The eccDNA replicon is a large extranuclear circular DNA that is composed of a sophisticated repetitive structure and harbors the EPSPS and several other genes that are transcribed during glyphosate stress.
Abstract
Gene copy number variation is a predominant mechanism used by organisms to respond to selective pressures from the environment. This often results in unbalanced structural variations that perpetuate as adaptations to sustain life. However, the underlying mechanisms that give rise to gene proliferation are poorly understood. Here, we show a unique result of genomic plasticity in Amaranthus palmeri: a massive, ∼400-kb extrachromosomal circular DNA (eccDNA) that harbors the 5-ENOYLPYRUVYLSHIKIMATE-3-PHOSPHATE SYNTHASE (EPSPS) gene and 58 other genes whose encoded functions traverse detoxification, replication, recombination, transposition, tethering, and transport. Gene expression analysis under glyphosate stress showed transcription of 41 of these 59 genes, with high expression of EPSPS, as well as genes coding for aminotransferases, zinc finger proteins, and several uncharacterized proteins. The genomic architecture of the eccDNA replicon is composed of a complex arrangement of repeat sequences and mobile genetic elements interspersed among arrays of clustered palindromes that may be crucial for stability, DNA duplication and tethering, and/or a means of nuclear integration of the adjacent and intervening sequences. Comparative analysis of orthologous genes in grain amaranth (Amaranthus hypochondriacus) and waterhemp (Amaranthus tuberculatus) suggests that higher order chromatin interactions contribute to the genomic origins of the A. palmeri eccDNA replicon structure.
INTRODUCTION
Barbara McClintock stated that “a sensing mechanism must be present in plants when experiencing unfavorable conditions to alert the cell to imminent danger and to set in motion the orderly sequence of events that will mitigate this danger” (McClintock, 1984). Amplification of genes and gene clusters is an example of one such stress-avoidance mechanism. It leads to altered physiology and is an intriguing example of genomic plasticity that confers genetic diversity that can rapidly lead to instances of adaptation. This phenomenon is conserved across kingdoms, and these amplified genes are often incorporated and maintained as extrachromosomal circular DNAs (eccDNAs). EccDNAs are found in human healthy somatic and cancerous tissues (Kumar et al., 2017; Møller et al., 2018) and cancer cell lines (van Loon et al., 1994). There have been few reports of eccDNAs in plants and other eukaryotes (Lanciano et al., 2017). A recent article reported that eccDNAs harbor bona fide oncogenes with massive expression levels driven by increased copy numbers (Wu et al., 2019). The same study also demonstrated that eccDNAs present in cancer cell lines contain highly accessible chromatin structure when compared with chromosomal DNA, which enables ultra-long-range chromatin contacts (Wu et al., 2019). Reported eccDNAs vary in size from a few hundred bp to Mbp and include double minutes and ring chromosomes (Storlazzi et al., 2010; Turner et al., 2017; Koo et al., 2018a, 2018b). The genesis and topological architecture of eccDNAs are not well understood. However, the formation of small eccDNAs (<2 kb) is thought to be a result of intramolecular recombination between adjacent repeats (telomeric, centromeric, satellites) initiated by a double-strand break (Mukherjee and Storici, 2012) and propagated using the breakage-fusion-bridge cycle (McClintock, 1941). Larger eccDNAs (>100 kb), such as those found in human tumors, most likely arise from a random mutational process involving all parts of the genome (Von Hoff et al., 1992). In yeast (Saccharomyces cerevisiae), some larger eccDNAs may have the capacity to self-replicate (Møller et al., 2015). Up to 80% of studied yeast eccDNAs contain consensus sequences for an autonomous origin of replication, which may partially explain their maintenance (Møller et al., 2015).

In plants, the establishment and persistence of weeds and other invasive species has been the focus of numerous investigations to understand the elements that contribute to their dominance (Thompson and Lumaret, 1992; Chandi et al., 2013). In the species Amaranthus palmeri, from the amaranth genus, amplification of the gene 5-ENOYLPYRUVYLSHIKIMATE-3-PHOSPHATE SYNTHASE (EPSPS) confers resistance to the herbicide glyphosate. The EPSPS gene may become amplified 40- to 100-fold in highly resistant populations (Gaines et al., 2010). Amplification of the EPSPS gene and the associated rise in EPSPS activity counterbalances the metabolic changes induced by glyphosate treatment of sensitive plants, such as shikimate accumulation and loss of aromatic amino acids (Gaines et al., 2010; Sammons and Gaines, 2014). Previous low-resolution fluorescence in situ hybridization analysis of glyphosate-resistant A. palmeri established that the amplified EPSPS gene was distributed over many chromosomes, suggesting a transposon-based mechanism of mobility. The amplification of EPSPS gene copy number was also concomitant with elevated EPSPS activity (Gaines et al., 2010). Partial sequencing of the genomic landscape flanking EPSPS from overlapping BACs revealed a large contiguous sequence of 297 kb that was termed the EPSPS cassette (Molin et al., 2017). The amplification of EPSPS also extended beyond the gene itself, with amplification of flanking genes and sequence. Flow cytometry revealed significant genome expansion (e.g., 11% increase in genome size with ∼100 extra copies of the cassette), suggesting that the amplification unit was large (Molin et al., 2017). A follow-up study reported that this unit was an intact eccDNA: high-resolution fiber extension microscopy identified various structural polymorphisms, including circular, dimerized circular, and linear forms (Koo et al., 2018a). A comprehensive analysis of eccDNA distribution in meiotic pachytene chromosomes also revealed a chromosome-tethering mechanism for inclusion in daughter cells during cell division rather than complete genome integration. Here, we present the reference sequence of the eccDNA replicon, its unique genomic content and structural organization, putative mechanism of genome persistence, and comparative analysis with waterhemp (Amaranthus tuberculatus) and grain amaranth (Amaranthus hypochondriacus). We anticipate that these findings will lead to a better understanding of adaptive evolution and, perhaps, to the development of stable artificial plant chromosomes carrying agronomically useful traits.
RESULTS
Sequence Composition and Structure of the eccDNA Plant Replicon
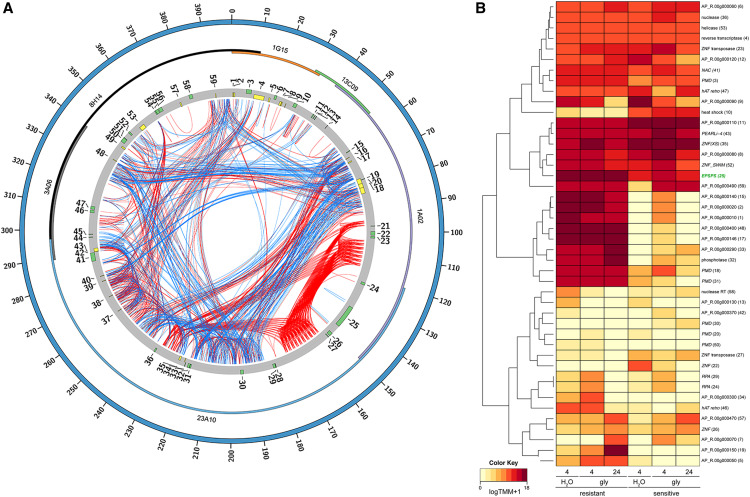
Earlier work had determined the partial genomic sequence and cytological verification of the presence of a massive eccDNA that tethers to mitotic and meiotic chromosomes for transgenerational maintenance in glyphosate-resistant A. palmeri (Molin et al., 2017; Koo et al., 2018a). Through single-molecule sequencing of the complete BAC tiling path, our sequence assembly further confirms that the circular structure is a massive extrachromosomal DNA element (Koo et al., 2018a). The circular assembly is composed of 399,435 bp, which is much larger than other eccDNAs reported to date in plants (Figure 1A). The lines at the center of the Circos plot connect direct (in red) and inverted (in blue) sequence repeats with their match(es) along the eccDNA, indicating extensive repetitive sequences within the replicon (Figure 1A).
Figure 1.
The EccDNA Replicon Structure and Gene Expression Profiles.
(A) Circos plot of the A. palmeri eccDNA replicon. The outermost solid blue ideogram represents the circular reference assembly of 399,435 bp. The multicolored inner ideogram displays the overlapping alignment of sequenced BAC clones (1G15, 13C09, 1A02, 23A10, 3A06, and 8H14) composing the 399-kb assembly. The next track (gray) depicts the 59 predicted genes with their direction of transcription (clockwise, green; counterclockwise, yellow). The internal links connect direct (red) and inverted (blue) sequence repeats with their respective internal match(es).
(B) A heatmap of gene expression of key genes under glyphosate (gly) and water controls in glyphosate-resistant and -sensitive biotypes 4 and 24 h after treatment with glyphosate. TMM, trimmed mean of M values.
The eccDNA replicon contains 59 predicted protein-coding genes, including the EPSPS gene (Supplemental Table 1). The EPSPS gene is flanked by two genes encoding putative AC transposases (Molin et al., 2017), which belong to a class of transcription factors known to interact with nucleic acid and other protein targets (Figure 1A; Supplemental Table 1). Near the EPSPS gene are two genes predicted to encode replication protein A (Figure 1A; Supplemental Table 1), which can be involved in the repair of double-strand DNA breaks induced by genotoxic stresses (Ishibashi et al., 2005). Downstream of EPSPS is a tandem array of genes coding for proteins of unknown functions but with a ribonuclease H-like domain and a C-terminal dimerization domain common to transposases of the hAT transposon superfamily (named after the founding elements hobo from fruit fly [Drosophila melanogaster], Ac from maize [Zea mays], and Tam3 from snapdragon [Antirrhinum sp]).
Many of the eccDNA replicon-encoded genes have predicted functional domains that may provide the critical cellular processes necessary for stress avoidance as well as the maintenance, stability, replication, and tethering of the eccDNA replicon. These processes include DNA transport and mobility, molecule sequestration, hormonal control, DNA replication and repair, heat shock, transcription regulation, DNA unwinding, and nuclease activity (Supplemental Table 1). For example, the eccDNA replicon encodes six proteins with aminotransferase domains. Three of the encoding genes are highly expressed in the glyphosate-resistant biotype, and they were shown to be crucial for proper cell division and differentiation (Ühlken et al., 2014). The eccDNA replicon also encodes a heat shock protein that belongs to the Hsp70 family, which is upregulated by heat stress and toxic chemicals. Hsp70 proteins also inhibit apoptosis in human cell lines (Beere et al., 2000). There are seven genes encoding proteins with plant mobile domains that function in posttranscriptional gene silencing, natural virus resistance, and the production of trans-acting small interfering RNAs (RNA interference machinery; Mourrain et al., 2000; Peragine et al., 2004). Another predicted and expressed gene encodes a protein with a No Apical Meristem (NAC) domain, found in a class of transcription factors that regulate plant defense (Xie et al., 1999) and abiotic stress responses (Hegedus et al., 2003). There are six genes coding for proteins with zinc finger domains, which are known to associate with DNA and protein, perhaps forming DNA-protein-DNA associations, and therefore with potential functions in chromosomal tethering. The AP_R.00g000430 gene product harbors a ZNF-SWIM domain, which has been shown to associate with RAD51 paralogues to promote homologous recombination (Makarova et al., 2002; Durrant et al., 2007). Furthermore, a recent study by Patterson et al. (2019) demonstrated that a RAD51 homologue was coduplicated with the EPSPS locus, which confers glyphosate resistance in Kochia scoparia. This may indicate parallel genomic recombination features. AP_R.00g000496 encodes a protein with a helicase domain, which is implicated in DNA replication and unwinding. The AP_R.00g000450 gene product contains a zinc binding reverse transcriptase domain and an integrase catalytic core, which are characteristic of a retroviral mechanism to integrate viral DNA into the host. This integrase is also found in various transposase proteins and belongs to the ribonuclease H-like superfamily involved in replication, homologous recombination, DNA repair, transposition, and RNA interference (Supplemental Table 1). In the assembled eccDNA replicon from the glyphosate-resistant biotype, the most highly expressed genes 24 h after glyphosate treatment code for a number of uncharacterized proteins, EPSPS (AP_R.00g000210), a zinc finger nuclease (AP_R.00g000492), and several proteins with plant mobile domains (Figure 1B).
Transcriptional Activity of the eccDNA Replicon under Glyphosate Stress
Forty-one of the 59 genes encoded by the eccDNA replicon are transcriptionally active when replicon-containing glyphosate-resistant plants are exposed to glyphosate at 4 or 24 h after treatment (Figure 1B; Supplemental Table 2). The most transcriptionally active genes encode two uncharacterized proteins, followed by the EPSPS gene itself (Supplemental Table 2). Additional genes include six unknown genes and two genes encoding proteins with similarity to aminotransferases that are mostly expressed in the glyphosate-resistant biotype. Stress treatments included surfactant and surfactant plus glyphosate. Surfactant treatments behaved exactly as the water-only controls, while the glyphosate treatment influenced unique gene expression profiles in the resistant biotype. Interestingly, at 4 h after glyphosate and water control treatments, the resistant and sensitive plants’ gene expression profiles were indistinguishable from their water controls (Supplemental Figure 1). At 24 h after glyphosate exposure, we detected 11 upregulated genes in the glyphosate-resistant biotype with at least a twofold increase when compared with the glyphosate-sensitive biotype (Supplemental Table 3). Their associated gene products include seven unknown proteins, two aminotransferase-like enzymes, EPSPS, and a protein with a zinc finger SWIM domain (Supplemental Table 3).
Repetitive Elements and Structural Organization
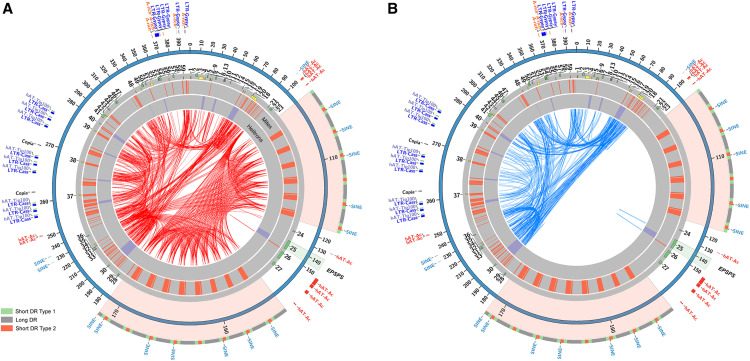
The eccDNA replicon sequence is composed of a complex arrangement of direct and indirect repeat motifs of variable lengths dispersed among retroelements composed of SINEs, LINEs, and LTR elements in addition to DNA transposons interspersed by predicted MITE and Helitron elements (Figures 2A and 2B). Flanking the EPSPS gene is an asymmetric set of direct repeats composed of arrays of clustered long and short interspersed palindromic repeat sequences (CLiSPrs), separated by identical MITES (Figure 2A). The complete palindromic array is bordered by LTR/ERVK, DNA/MULE MUDR, and DNA/TdMar Stowaway elements. The CLiSPr block regions are also composed of A/T-rich segments (up to 80%), which may serve as a mechanism for stability or nuclear recognition sites for tethering, integration into open chromatin, or transcriptional hotspots. Downstream of the CLiSPr arrays are repetitive triplicate clusters of LTR-Cassandra and DNA hAT-Ac elements, each bordered by MITE elements. Still further downstream are clustered A-rich and LTR/Gypsy elements. These arrangements may indicate functional relationships between clustered elements.
Figure 2.
The EccDNA Replicon Repeat Structure.
(A) Circos plot of the A. palmeri eccDNA replicon and key repeat content. The outer colored histograms with labels are predicted repetitive elements with organized arrangements. The highlighted repeat arrays are the CLiSPr arrays that flank the EPSPS gene. The two repeat blocks are large and asymmetric direct repeats. The inner tracks depict predicted MITE (red) and Helitron (purple) repetitive elements. The internal red links are direct repeats and their relationships within the replicon.
(B) As in (A), but the direct repeats are hidden and the indirect repeats are shown as blue links.
Genome Tethering
The first cytological evidence of an eccDNA tethering to chromosomes in plants as a mechanism of genome persistence was recently reported (Koo et al., 2018b). EccDNA maintenance has been extensively studied in DNA viruses that maintain their genomes as eccDNAs, such as Epstein-Barr, Rhadinovirus, Papillomavirus, and others (Feeney and Parish, 2009). A common theme among these viruses is genome tethering, which is facilitated by virus-encoded DNA binding proteins that associate with repeated sequences in the viral genome, themselves demonstrating affinity for host cell proteins that associate with mitotic chromatin to ensure nuclear retention (Feeney and Parish, 2009). For example, the Epstein-Barr virus anchors to the host genome through two possible mechanisms: (1) the viral genome is brought to mitotic chromosomes through the interaction of the viral protein Epstein-Barr Nuclear Antigen1 (EBNA1) and the cellular protein (and probable rRNA-processing protein) EBNA1 Binding Protein2 (EBP2); and (2) EBNA1 associates to chromatin directly via an AT-hook motif in its C terminus that binds to A/T-rich sequences on metaphase chromosomes (Wu et al., 2000; Sears et al., 2004). In Rhadinoviruses, a role has been suggested for the latency-associated nuclear antigen protein, which is thought to interact with terminal repeat regions in the viral genome (composed of long tandem repeats; Russo et al., 1996), where the C terminus of the protein attaches the terminal repeat regions while the N terminus tethers the episome to the chromosome (Piolot et al., 2001). In Papillomaviruses, the E2 regulatory protein is a multifunctional DNA binding protein that interacts with the E1 helicase for replication (Masterson et al., 1998) and facilitates association with the host genome through interaction between the N-terminal transactivation domain and host proteins (Skiadopoulos and McBride, 1998).
Computational analysis of the eccDNA replicon revealed several genes that may participate in tethering. The AP_R.00g000496 gene product contains two core AT-hook motifs (a three-amino acid motif, Gly-Arg-Pro) as well as a SWIM-type zinc finger domain known to bind DNA, proteins, and/or lipid structures (Supplemental Table 1). The optimal binding sites for the core AT-hook are AAAT and AATT, which when bound together form a concave DNA conformation for tight binding (Reeves, 2000). In the eccDNA replicon, we counted 143 (AAAT)2 and 186 (AATT)2 motifs (Figure 2A). There are consistent clusters of tandemly repeated motifs in the CLiSPr repeats. Proteins with AT-hooks may interact with the A/T-rich regions of the eccDNA replicon and other nuclear scaffold proteins. Furthermore, the helicase domain has recently been demonstrated to be an important regulator of the chromatin association, establishment, and maintenance of the Herpes virus (Harris et al., 2016). AP_R.00g000496 is predicted to encode a protein with a helicase domain that may also have a role in tethering the eccDNA to nuclear chromatin.
Synteny and Collinearity with Grain Amaranth and Waterhemp
It is unknown whether the genesis of the eccDNA replicon arose from a focal amplification surrounding the EPSPS gene or from recombination of distal genomic segments that combined multiple genes for simultaneous amplification. The high degree of synteny and collinearity between angiosperms can offer insights into the genomic origin(s) of the eccDNA replicon. The eccDNA replicon was aligned to the two closely related species grain amaranth and waterhemp, each with a haploid chromosome number (n = 16) and pseudochromosome-scale reference genome assemblies (Lightfoot et al., 2017; Kreiner et al., 2019). Both comparator genome assemblies have a single copy of the EPSPS gene located near the middle of scaffold 5 (Figure 3). Out of the 59 eccDNA replicon genes, 24 and 34 had reciprocal best hits (RBHs) to grain amaranth and waterhemp pseudochromosomes, respectively. The spatial topology of these orthologous matches was distributed across most of the pseudochromosomes in the assemblies (Figure 3; Supplemental Table 4). Eleven of the RBHs between the eccDNA replicon and the two Amaranthus genomes were organized in a collinear fashion on the same scaffolds in the same order (Figure 3, colored ideograms and links). Interestingly, the first orthologous eccDNA gene (AP_R.00g000030, encoding an aminotransferase-like enzyme) with synteny to scaffold 2 in both genomes is ∼254 kb from the next cluster of collinear genes on the eccDNA replicon (Figure 3, purple links), which are annotated in grain amaranth as encoding F-box family proteins with an additional domain of unknown function (Supplemental Table 4). Moreover, this cluster of genes is separated in the middle by a gene that resides on a completely different pseudochromosome (scaffold 3) and encodes a NAC-type transcription factor (Figure 3, blue link; Supplemental Table 4). These results suggest that eccDNA replicon genes likely originated from multiple genomic regions in A. palmeri, which supports the hypothesis of intragenomic recombination during the formation of the eccDNA replicon.
Figure 3.
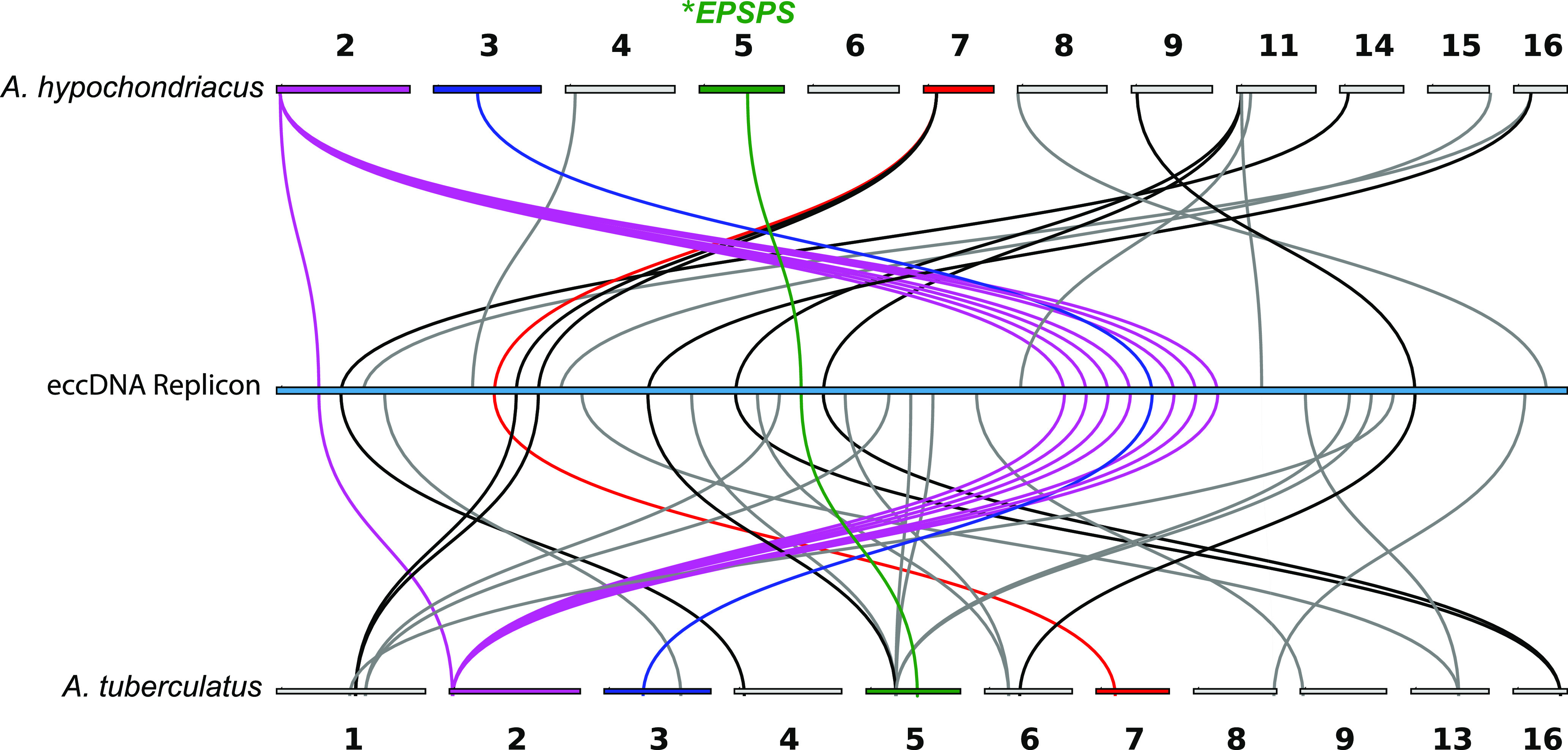
Collinear and Syntenic Arrangements in the Grain Amaranth and Waterhemp Genomes and Their Relationship to the EccDNA Replicon.
Gray lines indicate syntenic gene arrangements between one or the other genomes, and colored lines and ideograms indicate that the gene is collinear in both genomes. Purple lines, F-box family proteins with an additional domain of unknown function; blue line, NAC-type transcription factor.
The eccDNA replicon is a massive eccDNA vehicle for gene amplification, trait expression, and maintenance and transfer of genomic information. The genomic origin is unknown but likely a result of mobile element activation and extensive genome shuffling that may involve higher order chromatin interactions potentially influenced by xenobiotic pressures. It has various functional modalities for integration, stability, and maintenance to ensure genomic persistence. Furthermore, because of the functional implications of the putative genes in the eccDNA replicon, the presence of this unit could cause a general increase in abiotic stress resilience or, perhaps, an increased disposition to adapt. This vehicle may open new avenues in breeding and biotechnology through deeper understanding of its origin and function. Further investigations will be required to ascertain the essential components of eccDNA replicon proliferation and its compatibility in other species.
METHODS
Plant Material
The glyphosate-resistant Amaranthus palmeri genotype used here was collected in 2013 by W.T.M. from a soybean field in Washington County, Mississippi, that was exposed to extensive application of glyphosate during the previous decade. Sixteen plants were collected, and seeds collected from each plant were kept separate. For initial resistance assessment, seeds from each plant were sown in pots containing potting mix (Metro-Mix 360, Sun Gro Horticulture), lightly covered with 2 mm of potting mix, and watered from below. Pots were transferred to a greenhouse at the Jamie Whitten Delta States Research Center of the USDA-ARS in Stoneville, Mississippi, set to 25/20°C ± 3°C day/night temperature and a 15-h photoperiod under natural sunlight conditions supplemented with high-pressure sodium lights providing 400 µmol m−2 s−1. At the two-leaf stage, seedlings were sprayed with glyphosate at 0.84 kg of active ingredient (ai) ha−1 (16.7 mM) using an air-pressurized indoor spray chamber (DeVries Manufacturing) equipped with a nozzle mounted with 8002E flat-fan tip (Spraying Systems) delivering 190 L ha−1 at 220 kPa. All seedlings survived from a plant designated no. 13, and seedlings from this seed lot were used for all experiments. Plant no. 13 had a copy number as determined by qPCR of 34, using the single-copy gene ACETOLACTATE SYNTHASE for normalization (Molin et al., 2018).
BAC Isolation, Sequencing, and Analysis
BAC library construction, partial tile path isolation, sequencing, and analysis were described previously (Molin et al., 2017). Two additional BAC clones, 08H14 and 01G15, were determined by chromosome walking by hybridization with overgo probes designed from unique distal sequence on the terminal ends of the EPSPS cassette (clones 03A06 and 13C09). These two BAC clones were harvested and sequenced using Pacific Biosciences RSII sequencing to a depth greater than 100×, as described (Molin et al., 2017). Raw single-molecule sequence was self-corrected using the CANU Celera assembler (Koren et al., 2017) with corOutCoverage=1000 to increase the output of corrected sequences. BAC-end sequences were determined using standard Sanger sequencing methods and aligned to the reference assemblies with Phrap and opened in Consed (Gordon et al., 1998) for editing. BAC overlaps were identified using CrossMatch (Gordon et al., 1998), and ends were joined manually to form a circular structure. The consensus eccDNA replicon was annotated using a combination of the MakerP pipeline (Campbell et al., 2014) with RNA-seq (below) used as evidence with final manual curation. Functional domain scans and homology-based annotations were determined by BLAST to the UniprotKB database, InterproScan (http://www.ebi.ac.uk/interpro/search/sequence/), and hidden Markov models to the PfamA database (https://pfam.xfam.org). Repeat characterization and masking were conducted with the RepeatMasker software (http://www.repeatmasker.org). MITE and Helitron sequences were predicted with the detectMITE (Ye et al., 2016) and HelitronScanner (Xiong et al., 2014) tools. Circular figures were prepared using the Circos plotting toolset (Krzywinski et al., 2009).
RNA-Seq
Plants for RNA-seq were grown as previously described to the two-true-leaf stage under greenhouse conditions described above, at which time they were transplanted into 8-cm × 8-cm × 7-cm pots containing the same potting mixture. Plants were then watered as needed and fertilized once every 2 weeks after transplanting with a water-soluble fertilizer (Miracle-Gro, Scotts Miracle-Gro Products) at a half rate of 0.42 kg ai ha−1 (8.3 mM). When seedlings reached the six-leaf stage, they were sprayed with either water, water plus surfactant, or water plus surfactant plus glyphosate using the spray chamber. The surfactant was 0.5% (v/v) Tween 20, and glyphosate was applied at 0.42 kg ai ha−1 after neutralization with 0.1 N KOH solution. We harvested leaves from the third and fourth nodes for RNA extraction at 0, 4, and 24 h after treatment and in biological triplicates. Plants were held for 2 weeks post leaf harvest to verify survival following glyphosate treatment.
Total RNA was harvested using the RNeasy plant mini kit (Qiagen). Purified RNA was verified for intactness on a Bioanalyzer 2100 (Agilent) and subject to stranded mRNA-seq using standard TruSeq procedures and sequenced to a target depth of at least 15 million reads per sample. Raw sequence data were preprocessed for adapter and low-quality bases with the Trimmomatic tool (Krzywinski et al., 2009), and cleaned reads were aligned to the eccDNA replicon consensus assembly with Bowtie2 v.2.3.4.1 and the following arguments: –no-mixed–no-discordant–gbar 1000–end-to-end -k 200 -q -X 800. Trimmed mean of M values and fragments per kilobase million transcript quantification was determined with RSEM v1.3.0.
Synteny and Collinearity
Coding sequences and annotation (gff3) files for grain amaranth (Amaranthus hypochondriacus; v1.0) were downloaded from Phytozome (https://phytozome.jgi.doe.gov). Coding sequences and annotations for waterhemp (Amaranthus tuberculatus; v2, id54057) were downloaded from CoGe (https://genomevolution.org/coge/SearchResults.pl?s=amaranthusandp=genome). RBHs were determined using BLAST (Altschul et al., 1990) considering high-identity paralogous matches. The linear riparian plot (Figure 3) was created with custom modifications to the python version of MCscan [https://github.com/tanghaibao/jcvi/wiki/MCscan-(Python-version)].
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under BioProject ID PRJNA413471 Submission MT025716 and PRJNA626536 Submission SUB7291792.
Supplemental Data
Supplemental Figure. Multidimensional scaling plot of RNaseq data aligned to the eccDNA replicon reference derived from glyphosate resistant and sensitive biotypes sampled at 4 h and 24 h after exposure to glyphosate and water only controls.
Supplemental Table 1. Predicted coding genes and functional annotation of the eccDNA replicon.
Supplemental Table 2. Transcriptional abundance (log2 TMM+1) of eccDNA replicon genes under water and glyphosate exposure 4 h and 24 h after treatment in a glyphosate sensitive and resistant biotype.
Supplemental Table 3. Genes upregulated in the glyphosate-resistant biotype compared with the glyphosate-sensitive biotype 24 h after glyphosate exposure.
Supplemental Table 4. Replicon gene ortholog matches and locations in A. hypochondriacus and A. tuberculatus pseudochromosome assemblies.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by the USDA National Institute of Food and Agriculture (project number SC-1700530 and Technical Contribution 6758 of the Clemson University Experiment Station).
AUTHOR CONTRIBUTIONS
W.T.M. and C.A.S. designed, conducted, analyzed, and interpreted the eccDNA replicon capture, sequencing, and analysis components of the study, which include BAC screening, isolation, sequencing, assembly, annotation, and data analysis; W.T.M. and C.A.S. also prepared the initial draft of the article and edited subsequent versions; A.Y. and M.B. interpreted data and edited the article.
Footnotes
Articles can be viewed without a subscription.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.(1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Beere H.M., Wolf B.B., Cain K., Mosser D.D., Mahboubi A., Kuwana T., Tailor P., Morimoto R.I., Cohen G.M., Green D.R.(2000). Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat. Cell Biol. 2: 469–475. [DOI] [PubMed] [Google Scholar]
- Campbell M.S., et al. (2014). MAKER-P: A tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164: 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandi A., Milla-Lewis S.R., Jordan D.L., York A.C., Burton J.D., Zuleta M.C., Whitaker J.R., Culpepper A.S.(2013). Use of AFLP markers to assess genetic diversity in palmer amaranth (Amaranthus palmeri) populations from North Carolina and Georgia. Weed Sci. 61: 136–145. [Google Scholar]
- Durrant W.E., Wang S., Dong X.N. (2007). Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc. Natl. Acad. Sci. USA 104: 4223–4227. Erratum Proc. Natl. Acad. Sci. USA 104: 7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney K.M., Parish J.L.(2009). Targeting mitotic chromosomes: A conserved mechanism to ensure viral genome persistence. Proc. Biol. Sci. 276: 1535–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines T.A., et al. (2010). Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc. Natl. Acad. Sci. USA 107: 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D., Abajian C., Green P.(1998). Consed: A graphical tool for sequence finishing. Genome Res. 8: 195–202. [DOI] [PubMed] [Google Scholar]
- Harris L., McFarlane-Majeed L., Campos-León K., Roberts S., Parish J.L.(2016). The cellular DNA helicase ChlR1 regulates chromatin and nuclear matrix attachment of the human papillomavirus 16 E2 protein and high-copy-number viral genome establishment. J. Virol. 91: e01853-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus D., Yu M., Baldwin D., Gruber M., Sharpe A., Parkin I., Whitwill S., Lydiate D.(2003). Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol. Biol. 53: 383–397. [DOI] [PubMed] [Google Scholar]
- Ishibashi T., Koga A., Yamamoto T., Uchiyama Y., Mori Y., Hashimoto J., Kimura S., Sakaguchi K.(2005). Two types of replication protein A in seed plants. FEBS J. 272: 3270–3281. [DOI] [PubMed] [Google Scholar]
- Koo D.H., Jugulam M., Putta K., Cuvaca I.B., Peterson D.E., Currie R.S., Friebe B., Gill B.S.(2018b). Gene duplication and aneuploidy trigger rapid evolution of herbicide resistance in common waterhemp. Plant Physiol. 176: 1932–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo D.H., Molin W.T., Saski C.A., Jiang J., Putta K., Jugulam M., Friebe B., Gill B.S.(2018a). Extrachromosomal circular DNA-based amplification and transmission of herbicide resistance in crop weed Amaranthus palmeri. Proc. Natl. Acad. Sci. USA 115: 3332–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M.(2017). Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27: 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiner J.M., Giacomini D.A., Bemm F., Waithaka B., Regalado J., Lanz C., Hildebrandt J., Sikkema P.H., Tranel P.J., Weigel D., Stinchcombe J.R., Wright S.I.(2019). Multiple modes of convergent adaptation in the spread of glyphosate-resistant Amaranthus tuberculatus. Proc. Natl. Acad. Sci. USA 116: 21076–21084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A.(2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Dillon L.W., Shibata Y., Jazaeri A.A., Jones D.R., Dutta A.(2017). Normal and cancerous tissues release extrachromosomal circular DNA (eccDNA) into the circulation. Mol. Cancer Res. 15: 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciano S., Carpentier M.C., Llauro C., Jobet E., Robakowska-Hyzorek D., Lasserre E., Ghesquière A., Panaud O., Mirouze M.(2017). Sequencing the extrachromosomal circular mobilome reveals retrotransposon activity in plants. PLoS Genet. 13: e1006630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot D.J., Jarvis D.E., Ramaraj T., Lee R., Jellen E.N., Maughan P.J.(2017). Single-molecule sequencing and Hi-C-based proximity-guided assembly of amaranth (Amaranthus hypochondriacus) chromosomes provide insights into genome evolution. BMC Biol. 15: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K.S., Aravind L., Koonin E.V.(2002). SWIM, a novel Zn-chelating domain present in bacteria, archaea and eukaryotes. Trends Biochem. Sci. 27: 384–386. [DOI] [PubMed] [Google Scholar]
- Masterson P.J., Stanley M.A., Lewis A.P., Romanos M.A.(1998). A C-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase alpha-primase p68 subunit. J. Virol. 72: 7407–7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B.(1941). The stability of broken ends of chromosomes in Zea Mays. Genetics 26: 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B.(1984). The significance of responses of the genome to challenge. Science 226: 792–801. [DOI] [PubMed] [Google Scholar]
- Molin W.T., Wright A.A., Lawton-Rauh A., Saski C.A.(2017). The unique genomic landscape surrounding the EPSPS gene in glyphosate resistant Amaranthus palmeri: A repetitive path to resistance. BMC Genomics 18: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin W.T., Wright A.A., VanGessel M.J., McCloskey W.B., Jugulam M., Hoagland R.E.(2018). Survey of the genomic landscape surrounding the 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) gene in glyphosate-resistant Amaranthus palmeri from geographically distant populations in the USA. Pest Manag. Sci. 74: 1109–1117. [DOI] [PubMed] [Google Scholar]
- Møller H.D., Mohiyuddin M., Prada-Luengo I., Sailani M.R., Halling J.F., Plomgaard P., Maretty L., Hansen A.J., Snyder M.P., Pilegaard H., Lam H.Y.K., Regenberg B.(2018). Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat. Commun. 9: 1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller H.D., Parsons L., Jørgensen T.S., Botstein D., Regenberg B.(2015). Extrachromosomal circular DNA is common in yeast. Proc. Natl. Acad. Sci. USA 112: E3114–E3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542. [DOI] [PubMed] [Google Scholar]
- Mukherjee K., Storici F.(2012). A mechanism of gene amplification driven by small DNA fragments. PLoS Genet. 8: e1003119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson E.L., Saski C.A., Sloan D.B., Tranel P.J., Westra P., Gaines T.A.(2019). The draft genome of Kochia scoparia and the mechanism of glyphosate resistance via transposon-mediated EPSPS tandem gene duplication. Genome Biol. Evol. 11: 2927–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S.(2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18: 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piolot T., Tramier M., Coppey M., Nicolas J.C., Marechal V.(2001). Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75: 3948–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R.(2000). Structure and function of the HMGI(Y) family of architectural transcription factors. Environ. Health Perspect. 108 (suppl. 5): 803–809. [DOI] [PubMed] [Google Scholar]
- Russo J.J., Bohenzky R.A., Chien M.C., Chen J., Yan M., Maddalena D., Parry J.P., Peruzzi D., Edelman I.S., Chang Y., Moore P.S.(1996). Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93: 14862–14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammons R.D., Gaines T.A.(2014). Glyphosate resistance: State of knowledge. Pest Manag. Sci. 70: 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears J., Ujihara M., Wong S., Ott C., Middeldorp J., Aiyar A.(2004). The amino terminus of Epstein-Barr virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J. Virol. 78: 11487–11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiadopoulos M.H., McBride A.A.(1998). Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 72: 2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi C.T., et al. (2010). Gene amplification as double minutes or homogeneously staining regions in solid tumors: Origin and structure. Genome Res. 20: 1198–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Lumaret R.(1992). The evolutionary dynamics of polyploid plants: Origins, establishment and persistence. Trends Ecol. Evol. (Amst.) 7: 302–307. [DOI] [PubMed] [Google Scholar]
- Turner K.M., et al. (2017). Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 543: 122–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ühlken C., Horvath B., Stadler R., Sauer N., Weingartner M.(2014). MAIN-LIKE1 is a crucial factor for correct cell division and differentiation in Arabidopsis thaliana. Plant J. 78: 107–120. [DOI] [PubMed] [Google Scholar]
- van Loon N., Miller D., Murnane J.P.(1994). Formation of extrachromosomal circular DNA in HeLa cells by nonhomologous recombination. Nucleic Acids Res. 22: 2447–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hoff D.D., McGill J.R., Forseth B.J., Davidson K.K., Bradley T.P., Van Devanter D.R., Wahl G.M.(1992). Elimination of extrachromosomally amplified MYC genes from human tumor cells reduces their tumorigenicity. Proc. Natl. Acad. Sci. USA 89: 8165–8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Ceccarelli D.F., Frappier L.(2000). The DNA segregation mechanism of Epstein-Barr virus nuclear antigen 1. EMBO Rep. 1: 140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., et al. (2019). Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 575: 699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Sanz-Burgos A.P., Guo H., García J.A., Gutiérrez C.(1999). GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol. Biol. 39: 647–656. [DOI] [PubMed] [Google Scholar]
- Xiong W., He L., Lai J., Dooner H.K., Du C.(2014). HelitronScanner uncovers a large overlooked cache of Helitron transposons in many plant genomes. Proc. Natl. Acad. Sci. USA 111: 10263–10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C.T., Ji G.L., Liang C.(2016). detectMITE: A novel approach to detect miniature inverted repeat transposable elements in genomes. Sci. Rep. 6: 19688. [DOI] [PMC free article] [PubMed] [Google Scholar]