Abstract
Mesenchymal stem cell (MSC)-secreted factors have been shown to significantly promote oligodendrogenesis from cultured primary adult neural stem cells (aNSCs) and oligodendroglial precursor cells (OPCs). Revealing underlying mechanisms of how aNSCs can be fostered to differentiate into a specific cell lineage could provide important insights for the establishment of novel neuroregenerative treatment approaches aiming at myelin repair. However, the nature of MSC-derived differentiation and maturation factors acting on the oligodendroglial lineage has not been identified thus far. In addition to missing information on active ingredients, the degree to which MSC-dependent lineage instruction is functional in vivo also remains to be established. We here demonstrate that MSC-derived factors can indeed stimulate oligodendrogenesis and myelin sheath generation of aNSCs transplanted into different rodent central nervous system (CNS) regions, and furthermore, we provide insights into the underlying mechanism on the basis of a comparative mass spectrometry secretome analysis. We identified a number of secreted proteins known to act on oligodendroglia lineage differentiation. Among them, the tissue inhibitor of metalloproteinase type 1 (TIMP-1) was revealed to be an active component of the MSC-conditioned medium, thus validating our chosen secretome approach.
Keywords: neural stem cells, mesenchymal stem cells, transplantation, oligodendroglia, glial fate modulation, myelin, spinal cord, secretome, TIMP-1
1. Introduction
In demyelinating diseases of the central nervous system (CNS), such as multiple sclerosis, myelin repair activities can be observed on the basis of recruitment, activation, and differentiation of resident precursor and stem cells [1]. Besides oligodendroglial precursor cells (OPCs), multipotent adult neural stem cells (aNSCs) located in the subventricular zone (SVZ) or the dentate gyrus of the hippocampus serve as an additional source for the generation of myelinating oligodendrocytes [2,3,4,5,6,7]. Yet, their regeneration capacity and overall degree of remyelination remains limited, which is likely a result of multiple extrinsic and intrinsic inhibitory factors [8,9,10,11,12,13,14]. Moreover, immune reactions and damaged tissues create an overall hostile environment, which further limits regeneration activities [15]. Insights into the underlying mechanisms on how aNSCs can be stimulated to differentiate into a specific cell lineage could therefore pave the way for novel neuroregenerative treatment approaches, which are still representing a clinically unmet need.
Mesenchymal stem cells (MSCs) are known for their immune-modulatory capacity and can also mediate a number of regenerative responses [16]. Regarding the restricted repair potential of the adult CNS, rodent and human MSCs as well as their secreted factors have repeatedly been shown to promote oligodendrogenesis from aNSCs and OPCs [16,17,18,19,20]. The corresponding factors are likely secreted or shedded, and their identification might therefore provide important information on how existing limitations regarding successful myelin repair might be overcome. However, the nature of MSC-derived differentiation and maturation factors acting on the oligodendroglial lineage have not been disclosed thus far [21] and might therefore to be deciphered by sophisticated strategies such as mass spectrometry-based secretome analysis [22]. Although MSC secretome and proteome data already exist, most of them deal with their anti-inflammatory properties [23,24,25]. In addition to the missing information on active ingredients, it also remains to be demonstrated to what degree MSC-dependent lineage instruction is functional in vivo.
In the present study, we demonstrate that mesenchymal stem cell factors can indeed stimulate aNSC-derived oligodendrogenesis in different CNS regions and we furthermore provide insights into the underlying mechanisms as we conducted a comparative secretome and proteome analyses. We identified several secreted proteins, many of which have unknown roles in the glial context. However, the analysis also revealed a group of proteins that were previously shown to act on oligodendroglia.
2. Results
2.1. Survival of MSC-CM-Stimulated aNSC In Vivo
Mesenchymal stem cell secreted factors were shown to stimulate aNSCs towards an oligodendrogenic differentiation by yet unknown mechanisms [17,19,20]. To investigate the potential of rat MSC-conditioned medium (MSC-CM) stimulated aNSCs to generate oligodendroglial cells, we performed in vivo cell transplantations into different CNS tissues. Before transplantation experiments were initiated, we pre-treated green fluorescent protein (GFP)-expressing aNSCs were with either control α-minimum essential medium containing 10% fetal calf serum (α-MEM/10% FCS) or MSC-CM, and tested for the expression of oligodendroglial markers after 1, 3, and 7 days in vitro. These analyses revealed a significant increase (up to 15%) in the number of GFP-positive cells upon incubation in both media over time (Figure S1). In addition, a significantly increased number of cells expressing oligodendroglial markers was observed after MSC-CM stimulation, whereas astrocytic marker expression was decreased in MSC-CM-treated aNSCs compared to control medium-treated cells (Figure S1), thus reproducing MSC-CM-dependent effects previously shown in non-transfected cells [20].
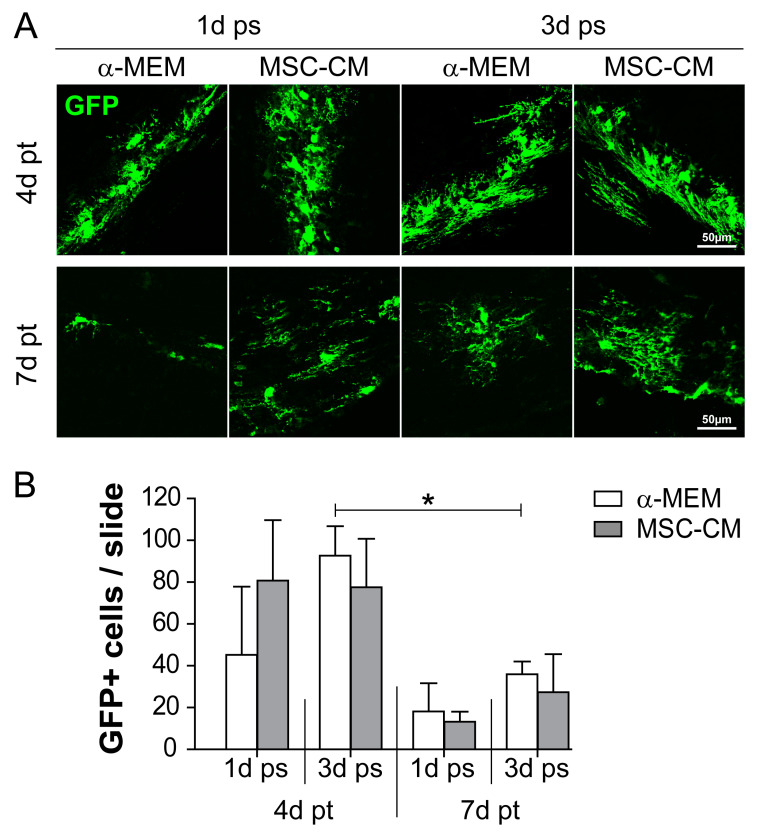
To investigate to what degree MSC-CM-stimulated aNSCs differentiate and integrate within different brain environments, we performed transplantations into either grey (GM) or white matter (WM) regions of young adult mouse brains as previously described [26]. Moreover, we tested different MSC-CM stimulation periods prior to transplantation. We first monitored to what extent GFP-expressing cells survive after transplantation and to what level stimulation with either control α-MEM or with MSC-CM affects cell survival in vivo. Transplantation into the grey (cortex) and white (corpus callosum) matter of the mouse brain revealed that transplanted aNSCs survived up to 7 days post-grafting, independent of their pre-stimulation (duration and condition). In addition, more GFP-positive cells were found after 4 days compared to 7 days post-transplantation (pt). A 3-day MSC-CM pre-stimulation period appeared to confer a slight survival benefit compared to the 1-day pre-treatment (Figure 1). Additionally, no significant differences were observed when cell survival between grey and white matter sites were compared (Figure S2). Our analysis also revealed that the survival rate of transplanted aNSCs decreased as nearly no GFP-positive cells were found at 14 days post-transplantation (data not shown).
Figure 1.
Survival rate of green fluorescent protein (GFP)-expressing cells after transplantation into the mouse brain. (A) Immunohistochemical staining was used to identify transplanted GFP-positive adult neural stem cells (aNSCs) (green) at 4 and 7 days post-transplantation (pt) into the mouse brain. aNSCs were pre-stimulated (ps) for either 1 or 3 days (1d ps and 3d ps, respectively) with α-minimum essential medium (α-MEM) or mesenchymal stem cell (MSC)-conditioned medium (MSC-CM). Representative pictures were taken from white (corpus callosum) / grey (cortex) matter implantation sites. (B) Quantitative evaluation of the total number of GFP-positive cells per slide including both grey and white matter. Statistical significance was calculated using a two-way ANOVA with Bonferroni posttest: * p ≤ 0.05. Data were generated on the basis of the following animal numbers (n): 1d ps and 4d pt, α-MEM and MSC-CM n = 4; 3d ps and 4d pt, α-MEM n = 5 and MSC-CM n = 4; 1d ps and 7d pt, α-MEM and MSC-CM n = 3; 3d ps and 7d pt, α-MEM and MSC-CM n = 4.
2.2. MSC-Derived Factors Promoted the Oligodendroglial Differentiation Process In Vivo
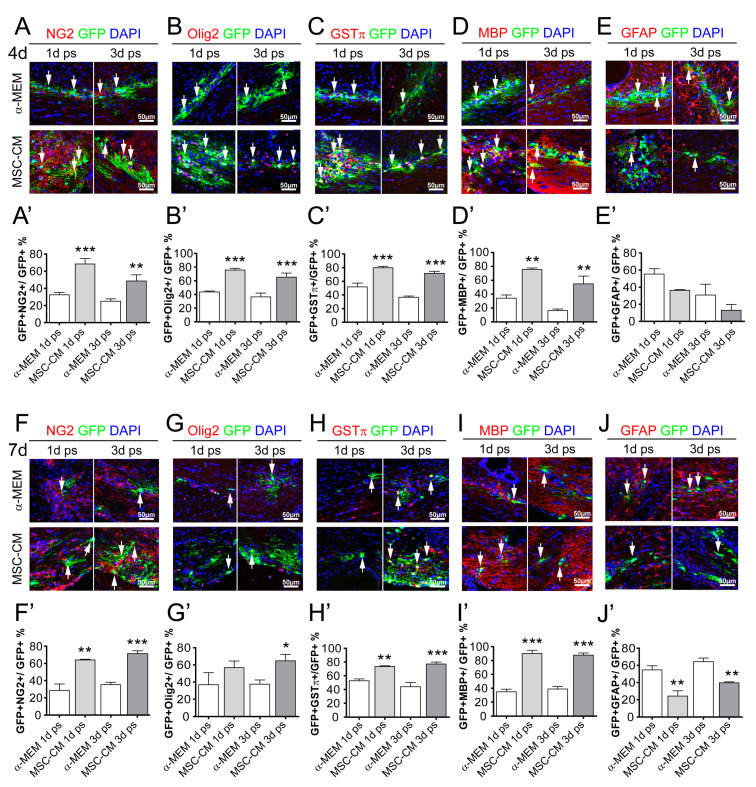
We next investigated the differentiation capacity of transplanted aNSCs after 4 and 7 days in the mouse brain. To this end, we examined expression of markers for OPCs, oligodendrocytes, and astrocytes in GFP-positive cells. MSC-CM pre-treatment led to an increase in neural/glial antigen 2 (NG2)-positive cells after 4 and 7 days compared to aNSCs that were pre-stimulated with α-MEM, (Figure 2A,A’,F,F’). Importantly, the degree of NG2 induction was similar between cells that were only stimulated for 1 day and for cells stimulated for 3 days (compare grey bars in Figure 2A’,F’). Likewise similar induction rates were observed for further markers along the oligodendroglial lineage such as oligodendrocyte transcription factor 2 (Olig2) (Figure 2B,B’,G,G’), glutathione-S-transferase-π (GSTπ) (Figure 2C,C’,H,H’), and myelin basic protein (MBP) (Figure 2D,D’,I,I’) at both investigated time points. Similar to the observed differentiation kinetics of cultured aNSCs [20], accumulation of oligodendroglial markers was accompanied by a MSC-CM dependent decrease of glial fibrillary acidic protein (GFAP)-positive cells (Figure 2E,E’,J,J’). When the differentiation capacity of transplanted cells in grey and white matter was compared, no significant difference was observed (Figure S3). Our data therefore indicate that a 1-day long pre-stimulation with MSC-derived factors is sufficient to boost the oligodendroglial differentiation process of aNSCs after transplantation in vivo. However, for practical reasons, we continued using a 3-day pre-stimulation period for aNSCs to be implanted into spinal cords.
Figure 2.
MSC-CM pre-stimulated aNSCs differentiated into mature myelin basic protein (MBP)-expressing oligodendrocytes after transplantation into the mouse brain. (A–E) Representative images of control (α-MEM) and MSC-CM pre-stimulated and transplanted GFP-positive aNSCs expressing neural/glial antigen 2 (NG2) (A), oligodendrocyte transcription factor 2 (Olig2) (B), glutathione-S-transferase-π (GSTπ) (C), MBP (D), or glial fibrillary acidic protein (GFAP) (E) 4 days after transplantation into the brain. Quantification of transplanted cells expressing NG2 (A’), Olig2 (B’), GSTπ (C’), and MBP (D’) revealed a significantly increased number of cells expressing oligodendroglial markers after 4 days, whereas the degree of GFAP-positive cells (E’) was significantly decreased. The same effects were observed at 7 days post-transplantation (F–J) with the corresponding quantifications shown for NG2 (F’), Olig2 (G’), GSTπ (H’), MBP (I’), and GFAP (J’). For statistical analysis, a two-way ANOVA with Bonferroni posttest was used: * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, MSC-CM compared to the respective α-MEM control (1- or 3-day pre-stimulation). Arrows point to GFP-positive cells (green) expressing the respective markers (red). Blue nuclei represent 4´,6-diamidin-2-phenylindol (DAPI) staining. Animal numbers (n): 3–5.
2.3. Extended Survival Time of MSC-CM Pre-Stimulated aNSCs Transplanted into the Adult Rat Spinal Cord
To evaluate the survival rate of MSC-CM pre-stimulated aNSCs and their differentiation potential after transplantation into other CNS tissues, we transplanted MSC-CM stimulated aNSCs into the grey or white matter of adult rat spinal cords. In this regard, 3-day pre-stimulated aNSCs were analyzed at 14 and 28 days after transplantation in order to monitor tissue integration, cell fate acquisition, maturation stages, and myelination capacities.
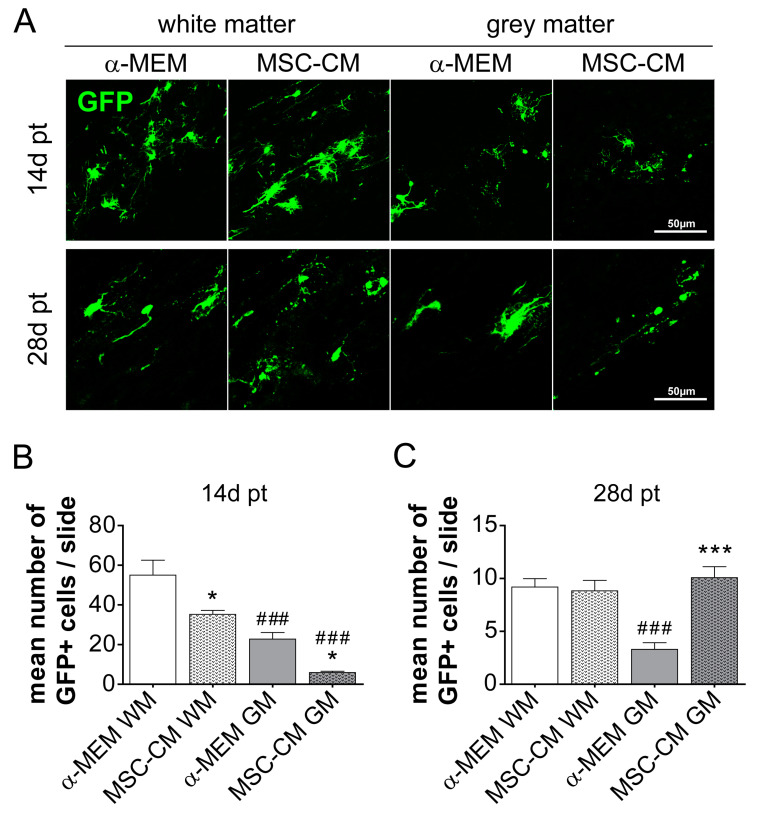
To evaluate the survival rate of transplanted cells, we counted the number of GFP-positive cells per slide and animal at both time points. Quantification of transplanted cells revealed that after 14 days the white matter environment exerted a general survival benefit on implanted cells. Furthermore, in both grey as well as white matter regions, fewer MSC-CM-stimulated cells compared to control (α-MEM)-stimulated cells were observed (Figure 3A,B). However, at the later time point (28 days), we observed no treatment difference in the white matter, while in the grey matter, MSC-CM pre-stimulated cells dominated over control (α-MEM)-treated cells.
Figure 3.
Survival rate of rat aNSCs after transplantation into the rat spinal cord. (A) Representative images of GFP-positive transplanted cells 14 and 28 days post-transplantation (pt) either in white or grey matter of the rat spinal cord. Corresponding quantifications of GFP-positive cells per slide (a mean of three sections per slide) and animal in white and grey matter at 14 days (B) and 28 days (C). Statistical significance was calculated using a two-way ANOVA with Bonferroni posttest: * p ≤ 0.05, *** p ≤ 0.001 (for comparison between control α-MEM to MSC-CM) and ### p ≤ 0.001 (for comparison between white and grey matter and respective transplantation paradigm). Animal numbers (n): 14 days pt α-MEM n = 4 and MSC-CM n = 5, 28 days pt α-MEM n = 6 and MSC-CM n = 4.
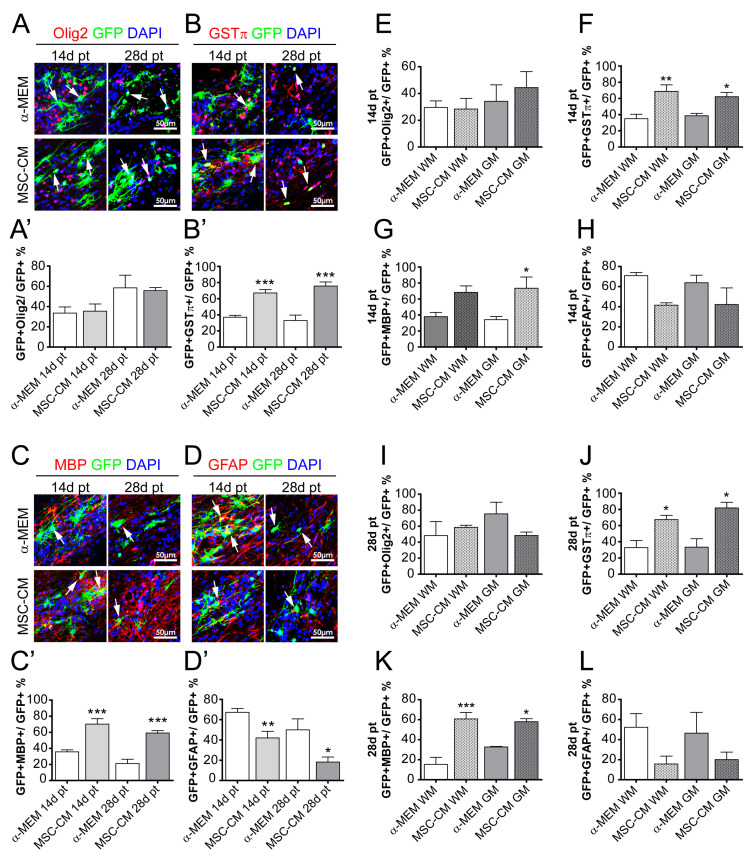
Investigating the differentiation capacity and maturation of spinal cord-transplanted cells confirmed an enhanced oligodendroglial cell fate upon MSC-CM pre-stimulation at 14 and 28 days post-transplantation. Although no impact of MSC-CM pre-stimulation on the degree of Olig2-positive cells was observed (Figure 4A,A’), the degree of GSTπ-positive and MBP-positive cells was strongly boosted upon MSC-CM pre-treatment at both time points (Figure 4B,B’,C,C’). Similar to the brain-transplanted cells, this increase in oligodendrogenesis was accompanied by an MSC-CM-dependent decrease in GFAP positivity among GFP-positive cells (Figure 4D,D’). This pro-oligodendroglial behavior did not substantially differ between WM- and GM-transplanted cells (Figure 4E–L). In addition, a potential neuronal differentiation capacity was tested by means of neuronal nuclei antigen (NeuN) and neurofilament (NF) immunohistochemical staining. No GFP/NeuN or GFP/NF-positive cells were found under both conditions (α-MEM or MSC-CM pre-stimulation) and at both time points (Figure S4). These data therefore confirmed a strong pro-oligodendroglial effect exerted by MSC-derived factors and at the same time revealed increased numbers of surviving and integrated cells in the spinal cord.
Figure 4.
Accumulation of oligodendroglial characteristics in spinal cord-transplanted aNSCs. Representative pictures of GFP-positive cells expressing Olig2 (A), GSTπ (B), MBP (C), and GFAP (D) at 14 and 28 days post-transplantation and following α-MEM or MSC-CM pre-treatment. The corresponding quantifications (percentage of expressing cells over total number of GFP-expressing cells) are depicted in (A’) for GFP/Olig2- in (B’) for GFP/GSTπ-, in (C’) for GFP/MBP-, and in (D’) for GFP/GFAP-positive cells. (E–L) Quantification and distribution of transplanted GFP-positive cells pre-treated with α-MEM and MSC-CM within white matter (WM) and grey (GM) at 14 and 28 days post-transplantation. Statistical significance was calculated using a two-way ANOVA with Bonferroni posttest: * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001 (for comparison between control α-MEM to MSC-CM). Arrows point to GFP-positive cells (green) expressing the respective marker proteins (red). Blue nuclei represent DAPI staining. Animal numbers (n): 4–6.
2.4. MSC-CM Pre-Stimulation Enabled aNSCs to Myelinate Axons In Vivo
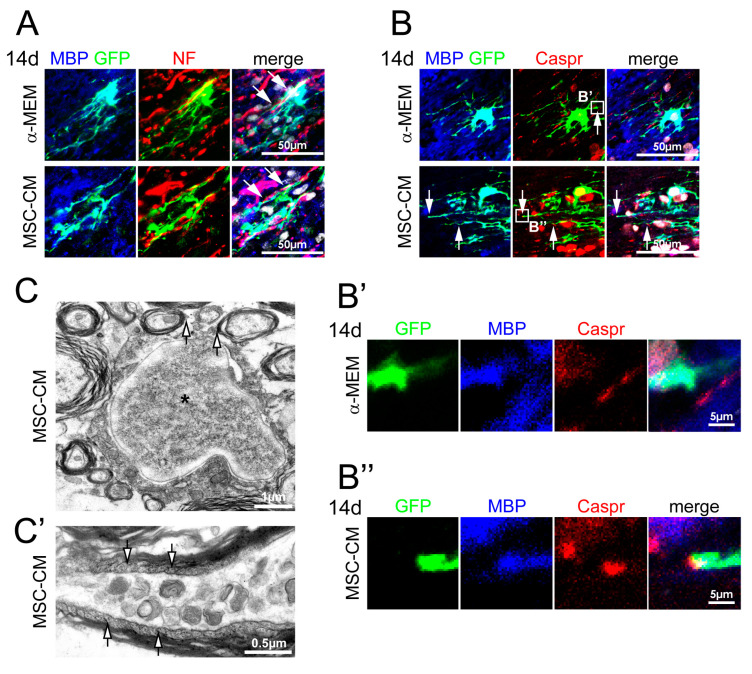
In order to analyze the functionality of MBP-expressing cells and to identify myelinated segments, we performed co-staining for neurofilament and contactin-associated protein (Caspr), a marker for paranodal axonal regions of nodes of Ranvier, after 14 days of cell transplantation into rat spinal cords as previously established for other stem cell transplantation approaches [27,28]. Immunohistochemical staining revealed that processes of transplanted GFP/MBP-double positive cells pre-stimulated with α-MEM or MSC-CM often aligned with neurofilament-positive axons (Figure 5A). Triple staining for GFP, MBP, and Caspr revealed that processes of GFP/MBP-positive cells abutted at Caspr-positive paranodal regions (Figure 5B,B’’). Importantly, such myelinated segments and nodal structures were only observed at MSC-CM pre-treated NSCs, thus providing strong evidence that mesenchymal factors contribute to the generation of myelinated segments in vivo. Moreover, ultrastructural analysis of transplanted MSC-CM pre-treated cells by means of immunoelectron microscopy (as established in [26]; transplanted cells were identified by chromogenic anti-GFP immunohistochemistry resulting in dark precipitates), demonstrated that these cells established myelin sheaths around axons (Figure 5C) and also revealed GFP-immunoreactive signals in paranodal loops (Figure 5C’).
Figure 5.
MSC-CM pre-treated aNSCs were able to generate myelinated segments. (A) Representative pictures of MBP-/GFP-/neurofilament-stained spinal cord sections at 14 days post-transplantation. For both α-MEM and MSC-CM pre-treated and transplanted cells, we observed an alignment of MBP-positive segments with neurofilament (NF)-positive axons (arrows). Nuclei are depicted in white. (B) MBP-positive processes of MSC-CM pre-stimulated cells often ended in Caspr-positive paranodes (arrows). Nuclei are depicted in white. Higher magnifications are depicted in (B’) for α-MEM and in (B’’) for MSC-CM pre-stimulated aNSCs. (C) Representative electron micrograph of a GFP-positive cells (asterisk: nucleus) 14 days post-transplantation. Chromogenic GFP-immunoreactivity (dark precipitate) identified myelinating processes (arrows). (C’) GFP expression was also detectable in the cytoplasm of paranodal loops (arrows).
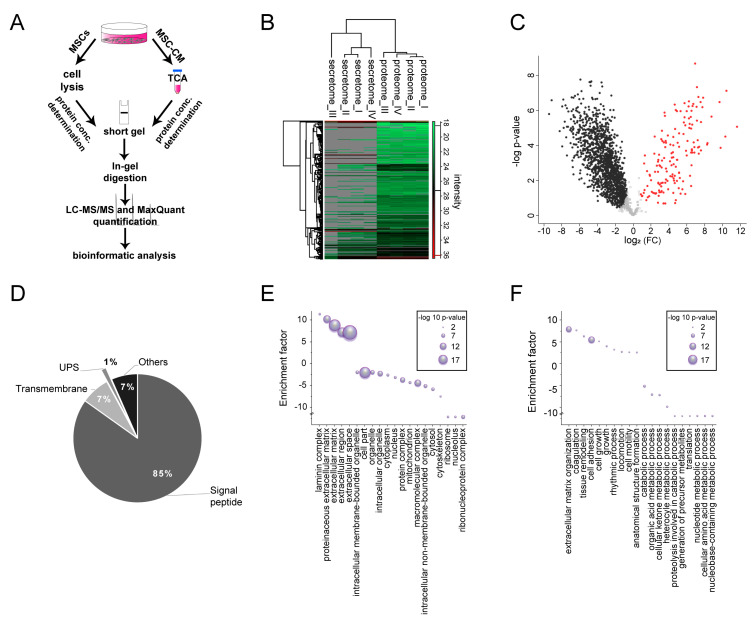
2.5. Characterization of Adult Rat Mesenchymal Stem Cell Secretome
Despite several approaches to identify active components, the underlying mechanism and acting factors of MSC-mediated pro-oligodendroglial effects is still unknown. In order to shed light onto the composition of active proteins in the MSC secretome and to identify MSC secreted pro-oligodendroglial proteins, we performed a mass spectrometry-based secretome analysis (Figure 6). For mass spectrometry analysis, MSCs and corresponding conditioned media were harvested after incubation with serum-free N2-medium for 48 h. Proteins of four independent replicates were processed and analyzed with liquid chromatography–mass spectrometry (LC–MS/MS; Figure 6A). Secretomes were compared to corresponding proteomes in order to identify bona fide secreted proteins and subtract potential contaminations from dying cells due to higher abundances in the secretome (Figure 6B; Table S1) (according to [29]). Thus, from the total number of proteins (691 proteins) identified in MSC-CM, we detected 152 proteins as bona fide secreted due to the comparison with the cellular proteome (Figure 6C; Table S1). From all secreted proteins, 85% of the proteins were predicted to have a signal peptide assigning them to the classical secretory pathway, 7% exhibited a transmembrane domain, and 1% were presumably unconventionally secreted proteins (Figure 6D). Categorical enrichment analysis revealed that bona fide secreted proteins were associated with extracellular components such as the laminin complex (enrichment factor (EF) 11.4, p-value 6.50 × 10−4), the proteinaceous extracellular matrix (EF 10.3, p-value 5.6 × 10−18), the extracellular matrix (EF 8.9, p-value 3.3 × 10−30), and extracellular space (EF 7.2, p-value 8.8 × 10−25) (Figure 6E), whereas categories related to intracellular compartments such as the nucleus (EF -3.0, p-value 4.7 × 10−6), the macromolecular complex (EF −4.3, p-value 1.2 × 10−16), and the ribonucleoprotein complex (EF −12.0, p-value 7.8 × 10−8) were under-represented (Figure 6E). Furthermore, MSC-secreted proteins were associated with biological processes related to extracellular compartments such as the extracellular matrix organization (EF 7.9, p-value 1.0 × 10−15), cell adhesion (EF 5.5, p-value 5.9 × 10−18), and locomotion (EF 2.7, p-value 7.4 × 10−4) (Figure 6F), whereas proteins related to intracellular processes such catabolic processes (EF −5.2, p-value 1.4 × 10−6) and translation (EF −12, p-value 2.6 × 10−3) were under-represented. We further analyzed these 152 proteins regarding a pro-oligodendroglial fate choice (Table 1). According to the already described functions, several proteins identified in the MSC secretome can either directly or indirectly be implicated in oligodendroglial differentiation or myelination. This includes, for example, extracellular matrix proteins such as laminin subunit beta-2 (Lamb2), tissue inhibitor of metalloproteinase inhibitor type 1 (TIMP-1), dystroglycan (Dag1), aggrecan (Acan), or bone morphogenetic protein (BMP)-associated proteins such as chordin (Chrd) and metalloendopeptidase (BMP-1). Moreover, growth factors acting as possible negative regulators such as connective tissue growth factor (CTGF) and insulin-like growth factor-binding proteins (IGFBP).
Figure 6.
Characterization of the mesenchymal stem cell secretome. (A) Workflow for preparation of MSC-derived proteomes and secretomes for quantitative LC–MS/MS analysis. (B) Heatmap of proteins quantified in the MSC secretome and proteome revealed a high similarity between the proteome samples that clearly differed from the secretomes (grey = proteins were not identified). (C) Volcano plot showing proteins found to be enriched in the MSC-derived secretome (152 secretome-enriched proteins, shown as red dots on right side) and the proteome (left side, black dots). (D) Out of the 152 proteins found to be enriched in the secretome, 85% of them were predicted to have a signal peptide, 7% were transmembrane proteins, and 1% were unconventionally secreted (UPS). (E) Gene Ontology (GO) enrichment analysis of cellular component revealed that these proteins were associated with extracellular space and matrix localization. (F) GO enrichment analysis of biological process revealed an association with extracellular matrix organization and cell adhesion (5% false discovery rate (FDR)).
Table 1.
Mesenchymal stem cell-secreted proteins associated with oligodendroglial lineage regulation.
| ID | Oligodendroglial Lineage-Promoting Factor | Gene Name | Description | Secretion Via/Localization | Reference |
|---|---|---|---|---|---|
| P07897 | Aggrecan core protein | Acn | Inhibits oligodendrocyte myelination | SP 1 | [30] |
| A0A140TA94 | Chordin | Chrd | Promotes oligodendrogenesis From subventricular zone derived stem cells in vitro and in vivo |
SP | [31] |
| Q9R1E9 | Connective tissue growth factor | Ctgf | Negatively regulates myelination | SP | [32] |
| F1M8K0 | Dystroglycan 1 | Dag1 | Intracellular portion of cleaved dag promotes oligodendroglial precursor cell proliferation/regulates myelin membrane production, growth, or stability | SP | [33,34] |
| A0A096P6L8 | Fibronectin | Fn | Maintenance and proliferation of OPCs | SP | [35] |
| Q63772 | Growth arrest-specific protein 6 | Gas6 | Protects oligodendrocytes from tumor necrosis factor-α-induced apoptosis | SP | [36] |
| P12843 | Insulin-like growth factor-binding protein 2 | Igfbp2 | Negative effector of oligodendrocyte survival and differentiation | SP | [37] |
| F1M9B2 | Insulin-like growth factor-binding protein 7 | Igfbp7 | Inhibits the differentiation of oligodendrocyte precursor cells via regulation of wingless-related integration site (Wnt) / β-catenin signaling | SP | [38] |
| P12843 | Laminin subunit beta-2 | Lamb2 | Extracellular matrix proteins known to be important for myelination | SP | [39,40] |
| F1M798 | Metalloendopeptidase | BMP1 | Inhibits oligodendrogenesis | SP | [41] |
| P30120 | Tissue inhibitor of metalloproteinase type 1 | Timp-1 | Promotes oligodendrocyte differentiation and central nervous system myelination | SP | [42,43] |
| P08721 | Osteopontin | Spp1 | Stimulates MBP synthesis and myelin sheath formation in vitro | SP | [44] |
| F7EPE0 | Sulfated glycoprotein 1/prosaposin | Psap | Known as a myelinotrophic factor protecting myelinating glial cells | SP | [45,46] |
| Q9JI92 | Syntenin-1 | Sdcbp | Plays a role in OPC migration | UPS 2 | [47] |
| M0R979 | Thrombospondin 1 | Thbs1 | Reduces oligodendrogenesis | SP | [48] |
| A0A0G2K1L0 | Tenascin c | Tnc | Inhibits OPC differentiation and myelination | SP | [49] |
1 SP: signal peptide; 2 UPS: unconventional protein secretion.
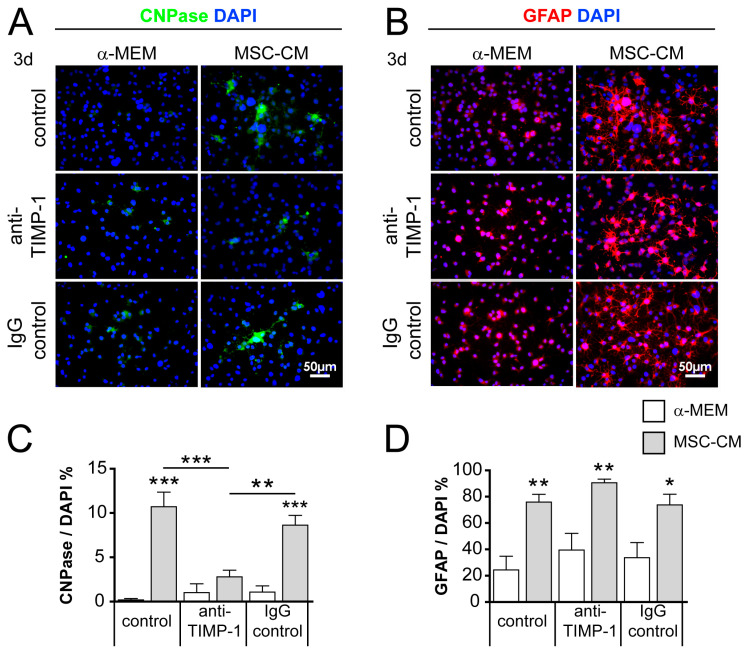
2.6. Neutralization of TIMP-1 Diminished the Pro-Oligodendroglial Effect of MSC-CM
Proteins found to be enriched in the MSC secretome included tissue inhibitor of metalloproteinase type 1 (TIMP-1), which represents a promising candidate protein with known pro-oligodendroglial properties [42,43,50]. In order to validate our protein identification approach, we performed specific TIMP-1 antibody blocking assays on cultured aNSCs. To this end, we applied the antibody to α-MEM and MSC-CM for 1 h prior to application of aNSCs. For further control, no antibody (normal control) and immunoglobulin G (IgG) isotype control antibody was applied. After 3 days of culture, we determined the degree of 2′,3′-cyclic-nucleotide 3′-phosphodiesterase (CNPase)-positivity among aNSCs. Neutralization of TIMP-1 in MSC-CM resulted in a significantly reduced number of CNPase-positive cells compared to untreated (control) MSC-CM, as well as to IgG antibody control-treated MSC-CM (Figure 7). After TIMP-1 neutralization, we further demonstrated that the number of GFAP-positive cells tended to increase compared to normal control and IgG antibody controls (Figure 7C). To exclude intrinsic TIMP-1 antibody effects, we also used a series of unconditioned α-MEM, which revealed no TIMP-1 or IgG control antibody effects on CNPase expression or GFAP-positive cells.
Figure 7.
Blocking the tissue inhibitor of metalloproteinase type 1 (TIMP-1) in MSC-CM reduced its pro-oligodendroglial activity on cultured aNSCs. Representative immunocytochemical staining of 2′,3′-cyclic-nucleotide 3′-phosphodiesterase (CNPase)- (A) and GFAP-positive (B) cells treated either with control (no antibody), anti-TIMP-1, or immunoglobulin G (IgG) control pre-incubated with α-MEM or MSC-CM after 3 days in culture. (C) Quantification of CNPase- and GFAP-positive (D) cell numbers. Number of experiments n = 4. Statistical significance was calculated using a two-way ANOVA with Bonferroni posttest: * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001.
3. Discussion
In demyelinating diseases, neural stem cells can be actively recruited for myelin repair [1,3,4,5,6,7]. However, despite of their ability to differentiate into oligodendrocytes, the extent of regeneration and remyelination in the adult CNS remains limited [11,12,13]. Thus, insights into the underlying mechanisms on how aNSCs can be fostered to differentiate into a specific cell lineage could help in establishing neuroregenerative treatment approaches, which are still a clinically unmet need. Forthcoming strategies could include the biological or pharmacological manipulation of endogenous aNSCs or, under specific circumstances, also the transplantation of aNSCs previously primed to acquire oligodendroglial cell fates. This second approach would nevertheless be restricted to well-described white matter lesions. However, in an experimental set-up, site-directed cell transplantation might indeed provide valuable information on how stem cells can be modulated and placed specifically in order to regenerate oligodendrocytes and white matter. Previous studies demonstrated that rodent and human MSC-secreted factors promote oligodendroglial differentiation of aNSCs in vitro [17,19,20,51]. However, the degree to which MSC-dependent lineage instruction is functional in vivo remains to be demonstrated. Moreover, the nature of MSC-derived differentiation factors regarding oligodendroglial lineage differentiation remain elusive thus far [21].
3.1. Enhanced Oligodendroglial Differentiation of MSC-CM-Stimulated aNSCs In Vivo
Our present study revealed that aNSC transplanted into different CNS tissues are able to survive at least up to 28 days post-grafting, independent of their pre-treatment. As expected, cell survival was diminished over time, which could depend on the immune response since the animals were not immune-suppressed as in other studies [52,53,54]. In comparison to the brain, the spinal cord environment appeared to provide a better survival rate. In this regard, it should be noted that this difference might also derive from rat cells being implanted into mouse brains, which could cause additional immune responses. Given that mouse neural stem cells have thus far not been shown to respond adequately to MSC-derived factors (own observations), we could not apply a direct comparison using a mouse-to-mouse transplantation scheme.
In the brain, grey and white matter can differently affect OPC differentiation [55], and we previously showed that white matter localization promotes differentiation towards myelinating oligodendrocytes at the expense of astrocyte generation when transplanted aNSC were depleted from the intrinsic oligodendrogenesis inhibitor p57kip2 [26]. However, in both studies, regional differences of the brain showed no impact on cell survival, which was confirmed by the here-described brain transplantation experiments. While transplantation of aNSCs into grey or white matter of the brain showed no difference in survival, most of the transplanted cells were found in white matter after spinal cord transplantation. It therefore appears that spinal cord white matter constitutes a preferred region for transplanted cells, which is in agreement with previous observations [56].
More importantly, our experiments clearly revealed that MSC-derived factors can indeed boost the generation of oligodendrocytes in vivo while preventing an astrocytic cell fate. Whereas such a directed stimulation depends on at least 3 days of pre-stimulation in cultured cells [20], our current results demonstrate that the pro-oligodendroglial effect in vivo can be equally well seen after a 1-day pre-treatment period. It thus appears that such an initial MSC-dependent stimulation of oligodendrogenesis is dominant and the corresponding impulse can be maintained in the CNS environment.
Besides the predominant expression of oligodendroglial markers, we only noticed myelin sheath generation in the spinal cord in response to pre-stimulation with mesenchymal-derived factors, indicating that not only quantitative but also qualitative effects are mediated. Myelin sheath generation was revealed by the presence of paranodal Caspr signals in close apposition to MBP-positive processes of MSC-CM pre-stimulated and transplanted cells [27,28]. Furthermore, representative examination of MSC-CM pre-stimulated cells by immunoelectron microscopy revealed myelinating processes ending in paranodal loops. This is furthermore supported by the observation that, at late time points, MSC-CM pre-treated aNSCs were more frequently observed in CNS tissues as these cells might have better survived due to successful tissue integration and axon ensheathment. Note that the generation of quantitative data on node formation and myelination is out of scope of this study and must be addressed in future investigations.
Although we detected more mature oligodendrocytes in response to MSC-CM, we found there was no difference in the number of Olig2-expressing cells, possibly reflecting the fact that Olig2-positivity, although differing in its subcellular distribution, is also featured by astrocytic cells [57].
The here-observed pro-oligodendroglial features within the almost healthy CNS (disregarding injection sites) suggest that similarly treated aNSCs might also be able to promote myelin repair upon injury and demyelination, also in light of the increased number of naked axons to be contacted. In addition, transplanted cells could also have an impact on endogenous progenitor or stem cells by direct cell–cell contact or by secretion of trophic factors, thus promoting endogenous remyelination capacity and functional regeneration. However, such experiments were out of the scope for the current analysis, but will be conducted in the future through applying toxin- as well as immune-mediated demyelination models.
3.2. MSCs Secreted a Number of Oligodendroglial Regulatory Proteins
Underlying pro-oligodendroglial mechanisms and proteins secreted by MSCs have not yet been deciphered in detail. Although some MSC secretome or proteome data already exist, most of them deal with anti-inflammatory, regenerative, and tumor-promoting properties of MSCs [23,24,25,58,59,60]. Therefore, we applied a quantitative secretome approach for the analysis of MSC-derived proteins with pro-oligodendroglial activity. From the initially identified 691 proteins, we determined 152 proteins (Table S1) as bona fide secreted due to the significant enrichment in MSC-CM. GO term enrichment analysis further confirmed that these proteins are mainly localized in the extracellular environment and predominantly involved in extracellular matrix (ECM) organization and cell adhesion. Moreover, in order to find pro-oligodendroglial factors, we further analyzed these 152 proteins regarding a pro-oligodendroglial fate choice (Table 1). On the basis of their previously shown impact on oligodendrogenesis, we identified first obvious promising candidates, among them ECM proteins such as Lamb2. Laminins were previously shown to be important for CNS myelination [61], and pericyte-dependent stimulation of aNSC-dependent oligodendrogenesis was recently found to depend on the related protein laminin subunit alpha 2 (Lama2) [62]. Laminins regulate CNS myelination by interacting with integrin receptors and by binding to the non-integrin ECM receptor dystroglycan [34], which was also enriched in the MSC secretome, possibly acting on OPC proliferation via its intracellular cleaved part [33]. Another bona fide secreted protein with a known pro-oligodendroglial function is chordin. As a BMP antagonist, it can promote oligodendrogenesis from SVZ neural stem cells [31], and we previously found chordin to be induced in response to the pro-oligodendroglial suppression of the p57kip2 inhibitor [63]. Furthermore, we detected syntenin-1, known to regulate OPC migration via interaction with NG2 [47]. While proteins such as dystroglycan and chordin promote proliferation and differentiation, prosaposin, also known as sulfated glycoprotein 1, promotes expression of myelin constituents and protects myelinating glial cells [45,46]. Protecting properties have also been assigned to the arrest-specific protein 6 (Gas6) [36], and its deficiency increases oligodendrocyte loss during cuprizone-induced demyelination [64]. Besides pro-oligodendroglial proteins, we also identified proteins inhibiting oligodendroglial differentiation such as metalloendopeptidase BMP 1 [41], thrombospondin 1 [48], and tenascin C [49]. An equilibrated action of positive and negative regulators might be necessary for a timed and successful cell fate acquisition, thereby contributing to successful establishment of new myelin sheaths. On the other hand, future studies will show whether blocking such proteins could also increase the efficiency of the MSC-CM treatment.
Finally, one of the most promising candidate proteins was TIMP-1, on the basis of its neuroprotective functions by means of matrix metalloproteinase (MMP) regulation [65] and its capacity to promote oligodendrocyte differentiation and myelination [42,43,50]. As we demonstrated that neutralization of TIMP-1 in MSC-CM results in a significantly reduced number of CNPase-positive cells and an increased (non-significant) degree of GFAP-positive cells in culture, we provide a proof-of-principle that the chosen secretome/proteome approach is valid. Moreover, we demonstrated that TIMP-1 is one of the key players in MSC-mediated oligodendrocyte differentiation and myelination.
Given the large number of differentiation-related proteins identified in the MSC secretome, we suggest that the MSC-CM effect depends on the interplay of both pro-oligodendroglial as well as anti-astrocytic factors possibly acting at different differentiation stages of which TIMP-1 constitutes one element. Upcoming experiments will therefore have to functionally investigate further protein candidates and also use combinations of neutralization antibodies or recombinant proteins for validation. Finally, given our observation that, in vivo, a much shorter instruction period by MSC-derived factors is sufficient for successful oligodendrogenesis, myelination, and cell integration, researchers should then test combinations of presumably active recombinant proteins on pre-stimulated aNSCs to be transplanted in vivo.
4. Materials and Methods
4.1. Animals
For the isolation of rat aNSCs and MSCs, female Wistar rats (8–10 weeks old, ≈210 g) were used throughout the study. Transplantation experiments into wild type female mice (C57Bl/6, 13–14 weeks old, ≈18–21g) and rats (Wistar, 10–12 weeks old, between 210 and 230 g) were all approved by the LANUV (Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen; Az.: 84-02.04.2015.A239; Az.: 84-02.04.2015.A525) and carried out in accordance with ethical care. All rodents were housed in a pathogen-free facility with 12 h light/dark cycle, and were supplied with food/water ad libitum.
4.2. Mesenchymal Stem Cell Culture
Preparation and cultivation of adult rat mesenchymal stem cells (MSCs) were performed as previously described [19,20]. For experiments, we cultured bone marrow-derived rat MSCs for 3 to 4 days in α-MEM/10% FBS (both Gibco Cell Culture, Life Technologies, Karlsruhe, Germany) containing 1% penicillin–streptomycin (Gibco) until reaching a confluent cell layer (90–100%). Afterwards, medium was changed and cells were cultured in α-MEM/10% FBS for another 3 to 4 days. The corresponding supernatant was filtered (0.2 µm filtropur S filter; Sarstedt AG and Co. KG, Nümbrecht, Germany) and used as rat mesenchymal stem cell-conditioned medium (MSC-CM) for aNSC stimulation. MSCs were used from passage 3 to 10.
4.3. Adult Rat Neural Stem Cell Culture and Transfection
Preparation and cultivation of aNSC derived from the rat subventricular zone (SVZ) were conducted as previously described [20,63]. SVZ-derived aNSCs were resuspended in neurobasal (NB) medium (Gibco) containing B27 (Gibco), 2 mM L-glutamine (Gibco), 1% penicillin–streptomycin (Gibco) supplemented with 2 mg/mL heparin (Sigma-Aldrich Chemie GmbH, Steinheim, Germany), 20 ng/mL fibroblast growth factor 2 (FGF-2; R&D Systems, Wiesbaden-Nordenstadt, Germany), and 20 ng/mL epidermal growth factor (EGF; R&D Systems, Wiesbaden-Nordenstadt, Germany), from here on referred to as NBall medium. For experiments, aNSCs were seeded with a cell number of 1 × 106 cells in 1 T75 culture flask in 10 mL NBall medium and cultured for 7 days at 37 °C in a humidified incubator with 5% CO2. The medium was exchanged twice a week, and on day 7, we passaged aNSCs using accutase (PAA Laboratories, Pasching, Austria) for separation (10–15 min at 37 °C). For differentiation, we seeded aNSC on acid-pretreated and poly-L-ornithine/laminin (100 µg/mL and 12 µg/mL, respectively, both Sigma-Aldrich Chemie GmbH)-coated 13 mm glass cover slips (6.3 × 104 cells per coverslip). Cells were cultured for 24 h in NBall medium before changing to differentiation control medium (α-MEM/10% FBS) or MSC-CM. For cell visualization of transplanted cells, we co-transfected aNSC with the UbC-StarTrack plasmids pCMV-hyPBase and UbC-EGFP [66] using the rat neural stem cell nucleofector kit (Lonza, Basel, Switzerland), as described previously [20]. Transfected cells were seeded with a concentration of 3 × 106 cells in 1 T75 culture flask in 10 mL NBall medium and were cultured for 7 days at 37 °C in a humidified incubator with 5% CO2. Two or four days before transplantation, we seeded aNSCs on poly-L-ornithine/laminin (100 µg/mL and 12 µg/mL, Sigma-Aldrich Chemie GmbH)-coated Petri dishes (35 × 10 mm) for 24 h in NBall medium before changing to control α-MEM/ 10% FBS or MSC-CM.
4.4. Antibody Blocking and Immunocytochemistry
For blocking experiments, we seeded aNSCs on acid-pretreated and poly-L-ornithine/laminin-coated glass cover slips (5 × 104 cells per coverslip) and cultured in NBall medium for 24 h. To neutralize metallopeptidase inhibitor 1 (TIMP-1), we incubated MSC-CM with an anti-rat TIMP-1 antibody (25 µg/mL, R&D System, AF580) for 1 h at 37 °C, prior to application to aNSCs. Isotype control IgG antibody (25 µg/mL, R&D System, AB-108-C) was used as control. In addition, we also applied both antibodies to normal control medium (α-MEM / 10% FCS) in order to exclude unspecific antibody effects. After 3 days (blocking experiments) or 1–7 days (PiggyBac-transfected aNSC differentiation) of incubation with α-MEM / 10% FCS or MSC-CM, we fixed aNSCs using 4% paraformaldehyde (PFA) and subjected them to immunocytochemical staining, as previously described [20]. After primary antibodies were blocked of unspecific binding sites and permeabilized for 45 min in 1% normal goat serum (NGS; Sigma-Aldrich Chemie GmbH) and 0.1% Triton in phosphate-buffered saline (PBS), we incubated them in 1% NGS (in PBS, 0.03% Triton) at 4 °C overnight. The following antibodies were used: rabbit anti-glial fibrillary acidic protein (GFAP; 1:4000, DAKO Agilent, Santa Clara, CA, USA; research resource identifiers (RRID): AB_10013382), mouse anti-2′,3′-cyclic-nucleotide 3′-phosphodiesterase (CNPase; 1:500, Biolegend, San Diego, CA, USA; RRID: AB_2565362), chicken anti-green fluorescent protein/citrine (GFP; 1:2000; Aves Labs, Tigard, OR, US; RRID: AB_10000240), rabbit anti-neural/glial antigen 2 (NG2; 1:100; Merck Millipore, Burlington, MA, USA; RRID: AB_11213678, 1 h RT), and rat anti-myelin basic protein (MBP; 1:250, Bio-Rad Laboratories, Inc., Hercules, CA, USA; RRID: AB_325004). Primary antibody incubation was followed by 3 washing steps with PBS and incubation with secondary (anti-mouse and anti-rabbit) antibodies conjugated with either Alexa Fluor594 or Alexa Fluor488 (1:500; Thermo Fisher Scientific, Darmstadt, Germany) in 1% NGS (in PBS, 0.03% Triton) supplemented with 4’,6-diamidin-2-phenylindol (DAPI, 0.02 µL/mL; Roche Diagnostic GmbH, Mannheim, Germany) at room temperature for 90 min. Cells were mounted with Citifluor (Cilifluor, Leicester, United Kingdom) and images were taken using a Zeiss Axioplan2 microscope (Carl Zeiss AG, Jena, Germany). For quantitative analyses, we used ImageJ Software (National Institute of Health (NIH) USA) [67,68] to count the positive cells that were normalized to the total cell number (DAPI-positive cells). For statistical analysis, we applied two-way analysis of variance (ANOVA) with Bonferroni posttest by using GraphPad Prism 5.0c software.
4.5. Stereotactic Cell Transplantations
Rat aNSCs were detached with accutase and kept at room temperature in their respective media. For transplantation into the mouse brain, aNSCs were centrifuged for 5 min at 140× g, washed once in PBS, and were finally resuspended in PBS to a concentration of 1 × 105 cells/μL. The transplantation procedure was performed as previously described [26]. Briefly, recipient C57Bl/6J mice were deeply anesthetized using isoflurane or sevoflurane inhalation. The skull was exposed using a scalpel, and holes were drilled at 0.7 mm (anterior–posterior), ± 1 mm (medial–lateral). Approximately 0.75 μL of the cell suspension was injected in the white (corpus callosum) and grey (cortex) matter of the mice brains (13–14 weeks old) according to [55]. Transplantations were performed with a Hamilton syringe (10 μL Neuros Model 1701 RN, ga 33, L 0–20 mm) at 0.7 mm (anterior–posterior), ± 1 mm (medial–lateral), 2.1 to 1.8 mm (dorsal–ventral) relative to Bregma using a motorized robot stereotaxic instrument and StereoDrivesoftware (Neurostar, Tubingen, Germany). Postoperative care comprised an analgesic treatment (Rimadyl, Pfizer Deutschland GmbH, Berlin, Germany; 5 mg/kg) for 3 days, starting on the day of operation. For tissue collection, mice were deeply anesthetized with isoflurane and transcardially perfused with 20 mL cold PBS followed by 20 mL 4% PFA. Mouse brains were post-fixed overnight in 4% PFA at 4 °C, followed by 24 to 48 h cryoprotective dehydration in 30% sucrose (in PBS) at 4 °C. Brains were embedded in Tissue-Tek OCT (Sakura Finetek Europe, The Netherlands), frozen, and stored at −80 °C until preparation of 10 μm sections using a cryostat (Leica CM3050S). Sections were stored at −80 °C.
For spinal cord transplantations, adult female rats were treated as previously described [26,69] with slight modifications. Briefly, rat aNSC transplantation was performed at thoracic level 8 (Th8) using a Hamilton syringe (10 μL Neuros Model 1701 RN, ga 33, L 0–20 mm) attached to a Small Animal Stereotaxic Instrument (David Kopf Instruments, Tujunga, CA, USA). Injections were performed at 0.1 mm lateral to the midline and 1.1 to 0.8 mm dorsal–ventral from the dural surface. At each transplantation site, we slowly and within 4 min injected 1 µL of either 1 × 105 control α-MEM pre-stimulated (3 day) or MSC-CM pre-stimulated (3 days) aNSCs in PBS. Postoperative care included prophylactic oral antibiotic treatment (Baytril, Bayer Health Care, Leverkusen, Germany; 0.4 mL/kg) for 7 days and analgesic treatment (Rimadyl, Pfizer, New York, NY, USA; 5 mg/kg) for 3 days starting, on the day of surgery. After 14 or 28 days post-operation, animals were deeply anesthetized using a mixed solution containing ketamine (100 mg/kg body weight) and xylazine (10 mg/kg body weight), and were transcardially perfused with 200 mL 4 °C ice-cold PBS followed by 400 mL 4% PFA. Spinal cords were harvested and post-fixed overnight in 4% PFA at 4 °C, followed by 24 h washing in PBS and 24 to 48 h cryoprotection in 30% sucrose (in PBS) at 4 °C. Spinal cords were then embedded in Tissue-Tek OCT (Sakura Finetek Europe, Netherlands), frozen, and stored at −30 °C until preparation of 14 μm sections using a cryostat (Leica, Wetzlar, Germany, CM3050S). Sections were stored at −30 °C.
4.6. Immunohistochemistry
Immunohistochemical staining was performed as previously described [26]. Briefly, defrosted brain and spinal cord sections were air-dried for at least 15 min at room temperature (RT), and were afterwards rehydrated for 5 min in distilled water, transferred to −20 °C acetone (5 min), and washed in 1x tris-buffered saline (TBS) (pH 7.6) and 1x TBS-T (TBS containing 0.02% Triton) for 5 min each. For blocking, we used 10% biotin-free bovine serum albumin (BSA; Carl Roth, Karlsruhe, Germany, in TBS-T) for 30 min at RT, followed by application of the following antibodies (in 5 to 10% BSA in TBS) and incubation overnight: rabbit anti-oligodendrocyte transcription factor 2 (Olig2; in brain 1:2000, in spinal cord 1:1000; Merck Millipore, Burlington, MA, USA; RRID: AB_570666, RT), rabbit anti-neural/glial antigen 2 (NG2; 1:100; Merck Millipore, Burlington, MA, USA; RRID: AB_11213678, RT), rabbit anti-glutathione-S-transferase-π (GSTπ; 1:4500; ENZO Life Sciences, GmbH, Lörrach, Germany; RRID: AB_10615079; 4 °C), rabbit GFAP (1:10000; DAKO Agilent, Santa Clara, CA, USA; RRID: AB_10013382; 4 °C), rat anti-MBP (1:500, Bio-Rad Laboratories, Inc., Hercules, CA, USA; RRID: AB_325004, RT or 4 °C), chicken anti-green fluorescent protein/citrine (GFP; 1:2000; Aves Labs, Tigard, OR, USA; RRID: AB_10000240; 4 °C or RT), mouse anti-Caspr (anti-Caspr/paranodin/neurexin IV, clone K65/35; 1:500; 75-001; UC Davis/NIH NeuroMab Facility, CA, USA; RRID: AB_10671175; 4 °C), rabbit anti-neurofilament (NF; 1:2500; Abcam, Cambridge, United Kingdom; RRID: AB_306298; 4°C), and rabbit anti-neuronal nuclei antigen (NeuN; 1:1000; Abcam, Cambridge, United Kingdom; RRID: AB_2532109; 4 °C).
Sections were washed 2 times in TBS for 5 min and incubated with appropriate secondary antibodies (goat anti-chicken; goat anti-rabbit; goat anti-rat conjugated with either Alexa Fluor 594, 488, 405, or 647; 1:200 in TBS; Thermo Fisher Scientific, Darmstadt, Germany) for 30 min at RT. DAPI (0.04 µl/mL; Roche Diagnostic GmbH) or RedDot 2 (1:200; Biotium, Fremont, CA, USA; cat. #: 40061) were included for nuclei labelling. Slices were mounted with Immu-Mount (Thermo Fisher Scientific, Darmstadt, Germany) and analyzed using a confocal confocal laser scanning microscopy (CLSM) microscope 510 (CLSM 510, Carl Zeiss AG, Oberkochen, Germany). Immunohistochemical staining on brain and spinal cord sections was performed using the corresponding centers of transplantation. An average of 25 brain sections and 13 spinal cord sections per marker and per time-point were analyzed, including 3 brain slices or 2 spinal cord slices per each slide. Fluorescently labeled cells were counted on each picture/section, and the mean number per slide (containing 3 brain or 2 spinal cord sections, respectively) was calculated, leading to an average value for each animal.
4.7. Immunoelectron Microscopy
Immunoelectron microscopy was performed as previously described [26], with slight modifications. Briefly, adult rats were perfused with 4% PFA in 1 M cacodylate buffer (Carl Roth GmbH + Co. KG, Karlsruhe, Germany), and spinal cords were harvested and post-fixed overnight in 4% PFA in 1M cacodylate buffer. After post-fixation, spinal cords were washed over night in PBS and embedded in 6% agarose in cacodylate buffer. Coronal sections (50 µm) were cut in PBS using a microtome (Microm HM 650 V, Thermo Fisher Scientific, Waltham, MA, USA). Free-floating sections were blocked with 1% BSA in PBS and incubated with 0.1 M NaIO3 in PBS and subsequently in 5% dimethyl sulfoxide in PBS. Sections were incubated with rabbit anti-GFP antibody (1:100; AB3080, Merck Millipore, Burlington, MA, USA; RRID: AB_91337) in 1% BSA in PBS overnight at 4 °C, and immune reactions were subsequently visualized using a biotinylated secondary antibody (biotinylated goat anti-rabbit IgG; 1:50; BA-1000, Vector Laboratories, Burlingame, CA, USA; RRID: AB_2313606) and streptavidin–biotin–peroxidase (PK-6100, Vector Laboratories, Burlingame, CA, USA; RRID: AB_2336819) complex using diaminobenzidine–HCl (SK-4105, Vector Laboratories, Burlingame, CA, USA; RRID: AB_2336520) and H2O2. After diaminobenzidine staining, appropriate transplantation regions were cut, and sections were osmicated and processed for light and electron microscopy by dehydration and embedding in Spurr’s medium. Ultrathin sections (70 nm) were mounted to copper grids, counterstained with lead citrate, and investigated using a ProScan Slow Scan CCD camera connected to a Leo 906 E electron microscope (Zeiss, Jena, Germany) and corresponding software iTEM (Soft Imaging System). Transplanted cells were identified on the basis of GFP immunoreactivity (indicated by dark precipitates).
4.8. Statistical Analysis of IHC and ICC
Statistical analyses and graphs were generated using Excel and Graph-Pad Prism 5.0 software. To determine statistical significance in graphs with more than two conditions, we applied ordinary two-way analysis of variance (ANOVA) with Bonferroni posttest. Statistical significance thresholds were set as follows: * p < 0.05; ** p < 0.01; *** p < 0.001. All data are shown as mean values ± SEM, and “n” represents the number of independent experiments performed.
4.9. Preparation of Mesenchymal Stem Cell Secretome and Proteome
Mesenchymal stem cells were prepared, as described previously, through seeding 4 × 105 cell/100 mm Ø culture dish in α-MEM-10% FBS until a confluent cell layer was achieved. For mass spectrometry analysis, we used four independent biological replicates of MSCs. Confluent MSCs were washed three times with PBS in order to remove the FBS and were then incubated for another 48h in serum-free N2-medium [22,70,71] consisting of Dulbecco’s modified Eagle’s medium (DMEM, Gibco Cell Culture, Thermo Fisher Scientific, Darmstadt, Germany) containing 5 mg/mL insulin, 30 nM sodium selenite, 100 µM putrescine, 20 nM progesterone, and 5 µg/mL transferrin (all Sigma-Aldrich Chemie GmbH). Medium was changed again after 48 h to further exclude FBS effects, and was then incubated for another 48 h in N2 medium.
To analyze secreted proteins within this second N2 medium preparation by mass spectrometry, we performed trichloroacetic acid (TCA) precipitation as previously described [70,71]. Briefly, conditioned medium was transferred to a 50 mL tube, centrifuged at 950× g at 4 °C for 10 min, and filtered through a 0.2 µm filter (Pall Acrodisc). The medium was mixed with 10% sodium lauroyl sarcosinate (Sigma-Aldrich Chemie GmbH) up to a final concentration of 0.1%. Ice-cold trichloroacetic acid (Sigma-Aldrich Chemie GmbH) was added (7.5% final concentration). The medium was incubated for 1 h on ice and then centrifuged at 7100× g at 4 °C for 10 min. After removing the supernatant, we resuspended the pellet in 1 mL ice-cold acetone and we then centrifuged it again. After removing the supernatant, we dried the pellet at room temperature and then dissolved it in lysis buffer consisting of 30 mM Tris base (Sigma-Aldrich Chemie GmbH), 7M urea (Sigma-Aldrich Chemie GmbH,), and 2 M thiourea (Sigma-Aldrich Chemie GmbH). The protein content was determined by Pierce 660 nm Protein Assay (Thermo Fisher Scientific).
To analyze the corresponding cell proteome, we washed MSCs 3 times with cold PBS after removing the conditioned medium for secretome analysis. Afterwards, 2 mL cold PBS was added and cells were detached using a cell scraper. Cell suspension was transferred to a 15 mL tube and centrifuged with 800× g at 4 °C for 5 min. The supernatant was removed, and cell pellets were immediately frozen and stored at −80 °C. MSCs were lysed by using the lysis buffer described above. The lysate was sonicated 6 × 10 s, centrifuged at 16,000× g for 15 min, and the protein concentration of the supernatant was then determined.
For in-gel digestion, 5 µg total protein of each sample (secretomes as well as proteomes) was used for short SDS gel electrophoresis (5 mm running distance, 10 min) and stained with Coomassie brilliant blue. The resulting lanes were excised, washed, reduced with 10 mM dithiothreitol (DTT; Serva Electrophoresis, Heidelberg, Germany) and alkylated with 55 mM iodacetamide (Sigma-Aldrich Chemie GmbH). Proteins were digested with 0.1 µg trypsin (Serva Electrophoresis) overnight at 37 °C, and peptides were extracted, dried, and finally resuspended in 0.1% trifluoroacetic acid.
4.10. Liquid Chromatography and Mass Spectrometric Analysis
Extracted peptides were separated by an Ultimate 3000 RSCLnano System (Thermo Fisher Scientific) with an Acclaim PepMap100 trap column (3 µm C18 particle size, 100 Å pore size, 75 µm inner diameter, 2 cm length, Thermo Fisher Scientific) as a precolumn using 0.1% trifluoroacetic acid (TFA) as a mobile phase and an Acclaim PepMapRSLC (2 µm C18 particle size, 100 Å pore size, 75 µm inner diameter, 25 cm length, Thermo Fisher Scientific) analytical column. The flow rate for analytical separation was constant with 300 nL/min, and a 2 h gradient of 0.1% formic acid (Fluka) to 0.1% formic acid/60% acetonitrile was used for peptide separation. Peptides were eluted via nano electrospray ionization into the mass spectrometer (Orbitrap Fusion Lumos mass spectrometer, Thermo Fisher Scientific) operated in positive mode with advanced peak determination enabled. Precursor mass spectra were recorded in the orbitrap analyzer within a mass range of 200–2000 m/z and a resolution of 120,000 (maximum ion time: 60 ms, automatic gain control target value: 250,000, profile mode). For a maximum of 2 s, we isolated precursors with charge states +2 to +7 and a minimum intensity of 5000 within a 1.6 m/z isolation window and fragmented via higher-energy collisional dissociation. MS/MS spectra were recorded in the linear ion trap in centroid mode with a maximal ion time of 50 ms and a target value for the automatic gain control was set to 10,000. The scan rate was “rapid”, and already fragmented precursors were excluded from further isolation for the next 60 s.
4.11. Analysis of Mass Spectrometric Data
To identify peptides and proteins, we used the MaxQuant environment (version 1.6.6.0, MPI for Biochemistry, Planegg, Germany). If not stated otherwise, the identification was done with standard parameters. Searches were performed using the rat UP000002494 proteome dataset (29,951 entries) downloaded on 10th April 2019 from the UniProt Knowledgebase using tryptic specificity (cleavage behind R and K) with a maximum of 2 missed cleavages sites. Carbamidomethylation at cysteines was considered as fixed, and methionine oxidation was set as variable modification. A first search was performed with 20 ppm precursor mass tolerance. Peptides identified with high confidence were then used for recalibration using the “software lock mass” feature of MaxQuant [72]. Thereafter, a main search was conducted with a precursor mass tolerance of 4.5 ppm. The mass tolerance or fragment spectrum was set to 0.5 Da. Peptides and proteins were accepted at a false discovery rate of 1%. The “match between runs” function was enabled as well as label-free quantification.
Corresponding secretomes and proteomes were compared to identify enriched proteins in the secretome, probably representing secreted proteins according to Grube et al. [29]. After a MaxQuant-based database search and quantification, we analyzed quantitative protein level data within the Perseus framework (version 1.6.6.0, MPI for Biochemistry, Planegg, Germany). Proteins identified only by site or marked as contaminant (from the MaxQuant contaminant list) as well as reverse hits were excluded. Proteins with at least 2 identified peptides and a minimum of 3 valid values in at least 1 group were considered. Calculations were done on normalized intensities (LFQ intensities), as provided by MaxQuant, and missing data were imputed before statistical analysis by values from a downshifted normal distribution (width 0.3 standard deviations, down-shift 1.8 standard deviations). The significance analysis of microarrays (SAM) method [73] was applied on log2-transformed values using a S0 constant of 0.8 and a 5% false discovery rate-based cutoff. Presented fold changes were calculated as difference from mean values of log2-transformed intensities.
To predict unconventionally secreted proteins, we used the OutCyte prediction tool [74], wherein we selected classically secreted proteins on the basis of the annotation from UniProtKB (signal peptide predicted, but neither a transmembrane domain nor a kdel sequence present). Gene Ontology (GO) biological process (GOBP) and cellular compartment (GOCC) were used for categorical annotations of proteins identified in the MSC-derived secretome, and annotation enrichments were calculated by Fisher’s exact tests. Enrichment associated p-values were adjusted via the method of Benjamini and Hochberg.
4.12. Data Availability
The mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE [75] partner repository with the dataset identifier PXD018231.
5. Conclusions
In the present study, we demonstrated that MSC-derived secreted factors promote oligodendrogenesis of aNSCs transplanted into different CNS regions. Furthermore, mass spectrometry-based secretome analysis provided insights into the underlying mechanisms. Our analysis identified several proteins previously shown to regulate oligodendrogenesis. As a promising protein candidate, we validated TIMP-1 to be an active component of the MSC-conditioned medium promoting oligodendrogenesis in vitro. Given that MSC-derived factors are sufficient to promote successful oligodendrogenesis, myelination and cell integration, transplantation of pre-stimulated aNSCs upon CNS injury, and demyelination could help to promote functional remyelination.
Acknowledgments
We thank Birgit Blomenkamp, Julia Jadasz, Zippora Kohne, Marion Hendricks, and Brigida Ziegler for technical assistance.
Abbreviations
| α-MEM | Alpha-minimum essential medium |
| Acan | Aggrecan |
| ANOVA | Analysis of variance |
| aNSCs | Adult neural stem cells |
| BMP | Bone morphogenetic protein |
| BSA | Bovine serum albumin |
| Caspr | Contactin-associated protein |
| Chrd | Chordin |
| CLSM | Confocal laser scanning microscopy |
| CNPase | 2′,3′-Cyclic-nucleotide 3′-phosphodiesterase |
| CNS | Central nervous system |
| CTGF | Connective tissue growth factor |
| Dag1 | Dystroglycan |
| DAPI | 4´,6-diamidin-2-phenylindol |
| DMEM | Dulbecco’s modified Eagle’s medium |
| DTT | Dithiothreitol |
| ECM | Extracellular matrix |
| EF | Enrichment factor |
| EGF | Epidermal growth factor |
| EM | Electron microscopy |
| FCS | Fetal calf serum |
| FDR | False discovery rate |
| FGF | Fibroblast growth factor |
| Fn | Fibronectin |
| Gas6 | Growth arrest-specific protein 6 |
| GFAP | Glial fibrillary acidic protein |
| GFP | Green fluorescent protein |
| GM | Grey matter |
| GSTπ | Glutathione-S-transferase-π |
| GO | Gene Ontology |
| GOBP | Gene Ontology biological process |
| GOCC | Gene Ontology cellular component |
| IGFBP | Insulin-like growth factor-binding protein |
| IgG | Immunoglobulin G |
| Lamb2 | Laminin subunit beta-2 |
| LC–MS/MS | Liquid chromatography–mass spectrometry/mass spectrometry |
| MBP | Myelin basic protein |
| MMP | Matrix metalloproteinase |
| MSCs | Mesenchymal stem cells |
| MSC-CM | Mesenchymal stem cell conditioned medium |
| NB | Neurobasal |
| NF | Neurofilament |
| NG2 | Neural/glial antigen 2 |
| NGS | Normal goat serum |
| Olig2 | Oligodendrocyte transcription factor 2 |
| OPCs | Oligodendrocyte progenitor cells |
| PBS | Phosphate-buffered saline |
| PFA | Paraformaldehyde |
| ps | Pre-stimulated |
| Psap | Prospaposin |
| pt | Post transplantation |
| Sdcbp | Syntenin-1 |
| SP | Signal peptide |
| Spp1 | Osteopontin |
| SVZ | Subventricular zone |
| TCA | Trichloroacetic acid |
| Thbs1 | Thrombospondin 1 |
| TIMP-1 | Tissue inhibitor of metalloproteinase type 1 |
| TM | Transmembrane |
| Tnc | Tenascin c |
| UPS | Unconventional protein secretion |
| WM | White matter |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/12/4350/s1.
Author Contributions
Conceptualization, I.S.A. and P.K.; data curation, I.S.A. and J.S.-H.; formal analysis, I.S.A., J.S.-H., and G.P.; funding acquisition, D.K., H.P.H., J.A., and P.K.; investigation, I.S.A., J.S.-H., F.B., J.G., C.B., V.E., and G.P.; methodology, I.S.A., J.S.-H., G.P., J.J.J., D.K., and K.S.; project administration, I.S.A. and P.K.; resources, A.B., L.L.-M., R.M., and H.W.M.; supervision, P.K.; validation, I.S.A., J.S.-H., and G.P.; visualization, I.S.A. and J.S.-H.; writing—original draft, I.S.A., J.S.-H., and P.K. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Christiane and Claudia Hempel Foundation for clinical stem cell research, iBrain, Stifterverband/Novartisstiftung and the James and Elisabeth Cloppenburg, Peek and Cloppenburg Düsseldorf Stiftung.
Conflicts of Interest
I.S.A, J.S.-H., F.B., J.G., C.B., V.E., G.P., A.B., J.J.J., L.L.-M., D.K., R.M. H.W.M., J.A., and K.S. report no conflict of interest. P. Küry performed consultancy work for Geneuro. H.-P. Hartung received compensation for consulting, speaking, and serving on steering committees from Bayer Healthcare, Biogen, Geneuro, MedImmune, Merck, Novartis, Opexa, Receptos Celgene, Roche, Sanofi Genzyme, and Teva, with approval by the Rector of Heinrich-Heine University.
References
- 1.Franklin R.J. Why does remyelination fail in multiple sclerosis? Nat. Rev. Neurosci. 2002;3:705–714. doi: 10.1038/nrn917. [DOI] [PubMed] [Google Scholar]
- 2.Jessberger S., Toni N., Clemenson G.D., Jr., Ray J., Gage F.H. Directed differentiation of hippocampal stem/progenitor cells in the adult brain. Nat. Neurosci. 2008;11:888–893. doi: 10.1038/nn.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akkermann R., Jadasz J.J., Azim K., Küry P. Taking Advantage of Nature’s Gift: Can Endogenous Neural Stem Cells Improve Myelin Regeneration? Int. J. Mol. Sci. 2016;17:1895. doi: 10.3390/ijms17111895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nait-Oumesmar B., Picard-Riera N., Kerninon C., Decker L., Seilhean D., Hoglinger G.U., Hirsch E.C., Reynolds R., Baron-Van Evercooren A. Activation of the subventricular zone in multiple sclerosis: Evidence for early glial progenitors. Proc. Natl. Acad. Sci. USA. 2007;104:4694–4699. doi: 10.1073/pnas.0606835104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nait-Oumesmar B., Picard-Riera N., Kerninon C., Baron-Van Evercooren A. The role of SVZ-derived neural precursors in demyelinating diseases: From animal models to multiple sclerosis. J. Neurol. Sci. 2008;265:26–31. doi: 10.1016/j.jns.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 6.Picard-Riera N., Decker L., Delarasse C., Goude K., Nait-Oumesmar B., Liblau R., Pham-Dinh D., Baron-Van Evercooren A. Experimental autoimmune encephalomyelitis mobilizes neural progenitors from the subventricular zone to undergo oligodendrogenesis in adult mice. Proc. Natl. Acad. Sci. USA. 2002;99:13211–13216. doi: 10.1073/pnas.192314199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xing Y.L., Roth P.T., Stratton J.A., Chuang B.H., Danne J., Ellis S.L., Ng S.W., Kilpatrick T.J., Merson T.D. Adult neural precursor cells from the subventricular zone contribute significantly to oligodendrocyte regeneration and remyelination. J. Neurosci. 2014;34:14128–14146. doi: 10.1523/JNEUROSCI.3491-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charles P., Reynolds R., Seilhean D., Rougon G., Aigrot M.S., Niezgoda A., Zalc B., Lubetzki C. Re-expression of PSA-NCAM by demyelinated axons: An inhibitor of remyelination in multiple sclerosis? Brain. 2002;125:1972–1979. doi: 10.1093/brain/awf216. [DOI] [PubMed] [Google Scholar]
- 9.Mi S., Miller R.H., Lee X., Scott M.L., Shulag-Morskaya S., Shao Z., Chang J., Thill G., Levesque M., Zhang M., et al. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat. Neurosci. 2005;8:745–751. doi: 10.1038/nn1460. [DOI] [PubMed] [Google Scholar]
- 10.Mi S., Pepinsky R.B., Cadavid D. Blocking LINGO-1 as a therapy to promote CNS repair: From concept to the clinic. CNS Drugs. 2013;27:493–503. doi: 10.1007/s40263-013-0068-8. [DOI] [PubMed] [Google Scholar]
- 11.De Castro F., Bribian A., Ortega M.C. Regulation of oligodendrocyte precursor migration during development, in adulthood and in pathology. Cell Mol. Life Sci. 2013;70:4355–4368. doi: 10.1007/s00018-013-1365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kremer D., Aktas O., Hartung H.P., Küry P. The complex world of oligodendroglial differentiation inhibitors. Ann. Neurol. 2011;69:602–618. doi: 10.1002/ana.22415. [DOI] [PubMed] [Google Scholar]
- 13.Kuhlmann T., Miron V., Cui Q., Wegner C., Antel J., Bruck W. Differentiation block of oligodendroglial progenitor cells as a cause for remyelination failure in chronic multiple sclerosis. Brain. 2008;131:1749–1758. doi: 10.1093/brain/awn096. [DOI] [PubMed] [Google Scholar]
- 14.Gruchot J., Weyers V., Göttle P., Förster M., Hartung H.P., Küry P., Kremer D. The Molecular Basis for Remyelination Failure in Multiple Sclerosis. Cells. 2019;8:825. doi: 10.3390/cells8080825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotter M.R., Stadelmann C., Hartung H.P. Enhancing remyelination in disease--can we wrap it up? Brain. 2011;134:1882–1900. doi: 10.1093/brain/awr014. [DOI] [PubMed] [Google Scholar]
- 16.Uccelli A., Moretta L., Pistoia V. Mesenchymal stem cells in health and disease. Nat. Rev. Immunol. 2008;8:726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 17.Rivera F.J., Couillard-Despres S., Pedre X., Ploetz S., Caioni M., Lois C., Bogdahn U., Aigner L. Mesenchymal stem cells instruct oligodendrogenic fate decision on adult neural stem cells. Stem Cells. 2006;24:2209–2219. doi: 10.1634/stemcells.2005-0614. [DOI] [PubMed] [Google Scholar]
- 18.Kassis I., Grigoriadis N., Gowda-Kurkalli B., Mizrachi-Kol R., Ben-Hur T., Slavin S., Abramsky O., Karussis D. Neuroprotection and immunomodulation with mesenchymal stem cells in chronic experimental autoimmune encephalomyelitis. Arch. Neurol. 2008;65:753–761. doi: 10.1001/archneur.65.6.753. [DOI] [PubMed] [Google Scholar]
- 19.Jadasz J.J., Kremer D., Göttle P., Tzekova N., Domke J., Rivera F.J., Adjaye J., Hartung H.P., Aigner L., Küry P. Mesenchymal stem cell conditioning promotes rat oligodendroglial cell maturation. PLoS ONE. 2013;8:e71814. doi: 10.1371/journal.pone.0071814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jadasz J.J., Tepe L., Beyer F., Samper Agrelo I., Akkermann R., Spitzhorn L.S., Silva M.E., Oreffo R.O.C., Hartung H.P., Prigione A., et al. Human mesenchymal factors induce rat hippocampal- and human neural stem cell dependent oligodendrogenesis. Glia. 2018;66:145–160. doi: 10.1002/glia.23233. [DOI] [PubMed] [Google Scholar]
- 21.Rivera F.J., Kandasamy M., Couillard-Despres S., Caioni M., Sanchez R., Huber C., Weidner N., Bogdahn U., Aigner L. Oligodendrogenesis of adult neural progenitors: Differential effects of ciliary neurotrophic factor and mesenchymal stem cell derived factors. J. Neurochem. 2008;107:832–843. doi: 10.1111/j.1471-4159.2008.05674.x. [DOI] [PubMed] [Google Scholar]
- 22.Schira-Heinen J., Grube L., Waldera-Lupa D.M., Baberg F., Langini M., Etemad-Parishanzadeh O., Poschmann G., Stühler K. Pitfalls and opportunities in the characterization of unconventionally secreted proteins by secretome analysis. Biochim. Biophys. Acta Proteins Proteom. 2019;1867:140237. doi: 10.1016/j.bbapap.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Maffioli E., Nonnis S., Angioni R., Santagata F., Cali B., Zanotti L., Negri A., Viola A., Tedeschi G. Proteomic analysis of the secretome of human bone marrow-derived mesenchymal stem cells primed by pro-inflammatory cytokines. J. Proteom. 2017;166:115–126. doi: 10.1016/j.jprot.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 24.Teixeira F.G., Carvalho M.M., Panchalingam K.M., Rodrigues A.J., Mendes-Pinheiro B., Anjo S., Manadas B., Behie L.A., Sousa N., Salgado A.J. Impact of the Secretome of Human Mesenchymal Stem Cells on Brain Structure and Animal Behavior in a Rat Model of Parkinson’s Disease. Stem Cells Transl. Med. 2017;6:634–646. doi: 10.5966/sctm.2016-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baberg F., Geyh S., Waldera-Lupa D., Stefanski A., Zilkens C., Haas R., Schroeder T., Stühler K. Secretome analysis of human bone marrow derived mesenchymal stromal cells. Biochim. Biophys. Acta Proteins Proteom. 2019;1867:434–441. doi: 10.1016/j.bbapap.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 26.Beyer F., Jadasz J., Samper Agrelo I., Schira-Heinen J., Groh J., Manousi A., Butermann C., Estrada V., Reiche L., Cantone M., et al. Heterogeneous fate choice of genetically modulated adult neural stem cells in gray and white matter of the central nervous system. Glia. 2020;68:393–406. doi: 10.1002/glia.23724. [DOI] [PubMed] [Google Scholar]
- 27.Samanta J., Grund E.M., Silva H.M., Lafaille J.J., Fishell G., Salzer J.L. Inhibition of Gli1 mobilizes endogenous neural stem cells for remyelination. Nature. 2015;526:448–452. doi: 10.1038/nature14957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koutsoudaki P.N., Papastefanaki F., Stamatakis A., Kouroupi G., Xingi E., Stylianopoulou F., Matsas R. Neural stem/progenitor cells differentiate into oligodendrocytes, reduce inflammation, and ameliorate learning deficits after transplantation in a mouse model of traumatic brain injury. Glia. 2016;64:763–779. doi: 10.1002/glia.22959. [DOI] [PubMed] [Google Scholar]
- 29.Grube L., Dellen R., Kruse F., Schwender H., Stühler K., Poschmann G. Mining the Secretome of C2C12 Muscle Cells: Data Dependent Experimental Approach To Analyze Protein Secretion Using Label-Free Quantification and Peptide Based Analysis. J. Proteome Res. 2018;17:879–890. doi: 10.1021/acs.jproteome.7b00684. [DOI] [PubMed] [Google Scholar]
- 30.Keough M.B., Rogers J.A., Zhang P., Jensen S.K., Stephenson E.L., Chen T., Hurlbert M.G., Lau L.W., Rawji K.S., Plemel J.R., et al. An inhibitor of chondroitin sulfate proteoglycan synthesis promotes central nervous system remyelination. Nat. Commun. 2016;7:11312. doi: 10.1038/ncomms11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jablonska B., Aguirre A., Raymond M., Szabo G., Kitabatake Y., Sailor K.A., Ming G.L., Song H., Gallo V. Chordin-induced lineage plasticity of adult SVZ neuroblasts after demyelination. Nat. Neurosci. 2010;13:541–550. doi: 10.1038/nn.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ercan E., Han J.M., Di Nardo A., Winden K., Han M.J., Hoyo L., Saffari A., Leask A., Geschwind D.H., Sahin M. Neuronal CTGF/CCN2 negatively regulates myelination in a mouse model of tuberous sclerosis complex. J. Exp. Med. 2017;214:681–697. doi: 10.1084/jem.20160446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leiton C.V., Aranmolate A., Eyermann C., Menezes M.J., Escobar-Hoyos L.F., Husain S., Winder S.J., Colognato H. Laminin promotes metalloproteinase-mediated dystroglycan processing to regulate oligodendrocyte progenitor cell proliferation. J. Neurochem. 2015;135:522–538. doi: 10.1111/jnc.13241. [DOI] [PubMed] [Google Scholar]
- 34.Colognato H., Galvin J., Wang Z., Relucio J., Nguyen T., Harrison D., Yurchenco P.D., Ffrench-Constant C. Identification of dystroglycan as a second laminin receptor in oligodendrocytes, with a role in myelination. Development. 2007;134:1723–1736. doi: 10.1242/dev.02819. [DOI] [PubMed] [Google Scholar]
- 35.Lourenco T., Paes de Faria J., Bippes C.A., Maia J., Lopes-da-Silva J.A., Relvas J.B., Graos M. Modulation of oligodendrocyte differentiation and maturation by combined biochemical and mechanical cues. Sci. Rep. 2016;6:21563. doi: 10.1038/srep21563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shankar S.L., O’Guin K., Kim M., Varnum B., Lemke G., Brosnan C.F., Shafit-Zagardo B. Gas6/Axl signaling activates the phosphatidylinositol 3-kinase/Akt1 survival pathway to protect oligodendrocytes from tumor necrosis factor alpha-induced apoptosis. J. Neurosci. 2006;26:5638–5648. doi: 10.1523/JNEUROSCI.5063-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuhl N.M., De Keyser J., De Vries H., Hoekstra D. Insulin-like growth factor binding proteins-1 and -2 differentially inhibit rat oligodendrocyte precursor cell survival and differentiation in vitro. J. Neurosci. Res. 2002;69:207–216. doi: 10.1002/jnr.10293. [DOI] [PubMed] [Google Scholar]
- 38.Li N., Han J., Tang J., Ying Y. IGFBP-7 inhibits the differentiation of oligodendrocyte precursor cells via regulation of Wnt/beta-Catenin signaling. J. Cell Biochem. 2018;119:4742–4750. doi: 10.1002/jcb.26654. [DOI] [PubMed] [Google Scholar]
- 39.Kim W.K., Kim D., Cui J., Jang H.H., Kim K.S., Lee H.J., Kim S.U., Ahn S.M. Secretome analysis of human oligodendrocytes derived from neural stem cells. PLoS ONE. 2014;9:e84292. doi: 10.1371/journal.pone.0084292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Relucio J., Menezes M.J., Miyagoe-Suzuki Y., Takeda S., Colognato H. Laminin regulates postnatal oligodendrocyte production by promoting oligodendrocyte progenitor survival in the subventricular zone. Glia. 2012;60:1451–1467. doi: 10.1002/glia.22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.See J., Zhang X., Eraydin N., Mun S.B., Mamontov P., Golden J.A., Grinspan J.B. Oligodendrocyte maturation is inhibited by bone morphogenetic protein. Mol. Cell Neurosci. 2004;26:481–492. doi: 10.1016/j.mcn.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 42.Moore C.S., Milner R., Nishiyama A., Frausto R.F., Serwanski D.R., Pagarigan R.R., Whitton J.L., Miller R.H., Crocker S.J. Astrocytic tissue inhibitor of metalloproteinase-1 (TIMP-1) promotes oligodendrocyte differentiation and enhances CNS myelination. J. Neurosci. 2011;31:6247–6254. doi: 10.1523/JNEUROSCI.5474-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicaise A.M., Johnson K.M., Willis C.M., Guzzo R.M., Crocker S.J. TIMP-1 Promotes Oligodendrocyte Differentiation Through Receptor-Mediated Signaling. Mol. Neurobiol. 2019;56:3380–3392. doi: 10.1007/s12035-018-1310-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selvaraju R., Bernasconi L., Losberger C., Graber P., Kadi L., Avellana-Adalid V., Picard-Riera N., Baron Van Evercooren A., Cirillo R., Kosco-Vilbois M., et al. Osteopontin is upregulated during in vivo demyelination and remyelination and enhances myelin formation in vitro. Mol. Cell Neurosci. 2004;25:707–721. doi: 10.1016/j.mcn.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 45.Meyer R.C., Giddens M.M., Coleman B.M., Hall R.A. The protective role of prosaposin and its receptors in the nervous system. Brain Res. 2014;1585:1–12. doi: 10.1016/j.brainres.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hiraiwa M., Campana W.M., Mizisin A.P., Mohiuddin L., O’Brien J.S. Prosaposin: A myelinotrophic protein that promotes expression of myelin constituents and is secreted after nerve injury. Glia. 1999;26:353–360. doi: 10.1002/(SICI)1098-1136(199906)26:4<353::AID-GLIA9>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 47.Chatterjee N., Stegmüller J., Schätzle P., Karram K., Koroll M., Werner H.B., Nave K.A., Trotter J. Interaction of syntenin-1 and the NG2 proteoglycan in migratory oligodendrocyte precursor cells. J. Biol. Chem. 2008;283:8310–8317. doi: 10.1074/jbc.M706074200. [DOI] [PubMed] [Google Scholar]
- 48.Lu Z., Kipnis J. Thrombospondin 1--a key astrocyte-derived neurogenic factor. FASEB J. 2010;24:1925–1934. doi: 10.1096/fj.09-150573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czopka T., von Holst A., ffrench-Constant C., Faissner A. Regulatory mechanisms that mediate tenascin C-dependent inhibition of oligodendrocyte precursor differentiation. J. Neurosci. 2010;30:12310–12322. doi: 10.1523/JNEUROSCI.4957-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang P., Chen C., Liu X.B., Pleasure D.E., Liu Y., Deng W. Human iPSC-Derived Immature Astroglia Promote Oligodendrogenesis by Increasing TIMP-1 Secretion. Cell Rep. 2016;15:1303–1315. doi: 10.1016/j.celrep.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steffenhagen C., Dechant F.X., Oberbauer E., Furtner T., Weidner N., Küry P., Aigner L., Rivera F.J. Mesenchymal stem cells prime proliferating adult neural progenitors toward an oligodendrocyte fate. Stem Cells Dev. 2012;21:1838–1851. doi: 10.1089/scd.2011.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fricker R.A., Carpenter M.K., Winkler C., Greco C., Gates M.A., Bjorklund A. Site-specific migration and neuronal differentiation of human neural progenitor cells after transplantation in the adult rat brain. J. Neurosci. 1999;19:5990–6005. doi: 10.1523/JNEUROSCI.19-14-05990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McGinley L.M., Kashlan O.N., Chen K.S., Bruno E.S., Hayes J.M., Backus C., Feldman S., Kashlan B.N., Johe K., Feldman E.L. Human neural stem cell transplantation into the corpus callosum of Alzheimer’s mice. Ann. Clin. Transl. Neurol. 2017;4:749–755. doi: 10.1002/acn3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karimi-Abdolrezaee S., Eftekharpour E., Wang J., Morshead C.M., Fehlings M.G. Delayed transplantation of adult neural precursor cells promotes remyelination and functional neurological recovery after spinal cord injury. J. Neurosci. 2006;26:3377–3389. doi: 10.1523/JNEUROSCI.4184-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vigano F., Möbius W., Götz M., Dimou L. Transplantation reveals regional differences in oligodendrocyte differentiation in the adult brain. Nat. Neurosci. 2013;16:1370–1372. doi: 10.1038/nn.3503. [DOI] [PubMed] [Google Scholar]
- 56.Enomoto M., Shinomiya K., Okabe S. Migration and differentiation of neural progenitor cells from two different regions of embryonic central nervous system after transplantation into the intact spinal cord. Eur. J. Neurosci. 2003;17:1223–1232. doi: 10.1046/j.1460-9568.2003.02555.x. [DOI] [PubMed] [Google Scholar]
- 57.Marshall C.A., Novitch B.G., Goldman J.E. Olig2 directs astrocyte and oligodendrocyte formation in postnatal subventricular zone cells. J. Neurosci. 2005;25:7289–7298. doi: 10.1523/JNEUROSCI.1924-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jungbluth P., Spitzhorn L.S., Grassmann J., Tanner S., Latz D., Rahman M.S., Bohndorf M., Wruck W., Sager M., Grotheer V., et al. Human iPSC-derived iMSCs improve bone regeneration in mini-pigs. Bone Res. 2019;7:32. doi: 10.1038/s41413-019-0069-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spitzhorn L.S., Rahman M.S., Schwindt L., Ho H.T., Wruck W., Bohndorf M., Wehrmeyer S., Ncube A., Beyer I., Hagenbeck C., et al. Isolation and Molecular Characterization of Amniotic Fluid-Derived Mesenchymal Stem Cells Obtained from Caesarean Sections. Stem. Cells Int. 2017;2017:5932706. doi: 10.1155/2017/5932706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Timaner M., Tsai K.K., Shaked Y. The multifaceted role of mesenchymal stem cells in cancer. Semin. Cancer Biol. 2020;60:225–237. doi: 10.1016/j.semcancer.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 61.Chun S.J., Rasband M.N., Sidman R.L., Habib A.A., Vartanian T. Integrin-linked kinase is required for laminin-2-induced oligodendrocyte cell spreading and CNS myelination. J. Cell Biol. 2003;163:397–408. doi: 10.1083/jcb.200304154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Silva M.E., Lange S., Hinrichsen B., Philp A.R., Reyes C.R., Halabi D., Mansilla J.B., Rotheneichner P., Guzman de la Fuente A., Couillard-Despres S., et al. Pericytes Favor Oligodendrocyte Fate Choice in Adult Neural Stem Cells. Front. Cell Neurosci. 2019;13:85. doi: 10.3389/fncel.2019.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jadasz J.J., Rivera F.J., Taubert A., Kandasamy M., Sandner B., Weidner N., Aktas O., Hartung H.P., Aigner L., Küry P. p57kip2 regulates glial fate decision in adult neural stem cells. Development. 2012;139:3306–3315. doi: 10.1242/dev.074518. [DOI] [PubMed] [Google Scholar]
- 64.Binder M.D., Cate H.S., Prieto A.L., Kemper D., Butzkueven H., Gresle M.M., Cipriani T., Jokubaitis V.G., Carmeliet P., Kilpatrick T.J. Gas6 deficiency increases oligodendrocyte loss and microglial activation in response to cuprizone-induced demyelination. J. Neurosci. 2008;28:5195–5206. doi: 10.1523/JNEUROSCI.1180-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tan H.K., Heywood D., Ralph G.S., Bienemann A., Baker A.H., Uney J.B. Tissue inhibitor of metalloproteinase 1 inhibits excitotoxic cell death in neurons. Mol. Cell Neurosci. 2003;22:98–106. doi: 10.1016/S1044-7431(02)00024-6. [DOI] [PubMed] [Google Scholar]
- 66.Figueres-Onate M., Garcia-Marques J., Lopez-Mascaraque L. UbC-StarTrack, a clonal method to target the entire progeny of individual progenitors. Sci. Rep. 2016;6:33896. doi: 10.1038/srep33896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schira J., Gasis M., Estrada V., Hendricks M., Schmitz C., Trapp T., Kruse F., Kögler G., Wernet P., Hartung H.P., et al. Significant clinical, neuropathological and behavioural recovery from acute spinal cord trauma by transplantation of a well-defined somatic stem cell from human umbilical cord blood. Brain. 2012;135:431–446. doi: 10.1093/brain/awr222. [DOI] [PubMed] [Google Scholar]
- 70.Schira J., Falkenberg H., Hendricks M., Waldera-Lupa D.M., Kögler G., Meyer H.E., Müller H.W., Stühler K. Characterization of Regenerative Phenotype of Unrestricted Somatic Stem Cells (USSC) from Human Umbilical Cord Blood (hUCB) by Functional Secretome Analysis. Mol. Cell Proteom. 2015;14:2630–2643. doi: 10.1074/mcp.M115.049312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schira J., Heinen A., Poschmann G., Ziegler B., Hartung H.P., Stühler K., Küry P. Secretome analysis of nerve repair mediating Schwann cells reveals Smad-dependent trophism. FASEB J. 2019;33:4703–4715. doi: 10.1096/fj.201801799R. [DOI] [PubMed] [Google Scholar]
- 72.Cox J., Michalski A., Mann M. Software lock mass by two-dimensional minimization of peptide mass errors. J. Am. Soc. Mass Spectrom. 2011;22:1373–1380. doi: 10.1007/s13361-011-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao L., Poschmann G., Waldera-Lupa D., Rafiee N., Kollmann M., Stuhler K. OutCyte: A novel tool for predicting unconventional protein secretion. Sci. Rep. 2019;9:19448. doi: 10.1038/s41598-019-55351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.