Abstract
The development of mutant BRAF inhibitors has improved the outcome for melanoma patients with BRAFV600E mutations. Although the initial response to these inhibitors can be dramatic, sometimes resulting in complete tumor regression, the majority of melanomas become resistant. To study resistance to BRAF inhibition, we developed a novel mouse model of melanoma using a tetracycline/doxycycline-regulated system that permits control of mutant BRAF expression. Treatment with doxycycline leads to loss of mutant BRAF expression and tumor regression, but tumors recur after a prolonged period of response to treatment. Vemurafenib, encorafenib and dabrafenib induce cell cycle arrest and apoptosis in BRAF melanoma cell lines; however, a residual population of tumor cells survive. Comparing gene expression in human cell lines and mouse tumors can assist with the identification of novel mechanisms of resistance. Accordingly, we conducted RNA sequencing analysis and immunoblotting on untreated and doxycycline-treated dormant mouse melanomas and human mutant BRAF melanoma cell lines treated with 2 μM vemurafenib for 20 days. We found conserved expression changes in histone methyltransferase genes ASH2, EZH2, PRMT5, SUV39H1, SUV39H2, and SYMD2 in P-ERK low, p-38 high melanoma cells following prolonged BRAF inhibition. Quantitative mass spectrometry, determined a corresponding reduction in histone Lys9 and Lys27 methylation and increase in Lys36 methylation in melanoma cell lines treated with 2 μM vemurafenib for 20 days. Thus, these changes as are part of the initiate response to BRAF inhibition and likely contribute to the survival of melanoma cells.
Keywords: BRAF, Histone, Melanoma, Mouse, Resistance
Introduction
Despite recent therapeutic advances in targeted therapy and immunotherapy, the 5-yr survival for patients with distant metastatic melanoma remains around 25% [1]. Therefore, there remains a need for a greater understanding of the disease and for new and improved therapeutic strategies. BRAFV600E/D/K mutations occur in ∼40% of malignant cutaneous melanomas [2]. These mutations result in constitutive kinase activity and elevated downstream MAPK signaling, which drives cell proliferation and survival [3]. Homozygous deletion of the cyclin-dependent kinase inhibitor 2A (CDKN2A) locus, is seen in ∼40% of sporadic melanomas and promoter methylation leading to silencing and loss of expression is observed in a high proportion of the remaining tumors [4]. The phosphatidylinositol 3-kinase (PI3K)/AKT pathway is active in most melanomas, due to the loss of PTEN expression, or PI3KCA or AKT mutation [5], [6], [7], [8], [9], [10], [11], [12]. In patients with BRAF mutant melanomas, the BRAF inhibitors, encorafenib, dabrafenib and vemurafenib, confer a survival advantage, demonstrating improvements in response-rates, progression-free survival, and overall survival [13], [14], [15]. The initial response to these inhibitors can be dramatic [16], sometimes causing complete tumor regression and dormancy; however, this response is often not durable, and patient relapse usually occurs within 6–7 months. This is normally associated with reactivation of MAPK signaling by pre-existing or newly acquired mutations [14], [17], [18], [19]. The use of concurrent BRAF and MEK inhibitors, such as binimetinib, cobimetinib, and trametinib, has been established as a synergistic treatment approach and one that has seen further improved response, as compared with monotherapy [20], [21]; however, after a period of response and dormancy the majority of patients develop resistance [22], [23].
Maximizing the effectiveness of targeted therapies requires greater knowledge of the molecular alterations that drive progression, maintenance, and resistance. However, the study of resistance to targeted therapy is highly complicated by tumor heterogeneity and melanoma has a mutation load that exceeds all other cancers [4]. Melanomas undergo significant changes during BRAF inhibitor-mediated tumor dormancy, and accumulating evidence suggests that dormant cells have cancer stem cell properties that confer faster and more aggressive melanoma relapse [24], [25]. The study of tumor dormancy is challenging because residual tumors that respond to BRAF inhibition are rarely biopsied. Based on this rational, models that permit regulated expression of oncogenes are particularly useful for understanding mechanisms of targeted therapies because abrogation of oncogene expression mimics pharmacological inhibition of the target. We have developed a mouse model of melanoma based on the targeted delivery of retroviruses to melanocytes in vivo [26]. This model recapitulates the human disease and allows for the temporal control of mutant BRAF expression with doxycycline [27]. In this model, tumors initially respond to loss of BRAF, but recur after a period of response and residual tumor dormancy, that models human tumor relapse following BRAF inhibition. An analysis of the recurrent and responding tumors has revealed new insights into melanoma tumor dormancy.
Epigenetic changes have been suggested to drive resistance to BRAF inhibition, as ∼40% of melanomas develop resistance to BRAF inhibition without any identifiable genetic changes[28].
Histone post translational modifications (PTMs), including methylation, play a key role in regulating gene expression programs to control cell differentiation, and aberrant histone H3 modification or mutation is associated with tumor formation. Broadly speaking, histone H3 Lys4 tri-methylation (H3K4me3) is enriched in transcriptionally active promoters. Histone H3 H3K36 tri-methylation (H3K36me3) is associated with gene activation. Histone H3 Lys9 tri-methylation (H3K9me3) and histone Lys27 tri-methylation (H3K27me3) are observed in repressed genes [29].
Epigenetic mechanism of acquired resistance to BRAF and MAPK inhibition in melanoma are known to include IGFBP2 expression via increased chromatin accessibility, H3 PTM including H3K56 acetylation at the IGFBP2 locus, and consequent activation of the IGF-1 receptor leading to increased downstream AKT signaling [30], [31], [32]. Here we demonstrate that an innate response to prolonged BRAF inhibition is loss of histone H3 K9 and H3 K27 methylation and increased H3.3 K36 methylation. These were the only constant changes to histone methylation in over 80 histone PTM examined. These changes are associated with specific changes in histone methyltransferase (HMT) gene expression in P-ERK low, p-38 high melanoma cells, following prolonged BRAF inhibition in melanoma cell lines and our novel somatic mouse model of melanoma.
Materials and methods
Vector constructs
The retroviral vectors used in this study were replication-competent avian leukosis virus long-terminal repeat, splice acceptor and bryan polymerase-containing vectors of envelope subgroup A, designated RCASBP(A) and replication-competent, avian leukosis long-terminal repeat, no splice acceptor designated RCANBP(A) [33]. RCASBP(A) CRE, RCASBP(A) BrafV600E and Tet-Off have been previously described. RCANBP(A)TRE-BRAF-KD V600E was created by PCR amplification from RCAS BRAFV600E template DNA [34], followed by TOPO cloning into the Gateway entry vector pCR8 (Thermo Fisher Scientific). This fragment was then cloned into an RCAN TRE vector by using LR Clonase (Thermo Fisher Scientific) for subsequent transfection and/or viral production, infection, and stable expression. Viral propagation and in vivo and in vitro infection were performed as previously described [33].
Animal model
Newborn male and female mice Dct::TVA; Cdkn2alox/lox; Ptenlox/lox mice [33] were injected with a Tet-regulated Braf viral vector, RCAN TRE BRAFVE, RCASBP(A) Tet-Off and RCASBP(A) CRE. Censored survival data was analyzed using a log-rank test of the Kaplan–Meier estimate of survival. All animal experiments were performed in compliance with ‘Care and Use of Animals’ in Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited facilities, and approved by the University of Minnesota IACUC. Mouse diets: Teklad global rodent diet formulated with doxycycline hyclate 625 mg/kg TD08541 and Teklad global rodent diet TD01306 (Envigo Indianapolis IN).
RNA-Seq
Melanoma tumor tissues were individually snap-frozen in liquid nitrogen, immersed in RNAlater-ICE RNA Stabilization Reagent (Thermo Fisher Scientific, Waltham, USA) and thereafter stored at −80 °C without thawing. Frozen tissues were homogenized in 1 mL of TRIzol® Reagent (15596-026, Thermo Fisher Scientific) using a sterile, autoclaved mortar, and pestle. RNA was extracted per the TRIzol® protocol and purified using Qiagen RNeasy Mini Kit (74104, Qiagen, Hilden, Germany) according to the manufacturer’s protocol. RNA sequencing (RNA-Seq) performed at the University of Minnesota Genomics Center [25 M reads per sample, paired-ends, a 50 bp read length, stranded library- Illumina HiSeq 2500]. The RNA-Seq analysis including mapping and gene FPKM (fragments per kilobase of exon model per million reads mapped) estimates (Supplemental tables 1–2) were carried out as previously described [35]. Bar graphs where generated in R using Tidyverse and Ggpubr packages to perfume student’s t-tests.
Western blotting
Immunoblotting assays were performed as described [33]. Primary antibodies were obtained from the following sources and used according to manufacturer’s recommendations: Cell Signaling Technology: anti-ERK (9102), phospho-eIF4E (9741), eIF4E (2067), P-4E-BP1 (9644), P-4E-BP1 (2855), anti-FRA (5281), phospho-FRA1 (5841), SPRY2 (14954), SOX2 (14962), MITF (12590), cyclin D1 (2978), P-ERK5 (3371), ERK5 (3372), p-p38 (4511), p38 (8690), p21 (2947), p27 (3686), EZH2 (5246), SUV39H1 (8729), H3K27me3 (9733), H3K9ac (9649), H3K27ac (8173), H3K4me3 (9751), H3 (4499), c-Met (8198); Abcam: histone H3.3-phospho S31 (ab92628), ATF3 (ab216569), SOX10 (ab216020), cystatin C (ab109508), SUV39H2 (ab190870); tubulin-HRP (21058). SOX17 (24903-1-AP); Thermo Fisher Scientific: SOX4 (PA5-40681); and Novus Biologicals: PML (NB100-59787). Covance: anti-HA monoclonal antibody HA.11. Secondary antibodies: Anti-mouse (7074) or rabbit IgG-HRP (7076) Cell Signaling.
Immunohistochemistry
Immunostaining for HA was performed using an anti-HA monoclonal antibody 1:1000 (HA.11, Covance, Berkeley, CA). Analysis of cytokeratin expression was performed using mouse cytokeratin-pan-antibody-cocktail (MA5-13203, Thermo Scientific). Analysis of S100 expression was performed using rabbit polyclonal antibody Z0331 (1:400) for S100 (Dako; Glostrup, Denmark). Detection of MAPK activation was performed using a 1:100 dilution of an antibody to phospho-ERK (4370, Cell Signaling). Cell proliferation was detected using a 1:250 dilution of a rabbit monoclonal antibody to Ki67 (RM-9106-R7 Thermo Fisher Scientific). Detection of HRP activity was performed using DAB (Cell Signaling). Sections were counterstained with hematoxylin.
Mass spectrometry
To analyze for changes in global histone PTM we used the MOD spec Quantitative mass spectrometry service from Active Motif (Carlsbad, CA). Histones were acid extracted, derivatized via propionylation, digested with trypsin, newly formed N-termini were propionylated as previously described [36], in triplicate using the Thermo Scientific TSQ Quantum Ultra mass spectrometer coupled with an UltiMate 3000 Dionex nano-liquid chromatography system. The data was quantified using Skyline [37] and represents the percent of each modification within the total pool of that tryptic peptide. Histone modification assessment was performed for H1.4: K25UN H1.4: K25AC H1.4: K25ME1 H1.4: K25ME2 H1.4: K25ME3 H2A: K5UN H2A: K5AC H2A: K9UN H2A: K9AC H2A: K36UN H2A: K36AC H2A1: K13UN H2A1: K13AC H2A1: K15UN H2A1: K15AC H2A1: K15UB H2A3: K13UN H2A3: K13AC H2A3: K15UN H2A3: K15AC H2A3: K15UB H3R2UN: K4UN H3R2UN: K4ME1 H3R2UN: K4ME2 H3R2UN: K4ME3 H3R2UN: K4AC H3R2UN: Q5UN H3R2UN: Q5ME1 H3: K9UN H3: K9AC H3: K9ME1 H3: K9ME2 H3: K9ME3 H3:K14UN H3: K14AC H3: K18UN H3: K18AC H3: K18ME1 H3: Q19UN H3: Q19ME1 H3: K23UN H3: K23AC H3: K23ME1 H3: R42UN H3: R42ME2 H3: R49UN H3: R49ME2 H3: Q55UN H3:Q55ME1 H3: K56UN H3: K56AC H3: K56ME1 H3: K64UN H3: K64AC H3: K79UN H3: K79AC H3: K79ME1 H3: K79ME2 H3: K79ME3 H3: K122UN H3: K122AC H3.1: K27UN H3.1: K27AC H3.1: K27ME1 H3.1: K27ME2 H3.1: K27ME3 H3.1: K36UN H3.1: K36AC H3.1: K36ME1 H3.1: K36ME2H3.1: K36ME3 H3.3: K27UN H3.3: K27AC H3.3: K27ME1 H3.3: K27ME2 H3.3: K27ME3 H3.3: K27M H3.3: K36UN H3.3: K36AC H3.3: K36ME1H3.3: K36ME2 H3.3: K36ME3 H4: K5UN H4: K5ACH4: K8UN H4: K8AC H4: K12UN H4: K12AC H4: K16UN H4: K16AC H4: K20UN H4: K20ME1 H4: K20ME2 H4: K20ME3 H4: K20AC (UN Unmodified, ME1/2/3 - Mono/di/tri methylation AC Acetylation UB, Ubiquitination H3R2UN).
Cell culture
Melanoma cell lines from the American Type Culture Collection (ATCC-A375s) or National Cancer Institute (M14s) were grown in Roswell Park Memorial Institute (RPMI) medium supplemented with 5% fetal bovine serum (FBS). DF-1 cells (ATCC) were grown in Dulbecco's Modified Eagle Medium (DMEM)-high glucose media supplemented with 10% FBS (Invitrogen), 5 mL penicillin–streptomycin, and maintained at 39 °C. Vemurafenib, dabrafenib, and entinostat were purchased from Selleckchem, (Houston, TX, USA). Clonogenic assays were performed as described [33].
Results
Delivery of BRAFV600E and Cre results in tumor formation in Dct::TVA;Cdkn2alox/lox; Ptenlox/lox mice
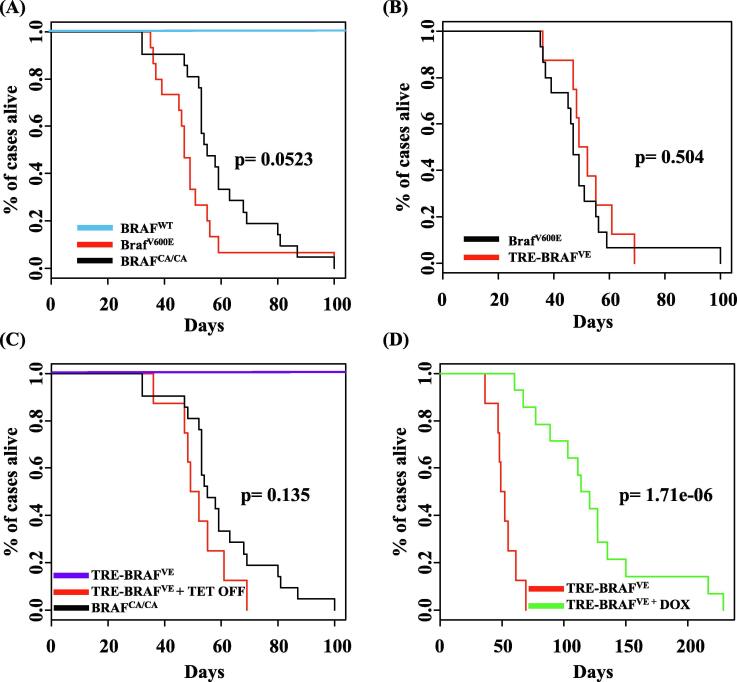
The RCAS/TVA retroviral vector system allows for tissue- and cell-specific targeted infection of mammalian cells through ectopic expression of the viral receptor [38]. The RCAS(A) receptor is encoded by the TVA gene that is normally expressed in avian cells. In this melanoma model, transgenic mice express the TVA receptor under the control of the dopachrome tautomerase (DCT) promoter, which allows targeting of the virus specifically to melanocytes in vivo [26]. Multiple genetic alterations can be introduced into the same cell, in the context of an unaltered microenvironment. We previously established the validity of the RCAS/TVA system to generate melanoma in vivo by delivering viral BRAF, NRAS, AKT, and MEK mutants in combination with Cre to melanocytes in Cdkn2alox/lox and Ptenlox/lox mice [33], [39], [40]. To allow for regulation of activated BRAF expression in vivo using the Tet-regulated system, we utilized the RCAN(A) vector as opposed to RCAS, wherein expression is decoupled from the viral long-terminal repeat (LTR) through the deletion of a key splice acceptor site, and is instead driven from a Tet-responsive element (TRE) [33]. Expression from the TRE requires the presence of a tetracycline transcriptional activator (tTA) such as Tet-Off. In the context of Tet-Off, the TRE-BRAF is expressed, but in the presence of doxycycline (Dox) it is repressed. An RCAS virus containing an influenza hemagglutinin (HA) epitope tagged human BRAF kinase domain with a V600E mutation [41] (hereafter BRAFVE) was introduced into immortalized DCT-TVA melanocytes with RCAN TRE-BRAFVE and RCAS Tet-Off and expression was assessed in the presence or absence of Dox. As expected, BRAF expression was present and suppressed by Dox (Supplementary Fig. 1, (3)). The deletion of the BRAF N-terminal auto-inhibitory domain leads to downstream activation of the MAPK pathway and is sufficient to drive tumorigenesis in glioma models [41]. Previously, viral producing cells harboring RCAS Cre were injected subcutaneously into newborn Dct::TVA;BrafCA/CACdkn2alox/lox;Ptenlox/lox mice (n = 15) and viruses containing mouse BrafV637E (the equivalent of human BRAFV600E) hereafter BrafV600E and Cre were injected subcutaneously into newborn Dct::TVA;Cdkn2alox/lox;Ptenlox/lox mice (n = 21) [33], [39]. Both of these models generated tumors in all animals with a mean survival of 50 days (range 36–100 days) and 60 days (range 32–100 days), respectively. No significant difference was observed between the BrafV600E and BrafCA/CA mouse cohorts (P = 0.523; Fig. 1A). Herein, to study resistance to BRAF inhibition in vivo, we delivered viruses containing Tet-Off, Cre, and/or TRE-BRAFVE to newborn Dct::TVA;Cdkn2alox/lox;Ptenlox/lox mice by subcutaneously injecting viral producing cells harboring the RCAS and RCAN viruses. Viruses containing TRE-BRAFVE and Cre were also delivered to newborn Dct::TVA; Cdkn2alox/lox; Ptenlox/lox mice as experimental controls. All mice were monitored for tumor growth and development. All of the Dct::TVA;Cdkn2alox/lox;Ptenlox/lox mice injected with viruses containing Tet-Off, Cre and TRE-BRAFVE developed melanomas (n = 35; Fig. 1B). To confirm that TRE-BRAF tumors have similar growth and survival rates as tumors driven by the viral LTR or endogenous BRAF promoter, we compared the survival to our previously published results using a log-rank test of the Kaplan–Meier estimate of survival. No significant difference was observed between the TRE-BRAFVE and BrafV600E or BrafCA/CA mouse cohorts (P = 0.504 and P = 0.135; Fig. 1B and C, respectively). Dct::TVA;Cdkn2alox/lox;Ptenlox/lox mice injected with viruses containing TRE- BRAFVE and Cre- (n = 11) in the absence of Tet-Off remained tumor-free for the duration of the study (Fig. 1C), demonstrating that the TRE-BRAFVE construct does not induce tumors in the absence of Tet-off.
Fig. 1.
The genetic suppression of BRAF increases survival in BRAFVE, Cdkn2a, and Pten mutant melanomas. (A) Kaplan–Meier percent survival curves for mice with BRAF tumors. Dct::TVA; BRAFCA/CACdkn2alox/lox; Ptenlox/lox mice were injected with viruses encoding Cre (black line, n = 21, tumor incidence 21/21) or Dct::TVA; Cdkn2alox/lox; Ptenlox/lox mice injected with viruses encoding BRAFV600E+Cre (red line, n = 15, tumor incidence 15/15). No significant difference was observed between the BRAFCA/CA and BRAFV600E tumors,P = 0.0523. No tumors developed in Dct::TVA; Cdkn2alox/lox; Ptenlox/lox mice injected with Cre alone as controls (Control A, blue line, n = 17, tumor incidence 0/17). (B) Kaplan–Meier percent survival curves comparing TRE-BRAFVE tumors with BRAFV600E tumors. Dct::TVA; Cdkn2alox/lox; Ptenlox/lox mice were injected with viruses encoding Cre (black line, n = 15 tumor incidence, 15/15) or Dct::TVA; Cdkn2alox/lox; Ptenlox/lox mice injected with viruses encoding BRAFV600E+Cre+Tet-Off (red line, n = 8, tumor incidence 8/8). No significant difference was observed between the TRE-BRAFVE and BRAFV600E tumors,P = 0.504. (C) Kaplan–Meier percent survival curves comparing TRE-BRAFVE tumors compared with BRAFCA/CA tumors. Dct::TVA; BRAFCA/CACdkn2alox/lox; Ptenlox/lox mice were injected with viruses encoding Cre (black line, n = 21 tumor incidence, 21/21); or Dct::TVA; Cdkn2alox/lox; Ptenlox/lox mice injected with viruses encoding TRE-BRAFV600E+Cre+Tet-Off (red line, n = 8, tumor incidence 8/18). No significant difference was observed between the BRAFCA/CAand BRAFV600E tumors,P = 0.135. No tumors developed in Dct::TVA; Cdkn2alox/lox; Ptenlox/lox mice injected with Cre+TRE-BRAFVE in the absence of Tet-Off as controls (n = 11, tumor incidence 0/11). (D) Kaplan–Meier survival curves demonstrating the effect of genetic BRAFVE inhibition. Dct::TVA;Cdkn2alox/lox; Ptenlox/lox mice were injected with viruses encoding Tet-Off+Cre+TRE-BRAFVE. Mice were monitored for tumor formation and randomized to receive either a Dox diet or control diet when tumors were measured at 1.0 cm3 (Dox diet dotted line, n = 14, control diet dashed line, n = 8). A significant increase in survival was found between Dox-treated and control mice (P = 0.0000017).
Suppression of BRAFVE expression promotes tumor regression and significantly increases survival
When the Dct::TVA;Cdkn2alox/lox; Ptenlox/lox Tet-Off, CRE and TRE-BRAFVE tumors reached 1000 mm3, 8/35 mice were randomly assigned to receive standard feed (untreated- controls), and 27/35 received Dox-containing food to suppress BRAF expression and determine whether down-regulation of BRAF expression results in tumor regression. Mice were sacrificed when the tumors exceeded 2500 mm3. In the Dox-treated cohort, tumors progressed for several days after initiation of treatment. This was then followed by rapid tumor regression in 85% (23/27) of the mice with the remaining 14% (4/27) only showing a partial response. In order to examine changes that occur in tumors that respond to Dox treatment, 13/27 mice in the Dox-treated cohort were sacrificed when the tumors regressed into a stable non-growing, dormant lesion, for 6 successive days. All of the remaining 10 tumor bearing mice that responded to Dox-treatment became resistant while on Dox-treatment after a highly variable period of response and tumor dormancy (Supplemental Fig. S2A). The mean time from the point the tumor regressed below 1000 mm3 until the tumor fully re-occurred and exceeded 1000 mm3, was 54.8 days (range 18–107 days). Survival rates, excluding the 13 dormant tumor mice (Supplemental Fig.S2B), were compared between the Dox-treated and untreated cohorts using a log-rank test of the Kaplan–Meier estimate of survival. The mean survival for the untreated TRE-BRAFVE+Tet-Off mice (n = 8) was 52 days (range 36–69 days) while the mean survival for the Dox-treated mice with recurrent tumors (n = 14) was 123 days (range 60–229 days). Thus, the administration of Dox and subsequent loss of TRE-BRAF expression significantly increased survival (P = 1.71 × 10−6; Fig. 1D).
Tumor histology
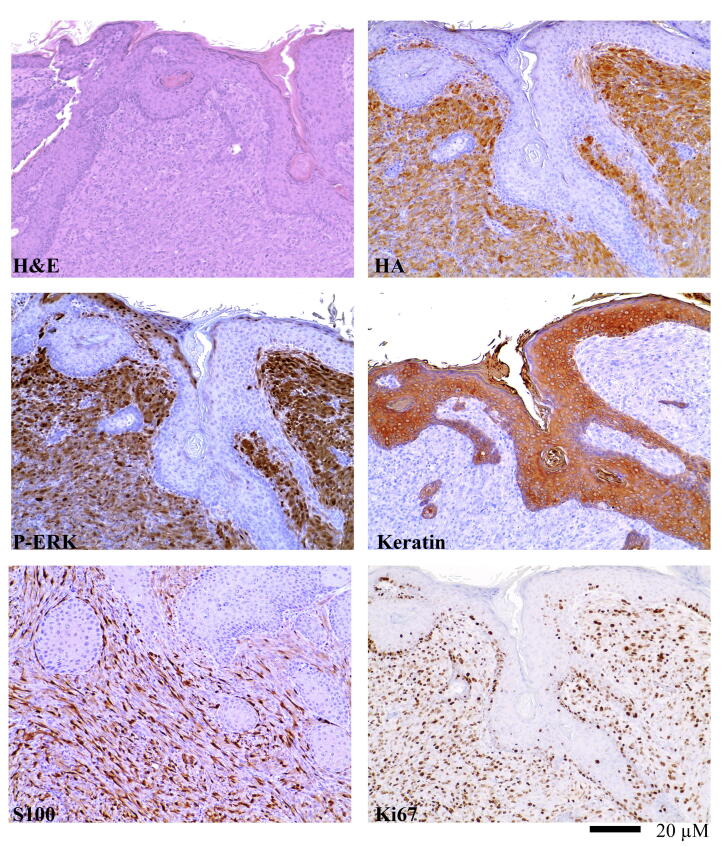
Tumors induced by expression of Tet-Off, TRE-BRAFVE, and Cre were indistinguishable from previously reported tumors induced by delivery of BRAF and Cre, where mutant expression was driven from the viral LTR, Cre, or the endogenous mouse BRAF promoter in BRAFCA/CA mice [27]. Microscopically, high-grade nuclear features were observed along with prominent nucleoli and numerous Ki67 positive mitotic figures (Fig. 2). The tumors consisted primarily of short spindle cells, although some possessed giant cells with epithelioid features. An HA epitope tag engineered on the N-terminus of mutant BRAF allowed for the distinction between virally delivered BRAFVE and endogenous Braf. Expression and activity of BRAFVE was verified by immunohistochemistry (IHC) for HA and phosphorylated ERK (P-ERK). IHC for S100 demonstrated melanocytic origin of the tumors (Fig. 2). Cytokeratin staining was used to demonstrate the tumors were not of epithelial origin.
Fig. 2.
Immunoprofile of a representative BRAFVE melanoma. BRAFVE melanomas are highly vascular and consist primarily of short spindle cells exhibiting high-grade nuclear features and prominent nucleoli. They invade into subcutaneous fat, muscle, and cartilage. IHC for S100 demonstrated the melanocytic origin of the tumors. IHC for P-ERK demonstrated canonical MAPK pathway activation. Assessment of the Ki67 cellular proliferation marker was performed using IHC on slides with uniform tumor cellularity and results demonstrated that all of the tumors were highly proliferative. IHC sections were counterstained with hematoxylin and a hematoxylin and eosin (H&E) stained tumor section is provided for comparison.
Analysis of resistance to BRAF inhibition
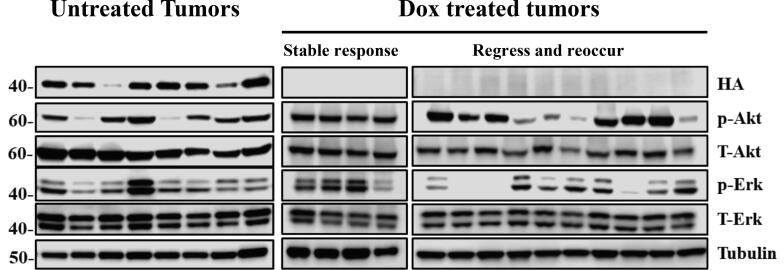
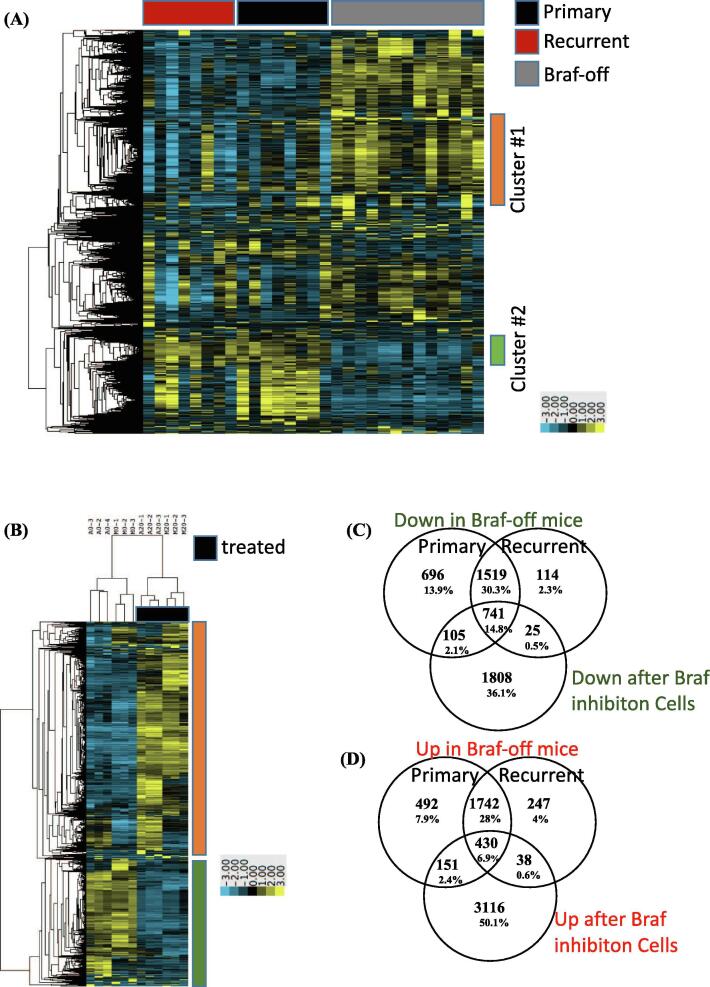
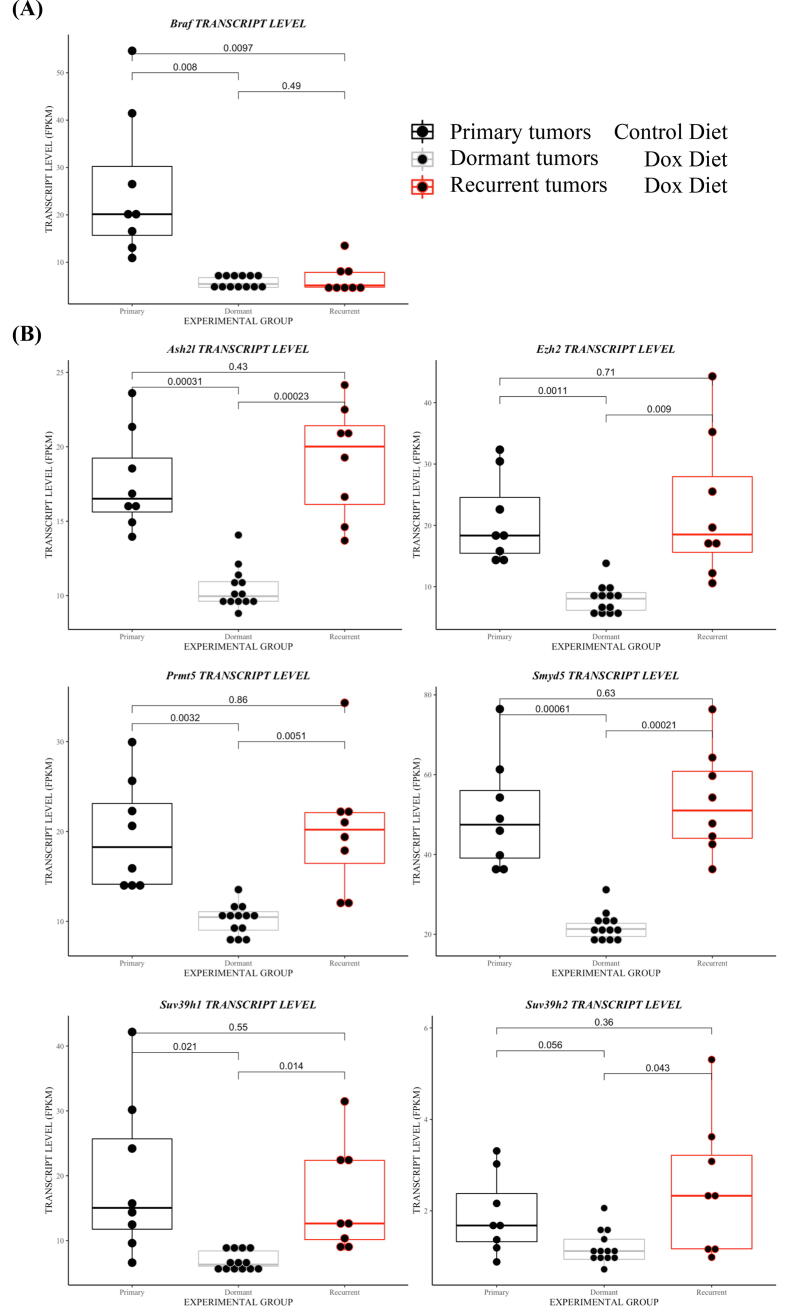
To delineate the mechanism(s) of resistance responsible for driving tumor recurrence, we first evaluated all tumor tissue for expression of virally delivered TRE-BRAFVE +/− DOX cohorts. Western Blot (WB) analysis of tumor samples for the HA epitope tag revealed that none of the Dox-treated tumors re-expressed TRE-BRAF (Fig. 3). These tumors were further evaluated for activation of the canonical MAPK pathway by immunoblotting for P-ERK. MAPK pathway activation was observed in 12/14 of the resistant tumors (Fig. 3). We previously reported c-Met amplification and overexpression can drive the escape of NRAS mutant melanoma from inhibition of RAS [40]. Accordingly, we evaluated tumor samples and controls for overexpression of Met; however, less than 3 of the resistant tumors overexpressed c-Met (Supplementary Fig. 3). To characterize the mechanisms of primary and acquired resistance to TRE-BRAF inhibition in an unbiased manner, we performed RNA sequencing (RNA-Seq) on primary tumors (n = 8), dormant tumor samples after Dox treatment (n = 13), and recurrent tumors resistant to Dox treatment (n = 8), (Supplemental table 1). We analyzed the resulting transcriptional patterns using both a statistical approach as well as an unsupervised clustering approach. We used a group-based T-test to identify genes, which significantly (p = 0.01) changed between tumors and dormant tumors, as well as to identify genes that significantly changed between dormant tumors and recurrent tumors (Supplemental table 3). A total of 2663 genes decreased between tumors and dormant tumor cells and 2976 genes increased between dormant tumors and recurrent tumors. A total of 2008 of these genes were on both lists. Expression of melanoma differentiation marker Melan A (Mlana-A) were significantly increased in dormant tumors compared to primary and recurrent melanomas. The mean FPKM values for Mlana-A were 0.28 in the primary, 11.5 in the dormant 0.22 in the recurrent tumors (P-value >0.005). Using the ENrichR tool [42], these gene lists (Supplemental Table 5) were independently highly enriched in E2F targets, genes involved in the canonical cell cycle pathway, and genes that are also repressed following exposure to anti-cancer drugs, including vemurafenib, which targets mutant BRAF (Supplemental Tables 6–9). Similarly, 2815 genes significantly increased in the dormant tumors relative to the primary tumors and 2743 genes significantly decreased in the recurrent tumors relative to the dormant tumors. Of these, 1981 were on both lists. These gene lists were independently highly enriched in transcripts associated with immune infiltrate (Supplemental Table 1, enrichment ENRICHR results). To visualize these changes and determine whether the primary changes are associated with these events, we selected all the transcripts with standard deviation (SD) greater than 1 and used hierarchical clustering to identify the patterns present in an unsupervised fashion. Visualization of the results show that generally a large set of genes change between primary tumors (BRAFVE on) and dormant tumors (BRAFVE off) and that these changes are reversed in the recurrent tumors (Fig. 4A). Analyses of the genes in cluster 1, which increases in dormant tumors (enriched in immune transcripts), and cluster 2, which decreases in dormant tumors (enriched in E2F cell cycle targets), showed very similar enrichment to the patterns observed in the direct statistically derived list (Fig. 4, Supplemental enrichment Table 1). The mean BRAF FPKM transcript levels were significantly reduced in Dox-treated mouse tumors compared to untreated tumor cohort (Dormant P = 0.008, Recurrent P = 0097, Fig. 5A). These results indicate that turning off BRAF is in part analogous to the pharmacological inhibition of BRAF and suggest that the dormant tumors are capable of escaping BRAFVE loss in a similar fashion as drug resistant tumors.
Fig. 3.
BRAFVE cooperates with Cdkn2a and Pten loss in the development of melanomas in vivo. Virally delivered BRAFVE expression was detected in proteins extracted from mouse melanomas by using an antibody for the HA epitope tag in tumors induced with RCAN TRE-BRAFVE+RCAS Tet-Off+Cre, but were absent from recurring Dox-treated tumors. MAPK pathway activity was evaluated by blotting for phosphorylated and total ERK 1/2. The PI3-K pathway was evaluated by blotting for phosphorylated and total Akt expression.
Fig. 4.
Expression Analysis of primary, dormant, and recurrent melanomas. RNA-Seq transcriptional response observed in (A) primary, recurrent and BRAFVE-off (dormant) mouse tumors and (B) human cell lines following treatment with vemurafenib. For inclusion in the Clustering heatmap Log base 2 transformed, mean centered FPKM values with standard deviation greater than 1 were clustered using average linkage clustering. T-tests were used to generate lists of genes with P-value >0.05 and absolute value of average fold change greater than 1.5 that were compared across human and mouse experiments to identify (C) transcripts that decreased following genetic removal of BRAFVE in murine tumors, and increased in recurrent tumors, as well as decreased after vemurafenib treatment in human cell lines and (D) increased following genetic removal of BRAFVE in murine tumors, and decreased in recurrent tumors, as well as increased after vemurafenib treatment in human cell lines.
Fig. 5.
Altered expression of histone modifying enzymes in response to suppression of BRAFVE. (A) Normalized gene expression (FPKMs) of virally delivered Dox-regulated BRAFVE in primary, dormant and recurrent tumors resistant to Dox treatment. (B) Normalized gene expression (FPKM) of Ezh2, Suv38h1, Suv38h2,Ash2l, ASH2L, SYMD2, and PRMT5 in primary, dormant and recurrent tumors. Students T-tests were performed to assess significance.
RNA-Seq analyses of BRAFV600E mutant cell lines following treatment with vemurafenib
Mutant BRAF inhibitors, encorafenib, vemurafenib, and dabrafenib, induce cell cycle arrest and high levels of apoptosis in BRAF mutant melanoma cell lines. However, a residual population of dormant tumor cells survive and over time can become resistant [27], [6]. Accordingly, we conducted RNA-seq analysis in triplicate on human melanoma cell lines with BRAFV600E mutations (A375 and M14) before and following 20 days of treatment with 2 µM vemurafenib (Supplemental table 2). We analyzed the resulting transcriptional patterns using both a statistical approach as well as an unsupervised clustering approach. Using the T-test approach (Supplemental table 4), 2679 genes decreased after vemurafenib treatment. The ENrichR results showed that this gene set is highly enriched in E2F4 targets, genes involved in canonical cell cycle and replication pathways, and genes that decrease following vemurafenib treatment–all changes that could be observed following removal of BRAF VE in the genetic model. Using the T-test approach, 3726 genes increase after vemurafenib treatment. Not surprisingly, these lists are highly enriched in genes that are increased following exposure to vemurafenib. As expected with in vitro analysis, genes associated with immune cells were not enriched. The T-test approach can also be obtained from the hierarchical clustering approaches, indicating that the primary differences in transcription resulted from the exposure of the cells to the drug (Fig. 4B). Overall, the changes between loss of BRAFVE in the mouse model and exposure of the cell lines to the BRAF inhibitor are consistent with a common biological mechanism. In additional support of this concept, many of the genes that were decreased after vemurafenib exposure were also decreased after BRAF removal relative to primary and recurrent tumors (n = 741, Fig. 4C). Similarly, many of the genes increased after vemurafenib exposure were also increased following BRAFVE suppression (n = 430, Fig. 4D). Differences likely stem from the presence of microenvironment transcripts in the mouse model as well as differing chromatin states.
Expression changes in histone methyltransferase are conserved in melanoma dormancy between human cell lines and mouse tumors
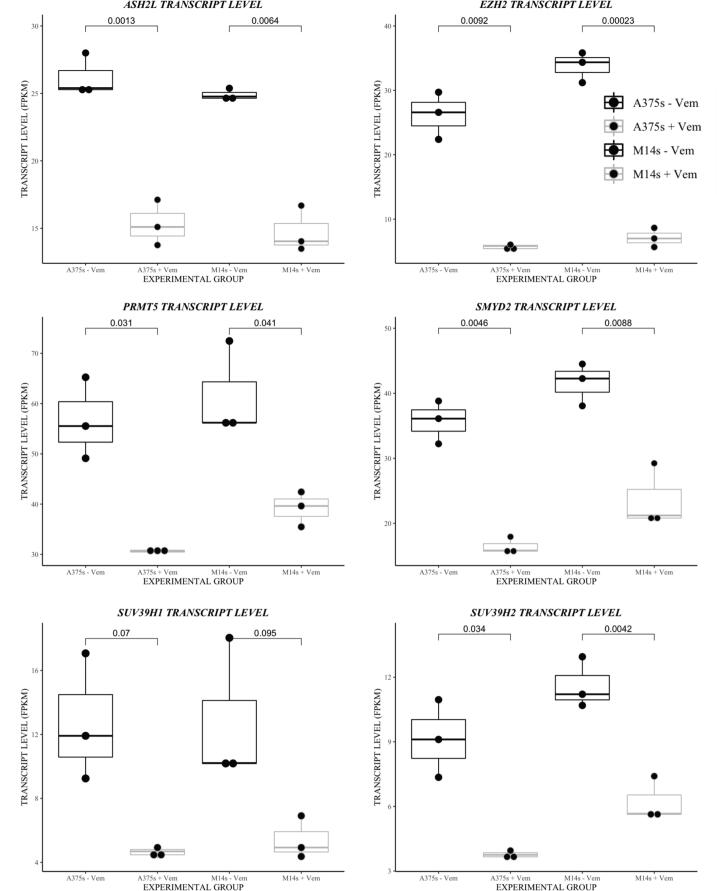
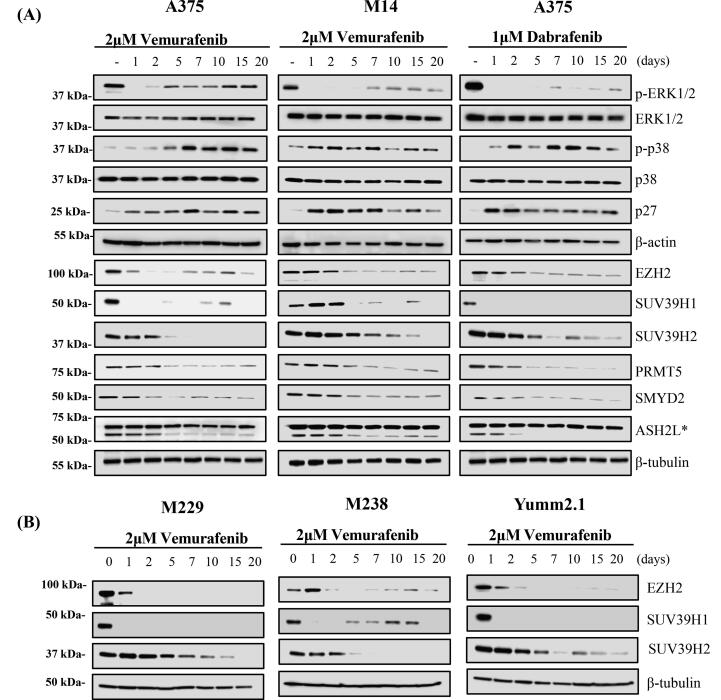
Our data revealed expression changes in specific histone methyltransferases and histone deacetylases in our dormant tumors in vivo and in pharmacologically dormant human melanoma cells treated with vemurafenib (Supplementary Fig. 4–response to BRAF inhibition). Despite the very different in vitro and in vivo conditions, expression of methyltransferase genes Enhancer Of Zeste 2 Polycomb Repressive Complex 2 Subunit (EZH2), Suppressor Of Variegation 3-9 Homolog 1(SUV39H1), Suppressor Of Variegation 3-9 Homolog 2 (SUV39H2), ASH2 Like, Histone Lysine Methyltransferase Complex Subunit (ASH2L), SET And MYND Domain Containing 2 (SYMD2), and Protein arginine methyltransferase 5 (PRMT5) decreased following BRAF inhibition (Figs. 5B, 6). To confirm the effect of altered histone methyltransferase and mRNA expression, we treated BRAF melanoma cells lines with 2 μM vemurafenib or 1 μM dabrafenib for 20 days. Immunoblotting of proteins from these cells confirmed that EZH2, SUV39H1, SUV39H2, ASH2L, SYMD2, and PRMT5 were downregulated in response to BRAF inhibition with vemurafenib or dabrafenib in the p-p38 high, p27 high, p-ERK1/2 low dormant A375 and M14 melanoma cells (Fig. 7A). The changes for EZH2 as well as for SUV39H1 and SUV39H2, which showed the least consistent significant change in gene mRNA expression, were further validated in two additional human BRAFV600E melanoma cell lines—M229 and M238—and in Yumm2.1s, a BRAFV600E mouse melanoma cell line (Fig. 7B).
Fig. 6.
Altered expression of histone modifying enzymes in response to BRAFV600E inhibition. Normalized gene expression (FPKMs) of EZH2, SUV39H1, SUV39H2, ASH2L, SYMD2, and PRMT5 in human melanoma cell lines following 20 days of BRAF inhibition with 2 µM vemurafenib or vehicle control. Students T-tests were performed to assess significance.
Fig. 7.
Prolonged BRAFV600E inhibition with vemurafenib leads to decreased methyltransferase in BRAF-expressing melanoma cells. (A) Melanoma cell lines with a BRAFV600E mutation were maintained in media containing 2 μM vemurafenib or 1 μM dabrafenib for 20 days. The expression, phosphorylation, and methylation of the indicated proteins were detected by immunoblotting. BRAF inhibition with vemurafenib led to rapid decreases in P-ERK and increased P-p38, p21, and p27 expression in A375 and M14 melanoma cells. The expression of EZH2, SUV39H1, SUV39H2, ASH2L, SYMD2 and PRMT5, decreased in response to BRAF inhibition with vemurafenib. (B) The effect of prolonged culture under BRAFV600E inhibition on EZH2, SUV39H1, SUV39H2 which demonstrated the most prominent response to BRAF inhibition expression were validated in three additional melanoma cell lines (M229, M238 and Yumm2.1s).
BRAF inhibition is associated with a loss of H3K9, H3K4 and H3K27-methylation and an increase in K36 methylation
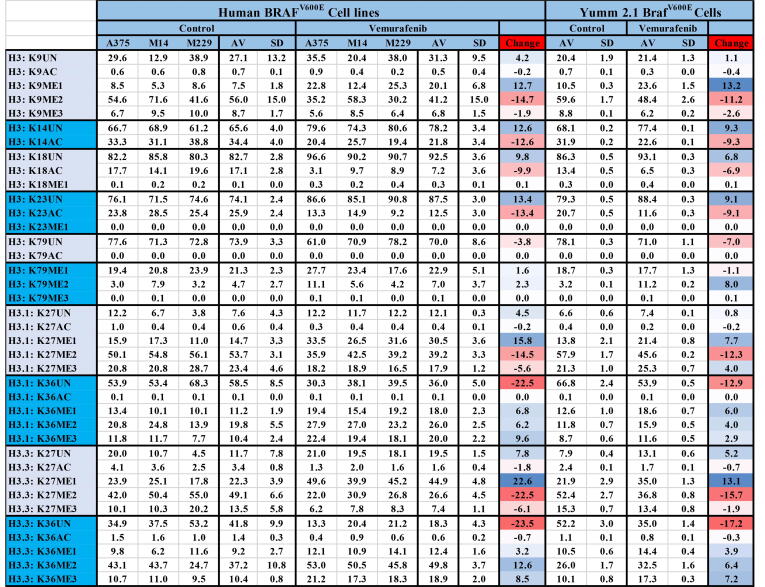
Immunoblotting and antibody-based protocols are not reliable for the assessment of histone PTMs due to antibody cross-reaction, the influence of neighboring PTMs, the inability to reliably distinguish between the modification states and similar histone variants; nor are they high throughput or fully quantitative [43], [44]. Therefore, to determine the actual effect of BRAF inhibition on histone PTM, we performed quantitative mass spectrometry to measure the relative abundance of the 80 most common histone PTMs in human A375, M14 and M229 melanoma cell lines treated with 2 μM vemurafenib or vehicle control for 20 days. The abundance of each peptide and the corresponding measurements from three separate experiments (Fig. 8, Supplemental table 10); as modification levels are below 0.5% can confounded by background signals from the instrumentation the results demonstrate that the loss of histone H3 K9, and K27 bi and tri methylation and an increase in H3 K36 bi and tri methylation and were the only consistent significant changes in histone PTM along with a small reduction in H3 K14, K18 and K23 acetylation.
Fig. 8.
Prolonged BRAFV600E inhibition with vemurafenib leads to specific changes to histone PTM in BRAF-expressing melanoma cells. The mean relative abundance of each form of a given tryptic peptide in BRAF melanoma cell lines treated with 2 μM vemurafenib or vehicle control for 20 days. A reduction in H3 K9 and K27 bi and tri-methylation and an increase in K36 bi and tri-methylation was seen in response to prolonged BRAF inhibition in human and mouse BRAF melanoma cell lines. A small but consistent reduction in acetylation was also seen at H3 K14, K18 and K23. No other significant changes to histone PTM were detected.
Discussion
Adaptation to therapy is a major factor that limits the long-term response of BRAF-mutant melanoma patients to BRAF inhibitors. To better understand resistance to BRAF inhibition, we have developed a targeted somatic cell gene delivery mouse model of melanoma that allows for temporal and spatial control of gene expression. This approach is useful as paired pre-biopsied samples of melanomas that are responding to treatment are nearly impossible to obtain. The targeted delivery of BRAFV600E, NRASQ61R, or MEK mutants in combination with Cre to Cdkn2lox/lox Ptenlox/lox melanocytes led to the development of melanomas that closely mimic the human disease [26], [39], [40]. Targeted delivery of Tet-Off, CRE, and TRE-BRAFVE, Dct::TVA; Cdkn2alox/lox; Ptenlox/lox to melanocytes caused mice to develop melanomas that regressed when they were fed doxycycline-containing food; however, after a long latency, the tumors re-occurred. Immunoblotting and RNA-Seq transcriptional analysis confirmed that BRAFVE remains under Dox control, and many of the resistant tumors reactivated the MAPK pathway.
RNA-Seq-based transcriptional analysis of tissue from pre-treated, responding, and recurrent melanomas revealed changes in gene expression potentially responsible for the survival of dormant melanoma cells. By comparing these gene expression changes to those in human cell lines treated with BRAF inhibitors, we were able to determine commonalties in suppressing BRAF genetically and the pharmacological inhibition of BRAF; these included the loss of differentiation markers such as Melan A [45], [46]. The inherent response in dormant mouse tumors and human melanoma cell lines to BRAF inhibition was the upregulation of HMTs, EZH2, SUV39H1, SUV39H2, ASH2L, SYMD2, and PRMT5.
EZH2 is the enzymatic subunit of polycomb repressive complex 2, which methylates H3 K27 to promote transcriptional silencing. EZH2 is overexpressed in melanoma, lymphoma, bladder cancer, glioma, liver, ovarian, and lung cancers, and gain-of-function mutations have also been identified in lymphoma and melanoma. Missense mutations at the H3 K27 site are common in pediatric brain stem gliomas [47]. Genes regulated by EZH2 in melanoma include p21/CDKN1A, RAF1, E-cadherin, NRAS and KRAS [48]. EZH2 also drives transformation by silencing genes relevant to the primary cilium, a key step in melanoma metastasis [49]. SUV39H1 and SUV39H2 are primarily responsible for di- and tri-methylation of H3K9. H3K9me3 leads to the silencing of E2F-responsive genes regulating cell cycle and cell differentiation. Overexpression of SUV39H1/H2 occurs in breast cancer, colorectal cancer, gastric cancer, lung cancer, liver cancer, leukemia, and lymphomas [50]. ASH2L is a core member of the SET1/ASH2 histone methyltransferase complex that specifically methylates H3K4. ASH2L is overexpressed in almost all human tumors and tumor cell lines [51]. PRMT5 regulates cellular functions, including apoptosis, pluripotency, and cell growth. ASH2L is, in turn, methylated by PRMT5. Overexpression of the PRMT5 protein is observed in leukemia, lymphoma, glioma, ovarian, breast, prostate, and lung cancer. Loss of PRMT5 expression reduces the expression of microphthalmia-associated transcription factor (MITF) and increases expression of the cell cycle regulator p27 (Kip1) in melanoma. The depletion of PRMT5 inhibits proliferation in melanoma cell lines, but accelerates the growth of other cancer cells [52]. SMYD2 specifically methylates H3K4 in complex with ASH2L and also methylates TP53, RB1, and PRAP1 proteins [53].
Histone methylation plays a key role in regulating gene expression programs to control cell differentiation and aberrant histone H3 modification or mutation is associated with tumor formation. Histone H3K4me3 is enriched in transcriptionally active promoters. H3K36me3 is associated with gene activation. H3K9me3 and H3K27me3 are observed in repressed genes [29]. An examination of histone modifications in the mouse melanoma cells and BRAF inhibitor-treated melanoma cell lines using quantitative mass spectrometry, determined a reduction in histone K9 and K27 methylation and increase in K36 methylation. These were the only changes to histone methylation in the melanoma cells out of 80 possible changes to histone PTM screened. Exposure of BRAFV600E melanoma cells to BRAF and MEK inhibitors, hypoxia or nutrient starvation has previously reported to induce the surviving dormant cells to adopt a less differentiated, drug-tolerant state characterized by the loss of differentiation marker Melan-A and the upregulation of HMT activity, and a loss of H3K4me3, H3K27me3 and gain of H3K9me3 [45], [46]. Our results demonstrated the loss of H3K9me3 and H3K27me3 in human and mouse BRAF melanoma cell lines as well as the novel discovery of increased H3K36me in response to BRAF inhibition; however, we did not detect a change in H3K4me3 and H3K4me2 in response to BRAF inhibition in human and mouse melanoma cell lines. This discrepancy may result from antibody-based approaches being unreliable for the detection of histone PTMs. Our use of mass spectrometry followed from past divergent results when examining histone PTMs with immunoblotting. The small size of the structural motifs makes the antibodies, which recognize PTM peptides/proteins independent of its surrounding sequences, unreliable [43], [44].
Conclusion
Almost 40% of melanomas develop resistance to BRAF inhibition without any identifiable genetic changes [28]. We have identified specific HMT and histone PTM that are dysregulated both in vitro and in vitro following the genetic suppression of vemurafenib inhibition BRAFV600E in melanomas. This suggest that one intrinsic response of melanoma cell to the challenge of BRAF inhibition is the induction of a transcriptionally permissive state through the global loss of repressive histone marks. Due to their key role in the regulation of gene expression, preventing H3 demethylation may be key to improving the long-term efficacy of BRAF inhibition. Histone deacetylase (HDAC) inhibitors are currently in Phase II trials in combination with pembrolizumab (anti PD-1) for patients with melanoma; however, HDAC inhibitors are known to indirectly prevent histone de-methylation [54]. Given that only minor changes in histone acetylation were seen in our study, the role of HDAC inhibitors in repressing histone methylation will require future investigation and needs to be decoupled from their role in deacetylation.
Author contributions
FG, HY, MVB, NH, AS, MVB, and JPR performed the experiments and prepared the manuscript.
Funding
This work was supported by a Research Scholar Grant ‘Defining resistance to BRAF inhibition in melanoma’ RSG-17-230-01-TBG from the American Cancer Society.
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2020.06.006. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE152723 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152723).
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020 doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Genomic T.C.G.A. Classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dankort D., Curley D.P., Cartlidge R.A., Nelson B., Karnezis A.N., Damsky W.E. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodis E., Watson I.R., Kryukov G.V., Arold S.T., Imielinski M., Theurillat J.P. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies M.A., Stemke-Hale K., Tellez C., Calderone T.L., Deng W., Prieto V.G. A novel AKT3 mutation in melanoma tumours and cell lines. Br J Cancer. 2008;99:1265–1268. doi: 10.1038/sj.bjc.6604637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nazarian R., Shi H., Wang Q., Kong X., Koya R.C., Lee H. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun C., Wang L., Huang S., Heynen G.J.J.E., Prahallad A., Robert C. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–122. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- 8.Johannessen C.M., Boehm J.S., Kim S.Y., Thomas S.R., Wardwell L., Johnson L.a. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villanueva J., Vultur A., Lee J.T., Somasundaram R., Fukunaga-Kalabis M., Cipolla A.K. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson T.R., Fridlyand J., Yan Y., Penuel E., Burton L., Chan E. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shull A.Y., Latham-Schwark A., Ramasamy P., Leskoske K., Oroian D., Birtwistle M.R. Novel somatic mutations to PI3K pathway genes in metastatic melanoma. PLoS One. 2012;7:1–10. doi: 10.1371/journal.pone.0043369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies M.a., Stemke-Hale K., Lin E., Tellez C., Deng W., Gopal Y.N. Integrated molecular and clinical analysis of AKT activation in metastatic melanoma. Clin Cancer Res. 2009;15:7538–7546. doi: 10.1158/1078-0432.CCR-09-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dummer R., Ascierto P.A., Gogas H.J., Arance A., Mandala M., Liszkay G. Encorafenib plus binimetinib versus vemurafenib or encorafenib in patients with BRAF-mutant melanoma (COLUMBUS): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2018 doi: 10.1016/S1470-2045(18)30142-6. [DOI] [PubMed] [Google Scholar]
- 14.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. 2011 doi:10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed]
- 15.Hauschild A., Grob J.J., Demidov L.V., Jouary T., Gutzmer R., Millward M. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 16.McArthur G.A., Puzanov I., Amaravadi R., Ribas A., Chapman P., Kim K.B. Marked homogeneous and early [18F]fluorodeoxyglucose-positron emission tomography responses to vemurafenib in BRAF-mutant advanced melanoma. J Clin Oncol. 2012;30:1628–1634. doi: 10.1200/JCO.2011.39.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McArthur G.A., Chapman P.B., Robert C., Larkin J., Haanen J.B., Dummer R. Safety and efficacy of vemurafenib in BRAFV600E and BRAFV600K mutation-positive melanoma (BRIM- 3): extended follow-up of a phase 3, randomised, open-label study. Lancet Oncol. 2014;15:323–332. doi: 10.1016/S1470-2045(14)70012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trunzer K., Pavlick A.C., Schuchter L., Gonzalez R., McArthur G.A., Hutson T.E. Pharmacodynamic effects and mechanisms of resistance to vemurafenib in patients with metastatic melanoma. J Clin Oncol. 2013;31:1767–1774. doi: 10.1200/JCO.2012.44.7888. [DOI] [PubMed] [Google Scholar]
- 19.Shtivelman E., Davies M.A., Hwu P., Yang J., Lotem M., Oren M. Pathways and therapeutic targets in melanoma. Oncotarget. 2014;5:1701–1752. doi: 10.18632/oncotarget.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larkin J., Ascierto P.a., Dréno B., Atkinson V., Liszkay G., Maio M. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med. 2014;371:1867–1876. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 21.Dummer R., Ascierto P.A., Gogas H.J., Arance A., Mandala M., Liszkay G. Overall survival in patients with BRAF-mutant melanoma receiving encorafenib plus binimetinib versus vemurafenib or encorafenib (COLUMBUS): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2018 doi: 10.1016/S1470-2045(18)30497-2. [DOI] [PubMed] [Google Scholar]
- 22.Flaherty K.T., Infante J.R., Daud A., Gonzalez R., Kefford R.F., Sosman J. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long G.V., Stroyakovskiy D., Gogas H., Levchenko E., de Braud F., Larkin J. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med. 2014;371:1877–1888. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 24.Touil Y., Segard P., Ostyn P., Begard S., Aspord C., El MacHhour R. Melanoma dormancy in a mouse model is linked to GILZ/FOXO3A-dependent quiescence of disseminated stem-like cells. Sci Rep. 2016;6 doi: 10.1038/srep30405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleffel S., Schatton T. Tumor dormancy and cancer stem cells: two sides of the same coin? Adv Exp Med Biol. 2013;734:145–179. doi: 10.1007/978-1-4614-1445-2_8. [DOI] [PubMed] [Google Scholar]
- 26.VanBrocklin M.W., Robinson J.P., Lastwika K.J., Khoury J.D., Holmen S.L. Targeted delivery of NRASQ61R and Cre-recombinase to post-natal melanocytes induces melanoma in Ink4a/Arflox/lox mice. Pigment Cell Melanoma Res. 2010;23:531–541. doi: 10.1111/j.1755-148X.2010.00717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang H., Kircher D.A., Kim K.H., Grossmann A.H., VanBrocklin M.W., Holmen S.L. Activated MEK cooperates with Cdkn2a and Pten loss to promote the development and maintenance of melanoma. Oncogene. 2017:1–10. doi: 10.1038/onc.2016.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kakadia S., Yarlagadda N., Awad R., Kundranda M., Niu J., Naraev B. Mechanisms of resistance to BRAF and MEK inhibitors and clinical update of us food and drug administration-approved targeted therapy in advanced melanoma. Onco Targets Ther. 2018 doi: 10.2147/OTT.S182721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyun K., Jeon J., Park K., Kim J. Writing, erasing and reading histone lysine methylations. Exp Mol Med. 2017 doi: 10.1038/emm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J., Huang S.K., Marzese D.M., Hsu S.C., Kawas N.P., Chong K.K. Epigenetic changes of EGFR have an important role in BRAF inhibitor-resistant cutaneous melanomas. J Invest Dermatol. 2015 doi: 10.1038/jid.2014.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emran A.Al, Marzese D.M., Menon D.R., Stark M.S., Torrano J., Hammerlindl H. Distinct histone modifications denote early stress-induced drug tolerance in cancer. Oncotarget. 2018 doi: 10.18632/oncotarget.23654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strub T., Ghiraldini F.G., Carcamo S., Li M., Wroblewska A., Singh R. SIRT6 haploinsufficiency induces BRAFV600E melanoma cell resistance to MAPK inhibitors via IGF signalling. Nat Commun. 2018 doi: 10.1038/s41467-018-05966-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang H., Kircher D.A., Kim K.H., Grossmann A.H., VanBrocklin M.W., Holmen S.L. Activated MEK cooperates with Cdkn2a and Pten loss to promote the development and maintenance of melanoma. Oncogene. 2017;36 doi: 10.1038/onc.2016.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson J.P., VanBrocklin M.W., Guilbeault A.R., Signorelli D.L., Brandner S., Holmen S.L. Activated BRAF induces gliomas in mice when combined with Ink4a/Arf loss or Akt activation. Oncogene. 2010;29:335–344. doi: 10.1038/onc.2009.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott M.C., Temiz N.A., Sarver A.E., LaRue R.S., Rathe S.K., Varshney J. Comparative transcriptome analysis quantifies immune cell transcript levels, metastatic progression, and survival in osteosarcoma. Cancer Res. 2018 doi: 10.1158/0008-5472.CAN-17-0576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia B.A., Mollah S., Ueberheide B.M., Busby S.A., Muratore T.L., Shabanowitz J. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc. 2007 doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010 doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahronian L.G., Lewis B.C. Using the RCAS-TVA system to model human cancer in mice. Cold Spring Harb Protoc. 2014;2014:1128–1135. doi: 10.1101/pdb.top069831. [DOI] [PubMed] [Google Scholar]
- 39.Cho J.H., Robinson J.P., Arave R.A., Burnett W.J., Kircher D.A., Chen G. AKT1 activation promotes development of melanoma metastases. Cell Rep. 2015;13 doi: 10.1016/j.celrep.2015.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson J.P., Rebecca V.W., Kircher D.A., Silvis M.R., Smalley I., Gibney G.T. Resistance mechanisms to genetic suppression of mutant NRAS in melanoma. Melanoma Res. 2017;27 doi: 10.1097/CMR.0000000000000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin CH, Grossmann AH, Holmen SL, Robinson JP. The BRAF kinase domain promotes the development of gliomas in vivo. 2015. [DOI] [PMC free article] [PubMed]
- 42.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016. doi:10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed]
- 43.Rothbart S.B., Dickson B.M., Raab J.R., Grzybowski A.T., Krajewski K., Guo A.H. An interactive database for the assessment of histone antibody specificity. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Y., Jensen O.N. Modification-specific proteomics: Strategies for characterization of post-translational modifications using enrichment techniques. Proteomics. 2009 doi: 10.1002/pmic.200900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menon D.R., Das S., Krepler C., Vultur A., Rinner B., Schauer S. A stress-induced early innate response causes multidrug tolerance in melanoma. Oncogene. 2015 doi: 10.1038/onc.2014.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khaliq M., Fallahi-Sichani M. Epigenetic mechanisms of escape from BRAF oncogene dependency. Cancers (Basel) 2019 doi: 10.3390/cancers11101480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larson J.D., Kasper L.H., Paugh B.S., Jin H., Wu G., Kwon C.H. Histone H3.3 K27M accelerates spontaneous brainstem glioma and drives restricted changes in bivalent gene expression. Cancer Cell. 2019 doi: 10.1016/j.ccell.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim K.H., Roberts C.W.M. Targeting EZH2 in cancer. Nat Med. 2016 doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zingg D., Debbache J., Peña-Hernández R., Antunes A.T., Schaefer S.M., Cheng P.F. EZH2-mediated primary cilium deconstruction drives metastatic melanoma formation. Cancer Cell. 2018 doi: 10.1016/j.ccell.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Li B., Zheng Y., Yang L. The oncogenic potential of SuV39H2: a comprehensive and perspective view. J Cancer. 2019 doi: 10.7150/jca.28254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wan M., Liang J., Xiong Y., Shi F., Zhang Y., Lu W. The trithorax group protein Ash2l is essential for pluripotency and maintaining open chromatin in embryonic stem cells. J Biol Chem. 2013 doi: 10.1074/jbc.M112.424515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nicholas C., Yang J., Peters S.B., Bill M.A., Baiocchi R.A., Yan F. PRMT5 Is Upregulated in malignant and metastatic melanoma and regulates expression of MITF and p27Kip1. PLoS One. 2013 doi: 10.1371/journal.pone.0074710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu L., Kimball S., Liu H., Holowatyj A., Yang Z.Q. Genetic alterations of histone lysine methyltransferases and their significance in breast cancer. Oncotarget. 2015 doi: 10.18632/oncotarget.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marinova Z., Leng Y., Leeds P., Chuang D.M. Histone deacetylase inhibition alters histone methylation associated with heat shock protein 70 promoter modifications in astrocytes and neurons. Neuropharmacology. 2011;60:1109–1115. doi: 10.1016/j.neuropharm.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.