Bodda et al. show that the deubiquitinase activity of the HSV1 VP1-2 protein inhibits induction of type I interferon in the brain. This is mediated through deubiquitination of the signaling protein STING, thus preventing activation of host defense after sensing of viral DNA by cGAS.
Abstract
Herpes simplex virus (HSV) is the main cause of viral encephalitis in the Western world, and the type I interferon (IFN) system is important for antiviral control in the brain. Here, we have compared Ifnb induction in mixed murine brain cell cultures by a panel of HSV1 mutants, each devoid of one mechanism to counteract the IFN-stimulating cGAS–STING pathway. We found that a mutant lacking the deubiquitinase (DUB) activity of the VP1-2 protein induced particularly strong expression of Ifnb and IFN-stimulated genes. HSV1 ΔDUB also induced elevated IFN expression in murine and human microglia and exhibited reduced viral replication in the brain. This was associated with increased ubiquitination of STING and elevated phosphorylation of STING, TBK1, and IRF3. VP1-2 associated directly with STING, leading to its deubiquitination. Recruitment of VP1-2 to STING was dependent on K150 of STING, which was ubiquitinated by TRIM32. Thus, the DUB activity of HSV1 VP1-2 is a major viral immune-evasion mechanism in the brain.
Introduction
HSV1 is a neurotropic alphaherpesvirus, and the cause of herpes simplex encephalitis (HSE), which is a devastating disease with high mortality if not treated (Whitley, 2006). Even if treatment is initiated early after onset of symptoms, HSE is associated with significant mortality and morbidity (Tyler, 2018). HSV1 infections are initiated at mucosal surfaces where the virus infects epithelial cells. This is followed by infection of innervating peripheral sensory nerves, where HSV1 replicates but also establishes latency. In rare cases, HSV1 can spread from the peripheral nerves and into the central nervous system (CNS) to cause HSE. This can occur during both primary infection and reactivation.
The innate immune system plays a central role in control of HSV1 infections, both in the periphery, and in the brain (Paludan et al., 2011). In particular, type I IFN is important for control of HSV1 in the brain of humans and mice (Conrady et al., 2013; Dupuis et al., 2003; Leib et al., 1999). We and the Carr laboratory reported that microglia are the key source of IFN in the HSV1-infected brain, while Leib and colleagues showed that IFN responsiveness in neurons is essential for antiviral activity (Conrady et al., 2013; Reinert et al., 2016; Rosato and Leib, 2015). This suggests that a microglia-neuron IFN circuit is important for control of HSV1 in the CNS. During a viral infection, type I IFN is induced primarily by viral nucleic acids, which are recognized by pattern recognition receptors. Endosomal viral RNA and DNA are detected by TLR3, TLR7, and TLR9 (Takeuchi and Akira, 2010); abnormal cytoplasmic RNA structures are detected by retinoic acid–inducible gene I–like receptors (Goubau et al., 2013); and cytoplasmic DNA is sensed by cyclic GMP-AMP synthase (cGAS; Paludan, 2015).
Human genetics data have revealed that loss of function of TLR3, or four downstream factors in the TLR3 signaling pathway, namely TIR domain–containing adaptor-inducing IFN-β (TRIF), TNF receptor-associated factor 3 (TRAF3), TANK-binding kinase 1 (TBK1), and IFN regulatory factor 3 (IRF3) lead to increased susceptibility to HSE (Andersen et al., 2015; Herman et al., 2012; Pérez de Diego et al., 2010; Sancho-Shimizu et al., 2011; Zhang et al., 2007). Moreover, mouse studies have shown an essential role for cGAS and its downstream adaptor protein stimulator of IFN genes (STING) in control of HSV1 in the brain (Ishikawa et al., 2009; Li et al., 2013; Reinert et al., 2016). Interestingly, TBK1 and IRF3 are also involved in signaling downstream of STING, and cells from a patient with HSE-associated mutations in IRF3 have defective signaling in both TLR3–TRIF and cGAS–STING (Andersen et al., 2015). This indicates that both the TLR3–TRIF and cGAS–STING pathways are involved in control of HSV1 infections. In further support of a role for the cGAS–STING pathway in control of HSV1, there are now reports of close to 10 HSV1-encoded proteins targeting this pathway (Christensen et al., 2016; Deschamps and Kalamvoki, 2017; Huang et al., 2018; Orzalli et al., 2012b; Su and Zheng, 2017; Verpooten et al., 2009; Wang et al., 2013b; Ye et al., 2017; Zhang et al., 2018). However, it remains unresolved how HSV1 evades the IFN-mediated immune response in the brain.
The cGAS–STING pathway is tightly regulated by posttranslational modifications, including phosphorylation (Liu et al., 2015), palmitoylation (Mukai et al., 2016), amidation (Zhang et al., 2018), glutamylation (Xia et al., 2016), and ubiquitination (Ni et al., 2017; Tsuchida et al., 2010; Wang et al., 2014; Zhang et al., 2012; Zhong et al., 2009). Regarding the latter, it is well established that K27- and K63-linked polyubiquitination of STING promotes recruitment of the kinase TBK1, which is an essential step in activation of the transcription factor activity of IRF3 (Paludan, 2015). The ubiquitin E3 ligase AMFR/INSIG1 is responsible for K27-linked polyubiquitination of STING (Wang et al., 2014), while MUL1 and tripartite motif-containing 32 (TRIM32) mediate K63-linked polyubiquitination of STING (Ni et al., 2017; Zhang et al., 2012). TRIM56 was proposed also to add K63-linked ubiquitin to STING (Tsuchida et al., 2010), but it was later demonstrated that monoubiquitination of cGAS is the TRIM56 target in the pathway (Seo et al., 2018).
In this work, we show that the deubiquitinase (DUB) activity of the HSV1 VP1-2 protein (also termed pUL36; Kattenhorn et al., 2005) is a potent inhibitor of HSV1-induced type I IFN expression in the brain. This is mediated through digestion of K63-linked ubiquitin chains from STING, thus preventing downstream activation of IRF3 and IFN expression. Deletion of HSV1 DUB activity leads to higher IFN production and reduced HSV1 replication in the brain following infection. Thus, HSV1 targets STING ubiquitination in the brain to promote viral infection and potentially progression to HSE.
Results
VP1-2 DUB activity is a major HSV1 IFN induction antagonist in mixed brain cell cultures
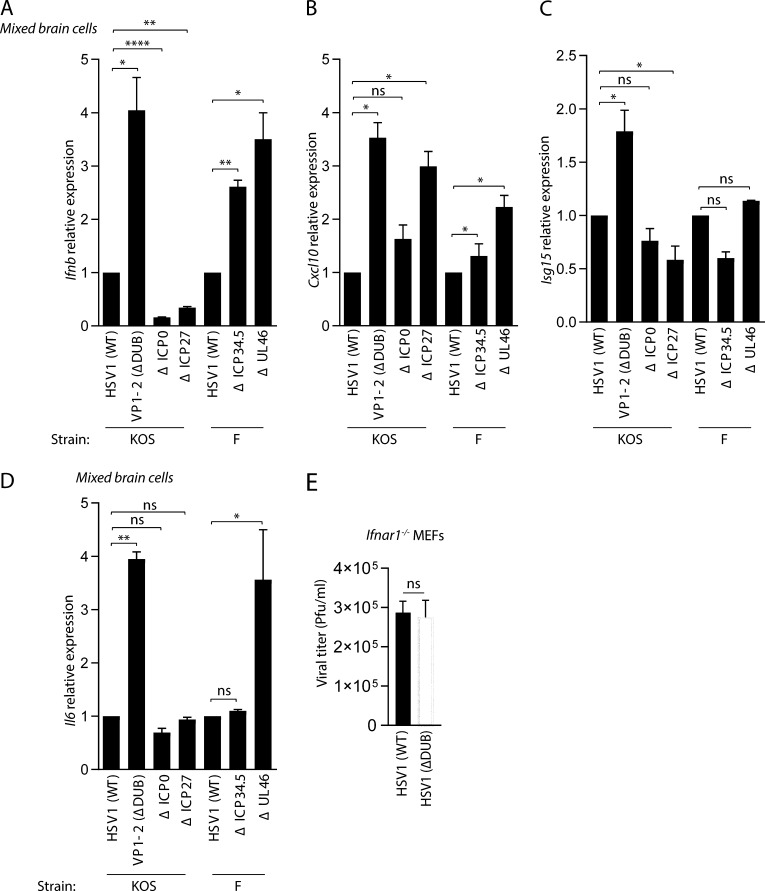
We wanted to evaluate the impact of different HSV1 evasion proteins by comparing IFN, IFN-stimulated genes (ISGs), and cytokine induction by viral mutants lacking reported cGAS–STING-targeting proteins/activities (Deschamps and Kalamvoki, 2017; Ma et al., 2012; Orzalli et al., 2012b; Ye et al., 2017). In our experiments, we used HSV1 mutants derived from the KOS and F strains. To focus on the CNS, we used mixed brain cell cultures from mice. The cells were infected with an equivalent inoculum of each virus at a multiplicity of infection (MOI) of 3, and virus-induced gene expression was assayed at 8 h. In this system, we observed that lack of ICP0 or ICP27 did not lead to elevated Ifnb expression. By contrast, virus mutants lacking the VP1-2 DUB activity (C65A, ΔDUB HSV), ICP34.5, or UL46 induced significantly more Ifnb expression than the respective WT viruses (Fig. 1 A). Brain cells infected with the ΔDUB HSV1 and to a lesser extent also the ΔUL46 mutant induced elevated levels of the two ISGs Cxcl10 and Isg15 (Fig. 1, B and C). These two viral deletion mutants also induced significantly elevated expression of the gene encoding the inflammatory cytokine interleukin 6. (Fig. 1 D). To test whether the observed elevated induction of Ifnb, ISGs, and Il6 by the ΔDUB HSV1 mutant was also seen on a different genetic background, we generated a ΔDUB HSV1 mutant in the strain 17+ (Fig. S1, A–C). As seen for the strain on the KOS background, the 17+-derived ΔDUB HSV1 mutant induced elevated IFN and ISG responses (Fig. S1, D and E). There is currently no knowledge on viral modulation of ubiquitin in the immune system in the brain. Moreover, the ΔDUB HSV1 mutant exhibited unaltered replication capacity in a cellular system devoid of type I IFN activity (Fig. 1 E), thus showing that the observed effects on host gene expression were not due to differential viral replication. Therefore, we decided to focus on the role of the HSV1 VP1-2 DUB activity in inhibition of type I IFN activity in brain cells.
Figure 1.
VP1-2 DUB activity is the major HSV1 IFN antagonist in mixed brain cell cultures. (A–D) Isolated mixed murine brain cells were infected with HSV1 (WT), VP1-2 (ΔDUB), ΔICP0, ΔICP27 (all KOS strain) and HSV1 (WT), ΔICP34.5, ΔUL46 (all F strain; MOI 3) for 8 h. Total RNA was isolated, and mRNA levels of Ifnb, Cxcl10, Isg15, and Il6 were analyzed by quantitative RT-PCR. The mRNA data were normalized to β-actin mRNA levels and are displayed as relative levels compared with HSV1 (WT) values (n = 3). (E) Ifnar1−/− MEFs were infected with WT and ΔDUB HSV1 (KOS strain) for 24 h at MOI 1. Viral loads in the supernatants were determined by plaque assay. P values were calculated using a two-tailed unpaired Student’s t test. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, not significant. Error bars represent standard deviation.
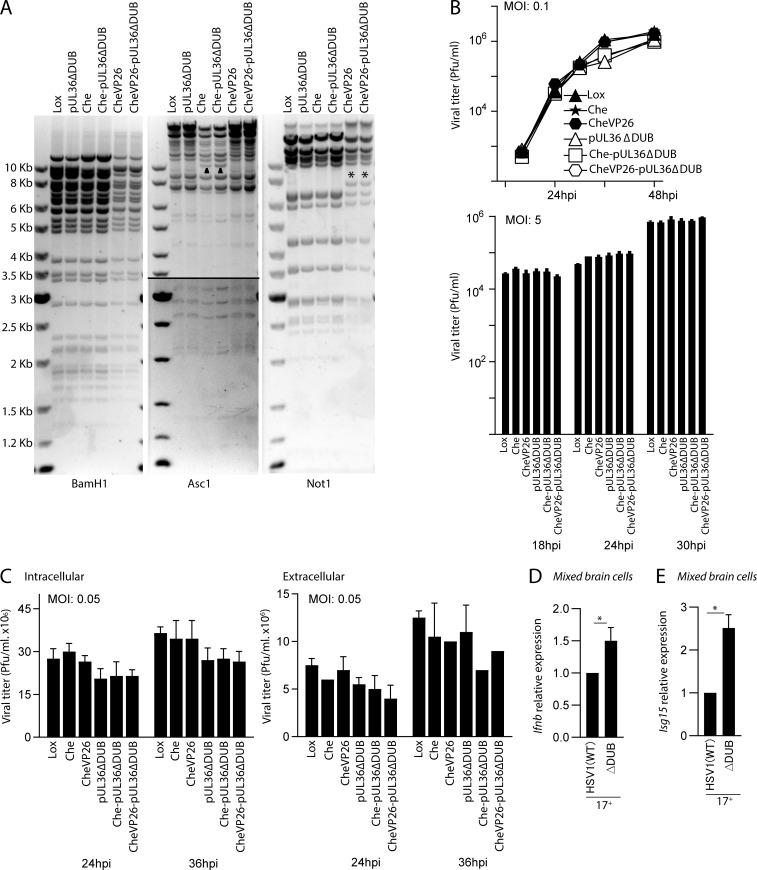
Figure S1.
Generation of new HSV1-pUL36ΔDUB strains, and effects of DUB deficiency on induction of IFN responses. (A) Characterization of HSV1-pUL36ΔDUB strains Restriction digest analysis with BamH1, Asc1, or Not1 of the parental BACs pHSV1(17+)Lox (Lox), pHSV1(17+)Lox-pMCMVmCherry (Che), and pHSV1(17+)Lox-CheVP26 (CheVP26) and the mutants pHSV1(17+)Lox-pUL36ΔDUB (pUL36ΔDUB), pHSV1(17+)Lox-pMCMVmCherry-pUL36ΔDUB (Che-pUL36ΔDUB), and pHSV1(17+)Lox-CheVP26-pUL36ΔDUB (CheVP26-pUL36ΔDUB). The agarose gels show the expected fragment sizes for HSV-1(17+; GenBank accession numbers NC001806 and JN555585) and pHSV1(17+)Lox. Asc1 fragments of ∼10 kb had shifted due to the insertion of pMCMVmCherry (triangle), and Not1 fragments of ∼8 kb due to the addition of mCherry to VP26 (asterisk). Kb, kilobase. (B) Vero cells were infected at MOI 0.1 or MOI 5. The combination of supernatants and cells was titrated at the respective time points. (C) HaCaT cells were infected at MOI 0.05, and the amount of infectious intracellular and extracellular virus was titrated at 24 and 36 h after infection (hpi). (D and E) Isolated mixed murine brain cells were infected with HSV1 (WT), VP1-2 (ΔDUB) on the 17+ background (MOI 3) for 8 h. Total RNA was isolated, and mRNA levels of Ifnb and Isg15 were analyzed by quantitative RT-PCR. The mRNA data were normalized to β-actin mRNA levels and are displayed as relative levels compared with HSV1 (WT) values (n = 3). P values were calculated using a two-tailed unpaired Student’s t test. *, P < 0.05. Error bars represent standard deviation.
The HSV1 VP1-2 DUB mutant exhibits elevated IFN induction and reduced replication in the brain
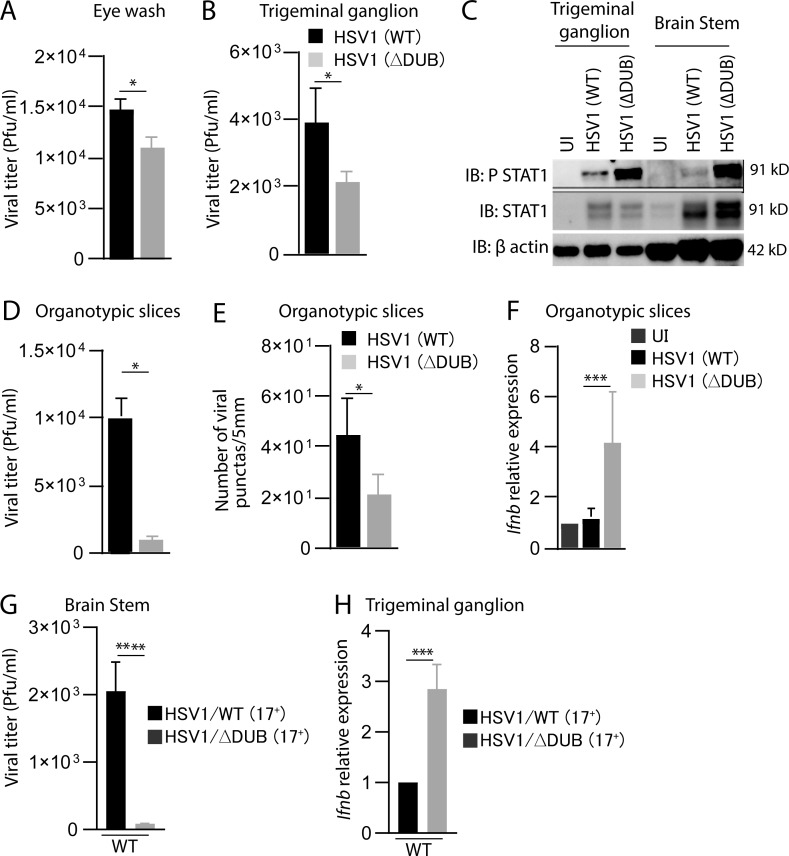
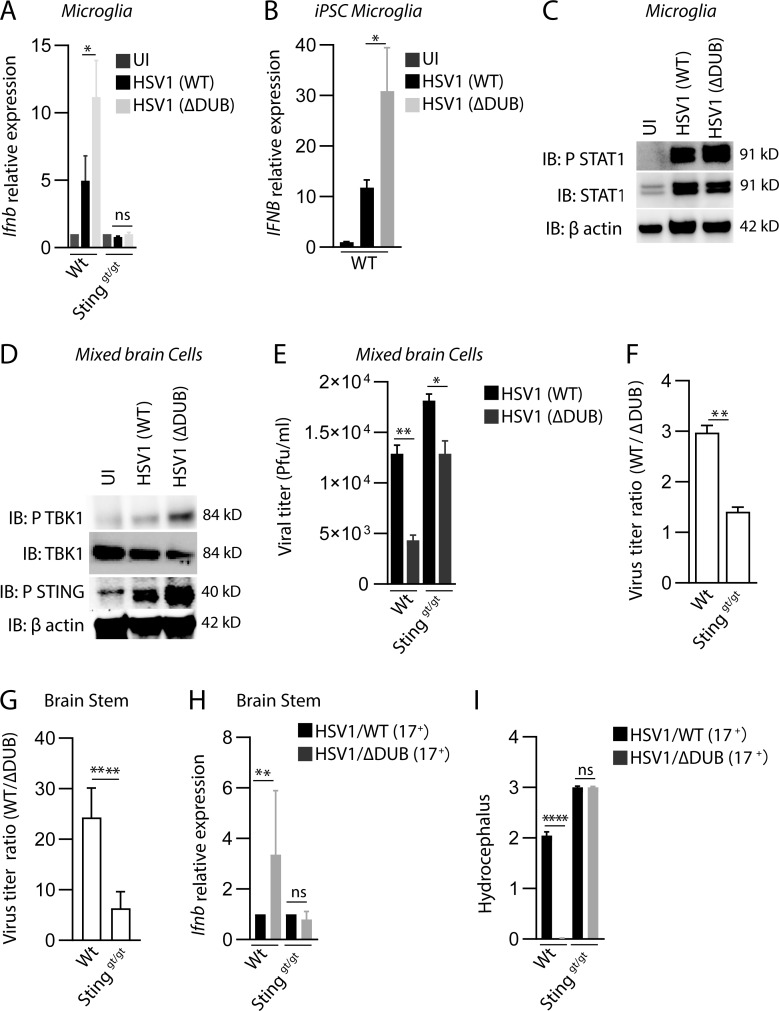
To examine whether VP1-2 DUB activity affects HSV1 replication and IFN induction in vivo, we first infected mice in the cornea with WT and ΔDUB HSV1 (KOS). We measured virus load in the eye washes and trigeminal ganglia, and observed a modest reduction in yields in mice infected with the mutant virus (Fig. 2, A and B). Interestingly, we found strongly elevated levels of the IFN-induced phosphorylation of STAT1 in trigeminal ganglion and brainstem from mice infected with the ΔDUB virus (Fig. 2 C). The HSV1 KOS strain is not very neuropathogenic in mice. Accordingly, we did not detect any virus in the brain tissue (data not shown). Therefore, we turned to organotypic brain slices to examine the role of the VP1-2 DUB activity in host control of HSV1 replication in the brain. Importantly, HSV1 replication in the organotypic brain slices was strongly impaired in the absence of viral DUB activity as measured by both viral titer and number of virus-producing cells (Fig. 2, D and E; and Fig. S2). In accordance with this, the levels of Ifnb transcripts were elevated in brain slices infected with HSV1 ΔDUB (Fig. 2 F). To test the role of DUB in infection in the CNS, we infected mice with HSV1 (17+)pUL36ΔDUB. Similar to the finding in organotypic brain slices, we observed reduced viral load and elevated Ifnb levels following infection with the DUB mutant (Fig. 2, G and H). Collectively, these data suggest that the HSV1 DUB activity inhibits Ifnb expression in the brain and hence facilitates viral replication.
Figure 2.
HSV1 VP1-2 ΔDUB exhibits elevated IFN induction and reduced growth in brain tissue. (A and B) C57BL/6 mice were infected in the cornea with HSV1 (WT) or HSV1 ΔDUB (KOS strain) as specified in Materials and methods. Eye washes were collected on day 2 after infection, and trigeminal ganglion and brain stem were isolated on day 6 after infection. Viral load was quantified using the plaque assay. n = 5 mice per group. (C) Mice were infected in the cornea with HSV1 (WT) or HSV1 (ΔDUB; KOS strain). Brain stem and trigeminal ganglion were isolated on day 6 after infection; homogenized and immunoblotted (IB) with the indicated antibodies. n = 5 mice per group. (D) P20 or P21 WT mice brain slices were cultured in semiporous membrane inserts and infected as specified in the methods section. After 6 d of infection, individual brain slices were homogenized and viral titers were determined by plaque assay. n = 5 mice per group. (E) Individual organotypic brain slices were sliced into 10 µm thickness after 6 d of infection with HSV1 (WT) or HSV1 (ΔDUB) virus and stained with anti–HSV1-2. The number of stained viral puncta were traced and analyzed by ImageJ. n = 4 mice per group. (F) Total RNA was isolated from brain slices on day 6 after infection. Ifnb mRNA levels were normalized to β-actin mRNA levels and showed as relative expression levels compared with uninfected controls. (G and H) C57BL/6 mice were infected in the cornea with HSV1/WT or HSV1 ΔDUB (17+ strain) as detailed in Materials and methods. Brain stems and trigeminal ganglia were isolated 5 d after infection for measurement of viral titer and Ifnb mRNA levels, respectively. n = 5 mice per group. P values were calculated using a two-tailed unpaired Student’s t test. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. Error bars represent standard deviation.
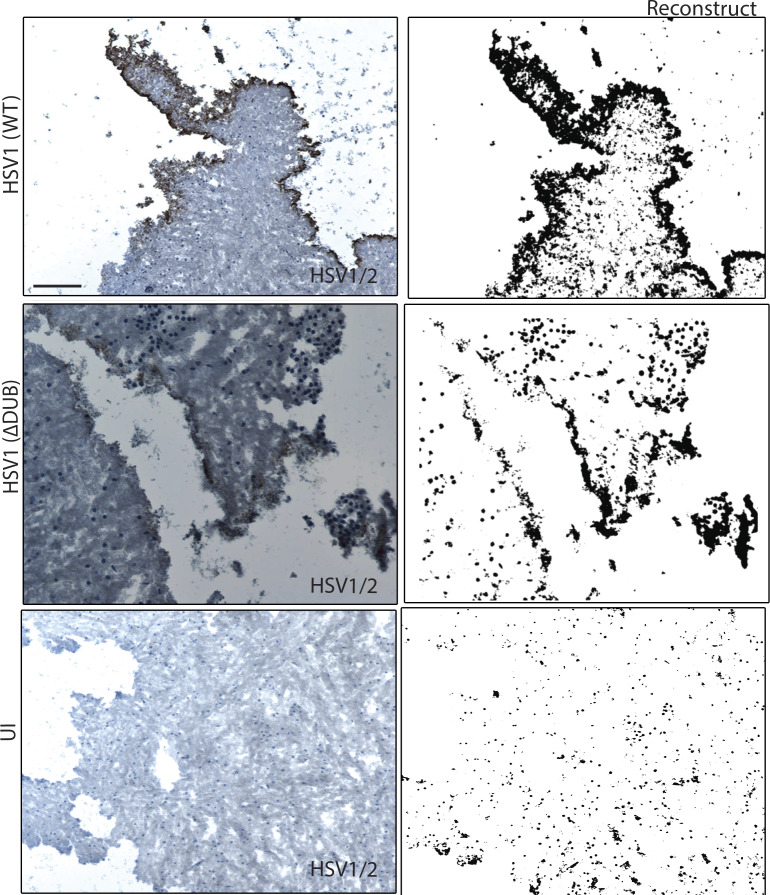
Figure S2.
Replication of DUB-deficient HSV1 in organotypic brain slices. Organotypic slices from WT mice brain were cultured in semiporous membrane inserts and infected with HSV1 (WT) or HSV1 ΔDUB, as specified in Materials and methods. After 6 d of infection, each organotypic brain slice was sectioned into 10-µm slices and stained with anti–HSV1-2. ImageJ was used to reconstruct the stained viral puncta. n = 4 mice per group. One representative image per group is shown (original and reconstructed images). Scale bar, 1 mm.
The HSV1 VP1-2 DUB targets the cGAS–STING pathway in microglia
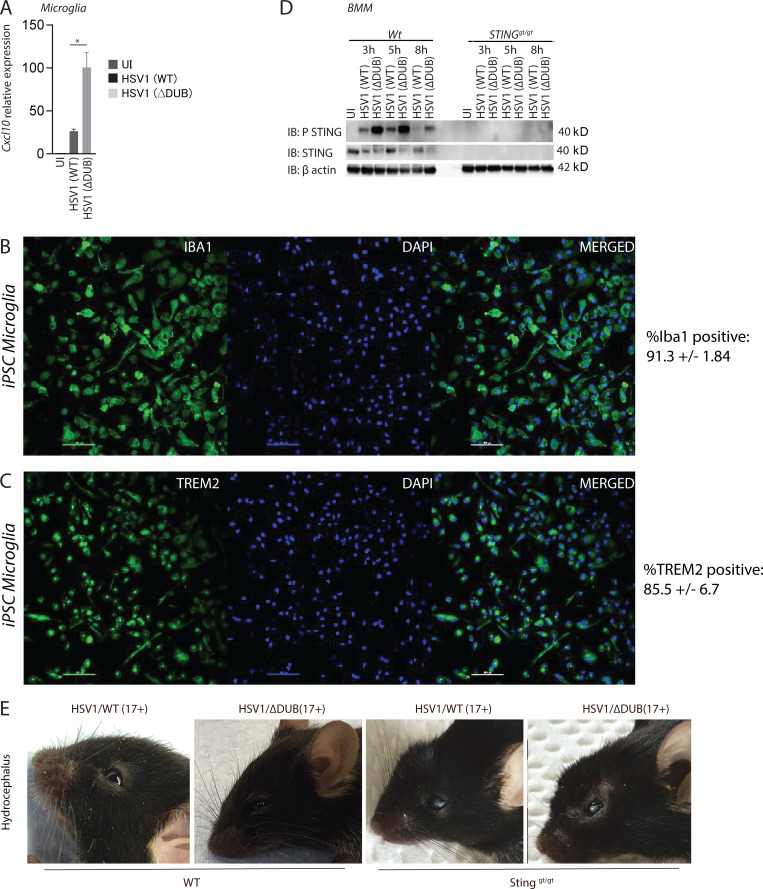
We previously reported that microglia are the predominant cell type producing type I IFN upon HSV1 infection in the brain of mice (Reinert et al., 2016). Therefore, we infected primary murine microglia from WT and STING-deficient mice with WT and HSV1 ΔDUB. Similar to what was observed in brain tissue, HSV1 ΔDUB induced higher levels of Ifnb and Cxcl10 than WT virus, and this Ifnb expression was entirely dependent on STING (Fig. 3 A and Fig. S3 A). To examine whether HSV1 ΔDUB also altered IFNB expression in human microglia, we generated microglia from human induced pluripotent stem cells (iPSCs). More than 85% of the differentiated cells expressed the microglia markers TREM2 and Iba1 (Fig. S3, B and C). Following infection with WT and HSV1 ΔDUB, we observed significantly higher IFNB expression in the cells infected with the mutant compared with WT (Fig. 3 B). The elevated Ifnb expression in murine microglia correlated with elevated levels of phospho-STAT1 in microglia infected with ΔDUB HSV1 as compared with WT HSV-1 (Fig. 3 C). When mixed brain cell cultures were infected with HSV1, the ΔDUB mutant virus induced stronger activation of the IFN-inducing pathway as measured by phosphorylation of TBK1 and STING (Fig. 3 D). Similar observations were made in bone marrow–derived macrophages, where we also observed the characteristic reduction of total STING levels after stimulation of the pathway (Prabakaran et al., 2018; Fig. S3 D). This suggests that the VP1-2 DUB generally targets STING in cells of the macrophage lineage. Accordingly, HSV1 ΔDUB replication was more impaired in mixed brain cell cultures from WT mice than STING-deficient mice (Fig. 3, E and F). Similar observations were obtained in vivo after infection of WT and Stinggt/gt mice with WT and ΔDUB HSV1 (Fig. 3, G and H), and the ΔDUB HSV1 virus also gained pathogenic activity in STING-deficient mice (Fig. 3 I and Fig. S3 E). However, HSV1 ΔDUB still exhibited some replication defects compared with the parental HSV1 in mixed brain cell cultures from STING-deficient mice and also in vivo. This argues for STING being the main, but not sole, target of the VP1-2 DUB and is consistent with previous studies suggesting that VP1-2 also deubiquitinates TRAF3 of the TLR3 pathway, which is known to be important for prevention of HSE (Pérez de Diego et al., 2010; Wang et al., 2013a; Zhang et al., 2007).
Figure 3.
The HSV1 VP1-2 DUB targets the cGAS–STING pathway in microglia. (A) WT and Stinggt/gt primary microglia were isolated, cultured, and infected with HSV1 (WT) or HSV1 ΔDUB virus (KOS strain, MOI 3) for 8 h. Total RNA as isolated and levels of Ifnb were measured by quantitative RT-PCR. The Ifnb mRNA levels were normalized to β-actin and shown relative levels compared with uninfected controls (n = 3). (B) iPSC-derived microglia were infected with HSV1 (WT) or HSV1 ΔDUB virus as above. IFNB mRNA levels were normalized to β-actin and are shown as relative levels compared with uninfected controls (n = 3). (C) WTmicroglia cultures were infected with HSV1 (WT) or HSV1 ΔDUB (KOS strain, MOI 3) for 8 h. Cell lysates were immunoblotted with the indicated antibodies. (D) Cultured mixed primary brain cells were infected with HSV1 (WT) or HSV1 ΔDUB (KOS strain) at MOI 3 for 16 h. Total cell lysates were cleared and immunoblotted with the indicated antibodies. (E) WT and Stinggt/gt mixed primary brain cell cultures were infected with HSV1 (WT) or HSV1 ΔDUB (KOS strain, MOI 10) for 24 h. Supernatants were collected, and the virus yield was quantified by plaque assay (n = 3). (F) Viral yield ratios between HSV1 (WT) and HSV1 ΔDUB in mixed brain cells from WT and Stinggt/gt mice. (G–I) C57BL/6 WT and Stinggt/gt mice were infected in the cornea with HSV1/WT or HSV1 ΔDUB (17+ strain) as described in Materials and methods. Viral load and Ifnb mRNA levels in the brain stem were quantified on day 5 after infection. Viral load is presented as ratios between WT and ΔDUB virus in the two mouse strains. (I) Degree of hydrocephalus was scored in the infected mice on day 5 after infection (n = 5). P values were calculated using a two-tailed unpaired Student’s t test. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, not significant. Error bars represent standard deviation.
Figure S3.
Activation of IFN responses by DUB-deficient HSV and disease development in vivo. (A) WT primary microglia were isolated and infected with HSV1 (WT) or HSV1 ΔDUB virus (MOI 3) for 16 h. Total RNA was isolated, and Cxcl10 levels were measured and normalized to β-actin by quantitative RT-PCR. (B and C) iPSCs were subjected to differentiation into microglia. The resulting cell population was stained for microglia markers Iba1 and TREM2 and visualized by immunofluorescence. (D) WT and Stinggt/gt bone marrow–derived macrophages (BMMs) were infected with HSV1 (WT) or HSV1 ΔDUB (MOI:10) for the indicated time intervals. Cleared lysates were immunoblotted with the indicated antibodies. (E) C57BL/6 WT and Stinggt/gt mice were infected in the cornea with HSV1 (WT) or HSV1 ΔDUB (17+ strain). The figure shows representative images of mice from each group. P value was calculated using a two-tailed unpaired Student’s t test. *, P < 0.05. Error bars represent standard deviation.
VP1-2 targets STING to prevent TBK1 activation
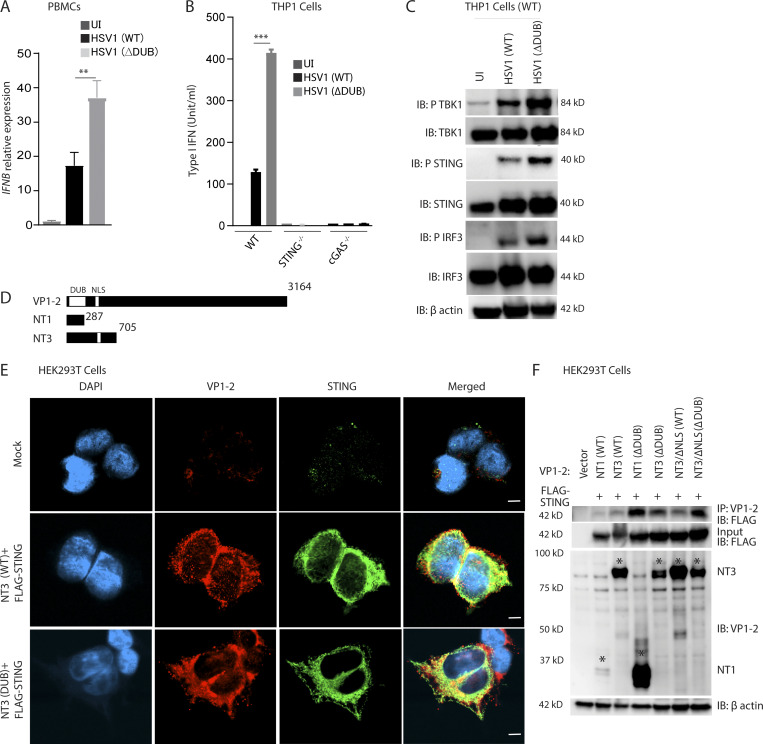
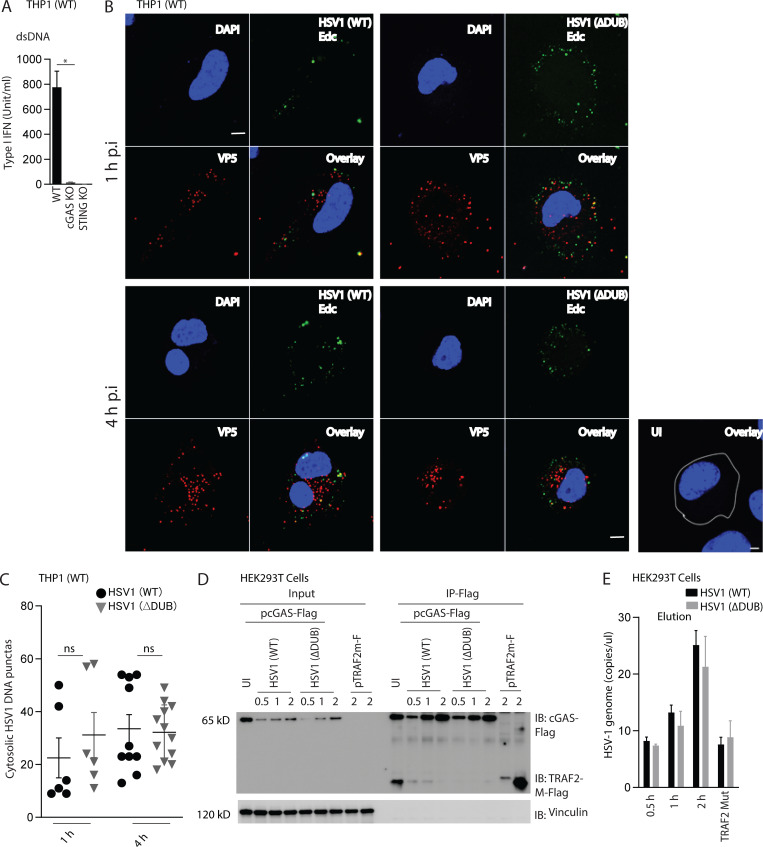
To gain mechanistic insight into how VP1-2 counteracts STING activity, we first infected peripheral blood mononuclear cells (PBMCs) and macrophage-like PMA-differentiated THP1 cells with WT and HSV1 ΔDUB. Similar to what was observed in microglia, HSV1 ΔDUB induced higher levels of IFNB and IFN bioactivity than WT virus in PBMCs and THP1 cells, respectively (Fig. 4, A and B). This was dependent on cGAS and STING (Fig. 4 B and Fig. S4 A) and was also observed at the signal transduction level, where the phosphorylated forms of STING, TBK1, and IRF3 were more abundant in lysates from cells infected with HSV1 ΔDUB (Fig. 4 C). The differential activation of the STING pathway by WT and ΔDUB was not due to differential release of viral DNA into the cytoplasm as evaluated by capsid-free viral DNA foci in the cytoplasm and coimmunoprecipitation of viral DNA with cGAS-FLAG (Fig. S4, B–E).
Figure 4.
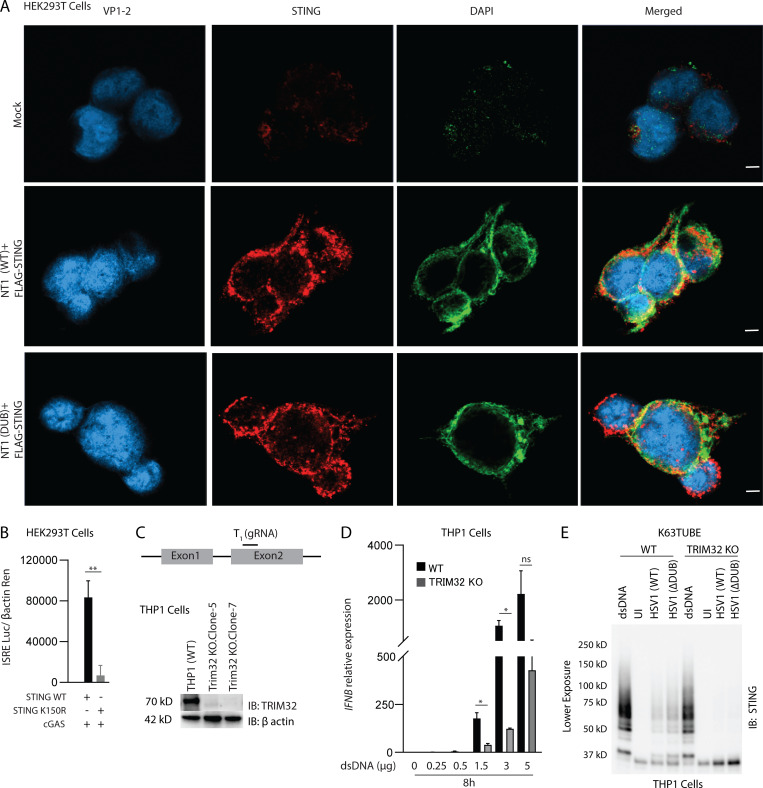
VP1-2 targets human STING to prevent TBK1 activation. (A) PBMCs were infected with HSV1 (WT) or HSV1 ΔDUB virus (KOS strain, MOI 3) for 8 h. IFNB mRNA levels were normalized with BACTIN, and relative gene expression levels were compared with uninfected controls. (B) Type I IFN bioactivity levels in supernatants from PMA-differentiated THP1 (WT), STING−/−, and cGAS−/− cells infected with HSV1 (WT) or HSV1 (ΔDUB; KOS strain, MOI 10) for 8 h (n = 3). (C) PMA-differentiated THP1 (WT) cells were infected with HSV1 (WT) or HSV1 ΔDUB (KOS strain, MOI 10) for 8 h. Cleared cell lysates were immunoblotted with the indicated antibodies. (D) Schematic representation of full-length VP1-2 and the two truncated versions, NT1 and NT, both encompassing the DUB domain. (E) HEK293T cells were transfected with FLAG-tagged STING (2 µg/ml) and NT1 (WT), NT3 (WT), NT1 (ΔDUB), or NT3 (ΔDUB) for 24 h. The cells were fixed and stained with anti–VP1-2, anti-FLAG, and DAPI. Scale bars, 5 µm. (F) HEK293T cells were transfected with FLAG-tagged STING (2 µg/ml) and one of the following constructs as indicated: NT1 (WT), NT3 (WT), NT1 (ΔDUB), NT3 (ΔDUB), NT3/ΔNLS (WT), or NT3/ΔNLS (ΔDUB; 2 µg/ml) for 36 h. Cell lysates were immunoprecipitated with anti–VP1-2 and immunoblotted with the indicated antibodies. NLS, nuclear localization signal. P values were calculated using a two-tailed unpaired Student’s t test. **, P < 0.01; ***, P < 0.001.
Figure S4.
DUB deficiency does not affect viral DNA release into the cytoplasm. (A) Type I IFN levels in supernatants from PMA-differentiated THP1 (WT), STING−/−, and cGAS−/− cells were transfected with dsDNA (4 µg/ml) for 8 h. (B) PMA differentiated THP1 (WT) cells were infected with 5-ethynyl-2′-deoxycytidine (EdC)-labeled HSV1(WT) or HSV1ΔDUB at indicated time points. The cells were fixed and stained for DNA and viral capsid using click chemistry and anti-VP5, respectively. Scale bars, 5 µm. (C) DNA puncta in 6–12 fields were counted using ImageJ. (D and E) HEK293T cells were transfected with cGAS-FLAG, TRAF2m-FLAG for 24 h and infected with HSV1 (WT) or HSV1 ΔDUB for the indicated time intervals. Lysates were immunoprecipitated with anti-FLAG and the eluate was analyzed by (D) immunoblotting for the quality of the precipitation or (E) PCR for viral DNA. Data are shown as HSV1 genome copies per microliter. n = 3. *, P < 0.05; ns, not significant. Error bars represent standard deviation.
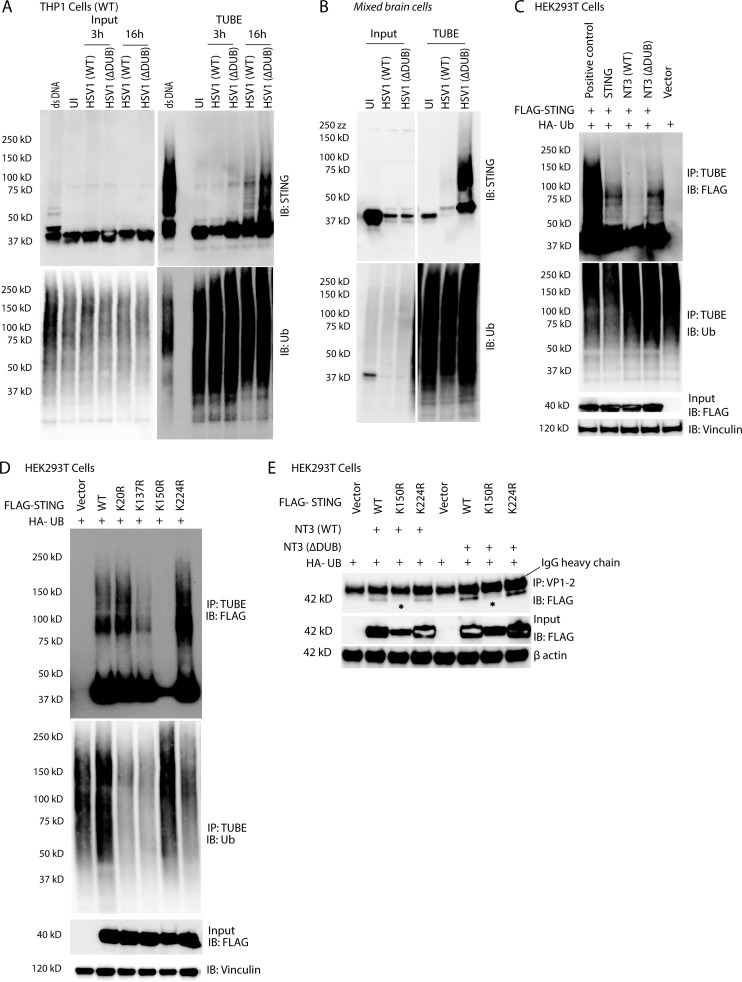
Next, we transfected HEK293T cells with FLAG-STING and truncated forms of the N-terminal part of VP1-2 harboring the DUB domain (Fig. 4 D). NT1 and NT3 were found to localize together with STING in the cytoplasm (Fig. 4 E and Fig. S5 A), and both NT1 and NT3 coprecipitated with STING; the association between VP1-2 and STING was increased for mutations lacking DUB activity (Fig. 4 F). This suggests that the N-terminal domain of VP1-2 is sufficient to mediate association with STING. The observation that the interaction was stabilized upon lack of DUB activity suggests, but does not unequivocally prove, that VP1-2 binds to ubiquitin chains on STING.
Figure S5.
Localization of VP1-2 NT1 and STING in HEK293T cells and role for STING K150 and TRIM32 for full activation of STING. (A) HEK293T cells were transfected with VP1-2 NT1 and STING. The cells were fixed 24 h after transfection and stained with anti–VP1-2 and anti-STING. Nuclear were visualized by staining with DAPI. Scale bars, 5 µm. (B) HEK293T cells were transfected with (50 ng) FLAG-tagged WT or K150R STING, cGAS, and IFNB1 promoter luciferase reporter and β-actin Renilla reporter. Reporter gene activity was measured 24 h after transfection (n = 3). (C) Illustration of targeting region for gRNA used to generate TRIM32 KO cells. Lysates from WT and two TRIM32 KO clones lysates were immunoblotted with anti-TRIM32 and anti–β-actin. (D) Shorter exposure of the anti-STING immunoblot shown to the right in Fig 6 F. (E) Type I IFN bioactivity levels in supernatants from PMA-differentiated THP1 (WT), and TRIM32 KO cells were transfected with dsDNA (0.25–5 µg) for 8 h. *, P < 0.05; **, P < 0.01; ns, not significant. Error bars represent standard deviation.
HSV1 VP1-2 deubiquitinates STING
To explore whether VP1-2 targets STING ubiquitination, endogenous ubiquitin-modified proteins (and associated proteins) were purified using the Tandem Ubiquitin Binding Entity (TUBE) technology. Strikingly, while polyubiquitinated forms of STING (Ub-STING) were detectable after infection with WT HSV1, Ub-STING accumulated to a much greater extent in cells infected with HSV1 ΔDUB (Fig. 5, A and B). To examine if the DUB activity of VP1-2 deubiquitnates STING, FLAG-tagged STING was coexpressed with the VP1-2 DUB domain. The WT, but not the enzymatically inactive, DUB domain diminished STING ubiquitination, suggesting that Ub-STING is a direct target of VP1-2 DUB activity (Fig. 5 C).
Figure 5.
HSV1 VP1-2 deubiquitinates STING. (A) PMA-differentiated THP1 (WT) cells were infected with HSV1 (WT) or HSV1 ΔDUB (KOS strain, MOI 10) for the indicated time intervals. Ubiquitin conjugates were purified by TUBE pull-down, and the precipitates were immunoblotted and probed with the indicated antibodies. (B) WT mixed primary murine brain cells were infected with HSV1 (WT) or HSV1 ΔDUB (KOS strain, MOI 3) for 16 h, and TUBE pull-downs were immunoblotted with the indicated antibodies. (C) HEK293T cells were transfected with FLAG-tagged STING (2 µg/ml), HA-tagged ubiquitin (Ub; 2 µg/ml), NT3 (WT), or NT1 (ΔDUB; 2 µg/ml) for 36 h. Ubiquitin conjugates were purified using TUBEs and immunoblotted with the indicated antibodies. (D) HEK293T cells were transfected with FLAG-tagged WT, K20R, K137R, K150R, or K224R STING (2 µg/ml) and HA-tagged ubiquitin (2 µg/ml) for 36 h. Total ubiquitin conjugates were purified using TUBE and immunoblotted with the indicated antibodies. (E) HEK293T cells were transfected with FLAG-tagged WT, K150R, or K224R STING, HA-tagged ubiquitin (2 µg/ml), and NT3 (WT or ΔDUB; 2 µg/ml) for 36 h. Total cell lysates were immunoprecipitated with anti–VP1-2 and immunoblotted as indicated.
It has previously been examined which lysines on STING are ubiquitinated upon activation of the pathway, but the published data are not consistent (Ni et al., 2017; Tsuchida et al., 2010; Zhang et al., 2012). We decided to focus on K20, K137, K150, and K224, given the previous publications. In our hands, overexpression of WT STING in HEK293T cells led to ubiquitination, and this was not significantly affected by the mutations K20R, K137R, or K224R (Fig. 5 D). By contrast, very little K150R STING was precipitated by TUBE despite similar expression of the WT and K-to-R mutation variants (Fig. 5 D). Accordingly, the K150R STING mutant was a poor inducer of IFN-β reporter gene activation (Fig. S5 B). Interestingly, the coimmunoprecipitation of the VP1-2 DUB domain and STING was abolished in lysates from the K150R STING mutant (Fig. 5 E). These data suggest that the VP1-2 DUB is recruited to STING in a manner dependent on ubiquitination of STING at K150, thus enabling deubiquitination STING by the viral DUB.
HSV1 VP1-2 digests K63-linked ubiquitin on STING to prevent activation of signaling
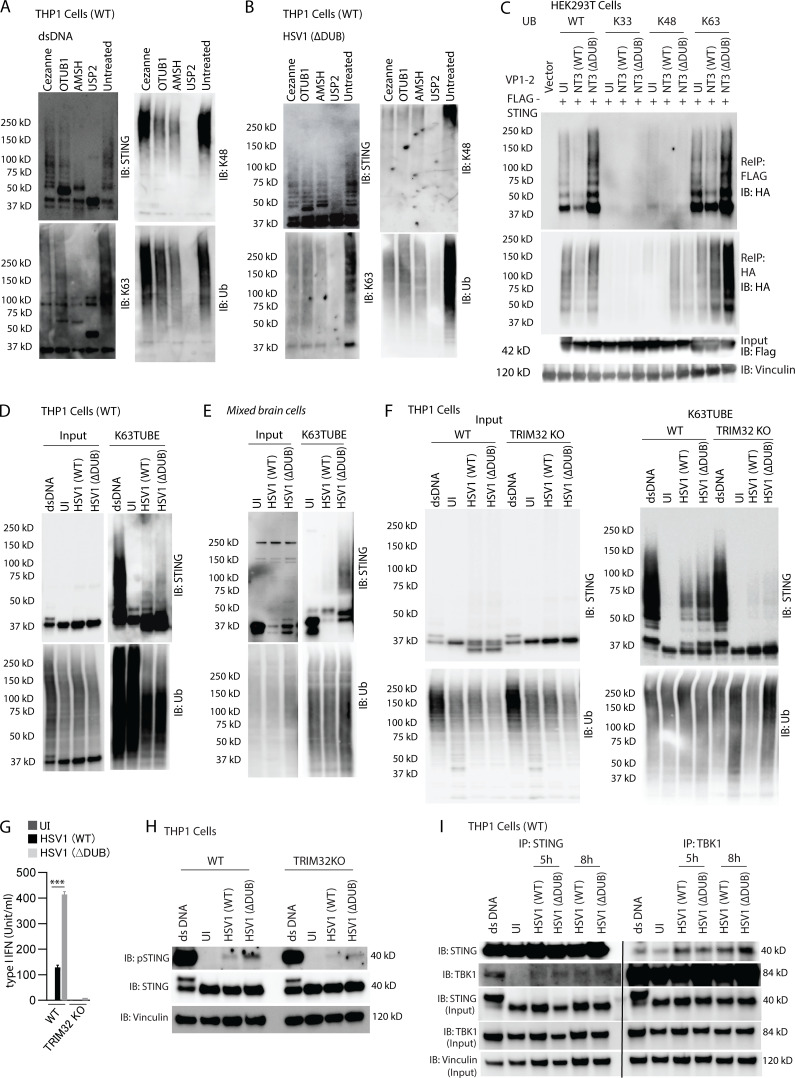
To get more detailed information on the type of ubiquitin linkage on STING after activation by synthetic DNA or HSV1, we performed UbiCrest, which takes advantage of the cleavage preference of specific DUBs to map the type of ubiquitin linkage on specific proteins (Hospenthal et al., 2015). In lysates from DNA-stimulated or HSV1 ΔDUB-infected cells, the K63-digesting DUB AMSH removed the majority of the STING ubiquitination, and a clear effect was also observed following treatment with the K48-digesting DUB OTUB1 (Fig. 6, A and B). Consistently, coexpression of STING, VP1-2 NT3, and single-lysine ubiquitin mutants in HEK293T cells showed similar levels of STING ubiquitination when WT and K63-only ubiquitin and these were both targeted by VP1-2 (Fig. 6 C). Expression of K48-only ubiquitin resulted in low levels of Ub-STING, whereas K33-only ubiquitin did not result in detectable Ub-STING. Finally, using K63-TUBE, we confirmed that K63-linked ubiquitination of STING was more pronounced in THP1 and mixed brain cells infected with HSV1 ΔDUB as compared with WT HSV1 (Fig. 6, D and E).
Figure 6.
VP1-2 targets K63-linked STING ubiquitination. (A and B) PMA-differentiated THP1 (WT) cells were transfected with dsDNA (4 µg/ml) for 5 h or infected with HSV1 ΔDUB (KOS strain, MOI 10) for 16 h. Endogenous polyubiquitin conjugates were precipitated using TUBE (1xUBA), and a ubiquitin chain restriction assay (UbiCRest) was performed and immunoblotted as indicated. (C) HEK293T cells were transfected with FLAG-tagged STING (2 µg/ml), HA-tagged WT, K33-only, K48-only, or K63-only ubiquitin (2 µg/ml) and NT3 (WT or ΔDUB; 2 µg/ml) for 36 h. STING was immunoprecipitated with anti-FLAG and eluted fractions were denatured and reimmunoprecipitated (ReIP) with either FLAG or HA and immunoblotted as shown. (D) THP1 (WT) cells were differentiated with PMA and infected with HSV1 (WT) or HSV1 ΔDUB (KOS strain, MOI 10) for 16 h. K63 TUBE was used to pull down endogenous K63 polyubiquitin conjugates from cell lysates and immunoblotted as indicated. (E) WT mixed primary mouse brain cells were infected with HSV1 (WT) or HSV1 ΔDUB (KOS strain, MOI 3) for 16 h. K63 polyubiquitin conjugates were precipitated using K63 TUBE pull-down and immunoblotted with the indicated antibodies. (F) PMA-differentiated THP1 (WT) and TRIM32 KO cells were infected with HSV1 (WT) or HSV1 ΔDUB (KOS strain, MOI 10) for 16 h. Endogenous K63 polyubiquitin conjugates were precipitated using TUBE and immunoblotted with the indicated antibodies. (G) Type I IFN bioactivity levels in supernatants from PMA-differentiated THP1 (WT) and TRIM32 KO cells infected with HSV1 (WT) or HSV1 (ΔDUB; KOS strain, MOI 10; n = 3). (H) PMA-differentiated THP1 (WT) and TRIM32 KO cells were infected with HSV1 (WT) or HSV1 ΔDUB (KOS strain, MOI 10) for 8 h. Cleared cell lysates were immunoblotted with the indicated antibodies. (I) PMA-differentiated THP1 (WT) cells were infected with HSV1 (WT) or HSV1 ΔDUB (KOS strain, MOI 10). Cells were lysed 5 and 8 h after infection, and cleared cell lysates were immunoprecipitated with either anti-STING or anti-TBK1 and immunoblotted with the indicated antibodies. P values were calculated using a two-tailed unpaired Student’s t test. ***, P < 0.001. Error bars represent standard deviation.
TRIM32, TRIM56, and Mul1 have been reported to exert K63-linked ubiquitination of STING (Ni et al., 2017; Tsuchida et al., 2010; Zhang et al., 2012). However, TRIM56 was later shown to actually ubiquitinate cGAS (Seo et al., 2018), whereas Mul1 ubiquitinates STING at K224 (Ni et al., 2017). Therefore, in order to get information on which E3 ligase that VP1-2 is counteracting, we first generated TRIM32-deficient THP1 cells (Fig. S5 C) and examined the impact of TRIM32 deficiency on STING ubiquitination and activation in response to HSV1 infection. Interestingly, THP1 cells lacking TRIM32 failed to accumulate K63-linked ubiquitin-modified STING after HSV infection, and showed partial defect after DNA stimulation (Fig. 6 F and Fig. S5 D). Accordingly, DNA- and HSV-induced expression of IFNB was severely affected by TRIM32 deficiency (Fig. 6 G and Fig. S5 E), and HSV-induced phosphorylation of STING was strongly reduced (Fig. 6 H). Immunoprecipitation (IP) of STING or TBK1 from cells infected with WT and ΔDUB HSV1 revealed that deletion of the HSV DUB did not affect recruitment of TBK1 to STING (Fig. 6 I), even though downstream TBK1 autophosphorylation and TBK1-mediated phosphorylation of STING and IRF3 were elevated (Fig. 4 C). Collectively, our data demonstrate that the HSV1 VP1-2 DUB is recruited to STING in a manner dependent on K150 on STING and that DUB activity is important for evasion of the IFN response in the brain. This viral evasion proceeds through deubiquitination of STING, thus reducing the signaling quality of the STING signalosome.
Discussion
HSE is a devastating disease with mortality rates of ∼70% for untreated cases (Whitley et al., 1977); even with acyclovir treatment, mortality rates are between 15% and 20%, and sequelae are significant (Jouan et al., 2015). The innate immune response is known to be a major determinant for control of HSV1 in the CNS, and studies from humans and mice demonstrate the type I IFN system of the innate immune system to be pivotal (Dupuis et al., 2003; Leib et al., 1999). Type I IFNs are induced following sensing of foreign or mislocalized nucleic acids by pattern recognition receptors, and inhibition of type I IFN expression is known to be a major immune-evasion strategy employed by most, if not all, viruses (Bowie and Unterholzner, 2008). Here, we report that the DUB activity of HSV1 VP1-2 is a central immune-evasion activity of HSV1 in the brain, and this impacts significantly on the ability of the virus to replicate in brain tissue. The VP1-2 DUB deubiquitinates STING with preference for K63-linked polyubiquitin, thus compromising STING signaling quality and downstream induction of IFN-β expression.
Like most DNA viruses, HSV1 has evolved multiple mechanisms to evade induction of type I IFN (Christensen and Paludan, 2017). This includes close to 10 known mechanisms, several of which specifically target the cGAS–STING pathway (Christensen et al., 2016; Deschamps and Kalamvoki, 2017; Huang et al., 2018; Su and Zheng, 2017; Ye et al., 2017; Zhang et al., 2018), and some inhibit more than one pathway by targeting proteins used by multiple pathways (Lin et al., 2004; Ma et al., 2012; Orzalli et al., 2012a; Wang et al., 2013b). HSV1 also inhibits autophagy through ICP34.5, and this contributes to the neuropathology of the virus (Orvedahl et al., 2007). By contrast, the mechanisms through which HSV1 evades the type I IFN response in the brain are not well understood. In brain cell cultures, we compared type I IFN production following infection with a panel of mutant viruses, each lacking one reported immune evasion protein or activity. A defect in the VP1-2 ΔDUB was found to lead to the highest virus-induced IFN/ISG levels, suggesting that this IFN evasion strategy is particularly important in the brain. We also observed elevated expression of TNF-α in agreement with the reported ability of VP1-2 to deubiquitinate IκBα downstream of STING (Ye et al., 2017). The elevated IFN/ISG levels in brain cell cultures were supported by data demonstrating elevated IFN induction and reduced viral replication in brain and organotypic brain slices following infection with HSV1 ΔDUB compared with WT HSV1. Interestingly, VP1-2 inhibits not only production of type I IFN, as we report in this study, but also the action of type I IFN through blockage of the interaction between IFNAR and JAK1 in a manner independent of DUB activity (Yuan et al., 2018). In fact, two other HSV proteins, ICP27 and UL41, which target the cGAS–STING pathway, have also been shown to inhibit type I IFN signaling (Duerst and Morrison, 2004; Johnson et al., 2008). The previously described VP1-2–mediated inhibition of type I IFN signaling was independent of VP1-2 DUB activity. In addition to the here-identified deubiquitination of STING, VP1-2 DUB has also been demonstrated to deubiquitinate TRAF3, which is essential for IFN induction through the TLR3 pathway (Wang et al., 2013a). In the mixed brain cell system, we observed that the defective replication of HSV1 VP1-2 ΔDUB in WT brain cell cultures was largely, but not fully, rescued in Sting−/− cultures. This suggests that DUB primarily targets STING but also other pathways, such as the TLR3–TRIF pathway. Collectively, these data demonstrate a central role for VP1-2 in antagonism of IFN induction through two key HSV1-sensing pathways in the brain.
Ubiquitination plays a central role in STING signaling (Paludan, 2015), and multiple types of ubiquitin linkages have been reported on STING, including K11, K27, K48, and K63. Among these, K27- and K63-linked polyubiquitination are directly associated with STING activation. The three E3 ligases TRIM32, TRIM56, and Mul1 have been reported to mediate K63-linked STING polyubiquitination (Ni et al., 2017; Tsuchida et al., 2010; Zhang et al., 2012), although TRIM56 was subsequently reported to ubiquitinate cGAS rather than STING (Tsuchida et al., 2010). TRIM32 mediates STING ubiquitination at multiple lysine residues, notably 20, 150, 224, and 236 (Zhang et al., 2012), whereas Mul1 acts specifically on lysine 224 (Ni et al., 2017). While TRIM32 is essential for both recruitment and activation of TBK1 as well as downstream IRF3 activation, Mul1 is specifically essential for IRF3 activation (Ni et al., 2017; Zhang et al., 2012). We found that HSV1 VP1-2 interacted with STING and mediated deubiquitination of STING with the strongest effect on K63-linked polyubiquitination, which was mediated by TRIM32. Consistently, the activity of DUB was associated with reduced phosphorylation of STING, TBK1, and IRF3 in cells infected with WT virus and consequently reduced activation of IFNB gene transcription. Thus, HSV1 VP1-2 counteracts TRIM32-mediated K63-linked polyubiquitination of STING, thus preventing signaling at the level of TBK1. The HSV1 DUB is conserved across herpesviruses and with protease activity specific for ubiquitin, since the ubiquitin-like molecules SUMO, Nedd8, and ISG15 are not cleaved (Kattenhorn et al., 2005). We found that the VP1-2 DUB activity is essential for HSV1 replication in vivo, and previous work has shown this activity not to be essential for virus replication in permissive cells in vitro (Bolstad et al., 2011). This suggests that the main function of VP1-2 DUB activity is to counteract ubiquitin-dependent virus–host interactions, including immune responses. Several immune evasion activities other than targeting of STING have been reported for the herpesvirus DUBs (van Gent et al., 2014; Inn et al., 2011; Sun et al., 2015), but it remains to be examined if the mechanism described in this work is shared by other herpesvirus DUBs or whether it is specific to HSV1.
HSV1 is able to enter into many cell types but replicates efficiently only in a limited number of cell types, including epithelial cells, fibroblasts, and neurons. It has generally been thought that accidental viral entry into a cell type not supporting replication was a waste of virus. However, one implication of the present results is that viruses that enter into, for instance, microglia can counteract STING-dependent antiviral activity via VP1-2–mediated deubiqutination. It is interesting to observe that only a subset of the reported mechanisms, through which HSV1 counteract the cGAS–STING pathway, have strong impact on IFN induction in brain cell cultures. This suggests that the relative contribution of the HSV1 immune-evasion mechanisms depends on cell types and potentially also various spatial and temporal factors, as well as the inflammatory microenvironment. At present, we do not have in-depth understanding of why VP1-2 DUB has particularly strong STING antagonistic activity in microglia and generally how, why, and when HSV1 employs its different immune evasion strategies in the most optimal manner to promote and sustain infection.
Collectively, in this work, we identify the DUB activity of the HSV1 tegument protein VP1-2 as a major viral mechanism to counteract induction of type I IFN expression through the cGAS–STING pathway in the brain. Together with the previous report on VP1-2 DUB also targeting the TLR3 pathway, this work identifies deubiquitination of innate immune signaling pathways as a key principle in HSV1-mediated inhibition of antiviral responses in the brain. One implication of our data are that pharmacological targeting of the viral DUB could unleash an efficient intrinsic antiviral response, which could benefit immunocompromised patients with severe HSV-1 infection in the CNS, particularly in cases with acyclovir-resistant HSV1 variants.
Materials and methods
Cell lines, reagents, and transfection
HEK293T cells, PBMCs, Ifnar1−/− MEFs, and mouse bone marrow–derived macrophages were maintained in DMEM 1640 supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 2 mM L-glutamine. THP1 cells were maintained in RPMI 1640 supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 2 mM L-glutamine. Media containing 150 nM PMA was used for the initial 24 h to differentiate THP1 cells and later was replaced with media containing no PMA. The converted macrophage-like cells were used for the experiments after 24 h. The 60mer double-stranded DNA (dsDNA) was transfected into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The 2′,3′-cGAMP was transfected into cells using digitonin according to a protocol described in a previous study (Luecke et al., 2017).
Mice
C57BL/6 and STINGgt/gt mice were bred at Taconic M&B. Mice were anesthetized using isoflurane (Abbott) or ketamine (MSD Animal Health) and xylazine (RompunVet). All information about mice treatment was blinded. No animals were barred from the analysis. The animals were closely monitored during HSV infection, and all efforts were put to minimize pain. Experimental protocols using mice have been reviewed and approved by Danish government authorities and comply with Danish laws (approval number 2016–15-0201-01085).
Virus propagation and generation of HSV-1 mutants
In this work, we used a panel of previously reported HSV1 mutant strains (Bolstad et al., 2011; Cai and Schaffer, 1989; Deschamps and Kalamvoki, 2017; Rice and Knipe, 1990). Virus titer was determined as described earlier (Reinert et al., 2016). Briefly, Vero cells were used to grow both WT HSV1 (strain KOS) and the derived VP1-2 C65A HSV1 (ΔDUB). Virus titers were measured by plaque assay on Vero cells. Beriglobin (CSL Behring) was used to neutralize extracellular HSV1 to calculate virus titers. A stock titer of 8–12 × 109 PFU/ml was used for all studies. In addition, we generated novel VP1-2 C65A mutants. As parental strains, we used HSV1(17+)Lox (Sandbaumhüter et al., 2013); HSV1(17+)Lox-pMCMVmCherry (Che for short), which expresses monomeric Cherry under the control of the mouse cytomegalovirus major immediate early promoter; and HSV1(17+)Lox-CheVP26 (CheVP26 for short) in which the first seven N-terminal residues of the small capsid protein VP26 have been replaced by mCherry (Sandbaumhüter et al., 2013). We mutated the core catalytic cysteine residue required for pUL36 DUB activity (Kattenhorn et al., 2005) in to alanine (C65A) using “en-passant mutagenesis.” We amplified a kanamycin-resistance cassette using the primers prW853 (5′-CATCAAATATGAGGCTGAGAAAGGACAGCGACGAGCGCATCGCCGATACCGACCCCCCCGGCTTAGGGATAACAGGGTAATCGATTT-3′) and prW855 (5′-CCAGTTCGCGCCCGACCTGGAGCCGGGGGGGTCGGTATCGGCGAT GCGCTCGTCGCTGTCCTTGTATATCTGGCCCGTACATCGATCT-3′) to introduce the mutated sequence (underlined). We transformed this cassette into the Escherichia coli GS1783 strains that carried the respective HSV-1 backbone as a bacterial artificial chromosome (BAC). After two-step Red-mediated recombination, we evaluated the integrity of the BAC DNA by restriction digestion with BamH1, Asc1, Not1 (Fig. S1 A), Xho1, EcoR1, or EcoRV (data not shown). The newly generated mutants HSV1(17+)Lox-pUL36ΔDUB, HSV1(17+)Lox-CheVP26-pUL36ΔDUB, HSV1(17+)Lox-pMCMVmCherry-pUL36ΔDUB were reconstituted by transfecting the respective BAC DNA into Vero cells as described before (Sandbaumhüter et al., 2013). We confirmed the point mutations by Sanger sequencing of the respective BAC DNA and next-generation sequencing of purified HSV-1 DNA after three passages of viral amplification. These viruses were used for infection of in vivo, ex vivo, and in vitro experiments.
Virus plaque assay
Dissected brain regions were homogenized with sterilized steel beads (Qiagen) in Tissuelyser II (Qiagen) for 2–3 min with 30-s intervals. One third of the homogenized tissue was pelleted by centrifugation at 1,600 g for 10 min at 4°C, and supernatants were collected and used for plaque assay with serial dilution on Vero cells in DMEM containing 2% FBS, 1% penicillin, and 1% streptomycin. After 1 h, human Beriglobin (CSL Behring) was added, and plates were incubated at 37°C in a humidified atmosphere at 5% CO2. After 2 d, the plaques were counted under the microscope.
Ocular HSV1 infection model
Ocular HSV1 infections in mice were performed as described previously (Reinert et al., 2016). Briefly, 6–10-wk-old male mice were anaesthetized with an i.p. injection combination of ketamine (100 mg/kg body weight) and xylazine (10 mg/kg body weight). Two corneas were scarified in a 10 × 10 notch pattern and infected with 5 µl HSV1 (108 PFUs for KOS strain and 8 × 106 PFUs for 17+ strain) with infection medium (DMEM 1640 with 200 U/ml penicillin and 200 µg/ml streptomycin) or mock infected with 5 μl medium. Eyewash was collected with a sterile cotton swab and stored in 500 µl DMEM medium at −80°C until the plaque assay was performed. Infected and control mice hydrocephalus were scored blinded at 5 d after infection. The hydrocephalus scores were indicated as no bump: 0, minor bump: 1, moderate bump: 2, or large bump: 3 (Reinert et al., 2016). 5–7 d after infection, brains were dissected, and the brainstem and trigeminal ganglia were dissected. One third of the brain was used for virus plaque assay as described and one third for RNA isolation, and the rest of the brain was used for immune blots.
Organotypic brain slices
Organotypic brain slice cultures were prepared from C57BL/6J mice postnatal day 20 (P20) to P21 (Bodda et al., 2013). Briefly, pups were gleaned by decapitation following protocols in line with Danish animal laws. Brains from pups were dissected and collected in ice-cold artificial cerebrospinal fluid containing 126 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 2.5 mM CaCl2, 13 mM MgCl2, 10 mM D-glucose, and 26 mM NaHCO3, pH 7.4. Brain slices (400 µm transversely) were cut on a Leica VT1200 S vibrating blade microtome (Leica Microsystems A/S) in ice-cold artificial cerebrospinal fluid. The slices were transferred to semiporous membrane insert (0.4-µm pore size; Millipore) with prewarm DMEM culture medium containing 25% FCS, 2 mM L-glutamine (Gibco), CaCl2 (Sigma), MgSO4 (Sigma), NaHCO3, Hepes, insulin (1 mg/ml), B-27 supplement (Gibco), 100 U/ml penicillin, and 100 µg/ml streptomycin. The organotypic brain slices were incubated at 37°C in 5% CO2 for 24 h, followed by HSV1 infection (108 PFUs per brain slice). The culture medium was changed every alternate day for 6 d after infection, and the brain slices were collected to determine viral loads, protein expression, and RNA preparation.
Isolation of primary microglia and mixed brain cells
Mixed brain cells and microglia were isolated from P0 to P2 mice. The brains were collected in HBSS (Gibco), and the meninges were lifted carefully. The brains were minced and washed in HBSS medium followed by 2.5% Trypsin (Gibco) treatment. After 10 min, the reaction was stopped by the addition of DMEM 1640 supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, 2 mM L-glutamine, and 0.5 μg/ml DNase1. Cells were triturated and filtered through a 70-µm pore size filter and seed on 75-cm2 culture flasks coated with poly-L-lysine (Gibco). The culture medium was replenished on days 2 and 5 after plating. The microglia were isolated after 5 d of mixed brain culture. The culture medium was supplemented with 33% L929 fibroblast-conditioned medium to stimulate proliferation of microglia. After 12 d of culture, microglia were harvested by 1-h shake off at 150 rpm in shaker and cultured with DMEM containing 10% FBS, 1% penicillin, 1% streptomycin, 2 mM L-glutamine, and filtered conditional medium from mixed glial cultures in a ratio of 50:50 at 37°C in a humidified atmosphere at 5% CO2 for 3 wk.
iPSC-derived microglia
The human iPSC line WTSIi015-A (EBiSC; Sigma) was maintained on Matrigel (Corning) in mTeSR1 (StemCell Technologies). iPSC colonies were dissociated into single cells using TrypLE Express (Thermo Fisher). 4 × 106 iPSCs were seeded per Aggrewell 800 (StemCell Technologies) in a 24-well plate in embryonic body medium. Cells were cultured for 4 d in Aggrewells to form embryonic bodies with half media change every day. Embryonic bodies were harvested using an inverted cell strainer (40 µm), and ∼15 embryonic bodies were plated per 6 wells in HM. 2 ml media were replaced every 7 d by fresh hematopoietic medium (HM). After ∼30 d, primitive macrophage precursors can be harvested during the media change and plated in microglia medium (MiM) into 48 wells at a density of 100,000 cells/cm2. Cells were differentiated in MiM for another 7–10 d with a full media change every second day. Embryonic body medium contained mTeSR1 supplemented with 10 µM ROCK inhibitor, 50 ng/ml BMP-4, 20 ng/ml SCF, and 50 ng/ml VEGF-121 (all from Peprotech). HM contained X-VIVO 15 (Lonza) supplemented with 2 mM Glutamax, 100 U/ml penicillin, 100 µg/ml streptomycin, 55 µM β-mercaptoethanol, 100 ng/ml M-CSF (Peprotech), and 25 ng/ml IL-3 (Cell Guidance Systems). MiM contained X-VIVO 15 (Lonza) supplemented with 2 mM Glutamax, 100 U/ml penicillin, 100 µg/ml streptomycin, 55 µM β-mercaptoethanol, 100 ng/ml IL-34 (Peprotech), and 10 ng/ml GM-CSF (Peprotech).
Ectopic protein expression in HEK293T cells
HEK293T cells were seeded into 6-cm dishes at ∼70% confluency. The cells were transfected with 2 µg FLAG-tagged-STING (Addgene) and 2 µg hemagglutinin (HA)-tagged ubiquitin (Addgene) and 2 µg NT3 (WT) or NT3 (ΔDUB) using Lipofectamine 2000 (Invitrogen). After 24 h of transfection, cells were lysed in IP lysis buffer containing 1× cOmplete mini, EDTA- free protease inhibitor (Sigma), 1× PhosSTOP (Sigma), 5 mM N-ethylmaleimide (Sigma), and 10 mM NaF and incubated for 2 h on ice and pelleted down. The supernatants were incubated overnight with FLAG M2 Magnetic Beads (Sigma) at 4°C. 3× FLAG tag peptide (Sigma) was used to elute the immunoprecipitated complexes. Two thirds of the eluted complexes were boiled for 5 min with 1% SDS and diluted in 1:10 with lysis buffer. The diluted probes were reimmunoprecipitated with HA Magnetic Beads (Invitrogen) for 2 h at room temperature. The immunoprecipitated complexes were boiled in 2× Laemmli sample buffer (Sigma) at 95°C and processed for immunoblot analysis.
IP
Experimentally stimulated cells were lysed in IP lysis buffer containing 1× cOmplete mini, EDTA-free protease inhibitor (Sigma), 1× PhosSTOP (Sigma), 5 mM N-ethylmaleimide (Sigma), and 10 mM NaF and incubated for 10 min on ice, and the cell lysates were pelleted at 8,000 g. Supernatants were incubated with sheep anti-STING or rabbit anti-TBK1 or mouse anti–VP1-2 overnight at 4°C. Dynabeads Protein G (Invitrogen) were added to the lysates and incubated for 2 h at 4°C followed by three washes with IP lysis buffer containing all the inhibitors. The immunoprecipitated complexes were boiled with 2× Laemmli sample buffer (Sigma) at 95°C to run immunoblot.
TUBEs
TUBEs were used to purify ubiquitin conjugates from HEK293T, PMA-differentiated THP1 cells, or mixed brain cell lysates. For isolation of total ubiquitin chains and K63-linked ubiquitin chains, we used Magnetic Beads TUBE1 and anti-K63 TUBE, respectively (both from Life Sensors), according to the manufacturer’s instructions with few modifications. 10 × 106 PMA-differentiated THP1 cells were lysed in ice-cold 600 µl TUBE lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 5 mM N-ethylmaleimide [Sigma], 5 mM sodium orthovanadate [Sigma], 1% NP 40, 0.5% Triton X-100, 10 mM NaF, 1× cOmplete mini, EDTA free protease inhibitor [Sigma], and 1× PhosSTOP [Sigma]). The lysates were incubated on ice and cleared by sonication and centrifuged 14,000 g for 20 min at 4°C. Supernatants were incubated with TUBE magnetic beads overnight at 4°C. The reaction mixture, along with magnetic beads, was washed three times with wash buffer containing 50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, and 0.05% NP-40. Samples were boiled in 2× Laemmli sample buffer (Sigma) at 95°C and processed for immunoblot analysis.
Immunoblotting
The whole-cell extracts, eluted proteins from TUBE’s, or IP samples were diluted in 2× Laemmli sample buffer (Sigma) containing 2-mercaptoethanol and analyzed on 4–20% SDS-PAGE gel (Bio-Rad). The resolved proteins in the gel were transferred to polyvinylidene difluoride membrane using Trans-Blot Turb Transfer System (Bio-Rad). The membrane was blocked in 3–5% nonfat skim milk (Sigma) or 3–5% BSA (Sigma). The antibodies used for immunoblotting were rabbit anti-STING (D2P2F/#13647, 1:1,000; Cell Signaling), sheep anti-STING (R&D Systems, AF6516, 1:500), rabbit anti-STING (rodent preferred; #50494, 1:1,000; Cell Signaling), rabbit anti-pSTING (S366; #85735, 1:500; Cell Signaling), rabbit anti-pSTING (S365; #72971, 1:500; Cell Signaling), rabbit anti-TBK1 (3504, 1:1,000; Cell Signaling), rabbit anti-pTBK1 (Ser172; D52C2/#5483, 1:500; Cell Signaling), mouse anti-FLAG M2 (F3165, 1:1,000; Sigma), rabbit anti-HA (#3274, 1:1,000; Cell Signaling), rabbit anti-VPS34 (#4263,1:1,000; MBL), rabbit anti-IRF3 (D83B9/#4302, 1:1,000; Cell Signaling), rabbit anti-pIRF3 (Ser396; D6O1M/ #29047, 1:1,000; Cell Signaling), mouse anti-ubiquitin (P4D1, 1:1,000; Cell Signaling), rabbit anti-K63 (#05-1307, 1:2500; Millipore), rabbit anti-K48 (#05-1307, 1:2,500; Millipore), rabbit anti–p-STAT1 (Tyr701; 9167, 1:1,000; Cell Signaling), rabbit anti-STAT1 (14994, 1:1,000; Cell Signaling), mouse anti–b-actin (Abcam, AC-15/ ab49900, 1:10,000), mouse anti-vinculin (#V9131, 1:10,000; Sigma).
Ubiquitin chain restriction assay
Ubiquitin chain restriction assay was performed following the protocol published by Komander and colleagues (Hospenthal et al., 2015). Endogenous polyubiquitin conjugates were isolated from PMA-differentiated THP1 cells using TUBE1 (Life Sensors) and the Ubiquitin chain restriction assay (UbiCRest) was performed immediately. Recombinant USP2, AMSH, OTUB1, and Cezanne (Boston Biochem) were used for the reaction according to the manufacturer’s instructions. The reaction was terminated by adding 2x Laemmli sample buffer (Sigma) at 95°C and processed for immunoblot analysis.
IFN bioassay
Cell supernatants were collected at indicated events after stimulation. Bioactive human type I IFN was measured using HEK-Blue IFN-α/β cells as reporter cells according to the manufacturer’s instructions (InvivoGen).
Immunofluorescence and confocal microscopy
All experiments were performed on indicated cells seeded on coverslips and stimulated as defined. Cells were fixed with ice-cold 100% methanol for 5 min at −20°C. Subsequently, they were blocked with 1% BSA, incubated with primary antibody overnight at 4°C, and then stained with secondary antibody (1:300; Alexa Fluor; Invitrogen) for 1 h at room temperature. DAPI (Invitrogen) was used to stain cell nuclei. Images were captured by Zeiss LSM 780 confocal microscope and processed with the Zen Blue software (Zeiss). The antibodies used were sheep anti-STING (1:50) and mouse VP1-2 (1:50). For the imaging of iPSC-derived microglia, cells were grown in chamber slides (Ibidi), fixed with cold 4% PFA (Histolab) for 10 min at room temperature, and washed with Tris-buffered saline (TBS) three times for 5 min each. Cells were permeabilized with 0.3% Triton X-100 (Sigma) in TBS for 15 min at room temperature, blocked in blocking buffer (5% donkey serum [Sigma] in 0.3% Triton X-100 in TBS) for 1 h at room temperature. Cells were incubated with primary antibodies against TREM2 (R&D Systems) and Iba-1 (Abcam), diluted 1:50 in blocking buffer, overnight at 4°C. The next day, cells were washed with TBS three times for 5 min each and incubated with conjugated secondary antibodies (Thermo Fisher), diluted in blocking buffer, for 1 h at room temperature. Cells were washed with TBS three times for 5 min each and incubated with DAPI (Thermo Fisher) for 5 min at room temperature. Cells were washed once with water and then mounted using Ibidi mounting media and kept at 4°C until analysis. Samples were analyzed using an Eclipse Ti-E inverted confocal microscope with a 20× objective and the NIS Element software (Nikon).
RNA extraction and quantitative RT-PCR
RNA was purified from various cell types using a High Pure RNA isolation kit (Roche) according to the manufacturer's protocol. TaqMan primers used to perform quantitative PCR are IFN-β (Mm00439552_s1), CXCL10 (Mm00445235_m1), HSV-1 gB forward primer (5′-CGCATCAAGACCACCTCCTC-3′), HSV-1 gB reverse primer (5′-AGCTTGCGGGCCTCGTT-3′), and probe 5′-CGGCCCAACATATCGTTGACATGGC-3′. The mRNAs levels were normalized to β-actin using the formula 2Ct(bactin)−Ct (mRNA X) and further normalized to ratio of untreated cells.
Luciferase assay
Cells were cotransfected with IFN-β promoter-luc (firefly) and β–actin promoter (Renilla), FLAG-tagged STING, NT3 (WT), or NT3 (ΔDUB) constructs. Dual-Glo luciferase assay was performed according to the manufacturer’s instructions (Promega).
CRISPR/Cas9-mediated genome editing
Trim32-deficient KOs in THP1 cells were generated by CRISPR/Cas9 technology as previously described (Holm et al., 2016). pSp Cas9n(BB) 2A-GFP all in one construct was used to generate Trim32-deficient THP1 cells. The gRNAs were designed to target the TRIM32 gene on the first exon (Trim32 T1: 5′-CACCCGGCAATTCTGCCGGAGCTG-3′; T2: 5′-CACCCGTCCCAAGCTTCTGCACTGTGG-3′). The cells were transfected and selected for GFP-positive single cells by FACS in aseptic conditions. These single clones were expanded and confirmed by sequencing of deleted fragment of the allele using primers 5′-CACCGACCTGTGATGAGGTTCCTG-3′ and 5′-AAACCAGGAACCTCATCACAGGTC-3′ of the PCR product and quantitative RT-PCR. Elimination of TRIM32 protein expression was confirmed by immunoblotting.
Statistical analysis
All statistical analyses were performed using Prism 8 (GraphPad Software). If the data exhibited normal distributions, they were calculated using two-tailed Student’s t test, and if they did not pass the standard distribution test, the Wilcoxon rank-sum test was employed. The data shown are from one individual experiment out of at least three (replicates). All experiments and samples were biological replicates.
Online supplemental material
Fig. S1 shows data related to the generation of new HSV1-pUL36ΔDUB strains and the effects of DUB deficiency on induction of IFN responses. Fig. S2 shows replication of DUB-deficient HSV1 in organotypic brain slices. Fig. S3 shows activation of IFN responses by DUB-deficient HSV1 in different cells types and disease development in vivo. Fig. S4 shows that DUB deficiency does not affect viral DNA release into the cytoplasm. Fig. S5 shows the localization of VP1-2 NT1 and STING in HEK293T cells and that STING K150 and TRIM32 are central for full activation of STING.
Acknowledgments
We thank Yonglun Luo for helping to designing the gRNA for CRISPRs and Thomas Hennig (Virology, Würzburg, Germany) for providing the BAC backbone pHSV1(17+)Lox-CheVP26-pUL36∆DUB.
This work was funded by the European Research Council (ERC-AdG ENVISION; 786602), the Novo Nordisk Foundation (NNF18OC0030274), the Lundbeck Foundation (R198-2015-171 and R268-2016-3927), and the EU FP7 Mobilex program (4092-00253). M. Gyrd-Hansen is supported by the Ludwig Institute for Cancer Research and a Wellcome Trust senior research fellowship (102894/Z/13/Z). Work in the laboratory of B. Sodeik was supported by the Deutsche Forschungsgemeinschaft (German Research Foundation) under Germany’s Excellence Strategy (EXC 2155 “RESIST”; project ID 39087428). H. Zetterberg is a Wallenberg Scholar supported by the Swedish Research Council (grant 2018-02532), the European Research Council (grant 681712), Swedish State Support for Clinical Research (grant ALFGBG-720931), the UK Dementia Research Institute, and the Alzheimer’s Drug Discovery Foundation.
Author contributions: C. Bodda and S.R. Paludan conceived the idea. C. Bodda, L.S. Reinert., S. Fruhwürth, T. Richardo, C. Sun, and B.-c. Zhang performed the experiments. M. Kalamvoki, A. Pohlmann, T.H. Mogensen, P. Bergström, L. Agholme, P. O'Hare, and B. Sodeik provided materials. M. Gyrd-Hansen, H. Zetterberg, and S.R. Paludan supervised experiments. C. Bodda and S.R. Paludan wrote the manuscript.
References
- Andersen L.L., Mørk N., Reinert L.S., Kofod-Olsen E., Narita R., Jørgensen S.E., Skipper K.A., Höning K., Gad H.H., Østergaard L., et al. . 2015. Functional IRF3 deficiency in a patient with herpes simplex encephalitis. J. Exp. Med. 212:1371–1379. 10.1084/jem.20142274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodda C., Tantra M., Mollajew R., Arunachalam J.P., Laccone F.A., Can K., Rosenberger A., Mironov S.L., Ehrenreich H., and Mannan A.U.. 2013. Mild overexpression of Mecp2 in mice causes a higher susceptibility toward seizures. Am. J. Pathol. 183:195–210. 10.1016/j.ajpath.2013.03.019 [DOI] [PubMed] [Google Scholar]
- Bolstad M., Abaitua F., Crump C.M., and O’Hare P.. 2011. Autocatalytic activity of the ubiquitin-specific protease domain of herpes simplex virus 1 VP1-2. J. Virol. 85:8738–8751. 10.1128/JVI.00798-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie A.G., and Unterholzner L.. 2008. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 8:911–922. 10.1038/nri2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W.Z., and Schaffer P.A.. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J. Virol. 63:4579–4589. 10.1128/JVI.63.11.4579-4589.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen M.H., and Paludan S.R.. 2017. Viral evasion of DNA-stimulated innate immune responses. Cell. Mol. Immunol. 14:4–13. 10.1038/cmi.2016.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen M.H., Jensen S.B., Miettinen J.J., Luecke S., Prabakaran T., Reinert L.S., Mettenleiter T., Chen Z.J., Knipe D.M., Sandri-Goldin R.M., et al. . 2016. HSV-1 ICP27 targets the TBK1-activated STING signalsome to inhibit virus-induced type I IFN expression. EMBO J. 35:1385–1399. 10.15252/embj.201593458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrady C.D., Zheng M., van Rooijen N., Drevets D.A., Royer D., Alleman A., and Carr D.J.. 2013. Microglia and a functional type I IFN pathway are required to counter HSV-1-driven brain lateral ventricle enlargement and encephalitis. J. Immunol. 190:2807–2817. 10.4049/jimmunol.1203265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps T., and Kalamvoki M.. 2017. Evasion of the STING DNA-Sensing Pathway by VP11/12 of Herpes Simplex Virus 1. J. Virol. 91 e00535-17 10.1128/JVI.00535-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerst R.J., and Morrison L.A.. 2004. Herpes simplex virus 2 virion host shutoff protein interferes with type I interferon production and responsiveness. Virology. 322:158–167. 10.1016/j.virol.2004.01.019 [DOI] [PubMed] [Google Scholar]
- Dupuis S., Jouanguy E., Al-Hajjar S., Fieschi C., Al-Mohsen I.Z., Al-Jumaah S., Yang K., Chapgier A., Eidenschenk C., Eid P., et al. . 2003. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 33:388–391. 10.1038/ng1097 [DOI] [PubMed] [Google Scholar]
- Goubau D., Deddouche S., and Reis e Sousa C.. 2013. Cytosolic sensing of viruses. Immunity. 38:855–869. 10.1016/j.immuni.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman M., Ciancanelli M., Ou Y.H., Lorenzo L., Klaudel-Dreszler M., Pauwels E., Sancho-Shimizu V., Pérez de Diego R., Abhyankar A., Israelsson E., et al. . 2012. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J. Exp. Med. 209:1567–1582. 10.1084/jem.20111316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C.K., Rahbek S.H., Gad H.H., Bak R.O., Jakobsen M.R., Jiang Z., Hansen A.L., Jensen S.K., Sun C., Thomsen M.K., et al. . 2016. Influenza A virus targets a cGAS-independent STING pathway that controls enveloped RNA viruses. Nat. Commun. 7:10680 10.1038/ncomms10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospenthal M.K., Mevissen T.E.T., and Komander D.. 2015. Deubiquitinase-based analysis of ubiquitin chain architecture using Ubiquitin Chain Restriction (UbiCRest). Nat. Protoc. 10:349–361. 10.1038/nprot.2015.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., You H., Su C., Li Y., Chen S., and Zheng C.. 2018. Herpes Simplex Virus 1 Tegument Protein VP22 Abrogates cGAS/STING-Mediated Antiviral Innate Immunity. J. Virol. 92 e00841-18 10.1128/JVI.00841-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inn K.S., Lee S.H., Rathbun J.Y., Wong L.Y., Toth Z., Machida K., Ou J.H., and Jung J.U.. 2011. Inhibition of RIG-I-mediated signaling by Kaposi’s sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J. Virol. 85:10899–10904. 10.1128/JVI.00690-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Ma Z., and Barber G.N.. 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 461:788–792. 10.1038/nature08476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K.E., Song B., and Knipe D.M.. 2008. Role for herpes simplex virus 1 ICP27 in the inhibition of type I interferon signaling. Virology. 374:487–494. 10.1016/j.virol.2008.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouan Y., Grammatico-Guillon L., Espitalier F., Cazals X., François P., and Guillon A.. 2015. Long-term outcome of severe herpes simplex encephalitis: a population-based observational study. Crit. Care. 19:345 10.1186/s13054-015-1046-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattenhorn L.M., Korbel G.A., Kessler B.M., Spooner E., and Ploegh H.L.. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell. 19:547–557. 10.1016/j.molcel.2005.07.003 [DOI] [PubMed] [Google Scholar]
- Leib D.A., Harrison T.E., Laslo K.M., Machalek M.A., Moorman N.J., and Virgin H.W.. 1999. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J. Exp. Med. 189:663–672. 10.1084/jem.189.4.663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.D., Wu J., Gao D., Wang H., Sun L., and Chen Z.J.. 2013. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 341:1390–1394. 10.1126/science.1244040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Noyce R.S., Collins S.E., Everett R.D., and Mossman K.L.. 2004. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J. Virol. 78:1675–1684. 10.1128/JVI.78.4.1675-1684.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y.T., Grishin N.V., et al. . 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 347 aaa2630 10.1126/science.aaa2630 [DOI] [PubMed] [Google Scholar]
- Luecke S., Holleufer A., Christensen M.H., Jønsson K.L., Boni G.A., Sørensen L.K., Johannsen M., Jakobsen M.R., Hartmann R., and Paludan S.R.. 2017. cGAS Is Activated by DNA in a Length-Dependent Manner. EMBO Rep. 18:1707–1715. 10.15252/embr.201744017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Jin H., Valyi-Nagy T., Cao Y., Yan Z., and He B.. 2012. Inhibition of TANK binding kinase 1 by herpes simplex virus 1 facilitates productive infection. J. Virol. 86:2188–2196. 10.1128/JVI.05376-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai K., Konno H., Akiba T., Uemura T., Waguri S., Kobayashi T., Barber G.N., Arai H., and Taguchi T.. 2016. Activation of STING requires palmitoylation at the Golgi. Nat. Commun. 7:11932 10.1038/ncomms11932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni G., Konno H., and Barber G.N.. 2017. Ubiquitination of STING at lysine 224 controls IRF3 activation. Sci. Immunol. 2 eaah7119 10.1126/sciimmunol.aah7119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A., Alexander D., Tallóczy Z., Sun Q., Wei Y., Zhang W., Burns D., Leib D.A., and Levine B.. 2007. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 1:23–35. 10.1016/j.chom.2006.12.001 [DOI] [PubMed] [Google Scholar]
- Orzalli M.H., DeLuca N.A., and Knipe D.M.. 2012a HSV-1 ICP0 redistributes the nuclear IFI16 pathogen sensor and promotes its degradation. Proc. Natl. Acad. Sci. USA. 109:E3008–E3017. 10.1073/pnas.1211302109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli M.H., DeLuca N.A., and Knipe D.M.. 2012b Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc. Natl. Acad. Sci. USA. 109:E3008–E3017. 10.1073/pnas.1211302109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan S.R. 2015. Activation and regulation of DNA-driven immune responses. Microbiol. Mol. Biol. Rev. 79:225–241. 10.1128/MMBR.00061-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan S.R., Bowie A.G., Horan K.A., and Fitzgerald K.A.. 2011. Recognition of herpesviruses by the innate immune system. Nat. Rev. Immunol. 11:143–154. 10.1038/nri2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez de Diego R., Sancho-Shimizu V., Lorenzo L., Puel A., Plancoulaine S., Picard C., Herman M., Cardon A., Durandy A., Bustamante J., et al. . 2010. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity. 33:400–411. 10.1016/j.immuni.2010.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran T., Bodda C., Krapp C., Zhang B.C., Christensen M.H., Sun C., Reinert L., Cai Y., Jensen S.B., Skouboe M.K., et al. . 2018. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 37 e97858 10.15252/embj.201797858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinert L.S., Lopušná K., Winther H., Sun C., Thomsen M.K., Nandakumar R., Mogensen T.H., Meyer M., Vægter C., Nyengaard J.R., et al. . 2016. Sensing of HSV-1 by the cGAS-STING pathway in microglia orchestrates antiviral defence in the CNS. Nat. Commun. 7:13348 10.1038/ncomms13348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice S.A., and Knipe D.M.. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J. Virol. 64:1704–1715. 10.1128/JVI.64.4.1704-1715.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato P.C., and Leib D.A.. 2015. Neuronal Interferon Signaling Is Required for Protection against Herpes Simplex Virus Replication and Pathogenesis. PLoS Pathog. 11 e1005028 10.1371/journal.ppat.1005028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho-Shimizu V., Pérez de Diego R., Lorenzo L., Halwani R., Alangari A., Israelsson E., Fabrega S., Cardon A., Maluenda J., Tatematsu M., et al. . 2011. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J. Clin. Invest. 121:4889–4902. 10.1172/JCI59259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandbaumhüter M., Döhner K., Schipke J., Binz A., Pohlmann A., Sodeik B., and Bauerfeind R.. 2013. Cytosolic herpes simplex virus capsids not only require binding inner tegument protein pUL36 but also pUL37 for active transport prior to secondary envelopment. Cell. Microbiol. 15:248–269. 10.1111/cmi.12075 [DOI] [PubMed] [Google Scholar]
- Seo G.J., Kim C., Shin W.J., Sklan E.H., Eoh H., and Jung J.U.. 2018. TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat. Commun. 9:613 10.1038/s41467-018-02936-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C., and Zheng C.. 2017. Herpes Simplex Virus 1 Abrogates the cGAS/STING-Mediated Cytosolic DNA-Sensing Pathway via Its Virion Host Shutoff Protein, UL41. J. Virol. 91 e02414-16 10.1128/JVI.02414-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Schattgen S.A., Pisitkun P., Jorgensen J.P., Hilterbrand A.T., Wang L.J., West J.A., Hansen K., Horan K.A., Jakobsen M.R., et al. . 2015. Evasion of innate cytosolic DNA sensing by a gammaherpesvirus facilitates establishment of latent infection. J. Immunol. 194:1819–1831. 10.4049/jimmunol.1402495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., and Akira S.. 2010. Pattern recognition receptors and inflammation. Cell. 140:805–820. 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- Tsuchida T., Zou J., Saitoh T., Kumar H., Abe T., Matsuura Y., Kawai T., and Akira S.. 2010. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 33:765–776. 10.1016/j.immuni.2010.10.013 [DOI] [PubMed] [Google Scholar]
- Tyler K.L. 2018. Acute Viral Encephalitis. N. Engl. J. Med. 379:557–566. 10.1056/NEJMra1708714 [DOI] [PubMed] [Google Scholar]
- van Gent M., Braem S.G.E., de Jong A., Delagic N., Peeters J.G.C., Boer I.G.J., Moynagh P.N., Kremmer E., Wiertz E.J., Ovaa H., et al. . 2014. Epstein-Barr virus large tegument protein BPLF1 contributes to innate immune evasion through interference with toll-like receptor signaling. PLoS Pathog. 10 e1003960 10.1371/journal.ppat.1003960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpooten D., Ma Y., Hou S., Yan Z., and He B.. 2009. Control of TANK-binding kinase 1-mediated signaling by the gamma(1)34.5 protein of herpes simplex virus 1. J. Biol. Chem. 284:1097–1105. 10.1074/jbc.M805905200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wang K., Li J., and Zheng C.. 2013a Herpes simplex virus 1 ubiquitin-specific protease UL36 inhibits beta interferon production by deubiquitinating TRAF3. J. Virol. 87:11851–11860. 10.1128/JVI.01211-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wang K., Lin R., and Zheng C.. 2013b Herpes simplex virus 1 serine/threonine kinase US3 hyperphosphorylates IRF3 and inhibits beta interferon production. J. Virol. 87:12814–12827. 10.1128/JVI.02355-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Liu X., Cui Y., Tang Y., Chen W., Li S., Yu H., Pan Y., and Wang C.. 2014. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity. 41:919–933. 10.1016/j.immuni.2014.11.011 [DOI] [PubMed] [Google Scholar]
- Whitley R.J. 2006. Herpes simplex encephalitis: adolescents and adults. Antiviral Res. 71:141–148. 10.1016/j.antiviral.2006.04.002 [DOI] [PubMed] [Google Scholar]
- Whitley R.J., Soong S.J., Dolin R., Galasso G.J., Ch’ien L.T., and Alford C.A.. 1977. Adenine arabinoside therapy of biopsy-proved herpes simplex encephalitis. National Institute of Allergy and Infectious Diseases collaborative antiviral study. N. Engl. J. Med. 297:289–294. 10.1056/NEJM197708112970601 [DOI] [PubMed] [Google Scholar]
- Xia P., Ye B., Wang S., Zhu X., Du Y., Xiong Z., Tian Y., and Fan Z.. 2016. Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat. Immunol. 17:369–378. 10.1038/ni.3356 [DOI] [PubMed] [Google Scholar]
- Ye R., Su C., Xu H., and Zheng C.. 2017. Herpes Simplex Virus 1 Ubiquitin-Specific Protease UL36 Abrogates NF-κB Activation in DNA Sensing Signal Pathway. J. Virol. 91 e02417-16 10.1128/JVI.02417-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., You J., You H., and Zheng C.. 2018. Herpes Simplex Virus 1 UL36USP Antagonizes Type I Interferon-Mediated Antiviral Innate Immunity. J. Virol. 92 e01161-18 10.1128/JVI.01161-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.Y., Jouanguy E., Ugolini S., Smahi A., Elain G., Romero P., Segal D., Sancho-Shimizu V., Lorenzo L., Puel A., et al. . 2007. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 317:1522–1527. 10.1126/science.1139522 [DOI] [PubMed] [Google Scholar]
- Zhang J., Hu M.M., Wang Y.Y., and Shu H.B.. 2012. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J. Biol. Chem. 287:28646–28655. 10.1074/jbc.M112.362608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhao J., Xu S., Li J., He S., Zeng Y., Xie L., Xie N., Liu T., Lee K., et al. . 2018. Species-Specific Deamidation of cGAS by Herpes Simplex Virus UL37 Protein Facilitates Viral Replication. Cell Host Microbe. 24:234–248.e5. 10.1016/j.chom.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B., Zhang L., Lei C., Li Y., Mao A.P., Yang Y., Wang Y.Y., Zhang X.L., and Shu H.B.. 2009. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 30:397–407. 10.1016/j.immuni.2009.01.008 [DOI] [PubMed] [Google Scholar]