Abstract
Salinity and drought are the most important abiotic stresses hampering crop growth and yield. It has been estimated that arid areas cover between 41% and 45% of the total Earth area worldwide. At the same time, the world’s population is going to soon reach 9 billion and the survival of this huge amount of people is dependent on agricultural products. Plants growing in saline/arid soil shows low germination rate, short roots, reduced shoot biomass, and serious impairment of photosynthetic efficiency, thus leading to a substantial loss of crop productivity, resulting in significant economic damage. However, plants should not be considered as single entities, but as a superorganism, or a holobiont, resulting from the intimate interactions occurring between the plant and the associated microbiota. Consequently, it is very complex to define how the plant responds to stress on the basis of the interaction with its associated plant growth-promoting bacteria (PGPB). This review provides an overview of the physiological mechanisms involved in plant survival in arid and saline soils and aims at describing the interactions occurring between plants and its bacteriome in such perturbed environments. The potential of PGPB in supporting plant survival and fitness in these environmental conditions has been discussed.
Keywords: salinity, aridity, microbial community, rhizosphere, PGPB, plant stress
1. Arid and Saline Soil
Drought and salinity are the most important abiotic stresses hampering crop growth and yield. Many environmental and anthropogenic factors influence the soil salinization and its aridity: the scarce vegetation cover associated with low evapotranspiration, drainage, irrigation water quality, agronomic practices (e.g., fertilization and pesticide application), saline groundwater intrusion, marine floods, and shallow aquifers that facilitate salt accumulation. In general, soil salinity is classified as “primary”, if salt is accumulated by natural process and “secondary” if the accumulation is a consequence of the management of natural resources [1,2]. Prolonged exposure to meteorological agents such as precipitations and winds can result in semi-arid and arid areas with naturally saline soils [3]. In these areas, rainfall is not enough to leach soluble salts from the soil [4]. This kind of salinity, called “transient salinity”, falls into one of the three types of the salinity examined by Rengasamy [2], together with “salinity associated to groundwater” (also known as “dryland salinity”) and “irrigation salinity”.
Reclamation of soil salinity and sodicity includes a combination of different physical, chemical (amendments as CaCl2·2H2O and CaSO4·2H2O), hydrological, and biological (biofertilizers and genetic engineering) methods aimed to remove Na+ from colloids and from the root zone, in order to allow plant cultivation in arid and semiarid areas [5].
Soil salinity is a global problem that severely affects more than 800 million hectares of arable land worldwide [6,7]. A soil is defined “saline” if the electrical conductivity of its saturation extract (ECe), measured at 25 °C, is over 4 dS m−1 [8], its salt concentration is ≥0.25 % and pH < 8.5; it must be considered that salinity of sea water ranges between 3 % and 4 % [9]. Salinity in soil is mainly due to NaCl, but also carbonates and sulfates of Ca, Mg, and K can be accumulated in large amounts. When soil clay particles absorb sodium until leading to ECe lower than 4 dS m−1 and pH > 8.5, many physico-chemical problems begin to occur, and the soil becomes “sodic” [9].
An excess of soluble salt accumulation in arable soils negatively affects agriculture, with a heavy economic impact due to low crop productivity, with repercussions on human nutrition and health [9]. Arid areas are considered the largest biome on Earth and cover 41% of the total Earth area in the world [10]. More recent estimates increase this percentage to 45% [11], highlighting the development of new vast and increasingly arid territories located in Alaska, Siberia, and towards lower latitudes, such as in central-Southern Asia, where only China represents the 55% of the lands newly exposed to aridity in Asia, or 31% worldwide. Southeast Africa and the North, including Morocco, Algeria, and Libya, have also seen an increase in drought. In North and Central America, the largest expansions of arid areas occur in the United States, Canada, and Mexico, while in South Argentina, shows an increase in dry areas equal to about 750,000 km2, or 27% of the national area, while in Chile, they exceed 50,000 km2. Australia has also been affected by an expansion of total arid lands of over 280,000 km2 (4% of the total area). In a study conducted in 2017, it emerged that the part of the territory with high or very high risk of desertification had increased, in less than a decade, by 177,000 km2 [12].
Studies carried out between 1999 and 2013 show that around 34% of European ecosystems have undergone significant alterations in their functioning. In Europe, salinization affects about 1 million hectares of lands, especially those of Mediterranean countries [1]. These include vast regions in Ireland, Germany, Poland, Greece, Italy, Spain, and Turkey. On average, 2.4% of European ecosystems have undergone a strong negative variation every year, corresponding to dry years and very dry seasons; 2005 was the year with the highest rate of sudden changes [13].
This work is an overview of the main processes occurring in the microbial community present in arid and saline perturbed soils. In particular, we focused our attention on arid environments, on the PGPB role, and on the bacterial communities occurring in saline soil. Moreover, the interaction between rhizobacteria and plants grown in arid soil and the plant responses to salt were explored.
2. Rhizosphere: A Scenario for Plant-Microbe Interaction
It was 1904 when Lorenz Hiltner [14] proposed the term “rhizosphere” for the first time, indicating the portion of soil attached to the root, in which the microbial density was higher than in uncultivated soil. Several definitions of rhizosphere were proposed afterwards, but the Hiltner one remained universally accepted [15].
The higher bacterial density observed by Hiltner in the rhizosphere is due to the release by the root of different compounds in solid, liquid, or gaseous forms through the secretion process known as root exudation or rhizodeposition. The flow of photosynthetic carbon that spreads from the root to the soil consists in the secretion activity of exudates such as sugars, organic acids, vitamins, mucilage, lysates, and many other substances [16]. There is a general consensus on the fact that about 11% of the total fixed C (or about 27% of the C allocated to roots) is released in the surrounding soil. However, these values, as well as the composition of the root exudates, vary according to the plant species, age, or plant nutritional status and to the exposition of environmental (biotic and abiotic) factors [16,17,18]. A direct link subsists between the composition of the root exudates and the composition of the associated microbiota. In fact, such molecules can play an important role as chemoattractants and repellents for bacteria, fungi, and viruses, which in turn, can affect plant growth, yield, health, and the nutritional value of seeds and fruits [19,20].
This concept is known as the rhizosphere feed-back loop and it is a dynamic process. The rhizodeposition modifies the environment surrounding the root, and the soil microbiota sense these variations. Then, different bacterial populations respond to the environmental modification, induced by the release of root exudates, modifying their own physiology. This leads to a selection favoring the bacterial populations best adapted to this new condition. This is at the base of the coevolution between the plant and its microbiota [21]. As a result, the induced shift in the microbial community composition and activity leads to different effects on plant development. Microorganisms can stimulate rhizodeposition and establish relationships for nutrient acquisition [21]. They can produce and consume trace gas, influence acidity, water availability, and hydrophobicity of soil. Moreover, they can degrade xenobiotic compounds and chelate metals [22]. On the other hand, there are also harmful parasitic interactions for plants and even fatal ones. Soil borne diseases in agricultural land reduce both the yield of the crop product and its quality [23], thus leading to lower financial returns. As an example, yield losses due to pathogenic organisms in rice ranged from 24% to 41% in Asia [24]; similarly, an amount ranging from 5% to 96% of potato production in France is lost due to plant diseases [25].
The plant selects the most suitable species-specific microbial composition, but the community structure and stability also depend on the type of soil [26]. In fact, different plant species host distinct microbial communities and specific microbial taxa are preferentially associated with specific plant species [26]. It is also true, however, that many soil bacterial taxa can be cosmopolitan and able to associate with a wide range of plant taxa [22].
As easily understood, the relation between plant development and its associated microbiota is so intimate that we referred to the plant as a superorganism or a holobiont, resulting directly from very complex plant-microorganisms and microbe-microbe interactions [27,28].
3. Plant Growth-Promoting Bacteria (PGPB)
As previously stated, following the release of root exudates, a large amount of microorganisms colonize plant roots with a different degree of intimacy (ranging from bacteria living in the rhizoplane to endophytic bacteria, who enter the root and occupy the internal plant tissues or even cells) and with different effect of plant growth and health. Among them, plant growth-promoting bacteria (PGPB) are typically defined as symbiotic or free-living bacteria in soil, able to efficiently colonize the root and induce a beneficial impact on the host plant. The major taxonomic groups of PGPB belong to Proteobacteria and Firmicutes [29,30]. Bacillus sp. is the most known genus of Firmicutes inducing plant growth promotion. Strains ascribed to the genera Rhizobium, Azospirillum, Burkholderia, Achromobacter, Pseudomonas, Acinetobacter, Serratia, Pantoea, Enterobacter, and Rahnella are the most representative of the phylum Proteobacteria reported to behave as PGPB [31].
The stimulation of plant growth occurs by direct and indirect mechanisms. While direct mechanisms include the facilitation of nutrient acquisition and the modulation of plant hormone levels, indirect mechanisms are based on the suppression of phytopathogenic microorganisms responsible for soil borne diseases, thereby acting as a biocontrol agent [32]. In agreement with the topic of the review, a brief overview of the direct mechanisms (Figure 1) is reported in the next paragraph.
Figure 1.
Overview of the main direct mechanisms used by PGPB. Improvement of mineral nutrition via nitrogen fixation, phosphate solubilization, and iron chelation, as well as the modulation of phytohormones levels.
3.1. Stimulation of Plant Growth Induced by Direct Mechanisms
PGPB-dependent plant growth promotion can occur through the improvement of plant mineral nutrition, especially as far as nitrogen (N), phosphate (P), and iron (Fe) uptake are concerned [33]. This enhancement of P, N, and potassium (K) uptake in plants, especially in the shoot, can reduce the accumulation of Na+ and Ca+ and this is an important process for the plants growing in saline/arid soil.
Nitrogen is one of the most important nutrients for plant growth and yield: this element is very abundant in the atmosphere as N2, but unavailable to plants in such form. However, N2 can be converted in ammonia with the help of specific groups of microorganisms able to carry out the biological N fixation (BNF) through the activity of the nitrogenase [34,35]. N-fixing microorganisms can be free living or symbiotic. Free-living microorganisms include genera as Azospirillum and Azotobacter [36]. Among the symbiotic bacteria, the members of the family Rhizobiaceae (Rhizobium, Bradyrhizobium, Sinorhizobium, Azorhizobium, and Mesorhizobium) can establish symbiotic relationships with legume, while bacteria belonging to Frankia genus can be associated with other plant families among which, Betulaceae (Alnus glutinosa for example) [37].
The close dialogue between rhizobia and the leguminous plants, mediated by the bacterial lipochitooligosaccharides (Nod factors) and plant flavonoids, leads to the formation of root nodules [36,38]. The different zones of the nodule are readily colonized by rhizobia cells that differentiate into bacteroids with diverse size and metabolic activity [38]. Inside the nodule, N fixation is driven by the nitrogenase enzyme that is composed by two subunits: the dinitrogenase reductase, with iron as cofactor, and dinitrogenase, having iron and molybdenum as cofactors [35]. Nitrogenase is oxygen sensitive, but rhizobia are strictly aerobic microorganisms: the trade-off solution is offered by the synthesis of leg-hemoglobin, a protein binding oxygen with high affinity, therefore ensuring nitrogenase activity, as well as bacterial viability [39]. Moreover, the nodule external structure, made by compact layers of cells, is characterized by a low permeability to O2 and is involved in reducing oxygen availability [40].
Nitrogen accumulation in crop plants can be reduced by salinity since an increase in the uptake of Cl− occurs together with the decrease in shoot NO−3 of many crops. Moreover, soil salinity may hamper symbiotic N2-fixation by reducing the density of rhizobia in the soil and their efficiency in colonizing root hairs. Even though the symbiosis is established, the stunted growth of the host plant in such condition leads to a reduction of the amount of photosynthate transferred to the nodule [41].
Non-symbiotic diazotrophic bacteria are able to fix N in non-leguminous plants: Azospirillum and Azotobacter are the most studied genera among them. Azospirillum is a Gram-negative, non-nodule-forming aerobic N-fixing bacterium belonging to the family Spirillaceae [42], while Azotobacter is free-living, aerobic, photoautotrophic non symbiotic bacterium assigned to the family Azotobacteriaceae. The most beneficial species are Azospirillum lipoferum/brasilense and Azotobacter chroococcum/vinelandii, respectively [43].
The mechanisms at the base of the positive effects on plant growth induced by strains of these two genera are not limited to N fixation. Azospirillum strains synthesize phytohormones (IAA, gibberellins, and cytokinin) and modulate root development and uptake of plant nutrients (N, P, and K). Similarly, Azotobacter releases vitamin B complex, different phytohormones such as gibberellins, naphthalene acetic acid, and other substances involved in biocontrol of root pathogens and promoting root growth and mineral uptake [44].
Similar to N, P is very abundant in soil, but its bioavailability is very low. P in soil is present in two main insoluble forms: mineral forms such as apatite, hydroxyapatite, and oxyapatite, and organic forms including inositol phosphate (soil phytate), phosphomonoesters, phosphodiesters, and phosphotriesters [45,46]. It has been estimated that the average P content of soil is about 0.05% (w/w): only 0.1% of this amount is available for plants [47]. As a consequence, P-based fertilizers are usually applied; however, right after the application, a fraction corresponding to 75–90% of the added P precipitates with metal-cation complexes, rapidly becomes fixed in soil [48,49,50].
In this context, phosphate solubilizing bacteria represent a sustainable and environment friendly solution to enhance P availability for plants especially in P poor soils. Inorganic P is solubilized by the synthesis of low molecular weight organic acids such as gluconic and citric acid or by the release of protons, and the production of chelating substances. On the other hand, the mineralization of organic phosphorus occurs through the synthesis of phosphomonoesterase, phosphodiesterase, and phosphotriesterase which catalyze the hydrolysis of phosphoric esters [51,52,53,54]. Phosphate solubilization tends to be more efficient in microorganisms isolated from extreme soils (such as saline/alkaline soils and soil with nutrient deficiency) than in those coming from soils characterized by moderate conditions [47].
Iron is the fourth most abundant element on earth [55], but in aerobic soils, Fe forms insoluble hydroxides and oxyhydroxides [56], which limit its bioavailability particularly in calcareous soils [57], that cover about 30% of cultivated soils in the world. For this reason, plant Fe nutrition in these soils is one of the most important agricultural problems worldwide [57,58]. Iron is crucial for all forms of life [59]: while microorganisms grow at an optimal Fe concentration of about 10−5 to 10−7 mol·L−1 [60], for plants, the optimal amount of Fe ranges between 10−9 to 10−4 mol·L−1 [61,62].
Usually, the low bioavailability of iron is faced with application of synthetic iron chelates, such as ethylenediaminetetraacetic acid (EDTA) or ethylenediamine-N, N’-bis(2-hydroxyphenylacetic) acid (EDDHA). This kind of approach is cheap and effective but, besides their high affinity for Fe, these chelators are not biodegradable and their accumulation in the environment is of great concern [63]. The availability of Fe in saline sodic soils is usually very low [64,65] and plant growth reduction is often reported to occur in such environmental condition. The high level of Fe starvation in saline soil leads to an altered chlorophyll content of young leaves and subsequently to the appearance of Fe chlorosis symptoms. Moreover, salt stress severely affects Fe transport to shoots, since its content is usually lower in aerial organs than in roots [65].
The Fe requirement is more accentuated in the rhizosphere, where the competition for this element among plants, bacteria, and fungi is higher than in bulk soil [61,66]. In order to cope with the low amount of available Fe, bacteria have evolved the synthesis of low weight molecules (between 500 and 1500 Da) called siderophores, having a high affinity for Fe3+. When the bacterial cell is in starvation for Fe, the synthesis of siderophores is induced and the chelators are released outside the cell. The siderophore binds Fe3+ and the Fe-siderophore complex, called ferrisiderophore, is recognized by a receptor localized on the cell membrane. The ferrisiderophore is taken up to a membrane receptor, allowing the internalization of this element that is subsequently transformed at the membrane level as Fe2+ and released inside the cell [67] (Figure 2).
Figure 2.
Bacterial Indole- 3-Acetic Acid (IAA) and 1-Amino-Cyclopropane-1-Carboxylic Acid (ACC) deaminase interplay: Plant ethylene is synthesized from S-adenosyl-L-methionine (SAM) and ACC through the enzymes SAM synthetase and ACC synthase, respectively. Then, ACC is converted to ethylene by ACC oxidase.
Microorganisms able to synthesize siderophores are present in a large amount in the rhizosphere [68]. As a consequence, the siderophore concentration is maximum in the rhizosphere, where it reaches 0.1 μmol·L−1 and 1 mmol·L−1 for bacterial and plant siderophore, respectively [69], while a very low level is found in bulk soil (10 nmol·L−1) [70]. Among rhizobacteria, the use of homologous siderophores, produced by members of the same genus, is very common, while the use of heterologous ones, synthesized by bacteria belonging to different genera, is also possible [71].
Plants assimilate Fe from bacterial siderophores by different mechanisms such as chelating and releasing Fe, direct uptake of ferrisiderophores, or by a ligand exchange reaction [72,73]. In plants, active Fe uptake occurs mainly through two strategies [74]. The first strategy (Strategy I) occurs typically in dicotyledonous and nongraminaceous monocotyledonous plants, and it is based on the acidification of the rhizosphere by H+ excretion, leading to the reduction of Fe3+ to Fe2+ by a plasma membrane-bound reductase and its transport inside root cells [75,76,77]. The second strategy (Strategy II), is mainly used by grasses and graminaceous plants, and it is based on the synthesis of Fe chelators (phytosiderophores) and on the uptake of the Fe–phytosiderophore complex in root cells mediated by specific transporter molecules [78,79]. Therefore, siderophores are involved both in suppressing the growth of phytopathogenic microorganisms via competition for the available Fe and in stimulation of plant growth by supplying the plant this nutrient.
Rhizobacteria can produce phytohormones and can modulate their endogenous amount in plants. It is well known that phytohormones play a crucial role in plant growth and development [80,81,82].
It has been estimated that about 80% of microorganisms living in the rhizosphere of various crops are able to synthesize and release auxins (IAA) by five biochemical pathways, depending or not on tryptophan as precursor [83,84]. IAA regulates both plant growth and defense responses: although its main role is the modulation of cell division, extension, and differentiation. IAA is also involved in seed germination, enhancement of root branching or elongation, regulation of responses to light, gravity, fluorescence, photosynthetic pigment biosynthesis, metabolite production, and resistance to environmental stresses [82,83,85]. In fact, when plants are exposed to stressful conditions, a regulation of plant phytohormones finalized to the adaptation to the stressor is often observed [86].
Plant reactions to exogenous IAA vary according to the endogenous concentration of this phytohormone, the plant species, its phenological stage, sensitivity, and the tissue involved [87]. For example, root elongation in Arabidopsis thaliana seedlings was stimulated only by exogenous concentrations ranging from 10−10 and 10−12 M [88]. Moreover, it should be considered that plants respond to a total pool of IAA that is composed by the endogenous fraction as well as by the IAA synthesized by soil microorganisms. The impact of bacterial IAA on plant development ranges from positive to negative effects according to the amount of IAA available to the plant and to the sensitivity of the host plant to the phytohormone. Therefore, the amount of IAA synthesized by the plant itself determines if the bacterial IAA will stimulate (optimal level) or suppress (suboptimal level) plant growth [84]. Therefore, the amount of IAA driving the growth of the plant has to be carefully regulated. In order to avoid inhibitory effects from over dosage, plants possess neutralization mechanisms to control IAA excess, conjugating IAA with sugars, amino acids, or peptides [89].
3.2. Increasing Plant Tolerance to Stress: 1-Amino-Cyclopropane-1-Carboxylic Acid (ACC) Deaminase and Exopolysaccharide Synthesis
Being sessile organisms, plants are characterized by a high level of physiological plasticity, which ensures a wide range of protective responses and enables them to survive a variety of different environmental stresses that is now called the “plant like-immune system” [90,91]. Ethylene is a gaseous plant hormone driving several aspects of plant growth, such as fruit ripening, flower senescence, leaf and petal abscission [92,93], and plays a key role in plant defense responses to stress and to symbiotic relationship [94,95,96].
In fact, the term “stress ethylene” [97] is usually used to indicate the rise in the ethylene level after plant exposure to environmental stresses such as: extremes of temperature, high light, flooding, drought, heavy metal, organic pollution, radiation, wounding, insect predation, high salt, and various pathogens including viruses, bacteria, and fungi [98].
Ethylene biosynthesis occurs in all higher plants via a methionine dependent pathway, in which methionine is converted to S-adenosyl-L-methionine (SAM) by the SAM synthetase. SAM become the substrate for ACC synthase that converts it in ACC, which is the immediate precursor of ethylene. Then, ACC oxidase catalyzes the conversion of ACC to ethylene, carbon dioxide, and cyanide (Figure 2) [99].
The amino acid tryptophan is exuded by plant cells, taken up by PGPB living around the roots, and used to synthesize IAA. This bacterial IAA is taken up by the plant cells and, together with the plant’s endogenous pool of IAA, stimulates an auxin signal transduction pathway which results in cell growth and proliferation, and in increased transcription of the gene for ACC synthase, eventually yielding an increased concentration of ACC. As a consequence, ACC is synthesized on SAM as substrate favoring the production of ethylene and leading to a two peaks plant stress response. The newly produced ACC is partly exuded from plant roots, taken up by the bacteria, and converted by ACC deaminase to ammonia and α-ketobutyrate. As a result, the amount of ethylene produced by the plant is reduced. Alleviation or exacerbation of stress depends on the plant species, its age, and the nature of the stress itself [100]. The answer of the plant to environmental stress involving ethylene has been explained by Glick et al. [101]. In this model, a few hours after the onset of a stress, an initial small peak of ethylene rises. This peak is considered to be beneficial since it triggers a protective response by the plant, such as the transcription of pathogenesis-related genes and induction of acquired resistance [100,102]. When the stress become chronic or more intense, the transcription of the ACC synthase genes occurs some days later, and another ACC is synthesized leading to a second, much larger ethylene peak. This second peak induces processes such as senescence, chlorosis, and abscission; therefore, the growth and the survival of the plant are hampered. A fraction of the ACC synthesized by this two-step process is released from seeds or plant roots, taken up by the bacteria, and converted to ammonia and α-ketobutyrate by the enzyme ACC deaminase [103]. As a consequence, the amount of ethylene produced by the plant after the stress is reduced and does not reach inhibitory levels. Various environmental stresses can also either increase the synthesis of IAA or stimulate the transcription of the gene for ACC synthase. As the amount of ethylene in a plant increases, the transcription of auxin response factors is inhibited. In the absence of bacterial ACC deaminase, by limiting transcription of auxin response factors, ethylene hampers both cell growth and IAA stimulation of the synthesis of additional ethylene. In the presence of bacterial ACC deaminase, less ethylene is formed because some of the ACC is degraded by ACC deaminase. When ACC deaminase is synthesized, transcription of auxin response factors is not suppressed, and IAA can stimulate cell growth and proliferation without inducing a rise of the ethylene level. In this model, ACC deaminase both decreases ethylene inhibition of plant growth and allows IAA to boost plant growth, both in the presence and absence of plant stress (Figure 2). Therefore, plants hosting bacteria able to synthesize ACC deaminase and exposed to stressful conditions result to be more tolerant to stress and their growth is comparable to that of plants grown in normal conditions. In the same way, bacteria synthesizing this enzyme favor the establishment of symbiosis with rhizobia (with an increased nodulation) [104,105,106] or mycorrhizal fungi [107,108,109], thus indirectly improving both plant growth and health.
The term exopolysaccharide (EPS) was coined in 1972 by Sutherland [110] in order to indicate the different types of layers occurring outside the bacterial cell wall and represents the 40–95% of the bacterial mass. Bacterial EPSs fall in two categories: slime EPSs and capsular EPSs both composed by a mixture of polysaccharides, proteins, nucleic acids, and lipids [110] whose weight reach 10–30 KDa [111]. The water content in the EPS reaches ∼97% and is considered as a sink of water involved in protection of bacteria from desiccation [112].
Due to their chemical and physical properties, EPSs found application in biotechnology industries as gelling, stabilizing, thickening, coagulating, film forming agents for detergents, textile, paper, paints, adhesive, beverages, and mainly food production [113,114]. Because of their impact on human health, bacterial exopolysaccharides, and especially those produced by lactic acid bacteria, are considered as beneficial since their modulation of the immune system, cholesterol-glucose lowering ability, antitumoral activities, and prebiotic effect [115]. Many PGPB belonging to the Pseudomonas, Azospirillum, and Rhizobium genera are reported to form strict association with plant roots through the formation of an EPS layer that is involved in biofilm building [116]. Moreover, during drought or salinity stress, EPS synthesis in bacteria is higher than in non-stressful conditions [117]. Inoculation of plants with EPS synthesizing PGPB increase plant resistance to drought and excess of salt in soil [118]. While both these stresses lead to severe change of the soil chemical and physical properties, making it unsuitable for an optimal crop productivity, the bacterial EPS produced in soil and all around the roots is crucial in maintenance of micro and macro aggregates and contributes to the formation of a site where the soil microbial process occurs, efficiently leading to an optimal flow of nutrient, water, and ions to the plant root systems, also in conditions of low water availability [119].
4. Effects of Salinity on Plants and Plant Responses
On the basis of their ability to grow in a saline environment, plants are classified into glycophytes and halophytes [120,121,122], the former are unable to grow in saline soils, the latter are salt tolerant. About 98% of the plants are glycophytes: they tend to exclude sodium, maintaining low levels of Na+ in their tissues (0.2–2.0 g Kg−1). Therefore, these plants are very salt-sensitive and are heavily affected by the presence of salt in the soil [122]. Some representative genera are Citrus and Persea, besides different species of the family Fabaceae (e.g., bean, chickpea and pea) and Poaceae (e.g., maize and rice). Only a very small number of glycophytes, among which Medicago sativa, A. thaliana, Gossypium spp., Solanum lycopersicum, and Hordeum vulgare are able to develop a certain degree of tolerance in response to the changing of soil salinity, showing reduced sensitivity to salt [123]. Halophytes represent only 2% of the total flora and generally tolerate high salt concentrations, therefore they are considered salt-resistant plants [121]. Halophytes evolved a series of adaptative traits that allow them to complete their life cycle under elevated salinity (more than 200 mM NaCl) [124]. Three main categories of halophytes are known: obligate, facultative, and preferential halophytes. Obligate halophytes require salt concentration in a range between 200–300 mM NaCl to reach optimal growth and can tolerate seawater salinity. The genus Salicornia, Suaeda, Halimione, Atriplex, Tamarix, and Petrosimonia are included in this group. Facultative halophytes do not necessarily require salt for their growth, therefore they can live also in soil devoid of salt [125]. Chenopodium quinoa has been classified as facultative halophyte, able to grow in marginal soils such as saline and arid ones; its ability to tolerate these extreme conditions could be improved by the use of halotolerant rhizobacteria as bioprotectors [126]. Preferential halophytes can grow at a relatively higher concentration of salt in soil, in respect to glycophytes. On the basis of anatomical adaptations in response to salinity, halophytes are also divided into extreme-halophytes (mainly succulent plants), able to store water in plant aerial tissues or organs and meso-halophytes, that include species with expanded endodermis, aerenchyma and bulliform cells, with intermediary anatomical adaptations between extreme-halophytes and glycophytes [127].
Similar to other important abiotic stresses, among which aridity, salinity induces a variety of responses in plants including morphological, physiological, biochemical, and molecular changes. It is well known that water deficit and salt stresses are strictly associated, since the negative effects of salt stress on plants have been mostly associated to the osmotic imbalance due to the decrease of water potential in soil, following by decreased water absorption by the plant root system. Therefore aridity and salinity can induce similar negative effects on plant growth and physiology [6,128,129,130,131], in particular, damaging membrane components (lipids and proteins), inducing seed dormancy, reducing germination rate, root elongation, shoot biomass, leaf expansion, stomatal conductance, and photosynthetic efficiency, leading to a substantial loss of crop productivity and yield, resulting in significant economic damage [3,9,130,132,133]. It has been demonstrated that plant fitness and productivity are closely related to the seed germination rate, a crucial phase of plant growth, strongly affected by many adverse environmental factors such as soil salinity. Cuartero and Fernandez-Muñoz [134] showed that tomato seed germination was strongly inhibited (from 50% to 100%) at 80 and 190 mM NaCl, respectively, if compared to that of seeds grown in not disturbed soil. Similar results were reported, at different salt concentrations, in many other plant families such as Poaceae [135,136], Fabaceae [137], and Brassicaceae [138,139]. Besides the seeds, other plant organs can be severely affected by salt toxicity. Roots are embedded in the soil and they are the first organ to face salinity [129]. For this reason, growth reduction is greater at root level than at shoot. An increased root to shoot ratio was reported under salt stress [7]. However, it was observed that the extent of salt-induced damage could depend also on plant/organ developmental stage [140]. In fact, it has been shown in some studies that high concentrations of salt can induce the expression of genes which, in addition to conferring a different ability of the plant to respond to stress, are also involved in regulating the development of the plant in different phenological stages such as flowering, seed germination, root growth /branching, and leaf morphology [7,141,142]. As suggested by Munns [143], mature leaves are more affected by salinity than young leaves: they present necrotic tissue (sign of premature senescence), probably due to the imbalance of the detoxification process in vacuole. Several anatomical changes (such as variation in palisade and spongy parenchyma thickness; increased intercellular spaces; damage of chloroplast ultrastructure and increased number of plastoglobules) were recorded in leaves of plants exposed to salt stress [7].
At the cellular level, the primary site of salt injury is the plasma membrane [144,145]. The so called “salt shock” induces changes in the membrane lipids and proteins due to the fact that salts are found in the solution in ionic form. The positive charges of the ions can break the electrostatic bonds present between the polypeptidic chains, increasing hydrophobic interactions and leading to loss of protein/membrane functionality. The alteration of the ionic flow across membrane, has a negative impact on important processes such as photosynthesis, respiration, and protein metabolism. High concentrations of Na+ and Cl− can also influence the membrane potential and the activities of membrane bound enzymes, besides the uptake of other minerals, therefore resulting in a nutritional imbalance for the plant. Different studies showed that salinity decreases the availability of nutrients and their assimilation in plants [3,146]: for example, Na+ ions can compete with other cations (such as K+) for membrane transporters, causing toxic accumulation of Na+ in the cell [147,148], decreasing the uptake of K+. The effects of salt stress can also be reflected in variations of membrane permeability, which has been reported as an effective selection criterion for salt tolerance in different crops [145,149].
Salt-induced osmotic stress is responsible for oxidative damage caused by reactive oxygen species (ROS) as superoxide anion (O2−), hydrogen peroxide (H2O2), and hydroxyl radical (HO•) [3,129]. The production sites of ROS in cell are various: they are produced in mitochondria during respiration, in peroxisomes during photorespiration, and in chloroplasts during photosynthesis. Therefore, the control of ROS intracellular levels is strictly necessary to avoid serious damage to all these cellular processes [3,150]. Moreover, ROS are interconnected with other molecules, including phytohormones (e.g., abscisic and gibberellic acids), and this connection is very important in modulating plant stress responses [150,151].
The plant photosynthetic apparatus is negatively affected by ROS, especially photosystem II (PSII) and the electron transport chain [152]: salt stress induces stomatal closure (due to a decreased uptake of CO2) and oxidative stress, resulting in a low photosynthetic rate and in an imbalance of photosynthetic pigment biosynthesis [3,153,154].
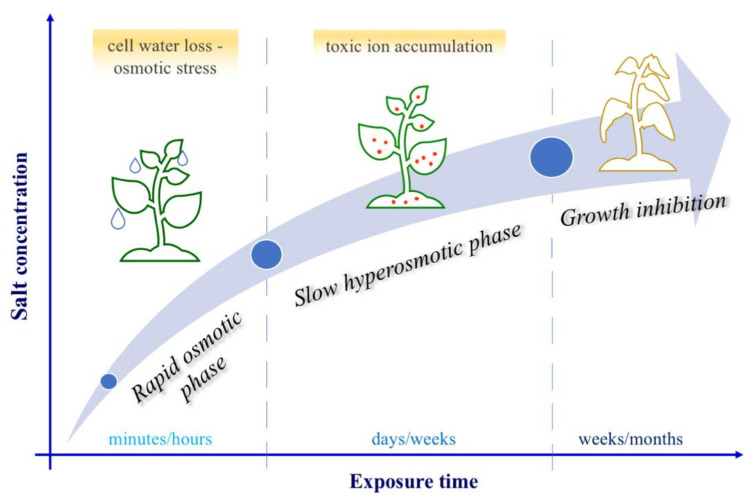
The above mentioned effects of salt stress on plants largely depend on the concentration of salt in soil, on the exposure time to stress (that can vary between minutes/days to days/weeks/months) [6,155] and on the plant’s ability to absorb salt before it becomes highly toxic, strongly affecting metabolic function and plant life itself [6]. An increase of salinity in soil leads cells, in a few minutes, to lose water due to osmotic stress: this phenomenon, known as “rapid osmotic phase” is followed by a “slower (days/weeks) ion toxicity phase” (or “hyperosmotic stress phase”) (Figure 3), that is characterized by a high accumulation of toxic ions, decreasing both cell division and expansion rate [6,156]. Therefore, over time (weeks/months), the vegetative development of the different plant organs is strongly altered.
Figure 3.
Plant responses to salt stress. Plants cope with the increase in salt concentration in different ways as the time of exposure to stress increases. Three phases are identified: a faster first reaction (minutes/hours) called rapid osmotic phase; one in the order of days/weeks called slow hyperosmotic phase; and a long term exposure (weeks/months) which leads to inhibition of growth and cell death.
Another factor to consider is plant tolerance/sensitiveness to salinity. It was observed that monocotyledonous species, especially those belonging to the family Poaceae, are more sensitive to salinity than dicotyledonous ones [157], but within each of these two large groups of plants, a wide range in salt stress responses is displayed. For example, among monocots, rice is the most sensitive and barley the most tolerant [158,159].
Plants have evolved different mechanisms to cope with salt stress. Munns and Tester [6] suggested three main mechanisms: osmotic stress tolerance, ion exclusion, and tissue tolerance. At first, plants respond to salt stress with osmotic adjustments in order to maintain cell turgor, which is necessary to the right functioning of the cell [130,160]. The increased soil salinity causes a decrease in the water potential, inducing root cells to release water into a hypertonic medium. In this context, plants synthesize osmolytes (such as proline, glycine-betaine, and soluble sugars) to mitigate the effects of high salt concentration, counterbalancing osmotic impairment [130,161,162]. Among these osmolytes, proline and glycine-betaine are the most efficient compatible solutes. Due to its ability to be accumulated at high concentrations in response to saline stress, proline can be used as a marker of salt toxicity [163]. Glycine-betaine acts mainly in protecting antioxidant enzymes and PSII [164]. In addition to their osmotic function, osmolytes have been reported to have also a ROS scavenging function [165,166]. During stress exposure, plants try to maintain also the turgor of mesophyll cells in leaves, avoiding the translocation of toxic ions from root to shoot and regulating leaf transpiration. Abscisic acid (ABA) plays a key role in reducing the transpiration rate within a few hours. Its production occurs in response to high salinity at the root level, then it is transported via xylem to leaves, also inducing stomatal closure [3,167]. A lower water loss has been found in species with a certain tolerance to salt. In fact, halophytes generally show a reduced transpiration, if compared to glycophytes [168].
The maintenance of the optimal cytosolic K+/Na+ ratio is of primary importance to protect cellular functions [169]. As reported before, an increasing uptake of Na+ due to salt stress induces an imbalance of K+ transport in root cells. Na+ ions in excess need to be extruded or compartmentalized in the vacuole not to become toxic [130,170]. Ion sequestration in the vacuole, or exclusion from the cytosol, are two tolerance mechanisms adopted by plants subjected to salinity stress. In order to allow the “normal” metabolic and enzymatic function, plant cells continue to store toxic ions in the vacuole until saturation level. This compartmentalization process effectively reduces the cytoplasmic concentration of Na+ [171]. On the other side, the plasma membrane Na+/H+ antiporters translocate the excess of Na+ outside the cell, into the extracellular region, in order to maintain a low concentration of Na+ in the cell [172]. The salt overly sensitive (SOS) pathway was discovered in A. thaliana and includes SOS1, SOS2, SOS3 genes. The SOS pathway has a key role in maintenance of homeostasis, in cytoskeleton reorganization, and in modification of root architecture [173]. The main gene involved in the transport of Na+ is SOS1, encoding for a Na+/H+ antiporter.
5. Influence of Salinity and Aridity on Microbial Communities and on Plant Microbial Interactions
Salinity negatively affects the physico-chemical properties of soil such as pH, nutrient availability for plants, cation exchange, and soil organic carbon input, strongly influencing the soil microbial communities and their activity [174,175]. The maintenance of soil organic carbon input is essential for soil biological and ecological functions. Soil salinization is a degradation mechanism that involves the accumulation of water-soluble salts in the upper part of the soil and affects the agricultural yield, the productivity of ecosystems, and therefore economic well-being. Strategies for maintaining agricultural productivity and alleviating water and salt stress include the development of salt tolerant cultivars, the implementation of alternative cultivation systems and specific management practices, such as drip irrigation or soil drainage. Stress can also be reduced by modifying soils with organic substances such as manure, compounds, or straw, which favor the promotion of the growth of microbial biomass. In fact, plants can benefit from the activity of microorganism communities that colonize their root systems: as previously explained, the bacterial production of EPS can help plants to resist drought and salinity, as well as enzymatic activities facilitate the formation of soil aggregates, the stabilization of organic carbon, and the production of hormones and volatile compounds that trigger defense mechanisms and promote growth [174]. The study of the microbiota associated with plants, its characterization and understanding of the role of both bacteria and fungi within the rhizosphere, is one of the main topics in current research.
Microbial communities are influenced by various abiotic and biotic factors. Greenhouse and field studies have shown that soil type, plant genotype, plant developmental stage, and cultivation practices determine the composition and structure of the microbiota [175,176,177,178,179]. Among these factors, the type of soil and, in particular, its physical-chemical properties, are the strongest determinants in the variation of the microbial community: pH is the most critical parameter. Soils with almost neutral pH generally have a greater bacterial diversity than more acidic or more basic soils, such as deserts.
Although the relationship between soil characteristics and its microbial communities present has been extensively studied, the effect of one on the other is largely unknown [180]. Furthermore, the microbiota is not static or passive: plants can actively modulate the construction of their microbiota, especially thanks to the deposition of extraradical exudates [181].
Autochthonous plants of the desert or semi-arid environments are constantly subjected to water and saline stress: these factors limit the number of species that manage to survive and shape the composition of the microbial community [182]. This idea is also confirmed by Fierer [183], based on the analysis of the microbiota in cold deserts, hot deserts, forests, prairies, and tundra. Fierer demonstrated that antibiotic resistance genes were three times less abundant in desert soils than other soils, suggesting that abiotic conditions are more important than competitive interactions in shaping desert microbial communities. Studies on various desert environments reveal both the antifungal [184] and the antibiotic properties [185] for the communities of Actinobacteria.
Investigations regarding bacterial diversity in the semi-arid zone of central Mexico have revealed the predominance of Actinobacteria, Proteobacteria, and Acidobacteria in the rhizosphere of cactus plants [186]. An analysis of the microbial communities in the Atacama desert (Chile) has shown a high number of Actinobacteria, Chloroflexi, low levels of Acidobacteria, and alpha and beta-Proteobacteria [187]. Acidobacteria are typically found in environments that vary in temperature, soil type, and pH. Basic soils such as desert generally have higher relative abundance of Actinobacteria than Acidobacteria [188]. This observation is confirmed by the study on the vineyards of Berlanas [189]: the vineyards have a more acidic soil than the desert, and the Acidobacteria (13.7% and 16.4%) immediately follow the Proteobacteria and the Actinobacteria (almost 50% of the total).
Both beta and alpha-proteobacteria are commonly associated with soils that receive high organic carbon input rates, a condition absent in the desert. Fierer [183] showed that all communities related to the different biomes (cold deserts, warm deserts, forests, grasslands, and tundra) were dominated by Acidobacteria, Actinobacteria, Bacteroidetes, Proteobacteria, Verrucomicrobia, and Gemmatimonadetes, bacterial phyla known to be relatively abundant and omnipresent in the soil. Actinobacteria and Bacteroidetes are generally more abundant in desert soils than in forests, grasslands, and tundra, while Verrucomicrobia and Acidobacteria have shown the opposite trend. Chloroflexi phyla, Cyanobacteria, Firmicutes, and Gemmatimonadetes have also been found in almost all soils, but their relative abundances are highly variable and represent less than 5% of the identified sequences. Firmicutes members, particularly Bacillus, are responsible for the production of a wide range of antimicrobial metabolites, enzymes, and surfactants that promote plant growth and induce systemic resistance in plants [190]. It is also possible that the low diversity of desert soils is not directly related to their very high pH levels, but rather to the high salinity. The desert soils are very dry, poor in nutrients, generally have a basic pH, higher than other biomes and the scarcity (or complete absence) of plant biomass reduces the inputs of organic carbon useful for bacterial metabolism. When the amount of organic matter is greater in soil, the stability and diversity of the microbiota is higher too [26]. For example, Gao and coworkers [188], in a study concerning the effect of aridity and dune type (in a desert of Northern China), showed that Firmicutes, Actinobacteria, Proteobacteria, and Acidobacteria were the dominant phyla in all samples of the rhizosphere of Caragana microphylla. The increased abundance of Actinobacteria in the rhizosphere soil was mainly caused by the decreased soil pH due to rhizodeposition [188]. The authors stated that the structure of the rhizospheric bacterial community was modulated mainly by soil total organic C, total N, Na+, and total P while total organic carbon, electronic conductivity, pH, and total phosphorus were the dominant factors to affect the microbial communities associated to the different dunes. Pereira [191] confirms that the low density of bacteria in the desert, compared to a soil not subjected to water stress, can be due to the previous reported physical-chemical characteristics of the arid soil.
Although the interest in the study of microbial communities occurring in arid environments is increasing, the number of works present in the literature and the results obtained do not yet allow to generalize a common trend. Furthermore, finding a common thread could become even more complicated taking into consideration the different species of plants.
6. Application of PGPB in Salt Stressed Soil
There is an increasing request for food worldwide: there are 7.4 billion people in the world, and this value is expected to reach 9.7 billion by 2050 [192]. As a consequence, the exploitation of chemical fertilizers in agriculture is considered as necessary regardless of its side effects on the environment and especially on soil. Even more, as previously explained, environmental abiotic stresses such as drought, high salinity, and high and low temperature have negative impact on agriculture productivity. As an example, it has been recently reported that soil salinization leads to a loss of US$ 27.3 billion [193]; in the same way, drought caused a 5% reduction of the yield in the primary food-producing nations and an annual loss of 17% in harvest yield in the tropics, arid, and semi-arid regions. In this context, in order to protect the sustainability of both the natural and agricultural ecosystem and, at the same time, obtain crop yields that meet the increased food demand, there is urgent need of new and eco-friendly approaches in agricultural practices.
PGPB offer a realistic opportunity in protecting crop yield also in stressed soils, due to their beneficial effect on soil fertility, using different strategy (as previously reported in the above paragraphs). In fact, in the event of environmental stress, rhizobacteria modulate their activities by interfering with the stress tolerance mechanisms in plants. The role of bacteria in the rhizosphere in a context of stress is really difficult to be fully understood. As already mentioned, each plant builds its own microbiota which will be different in relation to the type of soil, the environmental components, and the genetics of plant. Therefore, it is very complex to define how the plant responds to stress on the bases of interaction of the associated PGPB. Here are presented some positive rhizobacteria actions that help plants to adapt to stressful conditions.
Both fungi and bacteria can produce ACC-deaminase and a series of phytohormones to influence the plant endogenous hormone levels and eventually reprogram the plant hormonal signaling. Plants subjected to salinity, drought, and pathogenic stress are known to produce an excess of ethylene, which seriously slows down root development [194]. One of the features of PGPB is that they show the ability to produce ACC deaminase, which catalyzes the conversion of ACC (the precursor of ethylene biosynthesis) to ammonia and α-ketobutyrate. Therefore, plants with reduced ethylene levels would eventually overcome salt-induced growth inhibition by associating with the bacteria that produce ACC deaminase [108]. One of the subsequent effects of this particular type of plant-bacterial association would be the supply of a source of N (ammonia) to plants [195]. Little is known about how these PGPB influence this process, although there is some evidence that PGPB improve plant salt tolerance by altering the state of the endogenous hormone [196]. Studies carried out on pepper in the presence of water stress show that plants treated with ACC rhizobacteria recorded higher efficiency of photosynthetic processes and even higher tissue turgidity. These beneficial effects lead to an increase in biomass and root length of up to 50% compared to uninoculated plants. The most pronounced protection against drought has been obtained with strains of the genera Achromobacter, Klebsiella, and Citrobacter. Considering the ability to colonize the roots of these genera, it is conceivable that this protection activity can also be carried out in field conditions and not only in vitro [197].
PGPB can offer a realistic opportunity in increasing crop yields also in stressed soils due to their beneficial effect on soil fertility, improvement of nutrient uptake by plants, the expression of plant beneficial physiological traits, and reducing soil borne plant diseases [198].
In this paragraph, several examples of applications of PGPB in crop growing in salt or drought stressed soils are provided, referring to the most recent scientific papers.
In their work, Amna et al. [199] assessed that effects of Bacillus siamensis PM13, B. methylotrophicus PM19, and Bacillus sp. PM15, three halotolerant strains, able to synthesize ACC deaminase and EPS, on wheat seed germination and seedling growth under three level of salt stress (4, 8, and 16 dS m−1). Although a positive impact of bacteria was observed on both shoot and root development, this effect slowed down as the salinity level increased. The induced salt tolerance detected at moderate salinity stress may be based on EPS or ACC deaminase synthesis but also other physiological traits as P solubilization and IAA production could moderate the extent of the damages induced by NaCl [199].
The Burkholderia sp. strain P50 is able to produce ACC deaminase, IAA, EPS, and the osmolyte proline also under salinity stress. Rice seedlings inoculated with the strain P50 and grown in presence of 185 mM NaCl were improved in several morphological and biochemical characteristics, as well as in producing antioxidants able to scavenge ROS. Moreover, the amount of stress ethylene in seedlings exposed to salt stress and treated with Burkholderia sp. P50 was reduced compared to non-inoculated plants. The mutant of the strain P50 lacking the ACC deaminase genes was unable to reduce stress ethylene, indicating that the action of the Burkholderia sp. P50 strain was related to a decrease of ethylene level [200].
In their work, Chandra et al. [201] assessed the ability to alleviate drought stress in wheat by using strains of Variovorax paradoxus RAA3, Pseudomonas palleroniana DPB16, and Pseudomonas sp. UW4 in a field experiment under irrigated and rainfed conditions. The best response to bacterial inoculation was obtained under rainfed conditions, in which wheat was grown under extensive water stress. Moreover, all the three strains induced grain yield increase, but the most performer strain was V. paradoxus RAA3: all the plant growth and yield parameters considered (plant height and tillers/plant, spike length, number of grains/spike, total tillers, 1000 grain weight) in both irrigated and rainfed field conditions were increased after inoculation with the strain RAA3. Although the mechanisms at the base of this effect is not currently known, it is true that V. paradoxus RAA3 had the highest ACC deaminase activity among the three bacterial strains [201].
Tomato has a great economic impact worldwide, but its sensitivity to water stress can limit its yield [202]. In their recent study, Brilli et al. [203] inoculated tomato seeds with P. chlororaphis subsp. aureofaciens M71 and exposed plants to a mild water stress condition. The occurrence of this bacterial strain at the root level induced an increased production of osmolytes, such as proline, and enhanced antioxidant molecule levels in leaves, especially those of superoxide dismutase (SOD) and catalase (CAT) enzymes [203]. This kind of effect may boost the plant tolerance to osmotic stress and, at the same time, maintains low ROS levels for signaling purposes. This fact allows the plant to give a quick response to ROS produced by the plant itself during environmental stresses [204]. Therefore, it is suggested that P. chlororaphis acted as a ‘priming stimulus’ triggering in inoculated tomatoes enhanced tolerance to water stress [205]. Moreover, an increased content of ABA in leaves was observed in plants inoculated with M71 grown under water stress: this is an interesting effect induced by the bacterial strain since high ABA levels result in stomatal closure, that in turn, has a positive feedback on water use efficiency. As mentioned above, many microbial groups secrete phytohormones, such as IAA, ABA, cytokinins (Cyt), and gibberellic acids (GA) [206]. In the study of Bianco and Defez, it is highlighted how the Sinorhizobium meliloti (alpha Proteobacteria) strain produces IAA, leading to an improvement in the growth of Medicago truncatula in saline soils [207]. Salt tolerance probably increases in response to stimulation of root proliferation as suggested by Dodd and Pérez-Alfocea [202]. Under saline or normal conditions, PGPB inoculation often decreased the accumulation of ABA in roots and has even significantly altered the remote signaling of phloematic ABA shoot-to-root flows and xylematic root-to-shoot flows [208], flows that can mitigate the adaptation of plants to water deficit. Recently, two rhizospheric bacteria, Rhodococcus sp. (Actinobacteria) and Novosphingobium sp. (alpha Proteobacteria), have been shown to be able to metabolize ABA in vitro and have contributed at least in part to reducing ABA concentrations in plants [208]. Even more interesting, ABA homeostasis disruption influences the activity of the PGPB.
Furthermore, from Porcel’s study, it emerges how Bacillus megaterium can promote tomato growth by inhibiting mutant plants deficient in ABA [209]. In conclusion, these results suggest that the PGPB that produce and/or metabolize ABA in general will act differently in modulating the ABA state of plants and therefore could cause variable responses of plants to salinity stress.
PGPB can improve the absorption of nutrients in plants. Achromobacter piechaudii, when inoculated in Solanum lycopersicum, exposed to high salinity conditions, increases the acquisition of P and K and improves efficiency in the use of water [210]. Inoculation with Bacillus aquimaris significantly increase N, P, and K concentration in leaves of wheat, cultivated in field under high saline stress [211]. These results highlight PGPB ability to solubilize insoluble P and to fix atmospheric N in a non-symbiotic way. Similarly, PGPB are able to release siderophores in order to eliminate Fe, then ferri-siderophore can be easily accessible to plants [212].
A useful mechanism for maintaining ionic homeostasis in plants when subjected to saline conditions is expressed by the synthesis of EPS which binds toxic Na and limits its inflow into the roots. EPS can also promote biofilm formation on root surfaces, thereby limiting the flow of Na+. Recently, EPS production has been identified as involved in the beneficial effect of B. amyloliquefaciens FZB42 in protecting A. thaliana from drought stress [213]. The wild-type strain FZB42 promotes plant growth and drought tolerance by enhancing the plant survival rate, the plant biomass, root length, and branching. The EPS deficient mutant of this bacterial strain, once inoculated on the plant root, boosted cellular defense responses such as the level of proline and the activities of SOD and peroxidase and modulated the synthesis of ethylene and jasmonate.
Similar results were obtained in sunflower inoculated with the EPS producer Pseudomonas putida GAP-P45 [214], cowpea colonized by Azospirillum [215], and foxtail millet treated with P. fluorescens DR7 [216].
In case of prolonged exposure to high concentrations, Na+ moves from the roots to the breathable leaves where it can increase toxicity. At this point, volatile organic compounds (VOCs) produced by PGPB can be useful, inducing HKT1 gene expression in the shoots, which codes for a high affinity transporter protein of K, expressed in xylem parenchyma cells, that allow to maintain Na+ homeostasis. VOCs also induce the reduction of HKT1 in the roots, limiting the entry of Na+ and facilitating the recirculation of Na+ from bud to root. HKT1 then mediates the loading of Na+ into the phloem lymph in the shoots and the discharge into the roots, thus removing large quantities of this ion from the shoots [206].
PGPB positively regulate the expression of genes that code for the integral proteins of the plasma membrane (PIPs) and the activity of aquaporins, for an efficient absorption of water in stressed plants. Analysis of the gene expression of Pantoea agglomerans and Zea mays roots infected with B. megaterium showed that the PIP2 and ZmPIP1-1 genes were up-regulated under saline stress conditions, helping to increase the values of hydraulic conductance in inoculated plants [217]. In a parallel study, inoculation with Azospirillum brasilense triggered the transcription of HvPIP2-1 in the seedlings of Hordeum vulgare [218]. Furthermore, proteomic analysis revealed that the expression levels of different proteins involved in photosynthesis, antioxidant processes, membrane transport, and pathogenesis-related responses were altered in the presence of PGPB [219].
Almost all PGPB are able to boost antioxidant systems contributing to the scavenging of ROS produced in case of stress, which can alter fatty acids, amino acids, pigments, and other biomolecules of plant cells. The expression of ROS elimination genes, such as SOD, CAT, ascorbate peroxidase (APX), dehydroascorbate reductase (DHAR), and glutathione reductase (GR), increases in PGPB-treated Solanum tuberosum, under conditions of abiotic stress [220]. Even after short-term treatment with Enterobacter sp., the expression of saline stress sensitive genes related to proline biosynthesis has been up-regulated in Arabidopsis [221]. The production of some important osmolites, such as proline and polyamines, is promoted not only in plants subjected to saline and water stress, but also in those subject to microbial infection.
In the case of saline stress, Yuan’s work highlighted that there is an increase in the production of salicylic and jasmonic acid in the soil [222], which seems to be effective in producing tolerance. Many PGPB produce VOCs that interfere with normal hormonal production in the plant, allowing the activation of the defense mechanisms dependent on these hormones. The way in which VOCs work is partially unknown and the study is made difficult since they do not act as individual compounds, but often act synergistically. Endophytes also produce some effectors, including plant hormones, signaling molecules, and microRNAs, for the activation of signaling pathways and often significantly promote metabolic efficiency, altering the ratio between upregulated/downregulated genes. Other classes of endophytes produce some bioactive compounds (melanin, mannitol, and trehalose) to relieve the abiotic stress of plants [206].
In 2016, Tiwari [223] examined the role of Pseudomonas putida MTCC5279 inoculated in Cicer arietinum plants and showed positive actions against drought stress, including modulation and maintenance of membrane integrity, the accumulation of osmolites (proline, glycine betaine), the ability to wash ROS with transcription of the SOD, CAT genes, APX, and glutathione-S-transferase (GST), the differential expression of genes involved in the biosynthesis of ethylene, salicylic acid, the activation of jasmonic acid, and the greater production of late embryogenesis abundant (LEA) proteins and dehydrins (DHN).
The application of thuricin 17 produced by Bacillus thuringiensis NEB17 to soybeans (Glycine max) in conditions of water deficit has led to the modification of the root structures, an increase in the biomass of the roots and nodules, in the length of the roots, an increase in ABA, and the total N content in the roots [224].
Wang et al. [225] describe how pea plants inoculated with Variovorax paradoxus 5C-2, which produce ACC deaminases, resist saline stress. V. paradoxus involves an increase in photosynthetic rate, electron transport, balanced ionic homeostasis through an increase in K+ flow to the shoots and deposition of Na+ on the roots, a decrease in stomatal resistance and an equilibrium pressure of the xylem. In addition, there is also an increase in biomass at 70 and 130 mM NaCl [225].
Finally, wheat plants (Triticum aestivum) inoculated with Dietzia natronolimnaea showed an upregulation of the genes involved in the ABA signaling cascade, in the SOS pathway, in ion transporters and in antioxidant enzymes; stress tolerance is induced by the modulation of a complex network of gene families [226].
7. Conclusions
It has been estimated that the world’s population reached 7.4 billion and is promptly going to exceed 9 billion [192]. The survival of this huge amount of people is dependent on agricultural products. While the food demand is increasing, the soil quality is reducing due to the intensive use of chemicals and to other different stresses such as salinity and drought. Even more, there is a diffuse reluctance by consumers to embrace the use of genetically modified plants as foods. In this scenario, the exploitation of beneficial microorganisms able to improve plant tolerance and growth in stressed environments may represent an alternative whose aim is to reduce the worldwide dependence on chemicals in agricultural practices and at the same time, enhance the yield potential. Sustainable agriculture should take advantage of the potential of PGPB strains that can be used as biofertilizer, biostimulant, and biocontrol agents. However, we are not completely ready as we still need to make an effort in order to disentangle several variables as the best commercial formulation, the survival of the bacterial strains in field conditions, the specificity against the host plant. For these reasons, studies regarding the impact of salinity or drought on soil bacterial communities are essential in order to understand the mechanisms at the base of the bacterial and plant survival in these kinds of environments, as well as the possible interactions between the plant and its microbiota.
Author Contributions
The manuscript was written by all the authors. In particular, paragraphs 2, 3, 7 was written by E.G., 1 and 4 by V.T. and G.L., 5 and 6 by E.B. All the authors revised the complete version of the manuscript before the submission. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Daliakopoulos I.N., Tsanis I.K., Koutroulis A., Kourgialas N.N., Varouchakis A.E., Karatzas G.P., Ritsema C.J. The threat of soil salinity: A European scale review. Sci. Total Environ. 2016;573:727–739. doi: 10.1016/j.scitotenv.2016.08.177. [DOI] [PubMed] [Google Scholar]
- 2.Rengasamy P. World salinization with emphasis on Australia. J. Exp. Bot. 2006;57:1017–1023. doi: 10.1093/jxb/erj108. [DOI] [PubMed] [Google Scholar]
- 3.Polash M.A., Sakil M.A., Hossain M.A. Plants responses and their physiological and biochemical defense mechanisms against salinity: A review. Trop. Plant Res. 2019;6:250–274. doi: 10.22271/tpr.2019.v6.i2.35. [DOI] [Google Scholar]
- 4.Sparks D.L. Environmental Soil Chemistry. Academic Press; Cambridge, MA, USA: 2003. The chemistry of saline and sodic soil. [Google Scholar]
- 5.Zaman M., Shahid S.A., Heng L. Guideline for Salinity Sssessment, Mitigation and Adaptation Using Nuclear and Related Techniques. Springer; Berlin/Heidelberg, Germany: 2018. [Google Scholar]
- 6.Munns R., Tester M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 7.Acosta-Motos J., Ortuño M., Bernal-Vicente A., Diaz-Vivancos P., Sanchez-Blanco M., Hernandez J. Plant responses to salt stress: Adaptive mechanisms. Agronomy. 2017;7:18. doi: 10.3390/agronomy7010018. [DOI] [Google Scholar]
- 8.US Salinity Laboratory Staff . Diagnosis and Improvement of Saline and Alkali Soils. Volume 60 US Department of Agriculture, Agricultural Handbook, US Government Printer; Washington, DC, USA: 1954. [Google Scholar]
- 9.Rengasamy P. Soil processes affecting crop production in salt-affected soils. Funct. Plant Biol. 2010;37:613. doi: 10.1071/FP09249. [DOI] [Google Scholar]
- 10.Saffriel U., Adeel Z. Ecosystems and Human Well-Being: Current State and Trends. Volume 53. Island Press; Washington, DC, USA: 2005. Dryland System; pp. 623–662. [Google Scholar]
- 11.Prăvălie R., Bandoc G., Patriche C., Sternberg T. Recent changes in global drylands: Evidences from two major aridity databases. CATENA. 2019;178:209–231. doi: 10.1016/j.catena.2019.03.016. [DOI] [Google Scholar]
- 12.Prăvălie R., Patriche C., Bandoc G. Quantification of land degradation sensitivity areas in Southern and Central Southeastern Europe. New results based on improving DISMED methodology with new climate data. CATENA. 2017;158:309–320. doi: 10.1016/j.catena.2017.07.006. [DOI] [Google Scholar]
- 13.Horion S., Ivits E., De Keersmaecker W., Tagesson T., Vogt J., Fensholt R. Mapping European ecosystem change types in response to land-use change, extreme climate events, and land degradation. Land Degrad. Dev. 2019;30:951–963. doi: 10.1002/ldr.3282. [DOI] [Google Scholar]
- 14.Hiltner L. Über neuere erfahrungen und problem auf dem gebeit der bodenbakteriologie und unter besonderer berucksichtigung der grundungung und brache. Arb. Dtsch. Landwirschaft Ges. 1904:59–78. [Google Scholar]
- 15.Hartmann A., Rothballer M., Schmid M., Schmid M. Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil. 2008;312:7–14. doi: 10.1007/s11104-007-9514-z. [DOI] [Google Scholar]
- 16.Bais H.P., Weir T.L., Perry L.G., Gilroy S., Vivanco J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006;57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- 17.Jones D.L., Nguyen C., Finlay R.D. Carbon flow in the rhizosphere: Carbon trading at the soil–root interface. Plant Soil. 2009;321:5–33. doi: 10.1007/s11104-009-9925-0. [DOI] [Google Scholar]
- 18.Nakayama M., Tateno R. Solar radiation strongly influences the quantity of forest tree root exudates. Trees. 2018;32:871–879. doi: 10.1007/s00468-018-1685-0. [DOI] [Google Scholar]
- 19.Compant S., Samad A., Faist H., Sessitsch A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019;19:29–37. doi: 10.1016/j.jare.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bona E., Lingua G., Todeschini V. Effect of bioinoculants on the quality of crops. In: Arora N.K., Mehnaz S., Balestrini R., editors. Bioformulations: For Sustainable Agriculture. Springer; Berlin/Heidelberg, Germany: 2016. pp. 93–124. [Google Scholar]
- 21.Lemanceau P., Offre P., Mougel C., Gamalero E., Dessaux Y., Moënne-Loccoz Y., Berta G. Microbial ecology of the rhizosphere. In: Bloem J., Hopkins D.W., Benedetti A., editors. Microbiological Methods for Assessing Soil Quality. CAB International Wallingford; Oxfordshire, UK: 2005. pp. 228–230. [Google Scholar]
- 22.Fierer N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017;15:579–590. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 23.Savary S., Teng P.S., Willocquet L., Nutter F.W. Quantification and modelling of crop losses: A review of purposes. Annu. Rev. Phytopathol. 2006;44:89–112. doi: 10.1146/annurev.phyto.44.070505.143342. [DOI] [PubMed] [Google Scholar]
- 24.Savary S., Willocquet L., Elazegui F.A., Castilla N.P., Teng P.S. Rice pest constraints in tropical Asia: Quantification of yield losses due to rice pests in a range of production situations. Plant Dis. 2000;84:357–369. doi: 10.1094/PDIS.2000.84.3.357. [DOI] [PubMed] [Google Scholar]
- 25.Fiers M., Edel-Hermann V., Chatot C., Le Hingrat Y., Alabouvette C., Steinberg C. Potato soil-borne diseases. A review. Agron. Sustain. Dev. 2012;32:93–132. doi: 10.1007/s13593-011-0035-z. [DOI] [Google Scholar]
- 26.Tkacz A., Cheema J., Chandra G., Grant A., Poole P.S. Stability and succession of the rhizosphere microbiota depends upon plant type and soil composition. ISME J. 2015;9:2349–2359. doi: 10.1038/ismej.2015.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zilber-Rosenberg I., Rosenberg E. Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol. Rev. 2008;32:723–735. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 28.Vandenkoornhuyse P., Quaiser A., Duhamel M., Le Van A., Dufresne A. The importance of the microbiome of the plant holobiont. New Phytol. 2015;206:1196–1206. doi: 10.1111/nph.13312. [DOI] [PubMed] [Google Scholar]
- 29.Compant S., Clément C., Sessitsch A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010;42:669–678. doi: 10.1016/j.soilbio.2009.11.024. [DOI] [Google Scholar]
- 30.Ferreira C.M.H., Soares H.M.V.M., Soares E.V. Promising bacterial genera for agricultural practices: An insight on plant growth-promoting properties and microbial safety aspects. Sci. Total Environ. 2019;682:779–799. doi: 10.1016/j.scitotenv.2019.04.225. [DOI] [PubMed] [Google Scholar]
- 31.Batista B.D., Lacava P.T., Ferrari A., Teixeira-Silva N.S., Bonatelli M.L., Tsui S., Mondin M., Kitajima E.W., Pereira J.O., Azevedo J.L., et al. Screening of tropically derived, multi-trait plant growth- promoting rhizobacteria and evaluation of corn and soybean colonization ability. Microbiol. Res. 2018;206:33–42. doi: 10.1016/j.micres.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Gamalero E., Glick B.R. Plant growth-promoting bacteria in agriculture and stress environments. In: Van Elsas J.D., Trevors J.T., Rosado A.S., Nannipieri P., editors. Modern Soil Microbiology. 3rd ed. Taylor & Francis Inc.; Portland, OR, USA: 2019. pp. 361–380. [Google Scholar]
- 33.Pii Y., Mimmo T., Tomasi N., Terzano R., Cesco S., Crecchio C. Microbial interactions in the rhizosphere: Beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol. Fertil. Soils. 2015;51:403–415. doi: 10.1007/s00374-015-0996-1. [DOI] [Google Scholar]
- 34.Tairo E.V., Ndakidemi P.A. Possible benefits of rhizobial inoculation and phosphorus supplementation on nutrition, growth and economic sustainability in grain legumes. Am. J. Res. Commun. 2013;1:25. [Google Scholar]
- 35.Smith B.E., Richards R.L., Newton W.E. Catalysts for Nitrogen Fixation: Nitrogenases, Relevant Chemicals Models and Commercial Processes. Springer Science & Business Media; Berlin, Germany: 2013. p. 340. [Google Scholar]
- 36.Tao K., Kelly S., Radutoiu S. Microbial associations enabling nitrogen acquisition in plants. Curr. Opin. Microbiol. 2019;49:83–89. doi: 10.1016/j.mib.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Ben Tekaya S., Guerra T., Rodriguez D., Dawson J.O., Hahn D. Frankia diversity in host plant root nodules is independent of abundance or relative diversity of Frankia populations in corresponding rhizosphere soils. Appl. Environ. Microbiol. 2017;84:e02248-17. doi: 10.1128/AEM.02248-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prell J., Poole P. Metabolic changes of rhizobia in legume nodules. Trends Microbiol. 2006;14:161–168. doi: 10.1016/j.tim.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Gallon J.R. Reconciling the incompatible: N2 fixation and O2. New Phytol. 1992;122:571–609. doi: 10.1111/j.1469-8137.1992.tb00087.x. [DOI] [Google Scholar]
- 40.Clúa J., Roda C., Zanetti M., Blanco F. Compatibility between legumes and rhizobia for the establishment of a successful nitrogen-fixing symbiosis. Genes. 2018;9:125. doi: 10.3390/genes9030125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fageria N.K., Gheyi H.R., Moreira A. Nutrient bioavailability in salt affected soils. J. Plant Nutr. 2011;34:945–962. doi: 10.1080/01904167.2011.555578. [DOI] [Google Scholar]
- 42.Mehnaz S. Plant Microbes Symbiosis: Applied Faccets. Springer; Berlin/Heidelberg, Germany: 2015. Azospirillum: A biofertilizer for every crop; pp. 297–314. [Google Scholar]
- 43.Van Dommelen A., Vanderleyden J. Biology of the Nitrogen Cycle. Elsevier Science; Amsterdam, The Netherlands: 2007. Associative nitrogen fixation; p. 179. [Google Scholar]
- 44.Fukami J., Cerezini P., Hungria M. Azospirillum: Benefits that go far beyond biological nitrogen fixation. AMB Express. 2018;8:73. doi: 10.1186/s13568-018-0608-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan M.S., Zaidi A., Wani P.A. Role of phosphate-solubilizing microorganisms in sustainable agriculture—A review. Agron. Sustain. Dev. 2007;27:29–43. doi: 10.1051/agro:2006011. [DOI] [Google Scholar]
- 46.Mahdhi S.S., Hassan G.I., Hussain A., Rasol F. Phosphorus availability issue - its fixation and role of phosphate solubilizing bacteria in phosphate solubilization. Res. J. Agric. Sci. 2011;2:174–179. [Google Scholar]
- 47.Zhu F., Qu L., Hong X., Sun X. Isolation and characterization of a phosphate-solubilizing halophilic bacterium Kushneria sp. YCWA18 from Daqiao Saltern on the Coast of Yellow Sea of China. Evid. Based Complement. Alternat. Med. 2011;2011:615032. doi: 10.1155/2011/615032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng K., Lu H.M., Sheng H.J., Wang X.L., Mao J. Effect of organic ligands on biological availability of inorganic phosphorus in soils. Pedosphere. 2004;14:85–92. [Google Scholar]
- 49.Angus J.F. Sustainable Production. Springer; Berlin/Heidelberg, Germany: 2013. Fertilizer science and technology. [Google Scholar]
- 50.Stevenson F.J. Micronutrients. Wiley; New York, NY, USA: 1986. Cycles of soil carbon, nitrogen, phosphorus, sulfur. [Google Scholar]
- 51.Rodriguez H., Fraga R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999;17:319–339. doi: 10.1016/S0734-9750(99)00014-2. [DOI] [PubMed] [Google Scholar]
- 52.McComb R.B., Bowers G.N., Jr., Posen S. Alkaline Phosphatase. Springer Science and Business Media; Berlin/Heidelberg, Germany: 2013. pp. 79–436. [Google Scholar]
- 53.Gamalero E., Glick B.R. Bacteria in Agrobiology, Plant Nutrient Management. Volume 2. Springer; Berlin, Germany: 2012. Mechanisms used by plant growth-promoting bacteria; pp. 17–46. [Google Scholar]
- 54.Alori E.T., Glick B.R., Babalola O.O. Microbial phosphorus solubilization and Its potential for use in sustainable agriculture. Front. Microbiol. 2017;8:971. doi: 10.3389/fmicb.2017.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma J.F. Plant root responses to three abundant soil minerals: Silicon, aluminum and iron. Crit. Rev. Plant Sci. 2005;24:267–281. doi: 10.1080/07352680500196017. [DOI] [Google Scholar]
- 56.Colombo C., Palumbo G., He J.-Z., Pinton R., Cesco S. Review on iron availability in soil: Interaction of Fe minerals, plants, and microbes. J. Soils Sediments. 2014;14:538–548. doi: 10.1007/s11368-013-0814-z. [DOI] [Google Scholar]
- 57.Hansen N.C., Hopkins B.G., Ellsworth J.W., Jolley V.D. Iron Nutrition in Plants and Rhizosphere Microorganisms. Springer; Berlin/Heidelberg, Germany: 2006. Iron nutrition in field crops. [Google Scholar]
- 58.Barker A.V., Pilbeam D.J. Handbook of Plant Nutrition. CRC Press; Boca Raton, FL, USA: 2015. [Google Scholar]
- 59.Rajkumar M., Ae N., Prasad M.N.V., Freitas H. Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 2010;28:142–149. doi: 10.1016/j.tibtech.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 60.Saha M., Sarkar S., Sarkar B., Sharma B.K., Bhattacharjee S., Tribedi P. Microbial siderophores and their potential applications: A review. Environ. Sci. Pollut. Res. 2016;23:3984–3999. doi: 10.1007/s11356-015-4294-0. [DOI] [PubMed] [Google Scholar]
- 61.Guerinot M.L., Yiang Y. Iron: Nutritious, noxious, and not readily available. Plant Physiol. 1994;104:6. doi: 10.1104/pp.104.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lindsay W.L., Schwab A.P. The chemistry of iron in soils and its availability to plants. J. Plant Nutr. 1982;5:821–840. doi: 10.1080/01904168209363012. [DOI] [Google Scholar]
- 63.Adesemoye A.O., Torbert H.A., Kloepper J.W. Plant growth-promoting rhizobacteria allow reduced application rates of chemical fertilizers. Microb. Ecol. 2009;58:921–929. doi: 10.1007/s00248-009-9531-y. [DOI] [PubMed] [Google Scholar]
- 64.Abbas G., Saqib M., Akhtar J., Haq M.A. Interactive effects of salinity and iron deficiency on different rice genotypes. J. Plant Nutr. Soil Sci. 2015;178:306–311. doi: 10.1002/jpln.201400358. [DOI] [Google Scholar]
- 65.Rabhi M., Barhoumi Z., Ksouri R., Abdelly C., Gharsalli M. Interactive effects of salinity and iron deficiency in Medicago ciliaris. C. R. Biol. 2007;330:779–788. doi: 10.1016/j.crvi.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 66.Loper J.E., Buyer J.S. Siderophores in microbial interactions on plant surfaces. Mol. Plant. Microbe Interact. 1991;4:5–13. doi: 10.1094/MPMI-4-005. [DOI] [Google Scholar]
- 67.Neilands J.B. Iron absorption and transport in microorganisms. Annu. Rev. Nutr. 1981;1:27–46. doi: 10.1146/annurev.nu.01.070181.000331. [DOI] [PubMed] [Google Scholar]
- 68.Chauhan H., Bagyaraj D.J., Selvakumar G., Sundaram S.P. Novel plant growth promoting rhizobacteria—Prospects and potential. Appl. Soil Ecol. 2015;95:38–53. doi: 10.1016/j.apsoil.2015.05.011. [DOI] [Google Scholar]
- 69.Harrington J.M., Duckworth O.W., Haselwandter K. The fate of siderophores: Antagonistic environmental interactions in exudate-mediated micronutrient uptake. BioMetals. 2015;28:461–472. doi: 10.1007/s10534-015-9821-4. [DOI] [PubMed] [Google Scholar]
- 70.Ahmed E., Holmström S.J.M. Siderophores in environmental research: Roles and applications. Microb. Biotechnol. 2014;7:196–208. doi: 10.1111/1751-7915.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Butaitė E., Baumgartner M., Wyder S., Kümmerli R. Siderophore cheating and cheating resistance shape competition for iron in soil and freshwater Pseudomonas communities. Nat. Commun. 2017;8:414. doi: 10.1038/s41467-017-00509-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thomine S., Lanquar V. Transporters and Pumps in Plant Signalings. Springer; Berlin/Heidelberg, Germany: 2011. Iron transport and segnaling in plants; pp. 99–131. [Google Scholar]
- 73.Radzki W., Gutierrez Mañero F.J., Algar E., Lucas García J.A., García-Villaraco A., Ramos Solano B. Bacterial siderophores efficiently provide iron to iron-starved tomato plants in hydroponics culture. Antonie Van Leeuwenhoek. 2013;104:321–330. doi: 10.1007/s10482-013-9954-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Curie C., Briat J.-F. Iron transport and signalling. Annu. Rev. Plant Biol. 2003;54:183–206. doi: 10.1146/annurev.arplant.54.031902.135018. [DOI] [PubMed] [Google Scholar]
- 75.Robinson N.J., Procter C.M., Connolly E.L., Guerinot M.L. A ferric-chelate reductase for iron uptake from soils. Nature. 1999;397:694–697. doi: 10.1038/17800. [DOI] [PubMed] [Google Scholar]
- 76.Marschner H. Mineral Nutrition of Higher Plants. 2nd ed. Academic; London, UK: 1995. [Google Scholar]
- 77.Vert G., Grotz N., Dédaldéchamp F., Gaymard F., Guerinot M.L., Briat J.-F., Curie C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Altomare C., Tringovska I. Genetics, Biofuels and Local Farming Systems. Sustainable Agriculture Reviews. Springer; Berlin/Heidelberg, Germany: 2011. Beneficial soil microorganisms, an ecological alternative for soil fertility management; pp. 161–214. [Google Scholar]
- 79.Guerinot M. Cell Biology of Metals and Nutrients, Plant Cell Monographs. Springer; Berlin/Heidelberg, Germany: 2010. Iron; pp. 75–94. [Google Scholar]
- 80.Peleg Z., Blumwald E. Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 2011;14:290–295. doi: 10.1016/j.pbi.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 81.Korasick D.A., Enders T.A., Strader L.C. Auxin biosynthesis and storage forms. J. Exp. Bot. 2013;64:2541–2555. doi: 10.1093/jxb/ert080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weijers D., Nemhauser J., Yang Z. Auxin: Small molecule, big impact. J. Exp. Bot. 2018;69:133–136. doi: 10.1093/jxb/erx463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Spaepen S., Vanderleyden J. Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 2011;3:a001438. doi: 10.1101/cshperspect.a001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Duca D., Lorv J., Patten C.L., Rose D., Glick B.R. Indole-3-acetic acid in plant–microbe interactions. Antonie Van Leeuwenhoek. 2014;106:85–125. doi: 10.1007/s10482-013-0095-y. [DOI] [PubMed] [Google Scholar]
- 85.Gupta G., Parihar S.S., Ahirwar N.K., Snehi S.K., Sing V. Plant Growth Promoting Rhizobacteria (PGPR): Current and future prospects for development of sustainable agriculture. J. Microb. Biochem. Technol. 2015;7 doi: 10.4172/1948-5948.1000188. [DOI] [Google Scholar]
- 86.De Garcia Salamone I.E., Hynes R.K., Nelson L.M. Biocontrol and fertilization. Springer; Berlin/Heidelberg, Germany: 2006. Role of cytochinins in plant growth promotion; pp. 173–195. [Google Scholar]
- 87.Peck S.C., Kende H. Sequential induction of the ethylene biosynthetic enzymes by indole-3-acetic acid in ethiolated peas. Plant Mol. Biol. 1995;28:293–301. doi: 10.1007/BF00020248. [DOI] [PubMed] [Google Scholar]
- 88.Evans M.L., Hikawa H., Estelle M.A. Responses of Arabidopsis roots to auxin studied with high temporal resolution: Comparison of wild type and auxin-response mutants. Planta. 1994;194:215–222. doi: 10.1007/BF01101680. [DOI] [Google Scholar]
- 89.Sitbon F., Hennion S., Sundberg B., Little C.H.A., Olsson O., Sandberg G. Transgenic tobacco plants coexpressing the Agrobacterium tumefaciens iaaM and iaaH genes display altered growth and indoleacetic acid metabolism. Plant Physiol. 1992;99:1062–1069. doi: 10.1104/pp.99.3.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jones J.D.G., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 91.Han G. Origin and evolution of the plant immune system. New Phytol. 2019;222:70–83. doi: 10.1111/nph.15596. [DOI] [PubMed] [Google Scholar]
- 92.Van de Poel B., Smet D., Van Der Straeten D. Ethylene and hormonal cross talk in vegetative growth and development. Plant Physiol. 2015;169:61–72. doi: 10.1104/pp.15.00724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vanderstraeten L., Van Der Straeten D. Accumulation and transport of 1-Aminocyclopropane-1-Carboxylic Acid (ACC) in plants: Current status, considerations for future research and agronomic applications. Front. Plant Sci. 2017;8 doi: 10.3389/fpls.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Desbrosses G.J., Stougaard J. Root nodulation: A paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe. 2011;10:348–358. doi: 10.1016/j.chom.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 95.Guinel F.C. Ethylene, a hormone at the center-stage of nodulation. Front. Plant Sci. 2015;6 doi: 10.3389/fpls.2015.01121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tsang D.L., Edmond C., Harrington J.L., Nühse T.S. Cell wall integrity controls root elongation via a general 1-aminocyclopropane-1-carboxylic acid-dependent, ethylene-independent pathway. Plant Physiol. 2011;156:596–604. doi: 10.1104/pp.111.175372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Abeles F. Ethylene in Plant Biology. Academic Press; Cambridge, MA, USA: 1973. [Google Scholar]
- 98.Morgan P.W., Drew M.C. Ethylene and plant responses to stress. Physiol. Plant. 1997;100:620–630. doi: 10.1111/j.1399-3054.1997.tb03068.x. [DOI] [Google Scholar]
- 99.John P. How plant molecular biologists revealed a surprising relationship between two enzymes, which took an enzyme out of a membrane where it was not located, and put it into the soluble phase where it could be studied. Plant Mol. Biol. Rep. 1991;9:192–194. doi: 10.1007/BF02672067. [DOI] [Google Scholar]
- 100.Van Loon L.C., Glick B.R. Molecular Ecotoxicology of Plants. Springer; Berlin/Heidelberg, Germany: 2004. Increased plant fitness by rhizobacteria; pp. 177–205. [Google Scholar]
- 101.Glick B.R., Todorovic B., Czarny J., Cheng Z., Duan J., McConkey B. Promotion of plant growth by bacterial ACC deaminase. Crit. Rev. Plant Sci. 2007;26:227–242. doi: 10.1080/07352680701572966. [DOI] [Google Scholar]
- 102.Ciardi J.A., Tieman D.M., Lund S.T., Jones J.B., Stall R.E., Klee H.J. Response to Xanthomonas campestris pv. vesicatoria in tomato involves regulation of ethylene receptor gene expression. Plant Physiol. 2000;123:81–92. doi: 10.1104/pp.123.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bayliss C., Bent E., Culham D.E., MacLellan S., Clarke A.J., Brown G.L., Wood J.M. Bacterial genetic loci implicated in the Pseudomonas putida GR12-2R3–canola mutualism: Identification of an exudate-inducible sugar transporter. Can. J. Microbiol. 1997;43:809–818. doi: 10.1139/m97-118. [DOI] [PubMed] [Google Scholar]
- 104.Ma W., Guinel F.C., Glick B.R. Rhizobium leguminosarum biovar viciae 1-aminocyclopropane-1-carboxylate deaminase promotes nodulation of pea plants. Appl. Environ. Microbiol. 2003;69:4396–4402. doi: 10.1128/AEM.69.8.4396-4402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nascimento F.X., Rossi M.J., Glick B.R. Ethylene and 1-Aminocyclopropane-1-carboxylate (ACC) in plant–bacterial interactions. Front. Plant Sci. 2018;9:114. doi: 10.3389/fpls.2018.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nascimento F.X., Tavares M.J., Franck J., Ali S., Glick B.R., Rossi M.J. ACC deaminase plays a major role in Pseudomonas fluorescens YsS6 ability to promote the nodulation of Alpha- and Betaproteobacteria rhizobial strains. Arch. Microbiol. 2019;201:817–822. doi: 10.1007/s00203-019-01649-5. [DOI] [PubMed] [Google Scholar]
- 107.Gamalero E., Pivato B., Bona E., Copetta A., Avidano L., Lingua G., Berta G. Interactions between a fluorescent pseudomonad, an arbuscular mycorrhizal fungus and a hypovirulent isolate of Rhizoctonia solani affect plant growth and root architecture of tomato plants. Plant Biosyst. Int. J. Deal. Asp. Plant Biol. 2010;144:582–591. doi: 10.1080/11263504.2010.489315. [DOI] [Google Scholar]
- 108.Barnawal D., Bharti N., Maji D., Chanotiya C.S., Kalra A. ACC deaminase-containing Arthrobacter protophormiae induces NaCl stress tolerance through reduced ACC oxidase activity and ethylene production resulting in improved nodulation and mycorrhization in Pisum sativum. J. Plant Physiol. 2014;171:884–894. doi: 10.1016/j.jplph.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 109.Selvakumar G., Shagol C.C., Kim K., Han S., Sa T. Spore associated bacteria regulates maize root K+/Na+ ion homeostasis to promote salinity tolerance during arbuscular mycorrhizal symbiosis. BMC Plant Biol. 2018;18:109. doi: 10.1186/s12870-018-1317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Suttherland I.W. Bacterial exopolisaccharides. Adv. Microb. Physiol. 1972;8:143–213. doi: 10.1016/s0065-2911(08)60190-3. [DOI] [PubMed] [Google Scholar]
- 111.Kumar M.A., Anandapandian K.T.K., Parthiban K. Production and characterization of exopolysaccharides (EPS) from biofilm forming marine bacterium. Braz. Arch. Biol. Technol. 2011;54:259–265. doi: 10.1590/S1516-89132011000200006. [DOI] [Google Scholar]
- 112.Wingender J., Neu T.R., Flemming H.C. Microbial Extracellular Polymeric Substances. Springer; Berlin/Heidelberg, Germany: 1999. What are bacterial extracellular polymeric substances; pp. 1–9. [Google Scholar]
- 113.Kumar A.S., Kalpana M., Bhavanath J. Bacterial exopolysaccharides–a perception. J. Basic Microbiol. 2007;47:103–117. doi: 10.1002/jobm.200610203. [DOI] [PubMed] [Google Scholar]
- 114.Esbelin J., Santos T., Hébraud M. Desiccation: An environmental and food industry stress that bacteria commonly face. Food Microbiol. 2018;69:82–88. doi: 10.1016/j.fm.2017.07.017. [DOI] [PubMed] [Google Scholar]
- 115.Behare P., Singh R., Singh R.P. Exopolysaccharide-producing mesophilic lactic cultures for preparation of fat-free Dahi–an Indian fermented milk. J. Dairy Res. 2009;76:90–97. doi: 10.1017/S0022029908003865. [DOI] [PubMed] [Google Scholar]
- 116.Dragoš A., Kovács Á.T. The peculiar functions of the bacterial extracellular matrix. Trends Microbiol. 2017;25:257–266. doi: 10.1016/j.tim.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 117.Roberson E.B., Firestone M.K. Relationship between desiccation and exopolysaccharide production in a soil Pseudomonas sp. Appl. Environ. Microbiol. 1992;58:1284–1291. doi: 10.1128/AEM.58.4.1284-1291.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Naseem H., Ahsan M., Shahid M.A., Khan N. Exopolysaccharides producing rhizobacteria and their role in plant growth and drought tolerance. J. Basic Microbiol. 2018;58:1009–1022. doi: 10.1002/jobm.201800309. [DOI] [PubMed] [Google Scholar]
- 119.Costa O.Y.A., Raaijmakers J.M., Kuramae E.E. Microbial extracellular polymeric substances: Ecological function and impact on soil aggregation. Front. Microbiol. 2018;9:1636. doi: 10.3389/fmicb.2018.01636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Radyukina N.L., Kartashov A.V., Ivanov Y.V., Shevyakova N.I., Kuznetsov V.V. Functioning of defense systems in halophytes and glycophytes under progressing salinity. Russ. J. Plant Physiol. 2007;54:806–815. doi: 10.1134/S1021443707060131. [DOI] [Google Scholar]
- 121.Kosová K., Vítámvás P., Urban M.O., Prášil I.T. Plant proteome responses to salinity stress—Comparison of glycophytes and halophytes. Funct. Plant Biol. 2013;40:775. doi: 10.1071/FP12375. [DOI] [PubMed] [Google Scholar]
- 122.Green T.G.A., Sancho L.G., Pintado A., Saco D., Martín S., Arróniz-Crespo M., Angel Casermeiro M., de la Cruz Caravaca M.T., Cameron S., Rozzi R. Sodium chloride accumulation in glycophyte plants with cyanobacterial symbionts. AoB PLANTS. 2017;9 doi: 10.1093/aobpla/plx053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sanità di Toppi L., Bruno L., Bruni R., Ligrone R., Ferrarese A., Paoli L., Lingua G. Interazioni Piante-Ambiente. Piccin; Padova, Italy: 2018. [Google Scholar]
- 124.Flowers T.J., Colmer T.D. Salinity tolerance in halophytes. New Phytol. 2008;179:945–963. doi: 10.1111/j.1469-8137.2008.02531.x. [DOI] [PubMed] [Google Scholar]
- 125.Mishra A., Tanna B. Halophytes: Potential resources for salt stress tolerance genes and promoters. Front. Plant Sci. 2017;8:829. doi: 10.3389/fpls.2017.00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hinojosa L., Gonzalez J.A., Barrios-Masias F.H., Fuentes F., Murphy K.M. Quinoa abiotic stress responses: A review. Plants. 2018;7:106. doi: 10.3390/plants7040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Grigore M.N., Toma C. A proposal for a new halophytes classification, based on integrative anatomy observations. Olten. Stud. Şi Comunicări Ştiinţ. Nat. 2010;26:45–50. [Google Scholar]
- 128.Gupta B., Huang B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genomics. 2014;2014:701596. doi: 10.1155/2014/701596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.AbdElgawad H., Zinta G., Hegab M.M., Pandey R., Asard H., Abuelsoud W. High salinity induces different oxidative stress and antioxidant responses in maize seedlings organs. Front. Plant Sci. 2016;7 doi: 10.3389/fpls.2016.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liang W., Ma X., Wan P., Liu L. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018;495:286–291. doi: 10.1016/j.bbrc.2017.11.043. [DOI] [PubMed] [Google Scholar]
- 131.Isayenkov S.V., Maathuis F.J.M. Plant salinity stress: Many unanswered questions remain. Front. Plant Sci. 2019;10:80. doi: 10.3389/fpls.2019.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gamalero E., Berta G., Massa N., Glick B.R., Lingua G. Interactions between Pseudomonas putida UW4 and Gigaspora rosea BEG9 and their consequences for the growth of cucumber under salt-stress conditions: Salt affects bacteria–AM fungi interactions. J. Appl. Microbiol. 2010;108:236–245. doi: 10.1111/j.1365-2672.2009.04414.x. [DOI] [PubMed] [Google Scholar]
- 133.Baskin C.C., Baskin J.M. Seeds: Ecology, Biogeography, and Evolution of Dormancy and Germination. 2nd ed. Academic Press; San Diego, CA, USA: 2014. [Google Scholar]
- 134.Cuartero J. Fernandez-Munoz Tomato and salinity. Sci. Hortic. 1999;78:83–125. doi: 10.1016/S0304-4238(98)00191-5. [DOI] [Google Scholar]
- 135.Xu S., Hu B., He Z., Ma F., Feng J., Shen W., Yang J. Enhancement of salinity tolerance during rice seed germination by presoaking with hemoglobin. Int. J. Mol. Sci. 2011;12:2488–2501. doi: 10.3390/ijms12042488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhao Y., Lu Z., He L. Effects of saline-alkaline stress on seed germination and seedling growth of Sorghum bicolor (L.) Moench. Appl. Biochem. Biotechnol. 2014;173:1680–1691. doi: 10.1007/s12010-014-0956-5. [DOI] [PubMed] [Google Scholar]
- 137.Li H., Smith F.A., Dickson S., Holloway R.E., Smith S.E. Plant growth depressions in arbuscular mycorrhizal symbioses: Not just caused by carbon drain? New Phytol. 2008;178:852–862. doi: 10.1111/j.1469-8137.2008.02410.x. [DOI] [PubMed] [Google Scholar]
- 138.Bojovi B., Đelić G., Topuzović M., Stanković M. Effects of NaCl on seed germination. Kragujev. J. Sci. 2010:83–87. [Google Scholar]
- 139.Santo A., Mattana E., Frigau L., Marzo Pastor A., Picher Morelló M.C., Bacchetta G. Effects of NaCl stress on seed germination and seedling development of Brassica insularis Moris (Brassicaceae) Plant Biol. 2017;19:368–376. doi: 10.1111/plb.12539. [DOI] [PubMed] [Google Scholar]
- 140.Houle G., Morel L., Reynolds C.E., Siégel J. The effect of salinity on different developmental stages of an endemic annual plant, Aster laurentianus (Asteraceae) Am. J. Bot. 2001;88:62–67. doi: 10.2307/2657127. [DOI] [PubMed] [Google Scholar]
- 141.Trupkin S.A., Auge G.A., Zhu J.K., Sanchez R.A., Botto J.F. Salt Overly Sensitive 2 (SOS2) and interacting partners SOS3 and Abscisic Acid-Insensitive 2 (ABI2) promote red-light-dependent germination and seedling deetiolation in Arabidopsis. Int. J. Plant Sci. 2017;178:485–493. doi: 10.1086/692097. [DOI] [Google Scholar]
- 142.Yang Y., Guo Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018;217:523–539. doi: 10.1111/nph.14920. [DOI] [PubMed] [Google Scholar]
- 143.Munns R. Physiological processes limiting plant growth in saline soils: Some dogmas and hypotheses. Plant Cell Environ. 1993;16:15–24. doi: 10.1111/j.1365-3040.1993.tb00840.x. [DOI] [Google Scholar]
- 144.Mansour M.M.F., Salama K.H.A. Cellular basis of salinity tolerance in plants. Environ. Exp. Bot. 2004;52:113–122. doi: 10.1016/j.envexpbot.2004.01.009. [DOI] [Google Scholar]
- 145.Mansour M.M.F. Plasma membrane permeability as an indicator of salt tolerance in plants. Biol. Plant. 2013;57:1–10. doi: 10.1007/s10535-012-0144-9. [DOI] [Google Scholar]
- 146.Rogers M.E., Grieve C.M., Shannon M.C. Plant growth and ion relations in lucerne (Medicago sativa L.) in response to the combined effects of NaCl and P. Plant Soil. 2003;253:187–194. doi: 10.1023/A:1024543215015. [DOI] [Google Scholar]
- 147.Schachtman D.P. Molecular insights into the structure and function of plant K+ transport mechanisms. Biochim. Biophys. Acta BBA Biomembr. 2000;1465:127–139. doi: 10.1016/S0005-2736(00)00134-6. [DOI] [PubMed] [Google Scholar]
- 148.Flowers T.J., Galal H.K., Bromham L. Evolution of halophytes: Multiple origins of salt tolerance in land plants. Funct. Plant Biol. 2010;37:604. doi: 10.1071/FP09269. [DOI] [Google Scholar]
- 149.Flowers T.J., Flowers S.A. Why does salinity pose such a difficult problem for plant breeders? Agric. Water Manag. 2005;78:15–24. doi: 10.1016/j.agwat.2005.04.015. [DOI] [Google Scholar]
- 150.Mhamdi A., Van Breusegem F. Reactive oxygen species in plant development. Development. 2018;145:dev164376. doi: 10.1242/dev.164376. [DOI] [PubMed] [Google Scholar]
- 151.Noctor G., Lelarge-Trouverie C., Mhamdi A. The metabolomics of oxidative stress. Phytochemistry. 2015;112:33–53. doi: 10.1016/j.phytochem.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 152.Khan A.L., Asaf S., Al-Rawahi A., Lee I.-J., Al-Harrasi A. Rhizospheric microbial communities associated with wild and cultivated frankincense producing Boswellia sacra tree. PLoS ONE. 2017;12:e0186939. doi: 10.1371/journal.pone.0186939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Aarti P.D., Tanaka R., Tanaka A. Effects of oxidative stress on chlorophyll biosynthesis in cucumber (Cucumis sativus) cotyledons. Physiol. Plant. 2006;128:186–197. doi: 10.1111/j.1399-3054.2006.00720.x. [DOI] [Google Scholar]
- 154.Chutipaijit S., Cha-um S., Sompornpailin K. High contents of proline and anthocyanin increase protective response to salinity in Oryza sativa L. spp. indica. Aust. J. Crop Sci. 2011;9:1190. [Google Scholar]
- 155.Negrão S., Schmöckel S.M., Tester M. Evaluating physiological responses of plants to salinity stress. Ann. Bot. 2017;119:1–11. doi: 10.1093/aob/mcw191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Munns R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002;25:239–250. doi: 10.1046/j.0016-8025.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- 157.De Oliveira A.B., Mendes Alencar N.L., Gomes-Filho E. Comparison between the water and salt stress effects on plant growth and development. In: Akinci S., editor. Responses of Organisms to Water Stress. InTech; London, UK: 2013. [Google Scholar]
- 158.Landi S., Hausman J.-F., Guerriero G., Esposito S. Poaceae vs. abiotic stress: Focus on drought and salt stress, recent insights and perspectives. Front. Plant Sci. 2017;8:1214. doi: 10.3389/fpls.2017.01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chen M., Yang Z., Liu J., Zhu T., Wei X., Fan H., Wang B. Adaptation mechanism of salt excluders under saline conditions and its applications. Int. J. Mol. Sci. 2018;19:3668. doi: 10.3390/ijms19113668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sharp R.E., Hsiao T.C., Silk W.K. Growth of the maize primary root at low water potentials. Plant Physiol. 1990;93:1337–1346. doi: 10.1104/pp.93.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen B., Xiao X., Zhu Y.-G., Smith F.A., Xie Z.M., Smith S.E. The arbuscular mycorrhizal fungus Glomus mosseae gives contradictory effects on phosphorus and arsenic acquisition by Medicago sativa Linn. Sci. Total Environ. 2007;379:226–234. doi: 10.1016/j.scitotenv.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 162.Deinlein U., Stephan A.B., Horie T., Luo W., Xu G., Schroeder J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014;19:371–379. doi: 10.1016/j.tplants.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Toyooka K., Goto Y., Asatsuma S., Koizumi M., Mitsui T., Matsuoka K. A Mobile secretory vesicle cluster involved in mass transport from the Golgi to the plant cell exterior. Plant Cell. 2009;21:1212–1229. doi: 10.1105/tpc.108.058933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Raza S.H., Athar H.R., Ashraf M., Hameed A. Glycinebetaine-induced modulation of antioxidant enzymes activities and ion accumulation in two wheat cultivars differing in salt tolerance. Environ. Exp. Bot. 2007;60:368–376. doi: 10.1016/j.envexpbot.2006.12.009. [DOI] [Google Scholar]
- 165.Shen B., Jensen R.G., Bohnert H.J. Mannitol protects against oxidation by hydroxyl radicals. Plant Physiol. 1997;115:527–532. doi: 10.1104/pp.115.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chen C.-Y., Yeh K.-L., Aisyah R., Lee D.-J., Chang J.-S. Cultivation, photobioreactor design and harvesting of microalgae for biodiesel production: A critical review. Bioresour. Technol. 2011;102:71–81. doi: 10.1016/j.biortech.2010.06.159. [DOI] [PubMed] [Google Scholar]
- 167.Fricke W. Rapid and tissue-specific changes in ABA and in growth rate in response to salinity in barley leaves. J. Exp. Bot. 2004;55:1115–1123. doi: 10.1093/jxb/erh117. [DOI] [PubMed] [Google Scholar]
- 168.Lovelock C.E., Ball M.C. Salinity: Environment-Plants Molecules. Springer; Berlin/Heidelberg, Germany: 2002. Influence of salinity oh photosynthesis of halophytes; pp. 315–339. [Google Scholar]
- 169.Zhu J.K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003;6:441–445. doi: 10.1016/S1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- 170.Hasegawa P.M., Bressan R.A., Zhu J.-K., Bohnert H.J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- 171.Gaxiola R.A., Rao R., Sherman A., Grisafi P., Alper S.L., Fink G.R. The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. USA. 1999;96:1480–1485. doi: 10.1073/pnas.96.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhu J.K. Plant Salt Tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/S1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- 173.Ji H., Pardo J.M., Batelli G., Van Oosten M.J., Bressan R.A., Li X. The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol. Plant. 2013;6:275–286. doi: 10.1093/mp/sst017. [DOI] [PubMed] [Google Scholar]
- 174.Szoboszlay M., Näther A., Liu B., Carrillo A., Castellanos T., Smalla K., Jia Z., Tebbe C.C. Contrasting microbial community responses to salinization and straw amendment in a semiarid bare soil and its wheat rhizosphere. Sci. Rep. 2019;9:9795. doi: 10.1038/s41598-019-46070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bulgarelli D., Schlaeppi K., Spaepen S., van Themaat E.V.L., Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 176.Peiffer J.A., Spor A., Koren O., Jin Z., Tringe S.G., Dangl J.L., Buckler E.S., Ley R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA. 2013;110:6548–6553. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chaparro J.M., Badri D.V., Vivanco J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014;8:790–803. doi: 10.1038/ismej.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Schlaeppi K., Bulgarelli D. The plant microbiome at work. Mol. Plant. Microbe Interact. 2015;28:212–217. doi: 10.1094/MPMI-10-14-0334-FI. [DOI] [PubMed] [Google Scholar]
- 179.Edwards J., Johnson C., Santos-Medellín C., Lurie E., Podishetty N.K., Bhatnagar S., Eisen J.A., Sundaresan V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA. 2015;112:E911–E920. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lee S.A., Kim Y., Kim J.M., Chu B., Joa J.-H., Sang M.K., Song J., Weon H.-Y. A preliminary examination of bacterial, archaeal, and fungal communities inhabiting different rhizocompartments of tomato plants under real-world environments. Sci. Rep. 2019;9:9300. doi: 10.1038/s41598-019-45660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liu F., Hewezi T., Lebeis S.L., Pantalone V., Grewal P.S., Staton M.E. Soil indigenous microbiome and plant genotypes cooperatively modify soybean rhizosphere microbiome assembly. BMC Microbiol. 2019;19:201. doi: 10.1186/s12866-019-1572-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Martirosyan V., Unc A., Miller G., Doniger T., Wachtel C., Steinberger Y. Desert perennial shrubs shape the microbial-community miscellany in laimosphere and phyllosphere space. Microb. Ecol. 2016;72:659–668. doi: 10.1007/s00248-016-0822-9. [DOI] [PubMed] [Google Scholar]
- 183.Fierer N., Leff J.W., Adams B.J., Nielsen U.N., Bates S.T., Lauber C.L., Owens S., Gilbert J.A., Wall D.H., Caporaso J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA. 2012;109:21390–21395. doi: 10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Basil A., Strap J.L., Knotek-Smith H.M., Krawford D.L. Studies on the microbial population of the rhizosphere of big sagebrush (Artemisia tridentata) J. Ind. Microbiol. Biotechnol. 2004;31:278–288. doi: 10.1007/s10295-004-0140-y. [DOI] [PubMed] [Google Scholar]
- 185.Hozzein W.N., Ali M.I.A., Rabie W. A new preferential medium for enumeration and isolation of desert actinomycetes. World J. Microbiol. Biotechnol. 2008;24:1547–1552. doi: 10.1007/s11274-007-9641-y. [DOI] [Google Scholar]
- 186.Torres-Cortés G., Millán V., Fernández-González A.J., Aguirre-Garrido J.F., Ramírez-Saad H.C., Fernández-López M., Toro N., Martínez-Abarca F. Bacterial community in the rhizosphere of the cactus species Mammillaria carnea during dry and rainy seasons assessed by deep sequencing. Plant Soil. 2012;357:275–288. doi: 10.1007/s11104-012-1152-4. [DOI] [Google Scholar]
- 187.Neilson J.W., Quade J., Ortiz M., Nelson W.M., Legatzki A., Tian F., LaComb M., Betancourt J.L., Wing R.A., Soderlund C.A., et al. Life at the hyperarid margin: Novel bacterial diversity in arid soils of the Atacama Desert, Chile. Extremophiles. 2012;16:553–566. doi: 10.1007/s00792-012-0454-z. [DOI] [PubMed] [Google Scholar]
- 188.Gao J., Luo Y., Wei Y., Huang Y., Zhang H., He W., Sheng H., An L. Effect of aridity and dune type on rhizosphere soil bacterial communities of Caragana microphylla in desert regions of northern China. PLoS ONE. 2019;14:e0224195. doi: 10.1371/journal.pone.0224195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Berlanas C., Berbegal M., Elena G., Laidani M., Cibriain J.F., Sagües A., Gramaje D. The fungal and bacterial rhizosphere microbiome associated with grapevine rootstock genotypes in mature and young vineyards. Front. Microbiol. 2019;10:1142. doi: 10.3389/fmicb.2019.01142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bibi F. Diversity of antagonistic bacteria isolated from medicinal plant Peganum harmala L. Saudi J. Biol. Sci. 2017;24:1288–1293. doi: 10.1016/j.sjbs.2015.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Pereira L.B., Andrade G.S., Meneghin S.P., Vicentini R., Ottoboni L.M.M. Prospecting plant growth-promoting bacteria isolated from the rhizosphere of sugarcane under drought stress. Curr. Microbiol. 2019;76:1345–1354. doi: 10.1007/s00284-019-01749-x. [DOI] [PubMed] [Google Scholar]
- 192.Muller A., Schader C., El-Hage Scialabba N., Brüggemann J., Isensee A., Erb K.-H., Smith P., Klocke P., Leiber F., Stolze M., et al. Strategies for feeding the world more sustainably with organic agriculture. Nat. Commun. 2017;8:1290. doi: 10.1038/s41467-017-01410-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Meena M.D., Joshi P.K., Jat H.S., Chinchmalatpure A., Narjari B. Changes in biological and chemical properties of saline soil amended with municipal solid waste compost and chemical fertilizers in a mustard–pearl millet cropping system. Catena. 2016;140:1–8. doi: 10.1016/j.catena.2016.01.009. [DOI] [Google Scholar]
- 194.Mahajan S., Tuteja N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005;444:139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 195.Hardoim P.R., van Overbeek L.S., Elsas J.D. van Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008;16:463–471. doi: 10.1016/j.tim.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 196.Kang S.-M., Khan A.L., You Y.-H., Kim J.-G., Kamran M., Lee I.-J. Gibberellin production by newly isolated strain Leifsonia soli SE34 and its potential to promote plant growth. J. Microbiol. Biotechnol. 2014;24:106–112. doi: 10.4014/jmb.1304.04015. [DOI] [PubMed] [Google Scholar]
- 197.Marasco R., Rolli E., Ettoumi B., Vigani G., Mapelli F., Borin S., Abou-Hadid A.F., El-Behairy U.A., Sorlini C., Cherif A., et al. A drought resistance-promoting microbiome is selected by root system under desert farming. PLoS ONE. 2012;7:e48479. doi: 10.1371/journal.pone.0048479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Esitken A., Yildiz H.E., Ercisli S., Figen Donmez M., Turan M., Gunes A. Effects of plant growth promoting bacteria (PGPB) on yield, growth and nutrient contents of organically grown strawberry. Sci. Hortic. 2010;124:62–66. doi: 10.1016/j.scienta.2009.12.012. [DOI] [Google Scholar]
- 199.Din B.U., Sarfraz S., Xia Y., Kamran M.A., Javed M.T., Sultan T., Hussain Munis M.F., Chaudhary H.J. Mechanistic elucidation of germination potential and growth of wheat inoculated with exopolysaccharide and ACC- deaminase producing Bacillus strains under induced salinity stress. Ecotoxicol. Environ. Saf. 2019;183:109466. doi: 10.1016/j.ecoenv.2019.109466. [DOI] [PubMed] [Google Scholar]
- 200.Sarkar A., Pramanik K., Mitra S., Soren T., Maiti T.K. Enhancement of growth and salt tolerance of rice seedlings by ACC deaminase-producing Burkholderia sp. MTCC 12259. J. Plant Physiol. 2018;231:434–442. doi: 10.1016/j.jplph.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 201.Chandra D., Srivastava R., Gupta V.S.R., Franco C.M.M., Sharma A.K. Evaluation of ACC-deaminase-producing rhizobacteria to alleviate water-stress impacts in wheat (Triticum aestivum L.) plants. Can. J. Microbiol. 2019;65:387–403. doi: 10.1139/cjm-2018-0636. [DOI] [PubMed] [Google Scholar]
- 202.Dodd I.C., Perez-Alfocea F. Microbial amelioration of crop salinity stress. J. Exp. Bot. 2012;63:3415–3428. doi: 10.1093/jxb/ers033. [DOI] [PubMed] [Google Scholar]
- 203.Brilli F., Pollastri S., Raio A., Baraldi R., Neri L., Bartolini P., Podda A., Loreto F., Maserti B.E., Balestrini R. Root colonization by Pseudomonas chlororaphis primes tomato (Lycopersicum esculentum) plants for enhanced tolerance to water stress. J. Plant Physiol. 2019;232:82–93. doi: 10.1016/j.jplph.2018.10.029. [DOI] [PubMed] [Google Scholar]
- 204.Sewelam N., Kazan K., Schenk P.M. Global plant stress signaling: Reactive oxygen species at the cross-road. Front. Plant Sci. 2016;7 doi: 10.3389/fpls.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Balmer A., Pastor V., Gamir J., Flors V., Mauch-Mani B. The ‘prime-ome’: Towards a holistic approach to priming. Trends Plant Sci. 2015;20:443–452. doi: 10.1016/j.tplants.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 206.Qin Y., Druzhinina I.S., Pan X., Yuan Z. Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol. Adv. 2016;34:1245–1259. doi: 10.1016/j.biotechadv.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 207.Bianco C., Defez R. Medicago truncatula improves salt tolerance when nodulated by an indole-3-acetic acid-overproducing Sinorhizobium meliloti strain. J. Exp. Bot. 2009;60:3097–3107. doi: 10.1093/jxb/erp140. [DOI] [PubMed] [Google Scholar]
- 208.Belimov A.A., Dodd I.C., Safronova V.I., Dumova V.A., Shaposhnikov A.I., Ladatko A.G., Davies W.J. Abscisic acid metabolizing rhizobacteria decrease ABA concentrations in planta and alter plant growth. Plant Physiol. Biochem. 2014;74:84–91. doi: 10.1016/j.plaphy.2013.10.032. [DOI] [PubMed] [Google Scholar]
- 209.Porcel R., Zamarreño Á., García-Mina J., Aroca R. Involvement of plant endogenous ABA in Bacillus megaterium PGPR activity in tomato plants. BMC Plant Biol. 2014;14:36. doi: 10.1186/1471-2229-14-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mayak S., Tirosh T., Glick B.R. Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol. Biochem. 2004;42:565–572. doi: 10.1016/j.plaphy.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 211.Upadhyay S.K., Singh D.P. Effect of salt-tolerant plant growth-promoting rhizobacteria on wheat plants and soil health in a saline environment. Plant Biol. 2015;17:288–293. doi: 10.1111/plb.12173. [DOI] [PubMed] [Google Scholar]
- 212.Kloepper J.V., Leong J., Teintze M., Schroth M.N. Antiquity of the vertebrate pattern of activity metabolism and its possible relation to vertebrate origins. Nature. 1980;286:886–888. doi: 10.1038/286886a0. [DOI] [PubMed] [Google Scholar]
- 213.Lu X., Liu S.-F., Yue L., Zhao X., Zhang Y.-B., Xie Z.-K., Wang R.-Y. Epsc involved in the encoding of exopolysaccharides produced by Bacillus amyloliquefaciens FZB42 act to boost the drought tolerance of Arabidopsis thaliana. Int. J. Mol. Sci. 2018;19:3795. doi: 10.3390/ijms19123795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sandhya V.Z.A.S., Grover M., Reddy G., Venkateswarlu B. Alleviation of drought stress effects in sunflower seedlings by the exopolysaccharides producing Pseudomonas putida strain GAP-P45. Biol. Fertil. Soils. 2009;46:17–26. doi: 10.1007/s00374-009-0401-z. [DOI] [Google Scholar]
- 215.Skvortsov I.M., Ignatov V.V. Extracellular polysaccharides and polysaccharide-containing biopolymers from Azospirillum species: Properties and the possible role in interaction with plant roots. FEMS Microbiol. Lett. 1998;165:223–229. doi: 10.1111/j.1574-6968.1998.tb13150.x. [DOI] [PubMed] [Google Scholar]
- 216.Niu X., Song L., Xiao Y., Ge W. Drought-tolerant plant growth-promoting rhizobacteria associated with foxtail millet in a semi-arid agroecosystem and their potential in alleviating drought stress. Front. Microbiol. 2018;8:2580. doi: 10.3389/fmicb.2017.02580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Gond S.K., Torres M.S., Bergen M.S., Helsel Z., White J.F. Induction of salt tolerance and up-regulation of aquaporin genes in tropical corn by rhizobacterium Pantoea agglomerans. Lett. Appl. Microbiol. 2015;60:392–399. doi: 10.1111/lam.12385. [DOI] [PubMed] [Google Scholar]
- 218.Zawoznik M.S., Ameneiros M., Benavides M.P., Vázquez S., Groppa M.D. Response to saline stress and aquaporin expression in Azospirillum-inoculated barley seedlings. Appl. Microbiol. Biotechnol. 2011;90:1389–1397. doi: 10.1007/s00253-011-3162-1. [DOI] [PubMed] [Google Scholar]
- 219.Cheng Z., Woody O.Z., McConkey B.J., Glick B.R. Combined effects of the plant growth-promoting bacterium Pseudomonas putida UW4 and salinity stress on the Brassica napus proteome. Appl. Soil Ecol. 2012;61:255–263. doi: 10.1016/j.apsoil.2011.10.006. [DOI] [Google Scholar]
- 220.Gururani M.A., Upadhyaya C.P., Baskar V., Venkatesh J., Nookaraju A., Park S.W. Plant growth-promoting rhizobacteria enhance abiotic stress tolerance in Solanum tuberosum through inducing changes in the expression of ROS-scavenging enzymes and improved photosynthetic performance. J. Plant Growth Regul. 2013;32:245–258. doi: 10.1007/s00344-012-9292-6. [DOI] [Google Scholar]
- 221.Kim K., Jang Y.J., Lee S.M., Oh B.T., Chae J.C., Lee K. Alleviation of salt stress by Enterobacter sp. EJ01 in tomato and Arabidopsis is accompanied by up-regulation of conserved salinity responsive factors in plants. Mol. Cells. 2014;37:109–117. doi: 10.14348/molcells.2014.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Yuan J., Chaparro J.M., Manter D.K., Zhang R., Vivanco J.M., Shen Q. Roots from distinct plant developmental stages are capable of rapidly selecting their own microbiome without the influence of environmental and soil edaphic factors. Soil Biol. Biochem. 2015;89:206–209. doi: 10.1016/j.soilbio.2015.07.009. [DOI] [Google Scholar]
- 223.Tiwari S., Lata C., Chauhan P.S., Nautiyal C.S. Pseudomonas putida attunes morphophysiological, biochemical and molecular responses in Cicer arietinum L. during drought stress and recovery. Plant Physiol. Biochem. 2016;99:108–117. doi: 10.1016/j.plaphy.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 224.Prudent M., Salon C., Souleimanov A., Emery R.J.N., Smith D.L. Soybean is less impacted by water stress using Bradyrhizobium japonicum and thuricin-17 from Bacillus thuringiensis. Agron. Sustain. Dev. 2015;35:749–757. doi: 10.1007/s13593-014-0256-z. [DOI] [Google Scholar]
- 225.Wang D.-Z., Kong L.-F., Li Y.-Y., Xie Z.-X. Environmental microbial community proteomics: Status, challenges and perspectives. Int. J. Mol. Sci. 2016;17:1275. doi: 10.3390/ijms17081275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Bharti N., Pandey S.S., Barnawal D., Patel V.K., Kalra A. Plant growth promoting rhizobacteria Dietzia natronolimnaea modulates the expression of stress responsive genes providing protection of wheat from salinity stress. Sci. Rep. 2016;6:34768. doi: 10.1038/srep34768. [DOI] [PMC free article] [PubMed] [Google Scholar]