Abstract
Ovarian cancer is a common cancer worldwide. Epithelial ovarian cancer (EOC) is the most common subtype of ovarian cancer. This study was designed to explore the function of miR-362-3p in EOC. QRT-PCR analysis was used to test miR-362-3p levels in EOC tissues and cell lines. Cell viability was tested via MTT assay. Transwell systems were applied to assay cell migration. The target gene of miR-362-3p was evaluated using dual luciferase reporter assays. The MyD88 protein in EOC cells was tested via western blot. Our data showed that miR-362-3p was expressed at low levels in EOC tissues and cells. miR-362-3p inhibited cell proliferation and migration, bound the 3′-untranslated region (UTR) of MyD88, and inhibited MyD88 expression. MyD88 was inversely correlated with miR-362-3p in EOC, and MyD88 overexpression partly reduced the anti-proliferative effect of miR-362-3p in EOC cells. In conclusion, our data showed that miR-362-3p has an anti-proliferative effect on EOC.
Keywords: Cell biology, Bioinformatics, Cancer research, Oncology, Clinical research, Ovarian cancer, microRNAs, MyD88
Cell biology; Bioinformatics; Cancer research; Oncology; Clinical research; Ovarian cancer, microRNAs, MyD88.
1. Introduction
Ovarian cancer (OC) accounts for an estimated 239,000 new cases and 152,000 deaths worldwide annually [1]. Although China has a relatively low incidence rate (4.1 per 100,000), China's large population yielded an estimated 52,100 new cases and 22,500 related deaths in 2015 [2]. Epithelial ovarian cancer (EOC) is the most common subtype of ovarian cancer [3]. EOC responds temporarily to surgery and cytotoxic agents; however, EOC frequently persists and recurs [4]. New molecular targets must be identified to develop targeted therapies [5].
MicroRNA (miRNA) is a small endogenous noncoding RNA (containing approximately 22 nucleotides) that negatively regulates gene expression by inhibiting messenger RNA (mRNA) translation or by targeting mRNAs for degradation [6]. Aberrant miRNA expression is involved in tumorigenesis in various cancers [7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17].
The role miR-362-3p was investigated in various type of cancer. For example anti-miR-362-3p inhibits migration and invasion of human gastric cancer cells [18]; miR-362-3p functions as a tumor suppressor in cervical adenocarcinoma [19] and in renal cancer cell [20]. These data above indicate that miR-362-3p may played different roles in various types of cancer.
However, the function of miR-362-3p remains unknown. This study aimed to reveal the role of miR-362-3p in EOC. We hope our data will provide insight into the EOC pathogenesis.
2. Materials and methods
2.1. Tissues samples
For this study, twelve EOC tissue samples (serous subtype) were collected from the Department of Gynecology and Obstetrics, West China Second University Hospital, Sichuan University, Chengdu. The ethics committee of Sichuan University evaluated and approved the use of human tissues. Written informed consent was obtained from all patients enrolled in this study, and all specimens were handled and anonymized as required by the legal standards of China. The clinical and pathological characteristics of 12 EOC patients was listed as below:
| NO | AGE | HISTOLOGY | STAGE | GRADE |
|---|---|---|---|---|
| 1 | 72 | Serous | II | 3 |
| 2 | 46 | Serous | III | 3 |
| 3 | 59 | Serous | II | 3 |
| 4 | 68 | Serous | IV | 2 |
| 5 | 75 | Serous | III | 3 |
| 6 | 53 | Serous | II | 3 |
| 7 | 54 | Serous | IV | 3 |
| 8 | 65 | Serous | III | 3 |
| 9 | 61 | Serous | II | 2 |
| 10 | 64 | Serous | IV | 3 |
| 11 | 49 | Serous | III | 3 |
| 12 | 53 | Serous | II | 2 |
2.2. Cell lines
Two EOC cell lines, SKOV3 and HO8910, and the normal immortalized human ovarian surface epithelial cells, IOSE29, were acquired from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI-1640 medium with 10% fetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in 6-well plates in 37 °C and 5% CO2.
2.3. Detection of miR-362-3p and MyD88 in EOC samples and SKOV3 and HO8910 cells
The miR-362-3p levels in 12 EOC tissue samples and the SKOV3, HO8910 and IOSE29 cell lines were detected via qRT-PCR. In detail, total RNA (10ng is taken for next step) was extracted from each of the 12 EOC tissues using TRIzol reagent, per the manufacturer's protocol (Invitrogen, Carlsbad, CA, USA). The miR-362-3p levels were determined using a miRNA RT-PCR Kit (Kaifeng, Biotechnology, Kunshan, China) using a specific stem-loop primer. A diluted reverse transcription product that was a miRNA-specific cDNA was used for each real-time PCR reaction. Real-time PCR was performed using the ABI PRISM 7900 HT Sequence Detection System (Life Technologies, Carlsbad, CA, USA). The thermal PCR profile was as follows: 95 °C for 10 min, then 40 cycles at 95 °C for 15 s, and 60 °C for 1 min. Different combinations of U6 small nuclear RNAs were used as endogenous controls. The ΔΔCT method was used to calculate the expression ratios. The primer sequences used were as follows: miR-362-3p F: AACACACCTATTCAAGGATTCA, R: ACGTGA CACGTTCGGAGAATT; U6 F: CTCGCTTCGGCAGCACA, R: AACGCTTCACGAATTTGCGT; MyD88 F: TCATCGAAAAGAGGTTGGCT, R: GATGGGGATCAGTCGCTTCT; and GAPDH F: CGACAGTCAGCCGCATCTTC, R: CGTTCTCAGCCTTGACGGTG.
2.4. miRNAs and plasmid transfection
In the OC cell lines, miR-362-3p was overexpressed by miR-362-3p mimics and decreased with miR-362-3p antisense oligonucleotides (ASO). miR-362-3p mimics, miR-362-3p ASO and control miRNA were designed and purchased from Sangon Biotech Company (https://www.sangon.com/; Shanghai, China). The sequence were miR-362-3p mimics: CCUCUGGGCCCUUCCUCCAG and miR-362-3p ASO: UUAGGAACCUUGGAUCCAUCACAAAA. miRNAs were transfected into cells using Lipofectamine® 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA). Similarly, For MyD88 overexpression transfection, the pcDNA3.1-MyD88 was transfected into cells to up-regulate the MyD88 level in SKOV3 cells.
2.5. Cell viability assay
Cell viability was analyzed via MTT assay [21]. Briefly, SKOV3 and HO8910 were placed into 96-well plates at a density of 5×103/well. For cell viability assay, MTT reagent was added to the medium at a final concentration of 0.1 mg/ml, and 100 μl of DMSO was added. The optical density was measured on a microplate reader with a 570-nm filter. The measurement were performed in 24h, 48, 72h following miR-362-3p mimics or miR-362-3p transfection.
2.6. Migration
Transwell systems were applied to assay cell migration [22]. In detail, the transwell chambers (8.0-μm pore; Sigma, St. Louis, MI, USA) were placed in 24-well plates. The miR-362-3p mimics or ASO-transfected cells were fetal bovine serum (FBS)-deprived for 12 h, then subsequently added to the upper chambers. Medium containing 10% FBS was placed in the lower chambers, and the migrating cells were counted after 24 h. We count the number of migrating cells at least for three time.
2.7. Prediction of the putative miR-362-3p targets
Targetscan software (http://www.targetscan.org) was used to predict the putative targets of miR-362-3p [23, 24, 25].
2.8. Dual luciferase reporter assays
SKOV3 cells were seeded at 1×105 per well and were serum-starved for 6 h pretransfection. The 3′-untranslated region (UTR) of MyD88 and the mutated controls were cloned and inserted into the reporter plasmid (500 ng) and the pGL3-control (100 ng; Promega, USA). miR-362-3p mimics (50 nM) were then transfected into the SKOV3 cells containing wild-type or mutant 3′-UTR plasmids, using Lipofectamine 2000 (Invitrogen, USA). Cells were harvested, and luciferase activity was measured for each specimen after 24 h, using the Dual-Luciferase Reporter Assay System (Invitrogen, USA). MyD88 3′-UTR mutants were generated using the Site-Directed Mutagenesis kit (Invitrogen, USA).
2.9. Western blot analysis
SKOV3 cells were frozen and lysed in lysis buffer (150 mM NaCl, 50 mM Tris-HCI, 1% Triton X-100 and 0.1% SDS) with the protease inhibitor cocktail and the phosphatase inhibitor cocktail (Cat. No. P5726, Sigma, USA). For MyD88 analysis, an anti-MyD88 antibody (Cat. No, ab133739, Abcam, Cambridge, UK) was used at a dilution of 1:1000, then detect with a peroxidase-linked anti-rabbit IgG antibody (Cat. NO. ab6721, 1:2000 dilution, Abcam). Proteins were detected using ECL western blotting detection reagents (GE Healthcare, USA). The beta-actin antibody (Cat. NO. ab8226, 1:1000 dilution, Abcam) is applied to beta-actin (internal control), Goat anti-mouse IgG (HRP) (ab97040, Abcam) was used the secondary antibody.
2.10. Statistical analysis
All experiments were repeated three times. Data are shown as the means ± SD. A two-tailed Student's t-test was used to analyze the mean value between two groups; one-way ANOVA was used to test the mean value among 3 groups or more with post hoc contrasts by Student-Newman-Keuls test. P < 0.05 and P < 0.01 indicated statistical significance. All calculations were performed using SPSS software (version 16.0, IBM Armonk, New York, USA).
3. Results
3.1. Low miR-362-3p levels in OC tissue samples
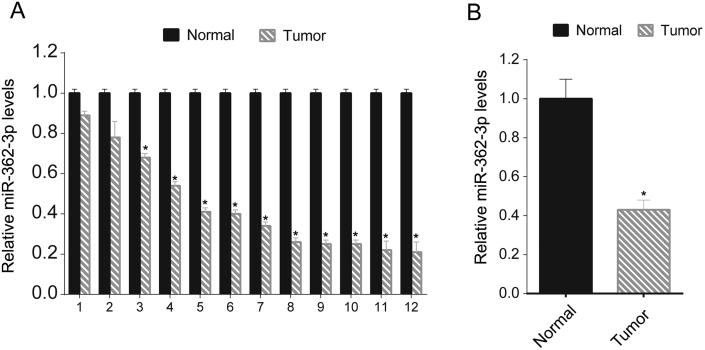
To investigate the role of miR-362-3p in OC, we firstly assayed the miR-362-3p levels in 12 OC tissues previously collected from the hospital via qRT-PCR. miR-362-3p levels were lower in tumor tissues than in adjacent normal tissues (Figure 1A). The mean values of the 12 OC tissue samples and matched adjacent tissue samples were compared, and the mean miR-362-3p value was lower in the OC tissues than in the normal tissues (Figure 1B).
Figure 1.
Relatively low levels of miR-362-3p in OC tissue samples. miR-362-3p levels in 12 OC tissues and their matched adjacent normal tissues were assessed via qRT-PCR. The relative miR-362-3p levels tumor/normal (log2) are listed (A); the mean miR-362-3p values of the OC tissues and their matched adjacent normal tissues were also recorded (B). These experiments were performed in triplicate ∗P < 0.05.
3.2. Forced miR-362-3p expression inhibited cell viability and migration
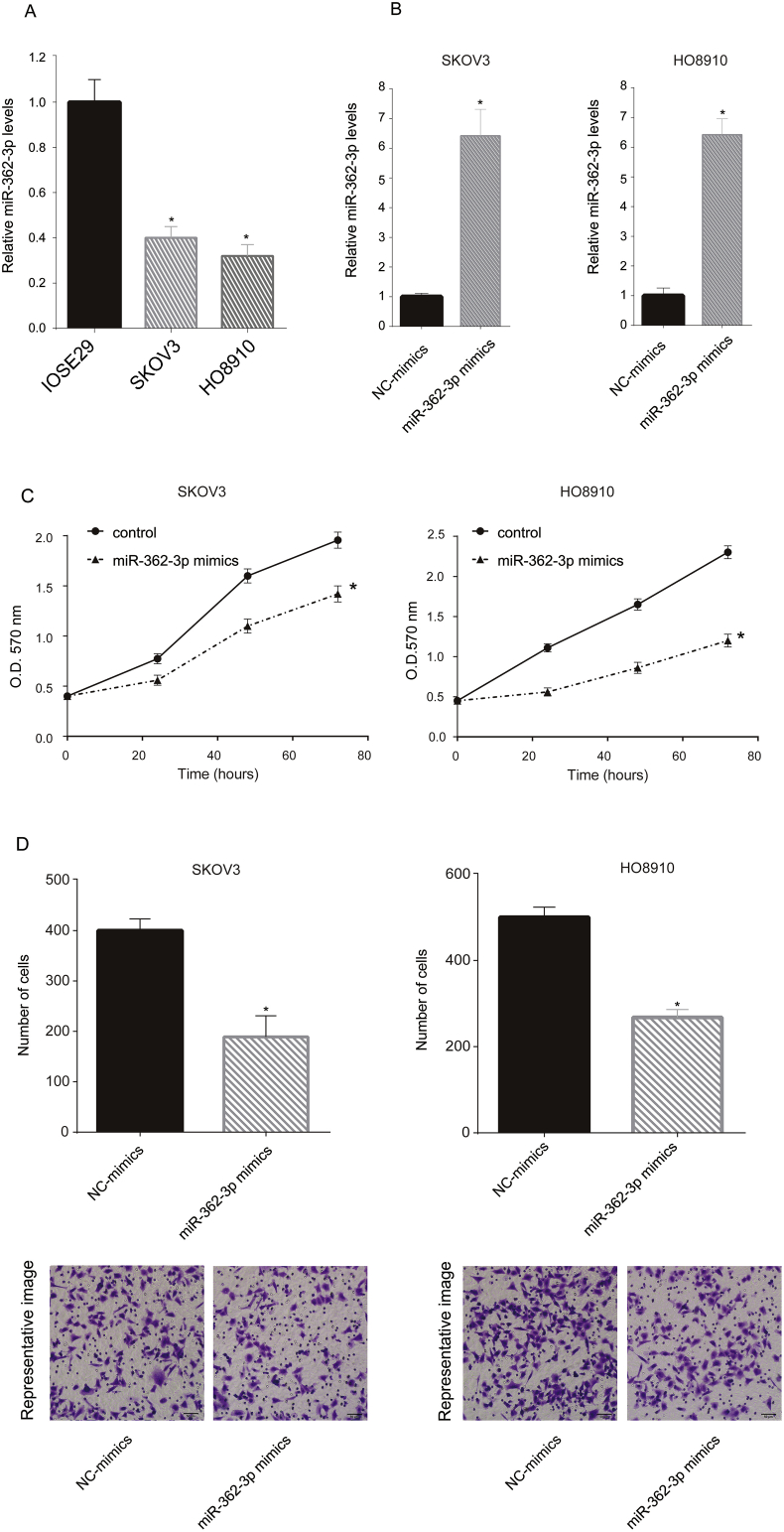
The in vitro function of miR-362-3p was tested in two OC cell lines (SKOV3 and HO8910). The cellular miR-362-3p levels were tested via qRT-PCR. Compared with IOSE29, the SKOV3 and HO8910 cells showed low miR-362-3p levels (Figure 2A). Next, miR-362-3p mimics were transfected into SKOV3 and HO8910 cells, which increased the miR-362-3p levels in these cells (Figure 2B). Next, we analyzed cell viability following the miR-362-3p mimic transfection and found that miR-362-3p mimics inhibited SKOV3 and HO8910 cell viability at the 72 h following transfection (Figure 2C). Subsequently, we investigated cell migration following transfection and found that miR-362-3p mimics decreased the number of migrating cells (Figure 2D).
Figure 2.
Overexpression of miR-362-3p inhibited SKOV3 and HO8910 proliferation and migration. miR-362-3p levels of normal human ovarian epithelial cells SKOV3 and HO8910 were assayed via qRT-PCR. The miR-362-3p level in normal human ovarian epithelial cells was arbitrarily defined as 1 (A). SKOV3 and HO8910 cells were transfected with miR-362-3p mimics, and miR-NC mimics were used as negative controls. After 24 h, the miR-362-3p levels in the transfected cells were assayed via qRT-PCR. The miR-362-3p level in the miR-NC mimic-transfected cells was arbitrarily defined as 1 (B). Following miR-362-3p mimic transfection, SKOV3 and HO8910 cell viability were tested via MTT assay at the indicated time point (0, 24, 48, 72h) (C). To assess cell migration, SKOV3 and HO8910 cells were added to the upper chamber with a noncoated membrane in each group. After 6 h, the cells in the lower chamber were counted (D). These experiments were performed in triplicate ∗P < 0.05.
3.3. Downregulation of miR-362-3p promoted cell viability and migration
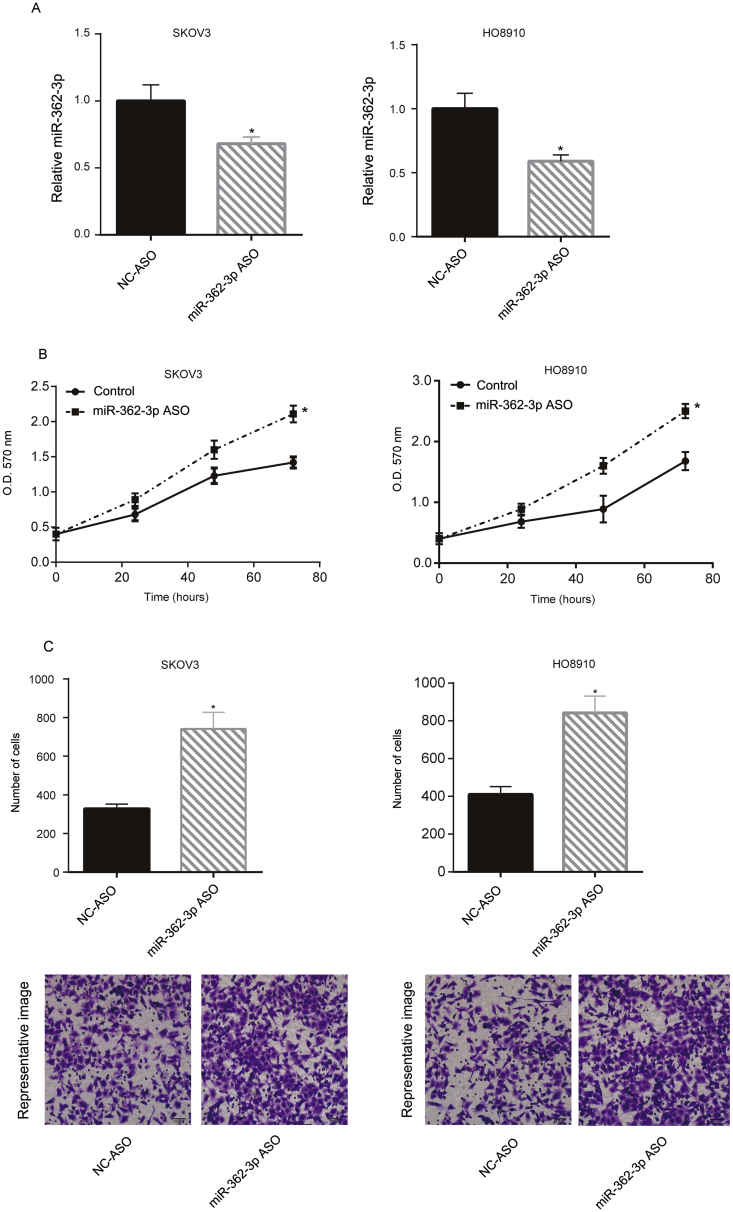
Next, we downregulated the miR-362-3p levels in SKOV3 and HO8910 cells by miR-362-3p ASO transfection. The miR-362-3p levels in these two cell lines were suppressed following miR-362-3p ASO transfection (Figure 3A). miR-362-3p ASO also promoted SKOV3 and HO8910 cell growth at the 72 h following transfection as shown via MTT assay (Figure 3B). Subsequently, we investigated cell migration following transfection, and miR-362-3p ASO increased the number of migrating cells (Figure 3C).
Figure 3.
Downregulation of miR-362-3p promoted SKOV3 and HO8910 proliferation and migration. SKOV3 and HO8910 cells were transfected with miR-362-3p ASO, and miR-NC ASO was used as the negative control. After 24 h, the miR-362-3p levels of the transfected cells were assayed via qRT-PCR. The miR-362-3p level in the miR-NC ASO transfected cells was arbitrarily defined as 1 (A). Following miR-362-3p ASO transfection, the cell growth of SKOV3 and HO8910 were tested via MTT assay at indicated time point (0, 24, 48, 72h) (B). To assess cell migration, SKOV3 and HO8910 cells were added to the upper chamber with a noncoated membrane in each group. After 6 h, the cells in the lower chamber were counted (C). These experiments were performed in triplicate ∗P < 0.05.
3.4. MyD88 is one of the target genes of miR-362-3p
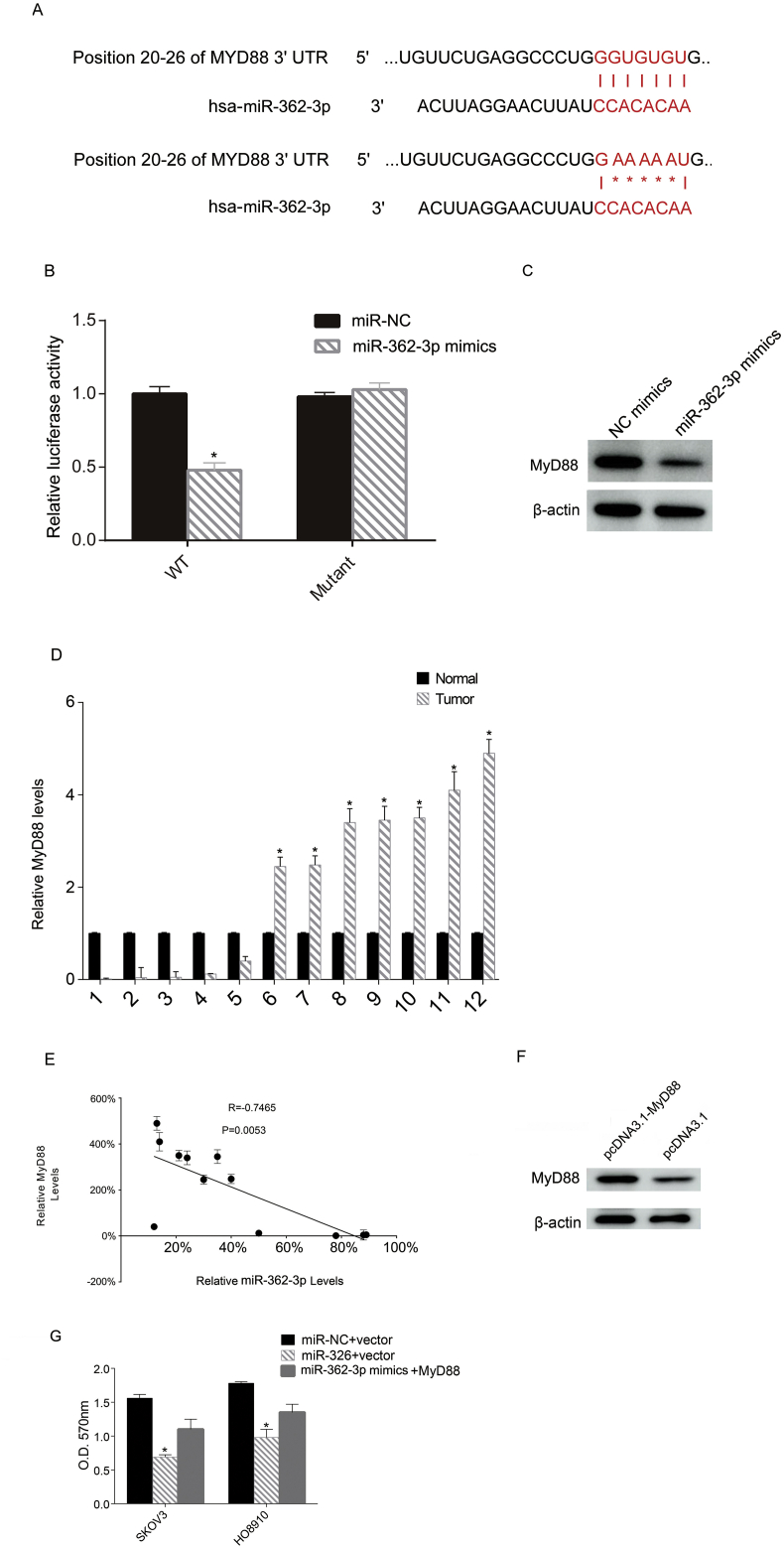
To investigate the underlying mechanism of miR-362-3p in OC, we tested the potential target gene of miR-362-3p, MyD88. A previous study showed strong MyD88 staining and poor survival in low-grade serous ovarian cancer [26]. The online software Targetscan showed that MyD88 is a potential targeted gene. Here, we tested the relationship between miR-362-3p and MyD88 by mutating the binding sites (Figure 4A). Next, the mutated sites were cloned into a luciferase reporter plasmid. miR-362-3p mimics and the reporter plasmid were cotransfected into SKOV3 cells. The activity of luciferase was assessed 12 h following transfection. Up-regulation of miR-362-3p inhibited luciferase activity, whereas the mutated binding site restored it, indicating that miR-362-3p targeted MyD88 in the SKOV3 cells (Figure 4B). Next, we determined the MyD88 protein levels following miR-362-3p mimic transfection and found that miR-362-3p transfection inhibited the MyD88 protein levels in SKOV3 cells (Figure 4C). We then tested the relative Myd88 mRNA levels in the 12 tumor tissues and adjacent normal tissues, and found that tumor tissues showed higher Myd88 levels (Figure 4D). Then we analyzed the correlation between relative miR-362-3p levels and MyD88 mRNA levels via Pearson's correlation coefficient analysis. The relative levels of miR-362-3p and MyD88 were calculated by comparing the miR-362-3p and MyD88 levels in tumor tissue with adjacent normal tissue. The data indicated that MyD88 was inversely correlated with miR-362-3p in epithelial ovarian cancer (Figure 4E). Furthermore, we up-regulated the expression levels of MyD88 in SKOV3 cells by transfection, and the transfection efficacy was confirmed by Western blot (Figure 4F). Then, proliferation analysis via MTT assay of cells overexpressing MyD88 after miR-362-3p mimic transfection revealed that MyD88 overexpression partly reduced the antiproliferative effect of miR-362-3p on these cells (Figure 4G).
Figure 4.
miR-362-3p targets MyD88. The binding sites and mutation location (positions 20–26) are listed (A). miR-362-3p mimics and a plasmid containing either a wild-type or mutated 3′-UTR sequence of MyD88 were transfected into SKOV3 cells. After 48 h, the luciferase activity was analyzed (B). miR-362-3p mimics were transfected into SKOV3 cells and the amount of MyD88 protein was determined via western blot (C). MyD88 levels in 12 OC tissues and their matched adjacent normal tissues were assessed via qRT-PCR. The relative MyD88 levels tumor/normal (log2) are listed (D). Pearson's correlation coefficient analysis revealed that miR-362-3p levels and MyD88 mRNA levels were inversely correlated in the OC tissue samples (E). SKOV3 and HO8910 cells (6×105 cells/well) were transfected with miR-362-3p or miR-NC (NC) separately, followed by transfection 12 h later with an empty plasmid or a MyD88-overexpression plasmid (pcDNA3.1-MyD88). The western blot was performed to test the MyD88 protein levels following transfection (F). An MTT assay was performed, and the OD value is presented as the relative cell number. The relative cell number in the miR-NC + vector (negative control) was defined as l00% (G). The full, not adjusted blot image for figure 4C and figure-4F were include as supplementary materials. (SM1: figure-4C β-actin, SM2: figure-4C MyD88, SM3: figure-4F β-actin, SM4: figure-4F MyD88). Each experiment was repeated at least three times. ∗P < 0.05.
4. Discussion
In this study, we found that miR-362-3p played a suppressive role in EOC. miR-362-3p showed lower expression levels in tumor tissues than in adjacent normal tissue. Forced miR-362-3p expression inhibited cell growth and migration, whereas downregulation of miR-362-3p promoted cell growth and migration. The MyD88 gene is the target of miR-362-3p.
To our knowledge, this is the first report of miR-362-3p function in EOC and miR-362-3p showed a suppressive role in EOC. However, miR-362-3p played a oncogenic role in gastric cancer, and miR-362-3p expression induce gastric cell metastasis capacity by suppression of CD82 expression [18]. We guess that the reason why miR-362-3p showed the opposite role in OC and gastric cancer is that miR-362-3p targeted different genes in various types of cancer.
Previous two studies have suggested that TLR4 and MyD88 expressions are associated with poorer survival in EOC [27, 28], indicating the importance of the tumor-associated inflammatory microenvironment. In another study, it showed that in high-grade serous ovarian carcinomas (HGSOCs). Interesting, the function of MyD88 seemed have related with the grade of serous ovarian cancer [26]. It showed that strong MyD88 expression was modestly associated with shortened overall survival, and in low-grade serous ovarian cancer (LGSOC), strong expression of both MyD88 and TLR4 was associated with favorable survival.
Classically, the TLR4-MyD88 pathway could be activated by pathogen-associated molecular patterns, such as LPS and HMGB1, and will regulate the inflammatory microenvironment [29, 30], TLR4-MyD88 also play unnoticeable roles in tumor or tumor-host response, such as the interaction between HMGB1 and TLR4 dictates the outcome of anticancer chemotherapy and radiotherapy [31] and TLR4-MyD88 could determines the metastatic potential of breast cancer cells [32]. Interestingly, it seemed that MyD88 play dual functional roles in colorectal cancer [33], which indicating the intricate relationship between inflammatory and tumor.
Here our data showed that miR-362-3p may inhibit MyD88 expression, thus it is possible that miR-362-3p may regulate inflammation response and host-tumor response.
MiRNAs may regulate other key factors in the TLR4-MyD88 pathway. MiR-21 is reported to regulate TLR4 by targeting the tumor suppressor protein, PDCD4 [34]. miR-146a is NF-κB-dependent and targets IRAK-1, IRAK-2 and TRAF-6, which are key MyD88 pathway components [35, 36].
A shortcoming of this study was the limited number of OC tumor tissue samples and limitation of the in vivo model. Because MyD88 is associated with poor patient survival in EOC [27], we hypothesized that the low expression of miR-362-3p was associated with poor patient survival in EOC.
In conclusion, miR-362-3p can inhibit cell proliferation and migration and may target MyD88.
Declarations
Author contribution statement
M. Hou: Conceived and designed the experiments; Wrote the paper.
J. Yuan: Performed the experiments.
T. Li: Performed the experiments; Analyzed and interpreted the data.
K. Yi: Contributed reagents, materials, analysis tools or data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
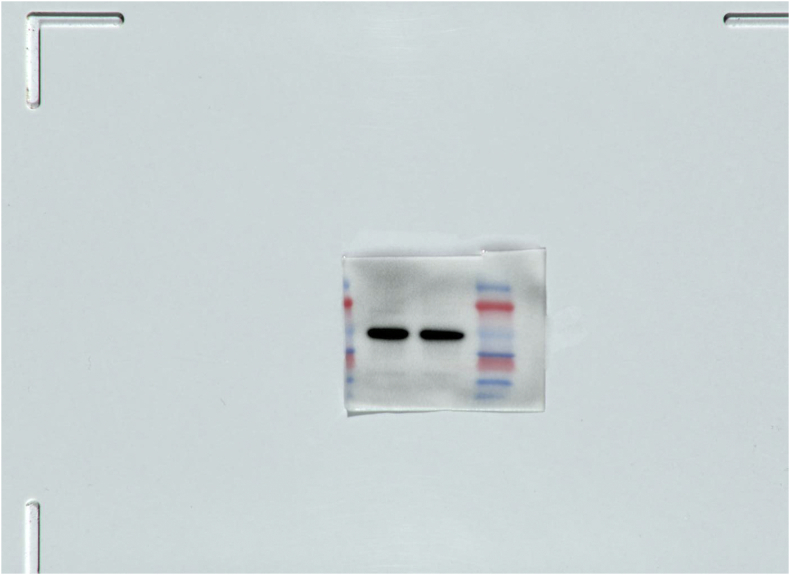
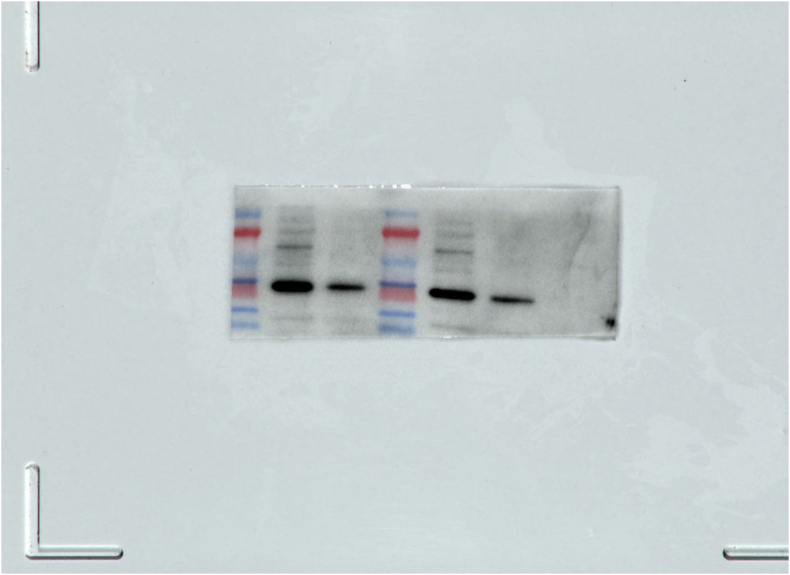
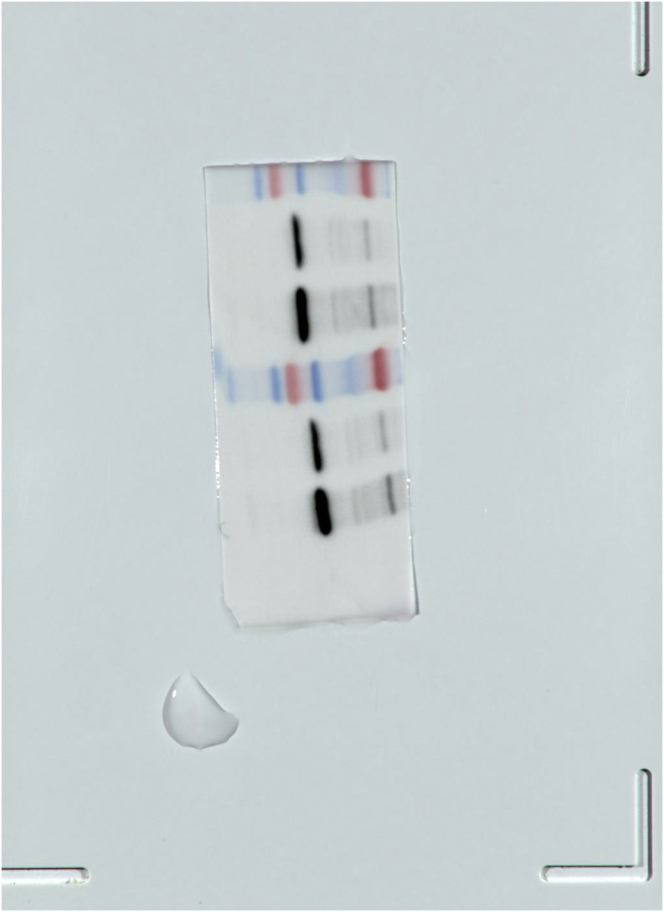
SM1.
The full, not adjusted blot image for figure 4C β-actin.
SM2.
The full, not adjusted blot image for figure 4C MyD88.
SM3.
The full, not adjusted blot image for figure 4F β-actin.
SM4.
The full, not adjusted blot image for figure 4F MyD88.
References
- 1.Ferlay J., Soerjomataram I., Ervik M., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D., Forman D., Bray F. International Agency for Research on Cancer; Lyon, France: 2014. Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. 2013. [Google Scholar]
- 2.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Reid B.M., Permuth J.B., Sellers T.A. Epidemiology of ovarian cancer: a review. Cancer Biol. Med. 2017;14:9–32. doi: 10.20892/j.issn.2095-3941.2016.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berek J.S., Bast R.C., Jr. 2003. Epithelial Ovarian Cancer. [Google Scholar]
- 5.Yap T.A., Carden C.P., Kaye S.B. Beyond chemotherapy: targeted therapies in ovarian cancer. Nat. Rev. Canc. 2009;9:167. doi: 10.1038/nrc2583. [DOI] [PubMed] [Google Scholar]
- 6.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calin G.A., Croce C.M. MicroRNA-cancer connection: the beginning of a new tale. Canc. Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 8.Xie B., Ding Q., Han H., Wu D. miRCancer: a microRNA–cancer association database constructed by text mining on literature. Bioinformatics. 2013;29:638–644. doi: 10.1093/bioinformatics/btt014. [DOI] [PubMed] [Google Scholar]
- 9.Farazi T.A., Hoell J.I., Morozov P., Tuschl T. Springer; 2013. MicroRNAs in Human Cancer, MicroRNA Cancer Regulation; pp. 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nugent M. Springer; 2015. microRNA and Bone Cancer, microRNA: Cancer; pp. 201–230. [DOI] [PubMed] [Google Scholar]
- 11.Del Vescovo V., Denti M.A. Springer; 2015. microRNA and Lung Cancer, microRNA: Cancer; pp. 153–177. [DOI] [PubMed] [Google Scholar]
- 12.Langhe R. Springer; 2015. microRNA and Ovarian Cancer, microRNA: Cancer; pp. 119–151. [DOI] [PubMed] [Google Scholar]
- 13.Volinia S., Calin G.A., Liu C.-G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng F., Henson R., Wehbe–Janek H., Ghoshal K., Jacob S.T., Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iorio M.V., Ferracin M., Liu C.-G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M. MicroRNA gene expression deregulation in human breast cancer. Canc. Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 16.Taylor D.D., Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 17.Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., Yi M., Stephens R.M., Okamoto A., Yokota J., Tanaka T. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Canc. Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q.H., Yao Y.L., Wu X.Y., Wu J.H., Gu T., Chen L., Gu J.H., Liu Y., Xu L. Anti-miR-362-3p inhibits migration and invasion of human gastric cancer cells by its target CD82. Dig. Dis. Sci. 2015;60:1967–1976. doi: 10.1007/s10620-015-3563-6. [DOI] [PubMed] [Google Scholar]
- 19.Wang D., Wang H., Li Y., Li Q. MiR-362-3p functions as a tumor suppressor through targeting MCM5 in cervical adenocarcinoma. Biosci. Rep. 2018;38 doi: 10.1042/BSR20180668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou X., Zhong J., Li J., Su Z., Chen Y., Deng W., Li Y., Lu S., Lin Y., Luo L., Li Z., Cai Z., Tang A. miR-362-3p targets nemo-like kinase and functions as a tumor suppressor in renal cancer cells. Mol. Med. Rep. 2016;13:994–1002. doi: 10.3892/mmr.2015.4632. [DOI] [PubMed] [Google Scholar]
- 21.Gerlier D., Thomasset N. Use of MTT colorimetric assay to measure cell activation. J. Immunol. Methods. 1986;94:57–63. doi: 10.1016/0022-1759(86)90215-2. [DOI] [PubMed] [Google Scholar]
- 22.Rüster B., Grace B., Seitz O., Seifried E., Henschler R. Induction and detection of human mesenchymal stem cell migration in the 48-well reusable transwell assay. Stem Cell. Dev. 2005;14:231–235. doi: 10.1089/scd.2005.14.231. [DOI] [PubMed] [Google Scholar]
- 23.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015:4. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia D.M., Baek D., Shin C., Bell G.W., Grimson A., Bartel D.P. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Block M.S., Vierkant R.A., Rambau P.F., Winham S.J., Wagner P., Traficante N., Toloczko A., Tiezzi D.G., Taran F.A., Sinn P., Sieh W., Sharma R., Rothstein J.H., Ramon Y.C.T., Paz-Ares L., Oszurek O., Orsulic S., Ness R.B., Nelson G., Modugno F., Menkiszak J., McGuire V., McCauley B.M., Mack M., Lubinski J., Longacre T.A., Li Z., Lester J., Kennedy C.J., Kalli K.R., Jung A.Y., Johnatty S.E., Jimenez-Linan M., Jensen A., Intermaggio M.P., Hung J., Herpel E., Hernandez B.Y., Hartkopf A.D., Harnett P.R., Ghatage P., Garcia-Bueno J.M., Gao B., Fereday S., Eilber U., Edwards R.P., de Sousa C.B., de Andrade J.M., Chudecka-Glaz A., Chenevix-Trench G., Cazorla A., Brucker S.Y., Australian Ovarian Cancer Study G., Alsop J., Whittemore A.S., Steed H., Staebler A., Moysich K.B., Menon U., Koziak J.M., Kommoss S., Kjaer S.K., Kelemen L.E., Karlan B.Y., Huntsman D.G., Hogdall E., Gronwald J., Goodman M.T., Gilks B., Garcia M.J., Fasching P.A., de Fazio A., Deen S., Chang-Claude J., Candido Dos Reis F.J., Campbell I.G., Brenton J.D., Bowtell D.D., Benitez J., Pharoah P.D.P., Kobel M., Ramus S.J., Goode E.L. MyD88 and TLR4 expression in epithelial ovarian cancer. Mayo Clin. Proc. 2018;93:307–320. doi: 10.1016/j.mayocp.2017.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z., Block M.S., Vierkant R.A., Fogarty Z.C., Winham S.J., Visscher D.W., Kalli K.R., Wang C., Goode E.L. The inflammatory microenvironment in epithelial ovarian cancer: a role for TLR4 and MyD88 and related proteins. Tumour Biol. 2016;37:13279–13286. doi: 10.1007/s13277-016-5163-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.d'Adhemar C.J., Spillane C.D., Gallagher M.F., Bates M., Costello K.M., Barry-O'Crowley J., Haley K., Kernan N., Murphy C., Smyth P.C., O'Byrne K., Pennington S., Cooke A.A., Ffrench B., Martin C.M., O'Donnell D., Hennessy B., Stordal B., Finn S., McCann A., Gleeson N., D'Arcy T., Flood B., O'Neill L.A., Sheils O., O'Toole S., O'Leary J.J. The MyD88+ phenotype is an adverse prognostic factor in epithelial ovarian cancer. PloS One. 2014;9 doi: 10.1371/journal.pone.0100816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y., Yin H., Zhao M., Lu Q. TLR2 and TLR4 in autoimmune diseases: a comprehensive review. Clin. Rev. Allergy Immunol. 2014;47:136–147. doi: 10.1007/s12016-013-8402-y. [DOI] [PubMed] [Google Scholar]
- 30.Takeda K., Akira S. TLR signaling pathways. Semin. Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Apetoh L., Ghiringhelli F., Tesniere A., Criollo A., Ortiz C., Lidereau R., Mariette C., Chaput N., Mira J.P., Delaloge S., Andre F., Tursz T., Kroemer G., Zitvogel L. The interaction between HMGB1 and TLR4 dictates the outcome of anticancer chemotherapy and radiotherapy. Immunol. Rev. 2007;220:47–59. doi: 10.1111/j.1600-065X.2007.00573.x. [DOI] [PubMed] [Google Scholar]
- 32.Wu K., Zhang H., Fu Y., Zhu Y., Kong L., Chen L., Zhao F., Yu L., Chen X. TLR4/MyD88 signaling determines the metastatic potential of breast cancer cells. Mol. Med. Rep. 2018;18:3411–3420. doi: 10.3892/mmr.2018.9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L., Yu K., Zhang X., Yu S. Dual functional roles of the MyD88 signaling in colorectal cancer development. Biomed. Pharmacother. 2018;107:177–184. doi: 10.1016/j.biopha.2018.07.139. [DOI] [PubMed] [Google Scholar]
- 34.Sheedy F.J., Palsson-McDermott E., Hennessy E.J., Martin C., O'Leary J.J., Ruan Q., Johnson D.S., Chen Y., O'Neill L.A. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 35.Liston A., Linterman M., Lu L.F. MicroRNA in the adaptive immune system, in sickness and in health. J. Clin. Immunol. 2010;30:339–346. doi: 10.1007/s10875-010-9378-5. [DOI] [PubMed] [Google Scholar]
- 36.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]