Abstract
Natural products (NPs) are important sources of human therapeutic agents and pesticides. To prevent self-harm from bioactive NPs, some microbial producers employ self-resistance genes to protect themselves. One effective strategy is to employ a self-resistance enzyme (SRE), which is a slightly mutated version of the original metabolic enzyme, and is resistant to the toxic NP but is still functional. The presence of a SRE in a gene cluster can serve as a predictive window to the biological activity of the NPs synthesized by the pathway. In this highlight, we summarize representative examples of NP biosynthetic pathways that utilize self-resistance genes for protection. Recent discoveries based on self-resistance gene identification have helped in bridging the gap between activity-guided and genome-driven approaches for NP discovery and functional assignment.
Graphic abstract
This review covers recent natural product research directed by self-resistance genes, which bridges the gap between activity-guided and genome-driven approaches.
1. Introduction
Natural products (NPs) are small organic molecules derived from secondary metabolism of living organisms, mostly found in microbes and plants. NPs have been evolutionally optimized to bind to specific but diverse macro-biomolecule targets, facilitated by their complex structures unrivalled by synthetic molecule libraries. As a result, NPs continue to inspire innovative discoveries in the field of chemistry, biology and medicine. From 1981 to 2014, among 1562 newly approved drugs, 710 were derived from NPs. Between 1997 and 2010, NPs and their derivatives made up approximately 36% of all new registered pesticide ingredients1, 2. NPs will remain an important source for discovering new human therapeutic agents or agrochemicals3.
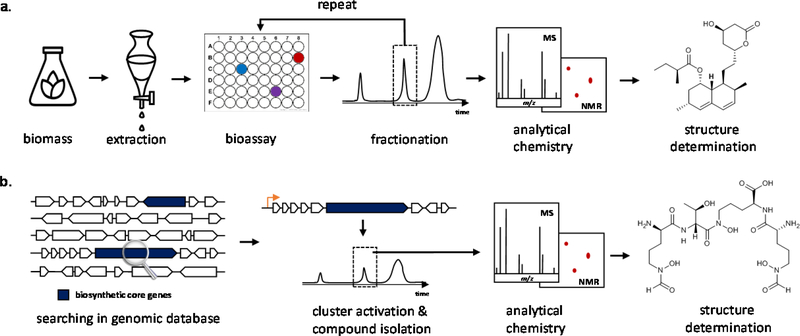
In the past century, the rapid development of biological assays and chemical separation techniques has led to the identification and isolation of many bioactive NPs1, 2. In activity-guided NP discovery, a biomass or extract with bioactivity is first identified using an activity screen. The desired activity is enriched and tracked by several rounds of fractionation and purification, until a pure bioactive compound is obtained4 (Fig. 1a). The most famous example is perhaps the serendipitous discovery of penicillin, identified through inhibition of bacterial growth. Such activity-guided discovery of NPs has remained a mainstay for almost a century, becoming more versatile with the development of higher-resolution and higher-throughput assays5.
Fig. 1.
Workflow of NP discovery. (a) activity-guided NP discovery approach. (b) genome-driven NP discovery approach.
The rapid development of genome sequencing technologies has revolutionized NP discovery6, 7. Extensive experimental studies of the biosynthetic genes of NPs showed that the genes encoding enzymes responsible microbial for biosynthesis of one NP are typically clustered in a genome as a biosynthetic gene cluster (BGC)8. This may facilitate co-regulation, co-expression and horizontal transfer of all genes within a BGC together. With the clustering of biosynthetic genes and powerful bioinformatics tools9, 10, 11, tens of thousands of BGCs have been identified from sequenced microbial genomes. Identification of a BGC is typically performed first through cataloguing of anchoring biosynthetic core enzymes, such as polyketide synthases (PKSs), nonribosomal peptide synthetases (NRPSs), terpene synthases (TSs), etc12, that build up the backbone of the molecules. Among identified BGCs, it is estimated that fewer than 10% are associated with characterized NPs, and the other >90% of gene clusters have unknown products13. This led to the common characterization of these BGCs as transcriptionally silent, cryptic and/or biosynthetic dark matter. The lack of production of NPs from these BGCs can be attributed to several reasons: 1) the production of many NPs in native strains is in responses to complex combinations of environmental and growth signals, which are difficult to recapitulate under axenic laboratory culturing conditions; 2) some NPs are only produced at very low concentrations, which could elude detection; and 3) the biological activities of the NPs are not targeted during activity-guided screens, and thereby overlooked during extract analysis. Uncovering the structures and activities of the >90% cryptic BGCs of unknown products is therefore a tantalizing new approach of drug discovery.
In the last decade, genome-driven NP discovery approaches have led to the effective mining of these cryptic BGCs14 (Fig. 1b). The application of modern cloning and synthetic biology tools, which include overexpression of pathway-specific transcription factors, epigenetic modification of chromatin structure, and heterologous expression of pathways in model hosts, have demonstrated there is significant potential to increase the NP structure space from microbes11, 15–17. Although many NPs with new structures have been discovered using genome mining approaches, the molecular targets and biological functions of these NPs are not easily identified. This is because genome mining efforts have been generally targeting the presence of interesting biosynthetic enzymes and from ecologically unique species, and are not activity-guided. As there are exponential increases in the number of cryptic BGCs, the challenge is to prioritize BGC mining with biological activity to identify the most therapeutically promising NPs18.
How can we predict the activity of a NP based on a genomic sequence? The answer to this question can help us pinpoint the untapped bioactive NPs from microbial secondary metabolism. In this review, we will summarize the occurrence of self-resistance genes in characterized BGCs. We will then highlight the use of the self-resistance gene as a predictive window for linking BGCs to NP activities, and to mine NPs with desired activity. In our opinion, this approach could be effective in leading the renaissance of NP discovery for novel human therapeutic agents and agricultural chemicals.
2. Self-resistance gene co-localized with BGC implies the molecular target of a NP
Many NPs are believed to be produced by organisms (the host) to kill or limit the growth of competitor organisms through the inhibition or inactivation of essential housekeeping enzymes. However, if the same enzyme target is conserved in the producing organism and is essential, the NP will also be toxic to the producing host. Therefore, self-protecting methods must be present in the host to confer resistance to the biosynthesized NPs. Several mechanisms of self-resistance are known, including efflux pumps that actively transport the metabolites to extracellular space; resistance proteins that are present to sequester or modify highly active NPs that remain in the cell; and enzymes that modify housekeeping enzymes in the host to evade NP inhibition19, 20. The accumulating knowledge of self-resistance mechanisms have facilitated NP research. For example, Wright and co-workers designed a resistance-based NP discovery platform for the discovery of novel antibiotics21. This platform can be summarized in the following steps: 1. Screening of environmental samples using desired antibiotic to enrich microbial producers of selected antibiotic scaffolds; 2. Determining the presence of a desired BGC in unique microbial isolates using polymerase chain reaction (PCR)-based screening; 3. Further phylogenetic analysis of the producers of new NPs
An additional strategy nature employs for self-resistance is to encode a functionally equivalent, self-resistance enzyme (SRE) that is a variant of the housekeeping enzyme. The SRE is highly similar in sequence to the housekeeping target, but contains mutations that render the enzyme insensitive to NP inhibition, while still maintaining activity22, 23. Because of the sequence similarities, the functions of SREs can be readily predicted through bioinformatic analysis. The SRE is often co-clustered with the BGC of the NP and is transcriptionally co-regulated with the biosynthetic genes during NP production. This ensures the SRE is present when the NP starts to accumulate intracellularly. The use of SRE strategy is widely adopted by NP producing microorganisms in both bacterial and fungal species22, 23.
From a genome mining perspective, the SRE effectively serves as a readout of the enzyme target of the associated NP. Here we summarize 36 examples of SRE co-localizations, which are grouped into different modes of action of NPs (Table 1). Among these 36 examples, 24 proposed SREs have been functionally validated by either in vivo or in vitro experiments (Table 1). The examples demonstrate co-localization of biosynthetic and SRE genes is prevalent for NP producing microorganisms.
Table 1.
Example of BGCs containing a self-resistance-gene as a duplicate copy of the housekeeping gene (molecular target)
| natural products | organism | molecular target | biosynthetic core gene | SRE verifieda | FDA approvedb | SRE inspiredc |
|---|---|---|---|---|---|---|
| inhibitors of DNA replication | ||||||
| novobiocin (1) | Streptomyces niveus | bacterial DNA gyrase | NRPS | V | Y | N |
| chlorobiocin (2) | Streptomyces roseochromogenes | bacterial DNA gyrase | NRPS | V | N | N |
| coumermycin A1 (3) | Streptomyces rishiriensis | bacterial DNA gyrase | NRPS | V | N | N |
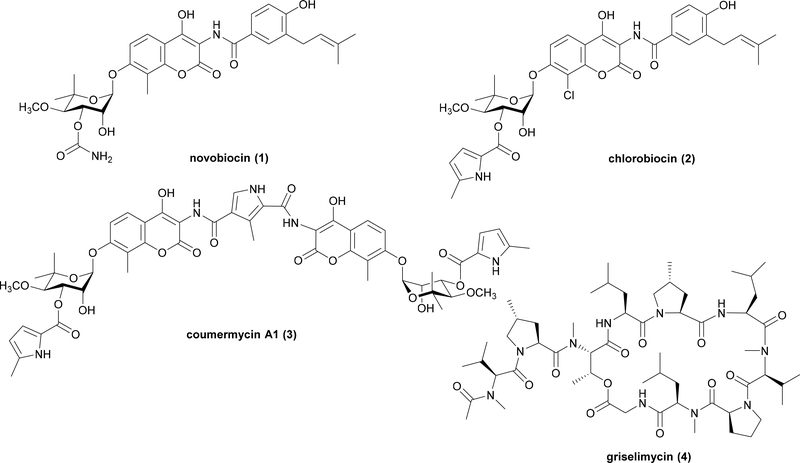
| griselimycin (4) | Streptomyces sp. DSM-40835 | DnaN | NRPS | V | N | Y |
| inhibitors of protein biosynthesis | ||||||
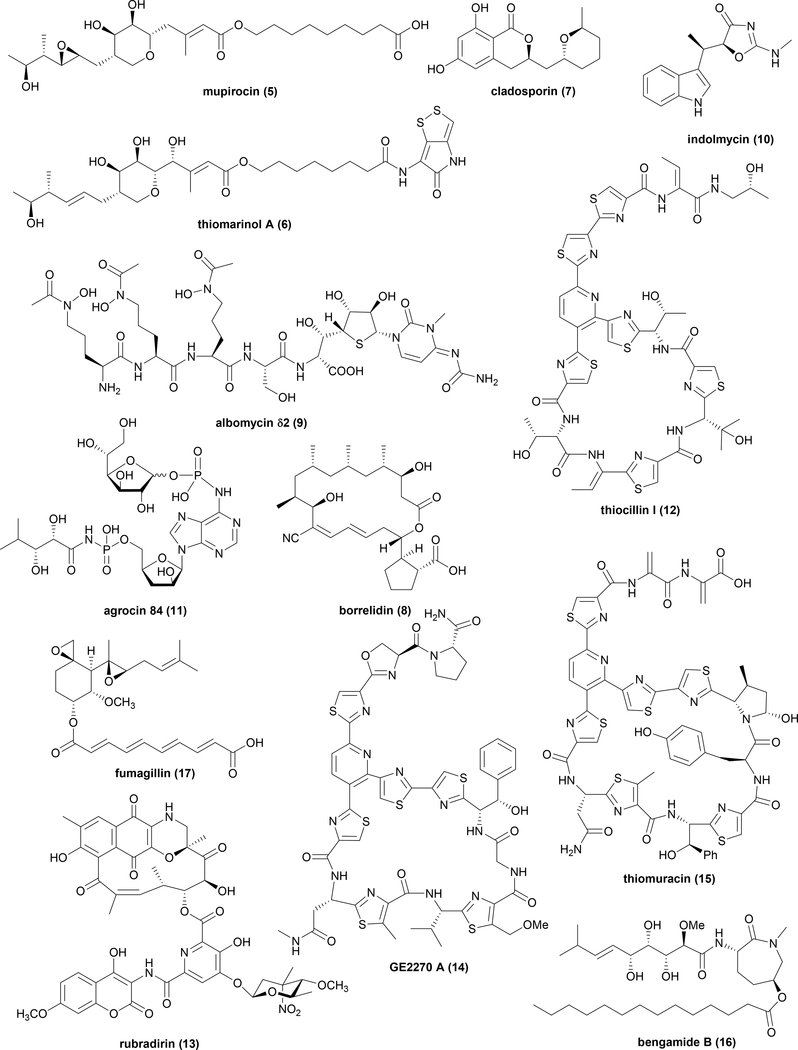
| mupirocin (5) | Pseudomonas fluorescens | isoleucyl-tRNA synthetase | PKS | V | Y | N |
| thiomarinol A (6) | Pseudoalteromonas sp. SANK 73390 | isoleucyl-tRNA synthetase | PKS | V | N | N |
| cladosporin (7) | Cladosporium cladosporioides | lysyl-tRNA synthetase | NRPS | V | Y | N |
| borrelidin (8) | Streptomyces parvulus | threonyl-tRNA synthetase | PKS | V | N | N |
| albomycin δ2 (9) | Streptomyces sp. ATCC 700974 | seryl-tRNA synthetase | NRPS | V | N | N |
| indolmycin (10) | Streptomyces griseus | tryptophanyl-tRNA synthetase | other | P | N | N |
| agrocin 84 (11) | Agrobacterium radiobacter | leucyl-tRNA synthetase | other | V | N | N |
| thiocillin (12) | Bacillus cereus | 50S ribosomal protein L11 | precursor peptide | P | N | N |
| rubradirin (13) | Streptomyces achromogenes | translation initiation factor | PKS | P | N | N |
| GE2270 (14) | Planobispora rosea | elongation factor Tu | precursor peptide | P | N | N |
| thiomuracin (15) | Thermobispora bispora | elongation factor Tu | P | N | N | |
| bengamide B (16) | Myxococcus virescens | methionine aminopeptidase, type I | NRPS | V | N | N |
| fumagillin (17) | Aspergillus fumigatus | methionine aminopeptidase, type II | PKS | P | N | N |
| inhibitors of protein degradation | ||||||
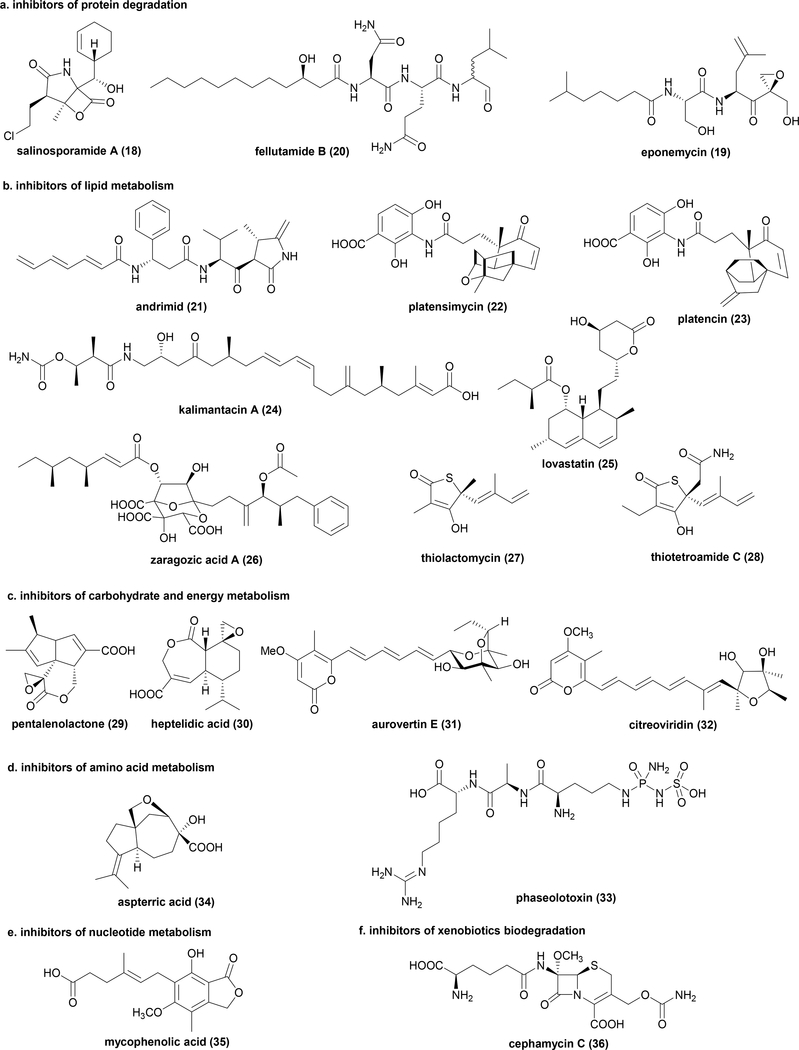
| salinosporamide A (18) | Salinispora tropica | proteasome subunit beta | PKS-NRPS hybrid | V | N | N |
| eponemycin (19) | Streptomyces hygroscopicus | proteasome subunit beta | NRPS | V | N | N |
| fellutamide B (20) | Aspergillus nidulans | proteasome component C5 | NRPS | V | N | Y |
| inhibitors of lipid metabolism | ||||||
| andrimid (21) | Pantoea agglomerans | acetyl-CoA carboxylase | NRPS | P | N | N |
| platensimycin (22) / platencin (23) | Streptomyces platensis | FabF | TS | V | N | N |
| kalimantacin (24) | Pseudomonas fluorescens | FabI | PKS | V | N | N |
| lovastatin (25) | Aspergillus terreus | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | PKS | V | Y | N |
| zaragozic acid (26) | Curvularia lunata | squalene synthase | PKS | P | N | N |
| thiolactomycin (27) | Salinispora pacifica | fatty acid synthase II | PKS | V | N | Y |
| thiotetroamide C (28) | Streptomyces afghaniensis | fatty acid synthase II | PKS | V | N | Y |
| inhibitors of carbohydrate and energy metabolism | ||||||
| pentalenolactone (29) | Streptomyces avermitilis | glyceraldehyde-3-phosphate dehydrogenase | TS | V | N | N |
| heptelidic acid (30) | Aspergillus oryzae | glyceraldehyde-3-phosphate dehydrogenase | TS | V | N | N |
| aurovertin E (31) | Metarhizium anisopliae | ATP synthase beta chain | PKS | P | N | N |
| citreoviridin (32) | Aspergillus terreus | ATP synthase beta chain | PKS | P | N | N |
| inhibitors of amino acid metabolism | ||||||
| phaseolotoxin (33) | Pseudomonas syringae | ornithine carbamoyltransferase | other | V | N | N |
| aspterric acid (34) | Aspergillus terreus | dihydroxy-acid dehydratase | TS | V | N | Y |
| inhibitors of nucleotide metabolism | ||||||
| mycophenolic acid (35) | Penicillium brevicompactum | inosine-5′-monophosphate dehydrogenase | PKS | V | Y | Y |
| inhibitors of xenobiotics biodegradation | ||||||
| cephamycin C (36) | Streptomyces clavuligerus | beta-lactamase | NRPS | P | N | N |
verified (V) or proposed (P).
approved (A) or not (N).
discovery is SRE inspired (Y) or not (N).
2.1. Inhibitors of DNA replication
DNA replication is a fundamental life process that produces identical copies of DNA molecules using the original chromosome in a template process. This process requires multiple enzyme complexes in coordination to achieve high efficiency and accuracy. This process and the enzymes involved are highly conserved among prokaryotes, while divergent in eukaryotes. Therefore, bacterial DNA replication enzymes are ideal targets for mining antibacterial agents24. The first NP identified to target DNA replication process was novobiocin (1), which was isolated in 1955 and used as an anti-infection agent in clinical treatment of Staphylococcus25 (Fig. 2). The enzyme target of 1 was determined to be DNA gyrase, which belongs to a subclass of type II topoisomerase26. This enzyme unwinds double-stranded DNA by reducing the topological strain in an ATP dependent manner27. Biosynthetic studies of 1 revealed the gyrB gene present within the BGC encodes a variant of the housekeeping DNA gyrase that is not sensitive to 128, 29 (Table 1). Co-localization of self-resistant DNA gyrase variants were also found in BGCs of other DNA gyrase inhibitors, such as chlorobiocin (2)30 and coumermycin A1 (3)31 (Fig. 2 and Table 1).
Fig. 2.
Example NP DNA replication inhibitors that employ a mutated target as SRE encoded in biosynthetic gene clusters.
2.2. Inhibitors of protein biosynthesis
Enzymes involved in protein biosynthesis are classic targets for the development of antibacterial agents because of the well-studied functional differences between prokaryotes and eukaryotes32. During protein biosynthesis, transfer RNAs (tRNAs) are first aminoacylated by the cognate amino acids by the 20 aminoacyl-tRNA synthetases (aaRSs). There are several NPs targeting different aaRSs including mupirocin (5) (IleRS) 33, 34, thiomarinol (6) (IleRS)35, cladosporin (7) (LysRS)36, borrelidin (8) (ThrRS)37, albomycin δ2 (9) (SerRS)38, indolmycin (10) (TrpRS)39 and agrocin 84 (11) (LeuRS)40, 41 (Fig. 3). Among these natural inhibitors, 5 is approved by the FDA to treat a skin infectious disease impetigo, and 7 is a FDA approved drug against malarial parasites. To prevent inhibition of housekeeping aaRSs by these potent inhibitors, the producing strains of these NPs encode mutated versions of corresponding aaRSs as SREs in the BGCs34–38, 41 (Table 1).
Fig. 3.
Example NP protein biosynthesis inhibitors that employ a mutated target as SRE encoded in biosynthetic gene clusters.
Many classic NPs inhibit the translation process at the ribosome, including tetracyclines, erythromycins, etc. The producers of these well-studied NPs use different mechanisms of self-resistance as noted earlier. The use of SRE for host protection of ribosomal inhibitors is also a widely adopted strategy. Thiopeptides such as thiocillin (12) perturb translation by binding within a cleft located between the ribosomal protein L11 and rRNA to block elongation factor G42. Study of 12 biosynthesis revealed that two identical copies of a variant of the housekeeping 50S ribosomal protein L11 are encoded in the BGC of 12, which are proposed to function as SREs to 1243 (Fig. 3 and Table 1). In another example, rubradirin (13) can selectively bind to one specific translation initiation factor to arrest protein biosynthesis44 (Fig. 3). Similar to 12, two identical copies of the translation initiation factor in the BGC are proposed to protect the host during biosynthesis of 1345 (Table 1). Similarly, identification of BGCs of NPs thiomuracin (15)46 and GE2270 (14)47 that inhibit elongation factors Tu (EF-Tu) revealed both clusters contain genes encoding variants of housekeeping EF-Tu as SREs (Fig. 3 and Table 1).
Most nascent proteins synthesized by ribosome require post-translational modifications to become mature and fully functional. One required modification in bacteria is removal of N-terminal methionine residue catalysed by methionine aminopeptidase (MetAP). A bacterial antibiotic bengamide B (16) produced by Myxococcus virescens is able to block type I MetAP48 (Fig. 3). Experimental studies of the type I MetAP variant encoded within the corresponding BGC confirmed it can confer self-resistance in the presence of 1649 (Table 1). The presence of MetAP variants as SREs is also observed in the BGC of fumagillin (17), which is a fungal MetAP inhibitor produced by Aspergillus fumigatus. Interestingly, two proposed SREs are encoded in the cluster, which correspond to additional copies of type I and type II MetAP50 (Fig. 3 and Table 1).
2.3. Inhibitors of protein degradation
Protein degradation is a cellular process that controls the intracellular concentration of proteins and enzymes, and is a key checkpoint for programmed cell death. Accurate and timely protein degradation is catalysed by protein complexes known as proteasomes. Disorder of a proteasome subunit is lethal to the organism. Salinosporamide A (18), isolated from a marine bacteria Salinispora tropica, is able to covalently inactivate proteasome subunit beta through a β-lactone ring opening step 51 (Fig. 4a). In the BGC of 18, an additional, mutated copy of the proteasome subunit beta was encoded. Functional assays confirmed the additional copy is indeed a SRE that confers resistance to 1852 (Table 1). Moore and coworkers showed that the SRE contains a single A49V mutation, which has been shown to constrict the S1 binding pocket and hinder 18 binding52. The biosynthesis of proteasome inhibitor eponemycin (19), produced by Streptomyces hygroscopicus, also involves the use of a mutated copy of proteasome subunit beta in the BGC that functions as SRE53 (Fig. 4a and Table 1). Although 19 itself is not further developed into a new drug, the epoxyketone warhead of 19 was used to design the FDA approved therapeutic agent Carfilzomib that targets the proteasome in the treatment of multiple myeloma54.
Fig. 4.
Example NP metabolic enzyme inhibitors that employ a mutated target as SRE encoded in biosynthetic gene clusters.
2.4. Inhibitors of lipid metabolism
Lipids are essential biomolecules involved in formation of cellular membranes, energy storage and signal transduction, etc. Therefore, most enzymes involved in the synthesis and degradation of lipids are essential to living organisms. A number of BGCs producing NPs targeting the fatty acid biosynthetic pathway encode SREs to confer self-resistance. For example, andrimid (21) targets acetyl-coenzyme A (acetyl-CoA) carboxylase, which catalyses the synthesis of malonyl-CoA via carboxylation of acetyl-CoA55 (Fig. 4b). A second copy of acetyl-CoA carboxylase in the BGC of 21 was validated to be the self-resistance-gene56, 57 (Table 1). Both platensimycin (22)58 and platencin (23)59 produced by Streptomyces platensis are potent inhibitors of FabF, the beta-ketoacyl-acyl carrier protein synthase II in fatty acid biosynthesis (Fig. 4b). The FabF homologs encoded within 22 and 23 BGCs have been demonstrated to be SREs60, 61 (Table 1). The bacterial polyketide kalimantacin (24) inhibits FabI, the enoyl-acyl carrier protein reductase in the fatty acid biosynthetic pathway62 (Fig. 4b). The second copy of fabI co-localized with 24 biosynthetic genes was verified to be the self-resistance gene62 (Table 1).
In addition to fatty acids, steroids and steroid alcohols are also lipids that function as vital components of cell membranes or signalling molecules in microorganisms. Both cholesterol (used by animals) and ergosterol (used by fungi and protozoa) are derived from triterpene precursors such as squalene, which in turn are polymerized from isopentenyl pyrophosphate build blocks derived from the mevalonate pathway. Whereas the ergosterol pathway in fungi is the target of antifungal drugs such as azoles63, the cholesterol biosynthetic pathway in humans has been a very successful target for treating hypercholesterolemia64. One of the most famous and successful fungal NPs, lovastatin (25), targets the rate limiting step of the mevalonate pathway, 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR). 25 is an approved drug (Mevacor) that treats high blood cholesterol levels65 (Fig. 4b), and has inspired the development of both semisynthetic and synthetic statin drugs. 25 is synthesized by the lov BGC from Aspergillus terreus, presumably to target the sterol biosynthetic pathways of competing fungal species. In the middle of the lov BGC is a second copy of the gene encoding a putative HMGR, and is demonstrated to confer self-resistance66, 67 (Table 1). Other NPs inhibiting the sterol biosynthetic pathway have been explored as potential cholesterol-lowering drugs. One notable example is zaragozic acid (26) that inhibits squalene synthase, the enzyme that dimerizes farnesyl diphosphate to afford squalene68 (Fig. 4b). As squalene synthase is vital to the producing fungus, a gene encoding a slightly mutated squalene synthase was found to be co-localized in the gene cluster, although the functional role as an SRE has not been verified69, 70 (Table 1).
2.5. Inhibitors of carbohydrate and energy metabolism
Not surprisingly, the central metabolism of microorganisms is heavily targeted by NPs in microbial warfare, including both anaerobic (glycolysis) and aerobic (respiration) processes. Because of the highly conserved enzymes in metabolism across species, SREs are commonly found in biosynthetic pathways of producing hosts. For example, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) catalyses the sixth step of glycolysis, which converts glyceraldehyde 3-phosphate to the high energy D-glycerate 1,3-bisphosphate in an NAD+ dependent fashion. There are currently two known NPs, the bacterial pentalenolactone (29)71 and fungal heptelidic acid (30)72, that can covalently inactivate GAPDH using an epoxide moiety as a warhead targeting the active site cysteine (Fig. 4c). In the BGC of both NPs, an additional copy of GAPDH has been found and is verified to serve as SRE73, 74 (Table 1). At the end of aerobic respiration, adenosine triphosphate (ATP) synthase is an essential enzyme to generate ATP driven by the electrochemical gradient. Two structurally similar fungal NPs, aurovertin E (31)75, 76 and citreoviridin (32)77, produced by Metarhizium anisopliae and A. terreus, target ATP synthase beta chain as noncompetitive inhibitors75 (Fig. 4c). An additional mutated copy of ATP synthase beta chain is encoded by the BGC of each molecule, presumably to protect the producing hosts76, 78 (Table 1).
2.6. Inhibitors of amino acid metabolism
Phaseolotoxin (33) isolated from plant pathogen Pseudomonas syringae is a potent irreversible inhibitor of ornithine carbamoyltransferase in the arginine biosynthetic pathway79 (Fig. 4d). 33 is a devastating toxin in agriculture, because it leads to halo blight disease that caused loss of beans production worldwide80. The producing organism is able to survive during NP production by co-expressing a mutated ornithine carbamoyltransferase SRE, which is confirmed to be insensitive to this toxin81, 82 (Table 1).
2.7. Inhibitors of nucleotide metabolism
Inosine-5’-monophosphate dehydrogenase (IMPDH) catalyses the first step of guanine biosynthesis. Inhibition of this pathway has been effective in arresting the development of lymphocytes due to de novo biosynthesis of guanine, and is the only way to supply both adenosine monophosphate (AMP) and guanosine monophosphate (GMP) in these cells. The fungal NP mycophenolic acid (35) targeting this enzyme was approved by FDA as an immunosuppressant to prevent rejection during organ transplantation83 (Fig. 4e). Biosynthesis studies of 35 in Penicillium brevicompactum revealed a second copy of IMPDH is encoded in the BGC as a SRE84 (Table 1).
2.8. Inhibitors of xenobiotics biodegradation
Xenobiotics are substances that are not naturally produced within the host cells such as some environmental toxins, external antibiotics or synthetic chemicals. Biodegradation of these xenobiotic compounds is often a necessary process to ensure the survival of the host. β-lactam antibiotics are commonly produced by microorganisms as cell wall biosynthesis inhibitors and are widely used as antibiotics. In nature, an evasive strategy employed by bacteria is to evolve β-lactamase that can cleave the β-lactam compounds and become resistant. Interestingly, nature has evolved NPs that can inhibit β-lactamase which renders the organisms sensitive again. This has led to the co-use of both β-lactams and β-lactamase inhibitors to overcome clinical resistance. Cephamycin C (36), produced by Streptomyces clavuligerus, is such a NP85 (Fig. 4f). In the BGC of 36, a mutated copy of β-lactamase is co-expressed and is not sensitive to 3686 (Table 1).
3. Recent studies on natural products using a resistance gene directed approach
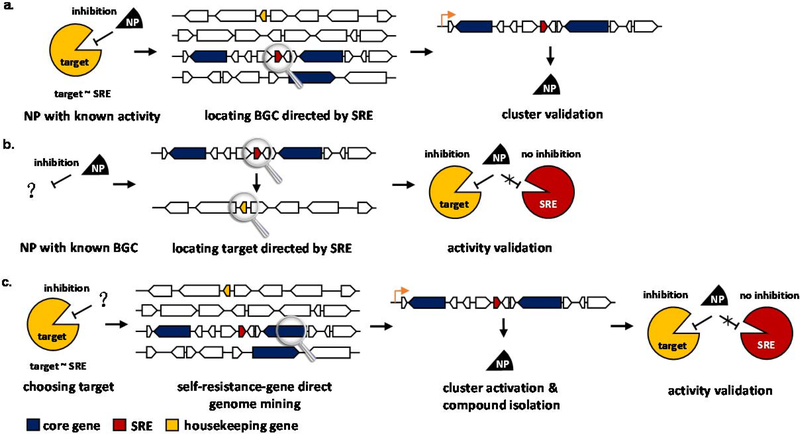
The above examples illustrate that the co-localizations of SRE genes within the BGC is a common occurrence. Most importantly, the identification of a putative SRE gene in the BGC through bioinformatic analysis or functional verification can connect the encoded NP to a potential target. The SRE, if predicted correctly, can therefore be a powerful predictive window to the bioactivity of a NP. This has several important implications to genome mining (Fig. 5): i) If the activity of the compound is already known, searching for a BGC with a potential resistance gene can help locate the gene cluster; ii) Many NPs are discovered without a molecular target. If a BGC is identified and contains a potential SRE, this can connect the molecule to its activity; and iii) Using a molecular target as a query, searching for BGCs that contain a corresponding SRE can lead to target-guided genome mining. Here we will highlight recent examples of using SREs in these three approaches.
Fig. 5.
NPs discovery inspired by a self-resistance gene. a. Locating the BGC of a NP with known activity. b. Rediscovering the activity of a known NP. c. Discovering NP with desired activity.
3.1. Locating the BGCs of NPs with known biomolecular targets
With both the structure and activity of a NP elucidated, one can use both information as probes to locate the BGCs (Fig. 5a). For example, the BGC of gyrase inhibitor 3 was located in the producing strain Streptomyces rishiriensis by identifying BGC that contains both the biosynthetic core gene dTDP-glucose 4,6-dehydratase and the self-resistance-gene gyrB20, 23. The latter was hinted from the presence of a SRE in the BGC of a related compound 1. In an example from fungi, the BGC of IMPDH inhibitor 35 was located within the producing strain Penicillium brevicompactum (Fig. 4e). The biosynthetic origin of 35 was not immediately clear at the onset of the study83, and core enzymes were not useful in pinpointing the BGC83, 84. Although there was no precedent of BGCs encoding IMPDH inhibitors requiring a SRE, the authors proposed that a homolog of IMPDH may be present to confer resistance to 3584. Based on this hypothesis, the BGC of 35 was successfully discovered using the IMPDH gene as a probe.
An example of using a SRE to locate a gene cluster from our lab is that of the fungal MetAP2 inhibitor 1750. It is known that the bis-epoxide cyclohexanol portion of 17 (fumagillol) is derived from a farnesyl diphosphate precursor87. When we searched through the sequenced and annotated A. fumigatus genome for BGCs containing canonical sesquiterpene cyclases, no suitable BGC could be identified. Suitability here refers to the presence of expected tailoring enzymes that match the structure of 17. An alternative strategy was to search for additional copies of MetAP, which was an elucidated target of 1788. In doing so, a BGC on the eighth chromosome was identified to contain additional copies of both MetAP1 and MetAP2. While the BGC encoded a multitude of tailoring enzymes, as well as a PKS that could synthesize the dioic acid, no terpene cyclase was found. Further detailed annotation of the gene cluster revealed that one gene was misannotated. Upon correction of the 5’ region assignment, the gene was reannotated as a putative membrane bound prenyltransferase with weak sequence homology to UbiA. Biochemical characterization of the enzyme demonstrated it is a new type of sesquiterpene cyclase that synthesizes the 17 precursor β-trans-bergamotene50. All the remaining genes in the BGC were characterized to be responsible for synthesizing 17 by both the Keller group and our group50, 89, 90. This example illustrates the utility of using the SRE hypothesis to find BGCs that may otherwise be difficult to identify using a core enzyme alone.
3.2. Discovering the biomolecular targets of known NPs
Besides locating the BGC of a NP of known bioactivity, the SRE presence in a BGCs can also reveal the bioactivity of discovered NPs (Fig. 5b). For example, 24 isolated from Pseudomonas fluorescens were identified to be promising novel antibacterial agents with a strong selective anti-staphylococcal activity (Fig. 4b). The mechanism of action of 24 however, had remained unknown62. When characterizing the function of each gene within the BGC, the Lavigne group observed a knockout of BatG, a predicted FabI isozyme, did not affect the production of this antibiotic. On the other hand, heterologous expression of BatG in a 24 sensitive host conferred this host resistance to 2462. FabI is an essential metabolic enzyme that functions as a trans-2-enoyl-ACP reductase in the fatty acid biosynthetic pathway. Thus they proposed the molecular target of 24 is FabI, and BatG is able to complement the function of FabI during production of 2462 (Table 1). One may ask how the original host can survive when batG is inactivated, since the SRE is no longer present. It may be that the housekeeping copy of FabI has already accumulated some mutations to gain resistance to 24, and the SRE is an additional safeguard. This may explain why there is not always an additional copy of the SRE in the BGCs of the bioactive NP. The ability of the housekeeping copy to be resistant may already be built in during evolution, and render SRE unnecessary. However, identification of these self-resistant housekeeping copies will need higher resolution bioinformatic analysis.
In another example, griselimycin (4) is a cyclic peptide isolated from Streptomyces strains in 1960s (Fig. 2). Although it has excellent broad spectrum antibacterial activity including against Mycobacterium tuberculosis that causes tuberculosis (TB), further development of this NP as an anti-TB drug was impeded because of its poor pharmacokinetics properties91. While analogues of 4 were synthesized and tested, however, efforts were abandoned when rifampin became available to treat TB92. Without knowledge of the mode of action, further medicinal chemical development for activity improvement was difficult. Recently, Kling et al. discovered the molecular target of 4 is DNA polymerase sliding clamp DnaN, which was hinted by a DnaN homolog encoded in the BGC that was confirmed to be the SRE93 (Table 1). This discovery unveiled a novel mode of action of 4, and resurrected its promising potential to be developed into a new anti-TB drug. This research outcome again indicated that the BGCs of NPs contain much more information than just the biosynthetic pathway: it is not just limited to how a NP is made, but also how that NP functions.
3.3. (Re)discovering NPs with desired biomolecular targets
While the above examples successfully leveraged the SRE presence to connect NPs to the biological activity, the more useful application of a SRE is to accurately connect biological activity and genomic information in target-guided mining of NPs. The concept is to use SREs as a predictive window to help prioritize and identify BGCs that encode a NP inhibiting the desired target. This effectively bridges the gap discussed earlier between activity-guided screening and untargeted genome mining. A workflow of resistance-gene directed NP discovery could be proposed as follows: 1) Identifying a desired drug or pesticide target that is also essential and conserved in microorganisms of interest; 2) Searching through a genome database for BGCs carrying duplicate copies of the target using desired biosynthetic core enzymes; 3) Activating the biosynthetic genes to produce the NP using various synthetic biology tools; 4) Isolating the NP and determining its structure via multiple analytical chemistry techniques. 5) Validating the bioactivity of NP using in vitro biochemical assays of housekeeping enzymes or in vivo growth inhibition assays of a susceptible heterologous host with or without expressing SRE (Fig. 5c). This can be a general workflow to perform resistance-gene directed NP discovery using a genomic database.
To identify BGCs associated with SREs for genome mining, new bioinformatic tools are being developed with more efficient search algorithms. Ziemert and co-workers developed a web-based genome mining tool Antibiotic Resistant Target Seeker (ARTS), which specifically uses SRE-directed approaches for NPs discovery in bacteria94. The user provides genomic data of an organism of interest as input, and the putative BGCs that could produce NPs to inhibit all possible targets will be displayed in a dynamic graphic as an output. This search algorithm processes data in the following steps: 1) The genome will first be processed by automated genome mining tools to detect BGCs based on biosynthetic core genes; 2) The putative resistance genes will be located according to known self-resistance models and duplication of essential housekeeping genes, which is validated by phylogenetic analysis of horizontal gene transfer; 3) The BGCs co-localized with a self-resistance gene will be screened and highlighted as an output, which are summarized into an interactive output table to rapidly visualize both known and putative targets. Although ARTS is currently focused on actinobacteria, the methodology of this automated pipeline can be extended to other organisms to prioritize genome mining based on potential bioactivity.
Another genome mining software, clusterTools, developed by Challis and co-workers also enables in silico resistance-gene directed NP discovery95. Compared to existing bioinformatics tools, clusterTools is designed to identify putative BGCs of interest using Hidden Markov Models (HMMs) of specific functional elements. A customized search can be performed with inputs of HMMs including not only functional domains of biosynthetic core enzymes and tailoring enzymes, but also other elements such as resistance or regulatory genes. This software enables a more precise biosynthetic-hypothesis driven and resistance-gene directed search of BGCs at the same time.
Recently, Andersen and co-workers developed a similar pipeline to perform resistance-gene directed genome mining from fungal species96. 72 unique putative resistance genes and clusters were identified by applying the search algorithm to 51 Aspergillus and Penicillium species.
The first detailed experimental methodology was performed by Moore’s group97. They successfully discovered a group of FASII inhibitors using their search pipeline to identify NPs that may inhibit parts of the fatty acid biosynthetic pathway. The pipeline can be summarized as follows: 1) Possible self-resistance gene were selected from duplicate members within delineated orthologous groups in the pan-genome of 86 Salinispora strains; 2) One PKS44 cluster was identified to contain a putative SRE, which is a homolog of fatty acid biosynthetic enzyme; 3) This PKS44 cluster was reconstituted in a heterologous host Streptomyces coelicolor, which produced a group of thiolactomycins (27) that were previously reported as FASII inhibitors98 (Fig. 4b and Table 1). Following a similar procedure, the thiotetroamide C (28) gene cluster was also identified in Streptomyces afghaniensis97, 99 (Fig. 4b and Table 1); and 4) The putative self-resistance genes ttmE and ttmJ were verified to aid the heterologous host to survive in the presence of 2897. The output of this pipeline demonstrated the practicality to perform a self-resistance-gene directed approach to find NPs with desired mode of action.
The proteasome inhibitor fellutamide B (20) was rediscovered by Yeh and co-workers guided by a putative resistance gene encoding a proteasome component C5 homolog100 (Fig. 4a and Table 1). First, the authors identified a BGC in Aspergillus nidulans that contains the gene inpE that encodes a putative proteasome subunit. This cluster was then activated by a serial promoter replacement to produce the NP 20, which was previously characterized to be a potent proteasome inhibitor101. Finally inpE was demonstrated to confer self-resistance to 20 by in vivo studies100.
Notwithstanding the above examples of SRE-directed mining, the rediscovery of 27, 28 and 20 led to known NPs with previously established modes of action. In the last few years, a couple of examples have emerged for the discovery of NPs with previously unknown targets. For example, Müller’s group discovered novel topoisomerase inhibitors from myxobacteria using topoisomerase-targeting pentapeptide repeat protein (PRP) as a probe102. This SRE was previously observed in BGCs of gyrase inhibitors albicidin and cystobactamid. A BGC encoding a polyketide metabolite was identified to contain a PRP in the genome of Pyxidicoccus fallax. Two new NPs, pyxidicycline A and B, were produced upon mutating the promoters of polyketide synthase genes. Bioactivity studies confirmed both pyxidicycline A and B are selective inhibitors of topoisomerases. Although the resistance gene here is not directly a mutated copy of the targeted housekeeping topoisomerase, this example validated the possibility of using SREs to discover new NPs with desired bioactivities.
We applied target-guiding genome mining to find herbicide leads with a new mode of action from a fungal genomic database. We targeted the enzyme dihydroxy-acid dehydratase (DHAD), the third enzyme in the branched chain amino acid biosynthesis pathway103. The first enzymes on this pathway, acetolactate synthase is the most targeted enzyme for herbicide development with over 50 commercialized agents. However, there is no inhibitor of DHAD that has been reported to work in planta. After confirming DHAD enzymes in plants are close homologs of the housekeeping fungal DHADs, we scanned fungal genomes in publicly available databases for a BGC that encodes a possible SRE copy of DHAD. We located a conserved gene cluster that encodes a duplicate copy of DHAD that is ~60% identical to the well-conserved housekeeping DHAD present in all fungi. The biosynthetic core genes from Aspergillus terreus were then reconstituted in a heterologous host to produce aspterric acid (34), a known sesquiterpene NP with unknown biological target104 (Fig. 4d, Table 1). We demonstrated that 34 is a competitive inhibitor of DHAD, and the SRE AstD is indeed resistant to 34. The broad spectrum herbicidal activities of 34 were then confirmed using a variety of model plant species. Furthermore, we introduced astD into Arabidopsis thaliana to construct a transgenic plant that is resistant to 34. The combination of the compound with new mode of action and a resistant plant makes 34 a promising herbicide lead. Finally, the structure of A. thaliana DHAD was solved and the homology structure of AstD was modelled, which showed the active site entrance in AstD may be significantly narrowed to prevent binding of 34. Our rediscovery of 34 with the target-guided mining demonstrates this approach can indeed lead to NPs with novel modes of action.
4. Conclusions and prospects
The rapid development of genome sequencing technologies has reinvigorated NP discovery using genome-mining approaches. The duplicated and insensitive SREs co-localized with NP BGCs likely co-evolved with the biosynthetic pathways of NPs. While study of the molecular mechanism of how SREs are insensitive can lead to new insights in how nature evolves resistance to powerful antibiotics, the SRE provides a unique window to predict the biological activities of NPs22, 23.
However, it is important to note that our limited biosynthetic knowledge means that prediction of a self-resistance gene in a BGC could sometimes be inaccurate. Using an example from our lab, when we tried to discover a new IMPDH inhibitor using the presence of a SRE, we recently discovered a BGC that synthesizes the PKS-NRPS product pyranonigrin A. However, pyranonigrin A was shown not to be an IMPDH inhibitor105. Therefore, a housekeeping enzyme homologue could be coincidently duplicated in the BGC of a NP and does not serve as a SRE.
In other cases, a housekeeping enzyme variant in a BGC could be bona fide biosynthetic enzyme that involved in the assembly of NPs rather than a SRE. For example, similar to that of 9, a second copy of seryl-tRNA synthetase is located within the valanimycin BGC. However, further characterization of the function revealed that this seryl-tRNA synthetase is responsible for catalysing seryl transfer in the valanimycin biosynthetic pathway106. In another recent example, Abe and coworkers discovered the BGC of an Ile-tRNA synthetase (IleRS) inhibitor SB-203208 in Streptomyces sp. NCIMB 40513 using the presence of an SRE copy of IleRS as a search criterion107. Surprisingly, the additional copy of IleRS (SbzA) in the BGC was demonstrated to participate in the biosynthetic steps of SB-203208. Therefore, it is challenging to accurately predict true SREs, especially for enzymes that may play functional roles in the biosynthetic pathway. However, we expect this to be less ambiguous as we build up more knowledge in biosynthetic enzymology. The accumulating knowledge of self-resistance mechanisms in the future will also facilitate new automated bioinformatics tools to prioritize cryptic BGCs with higher accuracy. We expect a self-resistance-gene guided approach to bridge the traditional and modern methods of NP discovery during this exciting period of NP renaissance.
6. Acknowledgements
This work was supported by the NIH (1R35GM118056) to Yi Tang.
Biography
 Nicholas Liu received his B.S. in Chemical and Biomolecular Engineering from Johns Hopkins University in 2014. He is currently working toward his Ph.D. under the guidance of Prof. Yi Tang at the University of California, Los Angeles. His research interests focus on self-resistance gene directed natural product discovery and biosynthesis, particularly on polyketide natural products.
Nicholas Liu received his B.S. in Chemical and Biomolecular Engineering from Johns Hopkins University in 2014. He is currently working toward his Ph.D. under the guidance of Prof. Yi Tang at the University of California, Los Angeles. His research interests focus on self-resistance gene directed natural product discovery and biosynthesis, particularly on polyketide natural products.
 Yi Tang received his undergraduate degree in Chemical Engineering and Material Science from Penn State University. He received his Ph.D. in Chemical Engineering from California Institute of Technology in 2002. After NIH postdoctoral training in Chemical Biology at Stanford University, he started his independent career at the University of California Los Angeles in 2004. He is currently Professor in the Department of Chemical and Biomolecular Engineering at UCLA, and in the Department of Chemistry and Biochemistry. His lab is interested in natural product biosynthesis, biocatalysis and protein engineering.
Yi Tang received his undergraduate degree in Chemical Engineering and Material Science from Penn State University. He received his Ph.D. in Chemical Engineering from California Institute of Technology in 2002. After NIH postdoctoral training in Chemical Biology at Stanford University, he started his independent career at the University of California Los Angeles in 2004. He is currently Professor in the Department of Chemical and Biomolecular Engineering at UCLA, and in the Department of Chemistry and Biochemistry. His lab is interested in natural product biosynthesis, biocatalysis and protein engineering.
 Yan Yan received his B.S. in Biology from China Agricultural University in 2008, and his M.S. in Organic Chemistry from Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences in 2013. After receiving his Ph.D. under the direction of Prof. Yi Tang at University of California, Los Angeles in 2019, he is currently a postdoctoral scholar in the same group. His research interests focus on self-resistance-gene directed natural products discovery, self-resistance mechanism of natural products, and natural products biosynthesis.
Yan Yan received his B.S. in Biology from China Agricultural University in 2008, and his M.S. in Organic Chemistry from Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences in 2013. After receiving his Ph.D. under the direction of Prof. Yi Tang at University of California, Los Angeles in 2019, he is currently a postdoctoral scholar in the same group. His research interests focus on self-resistance-gene directed natural products discovery, self-resistance mechanism of natural products, and natural products biosynthesis.
Footnotes
5. Conflicts of interest
Yi Tang is a cofounder of Hexagon Biosciences, a start-up biotechnology company using genome mining to discover new bioactive natural products.
7. References
- 1.Newman DJ and Cragg GM, J. Nat. Prod, 2016, 79, 629–661. [DOI] [PubMed] [Google Scholar]
- 2.Cantrell CL, Dayan FE and Duke SO, J. Nat. Prod, 2012, 75, 1231–1242. [DOI] [PubMed] [Google Scholar]
- 3.Yan Y, Liu Q, Jacobsen SE and Tang Y, EMBO Rep., 2018, 19, e46824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henke MT and Kelleher NL, Nat. Prod. Rep, 2016, 33, 942–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lobanovska M and Pilla G, Yale J Biol Med, 2017, 90, 135–145. [PMC free article] [PubMed] [Google Scholar]
- 6.Harvey AL, Edrada-Ebel R and Quinn RJ, Nat. Rev. Drug Discov, 2015, 14, 111–129. [DOI] [PubMed] [Google Scholar]
- 7.Van Lanen SG and Shen B, Curr. Opin. Microbiol, 2006, 9, 252–260. [DOI] [PubMed] [Google Scholar]
- 8.Nützmann H-W, Scazzocchio C and Osbourn A, Annu. Rev. Genet, 2018, 52, 159–183. [DOI] [PubMed] [Google Scholar]
- 9.Kim HU, Blin K, Lee SY and Weber T, Curr. Opin. Microbiol, 2017, 39, 113–120. [DOI] [PubMed] [Google Scholar]
- 10.Medema MH and Fischbach MA, Nat. Chem. Biol, 2015, 11, 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rutledge PJ and Challis GL, Nat. Rev. Microbiol, 2015, 13, 509–523. [DOI] [PubMed] [Google Scholar]
- 12.Cimermancic P, Medema Marnix H., Claesen J, Kurita K, Brown Laura C. Wieland, Mavrommatis K, Pati A, Godfrey Paul A., Koehrsen M, Clardy J, Birren Bruce W., Takano E, Sali A, Linington Roger G. and Fischbach Michael A., Cell, 2014, 158, 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walsh CT and Fischbach MA, J. Am. Chem. Soc, 2010, 132, 2469–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lautru S, Deeth RJ, Bailey LM and Challis GL, Nat. Chem. Biol, 2005, 1, 265–269. [DOI] [PubMed] [Google Scholar]
- 15.Clevenger KD, Bok JW, Ye R, Miley GP, Verdan MH, Velk T, Chen C, Yang K, Robey MT, Gao P, Lamprecht M, Thomas PM, Islam MN, Palmer JM, Wu CC, Keller NP and Kelleher NL, Nat. Chem. Biol, 2017, 13, 895–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang JJ, Tang X and Moore BS, Nat. Prod. Rep, 2019, DOI: 10.1039/c9np00025a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hopwood DA, Chem. Rev, 1997, 97, 2465–2498. [DOI] [PubMed] [Google Scholar]
- 18.Ziemert N, Alanjary M and Weber T, Nat. Prod. Rep, 2016, 33, 988–1005. [DOI] [PubMed] [Google Scholar]
- 19.Galm U, Hager MH, Van Lanen SG, Ju J, Thorson JS and Shen B, Chem. Rev, 2005, 105, 739–758. [DOI] [PubMed] [Google Scholar]
- 20.Weisblum B, Antimicrob. Agents Chemother, 1995, 39, 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thaker MN, Wang W, Spanogiannopoulos P, Waglechner N, King AM, Medina R and Wright GD, Nat. Biotechnol, 2013, 31, 922–927. [DOI] [PubMed] [Google Scholar]
- 22.Almabruk KH, Dinh LK and Philmus B, ACS Chem. Biol, 2018, 13, 1426–1437. [DOI] [PubMed] [Google Scholar]
- 23.O’Neill EC, Schorn M, Larson CB and Millán-Aguiñaga N, Crit. Rev. Microbiol, 2019, DOI: 10.1080/1040841x.2019.1590307, 1–23. [DOI] [PubMed] [Google Scholar]
- 24.van Eijk E, Wittekoek B, Kuijper EJ and Smits WK, J Antimicrob. Chemother, 2017, 72, 1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith CG, Dietz A, Sokolski WT and Savage GM, Antibiot. Chemother, 1956, 6, 135–142. [PubMed] [Google Scholar]
- 26.Maxwell A, Mol. Microbiol, 1993, 9, 681–686. [DOI] [PubMed] [Google Scholar]
- 27.Pommier Y, Leo E, Zhang H and Marchand C, Chem. Biol, 2010, 17, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steffensky M, Mühlenweg A, Wang Z-X, Li S-M and Heide L, Antimicrob. Agents Chemother, 2000, 44, 1214–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiara AS and Cundliffe E, Gene, 1989, 81, 65–72. [DOI] [PubMed] [Google Scholar]
- 30.Pojer F, Li S-M and Heide L, Microbiology, 2002, 148, 3901–3911. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z-X, Li S-M and Heide L, Antimicrob. Agents Chemother, 2000, 44, 3040–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCoy LS, Xie Y and Tor Y, Wiley Interdiscip. Rev. RNA, 2011, 2, 209–232. [DOI] [PubMed] [Google Scholar]
- 33.El-Sayed AK, Hothersall J, Cooper SM, Stephens E, Simpson TJ and Thomas CM, Chem. Biol, 2003, 10, 419–430. [DOI] [PubMed] [Google Scholar]
- 34.Yanagisawa T and Kawakami M, J. Biol. Chem, 2003, 278, 25887–25894. [DOI] [PubMed] [Google Scholar]
- 35.Fukuda D, Haines AS, Song Z, Murphy AC, Hothersall J, Stephens ER, Gurney R, Cox RJ, Crosby J, Willis CL, Simpson TJ and Thomas CM, PLoS One, 2011, 6, e18031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cochrane RVK, Sanichar R, Lambkin GR, Reiz B, Xu W, Tang Y and Vederas JC, Angew. Chem. Int. Ed. Engl, 2016, 128, 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olano C, Wilkinson B, Sánchez C, Moss SJ, Sheridan R, Math V, Weston AJ, Braña AF, Martin CJ, Oliynyk M, Méndez C, Leadlay PF and Salas JA, Chem. Biol, 2004, 11, 87–97. [DOI] [PubMed] [Google Scholar]
- 38.Zeng Y, Kulkarni A, Yang Z, Patil PB, Zhou W, Chi X, Van Lanen S and Chen S, ACS Chem. Biol, 2012, 7, 1565–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du Y-L, Alkhalaf LM and Ryan KS, Proc. Natl. Acad. Sci, 2015, 112, 2717–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slater SC, Goldman BS, Goodner B, Setubal JC, Farrand SK, Nester EW, Burr TJ, Banta L, Dickerman AW, Paulsen I, Otten L, Suen G, Welch R, Almeida NF, Arnold F, Burton OT, Du Z, Ewing A, Godsy E, Heisel S, Houmiel KL, Jhaveri J, Lu J, Miller NM, Norton S, Chen Q, Phoolcharoen W, Ohlin V, Ondrusek D, Pride N, Stricklin SL, Sun J, Wheeler C, Wilson L, Zhu H and Wood DW, J. Bacteriol, 2009, 191, 2501–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J-G, Park BK, Kim S-U, Choi D, Nahm BH, Moon JS, Reader JS, Farrand SK and Hwang I, Proc. Natl. Acad. Sci, 2006, 103, 8846–8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CMT and Fucini P, Mol. Cell, 2008, 30, 26–38. [DOI] [PubMed] [Google Scholar]
- 43.Wieland Brown LC, Acker MG, Clardy J, Walsh CT and Fischbach MA, Proc. Natl. Acad. Sci, 2009, 106, 2549–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reusser F, Biochemistry, 1973, 12, 4524–4528. [DOI] [PubMed] [Google Scholar]
- 45.Kim C-G, Lamichhane J, Song K-I, Nguyen VD, Kim D-H, Jeong T-S, Kang S-H, Kim K-W, Maharjan J, Hong Y-S, Kang JS, Yoo J-C, Lee J-J, Oh T-J, Liou K and Sohng JK, Arch. Microbiol, 2008, 189, 463–473. [DOI] [PubMed] [Google Scholar]
- 46.Hudson GA, Zhang Z, Tietz JI, Mitchell DA and van der Donk WA, J. Am. Chem. Soc, 2015, 137, 16012–16015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tocchetti A, Maffioli S, Iorio M, Alt S, Mazzei E, Brunati C, Sosio M and Donadio S, Chem. Biol, 2013, 20, 1067–1077. [DOI] [PubMed] [Google Scholar]
- 48.Towbin H, Bair KW, DeCaprio JA, Eck MJ, Kim S, Kinder FR, Morollo A, Mueller DR, Schindler P, Song HK, van Oostrum J, Versace RW, Voshol H, Wood J, Zabludoff S and Phillips PE, J. Biol. Chem, 2003, 278, 52964–52971. [DOI] [PubMed] [Google Scholar]
- 49.Wenzel SC, Hoffmann H, Zhang J, Debussche L, Haag-Richter S, Kurz M, Nardi F, Lukat P, Kochems I, Tietgen H, Schummer D, Nicolas J-P, Calvet L, Czepczor V, Vrignaud P, Mühlenweg A, Pelzer S, Müller R and Brönstrup M, Angew. Chem. Int. Ed, 2015, 54, 15560–15564. [DOI] [PubMed] [Google Scholar]
- 50.Lin H-C, Chooi Y-H, Dhingra S, Xu W, Calvo AM and Tang Y, J. Am. Chem. Soc, 2013, 135, 4616–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR and Fenical W, Angew. Chem. Int. Ed, 2003, 42, 355–357. [DOI] [PubMed] [Google Scholar]
- 52.Kale AJ, McGlinchey RP, Lechner A and Moore BS, ACS Chem. Biol, 2011, 6, 1257–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schorn M, Zettler J, Noel JP, Dorrestein PC, Moore BS and Kaysser L, ACS Chem. Biol, 2014, 9, 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim KB and Crews CM, Nat. Prod. Rep, 2013, 30, 600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freiberg C, Brunner NA, Schiffer G, Lampe T, Pohlmann J, Brands M, Raabe M, Häbich D and Ziegelbauer K, J. Biol. Chem, 2004, 279, 26066–26073. [DOI] [PubMed] [Google Scholar]
- 56.Jin M, Fischbach MA and Clardy J, J. Am. Chem. Soc, 2006, 128, 10660–10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X, Fortin PD and Walsh CT, Proc. Natl. Acad. Sci, 2008, 105, 13321–13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, Painter R, Parthasarathy G, Tang YS, Cummings R, Ha S, Dorso K, Motyl M, Jayasuriya H, Ondeyka J, Herath K, Zhang C, Hernandez L, Allocco J, Basilio Á, Tormo JR, Genilloud O, Vicente F, Pelaez F, Colwell L, Lee SH, Michael B, Felcetto T, Gill C, Silver LL, Hermes JD, Bartizal K, Barrett J, Schmatz D, Becker JW, Cully D and Singh SB, Nature, 2006, 441, 358–361. [DOI] [PubMed] [Google Scholar]
- 59.Wang J, Kodali S, Lee SH, Galgoci A, Painter R, Dorso K, Racine F, Motyl M, Hernandez L, Tinney E, Colletti SL, Herath K, Cummings R, Salazar O, González I, Basilio A, Vicente F, Genilloud O, Pelaez F, Jayasuriya H, Young K, Cully DF and Singh SB, Proc. Natl. Acad. Sci, 2007, 104, 7612–7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smanski MJ, Yu Z, Casper J, Lin S, Peterson RM, Chen Y, Wendt-Pienkowski E, Rajski SR and Shen B, Proceedings of the National Academy of Sciences, 2011, 108, 13498–13503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peterson Ryan M., Huang T, Rudolf Jeffrey D., Smanski Michael J. and Shen B, Chem. Biol, 2014, 21, 389–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mattheus W, Masschelein J, Gao L-J, Herdewijn P, Landuyt B, Volckaert G and Lavigne R, Chem. Biol, 2010, 17, 1067–1071. [DOI] [PubMed] [Google Scholar]
- 63.Sheehan DJ, Hitchcock CA and Sibley CM, Clin. Microbiol. Rev, 1999, 12, 40–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Charlton-Menys V and Durrington PN, Exp. Physiol, 2008, 93, 27–42. [DOI] [PubMed] [Google Scholar]
- 65.Stancu C and Sima A, J. Cell. Mol. Med, 2001, 5, 378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chamilos G, Lewis RE and Kontoyiannis DP, Antimicrob. Agents Chemother, 2006, 50, 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hutchinson CR, Kennedy J, Park C, Kendrew S, Auclair K and Vederas J, Antonie van Leeuwenhoek, 2000, 78, 287–295. [DOI] [PubMed] [Google Scholar]
- 68.Bergstrom JD, Kurtz MM, Rew DJ, Amend AM, Karkas JD, Bostedor RG, Bansal VS, Dufresne C, VanMiddlesworth FL and Hensens OD, Proc. Natl. Acad. Sci, 1993, 90, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu N, Hung Y-S, Gao S-S, Hang L, Zou Y, Chooi Y-H and Tang Y, Org. Lett, 2017, 19, 3560–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bonsch B, Belt V, Bartel C, Duensing N, Koziol M, Lazarus CM, Bailey AM, Simpson TJ and Cox RJ, Chem. Comm, 2016, 52, 6777–6780. [DOI] [PubMed] [Google Scholar]
- 71.Cane DE and Sohng J-K, Biochemistry, 1994, 33, 6524–6530. [DOI] [PubMed] [Google Scholar]
- 72.Kato M, Sakai K and Endo A, Biochim. Biophys. Acta 1992, 1120, 113–116. [DOI] [PubMed] [Google Scholar]
- 73.Tetzlaff CN, You Z, Cane DE, Takamatsu S, Omura S and Ikeda H, Biochemistry, 2006, 45, 6179–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shinohara Y, Nishimura I and Koyama Y, Biosci. Biotechnol. Biochem, 2018, DOI: 10.1080/09168451.2018.1549934, 1–8. [DOI] [PubMed] [Google Scholar]
- 75.van Raaij MJ, Abrahams JP, Leslie AG and Walker JE, Proc. Natl. Acad. Sci, 1996, 93, 6913–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mao X-M, Zhan Z-J, Grayson MN, Tang M-C, Xu W, Li Y-Q, Yin W-B, Lin H-C, Chooi Y-H, Houk KN and Tang Y, J. Am. Chem. Soc, 2015, 137, 11904–11907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gause EM, Buck MA and Douglas MG, J. Biol. Chem, 1981, 256, 557–559. [PubMed] [Google Scholar]
- 78.Lin T-S, Chiang Y-M and Wang CCC, Org. Lett, 2016, 18, 1366–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Langley DB, Templeton MD, Fields BA, Mitchell RE and Collyer CA, J. Biol. Chem, 2000, 275, 20012–20019. [DOI] [PubMed] [Google Scholar]
- 80.Mitchell RE and Bieleski RL, Plant Physiol, 1977, 60, 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Staskawicz BJ, Panopoulos NJ and Hoogenraad NJ, J. Bacteriol, 1980, 142, 720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aguilera S, López-López K, Nieto Y, Garcidueñas-Piña R, Hernández-Guzmán G, Hernández-Flores JL, Murillo J and Alvarez-Morales A, J. Bacteriol, 2007, 189, 2834–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bentley R, Chem. Rev, 2000, 100, 3801–3826. [DOI] [PubMed] [Google Scholar]
- 84.Regueira TB, Kildegaard KR, Hansen BG, Mortensen UH, Hertweck C and Nielsen J, Appl. Environ. Microbiol, 2011, 77, 3035–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Daoust DR, Onishi HR, Wallick H, Hendlin D and Stapley EO, Antimicrob. Agents Chemother, 1973, 3, 254–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Coque JJ, Liras P and Martín JF, EMBO J, 1993, 12, 631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Birch AJ and Hussain SF, J. Chem. Soc. C, 1969, DOI: 10.1039/j39690001473, 1473–1474. [DOI] [PubMed] [Google Scholar]
- 88.Liu S, Widom J, Kemp CW, Crews CM and Clardy J, Science, 1998, 282, 1324–1327. [DOI] [PubMed] [Google Scholar]
- 89.Lin H-C, Tsunematsu Y, Dhingra S, Xu W, Fukutomi M, Chooi Y-H, Cane DE, Calvo AM, Watanabe K and Tang Y, J. Am. Chem. Soc, 2014, 136, 4426–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wiemann P, Guo C-J, Palmer JM, Sekonyela R, Wang CCC and Keller NP, Proc. Natl. Acad. Sci, 2013, 110, 17065–17070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Toyohara M, Annales de l’Institut Pasteur. Microbiology, 1987, 138, 737–744. [DOI] [PubMed] [Google Scholar]
- 92.Hoagland DT, Liu J, Lee RB and Lee RE, Adv. Drug Deliv. Rev, 2016, 102, 55–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kling A, Lukat P, Almeida DV, Bauer A, Fontaine E, Sordello S, Zaburannyi N, Herrmann J, Wenzel SC, König C, Ammerman NC, Barrio MB, Borchers K, Bordon-Pallier F, Brönstrup M, Courtemanche G, Gerlitz M, Geslin M, Hammann P, Heinz DW, Hoffmann H, Klieber S, Kohlmann M, Kurz M, Lair C, Matter H, Nuermberger E, Tyagi S, Fraisse L, Grosset JH, Lagrange S and Müller R, Science, 2015, 348, 1106–1112. [DOI] [PubMed] [Google Scholar]
- 94.Alanjary M, Kronmiller B, Adamek M, Blin K, Weber T, Huson D, Philmus B and Ziemert N, Nucleic Acids Res, 2017, 45, W42–W48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.de los Santos ELC and Challis GL, bioRxiv, 2017, DOI: 10.1101/119214, 119214. [DOI] [Google Scholar]
- 96.Kjærbølling I, Vesth T and Andersen MR, mSystems, 2019, 4, e00085–00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tang X, Li J, Millán-Aguiñaga N, Zhang JJ, O’Neill EC, Ugalde JA, Jensen PR, Mantovani SM and Moore BS, ACS Chem. Biol, 2015, 10, 2841–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Price AC, Choi K-H, Heath RJ, Li Z, White SW and Rock CO, J. Biol. Chem, 2001, 276, 6551–6559. [DOI] [PubMed] [Google Scholar]
- 99.Li J, Tang X, Awakawa T and Moore BS, Angew. Chem. Int. Ed, 2017, 56, 12234–12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yeh H-H, Ahuja M, Chiang Y-M, Oakley CE, Moore S, Yoon O, Hajovsky H, Bok J-W, Keller NP, Wang CCC and Oakley BR, ACS Chem. Biol, 2016, 11, 2275–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin G, Li D, Chidawanyika T, Nathan C and Li H, Arch. Biochem. Biophys, 2010, 501, 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Panter F, Krug D, Baumann S and Müller R, Chem. Sci, 2018, 9, 4898–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yan Y, Liu Q, Zang X, Yuan S, Bat-Erdene U, Nguyen C, Gan J, Zhou J, Jacobsen SE and Tang Y, Nature, 2018, 559, 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tsuda Y, Kaneda M, Tada A, Nitta K, Yamamoto Y and Iitaka Y, J. Chem. Soc., Chem. Comm, 1978, 0, 160–161. [Google Scholar]
- 105.Tang M-C, Zou Y, Yee D and Tang Y, AIChE J, 2018, 64, 4182–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Garg RP, Qian XL, Alemany LB, Moran S and Parry RJ, Proc. Natl. Acad. Sci, 2008, 105, 6543–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hu Z, Awakawa T, Ma Z and Abe I, Nat. Commun, 2019, 10, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]