Abstract
Background
Bone marrow stem cell clonal dysfunction by somatic mutation is suspected to affect post-infarction myocardial regeneration after coronary bypass surgery (CABG).
Methods
Transcriptome and variant expression analysis was studied in the phase 3 PERFECT trial post myocardial infarction CABG and CD133+ bone marrow derived hematopoetic stem cells showing difference in left ventricular ejection fraction (∆LVEF) myocardial regeneration Responders (n=14; ∆LVEF +16% day 180/0) and Non-responders (n=9; ∆LVEF -1.1% day 180/0). Subsequently, the findings have been validated in an independent patient cohort (n=14) as well as in two preclinical mouse models investigating SH2B3/LNK antisense or knockout deficient conditions.
Findings
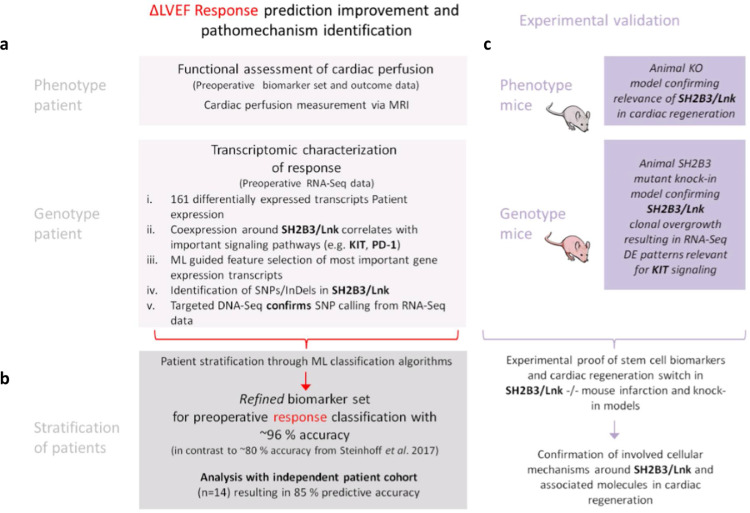
1. Clinical: R differed from NR in a total of 161 genes in differential expression (n=23, q<0•05) and 872 genes in coexpression analysis (n=23, q<0•05). Machine Learning clustering analysis revealed distinct RvsNR preoperative gene-expression signatures in peripheral blood acorrelated to SH2B3 (p<0.05). Mutation analysis revealed increased specific variants in RvsNR. (R: 48 genes; NR: 224 genes). 2. Preclinical:SH2B3/LNK-silenced hematopoietic stem cell (HSC) clones displayed significant overgrowth of myeloid and immune cells in bone marrow, peripheral blood, and tissue at day 160 after competitive bone-marrow transplantation into mice. SH2B3/LNK−/− mice demonstrated enhanced cardiac repair through augmenting the kinetics of bone marrow-derived endothelial progenitor cells, increased capillary density in ischemic myocardium, and reduced left ventricular fibrosis with preserved cardiac function. 3. Validation: Evaluation analysis in 14 additional patients revealed 85% RvsNR (12/14 patients) prediction accuracy for the identified biomarker signature.
Interpretation
Myocardial repair is affected by HSC gene response and somatic mutation. Machine Learning can be utilized to identify and predict pathological HSC response.
Funding
German Ministry of Research and Education (BMBF): Reference and Translation Center for Cardiac Stem Cell Therapy - FKZ0312138A and FKZ031L0106C, German Ministry of Research and Education (BMBF): Collaborative research center - DFG:SFB738 and Center of Excellence - DFG:EC-REBIRTH), European Social Fonds: ESF/IV-WM-B34-0011/08, ESF/IV-WM-B34-0030/10, and Miltenyi Biotec GmbH, Bergisch-Gladbach, Germany. Japanese Ministry of Health : Health and Labour Sciences Research Grant (H14-trans-001, H17-trans-002)
Trial registration
ClinicalTrials.gov NCT00950274
Keywords: Clonal hematopoiesis of indeterminate pathology, CHIP, SH2B3, Myocardial regeneration, Cardiac stem cell therapy, Angiogenesis induction, Post myocardial infarction heart failure, Coronary bypass surgery, CABG, Machine learning
Abbreviation: ANCOVA, Analysis of covariance; Ang-1, Angiopoeitin 1; AUR, Arch user repository; AUC, Area under curve; BM, Bone marrow; BMMNC, Bone marrow mononuclear cell; BMSC, Bone marrow stem cells; BrdU, Brome deoxyuridine; CABG, Coronary Artery Bypass Graft; CAP-EPC, Concentrated Ambient Particles – Endothelial Progenitor Cells; CD, Cluster of Differentiation; CEC, Circulating endothelial cells, CEC panel, CDs measured in PB; CFU, Colony-forming unit; CHIP, Clonal hematopoiesis of indeterminate potential; CI, Confidence interval; c-KIT/CD117, Stem Cell Factor Receptor c-KIT, CD117; CLARA, Clustering for Large Applications; CPC, cardiac progenitor cell; CSC, cardiac stem cell; DE, Differential gene expression; DNAseq, desoxyribonucleid acid sequencing; EGF, Epidermal growth factor; EGFR, Epidermal growth factor receptor; ELISA, Enzyme-Linked Immunosorbent Assay; EPC, Endothelial Progenitor Cells, EPC panel, CDs measured in PB; EPO, Erythropoietin; EPOR, Erythropoietin receptor; FACS, Fluorescence activated cell sorter; FDR, False discovery rate; GADPH, Glyceraldehyde 3 phosphate dehydrogenase; GATA4, Transcriptional activator that binds to the consensus sequence 5′-AGATAG-3’; GFP, Green fluorescent protein; GMP, Good Manufacturing Practice; GWAS, Genome wide association study; HR, Hazard ratio; HIF, Hypoxia-Inducible Factor, transcription factor; HSC, Hemopoeitic stem cell; hu, human; ICH GCP, Tripartite Guidelines Guideline for Good Clinical Practice; IGF-1, Insuline-like Growth Factor 1; IGFBP, Insuline-like growth factor binding proteine; IHG, Analysis performed in accordance with ISHAGE guidelines; IL, Interleukin; InDel, mutation insertion or deletion variant; KSL, mouse bone marrow stem cell subpopulation c-KIT+ Sca-I+ lin-; LAD, Left anterior descending coronary artery, RIVA; LAS, Longitudinal axis strain; LCRC, Loss of cardiac regeneration capacity; LNK, SH2B adapter protein 3 (lymphocyte adapter protein); LVEDV, Left Ventricular End Diastolic Volume; LVEF, Left Ventricular Ejection Fraction; LVESD, Left Ventricular End Systolic Dimension; m, mouse; MI, myocardial infarction; ML, Machine learning; MNC, Mononuclear cells; MRI, Magnetic resonance imaging; 6MWT, 6-Minute Walk Test; NGS, Next Generation Sequencing; NR, non-responder; PB, Peripheral blood; PBMNC, mononuclear cells isolated from peripheral blood; PCR, Polymerase chain reaction; PDGF, Platelet derived growth factor; PDFR, Platelet derived growth factor receptor; PEI, Paul-Ehrlich Institute; PI3K, Phosphoinositide-3-Kinase; PBMNC, Peripheral blood mononuclear cell; PPMC, Pearson Product Moment Correlation; qPCR, Quantitative polymerase chain reaction; R, responder; RFI, Reactome functional interaction; RNASeq, Ribonucleid acid sequencing; ROC, Receiver operating characteristics; RT-PCR, Reverse transcriptase polymerase chain reaction; RWMS, Regional wall motion score; SDF-1, Stromal Cell-derived Factor 1; SH2B3, LNK [Src homology 2-B3 (SH2B3)] belongs to a family of SH2-containing proteins with important adaptor functions; SCF, Stem Cell Factor; SNP, Single nucleotide polymorphism, variant; STEMI, ST- segment Elevation Infarction; SUSAR, Suspected Unexpected Serious Adverse Reaction; TiCoNE, Time course network enrichment; TNF, Tumor Necrosis Factor; t-SNE, t-distributed stochastic neighbour embedding; VCA, Virus-Capsid-Antigen; VEGF, Vascular Endothelial Growth Factor; VEGFR, Vascular Endothelial Growth Factor Receptor; WT, wild type; WGCNA, Weighted gene coexpression network analysis
Graphical abstract
Research in context.
Evidence before this study
The basis for this current work is the randomized double-blinded placebo controlled multicenter Phase 3 PERFECT-trial in which post myocardial infarction (MI) patients after coronary artery bypass graft (CABG) surgery have been treated with intramyocardial CD133+ bone marrow derived hematopoetic stem cells (BM-HSC) or Placebo. At the time we identified the correlation of myocardial regeneration with systemic bone marrow response characterized by a preoperative biomarker signature in peripheral blood (PB) of 20 angiogenesis and stem cell related factors [17]. An additional outcome prediction obtained by Machine Learning (ML) received an accuracy rate of 85% for responder (R) and 80% for non-responder (NR). Here, genetic dysregulation of BM-HSC was suspected and now followed up by gene expression and mutational dysregulation analysis. LNK is an adaptor protein coded by the gene SH2B3 and negatively regulates multiple essential signals in hematopoietic stem cells (HSC). Its regulatory role for BM-HSC in cardiovascular repair remains shallow and will be investigated throughout this underlying manuscript.
Added value of this study
In the present series of experiments, we clarified that HSC signaling adaptor gene mutations in SH2B3 contribute to a polygenic gene expression circuit switch including the genes PLCG1, LPCAT2, GRB2, AFAP1, AP1B1, KLF8, MARK3 favorable for the cardiac healing process in MI-patients undergoing cardiac recovery after CABG surgery. An integrative ML analysis of preoperative PB enables highly sensitive clinical diagnosis and prediction of cardiac regeneration response after CABG. It may be used for treatment monitoring for cardiac regeneration and give rise to a patient specific ML supported therapy in the future. Our findings in PERFECT about RvsNR and in SH2B3/LNK−/− mice suggest that the significantly reduced ischemic myocardial damage with preserved cardiac function following MI is mainly due to enhanced angiogenesis in ischemic myocardium.
Implications of all the available evidence
This novel approach of disease genotype/phenotype analysis combining gene expression, coexpression, and transcript variant calling in a randomized clinical trial led to the discovery of a polygenic circuit involved in HSC response associated to cardiac regeneration capacity. In the following, the findings were verified by animal studies and assisted by correlation analysis of human and mice. This comparison enabled new insights into adaptor proteins, proliferation signaling, and immune checkpoint regulation controlling for vasculogenesis/angiogenesis and cardiac tissue repair. Recovery of expedient cardiac function was observed through up-regulation of HSC/EPCs circulation and stimulation of immune progenitor cell (PC) proliferation. Our findings show that mutational changes in gene expression transcripts have important implications for formulations of new therapeutic strategies to diagnose and enhance cardiac repair by stem cells.
Alt-text: Unlabelled box
1. Introduction
The hematopoietic system has traditionally been considered as an organized, hierarchical system with multipotent, self-renewing stem cells at the top, lineage-committed progenitor cells in the middle, and lineage-restricted precursor cells, which give rise to terminally differentiated cells, at the bottom [[1], [2], [3]]. However, disorders of clonal hematopoiesis of indeterminate pathology (CHIP) has been described in hematological and cardiovascular disease patients and associated to congenital or somatic DNA mutations [4,5]. The question arises, which mutations in stem congenital or somatic cell regulatory genes cause hematopoietic clonal advantage and impact cardiovascular pathology [6,7]. SH2B3, which codes for the LNK adaptor protein, is one of the major mutated genes associated with hematopoietic stem cell (HSC) proliferation disorders, such as myelodysplasia, erythrocytosis or leukemia [8,9]. In genome wide association studies (GWAS) of cardiovascular patients, the SH2B3 phosphorylation related missense variant rs3184504 was found to be associated with increased platelet count, monocyte proliferation, hypertension, peripheral/coronary artery disease, autoimmune disease, and longevity [9], [10], [11], [12], [13], [14], [15]. SH2B3/LNK expression regulation is largely unknown, but expected to impact cardiovascular regeneration through c-KIT/CD117 expressing hematopoietic, myeloid, lymphocytic, endothelial, and mesenchymal progenitor cells in blood [9,10]. In contrast to this, intracardiac SH2B3/LNK expression was found to be associated with pressure overload cardiac hypertrophy regulation [16]. At present, the regulatory role of SH2B3 in stem cell proliferation and inflammation response remains unclear in patients with coronary artery disease, especially in post-myocardial infarction repair leading either to regeneration or inflammatory fibrosis of the myocardium [9,13]. Furthermore, it is unclear, if a monogenic switch of SH2B3 gene expression or SNP altered LNK protein function in bone marrow stem cells is able to control cardiac regeneration by altering bone marrow response [9]. Moreover, frequency and type of SH2B3 clonal mutations of HSC of patients with cardiac disease is unknown and may have impact on variable pathology. In the recent outcome analysis of the phase 3 clinical PERFECT trial we are investigating intramyocardial transplantation of c-KIT/CD117+/CD133+,/CD34+ bone marrow derived hematopoeitic stem cells (BM-HSC) in post-myocardial infarction (MI) coronary artery bypass graft (CABG) patients. We found striking differences in induction of cardiac regeneration in 60% of BM-HSC treated and placebo groups characterized by a preoperative Machine Learning (ML) signature in peripheral blood (PB) [17]. Responders (R) vs. non-responders (NR) were significantly different preoperatively, with R characterized by increased peripheral blood c-KIT/CD117+/CD133+/CD34+ circulating stem cells (EPC), increased thrombocytes, while NR had increased Erythropoeitin (EPO), Vascular endothelial growth factor (VEGF) and N-terminal pro b-type natriuretic peptide (NTproBNP) in preoperative serum [17]. Induced bone marrow stem cell proliferation responses in R was suspected to be due to adaptor protein SH2B3/LNK activity [17]. Based on this, we first performed variant and gene expression analyses in PERFECT responders vs. non-responders and compared diagnostic RvsNR signatures (Fig. 1A). Then we validated the effect on R/NR signature switch in SH2B3/LNK deficient mouse models to investigate the role of HSC dysfunction in cardiac repair . Final evaluation of the signatures was performed in an independent patient cohort and by mouse/man correlation analysis (Fig. 1B,C).
Fig. 1.
Overview of utilized integrative analysis approach integrating clinical patient data with murine pre-clinical models: Genotype/phenotype analysis in randomised clinical trial PERFECT cardiac regeneration outcome and knock-out animal disease model verification of regulatory genes.
2. Methods
2.1. Study Design
Peripheral blood bone marrow response was studied by whole transcriptome analysis in the randomized phase 3 PERFECT trial biomarker subgroup (total n=39, Rostock n=23, Hannover n=14, Leipzig n=2) [17]. Primary analysis was performed at the Rostock trial site with available biobank, clinical (per protocol), and biomarker data (n=23). CD133+ BMSC treated (n=13) and placebo controls (n=10) were equally distributed. In the biomarker patient cohort (n=23; Placebo/CD133+9/14), we investigated systemic bone marrow stem cell response in peripheral blood in Responders (R) classified by the difference in left ventricular ejection fraction (∆LVEF) ≥5 % after 180d (n=14; Placebo/CD133+ 7/7) and Non-responders (NR) classified by ∆LVEF <5 % after 180d (n=9; Placebo/CD133+ 5/4) in both treatment groups (intramyocardial Placebo vs. CD133+). For validation variant expression analysis, MRI, and clinical outcome was tested in 14 patients from the Hannover trial center with available clinical data.
2.2. Ethical approval and trial setting
RNA sequencing (RNA-Seq) analysis and mRNA RT-PCR in PB: Samples were taken from informed study patients who gave their written consent according to the Declaration of Helsinki (Approval by the Ethical committee, Rostock University Medical Center 2009; No. HV-2009-0012). Analyses and examinations were performed before unblinding of the trial and under careful adherence to the protection of data privacy (pseudonyms).
2.3. Transcriptome Analysis of EDTA blood samples using NGS
RNA of frozen EDTA blood samples was isolated in a three step procedure: First, the GeneJET Stabilized and Fresh Whole Blood RNA KIT (Thermo Scientific) was used following manufacturer's instructions. Second, isolated RNA was precipitated with 2.5 volumes ethanol under high salt conditions (10 % of 3 M sodium acetate, pH 5•2). After DNase digest (Thermo Scientific), the RNA was finally purified using Agencourt RNAClean XP beads (Beckman Coulter). Purified RNA was analyzed on a Bionalyzer (Agilent) using RNA 6000 Nano Chips (Agilent). Quality controlled RNA was used to construct sequencing libraries using the Universal Plus mRNA–Seq Technology (Nugen) according to manufacturer's instructions. Briefly, mRNA was selected by oligo d(T) beads, reverse transcribed and cDNA from Globin messengers was removed by the Globin depletion module (Nugen). Quality controlled and quantified libraries were sequenced on a HiSeq1500 system (Illumina) in single-end mode (100 nt read length). For RNA-Seq data analysis, adapter clipping and quality trimming procedures for data pre-processing were performed and aligned the reads to the hg19 genome (for patient data) and mm10 genome (for murine data) with the aid of kallisto, respectively. Differential expression analysis was performed using the likelihood ratio test of the DESeq2 package (genes with >2-fold change and a q-value < 0•05 are considered as significantly differentially expressed). The gene set enrichment analysis (GSEA), annotation, including functional annotation clustering and functional classification, was performed according to Enrichr [18].
2.4. Variant calling from transcriptomic data
The previously preprocessed human RNA-Seq datasets were realigned to the hg19 reference (Ensembl version 94) with Star (2-pass mode). The variant calling was applied by the Gatk toolkit [19] with specialized filters (e.g. variants are only considered, if they are confirmed with five independent reads - a comprehensive workflow is shown in Supplement Figure S1.
2.5. Experimental CRISPR-Cas9 induced SH2B3/LNK antisense silencing mouse model
2.5.1. Lentiviral vector production
The lentiviral vectors pRRL.U6.Lnk-sgRNA.EFS.dTomato.pre or pRRL.U6.NT-sgRNA.EFS.eBFP2.pre were packaged into viral particles by transfection of 10 µg vector, 12 µg pcDNA3.GP.4xCTE, 6 µg pRSV-Rev and 2 µg pMD2.G into HEK-293T cells in 10 cm plates using the calcium-phosphate method. Medium change was performed 6-8 h later and viral supernatants were harvested 30 h and 54 h post-transfection. The lentiviral supernatants were pooled and concentrated by ultracentrifugation. Vector titers were determined on lineage-negative mouse bone marrow cells.
2.5.2. Competitive bone marrow transplantation
Lineage-negative mouse bone marrow cells were isolated by flushing femurs, tibias and pelves of GFP+Cas9 mice (B6J.129(Cg)-Gt(ROSA)26Sortm1.1(CAG-cas9*,-EGFP)Fezh/J) (IMSR Cat# JAX:026179, RRID:IMSR_JAX:026179) followed by lineage depletion using the MojoSort Mouse Hematopoietic Progenitor Cell Isolation Kit (BioLegend). Cells were prestimulated for 24 hin StemSpan (Stem Cell Technologies) supplemented with 100 U/mL penicillin and 100 µg/mL streptomycin (PAA), 2 mM L-glutamine (Biochrom), 20 ng/mL mTPO, 20 ng/mL mIGF-2, 10 ng/mL mSCF, 10 ng/mL hFGF-1 (all cytokine: Peprotech), 20 µg/mL meropenem (Hexal) and 10 µg/mL heparin (Ratiopharm). Cells were transduced at a density of 1•5 × 106 cells/mL at an MOI of 30. For competitive transplantation, equal cell numbers of cells transduced with lentiviral vector pRRL.U6.Lnk-sgRNA.EFS.dTomato.pre or pRRL.U6.NT-sgRNA.EFS.eBFP2.pre were mixed and about 5 × 105 cells per mouse were transplanted into irradiated (2 × 4•5 Gy) GFP− CD45.2 B6 (C57BL/6) recipients. Cell mixtures were analyzed by flow cytometry 4-5 d after transduction to confirm equal distribution of both cell fractions. At week 4, 8, 12, and 18 after transplantation, blood counts were performed and peripheral blood was analyzed for donor cell engraftment and lineage distribution by flow cytometry. All experimental procedures were conducted in accordance with the German Animal Protection Law Guidelines for the Care and Use of Laboratory Animals and the study protocol was approved by the Ethics Committee of the LAVES (Lower Saxony State Department for Consumer and Food Safety Protection), Germany.
2.5.3. Experimental SH2B3/LNK knockout model
The SH2B3/LNK–/– mouse strain was generated as described previously [10]. C57BL/6 mice (CLEA Japan, Tokyo, Japan) were used as WT control mice. GFP transgenic mice (GFP-Tg mice; C57BL/ 6TgN [act EGFP] Osb Y01) were mated with WT mice or SH2B3/LNK–/– mice and generated WT/GFP mice or SH2B3/LNK–/–/GFP mice, respectively, for BM transplantation (BMT) studies. All experimental procedures were conducted in accordance with the Japanese Physiological Society Guidelines for the Care and Use of Laboratory Animals and the study protocol was approved by the Ethics Committee in RIKEN Center for Developmental Biology.
2.5.4. Statistical analysis
The results were statistically analyzed using a software package (Statview 5.0, Abacus Concepts Inc, Berkeley, CA). All values were expressed as mean±standard deviation (mean±SD). The comparisons among more than three groups were made using the one-way analysis of variances (ANOVA) in Prism 4 (GraphPad Software, San Diego, CA). Post hoc analysis was performed by Tukey's multiple comparison test, Mann-Whitney comparison test or Bonferroni post-hoc test. Differences of p<0•05 were considered to denote statistical significance.
2.5.5. Data analysis with machine learning
Identifying key features and classification of the comprehensive patient data was obtained by employing supervised and unsupervised Machine Learning (ML) algorithms . We preprocessed the data, while removing features with low variance and high correlation for dimension reduction, following best practices recommendations. We compared the following supervised algorithms: AdaBoost (AB), Gradient Boosting (GB), Support Vector Machines (SVM), and Random Forest (RF) [20]. We employed classifiers that are suitable for training on small data sets for a comparison of features given little training and chose the most appropriate algorithm according to accuracy and robustness towards overfitting [21]. Supervised ML models were 10-fold cross-validated and 100 times repeated. We then applied feature selection for the AB, GB, and RF classifiers to further reduce the number of features to <20. We employed principial component analysis (PCA), t-distributed Stochastic Neighbor Embedding (tSNE), and Uniform Manifold Approximation and Projection (UMAP, https://arxiv.org/abs/1802.03426) for unsupervised machine learning classification and nonlinear dimensionality reduction.
2.5.6. WGCN analysis
Weighted gene coexpression network analysis (WGCNA) was performed by applying the R package “WGCNA” to the human RNA-Seq count data. We first constructed the topological overlap matrix (TOM) of all investigated transcripts (~160,000) using the soft thresholding method. We calculated the eigenvalues of the transcripts and evaluated the adjacency based on distance. We subjected transcripts to hierarchical clustering (average linkage) and assigned transcripts with the dynamic hybrid method into groups. We computed the connectivity based on the interaction partners (k) and evaluated the gene significance, which represents the resulting module membership.
3. Results
In our analysis we integrated clinical genotype and phenotype data as well as experimental gene knockout animal modeling in which we aimed to unravel and validate diagnostic associations of blood, bone marrow, and heart tissue (Fig. 1). At the phenotypic level, left ventricular function measured in magnetic resonance imaging (MRI) showed recovery with a mean difference in primary endpoint outcome ∆LVEF (d.180/0) in Responders (R) +16% vs. Non-responders (NR) -1•1% (p<0•01, t-test; Mann-Whitney Rank Sum test) (Table 1). Significant difference was found in R for myocardial capillary perfusion measured in MRI with increased epicardial (p=0•038, t-test; Mann-Whitney Rank Sum test) and endocardial (p=0•024, t-test; Mann-Whitney Rank Sum test) maximal upslope velocity after 180 days (Table 1).
Table 1.
Left ventricular function and myocardial perfusion outcome analysis. MRI evaluation biomarker .subgroup (n=23) for primary endpoint (delta LVEF 180/0), myocardial function by long-axis-strain analysis, and myocardial perfusion by semiquantitative analysis (mean value of 16 segments). Responders (n=14) were classified according to primary endpoint outcome by delta LVEF >5% d. 180/0, non-responders (n=9) by delta LVEF <5% d.180/0. Long-axis-strain measurement was performed according to Giesdal O et al [22], myocardial perfusion was measured according to Mordini FE et al [23].
| Baseline (day 0) | SD | Primary endpoint (day 180) | SD | Delta (180/0) | P-value (t-test; Mann-Whitney Rank Sum test) | |
|---|---|---|---|---|---|---|
| LVEF (%) | ||||||
| Responder (n evaluable=14) | 33,3 | 5,0 | 49,3 | 6,7 | 16,0 | P≤0.001 |
| Non-Responder (n evaluable = 9) | 33,3 | 7,5 | 32,2 | 9,1 | -1,1 | P=0.781 |
| Responder – Non-responder | 0 | 17,1 | 17,1 | P≤0.001 | ||
| Long axis strain (LAS global) | ||||||
| Responder (n evaluable=14) | -7,6 | 2,2 | -9,4 | 2,2 | -1,8 | P=0.032 |
| Non-Responder (n evaluable = 9) | -8,4 | 2,7 | -9,5 | 2,7 | -1,1 | P=0.402 |
| Responder – Non-responder | +0,8 | +0,1 | -0,7 | P=0.416 | ||
| Maximal upslope epicardial | ||||||
| Responder (n evaluable=13) | 27,0 | 10,4 | 37,4 | 17,3 | 10,4 | P=0.018 |
| Non-Responder (n evaluable = 9) | 29,6 | 11,5 | 28,7 | 9,1 | -0,9 | P=0.895 |
| Responder – Non-responder | -2,6 | +8,7 | 11,3 | P=0.038 | ||
| Maximal upslope endocardial | ||||||
| Responder (n evaluable=13) | 29,7 | 12,6 | 42,4 | 19,5 | 12,7 | P=0.014 |
| Non-Responder (n evaluable = 9) | 33,6 | 11,5 | 33,8 | 9,7 | 0,2 | P=0.967 |
| Responder – Non-responder | -3,9 | 8,6 | 12,5 | P=0.024 | ||
3.1. A: Clinical phenotype and genotype of cardiac regeneration response
3.1.1. Gene expression analysis
In addition to previously identified correlating angiogenesis biomarkers and SH2B3/LNK RT-PCR analysis of PB [17], we performed an in-depth gene expression analysis. In order to study transcriptome profile patterns of R and NR signatures, the capture of polyadenylated RNA was conducted by high throughput sequencing. The experimental procedure included a depletion of cDNA derived from Globin messengers transcriptome to enable high resolution RNA-Seq in preoperative PB samples from 23 patients (14 R, 9 NR).
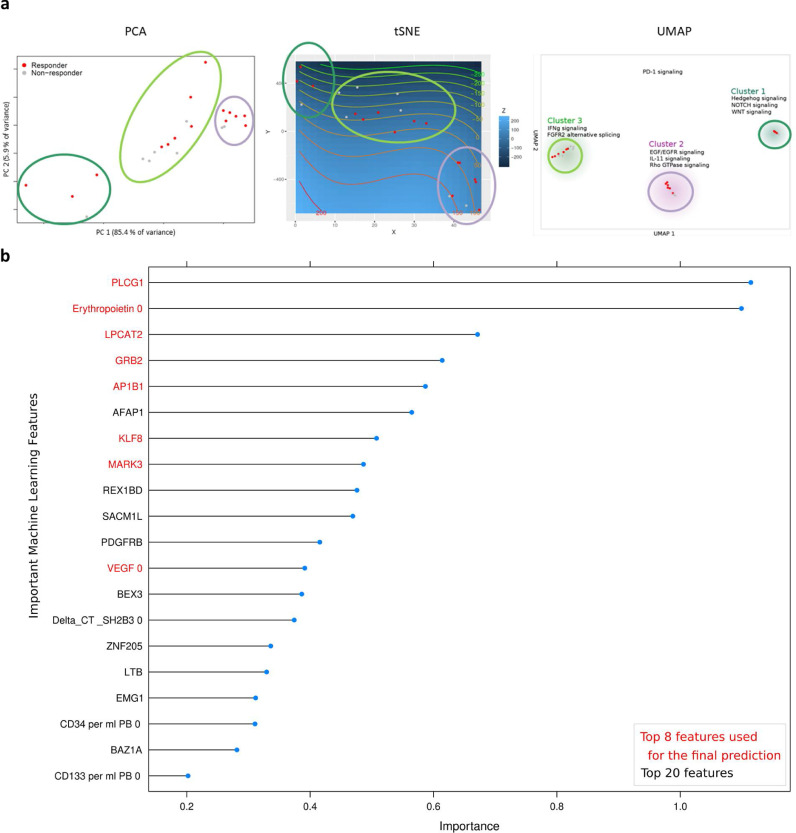
Differential gene expression analysis revealed distinct R/NR patterns consisting of 161 significant genes (q<0•05) out of ~160,000 transcripts. The highest significance was found for 122 unique genes (R/NR: q=0•02) (Supplementary Data SD1a). Clustering for all used methods examined potentially occuring patient subgroups. Three independent clustering analyses (PCA, tSNE, and UMAP) on all gene expression read counts showed a clear distinction between patients (Fig. 2a, Supplementary Figure 1c). All methods clustered the patients into the same defined subgroups, which did not change. Pathway analysis of differing genes was subsequently conducted on each of the three clusters to investigate the specific differences towards the gene signaling among these subgroups (Fig. 2a, Supplementary Data SD1b). Then, we performed the coexpression analysis by WGCNA, an so-called guilt-by-association approach, to be able to interconnect SH2B3/LNK with similarly regulated transcripts. SH2B3/LNK was identified to be coexpressed within a cluster of 872 genes (Supplementary Data SD1c). The corresponding pathways of the coexpressed genes were c-KIT receptor signaling pathway, as well as EGF, PDGF, TCR, IL6, and Interferon 1 signaling (Table 2).
Fig. 2.
a: ML subgroup clusters of cohort study (Responder, n=14, red points; Non-responder, n=9, grey points). b: Machine learning feature selection on clinical trial research data and RNA-Seq data. Accuracy comparison for the supervised prediction of the patient responsiveness using only preoperative data. Results are obtained after feature selection and subsequent prediction with two independent classifiers. The graph shows the true positive prediction weights of the ML model (RF for feature selection and SVM for final prediction). Combinations and subsets of these features have been subsequently used to train the final model. The importance indicates a hierarchy of the most relevant features needed for a classification.
Table 2.
Gene set enrichment pathway analysis utilized by Enrichr for differential gene expression, coexpression, and transcriptomic variants is based on preoperative RNA-Seq data . The 161 significantly differentially expressed transcripts identified by DESeq2 and 872 WGCNA transcripts have been applied to the pathway enrichment analysis of Enrichr for the WikiPathways and BioCarta database. The obtained pathways are significantly enriched according to the adjusted p-value < 0.05
| Type of data analysis | Database | Pathway Term | p-value | Adjusted p-value |
|---|---|---|---|---|
| Differential expression | BioCarta | Ras Signaling Pathway_Homo sapiens_h_rasPathway | 0,0004127 | 0,0235251 |
| AKT Signaling Pathway_Homo sapiens_h_aktPathway | 0,0076997 | 0,1097214 | ||
| Cyclin E Destruction Pathway_Homo sapiens_h_fbw7Pathway | 0,0018925 | 0,0447290 | ||
| E2F1 Destruction Pathway_Homo sapiens_h_skp2e2fPathway | 0,0023542 | 0,0447290 | ||
| Control of Gene Expression by Vitamin D Receptor_Homo sapiens_h_vdrPathway | 0,0169139 | 0,1722122 | ||
| Beta-arrestins in GPCR Desensitization_Homo sapiens_h_bArrestinPathway | 0,0181276 | 0,1722122 | ||
| WikiPathways | Hematopoietic Stem Cell Differentiation_Homo sapiens_WP2849 | 0,0050742 | 0,2572584 | |
| Translation Factors_Homo sapiens_WP107 | 0,0063783 | 0,2572584 | ||
| AMPK Signaling_Homo sapiens_WP1403 | 0,0145744 | 0,4408766 | ||
| RalA downstream regulated genes_Homo sapiens_WP2290 | 0,0034194 | 0,2572584 | ||
| EGFR1 Signaling Pathway_Mus musculus_WP572 | 0,0384020 | 0,4723207 | ||
| IL-6 signaling Pathway_Mus musculus_WP387 | 0,0353539 | 0,4723207 | ||
| Androgen receptor signaling pathway_Homo sapiens_WP138 | 0,0284065 | 0,4723207 | ||
| Striated Muscle Contraction_Mus musculus_WP216 | 0,0369516 | 0,4723207 | ||
| Striated Muscle Contraction_Homo sapiens_WP383 | 0,0321361 | 0,4723207 | ||
| Coexpression | BioCarta | EGF Signaling Pathway_Homo sapiens_h_egfPathway | 0,0001867 | 0,0148901 |
| PDGF Signaling Pathway_Homo sapiens_h_pdgfPathway | 0,0004153 | 0,0148901 | ||
| Control of Gene Expression by Vitamin D Receptor_Homo sapiens_h_vdrPathway | 0,0004653 | 0,0148901 | ||
| IL 6 signaling pathway_Homo sapiens_h_il6Pathway | 0,0023696 | 0,0440702 | ||
| Cell to Cell Adhesion Signaling_Homo sapiens_h_cell2cellPathway | 0,0020125 | 0,0440702 | ||
| Eukaryotic protein translation_Homo sapiens_h_eifPathway | 0,0027544 | 0,0440702 | ||
| T Cell Receptor Signaling Pathway_Homo sapiens_h_tcrPathway | 0,0037212 | 0,0486744 | ||
| Map Kinase Inactivation of SMRT Corepressor_Homo sapiens_h_egfr_smrtePathway | 0,0040712 | 0,0486744 | ||
| Internal Ribosome entry pathway_Homo sapiens_h_iresPathway | 0,0045632 | 0,0486744 | ||
| TPO Signaling Pathway_Homo sapiens_h_TPOPathway | 0,0080521 | 0,0515331 | ||
| Inhibition of Cellular Proliferation by Gleevec_Homo sapiens_h_gleevecpathway | 0,0067889 | 0,0515331 | ||
| Erk1/Erk2 Mapk Signaling pathway_Homo sapiens_h_erkPathway | 0,0067889 | 0,0515331 | ||
| Sprouty regulation of tyrosine kinase signals_Homo sapiens_h_spryPathway | 0,0056252 | 0,0515331 | ||
| How Progesterone Initiates the Oocyte Maturation_Homo sapiens_h_mPRPathway | 0,0074082 | 0,0515331 | ||
| mTOR Signaling Pathway_Homo sapiens_h_mTORPathway | 0,0080521 | 0,0515331 | ||
| WikiPathways | Kit Receptor Signaling Pathway_Mus musculus_WP407 | 0,0000385 | 0,0056566 | |
| mRNA processing_Mus musculus_WP310 | 0,0000874 | 0,0064241 | ||
| Interferon type I signaling pathways_Homo sapiens_WP585 | 0,0002457 | 0,0101753 | ||
| EGF/EGFR Signaling Pathway_Homo sapiens_WP437 | 0,0003291 | 0,0101753 | ||
| EPO Receptor Signaling_Homo sapiens_WP581 | 0,0004153 | 0,0101753 | ||
| EPO Receptor Signaling_Mus musculus_WP1249 | 0,0004153 | 0,0101753 | ||
| mRNA Processing_Homo sapiens_WP411 | 0,0007745 | 0,0162654 | ||
| PDGF Pathway_Homo sapiens_WP2526 | 0,0013840 | 0,0254306 | ||
| IL-6 signaling pathway_Homo sapiens_WP364 | 0,0018385 | 0,0270260 | ||
| IL-7 Signaling Pathway_Mus musculus_WP297 | 0,0018385 | 0,0270260 | ||
| Translation Factors_Mus musculus_WP307 | 0,0022338 | 0,0298523 | ||
| IL-3 Signaling Pathway_Homo sapiens_WP286 | 0,0026782 | 0,0325360 | ||
| EGFR1 Signaling Pathway_Mus musculus_WP572 | 0,0028773 | 0,0325360 | ||
| SNP-Responder | BioCarta | Calcium Signaling by HBx of Hepatitis B virus_Homo sapiens_h_HBxPathway | 0,0025401 | 0,0379514 |
| T Cell Receptor Signaling Pathway_Homo sapiens_h_tcrPathway | 0,0027108 | 0,0379514 | ||
| IL 4 signaling pathway_Homo sapiens_h_il4Pathway | 0,0025401 | 0,0379514 | ||
| Repression of Pain Sensation by the Transcriptional Regulator DREAM_Homo sapiens_h_dreampathway | 0,0025401 | 0,0379514 | ||
| Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription in carcinoma cells_Homo sapiens_h_rarrxrPathway | 0,0025401 | 0,0379514 | ||
| WikiPathways | mRNA Processing_Homo sapiens_WP411 | 0,0000037 | 0,0005582 | |
| Diurnally Regulated Genes with Circadian Orthologs_Homo sapiens_WP410 | 0,0001005 | 0,0037941 | ||
| Diurnally Regulated Genes with Circadian Orthologs_Mus musculus_WP1268 | 0,0001005 | 0,0037941 | ||
| Exercise-induced Circadian Regulation_Mus musculus_WP544 | 0,0001005 | 0,0037941 | ||
| mRNA processing_Mus musculus_WP310 | 0,0001861 | 0,0056192 | ||
| IL-2 Signaling Pathway_Homo sapiens_WP49 | 0,0012438 | 0,0268307 | ||
| Cytoplasmic Ribosomal Proteins_Homo sapiens_WP477 | 0,0010767 | 0,0268307 | ||
| IL-4 Signaling Pathway_Homo sapiens_WP395 | 0,0025723 | 0,0409333 | ||
| RANKL/RANK Signaling Pathway_Homo sapiens_WP2018 | 0,0027108 | 0,0409333 | ||
| Apoptosis-related network due to altered Notch3 in ovarian cancer_Homo sapiens_WP2864 | 0,0024383 | 0,0409333 | ||
| SNP-Non-responder | BioCarta | Mechanism of Protein Import into the Nucleus_Homo sapiens_h_npcPathway | 0,0017807 | 0,1887530 |
| Thrombin signaling and protease-activated receptors_Homo sapiens_h_Par1Pathway | 0,0187440 | 0,2862846 | ||
| Role of MEF2D in T-cell Apoptosis_Homo sapiens_h_mef2dPathway | 0,0248418 | 0,2862846 | ||
| ADP-Ribosylation Factor_Homo sapiens_h_arapPathway | 0,0227045 | 0,2862846 | ||
| Spliceosomal Assembly_Homo sapiens_h_smPathway | 0,0114323 | 0,2862846 | ||
| Cycling of Ran in nucleocytoplasmic transport_Homo sapiens_h_ranPathway | 0,0144950 | 0,2862846 | ||
| Role of Ran in mitotic spindle regulation_Homo sapiens_h_ranMSpathway | 0,0215374 | 0,2862846 | ||
| Erythropoietin mediated neuroprotection through NF-kB_Homo sapiens_h_eponfkbPathway | 0,0297088 | 0,2862846 | ||
| WikiPathways | Proteasome Degradation_Homo sapiens_WP183 | 0,0000001 | 0,0000161 | |
| Allograft Rejection_Homo sapiens_WP2328 | 0,0000055 | 0,0007073 | ||
| Proteasome Degradation_Mus musculus_WP519 | 0,0000215 | 0,0018437 | ||
| G13 Signaling Pathway_Mus musculus_WP298 | 0,0007868 | 0,0505532 |
3.1.2. Stratification of patients by ML selected features and correlation analysis
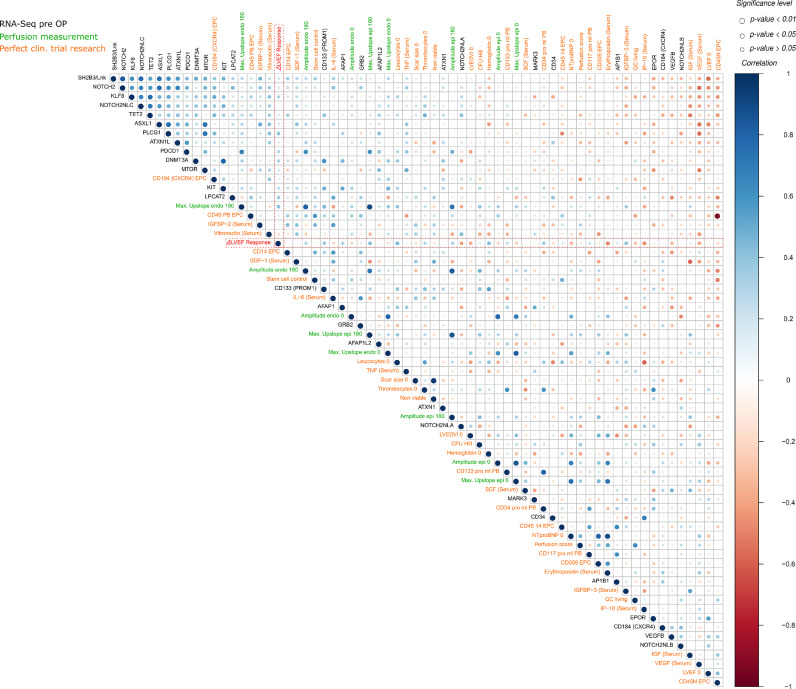
ML feature selection was applied as an independent method identifying the most important features among all gene expression and PERFECT trial outcome data. A first model achieved a prediction accuracy of 90% (ROC AUC 91•2%; CI: 89•4-93•0) when selecting the most important top 20 features as potential biomarkers (Fig. 2b). An integrational correlation analysis was applied to identify interrelations among the transcriptomic and phenotypic layer as well as between known angiogenesis and immune response markers. In particular, we correlated previously identified preoperative biomarkers for RvsNR found in the prior PERFECT trial analysis [17], ML top-selected genes (PLCG1, LPCAT2, GRB2, KLF8, AFAP1, MARK3, AP1B1), CHIP-related genes (TET2, ASXL-1, DNMT3A), previously identified adaptor protein LNK coding gene SH2B3, and related pathways (EPOR, KIT, KIT-L, PROM1/CD133, NOTCH2, PDCD1/PD-1, ATXN1L, MTOR genes) as well as myocardial perfusion parameters (Fig. 3). Top-listed correlations (p<0•05; Pearson correlation coefficient) were found for SH2B3 to the gene expression of NOTCH2, KLF8, NOTCH2NLC, TET2, ASXL1, PLCG1, and ATXN1L (Fig. 3). ML-top listed PLCG1, LPCAT2 were correlated to ∆LVEF response (p>0•05; Pearson correlation coefficient), PDCD1/PD-1 to ∆LVperfusion (p<0•05). Response was also correlated to increased PROM1/CD133 RNA, AFAP1 RNA, myocardial perfusion (∆ maximal upslope gradient epicardial after 180 days), preoperative leukocyte count, CD34 count, IGFBP3 serum protein, and hemoglobin (p>0•05; Pearson correlation coefficient; n=23). NR (negative correlation to ΔLVEF Response) correlated to preoperative LVEDV Index, VEGF-B, NOTCH2NLA gene expression, serum levels of NT proBNP, VEGF, Erythropoietin, and IP10 (p<0•05; Pearson correlation coefficient; n=23). Examplarily, an even higher complexity of differential gene transcript correlations to different genes were demonstrated for PROM1/CD133 and NOTCH2 on the isoform level(Supplemental Fig. S2).
Fig. 3.
Integration of RNA-Seq, perfusion, and clinical trial research data for Pearson correlation analysis. Comparison of peripheral blood (PB) circulating cells and biomarkers (orange), MRI myocardial perfusion parameters (green), and human PB gene expression data (RNA-Seq) (black). The ΔLVEF response (red) is highlighted for an improved visual analysis of important correlations. The color scale, ranging from 1 to -1 in the upper panel (blue to red), represents the correlation between the different factors. The size of the dots represents the significance (p<0,01, p<0,05, and p>0,05; Pearson correlation) of the respective correlation (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
3.1.3. Gene variant analysis
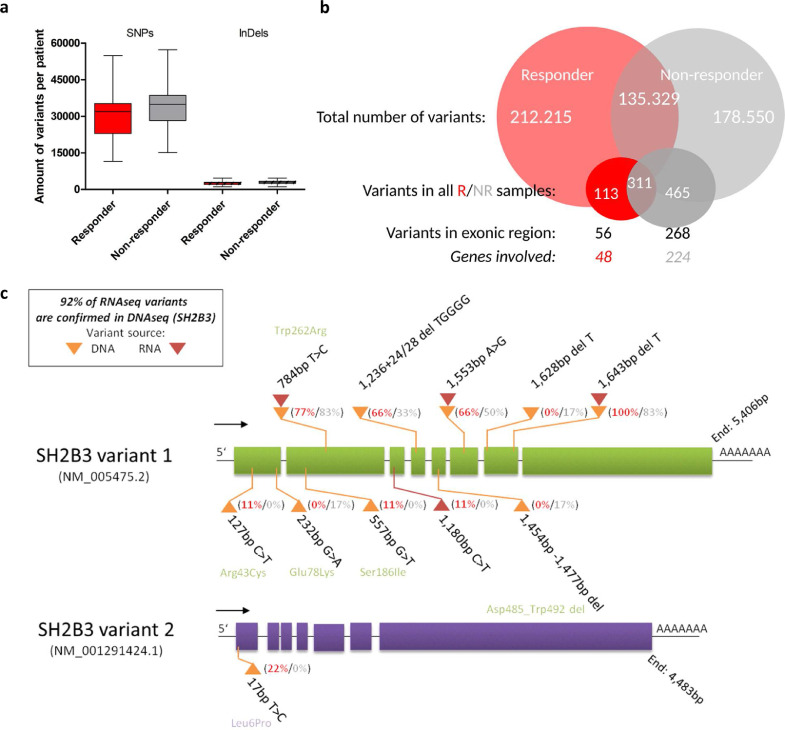
Transcriptomic mutation signature analysis performed on RNA-Seq data revealed increased specific variants in NR vs. R (NR: 465/178550 variants were contained in all NR, involved 224 genes with 268 exon regions; R: 113/212215 variants contained in all R, involved 48 genes with 56 exon regions (Supplemental data SD1d) (Fig. 4a, 4b). The DNA sequencing (DNA-Seq) confirmed more than 90% of the variants that were called in SH2B3 from RNA-Seq data (Fig. 4c, Supplemental Table 2). Total amount of variants, SNPs, and InDel mutations were not different in RvsNR (Fig. 4a, 4b). Main pathways possibly affected by variants were proteasome degradation (NR) and mRNA processing (R) (Table 3). Frequent mutations were present in all top-listed R/NR correlating genes including SH2B3/LNK, NOTCH2, PDCD1/PD-1, VEGF-B, PLCG1, GRB2, PROM1/CD133, mTOR, but also in the CHIP-related genes DNMT3, TET2, ASXL1 that were identified by RNA-Seq SNP calling (Supplementary Data SD1d). Moreover, variants in the reference gene GAPDH used for RT-PCR were observed with differences in ΔΔCT calculation as compared to Pol2a (Supplementary Data SD1e). Therefore, RT-PCR gene expression previously used for SH2B3 [17] was not used for the final outcome analysis. In addition, the Src-family adaptor protein coding gene SH2B3/LNK was found to be modified by SNP in DNA-Seq analysis of all patients (23/23) by deletions (100%) or nucleotide exchange (100%), with 83% of SNP resulting in amino acid substitution (Table 3). In RNA-Seq transcriptome analysis, the SNP variant rs3184504 p.Trp262Arg amino acid exchange in the pleckstrin binding domain was found in DNA-Seq (78%) and in RNA-Seq (83%) (Fig. 4c, supplementary Data SD1d, Table S2). Patients with rs3184504 SNPs in DNA-Seq (n=17/23) were distributed: R 12/14, NR 5/9. In order to validate the influence of single gene silencing on R/NR gene circuit and the resulting phenotype, we studied gene knockout effects of SH2B3 in mouse models.
Fig. 4.
Summary of genetic mutation signature analysis in PERFECT patients via sequencing analysis. a: Transcriptomic variants identified through RNA-Seq data analysis. Plot shows the average number of variants (SNPs and InDels) per patient that have been identified by applying our customized transcriptomic variant calling pipeline and filtering approaches. SNPs and InDels are considered as successfully called, if at least five independent reads support the individual variant. b: Venn diagram for the RvsNR variant comparison, exonic region association, and unique gene identification. c: Targeted DNA-Seq (yellow triangle) and RNA-Seq (red triangle) variant summary of SH2B3. The plot shows the ratio of SNP/del sites that are identified in Responders (red) and Non-responders (grey) as well as the possible amino acid transfer from its origin to its potential replacement (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Table 3.
Variant frequency of the SH2B3 Gene by DNA sequencing analysis. Variants, amino acid exchange, and frequency (homozygous > 50%, heterozygous > 25%). LVEF Responder (R) and non-responder (NR) are indicated. Sequencing analysis DNA was performed on peripheral blood by Centogene GmbH, Rostock. Validation was performed by mRNA sequencing performed by GeneCenter, LMU Munich.
| DNA seq SH2B3 | amino acid exchange | |||
|---|---|---|---|---|
| study ID | Results (SNP) | transcript | zygosity | |
| RTC106 | c.784T>C | p.Trp262Arg | NM_005475.2 | heterozygous |
| NR | c.1236+24_1236+28delTGGGG | NM_005475.2 | heterozygous | |
| c.*1553A>G | NM_005475.2 | heterozygous | ||
| c.*1643delT | NM_005475.2 | heterozygous | ||
| RTC133 | c.784T>C | p.Trp262Arg | NM_005475.2 | heterozygous |
| Resp | c.1236+24_1236+28delTGGGG | NM_005475.2 | heterozygous | |
| c.*1553A>G | NM_005475.2 | heterozygous | ||
| c.*1643delT | NM_005475.2 | heterozygous | ||
| RTC124 | c.127C>T | p.Arg43Cys | NM_005475.2 | heterozygous |
| Resp | c.784T>C | p.Trp262Arg | NM_005475.2 | heterozygous |
| c.1236+24_1236+28delTGGGG | NM_005475.2 | heterozygous | ||
| c.*1553A>G | NM_005475.2 | heterozygous | ||
| c.*1643delT | NM_005475.2 | heterozygous | ||
| RTC117 | c.784T>C | p.Trp262Arg | NM_005475.2 | heterozygous |
| Resp | c.1236+24_1236+28delTGGGG | NM_005475.2 | heterozygous | |
| c.*1553A>G | NM_005475.2 | heterozygous | ||
| c.*1643delT | NM_005475.2 | heterozygous | ||
| RTC146 | c.*1643delT | NM_005475.2 | heterozygous | |
| NR | ||||
| RTC137 | c.784T>C | p.Trp262Arg | NM_005475.2 | homozygous |
| NR | c.*1553A>G | NM_005475.2 | homozygous | |
| c.*1643delT | NM_005475.2 | heterozygous | ||
| RTC140 NR | c.784T>C | p.Trp262Arg | NM_005475.2 | homozygous |
| c.1236+24_1236+28delTGGGG | NM_005475.2 | heterozygous | ||
| c.*1553A>G | NM_005475.2 | heterozygous | ||
| c.*1643delT | NM_005475.2 | heterozygous | ||
| RTC099 NR | c.784T>C | p.Trp262Arg | NM_005475.2 | heterozygous |
| c.*1628delT | NM_005475.2 | heterozygous | ||
| RTC139 R | c.17T>C | p.Leu6Pro | NM_001291424.1 | heterozygous |
| c.784T>C | p.Trp262Arg | NM_005475.2 | heterozygous | |
| c.1236+24_1236+28delTGGGG | NM_005475.2 | heterozygous | ||
| c.*1553A>G | NM_005475.2 | heterozygous | ||
| c.*1643delT | NM_005475.2 | heterozygous | ||
| RTC145 R | c.*1643delT | NM_005475.5 | heterozygous | |
| RTC116 NR | c.1454_1477del | p.Asp485_Trp492del | NM_005475.2 | heterozygous |
| c.*1643delT | NM_005475.2 | heterozygous | ||
| RTC143 R | c.784T>C | p.Trp262Arg | NM_005475.2 | heterozygous |
| c.*1643delT | NM_005475.2 | heterozygous | ||
| RTC127 R | c.784T>C | p.Trp262Arg | NM_005475.2 | heterozygous |
| c.1236+24_1236+28delTGGGG | NM_005475.2 | heterozygous | ||
| c.*1553A>G | NM_005475.2 | heterozygous | ||
| c.*1643delT | NM_005475.2 | heterozygous | ||
| RTC115 R | c.*1643delT | NM_005475.2 | heterozygous | |
| RTC114 R | c.784T>C | p.Trp262Arg | NM_005475.2 | homozygous |
| c.1236+24_1236+28delTGGGG | NM_005475.2 | heterozygous | ||
| c.*1553A>G | NM_005475.2 | heterozygous | ||
| c.*1643delT | NM_005475.2 | heterozygous | ||
| RTC110 R | c.784T>C | p.Trp262Arg | NM_005475.2 | heterozygous |
| c.*1643delT | NM_005475.2 | heterozygous | ||
| RTC113 R | c.784T>C | p.Trp262Arg | NM_005475.2 | homozygous |
| c.1236+24_1236+28delTGGGG | NM_005475.2 | heterozygous | ||
| c.*1553A>G | NM_005475.2 | heterozygous | ||
| c.*1643delT | NM_005475.2 | heterozygous | ||
| RTC119 NR | c.17T>C | p.Leu6Pro | NM_001291424.1 | heterozygous |
| c.*1643delT | NM_005475.2 | heterozygous | ||
| RTC136 R | c.784T>C | p.Trp262Arg | NM_005475.2 | homozygous |
| c.*1643delT | NM_005475.2 | heterozygous | ||
| RTC134 NR | c.232G>A | p.Glu78Lys | NM_005475.2 | heterozygous |
| c.784T>C | p.Trp262Arg | NM_005475.2 | heterozygous | |
| c.*1643delT | NM_005475.2 | heterozygous | ||
| RTC130 R | c.557G>T | p.Ser186Ile | NM_005475.2 | heterozygous |
| c.784T>C | p.Trp262Arg | NM_005475.2 | homozygous | |
| c.1236+24_1236+28delTGGGG | NM_005475.2 | heterozygous | ||
| c.*1553A>G | NM_005475.2 | heterozygous | ||
| c.*1643delT | NM_005475.2 | heterozygous | ||
| RTC132 NR | c.*1643delT | NM_005475.2 | heterozygous | |
| RTC131 R | c.784T>C | p.Trp262Arg | NM_005475.2 | homozygous |
| c.*1643delT | NM_005475.2 | heterozygous | ||
3.2. B: Experimental phenotype and genotype validation of cardiac regeneration response switch
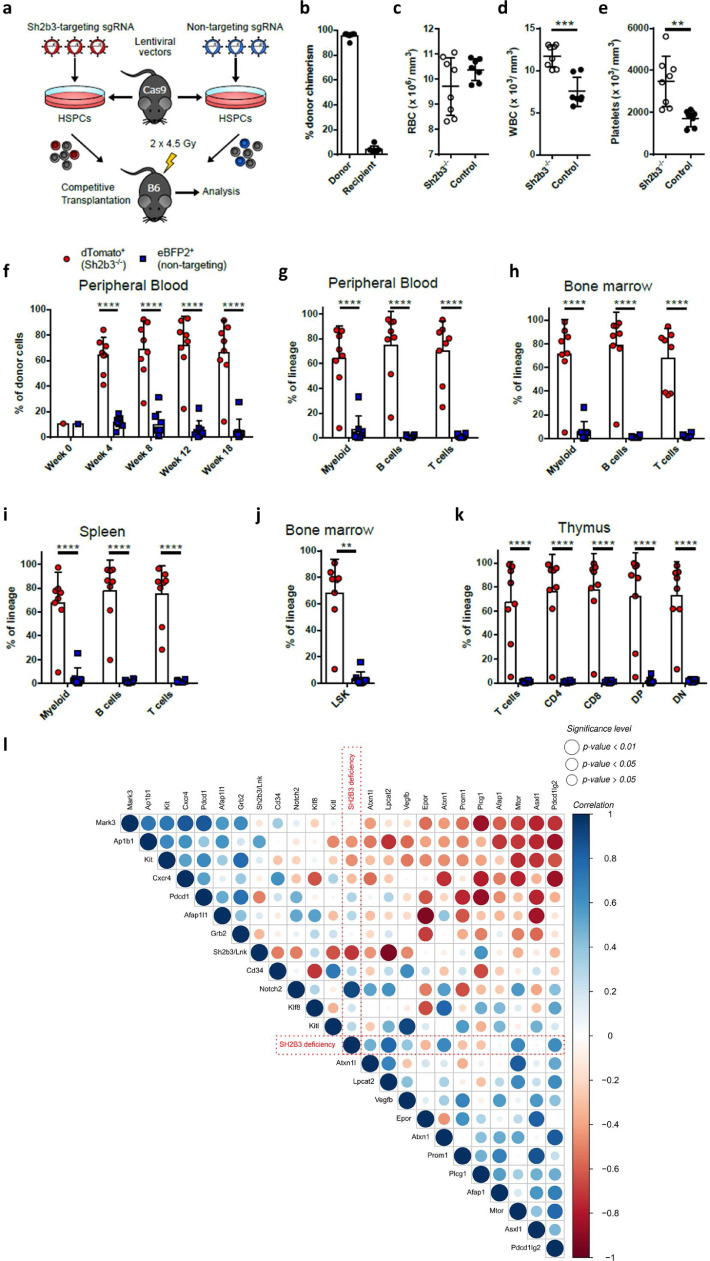
3.2.1. Clonal advantage of SH2B3/LNK knockout HSC in an experimental competitive transplantation assay
To show that SH2B3/LNK-deficient hematopoietic stem and progenitor cells (HSPCs) have an advantage to repopulate the bone marrow after partial bone marrow ablation, we performed a competitive transplantation assay. CRISPR-Cas9-mediated SH2B3−/- knockout cells (labeled with a dTomato fluorescent reporter) competed against eBFP2-marked SH2B3-intact competitor cells transduced with a non-targeting sgRNA (Fig. 5A). At week 18 post-transplantation, the bone marrow of transplanted mice consisted of 95•7% ± 2•4% donor cells, which indicated that despite partial bone marrow ablation, the remaining recipient cells were displaced by donor cells (Fig. 5B). Blood counts of transplanted mice revealed a normal red blood cell count compared to untreated control mice (Fig. 5C). However, we observed a significant increase of white blood cells and platelets in SH2B3−/− transplanted mice in comparison to control animals (Fig. 5D, E). Already four weeks after transplantation, the donor cells were dominant with 64•3% ± 13•8% by dTomato+ SH2B3−/- cells, which remained stable over time (Fig. 5F). Moreover, dTomato+ SH2B3−/− cells predominantly contributed to the hematopoiesis and outcompeted eBFP2+ competitor cells in myeloid, B-cell, and T-cell formation as quantified in the peripheral blood (Fig. 5G). Similar results were obtained for the different lineages (myeloidCD11b+, BB220+ and TCD3+ cells) in the bone marrow and spleen (Fig. 5H, I). Even 68•2% ± 25•5% of Lineage Sca1+ cKIT+ (LSK) HSPCs in the bone marrow were dTomato+ SH2B3−/− cells, which revealed a repopulating advantage also on the stem cell level (Fig. 5J). The selective advantage of SH2B3−/− was also present in the T cell compartment, including CD4+ and CD8+ single-positive, double-positive, and double-negative T cells in the thymus of transplanted mice (Fig. 5K). To compare gene expression profiles between WT and SH2B3−/− cells, we performed an in-depth gene expression analysis. In order to study transcriptome profile patterns of SH2B3−/- and WT peripheral blood signatures, the capture of polyadenylated RNA by high throughput sequencing was applied. To ensure a high comparability to the human samples, the cDNA derived from Globin messengers was also depleted. Correlation analysis was performed (Supplementary Figure 3; Supplementary Data SD1f) and showed a correlation (p<0•01) of SH2B3−/- with LPCAT2, NOTCH2, PDCD1LG2/PD-1, PROM1/CD133, ATXN1, and MTOR. SH2B3+/+ was negatively correlated to gene expression of LPCAT2, KITL, PDCD1/PD-1 and positively correlated to PLCG1 and AP1B1 (Fig. 5L). Affected pathways detected by GSEA include the Kit receptor, EGFR1, and IL-2 signaling pathways (p<0•05).
Fig. 5.
Influence of SH2B3 on HSC clonal overgrowth by using competitive bone marrow transplantation of Sh2b3−/− HSPCs. a: Scheme of the competitive transplantation assay is shown. HSPCs, which are derived from a SpCas9 transgenic mouse model (GFP+), were transduced with a lentiviral vector carrying a sgRNA against Sh2b3 and a dTomato fluorescent reporter. As competitor cells, HSPCs were transduced with a non-targeting sgRNA and an eBFP2 fluorescent reporter. After transduction, the Sh2b3−/− and Sh2b3-intact competitor cells were transplanted in a 1:1 mixture into irradiated C57BL/6 (B6, GFP−) recipient mice. Irradiation was performed using a fractionated dose of 2 × 4.5 Gy. b: Percentage of donor (GFP+) and recipient (GFP−) cells of total CD45+ cells in the bone marrow of mice at week 18 after transplantation. c: Red blood cell (RBC) count in Sh2b3−/− transplanted mice and untreated control animals at week 18 after transplantation. d: White blood cell (WBC) count in Sh2b3−/− transplanted mice and untreated control animals at week 18 after transplantation. e: Platelet count in Sh2b3−/− transplanted mice and untreated control animals at week 18 after transplantation. f: Presence of Sh2b3−/- (dTomato+) and competitor (eBFP2+) cells in the donor cell population in the peripheral blood at week 4, 8, 12, and 18 after transplantation. Week 0 shows the presence of dTomato+ and eBFP2+ cells in the transplanted cell population. g-k: Presence of Sh2b3−/- (dTomato+) and Sh2b3-intact competitor (eBFP2+) cells in the indicated lineage of donor cells in the peripheral blood g:, in the bone marrow h:, in the spleen i:, in Lineage− Sca1+ cKIT+ (LSK) HSPCs of the bone marrow j:, and in T cells of the thymus k: at week 18 after transplantation. l: Pearson correlation analysis of RNA-Seq data derived from murine Sh2b3 HSC clonal overgrowth model. The Sh2b3 deficiency (red) is highlighted for an improved visual analysis of important correlations. The color scale, ranging from 1 to -1 in the upper panel (blue to red), represents the correlation between the different factors. The size of the dots represents the significance (p<0.01, p<0.05, and p>0.05, Pearson correlation) of the respective correlation. Transplanted mice: n=8. Control mice: n=7. All graphs represent mean ± SD. Statistics: c-e: Unpaired t-test after normality test (D'Agostino & Pearson omnibus normality test) was passed; f-i:, k: Two-way ANOVA. j: Kolmogorov-Smirnov test. Significance level: ** p<0. 01, *** p<0.001, and **** p<0.0001 (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
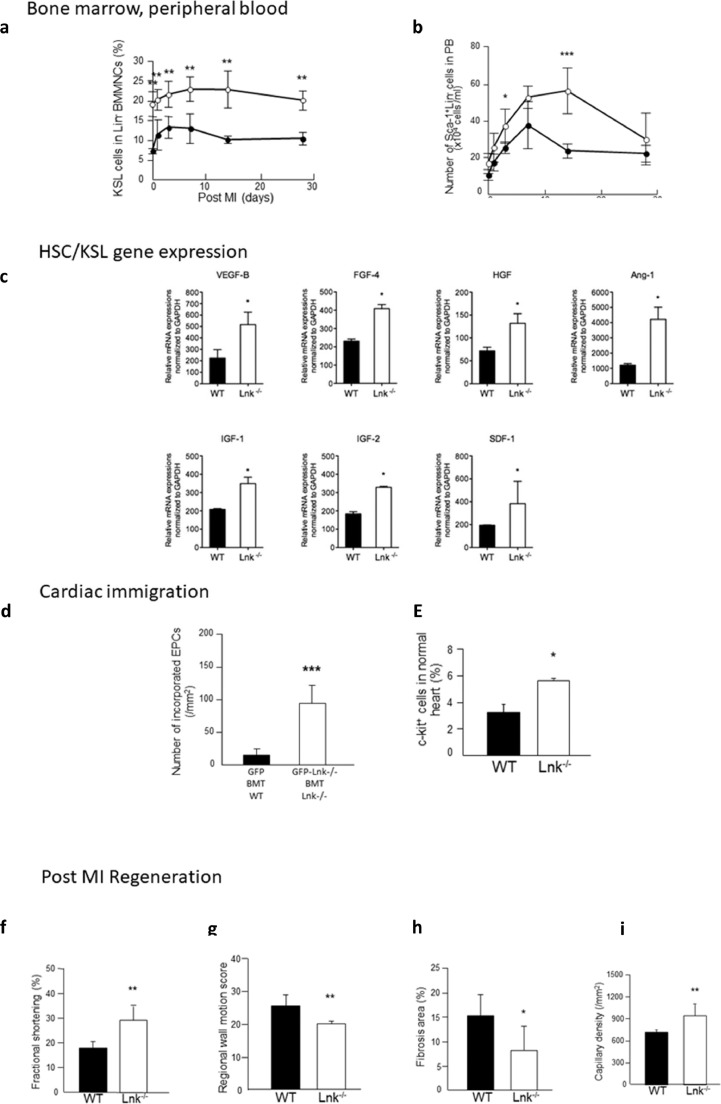
To elucidate the effect of SH2B3/LNK-deficiency on previously observed HSC/EPC kinetics in the setting of a mouse MI model, we used a second mouse model and examined the frequency of the kit+/sca+/lin− HSC (KSL) population.. Interestingly, the KSL population was significantly increased in SH2B3/LNK−/− mice compared to WT mice, i.e. on day 1, 3, 7, 14, and 28 (day 7: WT, 12•9 ± 3•7 vs. SH2B3/LNK−/−, 22•9±3•1 %, p<0•01; day 14: WT, 10•1±1•0 vs. SH2B3/LNK−/−, 22•9±4•8 %, p<0•01 two-way ANOVA followed by Tukey's multiple comparisons test) (Fig. 6A, Supplemental Data SD2). To examine the effect of SH2B3/LNK-deficiency on EPC mobilization into circulation post MI, we performed FACS analysis for Sca-1+/ lineage− cells, an EPC-enriched fraction in PB. The number of the cells pre-infarction was similar in SH2B3/LNK−/− and WT mice, whereas on days 3 and 14 post infarction, it was significantly greater in SH2B3/LNK−/− mice than WT mice (day 3: WT, 25•3±3.0 vs. SH2B3/LNK−/−, 37•0±9•4 × 104 cells/mL, p<0•05; Day 14: WT, 23•5±3•7 vs. SH2B3/LNK−/−, 56.0±12•4 × 104 cells/mL, p<0•001 two-way ANOVA followed by Tukey's multiple comparisons test) (Fig. 6B, Supplemental Data SD2). In isolated KSL cells, the mRNA expression of angiogenic factors (VEGF-B, FGF-4, HGF and Ang-1), survival factor (IGF-1), and stem/progenitor chemokines (IGF-2 and SDF-1) was significantly up-regulated in SH2B3/LNK−/− mice compared to WT mice (Fig. 6C; Supplemental Data SD2). Recruitment of BM-derived EPCs to infarcted myocardium was significantly increased in SH2B3/LNK−/- mice vs. WT mice (WT, 87•1±74•9 vs. SH2B3/LNK−/−, 647•1±174•7/mm2, p<0•001, Mann-Whitney comparison test) (Fig. 6D, Supplemental Data SD2). Moreover, incorporation of BM-derived EPCs into capillary vessels was significantly increased in SH2B3/LNK−/- EPCs than WT EPCs even at day 28 post MI (WT, 13•8±9•6 vs. SH2B3/LNK−/−, 92•0±27•0/mm2, p<0•001, Mann-Whitney comparison test). Frequency of c-KIT+ cells in whole heart analyzed by FACS was greater in SH2B3/LNK−/− than WT mice (WT, 3•2±0•7 vs. SH2B3/LNK−/−, 5•2±0•6 %, p<0•05, Mann-Whitney comparison test) (Fig. 6E, Supplemental Data SD2).
Fig. 6.
Experimental SH2B3 mouse MI-model
Bone marrow, peripheral blood kinetics of KSL cells in BM and SL cells in PB in WT vs. SH2B3/LNK−/- mice following MI. SH2B3/LNK−/− leads to increased EPC in bone marrow and circulation post MI a: Percent of KSL cells in Lin− BMMNCs before and after MI significantly increased in SH2B3/LNK−/− mice (open circles) compared with WT mice (closed circles). Two-way ANOVA followed by Tukey's multiple comparisons test **, p<0.01 vs. WT (n=3-4). b: Number of Sca-1+/Lin− (SL) cells in PB in SH2B3/LNK−/− mice (open circles) and WT mice (closed circles) before (Pre) and one day, 3, 7, 14, and 28 days after MI. Two-way ANOVA followed by Tukey's multiple comparisons test *, p<0.05 and ***, p<0.01 vs. WT (n=3-4).
c: HSC/KSL gene expression - Growth Factor and Chemokine mRNA expressions in WT BM-KSL cells vs. SH2B3/LNK−/− BM-KSL cells. KSL cells were sorted from freshly isolated BMMNCs by FACS, and were analysed the expressions of VEGF-B, FGF-4, HGF, Ang-1, IGF-1, IGF-2, and SDF-1 by quantitative real-time RT-PCR. Each relative mRNA expression was normalized to GAPDH and compared between WT BM-KSL cells (solid bar) and SH2B3/LNK−/− BM-KSL cells (open bar). Bonferroni post hoc test *, p<0.05. (n=3).
d: Effect of SH2B3/LNK gene deficiency on recruitment of BM-derived progenitors to ischemic myocardium. d: Double fluorescent immunostaining for GFP (green) and isolectin B4 (red) in heart sections in WT mice transplanted with GFP+ BM and in SH2B3/LNK−/− mice transplanted with GFP+- SH2B3/LNK−/− BM 7 days following MI. Number of recruited BM-derived cells into vasculature in ischemic myocardium 28 days following MI were counted and averaged. Mann-Whitney comparison test **, p<0.01 and ***, p<0.001 vs. WT mice transplanted with GFP BM. (n=3).
e: Assessment for proliferation activity in CSCs/CPCs and cardiomyocytes in ischemic myocardium. Double fluorescent immunostaining for BrdU (red) and c-KIT (green) in heart sections in WT mice and in SH2B3/LNK−/− mice 7 days following MI. Number of BrdU+/c-KIT+ cells in ischemic myocardium 7 days following MI were counted and averaged. Mann-Whitney comparison test *, p<0.05 vs. WT mice (WT: n=4 and SH2B3/LNK−/−: n=3).
f-i: Post MI regeneration: physiological and histological assessment for LV function in WT vs. SH2B3/LNK−/- mice following MI. M-mode echocardiography in WT mice and SH2B3/LNK−/− mice 28 days following MI. Fractional shortening (f) and regional wall motion score (g) were significantly great in SH2B3/LNK−/− mice than that in WT mice. Hemodynamic study using a micro-tip catheter in WT mice and SH2B3/LNK−/− mice 28 days following MI. +dP/dt, -dP/dt and EDP were significantly preserved in SH2B3/LNK−/− mice than those in WT mice. Mann-Whitney comparison test *, p<0.05 and **, p<0.01 vs. WT. (n=11) (+dP/dt: WT, 5,942.1±823.7 vs. SH2B3/LNK−/−, 8,901.6±1,147.9 mmHg/sec, p<0.01; -dP/dt: WT, -4,675.9±615.9 vs. SH2B3/LNK−/−, -6,201.4±875.4 mmHg/sec, p<0.01; EDP: WT, 8.6±2.1 vs. SH2B3/LNK−/−, 4.4±1.2 mmHg, p<0.05) (h) Representative Masson's trichrome stained heart sections in WT mice and SH2B3/LNK−/− mice 28 days following MI. Percent of fibrosis area in entire LV area on cross-sections. Histological analysis was performed on day 28 post MI. The percentage of fibrosis area was less in SH2B3/LNK−/− mice than WT mice (WT, 15.2±4.3 vs. SH2B3/LNK−/−, 8.0±5.0 %, p<0.05). Fibrosis area was significantly reduced in SH2B3/LNK−/− mice compared with WT mice. Bonferroni post hoc test * p<0,05 vs. WT. (WT: n=6 and SH2B3/LNK−/−: n=10) (i) Immunostaining for isolectin B4 (brown) in WT and SH2B3/LNK−/− mice 28 days following MI. Capillary density in ischemic border zone in infarcted myocardium of WT mice and SH2B3/LNK−/− mice. Bonferroni post hoc test **, p<0.01 vs. WT. (WT: n=5 and SH2B3/LNK−/−: n=9) capillary density in infarction border zone was significantly greater in SH2B3/LNK−/− mice than WT mice (WT, 713±28 vs. SH2B3/LNK−/−, 937±157/mm2, p<0.01). On the other hand, there was no significant difference in LV function and capillary density between WT mice and SH2B3/LNK−/− mice without ischemic injury.
j: Pearson correlation analysis between the mouse infarction model (SH2B3/LNK -/−vs. WT) and human phase 3 PERFECT trial (ΔLVEF Responder vs. Non-responder). The human ΔLVEF response is highlighted for an improved visual analysis of important correlations. The color scale, ranging from 1 to -1 in the upper panel (blue to red), represents the correlation between the different factors. The size of the dots represents the significance (p<0.01, p<0.05, and p>0.05; Pearson correlation) of the respective correlation. Comparison of peripheral blood (PB) circulating cells and biomarkers between mice (purple) and human (serum) (black) (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
3.2.2. Preserved left ventricular function and structural integrity in infarcted myocardium in SH2B3/LNK−/− mice
To investigate the effect on the phenotypic level in mice, LV function of WT mice and SH2B3/LNK−/− mice on day 28 post MI was analyzed by echocardiography. Fractional shortening (FS: WT, 17•7±2•6 vs. SH2B3/LNK−/−, 29•2±5•6 %, p<0,01, Mann-Whitney comparison test) (Fig. 6F), a parameter of global left ventricular contractility, and regional wall motion score (RWMS) were examined (RWMS: WT, 25•4±3•4 vs. SH2B3/LNK−/−, 19•9±0•9, p<0•01, Mann-Whitney comparison test) (Fig. 6G, Supplemental Data SD2). LV function was also examined using micromanometer-tipped catheters on day 28. +dP/dt, -dP/dt and endodiastric pressure (EDP), infarction borderzone fibrosis and capillary angiogenesis were significantly better preserved in SH2B3/LNK−/− mice (Fig. 6H, I, Supplemental Data SD2). These results suggest that gene deficiency of SH2B3/LNK contributes to the preservation of LV function and structural integrity of infarcted myocardium post MI.
3.3. C: Evaluation
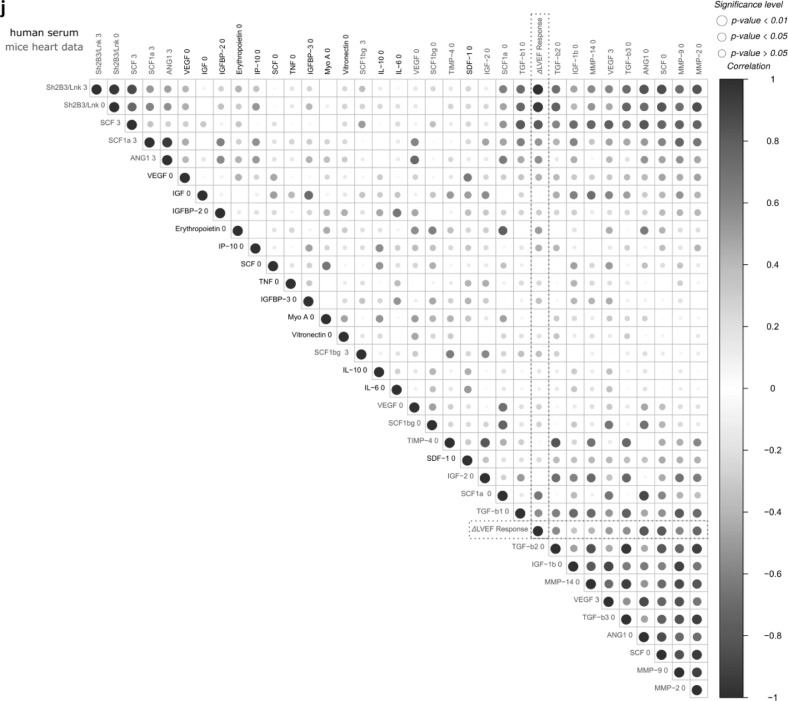
3.3.1. Bone marrow, peripheral blood, and heart tissue data in mice and man
We compared the previously investigated datasets of mice and man using correlation analyses visualised as heatmaps with respect to comparability of SH2B3/LNK−/− vs WT post MI cardiac regeneration biomarkers to post CABG/CD133+ cardiac regeneration (RvsNR) (JFig. 6J). Circulating EPCs in PB were correlated (p<0•05) to ∆LVEF response (RvsNR and SH2B3/LNK−/− vs. WT) for huCD34+(day 0), huCD45+14+(day 0), huCD133+(day 0), huCD45+CD184(CXCR4)+(day 0), huCD117(c-KIT)+(day 0), mSca-1+(day 3, 7), and mCD184(CXCR4)+(day 0, 14, 28) (Supplementary Figure S4). KSL bone marrow SC (day 1, 7, 14) in mice were positively correlated (p<0•05) to human PB CD133+ EPC, CFU-Hill, and ∆LVEF-response (Supplementary Figure S5). ∆LVEF-response was negatively correlated (p<0•01) to cardiac SH2B3/LNK expression in mice (day 0, 3) and positively correlated (p<0•05) to mouse cardiac remodeling factors MMP2, SCF1, and Angiopoietin 1 on day 0 (JFig. 6J).
Fig. 6.
(Continued).
3.3.2. ML- integrated stratification of patients
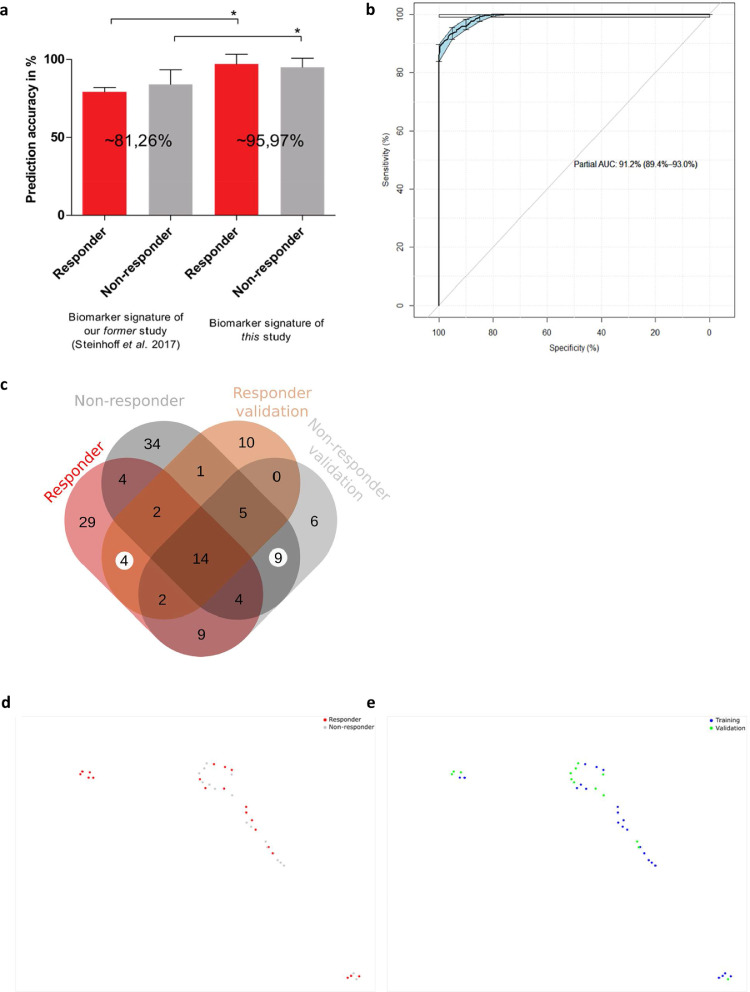
Combining gene expression features and biomarkers led to an improved patient signature for outcome prediction from 81% [17] to finally 96%, which was achieved by only using the top eight ML-selected features (Fig. 7A). Pathway analysis of most important ML features focusses on genes regulating hematopoietic stem cell receptor and proliferation signaling (Table 2).
Fig. 7.
Patient stratification to responder and non-responder. Clustering and SNP signature comparison for the analysis and validation cohort.
a: Machine learning accuracy comparison for the supervised prediction of the patient responsiveness using only preoperative data. Results are obtained after feature selection and subsequent prediction with two independent classifiers. The graph shows the true positive prediction results of two ML models (AdaBoost for feature selection and RF for final prediction for the former study and RF and SVM for the current study).The error bars indicate the respective accuracy standard deviation for the constructed models that have been obtained after 100 iterations. * indicates that the 100 model iterations are significant different according to Bonferroni post-hoc test (p<0.01). b: Receiver Operating Characteristics (ROC) curve for the random forest machine learning model. The plot represents the sensitivity (true positive rate) and the specificity (false positive rate) of the model. The area under the ROC curve (AUC) represents the entire area underneath the ROC curve and the confidence intervals (95%CI) are indicated in blue. c: Venn diagram summarizing the identified SNPs in RvsNR for the analysis and validation cohort.
d-e: Validation for the clustering with primary cohort (n=23, blue, Rostock trial center biomarker cohort) and independent validation cohort (n=14, green, Hannover center). UMAP representation with k=4 and 2,000 epochs (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
R/NR selective discriminative factors were identified by combining gene expression difference, coexpression analysis, SNP/variants, and Machine Learning (Fig. 7B; Supplementary Fig. S6). Validation of an independent patient data set revealed 85% accuracy (12/14 patients) for stratification (Fig. 7C) and classification concordance to ML-clusters (Fig. 7D, E). All missclassified patients in the primary biomarker cohort analysis (1/23, male R) and the validation cohort (2/14; male/R and female/NR) exclusively showed a single SNP (rs.56313931 G/A) mutation in the X-chromosome linked target biomarker KLF-8 (3/37) (Supplementary Data SD1D).
4. Discussion
In the randomized PERFECT phase 3 CABG/CD133+ stem cell trial, we observed a preoperative signature of circulating BM-HSC/EPC and angiogenesis parameters in peripheral blood that significantly correlated to postoperative response of myocardial regeneration in contrast to no difference in surgical CABG-procedure with complete coronary revascularization [17]. Using RNA sequencing analysis in blood, we have found for the first time a corresponding signature for a decreased (NR) or enhanced (R) bone marrow stem cell response upon ischemic/inflammatory response to CABG. Gene expression patterns are characteristic for a RvsNR preoperative steady-state and include pathways for proliferation control (EGFR, PDFR, TCR, c-KIT), inflammation (IL2, IFN1), and platelet activating factor synthesis (LPCAT2). Identified hub genes related to BM-HSC response were mainly genes coding for signaling, adaptor, and transcription regulation proteins.
Our independent ML-based feature selection of RvsNR discriminating the most important factors gives evidence for a gene circuit effecting myocardial repair by modified expression of signaling and adaptor protein gene transcription. Given the complexity of stem cell response to ischemic or inflammatory stimuli as induced by CABG-surgery, we found access to circuit pattern recognition in patients only by unbiased ML methods. Learning from failed reductionistic attempts to define single control factors of stem cell reactivity, we propose this approach as an independent and unbiased start. The association of genes such as PLCG1, LPCAT2, AP1B1, AFAP1, GRB2, KLF8, MARK3, with circulating CD133+/CD34+ cells, and serum proteins EPO/VEGF were initially identified using ML and sequentially validated experimentally in mice and in an additional patient cohort. Further clustering analysis of the overall patient cohort revealed subgroups representing the complexity of gene expression, coexpression, and mutational variants. Their functional interplay has to be studied in additional experimental models as exemplarily shown here for the Src adaptor protein SH2B3/LNK that regulates crosstalk between integrin and cytokine signaling pathways in BM-HSC/megakaryocyte proliferation.
Moreover, for the first time multiple somatic mutations involving stem cell functional genes as NOTCH2, PROM1/CD133, MTOR as well as SH2B3 gene were found in PERFECT patients with coronary arteriosclerotic disease. In this context, mutations of SH2B3/LNK were present in all patients with a majority of responders expressing the exon variant RS3184504 that is associated to coronary artery disease and increased thrombocyte count. Interestingly, human SH2B3 gene expression was not linked to thrombocyte count and circulating CD133+/CD34+ BM-HSC, whereas correlation was found to LVEF response. It is conceivable, that not reduced gene expression, but mutant related altered SH2B3/LNK protein signaling function impairs control of stem cell proliferation. Therefore, we compared gene expression patterns in a mouse model of CRISPR-Cas9-mediated SH2B3 knockout with human R/NR.
Similar to SH2B3/LNK in humans, LNK in the mouse model is an adaptor protein that negatively regulates multiple essential signals, including the SCF/c-KIT system, in stem/progenitor cells. We have previously reported that the deficiency accelerated hindlimb ischemia recovery and bone fracture healing [1], [24] is mainly achieved by restoring local blood perfusion with increased angiogenesis and osteogenesis, respectively. Following the series of SH2B3/LNK-related clinical observations in R/NR, we successfully demonstrated for the first time clonal dominance in hematopoiesis, lymphopoiesis, and myelopoiesis by reduced SH2B3/LNK signaling. To test the hypothesis of a clonal network switch found in silico, we subsequently performed a competitive syngeneic bone marrow transplantation model in mice transplanted with unmodified and SH2B3/LNK knockout HSC. SH2B3/LNK knockout HSC clones displayed significant overgrowth of myeloid and immune cells in bone marrow, peripheral blood, and tissue at day 160 after BMT. Moreover, the gene expression profile of peripheral blood was similar to the human R/NR signature (Table 4).
Table 4.
Comparison of whole blood RNA-Seq gene expression in human RvsNR in PERFECT trial in comparison to CRISPcas SH2B3 knock-out bone marrow transplantation (BMT) mouse model. Selected correlating genes to SH2B3 in PERFECT trial RvsNR and SH2B3 CRISPcas HSC knock-out mouse model. Depicted are RNA-Seq analyzed gene expression levels of selected genes with baseline PB expression in PERFECT RvsNR as compared to mouse CRISPcas SH2B3 knock-out BMT model. Identical pattern was observed for NOTCH2, PLCG1, LPCAT2, Prom1/CD133, MTOR, whereas VEGF-B was differently regulated.
| SH2B3/LNK correlating genes | RvsNR expression human PERFECT trial |
SH2B3 neg. vs pos. mouse BMT model |
|||||
| Abbreviation | Compartment | Protein function | SH2B3 GE correlation |
Responder Delta LVEF |
Non Responder dLVEF |
KO/CRISP CAS Mouse SH2B3 deficient |
WT Mouse SH2B3+ |
| NOTCH2 | Nucleus, ER, golgi, extracellular | Proliferation, differentiation, apoptosis |
P<0.01 Pos |
↑ | ↓ | ↑ | ↓ |
| PLCG1 (RNA) | Cytosol, Nucleus | Signal transduction TK growth receptors |
P<0.01 Pos |
↑ | ↓ | ↑ | ↓ |
| PDCD1/PD-1 (RNA) | Cytosol, extracellular | Immune checkpoint receptor ligand, apoptosis control |
P<0.05 Pos |
↑ | ↓ | ↑ | ↓ |
| LPCAT2 (RNA) | ER, Golgi | Phospholipid metabolism, PAF synthesis | P>0.05 Pos |
↑ | ↓ | ↑ | ↓ |
| Prom1/CD133 (RNA) | ER, Membrane | Differentiation, proliferation, apoptosis signaling | P<0.05 | ↑ | ↓ | ↑ | ↓ |
| MTOR (RNA) | Lysosome, Cytosol | Proliferation, metabolism signaling |
P<0.05 Pos |
↑ | ↓ | ↑ | ↓ |
| VEGF-B (RNA) | extracellular | Vascular growth factor |
P<0.05 Neg |
↓ | ↑ | ↑ | ↓ |
Furthermore, in ischemic myocardium using a SH2B3/LNK−/− mouse MI model, we demonstrated that: 1) SH2B3/LNK deficiency increased the number of HSC/KSL stem/progenitor cells (including EPCs) in BM and stem/progenitor cells in myocardium, 2) Angiogenic growth factor, survival factor, and stem/progenitor chemokine mRNA expressions were up-regulated in SH2B3/LNK deficient BM/PB HSC stem/progenitor cells, 3) Significant mobilization of BM/PB stem/progenitor cells occurred following myocardial ischemia in SH2B3/LNK-negative mice, 4) SH2B3/LNK deficiency reduced myocardial ischemic insult with increased angiogenesis to recruit BM-derived EPCs, 5) Resident stem/precursor cells in the heart proliferated and contributed to tissue regeneration in ischemic myocardium following MI.
Taken together, the obtained transcriptome data reveal that clonal HSC dysregulation led to specific disease phenotypes as reduced angiogenesis and sustained myocardial ischemia were observed to regulate cardiac recovery outcome in PERFECT trial coronary artery disease patients. Moreover, not specifically CHIP-gene mutations, but distinct regulatory angiogenesis and HSC proliferation pathway gene mutations were observed [5]. These observations underline the hypothesis of coronary artery disease (CAD) as a phenotype of hematological stem cell disease and acquired senescence by somatic mutations in the tissue repair gene circuit [4]. From this knowledge, treatment strategies for CAD with stem cells have to be reassessed [9,25]. We have to reconsider the current approach to diagnose and treat hematopoietic stem cell dysfunction as a truly cardiovascular and immune stem cell disease [9]. On this basis, temporary SH2B3 gene downregulation or functional abrogation of LNK protein would be one of the therapeutically effective modifications in currently ongoing autologous progenitor cell transplantation therapy [26]. Moreover, modulation of SH2B3 mutant gene expression in proliferative arteriosclerotic or hypoproliferative ischemic cardiovascular diseases may give rise to the next generation cell based therapy [27]. Temporary downregulation of adaptor gene SH2B3 in cKIT-CD117+/CD133+/CD34+ HSC can be a therapeutic switch to improve downregulated stem cell response in ischemia, tissue repair, and myocardial infarction. Using the R/NR signature response described here may also guide specific immune checkpoint or interferon based drug treatment interventions.
At this stage it is speculative, but it may be assumed that the myocardial repair response or non-response to stem cell therapy of coronary artery disease patients could be predicted on the basis of the gene expression signatures found in the PERFECT trial patients. This approach may complement the current evaluation in the CardiAMP Heart Failure Trial to predict treatment response from colony forming capacity of bone marrow stem cells [28]. Next phase studies should test the validity of our integrated data analysis approach combining whole transriptome, protein expression, cell function (colony forming units/CFU or Boyden chamber migration assay) with clinical MRI-imaging, laboratory, and symptom data for ML-based clinical outcome prediction accuracy. This may enable treatment of acquired stem cell senescence in a presymptomatic state and aim for restoration of normal bone marrow function. For selection and clonal overgrowth of healthy autologous HSC using ressources like cord blood HSC banking, iPSC-technology of non-mutated cell types as cardiomyocytes for HSC generation can be a desired option. Clonal selection and expansion biotechnology or allogenic BMT can be used to treat early or advanced stages of HSC senescence. In cases of advanced HSC senescence, the positive experience of autologous/allogenic BMT in multiple myeloma patients may be followed [29].
In conclusion, the proposed ML biomarker/gene signature, resulting in 96% classification accuracy, opens a perspective for the analysis of polygenic risk and cardiovascular disease pathomechanism profiling [30]. The first use of an integrative algorithm for gene expression, hub gene coexpression, and transcribed RNA variants derived from RNA-Seq datasets allows identification of patient-specific perturbations. This leads to an individualized pathomechanism switch and targeted treatment as shown for SH2B3/LNK. Moreover, validation of misclassifications can be enabled by whole genome variant analysis as shown here for the X-chromosome linked target gene KLF8. ML-based diagnosis of stem cell based cardiac regeneration capacity in tissue ischemia, infection or vascular repair can be applied and tested for differential diagnosis in heart and degenerative organ disease.
Role of the funding
The funding had no role in study design, in the collection, analysis, interpretation of data, in the writing of the report, and in the decision to submit the manuscript for publication. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Declaration of Competing Interest
All authors declare no competing interests.
Contributors
G Steinhoff contributed to study design, trial organization, medical controlling, enrolment and clinical follow-up of patients, research plan, analysis of clinical data, analysis of research data, data collection, data control, data analysis, and drafted the manuscript.
A Haverich, J Garbade, C Stamm, J Gummert , F M Wagner contributed to enrolment and clinical follow-up of patients, data collection and interpretation.
H Blum, S Krebs, J Philippou-Massier performed tissue processing and gene sequencing analysis, construction of cDNA libraries from polyA RNA with depletion of Globin cDNA and generation of sequencing data.
J Kowallick, CO Ritter, E Schrinner analysed MRI data.
J Nesteruk contributed to laboratory study design, follow-up analysis of patients, data collection, and statistical analyses.
R Gaebel performed laboratory analysis and examined data collection.
M Wolfien, H Hennig and O Wolkenhauer applied and investigated gene expression and machine learning data analysis.
T Asahara contributed to study design, experimental trial organization, research plan, analysis of experimental research data, data collection, data control, and drafted the manuscript.
A Kawamoto contributed to laboratory study design, data collection, and statistical analyses.
A Salybekov, M Ii, M Horii, H Iwasaki, H Akimaru, E Akimaru, A Yokoyama performed experimental animal model, laboratory analysis and examined data collection.
D Klatt and A Schambach performed experimental gene therapy, gene knock outs, competitive bone marrow transplantation model, and drafted the manuscript.
All authors contributed to final data interpretation, critically revised the manuscript, and approved the final version for submission.
Acknowledgments
We would like to thank the PERFECT study group and the safety monitoring board led by Prof Giulio Pompilio, MD PhD, (Milano) for their dedicated performance and support of the trial.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102862.
Contributor Information
Markus Wolfien, Email: markus.wolfien@uni-rostock.de.
Denise Klatt, Email: denise_klatt@DFCI.HARVARD.EDU.
Masaaki Ii, Email: masa0331@mac.com.
Miki Komatsu-Horii, Email: komatsu@fbri.org.
Ralf Gaebel, Email: ralf.gaebel@med.uni-rostock.de.
Julia Philippou-Massier, Email: massier@genzentrum.lmu.de.
Eric Schrinner, Email: eric.schrinner@web.de.
Hiroshi Akimaru, Email: akimaruh@cdb.riken.jp.
Erika Akimaru, Email: eakimaru@fbri.org.
Robert David, Email: robert.david@med.uni-rostock.de.
Jens Garbade, Email: jens.garbade@helios-kliniken.de.
Jan Gummert, Email: jan.gummert@ruhr-uni-bochum.de.
Axel Haverich, Email: haverich.axel@mh-hannover.de.
Holger Hennig, Email: holger.hen@gmail.com.
Hiroto Iwasaki, Email: hiroto@iwasakiclinic.net.
Alexander Kaminski, Email: kaminski.alexander@drguth.de.
Atsuhiko Kawamoto, Email: kawamoto@fbri.org.
Christian Klopsch, Email: christian.klopsch@med.uni-rostock.de.
Johannes T. Kowallick, Email: johannes.kowallick@med.uni-goettingen.de.
Stefan Krebs, Email: krebs@genzentrum.lmu.de.
Julia Nesteruk, Email: jullyn@yandex.ru.
Hermann Reichenspurner, Email: reichenspurner@uke.de.
Christian Ritter, Email: christian.ritter@med.uni-goettingen.de.
Christof Stamm, Email: stamm@dhzb.de.
Ayumi Tani-Yokoyama, Email: a-tani@fbri.org.
Helmut Blum, Email: blum@genzentrum.lmu.de.
Olaf Wolkenhauer, Email: olaf.wolkenhauer@uni-rostock.de.
Axel Schambach, Email: schambach.axel@mh-hannover.de.
Takayuki Asahara, Email: asa777@is.icc.u-tokai.ac.jp.
Gustav Steinhoff, Email: gustav.steinhoff@med.uni-rostock.de.
Appendix. Supplementary materials
References
- 1.Blau HM, Brazelton TR, Weimann JM. The evolving concept of a stem cell: entity or function? Cell. 2001;105:829–841. doi: 10.1016/s0092-8674(01)00409-3. [DOI] [PubMed] [Google Scholar]
- 2.Korbling M, Estrov Z. Adult stem cells for tissue repair - a new therapeutic concept? N Engl J Med. 2003;349:570–582. doi: 10.1056/NEJMra022361. [DOI] [PubMed] [Google Scholar]
- 3.Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- 4.Moehrle BM, Geiger H. Aging of hematopoietic stem cells: DNA damage and mutations? Exp Hematol. 2016; Oct;44(10):895–901. doi: 10.1016/j.exphem.2016.06.253. Epub 2016 Jul 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaiswal S, Natarajan P, Silver AJ. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N Engl J Med. 2017;377(2):111–121. doi: 10.1056/NEJMoa1701719. Epub 2017 Jun 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Machiela MJ, Chanock SJ. The ageing genome, clonal mosaicism and chronic disease. Curr Opin Genet Dev. 2017;42:8–13. doi: 10.1016/j.gde.2016.12.002. FebEpub 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elias HK, Bryder D, Park CY. Molecular mechanisms underlying lineage bias in aging hematopoiesis. Semin Hematol. 2017;54(1):4–11. doi: 10.1053/j.seminhematol.2016.11.002. Epub 2016. [DOI] [PubMed] [Google Scholar]
- 8.Baughn LB, Meredith MM, Oseth L, Smolarek TA, Hirsch B. SH2B3 aberrations enriched in iAMP21 B lymphoblastic leukemia. Cancer Genet. 2018:226–227. doi: 10.1016/j.cancergen.2018.05.004. 30-35Epub 2018. [DOI] [PubMed] [Google Scholar]
- 9.Steinhoff G, Nesteruk J, Wolfien M, Große J, Ruch U, Vasudevan P, Müller P. Stem cells and heart disease - Brake or accelerator? Adv Drug Deliv Rev. 2017;120:2–24. doi: 10.1016/j.addr.2017.10.007. Oct 1Epub 2017. [DOI] [PubMed] [Google Scholar]
- 10.Takaki S, Morita H, Tezuka Y, Takatsu K. Enhanced hematopoiesis by hematopoietic progenitor cells lacking intracellular adaptor protein, SH2B3/LNK. J Exp Med. 2002;195:151–160. doi: 10.1084/jem.20011170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siedlinski M, Jozefczuk E, Xu X, Teumer A, Evangelou E, Schnabel RB, Welsh P, Maffia P, Erdmann J, Tomaszewski M, Caulfield MJ, Sattar N, Holmes MV, Guzik TJ. White Blood Cells and Blood Pressure: A Mendelian Randomization Study. Circulation. 2020 doi: 10.1161/CIRCULATIONAHA.119.045102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Mo X, Zhang H, Zhang Y, Shen Y. Identification of Phosphorylation Associated SNPs for Blood Pressure, Coronary Artery Disease and Stroke from Genome-wide Association Studies. Curr Mol Med. 2019;19(10):731–738. doi: 10.2174/1566524019666190828151540. [DOI] [PubMed] [Google Scholar]
- 13.Wang W, Tang Y, Wang Y. SH2B3/LNK Loss of Function Promotes Atherosclerosis and Thrombosis. Circ Res. 2016;119(6):e91–e103. doi: 10.1161/CIRCRESAHA.116.308955. Epub 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mo X, Guo Y, Qian Q, Fu M, Zhang H. Phosphorylation-related SNPs influence lipid levels and rheumatoid arthritis risk by altering gene expression and plasma protein levels. Rheumatology (Oxford) 2019 doi: 10.1093/rheumatology/kez466. pii: kez466. [DOI] [PubMed] [Google Scholar]
- 15.Kuo CL, Joaquim M, Kuchel GA, Ferrucci L, Harries L, Pilling LC, Melzer D. The Longevity Associated Sh2b3 (LNK) Genetic Variant: Selected Aging Phenotypes in 379,758 Subjects. J Gerontol A Biol Sci Med Sci. 2019 doi: 10.1093/gerona/glz191. pii: glz191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu X, Fang J, Jiang DS, Zhang P, Zhao GN, Zhu X, Yang L, Wei X. Exacerbating Pressure Overload-Induced Cardiac Hypertrophy: Novel Role of Adaptor Molecule Src Homology 2-B3Hypertension2015; Sep;66(3):571-81. doi: 10.1161/HYPERTENSIONAHA.115.05183. Epub 2015. [DOI] [PubMed]
- Steinhoff G, Nesteruk J, Wolfien M. Cardiac function improvement and bone marrow response - Outcome analysis of the randomized PERFECT phase III clinical trial of intramyocardial CD133+Application after myocardial infarction. EBioMedicine. 2017;22:208–224. doi: 10.1016/j.ebiom.2017.07.022. AugEpub 2017 Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuleshov M V, Jones MR, Rouillard AD. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkw377. W90-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van der Auwera GA, Carneiro MO, Hartl C. Current Protocols in Bioinformatics. Jon Wiley&Sons Inc.; Hoboken, NJ, USA: 2013. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolit Best Practises Pipeline. p11.10.1-11.10.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn M. Building Predictive Models in R using the caret package. J Stat Softw. 2008;28(5):1–26. doi: 10.18637/jss.v028.i05. [DOI] [Google Scholar]
- 21.Forman G., Cohen I. Learning from little: comparison of classifiers given little training knowledge discovery in databases. PKDD. 2004 doi: 10.1007/978-3-540-30116-5_17. [DOI] [Google Scholar]
- 22.Gjesdal O, Almeida AL, Hopp E, Beitnes JO, Lunde K, Smith HJ, Lima JA, Edvardsen T. Long axis strain by MRI and echocardiography in a postmyocardial infarct population. J Magn Reson Imaging. 2014;40(5):1247–1251. doi: 10.1002/jmri.24485. NovEpub 2013 Nov 8. [DOI] [PubMed] [Google Scholar]
- Mordini FE, Haddad T, Hsu LY, Kellman P, Lowrey TB, Aletras AH, Bandettini WP, Arai AE. Diagnostic accuracy of stress perfusion CMR in comparison with quantitative coronary angiography: fully quantitative, semiquantitative, and qualitative assessment. JACC Cardiovasc Imaging. 2014;7(1):14–22. doi: 10.1016/j.jcmg.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto T, Ii M, Nishimura H. SH2B3/LNK-dependent axis of SCF-cKIT signal for osteogenesis in bone fracture healing. J Exp Med. 2010;207:2207–2223. doi: 10.1084/jem.20100321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murry CE, MacLellan WR. Stem cells and the heart—the road ahead. Science. 2020;367(6480):854–855. doi: 10.1126/science.aaz3650. [DOI] [PubMed] [Google Scholar]
- 26.Kawakami Y, Ii M, Matsumoto T, Kawamoto A, Kuroda R, Akimaru H. A small interfering RNA targeting Lnk accelerates bone fracture healing with early neovascularization. Lab Invest. 2013;93(9):1036–1053. doi: 10.1038/labinvest.2013.93. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Jin BJ, Chen Q, Yan BJ, Liu ZL. MicroRNA-29b upregulation improves myocardial fibrosis and cardiac function in myocardial infarction rats through targeting SH2B3. Eur Rev Med Pharmacol Sci. 2019;23(22):10115–10122. doi: 10.26355/eurrev_201911_19581. [DOI] [PubMed] [Google Scholar]
- 28.Johnston PV, Duckers HJ, Raval AN, Cook TD, Pepine CJ. Not all stem cells are created equal - the case for prospective assessment of stem cell potency in the CardiAMP Heart Failure Trial. Circulation Res. 2018;123:944–946. doi: 10.1161/CIRCRESAHA.118.313425. [DOI] [PubMed] [Google Scholar]
- 29.Fabre C, Koscielny S, Mohty M, Fegueux N, Blaise D, Maillard N, Tabrizi R, et al. Younger Donor's Age and Upfront Tandem Are Two Independent Prognostic Factors for Survival in Multiple Myeloma Patients Treated by Tandem Autologous-Allogeneic Stem Cell Transplantation: A Retrospective Study From the Société Française De Greffe De Moelle Et De Thérapie Cellulaire (SFGM-TC) Haematologica2012; 97 (4), 482-90 [DOI] [PMC free article] [PubMed]
- 30.Torkamani A, Wineinger NE, Topol E. The personal and clinical utility of polygenic risk scores. Nature Rev Genet. 2018;19:581–590. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.