Abstract
Transmembrane and ubiquitin-like domain-containing 1 (Tmub1) inhibits hepatocyte proliferation during liver regeneration, but its role in hepatocellular carcinoma (HCC) has yet to be revealed. In this study, we show that the levels of Tmub1 were significantly lower in HCC tissues and cells than they were in adjacent tissues and normal hepatic cells, and the low levels of Tmub1 indicated a poor prognosis in HCC patients. Xenograft growth assay revealed that Tmub1 represses HCC growth in vivo. In addition, Tmub1 formed a protein complex with apoptosis-associated protein tumor protein 63 (p63), especially with the ΔN isoforms (ΔNp63α, β, and γ). Further loss- and gain-of-function analyses indicated that Tmub1 promotes apoptosis of Hep3B and MHCC-LM3 cells. Tmub1 decreased the protein expression of ΔNp63, and the pro-apoptotic effect of Tmub1 can be reversed by ΔNp63 isoforms (α, β, and γ). Additionally, we report that Tmub1 promotes the ubiquitination and degradation of ΔNp63 proteins. Finally, we confirmed in HCC tissues that Tmub1 is negatively correlated with ΔNp63 and positively correlated with the level of apoptosis. Taken together, Tmub1 suppresses HCC by enhancing the ubiquitination and degradation of ΔNp63 isoforms to induce HCC cell apoptosis. These findings provide a potential strategy for the management of HCC.
Keywords: transmembrane and ubiquitin-like domain containing 1 protein, hepatocellular carcinoma, tumor suppressor, cell apoptosis, tumor protein 63, post-translational modification, ubiquitination
Graphical Abstract
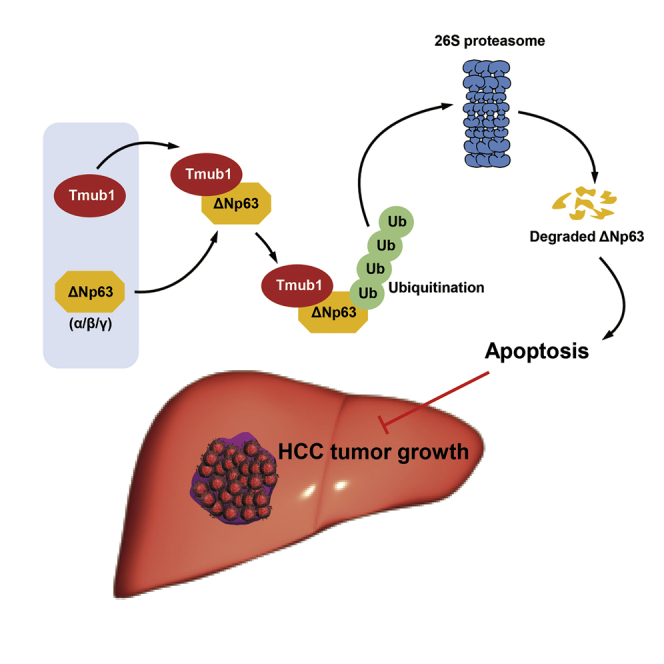
Chen and colleagues report that Tmub1 was a tumor suppressor in hepatocellular carcinoma. Mechanically, Tmub1 promotes apoptosis by binding to p53 family member ΔNp63 proteins and promotes their ubiquitination-mediated degradation.
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common cancer worldwide and the fourth most common cause of cancer mortality.1 HCC is difficult to treat because patients may be asymptomatic until the cancer has developed to an advanced stage. Although various treatment options are available, including surgical resection, chemotherapy, sorafenib, and combined immunotherapy, the 5-year survival rate of HCC patients still remains low.2 Since the precise molecular mechanisms responsible for HCC development have not been clarified, identifying HCC-related molecules may enable the development of effective efforts in improving the prognosis for HCC patients.
Transmembrane and ubiquitin-like domain-containing 1 (Tmub1), also known as hepatocyte odd protein shuttling (HOPS) or dendritic cell-derived ubiquitin-like protein (DULP), was first reported by Della Fazia et al.,3 and it is involved in liver regeneration and plays essential regulatory roles in the hepatocyte cell cycle. Our previous studies revealed that Tmub1 is a cell cycle-associated protein and a negative regulator of hepatocyte proliferation.4 Tmub1 can be induced by interleukin 6 (IL-6) and the transcriptional factor C/EBPβ, and it may form a negative feedback loop with STAT3 to regulate cell proliferation.5,6 Despite these data regarding the functions of Tmub1 in normal hepatocytes, the possible role of Tmub1 in HCC remains unknown, and other physiological functions of Tmub1 also need further explication. In 2009, Liu et al.7 mentioned that Tmub1 can induce apoptosis in 293T cells, but the specific role and the underlying mechanisms are yet to be revealed. In our preliminary studies, we found that Tmub1 may interact with an apoptosis-related protein, p63, indicating a possible relationship between Tmub1, p63, and apoptosis.
p63, a member of the tumor protein 53 (p53) family, shares DNA binding, oligomerization, and possible transactivation (TA) domains with p53 and p73. Using alternative promoters, p63 can be expressed as TAp63 and ΔNp63, which have opposite functions in transcription control. There are three major isoforms (α, β, and γ) for both TAp63 and ΔNp63 because of RNA splicing.8 Similar to p53, TAp63 promotes apoptosis and is often thought to function as a tumor suppressor. In contrast to TAp63, ΔNp63 isoforms can act as oncoproteins with anti-apoptotic activity. ΔNp63 isoforms lack the TA domain, and they prevent target gene activation by competing with TA isoforms, serving as dominant-negative forms of TAp63. Therefore, the ratios between TAp63 and ΔNp63 may be important in determining overall oncogenic or tumor-suppressive properties.9 Unlike p53, which is frequently mutated in HCC, p63 rarely acquires a loss of heterozygosity mutation. In fact, p63 is thought to be regulated predominantly at the protein level.10 p63 is commonly regulated by posttranslational modifications, particularly ubiquitination-proteasome-mediated degradation, which is a major pathway that regulates the cellular proteome by targeting specific proteins for proteasome-mediated degradation.11
In this study, we report that Tmub1 is downregulated in HCC, and that low levels of Tmub1 indicate a poor prognosis in HCC patients. Mechanistically, Tmub1 promotes apoptosis in HCC cells by enhancing the ubiquitination and degradation of ΔNp63 isoforms.
Results
The Poor Expression of Tmub1 Is Associated with the Malignancy of HCC
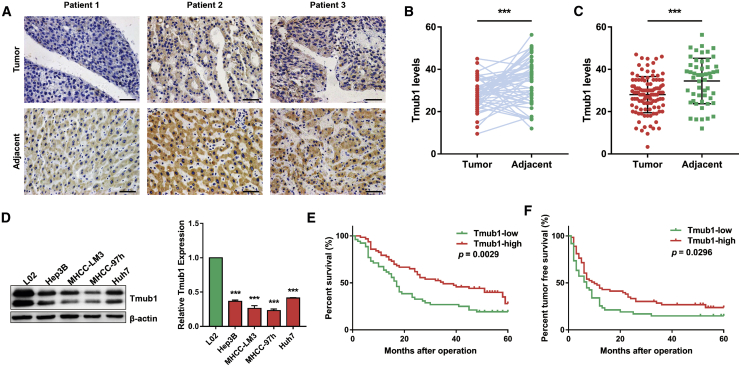
To characterize the expression of Tmub1 in HCC, Tmub1 immunohistochemistry was conducted on the tissue microarray, including 52 pairs of HCC tissue and matched adjacent paracancer tissue and 58 cases of only HCC tissue. As shown in Figures 1A–1C, a lower level of Tmub1 immunostaining was detected in the HCC tissue than what was observed in the adjacent normal tissue. Moreover, western blotting assays showed that the levels of Tmub1 in HCC cell lines (Hep3B, MHCC-LM3, MHCC-97h, and Huh7) were significantly lower than they were in the normal hepatocyte cell line L02 (Figure 1D). Then, we analyzed the effect of Tmub1 expression on HCC pathological characteristics and the prognosis of HCC patients. The results confirmed that Tmub1 staining was negatively correlated with tumor size, differentiation, and tumor, node, and metastasis (TNM) stage (Table 1). In addition, the overall survival rate (19.23% versus 27.95%, p = 0.0029) and the disease-free survival rate (14.88% versus 23.83%, p = 0.0296) of the HCC patients with high Tmub1 expression were significantly higher than they were in those patients with low expression, respectively (Figures 1E and 1F). These results show that the abnormal low expression of Tmub1 is associated with malignancy and leads to poor survival in HCC patients.
Figure 1.
Low Tmub1 Expression Is Associated with the Malignancy of HCC
(A) Representative images of Tmub1 immunostaining on the HCC-adjacent sample tissue microarray (original magnification, ×400). (B) Tmub1 levels in 52 pairs of HCC and adjacent tissues. (C) Tmub1 levels in 110 HCC tissues and 52 adjacent tissues. ∗∗∗p < 0.001. (D) Western blotting assays of Tmub1 levels in normal liver cell line L02 and HCC cell lines Hep3B, MHCC-LM3, MHCC-97h, and Huh7. Densitometric analysis showed significantly lower expression in HCC cells than in L02 cells. ∗∗∗p < 0.001 compared with L02 cells. (E) Overall survival of HCC patients with low and high expression of Tmub1. (F) Disease-free survival of HCC patients with low and high expression of Tmub1.
Table 1.
Relationship between the Expression of Tmub1 and the Clinicopathological Features of HCC Patients
| Characteristics | Cases (%) | Mean ± SEM | p Value |
|---|---|---|---|
| Sex | |||
| Male | 101 (91.8) | 28.48 ± 0.8206 | 0.6546 |
| Female | 9 (8.2) | 27.17 ± 3.516 | |
| Age | |||
| ≥50 | 58 (52.7) | 28.41 ± 1.202 | 0.8046 |
| <50 | 52 (47.3) | 28 ± 1.102 | |
| HBsAg | |||
| Positive | 98 (89.1) | 28.01 ± 0.8362 | 0.3325 |
| Negative | 12 (10.9) | 30.53 ± 2.883 | |
| AFP | |||
| ≥400 ng/mL | 60 (54.5) | 27.76 ± 1.144 | 0.4529 |
| <400 ng/mL | 50 (45.5) | 26.23 ± 1.759 | |
| Tumor size | |||
| ≥5 cm | 72 (65.5) | 26.92 ± 0.9886 | 0.0196 |
| <5 cm | 38 (34.5) | 30.86 ± 1.315 | |
| Differentiation | |||
| High/moderate | 89 (80.9) | 29.13 ± 0.8238 | 0.0048 |
| Low | 21 (19.1) | 23.37 ± 2.209 | |
| TNM stage | |||
| I/II | 60 (54.5) | 29.56 ± 1.211 | 0.0382 |
| III/IV | 50 (45.5) | 26.19 ± 0.9959 |
HBsAg, hepatitis B surface antigen; AFP, alpha-fetoprotein; TNM, tumor, node, metastasis.
Tmub1 Represses HCC Growth In Vivo
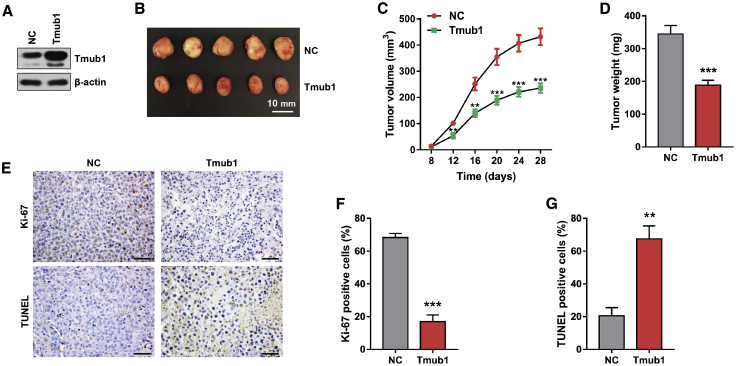
In view of the inhibitory role of Tmub1 in hepatocytes,5 we investigated the role of Tmub1 in tumor growth in vivo. The xenograft growth assays showed that Tmub1 significantly suppressed tumor growth, as reflected by tumor volume and weight, compared with the negative control group cells (Figures 2A–2D). Furthermore, TUNEL (terminal deoxynucleotidyltransferase-mediated deoxyuridine triphosphate nick end labeling) staining and immunohistochemical staining of Ki-67 showed that fewer proliferating cells and more apoptosis cells were found in Tmub1 overexpressing tumors (Figures 2E–2G). Taken together, Tmub1 negatively regulated the proliferation and facilitated the apoptosis of HCC cells in vivo.
Figure 2.
Tmub1 Inhibits Tumor Growth In Vivo
Nude mice (n = 5) were subcutaneously injected with Tmub1-overexpressing MHCC-LM3 cells (Tmub1) and negative control cells (NC) by lentivirus infection. Tumor sizes were measured after sizeable tumor formation (day 8). Inoculated mice were sacrificed on day 28 and the tumors were excised for analysis. (A) Western blotting analysis of Tmub1 levels. (B) Macroscopic appearance of the isolated tumor. (C and D) Tumor volume curves (C) and tumor weights (D) in Tmub1 and NC groups in xenograft nude mice. (E) Representative Ki-67 immunohistochemical staining and TUNEL staining of the xenograft tumor (original magnification, ×400). (F and G) Percentage of Ki-67-positive cells (F) and TUNEL-positive cells (G). ∗∗p < 0.01, ∗∗∗p < 0.001 compared with the NC group.
Tmub1 Forms a Protein Complex with p63
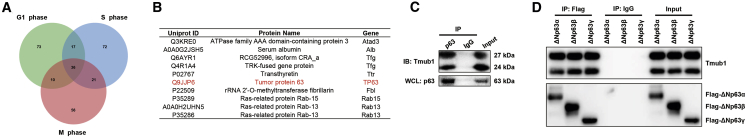
In our previous studies, we identified Tmub1 as a cell cycle-associated protein in rat hepatocytes.4 To identify the Tmub1-interaction proteins in the cell cycle, we conducted a preliminary immunoprecipitation (IP)-mass spectrometry (MS) experiment after cell cycle synchronization. However, we surprisingly found that Tmub1 interacts with the apoptosis protein p63 regardless of the cell cycle phase in hepatocytes (Figures 3A and 3B), which indicated a possible role of Tmub1 in apoptosis through p63.
Figure 3.
Tmub1 Forms a Protein Complex with p63
(A) IP-MS analysis of Tmub1-associated proteins in hepatocytes. Rat normal hepatocyte BRL-3A cells were synchronized at the G1, S, or M phases by serum starvation, double-thymidine, and nocodazole treatment, respectively. A Venn diagram shows the common Tmub1-interacting proteins within the three cell cycle phases. (B) The top 10 proteins from the 36 common Tmub1-interacting proteins identified by IP-MS assays are listed. (C) coIP assays for Tmub1 and p63 in Hep3B cells. (D) coIP assays for Tmub1 and ΔNp63 isoforms in 3× flag-ΔNp63α-, 3× flag-ΔNp63β-, and flag-ΔNp63γ transfected Hep3B cells.
Since p63 expression can be disrupted by p53, we chose a p53 null HCC cell line, Hep3B, to conduct the following experiments. First, we confirmed the Tmub1-p63 interaction with coimmunoprecipitation (coIP) assays (Figure 3C). Since the TAp63 and ΔNp63 isoforms perform opposite roles in apoptosis, we tested which p63 isoforms may be associated with Tmub1. Our immunohistochemistry assays on the HCC tissue microarray showed that the expression of TAp63 isoforms is very low in HCC tissues (Figure S1A). Moreover, after transfecting the six p63 isoforms into Hep3B cells, we found that TAp63 isoforms were much more unstable than the ΔNp63 isoforms (Figure S1B). Based on these results, we concentrated on the relationship between Tmub1 and the ΔNp63 isoforms in this study. coIP experiments showed that Tmub1 may nonselectively bind to ΔNp63 proteins (Figure 3D).
Tmub1 Promotes HCC Cell Apoptosis via ΔNp63
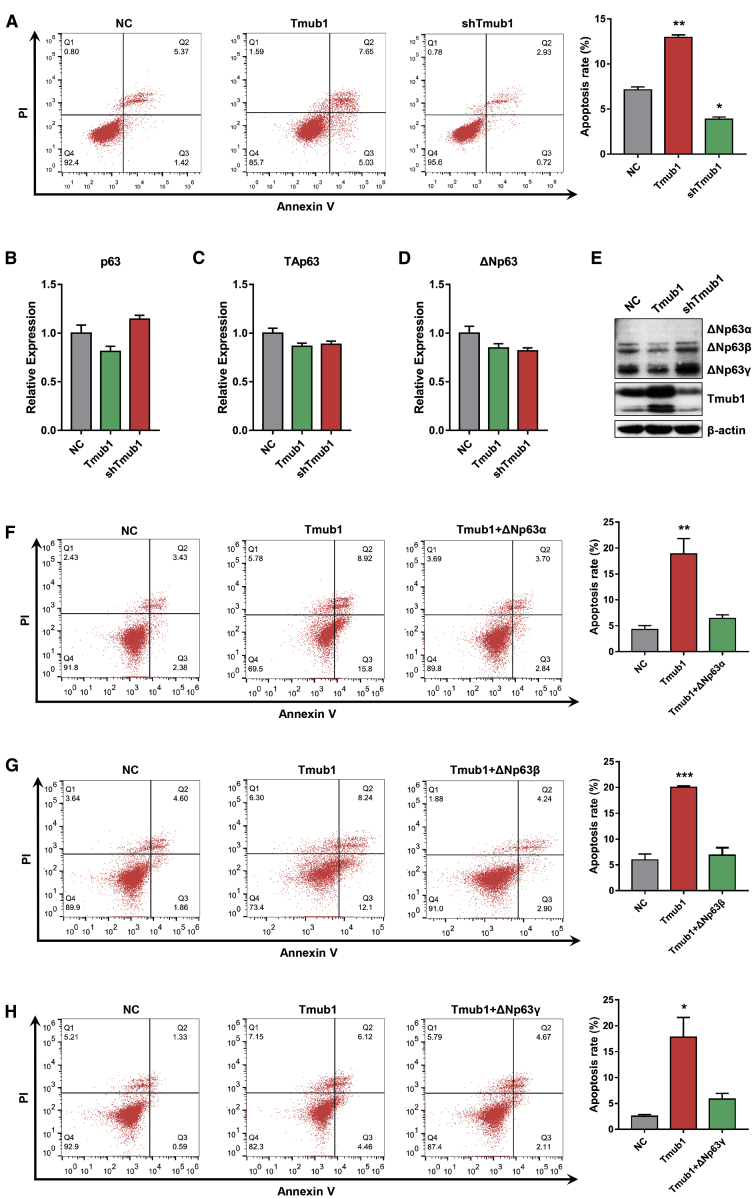
To investigate the role of Tmub1 in HCC cell apoptosis, we performed flow cytometry assays on Hep3B cells after manipulating the expression of Tmub1. The results showed that Tmub1 overexpression significantly increased the apoptosis rate of Hep3B cells, and knockdown of Tmub1 significantly reduced the apoptosis rate in those cells (Figure 4A). Similar results were also gathered from experiments using MHCC-LM3 cells (Figure S2).
Figure 4.
Tmub1 Promotes HCC Cell Apoptosis by Downregulating the Protein Levels of ΔNp63
(A) Flow cytometric analysis of Hep3B cells transfected with negative control (NC), Tmub1, or Tmub1 shRNA plasmids. Forty-eight hours after transfection, cells were collected and stained with annexin V/propidium iodide (PI), which was followed by flow cytometry assays to determine the apoptosis rate. (B–D) Quantitative real-time PCR assays for p63 (B), TAp63 (C), and ΔNp63 (D) in Hep3B cells transfected with negative control (NC), Tmub1, or Tmub1 shRNA plasmids. (E) Western blot assays for ΔNp63 on Hep3B cells after manipulating the expression of Tmub1. (F–H) The effect of Tmub1 on Hep3B cell apoptosis was reversed by (F) ΔNp63α, (G) ΔNp63β, and (H) ΔNp63γ. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 compared with NC cells.
Next, we questioned whether Tmub1 promotes Hep3B cell apoptosis by affecting the expression of ΔNp63. Although the mRNA levels of p63 may not be affected by manipulating Tmub1 expression (Figures 4B–4D), overexpression of Tmub1 significantly reduced the expression of ΔNp63 proteins, and knockdown of Tmub1 significantly increased the expression of ΔNp63 proteins (Figure 4E). Furthermore, the pro-apoptotic effect of Tmub1 was attenuated by transfection with ΔNp63α, ΔNp63β or ΔNp63γ (Figures 4F, 4G, and 4H, respectively). These results indicated that Tmub1 promotes HCC cell apoptosis by reducing ΔNp63 expression.
Tmub1 Promotes the Ubiquitination and Protein Degradation of ΔNp63 Isoforms
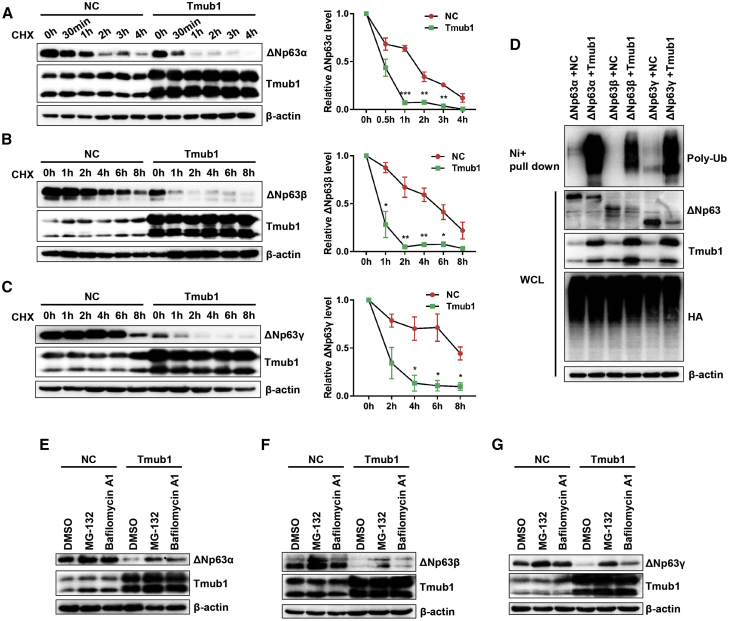
Considering that the ectopic expression of Tmub1 significantly reduced the expression of ΔNp63 proteins without affecting the mRNA levels, Tmub1 may control ΔNp63 expression at the posttranslational levels. Consistently, cycloheximide pulse-chase assays showed that ectopic expression of Tmub1 significantly shortened the half-life of ΔNp63α, ΔNp63β, and ΔNp63γ proteins in Hep3B cells (Figures 5A–5C). These data suggested that Tmub1 may promote the protein degradation of ΔNp63.
Figure 5.
Tmub1 Promotes the Ubiquitination and Degradation of ΔNp63 Proteins
(A) Measurement of ΔNp63α protein half-lives by cycloheximide (CHX) chase assays and western blotting. Hep3B cells were cotransfected with ΔNp63α and the indicated plasmids. Forty-eight hours after transfection, the cells were treated with 75 μg/mL CHX for different times (0–4 h), and the cells were then collected for western blot analysis. Quantitative results were obtained using ImageJ software. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 compared with NC cells. (B and C) Measurement of ΔNp63β (B) and ΔNp63γ (C) protein half-lives. The CHX treatment times ranged from 0 to 8 h. (D) Ubiquitination assay of ΔNp63 isoforms by Ni-NTA pull-down assays. Hep3B cells were cotransfected with HA-his-Ub and the indicated plasmids. Forty-eight hours after the transfection, cells were collected after additional treatment with MG132 for 6 h. (E–G) The pro-degradation effect of Tmub1 on (E) ΔNp63α, (F) ΔNp63β, and (G) ΔNp63γ proteins can be reversed by treatment with the proteasome inhibitor MG132 and the lysosome inhibitor bafilomycin A1. Hep3B cells were cotransfected with the ΔNp63 isoform and the indicated plasmids. Forty-eight hours after the transfection, cells were treated with MG132 or bafilomycin A1 for 6 h and were then collected.
Next, we tested whether Tmub1 controls ΔNp63 protein stability in HCC cells. In this regard, ΔNp63α, ΔNp63β, and ΔNp63γ protein levels were rescued by treatment with the proteasome inhibitor MG132 and the lysosome inhibitor bafilomycin A1 in Tmub1 overexpresing cells (Figures 5E–5G). We next examined whether Tmub1 affected the ubiquitination of ΔNp63 isoforms. The results showed that ectopic expression of Tmub1 increased the polyubiquitination of ΔNp63α, ΔNp63β, and ΔNp63γ (Figure 5D). In conclusion, Tmub1 promotes ΔNp63 protein degradation by enhancing its ubiquitination.
The Expression of Tmub1 Is Positively Correlated with Apoptosis in HCC Tissue
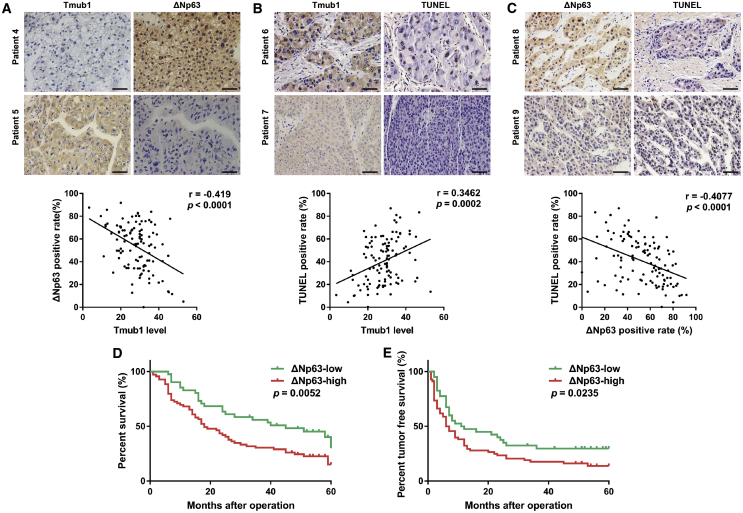
To study the relationships among Tmub1, ΔNp63, and apoptosis in HCC, immunohistochemical analysis of an HCC tissue microarray with serial sections was performed. The representative images showed that the cases with a low expression of Tmub1 had stronger ΔNp63 staining and weaker TUNEL staining than what was seen when there was high Tmub1 expression. Statistical analysis showed that the Tmub1 levels are negatively correlated with ΔNp63 levels and positively correlated with TUNEL-positive rates, while ΔNp63 was negatively correlated with TUNEL-positive rates (Figure 6A–6C). Moreover, survival analysis confirmed that the overall survival rate (30.14% versus 15.11%, p = 0.0052) and the disease-free survival rate (29.80% versus 13.87%, p = 0.0235) in the ΔNp63 underexpression group were significantly higher than those in the overexpression group (Figures 6D and 6E). These results suggested that downregulation of Tmub1 is closely related to the higher level of ΔNp63 and lower apoptosis rate in HCC tissues, which is involved in HCC development.
Figure 6.
The Expression of Tmub1 in HCC Tissues Is Correlated with Apoptosis and Negatively Correlated with ΔNp63
(A) Representative IHC images of samples with high and low expression levels of Tmub1 and ΔNp63. The Tmub1 level is negatively correlated with the ΔNp63 level. (B) Representative images of samples with high and low expression levels of Tmub1 and TUNEL staining. The Tmub1 level is positively correlated with the rate of TUNEL-positive staining. (C) Representative images of samples with high and low expression levels of ΔNp63 and TUNEL staining. The ΔNp63 level is negatively correlated with the rate of TUNEL-positive staining. n = 110. (D) Overall survival of HCC patients with low and high expression of ΔNp63. (E) Disease-free survival of HCC patients with low and high expression of ΔNp63.
Discussion
Because of the molecular links between apoptosis and tumorigenesis, current attempts to improve the survival of HCC patients will have to include strategies that specifically target tumor cell resistance to apoptosis. The observation that p53 function is lost in most cancers makes it a unique molecular target for cancer therapies, and it has been intensively studied during the last decades.12 For p53-deficient tumor cells, reconstitution of p53 activity has been demonstrated to be feasible and practical. The discovery of the p53-related genes p63 and p73 raised the possibility that they may be cancer-associated genes and, as a consequence, suggested that p53 is not the only component in predicting prognosis and the response to chemotherapy, and that the status of a network that contains p53, p63, and p73 should be considered.13,14
p63 plays a pivotal role in a wide range of biological processes, including cell proliferation, survival, apoptosis, differentiation, cell migration and invasion, and senescence. In contrast to TAp63, which may mimic the functions of p53, ΔNp63 exerts anti-apoptotic functions. Accumulating evidence indicates that the reduced tumor expression of TAp63 and the increased ΔNp63 expression may favor tumor development and may be associated with tumor recurrence and reduced survival.15, 16, 17 In the case of ΔNp63, ΔNp63α has been relatively intensively studied compared to the other two isoforms. ΔNp63α was correlated with cancer in human biopsy samples and is a common inhibitory target of oncogenic phosphatidylinositol 3-kinase (PI3K), Ras, and Her2.18 Additionally, ΔNp63α in HCC interferes with both the death receptor and the mitochondrial apoptosis activity of the TA isoforms:19 ΔNp63α may function as a critical integrator of oncogenic signaling in cancer by playing a causative role in promoting cell proliferation and metastasis, as well as by inhibiting apoptosis.20
Tmub1 is mainly regarded as a cell cycle-associated protein involved in the process of liver regeneration. After partial hepatectomy, Tmub1 is upregulated in the regenerating liver,3 and it plays a negative regulatory role in the process of hepatocyte proliferation.21 Additionally, Tmub1 regulates locomotor activity and wakefulness by interacting with CAMLG, and it facilitates the recycling of the AMPAR subunit GluR2 to the cell surface in the mouse brain,22,23 suggesting that it has additional roles in the central nervous system. Our previous studies on mice, rat hepatocytes, and human normal hepatocytes showed that Tmub1 significantly inhibits hepatocyte proliferation by decreasing the expression of cell cycle-related genes.4,5 However, cellular functions of Tmub1 in addition to hindering proliferation have yet to be revealed, and whether Tmub1 exerts similar inhibitory roles in HCC cells has not been studied.
In this study, we report that the expression of Tmub1 is downregulated in HCC cells compared to normal hepatocytes, and this decrease in Tmub1 expression is associated with HCC malignancy. By analyzing the Tmub1 protein complex, we found that p63 may be associated with the function of Tmub1. Further studies revealed that Tmub1 promotes HCC cell apoptosis by increasing the ubiquitination and degradation of ΔNp63 proteins.
As described previously, the stability of all p63 isoforms was proteasome-dependent,11,24,25 and ΔN isoforms were expressed at consistently higher intracellular levels than were the TA isoforms. Additionally, the expression of TA and ΔN isoforms are regulated by different mechanisms in liver cells.26 The TAp63 isoform is expressed at very low levels in the Hep3B cell line under basal conditions, and TAp63 isoforms are rapidly degraded and have a half-life of approximately 1 h; however, ΔNp63 isoforms are relatively more stable than the TAp63 isoforms.27 Our results were consistent with these studies, and we also propose that ΔNp63 proteins may be more useful than TAp63 isoforms in the diagnosis of HCC in biopsy samples, because of the low baseline expression of TAp63. Moreover, we found that the isoform most affected by Tmub1 was ΔNp63α. This may provide a deeper understanding into how Tmub1 affects apoptosis.
The ubiquitin-proteasome system (UPS) is an important ATP-dependent proteolytic system that targets specific proteins for proteasome-mediated degradation. In addition to the canonical ubiquitination pathway, proteins can also be modified through attachment to ubiquitin-like proteins (UBLs), such as NEDD8 and SUMO, which have the conserved globular β-grasp ubiquitin superfold and have demonstrated crosstalk with ubiquitination.28, 29, 30 In parallel to the single entities of UBLs, ubiquitin-like folds are now also recognized as important integral parts of proteins that form so-called ubiquitin-like domains (ULDs), which are present in a large variety of protein families. The most studied ULD proteins are the ubiquitin shuttle receptor families, which are involved in the targeting of polyubiquitinated proteins for proteasomal degradation.31 However, emerging roles indicate that in the wider family of UBL domain-containing proteins, the ULD is part of a larger polypeptide, but it is usually neither processed nor covalently attached to other proteins. Such ULDs confer properties to a protein that are similar to those acquired from a transferable UBL, including the ability to bind to specific target proteins.32 It has been adapted to a broad array of functions, ranging from a scaffold for various enzymatic activities and iron-sulfur cluster binding to an adaptor module for specific protein-protein interactions.33
As a ubiquitin-like domain-containing protein, the Tmub1 ULD only shares 26% identity and 34% similarity with ubiquitin, so it is not likely that Tmub1 can serve as a UBL to conjugate protein substrates through enzymatic cascades in ways that are similar to those of ubiquitin conjugation. So far, the only report about Tmub1 and ubiquitination is that Tmub1 may mediate the ubiquitination and degradation of the HMG-CoA (β-hydroxy β-methylglutaryl-coenzyme A) reductase HMGCR. In this process, Tmub1 links SPFH2 to a membrane-bound ubiquitin ligase gp78 in endoplasmic reticulum membranes.34 Our present work supports the notion that Tmub1 may affect the ubiquitination process by binding to specific proteins, but studies are needed to reveal the exact underlying molecular mechanisms. Another interesting phenomenon observed in this study is that the drop of DNp63 caused by Tmub1 can be partly rescued by the proteasome inhibitor MG-132 as well as the lysosome inhibitor bafilomycin A1. Considering that ubiquitinated proteins can be cleared from the cells by both proteasome- and lysosome-mediated degradation,35 DNp63 ubiquitination caused by Tmub1 may also lead to its degradation in the lysosome. The relationship between Tmub1-mediated ubiquitination and lysosome-mediated degradation will be studied in the future.
In conclusion, our study demonstrates that Tmub1 suppresses HCC growth by exerting a pro-apoptotic role in HCC cells, which is at least partially mediated by binding to ΔNp63 and promoting its ubiquitination and degradation. Our findings enrich the understanding of the p63-apoptosis regulatory network in HCC, and they support the use of Tmub1 as a potential therapeutic target for HCC.
Materials and Methods
Cases and Follow-up
In this study, we analyzed 110 HCC patients who received pathological liver resection at Southwest Hospital between January 2010 and December 2012. The clinicopathologic data for the patients were collected by acquiring patient records and performing pathological examinations. The patients were followed up until 5 years after surgery, and tumor recurrence and patient death were recorded. All subjects signed an informed consent form. The study was approved by the Institutional Research Ethics Committee of Southwest Hospital.
Immunohistochemical Staining
Patient pathological tissues were made into a tissue array that included 52 pairs of cancer and paracancer tissues, and another 58 cases of only HCC tissue. After dewaxing and hydration, the tissue arrays were heated in sodium citrate solution for antigen retrieval. After allowing to cool, the array was incubated in 3% H2O2 for 10 min and blocked in 10% goat serum at room temperature for 1 h. The arrays were incubated with anti-Tmub1 (1:50, Abcam) and anti-ΔNp63 (1:100, Abcam) antibodies overnight at 4°C. An anti-rabbit immunohistochemical detection kit (ZSGB-Bio) was used to detect the immunohistochemical reaction. Finally, the arrays were dehydrated, stained with hematoxylin, and mounted with neutral resin. The expression levels of Tmub1 were determined by counting mean gray value with ImageJ, and the positive nuclear staining rate for ΔNp63 of each tissue was calculated. A score higher than the mean was defined as high expression, while a score equal to or lower than the mean was categorized as low expression in tumor.
TUNEL Assay
TUNEL staining of the above tissue array was performed with a TUNEL staining kit according to the manufacturer’s instructions (Wanleibio). The positive staining of each tissue was identified and analyzed under a light microscope, and the staining rate was calculated.
Cell Culture and Transfection
The human HCC cell lines Hep3B, MHCC-LM3, MHCC-97h, and Huh7, the normal human hepatocyte cell line L02, and the rat hepatocyte cell line BRL-3A were purchased from the cell bank of Academia Sinica (Shanghai, China). The cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Excell Biotech) and were maintained at 37°C with 5% CO2. Constructs for human Tmub1 and its short hairpin RNA (shRNA) plasmids were purchased from Cyagen Biosciences. The shRNA sequence was 5′-GACACCATTGGCTCCTTGAAA-3′. Constructs for human 3× flag-TAp63α, 3× flag-TAp63β, 3× flag-ΔNp63α, and 3× flag-ΔNp63β were purchased from GeneCopoeia. Human flag-TAp63γ and flag-ΔNp63γ plasmids were purchased from Genscript, and hemagglutinin (HA)-histidine (His)-ubiquitin (Ub) plasmids were purchased from Sino Biological. Plasmids were transfected using Lipofectamine 3000 reagent (Invitrogen) according to the manufacturer’s instructions.
Xenograft Growth Assay
The antitumor activity of Tmub1 in HCC was assessed by the MHCC-LM3 cell line xenograft model. In detail, 5 × 106 cells from Tmub1 lentivirus-transfected cells and their negative control cells were implanted subcutaneously into 4-week-old nude mice. The subcutaneous tumor size was calculated and recorded every 4 days using a Vernier caliper as follows: tumor volume (mm3) = (length × width2)/2. Inoculated mice were sacrificed on day 28 and the tumors were excised for analysis. All animal experiments were approved by the Institutional Animal Care and Use Committees and performed in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care guidelines (https://www.aaalac.org).
Flow Cytometry Assay of Cell Apoptosis
Cell apoptosis assay were conducted with an annexin V-FITC apoptosis detection kit (Solarbio Life Sciences) according to the manufacturer’s instructions. Briefly, cells were seeded into a six-well plate for 24 h and then were transfected with the indicated plasmids. Forty-eight hours after transfection, cells were isolated in 1× annexin binding buffer and treated with both fluorescein isothiocyanate-annexin V stain and propidium iodide. After 30 min of incubation, the samples were subjected to flow cytometry analysis.
Western Blotting
Cell lysates were prepared with radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitor cocktail and phosphatase inhibitor cocktail (CoWin Biosciences). Protein concentrations were quantified with a bicinchoninic acid (BCA) protein assay kit (CoWin Biosciences) according to the manufacturer’s protocol. Protein extracts (20 μg) were denatured in Laemmli buffer, loaded onto a 10% SDS-polyacrylamide gel, and separated by electrophoresis. The proteins were transferred to nitrocellulose membranes and blocked with 5% nonfat dry milk buffer for 1 h. The membranes were incubated with the indicated primary antibodies at 4°C overnight and then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies at room temperature for 1 h. Anti-Tmub1 and anti-ΔNp63 antibodies were purchased from Abcam. An anti-flag antibody was purchased from Cell Signaling Technology. Anti-p63, anti-β-actin, HRP-conjugated goat anti-mouse, and goat anti-rabbit secondary antibodies were purchased from Proteintech.
Quantitative Real-Time PCR
Total RNA was extracted with an RNA extraction kit (BioTeke). Reverse transcription was performed according to a reverse transcription reagent protocol (Takara). Quantitative real-time PCR reactions were performed with SYBR Premix Ex Taq II (Takara) according to the manufacturer’s protocols. The sequences of the PCR primers used are as follows: p63, forward, 5′-CCCTCCAACACCGACTACCC-3′, reverse, 5′- CACCGCTTCACCACCTCCGT-3′; TAp63, forward, 5′-GACCTGAGTGACCCCATGTG-3′, reverse, 5′-CGGGTGATGGAGAGAGAGCA-3′; ΔNp63, forward, 5′-TGCCCAGACTCAATTTAGTGAG-3′, reverse, 5′-AGAGAGAGCATCGAAGGTGGGAG-3′;26 and β-actin, forward, 5′-CTCGCCTTTGCCGATCC-3′, reverse, 5′-TCTCCATGTCGTCCCAGTTG-3′.
CoIP
Cells lysate for western blot and IP experiments was generated by collecting the cells and lysing them in 500 μL of cell lysis buffer (Beyotime Biotechnology) containing protease inhibitor cocktail (1:50) and phosphatase inhibitor cocktail (1:50). Next, 60 μL of protein A/G magnetic beads (Bimake) was incubated with 4 μg of anti-flag (Cell Signaling Technology) antibody on a rotating platform for 30 min at room temperature; then the mix was incubated with 500 μL (1 mg) of cell lysate and rotated overnight at 4°C. Normal rabbit immunoglobulin G (IgG) was used as a negative control. The IP products were eluted with 40 μL of 1× Laemmli buffer and incubation for 10 min at 95°C. Western blot analysis was used for subsequent protein detection.
MS
Prior to MS analysis, Tmub1-binding proteins were enriched by IP from G1/S/M phase synchronized rat BRL-3A cells. Cell cycle synchronization was conducted as previously described.4 The MS analyses were performed at Shanghai Applied Protein Technology. Then, Mascot 2.2 software was used to assign the experimental mass values to specific peptide sequences from the UniProt database (https://www.uniprot.org/).
Protein Half-Life Assay
To examine the protein half-life of ΔNp63 proteins under different conditions, cycloheximide pulse-chase experiments were performed as previously described.27 Briefly, Hep3B cells were seeded in six-well plates and were then pretreated with 75 μg/mL of CHX for the indicated times. Whole-cell lysates were collected for immunoblotting.
Ubiquitination Assay
To detect ΔNp63 ubiquitination, nickel-nitrilotriacetic acid (Ni-NTA) pull-down assays were performed as described previously.36 In brief, Hep3B cells were transfected with HA-His-Ub and the indicated vectors, and then they were pretreated with MG132 for 6 h. Whole-cell lysates were collected using buffer A (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, and 10 mM imidazole [pH 8.0] in a 250-mL vol). After ultrasonication and centrifugation, lysates were purified by incubating with Ni-NTA (QIAGEN) 4°C overnight. Purification products were washed in buffer A, buffer A/buffer TI (1:3), and buffer TI (25 mM Tris-HCl, 20 mM imidazole [pH 6.8]). The pull-down products were then separated by SDS-PAGE for immunoblot analysis.
Statistical Analysis
Statistical analyses were conducted with SPSS 19.0 (SPSS, USA) for Windows and Prism 7 (GraphPad, USA). Continuous variables were expressed as the means ± standard deviation and compared using the independent sample t test and the Mann-Whitney U test. For categorical variables, comparisons were made using chi-square analysis or Fisher’s exact test. All statistical tests were two-way. p <0.05 was considered statistically significant.
Author Contributions
H.F. and P.C. conceived the project and designed the experiments. H.F. and Y.Z. performed and interpreted the majority of the experiments. H.F. wrote the manuscript. J.C. collected the clinical data. H.F., G.C., and B.Z. analyzed the data. G.C. supervised the project. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the Chongqing Technological Innovation and Application Project (cstc2018jscx-msybX0130) and the National Natural Science Foundation of China (grants 81270523 and 81570594). We thank the hepatobiliary surgery laboratory of the Southwest Hospital, Army Medical University for providing laboratory resources. We also thank Dr. Deng Huang, Dr. Ye Tan, and Dr. Xiaotong Lin (Institute of Hepatobiliary Surgery, Southwest Hospital, Army Medical University) for technical assistance.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.06.005.
Contributor Information
Geng Chen, Email: dr.chengeng@vip.126.com.
Ping Chen, Email: chenpingsyd@126.com.
Supplemental Information
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Zhu Q., Li N., Zeng X., Han Q., Li F., Yang C., Lv Y., Zhou Z., Liu Z. Hepatocellular carcinoma in a large medical center of China over a 10-year period: evolving therapeutic option and improving survival. Oncotarget. 2015;6:4440–4450. doi: 10.18632/oncotarget.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Della Fazia M.A., Castelli M., Bartoli D., Pieroni S., Pettirossi V., Piobbico D., Viola-Magni M., Servillo G. HOPS: a novel cAMP-dependent shuttling protein involved in protein synthesis regulation. J. Cell Sci. 2005;118:3185–3194. doi: 10.1242/jcs.02452. [DOI] [PubMed] [Google Scholar]
- 4.Fu H., Xu J., Chen J., Li G., Zhao X., Chen P. Microarray analysis reveals Tmub1 as a cell cycle-associated protein in rat hepatocytes. Mol. Med. Rep. 2018;17:4337–4344. doi: 10.3892/mmr.2018.8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu H., Dong R., Zhang Y., Xu J., Liu M., Chen P. Tmub1 negatively regulates liver regeneration via inhibiting STAT3 phosphorylation. Cell. Signal. 2019;55:65–72. doi: 10.1016/j.cellsig.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Liu M., Liu H., Wang X., Chen P., Chen H. IL-6 induction of hepatocyte proliferation through the Tmub1-regulated gene pathway. Int. J. Mol. Med. 2012;29:1106–1112. doi: 10.3892/ijmm.2012.939. [DOI] [PubMed] [Google Scholar]
- 7.Liu G.Y., Liu S.X., Li P., Tang L., Han Y.M., An H.Z., Li J.Y., Dai X.K., Li N., Cao X.T., Yu Y.Z. Cloning and characterization of DULP, a novel ubiquitin-like molecule from human dendritic cells. Cell. Mol. Immunol. 2009;6:27–33. doi: 10.1038/cmi.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Candi E., Agostini M., Melino G., Bernassola F. How the TP53 family proteins TP63 and TP73 contribute to tumorigenesis: regulators and effectors. Hum. Mutat. 2014;35:702–714. doi: 10.1002/humu.22523. [DOI] [PubMed] [Google Scholar]
- 9.Moll U.M., Slade N. p63 and p73: roles in development and tumor formation. Mol. Cancer Res. 2004;2:371–386. [PubMed] [Google Scholar]
- 10.Rossi M., Aqeilan R.I., Neale M., Candi E., Salomoni P., Knight R.A., Croce C.M., Melino G. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc. Natl. Acad. Sci. USA. 2006;103:12753–12758. doi: 10.1073/pnas.0603449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armstrong S.R., Wu H., Wang B., Abuetabh Y., Sergi C., Leng R.P. The Regulation of tumor suppressor p63 by the ubiquitin-proteasome system. Int. J. Mol. Sci. 2016;17:2041. doi: 10.3390/ijms17122041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flores E.R., Tsai K.Y., Crowley D., Sengupta S., Yang A., McKeon F., Jacks T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 13.Müller M., Schleithoff E.S., Stremmel W., Melino G., Krammer P.H., Schilling T. One, two, three—p53, p63, p73 and chemosensitivity. Drug Resist. Updat. 2006;9:288–306. doi: 10.1016/j.drup.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Seitz S.J., Schleithoff E.S., Koch A., Schuster A., Teufel A., Staib F., Stremmel W., Melino G., Krammer P.H., Schilling T., Müller M. Chemotherapy-induced apoptosis in hepatocellular carcinoma involves the p53 family and is mediated via the extrinsic and the intrinsic pathway. Int. J. Cancer. 2010;126:2049–2066. doi: 10.1002/ijc.24861. [DOI] [PubMed] [Google Scholar]
- 15.González R., De la Rosa A.J., Rufini A., Rodríguez-Hernández M.A., Navarro-Villarán E., Marchal T., Pereira S., De la Mata M., Müller-Schilling M., Pascasio-Acevedo J.M. Role of p63 and p73 isoforms on the cell death in patients with hepatocellular carcinoma submitted to orthotopic liver transplantation. PLoS ONE. 2017;12:e0174326. doi: 10.1371/journal.pone.0174326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patturajan M., Nomoto S., Sommer M., Fomenkov A., Hibi K., Zangen R., Poliak N., Califano J., Trink B., Ratovitski E., Sidransky D. ΔNp63 induces β-catenin nuclear accumulation and signaling. Cancer Cell. 2002;1:369–379. doi: 10.1016/s1535-6108(02)00057-0. [DOI] [PubMed] [Google Scholar]
- 17.Wu G., Nomoto S., Hoque M.O., Dracheva T., Osada M., Lee C.C., Dong S.M., Guo Z., Benoit N., Cohen Y. ΔNp63α and TAp63α regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res. 2003;63:2351–2357. [PubMed] [Google Scholar]
- 18.Hu L., Liang S., Chen H., Lv T., Wu J., Chen D., Wu M., Sun S., Zhang H., You H. ΔNp63α is a common inhibitory target in oncogenic PI3K/Ras/Her2-induced cell motility and tumor metastasis. Proc. Natl. Acad. Sci. USA. 2017;114:E3964–E3973. doi: 10.1073/pnas.1617816114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mundt H.M., Stremmel W., Melino G., Krammer P.H., Schilling T., Müller M. Dominant negative (ΔN) p63α induces drug resistance in hepatocellular carcinoma by interference with apoptosis signaling pathways. Biochem. Biophys. Res. Commun. 2010;396:335–341. doi: 10.1016/j.bbrc.2010.04.093. [DOI] [PubMed] [Google Scholar]
- 20.Ramalho F.S., Ramalho L.N., Della Porta L., Zucoloto S. Comparative immunohistochemical expression of p63 in human cholangiocarcinoma and hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2006;21:1276–1280. doi: 10.1111/j.1440-1746.2006.04309.x. [DOI] [PubMed] [Google Scholar]
- 21.Pieroni S., Della Fazia M.A., Castelli M., Piobbico D., Bartoli D., Brunacci C., Bellet M.M., Viola-Magni M., Servillo G. HOPS is an essential constituent of centrosome assembly. Cell Cycle. 2008;7:1462–1466. doi: 10.4161/cc.7.10.5882. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W., Savelieva K.V., Suwanichkul A., Small D.L., Kirkpatrick L.L., Xu N., Lanthorn T.H., Ye G.L. Transmembrane and ubiquitin-like domain containing 1 (Tmub1) regulates locomotor activity and wakefulness in mice and interacts with CAMLG. PLoS ONE. 2010;5:e11261. doi: 10.1371/journal.pone.0011261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang H., Takagi H., Konishi Y., Ageta H., Ikegami K., Yao I., Sato S., Hatanaka K., Inokuchi K., Seog D.H., Setou M. Transmembrane and ubiquitin-like domain-containing protein 1 (Tmub1/HOPS) facilitates surface expression of GluR2-containing AMPA receptors. PLoS ONE. 2008;3:e2809. doi: 10.1371/journal.pone.0002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osada M., Inaba R., Shinohara H., Hagiwara M., Nakamura M., Ikawa Y. Regulatory domain of protein stability of human P51/TAP63, a P53 homologue. Biochem. Biophys. Res. Commun. 2001;283:1135–1141. doi: 10.1006/bbrc.2001.4905. [DOI] [PubMed] [Google Scholar]
- 25.Okada Y., Osada M., Kurata S., Sato S., Aisaki K., Kageyama Y., Kihara K., Ikawa Y., Katoh I. p53 gene family p51(p63)-encoded, secondary transactivator p51B(TAp63α) occurs without forming an immunoprecipitable complex with MDM2, but responds to genotoxic stress by accumulation. Exp. Cell Res. 2002;276:194–200. doi: 10.1006/excr.2002.5535. [DOI] [PubMed] [Google Scholar]
- 26.Petitjean A., Cavard C., Shi H., Tribollet V., Hainaut P., Caron de Fromentel C. The expression of TA and ΔNp63 are regulated by different mechanisms in liver cells. Oncogene. 2005;24:512–519. doi: 10.1038/sj.onc.1208215. [DOI] [PubMed] [Google Scholar]
- 27.Petitjean A., Ruptier C., Tribollet V., Hautefeuille A., Chardon F., Cavard C., Puisieux A., Hainaut P., Caron de Fromentel C. Properties of the six isoforms of p63: p53-like regulation in response to genotoxic stress and cross talk with ΔNp73. Carcinogenesis. 2008;29:273–281. doi: 10.1093/carcin/bgm258. [DOI] [PubMed] [Google Scholar]
- 28.Watson I.R., Irwin M.S. Ubiquitin and ubiquitin-like modifications of the p53 family. Neoplasia. 2006;8:655–666. doi: 10.1593/neo.06439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staudinger J.L. The molecular interface between the SUMO and ubiquitin systems. Adv. Exp. Med. Biol. 2017;963:99–110. doi: 10.1007/978-3-319-50044-7_6. [DOI] [PubMed] [Google Scholar]
- 30.Ranieri M., Vivo M., De Simone M., Guerrini L., Pollice A., La Mantia G., Calabrò V. Sumoylation and ubiquitylation crosstalk in the control of ΔNp63α protein stability. Gene. 2018;645:34–40. doi: 10.1016/j.gene.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 31.Grabbe C., Dikic I. Functional roles of ubiquitin-like domain (ULD) and ubiquitin-binding domain (UBD) containing proteins. Chem. Rev. 2009;109:1481–1494. doi: 10.1021/cr800413p. [DOI] [PubMed] [Google Scholar]
- 32.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.May M.J., Larsen S.E., Shim J.H., Madge L.A., Ghosh S. A novel ubiquitin-like domain in IκB kinase β is required for functional activity of the kinase. J. Biol. Chem. 2004;279:45528–45539. doi: 10.1074/jbc.M408579200. [DOI] [PubMed] [Google Scholar]
- 34.Jo Y., Sguigna P.V., DeBose-Boyd R.A. Membrane-associated ubiquitin ligase complex containing gp78 mediates sterol-accelerated degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Biol. Chem. 2011;286:15022–15031. doi: 10.1074/jbc.M110.211326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reventun P., Alique M., Cuadrado I., Márquez S., Toro R., Zaragoza C., Saura M. iNOS-derived nitric oxide induces integrin-linked kinase endocytic lysosome-mediated degradation in the vascular endothelium. Arterioscler. Thromb. Vasc. Biol. 2017;37:1272–1281. doi: 10.1161/ATVBAHA.117.309560. [DOI] [PubMed] [Google Scholar]
- 36.Wang B., Jie Z., Joo D., Ordureau A., Liu P., Gan W., Guo J., Zhang J., North B.J., Dai X. TRAF2 and OTUD7B govern a ubiquitin-dependent switch that regulates mTORC2 signalling. Nature. 2017;545:365–369. doi: 10.1038/nature22344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.