We have identified five variable positions in response regulators that allow the rate constants of autophosphorylation and autodephosporylation reactions each to be altered over 3 orders of magnitude in CheY. The distributions of variable residue combinations across response regulator subfamilies suggest that distinct mechanisms associated with different variable positions allow reaction rates to be tuned independently during evolution for diverse biological purposes. This knowledge could be used in synthetic-biology applications to adjust the properties (e.g., background noise and response duration) of biosensors and may allow prediction of response regulator reaction kinetics from the primary amino acid sequence.
KEYWORDS: CheY, autodephosphorylation, autophosphorylation, receiver domain, response regulator, two-component systems
ABSTRACT
Microorganisms and plants utilize two-component systems to regulate adaptive responses to changing environmental conditions. Sensor kinases detect stimuli and alter their autophosphorylation activity accordingly. Signal propagation occurs via the transfer of phosphoryl groups from upstream kinases to downstream response regulator proteins. Removal of phosphoryl groups from the response regulator typically resets the system. Members of the same protein family may catalyze phosphorylation and dephosphorylation reactions with different efficiencies, exhibiting rate constants spanning many orders of magnitude to accommodate response time scales from milliseconds to days. We previously found that variable positions one or two residues to the C-terminal side of the conserved Asp phosphorylation site (D+2) or Thr/Ser (T+1/T+2) in response regulators alter reaction kinetics by direct interaction with phosphodonor or phosphoacceptor molecules. Here, we explore the kinetic effects of amino acid substitutions at the two positions immediately C-terminal to the conserved Lys (K+1/K+2) in the model Escherichia coli response regulator CheY. We measured CheY autophosphorylation and autodephosphorylation rate constants for 27 pairs of K+1/K+2 residues that represent 84% of naturally occurring response regulators. Effects on autodephosphorylation were modest, but autophosphorylation rate constants varied by 2 orders of magnitude, suggesting that the K+1/K+2 positions influence reaction kinetics by altering the conformational spectrum sampled by CheY at equilibrium. Additional evidence supporting this indirect mechanism includes the following: the effect on autophosphorylation rate constants is independent of the phosphodonor, the autophosphorylation rate constants and dissociation constants for the phosphoryl group analog BeF3− are inversely correlated, and the K+1/K+2 positions are distant from the phosphorylation site.
IMPORTANCE We have identified five variable positions in response regulators that allow the rate constants of autophosphorylation and autodephosporylation reactions each to be altered over 3 orders of magnitude in CheY. The distributions of variable residue combinations across response regulator subfamilies suggest that distinct mechanisms associated with different variable positions allow reaction rates to be tuned independently during evolution for diverse biological purposes. This knowledge could be used in synthetic-biology applications to adjust the properties (e.g., background noise and response duration) of biosensors and may allow prediction of response regulator reaction kinetics from the primary amino acid sequence.
INTRODUCTION
Two-component regulatory systems convert environmental stimuli into appropriate responses in bacteria, archaea, eukaryotic microorganisms, and plants (reviewed in references 1 and 2). A generic two-component system consists of a sensor histidine kinase protein (typically membrane bound) and a cytoplasmic response regulator protein. Stimuli detected by the sensor kinase modulate autophosphorylation of a conserved His residue in the cytoplasmic portion of the protein, thus capturing environmental information in the transferrable form of phosphoryl groups. The phosphoryl groups are passed from the kinase to a conserved Asp residue in the receiver domain of a response regulator. The phosphorylation state of the response regulator, in turn, controls the cellular response to the sensed stimuli (e.g., transcriptional regulation, binding to a target protein or RNA, or enzymatic activity). Two-component systems also incorporate mechanisms to remove phosphoryl groups from response regulators when stimuli subside. Thus, two-component systems are examples of a biological signal transduction pathway mediated by transient protein phosphorylation.
Receiver domains catalyze their own phosphorylation (3) and dephosphorylation (4) reactions using five conserved residues to form the active site (reviewed in references 5 and 6) (Fig. 1). Two acidic residues (DD) bind a divalent cation (typically Mg2+) that is essential for both reactions. The metal ion, a conserved Ser/Thr (T), and a conserved Lys (K) each coordinate one of the three oxygen atoms of the phosphoryl group. During phosphorylation, an oxygen atom from the side chain of the Asp (D) phosphorylation site executes a nucleophilic attack on the phosphorus atom of the phosphoryl group, whether the phosphodonor is the phosphohistidine of a sensor kinase or a small molecule, such as acetyl phosphate. During dephosphorylation, the oxygen atom of a water molecule carries out an analogous attack. In both cases, the reaction is believed to pass through a trigonal bipyramidal transition state with a planar phosphoryl group that undergoes a stereochemical inversion.
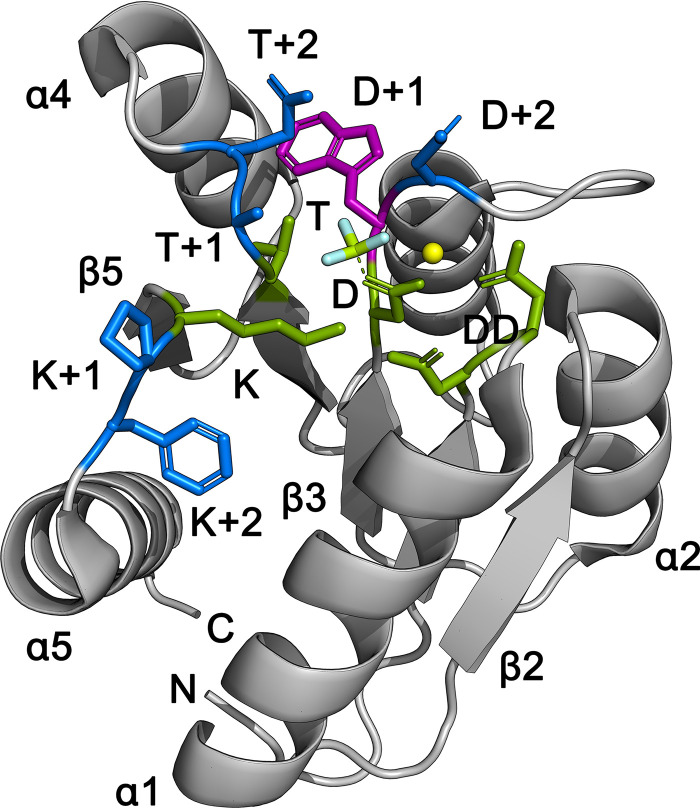
FIG 1.
Active site of E. coli CheY. Conserved active-site residues (green), Mn2+ (yellow), a BeF3− phosphoryl group analog (light blue), relevant variable residues (dark blue), and a unique Trp residue used for fluorescence assays (magenta) are shown. The N and C termini and selected secondary-structural elements (β strands and α helices) are also labeled. Structure from PDB 1fqw (55).
Response regulators exist in a spectrum of “inactive” and “active” conformations (7–11), where active is typically defined as being similar to the phosphorylated state. Unphosphorylated response regulators are believed to predominantly occupy inactive conformations. However, a small fraction of the unphosphorylated population can randomly sample more activated conformations with active-site geometries optimized for phosphorylation and hence preferentially react with phosphodonors (12, 13). Phosphorylation is believed to stabilize the more active conformations.
Receiver domains typically gain or lose phosphoryl groups in conjunction with partner proteins. Nevertheless, the autocatalytic reactions are of interest for multiple reasons. (i) Many examples exist of physiologically relevant self-catalyzed phosphorylation (reviewed in reference 14) or dephosphorylation (15) reactions. (ii) Kinases (16) and phosphatases (17–20) appear to accelerate the self-catalyzed reactions of receiver domains without altering the reaction mechanisms, emphasizing the fundamental importance of the underlying autocatalytic reactions. This modular design strategy is analogous to signal transduction by small GTPases, such as Ras, which can self-catalyze the exchange of GDP for GTP and hydrolysis of GTP to GDP but rely on guanine nucleotide exchange factors and GTPase-activating proteins to stimulate the intrinsic reaction rates (reviewed in reference 21). (iii) Partner proteins stimulate the rate of response regulator dephosphorylation from less than 1 order of magnitude (22–25) up to 1 or 2 orders of magnitude (19, 25–30). Similarly, proteins that inhibit response regulator dephosphorylation provide 1 to 2 orders of magnitude of phosphoryl group stabilization (31, 32). These effects are small with respect to the range of autodephosphorylation rate constants exhibited by wild-type response regulators (see below), suggesting that autodephosphorylation sets a physiologically relevant baseline rate of dephosphorylation that can be fine-tuned via accessory proteins.
A wide variety of cellular processes are controlled by two-component systems, including behavior, development, metabolism, and virulence. These responses operate over a wide range of time scales, from fractions of a second for chemotaxis (33) to days for multicellular development (34). To synchronize responses with stimuli, the kinetics of response regulator phosphorylation and dephosphorylation reactions must be faster than the processes they control. The reported range of autodephosphorylation rate constants for wild-type response regulators is 5 to 6 orders of magnitude (35–37). The reported range of rate constants for autophosphorylation of wild-type response regulators with phosphoramidate is only 2 orders of magnitude (13, 38), but this is almost certainly a lower bound because autophosphorylation kinetics have not been widely characterized. Indeed, the ranges of rate constants exhibited by mutants of a single response regulator for autophosphorylation with acetyl phosphate, phosphoramidate, or monophosphoimidazole are 2 to 5 orders of magnitude (39–42).
We seek to understand the mechanisms by which different wild-type response regulators that exhibit very similar tertiary structures, share conserved active-site machinery, and catalyze the same chemical reactions nevertheless support autophosphorylation and autodephosphorylation reactions with rate constants spanning many orders of magnitude. Variable residues within receiver domains provide a partial explanation. To identify equivalent variable positions across different response regulators, we note their positions relative to the landmarks provided by conserved active-site residues. Thus, D+2 signifies the residue located two positions C-terminal to the conserved Asp phosphorylation site. We previously showed that the residues located at variable positions D+2, T+1, and T+2 modulate autophosphorylation (39, 40, 42) and autodephosphorylation (36, 39, 42–44) rate constants by approximately 1 order of magnitude individually and 2 to 3 orders of magnitude collectively. Although these effects are substantial, they are not sufficient to account for the known range of rate constants (at least for autodephosphorylation) observed for wild-type response regulators. The search for additional factors that influence response regulator reaction kinetics led to the bioinformatic and biochemical characterization of positions K+1 and K+2 described here.
RESULTS
Rationale for investigation of receiver domain positions K+1 and K+2.
During the evolution of a protein family, disruptive changes at functionally important positions can be mitigated by compensatory changes at another position(s) within the same family member. Natural selection then leads to covariation between two correlated positions across family members. This coevolutionary relationship can be quantified using the concept of pairwise mutual information from information theory, providing a logical method to predict residues with functional significance. Receiver domain positions D+2, T+1, and T+2 are known to affect autophosphorylation and autodephosphorylation rate constants. The pairwise mutual information between positions D+2 and T+2 is in the top 0.1% of receiver domain pairs (36), and that between D+2/T+1 is in the top 1%. In turn, the mutual information between positions T+1/K+1 is in the top 1%, and that between K+1/K+2 is in the top 0.1% (data not shown). Thus, a chain of coevolutionary correlation connects three residues known to influence receiver domain reaction kinetics with variable positions K+1 and K+2, located on the β5α5 loop. We therefore experimentally investigated the effects of positions K+1 and K+2 on autophosphorylation and autodephosphorylation kinetics of the model response regulator Escherichia coli CheY.
Natural abundances of amino acids at receiver domain positions K+1 and K+2.
We used the natural abundances of amino acids at variable positions K+1 and K+2 to focus our investigation on the most physiologically relevant variants. In a nonredundant database of more than 33,000 receiver domains (36), Pro is found at K+1 in 82% of wild-type receivers, followed by 5% Asp and 3% Ser. Thus, just three amino acids at K+1 represent 90% of all receiver domains. The natural abundances of amino acids at position K+2 are 39% Phe, 15% Val, 13% Ile, 6% Leu, 5% Ala, and 5% Tyr. Thus, six hydrophobic residues account for 83% of all receiver domains. The relatively limited sequence space of positions K+1 and K+2 allowed us to experimentally evaluate the vast majority of biologically relevant amino acid combinations.
Among receiver domains with a Pro (the most common amino acid) at position K+1, the amino acids found at K+2 exhibit remarkable clustering by side chain physicochemical properties when ranked in order of abundance (Table 1). This correlation raises the possibility that, in the context of a Pro at K+1, amino acids at K+2 with similar chemical properties result in receiver domains with similar functional properties. In contrast, receiver domains with Asp or Ser (the second and third most common amino acids) at position K+1 exhibit no obvious correlation between the properties of the amino acids found at K+2 and natural abundance (data not shown). However, the high mutual information content between K+1 and K+2 is evident. If K+1 is a Pro, then there is an 85% chance that K+2 is Ile, Leu, Val, or Phe. If K+1 is Asp, then there is a 52% chance that K+2 is Ala, Thr, or Val. If K+1 is Ser, then there is a 59% chance that K+2 is Ala, Ser, Thr, Asp, or Glu. The mutual information content is even more striking in the other direction (see Table S1 in the supplemental material). If K+2 is Ile, Leu, Phe, Trp, Tyr, or Val (six of the nine most abundant amino acids at K+2), then there is a 91% to 99% chance that K+1 is Pro. In contrast, if K+2 is Ala, Ser, or Thr (the other three of the top nine amino acids at K+2), then there is a greater chance that K+1 is Asp or Ser than Pro.
TABLE 1.
Distribution of amino acids at K+2 in receiver domains with Pro at K+1a
| Amino acid at K+2 | % of PX pairs |
|---|---|
| Large branched/aromatic hydrophobic | |
| Phe | 47.4 |
| Val | 16.4 |
| Ile | 14.4 |
| Leu | 6.5 |
| Tyr | 5.6 |
| Trp | 2.0 |
| Small hydrophobic: Ala | 1.9 |
| Hydroxyl/sulfhydryl | |
| Cys | 1.3 |
| Ser | 0.90 |
| Thr | 0.69 |
| Proline: Pro | 0.61 |
| Large unbranched hydrophobic: Met | 0.50 |
| Small: Gly | 0.31 |
| Charged/amine | |
| Lys | 0.31 |
| Asp | 0.26 |
| Glu | 0.25 |
| Arg | 0.21 |
| Asn | 0.17 |
| Gln | 0.11 |
| His | 0.096 |
The total sample size was 27,177 sequences.
To investigate the potential impact(s) of various amino acids at positions K+1 and K+2 on reaction kinetics, we made specific amino acid substitutions in E. coli CheY based on the natural abundances described above. The 27 CheY variants (the wild type plus 26 mutants) characterized here make up only 9% of the 302 distinct K+1/K+2 amino acid pairs in our database of nonredundant receiver domain sequences but represent 84% of wild-type receiver domains and 13 of the 14 most abundant K+1/K+2 pairs.
Measurement of CheY reaction kinetics.
A Trp at position D+1 in E. coli CheY allows high-resolution measurement of autophosphorylation and autodephosphorylation kinetics via changes in intrinsic fluorescence intensity (3, 16, 45, 46) (Fig. 1). CheY consists of a single receiver domain, which avoids the inhibition of autophosphorylation by other domains observed in some response regulators (47–49).
To determine the autodephosphorylation rate constant, kdephos, we used the well-established pH jump method to selectively monitor the dephosphorylation reaction (45). We first incubated CheY in the presence of the small molecule phosphoramidate, which must be protonated to serve as a phosphodonor. We then measured changes in fluorescence intensity after raising the pH from 7.5 to 10.2, which selectively slows autophosphorylation by a factor of 500 but does not affect autodephosphorylation kinetics.
To determine the autophosphorylation rate constant, kphos/KS, we used a new method (see Materials and Methods). The traditional method measures the observed pre-steady-state rate constant, kobs, for approach to equilibrium between autophosphorylation and autodephosphorylation reactions (16, 40, 50). In contrast, the new method is based on a steady-state measurement and involves determination of K1/2, the concentration of phosphodonor required to phosphorylate half of the CheY population (51). The key insight is the following relationship: kdephos/K1/2 = kphos/KS (derived in Materials and Methods). The K1/2 method requires substantially less material and time than the kobs method but yields closely similar results (see Table S2 in the supplemental material).
Rate constants for autophosphorylation with phosphoramidate and autodephosphorylation with water are reported in Table 2. The values for wild-type CheY, which contains the most common K+1/K+2 pair (Pro Phe), are similar to previously reported values (40, 42). In general, kdephos values were very reproducible, with standard deviations of less than 10%. Most K1/2 values were similarly reproducible, but three CheY variants (Pro Arg, Leu Leu, and Asp Phe) exhibited high variation in spite of numerous measurements, which led to high variation in calculated kdephos/K1/2 values. Because these three K+1/K+2 combinations are very rare in wild-type receiver domains (<0.25% combined), we did not investigate the source of variation further.
TABLE 2.
Rate constants for E. coli CheY mutants altered at K+1 and/or K+2
| Mutant or wild type | Amino acid at: |
Natural abundance (%) | K1/2 PAM (mM)b | n | kdephos (min−1)b | n | kdephos/K1/2 PAM (M−1 s−1)a,b,c | |
|---|---|---|---|---|---|---|---|---|
| K+1 | K+2 | |||||||
| Wild type | Pro | Phe | 39 | 6.5 ± 0.8 | 11 | 3.5 ± 0.3 | 4 | 9.0 ± 1 |
| K+2 substitutions | ||||||||
| Branched hydrophobic | Pro | Val | 13 | 0.74 ± 0.1 | 4 | 7.6 ± 0.3 | 5 | 170 ± 20 |
| Pro | Ile | 12 | 0.57 ± 0.1 | 3 | 6.7 ± 0.5 | 6 | 200 ± 40 | |
| Pro | Leu | 5.3 | 0.60 ± 0.07 | 3 | 6.9 ± 0.4 | 2 | 190 ± 30 | |
| Aromatic | Pro | Tyr | 4.6 | 5.6 ± 0.4 | 3 | 9.1 ± 0.3 | 5 | 27 ± 2 |
| Pro | Trp | 1.7 | 8.1 ± 0.8 | 3 | 9.5 ± 0.7 | 6 | 20 ± 2 | |
| Small hydrophobic | Pro | Ala | 1.6 | 0.20 ± 0.006 | 3 | 8.7 ± 1 | 7 | 730 ± 90 |
| Hydroxyl/glycine | Pro | Ser | 0.73 | 2.2 ± 0.1 | 4 | 5.8 ± 0.3 | 6 | 44 ± 3 |
| Pro | Thr | 0.56 | 1.0 ± 0.1 | 3 | 6.2 ± 0.3 | 6 | 100 ± 10 | |
| Pro | Gly | 0.25 | 2.0 ± 0.07 | 2 | 7.5 ± 1 | 2 | 63 ± 10 | |
| Long unbranched | Pro | Met | 0.41 | 4.6 ± 0.4 | 2 | 3.6 ± 0.04 | 2 | 13 ± 1 |
| Pro | Asn | 0.14 | 4.0 ± 0.4 | 3 | 4.3 ± 0.1 | 3 | 18 ± 2 | |
| Pro | Gln | 0.09 | 3.2 ± 0.6 | 4 | 3.6 ± 0.2 | 3 | 19 ± 4 | |
| Charged | Pro | Lys | 0.25 | 8.1 ± 0.9 | 3 | 3.7 ± 0.1 | 6 | 7.6 ± 0.9 |
| Pro | Asp | 0.21 | 6.3 ± 0.4 | 3 | 3.7 ± 0.2 | 3 | 9.8 ± 0.8 | |
| Pro | Glu | 0.20 | 7.2 ± 2 | 4 | 3.5 ± 0.07 | 3 | 8.1 ± 2 | |
| Pro | Arg | 0.17 | 10 ± 8 | 6 | 3.6 ± 0.2 | 3 | 6.0 ± 5 | |
| K+1 K+2 substitutions | Asp | Ala | 1.3 | 2.6 ± 0.2 | 3 | 5.7 ± 0.1 | 3 | 37 ± 3 |
| Asp | Thr | 0.91 | 2.9 ± 0.2 | 3 | 5.8 ± 0.1 | 3 | 33 ± 3 | |
| Asp | Val | 0.62 | 5.2 ± 0.8 | 6 | 8.8 ± 0.9 | 6 | 28 ± 5 | |
| Ser | Ala | 0.59 | 3.3 ± 0.3 | 5 | 7.8 ± 0.0 | 6 | 39 ± 4 | |
| Gly | Gly | 0.33 | 2.5 ± 0.3 | 3 | 6.0 ± 0.03 | 5 | 40 ± 4 | |
| Ala | Ala | 0.08 | 6.3 ± 1 | 2 | 7.5 ± 0.4 | 2 | 20 ± 4 | |
| Leu | Leu | 0.01 | 18 ± 7 | 13 | 13 ± 4 | 6 | 12 ± 6 | |
| K+1 substitutions | Ser | Phe | 0.08 | 5.3 ± 1 | 3 | 4.6 ± 0.1 | 6 | 14 ± 4 |
| Asp | Phe | 0.04 | 17 ± 8 | 17 | 3.9 ± 0.5 | 6 | 3.8 ± 2 | |
| Ala | Phe | 0.01 | 23 ± 2 | 4 | 4.4 ± 0.1 | 2 | 3.2 ± 0.3 | |
kdephos/K1/2 PAM is equivalent to kphos/KS PAM. Subscript PAM indicates values determined with phosphoramidate.
Means and standard deviations of the mean are shown.
Calculation of standard deviation for kdephos/K1/2 recognizes propagation of error and incorporates the standard deviations of both numerator and denominator.
Position K+2 modulated CheY reaction kinetics.
We characterized all possible amino acid substitutions at K+2 except Cys, His, and Pro. The CheY variants listed in Table 2 reflect 98% of the receiver domains with a Pro at K+1, which in turn account for 82% of wild-type receiver domains. Changes at position K+2 affected both autophosphorylation and autodephosphorylation reactions, modulating rate constants approximately 3-fold and 100-fold, respectively. However, none of the substitutions at K+2 significantly diminished the rate constant for either reaction compared to Phe.
As might be anticipated from the data in Table 1, the effects of amino acids at K+2 appear to be closely linked to the chemical properties of the side chains and can be put into six groups based on comparison with the rate constants supported by Phe (a natural reference point, as the most common amino acid at K+2). (i) Branched hydrophobic residues (Ile, Leu, and Val) increased kdephos about 2-fold and kdephos/K1/2 about 20-fold. (ii) Aromatic residues (Trp and Tyr) doubled or tripled both kdephos and kdephos/K1/2. (iii) The small hydrophobic amino acid Ala doubled kdephos and increased kdephos/K1/2 80-fold, the largest increase seen in this study. (iv) Hydroxyl amino acids (Ser and Thr) and Gly doubled or tripled kdephos and increased kdephos/K1/2 about 5-fold. Thr, which has a side chain similar to that of Ser but includes a branched methyl group, increased kdephos/K1/2 another 2-fold compared to Ser. (v) Long neutral residues (Asn, Gln, and Met) had no effect on kdephos and increased kdephos/K1/2 about 2-fold. (vi) Charged residues (Arg, Asp, Glu, and Lys) had no effect on kdephos or kdephos/K1/2.
Asp is the second most common amino acid at K+1. Ala, Thr, and Val are the three most common amino acids at K+2 in the context of an Asp at K+1 and together account for 52% of such receiver domains. CheY variants with these three K+1/K+2 combinations each enhanced both autophosphorylation and autodephosphorylation (kdephos values, 2- to 3-fold; kdephos/K1/2 values, 3- to 4-fold) compared to the wild type. Ser is the third most common amino acid at K+1 and is most commonly paired with Ala (21%). CheY bearing Ser/Ala at K+1/K+2 exhibited rate constants similar to those of the three most common combinations with Asp at K+1.
We also characterized several variants that were not inspired by natural abundance. Due to the limited sequence space at K+1 (82% Pro) and K+2 (39% Phe), we explored the consequences of changing one or both positions. Changing K+1 alone to Asp or Ser (the second and third most common amino acids at K+1) or Ala (a minimal side chain) had no effect on autodephosphorylation and modest effects on autophosphorylation. Two of these pairs were the only combinations in this study to reduce autophosphorylation. An Ala Ala pair, with minimal side chains at both positions, increased the rate constants for both reactions about 2-fold. All four combinations described above are rare (<0.1%) in nature.
The effect of position K+2 on CheY autophosphorylation was independent of phosphodonors.
Response regulators can use phosphoramidates (N-P bond) or acyl phosphates (O-P bond) as phosphodonors. Variable positions D+2, T+1, and T+2 differentially affect autophosphorylation with various phosphodonors, likely due to direct interactions with the donor molecules (40, 42). In contrast, substitutions at K+2 affected the CheY autophosphorylation rate constants for phosphoramidate and acetyl phosphate similarly. The K1/2 values for acetyl phosphate were 7.0 ± 0.7 mM, 1.2 ± 0.1 mM, and 0.31 ± 0.08 mM (n = 3 for each) for CheY variants bearing Phe, Val, or Ala, respectively, at K+2. This gives a kdephos/K1/2 value for wild-type CheY of 8.3 ± 1 M−1 s−1 with acetyl phosphate, similar to previous kphos/KS measurements of 11 ± 0.8 M−1 s−1 (40). Changing K+2 from Phe to Val or Ala substantially increased the kdephos/K1/2 value with acetyl phosphate to 110 ± 10 M−1 s−1 or 470 ± 130 M−1 s−1, respectively. For both mutants, the fold increases in autophosphorylation rate constants for acetyl phosphate observed upon changing K+2 were 70% of those seen for phosphoramidate.
Position K+2 affected the conformational equilibria of CheY.
Similar effects on rate constants when using phosphodonors with different chemical and steric properties suggest that position K+2 influenced autophosphorylation kinetics through an indirect mechanism. The location of K+2 ∼9 Å from the site of phosphorylation (Fig. 1) is consistent with an indirect mechanism, because it is too far to directly participate in the in-line chemical attacks of phosphorylation or dephosphorylation reactions (52). The observation (Table 2) that altering position K+2 alone or in conjunction with K+1 generally enhanced autophosphorylation more than autodephosphorylation is reminiscent of previous reports involving the manipulation of response regulator conformational equilibria. In the presence of ompC1 target DNA, the autophosphorylation rate of OmpR increases ∼20-fold, whereas the autodephosphorylation rate increases only 2-fold (12). Similarly, in the presence of FliM target peptide, the autophosphorylation rate constant of CheY increases up to 30-fold. whereas the autodephosphorylation rate constant increases only up to 2-fold (13). A simple interpretation of these observations is that binding to a target molecule constrains the conformational space explored by an unphosphorylated response regulator and increases the chance of achieving an active conformation primed for phosphorylation. The experimentally observed autophosphorylation rate constant is the weighted average of a population of response regulators in inactive conformations that phosphorylate slowly and response regulators in active conformations that phosphorylate rapidly. The difference in autophosphorylation rate constants between active and inactive conformations of CheY is at least 2 orders of magnitude (13), which implies that shifting equilibria can have a substantial effect on observed rate constants. In contrast, phosphorylation constrains the conformational space explored by response regulators to be near an active conformation primed for dephosphorylation, so binding a target molecule provides little additional constraint and thus enhances observed autodephosphorylation kinetics modestly, if at all.
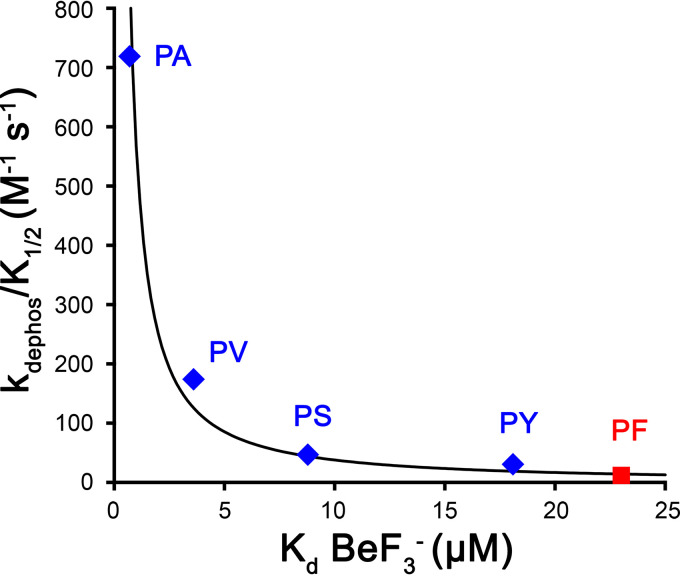
To determine whether altering the amino acid at K+2 increases the fraction of the CheY population in an active conformation, we measured the binding affinities of CheY variants for the stable phosphoryl group analog BeF3−. In the presence of the FliM target peptide, which increases the chance of achieving an active conformation, the population weighted average affinity of unphosphorylated CheY for BeF3− increased about 1 order of magnitude compared to that in the absence of FliM (13). This suggests that active conformations of CheY bind BeF3− substantially more tightly than inactive conformations. In the absence of FliM peptide, the dissociation constant (Kd) of unphosphorylated wild-type CheY for BeF3− was 23 μM (see Table S3 in the supplemental material), modestly weaker than the previously reported 7.7 μM (13). The difference is consistent with the higher-ionic-strength environment (230 versus 140 mM) used for our measurement. Among CheY mutants with substitutions at K+2, binding affinity for BeF3− increased with increasing autophosphorylation rate constants (see Table S3), as would be expected if the mutants have increased propensity to occupy an active conformation. The inverse variation of kdephos/K1/2 with the BeF3− Kd (Fig. 2), combined with no correlation between kdephos and Kd (see Table S3), is consistent with an effect on conformational sampling.
FIG 2.
Inverse relationship between autophosphorylation rate constants (Table 2) and binding constants for BeF3− (see Table S3) of select CheY variants at K+2. The best-fit line (r2 = 0.96) takes the following form, where c is a scaling constant: y = cx−1.2. A perfect inverse relationship would be of the following form: y = cx−1.
Positions K+1/K+2 were not sufficient to support extreme dephosphorylation kinetics.
Reported kdephos values for wild-type response regulators span a range of 5 to 6 orders of magnitude. Many response regulators near the reported extremes (and indeed throughout the range) contain the most frequent K+1/K+2 combination, Pro Phe, which is also found in CheY. This distribution suggests K+1/K+2 is not a primary determinant of autodephosphorylation kinetics, consistent with the finding that substitutions at K+1/K+2 have little effect on the kdephos value of CheY (Table 2). However, the response regulators with the most extreme dephosphorylation kinetics contain unusual combinations of K+1/K+2 residues, so we explored them further. The response regulators with the highest reported kdephos values of which we are aware are Rhodobacter sphaeroides CheY6 (37) and E. coli CheB (53). These response regulators contain the unusual K+1/K+2 combinations Pro Ser and Pro Gln, respectively. Neither combination significantly increased the autodephosphorylation rate of CheY (Table 2).
Myxococcus xanthus RedF has the lowest reported kdephos value (35) of which we are aware. Based on the amino acid sequence, RedF appears to have an unusual predicted structure. There are two Lys residues in the C-terminal portion of RedF that could serve as the conserved Lys residue, but one is closer and one is farther than the typical distance from the conserved Thr/Ser. The residues adjacent to the candidate lysines are Gly Gly and Leu Leu. A structural-homology model suggests that Gly Gly is adjacent to the catalytically important Lys in RedF (data not shown). We made and tested both combinations, which led to modest increases (rather than dramatic decreases) in the kdephos value of CheY (Table 2).
Effects of higher-order combinations of variable residues.
As noted previously, a chain of mutual information connects position K+2 to K+1 to T+1 to D+2 to T+2. Substitutions at each of these five variable positions affect response regulator reaction kinetics. We therefore explored whether combining K+1/K+2 substitutions (indirect effects) with D+2/T+1/T+2 substitutions (direct effects) might have larger effects than either alone. There are 205 (3.2 million) possible combinations of amino acids at these five positions, but the vast majority (>99%) of combinations do not naturally occur in response regulators. Our database of 33,253 nonredundant receiver domain sequences contains only 7,583 different combinations of the five positions, and the top 50 combinations represent a third of all wild-type receiver domain sequences.
We first created mimics of the response regulators with extreme kdephos values using the CheY backbone with substitutions at D+2/T+1/T+2/K+1/K+2. The combinations of five variable amino acids found in CheY6 and CheB are rare (0.02%) in nature but nevertheless occur 2 orders of magnitude more frequently than would be expected from the abundances of their constituent D+2/T+1/T+2 and K+1/K+2 combinations. One might anticipate such enrichment if the overall combination led to emergent properties of functional importance. However, in both the CheY6 and CheB mimics, introduction of the naturally occurring K+2 residue did not increase kdephos further than what was supported by the D+2/T+1/T+2 combinations (Table 3) in the context of CheY K+1/K+2 residues. The full CheY mimic of RedF was poorly soluble and unsuitable for data collection. We were able to examine the consequences of the D+2/T+1/T+2 combination found in RedF and observed modest effects on autodephosphorylation (4-fold decrease) and autophosphorylation (7-fold increase) rate constants in CheY compared to the wild type (Table 3). These differences are far from sufficient to account for the observed properties of RedF.
TABLE 3.
Rate constants for E. coli CheY mutants altered at D+2, T+1, T+2, K+1, and/or K+2
| Mutant or wild type | Amino acid atd
: |
K1/2 PAM (mM)c | n | kdephos (min−1)c | n | kdephos K1/2 PAM (M−1 s−1)a,c | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| D+2 | T+1 | T+2 | K+1 | K+2 | ||||||
| Wild type | Asn | Ala | Glu | Pro | Pheb | 6.5 ± 0.8 | 11 | 3.5 ± 0.3 | 4 | 9.0 ± 1 |
| R. sphaeroides CheY6 mimic | Asn | Ala | Glu | Pro | Serb | 2.2 ± 0.1 | 4 | 5.8 ± 0.3 | 6 | 44 ± 3 |
| Glu | Ser | Val | Pro | Phe | 8.6 ± 2 | 6 | 13 ± 0.7 | 6 | 25 ± 6 | |
| Glu | Ser | Val | Pro | Ser | 1.6 ± 0.05 | 5 | 12 ± 2 | 6 | 130 ± 20 | |
| E. coli CheB mimic | Asn | Ala | Glu | Pro | Glnb | 3.2 ± 0.6 | 4 | 3.6 ± 0.2 | 3 | 19 ± 3 |
| Glu | Ser | Leu | Pro | Phe | 1.9 ± 0.5 | 5 | 26 ± 0.6 | 3 | 230 ± 60 | |
| Glu | Ser | Leu | Pro | Gln | 0.73 ± 0.09 | 3 | 22 ± 0.6 | 3 | 500 ± 70 | |
| M. xanthus RedF mimic | Asn | Ala | Glu | Gly | Glyb | 2.5 ± 0.3 | 3 | 6.0 ± 0.03 | 5 | 40 ± 4 |
| Ser | Gly | His | Pro | Phe | 0.20 ± 0.006 | 3 | 0.80 0.04 | 3 | 67 ± 4 | |
| Ser | Gly | His | Gly | Gly | NDe | ND | ||||
| QAN common with PI/PL/PV | Asn | Ala | Glu | Pro | Valb | 0.74 ± 0.1 | 4 | 7.6 ± 0.3 | 5 | 170 ± 20 |
| Gln | Ala | Asn | Pro | Phe | 2.0 ± 0.03 | 2 | 36 ± 1f | |||
| Gln | Ala | Asn | Pro | Val | 3.2 ± 0.09 | 3 | 470 ± 20f | |||
| NTS common with PI/PL/PV | Asn | Ala | Glu | Pro | Valb | 0.74 ± 0.1 | 4 | 7.6 ± 0.3 | 5 | 170 ± 20 |
| Asn | Thr | Ser | Pro | Phe | 18 ± 5 | 3 | 3.3 ± 0.3 | 3 | 3.1 ± 0.9 | |
| Asn | Thr | Ser | Pro | Val | 2.0 ± 0.06 | 3 | 3.2 ± 0.1 | 3 | 27 ± 1 | |
| ESR most common with PY | Asn | Ala | Glu | Pro | Tyrb | 5.6 ± 0.4 | 3 | 9.1 ± 0.3 | 5 | 27 ± 2 |
| Glu | Ser | Arg | Pro | Phe | 0.55 ± 0.2 | 5 | 6.1 ± 0.4 | 3 | 180 ± 70 | |
| Glu | Ser | Arg | Pro | Tyr | 0.61 ± 0.08 | 4 | 10 ± 0.7 | 3 | 270 ± 40 | |
| RTF most common with DA | Asn | Ala | Glu | Asp | Alab | 2.6 ± 0.2 | 3 | 5.7 ± 0.1 | 3 | 37 ± 3 |
| Arg | Thr | Phe | Pro | Phe | 0.022 ± 0.006 | 3 | 0.70 ± 0.04 | 3 | 530 ± 100 | |
| Arg | Thr | Phe | Asp | Ala | ND | ND | ||||
kdephos/K1/2 PAM is equivalent to kphos/KS PAM. Subscript PAM indicates values determined with phosphoramidate.
Data for the first variant in each section are from Table 2.
Means and standard deviations of the mean are given.
For chimeras, italics indicate amino acids from wild-type E. coli CheY, boldface indicates amino acids from the target sequence, and regular type indicates amino acids that are the same in both.
ND, not determined because protein was poorly soluble.
Value is the kphos/KS PAM value from duplicate determinations of kobs. K1/2 values could not be determined because fluorescence changes did not monotonically increase with [PAM].
Half of the 20 most abundant D+2/T+1/T+2/K+1/K+2 combinations contain Pro Phe at K+1/K+2, as does the CheY model response regulator used in this study. We next investigated four combinations in the top 20 that did not contain Pro Phe. All were 2 to 20 times more abundant in wild-type receiver domains than would be expected based on the natural frequencies of their D+2/T+1/T+2 and K+1/K+2 sequences. The D+2/T+1/T+2 combinations Gln Ala Asn and Asn Thr Ser cooccur much more frequently with K+2 residues Val/Ile/Leu (second, third, and fourth most abundant, respectively, at K+2) than with Phe (most abundant at K+1). We therefore constructed and characterized CheY variants containing these two triple combinations with Val at K+2. Gln Ala Asn at D+2/T+1/T+2 and Val at K+2 each enhanced autophosphorylation compared to the wild type; the combination enhanced autophosphorylation even further (Table 3). In contrast to Val at K+2, Asn Thr Ser at D+2/T+1/T+2 diminished autophosphorylation, and the combination produced an intermediate effect. Gln Ala Asn and Asn Thr Ser at D+2/T+1/T+2 had little to no effect on autodephosphorylation, compared to the wild type, with either Phe or Val at K+2.
The fifth most common K+1/K+2 pair is Pro Tyr. The most common D+2/T+1/T+2 combination cooccurring with Pro Tyr is Glu Ser Arg. Both sets of substitutions enhanced autophosphorylation kinetics compared to the wild type. When they were combined, the enhancement was even greater.
The most common K+1/K+2 pair without a Pro is Asp Ala. The most common D+2/T+1/T+2 combination cooccurring with Asp Ala is Arg Thr Phe. Both sets of substitutions enhanced autophosphorylation kinetics compared to the wild type. CheY bearing all five substitutions was poorly soluble, so we were unable to determine rate constants for the quintuple mutant.
DISCUSSION
Effects of substitutions at K+1 and K+2 on CheY reaction kinetics.
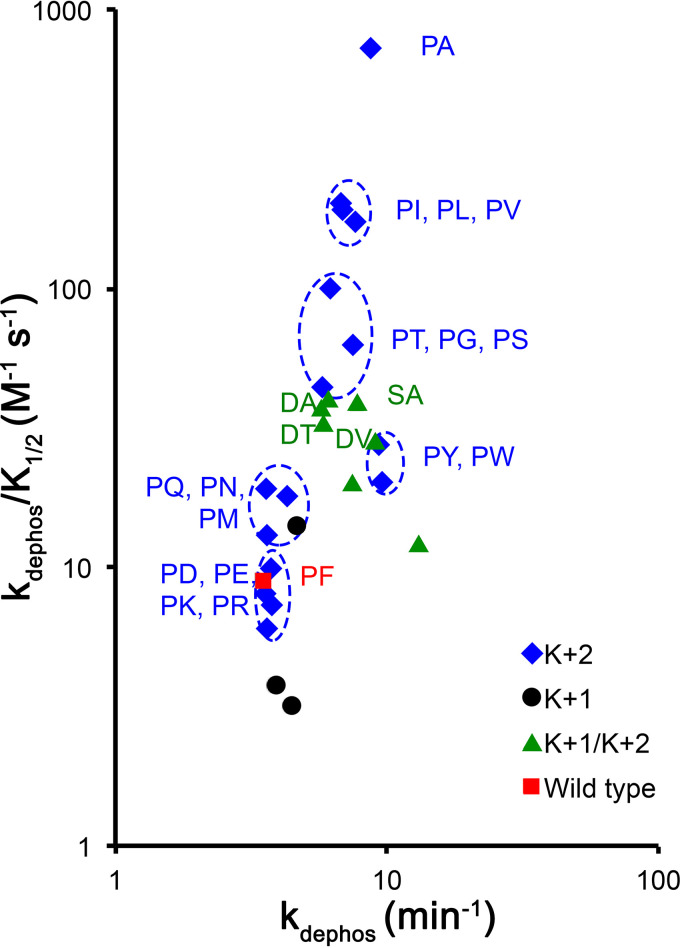
The rate constants for 27 CheY variants listed in Table 2 are plotted in Fig. 3. Recall that wild-type CheY contains by far the most common (39%) naturally occurring pair of amino acids at positions K+1/K+2 (Pro Phe) and therefore provides a biologically relevant reference point. Several conclusions are readily apparent. (i) The observed range of rate constants was more than 2 orders of magnitude for autophosphorylation but less than 1 order of magnitude for autodephosphorylation. No tested variants exhibited a decrease in kdephos, and almost all exhibited an increase in the autophosphorylation rate constant. (ii) Substitutions at K+2 were sufficient to have substantial effects, which clustered by physiochemical properties. After Phe, the most common amino acids at K+2 cooccuring with Pro at K+1 are Ile/Leu/Val (30%), Tyr/Trp (6%), Ala (2%), and Thr/Gly/Ser (1.5%). The two clusters (Gln/Asn/Met and Asp/Glu/Lys/Arg) of seven variants that resulted in kinetic characteristics similar to those of the wild type are quite rare in nature (no more than 0.5% individually and less than 2% collectively) (Table 1). (iii) Substitutions at K+1 alone (i.e., in the context of Phe at K+2) had remarkably modest effects, given the semiconserved nature of K+1 (Pro in 82% of receiver domains) and the unique properties of the Pro side chain. Because 99% of K+1/K+2 pairs with Phe at K+2 have Pro at K+1 (see Table S1 in the supplemental material), the tested K+1 variants are likely not physiologically relevant. (iv) The four most naturally abundant pairs that do not contain Pro at K+1 were created by changing K+2 in addition to K+1. Changing both positions increased both autophosphorylation and autodephosphorylation in all such combinations tested, whether compared to the wild type (Pro Phe) or to substitution mutants with Ser/Asp/Ala at K+1 and Phe at K+2 (Table 2). However, in three of the four cases, the physiologically relevant combination of residues had antagonistic effects on autophosphorylation, with the double substitution supporting a rate constant substantially less than expected from the effects of the single substitutions (see Table S4 in the supplemental material). In contrast, the kdephos values of the CheY double mutants were as expected from the single mutants.
FIG 3.
Autophosphorylation and autodephosphorylation rate constants for CheY variants with K+1 and/or K+2 substitutions. Data from Table 2.
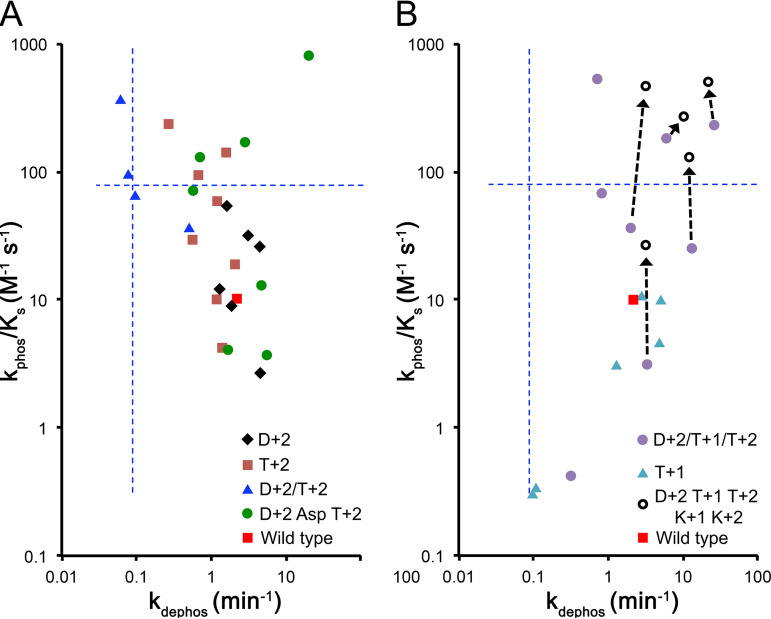
Effects of substitutions at D+2, T+1, and T+2 on CheY reaction kinetics.
We previously characterized the effects of substitutions at positions D+2, T+1, and T+2 on autodephosphorylation kinetics in multiple response regulators and on autophosphorylation kinetics in CheY (36, 39, 40, 42–44). To provide context for the K+1/K+2 results reported here, previous results with CheY D+2/T+1/T+2 mutants are plotted in Fig. 4 and can be summarized as follows. (i) The observed range of rate constants in the CheY backbone is 3 orders of magnitude for both autophosphorylation and autodephosphorylation. (ii) Substitutions at D+2 and/or T+2 generally have inverse effects on autophosphorylation and autodephosphorylation rate constants, with points falling around a trend line from upper left to lower right (Fig. 4A). This is likely because hydrophobic residues at these positions interact favorably with the leaving group portion of the phosphodonor (40), whereas hydrophilic residues interact favorably with attacking water molecules (36). (iii) Substitutions at T+1 (light-blue triangles and purple circles) generally have similar effects on autophosphorylation and autodephosphorylation rate constants, with points falling around a trend line from lower left to upper right (Fig. 4B). This is likely due to direct steric interactions between the residue at T+1 and the phosphodonor or phosphoacceptor molecules (42).
FIG 4.
Autophosphorylation and autodephosphorylation rate constants for CheY variants with D+2 and/or T+2 substitutions (A) and/or T+1 substitutions (B). All contain Pro at K+1. All contain Phe at K+2 except the open circles in panel B. The dashed arrows show the effects of combining D+2/T+1/T+2 with K+2 substitutions. The two triangles in panel A near the origin of the dashed lines are the two most common combinations of D+2/T+1/T+2/K+1/K+2 residues (MAKPF and MARPF; 11% of all wild-type receiver domains) in the context of a CheY backbone. (Data from references 36, 39, 40, and 42, to ,44 and Table 3.) kdephos values in Fig. 3 were determined by fluorescence change, whereas most values in this figure were determined by loss of 32P. Autophosphorylation rate constants in Fig. 3 were mostly determined by K1/2, whereas those in this figure were mostly determined by kobs. As noted in Materials and Methods, the two methods for determining each rate constant are reproducible and in close agreement (within ≤1.5-fold). Thus, the data in Fig. 3 and this figure can be compared directly (note the differences in the axis scales).
Substitutions at K+2 likely affect conformational equilibria of CheY.
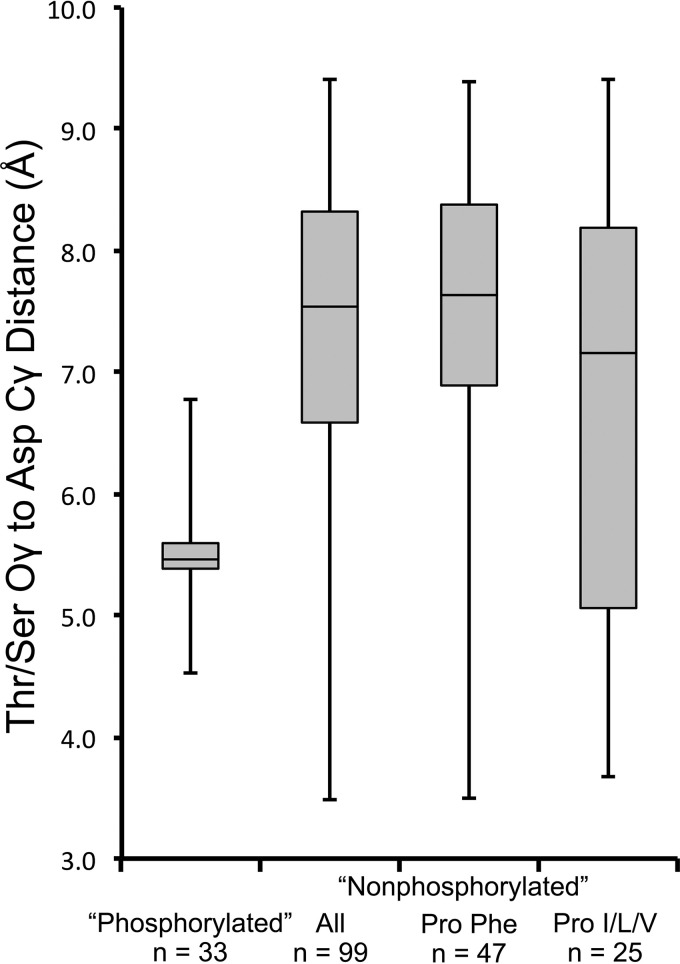
The pattern of kinetic effects caused by substitutions at K+2 (Fig. 3) is distinct from the D+2/T+1/T+2 results (Fig. 4) and likely arises from a different mechanism. Multiple lines of evidence suggest that amino acids at K+2 affect reaction kinetics by altering the conformational equilibria of CheY. (i) K+2 is distant from the Asp site of phosphorylation (Fig. 1), consistent with an indirect mechanism, such as allostery. (ii) Substitutions at K+2 have a larger effect on autophosphorylation than on autodephosphorylation (Table 2 and Fig. 3). This is analogous to the kinetic effects previously reported for binding of response regulators to their targets (12, 13), which in the case of CheY has been shown to directly affect conformation. (iii) Substitutions at D+2, T+1, and T+2 have differential effects on phosphodonors with different chemical and structural properties, consistent with direct interactions between the side chains and the phosphodonors (40, 42). In contrast, substitutions at K+2 had similar effects on autophosphorylation with acetyl phosphate and phosphoramidate, consistent with an indirect mechanism. (iv) A strong correlation exists between the fraction of the CheY population in an active conformation (BeF3− binding affinity) and the autophosphorylation rate constant (Fig. 2). (v) The outlier among CheY D+2/T+2 mutants (Fig. 4A, green circle at upper right) corresponds to an Asp Arg pair, a combination that is rare (∼0.2%) among receiver domains but common among the structurally related haloalkanoic acid dehalogenase enzymes (54). The kinetic effects of the D+2/T+2 Asp Arg mutant are likely due to an enhanced propensity to adopt the active conformation (39). The kinetic properties of CheY bearing an Ala substitution at K+2 (Fig. 3) are similar to those of the D+2/T+2 Asp Arg mutant, consistent with a conformational mechanism. (vi) A key distinction between active and inactive conformations of receiver domains is the location of the conserved Ser/Thr with respect to the Asp site of phosphorylation. In activated conformations, Ser/Thr is positioned to form a hydrogen bond with an oxygen atom of the phosphoryl group, whereas Ser/Thr is farther away from the Asp in inactive conformations (5, 6, 55). Snapshots of protein structures obtained by X-ray crystallography are imperfect probes of conformational accessibility, but enough receiver domain structures exist to provide valuable insight. In 33 wild-type receiver domains activated by phosphorylation or BeF3− binding, the distance between the Ser/Thr Oγ and Asp Cγ atoms was tightly clustered (range, 2.3 Å) around a median value of 5.5 Å (Fig. 5). In contrast, in 99 wild-type receiver domains lacking BeF3− or a phosphoryl group (nonphosphorylated), the observed distances spanned a much larger range (5.9 Å) and the median separation between Ser/Thr and Asp was substantially greater (7.6 Å). Both observations are as expected. The data suggest that receiver domains occupy a broader range of conformations when nonphosphorylated than when phosphorylated and that nonphosphorylated receiver domains rarely occupy an active conformation. To test the hypothesis that position K+2 affects conformational equilibria, we measured the distances from Ser/Thr to Asp in nonphosphorylated receiver domains as a function of the amino acid composition at K+1/K+2. Nearly half of the proteins in the nonphosphorylated data set contained Pro Phe at K+1/K+2 compared to the 40% expected from protein sequences (see Table S6 in the supplemental material). Not surprisingly, the median distance and range exhibited by the Pro Phe subset were similar to those of the entire nonphosphorylated set (Fig. 5). In contrast, the 25% of nonphosphorylated receiver domains that contained Pro at K+1 and Ile, Leu, or Val at K+2 (versus the 29% expected) exhibited a significant overlap with activated conformations. These structural differences are consistent with the increased autophosphorylation rate constants observed for CheY variants carrying Pro at K+1 and Ile/Leu/Val instead of Phe at K+2 (Table 2). There are insufficient structures to derive meaningful conclusions for response regulators with other combinations of amino acids at K+1/K+2.
FIG 5.
The amino acid at K+2 is correlated with the distance between the Oγ atom of the conserved Thr/Ser and the Cγ atom of the phosphorylatable Asp in X-ray crystal structures of wild-type receiver domains. During the phosphorylation reaction, the Thr/Ser orients toward and forms a hydrogen bond with one of the oxygen atoms of the phosphoryl group, neutralizing the negative charge and stabilizing the transition state. This results in a significant shift in the distance between the Thr/Ser and Asp side chains as a function of the activation state, which can be used as a convenient metric to characterize receiver domain structures. Phosphorylated structures contain either PO32− or BeF3− in the active site, whereas nonphosphorylated structures do not. Phosphorylated structures contain a divalent cation in the active site, whereas nonphosphorylated structures may or may not. Subsets of proteins with the indicated amino acids at positions K+1 and K+2 are represented by the two boxes on the right. Details of structure curation and distance calculation are given in Materials and Methods. The boxes show first quartile, median, and third quartile values, whereas the whiskers show minimum and maximum values.
Molecular mechanism of conformational influence.
The location of the β5α5 loop, which includes residues K+1/K+2, differs significantly between inactive and active conformations (6). To seek clues about the mechanism by which the amino acid at position K+2 might influence receiver domain conformation, we also examined the conformation of the K+2 side chain in structures of nearly 100 wild-type receiver domains that were neither complexed with other macromolecules (proteins or DNA) nor activated by phosphorylation or BeF3− binding. The amino acid at K+2 is almost always buried within the structure unless it is hydrophilic (data not shown). A potential mechanism consistent with kinetic and structural observations for response regulators with Pro at K+1 is that snugly burying a large Phe at K+2 could restrict sampling of the active conformation. Other large aromatic residues (Trp or Tyr) would be similarly constrained in the protein interior and result in similar autophosphorylation rate constants. The smaller hydrophobic residues Ile/Leu/Val might have more room to move while buried, thus allowing more conformational sampling (consistent with the data in Fig. 5) and yielding significantly higher autophosphorylation rate constants. The logical extension of this model would be that the small hydrophobic residue Ala is least restricted while buried and allows the most access to active conformations. The presumed preference for charged (Arg, Asp, Glu, and Lys) and polar (Asn and Gln) side chains to be solvent exposed rather than buried may make it difficult for the β5α5 loop to achieve an active conformation, resulting in slow autophosphorylation. Alternatively, if such residues (and the large Met) are instead buried, then their tolerated conformations in a hydrophobic environment would likely be very limited, again leading to relatively slow autophosphorylation. The proposed model does not easily account for the observed reaction kinetics of CheY mutants bearing the small/polar residues Gly/Ser/Thr at K+2, which will require additional investigation.
Binding of FliM increases CheY autophosphorylation kinetics substantially (13) by stabilizing a partially active conformation (56). In terms of the hypothetical model proposed above, FliM binding would help CheY overcome the conformational restrictions imposed by burying Phe at K+2. The rate constant for autophosphorylation with phosphoramidate by CheY bearing an Ala substitution at K+2 (720 M−1 s−1) (Table 2) is approximately three times greater than that achieved by FliM binding to 80% of wild-type CheY molecules (230 M−1 s−1) (13). This suggests that the K+2 Ala substitution allows CheY to adopt a more active conformation than FliM binding can achieve with wild-type CheY. The amino acids at K+2 may affect CheY conformation by a different mechanism than FliM binding.
Variable residues potentially modulate response regulator reaction kinetics.
The solid points in Fig. 4 represent the expansion of rate constants for CheY self-catalyzed reactions around a single K+1/K+2 combination (Pro Phe; red squares) by altering residue D+2, T+1, and/or T+2. The result is a range of 3 orders of magnitude in both autophosphorylation and autodephosphorylation rate constants.
We determined rate constants for five CheY variants with potentially interesting combinations of residues at variable positions D+2, T+1, T+2, and K+2. In each case, the effects of substitutions at D+2/T+1/T+2, and K+2 were exerted essentially independently of one another (see Table S5 in the supplemental material). There was no evidence of synergy and minor (approximately 2-fold difference between expected and observed rate constants) antagonism only in the variant containing a branched Thr side chain at T+1. These results are consistent with our earlier conclusion that position K+2 influences reaction kinetics by a different mechanism than positions D+2/T+1/T+2. It should be possible to alter the autophosphorylation kinetics of most variants by at least 1 order of magnitude simply by changing the residue at K+2 (Fig. 4B, dashed arrows), unless the consequences of conformational changes are thwarted by steric hindrance from the amino acid at T+1.
Multiple variable residues that affect phosphorylation and dephosphorylation rate constants via different mechanisms should make it possible to evolve response regulators with reaction kinetics suited for various biological purposes. Some examples are described below.
The two most common combinations of the five variable positions that we have investigated are represented by CheY variants in the upper left of Fig. 4A (intersection of the dashed lines). They are abundant in the OmpR/PhoB family of response regulators and represent ∼11% of all wild-type receiver domains. The combinations support fast phosphorylation and slow dephosphorylation, which appears suitable to generate a stable signal for regulating transcription. Background noise from autophosphorylation with small-molecule phosphodonors is suppressed by the phosphatase activity of the sensor kinase (57).
Chemotaxis operates on a much shorter time scale than transcription and so requires rapid dephosphorylation. In contrast to typical sensor kinases, whose DHp domains support opposing kinase and phosphatase activities, chemotaxis kinases utilize Hpt domains that lack phosphatase activity. Thus, chemotaxis systems require other means to dephosphorylate response regulators. The chemotaxis response regulator CheY is located in the centers of Fig. 4 (red squares) and exhibits faster autodephosphorylation (to enhance response time) and slower autophosphorylation (perhaps to reduce noise from intracellular small-molecule phosphodonors).
The combinations of amino acids at five variable positions, indicated by open circles in the upper right (fast phosphorylation and fast dephosphorylation) portion of Fig. 4B, represent two classes of receiver domains. (i) The chemotaxis proteins E. coli CheB and R. sphaeroides CheY6 exhibit fast kdephos. CheB does not have a partner phosphatase and relies entirely on autodephosphorylation. Similarly, the CheA3 phosphatase for CheY6 stimulates dephosphorylation only 3-fold (24) compared to 2 orders of magnitude for the CheZ phosphatase for E. coli CheY (28). (ii) The D+2/T+2 combinations Gln Asn and Glu Arg are primarily found in hybrid kinases (36), and the K+1/K+2 combinations Pro Val and Pro Tyr are enriched in hybrid kinases (see Table S6). The receiver domains of hybrid kinases may need fast autophosphorylation to accept phosphoryl groups from Hpt domains and fast autodephosphorylation to act as phosphate sinks that drain phosphoryl groups from the system (1). Finally, the green circle at the upper right of Fig. 4A represents a common D+2/T+2 combination in haloalkanoic acid dehalogenases, enzymes that need to cycle rapidly through phosphorylation and dephosphorylation reactions to serve as catalysts, whereas structurally related receiver domains require a somewhat stable phosphoprotein form to support signal transduction (39).
Consistent with the notion that variable positions D+2, T+1, T+2, K+1, and K+2 can tune response regulator reaction kinetics to suit biological purposes, the frequencies of amino acids at these positions vary substantially between different response regulator families (36, 42). There are three striking aspects of the amino acid distribution at K+1/K+2 (see Table S6). First, the OmpR/PhoB family contains almost exclusively Pro Phe. Second, all but one of the other major groups primarily have Pro at K+1 and Ile/Leu/Val in addition to Phe at K+2. If the results we have observed with CheY can be extrapolated to other response regulators, then this would suggest a need for faster phosphorylation in many family members than can be achieved with Pro Phe.
Variable residues potentially modulate response regulator interactions with partner proteins.
The third striking feature of the data in Table S6 is that FixJ/NarL is completely different from other response regulator families and contains many K+1/K+2 combinations that we did not experimentally characterize. In particular, Pro is uncommon at K+1 whereas Asp is relatively common at K+1. Although FixJ and NarL share an output domain and are included in the same family in Table S6, analysis of receiver domain sequences showed that FixJ and NarL belong to two phylogenetically distinct groups (42, 58). The divergent amino acid composition at K+1/K+2 is attributable to NarL-type response regulators, not FixJ type (data not shown). This is reminiscent of amino acid abundances at T+1. Among all receiver domains, T+1 is ∼75% Ala/Gly and ∼5% Ile/Val/Met, whereas in NarL family response regulators, T+1 is ∼15% Ala/Gly and ∼45% Ile/Val/Met (42). Bulky residues at T+1 allowed modulation of rate constants to the lower left quadrant of Fig. 4B.
The unusual variable amino acids in NarL-type response regulators may be a consequence of two type-specific protein/protein interactions. First, NarL-type response regulators partner with HisKA_3-type sensor kinases, a minor class that differs in some properties from the predominant HisKA sensor kinases (59). For example, in cocrystal structures (Protein Data Bank [PDB] IDs 5iuj, 5iuk, 5iul, and 5iun) of the HisKA_3 sensor kinase DesK complexed with the NarL-type response regulator DesR, the Asp at K+1 of DesR forms a salt bridge with an Arg on the DesR helical bundle (60). A Ser at DesK position H+8 is located nearby. These interactions are consistent with the abundances of polar amino acids at K+1 in NarL-type receiver domains. In contrast, cocrystal structures (PDB IDs 3dge and 5uht) of the HisKA sensor kinase HK853 with the response regulator RR468 show that the Pro at K+1 inserts into a hydrophobic portion of the HK853 helical bundle (61, 62). In the structure, the response regulator K+1 residue is immediately adjacent to the sensor kinase H+7 and H+11 residues identified as important for interaction specificity between HisKA sensor kinases and their partner response regulators (59). Second, NarL-type response regulators dimerize at the α1-α5 interface following phosphorylation, rather than the α4-β5-α5 interface utilized by OmpR-type response regulators or the α4-β5 interface used by NtrC-type response regulators (6). Key features of the NarL-type dimer interface include the polar residue at position K+1 and a Met at DD+3 on one monomer interacting with a polar residue at DD+1 and a hydrophobic cavity under the β5α5 loop (the location of K+1), respectively, on the other monomer (63–65). In contrast, the β5α5 loop is not part of the dimer interface in OmpR- and NtrC-type response regulators. Thus, the choice of amino acids at some response regulator variable positions may be evolutionarily constrained both to achieve appropriate reaction kinetics and to preserve interactions with partner proteins (66).
Relationship between response regulator autophosphorylation and phosphotransfer reactions.
Response regulators catalyze their own phosphorylation using either small-molecule or protein phosphodonors. The range of reported second-order rate constants for reaction of various phosphodonors with response regulators provides a useful perspective on potential relationships between the two reactions, which appear to use the same mechanism. The rate constants for autophosphorylation of CheY variants with phosphoramidate span 3 orders of magnitude (∼100 to ∼103 M−1 s−1) (Fig. 4) (39, 42). Monophosphoimidazole is a closer analog of phosphohistidine than phosphoramidate and results in a range of at least 5 orders of magnitude (∼10−1 to ∼104 M−1 s−1) for autophosphorylation of CheY variants (40, 41). For phosphotransfer from wild-type sensor kinases to wild-type response regulators, the four reported cases span 4 orders of magnitude (∼101 to ∼105 M−1 s−1) (67–69), which extensively overlaps the known range for CheY autophosphorylation with monophosphoimidazole. The rate constants for phosphotransfer between wild-type Hpt domains and response regulators are generally larger (∼104 to ∼108 M−1 s−1) and also span 4 orders of magnitude (68, 70–72).
To specifically connect stimuli to appropriate responses, two-component systems predominantly employ protein-protein interactions. As noted above, rate constants for response regulator phosphorylation are generally higher with protein phosphodonors than with small molecules. A protein phosphodonor can enhance reaction kinetics through two general types of effects. First, binding of the protein phosphodonor to the response regulator increases the local concentration of the phosphohistidine residue enormously compared to small molecules. In fact, binding of current small-molecule phosphodonors to CheY is so weak that it has not been experimentally possible to saturate the reaction kinetics (16). Second, binding between a protein phosphodonor and a response regulator can optimize the geometry of the reactants and hence speed the reaction. Additional investigation will be necessary to determine whether effects of variable amino acid substitutions on response regulator autophosphorylation kinetics persist or vanish in the context of reaction with a protein phosphodonor, but there are some suggestive hints. The considerable overlap between autophosphorylation and phosphotransfer rate constants makes it conceivable that variable residues could have similar effects on both reactions. A substitution at T+2 in the Brucella abortis NarX response regulator diminishes both autophosphorylation with acetyl phosphate and phosphotransfer from the NarY sensor kinase (73), but the latter effect could be a trivial consequence of reduced NarX/NarY binding. More intriguing are effects on conformation. Part of the mechanism for phosphotransfer from the DesK sensor kinase to the DesR response regulator is selection of active DesR conformations by DesK (65). Could substitutions at K+2 that enhance access to active response regulator conformations therefore speed phosphotransfer by enhancing binding to the sensor kinase? We already know that binding of target DNA to the output domain of the OmpR response regulator enhances both autophosphorylation with acetyl phosphate and phosphotransfer from the EnvZ sensor kinase, apparently by altering the conformation of the receiver domain (12). Furthermore, the rate constant for phosphotransfer from EnvZ to OmpR is only 100 M−1 s−1 (69), well within the range of autophosphorylation rate constants reported here for CheY variants bearing substitutions at K+2.
Potential predictive value of receiver domain variable positions.
Amino acid substitutions at positions D+2, T+1, and T+2 are believed to exert effects on reaction kinetics via direct interactions between side chains and phosphodonor or phosphoacceptor molecules (36, 40, 42). Substitutions at positions D+2, T+1, and T+2 have comparable effects on kdephos in different response regulators, consistent with interactions involving side chains, independent of the protein backbone (36, 42, 44). Effects on autophosphorylation kinetics have been examined only in CheY but similarly depend on phosphodonors in a manner consistent with direct involvement of residue side chains (40, 42). If side chain properties are the predominant determinant of kinetic effects, then the examination of receiver domain primary amino acid sequences may have predictive value.
The effects of substitutions at K+1 and K+2 on reaction kinetics have so far been systematically examined only in CheY. The K+1/K+2 positions appeared to mediate effects through conformation rather than direct interactions with reactants. Given the structural similarities shared by receiver domains, analogous differences between phosphorylated and unphosphorylated conformations, the limited sequence diversity at K+1 and K+2, and the observed correlation between CheY autophosphorylation rate constants (Table 2) and natural abundances of amino acids at K+2 in the presence of Pro at K+1 (Table 1), it is plausible that specific amino acids will have similar consequences in response regulators other than CheY.
Five variable positions are currently known to modulate the kinetics of CheY self-catalyzed reactions over 3 orders of magnitude. However, this is not sufficient to explain the 5- to 6-order-of-magnitude range reported for response regulator autodephosphorylation rate constants. Several nonexclusive mechanisms may contribute to the as yet unexplained range of rate constants. Additional variable positions with discrete, backbone-independent effects on autodephosphorylation kinetics may remain to be identified. Subtle structural differences between receiver domain backbones might cause differences in transition state bond lengths/energies and, hence, reaction kinetics. The multimeric state of at least one response regulator affects phosphoryl group stability (65). Finally, interactions between receiver and output domains substantially affect the autophosphorylation kinetics of some response regulators (47). We are not aware of cases in which interdomain interactions affect autodephosphorylation kinetics, but such an effect is conceivable. The relative contributions of backbone-independent and backbone-dependent factors will affect our ability to predict response regulator reaction kinetics from the amino acid sequence alone.
Potential applications to synthetic biology.
Two-component system proteins are used as building blocks for a variety of synthetic-biology applications, including biosensors (74–81). If substitutions at K+2 have effects in other response regulators similar to those reported here for CheY, then response regulators could be rationally engineered to optimize their properties for various purposes. For example, changing K+2 to Phe from other hydrophobic amino acids (in the context of Pro at K+1) could reduce background noise due to autophosphorylation with intracellular acetyl phosphate. If the phosphatase activity of the sensor kinase were selectively disabled (82, 83), then the duration of the response could be set to a desired value over a time scale of seconds to hours by using variable residues to adjust the autodephosphorylation rate.
MATERIALS AND METHODS
Bioinformatics.
Mutual information analysis to assess coevolution of residues within receiver domains was performed using multiple-sequence alignments of the Pfam (84) Response_reg family (PF00072) and the MISTIC server (http://mistic.leloir.org.ar/index.php) (85) in October 2015 and June 2019, with similar results. The five conserved active-site residues are present at frequencies of only 93% to 97% in the 2019 Pfam alignment, indicating the inclusion of pseudoreceiver domains (86, 87). The effect of minor contamination with pseudoreceiver domains on mutual information analysis results is unknown, but the results were used only to suggest positions K+1 and K+2 as candidates for experimental investigation.
To calculate amino acid abundances at various receiver domain positions and combinations thereof, we used a previously described database of 33,252 nonredundant wild-type receiver domain sequences (36) curated to exclude pseudoreceivers.
Strains and mutant construction.
Plasmid pKC1 is pET28a carrying E. coli cheY with an N-terminal His6 tag under the control of the T7 promoter (38). Three nonnative amino acids (Gly, Ser, and His) remain at the N terminus after thrombin cleavage to remove the affinity tag but are located on the opposite side of the protein from the active site and do not affect either the autophosphorylation (38) or autodephosphorylation (51) rate constants.
Site-directed mutations in cheY were created using a QuikChange kit (Agilent). The reaction products were transformed into E. coli DH5α cells. Transformants were streaked for single colonies to ensure homogeneous clonal populations. Plasmids were isolated, and the entire cheY gene was sequenced. Plasmids confirmed to contain only the desired mutations were transformed into E. coli BL21(DE3) cells for protein expression, and the resulting strains were stored at –80°C.
Protein purification.
To purify mutant CheY proteins, LB (10 g tryptone, 5 g yeast extract, 10 g NaCl per liter) plus 30 μg/ml kanamycin was inoculated from a frozen stock of BL21(DE3) cells containing the desired pKC1 variant. After overnight growth to saturation, 1 liter of the same medium was inoculated with a 1:100 dilution of the overnight culture and grown with shaking at 37°C. When the culture reached an optical density at 600 nm (OD600) of ∼0.5, 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added to induce expression of T7 RNA polymerase and hence CheY. Growth was continued overnight with shaking at room temperature. Cells were collected by centrifugation (30 min; 4,200 × g; 4°C), and the pellet was resuspended in 25 ml CheY lysis buffer (20 mM Tris [pH 7.5], 300 mM NaCl, 10 mM imidazole). The cells were lysed in an Avestin Emulsiflex C3 homogenizer. Debris was removed by ultracentrifugation (45 min; 145,000 × g; 4°C).
To separate tagged CheY from other soluble proteins, a 2-ml Ni-nitrilotriacetic acid (NTA) column was first equilibrated with 10 column volumes of water, followed by 10 column volumes of CheY lysis buffer. The supernatant from the ultracentrifugation was applied to the resin, and the column was washed with 10 column volumes of CheY wash buffer (20 mM Tris [pH 7.5], 300 mM NaCl, 20 mM imidazole). His6-tagged CheY was eluted with 10 column volumes of CheY elution buffer (20 mM Tris [pH 7.5], 300 mM NaCl, 250 mM imidazole). To remove the affinity tag, 40 U of human alpha-thrombin was added to the material eluted from the Ni-NTA column. To remove NaCl and imidazole that would interfere with measurement of reaction rate constants, overnight dialysis against TMG (25 mM Tris [pH 7.5], 5 mM MgCl2, 10% [vol/vol] glycerol) was performed simultaneously with thrombin cleavage at room temperature. To prepare for gel filtration, the sample volume was reduced in an Amicon Ultra 15 Centricon apparatus with 10-kDa molecular weight cutoff and passed through a 0.22-μm filter to remove any aggregates. The sample was injected onto a Sephacryl S-200 high-resolution column using a GE AKTA protein chromatography system and chromatographed in TMG to separate CheY from thrombin, the cleaved tag, and remaining contaminants. Fractions containing pure CheY were again concentrated and filtered before storage at –20°C.
Kinetic theory.
The rate at which a coupled system of CheY autophosphorylation and autodephosphorylation reactions comes to equilibrium is defined by the equilibrium dissociation constant for the phosphodonor substrate (KS), the autophosphorylation rate constant (kphos), and the autodephosphorylation rate constant (kdephos). Measurements of CheY reaction kinetics have traditionally used the observed rates of approach to equilibrium (kobs) at several concentrations of phosphodonor (e.g., the phosphoramidate concentration, [PAM]). Plotting kobs versus [PAM] with the following equation reveals kphos/KS from the slope and kdephos from the y intercept (40, 45): . We show here that it is also possible to determine the effective autophosphorylation rate constant, kphos/KS, from an equilibrium experiment. The rate of change in the concentration of phosphorylated CheY ([Yp]) is the difference between the rates of formation and destruction: d. For CheY, KS >> [PAM] (16), so the following equation is true: . We define K1/2 as the concentration of phosphodonor at which half the CheY molecules are phosphorylated and half are not. Therefore, when [PAM] is equal to K1/2, then [Y] is equal to [Yp] by definition. Furthermore, at equilibrium, d[Yp]/dt is equal to 0. Substituting and rearranging gives the following equations: and . kdephos can be determined separately by the pH jump method described below.
The advantages of determining kphos/KS from K1/2 rather than kobs are that the K1/2 method takes much less time and consumes far less protein and phosphodonor. The disadvantage is that the ratio of two separate experimental measurements (K1/2 and kdephos) has greater error than determining kphos/KS in a single experiment. To verify that autophosphorylation rate constants determined by the two methods were in agreement, they were determined by both methods for four different CheY variants (see Table S2 in the supplemental material). The values differed by ∼10% or less in three cases and ∼25% in the fourth. This agreement is more than sufficient for our purposes, given that we are interested in much larger effects and the observed range of kdephos/K1/2 values reported in Tables 2 and 3 is more than 2 orders of magnitude.
Determination of K1/2.
The small-molecule phosphodonor potassium phosphoramidate was synthesized as described previously (50, 88). The purity of phosphoramidate was determined by 31P nuclear magnetic resonance (NMR). The fluorescence intensity of Trp58 in E. coli CheY is quenched by phosphorylation (3). We used a PerkinElmer LS-50B fluorimeter with an excitation wavelength of 295 nm and an emission wavelength of 346 nm to monitor the extent of CheY phosphorylation. A solution of 5 μM CheY, 10 mM MgCl2, 100 mM HEPES (pH 7.0), and 100 mM KCl was placed in a quartz cuvette and stirred slowly at 25°C. A phosphodonor solution of 10 mM MgCl2, 100 mM HEPES (pH 7.0), and 100 mM potassium phosphoramidate was prepared and kept on ice until use. Because the rate of CheY autophosphorylation decreases with increasing ionic strength (16), a constant ionic strength was maintained in both the protein and phosphodonor solutions. For certain highly reactive CheY variants, the stock phosphoramidate concentration was decreased (20 to 100 mM) to bracket the K1/2 value, and the KCl concentration was increased to maintain 230 mM ionic strength. Multiple aliquots of phosphodonor solution were titrated into the protein solution using a Hamilton syringe. The reactions were allowed to come to equilibrium (i.e., no further fluorescence change) before the next addition. In a typical experiment, the concentrations of phosphoramidate in the cuvette ranged from 0.1 to 30 mM. For CheY variants that supported particularly fast autophosphorylation, the protein concentration was reduced 5- to 10-fold to prevent repeated cycles of phosphorylation and dephosphorylation from significantly changing the phosphodonor concentration during data collection.
To analyze the data, the raw fluorescence intensities of each plateau were first corrected for the effects of dilution using the following formula: .
The corrected fluorescence intensities were then used to calculate the net changes in fluorescence intensity (ΔI) from the initial sample with no phosphodonor. Finally, GraphPad Prism software was used to obtain the best fit of a hyperbolic equation to the ΔI data as a function of the phosphodonor concentration ([PAM]) by adjusting the variables K1/2 and Imax (maximum fluorescence intensity): .
Determination of kphos/KS.
The rate of approach to equilibrium between autophosphorylation and autophosphorylation kobs was determined at multiple phosphoramidate concentrations as previously described (50). The autophosphorylation rate constant, kphos/KS, is the slope of the best-fit line of a plot of kobs versus the phosphoramidate concentration.
Determination of kdephos.
Small-molecule phosphodonors with an N-P bond require a positively charged nitrogen to act as a good leaving group (46). At neutral pH, CheY catalyzes autophosphorylation in the presence of phosphoramidate, as well as subsequent autodephosphorylation. Raising the pH results in deprotonation of phosphoramidate and thus selectively slows autophosphorylation without affecting autodephosphorylation (45, 46). To determine kdephos, the same fluorimeter, wavelengths, and reaction temperature described for determination of K1/2 were used. CheY was continuously phosphorylated in a solution containing 10 μM CheY, 5 mM Tris (pH 7.5), 10 mM MgCl2, and phosphoramidate at a concentration of five times the K1/2 value for the particular CheY variant tested. For certain variants with solubility constraints, the protein concentration was decreased (5 to 10 μM). The phosphodonor concentration was high enough to phosphorylate most CheY molecules yet low enough to no longer support significant autophosphorylation when the fraction of protonated phosphoramidate molecules was reduced about 3 orders of magnitude by raising the pH by about 3 (51). The CheY solution was combined with an equal volume of 200 mM sodium carbonate, pH 10.2, in an Applied Photophysics RX2000 rapid mixer accessory (dead time, ∼8 ms), and fluorescence intensity data were collected at 50-ms intervals. The mixing was repeated five or six times, the data from each mixing were fit separately as described below, and the values were averaged to produce a single determination of kdephos.
GraphPad Prism software was used to obtain the best fit of an exponential-decay equation to the fluorescence intensity (It) data as a function of time by adjusting the variables kdephos, I0 (initial fluorescence intensity), and I∞ (final fluorescence intensity): . Because noise is larger than signal in the long exponential tail, we truncated the It data after the first 95% of the change in It (greater than four half-lives) to avoid distorting the fit.
Values of kdephos determined by changes in fluorescence at 25°C are up to ∼50% larger than those determined by loss of 32P at room temperature (51), perhaps due to the temperature difference between the two methods. Although results from the two methods are not exactly comparable, the differences are small compared to the range of kdephos values observed for different response regulators or mutants (36). All the values reported here were determined by fluorescence and therefore can be directly compared to one another.
The methods used to determine K1/2, kphos/KS, and kdephos exclusively monitor the fraction of the CheY population that reacts (changes fluorescence). The results are robust because they are independent of the protein concentration, purity, or specific activity.
Determination of BeF3− binding affinity.
Beryllium forms a series of complexes with one to four fluoride atoms, depending on the concentration of fluoride. BeF3− is the dominant species at 10 mM fluoride (89). For purposes of calculation, we made the generally accepted assumption that the concentration of BeF3− is equal to the concentration of Be2+ added. The Kd of CheY for BeF3− was determined in a manner analogous to that for K1/2, with suitable modifications. Because the Kd is similar to the protein concentration, the concentration of CheY was reduced to 1 μM to minimize changes in the BeF3− concentration due to binding. Thus, the cuvette contained 100 mM HEPES (pH 7.0), 10 mM MgCl2, 1 μM CheY, 10 mM NaF, and 90 mM KCl for a total ionic strength of 230 mM. A solution at the same ionic strength (100 mM HEPES [pH 7.0], 10 mM MgCl2, 1 mM BeSO4, and 96 mM KCl) was titrated into the cuvette, and changes in fluorescence were recorded. We have previously used BeCl2 to form BeF3−, but BeCl2 is no longer commercially available. We encountered no obvious difficulties using BeSO4 instead.
The fluorescence changes were corrected for dilution. At low concentrations of BeF3−, the ligand is not in large excess over CheY, so [BeF3−free] is not equal to [BeF3−total], and the concentration of BeF3− bound to CheY must be considered. GraphPad Prism software was used to obtain the best fit of a quadratic binding equation that accounts for ligand depletion to the ΔI data as a function of the Be2+ concentration by adjusting the variables Kd and S (a scaling factor): . The concentration of CheY was determined using an experimentally determined extinction coefficient of 10,200 M−1 cm−1 at 280 nm (90).
Statistical methods.
The values of all rate and equilibrium constants derived from best fits of equations to data were rounded to two significant figures prior to calculation of means and standard deviations. Standard deviations were rounded to one significant figure. To calculate the standard deviation of the quotient kdephos/K1/2, we used the following propagation-of-error formula, where σ is the standard deviation of the respective means: .
Analysis of wild-type receiver domain structures.
All entries featuring a receiver domain (Pfam PF00072) were obtained from the RCSB Protein Data Bank (current as of December 2019) (91). The data set was filtered to include only X-ray crystallographic models. To limit our analysis to wild-type receiver domains, pseudoreceiver domains (lacking any of the conserved DD, D, T, and K catalytic residues) and proteins that contained amino acid substitutions at any place other than the N or C terminus (remnants of purification tags that are typically known not to affect function) were excluded, as were chimeric or domain-swapped receiver domains. Because binding to biologically relevant partners can affect receiver domain conformation (6, 12, 13, 56), structures bound to other proteins, peptides, DNA, cyclic di-GMP, etc., were excluded. To avoid potential artifactual effects on conformation, structures with significant but nonrelevant ligands bound at the active site (Ca2+, K+, PO43−, or SO42−) were excluded. Structures with a BeF3− or PO32− moiety lacking tetrahedral geometry bound to the Asp were excluded.
Because we sought to measure the distance between the phosphorylatable Asp and the conserved Thr/Ser, structures lacking density for either side chain were excluded. Structures exhibiting multiple occupancies or ambiguous density for these side chains were examined manually. In almost all cases, conformers exhibited nearly identical distances, and one was arbitrarily chosen as representative for analysis. In models 1k68_A and 1k68_B, the side chain of Ser100 exhibits two conformations, one pointing toward BeF3− and one oriented away. The canonical orientation toward BeF3− was chosen for analysis. In model 1nxt_A, the secondary conformer of Asp52 lacking the phosphoryl mimic was excluded.
The remaining structures were classified as either “phosphorylated” (containing a divalent cation, such as Mg2+ or Mn2+, and a phosphoryl group or BeF3− in the active site) or “nonphosphorylated” (containing or not containing a divalent cation and not containing a phosphoryl group or BeF3−). PyMOL was used to calculate the distance between the Cγ atom of the catalytic Asp and the Oγ atom of the conserved Thr/Ser for every model. The Cγ atom of the Asp was utilized due to the equivalency of the side chain oxygen atoms and the relative flexibility of the Asp side chain. For entries with multiple protomers (chains), a single average measurement was calculated over all chains. To eliminate potential effects of redundancy, a final average measurement was calculated from the various entries for each unique receiver domain.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ruth Silversmith for inspiring the use of K1/2 analysis, Kailey Ezekiel and Ange Barasebwa for construction and initial characterization of some of the triple and quintuple variants, and Sarah Barr and Kailey Ezekiel for useful comments on the manuscript.
This work was funded by National Institutes of Health grant GM050860 to Robert B. Bourret.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Alvarez AF, Barba-Ostria C, Silva-Jiménez H, Georgellis D. 2016. Organization and mode of action of two component system signaling circuits from the various kingdoms of life. Environ Microbiol 18:3210–3226. doi: 10.1111/1462-2920.13397. [DOI] [PubMed] [Google Scholar]
- 2.Zschiedrich CP, Keidel V, Szurmant H. 2016. Molecular mechanisms of two-component signal transduction. J Mol Biol 428:3752–3775. doi: 10.1016/j.jmb.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lukat GS, McCleary WR, Stock AM, Stock JB. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci U S A 89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hess JF, Bourret RB, Oosawa K, Matsumura P, Simon MI. 1988. Protein phosphorylation and bacterial chemotaxis. Cold Spring Harbor Symp Quant Biol 53(Part 1):41–48. doi: 10.1101/sqb.1988.053.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Bourret RB. 2010. Receiver domain structure and function in response regulator proteins. Curr Opin Microbiol 13:142–149. doi: 10.1016/j.mib.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao R, Bouillet S, Stock AM. 2019. Structural basis of response regulator function. Annu Rev Microbiol 73:175–197. doi: 10.1146/annurev-micro-020518-115931. [DOI] [PubMed] [Google Scholar]
- 7.Bobay BG, Thompson RJ, Hoch JA, Cavanagh J. 2010. Long range dynamic effects of point-mutations trap a response regulator in an active conformation. FEBS Lett 584:4203–4207. doi: 10.1016/j.febslet.2010.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster CA, West AH. 2017. Use of restrained molecular dynamics to predict the conformations of phosphorylated receiver domains in two-component signaling systems. Proteins 85:155–176. doi: 10.1002/prot.25207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDonald LR, Boyer JA, Lee AL. 2012. Segmental motions, not a two-state concerted switch, underlie allostery in CheY. Structure 20:1363–1373. doi: 10.1016/j.str.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pontiggia F, Pachov DV, Clarkson MW, Villali J, Hagan MF, Pande VS, Kern D. 2015. Free energy landscape of activation in a signalling protein at atomic resolution. Nat Commun 6:7284. doi: 10.1038/ncomms8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stock AM, Guhaniyogi J. 2006. A new perspective on response regulator activation. J Bacteriol 188:7328–7330. doi: 10.1128/JB.01268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ames SK, Frankema N, Kenney LJ. 1999. C-terminal DNA binding stimulates N-terminal phosphorylation of the outer membrane protein regulator OmpR from Escherichia coli. Proc Natl Acad Sci U S A 96:11792–11797. doi: 10.1073/pnas.96.21.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuster M, Silversmith RE, Bourret RB. 2001. Conformational coupling in the chemotaxis response regulator CheY. Proc Natl Acad Sci U S A 98:6003–6008. doi: 10.1073/pnas.101571298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolfe AJ. 2010. Physiologically relevant small phosphodonors link metabolism to signal transduction. Curr Opin Microbiol 13:204–209. doi: 10.1016/j.mib.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hess JF, Oosawa K, Kaplan N, Simon MI. 1988. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell 53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 16.Da Re SS, Deville-Bonne D, Tolstykh T, Ron MV, Stock JB. 1999. Kinetics of CheY phosphorylation by small molecule phosphodonors. FEBS Lett 457:323–326. doi: 10.1016/S0014-5793(99)01057-1. [DOI] [PubMed] [Google Scholar]
- 17.Huynh TN, Noriega CE, Stewart V. 2010. Conserved mechanism for sensor phosphatase control of two-component signaling revealed in the nitrate sensor NarX. Proc Natl Acad Sci U S A 107:21140–21145. doi: 10.1073/pnas.1013081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parashar V, Mirouze N, Dubnau DA, Neiditch MB. 2011. Structural basis of response regulator dephosphorylation by Rap phosphatases. PLoS Biol 9:e1000589. doi: 10.1371/journal.pbio.1000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pazy Y, Motaleb MA, Guarnieri MT, Charon NW, Zhao R, Silversmith RE. 2010. Identical phosphatase mechanisms achieved through distinct modes of binding phosphoprotein substrate. Proc Natl Acad Sci U S A 107:1924–1929. doi: 10.1073/pnas.0911185107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao R, Collins EJ, Bourret RB, Silversmith RE. 2002. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat Struct Biol 9:570–575. doi: 10.1038/nsb816. [DOI] [PubMed] [Google Scholar]
- 21.Bos JL, Rehmann H, Wittinghofer A. 2007. GEFs and GAPs: critical elements in the control of small G proteins. Cell 129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 22.Jeong DW, Cho H, Jones MB, Shatzkes K, Sun F, Ji Q, Liu Q, Peterson SN, He C, Bae T. 2012. The auxiliary protein complex SaePQ activates the phosphatase activity of sensor kinase SaeS in the SaeRS two-component system of Staphylococcus aureus. Mol Microbiol 86:331–348. doi: 10.1111/j.1365-2958.2012.08198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keener J, Kustu S. 1988. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc Natl Acad Sci U S A 85:4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porter SL, Roberts MA, Manning CS, Armitage JP. 2008. A bifunctional kinase-phosphatase in bacterial chemotaxis. Proc Natl Acad Sci U S A 105:18531–18536. doi: 10.1073/pnas.0808010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willett JW, Kirby JR. 2011. CrdS and CrdA comprise a two-component system that is cooperatively regulated by the Che3 chemosensory system in Myxococcus xanthus. mBio 2:e00110-11. doi: 10.1128/mBio.00110-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albanesi D, Mansilla MC, de Mendoza D. 2004. The membrane fluidity sensor DesK of Bacillus subtilis controls the signal decay of its cognate response regulator. J Bacteriol 186:2655–2663. doi: 10.1128/jb.186.9.2655-2663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutu AD, Wayne KJ, Sham LT, Winkler ME. 2010. Kinetic characterization of the WalRKSpn (VicRK) two-component system of Streptococcus pneumoniae: dependence of WalKSpn (VicK) phosphatase activity on its PAS domain. J Bacteriol 192:2346–2358. doi: 10.1128/JB.01690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silversmith RE, Levin MD, Schilling E, Bourret RB. 2008. Kinetic characterization of catalysis by the chemotaxis phosphatase CheZ. Modulation of activity by the phosphorylated CheY substrate. J Biol Chem 283:756–765. doi: 10.1074/jbc.M704400200. [DOI] [PubMed] [Google Scholar]
- 29.Tzeng YL, Feher VA, Cavanagh J, Perego M, Hoch JA. 1998. Characterization of interactions between a two-component response regulator, Spo0F, and its phosphatase, RapB. Biochemistry 37:16538–16545. doi: 10.1021/bi981340o. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Y, Inouye M. 2002. The role of the G2 box, a conserved motif in the histidine kinase superfamily, in modulating the function of EnvZ. Mol Microbiol 45:653–663. doi: 10.1046/j.1365-2958.2002.03061.x. [DOI] [PubMed] [Google Scholar]
- 31.Janiak-Spens F, Sparling DP, West AH. 2000. Novel role for an HPt domain in stabilizing the phosphorylated state of a response regulator domain. J Bacteriol 182:6673–6678. doi: 10.1128/jb.182.23.6673-6678.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato A, Groisman EA. 2004. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev 18:2302–2313. doi: 10.1101/gad.1230804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segall JE, Manson MD, Berg HC. 1982. Signal processing times in bacterial chemotaxis. Nature 296:855–857. doi: 10.1038/296855a0. [DOI] [PubMed] [Google Scholar]
- 34.O'Connor KA, Zusman DR. 1991. Development in Myxococcus xanthus involves differentiation into two cell types, peripheral rods and spores. J Bacteriol 173:3318–3333. doi: 10.1128/jb.173.11.3318-3333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jagadeesan S, Mann P, Schink CW, Higgs PI. 2009. A novel “four-component” two-component signal transduction mechanism regulates developmental progression in Myxococcus xanthus. J Biol Chem 284:21435–21445. doi: 10.1074/jbc.M109.033415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Page SC, Immormino RM, Miller TH, Bourret RB. 2016. Experimental analysis of functional variation within protein families: receiver domain autodephosphorylation kinetics. J Bacteriol 198:2483–2493. doi: 10.1128/JB.00853-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porter SL, Armitage JP. 2002. Phosphotransfer in Rhodobacter sphaeroides chemotaxis. J Mol Biol 324:35–45. doi: 10.1016/s0022-2836(02)01031-8. [DOI] [PubMed] [Google Scholar]
- 38.Creager-Allen RL, Silversmith RE, Bourret RB. 2013. A link between dimerization and autophosphorylation of the response regulator PhoB. J Biol Chem 288:21755–21769. doi: 10.1074/jbc.M113.471763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Immormino RM, Starbird CA, Silversmith RE, Bourret RB. 2015. Probing mechanistic similarities between response regulator signaling proteins and haloacid dehalogenase phosphatases. Biochemistry 54:3514–3527. doi: 10.1021/acs.biochem.5b00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas SA, Immormino RM, Bourret RB, Silversmith RE. 2013. Nonconserved active site residues modulate CheY autophosphorylation kinetics and phosphodonor preference. Biochemistry 52:2262–2273. doi: 10.1021/bi301654m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Page SC, Silversmith RE, Collins EJ, Bourret RB. 2015. Imidazole as a small molecule analogue in two-component signal transduction. Biochemistry 54:7248–7260. doi: 10.1021/acs.biochem.5b01082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Immormino RM, Silversmith RE, Bourret RB. 2016. A variable active site residue influences the kinetics of response regulator phosphorylation and dephosphorylation. Biochemistry 55:5595–5609. doi: 10.1021/acs.biochem.6b00645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pazy Y, Wollish AC, Thomas SA, Miller PJ, Collins EJ, Bourret RB, Silversmith RE. 2009. Matching biochemical reaction kinetics to the timescales of life: structural determinants that influence the autodephosphorylation rate of response regulator proteins. J Mol Biol 392:1205–1220. doi: 10.1016/j.jmb.2009.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas SA, Brewster JA, Bourret RB. 2008. Two variable active site residues modulate response regulator phosphoryl group stability. Mol Microbiol 69:453–465. doi: 10.1111/j.1365-2958.2008.06296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayover TL, Halkides CJ, Stewart RC. 1999. Kinetic characterization of CheY phosphorylation reactions: comparison of P-CheA and small-molecule phosphodonors. Biochemistry 38:2259–2271. doi: 10.1021/bi981707p. [DOI] [PubMed] [Google Scholar]
- 46.Silversmith RE, Appleby JL, Bourret RB. 1997. Catalytic mechanism of phosphorylation and dephosphorylation of CheY: kinetic characterization of imidazole phosphates as phosphodonors and the role of acid catalysis. Biochemistry 36:14965–14974. doi: 10.1021/bi9715573. [DOI] [PubMed] [Google Scholar]
- 47.Barbieri CM, Mack TR, Robinson VL, Miller MT, Stock AM. 2010. Regulation of response regulator autophosphorylation through interdomain contacts. J Biol Chem 285:32325–32335. doi: 10.1074/jbc.M110.157164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Correa F, Gardner KH. 2016. Basis of mutual domain inhibition in a bacterial response regulator. Cell Chem Biol 23:945–954. doi: 10.1016/j.chembiol.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herrou J, Willett JW, Crosson S. 2015. Structured and dynamic disordered domains regulate the activity of a multifunctional anti-sigma factor. mBio 6:e00910. doi: 10.1128/mBio.00910-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silversmith RE, Bourret RB. 2018. Fluorescence measurement of kinetics of CheY autophosphorylation with small molecule phosphodonors. Methods Mol Biol 1729:321–335. doi: 10.1007/978-1-4939-7577-8_25. [DOI] [PubMed] [Google Scholar]
- 51.Bourret RB, Thomas SA, Page SC, Creager-Allen RL, Moore AM, Silversmith RE. 2010. Measurement of response regulator autodephosphorylation rates spanning six orders of magnitude. Methods Enzymol 471:89–114. doi: 10.1016/S0076-6879(10)71006-5. [DOI] [PubMed] [Google Scholar]
- 52.Lassila JK, Zalatan JG, Herschlag D. 2011. Biological phosphoryl-transfer reactions: understanding mechanism and catalysis. Annu Rev Biochem 80:669–702. doi: 10.1146/annurev-biochem-060409-092741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stewart RC. 1993. Activating and inhibitory mutations in the regulatory domain of CheB, the methylesterase in bacterial chemotaxis. J Biol Chem 268:1921–1930. [PubMed] [Google Scholar]
- 54.Lu Z, Dunaway-Mariano D, Allen KN. 2008. The catalytic scaffold of the haloalkanoic acid dehalogenase enzyme superfamily acts as a mold for the trigonal bipyramidal transition state. Proc Natl Acad Sci U S A 105:5687–5692. doi: 10.1073/pnas.0710800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee SY, Cho HS, Pelton JG, Yan D, Berry EA, Wemmer DE. 2001. Crystal structure of activated CheY. Comparison with other activated receiver domains. J Biol Chem 276:16425–16431. doi: 10.1074/jbc.M101002200. [DOI] [PubMed] [Google Scholar]
- 56.Dyer CM, Dahlquist FW. 2006. Switched or not? The structure of unphosphorylated CheY bound to the N terminus of FliM. J Bacteriol 188:7354–7363. doi: 10.1128/JB.00637-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Russo FD, Silhavy TJ. 1993. The essential tension: opposed reactions in bacterial two-component regulatory systems. Trends Microbiol 1:306–310. doi: 10.1016/0966-842X(93)90007-E. [DOI] [PubMed] [Google Scholar]
- 58.Pao GM, Saier MH. Jr. 1995. Response regulators of bacterial signal transduction systems: selective domain shuffling during evolution. J Mol Evol 40:136–154. doi: 10.1007/BF00167109. [DOI] [PubMed] [Google Scholar]
- 59.Buschiazzo A, Trajtenberg F. 2019. Two-component sensing and regulation: how do histidine kinases talk with response regulators at the molecular level?. Annu Rev Microbiol 73:507–528. doi: 10.1146/annurev-micro-091018-054627. [DOI] [PubMed] [Google Scholar]
- 60.Trajtenberg F, Imelio JA, Machado MR, Larrieux N, Marti MA, Obal G, Mechaly AE, Buschiazzo A. 2016. Regulation of signaling directionality revealed by 3D snapshots of a kinase:regulator complex in action. Elife 5:e21422. doi: 10.7554/eLife.21422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Casino P, Rubio V, Marina A. 2009. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 62.Liu Y, Rose J, Huang S, Hu Y, Wu Q, Wang D, Li C, Liu M, Zhou P, Jiang L. 2017. A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases. Nat Commun 8:2104. doi: 10.1038/s41467-017-02310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davlieva M, Tovar-Yanez A, DeBruler K, Leonard PG, Zianni MR, Arias CA, Shamoo Y. 2016. An adaptive mutation in Enterococcus faecium LiaR associated with antimicrobial peptide resistance mimics phosphorylation and stabilizes LiaR in an activated state. J Mol Biol 428:4503–4519. doi: 10.1016/j.jmb.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leonard PG, Golemi-Kotra D, Stock AM. 2013. Phosphorylation-dependent conformational changes and domain rearrangements in Staphylococcus aureus VraR activation. Proc Natl Acad Sci U S A 110:8525–8530. doi: 10.1073/pnas.1302819110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trajtenberg F, Albanesi D, Ruetalo N, Botti H, Mechaly AE, Nieves M, Aguilar PS, Cybulski L, Larrieux N, de Mendoza D, Buschiazzo A. 2014. Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation. mBio 5:e02105. doi: 10.1128/mBio.02105-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salazar ME, Laub MT. 2015. Temporal and evolutionary dynamics of two-component signaling pathways. Curr Opin Microbiol 24:7–14. doi: 10.1016/j.mib.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fisher SL, Kim SK, Wanner BL, Walsh CT. 1996. Kinetic comparison of the specificity of the vancomycin resistance VanS for two response regulators, VanR and PhoB. Biochemistry 35:4732–4740. doi: 10.1021/bi9525435. [DOI] [PubMed] [Google Scholar]
- 68.Grimshaw CE, Huang S, Hanstein CG, Strauch MA, Burbulys D, Wang L, Hoch JA, Whiteley JM. 1998. Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis. Biochemistry 37:1365–1375. doi: 10.1021/bi971917m. [DOI] [PubMed] [Google Scholar]
- 69.Groban ES, Clarke EJ, Salis HM, Miller SM, Voigt CA. 2009. Kinetic buffering of cross talk between bacterial two-component sensors. J Mol Biol 390:380–393. doi: 10.1016/j.jmb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Janiak-Spens F, Cook PF, West AH. 2005. Kinetic analysis of YPD1-dependent phosphotransfer reactions in the yeast osmoregulatory phosphorelay system. Biochemistry 44:377–386. doi: 10.1021/bi048433s. [DOI] [PubMed] [Google Scholar]
- 71.Stewart RC. 1997. Kinetic characterization of phosphotransfer between CheA and CheY in the bacterial chemotaxis signal transduction pathway. Biochemistry 36:2030–2040. doi: 10.1021/bi962261k. [DOI] [PubMed] [Google Scholar]
- 72.Stewart RC, Jahreis K, Parkinson JS. 2000. Rapid phosphotransfer to CheY from a CheA protein lacking the CheY-binding domain. Biochemistry 39:13157–13165. doi: 10.1021/bi001100k. [DOI] [PubMed] [Google Scholar]
- 73.Fernandez I, Otero LH, Klinke S, Carrica MDC, Goldbaum FA. 2015. Snapshots of conformational changes shed light into the NtrX receiver domain signal transduction mechanism. J Mol Biol 427:3258–3272. doi: 10.1016/j.jmb.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 74.Amin M, Kothamachu VB, Feliu E, Scharf BE, Porter SL, Soyer OS. 2014. Phosphate sink containing two-component signaling systems as tunable threshold devices. PLoS Comput Biol 10:e1003890. doi: 10.1371/journal.pcbi.1003890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Castillo-Hair SM, Baerman EA, Fujita M, Igoshin OA, Tabor JJ. 2019. Optogenetic control of Bacillus subtilis gene expression. Nat Commun 10:3099. doi: 10.1038/s41467-019-10906-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Daeffler KN, Galley JD, Sheth RU, Ortiz-Velez LC, Bibb CO, Shroyer NF, Britton RA, Tabor JJ. 2017. Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol Syst Biol 13:923. doi: 10.15252/msb.20167416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fernandez-Rodriguez J, Moser F, Song M, Voigt CA. 2017. Engineering RGB color vision into Escherichia coli. Nat Chem Biol 13:706–708. doi: 10.1038/nchembio.2390. [DOI] [PubMed] [Google Scholar]
- 78.Landry BP, Palanki R, Dyulgyarov N, Hartsough LA, Tabor JJ. 2018. Phosphatase activity tunes two-component system sensor detection threshold. Nat Commun 9:1433. doi: 10.1038/s41467-018-03929-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ninfa AJ. 2010. Use of two-component signal transduction systems in the construction of synthetic genetic networks. Curr Opin Microbiol 13:240–245. doi: 10.1016/j.mib.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ravikumar S, Baylon MG, Park SJ, Choi JI. 2017. Engineered microbial biosensors based on bacterial two-component systems as synthetic biotechnology platforms in bioremediation and biorefinery. Microb Cell Fact 16:62. doi: 10.1186/s12934-017-0675-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schmidl SR, Ekness F, Sofjan K, Daeffler KN, Brink KR, Landry BP, Gerhardt KP, Dyulgyarov N, Sheth RU, Tabor JJ. 2019. Rewiring bacterial two-component systems by modular DNA-binding domain swapping. Nat Chem Biol 15:690–698. doi: 10.1038/s41589-019-0286-6. [DOI] [PubMed] [Google Scholar]
- 82.Huynh TN, Stewart V. 2011. Negative control in two-component signal transduction by transmitter phosphatase activity. Mol Microbiol 82:275–286. doi: 10.1111/j.1365-2958.2011.07829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Willett JW, Kirby JR. 2012. Genetic and biochemical dissection of a HisKA domain identifies residues required exclusively for kinase and phosphatase activities. PLoS Genet 8:e1003084. doi: 10.1371/journal.pgen.1003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. 2019. The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simonetti FL, Teppa E, Chernomoretz A, Nielsen M, Marino Buslje C. 2013. MISTIC: mutual information server to infer coevolution. Nucleic Acids Res 41:W8–W14. doi: 10.1093/nar/gkt427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maule AF, Wright DP, Weiner JJ, Han L, Peterson FC, Volkman BF, Silvaggi NR, Ulijasz AT. 2015. The aspartate-less receiver (ALR) domains: distribution, structure and function. PLoS Pathog 11:e1004795. doi: 10.1371/journal.ppat.1004795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Desai SK, Kenney LJ. 2017. To ∼P or not to ∼P? Non-canonical activation by two-component response regulators. Mol Microbiol 103:203–213. doi: 10.1111/mmi.13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sheridan RC, McCullough JF, Wakefield ZT. 1972. Phosphoramidic acid and its salts. Inorg Synth 13:23–26. [Google Scholar]
- 89.Martin RB. 1988. Ternary hydroxide complexes in neutral solutions of Al3+ and F−. Biochem Biophys Res Commun 155:1194–1200. doi: 10.1016/S0006-291X(88)81266-X. [DOI] [PubMed] [Google Scholar]
- 90.Silversmith RE, Smith JG, Guanga GP, Les JT, Bourret RB. 2001. Alteration of a nonconserved active site residue in the chemotaxis response regulator CheY affects phosphorylation and interaction with CheZ. J Biol Chem 276:18478–18484. doi: 10.1074/jbc.M011418200. [DOI] [PubMed] [Google Scholar]
- 91.Burley SK, Berman HM, Bhikadiya C, Bi C, Chen L, Di Costanzo L, Christie C, Dalenberg K, Duarte JM, Dutta S, Feng Z, Ghosh S, Goodsell DS, Green RK, Guranovic V, Guzenko D, Hudson BP, Kalro T, Liang Y, Lowe R, Namkoong H, Peisach E, Periskova I, Prlic A, Randle C, Rose A, Rose P, Sala R, Sekharan M, Shao C, Tan L, Tao YP, Valasatava Y, Voigt M, Westbrook J, Woo J, Yang H, Young J, Zhuravleva M, Zardecki C. 2019. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res 47:D464–D474. doi: 10.1093/nar/gky1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.