Abstract
Clinical manifestations of coronavirus disease 2019 (COVID-19) vary from asymptomatic virus shedding, nonspecific pharyngitis, to pneumonia with silent hypoxia and respiratory failure. Dendritic cells and macrophages are sentinel cells for innate and adaptive immunity that affect the pathogenesis of severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS). The interplay between SARS-CoV-2 and these cell types remains unknown. We investigated infection and host responses of monocyte-derived dendritic cells (moDCs) and macrophages (MDMs) infected by SARS-CoV-2. MoDCs and MDMs were permissive to SARS-CoV-2 infection and protein expression but did not support productive virus replication. Importantly, SARS-CoV-2 launched an attenuated interferon response in both cell types and triggered significant proinflammatory cytokine/chemokine expression in MDMs but not moDCs. Investigations suggested that this attenuated immune response to SARS-CoV-2 in moDCs was associated with viral antagonism of STAT1 phosphorylation. These findings may explain the mild and insidious course of COVID-19 until late deterioration.
Keywords: COVID-19, SARS-CoV-2, coronavirus, dendritic cells, macrophages, moDCs, MDMs
MoDCs and MDMs were permissive to SARS-CoV-2 infection but not replication. The virus did not trigger IFN response in either cell type. The attenuated interferon and proinflammatory response in SARS-CoV-2–infected moDCs was associated with viral antagonism of STAT1 phosphorylation.
A novel human coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was recently identified as the causative agent of coronavirus disease 2019 (COVID-19) pandemic, affecting more than 4 million patients with over 250 000 deaths globally in less than 6 months [1–4]. Clinical features of COVID-19 in early reports included fever, myalgia, dry cough, dysphonia, fatigue, lymphopenia, and radiological evidence of multifocal peripheral ground-glass lung opacities. About 15% of COVID-19 cases progressed to acute respiratory distress syndromes and multiorgan failure [5]. Extrapulmonary manifestations included diarrhea, anosmia, dysgeusia, meningoencephalitis, thromboembolism, and multisystemic inflammatory syndrome. Despite sharing some clinical manifestations with that of SARS, a large proportion of COVID-19 cases were mildly symptomatic or asymptomatic, which may contribute to the high transmissibility of COVID-19 [6, 7].
Dendritic cells and macrophages are key sentinel cells of the host that play essential roles in the innate and adaptive immune defense. However, a number of viruses, including coronaviruses, are capable of infecting dendritic cells and macrophages to facilitate their replication and dissemination [8–10]. Importantly, we and others have reported that the highly pathogenic human coronaviruses, including SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV), triggered aberrant production of proinflammatory cytokines and chemokines in these cell types, which might contribute to viral pathogenesis [11–15]. In agreement with the in vitro findings, increasing monocyte, macrophage, and neutrophil infiltration was found in the lungs of SARS and MERS patients and was associated with elevated serum proinflammatory cytokines, contributing to disease severity and even multiorgan failure due to cytokine storm [16–18].
Similar to SARS, recent studies observed higher levels of cytokines and chemokines in severe COVID-19 cases as compared to mild cases, which is believed to contribute to COVID-19 disease progression and severity [19, 20]. We recently demonstrated that SARS-CoV-2 was capable of infecting the alveolar macrophages in ex vivo human lung tissues [21]. Similarly, SARS-CoV-2 also readily targeted the alveolar macrophages in the lungs of infected golden Syrian hamsters [22]. Additionally, CD68+CD169+ macrophages have been reported to be susceptible to SARS-CoV-2, which could contribute to viral spread, excessive inflammation, and activation-induced lymphocytic cell death during SARS-CoV-2 infection [23]. Despite the critical roles of dendritic cells and macrophages in coronavirus pathogenesis, the interplay between SARS-CoV-2 and these 2 important sentinel cell types remains elusive.
In this study, we comprehensively investigated the viral protein expression, replication kinetics, and host response in monocyte-derived dendritic cells (moDCs) and monocyte-derived macrophages (MDMs) upon SARS-CoV-2 infection. We found that despite efficient nucleocapsid (N) protein expression in SARS-CoV-2–infected moDCs or MDMs, infectious virus particles were not detected from the culture of both infected cell types. Interestingly, SARS-CoV-2 did not activate any interferon (IFN) gene upregulation, including that of IFN type I, type II, or type III, in both infected moDCs and MDMs. In addition, while SARS-CoV-2 triggered significant proinflammatory cytokines and chemokines expressions from infected MDMs, it did not activate the expression of these genes in the infected moDCs with the exception of IP-10. Further investigations suggested that the attenuated immune activation in SARS-CoV-2–infected moDCs was associated with viral antagonism of STAT1 phosphorylation.
METHODS
Cells and Viruses
All cell lines were purchased from the American Type Culture Collection. VeroE6 and Calu3 were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) and DMEM/F12, respectively, supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 μg/mL streptomycin. SARS-CoV-2 was isolated from the nasopharyngeal aspirate specimen of a laboratory-confirmed COVID-19 patient in Hong Kong [24]. SARS-CoV was a clinical isolate archived at Department of Microbiology, the University of Hong Kong. Both SARS-CoV-2 (GeneBank accession number: MT230904) and SARS-CoV GZ50 (AY304495) were titered in VeroE6 cells by plaque assays. All experiments involving live SARS-CoV-2 and SARS-CoV followed the approved standard operating procedures of the University of Hong Kong biosafety level-3 facility [24].
Preparation of moDCs and MDMs
Healthy volunteer blood samples were collected from the Hong Kong Red Cross Blood Transfusion Service according to a protocol approved by the institutional review board of the University of Hong Kong. Human peripheral blood mononuclear cells (PBMCs) were isolated from the buffy coats as previously described [25, 26]. MDMs were differentiated from PBMCs by providing RPMI-1640 supplemented with 10% FBS, 100 units/mL penicillin, 100 μg/mL streptomycin, 1% sodium pyruvate, 1% nonessential amino acids, and 1 U/mL granulocyte-macrophage colony-stimulating factor (Cell Sciences) [13]. MoDCs were differentiated and maintained as same condition as MDMs with interleukin-4 (IL-4; R&D systems) [14].
Immunofluorescence Staining and Confocal Microscopy
Immunostaining and confocal microscopy were performed as we previously described with slight modifications [27]. MoDCs and MDMs infected with SARS-CoV-2 or SARS-CoV were washed with phosphate-buffered saline and fixed in 4% paraformaldehyde. For immunostaining, the cells were permeabilized for 10 minutes with 0.2% Triton X-100 and blocked with Dako blocking buffer for 30 minutes at room temperature. The cells were incubated with an in-house rabbit antiserum against SARS-CoV-2 N protein [24] diluted with Dako antibody diluent overnight at 4°C, followed by incubation with Alexa Fluor secondary antibody (Thermo Fisher Scientific) diluted with Dako antibody diluent for 1 hour. The nuclei of cell were stained by 4′,6-diamidino-2-phenylindole (DAPI; Thermo Fisher Scientific) nucleic acid stain for 10 minutes. The slides were imaged with confocal microscopy using a Carl Zeiss LSM 880 system.
Virus Infection of moDCs and MDMs
MoDCs and MDMs were differentiated in complete RPMI-1640 with specific growth factors for 6–7 days before infection. For replication kinetics, cell viability, and host factor expressions (potential entry factors, cytokines, and chemokines) experiments, moDCs and MDMs were infected with SARS-CoV-2 or SARS-CoV at a multiplicity of infection (MOI) of 1 for 2 hours at 37°C. After 2 hours’ incubation, the virus inoculum was washed off and the cells were maintained with fresh RPMI-1640. For immunofluorescence microscopy experiment, moDCs and MDMs cultured in 24-well plates were digested by trypsin and reseeded into chamber slides. After 1 day of culture in full medium, the cells were infected with SARS-CoV-2 or SARS-CoV at an MOI of 10 for 2 hours at 37°C. After 2 hours’ incubation, the virus inoculum was washed off and the cells were maintained with fresh RPMI-1640.
Evaluation of Cell Viability
Cell viability of mock- or virus-infected moDCs and MDMs was determined using CellTiterGlo assays (Promega) according to manufacturer’s instruction. Briefly, the cells were infected with SARS-CoV-2 or SARS-CoV at an MOI of 1 for 2 hours at 37°C. The cells were lysed together with culture supernatant at a 1:1 ratio with CellTiterGlo reagent at the indicated hours post infection and placed on an orbital shaker for 10 minutes to induce cells lysis. The plates were read by measuring the luminescence signal with a Vector X3 multilabel plate reader (PerkinElmer) as we previously described [28].
RNA Extraction and Quantitative Reverse Transcription Polymerase Chain Reaction
To determine the viral genome copy number in cell lysate samples from infected cells, RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis were performed as previously described [24, 29]. In brief, real-time qRT-PCR was used to quantify SARS-CoV-2 and SARS-CoV genome copy number using QuantiNova Probe One Step RT-PCR kit (Qiagen) with LightCycler 96 Real-Time PCR System (Roche). The primers and probes sequences were designed to recognize viral RNA-dependent RNA polymerase/helicase regions. Forward primer for both SARS-CoV-2 and SARS-CoV: 5′-CGCATACAGTCTTRCAGGCT-3′; reverse primer for both SARS-CoV-2 and SARS-CoV: 5′-GTGTGATGTTGAWATGACATGGTC-3′; SARS-CoV-2–specific probe: 5′-FAM-TTAAGATGTGGTCTTGCATACGTAGAC-IABkFQ-3′; and SARS-CoV–specific probe: 5′-Cy5-CTTCGTTGCGGTGCCTGTGCCTGTATTAGGIAbRQSp-3′. Cellular RNA extraction, reverse transcription, and quantitative PCR were performed as we described previously [21]. For entry factors, cytokines, and chemokines analysis, the cells were lysed using the RLT buffer provided from the RNA extraction kit (Qiagen). The levels of cellular gene expression were normalized to glyceraldehyde-3-phosphate dehydrogenase and presented as fold change in gene expression of infected cells relative to that of mock-infected cells. Entry factors analyzed included angiotensin-converting enzyme 2 (ACE2), transmembrane protease serine 2 (TMPRSS2), FURIN, and dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin (DC-SIGN). Cytokines and chemokines analyzed included IFN-α, IFN-β, IFN-γ, IFN-λ1, tumor necrosis factor-α (TNF-α), IL-6, IL-8, IFN-γ inducible protein-10 (IP-10), monocyte chemoattractant protein-1 (MCP-1), macrophage-inflammatory protein-1α (MIP-1α), regulated upon activation normal T-cell expressed and secreted (RANTES), and IL-1β.
Western Blots and Analysis
Western blots were performed as we previously described with slight modifications [30]. MoDCs and MDMs were infected with SARS-CoV-2 or SARS-CoV at an MOI of 10 at 37°C. At 24 hours post infection, the cells were treated with 1000 IU/mL of recombinant human IFN-α (PBL Assay Science) for 40 minutes and then lysed with RIPA buffer supplemented with protease inhibitor cocktail and phosphatase inhibitor cocktail II (Thermo Fisher Scientific). The expression levels of STAT1, STAT1 phosphotyrosine residue 701 (pSTAT1), and β-actin were detected using anti-STAT1 (catalog number, 610186; BD Biosciences), anti-pSTAT1 (catalog number, 58D6; Cell Signaling Technology), and anti-β-actin (catalog number, A5441; Sigma-Aldrich) primary antibodies, respectively. The intensity of the bands was quantified using Image J software.
Statistical Analysis
Data represent mean and standard deviations. Statistical significance was calculated with 1-way ANOVA or 2-way ANOVA, and was considered significant when P < .05.
RESULTS
Infection of moDCs and MDMs by SARS-CoV-2
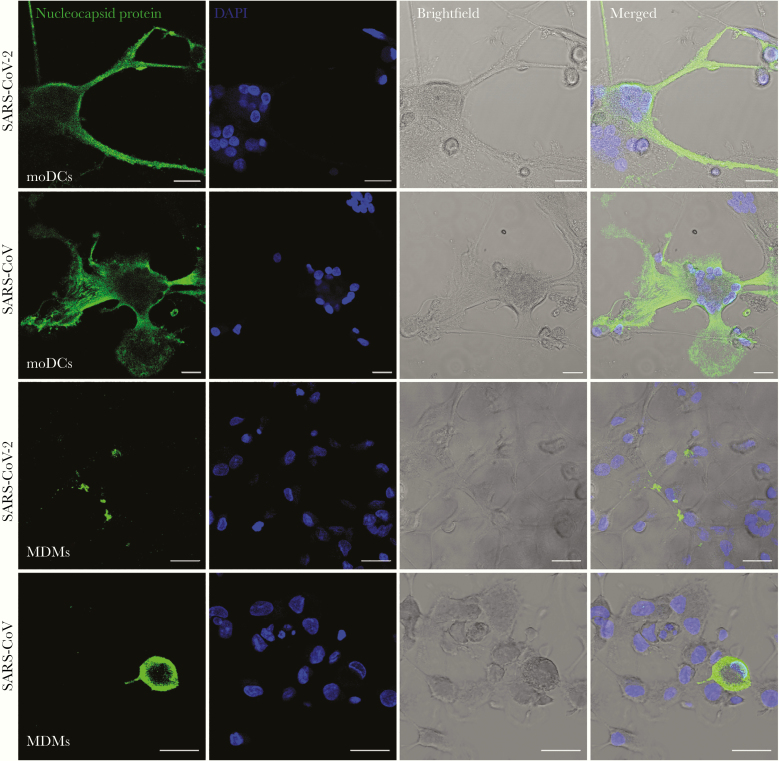
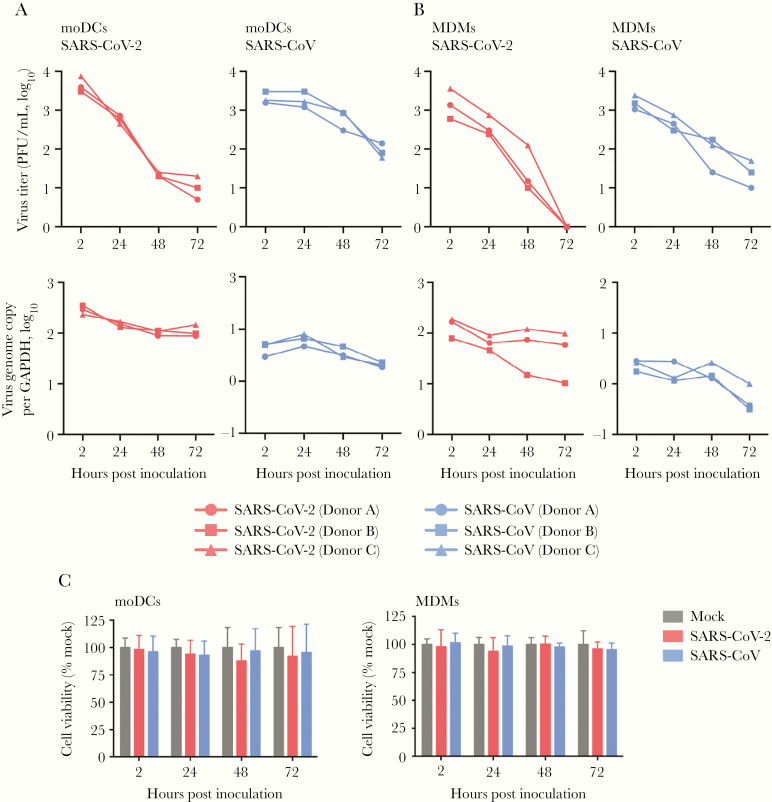
To investigate whether human dendritic cells and macrophages are susceptible to SARS-CoV-2 infection, we isolated and differentiated primary moDCs and MDMs from monocytes of healthy donors, and infected the cells with SARS-CoV-2. The expression of SARS-CoV-2 N protein in the infected moDCs and MDMs was evaluated using immunofluorescence staining and confocal microscopy. Our result demonstrated that SARS-CoV-2 N protein expression was readily detected from both infected moDCs and MDMs, suggesting that both cell types were permissive to SARS-CoV-2 infection (Figure 1). In particular, substantial cell-cell fusion was detected in SARS-CoV-2–infected moDCs (Figure 1). Next, we further evaluated the replication kinetics of SARS-CoV-2 in moDCs and MDMs with SARS-CoV included for comparison. As demonstrated in Figure 2, the virus titer in the supernatants and virus genome copy in the cell lysates did not increase over the 72-hour interval in both SARS-CoV-2–infected moDCs (Figure 2A) and MDMs (Figure 2B), suggesting that SARS-CoV-2 infection of moDCs and MDMs were abortive. We previously reported that MERS-CoV rapidly triggered substantial apoptosis in primary human T lymphocytes upon virus inoculation even in the absence of productive virus replication [31]. To investigate whether infection of SARS-CoV-2 induce cell death in moDCs and MDMs, we evaluated cell viability upon virus infection by the CellTiterGlo assays. As demonstrated in Figure 2C, SARS-CoV-2 infection resulted in a modest reduction in viability (10%–20%) in the infected moDCs between 24 and 72 hours post inoculation, which was not statistically significant due to sizable donor-to-donor variability. The viability of MDMs was largely unaffected upon SARS-CoV-2 and SARS-CoV infection (Figure 2C). Overall, our result demonstrated that moDCs and MDMs were susceptible to SARS-CoV-2 infection. Both cell types supported viral protein production but did not efficiently support virus replication and generation of infectious virus progenies.
Figure 1.
MoDCs and MDMs were susceptible to SARS-CoV-2 infection. MoDCs and MDMs were challenged with SARS-CoV-2 or SARS-CoV at a MOI of 10. At 24 hours post inoculation, the cells were fixed in 4% paraformaldehyde, permeabilized, and incubated with an in-house rabbit antiserum against SARS-CoV-2 nucleocapsid protein, followed by incubation with Alexa Fluor 488 secondary antibody for 1 hour. The slides were imaged with confocal microscopy using a Carl Zeiss LSM 880 system. Bars represent 20 μm. Abbreviations: MOI, multiplicity of infection; DAPI, 4′,6-diamidino-2-phenylindole; moDCs, monocyte-derived dendritic cells; MDMs, monocyte-derived macrophages; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Figure 2.
Infection of SARS-CoV-2 in moDCs and MDMs was not productive. MoDCs (A) and MDMs (B) were infected with SARS-CoV-2 or SARS-CoV at an MOI of 1. The live infectious virus particles in the supernatants and the viral genome copy in the cell lysates were determined by plaque assays and qRT-PCR, respectively. C, The cell viability of moDCs and MDMs upon SARS-CoV-2 or SARS-CoV infection at an MOI of 1 was quantified at the indicated hours post infection using CellTiterGlo assays. The mean cell viability of SARS-CoV-2– or SARS-CoV–infected cells was compared with that of mock-infected cells at each time point. The results represent mean and standard deviations from 3 individual donors in 3 independent experiments. Statistical significance between the groups was determined with 1-way ANOVA and was consider significant when P < .05. Abbreviations: MOI, multiplicity of infection; PFU, plaque-forming unit; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; moDCs, monocyte-derived dendritic cells; MDMs, monocyte-derived macrophages; qRT-PCR, quantitative reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
SARS-CoV-2 Infection Modulated the Expression of Entry-Related Host Factors in moDCs and MDMs
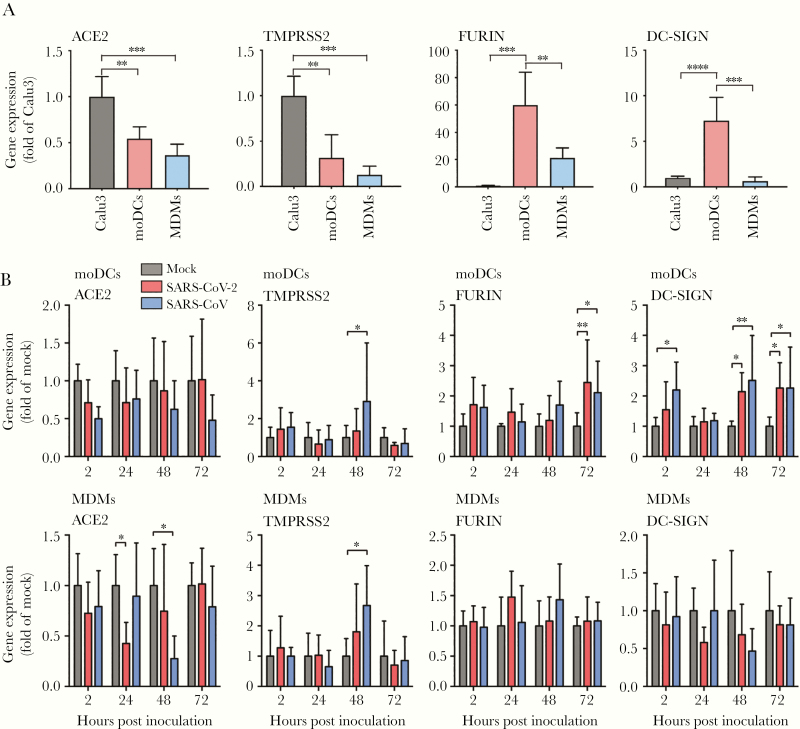
To dissect the reason behind the inefficient SARS-CoV-2 replication in moDCs and MDMs, we next assessed the endogenous expression of host factors that are potentially involved in SARS-CoV-2 infection and replication, which included ACE2, TMPRSS2, FURIN, and DC-SIGN. ACE2 was recently identified as the receptor of SARS-CoV-2 [32]. Efficient SARS-CoV-2 entry and replication also depend on host proteases, including TMPRSS2 and FURIN [32, 33]. DC-SIGN was previously suggested as an entry factor for SARS-CoV [34, 35]. Calu3 cells, which efficiently support SARS-CoV-2 replication, were included as a control group for comparison [24]. Our results demonstrated that while moDCs and MDMs expressed less ACE2 and TMPRSS2 than Calu3, they expressed 20- to 60-fold more FURIN than that of Calu3. In addition, moDCs expressed high level of DC-SIGN (Figure 3A). Upon infection, both SARS-CoV-2 and SARS-CoV had a trend to downregulate ACE2 expression in moDCs and MDMs, albeit the difference was only statistically significant in MDMs (Figure 3B). Interestingly, both viruses upregulated FURIN and DC-SIGN expression in moDCs upon infection. In addition, SARS-CoV but not SARS-CoV-2 upregulated the expression of TMPRSS2 in both moDCs and MDMs (Figure 3B). Taken together, our results suggested that the inefficient replication of SARS-CoV-2 in moDCs and MDMs was not a result of expression deficiency in receptor and host protease, and that SARS-CoV-2 infection modulated the expression of these entry-related host factors in the absence of productive virus replication.
Figure 3.
SARS-CoV-2 modulated the differential expression of entry-related host factors in moDCs and MDMs. A, The expression of endogenous ACE2, TMPRSS2, FURIN, and DC-SIGN was evaluated in moDCs and MDMs. Calu3 cells were included as a positive control. B, MoDCs and MDMs were infected with SARS-CoV-2 or SARS-CoV at an MOI of 1. The cell lysates were harvested to detect expression of entry-related host factors using qRT-PCR at the indicated hours post infection. The expression of entry-related host factors in SARS-CoV-2–infected or SARS-CoV–infected cells was compared with that of mock-infected cells at each time point. The results represent mean and standard deviations from 3 to 6 individual donors in 3 independent experiments. Statistical significance between groups was determined with 1-way ANOVA and was consider significant when P < .05. * P < .05, ** P < .01, *** P < .001, **** P < .0001. Abbreviations: MOI, multiplicity of infection; ACE2, angiotensin-converting enzyme 2; TMPRSS2; transmembrane protease serine 2; DC-SIGN, dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin; moDCs, monocyte-derived dendritic cells; MDMs, monocyte-derived macrophages; qRT-PCR, quantitative reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
SARS-CoV-2 Infection Did Not Activate IFN Response in moDCs and MDMs But Triggered Significant Proinflammatory Response in MDMs
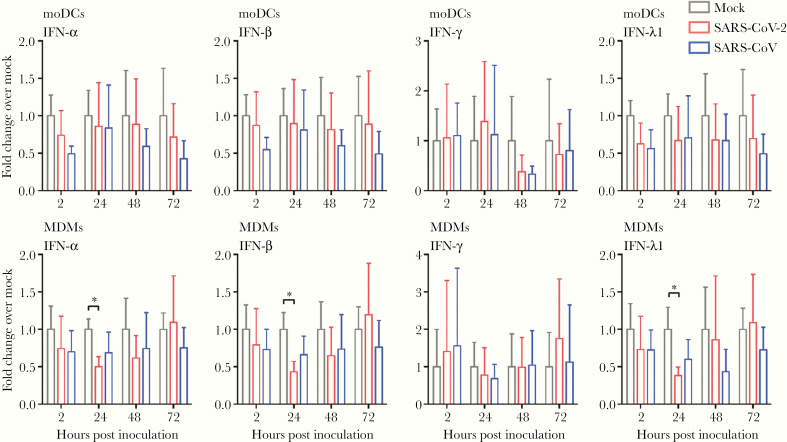
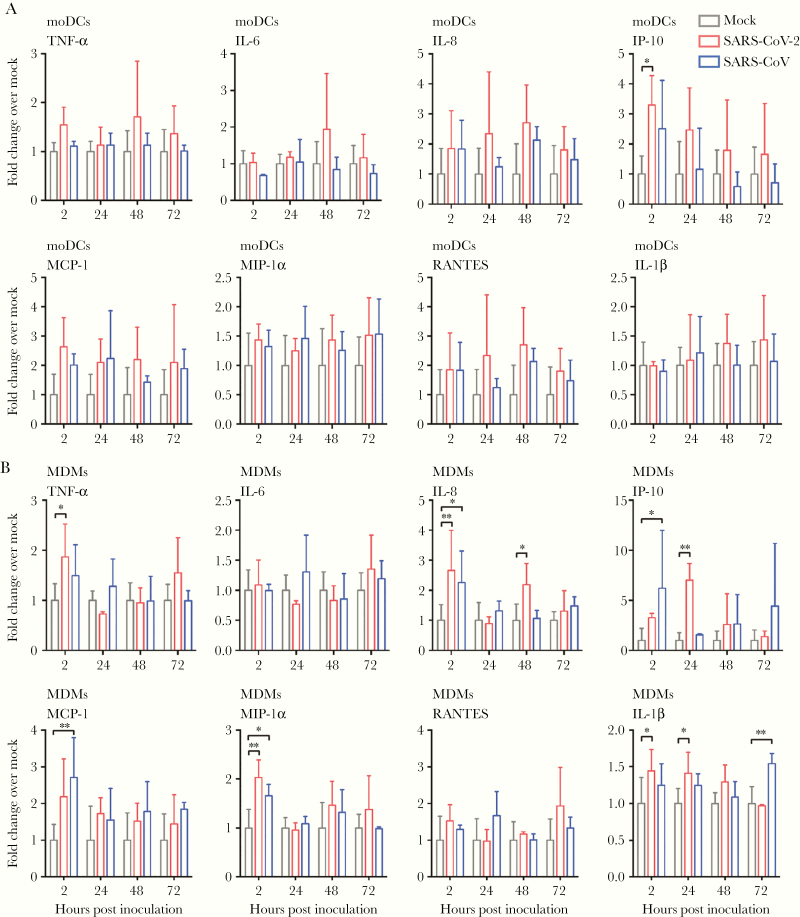
We next investigated the expression profile of representative IFN and proinflammatory cytokines and chemokines in moDCs and MDMs upon SARS-CoV-2 infection. Importantly, SARS-CoV-2 did not activate any IFN response, including that of IFN type I (IFN-α and IFN-β), IFN type II (IFN-γ), and IFN type III (IFN-λ1), from infected moDCs and MDMs (Figure 4). Intriguingly, in SARS-CoV-2–infected but not SARS-CoV–infected MDMs, the expression of IFN type I (IFN-α and IFN-β) and IFN type III (IFN-λ1) was even lower than that of the mock-infected MDMs at 24 hours post infection (Figure 4). In parallel, our data suggested that the expression of proinflammatory cytokines and chemokines was not substantially triggered by SARS-CoV-2 infection in moDCs. Specifically, among the 8 representative cytokine/chemokine evaluated, SARS-CoV-2 only significantly upregulated the expression of IP-10 (3.3-fold, P = .015) (Figure 5A). In contrast, SARS-CoV-2 significantly activated the expression of 5 of the 8 (5/8 or 62.5%) evaluated proinflammatory cytokine/chemokine in MDMs, including that of TNF-α, IL-8, IP-10, MIP-1α, and IL-1β. The most upregulated proinflammatory cytokine/chemokine in MDMs by SARS-CoV-2 infection was again IP-10 (7.0-fold, P = .003) (Figure 5B). Collectively, these findings demonstrated that SARS-CoV-2 infection did not activate IFN type I, II, or III response in moDCs and MDMs. In addition, while SARS-CoV-2 triggered significant proinflammatory cytokine/chemokine expressions in the infected MDMs, the proinflammatory response in moDCs upon SARS-CoV-2 infection was largely absent.
Figure 4.
Attenuated IFN response in SARS-CoV-2–infected moDCs and MDMs. MoDCs and MDMs were inoculated with SARS-CoV-2 or SARS-CoV at an MOI of 1. The cell lysates were harvested for qRT-PCR analysis of type I (IFN-α and IFN-β), II (IFN-γ), and III (IFN-λ1) IFN. The results represent mean and standard deviations from 3 to 6 individual donors in 3 independent experiments. Statistical significance between the groups was determined with 2-way ANOVA and was consider significant when P < .05. * P < .05. Abbreviations: MOI, multiplicity of infection; IFN, interferon; moDCs, monocyte-derived dendritic cells; MDMs, monocyte-derived macrophages; qRT-PCR, quantitative reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Figure 5.
SARS-CoV-2 induced significant proinflammatory response in MDMs but not moDCs. MoDCs (A) and MDMs (B) were inoculated with SARS-CoV-2 or SARS-CoV at an MOI of 1. The cell lysates were harvested for qRT-PCR analysis of representative proinflammatory cytokines and chemokines. The results represent mean and standard deviations from 3 to 6 individual donors in 3 independent experiments. Statistical significance between the groups was determined with 2-way ANOVA and was consider significant when P < .05. * P < .05, ** P < .01. Abbreviations: MOI, multiplicity of infection; TNF-α, tumor necrosis factor-α; IP-10, IFN-γ inducible protein-10; MCP-1, monocyte chemoattractant protein-1; MIP-1α, macrophage-inflammatory protein-1α; RANTES, regulated upon activation normal T-cell expressed and secreted; moDCs, monocyte-derived dendritic cells; MDMs, monocyte-derived macrophages; qRT-PCR, quantitative reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
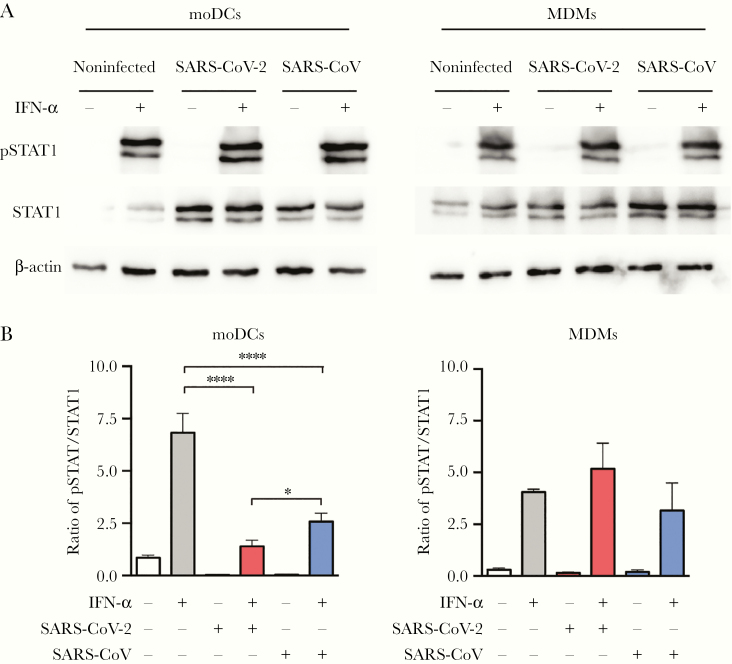
SARS-CoV-2 Antagonized STAT1 Phosphorylation in moDCs
Despite successful virus entry and protein production, antiviral IFN and proinflammatory response were not triggered by SARS-CoV-2 infection in moDCs, suggesting potential manipulation of the IFN signaling pathways by SARS-CoV-2 infection. To this end, we evaluated the protein expression level of STAT1 and examined the efficiency of STAT1 phosphorylation upon IFN-α treatment with or without SARS-CoV-2 infection. Our results demonstrated that SARS-CoV-2 infection upregulated STAT1 protein expression in moDCs (Figure 6A). We next evaluated the activation of STAT1 by measuring the ratio of phosphorylated STAT1 over STAT1 based on the intensity of individual bands. Interestingly, our data demonstrated that while IFN-α treatment substantially activated STAT1 phosphorylation, the extent of upregulation was significantly mitigated by SARS-CoV-2 infection in moDCs (Figure 6B). Importantly, while SARS-CoV also inhibited STAT1 phosphorylation in moDCs, the extent of inhibition by SARS-CoV-2 was significantly stronger than that by SARS-CoV (P = .035). In contrast, SARS-CoV-2 infection did not have a modulatory effect on STAT1 phosphorylation in MDMs (Figure 6B). Taken together, our results suggested that SARS-CoV-2 infection subverted IFN signaling in moDCs through antagonizing STAT1 phosphorylation.
Figure 6.
SARS-CoV-2 antagonized STAT1 phosphorylation in moDCs. MoDCs and MDMs were mock-infected or infected with SARS-CoV-2 or SARS-CoV at an MOI of 10. At 24 hours post infection, the cells were untreated or treated with 1000 U/mL of recombinant human IFN-α for 40 minutes. The cell lysates were collected for the detection of STAT1, pSTAT1, and β-actin by Western blots. A, Representative blots are shown from 3 donors in 3 independent experiments. B, Quantitation was calculated as the ratio of pSTAT1 over STAT1 protein. Statistical analysis was performed with 1-way ANOVA and the differences were considered significant when P < .05. *P < .05, ****P < .0001. Abbreviations: MOI, multiplicity of infection; IFN, interferon; moDCs, monocyte-derived dendritic cells; MDMs, monocyte-derived macrophages; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
DISCUSSION
Dendritic cells and macrophages play essential roles in innate and adaptive immunity against virus infection but are also potent producers of cytokines that may contribute to immunopathology [11, 36]. Both cell types could be infected by the highly pathogenic human coronaviruses, including SARS-CoV and MERS-CoV, and were suggested to play a role in viral pathogenesis [11–14]. However, the interplay between SARS-CoV-2 and these 2 sentinel cell types is currently unknown. In this study, we found that moDCs and MDMs were permissive to SARS-CoV-2 infection but not replication. Specifically, SARS-CoV-2 did not productively replicate in both cell types despite efficient N protein expression. In addition, SARS-CoV-2 infection downregulated the ACE2 expression in both infected moDCs and MDMs. Importantly, SARS-CoV-2 did not upregulate any IFN response in both cell types and only triggered significant proinflammatory response in the infected MDMs but not moDCs. Furthermore, we demonstrated that the largely silent infection of SARS-CoV-2 in moDCs was associated with viral antagonism of STAT1 phosphorylation. Overall, our study contributes to our understanding on the role of dendritic cells and macrophages in COVID-19 biology.
Importantly, despite efficient viral protein production, SARS-CoV-2 infection did not activate type I (IFN-α and IFN-β), II (IFN-γ), or III (IFN-λ1) IFN response in moDCs and MDMs. The attenuated host IFN response is consistent with our previous findings from SARS-CoV-2–infected ex vivo human lung tissues [21] as well as a recent study that examined the transcriptional response of SARS-CoV-2–infected samples [37]. Type I IFN is known to potently inhibit the replication of human coronaviruses, including hCoV-229E, SARS-CoV, and MERS-CoV, both in vitro and in vivo [38–41]. A recent study similarly suggested that SARS-CoV-2 was highly sensitive to IFN treatment [42]. In this regard, the deficiency of SARS-CoV-2–infected moDCs and MDMs in launching effective IFN response might delay viral clearance and contribute to COVID-19 pathogenesis. The treatment of COVID-19 by IFN may jump start and improve the antiviral response of the host to achieve better clinical outcome [43].
Coronaviruses have evolved a variety of mechanisms to intervene in IFN signaling [44], including inhibition of IRF phosphorylation, antagonizing mitochondrial antiviral-signaling (MAVS)/retinoic acid-inducible gene-1 (RIG-I) or TANK-binding kinase 1 (TBK1)/inhibitor of NF-κB kinase epsilon (IKKε) signaling, and blocking STAT1 nuclear import [45–47]. A recent report suggested that SARS-CoV-2 failed to activate the induction of TBK1 and STAT1 [37]. However, the detailed mechanism of SARS-CoV-2–mediated IFN antagonism has not been explored. We found that SARS-CoV-2 infection did not trigger IFN and proinflammatory response with the exception of IP-10 in moDCs. We further showed that this attenuated host response was associated with inhibition of STAT1 phosphorylation. STAT1 is a key modulator of type I, II, and III signaling and plays critical roles in the innate immune response in the clearance of SARS-CoV [48, 49]. A previous study demonstrated that the nsp1 nonstructural protein of SARS-CoV antagonized type I IFN by suppressing STAT1 phosphorylation [50]. Further investigations should be conducted to determine the SARS-CoV-2 protein that is associated with inhibition of STAT1 phosphorylation.
Dysregulated host immune response and excessive production of inflammatory cytokines are associated with disease severity in SARS, MERS, and COVID-19 [5, 19, 51]. Interestingly, despite a lack of IFN response, SARS-CoV-2 triggered potent expression of proinflammatory mediators including that of TNF-α, IL-8, IP-10, MIP-1α, and IL-1β in the infected MDMs. Among them, IP-10 was of particular interest as it was the only inflammatory mediator that was upregulated in both moDCs and MDMs upon SARS-CoV-2 infection. Importantly, excessive IP-10 could exacerbate the pathology of ARDS (acute respiratory distress syndrome) [52] and was recently suggested to play a critical role in the pathogenesis of SARS-CoV-2 [53]. Together, our data suggest that the SARS-CoV-2–infected dendritic cells and macrophages in the lungs of COVID-19 patients might be a source of proinflammatory cytokine production that exacerbates the clinical manifestation of COVID-19.
Overall, our study has provided the first evidence that demonstrates SARS-CoV-2 infection and the associated host responses in primary human dendritic cells and macrophages, which provides novel insights into the pathogenesis of COVID-19.
Notes
Acknowledgments. We sincerely thank the staff at the Core Facility, Li Ka Shing Faculty of Medicine, The University of Hong Kong, for facilitation of the study.
Disclaimer. The funding sources had no role in the study design, data collection, analysis, interpretation, or writing of the report.
Financial support. This work was supported in part by the May Tam Mak Mei Yin, Shaw Foundation of Hong Kong, Richard Yu and Carol Yu, Michael Seak-Kan Tong, Respiratory Viral Research Foundation Limited, Hui Ming, Hui Hoy and Chow Sin Lan Charity Fund Limited, Chan Yin Chuen Memorial Charitable Foundation, Marina Man-Wai Lee, Hong Kong Hainan Commercial Association South China Microbiology Research Fund, Jessie and George Ho Charitable Foundation, Perfect Shape Medical Limited, Lo Ying Shek Chi Wai Foundation, and Kai Chong Tong; and the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Diseases and Research Capability on Antimicrobial Resistance, Department of Health, Hong Kong Special Administrative Region; Research Grants Council, Hong Kong Special Administrative Region, Theme-Based Research Scheme (grant number T11/707/15); Sanming Project of Medicine in Shenzhen, China (grant number SZSM201911014); and the High Level-Hospital Program, Health Commission of Guangdong Province, China.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Zhu N, Zhang D, Wang W, et al. . A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chan JF, Yuan S, Kok KH, et al. . A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 2020; 395:514–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chan JF, Kok KH, Zhu Z, et al. . Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect 2020; 9:221–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. World Health Organization. Coronavirus disease (COVID-19) situation report–108 https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200507covid-19-sitrep-108.pdf?sfvrsn=44cc8ed8_2. Accessed 23 June 2020.
- 5. Guan WJ, Ni ZY, Hu Y, et al. . Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; 382:1708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bai Y, Yao L, Wei T, et al. . Presumed asymptomatic carrier transmission of COVID-19. JAMA 2020; 323:1406–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nishiura H, Kobayashi T, Miyama T, et al. . Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19). Int J Infect Dis 2020; 94:154–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bray M, Geisbert TW. Ebola virus: the role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int J Biochem Cell Biol 2005; 37:1560–6. [DOI] [PubMed] [Google Scholar]
- 9. Wu L, KewalRamani VN. Dendritic-cell interactions with HIV: infection and viral dissemination. Nat Rev Immunol 2006; 6:859–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simoni MK, Jurado KA, Abrahams VM, Fikrig E, Guller S. Zika virus infection of Hofbauer cells. Am J Reprod Immunol 2017; 77:10.1111/aji.12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheung CY, Poon LL, Ng IH, et al. . Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J Virol 2005; 79:7819–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Law HK, Cheung CY, Ng HY, et al. . Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood 2005; 106:2366–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou J, Chu H, Li C, et al. . Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J Infect Dis 2014; 209:1331–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chu H, Zhou J, Wong BH, et al. . Productive replication of Middle East respiratory syndrome coronavirus in monocyte-derived dendritic cells modulates innate immune response. Virology 2014; 454–455:197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scheuplein VA, Seifried J, Malczyk AH, et al. . High secretion of interferons by human plasmacytoid dendritic cells upon recognition of Middle East respiratory syndrome coronavirus. J Virol 2015; 89:3859–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nicholls JM, Poon LL, Lee KC, et al. . Lung pathology of fatal severe acute respiratory syndrome. Lancet 2003; 361:1773–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Page C, Goicochea L, Matthews K, et al. . Induction of alternatively activated macrophages enhances pathogenesis during severe acute respiratory syndrome coronavirus infection. J Virol 2012; 86:13334–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Faure E, Poissy J, Goffard A, et al. . Distinct immune response in two MERS-CoV-infected patients: can we go from bench to bedside? PLoS One 2014; 9:e88716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang C, Wang Y, Li X, et al. . Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xiong Y, Liu Y, Cao L, et al. . Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect 2020; 9:761–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chu H, Chan JF, Wang Y, et al. . Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19 [published online ahead of print 9 April 2020]. Clin Infect Dis doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chan JF, Zhang AJ, Yuan S, et al. . Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility [published online ahead of print 26 March 2020]. Clin Infect Dis doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen Y, Feng Z, Diao B, et al. . The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. medRxiv 20045427 [Preprint]. 27. March 2020. [cited 23 June 2020]. Available from: https:// 10.1101/2020.03.27.20045427. [DOI] [Google Scholar]
- 24. Chu H, Chan JFW, Yuen TTT, et al. . Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe 2020; 1:e14–e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chu H, Wang JJ, Qi M, et al. . Tetherin/BST-2 is essential for the formation of the intracellular virus-containing compartment in HIV-infected macrophages. Cell Host Microbe 2012; 12:360–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chu H, Wang JJ, Qi M, et al. . The intracellular virus-containing compartments in primary human macrophages are largely inaccessible to antibodies and small molecules. PLoS One 2012; 7:e35297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chu H, Chan CM, Zhang X, et al. . Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. J Biol Chem 2018; 293:11709–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chu H, Yuen T, Chik KKH, et al. . Targeting the inositol-requiring enzyme-1 pathway efficiently reverts Zika virus-induced neurogenesis and spermatogenesis marker perturbations [published online ahead of print 21 May 2020]. ACS Infect Dis doi: 10.1021/acsinfecdis.9b00526. [DOI] [PubMed] [Google Scholar]
- 29. Chan JF, Yip CC, To KK, et al. . Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J Clin Microbiol 2020; 58:e00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chan CM, Chu H, Wang Y, et al. . Carcinoembryonic antigen-related cell adhesion molecule 5 is an important surface attachment factor that facilitates entry of Middle East respiratory syndrome coronavirus. J Virol 2016; 90:9114–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chu H, Zhou J, Wong BH, et al. . Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J Infect Dis 2016; 213:904–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoffmann M, Kleine-Weber H, Schroeder S, et al. . SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; 181:271–80.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020; 181:281–92.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang ZY, Huang Y, Ganesh L, et al. . pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol 2004; 78: 5642–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jeffers SA, Tusell SM, Gillim-Ross L, et al. . CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci U S A 2004; 101:15748–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Steinman RM, Hemmi H. Dendritic cells: translating innate to adaptive immunity. Curr Top Microbiol Immunol 2006; 311:17–58. [DOI] [PubMed] [Google Scholar]
- 37. Blanco-Melo D, Nilsson-Payant BE, Liu WC, et al. . Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020; 181:1036–45.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hensley LE, Fritz LE, Jahrling PB, Karp CL, Huggins JW, Geisbert TW. Interferon-beta 1a and SARS coronavirus replication. Emerg Infect Dis 2004; 10:317–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Haagmans BL, Kuiken T, Martina BE, et al. . Pegylated interferon-alpha protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat Med 2004; 10:290–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chan JF, Yao Y, Yeung ML, et al. . Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J Infect Dis 2015; 212:1904–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sperber SJ, Hayden FG. Comparative susceptibility of respiratory viruses to recombinant interferons-alpha 2b and -beta. J Interferon Res 1989; 9:285–93. [DOI] [PubMed] [Google Scholar]
- 42. Lokugamage KG, Hage A, Schindewolf C, Rajsbaum R, Menachery VD. SARS-CoV-2 is sensitive to type I interferon pretreatment. BioRxiv 982264 [Preprint]. 9. April 2020. [cited 23 June 2020]. Available from: doi: 10.1101/2020.03.07.982264. [DOI]
- 43. Hung IF, Lung KC, Tso EY, et al. . Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet 2020; 395:1695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Totura AL, Baric RS. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol 2012; 2:264–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kamitani W, Huang C, Narayanan K, Lokugamage KG, Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat Struct Mol Biol 2009; 16:1134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Devaraj SG, Wang N, Chen Z, et al. . Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem 2007; 282:32208–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Siu KL, Kok KH, Ng MH, et al. . Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J Biol Chem 2009; 284:16202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Frieman MB, Chen J, Morrison TE, et al. . SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog 2010; 6:e1000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hogan RJ, Gao G, Rowe T, et al. . Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J Virol 2004; 78:11416–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wathelet MG, Orr M, Frieman MB, Baric RS. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J Virol 2007; 81:11620–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chan JF, Lau SK, To KK, Cheng VC, Woo PC, Yuen KY. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev 2015; 28:465–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ichikawa A, Kuba K, Morita M, et al. . CXCL10-CXCR3 enhances the development of neutrophil-mediated fulminant lung injury of viral and nonviral origin. Am J Respir Crit Care Med 2013; 187:65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang Y, Shen C, Li J, et al. . Exuberant elevation of IP-10, MCP-3 and IL-1ra during SARS-CoV-2 infection is associated with disease severity and fatal outcome. medRxiv 20029975 [Preprint]. 2. March 2020. [cited 23 June 2020]. Available from: doi: https:// 10.1101/2020.03.02.20029975. [DOI] [Google Scholar]