Abstract
Innate lymphoid cells (ILCs) reside in mucosal surfaces to potentiate immune responses, sustain mucosal integrity and maintain tissue homeostasis. However, how tumor infiltrating ILCs modulate tumor development and progression is unclear. Here we profiled tumor infiltrating ILCs during colorectal cancer (CRC) progression by single-cell RNA sequencing. We identified six clusters of tumor infiltrating ILCs with unique features. ILC1s expressed inhibitory receptors and underwent inhibitory functional conversion at the late stage of CRC. ILC2s were classified into three subsets (called ILC2-A, -B, -C), of which ILC2-C subset could facilitate tumor progression. HS3ST1 and PD1 were highly expressed in ILC2s of late stage CRC tumors and deficiency of HS3ST1 or PD1 in ILC2s suppressed tumor growth. Moreover, ILC3s transdifferentiated into ILCregs during CRC progression and ILCregs promoted tumor growth. Of note, TGF-β signaling initiated the conversion of ILC3s to ILCregs and blockade of TGF-β signaling could disrupt the ILCreg transdifferentiation and inhibited tumor growth. Thus, intervention of ILC conversions might be a potential strategy for CRC immunotherapy.
Subject terms: Tumour immunology, Tumour immunology
Introduction
The innate immune system provides first-line defence against invading pathogens by triggering inflammatory and antimicrobial responses. Although robust inflammation contributes to the clearance of pathogens, inflammatory disorders and chronic inflammation are also correlated with tumor development and progression.1–4 In addition, several cancers display characteristics of chronic inflammation during their progression. Colorectal cancer (CRC) is considered as a good example of a tumor correlated with chronic inflammation.2,5 Accumulating evidence shows that inflammatory disorder has substantial effect on CRC development and progression.4,6
Tumor infiltrating lymphocytes (TILs) are thought to be a sign of attacking transformed cells by the immune system at an early stage.7 Tumor cells can be recognized by TILs, which produce large amounts of IFN-γ (such as NK, NKT, and γδT cells) or directly kill transformed cells (such as NK and cytotoxic T lymphocyte (CTL cells)), leading to elimination of tumor cells.8,9 TILs withstand immune selection pressure against tumor cells and immunoevasive mutations might be acquired, which lead to tumor escape from immunosurveillance.10 Meanwhile, chronic inflammation caused by immune dysregulation promotes tumor development. Inflammatory microenvironment alters the fate of lymphocytes by various mediators such as cytokines and chemokines. For instance, tumor cells produce immunosuppressive cytokines such as vascular endothelial growth factor (VEGF), transforming growth factor-β (TGF-β), which convert effector T cells into regulatory T cells (Tregs). Tregs secrete IL-10 and produce PD-L1 and CTLA-4 to suppress CTL functions.11 In addition, continuous tumor antigen stimulation maintains the expression of inhibitory receptors on T cells, such as PD-1 and CTLA-4, which disarms effector T cells leading to tumor development and progression.12 At present, therapeutic applications targeting these immunosuppressive molecules become promising strategies for antitumor therapy.
Innate lymphoid cells (ILCs) are innate lymphocytes that lack other lineage markers and contribute to early pathogen defence.13 ILCs were previously categorized into three subgroups, namely group 1 ILC (ILC1s), group 2 ILC (ILC2s) and group 3 ILC (ILC3s), according to their featured cytokine profiles and unique transcription factors.14 ILC1s are characterized by expressing T-bet and producing IFN-γ, which is essential for clearance of intracellular microbial infections.15,16 ILC2s require Gata3 for their differentiation and maintenance.17 After activation by IL-25 and IL-33, ILC2s produce cytokines, such as IL-5 and IL-13, to promote resolution of helminth infections and to participate in pathogenesis of asthma.18,19 ILC3s are defined by RORγt expression and generate cytokines such as IL-22 and IL-17, which are extremely critical for resistance against bacterial infections.20,21 We recently defined a new regulatory subpopulation of ILCs named ILCregs, which contain Id3 and produce IL-10 to modulate inflammatory response.22 However, it is unknown about intratumoral heterogeneity of ILC populations and their associations with tumor progression. Here we used single-cell RNA sequencing (scRNA-seq) to profile tumor infiltrating ILCs in CRC at early and late stages. We identified six clusters of ILCs inside colon tumors and revealed conversions of their functions and subsets during CRC progression, and blocking ILC transdifferentiation could suppress tumor development.
Results
Profiling of tumor infiltrating ILCs in early and late stages of colorectal tumor
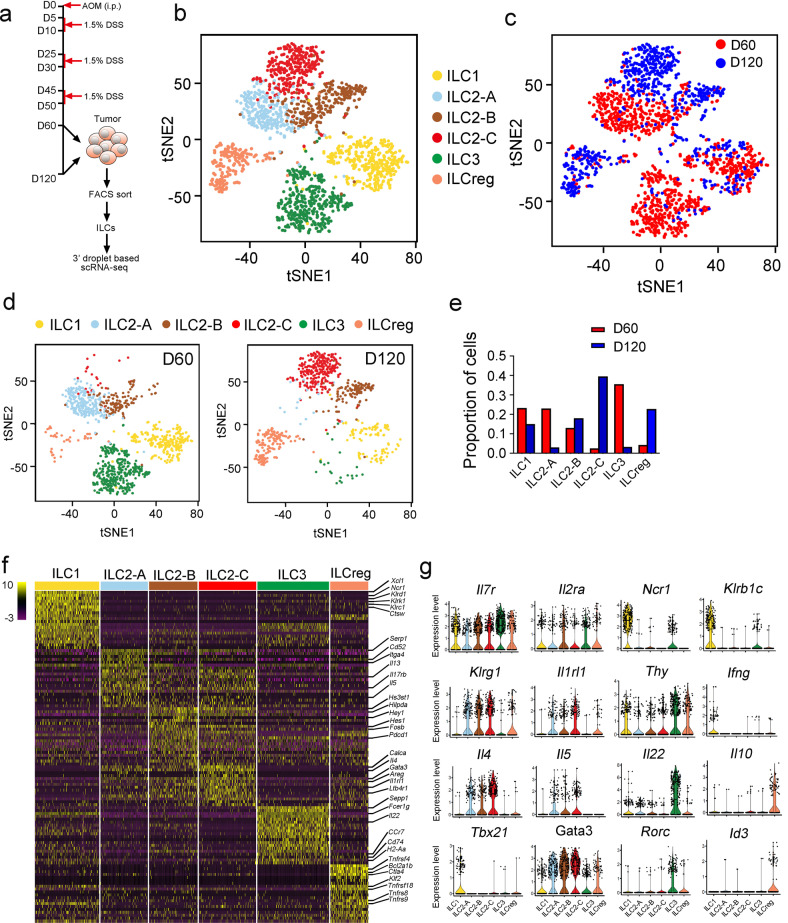
In order to explore dynamic regulation of tumor infiltrating ILCs during CRC progression, we used azoxymethane/dextran sodium sulfate (AOM/DSS)-induced colitis-associated CRC, which mimics tumor progression caused by chronic colitis as seen in inflammatory bowel disease (IBD).23,24 We isolated tumor infiltrating ILCs and conducted droplet-based scRNA-seq to profile ILCs inside colon tumors at day 60 (early stage) and day 120 (late stage) (Fig. 1a). We excluded contaminating lineage cells and low quality cells, and finally obtained 1,654 ILC cells (Supplementary information, Fig. S1a–f). Unsupervised graph clustering divided ILCs into six groups, and we labeled them with known ILC markers (Supplementary information, Fig. S1g).25,26 Six ILC subsets inside CRC tumors included ILC1s, three groups of ILC2s (termed ILC2-A, B, C), ILC3s and ILCregs (Fig. 1b). Based on scRNA-seq data, cell proportions were dynamically changed during tumor progression (Fig. 1c–e). ILC1 cells slightly reduced at the late stage of CRC tumors (Fig. 1c–e). ILC-2A cells were dominant ILC2 cells at the early stage of CRC tumors (Fig. 1c–e). ILC2-B cells distributed similarly at both early and late stages. However, ILC2-C cells mainly existed in colon tumors at the late stage (Fig. 1c–e). Of note, ILC3 cells mainly resided at the early stage of CRC tumors, whereas ILCregs appeared at the late stage of CRC tumors (Fig. 1c–e).
Fig. 1. Single-cell expression survey of tumor infiltrating ILCs.
a Strategy for scRNA-seq of tumor infiltrating ILCs. WT mice were i.p. injected with azoxymethane (AOM, 10 mg/kg body weight), and treated with 1.5% DSS in drinking water for three cycles as shown. Colon tumor infiltrating ILCs (Lin−CD45+CD127+) (Lin = CD3e,CD8a,CD19,CD11b,CD11c,Gr1,F4/80,Ter119) from tumors at day 60 (D60) or day 120 (D120) were isolated by flow cytometry and subjected to 3’ droplet-based scRNA-seq. Purity of ILC cells was over 95% for each sample that was determined by post sorting analysis of flow cytometry. b t-distributed stochastic neighbor embedding (tSNE) plots showed distribution of ILC clusters (ILC1 = 325 cells, ILC2-A = 289 cells, ILC2-B = 246 cells, ILC2-C = 239 cells, ILC3 = 363 cells, ILCreg = 192 cells). Low quality cells were removed as described in Materials and Methods. Contaminating lineage cell clusters were also removed according to the expression of lineage markers. c ILCs in tumor from different stages (D60 and D120) were shown by tSNE plot (D60 = 973 cells, D120 = 681 cells). d, e Tumor infiltrating ILC subsets from different stages were analyzed by tSNE (d) and proportions of each cell subset at D60 and D120 respectively were calculated and shown in (e). f Heat map of top 20 signature genes for indicated cell types were shown. g Distribution of expression levels of ILC marker genes in the indicated clusters were shown by violin plots.
We next assessed signature genes for each cell type and defined many known and newly identified markers (Fig. 1f). Tumor infiltrating ILC2 subsets shared several common ILC2 markers, and also harbored their unique signature expression patterns (Fig. 1g). All of the identified ILC subsets in CRC tumors expressed common ILC signature genes such as Il7r and Il2ra (Fig. 1g). Notably, three ILC2 subsets highly expressed Klrg1, Il1rl1 (encoding ST2), and Gata3. Thus, unbiased analyses of tumor infiltrating ILCs reveal dynamic changes of ILC subsets and different gene expression profiles inside CRC tumors during CRC progression.
Characterizing ILC1 cells in colorectal tumor
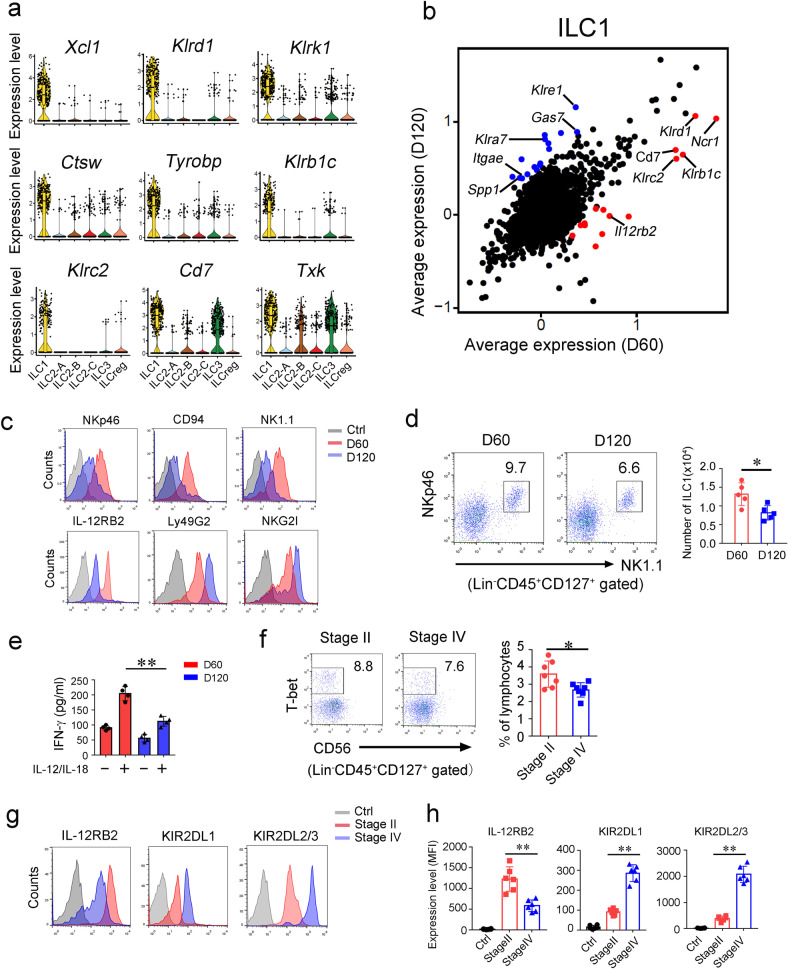
It has been reported that ILC1-like cells in mammary tumor exert tumor suppressive functions.27 The exact role of ILC1s inside CRC tumor is unknown. According to our scRNA seq data, we identified top 9 signature genes of tumor infiltrating ILC1s. Of these 9 genes, Tyrobp, Klrd1, Klrk1, Klrb1c and Klrc2 were previously identified as ILC1-related genes.25,28 In addition, Ctsw (encoding cathepsin W preproprotein), Txk (encoding RIk kinase), and Cd7 were also highly expressed in ILC1 cells (Fig. 2a). We next compared gene expression profiles of tumor infiltrating ILC1s at early stage versus late stage. Intriguingly, ILC1s at the early stage expressed high levels of activating receptors (Klrd1, Ncr1, Klrc2, Klrb1c), whereas they expressed inhibitory receptors (Klre1, Klra7) at the late stage (Fig. 2b).29 These observations were further validated by flow cytometry (Fig. 2c). However, expression levels of these receptors did not show significant changes on ILC1s of peri-tumor tissues (Supplementary information, Fig. S2a). We found that ILC1 cells inside CRC tumors slightly declined at the late stage (Fig. 2d), but no overt changes in peri-tumor tissues between the early stage and the late stage (Supplementary information, Fig. S2b). Furthermore, Il12rb2 was decreased in ILC1s in the late stage (Fig. 2b), suggesting impaired response of ILC1s to IL-12 stimulation. IFN-γ plays a critical role in eradication of early tumors.8,30 We isolated ILC1s from early and late stages of CRC tumors and stimulated with IL-12 plus IL-18 in vitro. We found that IFN-γ production was remarkably decreased (Fig. 2e). We also assessed tumor infiltrating ILC1s in CRC patients according to the defined markers on human ILC1s.16 We observed that tumor infiltrating ILC1s in advanced CRC patients showed lower frequencies with high levels of inhibitory receptors (Fig. 2f–h). In contrast, these receptors did not show apparent changes in peri-tumor tissues (Supplementary information, Fig. S2c). Taken together, ILC1s undergo functional change during CRC progression.
Fig. 2. ILC1s undergo functional conversion during CRC progression.
a Expression distributions of selected ILC1 signature genes were shown by violin plots. b Scatter plot displaying gene expression of ILC1s at early and late tumor stages. Scale data generated from Seurat analysis for the scatter plot. c Indicated surface markers of ILC1s were analyzed by flow cytometry. d Flow cytometry analysis of ILC1s (Lin−CD45+CD127+NK1.1+NKp46+) inside colon tumors of indicated stages. ILC1 numbers per cm3 colon tumor were calculated and shown as means ± SD (right panel). *P < 0.05 by Two-tailed unpaired Student’s t test. n = 5 for each group. e ILC1s (Lin−CD45+CD127+NK1.1+NKp46+) in tumors at indicated stages were isolated and treated with 10 ng/mL IL-12/IL-18 for 12 h. Secreted IFN-γ was analyzed by ELISA and shown as means ± SD. **P < 0.01 by Two-tailed unpaired Student’s t test. n = 4 for each group. f Tumor infiltrating ILC1s from CRC patient samples of indicated stages were analyzed by flow cytometry. The percentage of ILC1 in lymphocyte population was calculated and shown as means ± SD (right panel). *P < 0.05 by Two-tailed unpaired Student’s t test. n = 6 for each group. (human ILC1 = Lin−CD45+CD127+cKit−NKp44−CD56−Tbet+, Lin = CD2, CD3, CD14, CD16, CD19, CD56, CD235a). g, h Surface markers of ILC1s in tumors from CRC patient samples were analyzed by flow cytometry. Isotype Ig of each antibody was used as a negative control (Ctrl). Medians of fluorescence intensity (MFI) of each cell were calculated and shown as means ± SD. **P < 0.01 by One Way ANOVA. n = 6 for each group. For d–e, data are representative of at least four independent experiments. For f–h, data are representative of at least six independent experiments.
HS3ST1 and PD1 are highly expressed in tumor filtrating ILC2 cells of late stage CRC tumors
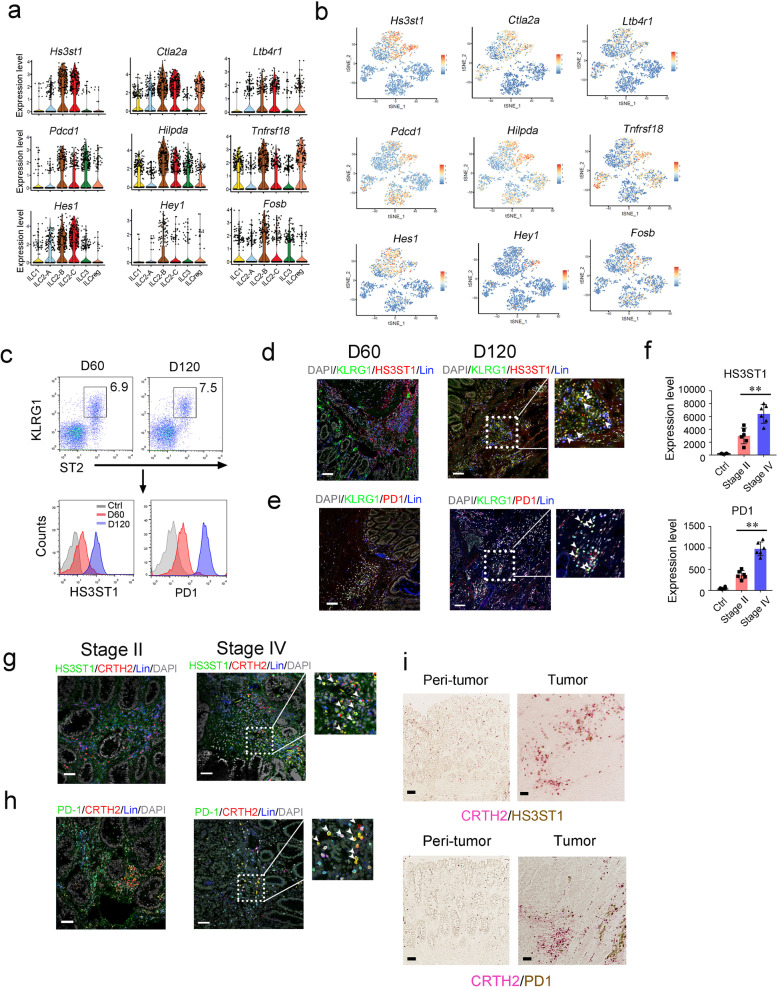
As shown in Fig. 1g, all three ILC2 subsets expressed ILC2 signature genes. Besides these signature genes, we also identified several genes in tumor infiltrating ILC2s that were gradually induced during CRC progression, including Hs3st1, Ctla2a, Ltb4r1, Pdcd1, Tnfrsf18, and Hes1 (Fig. 3a, b). These genes were indeed highly expressed in the late stage of CRC tumors (Supplementary information, Fig. S3a). Of these induced genes in ILC2s, we focused on Hs3st1 and Pdcd1. Hs3st1, encoding heparan sulfate 3-O-sulfotransferase 1 (HS3ST1), catalyzes biosynthesis of heparan sulfate.31,32 Hs3st1 was restrictedly expressed in ILC2s and substantially induced during CRC progression (Fig. 3a, b). Pdcd1, encoding PD1, is an immune checkpoint receptor,33,34 which was also highly expressed in ILC2 cells over CRC progression, suggesting a critical role of Pdcd1 for ILC2 function. Consistent with the mRNA levels, these two molecules were indeed highly expressed in ILC2s in late stage CRC tumors by flow cytometry (Fig. 3c). These observations were further validated by immunofluorescence staining (Fig. 3d, e). Intriguingly, HS3ST1 and PD1 were substantially expressed in ILC2s of human advanced CRC tumor tissues (Fig. 3f–h). Furthermore, these two molecules were restrictedly expressed in tumor tissues but not in peri-tumor tissues (Fig. 3i). Collectively, HS3ST1 and PD1 are highly expressed in tumor filtrating ILC2 cells of late stage CRC tumors.
Fig. 3. Characterization of ILC2 subsets in colorectal cancer.
a Violin plots showed 9 signature genes of ILC2s during tumor progression. b Scale data derived from Seurat analysis pipeline to plot the ILC2 signature genes on a tSNE plot. c Analysis of HS3ST1 and PD1 expression in mouse tumor infiltrating ILC2s (Lin−CD45+CD127+ST2+KLRG1+) by flow cytometry. d, e Immunofluorescence staining of HS3ST1 or PD1 (red) on ILC2 cells (KLRG1+, green) of early stage tumors (D60) or late stage tumors (D120). Scale bar, 50 μm. White arrowhead indicates ILC2s. f Expression level of indicated protein on tumor infiltrating ILC2s (Lin−CD45+CD127+CRTH2+) from colorectal cancer patients were analyzed by flow cytometry. Medians of fluorescence intensity (MFI) of each cell were calculated and shown as means ± SD. **P < 0.01 by One Way ANOVA. n = 6 for each group. g, h Immunofluorescence staining of HS3ST1 or PD1 (green) of ILC2s (CRTH2+, red) from stage II or stage IV CRC patient samples. Scale bar, 50 μm. i Sections of peri-tumor and tumor tissues from stage IV CRC patients were subjected to immunohistochemistry (IHC) staining of CRTH2 (red) and HS3ST1 or PD1 (brown). Scale bar, 50 μm. For c–e, data are representative of four independent experiments. For f, data are representative of six independent experiments. For g–i, data are representative of three independent experiments.
ILC2-C subset promotes CRC progression
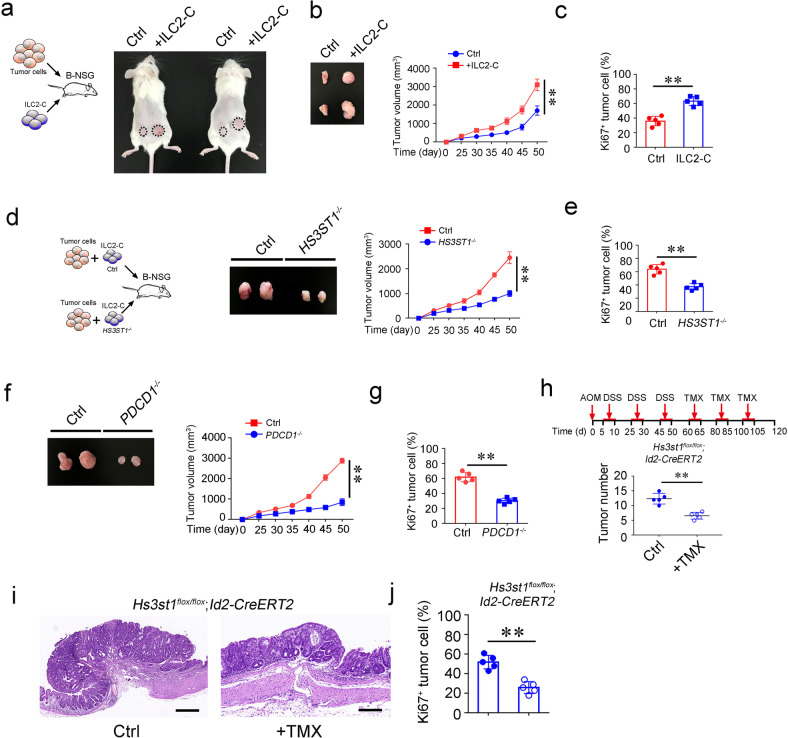
Tumor infiltrating ILC2s expressed high levels of PD1 in human advanced CRC tumors. ILC2s expressed differential levels of PD1 over CRC stages (Fig. 3f). We thus defined PD1low ILC2s as ILC2-A in human CRC tumors and PD1high ILC2s as ILC2-C. In order to determine the role of ILC2s in the tumor growth of human CRC, we isolated human PD1high ILC2-C cells from advanced CRC samples and engrafted them with tumor cells into B-NSG mice. We found that ILC2-C cells apparently promoted tumor growth, whereas ILC2-A cells had no such effect (Fig. 4a, b; Supplementary information, Fig. S3b). Additionally, tumor cells showed increased proliferation rates with ILC2-C cell engraftment (Fig. 4c). These data suggest that ILC2-C cells facilitate tumor growth. We next generated HS3ST1- or PD1-deficient ILC2-C cells by CRISRP-Cas9 technology and transferred them together with tumor cells into B-NSG mice (Fig. 4d; Supplementary information, Fig. S3c). Deletion of HS3ST1 or PD1 in ILC2-C cells did not apparently changed cell death (Supplementary information, Fig. S3d). Deletion of HS3ST1 or PD1 in human ILC2-C cells markedly suppressed tumor growth (Fig. 4d, f; Supplementary information, Fig. S3e), and inhibited tumor cell proliferation rates as well (Fig. 4e, g). In order to verify the role of HS3ST1 of ILC2s in CRC progression, we conditionally knocked out Hs3st1 in ILC2s (Supplementary information, Fig. S3f). Given that Hs3st1 was extremely highly expressed in ILC2s compared with other immune cells (Supplementary information, Fig. S3g), we thus crossed Hs3st1flox/flox mice with Id2-CreERT2 mice to delete Hs3st1 in ILCs. With administration of tamoxifen, HS3ST1 was efficiently depleted in ILC2s (Supplementary information, Fig. S3h). Of note, deficiency of HS3ST1 suppressed AOM/DSS-induced colon tumor development (Fig. 4h–i), and proliferation rates of tumor cells (Fig. 4j). Whereas tamoxifen did not affect tumor growth (Supplementary information, Fig. S3i). In addition, we used anti-PD1 antibody to inject into CRC patient-derived tumor cell (PDC) tumors once a week. Of note, anti-PD1 antibody treatment remarkably suppressed tumor growth (Supplementary information, Fig. S3j), suggesting that blocking PD1 on ILC2s might be a potential target against CRC patients. Collectively, ILC2-Cs promote tumor growth and HS3ST1 and PD1 in ILC2s are involved in the CRC tumor progression.
Fig. 4. ILC2-C subset facilitates colorectal cancer growth.
a ILC2-C cells promote colon tumor cell growth. Primary tumor cells (1 × 106) and PD1hi ILC2-C (ILC2-C = Lin−CD45+CD127+CRTH2+PD1high, 1 × 105 sorted by FACS) from stage IV CRC patients were isolated and subcutaneously injected into B-NSG (NOD-PrkdcscidIL2rgtm1/Bcgen) mice. ILC2s (Lin−CD45+CD127+CRTH2+, 1 × 105 sorted by FACS) from peri-tumor tissues were co-injected with tumor cells (1 × 106) into the other side of mice and served as negative control (Ctrl). After 50 days, tumor photos were shown. b Tumor volumes were measured at indicated time points and calculated as means ± SD. **P < 0.01 by Non parametric Mann–Whitney U-test. n = 5 for each group. Isolated tumors were shown in the left panel. c Proliferation of tumor cells was analyzed with Ki67 staining by flow cytometry. **P < 0.01 by One Way ANOVA. n = 5 for each group. d ILC2-Cs were isolated by FACS and transfected with virus carrying CRISPR/Cas9 elements against HS3ST1 gene. ILC2-C cells transfected with control element served as negative control (Ctrl). Control or HS3ST1−/− ILC2-C cells together with primary tumor cells from stage IV CRC patients were subcutaneously injected into B-NSG mice. Subcutaneous tumors were measured at the indicated time points. Tumor sizes were calculated as means ± SD. **P < 0.01 by Non parametric Mann–Whitney U-test. n = 5 for each group. After 50 days, tumors were isolated and shown as left panel. e Ki67 staining of tumor cells from indicated mice treated as (d) was analyzed by flow cytometry. **P < 0.01 by Two-tailed unpaired Student’s t test. n = 5 for each group. f PDCD1−/− ILC2-Cs were generated as in (d). Control or PDCD1−/− ILC2-Cs together with primary tumor cells were subcutaneously injected into B-NSG mice. Tumor sizes were calculated and shown as means ± SD. **P < 0.01 by Non parametric Mann–Whitney U-test. n = 5 for each group. g Analysis of tumor cell proliferation by Ki67 staining and shown as means ± SD. **P < 0.01 by Two-tailed unpaired Student’s t test. n = 5 for each group. h Hs3st1flox/flox;Id2-CreERT2 mice were subjected to AOM/DSS treatment. At day 60, mice were treated with tamoxifen (TMX) (50 mg/kg i.p. for six consecutive days every 20 days). Vehicle treatment served as negative control (Ctrl). At day 120, colon tumors of indicated mice were calculated and shown as means ± SD. **P < 0.01 by Two-tailed unpaired Student’s t test. n = 5 for each group. i H&E staining of colon tumors from Hs3st1flox/flox;Id2-CreERT2 mice with indicated treatment. Scale bar, 50 μm. j Ki67 staining of tumor cells from mice with indicated treatment was analyzed by flow cytometry and shown as means ± SD. Solid circles indicate control (Ctrl) and open circles indicate tamoxifen treatment (TMX). **P < 0.01 by Two-tailed unpaired Student’s t test. n = 5 for each group. For a–j, data are representative of five independent experiments.
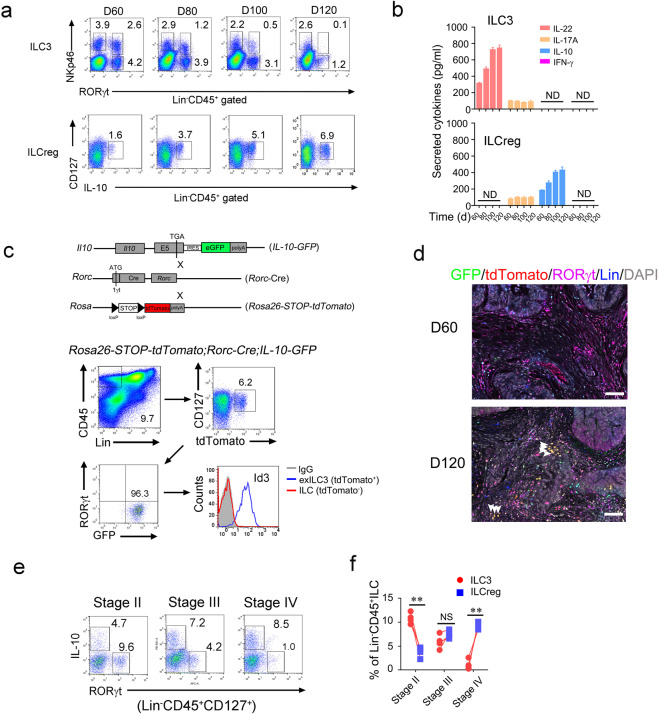
Transdifferentiation of ILC3 to ILCreg during CRC progression
Based on our scRNA seq data, the proportion of ILC3s was decreased with increased ILCregs over CRC tumor progression. We used diffusion map to analyze distributions of ILC3 and ILCreg. We observed that conversion of ILC3s to ILCregs at the late stage of CRC tumor (Supplementary information, Fig. S4a). In parallel, ILC3s decreased during CRC tumor progression, whereas ILCregs gradually increased (Fig. 5a). At day 120, ILC3s dramatically decreased and accompanied with a large number of ILCregs inside CRC tumors (Fig. 5a). In addition, tumor infiltrating ILC3s mainly produced IL-22, while ILCregs majorly secreted IL-10 (Fig. 5b). In order to trace the fate of lost ILC3s in CRC tumors, we generated Rosa26-STOP-tdTomato;Rorc-Cre;IL-10-GFP lineage tracing mice and followed by AOM/DSS treatment. We found that Lin−tdTomato+RORγt− cells used to be ILC3 (called exILC3) appeared in late stage CRC tumors, but not in early stage tumors (Fig. 5c and Supplementary information, Fig. S4b). Moreover, exILC3s expressed IL-10 and Id3 (Fig. 5c), which are signature markers of ILCregs.22 Moreover, tdTomato+RORγt- ILCregs only appeared at late stage tumors (Supplementary information, Fig. S4c), suggesting conversion of ILC3s to ILCregs over CRC progression. Consistently, exILC3s displayed in late stage tumors, but not in early stage tumors (Fig. 5d). In addition, tumor infiltrating ILC3s overtly decreased and ILCregs increased in human advanced CRC tumors (Fig. 5e, f). Notably, ILCregs in CRC tumors did not express signature markers of other ILCs (Supplementary information, Fig. S4d). Altogether, ILC3s transdifferentiate into ILCregs during CRC progression.
Fig. 5. Conversion of ILC3s to ILCregs during colorectal tumor progression.
a Analysis of ILC3s and ILCregs during progression of AOM/DSS-induced colorectal tumor by flow cytometry at the indicated time points. b Tumor infiltrating ILC3s (from RORγt-GFP mice) and ILCregs (from IL-10-GFP mice) were isolated for ELISA assay and secreted cytokines were shown as means ± SD. ND, not detectable. c Lineage tracing of ILC3 cells during tumor progression. Rosa26-STOP-tdTomato;Rorc-Cre;IL-10-GFP mice were treated with AOM/DSS as described previously. exILC3s (Lin−CD45+CD127+tdTomato+RORγt-) in colorectal tumor of the indicated mice were analyzed by flow cytometry. d Immunofluorescence staining of ILC3s and ILCregs in tumor section from Rosa26-STOP-tdTomato;Rorc-Cre;IL-10-GFP mice. Scale bar, 50 μm. White arrowheads denote tdTomato+ILCregs. e, f Analysis of ILC3s and ILCregs in CRC samples by flow cytometry (e). Lin−CD45+CD127+ cells were gated out for IL-10 and RORγt staining. Cell frequencies of ILCs were calculated and shown as means ± SD (f). **P < 0.01 by One Way ANOVA. NS, not significant by One Way ANOVA. n = 4 for each group. For a–d, data represent three independent experiments. For e–f, data represent four independent experiments.
TGF-β induces the conversion of ILC3 to ILCreg
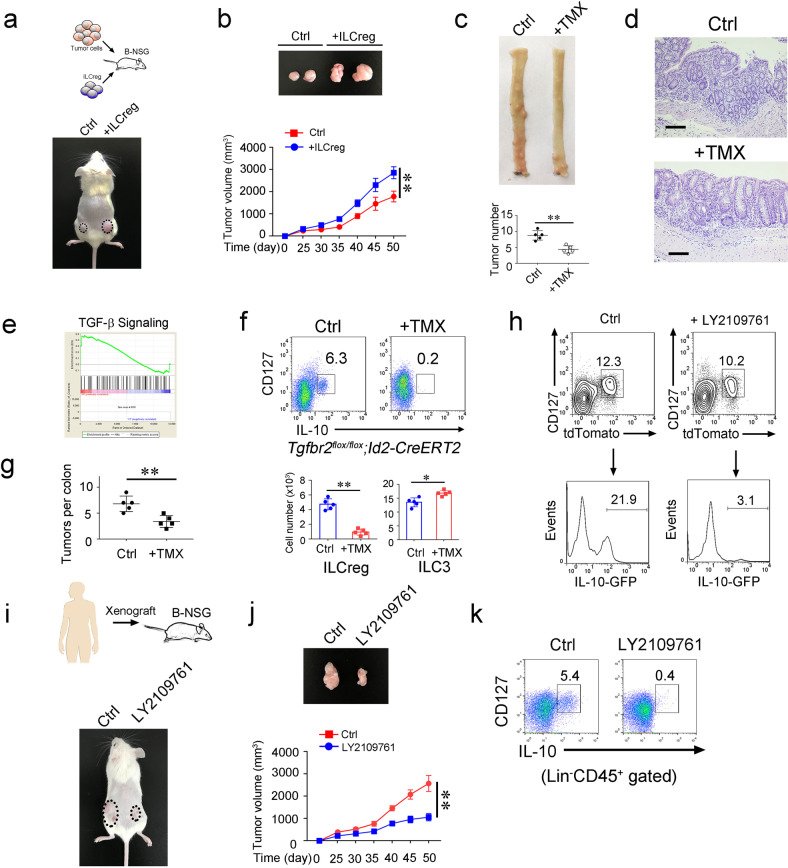
To explore the role of ILCregs in CRC progression, we isolated tumor infiltrating ILCregs from CRC tumors and transferred them together with tumor cells into B-NSG mice (Fig. 6a). We observed that ILCregs apparently promoted tumor growth (Fig. 6a, b). We previously showed that the fate decision factor Id3 drives progenitor CHILP cells toward ILCregs.22 We noticed that Id3 also existed in exILC3s (Fig. 5c). We established Id3flox/flox;Id2-CreERT2 mice to deplete ILCregs, followed by AOM/DSS treatment (Supplementary information, Fig. S5a). Deletion of Id3 indeed depleted ILCregs in CRC tumors, whereas ILC3s slightly increased (Supplementary information, Fig. S5a, b). By contrast, other ILCs and Tregs were not apparently changed (Supplementary information, Fig. S5b). Of note, depletion of ILCregs dramatically suppressed tumor growth (Fig. 6c–d). ILCregs are able to produce large amounts of IL-10 that participates in immunosuppression of tumor progression. We next used CRISRP/Cas9 technology to delete IL-10 in ILCregs as previously described,22 followed by adoptive transfer assays with IL-10-deleted ILCregs (Supplementary information, Fig. S5c). Notably, deletion of IL-10 in ILCregs dramatically suppressed tumor development (Supplementary information, Fig. S5d), suggesting that ILCregs promote tumor progression in an IL-10 dependent manner. These data indicate that ILCregs promote CRC tumor growth.
Fig. 6. ILCregs promote colorectal cancer progression.
a CT26 colon tumor cells (1 × 106) and ILCregs (1 × 105) sorted from AOM/DSS-induced colon tumors of IL-10-GFP mice subcutaneously injected into B-NSG mice. After 50 days, tumor photos were shown. b Tumor volumes were measured at indicated time points and calculated as means ± SD. After 50 days, tumors were isolated and shown (upper panel).**P < 0.01 by Non parametric Mann–Whitney U-test. n = 5 for each group. c Id3flox/flox;CreERT2 mice were treated with AOM/DSS as described previously. At day 60, mice were treated with TMX (50 mg/kg i.p. for six consecutive days every 20 days). Vehicle treatment served as negative control (Ctrl). At day 120, colon from indicated mice were isolated and cut open longitudinally. Colon tumor photo was shown in upper panel. Tumor numbers were calculated and shown as means ± SD. **P < 0.01 Two-tailed unpaired Student’s t test. n = 5 for each group. d H&E staining of colon from indicated mice treated as in (c). Scale bar, 50 μm. e Gene set enrichment analysis (GSEA) comparing TGF-β signaling of ILCreg and ILC3 subsets. NES (Normalized Enrichment Score) = 1.67, Nominal p = 0.00. f, g Tgfbr2flox/flox;Id2-CreERT2 mice were subjected to AOM/DSS treatment. After 60 days, mice were i.p. injected with TMX. Tumor infiltrating ILCreg and ILC3 cells were assessed by flow cytometry. Cell numbers of ILCregs and ILC3s were calculated and shown as means ± SD (f). Tumor numbers per colon were calculated and shown as means ± SD (g). *P < 0.05; **P < 0.01 by Two-tailed unpaired Student’s t test. n = 5 for each group. h tdTomato+ ILC3s from Rosa26-STOP-tdTomato;Rorc-Cre;IL-10-GFP mice were isolated and treated with 10 ng/mL TGF-β in complete cell culture in the presence or absence of 2 μM TGF-β inhibitor LY2109761. After 7 days, cells were analyzed by flow cytometry. i Human colon tumor xenografts were subcutaneously injected into B-NSG mice. After 25 days, LY2109761 was injected into xenograft tumors. 50 days after xenograft engraftment, tumors on mice were shown in the lower panel. j Xenograft tumor volumes were measured at the indicated time points and calculated as means ± SD. **P < 0.01 by Non parametric Mann–Whitney U-test. Tumors at day 50 were isolated and shown in the upper panel. n = 6 for each group. k Analysis of ILCregs (Lin−CD45+CD127+IL-10+) (Lin = CD2,CD3,CD14,CD16,CD19,CD56,CD235a) in xenograft tumors by flow cytometry. For a and b, data are representative of three independent experiments. For c–h, data are representative of five independent experiments. For i–k, data are representative of four independent experiments.
We next wanted to explore the mechanism by which ILC3s converted into ILCregs over CRC progression. Through analysis of scRNA seq data, we noticed that TGF-β signaling was elevated in ILCregs (Fig. 6e). We established Tgfbr2flox/flox;Id2-CreERT2 mice to delete TGF-β receptor on ILCs and treated with AOM/DSS. Of note, ILCregs were almost undetectable in CRC tumors, whereas ILC3s slightly increased (Fig. 6f). In addition, deletion of TGF-β receptor on ILCs remarkably suppressed tumor growth (Fig. 6g). Moreover, deletion of TGF-β receptor on ILC3s also suppressed tumor growth and decreased ILCreg numbers (Supplementary information, Fig. S5e, f). Consistently, TGF-β inhibitor treatment abrogated the conversion of ILC3s to ILCregs (Fig. 6h; Supplementary information, Fig. S5g), and consequently inhibited patient derived xenograft (PDX) tumor growth (Fig. 6i, j). Notably, TGF-β inhibitor treatment also blocked the conversion of ILC3s to ILCregs in PDX tumors (Fig. 6k). We conclude that TGF-β signaling is involved in the transdifferentiation of ILC3s to ILCregs over CRC progression. Altogether, TGF-β induces the conversion of ILC3 to ILCreg, and blocking the conversion of ILC3s to ILCregs can suppress tumor growth both in AOM/DSS-induced CRC tumors and in PDX tumors.
Discussion
Colorectal cancer is a common malignancy worldwide, which is highly related to chronic inflammation. Inflammatory microenvironment affects the fate and function of immune cells leading to immune evasion and tumor development. ILCs play critical roles in early antigen defence, allergy and inflammation. However, the physiological roles and regulation of ILCs in colorectal cancer progression are still unclear. Here we used scRNA-seq to profile colorectal tumor infiltrating ILCs and found ILCs are dynamically regulated during CRC progression. ILC1s undergo functional conversion during CRC progression, which exert inhibitory functions at the late stage of CRC. Three ILC2 subsets were identified inside CRC tumors and ILC2-Cs promote tumor development. HS3ST1 and PD1 are highly expressed in tumor filtrating ILC2s of late stage CRC tumors and deletion of HS3ST1 and PD1 in ILC2s suppress tumor growth. ILC3s convert to ILCregs under the stimulation of TGF-β during CRC progression. Importantly, ILCregs can promote tumor development and progression. Blocking TGF-β signaling can abrogate the transdifferention of ILC3s to ILCregs and suppress tumor development as well.
The roles of ILCs in regulation of tumorigenesis and progression still remain elusive. It has been reported that ILC1-like cells inside mammary tumors exert immunosurveillance function at an early stage.27 However, under TGF-β-rich tumor environment, NK cells convert to ILC1s leading to tumor escape from immunosurveillance.35 Herein we found that tumor infiltrating ILC1s express activating receptors at an early stage and inhibitory receptors at a late stage. Production of IFN-γ is suppressed in ILC1s of late stage CRC tumors, suggesting ILC1s undergo functional conversion during CRC progression. IFN-γ is considered as a critical cytokine in tumor immunosurveillance,30 and production of IFN-γ by innate lymphocytes such as NK cells apparently inhibit tumor development.36 It is possible that ILC1s and NK cells produce initial levels of IFN-γ and inhibit tumor development. However, inflammatory environment might affect ILC1 function and disarm their immunosurveillance function leading to CRC development. The mechanism for ILC1 conversion and regulation is needed to be further investigated.
Several reports show that ILC2s are associated with pulmonary and hepatic fibrosis, suggesting a potential role of ILC2s in the inflammation of lung and liver.37,38 In this study, we showed that tumor filtrating ILC2s are heterogeneous inside CRC tumors. The ratio of tumor cell/ILC2-C is 10:1 that can efficiently promote tumor growth. We tested tumor/ILC2-C ratios from 20:1 to 4:1 and successfully induced tumor growth (data not shown). ILC2-Cs predominate in the late stage of CRC and markedly promote tumor growth. Heparan sulfate 3-O-sulfotransferase 1 (HS3ST1) and PD1 are highly expressed in tumor filtrating ILC2s. HS3ST1 is a rate-limiting enzyme for heparan sulfate production. However, HS3ST1 deficient mice are lethal but do not display an obvious procoagulant phenotype.31 Heparan sulfate has been reported to be implicated in tumor proliferation, metastasis, angiogenesis.32 In T cells, PD1 recruits SHP2 and inhibits ZAP70 activation leading to T cell suppression.39 Deficiency of HS3ST1 or PD1 in ILC2s apparently suppresses tumor growth, but whose deletion does not affect cell death of ILC2s. In addition, conditional deletion of HS3ST1 remarkably inhibits AOM/DSS-induced CRC tumor development. These data suggest that targeting HS3ST1 and PD1 might be ideal strategies for CRC immunotherapy. However, how HS3ST1 and PD1 regulate ILC2 function requires to be further investigated. Besides HS3ST1 and PD1, we also identified several genes in tumor filtrating ILC2s, including Ltb4r1 (encoding leukotriene B4 receptor, BLT1), Hilpda (encoding HILPDA), and Tnfrsf18 (encoding Glucocorticoid-induced TNFR-related protein, GITR). The roles of these molecules in the regulation of ILC2s for CRC development are worthy of further investigation.
ILC3s in gut are related to chronic inflammation that might cause gut tumor.40 IL-23 signaling and high levels of IL-22 promote gut tumorigenesis.41,42 We previously identified a regulatory subpopulation of ILCs called ILCregs, which promote resolution of gut inflammation by secretion of IL-10.22 IL-10 is considered to be a negative indicator for clinical outcome in many cancers.43 We showed that ILC3s transdifferentiate into ILCregs during CRC progression by stimulation of TGF-β. Moreover, ILCregs can promote colon tumor growth and depletion of ILCregs dramatically inhibits tumor growth. Our results suggest that the conversion of ILC3s to ILCregs plays a critical role in the progression of CRC. The transdifferentiation of ILC3 to ILCreg is worthy to be analyzed by RNA velocity in our further analysis.44 In addition, TGF-β signaling induces the transdifferentiation of ILC3s to ILCregs during CRC progression. Blocking TGF-β signaling remarkably suppresses tumor growth of CRC, suggesting inhibitors against TGF-β signaling might be ideal agents for CRC treatment.
In summary, we define six clusters of tumor infiltrating ILCs during CRC progression. ILC2-Cs and ILCregs promote tumor development and growth of CRC. Our findings reveal that tumor infiltrating ILCs undergo conversions of their functions and subsets during CRC progression. The conversions of ILCs are related to CRC development and tumor progression. Blocking ILCreg transdifferentiation can suppress tumor development. Therefore intervention of ILC conversions may be a potential immunotherapy for CRC patient treatment.
Materials and methods
Antibodies and reagents
Antibodies used for flow cytometry: anti-mouse CD3(17A2), anti-mouse CD8a (53-6.7), anti-mouse CD19 (1D3), anti-mouse NK1.1 (PK136), anti-mouse CD11b (M1/70), anti-mouse CD11c (N418), anti-mouse Gr1 (RB6-8C5), anti-mouse F4/80 (BM8), anti-mouse Ter119 (TER-119), anti-mouse CD45 (30-F11), anti-mouse CD127 (A7R34), anti-mouse IL-10 (JES5-16E3), anti-mouse CD117 (2B8), anti-mouse NKp46 (29A1.4), anti-mouse ST2 (RMST2-33), anti-mouse KLRG1 (2F1), anti-mouse RORγt (AFKJS-9), anti-mouse Ly49G2 (4D11), anti-human IL-10 (JES3-9D7), anti-human CD127 (eBioRDR5), anti-human CD45 (2D1), Human Hematopoietic Lineage Monoclonal Antibody, anti-human T-bet (4B10), anti-human CD56 (TULY56), anti-human CRTH2 (BM16) were purchased from eBioscience; anti-mouse Id3 (S30-778) was from BD Pharmingen; anti-human/mouse IL-12RB2 was from R&D System; anti-mouse NKG2I was from Invitrogen; anti-human KIR2DL2/L3(DX27) was from Biolegend; anti-KIR2DL1 (REA284) was from Miltenyi Biotec. Anti-human PD1 antibody (nivolumab) was from invivogen. Recombinant murine IL-12, mIL-18, and mTGF-β1 were purchased from PeproTech. Brefeldin A and 7-AAD was from eBioscience. LY2109761 was from MedChemExpress.
Mouse and CRC patient samples
Id2-CreERT2, Rorc-Cre, and Tgfbr2flox/flox mice were from Jackson laboratory. Rosa26-STOP-tdTomato mice were from Shanghai Research Center for Model Organisms. B-NSG (NOD-PrkdcscidIL2rgtm1/Bcgen) mice were from Beijing Biocytogen Co., Ltd. IL-10-GFP mice were from Dr. Flavell (Yale University). Id3flox/flox mice were from Drs. Yuan Zhuang (Duke University) and Chen Dong (Tsinghua University). Lineage tracing mice Rosa26-STOP-tdTomato;Rorc-Cre were obtained by crossing Rosa26-STOP-tdTomato mice with Rorc-Cre mice. Both female and male mice were used in experiments. Age- and sex-matched littermates between 8 and 28 weeks of age were used. Mice were assigned randomly to experimental groups. To generate inducible TGF-β1 receptor deletion mouse model, Tgfbr2flox/+ mice were crossed with Id2-CreERT2 mice. To generate Id3 deletion mouse model, Id3flox/+ mice were crossed with Id2-CreERT2 mice for several rounds to obtain Id3flox/flox;CreERT2.
Hs3st1flox/+ mice were generated by inserting loxp sites flanking exon 2 of Hs3st1 gene by CRISPR/Cas9 technology (Supplementary information, Fig. S3f) as described previously.45 In short, a targeting construct containing loxP-flanked exon 2 of Hs3st1 was generated with CRISPR mediated long-chain single-stranded oligodeoxynucleotides (lsODNs). This construct was cloned into the pLSODN-1 plasmid and introduced by homologous recombination into the endogenous Hs3st1 gene. Targeted alleles were identified by PCR screening and DNA sequencing. Upstream sgRNA: 5’-CAGACAATAGACAGAGACAGGGG-3’; downstream sgRNA: 5’-CAGGGGTTAAACCCACATGTGGG-3’.
All the mice are C57BL/6 background and maintained under specific pathogen-free conditions with approval by the Institutional Committee of Institute of Biophysics, Chinese Academy of Sciences. The study is compliant with all relevant ethical regulations regarding animal research.
Human resected colon tissues of CRC patients were obtained from West China Hospital, Sichuan University (Chengdu, China) and Cancer Hospital Chinese Academy of Medical Sciences (Beijing, China) with informed consents, and approved by the Institutional Review Board of the Institute of Biophysics, Chinese Academy of Sciences. The study is compliant with all relevant ethical regulations regarding research involving human participants.
AOM/DSS-induced colitis-associated colorectal cancer
8–12 week-old mice of body weight > 20 g were i.p. injected of azoxymethane (AOM, 10 mg/kg body weight). After 4 days, mice were treated with 1.5% dextran sodium sulfate (DSS) in drinking water for six days. Then, DSS was replaced by drinking water. After two weeks, mice were treated with 1.5% DSS for another six days. Three cycles of DSS treatment were needed before tumor evaluation. Colon tumors were analyzed at early stage (60 days after AOM injection) or late stage (120 days after AOM injection).
Isolation of tumor infiltrating ILCs
Colon tumors from mice colons or CRC patient samples were isolated and cut into fine pieces. Tumor samples were digested twice for 40 min each at 37 °C with digestion buffer containing 5% FBS, Collagenase II and III (1 mg/mL; Worthington), DNase I (200 μg/mL; Roche) and dispase (4U/mL; Sigma). Mononuclear cells were isolated with 40%–80% Percol gradient, and washed twice with PBS. Isolated cells were blocked with anti-CD16/32 antibody for 30 min on ice and then stained with antibodies against CD45, CD127, lineage cocktail (Lin = CD3e,CD8a,CD19,CD11b,CD11c,Gr1,F4/80,Ter119) and ILC markers on ice for 1 h followed by 7AAD staining. Tumor infiltrating ILCs were isolated by flow cytometer (FACS Aria III, BD) for functional study or PDC experiments. Purity of ILC cells was over 95% for each assay that was determined by post sorting analysis of flow cytometry.
Flow cytometry
Tumor infiltrating cells were isolated and blocked with anti-CD16/32 antibody for 30 min on ice. Surface markers were then stained for 1 h on ice. For intracellular cytokine staining, cells were cultured in completed RPMI1640 media in the presence of Brefeldin A at 37 °C for 4 h. Next, cells were harvested for surface marker staining and fixed and permeablized by Intracellular Fixation & Permeablization buffer set (eBioscience), followed by intracellular antigen staining. Cell samples were analyzed by flow cytometer (FACS Aria III, BD) as described.46
Immunofluorescence assay
For in situ immunofluorescence of ILCs, mouse colons were treated as previously described.22 Briefly, colon tumors or intestines with longitudinally opened were fixed in 4% paraformaldehyde (PFA) (Sigma) fixative for 8 h, and rehydrated in 30% sucrose solution for 24 h and frozen in OCT for sectioning. Tumor or intestinal sections were rehydrated in PBS and blocked in 10% donkey serum and anti-CD16/32 antibody. Primary antibodies were used for section staining for 2 h at room temperature (RT). Next, we used Alex488-conjugated and Alex594-conjugated donkey secondary antibodies for further staining. Then, we used APC-conjugated lineage cocktails to stain slides at RT for 1 h. After washing for 3 times, nuclei were stained by DAPI if needed and the sections were subjected to dehydrating in EtOH gradient: 70%, 85%, 95%, and 100%. Mounted sections were analyzed by confocal microscopy (ZIESS LSM700). Antibodies for immunofluorescence staining were anti-mouse KLRG1 (2F1; eBioscience), anti-human/mouse HS3ST1 (Sigma), anti-human/mouse CD279 (PD1) (Invitrogen), anti-human CRTH2 (BM16, eBioscience), anti-GFP (GF28R, Invitrogen), anti-tdTomato (Biorbyt), anti-human/mouse RORγt (AFKJS-9, eBioscience). For 5-color staining, OpalTM 7-color fIHC kit (PerkinElmer) was used according to the manufacturer’s instructions. Briefly, paraffin sections of colon tumors were sequentially stained with primary antibodies and HRP conjugated secondary antibody. Microwave treatment was used after each Opal dye staining. Anti-GFP, anti-tdTomato, anti-RORγt, and anti-Lin antibodies were used as primary antibodies. Opal520, Opal570, Opal620, Opal690 dyes were used for secondary staining. Samples were observed using Vectra Automated Quantitative Pathology Imaging System (PerkinElmer).
Droplet-based scRNA-seq cDNA library preparation and sequencing
Tumor infiltrating ILCs from early (D60) or late (D120) stage of colon tumors were harvested and sorted by flow cytometry (n = 5 for each group). These mice shared the same background and similar distribution of ILCs according to our flow cytometry analysis (data not shown) and we merged them together for scRNA-seq. Tumor infiltrating ILCs (Lin−CD45+CD127+) (Lin = CD3e,CD8a,CD19,CD11b,CD11c,Gr1,F4/80,Ter119) were isolated and cell suspensions (300-500 living cells per microliter) were loaded on a Chromium Single Cell Controller (10x Genomics) to generate single-cell Gel Bead-In-Emulsions (GEM) by using Single Cell 3’ Library and Gel Bead Kit V2 (10x Genomics, 120237). mRNAs of captured cells were barcoded through reverse transcription. Barcoded cDNAs were pooled and cleaned up by using DynaBeads® MyOne Silane Beads (Invitrogen, 37002D). Single-cell RNA-seq cDNA libraries were prepared using Single Cell 3’ Library Gel Bead Kit V2 (10x Genomics, 120237) according to the manufacture’s protocol followed by sequencing on an Illumina HiSeq X Ten with pair end 150 bp (PE150). GSE number of scRNA sequencing is GSE142694.
Droplet-based scRNA-seq data preprocess
Raw sequencing data was checked by FastQC software. Next, fastq sequences were trimmed by FASTX-Toolkit. Gene counts were obtained by aligning reads to the mouse mm10 genome by using CellRanger software (version 2.0.1) to generate the single cell information. We abandoned cells with apparent lineage markers such as Cd3e, Cd19, Ly6g, Ly6c and Adgre1 which might be caused by a contamination during FACS-based cell separation. Next, we removed doublets and poor-quality cells by Seurat (v2.2.1) based on the number of unique molecular identifiers (UMIs) and percentage of mitochondrial genes. Briefly, raw cell counts were filtered in Seurat CreateSeuratObject function with the following criteria: one gene was expressed in at least 3 cells and at least 200 genes were detected in one cell. Mitochondrial genes inside one cell were also calculated with a percentage under 20%. Cells were further filtered with the following requirements: gene numbers inside on cell were no more than 8000 and no less than 200. “FindVariableGenes” function of Seurat was used to get the variable genes with the following parameters: x.low.cutoff 0.05, x.high.cutoff 8 and y.cutoff 0.5. 1226 variable genes were found as the input genes for PCA or DiffusionMap analysis. PCA analysis was conducted using the “RunPCA” function of Seurat and PCs 1–12 were chosen for dimension reduction analysis with Seurat function “RunTSNE” and DiffusionMap analysis with Seurat function “RunDiffusion”. For the clustering analysis, the first twelve PCs were used to calculate clusters with a resolution of 0.7 using Seurat function “FindClusters”. Differently expressed genes were calculated through Seurat “FindAllMarkers”. For heatmap and scatter plots, scaled data (scRNA@scale.data) from Seurat pipeline were used. For violin plots, normalized data (scRNA@data) from Seurat pipeline were used.
Diffusion map was drawn using the Seurat package command RunDiffusion with the following parameters: max.dim = 5, dims.use = 1:12 using top 10 marker genes of ILC3 and ILCreg. DM1, DM3, DM4 were used to plot the three-dimensional diffusion map using the R package “rgl” with the default parameters.
Gene knockout by CRISPR/Cas9 technology
ILC2-C cells from stage IV CRC patient samples were isolated and infected with lentivirus (lentiCRISPR-v2) carrying sgRNA against HS3ST1 or PDCD1 gene (Supplementary information, Table S2). Infected cells were selected with 0.5 μg/mL puromycin (Invivogen) and subcutaneously injected into B-NSG mice together with human colon tumor cells. After 3 days, expression of HS3ST1 or PD1 in ILC2-C cells was analyzed by flow cytometry. HS3ST1−/− or PDCD1−/− ILC2-C cells together with primary tumor cells from stage IV CRC patients were subcutaneously injected into B-NSG mice.
CRC patient-derived xenograft (PDX) and patient-derived cell (PDC) models
CRC PDX and PDC models were established as previously described.47 For PDX models, tumor samples from different stages of CRC patient samples were cut into 25–30 mm3 pieces in PBS with 100 mg/mL penicillin and 100 U/mL streptomycin. Tumor sample pieces were coated in Matrigel (BD Biosciences), followed by subcutaneous implantation into 6-week-old B-NSG mice (Biocytogen Co.). While the established tumors achieved 400 mm3, the mice were intratumorally injected with TGF-β inhibitor LY2109761. Tumor volumes were determined every 5 days. For PDC models, tumor samples from different stages of CRC patient samples were cut into small pieces and subjected with digestion by Collagenase II and III (1 mg/mL; Worthington). Tumor cells (EPCAM+) and ILC2 cells were isolated by flow cytometry and subcutaneously injected into 6-week-old B-NSG mice. Isolated ILC2 cells were transfected with virus carrying CRISPR/Cas9 elements against HS3ST1 or PDCD1 and subcutaneously injected into B-NSG mice together with tumor cells. For anti-PD1 antibody treatment, 1 mg/kg anti-human PD1 antibody (nivolumab, Invivogen) was injected into tumors at day 25 after tumor injection once a week. PBS was used as negative control. Tumor volumes were examined every 5 days.
Generation of mouse model with IL-10-deficient ILCregs
ILCregs were isolated from Rosa26-STOP-Cas9;Id2-ERT2-Cre;IL-10-GFP mice and infected with lentivirus (lentiGuide-Puro-sgRNA-Il10) carrying sgRNA against Il10 gene (#1: 5’-TATTGTCTTCCCGGCTGTAC-3’; #2: 5’-GCATGTGGCTCTGGCCGACT-3’). Infected cells were selected with 1 μg/mL puromycin (Invivogen) and adoptively transferred into Rag1−/−Il2rg−/− mice together with respective WT (CD45.1) lymphocytes. Expression of IL-10 in ILCregs (CD45.2) after treatment with TMX (50 mg/kg i.p. for five consecutive days) was detected prior to transplantation.
ELISA assay
Tumor-infiltrating ILCs from colon tumors were isolated and cultured in complete media for 24 h. Cytokines in supernatants were analyzed by ELISA kit (eBioscience) by manufacturer’s instructions. Detection thresholds of different cytokines were 50 pg/mL for IFN-γ, 50 pg/mL for IL-17A, 50 pg/mL for IL-22, and 50 pg/mL for IL-10. Values below detection thresholds were shown as ND (not detectable). ELISA data were normalized to total cell numbers and shown as means ± SD per 5 ×103 cells.
Real time PCR assay
Tumor infiltrating ILCs were isolated from colon tumors. mRNA was extracted by using Neasy Micro Kit (Qiagen) according to the manufacturer’s instructions. mRNA quality was determined by A260/A280 (between 1.8 and 2.2) and A260/230 (> 1.7) ratios. RNA integrity was assessed by agarose gel electrophoresis. cDNAs were synthesized by using oligo dT followed by real time PCR. Primers for real-time PCR in this study are as shown in Supplementary information, Table S1.
Statistical analysis
For statistical analysis, data were analyzed by using GraphPad Prism 7.0. Two-tailed unpaired Student’s t test, One way ANOVA and Non parametric Mann–Whitney U-test were used according to the type of experiments. P-values <0.05 were considered significant (*P < 0.05; **P < 0.01; P < 0.001); P > 0.05, non-significant (NS). All flow cytometry data were analyzed with FlowJo (Treestar). No statistical methods were used to predetermine sample sizes.
Supplementary information
Acknowledgements
We thank Junying Jia, Shuang Sun, Xiaoxiao Zhu, Yihui Xu, Shu Meng, Dongdong Fan and Yan Teng for technical support. We thank Drs Flavell (Yale University), Yuan Zhuang (Duke University) and Chen Dong (Tsinghua University) for providing genetic mouse strains. We thank Jing Li (Cnkingbio Company Ltd., Beijing, China) for technical support. We thank CapitalBio Technology Inc. for technical support. This work was supported by the Strategic Priority Research Programs of the Chinese Academy of Sciences (XDB29020000, XDB19030203), the National Natural Science Foundation of China (81722023, 81922031, 81921003, 31930036, 91640203, 31671531, 91940305, 31770939), Key Research Program of Frontier Sciences of Chinese Academy of Sciences (ZDBS-LY-SM025), National Key R&D Program of China (2019YFA0111800, 2019YFA0508501), CAMS Innovation Fund for Medical Sciences (2017-I2M-1-006), Beijing Natural Science Foundation (7181006) and Youth Innovation Promotion Association of CAS to S.W.
Author contributions
S.W. designed and performed experiments, analyzed data and wrote the paper; Y.Q., P.X., and Y.C. performed experiments and analyzed data; X.Z. and Y.T. generated animal models. G.W., J.Z. and J.Y provided human CRC samples. Z.F. initiated the study, organized, designed, and wrote the paper.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Shuo Wang, Yuan Qu, Pengyan Xia, Yi Chen.
Change history
6/15/2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Change history
5/25/2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Contributor Information
Shuo Wang, Email: wangshuo@im.ac.cn.
Yong Tian, Email: tiany@ibp.ac.cn.
Jianming Ying, Email: jmying@cicams.ac.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-020-0312-y.
References
- 1.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lasry A, Zinger A, Ben-Neriah Y. Inflammatory networks underlying colorectal cancer. Nat. Immunol. 2016;17:230–240. doi: 10.1038/ni.3384. [DOI] [PubMed] [Google Scholar]
- 3.Shalapour S, Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J Clin. Invest. 2015;125:3345–55. doi: 10.1172/JCI80007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feagins LA, Souza RF, Spechler SJ. Carcinogenesis in IBD: potential targets for the prevention of colorectal cancer. Nat. Rev. Gastroenterol. Hepatol. 2009;6:297–305. doi: 10.1038/nrgastro.2009.44. [DOI] [PubMed] [Google Scholar]
- 5.Terzic J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–2114. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 6.Arthur JC, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 8.Shankaran V, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 9.Sun JC, Lanier LL. NK cell development, homeostasis and function: parallels with CD8(+) T cells. Nat. Rev. Immunol. 2011;11:645–657. doi: 10.1038/nri3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 2011;29:235–271. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- 11.Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat. Rev. Immunol. 2006;6:295–307. doi: 10.1038/nri1806. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto M, et al. CD8 T cell exhaustion in chronic infection and cancer: opportunities for interventions. Annu. Rev. Med. 2018;69:301–318. doi: 10.1146/annurev-med-012017-043208. [DOI] [PubMed] [Google Scholar]
- 13.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 14.Serafini N, Vosshenrich CA, Di Santo JP. Transcriptional regulation of innate lymphoid cell fate. Nat. Rev. Immunol. 2015;15:415–428. doi: 10.1038/nri3855. [DOI] [PubMed] [Google Scholar]
- 15.Fuchs A, et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity. 2013;38:769–781. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernink JH, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 2013;14:221–229. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- 17.Hoyler T, et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634–648. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerbe F, et al. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature. 2016;529:226–230. doi: 10.1038/nature16527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallrapp A, et al. The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature. 2017;549:351–356. doi: 10.1038/nature24029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buonocore S, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ibiza S, et al. Glial-cell-derived neuroregulators control type 3 innate lymphoid cells and gut defence. Nature. 2016;535:440–443. doi: 10.1038/nature18644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang S, et al. Regulatory innate lymphoid cells control innate intestinal inflammation. Cell. 2017;171:201–216. doi: 10.1016/j.cell.2017.07.027. [DOI] [PubMed] [Google Scholar]
- 23.Neufert C, Becker C, Neurath MF. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat. Protoc. 2007;2:1998–2004. doi: 10.1038/nprot.2007.279. [DOI] [PubMed] [Google Scholar]
- 24.Man SM, et al. Critical role for the DNA sensor AIM2 in stem cell proliferation and cancer. Cell. 2015;162:45–58. doi: 10.1016/j.cell.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gury-BenAri M, et al. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell. 2016;166:1231–1246. doi: 10.1016/j.cell.2016.07.043. [DOI] [PubMed] [Google Scholar]
- 26.Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348:aaa6566. doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dadi S, et al. Cancer immunosurveillance by tissue-resident innate lymphoid cells and innate-like T ccells. Cell. 2016;164:365–377. doi: 10.1016/j.cell.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinette ML, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 2015;16:306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat. Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 30.Kaplan DH, et al. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc. Natl. Acad. Sci. USA. 1998;95:7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.HajMohammadi S, et al. Normal levels of anticoagulant heparan sulfate are not essential for normal hemostasis. J Clin. Invest. 2003;111:989–999. doi: 10.1172/JCI200315809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knelson EH, Nee JC, Blobe GC. Heparan sulfate signaling in cancer. Trends Biochem. Sci. 2014;39:277–288. doi: 10.1016/j.tibs.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin. Invest. 2015;125:3384–3391. doi: 10.1172/JCI80011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 35.Gao Y, et al. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 2017;18:1004–1015. doi: 10.1038/ni.3800. [DOI] [PubMed] [Google Scholar]
- 36.Martin-Fontecha A, et al. Induced recruitment of NK cells to lymph nodes provides IFN-gamma for T(H)1 priming. Nat. Immunol. 2004;5:1260–1265. doi: 10.1038/ni1138. [DOI] [PubMed] [Google Scholar]
- 37.Hams E, et al. IL-25 and type 2 innate lymphoid cells induce pulmonary fibrosis. Proc. Natl. Acad. Sci. USA. 2014;111:367–372. doi: 10.1073/pnas.1315854111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McHedlidze T, et al. Interleukin-33-dependent innate lymphoid cells mediate hepatic fibrosis. Immunity. 2013;39:357–371. doi: 10.1016/j.immuni.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okazaki T, Chikuma S, Iwai Y, Fagarasan S, Honjo T. A rheostat for immune responses: the unique properties of PD-1 and their advantages for clinical application. Nat. Immunol. 2013;14:1212–1218. doi: 10.1038/ni.2762. [DOI] [PubMed] [Google Scholar]
- 40.Ebbo M, Crinier A, Vely F, Vivier E. Innate lymphoid cells: major players in inflammatory diseases. Nat. Rev. Immunol. 2017;17:665–678. doi: 10.1038/nri.2017.86. [DOI] [PubMed] [Google Scholar]
- 41.Chan IH, et al. Interleukin-23 is sufficient to induce rapid de novo gut tumorigenesis, independent of carcinogens, through activation of innate lymphoid cells. Mucosal Immunol. 2014;7:842–856. doi: 10.1038/mi.2013.101. [DOI] [PubMed] [Google Scholar]
- 42.Jiang R, et al. IL-22 is related to development of human colon cancer by activation of STAT3. BMC Cancer. 2013;13:59. doi: 10.1186/1471-2407-13-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore KW, Malefyt deWaal, Coffman R, O’Garra RLA. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 44.La Manno G, et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia P, et al. The ER membrane adaptor ERAdP senses the bacterial second messenger c-di-AMP and initiates anti-bacterial immunity. Nat. Immunol. 2018;19:141–150. doi: 10.1038/s41590-017-0014-x. [DOI] [PubMed] [Google Scholar]
- 46.Wang S, et al. Natural killer-like B cells prime innate lymphocytes against microbial infection. Immunity. 2016;45:131–144. doi: 10.1016/j.immuni.2016.06.019. [DOI] [PubMed] [Google Scholar]
- 47.Zhu P, et al. LncGata6 maintains stemness of intestinal stem cells and promotes intestinal tumorigenesis. Nat. Cell. Biol. 2018;20:1134–1144. doi: 10.1038/s41556-018-0194-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.