Abstract
The human brain is composed of billions of cells, including neurons and glia, with an undetermined number of subtypes. During the embryonic and early postnatal stages, the vast majority of these cells are generated from neural progenitors and stem cells located in all regions of the neural tube. A smaller number of neurons will continue to be generated throughout our lives, in localized neurogenic zones, mainly confined at least in rodents to the subependymal zone of the lateral ventricles and the subgranular zone of the hippocampal dentate gyrus. During neurogenesis, a combination of extrinsic cues interacting with temporal and regional intrinsic programs are thought to be critical for increasing neuronal diversity, but their underlying mechanisms need further elucidation. In this review, we discuss the recent findings in Drosophila and mammals on the types of cell division and cell interactions used by neural progenitors and stem cells to sustain neurogenesis, and how they are influenced by glia.
Keywords: neurogenesis, neural stem cells, neural progenitors, niche, glia, intrinsic factors, extrinsic factors, adult neurogenesis
Neural Progenitors: Division Throughout Development and in Adult Neurogenic Niches
In all animals with a brain, from insects to humans, the complex functions the brain reliably carries out at every moment depend on its many neuronal and glial cell types being generated in the proper quantities and locations. Throughout the course of life, the production of new neurons that characterizes developmental stages also persists in two regions of the adult mammalian brain, the ventricular-subventricular or subependymal zone (SEZ) adjacent to the lateral ventricles and the subgranular zone (SGZ) of the hippocampal dentate gyrus. Recently there has been a great deal of controversy regarding the existence of adult neurogenesis in the human brain (Boldrini et al., 2018; Kempermann et al., 2018; Sorrells et al., 2018; Moreno-Jimenez et al., 2019), with confusion arising in part from technical problems and perhaps from interspecies differences in the dynamics of the process. Nevertheless, based on the accumulated evidence from previous work (Eriksson et al., 1998; Sanai et al., 2011; Spalding et al., 2013; Kempermann et al., 2018), it has become increasingly clear that adult humans probably generate new neurons only in the hippocampus and not in the SEZ.
For the purpose of this review, we will focus on the knowledge that deals with the neurogenic process in flies and mice. Here, we revisit recent findings on how neural stem cells (NSCs) divide to generate neuronal diversity during brain development and adulthood. We focus on the intrinsic and extrinsic mechanisms that explain the temporal and regional heterogeneity of neural progenitor and stem cells, and their progenies. We also summarize the role of niche glia in the early and late phases of neurogenesis and discuss their diversity. Whenever possible, we compare NSCs in Drosophila and rodents, at embryonic and larval stages and in adult neurogenic zones.
The vast cell diversity in adult brains is mostly generated during the embryonic and larval stages in Drosophila, and in the embryonic and early postnatal stages in mammals, from a pool of neural progenitor and stem cells (Doe, 2017; Holguera and Desplan, 2018; Ramon-Canellas et al., 2018; Cardenas and Borrell, 2019; Morales and Mira, 2019; Obernier and Alvarez-Buylla, 2019). This pool initially includes neuroepithelial cells (NECs), which later produce multipotent NSCs (neuroblasts in Drosophila) and apical radial glia (aRG) (Figure 1). These cells span the apical–basal axis of the developing brain in mammals and have been best characterized in the neocortex (Gotz and Huttner, 2005; Kriegstein and Alvarez-Buylla, 2009; Brand and Livesey, 2011). By mid-gestation, a fraction of cortical, striatal, and septal radial glia diverge from other progenitors and are set aside as relatively quiescent cells that will give rise to postnatal and adult NSCs in the SEZ (Fuentealba et al., 2015; Furutachi et al., 2015). The development of the hippocampal dentate gyrus is longer than in other brain areas. Progenitor cells from the embryonic dentate neuroepithelium migrate out of this zone through the dentate migratory stream and occupy several transient germinal niches before finally settling in a newly formed abventricular SGZ, transforming into quiescent SGZ NSCs mainly postnatally (Seki et al., 2014; Nicola et al., 2015; Matsue et al., 2018; Berg et al., 2019; Morales and Mira, 2019; Nelson et al., 2020). Adult NSCs share many features with embryonic aRGs, including a polarized morphology and the expression of common markers such as nestin, brain lipid-binding protein (BLBP), glutamate/aspartate transporter (GLAST) and the transcription factor Sox2 (Lagace et al., 2007; Ninkovic et al., 2007; Suh et al., 2007; Giachino et al., 2014), and are often referred to as radial glia-like NSCs. At the transcriptome level, single-cell RNA sequencing (scRNA-seq) and conventional RNA-seq studies show that adult NSCs are also closely related to, but distinct from, mature astrocytes (Beckervordersandforth et al., 2010; Llorens-Bobadilla et al., 2015; Hochgerner et al., 2018).
FIGURE 1.
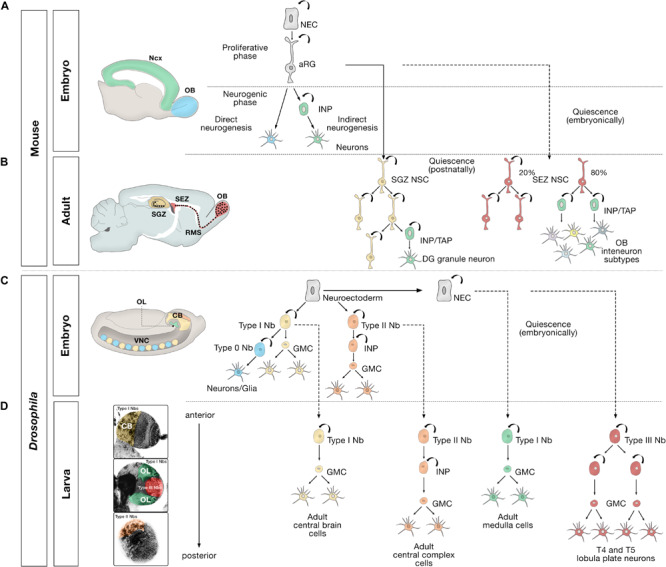
Embryonic, larval and adult neurogenesis in flies and mammals. (A) Overview of embryonic mammalian neurogenesis in the neocortex and olfactory bulb. In the developing dorsal pallium, the nervous system originates from neuroepithelial cells (NECs) that initially proliferate symmetrically before they transition to apical radial glial cells (aRG). aRGs give rise to neurons directly (blue), or indirectly (green) through intermediate neural progenitors (INP). Direct neurogenesis predominates in the olfactory bulb (OB, blue); indirect neurogenesis predominates in the neocortex (Ncx, green). (B) Adult mammalian neurogenesis in the subgranular zone (SGZ, yellow) and subependymal zone (SEZ, red). Quiescent postnatal neural stem cells in the SGZ (SGZ NSC) (yellow) undergo symmetric self-renewal before they give rise to transient amplifying cells, a type of intermediate neural progenitor (INP/TAP) (green) and differentiate into dentate gyrus granule neurons. Quiescent embryonic SEZ NSCs (red) are activated in the adult stage and undergo either symmetric self-renewing divisions (20%) or primarily produce INP/TAPs before differentiating into OB interneurons. (C) Different modes of division of neural progenitors in embryonic Drosophila. In the embryo, the nervous system originates from a neuroectoderm before they transit into neuroblasts (Nbs). Type 0 Nbs (blue) self-renew and produce a single ventral nerve cord (VNC) neuron at each division. Type I Nbs (yellow) self-renew and produce ganglion mother cells (GMC) that divide once to generate two cells in the central brain (CB, yellow) and VNC (yellow). Type II Nbs (orange) self-renew and produce intermediate neural progenitors (INP), which also self-renew multiple times before producing GMCs, which divide once and differentiate into central brain neurons (orange). Optic lobe cells (OL, green) originate from NECs. (D) Different modes of division of neural progenitors in the Drosophila larval brain. After the first, embryonic, wave of neurogenesis (shown in C), most of the remaining central brain and ventral nerve cord neuroblasts, and optic lobe NECs enter a quiescent state (dashed lines). In a second, larval, wave of neurogenesis, via ganglion mother cells (GMC), Type I Nbs in the central brain (CB, yellow region depicted in the larval brain) produce the majority of adult central brain cells, and Type II Nbs (orange region) produce the vast majority of central complex cells, an essential central brain region for sensorimotor integration (Pfeiffer and Homberg, 2014). Quiescent outer proliferation center (OPC) NECs are activated to transition into Type I Nbs (green region) and produce medulla cells in the OL. Type III Nbs (red) originate from NECs of the inner proliferation center (IPC), and undergo symmetric self-renewal to produce two identical progenies that retain the identity of neuroblasts and produce lobula plate cells in the OL.
Division Throughout Development
Cell division in neural progenitors and stem cells in the central nervous system has been elucidated using a combination of techniques. Key examples are selective lineage tracing; clonal analysis at single-cell resolution; and in vivo or whole-mount time-lapse imaging of Drosophila neuroblasts (Nbs), embryonic mammalian aRGs, and adult RG-like NSCs (Bossing et al., 1996; Schmidt et al., 1997; Urbach and Technau, 2004; Gao et al., 2014; Taverna et al., 2014; Doe, 2017; Cardenas et al., 2018; Cardenas and Borrell, 2019). Early during gestation, NECs first divide symmetrically and later asymmetrically to produce neuroblasts in the fly and aRGs in the mammalian brain (Figure 1; Gotz and Huttner, 2005; Kriegstein and Alvarez-Buylla, 2009; Brand and Livesey, 2011). In turn, aRGs initially divide symmetrically in the ventricular zone, generating more aRGs. They then switch to producing neurons either through direct neurogenesis, in which the aRG divides asymmetrically to self-renew and generate a neuron, or through indirect neurogenesis to generate various intermediate neural progenitors (INPs) with proliferative capacity, which amplifies neuronal production (Taverna et al., 2014; Cardenas and Borrell, 2019).
The orientation of the cleavage plane determines symmetric vs. asymmetric division (Gotz and Huttner, 2005) and is also important in the proper seeding of future adult NSCs during development (Falk et al., 2017). The indirect mode of asymmetric neurogenesis leads to the formation of an embryonic subventricular zone, where these INPs migrate before the neurons are ultimately produced (Haubensak et al., 2004; Miyata et al., 2004; Noctor et al., 2004). Indirect neurogenesis predominates in humans and other primates with expanded cortices, where additional types of progenitors are formed (Cardenas and Borrell, 2019). In the mouse, this mode is predominant in the neocortex but limited in the olfactory bulb (Cardenas et al., 2018; Cardenas and Borrell, 2019).
Similarly, Drosophila neuroblasts undergo distinct types of cell division to shape different areas of the fly brain (Figures 1C,D). Type I neuroblasts are the most abundant neuroblast in the embryonic central brain (CB) and ventral nerve cord, and in the CB and optic lobes (Figures 2A,A’) of larval Drosophila. Type II neuroblasts exist in sets of eight in each brain lobe. Type I and II neuroblasts have been broadly studied (Doe, 2017). Both types divide asymmetrically; the main difference between them is that Type I neuroblasts produce ganglion mother cells (GMCs) directly, whereas neurogenesis from Type II neuroblasts is mediated by INPs, which then produce GMCs, which ultimately divide symmetrically to generate two neurons or glia (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008). In the embryonic ventral nerve cord, the equivalent of the vertebrate spinal cord, most neuroblasts begin in Type I mode and then they switch to Type 0 mode, where each Type 0 neuroblast divides asymmetrically multiple times and produces progeny that differentiate directly into neurons (Baumgardt et al., 2014). Conversely, in the tip of the outer proliferation center (t-OPC), larval neuroblasts transit from Type 0 to Type I mode to generate diverse cell types in the adult optic lobe (Bertet et al., 2014). A Type III neuroblast has recently been described in the larval optic lobe (Mora et al., 2018). These distal inner proliferation center (d-IPC)-derived neuroblasts show the particularity that, like the SEZ NSCs (Obernier et al., 2018), they undergo symmetric self-renewal to produce two identical progenies that retain neuroblast markers and produce T4 and T5 lobula plate neurons. The identification of these Type III neuroblasts has generated some controversy, and their existence has not been corroborated in other studies (Apitz and Salecker, 2018; Pinto-Teixeira et al., 2018). Future research will be necessary to confirm the presence of this Type III novel neuroblast division mode.
FIGURE 2.
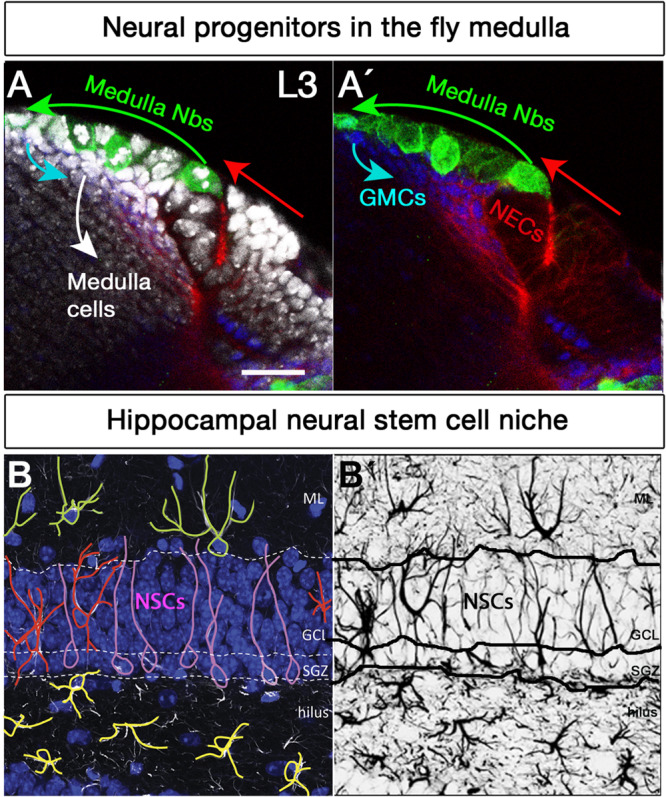
Neural stem cell niches in the Drosophila larval medulla and adult mouse hippocampus. (A,A’) Neural stem cell niche in the larval medulla: (A) neuroepithelial cells (NECs, DE-Cadherin, red) in the outer proliferation center, and their transition into medulla neuroblasts (Nbs, Miranda, green), ganglion mother cells (GMCs, Prospero, blue) and medulla postmitotic cells (DAPI, gray). Arrows indicate transition direction. Scale bar = 20 μm. (B,B’) Neural stem cell niche in the adult mouse hippocampus: (B) Neural stem cells with radial glia-like morphology (pink) are located in the hippocampal dentate gyrus (GFAP, white; DAPI, blue). Their soma sits at the border of the densely packed granule cell layer (GCL), the so-called subgranular zone (SGZ). Their primary process extends across the GCL and reaches the inner molecular layer (ML). NSCs express the markers Sox2, Prominin 1, Nestin (not shown) and glial fibrillary acidic protein GFAP (white), among others, and are mostly quiescent. Surrounding the NSCs a variety of highly branched niche astrocytes located in different layers are found. Those in the ML, GCL, and hilus are shown in green, red, and yellow, respectively. Mature astrocytes do not proliferate and express markers such as glial glutamate transporter 1 (GLT1), S100β (not shown) and GFAP (white), among others. Other niche elements such as blood vessels, INPs and neurons are not shown. (B’) GFAP immunostaining, marking both NSCs and astrocytes with distinctive morphologies.
Division in Adult Neurogenic Niches
Intermediate neural progenitor-mediated amplification of neuronal production also characterizes adult neurogenic niches (Figure 1B). Adult NSCs in the SEZ were long thought to behave like developmental aRGs, predominantly adopting an asymmetric neurogenic division mode as the main strategy to produce differentiated progeny while maintaining a pool of stem cells, before becoming depleted through a terminal symmetric division (Calzolari et al., 2015). However, this view has been recently challenged. Instead, it has been proposed that adult SEZ NSCs engage in two types of coexisting divisions, 20% of them undergoing symmetric self-renewal to contribute to the stem cell reservoir and the other 80% undergoing symmetric consuming divisions that produce transient amplifying progenitors, a type of intermediate neural progenitor (INP/TAP) (Obernier et al., 2018), which, in turn, generate a large variety of olfactory bulb interneurons (Merkle et al., 2007, 2014). This division mode allows the uncoupling of self-renewal and differentiation. The transition between the two NSC pools remains unclear.
In the adult SGZ niche (Figures 2B,B’), most divisions of radial glia-like NSCs are asymmetric (Figure 1B), giving rise to NSCs and dividing progenitors that will later become neurons (Encinas et al., 2011). However, symmetric self-renewing divisions have also been detected by in vivo clonal analysis with genetic marking (Bonaguidi et al., 2011). Recent live-imaging data suggests that radial glia-like NSCs follow a temporal developmental-like program upon activation, comprising an initial proliferative (symmetric) phase followed by a neurogenic (asymmetric) phase (Pilz et al., 2018). Active radial glia-like NSCs likely retain a molecular memory of their history and return to a less dormant quiescent state (Urban et al., 2016; Blomfield et al., 2019; Urban et al., 2019).
Adult NSCs in the SGZ will give rise to only one type of excitatory neuron (the dentate gyrus granule neuron) and, to a lesser extent, will produce local astroglial cells (Suh et al., 2007; Bonaguidi et al., 2011). After undergoing a series of neurogenic asymmetric divisions, radial glia-like NSCs become exhausted and terminally differentiate into mature astrocytes. This gliogenic process is poorly defined but is exacerbated during aging (Encinas et al., 2011; Gebara et al., 2016; Diaz-Moreno et al., 2018; Martin-Suarez et al., 2019) and in pathology (Sierra et al., 2015). Intriguingly, the terminal conversion of radial glia-like NSCs into astrocytes has not yet been captured by live imaging (Pilz et al., 2018), so additional studies are required to elucidate this pathway.
In summary, the presence of INPs is conserved in the fly and in mammalian adult neurogenic niches as a strategy to produce lineages with more neurons, in ways that resemble indirect neurogenesis in the developing mammalian brain. During adulthood, the dynamics of NSCs in the SGZ recapitulate the irreversible switch from a symmetrical self-renewing phase to an asymmetrical neurogenic division phase that characterizes cortical development. Conversely, in the SEZ, two subtypes of NSCs seem to coexist based on their symmetric mode of division. The molecular basis of these division modes, and the number of times adult NSCs divide before depletion, remains elusive.
Intrinsic Control of Neural Progenitor Fate: Temporal and Regional Patterns
Production of Cell Types in Drosophila
Neural stem cells can proliferate and differentiate into various cell types in response to both intrinsic factors and extrinsic cues from their stem cell niche. Spatial patterning plays a key role in acquiring NSCs identities in the developing nervous system. In the early embryo, the combined action of segment polarity, dorso-ventral, columnar and Hox genes act in gradients along the AP and DV axes and form an orthogonal grid that regionally divides the ventral nerve cord neuroectoderm and specifies the neuroblast identity (Table 1; Skeath and Thor, 2003; Technau et al., 2006; Urbach and Technau, 2008; Estacio-Gomez and Diaz-Benjumea, 2014). Interestingly, neuronal subtype specification in the vertebrate hindbrain and spinal cord relies on Hox-dependent regionalization of progenitor and postmitotic cells along the rostrocaudal axis as well in response to opposing morphogen gradients (Sagner and Briscoe, 2019), indicating that spatial colinearity is conserved among vertebrates and Drosophila (Philippidou and Dasen, 2013). In the larval brains, optic lobe neurons originate from two neuroepithelia, called the outer (OPC, Figures 2A,A’) and inner proliferation centers (IPC) (Hofbauer and Campos-Ortega, 1990). The OPC that gives rise to neuroblasts (Egger et al., 2007) is patterned into spatial domains along the anterior–posterior axis by expression of the transcription factors Visual system homeobox 1 (Vsx1), Optix and retinal homeobox (Rx), and the signaling molecules decapentaplegic (dpp), wingless (wg), and hedgehog (hh) (Table 1; Erclik et al., 2008, 2017; Gold and Brand, 2014).
TABLE 1.
Intrinsic factors and glial-derived extrinsic signals influencing cell decisions in Drosophila and mammalian neurogenic niches.
| Names/symbols | Human orthologs | Gene groups and pathways | Description |
| In Drosophila: | |||
| Intrinsic factors | |||
| Abdominal A (abd-A) | HOXA6 HOXC6 | Bithorax complex HOX-like homeobox TFs | Required for segmental identity of the second through eighth abdominal segments |
| Abdominal B (Abd-B) | HOXA11 HOXD11 | Bithorax complex HOX-like homeobox TFs | Specifies the identity of the posterior abdominal segments |
| Antennapedia (Antp) | HOXB7 | Antennapedia complex HOX-like homeobox TFs | Regulates segmental identity in the mesothorax |
| Asense (ase) | ASCL1 ACSL2 | Basic helix-loop-helix TFs | tTF in d-IPC Type I Nbs |
| Atonal (ato) | ATOH7 | Basic helix-loop-helix TFs | tTF in d-IPC Type I and III Nbs |
| Baboon (babo) | TGFβR1 | TGF-β type I receptors | Required for proliferation of Nbs |
| Broad (br) | BTBD18 | C2H2 zinc finger TFs | tTF in thoracic later-born neurons |
| Castor (cas) | CASZ1 | C2H2 zinc finger TFs | tTF in VNC Type I Nbs, thoracic Type I Nbs, CB Type II Nbs and INPs |
| Chronologically inappropriate morphogenesis (Chinmo) | BTBD18 | C2H2 zinc finger TFs | tTF in thoracic early-born neurons |
| Dachshund (dac) | DACH1 | Other DNA binding domain TFs | tTF in d-IPC Type I Nbs |
| Decapentaplegic (dpp) | BMP2 | Bone morphogenetic proteins signaling pathway core components | Patterns the dorsal surface of the embryo and is expressed in a subset of Rx+ tOPC NECs |
| Deformed (Dfd) | HOXC4 | Antennapedia complex HOX-like homeobox TFs | Involved in proper morphological identity of the maxillary segment and the posterior half of the mandibular segment |
| Dichaete (D) | SOX12 SOX14 SOX21 | High mobility group box TFs | tTF in Me, tOPC and d-IPC Type I Nbs, CB Type II Nbs and INPs |
| Distal-less (Dll) | DLX1 DLX6 | NK-like homeobox TFs | Expressed in Wg+ tOPC NECs and tTF in tOPC Type 0 Nbs |
| Dorsal (dl) | RELA RELB | Nuclear factor-κB | Patterns the ventral side of the embryo |
| Drop (Dr) | MSX2 | NK-like homeobox TFs | Specifies the dorsal portion of the neuroectoderm |
| Engrailed (en) | EN1 | NK-like homeobox TFs | Segment polarity gene involved in compartment identity and boundary formation |
| Epidermal growth factor receptor (EGFR) | EGFR | Receptor tyrosine kinases | Required for expansion of OPC NECs and patterns the ventral side of the embryo |
| Eyeless (ey) | PAX6 | Paired homeobox TFs | tTF in Me and tOPC Type I Nbs, CB Type II Nbs and INPs |
| Grainy head (grh) | GRHL1 | Polycomb group recruiters/DNA-binding proteins | tTF in CB Type II Nbs and INPs |
| Gooseberry (gsb) | PAX3 | Paired homeobox TFs | Expressed in segmentally repeating pattern to define the A/P polarity of embryonic segments |
| Hedgehog (hh) | SHH DHH | Hedgehog signaling pathway core component | Marks ventral half of the OPC NECs |
| Homothorax (hth) | MEIS1 MEIS2 | Tale homeobox TFs | tTF in Me Type I Nbs |
| Hunchback (hb) | IKZF5 | C2H2 zinc finger TFs | tTF in VNC Type I Nbs |
| IGF-II mRNA-binding protein (Imp) | IGF2BP1 IGF2BP2 IGF2BP3 | mRNA-binding protein | tTF in thoracic early-born neurons |
| Intermediate neuroblasts defective (ind) | GSX1 | HOX-like homeobox TFs | Specifies the intermediate portion of the neuroectoderm |
| Klumpfuss (Klu) | ZBTB7A | C2H2 zinc finger TFs | tTF in Me Type I Nbs |
| Kruppel (Kr) | BCL6 | C2H2 zinc finger TFs | tTF in VNC Type I Nbs |
| Labial (lab) | HOXA1 HOXB1 | Antennapedia complex HOX-like homeobox TFs | Specifies derivatives of gnathocephalic segments |
| Optix | SIX3 SIX6 | Six/Sine oculis homeobox TFs | Marks the adjacent ventral and dorsal main regions to Vsx1+ OPC NECs |
| Retinal Homeobox (Rx) | RAX | Paired-like homeobox TFs | Marks the tOPC NECs |
| POU domain protein 2 (Pdm2) | POU2F3 | POU homeobox TFs | tTF in VNC Type I Nbs |
| Proboscipedia (pb) | HOXA2 HOXB2 | Antennapedia complex HOX-like homeobox TFs | Required for the formation of labial and maxillary palps |
| Seven up (svp) | NR2F2 | Nuclear receptor TFs | tTF in CB Type II Nbs and INPs |
| Sex combs reduced (Scr) | HOXA5 | Antennapedia complex HOX-like homeobox TFs | Required for labial and first thoracic segment development |
| Sloppy paired 1 (slp 1) | FOXG1 | Fork head box TFs | tTF in Me and tOPC Type I Nbs |
| Syncrip (Syp) | HNRNPR SYNCRIP | mRNA-binding protein | tTF in thoracic later-born neurons |
| Tailless (tll) | NR2E1 | Nuclear receptor TFs | tTF in Me Type I Nbs |
| Ultrabithorax (Ubx) | HOXB6 | Bithorax complex HOX-like homeobox TFs | Controls development of the posterior thoracic and first abdominal segments |
| Ventral nervous system defective (vnd) | NKX2-2 | NK-like homeobox TFs | Specifies the ventral portion of the neuroectoderm |
| Visual system homeobox 1 (Vsx1) | VSX2 | Paired-like homeobox TFs | Expressed in central OPC NECs |
| Wingless (wg) | WNT1 | Wnt-TCF signaling pathway core component | Segment polarity gene involved in controlling the segmentation pattern of embryos by affecting the posterior cells of each parasegment and is expressed in a second subset of Rx+ tOPC NECs |
| Niche/glia-derived factors | |||
| Activin-β (Actβ) | INHBA INHBB | TGFβ superfamily ligand | Secreted from surface glia |
| Anachronism (ana) | – | Glycoprotein | Secreted from cortex glia |
| Dally-like (dlp) | GPC4 | Heparan sulfate proteoglycan Glypican (Membrane tethered) | Secreted from surface glia |
| Drosophila insulin-like peptides 1–8s (dILP1–8s) | IGF1/2 | Insulin-like peptides | Secreted from cortex and surface glia |
| Glass bottom boat (gbb) | BMP7 | Bone morphogenetic proteins signaling pathway ligand | Secreted from surface glia |
| Jelly belly (jeb) | – | Ligand of anaplastic lymphoma kinase | Secreted from glia |
| Spitz (Spi) | TGF-α | EGFR signaling pathway ligand | Secreted from cortex glia |
| Terribly reduced optic lobes (trol) | HSPG2 | Heparan sulfate proteoglycan Perlecan (ECM component) | Secreted from surface glia |
| Symbols/names | Drosophila orthologs | Gene groups and pathways | Description |
| In mammals: | |||
| Intrinsic factors | |||
| Castor zinc finger 1 (CasZ1) | Cas | C2H2 zinc finger TFs | tTF in the specification of late-born cell types in the retina |
| COUP-TF interacting protein 2/B cell leukemia/lymphoma 11B (Citp2/BCL11B) | CG9650 | C2H2 zinc finger TFs | Specification of Layer V neurons |
| Distal-less homeobox 2 (Dlx2) | Dll | NK-like homeobox TFs | Regional specification (embryonic subpallium (LGE and MGE) and lateral postnatal/adult SEZ) |
| Empty spiracles homeobox 1 (Emx1) | ems | NK-like homeobox TFs | Regional specification (embryonic pallium and dorsal postnatal/adult SEZ) |
| Empty Spiracles Homeobox 2 (Emx2) | ems | NK-like homeobox TFs | Dentate gyrus regional identity |
| Eomesodermin (Tbr2) | Doc1 | T-Box TFs | Specification of INPs |
| Eukaryotic translation initiation factor 4E nuclear import factor 1 (4E-T/EIF4ENIF1) | 4E-T | eIF4E/mRNA translation regulator | Translational repression of neuronal specification TFs |
| Fez family zinc finger 2 (Fezf2) | erm | C2H2 zinc finger TFs | Specification of Layer V neurons |
| Forkhead box G1 (Foxg1) | Slp2 | Fork head box TFs | Specification of deep-layer neurons |
| GS homeobox 2 (Gsh2/Gsx2) | ind | HOX-like homeobox TFs | Regional specification (embryonic subpallium (LGE and MGE) and dorsolateral postnatal/adult SEZ) |
| HOP homeobox (Hopx) | – | Homeobox TFs | Dentate gyrus regional identity |
| IKAROS family zinc finger 1 (Ikzf1) | Hb | C2H2 Zinc finger TFs | tTF in the specification of early-born cell types in the cortex and retina |
| Lysine (K)-specific methyltransferase 2A (Mll1/KMT2B) | trx | Trithorax complex | Preservation of regional identity |
| Lymphoid enhancer binding factor 1 (Lef1) | pan | High mobility group box TFs | Dentate gyrus regional identity |
| Neurogenic differentiation 1 (Neurod1) | amos ato | Proneural basic helix-loop-helix TFs | Required for neuronal differentiation |
| Neurogenin 2 (Neurog2) | tap | Proneural basic helix-loop-helix TFs | Drives differentiation of NSCs into neurons |
| NK2 homeobox 1 (Nkx2-1) | scro | NK-like homeobox TFs | Regional specification (embryonic subpallium (MGE), and ventrolateral and medial postnatal/adult SEZ) |
| Nuclear receptor subfamily 2, group F, member 1 (Nr2f1/COUP-TFI) | svp | Nuclear receptor TFs | Specification of upper-layer neurons |
| Paired box 6 (Pax6) | ey | Paired homeobox TFs | Expressed in Radial glia/NSCs; regional specification (embryonic pallium, dorsal postnatal/adult SEZ) |
| POU domain, class 3, transcription factor 3 (Brn1/POU3F3) | vvl | POU homeobox TFs | Specification of upper-layer neurons |
| Pumilio RNA-binding family member 2 (Pum2) | pum | RNA-binding family | Translational repression of neuronal specification TFs |
| Special AT-rich sequence binding protein 2 (Satb2) | dve | CUT homeobox TFs | Specification of upper-layer neurons |
| SRY (sex determining region Y)-box 5 (Sox5) | Sox102F | High mobility group box TFs | Specification of layer VI neurons |
| T-box brain transcription factor 1 (Tbr1) | Doc1 | T-Box TFs | Specification of layer VI neurons |
| Transducin-like enhancer of split 4 (Tle4) | gro | Transcriptional corepressor | Specification of deep-layer neurons |
| Zinc finger E-box binding homeobox 2 (Sip1/Zeb2) | zfh1 | C2H2 zinc finger TFs | Feedback signaling from neurons to progenitors |
| Zinc finger protein of the cerebellum 1 (Zic1) | opa | C2H2 zinc finger TFs | Regional specification (embryonic medial subpallium and septal postnatal/adult SEZ) |
| Niche/Astroglia-derived factors | |||
| Insulin-like growth factor binding protein 6 (IGFBP6) | – | Regulation of insulin-like growth factor receptor signaling pathway | Secreted by non-neurogenic astroglia |
| Insulin-like growth factor 1 (Igf1) | dilp2 | Insulin-like growth factor ligand | Systemic/niche factor |
| Interleukin 1 beta (IL-1β) | – | Cytokine activity | Secreted by SGZ niche astroglia |
| interleukin 6 (IL-6) | – | Cytokine activity | Secreted by SGZ niche astroglia |
| Jagged 1 (Jag1) | Ser | Notch signaling pathway membrane-bound ligand | Expressed by forebrain astroglia |
| Neurogenesin-1/Chordin-like protein 1 (Ng1/Chrdl1) | – | BMP antagonist | Secreted by SGZ niche astroglia |
| Secreted frizzled-related protein 4 (sFRP4) | – | Wnt antagonist | Secreted by OB astroglia |
| Thrombospondin 1 (Thbs1) | Tsp | Glycoprotein (ECM component) | Secreted by forebrain astroglia |
| Wingless-type MMTV integration site family (Wnt3,Wnt7a) | wg | Wnt pathway ligand | Secreted by SGZ/SEZ niche astroglia |
CB, central brain; d-IPC, distal inner proliferation center; ECM, extracellular matrix; LGE, lateral ganglionic eminence; Me, medulla; MGE, medial ganglionic eminence; Nbs, neuroblasts; NECs, neuroepithelial cells; NSCs, neural stem cells; OB, olfactory bulb; SEZ, subependymal zone; SGZ, subgranular zone; TFs, transcription factors; tTF, temporal transcription factor; t-OPC, tip of the outer proliferation center; VNC, ventral nerve cord.
Neural progenitors and NSCs also generate distinct neuronal and glial subtypes over time (Figure 3). This generation of diversity in the developing brain depends on the sequential expression of transcription factors, a phenomenon known as temporal patterning (Doe, 2017; Holguera and Desplan, 2018) that was first observed in the embryonic Drosophila ventral nerve cord (Kambadur et al., 1998; Brody and Odenwald, 2000; Grosskortenhaus et al., 2005; Baumgardt et al., 2009; Doe, 2017). Indeed, temporal patterning is how the neural progenitors in Drosophila generate cellular diversity in different areas of the brain (Morante and Desplan, 2008; Erclik et al., 2017; Konstantinides et al., 2018). For example, Type I neuroblasts of the larval central outer proliferation center (c-OPC) express six different transcription factors as they age: homothorax (hth), klumpfuss (klu), eyeless (ey), sloppy paired 1 (slp1), Dichaete (D), and tailless (tll) (Table 1; Morante et al., 2011; Li et al., 2013; Suzuki et al., 2013). These temporal series are not unique and, for example, larval neuroblasts at the t-OPC express Distal-less (Dll), ey, Slp1, and D (Bertet et al., 2014) while neuroblasts from the d-IPC express asense (ase), D, atonal (ato), and dachshund (dac) (Table 1; Apitz and Salecker, 2018; Mora et al., 2018; Pinto-Teixeira et al., 2018).
FIGURE 3.
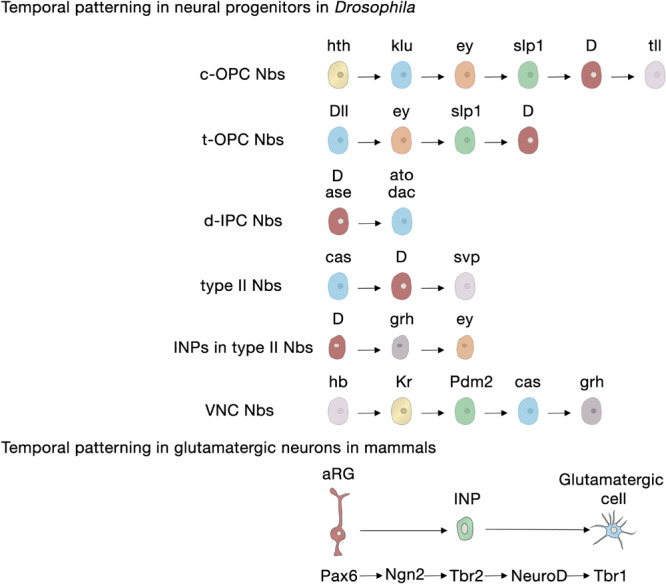
Temporal patterning in neural progenitors in Drosophila and in mammals. Upper panel, transcription factor sequences expressed in embryonic and larval Drosophila neural progenitors. Lower panel, core transcription factor sequence expressed in glutamatergic neurogenesis in the developing cerebral cortex, adult SGZ neurogenesis and adult OB glutamatergic juxtaglomerular interneuron neurogenesis. aRG, apical radial glia; c-OPC, central outer proliferation center; d-IPC, distal inner proliferation center; INP, intermediate neural progenitors; Nbs, neuroblasts; t-OPC, tip of the outer proliferation center; VNC, ventral nerve cord.
Embryonic and larval Type II neuroblasts, and their derived INPs, have adopted a different strategy to increase neural diversity in the adult central complex. In their case, both neuroblasts and INPs express their respective sequences of temporal transcription factors that remain identical as they age from the embryonic to the larval stage. These are castor (cas), D, and seven up (svp) in neuroblasts; and D, grainyhead (grh) and ey in INPs (Table 1; Bayraktar and Doe, 2013; Walsh and Doe, 2017; Alvarez and Diaz-Benjumea, 2018).
However, not all temporal transcription factor sequences remain identical during the embryonic and larval stages. Type I neuroblasts in the embryonic ventral nerve cord and thoracic larval neuroblasts, which delaminate from the embryonic neuroectoderm of the ventral nerve cord, also sequentially express transcription factors, but these sequences differ between the animals’ embryonic and larval lives. Embryonic ventral nerve cord neuroblasts express a complex series of transcription factors [hunchback (hb), krueppel (Kr), POU domain protein 2 (Pdm2), Cas and grh], but thoracic Type I neuroblasts only express Cas and give rise to a series of early-born neurons expressing the BTB transcription factor Chinmo and the RNA-binding protein Imp, and later-born neurons expressing broad (Br) and another RNA-binding protein, Syp (Zhu et al., 2006; Maurange et al., 2008). Svp, an orphan nuclear hormone belonging to the COUP-TF family, triggers this temporal transition from Imp and Chinmo expression to Syp and Br expression by terminating Cas expression (Table 1; Maurange et al., 2008; Ren et al., 2017; Syed et al., 2017; Kanai et al., 2018).
In summary, these studies show that combinatorial inputs from the temporal and spatial axes act together to promote neural diversity in the central nervous system (Figure 4).
FIGURE 4.
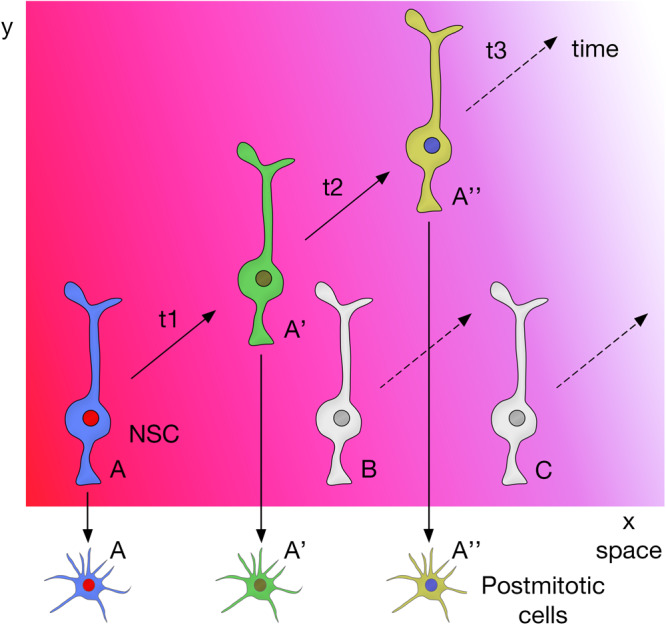
Spatial and temporal patterning on NSCs act together to promote neural diversity in the central nervous system.
Production of Neurons During Development in Mammals
In contrast to Drosophila neural progenitors, the temporal sequence of transcription factors in mammalian NSCs during development is less well understood. In the following section we will focus on the current understanding of cortical glutamatergic projection neuron production, a highly stereotyped process in which early-generated neurons occupy the deep layers of the cortex (V and VI) and late born neurons occupy the upper layers (II–IV) (Angevine and Sidman, 1961; Rakic, 1972). In this lineage, the transcription factor Pax6 specifically expressed in radial glia participates in the patterning of the telencephalon and in instructing neurogenesis through a cascade of transcription factors (Pax6 → Ngn2 → Tbr2 → Tbr1) that are sequentially expressed in radial glia cells, INPs and postmitotic glutamatergic projection neurons (Figure 3; Gotz et al., 1998; Stoykova et al., 2000; Heins et al., 2002; Englund et al., 2005). Tbr2, the transcriptional target of Ngn2, exerts key functions in this cascade: it directly represses Pax6 to favor the transition between cellular stages, from radial glia to INP; it promotes differentiation through the activation of projection neuron transcription factors including Tbr1, Ctip2, and Satb2 (Elsen et al., 2013; Kovach et al., 2013; Mihalas et al., 2016) and it regulates regional identity through the repression of ventral genes (Kovach et al., 2013). In addition, from the well-established gene repression network involved in the specification of deep neurons (Sox5, Tbr1, Fezf2, Ctip2) and upper-layer neurons (Satb2) within the mammalian cortex (Table 1; Britanova et al., 2008; Kwan et al., 2008; McKenna et al., 2011; Srinivasan et al., 2012; Leone et al., 2015), only Fezf2 has been detected in cortical progenitors (Hirata et al., 2004) and the rest are expressed in postmitotic neurons. Upper-layer competence has been linked to deep-layer neurogenesis. It is determined primarily through the repression of Tbr1 and derepression of Fezf2 by Foxg1 to acquire deep-layer identity, and the posterior feedback signal from deep-layer neurons to repress Fezf2/Ctip2 (Toma et al., 2014). Brn1 and Brn2 are also involved in this transition, as upper-layer neurons fail to be generated in Brn1/2 double mutants (Dominguez et al., 2013). Besides transcriptional regulation, another additional regulatory layer to ensure appropriate upper- versus deep-layer cortical neuron identities (Brn1 and Tle4) involves regulation at the translational level by the Pum2/4E-T repressor complex from aRGs that are transcriptionally primed to generate diverse types of neurons (Zahr et al., 2018).
Despite the lack of an unequivocal temporal transcription factor sequence in cortical neural progenitors, the COUP-TF family is still required for the temporal specification of mammalian neural progenitors. Knockdown of COUP-TFI/II in the mouse neocortex causes sustained neurogenesis and prolonged generation of early-born neurons, preventing the onset of gliogenesis (Naka et al., 2008). Other mammalian temporal factors include Ikaros, the ortholog of Hb, which specifies early-born neural identity in the cortex and retina (Elliott et al., 2008), and CasZ1, the ortholog of Cas, which specifies late-born neurons in the mammalian retina (Mattar et al., 2015). These studies reveal an overall conserved strategy regulating temporal identity transitions from flies to mammals and highlight the existence of very precise modes of gene expression control.
Production of New Neurons During Adulthood
The generation of neuronal diversity relies largely on the regional patterning experienced by the cells during development, which, in turn, depends on morphogen gradients. In mice, SEZ NSCs are found in the lateral, medial and dorsal walls of the lateral ventricles, in the rostral migratory stream that directs the new neurons to the olfactory bulb, and in the olfactory bulb core itself (Figure 1B; Merkle et al., 2007; Alonso et al., 2008; Moreno-Estelles et al., 2012; Mizrak et al., 2019). NSCs in these different locations produce a variety of interneuron subtypes in the olfactory bulb due to a mosaic of patterned progenitor domains: ventral NSCs are the source of deep granule neurons and calbindin-positive periglomerular neurons, whereas NSCs in dorsal domains generate superficial granule neurons and dopaminergic periglomerular neurons (Merkle et al., 2007). Ultimately, this complex organization depends on the positional information integrated by the NSCs during development that correlates with the expression of a transcription factor code (Merkle et al., 2007; Fuentealba et al., 2015). For instance, pallial markers such as Emx1 and Pax6 are expressed in the dorsal domains; subpallial markers such as Dlx2, Gsh2, and Nkx2-1 are expressed in the lateral and ventral domains, respectively; and septal ventricular markers like Zic1/2 are expressed in the medial domains (Table 1; Kohwi et al., 2007; Young et al., 2007; Winpenny et al., 2011; Azim et al., 2012; Lopez-Juarez et al., 2013; Merkle et al., 2014; Delgado and Lim, 2015; Fiorelli et al., 2015; Tiveron et al., 2017). Interestingly, a population of glutamatergic juxtaglomerular excitatory OB interneurons that end up in the external plexiform layer are produced in the SEZ through the conserved Pax6 → Ngn2 → Tbr2 → NeuroD → Tbr1 transcription factor sequence (Brill et al., 2009; Roybon et al., 2009a; Figure 3), highlighting the conservation of this cascade for the specification of glutamatergic cell fate.
Cell-intrinsic programs are maintained even when ventral SEZ progenitors are heterotopically grafted into the dorsal SEZ or when they are cultured in vitro (Merkle et al., 2007), although cells can still switch fate when a single dorsal or lateral SEZ-enriched transcription factor is overexpressed (Azim et al., 2015). This indicates that the positional identity acquired by SEZ NSCs during development becomes independent of morphogen signaling in the adult brain. Recent data show that the preservation of this positional identity during adulthood involves a cell-autonomous epigenetic memory mechanism that depends on the chromatin regulator mixed-lineage leukemia 1 (Mll1) (Delgado et al., 2020), the mammalian homolog of the Drosophila gene trithorax (trx). Trx proteins are a heterogeneous group with varied activities mainly related to chromatin modification and remodeling to maintain active states and, thus, counteract the silencing activity of the polycomb group proteins (Piunti and Shilatifard, 2016; Schuettengruber et al., 2017). Classical genetics approaches in Drosophila revealed that both groups of proteins preserve the expression of Hox genes that determine anterior–posterior identities, although they are not involved in their induction (Geisler and Paro, 2015). In ventral SEZ NSCs, Trx/Mll1 is similarly required for maintaining Nkx2-1 expression, yet it does not participate in the initial induction of this transcription factor, which ultimately depends on the ventral morphogen sonic hedgehog (Shh) (Delgado et al., 2020).
Intriguingly, embryonic progenitors and adult NSCs located at equivalent sites and patterned similarly produce different progenies; for instance, aRGs in the dorsal pallium generate excitatory cortical neurons, whereas adult dorsal SEZ NSCs, related to these aRGs, produce inhibitory olfactory bulb interneurons (Fuentealba et al., 2015). Furthermore, there is a temporal pattern in the production of different subtypes of olfactory bulb interneurons, suggesting that different NSC domains dominate neuronal production at specific time points (Batista-Brito et al., 2008). The molecular program underlying these temporal switches has not been completely defined yet and future lineage tracing and scRNA-seq studies are required to solve the intricate codes that define SEZ NSC heterogeneity in time and space.
For producing new granule neurons in the adult hippocampal SGZ, the patterning information would also be acquired early during embryogenesis and preserved across development and into adulthood (Morales and Mira, 2019). SGZ NSCs are a continuum derived from progenitor cells that migrate out of the dentate neuroepithelium expressing the homeodomain-only protein, Hopx (Li et al., 2015; Berg et al., 2019) and the transcription factors Pax6 and Sox9 (Nelson et al., 2020). Recent data suggest that an early cohort of Tbr2 INPs expressing Notch ligands pioneers the subsequent NSC migration toward the newly formed outer (abventricular) SGZ niche, keeping neighboring NSCs in an undifferentiated state through Notch signaling (Nelson et al., 2020). The regional identity of SGZ NSCs is markedly influenced by a number of other transcription factors (Hatami et al., 2018), including Emx2 and Lef1 (Pellegrini et al., 1996; Galceran et al., 2000; Oldekamp et al., 2004), as well as by Wnt and bone morphogenetic protein signaling from the adjacent cortical hem (Li and Pleasure, 2007). Interestingly, the transcription factor sequence Pax6 → Ngn2 → Tbr2 → NeuroD → Tbr1 observed in developmental glutamatergic neurogenesis in the cortex is conserved along the lineage progression of adult SGZ neurogenesis (Figure 3; Hodge et al., 2008; Roybon et al., 2009b). Tbr2 likely facilitates the progression from the NSC to the INP state by directly binding and repressing Sox2 (Hodge et al., 2012).
Hopx-positive dentate progenitors upregulate cell membrane genes over development, pointing to a transition from an intrinsic mode of regulation in embryonic radial glia to an extrinsic, niche-dependent mode in adult RG-like NSCs (Berg et al., 2019). Similarly, sc-RNAseq data of adult hippocampal quiescent NSCs confirm that adult NSCs are enriched in genes encoding membrane-related proteins, pointing to an enhanced niche signaling integration capacity (Shin et al., 2015; Artegiani et al., 2017; Hochgerner et al., 2018). Furthermore, at least for some signaling pathways such as Notch, there is a switch in the expression of receptor subtypes in NSCs during the transition from development to adulthood that could influence the outcome of the signaling (Nelson et al., 2020).
Extrinsic Control of Neural Progenitors
Neural progenitors and NSCs are also influenced by the local microenvironment where they reside, which determines their fate and self-renewal capacity (Morrison and Spradling, 2008; Siegenthaler et al., 2009; Lehtinen and Walsh, 2011; Lehtinen et al., 2011; Siegenthaler and Pleasure, 2011). The microenvironments in different brain regions and stages of development can be quite diverse, and this can be exploited as a strategy to generate cellular diversity.
Recent studies in the developing mouse forebrain have shown that transmission of temporal birthmarks from mother apical progenitors to their daughter cells fades with differentiation as environmental factors predominate (Vitali et al., 2018; Telley et al., 2019). A good example of this process occurs in developing layer IV neurons, whose final molecular identity and function is instructed by thalamocortical inputs (Pouchelon et al., 2014). Another remarkable illustration of microenvironmental influences is the production of signaling factors in postmitotic neurons. For example, neurotrophin-3 is regulated by the transcription repressor Sip1 (Zeb2), which feeds back to progenitors to modulate the timing of two cell fate switches during corticogenesis: neurogenesis to gliogenesis, and deep- to upper-layer neuron transitions (Seuntjens et al., 2009; Parthasarathy et al., 2014).
Classical transplantation experiments established that local environmental cues change over time and can control the competence of embryonic mammalian neural progenitors to produce neurons of different layers (McConnell, 1991). However, very few of these niches and their molecular signals have been characterized. An example of one that has is the extrinsic signaling from non-neural tissues, which has been proposed to co-ordinate neural progenitor and NSC proliferation in the developing mammalian forebrain (Siegenthaler and Pleasure, 2010; Lehtinen and Walsh, 2011; Lehtinen et al., 2011). In particular, retinoic acid signaling from the meninges was established to be important for switching from symmetric to asymmetric neurogenic proliferation in Foxc1-knockout mice (Siegenthaler et al., 2009). Additionally, meningeal cells organize the pial basement membrane, an extracellular matrix enriched in a variety of growth factors that covers the brain and might be involved in signaling at the basal side (Siegenthaler and Pleasure, 2011). Meanwhile, the apical side of embryonic and adult neural progenitors are in contact with the cerebrospinal fluid and the vascular system, and therefore might be influenced by extrinsic cues released from these non-neural tissues to regulate their self-renewal, differentiation and migratory capacity (Sawamoto et al., 2006; Tavazoie et al., 2008; Lehtinen and Walsh, 2011; Gato et al., 2019; Fame and Lehtinen, 2020). Indeed, isolated mouse apical progenitors cultured in vitro show only limited progression of temporal gene expression (Okamoto et al., 2016), suggesting that temporal progression in mammalian cortical progenitors may also require cell-extrinsic cues. This does not, however, seem to be the case with embryonic fruit fly neuroblasts. Isolated embryonic neuroblasts cultured in vitro express the same temporal sequences as observed in vivo (Grosskortenhaus et al., 2005), suggesting that the timing of temporal identity transitions in embryonic neuroblasts is regulated by an intrinsic mechanism (Doe, 2017), probably due to the short duration of the embryonic stage (24 h) and rapid divisions of embryonic neuroblasts compared to mammals.
Adult Neurogenic Niches in Rodents
Heterotopic transplantation experiments demonstrated that cell-intrinsic programs tightly regulate SEZ NSCs (Merkle et al., 2007). Yet, despite all the intrinsic determinants, SGZ NSCs are highly plastic and their fate can be redirected when exposed to an adequate milieu (Suhonen et al., 1996). For instance, when transplanted into the rostral migratory stream, their progeny migrates to the olfactory bulb and differentiates into dopaminergic neurons (a non-hippocampal subtype), but when grafted into a non-neurogenic area, such as the cerebellum, they do not generate neurons (Suhonen et al., 1996). Similarly, grafting clonally expanded non-neurogenic NSCs from the spinal cord to the dentate gyrus results in the cells differentiating into new neurons that resemble resident hippocampal granule neurons, whereas cells grafted into non-neurogenic areas of the hippocampus either remain undifferentiated or give rise primarily to NG2 oligodendrocyte precursors, but not to neurons (Shihabuddin et al., 2000). This indicates that the grafted cells are instructed by local signals emanating from the neurogenic niche. A variety of factors released form the niche vasculature, choroid plexus, cerebrospinal fluid, ependymal cells, and local interneurons influence adult NSCs. Their role falls beyond the scope of the next section, that will instead focus on glial-derived niche signals.
The Prevalent Role of Glial-Derived Signals
Glial-Derived Signaling in Drosophila
Extrinsic signals from glia play important roles in microenvironments where they can act directly on different biological processes (Figure 5A). In larval Drosophila brains, cortex glia are the source of Spitz, a homolog of transforming growth factor-alpha, which is required for the initial proliferation of NECs in the medulla through the activation of the EGFR (Morante et al., 2013). Other glial-derived signals that regulate neuroblast proliferation in the developing larval brain include Activin-β (Act-β), via its receptor baboon (babo) (Zhu et al., 2008), dally-like (dlp), a heparan sulfate proteoglycan, and glass-bottom boat (gbb), a BMP homolog (Kanai et al., 2018). Surface and cortex glia also provide Drosophila insulin-like peptides (dILPs) in response to systemic nutritional cues (Chell and Brand, 2010; Sousa-Nunes et al., 2011; Lanet et al., 2013; Otsuki and Brand, 2017, 2018), and components of the extracellular matrix, such as trol, a secreted heparan sulfate proteoglycan Perlecan (Voigt et al., 2002; Park et al., 2003) required for timely reactivation of quiescent larval neuroblasts in the ventral nerve cord and CB. Conversely, secretion of the anachronism (ana) glycoprotein also affects the initiation of neuroblast proliferation, but in the opposite way: in ana mutants, mitotically regulated neuroblasts begin cell division too early (Ebens et al., 1993). Glial cells are not only necessary to regulate the proliferation of neuroblasts, but also protect the proliferation of neuroblasts under conditions of hypoxia and oxidative stress (Bailey et al., 2015), or nutrient restriction, through positive regulation of the jelly belly (jeb) secretion to stimulate Anaplastic lymphoma kinase (Alk)-dependent PI3K signaling in neuroblasts and protects their proliferation (Cheng et al., 2011). Release of dILPs by wrapping glia also stimulates lamina precursor cells to differentiate into lamina neurons in the visual system (Fernandes et al., 2017). These examples show the important function of glial-derived signaling within the stem cell niche in flies. Recent studies have also highlighted that establishing the correct niche architecture is necessary for encasing neural progenitor cells and NSCs and allowing them to divide (Morante et al., 2013; Speder and Brand, 2018).
FIGURE 5.
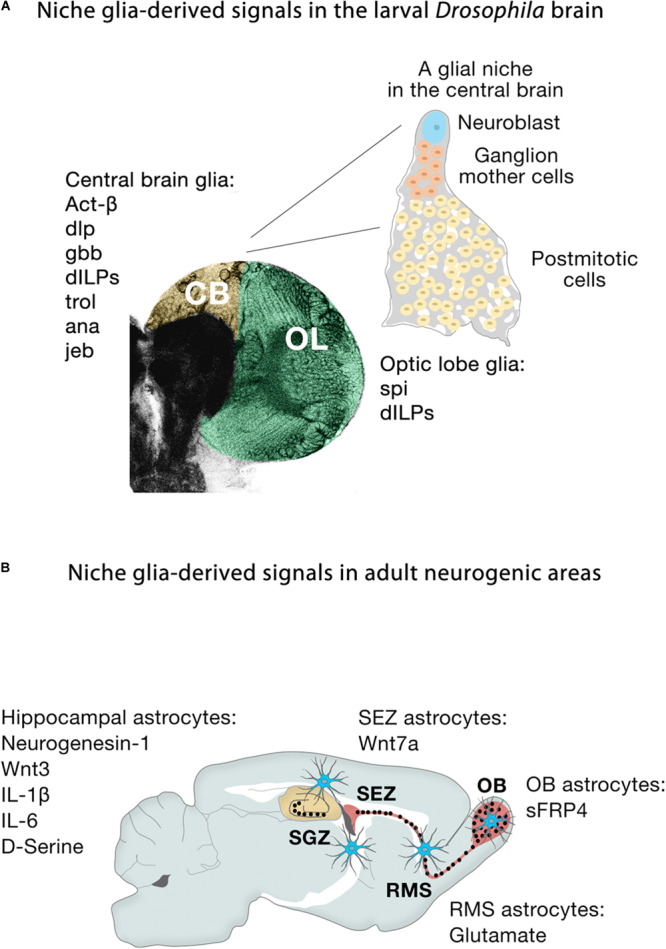
Niche glia-derived signals in the larval Drosophila brain and adult neurogenic areas. (A) Schematic representation of niche glia-derived signals in the larval central brain (CB, yellow) and optic lobe (OL, green), and schematic illustration of a GFP-labeled cortex glia Flp-out clone (gray) showing how the cortex glia ensheaths a Type I Nb (blue), ganglion mother cells (orange) and its neural progeny (yellow) in the central brain. (B) Schematic representation of niche astroglia-derived signals in the adult neurogenic areas. Astrocytes (blue cells) are regionally specified and secrete a variety of local signals to regulate neurogenesis. Astrocytes from the SEZ (the site of neuronal birth) secrete Wnt7a while olfactory bulb astrocytes (OB, the site of neuronal integration) express the Wnt antagonist sFRP4. Hippocampal astrocytes specifically release Wnt3, IL-1β, IL-6, and neurogenesin-1. Rostral migratory stream (RMS) astrocytes that ensheath migratory neuroblasts en route to the OB and hippocampal astrocytes also modulate adult neurogenesis through the supply of neurotransmitters (glutamate, D-serine).
Niche Astroglial-Derived Signaling and Cell Heterogeneity in Rodents
During embryonic development in mammals, neurogenesis precedes gliogenesis, so the new neurons are generated in environments devoid of mature astroglial cells. However, in the adult, astrocytes are distributed throughout the neurogenic niches (Figure 5B), where they play fundamental roles. Gene expression profiling shows that astrocytes are heterogeneous across, and even within, regions (Doyle et al., 2008; Morel et al., 2017, 2019; Boisvert et al., 2018; Batiuk et al., 2020; Bayraktar et al., 2020), in line with the remarkable morphological and functional heterogeneity of astroglia throughout the brain (Emsley and Macklis, 2006; Matyash and Kettenmann, 2010). Specifically, co-culture experiments demonstrate that niche astrocytes (but not those from non-neurogenic areas such as the adult spinal cord) are regulators of all the stages along the neurogenic cascade, supporting NSC self-renewal, proliferation and neuronal differentiation of precursor cells through the release of soluble and/or cell membrane-bound factors (Lim and Alvarez-Buylla, 1999; Song et al., 2002). For instance, the molecular signatures of astroglia from the SEZ (the site of neuronal birth) and the olfactory bulb (the site of neuronal differentiation and maturation) are remarkably different: Wnt7a secreted by SEZ astrocytes promotes symmetric NSC self-renewing divisions, whereas its antagonist sFRP4 expressed by olfactory bulb astrocytes presumably blocks the activation of local olfactory bulb NSCs (Moreno-Estelles et al., 2012). In the SGZ, Wnt3, IL-1β, and IL-6 and the BMP antagonist neurogenesin-1 released by hippocampal astrocytes enhance neuronal fate specification and differentiation (Ueki et al., 2003; Lie et al., 2005; Barkho et al., 2006; Figure 5B). Non-neurogenic astrocytes instead secrete IGFBP6 (Barkho et al., 2006), which negatively regulates insulin growth factor (IGF)-II, an important player (together with IGF-I) in adult hippocampal neurogenesis (Bracko et al., 2012; Nieto-Estevez et al., 2016).
Hippocampal astrocytes can also negatively affect neurogenesis possibly through the cell membrane-bound Notch ligand, Jagged1 as suggested from in vitro experiments performed with astrocytes and NSCs isolated from the postnatal forebrain (Wilhelmsson et al., 2012). Astrocytes modulate the late phases of adult neurogenesis through the supply of neurotransmitters such as glutamate and D-serine. Blocking their exocytotic vesicular release from astrocytes or knocking-out NMDA receptors in neuroblasts compromises neuroblast survival during migration from the SEZ toward the olfactory bulb (Platel et al., 2010) and reduces dendritic spine maturation and synaptic integration of adult-born hippocampal neurons (Sultan et al., 2015). The extracellular matrix protein trombospondin-1 secreted by astrocytes, involved in astrocyte-induced synaptogenesis, is probably an astrocyte-derived factor that affects several steps of the neurogenic process, although its expression is not restricted to the niches (Lu and Kipnis, 2010). Other astrocyte-related factors regulating neuronal function throughout the brain may also contribute to the adequate functionality of the adult-born neurons once these neurons become mature, fully integrated and indistinguishable from their embryonically born counterparts.
Astrocytes may be more diverse than anticipated, with differences not only between distant regions such as the SEZ niche and olfactory bulb (Moreno-Estelles et al., 2012), but also possibly even within regions (Figures 2B,B’), including the hippocampal niche (Beckervordersandforth et al., 2014; Schneider et al., 2019). The advent of transcriptomics is unveiling the molecular basis for the plurality of astrocytes (Matias et al., 2019). A large scRNA-seq study uncovered seven regionally restricted astrocyte subtypes in the brain that correspond to developmental boundaries (Zeisel et al., 2018), while a more recent scRNA-seq dataset identified five distinct astrocyte subtypes in the cortex and hippocampus that are distinguished on the basis of their gene expression signature and topographic distribution (Batiuk et al., 2020). Another recent single-nucleus RNA-seq study of the hippocampus confirmed the existence of a complex atlas of astroglial cells with a continuous range of profiles and revealed the existence of an additional astrocyte state associated to aging and Alzheimer’s (Habib et al., 2020). Other studies have also uncovered the existence of intra-cortical astroglial heterogeneity and highlight layer-specific interactions between neurons and astrocytes (Lanjakornsiripan et al., 2018; Morel et al., 2019; Bayraktar et al., 2020), so it is conceivable that this holds true for other zones. A putative enrichment of specific astrocytic subtypes in defined subdomains of adult neurogenic areas may have interesting implications for our understanding of the functional interactions taking place between astrocytes, NSCs, newly born neurons, and pre-existing neurons.
Future Perspectives of NSC Research
Single-cell RNA sequencing technology is revolutionizing how cell types are identified in developing and adult brains, providing astonishing insight into cellular diversity in specific regions including the optic lobe (Konstantinides et al., 2018), antennal lobe (Li et al., 2017), ventral nerve cord (Allen et al., 2020), and CB (Croset et al., 2018). In organisms with relatively simple brains, such as flies, whose brains consist of approximately 150,000 cells, the whole adult brain (Davie et al., 2018) can be investigated using this technology. And in animals with larger brains, like mice, the hypothalamus (Campbell et al., 2017; Romanov et al., 2017), lateral geniculate nucleus (Kalish et al., 2018), midbrain (La Manno et al., 2016), somatosensory cortex and hippocampus (Zeisel et al., 2015), visual cortex and anterior lateral motor cortex (Tasic et al., 2016, 2018); have been examined, among other areas. Some studies employ transgenic line–based sampling strategies and retrograde labeling of projection neurons to further assess the correspondence between the scRNA-seq identified cell types and specific cellular functions, including differential electrophysiological properties and long-range projection specificity (Zeisel et al., 2015; Tasic et al., 2016, 2018; Economo et al., 2018). Furthermore, spatiotemporal gene expression analysis of scRNA-seq datasets is revealing in unprecedented detail the intricate developmental trajectories that brain cells undergo through differentiation from embryonic neural progenitors (Telley et al., 2016, 2019; Nowakowski et al., 2017).
In the adult mouse, scRNA-seq studies have improved our understanding of the cellular composition of neurogenic niches. They have identified cellular states along the neurogenic lineage of the SEZ (Llorens-Bobadilla et al., 2015; Luo et al., 2015; Dulken et al., 2017; Zywitza et al., 2018; Mizrak et al., 2019) and the SGZ of the hippocampal dentate gyrus (Shin et al., 2015; Artegiani et al., 2017; Hochgerner et al., 2018). In Drosophila, a pioneer single-cell transcriptomic study has established a molecular cell atlas of the first instar larval brain (Brunet Avalos et al., 2019), identifying neurons expressing distinct neurotransmitters, neuromodulators and neuropeptides; neural progenitor cells; glial cells of different types; undifferentiated neurons; and non-neural cells.
Future studies will complement current knowledge and allow us to establish a detailed catalog of brain cell types (Ecker et al., 2017; Regev et al., 2017), as well as to fully map the cellular, molecular and spatial organization of the complex niche networks that maintain and regulate the division capacity of neural progenitors and stem cells. New data are already starting to shed light on the intrinsic epigenetic mechanisms that preserve regional identities in NSCs as the brain increases its complexity from development to adulthood. Additional studies are needed to clarify if there are glial subtypes in the niches and, if so, to analyze their possible role in regulating the different stages of the neurogenic cascade. It will be equally interesting to explore whether adult NSCs contribute to the intra-regional heterogeneity of astroglia and to generating their own local glial niche. Finally, we need to better understand how extrinsic cues received by neural progenitors are effectively interpreted to produce the correct intrinsic responses, as little is known about the specifics of these interactions.
Author Contributions
HM and JM conceived the structure and content, and wrote the manuscript. Both authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Sergio Javaloy for helping with illustrations and Lucia Casares for providing confocal images. We apologize to those whose work we did not cite due to focus and space limitations.
Abbreviations
- aRG
apical radial glia
- CB
central brain
- GMC
ganglion mother cell
- INP
intermediate neural progenitor
- IPC
inner proliferation center
- Nb
neuroblast
- NEC
neuroepithelial cell
- NSC
neural stem cell
- OPC
outer proliferation center
- scRNA-seq
single-cell RNA sequencing
- SEZ
subependymal zone
- SGZ
subgranular zone.
Footnotes
Funding. This work was supported by the Spanish State Research Agency (BFU2016-76295-R) co-financed by the European Regional Development Fund (ERDF), and the Severo Ochoa Program for Centres of Excellence in R&D (SEV-2017-0723) to JM, and by the Spanish MICINN grant (SAF2015-70433-R) and PROMETEO/2018/055 grant from Generalitat Valenciana to HM. We acknowledge support for the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
References
- Allen A. M., Neville M. C., Birtles S., Croset V., Treiber C. D., Waddell S., et al. (2020). A single-cell transcriptomic atlas of the adult Drosophila ventral nerve cord. eLife 9:e54074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso M., Ortega-Perez I., Grubb M. S., Bourgeois J. P., Charneau P., Lledo P. M. (2008). Turning astrocytes from the rostral migratory stream into neurons: a role for the olfactory sensory organ. J. Neurosci. 28 11089–11102. 10.1523/jneurosci.3713-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez J. A., Diaz-Benjumea F. J. (2018). Origin and specification of type II neuroblasts in the Drosophila embryo. Development 145:dev158394. 10.1242/dev.158394 [DOI] [PubMed] [Google Scholar]
- Angevine J. B., Jr., Sidman R. L. (1961). Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature 192 766–768. 10.1038/192766b0 [DOI] [PubMed] [Google Scholar]
- Apitz H., Salecker I. (2018). Spatio-temporal relays control layer identity of direction-selective neuron subtypes in Drosophila. Nat. Commun. 9:2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artegiani B., Lyubimova A., Muraro M., van Es J. H., van Oudenaarden A., Clevers H. (2017). A Single-Cell RNA sequencing study reveals cellular and molecular dynamics of the hippocampal neurogenic niche. Cell Rep. 21 3271–3284. 10.1016/j.celrep.2017.11.050 [DOI] [PubMed] [Google Scholar]
- Azim K., Fiorelli R., Zweifel S., Hurtado-Chong A., Yoshikawa K., Slomianka L., et al. (2012). 3-dimensional examination of the adult mouse subventricular zone reveals lineage-specific microdomains. PLoS One 7:e49087. 10.1371/journal.pone.0049087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azim K., Hurtado-Chong A., Fischer B., Kumar N., Zweifel S., Taylor V., et al. (2015). Transcriptional hallmarks of heterogeneous neural stem cell niches of the subventricular zone. Stem Cells 33 2232–2242. 10.1002/stem.2017 [DOI] [PubMed] [Google Scholar]
- Bailey A. P., Koster G., Guillermier C., Hirst E. M., MacRae J. I., Lechene C. P., et al. (2015). Antioxidant role for lipid droplets in a stem cell niche of Drosophila. Cell 163 340–353. 10.1016/j.cell.2015.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkho B. Z., Song H., Aimone J. B., Smrt R. D., Kuwabara T., Nakashima K., et al. (2006). Identification of astrocyte-expressed factors that modulate neural stem/progenitor cell differentiation. Stem Cells Dev. 15 407–421. 10.1089/scd.2006.15.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista-Brito R., Close J., Machold R., Fishell G. (2008). The distinct temporal origins of olfactory bulb interneuron subtypes. J. Neurosci. 28 3966–3975. 10.1523/jneurosci.5625-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batiuk M. Y., Martirosyan A., Wahis J., de Vin F., Marneffe C., Kusserow C., et al. (2020). Identification of region-specific astrocyte subtypes at single cell resolution. Nat. Commun. 11:1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgardt M., Karlsson D., Salmani B. Y., Bivik C., MacDonald R. B., Gunnar E., et al. (2014). Global programmed switch in neural daughter cell proliferation mode triggered by a temporal gene cascade. Dev. Cell 30 192–208. 10.1016/j.devcel.2014.06.021 [DOI] [PubMed] [Google Scholar]
- Baumgardt M., Karlsson D., Terriente J., Diaz-Benjumea F. J., Thor S. (2009). Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell 139 969–982. 10.1016/j.cell.2009.10.032 [DOI] [PubMed] [Google Scholar]
- Bayraktar O. A., Bartels T., Holmqvist S., Kleshchevnikov V., Martirosyan A., Polioudakis D., et al. (2020). Astrocyte layers in the mammalian cerebral cortex revealed by a single-cell in situ transcriptomic map. Nat. Neurosci. 23 500–509. 10.1038/s41593-020-0602-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar O. A., Doe C. Q. (2013). Combinatorial temporal patterning in progenitors expands neural diversity. Nature 498 449–455. 10.1038/nature12266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckervordersandforth R., Deshpande A., Schaffner I., Huttner H. B., Lepier A., Lie D. C., et al. (2014). In vivo targeting of adult neural stem cells in the dentate gyrus by a split-cre approach. Stem Cell Rep. 2 153–162. 10.1016/j.stemcr.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckervordersandforth R., Tripathi P., Ninkovic J., Bayam E., Lepier A., Stempfhuber B., et al. (2010). In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell 7 744–758. 10.1016/j.stem.2010.11.017 [DOI] [PubMed] [Google Scholar]
- Bello B. C., Izergina N., Caussinus E., Reichert H. (2008). Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 3:5. 10.1186/1749-8104-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. A., Su Y., Jimenez-Cyrus D., Patel A., Huang N., Morizet D., et al. (2019). A common embryonic origin of stem cells drives developmental and adult neurogenesis. Cell 177:e615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertet C., Li X., Erclik T., Cavey M., Wells B., Desplan C. (2014). Temporal patterning of neuroblasts controls notch-mediated cell survival through regulation of hid or reaper. Cell 158 1173–1186. 10.1016/j.cell.2014.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomfield I. M., Rocamonde B., Masdeu M. D. M., Mulugeta E., Vaga S., van den Berg D. L., et al. (2019). Id4 promotes the elimination of the pro-activation factor Ascl1 to maintain quiescence of adult hippocampal stem cells. eLife 8:e48561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert M. M., Erikson G. A., Shokhirev M. N., Allen N. J. (2018). The aging astrocyte transcriptome from multiple regions of the mouse brain. Cell Rep. 22 269–285. 10.1016/j.celrep.2017.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrini M., Fulmore C. A., Tartt A. N., Simeon L. R., Pavlova I., Poposka V., et al. (2018). Human hippocampal neurogenesis persists throughout aging. Cell Stem Cell 22 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaguidi M. A., Wheeler M. A., Shapiro J. S., Stadel R. P., Sun G. J., Ming G. L., et al. (2011). In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell 145 1142–1155. 10.1016/j.cell.2011.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone J. Q., Doe C. Q. (2008). Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev. Neurobiol. 68 1185–1195. 10.1002/dneu.20648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossing T., Udolph G., Doe C. Q., Technau G. M. (1996). The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev. Biol. 179 41–64. 10.1006/dbio.1996.0240 [DOI] [PubMed] [Google Scholar]
- Bowman S. K., Rolland V., Betschinger J., Kinsey K. A., Emery G., Knoblich J. A. (2008). The tumor suppressors brat and numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell 14 535–546. 10.1016/j.devcel.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracko O., Singer T., Aigner S., Knobloch M., Winner B., Ray J., et al. (2012). Gene expression profiling of neural stem cells and their neuronal progeny reveals IGF2 as a regulator of adult hippocampal neurogenesis. J. Neurosci. 32 3376–3387. 10.1523/jneurosci.4248-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Livesey F. J. (2011). Neural stem cell biology in vertebrates and invertebrates: more alike than different? Neuron 70 719–729. 10.1016/j.neuron.2011.05.016 [DOI] [PubMed] [Google Scholar]
- Brill M. S., Ninkovic J., Winpenny E., Hodge R. D., Ozen I., Yang R., et al. (2009). Adult generation of glutamatergic olfactory bulb interneurons. Nat. Neurosci. 12 1524–1533. 10.1038/nn.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova O., de Juan Romero C., Cheung A., Kwan K. Y., Schwark M., Gyorgy A., et al. (2008). Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron 57 378–392. 10.1016/j.neuron.2007.12.028 [DOI] [PubMed] [Google Scholar]
- Brody T., Odenwald W. F. (2000). Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Dev. Biol. 226 34–44. 10.1006/dbio.2000.9829 [DOI] [PubMed] [Google Scholar]
- Brunet Avalos C., Maier G. L., Bruggmann R., Sprecher S. G. (2019). Single cell transcriptome atlas of the Drosophila larval brain. eLife 8:e50354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzolari F., Michel J., Baumgart E. V., Theis F., Gotz M., Ninkovic J. (2015). Fast clonal expansion and limited neural stem cell self-renewal in the adult subependymal zone. Nat. Neurosci. 18 490–492. 10.1038/nn.3963 [DOI] [PubMed] [Google Scholar]
- Campbell J. N., Macosko E. Z., Fenselau H., Pers T. H., Lyubetskaya A., Tenen D., et al. (2017). A molecular census of arcuate hypothalamus and median eminence cell types. Nat. Neurosci. 20 484–496. 10.1038/nn.4495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas A., Borrell V. (2019). Molecular and cellular evolution of corticogenesis in amniotes. Cell Mol. Life Sci. 77 1435–1460. 10.1007/s00018-019-03315-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas A., Villalba A., de Juan Romero C., Pico E., Kyrousi C., Tzika A. C., et al. (2018). Evolution of cortical neurogenesis in amniotes controlled by robo signaling levels. Cell 174 590–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chell J. M., Brand A. H. (2010). Nutrition-responsive glia control exit of neural stem cells from quiescence. Cell 143 1161–1173. 10.1016/j.cell.2010.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L. Y., Bailey A. P., Leevers S. J., Ragan T. J., Driscoll P. C., Gould A. P. (2011). Anaplastic lymphoma kinase spares organ growth during nutrient restriction in Drosophila. Cell 146 435–447. 10.1016/j.cell.2011.06.040 [DOI] [PubMed] [Google Scholar]
- Croset V., Treiber C. D., Waddell S. (2018). Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. eLife 7:e34550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie K., Janssens J., Koldere D., De Waegeneer M., Pech U., Kreft L., et al. (2018). A single-cell transcriptome atlas of the aging Drosophila brain. Cell 174 982–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado R. N., Lim D. A. (2015). Embryonic Nkx2.1-expressing neural precursor cells contribute to the regional heterogeneity of adult V-SVZ neural stem cells. Dev. Biol. 407 265–274. 10.1016/j.ydbio.2015.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado R. N., Mansky B., Ahanger S. H., Lu C., Andersen R. E., Dou Y., et al. (2020). Maintenance of neural stem cell positional identity by mixed-lineage leukemia 1. Science 368 48–53. 10.1126/science.aba5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Moreno M., Armenteros T., Gradari S., Hortiguela R., Garcia-Corzo L., Fontan-Lozano A., et al. (2018). Noggin rescues age-related stem cell loss in the brain of senescent mice with neurodegenerative pathology. Proc. Natl. Acad. Sci. U.S.A. 115 11625–11630. 10.1073/pnas.1813205115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C. Q. (2017). Temporal patterning in the Drosophila CNS. Annu. Rev. Cell Dev. Biol. 33 219–240. 10.1146/annurev-cellbio-111315-125210 [DOI] [PubMed] [Google Scholar]
- Dominguez M. H., Ayoub A. E., Rakic P. (2013). POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb. Cortex 23 2632–2643. 10.1093/cercor/bhs252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. P., Dougherty J. D., Heiman M., Schmidt E. F., Stevens T. R., Ma G., et al. (2008). Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135 749–762. 10.1016/j.cell.2008.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulken B. W., Leeman D. S., Boutet S. C., Hebestreit K., Brunet A. (2017). Single-cell transcriptomic analysis defines heterogeneity and transcriptional dynamics in the adult neural stem cell lineage. Cell Rep. 18 777–790. 10.1016/j.celrep.2016.12.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebens A. J., Garren H., Cheyette B. N., Zipursky S. L. (1993). The Drosophila anachronism locus: a glycoprotein secreted by glia inhibits neuroblast proliferation. Cell 74 15–27. 10.1016/0092-8674(93)90291-w [DOI] [PubMed] [Google Scholar]
- Ecker J. R., Geschwind D. H., Kriegstein A. R., Ngai J., Osten P., Polioudakis D., et al. (2017). The BRAIN initiative cell census consortium: lessons learned toward generating a comprehensive brain cell atlas. Neuron 96 542–557. 10.1016/j.neuron.2017.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economo M. N., Viswanathan S., Tasic B., Bas E., Winnubst J., Menon V., et al. (2018). Distinct descending motor cortex pathways and their roles in movement. Nature 563 79–84. 10.1038/s41586-018-0642-9 [DOI] [PubMed] [Google Scholar]
- Egger B., Boone J. Q., Stevens N. R., Brand A. H., Doe C. Q. (2007). Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev. 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott J., Jolicoeur C., Ramamurthy V., Cayouette M. (2008). Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron 60 26–39. 10.1016/j.neuron.2008.08.008 [DOI] [PubMed] [Google Scholar]
- Elsen G. E., Hodge R. D., Bedogni F., Daza R. A., Nelson B. R., Shiba N., et al. (2013). The protomap is propagated to cortical plate neurons through an Eomes-dependent intermediate map. Proc. Natl. Acad. Sci. U.S.A. 110 4081–4086. 10.1073/pnas.1209076110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley J. G., Macklis J. D. (2006). Astroglial heterogeneity closely reflects the neuronal-defined anatomy of the adult murine CNS. Neuron Glia Biol. 2 175–186. 10.1017/s1740925x06000202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinas J. M., Michurina T. V., Peunova N., Park J. H., Tordo J., Peterson D. A., et al. (2011). Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell 8 566–579. 10.1016/j.stem.2011.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C., Fink A., Lau C., Pham D., Daza R. A., Bulfone A., et al. (2005). Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 25 247–251. 10.1523/jneurosci.2899-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erclik T., Hartenstein V., Lipshitz H. D., McInnes R. R. (2008). Conserved role of the Vsx genes supports a monophyletic origin for bilaterian visual systems. Curr. Biol. 18 1278–1287. 10.1016/j.cub.2008.07.076 [DOI] [PubMed] [Google Scholar]
- Erclik T., Li X., Courgeon M., Bertet C., Chen Z., Baumert R., et al. (2017). Integration of temporal and spatial patterning generates neural diversity. Nature 541 365–370. 10.1038/nature20794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson P. S., Perfilieva E., Bjork-Eriksson T., Alborn A. M., Nordborg C., Peterson D. A., et al. (1998). Neurogenesis in the adult human hippocampus. Nat. Med. 4 1313–1317. [DOI] [PubMed] [Google Scholar]
- Estacio-Gomez A., Diaz-Benjumea F. J. (2014). Roles of Hox genes in the patterning of the central nervous system of Drosophila. Fly 8 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S., Bugeon S., Ninkovic J., Pilz G. A., Postiglione M. P., Cremer H., et al. (2017). Time-specific effects of spindle positioning on embryonic progenitor pool composition and adult neural stem cell seeding. Neuron 93 777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fame R. M., Lehtinen M. K. (2020). Emergence and developmental roles of the cerebrospinal fluid system. Dev. Cell 52 261–275. 10.1016/j.devcel.2020.01.027 [DOI] [PubMed] [Google Scholar]
- Fernandes V. M., Chen Z., Rossi A. M., Zipfel J., Desplan C. (2017). Glia relay differentiation cues to coordinate neuronal development in Drosophila. Science 357 886–891. 10.1126/science.aan3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorelli R., Azim K., Fischer B., Raineteau O. (2015). Adding a spatial dimension to postnatal ventricular-subventricular zone neurogenesis. Development 142 2109–2120. 10.1242/dev.119966 [DOI] [PubMed] [Google Scholar]
- Fuentealba L. C., Rompani S. B., Parraguez J. I., Obernier K., Romero R., Cepko C. L., et al. (2015). Embryonic origin of postnatal neural stem cells. Cell 161 1644–1655. 10.1016/j.cell.2015.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutachi S., Miya H., Watanabe T., Kawai H., Yamasaki N., Harada Y., et al. (2015). Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat. Neurosci. 18 657–665. 10.1038/nn.3989 [DOI] [PubMed] [Google Scholar]
- Galceran J., Miyashita-Lin E. M., Devaney E., Rubenstein J. L., Grosschedl R. (2000). Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development 127 469–482.10631168 [Google Scholar]
- Gao P., Postiglione M. P., Krieger T. G., Hernandez L., Wang C., Han Z., et al. (2014). Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell 159 775–788. 10.1016/j.cell.2014.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gato A., Alonso M. I., Lamus F., Miyan J. (2019). Neurogenesis: a process ontogenically linked to brain cavities and their content, CSF. Semin. Cell Dev. Biol. 102 21–27. 10.1016/j.semcdb.2019.11.008 [DOI] [PubMed] [Google Scholar]
- Gebara E., Bonaguidi M. A., Beckervordersandforth R., Sultan S., Udry F., Gijs P. J., et al. (2016). Heterogeneity of radial glia-like cells in the adult hippocampus. Stem Cells 34 997–1010. 10.1002/stem.2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S. J., Paro R. (2015). Trithorax and polycomb group-dependent regulation: a tale of opposing activities. Development 142 2876–2887. 10.1242/dev.120030 [DOI] [PubMed] [Google Scholar]
- Giachino C., Basak O., Lugert S., Knuckles P., Obernier K., Fiorelli R., et al. (2014). Molecular diversity subdivides the adult forebrain neural stem cell population. Stem Cells 32 70–84. 10.1002/stem.1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold K. S., Brand A. H. (2014). Optix defines a neuroepithelial compartment in the optic lobe of the Drosophila brain. Neural Dev. 9:18. 10.1186/1749-8104-9-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M., Huttner W. B. (2005). The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6 777–788. [DOI] [PubMed] [Google Scholar]
- Gotz M., Stoykova A., Gruss P. (1998). Pax6 controls radial glia differentiation in the cerebral cortex. Neuron 21 1031–1044. 10.1016/s0896-6273(00)80621-2 [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus R., Pearson B. J., Marusich A., Doe C. Q. (2005). Regulation of temporal identity transitions in Drosophila neuroblasts. Dev. Cell 8 193–202. 10.1016/j.devcel.2004.11.019 [DOI] [PubMed] [Google Scholar]
- Habib N., McCabe C., Medina S., Varshavsky M., Kitsberg D., Dvir-Szternfeld R., et al. (2020). Disease-associated astrocytes in Alzheimer’s disease and aging. Nat. Neurosci. 23 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatami M., Conrad S., Naghsh P., Alvarez-Bolado G., Skutella T. (2018). Cell-biological requirements for the generation of dentate gyrus granule neurons. Front. Cell Neurosci. 12:402. 10.3389/fncel.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubensak W., Attardo A., Denk W., Huttner W. B. (2004). Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 101 3196–3201. 10.1073/pnas.0308600100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins N., Malatesta P., Cecconi F., Nakafuku M., Tucker K. L., Hack M. A., et al. (2002). Glial cells generate neurons: the role of the transcription factor Pax6. Nat. Neurosci. 5 308–315. 10.1038/nn828 [DOI] [PubMed] [Google Scholar]
- Hirata T., Suda Y., Nakao K., Narimatsu M., Hirano T., Hibi M. (2004). Zinc finger gene fez-like functions in the formation of subplate neurons and thalamocortical axons. Dev. Dyn. 230 546–556. 10.1002/dvdy.20068 [DOI] [PubMed] [Google Scholar]
- Hochgerner H., Zeisel A., Lonnerberg P., Linnarsson S. (2018). Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nat. Neurosci. 21 290–299. 10.1038/s41593-017-0056-2 [DOI] [PubMed] [Google Scholar]
- Hodge R. D., Kowalczyk T. D., Wolf S. A., Encinas J. M., Rippey C., Enikolopov G., et al. (2008). Intermediate progenitors in adult hippocampal neurogenesis: Tbr2 expression and coordinate regulation of neuronal output. J. Neurosci. 28 3707–3717. 10.1523/jneurosci.4280-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge R. D., Nelson B. R., Kahoud R. J., Yang R., Mussar K. E., Reiner S. L., et al. (2012). Tbr2 is essential for hippocampal lineage progression from neural stem cells to intermediate progenitors and neurons. J. Neurosci. 32 6275–6287. 10.1523/jneurosci.0532-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer A., Campos-Ortega J. A. (1990). Proliferation pattern and early differentiation of the optic lobes in Drosophila melanogaster. Rouxs. Arch. Dev. Biol. 198 264–274. 10.1007/bf00377393 [DOI] [PubMed] [Google Scholar]
- Holguera I., Desplan C. (2018). Neuronal specification in space and time. Science 362 176–180. 10.1126/science.aas9435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalish B. T., Cheadle L., Hrvatin S., Nagy M. A., Rivera S., Crow M., et al. (2018). Single-cell transcriptomics of the developing lateral geniculate nucleus reveals insights into circuit assembly and refinement. Proc. Natl. Acad. Sci. U.S.A. 115 E1051–E1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambadur R., Koizumi K., Stivers C., Nagle J., Poole S. J., Odenwald W. F. (1998). Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 12 246–260. 10.1101/gad.12.2.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai M. I., Kim M. J., Akiyama T., Takemura M., Wharton K., O’Connor M. B., et al. (2018). Regulation of neuroblast proliferation by surface glia in the Drosophila larval brain. Sci. Rep. 8:3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G., Gage F. H., Aigner L., Song H., Curtis M. A., Thuret S., et al. (2018). Human adult neurogenesis: evidence and remaining questions. Cell Stem Cell 23 25–30. 10.1016/j.stem.2018.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M., Petryniak M. A., Long J. E., Ekker M., Obata K., Yanagawa Y., et al. (2007). A subpopulation of olfactory bulb GABAergic interneurons is derived from Emx1- and Dlx5/6-expressing progenitors. J. Neurosci. 27 6878–6891. 10.1523/jneurosci.0254-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinides N., Kapuralin K., Fadil C., Barboza L., Satija R., Desplan C. (2018). Phenotypic convergence: distinct transcription factors regulate common terminal features. Cell 174 622–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach C., Dixit R., Li S., Mattar P., Wilkinson G., Elsen G. E., et al. (2013). Neurog2 simultaneously activates and represses alternative gene expression programs in the developing neocortex. Cereb. Cortex 23 1884–1900. 10.1093/cercor/bhs176 [DOI] [PubMed] [Google Scholar]
- Kriegstein A., Alvarez-Buylla A. (2009). The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32 149–184. 10.1146/annurev.neuro.051508.135600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan K. Y., Lam M. M., Krsnik Z., Kawasawa Y. I., Lefebvre V., Sestan N. (2008). SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc. Natl. Acad. Sci. U.S.A. 105 16021–16026. 10.1073/pnas.0806791105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G., Gyllborg D., Codeluppi S., Nishimura K., Salto C., Zeisel A., et al. (2016). Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167 566–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagace D. C., Whitman M. C., Noonan M. A., Ables J. L., DeCarolis N. A., Arguello A. A., et al. (2007). Dynamic contribution of nestin-expressing stem cells to adult neurogenesis. J. Neurosci. 27 12623–12629. 10.1523/jneurosci.3812-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanet E., Gould A. P., Maurange C. (2013). Protection of neuronal diversity at the expense of neuronal numbers during nutrient restriction in the Drosophila visual system. Cell Rep. 3 587–594. 10.1016/j.celrep.2013.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanjakornsiripan D., Pior B. J., Kawaguchi D., Furutachi S., Tahara T., Katsuyama Y., et al. (2018). Layer-specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nat. Commun. 9:1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen M. K., Walsh C. A. (2011). Neurogenesis at the brain-cerebrospinal fluid interface. Annu. Rev. Cell Dev. Biol. 27 653–679. 10.1146/annurev-cellbio-092910-154026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen M. K., Zappaterra M. W., Chen X., Yang Y. J., Hill A. D., Lun M., et al. (2011). The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron 69 893–905. 10.1016/j.neuron.2011.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone D. P., Heavner W. E., Ferenczi E. A., Dobreva G., Huguenard J. R., Grosschedl R., et al. (2015). Satb2 regulates the differentiation of both callosal and subcerebral projection neurons in the developing cerebral cortex. Cereb. Cortex 25 3406–3419. 10.1093/cercor/bhu156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Takeda N., Jain R., Manderfield L. J., Liu F., Li L., et al. (2015). Hopx distinguishes hippocampal from lateral ventricle neural stem cells. Stem Cell Res. 15 522–529. 10.1016/j.scr.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Pleasure S. J. (2007). Genetic regulation of dentate gyrus morphogenesis. Prog. Brain Res. 163 143–152. [DOI] [PubMed] [Google Scholar]
- Li H., Horns F., Wu B., Xie Q., Li J., Li T., et al. (2017). Classifying Drosophila olfactory projection neuron subtypes by single-cell RNA sequencing. Cell 171 1206–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Erclik T., Bertet C., Chen Z., Voutev R., Venkatesh S., et al. (2013). Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature 498 456–462. 10.1038/nature12319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie D. C., Colamarino S. A., Song H. J., Desire L., Mira H., Consiglio A., et al. (2005). Wnt signalling regulates adult hippocampal neurogenesis. Nature 437 1370–1375. 10.1038/nature04108 [DOI] [PubMed] [Google Scholar]
- Lim D. A., Alvarez-Buylla A. (1999). Interaction between astrocytes and adult subventricular zone precursors stimulates neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 96 7526–7531. 10.1073/pnas.96.13.7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E., Zhao S., Baser A., Saiz-Castro G., Zwadlo K., Martin-Villalba A. (2015). Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell 17 329–340. 10.1016/j.stem.2015.07.002 [DOI] [PubMed] [Google Scholar]
- Lopez-Juarez A., Howard J., Ullom K., Howard L., Grande A., Pardo A., et al. (2013). Gsx2 controls region-specific activation of neural stem cells and injury-induced neurogenesis in the adult subventricular zone. Genes Dev. 27 1272–1287. 10.1101/gad.217539.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Kipnis J. (2010). Thrombospondin 1–a key astrocyte-derived neurogenic factor. FASEB J. 24 1925–1934. 10.1096/fj.09-150573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Coskun V., Liang A., Yu J., Cheng L., Ge W., et al. (2015). Single-cell transcriptome analyses reveal signals to activate dormant neural stem cells. Cell 161 1175–1186. 10.1016/j.cell.2015.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Suarez S., Valero J., Muro-Garcia T., Encinas J. M. (2019). Phenotypical and functional heterogeneity of neural stem cells in the aged hippocampus. Aging Cell 18:e12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias I., Morgado J., Gomes F. C. A. (2019). Astrocyte heterogeneity: impact to brain aging and disease. Front. Aging Neurosci. 11:59 10.3389/fncel.2018.0059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsue K., Minakawa S., Kashiwagi T., Toda K., Sato T., Shioda S., et al. (2018). Dentate granule progenitor cell properties are rapidly altered soon after birth. Brain Struct. Funct. 223 357–369. 10.1007/s00429-017-1499-7 [DOI] [PubMed] [Google Scholar]
- Mattar P., Ericson J., Blackshaw S., Cayouette M. (2015). A conserved regulatory logic controls temporal identity in mouse neural progenitors. Neuron 85 497–504. 10.1016/j.neuron.2014.12.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyash V., Kettenmann H. (2010). Heterogeneity in astrocyte morphology and physiology. Brain Res. Rev. 63 2–10. 10.1016/j.brainresrev.2009.12.001 [DOI] [PubMed] [Google Scholar]
- Maurange C., Cheng L., Gould A. P. (2008). Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell 133 891–902. 10.1016/j.cell.2008.03.034 [DOI] [PubMed] [Google Scholar]
- McConnell S. K. (1991). The generation of neuronal diversity in the central nervous system. Annu. Rev. Neurosci. 14 269–300. 10.1146/annurev.ne.14.030191.001413 [DOI] [PubMed] [Google Scholar]
- McKenna W. L., Betancourt J., Larkin K. A., Abrams B., Guo C., Rubenstein J. L., et al. (2011). Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J. Neurosci. 31 549–564. 10.1523/jneurosci.4131-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle F. T., Fuentealba L. C., Sanders T. A., Magno L., Kessaris N., Alvarez-Buylla A. (2014). Adult neural stem cells in distinct microdomains generate previously unknown interneuron types. Nat. Neurosci. 17 207–214. 10.1038/nn.3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle F. T., Mirzadeh Z., Alvarez-Buylla A. (2007). Mosaic organization of neural stem cells in the adult brain. Science 317 381–384. 10.1126/science.1144914 [DOI] [PubMed] [Google Scholar]
- Mihalas A. B., Elsen G. E., Bedogni F., Daza R. A. M., Ramos-Laguna K. A., Arnold S. J., et al. (2016). Intermediate progenitor cohorts differentially generate cortical layers and require tbr2 for timely acquisition of neuronal subtype identity. Cell Rep. 16 92–105. 10.1016/j.celrep.2016.05.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T., Kawaguchi A., Saito K., Kawano M., Muto T., Ogawa M. (2004). Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development 131 3133–3145. 10.1242/dev.01173 [DOI] [PubMed] [Google Scholar]
- Mizrak D., Levitin H. M., Delgado A. C., Crotet V., Yuan J., Chaker Z., et al. (2019). Single-cell analysis of regional differences in adult V-SVZ neural stem cell lineages. Cell Rep. 26 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora N., Oliva C., Fiers M., Ejsmont R., Soldano A., Zhang T. T., et al. (2018). A temporal transcriptional switch governs stem cell division, neuronal numbers, and maintenance of differentiation. Dev. Cell 45 53–66. [DOI] [PubMed] [Google Scholar]
- Morales A. V., Mira H. (2019). Adult neural stem cells: born to last. Front. Cell Dev. Biol. 7:96 10.3389/fncel.2018.0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morante J., Desplan C. (2008). The color-vision circuit in the medulla of Drosophila. Curr. Biol. 18 553–565. 10.1016/j.cub.2008.02.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morante J., Erclik T., Desplan C. (2011). Cell migration in Drosophila optic lobe neurons is controlled by eyeless/Pax6. Development 138 687–693. 10.1242/dev.056069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morante J., Vallejo D. M., Desplan C., Dominguez M. (2013). Conserved miR-8/miR-200 defines a glial niche that controls neuroepithelial expansion and neuroblast transition. Dev. Cell 27 174–187. 10.1016/j.devcel.2013.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel L., Chiang M. S. R., Higashimori H., Shoneye T., Iyer L. K., Yelick J., et al. (2017). Molecular and functional properties of regional astrocytes in the adult brain. J. Neurosci. 37 8706–8717. 10.1523/jneurosci.3956-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel L., Men Y., Chiang M. S. R., Tian Y., Jin S., Yelick J., et al. (2019). Intracortical astrocyte subpopulations defined by astrocyte reporter Mice in the adult brain. Glia 67 171–181. 10.1002/glia.23545 [DOI] [PubMed] [Google Scholar]
- Moreno-Estelles M., Gonzalez-Gomez P., Hortiguela R., Diaz-Moreno M., San Emeterio J., Carvalho A. L., et al. (2012). Symmetric expansion of neural stem cells from the adult olfactory bulb is driven by astrocytes via WNT7A. Stem Cells 30 2796–2809. 10.1002/stem.1243 [DOI] [PubMed] [Google Scholar]
- Moreno-Jimenez E. P., Flor-Garcia M., Terreros-Roncal J., Rabano A., Cafini F., Pallas-Bazarra N., et al. (2019). Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med. 25 554–560. 10.1038/s41591-019-0375-9 [DOI] [PubMed] [Google Scholar]
- Morrison S. J., Spradling A. C. (2008). Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132 598–611. 10.1016/j.cell.2008.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka H., Nakamura S., Shimazaki T., Okano H. (2008). Requirement for COUP-TFI and II in the temporal specification of neural stem cells in CNS development. Nat. Neurosci. 11 1014–1023. 10.1038/nn.2168 [DOI] [PubMed] [Google Scholar]
- Nelson B. R., Hodge R. D., Daza R. A., Tripathi P. P., Arnold S. J., Millen K. J., et al. (2020). Intermediate progenitors support migration of neural stem cells into dentate gyrus outer neurogenic niches. eLife 9:e53777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola Z., Fabel K., Kempermann G. (2015). Development of the adult neurogenic niche in the hippocampus of mice. Front. Neuroanat. 9:53 10.3389/fncel.2018.0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Estevez V., Oueslati-Morales C. O., Li L., Pickel J., Morales A. V., Vicario-Abejon C. (2016). Brain insulin-like growth factor-i directs the transition from stem cells to mature neurons during postnatal/adult hippocampal neurogenesis. Stem Cells 34 2194–2209. 10.1002/stem.2397 [DOI] [PubMed] [Google Scholar]
- Ninkovic J., Mori T., Gotz M. (2007). Distinct modes of neuron addition in adult mouse neurogenesis. J. Neurosci. 27 10906–10911. 10.1523/jneurosci.2572-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor S. C., Martinez-Cerdeno V., Ivic L., Kriegstein A. R. (2004). Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7 136–144. 10.1038/nn1172 [DOI] [PubMed] [Google Scholar]
- Nowakowski T. J., Bhaduri A., Pollen A. A., Alvarado B., Mostajo-Radji M. A., Di Lullo E., et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358 1318–1323. 10.1126/science.aap8809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernier K., Alvarez-Buylla A. (2019). Neural stem cells: origin, heterogeneity and regulation in the adult mammalian brain. Development 146:dev156059. 10.1242/dev.156059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernier K., Cebrian-Silla A., Thomson M., Parraguez J. I., Anderson R., Guinto C., et al. (2018). Adult neurogenesis is sustained by symmetric self-renewal and differentiation. Cell Stem Cell 22 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M., Miyata T., Konno D., Ueda H. R., Kasukawa T., Hashimoto M., et al. (2016). Cell-cycle-independent transitions in temporal identity of mammalian neural progenitor cells. Nat. Commun. 7:11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldekamp J., Kraemer N., Alvarez-Bolado G., Skutella T. (2004). bHLH gene expression in the Emx2-deficient dentate gyrus reveals defective granule cells and absence of migrating precursors. Cereb. Cortex 14 1045–1058. 10.1093/cercor/bhh064 [DOI] [PubMed] [Google Scholar]
- Otsuki L., Brand A. H. (2017). The vasculature as a neural stem cell niche. Neurobiol. Dis. 107 4–14. 10.1016/j.nbd.2017.01.010 [DOI] [PubMed] [Google Scholar]
- Otsuki L., Brand A. H. (2018). Cell cycle heterogeneity directs the timing of neural stem cell activation from quiescence. Science 360 99–102. 10.1126/science.aan8795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y., Rangel C., Reynolds M. M., Caldwell M. C., Johns M., Nayak M., et al. (2003). Drosophila perlecan modulates FGF and hedgehog signals to activate neural stem cell division. Dev. Biol. 253 247–257. 10.1016/s0012-1606(02)00019-2 [DOI] [PubMed] [Google Scholar]
- Parthasarathy S., Srivatsa S., Nityanandam A., Tarabykin V. (2014). Ntf3 acts downstream of Sip1 in cortical postmitotic neurons to control progenitor cell fate through feedback signaling. Development 141 3324–3330. 10.1242/dev.114173 [DOI] [PubMed] [Google Scholar]
- Pellegrini M., Mansouri A., Simeone A., Boncinelli E., Gruss P. (1996). Dentate gyrus formation requires Emx2. Development 122 3893–3898. [DOI] [PubMed] [Google Scholar]
- Pfeiffer K., Homberg U. (2014). Organization and functional roles of the central complex in the insect brain. Ann. Rev. Entomol. 59 165–184. 10.1146/annurev-ento-011613-162031 [DOI] [PubMed] [Google Scholar]
- Philippidou P., Dasen J. S. (2013). Hox genes: choreographers in neural development, architects of circuit organization. Neuron 80 12–34. 10.1016/j.neuron.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilz G. A., Bottes S., Betizeau M., Jorg D. J., Carta S., Simons B. D., et al. (2018). Live imaging of neurogenesis in the adult mouse hippocampus. Science 359 658–662. 10.1126/science.aao5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto-Teixeira F., Koo C., Rossi A. M., Neriec N., Bertet C., Li X., et al. (2018). Development of concurrent retinotopic maps in the fly motion detection circuit. Cell 173 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piunti A., Shilatifard A. (2016). Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 352:aad9780. 10.1126/science.aad9780 [DOI] [PubMed] [Google Scholar]
- Platel J. C., Dave K. A., Gordon V., Lacar B., Rubio M. E., Bordey A. (2010). NMDA receptors activated by subventricular zone astrocytic glutamate are critical for neuroblast survival prior to entering a synaptic network. Neuron 65 859–872. 10.1016/j.neuron.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouchelon G., Gambino F., Bellone C., Telley L., Vitali I., Luscher C., et al. (2014). Modality-specific thalamocortical inputs instruct the identity of postsynaptic L4 neurons. Nature 511 471–474. 10.1038/nature13390 [DOI] [PubMed] [Google Scholar]
- Rakic P. (1972). Mode of cell migration to the superficial layers of fetal monkey neocortex. J. Comp. Neurol. 145 61–83. 10.1002/cne.901450105 [DOI] [PubMed] [Google Scholar]
- Ramon-Canellas P., Peterson H. P., Morante J. (2018). From early to late neurogenesis: neural progenitors and the glial niche from a fly’s point of view. Neuroscience 399 39–52. 10.1016/j.neuroscience.2018.12.014 [DOI] [PubMed] [Google Scholar]
- Regev A., Teichmann S. A., Lander E. S., Amit I., Benoist C., Birney E., et al. (2017). The human cell atlas. eLife 6:e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q., Yang C. P., Liu Z., Sugino K., Mok K., He Y., et al. (2017). Stem cell-intrinsic, seven-up-triggered temporal factor gradients diversify intermediate neural progenitors. Curr. Biol. 27 1303–1313. 10.1016/j.cub.2017.03.047 [DOI] [PubMed] [Google Scholar]
- Romanov R. A., Zeisel A., Bakker J., Girach F., Hellysaz A., Tomer R., et al. (2017). Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat. Neurosci. 20 176–188. 10.1038/nn.4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybon L., Deierborg T., Brundin P., Li J. Y. (2009a). Involvement of Ngn2, Tbr and NeuroD proteins during postnatal olfactory bulb neurogenesis. Eur. J. Neurosci. 29 232–243. 10.1111/j.1460-9568.2008.06595.x [DOI] [PubMed] [Google Scholar]
- Roybon L., Hjalt T., Stott S., Guillemot F., Li J. Y., Brundin P. (2009b). Neurogenin2 directs granule neuroblast production and amplification while NeuroD1 specifies neuronal fate during hippocampal neurogenesis. PLoS One 4:e4779 10.1371/journal.pone.004779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagner A., Briscoe J. (2019). Establishing neuronal diversity in the spinal cord: a time and a place. Development 146:dev182154. 10.1242/dev.182154 [DOI] [PubMed] [Google Scholar]
- Sanai N., Nguyen T., Ihrie R. A., Mirzadeh Z., Tsai H. H., Wong M., et al. (2011). Corridors of migrating neurons in the human brain and their decline during infancy. Nature 478 382–386. 10.1038/nature10487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamoto K., Wichterle H., Gonzalez-Perez O., Cholfin J. A., Yamada M., Spassky N., et al. (2006). New neurons follow the flow of cerebrospinal fluid in the adult brain. Science 311 629–632. 10.1126/science.1119133 [DOI] [PubMed] [Google Scholar]
- Schmidt H., Rickert C., Bossing T., Vef O., Urban J., Technau G. M. (1997). The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev. Biol. 189 186–204. 10.1006/dbio.1997.8660 [DOI] [PubMed] [Google Scholar]
- Schneider J., Karpf J., Beckervordersandforth R. (2019). Role of astrocytes in the neurogenic niches. Methods Mol. Biol. 1938 19–33. 10.1007/978-1-4939-9068-9_2 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Bourbon H. M., Di Croce L., Cavalli G. (2017). Genome regulation by polycomb and trithorax: 70 Years and counting. Cell 171 34–57. 10.1016/j.cell.2017.08.002 [DOI] [PubMed] [Google Scholar]
- Seki T., Sato T., Toda K., Osumi N., Imura T., Shioda S. (2014). Distinctive population of Gfap-expressing neural progenitors arising around the dentate notch migrate and form the granule cell layer in the developing hippocampus. J. Comp. Neurol. 522 261–283. 10.1002/cne.23460 [DOI] [PubMed] [Google Scholar]
- Seuntjens E., Nityanandam A., Miquelajauregui A., Debruyn J., Stryjewska A., Goebbels S., et al. (2009). Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nat. Neurosci. 12 1373–1380. 10.1038/nn.2409 [DOI] [PubMed] [Google Scholar]
- Shihabuddin L. S., Horner P. J., Ray J., Gage F. H. (2000). Adult spinal cord stem cells generate neurons after transplantation in the adult dentate gyrus. J. Neurosci. 20 8727–8735. 10.1523/jneurosci.20-23-08727.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Berg D. A., Zhu Y., Shin J. Y., Song J., Bonaguidi M. A., et al. (2015). Single-cell RNA-Seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell 17 360–372. 10.1016/j.stem.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegenthaler J. A., Ashique A. M., Zarbalis K., Patterson K. P., Hecht J. H., Kane M. A., et al. (2009). Retinoic acid from the meninges regulates cortical neuron generation. Cell 139 597–609. 10.1016/j.cell.2009.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegenthaler J. A., Pleasure S. J. (2010). There’s no place like home for a neural stem cell. Cell Stem Cell 7 141–143. 10.1016/j.stem.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Siegenthaler J. A., Pleasure S. J. (2011). We have got you ‘covered’: how the meninges control brain development. Curr. Opin. Genet. Dev. 21 249–255. 10.1016/j.gde.2010.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra A., Martin-Suarez S., Valcarcel-Martin R., Pascual-Brazo J., Aelvoet S. A., Abiega O., et al. (2015). Neuronal hyperactivity accelerates depletion of neural stem cells and impairs hippocampal neurogenesis. Cell Stem Cell 16 488–503. 10.1016/j.stem.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeath J. B., Thor S. (2003). Genetic control of Drosophila nerve cord development. Curr. Opin. Neurobiol. 13 8–15. 10.1016/s0959-4388(03)00007-2 [DOI] [PubMed] [Google Scholar]
- Song H., Stevens C. F., Gage F. H. (2002). Astroglia induce neurogenesis from adult neural stem cells. Nature 417 39–44. 10.1038/417039a [DOI] [PubMed] [Google Scholar]
- Sorrells S. F., Paredes M. F., Cebrian-Silla A., Sandoval K., Qi D., Kelley K. W., et al. (2018). Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 555 377–381. 10.1038/nature25975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Nunes R., Yee L. L., Gould A. P. (2011). Fat cells reactivate quiescent neuroblasts via TOR and glial insulin relays in Drosophila. Nature 471 508–512. 10.1038/nature09867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding K. L., Bergmann O., Alkass K., Bernard S., Salehpour M., Huttner H. B., et al. (2013). Dynamics of hippocampal neurogenesis in adult humans. Cell 153 1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speder P., Brand A. H. (2018). Systemic and local cues drive neural stem cell niche remodelling during neurogenesis in Drosophila. eLife 7:e30413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K., Leone D. P., Bateson R. K., Dobreva G., Kohwi Y., Kohwi-Shigematsu T., et al. (2012). A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proc. Natl. Acad. Sci. U.S.A. 109 19071–19078. 10.1073/pnas.1216793109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoykova A., Treichel D., Hallonet M., Gruss P. (2000). Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J. Neurosci. 20 8042–8050. 10.1523/jneurosci.20-21-08042.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H., Consiglio A., Ray J., Sawai T., D’Amour K. A., Gage F. H. (2007). In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell 1 515–528. 10.1016/j.stem.2007.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhonen J. O., Peterson D. A., Ray J., Gage F. H. (1996). Differentiation of adult hippocampus-derived progenitors into olfactory neurons in vivo. Nature 383 624–627. 10.1038/383624a0 [DOI] [PubMed] [Google Scholar]
- Sultan S., Li L., Moss J., Petrelli F., Casse F., Gebara E., et al. (2015). Synaptic integration of adult-born hippocampal neurons is locally controlled by astrocytes. Neuron 88 957–972. 10.1016/j.neuron.2015.10.037 [DOI] [PubMed] [Google Scholar]
- Suzuki T., Kaido M., Takayama R., Sato M. (2013). A temporal mechanism that produces neuronal diversity in the Drosophila visual center. Dev. Biol. 380 12–24. 10.1016/j.ydbio.2013.05.002 [DOI] [PubMed] [Google Scholar]
- Syed M. H., Mark B., Doe C. Q. (2017). Steroid hormone induction of temporal gene expression in Drosophila brain neuroblasts generates neuronal and glial diversity. eLife 6:e26287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B., Menon V., Nguyen T. N., Kim T. K., Jarsky T., Yao Z., et al. (2016). Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 19 335–346. 10.1038/nn.4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B., Yao Z., Graybuck L. T., Smith K. A., Nguyen T. N., Bertagnolli D., et al. (2018). Shared and distinct transcriptomic cell types across neocortical areas. Nature 563 72–78. 10.1038/s41586-018-0654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie M., Van der Veken L., Silva-Vargas V., Louissaint M., Colonna L., Zaidi B., et al. (2008). A specialized vascular niche for adult neural stem cells. Cell Stem Cell 3 279–288. 10.1016/j.stem.2008.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna E., Gotz M., Huttner W. B. (2014). The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 30 465–502. 10.1146/annurev-cellbio-101011-155801 [DOI] [PubMed] [Google Scholar]
- Technau G. M., Berger C., Urbach R. (2006). Generation of cell diversity and segmental pattern in the embryonic central nervous system of Drosophila. Dev. Dyn. 235 861–869. 10.1002/dvdy.20566 [DOI] [PubMed] [Google Scholar]
- Telley L., Agirman G., Prados J., Amberg N., Fievre S., Oberst P., et al. (2019). Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science 364:eaav2522. 10.1126/science.aav2522 [DOI] [PubMed] [Google Scholar]
- Telley L., Govindan S., Prados J., Stevant I., Nef S., Dermitzakis E., et al. (2016). Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science 351 1443–1446. 10.1126/science.aad8361 [DOI] [PubMed] [Google Scholar]
- Tiveron M. C., Beclin C., Murgan S., Wild S., Angelova A., Marc J., et al. (2017). Zic-proteins are repressors of dopaminergic forebrain fate in mice and C. elegans. J. Neurosci. 37 10611–10623. 10.1523/jneurosci.3888-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma K., Kumamoto T., Hanashima C. (2014). The timing of upper-layer neurogenesis is conferred by sequential derepression and negative feedback from deep-layer neurons. J. Neurosci. 34 13259–13276. 10.1523/jneurosci.2334-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki T., Tanaka M., Yamashita K., Mikawa S., Qiu Z., Maragakis N. J., et al. (2003). A novel secretory factor, Neurogenesin-1, provides neurogenic environmental cues for neural stem cells in the adult hippocampus. J. Neurosci. 23 11732–11740. 10.1523/jneurosci.23-37-11732.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach R., Technau G. M. (2004). Neuroblast formation and patterning during early brain development in Drosophila. Bioessays 26 739–751. 10.1002/bies.20062 [DOI] [PubMed] [Google Scholar]
- Urbach R., Technau G. M. (2008). Dorsoventral patterning of the brain: a comparative approach. Adv. Exp. Med. Biol. 628 42–56. 10.1007/978-0-387-78261-4_3 [DOI] [PubMed] [Google Scholar]
- Urban N., Blomfield I. M., Guillemot F. (2019). Quiescence of adult mammalian neural stem cells: a highly regulated rest. Neuron 104 834–848. 10.1016/j.neuron.2019.09.026 [DOI] [PubMed] [Google Scholar]
- Urban N., van den Berg D. L., Forget A., Andersen J., Demmers J. A., Hunt C., et al. (2016). Return to quiescence of mouse neural stem cells by degradation of a proactivation protein. Science 353 292–295. 10.1126/science.aaf4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali I., Fievre S., Telley L., Oberst P., Bariselli S., Frangeul L., et al. (2018). Progenitor hyperpolarization regulates the sequential generation of neuronal subtypes in the developing neocortex. Cell 174 1264–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt A., Pflanz R., Schafer U., Jackle H. (2002). Perlecan participates in proliferation activation of quiescent Drosophila neuroblasts. Dev. Dyn. 224 403–412. 10.1002/dvdy.10120 [DOI] [PubMed] [Google Scholar]
- Walsh K. T., Doe C. Q. (2017). Drosophila embryonic type II neuroblasts: origin, temporal patterning, and contribution to the adult central complex. Development 144 4552–4562. 10.1242/dev.157826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsson U., Faiz M., de Pablo Y., Sjoqvist M., Andersson D., Widestrand A., et al. (2012). Astrocytes negatively regulate neurogenesis through the Jagged1-mediated Notch pathway. Stem Cells 30 2320–2329. 10.1002/stem.1196 [DOI] [PubMed] [Google Scholar]
- Winpenny E., Lebel-Potter M., Fernandez M. E., Brill M. S., Gotz M., Guillemot F., et al. (2011). Sequential generation of olfactory bulb glutamatergic neurons by Neurog2-expressing precursor cells. Neural Dev. 6:12. 10.1186/1749-8104-6-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K. M., Fogarty M., Kessaris N., Richardson W. D. (2007). Subventricular zone stem cells are heterogeneous with respect to their embryonic origins and neurogenic fates in the adult olfactory bulb. J. Neurosci. 27 8286–8296. 10.1523/jneurosci.0476-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahr S. K., Yang G., Kazan H., Borrett M. J., Yuzwa S. A., Voronova A., et al. (2018). A translational repression complex in developing mammalian neural stem cells that regulates neuronal specification. Neuron 97 520–537. [DOI] [PubMed] [Google Scholar]
- Zeisel A., Hochgerner H., Lonnerberg P., Johnsson A., Memic F., van der Zwan J., et al. (2018). Molecular architecture of the mouse nervous system. Cell 174 999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A., Munoz-Manchado A. B., Codeluppi S., Lonnerberg P., La Manno G., Jureus A., et al. (2015). Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347 1138–1142. 10.1126/science.aaa1934 [DOI] [PubMed] [Google Scholar]
- Zhu C. C., Boone J. Q., Jensen P. A., Hanna S., Podemski L., Locke J., et al. (2008). Drosophila Activin- and the Activin-like product Dawdle function redundantly to regulate proliferation in the larval brain. Development 135 513–521. 10.1242/dev.010876 [DOI] [PubMed] [Google Scholar]
- Zhu S., Lin S., Kao C. F., Awasaki T., Chiang A. S., Lee T. (2006). Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell 127 409–422. 10.1016/j.cell.2006.08.045 [DOI] [PubMed] [Google Scholar]
- Zywitza V., Misios A., Bunatyan L., Willnow T. E., Rajewsky N. (2018). Single-cell transcriptomics characterizes cell types in the subventricular zone and uncovers molecular defects impairing adult neurogenesis. Cell Rep. 25 2457–2469. [DOI] [PubMed] [Google Scholar]


