Abstract
Niemann–Pick type C (NPC) disease is a lysosomal storage disorder arising from mutations in the cholesterol-trafficking protein NPC1 (95%) or NPC2 (5%). These mutations result in accumulation of low-density lipoprotein-derived cholesterol in late endosomes/lysosomes, disruption of endocytic trafficking, and stalled autophagic flux. Additionally, NPC disease results in sphingolipid accumulation, yet it is unique among the sphingolipidoses because of the absence of mutations in the enzymes responsible for sphingolipid degradation. In this work, we examined the cause for sphingosine and sphingolipid accumulation in multiple cellular models of NPC disease and observed that the activity of sphingosine kinase 1 (SphK1), one of the two isoenzymes that phosphorylate sphingoid bases, was markedly reduced in both NPC1 mutant and NPC1 knockout cells. Conversely, SphK1 inhibition with the isotype-specific inhibitor SK1-I in WT cells induced accumulation of cholesterol and reduced cholesterol esterification. Of note, a novel SphK1 activator (SK1-A) that we have characterized decreased sphingoid base and complex sphingolipid accumulation and ameliorated autophagic defects in both NPC1 mutant and NPC1 knockout cells. Remarkably, in these cells, SK1-A also reduced cholesterol accumulation and increased cholesterol ester formation. Our results indicate that a SphK1 activator rescues aberrant cholesterol and sphingolipid storage and trafficking in NPC1 mutant cells. These observations highlight a previously unknown link between SphK1 activity, NPC1, and cholesterol trafficking and metabolism.
Keywords: sphingosine kinase, cholesterol, Niemann–Pick type C, sphingolipids, lysosomal storage disease, genetic disorder, lipid metabolism, neurodegeneration, sphingolipid, sphingosine kinase (SphK), sphingosine-1-phosphate (S1P), NPC1
Introduction
NPC disease is caused by genetic mutations in the cholesterol transport proteins NPC1 and NPC2, with the vast majority of cases attributed to NPC1 mutations (95%) (1–4). Whereas NPC pathology often first presents systemically with hepatosplenomegaly, morbidity and mortality are almost entirely attributed to the development of a complex progressive neurodegeneration (5). It has been suggested that NPC proteins act sequentially in late endosomes/lysosomes (LE/L), where the soluble NPC2 protein binds unesterified cholesterol and transfers it to the luminal sterol-binding site of membrane-associated NPC1, which then facilitates its transport out of the LE/L compartment (6, 7). Aberrant functions of NPC proteins lead to excessive accumulation of unesterified cholesterol in the liver and spleen and also accumulation of sphingolipids, particularly in the brain (8). It was suggested that cholesterol accumulation can cause a secondary reduction in the activity of acidic sphingomyelinase that may explain accumulation of sphingomyelin (9). However, it is still not clear why sphingosine, the final breakdown product of all sphingolipids in the lysosome, also accumulates LE/L in NPC disease (10–15). Intriguingly, sphingosine is the first detectable lipid that accumulates upon pharmacological inhibition of NPC1 (10) and is the only lipid that can induce an NPC phenotype when added exogenously at the levels observed in NPC cell models (11). Sphingolipid catabolism depends on egress of sphingosine from lysosomes and phosphorylation to sphingosine-1-phosphate (S1P), and then it can be irreversibly degraded by S1P lyase (16) or dephosphorylated to sphingosine that is reutilized for synthesis of ceramide and more complex sphingolipids in the ER (17). SphK1 is a cytosolic enzyme, and multiple growth factors, including vascular endothelial growth factor (VEGF), activate and translocate it to the plasma membrane, leading to increased S1P formation (18). This S1P in turn is exported from cells by transporters and signals by binding to S1PR1-5 to regulate many physiologic responses (19). It was previously suggested that loss of Purkinje neurons in NPC is caused by defective SphK activity because of reduced levels of VEGF and implicated SphK1 in the neuropathogenesis of NPC (20). Inhibition or deletion of SphK1 has also been shown to impair endocytic trafficking and accumulation of sphingosine in the LE/L, similar to the cellular phenotype of NPC (21, 22).
Here, we showed that accumulation of sphingosine in several types of NPC1 mutant cells correlated with decreased SphK1 activity. Treating NPC1 mutant cells with a novel SphK1 activator that we identified reduced sphingosine accumulation, ameliorated autophagic defects, and restored the impaired cholesterol trafficking. Conversely, inhibition of SphK1 in WT cells with an isozyme-specific inhibitor increased accumulation of cholesterol. Our results highlight a link between lysosomal cholesterol accumulation and SphK1 activity and suggest that a SphK1 activator could be an attractive therapeutic option in NPC1 disease.
Results
NPC1MUTANT fibroblasts have defects in sphingoid base phosphorylation
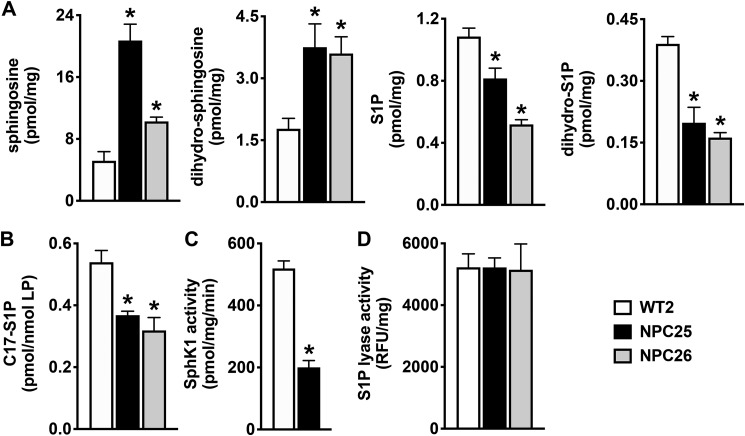
In agreement with previous studies demonstrating that in addition to cholesterol, sphingosine also accumulates in NPC disease (10, 11, 15), we found that sphingosine levels were elevated in NPC1 mutant fibroblasts derived from well-characterized patients with a wide disease spectrum (NPC25 and NPC26) compared with control fibroblasts from a healthy individual (Fig. 1A). In addition, dihydrosphingosine, an intermediate in de novo sphingolipid biosynthesis, also accumulated in these mutant fibroblasts. However, despite accumulation of these sphingoid bases, S1P and dihydro-S1P levels were reduced (Fig. 1A). As this could result from either decreased sphingoid base phosphorylation by sphingosine kinases (SphKs) or increased degradation by S1P lyase, we next determined their enzymatic activities. Sphingosine kinase activity was assessed by short incubation of cells with C17-sphingosine, and levels of C17-S1P were quantified by liquid chromatography, electrospray ionization–tandem MS (LC-ESI-MS/MS). Formation of C17-S1P was greatly reduced in the mutant fibroblasts compared with WT cells (Fig. 1B). Furthermore, an in vitro isoform-specific assay demonstrated that SphK1 enzymatic activity was significantly reduced in the mutant fibroblasts (Fig. 1C). In contrast, S1P lyase activity was not significantly different in the NPC1 mutant cells compared with WT (Fig. 1D). Taken together, these results suggest that impaired SphK1 activity leads to increased levels of sphingoid bases.
Figure 1.
Sphingosine kinase activity is attenuated in NPC1 mutant fibroblasts. A, sphingolipids were extracted from normal human fibroblasts (WT2) and NPC1 mutant fibroblasts (NPC25, NPC26), and levels of sphingosine, dihydrosphingosine, S1P, and dihydro-S1P measured by LC-ESI-MS/MS. B, in vivo SphK activity in WT and NPC1 mutant fibroblasts was determined by LC-ESI-MS/MS quantification of conversion of d17:1 sphingosine (1 μm) to C17-S1P after 30 min. Levels were normalized to total lipid phosphates. C, isotype-specific enzymatic activity of SphK1 was determined in fibroblast lysates. D, S1P lyase enzymatic activity was determined in fibroblast lysates by fluorometric assays and normalized to total protein. Data are representative of at least three independent experiments and presented as mean ± S.D. (error bars) of three biological replicates. *, p < 0.05 compared with WT2 by one-way ANOVA (A, B, and D) with Dunnett's post hoc test or by two-tailed Student's t test (C).
Identification of a novel SphK1 isoform-specific activator
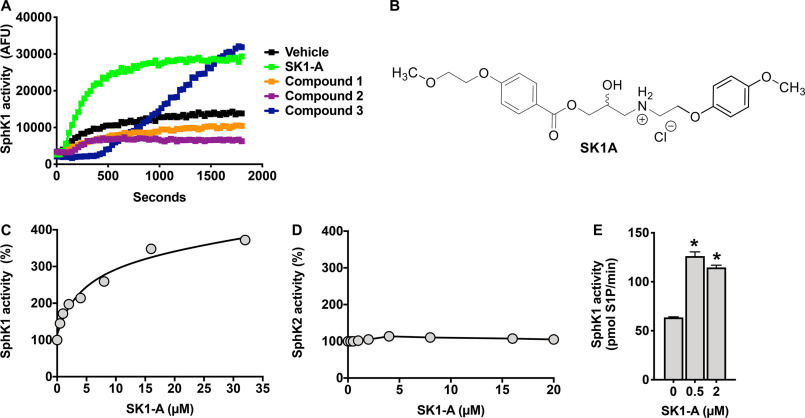
A previous study suggested that pathogenesis in NPC neurons resulted from defective SphK1 activity due to impaired VEGF levels and that correction of this activity by VEGF can reduce NPC pathological changes (20), suggesting that enhancing SphK1 activity could be a potential therapeutic intervention for this disorder. Therefore, it was important to identify a small molecule capable of increasing SphK1 activity. To this end, we used a high-throughput fluorescence assay to screen a chemical library with NBD-sphingosine as substrate that was used previously to identify a new inhibitor of SphK1 (23). In this screen, we discovered several compounds that increased SphK1 activity (data not shown; see Fig. 2A). However, only one compound, 2-hydroxy-3-{[4-(2-methoxyethoxy)benzoyl]oxy}-N-[2-(4-methoxyphenoxy)ethyl]propan-1-aminium, hereafter referred to as SK1-A (Fig. 2B), consistently stimulated SphK1 activity. In contrast, compound 3 in further assays with recombinant SphK1 did not increase its enzymatic activity. A more in-depth kinetic analysis of the effects of SK1-A resulted in an estimated half-maximal activation constant (AC50) of 4.5 µm for SphK1 (Fig. 2C), with a complete absence of any effects on SphK2 activity (Fig. 2D). Furthermore, with sphingosine as substrate, maximum activation of SphK1 was observed at a concentration as low as 0.5 µm (Fig. 2E). Moreover, SK1-A enhanced endogenous SphK activity in WT but not in SphK1 knockout mouse embryonic fibroblasts (Fig. S1). Thus, we next determined whether SK1-A could stimulate SphK1 activity in NPC1 mutant cells.
Figure 2.
Identification of a SphK1 activator. A, SK1-A was identified as an activator of SphK1 using a high-throughput fluorescent screening assay with recombinant SphK1 and NBD-sphingosine as a substrate as described previously (62). Data are time-resolved fluorescence emission of NBD-sphingosine (35 μm) in the absence or presence of the indicated compounds. AFU, arbitrary fluorescence units. B, structure of SK1-A. C and D, SK1-A concentration-dependent effect on recombinant SphK1 (C) and SphK2 (D) activity. C, SK1-A stimulated SphK1 with an estimated AC50 of 4.5 μm. SK1-A did not stimulate SphK2 activity. E, effect of SK1-A at the indicated concentrations on in vitro SphK1 activity with sphingosine (0.5 μm) as substrate. Data are representative of at least three independent experiments and presented as mean ± S.D. (error bars) of three biological replicates. *, p < 0.05 compared with vehicle, determined by one-way ANOVA with Dunnett's post hoc test.
SK1-A increases sphingosine phosphorylation and reduces autophagic defects in NPC1 mutant fibroblasts
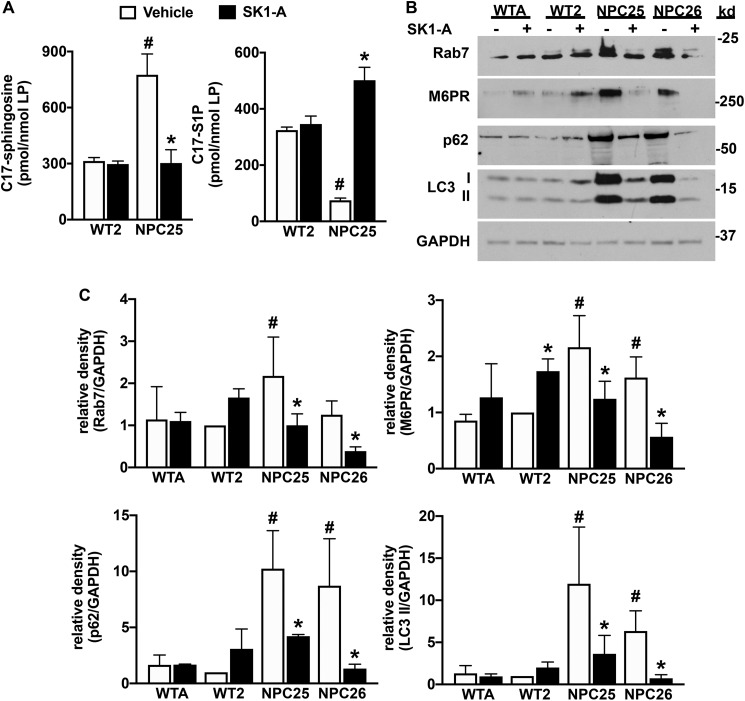
Next, cells were incubated with SK1-A, and cellular SphK activity was determined. As expected, C17-sphingosine accumulated in vehicle-treated NPC mutant fibroblasts, whereas C17-S1P formation was reduced compared with WT fibroblasts (Fig. 3A). Treatment of NPC mutant fibroblasts with SK1-A corrected this phosphorylation defect, reduced the high level of C17-sphingosine, and increased C17-S1P to a similar level observed in WT fibroblasts (Fig. 3A). Nevertheless, SK1-A treatment did not affect SphK1 protein expression in NPC1 mutant fibroblasts (data not shown).
Figure 3.
SK1-A increases sphingosine kinase activity and suppresses autophagic defects in NPC1 mutant fibroblasts. A, normal human fibroblasts (WT2) and NPC1 mutant fibroblasts (NPC25) were treated without or with SK1-A for 2 h and then incubated for 30 min with C17-sphingosine. Cellular levels of C17-sphingosine and C17-S1P were measured by LC-ESI-MS/MS. Data are representative of at least three independent experiments, normalized to total lipid phosphate, and presented as mean ± S.D. of three biological replicates (n = 3). #, p < 0.05 compared with WT2 vehicle; *, p < 0.05 compared with vehicle-treated NPC25 cells. Data were analyzed by two-way ANOVA with Tukey's post hoc test. B, normal human fibroblasts (WTA, WT2) and NPC1 mutant fibroblasts (NPC25, NPC26) were cultured for 2 h in the absence of serum and then treated without or with SK1-A (2 μm) for 3 h. Cell lysate proteins were immunoblotted with anti-M6PR, anti-Rab7, anti-p62, or anti-LC3 antibodies. Blots were stripped and reprobed with anti-GAPDH as a loading control. Images are representative of three independent experiments. C, relative density of the indicated immunopositive bands normalized to GAPDH. Data are mean ± S.D. (error bars) of three biological replicates. #, p < 0.05 compared with WT2 vehicle; *, p < 0.05 compared with vehicle-treated NPC1 mutant fibroblasts. p values were determined by two-way ANOVA with Fisher's LSD.
Autophagy, an essential process that ensures that misfolded or dysfunctional proteins are degraded by lysosomes, is known to be impaired in NPC and contributes to disease progression (24, 25). Because it was suggested that sphingosine accumulation due to impaired SphK activity causes defective endocytic and autophagic fluxes in NPC mutant cells (20), and we and others have shown that SphK1 is rapidly recruited to vesicles during endocytosis and autophagy to regulate these processes (21, 22, 26), it was of interest to determine the effects of SK1-A on autophagy. Consistent with previous results (24), in NPC1 mutant fibroblasts we found increased processing of the cytosolic soluble MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3) form (LC3-I) to its lipidated membrane-bound form (LC3-II), a marker of increased formation of autophagosomes or amphisomes. This increased LC3-II level was reduced by treatment with SK1-A (Fig. 3, B and C). Likewise, accumulation of p62, an adapter protein that recruits polyubiquitinated and aggregated proteins to autophagosomes by its interaction with LC3-II (27), was significantly higher in NPC1 mutant fibroblasts compared with WT cells (Fig. 3, B and C), as was reported previously (24, 28). The elevated levels of p62 in NPC1 mutant fibroblasts were also decreased by SK1-A (Fig. 3, B and C). Consistent with the accumulation of late endosomes in NPC1 mutant cells due impaired recruitment of components of the SNARE machinery to late endosomes (24, 29), Rab7 and other late endosome markers, such as cation-independent mannose 6-phosphate receptor (M6PR), were all increased compared with WT, and treatment with SK1-A reduced their levels (Fig. 3, B and C). Thus, increased phosphorylation of sphingosine and reduced accumulation of sphingosine correlates with improved autophagic and endocytic defects in NPC1 mutant cells.
Unexpected effects of SphK1 activation on cellular cholesterol trafficking
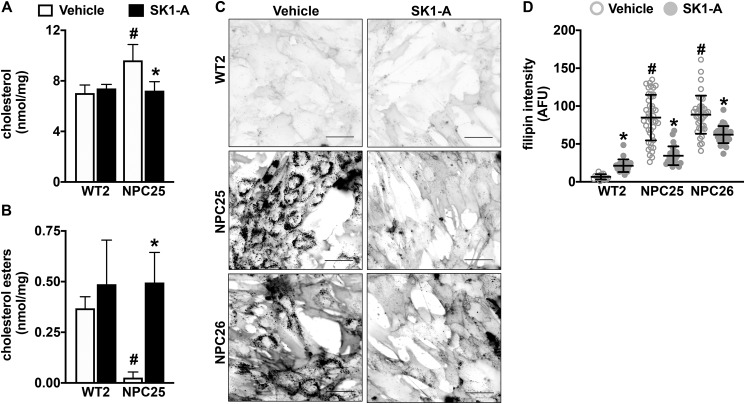
The accumulation of unesterified cholesterol inside the LE/L compartment is the hallmark cellular defect in NPC disease (1, 2, 5–7). Because autophagic defects can increase accumulation of cholesterol (24, 30) and up-regulation of autophagy is beneficial in several neurodegenerative diseases (31), we sought to examine the effect of SK1-A on cholesterol accumulation in NPC1 mutant fibroblasts. Remarkably, treatment with 2 µm SK1-A significantly decreased the levels of unesterified (free) cholesterol in NPC25 mutant fibroblasts (Fig. 4A). The reduction in unesterified cholesterol could result from increased trafficking from the LE/L compartment or a result of reduction in uptake. Thus, cholesterol ester formation was examined, as this requires delivery of cholesterol to acyl-CoA:cholesterol acyltransferase at the ER (32–34). SK1-A treatment dramatically increased cholesterol ester formation in NPC1 mutant fibroblasts (Fig. 4B), suggesting that activation of SphK1 by SK1-A increased delivery of cholesterol to the ER. To confirm these results, cells were stained with filipin, a fluorescent polyene antibiotic that specifically binds unesterified cholesterol (35), to visualize cholesterol levels inside the cells. Vehicle-treated NPC25 and NPC26 fibroblasts showed higher cholesterol levels compared with WT2 cells, yet after SK1-A treatment, this accumulation was greatly reduced (Fig. 4, C and D). These results suggest that activation of SphK1 can reduce the pathogenic accumulation of cholesterol in these cells.
Figure 4.
Activation of SphK1 with SK1-A decreases cholesterol accumulation in NPC1 mutant fibroblasts. A–D, normal human fibroblasts and NPC1 mutant fibroblasts were treated with vehicle or with SK1-A (2 μm) for 2 h. Levels of unesterified cholesterol (A) and esterified cholesterol (B) were quantified by Amplex Red assay and are representative of three independent experiments. Data are mean ± S.D. (error bars) of biological triplicates. #, p < 0.05 compared with WT2 vehicle; *, p < 0.05 compared with vehicle-treated NPC25 cells determined by two-way ANOVA with Tukey's post hoc test. C and D, cellular cholesterol was visualized by filipin staining (C). Representative images are shown. Scale bars, 50 μm. D, intensity of filipin staining was determined using ImageJ. Dots, fluorescent intensities of single cells. Means ± S.D. are indicated. #, p < 0.05 compared with WT2 vehicle; *, p < 0.05 compared with vehicle-treated NPC1 mutant fibroblasts, determined by two-way ANOVA followed by Tukey's post hoc analysis.
Inhibition of SphK1 increases cholesterol levels in NPC1 WT cells
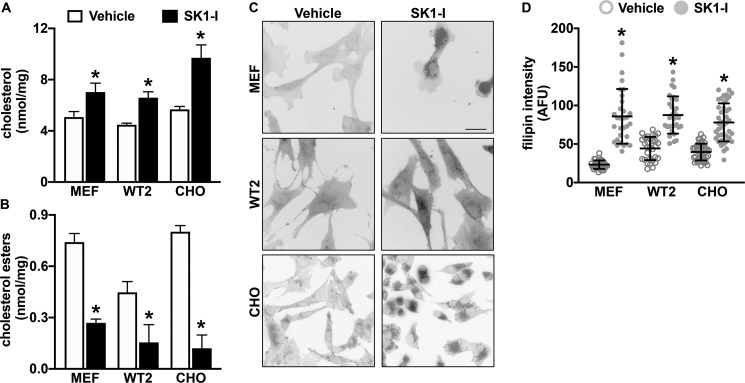
We have previously shown that inhibition of SphK1 with the isoform-specific SphK1 inhibitor SK1-I induces a stall in the late endosomal compartment, producing dilated Rab7a-positive vesicles containing sphingosine (22). As restoring SphK1 activity reduced cholesterol in NPC1mutant fibroblasts (Fig. 4), we sought to determine whether inhibition of SphK1 activity would have an inverse effect on cellular cholesterol in cells expressing WT NPC1 protein. Treatment of multiple cell types, including WT2 human fibroblasts, WT mouse embryonic fibroblasts (MEFs), and WT (JP17) CHO cells with SK1-I for 2 h significantly increased levels of free cholesterol with a decrease in cholesterol esters (Fig. 5, A and B). Similarly, increased filipin staining was observed after treatment of these cells with SK1-I (Fig. 5, C and D), supporting the notion that inhibition of SphK1 and sphingosine elevation reduced trafficking of cholesterol from lysosomes to the ER.
Figure 5.
Inhibition of SphK1 increases unesterified cholesterol accumulation. Normal human fibroblasts (WT2), MEFs, or CHO cells were treated with 7.5 μm SK1-I for 2 h. Cellular unesterified cholesterol (A) and esterified cholesterol levels (B) were quantified by Amplex Red assay and are representative of three independent experiments. Data are mean ± S.D. (error bars) of biological triplicates. C and D, cellular cholesterol was visualized by filipin staining. C, representative images are shown. Scale bar, 50 μm. D, intensity of filipin staining was determined using ImageJ. Dots, fluorescent intensities of single cells. Means ± S.D. are indicated. *, p < 0.05 compared with vehicle-treated cells as determined by two-tailed Student's t test.
Activation of SphK1 by SK1-A reduces accumulation of sphingoid bases and complex sphingolipids in NPC1 null cells
NPC is a complex disease in which many mutations have been identified, and affected individuals display a broad range of disease severity and age of onset of symptoms, ranging from infant to adult (5). Therefore, we sought to examine the effects of SK1-A in the most severe type of NPC disease where NPC1 is completely absent.
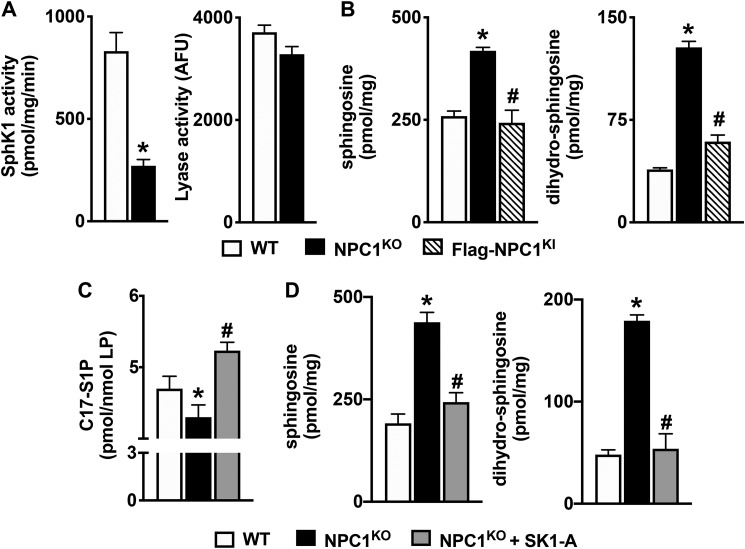
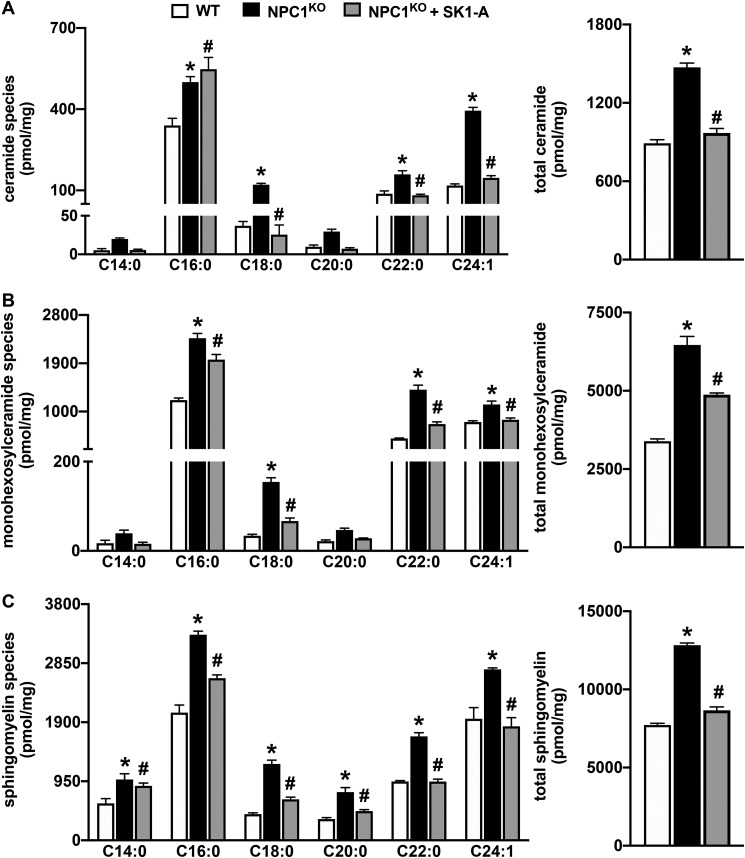
SphK1 activity in isogenic CHO cells with homozygous deletion of the NPC1 gene (NPC1KO) was greatly reduced (Fig. 6A), similar to the defect observed in NPC1MUTANT fibroblasts (Fig. 1C). Likewise, no major effects on S1P lyase activity were detected in NPC1KO cells (Fig. 6A). Similar to NPC1MUTANT fibroblasts, CHO NPC1KO cells accumulated sphingoid bases sphingosine and dihydrosphingosine (Fig. 6B). As expected, rescuing the expression of NPC1 in knockin cells that stably express FLAG-tagged human NPC1 in an endogenous NPC1 knockout background (FLAG-NPC1KI) reversed accumulation of the sphingoid bases (Fig. 6B). Moreover, the reduced conversion of C17-sphingosine to C17-S1P (Fig. 6C) in NPC1KO cells compared with WT CHO cells also supports the notion that this is the result of reduced SphK1 activity. Furthermore, treatment of NPC1KO cells with the SphK1 activator SK1-A, which increased conversion of C17-sphingosine to C17-S1P (Fig. 6C), also reduced levels of sphingosine and dihydrosphingosine to almost the same levels observed in WT CHO cells (Fig. 6D). Similarly, SK1-A significantly reduced accumulation of complex sphingolipids, including ceramides, monohexosylceramides, and sphingomyelins in NPC1KO cells (Fig. 7, A–C). Interestingly, an examination of the individual species of these complex sphingolipids, defined by the number of carbons present in their acyl chains, indicates that with the exception of C16:0 ceramide, all other acyl chain species of the complex sphingolipids were reduced by SK1-A treatment (Fig. 7, A–C).
Figure 6.
SK1-A decreases sphingoid base accumulation in NPC1KO cells. A, SphK1 and S1P lyase enzymatic activity in WT and NPC1KO CHO cells was measured and normalized to total protein. B, lipids were extracted from WT, NPC1KO, and NPC1KI CHO cells, and sphingosine and dihydrosphingosine were measured by LC-ESI-MS/MS. C and D, NPC1KO CHO cells were treated without or with SK1-A (10 μm) for 24 h. *, p < 0.05 compared with WT; #, p < 0.05 compared with NPC1KO. C, in vivo SphK activity was determined by LC-ESI-MS/MS quantification of conversion of C17-sphingosine (1 μm) to C17-S1P after 30 min and normalized to total lipid phosphates. *, p < 0.05 compared with WT. D, sphingosine and dihydrosphingosine were measured by LC-ESI-MS/MS. All data are means ± S.D. (error bars) and representative of three independent experiments, each with three biological replicates. *, p < 0.05 compared with WT; #, p < 0.05 compared with NPC1KO. p values were determined by two-tailed Student's t test (A) or one-way ANOVA followed by Tukey's post hoc analysis (B–D).
Figure 7.
Accumulation of complex sphingolipids in NPC1KO cells is reduced by SK1-A treatment. NPC1KO CHO cells were treated without or with SK1-A (10 μm) for 24 h. Lipids were extracted from the indicated CHO cells, and ceramide (A), monohexosylceramide (B), and sphingomyelin species (C) were measured by LC-ESI-MS/MS. Different chain-length species are shown; numbers indicate N-acyl chain length followed by the number of double bonds in the fatty acid. Data are means ± S.D. (error bars) of biological replicates and representative of three independent experiments. *, p < 0.05 compared with WT; #, p < 0.5 compared with NPC1KO as determined by one-way ANOVA (totals) or two-way ANOVA (species) followed by Tukey's post hoc analysis.
SK1-A corrects autophagic and cholesterol-trafficking defects in NPC1 null cells
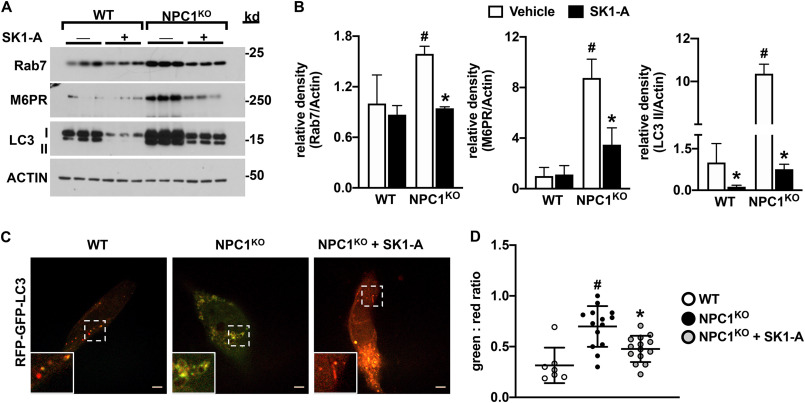
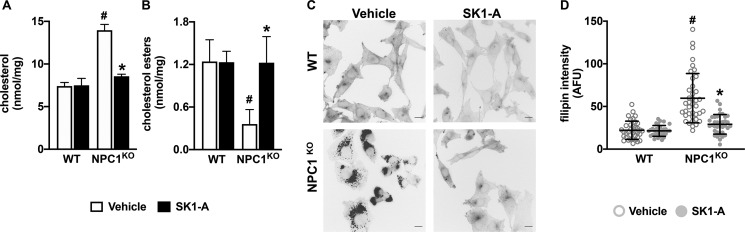
Similar to NPC1 mutant fibroblasts (Fig. 3), NPC1KO cells also have autophagic and endocytic defects, as shown by their increased levels of LC3-I and LC3-II as well as Rab7 and M6PR compared with WT cells (Fig. 8, A and B). Activation of SphK1 in these cells with SK1-A ameliorates these defects (Fig. 8, A and B), as assessed by a reduction in LC3-II, Rab7, and M6PR levels. Previous work suggested that autophagic flux is impaired in NPC mutant cells because of a loss in vesicular fusion between autophagosomes and late endosomes, inhibiting amphisome formation and cargo degradation (36, 37). Autophagic flux can be directly monitored using expression of a tandem monomeric RFP-GFP–tagged LC3 reporter, which emits both green and red fluorescence when localized to autophagosomes yet only emits red fluorescence when localized to autolysosomes because of fluorescence quenching of the GFP protein in the acidic environment of lysosomes (38). Consistent with previous reports (36, 37, 39), the majority of accumulated vesicles in NPC1KO CHO cells expressing RFP-GFP–tagged LC3 show increased yellow (red + green) fluorescence, indicating a block in autophagosome and lysosome fusion compared with WT cells (Fig. 8, C and D). Treatment with SK1-A resulted in the correction of impaired autophagosome maturation in NPC1KO cells (Fig. 8, C and D), suggesting that SK1-A can restore autophagic flux. Importantly, treatment of NPC1KO CHO cells with SK1-A decreased cellular unesterified (free) cholesterol levels (Fig. 9A) and increased cholesterol esters (Fig. 9B). In contrast, treatment with 100 nm S1P, a ligand for the S1PRs, had no effect on cholesterol accumulation in NPC1KO CHO cells (Fig. S2). Furthermore, visualization of unesterified cholesterol by filipin staining confirmed that SK1-A treatment of NPC1KO CHO cells decreased free cholesterol (Fig. 9, C and D). Taken together, these data indicate that activation of SphK1 with SK1-A restores cholesterol-trafficking defects in diverse NPC1 aberrant cells.
Figure 8.
SK1-A ameliorates autophagic defects in NPC1KO cells. A and B, WT and NPC1KO CHO cells were treated without or with 10 μm SK1-A for 2 h. A, cell lysate proteins were immunoblotted with anti-M6PR, anti-Rab7, or anti-LC3 antibodies. Blots were stripped and reprobed with anti-actin to show equal transfer and loading. Results are representative of three independent experiments, each with three biological replicates. B, relative density of the indicated immunopositive bands normalized to actin. Data are means ± S.D. of the blot presented in A. #, p < 0.05 compared with WT; *, p < 0.5 compared with NPC1KO, as determined by two-way ANOVA followed by Fisher's LSD analysis. C and D, confocal images of WT and NPC1KO CHO cells expressing RFP-GFP-LC3 treated with vehicle or 10 μm SK1-A for 2 h, as indicated. Scale bars, 5 μm. Inset box, ×2.4 magnification. D, GFP/RFP fluorescence ratio determined individually by ImageJ. Means ± S.D. (error bars) are indicated. Data and images are representative of three independent experiments. #, p < 0.05 compared with WT; *, p < 0.05 compared with vehicle-treated NPC1 mutant fibroblasts as determined by one-way ANOVA followed by Tukey's post hoc analysis.
Figure 9.
SK1-A treatment decreases cholesterol accumulation and enhances cholesterol esterification in NPC1KO cells. WT and NPC1KO CHO cells were treated with SK1-A (10 μm) for 2 h. Levels of unesterified cholesterol (A) and esterified cholesterol (B) were quantified by Amplex Red assay and are representative of three independent experiments. Data are mean ± S.D. (error bars) of biological triplicates. C and D, cellular cholesterol was visualized by filipin staining. C, representative images are shown. Scale bars, 20 μm. D, intensity of filipin staining was determined using ImageJ. Dots, fluorescent intensities of single cells. Means ± S.D. are indicated. #, p < 0.05 compared with WT; *, p < 0.5 compared with NPC1KO, as determined by two-way ANOVA followed by Tukey's post hoc analysis. AFU, arbitrary fluorescence units.
Discussion
Although the causative relationship between NPC mutations and cholesterol accumulation has been extensively studied, surprisingly little is known about the mechanism behind sphingosine and complex sphingolipid accumulation. Here, we report that the enzymatic activity of SphK1, one of the kinases that phosphorylate sphingosine to S1P, is reduced in diverse NPC1 mutant fibroblasts and in NPC1-deleted cells. As S1P cleavage by S1P lyase is the only exit point for complex sphingolipid degradation, we suggest that this decreased phosphorylation of sphingoid bases accounts for accumulation of sphingolipids in NPC. Consistent with our findings, a previous study showed that SphK activity is reduced in NPC mouse Purkinje neurons because of decreased levels of VEGF (20). Moreover, treatment with VEGF, which activates SphK1 by binding to VEGFR2, decreased sphingosine accumulation as well as improved neuronal survival and rota-rod scores in NPC1 knockout mice (20). Taken together, these results suggest that reversing defective SphK1 activity in NPC might be beneficial. In this study, we identified SK1-A as a potent and specific small-molecule activator of SphK1. SK1-A enhanced SphK activity and decreased accumulation of sphingoid bases in NPC1 mutant human fibroblasts and in NPC1 knockout cells.
Previous studies have shown that autophagic flux is impaired in NPC1 disease (40–42) because of defective amphisome formation caused by failure of the SNARE machinery (24). Furthermore, inhibition of autophagy caused accumulation of cholesterol (24, 30), suggesting that defective autophagic flux could be a causative factor in NPC1 disease. Methyl-b-cyclodextrin, which reduces lysosomal cholesterol accumulation in NPC disease restores impaired autophagy flux in NPC1-deficient cells through activation of AMP-activated protein kinase (43). We showed that activation of SphK1 with SK1-A suppressed defective autophagic flux and endocytic defects in NPC1 mutant and knockout cells. These results support recent findings that SphK1 regulates autophagic flux (44, 45) and cooperates with autophagy to maintain endocytic membrane trafficking (21).
Increasing autophagy has been suggested as a potential strategy for several neurodegenerative disorders (31). Unfortunately, although efforts to enhance autophagy using mTOR-dependent (rapamycin) and mTOR-independent (lithium) induction corrected the autophagic defects in NPC1 mutant cells and increased cell viability, they did not correct the hallmark cholesterol-trafficking defects (24). Remarkably, however, we found that treatment with SK1-A not only reduced autophagic defects but also reversed cholesterol accumulation in NPC1 mutant and deleted cells.
It has been elegantly established that low-density lipoprotein–derived free cholesterol is transferred by the soluble NPC2 protein in the lysosome to the membrane-bound NPC1 for subsequent egress from this compartment (7, 46, 47). Perhaps the most surprising discovery of our work is that SphK1 activation alone is sufficient to reduce lysosomal accumulation of free cholesterol and restore production of cholesterol esters even in NPC1 null cells. Thus, cholesterol egress can be restored in an NPC1-independent manner. There are several potential mechanisms that might explain NPC1-independent transport of cholesterol. We recently showed that expansion of membrane contact sites by overexpression of ORP1L-Δ-ORD mutant, which is known to act as a constitutively active tether between late endosomes and the ER without transporting cholesterol (48, 49), rescued the lysosomal cholesterol accumulation in NPC1-deficient cells (50). It is possible that when membrane contact sites are expanded, another cholesterol transporter, such as STARD3 (51), ORP1L (48, 49, 52–55), or Gramd1b (50), could compensate for loss of NPC1. It is also possible that cholesterol could diffuse along a concentration gradient across the expanded membrane contact sites without the need for a cholesterol transporter (56). It is tempting to speculate that conversion of sphingosine to S1P could regulate formation of membrane contact sites between late endosomes/lysosomes and the ER; however, other possibilities cannot be excluded. For example, it has been suggested that the ABC transporter A1 (ABCA1) has lysosomal cholesterol transport activity that requires functional NPC2 but not NPC1 (57). Furthermore, intracellular cholesterol trafficking can be enhanced by direct interaction of NPC2 with the phospholipid lysobisphosphatidic acid in the absence of NPC1 (58). It is still not known whether activation of SphK1 can increase ABCA1 transporter activity or levels of lysobisphosphatidic acid. Moreover, it was recently shown that reduction of the elevated sphingomyelin in NPC1 null cells decreased the cholesterol accumulation (59). Therefore, the dramatic decrease in sphingomyelin because of activation of SphK1 by SK1-A could be responsible for restoration of cholesterol trafficking via Rab9-dependent vesicular trafficking of cholesterol in NPC1-deficient cells (59). In addition, we cannot exclude the possibility that SK1-A also has effects on cholesterol metabolism and/or trafficking independent of its effects on SphK1.
Together these results demonstrate that impaired SphK1 activity in NPC cells contributes to accumulation of sphingosine and storage of complex sphingolipids and highlight a novel link between SphK1 activity, NPC1, and cholesterol trafficking and metabolism. Furthermore, the SphK1 activator SK1-A that we have characterized might lead to development of a new therapeutic strategy to reduce storage of lipids important in the pathogenesis of NPC disease.
Experimental procedures
Cell culture
NPC1MUTANT dermal fibroblasts (NPC25 and NPC26) and corresponding WT cells (WTA and WT2) were originally derived from NPC1 patients evaluated as part of an NICHD, National Institutes of Health, Institutional Review Board–approved protocol and obtained with consent. The following mutations were present: NPC25 (fs-exon 20, c.2979dupA, and p.N701K); NP-26 (c.3742_3745delCTCA fs-exon24 and p.R1059Q). WT (JP-17 parental), NPC1KO, and NPC1KO CHO cells stably expressing a FLAG-tagged NPC1 protein (referred to as NPC1KI) described previously (60) were a generous gift from Dr. Judith White (University of Virginia). Mouse embryonic fibroblasts (MEFs) were purchased from ATCC. Human fibroblasts were cultured in MEM (Thermo Fisher Scientific, Waltham, MA, USA), CHO cells were cultured in MEM-α (Thermo Fisher Scientific), and MEFs were cultured in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific), each supplemented with 10% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA, USA) containing 100 units/ml penicillin and 100 µg/ml streptomycin (Thermo Fisher Scientific) as described (34).
Sphingolipid analyses
Cells were washed three times with ice-cold PBS, lipids were extracted, and sphingolipids were quantified by LC-ESI-MS/MS (5500 QTrap; AB Sciex, Framingham, MA, USA) (61) and normalized to total protein.
Determination of SphK activity in cells
Cells were seeded on 6-well plates 24 h prior to a 30-min pulse with 1 µm C17-sphingosine (Avanti Polar Lipids). Cells were then washed three times with ice-cold PBS and then harvested directly by adding 1 ml of ice-cold methanol to each well. Sphingolipids were extracted, and C17-sphingosine and C17-S1P were quantified by LC-ESI-MS/MS and normalized to total lipid phosphates. SphK activity in mouse embryonic fibroblasts was determined with NBD-sphingosine as described previously (22).
Sphingosine kinase 1 assay
Recombinant SphK1 (15 nm) (62) was added to reaction mixtures containing 0.5 µm d-erythro-sphingosine (Avanti Polar Lipids), 30 mm Tris-HCl (pH 7.4), 0.05% Triton X-100, 150 mm NaCl, 10% glycerol, 1 mm Na3VO4, 10 mm NaF, and 10 mm β-glycerophosphate. Reactions were stopped by adding methanol, lipids were extracted, and S1P was quantified by LC-ESI-MS/MS.
Identification of the SphK1 activator SK1-A
Potential specific SphK1 inhibitors and activators were evaluated by high-throughput library screening fluorescence assays with recombinant SphK1 and SphK2 as described previously (23, 62). Briefly, compounds were dissolved in DMSO and initially screened by measuring effects on rates of production of NBD-S1P from NBD-sphingosine (Avanti) at a concentration of 650 µm. Candidates showing inhibition or stimulation of SphK1 activity in this assay were further characterized with NBD-sphingosine at lower concentrations to obtain IC50 values. SphK1 isoform specificity was determined using previously reported isoform-specific reaction conditions (63): SphK1 reaction buffer contained 30 mm Tris-HCl (pH 7.4), 0.5% Triton X-100 (MilliporeSigma), 150 mm NaCl, and 10% glycerol; SphK2 buffer contained 30 mm Tris-HCl (pH 7.4), 200 mm KCl, and 10% glycerol. Assays were carried out in 384‐well plates (Greiner Bio‐One, Frickenhausen, Germany) in the presence of recombinant SphKs (15 nm). Reactions were initiated by adding an ATP–Mg mixture (1 mm ATP, 2 mm MgCl2, 40 mm Tris‐HCl, pH 7.4) and followed in a TECAN Infinite M1000 fluorescence plate reader (Männedorf, Switzerland) at 37 °C. Excitation and emission wavelengths were 550 and 584 nm, respectively. All data were analyzed using Prism (GraphPad, La Jolla, CA, USA).
Synthesis of 2-hydroxy-3-{[4-(2-methoxyethoxy)benzoyl]oxy}-N-[2-(4-methoxyphenoxy)ethyl]propan-1-aminium chloride (SK1-A)
All reagents were purchased from Acros Organics (Geel, Belgium) or Sigma-Aldrich. 1H NMR and 13C NMR spectra were recorded at 25 °C with DMSO-d6 as solvents on a JNM-ECZ4OOR FT-NMR spectrometer 9.39 T (JEOL Resonance Inc., Tokyo, Japan; 399.78 MHz for 1H and 100.53 MHz for 13C). High-resolution mass spectra were determined with a high-performance liquid chromatograph Dionex UltiMate® 3000 (Thermo Scientific, West Palm Beach, FL, USA) coupled with a LTQ Orbitrap XL™ Hybrid Ion Trap-Orbitrap Fourier Transform Mass Spectrometer (Thermo Scientific) with injection into HESI II in the positive ion mode.
Oxirane intermediate, (oxiran-2-yl)methyl 4-(2-methoxyethoxy)benzoate, 1a (4 mmol) (23, 64) in isopropyl alcohol was added to 2-(4-methoxyphenoxy)ethylamine, intermediate 1b (4 mmol) (65)) in 15 ml of isopropyl alcohol (Fig. S3). The mixture was refluxed at 80 °C and then stirred for 72 h at room temperature and finally cooled at −18 °C for 48 h. The precipitate was collected and dissolved in a saturated ethereal solution of HCl. The hydrochloride salt was recrystallized from isopropyl alcohol (yield: 55%; MP: 121-122 °C; IR (ATR, cm−1): 3369w, 2948w, 2932w, 2890w, 2810w, 2404w, 1713s, 1606m, 1509s, 1467m, 1255m, 1226s, 1108m, 820m, 769m, 731m; 1H NMR (DMSO-d6) δ [ppm]: 9.39 (bs, 1H, ); 9.12 (bs, 1H, ); 7.95 (d, 3J = 8.9, 2H, HArCOO); 7.04 (d, 3J = 8.9, 2H, HArOEtO); 6.92 (d, 3J = 9.1, 2H, HArOEtN); 6.87 (d, 3J = 9.1, 2H, HArOMe); 5.97 (bs, 1H, OH); 4.28-4.24 (m, 5H, COOCH2CH + NCH2CH2O); 4.18-4.17 (m, 2H, CH2OAr); 3.70 (s, 3H, ArOCH3); 3.68-3.67 (m, 2H, CH2CH2OAr); 3.39-3.36 (m, 2H, NCH2CH2O); 3.31 (s, 3H, OCH3); 3.28-3.27 (m, 1H, CH2N); 3.11-3.08 (m, 1H, CH2N); 13 °C-NMR (DMSO-d6) δ [ppm]: 165.14, 162.47, 153.82, 151.70, 131.46, 121.69, 115.74, 114.60, 114.37, 70.14, 67.27, 65.86, 64.45, 63.84, 58.16, 55.34, 49.74, 46.16; HR-MS: C22H30NO7 [M + H]+ calculated 420.2016 m/z, found 420.2014 m/z.
Measurements of cholesterol and cholesterol esters
Cholesterol and cholesterol esters were quantified using the Amplex Red cholesterol assay kit as described previously (34). Briefly, cells in 6-well plates were treated as indicated in the figure legends, washed three times with ice-cold PBS, and lysed. To determine cholesterol esters, lysates were incubated without or with 0.2 units/ml cholesterol esterase. Lysates were subsequently incubated with 2 units/ml each of horseradish peroxidase and cholesterol oxidase and 300 µm Amplex Red reagent for 30 min at 37 °C in a 100 mm potassium phosphate buffer (pH 7.4) containing 50 mm NaCl, 5 mm cholic acid, and 0.1% Triton X-100. Fluorescence was measured with a TECAN Infinite M1000 fluorescent plate reader (TECAN, Männedorf, Switzerland). Levels of unesterified cholesterol and cholesterol esters were calculated from standard curves and normalized to total protein measured with the Bio-Rad protein assay kit.
Filipin staining
Cells were seeded on 22-mm coverslips in 6-well plates. After 24 h, cells were incubated with test compounds for 2 h. Cells were then washed and fixed in 4% paraformaldehyde for 15 min and washed. After incubating with 1.5 mg/ml glycine to quench the paraformaldehyde, cells were washed in PBS and stained with Filipin III (0.1 mg/ml; MiliporeSigma) for 2 h. Coverslips were then washed and mounted onto glass slides using Prolong Gold anti-fade reagent (Thermo Fisher Scientific) and imaged on an Axioimager A1 fluorescent microscope using Zen software (Zeiss, Jena, Germany). Images were quantified using ImageJ (National Institutes of Health) as described previously (34).
Immunoblotting
Cells were washed three times with ice-cold PBS and then scraped into lysis buffer containing 20 mm HEPES, pH 7.4 (Thermo Fisher Scientific), 250 mm NaCl, 1 mm DTT, 1 mm EDTA, 20% glycerol, 1% Triton X-100 (Thermo Fisher Scientific), and Halt protease and phosphatase inhibitor mixture (Thermo Fisher Scientific). After sonication, proteins were quantified, and equal amounts were separated by SDS-PAGE. Proteins were transferred onto polyvinylidene difluoride membranes (Bio-Rad), blocked with 2.5% blotting grade milk (Bio-Rad). The following primary antibodies were used for immunoblotting: Rab7 (#9367, 1:1000), GAPDH (#2118, 1:5000), actin (#3700, 1:5000), LC3 (#12741, 1:1000), or p62 (#5114, 1:1000) (Cell Signaling Technology, Danvers, MA, USA) or M6PR (#124767, 1:1000) (Abcam, Cambridge, MA, USA). Immunopositive bands were visualized by chemiluminescence with horseradish peroxidase–conjugated secondary antibodies (anti-rabbit #111035045 or anti-mouse #115035166, Jackson ImmunoResearch Laboratories, West Grove, PA, USA) and Super-Signal West Pico Plus (Thermo Fisher Scientific) substrate. Images were quantified using ImageJ as described previously (34).
S1P lyase activity assays
S1P lyase activity was measured as described (66). Briefly, cells (350,000–400,000) were seeded in 10-cm2 plates. The next day, cells were washed three times with cold PBS, scraped in 100 µl of 0.5 m potassium phosphate buffer (pH 7.4), and then lysed by brief sonication. An aliquot (75 µl) was added to 96-well plates with 15 µl of reaction mixture containing 0.5 m potassium phosphate buffer (pH 7.4), 25 µm Na3VO4, 0.25 mm pyridoxal 5′-phosphate, and 125 µm S1P lyase fluorogenic substrate (Cayman Chemical, Ann Arbor, MI, USA). Plates were incubated at 37 °C for 6 h in the dark, and reactions were stopped by adding 50 µl of methanol and incubating for 2 h. Fluorescence was measured with a TECAN Infinite M1000 fluorescent plate reader at excitation and emission wavelengths of 360 and 465 nm, respectively.
RFP-GFP-LC3 autophagic flux measurements
CHO cells were seeded on 96-well glass bottom plates (Cellvis, Mountain View, CA, USA) and infected with 80 viral particles (RFP-GFP-LC3; Life Technologies, Inc.) per cell for 48 h to express LC3 fused to a tandem green fluorescence– and red fluorescence–tagged protein. Autolysosomes are then displayed as red-only structures, as their acidic compartment causes the loss of the green fluorescent signal. After treatment without or with SK1-A, GFP-, and RFP-positive cells were visualized at ×100 in a Zeiss Cell Observer Spinning Disc (Zeiss, Germany) confocal microscope equipped with a growth chamber maintained at 37 °C and 5% CO2. RFP and GFP fluorescence was collected with identical settings across wells, and GFP/RFP ratios were quantified using ImageJ (45).
Statistics
Data were analyzed by Student's t test when comparing two groups or by ANOVA with Fisher's LSD, Dunnett's, or Tukey's post hoc analysis as appropriate using Prism. Results are expressed as means ± S.D. p ≤ 0.05 was considered statistically significant. Experiments were repeated three times with similar results.
Data availability
All data are contained within the article.
Supplementary Material
Acknowledgments
We thank Dr. Jeremy Allegood (Virginia Commonwealth University, Richmond, VA, USA) for skillful sphingolipid analyses, Drs. J. White (University of Virginia, Charlottesville, VA, USA) and Frances J. Sharom (University of Guelph, Guelph, Ontario, Canada) for the CHO cell line, and Dr. Forbes D. Porter (National Institutes of Health) for the human NPC mutant fibroblasts. The VCU Lipidomics and Microscopy Cores are supported in part by NCI, National Institutes of Health, Support Grant P30 CA016059.
This article contains supporting information.
Author contributions—J. N. and S. S. conceptualization; J. N. data curation; J. N. and S. S. funding acquisition; J. N. validation; J. N. writing-original draft; E. N. P., C. W., M. M., C. E. S., R. D. E., P. M., J. J., and S. L. investigation; M. H. G. software; C. E. S. methodology; S. M. and S. S. supervision; S. M. and S. S. writing-review and editing; S. S. resources; S. S. project administration.
Funding and additional information—This work was supported by National Institutes of Health Grants R01GM043880 (to S. S.) and K99HD096117 (to J. N.). S. L. was supported by National Institutes of Health Grant K22CA187314. The synthesis of SK1-A was supported by Slovak Research and Development Agency Grants APVV-17-0373 and APVV-17-0318 and by Slovak Grant Agency for Science Grant VEGA 1/0787/18. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- NPC
- Niemann–Pick type C
- ER
- endoplasmic reticulum
- RFP
- red fluorescent protein
- ESI
- electrospray ionization
- M6PR
- mannose 6-phosphate receptor
- MEM
- minimum essential medium
- NBD
- nitrobenzoxadiazole
- S1P
- sphingosine-1-phosphate
- SK1-A
- sphingosine kinase 1 activator
- SK1-I
- sphingosine kinase 1 inhibitor
- SphK
- sphingosine kinase
- VEGF
- vascular endothelial growth factor
- AC50
- half-maximal activation constant
- CHO
- Chinese hamster ovary
- MEF
- mouse embryo fibroblast
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ANOVA
- analysis of variance
- LSD
- least significant difference.
References
- 1. Carstea E. D., Morris J. A., Coleman K. G., Loftus S. K., Zhang D., Cummings C., Gu J., Rosenfeld M. A., Pavan W. J., Krizman D. B., Nagle J., Polymeropoulos M. H., Sturley S. L., Ioannou Y. A., Higgins M. E., et al. (1997) Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science 277, 228–231 10.1126/science.277.5323.228 [DOI] [PubMed] [Google Scholar]
- 2. Loftus S. K., Morris J. A., Carstea E. D., Gu J. Z., Cummings C., Brown A., Ellison J., Ohno K., Rosenfeld M. A., Tagle D. A., Pentchev P. G., and Pavan W. J. (1997) Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science 277, 232–235 10.1126/science.277.5323.232 [DOI] [PubMed] [Google Scholar]
- 3. Neufeld E. B., Wastney M., Patel S., Suresh S., Cooney A. M., Dwyer N. K., Roff C. F., Ohno K., Morris J. A., Carstea E. D., Incardona J. P., Strauss J. F. 3rd, Vanier M. T., Patterson M. C., Brady R. O., et al. (1999) The Niemann-Pick C1 protein resides in a vesicular compartment linked to retrograde transport of multiple lysosomal cargo. J. Biol. Chem. 274, 9627–9635 10.1074/jbc.274.14.9627 [DOI] [PubMed] [Google Scholar]
- 4. Pentchev P. G. (2004) Niemann-Pick C research from mouse to gene. Biochim. Biophys. Acta 1685, 3–7 10.1016/j.bbalip.2004.08.005 [DOI] [PubMed] [Google Scholar]
- 5. Vanier M. T. (2013) Niemann–Pick diseases. Handb. Clin. Neurol. 113, 1717–1721 10.1016/B978-0-444-59565-2.00041-1 [DOI] [PubMed] [Google Scholar]
- 6. Wang M. L., Motamed M., Infante R. E., Abi-Mosleh L., Kwon H. J., Brown M. S., and Goldstein J. L. (2010) Identification of surface residues on Niemann-Pick C2 essential for hydrophobic handoff of cholesterol to NPC1 in lysosomes. Cell Metab. 12, 166–173 10.1016/j.cmet.2010.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xie X., Brown M. S., Shelton J. M., Richardson J. A., Goldstein J. L., and Liang G. (2011) Amino acid substitution in NPC1 that abolishes cholesterol binding reproduces phenotype of complete NPC1 deficiency in mice. Proc. Natl. Acad. Sci. U.S.A. 108, 15330–15335 10.1073/pnas.1112751108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vanier M. T. (2015) Complex lipid trafficking in Niemann-Pick disease type C. J. Inherit. Metab. Dis. 38, 187–199 10.1007/s10545-014-9794-4 [DOI] [PubMed] [Google Scholar]
- 9. Oninla V. O., Breiden B., Babalola J. O., and Sandhoff K. (2014) Acid sphingomyelinase activity is regulated by membrane lipids and facilitates cholesterol transfer by NPC2. J. Lipid Res. 55, 2606–2619 10.1194/jlr.M054528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lloyd-Evans E., Morgan A. J., He X., Smith D. A., Elliot-Smith E., Sillence D. J., Churchill G. C., Schuchman E. H., Galione A., and Platt F. M. (2008) Niemann-Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nat. Med. 14, 1247–1255 10.1038/nm.1876 [DOI] [PubMed] [Google Scholar]
- 11. Lloyd-Evans E., and Platt F. M. (2010) Lipids on trial: the search for the offending metabolite in Niemann-Pick type C disease. Traffic 11, 419–428 10.1111/j.1600-0854.2010.01032.x [DOI] [PubMed] [Google Scholar]
- 12. Stein V. M., Crooks A., Ding W., Prociuk M., O'Donnell P., Bryan C., Sikora T., Dingemanse J., Vanier M. T., Walkley S. U., and Vite C. H. (2012) Miglustat improves Purkinje cell survival and alters microglial phenotype in feline Niemann-Pick disease type C. J. Neuropathol. Exp. Neurol. 71, 434–448 10.1097/NEN.0b013e31825414a6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Höglinger D., Haberkant P., Aguilera-Romero A., Riezman H., Porter F. D., Platt F. M., Galione A., and Schultz C. (2015) Intracellular sphingosine releases calcium from lysosomes. Elife 4, e10616 10.7554/eLife.10616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tharkeshwar A. K., Trekker J., Vermeire W., Pauwels J., Sannerud R., Priestman D. A., Te Vruchte D., Vints K., Baatsen P., Decuypere J. P., Lu H., Martin S., Vangheluwe P., Swinnen J. V., Lagae L., et al. (2017) A novel approach to analyze lysosomal dysfunctions through subcellular proteomics and lipidomics: the case of NPC1 deficiency. Sci. Rep. 7, 41408 10.1038/srep41408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Praggastis M., Tortelli B., Zhang J., Fujiwara H., Sidhu R., Chacko A., Chen Z., Chung C., Lieberman A. P., Sikora J., Davidson C., Walkley S. U., Pipalia N. H., Maxfield F. R., Schaffer J. E., and Ory D. S. (2015) A murine Niemann-Pick C1 I1061T knock-in model recapitulates the pathological features of the most prevalent human disease allele. J. Neurosci. 35, 8091–8106 10.1523/JNEUROSCI.4173-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aguilar A., and Saba J. D. (2012) Truth and consequences of sphingosine-1-phosphate lyase. Adv. Biol. Regul. 52, 17–30 10.1016/j.advenzreg.2011.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maceyka M., and Spiegel S. (2014) Sphingolipid metabolites in inflammatory disease. Nature 510, 58–67 10.1038/nature13475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Spiegel S., and Milstien S. (2003) Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol. 4, 397–407 10.1038/nrm1103 [DOI] [PubMed] [Google Scholar]
- 19. Spiegel S., and Milstien S. (2011) The outs and the ins of sphingosine-1-phosphate in immunity. Nat. Rev. Immunol. 11, 403–415 10.1038/nri2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee H., Lee J. K., Park M. H., Hong Y. R., Marti H. H., Kim H., Okada Y., Otsu M., Seo E. J., Park J. H., Bae J. H., Okino N., He X., Schuchman E. H., Bae J. S., and Jin H. K. (2014) Pathological roles of the VEGF/SphK pathway in Niemann-Pick type C neurons. Nat. Commun. 5, 5514 10.1038/ncomms6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Young M. M., Takahashi Y., Fox T. E., Yun J. K., Kester M., and Wang H. G. (2016) Sphingosine kinase 1 cooperates with autophagy to maintain endocytic membrane trafficking. Cell Rep. 17, 1532–1545 10.1016/j.celrep.2016.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lima S., Milstien S., and Spiegel S. (2017) Sphingosine and sphingosine kinase 1 involvement in endocytic membrane trafficking. J. Biol. Chem. 292, 3074–3088 10.1074/jbc.M116.762377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vettorazzi M., Angelina E., Lima S., Gonec T., Otevrel J., Marvanova P., Padrtova T., Mokry P., Bobal P., Acosta L. M., Palma A., Cobo J., Bobalova J., Csollei J., Malik I., et al. (2017) An integrative study to identify novel scaffolds for sphingosine kinase 1 inhibitors. Eur. J. Med. Chem. 139, 461–481 10.1016/j.ejmech.2017.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sarkar S., Carroll B., Buganim Y., Maetzel D., Ng A. H., Cassady J. P., Cohen M. A., Chakraborty S., Wang H., Spooner E., Ploegh H., Gsponer J., Korolchuk V. I., and Jaenisch R. (2013) Impaired autophagy in the lipid-storage disorder Niemann-Pick type C1 disease. Cell Rep. 5, 1302–1315 10.1016/j.celrep.2013.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schultz M. L., Krus K. L., Kaushik S., Dang D., Chopra R., Qi L., Shakkottai V. G., Cuervo A. M., and Lieberman A. P. (2018) Coordinate regulation of mutant NPC1 degradation by selective ER autophagy and MARCH6-dependent ERAD. Nat. Commun. 9, 3671 10.1038/s41467-018-06115-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shen H., Giordano F., Wu Y., Chan J., Zhu C., Milosevic I., Wu X., Yao K., Chen B., Baumgart T., Sieburth D., and De Camilli P. (2014) Coupling between endocytosis and sphingosine kinase 1 recruitment. Nat. Cell Biol. 16, 652–662 10.1038/ncb2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., and Johansen T. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- 28. Elrick M. J., Yu T., Chung C., and Lieberman A. P. (2012) Impaired proteolysis underlies autophagic dysfunction in Niemann-Pick type C disease. Hum. Mol. Genet. 21, 4876–4887 10.1093/hmg/dds324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wheeler S., Schmid R., and Sillence D. J. (2019) Lipid–protein interactions in Niemann–Pick type C disease: insights from molecular modeling. Int. J. Mol. Sci. 20, 717 10.3390/ijms20030717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., and Czaja M. J. (2009) Autophagy regulates lipid metabolism. Nature 458, 1131–1135 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rubinsztein D. C., Codogno P., and Levine B. (2012) Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 11, 709–730 10.1038/nrd3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rosenbaum A. I., Zhang G., Warren J. D., and Maxfield F. R. (2010) Endocytosis of β-cyclodextrins is responsible for cholesterol reduction in Niemann-Pick type C mutant cells. Proc. Natl. Acad. Sci. U.S.A. 107, 5477–5482 10.1073/pnas.0914309107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gelsthorpe M. E., Baumann N., Millard E., Gale S. E., Langmade S. J., Schaffer J. E., and Ory D. S. (2008) Niemann-Pick type C1 I1061T mutant encodes a functional protein that is selected for endoplasmic reticulum-associated degradation due to protein misfolding. J. Biol. Chem. 283, 8229–8236 10.1074/jbc.M708735200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Newton J., Hait N. C., Maceyka M., Colaco A., Maczis M., Wassif C. A., Cougnoux A., Porter F. D., Milstien S., Platt N., Platt F. M., and Spiegel S. (2017) FTY720/fingolimod increases NPC1 and NPC2 expression and reduces cholesterol and sphingolipid accumulation in Niemann-Pick type C mutant fibroblasts. FASEB J. 31, 1719–1730 10.1096/fj.201601041R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vanier M. T., and Latour P. (2015) Laboratory diagnosis of Niemann-Pick disease type C: the filipin staining test. Methods Cell Biol 126, 357–375 10.1016/bs.mcb.2014.10.028 [DOI] [PubMed] [Google Scholar]
- 36. Maetzel D., Sarkar S., Wang H., Abi-Mosleh L., Xu P., Cheng A. W., Gao Q., Mitalipova M., and Jaenisch R. (2014) Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann-Pick Type C patient-specific iPS cells. Stem Cell Reports 2, 866–880 10.1016/j.stemcr.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sarkar S., Maetzel D., Korolchuk V. I., and Jaenisch R. (2014) Restarting stalled autophagy a potential therapeutic approach for the lipid storage disorder, Niemann-Pick type C1 disease. Autophagy 10, 1137–1140 10.4161/auto.28623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kimura S., Noda T., and Yoshimori T. (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 10.4161/auto.4451 [DOI] [PubMed] [Google Scholar]
- 39. Campbell C. D., Mohajeri K., Malig M., Hormozdiari F., Nelson B., Du G., Patterson K. M., Eng C., Torgerson D. G., Hu D., Herman C., Chong J. X., Ko A., O'Roak B. J., Krumm N., et al. (2014) Whole-genome sequencing of individuals from a founder population identifies candidate genes for asthma. PLoS ONE 9, e104396 10.1371/journal.pone.0104396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ko D. C., Milenkovic L., Beier S. M., Manuel H., Buchanan J., and Scott M. P. (2005) Cell-autonomous death of cerebellar purkinje neurons with autophagy in Niemann-Pick type C disease. PLoS Genet. 1, 81–95 10.1371/journal.pgen.0010007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pacheco C. D., Kunkel R., and Lieberman A. P. (2007) Autophagy in Niemann-Pick C disease is dependent upon Beclin-1 and responsive to lipid trafficking defects. Hum. Mol. Genet. 16, 1495–1503 10.1093/hmg/ddm100 [DOI] [PubMed] [Google Scholar]
- 42. Ordonez M. P., Roberts E. A., Kidwell C. U., Yuan S. H., Plaisted W. C., and Goldstein L. S. (2012) Disruption and therapeutic rescue of autophagy in a human neuronal model of Niemann Pick type C1. Hum. Mol. Genet. 21, 2651–2662 10.1093/hmg/dds090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dai S., Dulcey A. E., Hu X., Wassif C. A., Porter F. D., Austin C. P., Ory D. S., Marugan J., and Zheng W. (2017) Methyl-β-cyclodextrin restores impaired autophagy flux in Niemann-Pick C1-deficient cells through activation of AMPK. Autophagy 13, 1435–1451 10.1080/15548627.2017.1329081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu H., Ma Y., He H. W., Zhao W. L., and Shao R. G. (2017) SPHK1 (sphingosine kinase 1) induces epithelial-mesenchymal transition by promoting the autophagy-linked lysosomal degradation of CDH1/E-cadherin in hepatoma cells. Autophagy 13, 900–913 10.1080/15548627.2017.1291479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lima S., Takabe K., Newton J., Saurabh K., Young M. M., Leopoldino A. M., Hait N. C., Roberts J. L., Wang H. G., Dent P., Milstien S., Booth L., and Spiegel S. (2018) TP53 is required for BECN1- and ATG5-dependent cell death induced by sphingosine kinase 1 inhibition. Autophagy 14, 1–957 10.1080/15548627.2018.1429875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kwon H. J., Abi-Mosleh L., Wang M. L., Deisenhofer J., Goldstein J. L., Brown M. S., and Infante R. E. (2009) Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell 137, 1213–1224 10.1016/j.cell.2009.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Infante R. E., Wang M. L., Radhakrishnan A., Kwon H. J., Brown M. S., and Goldstein J. L. (2008) NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc. Natl. Acad. Sci. U.S.A. 105, 15287–15292 10.1073/pnas.0807328105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rocha N., Kuijl C., van der Kant R., Janssen L., Houben D., Janssen H., Zwart W., and Neefjes J. (2009) Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J. Cell Biol. 185, 1209–1225 10.1083/jcb.200811005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Eden E. R., Sanchez-Heras E., Tsapara A., Sobota A., Levine T. P., and Futter C. E. (2016) Annexin A1 tethers membrane contact sites that mediate ER to endosome cholesterol transport. Dev. Cell 37, 473–483 10.1016/j.devcel.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Höglinger D., Burgoyne T., Sanchez-Heras E., Hartwig P., Colaco A., Newton J., Futter C. E., Spiegel S., Platt F. M., and Eden E. R. (2019) NPC1 regulates ER contacts with endocytic organelles to mediate cholesterol egress. Nat. Commun. 10, 4276–4290 10.1038/s41467-019-12152-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wilhelm L. P., Wendling C., Védie B., Kobayashi T., Chenard M. P., Tomasetto C., Drin G., and Alpy F. (2017) STARD3 mediates endoplasmic reticulum-to-endosome cholesterol transport at membrane contact sites. EMBO J. 36, 1412–1433 10.15252/embj.201695917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dong J., Du X., Wang H., Wang J., Lu C., Chen X., Zhu Z., Luo Z., Yu L., Brown A. J., Yang H., and Wu J. W. (2019) Allosteric enhancement of ORP1-mediated cholesterol transport by PI(4,5)P2/PI(3,4)P2. Nat. Commun. 10, 829 10.1038/s41467-019-08791-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhao K., and Ridgway N. D. (2017) Oxysterol-binding protein-related protein 1L regulates cholesterol egress from the endo-lysosomal system. Cell Rep. 19, 1807–1818 10.1016/j.celrep.2017.05.028 [DOI] [PubMed] [Google Scholar]
- 54. Wijdeven R. H., Janssen H., Nahidiazar L., Janssen L., Jalink K., Berlin I., and Neefjes J. (2016) Cholesterol and ORP1L-mediated ER contact sites control autophagosome transport and fusion with the endocytic pathway. Nat. Commun. 7, 11808 10.1038/ncomms11808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cianciola N. L., Greene D. J., Morton R. E., and Carlin C. R. (2013) Adenovirus RIDα uncovers a novel pathway requiring ORP1L for lipid droplet formation independent of NPC1. Mol. Biol. Cell 24, 3309–3325 10.1091/mbc.e12-10-0760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cheruku S. R., Xu Z., Dutia R., Lobel P., and Storch J. (2006) Mechanism of cholesterol transfer from the Niemann-Pick type C2 protein to model membranes supports a role in lysosomal cholesterol transport. J. Biol. Chem. 281, 31594–31604 10.1074/jbc.M602765200 [DOI] [PubMed] [Google Scholar]
- 57. Boadu E., Nelson R. C., and Francis G. A. (2012) ABCA1-dependent mobilization of lysosomal cholesterol requires functional Niemann-Pick C2 but not Niemann-Pick C1 protein. Biochim. Biophys. Acta 1821, 396–404 10.1016/j.bbalip.2011.11.013 [DOI] [PubMed] [Google Scholar]
- 58. McCauliff L. A., Langan A., Li R., Ilnytska O., Bose D., Waghalter M., Lai K., Kahn P. C., and Storch J. (2019) Intracellular cholesterol trafficking is dependent upon NPC2 interaction with lysobisphosphatidic acid. Elife 8, e50832 10.7554/eLife.50832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wanikawa M., Nakamura H., Emori S., Hashimoto N., and Murayama T. (2020) Accumulation of sphingomyelin in Niemann-Pick disease type C cells disrupts Rab9-dependent vesicular trafficking of cholesterol. J. Cell. Physiol. 235, 2300–2309 10.1002/jcp.29137 [DOI] [PubMed] [Google Scholar]
- 60. Higaki K., Ninomiya H., Sugimoto Y., Suzuki T., Taniguchi M., Niwa H., Pentchev P. G., Vanier M. T., and Ohno K. (2001) Isolation of NPC1-deficient Chinese hamster ovary cell mutants by gene trap mutagenesis. J. Biochem. 129, 875–880 10.1093/oxfordjournals.jbchem.a002932 [DOI] [PubMed] [Google Scholar]
- 61. Maczis M. A., Maceyka M., Waters M. R., Newton J., Singh M., Rigsby M. F., Turner T. H., Alzubi M. A., Harrell J. C., Milstien S., and Spiegel S. (2018) Sphingosine kinase 1 activation by estrogen receptor α36 contributes to tamoxifen resistance in breast cancer. J. Lipid Res. 59, 2297–2307 10.1194/jlr.M085191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lima S., Milstien S., and Spiegel S. (2014) A real-time high-throughput fluorescence assay for sphingosine kinases. J. Lipid Res. 55, 1525–1530 10.1194/jlr.D048132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Paugh S. W., Paugh B. S., Rahmani M., Kapitonov D., Almenara J. A., Kordula T., Milstien S., Adams J. K., Zipkin R. E., Grant S., and Spiegel S. (2008) A selective sphingosine kinase 1 inhibitor integrates multiple molecular therapeutic targets in human leukemia. Blood 112, 1382–1391 10.1182/blood-2008-02-138958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pospisilova S., Marvanova P., Treml J., Moricz A. M., Ott P. G., Mokry P., Odehnalova K., Sedo O., Cizek A., and Jampilek J. (2019) Activity of N-phenylpiperazine derivatives against bacterial and fungal pathogens. Curr. Protein Pept. Sci. 20, 1119–1129 10.2174/1389203720666190913114041 [DOI] [PubMed] [Google Scholar]
- 65. Tengler J., Kapustíková I., Peško M., Govender R., Keltošová S., Mokrý P., Kollár P., O'Mahony J., Coffey A., Král'ová K., and Jampílek J. (2013) Synthesis and biological evaluation of 2-hydroxy-3-[(2-aryloxyethyl)amino]propyl 4-[(alkoxycarbonyl)amino]benzoates. Sci. World J. 2013, 1–13 10.1155/2013/274570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bedia C., Camacho L., Casas J., Abad J. L., Delgado A., Van Veldhoven P. P., and Fabriàs G. (2009) Synthesis of a fluorogenic analogue of sphingosine-1-phosphate and its use to determine sphingosine-1-phosphate lyase activity. Chembiochem 10, 820–822 10.1002/cbic.200800809 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.