Summary
The yeast Hsp70 chaperone Ssb interacts with ribosomes and nascent polypeptides to assist protein folding. To reveal its working principle, we determined the nascent chain-binding pattern of Ssb at near-residue resolution by in vivo selective ribosome profiling. Ssb associates broadly with cytosolic, nuclear and hitherto unknown substrate classes of mitochondrial and ER nascent proteins, supporting its general chaperone function. Ssb engages most substrates by multiple binding-release cycles to a degenerate sequence enriched in positively charged and aromatic amino acids. Timely association with this motif upon emergence at the ribosomal tunnel exit requires ribosome-associated complex RAC, but not nascent polypeptide-associated complex NAC. Ribosome footprint densities along orfs reveal faster translation at times of Ssb binding, mainly imposed by biases in mRNA secondary structure, codon usage and Ssb action. Ssb thus employs substrate-tailored dynamic nascent chain associations to coordinate co-translational protein folding, facilitate accelerated translation and support membrane targeting of organellar proteins.
Introduction
Timely production of correctly folded and localized proteins is of fundamental importance for all organisms. Cells have evolved a multilayered machinery to secure and regulate all critical protein maturation steps, including N-terminal processing, membrane targeting and native folding (Kramer et al., 2009; Pechmann et al., 2013). This machinery is coordinated by the translating ribosome which serves as docking platform for the transient association of enzymes, targeting factors and chaperones, thereby allowing their privileged and ordered interaction with emerging polypeptide chains. Ribosomes furthermore exhibit local translation speed variations that facilitates co-translational folding and membrane targeting (Clarke and Clark, 2008; Gloge et al., 2014; Zhang and Ignatova, 2011). mRNA sequences imposing translational slow-down often occur at sites coinciding with the exposure of domain boundaries to facilitate membrane targeting and domain-wise folding (Pechmann and Frydman, 2013) thereby reducing unwanted inter-domain interactions and misfolding. How chaperones prevent non-productive intermolecular interactions and promote on-pathway folding trajectories of nascent proteins is unclear.
In eukaryotic cells the network of chaperones linked to protein synthesis (CLIPS) that engage nascent chains (Albanese et al., 2006) include ATP independent chaperones and members of the ATP dependent Hsp60, Hsp70 and Hsp90 chaperone families. In S. cerevisiae, the Hsp70 chaperone Ssb, the ribosome-associated complex (RAC) composed of the non-canonical Hsp70 Ssz and the J-protein Zuo1, and the heterodimeric nascent polypeptide-associated complex (NAC) directly bind ribosomes to promote early folding (Gautschi et al., 2001; Nelson et al., 1992; Yan et al., 1998). NAC's substrate pool is large (del Alamo et al., 2011), supporting its proposed roles in protein folding (Koplin et al., 2010), targeting of mitochondrial proteins (George et al., 1998) and regulation of SRP specificity (Gamerdinger et al., 2015; Wiedmann et al., 1994).
Ssb employs two positively charged regions in its substrate-binding domain to bind sites on the 60S ribosomal subunit (Gumiero et al., 2016; Hanebuth et al., 2016). Yeast encodes two isoforms Ssb1 and Ssb2, which differ by only four amino acids (aa). Ssb and RAC form a tripartite chaperone system (Gautschi et al., 2002) in which RAC stimulates Ssb's ATPase activity to facilitate stable substrate binding by Ssb (Huang et al., 2005; Willmund et al., 2013). Substrate release is stimulated by the nucleotide exchange factors (NEF) Sse1, Fes1 and Snl1 (Raviol et al., 2006; Shaner et al., 2005). Deletion of SSB1 or SSB2 is phenotypically silent, while deletion of both SSBs, or one RAC component, causes slow growth, sensitivity against cold, high salt and aminoglycosides and defects in ribosome biogenesis and protein folding (Koplin et al., 2010; Nelson et al., 1992). In accordance with these pleiotropic phenotypes Ssb is involved in many processes, including translation termination (Chiabudini et al., 2014), ribosome biogenesis (Albanese et al., 2010; Koplin et al., 2010), regulation of metabolism (von Plehwe et al., 2009) and prion propagation (Chernoff and Kiktev, 2016). Ssb binds many nascent chains, with preference for large proteins with complex folds and enriched in hydrophobic, aggregation-prone and intrinsically disordered regions (Willmund et al., 2013). Recent data suggest an additional role of RAC in coordinating mRNA translation with protein folding (Lee et al., 2016; Leidig et al., 2013; Zhang et al., 2014).
Many fundamental questions concerning the mechanistic principles of chaperone-assisted co-translational folding in eukaryotes remain unanswered: How and when do chaperones recognize nascent substrates and how does this affect the folding process? What rules govern the interplay of the factors engaging nascent chains? Do translation speed variations coordinate chaperone function? Here we reveal molecular features of chaperone action during translation in eukaryotes by providing proteome-wide Ssb interaction profiles with nascent chains at near-codon resolution. The Ssb interactome is broader than previously thought and includes nascent mitochondrial and ER-translocated proteins. Ssb operates through dynamic, nascent chain-specific cycles of binding to emerging recognition motifs near the peptide tunnel exit. Ssb binding coincides with a speed-up of translation, indicating an unexpected mode of coupling of chaperone recruitment and translation elongation kinetics.
Results
Profiling of Ssb bound ribosomes reveals in vivo interactome
To identify the nascent chain interaction profile of Ssb in vivo we employed selective ribosome profiling (SeRP, (Becker et al., 2013)) which compares ribosome protected mRNA footprints of all translating ribosomes (translatome) with those generated from the subset of Ssb bound ribosomes (Ssb-bound translatome). Forming the ratio of both datasets reveals codons translated when Ssb engages translating ribosomes (Data S1). The already translated codons reflect the sequence of the emerging polypeptide and, considering the ribosomal tunnel, the part of the chain that is available for Ssb interaction (Figure 1A). It should be noted that SeRP data represent averages over populations of ribosome-nascent chain complexes (RNCs) and therefore do not allow distinguishing between single or multiple Ssb binding/release events per binding peak and whether each RNC of a population is bound. The collated interaction profiles of Ssb with all nascent chains or subsets thereof (termed metagene analysis) instead reveal the averaged binding profiles of Ssb as function of nascent chain length.
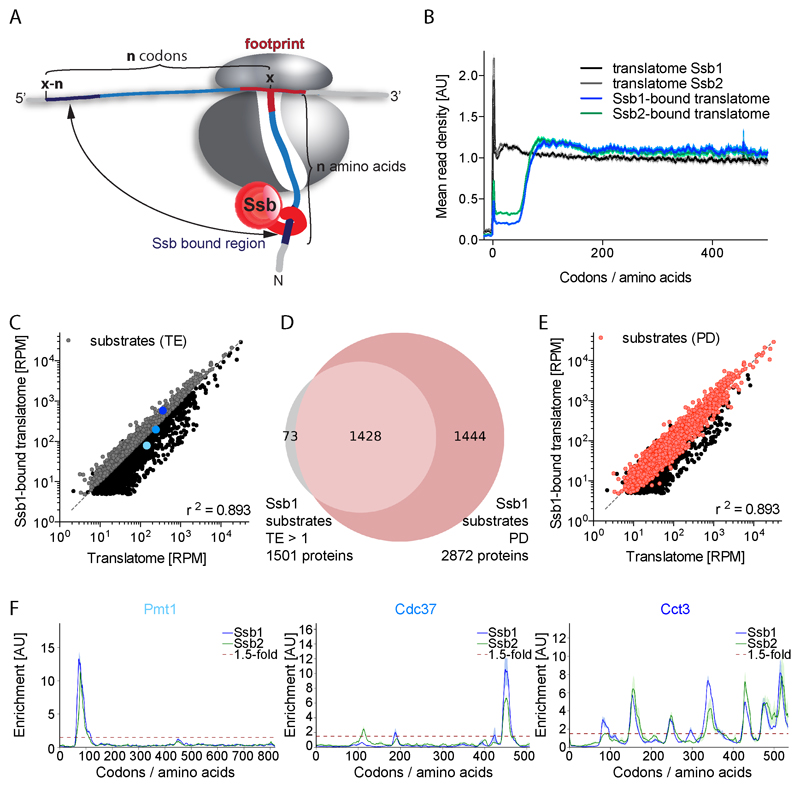
Figure 1. Nascent chain substrates of Ssb1 and Ssb2.
(A) Model illustrating the identification of Ssb binding positions. (B) Metagene profiles of translatomes and Ssb1- and Ssb2-bound translatomes. Shaded areas show the 95% confidence interval (CI). AU: arbitrary units. (C) and (E) Gene expression levels of translatome and Ssb1-bound translatome in reads per million (RPM). (C) Ssb1 substrates (grey) identified by total enrichment (TE >1). (E) Ssb1 substrates (red) identified by peak detection (PD). (D) Ssb1 substrate overlap detected by TE and PD. (F) Ssb1 and Ssb2 interaction profiles of proteins highlighted in (C). Shaded areas show the variation between replicates. n=2. See also Figures S1 and S2.
For purification of Ssb engaged RNCs, we chromosomally tagged Ssb with C-terminal GFP, which does not affect its in vivo function (Figure S1A) and ribosome binding (Figure S1B) and serves as specific affinity tag for SeRP (Shieh et al., 2015). To ensure selective purification of Ssb-RNC complexes formed in vivo, rather than after cell lysis, we developed a protocol that combines rapid cell harvest by filtration, flash freezing and lysis in frozen state with ATP depletion and dilution of the lysates upon thawing. This protocol was optimized to most efficiently suppress RNC interactions of purified Ssb1-GFP added to thawing lysates of either wild-type (wt) or ssb1∆ssb2∆ cells (Figure S1C) and stabilize existing Ssb-RNC complexes. Under the experimental conditions used, the half-life of Ssb-peptide complexes is 3.9 ± 0.2 hours and the half-life of Ssb-ribosome complexes is about 14 hours (Figure S2A and S2B). Our calculations of the association rate of Ssb to a binding peptide based on (Hanebuth et al., 2016) further suggest that potential post-lysis interactions of Ssb do not significantly affect the outcome of our SeRP experiments. Consistent with earlier findings (Pfund et al., 1998), purified Ssb-RNCs are stable under low and high salt conditions but resolved upon puromycin induced release of nascent chains (Figures S2C and S2D). These features indicate that the stability of Ssb-RNCs critically depends on Ssb interactions with nascent chains. We conclude our SeRP setup provides snapshots on physiologically relevant interactions of Ssb with RNCs that were established under non-equilibrium in vivo conditions.
Co-translational engagement of Ssb with RNCs
We performed SeRP of Ssb1 and Ssb2 separately, in wt cells or single SSB mutants. Ssb interaction profiles are independent on the salt conditions used during RNC preparations (Figures S3A and S3B).
The metagene translatome of cells encoding either Ssb1-GFP or Ssb2-GFP reveals the typical accumulation of reads near the start codon, indicative of slow translation initiation kinetics, and a nearly uniform footprint distribution along the first 500 codons (Figure 1B). The Ssb1- and Ssb2-bound translatomes are highly similar but differ from the total translatomes. The mean read density in Ssb-bound translatomes is low for the first 50 codons and later reaches a stable plateau. Ssb therefore, on average, starts to engage nascent chains beyond a length of approx. 50 residues and can bind/rebind throughout further synthesis (Figure 1B). Given that 25-30 aa are buried inside the ribosome, Ssb starts engaging nascent chains when approx. 20 residues are exposed.
Substrate interactions of Ssb1 and Ssb2
To assess the nascent chain interactome of both Ssbs, we initially calculated the total enrichment factor (TE) of each gene (ratio of footprints per gene of Ssb-bound translatome to translatome). We identified 1501 genes with a ratio >1 which qualifies them as Ssb1 substrates (Figure 1C). This quantification method however fails to detect transient Ssb binding, and has limited sensitivity if factors engage large substrate pools. Therefore we developed a peak detection (PD) algorithm, which scans all Ssb interaction profiles to identify local Ssb binding peaks (>1.5-fold enrichment of footprint density over a stretch of five codons, Figures 1D and 1E). The PD algorithm reliably detects even nascent chains that engage Ssb only for one short period during synthesis, which are often missed by TE detection (Figure 1F). Combining both analyses identified 2945 Ssb1 substrates and 2859 Ssb2 substrates (Figures 1D, 2A, and 2B; Table S1) out of 4152 and 4073 reliably detected genes, respectively.
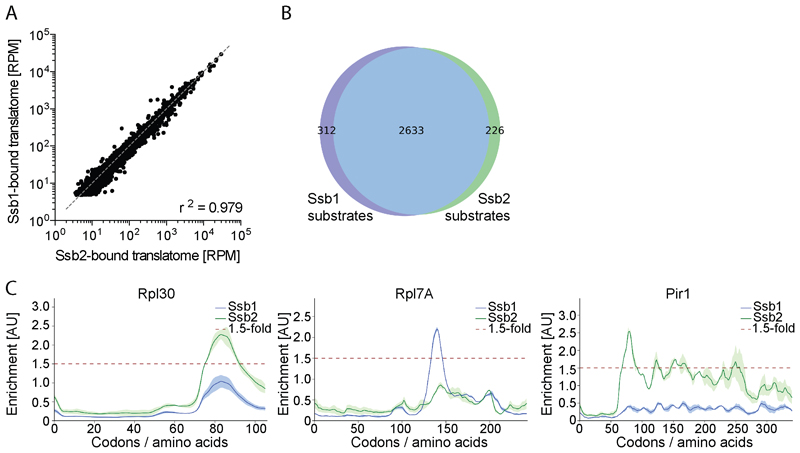
Figure 2. Ssb1 and Ssb2 nascent chain binding.
(A) Reads per gene of Ssb1- and Ssb2-bound translatomes. (B) Ssb1 and Ssb2 substrate overlap. (C) Ssb1 and Ssb2 binding profiles of three proteins. Shaded areas show the variation between replicates. n=2
The Ssb1 and Ssb2-bound translatomes highly correlate (Figure 2A and 2B) and most protein-specific interaction profiles are very similar (Figure 1F), in agreement with genetic data suggesting functional redundancy (Craig and Jacobsen, 1985). Gene-by-gene inspection revealed that the small discrepancy in substrate pools is largely due to subtle differences in enrichment factors that affect bioinformatic detection (e.g. Rpl30 in Figure 2C). For only a small subset of proteins, we find distinct interaction patterns of Ssb1 and Ssb2 (Figure 2C). These differences may be biologically relevant, as Ssb1 and Ssb2 specific functions were reported (Hatin et al., 2007). Of note three out of the four amino acid deviations are in the substrate-binding domain, suggesting slightly differing binding properties of Ssb1 and Ssb2.
Ssb engages nascent cytosolic, nuclear and organellar proteins
Consistent with previous data (Willmund et al., 2013), Ssb1 and Ssb2 (now referred to as Ssb) interact with nascent chains of more than 80% of the cytoplasmic and nuclear proteins (Figure 3A). We find Ssb also binds about 80% of nascent mitochondrial proteins and more than 40% of the ER-targeted proteins, suggesting a function of Ssb in membrane targeting and/or translocation of these proteins. Supporting this assumption, the SSB1,2 deletion causes the aggregation of 76 ER and 136 mitochondrial proteins (Willmund et al., 2013), many of which we identify as Ssb substrates (85% of the aggregated mitochondrial; 64% of the aggregated ER proteins). The metagene profiles of Ssb interaction with the different subclasses of substrates vary, suggesting distinct roles for Ssb in folding, sorting and translocation of nascent proteins (Figure 3B).
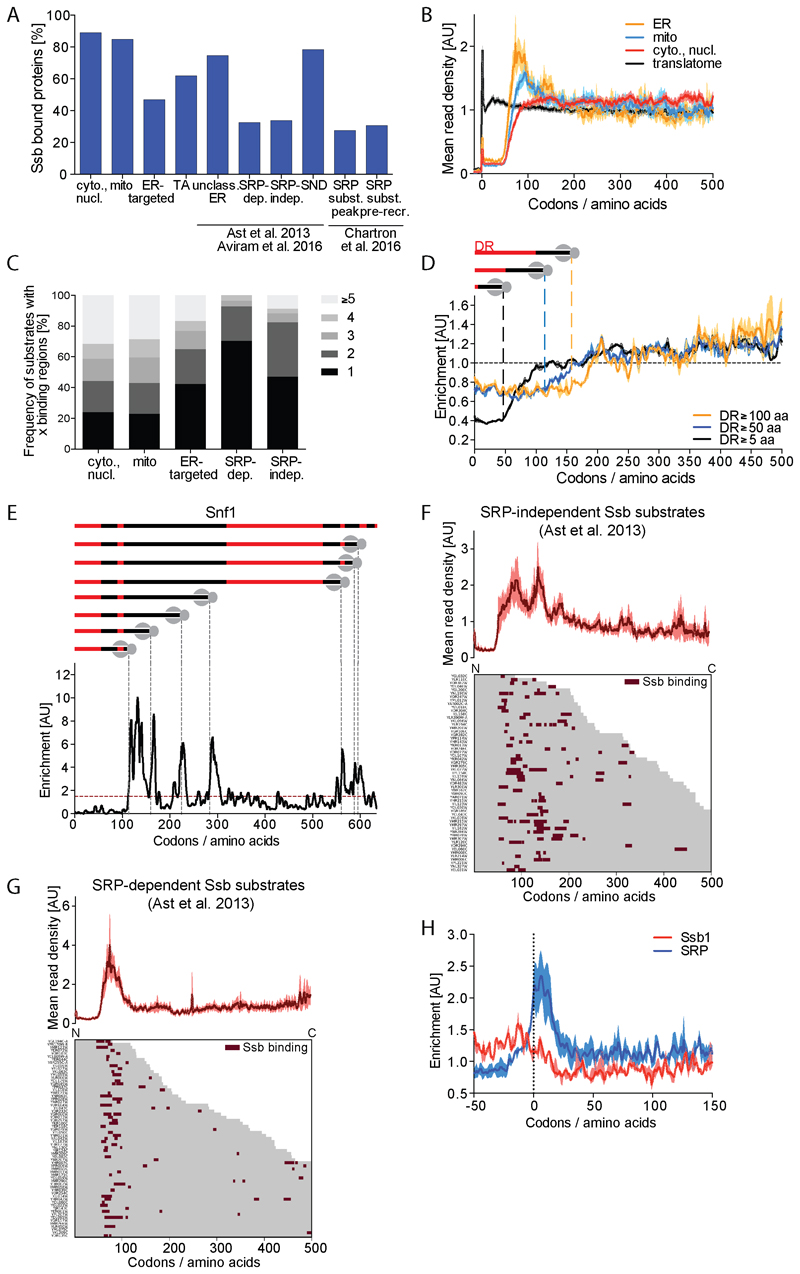
Figure 3. Subcellular destination of substrates affects Ssb interaction profiles.
(A) Fraction of Ssb substrates within indicated protein categories; cytoplasmic and nuclear, mitochondrial, ER-targeted, tail-anchored (TA), unclassified ER-targeted, SRP-dependent, SRP-independent, SND-pathway, SRP substrate (with defined SRP binding peak) and SRP substrate (with SRP pre-recruitment). (B) Metagene translatome and Ssb-bound translatomes with nascent proteins sorted by intracellular destination (averaged translatomes in black). Shaded areas indicate the 95% CI. (C) Number of Ssb binding regions per protein sorted by localization. (D) Metagene enrichment of Ssb binding to proteins containing disordered regions (DR) with indicated minimal lengths aligned to the start of first DR. Shaded areas indicate 95% CI. (E) Ssb binding to Snf1. Top: Position and length of DR (red) in Snf1. Gray dashed lines indicate onset of Ssb binding. Bottom: Ssb binding profile. (F, G) Heatmap of Ssb binding to ribosomes synthesizing SRP-independent (F) and SRP-dependent (G) nascent proteins. Corresponding metagene profile shown on top. (H) Metagene Ssb and SRP enrichment for genes with distinct SRP binding peak (Chartron et al., 2016) aligned to SRP peak position. Shaded areas show 95% CI. See also Figures S3, S4 and S7.
For the group of cytoplasmic and nuclear proteins the metagene profile implies Ssb frequently engages their nascent chains until the end of translation with no apparent clustering at defined chain lengths (Figure 3B). Almost 80% of the individual profiles show multiple binding peaks, consistent with repeated Ssb engagement during protein synthesis (Figure 3C). Binding often correlates with the emergence of peptide stretches that upon folding become buried in the hydrophobic core (Figures S3C and S3D). Considering that Ssb binds the emerging protein close to the ribosomal surface (see below), binding therefore must interfere with native folding. This raises the question whether Ssb binding is coordinated with the domain structure of the protein. To explore this possibility, we calculated meta profiles of Ssb binding to cytosolic and nuclear single and multi domain proteins with known domain structure (Figures S3E, S3F and S3G). Strikingly, Ssb binding to the 125 proteins containing ≥2 domains, and particularly the 42 proteins containing ≥3 domains, shows periodicity that follows the domain profile and specifically avoids linker regions. This binding pattern suggests Ssb may coordinate domain-wise co-translational folding by binding to core segments of partially exposed domains to keep them unfolded until domain synthesis is completed.
A significant fraction of the cellular proteome harbors intrinsically disordered regions involved in phase segregation and prion formation (Chernoff and Kiktev, 2016; Malinovska et al., 2013). Calculating meta profiles of Ssb binding to disordered regions of cytoplasmic proteins with minimal lengths of 5, 50 or 100 aa (van der Lee et al., 2014) we find Ssb strongly disfavors disordered regions (Figure 3D). This is further illustrated for Snf1 which has multiple intrinsically disordered regions of differing lengths, which are explicitly avoided by Ssb (Figure 3E). These findings are inconsistent with previous reports (Hubscher et al., 2016; Willmund et al., 2013) but support the proposed concept that Ssb is involved in the folding of protein domains, from which intrinsically disordered regions are exempted.
The association of Ssb with 80% of nascent mitochondrial proteins is unexpected. Metagene profiles indicate these proteins are frequently engaged at lengths of about 100 aa (Figure 3B). However, both number and position of binding peaks is diverse, similar to cytoplasmic Ssb substrates (Figures S4A and 3C). A recent study identified active ribosomes in proximity to mitochondria, implying some mitochondrial proteins are imported co-translationally (Williams et al., 2014). Co-translational import that involves ribosome docking to the translocon would exclude Ssb binding to longer nascent chains. A heatmap of Ssb binding to this subset of mitochondrial proteins does not support this assumption (Figure S4A), suggesting a more general role for Ssb in mitochondrial membrane targeting or import. However, we do not find a role for Ssb in these processes as the import kinetics of Ssb substrates in wt and ssb1∆ssb2∆ cells are unchanged (Figure S4B). Nonetheless fluorescence microscopic analysis revealed that ssb1∆ssb2∆ mutants contain more reticulated mitochondria than wt cells (Figure S4C). Furthermore, when comparing protein contents of mitochondria in wt and ssb1∆ssb2∆ cells, we repeatedly failed to isolate sufficient amounts of mitochondria from the mutant. These observations suggest a role of Ssb in maintaining structural integrity of mitochondria.
The Ssb metagene interaction profile of ER-targeted proteins displays two local maxima of Ssb binding at nascent chain lengths of 80 to 100 and 120 to 150 aa (Figure 3B). This pattern may result from distinct Ssb binding modes to substrates that use different ER targeting routes (Ast et al., 2013; Chartron et al., 2016). We therefore individually analyzed Ssb binding to substrates of the SRP and the newly discovered SND (SRP independent) pathway (Aviram et al., 2016), tail-anchored proteins and unclassified substrates.
Ssb interacts with 62% of the tail-anchored proteins (Figure 3A) (Jan et al., 2014) frequently with several binding sites per protein species (Figure S4D). This pattern agrees with the cytosolic synthesis and post-translational membrane insertion of these proteins, and a potential role for Ssb in postponing folding. Similarly, most SND substrates (78%) and the unclassified ER proteins (75%) can be engaged throughout translation, suggesting their translocation does not involve ribosome docking (Figures 3A, S4E, and S4F). Ssb furthermore binds approx. 30% each of SRP-dependent and SRP-independent proteins (according to (Ast et al., 2013)), with no particular preference for ER membrane versus luminal proteins (Figure 3A).
Metagene profiles indicate Ssb predominantly binds SRP-independent substrates early, with two local maxima at 80 to 100 and 130 to 160 aa (Figure 3F). The heatmap shows that the first possible Ssb engagement is not coordinated by an emerging signal sequence (Figure S4H).
About 70% of the Ssb bound SRP-dependent proteins engage Ssb only at nascent chain lengths of 80 to 100 aa (Figures 3C and 3G), consistent with a co-translational translocation mode. Binding is not coordinated by the emergence of transmembrane domains (TMD) or signal sequences, indicating that Ssb does not specifically recognize these hydrophobic stretches (Figure S4I). To understand the interplay of Ssb with SRP, we compared the position of Ssb binding regions with specific binding sites of SRP detected in a subpopulation of its substrates (Chartron et al., 2016). Ssb binds to 31% of this population, predominantly at lengths between 50 and 200 aa (Figure S4G). To reveal the order of Ssb and SRP binding in this subpopulation, we aligned all nascent chain sequences to the position of the SRP binding peak, and calculated the metagene Ssb binding profile relative to this position (Figure 3H). We find that 90% of the shared substrates engage Ssb before SRP.
Sequence recognition motif of Ssb
Considering Ssb engagement starts at nascent chain lengths of about 50 aa and the ribosomal tunnel covers the C-terminal 25-30 residues, Ssb should recognize a stretch of 20-25 exposed residues. To identify the Ssb binding motif within nascent chains, and to map its distance from the tunnel exit when Ssb binds, we bioinformatically analyzed the ribosome-proximal 60 residues of nascent chains. We could not identify a distinct sequence motif with precise amino acid specificity in the exposed peptide segments of Ssb bound substrates. However a multiple sequence analysis (WebLogo (Li et al., 2008), Figure 4A) revealed a degenerative Ssb binding motif positioned at a distance of 35 to 53 residues from the peptidyl-transferase center (PTC) that is enriched in positively charged (Arg and Lys) and hydrophobic residues (Val), similar to the sequence motif recognized by other Hsp70s (Rüdiger et al., 1997). Supporting this finding a metagene interaction profile of Ssb to proteins containing polybasic stretches composed of at least three Lys or Arg residues in random order (Figure S5A) shows a specific recruitment of Ssb once these stretches are positioned at a distance of about 50 aa from the PTC.
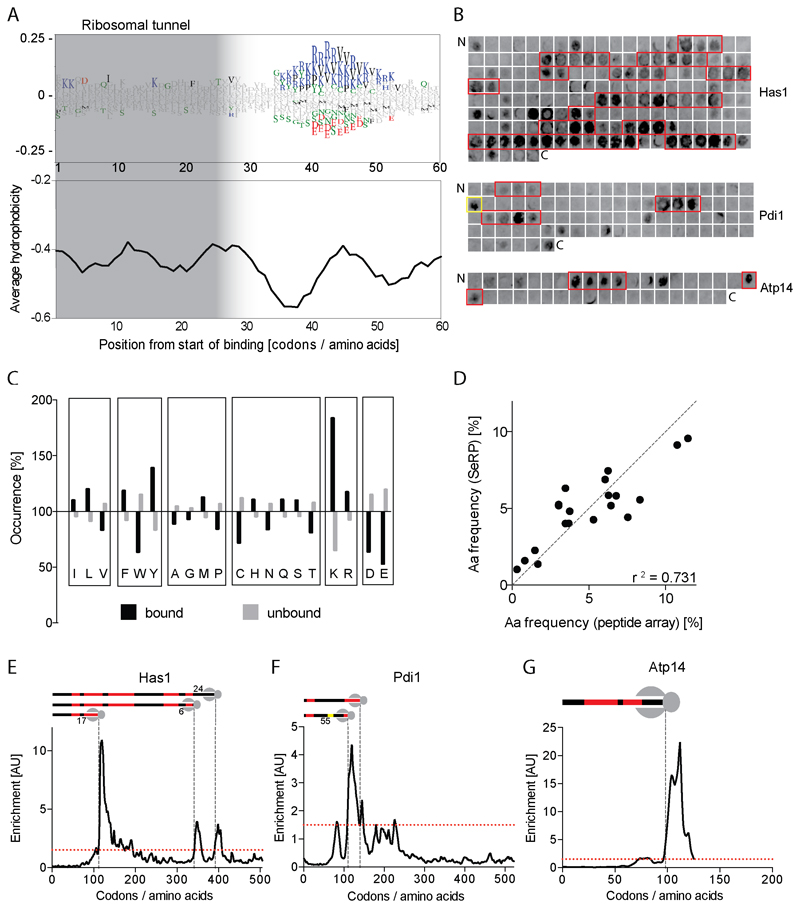
Figure 4. Substrate recognition motif of Ssb.
(A) Top: Aa enrichment (WebLogo) in the C-terminal 60 aa of nascent proteins at the time point of Ssb binding. Significantly enriched (>0) or depleted (<0) aa (chi-square test, p <0.05) are colored. Bottom: Average Kyte-Doolittle hydrophobicity plot (7 aa window). (B) Ssb1 binding to one peptide array scanning the sequences of Has1, Pdi1 and ATP14. Regions bound reproducibly are boxed. (C) Normalized occurrence of aa in Ssb bound and unbound peptides. (D) Correlation of aa frequencies in Ssb bound regions (SeRP) or peptides (array). (E-G) Comparison of SeRP and array data for Has1 (E), Pdi1 (F), Atp14 (G). Cartoons indicate areas bound by Ssb in vitro (red). Distances between ribosome and Ssb-bound peptides are indicated. Single Pdi1 peptide strongly bound by Ssb in yellow. See also Figures S5 and S7.
To independently verify the Ssb binding preference, we analyzed Ssb binding to peptide arrays representing the sequences of four proteins, among them three Ssb substrates identified by SeRP from different cellular compartments (Figure 4B). We find highly specific and reproducible peptide binding features of Ssb that are similar to those of E. coli DnaK, which we included for comparison (Figures S5B-S5E). Statistical analysis of the amino acid abundance in Ssb bound versus non-bound peptides fully supports the WebLogo analysis: Ssb prefers binding to peptides enriched with Lys and Arg, and avoids negatively charged peptides (Figures 4C, 4D, and S5F). Sorting the peptides by net charge reveals the importance of hydrophobic residues for Ssb binding. Ssb avoids binding positively charged peptides disenriched in Phe, Trp and Tyr (Figure S5G) but binds neutral and even negatively charged peptides enriched in aromatic (Phe, Tyr) or aliphatic (Ile, Val, Leu) residues (Figures S5H and S5I). Trp is not generally enriched in Ssb bound peptides, most likely because it is too rare to allow statistically meaningful conclusions. We conclude Ssb binds nascent chains close to the ribosomal surface with sequence preferences reminiscent to that of DnaK (Rüdiger et al., 1997). However, contrasting DnaK, the binding motif of Ssb does not indicate a clear separation into a hydrophobic core and positively charged flanking regions.
Connecting these findings we compared in vivo Ssb-nascent chain interaction profiles with in vitro peptide array data (Figures 4E-4G). For the three substrates analyzed, in vivo Ssb binding generally coincides with exposure of two overlapping peptides bound by Ssb in vitro. The single exception is the first Ssb binding region of nascent Pdi1, which correlates to only one peptide strongly bound by Ssb in vitro (Figure 4F). Importantly the exposure of an Ssb binding site in a nascent chain does not always trigger Ssb recruitment to the RNC. Skipping of emerging binding sites implies Ssb binding is additionally controlled, for example by coordinated action with RAC or NAC.
Impact of RAC on co-translational Ssb function
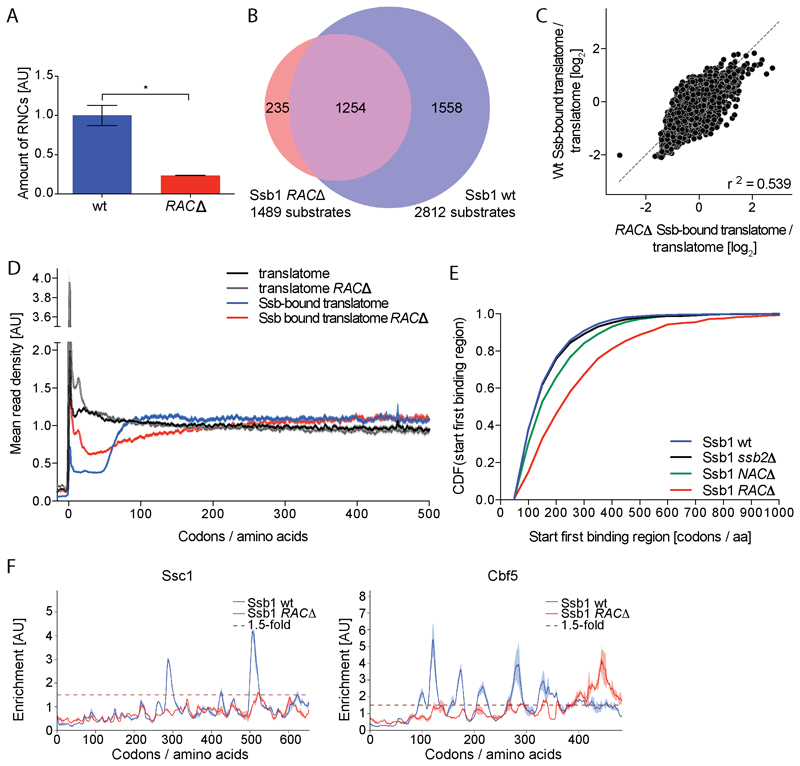
Genetic and biochemical evidence indicate that RAC and Ssb act synergistically in the same pathway (Gautschi et al., 2002; Willmund et al., 2013). We investigated the effect of RAC on Ssb binding to RNCs by SeRP of mutant cells lacking both RAC subunits (zuo1Δssz1Δ, referred to as RACΔ). RAC absence strongly reduces Ssb-RNC yields obtained by affinity purification, demonstrating RAC facilitates or stabilizes Ssb-RNC interactions (Figures 5A and 5B). Furthermore, RAC absence severely affects both, translatome and Ssb-bound translatome (Figures 5C and 5D). First, the metagene translatome reveals a strong accumulation of ribosomes near the translation initiation site, suggesting initiated ribosomes progress more slowly to the elongation phase (Figure 5D). Second, the metagene Ssb-bound translatome shows ribosome footprints are less disenriched in the mutant during translation of the first 50 codons, supporting the previous notion of impaired Ssb-ribosome interactions. Third, the sharp onset of Ssb binding at 50 aa nascent chains length is lost. Instead Ssb engagement very slowly increases, reaching a late plateau after translation of around 300 to 400 codons (Figure 5D). Accordingly, the average first Ssb engagement is much delayed in RAC∆ cells as compared to wt (Figure 5E). In the interaction profiles of individual proteins a general loss of distinct Ssb binding peaks is observed, and remaining binding events occur late during protein synthesis (Figure 5F).
Figure 5. RAC coordinates Ssb binding.
(A) Relative quantification of Ssb1 bound RNCs in wt and RACΔ cells (unpaired t-test, p <0.05). Error bars show standard deviation. (B) Ssb1 substrate overlap. (C) Comparison of TE in wt and RAC∆. (D) Metagene profiles of translatomes and Ssb1-bound translatomes. Shaded areas show 95% CI. (E) Cumulative distribution function (CDF) comparing the onsets of first Ssb binding regions (Wilcoxon rank-sum test, p <0.0001 for wt vs. RAC∆ and wt vs. NAC∆, not significant for wt vs. ssb2∆). (F) Ssb1 interaction profiles for exemplary proteins. Variation between replicates is shaded. n=2
Interplay of Ssb and NAC
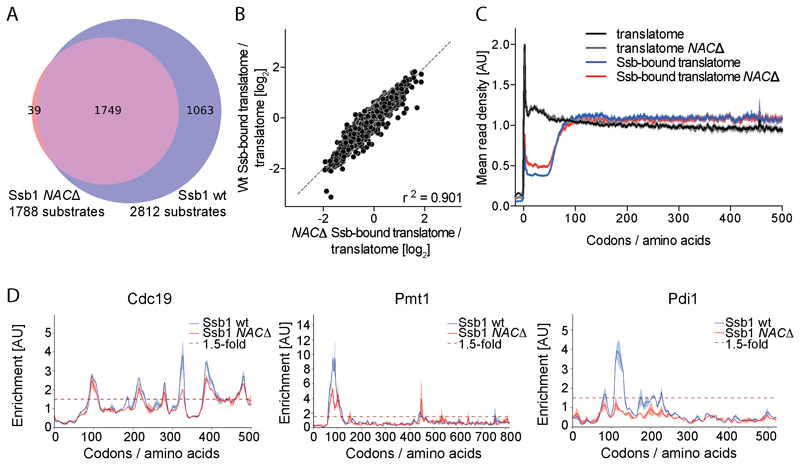
We next explored the impact of NAC on Ssb function, by Ssb-specific SeRP of mutant cells lacking all three NAC subunits (egd1∆btt1∆egd2∆, referred to as NAC∆). TE and PD analyses suggest less Ssb substrates in NAC∆ cells as compared to wt cells encoding both Ssbs (1788 compared to 2812, Figure 6A). However, comparing both strains reveals the Ssb-bound to total translatome ratios and the metagene Ssb interaction profiles are highly similar, demonstrating the coordination of Ssb binding with nascent chain length is the same in the NAC mutant (Figure 6B and 6C). We find only nine proteins that interact with Ssb exclusively in the presence of NAC (one is ER protein Pdi1, Figure 6D), while all other substrates are highly overlapping. Also, individual interaction profiles show that the reduced number of detected substrates is mostly caused by more equal distribution of footprints in the Ssb-bound translatome and reduced height of binding peaks (Figure 6D), which decreases the number of Ssb substrates detected. The position of binding peaks largely remains the same for wt and NAC∆ cells (Figure 6D) and also the WebLogo analysis shows the binding motif is preserved. These findings indicate a general synergistic effect of NAC in promoting Ssb binding to nascent chains, which affects the amount of Ssb substrates but not substrate selectivity.
Figure 6. Ssb acts independently of NAC.
(A) Ssb1 substrate overlap. (B) Comparison of TE in wt and NAC∆. (C) Metagene profiles of translatomes and Ssb1-bound translatomes. Shaded areas show 95% CI. (D) Ssb1 binding profiles for exemplary proteins. Variation between replicates is shaded. n=2
Coordination of Ssb binding with translation elongation rates
We next asked whether Ssb binding coincides with an altered speed of translation, e.g. ribosomal pausing, indicative of an additional level of coordination. For statistically robust data sets, we restricted our analysis to high-coverage genes (Figure S6A). We first determined the mRNA segments translated by Ssb-bound ribosomes and analyzed the footprint density in these segments of the translatome as a measure of local translation speed (Figures 7A, S6B, and S6C). Then we compared Ssb bound and unbound mRNA segments in independent RP datasets analyzing translation in wt cells. With high significance, mRNA segments translated by Ssb-bound ribosomes are generally less populated by ribosomes, and hence are translated with higher rates. We detect these differences at the metagene level (Figure 7B) and the level of individual genes. Dependent on the stringency of the selection criteria used to define bound and unbound segments (Figure S6D), the differences in local translation speed in wt cells vary by 10 to 38% (Figure 7B).
Figure 7. Altered translation kinetics of Ssb bound ribosomes.
(A) Average ribosome densities in translatomes and Ssb-bound translatomes related to Ssb binding. 95% CI is shaded. (B) Change in translation speed for Ssb bound and unbound ribosomes in wt and ssb1Δssb2Δ translatomes (Wilcoxon rank-sum test, p <0.0001 for all thresholds). Error bars show 95% CI. (C) Contribution of Ssb binding and mRNA features to faster translation. Error bars show 95% CI. (D) Enrichment of fast codons and depletion of slow codons in bound vs. unbound segments for indicated thresholds (Wilcoxon signed-rank test, p <0.05 for (P95, P5), p <0.0001 for other thresholds). Error bars show 95% CI. (E) Change in DMS reactivity of bound vs. unbound segments reflecting the probability of secondary structure formation with the indicated offsets from each nucleotide. Differences are significant up to an offset of 15 nt (paired t-test, p < 0.0001). See also Figures S6 and S7.
Two non-exclusive mechanisms may account for the change in translation speed. One is that Ssb binding confers the acceleration of translation. The second is that intrinsic features of the mRNA or the nascent chain accelerate translation. We first explored the impact of Ssb binding, by analyzing relative elongation speed in translatomes of ssb1∆ssb2∆ cells. Translation is accelerated also in the absence of Ssb (Figure 7B) although to a slightly lesser extent (Figure S6E). Independent of the stringency of our criteria to identify mRNA segments translated by Ssb bound or unbound ribosomes, the Ssb contribution to the translation speed-up is limited (up to 15%, Figure 7C). Testing for mRNA features that can slow translation, we find that mRNA segments translated by Ssb-engaged ribosomes are enriched for fast-translated codons, depleted for slowly translated codons and contain less proline codons (Figures 7D and S6F). In addition, mRNA segments located 1-15 nucleotides downstream of Ssb bound ribosomes have reduced mRNA secondary structure (Figure 7E). Finally, nascent chain segments located in the ribosomal tunnel of Ssb bound ribosomes have average numbers of positively charged residues but are enriched in negatively charged residues (Figure S6G).
Discussion
Our SeRP analysis identified approx. 2900 nascent chain substrates of Ssb out of 4200 detected translated proteins (Figure 1D). This large number of substrates qualifies Ssb as general chaperone for folding of newly synthesized proteins in yeast and substantially extends the known interactome list (Willmund et al., 2013) by mitochondrial and ER-targeted proteins (Figure 3A). Importantly, our approach revealed the Ssb binding profiles of the entire nascent proteome, identifying binding sites at near-residue resolution and for all nascent chain lengths. This uncovers molecular principles of the modus operandi of a eukaryotic Hsp70 chaperone. It furthermore discloses an unexpected connection of Ssb binding with translation speed enhancement (Figure S7).
Ssb binds translating ribosomes only after the nascent chain reached a length of about 50 aa, implying exposure of approx. 20 N-terminal aa (Figure 1B). WebLogo analysis revealed that Ssb binds RNCs when a degenerative recognition motif is positioned at about 5 aa from the tunnel exit (Figure 4A). Ssb thus provides one of the earliest encounters of nascent chains with the protein folding machinery.
The lack of Ssb binding to RNCs prior to the emergence of nascent chains parallels the behavior of the bacterial chaperone TF (Oh et al., 2011). TF however starts engaging nascent chains later, at about 110 aa chain length. This difference correlates with the distinct modes by which TF and Ssb associate with substrates. TF binds long sometimes discontinued substrate stretches involving binding surfaces spread over an extended cavity (Hoffmann et al., 2010; Merz et al., 2008; Saio et al., 2014). Ssb instead uses a single pocket to bind a short stretch positioned close to or even right above the tunnel exit (Gumiero et al., 2016). Interestingly the cellular concentrations of TF and Ssb and their in vitro affinities for vacant ribosomes should suffice to engage RNCs even earlier during translation (Hoffmann et al., 2010; Raue et al., 2007). The delayed chaperone engagement we observe in vivo may result from (i) lower effective concentrations of free chaperones, which restrict their association to RNCs exposing binding sites within nascent chains, (ii) occupancy of chaperone binding sites on ribosomes by other factors, or, for Ssb, (iii) regulatory effects exerted by RAC (see below). We envision late chaperone engagement increases the efficiency of chaperone utilization and generates a time window for other processes that require prioritization, such as enzymatic processing of nascent N-termini.
Our SeRP binding profiles indicate that Ssb dynamically engages nascent chains in typically multiple binding and release cycles (Figure 1F). The maximal life time of Ssb-RNC complexes is given by the width of Ssb binding peaks, which comprises a range of codons with a (median) stretch length of 11 translated codons. Assuming translation kinetics is 5 to 10 codons/second (Boehlke and Friesen, 1975), Ssb on average engages RNCs for 1-2 seconds. Shorter residence times for individual nascent chain molecules, which are blurred in our ensemble measurements, may exist. This rather short residence time, together with the fact that footprint densities do not increase towards the 3'-end of genes, suggests that a RNC is generally engaged by one Ssb at a time and that Ssb does not remain tethered to ribosome-associated RAC during chain elongation. The sharp decline of the Ssb binding peaks instead indicates coordinated RNC release, most likely mediated by one or several of the Ssb NEFs including Sse1, Fes1 and Snl1.
Number and position of Ssb binding sites vary strongly between nascent chain species. Despite this variability, Ssb binding follows clear rules, main parameters being sequence motifs of the nascent chains and the intracellular destination of the substrates (Figures 3B, 3C, and 4A).
SeRP and in vitro peptide array scanning show consistently that Ssb favors binding to positively charged, hydrophobic peptide stretches also enriched with aromatic residues (Figures 4A and 4C). This degenerate peptide motif occurs frequently in proteins, in agreement with Ssb´s role as general chaperone. While the binding motif shows similarities to that of canonical Hsp70 chaperones (Rüdiger et al., 1997), the strong preference for positively charged stretches is unique among Hsp70s, likely due to the more negatively charged surface near the substrate binding pocket of Ssb. This Ssb binding preference may serve to prevent non-productive contacts of positively charged nascent chain segments with negatively charged ribosomal RNA. Agreeing with this, highly positively charged ribosomal proteins are strongly enriched among the proteins that aggregate in cells lacking Ssb (Koplin et al., 2010).
Recognition of this motif allows Ssb to bind segments that will be surface exposed or form the hydrophobic core of the folded protein (Figures S3C and S3D). This implies Ssb binding will delay co-translational folding, very much like TF in bacteria (Hoffmann et al., 2012; Mashaghi et al., 2013). We speculate that upon Ssb release co-translational folding of a protein domain may either resume, or the nascent chain be handed over to another chaperone as part of a functional network. In agreement with the proposed function of Ssb to assist domain-wise folding by retarding premature folding, Ssb does not bind intrinsically disordered regions and disfavors linker regions connecting domains. Ssb instead binds emerging domains and the vast majority of mitochondrial precursors that must be kept unfolded for import into mitochondria (Figures 3A, 3D, S3E, and S7B).
It is intriguing that Ssb frequently ignores emerging binding motifs in nascent chains, suggesting a higher level of site-specific control (Figures 4E-4G). While this control is currently not understood, we suspect it may be exerted by RAC, which has a dominating effect on Ssb´s timing and specificity of substrate engagement. RAC absence severely impairs Ssb binding to emerging recognition motifs and delays RNC engagement (Figure 5D). RAC is suggested to target Ssb to its binding sites in nascent chain substrates by positioning the Ssb substrate binding site near the tunnel exit and promoting substrate enclosure by Zuo1-dependent stimulation of ATP hydrolysis by Ssb (Gumiero et al., 2016; Hanebuth et al., 2016; Huang et al., 2005). We propose skipping of selected binding sites is mediated by tuning this intricate RAC-Ssb interplay.
We also investigated the functional interplay between Ssb and NAC. NAC absence does not profoundly affect Ssb´s ability to interact with substrates (Figures 6C and 6D). Previous genetic analysis suggested partial functional overlap between Ssb and NAC since lack of both factors causes more severe protein misfolding than the lack of either Ssb or NAC. We note that NAC mutants have more subtle growth defects than mutants lacking both SSBs (del Alamo et al., 2011; Koplin et al., 2010; Willmund et al., 2013). Together these findings suggest that Ssb is the dominant chaperone in assisting co-translational protein folding, largely independent of NAC.
A large 80% fraction of all mitochondrial proteins are substrates of Ssb, suggesting a role of Ssb in protein targeting to this organelle. Ssb preferentially binds these nascent chains at lengths of about 100 residues and most substrates can be bound multiple times (Figures 3B and 3C). Together with the reported aggregation of mitochondrial proteins in ssb1∆ssb2∆ cells (Willmund et al., 2013), our results suggest Ssb may increase the targeting efficiency of protein precursors to mitochondria, potentially by preventing premature folding and misfolding.
Ssb also binds 46% of the ER-targeted proteins, with preference for proteins translocated without RNC docking to the translocon (Figures 3A and S4D-S4F). The co-translationally translocated, SRP-dependent Ssb substrates are frequently bound once, early during synthesis (Figure 3G) and binding is not correlated with the emergence of a signal sequence or a TMD (Figure S4I). Ssb generally binds prior to SRP (Figure 3H) which contrasts the current view of SRP being the first and only cytosolic interactor of nascent chains of the co-translational translocation pathway prior to RNC docking to the translocon. Our findings are consistent with two mutually non-exclusive scenarios: First, a hand-over of nascent chains from Ssb-RAC to SRP, ensuring faithful targeting. Second, substrate triage between Ssb and SRP, implicating two alternative targeting routes to the ER membrane. The plausibility of the latter scenario is suggested by a recent report showing that Ssb via its ATPase domain binds the Sec72 subunit of the ER translocation channel (Tripathi et al., 2017). Parallel pathways would provide functional redundancy, and hence biological robustness, to the ER targeting process, and explain the lack of a strong ER-specific phenotype of ssb1∆ssb2∆ mutants and the viability of yeast mutants lacking SRP.
An unexpected finding of our bioinformatics analysis is that Ssb binding to RNCs generally coincides with an acceleration of translation speed (Figure 7B). As a control, we examined if the same feature was present in nine other published Ribosome Profiling datasets from yeast that did not use CHX as a pretreatment (Figure S6H). We find that seven out of nine of the datasets exhibit this speedup with Ssb-bound regions (the origins of the inconsistency with two of the datasets is unknown). Thus, the speedup we observe in our data is largely reproducible across labs. Interestingly, canonical Hsp70 proteins in mammalian cells can overcome translational pausing around codon 65, at similar position where initial Ssb engagement peaks in yeast (Liu et al., 2013; Shalgi et al., 2013), potentially linking protein synthesis to the availability of Hsp70. However, our findings for Ssb are clearly different, as (i) intrinsic features of mRNA and nascent chains are largely responsible for faster translation, (ii) there is no indication for translation stalling at this position in SSB deletion mutants and (iii) Ssb binding contributes only up to 15% to the speed-up (Figure 7C).
What is the biological reason for evolving faster translation kinetics upon chaperone engagement? We speculate translation is allowed to occur faster because the folding-delaying function of Ssb binding generates a time window during which co-translational folding is uncoupled from translation kinetics. Faster translation during the periods of Ssb binding reduces the amount of ribosomes required to maintain protein synthesis and thereby reduces the costs of ribosome biogenesis in growing cells.
Taken together, by exploring proteome-wide in vivo interaction profiles we elucidate functional principles of Hsp70 chaperone action in co-translational protein folding and membrane targeting. Our findings reveal additional evidence for the intimate coordination of protein synthesis and folding, suggesting the genetic code and translation speed are directly adapted to the function of chaperones in co-translational protein folding.
Contact for Reagent and Resource Sharing
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Bernd Bukau (bukau@zmbh.uni-heidelberg.de).
Experimental Model
Recombinant proteins were expressed in Escherichia coli BL21 (DE3) cells grown in Lysogeny Broth.
Methods Details
Strain construction
GFP-tagged strains and deletion strains were constructed according to (Janke et al., 2004). For the GFP-tag, a cassette containing the monomeric GFP gene and a G418 resistance marker was amplified from the pYM12-mGFP plasmid. For gene deletions, a cassette containing only a selection marker was PCR amplified. All experiments were performed in the BY4741 strain background.
Purification of Ssb-RNCs for SeRP
200 ml of cells were grown to an OD600 of 0.5-0.6 in YPD, filtered and lysed by mixer milling (2 min, 30 Hz, MM400 Retsch) with 600 μl of lysis buffer (20 mM Tris-HCl pH 8.0, 140 mM KCl, 6 mM MgCl2, 0.1% NP-40, 0.2% glucose, 0.1 mg/ml CHX, 1 mM PMSF, 2x protease inhibitors (Complete EDTA-free, Roche), 0.02 U/μl DNaseI (recombinant DNaseI, Roche), 20 μg/mL leupeptin, 20 μg/mL aprotinin, 10 μg/mL E-64, 40 μg/mL bestatin). Lysates were thawed by adding the frozen powder stepwise to 100 μl hexokinase buffer (20 mM Tris-HCl pH 8.0, 40 mM NaPO4 pH 7.0, 0.2% glucose) containing 100 u of hexokinase while stirring with a magnetic flea to immediately deplete the ATP. Lysates were cleared by centrifugation (2 min at 30,000 g, 4 °C) and supernatants were digested using 60u/A260 nm of RNaseI for 5 min at 4 °C. 400 μl of digested lysates were loaded onto 800 μl of sucrose cushions (25% sucrose, 20 mM Tris-HCl pH 8.0, 140 mM KCl, 10 mM MgCl2, 0.1 mg/ml CHX, 1x protease inhibitors) and centrifuged in a TLA120-rotor for 90 min at 75,000 rpm, 4 °C. Pellets were resuspended in lysis buffer and transferred to non-stick tubes. 100-200 μg of total RNA were removed for ribosome profiling of the total translatome. To the residual RNA 500-750 μl of GFP-binder slurry (washed three-times with 1 ml of wash buffer I (20 mM Tris-HCl pH 8.0, 140 mM KCl, 10 mM MgCl2, 1 mM PMSF, 0.1% NP-40, 0.1 mg/ml CHX, 2x protease inhibitors)) was added and the suspension was rotated for 30 min, 4 °C. Beads were washed four-times in wash buffer I (20 min, 3x 5 min) and twice in wash buffer II (20 mM Tris-HCl, 140 mM KCl, 10 mM MgCl2, 1 mM PMSF, 0.1 mg/ml CHX, 0.01% NP-40, 10% glycerol, 2x protease inhibitors) (20 min, 5 min). Tubes were changed four-times during washing steps. The washed beads were subsequently used for RNA or protein extraction.
Deep sequencing library preparation
Library preparation was performed as described in (Becker et al., 2013). All steps were performed in non-stick, RNase free microfuge tubes (Ambion). In summary, RNA extraction was performed by mixing 0.75 ml pre-warmed acid phenol (Ambion) with either the purified monosomes resembling the total translatome or the monosomes bound to affinity beads for the Ssb-bound translatome and 40 μl 20% SDS (Ambion). After shaking at 1400 rpm for 5 min at 65°C, samples were incubated 5 min on ice and centrifuged at 20,000xg for 2 min. Top aqueous layers were transferred to fresh tubes and mixed again with 0.7 ml acid phenol. Samples were incubated for 5 min at room temperature with occasional vortexing and afterwards centrifuged for 2 min at 20,000xg. Top aqueous layers were transferred to fresh tubes and mixed with 0.6 ml chloroform, vortexed and centrifuged for 1 min at 20,000xg. Nucleic acids were precipitated by adding 78 μl 3 M NaOAc pH 5.5, 2 μl glycoblue and 0.75 ml isopropanol and incubating for 1 h to 16 h at -80°C. Samples were centrifuged for 30 min at 20,000xg and 4°C and pellets were washed with ice-cold 80% ethanol and resuspended in 10 mM Tris-HCl pH 7.0. Samples were heated at 80°C for 2 min and for total translatome 50 μg of RNA and for Ssb-bound translatome the entire sample was loaded onto a 15% TBE-Urea polyacrylamide gels (Invitrogen) in 1x TBE (Ambion) and run for 65 min at 200 V. Gels were stained for 20 min with SYBR gold (Invitrogen). To recover ribosomal footprints, the gel pieces were excised that contained RNA fragments with a size between 25 and 33 nt. Gel pieces were placed into 0.5 ml gel breaker tubes, nested into a 1.5 ml tube and centrifuged for 3 min at 20,000xg. 0.5 ml 10 mM Tris-HCl pH 7.0 was added and tubes were incubated at 70°C for 10 min with maximal shaking in an Eppendorf thermomixer. Gel pieces were removed using a Spin-X cellulose acetate column (Fisher) and the flow through was transferred to a new tube. 55 μl 3 M NaOAc pH 5.5, 2 μl glycoblue and 0.55 ml isopropanol were added. After mixing, tubes were frozen at -80°C for 1 h to 16 h. Samples were centrifuged for 30 min at 20,000xg and 4°C and pellets were washed with ice-cold 80% ethanol and resuspended in 15 μl of 10 mM Tris-HCl pH 7.0. For dephosphorylation, 2 μl 10x T4 polynucleotide kinase buffer without ATP (NEB), 1 μl murine RNase inhibitor and 2 μl T4 polynucleotide kinase (NEB) were added to each sample. Samples were incubated at 37°C for 1 h before heat inactivation of the enzyme for 10 min at 75°C and precipitation of nucleic acids by adding 0.5 ml 10 mM Tris-HCl pH 7.0, 55 μl 3 M NaOAc pH 5.5, 2 μl glycoblue and 0.55 ml isopropanol and incubating for 1 h to 16 h at -80°C. Samples were centrifuged for 30 min at 20,000xg and 4°C, pellets were washed with ice-cold 80% ethanol and resuspended in 6-11 μl of 10 mM Tris-HCl pH 7.0. For linker ligation, a maximum of 5 pmol RNA in 5 μl were denatured for 2 min at 80°C before 8 μl 50% sterile filtered PEG MW 8000, 2 μl DMSO, 2 μl 10x T4 RNA Ligase 2 buffer (NEB), 1 μl murine RNase inhibitor, 1 μl 1 μg/μl linker L1 and 1 μl truncated T4 RNA Ligase 2 (NEB) were added and incubated for 2.5 h at 37°C or 23°C. Nucleic acids were precipitated as described before and resuspended in 6 μl 10 mM Tris-HCl pH 7.0. Samples were run on a 10% TBE-Urea polyacrylamide gel (Invitrogen) in 1x TBE (Ambion) for 50 min at 200 V. Gels were stained for 20 min with SYBR gold and desired gel pieces were excised and RNA was extracted as described before. For reverse transcription, RNA was resuspended in 10 μl 10 mM Tris-HCl pH 7.0 and 1 μl 10 mM dNTP (NEB), 1 μl 25 linker L1’L2’ and 1.5 μl DEPC H20 were added to each sample. Samples were incubated at 65°C for 5 min followed by addition of 4 μl 5x FSB buffer (Invitrogen), 1 μl murine RNase inhibitor, 1 μl 0.1 M DTT (Invitrogen) and 1 μl Superscript III (Invitrogen). Samples were incubated at 50°C for 30 min and afterwards 2.3 μl 1 N NaOH was added to hydrolyze RNA and samples were further incubated at 95°C for 15 min. Samples were run on a 10% TBE-Urea polyacrylamide gel for 70 min at 200 V. Gels were stained as described before and desired bands were excised and nucleic acids were extracted as mentioned earlier but using Tris-HCl pH 8.0 and precipitating nucleic acids by adding 32 μl 5 M NaCl, 1 μl 0.5 M EDTA, 2 μl glycoblue and 0.55 ml isopropanol. For circularization, DNA was resuspended in 15 μl 10 mM Tris-HCl pH 8.0 and 2 μl 10x CircLigase buffer (EPICENTRE), 1 μl 1 mM ATP, 1 μl 50 mM MnCl2 and 1 μl CircLigase™ (EPICENTRE) were added. Samples were incubated at 60°C for 1 h. Addition of 1 μl CircLigase™ was repeated and samples were incubated for another hour at 60°C. Afterwards, the enzyme was inactivated by incubating 10 min at 80°C. 5 μl of circularized DNA was used for PCR amplification. Therefore, 16.7 μl 5x HF buffer, 1.7 μl 10 mM dNTPs, 0.4 μl 100 μM PCR primer L1’, 0.4 μl 100 μM barcoding primer, 59.2 μl DEPC H20 and 0.8 μl HF Phusion (NEB) were added. 17 μl PCR mix were aliquoted to 4 separate PCR tubes and the following PCR reaction cycles were run: 1.) 98°C, 30 sec; 2.) 98°C, 10 sec; 3.) 60°C, 10 sec; 4.) 72°C, 5 sec. Steps 2 through 4 were repeated ten times and one tube was removed after cycles 7, 8, 9, 10.
Samples were run on a 8% TBE polyacrylamide gel (Invitrogen) in 1x TBE (Ambion) for 45 min at 180 V. Gels were stained as mentioned before and desired bands were excised and DNA was extracted as described before. After a quality control step using a high sensitivity bioanalyzer chip (Agilent), samples were sequenced on a HiSeq 2000 (Illumina).
Purification of Ssb and Ssb-GFP
Ssb or Ssb-GFP was purified from E. coli cells encoding Ssb N-terminally fused with a cleavable His6-SUMO tag. Cells were lysed in buffer A (40 mM Hepes KOH pH 7.4, 150 mM KCl, 5 mM MgCl2, 5% glycerol, 1 mM PMSF, 10 mM β-mercaptoethanol, 1 mM ETDA, DNaseI) and cleared lysates were mixed with nickel–iminodiacetic acid (Ni-IDA, Protino, Macherey-Nagel). After extensive washing with buffer A, His6-SUMO-Ssb was eluted with buffer A containing 250 mM Imidazole. His6-SUMO was removed by treatment with His6-ULP protease during overnight dialysis against buffer A. His6-ULP, His6-SUMO and uncleaved His6-SUMO-Ssb were removed by passing the eluate through a second Ni-IDA column. Ssb was loaded onto a HiLoad 16/60 Superdex 75 gel filtration column (GE Healthcare Life Sciences) and gel filtration was performed in buffer B (40 mM Hepes KOH pH 7.4, 50 mM KCl, 5 mM MgCl2, 5% glycerol, 10 mM β-mercaptoethanol) containing 5 mM ATP. Aliquots were stored at - 80 °C.
In vitro binding studies
Two different cellulose-bound peptide arrays were used for the Ssb binding studies. PepSpots™ Peptide Arrays on cellulose were ordered from JPT. Has1, Pdi1, Atp14 and λ cI repressor protein sequences were subdivided into 13-mer peptides with 3 aa shift (= 10 aa overlap). C-termini of peptides were attached via ß-alanine PEG-linker to the cellulose membrane. Additional experiments were performed using CelluSpots™ Peptide Arrays obtained from Intavis that scanned the sequences of six proteins from bacteria and nematodes (subdivided into 14-mer peptides with 5 aa shift (= 9 aa overlap)). Experiments were performed according to (Rüdiger et al., 1997) with minor modifications. Peptide arrays were incubated with either 150 nM DnaK or 1 μM Ssb1 in MP buffer (31 mM Tris-HCl pH 7.6, 5 mM MgCl2, 100 mM KCl, 0.05% Tween-20, 5% glucose) for 40 min at RT with agitation and blotted six-times for 30 min at 0.8 mA/cm2 in a semi-dry blotter. Transferred proteins were detected with either Ssb-specific or DnaK-specific antibodies. The signal intensity of each peptide spot was quantified using Multi Gauge (FUJIFILM Co.) and normalized according to the highest signal obtained. Peptides classified as bound had a signal intensity of at least 24% and were visible in at least two independent experiments. Peptides showing a ring-like signal or artificially high background were excluded from the analysis.
Polysome profiles
Cells were grown and lysed as described for the purification of Ssb-RNCs for SeRP. After thawing (either with or without ATP depletion), lysates were cleared and 500-1000 μg of RNA were loaded onto a 10-50% linear sucrose gradient. To prepare gradients, 50% sucrose dissolved in sucrose gradient buffer (20 mM Tris-HCl pH 8.0, 140 mM KCl, 6 mM MgCl2, 0.1 mg/ml CHX) were layered below a 10% sucrose solution and solutions were mixed using the Gradient MasterTM (Biocomp). The gradients were centrifuged for 2.5 h at 35,000 rpm, 4 °C (SW40-rotor, Sorvall Discovery 100SE Ultracentrifuge). After centrifugation the polysome profiles were recorded with the Piston Gradient FractionatorTM (Biocomp). 300-600 μl fractions were collected and frozen in liquid nitrogen for later analysis.
Post-lysis binding control
Cells were grown and lysed as described for the purification of Ssb-RNCs for SeRP. Lysates were thawed in the presence of exogenously added Ssb1-GFP (1:1 ratio to endogenous Ssb, 25 ng of Ssb1-GFP for every μg of total protein in wt lysate) and hexokinase. Lysates were cleared by centrifugation (20,000xg for 2 min) and supernatants were digested with RNaseI for 5 min at 4°C. Following, 400 μl of digested lysates were loaded onto 800 μl of sucrose cushions (25% sucrose, 20 mM Tris-HCl pH 8.0, 140 mM KCl, 10 mM MgCl2, 0.1 mg/ml CHX, 1x protease inhibitors) and centrifuged in a TLA120-rotor for 90 min at 75,000 rpm, 4 °C. Pellets were resuspended in lysis buffer and lysate and pellet fractions were tested for the presence of endogenous and GFP-tagged Ssb.
In vitro peptide release measurements
Ssb2 was incubated for 2 hours at 30 °C with 2-(40-(iodoacetamido)anilino) naphthalene-6-sulfonic acid (IAANS) labeled peptide substrate σ32-Q132-Q144-C in HKM buffer (25 mM HEPES-KOH pH 7.5, 50 mM KCl, 5 mM MgCl2) before mixing with a 50-fold excess of unlabeled σ32-Q132-Q144-C peptide using stopped-flow instrumentation (Applied Photophysics SX20). Changes of fluorescence intensity (excitation wavelength 335 nm, emission filter 420 nm cutoff) were measured at 29°C (1000 s) and 2.9°C (2-12 h). The determined half-life of the complex was 7.7 ± 6.7 min (29°C) and 3.9 ± 0.2 hours at 2.9°C.
In vitro dissociation experiment
Cells lysates were prepared, thawed and digested as described for the purification of Ssb-RNCs for SeRP. RNase treated Ssb1-GFP ssb2Δ lysates and purified Ssb1-GFP (control, same amount as in the lysate) were separately loaded onto a 10-30% linear sucrose gradient and centrifuged for 4 h at 4°C and 35,000 rpm (SW40-rotor, Sorvall Discovery 100SE Ultracentrifuge). 10 fractions of the sucrose gradient were collected and the Ssb content of each fraction was determined by quantitative western blotting. The first three fractions of the gradient resembling free Ssb1-GFP present in the cell lysate were excluded from the analysis. Taking into account that purified Ssb-GFP migrated to the first fractions of the gradient, the Ssb1-GFP amount determined in the control run was subtracted from the Ssb1-GFP amount determined in the lysate centrifugation. Comparing the Ssb1-GFP amount in fractions 4-6 (dissociated Ssb1-GFP) with the amount in the ribosome fractions (7-10) showed that about 18.6% of Ssb dissociated within the 4 h centrifugation.
SILAC-coupled to mass spectrometry of purified mitochondria
For SILAC (stable isotope labeling with amino acids in cell culture) experiments, wt yeast cells were grown at 30 °C in SC-medium containing 2% galactose and isotope labeled lysine and arginine (13C6,15N2-L-Lysine HCl, 13C6,15N2-L-Arginine HCl) to an OD600 of 0.5 and mixed with same amounts of ssb1Δssb2Δ cells grown under the same conditions with unlabeled lysine and arginine. Mitochondria were purified according to published protocols (Altmann et al., 2007), proteins were separated by SDS-PAGE and identified by mass spectrometry.
Fluorescence microscopy
Cells were grown in SC-medium containing 2% glucose, harvested by centrifugation and resuspended in PBS. Optical sections of 0.2 μm were acquired to image the whole cell volume using a widefield system (xcellence IX81, Olympus) equipped with a Plan Apochromat 100x/NA 1.45 oil immersion objective and an EMCCD camera Hamamatsu EM-CCD (C9100-02). Acquired Z-stacks were deconvolved with olympus xcellence software using the Wiener Filter. All further processing of digital images was performed with ImageJ.
In vivo mitochondrial import assays
Yeast cells were grown for 2 days in SC-medium containing 2% galactose. Then, cells were diluted to an OD600 of 0.3 and grown for 4 h at 30 ºC. Cells were washed twice with medium lacking amino acids and an equivalent of 3 OD600 per sample was used for radioactive pulse chase experiment. Cells were pulse-labeled by addition of 200 °Ci/ml [35S]-methionine and an amino acid mix lacking methionine and cysteine for 10 min at 30 ºC. Labeling was stopped by addition of 20 mM non-radioactive methionine and 1 mM cysteine. Cells were pelleted, resuspended in SC-medium containing 2% galactose and chased for indicated times at 30 ºC. To dissipate the mitochondrial membrane potential, one sample with a cell equivalent of 6 OD600 was incubated with 10 μM CCCP throughout the experiment. The chase was stopped by trichloroacetic acid precipitation. The resulting protein pellet was dissolved in 50 μl buffer A (6 M Urea, 0.2 M Tris-HCl pH 7.5, 10 mM EDTA, 2% SDS, 100 mM β-mercaptoethanol) by sonication and incubation for 5 min at 55 ºC. Cell walls were disrupted by vortexing for 2 min with glass beads. The samples were subjected to denaturing immunoprecipitation against the HA-epitope tag.
Data analysis
Sequenced reads were processed as described previously (Becker et al., 2013) using standard analysis tools (Bowtie2, Tophat2) and python scripts adapted to S. cerevisiae. Further analyses were performed using customized python scripts. SeRP analyses are based on at least two independent biological replicates that were highly reproducible.
Metagene analysis
For metagene analyses genes were normalized to their expression level by dividing the read density of each nucleotide by the average read density per nucleotide of the respective gene. Analyses were performed for all samples using either all genes that were detected in the dataset or by splitting genes into pre-defined subsets, e.g. based on their cellular localization or according to Table S5 (Jan et al., 2014). Localization information was received from the SGD homepage (www.yeastgenome.org) or – for mitochondrial proteins – collected from several publications (Herrmann and Riemer, 2010; Sickmann et al., 2003; Vogtle et al., 2012; Vogtle et al., 2009; Zahedi et al., 2006). To equalize the length of domains / Ssb binding segments, the reads of all regions were normalized to the length of the shortest region included in the analysis (50 codons in case of domains and 15 nt in case of Ssb binding segments). Due to the delayed Ssb binding to translating ribosomes, the first 50 codons of genes were excluded from the analysis. Ssb binding was assessed considering the distance of 35 residues needed to span the ribosomal tunnel according to WebLogo analysis. Linker regions were defined as regions in between domains and were normalized to a length of 1. Proteins that lack linkers in between domains were not excluded from the analysis but not considered for analyzing Ssb binding to the linker region. For all normalized regions, reads from each gene were binned into the respective length and each bin was averaged. Differences in Ssb-binding between regions were assessed using the Wilcoxon rank-sum test and effect sizes were calculated using Cohen’s d formula. 95% CI of the medians were calculated using the Bootstrapping method.
Substrate identification
Substrates were detected by two methods, either by analyzing the total enrichment of genes between Ssb-bound translatome and total translatome (> 1 in both replicates) or by detecting transient binding peaks. For the peak detection, an algorithm based on the scanning of nucleotide sequences for the appearance of positions with an increased number of reads in the Ssb-bound translatome compared to the translatome was developed. To exclude genes that are expressed close to the background level or have a low read coverage, we defined minimal requirement thresholds that must be all passed before genes were considered for peak detection: i) at least 64 reads in both Ssb-bound translatome data sets; (ii) at least 8 RPKM in both translatome data sets; (iii) Pearson correlation coefficient of the Ssb-bound translatome replicates > 0.5; (iv) at least one position after the first 90 nucleotides in the Ssb-bound translatome that has a two-fold higher read number than the average of the first 90 nucleotides (designated 90 nt background giving the specific background signal for every gene; for genes lacking any read in the 90 nt background, the average read per nucleotide along the complete gene from the corresponding translatome is used).
Genes fulfilling all these requirements were then scanned for binding peaks in both replicates with a minimal width of at least 15 nucleotides. The minimal peak overlap between replicates was 8 nucleotides. We developed two independent selection protocols to identify binding sites of Ssb:
Protocol 1: Footprint enrichment (Ssb-bound translatome/translatome) > 1.5-fold for at least 5 consecutive codons
Protocol 2: Local footprint density over a sequence stretch of minimal 15 nucleotides must be at least three-fold enriched over the 90 nt background (see above) at every position AND every position of the selected sequence stretch must show a 1.5-fold increased footprint enrichment compared to the average footprint enrichment along the complete gene.
WebLogo analysis
WebLogo analyses to detect the binding motif were performed by extracting the sequences near Ssb peaks as indicated and by using the WebLogo online tool Version 2.8.2 (2005-09-08) (http://www.bioinformatics.org/blogo/cgi-bin/Blogo/Blogoform.pl) (Li et al., 2008).
TMD predictions
Transmembrane domains / signal sequences in proteins were predicted using the ΔG prediction server (full protein scan mode (Hessa et al., 2007)), http://dgpred.cbr.su.se.
Translation kinetics analysis
High coverage genes were obtained from list of substrates which have greater than zero reads at every codon position along the transcript. The first 40 codons as well as last 20 codons were excluded from the subsequent analyses of these transcripts since these regions can be influenced by initiation and termination, respectively. Ssb bound and unbound segments were initially defined using the peak detection algorithm in which a region is defined as Ssb bound if its Fold Enrichment (FE) value is greater than 1.5 over a stretch of at least 15 nt. To study the effect of translation rate at the extremities of Ssb-binding probabilities, varying stringency thresholds were set to define the Ssb bound and unbound segments. These thresholds are defined by the percentiles from the Cumulative Distribution Function of FE values (Figures S6B and S6C). Setting an initial threshold of P50, every nucleotide position with an FE value higher than P50 was classified as Ssb bound and every nucleotide position with an FE value lower than the P50 threshold was classified as Ssb unbound (Figure S6D). For all other pairs of thresholds, e.g. (P95, P5), all positions with FE values higher than the upper threshold (e.g. P95) were classified as Ssb bound while all values below the lower threshold (e.g. P5) were classified as Ssb unbound. The other positions with FE values between the thresholds were excluded from the analysis. Ssb bound and unbound segments were defined in the Ssb1-GFP strain background. These regions were then used to perform the relative translation speed analysis in independent translatomes (wt and ssb1∆ssb2∆).
Speed-up of translation
The translation rate was calculated as the inverse of the average number of ribosome reads per nucleotide and translation rate for the Ssb bound and unbound segments computed. To control for expression level differences across the genes, the percent change in translation rate was calculated for each gene separately using the equation where < RB >and < RUB > are the average number of reads per codon in the Ssb bound and unbound segments. The statistical significance of the speed-up across the gene dataset was calculated using the Wilcoxon rank-sum test. Error bars in the associated plots are 95% CI about the median calculated using the Bootstrapping method (Figure 7B).
Contribution of mRNA versus Ssb binding
For every gene in our dataset, we use the percent change calculation described above to estimate the contribution of mRNA and Ssb binding to the translation speed-up using the equation: and % contribution of Ssb binding = 100 – % contribution of mRNA % change ssb1∆ssb2∆ and % changewt correspond to the % change in the ssb1∆ssb2∆ and wt cells, respectively. The error bars in the associated plots are 95% CI about the median calculated using the Bootstrapping method (Figure 7C).
Enrichment/Depletion of Fast/Slow codons
The 61 sense codons were classified as being either Fast or Slow translating based on the local tAI values reported in (Tuller et al., 2010). The 31 codons with the highest tAI values were classified as 'Fast' and the remaining 30 codons as 'Slow'. The probability of finding Fast and Slow codons in the B and UB segments were then calculated and the percent change in these values between these segments computed. The statistical significance of this difference was computed using the paired Permutation test (Good, 2005). 95% CI for the percent change in probability were calculated using Bootstrapping. The enrichment/depletion of proline residues was determined in the same manner.
Upstream charged residues
To test for enrichment/depletion of charged residues in the exit tunnel, we defined a 30 residue window upstream of the Ssb bound and unbound segments along with the region itself. The probability of finding a positively charged residue (K, N, H) and negatively charged residue (D, E) were compared between the defined upstream regions of Ssb bound and unbound segments. We find the results do not change even if overlapping upstream positions of the Ssb bound and unbound segments are excluded from this analysis.
Downstream mRNA secondary structure
In vivo mRNA secondary structure information for all yeast genes was taken from (Rouskin et al., 2014). 'A' and 'C' bases react with DMS if they are not base-paired into the mRNA's secondary structure. Hence, DMS reactivity is inversely proportional to the probability of the nucleotide position forming secondary structure. DMS reactivities of ‘A’ and ‘C’ nucleotides within the Ssb bound and the unbound segments were compared as a function of nucleotide offset downstream of each nucleotide position. The significance of the change in DMS reactivity was assessed using the paired t-test.
Quantification and Statistical Analysis
For none of the experiments blinding or randomization was used. The number of independent biological replicates used for an experiment is indicated in the respective figure legends. The statistical tests and p-values used for the interpretation of data are mentioned in the figure legends and in the method details part of the STAR methods.
Supplementary Material
(A) Overlap of high coverage genes with Ssb bound mRNA segments in Ssb1 wt translatome in two biological replicates. (B) Probability distribution of Fold Enrichment (FE) values across the high coverage gene set. (C) Cumulative Distribution Function (CDF) of FE values. Percentiles were used to define the stringency thresholds to identify Ssb bound and unbound periods. P5, P50 and P95 are shown in red. (D) An example of an Ssb binding profile for the gene CYS3 using different thresholds for the bound and unbound segment definition. The first 120 nt and last 60 nt are excluded from the analysis as well as nucleotide positions exhibiting fold enrichment values between the thresholds (white). Ssb bound (green) and unbound (red) segments are either defined using the P50/P50 threshold (upper panel) or the P95/P5 threshold resulting in less nucleotide positions (i.e., more white space) that are used for the statistical analysis (lower panel). (E) The differences in speed-up between wt and ssb1Δssb2Δ translatomes. (Wilcoxon signed-rank test, p<0.0001 for all thresholds). (F) Depletion of proline residues in Ssb bound compared to unbound segments for the indicated thresholds. (Wilcoxon signed-rank test, p<0.0001 for all thresholds). Error bars show 95% CI. (G) Percent change of probability of finding positively and negatively charged residues in upstream regions of Ssb bound compared to upstream regions of unbound segments for the indicated thresholds. (Wilcoxon signed-rank test, p <0.0001 for negatively charged residues and p >0.05 for positively charged residues for all thresholds). (H) Change in translation speed for Ssb bound and unbound ribosomes in publicly available RP datasets. GEO: GSE63789 (Pop), GSE69414 (Young), GSE61011 (Williams), GSE61012 (Jan), GSE52968 (Guydosh), GSE75322 (Nissley), GSE67387 (Nedialkova), GSE51164 (Gardin), GSE53268 (Weinberg). Error bars show 95% CI. The samples from these datasets were chosen based on the criteria that they do not use CHX for pretreatment and hence capture in vivo translation dynamics reliably.
(A) Ssb binds to emerging recognition motifs (yellow) enriched in positively charged and hydrophobic amino acids. Binding is coordinated by RAC. Ssb bound ribosomes translate faster due to intrinsic features of both, the mRNAs and the nascent chain (enrichment of fast codons, depletion of slow codons, less secondary structures in the mRNA, less proline codons and enrichment of negatively charged amino acids within the ribosomal tunnel). (B) 1. Ssb postpones folding of cytoplasmic and nuclear proteins by rounds of binding to core segments of emerging domains. 2. Rounds of Ssb binding keep mitochondrial and non-SRP class ER-targeted substrates unfolded for translocation. Simultaneous Ssb binding and co-translational translocation is possible as long as ribosomes are not stably docked to the translocon. 3. Ssb binding to SRP class ER-targeted proteins can be explained by two alternative models. Model 1: Ssb binds to RNCs prior to SRP, and is later replaced by SRP for ribosome docking and translocation. Model 2: Ssb and SRP mediate two alternative pathways for targeting RNCs to the ER membrane. The Ssb-mediated targeting pathway would require a so far unknown Ssb binding partner in the ER membrane.
(A) Heatmap and corresponding metagene profile (top) of Ssb binding to RNCs synthesizing mitochondrial proteins and co- localizing with mitochondria (Williams et al., 2014). (B) Mitochondrial import assays in wt and ssb1Δssb2Δ cells analyzing import kinetics of HA-tagged Lpd1 (upper panel) or Pim1 (lower panel). Maturation of proteins was analyzed by radioactive pulse labeling followed by immunoprecipitation, SDS-PAGE and autoradiography. As control, CCCP (Carbonyl cyanide m-chlorophenyl hydrazone) is used to prevent import and specifically enrich the precursor protein. (C) Microscopic analysis of mitochondria in exponentially growing wt and ssb1Δssb2Δ cells. Om45-mcherry served as a mitochondrial marker. Maximum intensity projections of z-stack images are shown. Percentage number of cells exhibiting a given phenotype is indicated (n = 100). (D-G) Heatmaps and metagene binding profiles (top) of Ssb binding to ribosomes synthesizing subsets of ER-targeted proteins. Tail-anchored proteins (Jan et al., 2014) (D), unclassified proteins (E), SND-pathway proteins (Aviram et al., 2016) (F) and SRP substrates with distinct SRP binding peaks (Chartron et al., 2016) (G). (H-I) Heatmap indicating the position of TMDs and signal sequences (SS) upon start of the first Ssb binding region to nascent SRP-independent (H) and SRP-dependent (I) proteins.
(A) Metagene Ssb binding profile. The start of all mRNA segments encoding for three consecutive Arg or Lys amino acids (in arbitrary order) were aligned to codon position 0. (B) DnaK (E. coli) and Ssb1 binding to cellulose-coupled peptide arrays scanning the sequence of the λ cI repressor protein. (C-E) Quantification of Ssb1 binding to peptide arrays scanning the Has1 (C), Pdi1 (D) and Atp14 (E) sequence. Red dashed lines indicate the signal threshold. (F) Correlation of Ssb binding with overall charge of peptides. (G-I) Normalized occurrence of amino acids in Ssb bound or unbound peptides sorted by charge: peptides with a positive net charge (G), peptides with a negative net charge (H) and neutral peptides (I).
(A) Reads per gene in RPM in Ssb1-GFP-bound translatomes purified under low (LS, 140 mM) or high (HS, 500 mM KCl) salt conditions. n=2 (B) Metagene analyses of translatomes and Ssb-bound translatomes from LS and HS conditions. Shaded areas indicate the 95% CI. n=2 (C) Crystal structure of one octameric ring of the TRiC/CCT complex (PDB: 4v94). Individual subunits are shown in different grey shades and Ssb bound regions are indicated in red. Right structure shows the surface interacting with the second TRiC/CCT ring. (D) Cct3 subunit of TRiC/CCT (PDB: 4v94). The three domains are represented in different blue shades and Ssb bound regions are shown in red. (E) Ssb binding to length-normalized domains of single- or multi-domain-proteins as indicated. Shown are metagene Ssb binding profiles aligned to the start of the first domain. White areas indicate length-normalized linker regions. (F) Median of FE of domain and linker regions for proteins with more than three domains. Error bars show 95% CI (Wilcoxon rank-sum test, p<0.05). (G) Mean Cohen’s d effect size. Error bars show standard deviation.
(A) Dissociation kinetics of preformed Ssb complexes with fluorescently labeled peptide measured at 29 °C and 2.9 °C. (B) Western blotting to analyze the Ssb dissociation from ribosomes during sucrose gradient centrifugation. Top: Ssb1-GFP ssb2Δ lysate analyzed by sucrose-gradient centrifugation. Bottom: purified Ssb1-GFP analyzed by sucrose-gradient centrifugation (same amount as in lysate). Loading was as follows: fractions 1, 2 and 8: 1/1090; fractions 3-6: 1/100; fraction 7: 1/1071; fraction 9: 1/235; fraction 10: 1/168. Signals of fractions 4-10 were quantified and values of purified Ssb1-GFP fractions were subtracted from lysate fractions. Amount of Ssb in fractions 4-6 (dissociated Ssb) was compared to amount of ribosome-bound Ssb (fractions 7-10) to calculate the extent of dissociation of Ssb during the 4 h centrifugation. (C) Nascent chain dependent Ssb1-GFP binding to RNCs analyzed by immunoprecipitation and western blotting. (L: lysate, I: resuspended ribosomes used as input for IP, UB: unbound, supernatant of IP, W: first wash fraction, B: bound fraction of IP; CHX: cycloheximide, Puro: puromycin). (D) Quantification of ribosomes co-purified by Ssb-GFP immunoprecipitation analyzed by capillary gel electrophoresis (RNA nano chip, Bioanalyzer).
(A) Ssb-GFP complements the growth defects of a ssb1Δssb2Δ strain. (B) Ssb1-GFP binding to translating ribosomes analyzed by polysome profiling and western blotting using Ssb- and Rpl35-specific antisera. (C) Quantification of RNC binding of Ssb1-GFP added to cell lysates. Purified Ssb-GFP was added to either wt or ssb1Δssb2Δ lysates in the absence or presence of hexokinase. Lysates were loaded onto sucrose cushions, centrifuged and fractions were analyzed by western blotting using Ssb-specific antisera. As a control, a lysate containing endogenously expressed Ssb1-GFP and Ssb2 was treated identically.
Derivations demonstrating that the Fold Enrichment is directly proportional to the Ssb binding probability and that in the Fold Enrichment, the contribution of the elongation rate is eliminated. Related to STAR Methods.
Acknowledgments
We thank members of the Bukau laboratory and S. Anders (ZMBH) for insightful discussions; the DKFZ Genomics & Proteomics Core facility for sequencing; K. Richter and the DKFZ electron microscopy facility; the ZMBH mass spectrometry facility for experimental support; N. Altman for useful discussions on statistical analyses; U. Friedrich, C. Gläßer and D. Schibich for support with data analysis; Elizabeth Craig for providing antibodies. This work was supported by research grants from the HFSP to E.O. and B.B. and the Deutsche Forschungsgemeinschaft to M.P.M. and B.B. (SFB1036).
Footnotes
Contributions
K.D., T.R., G.K. and B.B. conceived the study and designed the experiments. K.D. and T.R. performed the experiments. H.G.S. performed the fluorescence microscopy.
M.H. and J.R. conducted the mitochondrial import assays. K.D., T.R., Y.V., M.P.M., G.K. and B.B. analyzed the data. N.A. and E.P.O. conceived and performed the translation speed analysis. K.D., G.K. and B.B. wrote the manuscript. All authors commented on the manuscript.
Data and Software Availability
The accession number for the data reported in this paper is GEO: GSE93830.
References
- Albanese V, Reissmann S, Frydman J. A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. The Journal of cell biology. 2010;189:69–81. doi: 10.1083/jcb.201001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese V, Yam AY, Baughman J, Parnot C, Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Altmann K, Durr M, Westermann B. Saccharomyces cerevisiae as a model organism to study mitochondrial biology: general considerations and basic procedures. Methods Mol Biol. 2007;372:81–90. doi: 10.1007/978-1-59745-365-3_6. [DOI] [PubMed] [Google Scholar]
- Ast T, Cohen G, Schuldiner M. A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell. 2013;152:1134–1145. doi: 10.1016/j.cell.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Aviram N, Ast T, Costa EA, Arakel EC, Chuartzman SG, Jan CH, Hassdenteufel S, Dudek J, Jung M, Schorr S, et al. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature. 2016;540:134–138. doi: 10.1038/nature20169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker AH, Oh E, Weissman JS, Kramer G, Bukau B. Selective ribosome profiling as a tool for studying the interaction of chaperones and targeting factors with nascent polypeptide chains and ribosomes. Nature protocols. 2013;8:2212–2239. doi: 10.1038/nprot.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehlke KW, Friesen JD. Cellular content of ribonucleic acid and protein in Saccharomyces cerevisiae as a function of exponential growth rate: calculation of the apparent peptide chain elongation rate. Journal of bacteriology. 1975;121:429–433. doi: 10.1128/jb.121.2.429-433.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron JW, Hunt KC, Frydman J. Cotranslational signal-independent SRP preloading during membrane targeting. Nature. 2016 doi: 10.1038/nature19309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Kiktev DA. Dual role of ribosome-associated chaperones in prion formation and propagation. Curr Genet. 2016;62:677–685. doi: 10.1007/s00294-016-0586-2. [DOI] [PubMed] [Google Scholar]
- Chiabudini M, Tais A, Zhang Y, Hayashi S, Wolfle T, Fitzke E, Rospert S. Release factor eRF3 mediates premature translation termination on polylysine-stalled ribosomes in Saccharomyces cerevisiae. Molecular and cellular biology. 2014;34:4062–4076. doi: 10.1128/MCB.00799-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TFt, Clark PL. Rare codons cluster. PloS one. 2008;3:e3412. doi: 10.1371/journal.pone.0003412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA, Jacobsen K. Mutations in cognate genes of Saccharomyces cerevisiae hsp70 result in reduced growth rates at low temperatures. Molecular and cellular biology. 1985;5:3517–3524. doi: 10.1128/mcb.5.12.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Alamo M, Hogan DJ, Pechmann S, Albanese V, Brown PO, Frydman J. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS biology. 2011;9:e1001100. doi: 10.1371/journal.pbio.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M, Hanebuth MA, Frickey T, Deuerling E. The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science. 2015;348:201–207. doi: 10.1126/science.aaa5335. [DOI] [PubMed] [Google Scholar]
- Gautschi M, Lilie H, Funfschilling U, Mun A, Ross S, Lithgow T, Rucknagel P, Rospert S. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:3762–3767. doi: 10.1073/pnas.071057198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M, Mun A, Ross S, Rospert S. A functional chaperone triad on the yeast ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4209–4214. doi: 10.1073/pnas.062048599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R, Beddoe T, Landl K, Lithgow T. The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proc Natl Acad Sci USA. 1998;95:2296–2301. doi: 10.1073/pnas.95.5.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloge F, Becker AH, Kramer G, Bukau B. Co-translational mechanisms of protein maturation. Current opinion in structural biology. 2014;24:24–33. doi: 10.1016/j.sbi.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Good P. Permutation, Parametric, and Bootstrap Tests of Hypotheses. Springer Series in Statistics. 2005 [Google Scholar]
- Gumiero A, Conz C, Gese GV, Zhang Y, Weyer FA, Lapouge K, Kappes J, von Plehwe U, Schermann G, Fitzke E, et al. Interaction of the cotranslational Hsp70 Ssb with ribosomal proteins and rRNA depends on its lid domain. Nature communications. 2016;7 doi: 10.1038/ncomms13563. 13563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanebuth MA, Kityk R, Fries SJ, Jain A, Kriel A, Albanese V, Frickey T, Peter C, Mayer MP, Frydman J, et al. Multivalent contacts of the Hsp70 Ssb contribute to its architecture on ribosomes and nascent chain interaction. Nature communications. 2016;7 doi: 10.1038/ncomms13695. 13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatin I, Fabret C, Namy O, Decatur WA, Rousset JP. Fine-tuning of translation termination efficiency in Saccharomyces cerevisiae involves two factors in close proximity to the exit tunnel of the ribosome. Genetics. 2007;177:1527–1537. doi: 10.1534/genetics.107.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JM, Riemer J. The intermembrane space of mitochondria. Antioxidants & redox signaling. 2010;13:1341–1358. doi: 10.1089/ars.2009.3063. [DOI] [PubMed] [Google Scholar]
- Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Becker AH, Zachmann-Brand B, Deuerling E, Bukau B, Kramer G. Concerted action of the ribosome and the associated chaperone trigger factor confines nascent polypeptide folding. Molecular cell. 2012;48:63–74. doi: 10.1016/j.molcel.2012.07.018. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Bukau B, Kramer G. Structure and function of the molecular chaperone Trigger Factor. Biochimica et biophysica acta. 2010;1803:650–661. doi: 10.1016/j.bbamcr.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Huang P, Gautschi M, Walter W, Rospert S, Craig EA. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat Struct Mol Biol. 2005;12:497–504. doi: 10.1038/nsmb942. [DOI] [PubMed] [Google Scholar]
- Hubscher V, Mudholkar K, Chiabudini M, Fitzke E, Wolfle T, Pfeifer D, Drepper F, Warscheid B, Rospert S. The Hsp70 homolog Ssb and the 14-3-3 protein Bmh1 jointly regulate transcription of glucose repressed genes in Saccharomyces cerevisiae. Nucleic acids research. 2016 doi: 10.1093/nar/gkw168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Williams CC, Weissman JS. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346 doi: 10.1126/science.1257521. 1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. The Journal of cell biology. 2010;189:57–68. doi: 10.1083/jcb.200910074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nature structural & molecular biology. 2009;16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- Lee K, Sharma R, Shrestha OK, Bingman CA, Craig EA. Dual interaction of the Hsp70 J-protein cochaperone Zuotin with the 40S and 60S ribosomal subunits. Nature structural & molecular biology. 2016 doi: 10.1038/nsmb.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidig C, Bange G, Kopp J, Amlacher S, Aravind A, Wickles S, Witte G, Hurt E, Beckmann R, Sinning I. Structural characterization of a eukaryotic chaperone--the ribosome-associated complex. Nature structural & molecular biology. 2013;20:23–28. doi: 10.1038/nsmb.2447. [DOI] [PubMed] [Google Scholar]
- Li W, Yang B, Liang S, Wang Y, Whiteley C, Cao Y, Wang X. BLogo: a tool for visualization of bias in biological sequences. Bioinformatics. 2008;24:2254–2255. doi: 10.1093/bioinformatics/btn407. [DOI] [PubMed] [Google Scholar]
- Liu B, Han Y, Qian SB. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Molecular cell. 2013;49:453–463. doi: 10.1016/j.molcel.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovska L, Kroschwald S, Alberti S. Protein disorder, prion propensities, and self-organizing macromolecular collectives. Biochimica et biophysica acta. 2013;1834:918–931. doi: 10.1016/j.bbapap.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Mashaghi A, Kramer G, Bechtluft P, Zachmann-Brand B, Driessen AJ, Bukau B, Tans SJ. Reshaping of the conformational search of a protein by the chaperone trigger factor. Nature. 2013;500:98–101. doi: 10.1038/nature12293. [DOI] [PubMed] [Google Scholar]
- Merz F, Boehringer D, Schaffitzel C, Preissler S, Hoffmann A, Maier T, Rutkowska A, Lozza J, Ban N, Bukau B, et al. Molecular mechanism and structure of Trigger Factor bound to the translating ribosome. The EMBO journal. 2008;27:1622–1632. doi: 10.1038/emboj.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 KDa heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- Oh E, Becker AH, Sandikci A, Huber D, Chaba R, Gloge F, Nichols RJ, Typas A, Gross CA, Kramer G, et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147:1295–1308. doi: 10.1016/j.cell.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Frydman J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nature structural & molecular biology. 2013;20:237–243. doi: 10.1038/nsmb.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Willmund F, Frydman J. The ribosome as a hub for protein quality control. Molecular cell. 2013;49:411–421. doi: 10.1016/j.molcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke BA, Lopez-Buesa P, Walter WA, Wiedmann M, Craig EA. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 1998;17:3981–3989. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raue U, Oellerer S, Rospert S. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. The Journal of biological chemistry. 2007;282:7809–7816. doi: 10.1074/jbc.M611436200. [DOI] [PubMed] [Google Scholar]
- Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. The EMBO journal. 2006;25:2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saio T, Guan X, Rossi P, Economou A, Kalodimos CG. Structural basis for protein antiaggregation activity of the trigger factor chaperone. Science. 2014;344 doi: 10.1126/science.1250494. 1250494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R, Hurt JA, Krykbaeva I, Taipale M, Lindquist S, Burge CB. Widespread regulation of translation by elongation pausing in heat shock. Molecular cell. 2013;49:439–452. doi: 10.1016/j.molcel.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner L, Wegele H, Buchner J, Morano KA. The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. The Journal of biological chemistry. 2005;280:41262–41269. doi: 10.1074/jbc.M503614200. [DOI] [PubMed] [Google Scholar]
- Shieh YW, Minguez P, Bork P, Auburger JJ, Guilbride DL, Kramer G, Bukau B. Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science. 2015;350:678–680. doi: 10.1126/science.aac8171. [DOI] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, et al. The proteome of Saccharomyces cerevisiae mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A, Mandon EC, Gilmore R, Rapoport TA. Two alternative binding mechanisms connect the protein translocation Sec71/Sec72 complex with heat shock proteins. The Journal of biological chemistry. 2017 doi: 10.1074/jbc.M116.761122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- van der Lee R, Lang B, Kruse K, Gsponer J, Sanchez de Groot N, Huynen MA, Matouschek A, Fuxreiter M, Babu MM. Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell reports. 2014;8:1832–1844. doi: 10.1016/j.celrep.2014.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogtle FN, Burkhart JM, Rao S, Gerbeth C, Hinrichs J, Martinou JC, Chacinska A, Sickmann A, Zahedi RP, Meisinger C. Intermembrane space proteome of yeast mitochondria. Mol Cell Proteomics. 2012;11:1840–1852. doi: 10.1074/mcp.M112.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogtle FN, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickmann A, Pfanner N, et al. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- von Plehwe U, Berndt U, Conz C, Chiabudini M, Fitzke E, Sickmann A, Petersen A, Pfeifer D, Rospert S. The Hsp70 homolog Ssb is essential for glucose sensing via the SNF1 kinase network. Genes & development. 2009;23:2102–2115. doi: 10.1101/gad.529409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmann B, Sakai H, Davis TA, Wiedmann M. A protein complex required for signal-sequence-specific sorting and translocation. Nature. 1994;370:434–440. doi: 10.1038/370434a0. [DOI] [PubMed] [Google Scholar]
- Williams CC, Jan CH, Weissman JS. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmund F, del Alamo M, Pechmann S, Chen T, Albanese V, Dammer EB, Peng J, Frydman J. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell. 2013;152:196–209. doi: 10.1016/j.cell.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Schilke B, Pfund C, Walter W, Kim S, Craig EA. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J. 1998;17:4809–4817. doi: 10.1093/emboj/17.16.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahedi RP, Sickmann A, Boehm AM, Winkler C, Zufall N, Schonfisch B, Guiard B, Pfanner N, Meisinger C. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Molecular biology of the cell. 2006;17:1436–1450. doi: 10.1091/mbc.E05-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Ignatova Z. Folding at the birth of the nascent chain: coordinating translation with co-translational folding. Current opinion in structural biology. 2011;21:25–31. doi: 10.1016/j.sbi.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ma C, Yuan Y, Zhu J, Li N, Chen C, Wu S, Yu L, Lei J, Gao N. Structural basis for interaction of a cotranslational chaperone with the eukaryotic ribosome. Nature structural & molecular biology. 2014;21:1042–1046. doi: 10.1038/nsmb.2908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Overlap of high coverage genes with Ssb bound mRNA segments in Ssb1 wt translatome in two biological replicates. (B) Probability distribution of Fold Enrichment (FE) values across the high coverage gene set. (C) Cumulative Distribution Function (CDF) of FE values. Percentiles were used to define the stringency thresholds to identify Ssb bound and unbound periods. P5, P50 and P95 are shown in red. (D) An example of an Ssb binding profile for the gene CYS3 using different thresholds for the bound and unbound segment definition. The first 120 nt and last 60 nt are excluded from the analysis as well as nucleotide positions exhibiting fold enrichment values between the thresholds (white). Ssb bound (green) and unbound (red) segments are either defined using the P50/P50 threshold (upper panel) or the P95/P5 threshold resulting in less nucleotide positions (i.e., more white space) that are used for the statistical analysis (lower panel). (E) The differences in speed-up between wt and ssb1Δssb2Δ translatomes. (Wilcoxon signed-rank test, p<0.0001 for all thresholds). (F) Depletion of proline residues in Ssb bound compared to unbound segments for the indicated thresholds. (Wilcoxon signed-rank test, p<0.0001 for all thresholds). Error bars show 95% CI. (G) Percent change of probability of finding positively and negatively charged residues in upstream regions of Ssb bound compared to upstream regions of unbound segments for the indicated thresholds. (Wilcoxon signed-rank test, p <0.0001 for negatively charged residues and p >0.05 for positively charged residues for all thresholds). (H) Change in translation speed for Ssb bound and unbound ribosomes in publicly available RP datasets. GEO: GSE63789 (Pop), GSE69414 (Young), GSE61011 (Williams), GSE61012 (Jan), GSE52968 (Guydosh), GSE75322 (Nissley), GSE67387 (Nedialkova), GSE51164 (Gardin), GSE53268 (Weinberg). Error bars show 95% CI. The samples from these datasets were chosen based on the criteria that they do not use CHX for pretreatment and hence capture in vivo translation dynamics reliably.
(A) Ssb binds to emerging recognition motifs (yellow) enriched in positively charged and hydrophobic amino acids. Binding is coordinated by RAC. Ssb bound ribosomes translate faster due to intrinsic features of both, the mRNAs and the nascent chain (enrichment of fast codons, depletion of slow codons, less secondary structures in the mRNA, less proline codons and enrichment of negatively charged amino acids within the ribosomal tunnel). (B) 1. Ssb postpones folding of cytoplasmic and nuclear proteins by rounds of binding to core segments of emerging domains. 2. Rounds of Ssb binding keep mitochondrial and non-SRP class ER-targeted substrates unfolded for translocation. Simultaneous Ssb binding and co-translational translocation is possible as long as ribosomes are not stably docked to the translocon. 3. Ssb binding to SRP class ER-targeted proteins can be explained by two alternative models. Model 1: Ssb binds to RNCs prior to SRP, and is later replaced by SRP for ribosome docking and translocation. Model 2: Ssb and SRP mediate two alternative pathways for targeting RNCs to the ER membrane. The Ssb-mediated targeting pathway would require a so far unknown Ssb binding partner in the ER membrane.
(A) Heatmap and corresponding metagene profile (top) of Ssb binding to RNCs synthesizing mitochondrial proteins and co- localizing with mitochondria (Williams et al., 2014). (B) Mitochondrial import assays in wt and ssb1Δssb2Δ cells analyzing import kinetics of HA-tagged Lpd1 (upper panel) or Pim1 (lower panel). Maturation of proteins was analyzed by radioactive pulse labeling followed by immunoprecipitation, SDS-PAGE and autoradiography. As control, CCCP (Carbonyl cyanide m-chlorophenyl hydrazone) is used to prevent import and specifically enrich the precursor protein. (C) Microscopic analysis of mitochondria in exponentially growing wt and ssb1Δssb2Δ cells. Om45-mcherry served as a mitochondrial marker. Maximum intensity projections of z-stack images are shown. Percentage number of cells exhibiting a given phenotype is indicated (n = 100). (D-G) Heatmaps and metagene binding profiles (top) of Ssb binding to ribosomes synthesizing subsets of ER-targeted proteins. Tail-anchored proteins (Jan et al., 2014) (D), unclassified proteins (E), SND-pathway proteins (Aviram et al., 2016) (F) and SRP substrates with distinct SRP binding peaks (Chartron et al., 2016) (G). (H-I) Heatmap indicating the position of TMDs and signal sequences (SS) upon start of the first Ssb binding region to nascent SRP-independent (H) and SRP-dependent (I) proteins.
(A) Metagene Ssb binding profile. The start of all mRNA segments encoding for three consecutive Arg or Lys amino acids (in arbitrary order) were aligned to codon position 0. (B) DnaK (E. coli) and Ssb1 binding to cellulose-coupled peptide arrays scanning the sequence of the λ cI repressor protein. (C-E) Quantification of Ssb1 binding to peptide arrays scanning the Has1 (C), Pdi1 (D) and Atp14 (E) sequence. Red dashed lines indicate the signal threshold. (F) Correlation of Ssb binding with overall charge of peptides. (G-I) Normalized occurrence of amino acids in Ssb bound or unbound peptides sorted by charge: peptides with a positive net charge (G), peptides with a negative net charge (H) and neutral peptides (I).
(A) Reads per gene in RPM in Ssb1-GFP-bound translatomes purified under low (LS, 140 mM) or high (HS, 500 mM KCl) salt conditions. n=2 (B) Metagene analyses of translatomes and Ssb-bound translatomes from LS and HS conditions. Shaded areas indicate the 95% CI. n=2 (C) Crystal structure of one octameric ring of the TRiC/CCT complex (PDB: 4v94). Individual subunits are shown in different grey shades and Ssb bound regions are indicated in red. Right structure shows the surface interacting with the second TRiC/CCT ring. (D) Cct3 subunit of TRiC/CCT (PDB: 4v94). The three domains are represented in different blue shades and Ssb bound regions are shown in red. (E) Ssb binding to length-normalized domains of single- or multi-domain-proteins as indicated. Shown are metagene Ssb binding profiles aligned to the start of the first domain. White areas indicate length-normalized linker regions. (F) Median of FE of domain and linker regions for proteins with more than three domains. Error bars show 95% CI (Wilcoxon rank-sum test, p<0.05). (G) Mean Cohen’s d effect size. Error bars show standard deviation.
(A) Dissociation kinetics of preformed Ssb complexes with fluorescently labeled peptide measured at 29 °C and 2.9 °C. (B) Western blotting to analyze the Ssb dissociation from ribosomes during sucrose gradient centrifugation. Top: Ssb1-GFP ssb2Δ lysate analyzed by sucrose-gradient centrifugation. Bottom: purified Ssb1-GFP analyzed by sucrose-gradient centrifugation (same amount as in lysate). Loading was as follows: fractions 1, 2 and 8: 1/1090; fractions 3-6: 1/100; fraction 7: 1/1071; fraction 9: 1/235; fraction 10: 1/168. Signals of fractions 4-10 were quantified and values of purified Ssb1-GFP fractions were subtracted from lysate fractions. Amount of Ssb in fractions 4-6 (dissociated Ssb) was compared to amount of ribosome-bound Ssb (fractions 7-10) to calculate the extent of dissociation of Ssb during the 4 h centrifugation. (C) Nascent chain dependent Ssb1-GFP binding to RNCs analyzed by immunoprecipitation and western blotting. (L: lysate, I: resuspended ribosomes used as input for IP, UB: unbound, supernatant of IP, W: first wash fraction, B: bound fraction of IP; CHX: cycloheximide, Puro: puromycin). (D) Quantification of ribosomes co-purified by Ssb-GFP immunoprecipitation analyzed by capillary gel electrophoresis (RNA nano chip, Bioanalyzer).
(A) Ssb-GFP complements the growth defects of a ssb1Δssb2Δ strain. (B) Ssb1-GFP binding to translating ribosomes analyzed by polysome profiling and western blotting using Ssb- and Rpl35-specific antisera. (C) Quantification of RNC binding of Ssb1-GFP added to cell lysates. Purified Ssb-GFP was added to either wt or ssb1Δssb2Δ lysates in the absence or presence of hexokinase. Lysates were loaded onto sucrose cushions, centrifuged and fractions were analyzed by western blotting using Ssb-specific antisera. As a control, a lysate containing endogenously expressed Ssb1-GFP and Ssb2 was treated identically.
Derivations demonstrating that the Fold Enrichment is directly proportional to the Ssb binding probability and that in the Fold Enrichment, the contribution of the elongation rate is eliminated. Related to STAR Methods.