Abstract
Background
African ancestry is associated with higher prevalence and greater severity of asthma. Yet genetic studies of the most significant childhood onset asthma locus on 17q12–21 in African Americans have been equivocal. The goal of this study was to leverage both the careful phenotyping in the CREW birth cohort consortium and the breakdown of linkage disequilibrium in African Americans to fine map this important asthma locus.
Methods
Using nine 17q12–21 tag SNPs, we performed association studies for childhood onset asthma in 1,613 European American and 870 African American CREW children, and meta-analyzed the results with publicly available summary data from the EVE consortium (n=4,343 European American, 3,035 African ancestry). We then performed eQTL studies of the SNPs associated with childhood asthma and the expression of 17q12–21 genes in resting blood-derived and upper airway epithelial cells from 85 and 246, respectively, African American children; and in lower airway epithelial cells from 44 European American and 72 African American adults from Chicago.
Findings
17q12–21 SNPs were broadly associated with asthma in European Americans, but only two SNPs (rs2305480 in GSDMB and rs8076131 in ORDML3) were associated with asthma in African Americans, at a Bonferroni-corrected P<0.0055. Genotype at rs2305480 was the most significant eQTL for GSDMB (P<0.0001), and to a lesser extent for PGAP3 (P<0.0001), in upper airway epithelial cells from African American children. No SNPs were eQTLs for ORMDL3. These patterns were replicated in lower airway cells from European and African American adults. The SNP that was the most significant eQTL (rs12936231) for GSDMB and ORMDL3 in blood cells (P<10−8) was not associated with asthma in African Americans.
Interpretations
Our study suggests that SNPs regulating GSDMB expression in airway epithelial cells play a major role in childhood onset asthma, while SNPs regulating the expression levels of 17q12–21 genes in resting blood cells are not central to asthma risk. Using both genetic and gene expression data in African Americans and European Americans, GSDMB, which induces cell death in response to intracellular pathogens, emerged as the leading candidate gene at this important asthma locus.
Disparities in the prevalence, severity and natural history of asthma have been well described, with African ancestry associated with increased asthma prevalence1,2 and poorer response to asthma therapies and suboptimal lung function.3,4 The relative contributions of genes, environment, or gene-environment interactions to these disparities remain unknown. In particular, the evidence for association with SNPs at the chromosome 17q12–21 locus, the most highly replicated childhood onset asthma locus in European ancestry populations,5–9 has been equivocal in African ancestry populations, with weak associations at best.7,8,10–14 Notably though, none of the studies in African Americans focused specifically on early childhood onset asthma and samples sizes have been small relative to studies of European ancestry populations.
A remarkable feature of the 17q12–21 asthma locus is extensive linkage disequilibrium (LD) that spans over 150 kb on European-derived chromosomes with lesser LD extending into the flanking regions, together encoding at least 10 genes. The asthma-associated alleles in European ancestry populations are on an extended “risk haplotype” that have been associated with the expression of two genes, ORMDL3 and GSDMB, primarily in blood cells. The risk and protective haplotypes at this locus occur at approximately equal frequencies in most European ancestry populations. The extensive LD makes it difficult to differentiate the effects of individual variants on asthma risk or on gene expression, or to identify causal variants, in European ancestry individuals. In contrast, there is significant breakdown of LD at this locus on African-derived chromosomes,15 yet this feature has so far not been used to fine map the independent effects of 17q12–21 SNPs on risk of childhood-onset asthma or on the expression of genes at this locus.
We hypothesized that the breakdown of LD in African Americans would facilitate fine mapping studies at this important locus, allowing us to narrow the associations with asthma and with the expression of genes at this locus to fewer SNPs. To this end, we leveraged the Children’s Respiratory and Environmental Workgroup (CREW) Consortium,16 a component study of the larger NIH-funded Environmental Influences on Child Health Outcomes Program (ECHO).17 CREW includes ethnically diverse subjects from 12 U.S. longitudinal birth cohorts with asthma outcome phenotypes. To first show that SNPs at this locus are associated with childhood onset asthma in the CREW children, we genotyped nine 17q12–21 tag SNPs in 1,613 European American and 870 African American CREW children. To increase power, we also performed a meta-analysis using summary statistics for 4,343 European American and 3,035 African ancestry individuals from the EVE consortium.7 We then sought to determine which SNPs are both associated with both childhood onset asthma and are eQTLs for 17q12–21 genes in 268 African American children from a CREW cohort, using unstimulated peripheral blood mononculear cells (PBMCs; n=85) and upper airway epithelium (n=246), two compartments that are central to the pathobiology of asthma. Finally, we validated the eQTL results in lower airway epithelial cells from 44 European American and 72 African American adults, which allowed us to generalize our findings to the lower airway and to European Americans. Using these orthogonal approaches, we show that patterns of association among asthma-associated SNPs and gene expression differ between airway epithelial cells and PBMCs, and that GSDMB, and possibly PGAP3, in airway epithelial cells likely underlie the risk for childhood onset asthma at the 17q12–21 locus.
Methods
Sample composition
Our study included 2,483 self-identified European American or African American unrelated children from nine CREW birth cohorts who were followed to at least age 6 years and provided DNA for genotyping (Table 1). Because the 17q12–21 locus is associated with early life wheezing illnesses and early diagnosis of asthma,9,18–20 we focused on asthma diagnoses by a health care provider by age 6 to maximize sample size and reduce heterogeneity (see appendix p. 5 for additional details). This work was approved by the institutional review boards at the participating institutions.
Table 1.
Description of the nine CREW cohorts included in this study. See Gern et al.16 for more details.
| Cohort | Recruitment Sites | Recruitment Years | High Risk* | European American (case/control) [%Female] |
African American (case/control) [%Female] |
|---|---|---|---|---|---|
| CAS | Detroit, MI | 1987 – 1989 | No | 294 (33/261) [42%/51%] |
3 (0/3) [−/33%] |
| CCAAPS | Cincinnati, OH | 2001 – 2003 | Yes | 324 (40/284) [35%/47%] |
89 (27/62) [37%/42%] |
| CCCEH | New York, NY | 1998 – 2006 | No | 0 | 71 (3/68) [100%/65%] |
| COAST | Madison, WI | 1998 – 2000 | Yes | 169 (56/113) [34%/50%] |
8 (5/3) [40%/67%] |
| EHAAS | Boston, MA | 1994 – 1996 | Yes | 238 (81/157) [26%/47%] |
21 (11/10) [55%/50%] |
| IIS | Tucson, AZ | 1997 – 2003 | No | 198 (34/164) [44%/57%] |
3 (2/1) [50%/0%] |
| TCRS | Tucson, AZ | 1980 – 1984 | No | 236 (23/213) [43%/49%] |
11 (1/10) [0%/70%] |
| URECA | Boston, MA/Baltimore, MD/New York, NY/St. Louis, MO | 2004 – 2006 | Yes | 3 (1/2) [0%/50%] |
328 (159/169) [47%/53%] |
| WHEALS | Detroit, MI | 2003 – 2007 | No | 151 (28/123) [46%/47%] |
336 (111/225) [45%/54%] |
| Total (case/control) |
1,613
(296/1,317) [36%/50%] |
870
(319/551) [45%/54%] |
High risk refers to families ascertained based on having at least one parent with asthma and/or allergy.
We also included two additional samples. First, to increase power in the association studies, asthma GWAS summary data were included from the EVE consortium7 for 6,141 (2,088 cases) European American and 3,976 (1,612 cases) African ancestry individuals from cases-control studies, in whom asthma was determined by a doctor’s diagnosis. Second, to provide validation of the eQTLs in the lower airway and in European Americans, we performed eQTL studies in bronchial epithelial cells from 44 (32 cases) European American and 72 (48 cases) African American participants in an case-control study in Chicago.21 Asthma was diagnosed based on a doctor’s diagnosis, current symptoms and asthma medication use (appendix p 5).
These studies were approved by the local Institutional Review Boards from each participating center.
Genotyping and quality control in CREW
DNA (500 ng at concentrations of 5 ng/μl) was shipped from the coordinating center of each CREW cohort to the University of Chicago for genotyping. Nine SNPs were selected based on LD patterns at the extended 17q12–21 locus and previously reported associations with asthma or gene expression (appendix p 9). Genotyping methods and quality control are described in the appendix (p 5).
SNP association tests
To avoid selection bias, we included all CREW cohorts, regardless of sample size. However, we repeated all analyses including cohorts with at least 20 cases to assure that the small samples were not skewing results. The genotype of each SNP was considered under an additive model (0, 1, or 2 effect alleles) and tested for association with asthma using logistic regression, including sex as a covariate. Because genome-wide SNPs were not available to estimate ancestry principal components (PCs), we also included the study site as a covariate to account for differences in allele and asthma frequencies among the cohorts. We considered the allele at each SNP that has been associated with increased asthma risk in European ancestry populations as the “effect” allele in all tests15. P-values were adjusted for nine tests; P<0.0055 was considered significant.
To increase power, we also included published summary data from the EVE Consortium.7 The majority of EVE cases had asthma onset in childhood (median age of onset <8 years in the European Americans and <7 years in the African ancestry subjects). We extracted summary statistics for seven of the nine SNPs that were available in EVE, and meta-analyzed across the two studies within each racial/ethnic group, using a fixed effect model in which we assumed each study’s log-transformed odds ratio (OR) estimate was centered around the true log-transformed OR with known study-specific variance. Statistical significance of the meta-analysis test statistics was assessed using standard normal approximations; analyses were performed in R (v3.3.3).
Gene expression studies
RNA-seq data were available in African American children from one of the CREW cohorts (URECA) in unstimulated PBMCs for 85 children (48 cases, 37 controls)22 and in upper airway (nasal) cells for 246 children (125 cases, 121 controls) at the age 11 visit (appendix p 5). We used an additive effects linear model to test for eQTL studies of the five 17q core region SNPs with expression of each of the 10 genes at the extended locus, including as covariates sex, sample collection site, and 10 latent factors23 in the PBMC studies and sex, study site, batch id, epithelial cell proportion and 12 latent factors in the epithelial cell studies. Latent factors were included to correct for unwanted variation.23 P-values were adjusted for 40 tests (5 SNPs x 8 genes); P<0.0012 was considered significant. Differential expression between cases and controls were tested in using a linear model, including the same covariates as described above.
We further examined associations between the same five SNPs with GSDMB and ORMDL3 expression in lower (bronchial) airway epithelial cells from 44 European American adults (32 cases, 12 controls) and African American adults (48 cases, 24 controls) who participated in a case-control study in Chicago (appendix pp 5–6, 10). Normalized gene expression counts were adjusted for age, sex, current smoking status, sequencing pool, the first three ancestry PCs, and cell composition (appendix p 5). Linear regression considering additive genotype effects on gene expression was performed using limma in R (v3.3.3). Differential expression between cases and controls were tested in the normalized data described above using a linear model.
Role of the funding source
Funding sources had no role in the study design, in the collection, analysis or interpretation of the data, in writing the report, or in decisions regarding choice of journals.
Results
LD patterns between 17q12–21 SNPs
Correlations between the nine SNPs in CREW children (Figure 1) are similar to those reported for European and African American populations (appendix p 12),11,15 both reflecting significant breakdown of LD in African Americans compared to European Americans. Throughout this paper we refer to the five SNPs within the approximately 150 kb LD block that includes the GSDMB and ORMDL3 genes as the core region; and the two SNPs in each of the flanking regions that show less LD with core region SNPs and less association with childhood onset asthma, as the proximal (encoding PGAP3 and ERBB2) and the distal (encoding GSDMA) regions, respectively, following Stein et al.15
Figure 1. Asthma association studies and LD nine SNPs at the 17q12–21 locus in CREW European American and African American children.
A) Pairwise LD between (see also appendix p 12). The top panel shows the location of genes at the locus, with the relative positions of the nine SNPs genotyped in the CREW children shown by vertical lines. The lower panel shows the LD plots in each population. The r2 value for pairs of SNPs are shown within the diamond shapes for all values <1.0. SNPs. Genes in the proximal region shown in blue, in the core region shown in orange, and in the distal region in green, as previously described.15 *rs3894194 was replaced in this study with rs8069202, which is in high LD with rs3894194 in both Europeans (r2=0.984) and African Americans (r2=0.953) (1000 Genomes). PGAP3, post-GPI attachment to proteins 3; ERBB2, erb-b2 receptor tyrosine kinase 2; MEN1, multiple endocrine neoplasia type 1; GRB7, growth factor receptor-bound protein 7; IKZF3, IKAROS family zinc finger 3; ZPBP2, zona pellucida binding protein 2; GSDMB, gasdermin B; ORMDL3, ORM1-like 3; LRRC3C, leucine-rich repeat-containing protein 3C; GSDMA, gasdermin A. B) Forest plots of association results in European Americans (EuAm) and African Americans (AfAm). Asthma odds ratio estimates (filled squares) and 95% confidence intervals (horizontal bars) are shown for the AfAm and EuAm samples in the CREW, EVE and CREW+EVE meta analyses. The size of the squares is proportional to the square root of the sample size. The bolded SNPs are the two that are associated with asthma in the AfAm meta-analysis (Bonferroni P <0.0055.
Associations between 17q12–21 SNPs and childhood onset asthma
The alleles at the nine SNPs were relatively common in CREW children (Table 2), as previously reported.15 All five SNPs in the core region were associated with asthma in the European American CREW children and in the meta-analysis with EVE (Table 2), reflecting the strong LD at this locus.
Table 2. Results of single SNP analysis of the 17q12–21 SNPs and asthma.
Different colors demarcate SNPs in the proximal, core, and distal regions of this locus (Figure 1A). Effect alleles are the asthma-associated alleles in Europeans, following 15. SNP numbers are from Figure 1A. The two rows in bolded font show the two SNPs associated with asthma in the meta-analysis of African American samples at the Bonferroni corrected P-value threshold (9 SNPs) = 0.005. The meta-analysis P-values were weighted by the number of cases. CI, confidence interval; EAF, effect allele frequency; n/a, SNP not available for EVE7.
| SNP Number | SNP_ Ref. Allele | Gene | European Americans | African Americans | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CREW (n = 1,613) | Meta-Analysis (CREW+EVE, n=5,916) | CREW (n = 870) | Meta-Analysis (CREW+EVE, n=3,904) | |||||||||||||
| RAF | OR | 95% CI | P | OR | 95% CI | P | RAF | OR | 95% CI | P | OR | 95% CI | P | |||
| 1 | rs2941504_A | PGAP3 | 0·31 | 1·15 | 0·94, 1·41 | 0·16 | 1·13 | 1·05, 1·22 | 0·0011 | 0·45 | 0·95 | 0·77, 1·17 | 0·67 | 1·038 | 0·97, 1·11 | 0·27 |
| 2 | rs2517955_C | PGAP3/ERBB2 | 0·35 | 1·11 | 0·91, 1·35 | 0·12 | 1·15 | 1·06, 1·25 | 0·0003 | 0·76 | 1·10 | 0·86, 1·40 | 0·41 | 1·059 | 0·98, 1·13 | 0·10 |
| 3 | rs12936231_C | ZPBP2 | 0·50 | 1·32 | 1·09, 1·59 | 0·0043 | 1·28 | 1·18, 1·38 | <0.0001 | 0·47 | 0·88 | 0·72, 1·08 | 0·24 | 1·013 | 0·91, 1·12 | 0·79 |
| 4 | rs2305480_G | GSDMB | 0·55 | 1·36 | 1·12, 1·65 | 0·0014 | 1·27 | 1·17, 1·38 | <0.0001 | 0·84 | 1·23 | 0·91, 1·66 | 0·16 | 1·17 | 1·05, 1·30 | 0·0035 |
| 5 | rs7216389_T | GSDMB | 0·51 | 1·38 | 1·11, 1·66 | 0·0008 | 1·28 | 1·18, 1·38 | <0.0001 | 0·79 | 1·15 | 0·89, 1·48 | 0·28 | 1·067 | 0·99, 1·14 | 0·059 |
| 6 | rs4065275_G | ORMDL3 | 0·52 | 1·31 | 1·09, 1·59 | 0·0043 | n/a | n/a | n/a | 0·62 | 0·81 | 0·66, 1·005 | 0·055 | n/a | n/a | n/a |
| 7 | rs8076131_A | ORMDL3 | 0·55 | 1·37 | 1·13, 1·67 | 0·0010 | 1·26 | 1·17, 1·34 | <0.0001 | 0·81 | 1·16 | 0·88, 1·52 | 0·26 | 1·17 | 1·06, 1·29 | 0·0018 |
| 8 | rs8069202_A | GSDMA | 0·45 | 1·29 | 1·07, 1·56 | 0·0077 | n/a | n/a | n/a | 0·32 | 0·87 | 0·70, 1·09 | 0·25 | n/a | n/a | n/a |
| 9 | rs3859192_T | GSDMA | 0·465 | 1·13 | 0·93, 1·36 | 0·12 | 1·12 | 1·03, 1·21 | 0·0040 | 0·35 | 0·85 | 0·69, 1·06 | 0·16 | 1·011 | 0·94, 1·07 | 0·73 |
None of the SNPs were significant in the African American CREW children, possibly due to the smaller sample size and the high frequency of the risk alleles compared to European American children. However, the overlapping confidence intervals for two SNPs in the core region (SNP4 [rs2305480] and SNP5 [rs7216389] in GSDMB and SNP7 [rs8076131] in ORMDL3) in the CREW samples suggest that the ORs are not different between these groups (Table 2; Figure 1B). Analyses including only CREW cohorts with ≥20 cases yielded similar results (appendix p 10). In the meta-analysis with EVE African American subjects, two SNPs were significantly associated with asthma: SNP4 (rs2305480) in GSDMB and SNP7 (rs8076131) in ORDML3 (Table 2; Figure 1B).
Gene expression studies at the 17q12–21 locus
The meta-analysis in African American subjects narrowed the association with asthma to two candidate SNPs. However, whether the effects of these SNPs are independently associated with asthma, or whether the associations are due to LD cannot be distinguished by genetic association studies. Therefore, to further investigate these possibilities, we performed eQTL fine mapping for the five core SNPs in African American CREW children and the expression of eight genes at this locus in two asthma-relevant cell compartments: PBMCs and upper airway epithelial cells.
Indeed, the breakdown of LD in the African American children allowed us to differentiate the eQTL effects at this locus (Figure 2, Table 3), which had not been possible in previous studies performed primarily European ancestry individuals (e.g. 24,25). In PBMCs, genotypes at the five core SNPs were associated only with expression of GSDMB and ORMDL3. The strongest associations were with SNP3 (rs12936231), an intronic SNP in the ZPBP2 gene (GSDMB: eQTL β=1·23 [95% CI 1·15, 1.32]; P<0.0001; ORMDL3: eQTL β = 1·18 [95% CI 1·12, 1·24], P<0.0001), with weaker associations at the other core region SNPs (Figure 2 upper panel, Table 3), consistent with earlier studies.26,27 The fact that SNP3 (rs12936231) genotype was most associated with GSDMB and ORMDL3 expression in PBMCs, and not associated with risk for childhood onset asthma in the African American CREW children or in the meta-analysis, suggests that expression of GSDMB and ORMDL3 in PBMCs may not underlie risk for asthma.
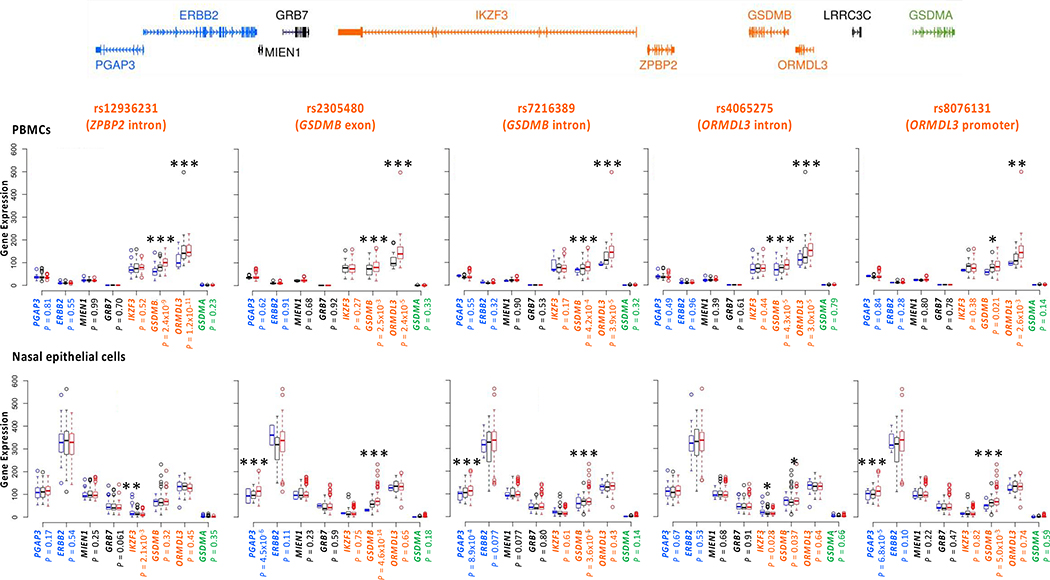
Figure 2. Associations between five core region SNPs and expression levels of genes at the extended 17q12–21 locus in African American children in A) PBMCs (n=85) and B) upper airway epithelial cells (n=246).
The relative location of genes at this locus are shown in the upper panel. ZPBP2 and LRRC3C were not detected as expressed in these cells. SNPs and the genes in which they reside are shown above each panel. Gene names are shown on the x-axes; normalized gene expression in counts per million are shown on the y-axis (see Methods). At each SNP, blue bars correspond to homozygotes for the non-effect (non-risk) alleles, black bars to heterozygotes, and red bars to homozygotes for the effect (risk) alleles. For SNP rs2305480, the AA and AG genotypes were combined in PBMCs because there was only one homozygote for the A allele. See Table 3 for beta values (95% CIs) and exact P-values. *P < 0.05, **P < 0.01, ***P ≤ 0.0012
Table 3. Effect sizes (beta), 95% confidence intervals (CI), and P-values for five core SNPs and expression of the eight 17q12–21 genes that are expressed in PBMCs (A) and upper airway epithelial cells (B).
The effect sizes (beta) reflect the expected increase in expression associated with each copy of the asthma risk allele. P-values were corrected for 40 tests (5 SNPs x 8 genes); P < 0.0012 are considered significant (shown in bolded font) (also see Figure 2).
|
A. PBMCs (n=85) | ||||||||||||
| SNP | PGAP3 | ERBB2 | MIEN1 | GRB7 | ||||||||
| Beta | 95% CI | P-value | Beta | 95% CI | P-value | Beta | 95% CI | P-value | Beta | 95% CI | P-value | |
| rs12936231 | 1·00 | 0·96, 1·041 | 0·81 | 0·976 | 0·91, 1·037 | 0·55 | 0·994 | 0·96, 1·024 | 0·99 | 1·02 | 0·82, 1·25 | 0·70 |
| rs2305480 | 1·01 | 0·95, 1·084 | 0·62 | 1.0 | 0·89, 1·11 | 0·91 | 0·987 | 0·93, 1·040 | 0·68 | 0·973 | 0·67, 1·40 | 0·92 |
| rs7216389 | 0·98 | 0·93, 1·042 | 0·55 | 0·945 | 0·86, 1·036 | 0·32 | 0·993 | 0·95, 1·038 | 0·90 | 0·888 | 0·65, 1·21 | 0·53 |
| rs4065275 | 0·98 | 0·95, 1·026 | 0·49 | 0·992 | 0·93, 1·057 | 0·96 | 1·008 | 0·97, 1·039 | 0·39 | 1·032 | 0·83, 1·27 | 0·61 |
| rs8076131 | 1·00 | 0·95, 1·064 | 0·84 | 0·942 | 0·85, 1·033 | 0·28 | 0·99 | 0·94, 1·035 | 0·8 | 0·943 | 0·69, 1·29 | 0·78 |
| SNP | IKZF3 | GSDMB | ORMDL3 | GSDMA | ||||||||
| Beta | 95% CI | P-value | Beta | 95% CI | P-value | Beta | 95% CI | P-value | Beta | 95% CI | P-value | |
| rs12936231 | 0·967 | 0·91, 1·027 | 0·52 | 1·238 | 1·15, 1·32 | <0.0001 | 1·186 | 1·12, 1·24 | <0.0001 | 1·202 | 0·91, 1·58 | 0·23 |
| rs2305480 | 0·938 | 0·84, 1·045 | 0·27 | 1·218 | 1·067, 1·39 | 0.0025 | 1·232 | 1·11, 1·35 | <0.0001 | 1·279 | 0·78, 2·094 | 0·33 |
| rs7216389 | 0·915 | 0·83, 1·001 | 0·17 | 1·239 | 1·11, 1·38 | 0.0004 | 1·203 | 1·10, 1·30 | <0.0001 | 1·279 | 0·84, 1·93 | 0·32 |
| rs4065275 | 1·003 | 0·94, 1·068 | 0·44 | 1·185 | 1·10, 1·27 | <0.0001 | 1·133 | 1·071, 1·19 | <0.0001 | 1·067 | 0·80, 1·42 | 0·79 |
| rs8076131 | 0·94 | 0·85, 1·029 | 0·38 | 1·158 | 1·034, 1·29 | 0·021 | 1·151 | 1·058, 1·25 | 0.0026 | 1·408 | 0·92, 2·13 | 0·14 |
|
B. Upper Airway Epithelial Cells (n=246) | ||||||||||||
| SNP | PGAP3 | ERBB2 | MIEN1 | GRB7 | ||||||||
| Beta | 95% CI | P-value | Beta | 95% CI | P-value | Beta | 95% CI | P-value | Beta | 95% CI | P-value | |
| rs12936231 | 1·026 | 0·989, 1·064 | 0·17 | 0·993 | 0·970, 1·016 | 0·54 | 1·009 | 0.993, 1.025 | 0·25 | 0·949 | 0.899, 1.002 | 0·061 |
| rs2305480 | 1·147 | 1.081, 1.216 | <0.0001 | 1·031 | 0.992, 1.071 | 0·12 | 1·016 | 0.991, 1.042 | 0·22 | 0·980 | 0.895, 1.074 | 0·67 |
| rs7216389 | 1·081 | 1.032, 1.133 | 0.0010 | 1·028 | 0.998, 1.060 | 0·070 | 1·018 | 0.998, 1.039 | 0·070 | 0·990 | 0.921, 1.064 | 0·78 |
| rs4065275 | 1·034 | 0.997, 1.072 | 0·74 | 1·008 | 0.985, 1.032 | 0·49 | 1·003 | 0.988, 1.019 | 0·67 | 1·003 | 0.949, 1.060 | 0·93 |
| rs8076131 | 1·107 | 1.052, 1.164 | <0.0001 | 1·028 | 0.995, 1.063 | 0·093 | 1·014 | 0.993, 1.036 | 0·20 | 0·971 | 0.898, 1.050 | 0·46 |
| SNP | IKZF3 | GSDMB | ORMDL3 | GSDMA | ||||||||
| Beta | 95% CI | P-value | Beta | 95% CI | P-value | Beta | 95% CI | P-value | Beta | 95% CI | P-value | |
| rs12936231 | 0·872 | 0.799, 0.951 | 0.0021 | 1·026 | 0.975, 1.080 | 0·32 | 0·989 | 0.962, 1.017 | 0·45 | 0·908 | 0.741, 1.112 | 0·35 |
| rs2305480 | 0·976 | 0.850, 1.119 | 0·73 | 1·353 | 1.251, 1.464 | <0.0001 | 0·990 | 0.945, 1.036 | 0·66 | 1·275 | 0.911, 1.785 | 0·16 |
| rs7216389 | 0·971 | 0.874, 1.078 | 0·58 | 1·144 | 1.073, 1.220 | <0.0001 | 1·015 | 0.979, 1.051 | 0·43 | 1·398 | 1.078, 1.813 | 0·11 |
| rs4065275 | 0·913 | 0.843, 0.989 | 0·026 | 1·054 | 1.003, 1.108 | 0·038 | 1·006 | 0.979, 1.035 | 0·65 | 0·962 | 0.786, 1.178 | 0·71 |
| rs8076131 | 0·985 | 0.877, 1.105 | 0·79 | 1·225 | 1.144, 1.312 | <0.0001 | 0·994 | 0.956, 1.034 | 0·78 | 1·328 | 0.998, 1.767 | 0·051 |
Very different patterns of association with gene expression were revealed in upper airway epithelial cells (Figure 2 lower panel, Table 3). The asthma-associated SNP in GSDMB (SNP4; rs2305480) was most strongly associated with expression of GSDMB (eQTL β=1·35 [95% CI 1·25, 1·46]; P<0.0001), and with PGAP3 to a lesser extent (eQTL β=1·22 [95% CI 1·14, 1·31]; P<0.0001), but not for any other genes at this locus, including ORMDL3 (eQTL P=0·66). The eQTL effects for SNP4 (rs2305480) were similar in children with and without asthma (appendix pp 10 and 14). The association between SNP4 (rs2305480) and PGAP3 expression may be due to LD between SNP4 and SNP2 (rs2517955) in PGAP3 (r2=0.40 in African American CREW children; Figure 1A). In fact, SNP2 (rs2517955) is a very strong eQTL for PGAP3 in upper airway epithelial cells (eQTL β=1·15 [95% CI 1·10, 1·20]; P<0.0001) (appendix p 15) in these children. Other SNPs (SNP5 [rs7216389] in GSDMB and SNP7 [rs8076131] in ORMDL3) showed orders of magnitude less significant association with GSDMB expression, likely reflecting LD with SNP4 (rs2305480). None of the SNPs were associated with the expression of any other genes. These combined data suggest that the effects of SNP4 (rs2305480), and/or other untyped SNPs that are in strong LD with SNP4, on airway epithelial cell expression of GSDMB may be responsible for modifying the risk for childhood onset asthma at this locus in African American children.
To assess the robustness of this finding, we performed parallel studies in lower airway epithelial cells from European American and African American adults. Similar to our observations in African American CREW children, SNPs in the core region were associated with the expression of GSDMB, but not with ORMDL3, in the European Americans (Figure 3), although no eQTLs reached significance after Bonferroni correction, possibly due to the small sample size (n=44). In the African American adults, SNP4 (rs2305480) was the most significant eQTL for GSDMB, but three other core region SNPs were also significant eQTLs for GSDMB. Four of the SNPs showed modest associations with ORMDL3, but none were significant. These results replicate our findings in CREW children indicating that asthma-associated variation at the 17q12–21 locus is most strongly associated with GSDMB expression levels in airway epithelial cells, and suggest that GSDMB expression in these cells underlies risk for childhood onset asthma in both European Americans and African Americans. The studies also raise the possibility that ORMDL3 expression in the lower airway may additionally contribute to risk in African American adults.
Figure 3. eQTL studies of five core region SNPs and expression levels of GSDMB and ORMDL3 in lower airway (bronchial) epithelial cells from European American (n=44) and African American (n=72) adults.
Upper panel: eQTL results with five core SNPs. The effect sizes (beta) reflect the expected increase in expression associated with each copy of the asthma risk allele. Bonferroni corrected P - value threshold (5 SNPs × 2 genes) = 0.0055. Lower panel: Boxplots showing associations between rs2305480 genotype and expression levels of GSDMB and ORMDL3.
Finally, we tested for differential expression of GSDMB and ORMDL3 between asthma cases and controls in resting blood-derived cells and upper airway epithelial cells from the African American children and in lower airway epithelial cells from the African American and European American adults. None of the analyses revealed differences in the overall abundance of transcripts from either gene between cases and controls, with and without including genotype for SNP4 (rs2305480) as a covariate (appendix pp 11–12).
Discussion
Asthma is the most common chronic disease of childhood, affecting about 8% of children in the U.S. this past decade.2 During this time, the prevalence of asthma was 15.7% among non-Hispanic black children, who also showed greater severity and poorer response to treatment compared to U.S. white populations.1–4 Among the more than 120 loci associated with childhood onset asthma in populations of European ancestry, a broad locus on chromosome 17q12–21 remains the most replicated to date.5–9 Yet, the effects of SNPs and genes at this locus on childhood onset asthma risk in African ancestry populations have been equivocal7,8,10–14. Moreover, the strong LD across more than 150 kb in European ancestry populations makes it particularly challenging to identify the SNPs and genes at this locus that underlie the strong associations with childhood onset asthma risk.
In this study, we used several approaches to identify the source of the asthma risk attributed to the 17q12–21 locus. First, the breakdown of LD in African ancestry populations allowed us to narrow the association signal to two tag SNPs. Second, eQTL studies in unstimulated PBMCs and airway epithelial cells from African American children revealed that the strongest eQTL in epithelial cells (SNP4; rs2305480), but not the strongest eQTL in PMBCs (SNP3; rs12936231), was also associated with asthma in African Americans. SNP3 (rs12936231) in the ZPBP2 gene is a known eQTL for ORMDL3 and GSDMB in immune cells,26,27 consistent with our findings. Yet, despite its profound effect on expression of ORMDL3 in immune cells from both European and African ancestry individuals,26,27 it has never been reported as the lead SNP at this locus in asthma GWASs.5–9 These combined observations suggest that expression of GSDMB in airway epithelial cells, but not ORMDL3 or GSDMB in PBMCs, modulates asthma risk at this locus. Moreover, we attribute the most of the association between SNP4 (rs2305480) with airway epithelial cell expression of PGAP3, which is located at the proximal end of the locus, to LD with a SNP in PGAP3 (SNP2, rs2517955; r2=0·40), which is a strong eQTL for PGAP3 in lung tissue15 and in the airway epithelial cells (P<0.0001) (appendix p 15). Third, our studies in lower airway epithelial cells from European American and African American adults replicated the finding that asthma-associated SNPs at this locus are strong eQTLs for GSDMB in airway cells and extended these observations to the lower airway, to adults, and to European Americans. Modest associations with ORMDL3 levels in lower airway cells raise the possibility that this gene may also contribute to asthma risk in African American adults. These combined data provide strong convergent lines of evidence that asthma risk at the 17q12–21 locus is modulated by genetic regulation of GSDMB expression in airway epithelial cells in African and European ancestry populations. The strength and specificity of association between 17q12–21 asthma-associated alleles with expression of GSDMB in airway epithelial cells further highlights the important role of epithelial function in the etiology of childhood onset asthma9.
GSDMB is an outstanding candidate asthma gene due to its critical role in pyroptosis, a form of necrotic and inflammatory cell death mediated by inflammatory caspases.28 Acting through a non-canonical pathway, the N-terminus of GSDMB binds to the CARD domain of caspase-4 in response to intracellular microbes, directly leading to activation of other caspases and cleavage of the N-terminus of another gasdermin, GSDMD, which then orchestrates pyroptosis by forming pores in the plasma membrane.29 SNP4 (rs2305480) is a missense variant (c. 892G>A; p.Pro298Ser) in GSDMB at a highly conserved amino acid. This SNP is in near perfect LD (r2∼1) in all populations with two GSDMB intronic SNPs,15 which were not genotyped in our study (appendix p 13). One intronic SNP, rs11078928 (c.662T>C), affects splicing efficiency of the GSDMB transcript by skipping exon 6,30 resulting in reduced expression of both the GSDMB transcript and GSDMB protein on the non-risk (C) allele. The second untyped SNP, rs11078927, is of unknown function. Thus, the lead candidate SNP in our studies represents the effects of three SNPs in GSDMB, including one with possible effects on protein function (SNP4; rs2305480) and one with known effects on transcript and protein levels (rs11078928).
The disparate frequencies of the asthma-associated alleles in GSDMB, ∼0.80 in African American and ∼0.50 in European American CREW children are notable. For example, only ∼4% of African American children are expected to be homozygous for the protective allele compared to nearly 25% of European American children. Among children with one or two copies of the risk allele, 19% of the European American and 36% of the African American children had a diagnosis of asthma by 6 years of age. On the one hand, the overall high penetrance of the risk genotypes in both groups may be due to inclusion of four CREW cohorts (accounting for 45% of European American children and 51% of African American children) in which participants were selected based on a family history of asthma or allergies (Table 1). On the other hand, the less than 100% penetrance of 17q12–21 genotypes in both groups, and the central role GSDMB in innate immune response to intracellular microbes, is consistent with the many interactions observed between 17q12–21 genotypes and early life exposures, including tobacco smoke,18,31 rhinovirus and wheezing illnesses,19,20 farm animals,20 and pets32 on childhood onset asthma risk.
There are several strengths and some caveats to this study. First, the CREW sample is relatively small for genetic association studies, particularly in the African American children. However, these children who were enrolled at birth and comprehensively monitored longitudinally improved the accuracy of the asthma phenotyping to an extent not achievable in the large samples needed for GWAS. In fact, prospectively collected “doctor-diagnosed asthma” definitions closely agree with other definitions that include lung function and other symptom-based criteria.33 Combining the CREW association results with published GWAS results increased power and revealed largely overlapping effect sizes at the associated loci in the CREW and EVE African Americans. This, together with the breakdown of LD in the African American children, allowed us to make several important observations that could not have been made in European ancestry samples. Second, because genome-wide genotypes for estimating ancestry were not available for the CREW children, we could not adjust for different levels of admixture. This potential limitation was minimized by focusing on a single locus and correcting for differences in asthma prevalence and allele frequencies by including study site as a covariate.
Third, this study included cells from different cellular compartments for gene expression studies, and the contrasting findings in airway and blood cells were informative. However, we cannot exclude that other genes at this locus, including ORMDL3, may be important in other tissues, in response to exposures, or at different stages of development. Lastly, we could not relate differential expression of GSDMB or ORMDL3 in these samples to asthma, similar to earlier studies (reviewed in Stein et al.15). This observation further suggests that expression differences of these genes between asthma cases and controls may only be evident during critical windows of development or in response to certain exposures, such as respiratory infections in early life.18,19
In conclusion, this study demonstrates the power of using populations of different ethnicities for association studies and eQTL studies in asthma-relevant cells to fine map the effects of SNPs on both disease risk and gene expression, and of using birth cohorts in which children have been carefully phenotyped during early life when disease risk trajectories are established. Our study highlights GSDMB as an important asthma gene at the 17q12–21 locus and airway epithelial cells as the most likely target for modulating the effects of variation at this locus on the development of childhood onset asthma. Combined with previous studies of GSDMB function, these results suggest a potential target for therapeutic intervention in children with the high-risk genotype at this locus.
Research in Context
Evidence before this study
We searched PubMed for all genetic and eQTL studies of the 17q12–21 locus since it was first reported as an asthma locus in 2007 through July 1, 2019, using the terms GSDMB, ORMDL3, 17q, and asthma. These studies supported the conclusion that this is the most highly replicated childhood onset asthma locus and is nearly always the most significant locus in GWASs of asthma in European ancestry populations. This locus has also been replicated in Latino populations, but much less so in African ancestry populations in whom associations with asthma are considerably weaker. SNPs at this locus have been reported as eQTLs for two genes, GSDMB and ORMDL3, primarily in blood-derived cells and primarily in individuals of European ancestry. Moreover, due to the extensive linkage disequilibrium (LD) at this locus in European ancestry populations, it has not been possible to distinguish the individual effects of SNPs spanning approximately 150 kb on asthma risk or on the expression of genes at this locus. No previous studies in African Americans focused on early childhood onset asthma or on eQTL studies in airway cells.
Added value of this study
Leveraging the breakdown of LD in African Americans, we were able to narrow the association with childhood onset asthma to two SNPs, we showed that one of the asthma-associated SNPs is also a strong eQTL for GSDMB, but not ORMDL3, in upper airway epithelial cells from African American children and in lower airway epithelial cells from African American adults. We further showed that 17q12–21 SNPs are eQTLs for GSDMB, but not ORMDL3, in lower airway epithelial cells from European American adults. The SNP that was the most significant eQTL for GSDMB and ORMDL3 in blood-derived cells from African American children was not associated with asthma.
Implications of all the available evidence
Our results indicate that GSDMB expression in airway epithelial cells, and the SNPs that regulate its expression, are the leading 17q12–21 asthma candidates, and identify GSDMB as a potential therapeutic target for childhood onset asthma.
Supplementary Material
Funding
Environmental influences on Child Health Outcomes (ECHO) is a nationwide research program supported by the National Institutes of Health (NIH), Office of the Director to enhance child health. This work was supported by UG3 OD023282, UL1 TR002373, UM1 AI114271, and U19 AI095230. The CREW cohorts are supported as follows: CAS: R01 AI024156, R03 HL067427, R01 AI051598; CCAAPS: R01 ES11170, R01 ES019890; CCCEH: P01 ES09600, R01 ES008977, P30ES09089, R01 ES013163, EPA R827027; COAST: P01 HL070831, U10 HL064305, R01 HL061879; EHAAS: R01 AI035786; IIS: HL56177, TCRS: R01 HL132523; URECA: NO1 AI25496, NO1 AI25482, HHS N272200900052C, HHS N272201000052I, NCRR/NIH RR00052, M01 RR00533, UL1 RR025771, M01 RR00071, UL1 RR024156, UL1 TR001079, UL1 RR024992, NCATS/NIH UL1TR000040; WHEALS: R01 AI050681, R56 AI050681, R01 AI061774, R21 AI059415, K01 AI070606, R21 AI069271, R01 HL113010, R21 ES022321, P01 AI089473, R21 AI080066, R01 AI110450, R01 HD082147, and the Fund for Henry Ford Health System.
Conflicts of Interest
Dr. Bacharier reports grants from NIH/NIAID,during the conduct of the study; personal fees from Aerocrine, personal fees from GlaxoSmithKline, personal fees from Genentech/Novartis, personal fees and non-financial support from Merck, personal fees from DBV Technologies, personal fees and non-financial support from Teva, personal fees and non-financial support from Boehringer Ingelheim, personal fees from AstraZeneca, personal fees from WebMD/Medscape, personal fees from Sanofi/Regeneron, personal fees from Vectura, personal fees from Circassia,outside the submitted work.
Dr. Gern reports grants from NIH, during the conduct of the study; personal fees from Regeneron, personal fees and stock options from Meissa Vaccines Inc, personal fees from MedImmune/AstraZeneca and Ena Therapeutics, outside the submitted work.
Dr. Gold reports grants from NIH, during the conduct of the study.
Dr. Hartert reports grants from NIH, during the conduct of the study; personal fees from Pfizer,outside the submitted work.
Dr. Hogarth reports personal fees and other from Auris, personal fees from Ambu, personal fees, non-financial support and other from Body Vision, personal fees and other from Eolo, other from Eon, other from Gravitas, personal fees and other from Noah Medical, personal fees and other from LX-Medical, other from Med-Opsys, other from Monogram Orthopedics, personal fees and other from Preora, other from VIDA, other from Viomics, personal fees from Boston Scientific, personal fees from Johnson and Johnson, personal fees from oncocyte, personal fees from veracyte, personal fees and other from Broncus, grants and personal fees from Gala, personal fees from Heritage Biologics, personal fees from IDbyDNA, personal fees from Level-Ex, personal fees from Medtronic, personal fees from Neurotronic, personal fees from olympus, personal fees from PulmonX, personal fees from Astra-Zeneca, personal fees from Biodesix, personal fees from Genetech, personal fees from Grifols, personal fees from Takeda, personal fees from CSL, personal fees from InhibRX, outside the submitted work. None of these COI have anything to do with this paper or my work on this paper.
Dr. Jackson reports grants from NIH, during the conduct of the study; personal fees from Sanofi-Regeneron, personal fees from AstraZeneca, personal fees from Novartis, personal fees from Vifor, personal fees from Pfizer, grants and personal fees from GlaxoSmithKline, personal fees from Merck, personal fees from Boehringer Ingelheim, personal fees from Commense, outside the submitted work.
Dr. Johnson reports grants from NIH, during the conduct of the study.
Dr. Kattan reports grants from NIH (NIAID), during the conduct of the study; personal fees from Sanofi/Regeron, outside the submitted work.
Dr. Lemanske reports grants from National Heart, Lung, and Blood Institute, grants from National Institutes of Health,during the conduct of the study; non-financial support from American Academy of Allergy, Asthma, and Immunology., grants from Clinical and Translational Science Award from NIH, personal fees from LSU, personal fees from Elsevier, personal fees from UpToDate, grants from Childhood Origins of ASThma (COAST), grants from AsthmaNet, personal fees from University of Kentucky, personal fees from Thermo Fischer, personal fees from Japanese Society of Allergy, personal fees from APAAACI & APAPARI, outside the submitted work.
Dr. Lynch reports grants from NIH/NIAID, grants from NIH/NICHD, during the conduct of the study; personal fees from Siolta Therapeutics, outside the submitted work.
Dr. Martinez reports grants from NIH/NHLBI, during the conduct of the study; grants from NIH/NHLBI, grants from NIH/NIEHS, grants from NIH/NIAID, grants from NIH/Office of Director, grants from Johnson & Johnson, personal fees from Copeval, outside the submitted work.
Dr. O’Connor reports grant support from NIH for the submitted work and personal fees from AstraZeneca, and grant support from Janssen Pharmaceuticals outside of the submitted work.
Dr. Ober reports grants from NIH, during the conduct of the study; grants and personal fees from NIH, personal fees from American Academy of Asthma, Allergy & Immunology, outside the submitted work.
Dr. White reports personal fees from Genentech, Inc, and grants from NHLBI outside the submitted work.
Dr. Wood reports grants from NIH, during the conduct of the study; grants from NIH, grants from Astellas, grants from DBV, grants from Regeneron, grants from Aimmune, personal fees from Up to Date, outside the submitted work.
Dr. Zoratti reports grants from U.S. National Institutes of Health, during the conduct of the study.
The other authors have nothing to disclose.
References
- 1.Urquhart A, Clarke P. US racial/ethnic disparities in childhood asthma emergent health care use: National Health Interview Survey, 2013–2015. J Asthma 2019: 1–11. [DOI] [PubMed] [Google Scholar]
- 2.Zahran HS, Bailey CM, Damon SA, Garbe PL, Breysse PN. Vital Signs: Asthma in Children - United States, 2001–2016. MMWR Morb Mortal Wkly Rep 2018; 67(5): 149–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naqvi M, Tcheurekdjian H, DeBoard JA, et al. Inhaled corticosteroids and augmented bronchodilator responsiveness in Latino and African American asthmatic patients. Ann Allergy Asthma Immunol 2008; 100(6): 551–7. [DOI] [PubMed] [Google Scholar]
- 4.Neophytou AM, White MJ, Oh SS, et al. Air Pollution and Lung Function in Minority Youth with Asthma in the GALA II (Genes-Environments and Admixture in Latino Americans) and SAGE II (Study of African Americans, Asthma, Genes, and Environments) Studies. Am J Respir Crit Care Med 2016; 193(11): 1271–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med 2010; 363(13): 1211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moffatt MF, Kabesch M, Liang L, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 2007; 448(7152): 470–3. [DOI] [PubMed] [Google Scholar]
- 7.Torgerson DG, Ampleford EJ, Chiu GY, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet 2011; 43(9): 887–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demenais F, Margaritte-Jeannin P, Barnes KC, et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet 2018; 50(1): 42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pividori M, Schoettler N, Nicolae DL, Ober C, Im HK. Shared and distinct genetic risk factors for childhood-onset and adult-onset asthma: genome-wide and transcriptome-wide studies. Lancet Respir Med 2019; 7(6): 509–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahlin A, Sordillo JE, Ziniti J, et al. Large-scale, multiethnic genome-wide association study identifies novel loci contributing to asthma susceptibility in adults. J Allergy Clin Immunol 2019; 143(4): 1633–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daya M, Rafaels N, Brunetti TM, et al. Association study in African-admixed populations across the Americas recapitulates asthma risk loci in non-African populations. Nature communications 2019; 10(1): 880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathias RA, Grant AV, Rafaels N, et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol 2010; 125(2): 336–46 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sleiman PM, Flory J, Imielinski M, et al. Variants of DENND1B associated with asthma in children. N Engl J Med 2010; 362(1): 36–44. [DOI] [PubMed] [Google Scholar]
- 14.White MJ, Risse-Adams O, Goddard P, et al. Novel genetic risk factors for asthma in African American children: Precision Medicine and the SAGE II Study. Immunogenetics 2016; 68(6–7): 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stein MM, Thompson EE, Schoettler N, et al. A decade of research on the 17q12–21 asthma locus: Piecing together the puzzle. J Allergy Clin Immunol 2018; 142(3): 749–64 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gern JE, Jackson DJ, Lemanske RF Jr., et al. The Children’s Respiratory and Environmental Workgroup (CREW) birth cohort consortium: design, methods, and study population. Respir Res 2019; 20(1): 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.https://echochildren.org/about/. (accessed January 2, 2020).
- 18.Bouzigon E, Corda E, Aschard H, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med 2008; 359(19): 1985–94. [DOI] [PubMed] [Google Scholar]
- 19.Caliskan M, Bochkov YA, Kreiner-Moller E, et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med 2013; 368(15): 1398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loss GJ, Depner M, Hose AJ, et al. The Early Development of Wheeze. Environmental Determinants and Genetic Susceptibility at 17q21. Am J Respir Crit Care Med 2016; 193(8): 889–97. [DOI] [PubMed] [Google Scholar]
- 21.Nicodemus-Johnson J, Myers RA, Sakabe NJ, et al. DNA methylation in lung cells is associated with asthma endotypes and genetic risk. JCI Insight 2016; 1(20): e90151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altman MC, Whalen E, Togias A, et al. Allergen-induced activation of natural killer cells represents an early-life immune response in the development of allergic asthma. J Allergy Clin Immunol 2018; 142(6): 1856–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKennan C, Nicolae DL. Accounting for unobserved covariates with varying degree of estimability in high dimensional experimental data. Biometrika 2019; 106(4): 823–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao K, Bosse Y, Nickle DC, et al. Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma. PLoS Genet 2012; 8(11): e1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Consortium G, Laboratory DA, Coordinating Center -Analysis Working G, et al. Genetic effects on gene expression across human tissues. Nature 2017; 550(7675): 204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verlaan DJ, Berlivet S, Hunninghake GM, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet 2009; 85(3): 377–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmiedel BJ, Seumois G, Samaniego-Castruita D, et al. 17q21 asthma-risk variants switch CTCF binding and regulate IL-2 production by T cells. Nature communications 2016; 7: 13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Man SM, Karki R, Kanneganti TD. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol Rev 2017; 277(1): 61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Q, Shi P, Wang Y, et al. GSDMB promotes non-canonical pyroptosis by enhancing caspase-4 activity. J Mol Cell Biol 2019; 11(6): 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison FS, Locke JM, Wood AR, et al. The splice site variant rs11078928 may be associated with a genotype-dependent alteration in expression of GSDMB transcripts. BMC Genomics 2013; 14: 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Valk RJ, Duijts L, Kerkhof M, et al. Interaction of a 17q12 variant with both fetal and infant smoke exposure in the development of childhood asthma-like symptoms. Allergy 2012; 67(6): 767–74. [DOI] [PubMed] [Google Scholar]
- 32.Stokholm J, Chawes BL, Vissing N, Bonnelykke K, Bisgaard H. Cat exposure in early life decreases asthma risk from the 17q21 high-risk variant. J Allergy Clin Immunol 2018; 141(5): 1598–606. [DOI] [PubMed] [Google Scholar]
- 33.O’Connor GT, Lynch SV, Bloomberg GR, et al. Early-life home environment and risk of asthma among inner-city children. J Allergy Clin Immunol 2018; 141(4): 1468–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.