Abstract
Various human pathogenic viruses employ envelope glycoproteins for host cell receptor recognition and binding, membrane fusion and viral entry. The spike (S) glycoprotein of betacoronavirus SARS-CoV-2 is a homotrimeric class I fusion protein that exists in a metastable conformation for cleavage by host cell proteases furin and TMPRSS2, thereby undergoing substantial structural rearrangement for ACE2 host cell receptor binding and subsequent viral entry by membrane fusion. The S protein is densely decorated with N-linked glycans protruding from the trimer surface that affect S protein folding, processing by host cell proteases and the elicitation of humoral immune response. Deep insight into the sophisticated structure of SARS-CoV-2 S protein may provide a blueprint for vaccination strategies, as reviewed herein.
Keywords: Coronavirus SARS-CoV-2, Spike protein, Protein structure, Receptor binding domain, Viral fusion protein, ACE2, Vaccination, Immune response
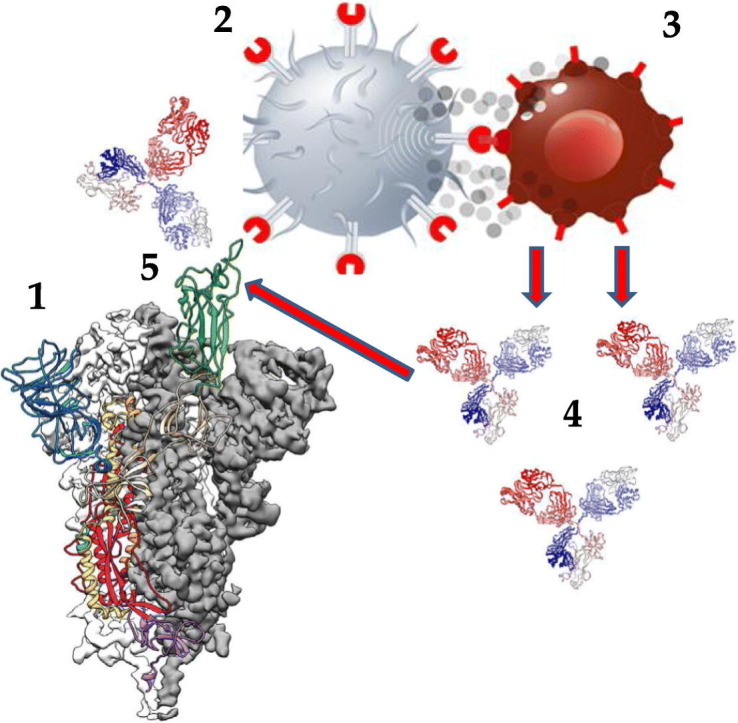
Graphical abstract
Immune response to SARS-CoV-2 spike protein (S):
The highly antigenic RBD (green) of S (1) is recognized by a T cell with a specific T cell receptor (2) that communicates to a B cell (3) to generate neutralizing antibodies (4) binding to the RBD (5).
1. Introduction
Coronaviruses, such as pandemic SARS-CoV and SARS-CoV-2, are highly pathogenic for humans and can induce a severe acute respiratory syndrome (SARS). Infection with SARS-CoV-2 which emerged in December 2019 in Wuhan, China, can progress to severe pneumonia, multi organ failure and death (COVID-19 disease) [[1], [2], [3]]. Like other human pathogenic enveloped viruses, coronaviruses use unique envelope protein complexes for host cell receptor recognition and binding, and subsequent viral and host cell membrane fusion, leading to cell entry [[4], [5], [6], [7], [8], [9], [10]]. Host cell entry of coronaviruses is mediated by a transmembrane homotrimeric class I fusion glycoprotein, the spike protein (S protein), which exists in a metastable prefusion conformation and comprises two functional subunits for binding to the host cell receptor angiotensin-converting enzyme 2 (ACE2) (S1 subunit) and for fusion of the viral and host cell membranes (S2 subunit) [11,12]. The S protein of SARS-CoV is cleaved by a host cell protease, the transmembrane protease/serine subfamily member 2 (TMPRSS2), an airway and alveolar cell serine protease preferentially expressed on epithelial cells of the respiratory tract, such as type II pneumocytes [[13], [14], [15]]. TMPRSS2-mediated cleavage and priming of SARS-CoV S protein is required for binding to ACE2, membrane fusion and cell entry that requires a concerted action of a viral and host cell machinery comprising S protein, TMPRSS2 and ACE2 [[13], [14], [15]]. SARS-CoV-2 also employs TMPRSS2 for priming of its S protein (S) and S-driven cell entry via ACE2 [[16], [17], [18], [19], [20], [21]]. Further, the S1/S2 boundary of SARS-CoV-2 S harbors multiple arginine residues not found in SARS-CoV and SARS-CoV-related S proteins. This S1/S2 boundary constitutes the cleavage site for the subtilisin-like host cell protease furin, which is ubiquitously expressed in humans [18,19,22].
The distal S1 subunit of S comprises the receptor-binding domains (RBDs) and contributes to stabilization of the prefusion state of the membrane-anchored S2 subunit that contains the fusion machinery [19]. For ACE2 receptor engagement, the RBDs located at the apex of S1 undergo hinge-like conformational movements that transiently expose (open status, “up”) or hide (closed status, “down”) the subdomains required for receptor binding, whereby the open status allows for receptor engagement, followed by shedding of S1 and refolding of S2 for membrane fusion [18,19]. Although the RBDs of the S1 subunit are more exposed on the viral surface than the S2 fusion machinery and are likely to be subject to selection pressure from immune surveillance, the S2 fusion machinery is densely decorated with heterogeneous N-linked glycans protruding from the S2 surface that may interfere with the elicitation of humoral immune responses and the accessibility to neutralizing antibodies [19]. In addition, the RBDs of S1 also contain N-linked glycans and unexpected O-linked glycans attached to the surface of S1 RBDs that also may interfere with the elicitation of neutralizing antibodies upon immune exposure or vaccination [23,24]. In individuals convalescent from COVID-19, the adaptive immunity to SARS-CoV-2 is largely mediated by CD4+ T cells with a T cell receptor repertoire specific for S epitopes, leading to the robust generation of neutralizing IgG, IgM and IgA antibodies against the RBDs and the ectodomain trimer of S1 [25,26]. Further, a recently designed human monoclonal IgG1 neutralizing antibody raised against and binding to a conserved epitope of the RBDs of S prevents infection of host cells [27], finally underscoring that understanding the structural features of S is key for vaccine design and development against SARS-CoV-2 infection.
2. Structural features of the SARS-CoV-2 S protein
Using sophisticated approaches, including high-resolution cryogenic electron microscopy (cryo-EM) at <4.0 Å, the labs of McLellan and Veesler recently uncovered the structural properties of SARS-CoV-2 S protein (S) [18,19] (Fig. 1A–C). S constitutes a tramsmembrane homotrimeric glycoprotein of ~180 kDa that belongs to the class I of trimeric fusion proteins found in other human pathogenic coronaviruses, including MERS-CoV and SARS-CoV. S is composed of two subunits, the apical V-shaped S1 ectotrimer subunit that harbors one ACE2-recognition motif per monomer (the receptor binding domain, RBD), and the S2 subunit required for fusion of the viral and cellular membranes (Fig. 2B, left) after being processed by the host cell protease furin at a polybasic cleavage site (with a four amino acid residue insertion, RRAR, at positions 681–684) that harbors multiple arginine residues and is located at the boundary between the S1 and S2 subunit [18,19,22]. Such polybasic cleavage sites are present in S proteins of human low pathogenic coronaviruses OC43 and HKU1, and in the S protein of the human high pathogenic coronavirus MERS.CoV [22], but are not present in SARS-CoV and SARS-CoV-related group 2b betacoronaviruses found in humans, civets, raccoon dog, pangolin and bats that possess a monobasic S1/S2 cleavage site processed upon entry of host cells [14,19,22,[28], [29], [30], [31], [32]]. The polybasic cleavage site of S may contribute to the high virulence of SARS-CoV-2, because furin and furin-like proteases required for proteolytic activation of S are ubiquitously expressed in humans, providing expanded tissue tropism of SARS-CoV-2 [18,19,22]. All 9 N-linked glycans protruding from the surface of one S2 monomer (Fig. 2A, right, Fig. 2B, right) are conserved among SARS-CoV and SARS-CoV-2, and the N-linked glycosylation sequons in S2 are mostly conserved across glycoproteins of SARS-CoV-related viruses [19], suggesting that these structures of S2 interfere with the elicitation of neutralizing antibodies and promote immune evasion [24].
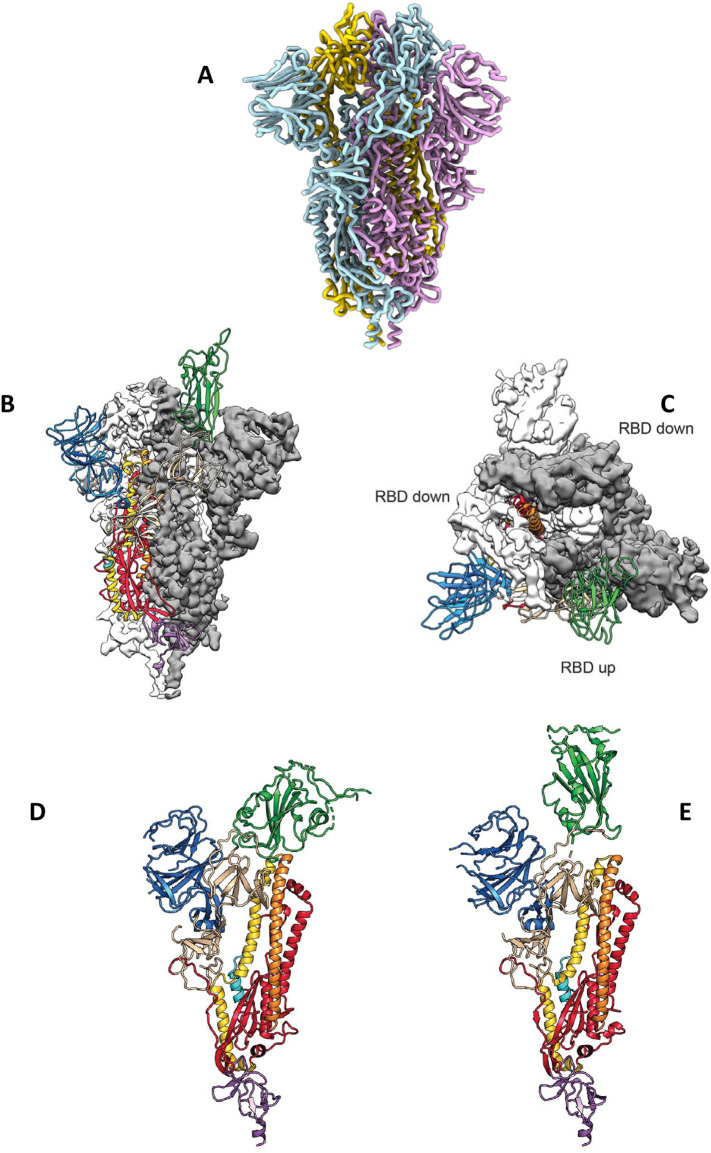
Fig. 1.
Structural features of the SARS-CoV-2 spike (S) protein. (A) Ribbon diagram of the homotrimeric S, adopted from [19], (with permission from Elsevier Inc.). (B) Side view of the prefusion structure of S, with a single RBD in open (“up”) conformation (green), adopted from [18], (with permission from Science.org). (C) Top view of the prefusion structure of S, with two single RBDs in closed (“down”) conformation (white and grey) and one single RBD in open (“up”) conformation (green), adopted from [18], (with permission from Science.org). (D) Single monomer of S, with the RBD in closed (“down”) conformation (green), adopted from [18], (with permission from Science.org). (E) Single monomer of S, with the RBD in open (“up”) conformation (green), adopted from [18], (with permission from Science.org). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
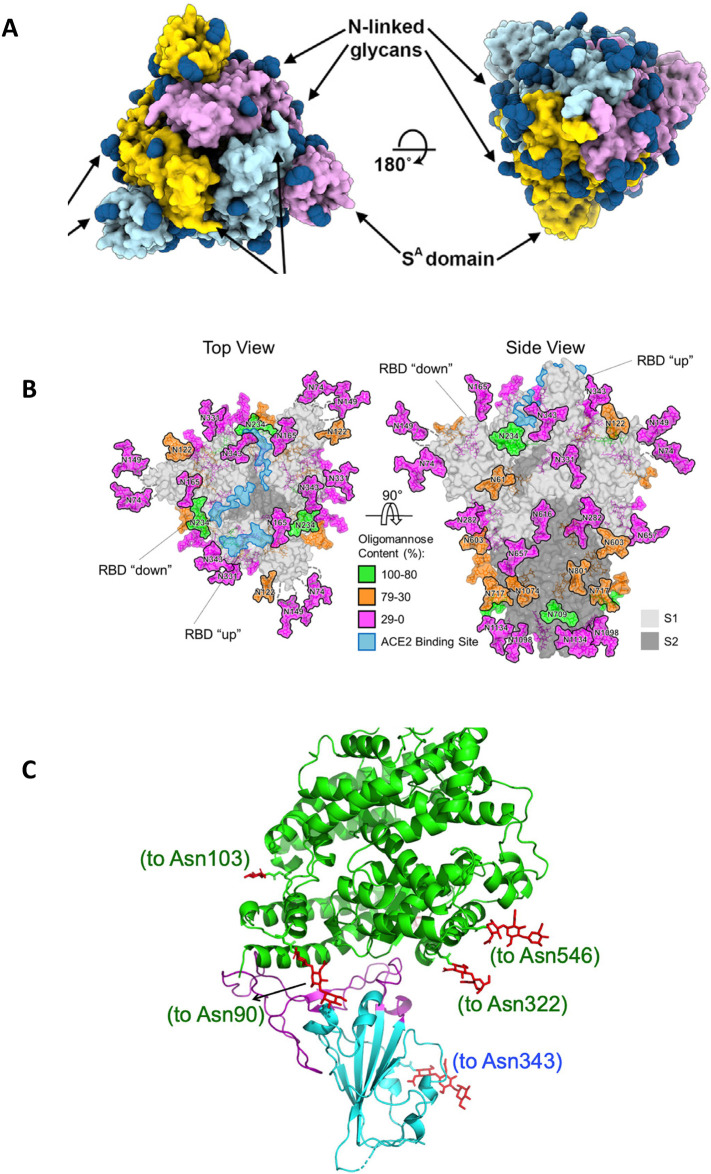
Fig. 2.
N-linked glycosylation of S, and the RBD of S binding to ACE2. (A) left: top view of the S1 homotrimer, with N-linked glycans as dark blue spheres; (A) right: bottom view of the S2 homotrimer, with N-linked glycans as dark blue spheres. Adopted from [19], (with permission from Elsevier Inc.). (B) Left: top view of the S1 homotrimer, with N-linked glycans colored according to their oligomannose content (green to pink), with the ACE2 binding site in light blue. (B) Right: side view of the S homotrimer, with N-linked glycans colored according to their oligomannose content (green to pink), with the ACE2 binding site in light blue, and the S1 (light grey) and S2 (dark grey) subunits. Adopted from [36], (with permission from Science.org). (C) The RBD (light blue), with its twisted five-stranded antiparallel β sheet of β1, β2, β3, β4 and β7 strands, binding to the bottom side of the small lobe of ACE2 (green helices). Adopted from [34], (with permission from Nature.com). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The S1 subunit of S is a 160-Å-long ectodomain trimer with a triangular cross-section [19] (Fig. 2A, left, Fig. 2B, left). At the apex of each S1 monomer, one RBD for ACE2 engagement is located. The RBDs undergo hinge-like conformational movements that transiently expose (open or “up” status) (Fig. 1B, C, E) or hide (closed or “down” status) (Fig. 1C, D) the determinants of receptor binding; the open status is required for ACE2 engagement [18,19].
The structural features of the RBD required for binding to ACE2 were recently determined using high-resolution X-ray crystallography [33,34] (Fig. 2C). Therefore, the RBD (residues Arg319-Phe541) and the N-terminal peptidase domain of ACE2 (residues Ser19-Asp615) were expressed in insect cells and subsequently purified. The structure of the complex was determined by molecular replacements using the RBD of SARS-CoV S and domains of ACE2 as search models, and was refined to a resolution of 2.45 Å [33]. The final complex contained residues Thr333-Gly526 of the RBD of SARS-CoV-2 S and residues Ser19-Asp615 of the ACE2 N-terminal peptidase domain [33]. This strategy revealed that the RBD of SARS-CoV-2 S contains a twisted five-stranded antiparallel β sheet of β1, β2, β3, β4 and β7 strands with connecting helices and loops that build the core of the RBD [33]. In the core, between the β4 and β7 strands, there is an extended insertion which contains short β5 and β6 strands and α4 and α5 helices and loops. This extended insertion constitutes the receptor-binding motif (RBM) of the RBD that contains the contacting residues that bind to ACE2 [33]. The extended RBM contacts the bottom side of a small lobe of ACE2, with a concave outer surface in the RBM that accommodates the N-terminal helix of ACE2 [33,34] (Fig. 2C, blue part). Compared to the RBM of SARS-CoV S, the RBM of SARS-CoV-2 S forms a larger binding interface and more contacts with ACE2 as well as higher binding affinity to ACE2 (equilibrium dissociation constant, K D, 4.7 nM versus 31 nM, respectively) [[33], [34], [35]], pointing out the higher infectivity and virulence of SARS-CoV-2 compared to SARS-CoV.
The S gene encodes 22 N-linked glycan sequons per monomer that affect S protein folding, processing by host cell proteases, elicitation of humoral immune response, and immune evasion [36]. Monomers of the S1 subunit are slightly more decorated with N-linked glycans (13) (Fig. 2A, left; Fig. B, left) than S2 monomers (9) (Fig. 2A, right; Fig. 2B, right), finally existing 66 N-linked glycans at N-linked glycosylation sites in one S homotrimer [19,36] (Fig. 2B, right). Compared to spike proteins of other human pathogenic coronaviruses, including HCoV-NL63 [37], MERS-CoV and SARS-CoV [38], and viral envelope glycoproteins, such as HIV-1 envelope glycoprotein [39] and Lassa virus GPC [40], S is less densely glycosylated [36] that may limit immune evasion and promote the elicitation of humoral immunity.
3. Humoral immunity against S
Extensive coronavirus S protein glycan shielding that obstructs the protein surface contributes to epitope masking and immune evasion by hiding specific epitopes from antibody neutralization [[36], [37], [38]]. Since S is less densely decorated with N-linked glycans compared to S proteins of other human pathogenic coronaviruses [[36], [37], [38]], S is likely to be highly immunogenic and a major target of neutralizing antibodies. Despite the high degree of structural homology between the S protein RBD of SARS-CoV and SARS-CoV-2, certain existing monoclonal antibodies raised against the SARS-CoV RBD (S230, m396, and 80R) failed to bind the RBD of SARS-CoV-2 S [18,33], suggesting that antibody cross-reactivity may be limited between the RBD of SARS-CoV and SARS-CoV-2. In contrast, sera obtained from patients convalescent from SARS-CoV infection, and rabbit sera raised against the S1 subunit of SARS-CoV S protein inhibited S-driven entry into simian Vero target cells [16]. Sera from mice immunized with a stabilized SARS-CoV S protein also significantly inhibited cell entry of SARS-CoV-2 into target cells [19], indicating that cross-neutralizing antibodies targeting conserved epitope of S proteins can be elicited upon vaccination. This is in line with the recent finding that CR3022, a human antibody isolated from a convalescent SARS-CoV patient and targeting the RBD of SARS-CoV S protein, binds to a highly conserved epitope distal from the ACE2 receptor binding site that enables cross-reactive binding between SARS-CoV-2 and SARS-CoV S proteins. Structural modeling approaches further demonstrate that the binding epitope can only be accessed by CR3022 when at least two RBDs of S protein are in the open conformation and slightly rotated [41]. Similar and extended results were obtained using a monoclonal antibody (S309) isolated from a convalescent SARS-CoV individual that potently neutralizes SARS-CoV-2 by engaging the RBD of S [42], indicating cross-neutralization by antibodies obtained from convalescent SARS-CoV individuals on SARS-CoV-2 by engagement of conserved S protein epitopes.
Consequently, monoclonal antibodies obtained from convalescent SARS-CoV-2 (COVID-19) individuals display neutralization activities against SARS-CoV-2 by targeting highly immunogenic epitopes of S, such as the RBD. In individuals convalescent from COVID-19, adaptive immune responses to SARS-CoV-2 are mediated by CD4+ T cells with a T cell receptor repertoire specific for S epitopes, leading to the robust generation of neutralizing IgG, IgM and IgA antibodies against the RBD and the ectodomain trimer of S [25,26]. By high-throughput single-cell RNA and VDJ sequencing of antigen-enriched B cells from 60 convalescent patients, various potent neutralizing antibodies were identified with the most potent one, BD-368-2, exhibiting strong neutralizing activity against SARS-CoV-2 [26]. Other monoclonal antibodies, B38 and H4, isolated from an individual convalescent from COVID-19 display neutralizing activity against SARS-CoV-2 by binding to the RBD-ACE2 interface [43], and RBD-specific monoclonal antibodies derived from single B cells of SARS-CoV-2 infected individuals exhibit potent neutralization activity that correlates with their competitive capacity with ACE2 for RBD binding [44]. Surprisingly, these monoclonal antibodies failed to bind the RBD of SARS-CoV and MERS-CoV S proteins [44], pointing out their specificity to the RBD of S.
A recently developed hybridoma-derived humanized monoclonal IgG1 neutralizing antibody (47D11) binds to the receptor-binding subdomain (residues 438–498) of S that loops out from the antiparallel β sheet core domain structure of the RBM of S that directly engages the binding domain of ACE2 [27]. Infection of simian VeroE6 cells with SARS-CoV-2 was effectively inhibited and neutralized at an IC50 value of 0.57 μg/ml [27], demonstrating high neutralizing activity of 47D11. Immunization of llama camelids, which are able to produce heavy-chain-only antibodies with a single variable domain (VHH) instead of two variable domains (VH and VL) that make up the equivalent antigen-binding fragment (Fab) of conventional immunoglobulin G (IgG) antibodies, with prefusion-stabilized SARS-CoV-1 S protein and MERS-CoV S protein in an alternating mode, resulted in the obtainment of cross-neutralizing VHHs targeting the RBD of S [45]. After engineering the VHH antibody into a bivalent Fc-fusion, the antibody construct was able to neutralize SARS-CoV-2 S pseudoviruses [45], revealing that S proteins of various coronaviruses including SARS-CoV-2 are highly immunogenic and can elicit effective humoral immune responses across mammalian species.
Recent work describes detection and isolation of potent neutralizing monoclonal antibodies from humans convalescent from SARS-CoV-2 infection and COVID-19 disease that may be engineered and used for passive immunization and therapeutic intervention [44,[46], [47], [48], [49]]. For instance, monoclonal antibodies derived from single B cells of SARS-CoV-2 infected individuals showed potent anti-SARS-CoV-2 neutralization activity that correlated with their competitive capacity with ACE2 for RDB binding [44]. SARS-CoV-2-neutralizing monoclonal antibodies isolated from infected patients hospitalized with severe COVID-19 disease displayed strong binding to the RBD and the N-terminal domain (NTD) of S, indicating that both of these S epitopes at the apex of S are highly immunogenic [46]. Another recent study demonstrated exclusive NTD specificity (epitope 4A8 of NTD) of neutralizing monoclonal antibodies isolated from convalescent COVID-19 patients [47], whereas predominant molecular targets of neutralizing monoclonal antibodies isolated from convalescent COVID-19 patients seem to be epitopes of the RBD of S [48,49] that correlates with the adaptive CD4+ T cell-mediated immune response to the RBD of S [25].
4. Vaccination strategies using S
Outbreak and pandemic of the betacoronaviruses SARS-CoV (2002/2003 in China) and MERS-CoV (2012 in Saudi Arabia) has led to the design and development of vaccination strategies mainly using recombinant viral S proteins as antigen [[50], [51], [52]].
Due to its high antigenicity and its proven ability to elicit robust humoral immune responses and neutralizing antibodies in individuals convalescent from SARS-CoV-2 infection and COVID-19 disease [25,26,43,44], S appears as an ideal candidate for vaccination against SARS-CoV-2 infection [[53], [54], [55], [56]], and constitutes an improved immunogen when stabilized in its prefusion conformation [18]. In a high-yield production approach, more than 100 structure-guided S variants based upon a previously determined cryo-EM structure of the prefusion S were designed, expressed and produced in Chinese hamster ExpiCHO cells [57]. The best prefusion S variant, termed HexaPro, was extremely stable in the prefusion state, retained the S2 subunit conformation and preserved its high antigenicity due to its stable prefusion conformation [57].
Recently, two synthetic DNA-based vaccine candidates expressing different forms of S were developed and investigated in rhesus macaques [58], and mice and guinea pigs [59]. A series of prototype DNA vaccines expressing six variants of the S: 1) full-length S, 2) deletion of the cytoplasmic tail of S, 3) deletion of the transmembrane domain and cytoplasmic tail reflecting the soluble ectodomain of S, 4) S1 domain with a foldon trimerization tag, 5) RBD of S with a foldon trimerization tag, and 6) a prefusion stabilized soluble ectodomain of S with deletion of the furin cleavage site, two proline mutations and a foldon trimerization tag, were produced [58]. Adult rhesus macaques in groups of 4 animals were immunized with one of the six prototype DNA vaccines, respectively, and each animal received 5 mg DNA vaccine by the intramuscular route without adjuvant at week 0 and week 3; ten animals not vaccinated served as control group [58]. After a boost immunization at week 5, S-specific binding antibodies and neutralizing antibodies (NAb) could be obtained from the animals, with median titers of the NAb comparable in the magnitude to NAb titers in a cohort of 9 convalescent macaques and in a cohort of 27 humans convalescent from SARS-CoV-2 infection [58]. Further, a Th1-biased cellular immune response of S-specific IFN-γ + CD4+ T cells to pooled S peptides was detected in the majority of vaccinated animals at week 5 [58]. Three weeks after the boost immunization, animals of the vaccine group and the control group were challenged with SARS-CoV-2, administered by the intranasal and the intratracheal route. In the broncho-alveolar lavage (BAL) and nasal swabs (NS) of the control group, high levels of viral RNA could be detected as compared to significant lower viral RNA levels in the vaccine group, and 8 of 25 vaccinated animals exhibited no detectable viral RNA in BAL and NS at any timepoint following the challenge [58], demonstrating high protective efficacy of the S-expressing DNA vaccine against intranasal and intratracheal SARS-CoV-2 infection in rhesus macaques.
In a similar study, a synthetic DNA plasmid, termed pGX9501/INO-4800, was designed to encode S that matches with >99.9% amino acid sequence identity of the recently published S sequences [59]. Intramuscular administration of INO-4800 in Balb/c mice on days 0 and 14 resulted in the elicitation of neutralizing IgG antibodies at day 21 that bind to S protein antigens, including S1 and S2 subunits, and RBD, as well as to the S-ACE2 interface, with limited cross-reactivity to SARS-CoV S protein antigens [59]. Similar results were obtained with INO-4800 in Hartley guinea pigs. Neutralizing antibodies were found in the BAL fluids of the animals at day 28 after vaccination, revealing strong lung tropism [56]. Moreover, a cellular immune response against S epitopes mediated by CD4+ and CD8+ IFN-γ + T cells was detected on day 14 after vaccination [59].
Furthermore, a vaccine candidate consisting in chimpanzee-derived adenoviral vector (ChAdOx1), expressing full-length S (GenBank accession number YP_009724390.1) and termed ChAdOx1 nCoV-19, has been shown to induce a robust humoral and cellular immune response in rhesus macaques after a single vaccination [60]. S-specific neutralizing antibodies and T-cell responses against full-length S could be detected 14 days post vaccination. A significantly reduced viral load in broncho-alveolar lavage fluid and respiratory tract tissue of vaccinated animals challenged with SARS-CoV-2 compared with control animals, and no pneumonia was observed in vaccinated rhesus macaques [60]. ChAdOx1 nCoV-19 is currently under investigation in a phase II/III clinical trial in the UK [60].
In a dose-escalation, open label, non-randomized, first-in-human trial, 108 healthy humans with mean age of 36 years received single low dose (n = 36), middle dose (n = 36) and high dose (n = 36) of a recombinant replication-defective adenovirus type-5 (Ad5) vectored vaccine expressing S, with full-length S gene based on Wuhan-Hu-1 strain (GenBank accession number YP_009724390) [61]. This resulted in the occurrence of frequent (in more than 80% of the participants) adverse reactions within the first 7 days after vaccination, including fever, fatigue, headache and muscle pain, but elicited robust cellular and humoral immune responses, with neutralizing antibodies binding to the RBD of S (from day 14, peaking at 28 after vaccination), and CD4+ and CD8+ T cells specific for S epitopes (from day 14 after vaccination) [61]. These inaugural results suggest that vaccination of humans using S as antigen will be successful. Consequently, a phase II clinical trial with Ad5 at low or middle dose has started in China [62], and Canada has approved a phase I/II clinical trial in humans with Ad5 [63]. However, pre-existing immunity against the Ad5 vector could reduce immunogenicity, potentially limiting efficacy in populations in which adenovirus type-5 is endemic, with a reported seroprevalence of 30–80% [64,65].
Currently, other vaccine candidates based on S or its RBD are in rapid development (Table 1 ), and different antigen delivery platforms, including recombinant protein vaccines, replicating or non-replicating viral-vector-based vaccines, and DNA or mRNA vaccines are under investigation [[53], [54], [55], [56],[66], [67], [68], [69]]. An overview of the current status of the pipeline of COVID-19 vaccine candidates is provided in Ref. [55] and Refs. [[66], [67], [68], [69]]. For example, phase I/II clinical first-in-humans trials using m-RNA vaccines encoding full-length S, the RBD of S [[70], [71], [72]] or a recombinant trimeric S subunit for vaccination [73] have been started recently (Table 1), pointing out a pivotal role of S and its RBD for vaccination against SARS-CoV-2.
Table 1.
Examples of vaccines targeting S that are in clinical development.
| Vaccine | Type | Target/Antigen | Clinical Status | Reference |
|---|---|---|---|---|
| ChAdOx1 nCoV-19 | Chimpanzee-derived adonoviral vectored vaccine expressing full-length S | Full-length S, (GenBank accession number YP_009724390.1) | Phase II/III clinical trial. UK, University of Oxford |
[60] |
| Ad5 | Recombinant replication-defective adenovirus type-5 (Ad5) vectored vaccine expressing full-length S | Full-length S, based on Wuhan-Hu-1 strain (GenBank accession number YP_009724390) | Phase I, dose-escalation, open label, non-randomized, first-in-human clinical trial. China |
[61] |
| Ad5 | Recombinant replication-defective adenovirus type-5 (Ad5) vectored vaccine expressing full-length S | Full-length S, based on Wuhan-Hu-1 strain (GenBank accession number YP_009724390) | Phase II, low and middle dose. China |
[62] |
| Ad5 | Recombinant replication-defective adenovirus type-5 (Ad5) vectored vaccine expressing full-length S | Full-length S, based on Wuhan-Hu-1 strain (GenBank accession number YP_009724390) | Phase I/II, low and middle dose. Canada |
[63] |
| mRNA-1273 | Lipid nanoparticle (LNP)-encapsulated mRNA-based vaccine encoding full-length, prefusion stabilized S | Full-length, prefusion stabilized S |
Phase II/III clinical trial. USA |
[70,71] |
| Different mRNAs: BNT 162a1, BNT 162b1, BNT 162b2, BNT 162c2 |
Lipid nanoparticle (LNP)-encapsulated mRNA-based vaccines encoding either full-length S or RBD of S | Full length S, or RBD of S |
Phase I/II, randomized, placebo-controlled, observer-blind, dose-finding clinical trial. Low, middle and high dose. USA, Germany |
[72] |
| SCB-2019 | Recombinant trimeric S subunit, with different adjuvants | Trimeric S subunit | Phase I, randomized, double blind, placebo controlled, first-in-human clinical trial. USA |
[73] |
5. Conclusion
The outbreak of coronavirus SARS-CoV-2 in Wuhan, China, in December 2019, the cause of COVID-19 disease, represents a pandemic threat to global health and has major consequences on global economy if SARS-CoV-2 spread and virulence is not contained, or effective treatments are not developed [74]. The pandemic has spread to more than 188 countries with more than 10,000,000 confirmed cases, more than 500,000 confirmed deaths and more than 5,000,000 total recoveries worldwide as of June 30th 2020 [75]. Since no specific drug against SARS-CoV-2 infection or COVID-19 disease is available or approved to date [76,77], it is mandatory to rapidly develop and provide successful vaccines against SARS-CoV-2 that should be available soon for large populations. Usually, it takes 10–15 years of vaccine development by the classical way, using inactivated or live attenuated vaccines after the generation of long-term safety and efficacy data [[78], [79], [80]]. Clinical development of vaccines begins with phase I trials to evaluate the safety of vaccine candidates, followed by phase II trials to establish doses and formulations to prove efficacy, and finally followed by phase III trials to prove and demonstrate safety and efficacy in larger human cohorts [53]. In an extraordinary situation like the COVID-19 pandemic, this procedure should be compressed, and an accelerated regulatory approval pathway might be developed [53].
The spike protein of SARS-CoV-2 (S) is a homotrimeric class I fusion protein protruding from the viral surface that is required for host cell receptor (ACE2) recognition and binding, and fusion of the viral and cellular membrane, leading to viral cell entry. Since S is highly exposed on the viral surface, it is likely to be subject to immune surveillance by T cells and professional antigen-presenting cells, leading to the elicitation of neutralizing antibodies against specific epitopes and domains of S. This is finally not surprising, because the human cellular and humoral immune system is capable of generating a response very specific to the structure of a foreign invading virus or protein, and the general adaptive immune response consisting of antigen capture and presentation by professional antigen-presenting cells, followed by T cell receptor-mediated antigen recognition and B cell-driven production of antibodies specific for the given antigen, is fortunately also operative in SARS-CoV-2 infection, obviously by choosing S epitopes as major antigens [25,81,82].
S is less densely decorated with N-linked glycans compared to S proteins of other human pathogenic coronaviruses, which display extensive S protein glycan shielding that obstructs the protein surface and thereby contributes to epitope masking and immune evasion. Therefore, S appears highly immunogenic and as a target for vaccination. In fact, a considerable number of recent studies discussed herein have demonstrated high immunogenicity of S and its RBD, leading to adaptive T cell-mediated immune responses and, finally, to the elicitation of neutralizing antibodies in humans and various mammals that can prevent SARS-CoV-2 infection and rechallenge.
In conclusion, deep understanding of the structural features of S will facilitate the design and development of successful vaccines against coronavirus SARS-CoV-2 for large populations.
Declaration of competing interest
None.
References
- 1.Liu J., Zheng X., Tong Q., Li W., Wang B., Sutter K., Trilling M., Lu M., Dittmer U., Yang D. Overlapping and dicrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J. Med. Virol. 2020;92:491–494. doi: 10.1002/jmv.25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang C., Yao X., Zhao Y., Wu J., Huang P., Pan C., Liu S., Pan C. Comparative review of respiratory diseases caused by coronaviruses and influenza a viruses during epidemic season. Microbes. Infect. May. 2020;13 doi: 10.1016/j.micinf.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y., Peng F., Wang R., Guan K., Jiang T., Xu G., Sun J., Chang C. The deadly coronaviruses: the 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greber U.F. Virus and host mechanics support membrane penetration and cell entry. J. Virol. 2016;90:3802–3805. doi: 10.1128/JVI.02568-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tong Y., Lavillette D., Li Q., Zhong J. Role of hepatitis C virus envelope glycoprotein E1 in virus entry and assembly. Front. Immunol. 2018;9:1411. doi: 10.3389/fimmu.2018.01411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arii J., Kawaguchi Y. The role of HSV glycoproteins in mediating cell entry. Adv. Exp. Med. Biol. 2018;1045:3–21. doi: 10.1007/978-981-10-7230-7_1. [DOI] [PubMed] [Google Scholar]
- 7.Malito E., Chandramouli S., Carfi A. From recognition to execution – the HCMV pentamer from receptor binding to fusion triggering. Curr. Opin. Virol. 2018;31:43–51. doi: 10.1016/j.coviro.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Azam K.D., Lee B. Differential features of fusion activation within the Paramyxoviridae. Viruses. 2020;12:161. doi: 10.3390/v12020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Millet J.K., Whittaker G.R. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology. 2018;517:3–8. doi: 10.1016/j.virol.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoeman D.J., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walls A.C., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J.M., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Tagushi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84 doi: 10.1128/JVI.01542-10. 12685-12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell Apr. 2020;27 doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:1–6. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shajahan A., Supekar N.T., Gleinich A.S., Azadi P. Deducing the N- and O- glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020 doi: 10.1093/glycob/cwaa042. May 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vigerust D.J., Shepherd V.L. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 2007;15:211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Rydyznski Moderbacher C., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell May. 2020;13 doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bosch B.J., Bartelink W., Rottier P.J. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 2008;82:8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam T.T., Shum M.H., Zhu H.C., Tong Y.G., Ni X.B., Liao Y.S., et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature Mar. 2020;26 doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 31.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARSCoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30:1346–1351. doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Zai J., Zhao Q., Nie Q., Li Y., Foley B.T., et al. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J. Med. Virol. Feb. 2020;27 doi: 10.1002/jmv.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 34.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020 doi: 10.1073/pnas.2003138117. May 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020 doi: 10.1126/science.abb9983. May 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walls A.C., Tortorici M.A., Frenz B., Snijder J., Li W., Rey F.A., et al. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016;23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walls A.C., Xiong X., Park Y.J., Tortorici M.A., Snijder J., Quispe J., et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Behrens A.J., Vasiljevic S., Pritchard L.K., Harvey D.J., Andev R.S., Krumm S.A., et al. Composition and antigenic effects of individual glycan sites of a trimeric HIV-1 envelope glycoprotein. Cell Rep. 2016;14:2695–2706. doi: 10.1016/j.celrep.2016.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe Y., Raghwani J., Allen J.D., Seabright G.E., Li S., Moser F., et al. Structure of the Lassa virus glycan shield provides a model for immunological resistance. Proc. Natl. Acad. Sci. U. S. A. 2018;115:7320–7325. doi: 10.1073/pnas.1803990115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan M., Wu N.C., Zhu X., Lee C.C.D., So R.T.Y., Lv H., et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020 doi: 10.1038/s41586-020-2349-y. May 18. [DOI] [PubMed] [Google Scholar]
- 43.Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020 doi: 10.1126/science.abc2241. May 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020 doi: 10.1038/s41586-020-2380-z. May 26. [DOI] [PubMed] [Google Scholar]
- 45.Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wand N., Van Breedam W., et al. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020 doi: 10.1016/j.cell.2020.04.031. May 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu L., Wang P., Nair M.S., Yu J., Huang Y., Rapp M.A., et al. 2020. Potent Neutralizing Monoclonal Antibodies Directed to Multiple Epitopes on the SARS-CoV-2 Spike. bioRxiv Jun 18. 06.17.153486. [DOI] [PubMed] [Google Scholar]
- 47.Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., et al. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020:eabc6952. doi: 10.1126/science.abc6952. Jun 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020:eabc5902. doi: 10.1126/science.abc5902. Jun 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi R., Chan C., Duan X., Chen Z., Liu P., Song J., et al. A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2381-y. May 26. [DOI] [PubMed] [Google Scholar]
- 50.Martin J.E., Louder M.K., Holman L.A., Gordon I.J., Enama M.E., Larkin B.D., et al. A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a phase I clinical trial. Vaccine. 2008;26:6338–6343. doi: 10.1016/j.vaccine.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong C.Y., Ong H.K., Yeap S.K., Ho K.L., Tan W.S. Recent advances in the vaccine development against middle east respiratory syndrome-coronavirus. Front. Microbiol. 2019;10:1781. doi: 10.3389/fmicb.2019.01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang F., Kream R.M., Stefano G.B. An evidence based perspective on mRNA-SARS-CoV-2 vaccine development. Med. Sci. Monit. 2020;26 doi: 10.12659/MSM.924700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mukherjee R. Global efforts on vaccines for COVID-19: since, sooner or later, we all will catch the coronavirus. J. Biosci. 2020;45:68. doi: 10.1007/s12038-020-00040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robson B. Computers and viral diseases. Preliminary bioinformatics studies on the design of a synthetic vaccine and a preventative peptidomimetic antagonist against the SARS-CoV-2 (2019-nCoV, COVID-19) coronavirus. Comput. Biol. Med. 2020;119 doi: 10.1016/j.compbiomed.2020.103670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsieh C.L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.C., Javanmardi K., et al. 2020. Structure-based Design of Prefusion-Stabilized SARS-CoV-2 Spikes. bioRxiv 2020 May 30. 05.30.125484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020 doi: 10.1126/science.abc6284. May 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., et al. 2020. ChAdOx1 nCoV-19 Vaccination Prevents SARS-CoV-2 Pneumonia in Rhesus Macaques. bioRxiv. 2020 May 13. 2020.05.13.093195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu F.C., Li Y.H., Guan X.H., Hou L.H., Wang W.J., Li J.X., et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020 doi: 10.1016/S0140-6736(20)31208-3. May 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. NCT04341389. Clinical Trials.gov. 2020. (Apr 10) [Google Scholar]
- 63. NCT04398147. ClinicalTrials.gov. 2020. (May 21) [Google Scholar]
- 64.Barouch D.H., Kik S.V., Weverling G.J., Dilan R., King S.L., Maxfield L.F., et al. International seroepidemiology of adenovirus serotypes 5, 26, 35, and 48 in pediatric and adult poulations. Vaccine. 2011;29:5203–5209. doi: 10.1016/j.vaccine.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu F.C., Wurie A.H., Hou L.H., Liang Q., Li Y.H., Russell J.B.W., et al. Safety and immunogenicity of a recombinant adenovirus type-5 vector-based Ebola vaccine in healthy adults in Sierra Leone: a single-centre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2017;389:621–628. doi: 10.1016/S0140-6736(16)32617-4. [DOI] [PubMed] [Google Scholar]
- 66.Lurie N., Saville M., Hatchett R., Halton J. Developing COVID-19 vaccines at pandemic speed. N. Engl. J. Med. 2020;382:1969–1973. doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- 67.Le T.T., Andreadakis Z., Kumar A., Román R.G., Tollefsen S., Saville M., et al. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 68.Graham B.S. Rapid COVID-19 vaccine development. Science. 2020;368:945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- 69.Chen W.H., Strych U., Hotez P.J., Bottazzi M.E. The SARS-CoV-2 vaccine pipeline: an overview. Curr. Trop. Med. Rep. 2020 doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. NCT04283461. ClinicalTrials.gov. 2020. (Feb 25) [Google Scholar]
- 71.Corbett K.S., Edwards D., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., et al. 2020. SARS-CoV-2 mRNA Vaccine Development Enabled by Prototype Pathogen Preparedness. bioRxiv Jun 11. 06.11.145920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. NCT04368728. ClinicalTrials.gov. 2020. (Apr 30) [Google Scholar]
- 73. NCT04405908. ClinicalTrials.gov. 2020. (May 28) [Google Scholar]
- 74.Lu R., Zhao X., Li J., Niu P., Wang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.https://coronavirus.jhu.edu/map.html, Johns Hopkins University, USA, (2020).
- 76.McKee D.L., Sternberg A., Stange U., Laufer S., Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol. Res. 2020;157 doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sternberg A., McKee D.L., Naujokat C. Novel drugs targeting the SARS-CoV-2/COVID-19 machinery. Curr.Top. Med. Chem. May. 2020;16 doi: 10.2174/1568026620999200517043137. [DOI] [PubMed] [Google Scholar]
- 78.Papaneri A.B., Johnson R.F., Wada J., Bollinger L., Jahrling P.B., Kuhn J.H. Middle east respiratory syndrome: obstacles and prospects for vaccine development. Expert Rev. Vaccines. 2015;14:949–962. doi: 10.1586/14760584.2015.1036033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kaiser J.A., Barrett A.D.T. Twenty years of progress toward West Nile virus vaccine development. Viruses. 2019;11:823. doi: 10.3390/v11090823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5(48):eabd2071. doi: 10.1126/sciimmunol.abd2071. Jun 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ni L., Ye F., Cheng M.L., Feng Y., Deng Y.Q., Zhao H., et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52(e3):971–977. doi: 10.1016/j.immuni.2020.04.023. Jun 16. [DOI] [PMC free article] [PubMed] [Google Scholar]