Summary
Dengue virus (DENV) and Zika virus (ZIKV), members of the Flavivirus genus, rearrange endoplasmic reticulum membranes to induce invaginations known as vesicle packets (VPs), which are the assumed sites for viral RNA replication. Mechanistic information on VP biogenesis has so far been difficult to attain due to the necessity of studying their formation under conditions of viral replication, where perturbations reducing replication will inevitably impact VP formation. Here, we report a replication-independent expression system, designated pIRO (plasmid-induced replication organelle formation) that induces bona fide DENV and ZIKV VPs that are morphologically indistinguishable from those in infected cells. Using this system, we demonstrate that sequences in the 3′ terminal RNA region of the DENV, but not the ZIKV genome, contribute to VP formation in a non-replicative manner. These results validate the pIRO system that opens avenues for mechanistically dissecting virus replication from membrane reorganization.
Keywords: flavivirus, vesicle packet, replication organelle, organelle biogenesis, membrane invagination, membranous organelle, viral replicase, replication complex
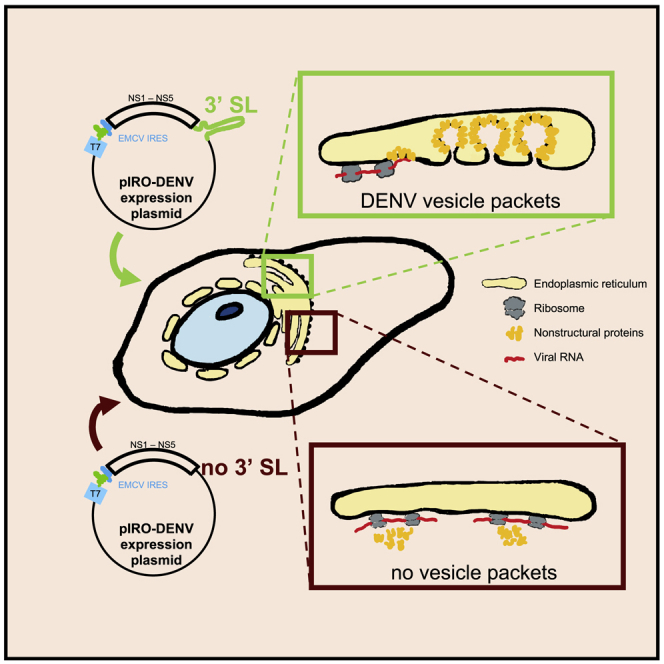
Graphical Abstract
Highlights
-
•
A replication-independent system to study DENV/ZIKV replication organelle formation
-
•
This system is called pIRO (i.e., plasmid-induced replication organelle formation)
-
•
Replication organelles induced with pIRO system analogous to those in infected cell
-
•
Non-replicative role of 3′ terminal RNA elements in organelle formation
Cerikan et al. devise an RNA replication-independent expression system designated pIRO (plasmid-induced replication organelle formation) phenocopying DENV/ZIKV-induced vesicle packets (VPs), the viral replication organelle. The authors find that RNA elements residing in the 3′ untranslated region of either virus genome are required for VP generation.
Introduction
Dengue virus (DENV) and Zika virus (ZIKV) belong to the Flaviviridae family, which consists of a large group of positive-strand RNA viruses (Neufeldt et al., 2018). Dengue is the most prevalent mosquito-borne viral disease, which, infecting up to 100 million people annually, is considered a global health problem (Stanaway et al., 2016). DENV has four distinct, but closely related, serotypes (DENV1–DENV4). Neutralizing antibodies for one serotype can exacerbate infection with a heterologous serotype, which can manifest as life-threatening dengue hemorrhagic fever or dengue shock syndrome (Bhatt et al., 2013; Rajapakse, 2011). ZIKV, first described to infect humans in 1954, has only recently become a global health concern following a large-scale epidemic that occurred in French Polynesia and the South Pacific in 2013 and 2014 (Cao-Lormeau et al., 2014; Faye et al., 2014; MacNamara, 1954). Importantly, these novel ZIKV strains were linked to the Guillain-Barré syndrome in adults and multiple neurodevelopmental defects, including microcephaly in neonates born to mothers infected during early stages of pregnancy (Wikan and Smith, 2016). Despite the high medical relevance, flavivirus-specific antiviral drugs are not available. Additionally, there is no vaccine for ZIKV, and the only DENV-approved vaccine has limited efficacy and depends on the baseline serostatus of the vaccine recipient (Sridhar et al., 2018).
Upon infection, flavivirus RNA genomes are released into the cytoplasm through a fusion event with endosomal membranes. For DENV and ZIKV, the viral genomic RNA shares a similar overall organization with a single long open reading frame, encoding for a polyprotein that is post- and co-translationally cleaved into structural and nonstructural (NS) proteins. The open reading frame is flanked by highly structured 5′ and 3′ untranslated regions (UTRs), which contribute to genome replication, protein production, and assembly of new virus particles (Wang et al., 2017). Within the 5′ UTR, there are several conserved stem-loop (SL) structures—including SL A (SLA), which serves as a viral polymerase binding site, and SL B (SLB), which contains the 5′ UAR (upstream of AUG region)—involved in long-range RNA-RNA interactions and genome replication (Figure 1A) (Alvarez et al., 2005a; Göertz et al., 2018; Yu et al., 2008). The 5′ UARs along with the 5′ cyclization sequence (CS) are responsible for genome circularization by hybridizing with their counterparts in the 3′ UTR, a process that is required for transferring the viral polymerase from the 5′ SLA to the 3′ end to initiate genome replication (Gebhard et al., 2011; Göertz et al., 2018; Villordo and Gamarnik, 2009).
Figure 1.
Expression of the Minimal DENV Replicase Does Not Suffice to Induce VP Formation
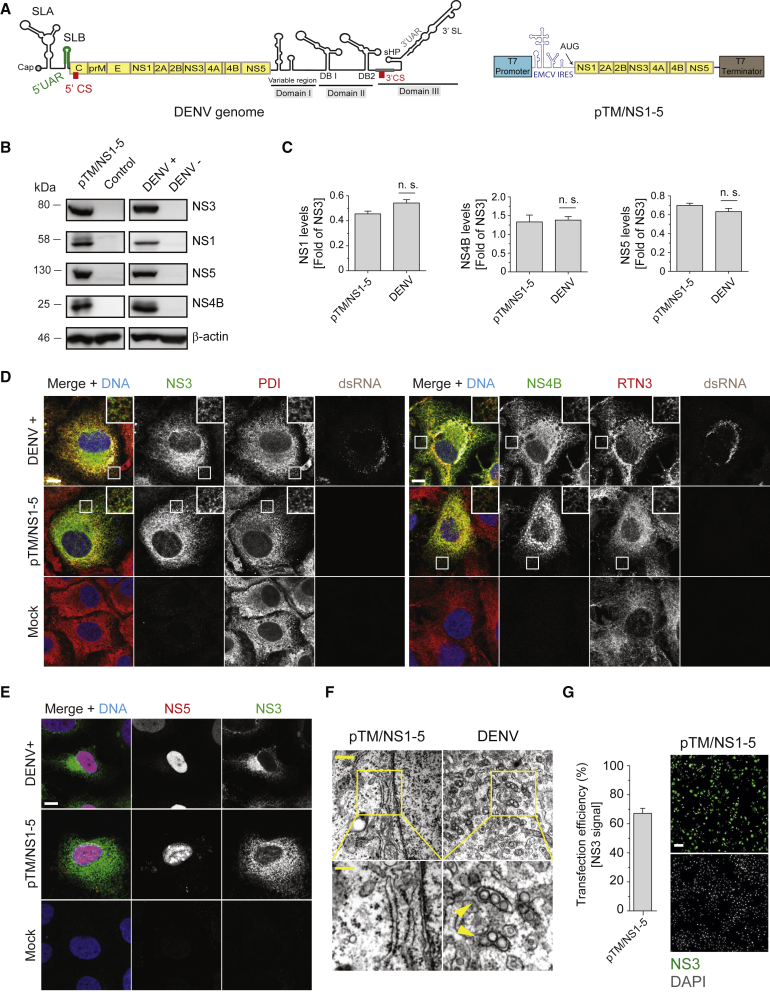
(A) Schematic representation of the DENV genome organization (left) and the T7 RNA polymerase-driven expression construct encoding the minimal DENV replicase NS1-5 (pTM/NS1-5; right panel). SL, stem-loop; UAR, upstream of AUG region; CS, cyclization sequence; DB, dumbbell; sHP, short-harpin.
(B) Huh7/Lunet-T7 cells were either transfected with pTM/NS1-5 for 20 h or infected with DENV (MOI = 5) for 48 h before being lysed and subjected to western blot analysis (left and right panels, respectively). β-actin was used as loading control.
(C) Relative abundance of viral proteins was determined by densitometry of the western blots, and values obtained for NS1, NS4B, or NS5 were normalized to NS3 expression levels. Values represent mean and standard error of three independent experiments. n.s., not significant.
(D) Cells were infected with DENV (upper row), transfected with the pTM/NS1-5 construct (middle row), or left untreated (bottom row) and fixed for immunofluorescence analysis after 48 h (infection) or 20 h (transfection and mock). PDI (protein disulfide isomerase) and RTN3 (reticulon 3) signals serve as ER makers. Scale bars: 10 μm.
(E) Nuclear localization of NS5 in DENV-infected or pTM/NS1-5-transfected cells. Scale bar: 10 μm.
(F) DENV-infected or pTM/NS1-5-transfected cells were fixed after 48 h or 20 h, respectively, and processed for TEM analysis. Lower panels are magnifications of areas highlighted with yellow squares in the upper panels. Yellow arrowheads show individual VPs. Scale bars: 500 and 200 nm in upper and lower panels, respectively.
(G) Transfection efficiency as determined by immunofluorescence staining of NS3. Error bars (left graph) represent the SEM of three independent experiments. Scale bar: 100 μm.
The ∼450-nucleotide-long 3′ UTRs of DENV and ZIKV can be divided into three main domains (Gebhard et al., 2011; Göertz et al., 2018) (Figure 1A). Domain I is the least conserved within the 3′ UTR, having significant variation in sequence and length among the four DENV serotypes (Aquino et al., 2006; Roche et al., 2007; Rossi et al., 2012; Shurtleff et al., 2001; Silva et al., 2008). Domain II consists of two RNA secondary structures, known as dumbbell elements (DB1 and DB2), which might play a role in viral RNA translation, genome replication, and/or species tropism (de Borba et al., 2019; Funk et al., 2010; Gritsun and Gould, 2006; Hahn et al., 1987; Manzano et al., 2011; Olsthoorn and Bol, 2001; Romero et al., 2006). RNA structures located within domains I and II were also shown to be non-essential enhancers of viral RNA replication (Alvarez et al., 2005b; Bredenbeek et al., 2003; Lo et al., 2003; Mandl et al., 1998; Men et al., 1996). Domain III, containing the 3′ CS element, a small hairpin (sHP) structure and the 3′ UAR (Figure 1A), is the most highly conserved region of the 3′ UTR and is indispensable for both DENV and ZIKV RNA replication (Brinton and Miller, 2015). The 3′ UAR and sHP are also part of the 3′ SL (3′ SL), which is essential for the initiation of viral genome replication (Filomatori et al., 2011; Gebhard et al., 2011; Ng et al., 2017; You et al., 2001).
Expression of the viral NS proteins, either in the context of virus infection or in cells containing sub-genomic replicons, leads to reorganization of the host endomembrane system to generate viral replication organelles (ROs). For flaviviruses, these ROs are composed of arrays of vesicle-like invaginations into rough endoplasmic reticulum (ER) membranes, originally designated vesicle packets (VPs) (Cortese et al., 2017; Mackenzie et al., 1996; Welsch et al., 2009). The detection of double-stranded RNA (dsRNA), an intermediate of viral genome replication, and viral replication components inside these vesicles suggest that VPs are the site of RNA genome replication (den Boon and Ahlquist, 2010; Cortese et al., 2017; Welsch et al., 2009). Each vesicle has a diameter of ∼85 nm and is surrounded by an outer ER membrane. The interior of these vesicles is connected to the cytosol through an ∼11-nm pore-like opening, presumably allowing the exchange of metabolites and other molecules required for RNA replication, as well as facilitating the release of newly synthesized viral RNA into the cytosol. In addition to VPs, in specific cell types, DENV and ZIKV induce bundled, smooth ER-derived structures known as convoluted membranes (CMs). These structures contain viral proteins, but no dsRNA, and are proposed to be involved in either polyprotein maturation or viral protein and lipid storage (Mackenzie et al., 1996; Neufeldt et al., 2018; Welsch et al., 2009).
While the morphology and architecture of the DENV and ZIKV ROs are well defined, the requisite host or virus factors, as well as the mechanisms governing the biogenesis of VPs, are poorly understood. Several viral proteins have been hypothesized to function in VP formation, but specifically evaluating their role in this process has not been possible because experimental perturbations of these factors cause replication defects in the context of virus infection or replicon cells. Additionally, studies using single viral protein expression have provided useful information about membrane-bending activity; however, these approaches fail to phenocopy VP formation. In order to study the mechanisms and factors involved in VP biogenesis, a replication-independent system is required. Here, we report a novel expression-based system for DENV and ZIKV that overcomes this limitation. We show that VPs formed with this system are indistinguishable from those generated in infected cells. In addition, we demonstrate the value of this system by showing that RNA elements in the 3′ UTR of the DENV and ZIKV genomes play non-replicative roles in RO biogenesis for these viruses.
Results
Expression of the DENV Replicase Polyprotein Is Not Sufficient to Induce VP Formation
With the goal of creating a DENV expression system that supports the formation of the viral RO in a replication-independent manner, we created a subgenomic DENV (sgDENV) expression construct, encoding the polyprotein region that encompasses the viral replicase (NS1-5). Considering that DENV replicates in the cytoplasm, we used the previously described T7 RNA polymerase-based system pTM (Moss et al., 1990) that allows RNA synthesis from the transfected plasmid DNA in the cytoplasm and avoids any undesired RNA modification such as splicing, which would impair viral protein production. To ensure robust, cap-independent RNA translation, we also added an encephalomyocarditis virus (ECMV)-derived internal ribosome entry site (IRES) (Palmenberg et al., 1984). This construct, designated pTM/NS1-5, (Figure 1A) was expressed in Huh7/Lunet-T7cells, which stably express the T7 RNA polymerase, and the resulting polyprotein expression and cellular membrane alterations were evaluated. Western blot analysis of transfected or DENV-infected cells, the latter serving as reference, revealed comparable polyprotein processing and DENV protein abundance (Figures 1B and 1C). Moreover, the subcellular localization of NS3, NS4B, and NS5 was comparable between pTM/NS1-5-transfected and DENV-infected cells (Figures 1D and 1E).
In order to determine if pTM/NS1-5 expression induced VPs, we analyzed the ultrastructure of transfected cells using thin-section transmission electron microscopy (TEM). Despite comparable levels and subcellular localization of DENV proteins, expression via pTM/NS1-5 failed to induce the formation of VPs that were abundant and readily detected in DENV-infected cells (Figure 1F). The absence of VPs in pTM/NS1-5-transfected cells was not due to transfection efficiency, which was >60% (Figure 1G). Therefore, we concluded that the sole expression of the NS1-5 polyprotein fragment is not sufficient to induce DENV ROs.
Formation of DENV ROs Requires Elements from the UTRs of the RNA Genome
The lack of VP formation in pTM/NS1-5-expressing cells suggested that in addition to viral proteins, viral RNA elements might be required for VP formation. Indeed, previous studies with Flock house virus (FHV) or alpha viruses have indicated that VP size and morphology can be affected by RNA length (Ertel et al., 2017; Kallio et al., 2013). In order to test the role of RNA elements in DENV VP production, we added the viral 5′ UTR and 3′ UTR sequences to the NS1-5 coding region, giving rise to the expression construct pTM/5′UTR/NS1-5/3′UTR DENV (Figure 2A). In addition, we created deletion mutations removing specific secondary structure elements in the 5′ UTR to identify possible RNA determinants for VP formation (Figure 2B). Each of these constructs was transfected into Huh7/Lunet-T7 cells, and protein expression as well as polyprotein processing was analyzed by western blot. In all 5′ UTR-mutant-expressing cells, the abundance of cleaved viral proteins was similar to that produced in infected cells (Figures 2C and 2D). As expected, the constructs lacking the polymerase binding SLA element (nucleotides 1–70 of the DENV genome) were replication incompetent (Figure S1). Interestingly, replication was also not detected following transfection with the 5′ wild-type (WT) construct that contains the complete 5′ UTR, which could be due to the heterologous IRES that might interfere with replication competence.
Figure 2.
Induction of VPs upon Expression of the Minimal DENV Replicase Requires Sequences in the 3′ UTR but Not the 5′ UTR
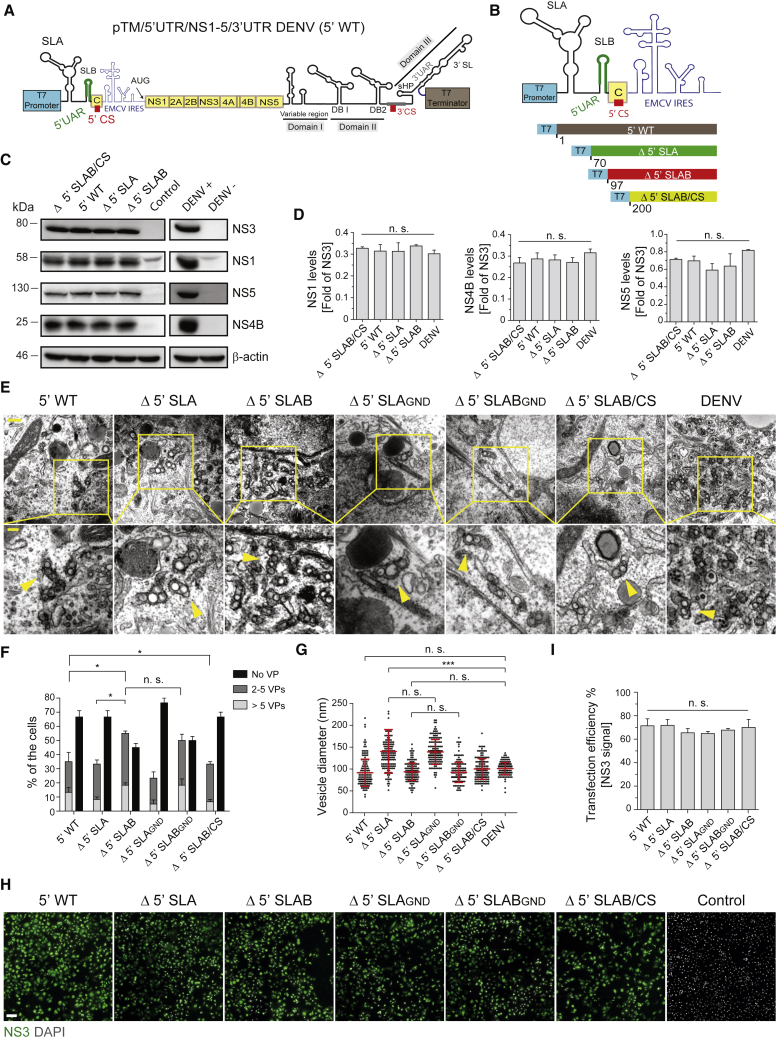
(A) Schematic of the pTM/5′UTR/NS1-5/3′UTR DENV expression plasmid (abbreviated 5′ WT).
(B) Schematic representation of 5′ UTR truncations introduced into the construct shown in (A). Numbers below each construct refer to the position in the DENV-2 genome (GenBank accession number NC_001474.2).
(C) Immunoblot showing the expression of NS3, NS1, NS5, and NS4B in cells 20 h after transfection with constructs specified on the top (left panel) or 48 h after DENV infection (right panel). β-actin served as sample loading control.
(D) Relative abundance of viral proteins was determined by densitometry and levels of NS1, NS4B, and NS5 normalized to NS3 signal. Values represent mean and standard error of three independent experiments. n.s., not significant.
(E) Thin-section TEM images of VPs induced upon transfection of indicated constructs. Cells were transfected or infected with DENV for 20 h and 48 h, respectively, before being processed for EM analysis. GND indicates the NS5 polymerase-inactivating mutation. Lower panels are magnified views of areas indicated with yellow squares in the upper panel images. Yellow arrowheads indicate individual VPs. Scale bars: 500 nm and 200 nm (upper and lower panels, respectively).
(F) For each condition, whole-cell sections of 20 cells were counted. Means ± SEM from three independent quantifications are given. n.s., not significant (∗p < 0.05).
(G) Vesicle diameters were measured manually using Fiji software. Means ± SEM were based on three independent measurements. For each condition, 50 vesicles were counted per experiment. n.s., not significant; ∗∗∗p < 0.001.
(H) Detection of NS3 by immunofluorescence microscopy to determine transfection efficiency for the indicated constructs. Scale bar: 100 μm.
(I) Transfection efficiencies were quantified according to NS3 staining. Error bars represent the standard error of three independent experiments. n.s., not significant.
Note that percentages given in (F) refer to all cells analyzed by TEM, but only ~70% of them had been transfected (I); therefore, not more than 70% of cells can contain VPs.
We next determined whether constructs containing the complete 5′ and 3′ UTRs were able to induce VPs and whether this property is affected by distinct deletions in the 5′ UTR (Figures 2E–2G). Huh7/Lunet-T7 cells were transfected with the different expression constructs and, 20 h later, were fixed and processed for immunofluorescence and TEM. Transfection efficiencies, determined by NS3 expression, were comparable between the constructs (Figures 2H and 2I). For VP quantification by TEM, we defined three classes of cells: cells that do not contain VPs, cells containing from two to five VPs, and cells containing more than five VPs. To avoid false positives, cells containing only one VP were not included in the quantifications. A VP was defined as either a single ER invagination vesicle or an array of ≥2 vesicles residing in the same ER lumen. Of note, VP formation was observed in cells expressing constructs with the complete 5′ and 3′ UTRs, suggesting that RNA elements in the viral genome facilitate VP formation (5′ WT in Figures 2E and 2F). Interestingly, each of the mutants, including the complete 5′ UTR deletion, was able to induce VP formation (Figures 2E and 2F), with the expression of the Δ 5′ SLAB construct inducing VP formation with the highest number of the cells (Figure 2F). Importantly, with the exception of the Δ 5′ SLA construct, the diameter of induced vesicles was comparable to the diameter of vesicles in DENV-infected cells (Figures 2E–2G).
To confirm that VP formation in this system was independent of RNA replication, we inserted a mutation converting the GDD active site of the RNA-dependent RNA polymerase to GND (Δ 5′ SLAGND and Δ 5′ SLABGND). We observed no significant differences in VP number between the constructs encoding WT NS5 and the corresponding GND mutation (Figures 2E–2G). Taken together, these results demonstrate that the expression of a DENV NS1-5 polyprotein translated from an RNA containing at least the 3′ UTR is sufficient to induce VP formation. Moreover, the data show that genome cyclization is not required for RO biogenesis because the construct lacking the 5′ CS element still supported VP formation, although the percentage of cells forming VPs after transfection of this construct was lower compared to cells transfected with the WT. Since the highest number of VPs per cell was obtained with the Δ 5′ SLAB construct (Figures 2B and 2F), we used it as parental construct for subsequent analyses of DENV RO biogenesis. This system was designated pIRO (plasmid-induced RO formation) and specifically for DENV will be referred as pIRO-D.
Involvement of ZIKV RNA Elements in RO Biogenesis and Generation of the pIRO-Z System
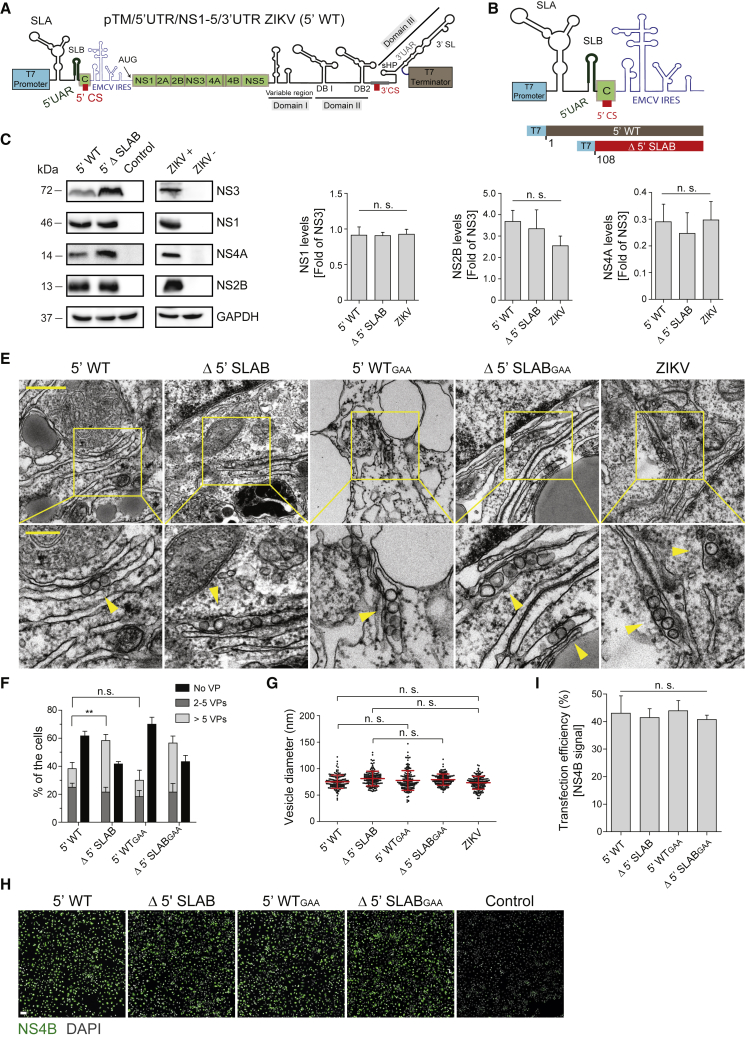
Given the role of RNA elements in DENV VP formation even under non-replicative conditions, we next aimed to determine whether this feature is conserved among flaviviruses. Employing a similar strategy to that used for DENV, we generated ZIKV constructs derived from the Asian H/PF/2013 strain genomic sequences (Münster et al., 2018). These constructs encoded for the ZIKV NS1-5 polyprotein and contained the 3′ UTR as well as the full-length or a truncated 5′ UTR, the latter corresponding to the most efficient DENV construct (5′ WT or Δ 5′ SLAB, respectively) (Figures 3A and 3B). Abundance and processing of the ZIKV polyprotein in transfected cells was comparable to ZIKV-infected cells (Figures 3C and 3D). Likewise, subcellular localization of NS3, NS4B, and NS5 was comparable between transfected and infected cells (Figures S2A and S2B). TEM analysis showed that 5′ WT and Δ 5′ SLAB constructs both induce VP formation (Figure 3E). Importantly, VP formation was not diminished by genetic ablation of NS5 polymerase activity in either construct (5′ WTGAA and Δ 5′ SLABGAA), demonstrating replication-independent VP formation in our system (Figures 3E and 3F). Diameters of vesicles induced by expression of 5′ WT, Δ 5′ SLAB, and the corresponding polymerase-dead mutants were similar to those in ZIKV-infected cells (Figure 3G). Comparable to the observations made for DENV (Figure 2F), expression of the ZIKV Δ 5′ SLAB construct yielded the highest percentage of cells forming VPs (Figure 3F), which was not due to higher transfection efficiencies (Figures 3H and 3I). Taken together, our data suggest that viral NS proteins, as well as RNA elements residing in the 3′ UTR, contribute to ZIKV and DENV RO formation in a replication-independent manner. Given that the highest number of cells forming VPs was observed upon transfection of the ZIKV Δ 5′ SLAB construct, it was used for all subsequent analyses and formed the basis of the pIRO-Z system.
Figure 3.
ZIKV 5′ UTR Is Dispensable for Expression-Based Induction of VPs
(A and B) Schematic of the pTM/5′UTR/NS1-5/3′UTR ZIKV expression plasmid (abbreviated as 5′ WT) (A) and the 5′ UTR deletion introduced into this construct (B). The number in the bottom refers to position in the ZIKV H/PF/2013 genome (GenBank accession number KJ776791.2).
(C) Immunoblot showing abundance of ZIKV NS3, NS1, NS4A, and NS2B in transfected and infected (MOI = 10) cells (left and right panel, respectively). GAPDH served as sample loading control.
(D) Relative abundance of ZIKV proteins was determined by densitometry and levels of NS1, NS2B, and NS4A normalized to NS3 signal. Values represent mean and standard error of three independent experiments. n.s., not significant.
(E) Thin-section TEM images showing VPs induced upon transfection of constructs specified on the top. Cells were transfected or infected with ZIKV for 18 h and 24 h, respectively, before being processed for EM analysis. GAA indicates the NS5 polymerase-inactivating mutation. Lower panels are magnified views of areas indicated with yellow squares in the upper panel images. Yellow arrowheads indicate individual VPs. Scale bars: 500 nm and 200 nm (upper and lower panels, respectively).
(F) For each condition, VPs contained in whole-cell sections from at least 20 cells were counted. Means ± SEM from three independent quantifications are given. n.s., not significant; ∗∗p < 0.01.
(G) Vesicle diameters were measured manually using Fiji software. Means ± SEM were calculated from three independent measurements. For each condition, 50 vesicles were counted per experiment. n.s., not significant.
(H) Detection of ZIKV NS4B by immunofluorescence microscopy to determine transfection efficiency for the constructs given on the top of each panel. Scale bar: 100 μm.
(I) Transfection efficiencies were quantified by counting NS4B-containing cells and normalization to the total number of cells. Error bars represent the standard error of three independent experiments. n.s., not significant.
Note that percentages given in (F) refer to all cells analyzed by TEM, but only a fraction of them had detectable amounts of ZIKV NS4B that were used to determine transfection efficiency (I). The higher percentage of cells with VPs attained with some constructs is most likely due to lower sensitivity of the immunofluorescence, allowing detection only of cells expressing high amounts of NS4B.
Comparable 3D Architecture of VPs Induced by pIRO-D and DENV Infection
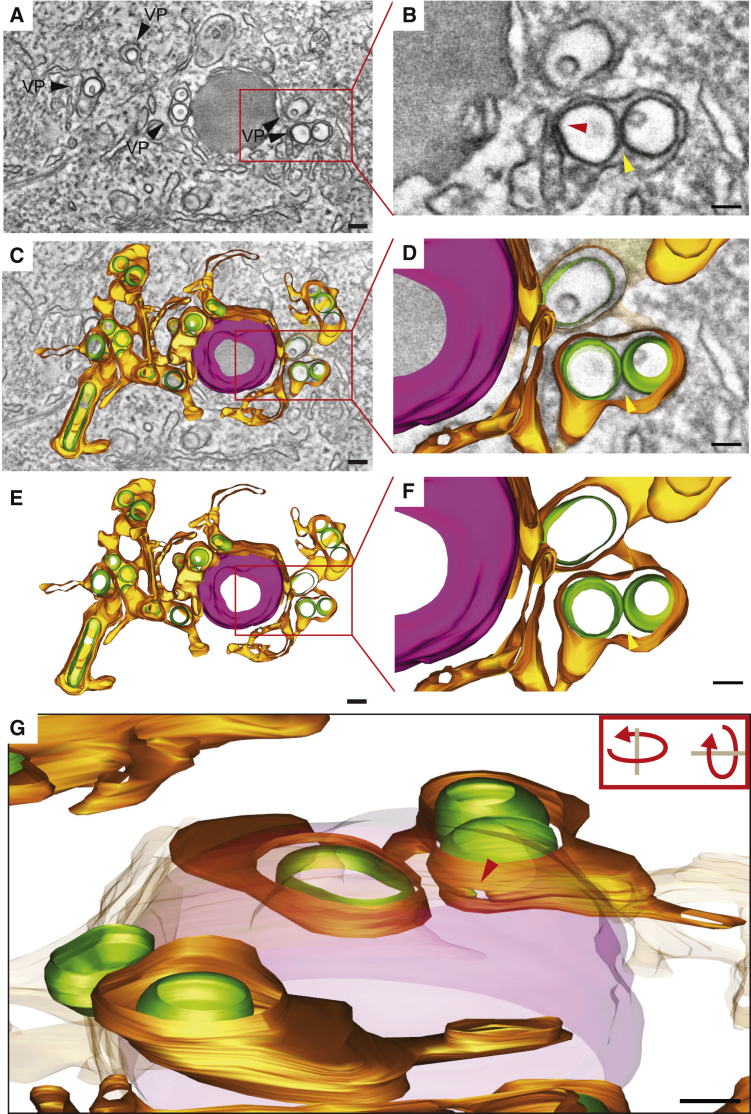
To determine whether VPs induced by the pIRO-D system are bona fide ER membrane invaginations as observed in DENV-infected cells, we performed electron tomography on 250-nm-thick sections of Huh7/Lunet-T7 cells transfected with the pIRO-D Δ 5′ SLAB construct (Figure 4). Comparable to DENV-infected cells, VPs induced in transfected cells also consisted of arrays of invaginated vesicles within dilated ER sheets (Welsch et al., 2009) (Figures 4A–4D). Moreover, similar to cells infected with DENV and other flaviviruses, such as ZIKV, tick-borne encephalitis virus (TBEV), or West Nile virus (WNV), the interior of the vesicles induced with the pIRO-D system was connected to the cytosol by a narrow pore-like structure, which was approximately 11 nm in diameter (Romero-Brey and Bartenschlager, 2014; Welsch et al., 2009) (Figures 4B and 4G). Also analogous to DENV-infection-induced VPs, no connection between adjacent vesicles was found in pIRO-D induced vesicles (Welsch et al., 2009) (Figures 4D–4F). These results show that VPs induced by transfection of the pIRO-D Δ 5′ SLAB construct are morphologically comparable to VPs formed in DENV-infected cells.
Figure 4.
Electron Tomography of Expression-Induced DENV VPs
(A–B) (A) Huh7/Lunet-T7 cells were fixed 24 h post-transfection with the pIRO-D Δ 5′ SLAB construct, embedded in epoxy resin, and analyzed by electron tomography. A tomographic slice showing pIRO-D-induced VPs within the ER is displayed. The red rectangle indicates the cropped section shown in (B), which is a magnified view of the ER lumen-containing vesicles. Two adjacent vesicles within the swollen ER lumen are shown (yellow arrowhead). Red arrowhead shows the pore-like opening to the cytosol.
(C–F) 3D surface model of the VPs next to a lipid droplet (magenta). The red rectangles in (C) and (E) indicate the cropped magnified sections shown in (D) and (F). Yellow arrowhead points to the adjacent vesicles shown in (B).
(G) Side view of the 3D model showing the pore-like opening (red arrowhead). Scale bars: 50 nm.
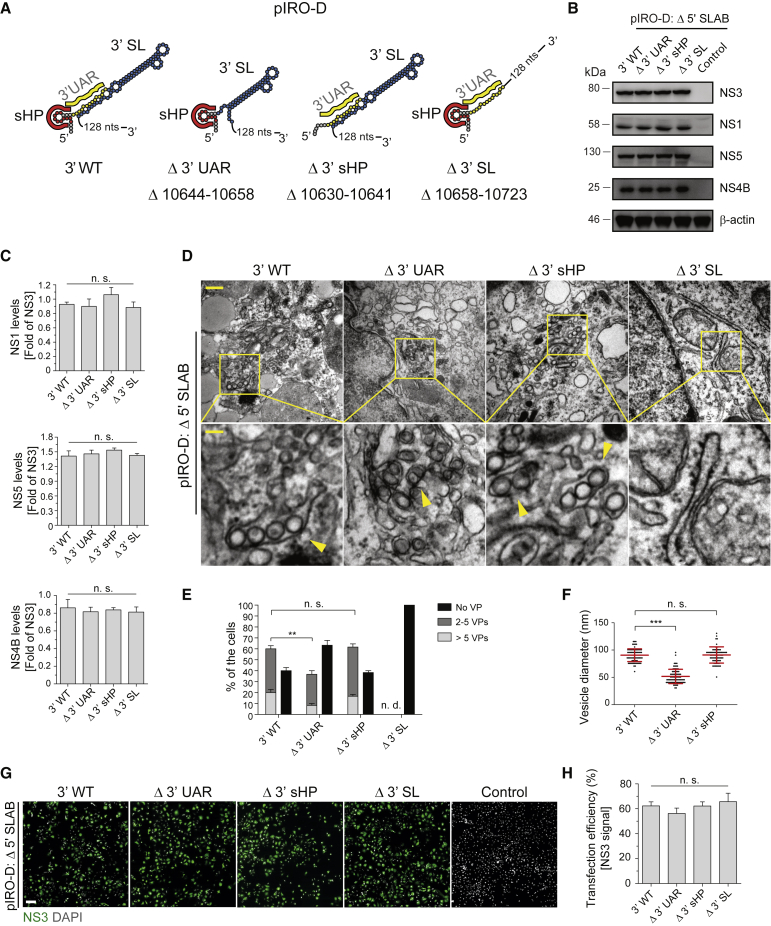
Essential Role of 3′ Terminal Sequences in the 3′ UTR for DENV RO Formation
The observation that the sole expression of the NS1-5 polyprotein is not sufficient to induce VP formation, together with our results showing that the 5′ UTR is largely dispensable for this process, pointed toward an important role of the 3′ UTR in DENV RO biogenesis. Therefore, we focused on evaluating the role of the highly conserved domain III of the 3′ UTR in VP formation through introduction of various deletions into the pIRO-D Δ 5′ SLAB construct (Figure 5A). None of the deletions altered polyprotein expression and processing (Figures 5B and 5C). TEM analysis and quantification of the number of VPs in transfected Huh7/Lunet-T7 cells revealed that disruption of the 3′ sHP had no effect, whereas deletion within the 3′ UAR significantly reduced VP formation (Figures 5D and 5E). Moreover, VPs induced by expression of the Δ 3′ UAR construct were smaller in diameter and morphologically different as compared to those induced by expression of the WT or the Δ 3′ sHP constructs (Figures 5D and 5F). Importantly, removal of most of the 3′ terminal SL (Δ 3′ SL) sequence abolished formation of VPs (Figures 5D and 5E). This was not due to poor transfection efficiency, as it was comparable between Δ 3′ SL and all the other constructs (Figures 5G and 5H). These results suggest that sequences in the 3′ UTR, most notably the 3′ SL, are required for VP formation in a non-replicative manner.
Figure 5.
Importance of the 3′ SL for DENV RO Formation
(A) Schematic representation of 3′ UTR truncations introduced into the pIRO-D Δ 5′ SLAB construct. Numbers refer to the deleted nucleotide positions of the DENV 2 genome (GenBank accession number NC_001474.2).
(B) Analysis of viral protein production in Huh7/Lunet-T7 cells 20 h after transfection with given constructs. β-actin served as sample loading control.
(C) Relative abundance of viral proteins was determined by densitometry, and levels of NS1, NS4B, and NS5 were normalized to NS3 signal. Values represent mean and standard error of three independent experiments. n.s., not significant.
(D) TEM images of Huh7/Lunet-T7 cells transfected with indicated 3′ UTR mutants or the 3′ WT reference construct. Cells were transfected and, after 20 h, fixed, processed, and embedded in resin for sectioning. Upper panel scale bar: 500 nm. Lower panels are magnifications of yellow squared areas in the upper panel images. Yellow arrowheads indicate individual VPs. Lower panel scale bar: 100 nm.
(E) For each condition, VPs contained in whole-cell sections from 20 cells or, in the case of the Δ 3′ SL mutant, 40 cells were counted. Means ± SEM from three independent experiments are shown. n.s., not significant; ∗∗p < 0.01.
(F) Vesicle diameters were measured by analyzing 25 vesicles per experimental condition using Fiji software. Means ± SEM from three independent experiments are given. n.s., not significant; ∗∗∗p < 0.001.
(G) Detection of NS3 by immunofluorescence microscopy to determine transfection efficiency for the indicated constructs. Nuclear DNA was stained with DAPI. Scale bar: 100 μm.
(H) Transfection efficiencies were quantified by counting NS3-containing cells and normalization to the total number of cells. Error bars represent the standard error of three independent experiments. n.s., not significant.
Note that percentages given in (E) refer to all cells analyzed by TEM, but only ~60% of them had been transfected (H); therefore, not more than 60% of cells can contain VPs.
Differential Requirements for RNA Elements in the 3′ UTR between DENV and ZIKV RO Formation
The requirement of certain RNA secondary structures in the DENV 3′ UTR for RO biogenesis raised the question of whether analogous RNA structures are also required for ZIKV RO formation. Therefore, we created pIRO-Z Δ 5′ SLAB-derived constructs lacking the 3′ sHP, the 3′ UAR, or the 3′ SL (Figure S3A). Abundance of viral proteins and polyprotein processing was comparable between all mutants and 3′ WT (Figures S3B and S3C). TEM analysis of these constructs showed that each mutant induced VP formation in transfected cells. However, in all cases, the number of cells containing VPs was lower than cells transfected with the parental (3′ WT) construct (Figures S3D and S3E). Vesicle diameters were unaltered (Figure S3F), and transfection efficiencies of constructs were comparable (Figures S3G and S3H). Importantly, the mutant lacking the complete sequence downstream of the 3′ UAR (construct Δ 3′ SL) was still able to induce formation of VPs. However, the number of VP-containing cells was significantly lower than cells transfected with the parental construct, arguing that also in the case of ZIKV, the 3′ UTR of the genome plays an important role in VP formation.
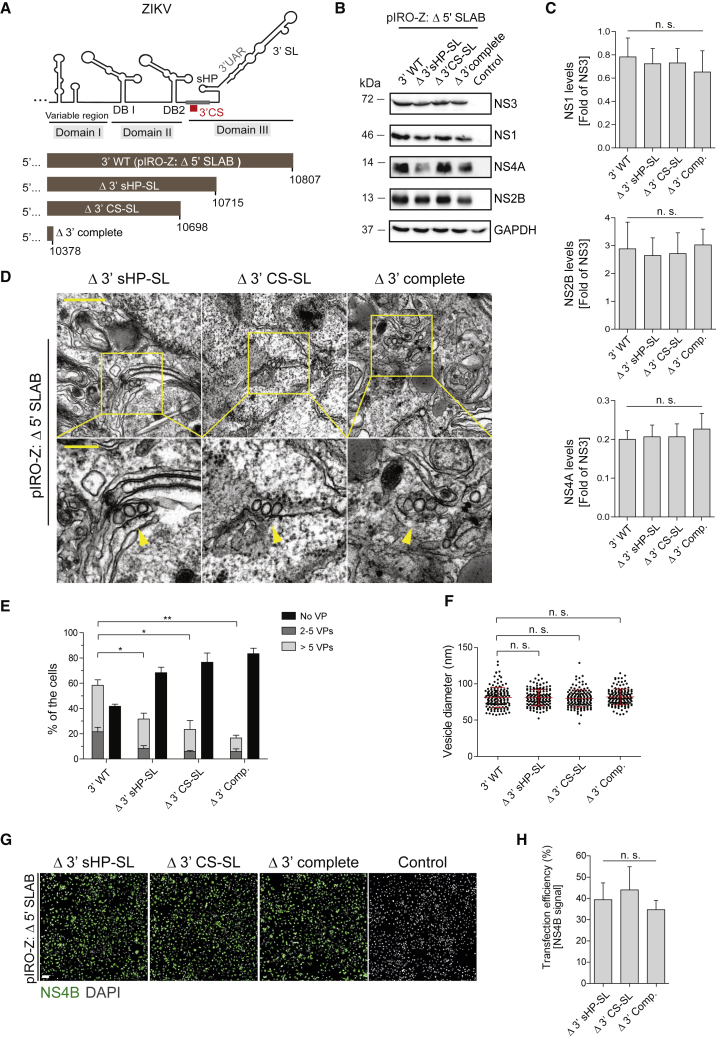
To determine the overall requirement for the 3′ UTR in ZIKV RO formation, we generated expression constructs with more extensive deletions in the 3′ UTR, including complete removal of the 3′ UTR (Δ 3′ complete; Figure 6A). Polyprotein expression and processing in cells transfected with these constructs were comparable (Figures 6B and 6C). TEM analysis of the mutants indicated that all the mutants were able to induce formation of VPs (Figure 6D). However, the number of cells containing VPs inversely correlated with the extent of the deletion introduced into the 3′ UTR. In fact, complete removal of the 3′ UTR caused the most pronounced reduction in VP abundance, compared to the parental construct, but had no effect on vesicle morphology and size (Figures 6E and 6F). Differences of VP abundance in cells expressing the various constructs were not due to different transfection efficiencies (Figures 6G and 6H).
Figure 6.
Elements in the ZIKV 3′ UTR Contribute to, But Are Not Essential for, VP formation
(A) Schematic representation of deletions introduced into the 3′ UTR of the parental pIRO-Z Δ 5′ SLAB construct. Numbers refer to the last nucleotide of the ZIKV genome contained in the expression construct (ZIKVH/PF/2013strain; GenBank accession number KJ776791.2).
(B) Huh7/Lunet-T7 cells were transfected with the parental construct (3′ WT), indicated mutants, or mock transfected (Control). After 18 h, cells were lysed and analyzed by western blot. GAPDH served as loading control.
(C) Relative ZIKV protein abundance was quantified by densitometry of western blots, and values for NS1, NS2B, or NS4A were normalized to NS3. Values represent mean and standard error of three independent experiments. n.s., not significant.
(D) TEM images of Huh7/Lunet-T7 cells 18 h after transfection with indicated 3′ UTR mutants. Cells were fixed, processed, and embedded in resin for sectioning. Lower panels are magnified views of areas indicated with yellow squares in the upper panel images. Yellow arrowheads indicate individual VPs. Scale bars: 500 nm and 200 nm (upper and lower panels, respectively).
(E) For each condition, VPs contained in whole-cell sections from 40 cells were counted. Means ± SEM from three independent quantifications are shown. ∗p < 0.05; ∗∗p < 0.01. Quantification of 3′ WT is taken from Figure 3F (Δ 5′ SLAB).
(F) Vesicle diameters were measured manually using Fiji software. Means ± SEM from three independent measurements (50 vesicles per experiment) are given. n.s., not significant. Vesicle diameter quantification of 3′ WT is taken from Figure 3F (parental construct pIRO-Z Δ 5′ SLAB containing an unaltered 3′ UTR).
(G) Detection of NS4B by immunofluorescence microscopy to determine transfection efficiency for the indicated constructs. Scale bar: 100 μm.
(H) Transfection efficiencies were quantified by counting NS4B-positive cells and normalization to the total cell number. Error bars represent the standard error of three independent experiments. n.s., not significant.
Note that percentages given in (E) refer to all cells analyzed by TEM, but only ~60% of them had been transfected (H); therefore, not more than 60% of cells can contain VPs.
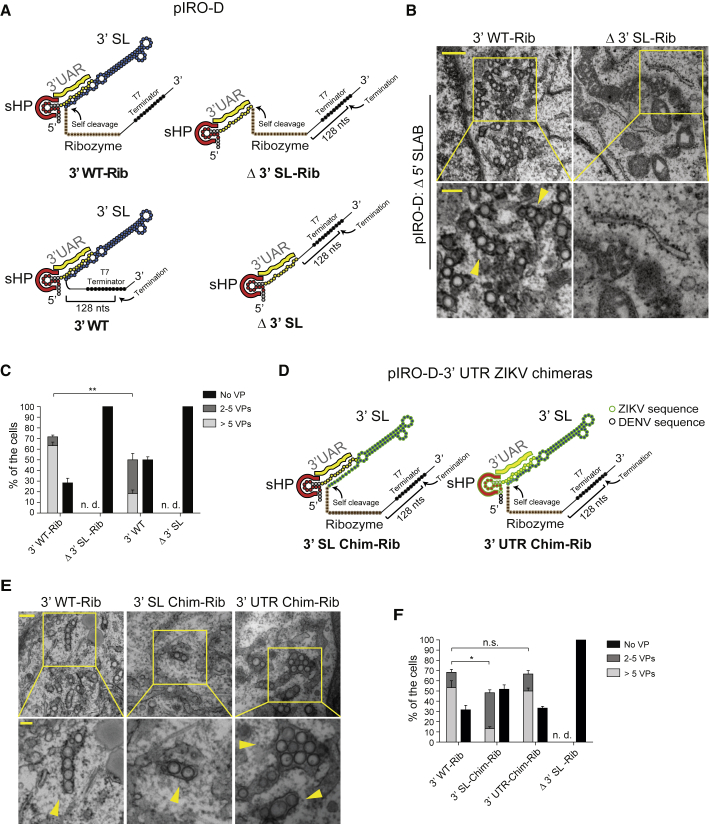
Given the essential role of the DENV 3′ UTR for RO biogenesis, we wondered whether a precise 3′ end is critical for VP formation. In the expression constructs described so far, the 3′ end was generated by the T7 terminator sequence (Macdonald et al., 1994); therefore, RNAs transcribed by the T7 RNA polymerase contain an extra 128-nucleotide-long sequence at their 3′ ends that is unrelated to the viral genome. To determine whether this heterologous sequence might impair VP formation, we inserted the hepatitis D virus (HDV) ribozyme downstream of the DENV sequence to allow the generation of viral RNAs with authentic 3′ ends (Figure 7A). We found that cells transfected with the parental ribozyme-containing construct (pIRO-D 3′ WT-Rib) contained significantly more VPs than cells expressing the ribozyme-less construct (pIRO-D 3′ WT; Figures 7B and 7C). In fact, ∼70% of cells transfected with the ribozyme-containing construct contained VPs (Figure 7C), which corresponds to the transfection efficiency (∼70%; Figures S5A and S5B), arguing that all cells expressing the RNA with the correct 3′ end also produce VPs. Importantly, even under these conditions, the construct lacking most of the 3′ end (Δ 3′ SL-Rib) did not induce VP formation (Figures 7B and 7C), even though transfection efficiency was comparably high (Figures S5A and S5B). In contrast, insertion of the HDV ribozyme into the analogous ZIKV construct had no effect, arguing again for different RNA requirements between DENV and ZIKV to induce VP formation (Figure S4).
Figure 7.
Critical Role of an Authentic 3′ UTR for VP Formation and Rescue of DENV Organelle Formation by the ZIKV 3′ SL
(A) Schematic representation of the 3′ terminal regions of used constructs containing or not containing the HDV ribozyme. Sites of RNA cleavage are indicated with arrows.
(B) TEM images of Huh7/Lunet-T7 cells transfected with the given pIRO-D constructs. Cells were transfected and, 20 h later, fixed, processed, and embedded in resin for sectioning. Upper panel scale bar: 500 nm. Lower panels are magnifications of yellow squared areas in the upper panel images. Yellow arrowheads indicate individual VPs. Lower panel scale bar: 200 nm.
(C) For each condition, VPs contained in whole-cell sections from 20 cells were counted. Means ± SEM from three independent quantifications are given. ∗∗p < 0.01. n.d., not detectable.
(D) Schematic representation of the 3′ terminal regions of the DENV constructs containing only the 3′ SL or the complete 3′ UTR of ZIKV (left and right, respectively).
(E) TEM images of Huh7/Lunet-T7 cells transfected with the constructs specified at the top of each panel. Cells were transfected and, 20 h later, fixed, processed, and embedded in resin for sectioning. Upper panel scale bar: 500 nm. Lower panels are magnifications of yellow squared areas in the upper panel images. Yellow arrowheads indicate individual VPs. Lower panel scale bar: 100 nm.
(F) For each condition, VPs contained in whole-cell sections from 20 cells were counted. Means ± SEM from three independent quantifications are given. ∗p < 0.1; n.s., not significant; n.d., non-detectable.
Rescue of DENV RO Formation by the ZIKV 3′ SL
The 3′ RNA element dependency differences between DENV and ZIKV for RO formation raised the question of whether swapping the DENV 3′ UTR elements with ZIKV 3′ elements would alter VP biogenesis. To address this question, we created two chimeric pIRO-D Δ 5′ SLAB constructs (Figure 7D). First, we fused the ZIKV 3′ SL to the 3′ end of the DENV mutant lacking the 3′ SL (construct 3′ SL Chim-Rib; Figure 7D); second, we replaced the 3′ UTR of DENV with the one of ZIKV in the context of the DENV polyprotein (construct 3′ UTR Chim-Rib; Figure 7D). Both chimeric constructs were able to induce VPs (Figures 7E and 7F), but a higher percentage of cells contained VPs upon transfection with the DENV construct containing the complete ZIKV 3′ UTR. This difference of VP abundance was not due to unequal transfection efficiency (Figures S5C and S5D). These results show that the ZIKV 3′ SL can functionally replace the DENV 3′ SL regarding RO formation. In addition, the higher percentage of VPs containing cells transfected with the DENV construct containing the complete ZIKV 3′ UTR suggested that cross-talk between RNA elements contained within the 3′ UTR might be of relevance for RO biogenesis.
In conclusion, our results suggest that RNA elements in the 3′ UTR of the DENV and ZIKV genome contribute to RO formation in a non-replicative manner. The magnitude of contribution differs between these close viruses with the 3′ terminal SL element of ZIKV being able to functionally replace the one of DENV.
Discussion
In this study, we report the establishment of a polyprotein expression system supporting the formation of flavivirus ROs independent from viral RNA replication. By using a cytoplasmic transcription-translation system based on the T7 RNA polymerase and a heterologous IRES, we achieved robust production and cleavage of viral polyproteins as well as VP formation (Jang et al., 1988; Studier and Moffatt, 1986).
The major advantage of the pIRO system reported here is the replication independence, enabling studies into the impact of replication-inactivating mutations in the viral genome or perturbations of the host cell on RO formation. In this respect, our system offers the opportunity to examine an important step in the viral life cycle that occurs after RNA translation and most likely before bulk RNA replication. For instance, past studies have suggested that NS4A and NS4B, which contain multiple membrane-spanning α helices, play a prominent role in the formation of DENV VPs, possibly by inducing lipid bilayer asymmetry through oligomerization (Paul and Bartenschlager, 2015; Zou et al., 2014). Additionally, NS1 is able to bind to and remodel liposomes in vitro, giving rise to positive membrane curvature, which might contribute to VP formation (Akey et al., 2014; Brown et al., 2016; Płaszczyca et al., 2019). Moreover, many studies have proposed roles for host factors in VP formation, which have yet to be confirmed experimentally (Aktepe et al., 2017; Neufeldt et al., 2019; Richardson et al., 2018). With the advent of our pIRO system, we can begin to dissect the specific functions of these proteins as well as many other host and viral factors in VP biogenesis.
We found that the sole expression of the DENV NS1-5 polyprotein failed to induce VPs in Huh7/Lunet-T7 cells, but the addition of the 3′ UTR of the viral genome rescued this defect. This result is contrary to observations made with HCV, where expression of the minimal viral replicase NS3-5B suffices to induce the HCV RO (Berger et al., 2014; Romero-Brey and Bartenschlager, 2014). However, HCV ROs consist primarily of double-membrane vesicles (DMVs) and thus are morphologically distinct from DENV and ZIKV ROs. Additionally, the mechanism of RO formation is fundamentally different between flaviviruses and HCV. For instance, HCV NS5A and NS5B recruit phosphatidylinositol 4-kinase-α (PI4Kα) to DMVs, where the kinase locally produces high amounts of phosphatidylinositol-4-phosphate (PtdIns4P), which in turn is required to alter membrane lipid composition (Paul and Bartenschlager, 2015; Reiss et al., 2011). In the case of the DENV, PtdIns4P was shown to be dispensable for RO formation, highlighting one important difference between HCV and DENV (Reiss et al., 2011; Wang et al., 2014).
Our experiments in determining the viral protein and genome elements needed for replication-independent VP production demonstrated a requirement for UTR elements in VP biogenesis, which prompted us to employ the pIRO system to further dissect the role of these RNA elements in this process. We found that the 5′ UTR is largely dispensable for DENV and ZIKV VP formation, which was even enhanced upon deletion of both SLA and SLB. In contrast, we observed a crucial role of the 3′ UTR for VP formation, as deduced from three observations. First, the DENV vesicle diameter and the percentage of cells forming VPs were significantly reduced when we removed the 3′ UAR (Figures 5D–5F); second, DENV VP formation was completely lost upon deletion of most of the 3′ SL (Figures 5D and 5E) but restored upon addition of the 3′ SL of ZIKV (Figures 7D–7F); and third, the number of cells with VPs were profoundly reduced when viral RNAs contained a 128-nucleotide-long heterologous sequence at their 3′ end (Figures 7B and 7C). Although these impairments were most pronounced in the case of DENV, an analogous reduction of VP formation was found in the case of ZIKV (Figures 6D and 6E). Taken together, these results provide compelling evidence for a role of the 3′ SL, especially in the DENV genome for VP biogenesis in a replication-independent manner.
The 3′ SL RNA sequences are highly conserved among DENV serotypes 1–3, whereas serotype 4 is somewhat distinct (Sievers et al., 2011) (Figure S6A). However, based on predicted RNA secondary structures, the 3′ SL of all DENV serotypes has a very similar fold (Mathews et al., 2004) (Figure S6B). Of note, the ZIKV 3′ SL could functionally rescue VP generation of DENV, although its homology is lowest compared to the four DENV serotypes (Figure S6). Therefore, we assume that viral or host cell factors binding to highly conserved regions within the 3′ SL are needed to induce VPs. Alternatively, a stable SL structure at the 3′ end per se is required for this process, with viral RNA acting as a structural component during vesicle formation. Along these lines, a recent study of the FHV, an unrelated positive-strand RNA virus belonging to the Nodaviridae family, proposed that formation of vesicular membrane invaginations of the outer mitochondrial membrane is driven by viral RNA replication, with nascent RNA exerting pressure onto the cytoplasmic side of this membrane (Ertel et al., 2017). In this way, the viral replicase, via newly synthesized RNA, provides mechanical force required for membrane bending and subsequent RO formation. Moreover, FHV spherule size was observed to depend on the length of the viral RNA (Ertel et al., 2017). A similar model has been suggested for two other positive-strand RNA viruses: the plant virus Tomato bushy stunt virus and Semliki Forest virus (SFV). In both cases, RO formation depends on viral RNA synthesis, with RNA template length affecting vesicle diameter (Kallio et al., 2013; Kovalev et al., 2016). Although these observations are consistent with a critical role of RNA replication in vesicle formation, a subsequent study conducted with SFV demonstrated that spherule formation can be induced by the sole expression of a partially processed replicase and, thus, is independent of RNA replication (Hellström et al., 2017). However, these spherules are less regular compared to those detected in SFV-replicating cells. Along the same lines, a recent study indicated that expression of a TBEV NS1-4B polyprotein fragment is sufficient to induce RO-like membrane invaginations, but these are morphologically distinct to those induced by TBEV infection (Miorin et al., 2013; Yau et al., 2019). In contrast, we found that an RNA encoding the minimal DENV replicase and containing the 3′ UTR is sufficient to induce vesicles that are morphologically indistinguishable to those found in infected cells.
The observation that the sole expression of the DENV polyprotein does not induce VP formation raises the question of whether the viral proteins form a replication complex outside VPs (e.g., at the ER that is the site of viral protein synthesis). Currently, we do not know the answer, but if a proper replicase complex would form, it might be able to trans-complement a truncated viral RNA lacking, for example, most of the coding region or containing replication-inactivating mutations. Alternatively, VPs might be required to orchestrate correct interaction between the viral proteins (e.g., by providing a proper scaffold for coordinated protein-protein or protein-RNA interaction). Moreover, subgenomic flavivirus RNA (sfRNA) generated in infected cells and containing parts or most of the DENV 3′ UTR, including the 3′ SL (Chapman et al., 2014a, 2014b; Pijlman et al., 2008), might promote VP formation in trans. Addressing these possibilities will require further studies for which the system described here will be instrumental.
In conclusion, we report a novel system, designated pIRO, supporting the biogenesis of DENV and ZIKV ROs independent from viral RNA replication. As shown here, this system is a valuable tool to dissect elements and mechanisms driving the biogenesis of the membranous ROs of these viruses. Insights into these aspects should help to devise novel approaches toward the development of antiviral drugs that are urgently required to treat these highly prevalent infections.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| α-Beta Actin | Sigma Aldrich | A5441; RRID: AB_476744 |
| α-RTN3 | Santacruz | sc374599; RRID: AB_10986405 |
| α-PDI | Thermofisher | P7496; RRID: AB_261952 |
| α-DENV NS3 | Genetex | GTX629477; RRID: AB_2801283 |
| α-DENV NS4B | Genetex | GTX124250; RRID: AB_11176998 |
| α-dsRNA | Scicons | 10010500; RRID: AB_2651015 |
| DENV NS1 | Welsch et al., 2009 | NA |
| DENV NS3 | Miller et al., 2006 | NA |
| DENV NS4B | Miller et al., 2006 | NA |
| DENV NS5 | Miller et al., 2006 | NA |
| ZIKV NS2B | Genetex | GTX133318 |
| ZIKV NS1 | Genetex | GTX634158 |
| ZIKV NS4B | Genetex | GTX133321 |
| ZIKV NS5 | Genetex | GTX133327; RRID: AB_2800435 |
| Goat anti–rabbit IgG-HRP | Sigma Aldrich | A6154; RRID: AB_258284 |
| Goat anti–mouse IgG-HRP | Sigma Aldrich | A4416; RRID: AB_258167 |
| Alexa Fluor 488 donkey anti-mouse IgG | Thermofisher | A-21202; RRID: AB_141607 |
| Alexa Fluor 488 donkey anti-mouse IgG2a | Thermofisher | A-21131; RRID: AB_2535771 |
| Alexa Fluor 568 donkey anti-rabbit IgG | Thermofisher | A-10042; RRID: AB_2534017 |
| Alexa Fluor 568 donkey anti-mouse IgG1 | Thermofisher | A-21124; RRID: AB_2535766 |
| Alexa Fluor 647 donkey anti-rabbit IgG | Thermofisher | A −31573; RRID: AB_2536183 |
| Virus Strains | ||
| DENV isolate 16681, synthetic genome | Fischl and Bartenschlager, 2013 | N/A |
| ZIKV strain H/PF/2013 | European Virus Archive (EVAg, France) | N/A |
| Chemicals | ||
| ProLong® Gold Antifade Reagent | Cell Signaling Technology | #9071 |
| DAPI-Fluoromount-G | Southern BioTech | Cat.#0100-20 |
| Transit LT-1 transfection reagent | Mirus | Cat.#MIR2304 |
| Experimental Models: Cell Lines | ||
| Huh7/Lunet T7 | Appel et al., 2005 | N/A |
| Oligonucleotides | ||
| Please see Table S1 for primers used in construct creation | This study | N/A |
| Recombinant DNA | ||
| pFK_sgDVR2A | Fischl and Bartenschlager, 2013 | N/A |
| pFK_sgDVR2A GND | Fischl and Bartenschlager, 2013 | N/A |
| Recombinant DNA (created using DENV isolate 16681 genomic sequence) | ||
| pTM/NS1-5 (pTM/NS1-5) | This Study | N/A |
| pTM-211 (Δ 5′ SLAB/CS) | This Study | N/A |
| pSM1 (5′ WT) | This Study | N/A |
| pSM2 (Δ 5′ SLA) | This Study | N/A |
| pSM3 (Δ 5′ SLAB) | This Study | N/A |
| pSM3 GND (Δ 5′ SLABGND) | This Study | N/A |
| pSM2 GND (Δ 5′ SLAGND) | This Study | N/A |
| pSM3 Δ 3′ UAR (Δ 5′ SLAB-Δ 3′ UAR) | This Study | N/A |
| pSM3 Δ 3′ sHP (Δ 5′ SLAB-Δ3′ sHP) | This Study | N/A |
| pSM3 Δ 3′ SL (Δ 5′ SLAB-Δ 3′ SL) | This Study | N/A |
| pSM3 WT-Rib (3′ WT-Rib) | This Study | N/A |
| pSM3 3′ Δ SL-Rib (Δ 3′ SL-Rib) | This Study | N/A |
| pSM3 3′ SL [ZIKV]-Rib (3′ SL Chim-Rib) | This Study | N/A |
| Recombinant DNA (created using ZIKV strain H/PF/2013 genomic sequence) | ||
| Δ 5′ SLAB | This Study | N/A |
| 5′ WTGAA | This Study | N/A |
| Δ 5′ SLABGAA | This Study | N/A |
| Δ 5′ SLAB-Δ 3′ UAR | This Study | N/A |
| Δ 5′ SLAB-Δ 3′ sHP | This Study | N/A |
| Δ 5′ SLAB-Δ 3′ SL | This Study | N/A |
| Δ 3′ sHP-SL, Δ 3′ CS-SL | This Study | N/A |
| Δ 3′ complete, 3′ WT-Rib | This Study | N/A |
| Software and Algorithms | ||
| ImageJ (FiJi) | Schindelin et al., 2012 | https://imagej.nih.gov/ij/ |
| Adobe Photoshop 5.5 | San Jose, CA, USA | N/A |
| GraphPad Prism 5.0 | LaJolla, CA, USA | N/A |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ralf Bartenschlager (ralf.bartenschlager@med.uni-heidelberg.de).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The published article includes all [datasets/code] generated or analyzed during this study.
Experimental Models and Subject details
Cell lines and culture conditions
Huh7/Lunet T7 cells (Appel et al., 2005) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with Glutamax (GIBCO), 10% fetal bovine serum, 100 U penicillin/ml, 100 μg streptomycin/ml, 2 mM L-glutamine and nonessential amino acids. Zeocin was added to the medium at a final concentration of 5 μg/ml to maintain stable expression of T7 RNA polymerase.
DNA plasmid constructs
The basic constructs pFK_sgDVR2A (wild-type replicon) and pFK_sgDVR2A GND (GND mutant replicon) were described elsewhere (Fischl and Bartenschlager, 2013). Additional DENV (pIRO-D) expression constructs generated in this study are as follows (plasmid names in parenthesis are abbreviations used in the main text): pTM/NS1-5 (pTM/NS1-5), pTM-211 (Δ 5′ SLAB/CS), pSM1 (5′ WT), pSM2 (Δ 5′ SLA), pSM3 (Δ 5′ SLAB), pSM3 GND (Δ 5′ SLABGND), pSM2 GND (Δ 5′ SLAGND), pSM3 Δ 3′ UAR (Δ 5′ SLAB-Δ 3′ UAR), pSM3 Δ 3′ sHP (Δ 5′ SLAB-Δ3′ sHP), pSM3 Δ 3′ SL (Δ 5′ SLAB-Δ 3′ SL), pSM3 WT-Rib (3′ WT-Rib), pSM3 3′ Δ SL-Rib (Δ 3′ SL-Rib), pSM3 3′ SL [ZIKV]-Rib (3′ SL Chim-Rib), pSM3 3′ UTR [ZIKV]-Rib (3′ UTR Chim-Rib).
In addition, the following ZIKV (pIRO-Z) expression constructs have been generated: Δ 5′ SLAB, 5′ WTGAA, Δ 5′ SLABGAA, Δ 5′ SLAB-Δ 3′ UAR, Δ 5′ SLAB-Δ 3′ sHP, Δ 5′ SLAB-Δ 3′ SL, Δ 3′ sHP-SL, Δ 3′ CS-SL, Δ 3′ complete and 3′ WT-Rib.
Antibodies and immunofluorescence reagents
Commercially available primary antibodies used in this study are as follows: α-Beta Actin (Sigma Aldrich: A5441), α-RTN3 (Santacruz: sc374599), α-PDI (Thermofisher: P7496), α-DENV NS3 (Genetex: GTX629477), α-DENV NS4B (Genetex: GTX124250), α-dsRNA (Scicons: 10010500), α-ZIKV NS2B (Genetex: GTX133318), α-ZIKV NS1 (Genetex: GTX634158), α-ZIKV NS3 (Genetex: GTX133320), α-ZIKV NS4A (Genetex: GTX133704), α-ZIKV NS4B (Genetex: GTX133321), α-ZIKV NS5 (Genetex: GTX133327). Commercially available secondary antibodies and IF reagents used in this study are as follows: Goat anti–rabbit IgG-HRP (Sigma Aldrich: A6154), Goat anti–mouse IgG-HRP (Sigma Aldrich: A4416), Alexa Fluor 488 donkey anti-mouse IgG (Thermofisher: A-21202), Alexa Fluor 488 donkey anti-mouse IgG2a (Thermofisher: A-21131), Alexa Fluor 568 donkey anti-rabbit IgG (Thermofisher: A-10042), Alexa Fluor 568 donkey anti-mouse IgG1 (Thermofisher: A-21124), Alexa Fluor 647 donkey anti-rabbit IgG (Thermofisher: A −31573), ProLong® Gold Antifade Reagent (Cell Signaling Technology: #9071) and DAPI (Sigma Aldrich: D9542). The DENV NS1-specific antibody was produced as described earlier (Welsch et al., 2009). DENV NS3-, NS4B- and NS5-specific antibodies were used for western blot analyses as described elsewhere (Miller et al., 2006).
Method Details
Plasmid DNA transfection
One day prior to transfection, 5 × 104 cells were seeded onto glass coverslips in a 24-well plate per well without zeocin antibiotic. On the next day, media was replaced by 500 μl per well fresh DMEM. Plasmid DNA (500 ng) was added to 100 μl of reduced serum OPTI-MEM media (Thermo Fisher: 31985070), mixed briefly and 1.5 μl of Trans-IT-LT1 transfection reagent (Mirus: MIR2304) was added into the DNA - OPTI-MEM mixture and mixed briefly again. After 20 min incubation at room temperature (RT), transfection mixture was added onto the cells in a dropwise manner. After 4 h incubation, medium was changed and cells were fixed 16 to 20 h after transfection for EM or immunofluorescence (IF) analysis. For western blot analysis, 2 × 105 Huh7/Lunet T7 cells were seeded per well of a 6-well plate and transfected as described above using 2 μg DNA and 6 μl Trans-IT-LT1 in 400 μl of OPTI-MEM. Cells were lysed in sample buffer 16 to 20 h after transfection.
Immunofluorescence analysis
Transfected cells were fixed with 4% paraformaldehyde (PFA) in Phosphate Buffered Saline (PBS) for 15 min at RT. After removal of PFA solution and washing with PBS cells were permeabilized with 0.2% Triton X-100 in PBS. Next, the Triton X-100 solution was replaced with 10% FBS solution (in PBS) and cells were blocked for 1 h at RT. Primary antibodies were diluted in 3% bovine serum albumin (BSA)-PBS solution and cells were incubated with primary antibodies for 1 h. After washing three times with PBS, cells were incubated with Alexa Fluorophore-conjugated secondary antibodies and DAPI, diluted in 3% BSA, for 30 min. Cells were washed three times with PBS and mounted in Prolong Gold solution. Microscopic analyses were conducted with a Nikon Eclipse Ti microscope (Nikon, Tokio, Japan) to determine the transfection efficiency, or a Leica SP8 confocal microscope (Leica) for the subcellular localization analyses.
Western blot analysis and imaging
Transfected cells in 6-well plates were washed with PBS and lysed with 100 μl of sample buffer (120 mM Tris-HCl [pH 6,8], 60 mM SDS, 100 mM DTT, 1.75% glycerol, 0.1% bromophenol blue) supplied with 1 μl of benzonase (Milipore: 70746-3) to digest contaminating nucleic acids. Samples were denatured by incubating at 95°C for 3 min. After SDS-PAGE, proteins were blotted onto PVDF (polyvinylidenfluorid) membranes that were blocked with 5% milk in PBS-T (PBS containing 1% Tween) for 1h at RT. Membranes were incubated with primary antibodies, diluted in 3% BSA in PBS, for 1h and washed three times for 10 min each with PBS-T. Horse radish peroxidase (HRP)-conjugated secondary antibodies were diluted in 5% milk in PBS-T and membranes were incubated for 1h at RT. After washing three times with PBS-T for 10 min each, membranes were developed using the Western Lightning Plus-ECL reagent (Perkin Elmer: NEL105001EA). Signals were visualized using a ChemoCam Imager 3.2 (Intas Science Imaging Instruments GmbH, Göttingen, Germany). Signals were quantified using the ImageJ (FiJi) software package (Schindelin et al., 2012). Values obtained for DENV or ZIKV NS1, NS4B or NS5 were normalized to the one obtained for NS3 of the respective virus. Same amount of lysate (based on equal cell numbers) was loaded onto each lane of the gel.
In vitro transcription and viral RNA transfection
Ten microgram of plasmid DNA per sample were linearized by XbaI restriction enzyme digestion and DNA was purified using the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel: 740609.250). For in vitro transcription of pFK_sgDVR2A (WT replicon) and pFK_sgDVR2A GND (GND replicon) SP6 RNA polymerase, and for 5′ WT, Δ 5′ SLA and Δ 5′ SLAB constructs T7 RNA polymerase were used according to the protocol described previously (Fischl and Bartenschlager, 2013). In vitro transcripts were purified with phenol-chloroform extraction and used for electroporation.Huh7/Lunet-T7 cells were trypsinized, washed with PBS, counted and resuspended in cytomix solution (120 mM KCl, 0.15 mM CaCl2,10 mM potassium phosphate buffer, 2 mM EGTA, 5 mM MgCl2, 25 mM HEPES [pH7.6], 2 mM ATP and 5 mM glutathione). ATP and glutathione were supplied freshly. Huh7/Lunet T7 cells (1x106) were mixed with 5 μg RNA in 400 μl of cytomix solution and transferred into a 0.2 cm gap-width electroporation cuvette (BioRad, Hercules). Mixtures were pulsed once with 166 V and 500 μF. Cells were diluted in media and seeded as duplicates for every time point as specified in the main text for further analysis.
Transmission electron microscopy and tomography
Cells on glass coverslips were fixed with EM fixation buffer (2% glutaraldehyde in 50 mM cacodylate buffer [pH7,2] containing 10 mM MgCl2, 10 mM CaCl2, 100 mM KCl and 2% sucrose) for 30 min at RT. Cells were washed with 50 mM cacodylatebuffer three times. Next, coverslips were incubated with 2% osmium tetroxide/50 mM cacodylate for 40 minutes on ice and washed three times with HPLC grade water. After treatment with 0.5% uranyl acetate for 30 minutes, cells were washed with water three times. Progressive dehydration was performed with increasing concentrations of ethanol (40% to 100%). Cells were embedded into an araldite-epon mixture (Araldite 502/Embed 812 kit; Electron Microscopy Sciences) and left two days at 60°C to achieve complete polymerization. Embedded samples were sectioned into 70-nm-thick slices using an Ultracut UCT microtome (Leica) and a diamond knife (Diatome). Sections were counterstained with 3% uranyl acetate in 70% methanol for 5 minutes and 2% lead citrate in water for 2 minutes. Imaging was performed with an EM-10 transmission electron microscope (Zeiss) equipped with a MegaView camera (Olympus).Electron tomography was performed as previously described (Cortese et al., 2017). In brief, 250 nm thick sections were prepared and10 nm-diameter protein A-gold was added to both sides of the grid. Gold labeled grids were placed in a high-tilt holder and digital images were recoded as single-axis tilt series over a −60° to +60° tilt range (increment 1°) on an FEI Tecnai TF20 microscope. Tomograms were reconstructed using the IMOD software package (Kremer et al., 1996) (https://bio3d.colorado.edu/imod).
Quantification and Statistical Analysis
Statistical analyses were performed using the GraphPad Prism 5.0 software package (LaJolla, CA, USA). Two-tailed paired Student’s t test with Welch’s correction or one-way ANOVA using a Tukey’s post analysis for multiple samples comparison was used to determine the statistical significance. Data are presented in the figures according to the following convention: n.s., not significant (p > 0.05); ∗, p ≤ 0.05; ∗∗, p ≤ 0.01; ∗∗∗, p ≤ 0.001; and ∗∗∗∗, p ≤ 0.0001
Acknowledgments
We thank Marie Bartenschlager and Micha Fauth for excellent technical support. We are grateful to the Electron Microscopy Core Facility at Heidelberg University, headed by Stefan Hillmer, and the Infectious Diseases Imaging Platform (IDIP) at the Center for Integrative Infectious Disease Research in Heidelberg, headed by Vibor Laketa, for expert support and access to their equipment. We are thankful to Hilmar Bading and Andrea Hellwig at the Interdisciplinary Center for Neuroscience of Heidelberg for providing access to their electron microscopy unit and for excellent technical support. This study was supported by the Deutsche Forschungsgemeinschaft (DFG), grants Ba1505/8-1 and 240245660-SFB 1129, both to R.B. C.J.N. was funded in part by a European Molecular Biology Organization (EMBO) long-term fellowship (ALTF 466-2016). K.M. was supported in part by a fellowship of the Hartmut Hoffmann-Berling International Graduate School of Molecular & Cellular Biology in Heidelberg.
Author Contributions
Conceptualization and Methodology, B.C., C.J.N., M.C., K.M., L.C.-C., and R.B.; Formal Analysis, B.C., M.C., S.G., and R.B.; Investigation, B.C., S.G., M.C., and U.H.; Writing – Original Draft, B.C. and R.B.; Writing – Review & Editing, B.C., M.C., C.J.N., and R.B; Funding Acquisition, R.B.
Declaration of Interests
The authors declare no competing interests.
Published: July 7, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107859.
Supplemental Information
References
- Akey D.L., Brown W.C., Dutta S., Konwerski J., Jose J., Jurkiw T.J., DelProposto J., Ogata C.M., Skiniotis G., Kuhn R.J. Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science. 2014;343:881–885. doi: 10.1126/science.1247749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktepe T.E., Liebscher S., Prier J.E., Simmons C.P., Mackenzie J.M. The Host Protein Reticulon 3.1A Is Utilized by Flaviviruses to Facilitate Membrane Remodelling. Cell Rep. 2017;21:1639–1654. doi: 10.1016/j.celrep.2017.10.055. [DOI] [PubMed] [Google Scholar]
- Alvarez D.E., Lodeiro M.F., Ludueña S.J., Pietrasanta L.I., Gamarnik A.V. Long-range RNA-RNA interactions circularize the dengue virus genome. J. Virol. 2005;79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez D.E., De Lella Ezcurra A.L., Fucito S., Gamarnik A.V. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005;339:200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Appel N., Pietschmann T., Bartenschlager R. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J. Virol. 2005;79:3187–3194. doi: 10.1128/JVI.79.5.3187-3194.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquino V.H., Anatriello E., Gonçalves P.F., DA Silva E.V., Vasconcelos P.F.C., Vieira D.S., Batista W.C., Bobadilla M.L., Vazquez C., Moran M., Figueiredo L.T. Molecular epidemiology of dengue type 3 virus in Brazil and Paraguay, 2002-2004. Am. J. Trop. Med. Hyg. 2006;75:710–715. [PubMed] [Google Scholar]
- Berger C., Romero-Brey I., Radujkovic D., Terreux R., Zayas M., Paul D., Harak C., Hoppe S., Gao M., Penin F. Daclatasvir-like inhibitors of NS5A block early biogenesis of hepatitis C virus-induced membranous replication factories, independent of RNA replication. Gastroenterology. 2014;147:1094–1105.e25. doi: 10.1053/j.gastro.2014.07.019. [DOI] [PubMed] [Google Scholar]
- Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., Drake J.M., Brownstein J.S., Hoen A.G., Sankoh O. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbeek P.J., Kooi E.A., Lindenbach B., Huijkman N., Rice C.M., Spaan W.J.M. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 2003;84:1261–1268. doi: 10.1099/vir.0.18860-0. [DOI] [PubMed] [Google Scholar]
- Brinton M.A., Miller W.A. Positive strand RNA virus replication: It depends on the ends. Virus Res. 2015;206:1–2. doi: 10.1016/j.virusres.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W.C., Akey D.L., Konwerski J.R., Tarrasch J.T., Skiniotis G., Kuhn R.J., Smith J.L. Extended surface for membrane association in Zika virus NS1 structure. Nat. Struct. Mol. Biol. 2016;23:865–867. doi: 10.1038/nsmb.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao-Lormeau V.-M., Roche C., Teissier A., Robin E., Berry A.-L., Mallet H.-P., Sall A.A., Musso D. Zika virus, French polynesia, South pacific, 2013. Emerg. Infect. Dis. 2014;20:1085–1086. doi: 10.3201/eid2006.140138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E.G., Costantino D.A., Rabe J.L., Moon S.L., Wilusz J., Nix J.C., Kieft J.S. The Structural Basis of Pathogenic Subgenomic Flavivirus RNA (sfRNA) Production. Science. 2014;344:307–310. doi: 10.1126/science.1250897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E.G., Moon S.L., Wilusz J., Kieft J.S. RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. eLife. 2014;3:e01892. doi: 10.7554/eLife.01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese M., Goellner S., Acosta E.G., Neufeldt C.J., Oleksiuk O., Lampe M., Haselmann U., Funaya C., Schieber N., Ronchi P. Ultrastructural Characterization of Zika Virus Replication Factories. Cell Rep. 2017;18:2113–2123. doi: 10.1016/j.celrep.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Borba L., Villordo S.M., Marsico F.L., Carballeda J.M., Filomatori C.V., Gebhard L.G., Pallarés H.M., Lequime S., Lambrechts L., Sánchez Vargas I. RNA Structure Duplication in the Dengue Virus 3′ UTR: Redundancy or Host Specificity? MBio. 2019;10:e02506-18. doi: 10.1128/mBio.02506-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J.A., Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 2010;64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- Ertel K.J., Benefield D., Castaño-Diez D., Pennington J.G., Horswill M., den Boon J.A., Otegui M.S., Ahlquist P. Cryo-electron tomography reveals novel features of a viral RNA replication compartment. eLife. 2017;6:e25940. doi: 10.7554/eLife.25940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye O., Freire C.C.M., Iamarino A., Faye O., de Oliveira J.V.C., Diallo M., Zanotto P.M.A., Sall A.A. Molecular evolution of Zika virus during its emergence in the 20(th) century. PLoS Negl. Trop. Dis. 2014;8:e2636. doi: 10.1371/journal.pntd.0002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomatori C.V., Iglesias N.G., Villordo S.M., Alvarez D.E., Gamarnik A.V. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J. Biol. Chem. 2011;286:6929–6939. doi: 10.1074/jbc.M110.162289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl W., Bartenschlager R. High-throughput screening using dengue virus reporter genomes. Methods Mol. Biol. 2013;1030:205–219. doi: 10.1007/978-1-62703-484-5_17. [DOI] [PubMed] [Google Scholar]
- Funk A., Truong K., Nagasaki T., Torres S., Floden N., Balmori Melian E., Edmonds J., Dong H., Shi P.-Y., Khromykh A.A. RNA structures required for production of subgenomic flavivirus RNA. J. Virol. 2010;84:11407–11417. doi: 10.1128/JVI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhard L.G., Filomatori C.V., Gamarnik A.V. Functional RNA elements in the dengue virus genome. Viruses. 2011;3:1739–1756. doi: 10.3390/v3091739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göertz G.P., Abbo S.R., Fros J.J., Pijlman G.P. Functional RNA during Zika virus infection. Virus Res. 2018;254:41–53. doi: 10.1016/j.virusres.2017.08.015. [DOI] [PubMed] [Google Scholar]
- Gritsun T.S., Gould E.A. Direct repeats in the 3′ untranslated regions of mosquito-borne flaviviruses: possible implications for virus transmission. J. Gen. Virol. 2006;87:3297–3305. doi: 10.1099/vir.0.82235-0. [DOI] [PubMed] [Google Scholar]
- Hahn C.S., Hahn Y.S., Rice C.M., Lee E., Dalgarno L., Strauss E.G., Strauss J.H. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 1987;198:33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- Hellström K., Kallio K., Utt A., Quirin T., Jokitalo E., Merits A., Ahola T. Partially uncleaved alphavirus replicase forms spherule structures in the presence and absence of RNA template. J. Virol. 2017;91:e00787-17. doi: 10.1128/JVI.00787-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S.K., Kräusslich H.-G., Nicklin M.J.H., Duke G.M., Palmenberg A.C., Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio K., Hellström K., Balistreri G., Spuul P., Jokitalo E., Ahola T. Template RNA length determines the size of replication complex spherules for Semliki Forest virus. J. Virol. 2013;87:9125–9134. doi: 10.1128/JVI.00660-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalev N., de Castro Martín I.F., Pogany J., Barajas D., Pathak K., Risco C., Nagy P.D. Role of Viral RNA and Co-opted Cellular ESCRT-I and ESCRT-III Factors in Formation of Tombusvirus Spherules Harboring the Tombusvirus Replicase. J. Virol. 2016;90:3611–3626. doi: 10.1128/JVI.02775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lo M.K., Tilgner M., Bernard K.A., Shi P.-Y. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 2003;77:10004–10014. doi: 10.1128/JVI.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald L.E., Durbin R.K., Dunn J.J., McAllister W.T. Characterization of two types of termination signal for bacteriophage T7 RNA polymerase. J. Mol. Biol. 1994;238:145–158. doi: 10.1006/jmbi.1994.1277. [DOI] [PubMed] [Google Scholar]
- Mackenzie J.M., Jones M.K., Young P.R. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;220:232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- MacNamara F.N. Zika virus: a report on three cases of human infection during an epidemic of jaundice in Nigeria. Trans. R. Soc. Trop. Med. Hyg. 1954;48:139–145. doi: 10.1016/0035-9203(54)90006-1. [DOI] [PubMed] [Google Scholar]
- Mandl C.W., Holzmann H., Meixner T., Rauscher S., Stadler P.F., Allison S.L., Heinz F.X. Spontaneous and engineered deletions in the 3′ noncoding region of tick-borne encephalitis virus: construction of highly attenuated mutants of a flavivirus. J. Virol. 1998;72:2132–2140. doi: 10.1128/jvi.72.3.2132-2140.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano M., Reichert E.D., Polo S., Falgout B., Kasprzak W., Shapiro B.A., Padmanabhan R. Identification of cis-acting elements in the 3′-untranslated region of the dengue virus type 2 RNA that modulate translation and replication. J. Biol. Chem. 2011;286:22521–22534. doi: 10.1074/jbc.M111.234302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews D.H., Disney M.D., Childs J.L., Schroeder S.J., Zuker M., Turner D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men R., Bray M., Clark D., Chanock R.M., Lai C.J. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 1996;70:3930–3937. doi: 10.1128/jvi.70.6.3930-3937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S., Sparacio S., Bartenschlager R. Subcellular localization and membrane topology of the Dengue virus type 2 Non-structural protein 4B. J. Biol. Chem. 2006;281:8854–8863. doi: 10.1074/jbc.M512697200. [DOI] [PubMed] [Google Scholar]
- Miorin L., Romero-Brey I., Maiuri P., Hoppe S., Krijnse-Locker J., Bartenschlager R., Marcello A. Three-dimensional architecture of tick-borne encephalitis virus replication sites and trafficking of the replicated RNA. J. Virol. 2013;87:6469–6481. doi: 10.1128/JVI.03456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B., Elroy-Stein O., Mizukami T., Alexander W.A., Fuerst T.R. Product review. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- Münster M., Płaszczyca A., Cortese M., Neufeldt C.J., Goellner S., Long G., Bartenschlager R. A Reverse Genetics System for Zika Virus Based on a Simple Molecular Cloning Strategy. Viruses. 2018;10:368. doi: 10.3390/v10070368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeldt C.J., Cortese M., Acosta E.G., Bartenschlager R. Rewiring cellular networks by members of the Flaviviridae family. Nat. Rev. Microbiol. 2018;16:125–142. doi: 10.1038/nrmicro.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeldt C.J., Cortese M., Scaturro P., Cerikan B., Wideman J.G., Tabata K., Moraes T., Oleksiuk O., Pichlmair A., Bartenschlager R. ER-shaping atlastin proteins act as central hubs to promote flavivirus replication and virion assembly. Nat. Microbiol. 2019;4:2416–2429. doi: 10.1038/s41564-019-0586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W.C., Soto-Acosta R., Bradrick S.S., Garcia-Blanco M.A., Ooi E.E. The 5′ and 3′ Untranslated Regions of the Flaviviral Genome. Viruses. 2017;9:137. doi: 10.3390/v9060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsthoorn R.C., Bol J.F. Sequence comparison and secondary structure analysis of the 3′ noncoding region of flavivirus genomes reveals multiple pseudoknots. RNA. 2001;7:1370–1377. [PMC free article] [PubMed] [Google Scholar]
- Palmenberg A.C., Kirby E.M., Janda M.R., Drake N.L., Duke G.M., Potratz K.F., Collett M.S. The nucleotide and deduced amino acid sequences of the encephalomyocarditis viral polyprotein coding region. Nucleic Acids Res. 1984;12:2969–2985. doi: 10.1093/nar/12.6.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D., Bartenschlager R. Flaviviridae Replication Organelles: Oh, What a Tangled Web We Weave. Annu. Rev. Virol. 2015;2:289–310. doi: 10.1146/annurev-virology-100114-055007. [DOI] [PubMed] [Google Scholar]
- Pijlman G.P., Funk A., Kondratieva N., Leung J., Torres S., van der Aa L., Liu W.J., Palmenberg A.C., Shi P.-Y., Hall R.A., Khromykh A.A. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Płaszczyca A., Scaturro P., Neufeldt C.J., Cortese M., Cerikan B., Ferla S., Brancale A., Pichlmair A., Bartenschlager R. A novel interaction between dengue virus nonstructural protein 1 and the NS4A-2K-4B precursor is required for viral RNA replication but not for formation of the membranous replication organelle. PLoS Pathog. 2019;15:e1007736. doi: 10.1371/journal.ppat.1007736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse S. Dengue shock. J. Emerg. Trauma Shock. 2011;4:120–127. doi: 10.4103/0974-2700.76835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss S., Rebhan I., Backes P., Romero-Brey I., Erfle H., Matula P., Kaderali L., Poenisch M., Blankenburg H., Hiet M.-S. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson R.B., Ohlson M.B., Eitson J.L., Kumar A., McDougal M.B., Boys I.N., Mar K.B., De La Cruz-Rivera P.C., Douglas C., Konopka G. A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nat. Microbiol. 2018;3:1214–1223. doi: 10.1038/s41564-018-0244-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche C., Cassar O., Laille M., Murgue B. Dengue-3 virus genomic differences that correlate with in vitro phenotype on a human cell line but not with disease severity. Microbes Infect. 2007;9:63–69. doi: 10.1016/j.micinf.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Romero T.A., Tumban E., Jun J., Lott W.B., Hanley K.A. Secondary structure of dengue virus type 4 3′ untranslated region: impact of deletion and substitution mutations. J. Gen. Virol. 2006;87:3291–3296. doi: 10.1099/vir.0.82182-0. [DOI] [PubMed] [Google Scholar]
- Romero-Brey I., Bartenschlager R. Membranous replication factories induced by plus-strand RNA viruses. Viruses. 2014;6:2826–2857. doi: 10.3390/v6072826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S.L., Nasar F., Cardosa J., Mayer S.V., Tesh R.B., Hanley K.A., Weaver S.C., Vasilakis N. Genetic and phenotypic characterization of sylvatic dengue virus type 4 strains. Virology. 2012;423:58–67. doi: 10.1016/j.virol.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff A.C., Beasley D.W.C., Chen J.J.Y., Ni H., Suderman M.T., Wang H., Xu R., Wang E., Weaver S.C., Watts D.M. Genetic variation in the 3′ non-coding region of dengue viruses. Virology. 2001;281:75–87. doi: 10.1006/viro.2000.0748. [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva R.L.A., de Silva A.M., Harris E., MacDonald G.H. Genetic analysis of Dengue 3 virus subtype III 5′ and 3′ non-coding regions. Virus Res. 2008;135:320–325. doi: 10.1016/j.virusres.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Sridhar S., Luedtke A., Langevin E., Zhu M., Bonaparte M., Machabert T., Savarino S., Zambrano B., Moureau A., Khromava A. Effect of Dengue Serostatus on Dengue Vaccine Safety and Efficacy. N. Engl. J. Med. 2018;379:327–340. doi: 10.1056/NEJMoa1800820. [DOI] [PubMed] [Google Scholar]
- Stanaway J.D., Shepard D.S., Undurraga E.A., Halasa Y.A., Coffeng L.E., Brady O.J., Hay S.I., Bedi N., Bensenor I.M., Castañeda-Orjuela C.A. The global burden of dengue: an analysis from the Global Burden of Disease Study 2013. Lancet Infect. Dis. 2016;16:712–723. doi: 10.1016/S1473-3099(16)00026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W., Moffatt B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Villordo S.M., Gamarnik A.V. Genome cyclization as strategy for flavivirus RNA replication. Virus Res. 2009;139:230–239. doi: 10.1016/j.virusres.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Perry J.W., Lauring A.S., Neddermann P., De Francesco R., Tai A.W. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology. 2014;146:1373–1385.e1-11. doi: 10.1053/j.gastro.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Thurmond S., Islas L., Hui K., Hai R. Zika virus genome biology and molecular pathogenesis. Emerg. Microbes Infect. 2017;6:e13. doi: 10.1038/emi.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S., Miller S., Romero-Brey I., Merz A., Bleck C.K.E., Walther P., Fuller S.D., Antony C., Krijnse-Locker J., Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikan N., Smith D.R. Zika virus: history of a newly emerging arbovirus. Lancet Infect. Dis. 2016;16:e119–e126. doi: 10.1016/S1473-3099(16)30010-X. [DOI] [PubMed] [Google Scholar]
- Yau W.-L., Nguyen-Dinh V., Larsson E., Lindqvist R., Överby A.K., Lundmark R. Model System for the Formation of Tick-Borne Encephalitis Virus Replication Compartments without Viral RNA Replication. J. Virol. 2019;93:e00292-19. doi: 10.1128/JVI.00292-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You S., Falgout B., Markoff L., Padmanabhan R. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 2001;276:15581–15591. doi: 10.1074/jbc.M010923200. [DOI] [PubMed] [Google Scholar]
- Yu L., Nomaguchi M., Padmanabhan R., Markoff L. Specific requirements for elements of the 5′ and 3′ terminal regions in flavivirus RNA synthesis and viral replication. Virology. 2008;374:170–185. doi: 10.1016/j.virol.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Xie X., Lee T., Chandrasekaran R., Reynaud A., Yap L., Wang Q.-Y., Dong H., Kang C., Yuan Z. Dimerization of flavivirus NS4B protein. J. Virol. 2014;88:3379–3391. doi: 10.1128/JVI.02782-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all [datasets/code] generated or analyzed during this study.