Abstract
The lack of tools for early detection of pancreatic ductal adenocarcinoma (PDAC) is directly correlated with the abysmal survival rates in patients. In addition to several potential detection tools under active investigation, we tested the gut microbiome and its metabolic complement as one of the earliest detection tools that could be useful in patients at high risk for PDAC. We used a combination of 16s rRNA pyrosequencing and whole-genome sequencing of gut fecal microbiota in a genetically engineered PDAC murine model (KRASG12DTP53R172HPdxCre or KPC). Metabolic reconstruction of microbiome was done using the HUMAnN2 pipeline. Serum polyamine levels were measured from murine and patient samples using chromogenic assay. Our results showed a Proteobacterial and Firmicutes dominance in gut microbiota in early stages of PDAC development. Upon in silico reconstruction of active metabolic pathways within the altered microbial flora, polyamine and nucleotide biosynthetic pathways were significantly elevated. These metabolic products are known to be actively assimilated by the host and eventually utilized by rapidly dividing cells for proliferation validating their importance in the context of tumorigenesis. In KPC mice, as well as PDAC patients, we show significantly elevated serum polyamine concentrations. Therefore, at the early stages of tumorigenesis, there is a strong correlation between microbial changes and release of metabolites that foster host tumorigenesis, thereby fulfilling the ‘vicious cycle hypothesis’ of the role of microbiome in health and disease states. Our results provide a potential, precise, noninvasive tool for early detection of PDAC, which may result in improved outcomes.
The gut microbiota changes during early stages of pancreatic ductal adenocarcinoma (PDAC) progression and potentially contributes toward host polyamine pool. Both changes may be a marker for early detection of the disease PDAC.
Introduction
Pancreatic cancer is the third most common cause of cancer related deaths in USA with a very poor 5 year survival rate of 9%. The disease is characterized by relatively late onset of symptoms and rapid progression with very limited treatment options. Lack of efficient biomarkers that can facilitate early detection of the disease is one of the primary challenges of the field. With effective tools for early detection, the survival rate would eventually increase, as more patients would be able to have their tumors resected (1). Thus, there is an urgent need for noninvasive, discriminatory biomarkers for early detection of this disease. During carcinogenesis, a pancreatic tumor progresses through the pre-cancerous pancreatic intraepithelial neoplasia (PanINs) lesions (2,3). However, even though these changes in the pancreas morphology are histologically distinct, imaging techniques used in the clinics are not able to distinguish the early PanINs from the normal pancreas. As a result, detection of a tumor at an early stage becomes difficult.
The role of gut microbiome, and specifically its dysbiosis in cancer development, is becoming more evident in recent years. It has become increasingly apparent that the gut microbiome plays an integral role in modulating host physiology (4). Although earlier focus on the microbial dysbiosis had been on identification of bacterial species that initiate tumorigenesis in the gastrointestinal tract, recent evidences have shown that this dysbiosis can induce systemic changes to affect tumor growth outside the gastrointestinal tract. Microbial dysbiosis has been associated with development of melanoma (5), lung cancer (6) and pancreatic ductal adenocarcinoma (PDAC) (7–14). It is now well established that the gut bacteria play an integral role in carcinogenesis by influencing host metabolic and inflammatory pathways (15,16).
Previous reports have suggested that oral microbiome dysbiosis occurs during pancreatic tumorigenesis (8,17). Bacterial species have been detected in pancreatic cysts, as well as in PDAC tumors (18). In addition, other studies have revealed that the microbiome plays an active role in stromal modulation (12) and depletion of the microbiome plays a profound role in reduction of pancreatic tumor burden (14). Most of these studies, however, have utilized the most common sequencing approach of an amplicon analysis of the 16s rRNA ribosomal RNA (rRNA) gene (19,20). This method is limited by the fact that the annotation is based on putative association of the 16s rRNA gene with an operational taxonomic unit (OTU). Because OTUs are analyzed at the phyla or genera level, the study is often unable to detect species within those taxa. An alternative approach to the 16s rRNA amplicon sequencing method is whole-genome shotgun sequencing (WGS) which uses sequencing with random primers to sequence overlapping regions of a genome allowing for the taxa to be more accurately defined at the species level (21).
Functional contribution of the gut microbial dysbiosis on cancer biology is increasingly being acknowledged. Among these, role of host metabolic pathways that may be influenced by the gut microbiota and promote cancer susceptibility has gained a lot of importance (16). Resident microbiota can promote differential metabolism of drugs in the host and confer differential effect to cancer chemotherapy (22). In addition, microbial antigens like LPS trigger inflammatory reactions in host and affect cancer progression (23). However, how microbial dysbiosis and the metabolic product from the dysbiotic microbiome may affect pancreatic carcinogenesis at the early stages have not been studied. In spite of the advances in microbiome research, most of the studies are still focused on the dysbiosis of the microbiome in itself in context of the disease (24–27). Not a lot of effort has been placed on whether microbial metabolites can be exploited as potential biomarker for early detection in pancreatic cancer.
In the present study, we analyzed the microbiome in a genetic mouse model for PDAC (KRASG12DTP53R172HPdxCre or KPC) and age-matched controls using WGS at very early time points of tumorigenesis. During these time points, the KPC mice do not show any detectable tumors in their pancreas. Our results show that at these early time points, the histological changes in the pancreas correspond to a significant change in certain gut microbial population. Our predictive metabolomic analysis on the identified bacterial species reveals that the primary microbial metabolites involved in progression and development of PDAC tumors are involved in polyamine metabolism. Consistent with our analysis in mice, estimation of polyamine from serum of PDAC patients show that total polyamine concentration in increased in PDAC patients compared with healthy volunteers. Furthermore, serum polyamine levels in KPC mice show an increased concentration of polyamines as tumors progress from PanINs to PDAC. Together these observations indicate that analysis of gut microbial flora along with an analysis of the microbial metabolites such as polyamines can be developed as potential biomarkers for detection of PDAC at early stages when histological changes are not yet grossly apparent.
Materials and methods
Ethics statement
All animal studies were performed according to the protocols approved by IACUC at University of Miami, USA (#16–066) in accordance with the principles of the Declaration of Helsinki. Serum from de-identified pancreatic cancer patients and healthy controls were obtained with informed consent, according to the approval from the IRB at University of Minnesota (1403M48826) accorded to Dr. Sulagna Banerjee as an author of the IRB proposal when she was in the Faculty of that University. All authors had access to all data and have reviewed and approved the final manuscript.
Animal models and experimental design
Spontaneous pancreatic animal model KRASG12D TP53R172HPdx-Cre (KPC) animals from both genders were enrolled in the study at 1 month of age. PDX-cre (Cre) animals that were age/gender matched with the KPC animals were used as control. Fecal samples were collected at 2, 3 and 4 months of age. Initially, eight to nine animals were kept in each group and randomized (group wise) to discount cage-effect in microbiome studies. At 4 months, all animals were killed according to protocols approved by University of Miami Animal Care Committee. Gut and pancreas samples were flash frozen in liquid nitrogen. Pancreas tissues were formalin fixed for paraffin embedding and histochemical analysis. Blood was collected by cardiac puncture prior to euthanizing the animals. Plasma and serum samples were stored for analysis of polyamines.
Isolation of DNA
DNA from the murine fecal samples was isolated using the Power Soil DNA Isolation Kit (Qiagen) according to manufacturer’s instructions. All samples were quantified using the Qubit® Quant-iT dsDNA High-Sensitivity Kit (Invitrogen, Life Technologies, Grand Island, NY) to ensure that they met minimum concentration and mass of DNA and were submitted to University of Minnesota Genomics Center for further analysis by 16s rRNA amplicon sequencing or WGS.
16s rRNA pyrosequencing and microbiome analysis
To enrich the fecal DNA for the bacterial 16s rRNA V5–V6 rDNA region, DNA was amplified utilizing fusion primers designed against the surrounding conserved regions which were tailed with sequences to incorporate Illumina (San Diego, CA) flow cell adapters and indexing barcodes. Each sample was PCR amplified with two differently bar coded V5–V6 fusion primers and advanced for pooling and sequencing. For each sample, amplified products were concentrated using a solid-phase reversible immobilization method for the purification of PCR products and quantified by electrophoresis using an Agilent 2100 Bioanalyzer®. The pooled 16s rRNA V5–V6 enriched, amplified, barcoded samples were loaded into the MiSeq® reagent cartridge and then onto the instrument along with the flow cell. After cluster formation on the MiSeq instrument, the amplicons were sequenced for 250 cycles with custom primers designed for paired-end sequencing.
Using QIIME 1.9.2 (Quantitative Insights into Microbial Ecology, version 1.9.2) (28), sequences were quality filtered and de-multiplexed using exact matches to the supplied DNA barcodes and primers. Resulting sequences were then searched against the Greengenes reference database of 16s rRNA sequences, clustered at 97% by uclust (closed-reference OTU picking) to obtain phylogenetic identities. Analysis for alpha and beta diversity was done with standardized QIIME 1.9.2 workflow or the ‘R’ statistical package, as we have shown before (29).
Metagenomic sequencing and microbiome analysis
Shotgun metagenomic library was constructed from fecal DNA with the Nextera DNA sample preparation kit (Illumina, San Diego, CA), as per manufacturer’s specification. Barcoding indices were inserted using Nextera indexing kit (Illumina). Products were purified using Agencourt AMpure XP kit (Beckman Coulter, Brea, CA) and pooled for sequencing. Samples were sequenced using MiSeq reagent kit V2 (Illumina).
Raw sequences were sorted using assigned barcodes and cleaned up before analysis (barcodes removed and sequences above a quality score, Q ≥ 30 taken forward for analyses). For assembly and annotation of sequences, MetAMOS (30) pipeline or Partek Flow software (Partek® Flow®, Partek, St. Louis, MO) were used. These softwares provide powerful tools to filter unique hits between human and mouse-specific genes versus microbial signatures. Alpha- and beta-diversity calculations were done using embedded programs within the metagenomic pipeline or using Stata15 (StataCorp LLC, College Station, TX) or EXPLICET software (31).
Functional profiling was performed using HUMAnN2-0.11.1 (32) with Uniref50 database to implement KEGG orthologies.
Microbial controls
Microbial contamination from tools and practices is a matter of concern in microbiota studies. Extreme rigor was exercised in our studies for both 16s rRNA and whole-genome bases sequencing. We had several controls that were run in parallel to the fecal samples:
(A) Power-soil sample vials (empty) carried through the entire process from DNA isolation to library preparation in triplicate.
(B) Typically, we collect the last time point fecal matter upon necropsy from the colonic area. For this, we use fresh set of implements (scissors, forceps etc.) for each animal. Prior to their use, we collect phosphate-buffered saline wash of these tools and they are also run through the entire process as a control.
(C) The sequencing center adds buffer blanks to the whole repertoire for the portion of the work they perform.
Any OTU or ID that appeared in the control samples were summarily rejected from the OTU/ID list before further analysis. For 16s rRNA pyrosequencing, we had 2–3 OTUs across study groups and one instance in the WGS groups that had to be deleted from the list for appearing in the controls.
Immunohistochemistry
For immunohistochemistry, 4 μm paraffin tissue sections were deparaffinized in xylene and rehydrated through graded ethanol. Hematoxylin and Eosin (H&E) staining were conducted to confirm histological features. Histological scoring was done by an arbitrary scoring method in which scores of 0–4 were assigned based on the dysplasia observed in the tissues. A score of 0 was assigned to the histologically normal pancreas, whereas a score of 4 was assigned to a fully dysplastic tissue (that observed in the late stages of tumor development in the KPC model). All scoring was done in a blinded manner by three separate investigators.
Polyamine estimation
Polyamine was estimated from the serum of KPC animals between the age groups of 2–8 months. The animals of 1–2 months old showed normal pancreas histology, those 3–4 months old showed various degrees of PanINs and those that were 4–8 months old had observable tumors. Polyamine was estimated by Total Polyamine Estimation Kit (Biovision, Milpitas, CA) according to manufacturers’ instruction after diluting the serum 1:25 in the assay buffer.
Statistical analyses
Microbiome analysis with QIIME and whole-genome analysis pipelines
OTU tables were rarefied to the sample containing the lowest number of sequences in each analysis. QIIME 1.9.2 was used to calculate alpha diversity (alpha_rarefaction.py) and to summarize taxa (summarize_taxa_through_plots.py). Principal co-ordinate analysis was done within this program using observation ID level. The Adonis test was utilized for finding significant compositional differences among discrete categorical or continuous variables with randomization/Monte Carlo permutation test (with Bonferroni correction). The fraction of permutations with greater distinction among categories (larger cross-category differences) than that observed with the non-permuted data was reported as the P-value. Relative abundance of species identified through the metagenomic pipelines were compared using non-parametric Mann–Whitney U test at P < 0.05 after FDR correction. Apart from statistical functions embedded within the metagenomic pipeline mentioned above, we have used GraphPad Prism (GraphPad, La Jolla, CA) or Stata15 (StataCorp LLC, College Station, TX) for different statistical analyses, mentioned in the text and figure legend as appropriate.
Data availability
Microbiome raw data sequences are available from EMBL ArrayExpress with the accession number E-MTAB-6921.
Results
Gut microbial profile in KPC mice diverges from age-matched control mice over time
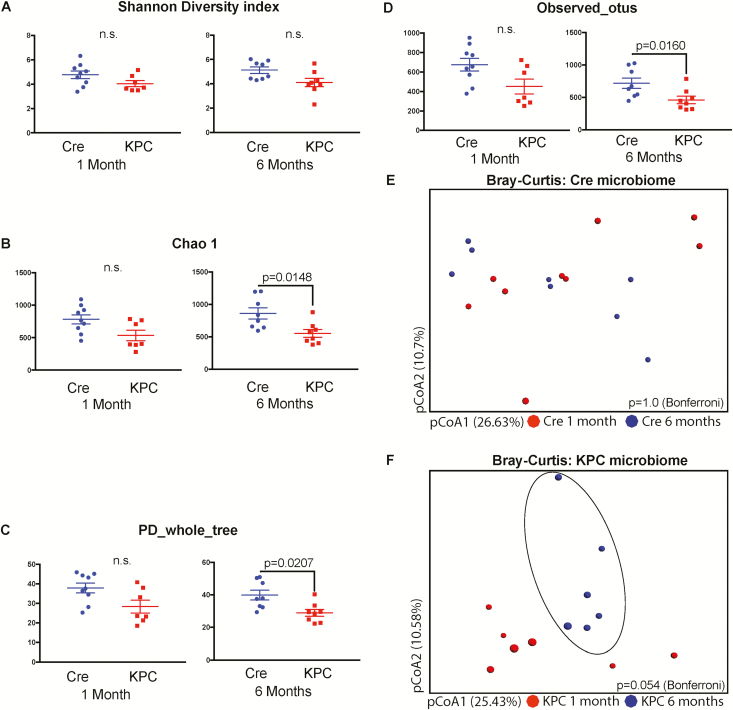
We employed a variety of indices to understand the composition and distribution of gut bacterial communities (species richness, evenness, distribution; alpha diversity) from the fecal matter of 1- and 6-month-old KPC and age-matched control mice. Although controversies exist in the implementation of various diversity indices on 16s rRNA microbial data, originally developed for ecological studies with macro-organisms, a combination of these indices for 16s rRNA phylogenetic studies provide an overall spectrum of the composition of each group (33). We started with the Shannon’s H index (34), which accounts for both the abundance and evenness (measure of species diversity and distribution in a community) in both age groups and genotypes (Figure 1A). The Shannon’s H index showed no difference between the overall composition of the two age groups (1- and 6-month-old control or KPC mice), implying that the numbers of different OTUs encountered (diversity), as well as the instances these unique OTUs were sampled (evenness) was relatively similar between the two groups. Because Shannon’s index is blinded to the identities of the OTUs, we also included Chao1 diversity (35), which accounts for the rarity or abundance of individual OTUs and provides a measure of species richness for individual groups which can then be compared (Figure 1B). The Chao1 diversity analysis showed significantly diminished OTU richness in KPC mice compared with control mice at 6 months of age. Faith’s phylogenetic diversity (PD_whole_tree) represents taxon richness of each sample expressed as the numbers of phylogenetic tree branches encountered for each sample based on sequenced OTUs (36). In a similar manner, as was seen in the Chao1 index, a significant reduction in OTU diversity was observed in KPC mice compared with control in 6-month age groups (Figure 1C).
Figure 1.
KPC microbiome exhibits clustering between 1 and 6 months, compared with control microbiome with 16s rRNA pyrosequencing. Various alpha-diversity indices were measured for the two genotypes and two time points. (A) Shannon’s H index was similar between the age groups and genotypes, whereas (B) Chao1 index, (C) Faith’s phylogenetic diversity and (D) observed OTU indices were significantly down in 6-month-old KPC animals, compared with age-matched control animals. Bray–Curtis principal co-ordinate analysis showed that (E) control animals did not form distinct clusters between 1 and 6 months of age, whereas (F) KPC animals for a distinct cluster at 6-month age, compared with 1-month-old animals. This difference, however, was not statistically significant (P = 0.056) with two-tailed test of significance with Bonferroni correction (n = 6–8 per group).
The disparity between Shannon index and Chao1/PD can be described through the ‘Observed OTU’ metric (Figure 1D), which accounts for an absolute presence/absence of the OTU irrespective of the number of instances it is represented in a sample. This index also exhibited a significant reduction of richness in 6-month-old KPC mice compared with the control animals. This disparity implies that some species of microbes are actually getting diminished with tumor development. Although there were differences in the Chao1 and PD indices, Shannon’s index was similar between the age groups. This is not uncommon, as described in other microbial studies (37), and ensures that the differences in the beta diversity (difference between groups) are not entirely due to differences in absolute OTU counts (37).
Next, we used Bray–Curtis principle co-ordinate analysis (pCoA; beta diversity) to qualitatively examine differences in microbial composition and associated changes due to age in control (Figure 1E) and KPC mice (Figure 1F). Microbial composition of control mice did not change over the course of 6 months, whereas a tendency toward clustering was observed in 6-month-old KPC mice. However, the differences were statistically insignificant (P = 0.054). A combined Bray–Curtis plot for all four groups is presented as Supplementary Figure 1A, available at Carcinogenesis online. Because there was a significant drop in phylogeny-informed diversity indices for KPC mice over time, we decided to study the major differences between the two KPC age groups, in the context of identified genera, family and class of microbes.
Because microbiome changes can be governed by age, sex and mutation status of the animals, we used PDX-Cre animals as our control. These animals have the same background as KPC animals and do not form tumors unless crossed with an animal having an LSL gene. Age- and gender-matched mice of PDX-Cre were also analyzed for the diversity of the species. These animals showed no difference between the time points (Figure 1E).
There are significant early differences in a small group of microbes at the class and genera level between 1- and 6-month-old KPC animals
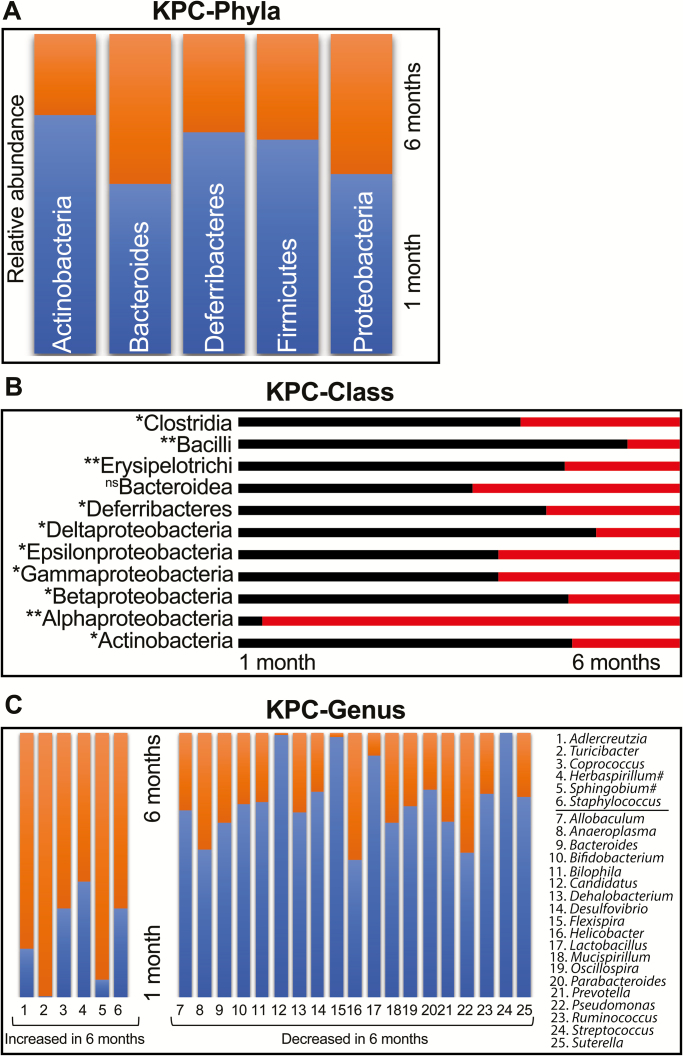
Analysis of the five major phyla revealed a relatively unchanged Bacteroides and a trend toward decrease in relative abundance in Actinobacteria, Deferribacteres, Firmicutes and Proteobacteria (Figure 2A). None of these phyla had statistically significant changes over time. However, when we looked at the class level in the hierarchy, several classes of bacteria showed significant changes in relative abundance between 1- and 6-month-old KPC animals (Figure 2B). Compared with non-significant changes in class Bacteroidea, there was a significant drop in the relative abundance for classes Clostridia, Bacilli, Erysipelotrichia (all phylum Firmicutes), Deferribacteres (phylum Deferribacteres), class Actinobacteria (phylum Actinobacteria) and delta-, epsilon-, gamma- and beta-proteobacteria (all phylum Proteobacteria). Within phylum Proteobacteria, however, there was a significant relative expansion of class Alphaproteobacteria in 6-month-old KPC animals. At the genus level, 6 genera exhibited significant relative expansion, whereas 19 genera showed diminished relative abundance from 1 to 6 months in KPC animals (Figure 2C). Furthermore, we also found Herbaspirilium and Sphingobium in significantly changing species list (with single-digit OTU counts), which are known environmental contaminants, despite not appearing in our control OTU list. Additionally, upon mapping all independent OTUs (n = 24) mapping to genus Bacteroides, several minor OTUs also have significantly elevated abundance in 6-month-old KPC animals (Supplementary Figure 1B, available at Carcinogenesis online) but are beyond the scope for any further identification due to the limitations of the 16s rRNA platform used. For mapping, microbial changes in spontaneous PDAC model and predicting functional attributes, it became abundantly clear that a better understanding of the early changes in genus/species level is of paramount importance. Unfortunately, phylogeny based on 16s rRNA amplicon sequencing does not provide sufficient genus-/species-level coverage, and, hence, we decided to restrict the time points and employ shotgun metagenomics for further studies.
Figure 2.
KPC animals show significant differences at the class and genera levels. In a head-to-head comparison between the OTUs representing the five major phyla, (A) Bacteroidetes did not show any difference between 1- and 6-month-old KPC mice. The other four phyla showed reduced relative abundance, which were statistically insignificant. At the class level (B) however, Alphaproteobacteria exhibited significantly high relative abundance in 6-month-old KPC. With Bacteroidea unchanged between the two age groups, all other Classes exhibited diminished relative abundance in 6-month-old KPC mice (test of significance—non-parametric Mann–Whitney U test. *P < 0.05, **P < 0.01). At the Genus level (C), six genera showed significant increase in relative abundance from 1 to 6 months of age, compensated by 19 genera with severely diminished relative abundance at the same time (all genera P < 0.05). Couple of potential environmental contaminants are marked with ‘#’ despite not appearing in our blank controls.
Microbial dysbiosis occurred during early processes of tumor development
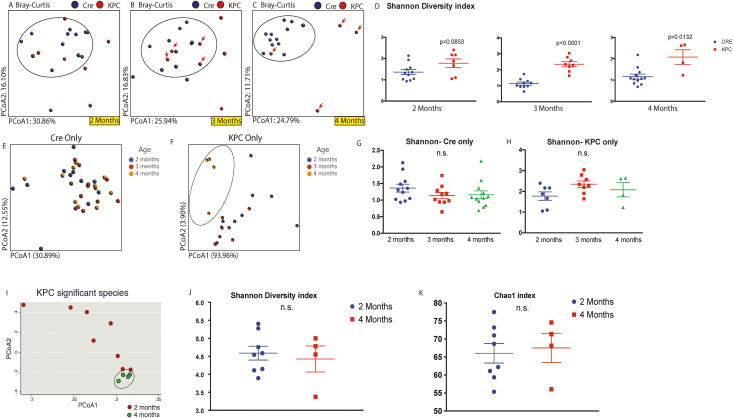
For the pre-neoplastic time points, in order to obtain functionally relevant data with species-level identification, we performed whole-genome sequencing on control and KPC animals at 2, 3, and 4 months of age in animals that did not have visible tumors. PDX-Cre mice were used as control. A total of 1359 genera were identified with 100% species-level coverage. Initial comparison of the overall microbial composition between control and KPC mice showed progressive microbial changes in the KPC mice from 2 to 4 months (Figure 3A–C). At 2 months of age, microbiome of control animals and majority of KPC animals form a single cluster (circled), with apparent changes in some of the KPC animals (Figure 3A). By the third month of age, control animals maintained the cluster, one KPC animal shifted out of the circled cluster, accompanied by mortality of another KPC mice, which was outside the cluster from 2 months of age (Figure 3B). Finally, at 4 months of age, all KPC animals, except one, were seen to acquire a distinct microbial composition compared with the control animals (Figure 3C). At this point however, all KPC animals outside the cluster at 3 months of age had not survived. This phenomenon is also evident from the class-/genus-level heatmap of top 50% representative bacteria at 2- and 4-month-old control and KPC mice, where we do not see any distinct pattern between the two genotypes at 2 months of age (Supplementary Figure 2, available at Carcinogenesis online), whereas a distinct pattern can be seen emerging in three out of four KPC animals at 4 months of age, consistent with the pCoA findings in Figure 3C (also see Supplementary Figure 3, available at Carcinogenesis online). In general, the KPC mice seem to maintain a higher species richness and evenness of distribution, which is significantly higher in the third and fourth months of age (Figure 3D).
Figure 3.
Whole-genome sequencing of control and KPC mice at ages 2, 3 and 4 months. As seen above, the microbial composition of KPC animals, which is similar to control (circled) animals at 2 months age (A), starts changing by 3-month age (B). By 4 months, there are significant differences in the control and KPC microbiome (C). In this experiment, only four KPC animals survived for 4 months (arrows point to the individual animals which survived from third to fourth month). This is accompanied by significantly increased Shannon’s H alpha-diversity index for 3- and 4-month-old KPC animals, compared with their control counterparts (D). Although control animals do not exhibit change in microbial composition in pCoA plot with age (E), four surviving members of 4-month-old KPC animals were seen to cluster separately from the 2 or 3 months of age collections (F). The Shannon index was not different within control (G) or KPC (H) animals over time. In KPC animals, between 2 and 4 months age, 82 species were found to be significantly different (see Supplementary Figure 4, available at Carcinogenesis online). Analysis of those 82 species with pCoA plot shows tight clustering of 4-month-old animals, compared with when they were 2 months old (I). Neither the Shannon (J) index nor Chao1 (K) index was different for the two age groups, when analyzed for the significantly changing species only.
This prompted us to analyze same samples from Figure 3A–C and render them according to their genotypes (Figure 3E and F), where the colors in each panel represent their age in months. The microbiota of control animals, as evident from Figure 3A–C, does not alter from the second to the fourth month (Figure 3E). Although there is some variability among individual animals across the time points, there were no distinct clusters within the defined time points. In KPC animals however, the microbiome at 4-month age was distinctly and significantly different from the other age groups (Figure 3F). These figures together show that over months, Cre microbiome remains similar in composition, whereas KPC animals have drastic compositional changes as they age. As mentioned earlier in Figure 3D, KPC microbiome maintains a higher level of species richness and evenness. However, there were no significant changes in control (Figure 3G) or KPC (Figure 3H) mice across the time points measured. Hence, the differences in the microbial composition with age are not influenced by the richness/evenness component of evaluation.
Next, we concentrated on the significantly changed species within the 2- and 4-month KPC animals (Supplementary Figure 4A, available at Carcinogenesis online). A total of 89 species, across all classes and phyla, were found to be significantly up- or down-regulated with age. Among the significantly changing species, only seven were found to lose abundance (marked with arrows), with four belonging to phylum Firmicutes and three from Proteobacteria (all gammaproteobacteria). The remaining 82 species exhibited significantly expanded relative abundance from 2 to 4 months of age, represented by 14 different bacterial phyla (combined phylum representation in Supplementary Figure 4B, available at Carcinogenesis online). As shown, approximately 40% of these species belong to phylum Proteobacteria, followed by Firmicutes (18%), Actinobacteria, Bacteroidetes, Cyanobacteria and Euryarchaeota (all 6–7%). The significantly altered species in 4-month-old KPC animals were compositionally close and formed a tight cluster in a pCoA plot, compared with 2-month-old KPC animals (Figure 3I). This is despite no significant differences seen in the Shannon (Figure 3J) or Chao1 (Figure 3K) indices, implying that the relative abundances of the significantly changing species in 4-month-old KPC animals are influenced by the disease and not the richness or evenness of the microbiome. Lack of change in the KC and PDX-Cre animals ruled out age-induced change in the microbiome.
The metabolic landscape of the microbiota shows a shift from dominance of energy metabolism to upregulation in polyamine and lipid metabolism with PDAC tumor progression
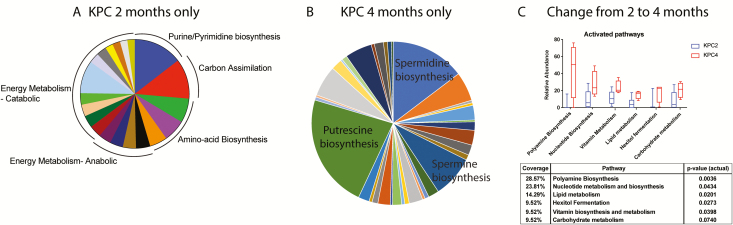
We then characterized the functional profile of the microbiota of 2- and 4-month-old KPC mice using the HUMAnN2 pipeline (32,38). The resulting pathways were classified and curated manually into (i) significant pathways in 2-month-old KPC only, (ii) significant pathways in 4-month-old KPC only and (iii) significantly changed pathways in 4-month-old KPC, compared with 2-month-old KPC animals (continuously valued relative abundance). We found that in 2 months old KPC, there is a distinct dominance of energy metabolism pathways in the 227 pathways identified (top 25 pathways depicted in Figure 4A). Within the top 25 pathways, apart from energy metabolism, pathways contributing to cell division (e.g., amino acid biosynthesis and nucleotide biosynthesis pathways) were adequately represented. However, in 4-month-old KPC animals, the landscape was found to be significantly dominated by polyamine biosynthesis (Figure 4B). Biosynthesis of polyamines Putrescine, Spermidine and Spermine dominated the metabolic landscape of the dysbiotic microbiome of 4-month-old KPC mice. The rest of the pathways were represented by metabolic shunt pathways and lipid metabolic pathways. Next, we looked into the differential upregulation of pathways between second- and fourth-month KPC animals (Figure 4C). We found a 29% upregulation of polyamine biosynthesis pathways in 4-month KPC animals, followed by 24% upregulation in nucleotide biosynthesis pathways. Essentially, >50% of bacterial metabolism seems to be promoting DNA synthesis/replication pathways, where the metabolic products are readily assimilated by the host (39–41). This is supported by a 14% upregulation of lipid biosynthetic pathways, important for de novo cell membrane synthesis/remodeling.
Figure 4.
Metabolic reconstruction of the microbiome. The HUMAnN2 pipeline generated the pathway abundance list from whole-genome sequencing input, and we manually curated it for predominantly active pathways found in (A) 2-month-old KPC only and (B) 4-month-old KPC only. While 2 months only was dominated by energy metabolism pathways among others, 4 months only was dominated by polyamine biosynthesis pathway. Overall, between the ages of 2 months and 4 months in KPC animals (C), the most significant metabolic pathways were dominated by biosynthetic pathways, where the majority of metabolites are exchanged between the host and the microbiota. Figure shows activated pathways in 4-month KPC animals compared with when they were 2 months old. Adjoining table shows the coverage and non-parametric 2-tailed P-value for each changing pathway.
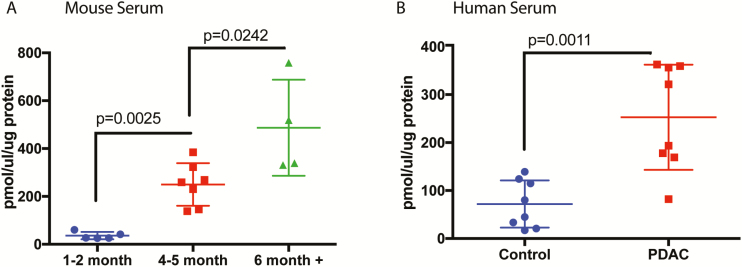
To validate our predictive metabolomics, we next estimated polyamines from serum samples of KPC animals at different stages of tumor progression. Our results showed that although there was low polyamine in the serum of the animals of 1- to 2-month age, there was a significant increase in the serum concentration of polyamines in 4-month animals that had PanIN2 and PanIN3 lesions but no observable tumor. Serum Polyamine concentration increased further in a full-tumor (≥6 months) mice (Figure 5A). To further validate this, we next estimated polyamines in the serum from PDAC patients and compared their concentration to that in serum from normal healthy volunteers. Our results showed that in PDAC patients, serum polyamine concentrations were significantly increased (Figure 5B).
Figure 5.
Serum polyamine levels are significantly higher in spontaneous murine model of PDAC and in PDAC patients. When we measured the actual total polyamine levels in KPC mice serum (A), significant elevation was seen with progressing age and cancer. Similarly, serum polyamines were found to be significantly elevated in PDAC patients, compared with healthy controls (B) (n = 5–8 for mice; n = 8 for human serum samples). Test of significance was two-tailed, non-parametric Mann–Whitney U test. P-values are exact and mentioned in the figure.
Discussion
The role of microbiome in disease development, progression and therapy is becoming increasingly clear. Although this became apparent in colon cancer and other colonic disorders such as inflammatory bowel disease where the bacterial population in the gut has direct influence on the inflammatory milieu of the disease, its influence in other diseases is only starting to be acknowledged (5,38,42). Changes in gut microbiome are now being associated with therapeutic response in melanoma (24,43), hepatocellular carcinoma (10) and even in PDAC (12,14). Although the current focus in microbiome research in PDAC has been primarily focused on therapy and its response with respect to microbial changes, there are no studies on the role of this dysbiosis with respect to disease development and progression. Earlier studies have revealed that changes in the salivary and oral microbiome may correlate with risk of PDAC (8). In a recent study by Ren et al., a miSeq analysis showed that some potential pathogens, such a Veillonella, Klebsiella and Selenomonas, and LPS-producing bacteria, including Prevotella, Hallella and Enterobacter, were enriched in pancreatic adenocarcinoma, whereas probiotics including Bifidobacterium and some butyrate-producing bacteria, such as Coprococcus, Clostridium IV, Blautia, Flavonifractor and Anaerostipes were reduced (44).
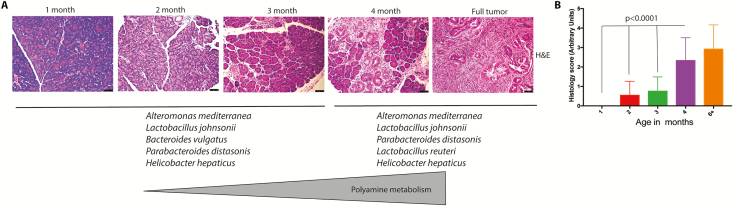
KPC mice have been a gold standard for understanding the progression of pancreatic tumors (45). In the present study, we found a distinct microbial dysbiosis between the microbiome of mice with no tumor at 1 month of age compared with mice with PDAC tumors at 6 months of age. To further resolve when the microbial dysbiosis occurs, we next collected and analyzed the fecal microbiome of 2- to 4-month-old mice that did not have overt tumors (but had histological indication of PanINs and some dysplasia). We gave an arbitrary dysplasia score in which histologically normal pancreases of 1-month-old mice were considered 0 and histological PDAC tumor-bearing mice were considered 4. Our results showed that microbial dysbiosis predominantly occurred at 4 months of age (mean histological score of 2.33), when there were no observable tumors in the animals (Figure 6). Our study corroborated the recent study by Pushalker et al. (12). We observed that there is a distinct difference in the microbial population as a pancreatic tumor develops (Figure 1). At this stage, a significant difference was observed at the class level, particularly for classes such as Clostridia, Bacilli and Erysipelotrichia, Actinobacteria, and Deferribacteres (Figure 2). Interestingly, when we further analyzed the microbiome of KPC mice at an early ‘pre-tumor’ stage in which there was no observable tumor (only histological indication), the microbial population seemed to emerge as a distinct pattern (Figure 3). The KPC animals appeared to maintain a higher species richness and evenness of distribution. Our analysis further showed that the significantly altered species in 4-month-old KPC animals were compositionally close and formed a tight cluster in a principal co-ordinate analysis despite no significant difference in the Shannon or Chao1 index, indicating that this difference was due to the progression of the disease (Figure 3). We also observed opposite trends in alpha-diversity matrices between initial 16s rRNA (Figure 1) and shotgun metagenomics analyses (Figure 3). This is due to differences in sequencing as well as analysis techniques. In 16s rRNA sequencing, the number of OTU hits are higher at 97% CI compared with shotgun sequencing; however, Genus-level ID is not >10–15% of all OTUs. Because alpha-diversity measures are mostly drawn from the sample-wise bacterial ID matrix, shotgun results tend to be more conservative than 16s rRNA pyrosequencing data and may have different outcomes. This has been discussed in refs (46) and (47), and it has even been contended that there is probably no linear comparability between the two measures (48). Although the alpha-diversity matrices between 16s rRNA sequencing and WGS are not reconcilable, qualitative compositional changes (beta diversity) were comparable between the sequencing methods. In this study, we can confidently say that WGS and WGS-derived metabolic reconstruction was more robust compared to 16s rRNA sequencing since the predicted outcomes were easily verifiable using biochemical assays (Figure 5, discussed below). Hence, it is perceivable that studies focusing on metabolic contribution of altered microbiome in disease pathogenesis should use WGS instead of 16s pyrosequencing.
Figure 6.
Progression to tumor and microbial/metabolic changes. With progressive cellular disorganization and tumor development in KPC mice (A; representative H&E staining), top bacterial species between 2 and 4 months are joined by Lactobacillus reuteri. Lactobacilli is known to actively participate in polyamine metabolism. (B) Blinded histological scoring of PDAC progression in KPC mice (n = 6/group with four fields per section]. Test of significance for (B) was non-parametric Mann–Whitney U test.
Microbial population is known to influence the host metabolism in a number of ways. Studies have shown that bile acid metabolism is one of the main metabolic pathways that are affected by microbial dysbiosis (29). Similarly, short-chain fatty acids that the exclusive products of microbial metabolism are known to affect the host epigenetic machinery (49). Furthermore, in the context of chemotherapy, microbes are known to differentially metabolize drugs to promote resistance of the tumor to cytotoxic agents (22). Our metagenomics analysis on the predictive metabolome of the microbial species shows that there is a distinct dominance of energy metabolism pathways (Figure 4). Interestingly, the major metabolic pathways that are deregulated from 2 to 4 months of age in KPC mice are those involved in polyamine biosynthesis and pyrimidine biosynthesis (Figure 4). It is well known that polyamine biosynthesis is critically regulated in a cell. This class of compounds is known to promote rapid proliferation by specifically contributing to the purine/pyrimidine biosynthesis in a cell and is thus considered a marker for neoplastic progression (50). Thus, the fact that our predictive metabolomics analysis shows this class of metabolites to be significantly deregulated in the pre-cancerous stage (prior to when invasive tumors start proliferating rapidly) is not surprising. This was further validated upon analysis of serum from KPC mice—mice that did not have observable tumors, but had PanINs, showed an increased serum polyamine concentration (Figure 5A and B). Interestingly, upon comparing the abundance of top five species as the KPC tumors progressed, we observed that the bacterial species, Lactobacillus reuteri, was detected in the 4-month sample, whereas it was below detection in the earlier time points (Figure 6). In our metabolomic reconstruction analysis, L.reuteri was associated with polyamine metabolism. It is reported that in gastric cancer Helicobacter pylori promotes DNA damage and neoplastic progression by deregulating polyamine metabolism and promoting oxidative stress in the gastric epithelial lining (51–53). Although members of Lactobacillus sp. are known to affect polyamine metabolism and growth inhibition in gastric cancer (52), their role in pancreatic cancer remains undefined. Consistent with this, we identified serum polyamine concentration was significantly higher in 4-month KPC samples (without observable tumors but with advanced PanINs) compared with the 2-month samples (Figure 5A).
In summary, early detection of PDAC has been an extremely challenging task. In this context, our results show for the first time that microbial dysbiosis and its altered metabolic pathways may potentially be exploited to develop a noninvasive biomarker panel. Furthermore, our results indicate a definitive shift in the microbial composition sufficiently early in the tumor development timeline. Thus, a comprehensive fecal analysis for the significantly changing bacterial species (as shown in Supplementary Figure 4, available at Carcinogenesis online), along with a serum analysis for pan-polyamines, can be further evaluated in the patient-derived samples.
Although based on spontaneous mouse models for PDAC, our study forms the first step toward understanding how the microbial dysbiosis during tumorigenesis can play a role in the metabolic regulation of active proliferation of tumor cells—by regulating polyamine metabolism and influencing purine/pyrimidine biosynthesis.
Funding
This study was partially funded by National Institutes of Health (NHLBI R21 HL125021 to Sa.B.), National Institutes of Health (NCI R01CA184274 to Su.B.) and University of Miami Institutional support to both. Sylvester Cancer center translational pilot award 2018 to Sa.B. The funders had no role in study design, collection, analysis or interpretation of data.
Supplementary Material
Acknowledgement
We would also acknowledge Prof. Ashok Saluja for infrastructural help and support.
Conflict of Interest Statement: University of Minnesota has a patent for Minnelide (WO/2010/129918/Triptolide Prodrugs), which has been licensed to Minneamrita Therapeutics, LLC. Dr. Sulagna Banerjee is a compensated consultant with Minneamrita Therapeutics LLC, and this relationship is managed by University of Miami. Rest of the authors declare no conflict of interest.
Glossary
Abbreviations
- OTU
operational taxonomic unit
- PanINs
pancreatic intraepithelial neoplasia
- pCoA
principle co-ordinate analysis
- PDAC
pancreatic ductal adenocarcinoma
- WGS
whole-genome shotgun sequencing
References
- 1. Chari S.T., et al. (2015) Early detection of sporadic pancreatic cancer: summative review. Pancreas, 44, 693–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hruban R.H., et al. (2000) Progression model for pancreatic cancer. Clin. Cancer Res., 6, 2969–2972. [PubMed] [Google Scholar]
- 3. Hruban R.H., et al. (2007) Precursors to pancreatic cancer. Gastroenterol. Clin. North Am., 36, 831–849, vi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fulbright L.E., et al. (2017) The microbiome and the hallmarks of cancer. PLoS Pathog., 13, e1006480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Salava A., et al. (2016) Skin microbiome in melanomas and melanocytic nevi. Eur. J. Dermatol., 26, 49–55. [DOI] [PubMed] [Google Scholar]
- 6. Mao Q., et al. (2018) Interplay between the lung microbiome and lung cancer. Cancer Lett., 415, 40–48. [DOI] [PubMed] [Google Scholar]
- 7. Ertz-Archambault N., et al. (2017) Microbiome and pancreatic cancer: a comprehensive topic review of literature. World J. Gastroenterol., 23, 1899–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fan X., et al. (2018) Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut, 67, 120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Farrell J.J., et al. (2012) Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut, 61, 582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mima K., et al. (2017) The microbiome and hepatobiliary-pancreatic cancers. Cancer Lett., 402, 9–15. [DOI] [PubMed] [Google Scholar]
- 11. Signoretti M., et al. (2017) Gut microbiota and pancreatic diseases. Minerva Gastroenterol. Dietol., 63, 399–410. [DOI] [PubMed] [Google Scholar]
- 12. Pushalkar S., et al. (2018) The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov., 8, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Riquelme E., et al. (2018) Immunotherapy for pancreatic cancer: more than just a gut feeling. Cancer Discov., 8, 386–388. [DOI] [PubMed] [Google Scholar]
- 14. Sethi V., et al. (2018) Gut microbiota promotes tumor growth in mice by modulating immune response. Gastroenterology, 155, 33–37.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zou S., et al. (2018) Dysbiosis of gut microbiota in promoting the development of colorectal cancer. Gastroenterol. Rep. (Oxf)., 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hullar M.A., et al. (2014) Gut microbes, diet, and cancer. Cancer Treat. Res., 159, 377–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Olson S.H., et al. (2017) The oral microbiota in patients with pancreatic cancer, patients with IPMNs, and controls: a pilot study. Cancer Causes Control, 28, 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li S., et al. (2017) Pancreatic cyst fluid harbors a unique microbiome. Microbiome, 5, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Human Microbiome Project Consortium (2012) Structure, function and diversity of the healthy human microbiome. Nature, 486, 207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qin J., et al. ; MetaHIT Consortium (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature, 464, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ranjan R., et al. (2016) Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun., 469, 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pouncey A.L., et al. (2018) Gut microbiota, chemotherapy and the host: the influence of the gut microbiota on cancer treatment. Ecancermedicalscience, 12, 868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cani P.D., et al. (2007) Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia, 50, 2374–2383. [DOI] [PubMed] [Google Scholar]
- 24. Alexander J.L., et al. (2017) Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol., 14, 356–365. [DOI] [PubMed] [Google Scholar]
- 25. Geller L.T., et al. (2018) Intratumoral bacteria may elicit chemoresistance by metabolizing anticancer agents. Mol. Cell. Oncol., 5, e1405139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Humphries A., et al. (2018) The gut microbiota and immune checkpoint inhibitors. Hum. Vaccin. Immunother., 14, 2178–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jaber D.F., et al. (2017) The effect of ciprofloxacin on the growth of B16F10 melanoma cells. J. Cancer Res. Ther., 13, 956–960. [DOI] [PubMed] [Google Scholar]
- 28. Caporaso J.G., et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat. Methods, 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Banerjee S., et al. (2016) Opioid-induced gut microbial disruption and bile dysregulation leads to gut barrier compromise and sustained systemic inflammation. Mucosal Immunol., 9, 1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Treangen T.J., et al. (2013) MetAMOS: a modular and open source metagenomic assembly and analysis pipeline. Genome Biol., 14, R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robertson C.E., et al. (2013) Explicet: graphical user interface software for metadata-driven management, analysis and visualization of microbiome data. Bioinformatics, 29, 3100–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abubucker S., et al. (2012) Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol., 8, e1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morgan X.C., et al. (2012) Chapter 12: human microbiome analysis. PLoS Comput. Biol., 8, e1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shannon C.E. (1997) The mathematical theory of communication. 1963. MD Comput., 14, 306–317. [PubMed] [Google Scholar]
- 35. Chiu C.H., et al. (2014) Distance-based functional diversity measures and their decomposition: a framework based on Hill numbers. PLoS One, 9, e100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Faith D.P. (2015) Phylogenetic diversity, functional trait diversity and extinction: avoiding tipping points and worst-case losses. Philos. Trans. R. Soc. Lond. B Biol. Sci., 370, 20140011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. He Y., et al. (2013) Comparison of microbial diversity determined with the same variable tag sequence extracted from two different PCR amplicons. BMC Microbiol., 13, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Golombos D.M., et al. (2018) The role of gut microbiome in the pathogenesis of prostate cancer: a prospective, pilot study. Urology, 111, 122–128. [DOI] [PubMed] [Google Scholar]
- 39. McKenna Iii J., et al. (2018) Metabolomic studies identify changes in transmethylation and polyamine metabolism in a brain-specific mouse model of tuberous sclerosis complex. Hum. Mol. Genet. 27, 2113–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhu Q., et al. (2012) Polyamine analogs modulate gene expression by inhibiting lysine-specific demethylase 1 (LSD1) and altering chromatin structure in human breast cancer cells. Amino Acids, 42, 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Johnson C.H., et al. (2016) Metabolite and microbiome interplay in cancer immunotherapy. Cancer Res., 76, 6146–6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petra C.V., et al. (2017) Gastric microbiota: tracing the culprit. Clujul Med., 90, 369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leslie M. (2017) Gut microbes may up PD-1 inhibitor response. Cancer Discov., 7, 448. [DOI] [PubMed] [Google Scholar]
- 44. Ren Z., et al. (2017) Gut microbial profile analysis by MiSeq sequencing of pancreatic carcinoma patients in China. Oncotarget, 8, 95176–95191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hingorani S.R., et al. (2005) Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell, 7, 469–483. [DOI] [PubMed] [Google Scholar]
- 46. Jovel J., et al. (2016) Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol., 7, 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hillmann B., Al-Ghalith G.A., Shields-Cutler R.R., Zhu Q., Gohl D.M., Beckman K.B., Knight R., Knights D (2018) Evaluating the information content of shallow shotgun metagenomics. mSystems, 3:e00069–18. doi:10.1128/mSystems.00069-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Clooney A.G., et al. (2016) Comparing apples and oranges? Next generation sequencing and its impact on microbiome analysis. PLoS One, 11, e0148028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Krautkramer K.A., et al. (2017) Metabolic programming of the epigenome: host and gut microbial metabolite interactions with host chromatin. Transl. Res., 189, 30–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Arruabarrena-Aristorena A., et al. (2018) Oil for the cancer engine: the cross-talk between oncogenic signaling and polyamine metabolism. Sci. Adv., 4, eaar2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Di Martino M.L., et al. (2013) Polyamines: emerging players in bacteria-host interactions. Int. J. Med. Microbiol., 303, 484–491. [DOI] [PubMed] [Google Scholar]
- 52. Russo F., et al. (2007) Effects of Lactobacillus rhamnosus GG on the cell growth and polyamine metabolism in HGC-27 human gastric cancer cells. Nutr. Cancer, 59, 106–114. [DOI] [PubMed] [Google Scholar]
- 53. Rasouli B.S., et al. (2017) In vitro activity of probiotic Lactobacillus reuteri against gastric cancer progression by downregulation of urokinase plasminogen activator/urokinase plasminogen activator receptor gene expression. J. Cancer Res. Ther., 13, 246–251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microbiome raw data sequences are available from EMBL ArrayExpress with the accession number E-MTAB-6921.