Abstract
Background and Aims
Intestinal inflammation in inflammatory bowel diseases [IBD] is thought to be T cell mediated and therefore dependent on the interaction between the T cell receptor [TCR] and human leukocyte antigen [HLA] proteins expressed on antigen presenting cells. The collection of all TCRs in one individual, known as the TCR repertoire, is characterised by enormous diversity and inter-individual variability. It was shown that healthy monozygotic [MZ] twins are more similar in their TCR repertoire than unrelated individuals. Therefore MZ twins, concordant or discordant for IBD, may be useful to identify disease-related and non-genetic factors in the TCR repertoire which could potentially be used as disease biomarkers.
Methods
Employing unique molecular barcoding that can distinguish between polymerase chain reaction [PCR] artefacts and true sequence variation, we performed deep TCRα and TCRβ repertoire profiling of the peripheral blood of 28 MZ twin pairs from Denmark and Germany, 24 of whom were discordant and four concordant for IBD.
Results
We observed disease- and smoking-associated traits such as sharing, diversity and abundance of specific clonotypes in the TCR repertoire of IBD patients, and particularly in patients with active disease, compared with their healthy twins.
Conclusions
Our findings identified TCR repertoire features specific for smokers and IBD patients, particularly when signs of disease activity were present. These findings are a first step towards the application of TCR repertoire analyses as a valuable tool to characterise inflammatory bowel diseases and to identify potential biomarkers and true disease causes.
Keywords: T cell receptor [TCR] repertoire, monozygotic twins, inflammatory bowel diseases [IBD]
1. Introduction
Inflammatory bowel diseases [IBD] are characterised by a chronic relapsing-remittent gastrointestinal inflammation, which is most often restricted to the colon in ulcerative colitis [UC] but it may involve the entire gastrointestinal tract in Crohn’s disease [CD]. Inflammation initiation and progression is thought to be driven by a dysregulated response of the immune system, and in particular of T cells, to the intestinal microbiota or other environmental factors in genetically susceptible individuals. IBD is therefore a complex, multifactorial disease affected by immunological, environmental, and genetic components.1
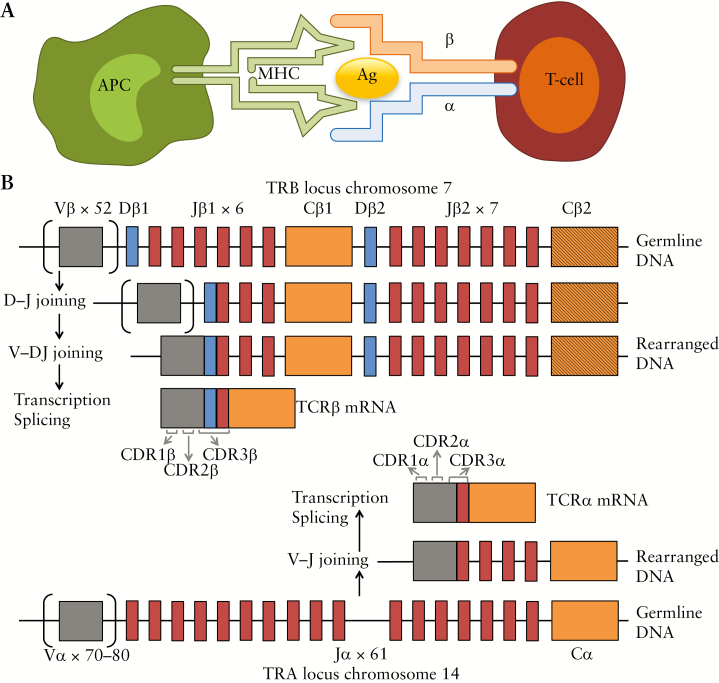
Activation of T cells by specific antigens is mediated by the interaction of their hypervariable T cell receptor [TCR] with the peptide-loaded major histocompatibility complex [pMHC] [Figure 1A]. The MHC is encoded in humans by the human leukocyte antigen [HLA] complex on chromosome 6p21.
Figure 1.
Interaction between an antigen presenting cell [APC] and a T cell, and V[D]J recombination, from Rosati et al. 2017. 12 [A] Interaction between the antigen–major histocompatibility complex [MHC] and the αβ T cell receptor [TCR]. [B] V[D]J recombination: During T cell development, the loci that encode T cell receptor α and β-chains are rearranged. For both loci, variable [V] and joining [J] gene segments, and an additional diversity [D] gene segment for the β-chain, are recombined to form the final rearranged TCR DNA sequence. This process also involves the deletion and insertion of nucleotides at the V-D, D-J, and V-J junctions [not shown]. Following transcription, the sequence between the recombined V[D]J regions and the gene encoding the constant [C] region is removed by splicing. The complementarity-determining region [CDR] 3 is encoded by the V[D]J junction, whereas the CDR1 and CDR2 loops are encoded within the germline V gene. V[D]J, variable diversity joining; TCR, T cell receptor.
Activated helper T cell subpopulations produce proinflammatory cytokines and are suspected to be the main drivers of inflammation in IBD.2 The TCR protein directly interacts with the antigens, mainly through its complementarity-determining region 3 [CDR3]. This region is characterised by great variability, acquired through the variable diversity joining [VDJ] recombination process undergone during T cell development in the thymus [Figure 1B].3,4 Throughout this manuscript we will refer to each unique CDR3 sequence, generated by a specific set of variable [V], diversity [D], and joining [J] genes of either the α [encoded on chromosome 14q11] or the β chain [encoded on chromosome 7q34] of the TCR, with the term TCRα or TCRβ clonotype, respectively.
The TCR repertoire, defined as the collection of different clonotypes of an individual, is known to be altered in a series of diseases, including cancer and infectious, autoimmune, and inflammatory diseases.5 Specifically, previous analyses of the TCR repertoire in IBD have shown repertoire alterations, and have aimed at identifying disease-associated traits and clonotypes.6–10 However, until the advent of next-generation sequencing technologies, technical limitations have prevented a comprehensive TCR repertoire analysis.11,12 Only recently, Chapman et al. published an article on the TCR repertoire in CD patients and compared the repertoire of blood and intestinal biopsies, identifying some disease-associated repertoire traits [i.e., gene usage] and at least one disease-associated clonotype.13 Also, Doorenspleet and colleagues analysed the intestinal repertoire of Crohn’s disease patients and controls14 and observed that T cell clones were more expanded in Crohn’s disease patients. By analysing the repertoire changes in response to therapy, the authors observed larger repertoire changes in individuals responding to therapy. However, specific disease-associated clonotypes could not be identified. Another related work was recently published by Allez et al.15 These studies represent a step towards the understanding of repertoire changes in IBD. However, interindividual repertoire variability coupled with multifactorial disease complexity make identification of disease traits extremely difficult in the absence of large sample numbers. For this reason, we aimed at identifying disease-associated traits in IBD patients’ blood by using a cohort of genetically matched individuals, such as monozygotic [MZ] twins. The analysis of MZ twins helps to reduce the degree of complexity of the study, both on the genetic—MZ twins are genetically identical16—and the environmental levels, as the majority of the twins grew up in the same environment.17
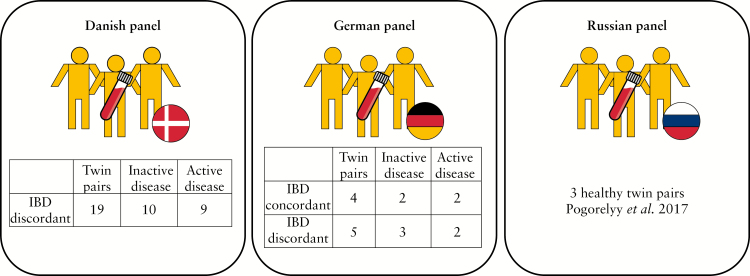
In the current study, we deeply profiled the α and β TCR repertoire of circulating T cells of 28 MZ twin pairs: nine German twin pairs, concordant [n = 4] and discordant [n = 5] for IBD, and 19 Danish twin pairs, discordant for IBD [Figure 2A]. For a comparison with healthy twins, we added previously published data of three healthy Russian MZ twins to our analysis.18 We were able to confirm previous findings regarding the TCR repertoire similarity in MZ twins, and then aimed at analysing if and how the repertoire differs in individuals with IBD. Additionally, we aimed at identifying repertoire-related disease markers by comparing the TCR repertoire of healthy and affected individuals in discordant twin pairs.
Figure 2.
Sample panel overview. Twin pair descriptions. In IBD-discordant twin pairs, twin number 1 is always the IBD one and twin number 2 is the healthy one in the pair. IBD, inflammatory bowel disease.
Indeed, we observed differences in IBD-discordant twin pairs. Particularly, individuals with active disease showed decreased repertoire sharing compared with individuals with inactive disease and with healthy individuals. Additionally, we identified specific TCRs at higher (or lower) frequency in IBD patients compared with their healthy twins. Finally, we observed that smoking seems to have an effect on the peripheral TCR repertoire too and in particular smokers seem to share less clonotypes compared with non-smokers.
2. Methods
2.1. Sample collection and ethics statement
German monozygotic twin pairs sequenced in this study were recruited as a part of the sample panel described in Spehlmann et al.17 Danish twin pairs were part of the twin cohort described in Moller et al.19 The study setup was approved by the Bioethical Committees of the University of Kiel and the Ethics Committee of the Region of Southern Denmark [S20120176], respectively. All patients gave written informed consent before data and biomaterials were collected. Basic phenotypic information is shown in Table 1.
Table 1.
Clinical characteristics. The table shows the different demographic and clinical traits of the twin pairs divided by ancestry. Medians are shown with interquartile range in brackets.
| Ancestry | Danish | German | Russian |
|---|---|---|---|
| Pair type | IBD | IBD | Healthy |
| N pairs | 19 | 9 | 3 |
| Of which IBD concordant | 0 | 4 | 0 |
| Of which IBD discordant | 19 | 5 | 0 |
| Pairs with CD | 4 | 4 | 0 |
| Pairs with UC | 15 | 5 | 0 |
| Median age [years] | 33 [29–40] | 34 [33–41] | 23 [23–24] |
| Males/females | 11/8 | 4/6 | 0/3 |
| Pairs with signs of disease activity [SDA] CD | 2 | 2 | 0 |
| Pairs with signs of disease activity [SDA] UC | 7 | 3 | 0 |
| Median HBI for CD | 4 [3–5.5] | na | - |
| Median HBI for CD with SDA | 7 [77] | na | - |
| Median SCCAI for UC | 3 [1–5] | na | - |
| Median SCCAI for UC with SDA | 6 [2–8] | na | - |
| Median CRP | 1.2 [1–4.15] | na | - |
| Median CRP for IBD with SDA | 3.3 [1–4.7] | na | - |
| Median FCal [ug/g] | 15 [15–116.5] | na | - |
| Median FCal [ug/g] for IBD with SDA | 134 [63–379] | na | - |
| Median Bristol stool scale | 4 [4–6] | na | - |
| Median Bristol stool scale for IBD with SDA | 5 [3.75–7] | na | - |
IBD, inflammatory bowel disease; CD, Crohn’s disease; UC, ulcerative colitis; SDA, signs of disease activity; na, data not available; -, not applicable; HDI, HarveyBradshaw Index; SCCAI, Simple Clinical Colitis Activity Index; CRP, C-reactive protein; FCal, faecal calprotectin.
For German patients, only self-reported disease activity information was available since the samples were not collected in a clinical environment. German patients were therefore defined as having signs of disease activity [SDA] when they did not have any disease manifestation for at least 1 year before sampling, and they were defined as having SDA when they had an acute disease manifestation at sampling time, or a maximum of 35 days before, or when their disease condition was chronically active at sampling time. For Danish twin pairs, information about C-reactive protein [CRP], faecal calprotectin [FCal] levels, the Bristol stool scale, the HarveyBradshaw Index [HBI] for CD patients, and the Simple Clinical Colitis Activity Index [SCCAI] for UC patients, was available at sampling time. Individuals that had HBI/SCCAI <5 and FCal <50 μg/g were considered as not having SDA and therefore to be in disease remission. All other patients had possible signs of disease activity including at least one of the following:
HBI >4 for CD patients and SCCAI >4 for UC patients
FCal >50 μg/g
Demographic and clinical details of all patients are summarised in Table 1.
Using PAXgene tubes [Qiagen], 2.5 ml of blood were drawn following the manufacturer’s instructions. Samples were stored at -80°C. Total RNA was isolated using the PAXgene blood miRNA kit [Qiagen] with automatic isolation using the Qiacube machine; 3500 ng of total RNA were used for TCR library preparation.
2.2. TCR library preparation and sequencing
Molecular-barcoded TCR complementary DNA [cDNA] libraries were prepared as previously described,18 with minor modifications for both TCRα and TCRβ chains. Briefly, cDNA synthesis was performed using SMARTScribe reverse transcriptase [Clontech, Takara] and eight cDNA synthesis reactions per sample were performed. A unique molecular identifier [UMI], also containing a sample barcode of 6 nucleotides, was introduced using the template-switching effect; cDNA synthesis was carried out for 60 min at 42°C and cDNA was then treated with Uracil DNA-Glycosylase [UDG, from New England Biolabs] and incubated for 30 min at 37°C. Samples were subsequently purified with the QIAquick PCR purification kit [Qiagen] and eluted in 100 µl H2O. Purified cDNA was then amplified with two consecutive nested PCRs, respectively 16 and 9 cycles, with purification after each PCR using MagSi-NGSprep Plus beads [MagnaMedics]. Illumina TruSeq compatible adapters and sample-specific barcodes were added during the second PCR.
Quality and concentration of the libraries were measured with Tapestation [Agilent] and Qubit [Thermo Fisher]. Libraries were pooled using 5 ng per library and were sequenced on Illumina HiSeq2500 with a single-index Rapid Run of 2 x 100 bp. Custom sequencing primers were added to the standard Illumina primers.
2.3. TCR data pre-processing
PCR and sequencing error correction was performed through identification and selection of unique molecular identifiers using the software MiGEC, version 1.2.6.20 Filtered sequences were aligned to a TCR gene reference, clonotypes were identified and grouped and CDR3 sequence was identified using the software MiXCR, version 2.1.1.21 Clonotype tables containing clonotype counts, frequencies, CDR3 nucleotide and amino acid sequence, and V[D]J genes were obtained and used for further analysis.
2.4. TCR data analysis
TCR gene usage, clonotype sharing, and abundance of mucosal associated invariant T cells [MAIT] TCRs and natural killer T cell [NKT] TCRs were analysed for the most abundant 1000, 10 000, and 20 000 sequences of each sample to normalise the samples [similar to Zvyagin et al.22]. Figures in the manuscript show results for the most abundant 10 000 clonotypes. TCR gene usage was analysed by calculating the number of clonotypes using specific V and J genes in each sample. Jensen-Shannon [JS] divergence23 was calculated for each possible pair of individuals in the sample panel, and median JS between IBD patients was compared with median JS between healthy controls.
Clonotype sharing was calculated as the number of clonotypes shared by two individuals, for each possible pair of individuals in the sample panel. The median number of clonotypes shared by two IBD patients was compared with the median number shared by two healthy individuals.
For analysis of differential clonotype sharing between IBD patients and healthy individuals, we calculated the number of times that each clonotype was found to be common in two individuals affected by IBD or in two healthy individuals, among the most abundant 10 000. We then checked which clonotypes were less shared by IBD patients and more shared by healthy individuals and vice versa, thus identifying candidates for which we assessed relative abundance differences between IBD patients and healthy individuals in the whole repertoire.
Additionally, the cumulative abundance of TCR sequences originating from known specific cell types (MAIT [mucosal associated invariant T cells] and NKT [natural killer T cells]) was analysed in each sample.24–27
Diversity analysis was performed by downsampling to the smallest sample [27 000 UMIs]. The inverse Simpson index28 was used and calculations were performed using the Vegan R package. TCR clonality was calculated as the inverse of the Shannon entropy as previously described.15
Statistical tests were performed using the non-parametric Wilcoxon-Mann-Whitney U test or the Kruskal-Wallis test, as indicated in the text. Statistical tests assessing differences between IBD patients and healthy individuals [gene usage, diversity, MAIT/NKT TCRs, specific clonotype abundance] were performed on IBD-discordant twin pairs using the Mann-Whitney U test for paired data.
Multiple comparison correction was performed using the Benjamini & Hochberg method.29 Analysis of presence/absence of specific TCRs in IBD individuals versus their healthy twins was conducted using the McNemar statistical test for paired data30 [on IBD-discordant twin pairs only].
TCR sequences were searched in VDJdb31 using the standalone VDJmatch software version 1.2.2 and allowing no mismatches, insertions, or deletions, and matching not only the CDR3 region sequence but also V and J genes.
Identification of disease-associated clonotypes was conducted using the VDJrec32 and ALICE33 software [on both IBD-concordant and -discordant twin pairs]. VDJrec uses a group of samples of interest, in this case IBD patients, to identify clonotypes that are present at a higher frequency in the group compared with what is expected by chance, following the TCR recombination probability model.34 In our analysis, only clonotypes present in at least three patients were considered. ALICE identifies groups of clonotypes which have the same CDR3 region length and only one mismatch of difference [called neighbour clonotypes] and, also using TCR generation probability, estimates which clonotypes have more neighbours than expected by chance, therefore identifying groups of clonotypes expanded in response to an antigen.
Plots were produced using the ggplot2 R package in the R programming environment version 3.4.0.
2.5. Analysis of smoking habits
Information on smoking habits was available for all of the twin pairs in our study [Table 2]. Individual twins were stratified into ‘current’, ‘former’, or ‘never’ smokers. An analysis distinguishing current from former smokers was attempted, but the sample size was not sufficient to run statistical tests; therefore current and former smokers were merged together to form the ‘ever-smoker’ group, which was compared with the ‘non-smokers’. The comparison of ever-smokers versus non-smokers was conducted only on UC patients and their healthy twins, since no concordantly smoker twin pair including a CD patient was present in our sample group.
Table 2.
Information on smoking habits. Only twin pairs concordant for smoking habits were used in the smoking analysis. Four twin pairs were excluded because one twin was a smoker and the other was not. Only twin pairs including UC patients were used for comparisons between smokers and non-smokers [16 pairs] due to the lack of twin pairs including CD patients and concordant smokers.
| CD | UC | |
|---|---|---|
| Twins both smokers | 0 | 5 |
| Twins both non-smokers | 4 | 11 |
| Twins discordant for smoking | 2 | 2 |
UC, ulcerative colitis; CD, Crohn’s disease.
Numbers in bold indicate twin pairs used in smoker VS non-smoker comparison.
3. Results
The investigated subjects included a total of 28 pairs of monozygotic twins, four concordant and 24 discordant for IBD, either UC or CD, nine from Germany and 19 from Denmark [Figure 2A]. Basic phenotypic information of all individuals is presented in Table 1. Additionally, we analysed the repertoire of three Russian healthy twin pairs previously published by Pogorelyy et al. in 2017.18
3.1. Data overview
On average, 151 000 unique TCRα and 181 000 unique TCRβ sequences for each individual were available for analysis, with a minimum of 22 000 and a maximum of 576 000 sequences per individual. A summary table on sequencing information is presented as Supplementary Data 1, including the number of reads, the number of UMIs used for TCR alignment, the number of sequences/UMIs used in each clonotype table, and the number of clonotypes per sample.
It was previously shown that MZ twins share specific TCR repertoire features. They share more highly abundant clonotypes compared with unrelated individuals and they have a similar VJ gene usage.22 We were able to replicate both twin-specific traits in our dataset [Supplementary Figure 1, available at ECCO-JCC online].
3.2. Individuals affected by IBD show specific repertoire traits and differ according todisease activity
In order to identify disease-associated traits, we focused our analyses mainly on case-control comparisons within IBD-discordant twin pairs. We compared all IBD patients against their healthy twins, as well as CD and UC patients separately, in order to identify possible existing differences between the two diseases.
We found differences between individuals affected by IBD and their healthy co-twins in TCR gene usage, diversity, MAIT TCRs and clonotype sharing [Mann-Whitney paired U test; all p-values are reported in figure legends].
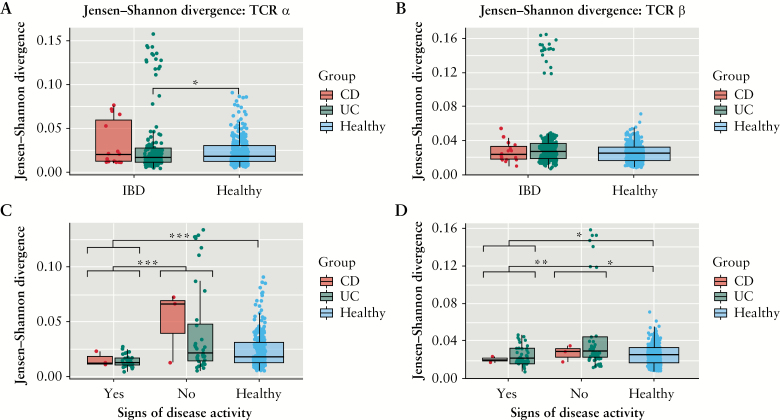
To this end, we investigated whether specific V genes were significantly more or less present in IBD patients compared with healthy individuals in IBD-discordant twin pairs. Particularly, we compared the number of clonotypes carrying specific genes among the most abundant clonotypes. None of the genes showed statistically significant differences after multiple comparison correction [Mann-Whitney paired U test]. However, among the different comparisons we made, a small number of genes showed a consistent trend of increment or decrement in individuals with IBD and particularly in those with signs of disease activity [SDA] [Supplementary Figure 2, available at ECCO-JCC online]. Analyses of the Jensen-Shannon divergence23 metric to measure gene usage similarity showed higher similarity in the TCRα V gene usage in the most abundant clonotypes of UC patients compared with healthy individuals [Figure 3A].This difference was not statistically significant for TCRβ V genes [Figure 3B]. Particularly, both CD and UC patients showing signs of disease activity displayed higher gene usage similarity compared with both CD and UC patients with inactive disease, and with healthy individuals, for both TCRα [Figure 3C] and TCRβ V genes [Figure 3D] [Mann-Whitney U test].
Figure 3.
Jensen-Shannon divergence [JS] for the 10 000 most abundant clonotypes. JS of healthy individuals and IBD patients divided in CD and UC patients for [A] TCRα [healthy-IBD p = 0.11, healthy-CD p = 0.55, healthy-UC p = 0.041, CD-UC p = 0.34]; and [B] TCRβ V gene usage [healthy-IBD p = 0.8, healthy-CD p = 0.8, healthy-UC p = 0.8, CD-UC p = 0.8]. In the bottom panels IBD patients are divided based on presence or absence of signs of disease activity [SDA] and by disease, for [C] TCRα [SDAyes-SDAno p = 0.0002, SDAyes-healthy p = 1.8 x 10-5, SDAno-healthy p = 0.09, UC: SDAyes-SDAno p = 0.007, CD: SDAyes-SDAno p = 0.2] and [D] TCRβ [SDAyes-SDAno p = 0.0005, SDAyes-healthy p = 0.007, SDAno-healthy p = 0.019, UC: SDAyes-SDAno p = 0.014, CD: SDAyes-SDAno p = 0.4]. Individuals showing signs of disease activity are characterised by significantly lower JS divergence particularly in TCRα V gene usage. [*] p-value <0.05; [**] p-value <0.005; [***] p-value <0.0005. IBD, inflammatory bowel disease; CD, Crohn’s disease; UC, ulcerative colitis; TCR, T cell receptor.
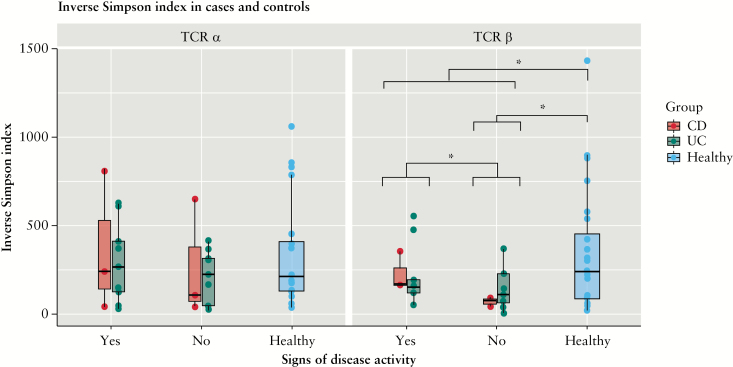
When analysing repertoire diversity in IBD-discordant twins, measured by the inverse Simpson index, we observed that diversity was decreased in individuals affected by IBD, particularly for the TCRβ chain [the difference for the TCRα chain was not significant] [Mann-Whitney paired U test] [Figure 4]. Interestingly, this diversity decrement was significantly different between individuals with and without signs of disease activity. Surprisingly, individuals with signs of disease activity had higher diversity compared with patients with inactive disease [Figure 4]. This observation is consistent with previous findings by others, showing a decrease in peripheral TCR repertoire diversity in patients affected by localised chronic inflammation.35,36
Figure 4.
Repertoire diversity comparison between IBD patients and their healthy twins. The plot is divided to show CD and UC patients separately [no significant difference was found between the two groups], as well as patients with or without signs of disease activity. TCRα [left] [IBD-healthy p = 0.3, SDAyes-SDAno p = 0.5, SDAyes-healthy p = 0.8, SDAno-healthy p = 0.2, UC: SDAyes-SDAno p = 0.6, CD: SDAyes-SDAno p = 0.7] and TCRβ [right] [IBD-healthy p = 0.01, SDAyes-SDAno p = 0.04, SDAyes-healthy p = 0.18, SDAno-healthy p = 0.04, UC: SDAyes-SDAno p = 0.3, CD: SDAyes-SDAno p = 0.1]. Diversity was assessed through the inverse Simpson diversity index, calculated on downsampled data. Diversity was decreased in IBD patients for TCRβ clonotypes and was significantly different between patients with and without signs of disease activity. [*] p-value <0.05. IBD, inflammatory bowel disease; CD, Crohn’s disease; UC, ulcerative colitis; TCR, T cell receptor; SDA, signs of disease activity.
It was previously shown that MAIT and NKT cells are less abundant in the blood of IBD patients compared with healthy controls, and they were also found to be increased and activated at the inflammation site in the gut.35–38
MAIT and NKT cells are characterised by a semi-invariant TCRα repertoire.24,25 Using previously published MAIT and NKT TCRα sequences,24–27 we assessed their abundance in our samples [the sequences are available in Supplementary Data 2, available at ECCO-JCC online]; estimation of MAIT/NKT cell abundance based on their TCRs is available in the Supplementary Materials, available at ECCO-JCC online]. We observed that MAIT TCRα sequences were reduced in twins affected by IBD compared with their healthy co-twins [Mann-Whitney paired Utest] [Supplementary Figure 3, available at ECCO-JCC online]
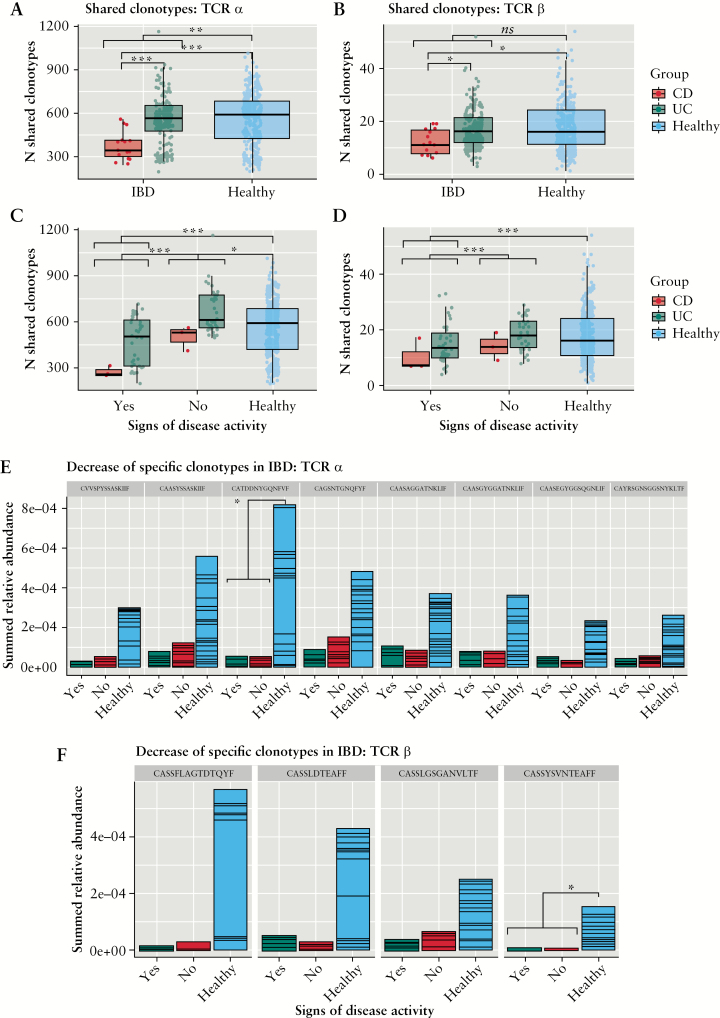
Furthermore, the number of clonotypes shared by each pair of individuals in our sample panel was decreased for IBD patients [Mann-Whitney U test]. Indeed, these shared fewer clonotypes with other IBD patients and also with healthy individuals, as compared with the average number of clonotypes shared by two healthy individuals. Of note, CD patients were shown to have significantly fewer shared clonotypes with one another compared with UC patients and controls [Figure 5A and B]. Most importantly, this characteristic was far more pronounced in individuals with signs of disease activity, for both CD and UC patients, whereas in individuals with inactive disease it was more similar to that of healthy individuals [Figure 5C and D].
Figure 5.
Clonotype sharing and clonotypes decreased in IBD patients. Number of shared clonotypes for each pair of IBD patients [CD and UC] and their healthy twins for the 10 000 most abundant clonotypes of [A] TCRα [healthy-IBD p = 0.002, healthy-CD p = 0.0002, healthy-UC p = 0.97, CD-UC p = 0.0002] and [B] TCRβ [healthy-IBD p = 0.064, healthy-CD p = 0.01, healthy-UC p = 0.96, CD-UC p = 0.01]. In the bottom panels, IBD patients are divided based on presence or absence of signs of disease activity [SDA] and by disease, for [C] TCRα [SDAyes-SDAno p = 1.4 x 10-11, SDAyes-healthy p = 1.8 x 10-8, SDAno-healthy p = 0.005, UC: SDAyes-SDAno p = 7 x 10-6, CD: SDAyes-SDAno p = 0.1, healthy-CD:SDAyes p = 0.01, healthy-UC:SDAyes p = 0.007, SDAyes: CD-UC p = 0.03, SDAno: CD-UC p = 0.03] and [D] TCRβ [SDAyes-SDAno p = 1.8x10-5, SDAyes-healthy p = 0.0001, SDAno-healthy p = 0.15, UC: SDAyes-SDAno p = 0.003, CD: SDAyes-SDAno p = 0.38, healthy-CD:SDAyes p = 0.12, healthy-UC:SDAyes p = 0.07, SDAyes: CD-UC p = 0.27, SDAno: CD-UC p = 0.12]. Clonotype sharing was decreased in between IBD patients and more in CD patients compared with UC patients. Particularly, individuals showing signs of disease activity were sharing less clonotypes compared with patients without signs of disease activity. Specific clonotypes whose abundance was found to be decreased in IBD patients are displayed for [E] TCRα [CVVSPYSSASKIIF p = 0.07, CAASYSSASKIIF p = 0.08, CATDDNYGQNFVF p = 0.02, CAGSNTGNQFYF p = 0.08, CAASAGGATNKLIF p = 0.07, CAASGYGGATNKLIF p = 0.08, CAASEGYGGSQGNLIF p = 0.07, CAYRSGNSGGSNYKLTF p = 0.08] and [F] TCRβ [CASSFLAGTDTQYF p = 0.17, CASSLDTEAFF p = 0.055, CASSLGSGANVLTF p = 0.09, CASSYSVNTEAFF p = 0.017] chain. [*] p-value <0.05; [**] p-value <0.005; [***] p-value <0.0005. IBD, inflammatory bowel disease; CD, Crohn’s disease; UC, ulcerative colitis; TCR, T cell receptor.
This observation is consistent with the decrease in TCR repertoire diversity in IBD patients, as described above. Of note, in the analyses about shared clonotypes described above, differences were statistically significant not only for IBD patients [CD and UC united] compared with healthy individuals, but also for UC or CD patients separately. However, often results for CD patients were not significant, probably due also to the low sample size, but a clear trend was present.
Following these observations, we investigated which of the 10 000 most abundant clonotypes were shared only or mainly by healthy individuals and not by IBD patients [in IBD-discordant twin pairs only]. Thirty TCRα sequences were shared by at least 28 pairs of healthy individuals and one or no pairs of IBD patients, whereas 16 TCRβ sequences were shared by at least 10 pairs of healthy individuals and by one or no pairs of IBD patients. Of note, TCRα sequences are less diverse compared with TCRβ and are therefore most commonly shared interindividually. Among these thus identified clonotypes, we investigated abundance differences between IBD twins and their healthy co-twins [Mann-Whitney paired U test]. Results are reported in Figure 5E and F. In particular, 12 TCRα [eight of which are shown in Figure 5E] and four TCRβ clonotypes showed a trend towards decreased abundance in IBD patients. This trend was significant after multiple comparison correction for one TCRα and one TCRβ sequence.
In comparison, 16 TCRα and 15 TCRβ sequences were shared most commonly among IBD patients, versus healthy individuals [Supplementary Data 3, available at ECCO-JCC online]. Among these, seven TCRα and five TCRβ sequences seemed to be more abundant in IBD patients compared with healthy individuals, and of these seven, five TCRα sequences were significantly more abundant in IBD patients [Supplementary Figure 4, available at ECCO-JCC online].
Interestingly, some of the clones identified to be more shared among IBD individuals compared with controls are known to be reactive to common viruses31 [Supplementary Figure 4; and Table 3].
Table 3.
CDR3 sequences with known antigen reactivity. These sequences were found at higher abundance in IBD patients compared with their healthy co-twins. Column 3 lists the pathogens which they were found to be reactive to, according to VDJdb.28 Column 4 indicates whether the V region of the antigen reactive clonotype was the same in our own data. Identical CDR3 regions with different V genes may or may not recognise the same antigen.
| CDR3 region | Chain | Antigen reactivity | V gene concordance with VDJdb |
|---|---|---|---|
| CAVSSNDYKLSF | alpha | Influenza A,39 yellow fever40 | No |
| CAVMNYGGSQGNLIF | alpha | CMV41 | No |
| CAVEDSNYQLIW | alpha | CMV41 | No |
| CAASGGGSYIPTF | alpha | CMV, EBV41 | No |
| CSARDRVGNTIYF | beta | EBV42 | Yes |
| CASSLGYEQYF | beta | CMV, EBV41,43 | No |
| CSVGAGGTNEKLFF | beta | EBV42 | Yes |
IBD, inflammatory bowel disease; V[D]J, variable diversity joining; CMV, cytomegalovirus; EBV, EpsteinBarr virus.
3.3. Smoking impacts on the peripheral TCR repertoire of healthy individuals and UC patients
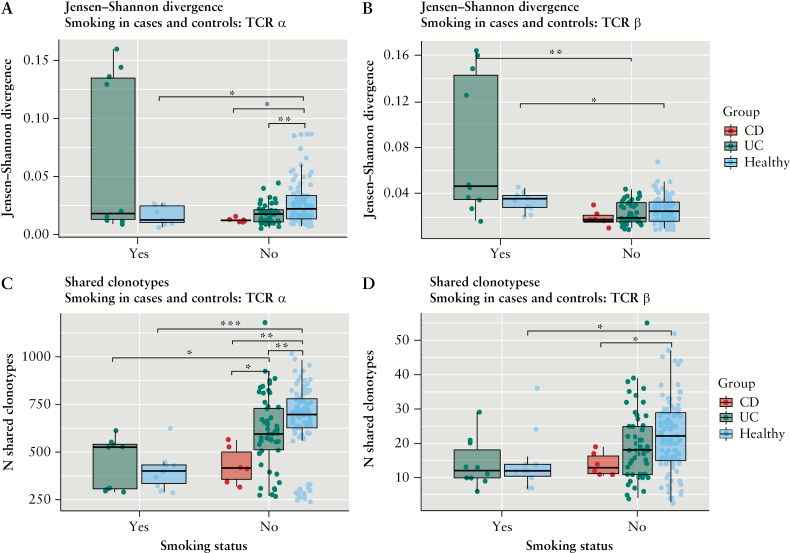
A recent work from Allez and colleagues15 showed how TCR clonality was higher in the ileum of smoking compared with non-smoking CD patients. We therefore analysed if and how smoking behaviour impacts on the peripheral TCR repertoire in our sample panel. We compared only twin pairs concordant for smoking behaviour. Therefore, we compared smoker and non-smoker UC patients but not CD patients, since no twin pair including CD patients had the same smoking habits. We did not observe significant differences between the TCR clonality and repertoire diversity of ever-smokers and non-smokers, both in healthy individuals and UC patients [Supplementary Figure 5, available at ECCO-JCC online]. We also compared gene usage similarity and clonotype sharing of smokers and non-smokers. In healthy individuals, TCRα gene usage is more similar among smokers compared with non-smokers, whereas TCRβ gene usage seems to diverge more among smokers. As previously mentioned, UC and CD patients seem to have more similar TCRα gene usage compared with healthy individuals; however this seems to be true only for non-smokers, and gene usage similarity seems to be more variable in smoker UC patients [Figure 6A and B] [Mann_Whitney paired U test].
Figure 6.
Impact of smoking behaviour on the peripheral TCR repertoire considering the 10 000 most abundant clonotypes. Smokers [yes] and non-smokers [no] are compared as well as healthy individuals and IBD patients. The Jensen-Shannon divergence between samples is depicted for [A] TCRα [healthy: smokers/non-smokers p = 0.03, UC: smokers/non-smokers p = 0.22, non-smokers: UC-healthy p = 0.0015, non-smokers: CD-healthy p = 0.01, non-smokers: CD-UC p = 0.21, smokers: UC-healthy p = 0.17] and [B] TCRβ [healthy: smokers/non-smokers p = 0.02, UC: smokers/non-smokers p = 0.0003, non-smokers: UC-healthy p = 0.31, non-smokers: CD-healthy p = 0.16, non-smokers: CD-UC p = 0.34, smokers: UC-healthy p = 0.09]. The number of shared clonotypes is shown for [C] TCRα [healthy: smokers/non-smokers p = 0.0001, UC: smokers/non-smokers p = 0.007, non-smokers: UC-healthy p = 0.003, non-smokers: CD-healthy p = 0.003, non-smokers: CD-UC p = 0.02, smokers: UC-healthy p = 0.5] and [D] TCRβ [healthy: smokers/non-smokers p = 0.01, UC: smokers/non-smokers p = 0.08, non-smokers: UC-healthy p = 0.06, non-smokers: CD-healthy p = 0.02, non-smokers: CD-UC p = 0.19, smokers: UC-healthy p = 0.85]. [*] p-value <0.05; [**] p-value <0.005; [***] p-value <0.0005. IBD, inflammatory bowel disease; CD, Crohn’s disease; UC, ulcerative colitis; TCR, T cell receptor.
Similarly, the number of shared clonotypes is higher in non-smokers compared with smokers in healthy individuals and also in UC patients. However, whereas non-smoker IBD patients share fewer clonotypes compared with healthy individuals, this does not seem to be the case among smokers [Mann-Whitney paired U test] [Figure 6C and D].
3.4. Towards the identification of disease-associated clonotypes
We also wanted to investigate whether disease-associated and peripherally enriched TCRs exist in IBD patients compared with healthy individuals, and to identify them. This is a highly complex task for such multifactorial diseases that exhibit high interindividual differences, and for which the antigenic triggers are still unknown.
We therefore employed different methodologies to identify such clonotypes, using methods which were recently used for similar purposes by Komech et al.44 for ankylosing spondylitis patients. VDJrec32 and ALICE33 are methods used for the identification of TCRβ clonotypes, which are condition-associated and involved in an active immune response, respectively. These methods are based on the TCR recombination probability model.34 Since they identify TCRs involved in immune responses in both patients and controls, these may also include responses to common pathogens such as influenza A, Epstein-Barr virus [EBV], and others. We therefore re-searched our candidate disease-associated sequences in the VDJdb database31 and were thus able to discriminate such known TCRs. To identify TCRs mostly present in IBD individuals, and not in their healthy co-twins, we used the McNemar statistical test30 for paired data. By employing these three methods to analyse our samples, we were able to identify a series of candidate disease-associated clonotypes. Results from all methods are available in the supplementary materials [Supplementary Data 3].
We then analysed the results overlapping between these three methods, in more detail. During this analysis we found that only one clonotype appeared to be enriched in IBD patients. This clonotype [CASSVRSSYEQYF, TRVB19, TRBJ2-7] is present in VDJdb and is known to be reactive to influenzaA virus.45 Three other clonotypes stood out in both the McNemar and ALICE analyses; however, these were enriched in healthy individuals compared with IBD patients.
Special emphasis needs to be put on the 10 clonotypes listed in Table 4A, since these were found to be enriched in IBD patients in both VDJrec and the McNemar test results, thus implying that these may be potentially disease-associated clonotypes. This table also includes the clonotype reactive for influenzaA mentioned above. Additionally, we have listed the clonotypes reaching the lowest p-value in the McNemar test for being potentially enriched in IBD patients [Table 4B]. These clonotypes were present in at least eight IBD patients, but not in their healthy co-twins, and they mainly used V genes TRBV5-1 and TRBV7-2.
Table 4.
Candidate disease-associated clonotypes. The table is divided into three parts. [A] TCRβ clonotypes which were identified as possibly disease-associated from both VDJrec and McNemar tests. [B] Possible disease-associated TCRβ clonotypes, top hits from McNemar test. [C] Possible disease-associated TCRα clonotypes, top hits from McNemar test. In addition to describing the clonotypes by their CDR3 region and V and J genes, the last four columns of the table describe the number of IBD discordant twin pairs in which these clonotypes where observed.
| CDR3 sequence | V gene | J gene | Present in both twins | Present only in IBD twin | Present only in healthy twin | Not present | |
|---|---|---|---|---|---|---|---|
| A | CASSFVAGTDTQYF | TRBV28 | TRBJ2-3 | 4 | 6 | 0 | 14 |
| CASSLAGGDQPQHF | TRBV5-5 | TRBJ1-5 | 0 | 8 | 0 | 16 | |
| CASSLDGTGPYNEQFF | TRBV7-2 | TRBJ2-1 | 0 | 6 | 0 | 18 | |
| CASSLGQGGTYEQYF | TRBV7-2 | TRBJ2-7 | 0 | 6 | 0 | 18 | |
| CASSPGTANSPLHF | TRBV5-1 | TRBJ1-6 | 1 | 8 | 0 | 15 | |
| CASSPPGQGGEKLFF | TRBV18 | TRBJ1-4 | 0 | 8 | 1 | 15 | |
| CASSPSGGRQPQHF | TRBV18 | TRBJ1-5 | 0 | 6 | 0 | 18 | |
| CASSQGQGAGQPQHF | TRBV3-1 | TRBJ1-5 | 0 | 8 | 1 | 15 | |
| CASSVRSSYEQYF | TRBV19 | TRBJ2-7 | 0 | 6 | 0 | 18 | |
| CSARRQGGTEAFF | TRBV20-1 | TRBJ1-1 | 1 | 9 | 1 | 13 | |
| B | CASSPGGTYEQYF | TRBV5-1 | TRBJ2-7 | 2 | 10 | 0 | 12 |
| CASSLVGGDTEAFF | TRBV5-1 | TRBJ1-1 | 3 | 9 | 0 | 12 | |
| CAWSVRGNTEAFF | TRBV30 | TRBJ1-1 | 0 | 9 | 0 | 15 | |
| CASSLAGGDQPQHF | TRBV5-5 | TRBJ1-5 | 0 | 8 | 0 | 16 | |
| CASSLALAGGTDTQYF | TRBV5-1 | TRBJ2-3 | 1 | 8 | 0 | 15 | |
| CASSLEGRSSYEQYF | TRBV5-1 | TRBJ2-7 | 0 | 8 | 0 | 16 | |
| CASSLGGGTYEQYF | TRBV7-2 | TRBJ2-7 | 1 | 8 | 0 | 15 | |
| CASSLSRNQPQHF | TRBV7-2 | TRBJ1-5 | 0 | 8 | 0 | 16 | |
| CASSLTGNTGELFF | TRBV7-2 | TRBJ2-2 | 1 | 8 | 0 | 15 | |
| CASSLVQGAYNEQFF | TRBV5-1 | TRBJ2-1 | 1 | 8 | 0 | 15 | |
| CASSPGTANSPLHF | TRBV5-1 | TRBJ1-6 | 1 | 8 | 0 | 15 | |
| CASSLRNTGELFF | TRBV12-3 | TRBJ2-2 | 1 | 10 | 1 | 12 | |
| C | CAASSLTGNQFYF | TRAV13-1 | TRAJ49 | 1 | 12 | 0 | 11 |
| CALLGGTSYGKLTF | TRAV9-2 | TRAJ52 | 4 | 11 | 0 | 9 | |
| CAYRSGYNNARLMF | TRAV38-2DV8 | TRAJ31 | 0 | 11 | 0 | 13 | |
| CAAKNDYKLSF | TRAV13-1 | TRAJ20 | 0 | 10 | 0 | 14 | |
| CAEGRDDKIIF | TRAV5 | TRAJ30 | 0 | 10 | 0 | 14 | |
| CAGDGGSQGNLIF | TRAV27 | TRAJ42 | 3 | 10 | 0 | 11 | |
| CALGGLTGGGNKLTF | TRAV6 | TRAJ10 | 0 | 10 | 0 | 14 | |
| CAMSSQGAQKLVF | TRAV12-3 | TRAJ54 | 4 | 10 | 0 | 10 | |
| CAVTPNQAGTALIF | TRAV8-1 | TRAJ15 | 5 | 10 | 0 | 9 | |
| CAVTPTGGFKTIF | TRAV8-6 | TRAJ9 | 1 | 10 | 0 | 13 | |
| CAYRITGNQFYF | TRAV38-2DV8 | TRAJ49 | 11 | 10 | 0 | 3 |
TCR, T cell receptor; IBD, inflammatory bowel disease; V[D]J, variable diversity joining
ALICE and VDJrec are currently available only for TCRβ analysis. However, we also ran the McNemar test for the TCRα chain. The clonotypes obtaining the lowest p-values are listed in Table 4C.
4. Discussion
The TCR repertoire composition is strongly influenced by the HLA type of the individuals,46,47 which should be taken into account when conducting standard case-control TCR repertoire studies. In our study design we therefore decreased the degree of complexity by comparing genetically identical individuals, making it easier to detect disease-associated traits.
By investigating the TCRα and TCRβ repertoire of the circulating lymphocytes of 28 MZ twin pairs concordant or discordant for IBD, we were able to replicate previous findings concerning the similarity in the repertoire of MZ twins and, most interestingly, we identified features associated with IBD, disease activity, and smoking habits. We also showed how the TCR repertoire of CD and UC patients differs in certain respects, such as clonotypes sharing.
We did not observe higher repertoire similarity in clonotype sharing or gene usage [data not shown] between twins concordant for IBD compared with twins discordant for IBD. However, this may also be due to a limited statistical power.
Our results regarding diversity, gene usage, clonotype sharing, and MAIT TCRs suggest that significant shifts occur in the peripheral TCR repertoire of IBD patients compared with healthy individuals. We observed that gene usage, particularly for TCRα chain, is more similar among UC patients compared with controls, and in both CD and UC patients showing signs of disease activity compared with healthy individuals [Figure 3]. This higher gene usage similarity did not translate into a higher number of shared clonotypes between patients. On the contrary, patients shared fewer clonotypes compared with healthy individuals. This effect was stronger for CD patients and patients showing signs of disease activity [Figure 5]. Additionally, repertoire diversity and MAIT TCRs were decreased in IBD patients [Figure 4; and Supplementary Figure 3]. These findings suggest that clonotypes normally present in the blood of healthy individuals may be decreased or absent in the blood of IBD patients. Others35,36,48 previously suggested that this may indicate a recruitment of certain circulating T cells to the inflammation site. However, it may also mean that different clonotypes arise in the peripheral repertoire of IBD patients. These two possible explanations are not mutually exclusive, and analyses of matched inflamed intestinal tissue and peripheral blood TCR repertoire will be necessary to better understand the dynamic of TCR repertoire changes in the blood and intestine of IBD patients.
We also analysed the impact of smoking habits on the peripheral TCR repertoire, and found that smokers shared fewer clonotypes compared with non-smokers, in both health and disease. Interestingly, although IBD patients showed lower gene usage divergence and clonotypes sharing compared with healthy individuals in the non-smoking group, this finding did not hold true among smokers [Figure 6]. Among smokers, we only analysed UC patients and their healthy twins, as we lacked CD patients in the smoker group. We did not observe higher TCR clonality in smoker patients compared with controls, as previously shown by Allez and colleagues.15 However, our analysis was limited to the peripheral blood. Additionally, smoking is known to have different effects in CD and UC,49 and therefore we cannot exclude that these may reflect on differences in the TCR repertoire, too. Both smoking and signs of IBD activity seem to lead to a lower number of shared clonotypes. These effects seem to be unrelated, since: [i] decrease in shared clonotypes was observed in healthy individuals who smoke and not only in patients; and [ii] only two patients among those showing signs of disease activity were smokers, and therefore there is a very small overlap between the two groups. It was shown that smoking leads to the relative increase of specific T cell populations in the blood,50 which may explain the decrease in clonotypes sharing between individuals. Simultaneously, smokers are exposed to a higher number of aero-antigens capable of triggering the immune system. This may potentially lead to the presence of different T cell clonotypes in the blood of smokers, which are not found in non-smokers.
In addition to investigating disease-associated repertoire traits, we also aimed at identifying specific candidate disease-associated clonotypes. We identified clonotypes with decreased or increased abundance in IBD patients’ blood. Most interestingly, one of the TCRβ sequences showing lower abundance in IBD patients’ blood [CASSLGSGANVLTF] was previously identified by Henriksen et al.51 to be specific for primary sclerosing cholangitis [PSC]. This sequence was found to be enriched in the liver of PSC patients and in the liver and gut of PSC-IBD patients, as compared with colorectal cancer [CRC] tissue. Since in this study the authors did not analyse blood from CRC patients, it is possible that this clonotype is normally present in the blood of healthy individuals, and that cells expressing this clonotype are recruited to the inflammation site in patients affected by chronic inflammatory conditions, therefore decreasing their relative frequency in the peripheral blood, as shown in our data.
In order to identify clonotypes enriched in IBD patients, we used different methodologies and analysed the best hits overlapping in all methods. We identified a number of clonotypes which seem to be enriched in IBD individuals, as compared with their healthy co-twins. The only clonotype [CASSVRSSYEQYF] that stood out, in the results of all three methods we used, was found to be reactive to influenza A. Interestingly, some of the clones identified to be more shared among IBD individuals compared with controls are known to be reactive to common viruses31 [Supplementary Figure 4; and Table 3]. For example, it is known that IBD patients have an increased risk of developing influenza.52 Therefore, they may also have more influenza-reactive T cells compared with controls. IBD patients’ higher risk of contracting or reactivating viral infections, also due to immunosuppressive treatment (as for EBV53 and cytomegalovirus [CMV]54), may be a possible explanation for the enrichment of these TCRs in IBD patients. However, it is important to keep in mind that TCRs are cross-reactive against different antigens. It is thus likely that these clonotypes are also reactive to other antigens that could be associated with IBD onset and/or progression. Also, TCRβ alone is not sufficient to define antigen specificity, which may differ depending on the employed TCRα chain and CD4/CD8 molecules.
Our findings do not seem to support the existence of a single strong signal or clonotype associated with IBD. Indeed, IBD is a highly complex disease, characterised by strong interindividual variability which is partly attributed to genetic differences, and for which the existence of a single antigenic trigger has not been demonstrated yet. Moreover different disease treatments, which often involve immunosuppressive drugs, may differently impact on the repertoire of the patients, potentially increasing the background noise of the data and the interindividual variability. Also, we were not able to distinguish between different T cell subtypes in our analysis. Taking these limitations into account, our TCR repertoire results, like others before us, 13, 15,55 rather seem to support the existence of multiple different antigens that trigger the response of different T cell clonotypes. Based on these considerations and on our results, we suggest that the search for disease-associated clonotypes [and indirectly antigens] should be conducted in selected patient subgroups matched, for example, by HLA type, medical treatment, or other meaningful clinical traits.
Even though the number of samples in our study may seem limited compared with huge genetics studies, our sample panel is the largest among TCR repertoire studies in twins. Repeated sampling of the same and of other genetically matched individuals may help to strengthen the findings and identify stronger signals.
We will replicate our findings and identify additional disease-associated traits in a large case-control sample collection, and will validate signatures observed in blood by comparing matched blood and intestinal TCR repertoires.
In conclusion, we think that TCR repertoire disease-associated traits and clonotypes may have the potential to be used as diagnostic or therapy response markers. Our results, showing differences not only between IBD and healthy twins but also between patients with and without signs of disease activity, represent, together with works by others,15 a first step towards this goal.
Supplementary Material
Acknowledgments
We thank Melanie Schlapkohl and Yewgenia Dolshanskaya for support with the sequencing procedures. We thank the Danish Twin Registry [DTR] for help with contact to the monozygotic twins and study design, and for storing the biological samples of the Danish twins. We thank Frank Hinrichs and Karin Eigner for personal care and support of the German twins.
Availability of Data and Materials
Raw sequencing data are available into the ENA database under accession number PRJEB27352. Sample barcode of each sample for demultiplexing is specified in the sample description.
Funding
This work was supported by the H2020 SYSCID program under the grant agreement No 733100, the DFG Excellence Clusters ExC306 ‘Inflammation at Interfaces’, the Research training group 1743 [RTG1743], the Kompetenznetz Darmerkrankungen e.V, Lundbeck Foundation, Region of Southern Denmark, University of Southern Denmark, and University Hospital of Southern Jutland. The work received further support from the Russian Science Foundation grant #15-15-00178 and RFBR #19-54-12011 for TCR cDNA library preparation and data analysis. The research work of Martina E. Spehlmann is supported by the Deutsches Zentrum für Herz-Kreislauf-Forschung e.V. [Grant 81X2700216] and the Medical Faculty of the Christian-Albrechts-University of Kiel,
Germany [Forschungsförderung 2019].
Conflict of Interest
The authors declare no competing interests.
Author Contributions
ER and MVP designed and performed the experiments. ER analysed the data with support from MVP and IZM. MES counselled the patients and the healthy siblings, collected the biomaterials, and provided the clinical data for the German twins. FTM, SBS, and VA provided the samples and metadata from Denmark and supported data analysis and manuscript writing. ER wrote the manuscript with support from MVP, IZM, CMD, and AF. CMD, YBL, SS, and NF contributed to supervising the study. AF and IZM supervised and coordinated the project. All authors discussed the results and commented on the manuscript. All authors read and approved the final manuscript.
References
- 1. Ellinghaus D, Bethune J, Petersen BS, Franke A. The genetics of Crohn’s disease and ulcerative colitis – status quo and beyond. Scand J Gastroenterol 2015;50:13–23. [DOI] [PubMed] [Google Scholar]
- 2. Chen ML, Sundrud MS. Cytokine networks and T cell subsets in inflammatory bowel diseases. Inflamm Bowel Dis 2016;22:1157–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nikolich-Zugich J, Slifka MK, Messaoudi I. The many important facets of T cell repertoire diversity. Nat Rev Immunol 2004;4:123–32. [DOI] [PubMed] [Google Scholar]
- 4. Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol 2015;33:169–200. [DOI] [PubMed] [Google Scholar]
- 5. Miles JJ, Douek DC, Price DA. Bias in the αβ T cell repertoire: implications for disease pathogenesis and vaccination. Immunol Cell Biol 2011;89:375–87. [DOI] [PubMed] [Google Scholar]
- 6. Shiobara N, Suzuki Y, Aoki H, et al. Bacterial superantigens and T cell receptor beta-chain-bearing T cells in the immunopathogenesis of ulcerative colitis. Clin Exp Immunol 2007;150:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chott A, Probert CS, Gross GG, Blumberg RS, Balk SP. A common TCR beta-chain expressed by CD8+ intestinal mucosa T cells in ulcerative colitis. J Immunol 1996;156:3024–35. [PubMed] [Google Scholar]
- 8. Shalon L, Gulwani-Akolkar B, Fisher SE, et al. Evidence for an altered T cell receptor repertoire in Crohn’s disease. Autoimmunity 1994;17:301–7. [DOI] [PubMed] [Google Scholar]
- 9. Saubermann LJ, Probert CS, Christ AD, et al. Evidence of T cell receptor beta-chain patterns in inflammatory and noninflammatory bowel disease states. Am J Physiol 1999;276:G613–21. [DOI] [PubMed] [Google Scholar]
- 10. Probert CS, Chott A, Saubermann LJ, Stevens AC, Balk SP, Blumberg RS. Prevalence of an ulcerative colitis-associated CD8+ T cell receptor beta-chain CDR3-region motif and its association with disease activity. J Clin Immunol 2001;21:126–34. [DOI] [PubMed] [Google Scholar]
- 11. Woodsworth DJ, Castellarin M, Holt RA. Sequence analysis of T cell repertoires in health and disease. Genome Med 2013;5:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rosati E, Dowds CM, Liaskou E, Henriksen EKK, Karlsen TH, Franke A. Overview of methodologies for T cell receptor repertoire analysis. BMC Biotechnol 2017;17:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chapman CG, Yamaguchi R, Tamura K, et al. Characterization of T cell receptor repertoire in inflamed tissues of patients with Crohn’s disease through deep sequencing. Inflamm Bowel Dis 2016;22:1275–85. [DOI] [PubMed] [Google Scholar]
- 14. Doorenspleet ME, Westera L, Peters CP, et al. Profoundly expanded T cell clones in the inflamed and uninflamed intestine of patients with Crohn’s disease. J Crohns Colitis 2017;11:831–9. [DOI] [PubMed] [Google Scholar]
- 15. Allez M, Auzolle C, Ngollo M, et al. ; REMIND Study Group T cell clonal expansions in ileal Crohn’s disease are associated with smoking behaviour and postoperative recurrence. Gut 2019;68:1961–70. [DOI] [PubMed] [Google Scholar]
- 16. Petersen BS, Spehlmann ME, Raedler A, et al. Whole genome and exome sequencing of monozygotic twins discordant for Crohn’s disease. BMC Genomics 2014;15:564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spehlmann ME, Begun AZ, Saroglou E, et al. Risk factors in German twins with inflammatory bowel disease: results of a questionnaire-based survey. J Crohns Colitis 2012;6:29–42. [DOI] [PubMed] [Google Scholar]
- 18. Pogorelyy MV, Elhanati Y, Marcou Q, et al. Persisting fetal clonotypes influence the structure and overlap of adult human T cell receptor repertoires. PLoS Comput Biol 2017;13:e1005572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moller FT, Knudsen L, Harbord M, et al. Danish cohort of monozygotic inflammatory bowel disease twins: clinical characteristics and inflammatory activity. World J Gastroenterol 2016;22:5050–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shugay M, Britanova OV, Merzlyak EM, et al. Towards error-free profiling of immune repertoires. Nat Methods 2014;11:653–5. [DOI] [PubMed] [Google Scholar]
- 21. Bolotin DA, Poslavsky S, Mitrophanov I, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods 2015;12:380–1. [DOI] [PubMed] [Google Scholar]
- 22. Zvyagin IV, Pogorelyy MV, Ivanova ME, et al. Distinctive properties of identical twins’ TCR repertoires revealed by high-throughput sequencing. Proc Natl Acad Sci U S A 2014;111:5980–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin J. Divergence measures based on the shannon entropy. IEEE T Inform Theory 1991;37:145–51. [Google Scholar]
- 24. Greenaway HY, Ng B, Price DA, Douek DC, Davenport MP, Venturi V. NKT and MAIT invariant TCRα sequences can be produced efficiently by VJ gene recombination. Immunobiology 2013;218:213–24. [DOI] [PubMed] [Google Scholar]
- 25. Gherardin Nicholas A, Keller Andrew N, Woolley Rachel E, et al. Diversity of T cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition. Immunity 2016;44:32–45. [DOI] [PubMed] [Google Scholar]
- 26. Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d-restricted antigens by natural killer T cells. Nat Rev Immunol 2012;12:845–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chamoto K, Guo T, Imataki O, et al. CDR3β sequence motifs regulate autoreactivity of human invariant NKT cell receptors. J Autoimmun 2016;68:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Simpson EH. Measurement of diversity. Nature 1949;163:688. [Google Scholar]
- 29. Benjamini Y, Hochberg Y.. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. London: Royal Statistical Society; 1995: 289–300. [Google Scholar]
- 30. Mcnemar Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 1947;12:153–7. [DOI] [PubMed] [Google Scholar]
- 31. Shugay M, Bagaev DV, Zvyagin IV, et al. VDJdb: a curated database of T cell receptor sequences with known antigen specificity. Nucleic Acids Res 2018;46:D419–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pogorelyy MV, Minervina AA, Chudakov DM, et al. Method for identification of condition-associated public antigen receptor sequences. Elife 2018;7:e33050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pogorelyy MV, Minervina AA, Shugay M, et al. Detecting T cell receptors involved in immune responses from single repertoire snapshots. PLoS Biol 2019;17:e3000314.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murugan A, Mora T, Walczak AM, Callan CG Jr. Statistical inference of the generation probability of T cell receptors from sequence repertoires. Proc Natl Acad Sci U S A 2012;109:16161–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Serriari NE, Eoche M, Lamotte L, et al. Innate mucosal-associated invariant T [MAIT] cells are activated in inflammatory bowel diseases. Clin Exp Immunol 2014;176:266–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Haga K, Chiba A, Shibuya T, et al. MAIT cells are activated and accumulated in the inflamed mucosa of ulcerative colitis. J Gastroenterol Hepatol 2016;31:965–72. [DOI] [PubMed] [Google Scholar]
- 37. Grose RH, Thompson FM, Baxter AG, Pellicci DG, Cummins AG. Deficiency of invariant NK T cells in Crohn’s disease and ulcerative colitis. Dig Dis Sci 2007;52:1415–22. [DOI] [PubMed] [Google Scholar]
- 38. van der Vliet HJ, von Blomberg BM, Nishi N, et al. Circulating V[alpha24+] Vbeta11+ NKT cell numbers are decreased in a wide variety of diseases that are characterized by autoreactive tissue damage. Clin Immunol 2001;100:144–8. [DOI] [PubMed] [Google Scholar]
- 39. Gil A, Yassai MB, Naumov YN, Selin LK. Narrowing of human influenza A virus-specific T cell receptor α and β repertoires with increasing age. J Virol 2015;89:4102–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee ES, Thomas PG, Mold JE, Yates AJ. Identifying T cell receptors from high-throughput sequencing: dealing with promiscuity in TCRα and TCRβ pairing. PLoS Comput Biol 2017;13:e1005313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.10xGenomics-resources. A new way of exploring immunity – linking highly multiplexed antigen recognition to immune repertoire and phenotype. https://www.10xgenomics.Com/resources/application-notes/a-new-wayof-exploring-immunity-linking-highly-multiplexed-antigen-recognitionto-immune-repertoire-and-phenotype/# Accessed September 01, 2019.
- 42. Cohen GB, Islam SA, Noble MS, et al. Clonotype tracking of TCR repertoires during chronic virus infections. Virology 2002;304:474–84. [DOI] [PubMed] [Google Scholar]
- 43. Chen G, Yang X, Ko A, et al. Sequence and structural analyses reveal distinct and highly diverse human CD8+ TCR repertoires to immunodominant viral antigens. Cell Rep 2017;19:569–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Komech EA, Pogorelyy MV, Egorov ES, et al. CD8+ T cells with characteristic T cell receptor beta motif are detected in blood and expanded in synovial fluid of ankylosing spondylitis patients. Rheumatology 2018;57:1097–104. [DOI] [PubMed] [Google Scholar]
- 45. Sant S, Grzelak L, Wang Z, et al. Single-cell approach to influenza-specific CD8+ T cell receptor repertoires across different age groups, tissues, and following influenza virus infection. Front Immunol 2018;9:1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sharon E, Sibener LV, Battle A, Fraser HB, Garcia KC, Pritchard JK. Genetic variation in MHC proteins is associated with T cell receptor expression biases. Nat Genet 2016;48:995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reed EF, Tugulea SL, Suciu-Foca N. Influence of HLA class I and class II antigens on the peripheral T cell receptor repertoire. Hum Immunol 1994;40:111–22. [DOI] [PubMed] [Google Scholar]
- 48. Hegazy AN, West NR, Stubbington MJT, et al. ; Oxford IBD Cohort Investigators Circulating and tissue-resident CD4+ T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology 2017;153:1320–37.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Parkes GC, Whelan K, Lindsay JO. Smoking in inflammatory bowel disease: impact on disease course and insights into the aetiology of its effect. J Crohns Colitis 2014;8:717–25. [DOI] [PubMed] [Google Scholar]
- 50. Bauer M, Linsel G, Fink B, et al. A varying T cell subtype explains apparent tobacco smoking induced single CpG hypomethylation in whole blood. Clin Epigenetics 2015;7:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Henriksen EK, Jørgensen KK, Kaveh F, et al. Gut and liver T cells of common clonal origin in primary sclerosing cholangitis-inflammatory bowel disease. J Hepatol 2017;66:116–22. [DOI] [PubMed] [Google Scholar]
- 52. Tinsley A, Navabi S, Williams ED, et al. Increased risk of influenza and influenza-related complications among 140,480 patients with inflammatory bowel disease. Inflamm Bowel Dis 2019;25:369–76. [DOI] [PubMed] [Google Scholar]
- 53. Li X, Chen N, You P, et al. The status of Epstein-Barr virus infection in intestinal mucosa of Chinese patients with inflammatory bowel disease. Digestion 2019;99:126–32. [DOI] [PubMed] [Google Scholar]
- 54. Römkens TE, Bulte GJ, Nissen LH, Drenth JP. Cytomegalovirus in inflammatory bowel disease: a systematic review. World J Gastroenterol 2016;22:1321–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Camus M, Esses S, Pariente B, et al. Oligoclonal expansions of mucosal T cells in Crohn’s disease predominate in NKG2D-expressing CD4 T cells. Mucosal Immunol 2014;7:325–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data are available into the ENA database under accession number PRJEB27352. Sample barcode of each sample for demultiplexing is specified in the sample description.