Listeria monocytogenes is a Gram-positive pathogen able to cause foodborne infections in humans and animals. Key virulence genes in L. monocytogenes are activated by the transcription regulator PrfA, a DNA binding protein belonging to the CRP/FNR family. Various signals from the environment are known to affect the activity of PrfA, either positively or negatively. Recently, we found that specific medium- and long-chain free fatty acids act as antimicrobial agents as well as signaling compounds in L. monocytogenes. Here, we show that both antimicrobial and nonantimicrobial free fatty acids inhibit PrfA-dependent activation of virulence gene transcription by interfering with the DNA binding activity of PrfA. Our findings suggest that free fatty acids could be candidates for alternative therapies against L. monocytogenes.
KEYWORDS: DNA-protein binding experiments, naturally occurring free fatty acids, antivirulence agents, antimicrobial agents, virulence regulators
ABSTRACT
Naturally occurring free fatty acids (FFAs) are recognized as potent antimicrobial agents that also affect the production of virulence factors in bacterial pathogens. In the foodborne pathogen Listeria monocytogenes, some medium- and long-chain FFAs act as antimicrobial agents as well as signaling compounds, causing a repression of transcription of virulence genes. We previously observed that the master virulence regulator PrfA is involved in both the antimicrobial and virulence-inhibitory response of L. monocytogenes to selected FFAs, but the underlying mechanisms are presently unknown. Here, we present a systematic analysis of the antimicrobial and PrfA-inhibitory activities of medium- and long-chain FFAs of various carbon chain lengths and degrees of saturation. We observed that exposure to specific antimicrobial and nonantimicrobial FFAs prevented PrfA-dependent activation of virulence gene transcription and reduced the levels of PrfA-regulated virulence factors. Thus, an antimicrobial activity was not compulsory for the PrfA-inhibitory ability of an FFA. In vitro binding experiments revealed that PrfA-inhibitory FFAs were also able to prevent the constitutively active variant PrfA* from binding to the PrfA box in the promoter region of the virulence gene hly, whereas noninhibitory FFAs did not affect its ability to bind DNA. Notably, the unsaturated FFAs inhibited the DNA binding activity of PrfA* most efficiently. Altogether, our findings support a model in which specific FFAs orchestrate a generalized reduction of the virulence potential of L. monocytogenes by directly targeting the key virulence regulator PrfA.
IMPORTANCE Listeria monocytogenes is a Gram-positive pathogen able to cause foodborne infections in humans and animals. Key virulence genes in L. monocytogenes are activated by the transcription regulator PrfA, a DNA binding protein belonging to the CRP/FNR family. Various signals from the environment are known to affect the activity of PrfA, either positively or negatively. Recently, we found that specific medium- and long-chain free fatty acids act as antimicrobial agents as well as signaling compounds in L. monocytogenes. Here, we show that both antimicrobial and nonantimicrobial free fatty acids inhibit PrfA-dependent activation of virulence gene transcription by interfering with the DNA binding activity of PrfA. Our findings suggest that free fatty acids could be candidates for alternative therapies against L. monocytogenes.
INTRODUCTION
Free fatty acids (FFAs) have diverse biological activities and are widely recognized as antimicrobial agents that can be bacteriostatic or even bactericidal (1–3). The mechanism behind the antimicrobial effect of FFAs is still not fully understood; nevertheless, the bacterial cell membrane seems to be the main target (1). For instance, large amounts of FFAs can solubilize the membrane and eventually cause cell lysis or interfere with the electron transport chain, as well as inhibit enzyme activity or impair nutrient uptake (1). Importantly, FFAs can also act as signaling molecules at subinhibitory concentrations, causing either repression or induction of gene expression. In pathogenic bacteria, FFAs are known to affect the production of virulence factors that are important for establishing a successful infection (4). In Vibrio cholerae, unsaturated FFAs inhibit the ToxT-dependent activation of genes encoding the cholera toxin and the toxin-coregulated pilus (5–7). Another example of virulence attenuation comes from Salmonella enterica, where unsaturated FFAs inhibit the activity of the HilD virulence regulator, consequently reducing the expression of the Salmonella pathogenicity island 1 type III secretion system (8).
In a recent study, we found that specific antimicrobial medium- and long-chain FFAs prevent PrfA-dependent activation of virulence genes in the Gram-positive foodborne pathogen Listeria monocytogenes (9). In this pathogen, the PrfA protein functions as a master virulence regulator (10, 11). PrfA belongs to the CRP/FNR family of regulators and binds as a homodimer to specific DNA sequences (PrfA boxes) placed in the promoter region of key virulence genes to activate their transcription (10). PrfA senses environmental signals, leading to the induction or repression of L. monocytogenes virulence machinery, as an unnecessary production of virulence factors represents a major fitness burden (12, 13). Accordingly, PrfA is kept in an inactive conformation when L. monocytogenes resides in the external environment (14). When crossing the intestinal barrier, PrfA becomes highly active and triggers an induced expression of the key virulence factors required for bacterial uptake into host cells, intracellular growth, and cell-to-cell transmission, namely, the internalins InlA and InlB, the pore-forming toxin LLO, the phospholipases PlcA and PlcB, and the surface protein ActA (10, 15). To become fully active, PrfA binds bacterium- and host-derived glutathione (GSH) (16), which results in a conformational change that promotes optimal PrfA-DNA interaction (17). Mutant variants of PrfA, called PrfA*, are known to lock the protein in its active conformation and constitutively express the PrfA-dependent virulence genes (10, 15). Several PrfA* mutants have been identified, all containing a single mutation within the prfA coding sequence (18–21). These mutations result in PrfA* proteins with different degrees of activity and DNA binding affinity, for example, the moderate-activity mutant proteins PrfA-E77K and PrfA-G155S and the high-activity mutant proteins PrfA-L140F and PrfA-G145S (14, 22–24). The prfA* mutants are hypervirulent in mouse infection models, clearly demonstrating that a high level of expression of PrfA-dependent virulence genes is beneficial for L. monocytogenes within the host environment (12). However, a high-level prfA* activity mutation (e.g., prfA-G145S), but not a midlevel prfA* activity mutation (e.g., prfA-G155S), confers a competitive disadvantage relative to the wild-type during growth in broth culture, as well as a greater sensitivity to stress conditions (12). These findings show that it is, indeed, critical for L. monocytogenes to tightly regulate PrfA activity throughout the environmental conditions it encounters (13, 20).
Due to its undeniable importance for listerial pathogenesis, PrfA became a desirable target for antivirulence agents, and therefore, recent studies have focused on identifying compounds that could act to prevent PrfA-dependent virulence gene expression (21, 25). We have recently shown that distinct FFAs can act as antimicrobial as well as virulence-inhibitory agents in L. monocytogenes (9). Three unsaturated long-chain FFAs and a saturated medium-chain FFA showed antimicrobial and virulence-inhibitory effects, whereas two saturated long-chain FFAs left L. monocytogenes unaffected (9). Curiously, PrfA seemed to play a role in the response of L. monocytogenes to the FFAs; first, the deletion of prfA resulted in an increased tolerance to antimicrobial FFAs, and second, subinhibitory concentrations of such FFAs could prevent PrfA-dependent activation of key virulence genes (9).
In the present study, we aimed to further uncover why some FFAs inhibit the growth and virulence potential of L. monocytogenes, while others do not. Therefore, we systematically analyzed the antilisteria activities of selected medium- and long-chain FFAs, various carbon chain lengths, and the number of double bonds, as well as the orientation of the double bonds (cis or trans). We performed a comparative analysis of the effect of these FFAs on bacterial growth and virulence gene expression and analyzed their impact on PrfA’s DNA binding activity.
RESULTS
We selected nine different medium- and long-chain FFAs of various carbon chain lengths and degrees of saturation (see Fig. S1 in the supplemental material) and examined their antimicrobial and PrfA-inhibitory properties against L. monocytogenes. Myristic acid (MA), which comprises 14 carbons in its chain, was compared with the previously tested C12:0 FFA lauric acid (LA), C16:0 FFA palmitic acid (PAL), and C18:0 FFA stearic acid (SA) (9) to understand how the number of carbons in saturated FFAs (SFFAs) affects their function. To address the importance of unsaturation, we varied the number of double bonds in FFAs all containing 18 carbons (C18 FFAs). We selected C18 FFAs with one (oleic acid [OA] and elaidic acid [EA]) or two (linoleic acid [LNA] and linolelaidic acid LLA]) double bonds, allowing a comparison to the previously tested C18 FFAs γ-linolenic acid (GLA) and SA, which contain three and zero double bonds, respectively (9). We also considered the configuration of the double bond, and as such, the C18:19 cis (OA) and trans (EA) FFAs, as well as the C18:29,12 cis (LNA) and trans (LLA) FFAs, were compared. The cis and trans isomers differ only in terms of conformation, meaning that the double bond of the cis isomer causes bending of the chain, whereas the trans isomer shows a configuration identical to its saturated counterpart (in this case SA) (see Fig. S1).
Table 1 shows the antimicrobial properties of the selected FFAs tested in this and in our previous study (9). To evaluate the role of PrfA in FFA tolerance, we examined the effect of FFAs on the L. monocytogenes EGD wild-type strain, as well as on a prfA* mutant strain (prfA-G155S) and on an EGD strain lacking prfA (ΔprfA). The strain encoding the midlevel constitutively active PrfA-G155S was chosen because it does not render a growth deficiency under stress conditions, as mentioned in the introduction. After 20 h of growth, we determined the inhibitory concentration (IC) of each specific FFA (see Fig. S2 in the supplemental material) (9). Our findings showed that the SFFA with the shortest carbon chain (LA; C12:0) was the only SFFA that exerted an antimicrobial effect on the L. monocytogenes wild-type and prfA-G155S strains (Table 1). Regarding the unsaturated C18 FFAs, we found that GLA (C18:3 cis) and LNA (C18:2 cis) were antimicrobial toward the wild-type and prfA-G155S strains, whereas OA (C18:1 cis) and SA (C18:0) were not, suggesting that the degree of unsaturation affects the antimicrobial activity of the FFAs. In addition, the results indicated that the configuration of the double bond also influences the antimicrobial properties of a certain FFA. Specifically, we found that LNA (C18:2 cis) was antimicrobial toward L. monocytogenes wild-type and prfA-G155S, whereas LLA (C18:2 trans) was not. For LA, GLA, and LNA, where an antimicrobial effect was observed against the wild-type and prfA-G155S strains, the ΔprfA strain was still able to grow at concentrations that were inhibitory for those strains, indicating a role for PrfA in the response to antimicrobial FFAs, as suggested earlier (9).
TABLE 1.
IC values of the various FFAs on L. monocytogenes EGD wild type, EGDΔprfA, and EGDprfA*
| Fatty acid | Abbreviation | Saturation | IC valuea
(μg/ml) for: |
||
|---|---|---|---|---|---|
| EGD | EGDΔprfA | EGDprfA* (prfA-G155S) | |||
| Lauric acid | LA | C12:0 | 50* | >75* | 50* |
| Myristic acid | MA | C14:0 | >160 | >160 | >160 |
| Palmitic acid | PAL | C16:0 | >150* | >150* | >150* |
| Stearic acid | SA | C18:0 | >75* | >75* | >75* |
| γ-Linolenic acid | GLA | C18:36,9,12 (cis) | 15* | 20* | 15* |
| Linoleic acid | LNA | C18:29,12 (cis) | ≤40 | >160 | 160 |
| Linolelaidic acid | LLA | C18:29,12 (trans) | >160 | >160 | >160 |
| Oleic acid | OA | C18:19 (cis) | >160 | >160 | >160 |
| Elaidic acid | EA | C18:19 (trans) | >160 | >160 | >160 |
The values represent the inhibitory concentration of a certain FFA determined for each strain, obtained either in this study (Fig. S2) or in our previous study (*) (9).
Nonantimicrobial FFAs can repress PrfA-dependent activation of virulence gene transcription.
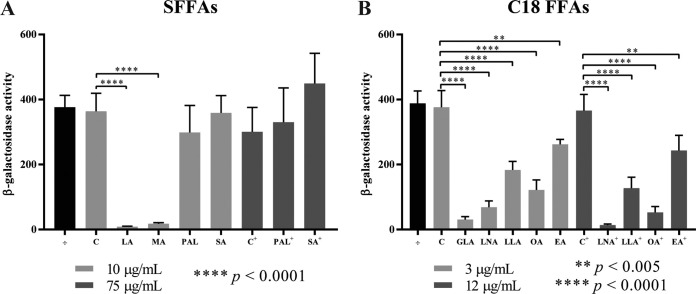
In our previous work, we noticed a putative correlation between the antimicrobial and virulence-inhibitory properties of the tested FFAs (9). To further investigate this, we measured the effect of the SFFAs and C18 FFAs on the expression of the PrfA-dependent virulence gene hly using a β-galactosidase assay. For these experiments, we used the L. monocytogenes prfA-G155S strain to ensure that PrfA-dependent activation of virulence genes could be observed during growth in brain heart infusion (BHI) medium. Briefly, exponentially growing cells of L. monocytogenes prfA-G155S, carrying the phly-lacZ fusion plasmid, were subjected either to subinhibitory concentrations of the FFAs or to the vehicle (C or C+). Alternatively, the cells were left untreated (÷). After 20 h of growth, cells were harvested, and the β-galactosidase activity was determined (Fig. 1). No differences in β-galactosidase activity were detected between untreated (÷) and vehicle-treated samples (C or C+). However, when the cultures were exposed to 10 μg/ml (35 to 50 μM) of each SFFA, corresponding to a subinhibitory concentration of the otherwise antimicrobial SFFA LA (Table 1), we observed that LA and MA could inhibit the β-galactosidase activity produced from phly-lacZ, whereas no effect was seen for the same concentration of PAL and SA (Fig. 1A). We tested a higher concentration to confirm that PAL and SA had, indeed, no effect on the PrfA activity (compare PAL+ and SA+ with C+), as previously reported (9) (Fig. 1A). To further verify that LA and MA affect PrfA activity, an identical experiment was done for the PrfA-independent gene lhrA (see Fig. S3A in the supplemental material). The results demonstrated a minor (<1.4-fold) reduction in β-galactosidase activity produced from plhrA36-lacZ-containing cells exposed to LA and MA, compared with the vehicle control (C) (Fig. S3A). Notably, the LA- and MA-mediated inhibition of phly-lacZ expression was substantially greater (42- and 20-fold reduction, respectively) (Fig. 1A). These results indicate that LA and MA both act to inhibit PrfA-dependent activation of hly; however, while LA is known to exhibit an antimicrobial effect on L. monocytogenes, MA clearly belongs to the group of nonantimicrobial FFAs (Table 1). These results represent the first evidence that an FFA (here, MA) does not have to exert an antimicrobial effect in order to effectively reduce PrfA-dependent activation in L. monocytogenes.
FIG 1.
Expression of phly-lacZ in response to FFA exposure. The promoter region of the PrfA-dependent hly gene cloned into vector pTCV-lac was transformed into EGDprfA*. The resulting strain was grown to an OD600 of 0.3 and exposed to SFFAs in a concentration of 10 μg/ml (50 μM LA, 44 μM MA, 39 μM PAL, or 35 μM SA) or 75 μg/ml (293 μM PAL+ or 264 μM SA+) (A) or to C18 FFAs in a concentration of 3 μg/ml (11 μM FFA) or 12 μg/ml (43 μM FFA+) (B). As controls, the cultures were left untreated (÷) or vehicle was added corresponding to the concentration present in the FFA-treated cultures (C or C+). Samples for the β-galactosidase assays were withdrawn after 20 h. Results are the average of three biological replicates, each carried out in technical duplicates.
In addition, we found that all five unsaturated C18 FFAs were able to significantly reduce the β-galactosidase activity produced from phly-lacZ, although to a different extent (Fig. 1B). For instance, when comparing the β-galactosidase activities of the cultures subjected to a subinhibitory concentration (3 μg/ml, corresponding to 11 μM) of the cis isomers GLA, LNA, and OA, the β-galactosidase activity clearly decreased with an increasing degree of unsaturation, with GLA being the most potent inhibitor. The trans isomers of LNA and OA, namely, LLA and EA, respectively, were still able to reduce the β-galactosidase activity but appeared to be less effective. A 4-fold higher subinhibitory concentration of LNA, LLA, OA, and EA corroborate the lower effectiveness of the trans isomers relative to the cis isomers of the C18:2 and C18:1 FFAs (Fig. 1B). Again, the inhibitory effect seen for the C18 FFAs seemed to specifically target PrfA activity, as L. monocytogenes prfA-G155S cells harboring plhrA36-lacZ were not affected by these FFAs (Fig. S3B). These results further substantiate that the growth inhibition property of an FFA (Table 1) does not correlate with its PrfA-inhibitory potential; clearly, the nonantimicrobial OA and, to a lesser extent, LLA and EA are capable of reducing the PrfA-dependent activation of phly-lacZ (Fig. 1B).
Altogether, our systematic analysis revealed that some FFAs (i.e., MA, OA, LLA, and EA) can repress the PrfA-dependent activation of hly transcription without exerting an antimicrobial effect on L. monocytogenes, whereas others, as exemplified by LA, GLA, and LNA, have antimicrobial as well as PrfA-inhibitory properties. Finally, as reported previously, SA and PAL do not affect L. monocytogenes. Thus, based on the present extended analysis, we conclude that the antimicrobial and PrfA-inhibitory properties of an FFA do not necessarily correlate.
Selected FFAs downregulate the virulence factors LLO and ActA.
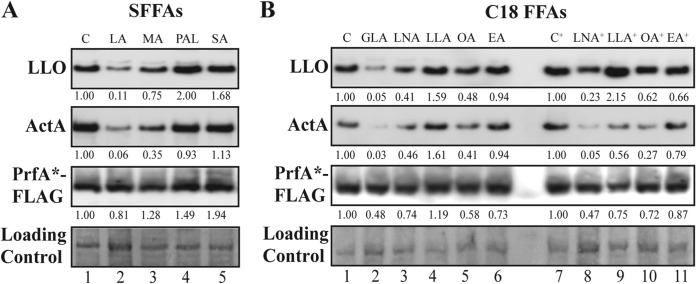
To further investigate whether the FFAs affect virulence gene expression through the repression of PrfA activity, the cellular protein levels of LLO and ActA, as well as FLAG-tagged PrfA*, were determined after exposure to the nine different FFAs. For Western blot analysis, the EGDprfA*-FLAG strain was grown in BHI until reaching an optical density at 600 nm (OD600) of 0.3; then, the culture was split, and the cells were treated with either SFFAs or C18 FFAs for 3 h, using the same concentrations as for the β-galactosidase assays. For the SFFA-treated cells, the levels of LLO and ActA decreased by more than 2.5-fold after exposure to LA and, to a lesser extent, MA (Fig. 2A, compare lanes C, LA, and MA). Regarding the unsaturated C18 FFAs, GLA had the most pronounced effect on LLO and ActA levels (Fig. 2B, compare lanes C and GLA), but also, exposure to LNA and, to a lesser extent, OA led to decreased levels of LLO and ActA at the highest concentration of FFA tested (Fig. 2B, compare lanes C+, LNA+, and OA+). Notably, 3 h of FFA exposure did not dramatically alter the levels of PrfA*-FLAG, supporting that the FFAs repress the levels of virulence factors through PrfA, most likely by inhibiting its transcription activator function.
FIG 2.
FFA-mediated downregulation of the virulence factors LLO and ActA. Western blot analyses of LLO, ActA, and PrfA*-FLAG. Samples were taken from EGDprfA*-FLAG grown to an OD600 of 0.3 and exposed for 3 h to SFFAs in a concentration of 10 μg/ml (corresponding to 50 μM LA, 44 μM MA, 39 μM PAL, or 35 μM SA) (A) or to C18 FFAs in a concentration of 3 μg/ml (corresponding to 11 μM FFA) or 12 μg/ml (corresponding to 43 μM FFA+) (B). As controls, vehicle was added corresponding to the concentration present in the FFA-treated cultures (C or C+). LLO and ActA were detected using anti-LLO and anti-ActA antibodies, whereas PrfA*-FLAG was detected using an anti-FLAG antibody. As a loading control, all proteins on the membrane were Coomassie stained. Relative levels of LLO, ActA, and PrfA*-FLAG (normalized to the loading control and relative to C or C+) are shown below each lane. Differences were considered significant if the relative amounts of normalized protein levels (FFA versus control) were consistently below 0.4 or above 2.5 in all replicates.
Selected FFAs rapidly reduce the mRNA levels of PrfA-activated genes.
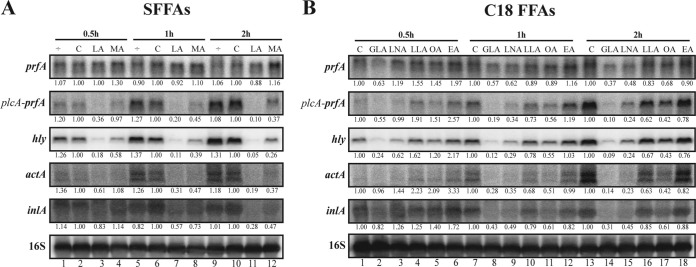
The downregulation of LLO and ActA by the SFFAs LA and MA, as well as the different levels of regulation seen for the C18 FFAs, led us to investigate how quickly the mRNA levels of several PrfA-regulated genes were reduced, following the exposure to the different FFAs. Total RNA was isolated from L. monocytogenes prfA-G155S cells grown to early exponential phase, and then, cells were subjected to subinhibitory concentrations of SFFAs or C18 FFAs for 0.5, 1, and 2 h. By Northern blotting, we found that exposure to 50 μM LA (Fig. 3A) reduced the mRNA levels of the virulence genes (except for the monocistronic prfA transcript). With a longer exposure time (2 h), the effect was more pronounced (>2.5-fold). As reported above (Fig. 1A and 2A), LA seems to be more effective than MA in reducing the transcript levels of the virulence genes. Exposure to the C18 FFAs (11 μM) (Fig. 3B) revealed that the degree of unsaturation is important for reducing the transcript levels, as GLA provided the fastest and most effective response of all five C18 FFAs. In addition, the cis isomer LNA also was more efficient in reducing the mRNA levels of the tested genes, compared with the trans isomer LLA. In fact, LLA had very little effect (less than 2.5-fold) on mRNA levels after 2 h of stress. To summarize, the Northern blots presented in Fig. 3 confirm the trends observed in both the β-galactosidase and Western blot assays; furthermore, we note that the virulence-inhibitory effect of selected FFAs (LA and GLA) can be clearly seen at the mRNA level for hly after just 0.5 h of FFA exposure.
FIG 3.
Time-dependent repression of PrfA-regulated virulence genes upon FFA exposure. Northern blot analyses of prfA, plcA (when cotranscribed with prfA), hly, actA, and inlA mRNAs. Samples were taken from EGDprfA* cultures exposed to 0.5, 1, and 2 h of SFFAs in a concentration of 50 μM (LA or MA) (A) or to C18 FFAs in a concentration of 11 μM (B). As controls, the cultures were left untreated (÷) or vehicle was added corresponding to the concentration present in the FFA-treated cultures (C). Northern blots were probed for prfA mRNA, hly mRNA, actA mRNA, inlA mRNA, and 16S rRNA (loading control). Relative levels of the transcripts (normalized to 16S and relative to C at each time point) are shown below each lane. Differences were considered significant if the relative amounts of normalized RNA levels (FFA versus control) were consistently below 0.4 or above 2.5 in all replicates.
The unsaturated C18 FFAs efficiently reduce the DNA binding activity of PrfA*.
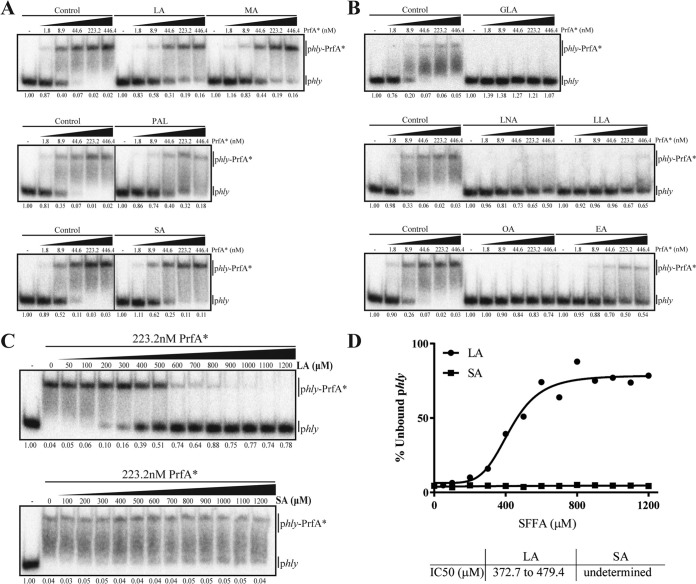
Based on the in vivo results obtained so far, specific FFAs seem to interfere with the regulatory activity of PrfA; however, it is not clear if these FFAs act directly on PrfA or if they act through a signaling pathway that leads to PrfA inactivation. To investigate if FFAs exert a direct effect on the DNA binding activity of PrfA, electrophoretic mobility shift assays (EMSAs) were performed. Reports have shown that the highly constitutively active PrfA* variant PrfA-G145S has a higher binding affinity to PrfA boxes than wild-type PrfA and other midlevel constitutively active PrfA* mutants (14, 19). Thus, the binding capacity of both PrfA-G145S and PrfA-G155S (the PrfA* mutant used for the in vivo experiments) was first tested. Increasing amounts of each of the purified PrfA* mutant proteins were added to individual binding reaction mixtures containing a constant concentration of 32P-labeled phly DNA probe, covering the PrfA box in the hly promoter region. A band shift showed that the PrfA-G145S-phly complex was clearly formed (see Fig. S4 in the supplemental material). In comparison, the PrfA-G155S-phly complex appeared to be highly unstable under the tested conditions (Fig. S4). Based on that, we decided to use PrfA-G145S for the following EMSAs.
Increasing amounts of PrfA* (PrfA-G145S) and a constant concentration of 32P-labeled phly DNA probe were mixed together with either bovine serum albumin (BSA) alone (control) or BSA complexed to the different FFAs. The resulting EMSAs showed that at the same micromolar concentration (i.e., 256 μM), the SFFAs, including LA and MA, hardly affected the PrfA*-phly complex formation (Fig. 4A), whereas the unsaturated C18 FFAs seemed very efficient at inhibiting the interaction between PrfA* and phly DNA (Fig. 4B). Since the SFFA LA inhibits PrfA-dependent activation very efficiently in vivo (Fig. 1A, 2A, and 3A), we speculated that a higher concentration of LA might be required to abolish the protein-DNA interaction in vitro. This led us to investigate in more detail the effect of LA, compared with the noninhibitory SA, on the DNA binding activity of PrfA*. In Fig. 4C, an in vitro titration experiment is shown, where increasing concentrations of LA or SA were added to constant amounts of PrfA* and 32P-labeled phly DNA. In the absence of FFAs, a PrfA*-phly complex was clearly formed. Notably, the addition of increasing amounts of LA progressively inhibited PrfA*-phly complex formation, whereas this was not the case for SA. The intensity of the bands was used to determine the percentage of unbound phly at each LA or SA concentration. Those values allowed us to calculate the concentration of LA or SA required to disrupt 50% (50% inhibitory concentration [IC50]) of the already formed protein-DNA complex (shown in Fig. 4D). Altogether, these data support the hypothesis that the PrfA inhibitory effect observed in vivo for LA, but not SA, may be explained by the specific ability of LA to inhibit the DNA binding activity of PrfA.
FIG 4.
FFA-mediated in vitro inhibition of the PrfA*-phly complex formation. (A and B) EMSAs of PrfA*-phly interaction. Labeled phly was tested for its ability to interact with increasing concentrations of purified PrfA* protein (His-tagged PrfA-G145S) in the absence (Control) or presence of 256 μM LA, MA, PAL, or SA (A) or 256 μM of GLA, LNA, LLA, OA, or EA (B). The fraction of unbound phly is shown below each lane. (C and D) Titration EMSAs of LA and SA. (C) Increasing concentrations of LA or SA were added to constant amounts of PrfA* and phly. The fraction of unbound phly is shown below each lane. (D) Unbound phly at each LA or SA concentration was used to draw a dose-response inhibition curve to obtain the concentration of LA or SA required to disrupt 50% of the already formed PrfA*-phly complex (IC50).
Among the C18 FFAs, GLA (C18:3 cis) is the most efficient inhibitor of PrfA activity, both in vivo and in vitro (compare Fig. 1B, 2B, and 3B with Fig. 4B). These findings indicate that GLA binds directly to PrfA*, hence preventing its ability to bind DNA. As shown above, the saturated counterpart to GLA, SA (C18:0) neither affects PrfA activity in vivo nor in vitro. Furthermore, the trans isomer EA is less efficient at inhibiting PrfA*-phly complex formation than the cis isomer OA (Fig. 4B). Collectively, our findings support that the unsaturated FFAs of the C18 series act as signaling molecules to inhibit PrfA activity, most likely by direct interaction with PrfA, which inhibits its ability to bind DNA.
DISCUSSION
Due to antibiotic overuse and misuse, antibiotic resistance has emerged as one of the biggest threats to global health. Bacteria are becoming resistant to the drugs currently used to treat infections at a much higher rate than novel antibiotics are discovered (26). Consequently, alternative therapies are gaining attention, such as antivirulence agents that can neutralize pathogenic bacteria by acting on their virulence factors (27–33). Multidrug-resistant Listeria species have been isolated from diverse sources, including food and clinical and external environments, and this acquired resistance has been associated with the presence of mutations within genes encoding efflux pumps or the acquisition of mobile genetic elements (34). L. monocytogenes is the third most common cause of death from food poisoning in humans, presenting a mortality rate of around 30% despite antibiotic treatment (34, 35). As a consequence, alternative therapies, such as plant products, have been tested lately for their antivirulent properties against L. monocytogenes. For example, thymoquinone (isolated from Nigella sativa seeds) was recently found to reduce motility, biofilm formation, secretion of LLO, and transcription of hly and other virulence-associated genes (36). Similar results were obtained with an essential oil extracted from Cannabis sativa L. and with the plant antimicrobials trans-cinnamaldehyde, carvacrol, and thymol, which additionally reduced L. monocytogenes adhesion to and invasion of Caco-2 enterocyte-like cells (37, 38). Effects on the hemolytic potential of L. monocytogenes were also observed upon exposure to subinhibitory concentrations of tea tree oil (39) and Stevia rebaudiana Bertoni (stevia), especially at lower temperatures (40). Notably, the mechanism by which exposure to such compounds reduces the expression of virulence genes is still unknown (36–40).
Naturally occurring FFAs have also been suggested as potential antivirulence agents against pathogenic bacteria (25, 30). Indeed, an earlier study reported a decreased invasiveness of L. monocytogenes into Caco-2 cells upon exposure to FFAs present in milk (41); however, the mechanism behind this effect was not unveiled. Previously, we suggested that selected antimicrobial FFAs could act through PrfA to reduce L. monocytogenes virulence potential (9). In the present study, we extended the number of FFAs tested and observed that 3 out of 9 FFAs, namely, LA, GLA, and LNA (Table 1), were antimicrobial toward L. monocytogenes. How the carbon chain length and degree of unsaturation affect the antimicrobial capacity of an FFA has been summarized by others (1), and the previously observed tendencies meet our findings, which are as follows: (i) unsaturated FFAs tend to have greater potency than saturated FFAs with the same length carbon chain; (ii) often a direct correlation exists between the number of double bonds in an unsaturated FFA and its antibacterial efficacy; (iii) FFAs with double bonds in a cis orientation tend to have greater antibacterial activity than the trans counterparts; and finally, (iv) for saturated FFAs, the most active ones have 10 or 12 carbons in the chain, and the antibacterial efficacy tends to decrease as the chain length gets longer or shorter (1). Previously, we suggested that antimicrobial FFAs also have PrfA inhibitory properties (e.g., LA and GLA) (9). Here, we corroborate those findings, as exposure to subinhibitory concentrations of the antimicrobial FFA LNA inhibits PrfA-mediated activation of virulence genes. We further demonstrate that an antimicrobial activity is not compulsory for the PrfA-inhibitory ability of an FFA, which is most clearly illustrated by the results obtained with the nonantimicrobial FFAs MA and OA (Fig. 1 to 3). Interestingly, the Western blots showed that the PrfA* protein was clearly present during exposure to these FFAs, supporting the idea that downregulation of the virulence factors is due to reduced PrfA activity.
Based on these findings, we hypothesized that FFAs act directly on PrfA to interfere with its DNA binding capacity. Accordingly, all nine FFAs were tested for their ability to interfere with the binding of PrfA* to the PrfA box in the hly promoter in vitro (Fig. 4). Curiously, we observed that at the same concentration, the unsaturated C18 FFAs promptly abolished the PrfA*-phly binding, whereas the SFFAs LA and MA, which appeared highly effective in vivo, barely affected the complex formation. It should be noted that for the Northern blot experiment, the concentration of SFFAs used was higher than the C18 FFAs (50 μM versus 11 μM) (Fig. 3), suggesting that higher concentrations of LA and MA might be necessary in order to disrupt PrfA*-DNA binding in vitro. This assumption was tested by the titration EMSAs performed for LA and SA, which showed that increasing concentrations of LA progressively disrupted PrfA*-DNA binding, whereas the addition of SA had no effect on complex formation. Altogether, the SFFA LA acts well in vivo and inhibits PrfA*-DNA binding in vitro. In contrast, SA neither affects virulence gene expression nor prevents binding of PrfA* to DNA under the conditions tested here. Regarding the unsaturated C18 FFAs, they were more efficient at interfering with PrfA*-phly complex formation, with GLA being the most efficient inhibitor of PrfA activity in vitro, as well as in vivo. In conclusion, the EMSAs support our hypothesis of a direct interaction between FFAs and PrfA, which prevents the protein from binding to DNA. Similar hypotheses have been investigated in other bacteria, including the intestinal pathogens V. cholera and S. enterica. In V. cholera, the unsaturated FFAs LNA and OA were able to reduce the expression of ToxT-regulated virulence genes, whereas the saturated FFAs PAL and SA had no significant effect (5). Interestingly, LNA and a conjugated form of LNA (CLA) were shown to reduce V. cholera ToxT binding to various virulence promoters in vitro (6, 7). In S. enterica, OA interacts directly with the virulence regulator HilD and affects its DNA binding ability, leading to a downregulation of virulence genes required for the expression of the Salmonella pathogenicity island 1 type III secretion system (8). Notably, ToxT and HilD belong to the AraC family of transcription regulators, whereas PrfA belongs to the CRP/FNR family (25). Altogether, these findings suggest that intestinal pathogens are using specific FFAs as signaling molecules to control the DNA binding activity of key virulence regulators.
Besides FFAs, other compounds acting to prevent PrfA-dependent virulence gene expression have been investigated (42–44). For instance, the purine analog 6-N-hydroxylaminopurine (6-N-HAP) has been shown to effectively reduce the expression of virulence genes by affecting both PrfA activity and PrfA protein levels (unlinked mechanisms) (43). Interestingly, 6-N-HAP did not affect the levels of the virulence factors in a strain expressing a PrfA* protein (43). These results demonstrated that while 6-N-HAP may prevent the activation of PrfA, a constitutively active PrfA* protein is able to overcome the inhibitory effect of the compound (43). Studies with ring-fused 2-pyridone molecules presented similar results, i.e., abolished expression of PrfA-regulated genes and a reduced uptake of L. monocytogenes by epithelial cells (42). Further in vitro experiments showed that the ring-fused 2-pyridone molecules could bind both PrfA and PrfA* (PrfA-G145S) (42); however, while binding these molecules to PrfA led to a lower affinity of PrfA for the virulence gene promoters, binding the compounds to PrfA* did not affect its DNA binding activity (42). Structurally, the C-terminal region of PrfA contains a winged helix-turn-helix (HTH) motif responsible for the binding to the DNA promoter regions (19). The amino acid substitution in PrfA* repositions the HTH motif to an active conformation with increased DNA binding affinity (19). Good et al. suggested that the inhibitory compounds are likely competing with a cofactor, like GSH, that stabilizes the HTH motif in the PrfA protein and that the binding of the inhibitor occurs prior to the stabilization of the DNA binding HTH motif, as the PrfA* protein with the stabilized HTH motif was not affected by the ring-fused 2-pyridone molecules (42). In a similar way, non-cysteine-containing peptides, such as Leu dipeptides, were shown to inhibit PrfA activity by binding the protein with high affinity and competing for the occupancy of the GSH binding site (44). Again, the inhibitory peptides were not able to inhibit the DNA binding capacity of a constitutively active PrfA* protein (44). Since our results demonstrate that specific FFAs can act on a constitutively active PrfA* protein (PrfA-G155S in vivo, and PrfA-G145S in vitro), we speculate that FFAs may act differently than ring-fused 2-pyridone molecules and inhibitory peptides and possibly do not bind to the GSH pocket. Alternatively, FFAs may bind to a different site on PrfA, which does not interfere with PrfA activation, but instead promotes a conformational change that hinders PrfA*-DNA binding. Further studies should focus on understanding where exactly FFAs bind in the PrfA protein and how a potential conformational change inhibits binding of PrfA to promoter regions and, consequently, prevents PrfA-dependent activation of virulence genes. Importantly, the inhibition of constitutively active PrfA* variants by specific FFAs suggests that these compounds could be feasible candidates for alternative and/or complementary therapies against pathogenic L. monocytogenes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Wild-type L. monocytogenes EGD serotype 1/2a was used in this study (obtained from W. Goebel, University of Wurzburg, Wurzburg, Germany). The isogenic mutant derivatives EGDΔprfA (45); EGDprfA*, expressing the midlevel constitutively active PrfA mutant derivative PrfA-G155S (9); and EGDprfA*-FLAG, expressing N-terminal 3× FLAG-tagged PrfA-G155S (9) have been described previously. L. monocytogenes was routinely grown at 37°C with aeration in brain heart infusion medium (BHI; Oxoid), and when required, cultures were supplemented with 50 μg/ml kanamycin.
To construct a plasmid expressing an N-terminal His-tagged PrfA-G145S, we performed 2-step PCR amplifications on EGD chromosomal DNA, where fragments containing the 6× His tag coding sequence inserted between codon 2 and 3 of prfA were amplified, as well as fragments containing the desired prfA* substitution. For construction of the N-terminal His-tagged PrfA-G155S, a 1-step PCR amplification was performed on EGDprfA* (prfA-G155S) chromosomal DNA to equally insert the 6× His tag coding sequence between codons 2 and 3 of prfA. Primers are listed in Table S1 in the supplemental material. The resulting DNA fragments were first inserted into pLITMUS28i (New England BioLabs) and then reinserted into the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression vector pUHE24-2 (46), resulting in the plasmids pUHE24-2-prfA-G145S-[His]6 and pUHE24-2-prfA-G155S-[His]6. For expression of His-tagged PrfA-G145S and PrfA-G155S (see below), Escherichia coli TOP10 (Invitrogen) containing the lacIq-carrying plasmid pMS421 (47) and the relevant pUHE24-2 construct were grown at 37°C with aeration in Luria-Bertani medium (LB) supplemented with 100 μg/ml spectinomycin and 100 μg/ml ampicillin.
The FFAs tested in this study were lauric acid (LA; C12:0; purity, ≥98%), myristic acid (MA; C14:0; purity, ≥99%), palmitic acid (PAL; C16:0; purity, ≥99%), stearic acid (SA; C18:0; purity, ≥98.5%), γ-linolenic acid (GLA; C18:36,9,12 cis; purity, ≥99%), linoleic acid (LNA; C18:29,12 cis; purity, ≥99%), linolelaidic acid (LLA; C18:29,12 trans; purity, ≥98%), oleic acid (OA; C18:19 cis; purity, ≥99%), and elaidic acid (EA; C18:19 trans; purity, ≥99%), and they were purchased from Sigma-Aldrich. All FFAs were solubilized and diluted in 96% ethanol, and thus, ethanol control samples were included as vehicle controls, corresponding to the highest concentration of FFA used.
FFA inhibitory concentration determination.
For the inhibitory concentration (IC) determination, overnight cultures of EGD wild type, EGDprfA* (prfA-G155S), or EGDΔprfA were diluted to an OD600 of 0.0002, and 4 ml was transferred to glass tubes containing increasing concentrations of MA, LNA, LLA, OA, or EA. After 20 h of growth, the OD600 measurements were recorded, and the lowest concentration with an OD600 of ≤0.1 was defined to be the IC. The experiment was performed in three biological replicates. For IC values presented as >X, the value X corresponds to the highest FFA concentration that could possibly be tested without precipitations being formed in the growth medium.
lacZ fusions and β-galactosidase assays.
To investigate the effect of FFAs on PrfA-G155S activity, the previously constructed L. monocytogenes EGDprfA* strains harboring the reporter plasmid phly-lacZ (48) or plhrA36-lacZ (49) were used; these strains contain the promoter region of a PrfA-dependent or PrfA-independent gene, respectively, fused to the promoterless lacZ in the transcriptional vector pTCV-lac (50). The strains were grown overnight, diluted to an OD600 of 0.02 into fresh BHI, and grown to an OD600 of 0.3. Cultures were split and FFAs were added to obtain a final concentration of 10 μg/ml (50 μM LA, 44 μM MA, 39 μM PAL, and 35 μM SA), 75 μg/ml (293 μM PAL and 264 μM SA), 3 μg/ml (11 μM GLA, LNA, LLA, OA, and EA), or 12 μg/ml (43 μM LNA, LLA, OA, and EA). As controls, the cultures were left untreated (÷) or vehicle was added corresponding to the concentration present in the FFA-treated cultures (C or C+). Samples (1 ml) were harvested after 20 h of growth. β-Galactosidase assays were conducted as previously described (51). The experiment was performed in three biological replicates, with each carried out in technical duplicates. The results were analyzed using a one-way analysis of variance (ANOVA) with Bonferroni’s multiple-comparison test. Only differences with at least 95% confidence were reported as statistically significant.
Preparation of protein extracts and Western blot analysis.
L. monocytogenes EGDprfA*-FLAG overnight cultures were diluted to an OD600 of 0.02 and grown to an OD600 of 0.3. Cultures were split and exposed to the same final concentrations of FFAs used for the β-galactosidase assays (except the highest concentrations of PAL and SA). Vehicle-treated cultures were included as controls. After 3 h of FFA exposure, 4 ml of culture was withdrawn and centrifuged at 3,000 relative centrifugal force (rcf) for 7 min at 4°C. The supernatant was discarded, and the pellets were resuspended in lysis buffer (0.1 M NaCl, 50 mM Tris-HCl [pH 7.5], 10 mM EDTA, and 0.5% SDS). Protein extract preparation, Western blot analysis, and antibody detection were conducted as described earlier (9). Coomassie staining of the membrane was achieved by washing the membrane three times in distilled water (dH2O) with shaking at 5 min each time, Coomassie staining (0.025% brilliant blue 250-G, 40% methanol, and 7% acetic acid) for 5 min, destaining (50% methanol and 7% acetic acid) for 5 to 10 min, and rinsing the membrane with dH2O several times before scanning (52). The protein bands were visualized using an Amersham Imager 680 (GE Healthcare), and the relative amounts of protein normalized to a representative band of the Coomassie-stained membrane were estimated using ImageJ. The Western blot analyses were repeated twice with similar results; representative examples are shown. Differences were considered significant if the relative amounts of normalized protein levels (FFA versus control) were consistently below 0.4 or above 2.5 in all replicates.
Total RNA extraction and Northern blot analysis.
To test the reduction on the mRNA levels of selected PrfA-dependent genes upon exposure to FFAs, L. monocytogenes EGDprfA* overnight cultures were diluted to an OD600 of 0.02 and grown to an OD600 of 0.3. Cultures were split, and the SFFAs LA and MA were added to a final concentration of 50 μM, whereas the C18 FFAs GLA, LNA, LLA, OA, and EA were added to a final concentration of 11 μM (corresponding to 3 μg/ml). Untreated and/or vehicle-treated cultures were included as controls. A total of 10 ml of each sample was taken from controls and FFA-exposed cultures after 0.5, 1, and 2 h, snap-cooled in liquid nitrogen, and centrifuged at 11,000 rcf for 3 min at 4°C. Cells were disrupted by the FastPrep instrument (Bio101; Thermo Scientific Corporation), and total RNA was extracted using the TRI reagent (Molecular Research Center, Inc.) as previously reported (53). The integrity, concentration, and purity of the RNA were confirmed by agarose gel electrophoresis and spectrophotometry on a DeNovix DS-11 Fx+ instrument. For Northern blot analysis (54, 55), 20 μg of total RNA was loaded on a formaldehyde-agarose gel and separated for 3 h and 15 min prior to capillarity blotting on a Zeta-Probe membrane (Bio-Rad). The membrane was hybridized with 32P-labeled single-stranded DNA probes (Table S1). A probe against 16S RNA was used for normalization of the RNA levels. RNA bands were visualized using a Typhoon FLA9000 instrument (GE Healthcare) and analyzed with IQTL 8.0 quantification software (GE Healthcare). The Northern blot experiments were performed in triplicates with similar results; representative examples are shown. Differences were considered significant if the relative amounts of normalized RNA levels (FFA versus control) were consistently below 0.4 or above 2.5 in all replicates.
Purification of His-tagged PrfA-G145S and PrfA-G155S.
The plasmids pUHE24-2-prfA-G145S-[His]6 and pUHE24-2-prfA-G155S-[His]6 were transformed into E. coli TOP10 containing pMS421 (47). The strains TOP10-pMS421-pUHE24-2-prfA-G145S-[His]6 and TOP10-pMS421-pUHE24-2-prfA-G155S-[His]6 were grown overnight in LB medium; diluted to an OD600 of 0.012 into fresh LB; and grown to an OD600 of ≈0.4, at which point 1 mM IPTG was added to induce protein expression. After 4 h, the cells were centrifuged at 10,000 rcf for 20 min at 4°C, and the pellets were resuspended in wash buffer (50 mM Na3PO4 [pH 7.0] and 300 mM NaCl). The cells were lysed twice using a French press at 1.8 kbar, and the debris was removed by centrifugation at 20,000 rcf for 1 h at 4°C. The cleared lysates were mixed with preequilibrated Talon metal affinity resins (Clontech laboratories, Inc.) and incubated with rotation for 20 min at 4°C. After incubation, the samples were centrifuged at 700 rcf for 5 min at 4°C, the supernatant was discarded, and the resins were washed twice in wash buffer. The resins were then resuspended in wash buffer and transferred to a gravity flow column. The column was washed with wash buffer containing 15 mM imidazole. Purified proteins were eluted in elution buffer (50 mM Na3PO4 [pH 7.0], 300 mM NaCl, and 150 mM imidazole) and dialyzed overnight against wash buffer. The protein samples were further dialyzed by use of Amicon Ultra centrifugal filter units (Ultra-15), with a cutoff at 10,000 kDa, by the addition of wash buffer and centrifugation at 5,000 rcf for 20 min at 4°C for a total of three times. Then, wash buffer with 10% glycerol was added twice to the filters and centrifuged. The remaining protein sample inside the filters was kept and stored at –20°C. The protein concentration was determined using the Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fischer Scientific) according to the manufacturer’s instructions.
Electrophoretic mobility shift assays (EMSAs).
The 54-bp DNA fragment containing the PrfA box of the hly promoter (phly) was prepared using 32P-labeled primers (Table S1) on EGD chromosomal DNA. A total of 0.0015 pmol of labeled phly was incubated with the indicated amounts of purified PrfA-G145S or PrfA-G155S to a total volume of 10 μl in the presence of binding buffer (20 mM HEPES [pH 8.0], 50 mM KCl, 1 mM EDTA, 3% Ficoll 400, and 1 mM dithiothreitol [DTT]) and unspecific DNA (10 μg/ml salmon sperm DNA; Invitrogen), as well as FFAs complexed with fat-free BSA (Sigma) to promote FFA solubility. Briefly, appropriate amounts of the FFAs dissolved in ethanol were dried under a stream of N2; 1,430 μM KOH was then added and the FFAs sonicated for 15 min; and finally, 256 μM of fat-free BSA dissolved in binding buffer was added to the FFAs. For the control conditions (no FFAs), BSA-KOH was used instead. The formation of the PrfA*-phly complexes was promoted by incubating the reaction mixtures at 37°C for 15 min and an additional 10 min on ice. Samples were loaded and separated at 4°C on a 5% nondenaturing polyacrylamide gel for 1.5 h. Bands were visualized and quantified as described for the Northern blotting experiments. For the titration experiments, curves based on the band quantification of unbound phly were drawn using GraphPad Prism 7.05 software (dose-response inhibition) to obtain the IC50 (concentration of the FFA required to free 50% of the complex formed). The EMSA experiments were repeated at least twice with similar results; representative examples are shown.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the Novo Nordisk Foundation (grant number NNF17OC0028528).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Desbois AP, Smith VJ. 2010. Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl Microbiol Biotechnol 85:1629–1642. doi: 10.1007/s00253-009-2355-3. [DOI] [PubMed] [Google Scholar]
- 2.Choi JS, Park NH, Hwang SY, Sohn JH, Kwak I, Cho KK, Choi IS. 2013. The antibacterial activity of various saturated and unsaturated fatty acids against several oral pathogens. J Environ Biol 34:673–676. [PubMed] [Google Scholar]
- 3.Das UN. 2018. Arachidonic acid and other unsaturated fatty acids and some of their metabolites function as endogenous antimicrobial molecules: a review. J Adv Res 11:57–66. doi: 10.1016/j.jare.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marques CN, Davies DG, Sauer K. 2015. Control of biofilms with the fatty acid signaling molecule cis-2-decenoic acid. Pharmaceuticals (Basel) 8:816–835. doi: 10.3390/ph8040816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterjee A, Dutta PK, Chowdhury R. 2007. Effect of fatty acids and cholesterol present in bile on expression of virulence factors and motility of Vibrio cholerae. Infect Immun 75:1946–1953. doi: 10.1128/IAI.01435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plecha SC, Withey JH. 2015. Mechanism for inhibition of Vibrio cholerae ToxT activity by the unsaturated fatty acid components of bile. J Bacteriol 197:1716–1725. doi: 10.1128/JB.02409-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Withey JH, Nag D, Plecha SC, Sinha R, Koley H. 2015. Conjugated linoleic acid reduces cholera toxin production in vitro and in vivo by inhibiting Vibrio cholerae ToxT activity. Antimicrob Agents Chemother 59:7471–7476. doi: 10.1128/AAC.01029-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golubeva YA, Ellermeier JR, Cott Chubiz JE, Slauch JM. 2016. Intestinal long-chain fatty acids act as a direct signal to modulate expression of the Salmonella pathogenicity island 1 type III secretion system. mBio 7:e02170-15. doi: 10.1128/mBio.02170-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sternkopf Lillebæk EM, Lambert Nielsen S, Scheel Thomasen R, Færgeman NJ, Kallipolitis BH. 2017. Antimicrobial medium- and long-chain free fatty acids prevent PrfA-dependent activation of virulence genes in Listeria monocytogenes. Res Microbiol 168:547–557. doi: 10.1016/j.resmic.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Scortti M, Monzó HJ, Lacharme-Lora L, Lewis DA, Vázquez-Boland JA. 2007. The PrfA virulence regulon. Microbes Infect 9:1196–1207. doi: 10.1016/j.micinf.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 11.de las Heras A, Cain RJ, Bielecka MK, Vázquez-Boland JA. 2011. Regulation of Listeria virulence: PrfA master and commander. Curr Opin Microbiol 14:118–127. doi: 10.1016/j.mib.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Bruno JC Jr, Freitag NE. 2010. Constitutive activation of PrfA tilts the balance of Listeria monocytogenes fitness towards life within the host versus environmental survival. PLoS One 5:e15138. doi: 10.1371/journal.pone.0015138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vasanthakrishnan RB, de Las Heras A, Scortti M, Deshayes C, Colegrave N, Vázquez-Boland JA. 2015. PrfA regulation offsets the cost of Listeria virulence outside the host. Environ Microbiol 17:4566–4579. doi: 10.1111/1462-2920.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miner MD, Port GC, Freitag NE. 2008. Functional impact of mutational activation on the Listeria monocytogenes central virulence regulator PrfA. Microbiology 154:3579–3589. doi: 10.1099/mic.0.2008/021063-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freitag NE, Port GC, Miner MD. 2009. Listeria monocytogenes—from saprophyte to intracellular pathogen. Nat Rev Microbiol 7:623–628. doi: 10.1038/nrmicro2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reniere ML, Whiteley AT, Hamilton KL, John SM, Lauer P, Brennan RG, Portnoy DA. 2015. Glutathione activates virulence gene expression of an intracellular pathogen. Nature 517:170–173. doi: 10.1038/nature14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall M, Grundstrom C, Begum A, Lindberg MJ, Sauer UH, Almqvist F, Johansson J, Sauer-Eriksson AE. 2016. Structural basis for glutathione-mediated activation of the virulence regulatory protein PrfA in Listeria. Proc Natl Acad Sci U S A 113:14733–14738. doi: 10.1073/pnas.1614028114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ripio MT, Domínguez-Bernal G, Lara M, Suárez M, Vazquez-Boland JA. 1997. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J Bacteriol 179:1533–1540. doi: 10.1128/jb.179.5.1533-1540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eiting M, Hageluken G, Schubert WD, Heinz DW. 2005. The mutation G145S in PrfA, a key virulence regulator of Listeria monocytogenes, increases DNA-binding affinity by stabilizing the HTH motif. Mol Microbiol 56:433–446. doi: 10.1111/j.1365-2958.2005.04561.x. [DOI] [PubMed] [Google Scholar]
- 20.Xayarath B, Freitag NE. 2012. Optimizing the balance between host and environmental survival skills: lessons learned from Listeria monocytogenes. Future Microbiol 7:839–852. doi: 10.2217/fmb.12.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansson J, Freitag NE. 2019. Regulation of Listeria monocytogenes virulence. Microbiol Spectr 7:GPP3-0064-2019. doi: 10.1128/microbiolspec.GPP3-0064-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shetron-Rama LM, Mueller K, Bravo JM, Bouwer HG, Way SS, Freitag NE. 2003. Isolation of Listeria monocytogenes mutants with high-level in vitro expression of host cytosol-induced gene products. Mol Microbiol 48:1537–1551. doi: 10.1046/j.1365-2958.2003.03534.x. [DOI] [PubMed] [Google Scholar]
- 23.Vega Y, Rauch M, Banfield MJ, Ermolaeva S, Scortti M, Goebel W, Vazquez-Boland JA. 2004. New Listeria monocytogenes prfA* mutants, transcriptional properties of PrfA* proteins and structure-function of the virulence regulator PrfA. Mol Microbiol 52:1553–1565. doi: 10.1111/j.1365-2958.2004.04052.x. [DOI] [PubMed] [Google Scholar]
- 24.Port GC, Freitag NE. 2007. Identification of novel Listeria monocytogenes secreted virulence factors following mutational activation of the central virulence regulator, PrfA. Infect Immun 75:5886–5897. doi: 10.1128/IAI.00845-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kallipolitis BH. 2017. How can naturally occurring fatty acids neutralize Listeria? Future Microbiol 12:1239–1241. doi: 10.2217/fmb-2017-0176. [DOI] [PubMed] [Google Scholar]
- 26.Ventola CL. 2015. The antibiotic resistance crisis: part 1: causes and threats. P T 40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 27.Ventola CL. 2015. The antibiotic resistance crisis: part 2: management strategies and new agents. P T 40:344–352. [PMC free article] [PubMed] [Google Scholar]
- 28.Clatworthy AE, Pierson E, Hung DT. 2007. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol 3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 29.Escaich S. 2010. Novel agents to inhibit microbial virulence and pathogenicity. Expert Opin Ther Pat 20:1401–1418. doi: 10.1517/13543776.2010.511176. [DOI] [PubMed] [Google Scholar]
- 30.Brown D. 2015. Antibiotic resistance breakers: can repurposed drugs fill the antibiotic discovery void? Nat Rev Drug Discov 14:821–832. doi: 10.1038/nrd4675. [DOI] [PubMed] [Google Scholar]
- 31.Dickey SW, Cheung GYC, Otto M. 2017. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat Rev Drug Discov 16:457–471. doi: 10.1038/nrd.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson BK, Abramovitch RB. 2017. Small molecules that sabotage bacterial virulence. Trends Pharmacol Sci 38:339–362. doi: 10.1016/j.tips.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fleitas Martinez O, Cardoso MH, Ribeiro SM, Franco OL. 2019. Recent advances in anti-virulence therapeutic strategies with a focus on dismantling bacterial membrane microdomains, toxin neutralization, quorum-sensing interference and biofilm inhibition. Front Cell Infect Microbiol 9:74. doi: 10.3389/fcimb.2019.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luque-Sastre L, Arroyo C, Fox EM, McMahon BJ, Bai L, Li F, Fanning S. 2018. Antimicrobial resistance in Listeria species. Microbiol Spectr 6:ARBA-0031-2017. doi: 10.1128/microbiolspec.ARBA-0031-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swaminathan B, Gerner-Smidt P. 2007. The epidemiology of human listeriosis. Microbes Infect 9:1236–1243. doi: 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 36.Miao X, Liu H, Zheng Y, Guo D, Shi C, Xu Y, Xia X. 2019. Inhibitory effect of thymoquinone on Listeria monocytogenes ATCC 19115 biofilm formation and virulence attributes critical for human infection. Front Cell Infect Microbiol 9:304. doi: 10.3389/fcimb.2019.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Upadhyay A, Johny AK, Amalaradjou MA, Ananda Baskaran S, Kim KS, Venkitanarayanan K. 2012. Plant-derived antimicrobials reduce Listeria monocytogenes virulence factors in vitro, and down-regulate expression of virulence genes. Int J Food Microbiol 157:88–94. doi: 10.1016/j.ijfoodmicro.2012.04.018. [DOI] [PubMed] [Google Scholar]
- 38.Marini E, Magi G, Ferretti G, Bacchetti T, Giuliani A, Pugnaloni A, Rippo MR, Facinelli B. 2018. Attenuation of Listeria monocytogenes virulence by Cannabis sativa L. essential oil. Front Cell Infect Microbiol 8:293. doi: 10.3389/fcimb.2018.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z, Meng R, Zhao X, Shi C, Zhang X, Zhang Y, Guo N. 2016. Inhibition effect of tea tree oil on Listeria monocytogenes growth and exotoxin proteins listeriolysin O and p60 secretion. Lett Appl Microbiol 63:450–457. doi: 10.1111/lam.12666. [DOI] [PubMed] [Google Scholar]
- 40.Sansano S, Rivas A, Pina-Perez MC, Martinez A, Rodrigo D. 2017. Stevia rebaudiana Bertoni effect on the hemolytic potential of Listeria monocytogenes. Int J Food Microbiol 250:7–11. doi: 10.1016/j.ijfoodmicro.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 41.Petrone G, Conte MP, Longhi C, di Santo S, Superti F, Ammendolia MG, Valenti P, Seganti L. 1998. Natural milk fatty acids affect survival and invasiveness of Listeria monocytogenes. Lett Appl Microbiol 27:362–368. doi: 10.1046/j.1472-765x.1998.00441.x. [DOI] [PubMed] [Google Scholar]
- 42.Good JA, Andersson C, Hansen S, Wall J, Krishnan KS, Begum A, Grundstrom C, Niemiec MS, Vaitkevicius K, Chorell E, Wittung-Stafshede P, Sauer UH, Sauer-Eriksson AE, Almqvist F, Johansson J. 2016. Attenuating Listeria monocytogenes virulence by targeting the regulatory protein PrfA. Cell Chem Biol 23:404–414. doi: 10.1016/j.chembiol.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krajewski SS, Isoz I, Johansson J. 2017. Antibacterial and antivirulence effect of 6-N-hydroxylaminopurine in Listeria monocytogenes. Nucleic Acids Res 45:1914–1924. doi: 10.1093/nar/gkw1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krypotou E, Scortti M, Grundström C, Oelker M, Luisi BF, Sauer-Eriksson AE, Vázquez-Boland J. 2019. Control of bacterial virulence through the peptide signature of the habitat. Cell Rep 26:1815–1827.e5. doi: 10.1016/j.celrep.2019.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Böckmann R, Dickneite C, Middendorf B, Goebel W, Sokolovic Z. 1996. Specific binding of the Listeria monocytogenes transcriptional regulator PrfA to target sequences requires additional factor(s) and is influenced by iron. Mol Microbiol 22:643–653. doi: 10.1046/j.1365-2958.1996.d01-1722.x. [DOI] [PubMed] [Google Scholar]
- 46.Jorgensen CI, Kallipolitis BH, Valentin-Hansen P. 1998. DNA-binding characteristics of the Escherichia coli CytR regulator: a relaxed spacing requirement between operator half-sites is provided by a flexible, unstructured interdomain linker. Mol Microbiol 27:41–50. doi: 10.1046/j.1365-2958.1998.00655.x. [DOI] [PubMed] [Google Scholar]
- 47.Grana D, Gardella T, Susskind MM. 1988. The effects of mutations in the ant promoter of phage P22 depend on context. Genetics 120:319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larsen MH, Kallipolitis BH, Christiansen JK, Olsen JE, Ingmer H. 2006. The response regulator ResD modulates virulence gene expression in response to carbohydrates in Listeria monocytogenes. Mol Microbiol 61:1622–1635. doi: 10.1111/j.1365-2958.2006.05328.x. [DOI] [PubMed] [Google Scholar]
- 49.Nielsen JS, Larsen MH, Lillebæk EMS, Bergholz TM, Christiansen MHG, Boor KJ, Wiedmann M, Kallipolitis BH. 2011. A small RNA controls expression of the chitinase ChiA in Listeria monocytogenes. PLoS One 6:e19019. doi: 10.1371/journal.pone.0019019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poyart C, Trieu-Cuot P. 1997. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galactosidase in gram-positive bacteria. FEMS Microbiol Lett 156:193–198. doi: 10.1111/j.1574-6968.1997.tb12726.x. [DOI] [PubMed] [Google Scholar]
- 51.Christiansen JK, Larsen MH, Ingmer H, Sogaard-Andersen L, Kallipolitis BH. 2004. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J Bacteriol 186:3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldman A, Harper S, Speicher DW. 2016. Detection of proteins on blot membranes. Curr Protoc Protein Sci 86:10.8.1–10.8.11. doi: 10.1002/cpps.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nielsen JS, Lei LK, Ebersbach T, Olsen AS, Klitgaard JK, Valentin-Hansen P, Kallipolitis BH. 2010. Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res 38:907–919. doi: 10.1093/nar/gkp1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheehan B, Klarsfeld A, Msadek T, Cossart P. 1995. Differential activation of virulence gene expression by PrfA, the Listeria monocytogenes virulence regulator. J Bacteriol 177:6469–6476. doi: 10.1128/jb.177.22.6469-6476.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sievers S, Lund A, Menendez-Gil P, Nielsen A, Storm Mollerup M, Lambert Nielsen S, Buch Larsson P, Borch-Jensen J, Johansson J, Kallipolitis BH. 2015. The multicopy sRNA LhrC controls expression of the oligopeptide-binding protein OppA in Listeria monocytogenes. RNA Biol 12:985–997. doi: 10.1080/15476286.2015.1071011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.