Abstract
The gut microbiome has been implicated in host metabolism, endocrinology, and pathophysiology. Furthermore, several studies have shown that gut bacteria impact host growth, partially mediated through the growth hormone (GH)/insulin-like growth factor 1 (IGF-1) axis. Yet, no study to date has examined the specific role of GH on the gut microbiome. Our study thus characterized the adult gut microbial profile and intestinal phenotype in GH gene-disrupted (GH-/-) mice (a model of GH deficiency) and bovine GH transgenic (bGH) mice (a model of chronic, excess GH action) at 6 months of age. Both the GH-/- and bGH mice had altered microbial signatures, in opposing directions at the phylum and genus levels. For example, GH-/- mice had significantly reduced abundance in the Proteobacteria, Campylobacterota, and Actinobacteria phyla, whereas bGH mice exhibited a trending increase in those phyla compared with respective controls. Analysis of maturity of the microbial community demonstrated that lack of GH results in a significantly more immature microbiome while excess GH increases microbial maturity. Several common bacterial genera were shared, although in opposing directions, between the 2 mouse lines (e.g., decreased in GH-/- mice and increased in bGH mice), suggesting an association with GH. Similarly, metabolic pathways like acetate, butyrate, heme B, and folate biosynthesis were predicted to be impacted by GH. This study is the first to characterize the gut microbiome in mouse lines with altered GH action and indicates that GH may play a role in the growth of certain microbiota thus impacting microbial maturation and metabolic function.
Keywords: gut microbiome, growth hormone, microbial maturity, bGH mice, GH-/- mice, short-chain fatty acid production
Two decades of research have radically shifted our understanding of the gut microbial community and its homeostatic role in maintaining human health. This microbial community, termed the gut microbiota, consists of trillions of microorganisms (mostly bacteria) that colonize the gastrointestinal tract, with the majority concentrated in the distal ileum and colon (1–3). The gut microbiome is defined as the collective gut microbiota, their genes, and how the expression of those genes influences the physiology of the host (2, 4). The gut microbiota and a subset of their by-products, such as short-chain fatty acids (SCFAs), have been implicated in numerous physiological functions, contributing to the intestinal barrier, host immune system, metabolism, production of several hormones (e.g., sex steroids, vitamin D, and gut hormones), and bone health (5–10). Conversely, microbial dysbiosis, or a lack of diversity in the gut, has been associated with metabolic and intestinal dysfunction (11), and microbial immaturity, or a delay in the expected development of the microbial community, has been associated with metabolic diseases, such as chronic undernutrition, anorexia nervosa, and acute malnutrition (12–15). Thus, altering the microbiome (either through administration of probiotics, SCFAs, or fecal microbiome transplant) has been studied as a means to treat various intestinal diseases, metabolic disorders, endocrine diseases, and growth failure secondary to malnutrition (5, 11, 16, 17).
Emerging evidence suggests a link between the gut microbiome and the growth hormone (GH)/insulin-like growth factor-1 (IGF-1) axis. That is, several studies point to microbial regulation of bone and linear growth by altering IGF-1 levels (18–21). In Drosophila, microbes (mainly Lactobacillus plantarum and Acetobacter pomonum) have been shown to increase growth factors, such as Drosophila insulin-like peptides, which are comparable to GH/IGF-1 signaling in mammals (19, 22). Interestingly, these findings have been translated in germ-free mice who experience restricted growth due to a nutrient-depleted diet, acting as a model of chronic undernutrition seen in children (18). Introduction of L. plantarum to the gut flora of these mice increases levels of serum IGF-1 and restores linear growth (18). Likewise, Yan et al demonstrated that colonization of commensal bacteria in the gut increases serum IGF-1 likely through SCFAs, whereas antibiotic treatment decreases serum IGF-1 levels and prevents bone formation (20, 21). Upstream of the GH/IGF-1 axis, full colonization of the gut microbiota has been associated with increasing ghrelin levels (23). The influence of the GH/IGF-1 axis on the gut microbiome has been minimally explored in 2 studies (13, 24); the first study focused on IGF-1 administration, which normalizes dysbiosis, independent of diet (13), while the second observed an altered gut microbiome in Ames mice, a model of hypopituitarism with deficiencies in GH, prolactin (PRL), and thyrotropin (TSH), also known as thyroid-stimulating hormone (24).
GH is a likely candidate to influence the microbiome. GH is well known to promote overall linear growth, has a catabolic effect on many organs (including intestines), increases lipolysis, and has a potent diabetogenic effect (25). GH action has also been associated with rebuilding the intestinal barrier and improving macronutrient and fat absorption (26–28). This pleiotropic effect of GH happens both synergistically with and independent of IGF-1, exemplified in linear and radial bone growth and metabolism (25, 29). Our laboratory has characterized the global and tissue-specific effect of GH in several mouse lines, 2 of which include the GH gene-disrupted (GH-/-) mice with absent GH action and bovine GH transgenic (bGH) mice with chronic, excess GH action. The GH-/- mice, like the GH receptor gene disrupted mice, are dwarf in size and have increased adiposity, decreased lean mass, and increased insulin sensitivity (30). On the other hand, the bGH mice are giant with decreased adiposity and increased lean mass and develop many comorbidities, including insulin resistance, diabetes, and accelerated aging (31, 32).
Although research points to a link between the GH/IGF-1 axis and the gut microbial community, the nature of the relationship between GH and the gut microbiome remains unclear, and to date, the specific impact of GH on the gut microbiome has not been explored. Therefore, this study aimed to examine the influence of GH on the mouse gut microbiome, characterizing the abundance, diversity, maturity, unique microbial signature, and predictive metabolic function in male GH-/- mice and bGH mice at 6 months of age. Moreover, the relationship between GH and the gut microbiome was analyzed in the context of the intestinal phenotype (i.e., intestinal gross anatomy, inflammation, morphology, and fecal output) of the 2 mouse lines compared with their respective littermate controls.
Materials and Methods
Mouse lines
Male mice from 2 different mouse lines were used: 1) GH gene disrupted (GH-/-) mice (absent GH) and littermate controls (n = 10 per group) and 2) bovine transgenic GH (bGH) mice (chronic, excess GH action) and their respective littermate controls (n = 10 per group). Both the GH-/- mice and the bGH mice have been previously characterized (30, 31). In brief, the GH-/- mouse line was generated by deleting the GH gene on chromosome 11 with the replacement of lacZ reporter and neomycin cassette originally in the C57BL/6N background at University of California (UC), Davis and then backcrossed 9 generations into a C57BL/6J background at Ohio University (30). The global deletion of the GH gene yields absent GH levels, conferring a phenotype similar to congenital GH deficiency. The bGH mice were generated using a metallothionein transcriptional regulatory element linked to the first exon and intron of the bGH cDNA that was injected into the pronucleus of C57BL/6J embryos. The bGH transgene in these mice allows for constitutively secreted GH, conferring chronic, excess GH signaling (approximately a 400-fold increase in endogenous GH levels and resultant 2-fold increase in endocrine IGF-1 levels) and a phenotype similar to patients with acromegaly (33). Mice used in this experiment were all genotyped at 4 weeks after birth from tail snips using PCR primers as described previously (30, 31).
Measurements on the microbiome in all 4 mouse groups occurred at 6 months of age. For intestinal gross anatomy, histology, and fecal markers, a separate cohort of 6-month-old male mice was used with the exception of intestinal histology in GH-/- mice, in which only older mice were available (12 months old). Male mice were chosen to eliminate the confounding factor of sex on the gut microbiome, and 6 months of age was chosen to ensure an adult microbiome. Separate controls for the GH-/- and bGH mice (and their respective littermate control mice) were used to eliminate the potential confounding factor of environmental, genetic, or phenotypic differences between the GH altered mouse lines. All 4 mouse groups (GH-/- mice, bGH mice, and their respective littermate controls) were housed in similar conditions, in a temperature-controlled (23°C) vivarium and exposed to a 14-hour light, 10-hour dark cycle. All mice were allowed access to chow (ProLab RMH 3000; PMI Nutrition International) and water ad libitum. All procedures performed with the mice were approved by the Ohio University Institutional Animal Care and Use Committee and are in accordance with all standards set forth by federal, state, and local authorities.
Fecal collections
Fecal pellets were freshly collected from GH-/- mice, bGH mice, and their respective littermate controls. Fecal pellets were collected at the same time of day to control for the circadian rhythm of the gut microbiome. Mice were allowed to naturally defecate or, after 10 minutes, encouraged to defecate through gentle massage in the hind-back region. Pellets were collected with sterilized forceps, weighed, measured for length, and immediately frozen on dry ice. Equipment and the bench surface were sterilized with diluted 1:10 bleach solution between each mouse collection. Frozen fecal pellets were stored at −80°C until used to measure fecal calprotectin or shipped on dry ice for 16S ribosomal RNA (rRNA) sequencing to the Microbiome and Host Response Core of the Mouse Metabolic Phenotyping Center (MMPC) at UC Davis.
Fecal calprotectin
Calprotectin, a biomarker for intestinal inflammation and typically used to diagnose and monitor Crohn’s disease, was assessed from fecal pellets of GH-/- mice, bGH mice, and their respective littermate controls. In brief, fecal pellets were homogenized, calprotectin was extracted in a 1:50 extraction buffer as provided by the manufacturer, and levels were quantified using an S100A8/S100A9 ELISA (Immundiagnostik AG; Bensheim, Germany, Cat#30–6936) (34).
Fecal DNA isolation
Total DNA from each individual pellet was extracted using Qiagen’s QIAamp PowerFecal DNA Isolation kit following the manufacturer’s protocol with minor modifications. In brief, each fecal pellet was homogenized and cells, both microbial and host, lysed by mechanical means (bead beater for 2 minutes) and chemical disruption of cell membranes (lysis solution and heating samples to 65°C and subsequent incubation for 10 minutes at 4°C). Each incubation step was set to 10 minutes rather than the protocol’s recommended 5 minutes to enhance the quality of the DNA. After several incubation steps, total genomic DNA was captured on a spin column, cleaned with an extra washing step with ethanol, and then eluted into 100 μL of 10 mM Tris. Quality of DNA was tested with a Qubit (Invitrogen). DNA was then stored in a −80°C freezer until 16S rRNA sequencing or quantitative polymerase chain reaction (PCR).
16S rRNA sequencing
Sample libraries were prepared and analyzed by barcoded amplicon sequencing as described previously (35). Purified DNA was amplified on the hypervariable V4 region of the 16S rRNA gene through PCR with primers F515 (5′-GTGCCAGCMGCCGCGGTAA-3′) and R806 (5′-GGACTACHVGGGTWTCTAAT-3′). The resultant amplicons were sequenced using pair-end, high-throughput sequencing with Illumina MiSeq (20 000 average sequence read depth) at the UC Davis Genomics Facility.
Quantitative PCR
Quantitative PCR (qPCR) was used to validate our microbial abundance findings with 16S rRNA sequencing. In brief, the same DNA isolated for 16S rRNA sequencing was used with several different primers at the kingdom (total microbial populations), phylum, and genus/species level (more details in Supplemental Table 1, available in an online data repository) (36). All primers were tested for both temperature and primer efficiency prior to testing experimental samples. Quantity and quality of DNA were checked, respectively, using Qubit 3.0 Fluorometer and NanoDrop 2000 Spectrophotometer. Sample DNA was diluted to 1:32 for the qPCR reactions, and qPCR assays were run using SYBR Green Master Mix and QuantStudio 3 system (Thermo Fisher Scientific). For quantification with qPCR, 2 approaches were used based upon other studies (37–39): (1) relative abundance against several reference genes (such as the V1V2 227F/2357R and Bac340F/515R primers that all target a conserved region of the 16S rRNA gene); and (2) absolute abundance against microbial standards (an even genomic mix of 12 known gut microbiota that were targeted by our bacterial primers; ATCC MSA-1006). Relative abundance was performed on genes targeting bacterial phyla and genera. The same interplate control was used across all plates for both mouse lines.
Microbiome data analysis
Microbial abundance analysis
Sequencing-derived data that were previously demultiplexed at UC, Davis Genomics Facility were then analyzed through the bioinformatics pipeline of QIIME 2 2019.1 version (40, 41). Sequences were first cleaned and denoised into amplicon sequence variants to reduce sequence errors and dereplicate sequences with dada2 function, creating both a frequency and sequence feature table for each mouse line with their respective controls. To quantify microbial abundance for each mouse group, taxonomical classification of features or operational taxonomical units (OTUs) with classify-sklearn function, a trained classifier, and the 99% SILVA 132 16S reference library were used (41, 42). Microbial abundance was analyzed from the phylum to genus level for each mouse and averaged for the 4 mouse groups: GH-/- mice, bGH mice, and both littermate control groups. All abundance data are reported mean ± standard error of the mean (SEM) with the effect size (Cohen d) between each mouse with altered GH action and their controls. The Welch’s t test was conducted due to unequal variance, and statistical significance was determined at P < 0.05 with a false-discovery rate (FDR) modification.
Microbial diversity and maturity
Diversity analyses were run on the resultant feature table sequences using QIIME 2 core diversity plugin. Rarefaction was used to ensure the same number of random reads from each sample for both alpha and beta diversity analysis. Alpha diversity was calculated with Faith’s phylogenetic diversity for microbial richness and with Pielou’s evenness. Beta diversity measures the differences within and between mouse groups on distance metric using principal coordinate analysis (PCoA) through nonphylogenetic (Bray-Curtis and Jaccard) and phylogenetic means (unweighted and weighted UniFrac). Permutation on a multivariate analysis of variance (PERMANOVA) was conducted with 1000 permutations with a post hoc Bonferroni test to determine statistical significance (P < 0.05) between GH-/- mice and bGH mice compared with their respective littermate controls.
Microbial maturity was assessed through q2-classifier on QIIME 2 (14). This method calculates the microbial maturity from a regression model on control group samples and then predicts the microbial age. A maturity index z-scores (MAZ) is calculated based upon the predicted microbial age compared with the actual chronological age of the mice.
Partial least squares-discriminant analysis—microbial signature
A partial least square–discriminant analysis (PLS-DA) using abundance at the genus level was performed using R to identify bacterial signatures unique to genotype differences. Data were first filtered to remove low abundance genera (mean of any group < 0.1%). Results were reported in a score and a loading values plot. The score plot demonstrates the difference between the 2 groups and their controls, whereas the loading values plot presents the top 20 bacterial genera responsible (both positive and negative) for the differentiation between those mouse groups. An additional analysis, variable influence on projection (VIP) scoring, was conducted using R, and a score higher than 1.0 was determined as a genus that differentiated the GH-altered mouse lines compared with respective controls.
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt)
The predictive metabolic function was assessed using the full Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2 (PICRUSt2) pipeline with EPA-NG. The full PICRUSt2 pipeline uses the input of the OTU feature table from QIIME 2 in both groups (GH-/- mice and bGH mice compared with their respective littermate controls) to calculate and differentiate the microbiome by MetaCyc abundance and coverage pathways. PLS-DA and VIP analyses were performed on the MetaCyc abundance pathways to identify which ones differentiated the GH-/- mice and bGH mice from their controls.
Intestinal gross anatomy and morphology
At dissection, both small and large intestines were extracted with weight and length measured as previously described (28). For histology, 6 cross-sections (2 each for the duodenum, jejunum, and ileum) of the small intestine and 4 longitudinal sections of the large intestine were collected and prepared with the Swiss-roll technique (43). Intestinal samples were then fixed in formalin for 24 hours and stored in 70% ethanol until further processing. Samples were embedded in paraffin, sectioned, and stained with hematoxylin and eosin for morphological assessment. Slides were visualized at 100× magnification with a Nikon Eclipse E600 microscope, and pictures of 10 nonoverlapping fields were taken per intestinal cross-section. An average of 20 measurements per mouse intestinal section for villus height, crypt length, and muscle thickness, respectively, were quantified using ImageJ and then averaged per mouse and group.
Other statistical measures
All values are reported as mean ± SEM, and statistics—other than microbial composition and beta diversity as stated above—were performed using R (44). Normality of data was tested plotting the data on a Q-Q plot and through the Shapiro-Wilks test (45). For all single time point measurements, equality of variance was tested using F-test of equal variance, or with Levene’s test of homogeneity of variance for data with an abnormal distribution (46). For data that passed assumptions of normality and variance between groups, a Student t test was performed. Due to unequal variance seen in the microbiome analysis, Welch’s t test was conducted between groups for abundance. For data that failed assumptions of normality, a Mann-Whitney U test was used. For all tests, statistical significance was set at P < 0.05. Effect size (calculated by Cohen d that takes into account the mean, standard deviation, and sample size of each mouse group) was used to describe the strength of the microbiome and intestinal findings and thus, more accurately compare findings between mouse lines. Cohen d is defined by the following cutoffs: a very small effect size is 0.01, small effect size 0.20, medium 0.50, large 0.80, and very large 1.20 (47, 48).
Correlations were used to examine the relationship between intestinal phenotype measurements (length and weight) and microbial richness, evenness, and abundance. Pearson’s correlation was used for intestinal length, whereas the nonparametric Spearman correlation was used for fecal weight and calprotectin due to failed Henze-Zirkler’s multivariate normality test (49). To correct for the number of correlations tested in each mouse line, the statistical significance was set at P < 0.0125. Beta diversity using PCoA was calculated looking at the effect of intestinal phenotype and caging on the microbiomes in both mouse lines, and statistical significance was determined through PERMANOVA and a post hoc Bonferroni test. Finally, to understand the context of intestinal anatomy and fecal output on the microbiome findings, linear mixed-effects models were created for microbial diversity and abundance with the variables of fecal weight, intestinal length, inflammation, and genotype, using R (50, 51).
Results
GH alters the gut microbial composition by 6 months of age
To determine the influence of GH on the gut microbial composition, abundance from the phylum to the genus level was characterized in adult GH-/- and bGH mice compared with their respective littermate controls through both 16S rRNA sequencing and qPCR (Fig. 1) (36). For all mouse groups, the most abundant phyla were Bacteroidetes, Firmicutes, Proteobacteria, and Campylobacterota. Compared with controls, both GH-/- and bGH mice exemplified a microbial shift at the phylum, family, and genus level (Fig. 1A–H and Table 1).
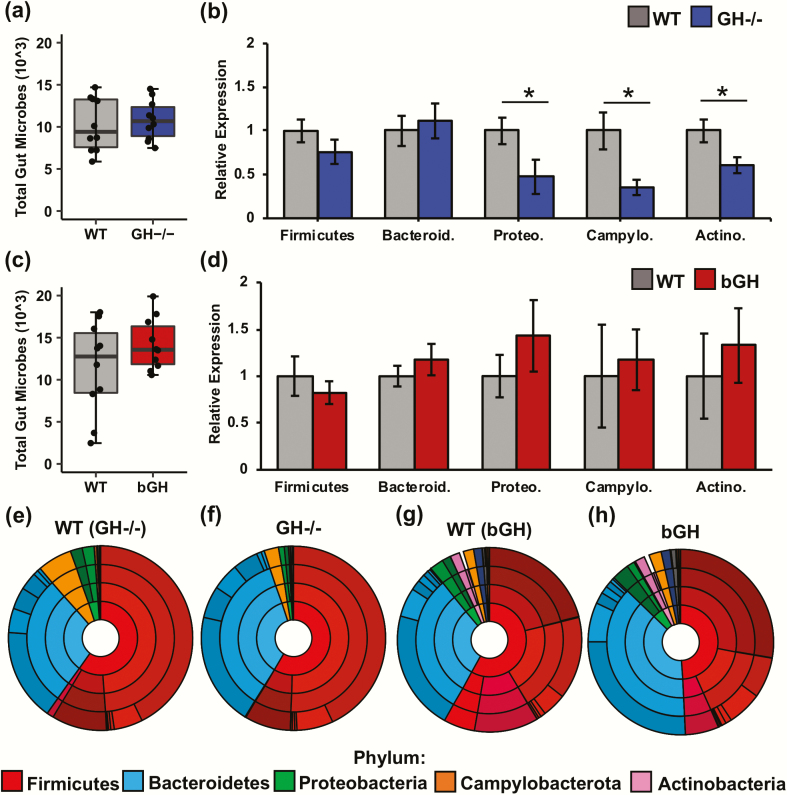
Figure 1.
Microbial abundance of the GH-/- and bGH mice at 6 months of age relative to their respective littermate controls. A. Total bacterial sequences in GH-/- mice and their littermate controls with a small effect size (Cohen d = 0.20). B. Abundance in GH-/- mice and controls at the phylum level seen through qPCR. GH-/- mice have a significant reduction in Proteobacteria (Proteo), Campylobacterota (Campylo), and Actinobacteria (Actino) (P = 0.048, P = 0.001, and P = 0.01, respectively). C. Total bacterial sequences in bGH mice and their respective littermate controls with a medium effect size (P = 0.18 and Cohen d = 0.62) D. Abundance in bGH mice and controls at the phylum level seen through qPCR. E-H: Microbial abundance from the phylum to the family level based on 16S rRNA sequencing (Table 1) in the following: E. GH-/- littermate control; F. GH-/- mice; G. bGH littermate control; and H. bGH mice.
Table 1.
Microbial Composition at the Phyla Level in GH-/- Mice, bGH Mice, and their Respective Littermate Controls
| Decreased GH Action | Increased GH Action | |||||
|---|---|---|---|---|---|---|
| Phylum | WT | GH-/- | Effect size | WT | bGH | Effect size |
| Firmicutes | 59.8% | 58.7% | 0.07 | 58.1% | 49.2% | 0.73 |
| Bacteroidetes | 28.8% | 36.1% | 0.44 | 31.0% | 37.9% | 0.61 |
| Proteobacteria | 4.14% | 1.73%* | 1.66 | 3.93% | 4.66% | 0.21 |
| Actinobacteria | 0.12% | 0.10% | 0.10 | 2.43% | 2.68% | 0.08 |
| Campylobacterota | 6.11% | 2.55%* | 1.65 | 1.79% | 2.10% | 0.10 |
| Cyanobacteria | 0.35% | 0.33% | 0.04 | 0.64% | 0.96% | 0.16 |
| Deferribacteres | 0.02% | 0.02% | 0.03 | 0.01% | 0.11%* | 0.96 |
*indicates statistical significance. Effect size was calculated by Cohen d between each mouse line and their respective littermate control group.
Overall, there was no significant difference in the total number of bacteria between the GH-/- mice and their littermate controls (Fig. 1A). At the phylum level, GH-/- mice exhibited a shift in the Firmicutes/Bacteroidetes ratio (2.4:1) compared with controls (3.1:1), with a small effect size (Cohen d = 0.33) (Fig. 1E and 1F; Table 1). Moreover, both Proteobacteria and Campylobacterota were significantly reduced in GH-/- mice (1.73% and 2.55%) compared with littermate controls (4.14% and 6.11%), with very large effect sizes (Fig. 1E and 1F; Table 1). The qPCR results confirmed these findings with a significant reduction in the Proteobacteria and Campylobacterota phyla relative to controls (Fig. 1B). Although the 16S rRNA sequencing did not show any differences in Actinobacteria between the GH-/- mice or controls, qPCR results demonstrated a significant decrease in the Actinobacteria phylum as well (Fig. 1B).
With GH excess, the bGH mice had a slightly higher total number of bacteria with a medium effect size than littermate controls, but this difference was not significant (Fig. 1C). Like GH-/- mice, the bGH mice exhibited a similar shift in the Firmicutes/Bacteroidetes ratio (1.4:1) compared with controls (2.5:1) (P = 0.09), with a large effect size (Cohen d = 0.83). That is, bGH mice had a decrease in Firmicutes and an increase in Bacteroidetes compared with controls (Fig. 1D, 1G, and 1H; Table 1). Moreover, the bGH microbiome demonstrated an inverse trend to the GH-/- microbiome with slightly increased abundance in most phyla compared with their respective controls, including Proteobacteria and Campylobacterota, and a significant increase in Deferribacteres (P < 0.05 and Cohen d = 0.96) (Fig. 1G and 1H; Table 1). The qPCR results demonstrated similar findings to the 16S rRNA sequencing analysis with a nonsignificant increase in Proteobacteria, Campylobacterota, and Actinobacteria in the bGH mice compared with controls (Fig. 1D).
GH shows minimal effect on microbial diversity yet significantly alters maturity
The diversity of the gut microbiota was measured in 2 different metrics, alpha and beta diversity. Alpha diversity measures the richness (or the number of bacterial populations) and evenness (or the proportionality of these bacterial populations) in an individual mouse microbiome (i.e., 1 fecal pellet). In contrast, beta diversity measures the microbial difference within and between groups on a taxonomical (nonphylogenetic) and phylogenetic basis.
For alpha diversity, richness was assessed through the Faith’s phylogenetic diversity (Fig. 2A and 2B), which assesses the biodiversity of a microbial community integrating the phylogenetic differences. Evenness was assessed through Pielou’s evenness index, which calculates the proportionality of the populations in the microbial community (Fig. 2C and 2D). There were no significant differences in richness or evenness between GH-/- mice and littermate controls nor between bGH mice and controls; all findings had small effect sizes.
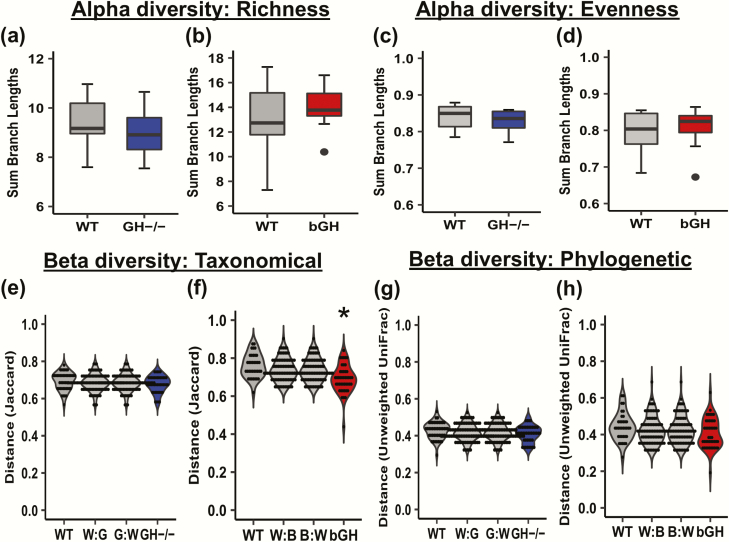
Figure 2.
Diversity in the GH-/- and bGH microbiome. A-D: Alpha diversity in GH-/- and bGH microbiome compared with respective littermate controls. Alpha diversity measures the richness (number of populations) and evenness (proportionality of the populations) in an individual mouse microbiome. A-B: Faith’s phylogenetic diversity. A. in GH-/- mice and littermate controls (Cohen d = 0.42). B. in bGH mice and controls (Cohen d = 0.32). C-D: Evenness alpha diversity. C. in the GH-/- mice and controls (Cohen d = 0.39). D. in bGH and control mice. (Cohen d = 0.30). E-H: Beta diversity in GH-/- and bGH microbiome compared with respective littermate controls. Beta diversity measures the microbial differences within and between groups on a distance matrix using a PCoA. E-F: Jaccard non-phylogenetic beta diversity measurements. E. GH-/- microbiome. F. bGH microbiome. *P = 0.025; determined by PERMANOVA with a post hoc Bonferroni test. G-H: Unweighted UniFrac phylogenetic diversity. G. GH-/- microbiome. H. bGH microbiome.
For beta diversity, a nonphylogenetic beta diversity (Jaccard diversity; Fig. 2E and 2F) test analyzes the microbial difference using a taxonomical basis, while a phylogenetic beta diversity (unweighted UniFrac diversity) (Fig. 2G and 2H) considers the phylogenetic relationship among bacteria. Overall, GH-/- mice and littermate controls were not significantly different in nonphylogenetic or phylogenetic beta diversity (Fig. 2E and 2G), as a wide variance within the GH-/- group and controls was seen, ablating any observable difference between groups. The bGH mice were significantly different at a taxonomical basis (Jaccard beta diversity; Fig. 2F). This significant difference was not observed in the phylogenetic beta diversity measurement (Fig.2H), suggesting that these differences were solely taxonomical in nature with a shared phylogenetic relationship.
Due to the shifts seen in microbial abundance, maturity was investigated by assessing the difference in biological and chronological age of the microbiome in the GH-/- mice and bGH mice compared with controls (Fig. 3). GH-/- mice exhibited a significantly immature microbiome compared with controls with a very large effect size (Fig. 3A). Inversely, the bGH mice showed a nonsignificant, trending increase in maturity compared with littermate controls with a large effect size (Fig. 3B).
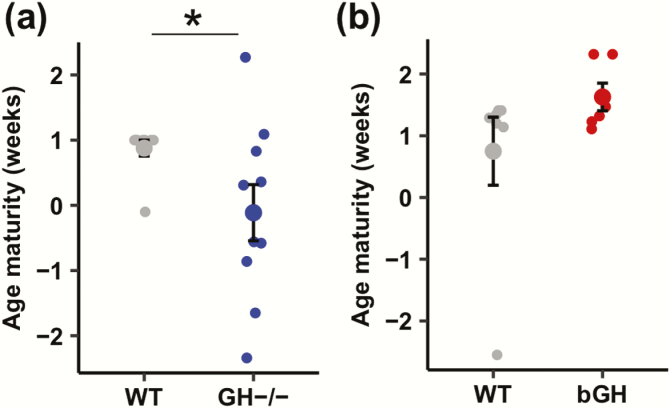
Figure 3.
Maturity of the microbiome in GH-/- (blue) and bGH mice (red) compared with respective littermate controls (gray). A. Maturity in the GH-/- microbiome compared with controls. GH-/- mice have a significant decrease with a very large effect size in age maturity between the expected microbial community and the actual microbial community (Cohen d effect size = 1.45). *P = 0.043, Mann Whitney U test. B. Maturity in the bGH microbiome compared with controls. bGH mice have a slight increase in maturity compared with littermate controls, but this finding is nonsignificant with a large effect size (Cohen d effect size = 0.79).
Unique microbial signature at the genus level observed in GH-/- and bGH mouse lines
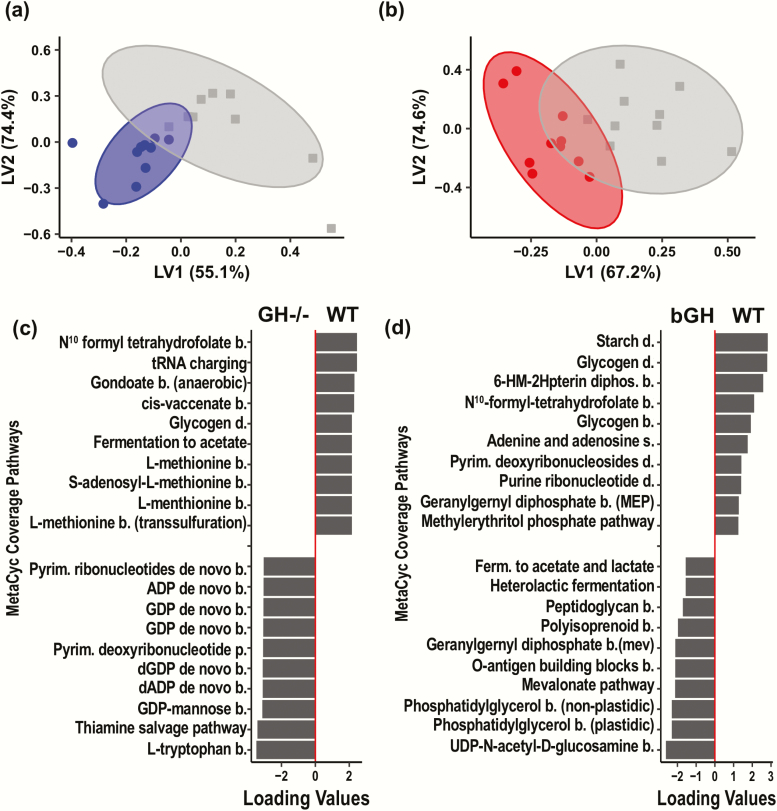
Two analyses were conducted to identify the specific microbial signatures that differentiated the 2 mouse lines versus controls based on abundance at the genus level (multivariate PLS-DA and VIP score analysis). The PLS-DA plot showed an overall distinct separation between each GH altered mouse line and their littermate controls, suggesting both bGH and GH-/- mice have a unique microbial signature compared with respective controls at the genus level (Fig. 4A and Fig. 4B).
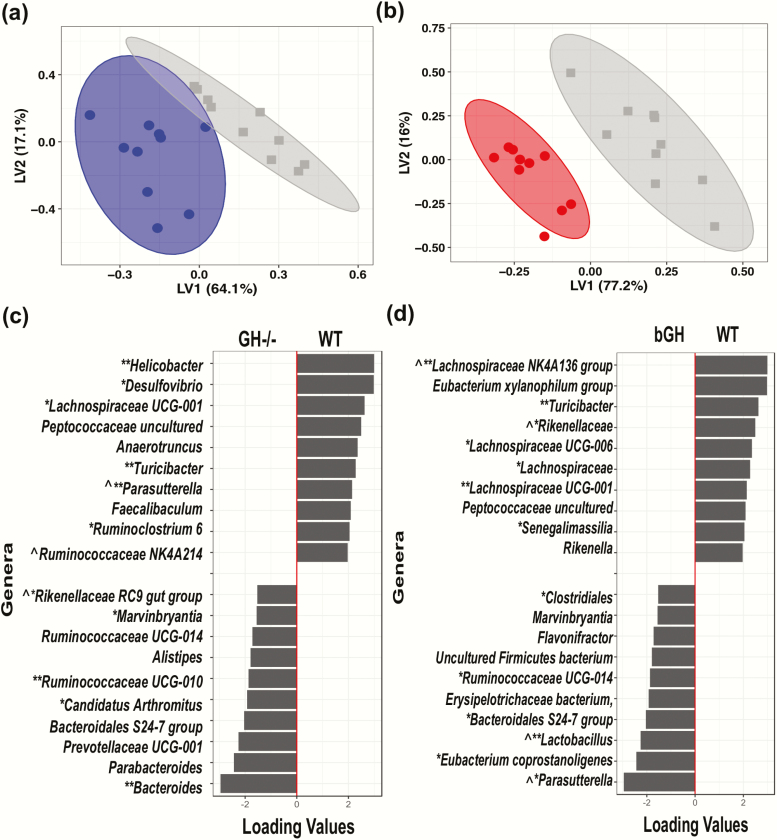
Figure 4.
Unique microbial signature in GH-/- and bGH microbiome determined by partial least squares-discriminant analysis (PLS-DA) at the genus level. A. Score plot of PLS-DA of GH-/- mice (blue) and controls (gray). B. Score plot of bGH mice (red) and controls (gray). C. Loading plot of the top 20 genera for GH-/- microbiome. Left column shows the top 10 genera increased in GH-/- mice while the right column represents the top 10 genera decreased in abundance in GH-/- mice. D. Loading plot for bGH microbiome. Left column shows the top 10 genera increased in abundance while the right column represents the top 10 genera decreased in abundance in bGH mice. *VIP bacteria; **Top 4 VIP genera in both mouse lines. ^Shared bacteria from PLS-DA and VIP analysis in present in GH-/- and bGH microbiomes in opposite directions.
For GH-/- mice and of the top 20 bacterial genera differentiated with the PLS-DA, 14 bacterial genera were also identified by the VIP analysis. The majority of the VIP bacterial genera belonged to the Firmicutes phylum (64.3%) and the remainder to Proteobacteria, Bacteroidetes, and Campylobacterota. Between both the PLS-DA analysis and VIP score, genera from the Proteobacteria and Campylobacterota phyla were identified as some of the most important players responsible for the differentiation between the 2 groups. In particular, Parasutterella, Helicobacter, and Desulfovibrio were all decreased in abundance in the GH-/- mice compared with controls (Fig. 4C). Conversely, the genera in the Bacteroidetes, such as Bacteroides, were increased in abundance in GH-/- mice compared with controls (Fig. 4C). Genera in the Firmicutes phylum were both upregulated (i.e., Marvinbryantia, Candidatus Arthromitus, and Ruminococcaceae UCG-010) and downregulated (i.e., Turicibacter, Ruminococcaceae, and Faecalibaculum) in the GH-/- mice (Fig. 4C).
For bGH mice, the PLS-DA identified the top 20 bacterial genera responsible for the differentiation between each genotype (Fig. 4D), most belonging to the Firmicutes, Bacteroidetes, and Proteobacteria phyla. There were 33 genera that were identified as VIP bacteria. Between the 2 analyses, 13 bacteria were identified as part of the unique microbial signature. The majority of the VIP bacteria belonged to the Firmicutes phylum (72%) with the remainder to Bacteroidetes, Proteobacteria, and Deferribacteres. The most differentially abundant candidates identified in both the PLS-DA and VIP included several bacteria from Firmicutes (Ruminococcaceae, Lachnospiraceae UCG-001, Turicibacter, Lachnospiraceae NK4A136 group, and Lactobacillus), Proteobacteria (Parasutterella and Bilophila), and Bacteroidetes (Rikenellaceae).
Several common bacterial genera were identified between the 2 mouse lines. A few microbes like Turicibacter, Marvinbryantia, and Lachnospiraceae (such as UCG-004 and UCG-001) were present in both GH altered microbiomes in the same direction. For instance, both Turicibacter and Lachnospiraceae were seen to be decreased in abundance in both GH-/- mice and bGH mice compared with littermate controls (Fig. 4) (Supplemental Figure 1) (36). Notably, several bacterial candidates appear to be associated with GH action as they were shared, albeit in opposite directions, between the 2 mouse lines (Table 2). Specifically, Parasutterella was decreased in the GH-/- microbiome while increased in the bGH microbiome (Fig. 4C and 4D). This trend was also confirmed by qPCR data, where we observed a significant reduction of 2 common strains of Parasutterella (P. secunda and P. excrementihomis) in GH-/- mice compared with respective controls (P = 0.02 and P < 0.001) and a trending increase in bGH mice of P. secunda (Supplemental Figure 1) (36). Similar trends were seen in Rikenellaceae and certain Ruminococcaceae and Lachnospiraceae (Fig. 4C and 4D) from the PLS-DA and VIP analysis. Interestingly, although Lactobacillus was only identified from the PLS-DA and VIP analysis in the bGH microbiome, there were significant opposing findings between the 2 groups in qPCR analysis (i.e., significantly decreased in GH-/- mice and increased in bGH mice [P < 0.05]).
Table 2.
Potential GH-Associated Bacterial Candidates Present in Opposite Directions between the GH-/- and bGH Microbiome Relative to Controls as Determined by PLS-DA and VIP Analysis
| GH-/- Microbiome | bGH Microbiome | ||||
|---|---|---|---|---|---|
| Bacterial Genus | Classification | Relative Abundance | VIP Score | Relative Abundance | VIP Score |
| Parasutterella | Proteobacteria; Betaproteobacteria; Burkholderiales; Alcaligenaceae | ↓ | 5.57 | ↑ | 1.365 |
| Ruminococcaceae NK4A214 group | Firmicutes; Clostridia; Clostridiales; Ruminococcaceae | ↓ | 0.85 | ↑ | 1.95 |
| Tyzzerella | Firmicutes; Clostridia; Clostridiales; Lachnospiraceae | ↓ | 1.08 | ↑ | 1.10 |
| Lactobacillus | Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae | ↓ | 0.75 | ↑ | 1.38 |
| Lachnospiraceae NKA214 group | Firmicutes; Clostridia; Clostridiales; Lachnospiraceae | ↑ | 0.90 | ↓ | 1.41 |
| Rikenellaceae | Bacteroidetes; Bacteroidia; Bacteroidales; Rikenellaceae | ↑ | 1.01 | ↓ | 1.11 |
Distinct predictive metabolic function associated with GH-/- and bGH mouse lines
Our previous analyses focused on identifying the gut bacteria associated with GH action. Next, we wanted to understand how GH impacts the predictive metabolic function of the total microbial community. A predictive metabolic function analysis was thereby performed through the platform PICRUSt2, focusing on the metabolic profile associated with microbial communities in the 4 groups (Fig. 5). Overall, both GH-/- and bGH mice exhibited a distinct predictive metabolic function relative to respective controls. To highlight a few pathways, the GH-/- microbiome is predicted to have upregulated aerobic respiration and branched chain amino acid biosynthesis and downregulated enterobactin biosynthesis. Meanwhile, the bGH microbiome was associated with a prediction of increased pyruvate fermentation to ethanol, lactate, and propionate along with downregulated aromatic compound degradation and vitamin B3 and B12 biosynthesis. More importantly, several pathways were predicted to be in opposite directions between the 2 mouse lines. Specifically, fermentation to the SCFAs acetate and butyrate was shown to be increased in the bGH microbiome and decreased in the GH-/- microbiome. Similar predicted trends were seen with folate biosynthesis and heme B biosynthesis seen in both the MetaCyc coverage pathways (Fig. 5) and MetaCyc abundance (Supplemental Figure 2) (36). These findings correspond with the shifts in the microbial populations at the phylum (i.e., Proteobacteria, Campylobacterota, and Actinobacteria) and genus level (i.e., Lactobacillus) observed in the GH-/- and bGH mice. Bacteria in the Firmicutes, Proteobacteria, and Campylobacterota phyla have been associated with heme B biosynthesis pathways, whereas specific bacteria from Firmicutes (Lactobacillus and Clostridium), Proteobacteria (Sutterella), and Actinobacteria (Bifidobacterium) have been associated with SCFA production.
Figure 5.
Distinct predictive metabolic function in GH-/- (blue) and bGH microbiome (red) using partial least squares-discriminant analysis (PLS-DA). A-B: Score plots of the GH-/- mice and bGH mice compared with respective littermate controls based on the MetaCyc coverage pathways. A. Score plot of GH-/- mice and controls. A significant difference was seen between these groups. B. Score plot of the bGH mice and controls. A significant difference was seen between groups. C-D: Top 20 metabolic pathways associated with GH-/- microbiome and bGH microbiome. (Left: top 10 upregulated; right: top 10 downregulated with altered GH action mouse lines). C. GH-/- microbiome compared with controls. D. bGH microbiome compared with controls.
GH significantly alters the intestinal phenotype
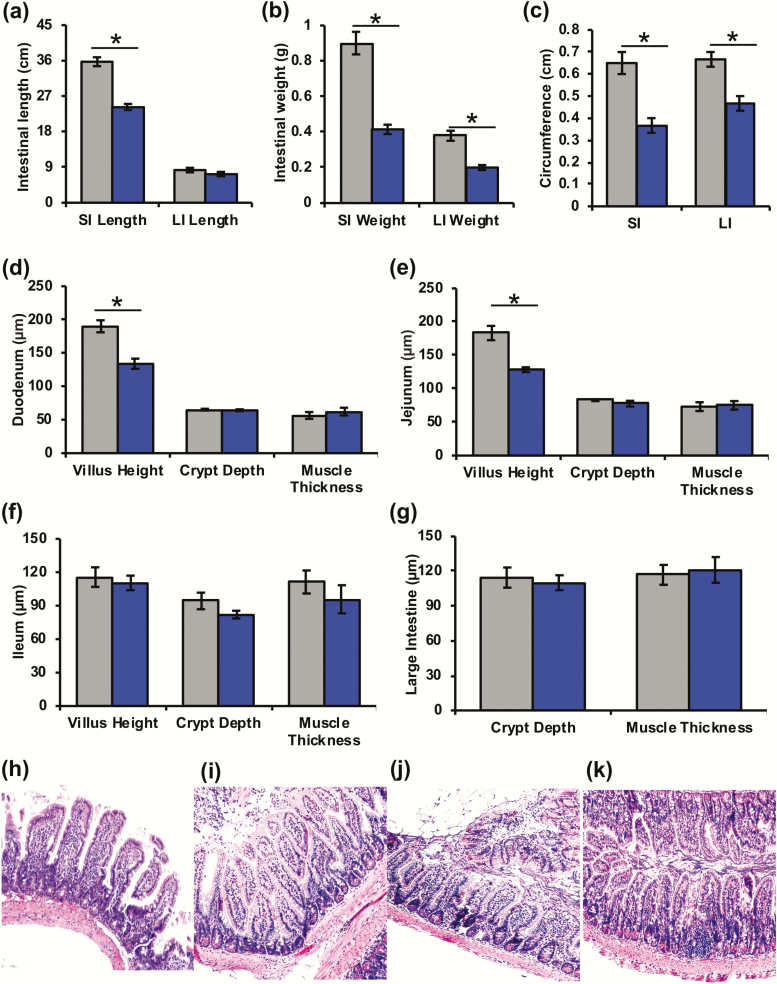
Since GH plays a role in intestinal homeostasis and gut functions, we evaluated the intestinal phenotype (i.e., gross anatomy, inflammation, histology, and fecal output) of the GH-/- and bGH mice. The data obtained demonstrate several opposing findings with large and very large effect sizes between the 2 mouse lines. The GH-/- mice at 6 months of age have significantly shorter small intestines (Fig. 6A), with both small and large intestines weighing significantly less compared with controls (Fig. 6B). At 12 months of age, GH-/- mice had a similar trend in shorter small and large intestines (P = 0.039 and 0.01) (data not shown) and had significantly smaller circumferences in jejunum and colon (Fig. 6C). Villus height in the duodenum and jejunum was significantly decreased in the 12-month-old GH-/- mice (Fig. 6D, 6E, and 6H–6K).
Figure 6.
Intestinal gross anatomy and histology in 6 month or 12 month old GH-/- mice compared (blue) to littermate controls (gray). A. Intestinal length of the small and large intestines (6 months). *P < 0.001 (Cohen d = 3.11). B. Intestinal weight of the small and large intestines (6 months). *P < 0.001 (Cohen d = 3.06 and 2.48, respectively). C. Intestinal circumference of the jejunum (SI) and colon (LI) in 12-month-old GH-/- mice compared with controls. *P = 0.01 and 0.03 (Cohen d = 4.05 and 3.33, respectively). D-F. Duodenum, jejunum and ileum morphology, respectively, including villus height, crypt depth, and muscle thickness of 12-month-old mice. D. GH-/- mice had significantly decreased villus height in duodenum. *P = 0.028 (Cohen d = 2.73). E. GH-/- had significantly decreased villus height in jejunum. *P = 0.049 (Cohen d = 2.08). G. Colon morphology in 12-month-old GH-/- mice compared to controls. H-K: Small intestinal histology in 12-month-old GH-/- mice and controls. H. Control jejunum. I. GH-/- jejunum. J. Control ileum. K. GH-/- Ileum.
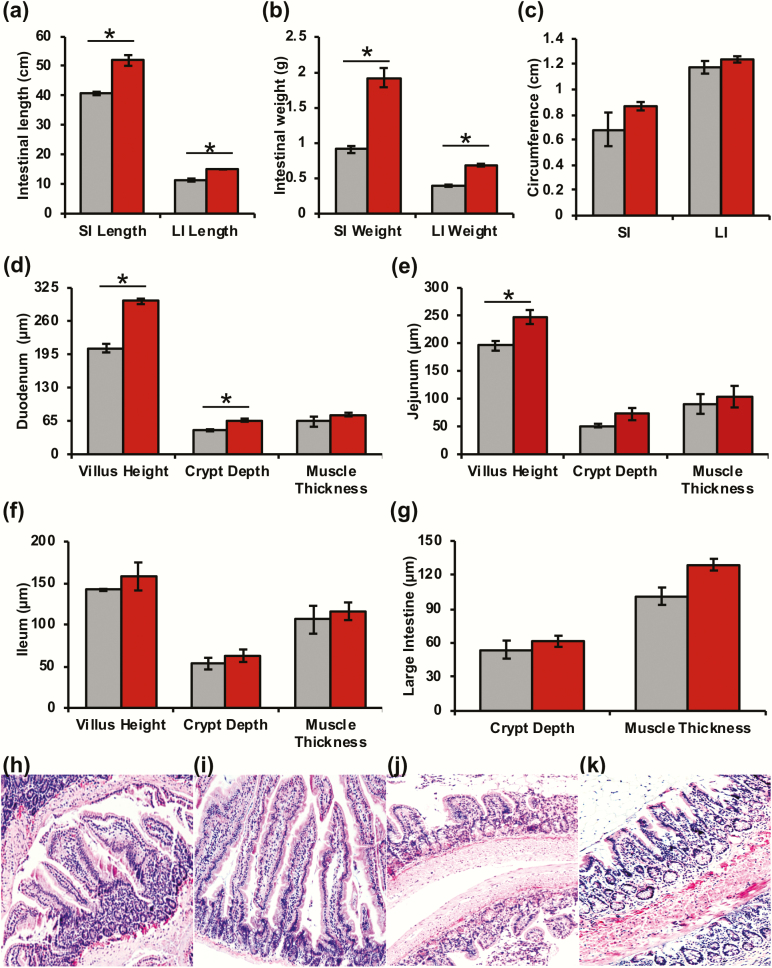
Inversely, bGH mice had significantly increased intestinal length and weight in the small and large intestines compared with littermate controls (Fig. 7A and 7B). Intestinal circumference also showed a trending increase in the jejunum and colon of bGH mice with a large effect size (Fig. 7C). Intestinal morphology was also altered with significantly increased villus height in the duodenum and jejunum, significantly increased crypt depth in the duodenum, and trending increase in villus height, crypt depth, and muscle thickness throughout the small and large intestines (Fig. 7D–K).
Figure 7.
Intestinal gross anatomy and histology in 6 month old bGH mice (red) and littermate controls (gray). A. Intestinal length of the small and large intestines. *P < 0.001 (Cohen d = 2.61 and 3.21, respectively). B. Intestinal weight of the small and large intestines *P < 0.001 (Cohen d = 4.4 and 7.19, respectively). C. Intestinal circumference in jejunum (SI) (Cohen d = 1.17) and colon (LI) (Cohen d = 1.01). D-F: Duodenum, jejunum, ileum morphology (i.e., villus height, crypt depth, and muscle thickness) in bGH mice and controls. D. The bGH mice have significantly increased villus height and crypt depth in the duodenum. *P < 0.001 and P = 0.002, respectively (Cohen d = 5.44 and 5.05, respectively). E. Villus height in the jejunum of bGH mice are significantly increased. *P = 0.02 (Cohen d = 2.62). G. Colon morphology. H-I: Small intestinal histology in bGH mice and controls. H. Control duodenum. I. bGH duodenum. J. Control ileum. K. bGH ileum.
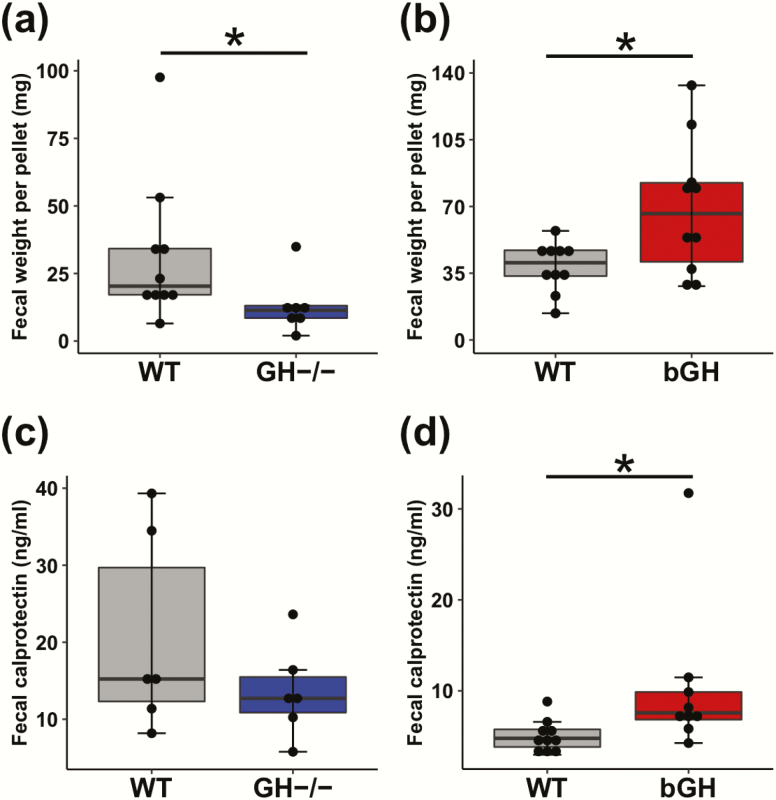
Likewise, fecal weight and calprotectin levels showed opposing trends in the 2 mouse lines relative to their respective controls. Fecal pellets weighed significantly less in GH-/- mice relative to controls (Fig. 8A). Inversely, fecal weight in bGH mice was significantly increased compared with controls (Fig. 8B). Fecal calprotectin levels, a measurement of intestinal inflammation, were not changed in our GH-/- mice with a medium effect size (Fig. 8C) and significantly increased in bGH mice compared with controls (Fig. 8D).
Figure 8.
Fecal output and intestinal inflammation in GH-/- (blue) and bGH mice (red) compared with littermate controls (gray). A-B. Fecal weight. A. in GH-/- mice and controls. *P = 0.033 (Cohen d = 0.92). B. in the bGH mice and controls. *P = 0.025 (Cohen d = 1.15). C-D. Fecal calprotectin levels. C. in GH-/- and control mice (Cohen d = 0.74). D. in bGH and control mice *P = 0.007 (Cohen d = 0.88).
Intestinal length and fecal weight minimally alter the gut microbial profile
The microbial community exists in the context of the host, influenced by environmental factors, genetics, and the intestinal phenotype. This study controlled for genetics and most environmental factors of the mice. However, as shown, GH does alter the intestinal gross anatomy, morphology, and fecal output, which could influence the microbiome. Thus, we attempted to decipher the impact of GH versus the intestinal environment on the microbial composition of the GH-/- and bGH mice relative to littermate controls through statistical means. First, we investigated the association between microbial findings and intestinal measurements (i.e., intestinal length, calprotectin levels, and fecal weight). In both GH-/- and bGH mouse lines, all intestinal phenotype measurements showed a nonsignificant, weak correlation to microbial richness and evenness (Supplemental Table 2) (36). In the GH-/- mouse line, the only significant correlations were observed between Proteobacteria and Campylobacterota abundance and small intestinal length and fecal weight (Supplemental Table 2) (36).
Second, we attempted to determine the collective impact of GH and the intestinal phenotype (i.e., fecal weight, intestinal length, and inflammation) on the gut microbiome using a linear mixed-effects model (Supplemental Table 3) (36). Specifically, linear mixed-effects models statistically identify which covariate(s)—in our case, genotype (i.e., GH level) and intestinal phenotype—are most responsible for microbial abundance and diversity. In GH-/- mice, genotype (i.e., lack of GH) exclusively explained microbial evenness (i.e., proportionality of the bacterial populations) and Proteobacteria abundance findings. Both genotype and large intestinal length collectively explained the microbial richness findings, whereas genotype and small intestinal length explained the Campylobacterota abundance findings. These findings suggest that GH is the major contributor to the gut microbial composition, at least in GH-/- mice.
In bGH mice, neither genotype nor the intestinal measurements fully explained the microbial richness findings, suggesting either a large variance in the groups or that there might be another contributing factor. Similar findings were seen with Campylobacterota, although independently genotype had a major influence on the abundance. Microbial evenness was explained exclusively by genotype and fecal weight, and intestinal length (small intestine and large intestine) and genotype all independently played a major influence on Proteobacteria. These findings paint a more complicated picture of how GH, intestinal length, and fecal weight contribute to the gut microbial composition; however, GH appears to still have a major influence on the microbiome.
Discussion
This study is the first to characterize the gut microbiome in both GH deficient (GH-/-) and GH excess (bGH) mouse lines. Importantly, both mouse lines have chronically altered GH levels with bGH mice having a 2-fold increase in endocrine IGF-1 (33), which needs to be considered when interpreting the results. We observed several interesting and opposing findings between the 2 mouse lines that provide insight into the role of GH on the gut microbial profile. Overall, both GH-/- and bGH mice have an altered microbial profile compared with their respective littermate controls, including differences in abundance in Proteobacteria, Campylobacterota, and to a smaller degree, Actinobacteria. Microbial maturity also differed between the 2 mouse lines with the GH-/- mice having an immature microbiome compared with controls. Additional multivariate analysis revealed that both mouse lines had a unique microbial signature at the genus level compared with their littermate controls, identifying several common bacterial candidates responsible for this differentiation. Some of the bacterial candidates, such as Turicibacter and certain Lachnospiraceae, exhibited similar abundance patterns in the 2 mouse lines. Interestingly, however, the abundance of several common bacterial genera (such as Parasutterella, Ruminococcaceae NK4A214, Rikenellaceae, and Lactobacillus) were opposed in the 2 mouse lines (e.g., decreased in GH-/- mice and increased in bGH mice). Similarly, the predictive function analysis demonstrated shared metabolic pathways regulated in opposite directions between the 2 mouse lines, suggesting that GH potentially plays a role in microbial fermentation of acetate and butyrate, as well as folate and heme B biosynthesis. Finally, we found opposing intestinal gross anatomy, histology, and fecal output between the 2 mouse lines, in which intestinal length might also contribute to the microbial profile associated with GH.
Although this study is the one of the first to report on GH-associated microbial abundance, other studies have evaluated (1) the impact of the gut microbiota—either the commensal microbial community or the addition of one bacteria (such as Lactobacillus plantarum)—on the GH/IGF-1 axis and host growth (i.e., Drosophila or mouse) (19–21, 23); or (2) the microbial composition seen with metabolic diseases (anorexia nervosa, chronic undernutrition, obesity, and diabetes) and intestinal disorders (pediatric Crohn’s disease) associated with abnormal GH action (5, 8, 52–55). To our knowledge, 2 published studies have shown an influence of the GH/IGF-1 axis on microbial composition (13). The first study focused on the direct influence of IGF-1 administration along with diet to BALB/c mice (with decreased IGF-1 levels due to nutrient restriction), resulting in increased Proteobacteria and significantly, Sutterella (a genus closely related to Parasutterella) (13). This positive correlation between IGF-1 and Sutterella and Proteobacteria corroborates the trend we see in our bGH mice and opposite trend observed in the GH-/- mice. The second study observed an altered gut microbial composition in Ames mice (who have GH, PRL, and TSH deficiencies) compared with controls (24). At 2 months of age, Ames mice showed an altered Firmicutes/Bacteroidetes ratio in a similar direction seen in our GH-/- and bGH mice. The Firmicutes/Bacteroidetes ratio is associated with metabolism, energy harvest, and more recently, longevity (8, 56, 57); however, studies still need to further explore the impact of an altered ratio on host health. A 2018 systemic review further suggests that a stronger indicator of metabolic diseases may be specific genera in these phyla rather than the ratio itself (56). Ames mice also had a differential abundance in fecal/colonic bacterial genera, including decreased Lactobacillus and increased Lachnospiraceae NKA214 group, among others (24). Although there were microbial differences observed between the findings of Wiesenborn et al (24) and ours—potentially due to the age of the mice and the additional hormonal deficiencies in Ames mice—the directionality of Lactobacillus and Lachnospiraceae confirm the trends seen in our GH-/-and bGH mouse lines at 6 months of age. Collectively, these findings suggest that GH action might play an important role in the Proteobacteria and Campylobacterota phyla and several genera, including Parasutterella and Lactobacillus. These changes in microbial abundance may influence the maturity and function of the gut microbiome and have an important role in the metabolism seen in these mouse lines.
GH appears to promote total microbial maturity, a finding that corresponds with studies that have examined the link between gut microbiota and growth. Both germ-free mice and antibiotic-treated mice exhibit decreased growth and are associated with decreased levels of ghrelin, GH, and IGF-1 (18, 20, 23). Moreover, lack of microbial maturity has been associated in mice and humans with restricted growth (with both chronic undernutrition and anorexia nervosa) (13–15). Although lack of microbial diversity has been traditionally associated with dysbiosis and intestinal and metabolic diseases, recent studies offer a more complex view of decreased microbial diversity. For instance, the richness and evenness of the developing microbiome is lower in infants, particularly those who breastfeed (58–61). Introduction of acarbose and a high-fiber diet decreases microbial diversity in individuals with type 2 diabetes, creating a niche for a guild of commensal microbes (62). Wilmanski et al (63) suggest an optimal range of microbial diversity with negative consequences with either too much or too little bacterial diversity. This complex interpretation of microbial diversity fits our microbial maturity findings in the GH-/- and bGH mice. Despite having increased adiposity and microbial immaturity, our GH-/- mice have no detectable intestinal disease or inflammation and are extremely insulin sensitive (30). Evidence has also shown that extended longevity (i.e., supercentenarians) is associated with a lack of microbial diversity (64, 65). Longevity has been associated with GH action in mouse lines—increased in mouse lines with decreased GH action (such as our GH receptor knockout mice) (66) and decreased in bGH mice (67). Microbial maturity then may reflect the differences in development and longevity between our GH altered mouse lines. These findings add to the debate on microbial maturity between the benefit of increased diversity/maturity (bGH mice) and decreased maturity with a beneficial guild of a few microbes (GH-/- mice).
The metabolic potential (e.g., acetate and butyrate production) of the GH-associated microbiome proves to be particularly important for endocrinology and host metabolism. The bacterial metabolic capacity is thought to significantly outweigh the host metabolic capacity with approximately 500 times more bacterial genes compared with human genes (68, 69). Microbial by-products–with the quintessence of SCFAs–have been shown to influence energy harvest, glucose and lipid metabolism, immunity, inflammation, and growth (68, 70, 71). Specifically, acetate has been shown to be important for lipid metabolism, and fermentation is thought to be conserved across bacterial groups (70, 71). Butyrate also has an important role in lipid metabolism and more so, acts as fuel for colonocytes; meanwhile, fermentation appears to be limited to fewer bacteria, including Lachnospiraceae, Ruminococcaceae, Lactobacillus, and Sutterella (20, 62, 70)—several of which were found to be associated with GH action. In part, the impact of SCFAs on metabolism may be mediated through altering hormones such as leptin, ghrelin, glucagon-like peptide 1, and peptide YY, and potentially, the GH/IGF-1 axis (7, 20, 68, 72). Yan et al (20) showed that SCFAs increase IGF-1 levels in serum, liver, and bone, while Wang et al (72) observed an inhibitory effect of SCFAs on GH transcription in bovine anterior pituitary cells. These observations, along with our findings, hint at a bidirectional relationship between GH and the gut microbiome, potentially through SCFAs (especially acetate and butyrate). Additional studies will be needed to confirm this association.
Another consideration to the link between GH and the gut microbiome is the intestinal environment, which was distinct in both of our mouse lines. Surprisingly, relatively few studies have examined GH’s impact on intestines, mainly focused on mouse lines with increased GH signaling and one intestinal specific GH receptor knockout (IntGHRKO) mouse line. With excess GH action, mice have increased colon length and weight, mucosal growth (villus height and crypt depth), dysplasia, lymphoid tissue, and muscle thickness (73–76); all of which concurs with our intestinal phenotype findings in bGH mice. Inversely, IntGHRKO mice tend to have decreased mucosal growth and sex-dependent effects on large intestinal length, glucose intolerance, and fat absorption (28). Acute administration of GH increases intestinal stem cell proliferation in vitro (77), has been shown to protect against colitis in rats (78), and improves the gut barrier, mucosal growth, and nutrient absorption in several animal models and clinical cohorts with intestinal diseases (such as short-bowel syndrome and intestinal obstruction) (26, 27, 79). Mucosal growth and muscle thickness are both crucial determinants of gut motility and absorption (80, 81); furthermore, mucosal growth, gut motility, absorption, and inflammation all influence the niche for the microbial community. Our linear mixed-effects models showed that the richness and evenness of the total microbial community was best explained by the combined effect of GH and intestinal length. This finding suggests that GH may alter the diversity of the microbial community through the intestinal environment. Yet interestingly, GH independent of the intestinal phenotype—at least of the factors measured—contributes to the abundance of Proteobacteria and Campylobacterota. Together, these statistical models propose that GH impacts microbial abundance and diversity through several different ways, one at least being its influence on the intestinal environment.
While this study confirmed a distinct microbial profile and intestinal phenotype associated with GH, there are additional questions that remain unanswered. First and foremost, we chose to focus on the effect of GH on a healthy adult gut microbiome and thus, only examined the microbiome at 6 months of age in male mice. This choice discounted the well-known differences in the microbiome seen between sexes and at different ages (82). It is also well-documented that males and females have distinct differences in total GH levels, GH troughs, and pulsatile secretion (83, 84). Moreover, Weger et al (23) has noted that germ-free mice have not only decreased GH levels and altered sexual maturation but consequentially, decreased sexual dimorphism in gene expression and metabolism of males and females, which highlights the importance of studying the relationship between GH and the gut microbiome in the context of sex. Subsequent studies on the microbiome in female and male mice with altered GH action at several age points will be conducted to afford a more thorough characterization. Another limitation of this study is that the wild-type littermate controls differed between mouse lines as seen in microbial abundance, maturity, and fecal calprotectin levels. Thus, we compared the GH altered mice to their respective littermate controls and then used effect size (Cohen d) to compare between mouse lines. Effect size allows us to compare the biological importance of the microbial findings between mouse lines and across studies in a standardized metric, independent of P values (which can be swayed by sample size). Finally, the technology to analyze the gut microbiome, such as next-generation sequencing and bioinformatics, is continuously evolving. For instance, the 16S rRNA sequencing (targeting the V4 region) and qPCR techniques (targeting more-specific sections in the hypervariable regions of the gene) yielded slightly different results for Actinobacteria and Lactobacillus, potentially due to different specificity of the targets. Thus, the use of both 16S rRNA sequencing and qPCR offered a more comprehensive view of the microbial composition in our mice.
In summary, this study is the first to report on the microbial profile (i.e., abundance, diversity, maturity, and predictive metabolic function) in mouse lines with altered GH action. Moreover, this study is the first to compare the intestinal gross anatomy, morphology, and inflammation in 6-month-old GH-/- and bGH mice. Our results demonstrate opposing findings in our 2 GH altered mouse lines in terms of microbial maturity and shifts in abundance and in intestinal phenotype (i.e., fecal output, intestinal size, morphology, and inflammation). This evidence suggests that GH may promote the growth of certain bacteria and thus, contribute toward microbial maturation and metabolic functions like SCFA, folate, and heme B biosynthesis. This study then highlights the potential of the bidirectional relationship between GH and the gut microbiome on intestinal and metabolic health. Although additional studies are needed to fully understand the direct or indirect mechanism of this relationship, our findings are significant in establishing a GH:microbiome connection.
Acknowledgments
This study was funded by the John J. Kopchick MCB/TBS Research Fellowship and a fellowship from Osteopathic Heritage Foundations, Dual Degree Program at Ohio University Heritage College of Osteopathic Medicine. Research was supported by NIH grant U24-DK092993 (Microbiome and Host Response core of Mouse Metabolic Phenotyping Center at UC Davis). We would like to acknowledge Trina Knotts and Helen Raybould (Microbiome and Host Response core at UC Davis) and William Broach (director of the Ohio University Genomics Facility) for consultation on microbiome analysis and the Ohio University Histology core for assistance in preparing intestinal samples. We would also like to acknowledge several students: Maria Onusko and Silvana Duran-Ortiz for their help with the qPCR experiments and Jaycie Kuhn for her help with fecal collection and intestinal histology.
Glossary
Abbreviations
- bGH
bovine growth hormone transgenic
- GH
growth hormone
- IGF-1
insulin-like growth factor 1
- OTU
operational taxonomical unit
- PCoA
principal coordinate analysis
- PERMANOVA
permutational multivariate analysis of variance
- PICRUSt2
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2
- PLS-DA
partial least square–discriminant analysis
- PRL
prolactin
- qPCR
quantitative polymerase chain reaction
- rRNA
ribosomal RNA
- SCFA
short-chain fatty acid
- SEM
standard error of the mean
- TSH
thyrotropin (thyroid-stimulating hormone)
- UC
University of California
Financial Support: This study was funded by the John J. Kopchick Molecular and Cellular Biology/Translational Biomedical Sciences Research Fellowship and a fellowship from Osteopathic Heritage Foundations, Dual Degree Program at Ohio University Heritage College of Osteopathic Medicine. Research was supported by NIH-NIDDK grant U24-DK092993 (Microbiome and Host Response core of Mouse Metabolic Phenotyping Center at UC Davis, RRID: SCR_015361). J.J.K., E.O.L., and D.E.B. were supported by NIH-NIA grant R01-AG059779.
Additional Information:
Disclosure Summary: The authors have nothing to disclose.
Data Availability. The datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
References
- 1. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. Plos Biol. 2016;14(8):e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bäckhed F, Fraser CM, Ringel Y, et al. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe. 2012;12(5):611–622. [DOI] [PubMed] [Google Scholar]
- 3. Ursell LK, Metcalf JL, Parfrey LW, Knight R. Defining the human microbiome. Nutr Rev. 2012;70(Suppl 1):S38–S44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sommer F, Bäckhed F. The gut microbiota — masters of host development and physiology. Nat Rev Micro. 2013;11(4):227–238. [DOI] [PubMed] [Google Scholar]
- 5. Aurigemma NC, Koltun KJ, VanEvery H, Rogers CJ, De Souza MJ. Linking the gut microbiota to bone health in anorexia nervosa. Curr Osteoporos Rep. 2018;16(1):65–75. [DOI] [PubMed] [Google Scholar]
- 6. Clarke G, Stilling RM, Kennedy PJ, Stanton C, Cryan JF, Dinan TG. Minireview: gut microbiota: the neglected endocrine organ. Mol Endocrinol. 2014;28(8):1221–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neuman H, Debelius JW, Knight R, Koren O. Microbial endocrinology: the interplay between the microbiota and the endocrine system. FEMS Microbiol Rev. 2015;39(4):509–521. [DOI] [PubMed] [Google Scholar]
- 8. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. [DOI] [PubMed] [Google Scholar]
- 9. Ximenez C, Torres J. Development of microbiota in infants and its role in maturation of gut mucosa and immune system. Arch Med Res. 2017;48(8):666–680. [DOI] [PubMed] [Google Scholar]
- 10. Novince CM, Whittow CR, Aartun JD, et al. Commensal gut microbiota immunomodulatory actions in bone marrow and liver have catabolic effects on skeletal homeostasis in health. Sci Rep. 2017;7(1):5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sanz Y, Santacruz A, Gauffin P. Gut microbiota in obesity and metabolic disorders. Proc Nutr Soc. 2010;69(3):434–441. [DOI] [PubMed] [Google Scholar]
- 12. Blanton LV, Barratt MJ, Charbonneau MR, Ahmed T, Gordon JI. Childhood undernutrition, the gut microbiota, and microbiota-directed therapeutics. Science. 2016;352(6293):1533. [DOI] [PubMed] [Google Scholar]
- 13. Chen J, Toyomasu Y, Hayashi Y, et al. Altered gut microbiota in female mice with persistent low body weights following removal of post-weaning chronic dietary restriction. Genome Med. 2016;8(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Subramanian S, Huq S, Yatsunenko T, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510(7505):417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tidjani Alou M, Million M, Traore SI, et al. Gut bacteria missing in severe acute malnutrition, can we identify potential probiotics by culturomics? Front Microbiol. 2017;8:899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med. 2018;24(4):392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lewis JD, Chen EZ, Baldassano RN, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe. 2015;18(4):489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schwarzer M, Makki K, Storelli G, et al. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science. 2016;351(6275):854–857. [DOI] [PubMed] [Google Scholar]
- 19. Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 2011;14(3):403–414. [DOI] [PubMed] [Google Scholar]
- 20. Yan J, Herzog JW, Tsang K, et al. Gut microbiota induce IGF-1 and promote bone formation and growth. Proc Natl Acad Sci U S A. 2016;113(47):E7554–E7563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yan J, Takakura A, Zandi-Nejad K, Charles JF. Mechanisms of gut microbiota-mediated bone remodeling. Gut Microbes. 2017;0(0):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shin SC, Kim SH, You H, et al. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334(6056):670–674. [DOI] [PubMed] [Google Scholar]
- 23. Weger BD, Gobet C, Yeung J, et al. The mouse microbiome is required for sex-specific diurnal rhythms of gene expression and metabolism. Cell Metab. 2019;29(2):362–382.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wiesenborn DS, Galvez EJC, Spinel L, et al. The role of Ames dwarfism and calorie restriction on gut microbiota. J Gerontol A Biol Sci Med Sci. 2019; glz236. doi:10.1093/gerona/glz236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Troike KM, Henry BE, Jensen EA, et al. Impact of growth hormone on regulation of adipose tissue. Compr Physiol. 2017;7(3):819–840. [DOI] [PubMed] [Google Scholar]
- 26. Byrne TA, Morrissey TB, Nattakom TV, Ziegler TR, Wilmore DW. Growth hormone, glutamine, and a modified diet enhance nutrient absorption in patients with severe short bowel syndrome. JPEN J Parenter Enteral Nutr. 1995;19(4):296–302. [DOI] [PubMed] [Google Scholar]
- 27. Kaymakci A, Guven S, Ciftci I, et al. Protective effects of growth hormone on bacterial translocation and intestinal damage in rats with partial intestinal obstruction. Bratisl Lek Listy. 2014;115(7):395–399. [DOI] [PubMed] [Google Scholar]
- 28. Young JA, Jensen EA, Stevens A, et al. Characterization of an intestine-specific GH receptor knockout (IntGHRKO) mouse. Growth Horm IGF Res. 2019;46-47:5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yakar S, Isaksson O. Regulation of skeletal growth and mineral acquisition by the GH/IGF-1 axis: Lessons from mouse models. Growth Horm IGF Res. 2016;28:26–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. List EO, Berryman DE, Buchman M, et al. GH knockout mice have increased subcutaneous adipose tissue with decreased fibrosis and enhanced insulin sensitivity. Endocrinology. 2019;160(7):1743–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Berryman DE, List EO, Coschigano KT, Behar K, Kim JK, Kopchick JJ. Comparing adiposity profiles in three mouse models with altered GH signaling. Growth Horm IGF Res. 2004;14(4):309–318. [DOI] [PubMed] [Google Scholar]
- 32. Householder LA, Comisford R, Duran-Ortiz S, et al. Increased fibrosis: a novel means by which GH influences white adipose tissue function. Growth Horm IGF Res. 2018;39:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Palmer AJ, Chung MY, List EO, et al. Age-related changes in body composition of bovine growth hormone transgenic mice. Endocrinology. 2009;150(3):1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. RRID: AB_10775454, http://antibodyregistry.org/AB_10775454.
- 35. Caporaso JG, Lauber CL, Walters WA, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jensen EA, Young JA, Jackson Z, et al. Data from: Growth Hormone Deficiency and Excess alter the Gut Microbiome in Adult Male Mice Dryad Digital Repository. Deposited January 23, 2020. doi:10.5061/dryad.73n5tb2sr [DOI] [PMC free article] [PubMed]
- 37. Tkacz A, Hortala M, Poole PS. Absolute quantitation of microbiota abundance in environmental samples. Microbiome. 2018;6(1):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rocha DJ, Santos CS, Pacheco LG. Bacterial reference genes for gene expression studies by RT-qPCR: survey and analysis. Antonie Van Leeuwenhoek. 2015;108(3):685–693. [DOI] [PubMed] [Google Scholar]
- 39. Kang C, Wang B, Kaliannan K, et al. Gut microbiota mediates the protective effects of dietary capsaicin against chronic low-grade inflammation and associated obesity induced by high-fat diet. mBio. 2017;8(3):e00470-17. doi:10.1128/mBio.00470-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bialkowska AB, Ghaleb AM, Nandan MO, Yang VW. Improved swiss-rolling technique for intestinal tissue preparation for immunohistochemical and immunofluorescent analyses. J Vis Exp. 2016;(113). doi: 10.3791/54161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Team RC. R: A Language and Environment for Statistical Computing, Vol. 2019 Vienna, Austria: R Foundation for Statistical Computing; 2018. [Google Scholar]
- 45. Wickham H, François R, Henry L, Muller K. Data from: dplyr: a grammar of data manipulation CRAN.R:Comprehensive R Archive Network Deposited January 31, 2020. https://CRAN.R-project.org/package=dplyr
- 46. Fox JW, Sanford. An {R} Companion to Applied Regression. 2nd ed. Thousand Oaks, CA: Sage; 2011. [Google Scholar]
- 47. Lakens D. Calculating and reporting effect sizes to facilitate cumulative science: a practical primer for t-tests and ANOVAs. Front Psychol. 2013;4:863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2nd ed. New York, NY: Routledge Academic; 1988. [Google Scholar]
- 49. Korkmaz S, Goksuluk D, Zararsiz G. MVN: An R package for assessing multivariate normality. R J. 2014;6(2):151–162. [Google Scholar]
- 50. Pinheiro J, Douglas B, DebRoy S, Sarkar D; R Core Team Data from: {nlme}: Linear and Nonlinear Mixed Effects Models CRAN.R:Comprehensive R Archive Network Deposited April 7, 2018 https://CRAN.R-project.org/package=nlme.
- 51. Barton K. Data from: MuMIn: Multi-Model Inference CRAN.R:Comprehensive R Archive Network Deposited July 23, 2018 https://CRAN.R-project.org/package=MuMIn.
- 52. Hoffman DJ, Campos-Ponce M, Taddei CR, Doak CM. Microbiome, growth retardation and metabolism: are they related? Ann Hum Biol. 2017;44(3):201–207. [DOI] [PubMed] [Google Scholar]
- 53. Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rieder F. The gut microbiome in intestinal fibrosis: environmental protector or provocateur? Sci Transl Med. 2013;5(190): 190ps10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hata T, Miyata N, Takakura S, et al. The gut microbiome derived from anorexia nervosa patients impairs weight gain and behavioral performance in female mice. Endocrinology. 2019;160(10):2441–2452. [DOI] [PubMed] [Google Scholar]
- 56. Indiani CMDSP, Rizzardi KF, Castelo PM, Ferraz LFC, Darrieux M, Parisotto TM. Childhood obesity and firmicutes/bacteroidetes ratio in the gut microbiota: a systematic review. Child Obes. 2018;14(8):501–509. [DOI] [PubMed] [Google Scholar]
- 57. Luo D, Chen K, Li J, et al. Gut microbiota combined with metabolomics reveals the metabolic profile of the normal aging process and the anti-aging effect of FuFang Zhenshu TiaoZhi(FTZ) in mice. Biomed Pharmacother. 2020;121:109550. [DOI] [PubMed] [Google Scholar]
- 58. Forbes JD, Azad MB, Vehling L, et al. ; Canadian Healthy Infant Longitudinal Development (CHILD) Study Investigators Association of exposure to formula in the hospital and subsequent infant feeding practices with gut microbiota and risk of overweight in the first year of life. JAMA Pediatr. 2018;172(7):e181161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Baumann-Dudenhoeffer AM, D’Souza AW, Tarr PI, Warner BB, Dantas G. Infant diet and maternal gestational weight gain predict early metabolic maturation of gut microbiomes. Nat Med. 2018;24(12):1822–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stewart CJ, Ajami NJ, O’Brien JL, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562(7728):583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vatanen T, Franzosa EA, Schwager R, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562(7728):589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhao L, Zhang F, Ding X, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359(6380):1151–1156. [DOI] [PubMed] [Google Scholar]
- 63. Wilmanski T, Rappaport N, Earls JC, et al. Blood metabolome predicts gut microbiome α-diversity in humans. Nat Biotechnol. 2019;37(10):1217–1228. [DOI] [PubMed] [Google Scholar]
- 64. Kong F, Hua Y, Zeng B, Ning R, Li Y, Zhao J. Gut microbiota signatures of longevity. Curr Biol. 2016;26(18):R832–R833. [DOI] [PubMed] [Google Scholar]
- 65. Biagi E, Rampelli S, Turroni S, Quercia S, Candela M, Brigidi P. The gut microbiota of centenarians: signatures of longevity in the gut microbiota profile. Mech Ageing Dev. 2017;165(Pt B):180–184. [DOI] [PubMed] [Google Scholar]
- 66. Junnila RK, List EO, Berryman DE, Murrey JW, Kopchick JJ. The GH/IGF-1 axis in ageing and longevity. Nat Rev Endocrinol. 2013;9(6):366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pendergrass WR, Li Y, Jiang D, Wolf NS. Decrease in cellular replicative potential in “giant” mice transfected with the bovine growth hormone gene correlates to shortened life span. J Cell Physiol. 1993;156(1):96–103. [DOI] [PubMed] [Google Scholar]
- 68. Rastelli M, Cani PD, Knauf C. The gut microbiome influences host endocrine functions. Endocr Rev. 2019;40(5):1271–1284. [DOI] [PubMed] [Google Scholar]
- 69. Li J, Jia H, Cai X, et al. ; MetaHIT Consortium; MetaHIT Consortium An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32(8):834–841. [DOI] [PubMed] [Google Scholar]
- 70. Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7(3):189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Oliphant K, Allen-Vercoe E. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome. 2019;7(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang JF, Fu SP, Li SN, et al. Short-chain fatty acids inhibit growth hormone and prolactin gene transcription via cAMP/PKA/CREB signaling pathway in dairy cow anterior pituitary cells. Int J Mol Sci. 2013;14(11):21474–21488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ohneda K, Ulshen MH, Fuller CR, D’Ercole AJ, Lund PK. Enhanced growth of small bowel in transgenic mice expressing human insulin-like growth factor I. Gastroenterology. 1997;112(2):444–454. [DOI] [PubMed] [Google Scholar]
- 74. Williams KL, Fuller CR, Dieleman LA, et al. Enhanced survival and mucosal repair after dextran sodium sulfate-induced colitis in transgenic mice that overexpress growth hormone. Gastroenterology. 2001;120(4):925–937. [DOI] [PubMed] [Google Scholar]
- 75. Chen T, Zheng F, Tao J, et al. Insulin-like growth factor-1 contributes to mucosal repair by β-arrestin2-mediated extracellular signal-related kinase signaling in experimental colitis. Am J Pathol. 2015;185(9):2441–2453. [DOI] [PubMed] [Google Scholar]
- 76. Michaylira CZ, Ramocki NM, Simmons JG, et al. Haplotype insufficiency for suppressor of cytokine signaling-2 enhances intestinal growth and promotes polyp formation in growth hormone-transgenic mice. Endocrinology. 2006;147(4):1632–1641. [DOI] [PubMed] [Google Scholar]
- 77. Chen Y, Tseng SH, Yao CL, Li C, Tsai YH. Distinct effects of growth hormone and glutamine on activation of intestinal stem cells. JPEN J Parenter Enteral Nutr. 2018;42(3):642–651. [DOI] [PubMed] [Google Scholar]
- 78. Christensen H, Flyvbjerg A, Orskov H, Laurberg S. Effect of growth hormone on the inflammatory activity of experimental colitis in rats. Scand J Gastroenterol. 1993;28(6):503–511. [DOI] [PubMed] [Google Scholar]
- 79. Scopa CD, Koureleas S, Tsamandas AC, et al. Beneficial effects of growth hormone and insulin-like growth factor I on intestinal bacterial translocation, endotoxemia, and apoptosis in experimentally jaundiced rats. J Am Coll Surg. 2000;190(4):423–431. [DOI] [PubMed] [Google Scholar]
- 80. Pereira-Fantini PM, Thomas SL, Taylor RG, et al. Colostrum supplementation restores insulin-like growth factor -1 levels and alters muscle morphology following massive small bowel resection. JPEN J Parenter Enteral Nutr. 2008;32(3):266–275. [DOI] [PubMed] [Google Scholar]
- 81. de Lima MB, Gama LA, Hauschildt AT, Dall’Agnol DJR, Corá LA, Americo MF. Gastrointestinal motility, mucosal mast cell, and intestinal histology in rats: effect of prednisone. Biomed Res Int. 2017;2017:4637621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Elderman M, Sovran B, Hugenholtz F, et al. The effect of age on the intestinal mucus thickness, microbiota composition and immunity in relation to sex in mice. Plos One. 2017;12(9): e0184274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hindmarsh PC, Dennison E, Pincus SM, et al. A sexually dimorphic pattern of growth hormone secretion in the elderly. J Clin Endocrinol Metab. 1999;84(8):2679–2685. [DOI] [PubMed] [Google Scholar]
- 84. Jessup SK, Dimaraki EV, Symons KV, Barkan AL. Sexual dimorphism of growth hormone (GH) regulation in humans: endogenous GH-releasing hormone maintains basal GH in women but not in men. J Clin Endocrinol Metab. 2003;88(10):4776–4780. [DOI] [PubMed] [Google Scholar]