Arabidopsis WRKY15 functions upstream of VND7, an important regulator of tracheary element (TE) differentiation, to suppress TE formation during plant vascular development.
Abstract
Formation of the vascular cylinder, a structure critical to water and nutrient transport in higher plants, is highly regulated. Here we identify WRKY15 as an important regulator that suppresses tracheary element (TE) differentiation in Arabidopsis (Arabidopsis thaliana). Overexpression of WRKY15 resulted in discontinuous protoxylem vessel files and TEs with reduced spiral wall thickening/lignification. Expression of a dominant-negative WRKY15 variant, WRKY15-EAR, led to extra protoxylem vessels and ectopic TEs with increased spiral wall thickening/lignification. Ectopic TE formation in the root cortex and hypocotyl/leaf epidermis reveals that the suppression of WRKY15 is sufficient to trigger the transdifferentiation of other types of cells to TEs. Expression profiling, RT-qPCR, and reporter analyses revealed that WRKY15 suppresses the expression of VASCULAR-RELATED NAC DOMAIN7 (VND7), a master transcriptional regulator that promotes TE differentiation. We propose that WRKY15 negatively regulates VND7 expression indirectly based on (1) the absence of a W-box in the promoter of VND7 and (2) the observation that WRKY15 and VND7 are expressed in different cells in the vascular cylinder, with WRKY15 expressed in the procambial cells and VND7 in the protoxylem poles of procambium and differentiating TEs. Future research is needed to reveal the details underlying the interaction of WRKY15 and VND7 in plant vascular development.
INTRODUCTION
Vascular plants have evolved xylem vessels to transport water and mineral nutrients, which enables plants to colonize the land and flourish (Caño-Delgado et al., 2010; Lucas et al., 2013; Xu et al., 2014; Cho et al., 2017; Ruonala et al., 2017). Xylem vessels consist of tracheary elements (TEs), a series of interconnected dead cells with perforations on both ends to form a continuous tubular structure. The development of TEs involves several cellular events including cell specification, patterned secondary wall deposition, and programmed cell death (PCD; Fukuda, 1997; Turner et al., 2007; Furuta et al., 2014). Secondary cell wall formation is a crucial step in the differentiation of xylem cells, and lignin is one of the characteristic components in secondary cell wall, which reinforces the mechanical strength and provides hydrophobic property of the xylem vessels (Zhao and Dixon, 2011; Oda and Fukuda, 2012; Voxeur et al., 2015; Zhong and Ye, 2015; Zhao, 2016; Ohtani and Demura, 2019). Based on the thickening patterns of secondary cell wall, we can identify protoxylem vessels, which have annular or spiral patterned secondary cell wall, and metaxylem vessels, which have pitted secondary cell wall, in Arabidopsis (Arabidopsis thaliana) roots (Kubo et al., 2005; Oda and Fukuda, 2012). Auxin, cytokinin, and brassinosteroids play pivotal roles in vascular development (Demura and Fukuda, 2007; Bishopp et al., 2011; Ursache et al., 2014; Smet and De Rybel, 2016; Cho et al., 2017).

A tremendous amount of work has focused on secondary cell wall formation during xylem cell differentiation (Kubo et al., 2005; Mitsuda et al., 2007; Soyano et al., 2008; Yamaguchi et al., 2008, 2010a; Zhong et al., 2008, 2010; Ohashi-Ito et al., 2010; Taylor-Teeples et al., 2015). These studies demonstrated that xylem cell fate is controlled by a hierarchical transcriptional regulatory network including several master regulators, mainly members of the NAC transcription factor family, and their downstream secondary regulators, mainly members of MYB transcription factor family. VND1-7 (Vascular-related NAC Domain) and NST1-3 (NAC Secondary Wall Thickening Promoting Factor), two subgroups of NAC transcription factors, play a leading role in the regulation of secondary wall formation (Kubo et al., 2005; Mitsuda et al., 2005; Zhong et al., 2006; Mitsuda et al., 2007; Endo et al., 2015; Taylor-Teeples et al., 2015).
The regulatory network with VND7 as the key player has been a hotspot of research for many years. VND7 functions as a transcriptional switch in the differentiation of protoxylem vessels in Arabidopsis roots (Kubo et al., 2005; Yamaguchi et al., 2010b). Overexpression of VND7 induced various cell types to transdifferentiate into protoxylem-like vessel elements. In addition, dominant-negative suppression of VND7 inhibited protoxylem vessel differentiation in roots (Kubo et al., 2005). Recently, two additional transcription factors, VND-INTERACTING2 (VNI2) and E2Fc, were identified as regulators in secondary cell wall formation (Yamaguchi et al., 2010a; Taylor-Teeples et al., 2015). VNI2 interacts with VND7 and other VND family proteins to negatively regulate TE differentiation (Yamaguchi et al., 2010a). E2Fc can activate or repress VND7 expression in a dose-dependent manner (Taylor-Teeples et al., 2015).
WRKY transcription factors, one of the largest transcription factor families unique to plants, are key transcriptional regulators in plant growth and development (Eulgem et al., 2000; Ulker and Somssich, 2004; Rushton et al., 2010; Yamasaki et al., 2013). Previous studies have shown that the pith parenchyma cells in the wrky12 mutant of Arabidopsis exhibited secondary cell wall thickening, and their homologous genes in Medicago truncatula and Populus trichocarpa also had similar functions (Wang et al., 2010; Yang et al., 2016). WRKY12 can restrict secondary wall thickening of pith parenchyma cells and maintain its primary cell wall characteristics, indicating its role in negatively regulating secondary wall formation in pith parenchyma cells, and this inhibitory mechanism seems to be evolutionarily conserved in dicotyledonous plants. In this study, we demonstrate that WRKY15 functions as a negative regulator of TE differentiation. Overexpression and dominant-negative suppression of WRKY15 resulted in opposite phenotypes in TE formation. In WRKY15 overexpression transgenic seedlings, protoxylem vessels become discontinuous, and the spiral wall thickening of the TEs was reduced. In contrast, expression of dominant-negative WRKY15-EAR led to the formation of extra protoxylem vessel files in the metaxylem positions, TEs with increased spiral wall thickening/lignification, and the transdifferentiation of nonvascular cells into ectopic TE cells. Ectopic TE formation in WRKY15-EAR plants was associated with ectopic overexpression of VND7, a master transcriptional regulator that promotes TE formation. Based on these findings, we propose that WRKY15 functions upstream of VND7 as a negative regulator, and the push-and-pull functions of VND7 and WRKY15 play an important role in regulating protoxylem vessel formation in the development of the root vascular cylinder.
RESULTS
WRKY15 Is Mainly Expressed In Root Procambial Cells and Root Caps
Previously, we demonstrated that the MPK3/MPK6 cascade functions downstream of the ERECTA receptor-like kinase (RLK) in regulating localized cell division. Loss of function of MPK3 and MPK6 or the upstream MAPKKs, MKK4 and MKK5, resulted in shortened pedicels and clustered inflorescences, phenocopying the erecta mutant (Meng et al., 2012). Expression profiling revealed that WRKY15 might be a downstream component in the ERECTA signaling pathway (Uchida et al., 2012). As a result, we became interested in the biological function of WRKY15. WRKY15 is a member of the WRKY IId subfamily, which includes seven members in the Arabidopsis genome. Of these, WRKY15 shares the highest similarity with WRKY7 followed by WRKY11 and WRKY17 (Supplemental Figure 1A; Supplemental Data Set 1). WRKY15 protein contains four putative domains including a C domain, a HARF domain, a nuclear localization signal (NLS), and a WRKY domain (Supplemental Figures 1B and 1C). The C domain interacts with calmodulin, which is dependent on calcium. The function of the HARF domain is unknown. The C-terminal WRKY domain, which is defined by the conserved amino acid sequence WRKYGQK, together with a C2H2 type of zinc finger motif, mediates its binding to the target DNA sequences.
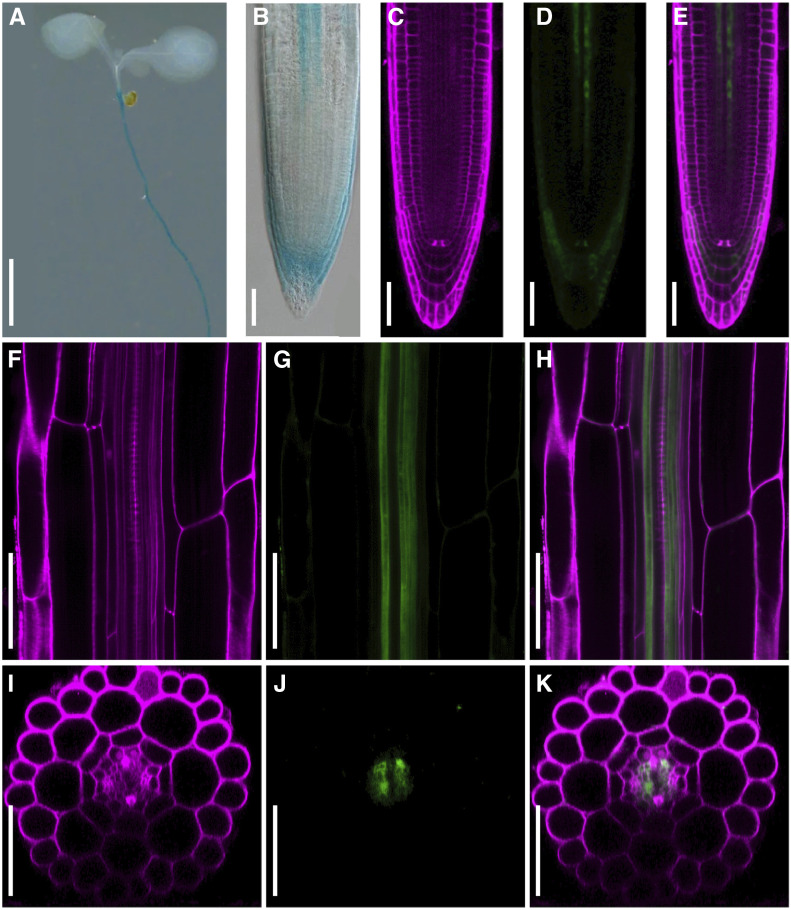
As a first step, we determined the spatial expression pattern of WRKY15 in Arabidopsis using a WRKY15 promoter-driven enhanced yellow fluorescent protein (eYFP) and β-glucuronidase (GUS) fusion reporter (WRKY15pro:eYFP-GUS). In 7-d-old Arabidopsis seedlings, GUS staining revealed high expression levels of WRKY15 in the hypocotyl-to-root transition zone, root vascular cylinder from root meristematic zone to the entire maturation zone, and root caps in cells below the quiescent center (Figures 1A and 1B). We also examined the eYFP fluorescence in roots using confocal microscopy. Again, the eYFP signal was detected in the root cap and root vascular cylinder from root meristematic zone to the entire maturation zone (Figures 1C to 1H).
Figure 1.
Localization of WRKY15-Promoter Reporter in Root Caps and Procambial Cells of the Root Vascular Cylinder.
(A) and (B) Expression pattern of WRKY15pro:eYFP-GUS reporter in 7-d-old Arabidopsis seedlings based on GUS staining. DIC images were taken using a Nikon microscope.
(C) to (K) Expression pattern of WRKY15pro:eYFP-GUS reporter from 7-d-old roots in the root tip [(C) to (E)] and the maturation zone [(F) to (K)] based on eYFP fluorescence detection. Images of eYFP and PI fluorescence were observed under a Nikon LSCM. (F), (G), and (H) are longitudinal section images through the vascular cylinder. (I), (J), and (K) are transverse section images built from Z-stack scanning of the longitudinal sections of roots. For (C), (F), and (I): PI fluorescence; (D), (G), and (J): eYFP fluorescence; (E), (H), and (K): merged images. Scale bar = 2.5 mm in (A) and 50 μm in (B) to (K).
To determine the cell types where the eYFP reporter was expressed, we performed longitudinal Z-stack scanning to generate transverse optical sections. In the root maturation zone, the eYFP signal was only detected in procambial cells (Figures 1F to 1K; Supplemental Movie 1). In the meristematic zone of the root tip, the eYFP signal was detected in several cell files within the procambial region (Supplemental Figure 2; Supplemental Movie 2). Collectively, these observations show that WRKY15 promoter activity is the highest in root procambial cells in the vascular cylinder, indicating a potential function of WRKY15 in root vascular tissue development. The broader localization detected using GUS staining is likely a result of the diffusion of the blue-colored product from the cells where the GUS enzyme was present to their neighboring cells.
Overexpression and Dominant-Negative Suppression of WRKY15 Have Opposite Effects on Root Elongation
To investigate the biological function of WRKY15, we overexpressed WRKY15 cDNA with a 4myc tag driven by the Cauliflower mosaic virus (CaMV) 35S promoter with a double enhancer. Transgenic lines with high transgene expression were identified by quantitative RT-PCR (RT-qPCR) and immunoblot analysis, and two independent lines (OE no. 8 and no. 43) were selected for further analyses (Figures 2A and 2B). For the loss-of-function study, we first used the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 approach to knock out the WRKY15 gene. Two independent deletion mutant lines were generated and examined. No growth or developmental phenotype was observed in these lines (Supplemental Figure 3), likely as a result of functional redundancy.
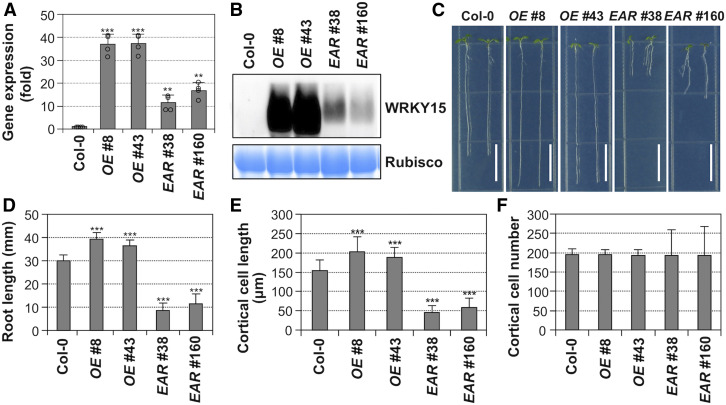
Figure 2.
Overexpression and Dominant-Negative Suppression of WRKY15 Transgenic Seedlings Have Opposite Effects on Root Elongation.
(A) Wild-type (Col-0), WRKY15 overexpression (WRKY15OE, lines no. 8 and no. 43), and WRKY15 dominant-negative suppression (WRKY15-EAR, lines no. 38 and no. 160) transgenic seedlings were cultured vertically on 1/2 MS plates under continuous light, and roots of 5-d-old seedlings (>50) were collected for total RNA preparation. Expression levels of transgenes were quantified by real-time qPCR and normalized to UBQ10. Error bars represent the standard deviations (n = 4).
(B) Total proteins from Col-0 and transgenic seedlings were extracted, levels of transgene expression were detected by immunoblot analysis using an anti-myc antibody (top), and equal loading was confirmed by Coomassie brilliant blue–stained gels (bottom).
(C) Root elongation phenotypes of WRKY15OE (lines no. 8 and no. 43) and WRKY15-EAR (lines no. 38 and no. 160) seedlings.
(D) Quantitation of primary root length of each genotype. Error bars indicate SD (n = 30).
(E) and (F) Root elongation phenotypes of WRKY15OE and WRKY15-EAR seedlings are associated with altered cell elongation (E), but not cell number (F). More than 500 mature root cortical cells from 20, 7-d-old seedlings were measured to determine the cell length and cell number per millimeter of root length. A significant difference is indicated by asterisks above the columns (One-way ANOVA, ***P < 0.001). Scale bar = 1 cm.
To circumvent this redundancy, we expressed a dominant-negative variant of WRKY15 by fusing it with the ETHYLENE RESPONSE FACTOR (ERF)-associated amphiphilic repression (EAR) motif (a repression domain) under the control of CaMV35S promoter, a method that previously allowed us to dissect successfully the function of the ERF transcription factor (Meng et al., 2013). Again, a 4myc tag was included in the N terminus of WRKY15-EAR fusion for easy protein detection. Two independent dominant-negative WRKY15-EAR transgenic lines (EAR no. 38 and no. 160) were selected for further analysis (Figures 2A and 2B).
Overexpression and dominant-negative suppression of WRKY15 transgenic seedlings showed opposite phenotypes in root elongation (Figures 2C and 2D). WRKY15 overexpression plants exhibited enhanced root growth/elongation. In contrast, the expression of dominant-negative WRKY15-EAR resulted in shorter roots. To investigate whether the altered root elongation was related to cell division and/or cell elongation rates, we assessed the cell number and cell length of the root cortex in 7-d-old seedlings. In WRKY15 overexpression transgenic seedlings, the number of cortical cells did not change, but the average cell length increased significantly (Figures 2E and 2F). By contrast, in WRKY15-EAR transgenic seedlings, cell number did not change either. However, the average cell length decreased significantly (Figures 2E and 2F). These results indicate that root elongation phenotypes of WRKY15 overexpression and dominant-negative suppression are due to altered cell elongation.
WRKY15 Negatively Regulates Root Protoxylem Vessel Differentiation and Secondary Cell Wall Thickening
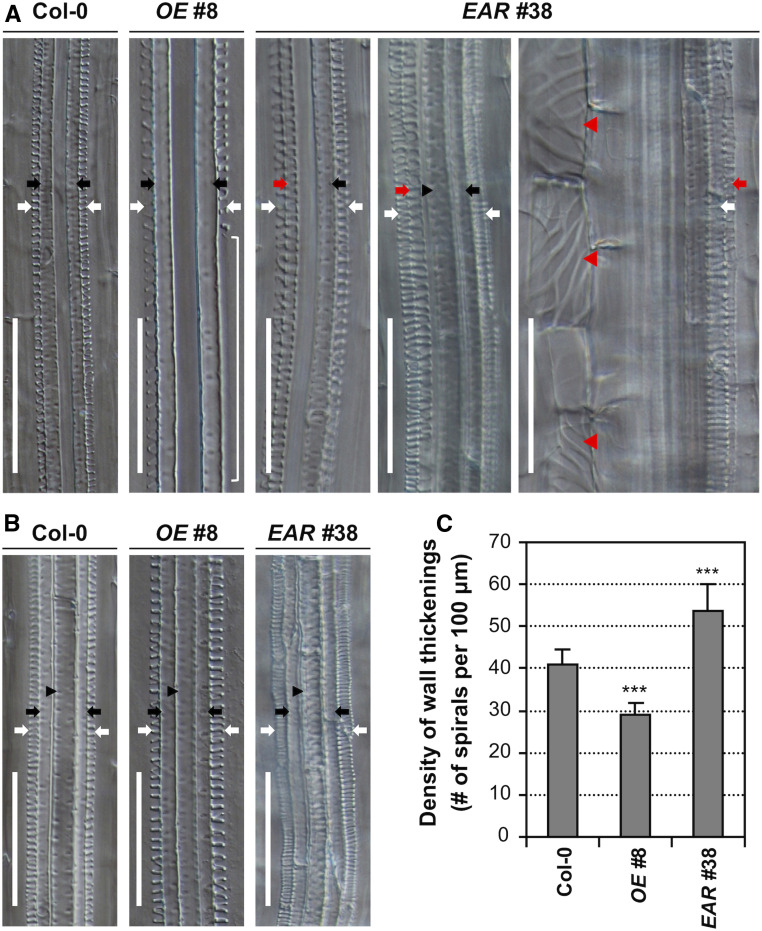
To understand the mechanism underlying the change of root elongation in WRKY15 overexpression and dominant-negative lines, we carefully examined the roots of Col-0, WRKY15OE, and WRKY15-EAR transgenic seedlings. As shown in Figure 3 and Supplemental Figure 4, the roots of WRKY15-EAR seedlings showed extra protoxylem cell files in the metaxylem positions. Moreover, the roots of WRKY15-EAR seedlings produced ectopic TE cells with spiral wall thickenings in the cortex, and these ectopic TE cells were similar to those in the protoxylem vessels but were larger in diameter and shorter in length (Figure 3A; Supplemental Figure 4A). In contrast, the roots of WRKY15OE seedlings were defective in protoxylem vessel formation, showing discontinuous protoxylem vessel files. In both WRKY15OE no. 8 and WRKY15OE no. 43 lines, ∼40% and 35% of the seedlings, respectively, displayed protoxylem gaps in roots (Supplemental Figure 5). To further characterize the root protoxylem vessels, we measured the density of spiral cell wall thickenings in protoxylem vessels. The roots of WRKY15OE seedlings exhibited looser spiral wall thickenings in the protoxylem vessels when compared with those in Col-0 roots. In contrast, the density of spiral wall thickenings in protoxylem vessels of WRKY15-EAR seedlings increased significantly (Figures 3B and 3C; Supplemental Figures 4B and 4C). These results suggest that WRKY15 plays a pivotal role in negatively controlling protoxylem vessel differentiation.
Figure 3.
WRKY15 Is Involved in Root Vascular Xylem Differentiation and Secondary Cell Wall Thickening.
(A) DIC images of longitudinal sections of roots from 7-d-old Col-0, WRKY15OE (line no. 8), and WRKY15-EAR (line no. 38) seedlings. Discontinuous protoxylem vessel strand can be observed in the roots of WRKY15OE seedlings (indicated by a white bracket). In the dominant-negative suppression WRKY15-EAR seedlings, formation of extra protoxylem vessels with spiral wall thickenings in the metaxylem position (red arrow) and ectopic formation of TEs in cortical cells (red arrowhead) are present.
(B) Decreased spiral wall thickenings in protoxylem vessels of WRKY15OE seedlings and increased spiral wall thickenings in protoxylem vessels of WRKY15-EAR seedlings.
(C) The density of spiral cell wall thickenings in protoxylem vessels of wild-type (Col-0), WRKY15OE, and WRKY15-EAR seedlings. A significant difference is indicated by asterisks above the columns (One-way ANOVA, ***P < 0.001). White arrows indicate protoxylem vessels. Black arrows and black arrowheads indicate primary and central metaxylem vessels, respectively. Red arrows and red arrowheads indicate ectopic protoxylem-like vessels and TE cells, respectively. Scale bar = 50 μm.
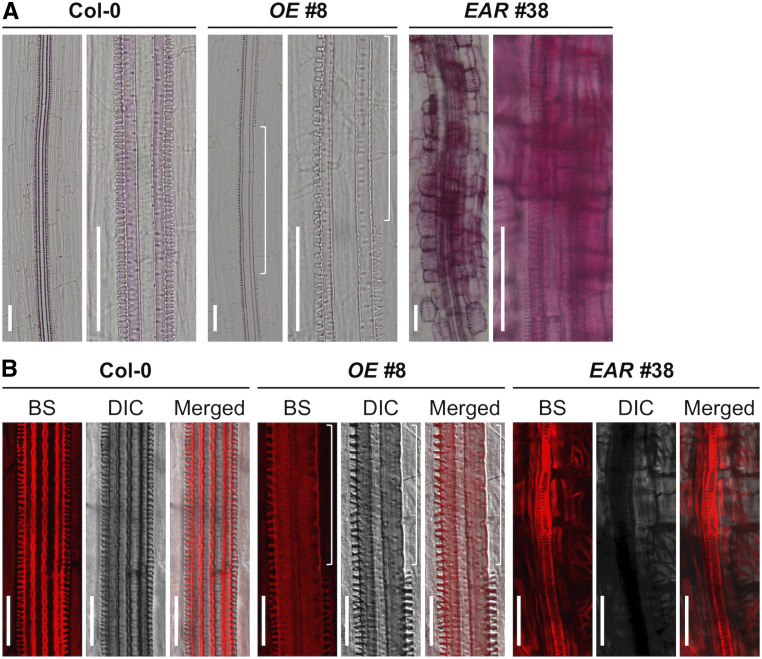
The formation of TEs is characterized by a series of events including cell expansion, patterned deposition of secondary cell walls, lignification, and PCD (Fukuda, 1997; Turner et al., 2007; Furuta et al., 2014). To further confirm the identity of the ectopic TE cells, we examined the lignin localization and deposition using two different histochemical staining methods, phloroglucinol-HCl staining and basic fuchsin staining. Consistent with the negative regulation of WRKY15 in cell wall lignification, both staining methods revealed an enhanced lignin deposition on the xylem vessels and ectopic TE cells of 7-d-old WRKY15-EAR roots (Figure 4; Supplemental Figure 6). In addition, ectopic lignin deposition was observed in epidermal and cortical cells that do not normally undergo lignification. Moreover, the lignin deposition on the xylem vessels was weakened and had gaps in the discontinuous protoxylem vessels of WRKY15OE roots (Figure 4; Supplemental Figure 6). Together, these findings demonstrate that WRKY15 plays an important role in the differentiation of TEs and secondary cell wall lignification/thickening. The ectopic lignin accumulation in epidermal and cortical cells of WRKY15-EAR seedlings was also observed (Figure 4; Supplemental Figure 6), which could inhibit cell wall expansion and contribute to the reduction of cell expansion/elongation and root length (Figure 2).
Figure 4.
Altered Lignin Deposition in the Roots of WRKY15OE and WRKY15-EAR Transgenic Seedlings.
(A) Lignin accumulation in 7-d-old Col-0, WRKY15OE (line no. 8), and WRKY15-EAR (line no. 38) roots was detected by phloroglucinol-HCl staining. Images were taken using a Nikon microscope.
(B) Basic fuchsin (BS) staining of lignin in the roots of 7-d-old Col-0, WRKY15OE (line no. 8), and WRKY15-EAR (line no. 38) seedlings. DIC and fluorescence images were photographed using a Nikon C2 Plus LSCM. The white long bracket represents a gap in protoxylem vessels. Scale bar = 50 μm.
In addition, the formation of TEs in WRKY15-EAR seedlings was early, starting in the meristematic/elongation zone before the completion of cell elongation, which may also cause the reduced cell expansion/elongation and root length. In wild-type roots, xylem vessel cell differentiation starts after cell elongation. In WRKY15OE seedlings, the reduced lignification and delayed formation of protoxylem (hence the formation of protoxylem gaps) are likely to allow the cells to elongate more, resulting in longer roots (Figure 2).
Despite the longer root phenotype of WRKY15OE seedlings on plates, WRKY15OE plants had shorter roots and were dwarf when grown in soil, possibly a result of defective transportation of water and nutrients due to the discontinuous protoxylem vessel files. In contrast, seedlings grown on vertically plates do not have these limitations because the seedlings are in contact with agar medium and can absorb water and nutrients readily throughout the body of the seedlings.
WRKY15 Is Important for the Proper Differentiation of TEs
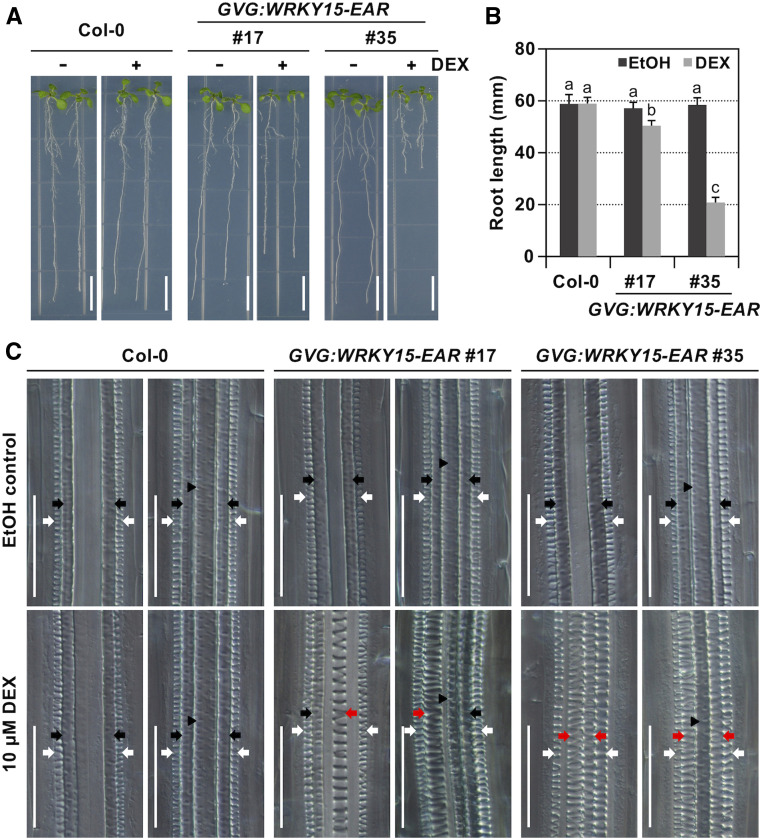
To confirm the role of WRKY15 in TE differentiation and secondary cell wall lignification, we also expressed the dominant-negative version of WRKY15 protein driven by dexamethasone (DEX)-inducible promoter and native WRKY15 promoter. In the DEX-inducible system, levels of transgene expression were determined using immunoblot analysis, and two independent transgenic lines (GVG:WRKY15-EAR no. 17 and no. 35) were selected for phenotypic characterizations (Supplemental Figure 7A). Seeds of GVG:WRKY15-EAR (lines no. 17 and no. 35) were germinated on vertically placed half-strength Murashige and Skoog (MS) plates. Five d later, seedlings were transferred onto plates supplemented with 10 μM DEX (+DEX) or ethanol solvent control (−DEX). Seedlings of 10 d old were collected for observation. Neither line showed an abnormal phenotype in the absence of DEX (Figure 5). However, in the presence of DEX, GVG:WRKY15-EAR transgenic seedlings exhibited inhibition of root elongation (Figures 5A and 5B).
Figure 5.
Conditional Expression of WRKY15-EAR Transgene Leads to Ectopic Differentiation of Protoxylem Vessel Files in the Metaxylem Positions.
(A) and (B) The induction of WRKY15-EAR expression inhibits root elongation. Col-0 and DEX-inducible promoter-driven WRKY15-EAR (GVG:WRKY15-EAR, lines no. 17 and no. 35) seeds were germinated on vertically placed 1/2 MS plates. Then 5 d later, seedlings were transferred onto plates supplemented with 10 μM DEX (+DEX) or ethanol solvent control (−DEX). Images were taken when seedlings were 10 d old (A), and primary root length was quantified (B). Error bars indicate SD (n = 25). Significant differences are indicated by different lowercase letters above the columns (One-way ANOVA, P < 0.01).
(C) Induction of WRKY15-EAR expression promotes protoxylem vessel development. DIC images of roots were obtained after clearing 10-d-old Col-0 and GVG:WRKY15-EAR (lines no. 17 and no. 35) seedlings. Images with roots formed two and three metaxylem vessels are displayed. White arrows indicate protoxylem vessels. Black arrows and black arrowheads indicate primary and central metaxylem vessels, respectively. Red arrows indicate ectopic differentiation of protoxylem files in the metaxylem positions. Scale bar = 1 cm in (A) and 50 μm in (C).
Roots of Col-0 and solvent-treated GVG:WRKY15-EAR no. 17 and no. 35 control seedlings developed the typical two files of protoxylem vessels at the outermost position of the vascular tissues with three to four files of metaxylem vessels in the center (Figure 5C). In DEX-treated GVG:WRKY15-EAR no. 17 and no. 35 seedlings, we did not observe any difference in the root region close to the hypocotyl, which developed before the exposure to DEX (d 0 to 5). In contrast, in the newly developed root region after the exposure to DEX (d 6 to 10), protoxylem vessel files formed in the metaxylem positions, resulting in more than two files of protoxylem vessels in the newly grown region close to the root tip (Figure 5C). Next, we performed lignin staining using phloroglucinol-HCl. Induction of WRKY15-EAR expression led to ectopic lignin deposition in roots of GVG:WRKY15-EAR (Supplemental Figure 7B), similar to that observed with constitutive overexpression of WRKY15-EAR (Figure 4A; Supplemental Figure 6A). In the native WRKY15 promoter-driven WRKY15-EAR transgenic plants, 9.7% (n = 62) of T1 seedlings generated an extra protoxylem strand in the root vascular tissues, and there was no other obvious defect in the roots (Supplemental Figure 8). The weak phenotype of the WRKY15pro:WRKY15-EAR plants is likely a result of relatively weak expression under the control of native WRKY15 promoter. These findings further confirmed the function of WRKY15 in protoxylem vessel formation and secondary cell wall lignification.
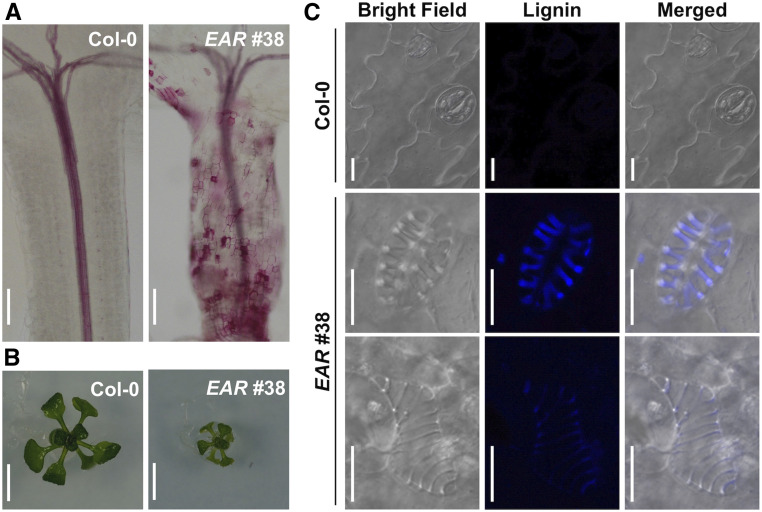
Expression of WRKY15-EAR led to severely dwarf seedlings, which was associated with ectopic TE cell formation in the epidermal layers of the hypocotyls and leaves (Figure 6; Supplemental Figure 9). The transdifferentiation of epidermal cells into protoxylem-like vessel elements was accompanied by ectopic lignin deposition, as detected by histological staining using phloroglucinol-HCl and lignin autofluorescence under UV light illumination (Figures 6A and 6C; Supplemental Figures 9A and 9C). Although the cell shape of the guard cells and pavement cells were maintained, they went through the cell wall lignification process and formed spiral cell wall thickenings. It was also observed that the cellular contents including chloroplasts were completed cleared from the ectopic guard cell-shaped TE cells (Figure 6C; Supplemental Figure 9C), suggesting the completion of autolysis/PCD as in normal TE differentiation. Conversion of only one of the two guard cells of a stoma into an ectopic TE cell (Supplemental Figure 9C) also indicates that the transdifferentiation of guard cells to ectopic TE cells could occur after stomata formation.
Figure 6.
Ectopic Differentiation of TEs in WRKY15-EAR Transgenic Plants.
(A) Hypocotyls of 7-d-old Col-0 and WRKY15-EAR no. 38 seedlings were stained by phloroglucinol-HCl. Lignin was ectopically deposited in WRKY15-EAR no. 38 hypocotyls.
(B) Images of 14-d-old Col-0 and WRKY15-EAR no. 38 seedlings. Seedlings of WRKY15-EAR no. 38 were severely dwarf.
(C) LSCM images of leaf epidermis in 14-d-old Col-0 and WRKY15-EAR no. 38 seedlings. Epidermal cells including guard cells in 14-d-old WRKY15-EAR no. 38 seedlings transdifferentiated into ectopic TE cells. Lignin autofluorescence (Lignin) were visualized under UV light illumination. Scale bar = 100 μm in (A); 500 μm in (B); 10 μm in (C).
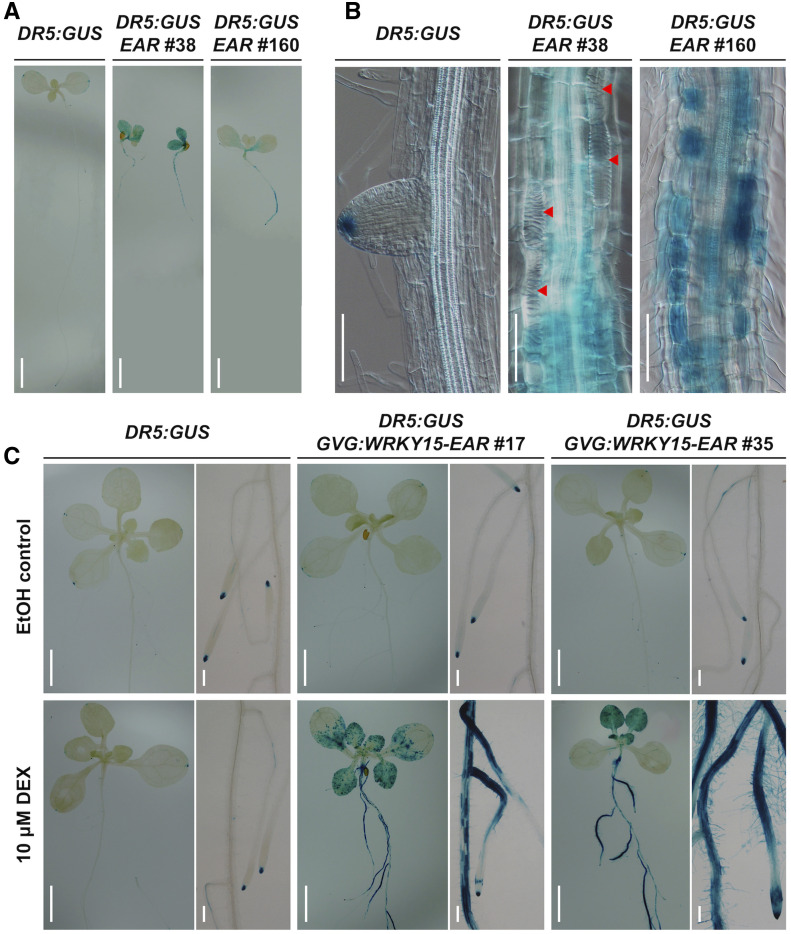
Auxin is known to play an important role in vascular development and TE formation (Kubo et al., 2005; Ursache et al., 2014; Smet and De Rybel, 2016; Cho et al., 2017). To investigate whether ectopic TE formation in WRKY15-EAR plants is associated with altered auxin activity, we introduced DR5:GUS into the WRKY15-EAR background and examined the DR5:GUS reporter activity. In both the constitutive and the inducible expression of WRKY15-EAR seedlings, DR5:GUS ectopically accumulated in roots, hypocotyls, and leaves (Figure 7). The ectopic and enhanced DR5:GUS reporter expression pattern in WRKY15-EAR seedlings was consistent with the pattern of TE formation (Figure 7B). Although DR5:GUS reporter activity was high in the immature ectopic TE cells that were undergoing differentiation, its activity decreased or disappeared completely in mature ectopic TE cells (Figure 7B). These results support a correlative relationship between the elevated auxin activity and the ectopic TE formation in WRKY15-EAR plants.
Figure 7.
WRKY15-EAR Transgenic Plants Enhance Auxin Reporter Activity.
(A) and (B) Ectopic and enhanced DR5:GUS reporter expression in WRKY15-EAR background. GUS staining of DR5:GUS reporter in 7-d-old seedlings of Col-0 and WRKY15-EAR (EAR, lines no. 38 and no. 160) backgrounds is shown. Red arrowheads indicate ectopic TE cells.
(C) Induction of WRKY15-EAR expression promotes DR5:GUS reporter activity. Seeds of DR5:GUS reporter in Col-0 and GVG:WRKY15-EAR (lines no. 17 and no. 35) backgrounds were vertically cultured for 5 d. Seedlings were then treated with 10 μM DEX or ethanol solvent control for 5 d before GUS staining. Scale bar = 250 μm in (A) and (C); 100 μm in (B).
Effects of WRKY15 on the Expression of Genes Involved in TE Differentiation
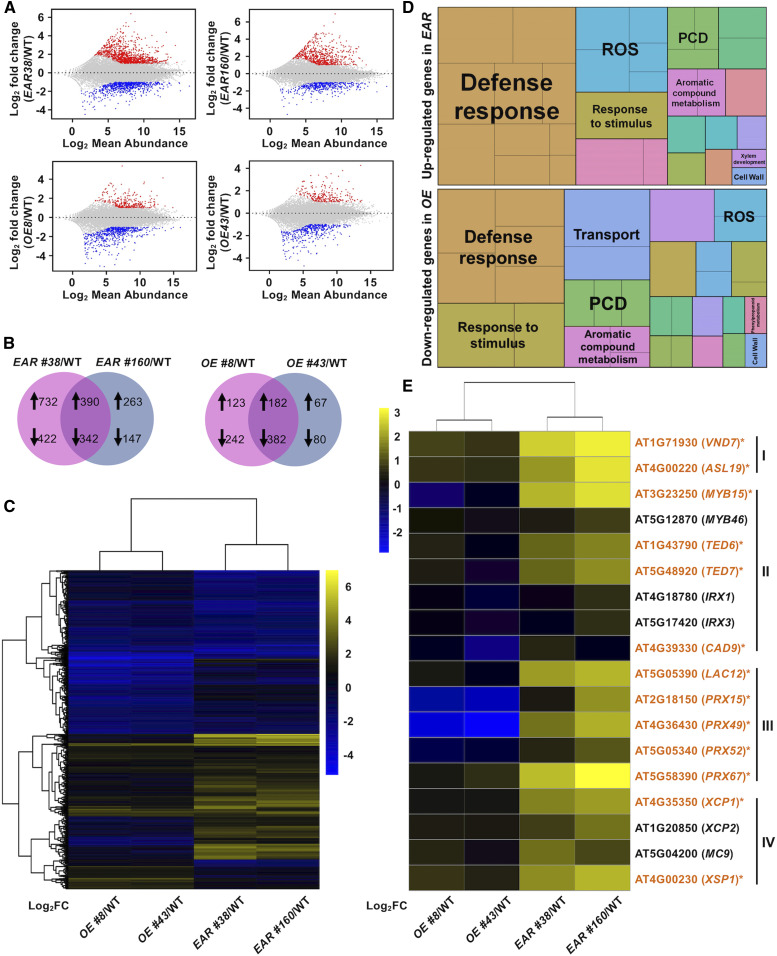
To identify the downstream target genes of WRKY15, we profiled gene expression in the roots of WRKY15OE and WRKY15-EAR seedlings (Supplemental Figure 10; Supplemental Table 1). In the roots from WRKY15-EAR no. 38 and no. 160, the expression of a common set of 390 genes was increased relative to wild type, and 342 genes reduced [padj < 0.05; |log2 fold change| > 1] (Figures 8A and 8B; Supplemental Data Set 2). In roots from WRKY15OE no. 8 and no. 43, the expression of a common set of 182 genes was increased relative to wild type, and 382 genes reduced [padj < 0.05; |log2 fold change| > 1] (Figures 8A and 8B; Supplemental Data Set 2). The logarithm to base 2 of the differentially expressed genes (DEGs) was shown as a heatmap in Figure 8C. Gene ontology enrichment analysis showed that the 390 genes significantly upregulated in two WRKY15-EAR transgenic lines were mainly associated with xylem differentiation such as lignin formation and PCD process, defense response, and response to stimulus (Figure 8D; Supplemental Data Sets 3 and 4). Consistent with the phenotype in WRKY15OE lines, the 382 downregulated genes in the two independent WRKY15OE transgenic lines relative to wild type were also associated with the above-mentioned processes (Figure 8D; Supplemental Data Sets 3 and 4). These results are consistent with the opposite phenotypes in WRKY15OE and WRKY15-EAR seedlings, and further indicate that WRKY15 regulates TE differentiation by modulating a wide range of genes.
Figure 8.
Genes Involved in Xylem Differentiation, including Lignin Formation and PCD Process, Are Significantly Enriched in WRKY15-EAR Transgenic Roots.
(A) The minus-average plot shows DEGs in four transgenic lines relative to wild type (WT). Mean values of the two independent experiments are shown. Genes that are significantly upregulated and downregulated are in red and blue, respectively (Cutoff values: padj < 0.05 and |log2 fold change| > 1). See also Supplemental Figure 10 and Supplemental Table 1.
(B) Venn diagrams illustrate common sets of DEGs. DEGs both in WRKY15EAR no. 38 and WRKY15-EAR no. 160 relative to Col-0 are designated EAR, and DEGs both in WRKY15OE no. 8 and WRKY15OE no. 43 relative to Col-0 are designated OE. Cutoff values: padj < 0.05 and |log2 fold change| > 1. Up arrows and down arrows indicate significantly upregulated and downregulated genes, respectively. See also Supplemental Data Set 2.
(C) Heatmap of expression of DEGs in EAR or OE. FC, fold change.
(D) Gene ontology analysis of DEGs both upregulated in WRKY15-EAR no. 38 and WRKY15EAR no. 160 relative to Col-0 (top), and DEGs both downregulated in WRKY15OE no. 8 and WRKY15OE no. 43 relative to Col-0 (bottom) using REVIGO. See also Supplemental Data Sets 3 and 4. ROS, reactive oxygen species.
(E) Heatmap showing gene expression levels in four transgenic lines relative to wild type based on RT-qPCR results (Supplemental Figure 11). Gene IDs highlighted with asterisks indicate those that are also identified as DEGs in expression profiling (A).
We selected 18 genes related to TE formation and re-examined their expression using RT-qPCR. All 18 genes were confirmed to be DEGs (Figure 8E; Supplemental Figure 11). This set of genes includes key regulators of TE differentiation (VND7 and ASL19/LBD30/JLO; Kubo et al., 2005; Soyano et al., 2008; Yamaguchi et al., 2010b), secondary cell wall-related genes [MYB15, MYB46, TED6, TED7, IRREGULAR XYLEM1 (IRX1)/CESA8, IRX3/CESA7, and CAD9; Zhong et al., 2007, 2010; Endo et al., 2009; Yamaguchi et al., 2011], genes encoding laccase and peroxidase responsible for lignin polymerization (LAC12, PRX15, PRX49, PRX52, and PRX62; Zhao et al., 2013), and genes involved in TE postmortem autolysis (XCP1, XCP2, MC9, and XSP1; Zhao et al., 2000; Funk et al., 2002; Avci et al., 2008; Bollhöner et al., 2013). Taking these results together, we can conclude that WRKY15 is likely to regulate a critical gene set required for TE formation.
Ectopic TE Formation in WRKY15-EAR Plants Is Associated with Ectopic Activation of VND7 Expression
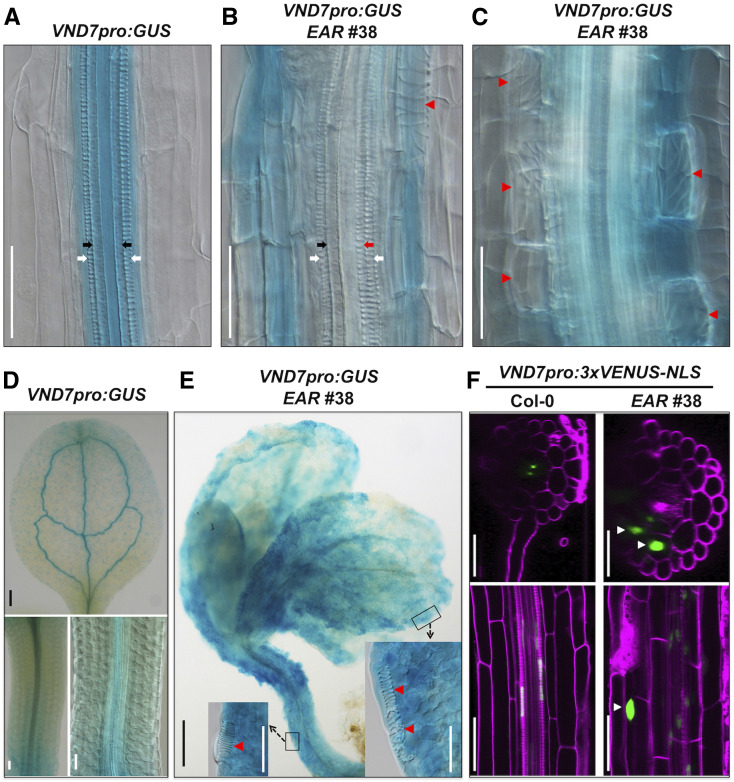
Expression of the dominant-negative WRKY15-EAR leads to the formation of ectopic TEs from nonvascular tissues, similar to the overexpression of VND7 (Kubo et al., 2005; Yamaguchi et al., 2010b; Endo et al., 2015). To further investigate the relationship between WRKY15 and VND7, we generated a VND7pro:eYFP-GUS reporter construct. After stable transgenic lines with single insertions were obtained, they were crossed into the WRKY15-EAR and WRKY15OE backgrounds. Histochemical GUS staining showed enhanced VND7 reporter activity and ectopic expression of VND7 reporter in nonvascular tissues of WRKY15-EAR plants while the expression pattern of VND7 in WRKY15OE plants was similar to that in Col-0 plants (Figures 9A to 9E; Supplemental Figures 12A and 12B). These results are consistent with the expression levels of VND7 in their corresponding backgrounds based on RNA sequencing (RNA-seq) and RT-qPCR analyses (Figure 8E; Supplemental Figure 11; Supplemental Data Set 2 ). Furthermore, reporter analysis revealed a more detailed spatiotemporal expression pattern of VND7. As shown in Figures 9B, 9C, and 9E and Supplemental Figure 12A, the expression of VND7pro:eYFP-GUS reporter decreased or disappeared in mature ectopic TE cells, likely a result of PCD associated with TE formation. In contrast, higher reporter activity was observed in the immature ectopic TE cells that were undergoing transdifferentiation. This is similar to the results of DR5:GUS reporter in WRKY15-EAR plants (Figure 7).
Figure 9.
Ectopic TE Formation in WRKY15-EAR Plants Is Associated with Ectopic Activation of VND7 Expression.
(A) to (C) Enhanced VND7pro:eYFP-GUS reporter expression in WRKY15-EAR background. GUS staining of VND7pro:eYFP-GUS reporter in Col-0 (A) and WRKY15-EAR no. 38 [(B) and (C)] backgrounds. White and black arrows indicate protoxylem vessels and metaxylem vessels, respectively. Red arrows and red arrowheads indicate ectopic protoxylem-like vessels and TE cells, respectively.
(D) and (E) Ectopic expression of VND7pro:eYFP-GUS reporter in WRKY15-EAR background. GUS staining of leaves and hypocotyls of VND7pro:eYFP-GUS transgenic plants in Col-0 (D) and WRKY15-EAR no. 38 (E) backgrounds. Insets are magnified regions in (E) to show the transdifferentiation of leaf and hypocotyl epidermal cells to ectopic TEs (indicated by red arrowheads).
(F) Ectopic VND7 expression in cortical cells in WRKY15-EAR background. LSCM images of 5-d-old VND7pro:3×VENUS-NLS seedlings in Col-0 (left) and WRKY15-EAR no. 38 (right) backgrounds. VENUS fluorescence (green) and PI fluorescence (magenta) are merged. At top, transverse optical sections through the root maturation zone are represented. At bottom, longitudinal optical sections through the root maturation zone are represented. Scale bar = 50 μm in white color, 250 μm in black color.
To further improve the resolution, we used a reporter that localized to the nucleus to avoid the diffusion of reporter protein to the neighboring cells. To this end, we generated VND7pro:3×VENUS-NLS reporter transgenic plants and crossed them into WRKY15-EAR plants. In the wild-type background, VND7 was predominantly expressed in immature TEs in the root maturation zone based on transverse and longitudinal optical sections using confocal microscopy (Figure 9F; Supplemental Figure 12C). In WRKY15-EAR plants, expression of VND7 was detected not only in immature TEs but also in the root cortex, where ectopic TE cells formed (Figure 9F; Supplemental Figure 12D). Secondary cell wall formation and PCD are two pivotal processes in TE maturation (Bollhöner et al., 2012; Escamez and Tuominen, 2014). We were able to capture root cortical cells during ectopic TE cell transdifferentiation and maturation, including the final autolysis of the cell contents (Supplemental Figure 13; Supplemental Movies 3 and 4). In the mature ectopic TE cells, we did not observe any nucleus-localized VENUS fluorescence in the Z-stack images/movies (Figure 9F; Supplemental Figures 11D and 12C; Supplemental Movies 3 and 4), suggesting that the nuclei in these cells have disintegrated and the cells have gone through the final autolysis process. These results demonstrate that the ectopic TE formation in WRKY15-EAR plants has all the hallmarks of normal TE formation, as well as the ectopic activation of VND7 expression. It is well established that high-level VND7 expression is sufficient for TE formation (Kubo et al., 2005; Yamaguchi et al., 2008, 2010b; Endo et al., 2015).
DISCUSSION
Xylem vessels are responsible for long-distance transportation of water and mineral nutrients in land plants, a process critical to plant survival. In this study, we demonstrate that WRKY15 is an important regulator of TE differentiation. The integration of WRKY15, a negative regulator, into the known VND7 regulatory network provides additional insights into the regulation of xylem vessel differentiation. WRKY15 is expressed mainly in root procambial cells (Figure 1; Supplemental Figure 2), suggesting a potential function in vascular differentiation. Similar to the overexpression of VND7 (Kubo et al., 2005; Yamaguchi et al., 2008, 2010b; Endo et al., 2015), expression of the dominant-negative WRKY15-EAR results in the transdifferentiation of nonvascular cells such as the root cortex and hypocotyl/leaf epidermis, including pavement cells and guard cells, into ectopic TE cells without disturbing cell division or changing cell shape (Figures 3, 4, 6, and 7; Supplemental Figures 4, 6, and 9). By contrast, in WRKY15-overexpressing transgenic seedlings, protoxylem vessel files become discontinuous (Figures 3 and 4; Supplemental Figures 4 and 6), similar to the overexpression of VND7-EAR (Kubo et al., 2005). In addition, WRKY15 also influences secondary cell wall formation/lignification (Figures 4 and 6; Supplemental Figures 6, 7, and 9). Based on these findings, we conclude that WRKY15 is an important regulator that negatively controls xylem vessel cell formation, which is in contrast with VDN7, a master regulator that positively influences xylem vessel cell fate.
Although we could not use live-cell imaging to follow the transdifferentiation process, we concluded that the ectopic TEs have previously acquired their initial cell identities such as epidermal cells, guard cells, and cortical cells, and later been converted to TEs in the WRKY15-EAR plants, similar to plants overexpressing VND7. These ectopic TEs have the morphology and anatomical location of their initial cell identities. For instance, ectopic TEs transdifferentiated from pavement cells still have the typical jigsaw puzzle piece shapes (Figure 6C). The ones from guard cells still maintain the kidney shape (Figure 6C; Supplemental Figure 9C). We also observed that sometimes only one of the two guard cells was converted to an ectopic TE cell (Supplemental Figure 9C), a stronger indication that they had already assumed guard cell identity because pairs of guard cells are always formed at the same time. Ectopic TEs from cortical cells all have the morphology of normal cortical cells and are located underneath the epidermal cell layer and outside of the vascular cylinder (Figures 3A and 4A; Supplemental Figures 4A and 6A). Their identity as ectopic TEs is based on the following evidence. First, they have the spiral cell wall thickening and lignification as the normal TEs in the vascular cylinder. Second, they express typical TE marker genes including VND7. Third, they have cleared all cellular contents, based on the lack of chloroplasts and nucleus. As shown in Figure 6C and Supplemental Figure 9C, normal guard cells contain chloroplasts that are easily visible. However, the chloroplasts are absent in the kidney-shaped ectopic TEs. When only one of the two guard cells is converted to the TE cell, chloroplasts are visible only in the remaining normal guard cell. Furthermore, when we examined the nucleus-localized Venus-NLS reporter (driven by the VND7 promoter) using Z-stack confocal imaging, no fluorescence could be observed in the mature ectopic TEs (Figure 9F; Supplemental Figures 12C and 13D; Supplemental Movies 3 and 4). Before the maturation of ectopic TEs, the nuclear localized Venus-NLS fluorescent protein first became cytosolically localized, an indication that the nuclear membrane has lost or partially lost its integrity. At this stage, we could still observe the large central vacuole where there was no fluorescence inside, an indication that the vacuolar membrane was still intact. Later, the fluorescence became weaker and diffuse, and filled up the whole cell, suggesting the disintegration of the nucleus and central vacuole. Finally, no fluorescence could be observed in the mature TEs, indicating the clearing of all cellular contents including the Venus-NLS fluorescent protein (Supplemental Figure 13). These findings suggest that the ectopic TEs have undergone a complete PCD process and cleared their cellular contents, similar to the normal TE differentiation.
In the wild-type background, we observed that the VND7pro:eYFP-GUS reporter is expressed in immature and mature TEs of roots, but the VND7pro:3×VENUS-NLS reporter is expressed in immature TEs of roots only (Figures 9A and 9F; Supplemental Figure 12C). The different expression patterns of VND7pro:eYFP-GUS reporter and VND7pro:3×VENUS-NLS reporter that we observed are consistent with previous reports (Kubo et al., 2005; Yamaguchi et al., 2008, 2010a). Mature TEs undergo the final autolysis process to clear the cellular contents including the fluorescent reporter proteins. The nuclear localization of VENUS-NLS produced from the VND7pro:3×VENUS-NLS reporter prevents the diffusion of the reporter protein into other mature TEs, making it a more precise reporter than VND7pro:eYFP-GUS. By crossing the VND7pro:3×VENUS-NLS reporter into the WRKY15-EAR transgenic background, we have found that VND7 ectopically overexpresses in WRKY15-EAR background (Figure 9; Supplemental Figures 12 and 13; Supplemental Movie 4), consistent with the ectopic TE formation. Furthermore, the disappearance of the nuclear-localized reporter fluorescent protein is associated with the maturation of ectopic TEs, which demonstrates the completion of the final autolysis of the cell contents in ectopic TEs in WRKY15-EAR plants (Figure 9; Supplemental Figures 12 and 13; Supplemental Movie 4). This event is also in concert with gene ontology enrichment analysis (Figure 8D; Supplemental Data Sets 3 and 4). We found that the upregulated genes in WRKY15-EAR transgenic lines relative to wild type are mainly associated with xylem differentiation (Figure 8D; Supplemental Data Sets 3 and 4). Moreover, the downregulated genes in WRKY15OE transgenic lines relative to wild type are mainly associated with TE formation including lignin formation and PCD process (Figure 8D; Supplemental Data Sets 3 and 4), illustrating the opposite effects of WRKY15 overexpression and negative suppression on TE formation at the gene expression level. These findings allow us to conclude that WRKY15 negatively regulates TE differentiation through modulating VND7 and other genes required for TE formation, and ectopic TE formation in WRKY15-EAR plants is associated with ectopic activation of VND7 expression.
Transcriptional upregulation of VND7 is important for its function as a master regulator of TE differentiation. However, the upstream regulator(s) are unknown. The identification and functional characterization of WRKY15 as an important regulator upstream of VND7 in TE formation shed some light on this important question. The negative regulatory role of WRKY15 and positive role of VND7 could, together, control the formation of protoxylem vessels in roots. Overexpression of WRKY15 also promotes root elongation by enhancing cell expansion (Figure 2), a process not associated with VND7 function, suggesting that WRKY15 may have additional downstream components in plant growth and development. Previously, WRKY15 was reported to function as a negative regulator of salt/osmotic stress tolerance and a positive regulator of growth. Its overexpression results in hypersensitivity to salt and osmotic stress, as well as enhanced growth in Arabidopsis (Vanderauwera et al., 2012). However, the underlying mechanisms were not clear.
Dominant-negative suppression of VND7 or VNI2 provided the loss-of-function evidence to support their roles in TE formation (Kubo et al., 2005; Yamaguchi et al., 2010a). Using the same approach, we demonstrated that the expression of WRKY15-EAR, a dominant-negative form of WRKY15, leads to extra protoxylem files and ectopic TE formation, suggesting that WRKY15 is a negative regulator of TE formation. In addition, conditional expression of WRKY15-EAR under an inducible promoter, which can avoid nonspecific secondary effects sometimes associated with constitutive overexpression, leads to the same phenotype, further supporting the negative regulatory role of WRKY15 in TE formation. However, single wrky15 mutant lines generated using the CRISPR-Cas9 approach had no difference in TE formation (Supplemental Figure 3), likely a result of functional redundancy. There are several close homologs of WRKY15 in the WRKY IId subfamily (Supplemental Figure 1A). Similarly, vnd7 mutants do not have defective TE formation, and overexpression of VND7 and VND7-EAR were used to acquire gain- and loss-of functional data to establish the function of VND7 in TE differentiation (Kubo et al., 2005; Yamaguchi et al., 2010a). Supporting the negative role of WRKY15 in TE formation, we found that overexpression of WRKY15 suppresses protoxylem vessel formation, leading to the formation of protoxylem gaps (Supplemental Figure 5).
VND7 is a positive master regulator of protoxylem vessel differentiation (Kubo et al., 2005; Yamaguchi et al., 2010b). We identified VND7 as a downstream gene of WRKY15 based on expression profiling and RT-qPCR analyses (Figure 8E; Supplemental Figure 11; Supplemental Data Set 2). GUS and eYFP reporter analyses of the WRKY15 promoter further affirmed this conclusion. WRKY15 is mainly expressed in procambial cells in the vascular cylinder starting from the meristematic zone of the root tip (Figure 1; Supplemental Figure 2; Supplemental Movies 1 and 2). Previous analysis of VND7 promoter-driven GUS/YFP reporter indicated its expression at the protoxylem poles of root procambium and differentiating vessels (Yamaguchi et al., 2008). Based on these two localization studies, WRKY15 is expressed in procambial cells surrounding the cells where VND7 is expressed (protoxylem poles of root procambium and differentiating vessels). Besides, the expression of WRKY15 can be detected in procambial cells in the meristematic zone, preceding the expression of VND7 during root development. This would be consistent with the function of WRKY15 being upstream of VND7 in regulating the formation of protoxylem. We propose that WRKY15 negatively regulates VND7 expression indirectly based on (1) the absence of a W-box in the promoter of VND7 and (2) the observation that WRKY15 and VND7 are expressed in different cells that are in close proximity in the vascular cylinder. There are precedents that a transcription factor can move to neighboring cells and suppress gene expression there. For instance, WUSCHEL RELATED HOMEOBOX 5 (WOX5), which is expressed in the root quiescent center (QC), can move to neighboring columella stem cells to recruits TPL/TPR corepressors and the histone deacetylase HDA19 to silence the expression of CYCLING DOF FACTOR 4 (CDF4), preventing the differentiation of columella stem cells (Pi et al., 2015). Consistent with this model, a previous study suggested that WRKY15 might function as a transcriptional repressor (Vanderauwera et al., 2012). Future research is needed to reveal the details underlying the interaction of WRKY15 and VND7 during plant vascular development.
METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Columbia (Col-0) ecotype was used as the wild type. For seedlings vertically grown in solid medium, seeds were surface sterilized. After stratification at 4°C for 3 to 5 d, seeds were sown in Petri dishes with solidified half-strength MS medium (0.8% [w/v] Phytoagar) and vertically cultured in a growth chamber at 22°C with continuous light (70 μmol m−2 s−1). The 5- or 7-d-old seedlings were used to do different experiments as stated in the figure legends. For conditional expression of WRKY15-EAR transgene, 5- or 9-d-old seedlings were transferred onto plates supplemented with 10 μM DEX or ethanol solvent control, and continued to grow under the same conditions for indicated days or hours. For transgenic selection, seeds were sown on solidified half-strength MS medium (0.45% [w/v] Phytoagar) containing corresponding antibiotics (glufosinate-ammonium, kanamycin, or hygromycin).
Generation of Transgenic Lines
For the generation of WRKY15pro:eYFP-GUS reporter construct, WRKY15 promoter (2-kb fragment upstream of the ATG start codon) and eYFP-GUS fragment were amplified by PCR using primers listed in Supplemental Table 2, and pCAMBIA3300 (pC3300) vector was linearized by Hind III and Sac I restriction digestion. Finally, the WRKY15 promoter and eYFP-GUS fragment were assembled into the linearized pC3300 vector using GBclonart Seamless Assembly Kit (Genebank Biosciences Inc) to generate pC3300-WRKY15pro:eYFP-GUS construct.
For the generation of pBId-Ω-4myc-WRKY15 construct, WRKY15 cDNA was amplified by PCR and inserted into the pBlueScript II KS (pBS) vector to generate pBS-WRKY15 cDNA construct. WRKY15 cDNA fragment was then cloned into pBS-Omega-4myc-NdeI vector using Nde I and EcoR I sites to generate the intermediate pBS-Ω-4myc-WRKY15 construct. The Ω-4myc-WRKY15 fragment was then cut out and inserted into the pBId vector using Xho I and Spe I sites to generate the final pBId-Ω-4myc-WRKY15 construct.
For the generation of pBId-Ω-4myc-WRKY15-EAR and DEX-inducible promoter-driven WRKY15-EAR transgenic plants, pBS-Ω-4myc-WRKY15 construct was used as template, and primers EAR-SpeI-pBS vector primer-L and WRKY15-pBS-R (Supplemental Table 2) were used to amplify and add the EAR sequence to generate pBS-Ω-4myc-WRKY15-EAR construct. The Ω-4myc-WRKY15-EAR fragment was inserted into the pBId vector and pTA7002 vector (with DEX-inducible promoter) using Xho I and Spe I sites to generate pBId-Ω-4myc-WRKY15-EAR construct and pTA7002-Ω-4myc-WRKY15-EAR construct, respectively.
For the generation of WRKY15pro:WRKY15-EAR transgenic plants, WRKY15 promoter (2-kb fragment upstream of the ATG start codon) was amplified by primers WRKY15-promoter-L2 and WRKY15-promoter-R2 (Supplemental Table 2), and cloned into pBS vector to generate pBS-WRKY15 promoter construct. Then the 35S promoter in pBId-Ω-4myc-WRKY15-EAR construct was replaced with the WRKY15 promoter cut out with Hind III and Xho I to generate the WRKY15pro:WRKY15-EAR construct.
For the generation of VND7pro:eYFP-GUS reporter construct, VND7 promoter (2-kb fragment upstream of the ATG start codon) was amplified using primers VND7-promoter-L1 and VND7-promoter-R (Supplemental Table 2), and inserted into pBS vector to generate pBS-VND7 promoter1 construct. The WRKY15 promoter in pC3300-WRKY15pro:eYFP-GUS construct was then replaced with VND7 promoter cut out using Stu I and BamH I to generate the pC3300-VND7pro:eYFP-GUS construct.
For the generation of VND7pro:3×VENUS-NLS reporter plants, VND7 promoter (2-kb fragment upstream of the ATG start codon) was amplified using primers VND7-promoter-L2 and VND7-promoter-R (Supplemental Table 2), and inserted into pBS vector to generate pBS-VND7 promoter2 construct. The VND7 promoter was then inserted into the pC3300-3×VENUS-NLS construct cut with Sma I and BamH I to generate the pC3300-VND7pro:3×VENUS-NLS construct.
For the generation of pYAO:hSpCas9-WRKY15 site 1-WRKY15 site 2-sgRNA construct, two target sites were designed and assembled into the vector as previously described by Yan et al. (2015). The two pairs of primers (Supplemental Table 2) with sticky ends for two target sites were annealed and cloned into the AtU6-26-sgRNA-SK construct using Bsa I site to generate AtU6-26-WRKY15 site 1-sgRNA and AtU6-26-WRKY15 site 2-sgRNA constructs, respectively. Then AtU6-26-WRKY15 site 2-sgRNA construct was digested by Spe I and Nhe I to get AtU6-26-WRKY15 site 2-sgRNA cassette, and AtU6-26-WRKY15 site 2-sgRNA cassette was inserted into the AtU6-26-WRKY15 site 1-sgRNA construct using Spe I site to generate AtU6-26-WRKY15 site 1-sgRNA-AtU6-26-WRKY15 site 2-sgRNA construct (Spe I and Nhe I are a pair of isocaudarner restriction enzymes). The AtU6-26-WRKY15 site 1-sgRNA-AtU6-26-WRKY15 site 2-sgRNA construct was digested by Spe I and Nhe I to get the cassette fragment, and the cassette fragment was inserted into the pYAO:hSpCas9 construct using Spe I site to generate pC1300-pYAO:hSpCas9-WRKY15 site 1-WRKY15 site 2-sgRNA construct.
These binary vectors were transformed into Agrobacterium trumefaciens strain GV3101 using electroporation. Arabidopsis transformation was performed by the floral dip method (Clough and Bent, 1998), and independent single insertion lines were identified for each transgene. The primers used in this study are listed in Supplemental Table 2.
Phylogenetic Analysis
Amino acid sequences were aligned by the Clustal W alignment program, which was shown in Supplemental Data Set 1. The phylogenetic tree was inferred using the Neighbor-Joining method by MEGA7 in Supplemental Figure 1A.
Quantitation of the Density of Spiral Wall Thickening
To quantity the density of cell wall thickness in protoxylem cells, we measured the protoxylem vessel length and counted the spiral wall thickenings in all protoxylem vessels in a microscopic field from the root maturation zone, where roots have formed three metaxylem vessels. A total of 20 roots were measured to calculate the density of the cell wall thickness.
GUS Staining
GUS staining in seedlings was detected by using X-Gluc as described previously (Meng et al., 2012). Seedlings were treated in precooled 90% (v/v) acetone for 20 min, and washed three times with precooled Millipore water for 5 min. Seedlings were then immersed in GUS staining solution (10 mM EDTA, 0.1% [v/v] Triton X-100, 2 mM potassium ferricyanide, 100 µg/mL chloramphenicol, 1 mg/mL X-Gluc in 50 mM sodium phosphate buffer, pH 7.0) at 37°C in the dark for 3 min to 3 h according to different materials. Seedlings were fixed in formaldehyde-alcohol-acetic acid solution (3.7% [w/v] formaldehyde, 50% [v/v] ethanol, and 5% [w/v] acetic acid in double distilled water) for 1 h before transferred to solution with 20% (w/v) lactic acid and 20% (v/v) glycerol. Finally, seedlings were mounted on glass slides and photographed on a Nikon Eclipse 80i microscope under differential interference contrast (DIC) settings.
Propidium Iodide Staining and eYFP Fluorescence Analysis
Seedlings were stained in 10 μg/mL propidium iodide (PI) solution for 3 min, and then they were mounted on glass slides with water and observed under a Nikon C2 Plus laser scanning confocal microscope (LSCM) with excitation and detection wavelengths as follows: PI (561 nm, 575–615 nm) and eYFP (or VENUS; 488 nm, 500–550 nm). To obtain the three-dimensional structures and transverse optical sections, z-stack procedure was adopted. All images were processed with NIS-Elements AR software (Nikon).
Clear, Lignin Staining, and Lignin Autofluorescence Analysis
Seedlings were fixed and destained in acetic acid:ethanol (1:3) solution for 1 h, and cleared in chloral hydrate:glycerol:H2O [8:1:2 (W:V:V)] for 3 min (Kubo et al., 2005; Endo et al., 2015). Seedlings were mounted in 50% (v/v) glycerol and observed on a Nikon Eclipse 80i microscope under DIC settings. For lignin staining, seedlings were first fixed for 1 h, and then stained in HCl-phloroglucinol solution [2% (w/v) phloroglucinol (dissolved in 95% [v/v] ethanol):2 M HCl = 1:1, fresh preparation] for 3 min (Taylor-Teeples et al., 2015). After being mounted in 50% (v/v) glycerol, they were observed on a Nikon Eclipse 80i microscope. Lignin staining in seedlings was also detected by using Basic fuchsin as previously described by Mahönen et al. (2006). After staining, seedlings were observed under LSCM with excitation and detection setup to 561 nm and 575–615 nm, respectively. For lignin autofluorescence analysis, leaves were cleared and destained in methanol (Zhong et al., 2007). They were then mounted with water and observed under LSCM with excitation and detection setup to 405 nm and 425–475 nm, respectively.
Protein Extraction and Immunoblot Analysis
Protein extraction and immunoblot were performed as previously described by Meng et al. (2013). Total protein was extracted from seedlings by grinding in liquid nitrogen to fine powders. Protein concentrations were determined by the Bradford assay with bovine serum albumin as the standard. WRKY15 and WRKY15-EAR (with a 4myc tag) transgene expression was detected using an anti-myc antibody (Millipore, dilution 1:2000).
RNA Extraction and RT-qPCR Analysis
Seedlings were cultured vertically on half-strength MS plates under continuous light, and roots of 5-d-old seedlings (>50) were collected for total RNA preparation using the Direct-zol RNA MicroPrep kit (Zymo Research). RNA quality was monitored using Agilent 2100 Bioanalyzer (Agilent RNA 6000 Nano Kit). Expression levels of selected genes were quantified by RT-qPCR as previously described (Xu et al., 2016) and normalized to UBQ10. Experiments were repeated independently at least three times with similar results. Primers used in RT-qPCR are listed in Supplemental Table 2.
Illumina RNA Sequencing and Analysis
Two sets of RNA samples were selected for profiling. RNA sequencing libraries were generated using the Tru-Seq RNA library preparation kit. Sequencing was performed on the HiSeq 2500 platform (Illumina). Dirty raw reads were filtered out using SOAPnuke software. After reads filtering, clean reads were mapped to The Arabidopsis Information Resource (TAIR10) reference genome using HISAT2 and to reference gene sequences using Bowtie 2. Gene expression levels were calculated using the FPKM method (fragments per kb per million reads). DESeq2 algorithms were used to detect the DEGs. Hierarchical clustering analysis for DEGs was performed by pheatmap, a function of R software. Library preparation and RNA sequencing were performed by the Shenzhen BGI (Beijing Genomics Institute). The raw Illumina reads generated from RNA-seq experiments were deposited at NCBI Sequence Read Archive (PRJNA558847).
Gene Ontology Enrichment
Gene ontology (GO) analysis was performed as previously described (Lee et al., 2018). Singular Enrichment Analysis of AgriGo (Du et al., 2010) was used with the Arabidopsis genome (TAIR10 assembly) as the input. GO terms with false discovery rate <0.05 were considered significantly enriched (Supplemental Data Set 3). GO term results were summarized and visualized by REVIGO (Supek et al., 2011). With use of SimRel semantic similarity measure, terms were clustered at a specified similarity cutoff of 0.4 (a ‘tiny’ REVIGO set) and were further manually modified to clarify the meaning of representative terms (Supplemental Data Set 4). Modified representative terms were used to generate treemaps shown in Figure 8D using treemap in R software.
Quantification and Statistical Analysis
Statistical details of experiments are described in Supplemental Data Set 5. The statistical analysis was performed by Student’s t test or one-way analysis of variance (ANOVA). P-values were shown in figure legends. ANOVA and t test results are shown in Supplemental Data Set 5.
Accession Numbers
Sequence data from this article can be found in TAIR or Genbank databases under the following accession numbers: ASL19 (AT4G00220), CAD9 (AT4G39330), IRX1 (AT4G18780), IRX3 (AT5G17420), LAC12 (AT5G05390), MC9 (AT5G04200), MYB15 (AT3G23250), MYB46 (AT5G12870), PRX15 (AT2G18150), PRX49 (AT4G36430), PRX52 (AT5G05340), PRX67 (AT5G58390), TED6 (AT1G43790), TED7 (AT5G48920), UBQ10 (AT4G05320), VND7 (AT1G71930), WRKY15 (AT2G23320), XCP1 (AT4G35350), XCP2 (AT1G20850), and XSP1 (AT4G00230).
Supplemental Data
Supplemental Figure 1. Phylogenetic analysis and structure characteristics of WRKY15.
Supplemental Figure 2. WRKY15 reporter expression in the procambium cells of the root meristematic zone (Related to Figure 1).
Supplemental Figure 3. Generation of wrky15 deletion mutant lines using the CRISPR-Cas9 approach.
Supplemental Figure 4. WRKY15 is involved in root vascular xylem differentiation and secondary cell wall thickening (Related to Figure 3, results from another independent line).
Supplemental Figure 5. Quantitation of protoxylem gaps in the WRKY15OE transgenic seedlings.
Supplemental Figure 6. Altered lignin deposition in the roots of the WRKY15OE and WRKY15-EAR transgenic seedlings (Related to Figure 4, results from another independent line).
Supplemental Figure 7. Conditional expression of WRKY15-EAR transgene leads to ectopic lignin deposition.
Supplemental Figure 8. T1 transgenic roots of WRKY15-EAR driven by native WRKY15 promoter generate an extra incomplete protoxylem strand.
Supplemental Figure 9. Ectopic differentiation of TEs in WRKY15-EAR transgenic plants (Related to Figure 6, results from another independent line).
Supplemental Figure 10. Correlation between samples from the RNA-Seq analysis. (Related to Figure 8).
Supplemental Figure 11. Effects of WRKY15 on the expression of genes involved in TE differentiation (Related to Figure 8).
Supplemental Figure 12. Ectopic TE formation in WRKY15-EAR plants is associated with ectopic activation of VND7 expression (Related to Figure 9).
Supplemental Figure 13. Changes of cellular morphology and Venus-NLS reporter during ectopic TE cell differentiation and maturation in WRKY15-EAR transgenic plants.
Supplemental Table 1. Gene mapping ratio and mapping statistics of RNA-seq reads.
Supplemental Table 2. List of primers used in this study.
Supplemental Data Set 1. Text file of alignment for the phylogenetic analysis in Supplemental Figure 1A.
Supplemental Data Set 2. List of differentially expressed genes in roots of WRKY15-EAR and WRKY15OE seedlings.
Supplemental Data Set 3. AgriGO analysis of the differentially expressed genes.
Supplemental Data Set 4. Clustering of DEGs based on gene ontology terms by REVIGO.
Supplemental Data Set 5. ANOVA test results showing variables, parameters, degrees of freedom, and test statistics.
Supplemental Movie 1. Expression pattern of WRKY15 in procambial cells of the root maturation zone.
Supplemental Movie 2. Expression pattern of WRKY15 in procambial cells of the root meristematic zone.
Supplemental Movie 3. Expression pattern of VND7 in the Col-0 roots.
Supplemental Movie 4. Expression pattern of VND7 in the WRKY15-EAR roots.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Wolfgang Lukowitz (University of Georgia) for pC3300-3×VENUS-NLS vector and Qi Xie (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for AtU6-26-sgRNA-SK and pYAO:hSpCas9 constructs. This research was supported by the Natural Science Foundation of Zhejiang Province (Zhejiang Provincial Natural Science Foundation; LR18C020001), the Young Elite Scientist Sponsorship Program by CAST (2018QNRC001), and 111 Project (B14027; to J.X.).
AUTHOR CONTRIBUTIONS
S.G., J.X., and S.Z. designed the project; S.G., X.H., X.X., Y.S., Q.Z., and Y.L. performed the experiments; S.G., J.D., J.X., and S.Z. analyzed the results; S.G. and S.Z. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Avci U., Earl Petzold H., Ismail I.O., Beers E.P., Haigler C.H.(2008). Cysteine proteases XCP1 and XCP2 aid micro-autolysis within the intact central vacuole during xylogenesis in Arabidopsis roots. Plant J. 56: 303–315. [DOI] [PubMed] [Google Scholar]
- Bishopp A., Help H., El-Showk S., Weijers D., Scheres B., Friml J., Benková E., Mähönen A.P., Helariutta Y.(2011). A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr. Biol. 21: 917–926. [DOI] [PubMed] [Google Scholar]
- Bollhöner B., Prestele J., Tuominen H.(2012). Xylem cell death: Emerging understanding of regulation and function. J. Exp. Bot. 63: 1081–1094. [DOI] [PubMed] [Google Scholar]
- Bollhöner B., Zhang B., Stael S., Denancé N., Overmyer K., Goffner D., Van Breusegem F., Tuominen H.(2013). Post mortem function of AtMC9 in xylem vessel elements. New Phytol. 200: 498–510. [DOI] [PubMed] [Google Scholar]
- Caño-Delgado A., Lee J.Y., Demura T.(2010). Regulatory mechanisms for specification and patterning of plant vascular tissues. Annu. Rev. Cell Dev. Biol. 26: 605–637. [DOI] [PubMed] [Google Scholar]
- Cho H., Dang T.V.T., Hwang I.(2017). Emergence of plant vascular system: Roles of hormonal and non-hormonal regulatory networks. Curr. Opin. Plant Biol. 35: 91–97. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Demura T., Fukuda H.(2007). Transcriptional regulation in wood formation. Trends Plant Sci. 12: 64–70. [DOI] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z.(2010). agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo H., Yamaguchi M., Tamura T., Nakano Y., Nishikubo N., Yoneda A., Kato K., Kubo M., Kajita S., Katayama Y., Ohtani M., Demura T.(2015). Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 56: 242–254. [DOI] [PubMed] [Google Scholar]
- Endo S., Pesquet E., Yamaguchi M., Tashiro G., Sato M., Toyooka K., Nishikubo N., Udagawa-Motose M., Kubo M., Fukuda H., Demura T.(2009). Identifying new components participating in the secondary cell wall formation of vessel elements in zinnia and Arabidopsis. Plant Cell 21: 1155–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamez S., Tuominen H.(2014). Programmes of cell death and autolysis in tracheary elements: When a suicidal cell arranges its own corpse removal. J. Exp. Bot. 65: 1313–1321. [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Robatzek S., Somssich I.E.(2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5: 199–206. [DOI] [PubMed] [Google Scholar]
- Fukuda H.(1997). Tracheary element differentiation. Plant Cell 9: 1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk V., Kositsup B., Zhao C., Beers E.P.(2002). The Arabidopsis xylem peptidase XCP1 is a tracheary element vacuolar protein that may be a papain ortholog. Plant Physiol. 128: 84–94. [PMC free article] [PubMed] [Google Scholar]
- Furuta K.M., Hellmann E., Helariutta Y.(2014). Molecular control of cell specification and cell differentiation during procambial development. Annu. Rev. Plant Biol. 65: 607–638. [DOI] [PubMed] [Google Scholar]
- Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., Mimura T., Fukuda H., Demura T.(2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19: 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., et al. (2018). A lignin molecular brace controls precision processing of cell walls critical for surface integrity in Arabidopsis. Cell 173: 1468–1480.e9. [DOI] [PubMed] [Google Scholar]
- Lucas W.J., et al. (2013). The plant vascular system: Evolution, development and functions. J. Integr. Plant Biol. 55: 294–388. [DOI] [PubMed] [Google Scholar]
- Meng X., Wang H., He Y., Liu Y., Walker J.C., Torii K.U., Zhang S.(2012). A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24: 4948–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Xu J., He Y., Yang K.Y., Mordorski B., Liu Y., Zhang S.(2013). Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25: 1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Iwase A., Yamamoto H., Yoshida M., Seki M., Shinozaki K., Ohme-Takagi M.(2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19: 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M.(2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17: 2993–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Fukuda H.(2012). Secondary cell wall patterning during xylem differentiation. Curr. Opin. Plant Biol. 15: 38–44. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K., Oda Y., Fukuda H.(2010). Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22: 3461–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani M., Demura T.(2019). The quest for transcriptional hubs of lignin biosynthesis: Beyond the NAC-MYB-gene regulatory network model. Curr. Opin. Biotechnol. 56: 82–87. [DOI] [PubMed] [Google Scholar]
- Pi L., Aichinger E., van der Graaff E., Llavata-Peris C.I., Weijers D., Hennig L., Groot E., Laux T.(2015). Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev. Cell 33: 576–588. [DOI] [PubMed] [Google Scholar]
- Ruonala R., Ko D., Helariutta Y.(2017). Genetic networks in plant vascular development. Annu. Rev. Genet. 51: 335–359. [DOI] [PubMed] [Google Scholar]
- Rushton P.J., Somssich I.E., Ringler P., Shen Q.J.(2010). WRKY transcription factors. Trends Plant Sci. 15: 247–258. [DOI] [PubMed] [Google Scholar]
- Smet W., De Rybel B.(2016). Genetic and hormonal control of vascular tissue proliferation. Curr. Opin. Plant Biol. 29: 50–56. [DOI] [PubMed] [Google Scholar]
- Soyano T., Thitamadee S., Machida Y., Chua N.H.(2008). ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 20: 3359–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F., Bošnjak M., Škunca N., Šmuc T.(2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Teeples M., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517: 571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S., Gallois P., Brown D.(2007). Tracheary element differentiation. Annu. Rev. Plant Biol. 58: 407–433. [DOI] [PubMed] [Google Scholar]
- Uchida N., Lee J.S., Horst R.J., Lai H.H., Kajita R., Kakimoto T., Tasaka M., Torii K.U.(2012). Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc. Natl. Acad. Sci. USA 109: 6337–6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker B., Somssich I.E.(2004). WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 7: 491–498. [DOI] [PubMed] [Google Scholar]
- Ursache R., Miyashima S., Chen Q., Vatén A., Nakajima K., Carlsbecker A., Zhao Y., Helariutta Y., Dettmer J.(2014). Tryptophan-dependent auxin biosynthesis is required for HD-ZIP III-mediated xylem patterning. Development 141: 1250–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S., Vandenbroucke K., Inzé A., van de Cotte B., Mühlenbock P., De Rycke R., Naouar N., Van Gaever T., Van Montagu M.C., Van Breusegem F.(2012). AtWRKY15 perturbation abolishes the mitochondrial stress response that steers osmotic stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 20113–20118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voxeur A., Wang Y., Sibout R.(2015). Lignification: different mechanisms for a versatile polymer. Curr. Opin. Plant Biol. 23: 83–90. [DOI] [PubMed] [Google Scholar]
- Wang H., Avci U., Nakashima J., Hahn M.G., Chen F., Dixon R.A.(2010). Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. USA 107: 22338–22343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Ohtani M., Yamaguchi M., Toyooka K., Wakazaki M., Sato M., Kubo M., Nakano Y., Sano R., Hiwatashi Y., Murata T., Kurata T., et al. (2014). Contribution of NAC transcription factors to plant adaptation to land. Science 343: 1505–1508. [DOI] [PubMed] [Google Scholar]
- Xu J., Meng J., Meng X., Zhao Y., Liu J., Sun T., Liu Y., Wang Q., Zhang S.(2016). Pathogen-responsive MPK3 and MPK6 reprogram the biosynthesis of indole glucosinolates and their derivatives in Arabidopsis immunity. Plant Cell 28: 1144–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Goué N., Igarashi H., Ohtani M., Nakano Y., Mortimer J.C., Nishikubo N., Kubo M., Katayama Y., Kakegawa K., Dupree P., Demura T.(2010b). VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 153: 906–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Kubo M., Fukuda H., Demura T.(2008). Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 55: 652–664. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Mitsuda N., Ohtani M., Ohme-Takagi M., Kato K., Demura T.(2011). VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 66: 579–590. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Ohtani M., Mitsuda N., Kubo M., Ohme-Takagi M., Fukuda H., Demura T.(2010a). VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22: 1249–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K., Kigawa T., Seki M., Shinozaki K., Yokoyama S.(2013). DNA-binding domains of plant-specific transcription factors: structure, function, and evolution. Trends Plant Sci. 18: 267–276. [DOI] [PubMed] [Google Scholar]
- Yan L., Wei S., Wu Y., Hu R., Li H., Yang W., Xie Q.(2015). High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 8: 1820–1823. [DOI] [PubMed] [Google Scholar]
- Yang L., Zhao X., Yang F., Fan D., Jiang Y., Luo K.(2016). PtrWRKY19, a novel WRKY transcription factor, contributes to the regulation of pith secondary wall formation in Populus trichocarpa. Sci. Rep. 6: 18643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Johnson B.J., Kositsup B., Beers E.P.(2000). Exploiting secondary growth in Arabidopsis. Construction of xylem and bark cDNA libraries and cloning of three xylem endopeptidases. Plant Physiol. 123: 1185–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q.(2016). Lignification: Flexibility, biosynthesis and regulation. Trends Plant Sci. 21: 713–721. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Dixon R.A.(2011). Transcriptional networks for lignin biosynthesis: More complex than we thought? Trends Plant Sci. 16: 227–233. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Nakashima J., Chen F., Yin Y., Fu C., Yun J., Shao H., Wang X., Wang Z.Y., Dixon R.A.(2013). Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25: 3976–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Demura T., Ye Z.H.(2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18: 3158–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Lee C., Ye Z.H.(2010). Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol. Plant 3: 1087–1103. [DOI] [PubMed] [Google Scholar]
- Zhong R., Lee C., Zhou J., McCarthy R.L., Ye Z.H.(2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Richardson E.A., Ye Z.H.(2007). The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19: 2776–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Ye Z.H.(2015). Secondary cell walls: Biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol. 56: 195–214. [DOI] [PubMed] [Google Scholar]