Abstract
Using the Allplex™ 2019-nCoV assay (Seegene, South Korea), 285 samples were tested; 49 (17%) were positive for 3 genes, 4 (1.4%) samples were positive for 2 genes (all N gene and RdRP gene), 8 (3%) samples were positive for 1 gene (all N gene only), and 224 (78.5%) samples were negative.
Keywords: SARS-CoV-2, Covid-19, PCR, Coronavirus
Highlights
-
•
All patients with detected PCR result were positive for all 3 gene targets.
-
•
12 of 61 positive staff were detected in only 1 or 2 of the 3 gene targets.
-
•
Positive results with high cycle threshold values were only seen in healthcare workers.
1. Introduction
SARS-CoV-2 is a novel coronavirus that has emerged in the last year, leading to a worldwide pandemic of Covid-19 disease (Ciotti et al., 2020; Kong et al., 2020). A significant volume of work has been invested in trying to provide rapid polymerase chain reaction (PCR) diagnostics to facilitate laboratory diagnosis of symptomatic patients and to develop an understanding of viral load dynamics (To KK et al., 2020; Xu et al., 2020). A feature of this pandemic has been the high numbers of healthcare workers who have acquired the disease (Chirico et al, 2020). Identifying and testing these healthcare workers have been essential to prevent nosocomial transmission of Covid-19 (Hoe Gan et al, 2020). Using PCR as a way to screen staff to ensure that they are no longer infectious has been proposed, but this is difficult as there can be variation in sampling technique and levels of detectable virus in the nasopharynx as the disease progresses, and also, it is unclear if the staff member remains infectious while low levels of virus are detected in the nasopharynx following clinical recovery (Ferrari et al., 2020; Xiao et al, 2020).
The aim of this retrospective study was to review SARS-CoV-2 PCR results of patients and staff using the Allplex™ 2019-nCoV assay (Seegene, South Korea) to increase knowledge around the expected cycle threshold value for PCR testing. This was not a diagnostic accuracy or validation study. The clinical details were sought from patients and staff if their result was considered a weak positive, i.e., only positive for 1 or 2 of the 3 genes tested.
2. Materials and methods
This was a retrospective review of SARS-CoV-2 PCR tests performed in the Rotunda Hospital. Only tests from staff and adult patients at the Rotunda Hospital and staff from Children's Health Ireland (CHI) at Temple Street taken between 24th March 2020 and 15th April 2020 were reviewed. All patients and staff were aged over 18 years. The Rotunda Hospital is a standalone tertiary-level care maternity hospital in the center of Dublin, Ireland, and the CHI at Temple Street is a standalone pediatric tertiary referral hospital. Testing was only performed on symptomatic patients and staff at the discretion of either occupational health department or the patient's clinician. At this time, there was no asymptomatic screening taking place for either staff or patients. As a consequence, a detected result at any cycle threshold value was considered to be positive, and the staff member was excluded for 14 days and contact tracing was performed. Confirmation on a second platform was not performed. Only the first PCR test per adult patient was included, and all samples were combined nasopharyngeal/oropharyngeal swabs. The eNAT (Copan, USA) flocked swabs for collection and preservation of nucleic acids were used for sampling. Extraction was performed using the Nimbus platform (Hamilton, USA), and then PCR was performed using the Allplex™ 2019-nCoV assay (Seegene, South Korea) on the CFX96 (Bio-Rad, USA) in line with manufacturer's instructions. The gene targets for the PCR assay were the N gene, the E gene, and the RdRP gene. The cycle threshold (Ct) value was recorded for each of the 3 genes. Samples with a detected result for all 3 genes, or a single target detected just the RdRP or N gene, were interpreted as SARS Cov2 PCR positive in line with manufacturer's guidance. For each sample, it was also recorded if the person was a patient or staff member, and for samples with only 1 or 2 of the genes detected, clinical symptoms were also recorded. This was approved by the Ethics committees of the Rotunda Hospital and the CHI at Temple Street as a retrospective audit (Rotunda ethics approval #RAG-2020-009; CHI at Temple Street approval #20.025).
3. Results
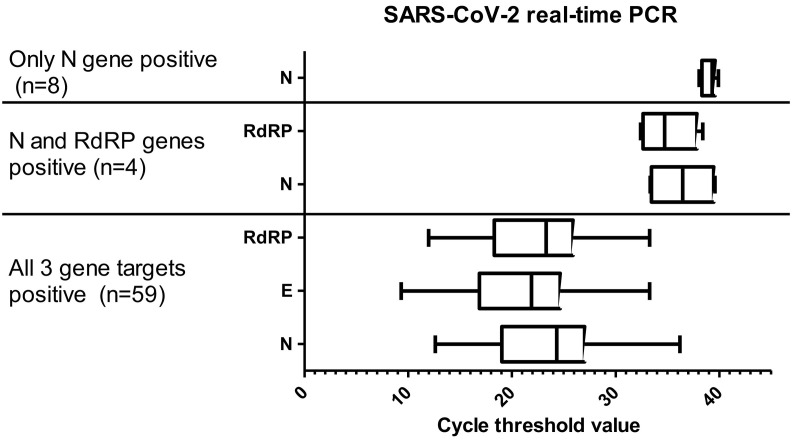
Over the 3-week period, 358 samples from the Rotunda Hospital and CHI at Temple Street were tested. Ten duplicate samples and 2 pediatric samples were removed, leaving a final list of 346 samples for analysis. Sixty-one (17%) were from adult patients with a median age of 35 years (range 15–52 years). The remaining 285 (83%) were from staff with a median age of 39 years (range 18–67 years). Of the 61 patients, 10 (16%) were positive for all 3 gene targets, and 51 (84%) were negative for all 3 gene targets. Of the 285 staff samples, 49 (17%) were positive for 3 gene targets, 4 (1.4%) samples were positive for 2 genes (all N gene and RdRP gene), 8 (3%) were positive for 1 gene (all N gene only), and 224 (78.5%) samples were negative. All positive healthcare workers were excluded from work for 14 days, and contact tracing was done in line with national guidelines. The available clinical details of these staff with 1 or 2 genes positive are shown in Table 1 . The median Ct value for the N gene was lower in samples that were positive for all 3 genes (median Ct 24, range 12–36) than in samples that were positive for 2 genes (median Ct 36) and just 1 gene (median Ct 39) (Fig. 1 ).
Table 1.
Available clinical details of healthcare workers that had only 1 or 2 genes detected in the Allplex™ 2019-nCoV assay (Seegene, South Korea).
| N gene (Ct) | RdRP gene (Ct) | Available clinical details of healthcare worker at time of testing |
|---|---|---|
| 2 genes positive (N and RdRP) | ||
| 39.1 | 36.15 | Previously positive Covid-19 case |
| 33.34 | 32.41 | Febrile day 10 post close Covid-19 contact |
| 39.58 | 38.39 | Fever and cough. Tested on day 6 of symptoms |
| 33.82 | 33.31 | Persistent cough for >7 days and known Covid-19 close contact |
| 1 gene positive (N gene only) | ||
| 38.23 | - | Previously positive Covid-19 case |
| 39.32 | - | Persistent cough for >7 days |
| 39.57 | - | Persistent cough for >7 days |
| 38.61 | - | Persistent cough for >7 days |
| 39.6 | - | Day 1 sore throat and fever, but day 12 post close Covid-19 contact |
| 39.29 | - | Day 2 chills, fatigue, and headache |
| 38.05 | - | Day 2 pyrexia, cough, and fatigue and day 4 post close Covid19 contact |
| 39.91 | - | Day 2 sore throat |
Fig. 1.
Cycle threshold value for detected results for SARS-CoV-2 PCR by number of genes detected in all positive patients and staff members.
4. Discussion
This study has shown that, overall, the positivity rate for SARS-CoV-2 by PCR in our population was 16% in patients and 17% in staff, which is a similar level to a study on staff in the United Kingdom (Keeley et al., 2020). All the patients had Ct values <36 in all 3 genes, likely reflecting that they are true positives as they were symptomatic as well. However, of the 61 staff with a detectable gene in the assay, 12 (20%) of them were positive in only 1 or 2 genes at a high Ct value. This 20% frequency of high Ct value results poses a difficulty when interpreting the potential infectivity of a staff member and whether or not they can return to work safely. As testing expands to include asymptomatic people presurgery, consideration should be given to having an indeterminate cutoff, after which positive results should be confirmed on a second platform.
The reason for these high Ct values is not clear, but in our study, we have detailed the symptoms recorded at the time of testing. For patients with 2 gene targets detected (N and RdRP genes), these seem to be clinically significant results as the patients were either known positives or symptomatic with a close contact history. For patients with just the N gene detectable, there appears to be 2 groups. The first has had symptoms for more than 7 days, and so the weak positive result may reflect degraded viral RNA and clearance of the virus. The second group was swabbed on the first or second day of their illness. Perhaps if they were retested 2 days later, the viral load may be higher and they may have had a lower Ct value result for each of the 3 genes. Further work is needed to determine if virus detected at these high cycle threshold values is viable and can lead to cross-transmission.
5. Conclusions
The interpretation of detected results with high Ct values needs to be done in the context of the clinical situation and timing of testing relative to symptoms or exposure.
Acknowledgments
The authors would like to thank the occupational health nurses, laboratory scientists, and the infection control midwives/nurses from the Rotunda Hospital and Temple Street for their help with data collection.
References
- Chirico F., Nucera G, Magnavita N. COVID-19: protecting healthcare workers is a priority. Infect Control Hosp Epidemiol. 2020:1–4. doi: 10.1017/ice.2020.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciotti M., Angeletti S., Minieri M., Giovannetti M., Benvenuto D., Pascarella S. COVID-19 outbreak: an overview. Chemotherapy. 2020;1-9 doi: 10.1159/000507423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari D., Motta A., Strollo M., Banfi GLocatelli M. Routine blood tests as a potential diagnostic tool for COVID-19. Clin Chem Lab Med. 2020;58(7):1095–1099. doi: 10.1515/cclm-2020-0398. [DOI] [PubMed] [Google Scholar]
- Hoe Gan W., Wah Lim J, Koh D. 2020. Preventing intra-hospital infection and transmission of COVID-19 in healthcare workers. Saf Health Work. [Google Scholar]
- Keeley A.J., Evans C., Colton H., Ankcorn M., Cope A., State A. Roll-out of SARS-oV-2 testing for healthcare workers at a large NHS Foundation Trust in the United Kingdom, March 2020. Euro Surveill. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.14.2000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W.H., Li Y., Peng M.W., Kong D.G., Yang X.B., Wang LLiu M.Q. 2020. SARS-CoV-2 detection in patients with influenza-like illness. Nat Microbiol. [DOI] [PubMed] [Google Scholar]
- To KK, Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A.T., Tong YX, Zhang S. False-negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: rather than recurrence. J Med Virol. 2020 doi: 10.1002/jmv.25855. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Chen C., Zhu Z., Cui M., Chen C., Dai HXue Y. Clinical features and dynamics of viral load in imported and non-imported patients with COVID-19. Int J Infect Dis. 2020;94:68–71. doi: 10.1016/j.ijid.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]