Abstract
Glioblastoma is a highly malignant tumor that contains stem-like cells known as glioma stem cells (GSCs), which lare associated with an increased risk of glioma occurrence, recurrence and poor prognosis. Circadian clock gene, period circadian clock 2 (PER2) expression has been revealed to be inhibited in various types of cancer. However, the precise role and potential mechanisms of PER2 in GSCs remains unclear. The present study demonstrated that PER2 mRNA and protein expression was downregulated in GSCs compared with non-stem glioma cells, which indicated that PER2 could be involved in the malignant process of glioma. Furthermore, functional studies revealed that PER2 overexpression could induce GSC arrest at the G0/G1 phase and suppress their proliferation, stemness and invasion ability in vitro and in vivo. Subsequently, the Wnt/β-catenin signaling pathway was identified as the target of PER2 in GSCs. These results indicated that PER2 plays a critical role in regulating the stemness of GSCs and provides a novel therapeutic target to overcome the effects of GSCs.
Keywords: PER2, glioma stem cells, stemness, Wnt/β-catenin
Introduction
Gliomas are the most common tumor type of the adult central nervous system (1). Although some progress has been made in recent years, regarding surgical resection and a number of adjuvant therapies, such as combined radiotherapy, chemotherapy and targeted therapy, the treatment of glioblastoma (GBM) is still unsatisfactory due to the high recurrence rate of glioma in patients and a median survival time of only 14 months (2–3).
Increasing evidence has revealed that some tumor-initiating cells with stem cell characteristics in glioma, also called glioma stem cells (GSCs), contribute to the high rates of therapeutic resistance and rapid recurrence (4–5). GSCs have self-renewal properties, high proliferation rates and high invasive growth characteristics, leading to increased resistance to radiotherapy and chemotherapy compared with non-stem glioma cells (NGSCs) (6–8). Consequently, GSCs are one of the main reasons of GBM treatment failure, and the treatment of GSCs has become the focus of current malignant glioma therapeutic strategies.
A previous study has shown that altered circadian regulation in cancer stem cells may contribute to disease progression (9). Endogenous circadian rhythms are maintained by two feedback loops. Clock circadian regulator (CLOCK) and brain and muscle Arnt-like protein-1 (BMAL1) form heterodimers to rhythmically activate the expression of transcriptional repressor proteins, such as period 1/2/3 and cryptochrome 1/2, which form a complex to inhibit CLOCK and BMAL1 transcriptional activity (9–11). In some types of cancer and model systems, period circadian clock 2 (PER2) serves as a tumor suppressor gene. Recent systems analysis revealed that changes in PER2 expression were correlated with lung cancer, gastric carcinoma, liver cancer, colon cancer, and head and neck squamous cell carcinomas (12–19). Our previous study has also revealed that PER1 and PER2 are significantly downregulated in glioma tissue compared with normal brain tissue and their expression is correlated with World Health Organization grading of glioma (20).
However, whether PER2 and PER1 play a critical role in GSCs and the circadian properties of human GSCs remains unclear. Based on the findings of previous studies, the present study investigated the potential role of PER2 on stemness, tumorigenesis, cell growth, cell cycle distribution, migration and invasion of GSCs in glioma. The present results revealed a novel mechanism of action underlying the role of PER2 in GSCs, as well as a new target to prevent glioma progression.
Materials and methods
Cell culture and patient samples
Normal human astrocytes (cat. no. 1800; http://www.sciencellonline.com/human-astrocytes.html) were obtained from ScienCell Research Laboratories, Inc., and last authenticated by the ScienCell in 2018. The human U87 (GBM of unknown origin) and U251 glioma cells were obtained from The Cell Bank Type Culture Collection of the Chinese Academy of Sciences and last authenticated with short tandem repeat (STR) profiling by an independent third-party company in 2019. All cells were grown in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a humidified atmosphere of 5% CO2. The glioma stem-like cell lines U251s and U87s were isolated from U251 and U87 cells using a serum-free clone formation method. Glioma cells were dispersed and resuspended in DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.) serum-free medium (SFM), B27 (20 mg/ml; Invitrogen; Thermo Fisher Scientific, Inc.), 20 ng/ml basic fibroblast growth factor (Sigma-Aldrich; Merck KGaA), 20 ng/ml epidermal growth factor (PeproTech, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.). All the cells used for analysis were within passage 10, generally in passages 3–7. To generate a PER2 overexpression model, the U87s and U251s cells were infected with lentiviruses packaged in pGMLV-PE1-PER2.
The present study consisted of 48 glioma tissue specimens, which were resected at the Department of Neurosurgery of the General Hospital of Ningxia Medical University between January 2017 and July 2019. None of the patients (17 female and 31 male patients; aged 12 to 73 years old) received preoperative treatment, such as irradiation or chemotherapy. All patients provided signed informed consent and the study was approved by the Ethics Committee of Ningxia Medical University. All research protocols for the present study were approved by the Ethics Committee of Ningxia Medical University.
Western blot analysis of protein expression
Cells were lysed in RIPA lysis buffer (Thermo Fisher Scientific, Inc.) and then centrifuged at 14,000 × g for 15 min at 4°C. Protein concentrations were determined using the BCA Protein assay (Gen5 software; Bio-Tek Instruments, Inc.). The supernatant was collected and denatured. Proteins (30 µg/lane) were separated via SDS-PAGE on a 10% gel and transferred onto a polyvinylidene difluoride (PVDF) membrane. The PVDF membrane was treated with TBS with Tween-20 containing 50 g/l skim milk at room temperature for 4 h, followed by incubation overnight at 4°C with the following primary antibodies: PER2 (product code ab94915; dilution 1:1,000; Abcam), CD133 (cat. no. NB120-16518; dilution 1:500; Bio-Techne), SOX2 (product code ab79351; dilution 1:1,000), NESTIN (product code ab6320; dilution 1:500), β-catenin (product code ab16051; dilution 1:500), Wnt7b (product code ab94915; dilution 1:200), matrix metallopeptidase (MMP)2 (product code ab97779; dilution 1:1,000), MMP9 (product code ab38898; dilution 1:2,000), c-Myc (product code ab32072; dilution 1:1,000), cyclin D1 (product code ab16663; dilution 1:200; all from Abcam) and GAPDH (cat. no. 10494-1-AP; dilution 1:2,000, ProteinTech Group, Inc.). Goat anti-rabbit IgG H&L IRDye® 800CW (product code ab216773; dilution 1:5,000) and goat anti-mouse IgG H&L IRDye® 680RD (product code ab216776; dilution 1:5,000; both from Abcam) were used as secondary antibodies incubated with the membrane for 2 h at room temperature. The western blot images were visualized on a LI-COR Odyssey Fc imaging system (LI-COR Biosciences).
RNA isolation and reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the standard protocols. cDNA was reverse transcribed from total RNA using a PrimeScript RT Master Mix Perfect Real-Time kit (Takara Bio, Inc.). mRNA expression was analyzed using SYBR Green PCR Master mix (Takara Biotechnology Co., Ltd.) and a 7300 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling program was as follows: 95°C for 1 min; (95°C for 15 sec; 60°C for 30 sec; 72°C for 30 sec) ×40 cycles. The primer sequences used were as follows: BMAL1 forward, 5′-CTCCTCCAATGTGGGCATCAA-3′, BMAL1 reverse, 5′-GGTGGCACCTCTTAATGTTTTCA-3′; CLOCK forward, 5′-TGCGAGGAACAATAGACCCAA-3′ and reverse, 5′-ATGGCCTATGTGTGCGTTGTA-3′; TIMELESS forward, 5′-TCTGATCCGCTATTTGAGGCA-3′ and reverse, 5′-GGCAGAAGGTCGCTCTGTAG-3′; NPAS2 forward, 5′-CGTGTTGGAAAAGGTCATCGG-3′ and reverse, 5′-TCCAGTCTTGCTGAATGTCAC-3′; Cry1 forward, 5′-CTCCTCCAATGTGGGCATCAA-3′ and reverse, 5′-CCACGAATCACAAACAGACGG-3′; Cry2 forward, 5′-TCCCAAGGCTGTTCAAGGAAT-3′ and reverse, 5′-TGCATCCCGTTCTTTCCCAAA-3′; PER1 forward, 5′-AGTCCGTCTTCTGCCGTATCA-3′ and reverse, 5′-AGCTTCGTAACCCGAATGGAT-3′; PER2 forward, 5′-GTGAAAGTGAGGAGAAAGGCAACC-3′ and reverse, 5′-CACCTCTTCCGAGCACCGTC-3′; PER3 forward, 5′-GCAGAGGAAATTGGCGGACA-3′ and reverse, 5′-GGTTTATTGCGTCTCTCCGAG-3′; and GAPDH forward, 5′-GGCTAGCACCAACCAAAAGTATAGTCATCCGG-3′ and reverse, 5′-GGAATTCGTCTTAAAATTCAATTTATTATAAA-3′. Fold changes in mRNA expression were quantified with the 2−∆∆Cq relative quantification method using GAPDH as a housekeeping control gene (21).
Neurosphere formation assay
GSCs were plated at 1,000 cells/well in 96-well plates. After culture for 7 days, the number of neurospheres that contained >20 cells was determined in each well. Wells containing no cells or >1 cell were excluded, and those with 1 cell were marked and monitored daily under a microscope. The number of tumor spheres per well was recorded, and the tumor sphere formation rate was calculated.
Cell proliferation assay and GSC flow cytometric assay
Cell proliferation was measured using a CCK-8 assay (Dojindo Molecular Technologies., Inc.) according to the manufacturer's protocol. The absorbance was then measured at 450 nm using a microplate reader (Tecan Group, Ltd.). Three independent experiments were carried out, and the proliferation fold was calculated as follows: (OD value at 24, 48 or 96 h-OD value at 0 h)/(OD value at 0 h).
To study the cell cycle, when GSCs reached ~70% confluency, they were detached with trypsin after washing twice with phosphate-buffered saline (PBS), centrifuged at 1,000 × g for 5 min, and then fixed with cold 70% alcohol and frozen at −20°C. In addition, cell cycle analysis was performed by standard PI staining (BD Biosciences). Briefly, cells were fixed, stained with PI solution for 30 min at 37°C, spun down and resuspended in PBS. After staining, the samples (5×104 cells/200 µl) were analyzed using a BD FACSCalibur™ Flow Cytometer (BD Biosciences). The data were analyzed with a BD FACS Diva (version 7.0; BD Biosciences).
Immunofluorescence and immunohistochemical staining
GSCs were fixed with 4% paraformaldehyde at room temperature, and PBS was used to wash the cells. Triton X-100 (0.1%) was used to permeabilize the cells, following which they were blocked with 5% BSA. Cells were incubated with the primary antibody for 20 h at 4°C (stem marker antibody concentrations were according to the manufacturer's instructions). Next, the samples were washed three times with PBS and incubated with fluorescent secondary antibodies at 37°C for 1 h in the dark: Goat anti-mouse IgG H&L (product code ab150117; dilution 1:500), goat anti-mouse IgG H&L (product code ab150115; dilution 1:1,000) and goat anti-rabbit IgG H&L (product code ab150079; dilution 1:1,000; all from Abcam). Then, the samples were washed three times with PBS, and DAPI was used to counterstain the cells at room temperature for 10 min. Finally, a fluorescence microscope (Leica Microsystems GmbH) at ×40 magnification was used to examine the cells.
For immunohistochemical (IHC) staining, brain tumor sections were incubated with the following primary antibodies: PER2 (product code ab94915; 1:500), Ki-67 (product code ab15580; 1:100; both from Abcam) and CD133 (cat. no. NB120-16518; 1:500, Bio-Techne), for 1 h at room temperature after deparaffinization, rehydration, antigen retrieval, endogenous peroxidase quenching and blocking. The sections were incubated with HRP-conjugated polymer (Dako; Agilent Technologies, Inc.) for 40 min and then with diaminobenzidine using an Ultravision DAB Plus Substrate Detection System (Thermo Fisher Scientific, Inc.) for 1–10 min at room temperature, followed by hematoxylin staining at room temperature for 2 min. Finally, a Leica DM6 light microscope (Leica Microsystems GmbH) at magnification ×10 and ×20 was used to examine the sections.
Migration and invasion assays
GSCs (1×105) in 200 µl of SFM were seeded into the top chamber of cell culture inserts coated with or without Matrigel (BD Biosciences), while the lower chamber was filled with 400 µl medium containing 8% FBS as a chemoattractant. After incubation at 37°C for 24 h, the cells that passed through the membrane were fixed in 100% methanol for 20 min at room temperature and stained with 0.1% crystal violet solution for 10 min at room temperature. Three independent fields of cells adhered to the lower side of the Transwell filters were photographed by an inverted light microscope (Nikon Corporation) at ×20 magnification for each well.
RNA-Sequencing (RNA-Seq)
Total RNA from U251s-control and U251s-PER2 overexpression cells was extracted using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) and treated with DEPC water. RNA-Seq was performed by Allwegene Technology Co., Ltd. (Beijing, China). The reference genome was from http://www.ensembl.org/Homo_sapiens/Info/Index. The data were analyzed on the online Majorbio I-Sanger Cloud Platform (www.i-sanger.com). The differential expression analysis was conducted with DEGseq 1.30.0 software. Genes with adjusted P-values (P-adjust) ≤0.05 were considered differentially expressed genes (DEGs). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) analyses of DEGs were analyzed with R package software (P<0.05).
Intracranial tumor formation in vivo
For intracranial implantation, U251 GSCs (1×105) transfected with lentiviral vectors expressing GFP-PER2 or GFP-control were implanted into the right frontal lobes of 6–8-week-old male BALB/c athymic nude mice (a total of 4 mice/group, Experimental Animals Center of Ningxia Medical University, Yinchuan, China) after anesthesia (1.5–2.5% isoflurane/oxygen inhalation for anesthetic maintenance). After 4, 10 and 15 days, a Xenogen in vivo imaging system (IVIS) was used to visualize the tumors, and photon measurement was defined around the tumor area and quantified using Living Image software (Caliper Life Science SA). The nude mice were raised under specific pathogen-free conditions in a temperature-controlled room (22±2°C; humidity, 50±10%) with a 12-h light/dark cycle, and with ad libitum access to sterilized water and mice food. The health and behavior of mice were monitored daily. After 2 weeks, tumor-bearing nude mice exhibited clinical signs of a brain tumor, such as spine curvature or dyskinesia, and thus the mice were sacrificed with transcardial perfusion under anesthesia (1.5–2.5% isoflurane/oxygen inhalation) on the 15th day after the implantation. The death of mice was verified by assessing cardiac arrest. Brain tumors in the mice were removed after transcardial perfusion with normal saline and then 4% paraformaldehyde (PFA) and were fixed in formalin or post-fixed in 4% PFA overnight at 4°C for OCT frozen tissue blocks. All mice included in the present study exhibited a single tumor, the maximum level of cachexia observed was a body weight loss <5%. The maximum tumor diameter was 6 mm, the volume was 78.45 mm3 (the tumor volume was calculated according to the formula: Tv=π/6 × tumor length × tumor width2) and the wet weight was 0.15 g, as <1% of total mice body weight (Fig. S1A). All mice experiments were performed with the approval of the Ningxia Medical University Experimental Animals Center IACUC.
Statistical analysis
The SPSS 20.0 software (IBM Corp.) was used for statistical analysis. All experiments were performed independently in triplicate, and all values were expressed as the mean ± standard deviation. The data were analyzed using two-tailed Student's t-test (for two-group comparisons) or one-way analysis of variance (ANOVA) with Tukey's post hoc test (for multiple comparisons). P<0.05 and P<0.01 were considered to indicate statistically significant differences.
Results
PER2 expression is downregulated in GSCs
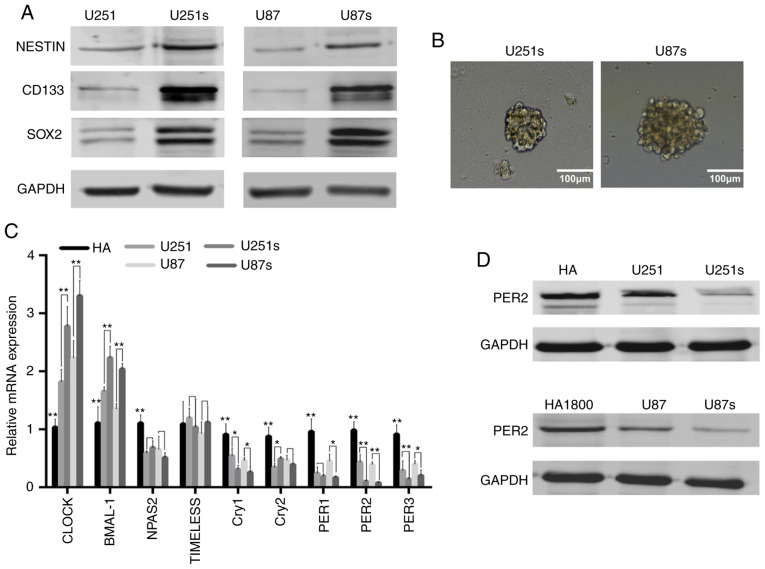
Based on previous study, GSCs could be enriched by sphere formation (22). Preliminary analysis of western blot data indicated that, as anticipated, the expression of critical GSC markers, such as CD133, NESTIN and SOX2, were markedly upregulated in U251 and U87 sphere-forming cells (Fig. 1A). The representative images of these GSCs are presented in Fig. 1B. These data confirmed that GSCs were enriched in U251 and U87 sphere-forming cells. Next, it was investigated whether circadian genes were ectopically expressed in GSCs. RT-qPCR and western blotting were performed to identify the mediator of GSCs stemness, it was demonstrated that PER2 was one of the most significantly downregulated core circadian genes in GSCs compared with NGSCs or human astrocyte cell line (Fig. 1C and D). These results indicated that PER2 expression was associated with GSCs, indicating that PER2 may be involved in the malignant process of glioma and the function of GSCs.
Figure 1.
PER2 expression is downregulated in GSCs. (A) Protein expression levels of NESTIN, CD133 and SOX2 increased in glioma sphere-forming cells line U251s and U87s. (B) Representative images of primary GSCs. (C) Relative mRNA expression of core circadian genes in normal human astrocyte cell, GSCs and matched non-GSCs glioma cell lines. (D) PER2 protein expression in those cell lines. One-way ANOVA with Tukey's post hoc test was used for statistical analysis of the results of B. *P<0.05, **P<0.01 vs. the control. PER2, period 2; GSCs, glioma stem cells.
PER2 overexpression reduces the stemness and self-renewal of GSCs
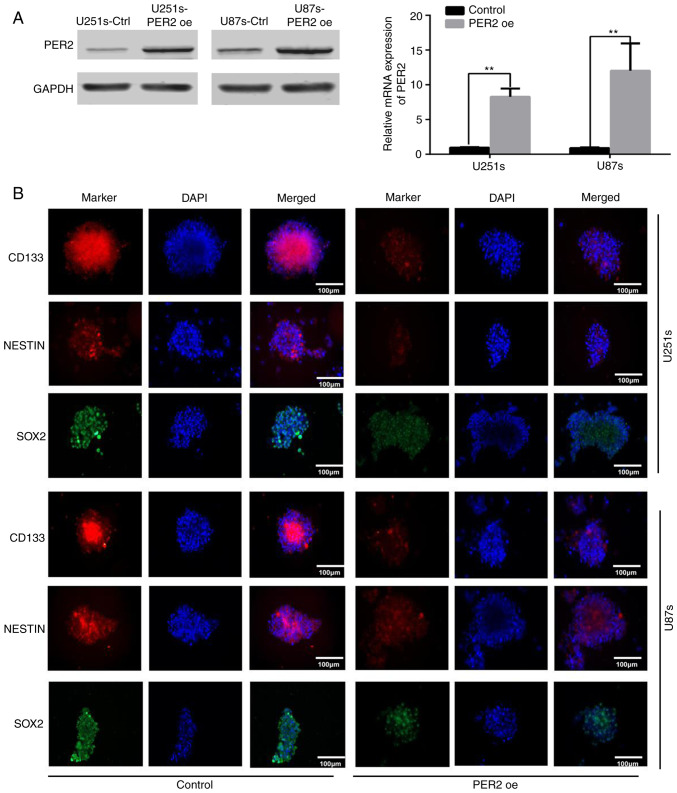
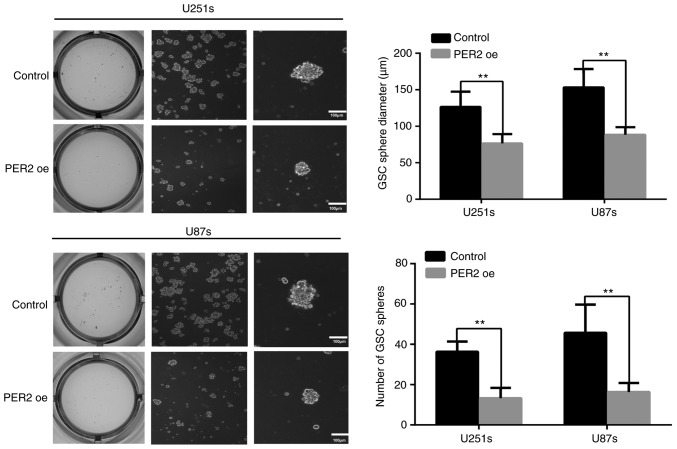
Maintenance of pluripotency is crucial for the proliferation and survival of GSCs during cancer development (4,7). To explore the functional significance of PER2 in inhibiting GSC stemness, GSCs were infected with lentivirus-PER2. Western blotting and qRT-PCR data indicated that, as anticipated, PER2 in these cell lines was overexpressed upon lentivirus-PER2 transfection (Fig. 2A). Following overexpression of PER2, the expression of stemness markers, such as CD133, NESTIN, and SOX2, was decreased in GSCs according to immunofluorescence (Fig. 2B) and western blot analysis (Fig. S1B). Furthermore, neurosphere formation revealed that PER2 could inhibit the self-renewal ability of GSCs. Both the neurosphere diameter and the number of GSC spheres were reduced after treatment with lentivirus-PER2 (Fig. 3). These results indicated that PER2 could inhibit the stemness and self-renewal capability of GSCs.
Figure 2.
PER2 overexpression reduces stemness and self-renewal of GSCs. (A) Overexpression of PER2 was observed in U251s and U87s transfected with lentivirus packaged pGMLV-per2 vector. (B) Representative immunofluorescence staining images of CD133 (red), NESTIN (red) and SOX2 (green) in GSCs; nuclei were counterstained with DAPI (blue) as a control, PER2 inhibited the expression of CD133, NESTIN and SOX2 in GSCs. Two-tailed Student's t-test for statistical analysis of the results of A. **P<0.01 vs. the control. PER2, period 2; GSCs, glioma stem cells.
Figure 3.
Self-renewal ability of GSCs was analyzed by tumor sphere formation assay. Diameters and number of PER2-overexpressing GSCs were both lower compared with the control group. **P<0.01 vs. the control (two-tailed Student's t-test). PER2, period 2; GSCs, glioma stem cells.
PER2 inhibits viability, cell cycle progression, migration, and invasion in GSCs
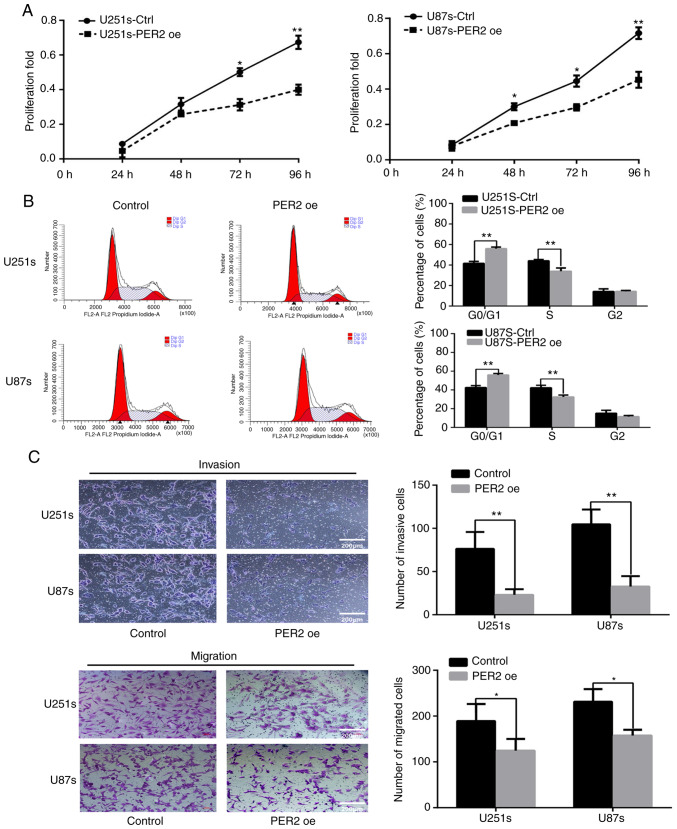
To investigate the role of PER2 in proliferation, CCK-8 assays were performed at different time-points (24, 48, 72 and 96 h) after transfection with an empty lentiviral vector (control) or lentiviral-PER2. The proliferation of GSCs was significantly inhibited by PER2 overexpression (Fig. 4A). Then, flow cytometry was performed to explore the function of PER2 in the cell cycle. The results indicated that the percentage of S-phase cells was significantly decreased after PER2 overexpression, and G0/G1 cycle arrest was observed in this group of cells (Fig. 4B). Previous research has suggested that cancer stem cells may possess higher invasive activity than differentiated cancer cells (8). Therefore, the effects of PER2 on the invasion and migration abilities of GCSs were detected. Transwell chamber assays revealed that overexpression of PER2 inhibited the invasion and migration abilities of GSCs (Fig. 4C).
Figure 4.
PER2 inhibits cell viability, the cell cycle, migration and invasion of GSCs. (A) The effect of PER2 overexpression on the proliferation of GSCs was evaluated using CCK-8 assay at the indicated time points. (B) Overexpression of PER2 led to a G0/G1 cycle arrest, as determined by flow cytometry. (C) Transwell assays revealed that the overexpression of PER2 inhibited the invasion and migration abilities of GSCs. One-way ANOVA with Tukey's post hoc test was used for statistical analysis of the results of A. Two-tailed Student's t-test was used for statistical analysis of the results of B and C. *P<0.05, **P<0.01 vs. the control. PER2, period 2; GSCs, glioma stem cells. **P<0.01.
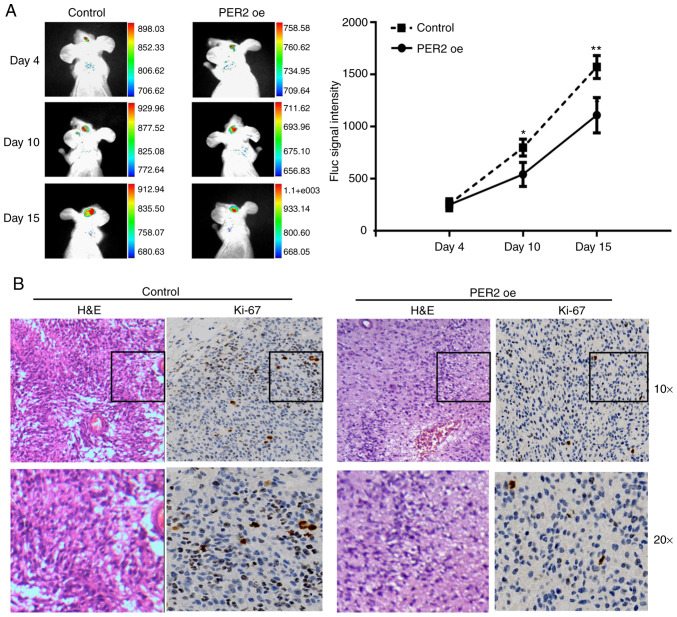
Based on the aforementioned data, it was hypothesized that PER2 could inhibit the malignant characteristics of GSCs in vivo. To test this hypothesis, the constructed U251-GSC line was implanted into nude mice by intracranial injection. As anticipated, IVIS (an imaging system) analysis and tumor volume assessment revealed that PER2 overexpression decreased tumor sizes at 4, 10, and 15 days after GSC transplantation (Fig. 5A) and the final tumor volume (Fig. S1A). Furthermore, fewer Ki-67-positive cells were observed in the PER2 overexpression group than in the control group (Fig. 5B).
Figure 5.
PER2 inhibited stemness in GSCs. (A) Tumor area measured by in vivo imaging system (IVIS) showed PER2 suppressed tumorigenic capacity of GSCs in vivo. (B) IHC of nude tumors showed a reduced number of Ki-67 positive cells in PER2 overexpression group compared with the control. One-way ANOVA with Tukey's post was used for statistical analysis of the results of A. *P<0.05, **P<0.01 vs. the control. PER2, period 2; GSCs, glioma stem cells; IHC, immunohistochemistry.
Next, it was determined whether PER2 also affected the phenotype of GSCs formed in humans. PER2 expression was analyzed in 48 paraffin-embedded glioma cancer tissue specimens. CD133 is the most well-accepted stemness marker of GSCs (22,23), and CD133 IHC revealed a significant negative association between PER2 expression and the stemness of glioma cells. High CD133-positive samples exhibited low or non-detectable PER2 staining. In contrast, samples with high PER2 staining did not have CD133 membrane staining (Fig. S1C).
PER2 suppresses the stem cell-like phenotype of GBM by dysregulating Wnt/β-catenin signaling
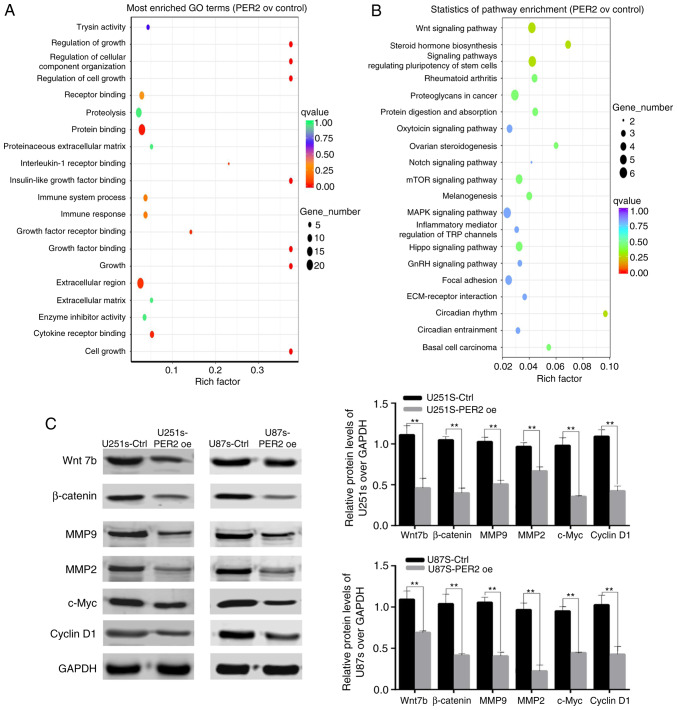
To characterize the molecular mechanisms of PER2 in GSCs, RNA-Seq analysis of control and PER2-overexpressed U251 GSCs was performed. Analysis of the RNA-Seq data indicated that PER2 overexpression decreased the expression of Wnt family genes, including Wnt7b and other Wnt family ligands and receptors (Fig. S2). To explore the molecular mechanisms underlying PER2 function, KEGG and GO analyses of the RNA-Seq data were also performed. The GO analysis revealed that PER2 altered the expression of genes involved in cell growth, metabolism and organismal systems, that aberrant signal transduction was closely related to the initiation and development of glioma (Fig. 6A). The results of the KEGG analysis also revealed that PER2 was significantly involved in the regulation of the Wnt signaling pathway (Fig. 6B; P=0.00694). Based on these data, it was hypothesized that PER2 may target the Wnt signaling pathway in order to regulate the development and progression of GSCs.
Figure 6.
PER2 suppresses the stem cell-like phenotype of GBM via dysregulation of Wnt/β-Catenin signaling. (A) GO and (B) KEGG analysis on the differential expression profiles. (C) Protein expression levels of Wnt/β-Catenin signaling pathway genes in the control and PER2-overexpressed GSCs. Two-tailed Student's t-test was used for statistical analysis of the results of C. **P<0.01 vs. the control. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PER2, period 2; GSCs, glioma stem cells.
To verify this hypothesis, western blotting was performed to assess the expression of some genes within the Wnt signaling pathway in the GSC cell lines. PER2 decreased the expression of Wnt7b and its downstream target genes, such as β-catenin, c-Myc, MMP2, MMP9, and cyclin D1. Consequently, PER2 suppressed the stemness of GSCs, at least partly, via the Wnt/β-catenin signaling pathway (Fig. 6C).
Discussion
Increasing evidence is showing that circadian genes play critical roles in the biological processes of cancer cells (12-14-24). Some of these genes also regulate cancer stem cell differentiation and cell stemness maintenance (25,26). Therefore, circadian genes may be potential targets for cancer stem cell treatment (27). Despite these insights into the role of circadian dysregulation in cancer, few novel circadian gene-targeting GSC therapies have emerged. Thus, nine core circadian gene expression profiles were detected in GSCs, normal glioma cells and human astrocyte cell lines, and PER2 was screened as the most significantly altered circadian gene involved in the malignant process of glioma. Consistent with data from our previous study, PER2 expression was notably downregulated in glioma samples compared with adjacent noncancerous tissues (20). These findings led to the development of the present study, which aimed to further explore the role of PER2 in GSCs and its potential mechanism of action.
In the present study, two GSC cell lines (U251s and U87s) were successfully established. Consistent with a previous study (6), our results confirmed that the stemness markers CD133, SOX2, and NESTIN were significantly increased in sphere-forming glioma cells compared with normal glioma cells. Next, it was demonstrated that PER2 overexpression led to the significant inhibition of stemness marker expression, decreased self-renewal capability and cell cycle arrest at the G0/G1 phase in GSCs. Furthermore, it was observed that PER2 suppressed the malignant characteristics of GSCs in vitro and in vivo. These results are consistent with a previous study revealing that the PER family gene PER3 could regulate self-renewal and chemoresistance in colorectal CSCs (28). Notably, we studied the malignant characteristics of GSCs in vivo with non-whole brain tumor pathology which could moderately demonstrate the result. However, pathological sections of the whole brain of tumor-bearing mice will provide relative position information of tumors in the brain which can further confirm that PER2 could inhibit the malignant characteristics of GSCs.
Previous research has demonstrated that PER2 acts as a potential anti-oncogene by targeting multiple genes, such as p53, AKT, OCT1 as well as several others (13,29–31). By analyzing the RNA-Seq results, it was determined that PER2 was involved in regulating the Wnt signaling pathway in GSCs. Griveau et al demonstrated that Wnt7b can affect glioma proliferation and invasion (32). As an important downstream component of the Wnt/β-catenin pathway, β-catenin can promote the uncontrolled proliferation and stemness of GBM cells (33). Tumor invasion is often due to the activities of metalloproteinases, which efficiently digest the extracellular matrix, such as MMP2 and MMP9 (34,35). It has been reported that c-Myc is required for the diverse features and pluripotency of GSCs (36). Therefore, in the present study, the downregulation of Wnt7b, β-catenin, MMP2, MMP9, and c-Myc induced by PER2 may reduce the invasive potential and stemness of GSCs. Cyclin D1 is a dominant cell cycle regulator and is involved in cell cycle control (37). which could explain the G0/G1 phase arrest observed in the PER2-overexpressing GSC cell lines.
In conclusion, the present study identified an important function of PER2 in regulating the stemness, cell growth, cell cycle progression and migration of GSCs in vitro and in vivo via the downregulation of Wnt/β-catenin signaling-associated gene expression. These findings indicated that PER2 may serve as a novel target in therapeutic strategies to overcome the effects of GSCs.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the National Natural Science Foundation of China (grant no. 81660419).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HX and HF conceived and designed the experiments. DM and LH performed the experiments. XJ and HL provided technical support, critical comments and suggestions. ZN analyzed the data. DM wrote the manuscript. All authors read and approved the final manuscript and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
All patients provided written informed consent, and the study was approved by the Ethics Committee of Ningxia Medical University. All mice experiments were performed with the approval of the Ningxia Medical University Experimental Animals Center IACUC.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cenciarini M, Valentino M, Belia S, Sforna L, Rosa P, Ronchetti S, D'Adamo MC, Pessia M. Dexamethasone in glioblastoma multiforme therapy: Mechanisms and controversies. Front Mol Neurosci. 2019;12:65. doi: 10.3389/fnmol.2019.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eisemann T, Costa B, Strelau J, Mittelbronn M, Angel P, Peterziel H. An advanced glioma cell invasion assay based on organotypic brain slice cultures. BMC Cancer. 2018;18:103. doi: 10.1186/s12885-018-4007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babu R, Komisarow JM, Agarwal VJ, Rahimpour S, Iyer A, Britt D, Karikari IO, Grossi PM, Thomas S, Friedman AH, Adamson C. Glioblastoma in the elderly: The effect of aggressive and modern therapies on survival. J Neurosurg. 2016;124:998–1007. doi: 10.3171/2015.4.JNS142200. [DOI] [PubMed] [Google Scholar]
- 4.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 5.Almenawer SA, Badhiwala JH, Alhazzani W, Greenspoon J, Farrokhyar F, Yarascavitch B, Algird A, Kachur E, Cenic A, Sharieff W, et al. Biopsy versus partial versus gross total resection in older patients with high-grade glioma: A systematic review and meta-analysis. Neuro Oncol. 2015;17:868–881. doi: 10.1093/neuonc/nou349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD, Rich JN. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006;66:7843–7848. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- 9.Kohler F, Rodriguez-Paredes M. DNA methylation in epidermal differentiation, aging, and cancer. J Invest Dermatol. 2020;140:38–47. doi: 10.1016/j.jid.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 10.Lowrey PL, Takahashi JS. Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikegami K, Refetoff S, Van Cauter E, Yoshimura T. Interconnection between circadian clocks and thyroid function. Nat Rev Endocrinol. 2019;15:590–600. doi: 10.1038/s41574-019-0237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shostak A. Circadian clock, cell division, and cancer: From molecules to organism. Int J Mol Sci. 2017;18:E873. doi: 10.3390/ijms18040873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang-Verslues WW, Chang PH, Jeng YM, Kuo WH, Chiang PH, Chang YC, Hsieh TH, Su FY, Lin LC, Abbondante S, et al. Loss of corepressor PER2 under hypoxia up-regulates OCT1-mediated EMT gene expression and enhances tumor malignancy. Proc Natl Acad Sci USA. 2013;110:12331–12336. doi: 10.1073/pnas.1222684110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiang R, Cui Y, Wang Y, Xie T, Yang X, Wang Z, Li J, Li Q. Circadian clock gene Per2 downregulation in nonsmall cell lung cancer is associated with tumour progression and metastasis. Oncol Rep. 2018;40:3040–3048. doi: 10.3892/or.2018.6704. [DOI] [PubMed] [Google Scholar]
- 15.Xiong H, Yang Y, Yang K, Zhao D, Tang H, Ran X. Loss of the clock gene PER2 is associated with cancer development and altered expression of important tumor-related genes in oral cancer. Int J Oncol. 2018;52:279–287. doi: 10.3892/ijo.2017.4180. [DOI] [PubMed] [Google Scholar]
- 16.Deng F, Yang K. Current status of research on the period family of clock genes in the occurrence and development of cancer. J Cancer. 2019;10:1117–1123. doi: 10.7150/jca.29212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu ML, Yeh KT, Lin PM, Hsu CM, Hsiao HH, Liu YC, Lin HY, Lin SF, Yang MY. Deregulated expression of circadian clock genes in gastric cancer. BMC Gastroenterol. 2014;14:67. doi: 10.1186/1471-230X-14-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazzoccoli G, Panza A, Valvano MR, Palumbo O, Carella M, Pazienza V, Biscaglia G, Tavano F, Di Sebastiano P, Andriulli A, Piepoli A. Clock gene expression levels and relationship with clinical and pathological features in colorectal cancer patients. Chronobiol Int. 2011;28:841–851. doi: 10.3109/07420528.2011.615182. [DOI] [PubMed] [Google Scholar]
- 19.Rahman S, Kraljević Pavelić S, Markova-Car E. Circadian (De)regulation in head and neck squamous cell carcinoma. Int J Mol Sci. 2019;20:E2662. doi: 10.3390/ijms20112662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia HC, Niu ZF, Ma H, Cao SZ, Hao SC, Liu ZT, Wang F. Deregulated expression of the Per1 and Per2 in human gliomas. Can J Neurol Sci. 2010;37:365–370. doi: 10.1017/S031716710001026X. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 23.Lv D, Ma QH, Duan JJ, Wu HB, Zhao XL, Yu SC, Bian XW. Optimized dissociation protocol for isolating human glioma stem cells from tumorspheres via fluorescence-activated cell sorting. Cancer Lett. 2016;377:105–115. doi: 10.1016/j.canlet.2016.04.022. [DOI] [PubMed] [Google Scholar]
- 24.Maiese K. Moving to the rhythm with clock (circadian) genes, autophagy, mTOR, and SIRT1 in degenerative disease and cancer. Curr Neurovasc Res. 2017;14:299–304. doi: 10.2174/1567202614666170718092010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Morita Y, Han B, Niemann S, Löffler B, Rudolph KL. Per2 induction limits lymphoid-biased haematopoietic stem cells and lymphopoiesis in the context of DNA damage and ageing. Nat Cell Biol. 2016;18:480–490. doi: 10.1038/ncb3342. [DOI] [PubMed] [Google Scholar]
- 26.Puram RV, Kowalczyk MS, de Boer CG, Schneider RK, Miller PG, McConkey M, Tothova Z, Tejero H, Heckl D, Järås M, et al. Core circadian clock genes regulate leukemia stem cells in AML. Cell. 2016;165:303–316. doi: 10.1016/j.cell.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ríos-Arrabal S, Muñoz-Gámez JA, Jiménez-Ruíz SM, Casado-Ruíz J, Artacho-Cordón F, León-López J. Circadian Regulation of Colon Cancer Stem Cells. Implications for Therapy. 2016 [Google Scholar]
- 28.Zhang F, Sun H, Zhang S, Yang X, Zhang G, Su T. Overexpression of PER3 inhibits self-renewal capability and chemoresistance of colorectal cancer stem-like cells via inhibition of notch and β-catenin signaling. Oncol Res. 2017;25:709–719. doi: 10.3727/096504016X14772331883976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miki T, Matsumoto T, Zhao Z, Lee CC. p53 regulates period 2 expression and the circadian clock. Nat Commun. 2013;4:2444. doi: 10.1038/ncomms3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X, He X, Yang Z, Jabbari E. Mammalian PER2 regulates AKT activation and DNA damage response. Biochem Cell Biol. 2012;90:675–682. doi: 10.1139/o2012-025. [DOI] [PubMed] [Google Scholar]
- 31.Gotoh T, Kim JK, Liu J, Vila-Caballer M, Stauffer PE, Tyson JJ, Finkielstein CV. Model-driven experimental approach reveals the complex regulatory distribution of p53 by the circadian factor period 2. Proc Natl Acad Sci USA. 2016;113:13516–13521. doi: 10.1073/pnas.1607984113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griveau A, Seano G, Shelton SJ, Kupp R, Jahangiri A, Obernier K, Krishnan S, Lindberg OR, Yuen TJ, Tien AC, et al. A glial signature and Wnt7 signaling regulate glioma-vascular interactions and tumor microenvironment. Cancer Cell. 2018;33:874–889.e7. doi: 10.1016/j.ccell.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, Huang H, Xue J, Liu M, Wang Y, et al. FoxM1 promotes β-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell. 2011;20:427–442. doi: 10.1016/j.ccr.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guichet PO, Guelfi S, Teigell M, Hoppe L, Bakalara N, Bauchet L, Duffau H, Lamszus K, Rothhut B, Hugnot JP. Notch1 stimulation induces a vascularization switch with pericyte-like cell differentiation of glioblastoma stem cells. Stem Cells. 2015;33:21–34. doi: 10.1002/stem.1767. [DOI] [PubMed] [Google Scholar]
- 35.Mikheev AM, Mikheeva SA, Severs LJ, Funk CC, Huang L, McFaline-Figueroa JL, Schwensen J, Trapnell C, Price ND, Wong S, Rostomily RC. Targeting TWIST1 through loss of function inhibits tumorigenicity of human glioblastoma. Mol Oncol. 2018;12:1188–1202. doi: 10.1002/1878-0261.12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di J, Duiveman-de Boer T, Zusterzeel PL, Figdor CG, Massuger LF, Torensma R. The stem cell markers Oct4A, Nanog and c-Myc are expressed in ascites cells and tumor tissue of ovarian cancer patients. Cell Oncol (Dordr) 2013;36:363–374. doi: 10.1007/s13402-013-0142-8. [DOI] [PubMed] [Google Scholar]
- 37.Kim JK, Diehl JA. Nuclear cyclin D1: An oncogenic driver in human cancer. J Cell Physiol. 2009;220:292–296. doi: 10.1002/jcp.21791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.