Abstract
Recently a novel subtype of endometrial stromal sarcoma (ESS) defined by recurrent genomic alterations involving BCOR has been described (HGESS‐BCOR). We identified a case of HGESS‐BCOR with a ZC3H7B‐BCOR gene fusion, which harbored an amplification of the MDM2 locus. This index case prompted us to investigate MDM2 amplification in four additional cases of HGESS‐BCOR. Tumors were analyzed for MDM2 amplification by array‐based profiling of copy number alterations (CNAs) and fluorescence in situ hybridization (FISH), as well as for MDM2 expression by immunohistochemistry (IHC). Additionally, a cohort of other mesenchymal uterine neoplasms, including 17 low‐grade ESS, 6 classical high‐grade ESS with YWHAE‐rearrangement, 16 uterine tumors resembling ovarian sex cord tumors, 7 uterine leiomyomas and 8 uterine leiomyosarcomas, was analyzed for CNAs in MDM2. Copy number profiling identified amplification of the 12q15 region involving the MDM2 locus in all five HGESS‐BCOR. Subsequent validation analyses of three tumors confirmed MDM2 amplification using MDM2 FISH. Accordingly, IHC showed MDM2 overexpression in all analyzed cases. None of the other uterine neoplasms in our series, including tumors that are in the histopathological differential diagnoses of HGESS‐BCOR, showed copy number gains of MDM2. Together, our results indicate that HGESS‐BCOR carries MDM2 amplifications, which has diagnostic implications and could potentially be used for targeted therapies in these clinically aggressive tumors.
Keywords: endometrial stromal sarcoma, MDM2, amplification, BCOR, YWHAE, uterine, neoplasm
Introduction
Endometrial stromal tumors (EST) represent rare uterine neoplasms of mesenchymal origin [1]. The 2014 WHO classification of tumors of the female reproductive tract distinguishes four categories of EST: endometrial stromal nodule (ESN), low‐grade endometrial stromal sarcoma (LGESS), high‐grade endometrial stromal sarcoma (HGESS), and undifferentiated uterine sarcoma (UUS) [2]. While ESN and LGESS histologically resemble stromal cells of the proliferating endometrium and harbor recurrent chromosomal translocations most frequently associated with a JAZF1‐SUZ12 gene fusion, UUS comprises endometrial and myometrial sarcomas which lack specific mesenchymal differentiation and are molecularly heterogenous [3, 4]. As defined by the 2014 WHO classification, HGESS harbor a t(10;17)(q22;p13) chromosomal translocation resulting in a YWHAE‐NUTM2 fusion [2, 5]. Such tumors represent a clinically more aggressive entity with patients diagnosed at higher stages and more likely to die of disease when compared to LGESS [6].
Recently, a rare subtype of ESS with high‐grade features and BCOR alterations, caused by either a gene fusion between BCOR and ZC3H7B or a mutually exclusive somatic internal tandem duplication (ITD) of exon 15 of BCOR, has been described (HGESS‐BCOR) [7, 8]. Although such BCOR alterations may well be the molecular driver in these tumors, little is known about their biology, or about potential cooperative and co‐occurring genetic events.
We recently identified a case of HGESS‐BCOR that carried an amplification of the 12q15 region involving the MDM2 locus. This observation prompted us to compile a multicenter cohort to investigate MDM2 amplification in HGESS‐BCOR.
Material and methods
Study cohort
A study cohort including HGESS‐BCOR, LGESS, HGESS, uterine tumors resembling ovarian sex cord tumors (UTROSCT), uterine leiomyomas (ULMO), and uterine leiomyosarcomas (ULMS) was collected from the referral center archives of two of the authors (DS and FK), the Department of Pathology, University of Heidelberg, and the KK Women's and Children's Hospital, Singapore. All cases were subject to expert pathology review including molecular pathology [9]. Fusion status of HGESS‐BCOR has previously been reported in part [10, 11]. This study was performed in accordance with the ethical standards of the institutional research committee and the Declaration of Helsinki.
Genomic DNA extraction and quantification
DNA of all tumors was extracted from formalin‐fixed paraffin‐embedded (FFPE) tissue samples. Extracted DNA was quantified using the QuantiFast SYBR Green PCR Kit (Qiagen, Duesseldorf, NW, Germany).
Copy number‐profile generation
A total of >100 ng DNA was available for array‐based DNA methylation analysis in all cases. Samples were analyzed using the Illumina Infinium HumanMethylation450 (450k) or EPIC (850k) BeadChip (Illumina, San Diego, IL, USA), according to the manufacturer's instructions at the Genomics and Proteomics Core Facility of the German Cancer Research Center (DKFZ), Heidelberg. DNA methylation data were normalized by performing background correction and dye bias correction as previously described [11]. Probes targeting sex chromosomes, probes containing multiple single nucleotide polymorphisms, and those which could not be uniquely mapped, were removed.
MDM2‐specific fluorescence in situ hybridization
MDM2‐specific fluorescence in situ hybridization (FISH) analysis was performed on whole tissue sections using the ZytoLight® SPEC MDM2/CEN 12 Dual Color Probe (ZytoVision GmbH, Bremerhaven, Germany) as previously described [12]. Amplification of MDM2 was defined as an MDM2/centromere 12 (CEN12) ratio ≥2.0 or an average number of MDM2 signals per tumor cell nucleus ≥6 or large clusters of MDM2 signals in ≥10% of tumor cells.
MDM2 immunohistochemistry
Four micrometer sections were cut and mounted on StarFrost Advanced Adhesive slides (Engelbrecht, Kassel, Germany) followed by heat induced antigen retrieval in high pH buffer. MDM2 immunohistochemistry was performed using a monoclonal mouse antibody (dilution 1:100, clone IF2, Invitrogen by Thermo Fisher Scientific Inc., Waltham, MA, USA) as previously described [12]. Specimens were examined according to their nuclear staining for MDM2.
Results
Clinicopathologic characteristics of HGESS‐BCOR
Five HGESS‐BCOR were available for analysis. All tumors showed a hypercellular appearance with haphazard fascicular architecture. Tumor cells were spindled with irregular nuclear contours and an even chromatin pattern (Figure 1A,B). More notable atypia was only seen in one tumor (case 2; Figure 1C). Island‐like myxoid stromal change was present in three tumors (cases 1, 3, and 4; Figure 1D). RNA‐seq analysis identified a ZC3H7B‐BCOR gene fusion in four cases (cases 1, 2, 4, and 5). In case 3, the fusion detection algorithm identified a rearrangement between BCOR and the LPP gene, including RNA reads spanning parts of both genes. This event, however, appears to result in overexpression of a C‐terminally truncated BCOR protein, without generation of a fusion protein (no coding part of LPP is included). Thus, while RNA‐seq data support the presence of a BCOR alteration, the precise functional consequences of this aberration remain unclear. Clinicopathological data of all HGESS‐BCOR cases are summarized in Table 1.
Figure 1.
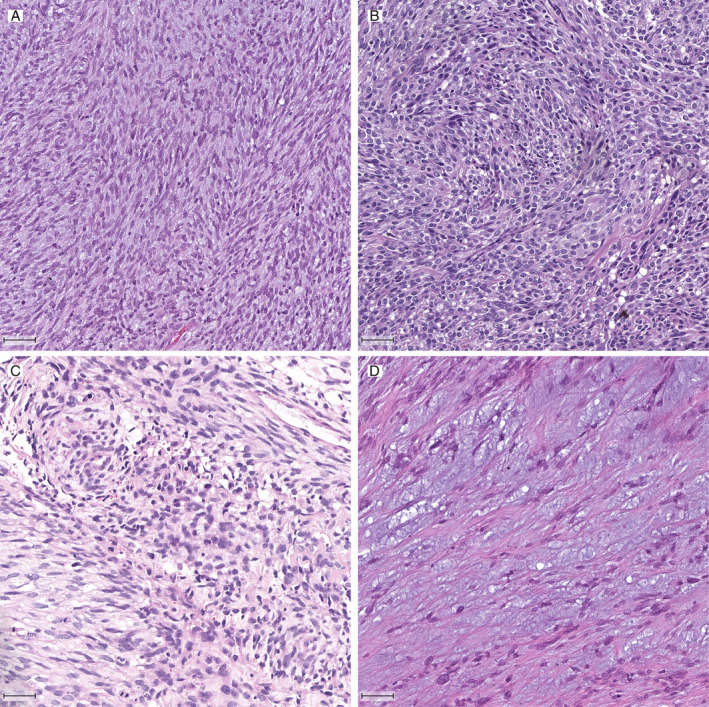
Histologic features of HGESS‐BCOR: sarcomatous proliferation of atypical and spindled neoplastic tumor cells with fascicular architecture (A and B), notable atypia (C) and prominent myxoid stroma (D). H&E stain; bar equals 50 μm.
Table 1.
Clinicopathological data of HGESS‐BCOR cases
| Case ID | Age | Diagnosis | Specimen type | Myxoid change | Molecular type |
|---|---|---|---|---|---|
| Case 1 | 48 | HGESS‐BCOR | Hysterectomy | Yes | ZC3H7B‐BCOR |
| Case 2 | 31 | HGESS‐BCOR | Hysterectomy | No | ZC3H7B‐BCOR |
| Case 3 | 50 | HGESS‐BCOR | Hysterectomy | Yes | BCOR alteration |
| Case 4 | 41 | HGESS‐BCOR | Hysterectomy | Yes | ZC3H7B‐BCOR |
| Case 5 | 34 | HGESS‐BCOR | Hysterectomy | No | ZC3H7B‐BCOR |
MDM2 amplification in HGESS‐BCOR
Copy number analysis of HGESS‐BCOR showed an amplification of the 12q15 region involving the MDM2 locus in all five cases (Figure 2A). For subsequent MDM2 validation analyses sufficient material was only available for three tumors (cases 1–3). FISH and IHC confirmed the amplification of MDM2 and overexpression (Figure 2B) of MDM2 in all three tumors. MDM2 amplification by FISH was present diffusely across the tumors analyzed. In addition to an MDM2 amplification, copy number analyses also revealed amplification of CDK4 in three tumors (cases 1–3) and a deletion of CDKN2A in one tumor (case 5).
Figure 2.

MDM2 amplification in HGESS‐BCOR: (A) copy number profiles (cases 1–5). (B) MDM2 FISH (MDM2: green, CEN12: red) and immunohistochemical staining for MDM2 (cases 1–3). Bar equals 50 μm.
MDM2 amplification is rare in other mesenchymal uterine neoplasms
Next, we analyzed copy number profiles of potential differential diagnoses of HGESS‐BCOR including 17 LGESS, 6 HGESS with YWHAE‐NUTM2 gene fusion, 16 UTROSCT, 7 ULMO, and 8 ULMS. None of these cases carried an amplification of the 12q15 region involving the MDM2 locus (Table 2).
Table 2.
12q15 (MDM2) amplification status of mesenchymal uterine neoplasms including LGESS, HGESS‐BCOR, HGESS with YWHAE‐rearrangement, UTROSCT, ULM, and ULMS
| Diagnosis | 12q15 amplification (MDM2) |
|---|---|
| Low‐grade endometrial stromal sarcoma | 0/17 (0%) |
| High‐grade endometrial stromal sarcoma (BCOR) | 5/5 (100%) |
| High‐grade endometrial stromal sarcoma (YWHAE‐NUTM2) | 0/6 (0%) |
| Uterine tumor resembling ovarian sex cord tumor | 0/15 (0%) |
| Uterine leiomyoma | 0/8 (0%) |
| Uterine leiomyosarcoma | 0/7 (0%) |
Discussion
HGESS‐BCOR represents a new subtype in the spectrum of EST, defined by genetic alterations involving BCOR. ESS with BCOR alteration show a greater degree of atypia when compared to LGESS, and available clinical data suggest an aggressive clinical course similar to that of classical HGESS [13]. Thus, it has been proposed to place such tumors in the HGESS category. We have recently shown BCOR‐ and YWHAE‐rearranged HGESS to share similar DNA methylation profiles, distinct from LGESS and other high‐grade uterine sarcomas such as ULMS, further supporting the proposed classification [11]. In the current study, we show HGESS‐BCOR to harbor MDM2 amplifications, while all HGESS with YWHAE‐NUTM2 gene fusion in our series were MDM2‐balanced.
MDM2 is the primary negative regulator of p53 and is overexpressed in cancers, e.g. certain subtypes of sarcomas [14]. Mechanistically, MDM2 overexpression functions as a powerful oncogene by negatively regulating TP53 transcriptional activity and therefore has been suggested as a druggable target for small molecule inhibitors [15]. Given the above, p53 immunohistochemistry in a subset of HGESS‐BCOR of our cohort did not exhibit strong or diffuse p53 expression (n = 3). In line with this staining pattern, additional DNA sequencing did not identify pathogenic TP53 mutations in any HGESS‐BCOR of our series (n = 5; data not shown). Thus, detecting MDM2 amplification in HGESS‐BCOR might have clinical implications in affected patients. To date, therapeutic strategies in cases of high‐grade uterine sarcoma are usually limited to surgery and radio‐/chemotherapy, and prognosis is generally dismal [16]. Targeting MDM2 might therefore be a promising way to extend the spectrum of therapeutic options for this aggressive neoplasm.
MDM2 has previously been investigated in EST (n = 43) including ESN, LGESS, HGESS with YWHAE‐rearrangement, and UUS [17]. Schoolmeester et al reported MDM2 amplification in two of 43 (5%) tumors, both of which showed an adverse clinical course. One of the latter tumors showed morphological features consistent with LGESS, and it was reported to harbor a JAZF1‐rearrangement. The second tumor harboring a MDM2 amplification had high‐grade morphology, polysomy of JAZF1, PHF1, and YWHAE but no rearrangements were reported, leading to a diagnosis of UUS. While the latter data imply that MDM2 amplification is also present in a subset of LGESS and UUS, we did not identify copy number gains of MDM2 in any other mesenchymal uterine tumor of our series including 17 LGESS. Interestingly, a recent study suggests that UUS often represents under‐recognized HGESS including tumors with BCOR alteration [18]. Moreover, ESS may rarely present as a primary intrabdominal soft tissue tumor or arise at distant sites in extragenital endometriosis. This is important to know when encountering undifferentiated mesenteric or retroperitoneal soft tissue tumors. While MDM2 amplification in such tumors usually supports a diagnosis of dedifferentiated liposarcoma, HGESS‐BCOR should also be considered in the differential diagnoses in such cases [19]. Notably, MDM2 positivity by immunohistochemistry has been reported in cases of uterine leiomyosarcoma and primary liposarcoma of the uterus [20, 21]. However, to date no systematic analysis of MDM2 amplification has been performed in such cases.
As a limitation of our study, it is important to mention that our study cohort did not include cases of Müllerian adenosarcoma, a lesion known to harbor MDM2 amplifications in up to 28% of cases [22]. Further, our study did not investigate tumors with BCOR‐ITD, which are exceedingly rare with only few cases reported in the literature to date [8]. Thus, future studies of larger cohorts of mesenchymal uterine neoplasms are needed to confirm, and to expand on, our results.
In conclusion, our data suggest that detection of MDM2 amplicons could be a useful tool in uterine sarcoma pathology and implicate MDM2 as a potential therapeutic target in HGESS‐BCOR.
Author contributions statement
FKFK and CK conceptualized the project and wrote the original draft. FKFK and CK coordinated data generation. FS and AvD supervised array‐based analysis. DSt, DTWJ, and AB analyzed array data. WH supervised the FISH and IHC analysis. FKFK, KTC, CEH, SF, AS, GM, HPS, DS, and FK provided tumor samples and corresponding metadata. All authors contributed to and approved of the final manuscript.
Acknowledgements
We want to thank the Microarray Unit of the Genomics and Proteomics Core Facility, German Cancer Research Center (DKFZ) for providing excellent technical support. This work was supported by the German Cancer Aid (Grant: 70112499). FKFK is funded by the Physician Scientist‐Program of Heidelberg University.
No conflicts of interest were declared.
References
- 1. Hoang L, Chiang S, Lee CH. Endometrial stromal sarcomas and related neoplasms: new developments and diagnostic considerations. Pathology 2018; 50: 162–177. [DOI] [PubMed] [Google Scholar]
- 2. Oliva E, Carcangiu ML, Carinelli S, et al Mesenchymal tumors In WHO Classification of Tumours of Female Reproductive Organs (4th edn), Kurman RJ, Carangiu ML, Herrington CS, et al (Eds). International Agency for Research on Cancer: Lyon, France, 2014; 135–147. [Google Scholar]
- 3. Koontz JI, Soreng AL, Nucci M, et al Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. Proc Natl Acad Sci U S A 2001; 98: 6348–6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Halbwedl I, Ullmann R, Kremser ML, et al Chromosomal alterations in low‐grade endometrial stromal sarcoma and undifferentiated endometrial sarcoma as detected by comparative genomic hybridization. Gynecol Oncol 2005; 97: 582–587. [DOI] [PubMed] [Google Scholar]
- 5. Lee CH, Ou WB, Marino‐Enriquez A, et al 14‐3‐3 fusion oncogenes in high‐grade endometrial stromal sarcoma. Proc Natl Acad Sci U S A 2012; 109: 929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee CH, Marino‐Enriquez A, Ou W, et al The clinicopathologic features of YWHAE‐FAM22 endometrial stromal sarcomas: a histologically high‐grade and clinically aggressive tumor. Am J Surg Pathol 2012; 36: 641–653. [DOI] [PubMed] [Google Scholar]
- 7. Panagopoulos I, Thorsen J, Gorunova L, et al Fusion of the ZC3H7B and BCOR genes in endometrial stromal sarcomas carrying an X;22‐translocation. Genes Chromosomes Cancer 2013; 52: 610–618. [DOI] [PubMed] [Google Scholar]
- 8. Mariño‐Enriquez A, Lauria A, Przybyl J, et al BCOR internal tandem duplication in high‐grade uterine sarcomas. Am J Surg Pathol 2018; 42: 335–341. [DOI] [PubMed] [Google Scholar]
- 9. Stichel D, Schrimpf D, Casalini B, et al Routine RNA sequencing of formalin‐fixed paraffin‐embedded specimens in neuropathology diagnostics identifies diagnostically and therapeutically relevant gene fusions. Acta Neuropathol 2019; 138: 827–835. [DOI] [PubMed] [Google Scholar]
- 10. Mansor S, Kuick CH, Lim SL, et al ZC3H7B‐BCOR‐rearranged endometrial stromal sarcomas: a distinct subset merits its own classification? Int J Gynecol Pathol 2018; 38: 420–425. [DOI] [PubMed] [Google Scholar]
- 11. Kommoss FK, Stichel D, Schrimpf D, et al DNA methylation‐based profiling of uterine neoplasms: a novel tool to improve gynecologic cancer diagnostics. J Cancer Res Clin Oncol 2020; 146: 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grunewald I, Trautmann M, Busch A, et al MDM2 and CDK4 amplifications are rare events in salivary duct carcinomas. Oncotarget 2016; 7: 75261–75272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoang LN, Aneja A, Conlon N, et al Novel high‐grade endometrial stromal sarcoma – a morphologic mimicker of Myxoid Leiomyosarcoma. Am J Surg Pathol 2017; 41: 12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ray‐Coquard I, Blay JY, Italiano A, et al Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2‐amplified, well‐differentiated or dedifferentiated liposarcoma: an exploratory proof‐of‐mechanism study. Lancet Oncol 2012; 13: 1133–1140. [DOI] [PubMed] [Google Scholar]
- 15. Vassilev LT, Vu BT, Graves B, et al In vivo activation of the p53 pathway by small‐molecule antagonists of MDM2. Science 2004; 303: 844–848. [DOI] [PubMed] [Google Scholar]
- 16. Tanner EJ, Garg K, Leitao MM Jr, et al High grade undifferentiated uterine sarcoma: surgery, treatment, and survival outcomes. Gynecol Oncol 2012; 127: 27–31. [DOI] [PubMed] [Google Scholar]
- 17. Schoolmeester JK, Sciallis AP, Greipp PT, et al Analysis of MDM2 amplification in 43 endometrial stromal tumors: a potential diagnostic pitfall. Int J Gynecol Pathol 2015; 34: 576–583. [DOI] [PubMed] [Google Scholar]
- 18. Cotzia P, Benayed R, Mullaney K, et al Undifferentiated uterine sarcomas represent under‐recognized high‐grade endometrial stromal sarcomas. Am J Surg Pathol 2019; 43: 662–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Binh MB, Sastre‐Garau X, Guillou L, et al MDM2 and CDK4 immunostainings are useful adjuncts in diagnosing well‐differentiated and dedifferentiated liposarcoma subtypes: a comparative analysis of 559 soft tissue neoplasms with genetic data. Am J Surg Pathol 2005; 29: 1340–1347. [DOI] [PubMed] [Google Scholar]
- 20. Hall KL, Teneriello MG, Taylor RR, et al Analysis of Ki‐ras, p53, and MDM2 genes in uterine leiomyomas and leiomyosarcomas. Gynecol Oncol 1997; 65: 330–335. [DOI] [PubMed] [Google Scholar]
- 21. McDonald AG, Dal Cin P, Ganguly A, et al Liposarcoma arising in uterine lipoleiomyoma: a report of 3 cases and review of the literature. Am J Surg Pathol 2011; 35: 221–227. [DOI] [PubMed] [Google Scholar]
- 22. Howitt BE, Sholl LM, Dal Cin P, et al Targeted genomic analysis of Mullerian adenosarcoma. J Pathol 2015; 235: 37–49. [DOI] [PubMed] [Google Scholar]


