Abstract
The ubiquitin family member SUMO is a covalent regulator of proteins that functions in response to various stresses, and defects in SUMO-protein conjugation or deconjugation have been implicated in multiple diseases. The loss of the Ulp2 SUMO protease, which reverses SUMO-protein modifications, in the model eukaryote Saccharomyces cerevisiae is severely detrimental to cell fitness and has emerged as a useful model for studying how cells adapt to SUMO system dysfunction. Both short-term and long-term adaptive mechanisms are triggered depending on the length of time cells spend without this SUMO chain-cleaving enzyme. Such short-term adaptations include a highly specific multichromosome aneuploidy and large changes in ribosomal gene transcription. While aneuploid ulp2Δ cells survive, they suffer severe defects in growth and stress resistance. Over many generations, euploidy is restored, transcriptional programs are adjusted, and specific genetic changes that compensate for the loss of the SUMO protease are observed. These long-term adapted cells grow at normal rates with no detectable defects in stress resistance. In this review, we examine the connections between SUMO and cellular adaptive mechanisms more broadly.
Subject terms: Biochemistry, Molecular biology
Stress response: how cells wrestle with SUMO disruption
Cellular stress caused by disrupting attachment of the ubiquitous small ubiquitin-like modifier (SUMO) proteins, which are present in most organisms and regulate numerous DNA processes and stress responses by attaching to key proteins, results in some remarkable adaptations. Mark Hochstrasser at Yale University, New Haven, USA, and co-workers review how this “sumoylation” is reversed by protease enzymes, and how imbalances between sumoylation and desumoylation may be linked to diseases including cancer. When certain SUMO proteases are deliberately disrupted, the cells quickly become aneuploid, i.e., carry an abnormal number of chromosomes. These cells show severe growth defects, but over many generations they regain the normal number of chromosomes. They also undergo genetic changes that promote alternative mechanisms that compensate for losing the SUMO protease and facilitate the same efficient stress responses as the original cells.
Introduction
The small ubiquitin-like modifier (SUMO) protein is evolutionarily conserved and found in all eukaryotes1. While humans have four genes encoding SUMO proteins, SUMO1, 2, 3, and 4, the budding yeast Saccharomyces cerevisiae has only a single SUMO gene, SMT3, whose protein product shares 48% identity and 75% similarity with human SUMO12. In the sumoylation pathway, SUMO is translated as a C-terminally extended precursor, which is subsequently trimmed by SUMO-specific proteases to release the mature form. The protein is conjugated to lysine side chains of target proteins via an enzyme cascade similar to that used for ubiquitin-protein conjugation. A heterodimeric SUMO-activating enzyme (E1) first forms a thioester linkage through its active-site cysteine with the carboxy terminus of SUMO, and the SUMO moiety is then transferred to the active-site cysteine of a SUMO-conjugating enzyme (E2). Typically, SUMO is then conjugated to a lysine side chain(s) of a substrate protein, which is mediated by one of the several SUMO ligases (E3s)3. Chains of SUMO can also assemble on substrates. Sumoylation is reversed by site-specific proteases; nine SUMO proteases have been reported in humans and two, Ulp1 and Ulp2, in S. cerevisiae4.
The SUMO protein posttranslationally modifies diverse substrates involved in various cellular processes, including transcription, DNA replication, cell-cycle progression, nucleo-cytoplasmic transport, apoptosis, and genome integrity and stability5. SUMO is an essential regulator of cell homeostasis when cells encounter environmental stresses such as osmotic shock, hypoxia, heat, oxidative stress, nutrient deprivation, or genotoxic stresses, and protein sumoylation levels increase sharply in response to stress6. Although the SUMO stress response is still not fully understood, it was previously reported that the Siz1 E3 ligase and Ulp2 SUMO protease are major factors involved in the SUMO stress response in S. cerevisiae7.
Recently, our group reported that distinct adaptive mechanisms counter a dysregulated SUMO system upon loss of the Ulp2 protease8,9. To overcome the stress caused by the acute loss of Ulp2, mutant yeast cells become aneuploid (i.e., they carry an abnormal number of chromosomes), which promotes compensatory mechanisms for rapid adaptation to Ulp2 loss. However, because aneuploidy is usually deleterious to cell fitness10 and such ulp2Δ cells exhibit severely impaired growth, long-term adaptation restores euploidy and leads to countervailing mutations in SUMO conjugation enzymes and regulatory shifts in ribosome biogenesis. The stepwise employment of these mechanisms in response to disturbed SUMO conjugation dynamics is likely relevant to the robust adaptive fitness gains of cells following the loss of the quasi-essential Ulp2 protease. The present review provides an overview of the mechanisms of adaptation to environmental stress and particularly how the perturbation of the SUMO system is countered.
First-line adaptive mechanisms
The types of adaptation that occur in response to exogeneous or endogenous stress stimuli generally depend on the severity, duration, and reversibility of the stress conditions. If they do not exceed a certain threshold, stress effects are counterbalanced by transient protective mechanisms that promote cell survival11. Rapid “first-line” cellular responses include changes in metabolism, gene expression, cell-cycle progression, protein homeostasis, cytoskeletal organization, vesicular trafficking, and/or enzyme activity, which can re-establish homeostasis and maintain viability. For example, many heat shock proteins function as molecular chaperones that ensure the proper refolding of misfolded proteins and prevent or reverse protein aggregation under multiple environmental stress conditions12. Another example is the unfolded protein response, a highly conserved eukaryotic signaling pathway that responds to the accumulation of unfolded or misfolded proteins in the endoplasmic reticulum (ER)13. The accumulation of such aberrant proteins is sensed by transmembrane proteins in the ER that activate specific transcriptional programs; these include genes for ER molecular chaperones and ER-associated degradation pathways to help clear the ER of damaged proteins. In many species, protein translation is also decreased, and ER-associated mRNAs are selectively destroyed to reduce the protein client load of the ER protein folding machinery.
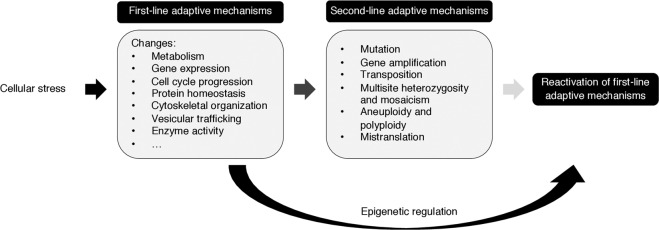
Other examples include disturbances in the balance between pro- and antioxidant factors, which cause an oxidative stress response that upregulates many antioxidant genes14, and DNA double-strand breaks (DSBs) and single-strand breaks induced by genotoxic stressors, which require the DNA lesions to be detected and repaired by the DNA damage response15. All of these first-line protective responses involve stresses that are reversible or short term. If a stress is persistent or irreversible, cells often engage a set of genetic, “second-line” adaptive mechanisms, which allow them to survive in the continued presence of the stress (Fig. 1)16.
Fig. 1. Stress-induced cellular adaptive strategies.
First-line adaptive mechanisms maximize cell survival under acute stress conditions. When these protective responses are not sufficient to protect cells from stress, the cells activate second-line adaptive mechanisms, which primarily consist of genetic changes that confer resistance to stress. However, because some second-line responses are deleterious to cell fitness, they may evolve or reactivate other adaptive mechanisms through genetic or epigenetic changes.
Second-line adaptive mechanisms
Mutation
Unlike spontaneous mutations, so-called adaptive mutations are genetic variations that are apparently induced specifically by exposure to an environment in which the mutation provides a selective advantage17. This contrasts with natural selection, in which cells bearing pre-existing genetic variants are selected if these mutations provide a reproductive advantage. Most studies of directed adaptive mutation, which remains a highly controversial concept (for example, see ref. 18), have been performed in Escherichia coli, but additional studies have been reported in other bacteria and eukaryotes, including yeast and mammals17,19,20. While there is no consensus regarding whether directed mutation can occur, there is a range of reported evidence for stress-induced hypermutation, which can increase the probability of creating favorable mutations that can be acted upon by natural selection21.
Stress-induced mutations include base substitutions, deletions, and insertions, and they are generated by evolutionarily conserved mechanisms. For instance, high rates of transcription have been associated with higher mutation frequencies due to exceeding the capacity of transcription-coupled DNA repair, which normally represses elevated rates of recombination and mutagenesis during transcription in bacteria and yeast22–24. The accumulation of genetic alterations in certain liver cancer cells also depends on active transcription25. Error-prone DNA polymerases are responsible for stress-induced mutations in bacterial lac, yeast CAN1, and human androgen receptor genes26–28. On the other hand, defects or limitation of DNA-repair processes caused by particular stress conditions in bacteria and mammalian cells or aneuploidy stress in yeast leads to increased mutation rates29–31. There is a switch from homologous recombination-mediated DSB repair to the more error-prone nonhomologous end-joining DNA-repair pathway under specific stress conditions in yeast32, and similar mechanisms have been reported in human cancer cells33.
Gene amplification
A study involving the Cairns and Foster lac frameshift system revealed that gene amplification could also contribute to Lac+ colonies following long periods of incubation on lactose medium34. Because the DNA junctions of amplified units are located in regions including nonhomologous joints in Lac+ cells34, a plausible hypothesis is that adaptive point mutations and gene amplification are both commonly initiated from DSBs, but amplification might primarily occur when no homologous sequence is available during repair. In yeast, long-term adaption to a glucose-limited environment also leads to the amplification of genes encoding the high-affinity hexose transporter35, and in human glioblastomas, double minutes, another manifestation of gene amplification, develop in response to hypoxia, a tumor microenvironment factor36.
Transposition
Transposons are DNA elements that autonomously move to distal locations within the genome, often inducing or reverting existing mutations37. Transposon mobility contributes to diverse regulatory mechanisms38. For example, glycerol limitation triggers the site-specific insertion of the IS5 transposon into a site upstream of the E. coli glpFK operon39, activating the expression of the operon to promote glycerol utilization. Similarly, in glucose-limited yeast cultures, specific chromosomal rearrangements may bring an activating transposon sequence to a position next to the CIT1 gene, which encodes citrate synthase, a key enzyme in the tricarboxylic acid cycle35. In human neuroblastoma cells, oxidative stress induces an increase in the retrotransposition of long interspersed element-1 and the upregulation of its transcript40.
Multisite heterozygosity and mosaicism
Multisite heterozygosity is defined as the coexistence of differences in the genetic composition at multiple loci in the same population and is often increased by outcrossing between different populations under environmental stress conditions41. Outcrossing between two or more populations gives rise to a larger proportion of allelic variation or mosaicism in the genome. The DNA sequencing of natural isolates of yeast strains, which tend to preferentially undergo asexual reproduction, revealed ~63% heterozygosity, suggesting some fitness advantage for this condition42. Higher levels of heterozygosity in yeast are observed under diverse stress conditions, such as exposure to the antifungal drug fluconazole, growth at high temperature, or incompatibility of the mismatch repair genes MLH1-PMS143,44.
Aneuploidy and polyploidy
Aneuploidy (an abnormal number of chromosomes) is usually acquired through unequal chromosome segregation during cell division, resulting in an imbalance in chromosome copy number, accompanied by parallel changes in both mRNA and protein levels10,45. An extra or missing chromosome has detrimental effects on cellular fitness and causes genomic instability in various organisms, including Drosophila, C. elegans, mice, plants, and humans46. In addition, almost all cancer cells, including 90% of solid tumors and 75% of hematopoietic cancers, have an aberrant number of chromosomes47; however, it is still debated whether such aneuploidies are a cause or consequence of cancer.
Although aneuploidy generally reduces cell fitness, it can provide a selective advantage relative to euploid cells under certain circumstances10. In the case of such beneficial aneuploidy, the increased dosage of a specific gene(s) on the duplicated chromosome(s) can mitigate the deleterious effects of the stress that induced the aneuploidy. Yeast has emerged as a versatile model organism for studying the adaptive effects of aneuploidy48. Adaptive aneuploidies are triggered in response to various stresses, including the application of the drugs fluconazole, radicicol, and 4-NQO; the deletion of genes such as MYO1, RPS24A, or RNR1; nutrient limitation; high temperature; or high pH. Since aneuploidy is an acute compensation mechanism with long-term fitness costs, continued encounters with the same stress often activate alternative adaptive mechanisms that can restore euploidy49. For example, specific aneuploidies induced by heat or high pH have been found to be eliminated and replaced with (unidentified) gene mutations and alterations in gene expression after long-term exposure to these conditions50.
Adaptive polyploidy, usually resulting from whole-genome duplication, has been reported in some cases. In vitro laboratory evolution experiments with baker’s yeast revealed that tetraploids experience accelerated adaptation compared with haploids and diploids51. In the pathogenic yeast Cryptococcus neoformans grown in the presence of antifungal azole drugs, transient polyploid states appear to give rise to aneuploid progeny with heightened drug resistance; this situation increases the dosage of specific genes, including ERG11, which encodes the azole drug target52. The treatment of human HL-60 cells with SKF 10496, which targets the ERG11 homolog that functions in cholesterol biosynthesis, has been found to lead to polyploidy53. Therefore, both polyploidy and aneuploidy can provide selective advantages under specific stress conditions.
Mis-translation
An increased translation error rate is a less established mechanism for adaptation to cell stress. The fidelity of translation is normally quite high, at approximately one error per 103–104 amino acids incorporated into proteins; fidelity is maintained through highly accurate tRNA aminoacylation and mRNA codon-cognate tRNA anticodon pairing by the ribosome54. The error rate of protein synthesis is typically increased under stressful conditions, which provoke tRNA misacylation, ribosome miscoding, frameshift errors, and translational readthrough, but cells can tolerate substantial decreases in translation fidelity55. Although the synthesis of mutant proteins frequently leads to protein misfolding or aggregation, some mutant protein forms can enhance specific stress responses and adaptations55. For instance, misfolded proteins are preferentially directed to the proteasome, and the peptide fragments generated by proteasomes can serve as ligands in antigen presentation56. An intriguing model has been proposed in which methionine-substituted mutant proteins resulting from tRNA misacylation with Met during oxidative stress could potentially provide sites for reversible modification by reactive oxygen species, sparing oxidizable active-site residues in these proteins55. Met-enriched proteins tend to remain active longer than the equivalent Met-depleted versions under oxidative stress conditions57,58. In the human commensal yeast Candida albicans, translation of CUG codons as either Ser or Leu can result in greater diversity in cell surface proteins that are normally recognized by the host immune system or can lead to resistance to the antifungal agent fluconazole59,60.
Return to first-line or other adaptive mechanisms
As noted above, most first-line responses to stress stimuli are rapidly reversible when the stress is removed or ameliorated, allowing the cell to return to its basal state16. When exposed to persistent stresses, second-line defense mechanisms are triggered, but cells often reactivate first-line mechanisms that impose a lower long-term fitness cost (Fig. 1). A clear example was reported by Dahan and colleagues, who showed that the aneuploidy-based adaptation of yeast to high temperature was replaced by refinements in gene expression during prolonged evolution at 39 °C50.
Epigenetic regulation appears to play a critical role in these adjustments of the transcriptional program. Such epigenetic mechanisms allow rapid and reversible, but durable adaptations through histone or DNA modifications that mediate changes in transcription, chromatin structure, or pre-mRNA processing61. Prion-mediated changes in protein states, which can be inherited without changes at the genetic level, may provide another mechanism of adaptation62,63. Cells can therefore switch or combine distinct adaptive strategies to maximize survival under various selective pressures.
SUMO and stress responses
SUMO is an essential modulator of cellular responses to various environmental stresses, such as heat, oxidative, osmotic, or genotoxic stresses, which lead to increased global levels of sumoylation in yeast and mammals6. The sumoylation level of target protein(s) depends on the nature, duration, and intensity of the stress64. The conjugation of SUMO to individual targets can be dramatically altered in response to specific stresses. For instance, heat shock induces a large increase in the SUMO conjugation of targets such as the heat shock factor-1 (HSF1) and c-Myb transcription factors65,66, whereas the sumoylation of the c-Fos, topoisomerase 1, and promyelocytic leukemia proteins decreases upon heat shock67–69. An interesting response is observed under different levels of oxidative stress. Low doses of H2O2 (below 10 μM) have minimal effects on SUMO conjugation, but moderate amounts of H2O2 (below 1 mM) lead to the inhibition of global sumoylation due to the formation of disulfide bond(s) between the catalytic cysteines of the SUMO E1 and E2 enzymes70–72. High doses of oxidants actually increase bulk SUMO-2/3 conjugate levels, probably through the attenuation of SUMO protease activity. Hypoxic stress induces increased global protein sumoylation through the upregulation of the expression of SUMO161,73. SUMO is crucial for the response to genotoxic stress as well, and the inhibition of the SUMO pathway leads to increased sensitivity to a wide range of genotoxic agents in both yeast and human cells74,75.
A key element in the SUMO stress response is the regulation of transcription. SUMO suppresses the transcriptional activity of diverse activators, including c-Myb, a major regulator of cell proliferation65. Conversely, transcription can be activated by the sumoylation of proteins such as HSF1, resulting in increased DNA binding and activity66, or NEMO, which activates NFκB in response to genotoxic stress76. Other targets of the SUMO stress response include basal components of the transcription machinery such as the TFIID and Mediator complexes, chromatin remodeling factors, the transcriptional corepressor Tup1-Cyc8, and subunits of the Set3 and Rpd3 histone deacetylase complexes7. A common feature of various types of stress is the desumoylation of RNA polymerase III subunits, which correlates with a decrease in tRNA transcription77. Histone sumoylation is also generally associated with transcriptional repression78, and its removal by the Ulp2 SUMO protease is required for the promotion of RNA polymerase II transcription elongation in yeast79.
The Ulp2 SUMO protease and adaptations to its loss
Although the SUMO system is implicated in the regulation of many cellular processes, how cells adapt to inhibition or dysfunction of the system is still largely unknown. As a first step in analyzing such mechanisms, our group examined how cells adapt to the loss of the yeast Ulp2 SUMO protease, a polySUMO chain-depolymerizing enzyme (Fig. 2). The deletion of the ULP2 gene causes severe growth defects even under optimal conditions80. Whole-genome RNA sequencing revealed that cells that survived the elimination of Ulp2 display a twofold increase in transcript levels across two specific chromosomes, chromosome I and ChrXII; this was traced to the duplication of these chromosomes9. Ulp2 plays roles in chromosome segregation and the maintenance of centromere cohesion81, so its loss may also facilitate the generation of cells with this very specific but aberrant chromosome complement by increasing chromosome segregation errors.
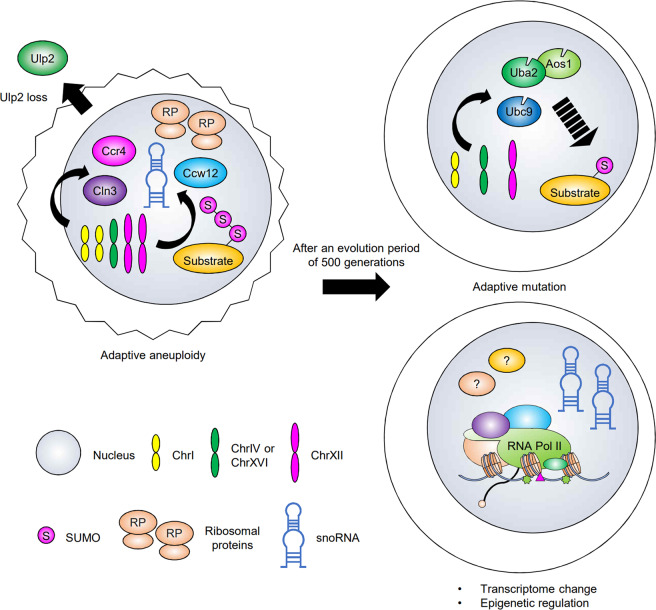
Fig. 2. Evolution of adaptive mechanisms upon the loss of Ulp2.
The loss of Ulp2 in yeast leads to the accumulation of polySUMO-conjugated proteins, increased expression of ribosomal proteins and reduced cell fitness (depicted by the irregular cell outline). Disomies of ChrI and ChrXII provide a transient adaptive solution by virtue of an increased dosage of three protein-coding genes, CCR4, CLN3, and CCW12, and a snoRNA gene cluster consisting of SNR61, SNR55, and SNR57. Following evolution over many cell generations, disomies of both ChrI and XII are replaced with two other adaptive mechanisms: mutations of SUMO-ligating enzymes and specific transcriptome changes. Point mutations in UBC9 or UBA2, on ChrIV, or AOS1, on ChrXVI, reduce SUMO conjugation and suppress the growth defects of ulp2Δ cells. In parallel, the upregulation of numerous snoRNA genes, which can repress the transcription of RP genes, and refined transcriptome alterations concomitant with epigenetic changes (unpublished data) appear to facilitate further adaptation.
The two-chromosome aneuploidy in ulp2Δ cells is an essential adaptation due to the increased dosage of three specific protein-coding genes, CCR4, CLN3, and CCW12, and a cluster of small nucleolar RNA (snoRNA) genes, SNR61, SNR55, and SNR578,9, which are all carried by these two chromosomes. Increased levels of Ccr4, a catalytic deadenylase subunit of the Ccr4–Not complex, in ulp2Δ mutant cells limit the expression of snoRNA and ribosomal RNA (rRNA) and thereby controls the synthesis of ribosomes, which is usually tightly coupled to growth. This compensates for the absence of Ulp2, which would normally check ribosome levels by inhibiting the transcription of the ribosomal protein (RP), snoRNA, and rRNA genes8. Increased levels of the three snoRNAs expressed from ChrXII also reduce the transcription of RP genes; interestingly, a previous report showed that a multiple myeloma-associated snoRNA, ACA11, downregulates RP gene expression in human cells82. An increased dosage of CLN3, encoding a G1 cyclin, likely prevents the development of aneuploidy by promoting cell-cycle progression. The adaptive role of elevated Ccw12, a mannoprotein required for cell wall integrity, is unknown.
Although these aneuploidy-mediated compensatory mechanisms help to overcome the acute stress caused by a dysregulated SUMO system, aneuploidy is deleterious, and aneuploid ulp2Δ cultures grow poorly and are highly sensitive to many stresses50,83. However, in vitro evolution over 250–500 generations restores euploidy8. Remarkably, despite the complete absence of the ULP2 gene, these evolved cells exhibit nearly normal growth and cell-cycle characteristics and do not present any obvious stress sensitivities. The evaluation of independent ulp2Δ cultures that evolved in parallel showed that the strains accrue different mutations in the genes encoding the SUMO-ligating enzymes Uba2/Aos1 (either subunit) or Ubc9. This is accompanied by a reduction in polySUMO-conjugate accumulation in most of the isolates. These results indicate that partial loss-of-function mutations in the essential SUMO ligation pathway can counter the hypersumoylation phenotype caused by Ulp2 loss8 and might affect the gene expression profile in a direction that increases cell fitness.
Interestingly, several of the evolved strains continued to maintain high levels of polySUMO conjugates, and no additional mutations in the SUMO pathway were found in these cells8. This indicates that additional adaptive mechanisms are possible. One such potential alternative mechanism might be the upregulation of snoRNA expression, which can repress the transcription of RPs and may help to resolve chromosome imbalances and restore stress resistance in ulp2Δ cells. However, additional copies of single snoRNAs do not suppress the growth defects caused by the loss of Ulp28. In addition to the evolutionary trajectories described above, we recently identified altered transcriptional profiles of certain genes in evolved ulp2Δ cells and likely epigenetic changes; we are now analyzing their potential adaptive advantages (unpublished data).
Eukaryotic ribosome biogenesis is critically linked to the SUMO pathway. The mammalian SUMO protease SENP3 physically interacts with multiple proteins involved in ribosome maturation, including NPM1 and the PELP1-WDR18-TEX10 complex, which are required for rRNA processing and the transit of the 60S ribosomal subunit from the nucleolus84. Several SUMO pathway mutants have been shown to exhibit defects in rRNA processing in S. cerevisiae as well85. SUMO and the Ulp2 protease localize to RP genes, reflecting the regulation of RP gene expression by balancing the sumoylation and desumoylation of the Rap1 transcription factor8,79,86. Consistent with these close ties between protein sumoylation and ribosome formation, regulatory shifts in ribosome biogenesis are an important factor in the response of cells to Ulp2 loss8.
Concluding remarks
Although SUMO is known to play essential roles in various stress responses, the adaptive mechanisms that allow cells to survive and flourish when elements of the essential SUMO system are altered have not been extensively explored. Here, we have briefly summarized various genetic or second-line adaptive responses to severe physiological stress or environmental insult, and we have described recent observations of the adaptation of cells to SUMO system dysfunction in particular. When the Ulp2 SUMO protease is lost, a complex adaptive aneuploidy is rapidly established; over longer periods, normal growth is usually restored by compensatory mutations in SUMO-ligating enzymes and the restoration of euploidy8,9. In response to severe or chronic stress, progression from rapid but nonideal adaptations to different, longer-term changes that boost fitness is likely to occur in association with many other pathways50. For instance, similar dynamic evolutionary trajectories may characterize a large fraction of tumors47.
The SUMO system is phylogenetically well conserved in eukaryotes, although it is not essential for viability in all organisms87. An interesting case relevant to the evolution of the SUMO system involves microsporidial Encephalitozoon spp. These obligate intracellular parasites have undergone extreme genome reduction, and their genome does not appear to encode a ULP2 ortholog, although it still harbors genes encoding a rudimentary SUMO pathway based on available genome sequences88. These organisms might have experienced a parallel deterioration in SUMO conjugation and deconjugation activities to maintain an optimal balance between them, similar to what we observed in our in vitro evolution studies in ulp2Δ cells. Analogous high-throughput in vitro evolution experiments hold promise for deciphering other aspects of SUMO system physiology.
Imbalances between sumoylation and desumoylation have been suggested to be important in the development of multiple diseases, such as cancer and neurodegenerative disorders89. Several SUMO enzymes appear to be upregulated in different cancers, where they seem to protect the stability and functionality of gene expression programs and signaling pathways in the face of cancer-induced changes90. Conversely, protein sumoylation contributes to certain pathological conditions and neurological disorders by promoting the formation of toxic protein aggregates89. The study of SUMO function in adaptive mechanisms may provide clues for the development of new therapeutic agents for these different disorders.
Acknowledgements
This study was supported by NIH grant GM053756 to M.H. and a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (no. 2020R1C1C1009367) to H.-Y.R and (no. NRF-2016R1A2B2008217) to S.H.A.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Johnson ES. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 2.Huang WC, Ko TP, Li SS, Wang AH. Crystal structures of the human SUMO-2 protein at 1.6 A and 1.2 A resolution: implication on the functional differences of SUMO proteins. Eur. J. Biochem. 2004;271:4114–4122. doi: 10.1111/j.1432-1033.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 3.Hendriks IA, Vertegaal AC. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 2016;17:581–595. doi: 10.1038/nrm.2016.81. [DOI] [PubMed] [Google Scholar]
- 4.Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 2012;13:755–766. doi: 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev. Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 6.Enserink JM. Sumo and the cellular stress response. Cell Div. 2015;10:4. doi: 10.1186/s13008-015-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewicki MC, Srikumar T, Johnson E, Raught B. The S. cerevisiae SUMO stress response is a conjugation-deconjugation cycle that targets the transcription machinery. J. Proteom. 2015;118:39–48. doi: 10.1016/j.jprot.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Ryu HY, et al. Distinct adaptive mechanisms drive recovery from aneuploidy caused by loss of the Ulp2 SUMO protease. Nat. Commun. 2018;9:5417. doi: 10.1038/s41467-018-07836-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryu HY, Wilson NR, Mehta S, Hwang SS, Hochstrasser M. Loss of the SUMO protease Ulp2 triggers a specific multichromosome aneuploidy. Genes Dev. 2016;30:1881–1894. doi: 10.1101/gad.282194.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheltzer JM, Amon A. The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends Genet. 2011;27:446–453. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulda S, Gorman AM, Hori O, Samali A. Cellular stress responses: cell survival and cell death. Int J. Cell Biol. 2010;2010:214074. doi: 10.1155/2010/214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kregel KC. Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J. Appl Physiol. (1985) 2002;92:2177–2186. doi: 10.1152/japplphysiol.01267.2001. [DOI] [PubMed] [Google Scholar]
- 13.Hetz C, Papa FR. The unfolded protein response and cell fate control. Mol. Cell. 2018;69:169–181. doi: 10.1016/j.molcel.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 14.Trachootham D, Lu W, Ogasawara MA, Nilsa RD, Huang P. Redox regulation of cell survival. Antioxid. Redox Signal. 2008;10:1343–1374. doi: 10.1089/ars.2007.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 2006;12:440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Cipponi A, Thomas DM. Stress-induced cellular adaptive strategies: ancient evolutionarily conserved programs as new anticancer therapeutic targets. Bioessays. 2014;36:552–560. doi: 10.1002/bies.201300170. [DOI] [PubMed] [Google Scholar]
- 17.Rosenberg SM. Evolving responsively: adaptive mutation. Nat. Rev. Genet. 2001;2:504–515. doi: 10.1038/35080556. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen A, Maisnier-Patin S, Yamayoshi I, Kofoid E, Roth JR. Selective Inbreeding: genetic crosses drive apparent adaptive mutation in the Cairns-foster system of Escherichia coli. Genetics. 2020;214:333–354. doi: 10.1534/genetics.119.302754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster PL. Adaptive mutation: implications for evolution. Bioessays. 2000;22:1067–1074. doi: 10.1002/1521-1878(200012)22:12<1067::AID-BIES4>3.0.CO;2-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heidenreich E. Adaptive mutation in Saccharomyces cerevisiae. Crit. Rev. Biochem Mol. Biol. 2007;42:285–311. doi: 10.1080/10409230701507773. [DOI] [PubMed] [Google Scholar]
- 21.Foster PL. Mechanisms of stationary phase mutation: a decade of adaptive mutation. Annu Rev. Genet. 1999;33:57–88. doi: 10.1146/annurev.genet.33.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datta A, Jinks-Robertson S. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]
- 23.Reimers JM, Schmidt KH, Longacre A, Reschke DK, Wright BE. Increased transcription rates correlate with increased reversion rates in leuB and argH Escherichia coli auxotrophs. Microbiology. 2004;150:1457–1466. doi: 10.1099/mic.0.26954-0. [DOI] [PubMed] [Google Scholar]
- 24.Pybus C, et al. Transcription-associated mutation in Bacillus subtilis cells under stress. J. Bacteriol. 2010;192:3321–3328. doi: 10.1128/JB.00354-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SK, et al. A model of liver carcinogenesis originating from hepatic progenitor cells with accumulation of genetic alterations. Int J. Cancer. 2014;134:1067–1076. doi: 10.1002/ijc.28445. [DOI] [PubMed] [Google Scholar]
- 26.Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 27.Shor E, Fox CA, Broach JR. The yeast environmental stress response regulates mutagenesis induced by proteotoxic stress. PLoS Genet. 2013;9:e1003680. doi: 10.1371/journal.pgen.1003680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara T, Kouno J, Nakamura K, Kusaka M, Yamaoka M. Possible role of adaptive mutation in resistance to antiandrogen in prostate cancer cells. Prostate. 2005;65:268–275. doi: 10.1002/pros.20282. [DOI] [PubMed] [Google Scholar]
- 29.Harris RS, et al. Mismatch repair protein MutL becomes limiting during stationary-phase mutation. Genes Dev. 1997;11:2426–2437. doi: 10.1101/gad.11.18.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheltzer JM, et al. Aneuploidy drives genomic instability in yeast. Science. 2011;333:1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mihaylova VT, et al. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol. Cell Biol. 2003;23:3265–3273. doi: 10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heidenreich E, Novotny R, Kneidinger B, Holzmann V, Wintersberger U. Non-homologous end joining as an important mutagenic process in cell cycle-arrested cells. EMBO J. 2003;22:2274–2283. doi: 10.1093/emboj/cdg203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bindra RS, Crosby ME, Glazer PM. Regulation of DNA repair in hypoxic cancer cells. Cancer Metastasis Rev. 2007;26:249–260. doi: 10.1007/s10555-007-9061-3. [DOI] [PubMed] [Google Scholar]
- 34.Hastings PJ, Bull HJ, Klump JR, Rosenberg SM. Adaptive amplification: an inducible chromosomal instability mechanism. Cell. 2000;103:723–731. doi: 10.1016/s0092-8674(00)00176-8. [DOI] [PubMed] [Google Scholar]
- 35.Dunham MJ, et al. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. U.S.A. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer U, Radermacher J, Mayer J, Mehraein Y, Meese E. Tumor hypoxia: Impact on gene amplification in glioblastoma. Int J. Oncol. 2008;33:509–515. [PubMed] [Google Scholar]
- 37.Munoz-Lopez M, Garcia-Perez JL. DNA transposons: nature and applications in genomics. Curr. Genomics. 2010;11:115–128. doi: 10.2174/138920210790886871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bucher E, Reinders J, Mirouze M. Epigenetic control of transposon transcription and mobility in Arabidopsis. Curr. Opin. Plant Biol. 2012;15:503–510. doi: 10.1016/j.pbi.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z, Saier MH., Jr. Transposon-mediated adaptive and directed mutations and their potential evolutionary benefits. J. Mol. Microbiol Biotechnol. 2011;21:59–70. doi: 10.1159/000333108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giorgi G, Marcantonio P, Del Re B. LINE-1 retrotransposition in human neuroblastoma cells is affected by oxidative stress. Cell Tissue Res. 2011;346:383–391. doi: 10.1007/s00441-011-1289-0. [DOI] [PubMed] [Google Scholar]
- 41.Raghavan V, Aquadro CF, Alani E. Baker’s yeast clinical isolates provide a model for how pathogenic yeasts adapt to stress. Trends Genet. 2019;35:804–817. doi: 10.1016/j.tig.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peter J, et al. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature. 2018;556:339–344. doi: 10.1038/s41586-018-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raghavan V, et al. Incompatibilities in mismatch repair genes MLH1-PMS1 contribute to a wide range of mutation rates in human isolates of Baker’s yeast. Genetics. 2018;210:1253–1266. doi: 10.1534/genetics.118.301550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magwene PM. Revisiting Mortimer’s genome renewal hypothesis: heterozygosity, homothallism, and the potential for adaptation in yeast. Adv. Exp. Med Biol. 2014;781:37–48. doi: 10.1007/978-94-007-7347-9_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavelka N, Rancati G, Li R. Dr Jekyll and Mr Hyde: role of aneuploidy in cellular adaptation and cancer. Curr. Opin. Cell Biol. 2010;22:809–815. doi: 10.1016/j.ceb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 47.Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 48.Mulla W, Zhu J, Li R. Yeast: a simple model system to study complex phenomena of aneuploidy. FEMS Microbiol. Rev. 2014;38:201–212. doi: 10.1111/1574-6976.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galluzzi L, Pedro JMBS, Kepp O, Kroemer G. Regulated cell death and adaptive stress responses. Cell Mol. Life Sci. 2016;73:2405–2410. doi: 10.1007/s00018-016-2209-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yona AH, et al. Chromosomal duplication is a transient evolutionary solution to stress. Proc. Natl. Acad. Sci. U.S.A. 2012;109:21010–21015. doi: 10.1073/pnas.1211150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selmecki AM, et al. Polyploidy can drive rapid adaptation in yeast. Nature. 2015;519:349–352. doi: 10.1038/nature14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwon-Chung KJ, Chang YC. Aneuploidy and drug resistance in pathogenic fungi. PLoS Pathog. 2012;8:e1003022. doi: 10.1371/journal.ppat.1003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandez C, Lobo Md Mdel V, Gomez-Coronado D, Lasuncion MA. Cholesterol is essential for mitosis progression and its deficiency induces polyploid cell formation. Exp. Cell Res. 2004;300:109–120. doi: 10.1016/j.yexcr.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 54.Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan T. Adaptive translation as a mechanism of stress response and adaptation. Annu Rev. Genet. 2013;47:121–137. doi: 10.1146/annurev-genet-111212-133522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yewdell JW, Reits E, Neefjes J. Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat. Rev. Immunol. 2003;3:952–961. doi: 10.1038/nri1250. [DOI] [PubMed] [Google Scholar]
- 57.Luo S, Levine RL. Methionine in proteins defends against oxidative stress. FASEB J. 2009;23:464–472. doi: 10.1096/fj.08-118414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. U.S.A. 1996;93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miranda I. et al. Candida albicans CUG mistranslation is a mechanism to create cell surface variation. mBio4, 10.1128/mBio.00285-13 (2013). [DOI] [PMC free article] [PubMed]
- 60.Weil T. et al. Adaptive mistranslation accelerates the evolution of fluconazole resistance and induces major genomic and gene expression alterations in Candida albicans. mSphere2, 10.1128/mSphere.00167-17 (2017). [DOI] [PMC free article] [PubMed]
- 61.Comerford KM, et al. Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc. Natl. Acad. Sci. U.S.A. 2003;100:986–991. doi: 10.1073/pnas.0337412100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fabrizio, P., Garvis, S. & Palladino, F. Histone methylation and memory of environmental stress. Cells8, 10.3390/cells8040339 (2019). [DOI] [PMC free article] [PubMed]
- 63.Westergard L, True HL. Wild yeast harbour a variety of distinct amyloid structures with strong prion-inducing capabilities. Mol. Microbiol. 2014;92:183–193. doi: 10.1111/mmi.12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tempe D, Piechaczyk M, Bossis G. SUMO under stress. Biochem. Soc. Trans. 2008;36:874–878. doi: 10.1042/BST0360874. [DOI] [PubMed] [Google Scholar]
- 65.Sramko M, Markus J, Kabat J, Wolff L, Bies J. Stress-induced inactivation of the c-Myb transcription factor through conjugation of SUMO-2/3 proteins. J. Biol. Chem. 2006;281:40065–40075. doi: 10.1074/jbc.M609404200. [DOI] [PubMed] [Google Scholar]
- 66.Hong Y, et al. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 2001;276:40263–40267. doi: 10.1074/jbc.M104714200. [DOI] [PubMed] [Google Scholar]
- 67.Bossis G, et al. Down-regulation of c-Fos/c-Jun AP-1 dimer activity by sumoylation. Mol. Cell. Biol. 2005;25:6964–6979. doi: 10.1128/MCB.25.16.6964-6979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mo YY, Yu Y, Shen Z, Beck WT. Nucleolar delocalization of human topoisomerase I in response to topotecan correlates with sumoylation of the protein. J. Biol. Chem. 2002;277:2958–2964. doi: 10.1074/jbc.M108263200. [DOI] [PubMed] [Google Scholar]
- 69.Nefkens I, et al. Heat shock and Cd2+ exposure regulate PML and Daxx release from ND10 by independent mechanisms that modify the induction of heat-shock proteins 70 and 25 differently. J. Cell Sci. 2003;116:513–524. doi: 10.1242/jcs.00253. [DOI] [PubMed] [Google Scholar]
- 70.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol. Cell. 2006;21:349–357. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 71.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 72.Xu Z, et al. Molecular basis of the redox regulation of SUMO proteases: a protective mechanism of intermolecular disulfide linkage against irreversible sulfhydryl oxidation. FASEB J. 2008;22:127–137. doi: 10.1096/fj.06-7871com. [DOI] [PubMed] [Google Scholar]
- 73.Shao R, et al. Increase of SUMO-1 expression in response to hypoxia: direct interaction with HIF-1alpha in adult mouse brain and heart in vivo. FEBS Lett. 2004;569:293–300. doi: 10.1016/j.febslet.2004.05.079. [DOI] [PubMed] [Google Scholar]
- 74.Jacquiau HR, et al. Defects in SUMO (small ubiquitin-related modifier) conjugation and deconjugation alter cell sensitivity to DNA topoisomerase I-induced DNA damage. J. Biol. Chem. 2005;280:23566–23575. doi: 10.1074/jbc.M500947200. [DOI] [PubMed] [Google Scholar]
- 75.Mo YY, Yu Y, Ee PL, Beck WT. Overexpression of a dominant-negative mutant Ubc9 is associated with increased sensitivity to anticancer drugs. Cancer Res. 2004;64:2793–2798. doi: 10.1158/0008-5472.can-03-2410. [DOI] [PubMed] [Google Scholar]
- 76.Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell. 2003;115:565–576. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- 77.Nguea PA, Robertson J, Herrera MC, Chymkowitch P, Enserink JM. Desumoylation of RNA polymerase III lies at the core of the Sumo stress response in yeast. J. Biol. Chem. 2019;294:18784–18795. doi: 10.1074/jbc.RA119.009721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nathan D, et al. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 2006;20:966–976. doi: 10.1101/gad.1404206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ryu HY, et al. The Ulp2 SUMO protease promotes transcription elongation through regulation of histone sumoylation. EMBO J. 2019;38:e102003. doi: 10.15252/embj.2019102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li SJ, Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 2000;20:2367–2377. doi: 10.1128/mcb.20.7.2367-2377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baldwin ML, Julius JA, Tang XY, Wang YC, Bachant J. The yeast SUMO isopeptidase Smt4/Ulp2 and the polo kinase Cdc5 act in an opposing fashion to regulate sumoylation in mitosis and cohesion at centromeres. Cell Cycle. 2009;8:3406–3419. doi: 10.4161/cc.8.20.9911. [DOI] [PubMed] [Google Scholar]
- 82.Chu L, et al. Multiple myeloma-associated chromosomal translocation activates orphan snoRNA ACA11 to suppress oxidative stress. J. Clin. Invest. 2012;122:2793–2806. doi: 10.1172/JCI63051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ryu HY, Hochstrasser M. Adaptive aneuploidy counters a dysregulated SUMO system. Cell Cycle. 2017;16:383–385. doi: 10.1080/15384101.2016.1256154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Finkbeiner E, Haindl M, Raman N, Muller S. SUMO routes ribosome maturation. Nucleus. 2011;2:527–532. doi: 10.4161/nucl.2.6.17604. [DOI] [PubMed] [Google Scholar]
- 85.Panse VG, et al. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic. 2006;7:1311–1321. doi: 10.1111/j.1600-0854.2006.00471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chymkowitch P, et al. Sumoylation of Rap1 mediates the recruitment of TFIID to promote transcription of ribosomal protein genes. Genome Res. 2015;25:897–906. doi: 10.1101/gr.185793.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2000;2:E153–E157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- 88.Haag KL, et al. Evolution of a morphological novelty occurred before genome compaction in a lineage of extreme parasites. Proc. Natl Acad. Sci. U.S.A. 2014;111:15480–15485. doi: 10.1073/pnas.1410442111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang Y. et al. Protein SUMOylation modification and its associations with disease. Open Biol. 7, 10.1098/rsob.170167 (2017). [DOI] [PMC free article] [PubMed]
- 90.Seeler JS, Dejean A. SUMO and the robustness of cancer. Nat. Rev. Cancer. 2017;17:184–197. doi: 10.1038/nrc.2016.143. [DOI] [PubMed] [Google Scholar]