Abstract
Background/purpose
TrkA overexpression occurs in over 20% of breast cancers, including triple-negative breast cancers (TNBC), and has recently been recognized as a potential driver of carcinogenesis. Recent clinical trials of pan-Trk inhibitors have demonstrated targeted activity against tumors harboring NTRK fusions, a relatively rare alteration across human cancers. Despite this success, current clinical trials have not investigated TrkA overexpression as an additional therapeutic target for pan-Trk inhibitors. Here, we evaluate the cancerous phenotypes of TrkA overexpression relative to NTRK1 fusions in human cells and assess response to pharmacologic Trk inhibition.
Experimental design/methods
To evaluate the clinical utility of TrkA overexpression, a panel of TrkA overexpressing cells were developed via stable transfection of an NTRK1 vector into the non-tumorigenic breast cell lines, MCF10A and hTERT-IMEC. A panel of positive controls was generated via stable transfection with a CD74-NTRK1 fusion vector into MCF10A cells. Cells were assessed via various in vitro and in vivo analyses to determine the transformative potential and targetability of TrkA overexpression.
Results
TrkA overexpressing cells demonstrated transformative phenotypes similar to Trk fusions, indicating increased oncogenic potential. TrkA overexpressing cells demonstrated growth factor-independent proliferation, increased PI3Kinase and MAPKinase pathway activation, anchorage-independent growth, and increased migratory capacity. These phenotypes were abrogated by the addition of the pan-Trk inhibitor, larotrectinib. In vivo analysis demonstrated increased tumorgenicity and metastatic potential of TrkA overexpressing breast cancer cells.
Conclusions
Herein, we demonstrate TrkA overexpressing cells show increased tumorgenicity and are sensitive to pan-Trk inhibitors. These data suggest that TrkA overexpression may be an additional target for pan-Trk inhibitors and provide a targeted therapy for breast cancer patients.
Keywords: Breast cancer, TrkA, NTRK1, Metastasis
Introduction
Triple-negative breast cancer (TNBC) is defined by the lack of ER (estrogen receptor-alpha), PR (progesterone receptor), or HER2 (human epidermal growth factor 2) expression and represents approximately 15–20% of all breast cancers [1]. Due to the lack of receptor expression, TNBCs are not suitable candidates for targeted therapies such as endocrine therapies or HER2 inhibitors [2–4]. As a result, TNBC cancers have higher rates of recurrence, metastasis, and mortality. Furthermore, TNBCs have a worse disease-free survival (DFS) and overall survival (OS) than other breast cancer subtypes [2]. Currently, standard of care options are limited to surgical intervention, radiation, and cytotoxic chemotherapies. Although local management and adjuvant chemotherapies of earlystage TNBC have demonstrated improved DFS/OS, those who develop metastatic disease have a median OS of 13–18 months with current treatment options [1, 5]. To date, there are no targeted therapies for TNBC and the establishment of treatment-resistant tumor populations leaves patients with few options.
Tropomyosin receptor kinases (Trk receptors) are a family of membrane-bound tyrosine kinases that regulate components of the mammalian nervous system. The three most common types of Trk receptors are TrkA, TrkB, and TrkC, which are encoded by the genes NTRK1, NTRK2, and NTRK3, respectively. Classification of Trk receptors is determined by distinct affinities for different neurotrophin ligands. TrkA serves as a high affinity receptor for nerve growth factor (NGF), while TrkB and TrkC serve as high affinity receptors for brain-derived neurotrophic factor (BDNF) and NT-3, respectively (5). Binding of ligand results in downstream kinase signaling and activation of the Ras/MAP Kinase and PI3 Kinase pathways, two pathways closely associated with several cancers [6]. The oncogenic role of Trk was first identified in colorectal cancer as a fusion protein due to an NTRK1 rearrangement, which resulted in upregulation of downstream pathways [7]. Recently, next-generation sequencing efforts confirmed the presence of NTRK1 rearrangements in multiple cancers and the subsequent upregulation of oncogenic pathways [7, 8]. Although the frequency of NTRK1 rearrangements in most cancers is rare, pan-Trk inhibitors have shown promising results with high rates of efficacy and low toxicity [9, 10]. Furthermore, a recent clinical study demonstrated the majority of breast cancer-associated NTRK1 rearrangements were found in TNBCs and may offer an option for targeted therapy for a small subset of TNBC patients [11].
Although NTRK1 rearrangements are relatively rare, NTRK1 amplification (and subsequent TrkA overexpression) is fairly frequent in a variety of human cancers, with reported rates as high as 20% in breast cancers (Supp. Fig. S1) [12]. In 2009, Lagadec et al. found that breast tumors demonstrated higher levels of TrkA and phospho-TrkA when compared to normal tissue. Furthermore, in vitro studies demonstrated that NTRK1 amplification exhibited similar pathway activation and oncogenic phenotypes to those observed with NTRK1 rearrangements [13]. Based on the recent success of Trk inhibitors for cancers with NTRK gene rearrangements [7, 8, 14, 15], we hypothesized that overexpression of TrkA may offer an alternative target in treating cancers with these inhibitors. Of specific interest, the ability to use these therapies for treating TNBC with NTRK1 amplification could be a potential strategy for targeted therapies against a particularly recalcitrant group of cancers.
In this study, we developed a panel of TrkA overexpressing human breast cell lines to determine the potential targetability of TrkA amplification in TNBC. Our results demonstrate that overexpression of NTRK1 in non-tumorigenic human breast epithelial cells results in transformative characteristics consistent with increased metastatic potential, which can be reversed using a pan-Trk inhibitor. Our studies suggest that NTRK1 amplification may provide a targetable biomarker for TNBC.
Results
Generation of TrkA overexpressing non-tumorigenic human breast epithelial cell lines
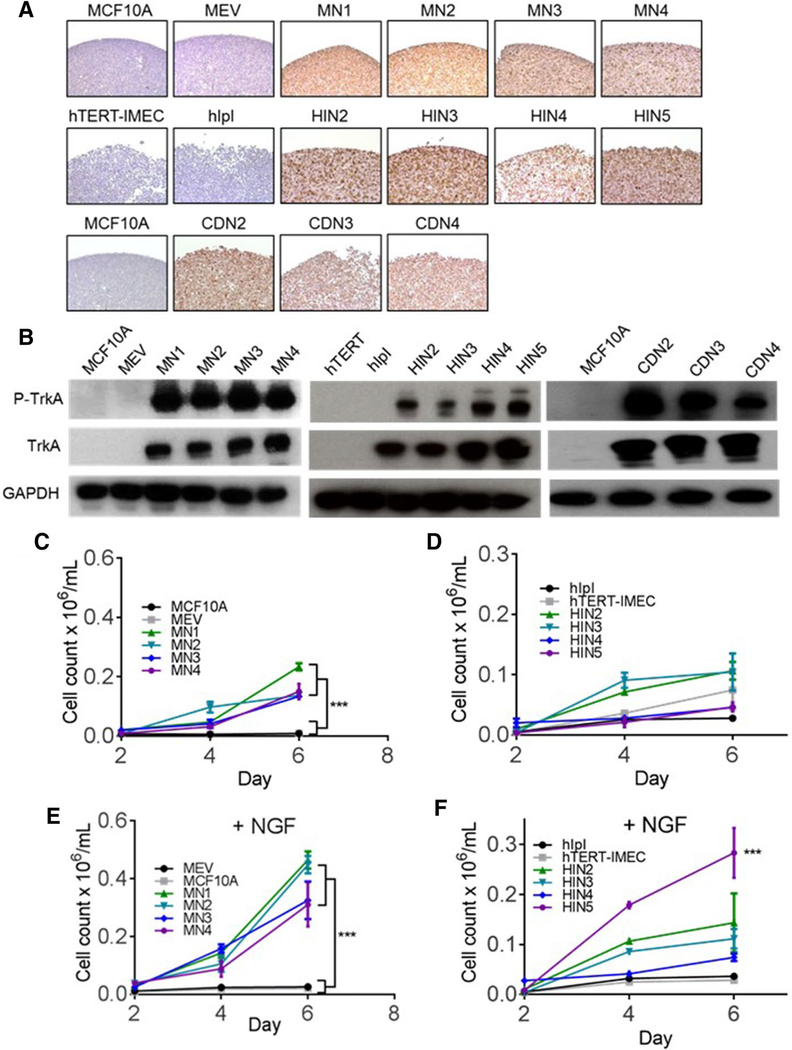
To determine the oncogenic potential of TrkA overexpression in normal breast cells, two non-tumorigenic human breast epithelial cell lines, MCF10A and hTERT-IMEC, were transfected with TrkA overexpression vectors (17). MCF10A is a spontaneously immortalized human non-tumorigenic breast epithelial cell line that requires exogenous growth factors and hormones, including epidermal growth factor (EGF), for normal propagation. Similarly, hTERT-IMEC is a non-tumorigenic human mammary cell line immortalized by TERT protein expression. Neither parental cell line expresses the TrkA protein at significant levels (Fig. 1a, b). Both cell lines were transfected with a pIRESneo3 plasmid containing a human full length NTRK1 cDNA. Four individual clones (referred to as MN1, 2, 3, 4 for MCF10A and HIN2, 3, 4, 5 for hTERT-IMEC) were isolated for each parental line and TrkA expression was confirmed by immunohistochemistry and immunoblot analyses (Fig. 1a, b). Empty vector controls expressing the pIRESneo3 plasmid containing no cDNA were developed as well (MEV for MCF10A and hIpI for hTERT-IMEC). To effectively compare the TrkA overexpression panel to a clinically druggable NTRK1 gene fusion, MCF10A cells were transfected with a plasmid containing a cDNA fusion CD74-NTRK1 as previously described [16]. Three clones were isolated (CDN2, 3, 4) and served as positive controls in subsequent studies (Fig. 1a, b).
Fig. 1.
Overexpression of TrkA leads to growth factor-independent proliferation. a Overexpression of TrkA in MCF10A (MN1, 2, 3, 4, top row) and hTERT-IMEC (HIN2, 3, 4, 5, middle row) were confirmed by immunohistochemistry. In addition to the overexpression cell models, CD74-NTRK1 fusions were expressed in MCF10As (CDN2,3,4, bottom row) as positive controls. Empty vector controls (MEV for MCF10A and hIpI for hTERT-IMEC) were used as negative controls. b Immunoblot analysis of TrkA overexpression panels in MCF10A (left) and hTERT-IMEC (middle), and CD74-NTRK1 fusions (right). c Proliferation analysis of the MCF10A TrkA overexpression panel and d hTERT-IMEC panel in growth factor-reduced media. Cells were plated at a density of 30,000 cells/well in 24-well plates and cell counted on 2, 4, and 6 days. e Proliferation analysis of the MCF10A TrkA overexpression panel and f hTERT-IMEC panel in the presence of 0.2 ng/mL neuronal growth factor (NGF) as described in (c). Mean ± SEM shown, ***P ≤ 0.001, by ANOVA analysis at 6-day time point
TrkA overexpression leads to growth factor-independent proliferation
Growth factor-independent cell proliferation in MCF10A is a characteristic of transformation and was previously demonstrated in MCF10A-derived cells containing oncogenic PIK3CA mutations [17]. To determine whether TrkA overexpression resulted in EGF independence, cell proliferation assays with TrkA overexpressing and CD74-NTRK1 fusion clones were carried out in growth factor-reduced conditions (16). Due to the inability of parental MCF10As to propagate in the absence of EGF, cell proliferation assays were also carried out at physiologic levels of EGF (0.2 ng/ mL) to compare changes in growth rate. In the absence of EGF, TrkA overexpression in both MCF10A and hTERT-IMEC cells resulted in EGF-independent growth (Fig. 1c, d respectively). Furthermore, the presence of physiologic levels of EGF led to a significant increase in proliferation of the TrkA overexpression lines when compared to parental and empty vector controls (Supp. Fig. S2A). As previously mentioned, TrkA is activated specifically by neuronal growth factor (NGF). To determine the proliferative impact of NGF on TrkA overexpression lines, both cell line panels were grown in physiologic doses of NGF (100 ng/mL for MCF10A-derived cells and 12.5 ng/mL for hTERT-IMEC). In both cell line panels, the addition of NGF increased cell proliferation rates in the TrkA-expressing clones but not in the parental cells or empty vector controls (Fig. 1e, f). The MCF10As demonstrated a significant increase across all clones consistent with the higher TrkA expression levels demonstrated in the western blots.
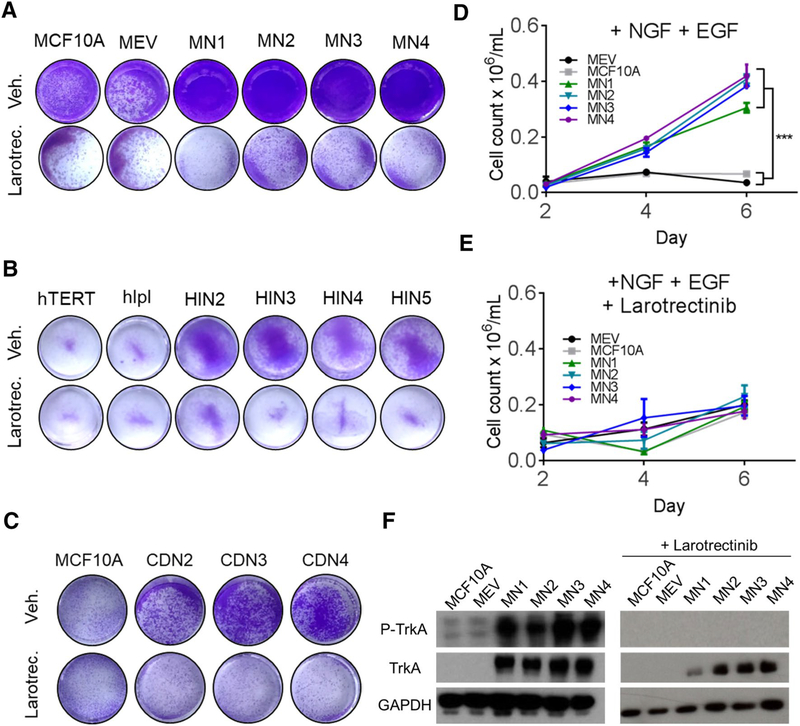
TrkA overexpressing cells are growth inhibited by TrkA pan-inhibitors
Recent basket trials have demonstrated the success of the pan-Trk inhibitor, larotrectinib, in targeting Trk fusions (NCT02576431). To determine if larotrectinib could effectively target TrkA overexpressing cells, cell line panels were grown in the presence of both physiologic NGF and EGF ± 1.5 μM larotrectinib. Despite the significant growth advantage of TrkA overexpressing cells in the presence of growth factor, the addition of larotrectinib resulted in nearly complete growth inhibition (Fig. 2a, b, Supp. Fig S2). The MCF10A CD74-NTRK1 fusion cells demonstrated similar growth inhibition further suggesting TrkA overexpression may be a comparable target for Trk inhibitors (Fig. 2c, Supp. Fig. S3). Quantification of the MCF10A-TrkA panel demonstrated a significant decrease in growth in the presence of larotrectinib despite the addition of both EGF and NGF (Fig. 2d, e). Immunoblot analysis confirmed overexpression of TrkA in MCF10A cells and demonstrated an increase in phospho-TrkA, consistent with increased receptor activation while the addition of larotrectinib resulted in the complete inhibition of phospho-TrkA (Fig. 2f).
Fig. 2.
TrkA inhibitions leads to decreased growth in TrkA overexpressing cells. a Proliferation analysis of the MCF10A TrkA overexpression panel in the presence of 0.2 ng/mL neuronal growth factor (NGF) and 0.2 ng/mL epidermal growth factor (+ EGF) and b hTERT-IMEC panel in the presence of 0.2 ng/mL neuronal growth factor (NGF). Cells were plated at a density of 30,000 cells/well in 24-well plates and stained with crystal violet at 5 days. Vehicle (Veh.) or 1.5 μM Larotrectinib (Larotrect.) were added as specified. c Growth analysis of CD74-NTRK1 fusion panel as described in (a). Larotrectinib (Larotrect.) was added at 2 μM as specified. d Quantification of proliferation in the MCF10A TrkA overexpression panel in the presence of 0.2 ng/mL neuronal growth factor (+ NGF) and 0.2 ng/mL epidermal growth factor (+ EGF), and e 1.5 μM Larotrectinib. Mean ± SEM shown, ***P ≤ 0.001, by ANOVA at 6-day time point. f Immunoblot analysis of the MCF10A TrkA overexpression panel ± 1.5 μM Larotrectinib in the presence of 0.2 ng/mL neuronal growth factor and 0.2 ng/mL epidermal growth factor
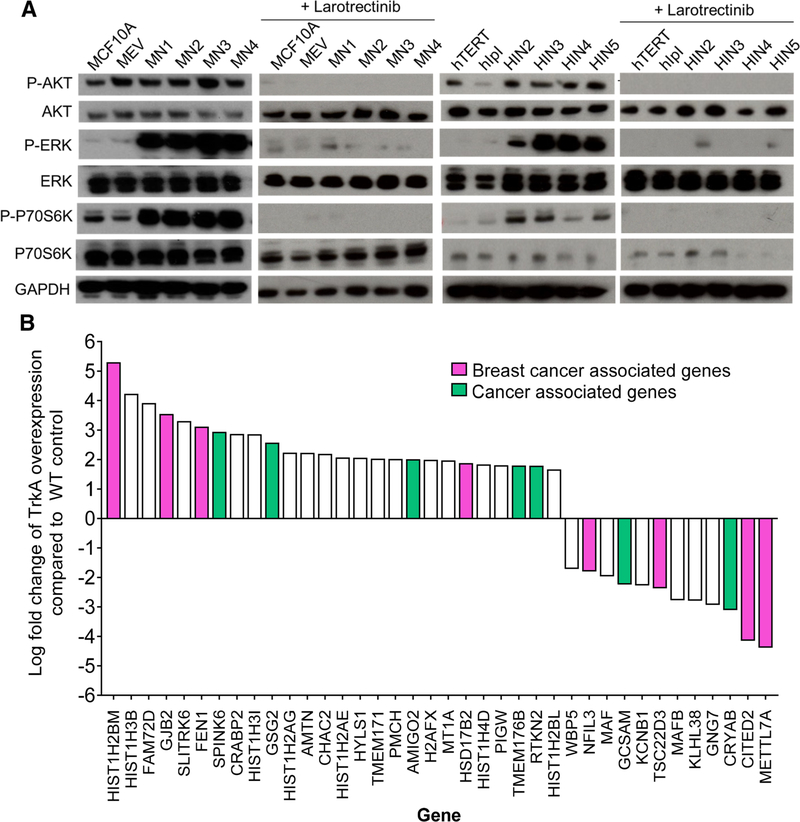
TrkA overexpression leads to increased MAPK and PI3Kinase signaling and altered gene expression of cancer pathway genes
Previous in vitro studies demonstrated NTRK1 rearrangements, as well as TrkA activation, led to upregulation of the MAPKinase and PI3Kinase pathways. Immunoblot analysis was carried out for both the MAPKinase and PI3Kinase-signaling pathways to determine if MCF10A and hTERT-IMEC TrkA overexpression demonstrated similar activation to NTRK1 rearrangements. Both cell line panels were grown in complete growth media to ensure exponential growth of the controls. As seen with NTRK1 rearrangements, overexpression of TrkA in both MCF10A and hTERT-MEC led to an increase in p-AKT, p-ERK, and p-S6K (or p-p70S6K) when compared to controls (Fig. 3a). Furthermore, in the absence of growth factors TrkA overexpression cells demonstrated moderate pathway activation suggesting a transformative phenotype similar to NTRK1 rearrangements (Supp. Fig. S4A). Interestingly, in the absence of EGF, the addition of NGF resulted in robust growth of the TrkA overexpressing cells when compared to empty vector controls (Supp. Fig. S4B). Addition of larotrectinib resulted in complete loss of AKT, ERK, and S6K activation. Taken together, these results suggest that TrkA overexpression is a driver for dysregulated proliferation in normal breast epithelia and may be a target for pharmacologic Trk inhibition (Fig. 3a). It is important to note the downregulation of the MAPK and PI3K kinase pathways in the controls may be attributed to the relatively high concentration of larotrectinib.
Fig. 3.
TrkA overexpression leads to increased MAPK/PI3K signaling and dysregulation of genes in oncogenic pathways. a Immunoblot analysis of the MCF10A and hTERT-IMEC TrkA overexpression panels. Protein lysates were collected after 24-h exposure to arrest media conditions supplemented with EGF and NGF for MCF10A (left) and hTERT-IMEC (right) cell line panels. 1.5 μM larotrectinib was added as indicated. b Microarray analysis of TrkA overexpression panels. Log fold change was calculated and averaged among respective parental controls. Log fold change > 2 shown. List of additional genes in Supplemental Table 2. Breast cancer-associated genes (pink) and cancer-associated genes (green) indicated
Previous in vitro studies have demonstrated oncogenic alterations result in distinct gene expression profiles that are indicative of transformation [18]. To gain further insight into signaling pathways that were dysregulated with TrkA overexpression, gene expression was analyzed using a microarray of approximately 20,000 genes. Both MCF10A and hTERT-IMEC TrkA overexpressing cells exhibited similar altered gene expression in over 100 genes. A complete list of genes exhibiting > 1 log fold change is listed in Supplemental Tables S2. Among genes demonstrating a > 2 log fold change, 40% have been previously implicated in cancer, with over half being specifically associated with breast cancer (Fig. 3b). Of note, the most dramatically up and downregulated genes have been previously associated with breast cancer [19–21]. These results suggest TrkA overexpression leads to the activation of oncogenic pathways that may play a role in breast carcinogenesis.
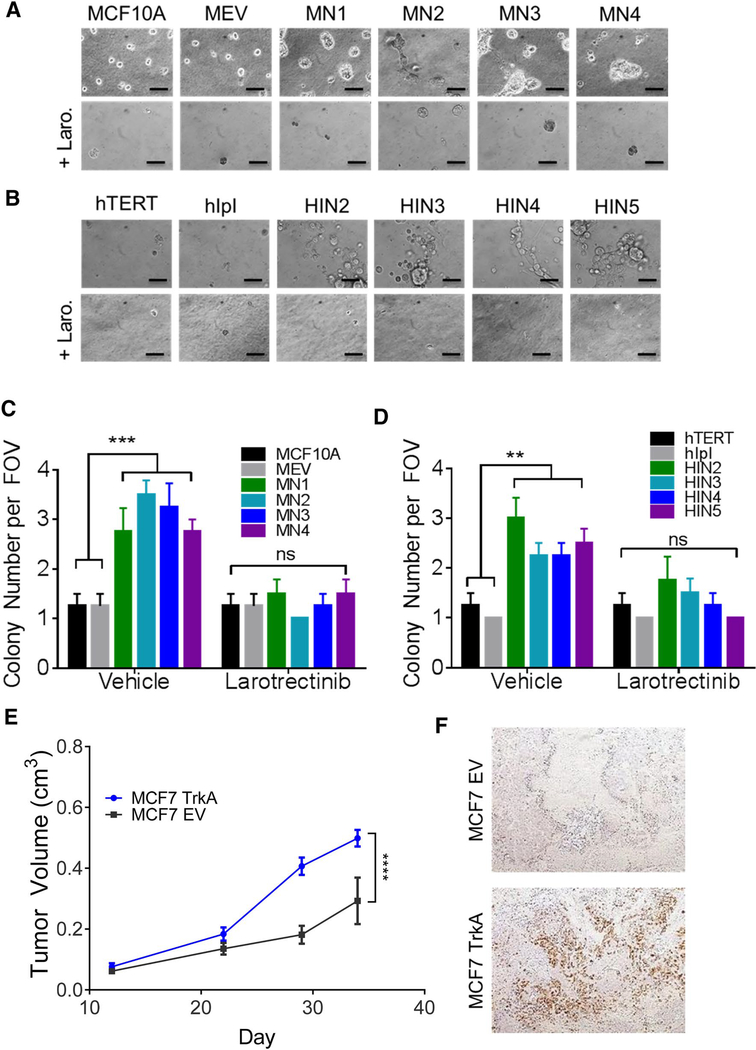
TrkA overexpression leads to dysregulated 3D growth and increased tumorigenicity
Previous studies have demonstrated dysregulation of proliferative cell survival pathways results in changes in 3D morphology, indicative of cellular transformation (17). Normal breast epithelial cells form hollow spherical structures termed acini. However, the presence of oncogenic alterations results in protrusions at the borders of acini, bridging between neighboring acini, and loss of structural integrity [22]. To determine the transformative capabilities of TrkA overexpression, cells were grown in a Matrigel matrix in both the presence and absence of growth factor. In the absence of growth factor, normal breast epithelial cells are unable to proliferate or form acini. However, as seen with proliferation assays, overexpression of TrkA in MCF10A and hTERT-IMEC conferred the ability to form acini in the absence of growth factors (Supp. Figure S5). Consistent with western blot and proliferation data, the addition of NGF augmented the transformative properties of TrkA overexpression and resulted in increased size and loss of structural integrity when compared to parental controls (Fig. 4a, b, top rows). The addition of larotrectinib resulted in complete inhibition of acini formation (Fig. 4a, b, bottom rows).
Fig. 4.
TrkA overexpression leads to dysregulated 3D growth and increased tumorigenicity. a MCF10A TrkA overexpressing panel and b hTERT-IMEC overexpressing panel were cultured at low density in Matrigel in the presence of EGF and NGF. 1.5 μM of Larotrectinib was added as indicated (Scale bar, 50 μm). c MCF10A TrkA overexpressing panel and d hTERT-IMEC overexpressing panel were cultured in low growth factor conditions at low density in 0.8% agar for two weeks, stained with crystal violet. Colonies were quantified per field of view (FOV) (n = 4). Mean ± SEM shown, ***P ≤ 0.001; **P ≤ 0.01, ns = not significant, as determined by ANOVA. e MCF7 TrkA (TrkA overexpression) and MCF7 EV (empty vector) xenografts were established subcutaneously in athymic nude mice and supplemented with a 17β-estradiol pellet (n = 5 per arm). Each data point represents the mean tumor volume in mm3 ± SD. ***P ≤ 0.001, as determined by ANOVA. f Tumors were harvested and FFPE tumor sections were prepared and subjected to IHC with TrkA as described in the methods. Representative images shown
Anchorage-independent growth in soft agar is a property of transformed cells that best correlates with in vivo tumorgenicity [23]. As non-tumorigenic breast epithelial cells, both MCF10A and hTERT-IMEC lack the capacity for anchorage-independent growth. However, TrkA overexpression in both non-tumorigenic cell lines resulted in a significant increase in colony formation and a higher propensity for anchorage-independent growth (Fig. 4c, d). These effects were ablated in the presence of the inhibitor larotrectinib (Fig. 4c, d). To determine if this correlated to an increase in potential for tumor growth in vivo, TrkA was overexpressed in the tumorigenic breast cell line, MCF7. Athymic nude mice were injected subcutaneously with MCF7 transfected with empty vector controls (MCF7 EV) or TrkA overexpressing MCF7 (MCF7 TrkA). TrkA overexpressing tumors demonstrated a marked increase in tumor volume when compared to the empty vector controls (Fig. 4e). TrkA expression was confirmed in xenografts using immunohistochemistry (Fig. 4f). Taken together, these studies suggest TrkA overexpression leads to anchorage-independent growth and increased tumorigenic potential.
TrkA overexpressing cells demonstrate increased migratory capabilities
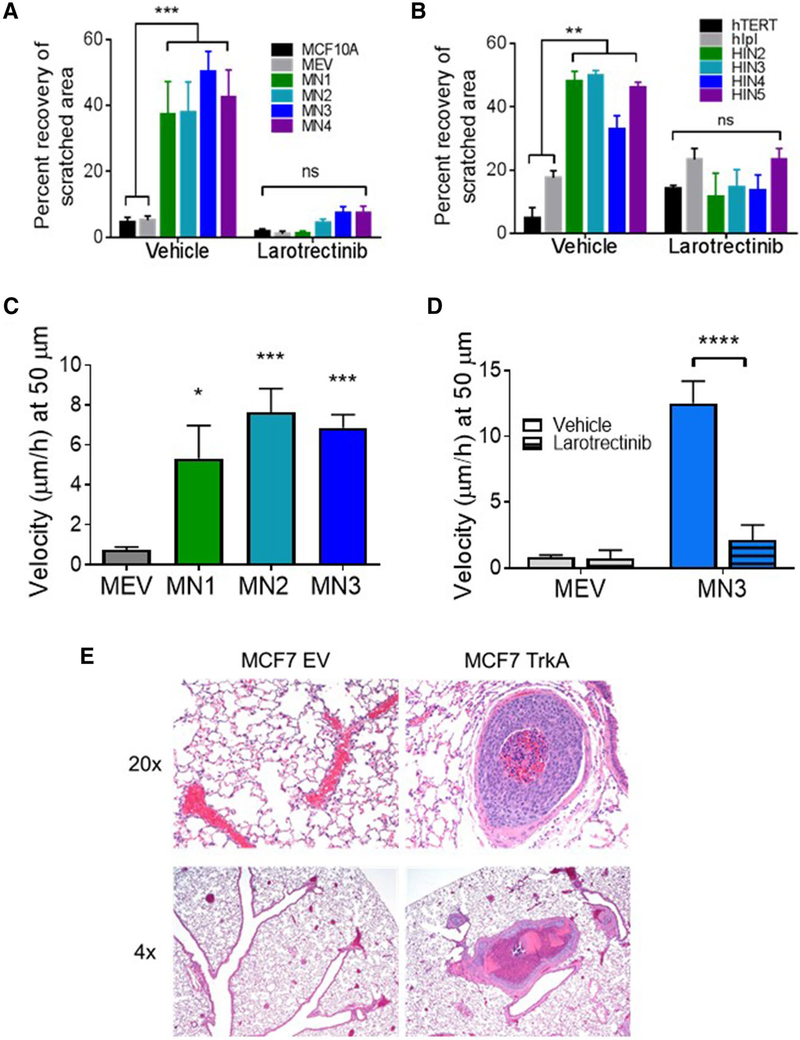
Migratory capacity has been associated in vitro and in vivo with a metastatic phenotype and is commonly linked with anchorage-independent growth. The migratory capacity of TrkA overexpressing clones was analyzed using scratch wound-healing and microchannel migration assays as previously described [24]. The TrkA overexpressing cell lines exhibited a significant increase in scratch wound healing, a direct measure of migratory propensity, when compared to control cell lines (Fig. 5a, b, Supp. Fig. S6). In concordance with other transformation assays, addition of larotrectinibmitigated migratory capabilities and further suggests TrkA overexpression is a carcinogenic driver that may be a therapeutic target for pan-Trk inhibitors (Fig. 5a, b, Supp. Fig. S6).
Fig. 5.
TrkA overexpression leads to increased migratory capabilities. a MCF10A TrkA overexpression and b hTERT-IMEC TrkA overexpression panels were plated in growth factor-supplemented media to a confluent monolayer, scratched, and imaged at 0 and 16 h time points ± 2 μM of larotrectinib; percent healing calculated from scratch dimensions quantified with ImageJ MiToBo software. ***P ≤ 0.001, **P ≤ 0.01, as determined by ANOVA. c Microchannel migration assays were performed in 50 μm channels. Individual cells from the MCF10A TrkA overexpression were tracked along a growth factor gradient. Cellular velocity of migrating cells was calculated as the product of speed and persistence (shown individually in Supplemental Fig. S6). ***P ≤ 0.001, *P ≤ 0.05, as determined by ANOVA. d Representative clone was tracked across growth factor gradient in the presence of 1.5 μM larotrectinib. ****P ≤ 0.0001 as determined by unpaired Student’s t test. e Tail vein injection of MCF7-TrkA (TrkA overexpression) and MCF7-EV (empty vector) cells in athymic nude mice (n = 5 per cell line). 2 of 5 mice injected with TrkA overexpression formed tumors. Mice were supplemented with a 17β-estradiol pellet. Representative images of lung sections at ×4 and ×20
To determine if increased migratory capacity of TrkA overexpressing cells was independent of proliferation rates, a microchannel migratory assay was performed. MCF10A overexpressing cells were seeded in microfluidic devices with growth factor-reduced media separated by microchannels from a channel containing media supplemented with NGF. Time lapse imaging allowed the tracking of individual migratory cells through the channel toward the growth factor attractant. The migratory capacity of a cell was measured based on the velocity of individual cells [24]. The TrkA panel demonstrated increased persistence and speed when compared to the empty vector control (Supp. Fig. S7). As a result, velocity was significantly higher in the TrkA overexpressing cells (Fig. 5c). It is important to note the TrkA’s demonstrated increased migratory capabilities across channel sizes smaller than 50 μm (Supp. Fig. S8). However, for statistical comparisons, a channel size that allowed migration of all cell types was selected. Notably, although the empty vector control cells rarely entered narrower (10 μm wide) microchannels, cells overexpressing TrkA were consistently able to squeeze into these channels, in line with their enhanced migratory potential (Supp. Fig. S8). As with other transformation assays, the addition of the inhibitor larotrectinib resulted in a significant decrease in the velocity of the TrkA overexpressing cells, suggesting the migratory capacity was a direct result of TrkA overexpression (Fig. 5d).
Increased migratory capabilities have been shown to correlate with increased metastatic potential in vivo. To determine the metastatic potential of TrkA overexpression, tail vein injections of MCF7 TrkA overexpression panel were carried out in athymic nude mice. Although tail vein injection of cancer cells is not a measure of metastasis, the ability of cancer cells to extravasate from the circulation and grow in distant sites has been considered a step that is necessary for metastatic potential. MCF7 cells with TrkA overexpression formed microscopic and macroscopic tumors in the lungs (Fig. 5e). In contrast, control cells did not form tumors. When considered with the increased migratory capabilities, the increased propensity of tumor development via tail vein injections suggest that TrkA overexpression may enhance cancer cells’ ability to disseminate to distant sites.
Discussion
In the past few years, there has been a concerted effort to develop individualized therapy for breast cancer patients. The ability to target key oncogenic drivers with effective therapies has led to substantial improvements in overall survival of cancer patients. Despite significant advancements in treatments targeting ER/PR and HER2-receptor positive subtypes, chemotherapy remains the standard of care for patients with triple-negative breast cancer (TNBC). Currently, there is a high demand to identify molecular markers and evaluate targeted therapies in TNBC. Interrogation of online databases (cbioportal) revealed that NTRK1 amplification occurs in over 20% of breast cancers, regardless of receptor status.
Herein, we provide the first direct evidence that TrkA overexpression alone in normal breast epithelial cells is sufficient to drive the oncogenic transformation of cells. Our in vitro data demonstrate TrkA overexpression is actively involved in breast carcinogenesis through oncogenic pathway activation. We determined that TrkA overexpression resulted in increased cell proliferation and migratory/invasive capacity, both hallmarks of cancer development, as well as increased MAP Kinase and PI3 Kinase signaling, two pathways heavily involved in cancer development. Previous studies have shown that NGF is ubiquitously expressed in breast tumor cells, regardless of subtype [25]. Our in vitro data demonstrated NGF alone was sufficient for transformation in TrkA overexpressing cells. This suggests TrkA may be a significant driver in breast carcinogenesis and inhibition of NGF/TrkA activation may offer a novel target for TNBC.
Recent clinical trials have demonstrated the remarkable success of pan-Trk inhibitors in rare cancers with NTRK1 rearrangements. Tumors harboring these rearrangements have upregulated TrkA activity and demonstrate remarkable responses to pan-Trk inhibitors [26]. Our studies demonstrate human breast cells with TrkA overexpression show a significant increase in transformation and metastatic potential in vitro and in vivo, comparable to targetable NTRK1 rearrangements. However, despite the high prevalence of TrkA overexpression and similar phenotypes to NTRK1 rearrangements in vitro, tumors with TrkA overexpression have been largely excluded in trials of Trk inhibitors. Our in vitro studies demonstrate TrkA overexpression in breast cells are highly susceptible to pan-Trk inhibitors and may offer a viable therapeutic target. The addition of the pan-Trk inhibitor, larotrectinib, results in the complete abrogation of these transformative phenotypes further suggesting these alterations act as a significant driver in breast carcinogenesis.
Our studies demonstrate TrkA overexpression is sufficient to drive oncogenic transformation in normal breast epithelial cells and is highly sensitive to pan-Trk inhibitors. A significant portion of breast cancers demonstrate TrkA overexpression, regardless of subtype and may offer an alternative or novel target for current clinical trials. Despite the promising preclinical evidence presented in this paper, further analysis determining the utility of pan-Trk inhibitors in TrkA overexpressing cancers are needed.
Materials and Methods
Cell Culture
The non-tumorigenic human breast epithelial cell lines MCF10A and hTERT-IMEC and their derivatives were cultured in DMEM/F12 (1:1) supplemented with 1% Penicillin–Streptomycin, 0.1 ng/μL cholera toxin, 10 μg/mL insulin, 20 ng/mL EGF, 0.5 μg/mL hydrocortisone and 5% horse serum for MCF10A or 10% charcoal–dextranstripped fetal bovine serum (FBS) for hTERT-IMEC. NTRK1-overexpressing clones and CD74-NTRK1 fusion expression clones were continuously cultured under neomycin selection. MCF10A parental, hTERT-IMEC parental (a generous gift from Myles Brown), and MCF7 parental were originally purchased from ATCC. Parental cell lines were authenticated by short tandem repeat profiling analysis at the Johns Hopkins Genetic Resources Core Facility. For assay with growth factor-reduced media, cells were conditioned in DMEM/F12 with 5% charcoal/ dextran-stripped FBS (C/D FBS), 1% Pen/Strep, hydrocortisone, and cholera toxin for 48 h.
Overexpression of NTRK1 or CD74-NTRK1 cDNA in human cells
NTRK1 cDNA was purchased as pCMV6-AC-GFPNTRK1 (RC213091, Origene) and subcloned into the pIRESneo3 vector (Clontech). Cells were transfected with pIRESneo3-NTRK1 using the Fugene 6 system, as directed. CD74-NTRK1 fusion cDNA was generously provided by Dr. Pasi Janne [8]. For in vivo studies, the tumorigenic breast cell line, MCF7 was transfected with the NTRK1 or empty vector and xenograft studies were carried out as discussed below.
Immunohistochemistry
TrkA overexpression was confirmed using immunohistochemistry as previously described [27]. Slides were stained with the anti-TrkA rabbit monoclonal antibody (2505S; Cell Signaling).
Cell proliferation assays
Cells were seeded at 30,000 cells/well in 12-well plates and assay media was added after 24 h. Assay media included the following modifications: 1% C/D FBS, 0.2 ng/mL EGF and 100 ng/mL or 12.5 ng/mL NGF. Cells were counted using a Beckman–Coulter cell counter (17). For crystal violet staining, cells were stained with 0.05% crystal violet solution in 70% ethanol at 1 week [24].
Immunoblot Analysis
Cells were seeded 24 h prior to the addition of specified assay media. Protein lysates were made using Laemmli buffer and immunoblot analysis was performed using the NuPAGE system. The primary antibodies included: TrkA (Cell Signaling 2505), p-TrkA (Cell Signaling 9141S), p-AKT (Cell Signaling 9271), total AKT (Cell Signaling 9272), p-ERK (Cell Signaling 4370), total ERK (Cell Signaling 9102), p-p70S6K (Cell Signaling 9205), Total p70S6k (Cell Signaling 9202), GAPDH (Cell Signaling 5174).
Microarray Analysis
Gene expression profiling was performed using the Affymetrix GeneChip System and the JHMI microarray core service as previously described [28]. Relative gene expression was determined using the log fold change (LogFC) of TrkA overexpression cells compared to parental controls. Significant genes for both panels were selected.
Colony formation in semisolid media
Cells were prepared in 1:1 mix of indicated media and 1.2% UltraPure Agarose for a final concentration of 0.6% agarose and soft agar assays were carried out as previously described [24]. Colonies were counted per field (n = 4) and photographs were taken with a Nikon SMZ 1500 stereoscopic zoom microscope.
Acinar morphogenesis assay
Acinar morphogenesis assays were performed with growth factor-reduced Matrigel as previously described [24].
Wound-healing assay
Wound-healing assays were performed as previously described [24]. Scratch Assay Analyzer tool in the MiToBo extension in ImageJ 2.0 with parameters sigma = 2 and entropy filter = 25.
Microchannel migration assays
A polydimethylsiloxane (PDMS) microfluidic device was produced and migration assays were performed as previously described [24]. NGF was used as a growth factor attractant.
Xenograft studies
For each group, a minimum of five 8- to 10-week old female athymic nude mice (Harlan Laboratories) were provided with estrogen pellet supplementation prior to cell injection. For solid tumor-formation studies, cell solutions were prepared in a 20% sterile PBS, 80% growth factor-reduced Matrigel (BD Biosciences) containing 1 million cells/200 μL and injected subcutaneously. Tumor volumes were measured weekly and calculated by multiplying width, length and height for each individual tumor. For tail vein injection assays, cell suspensions were prepared in sterile PBS at 0.5 million cells/200 μL and injected into the tail vein. Mice were monitored for signs of disseminated tumor formation including palpable mass and discomfort. Mice were euthanized as appropriate. All animal experiments were performed in accordance with institutional and NIH guidelines.
Statistics
All statistical analyses were performed using GraphPad Prism 5 software. Unpaired Student’s T tests, 1-way ANOVA, and 2-way ANOVA tests were used to compare experimental groups, as indicated. Significance levels are indicated by asterisks: P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***). Error bars represent ± SEM.
Supplementary Material
Acknowledgements
The authors would like to thank Loxo Oncology for providing larotrectinib for the studies presented in this article and Dr. Pasi Janne for providing reagents.
Funding This study was funded by NIH CA214494 and CA194024 (B.H.P.). We would also like to thank and acknowledge the support of Susan G. Komen, the Canney Foundation, the Breast Cancer Research Foundation, and the Marcie & Ellen Foundation. None of the funding sources influenced the design, interpretation or submission of this manuscript.
Footnotes
Compliance with ethical standards
Conflict of interest B.H.P. had ownership interest and was a paid member of the scientific advisory board of Loxo Oncology and was a paid consultant for Foundation Medicine, Inc, Jackson Laboratories, Casdin Capital and Roche. Under separate licensing agreements between Horizon Discovery, LTD and The Johns Hopkins University, B.H.P. is entitled to a share of royalties received by the University on sales of products. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies. All other authors declare no potential conflicts.
Ethical approval All studies were performed in compliance with institutional ethical standards. All animal experiments were performed in accordance with institutional and The National Institutes of Health Guide for the Care and Use of Laboratory Animals guidelines.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s10549-019-05506-3) contains supplementary material, which is available to authorized users.
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Garrido-Castro AC, Lin NU, Polyak K (2019) Insights into molecular classifications of triple-negative breast cancer: improving patient selection for treatment. Cancer Discov 9(2):176–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badve S, Dabbs DJ, Schnitt SJ, Baehner FL, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR et al. (2017) Electronic and optical properties of two-dimensional GaN from first principles. Nano Lett 17(12):7345–167 [DOI] [PubMed] [Google Scholar]
- 3.Hutchinson L (2014) Breast cancer: TNBC: can we treat the untargetable? Nat Rev Clin Oncol 11(7):379. [DOI] [PubMed] [Google Scholar]
- 4.Jamdade VS, Sethi N, Mundhe NA, Kumar P, Lahkar M, Sinha N (2015) Therapeutic targets of triple-negative breast cancer: a review. Br J Pharmacol 172(17):4228–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andre F, Zielinski CC (2006) Gradient doping negative electron affinity GaAs photocathodes. Opt Eng 23(Suppl 6):vi46–vi51 [Google Scholar]
- 6.Descamps S, Toillon RA, Adriaenssens E, Pawlowski V, Cool SM, Nurcombe V, Le Bourhis X, Boilly B, Peyrat JP, Hondermarck H (2001) Nerve growth factor stimulates proliferation and survival of human breast cancer cells through two distinct signaling pathways. J Biol Chem 276(21):17864–17870 [DOI] [PubMed] [Google Scholar]
- 7.Khotskaya YB, Holla VR, Farago AF, Mills Shaw KR, Meric-Bernstam F, Hong DS (2017) Targeting TRK family proteins in cancer. Pharmacol Ther 173:58–66 [DOI] [PubMed] [Google Scholar]
- 8.Doebele RC, Davis LE, Vaishnavi A, Le AT, Estrada-Bernal A, Keysar S, Jimeno A, Varella-Garcia M, Aisner DL, Li Y et al. (2015) An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer Discov 5(10):1049–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian Q, Du P, Li S, Bai Z, Yang Y, Zeng J (2017) Effect of antitumor treatments on triple-negative breast cancer patients: a PRISMA-compliant network meta-analysis of randomized controlled trials. Medicine (Baltimore) 96(45):e8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, Nathenson M, Doebele RC, Farago AF, Pappo AS et al. (2018) Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med 378(8):731–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ross JS, Chung J, Elvin JE, Vergilio J-A, Ramkissoon S, Suh J, Severson E, Daniel S, Frampton GM, Fabrizio D, Hartmaier RJ, Albacker LA, Ali SM, Schrock AB, Miller VA, Stephens PJ, Gay LM (2018) Abstract P2–09–15: NTRK fusions in breast cancer: Clinical, pathologic and genomic findings. In: Proceedings of the 2017 San Antonio Breast Cancer Symposium: 2017; San Antonio, TX: AACR. Cancer Res. 10.1158/1538-7445.SABCS17-P2-09-15 [DOI] [Google Scholar]
- 12.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6(269):pl 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagadec C, Meignan S, Adriaenssens E, Foveau B, Vanhecke E, Romon R, Toillon RA, Oxombre B, Hondermarck H, Le Bourhis X (2009) TrkA overexpression enhances growth and metastasis of breast cancer cells. Oncogene 28(18):1960–1970 [DOI] [PubMed] [Google Scholar]
- 14.Sabari JK, Santini F, Bergagnini I, Lai WV, Arbour KC, Drilon A (2017) Changing the therapeutic landscape in non-small cell lung cancers: the evolution of comprehensive molecular profiling improves access to therapy. Curr Oncol Rep 19(4):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaishnavi A, Capelletti M, Le AT, Kako S, Butaney M, Ercan D, Mahale S, Davies KD, Aisner DL, Pilling AB et al. (2013) Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med 19(11):1469–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chong CR, Bahcall M, Capelletti M, Kosaka T, Ercan D, Sim T, Sholl LM, Nishino M, Johnson BE, Gray NS et al. (2017) Identification of existing drugs that effectively target NTRK1 and ROS1 rearrangements in lung cancer. Clin Cancer Res 23(1):204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang GM, Wong HY, Konishi H, Blair BG, Abukhdeir AM, Gustin JP, Rosen DM, Denmeade SR, Rasheed Z, Matsui W et al. (2013) Single copies of mutant KRAS and mutant PIK3CA cooperate in immortalized human epithelial cells to induce tumor formation. Cancer Res 73(11):3248–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai KV, Xiao N, Wang W, Gangi L, Greene J, Powell JI, Dickson R, Furth P, Hunter K, Kucherlapati R et al. (2002) Initiating oncogenic event determines gene-expression patterns of human breast cancer models. Proc Natl Acad Sci USA 99(10):6967–6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choong LY, Lim S, Chong PK, Wong CY, Shah N, Lim YP (2010) Proteome-wide profiling of the MCF10AT breast cancer progression model. PLoS ONE 5(6):e11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narayan S, Jaiswal AS, Law BK, Kamal MA, Sharma AK, Hromas RA (2016) Interaction between APC and Fen1 during breast carcinogenesis. DNA Repair (Amst) 41:54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hilborn E, Stal O, Jansson A (2017) Estrogen and androgen-converting enzymes 17beta-hydroxysteroid dehydrogenase and their involvement in cancer: with a special focus on 17beta-hydroxysteroid dehydrogenase type 1, 2, and breast cancer. Oncotarget 8(18):30552–30562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Croessmann S, Wong HY, Zabransky DJ, Chu D, Rosen DM, Cidado J, Cochran RL, Dalton WB, Erlanger B, Cravero K et al. (2017) PIK3CA mutations and TP53 alterations cooperate to increase cancerous phenotypes and tumor heterogeneity. Breast Cancer Res Treat 162(3):451–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin SI, Freedman VH, Risser R, Pollack R (1975) Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growth in vitro. Proc Natl Acad Sci USA 72(11):4435–4439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zabransky DJ, Yankaskas CL, Cochran RL, Wong HY, Croessmann S, Chu D, Kavuri SM, Red Brewer M, Rosen DM, Dalton WB et al. (2015) HER2 missense mutations have distinct effects on oncogenic signaling and migration. Proc Natl Acad Sci USA 112(45):E6205–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adriaenssens E, Vanhecke E, Saule P, Mougel A, Page A, Romon R, Nurcombe V, Le Bourhis X, Hondermarck H (2008) Nerve growth factor is a potential therapeutic target in breast cancer. Cancer Res 68(2):346–351 [DOI] [PubMed] [Google Scholar]
- 26.Cocco E, Scaltriti M, Drilon A (2018) NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol 15(12):731–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong HY, Wang GM, Croessmann S, Zabransky DJ, Chu D, Garay JP, Cidado J, Cochran RL, Beaver JA, Aggarwal A et al. (2015) TMSB4Y is a candidate tumor suppressor on the Y chromosome and is deleted in male breast cancer. Oncotarget 6(42):44927–44940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes RM, Simons BW, Khan H, Miller R, Kugler V, Torquato S, Theodros D, Haffner MC, Lotan T, Huang J et al. (2019) Asporin restricts mesenchymal stromal cell differentiation, alters the tumor microenvironment, and drives metastatic progression. Cancer Res 79(14):3636–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.