Abstract
Food safety is of significant concern to consumers and is a major issue for the food industry. As such, the industry is aware of the importance of safety assessments of starters used in the production of fermented foods. Coagulase-negative staphylococci (CNS) are the predominant bacteria found in fermented foods worldwide. Because of this, food-derived CNS are used as starters for cheese and meat fermentation, and have been investigated for use as starters in soybean fermentation. Although food-derived CNS are generally considered non-pathogenic, their safety remains uncertain following the isolation of CNS from skin infections in humans and animals, and because they belong to the same genus as the highly pathogenic species Staphylococcus aureus. This review explores what is known about the safety of food-derived CNS, focusing on antibiotic resistance, enterotoxin genes, and biogenic amine production, to aid in the selection of starter candidates.
Keywords: Food-derived coagulase negative staphylococci, Starter culture, Safety, Diversity
Introduction
Staphylococci, which are identified under the microscope by the formation of irregular grape-like clusters of cells, are Gram-positive, facultatively anaerobic, catalase-positive, non-motile, and non-spore-forming bacteria with a high tolerance for salt (most strains survive in the presence of 10% NaCl) (Chapman, 1945; Götz et al., 2006). They are part of the normal microbiota of the skin and mucous membranes of both humans and animals, and are also ubiquitously distributed in a variety of niches, including soil, water, and air, as well as various foodstuffs (Coton et al., 2010b; Götz et al., 2006; Irlinger, 2008).
Staphylococci are classified as either coagulase-positive or coagulase-negative based on their ability to produce coagulase. Coagulase production is considered a virulence factor in staphylococci because it is associated with escape from phagocytic cells. Generally, coagulase-positive staphylococci (CPS) are pathogenic and/or toxigenic and are capable of causing infections such as pneumonia and food poisoning (Archer, 1998). Staphylococcus aureus is the representative CPS species and is a well-known virulent pathogen (Tong et al., 2015). In contrast, coagulase-negative staphylococci (CNS) are generally considered benign, despite some species (Staphylococcus epidermidis, Staphylococcus haemolyticus, and Staphylococcus saprophyticus) occasionally causing opportunistic infections (Kacica et al., 1994; Natoli et al., 2009; Widerstrom et al., 2012). As a result, few studies have examined the virulence of CNS species associated with fermented foods, such as Staphylococcus carnosus, Staphylococcus equorum, and Staphylococcus xylosus.
CNS reportedly contribute to the sensory properties of fermented foods, including color, aroma, and taste. As such, they are often used as starter cultures for meat and cheese fermentation to enhance color and flavor development (Brigante et al., 2008; Hugas and Monfort, 1997; Irlinger, 2008; Leroy et al., 2006; Schleifer et al. 2009). CNS have also been isolated from seafood- and soybean-based foods fermented at high-salt concentrations (Guan et al., 2011; Jeong et al., 2014a, 2014b), and Staphylococcus succinus was reported to produce species-specific volatile compounds during soybean fermentation when used as a starter culture (Jeong et al., 2017a, 2019). Despite their contribution to the sensory properties of fermented foods, CNS have not gained the qualified presumption of safety (QPS) status for use in food and feed in the European Union (EFSA, 2016). This lack of acknowledgement as food-safe microorganisms in the European Union and undocumented usage history in Korea hinders the introduction of CNS for use in fermentation in Korean food production. In this review, we explore the possibility of using CNS as starter cultures by comparing the safety of CNS species from different origins, and also examine the roles of CNS during fermentation.
Classification, habitat, and virulence of CNS
As of 2019, the genus Staphylococcus comprised 53 species and 27 subspecies in the List of Prokaryotic Names with Standing in Nomenclature (Parte, 2018). Of these, 40 species and 24 subspecies were CNS, which were classified into four representative groups by colonization: S. epidermidis group, Staphylococcus lugdunensis, S. saprophyticus subsp. saprophyticus, and other CNS (Becker et al., 2014). S. epidermidis group species colonize human body surfaces, S. lugdunensis colonizes human skin, and S. saprophyticus colonizes the human rectum and genitourinary tract. In comparison, bacteria belonging to the other CNS species group are only found in fermented foods and used as starter cultures (Becker et al., 2014).
Becker et al. (2014) reported that S. epidermidis, S. haemolyticus, Staphylococcus capitis, Staphylococcus pettenkoferi, and Staphylococcus auricularis belonged to the S. epidermidis group, which accounts for a substantial number of foreign body-related infections and infections in preterm newborns. S. epidermidis-group staphylococci have been isolated from various human and animal body surfaces, both healthy and infected. Despite generally having a benign relationship with its host, S. epidermidis is the representative opportunistic pathogen among the CNS (Otto, 2009). Yet unlike S. aureus, the S. epidermidis toxins responsible for infection in humans have not been clearly determined (Otto, 2012). S. haemolyticus, S. capitis, and S. pettenkoferi, all members of the S. epidermidis group, have been isolated from clinical infection patients, a prosthetic joint infection, and a bacteremia case, respectively (Barros et al., 2012; Hashi et al., 2015; Tevell et al., 2017). Interestingly, approximately 55% of clinical S. haemolyticus isolates contain methicillin resistance genes (Barros et al., 2012), although the toxin genes remain ambiguous. Some S. epidermidis strains have also been shown to contain the staphylococcal cassette chromosome mec (SSCmec), coding for methicillin resistance (Otto, 2013). However, antibiotic resistance is not directly associated with virulence. Although these species have been isolated from clinical infection cases and were shown to contain virulence factors such as adhesion factors and immune effectors (Sabate Bresco et al., 2017), their direct clinical impact might be underreported. In addition, given the reports of infection caused by S. epidermidis group species, these bacteria may have a higher clinical burden than other CNS.
Staphylococcus lugdunensis, a normal inhabitant of human skin, was first described by Freney et al. (1988) and has never been isolated from fermented foods. S. lugdunensis-associated skin infections have been reported, causing symptoms similar to those of S. aureus infection (Bieber and Kahlmeter, 2010). However, it is not clear what virulence factors S. lugdunensis uses for infection. In Greece, S. lugdunensis was isolated from various infection sites in hospitalized patients (Giormezis et al., 2015). Bierowiec et al. reported that while most tested S. lugdunensis isolates were susceptible to nine antibiotics (penicillin G, gentamicin, rifampicin, cefoxitin, fusidic acid, trimethoprim/sulfamethoxazole, norfloxacin, clindamycin, and erythromycin), some showed resistance to erythromycin and/or tetracycline via strain-specific carriage of ermC and/or tetK (Bierowiec et al., 2019). In addition, a very small number of S. lugdunensis isolates carried SSCmec type IVa. In contrast, a survey of clinical S. lugdunensis isolates by Giormezis et al. (2015) found high rates of resistance to ampicillin (50%), erythromycin (18.4%), and clindamycin (18.4%).
Staphylococcus saprophyticus subsp. saprophyticus is frequently isolated from young female outpatients with uncomplicated urinary tract infections but is rarely detected in healthy individuals, and has occasionally been implicated in cases of pyelonephritis, septicemia, nephrolithiasis, and endocarditis (Raz et al., 2005). Genome analysis revealed that S. saprophyticus ATCC 15305T does not possess genes coding for common S. aureus virulence factors such as coagulase and enterotoxins but does contain genes responsible for urease production and adhesion to uroepithelial cells, which might be involved in establishing uncomplicated urinary tract infections (Kuroda et al., 2005). While S. saprophyticus surface-associated protein and lipase have been suggested as potential virulence factors (Sakinc et al., 2005, 2007; Szabados et al., 2013; Tang et al., 2009), no investigations into the pathogenicity of S. saprophyticus in the urinary tract or the expression of potential virulence genes in this environment have been published as yet.
Staphylococcus saprophyticus is also commonly detected in fermented foods such as Greek and Italian fermented sausages (Drosinos et al., 2005; Mauriello et al., 2004; Papamanoli et al., 2002), Taiwanese naturally-fermented dry ham (Tu et al., 2010), and Korean fermented soybeans (Jeong et al., 2014b). It was also associated with the improvement of sausage aromatization during ripening (Mauriello et al., 2004; Samelis et al., 1998). Staphylococcus saprophyticus has been isolated and assessed for use as a starter culture for fermentation of French and Spanish dry sausages (Fonseca et al., 2013; Montel et al., 1993), as well as Korean fermented soybeans (Jeong et al., 2016a). Multilocus sequence typing (MLST)-based analysis of S. saprophyticus subsp. saprophyticus strains from fermented foods and clinical specimens revealed a correlation between the genetic backgrounds of the strains and their origins, and identified a link between genetic background and acquisition of lincomycin resistance (Lee et al., 2015). The results suggested that functional properties such as virulence and antimicrobial susceptibility might differ in an origin-dependent manner among S. saprophyticus subsp. saprophyticus strains. However, the notoriety of S. saprophyticus as an uropathogen hinders its application as a starter for fermented foods despite its documented contribution to the sensory properties of various foods during fermentation. Therefore, more evidence is needed to confirm the origin-dependence of the genetic backgrounds of S. saprophyticus strains before they can be used in food fermentation.
Other CNS classified by Becker et al. include S. carnosus, Staphylococcus condimenti, S. equorum, Staphylococcus piscifermentans, S. succinus, and S. xylosus (Becker et al., 2014), all of which are typically associated with fermented foods (Irlinger, 2008). Irlinger suggested that food-derived CNS from milk or dairy products should be classified as exceptional opportunistic pathogens (Irlinger, 2008) because although they are primarily nonpathogenic, there are isolated reports of these CNS species being recovered from clinical samples (Blaiotta et al., 2004; Novakova et al., 2006). Coton et al. (2010a) investigated 297 clinical CNS isolates and showed that they predominantly belonged to the species S. epidermidis, S. capitis, Staphylococcus hominis, Staphylococcus warneri, and S. haemolyticus, and none were identified as food-associated species S. carnosus, S. condimenti, S. equorum, S. piscifermentans, S. succinus, or S. xylosus. However, the relationship between food-derived CNS and human infection is still unclear.
Safety of CNS derived from fermented foods
The Food Safety Authority of the European Union has introduced the QPS approach for safety assessment of microorganisms throughout the food chain, with emphasis on acquired antibiotic resistance, presence of toxin genes (including hemolysin genes), and production of biogenic amines (EFSA, 2004). Four species, S. carnosus, S. equorum, S. succinus, and S. xylosus, account for the majority of food-derived CNS isolates from fermented meat, sausage, soybean products, and fish sauce (Becker et al., 2014; Guan et al., 2011; Jeong et al., 2014b; Mauriello et al., 2004; Sondergaard and Stahnke, 2002). As such, several studies including safety assessments of these four CNS species using the QPS approach have been undertaken.
Staphylococcus carnosus
Staphylococcus carnosus has been used as a starter culture for sausage fermentation since the 1950s (Lofblom et al., 2017). This long usage history has shown it to be a benign food-grade species, with complete genome sequencing of S. carnosus TM300 revealing that it lacks genes required for pathogenicity (Rosenstein et al., 2009).
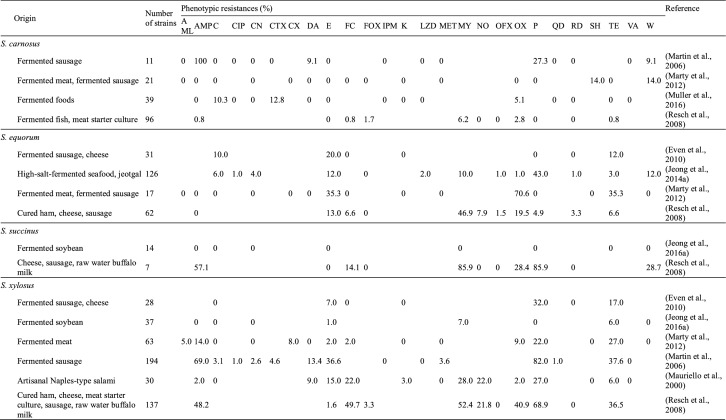
Most S. carnosus isolates from various fermented foods are sensitive to antibiotics; however, a few strains have shown resistance to a small number of antibiotics (Martin et al., 2006; Marty et al., 2012; Muller et al., 2016; Resch et al., 2008) (Table 1). For example, Muller et al. (2016) studied the antibiotic resistance profiles of 39 S. carnosus strains isolated from fermented sausage meat and found that while all 39 strains were susceptible to ampicillin, amoxicillin/clavulanic acid, ciprofloxacin, clindamycin, erythromycin, gentamicin, imipenem, kanamycin, linezolid, quinupristin/dalfopristin, rifampicin, tetracycline, and vancomycin), four and two strains showed resistance or intermediate resistance to chloramphenicol and oxacillin, respectively. Similarly, Marty et al. (2012) showed that while all 21 S. carnosus strains from fermented meats were susceptible to ampicillin, amoxicillin, clindamycin, cloxacillin, erythromycin, fusidic acid, methicillin, oxacillin, penicillin G, and tetracycline, 14% of the strains were resistant to streptomycin and trimethoprim (Marty et al., 2012). Interestingly, and unusually for S. carnosus strains, another study showed that while all tested S. carnosus strains from fermented sausages were sensitive to 12 different antibiotics, all 11 strains were resistant to ampicillin (Martin et al., 2006). Together, these findings suggest that S. carnosus has low overall rates of antibiotic resistance, and that any antibiotic resistance that does occur appears to be strain specific.
Table 1.
Antibiotic resistance profiles of food-derived coagulase-negative staphylococci
Blank box indicates that the antibiotic was not assessed
Antibiotics: AML, amoxicillin; AMP, ampicillin; C, chloramphenicol; CIP, ciprofloxacin; CN, gentamicin; CTX, cefotaxime; CX, cloxacillin; DA, clindamycin; E, erythromycin; FC, fusidic acid; FOX, cefoxitin; IPM, imipenem; K, kanamycin; LZD, linezolid; MET, methicillin; MY, lincomycin; NO, novobiocin; OFX, ofloxacin; OX, oxacillin; P, penicillin G; QD, quinupristin/dalfopristin; RD, rifampin; SH, streptomycin; TE, tetracycline; VA, vancomycin; W, trimethoprim
Staphylococcal enterotoxin genes, exfoliative toxin genes, and the toxic shock syndrome toxin-encoding gene have not been amplified from any of the S. carnosus strains examined to date (Table 2) (Martin et al., 2006; Muller et al., 2016). Only one among the 39 strains examined by Muller et al. in 2016 showed weak hemolysis activity (Muller et al., 2016), while a hemolysin gene was amplified from a second strain. Thus, the phenotype and genotype did not match for either strain. To date, there have been several reports of S. carnosus isolates showing hemolysis on solid media and successful PCR-based amplification of a partial hemolysin gene from some strains; however, the in vitro and in vivo virulence of S. carnosus has not been reported (Muller et al., 2016). Importantly though, the genome sequence of S. carnosus TM300 lacks virulence genes encoding the enterotoxin and hemolysin (Rosenstein et al., 2009).
Table 2.
Prevalence of hemolysis, hemolysin, and toxin genes among food-derived coagulase-negative staphylococci
| S. carnosus | S. equorum | S. succinus | S. xylosus | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hemolysis | ||||||||||
| Horse blood | 0 | 0 | 11.1 | |||||||
| Sheep blood | 2.4 | 0 | 16.7 | |||||||
| Human blood | 60.0 | 2.6 | 88.2 | 3.0 | ||||||
| Hemolysin genes | ||||||||||
| hla | 0 | 2.6 | 0 | 0 | 0 | 0 | ||||
| hlb | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| hld | 0 | 2.6 | 23.5 | 0 | 0 | 0 | 3.6 | |||
| SE_0613 | 0 | 0 | 3.2 | 0 | 0 | 0 | ||||
| SE_1760 | 0 | 0 | 3.2 | 0 | 0 | 0 | ||||
| Enterotoxin genes | ||||||||||
| sea | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| seb | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| sec | 0 | 0 | 0 | 0 | 0 | 1.5 | 0 | |||
| sed | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| see | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| seg | 0 | 5.9 | 0 | |||||||
| seh0 | 0 | 0 | 0 | |||||||
| sei | 0 | 0 | 0 | |||||||
| sej | 5.9 | 0 | 0 | |||||||
| selj | 0 | 0 | ||||||||
| selr | 0 | 0 | ||||||||
| set9 | 0 | 0 | ||||||||
| Exfoliative toxin genes | ||||||||||
| eta | 0 | 0 | 0 | 0 | 0 | |||||
| etb | 0 | 0 | 0 | 0 | ||||||
| Toxic shock syndrome toxin gene | ||||||||||
| tst-1 | 0 | 0 | 0 | 0 | 0 | |||||
| Number of strains | 11 | 5 | 39 | 17 | 31 | 126 | 14 | 194 | 28 | 18 |
| References | Martin et al. (2006) | Marty et al. (2012) | Muller et al. (2016) | Marty et al. (2012) | Even et al. (2010) | Jeong et al. (2014a) | Jeong et al. (2016a) | Martin et al. (2006) | Even et al. (2010) | Jeong et al. (2016a) |
Blank box indicates an assay was not performed
SE_0613 and SE_1760 are the hemolysin- and hemolysin III-encoding genes, respectively, from S. epidermidis ATCC 12228
hylIII, hemolysin III-encoding gene from Bacillus cereus
Staphylococcus carnosus does not produce the biogenic amines cadaverine, putrescine, tryptamine, tyramine, or histamine. However, some strains do produce phenethylamine, which is derived from phenylalanine via decarboxylation (Table 3) (Martin et al., 2006; Muller et al., 2016), although the amounts produced, even with an abundance of the precursor amino acid, are below the allowable concentration listed in Codex (100–200 μg/mL). In addition, the phenylalanine decarboxylase gene, which is required for phenethylamine production, was not detected in the genome of S. carnosus TM300 (Rosenstein et al., 2009).
Table 3.
Prevalence of in vitro production of biogenic amines and associated genes in coagulase-negative staphylococci
| Biogenic amines | Number of strains | Method | References | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cadaverine | Histamine | Phenyl-ethylamine | Putrescine | Tryptamine | Tyramine | ||||||||||
| Pa | Gb | Pa | Gb | Pa | Gb | Pa | Gb | Pa | Gb | Pa | Gb | ||||
| S. carnosus | |||||||||||||||
| 0 | 0 | 90.9 | 0 | 0 | 0 | 11 | HPLC | Martin et al. (2006) | |||||||
| 0 | 0 | 63.2 | 0 | 39 | HPLC | Muller et al. (2016) | |||||||||
| S. equorum | |||||||||||||||
| – | 3.2 | – | 0 | (116 μg/mL)c | 0 | (23 μg/mL)c | 0 | 31 | LC | Even et al. (2010) | |||||
| (29.6 ppm) d | (40.0 ppm) d | (22.6 ppm) d | (29.7 ppm) d | 39 | HPLC | Jeong et al. (2014a) | |||||||||
| 85.7 | 0 | 14.3 | 0 | 0 | 0 | 7 | Medium | Bonomo et al. (2009) | |||||||
| S. succinus | |||||||||||||||
| 100 | 0 | 0 | 0 | 0 | 2 | Medium | Bonomo et al. (2009) | ||||||||
| 76.2 | 0 | 32 | 76.2 | 41.3 | 10.5 | 9 | HPLC | Jeong et al. (2016a) | |||||||
| S. xylosus | |||||||||||||||
| 49.0 | 49.0 | 51 | HPLC | Martuscelli et al. (2000) | |||||||||||
| 1 | 0.5 | 6.7 | 1.5 | 0.5 | 3.6 | 194 | HPLC | Martin et al. (2006) | |||||||
| 0 | 0 | 0 | 0 | 0 | 28 | LC | Even et al. (2010) | ||||||||
| 41.2 | 5.9 | 0 | 5.9 | 0 | 17 | Medium | Bonomo et al. (2009) | ||||||||
| 72.1 | 0 | 32.8 | 72.1 | 42.8 | 11 | 2 | HPLC | Jeong et al. (2016a) | |||||||
Blank box indicates an assay was not conducted
–Concentration below the limit of detection
aPhenotype
bGenotype
cBiogenic amines produced by only 1 out of 31strains
dConcentration of biogenic amines produced by the highest producers
Staphylococcus equorum
Staphylococcus equorum is a common component of the microbial communities of high-salt-fermented foods produced in Europe such as meat products and smear-ripened and semi-hard cheeses (Blaiotta et al., 2004; Bockelmann et al., 2005; Corbiere Morot-Bizot et al., 2006; Mauriello et al., 2004; Place et al., 2003). In addition, S. equorum has been developed as a starter culture for fermented foods such as sausage and jeotgal (Jeong et al., 2014a; Mauriello et al., 2004; Place et al., 2003; Sondergaard and Stahnke, 2002). The genome sequence of S. equorum Mu2 from a French smear-ripened cheese revealed that it does not contain any of the virulence factors found in S. aureus (Irlinger et al., 2012), and genomic analysis of S. equorum KS1039, which has been selected as a starter candidate for jeotgal, confirmed that it is also missing the S. aureus virulence genes (Jeong et al., 2016b).
In a study by Even et al. (2010), all 31 tested S. equorum strains were susceptible to fusidic acid, kanamycin, penicillin G, and rifampicin, with fewer than 20% of strains found to be resistant to chloramphenicol, erythromycin, and tetracycline (Table 1). Marty et al. (2012) reported similar findings, with 17 strains isolated from fermented meats showing susceptibility to ampicillin, amoxicillin, chloramphenicol, clindamycin, cloxacillin, gentamycin, fusidic acid, kanamycin, methicillin, penicillin G, streptomycin, and trimethoprim, and approximately 20% of strains showing resistance to erythromycin, oxacillin, and tetracycline. In comparison, Resch et al. (2008) found that most S. equorum isolates were resistant to at least one antibiotic, with 46.9% of the tested isolates demonstrating resistance to lincomycin. As well as being common in fermented foods in Europe, S. equorum has also been identified as the dominant microbial species in jeotgal, a high-salt-fermented seafood product from Korea (Guan et al., 2011; Jeong et al., 2014a). One study found that 66/126 ampicillin-sensitive S. equorum strains from jeotgal exhibited phenotypic resistance to at least one antibiotic, with the highest rates of resistance observed for penicillin G (34.1%), erythromycin (9.5%), and trimethoprim (9.5%). Therefore, among the food-derived CNS, the antibiotic resistance of S. equorum appears to be unrivaled. Comparative genomic analysis of six S. equorum strains revealed the presence of several antibiotic resistance genes, although four of the strains were sensitive to the corresponding antibiotics (Jeong et al., 2017a). In the two remaining strains, antibiotic resistance was conferred by acquired antibiotic resistance genes.
Safety assessments of 17 S. equorum isolates from spontaneously-fermented Swiss meat products showed no antibiotic resistance and produced negative results for other safety concerns such as hemolysis, hemagglutination, virulence factors, and biogenic amines (Marty et al., 2012) (Table 2). In addition, while staphylococcal enterotoxin genes were not present in most of the isolates, partial seg and sej gene sequences were amplified from one isolate (Table 2) (Marty et al., 2012). Further screening of S. equorum isolates from jeotgal (n = 126) and fermented sausages and cheese (n = 31) also confirmed the absence of staphylococcal enterotoxin genes (Even et al., 2010; Jeong et al., 2014a). However, a complete enterotoxin gene sequence and the contribution of enterotoxins to virulence in S. equorum have not been reported. Weak hemolysis and moderate hemolytic activity were frequently observed among S. equorum isolates in a study by Marty et al. (2012), and a partial δ-hemolysin-encoding gene (hld) was amplified from one isolate. Similarly, S. equorum isolates from jeotgal commonly demonstrated δ-hemolysis, although α- and β-hemolysis were infrequently detected (Jeong et al., 2014a). Comparative genomic analysis of six S. equorum strains failed to identify α-, β-, and δ-hemolysin genes with homology to those observed in S. aureus (Jeong et al., 2017b), although several other annotated hemolysin genes were present. Interestingly, these other hemolysin genes are often found in non-hemolytic S. equorum strains, while hemolytic strains show strain-specific carriage of a gene encoding a hemolysin activation protein and a hemolysin-family calcium-binding region (Jeong et al., 2017b). These results suggest that S. equorum does not generally carry enterotoxin or hemolysin genes but some specific strains may have acquired them.
Most S. equorum isolates from traditionally-fermented sausages produced in the Basilicata region of Italy tested positive for decarboxylation of lysine and phenylalanine based on a color change on indicator media (Bonomo et al., 2009). However, 78% of the isolates did not carry any genes involved in biogenic amine production (Even et al., 2010). High-performance liquid chromatography (HPLC) analysis showed that some S. equorum isolates from jeotgal produced low concentrations of cadaverine, histamine, putrescine, and tyramine in media supplemented with amino acid precursors (Table 3)(Jeong et al., 2014a). However, comparative genomic analysis of six different strains revealed that while the S. equorum strains carried genes involved in cadaverine production, none of the genes required for production of histamine, putrescine, or tyramine were present in any of the genomes (Lee et al., 2018). These results confirm that S. equorum is fundamentally a benign species.
Staphylococcus succinus
Staphylococcus succinus is one of the predominant bacterial species in fermented sausage (Corbiere Morot-Bizot et al., 2006; Mauriello et al., 2004; Talon et al., 2008) and is used as a starter culture in various meat fermentation processes (Talon et al., 2008). Resch et al. (2008) found that S. succinus has high levels of antibiotic resistance against ampicillin, lincomycin, and penicillin G. However, safety assessments of 81 CNS isolates from fermented soybeans showed that none of the S. succinus isolates exhibited antibiotic resistance or hemolytic activity (Jeong et al., 2016a). Among them, strain 14BME20 was selected as a starter candidate. Genomic analysis confirmed that strain 14BME20 did not contain any of the known S. aureus virulence factor-encoding genes but did carry strain-specific genes for lipid degradation, which may contribute to the production of volatile compounds (Jeong and Lee, 2017).
Staphylococcus succinus isolates from doenjang, a high-salt-fermented soybean product, did not exhibit hemolysis (Jeong et al., 2016a) and attempts to amplify enterotoxin genes were unsuccessful (unpublished results). However, the isolates did produce cadaverine, putrescine, and tyramine, but not histamine, in a laboratory setting (Jeong et al., 2016a). Additionally, genome sequencing of the S. succinus isolates suggested that the genes required for histamine, putrescine, and tyramine production were missing from all isolates, although a lysine decarboxylase-encoding gene, which is needed for cadaverine production, was identified (Jeong and Lee, 2017; Megaw and Gilmore, 2016; Zhou et al., 2017). Cadaverine production by the isolates averaged 75.1 ppm in medium supplemented with excess precursors, suggesting that if precursor concentrations were restricted, cadaverine production should be very low.
Staphylococcus xylosus
Like S. succinus, S. xylosus is a predominant species in fermented sausages (Blaiotta et al., 2004; Corbiere Morot-Bizot et al., 2006; Mauriello et al., 2004; Talon et al., 2008) and is used as a starter culture in fermented sausage production because of its contribution to color and flavor development (Fiorentini et al., 2009; Martin et al., 2006; Talon et al., 2008). Genome sequence analysis of a S. xylosus strains used as a meat starter culture revealed that all known S. aureus genes coding for enterotoxins and virulence factors were missing from the genome (Labrie et al., 2014).
However, S. xylosus isolates have shown high levels of resistance to ampicillin, penicillin G, and tetracycline (Even et al., 2010; Martin et al., 2006; Marty et al., 2012; Mauriello et al., 2000; Resch et al., 2008), although the mecA gene, which is responsible for methicillin resistance, has only rarely been identified in S. xylosus (Martin et al., 2006; Resch et al., 2008). Microarray analysis of several S. xylosus isolates by Kastner et al. (2006) showed that while most antibiotic resistance genes were not present, partial tetK genes, responsible for tetracycline resistance, were detected in all of the tested isolates. However, other studies have not detected the tetK gene in S. xylosus. S. xylosus is a predominant species in fermented soybean along with S. succinus and S. saprophyticus (Jeong et al., 2014b, 2016a). A study by Jeong et al. (2014b) revealed that all 37 examined S. xylosus isolates from doenjang were susceptible to ampicillin, chloramphenicol, gentamycin, penicillin G, and trimethoprim, although 7% of isolates were resistant to erythromycin, lincomycin, and streptomycin.
Safety assessments of 18 S. xylosus isolates from doenjang showed that two and three isolates displayed weak α-hemolytic and β-hemolytic activity, respectively. However, α-hemolysin and β-hemolysin genes were not detected in the genome sequences of the S. xylosus isolates (Labrie et al., 2014). Similarly, approximately 3% of S. xylosus isolates from fermented sausages in a study by Martin et al. (2006) showed δ-hemolysis activity despite none of the isolates containing hld (Table 2). Amplification of staphylococcal enterotoxin genes has also been unsuccessful from most S. xylosus isolates (Table 2) (Even et al., 2010; Martin et al., 2006).
According to Ansorena et al. (2002), biogenic amines 2-phenylethylamine, putrescine, histamine, and tryptamine were not detected in sausages produced using a S. xylosus starter, although the S. carnosus starter cultures showed production of 2-phenylethylamine and tryptamine. Martuscellin et al. reported that approximately 80% of S. xylosus strains from artisanal fermented sausages did not demonstrate amino acid decarboxylase activity, which is required for biogenic amine production from amino acids, and 52% of strains were not able to produce biogenic amines (Martuscelli et al., 2000). Therefore, the production of biogenic amines by food-derived CNS appears to strain-specific (Table 3) (Ansorena et al., 2002; Bonomo et al., 2009; Even et al., 2010; Jeong et al., 2014a; Martin et al., 2006; Martuscelli et al., 2000). Additionally, genome sequence analysis of several S. carnosus, S. equorum, S. succinus, and S. xylosus isolates showed that none of the genomes contained genes involved in histamine and tyramine production; however, a lysine decarboxylase-encoding gene, which is required for cadaverine production, was identified (Table 4). These results again suggest that biogenic amines production might be strain-specific in S. xylosus.
Table 4.
Potential genes associated with biogenic amine production in five food-derived coagulase-negative staphylococci
| Product | S. carnosus | S. equorum | S. succinus | S. xylosus | S. saprophyticus |
|---|---|---|---|---|---|
| TM300 | KS1039 | 14BME20 | C2a | ATCC 15305 | |
| Lysine decarboxylase | SCA_RS00595 | SE1039_RS01180 | BK815_RS08125 | SXYL_RS11985 | SSP_RS11450 |
| SCA_RS01670 | SE1039_RS02575 | BK815_RS06925 | SXYL_RS10610 | SSP_RS10205 |
Food-derived CNS species are occasionally isolated from human skin infections (Blaiotta et al., 2004; Novakova et al., 2006). However, direct evidence of the involvement of food-derived CNS species in human disease is insufficient, and the virulence mechanism has yet to be determined. Moreover, genomic analysis revealed that food-derived CNS do not produce the adhesion proteins, invasion proteins, and enterotoxins that are necessary for the virulence of S. aureus (Heo et al., 2020; Jeong et al., 2017b). Therefore, we suggest that food-derived CNS are likely to be suitable for use in food production.
Contribution of food-derived CNS to food fermentation
CNS contribute to the sensory qualities of sausages via the catabolism of carbohydrates and amino acids, the formation of esters, and their interaction with fatty acids (Montel et al., 1998). CNS produce small molecule flavor compounds through proteolysis and lipolysis reactions (Berdague et al., 1993; Stahnke, 1994; Sondergaard and Stahnke, 2002). For example, S. equorum from jeotgal exhibited protease and lipase activity, while S. saprophyticus, S. succinus, and S. xylosus from doenjang showed protease and lipase activity (Jeong et al., 2014a, 2016a). During in vitro fermentation, S. carnosus, S. equorum, and S. xylosus were responsible for different aroma profiles, with S. carnosus producing higher concentrations of leucine-, isoleucine-, and valine-derived volatile compounds (Sondergaard and Stahnke, 2002). S. equorum from surface-ripened French cheese produced volatile compounds such as 3-methyl-3-buten-1-ol and 4-methyl-2-pentanone, which are responsible for fruity and sweet descriptors (Deetae et al., 2007), while S. succinus produced 3-mehylbutyl acetate, which is responsible for banana and pear descriptors, during soybean fermentation (Jeong et al., 2017a, 2019). CNS also contributed the volatile profile during fermentation of dry sausages (Ravyts et al., 2010). Additionally, S. succinus affected the volatile compounds detected in fermented soybean during pilot plant fermentation. These volatile compounds were derived from the conversion of amino acids (Jeong et al., 2019). Jeong et al. (2017a) also confirmed that the profile of volatile compounds produced by S. succinus was different from that of S. saprophyticus during soybean fermentation.
Unlike most lactic acid bacteria, genome sequence analysis of six strains revealed that S. equorum contains genes for the biosynthesis of all amino acids except asparagine and the conversion of branched chain fatty acids (Lee et al., 2018). S. equorum also carries genes required for the production of butane-2,3-diol, diacetyl, and acetoin via glycolysis, and ester compounds via protein degradation (Lee et al., 2018). In fermented sausage, S. succinus and S. xylosus produced acetoin diacetyl (Ravyts et al., 2010), while Flores and Toldra (2011) showed that CNS enzymatically converted free amino acids from peptides. Together, these results show that CNS contribute to flavor development in fermented foods via carbohydrate fermentation, proteolysis, lipolysis, and amino acid conversions. CNS can therefore be selected based on these various properties for use as starter cultures for fermentation.
Multilocus sequence typing and diversity of food-derived CNS
Food-derived CNS have several advantages for use as potential starter cultures for meat, seafood, and soybean fermentation. However, they also belong to the same genus as important human pathogen S. aureus, and have occasionally been isolation from human skin infections. In addition, food-derived CNS are still not included on the Pan American Health Organization list of safe organisms, despite the assertions of Irlinger (2008) that no CNS isolated from milk or dairy products has ever been involved in case of food poisoning or human pathology. Therefore, genetic and phenotypic differences between food-derived and clinical CNS isolates needs to be explored, along with the genetic diversity among food-derived CNS.
MLST based on internal fragments of five to seven housekeeping genes is an efficient tool for characterizing bacterial species in short-term epidemiological studies. As such, MLST has been used for strain characterization of food-derived CNS including S. carnosus (Buckle et al., 2017), S. equorum (Jeong et al., 2014c), and S. saprophyticus (Lee et al., 2015).
To examine the genetic diversity and population structure of S. carnosus strains, MLST using seven housekeeping genes (glpK (encoding glycerol kinase), tpiA (triosephosphate isomerase), dat (D-amino acid aminotransferase), xprT (xanthine phosphoribosyltransferase), gmk (guanylate kinase), narG (respiratory nitrate reductase alpha chain), and cstA (carbon starvation protein)) was conducted (Buckle et al., 2017). A total of 44 S. carnosus isolates from fish sauce, fish brine, sausage, smoked raw ham, salami, beef, and starter cultures were examined using the MLST assays. The results showed no correlation between sequence type and/or isolation year or source.
A MLST scheme was developed for S. saprophyticus to evaluate the genetic diversity between isolates from food and clinical origins based on seven housekeeping genes: aroE (encoding shikimate 5-dehydrogenase), dnaJ (chaperone protein), glpF (glycerol 3-phosphate dehydrogenase), gmk (guanylate kinase), hsp60 (heat shock protein 60), mutS (DNA mismatch repair protein), and pta (phosphotransacetylase) (Lee et al., 2015). A total of 48 isolates from human urine (n = 21), doenjang (n = 9), myeolchi-jeotgal (n = 6), saeu-jeotgal (n = 3), and sausage (n = 9) samples were subjected to MLST. Again, the results showed no correlation between sequence type and isolate origin, but clustering of the sequence types using the eBURST algorithm (Feil et al., 2004) revealed a correlation between the genetic backgrounds and origins of the isolates.
Another study developed a MLST scheme to evaluate the genetic diversity and background of S. equorum based on seven housekeeping genes: aroE, dnaJ, glpF, gmk, hsp60, mutS, and pta (Jeong et al., 2014c). A total of 117 S. equorum strains from saeu-jeotgal, myeolchi-jeotgal, sausage, cheese, and horse skin were examined. Results confirmed that MLST could not accurately identify correlations between the origins of the isolates and their sequence types.
Therefore, there is little information that can be used to accurately discriminate differences between CNS isolates of food and clinical origin. Complicating the issue is the lack of clinical isolates given the very rare occurrence of food-derived CNS-associated human infection. Additionally, CNS identification using commercial identification kits and ribotyping is difficult because of the variability within species (Carretto et al., 2005; Dupont et al., 2010; Sivadon et al., 2004), increasing the possibility of CNS misidentification. In addition, there is no consensus on the identification methods used. For example, S. xylosus isolates from patients with urinary tract infections were identified using phenotypic analysis and API 20 staph strips (Al-Mathkhury et al., 2008), while S. xylosus from bacteremia patients was identified using phenotypic methods only (Tselenis-Kotsowilis et al., 1982). Therefore, we suggest that species identification should be confirmed using several methods, especially food-derived CNS clinical isolates.
Food-derived CNS as starter candidates
Fermented foods are reservoirs and vehicles for large populations of living bacteria; therefore, the safety of starter cultures has become a significant issue. Prospective starters must be assessed for safety as well as functional properties such as the enhancement of sensory properties including color and volatile compound improvement. The safety of a starter is mainly assessed by two approaches: the Generally Recognized As Safe (GRAS) notification system in the United States and QPS in the European Union. The QPS system is based on four pillars of safety assessment dealing with taxonomy (establishing identity), familiarity (history of use, scientific literature, clinical aspects, industrial application, and ecology), pathogenicity (identification of safety concerns), and end use. Among the food-derived CNS, S. carnosus and S. xylosus have been nominated to achieve QPS status because of their extensive usage history in traditional meat fermentation (Talon and Leroy, 2011). However, further research is needed to obtain sufficient knowledge about the safety of food-derived CNS before they obtain QPS status. Food-derived CNS require case-by-case safety evaluations before they are cleared for use as starter cultures.
This review highlights the remaining safety concerns surrounding the use of food-derived CNS as starter cultures, particularly the small number of clinical infection cases attributed to these species. However, there have been no reported cases of food poisoning caused by food-derived CNS. In addition, comparative genomic analysis revealed that food-derived CNS do not carry the genes required for histamine, tyramine, putrescine, or cadaverine production. In terms of safety, the main hazard of bacteria used in food fermentation is their ability to acquire antibiotic resistance determinants, rather than their intrinsic resistance (EFSA, 2005). Intrinsic resistance is conferred by chromosomally-encoded genes that have been altered by mutation, and there is a very low risk of horizontal transmission of these genes. In contrast, acquired antibiotic resistance genes are located on mobile elements, which are readily transferred among strains. Given the lack of evidence of acquired antibiotic resistance genes and virulence genes in food-derived CNS, we propose that food-derived S. carnosus, S. equorum, S, succinus, and S. xylosus strains should be recognized as safe for use as starter cultures in food fermentation and they are should be contributed the enhancement of sensory properties including color, flavor, and taste in fermented food.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) [NRF-2019R1A2C1003639].
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
The original version of this article was revised due to a retrospective Open Access order.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
9/5/2020
The article “Food-derived coagulase-negative <Emphasis Type="Italic">Staphylococcus</Emphasis> as starter cultures for fermented foods”, written by Sojeong Heo, Jong-Hoon Lee, and Do-Won Jeong, was originally published Online First without Open Access.
Contributor Information
Sojeong Heo, Email: hsjeong32@daum.net.
Jong-Hoon Lee, Email: jhl@kgu.ac.kr.
Do-Won Jeong, Email: jeongdw@dongduk.ac.kr.
References
- Al-Mathkhury HJF, Flaih MT, Abdullah Z. Pathological study on Staphylococcus xylosus isolated from patients with urinary tract infections. Al-Nahrain Univ. J. Sci. 2008;11:123–130. [Google Scholar]
- Ansorena D, Montel MC, Rokka M, Talon R, Eerola S, Rizzo A, Raemaekers M, Demeyer D. Analysis of biogenic amines in northern and southern European sausages and role of flora in amine production. Meat Sci. 2002;61:141–147. doi: 10.1016/s0309-1740(01)00174-7. [DOI] [PubMed] [Google Scholar]
- Archer GL. Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis. 1998;26:1179–1181. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- Barros EM, Ceotto H, Bastos MC, Dos Santos KR, Giambiagi-Demarval M. Staphylococcus haemolyticus as an important hospital pathogen and carrier of methicillin resistance genes. J. Clin. Microbiol. 2012;50:166–168. doi: 10.1128/JCM.05563-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 2014;27:870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdague JL, Monteil P, Montel MC, Talon R. Effects of starter cultures on the formation of flavour compounds in dry sausage. Meat Sci. 1993;35:275–287. doi: 10.1016/0309-1740(93)90033-E. [DOI] [PubMed] [Google Scholar]
- Bieber L, Kahlmeter G. Staphylococcus lugdunensis in several niches of the normal skin flora. Clin. Microbiol. Infect. 2010;16:385–388. doi: 10.1111/j.1469-0691.2009.02813.x. [DOI] [PubMed] [Google Scholar]
- Bierowiec K, Korzeniowska-Kowal A, Wzorek A, Rypula K, Gamian A. Prevalence of Staphylococcus species colonization in healthy and sick Cats. Biomed Res. Int. 2019;2019:4360525. doi: 10.1155/2019/4360525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaiotta G, Pennacchia C, Villani F, Ricciardi A, Tofalo R, Parente E. Diversity and dynamics of communities of coagulase-negative staphylococci in traditional fermented sausages. J. Appl. Microbiol. 2004;97:271–284. doi: 10.1111/j.1365-2672.2004.02298.x. [DOI] [PubMed] [Google Scholar]
- Bockelmann W, Willems KP, Neve H, Heller KH. Cultures for the ripening of smear cheeses. Int. Dairy J. 2005;15:719–732. [Google Scholar]
- Bonomo MG, Ricciardi A, Zotta T, Sico MA, Salzano G. Technological and safety characterization of coagulase-negative staphylococci from traditionally fermented sausages of Basilicata region (Southern Italy) Meat Sci. 2009;83:15–23. doi: 10.1016/j.meatsci.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Brigante G, Menozzi MG, Pini B, Porta R, Somenzi P, Sciacca A, Spanu T, Stefani S. Identification of coagulase-negative staphylococci by using the BD phoenix system in the low-inoculum mode. J. Clin. Microbiol. 2008;46:3826–3828. doi: 10.1128/JCM.00877-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckle A, Kranz M, Schmidt H, Weiss A. Genetic diversity and population structure of food-borne Staphylococcus carnosus strains. Syst. Appl. Microbiol. 2017;40:34–41. doi: 10.1016/j.syapm.2016.11.005. [DOI] [PubMed] [Google Scholar]
- Carretto E, Barbarini D, Couto I, De Viti D, Marone P, Verhoef J, De Lencastre H, Brisse S. Identification of coagulase-negative staphylococci other than Staphylococcus epidermidis by automated ribotyping. Clin. Microbiol. Infect. 2005;11:177–184. doi: 10.1111/j.1469-0691.2004.01052.x. [DOI] [PubMed] [Google Scholar]
- Chapman GH. The significance of sodium chloride in studies of staphylococci. J. Bacteriol. 1945;50:201–203. doi: 10.1128/JB.50.2.201-203.1945. [DOI] [PubMed] [Google Scholar]
- Corbiere Morot-Bizot S, Leroy S, Talon R. Staphylococcal community of a small unit manufacturing traditional dry fermented sausages. Int. J. Food Microbiol. 2006;108:210–217. doi: 10.1016/j.ijfoodmicro.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Coton E, Desmonts MH, Leroy S, Coton M, Jamet E, Christieans S, Donnio PY, Lebert I, Talon R. Biodiversity of coagulase-negative Staphylococci in French cheeses, dry fermented sausages, processing environments and clinical samples. Int. J. Food. Microbiol. 2010;137:221–229. doi: 10.1016/j.ijfoodmicro.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Coton E, Mulder N, Coton M, Pochet S, Trip H, Lolkema JS. Origin of the putrescine-producing ability of the coagulase-negative bacterium Staphylococcus epidermidis 2015B. Appl. Environ. Microbiol. 2010;76:5570–5576. doi: 10.1128/AEM.00441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deetae P, Bonnarme P, Spinnler HE, Helinck S. Production of volatile aroma compounds by bacterial strains isolated from different surface-ripened French cheeses. Appl. Microbiol. Biotechnol. 2007;76:1161–1171. doi: 10.1007/s00253-007-1095-5. [DOI] [PubMed] [Google Scholar]
- Drosinos EH, Mataragas M, Xiraphi N, Moschonas G, Gaitis F, Metaxopoulos J. Characterization of the microbial flora from a traditional Greek fermented sausage. Meat Sci. 2005;69:307–317. doi: 10.1016/j.meatsci.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Dupont C, Sivadon-Tardy V, Bille E, Dauphin B, Beretti JL, Alvarez AS, Degand N, Ferroni A, Rottman M, Herrmann JL, Nassif X, Ronco E, Carbonnelle E. Identification of clinical coagulase-negative staphylococci, isolated in microbiology laboratories, by matrix-assisted laser desorption/ionization-time of flight mass spectrometry and two automated systems. Clin. Microbiol. Infect. 2010;16:998–1004. doi: 10.1111/j.1469-0691.2009.03036.x. [DOI] [PubMed] [Google Scholar]
- EFSA. EFSA scientific colloquium. Summary Report. QPS. Qualified Presumption of Saferty of microorganisms in Food and Feed. European Food Safety Authority, Bruessels, Belgium (2004)
- EFSA Opinion of the Scientific Panel on Additives and Products or Substances used in Animal Feed on the updating of the criteria used in the assessment of bacteria for resistance to antibiotics of human or veterinary importance. EFSA J. 2005;223:1–12. [Google Scholar]
- EFSA Update of the list of QPS-recommended biological agentsintentionally added to food or feed as notified to EFSA. EFSA J. 2016;15:4663. [Google Scholar]
- Even S, Leroy S, Charlier C, Zakour NB, Chacornac JP, Lebert I, Jamet E, Desmonts MH, Coton E, Pochet S, Donnio PY, Gautier M, Talon R, Le Loir Y. Low occurrence of safety hazards in coagulase negative staphylococci isolated from fermented foodstuffs. Int. J. Food Microbiol. 2010;139:87–95. doi: 10.1016/j.ijfoodmicro.2010.02.019. [DOI] [PubMed] [Google Scholar]
- Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentini AM, Sawitzki MC, Bertol TM, Sant’anna ES. Viability of Staphylococcus xylosus isolated from artisanal sausages for application as starter cultures in meat products. Braz. J. Microbiol. 2009;40:129–133. doi: 10.1590/S1517-838220090001000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores M, Toldra F. Microbial enzymaticactivities forimproved fermentedmeats. Trends Food Sci. Technol. 2011;22:81–90. [Google Scholar]
- Fonseca S, Cachaldora A, Gomez M, Franco I, Carballo J. Monitoring the bacterial population dynamics during the ripening of Galician chorizo, a traditional dry fermented Spanish sausage. Food Microbiol. 2013;33:77–84. doi: 10.1016/j.fm.2012.08.015. [DOI] [PubMed] [Google Scholar]
- Freney J, Brum Y, Bes M, Meugnier H, Grimont F, GrimontPatrick AD, Nervi C, Fleurette J. Staphylococcus lugdunensis sp. nov. and Staphylococcus schleiferi sp. nov., Two Species from Human Clinical Specimens. Int. J. Syst. Bacteriol. 1988;38:168–172. [Google Scholar]
- Giormezis N, Kolonitsiou F, Makri A, Vogiatzi A, Christofidou M, Anastassiou ED, Spiliopoulou I. Virulence factors among Staphylococcus lugdunensis are associated with infection sites and clonal spread. Eur. J. Clin. Microbiol. Infect. Dis. 2015;34:773–778. doi: 10.1007/s10096-014-2291-8. [DOI] [PubMed] [Google Scholar]
- Guan L, Cho KH, Lee JH. Analysis of the cultivable bacterial community in jeotgal, a Korean salted and fermented seafood, and identification of its dominant bacteria. Food Microbiol. 2011;28:101–113. doi: 10.1016/j.fm.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Götz F, Bannerman T, Schleifer K-H. The genera Staphylococcus and Macrococcus. Prokaryotes. 2006;4:5–75. [Google Scholar]
- Hashi AA, Delport JA, Elsayed S, Silverman MS. Staphylococcus pettenkoferi bacteremia: A case report and review of the literature. Can. J. Infect. Dis. Med. Microbiol. 2015;26:319–322. doi: 10.1155/2015/748154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo S, Lee JS, Lee JH, Jeong DW. Comparative genomic analysis of food-originaged coagulase-negative Staphylococcus: Analysis of conserved core genes and diversity of the pan-genome. J. Microbiol. Biotechnol. 2020;30:341–351. doi: 10.4014/jmb.1910.10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugas M, Monfort JM. Bacterial starter cultures for meat fermentation. Food Chem. 1997;59:547–554. [Google Scholar]
- Irlinger F. Safety assessment of dairy microorganisms: coagulase-negative staphylococci. Int. J. Food Microbiol. 2008;126:302–310. doi: 10.1016/j.ijfoodmicro.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Irlinger F, Loux V, Bento P, Gibrat JF, Straub C, Bonnarme P, Landaud S, Monnet C. Genome sequence of Staphylococcus equorum subsp. equorum Mu2, isolated from a French smear-ripened cheese. J. Bacteriol. 2012;194:5141–5142. doi: 10.1128/JB.01038-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DW, Lee JH. Complete genome sequence of Staphylococcus succinus 14BME20 isolated from a traditional Korean fermented soybean food. Genome Announc. 2017;5:e01731-16. doi: 10.1128/genomeA.01731-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DW, Han S, Lee JH. Safety and technological characterization of Staphylococcus equorum isolates from jeotgal, a Korean high-salt-fermented seafood, for starter development. Int. J. Food Microbiol. 2014;188:108–115. doi: 10.1016/j.ijfoodmicro.2014.07.022. [DOI] [PubMed] [Google Scholar]
- Jeong DW, Kim HR, Jung G, Han S, Kim CT, Lee JH. Bacterial community migration in the ripening of doenjang, a traditional Korean fermented soybean food. J. Microbiol. Biotechnol. 2014;24:648–660. doi: 10.4014/jmb.1401.01009. [DOI] [PubMed] [Google Scholar]
- Jeong DW, Kim HR, Lee JH. Genetic diversity of Staphylococcus equorum isolates from Saeu-jeotgal evaluated by multilocus sequence typing. Anton. Leeuw. Int. J. G. 2014;106:795–808. doi: 10.1007/s10482-014-0249-6. [DOI] [PubMed] [Google Scholar]
- Jeong DW, Lee B, Her JY, Lee KG, Lee JH. Safety and technological characterization of coagulase-negative staphylococci isolates from traditional Korean fermented soybean foods for starter development. Int. J. Food Microbiol. 2016;236:9–16. doi: 10.1016/j.ijfoodmicro.2016.07.011. [DOI] [PubMed] [Google Scholar]
- Jeong DW, Na H, Ryu S, Lee JH. Complete genome sequence of Staphylococcus equorum KS1039 isolated from Saeu-jeotgal. Korean high-salt-fermented seafood. J. Biotechnol. 2016;219:88–89. doi: 10.1016/j.jbiotec.2015.12.025. [DOI] [PubMed] [Google Scholar]
- Jeong DW, Heo S, Lee B, Lee H, Jeong K, Her JY, Lee KG, Lee JH. Effects of the predominant bacteria from meju and doenjang on the production of volatile compounds during soybean fermentation. Int. J. Food Microbiol. 2017;262:8–13. doi: 10.1016/j.ijfoodmicro.2017.09.011. [DOI] [PubMed] [Google Scholar]
- Jeong DW, Heo S, Ryu S, Blom J, Lee JH. Genomic insights into the virulence and salt tolerance of Staphylococcus equorum. Sci. Rep. 2017;7:5383. doi: 10.1038/s41598-017-05918-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DW, Lee H, Jeong K, Kim CT, Shim ST, Lee JH. Effects of starter candidates and NaCl on the production of volatile compounds during soybean fermentation. J. Microbiol. Biotechnol. 2019;29:191–199. doi: 10.4014/jmb.1811.11012. [DOI] [PubMed] [Google Scholar]
- Kacica MA, Horgan MJ, Preston KE, Lepow M, Venezia RA. Relatedness of coagulase-negative staphylococci causing bacteremia in low-birthweight infants. Infect. Cont. Hosp. Ep. 1994;15:658–662. doi: 10.1086/646829. [DOI] [PubMed] [Google Scholar]
- Kastner S, Perreten V, Bleuler H, Hugenschmidt G, Lacroix C, Meile L. Antibiotic susceptibility patterns and resistance genes of starter cultures and probiotic bacteria used in food. Syst. Appl. Microbiol. 2006;29:145–155. doi: 10.1016/j.syapm.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Kuroda M, Yamashita A, Hirakawa H, Kumano M, Morikawa K, Higashide M, Maruyama A, Inose Y, Matoba K, Toh H, Kuhara S, Hattori M, Ohta T. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc. Natl. Acad. Sci. USA. 2005;102:13272–13277. doi: 10.1073/pnas.0502950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie SJ, El Haddad L, Tremblay DM, Plante PL, Wasserscheid J, Dumaresq J, Dewar K, Corbeil J, Moineau S. First complete genome sequence of Staphylococcus xylosus, a meat starter culture and a host to propagate Staphylococcus aureus phages. Genome Announc. 2014;2(4):e00671-14. doi: 10.1128/genomeA.00671-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Jeong DW, Lee JH. Genetic diversity and antibiotic resistance of Staphylococcus saprophyticus isolates from fermented foods and clinical samples. J. Korean Soc. Appl. Biol. Chem. 2015;58:659–668. [Google Scholar]
- Lee JH, Heo S, Jeong DW. Genomic insights into Staphylococcus equorum KS1039 as a potential starter culture for the fermentation of high-salt foods. BMC Genomics. 2018;19:136. doi: 10.1186/s12864-018-4532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy F, Verluyten J, De Vuyst L. Functional meat starter cultures for improved sausage fermentation. Int. J. Food Microbiol. 2006;106:270–285. doi: 10.1016/j.ijfoodmicro.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Lofblom J, Rosenstein R, Nguyen MT, Stahl S, Gotz F. Staphylococcus carnosus: from starter culture to protein engineering platform. Appl. Microbiol. Biotechnol. 2017;101:8293–8307. doi: 10.1007/s00253-017-8528-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B, Garriga M, Hugas M, Bover-Cid S, Veciana-Nogues MT, Aymerich T. Molecular, technological and safety characterization of Gram-positive catalase-positive cocci from slightly fermented sausages. Int. J. Food Microbiol. 2006;107:148–158. doi: 10.1016/j.ijfoodmicro.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Martuscelli M, Crudele MA, Gardini F, Suzzi G. Biogenic amine formation and oxidation by Staphylococcus xylosus strains from artisanal fermented sausages. Lett. Appl. Microbiol. 2000;31:228–232. doi: 10.1046/j.1365-2672.2000.00796.x. [DOI] [PubMed] [Google Scholar]
- Marty E, Bodenmann C, Buchs J, Hadorn R, Eugster-Meier E, Lacroix C, Meile L. Prevalence of antibiotic resistance in coagulase-negative staphylococci from spontaneously fermented meat products and safety assessment for new starters. Int. J. Food Microbiol. 2012;159:74–83. doi: 10.1016/j.ijfoodmicro.2012.07.025. [DOI] [PubMed] [Google Scholar]
- Mauriello G, Casaburi A, Blaiotta G, Villani F. Isolation and technological properties of coagulase negative staphylococci from fermented sausages of Southern Italy. Meat Sci. 2004;67:149–158. doi: 10.1016/j.meatsci.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Mauriello G, Moschetti G, Villani F, Blaiotta G, Coppola S. Antibiotic resistance of coagulase-negative staphylococci isolated from artisanal Naples-type salami. Int. J. Food Sci. Nutr. 2000;51:19–24. doi: 10.1080/096374800100868. [DOI] [PubMed] [Google Scholar]
- Megaw J, Gilmore BF. Draft genome sequence of Staphylococcus succinus strain CSM-77, a moderately halophilic bacterium isolated from a triassic salt mine. Genome Announc. 2016;4:e00532-00516. doi: 10.1128/genomeA.00532-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montel MC, Masson F, Talon R. Bacterial role in flavour development. Meat Sci. 1998;49:S111–S123. [PubMed] [Google Scholar]
- Montel MC, Talon R, Berdague JL, Cantonnet M. Effects of starter cultures on the biochemical characteristics of French dry sausages. Meat Sci. 1993;35:229–240. doi: 10.1016/0309-1740(93)90053-K. [DOI] [PubMed] [Google Scholar]
- Muller A, Reichhardt R, Fogarassy G, Bosse R, Gibis M, Weiss J, Schmidt H, Weiss A. Safety assessment of selected Staphylococcus carnosus strains with regard to their application as meat starter culture. Food Control. 2016;66:93–99. [Google Scholar]
- Natoli S, Fontana C, Favaro M, Bergamini A, Testore GP, Minelli S, Bossa MC, Casapulla M, Broglio G, Beltrame A, Cudillo L, Cerretti R, Leonardis F. Characterization of coagulase-negative staphylococcal isolates from blood with reduced susceptibility to glycopeptides and therapeutic options. BMC Infect. Dis. 2009;9:83. doi: 10.1186/1471-2334-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakova D, Sedlacek I, Pantucek R, Stetina V, Svec P, Petras P. Staphylococcus equorum and Staphylococcus succinus isolated from human clinical specimens. J. Med. Microbiol. 2006;55:523–528. doi: 10.1099/jmm.0.46246-0. [DOI] [PubMed] [Google Scholar]
- Otto M. Staphylococcus epidermidis-the ‘accidental’ pathogen. Nat. Rev. Microbiol. 2009;7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. Molecular basis of Staphylococcus epidermidis infections. Semin. Immunopathol. 2012;34:201–214. doi: 10.1007/s00281-011-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: Staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. Bioessays. 2013;35:4–11. doi: 10.1002/bies.201200112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamanoli E, Kotzekidou P, Tzanetakis N, Litopoulou-Tzanetaki E. Characterization of micrococcaceae isolated from dry fermented sausage. Food Microbiol. 2002;19:441–449. doi: 10.1016/S0309-1740(02)00292-9. [DOI] [PubMed] [Google Scholar]
- Parte AC. LPSN—List of Prokaryotic names with Standing in Nomenclature (bacterio.net), 20 years on. Int. J. Syst. Evol. Microbiol. 2018;68:1825–1829. doi: 10.1099/ijsem.0.002786. [DOI] [PubMed] [Google Scholar]
- Place RB, Hiestand D, Gallmann HR, Teuber M. Staphylococcus equorum subsp. linens, subsp. nov., a starter culture component for surface ripened semi-hard cheeses. Syst. Appl. Microbiol. 2003;26:30–37. doi: 10.1078/072320203322337281. [DOI] [PubMed] [Google Scholar]
- Ravyts F, Steen L, Goemaere O, Paelinck H, De Vuyst L, Leroy F. The application of staphylococci with flavour-generating potential is affected by acidification in fermented dry sausages. Food Microbiol. 2010;27:945–954. doi: 10.1016/j.fm.2010.05.030. [DOI] [PubMed] [Google Scholar]
- Raz R, Colodner R, Kunin CM. Who are you-Staphylococcus saprophyticus? Clin. Infect. Dis. 2005;40:896–898. doi: 10.1086/428353. [DOI] [PubMed] [Google Scholar]
- Resch M, Nagel V, Hertel C. Antibiotic resistance of coagulase-negative staphylococci associated with food and used in starter cultures. Int. J. Food Microbiol. 2008;127:99–104. doi: 10.1016/j.ijfoodmicro.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Rosenstein R, Nerz C, Biswas L, Resch A, Raddatz G, Schuster SC, Gotz F. Genome analysis of the meat starter culture bacterium Staphylococcus carnosus TM300. Appl. Environ. Microbiol. 2009;75:811–822. doi: 10.1128/AEM.01982-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabate Bresco M, Harris LG, Thompson K, Stanic B, Morgenstern M, O’Mahony L, Richards RG, Moriarty TF. Pathogenic mechanisms and host interactions in Staphylococcus epidermidis device-related infection. Front. Microbiol. 2017;8:1401. doi: 10.3389/fmicb.2017.01401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakinc T, Woznowski M, Ebsen M, Gatermann SG. The surface-associated protein of Staphylococcus saprophyticus is a lipase. Infect. Immun. 2005;73:6419–6428. doi: 10.1128/IAI.73.10.6419-6428.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakinc T, Kleine B, Gatermann SG. Biochemical characterization of the surface-associated lipase of Staphylococcus saprophyticus. FEMS Microbiol. Lett. 2007;274:335–341. doi: 10.1111/j.1574-6968.2007.00857.x. [DOI] [PubMed] [Google Scholar]
- Samelis J, Metaxopoulos J, Vlassi M, Pappa A. Stability and safety of traditional Greek salami—a microbiological ecology study. Int. J. Food Microbiol. 1998;44:69–82. doi: 10.1016/s0168-1605(98)00124-x. [DOI] [PubMed] [Google Scholar]
- Schleifer KH, Bell JA. Staphylococcaceae family nov vol three. In: De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB, editors. Bergey’s manual of systemic bacteriology. New York: Springer; 2009. pp. 392–433. [Google Scholar]
- Sivadon V, Rottman M, Quincampoix JC, Avettand V, Chaverot S, de Mazancourt P, Trieu-Cuot P, Gaillard JL. Use of sodA sequencing for the identification of clinical isolates of coagulase-negative staphylococci. Clin. Microbiol. Infect. 2004;10:939–942. doi: 10.1111/j.1469-0691.2004.00939.x. [DOI] [PubMed] [Google Scholar]
- Sondergaard AK, Stahnke LH. Growth and aroma production by Staphylococcus xylosus, S. carnosus and S. equorum—a comparative study in model systems. Int. J. Food Microbiol. 2002;75:99–109. doi: 10.1016/s0168-1605(01)00729-2. [DOI] [PubMed] [Google Scholar]
- Stahnke LH. Aroma components from dried sausages fermented with Staphylococcus xylosus. Meat Sci. 1994;38:39–53. doi: 10.1016/0309-1740(94)90094-9. [DOI] [PubMed] [Google Scholar]
- Szabados F, Mohner A, Kleine B, Gatermann SG. Staphylococcus saprophyticus surface-associated protein (Ssp) is associated with lifespan reduction in Caenorhabditis elegans. Virulence. 2013;4:604–611. doi: 10.4161/viru.25875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon R, Leroy S. Diversity and safety hazards of bacteria involved in meat fermentations. Meat Sci. 2011;89:303–309. doi: 10.1016/j.meatsci.2011.04.029. [DOI] [PubMed] [Google Scholar]
- Talon R, Leroy S, Lebert I, Giammarinaro P, Chacornac JP, Latorre-Moratalla M, Vidal-Carou C, Zanardi E, Conter M, Lebecque A. Safety improvement and preservation of typical sensory qualities of traditional dry fermented sausages using autochthonous starter cultures. Int. J. Food Microbiol. 2008;126:227–234. doi: 10.1016/j.ijfoodmicro.2008.05.031. [DOI] [PubMed] [Google Scholar]
- Tang Y, Lu Y, Lu F, Bie X, Guo Y, Lu Z. Cloning and expression of organic solvent tolerant lipase gene from Staphylococcus saprophyticus M36. Sheng Wu Gong Cheng Xue Bao. 2009;25:1989–1995. [PubMed] [Google Scholar]
- Tevell S, Hellmark B, Nilsdotter-Augustinsson A, Soderquist B. Staphylococcus capitis isolated from prosthetic joint infections. Eur. J. Clin. Microbiol. Infect. Dis. 2017;36:115–122. doi: 10.1007/s10096-016-2777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG., Jr Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tselenis-Kotsowilis AD, Koliomichalis MP, Papavassiliou JT. Acute pyelonephritis caused by Staphylococcus xylosus. J. Clin. Microbiol. 1982;16:593–594. doi: 10.1128/jcm.16.3.593-594.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu RJ, Wu HY, Lock YS, Chen MJ. Evaluation of microbial dynamics during the ripening of a traditional Taiwanese naturally fermented ham. Food Microbiol. 2010;27:460–467. doi: 10.1016/j.fm.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Widerstrom M, Wistrom J, Sjostedt A, Monsen T. Coagulase-negative staphylococci: update on the molecular epidemiology and clinical presentation, with a focus on Staphylococcus epidermidis and Staphylococcus saprophyticus. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:7–20. doi: 10.1007/s10096-011-1270-6. [DOI] [PubMed] [Google Scholar]
- Zhou H, Yao Z, Shi H, Wang B, Li D, Hou J, Ma S. Draft Genome Sequence of Staphylococcus succinus subsp. succinus type strain DSM 14617, isolated from plant and soil inclusions within 25- to 35-million-year-old dominican amber. Genome Announc. 2017;5:16. doi: 10.1128/genomeA.01521-16. [DOI] [PMC free article] [PubMed] [Google Scholar]