Abstract
We report a label-free nanopore sensor for the detection of Zn2+ ion. By taking advantage of the cleavage of a substrate peptide by zinc dependent enzyme, nanomolar concentrations of Zn2+ ion could be detected within minutes. Furthermore, structurally similar transition metals such as Ni2+, Co2+, Hg2+, Cu2+, and Cd2+ did not interfere with its detection. The enzymatic reaction-based nanopore sensing strategy developed in this work may find potential applications in environmental monitoring and medical diagnosis.
Introduction
Anthropological activities have led to ubiquitous contamination by metals, some of which are toxic and some of which are essential to life. Although trace metal ions play important roles in many biological processes,1 long-term exposure to high levels of heavy metals could cause various serious health issues, including cancer and neurodegenerative diseases.2 For example, mercury bioaccumulation may induce the production of free radicals and oxidative stress in nervous system, causing DNA instability and thus leading to chronic degenerative dementias including Alzheimer’s and Parkinson’s diseases,3,4 whereas overload or deficiency of copper is associated with Wilson disease (WD) and Menkes disease (MD),5 respectively. Therefore, the ability to sensitively and selectively detect metal ions is an important aspect of environmental, medical, and biochemical research. Thus far, a variety of approaches have been developed for metal ion detection, including traditional methods such as spectroscopy, electrochemistry, and chromatography, as well as nanotechniques including nanopore sensor.6–11 Nanopore sensing can detect analytes at the single-molecule level, where the interactions between analyte molecules and a single nanoscale-sized pore are detected as transient ionic current modulations.12–17 In addition to its potential utility as the fourth generation DNA sequencer,18–20 nanopore technology has been used to investigate various other applications, especially biosensing.8,21
In terms of metal ion detection, two major strategies have been developed thus far. One relies on construction of a binding site in the nanopore interior,22,23 while the other uses peptides and DNA molecules as an external selective molecular probe to detect analytes based on chelation / coordination interaction.24–27 Although highly sensitive and selective detection of target analytes could be achieved by introducing bind sites in the nanopore, one major disadvantage of this sensing strategy is time-consuming and inconvenient because different nanopores having different binding sites need to be produced in order to detect different analytes. To this end, the molecular probe nanopore sensing strategy is flexible (only needs to use a different probe molecule for a different target analyte) and hence more advantageous.
Herein, we demonstrate a new enzymatic reaction-based nanopore sensing method for sensitive and selective detection of metal ions, which could be used as an alternative to the molecule probe strategy in the situation that a sensitive and selective probe molecule for the target analyte is difficult to obtain. Proteases, also known as peptidases, are enzymes that catalyze the hydrolysis of the peptide bonds that join amino acids within proteins. They occur naturally in all living organisms, and play key roles in diverse biological processes, including cell growth and migration, metastasis, cell death, tissue regeneration, angiogenesis, neurogenesis, immune defense, and so on.28 For some proteases, the presence of metal ions as cofactors is crucial for their activity.
Experimental
Materials and Reagents
ADAM17 was purchased from R&D Systems (Minneapolis, MN). Peptides LAQAVRSSSARLVFF, LAQAV, and RSSSARLVFF were synthesized by WatsonBio (Houston, TX). All the other chemicals used in this study, including Zn(NO3)2 (99.999%), Ni(NO3)2 (99.999%), Co(NO3)2 (99.999%), Cu(NO3)2 (99.999%), Hg(NO3)2 (99.999%), Cd(NO3)2 (99.999%), NaCl (99.999%), Trizma base (BioXtra grade, ≥99.9%), and HCl (ACS reagent, ≤ 1 ppm heavy metals), were obtained from Sigma-Aldrich (St. Louis, MO). Stock solutions of the peptides (10 mM each) and the ADAM17 stock solution (100 μg/mL) were prepared in nuclease-free water. Peptides and ADAM17 were kept at −20 °C, and −80 °C, respectively, before and immediately after use.
Bilayer Experiment and Electrical Recording
The detailed procedure for the formation of lipid bilayer (from 1,2-diphytanoylphosphatidylcholine, Avanti Polar Lipids, Alabaster, AL) and single channel recordings has been provided in our previous work.29 Briefly, the experiments were performed with an Axopatch 200B amplifier (Molecular Devices, Sunnyvale, CA) in a two-compartment nanopore sensing chamber under asymmetric electrolyte conditions, where the trans compartment was filled with an electrolyte buffer solution containing 1 M NaCl and 10 mM tris (pH 7.5), while the cis compartment was filled with 3 M NaCl and 10 mM tris (pH 7.5). The wild-type α-hemolysin protein was utilized as the nanopore sensing element, and was added to the cis compartment of the sensing chamber. The experimental temperature was 25 ± 1 °C, and the applied potential bias was +120 mV. Data analysis was carried out using Clampfit 10.5 software (Molecular Device).
Enzyme Digestion
Unless otherwise noted, enzymatic reactions were performed by incubating 5 μM peptide substrate, 0.25 μM ADAM17, and Zn2+ (ranging from 0 to 1 μM) at 37 °C for 120 minutes.
Preparation of Simulated Water Samples
Simulated Zn2+-contaminated water samples were prepared by spiking 4 μL of Zn(NO3)2 solutions (with the final Zn2+ concentrations ranging from 0.25 to 1 μM) into 40 μL of tap water (obtained from our life science building), bottled spring water (Ice Mountain brand), or lake water (from Lake Michigan) without further treatment.
Results and Discussions
Metal ion sensing strategy
The principle for the enzymatic reaction-based nanopore detection of metal ions is shown in Figure 1. In the absence of the target metal ion, the protease is not able to cleave the substrate peptide so that the current modulations are caused only by the substrate. However, in the presence of the target analyte, the protease is activated, and two peptide fragments will be produced due to the degradation of the substrate by the protease. Hence, new type of blockage events having smaller residence time (τoff) and/or amplitude values than those of the substrate peptide can be observed. Accordingly, the frequency (or number of counts) of these new events could be used to quantitate the concentration of metal ions in the electrolyte solution.
Fig. 1.
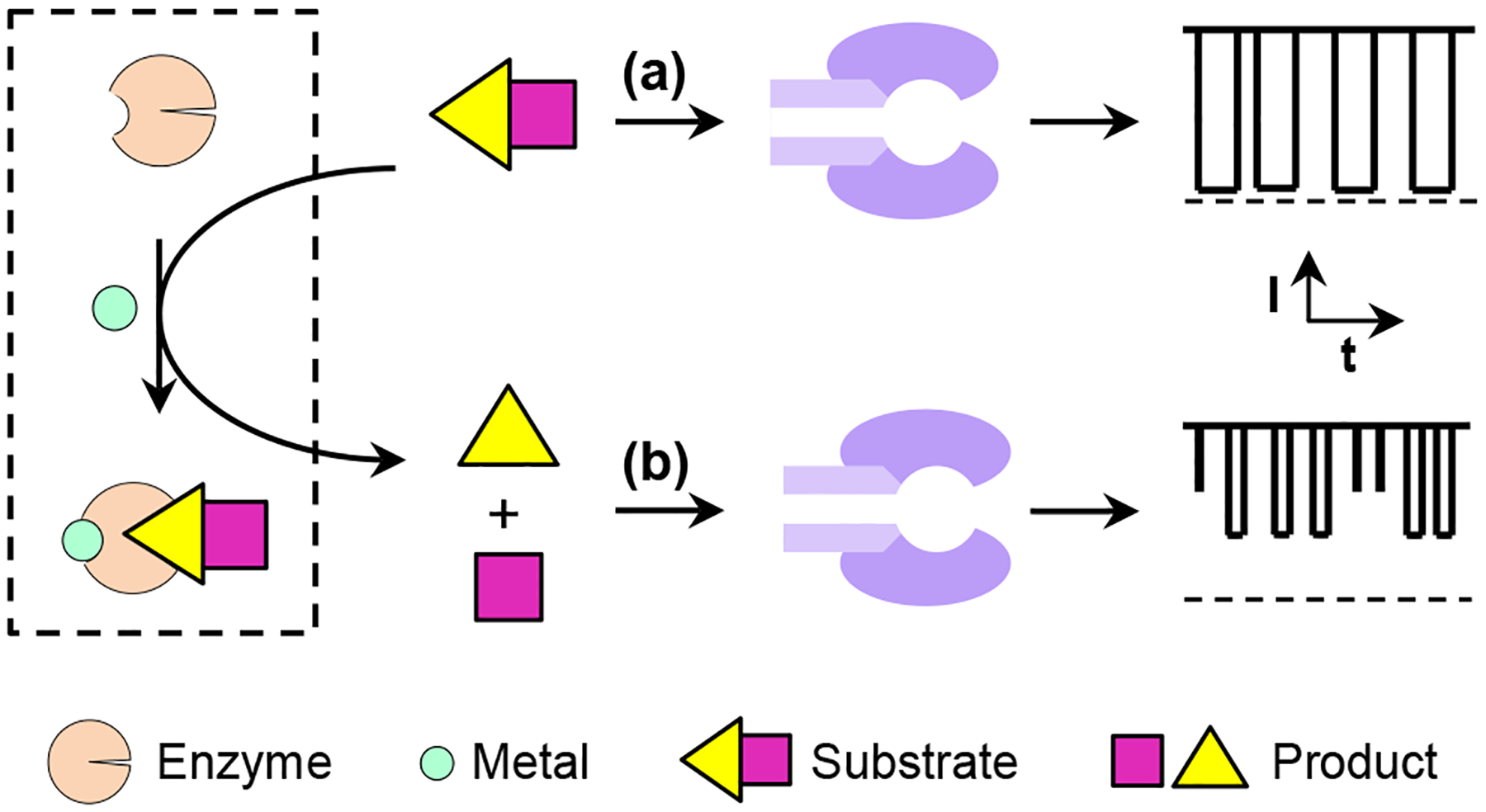
Schematic representation of the enzymatic reaction-based nanopore detection of metal ions: (a) without the target metal ions, the enzyme is inactive so that the current modulations are caused only by the peptide substrate; and (b) with the analyte in the solution, the enzyme is activated and the cleavage of the peptide substrate by its corresponding enzyme produces new types of blockage events with different residence time and/or amplitude values in the nanopore.
Detection of Zn2+ ions
To demonstrate this concept, ADAM17-based Zn2+ ion detection was utilized as a model system. ADAM17 (short for a disintegrin and metalloproteinase 17), also known as a tumor necrosis factor-α-converting enzyme, is a member of ADAM family. ADAMs are Zn2+ dependent endopeptidases that proteolytically cleave a wide variety of membrane-bound proteins, and are involved in cell-cell, cell-matrix interface-related processes.30,31 The experiments were initially carried out at +120 mV under an asymmetric electrolyte condition of 3 M/1 M NaCl (cis/trans) with the α-hemolysin protein pore and in the presence of ADAM17 and its well-known substrate, peptide LAQAVRSSSARLVFF.31,32 The reason why an asymmetric electrolyte condition was used in the nanopore sensing experiment instead of the commonly used symmetric electrolyte solutions was because the previous studies have shown that a better sensor resolution and sensitivity could be achieved under a salt gradient.25,31 The experimental results were summarized in Figure 2. As we expected, if there was no presence of Zn2+ in the solution, the substrate peptide produced only one major type of events (mean residence time: 3.00 ± 0.15 ms; event residual current: 27.9 ± 0.1% of full channel block). In contrast, after addition of Zn2+ ions to the peptide solution, a new type of current modulation events was observed. These new events had a shorter residence time (0.78 ± 0.15 ms) and a larger event residual current (42.4 ± 0.1% of full channel block) than those of the substrate peptide, which should be attributed to the RSSSARLVFF peptide fragment. Its identity was confirmed by direct measurement of its current blockages using the peptide RSSSARLVFF standard (event mean residence time: 0.65 ± 0.10 ms; block residual current: 43.5 ± 0.5% of full channel block, Electronic Supplementary Information, Figure S1). In theory, cleavage of the substrate peptide by ADAM17 would produce two fragments (LAQAV and RSSSARLVFF). However, we did not observe any significant events for LAQAV (Electronic Supplementary Information, Figure S2). One likely interpretation is that the interaction between the short-sequence peptide and the α-hemolysin pore was so weak that our nanopore sensing system (with ~200 μs resolution) could not capture its rapid translocation in the nanopore. Furthermore, we noticed that, with an increase in the concentration of the added zinc ions, the frequency of the new events increased, while that of the substrate events decreased, a clear indication of the feasibility of utilizing these events to quantify zinc ions. As an added control, the interaction between the peptide substrate LAQAVRSSSARLVFF and ADAM17 was further investigated in the presence of a mixture (1:1 molar ratio) of Zn2+ and EDTA (a well-known metal ion chelating agent). As a result, only one major type of (i.e., the peptide substrate) events were identified (Electronic Supplementary Information, Figure S3), indicating the necessity of the presence of free metal ions in the solution for ADAM17 to be activated.
Fig. 2.
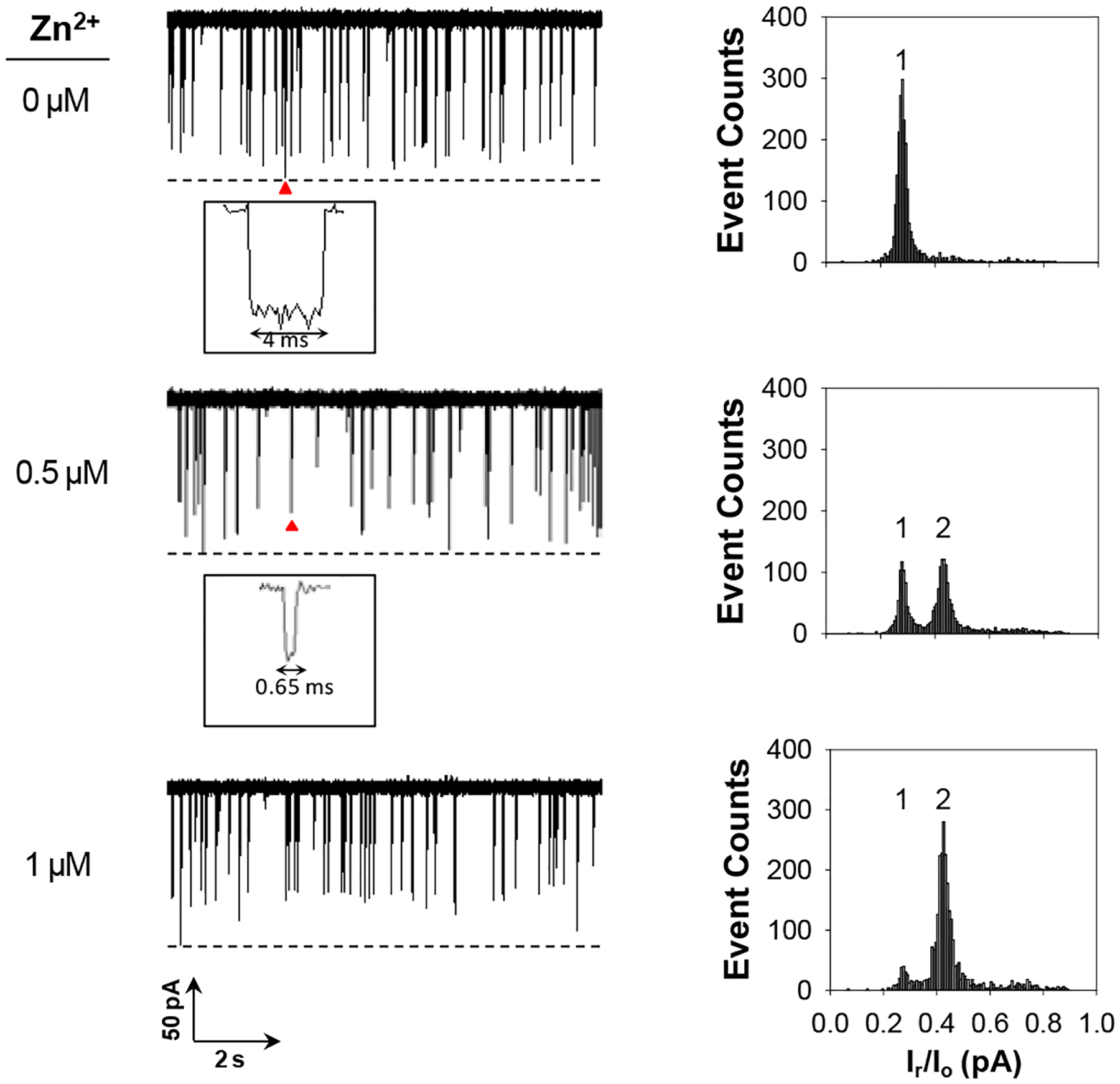
Nanopore detection of Zn2+ ions based on ADAM17-peptide LAQAVRSSSARLVFF cleavage reaction. (Left) Typical single-channel recording trace segments. Dashed lines represent the levels of zero current. Insets show enlarged images of arrow-marked events; (Right) Corresponding event amplitude histograms. Events attributed to peptides LAQAVRSSSARLVFF and RSSSARLVFF were marked as 1 and 2, respectively. The experiments were performed at +120 mV and with 5 μM LAQAVRSSSARLVFF and 0.25 μg/mL ADAM17 added into the trans compartment of the nanopore sensing chamber.
Effect of incubation time on Zn2+ ion detection
It should be noted that, in a cofactor-enzyme system, a variety of factors will affect the substrate cleavage by the enzyme. These include solution pH, temperature, and incubation time, as well as the concentrations of the substrate, enzyme, and cofactor. Clearly, under experimental conditions of constant solution pH and temperature, as well as given concentrations of substrate, enzyme and cofactor, the enzymatic reaction are only depends on the incubation time. To determine an appropriate incubation time for Zn2+ detection, a series of experiments was carried out, where digestion of the peptide substrate by ADAM17 was kept at 37 °C for a period from 15 min to 120 min. The results were summarized in Figure 3. We found that the percentage of the cleavage product events out of the total events increased rapidly with an increase in the incubation time until 60 min, after which the percentage increased slowly. To obtain larger sensor sensitivity, 120 min incubation time was used for subsequent experiments although a more rapid detection could be achieved with 60 min incubation.
Fig. 3.
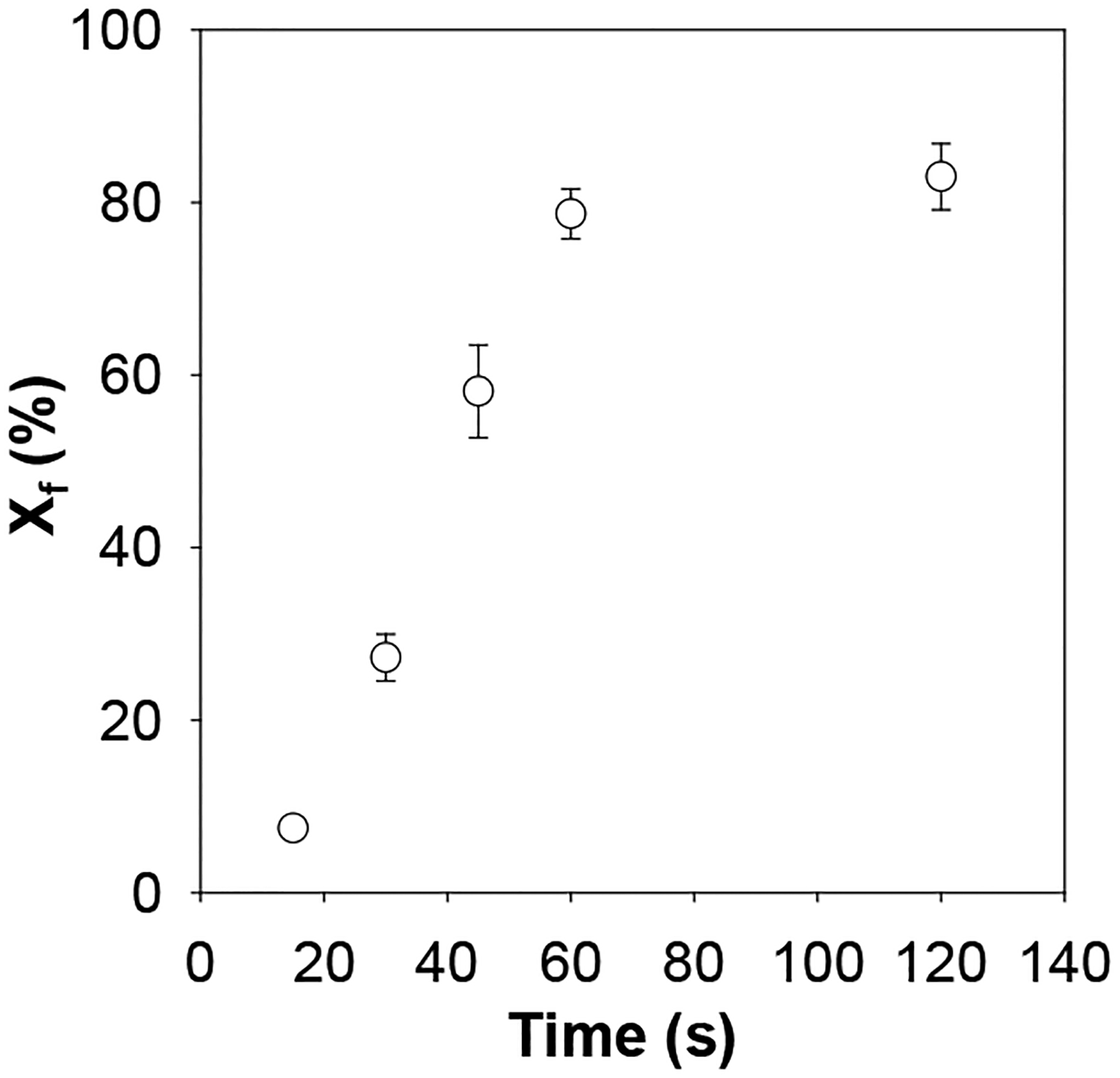
The effect of incubation time on the percentage of the cleavage product events out of the total events. The experiments were performed at +120 mV in a salt gradient of 3 M/1 M NaCl (cis/trans) and with 5 μM peptide substrate, 0.25 μg/mL ADAM17 and 1 μM Zn2+ added into the trans compartment of the nanopore sensing chamber.
Sensor sensitivity and selectivity
To determine the sensitivity of our nanopore sensor, the dose response curve for Zn2+ ion detection was constructed by monitoring the cleavage of the peptide substrate by ADAM17 in the presence of zinc ions at various concentrations, ranging from 0 to 1 μM. As shown in Figure 4a, the percentage of the cleavage product events out of the total events linearly increased with the zinc concentration. The limit of detection (LOD) of the nanopore sensor in a 10 min electrical recording was 100 nM, where LOD was defined as the concentration of Zn2+ corresponding to three times the standard deviation of the blank signal. Note that, although our Zn2+ sensor was not optimized, its current detection limit is comparable to those of most other Zn2+ detection methods33,34 reported thus far, which is far below the EPA standard concentration (76 μM) for zinc in drinking water. In addition, the selectivity of the nanopore zinc sensor was investigated. Divalent metal ions such as Ni2+, Co2+, Hg2+, Cu2+, and Cd2+ were selected as potential interfering species since they have similar chemical properties to Zn2+. As shown in Figure 4b, our single-channel recording experiments showed that the mixtures of zinc ion and these metals produced similar results to that of the single zinc ion standard, suggesting that other metal ions would not interfere with Zn2+ ion detection significantly.
Fig. 4.
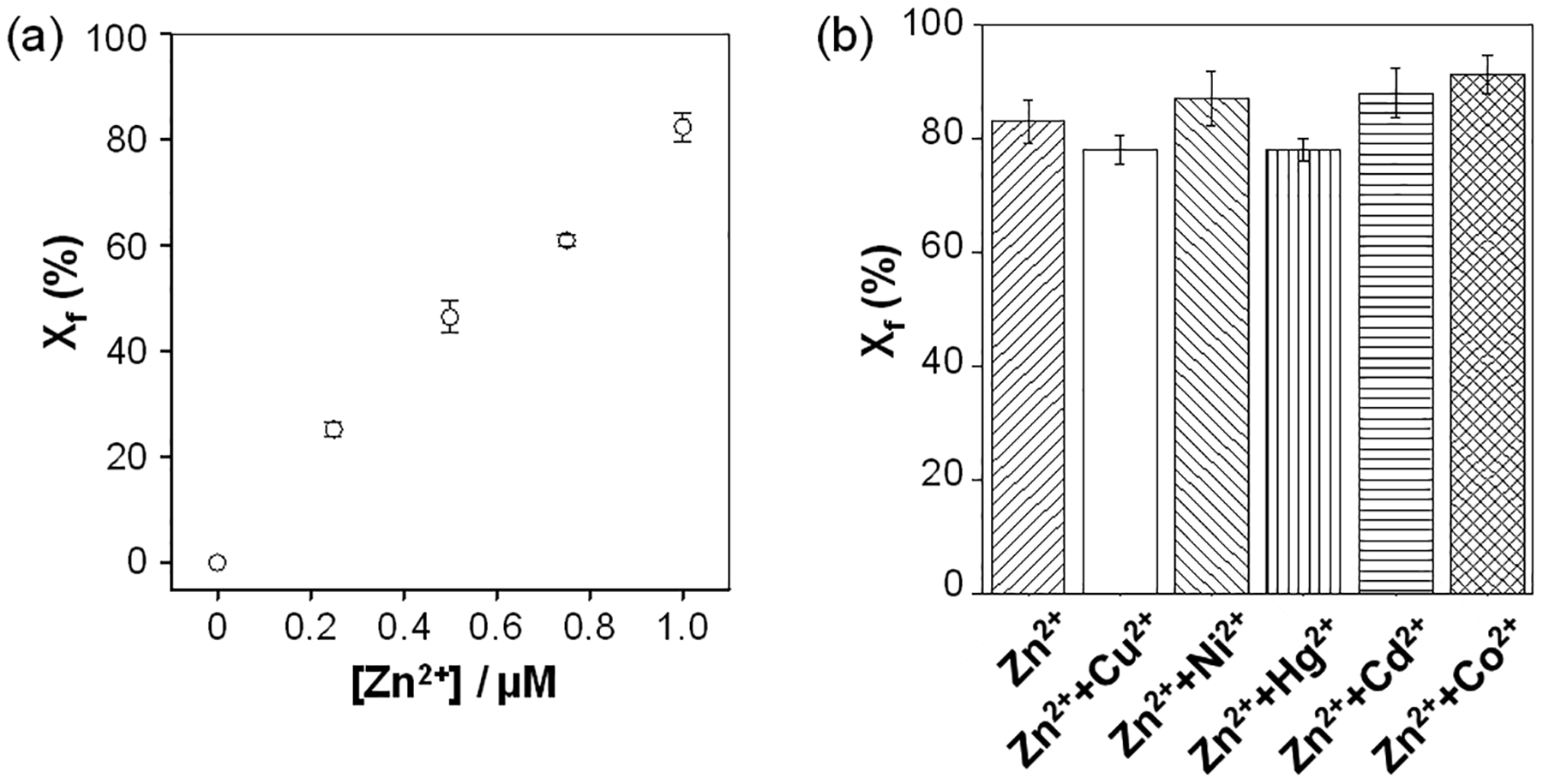
(a) Plot of the percentage of the cleavage product events out of the total events versus Zn2+ ion concentration. (b) Selectivity study. The experiments were performed at +120 mV in a salt gradient of 3 M/1 M NaCl (cis/trans) and with 5 μM peptide substrate and 0.25 μg/mL ADAM17 added into the trans compartment of the nanopore sensing chamber. The concentrations of metal ions used in Figure 4b were 1 μM each.
Simulated water sample analysis
To show our nanopore sensor has potential application for environmental sample analysis, five simulated Zn2+-contaminated water samples were analyzed. The five water samples were prepared by spiking Zn2+ ions (with concentrations ranging from 0.25 to 1 μM) into the tap water, bottled spring water, and lake water, respectively. Note that, these samples could be grouped into two categories. One category had the same matrix components but with different concentrations of Zn2+, while the other contained the same concentration of Zn2+ ions but having different matrices. The results were summarized in Table 1. From the table, we could see that, the recoveries (ranging from 99% to 116%) of Zn2+ from the water samples determined by use of the nanopore sensor were satisfactory, indicating that our developed Zn2+ sensing system has a great potential for application in the analysis of environmental samples.
Table 1.
Recovery of Zn2+ ions from water samples by use of the nanopore stochastic sensing method. Each experimental value represents the mean of three replicate analyses ± one standard deviation. The experiments were performed at +120 mV in the presence of 5 μM LAQAVRSSSARLVFF and 0.25 μg/mL ADAM17.
| Sample type | Theoretical value (μM) | Experimental value (μM) |
|---|---|---|
| Tap | 1 | 0.99 ± 0.04 |
| Spring | 1 | 0.99 ± 0.03 |
| Lake | 1 | 1.04 ± 0.01 |
| Lake | 0.25 | 0.28 ± 0.01 |
| Lake | 0.5 | 0.58 ± 0.05 |
Conclusions
In summary, by taking advantage of enzymatic reactions, we developed a selective and sensitive nanopore sensing strategy for metal ions. As discussed in the introduction section, thus far, two major nanopore strategies have been developed for metal ion detection. One relies on construction of a binding site in the nanopore interior, while the other uses peptides and DNA molecules as an external selective molecular probe. It should be noted that, in the well-established molecular probe approach, analyte detection was achieved based on chelation / coordination interaction, in which metal ions and probes formed stable metal-biomolecule chelates. The difference between the biomolecules and the produced metal-biomolecule chelates in terms of the structure, conformation, and/or net charge leads to the detection of the metal ion by the nanopore sensor. In contrast, in the present enzymatic reaction-based sensing strategy developed in this work, metal ions didn’t interact with the biomolecule, but instead activated the enzyme to speed up the cleavage of the biomolecule substrate. Although the reported work was focused on zinc ion detection by ADAM17, the strategy we developed is applicable to a variety of other cofactor-enzyme and coenzyme-enzyme systems. As a noted example, type II DNA topoisomerases are enzymes that remove knots and tangles from the genome.35 All type II topoisomerases require divalent metal ions to cleave and ligate DNA. Using an appropriately designed DNA substrate instead of the peptide substrate used in this work, we can study the effects of metal ions on the cleavage of DNA by topoisomerases and develop highly sensitive and selective nanopore sensors for metal ions. Given the advantages of label-free detection, high sensitivity and selectivity, our developed enzymatic reaction-based nanopore metal ion sensing strategy should find useful application in many fields, especially environmental monitoring and medical diagnosis.
Supplementary Material
Acknowledgements
We thank the National Institutes of Health (2R15GM110632-02) and National Science Foundation (1708596) for supporting this work.
Footnotes
Conflicts of interest
“There are no conflicts to declare”.
Electronic Supplementary Information (ESI) available: Residence time and amplitude histograms of peptide RSSSARLVFF events; typical single-channel recording trace segment of peptide fragment LAQAV.
Notes and references
- 1.Holm RH, Kennepohl P and Solomon EI, Chem. Rev, 1996, 96, 2239–2314. [DOI] [PubMed] [Google Scholar]
- 2.Jomova K and Valko M, Toxicology, 2011, 283, 65–87. [DOI] [PubMed] [Google Scholar]
- 3.Ronnback L and Hansson E, Occup. Environ. Med, 1992, 49, 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palmiter RD, Proc. Natl. Acad. Sci, 1998, 95, 8428–8430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tisato F, Marzano C, Porchia M, Pellei M and Santini C, Med. Res. Rev, 2010, 30, 708–49. [DOI] [PubMed] [Google Scholar]
- 6.Bond AM and Wallace GG, Anal. Chem, 1984, 56, 2085–2090. [DOI] [PubMed] [Google Scholar]
- 7.Nolan EM and Lippard SJ, J. Am. Chem. Soc, 2003, 125, 14270–14271. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Yao F and Kang X, Anal. Chem, 2017, 89, 7958–7965. [DOI] [PubMed] [Google Scholar]
- 9.Deo S and Godwin HA, J. Am. Chem. Soc, 2000, 122, 174–175. [Google Scholar]
- 10.Gumpu MB, Sethuraman S, Krishnan UM and Rayappan JBB, Sensors Actuators B Chem, 2015, 213, 515–533. [Google Scholar]
- 11.Roozbahani GM, Chen X, Zhang Y, Juarez O, Li D and Guan X, Anal. Chem, 2018, 90, 5938–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang G, Zhao Q, Kang X and Guan X, J. Phys. Chem. B, 2013, 117, 4763–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Z-L, Li M-Y, Liu S-C, Ying Y-L and Long Y-T, Chem. Sci, 2019, 10, 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stefureac RI, Madampage CA, Andrievskaia O and Lee JS, Biochem. Cell Biol, 2010, 88, 347–358. [DOI] [PubMed] [Google Scholar]
- 15.Verschueren DV, Pud S, Shi X, De Angelis L, Kuipers L and Dekker C, ACS Nano, 2019, 13, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao S, Restrepo-Pérez L, Soskine M, Maglia G, Joo C, Dekker C and Aksimentiev A, ACS Nano, 2019, 13, 2398–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meller A, Nivon L, Brandin E, Golovchenko J and Branton D, Proc. Natl. Acad. Sci, 2000, 97, 1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider GF and Dekker C, Nat. Biotechnol, 2012, 30, 326–328. [DOI] [PubMed] [Google Scholar]
- 19.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, V Pershin Y, Ramsey JM, Riehn R, Soni GV, Cossa VT, Wanunu M, Wiggin M and Schloss JA, in Nanoscience and Technology, Co-Published with Macmillan Publishers Ltd, UK, 2009, pp. 261–268. [Google Scholar]
- 20.Howorka S, Cheley S and Bayley H, Nat. Biotechnol, 2001, 19, 636–639. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Chen X, Zhou S, Roozbahani GM, Zhang Y, Wang D and Guan X, Chem. Commun, 2018, 54, 13977–13980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braha O, Gu L-Q, Zhou L, Lu X, Cheley S and Bayley H, Nat. Biotechnol, 2000, 18, 1005–1007. [DOI] [PubMed] [Google Scholar]
- 23.Braha O, Walker B, Cheley S, Kasianowicz JJ, Song L, Gouaux JE and Bayley H, Chem. Biol, 1997, 4, 497–505. [DOI] [PubMed] [Google Scholar]
- 24.Asandei A, Schiopu I, Iftemi S, Mereuta L and Luchian T, Langmuir, 2013, 29, 15634–15642. [DOI] [PubMed] [Google Scholar]
- 25.Roozbahani GM, Chen X, Zhang Y, Xie R, Ma R, Li D, Li H and Guan X, ACS Sensors, 2017, 2, 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stefureac RI and Lee JS, Small, 2008, 4, 1646–1650. [DOI] [PubMed] [Google Scholar]
- 27.Wang G, Wang L, Han Y, Zhou S and Guan X, Acc. Chem. Res, 2013, 46, 2867–2877. [DOI] [PubMed] [Google Scholar]
- 28.Neumann U, Kubota H, Frei K, Ganu V and Leppert D, Anal. Biochem, 2004, 328, 166–173. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Q, Jayawardhana DA and Guan X, Biophys. J, 2008, 94, 1267–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein T and Bischoff R, J. Proteome Res, 2011, 10, 17–33. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, Zhang Y, Mohammadi Roozbahani G and Guan X, ACS Appl. Bio Mater, 2019, 2, 504–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Chen X, Roozbahani GM and Guan X, Analyst, 2019, 144, 1825–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chae JB, Yun D, Kim S, Lee H, Kim M, Lim MH, Kim K-T and Kim C, Spectrochim. Acta Part A Mol. Biomol. Spectrosc, 2019, 219, 74–82. [DOI] [PubMed] [Google Scholar]
- 34.Du K, Liu J, Shen R and Zhang P, Luminescence, 2019, 34, 407–414. [DOI] [PubMed] [Google Scholar]
- 35.Wang JC, Nat. Rev. Mol. Cell Biol, 2002, 3, 430–440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


