Abstract
The coronavirus disease (COVID-19) belongs to the family Severe Acute Respiratory Syndrome (SARS-CoV). It can be more severe for some persons and can lead to pneumonia or breathing difficulties resulting in the death of immune-compromised patients. We performed a phylogenomic and phylogeographic tree from the collected datasets. Phylogenomic analysis or sequence-based phylogeny showed an evolutionary relationship between the geographical strains. The phylogenomic tree grouped into two major clades consists of various isolates of SARS-CoV-2 and Bat SARS-like coronavirus, Bat coronavirus, and Pangolin coronavirus. The phylogenetic neighbor of newly sequenced Indian strains (Accession: MT012098.1, MT050493.1) was revealed to identify the variations between the nCoV-19 strains. The results showed keen evidence that SARS-CoV-2 has evolved from Bat SARS-like coronavirus. The evolutionary history and comparative proteomic analysis provide a new avenue for the current scientific research related to the coronavirus.
Keywords: SARS-CoV-2, Bat SARS-CoV, Coronavirus, Bat-CoV, Pangolin-CoV, Evolutionary imprints, Genetic diversity
Abbreviations
1. Introduction
The nCoV-19 is potentially contagious, which can transmit quickly from one human to another human (Heymann and Shindo, 2020; Zheng, 2020). This is one of the viruses that infect the human population in almost all countries recently (Mackenzie and Smith, 2020). The nCoV-19 was taxonomically classified as SARS-CoV-2 which belongs to the Sarbecovirus subgenus (Beta-Coronavirus genus) (Chan et al., 2020) owing to its phylogenetic affiliation with Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) (Paraskevis et al., 2020). These belong to the family of RNA virus transmitted from animals (Fehr and Perlman, 2015). In this study, we infer the genome-based molecular evolutionary imprints of SARS-CoV-2 to identify its origin and replication mechanism and compare its proteomic regions with two new isolates of nCoV-19 from India and its ancestral species.
2. Materials and methods
The complete genome sequence of Wuhan seafood market pneumonia virus Wuhan-Hu-1 named as COVID-19 (ACC. No.: NC_045512.2) was retrieved from NCBI (https://www.ncbi.nlm.nih.gov/). The sequence similarity search was performed using the NCBI-BLAST (Boratyn et al., 2013) to collect 250 complete genome sequences of SARS-CoV-2 with high similarity query coverage along with ancestral species (Supplementary Table S1). The phylogenomic tree was manually condensed and constructed with 1000 bootstrapping replicates to identify the origin and replication of COVID-19 using Mega X 10.1.7 Version (Kumar et al., 2018). The phylogeographic large-scale supertree was constructed among the selected members using the Interactive Tree of Life (iTOL) v4 (Letunic and Bork, 2019). Based on the appearance of the phylogenomic cluster, a single isolate was chosen for each cluster along with two newly sequenced Indian strains (Accession: MT012098.1, MT050493.1) to study the comparative proteomic analysis. Furthermore, a statistical genomic diversity was computed for the selected species.
The nucleotide and amino acid base pair compositional variations in mmol fraction unit (Supplementary Table S2) were calculated in R statistical package. The phylogenetic hierarchical clustering was carried out with a library called gplots and Rcolourbrowsers packages for plotting the heatmap and color key representation. The principal component analysis (PCA) was performed in ggplots library to find the convergence and divergence between selected species. A dot plot comparison was carried out for functional coding regions using EMBOSS Dotmatcher (https://www.ebi.ac.uk/Tools/seqstats/emboss_dotmatcher/).
3. Results and discussion
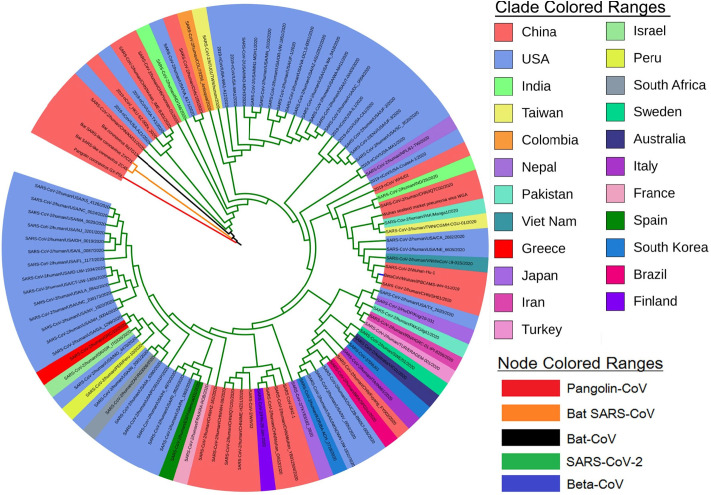
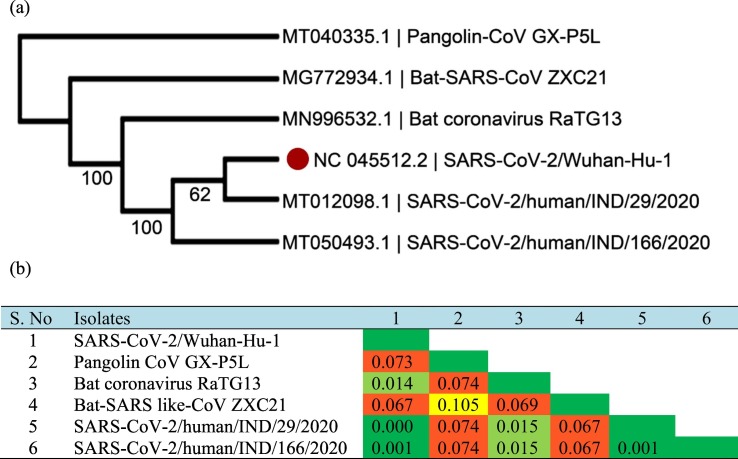
We performed a phylogenomic and phylogeographic tree for 250 isolates of SARS-CoV-2 genomes along with the out-groups. Phylogenomic analysis or sequence-based phylogeny reveals the understanding of the evolutionary relationship between the biological species (Yuan et al., 2014). The phylogenomic tree was separated and grouped according to country wise isolates (color representation in clade) and inter- and intra-specific species variation (color representation in nodes) as shown in Fig. 1 . The phylogenomic tree of our study indicates that the target Wuhan-Hu-1 genome was closely related to different isolates of SARS-COV-2 genomes. Bat coronavirus RaTG13, Bat-SARS-CoV ZXC21, and Pangolin-CoV GX-P5L were found to be an ancestor. A separate phylogenomic tree of various isolates of SARS-CoV-2 was represented in Supplementary Fig. S1. Bat SARS-like coronavirus was found to be an ancestor of newly identified COVID-19. Dataset used for the phylogenomic and phylogeographic tree construction and its closely related species are available in Supplementary Table S1. Based on the constructed genomic tree, we further summarized and select an isolate of phylogenetic neighbor from each cluster along with two newly sequenced Indian isolates (Accession: MT012098.1, MT050493.1), to identify proteomic differentiation between them. Based on these criteria, a proteome-based phylogenetic analysis was performed. It also provides support evidence to the zoonotic evolution theory that SARS-CoV-2 might have evolved from Bat SARS-like coronavirus and Pangolin coronavirus (as represented in Fig. 2a). When compared with the previous study, the genome of SARS-CoV-2 was found to be about 82%, 96%, and 86.9% identical to the SARS-CoV, Bat-CoV-RaTG13, and Bat-SARS like-CoVZC45, respectively (Zhou et al., 2020).
Fig. 1.
A circular view of phylogenomic and phylogeographic tree of SARS-CoV-2 genome and its closely related species and isolates. Each color of nodes and clades are represents a corresponding genomic features (refer, figure legend).
Fig. 2.
Proteome-based phylogenetic analyses (a) and calculation of genetic diversity (b) for selected species from each cluster in the phylogenomic tree.
The genetic diversity analysis of our study revealed that Wuhan-Hu-1 was closely related to new isolates of two Indian SARS-coronavirus 2 with 100% query coverage and 99.98% sequence similarity respectively. The pair-wise genetic distance occurred in the range between 0.0 and 0.001. However, the SARS-CoV-2 genome was genetically distinct from SARS-CoV and has a relatively long branch length to the Bat-CoV RaTG13 (0.014), Bat-SARS like-CoV ZXC21 (0.067), and Pangolin-CoV GX-P5L (0.073) (Fig. 2b). A total of 99.98% similarity was found across the SARS-CoV-2 genomes obtained from different patients (Lu et al., 2020). Genetic recombination events are complex and more likely occurring in Bat-CoV RaTG13 and Bat-SARS like-CoV ZXC21than in SARS-CoV-2 (Jaimes et al., 2020; Lu et al., 2020; Paraskevis et al., 2020). The Tajima's neutrality test (Tajima, 1989) was estimated to identify the mutations by DNA polymorphism. It shows that the nucleotide diversity appears in the range 0.045977 with a high number of segregating sites (1123). It further confirms that the rate of gene duplication events may occur due to radical changes in the segregating sites with a high recombination rate, as described by Clark et al. (2012). The fluctuation of amino acid frequencies was estimated using Jones et al. (1992) evolutionary model (Jones et al., 1992) as represented in Fig. 2c. Comparing to other amino acids, leucine (9.11%) is a highly enriched amino acid found in this group. As per the earlier investigation by Matsushima et al. (2019), the diverse effects of the mutations or recombinant events are occurred due to leucine-rich repeat, and it leads to the origin of several human diseases.
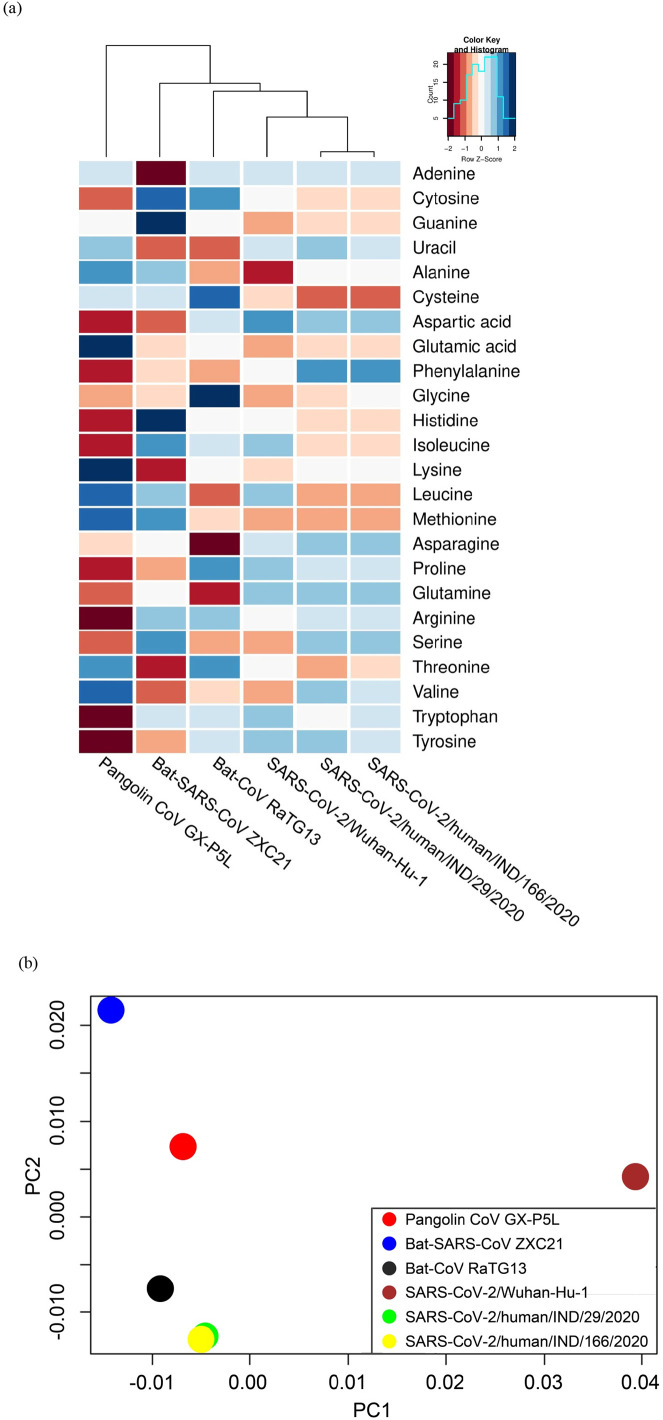
These conversions provide keen evidence of variance in chemical composition (ATP/UTP/GTP/CTP) between the species. The hierarchical clustering results show that the two Indian isolates SARS-CoV-2/human/IND/29/2020 and SARS-CoV-2/human/IND/166/2020 are closely related to each other. And it can deviate when SARS-CoV-2/Wuhan-Hu-1 is compared to other ancestral species (Fig. 3a). A low abundance of amino acid was found in Pangolin CoV GX-P5L and Bat-SARS-CoV ZXC21. It also indicates that several compositional variances are found in amino acid bases between the selected species. It was further proved by principal component analysis. It shows that the two Indian isolates and Bat-CoV RaTG13 were closely related to each other while compared to Wuhan-Hu-1 (Fig. 3b). Therefore, these variations may refer that the SARS-CoV-2 genome has been expanded in human hosts due to the establishment of a hyper-variable genomic hotspot population (Wen et al., 2020). As per the previous investigation, Magiorkinis et al. (2004) and Paraskevis et al., 2020 stated that these may occur due to altered diverse lineages in the evolutionary rates or either by ancient recombination events.
Fig. 3.
Representation of compositional variations of nucleotide and amino acid base pair (a) and principle component analysis (b) for selected species.
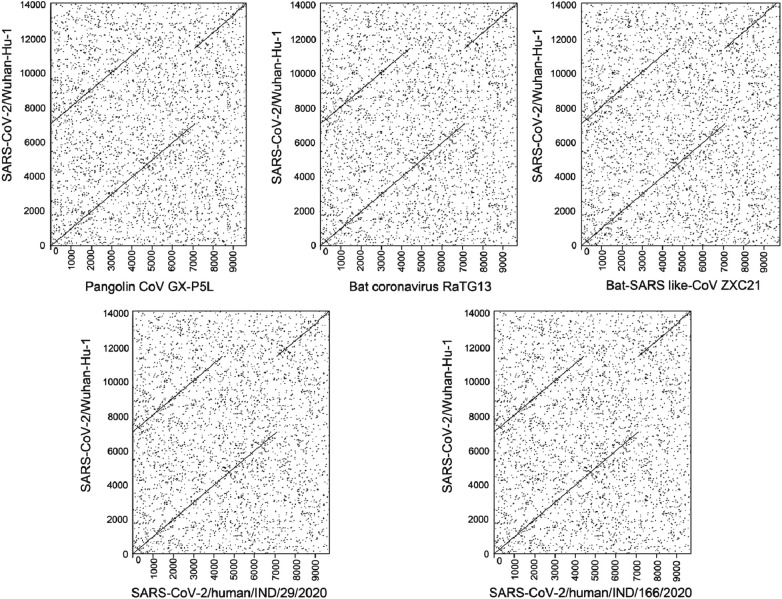
A dot plot comparison was performed for the function coding region between Wuhan-Hu-1 and selected phylogenetic neighbors and ancestors (Fig. 4 ). It shows that 70% of sequences from the SARS-CoV-2 Wuhan-Hu-1 match with selected species (represented in the colored block as aligned regions 1, 2 and 3). Interestingly, a few blocks were identified in Wuhan-Hu-1 isolates in the position 7096–11,500 bp (stand for a white block as mismatch or gapped region). A total of 4404 amino acid bases were uniquely found in the Wuhan-Hu-1. While comparing to our analysis, approximately 30% of Wuhan-Hu-1 doesn't match with any isolates and it may offer new ancestry relationships within the subgenus of sarbecovirus (Paraskevis et al., 2020).
Fig. 4.
A dotplot comparison of functional coding regions (proteomic features) for selected species. X-and Y-axis determines the base pair position of corresponding genomes.
4. Conclusion
We conducted a detailed genomic survey to understand molecular and functional diversity between SARS-CoV-2 and its phylogenomic neighbors. From our analysis, it is inferred that SARS-CoV-2 of Indian origin has highly diverged from the SARS-CoV-2 of Wuhan-Hu-1 which might due to the high mutagenicity rate of COVID. Further high mutagenicity rate was occurred due to the variations of transition/transversion genetic ratio between the selected pairs. It is speculated that this divergence in the evolution of Indian origin SARS-CoV-2 is responsible for the lower death rate scenario in India. Hence, the present genomic and proteomic impending of this virus will provide a lead to the researchers, in turn to understand the mutation rate among Indian strains and helps to unravel the mechanism behind its emergence and also in drug discovery.
Funding
Authors are thankful to Ministry of Science and Technology and Science and Engineering Research Board, New Delhi, India under NPDF scheme (Sanction No. PDF/2019/002762/Dated: 23/12/2019), India.
CRediT authorship contribution statement
R. Prathiviraj: Software, Data curation, Formal analysis, Investigation, Methodology, Writing - original draft. George Seghal Kiran: Conceptualization, Validation, Visualization, Writing - review & editing. Joseph Selvin: Funding acquisition, Project administration, Resources, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.genrep.2020.100777.
Appendix A. Supplementary data
Supplementary material
References
- Boratyn G.M., Camacho C., Cooper P.S., Coulouris G., Fong A., Ma N., Madden T.L., Matten W.T., McGinnis S.D., Merezhuk Y., Raytselis Y., Sayers E.W., Tao T., Ye J., Zaretskaya I. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013;41:W29–W33. doi: 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark B.K., Wabick K.J., Weidner J.G. Inversion and crossover recombination contributions to the spacing between two functionally linked genes. Biosystems. 2012;109(2):169–178. doi: 10.1016/j.biosystems.2012.04.013. [DOI] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann D.L., Shindo N. WHO scientific and technical advisory group for infectious hazards. COVID-19: what is next for public health? Lancet. 2020;395(10224):542–545. doi: 10.1016/S0140-6736(20)30374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J.A., André N.M., Chappie J.S., Millet J.K., Whittaker G.R. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J. Mol. Biol. 2020;432(10):3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie J.S., Smith D.W. COVID-19: a novel zoonotic disease caused by a coronavirus from China: what we know and what we don't. Microbiol Aust. 2020:MA20013. doi: 10.1071/MA20013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorkinis G., Magiorkinis E., Paraskevis D., Vandamme A.M., Van Ranst M., Moulton V., Hatzakis A. Phylogenetic analysis of the full-length SARS-CoV sequences: evidence for phylogenetic discordance in three genomic regions. J. Med. Virol. 2004;74(3):369–372. doi: 10.1002/jmv.20187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima N., Takatsuka S., Miyashita H., Kretsinger R.H. Leucine rich repeat proteins: sequences, mutations, structures and diseases. Protein Pept Lett. 2019;26(2):108–131. doi: 10.2174/0929866526666181208170027. [DOI] [PubMed] [Google Scholar]
- Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020;79 doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3):585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F., Yu H., Guo J., Li Y., Luo K., Huang S. Identification of the hyper-variable genomic hotspot for the novel coronavirus SARS-CoV-2. J. Inf. Secur. 2020;80(6):671–693. doi: 10.1016/j.jinf.2020.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Zhu Q., Liu B. Phylogenetic and biological significance of evolutionary elements from metazoan mitochondrial genomes. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0084330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020;16(10):1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material