Abstract
Bacterial dormancy can take many forms, including formation of Bacillus endospores, Streptomyces exospores, and metabolically latent Mycobacterium cells. In the actinobacteria, including the streptomycetes and mycobacteria, the rapid resuscitation from a dormant state requires the activities of a family of cell-wall lytic enzymes called resuscitation-promoting factors (Rpfs). Whether Rpf activity promotes resuscitation by generating peptidoglycan fragments (muropeptides) that function as signaling molecules for spore germination or by simply remodeling the dormant cell wall has been the subject of much debate. Here, to address this question, we used mutagenesis and peptidoglycan binding and cleavage assays to first gain broader insight into the biochemical function of diverse Rpf enzymes. We show that their LysM and LytM domains enhance Rpf enzyme activity; their LytM domain and, in some cases their LysM domain, also promoted peptidoglycan binding. We further demonstrate that the Rpfs function as endo-acting lytic transglycosylases, cleaving within the peptidoglycan backbone. We also found that unlike in other systems, Rpf activity in the streptomycetes is not correlated with peptidoglycan-responsive Ser/Thr kinases for cell signaling, and the germination of rpf mutant strains could not be stimulated by the addition of known germinants. Collectively, these results suggest that in Streptomyces, Rpfs have a structural rather than signaling function during spore germination, and that in the actinobacteria, any signaling function associated with spore resuscitation requires the activity of additional yet to be identified enzymes.
Keywords: bacteria; peptidoglycan; cell surface enzyme; enzyme mechanism; serine/threonine protein kinase; dormancy; lytic transglycosylase; resuscitation-promoting factor (Rpf,); LysM; LytM; cell wall; actinobacteria; enzyme
Introduction
Bacteria are masters of survival. When faced with unfavorable growth conditions, many bacteria have evolved the ability to enter a nonreplicative state, allowing them to survive a wide range of adverse conditions. These nonreplicating states include everything from persister cells and viable but not culturable (VBNC) bacteria, through to specialized dormant spores (1–3). Despite the different forms adopted by these nonreplicating cells, they all share reduced metabolic activity compared with their vegetative counterparts, and often have an altered (thicker) cell wall.
A major constituent of the cell wall, in both vegetative and dormant cells, is peptidoglycan. Peptidoglycan polymers are defined by their glycan backbones, composed of alternating N-acetylmuramic acid (MurNAc) and N-acetylglucosamine (GlcNAc) residues, and by short peptides extending from the lactyl groups of the MurNAc residues. The peptide stems of different glycan strands can in turn be joined together either directly, or by amino acid linkers of varying lengths. These peptide bridges cross-link parallel strands together, yielding a rigid structure that maintains the integrity of the cell membrane (4). In dormant cells, the peptidoglycan is relatively inert, whereas in actively growing cells, it is highly dynamic (5).
Cell wall cleavage is a critical component of cell growth, being required for the insertion of new peptidoglycan. Muralytic enzymes target the glycan strands of peptidoglycan, and are classified as either hydrolases or lytic transglycosylases. Hydrolases, including the lysozymes and β-N-acetylglucosaminidases, hydrolyze β-(1–4) linkages in the glycan strands (6). In contrast, lytic transglycosylases cleave the same bond as lysozymes (between MurNAc and GlcNAc), but they do not require water and instead generate GlcNAc and 1-6–anhydroMurNAc products (7). All of these enzymes can be further subdivided into exo- or endo-acting enzymes, depending on whether they cleave at the ends of glycan strands, or within strands, respectively.
For many dormant cells, a return to active growth requires the breakdown of the thick protective cell wall, and different bacteria have evolved distinct strategies to achieve this. Within the actinobacteria, dormant cells employ a common degradative enzyme that promotes the resumption of vegetative growth. The so-called “resuscitation promoting factor”' (Rpf) enzymes share structural homology with lysozyme and lytic transglycosylases (8), and have muralytic activity (9–15). In Micrococcus luteus, a single Rpf enzyme is required for the resuscitation of metabolically quiescent cells (16). Most other actinobacteria encode multiple Rpfs (17, 18), and these collectively stimulate the growth of dormant cells (13, 19–22). In Streptomyces coelicolor, the products of five rpf genes (rpfA-E) promote the rapid germination of dormant spores, and can influence both vegetative growth and sporulation (11, 13). Deleting individual rpf genes results in modest germination defects in some instances (11, 13), whereas the loss of all five has the greatest impact on germination (13).
Resuscitation is a complex process, and how the Rpfs promote resuscitation is not fully understood. Two models have been put forth to explain Rpf function during the escape from dormancy: 1) Rpf activity liberates peptidoglycan-derived signaling molecules that activate a regulatory cascade needed for growth resumption, and 2) Rpf activity relieves the physical constraints imposed by dormant cell walls, allowing cell growth to resume (23). Although these proposals are not mutually exclusive, investigations to date appear to favor a signaling-based mechanism (24).
Resuscitation from dormancy has been best studied in Bacillus, which forms highly resistant endospores (25, 26) and encodes an Rpf-like enzyme (27). Bacillus spore germination can be promoted by the addition of peptidoglycan fragments (muropeptides) (28). These muropeptides bind to PrkC, a eukaryotic-like Ser/Thr kinase located in the spore membrane, initiating a signaling cascade that triggers spore germination (28). PrkC contains tandem PASTA (penicillin-binding protein and Ser/Thr kinase associated) domain repeats, and these domains recognize both nascent peptidoglycan and muropeptides (28–32). A similar situation may exist in Mycobacterium, where emergence from latency can be stimulated by muropeptide binding to PknB, a PrkC homologue (32). It is worth noting, however, that mycobacterial resuscitation via this route is not robust, and the major function of muropeptide binding appears to be in directing the subcellular localization of PknB (32). The molecular basis for Streptomyces resuscitation, and the contribution made by the Rpf proteins to this process, remains to be determined.
There is considerable diversity in Rpf enzyme architecture, and a clear understanding of Rpf function requires not only a full characterization of the enzymes themselves, but also a systematic assessment of the contributions made by the different domains. Here, we show that Rpf accessory domains make critical contributions to enzyme activity. We establish that the Rpfs function as endo-acting lytic transglycosylases, and further demonstrate that their activity is independent of known signaling cascades associated with germination in other systems. Unlike most systems investigated to date, our data are most consistent with a cell wall remodeling role for the Rpfs in Streptomyces spore germination.
Results
Rpf domain diversity in the actinobacteria
The Rpf domain is found in proteins throughout the actinobacteria, in association with a variety of different protein domains (17, 18). How these accessory domains influence the biological and biochemical function of different Rpfs remains unclear. To prioritize different architectures for investigation, we searched for Rpf domain-containing proteins in the streptomycetes, mycobacteria, micrococci, and other actinobacteria (Table 1). We found the RpfASC (protein bearing a signal peptide, and Rpf and LysM domains) class to be the most widespread in the actinobacteria, followed closely by the short Rpf class, which have no obvious functional domains beyond their Rpf domain (and signal peptide).
Table 1.
Prevalence of distinct Rpf configurations in the actinobacteria
Based on bioinformatics analyses conducted on March 6, 2019.
| Class and associated domains1 | Number in streptomycetes2 | Number in mycobacteria2 | Number in micrococci2 | All actinobacteria2 |
|---|---|---|---|---|
| RpfASC (Rpf domain and LysM; e.g. RpfA, RpfC) | 1624 (65.2%) | 4 (0.2%) | 211 (63%) | 2336 (32.8%) |
| RpfB (Rpf, G5, DUF348) | 58 (2.3%) | 410 (24.5%) | 124 (37%) | 1547 (21.7%) |
| RpfD (Rpf, LysM, LytM) | 538 (21.6%) | 0 | 0 | 546 (7.7%) |
| Short Rpfs (Rpf; e.g. RpfE) | 236 (9.5%) | 1260 (75.3%)3 | 0 | 2287 (32.1%) |
| Rpf, DUF3235 | 0 | 0 | 0 | 286 (4%)4 |
| Rpf, PG binding 1 | 0 | 0 | 0 | 89 (1.2%) |
| Rpf, Peptidase, SLT/GEWL | 11 (0.4%) | 0 | 0 | 11 (0.1%) |
| Rpf, VCBS | 26 (1%) | 0 | 0 | 26 (0.4%) |
| Total | 2493 | 1674 | 335 | 7129 |
| Entries in UniProtKB | 543 | 408 | 187 | 2888 |
1The abbreviations used are: DUF, domain of unknown function; SLT/GEWL, soluble lytic transglycosylase/goose egg white lysozyme; VCBS, repeat domain in Vibrio, Colwellia, Bradyrhizobium, and Shewanella.
2Numbers in parentheses are % of all Rpfs in each genera.
3Includes those Rpf proteins with N- and C-terminal extensions lacking obvious domains.
4Confined to the corynebacteria.
There were interesting phylogenetic distributions associated with each of the Rpf domain architectures. Within the mycobacteria, Rpf domains were most frequently found in conjunction with uncharacterized N- or C-terminal extensions (17, 18) (Table 1). These extended regions lacked any obvious functional domains, and were confined to Rpf-associated proteins in the mycobacteria. The corynebacteria also encoded a distinct subset of Rpf proteins associated with an uncharacterized DUF3235 domain (17) (Table 1).
In addition to these Genus-specific subsets, Rpfs were also associated with other functional domains, with two configurations being highly represented: the RpfB subgroup and the LysM-containing groups (Table 1). Members of the RpfB group were found in a range of actinobacterial species, and contained a G5 domain and tandem repeats of the DUF348 domain. Recent structural studies on an RpfB variant from Mycobacterium tuberculosis revealed an interesting ubiquitin-like fold for the DUF348 domain, and a close physical association between these domains and the G5 domain (33, 34). G5 domains bind to GlcNAc residues and are thought to promote peptidoglycan binding (35), whereas the DUF348 domains facilitate RpfB dimerization, and appear to negatively affect RpfB cleavage activity (13). In contrast to the RpfB group, the LysM-containing groups of Rpfs are the predominant form in the streptomycetes and micrococci. The LysM domain, like the G5 domain from the RpfB subfamily, binds GlcNAc residues (36), and is proposed to enhance Rpf binding to its peptidoglycan substrate. In the streptomycetes, many LysM domain-containing Rpfs also possess a LytM domain (Pfam: M23 metallopeptidase), which is expected to have endopeptidase activity and thus the potential to cleave either within peptide stems or peptide cross-bridges (37, 38).
LysM and LytM domains enhance Rpf activity
There is currently nothing known about how the Rpf-associated LysM and LytM domains influence Rpf activity. Given that the vast majority (>85%) of Streptomyces Rpf proteins possess one or both of these domains (Fig. 1A), we sought to determine how they influenced the biochemical activity of the Rpfs. To probe the functional contributions made by these domains, we created a truncated version of RpfA lacking the LysM domain (RpfAΔLysM), alongside two RpfD variants: one missing the LysM domain (RpfDΔLysM), and one lacking both the LysM and LytM domains (RpfDΔLysMΔLytM). We overexpressed and purified these proteins, along with their full-length counterparts (minus their SignalP-predicted secretion signals (39)), and evaluated the enzyme activity of each using a fluorescence-based peptidoglycan cleavage assay. The assay employs fluorescein-labeled M. luteus peptidoglycan as a substrate, where the fluorescein labeling is sufficiently dense so as to quench the fluorescent signal. Peptidoglycan cleavage results in the release of fluorescein molecules, leading to increased fluorescence.
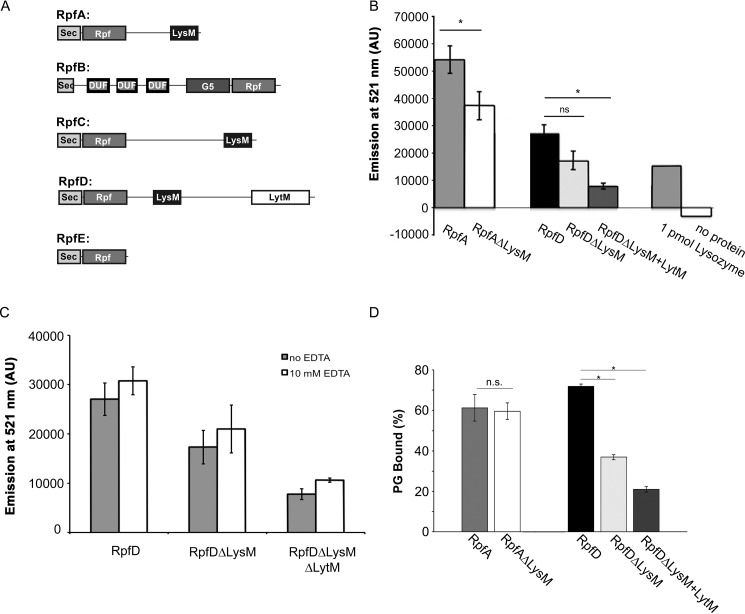
Figure 1.
LysM and LytM domains contribute to Rpf activity and peptidoglycan binding. A, schematic of the Rpf domain architectures in S. coelicolor. DUF, DUF348. B, a fluorescence-based assay was used to quantify the activity of Rpf proteins. Purified protein was mixed with fluorescein-labeled cell walls and incubated at 37 °C for 1 h. Cleavage of the substrate, and subsequent release of fluorescein, was detected by measuring the emission at 521 nm. One nanomole of each protein (or 1 pmol of lysozyme) was mixed with fluorescein-labeled M. luteus cell wall material. C, as described for B, purified protein was incubated in storage buffer with or without 10 mm EDTA for 1 h at room temperature prior to setting up the peptidoglycan cleavage assays. For both B and C, bars represent the average emission at 521 nm, for three independent biological replicates (two for RpfA), each of which is the average of three technical replicates, mean ± S.E. D, proportion of each protein bound to S. coelicolor peptidoglycan (PG), relative to input protein. Results presented are the average of two or three independently isolated protein preparations, mean ± S.E. Statistical significance was assessed using the Student's t test (*, p < 0.01; ns, not significant).
Mature versions of the full-length and truncated RpfA and RpfD enzymes were added in equimolar concentrations to the fluorescein-labeled substrate. For both RpfA and RpfD, we found that enzymes lacking the LysM domain had 65–70% of the activity of the full-length variants (Fig. 1B). This suggested that peptidoglycan targeting by the LysM domain may help position the Rpfs on their substrate and enhance their cleavage capabilities. We observed that removal of the LytM domain from RpfD led to a further decrease in activity; RpfD lacking both LysM and LytM domains had only ∼30% of the activity of the full-length enzyme (Fig. 1B).
The contribution of the LytM domain to RpfD activity may be enzymatic, as this domain typically has metallopeptidase activity, or it could function as an additional substrate specificity determinant. LytM peptidase activity requires a Zn2+ co-factor (40), and thus we tested the activity of all RpfD variants in the presence and absence of EDTA, which would be expected to chelate any associated Zn2+ ions. We found that EDTA had no effect on RpfD activity, irrespective of whether the LytM domain was present or not (Fig. 1C). This suggested that the RpfD-associated LytM domain may not function as an enzyme, and may instead provide additional targeting specificity for RpfD; although we cannot formally exclude the possibility that 10 mm EDTA was insufficient to remove any associated metal ions from RpfD. We examined the sequence of the LytM domain to determine whether it was lacking any key Zn2+-binding or active site residues, as is the case for EnvC and NlpD in Escherichia coli (41). However, all critical residues appeared to be present (Fig. S1), suggesting that the lack of enzyme activity was not due to the inability to bind the Zn2+ co-factor, nor to a degenerate active site.
To determine whether the difference in activity was due to a reduced ability to bind to peptidoglycan, we performed peptidoglycan-binding assays using peptidoglycan isolated from Streptomyces coelicolor. Surprisingly, we found that removing the LysM domain from RpfA had little impact on the peptidoglycan-binding capabilities of this enzyme (Fig. 1D, Fig. S2). In contrast, loss of the LysM domain from RpfD significantly reduced peptidoglycan binding. Removing the LytM domain from the LysM-deficient RpfD further reduced peptidoglycan binding (Fig. 1D, Fig. S2), supporting the proposal that the LytM domain may function to enhance peptidoglycan binding by RpfD.
Rpf domain functions as an endolytic transglycosylase
Having established that the LysM and LytM domains impacted Rpf activities in vitro, we next set out to investigate the mechanistic basis underlying peptidoglycan cleavage by the various Rpf enzymes. We opted to assess the activity of all five Rpfs from S. coelicolor (RpfA–E), as these enzymes represented four different structural classes (Rpf alone, RpfB, Rpf+LysM, and Rpf+LysM+LytM). For this, we used the assay of Herlihey et al. (42), taking advantage of the fact that hydrolases require water to break their cognate glycosidic bonds. Thus, in the presence of water labeled with the stable isotope 18O, hydrolase products would have an [18O]OH− incorporated at the C-1 position, altering the isotopic distribution of products detected using MS, relative to those produced in the presence of unlabeled water. In contrast, lytic transglycosylases do not use water in breaking the glycosidic bond in the peptidoglycan backbone, and as a result their 1,6-anhydromuroglycan products would have an unaltered isotopic distribution.
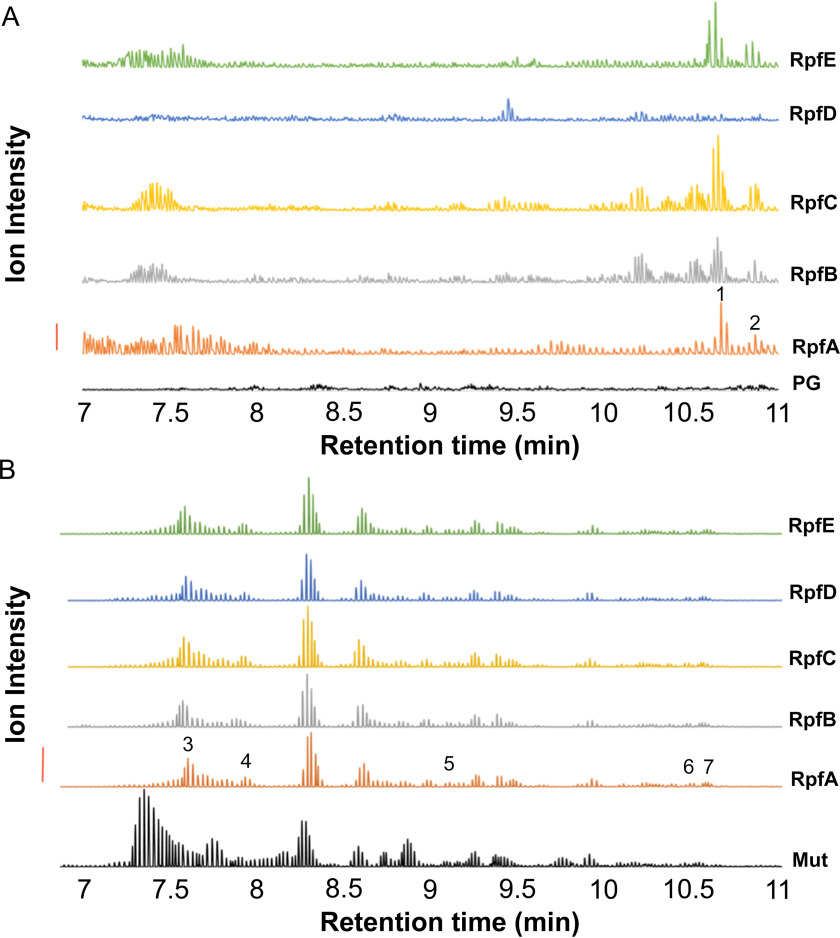
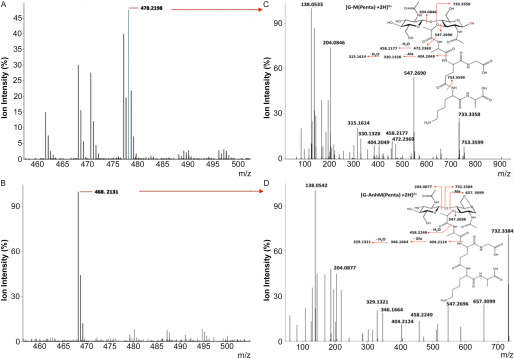
M. luteus peptidoglycan, suspended in 18O-labeled water, was initially used as a substrate for RpfA-E, and for the control hydrolytic enzyme mutanolysin. Being a highly active and efficient hydrolase, mutanolysin completely solubilized the peptidoglycan and we detected a variety of muroglycans by LC–MS analysis (Fig. 2). As expected, a number of these muroglycans were enriched with 18O. Subsequent MS/MS analyses confirmed the association of this 18O with only muramoyl residues (Fig. 3). In contrast, we detected very few soluble muroglycan products from each of the reactions involving RpfA, RpfB, RpfC, and RpfE, whereas none were produced by RpfD (Fig. 2A). MS analyses indicated that the few soluble muroglycans released did not contain 18O. These data suggested that the Rpfs did not function as either muramidases or β-N-acetylglucosaminidases. Instead, tandem MS analyses revealed that the released soluble muroglycans contained GlcNAc-1,6-anhydroMurNAc (peptides) (Table 2), indicating that each of RpfA/B/C/E functioned as lytic transglycosylases.
Figure 2.
Characterization of RpfA-E as endo-lytic transglycosylases by LC-Q-TOF MS analysis of their reaction products. Samples of M. luteus peptidoglycan suspended in [18O]H2O to a final concentration of 1.4 mg/ml were incubated separately with 1 nmol Rpf or 1.1 nmol mutanolysin (positive control). After incubation at 37 °C for 9.5 h, soluble reaction products were separated from insoluble material by centrifugation. The insoluble peptidoglycan pellets from the Rpf digestions were washed with water and then resuspended in 0.1 mm potassium phosphate buffer, pH 6.2, for solubilization by 1.1 μm mutanolysin. Soluble muropeptides from this secondary digestion were recovered by centrifugation. Each soluble and secondary-soluble fraction was subjected to LC-Q-TOF MS analysis. A, analysis of soluble fraction from peptidoglycan alone (PG), and reactions with 1 nmol RpfA–E, as indicated. B, analysis of insoluble products following secondary mutanolysin digestion from reaction with 1.1 μmol mutanolysin (Mut; positive control); or 1 nmol RpfA–E, as indicated. The identities of the numbered muropeptide fractions are listed in Table 2. The solid vertical bar to the left denotes 10,000 and 200,000 intensity units, for panels A and B, respectively.
Figure 3.
Tandem Q-TOF MS analysis of select muropeptides. Example of MS analysis of parent ions for muropeptides recovered from (A) mutanolysin (positive control) and (B) soluble RpfA digests of M. luteus peptidoglycan, by LC–MS as described in the legend of Fig. 2. C and D, tandem Q-TOF MS analysis of denoted parent ions from corresponding panels A and B. The blue spectral line in the MS spectrum of panel A denotes the 18O-containing isotope of the respective muropeptide. The monoisotopic masses (M + 2H)2+ are presented for each of the identified fragments.
Table 2.
LC-Q-TOF analysis of select muropeptides released from insoluble M. luteus peptidoglycan by RpfA-E
| Fraction No.1 | Annotation2 | Expected | Observed (m/z) |
z | ||||
|---|---|---|---|---|---|---|---|---|
| RpfA | RpfB | RpfC | RpfD | RpfE | ||||
| Rpf-soluble reaction products | ||||||||
| 1 | G-anhM(Penta) | 468.2050 | 468.2131 | 468.2112 | 468.2103 | NA3 | 468.2121 | 2 |
| 2 | G-anhM(Penta-Ala) | 503.7250 | 503.7322 | 503.7326 | 503.7330 | NA | 503.7309 | 2 |
| Rpf-insoluble reaction products | ||||||||
| 3 | G-anhM(Penta) | 468.2050 | 468.2118 | 468.2117 | 468.2120 | 468.2137 | 468.2123 | 2 |
| 4 | G-anhM(Penta-Ala) | 503.7250 | 503.7300 | 503.7309 | 503.7300 | 503.7320 | 503.7306 | 2 |
| 5 | G-M*-G-M(Penta)-G-anhM | 616.6200 | 616.9644 | 616.6293 | 616.9655 | 616.6328 | 616.6308 | 3 |
| 6 | G-M-G-M(Penta)-G-anhM(Penta) | 782.0300 | 782.O290 | 782.O266 | 782.O266 | 782.O297 | 782.O269 | 3 |
| 7 | G-M-G-M-G-anhM(Penta-Ala) | 979.9350 | 979.9418 | 979.9393 | 979.9386 | 979.9434 | 979.9394 | 2 |
1The muropeptide fractions correspond to those of the RP-HPLC separation presented in Fig. 2, A and B.
2Identification of each muropeptide was made by tandem Q-TOF-MS analysis of each parent ion (data not shown); G, GlcNAc; M, MurNAc; anhM, 1,6-anhydroMurNAc; penta, l-Ala-d-Glu-(Gly)-l-Lys-d-Ala; *, O-acetylation.
3NA, not observed.
We wondered if the lack of soluble products following reaction with RpfD, and the minimal amount produced by the other Rpf proteins, was due to a predominant endo-type lytic activity associated with each, where reaction products would remain cross-linked to the insoluble peptidoglycan sacculus. To analyze the insoluble fraction for any evidence of lysis, we washed the recovered insoluble peptidoglycan products and digested them with mutanolysin; any initial hydrolytic products of Rpf activity would retain their 18O enrichment, if present, following this secondary digestion. As seen in Fig. 2B, the muroglycan profiles of peptidoglycan incubated with each of the Rpfs, including RpfD, followed by mutanolysin digestion were distinct from the control reaction with mutanolysin alone. Tandem MS analysis of the unique muroglycan fractions revealed that none were enriched with 18O and that the majority were linear oligomers terminating with an anhydromuramoyl residue (Table 2). These data thus suggested that each of the Rpfs function as endo-acting lytic transglycosylases.
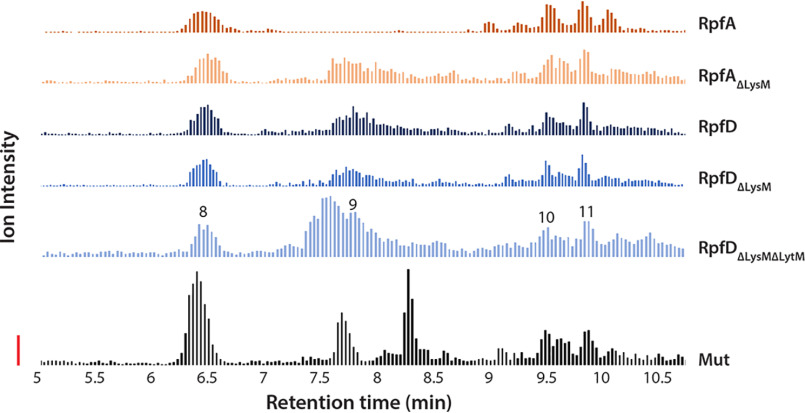
Unexpectedly, the muroglycan profiles for each of the five Rpfs were similar, and any specificity for glycan chain length, peptide stem composition, or cross-linking was not observed in the soluble or insoluble fractions. We noted, however, that the peptidoglycan composition of M. luteus differed slightly from that of S. coelicolor, specifically in the third amino acid and interpeptide bridge positions (43). Consequently, we sought to test whether any differences could be detected when using S. coelicolor peptidoglycan as substrate. As we saw for the cleavage profiles of the five WT Rpfs using M. luteus peptidoglycan, the S. coelicolor muroglycan profiles for RpfA, RpfD, and their LysM/LytM mutant derivatives were all similar (Fig. 4, Table 3), although the relative intensity units were different for the two substrates, suggesting greater Rpf affinity for/activity against the S. coelicolor peptidoglycan. These data collectively suggested that the different domains associated with the Rpf proteins did not confer any obvious substrate specificity with respect to peptidoglycan cleavage. Given the potential peptidase activity associated with the LytM domain of RpfD, we closely examined the cleavage products of both the M. luteus and S. coelicolor peptidoglycan for evidence of any hydrolytic activity not associated with the muroglycan backbone, but none were detected. These findings were consistent with our in vitro analyses, suggesting that the LytM domain enhanced RpfD function, not through peptidase activity, but instead by promoting peptidoglycan binding.
Figure 4.
LC-Q-TOF-MS analysis of insoluble reaction products generated by RpfA and RpfD variants during incubation with S. coelicolor peptidoglycan. Samples of S. coelicolor peptidoglycan were suspended in [18O]H2O to a final concentration of 1.4 mg/ml and incubated with 1 nmol of Rpf. Mutanolysin (1 nmol) was used as a positive control. Reactions were incubated at 37 °C for 9.5 h after which the soluble and insoluble fractions were isolated via centrifugation. Soluble fractions were prepared for LC-Q-TOF-MS analysis after this step, whereas insoluble fractions required further preparation. Insoluble pellets were washed with water and recovered each time via centrifugation. Washed samples were then solubilized with 1 nmol of mutanolysin prior to being subjected to LC-Q-TOF-MS analysis. A chromatogram of the combined extracted ion chromatograms (EICs) of each target ion species was generated for each enzyme variant using Agilent MassHunter Qualitative Analysis Software (version B.06.00). The identities of the numbered muropeptide species are as listed in Table 3. The solid vertical bar denotes 100 ion intensity units. Due to an overlapping of spectra involving an unidentified singly charged species (likely originating in the enzymatic preparation), an EIC could not be extracted for species 9 in the RpfA sample. However, the presence of this species was confirmed using tandem MS and was successfully identified, as listed in Table 3.
Table 3.
LC-Q-TOF analysis of select muropeptides released from insoluble S. coelicolor peptidoglycan by RpfA and D variants
| Annotation1 | Expected | Observed (m/z) |
z | ||||
|---|---|---|---|---|---|---|---|
| RpfD WT | RpfD ΔLysM | RpfD ΔLysM ΔLytM | RpfA WT | RpfA ΔLysM | |||
| Rpf soluble reaction products | |||||||
| G-anhM(Tri) | 426.1801 | 426.1839 | 426.1788 | 426.1873 | 426.1783 | 2 | |
| G-anhM(Tetra) | 461.69865 | 461.6976 | 461.6979 | 461.69 | 461.6965 | 461.697 | 2 |
| Rpf insoluble reaction products | |||||||
| G-anhM(Tri) | 426.1801 | 426.181 | 426.1893 | 426.1803 | 426.1793 | 426.1891 | 2 |
| G-anhM(Tetra) | 461.69865 | 461.6975 | 461.6963 | 461.6976 | 461.7062 | 461.6976 | 2 |
| G-M-G-anhM(Tetra-Tri) | 886.8748 | 886.8746 | 886.8794 | 886.8743 | 886.8799 | 886.8766 | 2 |
| G-M(Tetra)-G-anhM(Tetra) | 922.3894 | 922.3862 | 922.3927 | 922.387 | 922.3894 | 922.3865 | 2 |
1The muropeptide fractions correspond to those of the RP-HPLC separation presented in Fig. 2, C and D. Identification of each muropeptide was made by tandem Q-TOF-MS analysis of each parent ion (data not shown); G, GlcNAc; M, MurNAc; anhM, 1,6-anhydroMurNAc; Tri, l-Ala-d-Glu- l,l-DAP; Tetra l-Ala-d-Glu- l,l-DAP-d-Ala.
PASTA domain-containing Ser/Thr kinases in S. coelicolor inhibit germination and vegetative outgrowth
One hypothesis put forward to explain the role of Rpfs in cell resuscitation involves the release of muropeptide signals, which activate a regulatory cascade leading to the reactivation of metabolism. Such a model would be most consistent with exo-activity of the Rpfs, as this would promote the release of muropeptides; however, our results indicated that the Rpfs were endo-acting lytic transglycosylases.
In Bacillus, and to a lesser extent in Mycobacterium, the resuscitation-promoting signaling cascade is mediated through PASTA domain-containing Ser/Thr kinases (28, 32). We considered two possibilities that could accommodate both the endo-activity of the Rpfs and a role for Ser/Thr kinase signaling. In one, the PASTA domain-containing kinases in S. coelicolor may recognize the ends of cleaved peptidoglycan rather than a defined muropeptide. The second involved Rpf-cleaved products serving as a substrate for other cell wall lytic enzymes, resulting in the release of germination-stimulating muropeptides that are recognized by these kinases.
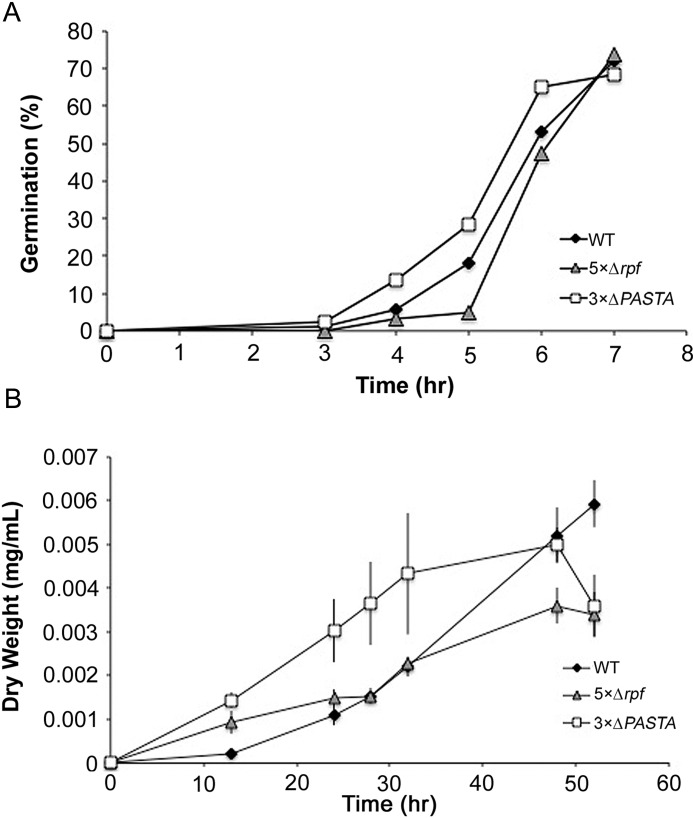
S. coelicolor encodes three PASTA domain-containing protein kinases, and we obtained a triple mutant strain (44), here dubbed the 3×ΔPASTA strain (Table S1). We expected that this strain would have similar germination rates to that of an rpf null mutant if the Rpfs were involved in generating appropriate peptidoglycan ends or germination-promoting muropeptides that were recognized by these kinases. We measured germination rates of the triple mutant strain, and compared these to the WT and rpf null strains. We found that germination of the 3×ΔPASTA strain was consistently more rapid than either the WT or the rpf null strain (Fig. 5A). This suggested that the three PASTA domain-containing Ser/Thr kinases in S. coelicolor were not involved in recognizing a product produced directly or indirectly by the Rpfs. Instead, the rapid germination of these strains implied that the activity of these kinases might inhibit germination. We also assessed the growth of the 3×ΔPASTA strain in liquid minimal medium, to determine whether it exhibited defects in vegetative growth compared with WT and the rpf null strain. Consistent with our germination results, growth of the 3×ΔPASTA strain was faster than either comparator strain (Fig. 5B). These results suggested that these Ser/Thr kinases may function to delay germination/growth, given the enhanced rates of both processes in the absence of these enzymes, and further indicated that the effect of the Rpfs on spore germination and vegetative growth was not a result of Rpf-dependent muropeptide signaling, at least through the PASTA domain-containing Ser/Thr kinases.
Figure 5.
PASTA domain Ser/Thr protein kinases function independently of the Rpfs to affect spore germination and growth. A, WT, rpf null (5×Δrpf), and PASTA Ser/Thr kinase null (3×ΔPASTA) spores were monitored for germination using light microscopy over the course of 8 h. Data are representative of three independent replicates (n ≥ 200 spores per strain per time point). B, growth profiles of the WT, rpf null, and Ser/Thr PASTA kinase null strains in new minimal medium with phosphate liquid medium. Data presented are the average of three independent replicates ± S.E.
Rpf activity is required for germination with alternative germinants
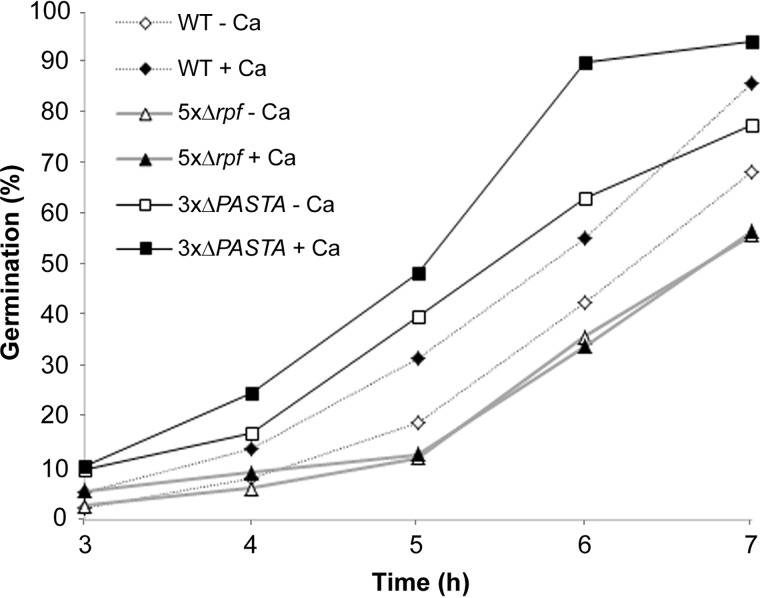
An alternative hypothesis to explain how Rpf enzymes promote germination is that their cell wall cleavage activities provide cells with an opportunity to insert new peptidoglycan, thus permitting cell expansion and growth. We predicted that if the role of the Rpfs was a physical one, we should be able to stimulate germination of the WT strain, but not an rpf mutant, by adding a known germinant. To test this, we incubated spores on minimal medium in the presence or absence of germination-promoting calcium chloride (45). We followed germination over a 7-h time course, and found that calcium chloride effectively stimulated germination of WT spores, but had little effect on the rpf null strain (Fig. 6). This supported the proposal that germination was inhibited at the outgrowth step in the rpf null strain (although we cannot exclude the possibility that calcium promotes germination by stimulating Rpf activity).
Figure 6.
Germination of the rpf null strain cannot be stimulated by a known germinant. Germination of the WT, rpf null (5×Δrpf), and 3×ΔPASTA mutant strains on minimal medium with and without 10 mm calcium chloride, was monitored over the course of 7 h using light microscopy. Data presented are representative of three independent replicates.
We also tested the effect of calcium chloride on the 3×ΔPASTA mutant strain, and found that like WT, spore germination was enhanced in the presence of calcium chloride (Fig. 6). This further implied that the activity of the PASTA-domain–containing Ser/Thr kinases was independent of Rpf activity in S. coelicolor.
Discussion
Here, we demonstrated that Rpfs were endo-acting lytic transglycosylases. We found that Rpf function was enhanced by their associated domains, with LytM domains appearing to promote peptidoglycan association but not peptidoglycan cleavage, and LysM domains increasing enzyme activity, either by enhancing peptidoglycan binding, as appears to be the case for RpfD, or by some other means, in the case of RpfA. Unexpectedly, we found Rpf function was not tied to an obvious signaling cascade in the streptomycetes. Instead our data were most consistent with a physical role for the Rpfs, where these enzymes functioned to structurally alter the germinating spore wall.
Role of LysM and LytM domains in Rpf activity
LysM-containing Rpfs represent one of the largest Rpf protein configurations in the actinobacteria, but the role of LysM domains in cell wall lytic enzymes is not universally conserved: some enzymes require these domains for activity, whereas others do not (46–49). We found that for RpfA and RpfD, deleting their LysM domains decreased their cell wall lytic activity. Interestingly, however, this domain only seemed to promote peptidoglycan association for RpfD, not RpfA, at least in vitro. LysM domains do not have catalytic activity of their own, and thus we suggest that the LysM domain may function to either increase the affinity of Rpf proteins for peptidoglycan (as for RpfD), or perhaps impact substrate orientation relative to the Rpf catalytic domain (RpfA). In their natural environments, LysM domains may further serve to anchor the Rpf proteins to the cell wall, thereby preventing indiscriminate peptidoglycan cleavage by these enzymes, and ensuring that the Rpfs remain a “private good” rather than a shared product in mixed microbial communities. In the mycobacteria, the Rpfs do not typically contain a peptidoglycan-binding domain. It is conceivable that the mycolic acid-based outer membrane encasing the peptidoglycan in these organisms may prevent the broad dispersal of the Rpfs, obviating the need for this additional LysM domain.
In contrast to the LysM domains, LytM domains are expected to have catalytic activity, and function in cleaving stem/cross-bridge peptides. Intriguingly, the RpfD-associated LytM domain does not appear to have peptidoglycan cleavage capabilities, at least in the assays conducted here. Despite the lack of LytM enzyme activity, removing the LytM domain, together with the LysM domain, significantly reduced the activity and peptidoglycan binding of RpfD beyond what had been observed by simply deleting the LysM domain. This suggests that the LytM domain further enhanced the affinity of RpfD for peptidoglycan through its peptide binding, although it is also possible that this domain increased Rpf enzyme activity (and/or peptidoglycan binding) through allosteric activation, as has been observed for EnvC and NlpD in their activation of AmiA, AmiB, and AmiC in E. coli (41).
Unlike EnvC and NlpD, however, the RpfD LytM domain has retained all active site and Zn2+-binding residues, suggesting that it may still be enzymatically competent. Some LytM metallopeptidases require additional processing for activation (40, 50). Such processing may occur upon secretion of RpfD to the Streptomyces cell surface, where the LytM domain then makes an enzymatic contribution to RpfD activity. Alternatively, this domain may be functionally silent in the context of the RpfD polypeptide, but may be processed in such a way that it acts independently of RpfD. RpfD is unusual among the Rpfs in S. coelicolor, in that rpfD transcript levels expression peaks later in development, as opposed to during germination as is the case for all other rpf genes with detectable transcription (13). It is therefore possible that RpfD function, and that of its LytM domain, may be more important at later stages of development than the other Rpfs in S. coelicolor. This would be consistent with the observation that LytM-containing Rpf proteins are found exclusively in the streptomycetes, and thus may function in aspects of development unique to these bacteria.
Specificity and redundancy in Rpf function
A key question is why multiple Rpfs are required for cellular resuscitation in many actinobacteria (13, 21). Our peptidoglycan cleavage assays did not reveal unique specificity for any individual Rpf class, although it is notable that the muropeptides released during cleavage of S. coelicolor peptidoglycan represented those that are most abundant in spore peptidoglycan (the relative proportion of different monomer and dimer products changes throughout development) (51). At this stage, however, we cannot exclude the possibility that these enzymes have differential specificity in vivo. Germination of Streptomyces spores occurs at the spore poles, and it is possible that the peptidoglycan architecture in these regions has specific modifications that were not captured at high levels in our peptidoglycan preparations. It is also possible that other proteins function to localize the Rpfs to specific sites within the cell wall of dormant spores. Future protein interaction and localization studies will help to resolve these questions.
Revising the model of Rpf function during germination
How the Rpfs promote resuscitation/germination is still being debated, 20 years after the discovery of these proteins. Do they function to liberate a signaling molecule that acts as a germinant? Or is their role more structural, where they permit cell expansion and new peptidoglycan incorporation through their cell wall cleavage activities?
Three lines of evidence support a peptidoglycan remodeling role for the Streptomyces Rpfs. One, we demonstrated that peptidoglycan-binding kinases, known to influence germination through a muropeptide-mediated signal transduction cascade in Bacillus (28) and to a lesser extent in Mycobacterium (32), are not associated with Rpf function in S. coelicolor. Instead, these kinases appear to negatively influence germination, based on the rapid germ tube outgrowth observed in their absence. Two, the endo-acting lytic transglycosylase activity of the Rpfs is more compatible with an architectural role than with a signaling role. Finally, a known germinant for Streptomyces (calcium chloride) stimulated germination of WT spores, but had no effect on the germination of an rpf null strain. The fact that alternative germinants could not substitute for the lack of Rpfs, suggests that the Rpfs may be universally required for efficient germination. It is, however, worth noting that it is not clear how calcium chloride promotes germination, and it will be interesting to determine whether its effects are mediated through Rpf activity, through other cell wall lytic enzyme(s) that act in conjunction with the Rpfs, or through some other means altogether.
In M. tuberculosis, an equivalent germination-promoting experiment (treating dormant cells with both an Rpf inhibitor and oleic acid, a known germinant) led to metabolic reactivation but delayed cellular outgrowth, again suggesting that the Rpf role may be more structural (52, 53). Taken together, the simplest explanation for these results would be that the Rpfs function to remodel the cell wall and promote cell expansion and growth after metabolic reactivation.
Our findings do not, however, definitively rule out an additional signaling role for the Rpfs. Indeed, a wide variety of muropeptides clearly enhance the resuscitation of Mycobacterium (24, 29, 54). In M. tuberculosis, RpfB acts synergistically in association with the endopeptidase RipA, and a product of their activity (a peptidoglycan-derived disaccharide-dipeptide) has been proposed to promote mycobacterial resuscitation (10, 54). In the streptomycetes, such a signaling phenomenon would require additional glycosidic enzymes, as the endo-lytic activity of the Rpfs would not allow for the generation of a disaccharide molecule. We suggest that muropeptide release may be a secondary effect of Rpf activity, and this could be accomplished either directly through the cleavage activity of Rpfs and any associated enzymes, or indirectly through cell growth and the associated peptidoglycan shedding that accompanies this process.
Experimental procedures
Bioinformatic analysis
The HMMER webserver (55) was used to search the UniProtKB database using the Rpf domain from RpfE in S. coelicolor to identify homologues of each Rpf configuration and their taxonomic distribution.
Bacterial strains and growth conditions
Bacterial strains used or created in this work are outlined in Table S1. S. coelicolor A3(2) strain M145 and its derivatives were grown at 30 °C on solid minimal medium (MM) or mannitol soya flour (MS) agar, with antibiotics to maintain plasmid selection where appropriate, or in new minimal medium with phosphate, as described by Kieser et al. (56). Growth curves were generated using dry weight. This involved transferring 1 ml of culture at a given time point, into pre-weighed tubes. The cells were then collected by centrifugation at full speed in a microcentrifuge for 5 min, after which the supernatant was removed. The remaining cell pellet was then dried at 60 °C for ∼2 days, after which the tubes were weighed again, allowing for calculation of the cell dry weight. All E. coli strains were grown at 37 °C on LB or nutrient agar (NA) plates (56), or in LB or super optimal broth liquid medium (57, 58) supplemented with antibiotics where appropriate to maintain plasmid selection.
Spore germination assay
To assess the germination efficiency of the different strains, spores were plated on MS agar overlaid with cellophane discs and incubated at 30 °C for up to 8 h. At the indicated time points, a 1 × 1-cm square was excised from the cellophane disc and examined using light microscopy. Images were acquired at ×1000 magnification using a Nikon Eclipse E600 microscope fitted with DS-Fi1 camera. Image capture was performed using Nikon NIS-Elements software. Spore germination was then assessed, scoring germinated spores (those possessing at least one germ tube) versus nongerminated spores, with a minimum of 200 spores being assessed per strain, at each time point, in at least three independent trials. Spore scoring was performed using the cell counter plug-in for ImageJ (59). To test the effects of Ca2+ on germination, spores were plated on MM agar with and without 10 mm CaCl2. Spore germination assays were then conducted as described above.
Protein overexpression and purification
RpfsA–E, excluding their SignalP-predicted signal peptide, were overexpressed and purified as N terminally His-tagged proteins, as described by Sexton et al. (13). The equivalent region for each RpfA and RpfD variant (coding sequence, minus signal peptide) was amplified using the primers outlined in Table S2. Overlap extension PCR (60) with the primers described in Table S2 was used to generate rpfDΔLysM. Other mutants were truncations of either RpfA or RpfD, and were generated using the primers indicated in Table S2. Digested PCR products were cloned into the BamHI and NdeI restriction sites of pET15b (Novagen) (Table S1). Construct integrity was confirmed by sequencing using the T7 promoter and terminator primers (Table S2). Each plasmid was freshly transformed into E. coli Rosetta 2 cells (Table S2) prior to overexpression. Transformants were grown overnight at 37 °C in 5 ml of LB liquid medium supplemented with ampicillin and chloramphenicol. These overnight cultures were used to inoculate 500 ml of LB medium, again supplemented with ampicillin and chloramphenicol. Cultures were grown at 37 °C until they reached an optical density at 600 nm (OD600) of 0.6-1.0 (depending on the Rpf variant), at which point 1 mm isopropyl β-d-thiogalactopyranoside was added to induce protein overexpression. Conditions for overexpression are summarized in Table S3. Overexpression of RpfDΔLysM was attempted at an initial OD600 of 0.4-1.2, using 0.25-2 mm isopropyl β-d-thiogalactopyranoside, and induced cultures were grown for 1.5 h to overnight at 16, 30, or 37 °C. Overexpression was also attempted using in vitro translation with the PURExpress kit (New England Biolabs) following the manufacturer's recommendations. None of the conditions tested yielded the desired protein.
For those proteins where overexpression was observed, cell pellets were resuspended in 5 ml of lysis buffer (50 mm NaH2PO4, 300 mm NaCl, 10 mm imidazole, pH 8.0) containing cOmplete Mini EDTA-free protease inhibitor (Roche Applied Science) and lysed using the Constant Systems TS-2 0.75 kW cell disruptor. The lysate was centrifuged at 10,000 × g for 20 min at 4 °C to remove insoluble debris. The clarified lysate was incubated with 1 ml of nickel-nitriloacetic acid-agarose (Thermo) for 1 h at 4 °C before being applied to a chromatography column. The column was washed twice with 5 ml of buffer (50 mm NaH2PO4, 300 mm NaCl, pH 8.0) containing 20 and 50 mm imidazole before His6-tagged proteins were eluted sequentially with buffers containing 100 mm, 250 mm, 500 mm, and 1 m imidazole. The success of protein overexpression and the quality of protein purification was assessed by separating purified proteins (and their accompanying washes and crude soluble and insoluble fractions) on a 10% Tricine polyacrylamide gel (61) and staining with Coomassie Brilliant Blue. Protein concentrations were determined using a Bradford assay (62), with BSA as a standard. Each Rpf was dialyzed into storage buffer (50 mm NaH2PO4, 10% glycerol, pH 8) overnight to remove imidazole. Proteins were stored at 4 °C for a maximum of 24 h before all assays.
Enzyme activity assays
Quantitative Rpf activity assays
The EnzChek lysozyme assay kit (Molecular Probes) was used to assess the ability of the different RpfA and RpfD variants to cleave fluorescein-labeled M. luteus cell walls, as described previously (13). Briefly, 1 nmol of purified Rpf protein was added to each reaction, and the volume was brought to 50 μl with storage buffer before adding 50 μl of fluorescein-labeled M. luteus cell wall substrate. One pmol of lysozyme was used as a positive control, whereas a reaction without protein served as a negative control. Reactions were set up in black 96-well–plates (Thermo). Fluorescein release was measured after 1 h using a Cytation 5 plate reader (BioTek) with an excitation wavelength of 494 nm and emission wavelength of 521 nm. Assays were conducted in technical triplicate, using at least two independent protein preparations.
Isolation and purification of peptidoglycan
Insoluble peptidoglycan for use in the enzymatic assays was isolated from overnight cultures of S. coelicolor using the boiling SDS protocol and purification by enzyme treatment (amylase, DNase, RNase, and Pronase), as described by Clarke (63); as Gram-positive bacteria, both M. luteus and S. coelicolor produce peptidoglycan with limited 1,6-anhydromuramoyl content (64). O-Acetyl groups were removed by incubating peptidoglycan in 20 mm NaOH at room temperature overnight, and insoluble peptidoglycan was isolated by centrifugation (9,000 × g, 30 min, room temperature) and washed with water at least three times. Teichoic acids were removed by extracting the peptidoglycan with 10% TCA overnight at room temperature and peptidoglycan was washed four times in water, frozen, lyophilized, and stored at −20 °C.
Peptidoglycan-binding assays
A solution containing 1 mg/ml of purified S. coelicolor peptidoglycan in 10 mm Tris, pH 7, was sonicated continuously on ice for 5 min (to provide a relatively even suspension). Two nanomoles of purified Rpf protein were mixed with 150 μl of the peptidoglycan solution and the volume was brought to 300 μl using 10 mm Tris, pH 7. Reactions were incubated at 4 °C for 2 h with gentle shaking. The peptidoglycan-bound protein was separated from unbound protein by centrifugation at 15,000 × g at 4 °C for 30 min, with the peptidoglycan-bound protein associated with the pellet, and the unbound protein present in the supernatant. The supernatant was then transferred to a new tube. All reactions were brought to 400 μl using sample loading buffer. Twenty microliters of each fraction were separated using 10% Tricine-PAGE and stained with Coomassie Brilliant Blue. ImageJ (59) was used to quantify band intensity.
[18O]H2O-based assay to differentiate between hydrolases and lytic transglycosylases
The [18O]H2O-based assays were conducted as described by Herlihey et al. (42) using M. luteus and S. coelicolor peptidoglycan as substrates. For each, peptidoglycan was resuspended to a final concentration of 1.4 mg/ml in [18O]H2O and briefly sonicated to homogenize the suspension. To start reactions, 1 nmol of purified Rpf protein was mixed with 100 μl of resuspended peptidoglycan in [18O]H2O, and the reaction was brought to 200 μl with Rpf storage buffer. Reactions were incubated at 37 °C for 9.5 h with gentle shaking and then stopped by rapid freezing. Mutanolysin (1.1 nmol) was used as a positive control, whereas reactions without added protein were used as negative controls. Reaction mixtures were thawed and soluble reaction products were separated from insoluble peptidoglycan by centrifugation (15,000 × g, 15 min, 4 °C) prior to analysis by LC-Q-TOF-MS. The insoluble fractions were washed four to five times with 200-μl volumes of water and recovered each time by centrifugation (15,000 × g, 6 min, room temperature). The washed peptidoglycan pellets were resuspended in 0.1 mm potassium phosphate buffer, pH 6.2, and solubilized by mutanolysin (1.1 µm, final concentration) prior to LC-Q-TOF-MS analysis. LC-Q-TOF-MS was performed by injecting samples into an Agilent 1260 Infinity liquid chromatograph interfaced to an Agilent 6540 UHD accurate Mass Q-TOF mass spectrometer as described previously (42). MS analyses were conducted at the Mass Spectrometry Facility at the University of Guelph.
Data availability
All data are shown, apart from the tandem Q-TOF-MS analysis of each parent ion (Tables 2 and 3), the data for which are available upon request, by emailing Anthony Clarke (ajclarke@wlu.ca) at the University of Guelph/Wilfred Laurier University.
Supplementary Material
Acknowledgments
We thank Dr. Mark Buttner for the gift of the 3×PASTA mutant strain, and Dr. Dyanne Brewer of the University of Guelph Mass Spectrometry Facility for helpful discussions.
This article contains supporting information.
Author contributions—D. L. S., F. A. H., A. S. B., D. A. C., E. S., A. J. C., and M. A. E. formal analysis; D. L. S., F. A. H., A. S. B., D. A. C., and E. S. investigation; D. L. S., F. A. H., A. S. B., and M. A. E. visualization; D. L. S., F. A. H., A. S. B., D. A. C., and E. S. methodology; D. L. S., A. J. C., and M. A. E. writing-original draft; D. L. S., F. A. H., A. S. B., D. A. C., E. S., A. J. C., and M. A. E. writing-review and editing; F. A. H., A. S. B., and E. S. validation; A. J. C. and M. A. E. supervision; A. J. C. and M. A. E. project administration; M. A. E. conceptualization; M. A. E. funding acquisition.
Funding and additional information—This work was supported by Canadian Institutes for Health Research Grant MOP-93635 (to M. A. E.), Natural Sciences and Engineering Research Council Grant RGPIN 3215-11 (to A. J. C.), and Canada Graduate Scholarship and Ontario Graduate Scholarships (to D. L. S. and A. S. B., respectively).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- VBNC
- viable but not culturable
- LC/MS
- liquid chromatography/mass spectrometry
- MurNAc
- N-acetylmuramic acid
- OD
- optical density
- Rpf
- resuscitation promoting factor
- PASTA
- penicillin-binding protein and Ser/Thr kinase associated
- NA
- nutrient agar
- MM
- minimal medium
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
References
- 1. Wood T. K., Knabel S. J., and Kwan B. W. (2013) Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 79, 7116–7121 10.1128/AEM.02636-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Swiercz J. P., and Elliot M. A. (2011) Streptomyces sporulation. in Bacterial spores: current research and applications, 1–18, Horizon Scientific Press, Poole, United Kingdom [Google Scholar]
- 3. Oliver J. D. (2010) Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 34, 415–425 10.1111/j.1574-6976.2009.00200.x [DOI] [PubMed] [Google Scholar]
- 4. Vollmer W., Blanot D., and De Pedro M. A. (2008) Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167 10.1111/j.1574-6976.2007.00094.x [DOI] [PubMed] [Google Scholar]
- 5. Cava F., and de Pedro M. A. (2014) Peptidoglycan plasticity in bacteria: Emerging variability of the murein sacculus and their associated biological functions. Curr. Opin. Microbiol. 18, 46–53 10.1016/j.mib.2014.01.004 [DOI] [PubMed] [Google Scholar]
- 6. Vollmer W., Joris B., Charlier P., and Foster S. (2008) Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32, 259–286 10.1111/j.1574-6976.2007.00099.x [DOI] [PubMed] [Google Scholar]
- 7. Scheurwater E., Reid C. W., and Clarke A. J. (2008) Lytic transglycosylases: bacterial space-making autolysins. Int. J. Biochem. Cell Biol. 40, 586–591 10.1016/j.biocel.2007.03.018 [DOI] [PubMed] [Google Scholar]
- 8. Cohen-Gonsaud M., Barthe P., Bagnéris C., Henderson B., Ward J., Roumestand C., and Keep N. H. (2005) The structure of a resuscitation-promoting factor domain from Mycobacterium tuberculosis shows homology to lysozymes. Nat. Struct. Mol. Biol. 12, 270–273 10.1038/nsmb905 [DOI] [PubMed] [Google Scholar]
- 9. Mukamolova G. V., Murzin A. G., Salina E. G., Demina G. R., Kell D. B., Kaprelyants A. S., and Young M. (2006) Muralytic activity of Micrococcus luteus Rpf and its relationship to physiological activity in promoting bacterial growth and resuscitation. Mol. Microbiol. 59, 84–98 10.1111/j.1365-2958.2005.04930.x [DOI] [PubMed] [Google Scholar]
- 10. Hett E. C., Chao M. C., Deng L. L., and Rubin E. J. (2008) A mycobacterial enzyme essential for cell division synergizes with resuscitation-promoting factor. PLoS Pathog. 4, e1000001 10.1371/journal.ppat.1000001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haiser H. J., Yousef M. R., and Elliot M. A. (2009) Cell wall hydrolases affect germination, vegetative growth, and sporulation in Streptomyces coelicolor. J. Bacteriol. 191, 6501–6512 10.1128/JB.00767-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ruggiero A., Tizzano B., Pedone E., Pedone C., Wilmanns M., and Berisio R. (2009) Crystal structure of the resuscitation-promoting factor ΔDUFRpfB from M. tuberculosis. J. Mol. Biol. 385, 153–162 10.1016/j.jmb.2008.10.042 [DOI] [PubMed] [Google Scholar]
- 13. Sexton D. L., St-Onge R. J., Haiser H. J., Yousef M. R., Brady L., Gao C., Leonard J., and Elliot M. A. (2015) Resuscitation-promoting factors are cell wall-lytic enzymes with important roles in the germination and growth of Streptomyces coelicolor. J. Bacteriol. 197, 848–860 10.1128/JB.02464-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cohen-Gonsaud M., Keep N. H., Davies A. P., Ward J., Henderson B., and Labesse G. (2004) Resuscitation-promoting factors possess a lysozyme-like domain. Trends Biochem. Sci. 29, 7–10 10.1016/j.tibs.2003.10.009 [DOI] [PubMed] [Google Scholar]
- 15. Telkov M. V., Demina G. R., Voloshin S. A., Salina E. G., Dudik T. V., Stekhanova T. N., Mukamolova G. V., Kazaryan K. A., Goncharenko A. V., Young M., and Kaprelyants A. S. (2006) Proteins of the Rpf (resuscitation promoting factor) family are peptidoglycan hydrolases. Biochem. 71, 414–422 10.1134/s0006297906040092 [DOI] [PubMed] [Google Scholar]
- 16. Mukamolova G. V., Kaprelyants A. S., Young D. I., Young M., and Kell D. B. (1998) A bacterial cytokine. Proc. Natl. Acad. Sci. U. S. A. 95, 8916–8921 10.1073/pnas.95.15.8916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ravagnani A., Finan C. L., and Young M. (2005) A novel firmicute protein family related to the actinobacterial resuscitation-promoting factors by non-orthologous domain displacement. BMC Genomics 6, 1–14 10.1186/1471-2164-6-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Machowski E. E., Senzani S., Ealand C., and Kana B. D. (2014) Comparative genomics for mycobacterial peptidoglycan remodelling enzymes reveals extensive genetic multiplicity. BMC Microbiol. 14, 75 10.1186/1471-2180-14-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hartmann M., Barsch A., Niehaus K., Pühler A., Tauch A., and Kalinowski J. (2004) The glycosylated cell surface protein Rpf2, containing a resuscitation-promoting factor motif, is involved in intercellular communication of Corynebacterium glutamicum. Arch. Microbiol. 182, 299–312 10.1007/s00203-004-0713-1 [DOI] [PubMed] [Google Scholar]
- 20. Downing K. J., Mischenko V. V., Shleeva M. O., Young D. I., Young M., Kaprelyants A. S., Apt A. S., and Mizrahi V. (2005) Mutants of Mycobacterium tuberculosis lacking three of the five rpf-like genes are defective for growth in vivo and for resuscitation in vitro. Infect. Immun. 73, 3038–3043 10.1128/IAI.73.5.3038-3043.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kana B. D., Gordhan B. G., Downing K. J., Sung N., Vostroktunova G., Machowski E. E., Tsenova L., Young M., Kaprelyants A., Kaplan G., and Mizrahi V. (2008) The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol. Microbiol. 67, 672–684 10.1111/j.1365-2958.2007.06078.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Russell-Goldman E., Xu J., Wang X., Chan J., and Tufariello J. A. M. (2008) A Mycobacterium tuberculosis Rpf double-knockout strain exhibits profound defects in reactivation from chronic tuberculosis and innate immunity phenotypes. Infect. Immun. 76, 4269–4281 10.1128/IAI.01735-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Keep N. H., Ward J. M., Robertson G., Cohen-Gonsaud M., and Henderson B. (2006) Bacterial resuscitation factors: revival of viable but non-culturable bacteria. Cell. Mol. Life Sci. 63, 2555–2559 10.1007/s00018-006-6188-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nikitushkin V. D., Demina G. R., Shleeva M. O., and Kaprelyants A. S. (2013) Peptidoglycan fragments stimulate resuscitation of “non-culturable” mycobacteria. Antonie van Leeuwenhoek 103, 37–46 10.1007/s10482-012-9784-1 [DOI] [PubMed] [Google Scholar]
- 25. Dworkin J., and Shah I. M. (2010) Exit from dormancy in microbial organisms. Nat. Rev. Microbiol. 8, 890–896 10.1038/nrmicro2453 [DOI] [PubMed] [Google Scholar]
- 26. Setlow P. (2014) Germination of spores of Bacillus species: what we know and do not know. J. Bacteriol. 196, 1297–1305 10.1128/JB.01455-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Keep N. H., Ward J. M., Cohen-Gonsaud M., and Henderson B. (2006) Wake up! peptidoglycan lysis and bacterial non-growth states. Trends Microbiol. 14, 271–276 10.1016/j.tim.2006.04.003 [DOI] [PubMed] [Google Scholar]
- 28. Shah I. M., Laaberki M. H., Popham D. L., and Dworkin J. (2008) A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135, 486–496 10.1016/j.cell.2008.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Turapov O., Loraine J., Jenkins C. H., Barthe P., McFeely D., Forti F., Ghisotti D., Hesek D., Lee M., Bottrill A. R., Vollmer W., Mobashery S., Cohen-Gonsaud M., and Mukamolova G. V. (2015) The external PASTA domain of the essential serine/threonine protein kinase PknB regulates mycobacterial growth. Open Biol. 5, 150025 10.1098/rsob.150025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Squeglia F., Marchetti R., Ruggiero A., Lanzetta R., Marasco D., Dworkin J., Petoukhov M., Molinaro A., Berisio R., and Silipo A. (2011) Chemical basis of peptidoglycan discrimination by PrkC, a key kinase involved in bacterial resuscitation from dormancy. J. Am. Chem. Soc. 133, 20676–20679 10.1021/ja208080r [DOI] [PubMed] [Google Scholar]
- 31. Yeats C., Finn R. D., and Bateman A. (2002) The PASTA domain: A β-lactam-binding domain. Trends Biochem. Sci. 27, 438 10.1016/S0968-0004(02)02164-3 [DOI] [PubMed] [Google Scholar]
- 32. Mir M., Asong J., Li X., Cardot J., Boons G. J., and Husson R. N. (2011) The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog. 7, e1002182 10.1371/journal.ppat.1002182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ruggiero A., Squeglia F., Romano M., Vitagliano L., De Simone A., and Berisio R. (2017) Structure and dynamics of the multi-domain resuscitation promoting factor RpfB from Mycobacterium tuberculosis. J. Biomol. Struct. Dyn. 35, 1322–1330 10.1080/07391102.2016.1182947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ruggiero A., Squeglia F., Romano M., Vitagliano L., De Simone A., and Berisio R. (2016) The structure of resuscitation promoting factor B from M. tuberculosis reveals unexpected ubiquitin-like domains. Biochim. Biophys. Acta 1860, 445–451 10.1016/j.bbagen.2015.11.001 [DOI] [PubMed] [Google Scholar]
- 35. Bateman A., Holden M. T. G., and Yeats C. (2005) The G5 domain: a potential N-acetylglucosamine recognition domain involved in biofilm formation. Bioinformatics. 21, 1301–1303 10.1093/bioinformatics/bti206 [DOI] [PubMed] [Google Scholar]
- 36. Mesnage S., Dellarole M., Baxter N. J., Rouget J. B., Dimitrov J. D., Wang N., Fujimoto Y., Hounslow A. M., Lacroix-Desmazes S., Fukase K., Foster S. J., and Williamson M. P. (2014) Molecular basis for bacterial peptidoglycan recognition by LysM domains. Nat. Commun. 5, 4269 10.1038/ncomms5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Firczuk M., and Bochtler M. (2007) Folds and activities of peptidoglycan amidases. FEMS Microbiol. Rev. 31, 676–691 10.1111/j.1574-6976.2007.00084.x [DOI] [PubMed] [Google Scholar]
- 38. Grabowska M., Jagielska E., Czapinska H., Bochtler M., and Sabala I. (2015) High resolution structure of an M23 peptidase with a substrate analogue. Sci. Rep. 5, 14833 10.1038/srep14833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 8, 785–786 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 40. Odintsov S. G., Sabala I., Marcyjaniak M., and Bochtler M. (2004) Latent LytM at 1.3 Å resolution. J. Mol. Biol. 335, 775–785 10.1016/j.jmb.2003.11.009 [DOI] [PubMed] [Google Scholar]
- 41. Uehara T., Parzych K. R., Dinh T., and Bernhardt T. G. (2010) Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 29, 1412–1422 10.1038/emboj.2010.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Herlihey F. A., Moynihan P. J., and Clarke A. J. (2014) The essential protein for bacterial flagella formation FlgJ functions as a β-N-acetylglucosaminidase. J. Biol. Chem. 289, 31029–31042 10.1074/jbc.M114.603944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schleifer K. H., and Kandler O. (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477 10.1128/MMBR.36.4.407-477.1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hempel A. M., Cantlay S., Molle V., Wang S.-B., Naldrett M. J., Parker J. L., Richards D. M., Jung Y.-G., Buttner M. J., and Flardh K. (2012) The Ser/Thr protein kinase AfsK regulates polar growth and hyphal branching in the filamentous bacteria Streptomyces. Proc. Natl. Acad. Sci. U.S.A. 109, E2371–E2379 10.1073/pnas.1207409109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eaton D., and Ensign J. C. (1980) Streptomyces viridochromogenes spore germination initiated by calcium ions. J. Bacteriol. 143, 377–382 10.1128/JB.143.1.377-382.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mukamolova G. V., Turapov O. A., Kazarian K., Telkov M., Kaprelyants A. S., Kell D. B., and Young M. (2002) The rpf gene of Micrococcus luteus encodes an essential secreted growth factor. Mol. Microbiol. 46, 611–621 10.1046/j.1365-2958.2002.03183.x [DOI] [PubMed] [Google Scholar]
- 47. Steen A., Buist G., Horsburgh G. J., Venema G., Kuipers O. P., Foster S. J., and Kok J. (2005) AcmA of Lactococcus lactis is an N-acetylglucosaminidase with an optimal number of LysM domains for proper functioning. FEBS J. 272, 2854–2868 10.1111/j.1742-4658.2005.04706.x [DOI] [PubMed] [Google Scholar]
- 48. Wong J. E. M. M., Alsarraf H. M. A. B., Kaspersen J. D., Pedersen J. S., Stougaard J., Thirup S., and Blaise M. (2014) Cooperative binding of LysM domains determines the carbohydrate affinity of a bacterial endopeptidase protein. FEBS J. 281, 1196–1208 10.1111/febs.12698 [DOI] [PubMed] [Google Scholar]
- 49. Eckert C., Lecerf M., Dubost L., Arthur M., and Mesnage S. (2006) Functional analysis of AtlA, the major N-acetylglucosaminidase of Enterococcus faecalis. J. Bacteriol. 188, 8513–8519 10.1128/JB.01145-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Firczuk M., Mucha A., and Bochtler M. (2005) Crystal structures of active LytM. J. Mol. Biol. 354, 578–590 10.1016/j.jmb.2005.09.082 [DOI] [PubMed] [Google Scholar]
- 51. van der Aart L. T., Spijksma G. K., Harms A., Vollmer W., Hankemeier T., and van Wezel G. P. (2018) High-resolution analysis of the peptidoglycan composition in Streptomyces coelicolor. J. Bacteriol. 200, e00290–18 10.1128/JB.00290-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Demina G. R., Makarov V. A., Nikitushkin V. D., Ryabova O. B., Vostroknutova G. N., Salina E. G., Shleeva M. O., Goncharenko A. V., and Kaprelyants A. S. (2009) Finding of the low molecular weight inhibitors of resuscitation promoting factor enzymatic and resuscitation activity. PLoS ONE 4, e8174 10.1371/journal.pone.0008174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shleeva M., Goncharenko A., Kudykina Y., Young D., Young M., and Kaprelyants A. (2013) Cyclic AMP-dependent resuscitation of dormant mycobacteria by exogenous free fatty acids. PLoS ONE 8, e82914 10.1371/journal.pone.0082914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nikitushkin V. D., Demina G. R., Shleeva M. O., Guryanova S. V., Ruggiero A., Berisio R., and Kaprelyants A. S. (2015) A product of RpfB and RipA joint enzymatic action promotes the resuscitation of dormant mycobacteria. FEBS J. 282, 2500–2511 10.1111/febs.13292 [DOI] [PubMed] [Google Scholar]
- 55. Finn R. D., Clements J., Arndt W., Miller B. L., Wheeler T. J., Schreiber F., Bateman A., and Eddy S. R. (2015) HMMER web server: 2015 update. Nucleic Acids Res. 43, W30–W38 10.1093/nar/gkv397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kieser T., Bibb M. J., Buttner M. J., Chater K. F., and Hopwood D. A. (2000) Practical Streptomyces Genetics, The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 57. Bertani G. (1951) Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62, 293–300 10.1128/JB.62.3.293-300.1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580 10.1016/S0022-2836(83)80284-8 [DOI] [PubMed] [Google Scholar]
- 59. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Heckman K. L., and Pease L. R. (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2, 924–932 10.1038/nprot.2007.132 [DOI] [PubMed] [Google Scholar]
- 61. Schägger H. (2006) Tricine–SDS-PAGE. Nat. Protoc. 1, 16–22 10.1038/nprot.2006.4 [DOI] [PubMed] [Google Scholar]
- 62. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 63. Clarke A. J. (1993) Compositional analysis of peptidoglycan by high-performance anion-exchange chromatography. Anal. Biochem. 212, 344–350 10.1006/abio.1993.1339 [DOI] [PubMed] [Google Scholar]
- 64. Desmarais S. M., De Pedro M. A., Cava F., and Huang K. C. (2013) Peptidoglycan at its peaks: How chromatographic analyses can reveal bacterial cell wall structure and assembly. Mol. Microbiol 89, 1–13 10.1111/mmi.12266 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are shown, apart from the tandem Q-TOF-MS analysis of each parent ion (Tables 2 and 3), the data for which are available upon request, by emailing Anthony Clarke (ajclarke@wlu.ca) at the University of Guelph/Wilfred Laurier University.