The emergence of multidrug-resistant Staphylococcus aureus makes the treatment of staphylococcal infections much more difficult. S. aureus can acquire a drug resistance gene from other bacteria, such as Enterococcus faecalis. Intriguingly, S. aureus produces a sex pheromone for the E. faecalis plasmid pAM373, raising the possibility that S. aureus actively promotes plasmid conjugation from E. faecalis. In this study, we found that the staphylococcal Eep protein is responsible for sex pheromone processing and contributes to the survival of the bacteria in the host. These results will enhance future research on the drug resistance acquisition of S. aureus and can lead to the development of novel antivirulence drugs.
KEYWORDS: S. aureus, protease, virulence, innate immunity, translocation
ABSTRACT
In Enterococcus faecalis, the site 2 protease Eep generates sex pheromones, including cAM373. Intriguingly, in Staphylococcus aureus, a peptide similar to cAM373, named cAM373_SA, is produced from the camS gene. Here, we report that the staphylococcal Eep homolog is not only responsible for the production of cAM373_SA but also critical for staphylococcal virulence. As with other Eep proteins, the staphylococcal Eep protein has four transmembrane (TM) domains, with the predicted zinc metalloprotease active site (HEXXH) in the first TM domain. eep deletion reduced the cAM373_SA activity in the culture supernatant to the level of the camS deletion mutant. It also markedly decreased the cAM373 peptide peak in a high-performance liquid chromatography (HPLC) analysis. Proteomics analysis showed that Eep affects the production and/or the release of diverse proteins, including the signal peptidase subunit SpsB and the surface proteins SpA, SasG, and FnbA. eep deletion decreased the adherence of S. aureus to host epithelial cells; however, the adherence of the eep mutant was increased by overexpression of the surface proteins SpA, SasG, and FnbA. eep deletion reduced staphylococcal resistance to killing by human neutrophils as well as survival in a murine model of blood infection. The overexpression of the surface protein SpA in the eep mutant increased bacterial survival in the liver. Our study illustrates that in S. aureus, Eep not only generates cAM373_SA but also contributes to the survival of the bacterial pathogen in the host.
IMPORTANCE The emergence of multidrug-resistant Staphylococcus aureus makes the treatment of staphylococcal infections much more difficult. S. aureus can acquire a drug resistance gene from other bacteria, such as Enterococcus faecalis. Intriguingly, S. aureus produces a sex pheromone for the E. faecalis plasmid pAM373, raising the possibility that S. aureus actively promotes plasmid conjugation from E. faecalis. In this study, we found that the staphylococcal Eep protein is responsible for sex pheromone processing and contributes to the survival of the bacteria in the host. These results will enhance future research on the drug resistance acquisition of S. aureus and can lead to the development of novel antivirulence drugs.
INTRODUCTION
The site 2 proteases (S2Ps) are a class of intramembrane metalloproteases widely distributed in bacteria (1, 2). S2Ps have a conserved zinc metalloprotease active site (HEXXH) in a transmembrane (TM) domain and cleave the target proteins in the membrane (2). They also have an XDG motif in another transmembrane domain, which forms the catalytic center with the active site (3). Many S2Ps also have a centrally located PDZ domain, which is named after three eukaryotic proteins, PSD-95, DLG, and ZO-1 (4, 5). The PDZ domain is important for recognizing target proteins by protein-protein interaction (2, 6, 7).
The Eep protein is an S2P involved in the processing of the signal peptide of lipoproteins in various Gram-positive bacteria, including Enterococcus faecalis, Listeria monocytogenes, and Streptococcus uberis (8–10). In particular, in E. faecalis, the Eep protein cleaves signal peptides of lipoproteins to produce various sex pheromones, such as cAD1, pPD1, and cCF10 (11–14). Sex pheromones are peptides of 7 or 8 amino acids (aa) and induce the conjugation of responsive plasmids (15). When sensed by a donor cell carrying a responsive plasmid, the peptide elicits the production of surface proteins from the responsive plasmid, and the surface protein mediates cell aggregation, a required step for plasmid conjugation (16). In addition to sex pheromone generation, the enterococcal Eep is known to contribute to biofilm formation and virulence (17).
Staphylococcus aureus is a Gram-positive pathogen causing diverse diseases from skin and soft tissue infections to life-threatening infections such as sepsis, pneumonia, endocarditis, and toxic shock syndrome (18). Intriguingly, the bacterium produces a peptide similar to the enterococcal sex pheromone cAM373 (12). Unlike other enterococcal sex pheromones, cAM373 is not processed by Eep, and its processing protease is not known (11). In S. aureus, the cAM373-like peptide, named cAM373_SA, is encoded by a putative lipoprotein gene (SAUSA300_1884, camS) (19, 20). As with the cAM373 peptide in E. faecalis, the enzyme responsible for the production of cAM373_SA has not been identified. In the S. aureus genome, we identified a gene highly homologous to the enterococcal eep gene. Although the enterococcal Eep does not process cAM373, Eep processes the peptide pheromone pPpIA in L. monocytogenes (8). Therefore, in this study, we examined whether the staphylococcal Eep is involved in the production of cAM373_SA. Our results indicate that the staphylococcal Eep not only is responsible for the production of cAM373_SA but also contributes to the virulence of the bacterium.
RESULTS
Staphylococcal Eep is a putative zinc metalloprotease in the membrane.
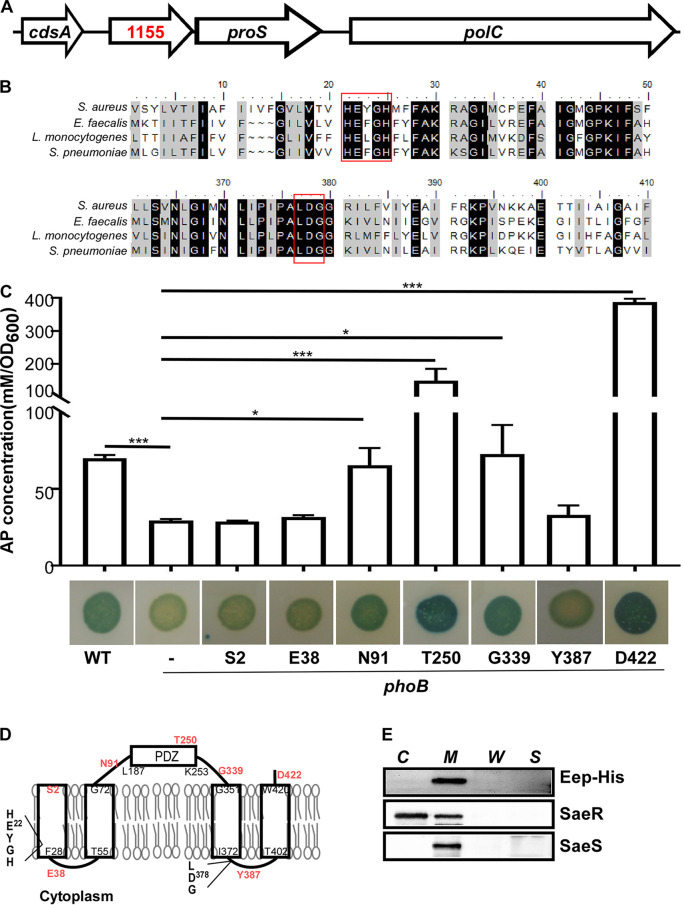
The S. aureus genome has a gene (SAUSA300_1155) encoding a putative membrane-associated zinc metalloprotease (Fig. 1A). The gene forms an operon with proS, encoding prolyl-tRNA synthetase. The gene product shows 40% to 43% identity to the Eep proteins from various Gram-positive bacteria (Fig. 1B). As with other Eep proteins, the staphylococcal Eep protein has a putative zinc metalloprotease active site (HEXXH) in the 1st transmembrane domain and an additional coordinating site (LDG) (Fig. 1B).
FIG 1.
Staphylococcal Eep is a membrane-associated zinc metalloprotease. (A) The genome map for eep in the chromosome in S. aureus USA300_FPR3757. The numbers are gene identifiers (IDs). (B) Sequence alignment of staphylococcal Eep to the Eep proteins of other Gram-positive bacteria with Clustal W2. The conserved HEXXH and LDG motifs are highlighted in red boxes. (C) phoB fusion analysis of Eep. In the single-copy plasmid peep, the phoB gene was inserted into eep at the positions indicated. The alkaline phosphatase (AP) activity was measured with p-nitrophenylphosphate (pNPP). The data were collected from two biological repeats. The statistical significance was measured by unpaired, two-tailed Student’s t test. *, P < 0.05; ***, P < 0.001. For the plate agar assay, the test strains were inoculated on a tryptic soy agar plate containing XP (5-bromo-4-chloro-3-indolylphosphate, toluidine salt, 100 μg/ml; Sigma). (D) Predicted topology of Eep. Rectangles represent transmembrane helices. The conserved HEXXH and LDG motifs are indicated. The amino acid positions for the phoB fusion are shown in red. (E) Cell localization analysis of Eep. Cells were lysed and fractionated into the cytoplasm (C), cytoplasmic membrane (M), cell wall (W), and culture supernatant (S). The proteins were detected by Western blot analysis with cognate antibodies. As fractionation controls, the following proteins were used: SaeR, a cytoplasmic protein, and SaeS, a membrane protein. The full-length blots are presented in Fig. S8.
Sequence analysis (SMART [http://smart.embl-heidelberg.de/]) predicted that staphylococcal Eep is a membrane protein with four transmembrane helices (aa 4 to 28, aa 55 to 72, aa 351 to 372, and aa 402 to 420). To examine the predicted topology, we fused the phoB gene, encoding staphylococcal alkaline phosphatase, to eep at S2, E38, N91, T250, G339, Y387, and D422 and assessed the alkaline phosphatase activities of the fusion proteins. Since alkaline phosphatase requires an oxidizing environment for activity, the protein is functional only in the extracytoplasmic environment (21). As shown in Fig. 1C, only the fusions at the predicted extracytoplasmic domain (i.e., the N91, T250, G339, and D422 fusions) showed substantial alkaline phosphatase activity, confirming the predicted topology (Fig. 1D). Cell fractionation further showed that Eep is localized in the membrane (Fig. 1E). Therefore, we concluded that staphylococcal Eep is a putative zinc metalloprotease residing in the membrane.
Eep is required for the processing of cAM373_SA.
cAM373 (AIFILAS) is an enterococcal sex pheromone inducing the conjugation of the pAM373 plasmid (19). Intriguingly, although S. aureus does not contain pAM373-like plasmid, it produces a cAM373-like sex pheromone (cAM373_SA; AIFILAA), which has Ala instead of Ser at the C terminus (12). cAM373_SA resides in the signal peptide of the CamS lipoprotein (12). Although in E. faecalis, cAM373 is not processed by Eep, other sex pheromone peptides are generated by Eep (8). Therefore, we hypothesized that staphylococcal Eep generates cAM373_SA by processing the signal peptide of CamS. To test this hypothesis, we generated eep and camS deletion mutants. The resulting mutants exhibited growth patterns similar to that of wild-type (WT) strain (see Fig. S1 in the supplemental material), showing that neither the Eep protease nor the CamS lipoprotein is essential for growth. To examine whether Eep is involved in the production of cAM373_SA, the culture supernatants of the WT and mutant strains were collected and subjected to an E. faecalis clumping assay. In this assay, the culture supernatants were serially diluted and mixed with the indicator strain E. faecalis OG1X, carrying pAM373. If the indicator strain detects cAM373_SA, it produces a surface protein that can cause the clumping of bacterial cells. In this assay, the pheromone titer was defined as the maximum dilution fold where the culture supernatant still retains the bacterial clumping activity (22). As shown in Fig. 2A, the WT culture supernatant showed a pheromone titer of 32, whereas the culture supernatants from the eep and the camS deletion mutants showed a pheromone titer of 8, indicating that Eep is required for the production of cAM373_SA from CamS. The clumping activity of the eep mutant was restored by the complementation plasmid peep but not by the vector itself (V) or by the plasmid producing the mutant Eep proteins either in the active site (E22A) or in the LDG motif (D378A), demonstrating that the Eep protease activity is required for peptide-pheromone processing. The culture medium had no clumping activity (TSB in Fig. 2A). Also, all of the culture supernatants failed to induce bacterial clumping in the control strain C1160, which does not have the pAM373 plasmid (Fig. 2A), confirming the specificity of cAM373_SA to the pAM373 plasmid.
FIG 2.
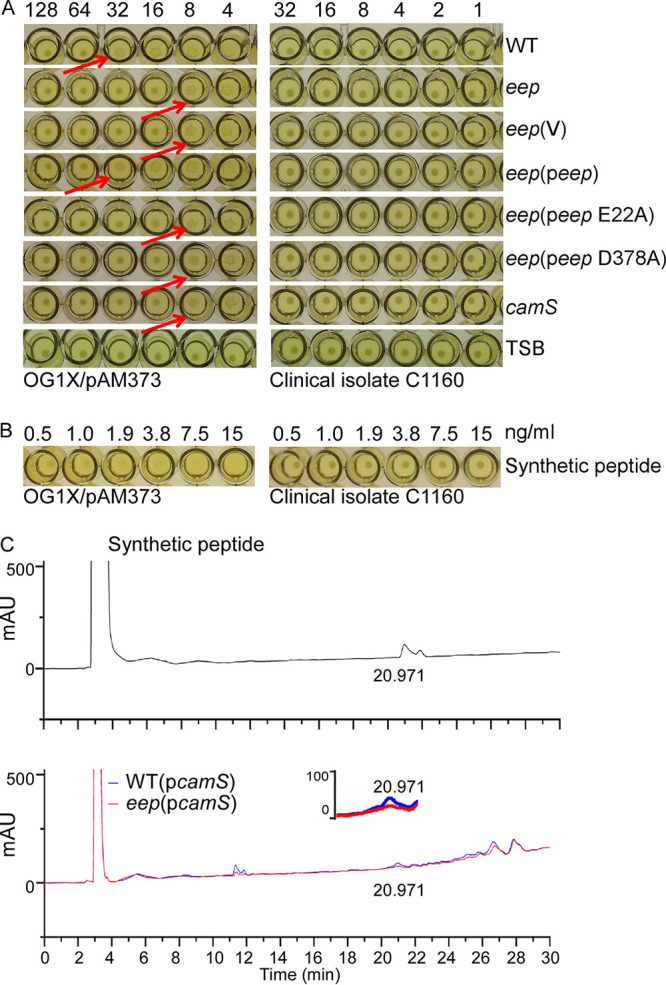
Eep generates cAM373_SA from CamS. (A) The bacterial clumping assay for cAM373_SA. The culture supernatants of test strains were serially diluted 2-fold and added to the bacterial indicator strain (E. faecalis OG1X carrying pAM373) or control strain C1160, a clinical isolate without pAM373. The arrows point to the clumped bacterial cells at the bottom of the well. WT, S. aureus USA300; eep, the eep deletion mutant; V, the vector pCL55; peep, pCL55 containing the eep gene; peep E22A, peep containing the E22A mutation; peep D378A, peep containing the D378A mutation; camS, the camS deletion mutant. Dilution folds are shown at the top. (B) The bacterial clumping assay for the synthetic peptide cAM373_SA (AIFILAA). (C) HPLC analysis of cAM373_SA peptide in the culture supernatants. WT (pcamS), S. aureus USA300 containing pOS1-camS-his; eep (pcamS), the eep deletion mutant containing pOS1-camS-his. The inset showed the enlarged chromatogram from 20 to 22 min. mAU, milli-absorbance unit.
To further verify that Eep is responsible for the production of cAM373_SA from CamS, we analyzed the culture supernatants by high-performance liquid chromatography (HPLC). As a control, the chemically synthesized cAM373_SA peptide was used. The synthetic peptide elicited E. faecalis clumping activity at as low as a 1-ng/ml concentration (Fig. 2B) and in HPLC analysis was eluted at 20.97 min as a somewhat broad peak (Fig. 2C). However, no such peak was detected from the WT culture supernatant (data not shown), indicating that the cAM373_SA concentration was too low to be detected by HPLC. Therefore, using the multicopy plasmid pOS1-camS-his, we overexpressed CamS in the WT and eep mutant strains (Fig. S2). Then culture supernatants were collected, concentrated 1,000-fold, and analyzed by HPLC. Although the culture supernatant of the WT (pOS1-camS-his) showed a small peak at the position of the synthetic cAM373_SA, the peak was missing in the culture supernatant of the eep mutant carrying pOS1-camS-his (Fig. 2C). Based on the results of the clumping assay and the HPLC analysis, we concluded that Eep is responsible for the production of cAM373_SA in S. aureus.
Identification of Eep-regulated protein candidates.
It is possible that as a membrane-bound protease, Eep not only processes the CamS signal peptide but also directly degrades membrane proteins. To examine this possibility, we determined by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis the cell-associated proteins whose abundance is affected by eep deletion. Interestingly, eep deletion changed the abundance of only five cell-associated proteins (4 decreased and 1 increased) (Table 1). Of the 4 decreased proteins, ProS is encoded by the gene in the same operon as eep (Fig. 1A) and is involved in the synthesis of proline-tRNA, whereas LysA synthesizes lysine-tRNA (23). SrrB is a sensor histidine kinase in the SrrAB two-component system (24). SpA is an IgG-binding protein contributing to immune evasion (25). On the other hand, the SpsB protein, a subunit of signal peptidase I, was increased by eep deletion (26). To confirm the effect of eep deletion on the expression of SpsB, we overexpressed His-tagged SpsB in the WT and eep deletion mutant strains and compared the SpsB levels by Western blot analysis. Indeed, the abundance of SpsB was significantly higher in the eep deletion mutant (Fig. 3), confirming the LC-MS/MS results.
TABLE 1.
Cell-associated proteins affected in abundance by eep deletion in USA300
| Gene ID | Protein name | Spectral count |
Fold change (eep/WT) | P value | Protein function | |
|---|---|---|---|---|---|---|
| WT | eep | |||||
| SAUSA300_1293 | LysA | 16.3 | 6.7 | 0.40 | 0.0125 | Diaminopimelate decarboxylase |
| SAUSA300_1441 | SrrB | 15.0 | 6 | 0.42 | 0.0263 | Staphylococcal respiratory response protein |
| SAUSA300_1156 | ProS | 135.3 | 55.3 | 0.42 | 0.0163 | Proline-tRNA ligase |
| SAUSA300_0113 | SpA | 193.3 | 79 | 0.46 | 0.0317 | Immunoglobulin G binding protein A |
| SAUSA300_0868 | SpsB | 11.3 | 19.6 | 1.77 | 0.0091 | Signal peptidase I |
FIG 3.
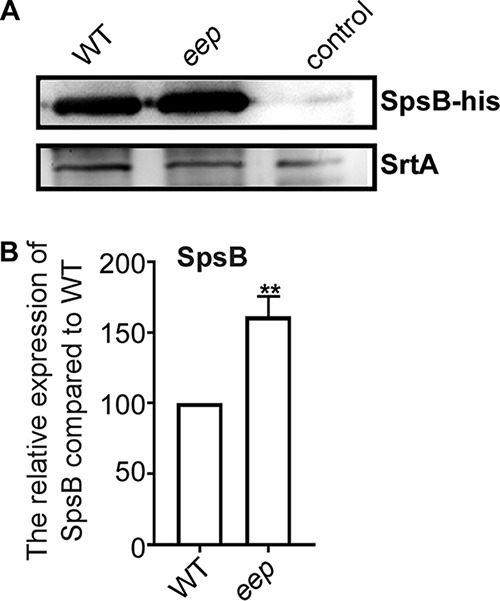
Effect of eep deletion on the expression of SpsB. (A) Western blot analysis for SpsB. Equal numbers of cells were used for the analysis (see Materials and Methods). SpsB protein was detected by the anti-His tag antibody. WT, S. aureus USA300 containing pOS1-spsB-His; eep, eep deletion mutant containing pOS1-spsB-His; control, S. aureus USA300 containing the pOS1 vector. The membrane protein sortase A (SrtA) was used as a loading control. The full-length blots are presented in Fig. S8. (B) Quantification of Western blot results. The protein bands were quantified by ImageJ. The statistical significance was measured by unpaired, two-tailed Student’s t test. **, P < 0.01.
Since SpsB is involved in the signal peptide process and translocation of extracellular proteins, we further examined whether eep deletion can affect the translocation of extracellular proteins. When the overall expression levels of secreted proteins were analyzed by SDS-PAGE, no apparent change was observed (Fig. S3), indicating that the increase of SpsB in the eep deletion mutant does not significantly affect protein secretions.
The sensitivity of the SDS-PAGE is relatively low and cannot detect small changes in secreted proteins. Therefore, we further carried out secretome analysis for the WT and the eep deletion mutant by LC-MS/MS. To our surprise, in this analysis, the deletion of eep affected the secretion and release of 57 proteins (30 increased and 27 decreased) (Fig. 4 and Table 2). Among them, 26 proteins (15 increased and 11 decreased) have a predicted signal peptide sequence. Of the 26 proteins with a signal peptide, 8 were lipoproteins whose signal peptide is cleaved by Lsp, whereas the rest are expected to be targeted by the SpsAB signal peptidase I (Table 3). Intriguingly, 9 of the 18 nonlipoproteins showed reduced secretion in the eep mutant (Table 3 and Fig. 4A), which cannot be explained by the increased level of SpsB. Therefore, it is likely that SpsB plays, at best, a minor role in the alteration in protein secretion.
FIG 4.
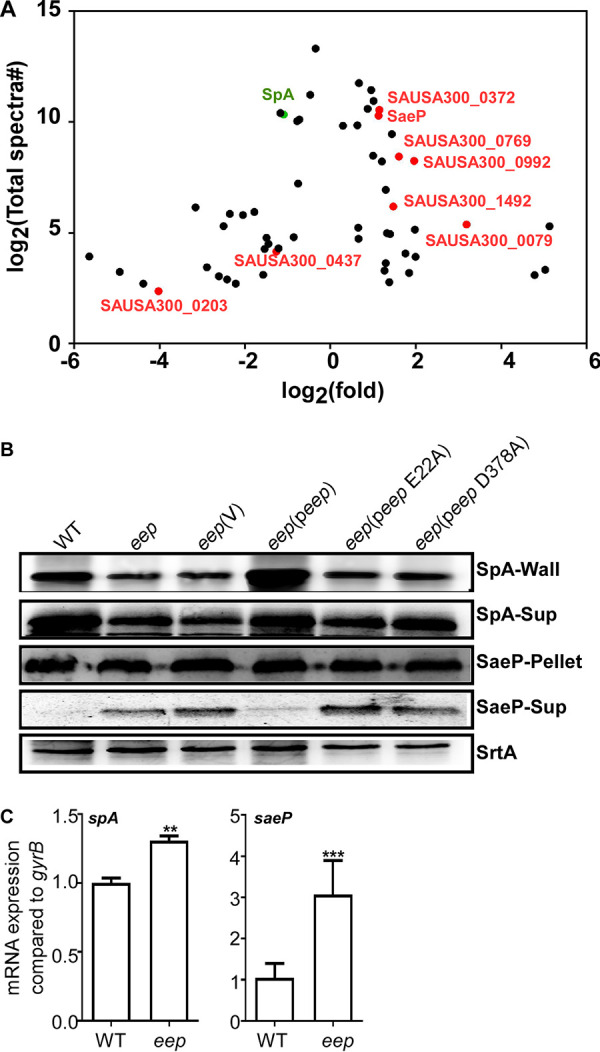
Identification of the staphylococcal proteins whose release or secretion was affected by eep deletion. (A) Proteins whose abundance in culture medium was significantly affected by eep deletion. The x axis represents the average fold changes of the proteins (Δeep/WT), while the y axis shows the total spectral counts of the corresponding proteins. Red color indicates lipoproteins. Staphylococcal protein A (SpA) is shown in green. The data were collected from three biological repeats. (B) Role of Eep in expression of SpA and SaeP. Wall, cell wall; Sup, culture supernatant; Pellet, cell pellets. The membrane protein sortase A (SrtA) was used as a loading control. WT, S. aureus USA300; eep, the ee deletion mutant; V, the vector pCL55; peep, pCL55 containing the eep gene; peep E22A, peep containing the E22A mutation; peep D378A, peep containing the D378A mutation. The full-length blots are presented in Fig. S8. (C) Effect of eep deletion on transcription of spA and saeP. The transcript levels were measured by qRT-PCR, in which gyrB was used as a reference. The data were collected from three biological repeats. The statistical significance was measured by unpaired, two-tailed Student’s t test. **, P < 0.01; ***, P < 0.001.
TABLE 2.
Secreted or released proteins affected by eep deletion in USA300
| Gene ID | Gene name | Spectral count |
Fold change (eep/WT) | P value | Protein function | |
|---|---|---|---|---|---|---|
| WT | eep | |||||
| Upregulated | ||||||
| SAUSA300_0079 | 1.4 | 12.5 | 9.10 | 0.0015 | Putative lipoprotein | |
| SAUSA300_0220 | pflB | 1.0 | 2.3 | 2.41 | 0.0381 | Formate acetyltransferase |
| SAUSA300_0278 | esxA | 11.8 | 29.1 | 2.46 | 0.0045 | ESAT-6-like protein |
| SAUSA300_0372 | 130.7 | 285.6 | 2.19 | 0.0076 | Putative lipoprotein | |
| SAUSA300_0693 | saeP | 155.3 | 343.5 | 2.21 | 0.0004 | Putative lipoprotein |
| SAUSA300_0759 | gpmI | 1.3 | 4.3 | 3.39 | 0.0471 | 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase |
| SAUSA300_0769 | 28.7 | 87.3 | 3.04 | 0.0015 | Putative lipoprotein | |
| SAUSA300_0793 | 0.1 | 2.8 | 27.55 | 0.0007 | Uncharacterized protein | |
| SAUSA300_0897 | trpS | 0.7 | 2.4 | 3.59 | 0.0242 | Tryptophan-tRNA ligase |
| SAUSA300_0950 | sspB | 315.6 | 612.2 | 1.94 | 0.0118 | Cysteine protease |
| SAUSA300_0951 | sspA | 182.8 | 334.3 | 1.83 | 0.0113 | V8 protease |
| SAUSA300_0992 | 20.7 | 80.4 | 3.89 | 0.0016 | Putative lipoprotein | |
| SAUSA300_1044 | trxA | 2.8 | 7.4 | 2.64 | 0.0019 | Thioredoxin |
| SAUSA300_1058 | hla | 444.3 | 707.6 | 1.59 | 0.0147 | Alpha-hemolysin |
| SAUSA300_1068 | 3.0 | 7.6 | 2.51 | 0.0253 | Antibacterial protein | |
| SAUSA300_1247 | 0.6 | 1.7 | 2.60 | 0.0464 | Uncharacterized protein | |
| SAUSA300_1295 | cspA | 4.9 | 7.7 | 1.57 | 0.0486 | Cold shock protein CspA |
| SAUSA300_1327 | ebh | 2.5 | 4.1 | 1.64 | 0.0501 | Extracellular matrix-binding protein |
| SAUSA300_1382 | lukS-PV | 218.3 | 439.8 | 2.01 | 0.0116 | Panton-Valentine leukocidin |
| SAUSA300_1440 | 0.4 | 12.8 | 35.04 | 0.0011 | Uncharacterized protein | |
| SAUSA300_1492 | 6.4 | 17.9 | 2.78 | 0.0021 | Putative lipoprotein | |
| SAUSA300_1704 | leuS | 1.2 | 2.9 | 2.46 | 0.0499 | Leucine-tRNA ligase |
| SAUSA300_1757 | splB | 120.9 | 187.3 | 1.55 | 0.0148 | Serine protease |
| SAUSA300_1759 | 2.4 | 9.4 | 3.95 | 0.0103 | Uncharacterized protein | |
| SAUSA300_1790 | prsA | 62.8 | 170.5 | 2.71 | 0.0002 | Foldase protein |
| SAUSA300_2161 | hysA | 39.6 | 79.2 | 2.00 | 0.0131 | Hyaluronate lyase |
| SAUSA300_2253 | ssaA | 136.0 | 167.0 | 1.23 | 0.0197 | Secretory antigen |
| SAUSA300_2299 | 3.4 | 5.4 | 1.58 | 0.0242 | Multidrug resistance protein A, drug resistance transporter | |
| SAUSA300_2546 | betB | 0.1 | 3.3 | 32.64 | 0.0005 | Glycine betaine aldehyde dehydrogenase |
| SAUSA300_2572 | aur | 30.0 | 69.3 | 2.31 | 0.0043 | Zinc metalloproteinase aureolysin |
| Downregulated | ||||||
| SAUSA300_0113 | spa | 293.7 | 137.6 | 0.47 | 0.0198 | Immunoglobulin G binding protein A |
| SAUSA300_0203 | 1.6 | 0.1 | 0.06 | 0.0009 | Putative lipoprotein | |
| SAUSA300_0224 | coa | 6.0 | 3.3 | 0.55 | 0.0232 | Staphylocoagulase |
| SAUSA300_0235 | ldh1 | 15.0 | 3.6 | 0.24 | 0.0079 | l-Lactate dehydrogenase 1 |
| SAUSA300_0437 | 4.2 | 1.7 | 0.42 | 0.0214 | Lipoprotein | |
| SAUSA300_0504 | pdxS | 11.2 | 2.0 | 0.18 | 0.0025 | Pyridoxal 5-phosphate synthase subunit |
| SAUSA300_0536 | hchA | 31.2 | 18.5 | 0.59 | 0.0285 | Molecular chaperone Hsp31 and glyoxalase 3 |
| SAUSA300_0547 | sdrD | 16.1 | 3.2 | 0.20 | 0.0135 | Serine-aspartate repeat-containing protein D |
| SAUSA300_0569 | 2.4 | 0.4 | 0.16 | 0.0034 | Putative heme-dependent peroxidase | |
| SAUSA300_0594 | adh | 5.0 | 0.1 | 0.02 | 0.0008 | Alcohol dehydrogenase |
| SAUSA300_0681 | 3.2 | 0.4 | 0.13 | 0.0036 | Uncharacterized protein | |
| SAUSA300_0717 | 4.8 | 1.7 | 0.34 | 0.0079 | Ribonucleoside-diphosphate reductase, beta subunit | |
| SAUSA300_0753 | 4.6 | 2.0 | 0.43 | 0.0218 | Epimerase family protein | |
| SAUSA300_0812 | 14.9 | 8.4 | 0.56 | 0.0074 | Uncharacterized protein | |
| SAUSA300_0871 | 6.7 | 2.4 | 0.35 | 0.0166 | Uncharacterized protein | |
| SAUSA300_0955 | atl | 1895.2 | 1489.6 | 0.79 | 0.0326 | Autolysin |
| SAUSA300_0973 | purM | 2.1 | 0.1 | 0.05 | 0.0004 | Phosphoribosylformylglycinamidine cycloligase |
| SAUSA300_1236 | 21.3 | 2.4 | 0.11 | 0.0029 | Uncharacterized protein | |
| SAUSA300_1491 | 1.8 | 0.4 | 0.22 | 0.0146 | Proline dipeptidase | |
| SAUSA300_1512 | pbp3 | 3.1 | 0.1 | 0.03 | 0.0002 | Penicillin-binding protein 3 |
| SAUSA300_1525 | glyQS | 2.2 | 0.7 | 0.34 | 0.0381 | Glycine-tRNA ligase |
| SAUSA300_1972 | int | 2.1 | 0.4 | 0.19 | 0.0141 | Integrase |
| SAUSA300_1974 | lukG | 463.6 | 334.2 | 0.72 | 0.0328 | Uncharacterized leukocidin-like protein |
| SAUSA300_2249 | ssaA | 230.0 | 139.0 | 0.60 | 0.0010 | Secretory antigen SsaA |
| SAUSA300_2364 | sbi | 222.2 | 129.5 | 0.58 | 0.0034 | Immunoglobulin-binding protein |
| SAUSA300_2436 | sasG | 312.8 | 139.0 | 0.44 | 0.0017 | Putative cell wall surface anchor family protein |
| SAUSA300_2441 | fnbA | 5.5 | 2.0 | 0.37 | 0.0026 | Fibronectin-binding protein A |
TABLE 3.
Signal peptide-containing proteins affected by eep deletion in USA300
| Gene ID | Gene name | Fold change (eep/WT) | P value | Signal peptidea |
|---|---|---|---|---|
| Lipoproteins | ||||
| SAUSA300_0079 | 9.10 | 0.0015 | MIKKLFFMILGSLLILSAC | |
| SAUSA300_0203 | 0.06 | 0.0009 | MKKIISIAIIVLALVLSGC | |
| SAUSA300_0372 | 2.19 | 0.0076 | MKLKSLAVLSMSAVVLTAC | |
| SAUSA300_0437 | 0.42 | 0.0214 | MKRLIGLVIVALVLLAAC | |
| SAUSA300_0693 | saeP | 2.21 | 0.0004 | MNTKYFLAAGAVITTLALGAC |
| SAUSA300_0769 | 3.04 | 0.0015 | MKKVMGILLASTLILGAC | |
| SAUSA300_0992 | 3.89 | 0.0016 | MKFGKTIAVVLASSVLLAGC | |
| SAUSA300_1492 | 2.78 | 0.0021 | MKKLVSIVGATLLLAGC | |
| Nonlipoproteins | ||||
| SAUSA300_0113 | spa | 0.47 | 0.0198 | MKKKNIYSIRKLGVGIASVTLGTLLISGGVTPAANA |
| SAUSA300_0224 | coa | 0.55 | 0.0232 | MKKQIISLGALAVASSLFTWDNKADA |
| SAUSA300_0547 | sdrD | 0.20 | 0.0135 | MLNRENKTAITRKGMVSNRLNKFSIRKYTVGTASI |
| SAUSA300_0950 | sspB | 1.94 | 0.0118 | MNSSCKSRVFNIISIIMVSMLILSLGAFANNNKAKADSHSKQLEIN |
| SAUSA300_0951 | sspA | 1.83 | 0.0113 | MKGKFLKVSSLFVATLTTATLVSSPAANA |
| SAUSA300_0955 | atl | 0.79 | 0.0326 | MAKKFNYKLPSMVALTLVGSAVTAHQVQA |
| SAUSA300_1058 | hla | 1.59 | 0.0147 | MKTRIVSSVTTTLLLGSILMNPVANA |
| SAUSA300_1327 | ebh | 1.64 | 0.0501 | MNYRDKIQKFSIRKYTVGTFSTVIATLVFLGFNTSQAHA |
| SAUSA300_1382 | lukS-PV | 2.01 | 0.0116 | MVKKRLLAATLSLGIITPIATSFHESKA |
| SAUSA300_1757 | splB | 1.55 | 0.0148 | MNKNVVIKSLAALTILTSVTGIGTTLVEEVQQTAKA |
| SAUSA300_1790 | prsA | 2.71 | 0.0002 | MKMINKLIVPVTASALLLGA |
| SAUSA300_1974 | lukG | 0.72 | 0.0328 | MIKQLCKNITICTLALSTTFTVLPATSFA |
| SAUSA300_2249 | ssaA | 0.60 | 0.0010 | MKKIATATIATAGFATIAIASGNQAHA |
| SAUSA300_2253 | ssaA1 | 1.23 | 0.0197 | MKKLVTATTLTAGIGTALVGQAHHADA |
| SAUSA300_2364 | sbi | 0.58 | 0.0034 | MKNKYISKLLVGAATITLATMISNGEAKA |
| SAUSA300_2436 | sasG | 0.44 | 0.0017 | MRDKKGPVNKRVDFLSNKLNKYSIRKFTVGTASILIGSLMYLGTQQEAEA |
| SAUSA300_2441 | fnbA | 0.37 | 0.0026 | MKNNLRYGIRKHKLGAASVFLGTMIVVGMGQDKEAA |
| SAUSA300_2572 | aur | 2.31 | 0.0043 | MRKFSRYAFTSMAALTLLSTLSPAAL |
The lipobox is in boldface type.
Confirmation of the MS analysis results.
To confirm the MS analysis results for selected proteins, we examined the effect of eep deletion on the expression and release of SpA, a cell wall protein, and SaeP, a lipoprotein. Although SpA is anchored to the cell wall, it can be released into the culture supernatant by murein hydrolase (27). As shown by the LC-MS/MS results, eep deletion significantly decreased the abundance of SpA in the culture supernatant (SpA-Sup in Fig. 4B and Fig. S4). Interestingly, eep deletion decreased the SpA abundance in the cell wall too (SpA-Wall in Fig. 4B and Fig. S4). In the transcriptional analysis, however, eep deletion rather modestly increased the level of the spA transcripts (Fig. 4C), suggesting that the effect of Eep on SpA expression is posttranscriptional. In the SaeP analysis, eep deletion did not affect the abundance of SaeP in cell pellets; however, it significantly increased the abundance of SaeP in the culture supernatant (Fig. 4B and Fig. S4). Quantitative reverse transcription-PCR (qRT-PCR) analysis showed that eep deletion significantly increased the transcription of saeP (Fig. 4C). The abundance of both SpA and SaeP was restored by the WT eep gene but not by the E22A or the D378A mutant gene (Fig. 4 and Fig. S4), confirming the involvement of Eep protease activity in those phenotypes. Since Eep is not a transcriptional factor, these results indicate that Eep can affect transcription of some proteins indirectly.
In S. aureus, Eep promotes bacterial adhesion to a human epithelial cell line by regulating the expression of adhesins.
In E. faecalis, Eep is reported to be a biofilm-associated virulence factor (17). To test whether Eep contributes to biofilm formation in S. aureus too, we compared biofilm formation by the WT with that by the eep deletion mutant. No significant difference was observed (Fig. S5), suggesting that unlike in E. faecalis, Eep is not required for biofilm formation in S. aureus.
Next, we examined whether Eep plays a role in bacterial adhesion and invasion into host epithelial cells. The WT, the eep mutant, and the complemented strains were incubated with human alveolar basal epithelial A549 cells for 2 h. After eliminating loosely associated bacterial cells by multiple washings, the host cells were lysed, and CFU of the bacterial cells, either tightly associated with or internalized into the A549 cells, were counted. As shown in Fig. 5A, the deletion of eep reduced the CFU counts 5-fold. The CFU counts were fully restored by the complement plasmid peep but not by the plasmid producing the inactive Eep proteins (i.e., E22A and D378A mutants) (Fig. 5A).
FIG 5.
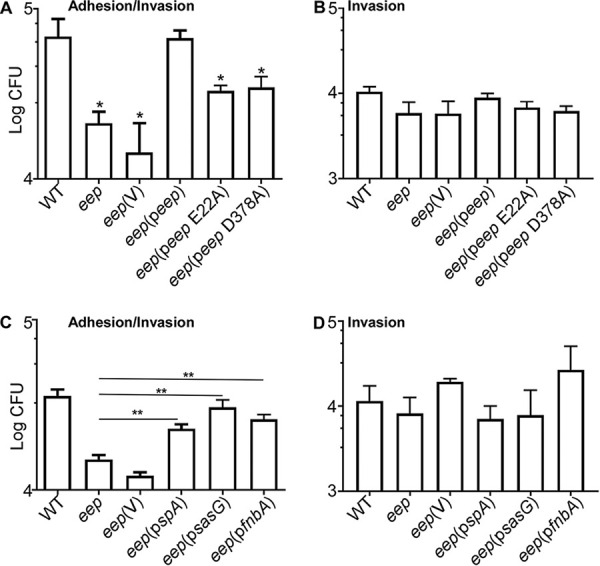
Eep promotes staphylococcal adhesion to human epithelial cells. (A) A549 epithelial cells were incubated with the bacterial strains for 2 h; then the CFU associated with the epithelial cells were measured. (B) Effect of eep deletion on bacterial invasion into A549 cells. In this assay, before CFU enumeration, bacterial cells outside the epithelial cells were eliminated by lysostaphin treatment. (C and D) Effect of the additional expression of three surface proteins on the adhesion/invasion (C) and the invasion (D) of the eep mutant. WT, S. aureus USA300; eep, the eep deletion mutant; V, the vector pCL55; peep, pCL55 containing the eep gene; peep E22A, peep containing the E22A mutation; peep D378A, peep containing the D378A mutation; pspA, pCL55 containing the spA gene; psasG, pOS1 containing the sasG gene; pfnbA, pOS1 containing the fnbA gene. The data were collected from three biological repeats. The statistical significance was measured by unpaired, two-tailed Student’s t test. *, P < 0.05; **, P < 0.01.
The assay described above did not distinguish the bacteria attached to the host cells from the ones inside the host cells. Therefore, to enumerate only the bacteria inside the host cells, after incubation of A549 cells with bacterial cells for 2 h, we washed the A549 cells twice and eliminated the bacterial cells outside A549 cells by lysostaphin treatment; then the bacterial CFU were measured. As shown in Fig. 5B, the deletion of eep did not affect the intracellular CFU counts. These results suggest that Eep plays a positive role in staphylococcal adhesion but not in the invasion of the bacterium into host cells.
Our secretome data showed that eep deletion reduced the release of several cell surface proteins, such as SpA, SasG, and FnbA (Table 2). Since surface proteins are involved in bacterial adhesion to the host cells, we hypothesized that the decreased expression/release of those surface proteins might contribute to the reduced adhesion of the eep deletion mutant. To test this hypothesis, we additionally expressed those surface proteins in the eep mutant using a plasmid and compared the bacterial adhesions. SpA was produced in a native form, whereas SasG and FnbA were expressed as a His-tagged protein. Western blot analysis confirmed the additional expression of those proteins in the eep mutant (Fig. S6). The additional expression of those surface proteins in eep mutants significantly increased the bacterial adhesion to the A549 cells without an effect on bacterial invasion (Fig. 5C and D), indicating that the decreased expression/release of those surface proteins, at least in part, explains the reduced adhesion of the eep mutant to the host cells.
Eep contributes to resistance to human neutrophil-mediated killing.
Neutrophils are a crucial component of the innate immune defense against staphylococcal infection (28). We further investigated whether Eep impacts S. aureus interaction with neutrophils. We incubated the WT and the eep mutant with purified human neutrophils and analyzed bacterial survival and neutrophil lysis. The eep mutant showed a significantly lower survival rate than the WT (Fig. 6A). Also, the neutrophil lysis was markedly lower with the eep mutant (Fig. 6B). The complementation test confirmed that the protease activity of Eep is critical for bacterial survival and the lysis of neutrophils (Fig. 6). These results strongly suggest that Eep is required for S. aureus to cope with attack by human neutrophils.
FIG 6.
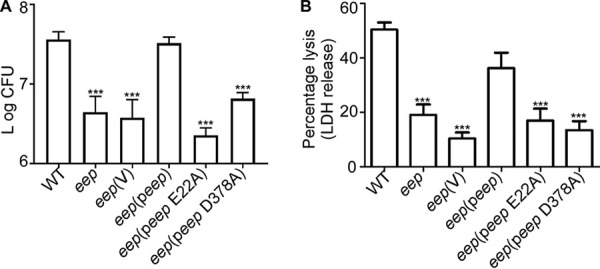
Eep contributes to staphylococcal defense against the attacks of human neutrophils. Bacterial cells at exponential growth phase were collected, washed, and incubated with purified human neutrophils for 3 h. (A) Role of Eep in the survival of bacteria measured by CFU counting. (B) Role of Eep in the lysis of neutrophils measured by the release of lactate dehydrogenase (LDH). WT, S. aureus USA300; eep, the eep deletion mutant; V, the vector pCL55; peep, pCL55 containing the eep gene; peep E22A, peep containing the E22A mutation; peep D378A, peep containing the D378A mutation. The results are representative of those from three independent experiments. The statistical significance was measured by unpaired, two-tailed Student’s t test. ***, P < 0.001.
Eep is required for staphylococcal survival during host infection.
So far, we found that Eep is required for staphylococcal adhesion into the host cells and the defense against neutrophil attacks. Next, using a murine model of a blood infection, we examined whether Eep is required for bacterial virulence and survival during host infection. Mice were infected with the WT and the eep deletion mutant via the retro-orbital route; then the mice were observed for 14 days. As shown in Fig. 7A, WT S. aureus killed 80% of the infected mice, whereas the eep mutant killed only 40%. The eep deletion mutant showed a CFU count approximately 10 to 100 times lower than that of the WT in the murine kidneys and livers (Fig. 7B), suggesting that Eep is required for staphylococcal survival during host infection. Furthermore, the kidneys of mice infected with the eep mutant strain showed lower abscess formation (Fig. 7C) and a decreased infiltration of inflammatory cells (Fig. 7D). These results demonstrate that Eep contributes to the virulence and survival of S. aureus during host infection.
FIG 7.
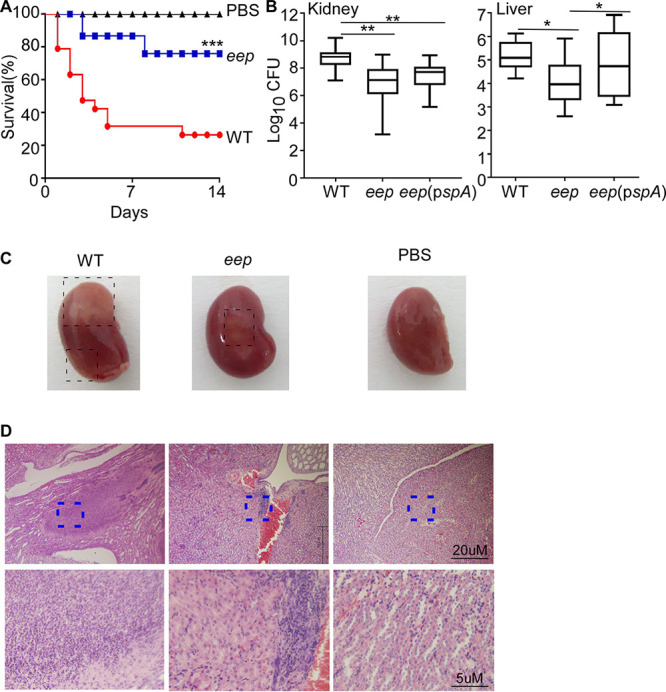
Eep contributes to staphylococcal virulence and survival during host infection. (A) Effect of eep deletion on survival of infected mice. USA300 WT and eep deletion mutant (eep) cells were collected at the exponential growth phase, washed, and administered into eight mice via the retro-orbital route. The survival difference between the mice infected with the WT and the eep mutant was analyzed by log rank (Mantel-Cox) test. ***, P < 0.001. The experiment was repeated, and the results were pooled. PBS, phosphate-buffered saline. (B) Role of Eep in survival of S. aureus during murine infection. At day 4 postinfection, eight mice in each group were killed, and the kidneys and livers were harvested and ground. The CFU of S. aureus in the ground kidneys and livers were measured by the serial dilution method. Statistical significance was measured by unpaired, two-tailed Student’s t test. The data were collected from two biological repeats. *, P < 0.05; **, P < 0.01. (C) Effect of eep deletion on kidney abscess formation. The kidney was harvested at day 4 postinfection. The dashed rectangles show the abscessed regions. (D) The H&E staining of the kidney section. The dashed squares show stronger infiltration of inflammatory cells in the kidney infected with WT S. aureus. WT, S. aureus USA300; eep, the eep deletion mutant; eep(pspA), the eep deletion mutant carrying the SpA expression plasmid pspA; PBS, the PBS control.
eep deletion lowered the expression of the surface protein SpA, an important virulence factor (Fig. 4) (25). To test whether the lower expression of SpA is, at least in part, responsible for the reduced survival of the eep mutant, we subjected the eep mutant producing extra SpA protein to the murine blood infection model. Although the increased expression of SpA did not affect staphylococcal survival in the kidney, it increased the bacterial CFU to the WT level in the liver (Fig. 7B), suggesting that the reduced SpA expression, at least in part, explains the lower survival of the eep mutant in the host.
DISCUSSION
As proteases widely distributed in bacteria, site 2 proteases (S2Ps) are involved in diverse molecular pathways (2). In S. aureus, the function of S2P Eep has been unknown. In this study, we showed that in S. aureus, Eep is responsible for the processing of cAM373_SA and contributes to bacterial adhesion into human epithelial cells. The protein was also required for resistance to killing by human neutrophils and bacterial survival during host infection. These results indicate that Eep is a new virulence factor in S. aureus that can be a target of novel therapeutic development.
Unlike the enterococcal sex pheromone cAM373 (29), which is not processed by Eep, the staphylococcal peptide cAM373_SA appeared to be processed by the Eep protein. First, the deletion of eep reduced cAM373_SA in the culture supernatant to the level of the camS deletion mutant (Fig. 2A). Second, eep deletion almost abolished the cAM373_SA peptide peak at 20.97 min in the HPLC analysis (Fig. 2C). Also, during the review of our study, Schilcher et al. reported that in S. aureus, Eep is responsible for the secretion of small linear peptides, including cAM373_SA (30), confirming our clumping assay and HPLC analysis results. Intriguingly, neither the eep deletion nor the camS deletion abolished the clumping activity of culture supernatant (Fig. 2A), indicating that S. aureus produces another cAM373-like sex pheromone, whose production is independent of Eep. When the protein sequences of the S. aureus USA300_FPR3757 genome were searched for peptide sequences similar to cAM373 or cAM373_SA, with one amino acid substitution allowed, the following three peptide sequences were found: AIFILAT in DnaX (SAUSA300_0452; aa 142 to 148), FIFILAS in PgsA (SAUSA300_1176; aa 48 to 54), and AIFIIAA in FmtC (SAUSA300_1255; aa 232 to 238), where the disparate amino acids are underlined. However, unlike CamS, all proteins are cytoplasmic proteins. Therefore, it is unlikely that those peptides are processed and released into the extracellular environment. Therefore, more work is needed to identify the source of the remaining pheromone activity.
Why does S. aureus produce cAM373-like peptide? In Gram-positive bacteria, small peptides play diverse roles from quorum sensing and competence development to plasmid conjugation, biofilm formation, and bacterial virulence (15, 31–34). One of the interesting aspects of the peptide pheromone system is that the released peptide pheromones are imported into the cytoplasm to bind their target molecules (8, 35). So far, except for the clumping activity toward E. faecalis carrying pAM373, no physiological role for cAM373_SA has been identified. Since the camS deletion mutant showed normal growth (see Fig. S1 in the supplemental material), the peptide is not required for bacterial growth. One possible function of the peptide is to facilitate the acquisition of useful genetic elements, such as antibiotic resistance genes from enterococci, by inducing the conjugation process. Indeed, the first vancomycin-resistant methicillin-resistant S. aureus (MRSA) isolate acquired Tn1546 carrying the vanA-type vancomycin resistance genes from E. faecalis (36). Since pAM373 cannot stably replicate in S. aureus, such useful genes acquired via pAM373 conjugation should be incorporated into an S. aureus chromosome or plasmid via illegitimate recombination or, if the acquired genes are in a transposon, by transposition (37). Also, it is possible that cAM373_SA has a receptor in S. aureus and carries out hitherto-unidentified functions. Finally, we cannot rule out the possibility that the staphylococcal production of cAM373_SA is an accident and cAM373_SA does not have any physiological roles in S. aureus. These possibilities are currently being tested in our laboratory.
In Gram-positive bacteria, lipoproteins are key players for bacterial virulence (38). As Toll-like receptor (TLR2) agonists, bacterial lipoproteins can play an important role in innate immune activation (38). Intriguingly, the abundance of eight lipoproteins in the culture supernatant was altered by eep deletion (Table 2). To examine whether the altered abundance of the lipoproteins affects the innate immune response, we transfected the HEK293 human embryonic kidney cell line with TLR2 and stimulated the cells with the WT and the eep mutant strains. When interleukin 8 (IL-8) was measured as an indicator for TLR2 activation, no significant difference was observed (Fig. S7), indicating that altered release of the lipoproteins does not bring about substantial changes in the innate immune response.
The secretome data showed that eep deletion lowered the release of several surface proteins, such as SpA, SasG, and FnbA (Table 2). When those surface proteins were additionally expressed from a plasmid in the eep deletion mutant, the adhesion of the eep mutant was significantly increased (Fig. S6), indicating the involvement of those surface proteins in the bacterial adhesion to the host cells. In particular, the additional expression of SpA also significantly increased bacterial survival in the murine liver (Fig. 7B), demonstrating the contribution of the surface protein to bacterial survival in the host. The positive role of SpA in staphylococcal virulence has been well documented (39). With its ability to bind to the Fcγ portion of human and animal immunoglobulin, it protects S. aureus from opsonophagocytic killing (40). It has a superantigen activity toward B cells and interferes with host adaptive immunity (41, 42). The protein is also required for staphylococcal abscess formation (43). SpA, in concert with Sbi, is a key factor in neutrophil extracellular trap (NET) formation (44). Indeed, monoclonal antibody against an SpA variant devoid of Fcγ binding and superantigen activity promoted opsonophagocytic killing of MRSA and reduced abscess formation in the murine kidney (45). It is possible that the decreased expression of SpA from the eep mutant contributes to the reduced survival and virulence of the mutant (Fig. 7). It should be noted that the abundance of many toxins and enzymes (e.g., SspAB, Hla, LukS-PV, SplB, and Aur) was increased in the culture medium of the eep mutants, implying that the increased secretion of those toxins and enzymes failed to compensate the adverse effect of the eep deletion on staphylococcal virulence.
In summary, we have shown that the staphylococcal Eep protein generates cAM373_SA from the lipoprotein CamS and plays a critical role in bacterial survival and virulence. The reduced production/release of surface proteins such as SpA, SasG, and FnbA partly explains the lower survival rate of the eep mutant in the host. However, it remains unknown how Eep, with very limited direct targets (Table 1), affects the production/release of 57 proteins, including the surface proteins. Finally, the physiological role of cAM373_SA in S. aureus remains to be determined. Thus, future research is warranted to answer those questions.
MATERIALS AND METHODS
Ethics statement.
The animal experiment was performed by following the Guide for the Care and Use of Laboratory Animals (46). The animal protocol was approved by the ethics committee of Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University. Human heparinized venous blood was taken from healthy individuals in accordance with a protocol approved by the ethics committee of Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University. All individuals gave written informed consent prior to donating blood.
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. Escherichia coli and S. aureus were grown in lysogeny broth (LB) and tryptic soy broth (TSB), respectively. For transduction of mutations and plasmids, heart infusion broth (HIB) supplemented with 5 mM CaCl2 was used. When necessary, antibiotics were added to the growth media at the following concentrations: ampicillin, 100 μg/ml; erythromycin, 10 μg/ml; and chloramphenicol, 5 μg/ml.
DNA manipulation.
Unless stated otherwise, all restriction enzymes and DNA modification enzymes were purchased from New England BioLabs. Plasmids and genomic DNA were extracted with a plasmid miniprep kit (Zymo Research). Plasmid DNA was introduced into E. coli by the method of Hanahan (47) and electroporated into S. aureus RN4220 with Gene Pulser (Bio-Rad).
Generation of mutant and complementation strains.
To construct plasmids for deletion of eep and camS, we used the ligation-independent cloning (LIC) method. Vector DNA was PCR amplified from pKOR1 with the primers P236/237 or from pIMAY with the primers P1986/1987 and PrimSTAR (TaKaRa) (Table S2). Two 1-kb DNA fragments, upstream and downstream of eep and camS, were PCR amplified from the chromosomal DNA with the following primer pairs: P474/475 and P476/477 for eep and P2481/2482 and P2483/2484 for camS. The PCR products were treated with T4 DNA polymerase in the presence of dGTP (vector) or dCTP (insert DNA) and mixed together. The DNA mixture was transformed into E. coli DH5α. The resulting plasmids, pIMAYΔeep and pKOR1ΔcamS, were electroporated into S. aureus strain RN4220 and subsequently transduced into USA300 with ϕ85. The eep and camS deletions were carried out as described previously (48, 49).
For the complementation test for eep, the single-copy plasmid pCL55 was used. The vector DNA was PCR amplified with the primers P35/80, whereas the eep gene, including the promoter sequence, was amplified with the primers P78/249 (for the His tag at the C terminus) (Table S2). The plasmid was assembled by LIC as described above.
Site-directed mutagenesis of the eep complementation plasmids was carried out as described by Ho et al. (50) with the following primers: PL85/86 for eep E22A and PL87/88 for eep D378A (Table S2). The mutations were verified by DNA sequencing.
For overexpression of CamS, the multicopy plasmid pOS1 was used. The camS gene was amplified with the primers PL351/PL352 (for the His tag at the C terminus) (Table S2). The amplified fragment was digested with SmaI/BamHI and inserted into the multicopy plasmid pOS1, resulting in pOS1-camS-his. The plasmid was inserted into E. coli DH5α and then into S. aureus RN4220. Finally, the plasmid was transduced by ϕ85 into the wild type and the eep mutants of S. aureus.
Construction of PhoB fusions.
To generate PhoB fusions at Ser2 (S2), Glu38 (E38), Asn91 (N91), Thr250 (T250), Gly339 (G339), Tyr387 (Y387), and Asp422 (D422) of Eep, the phoB fragment lacking the signal peptide sequence was PCR amplified with the following primer pairs: PL24/25 for S2-PhoB, PL82/83 for E38-PhoB, PL28/29 for N91-PhoB, PL32/33 for T250-PhoB, PL36/37 for G339-PhoB, PL40/41 for Y387-PhoB, and PL44/45 for D422-PhoB (Table S2). The target vector pCL55-eep was PCR amplified with primer pairs PL22/23 for S2-PhoB, PL80/81 for E38-PhoB, PL26/27 for N91-PhoB, PL30/31 for T250-PhoB, PL34/35 for G339-PhoB, PL38/39 for Y387-PhoB, and PL42/43 for D422-PhoB (Table S2). All resulting PCR products were treated with T4 DNA polymerase for 30 min at room temperature. The insert phoB fragment and its corresponding vector DNA were mixed and incubated at 37°C for 30 min; then the mixture was transformed first into E. coli and subsequently into RN4220 and its target strain, NMΔphoB.
Alkaline phosphatase assay.
Alkaline phosphatase activity was measured according to the manufacturer’s instructions (Yeasen, China). Briefly, bacteria were cultured in TSB for 4 h and collected by centrifugation. The collected cells were treated with lysostaphin (50 μg/ml) for 30 min. The samples were incubated with p-nitrophenylphosphate (pNPP) for 30 min, and OD at 405 nm (OD450) was measured with a Synergy 2 microplate reader (BioTek) and normalized by OD600. For agar plate assay, the test strains were inoculated on a tryptic soy agar plate containing XP (5-bromo-4-chloro-3-indolylphosphate, toluidine salt, 100 μg/ml; Sigma) and incubated at 37°C overnight (51).
Fractionation of cell components.
Bacterial cells were grown in TSB to the exponential growth phase (OD600 ≈ 1.0). Cells were collected by centrifugation, suspended in TSM (50 mM Tris-HCl, 0.5 M sucrose, 10 mM MgCl2 [pH 8.0]) containing lysostaphin (50 μg/ml), and incubated at 37°C for 30 min. The protoplasts were collected by centrifugation (4,600 × g, 5 min) and suspended in membrane buffer (100 mM Tris-HCl, 100 mM NaCl, 10 mM MgCl2 [pH 8.0]). After sonication, the membrane fraction was recovered by ultracentrifugation (120,000 × g) at 4°C for 30 min and suspended in 1× TKMG buffer (50 mM Tris-HCl, 50 mM KCl, 1 mM MgCl2, 25% glycerol [pH 8.0]). The supernatant was designated the cytoplasmic fraction. All samples were subjected to SDS-PAGE followed by Western blot analysis.
Western blot analysis.
Western blot analysis of proteins was carried out as described previously (52). Briefly, the test strains were grown to exponential phase. Equal numbers of cells were collected and treated with lysostaphin (50 μg/ml) for 30 min. The samples were mixed with protein loading buffer, boiled for 10 min, and subjected to 8% to 15% SDS-PAGE. In Western blot analysis, the Eep, SpsB, SasG, and FnbA proteins were detected with anti-His tag antibody, whereas other proteins were detected with their cognate antibodies. The His tag antibody was purchased from Yeasen Co. (China). The SpA antibody was purchased from Sigma. The SaeP, SaeR, SaeS, and SrtA antibodies were generated by our laboratory.
Synthesis of cAM373_SA.
cAM373_ SA (AIFILAA) was synthesized by GenScript. The synthesized peptide showed over 98% purity by high-performance liquid chromatography (HPLC).
Clumping assay.
The clumping assay for the peptide was carried out as previously described (22). Bacterial cells were grown in TSB to the exponential growth phase. The supernatant was collected by centrifugation and passed through a 0.22-μm filter to eliminate the remaining bacterial cells. The supernatant was then boiled for 15 min and stored at 4°C. The supernatant (100 μl) was added to 96-well microtiter plates and serially diluted (2-fold) into fresh TSB. Responder cells (E. faecalis OG1X carrying pAM373) were cultured in HIB and collected at the stationary growth phase. The cell density was adjusted to an OD600 of 0.5. The cell suspension (100 μl) was added to the culture supernatants, and the samples were incubated at 37°C for 4 h. The clumping of cells was visually examined.
HPLC identification of cAM373_SA.
USA300 (pOS1-camS-his) and the eep(pOS1-camS-his) strains were grown in TSB at 37°C overnight. Next day, the resulting cultures were diluted 100 times in fresh TSB and further incubated at 37°C for 4 h. After centrifugation, the culture supernatants were collected, boiled for 15 min, and passed through a 0.22-μm filter. The filtrated culture supernatants were precipitated by 10% trichloroacetic acid (TCA) at –20°C overnight. After centrifugation (13,000 × g, 10 min), the supernatant was passed through a Welchrom C18E column (Welch; WS18190923). The column was washed with water twice and eluted with 2 ml of dimethyl sulfoxide (DMSO) three times. The eluents were combined and concentrated 1,000-fold with a nitrogen-blowing instrument (MD200-1; Allsheng, China). The concentrated samples (20 μl) were subjected to the C18 column (Eclipse Plus; 5-μm particle size; 4.6 by 250 mm) in HPLC (Agilent Associates). In the HPLC analysis, two solutions were used: solution A (0.1% formic acid in 100% water) (vol/vol) and solution B (0.1% formic acid in 100% acetonitrile) (vol/vol). Peptides were eluted by a linear gradient of solution B from 5% to 40% in 40 min. The flow rate was 1 ml/min, and the column temperature was maintained at 30°C. The peptides were detected at 220 nm.
Mass spectrometry analysis.
S. aureus was grown to the exponential growth phase in TSB (OD600 ≈ 1.0). The resulting bacterial culture was centrifuged at 8 000 × g for 10 min, and the bacterial pellets and the supernatant were collected for further bacterial proteome and secretome analyses. The cell pellets were washed twice with phosphate-buffered saline (PBS) and then treated with lysostaphin (50 μg/ml) at 37°C for 30 min. For secretome analysis, the supernatant was passed through a 0.22-μm filter, precipitated with 10% TCA, washed with ice-cold acetone, and then suspended in TS buffer (50 mM Tris-HCl [pH 8.0], 4% sodium dodecyl sulfate). All samples were mixed with protein loading buffer, boiled for 10 min, and subjected to 12% SDS-PAGE and Coomassie staining. The gel was sliced into 10 pieces. Each piece was cut into small cubes and destained with 50% acetonitrile (ACN) in 50 mM NH4HCO2; then the cubes were dehydrated with 100% ACN before digestion with sequencing-grade trypsin (10 ng/μl of trypsin, 50 mM ammonium bicarbonate [pH 8.0]) at 37°C overnight. The peptides were vacuum dried prior to LC-MS/MS analysis on a nanoflow liquid chromatography instrument (EASY-nLC 1000; Thermo Scientific) coupled to an ion trap mass spectrometer (LTQ Velosro; Thermo Scientific). The outlet of the LC system was comprised of solvent A (97% H2O, 3% ACN, 0.1% formic acid) and solvent B (100% ACN, 0.1% formic acid). Peptides were eluted using a 55-min gradient: the gradient was started at 7% solvent B for 3 min and then raised to 35% solvent B for 40 min, and then solvent B was ramped to 90% for 2 min and maintained for 10 min for column wash. The eluted peptides were introduced into mass spectrometry for MS1 and MS2 analysis in a data-dependent mode. Full-scan MS spectra were obtained with m/z 350 to 1,500; the most intense ions (top 10) with multiple charges (+2 and +3) were required for MS2 analysis. Dynamic exclusion was set with a maximum repeat duration of 24 s and exclusion duration of 12 s. The data were collected with a centroid mode. Detailed LC-MS/MS settings have been described elsewhere (53).
Expression of Eep-regulated proteins in S. aureus.
To express SpsB, SasG, and FnbA in S. aureus, the respective gene with its own promoter was PCR amplified with the primers PL428/429 (for spsB), PL503/504 (for sasG), and PL501/502 (for fnbA), which insert the His tag sequence at the C terminus (Table S2). The amplified fragment was digested with SmaI/SalI (for spsB) or SmaI/BamHI (for sasG and fnbA) and inserted into the multicopy plasmid pOS1. For unknown reasons, the expression of SpA was not successful with pOS1. Therefore, we expressed SpA with the single-copy plasmid pCL55 (54). The spA gene with its own promoter was PCR amplified with the primers P221/222 (Table S2). The amplified fragment was digested with SmaI/BamHI and inserted into pCL55. The resulting plasmid was inserted into E. coli DH5α and then into S. aureus RN4220. Finally, the plasmids were transduced by φ85 into the wild type and the eep deletion mutant of S. aureus.
Real-time quantitative reverse transcription-PCR (qRT-PCR).
Cells were grown as described above in 3 ml of TSB, harvested, and broken with a mini-bead beater (Biospec Products) at maximum speed for 30 s. After incubation on ice for 5 min, the samples were centrifuged. The supernatant was collected and used to isolate total RNA according to the manufacturer’s instructions (Qiagen). After DNase treatment with a TRUBO DNA-free kit (Ambion), 1 μg of total RNA was reverse transcribed with a Prime Script RT reagent kit (Qiagen). The cDNA was used as a template for real-time PCR with SYBR green PCR reagents (Roche). Reactions were performed in a Micro Amp optical 96-well reaction plate with a 7500 sequence detector (Applied Biosystems). Primers used in this analysis are listed in Table S2. All RT-PCR experiments were carried out in triplicate with gyrB as an internal control. All experiments were repeated at least three times independently.
Semiquantitative biofilm assay.
The semiquantitative biofilm assay was performed as described previously (55). Briefly, overnight cultures of S. aureus strains were diluted 1:100 with fresh TSB containing 0.5% glucose. The diluted cultures were pipetted into sterile 96-well flat-bottom plates and incubated at 37°C for 24 h. Culture supernatants were gently removed, and wells were washed with PBS. The adherent organisms at the bottoms of the wells were fixed by Bouin fixative over 1 h. The fixative was removed gently, and wells were washed with PBS. The organisms in the wells were stained with 0.4% (wt/vol) crystal violet. Biofilm formation was measured with a Synergy 2 microplate reader (BioTek) at 570 nm.
Adhesion and invasion assay.
S. aureus was grown to the exponential growth phase in TSB (OD600 ≈ 1.0) and washed twice with F-12K medium. The A549 cells (i.e., adenocarcinomic human alveolar basal epithelial cells) were cultured at 37°C in F-12K medium supplemented with fetal bovine serum (FBS; 10%) and 5% CO2. The cells were infected with S. aureus at a multiplicity of infection (MOI) of 100.
For the adhesion and invasion assay, the cells were incubated for 2 h and then were collected, washed twice with PBS, and lysed by the addition of 0.1% deoxysodium cholate solution (500 μl). Bacterial CFU were enumerated by serial dilutions of the epithelial cell lysates and spreading of the diluted lysates onto tryptic soy agar (TSA) plates.
The invasion assay was carried out as described by Kim et al. (56). Briefly, after incubation with the bacteria for 2 h, the cells were washed with PBS twice and then incubated in F-12K medium supplemented with lysostaphin (8.8 nM) for 2 h. Then the lysostaphin was quenched by 50 mM EDTA (pH 8.7). The cells were washed twice with PBS and lysed by the addition of 0.1% deoxysodium cholate solution (500 μl). Bacterial CFU were enumerated by serial dilutions of epithelial cell lysates and spreading of the lysates onto TSA plates.
Overexpression of TLR2 in HEK293 cells.
To generate TLR2 overexpression plasmid in human cells, the TLR2 fragment was PCR amplified with primer pair PL464/465 (Table S2). The amplified fragment was digested with KpnI/XbaI and inserted into the pCMV3.0 vector. The resulting plasmid was inserted into E. coli DH5α, and the plasmid was extracted from E. coli using the Endo-free plasmid minikit (Omega). Human embryonic kidney 293 (HEK293) cells were cultured in 35-mm tissue culture plate with Dulbecco modified Eagle medium (DMEM) with 10% FBS at 37°C and 5% CO2 overnight. A total of 4 μg of plasmid DNA was transfected for 6 h. Then the transfected cells were stimulated with either WT or eep deletion mutant S. aureus at an MOI of 10 for 18 h. The culture supernatant was collected by centrifugation. Human IL-8 secretion was measured in cellular supernatants using CUSABIO enzyme-linked immunosorbent assay (ELISA) kits according to the manufacturer’s instructions.
Neutrophil lysis assay.
Neutrophil lysis was measured with a lactate dehydrogenase (LDH) cytotoxicity detection kit according to the manufacturer’s protocol (Roche). Bacteria were grown to exponential growth phase (OD600 ≈ 1.0). Neutrophils were infected by bacteria at an MOI of 10. After incubation for 3 h, the supernatant was collected for LDH activity. To determine the bacterial survival rates, 100 μl of the culture was serially diluted and spread on TSA plates for CFU counting.
Animal experiment.
Bacterial cells were grown in TSB to exponential growth phase (OD600 ≈ 1.0) and then washed with PBS. For the survival curve, the bacteria were suspended in PBS to an OD600 of 0.7 and the bacterial suspension (100 μl; ∼1 × 107 CFU) was administered into eight female BALB/c mice (6 weeks old) via retro-orbital injection. The infected mice were observed for 14 days. The survival was compared by log rank (Mantel-Cox) test with Prism 5 (GraphPad). To analyze the bacterial survival, the bacterial cells were suspended in PBS to an OD600 of 0.4, and the bacterial suspension (100 μl; ∼0.5 × 107 CFU) was administered into eight female BALB/c mice (6 weeks old) via retro-orbital injection. At day 4 postinfection, all mice were euthanized, and kidneys and livers were harvested. For each set of kidneys harvested, one was used for bacterial CFU counting and the other for histology analysis. For bacterial CFU counting, one of the kidneys and the liver were ground, diluted, and spread on TSA blood agar. The plates were incubated at 37°C overnight; then colonies were enumerated. For histology analysis, the remaining kidney was fixed in 4% formalin (Sigma). Then paraffin embedding and hematoxylin and eosin (H&E) staining were performed as previously described (57).
Supplementary Material
ACKNOWLEDGMENTS
We thank Gary M. Dunny at the University of Minnesota for providing the pheromone indicator strain (E. faecalis OG1X carrying pAM373). S. aureus NRS384 (USA300-0114) was obtained through the Network on Antimicrobial Resistance in S. aureus (NARSA) program.
This study was supported by the National Natural Science Foundation of China (grants 81772139 and 81501803) to Q.L., Innovative Research Team of High-Level Local Universities in Shanghai (SSMU-ZLCX20180701) to M.L. and Q.L., the Cultivation Fund from Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University (grant PYIII-17-001) to Q.L., and the Research Enhancement Grant of Indiana University School of Medicine to T.B.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Kinch LN, Ginalski K, Grishin NV. 2006. Site-2 protease regulated intramembrane proteolysis: sequence homologs suggest an ancient signaling cascade. Protein Sci 15:84–93. doi: 10.1110/ps.051766506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider JS, Glickman MS. 2013. Function of site-2 proteases in bacteria and bacterial pathogens. Biochim Biophys Acta 1828:2808–2814. doi: 10.1016/j.bbamem.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudner DZ, Fawcett P, Losick R. 1999. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci U S A 96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krojer T, Pangerl K, Kurt J, Sawa J, Stingl C, Mechtler K, Huber R, Ehrmann M, Clausen T. 2008. Interplay of PDZ and protease domain of DegP ensures efficient elimination of misfolded proteins. Proc Natl Acad Sci U S A 105:7702–7707. doi: 10.1073/pnas.0803392105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanehara K, Ito K, Akiyama Y. 2003. YaeL proteolysis of RseA is controlled by the PDZ domain of YaeL and a Gln-rich region of RseA. EMBO J 22:6389–6398. doi: 10.1093/emboj/cdg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider JS, Sklar JG, Glickman MS. 2014. The Rip1 protease of Mycobacterium tuberculosis controls the SigD regulon. J Bacteriol 196:2638–2645. doi: 10.1128/JB.01537-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schobel S, Zellmeier S, Schumann W, Wiegert T. 2004. The Bacillus subtilis sigmaW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol Microbiol 52:1091–1105. doi: 10.1111/j.1365-2958.2004.04031.x. [DOI] [PubMed] [Google Scholar]
- 8.Xayarath B, Alonzo F III, Freitag NE. 2015. Identification of a peptide-pheromone that enhances Listeria monocytogenes escape from host cell vacuoles. PLoS Pathog 11:e1004707. doi: 10.1371/journal.ppat.1004707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denham EL, Ward PN, Leigh JA. 2008. Lipoprotein signal peptides are processed by Lsp and Eep of Streptococcus uberis. J Bacteriol 190:4641–4647. doi: 10.1128/JB.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito A, Hizukuri Y, Matsuo E, Chiba S, Mori H, Nishimura O, Ito K, Akiyama Y. 2011. Post-liberation cleavage of signal peptides is catalyzed by the site-2 protease (S2P) in bacteria. Proc Natl Acad Sci U S A 108:13740–13745. doi: 10.1073/pnas.1108376108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An FY, Sulavik MC, Clewell DB. 1999. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J Bacteriol 181:5915–5921. doi: 10.1128/JB.181.19.5915-5921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clewell DB, Francia MV, Flannagan SE, An FY. 2002. Enterococcal plasmid transfer: sex pheromones, transfer origins, relaxases, and the Staphylococcus aureus issue. Plasmid 48:193–201. doi: 10.1016/s0147-619x(02)00113-0. [DOI] [PubMed] [Google Scholar]
- 13.An FY, Clewell DB. 2002. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J Bacteriol 184:1880–1887. doi: 10.1128/jb.184.7.1880-1887.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandler JR, Flynn AR, Bryan EM, Dunny GM. 2005. Specific control of endogenous cCF10 pheromone by a conserved domain of the pCF10-encoded regulatory protein PrgY in Enterococcus faecalis. J Bacteriol 187:4830–4843. doi: 10.1128/JB.187.14.4830-4843.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunny GM, Berntsson RP. 2016. Enterococcal sex pheromones: evolutionary pathways to complex, two-signal systems. J Bacteriol 198:1556–1562. doi: 10.1128/JB.00128-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunny GM. 2013. Enterococcal sex pheromones: signaling, social behavior, and evolution. Annu Rev Genet 47:457–482. doi: 10.1146/annurev-genet-111212-133449. [DOI] [PubMed] [Google Scholar]
- 17.Frank KL, Guiton PS, Barnes AM, Manias DA, Chuang-Smith ON, Kohler PL, Spaulding AR, Hultgren SJ, Schlievert PM, Dunny GM. 2013. AhrC and Eep are biofilm infection-associated virulence factors in Enterococcus faecalis. Infect Immun 81:1696–1708. doi: 10.1128/IAI.01210-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Archer GL. 1998. Staphylococcus aureus: a well-armed pathogen. Clin Infect Dis 26:1179–1181. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- 19.Flannagan SE, Clewell DB. 2002. Identification and characterization of genes encoding sex pheromone cAM373 activity in Enterococcus faecalis and Staphylococcus aureus. Mol Microbiol 44:803–817. doi: 10.1046/j.1365-2958.2002.02922.x. [DOI] [PubMed] [Google Scholar]
- 20.Clewell DB, An FY, White BA, Gawron-Burke C. 1985. Sex pheromones and plasmid transfer in Streptococcus faecalis: a pheromone, cAM373, which is also excreted by Staphylococcus aureus. Basic Life Sci 30:489–503. doi: 10.1007/978-1-4613-2447-8_35. [DOI] [PubMed] [Google Scholar]
- 21.Manoil C, Beckwith J. 1986. A genetic approach to analyzing membrane protein topology. Science 233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 22.Dunny GM, Craig RA, Carron RL, Clewell DB. 1979. Plasmid transfer in Streptococcus faecalis: production of multiple sex pheromones by recipients. Plasmid 2:454–465. doi: 10.1016/0147-619x(79)90029-5. [DOI] [PubMed] [Google Scholar]
- 23.Oogai Y, Yamaguchi M, Kawada-Matsuo M, Sumitomo T, Kawabata S, Komatsuzawa H. 2016. Lysine and threonine biosynthesis from aspartate contributes to Staphylococcus aureus growth in calf serum. Appl Environ Microbiol 82:6150–6157. doi: 10.1128/AEM.01399-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ulrich M, Bastian M, Cramton SE, Ziegler K, Pragman AA, Bragonzi A, Memmi G, Wolz C, Schlievert PM, Cheung A, Doring G. 2007. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol Microbiol 65:1276–1287. doi: 10.1111/j.1365-2958.2007.05863.x. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi SD, DeLeo FR. 2013. Staphylococcus aureus protein A promotes immune suppression. mBio 4:e00764-13. doi: 10.1128/mBio.00764-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schallenberger MA, Niessen S, Shao C, Fowler BJ, Romesberg FE. 2012. Type I signal peptidase and protein secretion in Staphylococcus aureus. J Bacteriol 194:2677–2686. doi: 10.1128/JB.00064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker S, Frankel MB, Schneewind O, Missiakas D. 2014. Release of protein A from the cell wall of Staphylococcus aureus. Proc Natl Acad Sci U S A 111:1574–1579. doi: 10.1073/pnas.1317181111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicolas-Avila JA, Adrover JM, Hidalgo A. 2017. Neutrophils in homeostasis, immunity, and cancer. Immunity 46:15–28. doi: 10.1016/j.immuni.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 29.Mori M, Tanaka H, Sakagami Y, Isogai A, Fujino M, Kitada C, White BA, An FY, Clewell DB, Suzuki A. 1986. Isolation and structure of the Streptococcus faecalis sex pheromone, cAM373. FEBS Lett 206:69–72. doi: 10.1016/0014-5793(86)81342-4. [DOI] [PubMed] [Google Scholar]
- 30.Schilcher K, Caesar LK, Cech NB, Horswill AR. 2020. Processing, export, and identification of novel linear peptides from Staphylococcus aureus. mBio 11:e00112-20. doi: 10.1128/mBio.00112-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji G, Beavis R, Novick RP. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 32.Havarstein LS, Coomaraswamy G, Morrison DA. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci U S A 92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moubareck C, Meziane-Cherif D, Courvalin P, Périchon B. 2009. VanA-type Staphylococcus aureus strain VRSA-7 is partially dependent on vancomycin for growth. Antimicrob Agents Chemother 53:3657–3663. doi: 10.1128/AAC.00338-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang JC, LaSarre B, Jimenez JC, Aggarwal C, Federle MJ. 2011. Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS Pathog 7:e1002190. doi: 10.1371/journal.ppat.1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leonard BA, Podbielski A, Hedberg PJ, Dunny GM. 1996. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc Natl Acad Sci U S A 93:260–264. doi: 10.1073/pnas.93.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shetty J, Killgore GE, Tenover FC. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 37.Perichon B, Courvalin P. 2009. VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 53:4580–4587. doi: 10.1128/AAC.00346-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen MT, Gotz F. 2016. Lipoproteins of Gram-positive bacteria: key players in the immune response and virulence. Microbiol Mol Biol Rev 80:891–903. doi: 10.1128/MMBR.00028-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomez MI, Lee A, Reddy B, Muir A, Soong G, Pitt A, Cheung A, Prince A. 2004. Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat Med 10:842–848. doi: 10.1038/nm1079. [DOI] [PubMed] [Google Scholar]
- 40.Forsgren A. 1970. Significance of protein A production by staphylococci. Infect Immun 2:672–673. doi: 10.1128/IAI.2.5.672-673.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodyear CS, Silverman GJ. 2003. Death by a B cell superantigen: in vivo VH-targeted apoptotic supraclonal B cell deletion by a staphylococcal toxin. J Exp Med 197:1125–1139. doi: 10.1084/jem.20020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falugi F, Kim HK, Missiakas DM, Schneewind O. 2013. Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. mBio 4:e00575-13. doi: 10.1128/mBio.00575-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. 2009. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J 23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoppenbrouwers T, Sultan AR, Abraham TE, Lemmens-den Toom NA, Hansenová Maňásková S, van Cappellen WA, Houtsmuller AB, van Wamel WJB, de Maat MPM, van Neck JW. 2018. Staphylococcal protein A is a key factor in neutrophil extracellular traps formation. Front Immunol 9:165. doi: 10.3389/fimmu.2018.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim HK, Emolo C, DeDent AC, Falugi F, Missiakas DM, Schneewind O. 2012. Protein A-specific monoclonal antibodies and prevention of Staphylococcus aureus disease in mice. Infect Immun 80:3460–3470. doi: 10.1128/IAI.00230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 47.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 48.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 49.Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3:e00277-11. doi: 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 51.Payne MS, Jackson EN. 1991. Use of alkaline phosphatase fusions to study protein secretion in Bacillus subtilis. J Bacteriol 173:2278–2282. doi: 10.1128/jb.173.7.2278-2282.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Q, Cho H, Yeo WS, Bae T. 2015. The extracytoplasmic linker peptide of the sensor protein SaeS tunes the kinase activity required for staphylococcal virulence in response to host signals. PLoS Pathog 11:e1004799. doi: 10.1371/journal.ppat.1004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu M, Liu Y, Yu K, Liu X. 2014. Decreasing the amount of trypsin in in-gel digestion leads to diminished chemical noise and improved protein identifications. J Proteomics 109:16–25. doi: 10.1016/j.jprot.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 54.Jeong DW, Cho H, Lee H, Li C, Garza J, Fried M, Bae T. 2011. Identification of the P3 promoter and distinct roles of the two promoters of the SaeRS two-component system in Staphylococcus aureus. J Bacteriol 193:4672–4684. doi: 10.1128/JB.00353-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Q, Fan J, Niu C, Wang D, Wang J, Wang X, Villaruz AE, Li M, Otto M, Gao Q. 2011. The eukaryotic-type serine/threonine protein kinase Stk is required for biofilm formation and virulence in Staphylococcus epidermidis. PLoS One 6:e25380. doi: 10.1371/journal.pone.0025380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim JH, Chaurasia AK, Batool N, Ko KS, Kim KK. 2019. Alternative enzyme protection assay to overcome the drawbacks of the gentamicin protection assay for measuring entry and intracellular survival of staphylococci. Infect Immun 87:e00119-19. doi: 10.1128/IAI.00119-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y, Li N, Yu X, Huang K, Zheng T, Cheng X, Zeng S, Liu X. 2018. Hematoxylin and eosin staining of intact tissues via delipidation and ultrasound. Sci Rep 8:12259. doi: 10.1038/s41598-018-30755-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.