Abstract
Glioblastoma is a difficult disease to diagnose. Proteomic techniques are commonly applied in biomedical research, and can be useful for early detection, making an accurate diagnosis and reducing mortality. The relevance of mitochondria in brain development and function is well known; therefore, mitochondria may influence the development of glioblastoma. The T98G (with oxidative metabolism) and U87MG (with glycolytic metabolism) cell lines are considered to be useful glioblastoma models for studying these tumors and the role of mitochondria in key aspects of this disease, such as prognosis, metastasis and apoptosis. In the present study, principal component analysis of protein abundance data identified by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) and matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF) from 2D gels indicated that representative mitochondrial proteins were associated with glioblastoma. The selected proteins were organized into T98G- and U87MG-specific protein-protein interaction networks to demonstrate the representativeness of both proteomic techniques. Gene Ontology overrepresentation analysis based on the relevant proteins revealed that mitochondrial processes were associated with metabolic changes, invasion and metastasis in glioblastoma, along with other non-mitochondrial processes, such as DNA translation, chaperone responses and autophagy. Despite the lower resolution of 2D electrophoresis, principal component analysis yielded information of comparable quality to that of LC-MS/MS. The present analysis pipeline described a specific and more complete metabolic status for each cell line, defined a clear mitochondrial performance for distinct glioblastoma tumors, and introduced a useful strategy to understand the heterogeneity of glioblastoma.
Keywords: brain cancer, metabolic change, electrophoresis, proteomics, multivariate analysis
Introduction
Pediatric solid brain tumors are the most common central nervous system neoplasia in children and the second most common in individuals <20 years old (1). Glioblastoma multiforme (GBM), or grade IV astrocytoma, is the most common and lethal adult malignant brain tumor, whereas it only occurs in 8–12% of the pediatric population (2). Nevertheless, glioblastoma in both populations is characterized by an aggressive medical behavior, as well as high mortality and morbidity rates, with an incidence of 3.19 cases per 100,000 individuals and a 5-year survival rate of 5% (3,4). GBM has a high diversity in terms of morphology, localization and genetic alterations; therefore, GBM has been poorly characterized, which makes glioblastoma difficult to diagnose (4). Understanding glioblastoma heterogeneity should be a priority for developing improved therapies and searching for novel biomarkers (5).
Mitochondria, which are the ‘power houses’ of the cell, are abundant in the brain. Biogenesis, mitophagy, migration and morphogenesis are crucial in brain development and synaptic pruning (6,7). Therefore, mitochondria can affect the susceptibility of the brain to injury, and they serve a role in innate immunity, in inflammation in response to infection and acute damage, and in antiviral and antibacterial defense (6,8). As mitochondria serve critical roles in numerous bioenergetic, anabolic and cell biochemical pathways (9,10), their genetic and metabolic alterations have been suggested to be a pathogenic cause of, or contributing factor to, a broad range of human diseases, including cancer (11,12). Several common tumor cell features can result from mitochondrial dysregulation, because, the biology of mitochondria supports cell transformation during carcinogenesis (11,13,14), which suggests that the mitochondrial proteome is versatile and can sense the spatial and temporal dynamics of cell biological processes from the onset to the end of cancer. However, the specific role of mitochondria in cancer has not been completely uncovered, mainly due to the large amount of information regarding mitochondrial processes in cancer not having been properly integrated.
Proteomic analysis can be applied in GBM research for early detection, for making a reliable diagnosis and for performing an accurate risk assessment. However, Petrak et al (15), Deighton et al (16) and Valledor and Jorrín (17) agree that, despite the utility of proteomic research to obtain insights into cancer-associated biological processes and the knowledge of neuro-oncology, the glioblastoma proteomic studies performed to date have focused on how proteins are up- or downregulated, and these results have been generated without any specific approach to establish the existence of key proteins and/or specific signaling pathways in cancer development or regulation. To the best of our knowledge, most of the generated data lack reproducibility, validity and comparability, mainly due to methodological and analytical constraints. The identified proteins in these studies are diverse and make it hard to understand the nature of the disease or its background.
Mitochondria compose a biological system that interacts in, with or between other living systems, and that maintains physiological associations with other subsystems in cells, such as organelles, genes and proteins. In a complex disease, the mitochondrion and its environment are altered (14). Therefore, systemic questions in the context of cancer are: How do the mitochondria interact with other components and their environment? Additionally, what are the roles of the mitochondria within the cell? (18) In this regard, analyzing protein abundance data according to fold-change, univariate statistics (such as Student's t-test or ANOVA) or analog nonparametric tests do not answer these questions due to their nature; on the other hand, multivariate statistical analysis involves >2 variables in ≥2 conditions simultaneously for any possible association or empirical relationship, so the description of the interactions between cellular components can be improved.
Proteomic data enclose information about the whole cellular system, bringing an improved description through multivariate statistical approaches. Additionally, principal component analysis (PCA) applied to GBM mitochondrial proteomic data could reveal that mitochondria metabolism acts as a cellular sensor of specific isolated cancer states, which could result in reliable and useful information to help improve diagnosis and risk assessment, as well as to understand the role of mitochondria in GBM.
In the present study, a proteomic functional analysis based on PCA of liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) and 2D isoelectric focusing (IEF)/SDS-PAGE intensity data was conducted. A specific mitochondrial proteomic landscape was obtained from the glioblastoma T98G and U87MG cell lines associated with biological processes that characterize ‘oxidative’ and ‘glycolytic’ types of tumor. Additionally, the present cell model resembles the metabolic transition from mitochondrial oxidative phosphorylation (OXPHOS) to glycolysis (a process known as the Warburg effect), as reported during tumorigenesis (19). Finally, protein-protein interaction networks (PPIns) and Gene Ontology (GO) overrepresentation based on PCA revealed that LC-MS/MS and 2D IEF/SDS-PAGE analysis were comparable and complementary with each other, indicating that mitochondria may act as key sensing organelles for GBM tumor characterization and serve as valuable tools for therapeutic targets.
Materials and methods
Cell culture
The T98G (CRL-1690™) and U87MG (glioblastoma of unknown origin; HTB-14™) cell lines were purchased from the American Type Culture Collection and cultured in 175-cm2 plastic flasks at 37°C with 5% CO2 in Eagle's Minimum Essential Medium (In Vitro S. A.) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc). Cells were grown to 80–90% confluence, harvested with trypsin, washed twice in PBS and used for mitochondrial isolation.
Mitochondrial isolation
Mitochondria were isolated by differential centrifugation. Cells were separately disrupted in 250 mM sucrose, 1 mM EGTA and 10 mM HEPES (pH 7.4) at 4°C and centrifuged for 10 min at 1,500 × g and 4°C to recover the supernatant. This step was repeated three times. Subsequently, all of the supernatants were pooled and centrifuged for 10 min at 12,000 × g and 4°C to obtain a mitochondrial pellet. The pellets were used immediately or kept at −80°C until use.
LC-MS/MS
Mitochondrial proteome extraction
A total of six mitochondrial pellets (3 biological replicates each from T98G and U87MG cells) from the same passages were lysed, incubated and sonicated at 4°C (5 cycles of 20 pulses) in lysis buffer (4% SDS, 0.1 M DTT and 0.1 M Tris pH 8.6). To reduce disulfide bridges, samples were incubated at 40°C for 30 min, and cysteine residues were alkylated with 100 mM iodoacetamide for 30 min in the dark. The protein content was estimated by 1D SDS-PAGE scanned in a GS-800 densitometer (Bio-Rad Laboratories, Inc.), stained overnight at room temperature with colloidal Coomassie brilliant blue R-250 and quantified using the Quantity One software v4.6.9 (Bio-Rad Laboratories, Inc.).
Peptide separation and spectrometry
The peptide mixture was subjected to reverse phase chromatography on a Dionex Ultimate 3,000 RSLC nano-UPLC system (Thermo Fisher Scientific, Inc.) in-line coupled to a Q-Exactive Plus high-resolution mass spectrometer (Thermo Fischer Scientific, Inc.). Peptides (2 µg) resuspended were first trapped on a precolumn (C18 PepMap 100, 5 µm, 100 A, 300 µm inner diameter × 5 mm; Thermo Fisher Scientific, Inc), then separated using an EASY-Spray PepMap RSLC C18 capillary column (2 µm, 15 cm × 50 µm; Thermo Fischer Scientific, Inc.) with a 250-min elution gradient at 250 nl/min. The mobile phases were: A) 2% acetonitrile and 0.1% formic acid in water; and B) 90:10 (v:v) acetonitrile:water and 0.1% formic acid in water. The mass spectrometer was operated in positive data-dependent acquisition mode and the full MS range was 300-1,800 m/z. A total of 10 of the most intense ions were isolated in the quadrupole and fragmented under higher-energy collisional dissociation with a normalized collision energy of 27%. Precursor ions were measured at a resolution of 70,000 (at 200 m/z) and the fragments were measured at 17,500. Only ions with charge states ≥2 were fragmented with an isolation window of 2 m/z.
Protein identification and quantification
Protein identification and label-free quantification were performed using MaxQuant v1.6.2.3 (20). The parameters included trypsin/P as the digestion enzyme, carbamidomethyl-cysteine as a fixed modification, and N-terminal protein acetylation and methionine oxidation as variable modifications. Proteins were identified with a 1% false discovery rate (FDR) based on the target-decoy strategy provided by MaxQuant, setting Arg-C as the digestion enzyme and carbamidomethylcysteine as a fixed modification. Proteins were identified with an FDR of 1% based on the target-decoy strategy provided by MaxQuant. Protein identification was performed according to the human reference proteome UP000005640 from the UniProt repository downloaded on August 03, 2018 (https://www.uniprot.org/proteomes/UP000005640). Label-free quantification was performed using proteins with ≥2 razor-unique peptides identified by LC-MS/MS.
Multivariate analysis of protein intensities
Statistical analysis of protein abundances was performed only with proteins with ≥2 intensity values in each cell. The protein abundance was normalized between conditions and missing values were imputed with the Random Forest method (missForest v.4; R package v3.5) (21). PCA was carried out on the protein intensity correlation matrix (FactoMiner v2.3; R package v3.5) (22) to generate a protein abundance pattern for the cell lines (15). To determine whether any component could distinguish between the cell lines, the sample scores for each component were plotted. After finding the component, the significant proteins with discriminatory capacity in that component were identified using the cos2 of the correlation matrix between the components and the proteins (23).
To evaluate the PCA performance on LC-MS/MS data, a t-test (significance level, 0.05) and a fold-change analysis were conducted to compare the abundance spots of proteins between cell lines.
2D SDS-PAGE
Mitochondrial proteome extraction
T98G and U87MG mitochondrial-associated proteins were obtained according to Hurkman's protocol (24), which was modified as follows: Six mitochondrial pellets (three biological replicates each from T98G and U87MG cells) from the same passage were resuspended in 500 µl extraction buffer (0.7 M sucrose, 0.5 M Tris-Base, 0.1 M KCI, 0.03 M HCI, 0.05 M EDTA and 2% β-mercaptoethanol) and 500 µl saturated phenol, and incubated for 20 min at −20°C. Subsequently, mitochondrial samples were centrifuged for 10 min at 400 × g and 4°C, and the phenolic phase was recovered after the addition of 0.1 M ammonium acetate for 12–15 h at −20°C. Subsequently, mitochondrial samples were washed twice with 0.1 M ammonium acetate and centrifuged for 10 min at 4,000 × g and 4°C. Pellets with mitochondrial proteins were washed with 1 ml 80% acetone and centrifuged for 10 min at 4,000 × g and 4°C. The supernatants were discarded, and the pellets were resuspended in IEF buffer (7 M urea, 2 M thiourea, 0.06 M DTT, 2% ampholytes at pH 3–10 and 4% CHAPS) and centrifuged for 30 min at 8,000 × g and 4°C. The obtained supernatants were recovered and frozen at −80°C until use in 2D electrophoresis (2DE).
2-DE gels
IEF was performed in acrylamide gel tubes as previously described (25). Briefly, gel tubes were pre-focused (2500 V; 110 mA; 1 h and 250/h per gel) before IEF (22 h at 125 V). Each gel (three for T98G and three for U87MG cells) was loaded with 500 µg protein, which was quantified via the Bradford method. Electrofocused gels were used for 12% 2D SDS-PAGE for additional spot separation. 2D gels were fixed for 30 min and stained for 1 h with colloidal Coomassie brilliant blue R-250 for image acquisition; fixing and staining processes were carried out at room temperature.
Image pre-processing
The gels were scanned in a GS-800 densitometer (Bio-Rad Laboratories, Inc.), and six images were acquired, wrapped and overlapped with PdQuest v8.0.1 software (Bio-Rad Laboratories, Inc.). Subsequently, with all six images combined, a master gel was created by the default PdQuest algorithm from the intensity sum of all of the spots in the gel images.
Random spot selection in the master gel
To increase the protein representativeness of the cellular processes carried out in the T98G and U87MG cell lines, 400 spots (of the 1,274 detected by PdQuest) were randomly selected from the master gel regardless of their size, intensity or abundance differences between cell lines. This process ensures that every spot in the master gel had an equal chance of being selected and allowed to obtain a representative mitochondrial proteome sample (26,27). The generated spot sample was rematched in all gel images to allow for a more reliable abundance analysis (17).
Multivariate analysis of spot intensities
To select the spots to be identified, a spreadsheet with the normalized intensity of the 400 spots sampled was exported from PdQuest. The abundance of the spots was logarithmically transformed, and missing values were imputed with the Random Forest method (missForest; R package) (21) to perform multivariate analysis.
Abundance analysis was performed using PCA of the spot intensity correlation matrix (FactoMineR; R package) (22) to generate a spot abundance pattern for the cell line gels (17). To understand whether any component could distinguish between the cell lines, the gel scores for each component were plotted. Subsequently, significant spots were identified using the cos2 of the correlation matrix between the components and the spots with discriminatory capacity (23).
To evaluate the PCA performance on 2DE-MALDI-TOF data, a t-test (significance level, 0.05) and a fold-change analysis were conducted to compare the abundance spots of proteins between cell lines.
Mass spectrometry
Each selected spot was cut from the gel, alkylated, reduced, digested and automatically transferred to a matrix-assisted laser desorption/ionization (MALDI) analysis target via a Proteineer SPII and SP robot using the SP control v3.1.48.0 software (Bruker Corporation) with the aid of a DP Chemicals 96 gel digestion kit (Bruker Corporation) and processed in a MALDI-time of flight (TOF) Autoflex spectrometer (Bruker Corporation) to obtain the peptide mass fingerprints. A total of 100 satisfactory shots in 20 short steps were performed, the peak resolution threshold was set at 1,500, the signal/noise ratio of tolerance was 6 and contaminants were not excluded. The spectrum was annotated using flexAnalysis 1.2 vSD1 Patch 2 (Bruker Corporation). The search engine MASCOT (28) was used to compare the fingerprints against the SwissProt (29) 2016 release database with the following parameters: Taxon-Human, mass tolerance of ≤200 ppm, one missed cleavage allowed, and fixed modification of carbamidomethyl and oxidation of methionine as the variable modifications.
PPIn construction
A PPIn network for T98G and another for U87MG cell lines were built with overexpressed and specific proteins obtained from the LC-MS/MS abundance data employing the GeneMANIA application v3.4.1 (30) in Cytoscape v3.3 (31). The networks were constructed only using the experimental evidence of physical interactions between proteins without added nodes. Subsequently, to compare the consistency between the PCA results from the LC-MS/MS and the 2DE data, overexpressed and specific proteins obtained from 2DE were localized to the LC-MS/MS PPIn.
Representative biological processes identification
To further understand the critical biochemical processes taking place in each PPIn, only the connected proteins in the networks were taken into account for comparative overrepresentation analysis based on GO (32). Overrepresentation was performed online employing the Gene List Analysis tool on the PANTHER Classification system site (33) (http://www.pantherdb.org/). As inputs, the official gene symbols were uploaded as identifiers. The overrepresented biological processes were clustered with REViGO web tool (http://revigo.irb.hr/) (34) and R software v3.4 (35).
Western blot analysis
For OXPHOS system comparison, 20 µg of mitochondrial extracts were separated via 12% SDS-PAGE (36) and transferred to a PVDF membrane (37) at 100 V for 1 h (36,37). The membrane was blocked with 5% non-fat milk in PBS-0.05% Tween-20 (PBST) for 1 h at room temperature, incubated with primary antibodies (overnight at 4°C) purchased from Abcam against each subunit of the OXPHOS complex, which is one of the most affected systems: NADH dehydrogenase [uniquinone] 1α subunit 10 or complex (CI; cat. no. ab14713; 1:2,000); subunit 70 kDa (CII; cat. no. ab14715; 1:10,000); core 2 (CIII; cat. no. ab14745; 1:4,000); subunit IV (CIV; cat. no. ab14744; 1:1,000) and β-subunit ATP synthase (CV; cat. no. ab110273; 1:1,000). Next, the membrane was washed three times with PBST, each for 10 min at room temperature, and incubated with HRP-conjugated goat anti-mouse IgG secondary antibody (cat. no. NB7539; Novus Biological; 1:5,000). After membrane washing as described, the reaction bands were detected via chemiluminescence (EMD Millipore) and read with a C-Digit Blot Scanner (LI-COR Biosciences). To compare the densitometry values [(pixels/cm2) ×104] between cell lines, a Mann-Whitney test was performed for 3 independent gels for each cell line using R software v3.4 (35). Comparisons are shown as 95% confidence intervals of the median.
To obtain bioenergetic signatures, total protein from the two cell lines was extracted using RIPA buffer (10 mM Tris-Cl pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS and 140 mM NaC) and protein concentration was determined using the Lowry method (38). As GAPDH and β-ATPase exhibited differences in abundance, western blotting was performed using both 10 and 20 µg of total protein to avoid artefacts due to protein quantity. Total protein was separated and transferred in the same way as mitochondrial extracts. The membrane was blocked, incubated and bands detected using the same protocol as for OXPHOS western blot analysis with primary antibodies purchased from Abcam against GAPDH (cat. no. ab8245; 1:1,000) and β-subunit ATP synthase (1:1,000), with an anti-mouse secondary antibody used (1:5,000). The experimental conditions, reagents, equipment and software used are those mentioned for mitochondrial extracts.
Results
PCA of LC-MS/MS identified proteins
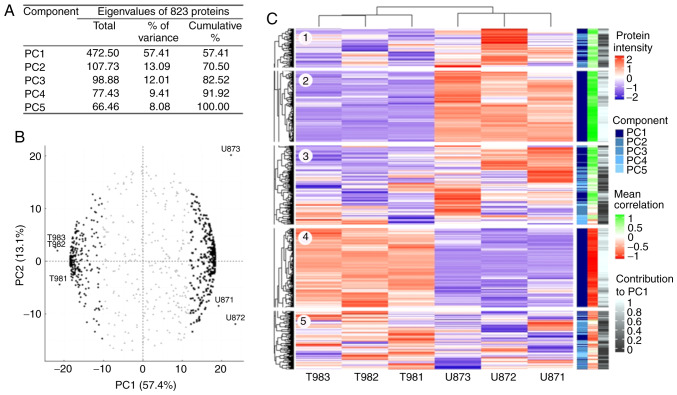
The LC-MS/MS process identified 1,805 proteins, and 1,069 proteins were identified with ≥2 unique peptides and had ≥2 intensity values for each cell line (Table SI and Fig. S1). However, 161 proteins were specific for T98G and 82 proteins for U87MG; these proteins were considered for the overrepresentation analysis (Fig. S1). Additionally, three proteins were eliminated during the imputation data process as they were identified as outliers. PCA was performed on 823 shared proteins (Table SII and Fig. S1). The total protein abundance variation was explained via five principal components (PCs; Fig. 1A). PC1 embraces 57% of the whole abundance variability, while the other four components only explain 43% of the remaining variability. The association of proteins and samples with PC1 and PC2 (Fig. 1B) revealed 235 proteins (black dots), with negative PC1 values, associated with T98G samples, and 308 proteins (black squares) with positive PC1 values, associated with U87MG samples. These 543 proteins (dark blue on the component scale in Fig. 1C) had a homogeneous intensity pattern within the cell lines (blocks 2 and 4 on the heatmap; Fig. 1C) and significant (r<-0.5 or r>0.5) correlation values with PC1 (light red and light green in the mean correlation scale in Fig. 1C and Table SII), and their contribution to explain this intensity pattern was significant (white color in the contribution scale in Fig. 1C and Table SII). The proteins grouped on the left had a greater abundance in T98G and a lower abundance in U87MG (block 4 on the heatmap; Fig. 1C), and those grouped on the right were more abundant in U87MG (block 2 on the heatmap; Fig. 1C). The proteins in the center of the circular biplot (grey triangles; Fig. 1B) had heterogeneous intensity values within the cell lines (blocks 1, 3 and 5 on the heatmap; Fig. 1C), as well as low correlation and contribution values (Fig. 1C and Table SII).
Figure 1.
Multivariate analysis of the protein abundance from LC-MS/MS. (A) PC analysis was performed using the protein abundance values from LC-MS/MS to obtain PCs that explained the total data variation. (B) Circular biplot showing the association between proteins and samples with PC1 and PC2. Black dots (left) represent proteins with negative correlation with PC1, black squares (right) proteins with positive correlation with PC1 and grey triangles (center) proteins not correlated with PC1. (C) Protein abundance patterns (blue-red scale into heatmap) showing that where PC1 proteins are present (dark blue in component scale), the protein expression values show homogeneous patterns with mean correlation values (red-green scale) and protein contribution to PC1 (white-black scale). PC, principal component; LC-MS/MS, liquid chromatography coupled to tandem mass spectrometry.
PCA for 2DE gel spots
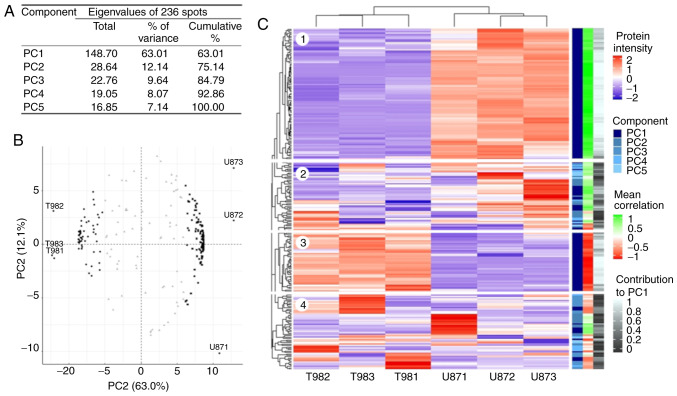
Overall, 400 protein spots were selected across all gel surfaces regardless of size, intensity or difference in abundance between the cell lines. A total of three spots did not pass quality control, and 161 protein spots were specific for either T98G or U87MG (Fig. S1). Finally, PCA was performed on 236 spots shared by both cell lines (Table SIII and Fig. S1). The 2DE-PCA results were close to those obtained via LC-MS/MS, as five PCs explained the gel intensity behavior (Fig. 2A). Similarly, to LC-MS/MS, PC1 accounted for 63% of the whole explained variance, while the other four components only explained 37%. The circular biplot (Fig. 2B) exhibits the same spatial arrangement between gels (proteins visualized as spots) as for the LC-MS/MS data. A total of 165 spots strongly correlated with PC1; 51 spots (black dots), associated with T98G gels, had negative values, and 114 spots (black squares) associated with U87MG gels, had positive PC1 values. Additionally, the 2DE intensity heatmap (Fig. 2C) replicated that obtained for LC-MS/MS. As expected, protein spots that differed between cell lines (dark blue on the component scale) had a homogeneous intensity within cell lines (blocks 1 and 3 on the heatmap; Fig. 2C), with significant correlation and contribution values (light red and light green in the mean correlation scale, and white in the contribution scale in Fig. 2C and Table SIII) with PC1. Likewise, the protein spots grouped on the left in the circular biplot (Fig. 2B), had a larger abundance in T98G cells and a lower abundance in U87MG cells (block 3 on the heatmap; Fig. 2C), and those grouped on the right were more abundant in U87MG cells (block 1 on the heatmap; Fig. 2C). Additionally, the spots in the center of the circular biplot (grey triangles in Fig. 2B) had very heterogeneous intensity values within the cell lines (blocks 2 and 4 on the heatmap; Fig. 2C), as well as low correlation and contribution values (Fig. 2C and Table SIII). According to the present results, these 165 spots, and 20 specific spots for T98G and 20 specific spots for U87MG cells (randomly selected) were selected for MALDI-TOF identification.
Figure 2.
Multivariate analysis of the spot abundance from 2D gels. (A) PC analysis was performed with the spot abundance values to obtain PCs that explained the total data variation. (B) Circular biplot showing the association between proteins and samples with PC1 and PC2. Black dots (left) represent proteins with negative correlation with PC1, black squares (right) proteins with positive correlation with PC1 and grey triangles (center) proteins not correlated with PC1. (C) Protein abundance patterns (blue-red scale into heatmap) show that where PC1 proteins are present (dark blue in component scale), the protein expression values show homogeneous patterns with mean correlation values (red-green scale) and protein contribution to PC1 (white-black scale). PC, principal component.
MALDI-TOF protein identification
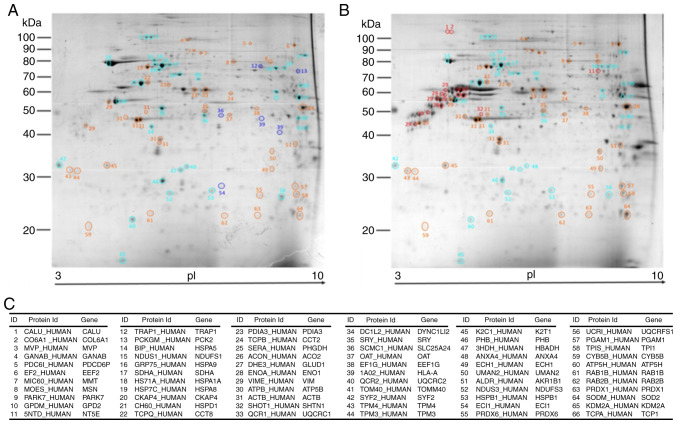
As a result of random sampling and PCA, 66 proteins exhibited a homogeneous distribution in the T98G and U87MG gels (Fig. 3A and B), which assured whole mitochondrial proteins were represented. The T98G cell line was represented by 33 proteins (4 specific and 29 upregulated), and the U87MG cell line was represented by 33 proteins (5 specific and 28 more abundant; Fig. 3).
Figure 3.
2D electrophoresis-matrix-assisted laser desorption/ionization-time of flight-identified protein distribution in T98G and U87MG gels. Mitochondria-enriched protein extracts from (A) T98G and (B) U87MG cells were analyzed via 2D PAGE; protein spots were randomly selected, and the density was measured for principal component analysis. Blue and light-blue circles represent specific and upregulated proteins, respectively, in T98G cells. Red and orange circles represent specific and upregulated proteins, respectively, in U87MG cells. (C) UniProt ID and gene symbol were provided for each of the 66 identified proteins.
PPIns from T98G and U87MG cell lines
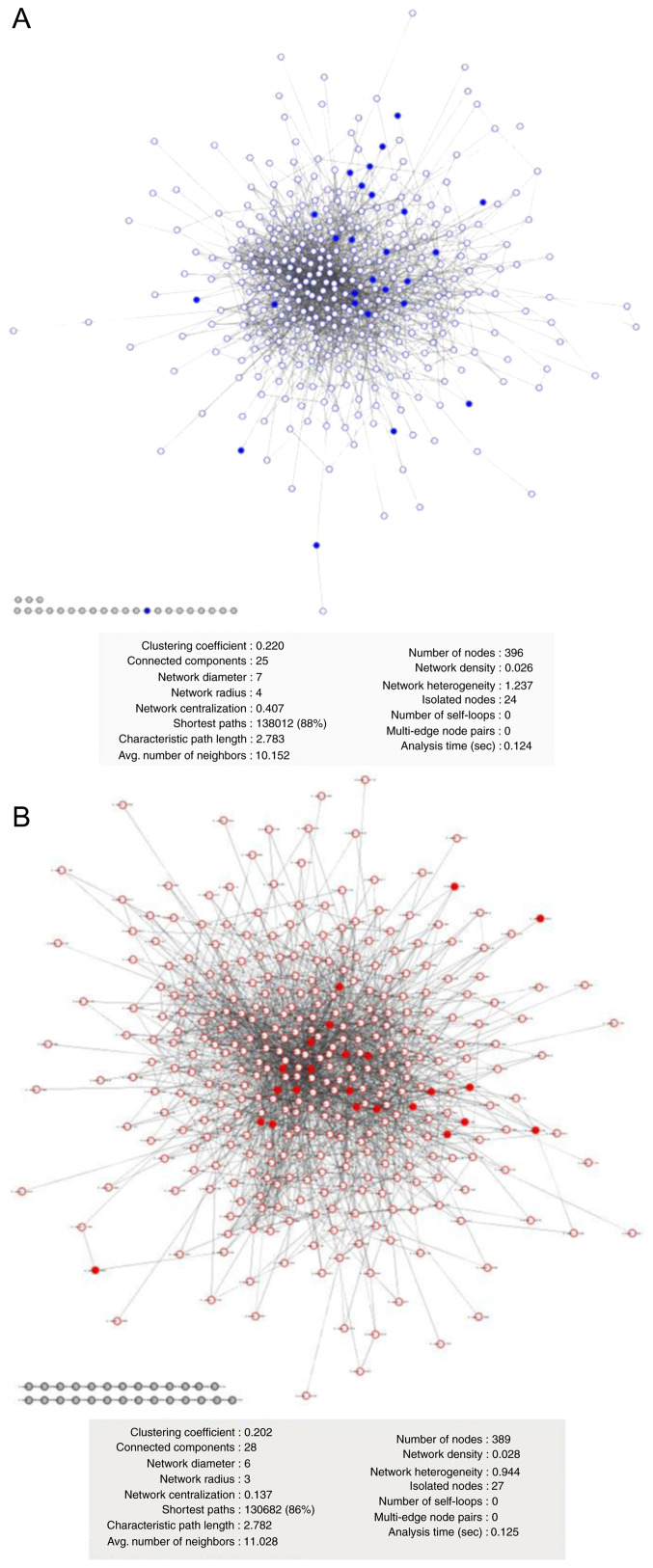
According to LC-MS/MS data analysis, a specific T98G PPIn (Fig. 4A) was built composed of 396 proteins (235 from PC1 and 161 specific). The U87MG PPIn (Fig. 4B) included 389 proteins (307 from PC1 and 82 specifics). The T98G PPIn had 24 no-interaction nodes, while U87MG had 27; their heterogeneity and connectivity were similar (Fig. 4A and B). For 2DE-identified proteins, 51 (77%) were found in PC1 from the LC-MS/MS data and mapped onto the T98G or U87MG LC-MS/MS PPIn (Fig. 4). As shown in Fig. 4, 2DE proteins were distributed throughout the T98G (solid blue dots in Fig. 4A) and U87MG (solid red dots in Fig. 4B) LC-MS/MS networks. The present results suggested that the proteins obtained from 2DE-PCA from randomly selected spots were comparable with the proteins from PCA applied to the LC-MS/MS label-free data.
Figure 4.
T98G and U87MG PPIns. PPIns were built using GeneMANIA v3.4.1 with PC1-proteins from the liquid chromatography coupled to tandem mass spectrometry data without adding more nodes. The edges represent only experimentally tested physical interactions between proteins. The solid blue and red dots represent 2D electrophoresis-matrix-assisted laser desorption/ionization-time of flight PC1 weighted proteins in (A) T98G and (B) U87MG PPIns. PPIns, protein-protein interaction networks; PC, principal component.
Overrepresentation analysis
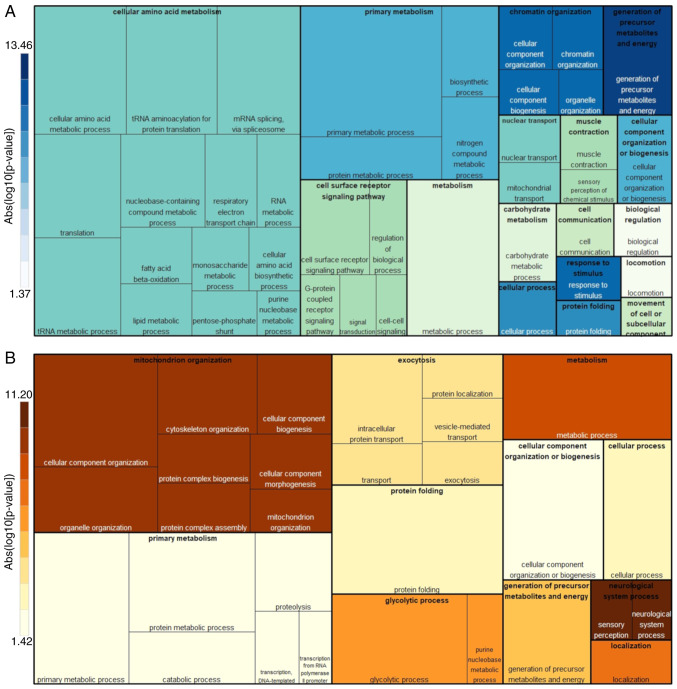
To determine whether 2DE-MALDI-TOF and LC-MS/MS data were biologically comparable, GO overrepresentation analysis was performed on the PC1 significant proteins from LC-MS/MS and 2DE. Proteins identified in T98G cells exhibited typical mitochondrial functions (Fig. 5A): ‘Generation of precursor metabolites and energy process’ was the main enriched cellular process, followed by ‘primary metabolism’ and ‘cellular amino acid metabolism’, which together involve OXPHOS (UCRI, QCR1, QCR2, NUDS1 and NUDS3 proteins identified by MALDI-TOF), ‘β-oxidation’ (ECI1), and ‘tricarboxylic acid (TCA) cycle’ (ACON, PCKGM, SDHA, DHE3, SERA and 3HIDH). Notably, other cellular processes that are less reported for mitochondrial function were also present: ‘Protein metabolic process’, ‘protein folding’ (TCPQ, TCPB and HSP7C), ‘translation’ (EF2 and EF1G) and ‘cellular component organization or biogenesis’ (TOMM40, UQCRC2 and NDUS1). The minor represented biological processes included ‘cell surface receptor signaling pathways’, ‘carbohydrate metabolism’ and ‘locomotion’. The present protein ranking provided a close picture of the mitochondrial function in T98G cells.
Figure 5.
Tree-maps of biological processes for glioblastoma cell lines. Overrepresented biological processes are shown according to liquid chromatography coupled to tandem mass spectrometry and 2D-matrix-assisted laser desorption/ionization-time of flight PC1 relevant proteins from the (A) T98G and (B) U87MG cell lines. Each tree-map groups all Gene Ontology biological processes mapped to a high hierarchical level obtained from the PANTHER analysis tool website. The box size is associated with the number of the proteins in that process, and the color intensity with the relative process representativeness (false discovery rate value).
On the other hand, U87MG protein classification in cellular processes (Fig. 5B) was different compared with the T98G results. The more notable biological processes identified were associated with cancer. One of the most obvious was associated with energy metabolism change, ‘glycolytic process’ (ENOA, PGAM1 and TPIS); however, this was ranked below mitochondrial organization issues such as ‘cytoskeleton organization’ (LMAN2, DYNC1 L12, TPM3 and TPM4) and ‘cellular component morphogenesis’ (CCT2 and CCT8). Notably, ‘exocytosis’ (ACTB, RAB1B and RAB2B) was another cancer-associated process identified in these cells, and a clear downregulation of the T98G cellular processes was evident (such as ‘primary metabolism’ and ‘cellular component organization or biogenesis’), indicating good biological congruence for the present analysis.
The aforementioned cellular processes were corroborated through protein spot identification by MALDI-TOF (Fig. 3), as the proteins associated with ‘glycolytic process’, ‘vesicle-mediated transport’, ‘protein translation and biomass’ (EF2 and EF1G) or molecular chaperones (HSP7C, TCPB and TPCQ) were upregulated. The present U87MG landscape presents mitochondria with modified cellular and metabolic functions, suggesting that mitochondria readjust their cellular processes according to different cancer states.
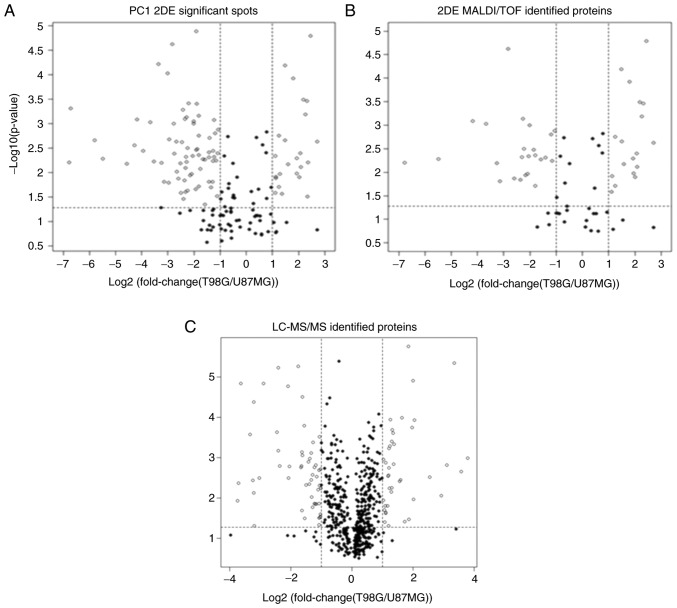
PCA versus fold-change and t-test
Through selection of random 2DE spots followed by determination of their PCA abundance, 165 spots were obtained as candidates for identification (Fig. 6A). Ultimately, the log2(fold-change) between −1 and 1 led to discard 31% of these 165 spots (black dots; Fig. 6A), and a t-test removed 30% (black dots below the horizontal dotted line; Fig. 6A). Following the log2(fold-change) and the t-test, as commonly performed to select differentially expressed spots, 43% of the spots were discarded (black dots; Fig. 6A).
Figure 6.
Comparison of the significant proteins between PCA, t-test and log2(fold-change). Random sampling and PCA show advantages over log2(fold-change) and the t-test for the description of T98G and U87MG cells. Volcano plots for (A) PC1 significant 2DE spots, (B) 2DE-MALDI-TOF and (C) LC-MS/MS significant identified proteins. Graphs show spots and identified proteins discarded by log2(fold-change) represented by black dots between −1 and 1 values of log2(fold-change) (vertical dotted lines) and/or by t-test, represented by black dots under the horizontal dotted line (P=0.05 boundary). PCA, principal component analysis; 2DE, 2D electrophoresis; MALDI-TOF, matrix-assisted laser desorption/ionization-time of flight; LC-MS/MS, liquid chromatography coupled to tandem mass spectrometry.
A similar behavior was observed for 89 MALDI-TOF- identified proteins (Fig. 6B), for which the log2(fold-change) comparison removed ~35% of the spots, while the t-test removed 30%. When considering both tests, 44% of the identified proteins could be discarded (black dots; Fig. 6B). With respect to the 543 proteins obtained from LC-MS/MS PC1, there was a high number of removed proteins (black dots; Fig. 6C): The log2(fold-change) and the t-test removed 80 and 35% of proteins, respectively. A total of 18% of proteins were significant according to the log2(fold-change) and the t-test, indicating a substantial loss of information and representativeness.
Warburg metabolism
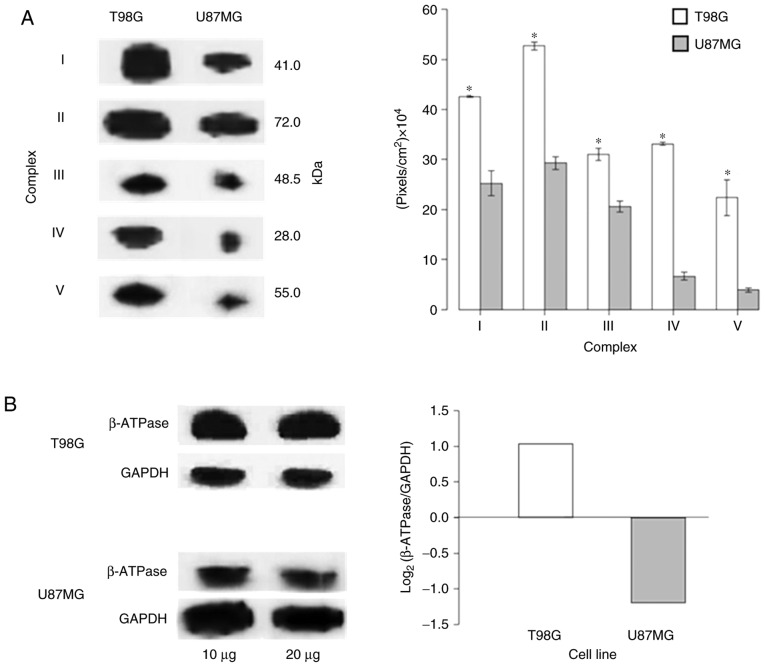
As the ‘generation of precursor metabolites and energy’ and ‘metabolism’ processes were amongst the most significantly enriched processes in the T98G (Fig. 5A) and U87MG (Fig. 5B) cell lines, respectively, the protein expression levels of OXPHOS complexes (CI–V and β-subunit ATP synthase) were verified on both cell lines via western blot analysis. The expression levels of the OXPHOS complexes in U87MG cells were significantly decreased compared with those in T98G cells (Fig. 7A and Fig. S2). Additionally, as the ‘glycolytic process’ was one of the most enhanced processes in U87MG (Fig. 5B), the bioenergetic signature (39) was investigated (Fig. 7B). The results were in accordance with Warburg's effect, as U87MG cells expressed more glycolytic proteins compared with T98G cells, where the OXPHOS system was predominant. The present results suggested that mitochondria may act as sensor organelles that change with biological states.
Figure 7.
OXPHOS and bioenergetic signature comparison between T98G and U87MG cell lines. (A) OXPHOS system is decreased in U87MG cells compared with T98G cells according to western blot analysis (mitochondrial protein, 20 µg; left panel) and mean densitometry from 3 gels (right panel); T98G cells have a well-represented OXPHOS system. The error bars represent the 95% confidence interval for the mean intensity. A loading control for these experiments is provided in Fig. S2. *P<0.05. (B) Western blot analysis from whole cell lysates (left panel) showing that GAPDH is upregulated in U87MG cells compared with β-ATPase, while in T98G cells, β-ATPase is upregulated compared with GAPDH. The bioenergetic signature log2(β-ATPase/GAPDH) (right panel) of U87MG produced a negative value, while that of T98G was positive, suggesting that T98G cells mainly maintained an OXPHOS metabolism, whereas U87MG cells maintain a glycolytic metabolism. As cells with an oxidative or glycolytic metabolism were compared, GAPDH and β-ATPase exhibited differences in abundance; thus, western blotting was performed using both 10 and 20 µg of total protein to avoid artefacts due to protein quantity. OXPHOS, oxidative phosphorylation.
Discussion
Previous studies have stated that cancer proteomics results have an unclear association with various diseases (15–17). In regard to cancer, there can be a number of reasons for this unclear association; one of these may be associated with ‘custom’ data analysis focused on protein abundance fold-changes and/or univariate hypothesis tests. However, numerous proteins exhibit multiple or moonlighting functions and are involved in different biological pathways, or as bidirectional enzymes, they are involved in synthesizing or hydrolyzing according to cell duties. Therefore, a more sensible statistical approach consistent with the biological systems under study is required.
Unlike the log2(fold-change) and t-test approach, PCA does not compare the protein mean abundance by multiple independent hypothesis tests between groups. Instead, the PCA summarizes the abundance behavior of whole proteins simultaneously in all samples to determine abundance patterns, i.e. proteins changing simultaneously in specific signaling pathways under certain conditions (40,41). On the other hand, random sampling on 2D gels is able to obtain enough representativeness to make reliable inferences (27,42).
LC and 2DE data analysis are in line with biochemical and proteomic evidence (15,16), providing an accurate description of T98G and U87MG cells with the representative biological processes according to each cell line (19,43–45). In the present study, mitochondrial proteome PCA of LC-MS/MS and 2DE random spot selection data revealed specific PPIns for each cell line in which 2D-selected and identified proteins were included in a larger and improved LC PPIn.
According to the current results, PPIns revealed that T98G protein groups belonged to well-characterized cellular processes closer to those of typical mitochondria. The most represented protein groups according to GO overrepresentation were associated with ‘generation of precursor metabolites and energy’, followed by ‘chromatin organization’, ‘primary metabolism’, ‘protein folding’, ‘cellular component organization or biogenesis’ and the major group of proteins for ‘cellular amino acid metabolism’, where OXPHOS and TCA are implicitly represented. Additionally, some less reported mitochondrial processes (‘carbohydrate metabolism’ or ‘locomotion’) or non-typical mitochondrial functions (‘cell surface receptor signaling pathway’ and ‘cell communication’) associated with cancer were identified. Therefore, PCA analysis appears to be sufficiently powerful to detect this ‘extra’ information. All the aforementioned processes were identified according to 2DE data, which demonstrated enough resolution power to build a reliable PPIn network skeleton; LC data detected the same pathways but in more detail.
U87MG cells displayed a more heterogeneous molecular landscape with more non-mitochondrial processes detected. In these cells, mitochondrial organization processes (including ‘cytoskeleton organization’, ‘cellular component morphogenesis’, ‘protein complex assembly’ and ‘protein complex biogenesis’) occupied a central role. Additionally, the canonic energy metabolic shift was clear, as ‘glycolytic process’ proteins were well represented and OXPHOS was dysregulated. Notably, other non-mitochondrial cancer-associated processes were observed, including ‘exocytosis’ (such as ‘intracellular protein transport’ and ‘vesicle-mediated transport’) and ‘protein folding’. Some of these cellular processes were also present in T98G cells but its commitment is different since in U87MG cells, i.e. the ‘generation of precursor metabolites and energy’ and ‘primary metabolism’ abundance pattern changes are represented mainly by ‘catabolic process’ or ‘proteolysis’.
PC1 of LC-MS/MS label-free quantification and 2DE spot abundance data explained 57 and 63% of the total abundance variation, respectively. Consequently, the proteins with the greatest contribution values in PC1 could be distinguished between the T98G and U87MG cell lines due to their abundance patterns. According to GO overrepresentation analysis of PC1-PPIns, the biological processes were mainly involved in ‘energy metabolism shift’ or Warburg effect, with U87MG cells representing an advanced, invasive and malignant cancer state with promiscuous interactions between the ER and nucleus, a maintained ‘chaperone response’, ‘DNA translation to proteins’ and invasion (vesicle formation, cytoskeleton proteins and proteolysis) compared with T98G cells. Conversely, T98G cells exhibited typical mitochondrial functions (‘OXPHOS’, ‘TCA cycle’ and ‘lipid metabolism’), as well as other cancer-associated processes (‘proliferation’, ‘amino acids metabolism’ or ‘chaperone response’).
PCA may reveal different cancer states or intervals of GBM, and may define processes for other types of cancer. In this way, T98G cells may represent a different cancer process or an earlier state with a molecular landscape similar to that of ‘oxidative tumors’, where ATP comes from an OXPHOS system fueled by glutamine synthesis (e.g. ‘lipid and amino acid metabolism’), as has been reported in glioblastoma (19,46). U87MG exhibited a different state, where ‘glycolytic process’ was well represented probably due to the Warburg effect, and a number of non-mitochondrial but cancer-associated proteins (16,47,48) were also present. U87MG mitochondria were associated with mobility or migration events (cytoskeleton and vesicle-associated proteins). Other less frequent processes associated with a biomass increase or metabolic energy source (DNA translation proteins and folding chaperones) were observed.
A notable GBM feature is the chaperone response, where potential biomarkers (49) or therapy targets (50) have been identified. Chaperones such as tumor necrosis factor receptor-associated protein 1 [heat shock protein (HSP)90 homologous], glucose-regulated protein (GRP)78, GRP75 and HSPB1, identified in gel spots in the present study, regulate certain mitochondrial metabolic pathways and stabilize cancer cells through apoptosis evasion (51) or can be involved in drug surveillance (44).
Non-mitochondrial protein presence is unsurprising, as mitochondrial interactions with the nucleus and the endoplasmic reticulum occur under normoxic conditions. Notably, U87MG mitochondria are more prone to these phenomena, suggesting a more promiscuous or heterogenous environment, as expected from an advanced cancer (14,52). This landscape resembles autophagy, which is a central process in advanced cancer that enables cell surveillance due to metabolite and nutrient recycling, such as amino acid generation by proteolysis, recycling (formation of metabolic precursors, RAB GTPases) and protein synthesis for fueling other pathways, such as the TCA cycle when basal or other metabolites are not available (53,54). In addition, there are proteins for amino acid and purine metabolism that could enable phagocytic structures such as phagosomes (55).
In addition to a metabolic shift, PCA can determine simultaneous cell processes, unlike other proteomic approaches based on proteins surpassing significant abundance changes (56,57). The present data analysis approach identified a specific proteomic landscape for T98G cells and another one for U87MG cells that defined concrete cell processes and temporality. This may allow the identification of targets or therapeutic tools that may result in reliable and useful information to help improve diagnosis and risk assessment.
The present data supported the hypothesis of mitochondria acting as dynamic organelles following and sensing the molecular events that take place during carcinogenesis (19,58–60). In conclusion, PCA applied to LC-MS/MS label-free quantified data was able to describe the most relevant biological processes in each cell type. Similarly, random sampling of spots and their abundance PCA from 2DE before protein identification identified proteins that exhibited the same information as LC, albeit with less resolution; with this information, a representative mitochondrial proteomic landscape was built specifically for the T98G and U87MG cell lines, in which overrepresented biological processes were highlighted with the identified mitochondrial proteins.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- GO
Gene Ontology
- LC-MS/MS
liquid chromatography coupled to tandem mass spectrometry
- m/z
mass/charge
- MALDI-TOF
matrix-assisted laser desorption/ionization- time of flight
- PC
principal component
- PCA
principal component analysis
- PPIn
protein-protein interaction network
Funding
The present study was supported by the Mexican Social Security Institute (IMSS) and National Autonomous University of Mexico (UNAM), by a grant from CONACyT (grant no. 220790), by a fellowship from the National Council of Science and Technology (grant no. 375815) and by General Directorate of Academic Staff Affairs-Support Program for Research and Technological Innovation Projects (grant nos. IN-207519 and IN-213216).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding authors on reasonable request.
Authors' contributions
LGC, FMS and SEG conceptualized the research. LGC, FMS, SEG, AJOL, HRV and AGMB conceived and designed the experiments. LGC, AJOL and AGMB performed the experiments. LGC analyzed the data. FMS, SEG and HRV contributed to the design of the study, acquisition of resources and equipment, and discussion of alternative experimental approaches. LGC and FMS wrote the paper. LGC, FMS and SEG revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Luna B, Bhatia S, Yoo C, Felty Q, Sandberg DI, Duchowny M, Khatib Z, Miller I, Ragheb J, Prasanna J, Roy D. Proteomic and mitochondrial genomic analyses of pediatric brain tumors. Mol Neurobiol. 2015;52:1341–1363. doi: 10.1007/s12035-014-8930-3. [DOI] [PubMed] [Google Scholar]
- 2.Pooladi M, Rezaei-Tavirani M, Hashemi M, Hesami-Tackallou S, Khaghani-Razi-Abad S, Moradi A, Zali AR, Mousavi M, Firozi-Dalvand L, Rakhshan A, Zamanian Azodi M. Cluster and Principal Component Analysis of Human Glioblastoma Multiforme (GBM) Tumor Proteome. Iran J cancer Prev. 2014;7:87–95. [PMC free article] [PubMed] [Google Scholar]
- 3.Fangusaro J. Pediatric high grade glioma: A review and update on tumor clinical characteristics and biology. Front Oncol. 2012;2:105. doi: 10.3389/fonc.2012.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batash R, Asna N, Schaffer P, Francis N, Schaffer M. Glioblastoma multiforme, diagnosis and treatment; recent literature review. Curr Med Chem. 2017;24:3002–3009. doi: 10.2174/0929867324666170516123206. [DOI] [PubMed] [Google Scholar]
- 5.Kafka A, Tomas D, Lechpammer M, Gabud T, Pažanin L, Pećina-Šlaus N. Expression Levels and Localizations of DVL3 and sFRP3 in Glioblastoma. Dis Markers. 2017;2017 doi: 10.1155/2017/9253495. 9253495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagberg H, Mallard C, Rousset CI, Thornton C. Mitochondria: Hub of injury responses in the developing brain. Lancet Neurol. 2014;13:217–232. doi: 10.1016/S1474-4422(13)70261-8. [DOI] [PubMed] [Google Scholar]
- 7.Raefsky SM, Mattson MP. Adaptive responses of neuronal mitochondria to bioenergetic challenges: Roles in neuroplasticity and disease resistance. Free Radic Biol. 2017;102:203–216. doi: 10.1016/j.freeradbiomed.2016.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Son G, Han J. Roles of mitochondria in neuronal development. BMB Rep. 2018;51:549–556. doi: 10.5483/BMBRep.2018.51.11.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Hallmarks of cancer. The next generation. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Floor SL, Dumont JE, Maenhaut C, Raspe E. Hallmarks of cancer: Of all cancer cells, all the time? Trends Mol Med. 2012;18:509–515. doi: 10.1016/j.molmed.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Czarnecka AM, Czarnecki JS, Kukwa W, Cappello F, Ścińska A, Kukwa A. Molecular oncology focus-is carcinogenesis a ‘mitochondriopathy’? J Biomed Sci. 2010;17:31. doi: 10.1186/1423-0127-17-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kroemer G, Pouyssegur J. Tumor cell metabolism: Cancer's Achilles' heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Galluzzi L, Morselli E, Kepp O, Vitale I, Rigoni A, Vacchelli E, Michaud M, Zischka H, Castedo M, Kroemer G. Mitochondrial gateways to cancer. Mol Aspects Med. 2010;31:1–20. doi: 10.1016/j.mam.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Vyas S, Zaganjor E, Haigis MC. Mitochondria and Cancer. Cell. 2016;166:555–566. doi: 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrak J, Ivanek R, Toman O, Cmejla R, Cmejlova J, Vyoral D, Zivny J, Vulpe CD. Déjà vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics. 2008;8:1744–1749. doi: 10.1002/pmic.200700919. [DOI] [PubMed] [Google Scholar]
- 16.Deighton RF, McGregor R, Kemp J, McCulloch J, Whittle IR. Glioma pathophysiology: Insights emerging from proteomics. Brain Pathol. 2010;20:691–703. doi: 10.1111/j.1750-3639.2010.00376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valledor L, Jorrín J. Back to the basics: Maximizing the information obtained by quantitative two dimensional gel electrophoresis analyses by an appropriate experimental design and statistical analyses. J Proteomics. 2011;74:1–18. doi: 10.1016/j.jprot.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Meehan MC. General system theory: Foundations, development, applications. JAMA. 1969;208((5)):8701969. [Google Scholar]
- 19.Obre E, Rossignol R. Emerging concepts in bioenergetics and cancer research: Metabolic flexibility, coupling, symbiosis, switch, oxidative tumors, metabolic remodeling, signaling and bioenergetic therapy. Int J Biochem Cell Biol. 2015;59:167–181. doi: 10.1016/j.biocel.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 21.Stekhoven DJ, Buhlmann P. MissForest-non-parametric missing value imputation for mixed-type data. Bioinformatics. 2012;28:112–118. doi: 10.1093/bioinformatics/btr597. [DOI] [PubMed] [Google Scholar]
- 22.Lê S, Josse J, Husson F. FactoMineR: An R package for multivariate analysis. J Stat Softw. 2008:1–18. doi.org/10.18637/jss.v025.i01 PMID: 18676020. [Google Scholar]
- 23.Abdi H, Williams LJ. Principal component analysis. Wiley Interdiscip Rev Comput Stat. 2010;2:433–459. doi: 10.1002/wics.101. [DOI] [Google Scholar]
- 24.Hurkman WJ, Tanaka CK. Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol. 1986;81:802–806. doi: 10.1104/pp.81.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salazar E, Díaz-Mejía JJ, Moreno-Hagelsieb G, Martínez- Batallar G, Mora Y, Mora J, Encarnación S. Characterization of the Nif A-RpoN regulon in rhizobium etli in free life and in symbiosis with phaseolus vulgaris. Appl Environ Microbiol. 2010;76:4510–4520. doi: 10.1128/AEM.02007-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mead R, Curnow RN, Hasted AM, editors. 3rd. Chapman and Hall/CRC Press; New York, NY: 2003. Statistical Methods in Agriculture and Experimental Biology; p. 472. [Google Scholar]
- 27.Cochran WG, editor. 3rd. Wiley; New Jersey: 1977. Sampling Techniques; p. 428. [Google Scholar]
- 28.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 29.The UniProt Consortium. UniProt: The universal protein knowledge base. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, Morris Q, Bader GD. GeneMANIA Cytoscape plugin: Fast gene function predictions on the desktop. Bioinformatics. 2010;26:2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashkenazi M, Bader GD, Kuchinsky A, Moshelion M, States DJ. Cytoscape ESP: Simple search of complex biological networks. Bioinformatics. 2008;24:1465–1466. doi: 10.1093/bioinformatics/btn208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Supek F, Bošnjak M, Škunca N, Šmuc T. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R Core Team, corp-author. Vienna, Austria: 2017. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 36.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 39.Cuezva JM, Krajewska M, de Heredia ML, Krajewski S, Santamaría G, Kim H, Zapata JM, Marusawa H, Chamorro M, Reed JC. The Bioenergetic Signature of Cancer: A Marker of Tumor Progression. Cancer Res. 2002;62:6674–6681. [PubMed] [Google Scholar]
- 40.Hair JF, Anderson RE, Tatham RL, Black WC. Multivariate Data Analysis. Int J Pharmaceutics. 1998;816 21335075. [Google Scholar]
- 41.Everitt B, Hothorn T, editors. Springer-Verlag; New York, NY: 2011. An Introduction to Applied Multivariate Analysis with R. [DOI] [Google Scholar]
- 42.Heinisch O. An introduction to sampling theory with application to agriculture. Biom Z. 2017;5:212. doi: 10.1002/bimj.19630050309. [DOI] [Google Scholar]
- 43.Cuezva JM, Ortega AD, Willers I, Sánchez-Cenizo L, Aldea M, Sánchez-Aragó M. The tumor suppressor function of mitochondria: Translation into the clinics. Biochim Biophys Acta. 2009;1792:1145–58. doi: 10.1016/j.bbadis.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Siegelin MD, Plescia J, Raskett CM, Gilbert CA, Ross AH, Altieri DC. Global targeting of subcellular heat shock protein-90 networks for therapy of glioblastoma. Mol Cancer Ther. 2010;9:1638–1646. doi: 10.1158/1535-7163.MCT-10-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolf DA. Is reliance on mitochondrial respiration a ‘chink in the armor’ of therapy-resistant cancer? Cancer Cell. 2014;26:788–795. doi: 10.1016/j.ccell.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jose C, Bellance N, Rossignol R. Choosing between glycolysis and oxidative phosphorylation: A tumor's dilemma? Biochim Biophys Acta. 2011;1807:552–561. doi: 10.1016/j.bbabio.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 47.Pooladi M, Abad SK, Hashemi M. Proteomics analysis of human brain glial cell proteome by 2D gel. Indian J Cancer. 2014;51:159–162. doi: 10.4103/0019-509X.138271. [DOI] [PubMed] [Google Scholar]
- 48.Ramão A, Gimenez M, Laure HJ, Izumi C, Vida RC, Oba-Shinjo S, Marie SK, Rosa JC. Changes in the expression of proteins associated with aerobic glycolysis and cell migration are involved in tumorigenic ability of two glioma cell lines. Proteome Sci. 2012;10:53. doi: 10.1186/1477-5956-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banerjee HN, Mahaffey K, Riddick E, Banerjee A, Bhowmik N, Patra M. Search for a diagnostic/prognostic biomarker for the brain cancer glioblastoma multiforme by 2D-DIGE-MS technique. Mol Cell Biochem. 2012;367:59–63. doi: 10.1007/s11010-012-1319-6. [DOI] [PubMed] [Google Scholar]
- 50.Karpel-Massler G, Ishida CT, Bianchetti E, Shu C, Perez-Lorenzo R, Horst B, Banu M, Roth KA, Bruce JN, Canoll P, Altieri DC, Siegelin MD. Inhibition of mitochondrial matrix chaperones and antiapoptotic bcl-2 family proteins empower antitumor therapeutic responses. Cancer Res. 2017;77:3513–3526. doi: 10.1158/0008-5472.CAN-16-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chae JI, Jeon YJ, Shim JH. Downregulation of Sp1 is involved in honokiol-induced cell cycle arrest and apoptosis in human malignant pleural mesothelioma cells. Oncol Rep. 2013;29:2318–2324. doi: 10.3892/or.2013.2353. [DOI] [PubMed] [Google Scholar]
- 52.Porporato PE, Filigheddu N, Pedro JMB, Kroemer G, Galluzzi L. Mitochondrial metabolism and cancer. Cell Res. 2018;28:265–280. doi: 10.1038/cr.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giatromanolaki A, Sivridis E, Mitrakas A, Kalamida D, Zois CE, Haider S, Piperidou C, Pappa A, Gatter KC, Harris AL, Koukourakis MI. Autophagy and lysosomal related protein expression patterns in human glioblastoma. Cancer Biol Ther. 2014;15:1468–1478. doi: 10.4161/15384047.2014.955719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sica V, Galluzzi L, Bravo-San Pedro JM, Izzo V, Maiuri MC, Kroemer G. Organelle-Specific Initiation of Autophagy. Mol Cell. 2015;59:522–539. doi: 10.1016/j.molcel.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 56.Gautam P, Nair SC, Gupta MK, Sharma R, Polisetty RV, Uppin MS, Sundaram C, Puligopu AK, Ankathi P, Purohit AK, et al. Proteins with altered levels in plasma from glioblastoma patients as revealed by iTRAQ-based quantitative proteomic analysis. PLoS One. 2012;7:e46153. doi: 10.1371/journal.pone.0046153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Locasale JW, Melman T, Song S, Yang X, Swanson KD, Cantley LC, Wong ET, Asara JM. Metabolomics of human cerebrospinal fluid identifies signatures of malignant glioma. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014688. M111.014688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iwadate Y, Sakaida T, Hiwasa T, Nagai Y, Ishikura H, Takiguchi M, Yamaura A. Molecular classification and survival prediction in human gliomas based on proteome analysis. Cancer Res. 2004;64:2496–2501. doi: 10.1158/0008-5472.CAN-03-1254. [DOI] [PubMed] [Google Scholar]
- 59.Ordys BB, Launay S, Deighton RF, McCulloch J, Whittle IR. The role of mitochondria in glioma pathophysiology. Mol Neurobiol. 2010;42:64–75. doi: 10.1007/s12035-010-8133-5. [DOI] [PubMed] [Google Scholar]
- 60.Collet B, Guitton N, Saïkali S, Avril T, Pineau C, Hamlat A, Mosser J, Quillien V. Differential analysis of glioblastoma multiforme proteome by a 2D-DIGE approach. Proteome Sci. 2011;9:16. doi: 10.1186/1477-5956-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding authors on reasonable request.