Abstract
Expression of olfactory receptors (ORs) in non-olfactory tissues has been widely reported over the last 20 years. Olfactory marker protein (OMP) is highly expressed in mature olfactory sensory neurons (mOSNs) of the olfactory epithelium. It is involved in the olfactory signal transduction pathway, which is mediated by well-conserved components, including ORs, olfactory G protein (Golf), and adenylyl cyclase 3 (AC3). OMP is widely expressed in non-olfactory tissues with an apparent preference for motile cells. We hypothesized that OMP is expressed in compartment-specific locations and co-localize with an OR, Golf, and AC3 in rat epididymal and human-ejaculated spermatozoa. We used immunocytochemistry to examine the expression patterns of OMP and OR6B2 (human OR, served as positive olfactory control) in experimentally induced modes of activation and determine whether there are any observable differences in proteins expression during the post-ejaculatory stages of spermatozoal functional maturation. We found that OMP was expressed in compartment-specific locations in human and rat spermatozoa. OMP was co-expressed with Golf and AC3 in rat spermatozoa and with OR6B2 in all three modes of activation (control, activated, and hyperactivated), and the mode of activation changed the co-expression pattern in acrosomal-reacted human spermatozoa. These observations suggest that OMP expression is a reliable indicator of OR-mediated chemoreception, may be used to identify ectopically expressed ORs, and could participate in second messenger signaling cascades that mediate fertility.
Keywords: chemoreception, fluorescence microscopy, interstitial tissue, olfactory epithelium, olfactory marker protein, reproduction, sperm
Introduction
Chemoreception is fundamental to all living organisms. It is the process by which a cell or an organism responds to a chemical substance in its environment through chemoreceptors often in the form of olfactory receptors (ORs).1,2 ORs have been shown to be expressed within and outside of the olfactory system.3 Within the olfactory system, they are expressed by olfactory sensory neurons (OSNs) where they detect and distinguish a diversity of volatile chemicals (odorants) of diverse structures.1,2,4–6 OR-mediated chemoreception begins when an odorant binds to an OR on the cilia of an OSN, initiating an intracellular second messenger pathway mediated by an olfactory G protein, Golf, and involving AC3. ORs also play a crucial role in axon guidance to the proper glomeruli by recognizing internal cues in the olfactory bulb (OB).5,7–9 Signals from the OSNs travel to the OB and higher centers where odor perceptions are processed.4,10–13 Each OR is expressed by only one of many hundreds of OR genes, and each OSN expresses only one type of OR.14–16 In spite of this specificity, individual ORs can recognize more than one odorant, and individual odorants can bind to more than one OR.5,17
ORs have also been found in non-olfactory tissues of different mammalian species.18–20 They are present in the tongue,21–23 spleen, pancreas,24,25 placenta,26 enterochromaffin cells of the gut,27 and spermatozoa of various species, including human, dog, mouse, hamster, and rat.28–32 Canine germ cells were shown to contain approximately 20 human OR transcripts.30 Mouse testes were found to express 66 OR genes.33 Human testes were shown to contain the highest number of ectopically expressed OR transcripts (at least 90 different OR transcripts) for multiple human tissues investigated.20
Olfactory marker protein (OMP) is notable because immunological studies have demonstrated it as phylogenetically conserved, widely distributed, and expressed in the olfactory system of virtually every vertebrate species, including humans.34–38 OMP characterizes mature olfactory sensory neurons (mOSNs) in the olfactory epithelium (OE), olfactory septal organ, and vomeronasal organ. In mOSNs, OMP is localized within cilia, dendrites, dendritic knobs (only in those with cilia expressing ORs), in the cytoplasm of cell bodies, and along axons targeting the OB.35,37,39 OMP is associated with the olfactory signal transduction cascade,40–46 is involved in clearing Ca2+ influx during olfactory transduction,47,48 may regulate neurogenesis in the olfactory system,49 and participates in axon targeting into the OB.50 OMP-knockout mice exhibit reduced odorant sensitivity, slower odorant response kinetics, and impaired odor discrimination.41–43,45,47,48,51 Expression of OMP is a key indicator for normal function in the olfactory system.45
In addition to the olfactory system, OMP is expressed in brain and spinal cord52 and in variety of non-olfactory tissues.18,53,54 OMP genes were detected in the liver, skeletal muscle, bladder, pancreas, stomach, duodenum, testes, spleen, heart, thymus, kidney, thyroid, and lung of mice, although conventional Western blot analysis using a specific OMP antibody failed to detect OMP expression.18 The more sensitive, high-resolution double immunoassay, however, showed different levels of expression in various tissues, with the highest levels of OMP expression occurring in skeletal muscle, heart, thymus, and thyroid, whereas the lowest levels of expression in liver, bladder, pancreas, stomach, duodenum, testes, spleen, and lung.18 Immunohistochemistry (IHC) confirmed the expression of OMP in five non-olfactory tissues, bladder, thyroid, thymus, heart, and testes,18 and double-labeling IHC using specific marker proteins showed that OMP was differentially expressed within specific cell types, including the interstitial cells of Cajal of the bladder, parafollicullar cells of the thyroid, medullary epithelial cells of the thymus, Leydig-like cells in the interstitial tissue (IT) of the testes, and in the heart. This widespread expression of OMP suggests a biological economy in the animal kingdom for detecting extracellular chemical cues and argues that OMP immunoreactivity might be a powerful indicator of non-olfactory OR-mediated chemosensing.18
We hypothesized that OMP would be expressed in human and rat spermatozoa in compartment-specific locations and that it would also be co-expressed with OR6B2, Golf, and AC3. The co-expression of these molecules in specific compartments of spermatozoa could indicate the presence of an intracellular signaling pathway that activates functional processes in the non-olfactory cells. We used IHC analysis and bright-field and fluorescent microscopy to identify OMP immunoreactivity in compartment-specific locations in human and rat spermatozoa under the different conditions of spermatozoal activation. These findings suggest a role for ectopically expressed ORs in spermatozoa that might have an impact on the fertilization process in mammals.
Materials and Methods
Animals
Adult male Wistar rats (n=16) were used in accordance with the NHMRC (National Health and Medical Research Council) Animal Experimentation Guidelines and the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (1990) and with the approval of the Animal Care and Experimentation Committee for the University of New South Wales (UNSW). Animal tissues and spermatozoa were obtained from the School of Optometry and Vision Science and from the Victor Chang Research Institute, UNSW.
Tissue Processing
Tissues of testes and olfactory turbinates containing OE and OBs were post-fixed (12–14 hr) in 10% neutral buffered formalin solution (Sigma-Aldrich Corp.; Sydney, Australia). Tissues were immersed overnight in 70% (v/v) ethanol and embedded in paraffin using standard embedding procedures. Tissues were sectioned at 5 μm thickness on a motorized microtome (Leica RM 2155; Leica Microsystems; Wetzlar, Germany), serially collected, and mounted in consecutive order on electrostatic slides (Menzel-Glaser; Braunschweig, Germany). Slides were randomly selected from each experimental animal from different points in the numbering spectrum. The exception was for experiments with negative control staining, for which adjacent slides were selected. Hematoxylin and eosin staining was performed to evaluate tissue structure and quality.
Rat Spermatozoa Collection and Preparation
Spermatozoa were obtained from the epididymis. The epididymis was separated from the testes, cut along its length, and placed in phosphate-buffered saline (PBS; D8537; Sigma-Aldrich Corp.). Spermatozoa were allowed to diffuse into PBS at room temperature for 10 min and then aliquoted into 10 to 20 µl batches, smeared, and air-dried on microscope slides (4951PLUS4; Menzel-Glaser). Spermatozoa preparations were then fixed by immersion in methanol (AR grade; Fronine; Sydney, Australia) at –20C for 10 min, then washed three times, and stored in PBS at 4C until staining.
Human Spermatozoa Collection and Preparation
Discarded anonymous samples for the current study were provided by consenting men attending the NSW Health Pathology–Andrology Laboratory at the Royal Hospital for Women, UNSW, for semen analysis. Clean samples were treated in accordance with the Declaration of Helsinki. Human ethics approval was not required because our study used only surplus anonymized clinical samples.
Samples were placed into a non-toxic specimen container and processed within 1 hr of collection. A total of 15 samples from 15 individual donors were analyzed according to the World Health Organization criteria.55 For semen analysis, only samples with normal sperm morphology and semen parameters were included in the study. Normal sperm morphology was considered to include a smooth, oval head, approximately 3.7–4.7 µm long, and 2.5–3.2 µm wide with a well-defined acrosomal area occupying 40–70% of the head; a middle piece, approximately 3.3–5.2 µm long and 0.6 µm wide; and a tail, approximately 45 µm long, without kinks or coils.55–57 Sperm concentration ranged from 42.0 × 106 to 68.9 × 106 ml–1 (mean, 57.4 × 106 ml–1) and sperm motility (grade a: progressive motility) from 36% to 65% (mean, 50.3%).55
Experimental Design to Test Functional Modes of Spermatozoa
Three functional modes of spermatozoa were prepared. Initially, a general procedure was used for all three modes of spermatozoa preparation. Following this, an additional procedure was used for each of the three modes.
Initial Procedure
Spermatozoa from 15 semen samples were separated from the seminal plasma by centrifugation using SAGE PureCeption Media (SAGE IVF Inc.; CooperSurgical; Trumbull, CT). In an 11 ml Nunc IVF centrifuge tube (137860; Thermo Fisher Scientific; Roskilde, Denmark), 2 ml of liquefied semen was layered in a single column over upper phase gradient ([40%, v/v, 2 ml]; silane-coated colloidal silica in HEPES-buffered human tubal fluid (HTF; ART-2040) and lower phase gradient ([80%, v/v, 2 ml]; ART-2080), and centrifuged for 18 min at 244 × g force at 30C. The seminal plasma and waste phase gradients were removed. The lower pellet of spermatozoa was removed and washed in a clean 11 ml Nunc tube by inversion in 10 ml Quinn’s Advantage Medium (QAM) with HEPES (ART-1024; SAGE IVF Inc.; CooperSurgical) supplemented with 10 mg/ml human serum albumin (HSA; ART-3003; SAGE IVF Inc.; CooperSurgical). The spermatozoa were then centrifuged for 8 min at 244 × g force at 30C. The wash solution was removed, and the remaining spermatozoa pellet resuspended in 0.5 ml in the same QAM wash solution.
Preparation of Mode 1: Control Spermatozoa
Aliquots (2 µl) of washed spermatozoa from all samples were placed inside a defined circle on a plain microscope slides (Menzel-Glaser) and the slides were labeled. Spermatozoa were air-dried and fixed with cold methanol (AR; Fronine) for 10 min at −20C.
Preparation of Mode 2: Activated (AC) Spermatozoa
The control spermatozoa from all samples were activated by incubating in the fresh QAM wash solution for 1.5 hr at 30C. On a plain microscope slide (Thermo Fisher Scientific), a circular area was defined into which 2 μl aliquots of the AC spermatozoa were placed. They were air-dried and fixed with cold methanol (AR grade; Fronine) for 10 min at −20C. The slides with fixed-spermatozoa were labeled and stored in PBS at 4C until immunostaining.
Preparation of Mode 3: Hyperactivated (HAC) Spermatozoa
Samples (0.5 ml) of control spermatozoa were placed in an 11 ml Nunc IVF centrifuge tube, and 0.5 ml of HAmax reagent was added. The HAmax reagent contained progesterone ([2 μg/ml]; P6149; Sigma-Aldrich Corp.), pentoxifylline ([7.2 mM]; P1784; Sigma-Aldrich Corp.) and HSA, and was dissolved in bicarbonate-buffered Earle’s Balanced Salt Solution (E2888; Sigma-Aldrich Corp.), a spermatozoa medium that supports human spermatozoa capacitation. The 11 ml Nunc tubes were then incubated for 1 hr at 37C in a CO2 incubator. On a plain microscope slide (Menzel-Glaser), a circular area was defined into which 2 μl aliquots of the HAC spermatozoa were placed. They were air-dried and fixed with cold methanol (AR grade; Fronine) for 10 min at −20C. The slides with fixed-spermatozoa were labeled and stored in PBS at 4C until immunostaining.
IHC
Immunoperoxidase (IP) Histochemistry
Paraffin sections (rat OE and testes) were dewaxed in xylene twice for 5 min, rehydrated in a graded series of alcohol dilutions (100% and 70% ethanol), and placed in water for 1 min. Sections were subjected to heat-induced antigen retrieval with 10 mM citrate buffer (CB; pH 6.0), using a microwave oven for 5 min. Sections were then rinsed three times for 5 min each in Tris buffer, pH 7.4. Endogenous peroxidase activity was blocked by exposing sections to 0.3% hydrogen peroxide (H2O2; Millipore Australia; Sydney, Australia) for 5 min. After rinsing in the buffer solution, sections were incubated for 1 hr at room temperature with primary antibody (Table 1). Sections were then rinsed three times in the buffer solution for 2 min each and left in a post-primary solution (DS 9800; Bond Polymer Refine; Leica Biosystems; Newcastle, UK) for 8 min. They were rinsed three times in the buffer solution and placed in polymer (DS 9800; Bond Polymer Refine; Leica Biosystems) for 8 min. Sections were then rinsed twice in the buffer solution for 2 min each and again in water once. Colorimetric visualization was performed using 3,3′-diaminobenzidine (DAB; DAKO liquid DAB+ Substrate Chromogen System; DAKO; Sydney, Australia). Reactions were stopped by washing slides in water. Sections were counterstained with hematoxylin for 5 min, and then rinsed once in water, once in the buffer solution, and again in water. They were dehydrated in four changes of 100% ethanol, cleared in xylene, and cover-slipped with Aqua mount (BDH Laboratories Supplies; Poole, UK).
Table 1.
Primary Antibodies and Their Specificity.
| Primary Antibody | Antibody Dilution |
Specificity | |||
|---|---|---|---|---|---|
| Rat |
Human |
||||
| OE | IT | Spermatozoa | |||
| OMP, goat antiserum (019-22291; Wako Pure Chemical Industries; Osaka, JP) | 1:1000IF1:25IP | 1:1000IF1:25IP | 1:800IF | 1:800IF | mOSNs |
| Golf, goat pAb (P-15) (sc-26763; Santa Cruz Biotechnology; Dallas, TX) | NA | NA | 1:250IF | NA | OSNs |
| AC3, rabbit pAb (C-20) (sc-588; Santa Cruz Biotechnology; Dallas, TX) | NA | NA | 1:250IF | NA | OSNs |
| OR6B2, rabbit pAb (NBP2-13711; Novus Biologicals; Littleton, CO) | NA | NA | NA | 1:100IF | OR6B2 |
| Calretinin, rabbit pAb (ab702; Abcam; Sydney, Australia) | NA | 1:400IF | NA | NA | Leydig cells |
Abbreviations: AC3, adenylyl cyclase 3; IT, interstitial tissue; IF, immunofluorescent histochemistry; IP, immunoperoxidase histochemistry; mOSNs, mature OSNs; OE, olfactory epithelium; OMP, olfactory marker protein; OR6B2, specific human olfactory receptor; OSNs, olfactory sensory neurons; NA, not applicable; pAb, polyclonal primary antibody.
Single-labeling Immunofluorescent (IF) Histochemistry
Paraffin sections (rat OE and testes) for IF histochemistry were dewaxed in Histo-Clear (National Diagnostic Products; Sydney, Australia), rehydrated in a descending series of alcohol dilutions (100%, 75%, 50%, H2O), and subjected to heat-induced antigen retrieval with CB. After cooling, sections were washed three times in PBS, and nonspecific staining was blocked with 10% normal donkey serum (NDS; D9663; Sigma-Aldrich Corp.) for 20 min at room temperature in a humidified chamber. Tissue sections were incubated in 2% NDS in PBS containing appropriate primary antibody (Table 1) and then placed in a humidified chamber for either 1 hr at room temperature or overnight at 4C. Sections were rinsed three times with PBS and incubated with appropriate Alexa secondary antibodies (Invitrogen Molecular Probes; Carlsbad, CA). Sections were left for 30 min at room temperature in the dark and then washed three times with PBS. Nuclei were stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) using an anti-fading fluorescence mounting medium (Vectashield Hard+Set Mounting Medium with DAPI; Vector Labs; Fluoroshield with DAPI; Sigma-Aldrich Corp.) and cover-slipped. Negative control slides were incubated with 1% normal serum (NS) without the primary antibody and subjected to the IHC protocol as described above.
Immunocytochemistry (ICC)
Single-labeling IF Cytochemistry
Fixed samples of human and rat spermatozoa were washed three times in PBS. Nonspecific staining was blocked with 10% NDS (Sigma-Aldrich Corp.) for 20 min at room temperature in a humidified chamber. Spermatozoa were incubated in 2% NDS in PBS containing appropriate primary antibody (Table 1) and then placed in a humidified chamber for overnight at 4C. The next morning, slides were rinsed three times with PBS and then incubated with appropriate Alexa secondary antibodies (Invitrogen Molecular Probes). Sections were left for 30 min at room temperature in the dark and then washed three times with PBS. Nuclei were stained with DAPI using an anti-fading fluorescence mounting media (Sigma-Aldrich Corp.). In the negative control slides, spermatozoa were incubated with 1% NS without the primary antibody and subjected to our ICC protocol, as described above.
Double-labeling IHC and ICC
Double-labeling staining was performed, using the same protocol as described for single-labeling staining, with the exception that two primary antibodies were mixed in PBS containing 2% NDS. The same method was used with the Alexa-labeled secondary antibodies (Invitrogen Molecular Probes).
Antibodies
The goat antiserum recognizing OMP, developed with rodent OMP as the immunogen, was used to identify mOSNs in the OE of rat, OMP expression in the IT of rat, and in spermatozoa of rat and human (019-22291; Wako Pure Chemical Industries; Osaka, Japan). OMP is a known marker for mOSNs, their axons, and terminals.34,36,52,58 The goat polyclonal primary antibody (pAb) recognizing Gαolf (Golf; P-15), developed against a peptide mapping near the N-terminus of Gαolf of human origin, was used to detect OSNs in the OE and Golf expression in the spermatozoa of rats (sc-26763; Santa Cruz Biotechnology; Dallas, TX) as recommended for detecting Gαolf in mice, rats, and humans.12,59–61 The rabbit pAb recognizing adenylyl cyclase III (AC3; C-20), developed against a peptide mapping at the C-terminus of AC3 of mouse, rat, and human origin, was used to detect OSNs in the OE and AC3expression in the spermatozoa of rats (sc-588; Santa Cruz Biotechnology). This pAb detected AC3 in mice, rats, and humans.11,62–64 The rabbit pAb OR6B2 (gene ID: 389090, gene symbol OR6B2), was developed against recombinant protein corresponding to amino acids ENVTKVSTFILVGLPTAPGLQYLL (NBP2-13711; Novus Biologicals; Littleton, CO). IHC data showed its specificity for OR6B2 in the human spermatozoa. OR6B2 protein was localized in two particular positions relevant to our study—on the equatorial segment of the head and from the annulus down the tail.65 The rabbit pAb recognizing calretinin (full-length protein, corresponding to calretinin) was used to identify Leydig cells in the IT of rat testes (ab702; Abcam; Sydney, Australia). Calretinin (CR), a 29-kDa neuronal calcium-binding protein, has been found in distinct subsets of neurons and fibers in the central and peripheral nervous systems.66–68 Expression of CR is found in all Leydig cells of the IT in human testes, and is a valuable marker for detecting Leydig cells in the testes.69,70 Primary antibodies used for IHC and ICC are summarized in Table 1.
Fluorescent secondary antibodies were Alexa Fluor 488 donkey anti-rabbit (DaR), Alexa Fluor 594 DaR, Alexa Fluor 488 donkey anti-goat (DaG), and Alexa Fluor 594 DaG (in dilution:1:500; Invitrogen Molecular Probes).
Digital Microscopy, Image Processing, and Photography
Bright-field and fluorescent images were viewed and analyzed using a motorized Axioplan 2ie microscope (Carl Zeiss; Gottingen, Germany). Images were captured with Axiocam HRc and MRm digital cameras using AxioVision microscopy software that allowed multichannel and Z stack image acquisition and processing. Stained slides of rat spermatozoa and slides of human spermatozoa from each of the three modes (control, AC and HAC) were examined using 40× (Zeiss; EC Plan-NEOFLUAR, 40×/0.75) and 63× (Zeiss; Plan-APOCHROMAT, 63×/1.4 oil DIC) objectives. Spermatozoa were randomly selected for analysis and photographed. Confocal Z stack image analysis was performed on control (Mode 1) and AC (Mode 2) spermatozoa. Deconvolution (constrained iterative algorithm) was performed on the fluorescence Z stacks to allow three-dimensional (3D) reconstructions and display. Leydig cells were also examined and photographed at 100× (Olympus Plan, 100×/1.25 oil) objective with Axioplan 2ie microscope (Carl Zeiss; Gottingen, Germany) and ProgRes C5 CCD 5.0 camera (Jenoptik; Jena, Germany). Stained tissues were scanned (Aperio ScanScope AT Slide Scanner; Leica Biosystems, Germany), viewed, and analyzed with Aperio ImageScope software. Representative images were selected and photographed. Stained slides were evaluated using light microscope, and the presence/absence of expression was assessed as “+” (staining observed) or “–” (no staining observed). Final images were imported into Adobe Photoshop (CS6; Adobe Systems; San Jose, CA) for adjusting brightness, contrast, and size.
Identification and Quantitative Analysis of CR-expressing and OMP-expressing Cells in IT
Double-labeling IHC with OMP and CR was performed on testes tissues from three different rats. Nuclei of the cells were counterstained with DAPI (blue fluorescence). Stained testes slides were inspected using the 40× objective lens. The randomly selected IT sections were magnified and photographed for cell counting using the 63× oil immersion lens. Images were opened in Adobe Photoshop (CS6; Adobe Systems). Each image was separated into three channels: red for OMP-expressing (OMP+) cells, green for CR-expressing (CR+) cells, and blue for nuclei (DAPI+). Images were then converted from color to black/white to emphasize cells, and were counted manually. To identify and count Leydig cells in the IT, we calculated the percentage of CR+ cells and cells that did not express CR (CR–) cells from the total number of DAPI-stained cell nuclei. Cell density of CR+ and CR– cells was expressed as a percentage. The chi-square test and Wald 95% confidence interval (CI) were applied for this calculation. To count OMP+ Leydig cells in the IT, we calculated the percentage of Leydig cells co-expressing CR and OMP (Double: CR+OMP+), percentage of Leydig cells expressing only OMP (OMP Single+), and percentage of Leydig cells expressing only CR (CR Single+) and called these percentage’s “cell density.” The chi-square test, Wald 95% CI, and Fisher’s exact (F) test were applied for these calculations.71–75 Values of p<0.05 were considered significant.
Results
Expression of OMP in Rat OE
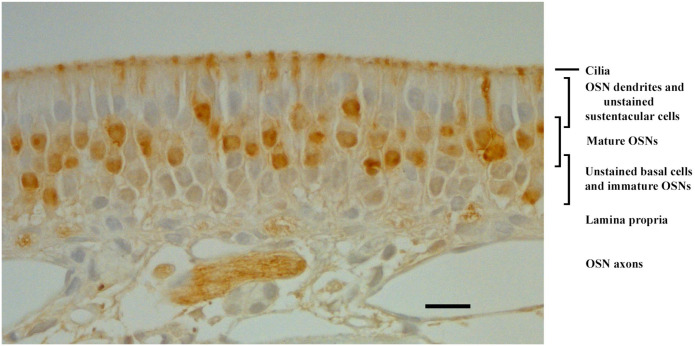
OMP was expressed in the mOSNs of the OE. The application of goat antiserum to OMP (Table 1) showed that OMP expression was restricted to the upper band of neuronal cell populations of the OE (Fig. 1). OMP specifically labeled mOSNs located above the lamina propria and above rows of OMP-negative (OMP–) cells at the base of the OE. The cytoplasm of mOSNs showed diffuse patterns of OMP expression, with occasional dense accumulations (Fig. 1). OMP was also expressed in dendrites of mOSNs and throughout most of the olfactory cilia. Intense OMP expression was observed along axons and within axon bundles (Fig. 1). No expression of OMP was observed in negative control slide (data not shown). We found that OMP was expressed in mOSNs in the rat OE, and this expression was used as a positive control for studies of OMP expression in non-olfactory tissues.
Figure 1.
Bright-field photomicrograph of OMP distribution in the rat OE as revealed by immunohistochemistry. The OMP brown reaction product is distributed in the somata of mature olfactory sensory neurons, in their dendrites and knobs, the cilia, and their axons. Sustentacular cells, basal cells, and immature OSNs are unlabeled. This pattern is as previously reported34,36,76 and thus serves as positive control for other rat tissues. Scale bar = 20 µm. Abbreviations: OMP, olfactory marker protein; OE, olfactory epithelium; OSNs, olfactory sensory neurons.
Expression of OMP in Rat Testes: IT
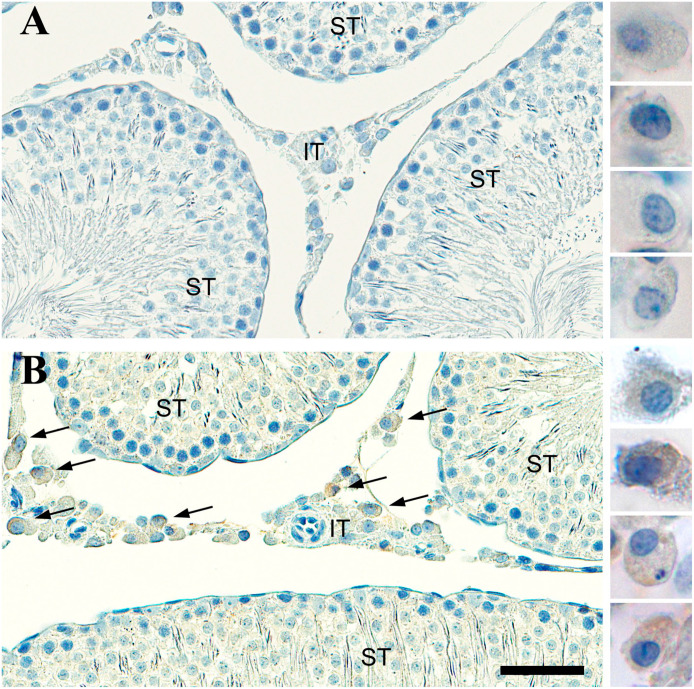
OMP was expressed in the Leydig cells of the IT. Sections of rat testes were immunostained with antibody to OMP and analyzed using digital microscopy at high magnification. We found that OMP was localized to specific cell type within the IT. No OMP expression was seen in cells of seminiferous tubules (Figs. 2B and 3B). We observed that immunostained OMP (OMP+) cells in the IT had distinct morphological features and were similar to those described for the Leydig cell-like population.77–80 The OMP+ Leydig-like cells were located singly or in clumps within the IT and generally grouped near blood and lymph capillaries (Fig. 2B). Cell bodies were polygonal or oval-round in shape and had prominent nucleoli in nuclei that tended to be eccentrically located (Fig. 2B). We observed diffuse patterns of OMP staining in the cytoplasm of Leydig-like cells at high magnification (Figs. 2B and 3B). No immunostaining of OMP was observed in negative control slides (Fig. 2A).
Figure 2.
Bright-field photomicrograph of olfactory marker protein (OMP) staining in the rat testes. (A) Control tissue containing seminiferous tubules (ST) and interstitial tissue (IT) is stained by cresyl violet but there is no immunostaining when the OMP antibody is left out. Leydig-like cells are shown on the right at higher magnification and are void of OMP reaction product. (B) Tissue stained by OMP antibodies reveal Leydig-like cells express OMP (brown) and these are only found in the IT. OMP+ Leydig-like cells are shown on the right at higher magnification. Scale bar = 50 µm for the tissue and 18 µm for the panel of individual cells.
Figure 3.
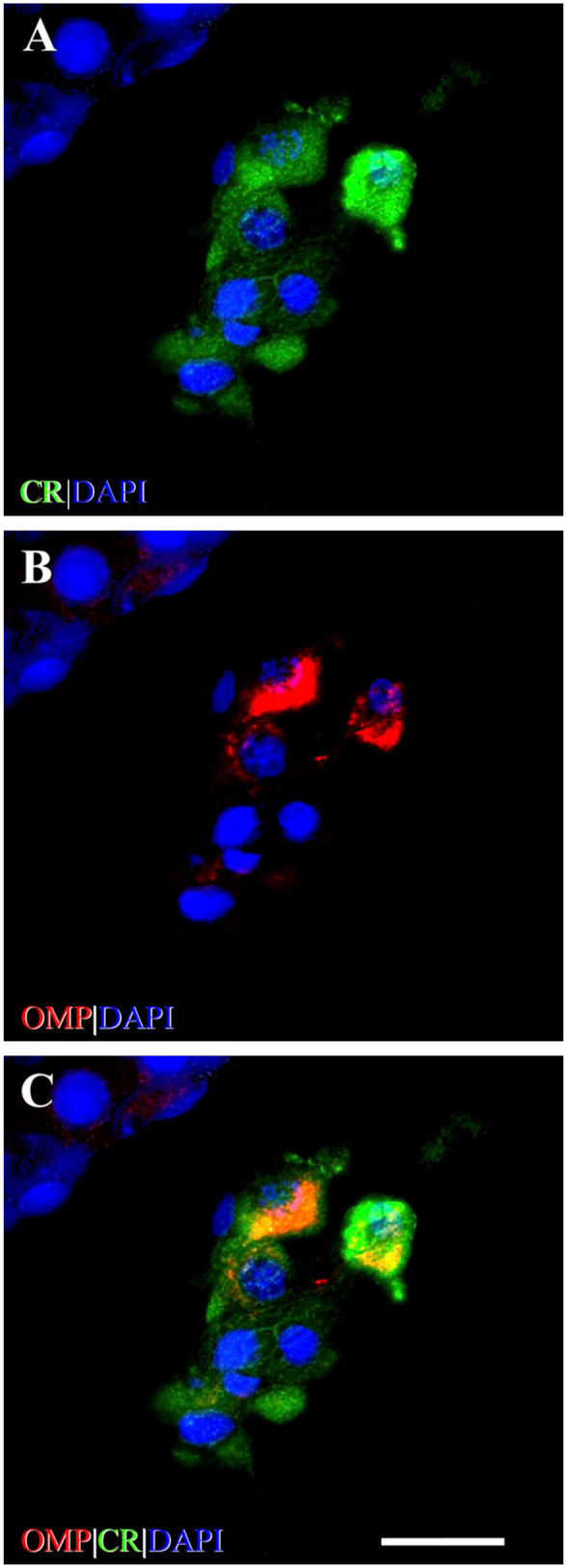
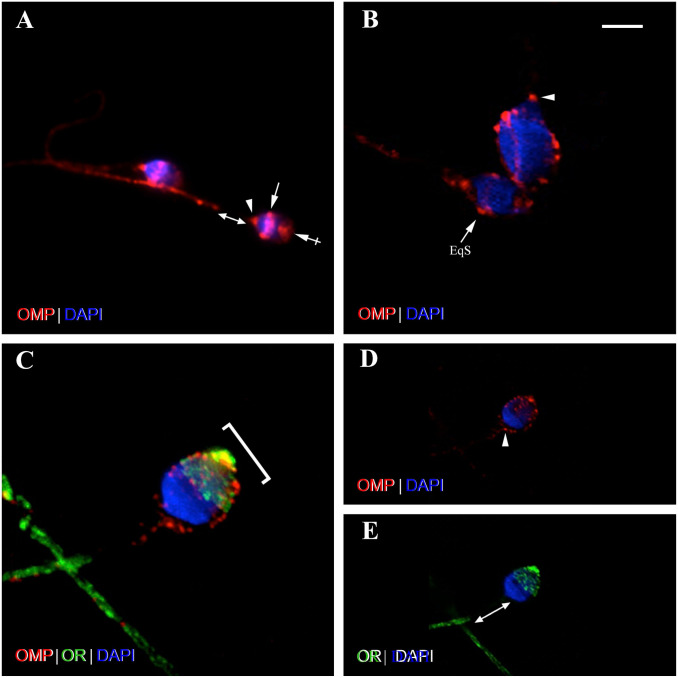
Identification of OMP+ staining of Leydig cells in the rat testes. (A) Photomicrograph of calretinin (CR) immunostaining (green) of cells of IT. CR is a marker for Leydig cells.69,70 The somata of Leydig cells are polygonal or round-oval in shape and have positively stained cytoplasm and prominent round nuclei. (B) Photomicrograph shows Leydig cells stained with OMP (red). OMP immunoreactivity varies in intensity between individual OMP+ cells but no OMP+ staining is observed in cells of the ST. (C) OMP is co-expressed with CR using double-labeling IHC as revealed by yellow-orange cytoplasm in merged image. CR immunoreactivity can be observed singly in some IT cells but all OMP+-stained cells are co-stained with CR. DAPI (blue) stains cell nuclei. Scale bar = 20 µm. Abbreviations: OMP, olfactory marker protein; IT, interstitial tissue; ST, seminiferous tubule; IHC, immunohistochemistry; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride.
Immunohistochemical techniques and quantitative analysis were performed to identify OMP+ cells in the rat IT. OMP+ cells in the IT were co-labeled using the antibody to CR (Table 1), a cell-specific marker for Leydig cells in human and rat testes.69,70 First, we identified the Leydig cell population and found that 79.13% (95% CI, 71.70–86.56) of all IT cells were immunostained by CR (CR+); other CR-negative (CR–) cells represent 20.87% (95% CI, 13.44–28.30, *p<0.001) (Figs. 3A and 4A). Then, double-labeling IHC showed that 81.32% (95% CI, 73.31–89.33) of Leydig cells were co-labeled by CR and OMP (CR+OMP+) with only 18.68% (95% CI, 10.67–26.69, *p<0.001) of cells expressing CR singly and no cells (0%; 95% CI, 0.0–1.05, *p<0.001) expressing OMP alone (Figs. 3C and 4B). Co-expression CR with OMP indicates that OMP was expressed in Leydig cells, and 81% of Leydig cells were OMP+ (Fig. 4B). These data demonstrated that OMP was expressed in Leydig cells of IT of rat testes.
Figure 4.
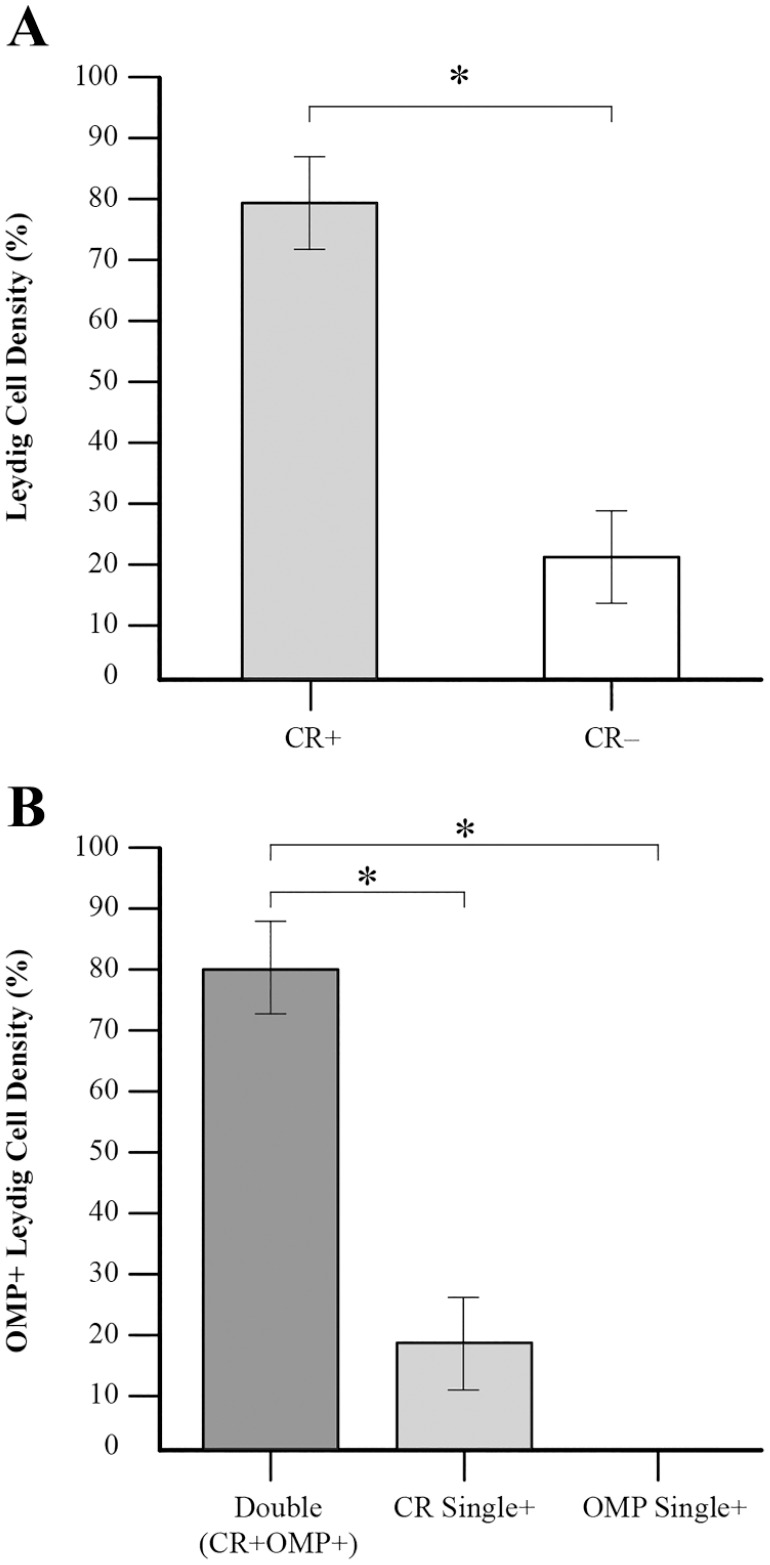
Graph of Leydig and OMP+ cells representation in the IT of the rat testes. (A) Leydig cell population, as defined by immunolabeling by CR (CR+), represent the majority—79% of all cells (n=115) in the IT of rat testes. The CR-negative (CR–) cells represent the remaining 21%. (B) 81% of the Leydig cell population (n=91) was OMP+ (Double: CR+OMP+). No cells are labeled by OMP alone (OMP Single+) and CR+ only cells (CR Single+) are represented 19% of the Leydig cell population. Statistical significance between groups is indicated by asterisks. CR+ vs CR– (χ2 test, *p<0.001); double vs CR single+ (χ2 test, *p<0.001); double vs OMP single+ (F test, *p<0.001). Abbreviations: OMP, olfactory marker protein CR, calretinin; IT, interstitial tissue.
OR-associated Signaling Molecules in Rat Spermatozoa
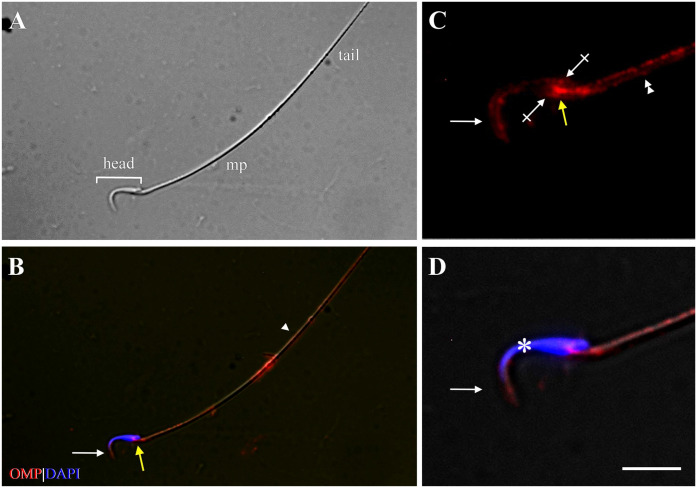
Expression of OMP, Golf, and AC3 was observed in spermatozoa collected from the rat epididymis with the use of IF cytochemistry. OMP was expressed on the head, connecting piece, and tail of rat spermatozoa (Fig. 5). The anatomic location (Fig. 5A) of OMP expression was determined by comparison with published images of rat spermatozoa.32,81–83 On the head, OMP was expressed on the acrosomal region (located on the apical part of the head) and on the equatorial segment. OMP was expressed prominently on the connecting piece where the centriole is located and in a discontinuous, punctate fashion along the plasma membrane of the tail. Strongest labeling was observed along the middle piece (Fig. 5B), where the mitochondria is situated.83 No expression of OMP was observed in negative control slides (data not shown).
Figure 5.
Expression of OMP showing compartment-specific distribution in the rat spermatozoa. (A) Phase-contrast photomicrograph shows the typical structure of rat spermatozoa: head (bracket), mp, and tail. (B) Immunofluorescence image shows expression of OMP (red) on the head (white arrow), cp (yellow arrow), and tail of spermatozoa, with strongest expression along the mp. The arrowhead marks the border of the mp and the tail. (C) High-power photomicrograph shows OMP expression on the acrosomal region (arrow) and EqS (crossed arrows) of the head, the cp (yellow arrow), and the mp (double arrowhead). (D) DAPI (blue) stains the spermatozoa nucleus (asterisk). Scale bar = 20 µm for low-power photomicrographs (A and B) and 8 µm for high-power photomicrographs of head region (C and D). Abbreviations: OMP, olfactory marker protein; mp, middle piece; cp, connecting piece; EqS, equatorial segment; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride.
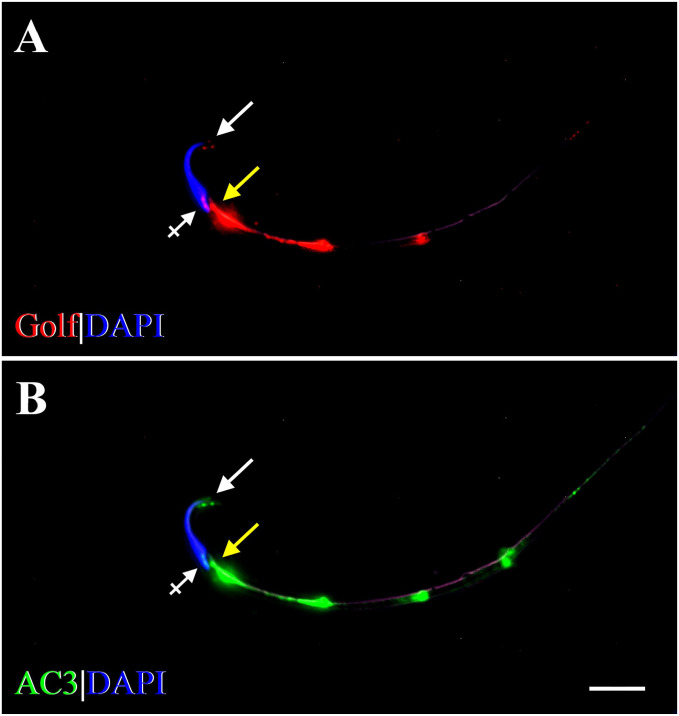
Double-labeling ICC with Golf and AC3 showed that they were co-expressed on the head, connecting piece, and tail of rat spermatozoa (Fig. 6). On the head, Golf and AC3 were expressed on the apical part of the acrosomal region and on the equatorial segment (Fig. 6A and B). Along the tail, Golf and AC3 were expressed in a dot-like, discontinuous fashion, with prominent expression occurring on the middle piece and gradually diminishing along the principal piece (Fig. 6A and B). These patterns of Golf and AC3 expression were similar to those of OMP expression on the head, connecting piece, and tail (Fig. 5B).
Figure 6.
Fluorescent photomicrographs showing compartment-specific expression of Golf and AC3 in the rat spermatozoa. (A) Expression of Golf (red) is seen on the acrosomal region (arrow) and EqS (crossed arrow) of the head, on the cp (yellow arrow), and along the tail in a discontinuous pattern. (B) Expression of AC3 (green) is seen in similar compartments: on the head (arrow, crossed arrow), on the cp (yellow arrow), and along the tail in a discontinuous pattern. Strong expression of Golf and AC3 is seen on the mp, gradually diminishing along the tail. DAPI (blue) stains spermatozoal nuclei. Scale bar = 10µm. Abbreviations: AC3, adenylyl cyclase 3; Golf, olfactory G protein; EqS, equatorial segment; cp, connecting piece; mp, middle piece; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride.
We found that OMP, Golf, and AC3 were expressed in the same compartment-specific locations in rat epididymal spermatozoa: on the head (acrosomal region and equatorial segment), connecting piece, and along the tail in a discontinuous pattern, with strongest expression occurring on the middle piece.
Expression of OMP in Human Spermatozoa
OMP was expressed in human-ejaculated spermatozoa. IF cytochemistry showed that OMP was expressed on the head, connecting piece, and tail of human spermatozoa (Fig. 7). On the head, OMP was expressed on the equatorial segment and the apical part of the acrosomal region. OMP expressed prominently on the equatorial segment—the membranous structure located in the middle of the head—that underlies the domain of the sperm that fuses with the oocyte membrane during fertilization.84–87 This expression was observed as ring-like staining around the head with dense accumulations on the periphery that often appeared as a pair of lateral blotches (Fig. 7B). OMP was also expressed, faintly, on the apical part of the acrosomal region of the head. This is a cap-like structure overlying the anterior half of the spermatozoa head (Fig. 7B) that is essential for fertilization. Agenesis of the acrosome produces infertility.88 OMP was prominently expressed on the connecting piece, where the centriole is located (Fig. 7B). Along the tail, OMP was expressed in discontinuous and punctate patterns along the entire length of tail, except for the middle piece (Fig. 7B and C). No OMP expression was observed in negative control slide (data not shown). Confocal Z stack image analysis showed that OMP was expressed along the equatorial segment and on the anterior part of the head (acrosomal region) in a dot-like pattern (Fig. 10A and B). OMP was prominently expressed on the connecting piece and in discontinuous and punctate pattern along the tail. However, the absence of OMP immunoreactivity on the middle piece is clearly visible (Fig. 10A).
Figure 7.
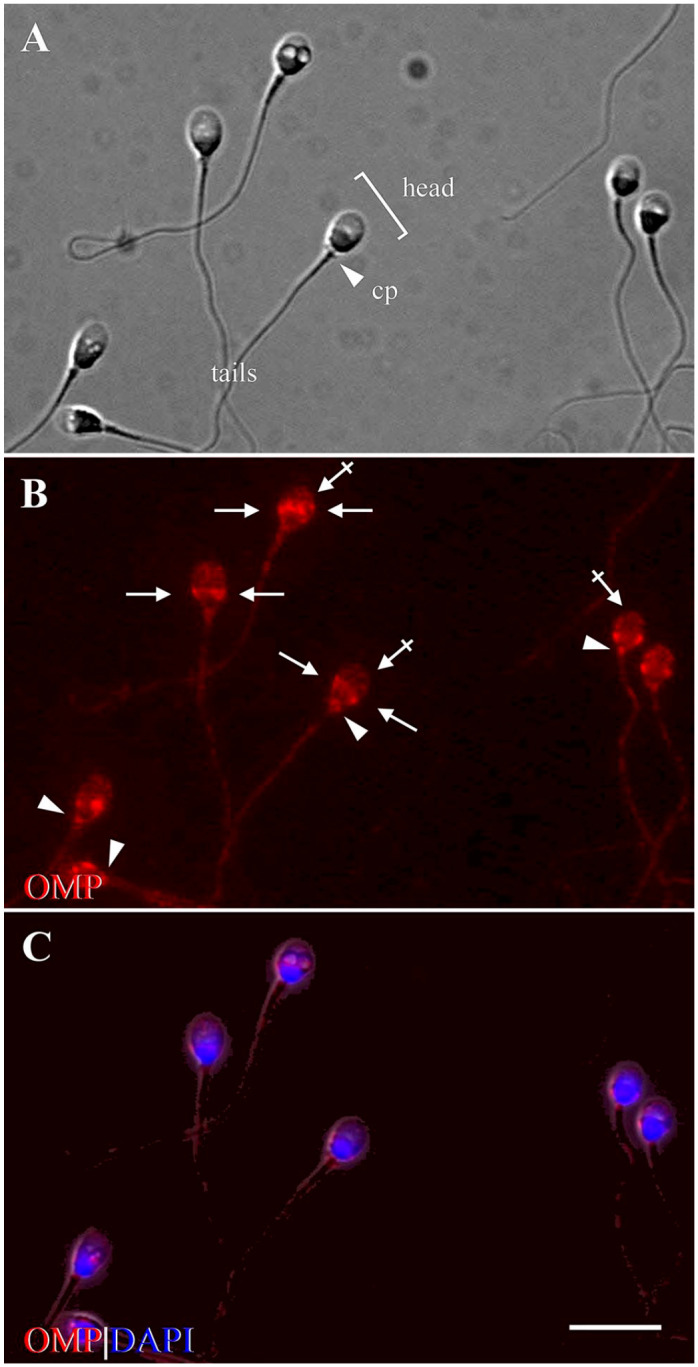
Photomicrographs show compartment-specific expression of OMP in ejaculated human spermatozoa (control). (A) Phase-contrast image shows the structure of human spermatozoa, consisting of the head (bracket), cp (arrowhead), and tail. (B) Immunofluorescence shows expression of OMP (red) on the apical part of the acrosomal region (crossed arrows) and along the EqS of the head (arrows), on the cp (arrowheads), and along the tail, except for the mp. (C) DAPI (blue) shows spermatozoal nuclei, surrounded by a thin rim of cytoplasm. Acrosome is seen above nuclei, on the anterior part of the head (crossed arrows in B). Scale bar = 10 µm. Abbreviations: OMP, olfactory marker protein; cp, connecting piece; EqS, equatorial segment; mp, middle piece; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride.
Figure 10.
Photomicrographs show confocal Z stack images of human spermatozoa in control (A and B) and activated (C to E) modes. Activation of spermatozoa affects OMP and OR6B2 expression pattern on the head. Photomicrographs (A and B) show control (unactivated) spermatozoa stained with OMP using single-labeling ICC. (A) Similar patterns of OMP expression (red) are seen in two control spermatozoa. OMP expression is seen on the EqS (arrow) and on the apical part of the acrosomal region (crossed arrow) of the head, on the cp (arrowhead), and, in a discontinuous pattern, along entire tail. No OMP expression is seen on the mp (double head arrow). (B) Photomicrograph shows heads of two control spermatozoa in two rotations. Dot-like expression of OMP (red) is seen on the EqS (arrow) and on the apical part of the acrosomal region of the head, and on the cp (arrowhead). Photomicrographs (C to E) show activated spermatozoon co-stained with OMP and OR6B2 using double-labeling ICC. Representative co-expression is shown in merged image C, and representative expression of each antibody is displayed in image D (OMP) and image E (OR6B2). (C) Three-dimensional image stack reconstruction of spermatozoon shows co-expression of OMP with OR6B2 (yellow) in dot-like patterns across the entire acrosomal region of the head (bracket), and along the tail. On the head, strongest co-expression is visible on the apical tip. (D) Single-image slice shows OMP (red) expression across the entire acrosomal region of the head in defined dot-like pattern, and along the tail. OMP expression is seen on the cp (arrowhead), and a few spots of OMP staining are seen in near proximity. (E) Single-image slice shows OR6B2 (OR, green) expression across the entire acrosomal region of the head in a dot-like pattern, and along the tail. No OR6B2 expression is visible on the cp or mp (double head arrow). DAPI (blue) stains spermatozoa nuclei. Scale bar = 5 µm for B, C and 10µm for A, D, E. Abbreviations: OMP, olfactory marker protein; OR, olfactory receptor (OR6B2); ICC, immunocytochemistry; EqS, equatorial segment; cp, connecting piece; mp, middle piece; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride.
We observed that OMP expression in human spermatozoa took place in compartment-specific locations that were similar to those in which OMP was expressed in rats. The main difference in expression locations was observed on the middle piece. In human spermatozoa, we did not observe OMP expression on the middle piece; however, in rat spermatozoa, OMP expression was prominent on the middle piece together with Golf and AC3.
Co-expression of OMP With OR6B2 in Human Spermatozoa: Effect of Functional Modes (AC, HAC) on Expression Pattern
We performed double-labeling ICC to test whether OMP and OR6B2 would co-express and whether modes of spermatozoal activation would affect this co-expression pattern. In a previous study on human spermatozoa, OR6B2 was the only OR to express in the equatorial segment of the head—where it express strongly—and in the principal piece of the tail.65 Because the OMP expression was observed on the equatorial segment of the head, the connecting piece, and along the tail of control spermatozoa, we used OR6B2 as our positive olfactory control. We found that OMP co-expressed with OR6B2 in control, AC, and HAC human spermatozoa in compartment-specific locations. In control spermatozoa, OMP and OR6B2 co-expressed on the head and along the tail, except for the middle piece (Fig. 8A). On the head, co-expression occurred on the equatorial segment of all control spermatozoa (Fig. 8A). No co-expression occurred on the apical part of the acrosomal region of the head or on the connecting piece, where single OMP expression was observed (Fig. 8A and B). OR6B2 was expressed on the equatorial segment of the head and along the tail, but not on the middle piece (Fig. 8C), and this expression pattern was consistent with that reported previously.65
Figure 8.
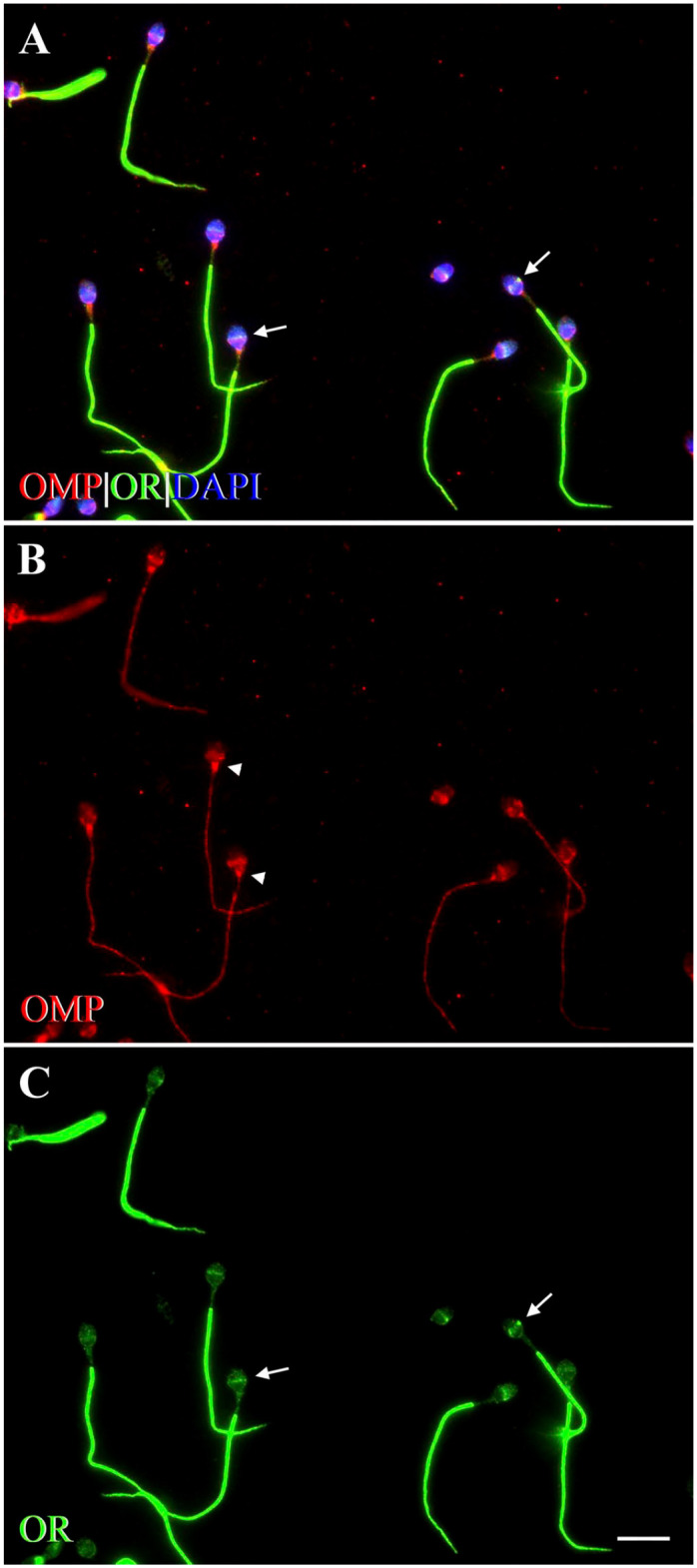
Fluorescent photomicrographs showing co-expression of OMP with OR6B2 in control human spermatozoa. OR6B2, a marker for a human OR,65 was co-labeled with OMP using double-labeling ICC. (A) OMP co-expresses with OR6B2 (yellow) on the EqS (arrows) of the head, and along the tail, except for the mp. DAPI (blue) stains spermatozoal nuclei. (B) Immunofluorescence shows expression of OMP (red) on the head, cp (arrowheads), and along the tail. On the head, expression of OMP is seen prominently on the EqS and faintly on the apical part of acrosomal region. OMP is also expressed in a discontinuous and punctate pattern along the tail, except for the mp. (C) Expression of OR6B2 (OR, green) is seen on the EqS of the head (arrows) and evenly along the tail, but not on the cp or mp. Scale bar = 10µm. Abbreviations: OMP, olfactory marker protein; OR, olfactory receptor (OR6B2); ICC, immunocytochemistry; EqS, equatorial segment; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride; cp, connecting piece; mp, middle piece.
In AC spermatozoa, OMP and OR6B2 co-expressed on the head and tail, except for the middle piece (Fig. 9A). In the majority of AC spermatozoa, co-expression occurred on the equatorial segment of the head and along the tail in a dot-like pattern, with OR6B2 expressing more strongly on the tail and OMP expressing more strongly on the equatorial segment (Fig. 9A to C). These patterns of co-expression were similar to those observed in control spermatozoa (Fig. 8A). In some AC spermatozoa, we observed prominent OMP and OR6B2 co-expression across the entire acrosomal region of the head (acrosomal-expressed AC spermatozoa), rather than on the equatorial segment (Fig. 9A). In these acrosomal-expressed AC spermatozoa, we also observed co-expression of OMP with OR6B2 along the tail. However, the intensity of OMP expression was less than that observed in controls, with no changes in intensity of OR6B2 expression along the tail (Fig. 9B and C). Confocal Z stack image analysis with 3D reconstruction of an acrosomal-expressed AC spermatozoon showed that OMP and OR6B2 prominently co-expressed across the entire acrosomal region of the head. This co-expression also occurred along the tail (Fig. 10C). No OR6B2 expression was observed on the connecting and middle piece (Fig. 10E). OMP expressed singly and densely on the connecting piece and in a few spots along the tail in close proximity to the connecting piece (Fig. 10D).
Figure 9.
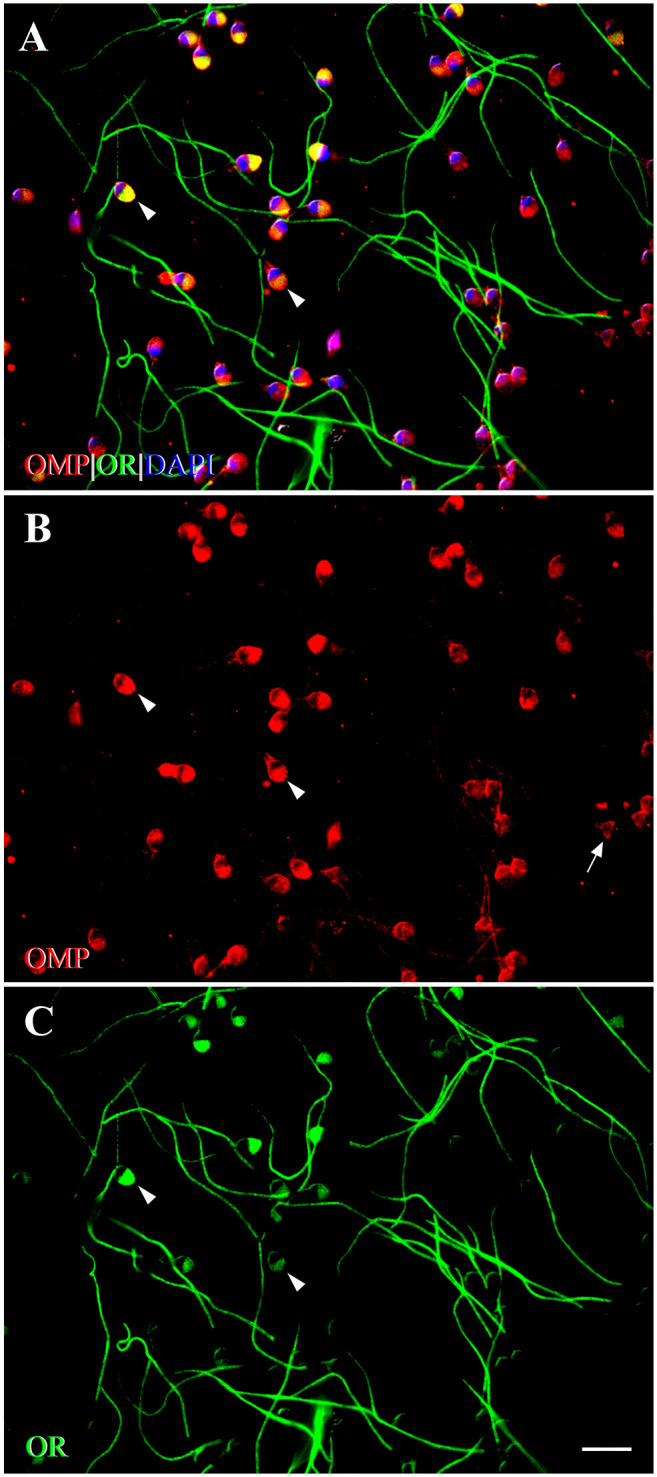
Fluorescent photomicrographs showing co-expression of OMP with OR6B2 in activated human spermatozoa using double-labeling ICC. (A) OMP co-expresses with OR6B2 (yellow) on the head and tail, but not on the mp. On the head, prominent co-expression is seen across the entire acrosomal region (arrowheads). DAPI (blue) stains spermatozoal nuclei. (B) Expression of OMP (red) is observed on the head, the cp, and along the tail. In some spermatozoa, OMP expression is seen across the entire acrosomal region (arrowheads). In other spermatozoa, OMP expression is seen primarily on the EqS (arrow). OMP is expressed along the tail in a discontinuous pattern. (C) Expression of OR6B2 (OR, green) is seen on the head (arrowheads), and tail in some spermatozoa, whereas in other spermatozoa, OR6B2 expression is seen only along the tail. No single expression of OMP or OR6B2 is seen on the mp. The pattern of staining in activated spermatozoa is distinct from that of control (unactivated) spermatozoa. Scale bar = 10µm. Abbreviations: OMP, olfactory marker protein; OR, olfactory receptor (OR6B2); ICC, immunocytochemistry; mp, middle piece; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride; cp, connecting piece; EqS, equatorial segment.
In HAC spermatozoa, we observed that OMP and OR6B2 co-expressed on the head and tail, except for the middle piece. On the head, we observed prominent co-expression only in a single location—across the entire acrosomal region of all HAC spermatozoa, and this pattern of co-expression was identical to the pattern of OMP and OR6B2 co-expression observed in acrosomal-expressed AC spermatozoa, shown in Fig. 10C.
We found that the co-expression of OMP with OR6B2 in control, AC, and HAC spermatozoa occurred in compartment-specific locations on the head and tail, but not on the middle piece. Activation and hyperactivation triggered a change in expression location of OMP and OR6B2 from the equatorial segment to the acrosomal region of the head (Fig. 10A–E).
Discussion
Fertilization remains a complex biological process with much still to be learned.89–91 Spermatozoa require chemosensory properties to respond to chemical cues as they pass through the epididymis and female genital tract and to communicate with the oocyte before and during fertilization.65,92–95 The connection between olfaction and reproduction in mating behavior is well known, so it seems remarkable that at a cellular level, an analogous chemical system seems involved in triggering sperm to migrate, identify, depolarize, and activate its target.96,97 Many signaling molecules such as ORs, G proteins, isoforms of adenylyl cyclase, protein kinases (GRK3/βARK2), β-arrestin2, β1 isoform of phospholipase C (PLC), inositol 1,4,5-trisphosphate (IP3), CNGA3, and a CNGA2 subunit of a cyclic nucleotide-gated (CNG) channel are conserved between the olfactory and reproductive systems.32,64,98–108 Given that only a small fraction of spermatozoa succeed in fertilizing an egg, there must be strong selective pressure for each spermatozoon to be equipped with a sensitive guidance system. Our observations are relevant to understanding this process in reproduction.
Despite intensive research, the identification of ORs and OR-associated proteins in non-olfactory tissues has been difficult. First, non-olfactory tissues express lower levels of OR transcripts than olfactory tissue.19 Second, there is a dearth of high-quality, commercially available OR antibodies due to the high homology between ORs (40–90%).109 Third, less than 100 OR ligands have been isolated.110–113 OMP is a protein that is intimately involved in olfactory signal transduction and is highly expressed in mOSNs.34,45,48,51,114 Ectopic expression of OMP has been found in some non-olfactory tissues of mice, suggesting that it could play a role in chemosensing.18,19 We confirmed and extended this finding in rats where OMP was expressed not only in mOSNs of the olfactory system, but also in Leydig cells of the reproductive system. These cells are involved in the production of testicular androgen, which is important for the paracrine regulation of spermatogenesis within the testes and for a multitude of endocrine effects, outside the testes.80 We applied OMP immunostaining as a surrogate representative of natural ORs and observed its relationship with Golf and AC3 in rat spermatozoa that indicate the presence of ORs and an OR-like signaling pathway.20 This expands knowledge about the ectopically expressed ORs and plays an important role in understanding the chemosensing in non-olfactory systems. Moreover, we co-localized OMP with OR6B2 which has been shown to be part of an olfactory phenotype with odorant’s perceived intensity and pleasantness.115 The OR6B2 gene is highly expressed in human trigeminal ganglia (TG), dorsal root ganglia (DRG), and neuronal retina, all of which perform sensory functions. The OR6B2 antibody detects the corresponding OR protein that is thought to be chemosensory in nature. OR6B2 stains satellite glial cells that envelop and form a discrete anatomical unit with somatosensory neurons—sensors for various innocuous and noxious physical and chemical stimuli—of the TG and DRG.20,116–118 These diverse observations are consistent with OMP being a reliable marker for OR proteins.
OMP Expression in Human and Rat Spermatozoa
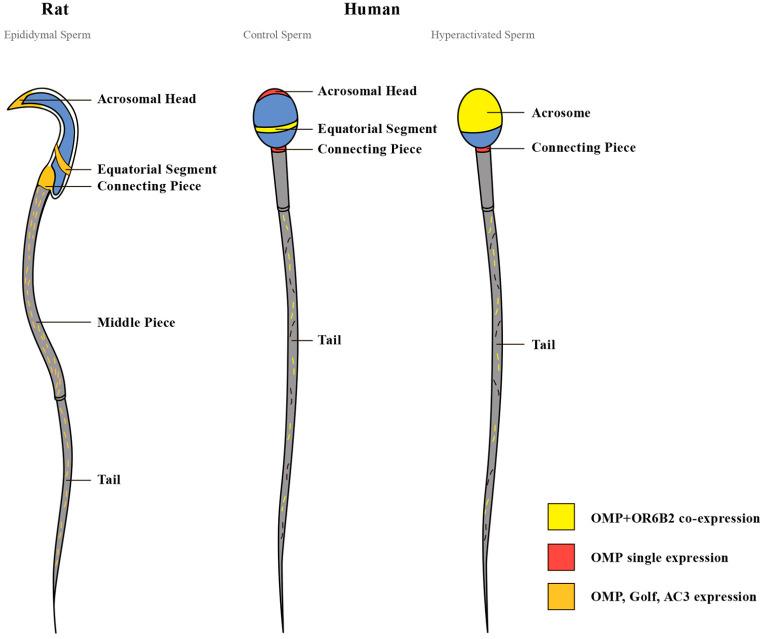
Our study is the first to demonstrate OMP expression in ejaculated human spermatozoa. OMP was expressed on the equatorial segment and the anterior acrosomal region of the head, on the connecting piece, and along the tail, except for the middle piece. We also showed OMP expression in rat epididymal spermatozoa. OMP was expressed on the equatorial segment and the acrosomal region of the head, on the connecting piece, and along the tail. The strongest OMP expression was observed on the middle piece. Expression of OMP on the tail of both human and rat spermatozoa occurred in a discontinuous punctate pattern. A clear difference in the pattern of OMP expression between human and rat spermatozoa was observed on the middle piece of the tail. In rats, OMP was expressed prominently together with Golf and AC3. In humans, however, no OMP was expressed on the middle piece (Fig. 11). The conserved molecular presence but variable distribution across species merits further consideration. Species-specific difference in OMP expression on the middle piece of the tail may be explained in several ways. The differences may be due to the maturational status of the spermatozoa studied. Whereas the human spermatozoa in our study were fully mature, rat spermatozoa were collected from different parts of the epididymis and were undoubtedly in different stages of maturation. Additional studies of ejaculated rat spermatozoa that mapped epididymal location could clarify the pattern of tail staining as it relates to maturational status and need to be conducted. Another explanation for the species-specific differences in OMP expression on the middle piece may be due simply to dissimilarity in the morphology of human and rat spermatozoa. In many species, including rats, cytoplasmic droplets (CDs) are shed from spermatozoa after ejaculation. However, in humans, CDs remain attached to spermatozoa, where they have a role in motility development during spermatozoa epididymal maturation, and in spermatozoa volume adaptation during movement through the female reproductive tract.119–122 Differences in OMP expression may reflect the divergent challenges that spermatozoa face as they negotiate species-specific variations in the female genital tracts across mammals. Species-specific differences have been already observed in the expression of ion channels and signaling molecules in the spermatozoa between humans and rodents.123–126
Figure 11.
OMP, OR6B2, Golf, and AC3 are localized in compartment-specific locations in human and rat spermatozoa. In rat spermatozoa, OMP, Golf, and AC3 are simultaneously expressed on the head (acrosomal region and equatorial segment), the connecting piece, and along the tail, with strongest expression on the middle piece. In control human spermatozoa, OMP co-expresses with OR6B2 on the head (equatorial segment), and along the tail, except for the middle piece. In the hyperactivated spermatozoa, OMP co-expresses with OR6B2 on the head (acrosome), and along the tail, except for the middle piece. In control and hyperactivated spermatozoa, OMP is singly expressed on the connecting piece. A clear difference in the pattern of OMP expression between human (both control and hyperactivated) and rat spermatozoa is observed on the middle piece. The differences in the pattern of OMP and OR6B2 co-expression between the control and hyperactivated human spermatozoa are observed on the head, with no changes of OMP single expression on the connecting piece. Abbreviations: OMP, olfactory marker protein; OR, olfactory receptor (OR6B2); AC3, adenylyl cyclase 3; Golf, olfactory G protein.
We found that OMP was expressed in compartment-specific locations in both human-ejaculated and rat epididymal spermatozoa. Such compartment-specific expression suggests that OMP may perform a number of different physiological processes. For example, expression of OMP on the head suggests the existence of a guidance mechanism to direct spermatozoa to the oocyte. Expression of OMP on the equatorial segment suggests that OMP may be involved in sperm–egg binding and fusion during fertilization.84,127 Expression of OMP on the acrosomal region of the head suggests that OMP may be involved in exocytotic release of acrosomal enzymes that help the sperm penetrate the zona pellucida of the egg.128,129 Expression of OMP on the head and on the tail could also contribute to spermatozoal development and maturation by recognizing chemicals, such as hormones, proteins, or lipids in the luminal environment as spermatozoa pass through the epididymis. The guided and timed movement of spermatozoa through the epididymis may involve epididymosomes—small membrane–bound vesicles released by principal epithelial cells and located in the luminal fluid of the epididymis in humans, mice, sheep, rats, hamsters, and bulls.130–138 It is thought that they may deliver a broad range of enzymes, proteins, chaperons, cytokines, and small non-coding RNA.139–141 The content of epididymosomes varies from one epididymal segment to another and is selectively transferred to spermatozoa to contribute to sperm function, protection, and maturation.130,142
Co-expression of OMP With an OR (OR6B2), Golf, and AC3 in Spermatozoa
We examined the co-expression of OMP with OR6B2 in human spermatozoa and of OMP with Golf and AC3 in rat spermatozoa. In all control human-ejaculated spermatozoa, OMP strongly co-expressed with OR6B2 in compartment-specific locations—on the head, and along the tail, except for the middle piece. In the rat epididymal spermatozoa, OMP, Golf, and AC3 were localized to similar compartment-specific locations: on the head, connecting piece, and along the tail, with strong expression on the middle piece. OMP co-localizes with OR6B2 in human spermatozoa in a pattern resembling that observed for OMP, Golf, and AC3 in rats with the exception of the middle piece.
The co-expression in spermatozoa of OMP with olfactory-signaling molecules, such as ORs, Golf, and AC3, implies a chemosensing role for OMP in the reproductive system that is similar to that in the olfactory system. Human and rat spermatozoa might use these molecules as an intracellular signaling pathway resembling that described in the bladder and thyroid of mice, suggesting a broad application of OR-mediated chemosensory signaling in the body.18
Co-expression OMP With OR6B2 in Control, AC, and HAC Human Spermatozoa
The co-expression of OMP with OR6B2 in control, AC, and HAC human spermatozoa was consistent with other data that OMP marked ORs in both olfactory and non-olfactory systems.18 This shared labeling was generally replicated in spatial location and for different functional modes of spermatozoa for both markers. Two types of co-expression were observed on the head of AC spermatozoa. In the majority of AC spermatozoa, OMP co-expressed with OR6B2 on the equatorial segment of the head, whereas, in a smaller proportion, OMP and OR6B2 co-expressed prominently across the entire acrosomal region of the head. The different co-expression patterns on the head may be due to the acrosomal status of the spermatozoa, especially as co-expression on the acrosomal region occurred only in acrosome-reacted spermatozoa.103,143 In HAC spermatozoa, OMP strongly co-expressed with OR6B2 only across the acrosomal region of the head. Whereas this type of co-expression occurred only in a small number of AC spermatozoa, it occurred in all HAC spermatozoa. This result may be because all HAC spermatozoa were acrosomal-reacted due to the presence of progesterone and pentoxifylline in HAmax reagent (Fig. 11). It has been reported that progesterone, a steroid hormone released post-ovulation by the corpus luteum, forms a gradient along the cumulus and beyond that may attract the spermatozoa.144,145 Progesterone also induces robust Ca2+ influx into human spermatozoa,146,147 triggers sperm hyperactivation, and initiates the acrosomal reaction—the preparative processes of spermatozoa essential for fertilization.125,144,148,149 Pentoxifylline has been found to enhance spermatozoa motility and induce hyperactivation and acrosome reaction in spermatozoa.150,151
OMP did not co-express with OR6B2 in only one location: the connecting piece of control, AC, and HAC human spermatozoa. This finding was consistent with a previous study that found that a small number of OMP+ cells did not co-express with ORs in the bladder and thyroid of mice.18 Although we found that OMP did not co-express with OR6B2 in the connecting piece, OMP may co-express with other ORs, such as OR3A2, OR2H1/2, OR10J1, or OR51E2,65 in this location, and this could be the subject of future investigations.
In conclusion, our study demonstrates that OMP is expressed in specific cells of the reproductive system in humans and rats. OMP co-expressed with the unique OR (OR6B2), Golf, and AC3 in compartment-specific locations in spermatozoa, suggesting that OMP expression is a reliable indicator of OR-mediated chemoreception in human and rat spermatozoa. Co-expression differences in the location and distribution of OMP and OR6B2 are evident with respect to activation and hyperactivation of spermatozoa and may reflect signaling processes involved in the different stages of fertilization. Further studies involving endogenous ligands from oocyte cumulus cells and follicular fluid are needed to confirm these associations. These data emphasize the significance of determining individual ORs and their specific ligands for advancing our knowledge of chemoreception. They also argue for human studies using a large cohort of fertile and infertile males to determine differences in expression patterns of olfactory-signaling proteins (ORs, OMP, Golf, and AC3). The resulting knowledge could be useful in developing clinical strategies for combating infertility.
Acknowledgments
The authors thank the following people for invaluable technical assistance and/or discussions about the data: Drs Nesli Avgan, Anne Cunningham, Harleen Kaur, Stephanie Kong, Michael Muniak, Roland Stocker and Kiera Grierson, and Lyn McLean. Special thanks to Arne Muller (Carl ZEISS Australia and New Zealand) for assistance with setting up the Axioplan 2ie microscope system for high-resolution fluorescence imaging.
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: The project was conceived by YM and DKR. YM performed experiments and analyzed data under the supervision of DKR. YM and DKR wrote the first draft of the paper and prepared all the figures. WLL and CN afforded access to human samples and provided critical discussions throughout the project. All authors read, edited, and approved the final manuscript before submission.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was jointly supported by an Australian Postgraduate Award, a Brain Top-Up scholarship, funding from the School of Women’s and Children’s Health, an NHMRC (National Health and Medical Research Council) grant 1080652, as well as donations from Alan and Lynne Rydge, the Walker Family Foundation, Haydn and Sue Daw, and Charlene and Graham Bradley.
Contributor Information
Yuliya Makeyeva, Garvan Institute of Medical Research, Royal Hospital for Women, Sydney, NSW, Australia; Westfield Research Laboratories, School of Women’s and Children’s Health, Royal Hospital for Women, Sydney, NSW, Australia.
Christopher Nicol, UNSW Sydney, Sydney, NSW, Australia, and Andrology Laboratory, NSW Health Pathology, Royal Hospital for Women, Sydney, NSW, Australia.
William L. Ledger, Fertility & Research Centre, Royal Hospital for Women, Sydney, NSW, Australia
David K. Ryugo, Garvan Institute of Medical Research, Royal Hospital for Women, Sydney, NSW, Australia School of Medical Sciences, UNSW, Royal Hospital for Women, Sydney, NSW, Australia; Department of Otolaryngology, Head, Neck & Skull Base Surgery, St. Vincent’s Hospital, Royal Hospital for Women, Sydney, NSW, Australia.
Literature Cited
- 1. Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65(1):175–87. [DOI] [PubMed] [Google Scholar]
- 2. Shepherd G. Discrimination of molecular signals by the olfactory receptor neuron. Neuron. 1994;13(4):771–90. [DOI] [PubMed] [Google Scholar]
- 3. Giorgi F, Maggio R, Bruni L. Are olfactory receptors really olfactive? Biosemiotics. 2011;4(3):331–47. [Google Scholar]
- 4. Buck L. Information coding in the vertebrate olfactory system. Annu Rev Neurosci. 1996;19(1):517–44. [DOI] [PubMed] [Google Scholar]
- 5. Malnic B, Hirono J, Sato T, Buck L. Combinatorial receptor codes for odors. Cell. 1999;96(5):713–23. [DOI] [PubMed] [Google Scholar]
- 6. Zhao H, Ivic L, Otaki J, Hashimoto M, Mikoshiba K, Firestein S. Functional expression of a mammalian odorant receptor. Science. 1998;279(5348):237–42. [DOI] [PubMed] [Google Scholar]
- 7. Mombaerts P. Axonal wiring in the mouse olfactory system. Annu Rev Cell Dev Biol. 2006;22:713–37. [DOI] [PubMed] [Google Scholar]
- 8. Feinstein P, Mombaerts P. A contextual model for axonal sorting into glomeruli in the mouse olfactory system. Cell. 2004;117(6):817–31. [DOI] [PubMed] [Google Scholar]
- 9. Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the β2 adrenergic receptor. Cell. 2004;117(6):833–46. [DOI] [PubMed] [Google Scholar]
- 10. Schild D, Restrepo D. Transduction mechanisms in vertebrate olfactory receptor cells. Physiol Rev. 1998;78(2):429–66. [DOI] [PubMed] [Google Scholar]
- 11. Bakalyar H, Reed R. Identification of a specialized adenylyl cyclase that may mediate odorant detection. Science. 1990;250(4986):1403–6. [DOI] [PubMed] [Google Scholar]
- 12. Jones D, Reed R. Golf: an olfactory neuron specific-G protein involved in odorant signal transduction. Science. 1989;244(4906):790–5. [DOI] [PubMed] [Google Scholar]
- 13. Restrepo D, Teeter J, Schild D. Second messenger signaling in olfactory transduction. J Neurobiol. 1996;30(1):37–48. [DOI] [PubMed] [Google Scholar]
- 14. Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78(5):823–34. [DOI] [PubMed] [Google Scholar]
- 15. Ressler K, Sullivan S, Buck L. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79(7):1245–55. [DOI] [PubMed] [Google Scholar]
- 16. Serizawa S, Miyamichi K, Sakano H. One neuron-one receptor rule in the mouse olfactory system. Trends Genet. 2004;20(12):648–53. [DOI] [PubMed] [Google Scholar]
- 17. Buck LB. Olfactory receptors and odor coding in mammals. Nutr Rev. 2004;62(11 Pt 2):S184–8. [DOI] [PubMed] [Google Scholar]
- 18. Kang N, Kim H, Jae Y, Lee N, Ku C, Margolis F, Lee EJ, Bahk YY, Kim MS, Koo J. Olfactory marker protein expression is an indicator of olfactory receptor-associated events in non-olfactory tissues. PLoS ONE. 2015;10(1):e0116097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kang N, Koo J. Olfactory receptors in non-chemosensory tissues. BMB Rep. 2012;45(11):612–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Flegel C, Manteniotis S, Osthold S, Hatt H, Gisselmann G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS ONE. 2013;8(2):e55368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gaudin J-C, Breuils L, Haertlé T. Mouse orthologs of human olfactory-like receptors expressed in the tongue. Gene. 2006;381:42–8. [DOI] [PubMed] [Google Scholar]
- 22. Durzyński Ł, Gaudin J-C, Myga M, Szydłowski J, Go�dzicka-Józefiak A, Haertlé T. Olfactory-like receptor cDNAs are present in human lingual cDNA libraries. Biochem Biophys Res Commun. 2005;333(1):264–72. [DOI] [PubMed] [Google Scholar]
- 23. Thomas M, Haines S, Akeson R. Chemoreceptors expressed in taste, olfactory and male reproductive tissues. Gene. 1996;178(1):1–5. [DOI] [PubMed] [Google Scholar]
- 24. Blache P, Gros L, Salazar G, Bataille D. Cloning and tissue distribution of a new rat olfactory receptor-like (OL2). Biochem Biophys Res Commun. 1998;242(3):669–72. [DOI] [PubMed] [Google Scholar]
- 25. Nakagawa Y, Nagasawa M, Yamada S, Hara A, Mogami H, Nikolaev V, Lohse MJ, Shigemura N, Ninomiya Y, Kojima I. Sweet taste receptor expressed in pancreatic β-cells activates the calcium and cyclic AMP signaling systems and stimulates insulin secretion. PLoS ONE. 2009;4(4):e5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Itakura S, Ohno K, Ueki T, Sato K, Kanayama N. Expression of Golf in the rat placenta: possible implication in olfactory receptor transduction. Placenta. 2006;27(1):103–8. [DOI] [PubMed] [Google Scholar]
- 27. Braun T, Voland P, Kunz L, Prinz C, Gratzl M. Enterochromaffin cells of the human gut: sensors for spices and odorants. Gastroenterology. 2007;132(5):1890–901. [DOI] [PubMed] [Google Scholar]
- 28. Spehr M, Gisselmann G, Poplawski A, Riffell J, Wetzel C, Zimmer R, Hatt H. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science. 2003;299(5615):2054–8. [DOI] [PubMed] [Google Scholar]
- 29. Fukuda N, Yomogida K, Okabe M, Touhara K. Functional characterization of a mouse testicular olfactory receptor and its role in chemosensing and in regulation of sperm motility. J Cell Sci. 2004;117(24):5835–45. [DOI] [PubMed] [Google Scholar]
- 30. Parmentier M, Libert F, Schurmans S, Schiffmann S, Lefort A, Eggerickx D, Ledent C, Mollereau C, Gérard C, Perret J, Grootegoed A, Vassart G. Expression of members of the putative olfactory receptor gene family in mammalian germ cells. Nature. 1992;355(6359):453–5. [DOI] [PubMed] [Google Scholar]
- 31. Vanderhaeghen P, Schurmans S, Vassart G, Parmentier M. Olfactory receptors are displayed on dog mature sperm cells. J Cell Biol. 1993;123(6):1441–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Walensky L, Roskams A, Lefkowitz R, Snyder S, Ronnett G. Odorant receptors and desensitization proteins colocalize in mammalian sperm. Mol Med. 1995;1(2):130–41. [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang X, Rogers M, Tian H, Zhang X, Zou D-J, Liu J, Ma M, Shepherd GM, Firestein SJ. High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc Natl Acad Sci U S A. 2004;101(39):14168–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Farbman A, Margolis F. Olfactory marker protein during ontogeny: immunohistochemical localization. Dev Biol. 1980;74(1):205–15. [DOI] [PubMed] [Google Scholar]
- 35. Johnson E, Eller P, Jafek B. An immuno-electron microscopic comparison of olfactory marker protein localization in the supranuclear regions of the rat olfactory epithelium and vomeronasal organ neuroepithelium. Acta Otolaryngol. 1993;113(6):766–71. [DOI] [PubMed] [Google Scholar]
- 36. Keller A, Margolis F. Immunological studies of the rat olfactory marker protein. J Neurochem. 1975;24(6):1101–6. [DOI] [PubMed] [Google Scholar]
- 37. Menco B. Electron-microscopic demonstration of olfactory-marker protein with protein G-gold in freeze-substituted, Lowicryl K11M-embedded rat olfactory-receptor cells. Cell Tissue Res. 1989;256(2):275–81. [DOI] [PubMed] [Google Scholar]
- 38. Weiler E, Benali A. Olfactory epithelia differentially express neuronal markers. J Neurocytol. 2005;34(3):217–40. [DOI] [PubMed] [Google Scholar]
- 39. Wensley C, Stone D, Baker H, Kauer J, Margolis F, Chikaraishi D. Olfactory marker protein mRNA is found in axons of olfactory receptor neurons. J Neurosci. 1995;15(7):4827–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Buiakova O, Krishna N, Getchell T, Margolis F. Human and rodent OMP genes: conservation of structural and regulatory motifs and cellular localization. Genomics. 1994;20(3):452–62. [DOI] [PubMed] [Google Scholar]
- 41. Youngentob S, Margolis F. OMP gene deletion causes an elevation in behavioral threshold sensitivity. Neuroreport. 1999;10(1):15–9. [DOI] [PubMed] [Google Scholar]
- 42. Ivic L, Pyrski M, Margolis J, Richards L, Firestein S, Margolis F. Adenoviral vector-mediated rescue of the OMP-null phenotype in vivo. Nat Neurosci. 2000;3(11):1113–20. [DOI] [PubMed] [Google Scholar]
- 43. Youngentob S, Kent P, Margolis F. OMP gene deletion results in an alteration in odorant-induced mucosal activity patterns. J Neurophysiol. 2003;90(6):3864–73. [DOI] [PubMed] [Google Scholar]
- 44. Lee A, He J, Ma M. Olfactory marker protein is critical for functional maturation of olfactory sensory neurons and development of mother preference. J Neurosci. 2011;31(8):2974–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reisert J, Yau KW, Margolis F. Olfactory marker protein modulates the cAMP kinetics of the odour-induced response in cilia of mouse olfactory receptor neurons. J Physiol. 2007;585(3):731–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith P, Firestein S, Hunt J. The crystal structure of the olfactory marker protein at 2.3 Å resolution. J Mol Biol. 2002;319(3):807–21. [DOI] [PubMed] [Google Scholar]
- 47. Buiakova O, Baker H, Scott J, Farbman A, Kream R, Grillo M, Franzen L, Richman M, Davis LM, Abbondanzo S, Stewart CL, Margolis FL. Olfactory marker protein (OMP) gene deletion causes altered physiological activity of olfactory sensory neurons. Proc Natl Acad Sci U S A. 1996;93(18):9858–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kwon H, Koo J, Zufall F, Leinders-Zufall T, Margolis F. Ca2+ extrusion by NCX is compromised in olfactory sensory neurons of OMP−/− Mice. PLoS ONE. 2009;4(1):e4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carr V, Walters E, Margolis F, Farbman A. An enhanced olfactory marker protein immunoreactivity in individual olfactory receptor neurons following olfactory bulbectomy may be related to increased neurogenesis. J Neurobiol. 1998;34(4):377–90. [PubMed] [Google Scholar]
- 50. St John J, Key B. Olfactory marker protein modulates primary olfactory axon overshooting in the olfactory bulb. J Comp Neurol. 2005;488(1):61–9. [DOI] [PubMed] [Google Scholar]
- 51. Youngentob S, Margolis F, Youngentob L. OMP gene deletion results in an alteration in odorant quality perception. Behav Neurosci. 2001;115(3):626–31. [DOI] [PubMed] [Google Scholar]
- 52. Baker H, Grillo M, Margolis F. Biochemical and immunocytochemical charaterization of olfactory marker protein in the rodent central nervous system. J Comp Neurol. 1989;285(2):246–61. [DOI] [PubMed] [Google Scholar]
- 53. Budanova E, Bystrova M. Immunohistochemical detection of olfactory marker protein in tissues with ectopic expression of olfactory receptor genes. Biochem Moscow Suppl Ser A. 2010;4(1):120–3. [Google Scholar]
- 54. Pronin A, Levay K, Velmeshev D, Faghihi M, Shestopalov V, Slepak V. Expression of olfactory signaling genes in the eye. PLoS ONE. 2014;9(4):e96435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. World Health Organization. WHO laboratory manual for the examination and processing of human semen. Geneva: World Health Organization; 2010. [Google Scholar]
- 56. James P, Hennessy C, Berge T, Jones R. Compartmentalisation of the sperm plasma membrane: a FRAP, FLIP and SPFI analysis of putative diffusion barriers on the sperm head. J Cell Sci. 2004;117(26):6485–95. [DOI] [PubMed] [Google Scholar]
- 57. Coward K, Wells D. Textbook of clinical embryology. Cambridge: Cambridge University Press; 2013. [Google Scholar]
- 58. Koo J, Gill S, Pannell L, Menco B, Margolis J, Margolis F. The interaction of Bex and OMP reveals a dimer of OMP with a short half-life. J Neurochem. 2004;90(1):102–16. [DOI] [PubMed] [Google Scholar]
- 59. Jones D, Masters S, Bourne H, Reed R. Biochemical characterization of three stimulatory GTP-binding proteins. The large and small forms of Gs and the olfactory-specific G-protein, Golf. J Biol Chem. 1990;265(5):2671–76. [PubMed] [Google Scholar]
- 60. Simon M, Strathmann M, Gautam N. Diversity of G proteins in signal transduction. Science. 1991;252(5007):802–8. [DOI] [PubMed] [Google Scholar]
- 61. Conklin B, Bourne H. Structural elements of Gα subunits that interact with Gβγ, receptors, and effectors. Cell. 1993;73(4):631–41. [DOI] [PubMed] [Google Scholar]
- 62. Lee Y, Li P, Huh J, Hwang I, Lu M, Kim J, Ham M, Talukdar S, Chen A, Lu WJ, Bandyopadhyay GK, Schwendener R, Olefsky J, Kim JB. Inflammation is necessary for long-term but not short-term high-fat diet-induced insulin resistance. Diabetes. 2011;60(10):2474–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Belluscio L, Gold G, Nemes A, Axel R. Mice deficient in G(olf) are anosmic. Neuron. 1998;20(1):69–81. [DOI] [PubMed] [Google Scholar]
- 64. Gautier-Courteille C, Salanova M, Conti M. The olfactory adenylyl cyclase III is expressed in rat germ cells during spermiogenesis 1. Endocrinology. 1998;139(5):2588–99. [DOI] [PubMed] [Google Scholar]
- 65. Flegel C, Vogel F, Hofreuter A, Schreiner B, Osthold S, Veitinger S, Becker C, Brockmeyer NH, Muschol M, Wennemuth G, Altmüller J, Hatt H, Gisselmann G. Characterization of the olfactory receptors expressed in human spermatozoa. Frontiers Mol Biosci. 2015;2:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Winsky L, Nakata H, Martin B, Jacobowitz D. Isolation, partial amino acid sequence, and immunohistochemical localization of a brain-specific calcium-binding protein. Proc Natl Acad Sci U S A. 1989;86(24):10139–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Arai R, Winsky L, Arai M, Jacobowitz D. Immunohistochemical localization of calretinin in the rat hindbrain. J Comp Neurol. 1991;310(1):21–44. [DOI] [PubMed] [Google Scholar]
- 68. Brookes S, Steele P, Costa M. Calretinin immunoreactivity in cholinergic motor neurones, interneurones and vasomotor neurones in the guinea-pig small intestine. Cell Tissue Res. 1991;263(3):471–81. [DOI] [PubMed] [Google Scholar]
- 69. Strauss KI, Isaacs KR, Ha QN, Jacobowitz DM. Calretinin is expressed in the Leydig cells of rat testis. Biochim Biophys Acta. 1994;1219(2):435–40. [DOI] [PubMed] [Google Scholar]
- 70. Augusto D, Leteurtre E, De La Taille A, Gosselin B, Leroy X. Calretinin: a valuable marker of normal and neoplastic Leydig cells of the testis. Appl Immunohistochem Mol Morphol. 2002;10(2):159–62. [DOI] [PubMed] [Google Scholar]
- 71. Newcombe RG. Two-sided confidence intervals for the single proportion: comparison of seven methods. Statist Med. 1998;17(8):857–72. [DOI] [PubMed] [Google Scholar]
- 72. Fay MP. Two-sided exact tests and matching confidence intervals for discrete data. R J. 2010;2(1):53–8. [Google Scholar]
- 73. McDonald JH. Handbook of biological statistics. Baltimore, MD: Sparky House Publishing; 2009. [Google Scholar]
- 74. Mann H, Wald A. On the choice of the number of class intervals in the application of the chi square test. Ann Math Statist. 1942;13(3):306–17. [Google Scholar]
- 75. Moore DS. Generalized inverses, Wald’s method, and the construction of chi-squared tests of fit. J Am Stat Assoc. 1977;72(357):131–7. [Google Scholar]
- 76. Graziadei GM, Stanley R, Graziadei P. The olfactory marker protein in the olfactory system of the mouse during development. Neuroscience. 1980;5(7):1239–52. [DOI] [PubMed] [Google Scholar]
- 77. Schulze W, Rehder U. Organization and morphogenesis of the human seminiferous epithelium. Cell Tissue Res. 1984;237(3):395–407. [DOI] [PubMed] [Google Scholar]
- 78. Saez JM. Leydig cells: endocrine, paracrine, and autocrine regulation. Endocr Rev. 1994;15(5):574–626. [DOI] [PubMed] [Google Scholar]
- 79. Russell L. Mammalian Leydig cell structure. In: Payne AH, Hardy MP, Russell LD, editors. The Leydig cell. Vienna, IL: Cache River Press; 1996. p. 43–96. [Google Scholar]
- 80. Teerds KJ, Huhtaniemi IT. Morphological and functional maturation of Leydig cells: from rodent models to primates. Hum Reprod Update. 2015;21(3):310–28. [DOI] [PubMed] [Google Scholar]
- 81. Oko R, Hermo L, Chan P, Fazel A, Bergeron J. The cytoplasmic droplet of rat epididymal spermatozoa contains saccular elements with Golgi characteristics. J Cell Biol. 1993;123(4):809–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Darszon A, Nishigaki T, Beltran C, Treviño C. Calcium channels in the development, maturation, and function of spermatozoa. Physiol Rev. 2011;91(4):1305–55. [DOI] [PubMed] [Google Scholar]
- 83. Clermont Y, Oko R, Hermo L. Cell biology of mammalian spermiogenesis. In: Desjardins C, Ewing LL, editors. Cell and molecular biology of the testis. vol. 1 New York: Oxford University Press; 1993. p. 332–76. [Google Scholar]
- 84. Wolkowicz M, Digilio L, Klotz K, Shetty J, Flickinger C, Herr J. Equatorial segment protein (ESP) is a human alloantigen involved in sperm-egg binding and fusion. J Androl. 2008;29(3):272–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Fujihara Y, Murakami M, Inoue N, Satouh Y, Kaseda K, Ikawa M, Okabe M. Sperm equatorial segment protein 1, SPESP1, is required for fully fertile sperm in mouse. J Cell Sci. 2010;123(9):1531–6. [DOI] [PubMed] [Google Scholar]
- 86. Nimlamool W, Bean BS, Lowe-Krentz LJ. Human sperm CRISP2 is released from the acrosome during the acrosome reaction and re-associates at the equatorial segment. Mol Reprod Dev. 2013;80(6):488–502. [DOI] [PubMed] [Google Scholar]
- 87. Nimlamool W. The sperm equatorial segment: an organizing center for sperm protein relocalization and facilitation of fertilization [doctoral thesis]; 2013. Available from: https://preserve.lehigh.edu/cgi/viewcontent.cgi?article=2575&context=etd
- 88. Wolkowicz MJ, Shetty J, Westbrook A, Klotz K, Jayes F, Mandal A, Flickinger CJ, Herr JC. Equatorial segment protein defines a discrete acrosomal subcompartment persisting throughout acrosomal biogenesis. Biol Reprod. 2003;69(3):735–45. [DOI] [PubMed] [Google Scholar]
- 89. Okabe M. Mechanism of fertilization: a modern view. Exp Anim. 2014;63(4):357–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Krauchunas A, Marcello M, Singson A. The molecular complexity of fertilization: introducing the concept of a fertilization synapse. Mol Reprod Dev. 2016;83(5):376–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Georgadaki K, Khoury N, Spandidos D, Zoumpourlis V. The molecular basis of fertilization. Intl J Mol Med. 2016;38(4):979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Veitinger T, Riffell J, Veitinger S, Nascimento J, Triller A, Chandsawangbhuwana C, Schwane K, Geerts A, Wunder F, Berns MW, Neuhaus EM, Zimmer RK, Spehr M, Hatt H. Chemosensory Ca2+ dynamics correlate with diverse behavioral phenotypes in human sperm. J Biol Chem. 2011;286(19):17311–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sun F, Bahat A, Gakamsky A, Girsh E, Katz N, Giojalas L, Tur-Kaspa I, Eisenbach M. Human sperm chemotaxis: both the oocyte and its surrounding cumulus cells secrete sperm chemoattractants. Hum Reprod. 2005;20(3):761–67. [DOI] [PubMed] [Google Scholar]
- 94. Spehr M, Schwane K, Heilmann S, Gisselmann G, Hummel T, Hatt H. Dual capacity of a human olfactory receptor. Curr Biol. 2004;14(19):R832–3. [DOI] [PubMed] [Google Scholar]
- 95. Teves M, Guidobaldi H, Uñates D, Sanchez R, Miska W, Publicover S, Morales Garcia AA, Giojalas LC. Molecular mechanism for human sperm chemotaxis mediated by progesterone. PLoS ONE. 2009;4(12):e8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kaupp U, Kashikar N, Weyand I. Mechanisms of sperm chemotaxis. Annu Rev Physiol. 2008;70:93–117. [DOI] [PubMed] [Google Scholar]
- 97. Meizel S. The sperm, a neuron with a tail: “neuronal” receptors in mammalian sperm. Biol Rev. 2004;79(4):713–32. [DOI] [PubMed] [Google Scholar]
- 98. Defer N, Best-Belpomme M, Hanoune J. Tissue specificity and physiological relevance of various isoforms of adenylyl cyclase. Am J Physiol Renal Physiol. 2000;279(3):F400–16. [DOI] [PubMed] [Google Scholar]
- 99. Hanoune J, Defer N. Regulation and role of adenylyl cyclase isoforms. Annu Rev Pharmacol Toxicol. 2001;41(1):145–74. [DOI] [PubMed] [Google Scholar]
- 100. Lee C, Park D, Wu D, Rhee S, Simon M. Members of the Gq alpha subunit gene family activate phospholipase C beta isozymes. J Biol Chem. 1992;267(23):16044–7. [PubMed] [Google Scholar]
- 101. Livera G, Xie F, Garcia M, Jaiswal B, Chen J, Law E, Storm DR, Conti M. Inactivation of the mouse adenylyl cyclase 3 gene disrupts male fertility and spermatozoon function. Mol Endocrinol. 2005;19(5):1277–90. [DOI] [PubMed] [Google Scholar]
- 102. Neuhaus EM, Mashukova A, Barbour J, Wolters D, Hatt H. Novel function of beta-arrestin2 in the nucleus of mature spermatozoa. J Cell Sci. 2006;119(Pt 15):3047–56. [DOI] [PubMed] [Google Scholar]
- 103. Spehr M, Schwane K, Riffell J, Barbour J, Zimmer R, Neuhaus E, Hatt H. Particulate adenylate cyclase plays a key role in human sperm olfactory receptor-mediated chemotaxis. J Biol Chem. 2004;279(38):40194–203. [DOI] [PubMed] [Google Scholar]
- 104. Vanderhaeghen P, Schurmans S, Vassart G, Parmentier M. Specific repertoire of olfactory receptor genes in the male germ cells of several mammalian species. Genomics. 1997;39(3):239–46. [DOI] [PubMed] [Google Scholar]
- 105. Walensky L, Ruat M, Bakin R, Blackshaw S, Ronnett G, Snyder S. Two novel odorant receptor families expressed in spermatids undergo 5’-splicing. J Biol Chem. 1998;273(16):9378–87. [DOI] [PubMed] [Google Scholar]
- 106. Walensky L, Snyder S. Inositol 1, 4, 5-trisphosphate receptors selectively localized to the acrosomes of mammalian sperm. J Cell Biol. 1995;130(4):857–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wiesner B, Weiner J, Middendorff R, Hagen V, Kaupp U, Weyand I. Cyclic nucleotide-gated channels on the flagellum control Ca2+ entry into sperm. J Cell Biol. 1998;142(2):473–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Weyand I, Godde M, Frings S, Weiner J, Muller F, Altenhofen W, Hatt H, Kaupp UB. Cloning and functional expression of a cyclic-nucleotide-gated channel from mammalian sperm. Nature. 1994;368(6474):859–63. [DOI] [PubMed] [Google Scholar]
- 109. Buettner A. Springer handbook of odor. New York: Springer; 2017. [Google Scholar]
- 110. Saito H, Chi Q, Zhuang H, Matsunami H, Mainland J. Odor coding by a Mammalian receptor repertoire. Sci Signal. 2009;2(60):ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Nara K, Saraiva L, Ye X, Buck L. A large-scale analysis of odor coding in the olfactory epithelium. J Neurosci. 2011;31(25):9179–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sanz G, Schlegel C, Pernollet J-C, Briand L. Comparison of odorant specificity of two human olfactory receptors from different phylogenetic classes and evidence for antagonism. Chem Senses. 2005;30(1):69–80. [DOI] [PubMed] [Google Scholar]
- 113. Matarazzo V, Clot-Faybesse O, Marcet B, Guiraudie-Capraz G, Atanasova B, Devauchelle G, Cerutti M, Etiévant P, Ronin C. Functional characterization of two human olfactory receptors expressed in the baculovirus Sf9 insect cell system. Chem Senses. 2005;30(3):195–207. [DOI] [PubMed] [Google Scholar]
- 114. Monti-Graziadei G, Margolis F, Harding J, Graziadei P. Immunocytochemistry of the olfactory marker protein. J Histochem Cytochem. 1977;25(12):1311–6. [DOI] [PubMed] [Google Scholar]
- 115. Trimmer C, Keller A, Murphy NR, Snyder LL, Willer JR, Nagai M, Katsanis N, Vosshall LB, Matsunami H, Mainland JD. Genetic variation across the human olfactory receptor repertoire alters odor perception. Proc Natl Acad Sci U S A. 2019;116(19):9475–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Verbeurgt C, Wilkin F, Tarabichi M, Gregoire F, Dumont JE, Chatelain P. Profiling of olfactory receptor gene expression in whole human olfactory mucosa. PLoS ONE. 2014;9(5):e96333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Flegel C, Schöbel N, Altmüller J, Becker C, Tannapfel A, Hatt H, Gisselmann G. RNA-Seq analysis of human trigeminal and dorsal root ganglia with a focus on chemoreceptors. PLoS ONE. 2015;10(6):e0128951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Jovancevic N, Wunderlich KA, Haering C, Flegel C, Maßberg D, Weinrich M, Weber L, Tebbe L, Kampik A, Gisselmann G, Wolfrum U, Hatt H, Gelis L. Deep sequencing of the human retinae reveals the expression of odorant receptors. Front Cell Neurosci. 2017;11:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Mortimer D, Leslie E, Kelly R, Templeton A. Morphological selection of human spermatozoa in vivo and in vitro. J Reprod Fertil. 1982;64(2):391–9. [DOI] [PubMed] [Google Scholar]
- 120. Abraham-Peskir J, Chantler E, Uggerhøj E, Fedder J. Response of midpiece vesicles on human sperm to osmotic stress. Hum Reprod. 2002;17(2):375–82. [DOI] [PubMed] [Google Scholar]
- 121. Cooper T, Yeung C-H, Fetic S, Sobhani A, Nieschlag E. Cytoplasmic droplets are normal structures of human sperm but are not well preserved by routine procedures for assessing sperm morphology. Hum Reprod. 2004;19(10):2283–8. [DOI] [PubMed] [Google Scholar]
- 122. Xu H, Yuan S-Q, Zheng Z-H, Yan W. The cytoplasmic droplet may be indicative of sperm motility and normal spermiogenesis. Asian J Androl. 2013;15(6):799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Strünker T, Goodwin N, Brenker C, Kashikar N, Weyand I, Seifert R, Kaupp UB. The CatSper channel mediates progesterone-induced Ca2+ influx in human sperm. Nature. 2011;471(7338):382–6. [DOI] [PubMed] [Google Scholar]
- 124. Brenker C, Goodwin N, Weyand I, Kashikar N, Naruse M, Krähling M, Müller A, Kaupp UB, Strünker T. The CatSper channel: a polymodal chemosensor in human sperm. EMBO J. 2012;31(7):1654–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Lishko PV, Kirichok Y, Ren D, Navarro B, Chung J-J, Clapham DE. The control of male fertility by spermatozoan ion channels. Ann Review of Physiol. 2012;74:453–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Strünker T, Alvarez L, Kaupp U. At the physical limit—chemosensation in sperm. Curr Opin Neurobiol. 2015;34:110–6. [DOI] [PubMed] [Google Scholar]
- 127. Evans J. Sperm-egg interaction. Annu Rev Physiol. 2012;74:477–502. [DOI] [PubMed] [Google Scholar]
- 128. Sieme H. Semen evaluation. In: Samper JC, editor. Equine breeding management and artificial insemination. Amsterdam: Elsevier; 2009. p. 57–74. [Google Scholar]
- 129. Moreno RD, Ramalho-Santos J, Sutovsky P, Chan EK, Schatten G. Vesicular traffic and golgi apparatus dynamics during mammalian spermatogenesis: implications for acrosome architecture. Biol Reprod. 2000;63(1):89–98. [DOI] [PubMed] [Google Scholar]
- 130. Sullivan R, Frenette G, Girouard J. Epididymosomes are involved in the acquisition of new sperm proteins during epididymal transit. Asian J Androl. 2007;9(4):483–91. [DOI] [PubMed] [Google Scholar]
- 131. Frenette G, Girouard J, Sullivan R. Comparison between epididymosomes collected in the intraluminal compartment of the bovine caput and cauda epididymidis. Biol Reprod. 2006;75(6):885–90. [DOI] [PubMed] [Google Scholar]
- 132. Aumüller G, Wilhelm B, Seitz J. Apocrine secretion—fact or artifact? Ann Anat. 1999;181(5):437–46. [DOI] [PubMed] [Google Scholar]
- 133. Fornes M, Barbieri A, Cavicchia J. Morphological and enzymatic study of membrane-bound vesicles from the lumen of the rat epididymis. Andrologia. 1995;27(1):1–5. [DOI] [PubMed] [Google Scholar]
- 134. Frenette G, Légaré C, Saez F, Sullivan R. Macrophage migration inhibitory factor in the human epididymis and semen. Mol Hum Reprod. 2005;11(8):575–82. [DOI] [PubMed] [Google Scholar]
- 135. Gatti J-L, Métayer S, Moudjou M, Andréoletti O, Lantier F, Dacheux J-L, Sarradin P. Prion protein is secreted in soluble forms in the epididymal fluid and proteolytically processed and transported in seminal plasma. Biol Reprod. 2002;67(2):393–400. [DOI] [PubMed] [Google Scholar]
- 136. Gatti J-L, Métayer S, Belghazi M, Dacheux F, Dacheux J-L. Identification, proteomic profiling, and origin of ram epididymal fluid exosome-like vesicles. Biol Reprod. 2005;72(6):1452–65. [DOI] [PubMed] [Google Scholar]
- 137. Dacheux JL, Gatti J, Dacheux F. Contribution of epididymal secretory proteins for spermatozoa maturation. Microsc Res Tech. 2003;61(1):7–17. [DOI] [PubMed] [Google Scholar]
- 138. Yanagimachi R, Kamiguchi Y, Mikamo K, Suzuki F, Yanagimachi H. Maturation of spermatozoa in the epididymis of the Chinese hamster. Dev Dynam. 1985;172(4):317–30. [DOI] [PubMed] [Google Scholar]
- 139. Kirchhoff C, Hale G. Cell-to-cell transfer of glycosylphosphatidylinositol-anchored membrane proteins during sperm maturation. Mol Hum Reprod. 1996;2(3):177–84. [DOI] [PubMed] [Google Scholar]
- 140. Reilly JN, McLaughlin EA, Stanger SJ, Anderson AL, Hutcheon K, Church K, Mihalas BP, Tyagi S, Holt JE, Eamens AL, Nixon B. Characterisation of mouse epididymosomes reveals a complex profile of microRNAs and a potential mechanism for modification of the sperm epigenome. Sci Rep. 2016;6:31794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Hutcheon K, McLaughlin EA, Stanger SJ, Bernstein IR, Dun MD, Eamens AL, Nixon B. Analysis of the small non-protein-coding RNA profile of mouse spermatozoa reveals specific enrichment of piRNAs within mature spermatozoa. RNA Biol. 2017;14(12):1776–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zhou W, Stanger SJ, Anderson AL, Bernstein IR, De Iuliis GN, McCluskey A, McLaughlin EA, Dun MD, Nixon B. Mechanisms of tethering and cargo transfer during epididymosome-sperm interactions. BMC Biol. 2019;17(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Mortimer S, Mortimer D. Kinematics of human spermatozoa incubated under capacitating conditions. J Androl. 1990;11(3):195–203. [PubMed] [Google Scholar]
- 144. Teves M, Barbano F, Guidobaldi H, Sanchez R, Miska W, Giojalas L. Progesterone at the picomolar range is a chemoattractant for mammalian spermatozoa. Fertil Steril. 2006;86(3):745–49. [DOI] [PubMed] [Google Scholar]
- 145. Guidobaldi H, Teves M, Uñates D, Giojalas L. Sperm transport and retention at the fertilization site is orchestrated by a chemical guidance and oviduct movement. Reproduction. 2012;143(5):587–96. [DOI] [PubMed] [Google Scholar]
- 146. Blackmore P, Beebe S, Danforth D, Alexander N. Progesterone and 17 alpha-hydroxyprogesterone. Novel stimulators of calcium influx in human sperm. J Biol Chem. 1990;265(3):1376–80. [PubMed] [Google Scholar]
- 147. Thomas P, Meizel S. Phosphatidylinositol 4,5-bisphosphate hydrolysis in human sperm stimulated with follicular fluid or progesterone is dependent upon Ca2+ influx. Biochem J. 1989;264(2):539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Yanagimachi R. Mammalian fertilization. In: Knobil E, Neill J, editors. The physiology of reproduction. New York: Raven Press; 1994. p. 189–317. [Google Scholar]
- 149. Kopf GS. The mammalian sperm acrosome reaction and the acrosome reaction. In: Wassarman PM, editor. Elements of mammalian fertilization. Boca Raton: CRC Press; 1991. p. 153–203. [Google Scholar]
- 150. Jayaprakash D, Kumar KS, Shivaji S, Seshagiri P. Pentoxifylline induces hyperactivation and acrosome reaction in spermatozoa of golden hamsters: changes in motility kinematics. Hum Reprod. 1997;12(10):2192–9. [DOI] [PubMed] [Google Scholar]
- 151. Nassar A, Morshedi M, Mahony M, Srisombut C, Lin M, Oehninger S. Pentoxifylline stimulates various sperm motion parameters and cervical mucus penetrability in patients with asthenozoospermia. Andrologia. 1999;31(1):9–15. [DOI] [PubMed] [Google Scholar]