Abstract
The balance between cell death and survival is a critical parameter in the regulation of cells and the maintenance of homeostasis in vivo. Three major mechanisms for cell death have been identified in mammalian cells: apoptosis (type I), autophagic cell death (type II), and necrosis (type III). These three mechanisms have been suggested to engage in cross talk with each other. Among them, autophagy was originally characterized as a cell survival mechanism for amino acid recycling during starvation. Whether autophagy functions primarily in cell survival or cell death is a critical question yet to be answered. Here, we present a comprehensive review of the cell death-related events that take place during autophagy and their underlying mechanisms in cancer and autoimmune disease development.
Subject terms: Kinases, Macroautophagy
Facts
Autophagy is a conserved catabolic process essential for homeostasis, however, its role as a protective mechanism in cancer remains controversial.
The complex cross talk between apoptosis, autophagic cell death, and necrosis underlies the pathogenesis of multiple diseases.
The serine/threonine kinase Akt and its downstream signaling molecules at the lysosome may have a role in the interaction between the autophagic and apoptotic pathways.
Autophagy may underly the pathological conditions of cancer and autoimmune diseases by altering metabolic conditions.
Open questions
Whether and how autophagy has a role in the cell death machinery remain unclear.
How the three forms of cell death, autophagy, apoptosis, and necrosis are mechanistically interconnected each other remain unclear.
How autophagy could underlie the pathogenesis of cancer and/or the carcinogenesis needs to be clarified.
How autophagy mechanistically is involved in the pathogenesis of autoimmune diseases needs to be clarified.
Understanding the precise modulation of autophagy may lead to novel therapeutic approaches for several diseases, including cancer and autoimmune diseases.
Autophagy is considered to be a conserved survival process prompted by several cellular stress conditions, which prevents cell damage, promotes cell survival in the event of nutrient scarcity, and responds to cytotoxic stimuli. Thus, autophagy serves as a regulator of homeostasis, effectively preserving the balance between the production of cellular components, and the breakdown of damaged or unnecessary organelles and other cellular constituents1–4. Although autophagy was originally characterized as a cytoprotective process in yeast under starvation conditions, it is now believed to be a form of cell death in mammalian cells5–7 along with the two other major cell death mechanisms—apoptosis and necrosis8.
Roles of lysosome and induction of autophagy
The molecular events associated with apoptosis, including caspase activation, chromatin condensation, DNA cleavage, and plasma membrane degeneration, are well characterized8–10. In contrast, the molecular processes that occur between the lysosomal degradation of cytosolic components and autophagic cell death are poorly understood11. Following engulfment of endocytosed and autophagocytosed material into the lysosomal/autolysosomal compartment, lysosomal hydrolases participate in the digestion of endocytosed and autophagocytosed material during acute cell death and cancer progression. The increased expression and altered trafficking of lysosomal enzymes participate in tissue invasion, angiogenesis and sensitization to the lysosomal death pathway12. Furthermore, the biological roles of lysosomes in specific cellular processes, including nutrient sensing, signaling, and metabolism have been established4,13. Lysosomes contain over 20 hydrolytic enzymes that become activated in the acidic lysosomal compartment6. During the later stages of autophagy (i.e., macroautophagy) induction, lysosomal acidification is important for maintaining the hydrolytic activity of lysosomal enzymes, such as cathepsin D. Recent studies suggest that the Akt-mammalian target of rapamycin complex (mTORC1) pathway regulates the proton pump [vacuolar H(+)-adenosine triphosphatase ATPase (v-ATPase)] in the lysosomal membrane, which transports protons from the cytosol, thereby regulating the lysosomal acidification process required for autophagy induction1–3,14,15. In addition, lysosomal interactions between the vaccinia-related kinase 2 (VRK2)-Akt complexes also regulates lysosomal acidification, and subsequent enzymatic hydrolytic activities16. Notably, lysosomal membrane permeabilization leads to hydrolytic activation and release of proteolytic enzymes into the cytosol, resulting in cell death—a process often referred to as lysosomal cell death17,18, which may differ from autophagy. In this process, lysosomal hydrolytic enzymes and their activation by the acidified state may have a role under various cellar conditions in human diseases1–3,19.
Cross talk between autophagy and apoptosis
Although autophagy was originally described in yeast, it is now being recognized as a mammalian cell survival/prosurvival mechanism. Research has specifically focused on the cross talk that occurs between autophagy and apoptosis as a means to understand how these two types of cell death are interconnected5,20–23. Inhibition of macroautophagy following treatment with small interfering RNAs (targeting autophagy-related protein (Atg)5, Atg6/Beclin-1, Atg10, or Atg12), or via pharmacological agents (3-methyladenine, hydroxychloroquine, bafilomycin A1, or monensin), enhances apoptosis-induced cell death as a consequence of reduced mitochondrial membrane stabilization or caspase inhibition24. Autophagy-related proteins (Atg)s have been described as regulators of apoptosis. For example, calpain-mediated cleavage of Atg5,which is known to induce autophagy, switches autophagy to apoptosis25.
Similarly, Atg12 is a positive mediator of mitochondrial apoptosis, it regulates the apoptotic pathway by binding and inactivating prosurvival B cell lymphoma (Bcl)-2 family members, including Bcl-2 and Mcl-125. Moreover, in the absence of Atg12-Atg3 complex formation, an expansion in mitochondrial mass and simultaneous inhibition of mitochondria-mediated cell death is observed26. These results support the notion that not only do autophagic and apoptotic pathways converge in mitochondria, but also that mitochondria provide and ideal molecular platform for counter regulation of autophagic versus apoptotic cell death5,9,27,28.
In growth factor-dependent Bax/Bak-deficient cells, autophagy is activated after growth factor withdrawal. These cells activate autophagy with progressive atrophy leading to cell death. This observation supports the notion that apoptosis is not essential to limit cell autonomous survival. Autophagy is essential for maintaining cell survival following growth factor withdrawal and can sustain viability21. Tumor necrosis factor (TNF)-related apoptosis and autophagy has been shown to cross talk in various physiological settings28–30. Blocking the p62(sequestosome1, SQSTM1)-dependent recruitment of the necrosome to participate in autophagy can activate apoptosis supporting that the machinery of autophagy can control programed cell death31,32.
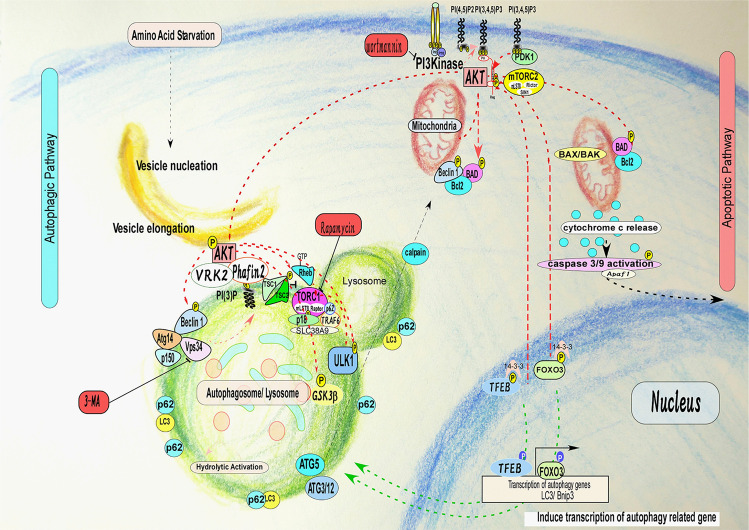
A critical question that remains to be answered is whether autophagy serves primarily as a cell survival mechanism, or rather as an active killing mechanism in mammalian cells12,29,30 (Fig. 1). If autophagy is solely a cell survival mechanism, it can protect against cell death induced by either apoptosis or necrosis. However, depending on the relative strength of the cell survival signal elicited through autophagy, cells can either survive or die with a combination of morphological changes associated with autophagic, apoptotic-, or necrotic- cell death. It is possible that depending on the circumstances, typical morphological features of cell death represent those of apoptosis, autophagy, or even necrosis. In contrast what happens in autophagy, the molecular regulation of the cell death cascade is well defined in apoptotic pathways from the death receptor to its downstream signal31. However, it remains unclear how lysosomal degradation of intracellular proteins can unavoidably induce cell death12,20,33.
Fig. 1. Cross talk regulation between autophagy and apoptosis.
Autophagy may contribute to enhanced cell death induced by apoptosis, depending on the relative potency of the cell death-inducing stimuli. It is also possible that apoptosis, autophagy, and necrosis, individually or conjointly, act to induce cell death8,9,35. This complex cross talk underlies the pathogenesis and manifestation of cancer likely through alteration of metabolic status via autophagy9,33,35. Although autophagy acts as a cytoprotective effect under starvation and/or stress conditions, it is now thought that autophagy also regulates metabolic conditions, which could differentially contribute to the various human diseases such as cancer and autoimmune diseases.
There is accumulating evidence regarding the deep connection between apoptosis regulation and the induction of autophagy7,9,17,18,29,34, and this cross talk underlies the pathogenesis of, for instance, cancer and its metabolism9,33,35. Autophagy is primarily a survival mechanism that maintains the balance between the creation of cellular components and the breakdown of damaged or unnecessary organelles and other cellular constituents. However, autophagy is also known to kill cells under certain conditions, in a process called autophagic cell death, which involves pathways and mediators that differ from those of apoptosis5,6,23. It is, therefore, plausible that autophagy enhances cell death caused by apoptosis; alternatively, it may also induce cell death in an apoptosis- or necrosis-independent manner. To support this notion, several experiments have shown that in the molecular process of apoptosis31, pro-apoptotic signals, such as TNF-related apoptosis-inducing ligand (TRAIL)36, TNF37, and FADD38 induce autophagy. Ectopic expression of Beclin-1 (Atg6) suppresses cell death, whereas reduced Beclin-1 levels induced by siRNA, sensitizes cells to TRAIL-induced cell death39.
Akt and downstream signaling molecules localize at the lysosome
Akt is a serine/threonine kinase and a major downstream effector that regulates diverse cellular processes through PI3K, and has reported anti-apoptotic, cell proliferative, cell cycle, cytoskeletal organization, vesicle trafficking, and glucose transporting properties23,40,41. Furthermore, Akt activation can inhibit autophagy induction in mammalian cells21,42–45. To support the roles of Akt in autophagy regulation, Sch9 kinase, a putative yeast orthologue of mammalian Akt and possibly S6K1, is suggested to have a role in the regulation of autophagy46,47.
Although the mTORC is considered to be a major factor in the control of autophagy induction, Akt activation can inhibit autophagy in mammalian cells21,42. In fact, recent studies have reported that various Akt effector molecules, including mTORC1, mTORC2, glycogen synthase kinase 3β (GSK3β), and the tuberous sclerosis complex (TSC) are present at the lysosome, a major locus for executing autophagy23,48–52. The level of phosphorylated Akt, and its substrates at the lysosome remained high even after Hanks Balanced Salt Solution (HBSS) treatment (essentially amino acid deprivation) to induce autophagy, which was sufficient to inhibit general intracellular activation of Akt16. Characterization of the protein complexes associated with Akt at the lysosomal membrane after induction of autophagy, showed that Akt forms a complex with Phafin2-vaccinia-related kinase 2 (VRK2), and this complex is important for determining the fate of autophagy in mammalian cells16,48.
Akt phosphorylates and inhibits TSC1/2, which leads to the stabilization of Rheb GTPase40,53. This in turn activates mTORC1, resulting in inhibition of autophagy40. Akt also directly phosphorylates ULK1 (ATG1) and Beclin-1, which control autophagy23,54,55. In Myr-Akt-expressing cells, in which Akt is constitutively activated, Beclin-1 is phosphorylated at Ser234 and Ser295 in the presence of Torin 1, an inhibitor of mTORC1, suggesting that Akt controls autophagy in a manner independent of mTORC155. The observation suggests that Akt signaling may be mechanistically linked to autophagy inhibition and tumorigenesis through regulation of the Beclin-1 complex56, possibly independent of mTOR55.
The involvement of Akt in regulating autophagy is also supported by the fact that Akt directly phosphorylates a wide variety of autophagy regulatory molecules localized at either mitochondria or autophagosomes, including ULK1 (Atg1) at Ser774 through insulin signaling54, as well as Beclin-1, and TSC223,49. Akt can also phosphorylate the anti-apoptotic molecule, Bcl-associated death (BAD), at mitochondria57, which then releases activated forms of Bcl-2 at the outer membrane to prevent subsequent cytochrome c release for triggering downstream caspase activation56.
In addition, transcription factor EB (TFEB), a transcriptional regulator of autophagy, is targeted by Akt to inhibit autophagy induction independent of mTORC119,58. These observations provide further support for the involvement of Akt in the autophagy process23,54,55.
A recent study reported that Beclin-1 is a substrate of Akt and can regulate tumorigenesis55. Akt-mediated phosphorylation of Beclin-1 inhibits autophagy by forming an autophagy-inhibitory complex composed of Beclin-1(Atg6), 14-3-3 proteins, vimentin, and the intermediate filament complex55. Beclin-1 also regulates PtdIns(3)P production in response to growth factor stimulation to control the residency time of growth factor receptors. As a consequence, suppressing Beclin-1 sustains growth factor-stimulated Akt, and extracellular signal-regulated kinase activation59. Thus, phosphorylation of Atgs is an additional aspect of autophagy regulation60.
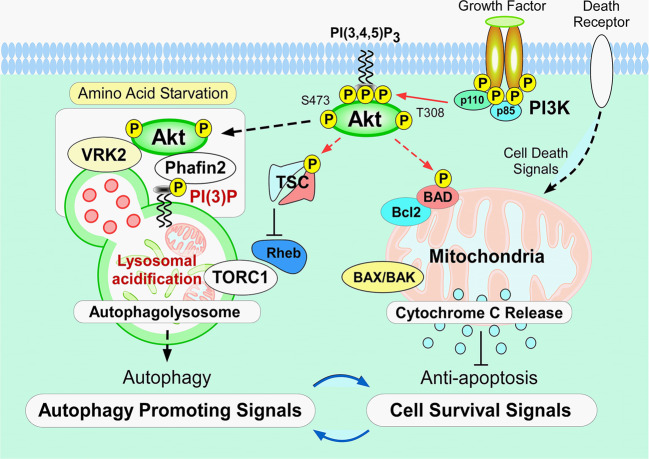
Akt–Phafin2–VRK2 forms a complex at the lysosome
PtdIns(3)P-dependent lysosomal accumulation of the Akt–Phafin2 complex is a critical step in inducing autophagy48. Yeast two-hybrid studies revealed an interaction between Akt and Phafin2 (EAPF or PLEKHF2), a lysosomal protein with unique N-terminal pleckstrin homology (PH) and C-terminal FYVE domains48. These conserved motifs place Phafin2 in a family of proteins known to induce caspase-independent apoptosis via the lysosomal-mitochondrial pathway61. Phafin2 controls receptor trafficking through the endosome62. However, the exact physiological roles of Phafin2 at the lysosome during the course of autophagy induction have not been thoroughly investigated. Following autophagy induction, Akt translocates with Phafin2 to the lysosome in a PtdIns(3)P-dependent manner following autophagy induction to form the Akt–Phafin2 complex, a critical step in inducing autophagy via interaction with PtdIns (3)P48. In this regard, a PtdIns(3)P-specific RNA aptamer has been identified, and its expression inhibits induction of autophagy as determined by LC3II/I conversion, p62 degradation, formation of LC3 puncta, lysosomal accumulation of Phafin2, lysosomal acidification, and subsequent inhibition of the hydrolytic activity of cathepsin D63. Given the roles of autophagy in cancer, infectious diseases, and autoimmune diseases, this RNA aptamer may have possible implications for the design of novel therapeutics63.
Through time-of-flight/mass spectrometry analysis of lysosomal Akt protein complexes, VRK2 was found to interact with Akt and to be enriched at lysosomes following autophagy induction16. VRK2 interacts with Akt in a kinase-dependent manner to maintain the phosphorylation of Akt at the lysosome. Ectopic expression of the active form of Akt, but not kinase-dead Akt, can induce autophagy induction suggesting the kinase activity of Akt is critical for induction of autophagy.
VRK2 facilitates lysosomal localization of activated Akt and its substrates after HBSS treatment to induce autophagy. Functional characterization of the VRK2–Akt interaction clarified the roles of VRK2 in sustaining the kinase activity of Akt at the lysosomes, which controls the size, acidification, and induction of autophagy in mammalian cells. Overall, these findings support a model in which spatial control of Akt and its kinase activity toward the lysosome serve as functional modulators of autophagy induction via VRK2A. Moreover, the functional interaction between Akt–Phafin2–VRK2 complex at the lysosome supports the hypothesis that spatial localization of activated Akt at the lysosomes has important roles in the process of autophagy induction, which many contribute to the regulation of autophagic versus apoptotic cell death16,23 (Fig. 2).
Fig. 2. Lysosomal Akt–Phafin2–VRK2 complex as a functional modulator of autophagy.
Regulatory roles of Akt at the lysosome in the course of autophagy are suggested by Beclin-1 (Atg6), transcription factor EB (TFEB), and Unc-51 like autophagy activating kinase 1 (ULK1) functioning as direct substrates23,54,55. Recent investigations revealed that target molecules of Akt, such as the tuberous sclerosis complex (TSC), Beclin-1 (ATG6), ULK1, and BAD, are present in the lysosomes23,48–52. Lysosomes are intracellular organelles that participate in intracellular protein degradation through activating hydrolytic enzymes in the process of autophagy for amino acid recycling8–10. Autophagy, a cell survival mechanism during starvation conditions, participates in the regulation of cell death/survival machinery5–7. Searching for the binding partner of Akt at the lysosome using yeast two-hybrid screening and time-of-flight/mass spectrometry analyses, we identified the formation of a Akt–Phafin2–VRK2 protein complexes at the lysosome in a phosphatidylinositol 3-phosphate (PtdIns(3)P)-dependent manner16,48. A functional study demonstrated the regulatory roles of the Phafin2–VRK2–Akt protein complex at the lysosome in the course of autophagy induction. These findings support the notion that the intersection between autophagic cell death and apoptosis may be at the mitochondria. The presence of the Akt–Phafin2–VRK2 at the lysosome sheds light on the roles of cell death and survival machinery in mammalian cells that may underlie various human diseases including cancer and autoimmune diseases.
Mitochondria at the crossroad of cell death
Mitochondria-associated proteins may also be responsible for the cross talk between the autophagic and the apoptotic pathways5,9,27,28. Bcl-2 inhibits Beclin-1-dependent autophagy64,65 and phosphorylation of Beclin-1 on T119 via death-associated protein kinases, inhibits the Bcl-2–Beclin-1 interaction and activates autophagy66. The protein complex composed of Beclin-1 and Bcl-2, which physically interacts at the mitochondrial outer membrane, is regulated by calpain-mediated cleavage of Atg5, a key regulator of apoptosis67. Ayg3/12 and Bim, another member of the Bcl-2 family of proteins, have also been shown to inhibit apoptosis via mitochondria26. In this regard, it is worth noting that mitochondria are also targeted for lysosomal-mediated degradation in a process known as mitophagy, a selective degradation of mitochondria by autophagy that often occurs to remove and digest defective mitochondria following damage or stress68. Decreased expression of genes that regulate autophagy or mitophagy can cause degenerative diseases in which deficient quality control results in inflammation and the death of cell populations. Thus, the altering of mitochondrial functions is suggested to be associated with multiple degenerative diseases5.
Beclin-1 and ULK1 (Atg1) are both involved in autophagosome formation and are direct substrates of Akt54,55. Bcl-2 is an anti-apoptotic protein that acts as a major effector of Akt signaling through BAD phosphorylation, which maintains the mitochondrial outer membrane potential to modulate cell survival69–71, in part by inhibiting Beclin-1-dependent autophagy64,71,72. Therefore, it is possible that mTORC1 (via TRAF6-p62-mediated ubiquitination)73 and Akt (via Phafin2–VRK2-induced autophagy) serve as molecular links between autophagy and apoptosis23,48.
Autophagic cell death
Three mechanisms for cell death are reported in mammalian cells; apoptosis (type I), autophagic cell death (type II), and necrosis (type III)6,74. To date, it is not entirely clear whether autophagy is indeed a cell survival mechanism or rather a cell death-inducing mechanism in mammalian cells8,27,75. Autophagy is believed to be a conserved mechanism, in diverged organisms, for the gross disposal and recycling of targeted intracellular proteins13,43,76. However, it also induces cell death with morphological findings that differ from those of apoptosis or necrosis; referred to as “autophagic cell death” or “type II cell death”8,74,75,77.
Apoptotic cell death (type I cell death) is characterized by morphologic changes including plasma membrane blebbing, apoptotic bodies, and nuclear condensation with reduction of cell volume7. Alternatively, necrotic cell death (type III cell death) is characterized by a gain of cell volume or swelling of organelles often associated with plasma membrane rupture without blebbing and subsequent loss of intracellular content. Notably, necrosis can occur as a consequence of apoptosis7,74. Autophagic cell death (type II cell death) is characterized by the appearance of cellular vacuoles without extensive condensation of the nucleus or caspase activation7,74. Morphologically, autophagic cell death differs considerably from cells undergoing apoptosis or necrosis, and is characterized as a form of cell death that mechanistically depends on the autophagic machinery with extensive cytoplasmic vacuolization, phagocytic uptake, and lysosomal degradation78.
Lysosomes can initiate a cell death pathway, designated lysosomal cell death, which is distinct from autophagic cell death8. Lysosomal cell death is mediated by the release of cathepsins into the cytoplasm following lysosomal damage79. However, the molecular processes that occur between the lysosomal degradation of cytosolic components and autophagic cell death are not well characterized11. It is also possible that the three biological mechanisms of cell death (apoptosis, autophagic cell death, and necrosis) act independently or interactively cross regulate each other in cell death and survival machinery8,9,35.
Inactivation of lysosomal membrane-associated protein (LAMP)2 by RNA interference or by homologous recombination leads to autophagic vacuolization in nutrient-depleted cells. Furthermore, cells that lack LAMP2 expression showed an enhanced accumulation of vacuoles carrying the marker LC3, yet a decreased colocalization of LC3 and lysosomes. Thus, supporting the complex relationship between localization of LC3 at the lysosome and enhanced vacuole formation in the course of autophagic cell death and apoptosis80.
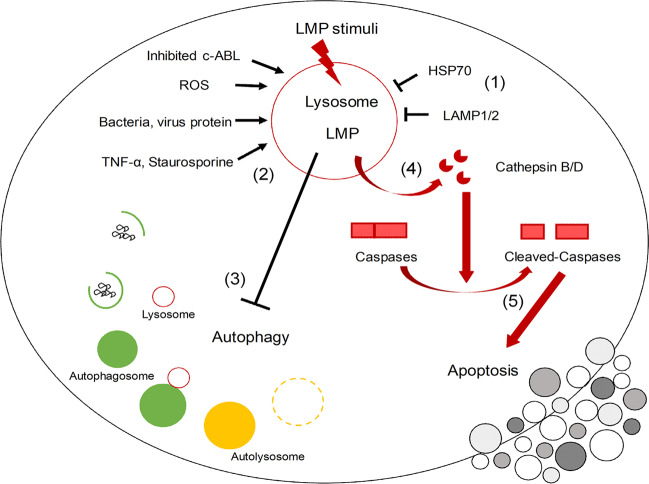
Recent papers further indicate that LAMP3 has a role in autophagy81,82. In these studies, LAMP3 knockdown cells exhibit a suppressed ability to complete the autophagic process. Furthermore, although death by lysosomal membrane permeabilization (LMP) and subsequent release of cathepsins into the cytoplasm has been previously reported, the mechanism of action is not well characterized and the inducing agents have only been described as compounds capable of sensitizing cancer cells18,83 (Fig. 3). Furthermore, LMP can induce cell death via several mechanisms including both caspase-dependent and independent pathways, as well as via activation of the inflammasome to release cytokines84. A common characteristic of LMP-associated cell death is sensitivity to cathepsin inhibitors such as pepstatin A85. In fact, cytoplasmic cathepsins are reported to cleave Bid and induce translocation of the pro-apoptotic proteins Bax and Bak, which in turn induce mitochondrial membrane permeabilization and caspase-dependent apoptosis86.
Fig. 3. The lysosome and lysosomal membrane permeability are central to the balance of autophagy and apoptosis.
(1) LAMP1, LAMP2, and HSP70 protect lysosomal membrane integrity from lysosomal membrane permeability (LMP) stimuli. (2) Lack of these protein, or increased LMP stimuli such as reactive oxide species (ROS), c-ABL dysfunction, bacteria or virus infection, TNF-α, and staurosporine treatment can disrupt lysosomal membrane integrity resulting in LMP. (3) The decrease in matured lysosomes induced by LMP leads to inhibition of autophagy-dependent protein degradation. (4) LMP also initiates relocalization of lysosomal enzyme such as cathepsins into cytoplasm. (5) Released cathepsins activate caspases resulting in apoptosis.
Autophagy as an underlying mechanisms of cancer
Apoptotic cell death and its evasion are shown to be directly involved in pathogenetic conditions of human cancer87–90. For example, activation of Bcl-2, an anti-apoptotic factor, via chromosomal translocation serves to enhance cell survival, which contributes to development of human B cell lymphoma. Furthermore, we previously reported that TCL1 functions as an Akt coactivator by enhancing transphosphorylation within the TCL1-Akt high molecular weight protein complex to enhance cell survival, thereby serving as an underlying molecular mechanism of human T cell prolymphocytic leukemia91–94. Alternatively, the tumor-suppressor gene PTEN dephosphorylates phosphatidylinositol (3,4,5) triphosphate (PIP3) and phosphatildylinositol (3,4) bisphosphate to inhibit Akt activation95,96. Loss of PTEN function leads to an elevated concentration of the PIP3 substrate, and consequent constitutive activation of the downstream components of the PI3K pathway including Akt and its downstream effector mTOR, underlying development of human glioblastoma, melanoma, prostate cancers, lung cancers, and lymphomas97,98. Consistently, PTEN knock-out mice revealed a high incidence of neoplastic diseases including T cell lymphomas, gonadostromal tumor, germline cancers, and, thyroid, prostate, breast, liver, and intestinal tract cancers96,99.
Metabolic regulations are important for controlling cell death27,100. The PI3K–Akt signaling pathways40,95,97,101, originally characterized to mediate insulin signaling, are critical metabolic pathways in mammalian cells required to divert nutrients toward anabolic processes to facilitate enhanced growth and proliferation102–104. Altering the metabolic state in cancer cells, which was originally described as the Warburg effect, increases aerobic glycolysis and is considered to be metabolic hallmarks of cancer cells through both post-translational and transcriptional regulation105,106. The PI3K–Akt–mTOR pathway coordinates the uptake and utilization of multiple nutrients, including glucose, glutamine, nucleotides, and lipids, in a manner best suited for supporting the enhanced growth and proliferation of cancer cells107,108.
Autophagy degrades and recycles proteins and organelles to support metabolism and survival in starvation. Given the possible roles of autophagy in the regulation of cell death machinery, it is possible that it may underlie molecular mechanisms of altered metabolism in human cancer100,109,110. Indeed, autophagy can act as a suppressor of early tumorigenesis through controlling cancer metabolism27,109.
Autophagy was initially considered a tumor suppressive mechanism based on indirect evidence from oncogene and tumor-suppressor gene alteration studies100. In addition autophagy can support tumor growth in multiple tumor types111,112, as well as promote resistance to a variety of therapies113,114. Amplification, or gain of function mutations in PI3K, AKT, or loss of function PTEN mutations, serve to activate mTOR thus inhibiting autophagy, and have, therefore, been defined as underlying mechanisms for oncogenicity, suggesting a potential importance of suppressing autophagy during tumor initiation112,115. To support this notion mice lacking the atg7 gene were created concurrently with K-rasG12D activation to investigate non-small-cell lung cancer. Results found that Atg7-deficient tumors accumulated dysfunctional mitochondria and prematurely induced p53 and proliferative arrest, which reduced tumor burden that was partly relieved by p53 deletion116. In some contexts, autophagy suppresses tumorigenesis, however, in most contexts autophagy facilitates tumorigenesis100,111. Autophagy is shown to promote tumor growth by suppressing the p53 response, maintaining mitochondrial function, sustaining metabolic homeostasis and survival in stress, and preventing diversion of tumor progression to benign oncocytomas suggesting that cancers require autophagy for distinct roles in metabolism that are oncogene- and tumor-suppressor gene-specific111. In this scenario, cancer cells may upregulate autophagy to survive microenvironmental stress and to increase growth and aggressiveness100,111. The signaling link between p62 (sequestosome1, SQSTM1), a core autophagy regulator, and the apoptosis pathway was suggested as a critical decision point that controls cell death and survival in tumorigenesis32. Mice with systemic mosaic deletion of Atg5 and liver-specific Atg7(−)/(−) mice develop benign liver adenomas with mitochondrial swelling, p62 accumulation, and oxidative stress and genomic damage responses. The size of the Atg7(−)/(−) liver tumors was reduced by simultaneous deletion of p62117. These observations together suggest that, analogous to the role of autophagy in normal tissues, that in tumor tissues appear to be complex and context-dependent9,100,109,118.
In addition to the fundamental roles of autophagy as a lysosomal degradation pathway for nutrient/amino acid recycling, autophagy can mediate various autophagy-independent functions, such as metabolic adaptation, cell proliferation, exosome secretion, pathogen control, centrosome functions, endocytosis, phagocytosis, inflammation, and innate immunity11,74. Thus, the involvement of autophagy in cancer pathogenesis may not be solely through controlling cell death machinery, but also through alteration of various cellular conditions. Therefore, attention has turned to autophagy as a pathological survival-promoting pathway for cancer metabolism5,6,27,28,33,75,100 underlying the mechanisms responsible for initiation and maintenance of various types of cancers55,100,117,119–121. Mechanisms by which autophagy promotes cancer include suppressing induction of the p53 tumor-suppressor protein and maintaining metabolic function of mitochondria100. Autophagy can impede early cancer development while facilitating advanced tumor progression33,109. Autophagy can impede early cancer development while facilitating advanced tumor progression33,109. Indeed, autophagy is suggested to underlie the conditions of various aspect of cancer and its pathological conditions including alteration of metabolic condition33,109.
A primary cilium is a microtubule-based sensory organelle that has an important role in human development and disease122. Loss of cilia has been observed in multiple malignant tumors, suggesting a potential suppressive role of cilia in cancer development through the dual interaction between autophagy and ciliogenesis123,124. Akt, primary regulator of cancer development, physically interacts with Inversin at the primary cilia controls normal cilial development125. Therefore, it is possible that Akt may have a regulatory role through the interplay between cilia and autophagy to alter metabolic status for cancer development.
Furthermore, chaperon-mediated autophagy (CMA), the direct delivery of cytosolic proteins targeted for degradation to the lysosome126, is suggested to exert a major regulatory effect on cell metabolism127. Therefore, it is possible that autophagy-induced alteration of energy homeostasis and cell metabolism may underlie the manifestation of cell death associated with cancer. Thus, autophagy inhibition can limit the growth of established tumors to potentially improve responses to cancer therapeutics114.
Autophagy and autoimmune diseases
Dysregulation of autophagy has been implicated in a variety of autoimmune diseases. Several lines of evidence from animal models, genome‐wide association studies (GWAS), and human patients are emerging to support the role of autophagy in progression and pathogenesis of systemic lupus erythematosus (SLE), a prototypic autoimmune disease characterized by the production of multiple autoantibodies in association with systemic inflammation and organ damage128. GWAS have identified several autophagy‐related genes (ATG5, ATG16L2, CDKN1B, CLEC16A, and DRAM1) that are associated with susceptibility to SLE (reviewed in ref. 129). In addition, several variants located in other autophagy‐related genes including ATG7, APOL1, IRGM, LRRK2, MAP1LC3B, and MTMR3 are associated with susceptibility to SLE129. This study strongly supports the notion that autophagy has an important role in the genetic etiology of SLE.
In the pathogenesis of SLE, immune complexes of multiple autoantibodies with nuclear autoantigens can deposit in various tissues and stimulate cytokine production to cause inflammation130. The extracellular release of nuclear autoantigens is likely to be a critical step in the pathway. Although the release is considered to be the consequence of apoptotic cell death, recent findings show that other forms of cell death, including autophagy may be similarly involved. Indeed, enhanced autophagy has been reported in T and B cells from a lupus mouse model, as well as in PBMCs from patients with SLE131. In addition, autophagy-related genes, including mTOR, Beclin-1 (Atg6), LC3, and p62, have been shown to be differentially expressed in PBMCs in SLE patients132. Importantly, mice lacking components of the LC3-associated phagocytosis (LAP) pathway show increased serum levels of inflammatory cytokines and autoantibodies, as well as kidney damage with glomerular immune complex deposition133. Moreover, repeated injection of dying cells into LAP-deficient mice accelerates the development of SLE-like disease133, suggesting the defects in LAP rather than canonical autophagy, may contribute to the pathogenesis of SLE (Fig. 4).
Fig. 4. LC3-associated phagocytosis (LAP) in the clearance of dying cells.
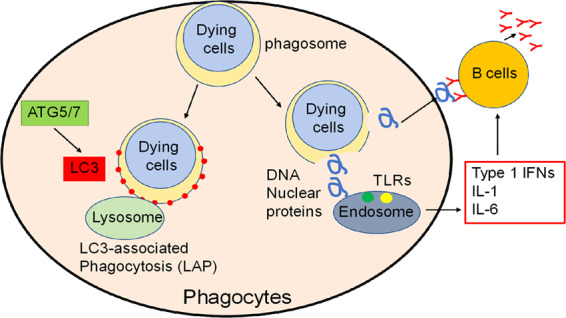
Generally, phagosomes become decorated with LC3 and are then trafficked to the lysosome for degradation (LC3-associated phagocytosis (LAP)). LAP uses a subset of the autophagy machinery distinct from canonical autophagy. As a result, dying cells are rapidly ingested and degraded and do not induce an autoimmune response. If components of the LAP pathway are lacking, dying cells are not degraded and, hence, the nuclear material escapes, which leads to autoimmune responses. Consequently, a lupus-like systemic inflammatory disease develops.
Dysfunction of the immune systems, including B cells and T cells of the adaptive immune systems as well as macrophages, dendritic cells (DCs), innate lymphoid cells (ILCs), and neutrophils of the innate immune systems, are considered to be involved in the pathogenesis of SLE130. Among them, dysfunction of regulatory T cells (Tregs) is likely to be deeply involved134. Indeed, in a murine model of SLE, the number of Tregs decreased along with disease progression135. In addition, the suppressive function of Tregs decreased in patients with active SLE136. Consistent with an earlier mouse study by Wei et al.137, autophagy has been shown to be augmented in Tregs over effector T cells in healthy controls134. However, the difference in autophagy is abolished in SLE patients as autophagy is enhanced in naive and proinflammatory effector CD4+ T cells yet profoundly diminished in Tregs in SLE138. Importantly, IL-21, a key cytokine produced by follicular helper T cells, induces mTOR activation and eliminates autophagy and differentiation of Tregs138 (Fig. 5). Consistently, dual blockade of mTORC1 and mTORC2 by rapamycin treatment induces autophagy, which restores the expression of GATA-3 and CTLA-4, and recovers the function of Tregs138. These findings suggest that increased activity of the IL-21–mTORC1 axis in SLE results in the blockade of autophagy in Tregs, leading to the development of SLE. Consistently, rapamycin has been shown to be efficient in treating SLE patients as well as lupus mice139.
Fig. 5. IL-21-mTOR axis in the regulation of autophagy in Tregs in SLE.
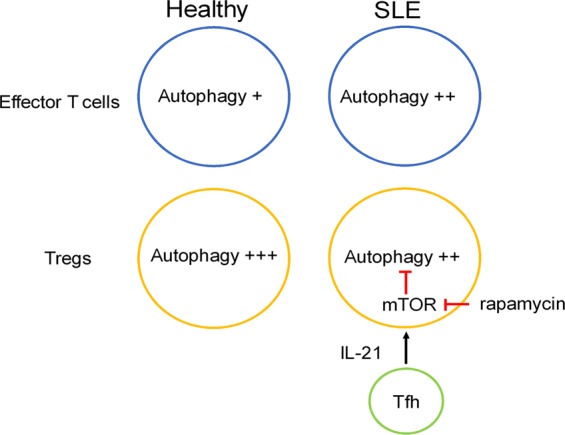
Autophagy is profoundly diminished in Tregs in SLE. IL-21 induces mTOR activation and eliminates autophagy in Tregs. The blockade of mTOR by rapamycin induces autophagy and recovers the function of Tregs.
Recent reports also suggest a role for autophagy in Sjögren’s syndrome, a complex autoimmune disease of secretory glands. Markers of macroautophagy are reported to be increased in tears, conjunctival cells, and salivary glands of primary Sjögren’s patients140,141. Investigation in MRL/lpr mice, which spontaneously develop systemic autoimmune disease that is similar to human SLE and secondary Sjögren’s syndrome, showed that markers of macroautophagy flux and CMA are reduced142. Although these findings seem contradictory, given the role of autophagy in protein degradation and release, markers of autophagy may be expected in gland secretions if the natural progression of protein degradation is affected. Additional studies of the minor salivary glands of Sjögren’s syndrome patients suggest that LAMP3 is upregulated in a subset of patients and contributes to a stalling of autophagy and an increase in LMP. This increase in LAMP3 also results in the release of autoantigens via a caspase-independent process, which may account for the release of other marker of autophagy by exocrine cells. Further studies will be required to more accurately describe whether the role of autophagy is central to the development of Sjögren’s syndrome or if this is a byproduct of cellular dysregulation.
In addition to SLE and Sjögren’s syndrome, the combined data from GWAS and analyses in murine models have implicated autophagy in other autoimmune diseases, including rheumatoid arthritis (RA), multiple sclerosis (MS), and Crohn’s disease. RA is an inflammatory autoimmune disease that primarily affects the joints. Although the role of autophagy in the survival of synovial fibroblasts remains controversial, increased levels of autophagy have been shown in the synovial tissues of active RA patients143. MS is an inflammatory disorder that is characterized by immune system reactivity against myelin in the central nervous system, resulting in relapsing or progressive neurological degeneration. Inhibition of autophagy in CD4+ T cells has been shown to exhibit a protective role in experimental autoimmune encephalomyelitis (EAE)144, a murine model of MS. Consistently, rapamycin treatment reduces the disease activity of EAE via inhibition of autophagy145. Moreover, GWAS have identified Atg16L1 and immunity-related GTPase family M (IRGM) in Crohn’s disease146, indicating a role of autophagy in the pathogenesis of Crohn’s disease.
Conclusion and perspective
Autophagy, originally described as a cellular survival mechanism initiated upon starvation conditions, underlies various aspect of cellular events by recycling intracellular proteins at the lysosome1–4. However, under certain conditions such as cancer pathogenesis, autophagy can induce cell death both directly and indirectly5–7. Experimental approaches including targeted deletion Atg genes in mice111, support a biphasic complex role for autophagy in the context of cell death109. Among the wide variety of cellular responses controlled by autophagy, metabolic function may have a key role in the progression of autophagic cell death8. Clarification of the roles played by autophagy in the cell death machinery will open a new avenue for the treatment of disease in which autophagy modulation may be a novel and impactful molecular target for cancer therapy and autoimmune diseases7,147.
Acknowledgements
M.N. is supported by Takeda and Naito Foundation. N.H. from JSPS (16K20872), F.S. from JSPS 16K08705, AMED 19gm5910024h0003. T.T. from JSPS Research Fellowship for Japanese Biomedical and Behavioral Researchers at NIH (71713). This study was supported by an NIDCR, NIH intramural research grant to J.A.C. (1ZIADE000695).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by G. M. Fimia
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.de Duve C. The lysosome turns fifty. Nat. Cell Biol. 2005;7:847–849. doi: 10.1038/ncb0905-847. [DOI] [PubMed] [Google Scholar]
- 2.Mindell JA. Lysosomal acidification mechanisms. Annu. Rev. Physiol. 2012;74:69–86. doi: 10.1146/annurev-physiol-012110-142317. [DOI] [PubMed] [Google Scholar]
- 3.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 4.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109–1112. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kroemer G, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green DR. The coming decade of cell death research: five riddles. Cell. 2019;177:1094–1107. doi: 10.1016/j.cell.2019.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galluzzi L, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galluzzi L, Green DR. Autophagy-independent functions of the autophagy machinery. Cell. 2019;177:1682–1699. doi: 10.1016/j.cell.2019.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elmore S. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryter SW, Mizumura K, Choi AM. The impact of autophagy on cell death modalities. Int. J. Cell Biol. 2014;2014:502676. doi: 10.1155/2014/502676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nat. Rev. Cancer. 2005;5:886–897. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- 13.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang B, et al. Proton pump inhibitor pantoprazole abrogates adriamycin-resistant gastric cancer cell invasiveness via suppression of Akt/GSK-beta/beta-catenin signaling and epithelial-mesenchymal transition. Cancer Lett. 2015;356:704–712. doi: 10.1016/j.canlet.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Zoncu R, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirata N, et al. Functional characterization of lysosomal interaction of Akt with VRK2. Oncogene. 2018;37:5367–5386. doi: 10.1038/s41388-018-0330-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aits S, Jaattela M. Lysosomal cell death at a glance. J. Cell Sci. 2013;126:1905–1912. doi: 10.1242/jcs.091181. [DOI] [PubMed] [Google Scholar]
- 18.Boya P, Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene. 2008;27:6434–6451. doi: 10.1038/onc.2008.310. [DOI] [PubMed] [Google Scholar]
- 19.Settembre C, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat. Rev. Mol. Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lum JJ, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 22.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J. Clin. Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noguchi M, Hirata N, Suizu F. The links between AKT and two intracellular proteolytic cascades: ubiquitination and autophagy. Biochim. Biophys. Acta. 2014;1846:342–352. doi: 10.1016/j.bbcan.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Boya P, et al. Inhibition of macroautophagy triggers apoptosis. Mol. Cell. Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubinstein AD, Eisenstein M, Ber Y, Bialik S, Kimchi A. The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol. Cell. 2011;44:698–709. doi: 10.1016/j.molcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 26.Radoshevich L, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142:590–600. doi: 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green DR, Galluzzi L, Kroemer G. Cell biology. Metabolic control of cell death. Science. 2014;345:1250256. doi: 10.1126/science.1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noguchi M, Hirata N, Edamura T, Ishigaki S, Suizu F. Intersection of apoptosis and autophagy cell death pathways. Austin J. Mol. Cell. Biol. 2015;2:1004–1007. [Google Scholar]
- 29.Gump JM, Thorburn A. Autophagy and apoptosis: what is the connection? Trends Cell Biol. 2011;21:387–392. doi: 10.1016/j.tcb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Debnath J, Baehrecke EH, Kroemer G. Does autophagy contribute to cell death? Autophagy. 2005;1:66–74. doi: 10.4161/auto.1.2.1738. [DOI] [PubMed] [Google Scholar]
- 31.Edinger AL, Thompson CB. Death by design: apoptosis, necrosis and autophagy. Curr. Opin. Cell Biol. 2004;16:663–669. doi: 10.1016/j.ceb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 32.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy. Apoptosis Cancer Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimmelman AC, White E. Autophagy and tumor metabolism. Cell Metab. 2017;25:1037–1043. doi: 10.1016/j.cmet.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen HM, Codogno P. Autophagic cell death: Loch Ness monster or endangered species? Autophagy. 2011;7:457–465. doi: 10.4161/auto.7.5.14226. [DOI] [PubMed] [Google Scholar]
- 35.Tang D, Kang R, Berghe TV, Vandenabeele P, Kroemer G. The molecular machinery of regulated cell death. Cell Res. 2019;29:347–364. doi: 10.1038/s41422-019-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mills KR, Reginato M, Debnath J, Queenan B, Brugge JS. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is required for induction of autophagy during lumen formation in vitro. Proc. Natl Acad. Sci. USA. 2004;101:3438–3443. doi: 10.1073/pnas.0400443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prins J, et al. Tumour necrosis factor induced autophagy and mitochondrial morphological abnormalities are mediated by TNFR-I and/or TNFR-II and do not invariably lead to cell death. Biochem. Soc. Trans. 1998;26:S314. doi: 10.1042/bst026s314. [DOI] [PubMed] [Google Scholar]
- 38.Thorburn J, et al. Selective inactivation of a Fas-associated death domain protein (FADD)-dependent apoptosis and autophagy pathway in immortal epithelial cells. Mol. Biol. Cell. 2005;16:1189–1199. doi: 10.1091/mbc.E04-10-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi KC, Kim SH, Ha JY, Kim ST, Son JH. A novel mTOR activating protein protects dopamine neurons against oxidative stress by repressing autophagy related cell death. J. Neurochem. 2010;112:366–376. doi: 10.1111/j.1471-4159.2009.06463.x. [DOI] [PubMed] [Google Scholar]
- 40.Manning BD, Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lien EC, Dibble CC, Toker A. PI3K signaling in cancer: beyond AKT. Curr. Opin. Cell Biol. 2017;45:62–71. doi: 10.1016/j.ceb.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arico S, et al. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J. Biol. Chem. 2001;276:35243–35246. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- 43.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindmo K, Stenmark H. How a RING finger protein and a steroid hormone control autophagy. Autophagy. 2006;2:321–322. doi: 10.4161/auto.3091. [DOI] [PubMed] [Google Scholar]
- 45.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat. Rev. Mol. Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 46.Madia F, et al. Oncogene homologue Sch9 promotes age-dependent mutations by a superoxide and Rev1/Polzeta-dependent mechanism. J. Cell Biol. 2009;186:509–523. doi: 10.1083/jcb.200906011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaeberlein M, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 48.Matsuda-Lennikov M, et al. Lysosomal interaction of Akt with Phafin2: a critical step in the induction of autophagy. PLoS ONE. 2014;9:e79795. doi: 10.1371/journal.pone.0079795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Menon S, et al. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arias E, et al. Lysosomal mTORC2/PHLPP1/Akt regulate chaperone-mediated autophagy. Mol. Cell. 2015;59:270–284. doi: 10.1016/j.molcel.2015.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, et al. Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat. Cell Biol. 2016;18:1065–1077. doi: 10.1038/ncb3407. [DOI] [PubMed] [Google Scholar]
- 53.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 54.Bach M, Larance M, James DE, Ramm G. The serine/threonine kinase ULK1 is a target of multiple phosphorylation events. Biochem. J. 2011;440:283–291. doi: 10.1042/BJ20101894. [DOI] [PubMed] [Google Scholar]
- 55.Wang RC, et al. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338:956–959. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang E, et al. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 57.Datta SR, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 58.Palmieri M, et al. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat. Commun. 2017;8:14338. doi: 10.1038/ncomms14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rohatgi RA, et al. Beclin 1 regulates growth factor receptor signaling in breast cancer. Oncogene. 2015;34:5352–5362. doi: 10.1038/onc.2014.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen W, et al. The lysosome-associated apoptosis-inducing protein containing the pleckstrin homology (PH) and FYVE domains (LAPF), representative of a novel family of PH and FYVE domain-containing proteins, induces caspase-independent apoptosis via the lysosomal-mitochondrial pathway. J. Biol. Chem. 2005;280:40985–40995. doi: 10.1074/jbc.M502190200. [DOI] [PubMed] [Google Scholar]
- 62.Pedersen NM, et al. The PtdIns3P-binding protein phafin 2 mediates epidermal growth factor receptor degradation by promoting endosome fusion. Traffic. 2012;13:1547–1563. doi: 10.1111/j.1600-0854.2012.01400.x. [DOI] [PubMed] [Google Scholar]
- 63.Donia T, et al. Identification of RNA aptamer which specifically interacts with PtdIns(3)P. Biochem. Biophys. Res. Commun. 2019;517:146–154. doi: 10.1016/j.bbrc.2019.07.034. [DOI] [PubMed] [Google Scholar]
- 64.Pattingre S, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 65.Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008;27(Suppl 1):S137–148. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sinha S, Colbert CL, Becker N, Wei Y, Levine B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy. 2008;4:989–997. doi: 10.4161/auto.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yousefi S, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 68.Du Toit A. Protein degradation: An alternative route for mitochondrial quality control. Nat. Rev. Mol. Cell Biol. 2014;15:150–151. doi: 10.1038/nrm3755. [DOI] [PubMed] [Google Scholar]
- 69.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 70.Moldoveanu T, Follis AV, Kriwacki RW, Green DR. Many players in BCL-2 family affairs. Trends Biochem. Sci. 2014;39:101–111. doi: 10.1016/j.tibs.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol. Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Linares JF, et al. K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol. Cell. 2013;51:283–296. doi: 10.1016/j.molcel.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Green, D. Means to an End: Apoptosis and Other Cell Death Mechanisms (Cold Spring Harbor Laboratory Press, 2010).
- 75.Green DR, Levine B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell. 2014;157:65–75. doi: 10.1016/j.cell.2014.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tasdemir E, et al. Methods for assessing autophagy and autophagic cell death. Methods Mol. Biol. 2008;445:29–76. doi: 10.1007/978-1-59745-157-4_3. [DOI] [PubMed] [Google Scholar]
- 78.Galluzzi L, et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811–1836. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gomez-Sintes R, Ledesma MD, Boya P. Lysosomal cell death mechanisms in aging. Ageing Res. Rev. 2016;32:150–168. doi: 10.1016/j.arr.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 80.Gonzalez-Polo RA, et al. The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J. Cell Sci. 2005;118:3091–3102. doi: 10.1242/jcs.02447. [DOI] [PubMed] [Google Scholar]
- 81.Dominguez-Bautista JA, et al. Loss of lysosome-associated membrane protein 3 (LAMP3) enhances cellular vulnerability against proteasomal inhibition. Eur. J. Cell Biol. 2015;94:148–161. doi: 10.1016/j.ejcb.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 82.Nagelkerke A, et al. LAMP3 is involved in tamoxifen resistance in breast cancer cells through the modulation of autophagy. Endocr. Relat. Cancer. 2014;21:101–112. doi: 10.1530/ERC-13-0183. [DOI] [PubMed] [Google Scholar]
- 83.Gonzalez P, et al. Impairment of lysosomal integrity by B10, a glycosylated derivative of betulinic acid, leads to lysosomal cell death and converts autophagy into a detrimental process. Cell Death Differ. 2012;19:1337–1346. doi: 10.1038/cdd.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Heid ME, et al. Mitochondrial reactive oxygen species induces NLRP3-dependent lysosomal damage and inflammasome activation. J. Immunol. 2013;191:5230–5238. doi: 10.4049/jimmunol.1301490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Serrano-Puebla A, Boya P. Lysosomal membrane permeabilization as a cell death mechanism in cancer cells. Biochem. Soc. Trans. 2018;46:207–215. doi: 10.1042/BST20170130. [DOI] [PubMed] [Google Scholar]
- 86.Oberle C, et al. Lysosomal membrane permeabilization and cathepsin release is a Bax/Bak-dependent, amplifying event of apoptosis in fibroblasts and monocytes. Cell Death Differ. 2010;17:1167–1178. doi: 10.1038/cdd.2009.214. [DOI] [PubMed] [Google Scholar]
- 87.Fernald K, Kurokawa M. Evading apoptosis in cancer. Trends Cell Biol. 2013;23:620–633. doi: 10.1016/j.tcb.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Benedict CA, Norris PS, Ware CF. To kill or be killed: viral evasion of apoptosis. Nat. Immunol. 2002;3:1013–1018. doi: 10.1038/ni1102-1013. [DOI] [PubMed] [Google Scholar]
- 89.Letai A. Apotosis and cancer. Ann. Rev. Cancer Biol. 2017;1:275–294. [Google Scholar]
- 90.Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 91.Laine J, Kunstle G, Obata T, Sha M, Noguchi M. The protooncogene TCL1 is an Akt kinase coactivator. Mol. Cell. 2000;6:395–407. doi: 10.1016/s1097-2765(00)00039-3. [DOI] [PubMed] [Google Scholar]
- 92.Kunstle G, et al. Identification of Akt association and oligomerization domains of the Akt kinase coactivator TCL1. Mol. Cell. Biol. 2002;22:1513–1525. doi: 10.1128/mcb.22.5.1513-1525.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Laine J, Kunstle G, Obata T, Noguchi M. Differential regulation of Akt kinase isoforms by the members of the TCL1 oncogene family. J. Biol. Chem. 2002;277:3743–3751. doi: 10.1074/jbc.M107069200. [DOI] [PubMed] [Google Scholar]
- 94.Noguchi M, Ropars V, Roumestand C, Suizu F. Proto-oncogene TCL1: more than just a coactivator for Akt. FASEB J. 2007;21:2273–2284. doi: 10.1096/fj.06-7684com. [DOI] [PubMed] [Google Scholar]
- 95.Eng C. PTEN: one gene, many syndromes. Hum. Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- 96.Lian Z, Di Cristofano A. Class reunion: PTEN joins the nuclear crew. Oncogene. 2005;24:7394–7400. doi: 10.1038/sj.onc.1209089. [DOI] [PubMed] [Google Scholar]
- 97.Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc. Natl Acad. Sci. USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 99.Carracedo A, Alimonti A, Pandolfi PP. PTEN level in tumor suppression: how much is too little? Cancer Res. 2011;71:629–633. doi: 10.1158/0008-5472.CAN-10-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.White E. The role for autophagy in cancer. J. Clin. Invest. 2015;125:42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fruman DA, et al. The PI3K pathway in human disease. Cell. 2017;170:605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Whiteman EL, Cho H, Birnbaum MJ. Role of Akt/protein kinase B in metabolism. Trends Endocrinol. Metab. 2002;13:444–451. doi: 10.1016/s1043-2760(02)00662-8. [DOI] [PubMed] [Google Scholar]
- 104.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 105.Lien EC, Lyssiotis CA, Cantley LC. Metabolic reprogramming by the PI3K-Akt-mTOR pathway in cancer. Recent Results Cancer Res. 2016;207:39–72. doi: 10.1007/978-3-319-42118-6_3. [DOI] [PubMed] [Google Scholar]
- 106.Morita M, et al. PKM1 confers metabolic advantages and promotes cell-autonomous tumor cell growth. Cancer Cell. 2018;33:355–367 e357. doi: 10.1016/j.ccell.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 107.Robey RB, Hay N. Is Akt the “Warburg kinase”?-Akt-energy metabolism interactions and oncogenesis. Semin. Cancer Biol. 2009;19:25–31. doi: 10.1016/j.semcancer.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hoxhaj G, Manning BD. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer. 2020;20:74–88. doi: 10.1038/s41568-019-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Santana-Codina N, Mancias JD, Kimmelman AC. The roles of autophagy in cancer. Annu. Rev. Cancer Biol. 2017;1:19–39. doi: 10.1146/annurev-cancerbio-041816-122338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 2018;20:521–527. doi: 10.1038/s41556-018-0092-5. [DOI] [PubMed] [Google Scholar]
- 111.Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell. 2013;155:1216–1219. doi: 10.1016/j.cell.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913–1930. doi: 10.1101/gad.287524.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Amaravadi RK, Kimmelman AC, Debnath J. Targeting autophagy in cancer: Recent advances and future directions. Cancer Discov. 2019;9:1167–1181. doi: 10.1158/2159-8290.CD-19-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Maiuri MC, et al. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ. 2009;16:87–93. doi: 10.1038/cdd.2008.131. [DOI] [PubMed] [Google Scholar]
- 116.Guo JY, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Takamura A, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 119.Santanam U, et al. Atg7 cooperates with Pten loss to drive prostate cancer tumor growth. Genes Dev. 2016;30:399–407. doi: 10.1101/gad.274134.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Poillet-Perez L, White E. Role of tumor and host autophagy in cancer metabolism. Genes Dev. 2019;33:610–619. doi: 10.1101/gad.325514.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kenific CM, Debnath J. Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol. 2015;25:37–45. doi: 10.1016/j.tcb.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 123.Cao M, Zhong Q. Cilia in autophagy and cancer. Cilia. 2015;5:4. doi: 10.1186/s13630-016-0027-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pampliega O, Cuervo AM. Autophagy and primary cilia: dual interplay. Curr. Opin. Cell Biol. 2016;39:1–7. doi: 10.1016/j.ceb.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Suizu F, et al. Phosphorylation-dependent Akt-Inversin interaction at the basal body of primary cilia. EMBO J. 2016;35:1346–1363. doi: 10.15252/embj.201593003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schneider JL, Suh Y, Cuervo AM. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 2014;20:417–432. doi: 10.1016/j.cmet.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dorner T, Furie R. Novel paradigms in systemic lupus erythematosus. Lancet. 2019;393:2344–2358. doi: 10.1016/S0140-6736(19)30546-X. [DOI] [PubMed] [Google Scholar]
- 129.Qi YY, Zhou XJ, Zhang H. Autophagy and immunological aberrations in systemic lupus erythematosus. Eur. J. Immunol. 2019;49:523–533. doi: 10.1002/eji.201847679. [DOI] [PubMed] [Google Scholar]
- 130.Tsokos GC. Systemic lupus erythematosus. N. Engl. J. Med. 2011;365:2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 131.Clarke AJ, et al. Autophagy is activated in systemic lupus erythematosus and required for plasmablast development. Ann. Rheum. Dis. 2015;74:912–920. doi: 10.1136/annrheumdis-2013-204343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wu ZZ, et al. Expression of autophagy related genes mTOR, Becline-1, LC3 and p62 in the peripheral blood mononuclear cells of systemic lupus erythematosus. Am. J. Clin. Exp. Immunol. 2017;6:1–8. [PMC free article] [PubMed] [Google Scholar]
- 133.Martinez J, et al. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature. 2016;533:115–119. doi: 10.1038/nature17950. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 134.Mizui M, Tsokos GC. Targeting regulatory T cells to treat patients with systemic lupus erythematosus. Front. Immunol. 2018;9:786. doi: 10.3389/fimmu.2018.00786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Humrich JY, et al. Homeostatic imbalance of regulatory and effector T cells due to IL-2 deprivation amplifies murine lupus. Proc. Natl Acad. Sci. USA. 2010;107:204–209. doi: 10.1073/pnas.0903158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bonelli M, et al. Quantitative and qualitative deficiencies of regulatory T cells in patients with systemic lupus erythematosus (SLE) Int. Immunol. 2008;20:861–868. doi: 10.1093/intimm/dxn044. [DOI] [PubMed] [Google Scholar]
- 137.Wei J, et al. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat. Immunol. 2016;17:277–285. doi: 10.1038/ni.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kato H, Perl A. Blockade of Treg cell differentiation and function by the interleukin-21-mechanistic target of rapamycin axis via suppression of autophagy in patients with systemic lupus erythematosus. Arthritis Rheumatol. 2018;70:427–438. doi: 10.1002/art.40380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lai ZW, et al. Sirolimus in patients with clinically active systemic lupus erythematosus resistant to, or intolerant of, conventional medications: a single-arm, open-label, phase 1/2 trial. Lancet. 2018;391:1186–1196. doi: 10.1016/S0140-6736(18)30485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Alessandri C, et al. CD4 T lymphocyte autophagy is upregulated in the salivary glands of primary Sjogren’s syndrome patients and correlates with focus score and disease activity. Arthritis Res. Ther. 2017;19:178. doi: 10.1186/s13075-017-1385-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Byun YS, Lee HJ, Shin S, Chung SH. Elevation of autophagy markers in Sjogren syndrome dry eye. Sci. Rep. 2017;7:17280. doi: 10.1038/s41598-017-17128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li B, Wang F, Schall N, Muller S. Rescue of autophagy and lysosome defects in salivary glands of MRL/lpr mice by a therapeutic phosphopeptide. J. Autoimmun. 2018;90:132–145. doi: 10.1016/j.jaut.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 143.Zhu L, et al. The autophagy level is increased in the synovial tissues of patients with active rheumatoid arthritis and is correlated with disease severity. Mediators Inflamm. 2017;2017:7623145. doi: 10.1155/2017/7623145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kovacs JR, et al. Autophagy promotes T-cell survival through degradation of proteins of the cell death machinery. Cell Death Differ. 2012;19:144–152. doi: 10.1038/cdd.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liang P, Le W. Role of autophagy in the pathogenesis of multiple sclerosis. Neurosci. Bull. 2015;31:435–444. doi: 10.1007/s12264-015-1545-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.McCarroll SA, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat. Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat. Rev. Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]