Abstract
Lactobacillus reuteri, a symbiotic inhabitant of the gastrointestinal tract in humans and animals, is marketed as a probiotic. The ability to adhere to intestinal epithelial cells and mucus is an interesting property with regard to probiotic features such as colonization of the gastrointestinal tract and interaction with the host. Here, we present a study performed to elucidate the role of sortase (SrtA), four putative sortase-dependent proteins (SDPs), and one C-terminal membrane-anchored cell surface protein of Lactobacillus reuteri ATCC PTA 6475 in adhesion to Caco-2 cells and mucus in vitro. This included mutagenesis of the genes encoding these proteins and complementation of mutants. A null mutation in hmpref0536_10255 encoding srtA resulted in significantly reduced adhesion to Caco-2 cells and mucus, indicating involvement of SDPs in adhesion. Evaluation of the bacterial adhesion revealed that of the five putative surface protein mutants tested, only a null mutation in the hmpref0536_10633 gene, encoding a putative SDP with an LPxTG motif, resulted in a significant loss of adhesion to both Caco-2 cells and mucus. Complementation with the functional gene on a plasmid restored adhesion to Caco-2 cells. However, complete restoration of adhesion to mucus was not achieved. Overexpression of hmpref0536_10633 in strain ATCC PTA 6475 resulted in an increased adhesion to Caco-2 cells and mucus compared with the WT strain. We conclude from these results that, among the putative surface proteins tested, the protein encoded by hmpref0536_10633 plays a critical role in binding of Lactobacillus reuteri ATCC PTA 6475 to Caco-2 cells and mucus. Based on this, we propose that this LPxTG motif containing protein should be referred to as cell and mucus binding protein A (CmbA).
Introduction
Strains of Lactobacillus reuteri have been isolated from the gastrointestinal tract, mother’s milk and vagina of humans and animals (Oh et al., 2010; Walter et al., 2011). However, the primary habitat of the species appears to be the gastrointestinal tract, and the species has been designated a universal entero-Lactobacillus (Casas & Dobrogosz, 2000) and a vertebrate symbiont of the gastrointestinal tract (Walter et al., 2011). Lactobacillus reuteri is marketed as a probiotic for humans and several clinical studies indicate positive effects (Weizman et al., 2005; Agustina et al., 2012; Hunter et al., 2012; Szajewska et al., 2013). Lactobacillus reuteri ATCC PTA 6475, isolated from human mother’s milk, is a candidate probiotic with anti-inflammatory properties (Lin et al., 2008; Jones et al., 2011; Thomas et al., 2012), which has shown promising effects in animal studies (Eaton et al., 2011; Preidis et al., 2012; McCabe et al., 2013).
Bacterial adherence to intestinal epithelial cells (IECs) and/or mucus is frequently considered to be a desirable feature for a probiotic strain as it can promote the gut residence time and interaction with host epithelial and immune cells (Lebeer et al., 2008; Kleerebezem et al., 2010; Juge, 2012). In general, adhesins of lactobacilli can be classified according to their targets in the intestine (e.g. mucus components, extracellular matrix proteins), according to their localization on the bacterial surface (e.g. surface layer proteins) or according to how they are anchored to the bacterial surface [e.g. sortase-dependent proteins (SDPs)] (Vélez et al., 2007). The mechanisms of adhesion are not fully understood; however, several reports have shed light on the mechanisms for a variety of Lactobacillus species (Roos et al., 1996; Rojas et al., 2002; Roos & Jonsson, 2002; Granato et al., 2004; Buck et al., 2005, 2009; Pretzer et al., 2005; Bergonzelli et al., 2006; van Pijkeren et al., 2006; Kankainen et al., 2009; Vélez et al., 2010; Sánchez et al., 2011; von Ossowski et al., 2011). In addition to specific bacterial adhesins, other cell surface molecules, such as S-layer proteins, lipoteichoic acid and exopolysaccharides (Lebeer et al., 2008), and extracellular appendages, such as flagella, fimbriae and pili (Juge, 2012), can also contribute to adhesion to host epithelial cells and mucus. See Vélez et al. (2007), Juge (2012) or Lebeer et al. (2008) for comprehensive reviews on adherence factors.
Several adhesins of Lactobacillus reuteri have been described. The first Lactobacillus reuteri surface protein involved in adhesion to be described was CnBP of Lactobacillus reuteri NCIB 11951 (Aleljung et al., 1994; Roos et al., 1996), which binds to collagen type I. Later, the adhesion-promoting protein MapA of Lactobacillus reuteri 140R (Rojas et al., 2002) was described. This protein binds to both mucus (Rojas et al., 2002) and Caco-2 cells (Miyoshi et al., 2006), and two receptor-like molecules for MapA have been identified on Caco-2 cells (Miyoshi et al., 2006). CnBP and MapA are considered homologues in light of their similarity at the amino acid level (94 %). Roos & Jonsson (2002) were the first to describe a mucus-binding protein of Lactobacillus reuteri when they described an extracellular mucus-binding protein (Mub) in Lactobacillus reuteri 1063. The crystal structure of the Mub protein has been determined, which revealed an unexpected immunoglobulin binding activity (MacKenzie et al., 2009). Other surface proteins of Lactobacillus reuteri that contribute to adhesion include a high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB), which both contribute to adherence and ecological performance of Lactobacillus reuteri 100-23 in the murine gut (Walter et al., 2005).
SDPs are a group of surface-associated proteins in Gram-positive bacteria, many of which have been shown to impact the adhesive ability of several lactobacilli. SDPs have a common molecular structure that includes an N-terminal signal peptide, often with an YSIRK-G/S motif that promotes secretion (Bae & Schneewind, 2003) and directs the protein to a specific surface localization (DeDent et al., 2008), a C-terminal LPxTG motif, followed by a C-terminal transmembrane helix and a positively charged tail (Lebeer et al., 2008; Call & Klaenhammer, 2013). Examples are the mannose-specific adhesin (Msa) in Lactobacillus plantarum WCFS1 (Pretzer et al., 2005), Mub in Lactobacillus reuteri ATCC 53608 (strain 1063) (Roos & Jonsson, 2002) and Lactobacillus epithelium adhesin of Lactobacillus crispatus ST1 (Edelman et al., 2012). Sortase A (SrtA) cleaves the LPxTG motif between the threonine and glycine residues, and covalently links the threonine carboxyl group to amino groups provided by the cell wall cross-bridges of peptidoglycan precursors (Marraffini et al., 2006). Thus, a SDP is linked to the cell wall and displayed on the bacterial surface.
Lactobacillus reuteri ATCC PTA 6475 has a single gene encoding SrtA, five putative SDPs and one putative C-terminal membrane-anchored cell surface protein with similarities to SDPs but lacking the LPxTG motif. Among the human Lactobacillus reuteri strains, ATCC PTA 6475 is highly adherent to mucus (MacKenzie et al., 2010) and various intestinal human cell lines (Wang et al., 2008; Jensen et al., 2012). Here, we present a study performed to elucidate whether SrtA and five of the six above-mentioned putative surface proteins of Lactobacillus reuteri ATCC PTA 6475 play a role in the ability of the strain to adhere to IECs and mucus. The characterization of the functionality of these proteins includes adhesion to IECs and mucus in vitro, mutagenesis of specific genes, and complementation of mutants. We found that the putative SDP encoded by the gene hmpref0563_10633 plays a significant role in the ability to adhere to IECs and mucus, and propose that this protein should be referred to as cell and mucus-binding protein A (CmbA).
Methods
Bacterial strains and growth conditions.
The strains used in this study are shown in Table 1. Strains were maintained at –80 °C in 20 % (v/v) glycerol. Lactobacillus reuteri strains were grown at 37 °C in Man–Rogosa–Sharpe (MRS) broth or on MRS agar. Lactococcus lactis MG1363 was grown at 30 °C in M17 supplemented with glucose [0.5 % (w/v)]. Strains harbouring the pSIP411 vector or its derivatives were cultured in the presence of 10 µg erythromycin ml−1 (Sigma-Aldrich). All culture media were from Oxoid.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference |
| Strains | ||
| Lactobacillus reuteri | ||
| 6475 | ATCC PTA 6475 (earlier designated MM4-1A); WT, host strain; human breast milk (a kind gift from BioGaia AB, Stockholm, Sweden) | Oh et al. (2010) |
| 6475 cmbA− | Derivative of 6475; nonsense mutation in cmbA (hmpref0536_10633) encoding a hypothetical LPxTG motif containing protein* | This work |
| 6475 10146 − | Derivative of 6475; nonsense mutation in hmpref0536_10146 encoding a hypothetical LPxTG motif containing protein* | This work |
| 6475 11993 − | Derivative of 6475; nonsense mutation in hmpref0536_11993 encoding a putative C-terminal membrane-anchored cell surface protein* | This work |
| 6475 10802 − | Derivative of 6475; nonsense mutation in hmpref0536_10802 encoding a LPxTG motif containing amidase* | This work |
| 6475 10154 − | Derivative of 6475; nonsense mutation in hmpref0536_10154 encoding a LPxTG motif containing Ser/Thr protein phosphatase* | This work |
| 6475 srtA− | Derivative of 6475; nonsense mutation in srtA (hmpref0536_10255) encoding sortase A* | This work |
| 6475(pSIPΔ) | Derivative of 6475 containing pSIPΔ | This work |
| 6475(pSIP-cmbA) | Derivative of 6475 containing pSIP-cmbA | This work |
| 6475 cmbA−(pSIPΔ) | Derivative of 6475 cmbA− containing pSIPΔ | This work |
| 6475 cmbA−(pSIP-cmbA) | Derivative of 6475 cmbA− containing pSIP-cmbA | This work |
| 6475 srtA−(pSIPΔ) | Derivative of 6475 srtA− containing pSIPΔ | This work |
| 6475 srtA−(pSIP-srtA) | Derivative of 6475 srtA− containing pSIP-srtA | This work |
| Lactococcus lactis | ||
| MG1363 | Intermediate cloning host | Gasson (1983) |
| Plasmids | ||
| pSIP411 | Emr; SppIP-based expression vector with PsppQ : : gusA | Sørvig et al. (2005) |
| pSIPΔ | Emr; pSIP411 derivative without gusA | This work |
| pSIP-cmbA | Emr; pSIP411 derivative containing cmbA under control of PsppQ | This work |
| pSIP-srtA | Emr; pSIP411 derivative containing srtA under control of PsppQ | This work |
Details given in Table S1.
Cell culture.
The human colorectal adenocarcinoma cell line Caco-2 (HTB-37) was obtained from the American Type Culture Collection. Caco-2 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 20 % heat-inactivated FBS, 1 % non-essential amino acids, 100 U penicillin ml−1 and 100 µg streptomycin ml−1. All solutions were obtained from Invitrogen. The cells were maintained at 37 °C in a humidified atmosphere of 5 % CO2 and subcultivated at 70–90 % confluence.
Sequence analysis of Lactobacillus reuteri ATCC PTA 6475.
In order to identify genes encoding cell wall anchor domain proteins (Marraffini et al., 2006), the genome sequence and identified proteins of strain ATCC PTA 6475, hereafter called 6475 (previously named MM4-1A; GenBank accession number ACGX02000000, sequences ACGX02000001–ACGX02000007), were reanalysed after the preliminary analysis made by Saulnier et al. (2011). The sorting motif LPxTG was searched for manually in the protein sequences, and YSIRK-G/S signal sequences (pfam04650), cell wall anchor domains (TIGR01167) and other protein domains were searched for in GenBank and with blastp at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov). Secretion signal peptides were predicted with SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP) (Petersen et al., 2011) and transmembrane helices were predicted with tmhmm 2.0 (http://www.cbs.dtu.dk/services/TMHMM). Repeats in the protein sequences were identified using radar (http://www.ebi.ac.uk/Tools/pfa/radar).
Construction of Lactobacillus reuteri mutants.
Mutagenesis in Lactobacillus reuteri 6475 was performed using ssDNA recombineering as described previously (van Pijkeren & Britton, 2012). Defined nonsense mutations were established within the Lactobacillus reuteri 6475 genes hmpref0536_10633, hmpref0536_10255, hmpref0536_10146, hmpref0536_11993, hmpref0536_10154 and hmpref0536_10802 (Table S1, available in the online Supplementary Material); hereafter referred to as cmbA, srtA, 10146, 11993, 10154 and 10802, respectively.
Reagents and enzymes.
Total DNA was isolated using the DNeasy Tissue kit (Qiagen) and the Bacterial Genomic DNA Purification kit (EdgeBio). Plasmid DNA was isolated using the QIAprep Miniprep kit (Qiagen). Lysozyme (20 mg ml−1) and mutanolysin (40 U ml−1) (Sigma-Aldrich) were used in all lysis steps. Restriction endonucleases NcoI, XbaI and XhoI, and T4 DNA ligase were obtained from New England Biolabs. Phusion High-Fidelity DNA Polymerase (Finnzymes/Thermo Fisher Scientific) and Maxima Hot Start polymerase (Thermo Fisher Scientific) were used for PCR. PCR fragments required for cloning were recovered from 0.7 % agarose gels using the QIAquick Gel Extraction kit (Qiagen). All kits were used according to the manufacturers’ instructions.
Cloning of Lactobacillus reuteri ATCC PTA 6475 cmbA and srtA.
Plasmid pSIP411 (Sørvig et al., 2005), known to yield inducible and titratable gene expression in Lactobacillus reuteri 6475 (van Pijkeren & Britton, 2012) through the PsppQ (previously PorfX) promoter, was used as cloning vector. Plasmid constructions were performed with Lactococcus lactis MG1363 as an intermediate host. Lactococcus lactis MG1363 and Lactobacillus reuteri were transformed by electroporation using the Gene Pulser system (Bio-Rad) as described previously (Holo & Nes, 1989; Ahrné et al., 1992), except that 0.5 % sucrose in 10 % glycerol was used as electroporation buffer for Lactobacillus reuteri. The Lactobacillus reuteri 6475 cmbA gene contains an internal NcoI site. In order to clone the gene at the NcoI/XbaI sites in pSIP411 (at the ATG start codon), replacing the reporter gene gusA, the internal NcoI site was removed and the fourth base of the gene was changed from C to G to create an NcoI site at the 5′ end of the gene. This was accomplished by fusion PCR (Horton & Pease, 1991), where the complete coding region of cmbA was amplified in two parts and assembled, including the base changes, before ligation into the pSIP411 vector at the NcoI and XbaI sites (primers used for fusion PCR are listed in Table S2). The base changes resulted in codon changes at the positions corresponding to aa 2 (CTA to GTA) and aa 407 (TCC to TCG) in cmbA. The former results in an amino acid residue change (Leu to Val), which was anticipated not to affect the function of the protein. Cloning of the Lactobacillus reuteri 6475 srtA gene was performed using synthetically manufactured DNA (GenScript). The synthetic fragment consisted of the PsppQ promoter and ribosome binding site, as present in pSIP411, and the srtA gene with the following base changes: A instead of G at position 1 (changing the start codon from GTG to ATG to ensure expression in the pSIP system) and T instead of C at positions 202 and 405 (removing internal BglII and NcoI sites to minimize problems in future cloning in the pSIP plasmids). None of the codon changes resulted in an amino acid residue change in the corresponding protein. The synthetic fragment was flanked by BglII and XhoI sites, which were used for cloning in pSIP411, replacing the PsppQ : : gusA region. pSIP411 with the gusA reporter gene deleted, designated pSIPΔ, was also used as a control. Sanger sequencing of the final constructions was performed to ensure correct sequences. Molecular cloning and gel electrophoresis procedures were performed using standard procedures. All primers used in this study are listed in Table S2.
Adhesion assays.
The initial adhesion assays (for strain 6475 and mutants) were performed as follows. Before adhesion experiments, strains were taken from frozen stock and grown in MRS broth at 37 °C. The strains were subcultured once before experiments. On the day of experiment, the strains were inoculated to OD600 0.1 in MRS broth and grown at 37 °C. At OD600 1.0±0.1, the cells were harvested by centrifugation at 4000 r.p.m. for 10 min, washed in Dulbecco’s PBS (DPBS) and resuspended in 1 vol. DPBS (Sigma-Aldrich). In the complementation/overexpression adhesion assays, where the pSIP-inducible gene expression system (Sørvig et al., 2005) was used, the strains were inoculated to OD600 0.1 in two separate tubes for each strain. At OD600 0.3, SppIP-inducing peptide (Molecular Biology Unit, University of Newcastle, UK) was added to one set of cultures. For Lactobacillus reuteri 6475, 6475 cmbA – and their derivative strains, 50 ng SppIP ml−1 was used. For 6475 srtA – and derivative strains, 1 ng SppIP ml−1 was used due to severe growth inhibition at higher concentrations for the strain containing pSIP-srtA. At 1.5 h after induction (OD600 1.0±0.1), both induced and non-induced cultures were harvested as described above. Lactobacillus reuteri adhesion to Caco-2 cells was tested as described previously (Jensen et al., 2012). Briefly, Lactobacillus reuteri strains were added (~5×106 c.f.u.) to confluent cell layers in 1 ml antibiotic-free cell media per well. After 1 h incubation, the cell layer was washed to remove non-adherent bacteria and lysed by addition of 0.1 % Triton X-100 (Sigma-Aldrich) in DPBS. The remaining suspensions with viable adhered bacteria were plated onto MRS agar and the number of c.f.u. was counted after 48 h incubation. Adhesion to Caco-2 cells was calculated as per cent of adhered bacteria in relation to the total number of bacteria added. Experiments were performed with triplicate determinations and repeated three to six times. The bacterial adhesion to mucus for Lactobacillus reuteri strains (Table 1) was tested as described previously (Roos & Jonsson, 2002). Briefly, mucus from pig small intestine (obtained from slaughterhouse material at Uppsala, Sweden) was prepared and coated in microtitre wells (MaxiSorp; Nunc). Strains were prepared as described above and washed once in PBS pH 6.0 (PBS) supplemented with 0.05 % Tween 20 (PBST) and resuspended in an equal volume of the same buffer. An aliquot of 100 µl bacterial suspension was added to each well and incubated overnight at 4 °C. The wells were washed three times with PBST, the buffer was poured off and OD405 was measured using a plate reader after the wells had dried. Wells coated with BSA were used as control. Experiments were performed with triplicate determinations and repeated three times.
Statistics.
ANOVA of the bacterial adhesion to Caco-2 cells was performed in Minitab version 16 (Minitab) using the General Linear Model and Tukey’s or Dunnet’s post-hoc test. Differences were considered statistically significant at P≤0.05. Illustrations were created in Prism version 5.0 (GraphPad).
Results
Sequence analysis of Lactobacillus reuteri ATCC PTA 6475
The sequences of the proteins encoded by the draft genome sequence of Lactobacillus reuteri ATCC PTA 6475 were analysed, and one sortase and five cell wall anchor domain proteins with LPxTG motifs were identified. One protein with a similar architecture, but lacking the actual LPxTG motif, thus predicted to be anchored to the cell envelope by a C-terminal membrane anchor, was also found. Furthermore, two putative pseudogenes encoding domains related to cell wall anchoring were identified. The genetic loci in strain 6475, the corresponding loci in the fully sequenced and highly related strain JCM 1112 (Walter et al., 2011), the identity of the proteins, and some of their features are shown in Table 2.
Table 2.
Genes encoding cell wall anchor domain proteins and related proteins in Lactobacillus reuteri 6475
| Locus tag HMPREF0536 | Homologue in strain JCM 1112 | Annotation/features/comments | Size (aa) | Mutant analysed |
| 10255 | LAR_0227 | Sortase | 234 | Yes |
| 10146 | LAR_0989 | LPxTG protein, Pilus_PilP and Rib regions | 630 | Yes |
| 10154 | LAR_0983 | LPxTG protein, 5′-nucleotidase/2′,3′-cyclic phosphodiesterase | 752 | Yes |
| 10633 | LAR_0958 | LPxTG protein, YSIRK-G/S signal sequence | 1030 | Yes |
| 10706 | LAR_0903 | LPxTG protein, a second putative start 87 bp downstream (size 272 aa) | 301 | NA |
| 10802 | LAR_0813 | LPxTG protein, amidase, a second putative start 25 bp downstream (size 637 aa) | 645 | Yes |
| 11242-11241 | LAR_1193-1192 | 11242: YSIRK-G/S signal sequence; 11241: LPxTG motif; ORFs separated by frame shift, putative pseudogene | 328* | NA |
| 12042 | LAR_0089 | YSIRK-G/S signal sequence, truncated, no LPxTG motif, putative pseudogene | 607* | NA |
| 11993 | LAR_0044 | C-terminal membrane-anchored cell surface protein | 951 | Yes |
na, Not available.
Truncated genes.
Adhesion of Lactobacillus reuteri mutants to Caco-2 cells and mucus
Bacterial adhesion to the human colorectal cell line Caco-2 and mucus was investigated initially for Lactobacillus reuteri 6475, the srtA mutant and the five mutants for genes encoding putative surface proteins (Table 1). The growth of the strains was followed as a part of the Caco-2 cell adhesion assay (see Methods). The growth rates were very similar (results not shown). Lactobacillus reuteri 6475 srtA− revealed a significantly lower adhesion to Caco-2 cells and mucus compared with 6475 (P = 0.0057 and P = 0.00017, respectively) (Fig. 1), indicating involvement of SDPs in adhesion. Of the LPxTG protein mutants, Lactobacillus reuteri 6475 cmbA− revealed a significant loss of adhesion to Caco-2 cells, whereas the other mutants did not show significantly reduced adhesion compared with 6475 (1.3 vs 4.8 %, P<0.0001) (Fig. 1a). Lactobacillus reuteri 6475 cmbA− also showed a total loss of adhesion to mucus (P<0.0001). Lactobacillus reuteri 6475 10802 − had a significant loss of adhesion to mucus compared with 6475 (P = 0.0161), whereas the other mutants did not show significantly reduced adhesion to mucus (Fig. 1b). Only Lactobacillus reuteri 6475 srtA− and 6475 cmbA− thus revealed significantly reduced adhesion to both Caco-2 cells and mucus, and only those strains were selected for complementation.
Fig. 1.
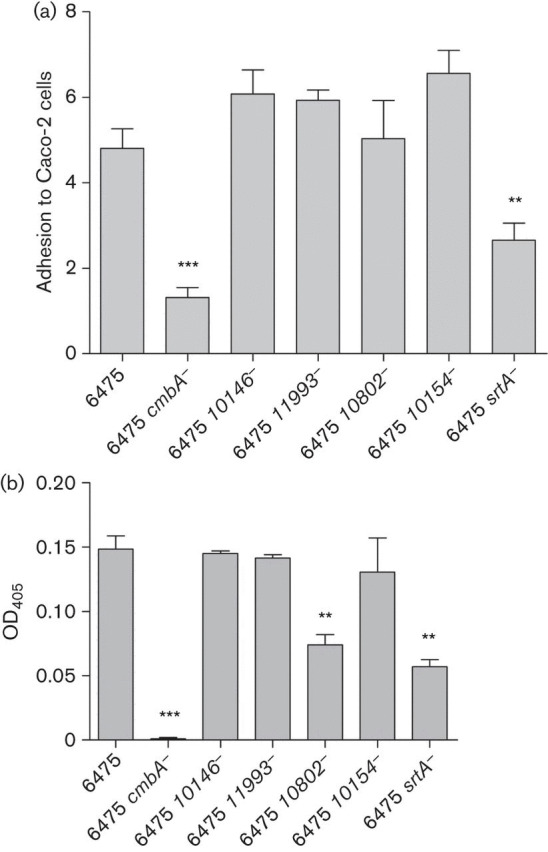
(a) Adhesion of Lactobacillus reuteri 6475 and mutant strains to Caco-2 cells. Lactobacillus reuteri strains were added (~5×106 c.f.u.) to confluent cell layers in 1 ml antibiotic-free cell media per well. After 1 h incubation, the cell layer was washed to remove non-adherent bacteria and lysed by addition of 0.1 % Triton X-100 (Sigma-Aldrich) in DPBS. The remaining suspensions with viable adhered bacteria were plated onto MRS agar and the number of c.f.u. counted after 48 h incubation. Adhesion to Caco-2 cells was calculated as per cent of adhered bacteria in relation to the total number of bacteria added. At least three independent biological replicates were performed with each strain in triplicate. (b) Adhesion of Lactobacillus reuteri 6475 and mutant strains to mucus. Mucus from pig small intestine was prepared and coated in microtitre wells. An aliquot of 100 µl bacterial suspension was added to each well and incubated overnight at 4 °C. The wells were washed three times with PBST, the buffer was poured off and OD405 was measured using a plate reader after the wells had dried. At least three independent biological replicates were performed with each strain in triplicate. All results are expressed as means; error bars, sem. Lactobacillus reuteri mutants were compared with Lactobacillus reuteri 6475 (ANOVA, General Linear Model, Dunnet’s post-hoc test), *P≤0.05, **P≤0.01, ***P≤0.001.
Cloning of cmbA
The annotated hmpref0536_10633 (cmbA) gene of strain 6475 is reported to be 3093 bp (Fig. S1). However, the cloning procedure yielded a gene of 2517 bp. Sequencing of the cloned gene revealed that the difference was due to a part where the annotated gene shows the presence of three identical tandem repeat regions of 288 bp (Fig. S1), whereas the cloned gene only has one such region. Several control PCRs with different primer pairs were performed, verifying that the cmbA gene in our culture of strain 6475 was ~0.6 kbp shorter than expected from the annotated genome sequence (not shown). Whether this represents a sequencing error and/or artefact in the reported 6475 genome sequence, or that variants of the gene exist, is not known at present.
Complementation of mutants: adhesion to Caco-2
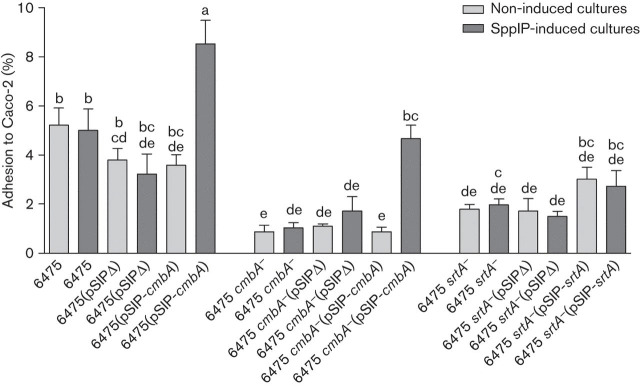
Based on the initial adhesion experiments (Fig. 1), Lactobacillus reuteri 6475 cmbA− and 6475 srtA− were complemented with the corresponding functional gene using the pSIP411 vector and its inducible gene expression system. Gene expression in complemented strains was first validated for cmbA using quantitative real-time PCR. This showed that cmbA expression increased in the complemented strain ~400-fold during induction compared with the WT expression from the chromosome (Table S3 and accompanying text). Together with controls, the adhesion of the complemented strains to Caco-2 cells was tested (Fig. 2). Similar to the initial experiments with the original mutants (see above), the growth was monitored for the different variant strains as a part of the adhesion assay. The growth rates of the strains again appeared similar (not shown). After SppIP induction of cmbA expression in Lactobacillus reuteri 6475 cmbA−(pSIP-cmbA) the adhesion was restored to that of Lactobacillus reuteri 6475 (4.7 vs 5.2 %), significantly higher than the corresponding non-induced strain (4.7 vs 0.9 %, P = 0.0017) (Fig. 2). Furthermore, SppIP induction of vector cmbA in Lactobacillus reuteri 6475(pSIP-cmbA), i.e. overexpression of cmbA in the WT strain, resulted in a significantly higher adhesion compared with the corresponding non-induced strain (8.5 vs 3.6 %, P = 0.0001) and the WT strain (8.5 vs 5.2 %, P = 0.0095) (Fig. 2). The SppIP-induced Lactobacillus reuteri 6475 srtA−(pSIP-srtA) did not show a higher adhesion than the corresponding non-induced strain (2.7 vs 3.0 %). However, both these strains showed a somewhat higher adhesion than Lactobacillus reuteri 6475 srtA− and 6475 srtA−(pSIPΔ) (~1.7 %) (Fig. 2), although the difference was not statistically significant. The presence of the pSIPΔ vector alone did not influence the adhesion with statistical significance. However, non-induced Lactobacillus reuteri 6475(pSIPΔ) and 6475(pSIP-cmbA) did show a somewhat poorer adhesion compared with Lactobacillus reuteri 6475 (3.8 and 3.6 vs 5.2 %, respectively) (Fig. 2). Furthermore, the induction peptide SppIP (1 or 50 ng ml−1) did not influence the growth of the strains (results not shown) or the adhesion to Caco-2 cells (Fig. 2).
Fig. 2.
Adhesion of Lactobacillus reuteri strains to Caco-2 cells with and without SppIP induction. Lactobacillus reuteri strains, either induced or non-induced with SppIP, were added (~5×106 c.f.u.) to confluent cell layers in 1 ml antibiotic-free cell media per well. After 1 h incubation, the cell layer was washed to remove non-adherent bacteria and lysed by addition of 0.1 % Triton X-100 (Sigma-Aldrich) in DPBS. The remaining suspensions with viable adhered bacteria were plated onto MRS agar and the number of c.f.u. was counted after 48 h incubation. Adhesion to Caco-2 cells was calculated as per cent of adhered bacteria in relation to the total number of bacteria added. At least three independent biological replicates were performed with each strain in triplicate. The results are expressed as means; error bars, sem. Letters above columns refer to the ANOVA: means that do not share a letter are significantly different (ANOVA, General Linear Model, Tukey’s post-hoc test accounting for the interaction between strains and level of induction), P≤0.05.
Complementation of mutants: adhesion to mucus
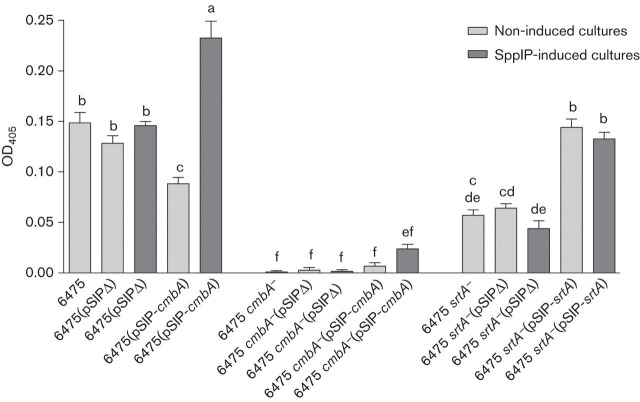
Bacterial adhesion to immobilized mucus was investigated for Lactobacillus reuteri 6475, 6475 cmbA−, 6475 srtA− and their derivative strains (Fig. 3). SppIP induction of vector cmbA in 6475(pSIP-cmbA) resulted in a significantly higher adhesion compared with both the corresponding non-induced strain (P<0.0001) and 6475 (P<0.0001) (Fig. 3). On the contrary, SppIP induction of vector cmbA in 6475 cmbA−(pSIP-cmbA), i.e. complementation of the cmbA mutant, did not restore fully the adhesion to that of 6475. A clear trend towards increased adhesion for the complemented strain compared with 6475 cmbA− was observed (Fig. 3), although the difference was not statistically significant in the ANOVA. After complementation with vector srtA in Lactobacillus reuteri 6475 srtA−, the adhesion was restored to that of 6475 for both SppIP-induced and non-induced cultures, and was significantly higher than 6475 srtA − (P<0.0001 for both SppIP-induced and non-induced cultures). Although showing clearly higher adhesion than the mutant, non-induced Lactobacillus reuteri 6475(pSIP-cmbA) revealed, for unknown reasons, a significantly lower adhesion compared with non-induced 6745 (P = 0.0002), whereas Lactobacillus reuteri 6475 harbouring pSIPΔ did not show reduced adhesion under these conditions (Fig. 3).
Fig. 3.
Adhesion of Lactobacillus reuteri strains to mucus with and without SppIP induction. Mucus from pig small intestine was prepared and coated in microtitre wells. An aliquot of 100 µl bacterial suspension, either induced or non-induced with SppIP, was added to each well and incubated overnight at 4 °C. The wells were washed three times with PBST, the buffer was poured off and OD405 was measured using a plate reader after the wells had dried. At least three independent biological replicates were performed with each strain in triplicate. The results are expressed as means; error bars, sem. Letters above columns refer to the ANOVA: means that do not share a letter are significantly different (ANOVA, General Linear Model, Tukey’s post-hoc test), P≤0.05.
Discussion
In this study, we addressed the effect of inactivating sortase (SrtA), four putative SDPs and one C-terminal membrane-anchored cell surface protein of Lactobacillus reuteri ATCC PTA 6475 on adhesion to Caco-2 cells and mucus. The putative SDP, encoded by the gene hmpref0536_10633, proved to be highly important for adhesion to both Caco-2 cells and mucus in vitro as shown by various mutant and complemented strains (Figs 1, 2 and 3). We therefore propose the protein encoded by this gene to be named and referred to as cell and mucus binding protein A (CmbA).
According to the genome sequence of Lactobacillus reuteri 6475, CmbA encodes a polypeptide of 1030 aa containing an N-terminal YSIRK-G/S type signal peptide and a C-terminal LPxTG motif followed by a hydrophobic region predicted to be a transmembrane helix and a positively charged tail (Fig. S1). The LPxTG motif is recognized by SrtA (Marraffini et al., 2006; Spirig et al., 2011; Call & Klaenhammer, 2013), which in turn is responsible for anchoring of the protein to the cell wall. When using the C-terminal 42 aa of the predicted protein (including the LPxTG motif, the hydrophobic region and the positively charged tail) as the search string in a Pfam search (http://pfam.sanger.ac.uk), a very significant similarity for this region was obtained with the protein family ‘Gram positive anchor’ (Pfam: PF00746). CmbA thus has all the features that define SDPs (Schneewind et al., 1992; Navarre & Schneewind, 1994, 1999; Bae & Schneewind, 2003; van Pijkeren et al., 2006; Schneewind & Missiakas, 2012; Call & Klaenhammer, 2013; Remus et al., 2013). CmbA is not classified as a ‘MucBP’ protein (Pfam: PF06458); however, blastp searches revealed that the best similarity scores were obtained with mucus binding proteins and LPxTG proteins in other Lactobacillus species, especially those of the so-called ‘acidophilus complex’ (Kullen et al., 2000), e.g. Lactobacillus gasseri MV-22 mucus binding protein (GenBank accession number ZP_07711536), Lactobacillus crispatus ST1 mucus binding protein (GenBank accession number YP_003602126), Lactobacillus johnsonii NCC 533 MucBP region protein (GenBank accession number NP_964406) and Lactobacillus acidophilus NCFM mucus binding protein (GenBank accession number YP_194552). The homologies to these proteins were in the range of 30–35 % identity (45–55 % similarity considering conservative amino acid changes). Among analysed and sequenced Lactobacillus reuteri strains, CmbA is unique to Lactobacillus reuteri 6475, JCM 1112 (DSM 20016), ATCC PTA 4659 (MM2-3) and ATCC PTA 5289 (FJ1). These strains are almost identical at the genome level with only a maximum of nine SNP differences between them (Walter et al., 2011) and are all clustered in one phylogenetic group, lineage II, of Lactobacillus reuteri (Oh et al., 2010), which essentially contains Lactobacillus reuteri strains of human origin. In addition, and notably, the cmbA gene is differently reported in GenBank for some of these strains with regard to the tandem repeat region (Fig. S1): the JCM 1112 sequence (GenBank accession number NC_010609.1) contains five repeats, the 6475 draft sequence (GenBank accession number ACGX02000000) reports three (with a comment: ‘unresolved tandem repeat’), and the ATCC PTA 4659 draft sequence (GenBank accession number ACLB01000000) has the repeat region as a separate contig. The cloning of cmbA from the 6475 strain in our collection yielded a gene with only one of these repeat regions. This might indicate that variants of the gene exist. Whether this is the case and if this has any bearing on the function of CmbA remains to be investigated. Worthy of note is that repeat regions, with variable numbers in different strains, have been found in other adhesins from lactobacilli (Boekhorst et al., 2006; Gross et al., 2010). Human Lactobacillus reuteri strains are also found in another phylogenetic group (lineage VI). One representative of this latter group is the commercial strain DSM 17938, which lacks CmbA. This strain has been shown to adhere significantly less to IECs than the strongly adhering strains 6475, DSM 20016 and ATCC PTA 5289 (FJ1) in the same assay system as used here (Jensen et al., 2012). One might therefore speculate that CmbA has a specific interaction with structures on human IECs that renders the strains possessing the protein to be highly adherent.
The cmbA mutant showed a significant reduction in adhesion to Caco-2 cells and a total loss of adhesion to mucus. This effect was reversible for adhesion to Caco-2 cells upon complementation with cmbA expressed from a vector. In addition, overexpression of cmbA in the WT strain [i.e. strain 6475(pSIP-cmbA)] did increase adhesion to both Caco-2 cells and mucus. In light of this, it was somewhat surprising that the adhesion to mucus was poorly restored with complemented cmbA. It should be noted, however, that there was a relatively clear trend towards increased adhesion for the complemented strain compared with the mutant. The lack of statistical significance in the ANOVA may in part be a result of uncertainties in the measurements at these low levels of adhesion (in the case of the cmbA mutant, no adhesion could be measured). Lack of full complementation cannot be copy number or pSIP system related, as such effects would also have been evident in studies of the same culture in adhesion to Caco-2 cells. The effect must therefore be specific for the mucus binding property of CmbA. Some possibilities exist for obtaining poor complementation, e.g. improper folding of the overexpressed protein that specifically affects mucus binding or improper co-expression of CmbA in relation to additional unidentified factors specifically involved in adhesion to mucus. This remains to be investigated, and has to await a more thorough characterization of CmbA and possible interactions with other proteins. Other adhesion factors to IECs and mucus have been described previously for Lactobacillus reuteri (Miyoshi et al., 2006; MacKenzie et al., 2010), and the present study also indicated that the putative SDP encoded by the gene hmpref0536_10802 was involved in mucus binding (Fig. 1b), but not in IEC binding. Nevertheless, the results indicate a significant role of CmbA in the adhesive properties of Lactobacillus reuteri 6475.
The srtA mutant showed a significantly reduced adhesion compared with Lactobacillus reuteri 6475, but a somewhat higher adhesion than the cmbA mutant. This supports the strong in silico evidence that CmbA is an SDP, and is in line with previous studies on SDPs and sortase-deficient mutants. SDPs may still be found as surface proteins in srtA mutants (Bierne et al., 2002; Nobbs et al., 2007; Remus et al., 2013), although significantly decreased in abundance and possibly displayed in a non-optimal fashion. The reason for the increased adhesion for the srtA mutant compared with the cmbA mutant may thus be explained by the physical properties of the C-terminal end of the CmbA protein and the fact that cmbA is still expressed in the srtA mutant. In the srtA mutant, the protein will be exported through the Sec machinery by way of the signal sequence (Ton-That et al., 2004). It is, however, no longer cleaved at the LPxTG site and therefore not covalently bound to the peptidoglycan, but the C-terminal transmembrane helix and the positively charged tail may anchor some of the expressed protein to the membrane. Thus, it is likely that whilst CmbA is not coupled to the peptidoglycan in the absence of SrtA, and therefore not optimally displayed, it remains surface associated in the cytoplasmic membrane by the C-terminal anchor and promotes some adhesion of Lactobacillus reuteri to IECs and mucus. The adhesion of the srtA mutant was essentially restored by introducing pSIP-srtA without induction. This was particularly evident with regard to mucus adhesion. Induction with 1 ng SppIP ml−1 did not increase adhesion further. This level of SppIP is well below the saturation level, but still promotes measurable and higher expression in Lactobacillus reuteri compared with the non-induced state (unpublished observations). Higher induction levels were tested, but this resulted in severe growth inhibition of the strain. This may indicate that srtA expression in the WT is at a relatively low level due to possible toxic effects of high SrtA concentrations, consistent with the protein being membrane located. The GTG start codon, used in the native srtA gene, but changed to ATG in the cloned version, is generally also an indication of downregulation of expression (Vellanoweth & Rabinowitz, 1992; O’Donnell & Janssen, 2001). The PsppQ promoter in the pSIP411 vector was shown previously to be not inactive completely in the non-induced state, i.e. a very minor, but still detectable, degree of gene expression occurred also without the presence of the induction peptide SppIP (Sørvig et al., 2005). This minor expression of srtA, together with a more effective translational start, could thus be enough to complement the mutant. Due to the sensitivity of srtA expression in Lactobacillus reuteri 6475 as described above, overexpression of srtA in the WT strain was anticipated to also give severe growth inhibition and therefore not tested.
Of the five putative surface protein mutants tested, only a null mutation in the hmpref0536_10633 gene (cmbA), encoding a putative surface protein with an LPxTG motif, had a significant loss of adhesion to both Caco-2 cells and mucus. The hmpref0536_10802 gene might play a role in adhesion of Lactobacillus reuteri 6475 to mucus, although the effect of inactivation of this gene was not as large as inactivation of cmbA. None of the other putative cell wall/membrane-anchored proteins investigated in the present study appeared to be important for adhesion to Caco-2 cells or mucus. Whether these proteins are expressed and, if so, their role in the surface properties of Lactobacillus reuteri 6475 remain to be investigated. The automatic bioinformatic analysis initially did not designate hmpref0536_10706 as SDP encoding (due to a possible start codon 87 bp downstream, leading to a protein without a signal sequence; Table 2). A mutant for this remaining putative SDP gene was therefore not available for this study. This SDP also remains to be evaluated for any role in the adhesive properties of strain 6475.
The importance of CmbA in adhesion to both IECs and mucus adds to the complexity of the interactions that mediate the adhesion of gut bacteria to the intestine. As the mucus layer of the intestine is renewed continuously, it would probably be advantageous for probiotic bacteria to have the ability to bind to various intestinal surfaces as this would allow for prolonged time in the intestine for interactions with IECs and immune cells. The importance of CmbA in colonization of Lactobacillus reuteri 6475 in the intestine in vivo will have to be validated in future studies, but as this strain of Lactobacillus reuteri most probably does not bind to intestinal surfaces of mice (Oh et al., 2010; Frese et al., 2011; Walter et al., 2011), such studies also require the development of suitable models.
In conclusion, we have identified a novel cell and mucus-binding protein, CmbA, of Lactobacillus reuteri 6475. Other surface proteins of Lactobacillus reuteri 6475 may contribute to the adhesion properties of the strain. However, since the cmbA mutant had a very significant loss of adhesion to Caco-2 cells and a total loss of adhesion to mucus, effects that were partially reversible by complementation of mutants, CmbA is clearly a highly important protein for the adhesive properties of Lactobacillus reuteri 6475.
Supplementary Data
Acknowledgements
This work was supported by the Fund for the Research Levy on Agricultural Products (Norway). The authors wish to thank Signe Marie Drømtorp for excellent technical assistance and Ingunn Berget for help with the statistical analysis.
Footnotes
Abbreviations: IEC, intestinal epithelial cell; SDP, sortase-dependent protein.
One supplementary figure and three supplementary tables are available with the online version of this paper.
Edited by: P. O'Toole
References
- Agustina R., Kok F. J., van de Rest O., Fahmida U., Firmansyah A., Lukito W., Feskens E. J., van den Heuvel E. G., Albers R., Bovee-Oudenhoven I. M.(2012).Randomized trial of probiotics and calcium on diarrhea and respiratory tract infections in Indonesian children Pediatrics 129e1155–e1164. 10.1542/peds.2011-1379 [DOI] [PubMed] [Google Scholar]
- Ahrné S., Molin G., Axelsson L.(1992).Transformation of Lactobacillus reuteri with electroporation: studies on the erythromycin resistance plasmid pLUL631 Curr Microbiol 24199–205. 10.1007/BF01579282 [DOI] [Google Scholar]
- Aleljung P., Shen W., Rozalska B., Hellman U., Ljungh A., Wadström T.(1994).Purification of collagen-binding proteins of Lactobacillus reuteri NCIB 11951 Curr Microbiol 28231–236. 10.1007/BF01575966 [DOI] [PubMed] [Google Scholar]
- Bae T., Schneewind O.(2003).The YSIRK-G/S motif of staphylococcal protein A and its role in efficiency of signal peptide processing J Bacteriol 1852910–2919. 10.1128/JB.185.9.2910-2919.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergonzelli G. E., Granato D., Pridmore R. D., Marvin-Guy L. F., Donnicola D., Corthésy-Theulaz I. E.(2006).GroEL of Lactobacillus johnsonii La1 (NCC 533) is cell surface associated: potential role in interactions with the host and the gastric pathogen Helicobacter pylori Infect Immun 74425–434. 10.1128/IAI.74.1.425-434.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H., Mazmanian S. K., Trost M., Pucciarelli M. G., Liu G., Dehoux P., Jänsch L., Garcia-del Portillo F., Schneewind O., Cossart P.(2002).Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence Mol Microbiol 43869–881. 10.1046/j.1365-2958.2002.02798.x [DOI] [PubMed] [Google Scholar]
- Boekhorst J., Helmer Q., Kleerebezem M., Siezen R. J.(2006).Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria Microbiology 152273–280. 10.1099/mic.0.28415-0 [DOI] [PubMed] [Google Scholar]
- Buck B. L., Altermann E., Svingerud T., Klaenhammer T. R.(2005).Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM Appl Environ Microbiol 718344–8351. 10.1128/AEM.71.12.8344-8351.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck B. L., Azcarate-Peril M. A., Klaenhammer T. R.(2009).Role of autoinducer-2 on the adhesion ability of Lactobacillus acidophilus J Appl Microbiol 107269–279. 10.1111/j.1365-2672.2009.04204.x [DOI] [PubMed] [Google Scholar]
- Call E. K., Klaenhammer T. R.(2013).Relevance and application of sortase and sortase-dependent proteins in lactic acid bacteria Front Microbiol 473. 10.3389/fmicb.2013.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas I. A., Dobrogosz W. J.(2000).Validation of the probiotic concept: Lactobacillus reuteri confers broad-spectrum protection against disease in humans and animals Microb Ecol Health Dis 12247–285. 10.1080/08910600050216246-1 [DOI] [Google Scholar]
- DeDent A., Bae T., Missiakas D. M., Schneewind O.(2008).Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus EMBO J 272656–2668. 10.1038/emboj.2008.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton K. A., Honkala A., Auchtung T. A., Britton R. A.(2011).Probiotic Lactobacillus reuteri ameliorates disease due to enterohemorrhagic Escherichia coli in germfree mice Infect Immun 79185–191. 10.1128/IAI.00880-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman S. M., Lehti T. A., Kainulainen V., Antikainen J., Kylväjä R., Baumann M., Westerlund-Wikström B., Korhonen T. K.(2012).Identification of a high-molecular-mass Lactobacillus epithelium adhesin (LEA) of Lactobacillus crispatus ST1 that binds to stratified squamous epithelium Microbiology 1581713–1722. 10.1099/mic.0.057216-0 [DOI] [PubMed] [Google Scholar]
- Frese S. A., Benson A. K., Tannock G. W., Loach D. M., Kim J., Zhang M., Oh P. L., Heng N. C., Patil P. B.& other authors (2011).The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri PLoS Genet 7e1001314. 10.1371/journal.pgen.1001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M. J.(1983).Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing J Bacteriol 1541–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato D., Bergonzelli G. E., Pridmore R. D., Marvin L., Rouvet M., Corthésy-Theulaz I. E.(2004).Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins Infect Immun 722160–2169. 10.1128/IAI.72.4.2160-2169.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross G., Snel J., Boekhorst J., Smits M. A., Kleerebezem M.(2010).Biodiversity of mannose-specific adhesion in Lactobacillus plantarum revisited: strain-specific domain composition of the mannose-adhesin Benef Microbes 161–66. 10.3920/BM2008.1006 [DOI] [PubMed] [Google Scholar]
- Holo H., Nes I. F.(1989).High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media Appl Environ Microbiol 553119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R. M., Pease L. R.(1991).Recombination and mutagenesis of DNA sequences using PCR Directed Mutagenesis: A Practical Approach 217–247.McPherson M. J.Oxford: IRL Press [Google Scholar]
- Hunter C., Dimaguila M. A., Gal P., Wimmer J. E., Jr, Ransom J. L., Carlos R. Q., Smith M., Davanzo C. C.(2012).Effect of routine probiotic, Lactobacillus reuteri DSM 17938, use on rates of necrotizing enterocolitis in neonates with birthweight < 1000 grams: a sequential analysis BMC Pediatr 12142. 10.1186/1471-2431-12-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen H., Grimmer S., Naterstad K., Axelsson L.(2012).In vitro testing of commercial and potential probiotic lactic acid bacteria Int J Food Microbiol 153216–222. 10.1016/j.ijfoodmicro.2011.11.020 [DOI] [PubMed] [Google Scholar]
- Jones S. E., Whitehead K., Saulnier D., Thomas C. M., Versalovic J., Britton R. A.(2011).Cyclopropane fatty acid synthase mutants of probiotic human-derived Lactobacillus reuteri are defective in TNF inhibition Gut Microbes 269–79. 10.4161/gmic.2.2.15282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juge N.(2012).Microbial adhesins to gastrointestinal mucus Trends Microbiol 2030–39. 10.1016/j.tim.2011.10.001 [DOI] [PubMed] [Google Scholar]
- Kankainen M., Paulin L., Tynkkynen S., von Ossowski I., Reunanen J., Partanen P., Satokari R., Vesterlund S., Hendrickx A. P. A.& other authors (2009).Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein Proc Natl Acad Sci U S A 10617193–17198. 10.1073/pnas.0908876106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem M., Hols P., Bernard E., Rolain T., Zhou M., Siezen R. J., Bron P. A.(2010).The extracellular biology of the lactobacilli FEMS Microbiol Rev 34199–230. 10.1111/j.1574-6976.2009.00208.x [DOI] [PubMed] [Google Scholar]
- Kullen M. J., Sanozky-Dawes R. B., Crowell D. C., Klaenhammer T. R.(2000).Use of the DNA sequence of variable regions of the 16S rRNA gene for rapid and accurate identification of bacteria in the Lactobacillus acidophilus complex J Appl Microbiol 89511–516. 10.1046/j.1365-2672.2000.01146.x [DOI] [PubMed] [Google Scholar]
- Lebeer S., Vanderleyden J., De Keersmaecker S. C.(2008).Genes and molecules of lactobacilli supporting probiotic action Microbiol Mol Biol Rev 72728–764. 10.1128/MMBR.00017-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. P., Thibodeaux C. H., Peña J. A., Ferry G. D., Versalovic J.(2008).Probiotic Lactobacillus reuteri suppress proinflammatory cytokines via c-Jun Inflamm Bowel Dis 141068–1083. 10.1002/ibd.20448 [DOI] [PubMed] [Google Scholar]
- MacKenzie D. A., Tailford L. E., Hemmings A. M., Juge N.(2009).Crystal structure of a mucus-binding protein repeat reveals an unexpected functional immunoglobulin binding activity J Biol Chem 28432444–32453. 10.1074/jbc.M109.040907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie D. A., Jeffers F., Parker M. L., Vibert-Vallet A., Bongaerts R. J., Roos S., Walter J., Juge N.(2010).Strain-specific diversity of mucus-binding proteins in the adhesion and aggregation properties of Lactobacillus reuteri Microbiology 1563368–3378. 10.1099/mic.0.043265-0 [DOI] [PubMed] [Google Scholar]
- Marraffini L. A., Dedent A. C., Schneewind O.(2006).Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria Microbiol Mol Biol Rev 70192–221. 10.1128/MMBR.70.1.192-221.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe L. R., Irwin R., Schaefer L., Britton R. A.(2013).Probiotic use decreases intestinal inflammation and increases bone density in healthy male but not female mice J Cell Physiol 2281793–1798. 10.1002/jcp.24340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi Y., Okada S., Uchimura T., Satoh E.(2006).A mucus adhesion promoting protein, MapA, mediates the adhesion of Lactobacillus reuteri to Caco-2 human intestinal epithelial cells Biosci Biotechnol Biochem 701622–1628. 10.1271/bbb.50688 [DOI] [PubMed] [Google Scholar]
- Navarre W. W., Schneewind O.(1994).Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria Mol Microbiol 14115–121. 10.1111/j.1365-2958.1994.tb01271.x [DOI] [PubMed] [Google Scholar]
- Navarre W. W., Schneewind O.(1999).Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope Microbiol Mol Biol Rev 63174–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs A. H., Vajna R. M., Johnson J. R., Zhang Y., Erlandsen S. L., Oli M. W., Kreth J., Brady L. J., Herzberg M. C.(2007).Consequences of a sortase A mutation in Streptococcus gordonii Microbiology 1534088–4097. 10.1099/mic.0.2007/007252-0 [DOI] [PubMed] [Google Scholar]
- O’Donnell S. M., Janssen G. R.(2001).The initiation codon affects ribosome binding and translational efficiency in Escherichia coli of cI mRNA with or without the 5′ untranslated leader J Bacteriol 1831277–1283. 10.1128/JB.183.4.1277-1283.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh P. L., Benson A. K., Peterson D. A., Patil P. B., Moriyama E. N., Roos S., Walter J.(2010).Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution ISME J 4377–387. 10.1038/ismej.2009.123 [DOI] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., Nielsen H.(2011).SignalP 4.0: discriminating signal peptides from transmembrane regions Nat Methods 8785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Preidis G. A., Saulnier D. M., Blutt S. E., Mistretta T. A., Riehle K. P., Major A. M., Venable S. F., Barrish J. P., Finegold M. J.& other authors (2012).Host response to probiotics determined by nutritional status of rotavirus-infected neonatal mice J Pediatr Gastroenterol Nutr 55299–307. 10.1097/MPG.0b013e31824d2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretzer G., Snel J., Molenaar D., Wiersma A., Bron P. A., Lambert J., de Vos W. M., van der Meer R., Smits M. A., Kleerebezem M.(2005).Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum J Bacteriol 1876128–6136. 10.1128/JB.187.17.6128-6136.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D. M., Bongers R. S., Meijerink M., Fusetti F., Poolman B., de Vos P., Wells J. M., Kleerebezem M., Bron P. A.(2013).Impact of Lactobacillus plantarum sortase on target protein sorting, gastrointestinal persistence, and host immune response modulation J Bacteriol 195502–509. 10.1128/JB.01321-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M., Ascencio F., Conway P. L.(2002).Purification and characterization of a surface protein from Lactobacillus fermentum 104R that binds to porcine small intestinal mucus and gastric mucin Appl Environ Microbiol 682330–2336. 10.1128/AEM.68.5.2330-2336.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos S., Jonsson H.(2002).A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components Microbiology 148433–442. [DOI] [PubMed] [Google Scholar]
- Roos S., Aleljung P., Robert N., Lee B., Wadström T., Lindberg M., Jonsson H.(1996).A collagen binding protein from Lactobacillus reuteri is part of an ABC transporter system? FEMS Microbiol Lett 14433–38. 10.1111/j.1574-6968.1996.tb08505.x [DOI] [PubMed] [Google Scholar]
- Sánchez B., González-Tejedo C., Ruas-Madiedo P., Urdaci M. C., Margolles A.(2011). Lactobacillus plantarum extracellular chitin-binding protein and its role in the interaction between chitin, Caco-2 cells, and mucin Appl Environ Microbiol 771123–1126. 10.1128/AEM.02080-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulnier D. M., Santos F., Roos S., Mistretta T. A., Spinler J. K., Molenaar D., Teusink B., Versalovic J.(2011).Exploring metabolic pathway reconstruction and genome-wide expression profiling in Lactobacillus reuteri to define functional probiotic features PLoS ONE 6e18783. 10.1371/journal.pone.0018783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneewind O., Missiakas D. M.(2012).Protein secretion and surface display in Gram-positive bacteria Phi Trans R Soc Lond B Biol Sci 3671123–1139. 10.1098/rstb.2011.0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneewind O., Model P., Fischetti V. A.(1992).Sorting of protein A to the staphylococcal cell wall Cell 70267–281. 10.1016/0092-8674(92)90101-H [DOI] [PubMed] [Google Scholar]
- Sørvig E., Mathiesen G., Naterstad K., Eijsink V. G., Axelsson L.(2005).High-level, inducible gene expression in Lactobacillus sakei and Lactobacillus plantarum using versatile expression vectors Microbiology 1512439–2449. 10.1099/mic.0.28084-0 [DOI] [PubMed] [Google Scholar]
- Spirig T., Weiner E. M., Clubb R. T.(2011).Sortase enzymes in Gram-positive bacteria Mol Microbiol 821044–1059. 10.1111/j.1365-2958.2011.07887.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajewska H., Gyrczuk E., Horvath A.(2013). Lactobacillus reuteri DSM 17938 for the management of infantile colic in breastfed infants: a randomized, double-blind, placebo-controlled trial J Pediatr 162257–262. 10.1016/j.jpeds.2012.08.004 [DOI] [PubMed] [Google Scholar]
- Thomas C. M., Hong T., van Pijkeren J. P., Hemarajata P., Trinh D. V., Hu W., Britton R. A., Kalkum M., Versalovic J.(2012).Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling PLoS ONE 7e31951. 10.1371/journal.pone.0031951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-That H., Marraffini L. A., Schneewind O.(2004).Protein sorting to the cell wall envelope of Gram-positive bacteria Biochim Biophys Acta 1694269–278. 10.1016/j.bbamcr.2004.04.014 [DOI] [PubMed] [Google Scholar]
- van Pijkeren J. P., Britton R. A.(2012).High efficiency recombineering in lactic acid bacteria Nucleic Acids Res 40e76. 10.1093/nar/gks147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Pijkeren J. P., Canchaya C., Ryan K. A., Li Y., Claesson M. J., Sheil B., Steidler L., O’Mahony L., Fitzgerald G. F.& other authors (2006).Comparative and functional analysis of sortase-dependent proteins in the predicted secretome of Lactobacillus salivarius UCC118 Appl Environ Microbiol 724143–4153. 10.1128/AEM.03023-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vélez M. P., De Keersmaecker S. C., Vanderleyden J.(2007).Adherence factors of Lactobacillus in the human gastrointestinal tract FEMS Microbiol Lett 276140–148. 10.1111/j.1574-6968.2007.00908.x [DOI] [PubMed] [Google Scholar]
- Vélez M. P., Petrova M. I., Lebeer S., Verhoeven T. L., Claes I., Lambrichts I., Tynkkynen S., Vanderleyden J., De Keersmaecker S. C.(2010).Characterization of MabA, a modulator of Lactobacillus rhamnosus GG adhesion and biofilm formation FEMS Immunol Med Microbiol 59386–398. [DOI] [PubMed] [Google Scholar]
- Vellanoweth R. L., Rabinowitz J. C.(1992).The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo Mol Microbiol 61105–1114. 10.1111/j.1365-2958.1992.tb01548.x [DOI] [PubMed] [Google Scholar]
- von Ossowski I., Satokari R., Reunanen J., Lebeer S., De Keersmaecker S. C., Vanderleyden J., de Vos W. M., Palva A.(2011).Functional characterization of a mucus-specific LPXTG surface adhesin from probiotic Lactobacillus rhamnosus GG Appl Environ Microbiol 774465–4472. 10.1128/AEM.02497-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J., Chagnaud P., Tannock G. W., Loach D. M., Dal Bello F., Jenkinson H. F., Hammes W. P., Hertel C.(2005).A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut Appl Environ Microbiol 71979–986. 10.1128/AEM.71.2.979-986.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J., Britton R. A., Roos S.(2011).Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm Proc Natl Acad Sci U S A 108Suppl. 14645–4652. 10.1073/pnas.1000099107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Wei H., Yuan J., Li Q., Li Y., Li N., Li J.(2008).Identification of a surface protein from Lactobacillus reuteri JCM1081 that adheres to porcine gastric mucin and human enterocyte-like HT-29 cells Curr Microbiol 5733–38. 10.1007/s00284-008-9148-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weizman Z., Asli G., Alsheikh A.(2005).Effect of a probiotic infant formula on infections in child care centers: comparison of two probiotic agents Pediatrics 1155–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.