Abstract
The CAGI-5 intellectual disability challenge asked to use computational methods to predict patient clinical phenotypes and the causal variant(s) based on an analysis of their gene panel sequence data. Sequence data for 74 genes associated with intellectual disability (ID) and/or Autism spectrum disorders (ASD) from a cohort of 150 patients with a range of neurodevelopmental manifestations (i.e. ID, autism, epilepsy, microcephaly, macrocephaly, hypotonia, ataxia) have been made available for this challenge. For each patient, predictors had to report the causative variants and which of the seven phenotypes were present. Since neurodevelopmental disorders are characterized by strong comorbidity, tested individuals often present more than one pathological condition. Considering the overall clinical manifestation of each patient, the correct phenotype has been predicted by at least one group for 93 individuals (62%). ID and ASD were the best predicted among the seven phenotypic traits. Also, causative or potentially pathogenic variants were predicted correctly by at least one group. However, the prediction of the correct causative variant seems to be insufficient to predict the correct phenotype. In some cases, the correct prediction has been supported by rare or common variants in genes different from the causative one.
Keywords: Critical assessment, community challenge, genetic testing, phenotype prediction, variant interpretation
Introduction
Neurodevelopmental disorders (NDDs) are a spectrum of disease conditions affecting brain development. Affected patients have increased manifestations as their childhood progresses, as the pathogenic conditions disturb normal brain development. Manifestations usually start with a non-specific form of intellectual disability (ID), characterized by limitations both in intellectual functioning (reasoning, learning, problem solving) and in adaptive behaviour, which covers a range of everyday social and practical skills. However, additional manifestations, such as autistic spectrum disorders (ASD) and epileptic seizures, can arise (Bowley and Kerr 2000; Tonnsen et al. 2016). Structural abnormalities of the cranium (i.e. microcephaly, macrocephaly) may also be present at birth or appear postnatally. People with ID show also a delayed motor development, which become evident with abnormalities in gait, such as ataxic gait (i.e. a lack of coordination in movement with a tendency to fall), hypotonia (general muscle weakness), or with ‘unconscious’ active motor behaviours (e.g. dyskinetic–dystonic movements or stereotypies) (Almuhtaseb, Oppewal, and Hilgenkamp 2014). NDDs are clinically and phenotypically diverse, but driven by a substantial and overlapping genetic component, with numerous shared risk genes underlying these conditions (Mitchell 2011). In particular, complex conditions such as ID and ASD have already been associated to hundreds of different genes. Next Generation sequencing (NGS) has led to the identification of many new NDDs genes with an excess of de novo mutations when compared to controls (Iossifov et al. 2014). Despite remarkable genetic heterogeneity, the findings from NGS and improvement in systems biology approaches, unraveled convergent biological pathways involved in brain development and help our understanding of disease pathophysiology (Pinto et al. 2014; Krumm et al. 2014; Barabási, Gulbahce, and Loscalzo 2011; An et al. 2014).
As NDDs can in principle be diagnosed even before birth by genetic tests, this has led to an increasing application of next-generation sequencing in clinical practice. Medical laboratories are routinely asked to screen hundreds of patients, which are either affected by NDDs or at risk of developing the condition. The limiting factor for successful diagnosis has therefore become the identification of causative mutations to associate to given pathogenic phenotypes. As most of these mutations are extremely rare or private, the problem is one of interpreting the effects of scores of variants of unknown significance on a wide range of candidate genes. This background fits well into the framework of the Critical Assessment of Genome Interpretation (CAGI) experiment, which has a declared goal of assessing methods to help interpret the effects of variants of unknown significance. A similar challenge was present in the CAGI-4 experiment with the Hopkins gene panel, where predictors were asked to predict phenotypes based on the results of a genetic screening performed on a set of 83 genes associated to 14 different conditions (Chandonia et al. 2017).
The setup of the CAGI-5 ID challenge starts from a similar background. The Padua Genetics of Neurodevelopmental Disorders Lab at the Department of Woman and Child Health, University of Padua (henceforth, Padua NDD lab) has been using a gene panel to diagnose different NDD subtypes for the past couple of years. For the purpose of the CAGI-5 challenge, a dataset of 150 unpublished pediatric patients was released. Starting from the gene panel sequencing data, predictors were asked to predict (a) the phenotypes and (b) their causative or potentially causative variations for each patient. Phenotypes have been derived from the clinical notes collected by geneticists visiting the patients. Candidate variants have been validated by segregation analysis, i.e. verifying their absence in the parents according to the de novo paradigm, inherited from affected parents. It should be noted that this is a difficult “open world” CAGI challenge, as clinical notes may be somewhat subjective and only a subset of genes have been screened. Furthermore, the phenotypic traits to predict are pathophysiology conditions that can be present in different NDDs, thus, in contrast to the CAGI-4 Hopkins challenge, patients may manifest more than one of these phenotypes, in different combinations.
The challenge is realistic as it well represents the difficult of assigning causative mutations to complex neurological diseases in clinical practice. In a few selected cases, consistent predictions were used to challenge previous assumptions and have led to a revised molecular diagnosis.
Materials and Methods
Sequencing, variant nomenclature and analysis by the Padua NDD lab
Coding sequences and nearest flanking regions of 74 genes were targeted for deep sequencing with a custom Ampliseq panel assay using a mixture of oligonucleotides generating 1,834 amplicons covering 520 kb. Multiple indexed libraries were pooled and sequenced on the Ion PGM platform (Thermo Fisher Scientific). Alignment and variant calling were performed with the Ion Torrent Suite Software v 5.02 (Thermo Fisher Scientific). The panel of 74 genes was sequenced in 150 individuals referred to the Padua NDD lab for intellectual disability with or without autistic features. VCF files of the 150 patients were provided to the CAGI-5 organizers with clinical information regarding the presence of seven ‘phenotypic traits’ for each patient (Suppl. Table S1). The clinical information was provided by the patient’s physician, which were asked to fill a clinical record for each patient. When the clinician leaved a field empty, we indicated information about the specific trait as not available, although we cannot exclude that some patients may present it. The Padua NDD lab also indicated the identified variants of the sequenced genes that have been classified as causative, putative, or contributing factors (see Suppl. Table S2). Causative variants are supported by segregation analysis and genotype-phenotype correlation, while “putative” ones are rare or novel variants predicted as pathogenic for which segregation analysis is not available. Contributing factors are rare or novel variants predicted as pathogenic, inherited from apparently healthy parents, mapping on genes that confer a risk but are not sufficient to cause the disease, mapping on genes causing ASD susceptibility, or found mutated in individuals with very mild phenotypes. Table 1 summarizes the amount of patients with variants associated to each phenotype.
Table 1.
Patients for whom Padua NDD lab identified at least one causative or potentially disease variant in the answer key, summarized by phenotype. Each variant is specific for each patient and one patients can be associated to more than one phenotype.
| Phenotype | Patients | Disease causing | Putative | Contributing factor | All variants |
|---|---|---|---|---|---|
| ID | 49 | 25 | 18 | 12 | 55 |
| ASD – Autistic traits | 31 | 14 | 12 | 10 | 36 |
| Epilepsy | 18 | 9 | 8 | 2 | 19 |
| Microcephaly | 8 | 5 | 2 | 1 | 8 |
| Macrocephaly | 4 | 4 | 0 | 0 | 4 |
| Hypotonia | 6 | 4 | 1 | 1 | 6 |
| Ataxia | 3 | 1 | 2 | 1 | 4 |
To evaluate the putative clinical impact of the variants, the following criteria were applied: 1) allele frequency <0.002% in the Gnomad database, or <0.45% for variants in autosomal-recessive genes, as indicated by (Whiffin et al. 2019; Piton, Redin, and Mandel 2013) 2) absence of the variant in other samples (in-house database), 3) stop gain, frameshift and splicing variants were a priori considered to be most likely pathogenic, 4) for missense mutations, amino acid conservation and consensus of pathogenicity predictions were evaluated, 5) inheritance mode, 6) phenotypic consistency with the clinical signs associated to mutations in the same gene.
It is important to note, that for a diagnostic purpose, the thresholds used by the Padua NDD lab to filter candidate variants, have been calculated based on the assumption that the patient phenotype follow a Mendelian transmission.
Whiffin and colleagues demonstrated that for human Mendelian disease clinical genome interpretation is empowered by using high-resolution variant frequencies (Whiffin et al. 2019). To select candidate variants responsible for ID, Piton and colleagues suggested to filter variants with a frequency compatible with the incidence of the disease (i=2% in the general population) (Piton, Redin, and Mandel 2013). Since the repeat expansion on FMR1 gene remains the most frequent cause of X-linked forms of ID and given the genetic heterogeneity of NDDs, we expect that mutations in other genes account for less than 0.1% of all ID cases, resulting in a disease frequency <0.002% (i= 0.02 × 0.001). Variants in genes associated to recessive disorders should not exceed the threshold of 0.45% (√0.002%).
Challenge format
Participants were provided with 150 VCF files, one per patient, a detailed description of the seven disease phenotypes given in Suppl. Table S1, the 74 gene identifiers, the gene captured regions used in sequencing the patients in Browser Extensible Data (BED) format, a submission template, and a submission validation script. Furthermore, participants were informed that each patient may have more than one phenotypic trait, and all have at least one.
Participants were asked to submit the predictions of phenotypic traits and causative variants for each patient, based on their gene panel sequences. For each submission, participants were required to predict the probability that a patient has a referring phenotypic trait in each of the 7 phenotypic classes provided, as well as the predicted causal variant(s) from the gene panel sequence dataset for every disease class with a non-zero probability. Each predicted disease class probability also included a mandatory standard deviation (SD) field indicating the confidence prediction, with low SD indicating high confidence and high SD indicating low confidence.
Assessment
The prediction assessment was focused on evaluating the predictive ability of the different submissions, considering their performance on each disease phenotype. This approach has been successfully used for the analysis of multilabel classifier performance, since it focuses on a set of two-class prediction problems (Fawcett 2006). It also simplifies the assessment procedure, allowing to compare and highlight different method performances on each single phenotype, instead of evaluating them considering the whole predicted class matrix (150 × 7, one prediction for each patient and phenotype).
Predicted disease classes for each submission were assessed against the clinical phenotype given in the Padua NDD lab answer key, using the procedure described below. When the predictors did not provide a probability value leaving the asterisk on the template file, it was treated as probability zero in the assessment.
The first phase of the assessment procedure was the conversion of submitted probability values to positive (1) or negative (0) classes. The conversion was done by each phenotype column, considering as threshold the probability value which maximizes the Matthew correlation coefficient (MCC) for that phenotype. We compared all probability values of each phenotype with the corresponding threshold and assign 0 or 1 if the value is lower or higher, respectively. In addition, different performance measures were used to assess the predictions for each phenotype. Sensitivity and specificity have been used to evaluate model capability to detect positive cases and discriminate between positive and negative classes. The MCC, accuracy (ACC) and F1 measures have been used to evaluate both negative and positive predictions at the same time (see Suppl. Material for details). Particularly, MCC has been proven to be less influenced by an unbalanced dataset (Vihinen 2012), as is the case of this challenge where some phenotypes are completely unbalanced (Figure 1C). ROC curves have been produced comparing experimental and predicted probability values for each phenotype. The Area Under ROC curve (AUC) was calculated for these.
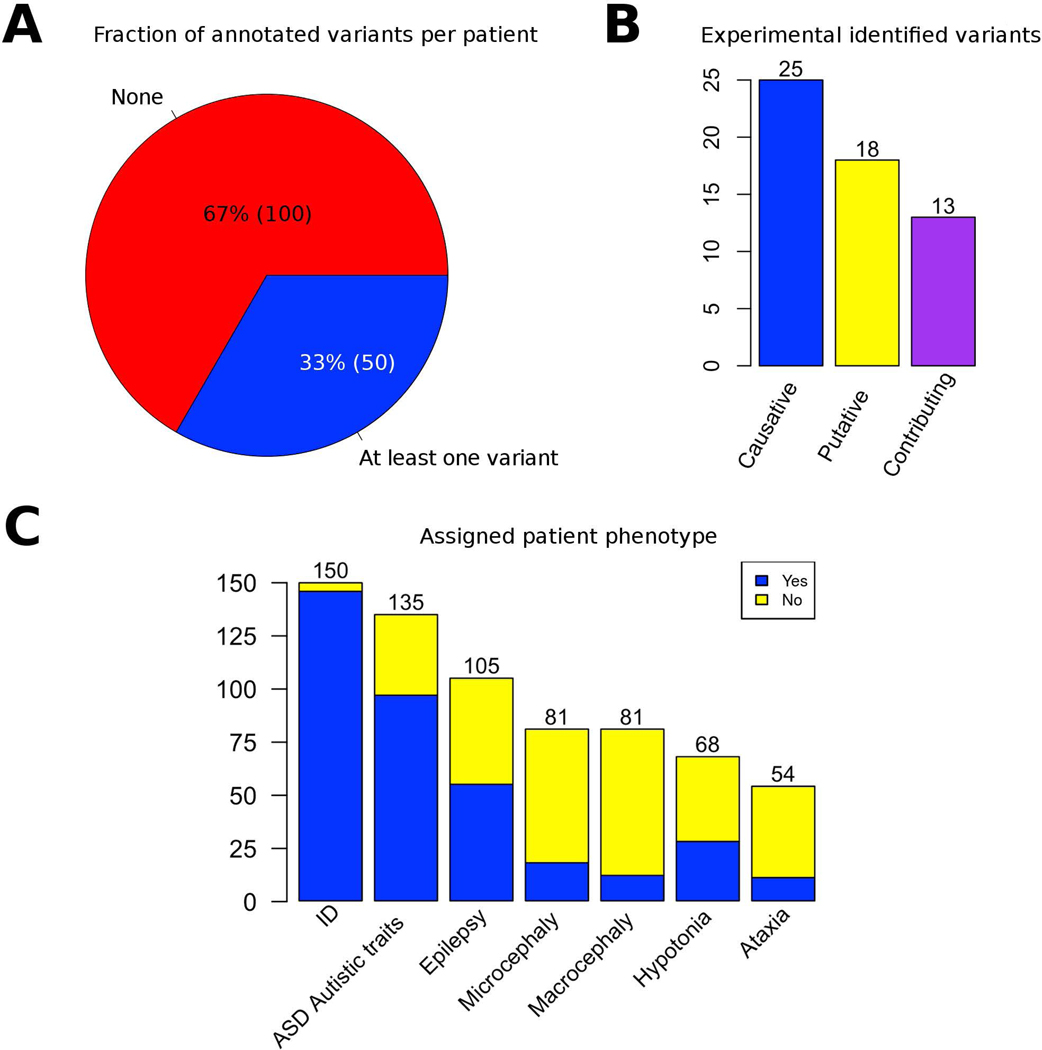
Figure 1.
Summary of CAGI-5 intellectual disability challenge experimental data. A) For the 150 patients included in the study, the Padua NDD lab noted at least one mutation relevant to the phenotype in the 33% of the patients B) Variant classes distribution. c) Number of patients where the presence or absence of the phenotype was ascertained by a clinician.
The R scripts used to perform the assessment are publicly available from the GitHub repository at URL: https://github.com/BioComputingUP/CAGI-ID-assessment.
Prediction methodology
A total of four groups, plus a late prediction (which can be found in the Suppl. Material), submitted predictions for the ID challenge. The group prediction approaches are summarized in Table 2 and described in detail below.
Table 2.
Computational approaches adopted by different groups.
| Group | Annotation | Gene-Phenotype | Variant impact | Filters | Inheritance model | |||
|---|---|---|---|---|---|---|---|---|
| ID | Submission | Name | Frequency | Low quality | ||||
| 1 | 1.1 | Mooney-Radivojac | ANNOVAR | HGMD, PhenoPred, and PPI for network propagation | MutPred2 and MutPredLOF | - | - | - |
| 2 | 2.1 | Moult Lab | Varant | OMIM+GHR, OMIM+HPO |
13 levels of variant impact | SNVs >1%, SNVs in LCR low complexity region | yes | yes |
| 2.2 | ||||||||
| 2.3 | ||||||||
| 2.4 | ||||||||
| 2.5 | ||||||||
| 2.6 | ||||||||
| 3 | 3.1 | Lichtarge Lab | ANNOVAR | Diffusion on CTD (Comparative Toxicogenomics Database) associations | Evolutionary Action | No | yes | no |
| 3.1 | ||||||||
| 3.3 | ||||||||
| 4 | 4.1 | Brenner Lab | CHESS v0.1 | Phenolyzer | VEP, REVEL score | SNVs MAF>5% | yes | yes |
| 4.2 | ||||||||
| 4.3 | ||||||||
Group 1 (Mooney - Radivojac Lab):
Annotation of the protein coding variant in the raw VCF files was performed using ANNOVAR [http://annovar.openbioinformatics.org/en/latest/], including extraction of wild type and mutant protein sequences (Wang, Li, and Hakonarson 2010). Pathogenicity prediction scores were assigned to missense, stop gain, and frameshifting indel variants with Mutpred2 [http://mutpred.mutdb.org/] (Pejaver et al. 2017) and Mutpred-LOF [http://mutpredlof.cs.indiana.edu/] (Pagel et al. 2017). In each individual, phenotypic trait risk was determined based only upon the variant with the highest pathogenicity prediction score across a set of phenotype-specific risk genes. For each phenotypic trait, a list of risk genes that are known to harbor disease-causing variants associated with that phenotypic trait was compiled from the Human Gene Mutation Database (HGMD) (Stenson et al. 2017).
Gene lists were extended, particularly those with fewer known risk genes (macrocephaly, hypotonia and ataxic gait), with the PhenoPred [https://www.phenopred.org/] web tool (Radivojac et al. 2008) and a gene prioritization algorithm. Confirmed risk genes have been used as “seed” genes on the human protein-protein interaction network for running a network propagation algorithm (Nabieva et al. 2005). The propagation algorithm was performed in a 5-fold cross validation manner so as to get an initial score between [0, 1] for all the genes. The AlphaMax algorithm (Jain, White, and Radivojac 2016) was used to estimate the positive proportion of the risk genes and calibrate those initial scores to be proper probability scores measuring the likelihood of a gene being associated with the disease. For each phenotypic trait, the probability was MutPred2 or MutPred-LOF score of the highest scoring variant in the associated risk genes.
Group 2 (Moult Lab):
The 150 VCF files (one VCF file per patient) provided for the challenge were annotated using the Varant tool [http://compbio.berkeley.edu/proj/varant/Home.html], including region of occurrence (intron, exon, splice site or intergenic), observed minor allele frequencies (MAF), mutation type, predicted impact on protein function, and previously established associated phenotypes reported in ClinVar (Landrum et al. 2014). The RefGene (Pruitt et al. 2014) gene definition file was used for gene and transcript annotations in Varant. In addition, in-house scripts were written to further annotate the VCF files with HGMD (Stenson et al. 2003) disease-related variants, with dbscSNV (Jian, Boerwinkle, and Liu 2014) and SPIDEX [http://tools.genes.toronto.edu/] (Xiong et al. 2015) variants that potentially alter splicing, and with REVEL (Ioannidis et al. 2016) scores for missense variants. A quality control (QC) analysis were performed to exclude outlier samples (see Suppl. Material). The transition/transversion ratio (Ts/Tv) and heterozygous/homozygous ratio were compared to the 1000 Genomes dataset for the genomic regions captured for sequencing in the challenge dataset. Comparison of common, rare, and novel variant counts across samples was also performed. The 74 genes were mapped to one or more of the seven phenotype traits using two independent approaches generating two different gene-phenotype mapped files. In addition to the OMIM database, the Genetic Home Reference (https://ghr.nlm.nih.gov/) or Human Phenotype Ontology (https://hpo.jax.org/app/) databases, respectively, were used to map the phenotypes to the genes. The variant prioritization procedure was performed on each of these phenotype lists. Only rare variants (MAF less than or equal to 1% in Exac (http://exac.broadinstitute.org) or novel variants (not reported in ExAC), flagged as PASS in the VCF files, were considered. Indels in low complexity regions (LCR) were excluded from the analysis, based on the LCR dataset pre-computed for the human genome by Heng Li (Li 2014). A strand bias filter was used to remove variants whose alternate allele was present only on one strand of the reads mapped to the variant position. Variant prioritization was based on two main criteria, variant quality and variant impact, that were applied in a sequential manner to each sample. For each criterion, five different levels of variant quality and 13 different types of variant impact were defined respectively (for more details see Suppl. Material). Putative causative variants identified were further filtered for inheritance model associated with the gene, according to the available information for the gene concerned in OMIM and Genetic Home Reference database.
To compute a probability score, i.e. the probability of a variant causing a disease phenotype, a number of ad hoc procedures were used. An exception was for missense variants, where the probability was assigned using the extent of consensus among the four missense-analysis methods, previously calibrated from HGMD data and a control set of inter-species variants. Other variant types were subjectively assigned probabilities depending on the severity of the impact. Furthermore, depending on the considered mode of inheritance, the probability score was adjusted. Ad hoc probabilities of a correct variant call were also assigned to each variant based on the variant quality filters (see Suppl. Material). Six different predictions were performed based on the two different gene-phenotype lists and different combination of probabilities.
Group 3 (Lichtarge Lab):
Variants of poor sequencing quality (QUAL<80) were excluded from the analysis and the rest variants were annotated with ANNOVAR [http://annovar.openbioinformatics.org/en/latest/] (Wang, Li, and Hakonarson 2010). There were three submissions that used i) only missense, ii) missense and nonsense, and iii) all variations. The effect of each variant was estimated with the Evolutionary Action (EA) [http://mammoth.bcm.tmc.edu/uea/] equation (Katsonis and Lichtarge 2014) and the function loss of each gene was calculated as: LOFg=1-∏(1-EAi/100), where ∏ indicates the product for all mutations i in that gene. Nonsense and fs-indel variants were given EA of 100, while silent variants were given EA of 0. Genes were also weighted for their ability to tolerate mutations (wg), calculated as the fractional rank of the average EA score of mutations seen in the gnomAD data (Lek et al., 2016). The weighted loss of function of each gene (wg*LOFg) was used as starting value for diffusion across the CTD gene-disease network (Mattingly et al. 2003). Diffusion scores were calculated for each disease (Lin et al. 2018) and a collective burden was calculated for each of the seven disease categories (normalized between 0–1). The relative ratios of the collective burden of the disease categories was used as the probability that a patient belongs to that disease category. The variants that contributed most to the collective burden of each disease category were reported as the causal variants.
Group 4 (Brenner Lab):
This group used their software CHESS v0.1 adjusting some parameters to perform predictions for the CAGI-5 ID challenge. Public data used on CHESS are variant frequency data from GNOMAD v2.0.2 [https://gnomad.broadinstitute.org/] (Lek et al. 2016), pre-calculated variant deleterious scores by REVEL [https://sites.google.com/site/revelgenomics/] (Ioannidis et al. 2016), and clinical evidence data from ClinVar (Landrum et al. 2016) (downloaded on 2017–10-02). Phenotype matching scores for all genes were calculated using Phenolyzer (Yang, Robinson, and Wang 2015). Pre-called variants from the case exome were annotated with data using VEP (McLaren et al. 2016), GNOMAD variant frequency data, ClinVar evidence, and the pre-calculated REVEL scores. To reduce the computing burden, common (variants with MAF >= 5%) and non-protein-altering variants have been excluded from the analysis. The selected variants were scored based on quality of data, impact severity, phenotype-match score (see Suppl. Material). Different scoring adjustments were also performed based on the inheritance mode considered. The three submissions correspond to three models with different stringency in the final decision, based on variant frequency in the 150 patient cohort and the probability score threshold used for each prediction.
Results
Summary of experimental data and submissions
Four groups submitted a total of 13 predictions for the CAGI-5 intellectual disability (ID) challenge. Group 2 submitted 6 predictions, groups 2 and 3 submitted 3 predictions each. In addition, a late submission (Group 5) was not considered for the general assessment but can be found in the Suppl. Material. Table 2 summarizes the participating groups, computational methods and their submissions.
An overview of the genetic and clinical data used in the ID challenge is shown in Figure 1. The 150 patients in the challenge can be divided into two groups: (1) patients for whom the Padua NDD lab identified at least one causative or potentially disease variant in the answer key (50 patients, 33%) and (2) patients for whom the Padua NDD lab excluded the presence of potentially pathogenic variants (100 patients, 67%). The total number of variants associated to at least one phenotype is 56 and variants are unique of each patient. In Table 1 is shown how variants are distributed in the different phenotypes. These variants were classified by the Padua NDD lab according to their possible effect as follows (Figure 1B): causative (25 variants), putative (18 variants) and contributing factor (13 variants). However, all variants were treated equivalently for purposes of assessing and ranking predictions.
Most of the patients with identified variants have at least one causative, and 16 and 13 patients show at least one putative or contributing factor variant, respectively. Combinations of different variant types in the same patient were observed only in a limited number of cases.
Phenotypic features were associated to each patient by a clinician. Although all patients have at least one feature assigned, the phenotypes were not equally represented in all patients. Figure 1C shows that most of the patients have ID, ASD, or Epilepsy. Other phenotypes (Microcephaly, Macrocephaly, Hypotonia and Ataxia) were less frequently observed in these patients. Nevertheless, for many patients no information was available about the presence or absence of a phenotype. Analyzing the overlap among phenotypes in patients, most patients have in common the phenotypes ID and ASD (39 patients), and ID, ASD and Epilepsy (21 patients) (Suppl. Figure S2).
Phenotype prediction assessment
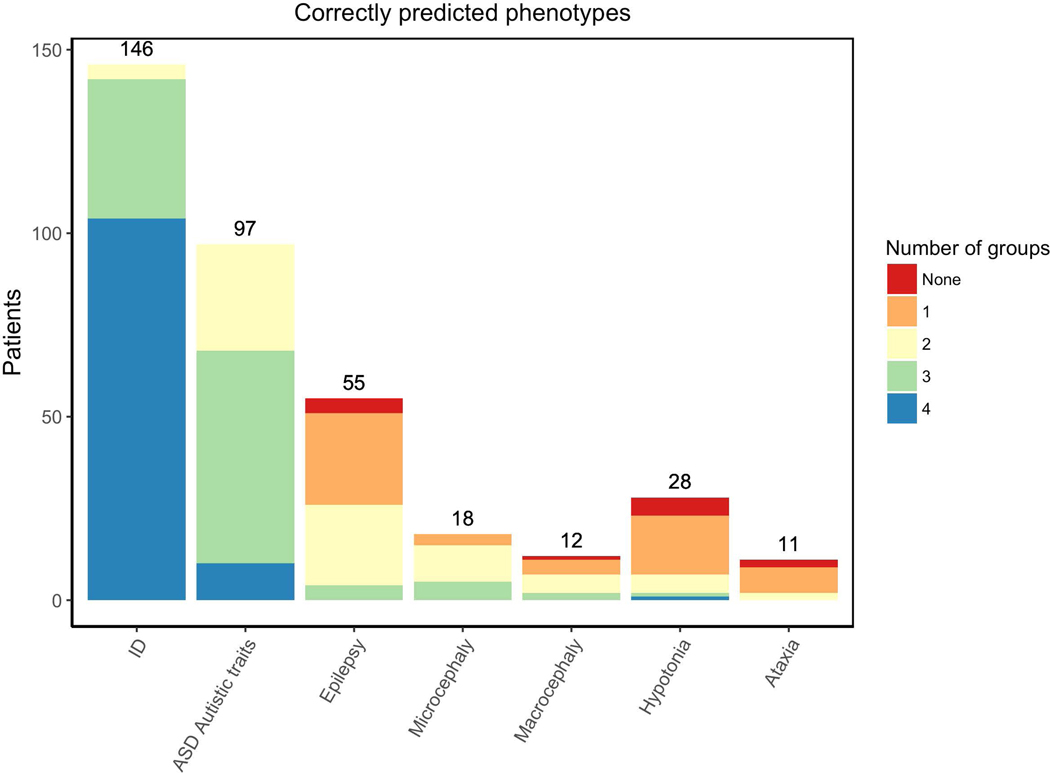
In this CAGI-5 challenge, the phenotype assessment was performed individually for each of the seven phenotypic traits assigned by the Padua NDD lab. Figure 2 shows the number of groups predicting correctly a patient phenotype when it was present. The ID phenotype was best predicted by most of the groups for about 90% of ID patients. ASD was the second best phenotype predicted among the patients. Despite the limited number of patients with Microcephaly (18 patients), this phenotype was correctly predicted in about 60% of cases by most of the groups. Other phenotypes, such as Epilepsy, Macrocephaly, Hypotonia and Ataxia, were poorly predicted by the different groups.
Figure 2.
Number of patients with the phenotype. Colors represent the proportion and number of groups which correctly predicted the phenotype.
Considering only patients for whom the presence or absence of a phenotype was ascertained by a clinician (Figure 1C), it is important to observe the patient coverage by each submission. Suppl. Figure S1 shows the number of patients with predictions in each submission for the different phenotypes; at least one prediction was made for all possible patients. However, group 3 and 5 submissions did not predict any probability values for many patients, particularly in ASD, Epilepsy, Macrocephaly, Hypotonia and Ataxia phenotype. Suppl. Figure S1 also shows the number of patients for whom the phenotype was correctly predicted.
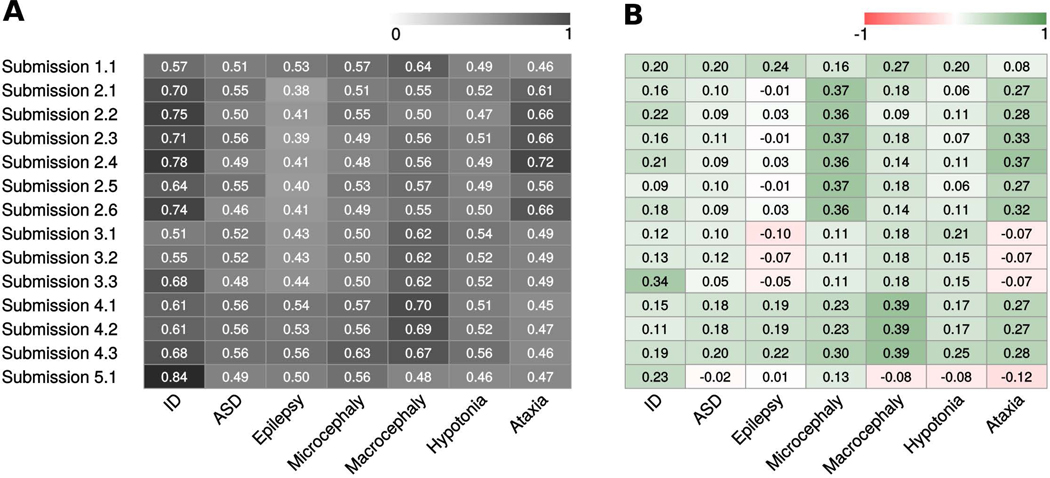
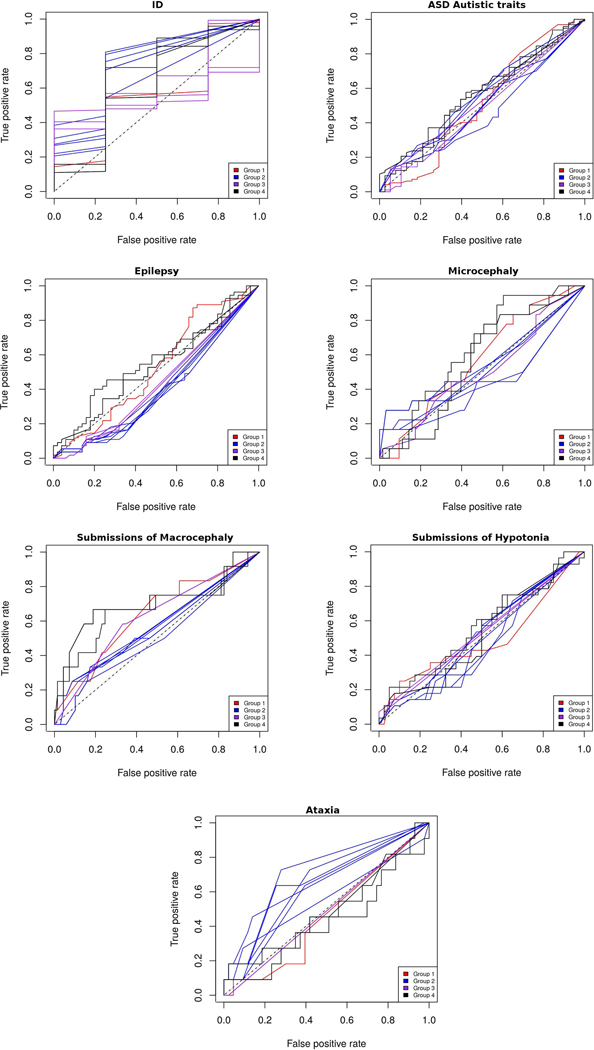
The overall submission performance was assessed using AUC for each phenotype, with MCC, ACC, and F1 measures used to better evaluate predictions. Figure 3 shows a summary of the AUC values obtained by each submission in the different phenotypes. In addition, Figure 4 and Suppl. Table S3 show the ROC curves and performance measures, respectively, for all submissions in each phenotype. Since the overall predictions are far from perfect performance, the prediction assessment for each phenotypic trait was performed also in the group of patients where the Padua NDD lab noted a potentially causative variants like previous CAGI challenge assessments (data not shown) (Chandonia et al. 2017). However, this did not show any improvement of predictor performance.
Figure 3.
Overall performance for each submission on phenotype prediction. A) Each cell represents the AUC values. The color scale ranges from dark (+1, perfect performance) to white (0, bad performance). White means random performance. B) Each cell represents the MCC values. The color scale ranges from green (+1, perfect correlation) to red (−1, negative correlation). White means no better than random prediction.
Figure 4.
ROC curves for each phenotype. Submissions are colored by predictor group.
For the ID phenotype, submission 4 of group 2 achieved the highest AUC value (0.78), followed by submissions 2, 6 and 3 of same group and submission 3 of group 3. Indeed, submission 3.3 obtained the highest overall performance considering all measures. They correctly predicted 146 patients out 150, and a moderate correlation with patient clinical data. ASD was the second most noted phenotype in patients by the Padua NDD lab. While all group 4 submissions and submission 2.3 reached higher AUC values than other groups, AUC values (average 0.56) and ROC curves remain close to random. Submission 4.3 and 1.1 achieved the best performance considering the other measures, with submission 1.1 equal in ACC, MCC and F1. Both submissions well identified patient phenotype in almost 100% of the cases.
Despite the rather good AUC, ACC and F1 values reached by some groups for the ID phenotype, and also for ASD, the MCC values remain quite low. Since MCC is not influenced by unbalanced categories, it shows a more realistic picture of prediction performance. As most of patients have the ID and ASD phenotypes, the confusion matrix is completely biased towards true positive values due to the highly imbalanced classes. This causes the ROC curve and consequent AUC not to reflect correctly the real predictor performance.
The presence or absence of the Epilepsy phenotype was poorly predicted by most groups, with an average MCC value of 0.05. This phenotype was particularly difficult, as roughly half of the patients had the disease. The best performances were achieved by group 4 and submission 1.1, predicting adequately more than 60% of patients. While performance measures show modest values, group 4 obtained the highest AUC (0.56) and group 1 the best MCC, AUC, and F1 values (see Suppl. Table S3).
Information about the presence or absence of Microcephaly and Macrocephaly was available for about half (81) of the patients. Microcephaly was reported in 18 patients and Macrocephaly in 12. Predictions for Microcephaly performed modestly, the best AUC being reached by submission 4.3, which correctly predicted 42 patients. Group 1 also predicted most of the patients with the phenotype (15 correct). In addition, most group 2 submissions obtained the best MCC and ACC values compared to other groups, predicting correctly 66 patients. However, best MCC values are again poor compared to other measures, denoting the effect of unbalanced categories in the predictions. Group 2 predictions were biased to identify patients without the phenotype (63 out of 63 patients) and just three patients with the disease. On the contrary, submission 4.3 was biased to predict patients with the disease (17 out of 18 patients) and 25 patients without the phenotype.
Performance assessment for Macrocephaly shows similar results as Microcephaly. Group 4 submissions performed better than other groups in terms of AUC. Submission 4.3 predicted the highest number of patients correctly (68 out 69 patients without the phenotype and 3 out 12 patients with the phenotype). Submissions 4.1 and 4.2 predicted correctly the highest number of patients with the phenotype (8 out 12 patients). Group 2 scored quite well in the prediction of patients without the phenotype, their submissions mostly predicting most of the patients where the phenotype was not noted. MCC values among submissions are again low, meaning that predictions were significantly biased.
The Hypotonia phenotype was positively or negatively noted in 68 patients by the Padua NDD lab. AUC values reached by different groups are poor, averaging around 0.5. Indeed, performance measures such as MCC and ACC are lower than in other phenotypes. Submission 4.3 obtained the best AUC, MCC and ACC values compared to other groups, correctly predicting 44 patients (6 out 28 with the phenotype and 38 out 40 without the phenotype). Submissions 2.1 and 2.3 predicted most of the patients with the phenotype (17 and 16, respectively).
The Ataxia phenotype was noted positively and negatively in 54 patients and only 11 patients had the disease. Submissions 4.1, 4.3, 2.1, 2.2 and 2.5 predicted well most of the patients but were biased to detect patients without the phenotype. Submissions 2.3 and 2.4 correctly predicted the presence of the disease in 7 and 8 patients, respectively. Consequently, the best AUC and MCC values were obtained by submission 2.4.
The overall submission ranking of this challenge was made considering the average AUC rankings for each phenotype. Table 3 shows the position reached by each submission in the different phenotypes. The best average ranked was submission 4.3, followed by other submissions of the same group.
Table 3.
Overall ranking among phenotypes by each submission. Individual phenotype ranking for each submission was made considering the performance measured by AUC.
| Submission | ID | ASD | Epilepsy | Microcephaly | Macrocephaly | Hypotonia | Ataxia | Avg. Ranking | Final |
|---|---|---|---|---|---|---|---|---|---|
| 4.3 | 6 | 4 | 1 | 1 | 3 | 1 | 11 | 3.86 | 1 |
| 4.1 | 10 | 1 | 2 | 3 | 1 | 7 | 13 | 5.29 | 2 |
| 4.2 | 9 | 3 | 4 | 4 | 2 | 5 | 10 | 5.29 | 2 |
| 3.3 | 7 | 12 | 5 | 9 | 6 | 3.5 | 8 | 7.21 | 4 |
| 2.3 | 4 | 2 | 12 | 12 | 10 | 8 | 3 | 7.29 | 5 |
| 3.2 | 12 | 7 | 6 | 9 | 6 | 3.5 | 8 | 7.36 | 6 |
| 1.1 | 11 | 9 | 3 | 2 | 4 | 11 | 12 | 7.43 | 7 |
| 2.1 | 5 | 5 | 13 | 7 | 11 | 6 | 5 | 7.43 | 7 |
| 2.4 | 1 | 11 | 8 | 13 | 9 | 10 | 1 | 7.57 | 9 |
| 3.1 | 13 | 8 | 7 | 9 | 6 | 2 | 8 | 7.57 | 9 |
| 2.2 | 2 | 10 | 9 | 5 | 13 | 13 | 4 | 8 | 11 |
| 2.5 | 8 | 6 | 11 | 6 | 8 | 12 | 6 | 8.14 | 12 |
| 2.6 | 3 | 13 | 10 | 11 | 12 | 9 | 2 | 8.57 | 13 |
For the CAGI-5 challenge, the assessment was performed also for each patient considering their overall clinical manifestations (Suppl. Table S4). Only 39 patients have the seven phenotypes negatively or positively assigned by Padua NDD lab. Among them, 13 patients (33%) were correctly predicted by at least one group, two patients were correctly predicted by two groups and three patients were correctly predicted by three groups. Four of these individuals have at least one variant. Group 2 was the best and predicted correctly the phenotype of 8 patients taking into account all their submissions (six submissions). Particularly, submission 2.4 predicted well 7 patients. Sixty-three patients (53%), among 119 with information about at least three phenotypes, were correctly predicted by at least one group. Group 1 and group 4 submission 3 were the best, correctly predicting 24 out 63 patients.
Furthermore, in this challenge we performed the assessment in predicting the overall clinical manifestation only for the 38 patients where the Padua NDD lab noted a causative or putative variant. For 22 of them (58%) the whole phenotype was correctly predicted by at least one group. Submission 3 group 4 and group 1 predicted correctly the phenotype in the same number of patients (13; 34.2%), 7 with causative and 6 with putative variants. On the other side, among the 100 patients with at least one assigned phenotype and where the Padua NDD lab did not report either a causative, putative or contributing factor variant; sixty-three (63%) were correctly predicted by at least one group. Considering only the patients with at least three assigned phenotypes (80), 22 (27%) of them were correctly predicted by at least one group. Again submission 4.3 and group 1 were the best groups in this subset of patients without pathogenic variants.
Variant prediction assessment
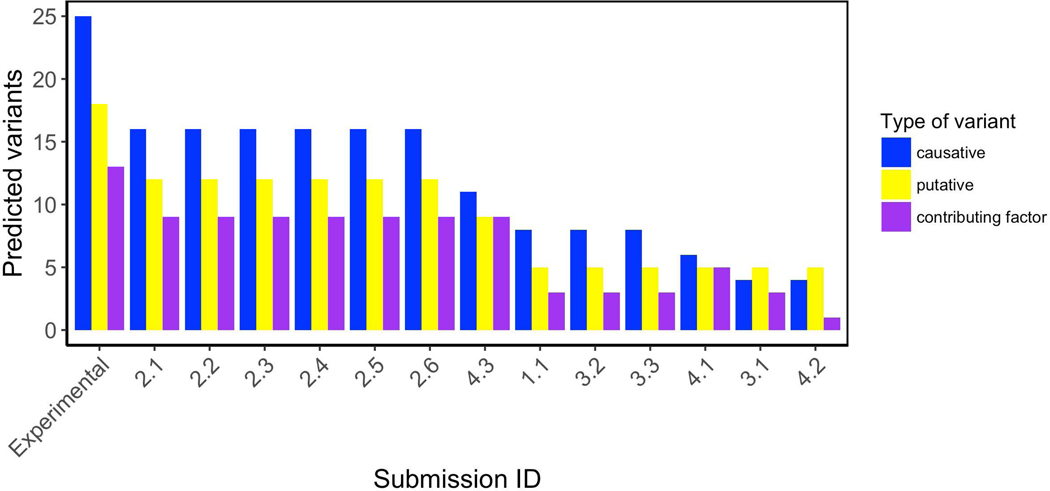
Predictors have been also assessed for their ability to detect variants in patients where clinicians have noted at least one variant probably associated to the phenotype. Figure 5 shows variant predictions for all patients and phenotypes by the different submissions. The amount of experimental variants (EV) with their corresponding classification are shown in the first three bars on the plot. Submissions of group 2 show the highest amount of well predicted variants associated to the different patient phenotypes (37 out of 56). Indeed, Group 2 outperformed other groups for causative (16 out of 25), putative causative (12 out of 18) and contributing factor (9 out of 13) variants. Submission 3 of group 4 was the second group predicting most of the variants. They correctly predicted 29 variants (11 causative, 9 putative causative and 9 contributing factor variants).
Figure 5.
Predicted variants distribution. Category “Experimental” is the amount of variants which were identified and classified by the Padua NDD lab. Each bar represents the amount of variants and type predicted by each submission.
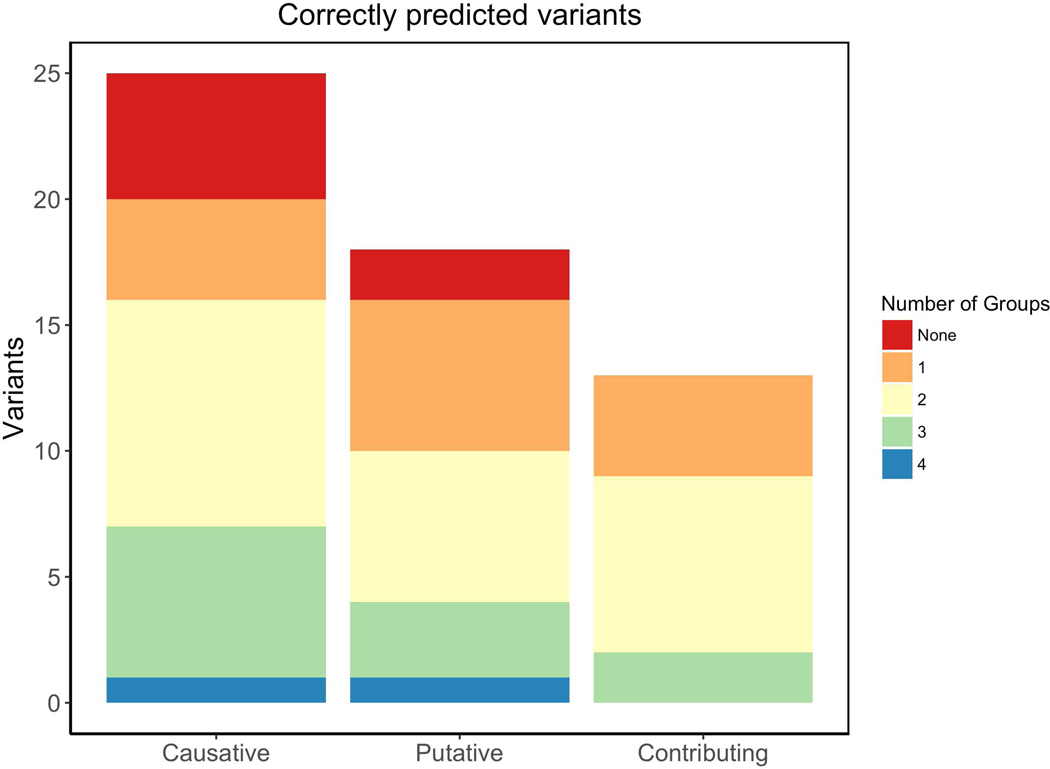
In addition, Figure 6 shows the fraction of each mutation type well predicted by the different groups. It is possible to see that just a small amount of variants were well predicted by all groups. The 28% of causative and 15% of contributing variants were correctly identified by at least 3 groups. On the other hand, 17% of putative variants were well predicted by at least 3 groups. Table 4 contains the fraction of well predicted variants by each group submission. Group 2 did not only predict most variants but also obtained the highest fraction of correctly predicted variants, calculated as the amount of variants well predicted divided by all the predicted variants for all patients and phenotypes Suppl. Table S2 summarizes all variants noted by the Padua NDD lab and the groups which predicted them correctly. All 25 causative variants, except the SHANK3 frameshift indel chr22:51159830:A:TTC in patient MR1970.01, were correctly predicted by at least one group. After the initial assessment, we realized that this complex genetic event (nucleotide substitution chr22:51159830:A:C plus a TT insertion) was molecularly characterized by Sanger validation of the chr22:51159830:A:C variant, but the variant caller plugin failed to call the insertion at near position of the same reads. However, group 4 correctly predicted chr22:51159830:A:C as a potentially pathogenic variant.
Figure 6:
Amount of variants classified by their effect. Colors indicate the proportion and number of groups which correctly predicted those variants.
Table 4:
Summary of variants prediction assessment by each submission.
| Submission | Correctly pred. Variants | Total pred. Variants | Correctly pred. Variants / Exp. Variants | Correctly pred. Variants / Total pred. Variants |
|---|---|---|---|---|
| 1.1 | 16 | 228 | 0.29 | 0.07 |
| 2.1 | 37 | 174 | 0.66 | 0.21 |
| 2.2 | 37 | 171 | 0.66 | 0.21 |
| 2.3 | 37 | 174 | 0.66 | 0.21 |
| 2.4 | 37 | 171 | 0.66 | 0.21 |
| 2.5 | 37 | 174 | 0.66 | 0.21 |
| 2.6 | 37 | 171 | 0.66 | 0.21 |
| 3.1 | 12 | 129 | 0.21 | 0.09 |
| 3.2 | 16 | 135 | 0.29 | 0.12 |
| 3.3 | 16 | 148 | 0.29 | 0.11 |
| 4.1 | 16 | 157 | 0.29 | 0.10 |
| 4.2 | 10 | 113 | 0.18 | 0.09 |
| 4.3 | 29 | 290 | 0.52 | 0.10 |
The Padua NDD lab considered some causative missense variants difficult to interpret (ATRX: p.N1377S; RAB39B p.F193L; GRIA3 p.R216Q; MED13L p.G706E), since pathogenicity predictions were discordant, allele frequency in control cohorts higher than expected, or proband phenotype partially consistent with those associated to the gene. However, the majority of the groups was able to predict these correctly. One example is the maternally inherited X-linked p.F193L of the RAB39B gene associated to recessive X-linked Mental Retardation syndrome (MR-XL72, OMIM 300271) or to Waissman syndrome, which is characterized by ID and early-onset Parkinson disease (OMIM 311510). This variant is predicted damaging by three out of twelve computational tools provided by ANNOVAR (LRT, Mutation Taster, and fathmmMKL), is moderately conserved during evolution, and present in a hemizygous state in two control cohort individuals. However, the variant maps to the C-terminal hypervariable tail of RAB39B which is relevant for protein interactions involved in protein targeting. The mother transmitting the p.F193L variant has a mild phenotype, consistent with those reported in the literature associated to a missense mutation at the close p.Gly192Arg position (Mata et al. 2015).
At least one group correctly predicted 16 out of 18 putative mutations. In particular, 7 variants were indicated by the majority of the groups. Three of these 7 variants were inherited and suspected to contribute to the disease together with other genetic or environmental factors. For the other four cases, after the CAGI-5 assessment, we contacted the families to follow up the molecular finding carrying out segregation analysis of the identified variants. Only one family answered our call, which allowed us to characterize the de novo status of the p.Y381H variant in the CASK gene. Even if the pathogenicity predictions were discordant, this variant was absent from control cohorts and in silico analysis suggested a structural role of this residue in the homo and hetero-dimerization of the CASK protein (Aspromonte et al. 2019). The proband phenotype is also consistent with those associated with CASK-related disorders.
In addition, at least one group correctly predicted the 13 variants classified as contributing factor, of which seven were indicated by the majority of the groups. This variant class is particularly relevant for autism susceptibility.
Novel variant predictions
Commonly predicted variants were also used to support those variants which were not considered by the Padua NDD lab. In order to check whether some relevant variant may be lost in the filtering process, the Padua NDD lab revised all of these 615 variants, which include 492 exonic (80%), 75 intronic (12.5%), 9 splicing (1.5%), and 6 5’/3’-UTR (untranslated region) (1%) variants.
Among the exonic variants, 80 (16.2%) were excluded for high allele frequency in the general population (MAF>0.002%). 150 variants with MAF<0.002% were excluded due to being present more than once in the cohort. 118 predicted to be likely gene disrupting variants (frameshift insertion, frameshift deletion, stop gain) were classified as sequencing errors after visual check of the raw data.
Focusing on variants indicated by the majority of the groups, we selected some variants to be reconsidered for Sanger validation and may be involved in the proband phenotype. In particular, for patient MR2001.01, three different groups (2, 3, and 4) predicted as potentially pathogenic two variants in NRXN1, p.L708I and p.I649V (NM_004801; 2:50765412 and 2:50765589 rs200074974). These variants were not reported to the patient due to being predicted neutral by the majority of the used computational methods. Variants of the NRXN1 gene are associated to schizophrenia, autism spectrum disorders, or the autosomal recessive Pitt-Hopkins-like syndrome 2, which is characterized by severe ID, developmental regression, hyper breathing, autistic behavior, and dysmorphic features. One of the putative variants found in MR2001.01, p.I649V, was reported in the literature in a patient with schizophrenia, inherited from the affected mother (Gauthier et al. 2011). Patient MR2001.01 has borderline ID with autistic traits and other behavioral psychiatric manifestations, such as depression and anxiety. Thus, its phenotype is not fully consistent with the recessive Pitt Hopkins-like disorder. However, we performed segregation analysis and found that the two NRXN1 variants were absent from the DNA of the mother and the healthy sister. This suggests that the two rare variants might be transmitted in cis from the father, who is not available for further investigation.
Two other variants have been reconsidered for Sanger validation and segregation analysis, a non-frameshift deletion and a synonymous variant, belonging to a class of variants for whom pathogenicity prediction is difficult to obtain. In patient MR1289_01, an in-frame deletion in the CC2D1A gene (NM_017721:exon1:c.27_35del:p.10_12del) was indicated as potentially pathogenic by groups 4 and 5. Mutations in CC2D1A are associated to ID, autosomal recessive 3 (MR-AR3), which is partially consistent with the proband phenotype. This position had a coverage of 74x, alternative allele frequency of 100% with a genotype quality of 24. At that position, which is part of a repeat sequence, other patients analyzed in the same experiment present sequence and alignment errors. However, Sanger sequencing of this amplicon revealed that the MR2001.01 case carries this variant in a heterozygous state. No other variants have been identified in the CC2D1A gene, which is completely covered by gene panel sequencing. As alterations of the CC2D1A have been implicated in NDDs only in recessive conditions, the final outcome for the patient does not change.
In patient MR1769_01, one hemizygous synonymous variant (NM_005120, c.3600C>T p.(Arg1200Arg) in the MED12 gene, has been indicated as pathogenic by group 5 (late predictor). The variant is predicted to potentially alter splicing by Human Splicing Finder (Desmet et al. 2009). For this patient, we reported two variants in the CNTNAP2 and FOXP1 genes, which we hypothesized to act in a two-hit model to determine the disorder, as previously described by (O’Roak et al. 2011). Nevertheless, a mutation in the MED12 gene could explain the family history evocative of an X-linked disorder, since the maternal uncle presents ID and ASD. However, segregation analysis revealed that the mother transmitted the MED12 variant to the two sons. Since the phenotype of the MR1769.01 brother is not consistent with MED12-related disorders, we can exclude this variant as a main molecular cause of the MR1769.01 phenotype.
Assessment by group summary
Group 1 (Mooney-Radivojac Lab):
Considering individually each phenotypic trait, group 2 predicted correctly all individuals with ASD, 15 out of 18 individuals presenting microcephaly, and 60% of the epileptic patients. Furthermore, group 1 predicted correctly the overall phenotype of 11 patients that other groups did not predict correctly. Although, their method was less accurate on the prediction of causative or putative variants indicated by the Padua NDD Lab, group 1 was one of the two best at predicting the correct combination of phenotype traits in cases where the Padua NDD Lab indicated a causative or putative variant. They also performed better in the phenotype prediction for patients with at least three phenotypic traits available. Given the discrepancy between the accuracy at predicting causal variants and phenotypic traits, the Padua NDD Lab checked the variants that they indicated supporting phenotype predictions. Although some supportive variants were classified as sequencing errors, many others were rare variants in genes associated to the specific phenotype trait. The Padua NDD Lab did not report these variants to the patient due to their relatively high frequency and discordant predictions among pathogenicity predictors. The group 1 method differs from that of other groups in the approach used to identify gene-phenotype association, particularly for traits with fewer known risk genes, such as macrocephaly, hypotonia, and ataxic gait. The use of protein-protein interaction networks to expand genetic association with the disease has been useful to select relatively low frequency variants with less functional impact that may contribute to the disease expression. This is in line with the emergent model explaining the genetic architecture of NDDs.
Group 2 (Moult Lab):
their method was the most accurate in predicting the correct combination of phenotypic traits in patients for whom the Padua NDD Lab provided information about all seven traits. Among these 13 individuals, group 2 correctly predicted the phenotype of 8 patients considering all their submissions (six in total). In particular, submission 2.4 predicted well 7 patients and predicted correctly the phenotype of 12 individuals that other groups did not. Group 2 was also one of the best in predicting the ID and ASD phenotype in all 150 individuals. Furthermore, they predicted correctly the individuals presenting ataxia and hypotonia, reaching the best AUC for ataxia. Interestingly, their method was the most accurate in predicting patients that did not present microcephaly or macrocephaly (together with group 4), reaching the best MCC and ACC scores. Furthermore, group 2 was the most accurate in the prediction of causal or likely pathogenic variants indicated by the Padua NDD Lab, with 66% correctly predicted. However, other groups performed better in the phenotype prediction for patients with causative/putative variants. This suggests that the identification of the correct causative or likely pathogenic variant is not sufficient to be able to predict all clinical manifestations in patients.
Group 3 (Lichtarge Lab):
Their predictions were based on the evolutionary burden of variations, thus the disease-gene association was supported by rare and common variants. Among patients with causative variants, group 3 predicted correctly the overall clinical manifestations for patient MR1974.01 that other groups, despite having identified the correct variants, did not predicted correctly. The Padua NDD lab checked the supporting variants indicated by group 3 and, besides variants classified as sequencing errors, they reported rare and common variants clustering in genes associated to the specific traits. Probably in some patients the phenotype prediction could be inferred if the set of analyzed genes contains rare and common variants associated to the phenotypic trait. This is in line with the recent finding that inherited common and rare variants cluster together with de novo variants in convergent pathways to determine the disease.
Group 4 (Brenner Lab):
Their prediction method obtained the best results considering the average overall performance for the prediction of each phenotypic trait. In addition, submission 3 of this group obtained the best MCC values for ASD, Macrocephaly and Hypotonia, and best AUC values for ASD, Epilepsy, Microcephaly and Hypotonia. Taking into account those patients where the Padua NDD Lab provided information about all seven traits, this group correctly predicted only three patients. However, their method was one of the best in correctly predicting patient phenotypes where the Padua NDD Lab provided at least three phenotypic traits. Despite their good performance to predict patient phenotypic traits, group 4 identified less causative or putative variants than groups 2 and 5. However, group 4 predicted correctly the phenotype of 8 individuals that other groups did not, two of them carrying likely pathogenic mutations of the CASK gene and one with a putative variant in PHF8 gene. This suggests that group 4 performed better for patients where they predicted correctly the causative or putative variant, thus they were good in the association of the gene with phenotypic traits. Group 4 used Phenolyzer to calculate the gene-phenotype matching score. Phenolyzer has been demonstrated to perform better than other available tools in the prioritization of candidate genes for complex disease (Yang, Robinson, and Wang 2015).
Discussion
We have described the assessment of the CAGI-5 ID challenge. This challenge is based on the phenotype evaluation of patients using gene panel sequences, in analogy to the CAGI-4 Hopkins panel challenge (Chandonia et al. 2017). Where the Hopkins panel was testing for different monogenic diseases with Mendelian inheritance, the ID challenge focuses on complex disorders. Neurodevelopmental conditions are characterized by strong clinical comorbidity and a complex genetic architecture (Mitchell 2011). The genetic information for each patient can at best be considered partial, as compounded by the rather limited fraction of patients (33%) where a putative or causative variant has been detected by the Padua NDD lab. As such, the CAGI-5 ID challenge can be expected to be more difficult that the CAGI-4 Hopkins panel. However, due to the genetic heterogeneity seen in NDDs, the presence of negative cases in the data set reflects the clinical practice, where the sequenced genes cannot explain the phenotype of all tested individuals. This implies that the identified rare variants should be interpreted with caution.
The phenotype prediction component of the ID challenge makes it also similar to the Personal Genome Project (PGP) challenge from previous rounds of CAGI (Cai et al. 2017). In the CAGI-2 PGP challenge, participants were initially asked to predict the presence of a set of phenotypic traits. Later CAGI editions turned the challenge into a matching game between sets of phenotypic profiles and genetic data. The ID challenge is similar to the original PGP challenge, but with a narrower focus on NDDs. Like PGP, it emphasizes complex disease conditions whose genetical bases are not fully understood. It is indeed increasingly accepted that the genetic architecture of NDDs involves the interplay of de novo, rare, and many common (>1% frequency) variants, which have a potential role in phenotype variability and severity of the disease. Furthermore, besides some well known monogenic conditions there are oligo- or polygenic forms with multiple gene-gene or gene-environment interactions (Lesch 2016; Mitchell 2011).
Despite these difficulties, several predictors participating in the CAGI-5 ID challenge were able to achieve AUC > 0.6 for three non-trivial phenotypes (microcephaly, macrocephaly and ataxia) and also for the ID phenotype which was heavily biased to the positive case. Intriguingly, group 4 (Brenner lab) has been able to make acceptable predictions for most of the individual phenotypic traits, except ataxia (Table 3). Furthermore, considering the overall clinical manifestations of each patient, for 93 individuals (62%) the correct phenotype has been predicted by at least one group. In particular, group 1 predicted 49 of them (52%), group 4 (submission 3) predicted 46 of them (50%) and 57 (61%) considering their three submissions. Finally, group 2 correctly predicted 43 of them (46%) considering all their six submissions. Group 2 in particular accurately predicted each of the seven phenotypic traits in 8 individuals, and the overall phenotype in 12 patients that were not correctly predicted by other groups. Even though this performance is not promising, we have to consider the extreme difficulty to predict a combination of several pathological conditions that often occur in comorbidity with variable expression and severity.
The assessment on phenotype prediction has also been performed considering only the patients with variants noted by the Padua NDD Lab, both considering each phenotypic trait individually and for the combination of the seven traits. We hypothesized that the phenotype of the individuals carrying a disease mutation must be easier to predict. Furthermore, the Hopkins challenge in CAGI-4 noted a higher performance of the prediction methods in phenotype prediction of cases where the Hopkins lab reported a variant, with at least one group correctly identifying the disease class in 84% of these patients. However, in the CAGI-5 ID challenge there were no improvements in the performance of methods. Surprisingly, group 2, which performed better in the causative or putative variants prediction, was less accurate in predicting phenotypic traits. Something similar occurs when we tried to remove patients for whom no method was able to correctly predict the phenotype (e.g. correctly predict the presence or absence of each class). We again observed that while some methods improved their performance, others decreased it.
In contrast to the Hopkins challenge, the Padua NDDs lab participated in the assessment of the challenge and provided feedback on predicted variants by the groups. This allowed us to observe that variants supporting the predictions of some groups, in particular group 1 and group 3, are rare or common variants with weak pathogenic predictions. Some of these variants were previously excluded by the Padua NDD lab as inherited from healthy parents (Aspromonte et al. 2019). However, it seems that taking into account the contribution of these inherited rare or common variants may help in the phenotype prediction. This can be explained by the complex genetic architecture of NDDs and the recent findings that different variants cluster in common pathways to determine the expression of the disease (Mitchell 2011). Thus, particularly for phenotypic traits with little genetic information, group 1 used protein-protein interaction networks to expand the gene-phenotype association, which has been useful to select relatively low frequency variants with less functional impact that may contribute to the expression of the phenotypic trait. Moreover, group 4 created their gene-phenotype association list using a well establish tool for the prioritization of risk genes in complex diseases.
However, no less important is that some groups made correct predictions based on variants that were excluded by the Padua NDD lab as sequencing errors. Methods using good quality filters, such as groups 2 and 4, are more reliable than others. Nonetheless, the Padua NDD lab reconsidered some of these predicted variants and validated them with Sanger sequencing and segregation analysis. Even if many of the reconsidered variants did not change the molecular diagnosis of the tested patients, the re-assessment of the interpreted data allowed to fix some rules in filtering sequencing errors and interpretation of variants, such as synonymous variants, that can be missed as causative. In particular, re-assessing putative variants that were predicted by the majority of groups as pathogenic, allowed us to select a limited set of putative variants for further investigation. The re-evaluation by segregation analysis was possible only for one family that answered our call. The variant resulted de novo, supporting the causative role of a probably hypomorphic CASK mutation in a male with a phenotype consistent with a CASK-related disorder.
This CAGI-5 challenge has provided a realistic framework to assess the performance of prediction methods in clinical practice. Despite all its inherent limitations, we believe it has demonstrated promising results and avenues for possible future improvements. We will hopefully be able to measure improvement over the next editions of the CAGI experiment.
Supplementary Material
Acknowledgments
The CAGI experiment coordination is supported by NIH U41 HG007346 and the CAGI conference by NIH R13 HG006650. The authors are grateful to all the italian proband families and clinical Institutions that referred the patients to the Laboratory of Molecular Genetics of NDDs, as well as to members of the BioComputing UP group and Molecular Genetics of NDDs, for insightful discussions. This work was supported by Italian Ministry of Health Young Investigator Grant GR-2011–02347754 to E.L and S.C.E.T.; Fondazione Istituto di Ricerca Pediatrica - Città della Speranza, Grant 18–04 to E.L; Italian Ministry of Health grant GR-2011–02346845 to S.C.E.T.
S.J.W., P.K., and O.L. were supported by the National Institutes of Health (GM079656 and GM066099).
References
- Almuhtaseb Sanaa, Oppewal Alyt, and Hilgenkamp Thessa I. M.. 2014. “Gait Characteristics in Individuals with Intellectual Disabilities: A Literature Review.” Research in Developmental Disabilities 35 (11): 2858–83. [DOI] [PubMed] [Google Scholar]
- An JY, Cristino AS, Zhao Q, Edson J, Williams SM, Ravine D, Wray J, et al. 2014. “Towards a Molecular Characterization of Autism Spectrum Disorders: An Exome Sequencing and Systems Approach.” Translational Psychiatry 4 (June): e394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspromonte Maria Cristina, Bellini Mariagrazia, Gasparini Alessandra, Carraro Marco, Bettella Elisa, Polli Roberta, Cesca Federica, et al. 2019. “Characterization of Intellectual Disability and Autism Comorbidity through Gene Panel Sequencing.” 10.1101/545772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási Albert-László, Gulbahce Natali, and Loscalzo Joseph. 2011. “Network Medicine: A Network-Based Approach to Human Disease.” Nature Reviews. Genetics 12 (1): 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowley C, and Kerr M. 2000. “Epilepsy and Intellectual Disability.” Journal of Intellectual Disability Research: JIDR 44 ( Pt 5) (October): 529–43. [DOI] [PubMed] [Google Scholar]
- Cai Binghuang, Li Biao, Kiga Nikki, Thusberg Janita, Bergquist Timothy, Chen Yun-Ching, Niknafs Noushin, et al. 2017. “Matching Phenotypes to Whole Genomes: Lessons Learned from Four Iterations of the Personal Genome Project Community Challenges.” Human Mutation 38 (9): 1266–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandonia John-Marc, Adhikari Aashish, Carraro Marco, Chhibber Aparna, Cutting Garry R., Fu Yao, Gasparini Alessandra, et al. 2017. “Lessons from the CAGI-4 Hopkins Clinical Panel Challenge.” Human Mutation 38 (9): 1155–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet François-Olivier, Hamroun Dalil, Lalande Marine, Gwenaëlle Collod-Béroud, Mireille Claustres, and Christophe Béroud. 2009. “Human Splicing Finder: An Online Bioinformatics Tool to Predict Splicing Signals.” Nucleic Acids Research 37 (9): e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett Tom. 2006. “An Introduction to ROC Analysis.” Pattern Recognition Letters 27 (8): 861–74. [Google Scholar]
- Gauthier Julie, Siddiqui Tabrez J., Huashan Peng, Yokomaku Daisaku, Hamdan Fadi F., Champagne Nathalie, Lapointe Mathieu, et al. 2011. “Truncating Mutations in NRXN2 and NRXN1 in Autism Spectrum Disorders and Schizophrenia.” Human Genetics 130 (4): 563–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis Nilah M., Rothstein Joseph H., Pejaver Vikas, Middha Sumit, McDonnell Shannon K., Baheti Saurabh, Musolf Anthony, et al. 2016. “REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants.” American Journal of Human Genetics 99 (4): 877–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov Ivan,, O’Roak Brian J., Sanders Stephan J., Michael Ronemus Niklas Krumm, Dan Levy, Stessman Holly A., et al. 2014. “The Contribution of de Novo Coding Mutations to Autism Spectrum Disorder.” Nature 515 (7526): 216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain Shantanu, White Martha, and Radivojac Predrag. 2016. “Estimating the Class Prior and Posterior from Noisy Positives and Unlabeled Data.” arXiv [stat.ML]. arXiv. http://arxiv.org/abs/1606.08561. [Google Scholar]
- Jian Xueqiu, Boerwinkle Eric, and Liu Xiaoming. 2014. “In Silico Prediction of Splice-Altering Single Nucleotide Variants in the Human Genome.” Nucleic Acids Research 42 (22): 13534–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsonis Panagiotis, and Lichtarge Olivier. 2014. “A Formal Perturbation Equation between Genotype and Phenotype Determines the Evolutionary Action of Protein-Coding Variations on Fitness.” Genome Research 24 (12): 2050–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm Niklas, O’Roak Brian J., Jay Shendure, and Eichler Evan E.. 2014. “A de Novo Convergence of Autism Genetics and Molecular Neuroscience.” Trends in Neurosciences 37 (2): 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum Melissa J., Lee Jennifer M., Benson Mark, Brown Garth, Chao Chen, Chitipiralla Shanmuga, Gu Baoshan, et al. 2016. “ClinVar: Public Archive of Interpretations of Clinically Relevant Variants.” Nucleic Acids Research 44 (D1): D862–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum Melissa J., Lee Jennifer M., Riley George R., Jang Wonhee, Rubinstein Wendy S., Church Deanna M., and Maglott Donna R.. 2014. “ClinVar: Public Archive of Relationships among Sequence Variation and Human Phenotype.” Nucleic Acids Research 42 (Database issue): D980–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek Monkol, Karczewski Konrad J., Minikel Eric V., Samocha Kaitlin E., Banks Eric, Fennell Timothy, O’Donnell-Luria Anne H., et al. 2016. “Analysis of Protein-Coding Genetic Variation in 60,706 Humans.” Nature 536 (7616): 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch Klaus-Peter. 2016. “Maturing Insights into the Genetic Architecture of Neurodevelopmental Disorders - from Common and Rare Variant Interplay to Precision Psychiatry.” Journal of Child Psychology and Psychiatry, and Allied Disciplines 57 (6): 659–61. [DOI] [PubMed] [Google Scholar]
- Li Heng. 2014. “Toward Better Understanding of Artifacts in Variant Calling from High-Coverage Samples.” Bioinformatics 30 (20): 2843–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Chih-Hsu, Konecki Daniel M., Liu Meng, Wilson Stephen J., Nassar Huda, Wilkins Angela D., Gleich David F., and Lichtarge Olivier. 2018. “Multimodal Network Diffusion Predicts Future Disease-Gene-Chemical Associations.” Bioinformatics, October 10.1093/bioinformatics/bty858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata Ignacio F., Jang Yongwoo, Kim Chun-Hyung, Hanna David S., Dorschner Michael O., Samii Ali, Agarwal Pinky, et al. 2015. “The RAB39B p.G192R Mutation Causes X-Linked Dominant Parkinson’s Disease.” Molecular Neurodegeneration 10 (September): 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly Carolyn J., Colby Glenn T., Forrest John N., and Boyer James L.. 2003. “The Comparative Toxicogenomics Database (CTD).” Environmental Health Perspectives 111 (6): 793–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren William, Gil Laurent, Hunt Sarah E., Riat Harpreet Singh, Ritchie Graham R. S., Thormann Anja, Flicek Paul, and Cunningham Fiona. 2016. “The Ensembl Variant Effect Predictor.” Genome Biology 17 (1): 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell Kevin J. 2011. “The Genetics of Neurodevelopmental Disease.” Current Opinion in Neurobiology 21 (1): 197–203. [DOI] [PubMed] [Google Scholar]
- Nabieva Elena, Jim Kam, Agarwal Amit, Chazelle Bernard, and Singh Mona. 2005. “Whole-Proteome Prediction of Protein Function via Graph-Theoretic Analysis of Interaction Maps.” Bioinformatics 21 Suppl 1 (June): i302–10. [DOI] [PubMed] [Google Scholar]
- O’Roak Brian J., Deriziotis Pelagia, Lee Choli, Vives Laura, Schwartz Jerrod J., Girirajan Santhosh, Karakoc Emre, et al. 2011. “Exome Sequencing in Sporadic Autism Spectrum Disorders Identifies Severe de Novo Mutations.” Nature Genetics 43 (6): 585–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel Kymberleigh A., Pejaver Vikas, Guan Ning Lin Hyun-Jun Nam, Mort Matthew, Cooper David N., Sebat Jonathan, Iakoucheva Lilia M., Mooney Sean D., and Radivojac Predrag. 2017. “When Loss-of-Function Is Loss of Function: Assessing Mutational Signatures and Impact of Loss-of-Function Genetic Variants.” Bioinformatics 33 (14): i389–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejaver Vikas, Urresti Jorge, Jose Lugo-Martinez Kymberleigh A. Pagel, Guan Ning Lin Hyun-Jun Nam, Mort Matthew, et al. 2017. “MutPred2: Inferring the Molecular and Phenotypic Impact of Amino Acid Variants.” bioRxiv. 10.1101/134981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto Dalila, Delaby Elsa, Merico Daniele, Barbosa Mafalda, Merikangas Alison, Klei Lambertus, Thiruvahindrapuram Bhooma, et al. 2014. “Convergence of Genes and Cellular Pathways Dysregulated in Autism Spectrum Disorders.” American Journal of Human Genetics 94 (5): 677–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piton Amélie, Redin Claire, and Mandel Jean-Louis. 2013. “XLID-Causing Mutations and Associated Genes Challenged in Light of Data from Large-Scale Human Exome Sequencing.” American Journal of Human Genetics 93 (2): 368–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter John. 1978. “Handbook of Clinical Neurology, Vol. 30 (congenital Malformations of the Brain and Skull, Part I): By P. J. Vinken and G. W. Bruyn (Eds.), in Collaboration with N.C. Myrianthopoulos, Xii + 708 Pages, 391 Illustrations, 44 Tables, North-Holland Publishing Company, Amsterdam, 1977, US 121.75, Dfl 280.00, Subscription Price US 103.50, Dfl 238.00.” Journal of the Neurological Sciences 38 (3): 442. [Google Scholar]
- Pruitt Kim D., Brown Garth R., Hiatt Susan M., Françoise Thibaud-Nissen Alexander Astashyn, Ermolaeva Olga, Farrell Catherine M., et al. 2014. “RefSeq: An Update on Mammalian Reference Sequences.” Nucleic Acids Research 42 (Database issue): D756–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac Predrag, Peng Kang, Clark Wyatt T., Peters Brandon J., Mohan Amrita, Boyle Sean M., and Mooney Sean D.. 2008. “An Integrated Approach to Inferring Gene-Disease Associations in Humans.” Proteins 72 (3): 1030–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson Peter D., Ball Edward V., Mort Matthew, Phillips Andrew D., Shiel Jacqueline A., Thomas Nick S. T., Abeysinghe Shaun, Krawczak Michael, and Cooper David N.. 2003. “Human Gene Mutation Database (HGMD): 2003 Update.” Human Mutation 21 (6): 577–81. [DOI] [PubMed] [Google Scholar]
- Stenson Peter D., Mort Matthew, Ball Edward V., Evans Katy, Hayden Matthew, Heywood Sally, Hussain Michelle, Phillips Andrew D., and Cooper David N.. 2017. “The Human Gene Mutation Database: Towards a Comprehensive Repository of Inherited Mutation Data for Medical Research, Genetic Diagnosis and next-Generation Sequencing Studies.” Human Genetics 136 (6): 665–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonnsen Bridgette L., Boan Andrea D., Bradley Catherine C., Charles Jane, Cohen Amy, and Carpenter Laura A.. 2016. “Prevalence of Autism Spectrum Disorders Among Children With Intellectual Disability.” American Journal on Intellectual and Developmental Disabilities 121 (6): 487–500. [DOI] [PubMed] [Google Scholar]
- Vihinen Mauno. 2012. “How to Evaluate Performance of Prediction Methods? Measures and Their Interpretation in Variation Effect Analysis.” BMC Genomics 13 Suppl 4 (June): S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Kai, Li Mingyao, and Hakonarson Hakon. 2010. “ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data.” Nucleic Acids Research 38 (16): e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiffin Nicola, Roberts Angharad M., Minikel Eric, Zappala Zach, Walsh Roddy, Anne H. O’Donnell-Luria, Konrad J. Karczewski, et al. 2019. “Using High-Resolution Variant Frequencies Empowers Clinical Genome Interpretation and Enables Investigation of Genetic Architecture.” American Journal of Human Genetics 104 (1): 187–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Hui Y., Alipanahi Babak, Lee Leo J., Bretschneider Hannes, Merico Daniele, Yuen Ryan K. C., Hua Yimin, et al. 2015. “RNA Splicing. The Human Splicing Code Reveals New Insights into the Genetic Determinants of Disease.” Science 347 (6218): 1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Hui, Robinson Peter N., and Wang Kai. 2015. “Phenolyzer: Phenotype-Based Prioritization of Candidate Genes for Human Diseases.” Nature Methods 12 (9): 841–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.