Abstract
Reduction of fitness due to deleterious mutations imposes a limit to adaptive evolution. By characterizing features that influence this genetic load we may better understand constraints on responses to both natural and human-mediated selection. Here, using whole-genome, transcriptome, and methylome data from >600 Arabidopsis thaliana individuals, we set out to identify important features influencing selective constraint. Our analyses reveal that multiple factors underlie the accumulation of maladaptive mutations, including gene expression level, gene network connectivity, and gene-body methylation. We then focus on a feature with major effect, nucleotide composition. The ancestral vs. derived status of segregating alleles suggests that GC-biased gene conversion, a recombination-associated process that increases the frequency of G and C nucleotides regardless of their fitness effects, shapes sequence patterns in A. thaliana. Through estimation of mutational effects, we present evidence that biased gene conversion hinders the purging of deleterious mutations and contributes to a genome-wide signal of decreased efficacy of selection. By comparing these results to two outcrossing relatives, Arabidopsis lyrata and Capsella grandiflora, we find that protein evolution in A. thaliana is as strongly affected by biased gene conversion as in the outcrossing species. Last, we perform simulations to show that natural levels of outcrossing in A. thaliana are sufficient to facilitate biased gene conversion despite increased homozygosity due to selfing. Together, our results show that even predominantly selfing taxa are susceptible to biased gene conversion, suggesting that it may constitute an important constraint to adaptation among plant species.
Keywords: deleterious mutations, biased gene conversion, machine-learning, DFE-alpha, evolutionary simulations, Arabidopsis
THE reduction of fitness due to recurrent deleterious mutations can constrain adaptive evolution (Haldane 1937; Muller 1950; Crow 1970; Charlesworth and Charlesworth 1998; Agrawal and Whitlock 2012). The extent of this genetic load depends on the efficacy of purifying selection, which may differ between species and populations due to factors such as mating-system and demographic history (Muller 1964; Ohta 1973; Lynch and Gabriel 1990; Charlesworth et al. 1993; Nordborg 2000). However, the strength of purifying selection also varies within individual genomes, so that some chromosomal regions are more prone to accumulate deleterious variants than others (Hill and Robertson 1966; Felsenstein 1974; Chun and Fay 2011; Hartfield and Otto 2011). Identifying features that influence this variation not only informs about the limits of adaptation but also may provide insight into processes such as the evolution of sexual reproduction (Charlesworth et al. 1993; Peck 1994; Charlesworth and Charlesworth 1998; Keightley and Otto 2006).
Features influencing genetic load within a genome are not well resolved, although some common patterns have been established. For instance, in nearly all studied taxa, the expression level of a gene is positively associated with the strength of purifying selection, suggesting that highly expressed genes tend to have essential roles in physiology and development (Koonin 2011). The same likely holds true for genes occupying central positions within gene networks, as multiple studies have found evidence that highly connected genes are under strong selective constraint (Rausher et al. 1999; Fraser et al. 2002; Papakostas et al. 2014; Josephs et al. 2017). Both theoretical (Muller 1964; Hill and Robertson 1966; Felsenstein 1974; Hartfield and Otto 2011) and empirical (Chun and Fay 2011; Zhang et al. 2016) studies also have established that recombination rate can modulate the strength of purifying selection, so that deleterious alleles are more efficiently removed in regions of high recombination. Moreover, features such as gene-body methylation and chromatin remodeling may be associated with genetic load, as methylated cytosines are known to have elevated mutation rate (Bird 1980; Weng et al. 2019), and accessible chromatin regions, indicative of cis-regulatory elements (Klemm et al. 2019), show high sequence conservation between species (Rodgers-Melnick et al. 2016; Lu et al. 2019).
GC-biased gene conversion (gBGC) also is expected to influence genetic load (Bengtsson 1990). gBGC takes place during meiotic recombination, when GC/AT heterozygotes occurring within a heteroduplex DNA are preferentially fixed to GC (as opposed to AT) nucleotides (Marais 2003; Galtier and Duret 2007; Mugal et al. 2015). gBGC increases the frequency of GC alleles regardless of their fitness effects, which can lead to an accumulation of deleterious mutations (Bengtsson 1990; Glémin 2010). Indeed, evidence for increased genetic load due to gBGC has been found in the human genome (Necşulea et al. 2011; Lachance and Tishkoff 2014). The increased frequency of GC alleles, as well as the decreased frequency of AT alleles, may also give the appearance of selection, leading to a biased view of the selection landscape (Galtier and Duret 2007; Galtier et al. 2009; Ratnakumar et al. 2010; Corcoran et al. 2017; Bolívar et al. 2018; Rousselle et al. 2019). The widespread occurrence of gBGC is well-established in animals (Galtier et al. 2001, 2009, 2018; Duret and Galtier 2009; Wallberg et al. 2015; Glémin et al. 2015; Mugal et al. 2015; Smeds et al. 2016; Corcoran et al. 2017; Bolívar et al. 2018; Rousselle et al. 2019) and yeast (Mancera et al. 2008; Lesecque et al. 2013), but less is known about it in plants (Glémin et al. 2014; Clément et al. 2017). The extent of gBGC is thought to be directly related to the outcrossing rate, so that it is either weak or absent in highly homozygous selfing species (Marais et al. 2004; Glémin 2010). Empirical data support this notion, as selfing species of the genus Oryza and Collinsia have shown weaker footprints of gBGC than outcrossing species (Muyle et al. 2011; Hazzouri et al. 2013). However, by examining the association between recombination rate and the nucleotide composition of segregating sites, Günther et al. (2013) found that gBGC may shape sequence variation in a predominantly selfing species, Arabidopsis thaliana. These results raise questions about the extent of gBGC in selfing species, and whether the accumulation of deleterious mutations and apparent signal of selection due to gBGC are limited to outcrossing taxa.
In this study, we first perform a comprehensive analysis of genomic features that likely underlie the accumulation of maladaptive mutations in A. thaliana. This species has recently switched from outcrossing to selfing (Bechsgaard et al. 2006; Tang et al. 2007; Bomblies et al. 2010; Durvasula et al. 2017), which has important implications for the dynamics of deleterious variants (Charlesworth et al. 1993). Specifically, selfing reduces the effective population size (Ne), thereby weakening the efficacy of purifying selection (Bustamante et al. 2002). By increasing homozygosity, selfing also weakens the effects of recombination on allelic diversity (Nordborg 2000), which may run counter to the expectation that regions of high recombination accumulate few deleterious mutations (Hartfield and Otto 2011). Here, by combining whole-genome, transcriptome, and methylome data from >600 individuals, we leverage the considerable genomic and functional information available for A. thaliana to identify important factors associated with maladaptive mutations. We then focus on a feature with major effect—nucleotide composition. Our results suggest that gBGC has a sizable effect on sequence variation in A. thaliana despite selfing. We present evidence that gBGC decreases the efficacy of purifying selection by increasing the frequency of slightly deleterious mutations, which intensifies genome-wide signals of relaxed selection. Comparisons with two outcrossing species, Arabidopsis lyrata and Capsella grandiflora, suggest that gBGC leads to a footprint of relaxed purifying selection in all three species, but weakens signals of positive selection only in the two Arabidopsis species. Moreover, the simulations we perform demonstrate that natural levels of outcrossing are sufficient to facilitate gBGC in A. thaliana. Together, our results suggest that the importance of gBGC on sequence evolution is not limited to outcrossing taxa, but can have considerable genome-wide impact also in predominantly selfing species—a group that includes many of the most important crop species (Ross-Ibarra et al. 2007).
Materials and Methods
Data acquisition
Our main analyses are based on publicly available genome, transcriptome, and methylome data from the model-species A. thaliana. We focus on 645 genotypes, for which these data were collected by The 1001 Genomes Consortium (1001 Genomes Consortium 2016; Kawakatsu et al. 2016). These individuals represent ecotypes collected across Eurasia, North Africa, and North America.
A VCF file containing both variant and invariant sites for 1135 A. thaliana genotypes was downloaded from The 1001 Genomes database (https://1001genomes.org/data/GMI-MPI/releases/v3.1/, last accessed September 20, 2019) and filtered with VCFtools (Danecek et al. 2011). We retained only genotypes for which transcriptomes were sequenced by Kawakatsu et al. (2016). We then removed individuals identified as “relics” in the original publications, as they exhibited genetic and expression profiles that were distinct from the other individuals, leaving 645 genotypes. All indels and SNPs with more than two alleles were removed. We also removed sites with >20% missing data, and imputed the missing genotypes with Beagle 5 (Browning et al. 2018). Out of 120 M sites, we retained 79 M (6.3 M variable) after filtering.
Expression data for the 645 individuals were downloaded from NCBI GEO: GSE80744. According to the original publication (Kawakatsu et al. 2016), leaf samples were collected from plants grown in a common greenhouse environment, the RNA-seq reads aligned against the TAIR10 reference genome (Lamesch et al. 2012), and the per-gene read counts batch-corrected and size-normalized.
Methylation calls for the 645 individuals were downloaded from GEO: GSE43857 (Kawakatsu et al. 2016). For each individual, we counted the proportion of methylated cytosines (mCG, mCHG, and mCHH contexts, where H is A, T, or C) per gene. Sites with coverage <5 were removed. We estimated two features of the methylation data: the average proportion of methylated cytosines and methylation variability (measured as coefficient of variation) across the individuals.
Genomic features
We characterized multiple features that may play a role in defining the rate of sequence evolution at different regions of the A. thaliana genome. First, we used the TAIR10 annotation to count the number of exonic and intronic base-pairs, number of splice variants, distance to the centromere, and distance to the nearest transposable element (TE) for each gene. We then used invariant sites from the SNP-calls to calculate the percentage of guanine and cytosine bases per gene (GC%). We used ENCprime (https://github.com/jnovembre/ENCprime, last accessed October 11, 2019) to estimate the effective number of codons, as reflected in the statistic (Novembre 2002). Using data from Lu et al. (2019), we defined accessible chromatin regions (ACRs), indicative of cis-regulatory elements (Klemm et al. 2019). Around half of the ACRs in A. thaliana are found in genic regions (Lu et al. 2019), so we utilized two features of the data: distance to the nearest ACR and the percentage of ACR-base-pairs (ACR%) per gene.
Recombination rates (r) for genes in the A. thaliana genome were estimated from a crossover map covering >17,000 meiotic crossover events [based on 1920 F2 progeny (Col-0 and Ler-0), Rowan et al. 2019]. The density of de novo mutations (n = 2023) from 107 mutation accumulation lines (maintained for 25 generations as single-seed descent, Weng et al. 2019), were used to define mutation rates (μ) for the genes. We used machine-learning-based regression modeling to predict per-gene estimates of r and μ, given their chromosomal locations. The Extremely Randomized Trees (Extra-Trees) method (Geurts et al. 2006), as implemented in the R package ranger (Wright and Ziegler 2017), was used for the prediction. For more information, see section Machine-learning.
Co-expression Network
We used the R package WGCNA (Langfelder and Horvath 2008) to identify modules of co-expressed genes within the transcriptome data, as well as to estimate among-gene connectivity. A soft-thresholding power of 12 was used to calculate adjacencies for a signed co-expression network. Topological overlap matrix (TOM) and dynamic-cut tree algorithm were used to define network modules. Modules with ≥90% identical expression profiles were merged. Connectivity was defined as the sum of adjacencies between the focal-gene and other genes in the network.
Quantification Of Selective Constraint
To identify putatively harmful mutations, we predicted mutational effects with SIFT4G (Vaser et al. 2016). SIFT predictions are based on protein conservation among homologous sequences, with rare nonsynonymous mutations assigned lower (i.e., more harmful) scores. Based on analysis of known deleterious variants, this method was found to perform well in A. thaliana (Kono et al. 2018). We used the existing A. thaliana database (https://sift.bii.a-star.edu.sg/sift4g/, last accessed October 4, 2019) to annotate SNPs with MAF ≥0.01 among the 645 individuals, and calculated an average SIFT-score for each gene (averaged across sites at which mutational effects were predicted). High SIFT-scores indicate a low average impact of segregating mutations, reflecting strong purifying selection, whereas low SIFT-scores are due to high average impact of segregating mutations, reflecting relaxed selective constraint.
As a comparison to SIFT-scores, we estimated two statistics reflecting the efficacy of purifying selection: the ratio of nonsynonymous to synonymous nucleotide divergence (dN/dS) between A. thaliana and A. lyrata, and the ratio of nonsynonymous to synonymous nucleotide diversities (πN/πS) within A. thaliana (Nielsen 2005; Chen et al. 2017). For each gene, we also estimated pairwise nucleotide diversity across all sites, which is sensitive to factors besides purifying selection (Cutter and Payseur 2013). For dN/dS, orthologous gene-pairs were identified with reciprocal BLAST (Camacho et al. 2009) and coding sequences aligned at the codon-level with PRANK (Löytynoja and Goldman 2008). dN and dS were then estimated with the R package SeqinR (Charif and Lobry 2007). We used the full VCF-file to estimate pairwise nucleotide diversity (Tajima 1983) across each callable site (variant and invariant).
Machine-learning
We explored what factors best predict gene-specific measures of sequence evolution using machine-learning based regression modeling. The following features were used in the models: GC%, number of exonic and intronic base pair, number of splice variants, distance to the centromere, effective number of codons, r, μ, distance to the nearest TE, distance to the nearest ACR, ACR%, methylation level, methylation variability, connectivity, expression level, expression variability, and the co-expression module assignment. The R package ranger (Wright and Ziegler 2017) was used to train Extra-Trees (Geurts et al. 2006) forests to estimate the relative importance of each predictor variable. To this end, settings -splitrule “extratrees” -replace = F and -sample.fraction = 1 were used in ranger. Extra-Trees is an extension of the popular ensemble learning method, Random Forest (Breiman 2001), in which a random selection of data is used to train decision trees, and the response variable predicted based on the resulting forest. In contrast to Random Forest, which trains trees on a subset of the learning sample and defines optimal cut-points for each node, Extra-Trees are trained on the whole sample and the cut-points are chosen randomly. This approach generally reduces the risk of overfitting, potentially leading to more accurate prediction (Geurts et al. 2006). Indeed, with our data, Extra-Trees outperformed Random Forest by consistently yielding ∼1.3× more accurate predictions (70% used for training and 30% used for testing). A total of 500 trees were trained in each model, and the best tuning parameters (number of variables split at each node and minimum node size) were chosen based on fivefold cross-validation, conducted with the R package caret (Kuhn 2008). Variable importance was estimated using a corrected Gini importance measure, which is not biased by the number or frequency of categories (Nembrini et al. 2018). Deviations from random expectations were assessed by permuting each predictor variable across genes. However, as training a large number of machine-learning models to estimate accurate permutation P-values is computationally intensive, we used a smaller number of repeats (n = 100) to establish a null distribution for each predictor. These empirical nulls approximately follow a normal distribution (Billingsley 2008), so we used the mean and SD to define P-values with the R function pnorm.
Derived Allele Frequency Estimation
To better understand how nucleotide composition influences selective constraint, we partitioned segregating alleles based on their ancestral vs. derived status. We utilized three species from the family Brassicaceae as outgroups: A. lyrata (Hu et al. 2011), Capsella rubella (Slotte et al. 2013), and Arabis alpina (Willing et al. 2015). The three reference genomes were aligned against the A. thaliana genome with MUMmer4 (Marçais et al. 2018) and variants in regions showing one-to-one alignments in at least two of the three comparisons were used to estimate derived allele frequencies (DAF). First, we used a method by Keightley and Jackson (2018) to infer probabilities for derived alleles based on polymorphism data and the outgroup species. Substitutions were assumed to follow a six-parameter (R6) model, which allows for variable mutation probabilities between different nucleotides (Keightley and Jackson 2018). The uncertainty in the assignment of derived alleles was then directly incorporated into the DAF estimation:
where xAi and xai are the counts of alleles Ai and ai in a site i, and n is the number of segregating sites in a gene. We employed the common division of nucleotides based on their number of hydrogen bonds, strong (S: G or C) and weak (W: A or T), to estimate DAF for classes: WS, SW, and SS+WW (the first letter or each pair corresponds to the ancestral allele and the second letter corresponds to the derived allele). gBGC tends to increase the frequency of derived S alleles and decrease the frequency of derived W alleles, and therefore the relative frequencies of WS, SW, and SS+WW alleles provide insight into the strength of gBGC at different regions of the genome. As a more specific measure of gBGC, we estimated the ratio of WS to SW (WS/SW) for each gene, with estimates >1 indicating an excess of segregating S alleles, potentially caused by gBGC. We note that mutation probabilities in the R6 substitution model are symmetric (e.g., C → T and T → C are represented by a single parameter) and therefore asymmetries in mutation rates, caused e.g., by the hypermutability of methylated cytosines, are not directly accounted for. To examine to what extent such asymmetries might bias the derived-allele probabilities, we used a parsimony-based approach (all three outgroup-species were required to carry the same allele) to estimate mutation proportions between each of the four nucleotides. Our results indicate that C → T (0.18) and G → A (0.18) transitions have been the most common mutations, followed by the opposite T → C (0.11) and A → G (0.11) transitions. By contrast, transversion have been less common and more symmetric (Supplemental Material, Figure S1). The observed transitional asymmetry may therefore cause the derived-allele probabilities to be overestimated at SW sites, leading to an increase of SW alleles being sampled, and underestimated at WS sites, leading to a decrease of WS alleles being sampled. Given that this pattern is opposite to what is expected under gBGC, the asymmetry should, on average, make our results conservative.
Dfe and α
We estimated the distribution of fitness effects (DFE) using DFE-alpha (Keightley and Eyre-Walker 2007). DFE-alpha models the DFE as a gamma distribution governed by the mean strength of selection (Nes) and the shape parameter β. The DFE can range from leptokurtic (L-shaped) to platykurtic (spike-shaped), providing insight into the strength of purifying selection (Eyre-Walker and Keightley 2007). An extension of the McDonald-Kreitman test (McDonald and Kreitman 1991) was used to estimate the rate of positive selection, while taking into account the number of nearly neutral mutations derived from the DFE (Eyre-Walker and Keightley 2009). Here, the ratio of nonsynonymous to synonymous polymorphisms (pN/pS) within species is compared against the ratio of nonsynonymous to synonymous divergence (dN/dS) between species. The proportion of adaptive substitutions is then estimated as: α = 1 – (pN/pS)/(dN/dS), and the rate of adaptive substitutions relative to the neutral mutation rate as: ωA = α(dN/dS).
We used the whole-genome alignments to count the number of 0-fold and fourfold substitutions in A. thaliana. All variant and invariant sites from the aligned regions were then used to estimate unfolded nonsynonymous and synonymous site frequency spectra (SFS), which were subsequently folded by DFE-alpha. The A. thaliana accessions used here are fully homozygous, so we treated them as haploid when estimating the SFS. To account for the uncertainty in the assignment of ancestral vs. derived alleles, which is important for the WS and SW sites, we sampled derived alleles based on their individual probabilities using the same approach as with DAF. To account for nonequilibrium population histories, two-step Ne change was included into the DFE models. Confidence intervals for DFE, α, and ωA were estimated by fitting the models to 500 parametric bootstrap SFS. We assumed that counts in the bootstrap replicates were distributed multinomially, with number of trials corresponding to total number of sites in the SFS and the probability of success corresponding to proportion of sites in a given derived allele group. The bootstrap SFS were generated with the R function rmultinom.
Arabidopsis Lyrata and Capsella Grandiflora
To better assess how selfing affects gBGC, we repeated part of our analyses using whole-genome data from two outcrossing Brassicaceae species, A. lyrata ssp. petraea and Capsella grandiflora. For A. lyrata, we used 21 individuals from Jotunheimen, Norway, published as part of two studies: Mattila et al. (2017) and Hämälä et al. (2018). For C. grandiflora, we used 21 individuals from Zagori, Greece, published by Steige et al. (2017). With the C. grandiflora data, we followed the approach of Steige et al. (2017) and aligned reads against the genome of a recently (<200 KYA, Koenig et al. 2019) diverged species C. rubella, which is more contiguous than the currently available C. grandiflora genome. For estimation of recombination rates, we used linkage maps constructed for both species. The A. lyrata map consists of 1515 markers, genotypes for 354 F2 progeny (Hämälä et al. 2017), and the C. grandiflora map consists of 890 markers, genotyped for 550 F2 progeny (Slotte et al. 2012).
For both A. lyrata and C. grandiflora, low quality reads and sequencing adapters were first removed with Trimmomatic (Bolger et al. 2014) and the surviving reads aligned against their respective reference genomes (A. lyrata v1.0, Hu et al. 2011; C. rubella v1.0, Slotte et al. 2013) with BWA-MEM (Li 2013). SAMtools (Li et al. 2009) was used to sort the alignments and remove duplicated reads. Calling of variant and invariant sites was done with BCFtools (Li 2011), using only reads with mapping quality ≥30 and base quality ≥20. The resulting VCF-files were filtered with the following requirements: site quality ≥20, genotype quality ≥20, read coverage ≥6, and missing data in <20% of individuals. All indels and SNPs with more than two alleles were further removed. For A. lyrata, we retained 110 M (4.9 M variable) out of 150 M sites, and, for C. grandiflora, we retained 100 M (7.3 M variable) out of 120 M sites. These data were used to estimate GC%, πN/πS, DFE, α, and ωA using the same methods as for A. thaliana.
Forward Simulations
The A. thaliana data are derived from accessions that were selfed multiple times between collection and sequencing, so they cannot be used to estimate the expected number of heterozygous sites that are susceptible to gBGC. We therefore conducted forward simulations with SLiM 3 (Haller and Messer 2019) to estimate to what extent gBGC could be expected in natural populations. Selfing in A. thaliana has evolved relatively recently, likely between 500 K and 1 M generations ago (Bechsgaard et al. 2006; Tang et al. 2007; Durvasula et al. 2017). For this reason, we started by establishing a single fully outcrossing population of N = 50,000 individuals, approximately corresponding to twice the current Ne estimate of European A. thaliana (Durvasula et al. 2017). After a burn-in of 10N generations, the population switched to (predominant) selfing, at which time N was reduced to 25,000 (as expected under selfing; Pollak 1987). We considered four rates of outcrossing: 0, 5, 10, and 15%, approximately corresponding to outcrossing rates estimated for natural A. thaliana populations from Germany (Bomblies et al. 2010; Sellinger et al. 2020). Mutation rate (μ) was set to 6.95 × 10−9 (Weng et al. 2019), with nucleotide replacements following a Jukes-Cantor model (Jukes and Cantor 1969) without any GC/AT bias. We considered three crossover rates (r) based on our gene-specific estimates: weak 2.3 × 10−8 (minimum estimate), moderate 4.0 × 10−8 (average estimate), and high 7.3 × 10−8 (maximum estimate). The N, μ, and r parameters were rescaled by a factor 10 to reduce computation time, while retaining the same product of Nμ and Nr as the unscaled data (Kim and Wiehe 2008). Based on Yang et al. (2012), gene conversions were assumed to outnumber crossovers by a factor of 50 and to have an average track length of 553 bp. For parameter governing the GC over AT repair bias, we considered three values: 5, 10, and 20%, approximately corresponding to different levels of bias estimated for A. thaliana (Yang et al. 2012; Wijnker et al. 2013; Liu et al. 2018). In total, we simulated 100 × 50 kb regions with each parameter combination. Following the switch to selfing, simulations were run 100 K generations (1 M/10, the rescaling parameter), during which the extent of gBGC was defined by estimating WS/SW and GC% as with the observed data. We note that the efficacy of gBGC can be dependent on the effective population size (Duret and Galtier 2009), so the rescaling of N, μ, and r might diminish the effects of gBGC. However, by conducting a subset of the simulations with unscaled parameters, we found that although rescaling has a slight effect on the absolute values of WS/SW and GC%, it does not influence the relative patterns arising from selfing (Figure S2). Therefore, conclusions drawn from these results should not be greatly affected by the parameter-scaling.
Data Availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental figures and tables, a compiled table of genomic features, and a code for running the simulations are available at figshare: https://doi.org/10.25386/genetics.12284174.
Results
Genomic Features Influence Selective Constraint
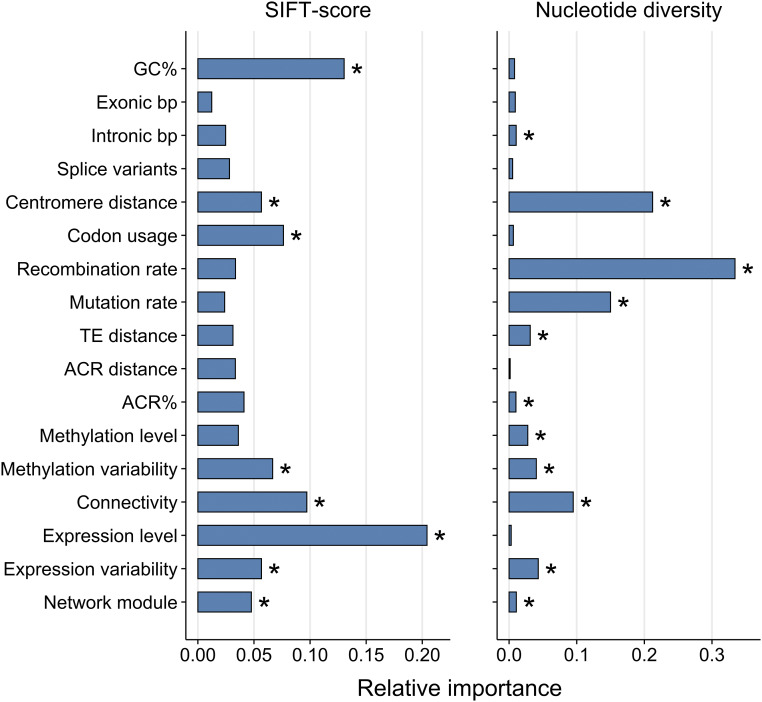
We used machine-learning based modeling to examine how genomic features influence the accumulation of deleterious mutations at different regions of the A. thaliana genome. To this end, we estimated average SIFT-scores for 24,855 protein-coding genes, of which 18,070 had complete data for the 16 features used as predictors in our Extra-Trees models (Figure S3). The two other statistics reflecting the strength of purifying selection, dN/dS and πN/πS, produced similar results as SIFT (Figure S4 and Table S1), so here we focus on identifying factors affecting SIFT-scores. Multiple features had an influence on SIFT-scores (Figure 1), with expression level and GC% having largest effects. Both features were positively correlated with SIFT-scores (Table 1), indicating that mutations segregating at >0.01 frequency in highly expressed genes and GC-rich genes were less deleterious than the genome-wide average. Genes with greater connectivity in a co-expression network also had higher than average SIFT-scores, whereas increase in the effective number of codons (i.e., lower codon-usage bias, Novembre 2002), variability in gene-body methylation, and distance from the centromere had the opposite effects (Figure 1 and Table 1). An additional important factor was the module assignment from a co-expression network (30 modules in total), indicating that genes from each module have more similar SIFT-scores than expected by chance. This similarity suggests that genes within the modules respond to correlated selection pressures, possibly due to shared biological function (Hämälä et al. 2020). Interestingly, effect of recombination rate on average SIFT-scores was minor and did not exceed values from random permutations. This pattern is in stark contrast to nucleotide diversity, for which recombination rate and the distance from the centromere (arguably, a proxy for recombination rate) were clearly the best predictors, while expression level and GC% were of minor importance (Figure 1 and Table 1).
Figure 1.
Variable importance from Extra-Trees models for SIFT-scores and nucleotide diversity. *P < 0.05 (Bonferroni corrected).
Table 1. Spearman’s rank correlation between genomic features and two measures of sequence evolution.
| Feature | SIFT-score | Nucleotide diversity | ||
|---|---|---|---|---|
| Pairwise | Partiala | Pairwise | Partiala | |
| GC% | 0.17* | 0.10* | −0.05* | −0.03* |
| Exonic bp | 0.01 | ∼0 | ∼0 | −0.03* |
| Intronic bp | −0.01 | ∼0 | −0.08* | −0.01 |
| Splice variants | −0.05* | −0.04* | −0.01 | ∼0 |
| Centromere distance | −0.06* | −0.05* | −0.27* | −0.11* |
| Codon usage | −0.08* | −0.03* | ∼0 | 0.03* |
| Recombination rate | 0.05* | 0.02* | 0.29* | 0.13* |
| Mutation rate | 0.01 | ∼0 | 0.10* | 0.04* |
| TE distance | −0.02* | ∼0 | −0.14* | −0.02 |
| ACR distanceb | −0.09* | 0.01 | −0.10* | ∼0 |
| ACR%b | 0.11* | 0.01 | 0.11* | 0.04* |
| Methylation level | −0.02 | ∼0 | −0.05* | 0.06* |
| Methylation variability | 0.01 | 0.01 | 0.12* | 0.08* |
| Connectivity | 0.12* | 0.02 | −0.17* | −0.12* |
| Expression level | 0.25* | 0.16* | −0.06* | −0.06* |
| Expression variability | −0.10 | −0.04* | 0.18* | 0.09* |
Partial correlation after controlling for all other features (Kim 2015).
Accessible chromatin region.
P < 0.05 (Bonferroni corrected).
Gc-Content is a Major Predictor of Selective Constraint
The positive association between expression level and the strength of purifying selection (Table 1) is well-established in multiple taxa (Koonin 2011). By contrast, the relationship between nucleotide composition and selection is less explored, particularly in plants (Glémin et al. 2014). We therefore conducted analyses to identify factors that might explain the relationship between GC% and the accumulation of deleterious mutations. In mammals, GC% is positively correlated with recombination rate, which is thought to arise from gBGC increasing the fixation probability of GC alleles in regions of high recombination (Duret and Galtier 2009). gBGC also can drive the spread of deleterious AT → GC alleles and inhibit the spread of deleterious GC → AT alleles. If gBGC is acting in A. thaliana, we would expect a positive correlation between GC% and recombination rate. However, like previous studies in A. thaliana (Giraut et al. 2011; Wijnker et al. 2013), we found this correlation to be negative (Spearman’s ρ = −0.12, P < 2 × 10−16). The Extra-Trees model also revealed that recombination rate is of minor importance in explaining variation in GC% (Table S2), being far less important than methylation variability, methylation level, expression level, codon usage, and intron length.
The minor importance of recombination rate in explaining GC% suggests that gBGC may not have an important effect on nucleotide composition in A. thaliana. However, it also is possible that GC% is a poor proxy for gBGC in predominantly selfing species. For this reason, we estimated DAF for each of three groups: WS (ancestral allele A or T, derived allele G or C; AT → GC), SW (ancestral allele G or C, derived allele A or T; GC → AT), and WW+SS (ancestral and derived A or T, and ancestral and derived G or C; AT → AT and GC → GC). If gBGC is affecting nucleotide composition in A. thaliana, we would expect gBGC to contribute to the spread of WS alleles, inhibit the spread of SW alleles, and not affect the evolution of WW+SS alleles. Consistent with this notion, we found that average DAF was highest for WS alleles (DAF = 0.10), lowest for SW alleles (DAF = 0.08), and intermediate for WW+SS alleles (DAF = 0.09; P < 2 × 10−16, Wilcoxon rank-sum test). Moreover, by estimating synonymous Tajima’s D (Tajima 1989) for sites with derived-allele probability >0.8, we found that, compared to the unbiased WW+SS sites (D = −0.95, 95% CI: −0.96 to −0.94), the SFS was shifted toward common variants at WS sites (D = −0.84, 95% CI: −0.85 to −0.83) and shifted toward rare variants at SW sites (D = −1.00, 95% CI: −1.01 to −0.99); a pattern indicative of gBGC (Lachance and Tishkoff 2014). The frequency of segregating alleles thus suggests that gBGC may shape nucleotide variation in A. thaliana despite it only having a minor role in the genome-wide GC%, which is more strongly affected by factors such as gene-body methylation, expression level, and gene structure (Table S2).
gBGC Affects the Efficacy of Purifying Selection
To assess whether gBGC can lead to an accumulation of deleterious mutations in A. thaliana, we examined the relationship between DAF and SIFT-scores. Overall, there was a positive correlation between the two measures (Figure 2A), indicating that high frequency derived alleles have, on average, lower negative impact on fitness. However, by comparing the DAF at each of the three allelic classes, we saw that the frequency of SW alleles was a better predictor of SIFT-scores than the frequency of either WS or WW+SS alleles (Figure 2A). The correlation between SW-allele frequency and SIFT-scores also was more highly positive than in the other DAF classes, indicating that genes with predominantly strong ancestral alleles tend to have segregating mutations that are less harmful. gBGC could reduce the impact of segregating mutations at these genes by decreasing the frequency of derived alleles, most of which are deleterious (Eyre-Walker and Keightley 2007). Under this model, the opposite pattern is expected at genes with predominantly weak ancestral alleles, as gBGC can increase the frequency of slightly deleterious mutations (Bengtsson 1990; Glémin 2010).
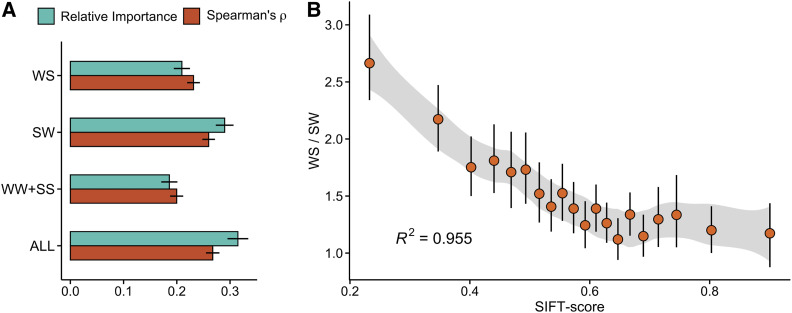
Figure 2.
The effect of gBGC on selective constraint. (A) Importance from an Extra-Trees model with SIFT-scores as the response and the four DAF classes as predictors, and Spearman’s rank-correlation ρ between SIFT-scores and DAF classes. Error bars show 95% CIs. (B) Relationship between WS/SW, a measure of gBGC, and SIFT-scores. Data were split into 20 bins of equal size based on their SIFT-scores. Figure shows means (circles) and 95% CIs (error bars) estimated for each bin. Also shown are 95% CI and R2 for a loess-model (shaded area) fit on the binned data.
To test this hypothesis, we examined the relationship between WS/SW and SIFT-scores. We found a clear negative trend between the two measures; genes with low average SIFT-scores (i.e., more harmful mutations) had a greater excess of derived S alleles (Figure 2B; similar associations were also found for dN/dS and πN/πS, Figure S5). Although the derived allele probabilities at WS and SW sites are likely influenced by asymmetric mutation rates (Figure S1), on average this bias would increase SW alleles and decrease WS alleles, making the trend observed here conservative. We further found that mutation rate and WS/SW are not correlated (Spearman’s ρ ≈ 0), indicating that variation in WS/SW is not driven by mutation bias. On the other hand, these results might be affected by the presence of methylated cytosines, which have the highest SW mutation rates in the A. thaliana genome (Weng et al. 2019). Polarization errors at such sites are more likely, which could lead to an apparent excess of high frequency WS mutations (Glémin et al. 2015), and thus inflate the WS/SW at genes with more hypermutable sites. To address this potential issue, we fit the following linear model to the data: SIFT-score = WS/SW + mC%, where mC% is the average density of methylated cytosines within a gene. The model showed that mC% has little effect on the association between WS/SW and SIFT-scores (without mC% as a cofactor: βWS/SW = −0.087, with mC% as a cofactor: βWS/SW = −0.085; P < 2 × 10−16 for both), indicating that the signal of gBGC is not biased by hypermutable sites. Our results are therefore consistent with gBGC preventing the purging of deleterious mutations at genes with predominantly weak ancestral alleles, while facilitating their removal at genes with predominantly strong ancestral alleles. In fact, including WS/SW into our Extra-Trees model revealed that it is among the best predictors of SIFT-scores, exceeded only by expression level and GC% (relative importance: expression level = 0.19, GC% = 0.12, WS/SW = 0.11). These results lead us to conclude that gBGC is strong enough in A. thaliana to influence the efficacy of purifying selection.
gBGC Leads to a Signal of Relaxed Selection in A. Thaliana
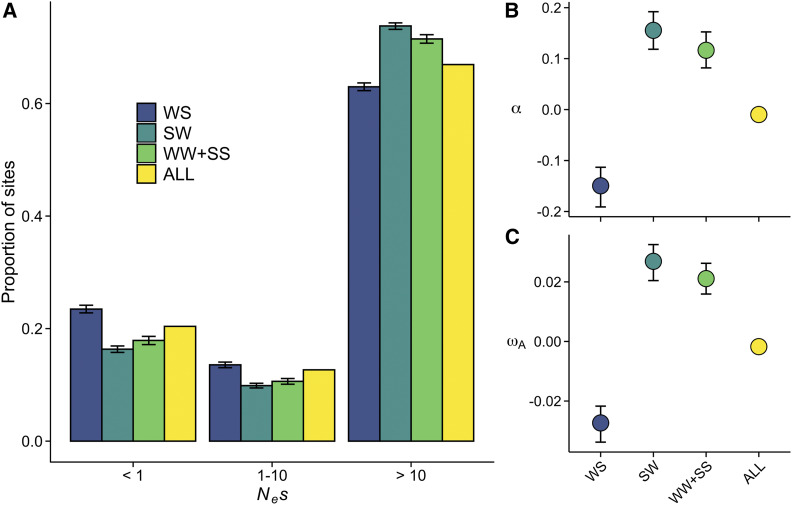
A characteristic feature of gBGC is that the increased frequency of S alleles and the decreased frequency of W alleles may give the appearance of selection (Galtier and Duret 2007). We therefore examined whether gBGC in A. thaliana is prominent enough to alter the estimates of selection. DFE estimated for different DAF classes revealed that WS had more, and SW fewer, nonsynonymous sites in the nearly neutral category (Nes < 1) than the genome-wide average, indicating relaxed purifying selection at WS sites and stronger than average selective constraint at SW sites (Figure 3A). Consistent with previous estimates for A. thaliana (Fay 2011; Slotte et al. 2011; Gossmann et al. 2012), the genome-wide α and ωA were close to zero, suggesting that positive selection has little effect on shaping nucleotide diversity in A. thaliana. By contrast, the α and ωA estimates for WS sites were clearly negative, and the estimates for SW sites were clearly positive (Figure 3, B and C). These results give the appearance of WS sites evolving slower than average rate and SW sites evolving faster than average rate. The DFE, α, and ωA results stay unchanged when sites that are most susceptible to gene-body methylation (mCG) were removed (Figure S6), indicating that hypermutable sites have little effect on the estimates of selection. Moreover, by conducting this analysis separately for four largest admixture groups defined by The 1001 Genomes Consortium (2016), we confirmed that our results are not biased by population structure, as each group showed patterns similar to those of the complete dataset (Figure S7).
Figure 3.
Apparent strength of negative and positive selection in A. thaliana (n = 645). (A) The distribution of fitness effects (DFE). Nonsynonymous sites were divided into three bins based on the strength of purifying selection (Nes): nearly neutral, intermediate, and highly deleterious, respectively. (B) The proportion of sites fixed by positive selection (α). (C) The rate of adaptive substitutions relative to the neutral mutation rate (ωA). For all three figures, error bars show 95% CIs (CIs are too narrow to show for ALL-sites).
The α and ωA are based on the ratios of nonsynonymous to synonymous divergence (dN/dS) and nonsynonymous to synonymous polymorphisms (pN/pS). In general, positive estimates are the result of dN/dS exceeding pN/pS, whereas the reverse is true for negative estimates. The contrasting estimates at WS and SW sites can therefore arise if gBGC has different effects on divergence and polymorphism rates at these sites. We found that, compared to the unbiased WW+SS sites, both dN/dS and pN/pS (excluding polymorphisms with frequency <15%; Charlesworth and Eyre-Walker 2008) were increased at WS sites and decreased at SW sites (Figure S8). However, the difference between the pN/pS estimates (WS = 0.22, SW = 0.14) was greater than between the dN/dS estimates (WS = 0.21, SW = 0.17), consistent with the notion that gBGC can prevent the removal of slightly deleterious polymorphisms at WS sites, while facilitating their removal at SW sites. Overall, our results suggest that gBGC contributes to the signal of decreased selection-efficacy in A. thaliana (genome-wide α and ωA ≈ 0). More accurate estimates of selection may be obtained by examining WW+SS sites, which should not be affected by gBGC. Estimates of α and ωA at WW+SS sites were clearly greater than zero (α = 0.12, 95% CI: 0.09 to 0.14; ωA = 0.022, 95% CI: 0.017 to 0.025), suggesting that positive selection has been more important in shaping nucleotide diversity in A. thaliana than previously thought.
Evidence of gBGC in Outcrossing Relatives A. Lyrata and C. Grandiflora
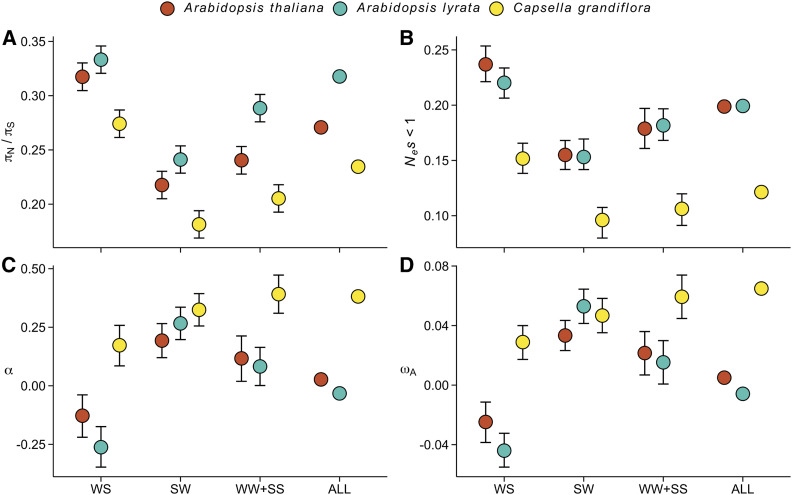
To examine the role of selfing in gBGC, we estimated πN/πS, DFE, α, and ωA for two related outcrossing species, A. lyrata and C. grandiflora. We used population data originating from Norway (A. lyrata, n = 21) and Greece (C. grandiflora, n = 21) for the two species. All else being equal, the increased heterozygosity due to outcrossing should result in stronger footprints of gBGC in A. lyrata and C. grandiflora than in A. thaliana. We note, however, that the two outcrossing species have very different demographic histories, with population size decline in A. lyrata (current Ne < 10 K, Hämälä et al. 2018; Hämälä and Savolainen 2019; Mattila et al. 2019) and population size increase in C. grandiflora (current Ne > 500 K, Douglas et al. 2015; Mattila et al. 2019). To test for an effect of the mating-system, we compared results from A. lyrata and C. grandiflora to a set of 42 A. thaliana individuals from Germany, yielding the same number of sampled chromosomes (due to full homozygosity) as the outcrossing species (note that estimates in Figure 3 are based on all 645 individuals).
Unlike in A. thaliana, we found a positive correlation between GC% and recombination rate in both A. lyrata (Spearman’s ρ = 0.09, P < 2 × 10−16) and C. grandiflora (Spearman’s ρ = 0.07, P = 2 × 10−16), suggesting that gBGC may more strongly increase the fixation probability of GC alleles in these species. Patterns of πN/πS (Figure 4A) and DFE (Figure 4B) were similar in the two outcrossing species and A. thaliana, with a signal of relaxed purifying selection at WS sites and a signal of stronger than average selective constraint at SW sites. The genome-wide estimates of α (Figure 4C) and ωA (Figure 4D) supported previous findings by showing weak signs of positive selection in A. lyrata (Gossmann et al. 2010; Mattila et al. 2019) and strong signs in C. grandiflora (Slotte et al. 2010; Williamson et al. 2014). The estimates of α and ωA for each of the DAF classes were similar in the two Arabidopsis species; compared to the genome-wide average, measures of positive selection were lower for WS sites and higher for SW sites. The unbiased WW+SS sites also had higher α and ωA than the genome-wide average. In contrast to the Arabidopsis species, α and ωA estimates showed weaker footprints (smaller deviations of WS and SW sites from the overall average) of gBGC in C. grandiflora. Overall, our results suggest that gBGC leads to a signal of relaxed efficacy of selection, particularly in the small-Ne species A. thaliana and A. lyrata. However, contrary to our initial expectation, the intensity of the signal was not stronger in the outcrossing species.
Figure 4.
The effect of gBGC on measures of protein evolution in three Brassicaceae species. Analyses were conducted using the same number of chromosomes (n = 42) from each species. (A) The ratio of nonsynonymous to synonymous nucleotide diversities (πN/πS). (B) The proportion of nearly neutral mutations (Nes < 1). (C) The proportion of sites fixed by positive selection (α). (D) The rate of adaptive substitutions relative to the neutral mutation rate (ωA). For all four figures, error bars show 95% CIs.
gBGC is expected in A. thaliana despite selfing
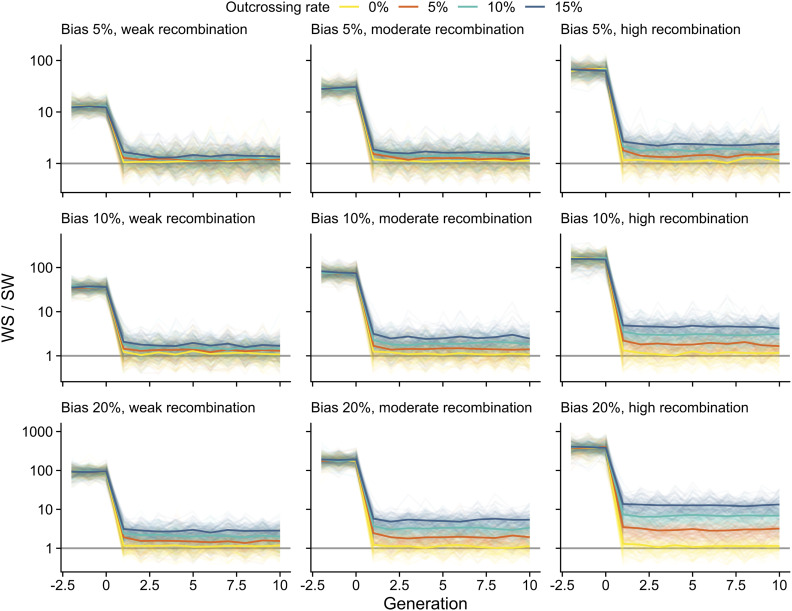
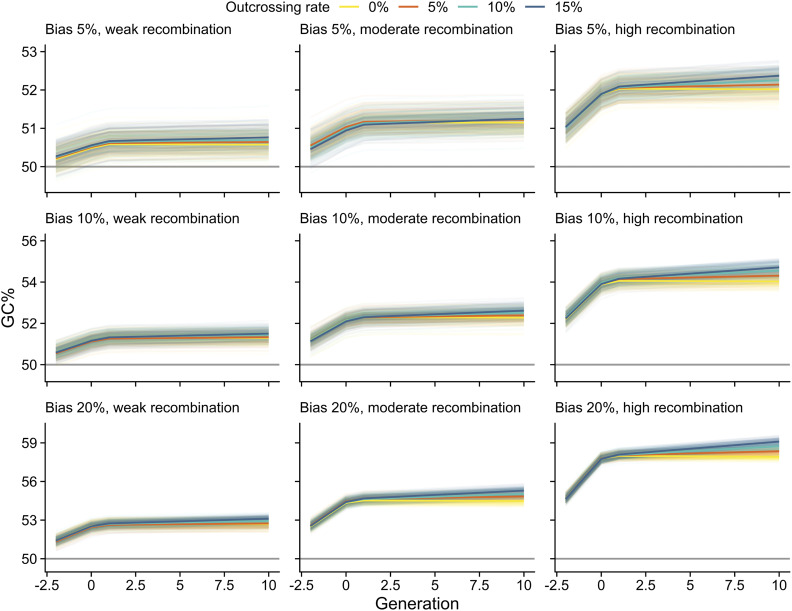
Contrary to the view that gBGC should be weak or absent in predominantly selfing species (Marais et al. 2004; Glémin 2010), our results suggest that it has a sizeable effect on sequence variation in A. thaliana. This seemingly unexpected result could arise from residual effects of recent outcrossing, and/or if natural levels of continuing outcrossing are high enough to facilitate gBGC. To evaluate this, we used forward simulations to explore the effects of a recent mating-system shift and low outcrossing rates on signals of gBGC. These simulations revealed that both WS/SW and GC% were affected by the switch to selfing; WS/SW rapidly dropped from initial high values and the increase of GC% slowed shortly after the end of full outcrossing. As expected, stronger repair bias and higher recombination rate led to clearer footprints of gBGC (Figure 5 and Figure 6).
Figure 5.
The simulated extent of WS/SW under gBGC. Time in number of generations is shown in the horizontal axes (×104 scaled, ×105 unscaled). At time zero, population switches from full outcrossing to predominant selfing. WS/SW estimates are shown for three levels of GC over AT repair bias, three recombination rates, and four outcrossing rates. Solid colors show average estimates. Results from each of 100 simulations are shown in transparent colors. Gray horizontal lines mark the expected estimates in the absence of gBGC.
Figure 6.
The simulated extent of GC% under gBGC. Time in number of generations is shown in the horizontal axes (×104 scaled, ×105 unscaled). At time zero, population switches from full outcrossing to predominant selfing. GC% estimates are shown for three levels of GC over AT repair bias, three recombination rates, and four outcrossing rates. Solid colors show average estimates. Results from each of 100 simulations are shown in transparent colors. Gray horizontal lines mark the expected estimates in the absence of gBGC.
The composition of segregating alleles, as measured by WS/SW, was highly sensitive to the mating-system shift, and 100 K generations of selfing was enough to remove any residual effects of full outcrossing. However, despite the radical drop in WS/SW after the mating-system shift, equilibrium values were clearly affected even by low levels of ongoing outcrossing. Under complete selfing, gBGC had little influence on segregating alleles, but the effects increased rapidly with increasing outcrossing rate. At 5% outcrossing, close to an average estimated by Bomblies et al. (2010), WS/SW estimates deviated from the expected values under most parameter combinations, and at 15% outcrossing, close to an estimate by Sellinger et al. (2020), WS/SW estimates were clearly higher than in the absence of gBGC (Figure 5). The increase in GC% that was driven by gBGC during outcrossing slowed sharply, but did not decrease, following the transition to selfing. After the mating-system shift, the ongoing outcrossing had little effect on GC% during the 1 M generations examined here (Figure 6). Our simulations are therefore consistent with continuing effects of gBGC on segregating sites (WS/SW) in A. thaliana, whereas fixed sites (GC%) may mostly reflect patterns established during the recent outcrossing.
Discussion
By using whole-genome, transcriptome, and methylome data from A. thaliana, we have gained new insights into factors influencing selective constraint in this model-species. As expected, segregating harmful mutations are not uniformly distributed among genes, but are influenced by variation in features such as expression level, connectivity, codon usage bias, and gene-body methylation. Interestingly, we found strong evidence that GC-biased gene conversion (gBGC) shapes sequence variation in A. thaliana, despite the high rate of self-fertilization. As gBGC is expected to be weak or absent in selfing species (Marais et al. 2004; Glémin 2010), we conducted simulations to assess whether natural levels of outcrossing in A. thaliana can facilitate gBGC. Our results are consistent with previous findings in showing that gBGC may be undetectable under complete selfing. However, even relatively weak outcrossing of ∼5% could result in noticeable effect, suggesting that the mating-system does not prevent gBGC from shaping nucleotide variation in predominantly selfing species such as A. thaliana.
A general pattern observed in mammals, birds, yeast, and grasses is that GC-content (GC%) is positively correlated with recombination rate (Eyre-Walker 1993; Duret and Galtier 2009; Pessia et al. 2012; Glémin et al. 2014; Mugal et al. 2015). Here, we also found such a correlation in the outcrossing species A. lyrata and C. grandiflora. This correlation is thought to arise from gBGC, as the frequency of derived GC alleles is more strongly increased in regions of high recombination (Eyre-Walker 1993; Marais 2003). A lack of positive correlation between GC% and recombination, as in A. thaliana, has been interpreted as evidence against gBGC (Marais et al. 2004; Pessia et al. 2012). However, there is direct evidence for GC-biased gene conversion in A. thaliana (Yang et al. 2012), and population genetic analyses have suggested that this leads to noticeable genome-wide effects on nucleotide variation (Cao et al. 2011; Günther et al. 2013). Why then does recombination rate appear to be weakly correlated with the genomic signatures of gBGC in A. thaliana? Although our simulations suggest that the residual effects of outcrossing could still be seen in GC%, this association only holds if the recombination landscape has remained largely unchanged since A. thaliana switched to selfing. Given the variable nature of the recombination landscape (Stapley et al. 2017; Lloyd et al. 2018), this condition could be easily violated. We further found that GC% is strongly influenced by many factors, potentially confounding the signal arising from recombination and gBGC. Our results therefore suggest that the correlation between GC% and recombination rate is not an appropriate proxy for gBGC in all organisms.
Theory predicts that gBGC increases genetic load by driving the frequencies of slightly deleterious mutations (Bengtsson 1990; Glémin 2010). Support for this model has been found in humans, where S alleles at disease causing loci tend to segregate at a higher than expected frequency (Necşulea et al. 2011; Lachance and Tishkoff 2014). By predicting mutational effects with SIFT4G (Vaser et al. 2016), we found that gBGC is associated with selective constraint in A. thaliana. Genes with low average SIFT-scores, indicative of increased density of segregating harmful mutations, had a greater excess of derived S alleles (high WS/SW). The observed pattern is consistent with gBGC preventing slightly deleterious mutations from being purged by purifying selection, potentially leading to increased genetic load at genes with predominantly weak (A,T) ancestral alleles. However, our results also indicate that genes with predominantly strong (G,C) ancestral alleles may have decreased genetic load, because gBGC can facilitate the removal of new mutations, most of which are deleterious (Eyre-Walker and Keightley 2007).
Consistent with studies in mammals and birds (Galtier and Duret 2007; Galtier et al. 2009; Ratnakumar et al. 2010; Corcoran et al. 2017; Bolívar et al. 2018; Rousselle et al. 2019), we found evidence that gBGC influences the rate of protein evolution in A. thaliana, thus affecting inferences of selection that are based on patterns of nucleotide divergence and diversity. However, the increased frequency of S alleles did not lead to a false signal of positive selection, but it appears that gBGC mainly masks selection in A. thaliana. By comparing results from A. thaliana to outcrossing relatives A. lyrata and C. grandiflora, we found that measures of negative and positive selection were similarly affected in the two Arabidopsis species, both with small Ne, and more weakly affected in C. grandiflora, a species with large Ne. Although gBGC is expected to be more efficient in species with large Ne (Duret and Galtier 2009; Glémin 2010; Mugal et al. 2015), the association between the two parameters is not always monotonous (Galtier et al. 2009, 2018), particularly in protein-coding genes at which large-Ne species may be more efficient at counteracting the harmful effects of gBGC. The strength of gBGC has been decoupled from Ne in other species groups (Clément et al. 2017; Galtier et al. 2018). For instance, strong evidence of gBGC was found in the small-Ne species honey bee (Wallberg et al. 2015), whereas only weak evidence has been found in the large-Ne species Drosophila melanogaster (Galtier et al. 2006; Williamson et al. 2014). Together, these results suggest that the effects of gBGC on protein evolution cannot be simply predicted based on the mating-system and Ne. This complicates the inference of gBGC, given that estimates of the underlying parameters (gene conversion rate, track length, and repair bias) are available only for a handful of well-studies species (Galtier et al. 2018). In any case, we found that gBGC results in the proportion of sites fixed by positive selection to be underestimated, at least in A. thaliana and A. lyrata. For these species, the unbiased WW+SS sites could more accurately reflect the rate of adaptive evolution. This puts the α estimate for A. thaliana (α = 0.12) in level with some other small-Ne species, such as humans (α = 0.14, Uricchio et al. 2019), but still considerably lower than estimates for large-Ne species, such as D. melanogaster (α > 0.5, Kousathanas and Keightley 2013).
Conclusions
We have shown that selective constraint is modulated by multiple genomic features in A. thaliana, with expression level, GC-content, and connectivity being the most influential. Importantly, our analyses provide strong evidence that gBGC shapes protein evolution in A. thaliana, despite the predominantly selfing mating-system. Both the increasing frequency of S alleles and the decreasing frequency of W alleles influence segregating deleterious mutations, while leading to a genome-wide signal of reduced selection-efficacy. By finding evidence that even weak outcrossing can facilitate gBGC, we have shown that it has more far-reaching consequences than previously appreciated. Based on these results, we propose that the importance of accounting for gBGC in genomic analyses should not be limited to outcrossing taxa, but it ought to be done regardless of the mating system.
Acknowledgments
We thank the associate editor, S.I. Wright, and three anonymous reviewers for their comments on improving the manuscript. Computational resources were provided by the Minnesota Supercomputing Institute (MSI) at the University of Minnesota. This work was supported by the National Science Foundation (NSF) grant IOS-1546863. Any opinions, findings, conclusions, or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the NSF.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12284174.
Communicating editor: S. Wright
Literature Cited
- 1001 Genomes Consortium 2016. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491. 10.1016/j.cell.2016.05.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A. F., and Whitlock M. C., 2012. Mutation load: the fitness of individuals in populations where deleterious alleles are abundant. Annu. Rev. Ecol. Evol. Syst. 43: 115–135. 10.1146/annurev-ecolsys-110411-160257 [DOI] [Google Scholar]
- Bechsgaard J. S., Castric V., Charlesworth D., Vekemans X., and Schierup M. H., 2006. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-Haplotypes over 10 Myr. Mol. Biol. Evol. 23: 1741–1750. 10.1093/molbev/msl042 [DOI] [PubMed] [Google Scholar]
- Bengtsson B. O., 1990. The effect of biased conversion on the mutation load. Genet. Res. 55: 183–187. 10.1017/S0016672300025519 [DOI] [PubMed] [Google Scholar]
- Billingsley P., 2008. Probability and Measure. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- Bird A. P., 1980. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 8: 1499–1504. 10.1093/nar/8.7.1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., and Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolívar P., Mugal C. F., Rossi M., Nater A., Wang M. et al. , 2018. Biased inference of selection due to GC-biased gene conversion and the rate of protein evolution in flycatchers when accounting for it. Mol. Biol. Evol. 35: 2475–2486. 10.1093/molbev/msy149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K., Yant L., Laitinen R. A., Kim S. T., Hollister J. D. et al. , 2010. Local-scale patterns of genetic variability, outcrossing, and spatial structure in natural stands of Arabidopsis thaliana. PLoS Genet. 6: e1000890 10.1371/journal.pgen.1000890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L., 2001. Random forests. Mach. Learn. 45: 5–32. 10.1023/A:1010933404324 [DOI] [Google Scholar]
- Browning B. L., Zhou Y., and Browning S. R., 2018. A one-penny imputed genome from next-generation reference panels. Am. J. Hum. Genet. 103: 338–348. 10.1016/j.ajhg.2018.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C. D., Nielsen R., Sawyer S. A., Olsen K. M., Purugganan M. D. et al. , 2002. The cost of inbreeding in Arabidopsis. Nature 416: 531–534. 10.1038/416531a [DOI] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J. et al. , 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Schneeberger K., Ossowski S., Günther T., Bender S. et al. , 2011. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43: 956–963. 10.1038/ng.911 [DOI] [PubMed] [Google Scholar]
- Charif D., and Lobry J. R., 2007. SeqinR 1.0–2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis, pp. 207–232 in Structural Approaches to Sequence Evolution. Springer, Berlin, Heidelberg. [Google Scholar]
- Charlesworth B., and Charlesworth D., 1998. Some evolutionary consequences of deleterious mutations. Genetica 102–103: 3–19. 10.1023/A:1017066304739 [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Morgan M. T., and Charlesworth B., 1993. Mutation accumulation in finite outbreeding and inbreeding populations. Genet. Res. 61: 39–56. 10.1017/S0016672300031086 [DOI] [Google Scholar]
- Charlesworth J., and Eyre-Walker A., 2008. The McDonald-Kreitman test and slightly deleterious mutations. Mol. Biol. Evol. 25: 1007–1015. 10.1093/molbev/msn005 [DOI] [PubMed] [Google Scholar]
- Chen J., Glémin S., and Lascoux M., 2017. Genetic diversity and the efficacy of purifying selection across plant and animal species. Mol. Biol. Evol. 34: 1417–1428. 10.1093/molbev/msx088 [DOI] [PubMed] [Google Scholar]
- Chun S., and Fay J. C., 2011. Evidence for hitchhiking of deleterious mutations within the human genome. PLoS Genet. 7: e1002240 10.1371/journal.pgen.1002240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément Y., Sarah G., Holtz Y., Homa F., Pointet S. et al. , 2017. Evolutionary forces affecting synonymous variations in plant genomes. PLoS Genet. 13: e1006799 10.1371/journal.pgen.1006799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran P., Gossmann T. I., Barton H. J., Slate J., Zeng K. et al. , 2017. Determinants of the efficacy of natural selection on coding and noncoding variability in two passerine species. Genome Biol. Evol. 9: 2987–3007. 10.1093/gbe/evx213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J. F., 1970. Genetic loads and the cost of natural selection , pp 128–177 in Mathematical Topics in Population Genetics. Springer, Berlin, Heidelberg: 10.1007/978-3-642-46244-3_5 [DOI] [Google Scholar]
- Cutter A. D., and Payseur B. A., 2013. Genomic signatures of selection at linked sites: unifying the disparity among species. Nat. Rev. Genet. 14: 262–274. 10.1038/nrg3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E. et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas G. M., Gos G., Steige K. A., Salcedo A., Holm K. et al. , 2015. Hybrid origins and the earliest stages of diploidization in the highly successful recent polyploid Capsella bursa-pastoris. Proc. Natl. Acad. Sci. USA 112: 2806–2811. 10.1073/pnas.1412277112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., and Galtier N., 2009. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu. Rev. Genomics Hum. Genet. 10: 285–311. 10.1146/annurev-genom-082908-150001 [DOI] [PubMed] [Google Scholar]
- Durvasula A., Fulgione A., Gutaker R. M., Alacakaptan S. I., Flood P. J. et al. , 2017. African genomes illuminate the early history and transition to selfing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 114: 5213–5218. 10.1073/pnas.1616736114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., 1993. Recombination and mammalian genome evolution. Proc. Biol. Sci. 252: 237–243. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., and Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618. 10.1038/nrg2146 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., and Keightley P. D., 2009. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol. Biol. Evol. 26: 2097–2108. 10.1093/molbev/msp119 [DOI] [PubMed] [Google Scholar]
- Fay J. C., 2011. Weighing the evidence for adaptation at the molecular level. Trends Genet. 27: 343–349. 10.1016/j.tig.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J., 1974. The evolutionary advantage of recombination. Genetics 78: 737–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H. B., Hirsh A. E., Steinmetz L. M., Scharfe C., and Feldman M. W., 2002. Evolutionary rate in the protein interaction network. Science 296: 750–752. 10.1126/science.1068696 [DOI] [PubMed] [Google Scholar]
- Galtier N., and Duret L., 2007. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 23: 273–277. 10.1016/j.tig.2007.03.011 [DOI] [PubMed] [Google Scholar]
- Galtier N., Piganeau G., Mounchiroud D., and Duret L., 2001. GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics 159: 907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N., Bazin E., and Bierne N., 2006. GC-biased segregation of noncoding polymorphisms in Drosophila. Genetics 172: 221–228. 10.1534/genetics.105.046524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N., Duret L., Glémin S., and Ranwez V., 2009. GC-biased gene conversion promotes the fixation of deleterious amino acid changes in primates. Trends Genet. 25: 1–5. 10.1016/j.tig.2008.10.011 [DOI] [PubMed] [Google Scholar]
- Galtier N., Roux C., Rousselle M., Romiguier J., Figuet E. et al. , 2018. Codon usage bias in animals: disentangling the effects of natural selection, effective population size, and GC-biased gene conversion. Mol. Biol. Evol. 35: 1092–1103. 10.1093/molbev/msy015 [DOI] [PubMed] [Google Scholar]
- Geurts P., Ernst D., and Wehenkel L., 2006. Extremely randomized trees. Mach. Learn. 63: 3–42. 10.1007/s10994-006-6226-1 [DOI] [Google Scholar]
- Giraut L., Falque M., Drouaud J., Pereira L., Martin O. C. et al. , 2011. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 7: e1002354 10.1371/journal.pgen.1002354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S., 2010. Surprising fitness consequences of GC-biased gene conversion: I. Mutation load and inbreeding depression. Genetics 185: 939–959 [corrigenda: Genetics 190: 1585 (2012)]. 10.1534/genetics.110.116368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S., Clément Y., David J., and Ressayre A., 2014. GC content evolution in coding regions of angiosperm genomes: a unifying hypothesis. Trends Genet. 30: 263–270. 10.1016/j.tig.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Glémin S., Arndt P. F., Messer P. W., Petrov D., Galtier N. et al. , 2015. Quantification of GC-biased gene conversion in the human genome. Genome Res. 25: 1215–1228. 10.1101/gr.185488.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossmann T. I., Song B. H., Windsor A. J., Mitchell-Olds T., Dixon C. J. et al. , 2010. Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol. Biol. Evol. 27: 1822–1832. 10.1093/molbev/msq079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossmann T. I., Keightley P. D., and Eyre-Walker A., 2012. The effect of variation in the effective population size on the rate of adaptive molecular evolution in eukaryotes. Genome Biol. Evol. 4: 658–667. 10.1093/gbe/evs027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther T., Lampei C., and Schmid K. J., 2013. Mutational bias and gene conversion affect the intraspecific nitrogen stoichiometry of the Arabidopsis thaliana transcriptome. Mol. Biol. Evol. 30: 561–568. 10.1093/molbev/mss249 [DOI] [PubMed] [Google Scholar]
- Haldane J. B. S., 1937. The effect of variation on fitness. Am. Nat. 71: 337–349. 10.1086/280722 [DOI] [Google Scholar]
- Haller B. C., and Messer P. W., 2019. SLiM 3: forward genetic simulations beyond the Wright–Fisher model. Mol. Biol. Evol. 36: 632–637. 10.1093/molbev/msy228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hämälä T., and Savolainen O., 2019. Genomic patterns of local adaptation under gene flow in Arabidopsis lyrata. Mol. Biol. Evol. 36: 2557–2571. 10.1093/molbev/msz149 [DOI] [PubMed] [Google Scholar]
- Hämälä T., Mattila T. M., Leinonen P. H., Kuittinen H., and Savolainen O., 2017. Role of seed germination in adaptation and reproductive isolation in Arabidopsis lyrata. Mol. Ecol. 26: 3484–3496. 10.1111/mec.14135 [DOI] [PubMed] [Google Scholar]
- Hämälä T., Mattila T. M., and Savolainen O., 2018. Local adaptation and ecological differentiation under selection, migration and drift in Arabidopsis lyrata. Evolution. 72: 1373–1386. 10.1111/evo.13502 [DOI] [PubMed] [Google Scholar]
- Hämälä T., Guiltinan M. J., Marden J. H., Maximova S. N., DePamphilis C. W. et al. , 2020. Gene expression modularity reveals footprints of polygenic adaptation in Theobroma cacao. Mol. Biol. Evol. 37: 110–123. 10.1093/molbev/msz206 [DOI] [PubMed] [Google Scholar]
- Hartfield M., and Otto S. P., 2011. Recombination and hitchhiking of deleterious alleles. Evolution. 65: 2421–2434. 10.1111/j.1558-5646.2011.01311.x [DOI] [PubMed] [Google Scholar]
- Hazzouri K. M., Escobar J. S., Ness R. W., Killian Newman L., Randle A. M. et al. , 2013. Comparative population genomics in Collinsia sister species reveals evidence for reduced effective population size, relaxed selection, and evolution of biased gene conversion with an ongoing mating system shift. Evolution. 67: 1263–1278. [DOI] [PubMed] [Google Scholar]
- Hill W. G., and Robertson A., 1966. The effects of linkage and the limits to artificial selection. Genet. Res. 8: 269–294. 10.1017/S0016672300010156 [DOI] [PubMed] [Google Scholar]
- Hu T. T., Pattyn P., Bakker E. G., Cao J., Cheng J.-F. et al. , 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43: 476–481. 10.1038/ng.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs E. B., Wright S. I., Stinchcombe J. R., and Schoen D. J., 2017. The relationship between selection, network connectivity, and regulatory variation within a population of Capsella grandiflora. Genome Biol. Evol. 9: 1099–1109. 10.1093/gbe/evx068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes T. H., and Cantor C. R.. 1969. Evolution of protein molecules. Mamm. Protein Metab. 3: 21– 132 10.1016/B978-1-4832-3211-9.50009-7 [DOI] [Google Scholar]
- Kawakatsu T., Carol Huang S.-S., Jupe F., Sasaki E., Schmitz R. J. et al. , 2016. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166: 492–505. 10.1016/j.cell.2016.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., and Otto S. P., 2006. Interference among deleterious mutations favours sex and recombination in finite populations. Nature 443: 89–92. 10.1038/nature05049 [DOI] [PubMed] [Google Scholar]
- Keightley P. D., and Eyre-Walker A., 2007. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics 177: 2251–2261. 10.1534/genetics.107.080663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., and Jackson B. C., 2018. Inferring the probability of the derived vs. the ancestral allelic state at a polymorphic site. Genetics 209: 897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., 2015. ppcor: an R package for a fast calculation to semi-partial correlation coefficients. Commun. Stat. Appl. Methods 22: 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., and Wiehe T., 2008. Simulation of DNA sequence evolution under models of recent directional selection. Brief. Bioinform. 10: 84–96. 10.1093/bib/bbn048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm S. L., Shipony Z., and Greenleaf W. J., 2019. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20: 207–220. 10.1038/s41576-018-0089-8 [DOI] [PubMed] [Google Scholar]
- Koenig D., Hagmann J., Li R., Bemm F., Slotte T. et al. , 2019. Long-term balancing selection drives evolution of immunity genes in Capsella. eLife 8: e43606 10.7554/eLife.43606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono T. J. Y., Lei L., Shih C. H., Hoffman P. J., Morrell P. L. et al. , 2018. Comparative genomics approaches accurately predict deleterious variants in plants. G3 Genes, Genomes. Genet. 8: 3321–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., 2011. Are there laws of genome evolution? PLoS Comput. Biol. 7: e1002173 10.1371/journal.pcbi.1002173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousathanas A., and Keightley P. D., 2013. A comparison of models to infer the distribution of fitness effects of new mutations. Genetics 193: 1197–1208. 10.1534/genetics.112.148023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M., 2008. Building predictive models in R using the caret package. J. Stat. Softw. 28: 1–26. 10.18637/jss.v028.i0527774042 [DOI] [Google Scholar]
- Lachance J., and Tishkoff S. A., 2014. Biased gene conversion skews allele frequencies in human populations, increasing the disease burden of recessive alleles. Am. J. Hum. Genet. 95: 408–420. 10.1016/j.ajhg.2014.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P., Berardini T. Z., Li D., Swarbreck D., Wilks C. et al. , 2012. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40: D1202–D1210. 10.1093/nar/gkr1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., and Horvath S., 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesecque Y., Mouchiroud D., and Duret L., 2013. GC-biased gene conversion in yeast is specifically associated with crossovers: molecular mechanisms and evolutionary significance. Mol. Biol. Evol. 30: 1409–1419. 10.1093/molbev/mst056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN].
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Huang J., Sun X., Li J., Hu Y. et al. , 2018. Tetrad analysis in plants and fungi finds large differences in gene conversion rates but no GC bias. Nat. Ecol. Evol. 2: 164–173. 10.1038/s41559-017-0372-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A., Morgan C., Franklin F. C. H., and Bomblies K., 2018. Plasticity of meiotic recombination rates in response to temperature in arabidopsis. Genetics 208: 1409–1420. 10.1534/genetics.117.300588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A., and Goldman N., 2008. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science 320: 1632–1635. 10.1126/science.1158395 [DOI] [PubMed] [Google Scholar]
- Lu Z., Marand A. P., Ricci W. A., Ethridge C. L., Zhang X. et al. , 2019. The prevalence, evolution and chromatin signatures of plant regulatory elements. Nat. Plants 5: 1250–1259. 10.1038/s41477-019-0548-z [DOI] [PubMed] [Google Scholar]
- Lynch M., and Gabriel W., 1990. Mutation load and the survival of small populations. Evolution 44: 1725–1737. 10.1111/j.1558-5646.1990.tb05244.x [DOI] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., and Steinmetz L. M., 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485. 10.1038/nature07135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G., 2003. Biased gene conversion: implications for genome and sex evolution. Trends Genet. 19: 330–338. 10.1016/S0168-9525(03)00116-1 [DOI] [PubMed] [Google Scholar]
- Marais G., Charlesworth B., and Wright S. I., 2004. Recombination and base composition: the case of the highly self-fertilizing plant Arabidopsis thaliana. Genome Biol. 5: R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G., Delcher A. L., Phillippy A. M., Coston R., Salzberg S. L. et al. , 2018. MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol. 14: e1005944 10.1371/journal.pcbi.1005944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila T. M., Tyrmi J., Pyhäjärvi T., and Savolainen O., 2017. Genome-wide analysis of colonization history and concomitant selection in Arabidopsis lyrata. Mol. Biol. Evol. 34: 2665–2677. 10.1093/molbev/msx193 [DOI] [PubMed] [Google Scholar]
- Mattila T. M., Laenen B., Horvath R., Hämälä T., Savolainen O. et al. , . 2019. Impact of demography on linked selection in two outcrossing Brassicaceae species. Ecol. Evol. 9: 9532–9545. 10.1002/ece3.5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J. H., and Kreitman M., 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. 10.1038/351652a0 [DOI] [PubMed] [Google Scholar]
- Mugal C. F., Weber C. C., and Ellegren H., 2015. GC-biased gene conversion links the recombination landscape and demography to genomic base composition: GC-biased gene conversion drives genomic base composition across a wide range of species. BioEssays 37: 1317–1326. 10.1002/bies.201500058 [DOI] [PubMed] [Google Scholar]
- Muller H., 1950. Our load of mutations. Am. J. Hum. Genet. 2: 111–176. [PMC free article] [PubMed] [Google Scholar]
- Muller H., 1964. The relation of recombination to mutational advance. Mutat. Res. 1: 2–9. 10.1016/0027-5107(64)90047-8 [DOI] [PubMed] [Google Scholar]
- Muyle A., Serres-Giardi L., Ressayre A., Escobar J., and Glémin S., 2011. GC-biased gene conversion and selection affect GC content in the Oryza genus (rice). Mol. Biol. Evol. 28: 2695–2706. 10.1093/molbev/msr104 [DOI] [PubMed] [Google Scholar]
- Necşulea A., Popa A., Cooper D. N., Stenson P. D., Mouchiroud D. et al. , 2011. Meiotic recombination favors the spreading of deleterious mutations in human populations. Hum. Mutat. 32: 198–206. 10.1002/humu.21407 [DOI] [PubMed] [Google Scholar]
- Nembrini S., König I. R., and Wright M. N., 2018. The revival of the Gini importance? Bioinformatics 34: 3711–3718. 10.1093/bioinformatics/bty373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., 2005. Molecular signatures of natural selection. Annu. Rev. Genet. 39: 197–218. 10.1146/annurev.genet.39.073003.112420 [DOI] [PubMed] [Google Scholar]
- Nordborg M., 2000. Linkage disequilibrium, gene trees and selfing: an ancestral recombination graph with partial self-fertilization. Genetics 154: 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J. A., 2002. Accounting for background nucleotide composition when measuring codon usage bias. Mol. Biol. Evol. 19: 1390–1394. 10.1093/oxfordjournals.molbev.a004201 [DOI] [PubMed] [Google Scholar]
- Ohta T., 1973. Slightly deleterious mutant substitutions in evolution. Nature 246: 96–98. 10.1038/246096a0 [DOI] [PubMed] [Google Scholar]
- Papakostas S., Vøllestad L. A., Bruneaux M., Aykanat T., Vanoverbeke J. et al. , 2014. Gene pleiotropy constrains gene expression changes in fish adapted to different thermal conditions. Nat. Commun. 5: 4071 10.1038/ncomms5071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck J. R., 1994. A ruby in the rubbish: beneficial mutations, deleterious mutations and the evolution of sex. Genetics 137: 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessia E., Popa A., Mousset S., Rezvoy C., Duret L. et al. , 2012. Evidence for widespread GC-biased gene conversion in eukaryotes. Genome Biol. Evol. 4: 675–682. 10.1093/gbe/evs052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak E., 1987. On the theory of partially inbreeding finite populations. I. Partial selfing. Genetics 117: 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnakumar A., Mousset S., Glémin S., Berglund J., Galtier N. et al. , 2010. Detecting positive selection within genomes: the problem of biased gene conversion. Philos. Trans. R. Soc. B Biol. Sci. 365: 2571–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausher M. D., Miller R. E., and Tiffin P., 1999. Patterns of evolutionary rate variation among genes of the anthocyanin biosynthetic pathway. Mol. Biol. Evol. 16: 266–274. 10.1093/oxfordjournals.molbev.a026108 [DOI] [PubMed] [Google Scholar]
- Rodgers-Melnick E., Vera D., Bass H., and Buckler E., 2016. Open chromatin reveals the functional maize genome. Proc. Natl. Acad. Sci. USA 113: E3177–E3184. 10.1073/pnas.1525244113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Ibarra J., Morrell P. L., and Gaut B. S., 2007. Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc. Natl. Acad. Sci. USA 104: 8641–8648. 10.1073/pnas.0700643104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousselle M., Laverré A., Figuet E., Nabholz B., and Galtier N., 2019. Influence of recombination and GC-biased gene conversion on the adaptive and nonadaptive substitution rate in mammals vs. birds. Mol. Biol. Evol. 36: 458–471. 10.1093/molbev/msy243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan B. A., Heavens D., Feuerborn T. R., Tock A. J., Henderson I. R. et al. , 2019. An ultra high-density Arabidopsis thaliana crossover map that refines the influence of structural variation and epigenetic features. Genetics 213: 771–787. 10.1534/genetics.119.302406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellinger T. P. P., Awad D. A., Moest M., and Tellierm A., 2020. Inference of past demography, dormancy and self-fertilization rates from whole genome sequence data. PLoS Genet. 16: e1008698 10.1371/journal.pgen.1008698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., Foxe J. P., Hazzouri K. M., and Wright S. I., 2010. Genome-wide evidence for efficient positive and purifying selection in capsella grandiflora, a plant species with a large effective population size. Mol. Biol. Evol. 27: 1813–1821. 10.1093/molbev/msq062 [DOI] [PubMed] [Google Scholar]
- Slotte T., Bataillon T., Hansen T. T., St. Onge K., Wright S. I. et al. , 2011. Genomic determinants of protein evolution and polymorphism in Arabidopsis. Genome Biol. Evol. 3: 1210–1219. 10.1093/gbe/evr094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., Hazzouri K. M., Stern D., Andolfatto P., and Wright S. I., 2012. Genetic architecture and adaptive significance of the selfing syndrome in Capsella. Evolution. 66: 1360–1374. 10.1111/j.1558-5646.2011.01540.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., Hazzouri K. M., Ågren J. A., Koenig D., Maumus F. et al. , 2013. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 45: 831–835. 10.1038/ng.2669 [DOI] [PubMed] [Google Scholar]
- Smeds L., Mugal C. F., Qvarnström A., and Ellegren H., 2016. High-resolution mapping of crossover and non-crossover recombination events by whole-genome re-sequencing of an avian pedigree. PLoS Genet. 12: e1006044 10.1371/journal.pgen.1006044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley, J., P. G. D. Feulner, S. E. Johnston, A. W. Santure, and C. M. Smadja, 2017 Variation in recombination frequency and distribution across eukaryotes: Patterns and processes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372: 20160455 (erratum: Philos. Trans. R. Soc. Lond. B Biol. Sci. 373: 2017.0360). [DOI] [PMC free article] [PubMed]
- Steige K. A., Laenen B., Reimegård J., Scofield D. G., and Slotte T., 2017. Genomic analysis reveals major determinants of cis-regulatory variation in Capsella grandiflora. Proc. Natl. Acad. Sci. USA 114: 1087–1092. 10.1073/pnas.1612561114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C., Toomajian C., Sherman-Broyles S., Plagnol V., Guo Y. L. et al. , 2007. The evolution of selfing in Arabidopsis thaliana. Science 317: 1070–1072. 10.1126/science.1143153 [DOI] [PubMed] [Google Scholar]
- Uricchio L. H., Petrov D. A., and Enard D., 2019. Exploiting selection at linked sites to infer the rate and strength of adaptation. Nat. Ecol. Evol. 3: 977–984. 10.1038/s41559-019-0890-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R., Adusumalli A., Leng S. N., Sikic M., and Ng P. C., 2016. SIFT missense predictions for genomes. Nat. Protoc. 11: 1–9. 10.1038/nprot.2015.123 [DOI] [PubMed] [Google Scholar]
- Wallberg A., Glémin S., and Webster M. T., 2015. Extreme recombination frequencies shape genome variation and evolution in the honeybee, Apis mellifera. PLoS Genet. 11: e1005189 10.1371/journal.pgen.1005189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M. L., Becker C., Hildebrandt J., Neumann M., Rutter M. T. et al. , 2019. Fine-grained analysis of spontaneous mutation spectrum and frequency in Arabidopsis thaliana. Genetics 211: 703–714. 10.1534/genetics.118.301721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnker E., James G. V., Ding J., Becker F., Klasen J. R. et al. , 2013. The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. eLife 2013: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson R. J., Josephs E. B., Platts A. E., Hazzouri K. M., Haudry A. et al. , 2014. Evidence for widespread positive and negative selection in coding and conserved noncoding regions of capsella grandiflora. PLoS Genet. 10: e1004622 10.1371/journal.pgen.1004622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing E. M., Rawat V., Mandáková T., Maumus F., James G. V. et al. , 2015. Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nat. Plants 1: 14023 10.1038/nplants.2014.23 [DOI] [PubMed] [Google Scholar]
- Wright M. N., and Ziegler A., 2017. ranger: a fast implementation of random forests for high dimensional data in C++ and R. J. Stat. Softw. 77: 1–17. 10.18637/jss.v077.i01 [DOI] [Google Scholar]
- Yang S., Yuan Y., Wang L., Li J., Wang W. et al. , 2012. Great majority of recombination events in Arabidopsis are gene conversion events. Proc. Natl. Acad. Sci. USA 109: 20992–20997. 10.1073/pnas.1211827110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Zhou L., Bawa R., Suren H., and Holliday J. A., 2016. Recombination rate variation, hitchhiking, and demographic history shape deleterious load in poplar. Mol. Biol. Evol. 33: 2899–2910. 10.1093/molbev/msw169 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental figures and tables, a compiled table of genomic features, and a code for running the simulations are available at figshare: https://doi.org/10.25386/genetics.12284174.