Abstract
The orientation of microtubule (MT) networks is exploited by motors to deliver cargoes to specific intracellular destinations and is thus essential for cell polarity and function. Reconstituted in vitro systems have largely contributed to understanding the molecular framework regulating the behavior of MT filaments. In cells, however, MTs are exposed to various biomechanical forces that might impact on their orientation, but little is known about it. Oocytes, which display forceful cytoplasmic streaming, are excellent model systems to study the impact of motion forces on cytoskeletons in vivo. Here we implement variational optical flow analysis as a new approach to analyze the polarity of MTs in the Drosophila oocyte, a cell that displays distinct Kinesin-dependent streaming. After validating the method as robust for describing MT orientation from confocal movies, we find that increasing the speed of flows results in aberrant plus end growth direction. Furthermore, we find that in oocytes where Kinesin is unable to induce cytoplasmic streaming, the growth direction of MT plus ends is also altered. These findings lead us to propose that cytoplasmic streaming - and thus motion by advection – contributes to the correct orientation of MTs in vivo. Finally, we propose a possible mechanism for a specialized cytoplasmic actin network (the actin mesh) to act as a regulator of flow speeds to counteract the recruitment of Kinesin to MTs.
INTRODUCTION
Eukaryotic life depends on many dynamic processes, including, for example, cell division, cell migration, and cell polarization. These processes in turn strongly rely on highly organized microtubule (MT) arrays. All MT networks are polarized, with the minus end of each filament linked to a nucleating center (MT organizing center or MTOC) and the plus end growing away from these centers. This polarity is exploited by specific motor proteins to transport cargoes along MTs in a defined direction and is thus essential for the function of MT networks.
A number of biophysical studies, using reconstituted in vitro systems, have helped to understand the mechanical properties of MTs, setting the stage for deciphering the behavior of MTs in vivo. However, much needs to be learned about the properties of MTs in their natural intracellular environment. For example, a rather new concept emanating from in vivo experiments is that controlling nucleation and the position of minus ends of single filaments alone is not always sufficient to establish the proper polarity of the entire MT network. MT plus ends must be controlled as well in order to allow motor proteins to deliver their cargoes to the correct destination. Plus ends can be regulated at various levels including dynamic instability, capturing, and direction of growth. Dynamic instability describes a process in which MT polymerization is interrupted by a rapid depolymerization phase, followed by a rescue process (Mitchison and Kirschner, 1984). Various MT-associated proteins, such as motors and MT plus end-tracking proteins (+TIPs), are known to regulate dynamic instability (Akhmanova and Steinmetz, 2015). MT plus ends can also be captured at the cell cortex, a process also involving +TIPs and motors, such as the Dynein/Dynactin complex (Nieuwburg et al., 2017 and reviewed in Akhmanova and Steinmetz, 2015). However, very little is known about how the direction of growth of plus ends, and therefore the orientation of MTs, is controlled in cells. In axons, adenomatous polyposis coli seems to regulate plus end direction (Purro et al., 2008), while Fidgetin-like1, a MT-associated ATPase, controls both dynamics and plus ends direction (Fassier et al., 2018). MT bending also impacts the direction of plus tip growth, as the MT tip rotates due to local bend formation (Kent et al., 2016). Furthermore, +TIPs that contain actin-binding motifs can influence MT growth direction by guiding dynamic plus ends along actin bundles (Kodama et al., 2003; Jiang et al., 2012).
In the midoogenesis stage 9 (st9) oocyte of Drosophila melanogaster, MTs are nucleated from the antero-lateral cortex in a gradient of diminishing abundance toward the posterior pole, where nucleation is absent. In addition, the growing plus ends exhibit a weak posterior orientation bias (Doerflinger et al., 2006; Parton et al., 2011; Nashchekin et al., 2016). This weak polarization bias of plus ends toward the posterior pole of the oocyte is sufficient and necessary for the localization of body plan determinants to the very “posterior tip,” as well as the formation of the pole plasm (needed for germ cell development) in this region. For example, the plus end motor Kinesin-1 (Kin from here on) localizes oskar mRNA to the posterior tip of the st9 oocyte, an essential step in the establishment of the anterior-posterior (A-P) axis and the formation of the pole plasm (Brendza et al., 2000).
In addition to a polarized MT cytoskeleton, the oocyte displays a Kin-dependent bulk motion of the cytoplasm called cytoplasmic streaming or flows (Palacios and St Johnston, 2002; Serbus et al., 2005; Ganguly et al., 2012; Drechsler et al., 2017). Compared with st9, late stage oocytes (st11) exhibit faster and larger scale flows that are induced not only by Kin-mediated viscous drag of transported cargo (similar to st9) but also by Kin-mediated MT-sliding (Lu et al., 2016; Monteith et al., 2016). At these later developmental stages, these faster flows are important for mixing the cytoplasm of the large oocyte (Ganguly et al., 2012) and aiding the asymmetric localization of developmental determinants and mitochondria (Forrest and Gavis, 2003; Hurd et al., 2016; Lu et al., 2018). In addition, mathematical modeling has suggested that faster flows can induce parallel MT arrays when the MTs are near a barrier (such as the cortex) (Monteith et al., 2016). However, in st9 oocytes, when posterior cargoes such as oskar mRNA are first localized, MT sliding has been excluded as the source of cytoplasmic streaming (Lu et al., 2016). Cytoplasmic flows at st9 are slowed down by a cytoplasmic actin mesh. When this actin mesh dissolves during midoogenesis, the flows become faster and more organized, and the MTs arrange into subcortical bundles (Dahlgaard et al., 2007; Quinlan, 2013). However, the mechanism by which the actin mesh regulates flows is unknown. Furthermore, it is unclear whether cytoplasmic flows at st9 have an impact on the organization of the MT cytoskeleton. Recently, we found that st9 flows constitute a key force driving the persistent motion of vesicles and actin filaments (Drechsler et al., 2017). These observations prompted us to address the question of how advection (active transport induced by fluid flows) impacts on the polarization of the MT network in st9 oocytes. In this way, we aim to contribute to the insufficient knowledge of how the direction of MT growth is controlled in vivo.
To assess the global MT orientation in st9 oocytes, and to investigate the growth direction of MT plus ends in vivo, we used EB1::GFP (EB1 from here on). EB1 exclusively decorates the growing plus end of MTs, resulting in dynamic comets moving through the cytoplasm at a speed of ∼100–600 nm/s across various cell types, including the oocyte (at an average and maximum speeds of 230 and 600 nm/s, respectively; Parton et al., 2011; Nieuwburg et al., 2017). Analyzing the orientation of MTs in complex networks has proven technically challenging and requires suitable imaging and image analysis tools. Especially for the Drosophila oocyte, we found the published method too demanding on the imaging level, requiring state of the art widefield deconvolution microscopy and elaborate image processing (Parton et al., 2011) and thus unfeasible for the various experimental conditions that our study required. Here, we developed an image analysis strategy that allows an efficient characterization of direction and distribution of EB1 comets from confocal image series by an optical flow (OF)-based motion estimation approach. In general, OF allows to estimate the apparent motion of objects or other intensity variations in a sequence of images (Horn and Schunck, 1981). In addition, variational OF methods constitute a well-established framework for dense motion estimation and do not require elaborate segmentation or tracking of the studied structures. OF methods outperform popular methods, such as particle image velocimetry (PIV), for motion analysis in certain settings and, in particular, in the presence of noise (Ruhnau et al., 2005; Vig et al., 2016). While variational OF methods have been used predominantly to investigate the dynamics of entire cells (Amat et al., 2013; Boric et al., 2013; Guo and Zhou, 2014), recent work focused also on intracellular motility (Delpiano et al., 2012; Frerking et al., 2014; Vig et al., 2016; Boquet-Pujadas et al., 2017; Huang et al., 2017).
In this study, we investigated how cytoplasmic streaming influences the polarization of the MT cytoskeleton. For this, we introduce a two-step image analysis approach that is based on image denoising and variational OF and is able to estimate approximate velocities (speed and direction) of EB1 motion in confocal image sequences. This approach will assist many researchers interested in characterizing MT polarity from confocal images of their tissue of choice. Our findings revealed that cytoplasmic streaming is necessary and sufficient to regulate the polarity of the MT network in st9 oocytes. Furthermore, our data suggest that the actin mesh regulates the recruitment of Kin to MTs, indicating a new mechanism by which the actin cytoskeleton influences MT-based transport and advection. With this work, we further contribute to understanding the in vivo properties of MTs and the interactions of MTs with their natural environment.
RESULTS
Quantification of MT plus tip directionality by OF analysis
To study the spatial orientation of MT filaments in vivo, we used oocytes expressing EB1 (Figures 1 and 3–5). EB1 constitutes a marker for growing MT plus tips and has been used in fly oocytes before (Parton et al., 2011; Nieuwburg et al., 2017). In these previous studies, widefield deconvolution microscopy was used to image EB1 dynamics in various areas of st9 oocytes. However, widefield microscopy has a limited focus depth, only able to image EB1 dynamics close to the cortex of the relatively large oocyte, and requires a complex acquisition procedure. To improve imaging depth and to simplify the acquisition procedure, we implemented a strategy that combines conventional confocal microscopy with image denoising and motion analysis by variational OF analysis.
FIGURE 1:
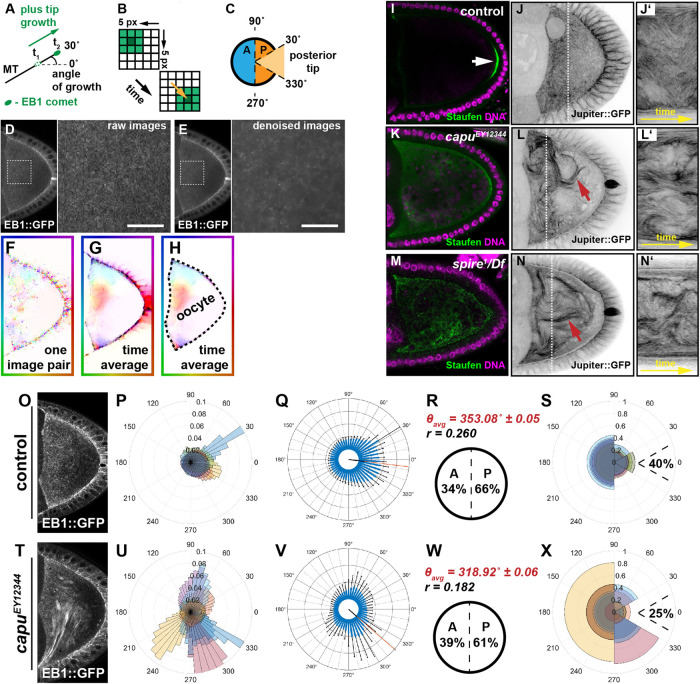
MT orientation is aberrant in oocytes with faster flows. (A) Schematic representation of MT orientation. EB1 specifically associates with the growing end (plus end) of MTs and therefore serves as readout of their spatial orientation. The orientation of MT growth is represented as an angle, deviating from an imaginary anterior (180°) to posterior axis (0°). (B) Simplified scheme illustrating the underlying principle of OF-based motion estimation. Shown are two consecutive frames of a 5 × 5 pixel-wide image sequence that contains a rectangular object of different pixel intensities—from light green (little signal) to dark green (maximum signal). OF assumes that the intensities of a signal do not change along its trajectory. Based on this assumption, variational OF allows to estimate a displacement vector for each pixel (the yellow arrow shows the displacement vector of the center pixel of the object). (C) Definition of growth directions based on OF-estimated velocities. Angles of velocities between 90° and 270° are regarded as anterior (A, blue hemicircle) and the complementary set of angles as posterior (P, orange hemicircle). The subpopulation of all posterior signals (orange) that fall between 330° and 30° (light orange sector) are considered to grow toward the posterior tip. (D) Single frame of an unprocessed image sequence (raw data) showing an oocyte expressing EB1. The magnified area is indicated by a dashed box. (E) Same frame as shown in D after applying the denoising step (Supplemental Movie S2). Scale bars are 10 µm. (F) Shown is the OF (displacement vector) field between two frames of the image sequence in D. (G) Average OF (over the entire sequence). (H) Hand-drawn segmentation mask of the oocyte. For the analysis, only the displacement vectors within this segmentation were considered. (I) Staufen (green) localization in control oocytes (arrow). The protein localizes in a tight posterior crescent by the end of st9. (J) Living control oocyte expressing the MT-binding protein Jup (Supplemental Movie S5). (J’) Kymograph/space-time plot (along the indicated dotted line in C showing dynamic bending of MTs over time. (K, M) Staufen fails to localize in capu or spire mutants and distributes throughout the cytoplasm of the cell. (L, N) Living capu (L) or spire (N) mutant oocytes expressing Jup. Fast cytoplasmic flows lead to the formation of dense and long MT bundles (red arrows), as well as increased MT bending (Supplemental Movie S5). (L’, N’) Kymograph (along the dashed line in L and N) indicate a stronger displacement of MT bundles over time in capu (L’) or spire (N’) mutants. (O) Standard deviation (temporal) projection of EB1 comets in a control image sequence (in total 65 s). (P) Rose diagram (angular histogram) with 50 bins depicting the distribution of EB1 directions in individual control cells within the corresponding segmented oocyte. Each color represents the angular histogram of the directions from one oocyte. (Q) Same data as shown in P with angular histograms averaged over all cells (n = 8). Error bars (in black) indicate the SD for each bin (in blue). (R) Mean angular direction θavg of the histogram shown in Q (also indicated by a red line in Q) and the length r (between 0 and 1) of the mean resultant vector (length of black line in Q originating from the center), which relates to the circular variance S = 1–r of the distribution shown in Q. Anterior-posterior bias of all EB1 growth directions. (S) Rose diagram similar to P for control cells but with growth directions binned into four bins (30°–90°, 90°–270°, 270°–330°, and 330°–30°). Moreover, the fraction of posterior-growing EB1 comets pointing toward the posterior tip (330°–30°) is indicated. (T) Standard deviation (temporal) projection of EB1 comets in an image sequence of a capu mutant oocyte (in total 65 s). (U–X) Quantified EB1 directions in capu cells following the same experimental pipeline described for the control cell.
FIGURE 3:
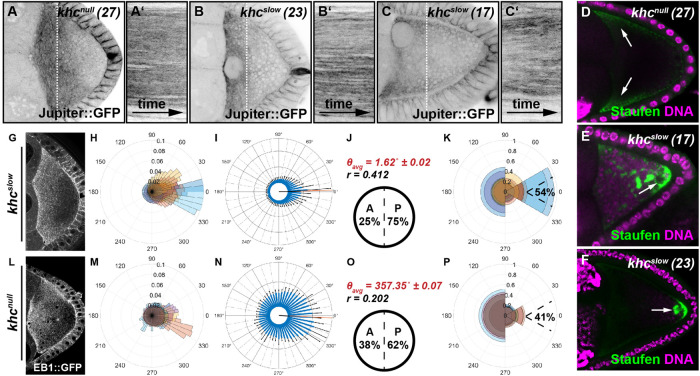
Kin activity impacts on the spatial orientation of MTs in the oocyte. (A–C) Still frames and kymographs of live oocytes expressing Jup. Cells harbor a null mutation (khcnull (27)), or single point mutations in the motor domain (khcslow (23) and khcslow (17)), rendering the motor slower. Compared with controls, all khc mutant cells exhibit no cytoplasmic flows and thus no MT bulk motion (Supplemental Movie S6). (D–F) Posterior cargo localization in different khc mutant alleles. Staufen (green) is not transported to the posterior in cells lacking Kin (khcnull (27)) and is found in the anterior corners of the cell (arrows). In contrast, in both of the slow Kin alleles (khcslow (23) and khcslow (17)) a considerable amount of Staufen becomes transported to the posterior. However, compared with controls (Figure 2A), Staufen is not only localized in a posterior crescent but also in dots within the posterior cytoplasm (arrows). (G–P) OF analysis of EB1 growth directionality in khcslow(G–K, n = 10) and khcnull(L–P, n = 10) oocytes, following the experimental pipeline described in Figure 1.
FIGURE 5:
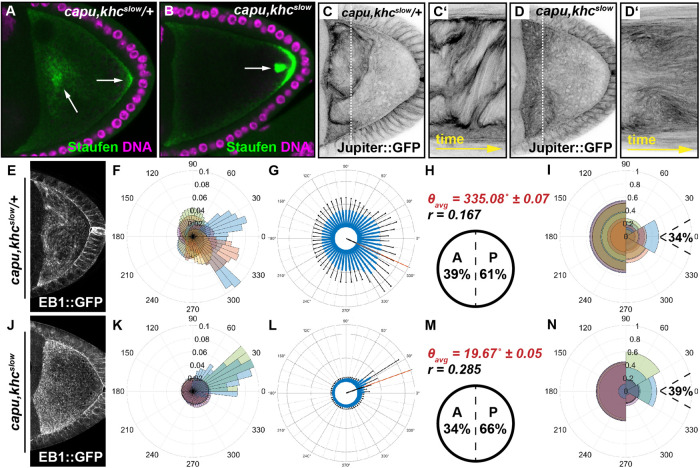
Cytoplasmic streaming constitutes a major contributor to MT orientation. (A, B) Posterior cargo localization in capu,khcslow/+ (A) and capu,khcslow double (B) mutant cells. While Staufen (green) partially localizes to the posterior of the cell in capu,khcslow/+ oocytes (right arrow in A), the majority of the protein localizes in cytoplasmic clouds (left arrow in A). In capu,khcslow double mutant cells, Staufen localizes in a posterior crescent but also accumulates in dots close to the posterior (arrow in B). (C, D) Still frames and kymographs of live capu,khcslow/+ (C, C’) or capu,khcslow double (D, D’) mutant oocytes, expressing Jup. (E–N) OF analysis of EB1 growth directionality in capu,khcslow/+ (E–I, n = 10) and capu,khcslow (J–N, n = 8) oocytes.
OF-based motion estimation relies on the assumption that particles approximately maintain their pixel intensity as they move. In thick biological samples, the intensity of fluorescent signals declines with the depth of the imaging plane. To minimize the effects of tissue thickness on signal intensity, we imaged within a single focal plane in the middle of the oocyte over the course of at least 1 min (for details, see Materials and Methods). With this approach, we avoided measurable bleaching and ensured that EB1 comets would only be detected within a thin focal plane of ∼1.3 µm. Thus, comets moving orthogonally to the focal plane would be lost instead of getting dimmer or brighter. As a consequence, our analysis captures the 2D motion of EB1 comets within a 3D intracellular environment. However, the orientation of individual oocytes on the microscope slide is random. Together with averaging across several cells, this results in 2D data obtained from various angles (one precise orientation per cell), which indirectly allows us to acquire information about the 3D behavior. As OF is formulated on a per-pixel basis, this method allows inferring a displacement vector for each pixel and does not require sophisticated tracking of individual particles (Figure 1, A and B). Since MTs grow one order of magnitude faster compared with the speed of cytoplasmic flows in st9 oocytes, we took one image every 650 ms, resulting in a loss of spatial resolution and causing the raw data to exhibit considerable high noise (Figure 1D). Due to this high noise level, preceding denoising of the image data was required prior to applying the motion estimation. We found that total variation-based (Rudin et al., 1992) image denoising with additional temporal regularization was sufficient to improve the signal-to-noise ratio and the visibility of EB1 comets (Figure 1E, Supplemental Figures S1–S3, and Supplemental Movie S1).
Movie S1.
MT bulk motion in control and capu mutant oocytes (related to Fig. 1). Dynamic bulk motion of Jup-labelled MTs in control (left) and capuEY12344 mutant oocytes. Images have been acquired at one frame every 10.4 s. Playback speed is 20 fps (∼ 200× real speed).
In the next step, displacement vector fields were computed with a variational OF method (Supplemental Figure S4). After accounting for the pixel size and the time interval between consecutive frames, these displacements can be regarded as approximate velocities of EB1 motion (Figure 1, F–H). Importantly, the acquired velocities are a sum of all motion experienced by the imaged EB1 signal, including filament growth and MT displacement by advection and transport. A typical image sequence of 100 frames and a 512 × 256 pixel resolution results in a total number of approximately 13 million computed vectors that require appropriate interpretation. The computation time for processing one typical sequence amounted to less than 25 min on average.
We visualized the estimated vector fields with the help of standard color coding (Baker et al., 2011). The vector at each pixel and at a certain frame is represented by a color that is determined by the direction of the signal’s movement (see the color coding at the boundary of the images in Figure 1, F–H). The color’s intensity is determined by the relative velocity of the signal. The stronger the intensity, the higher the velocity. For our statistical analyses, we considered only vectors located within a hand-drawn segmentation mask that outlines the oocyte in each sequence (Figure 1H). The direction of each vector could then be represented in polar coordinates. The distribution of the angles obtained from each individual cell is visualized in a rose diagram (Figure 1, O and P). In addition, we show the histogram of the distributions averaged over all cells of a given genotype (Figure 1Q).
We then used OF to describe the MT orientation in control oocytes (Figure 1, O–S; Supplemental Figure S5). Since all cells were orientated with the posterior pole to the right during imaging, the angles given in the rose diagram reflect the directional movement of EB1 comets. For a quantitative description of MT orientation, we determined the mean angular direction of the estimated motion of EB1 signals (θavg, considering all recorded cells of one genotype; Figure 1R; Table 1) and the frequency of movements directed toward the anterior (all within the region 270°–90°, blue hemicircle in Figure 1C) or the posterior (all within the region 90°–270°, orange hemicircle in Figure 1C). As a descriptor of how focused MTs grow toward the very posterior tip, we also determined the relative frequency of posterior-directed EB1 signals that fall within a sector of 60° (from 30° to 330°), called the posterior tip, inspired by the region where determinants localize and pole plasm forms (light orange sector in Figure 1C). This posterior tip frequency only considers EB1 signals that were previously found to grow toward the posterior (orange hemicircle in Figure 1C), and thus the posterior tip value represents a subpopulation of all posterior signals that grow with an angle between 30° and 330°. In summary, we find that all growing MTs exhibit a global (over the entire oocyte) posterior orientation bias, with 66% of all comets growing toward the posterior of control cells (Figure 1R). These findings are in agreement with previous reports of biased MT orientation in st9 oocytes (Parton et al., 2011) and provide a first level of validation to establish OF as a reliable way to investigate the orientation of MTs. In addition, we find that 40% of the 66% of comets that grow toward the posterior are oriented toward the posterior tip, where determinants localize and the pole plasm forms (Figure 1S).
TABLE 1:
Summary of EB1 orientation data extracted from confocal time series by variational OF analysis.
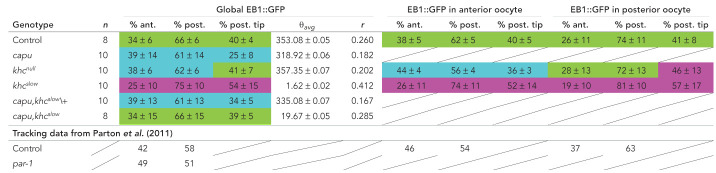
Anterior-posterior orientation bias of microtubule growth in percent (±95% confidence interval); n represents the number of cells analyzed. θavg gives the average angle of all EB1 orientations of all cells of a given genotype. r constitutes a measure of variance and lies between 0 and 1. The larger r, the less variable the data set. Green, similar to control; purple, stronger posterior bias; blue, weaker posterior bias.
OF is sufficient to detect changes in microtuble network orientation
To test whether OF-based approaches are able to capture EB1 directionality in other cell types, we investigated EB1 comets in the follicular epithelium surrounding the egg chamber. In these epithelial cells, MTs are nucleated apically and grow predominantly toward the basal membrane, parallel to the lateral membranes of the cell. Notably, single comets can easily be identified and followed by eye, which allowed us to assess the accuracy of the OF results. OF analysis captures the directionality of MTs in these follicle cells accurately, supporting the suitability of the approach to estimate MT orientation in vivo (Supplemental Movie S2 and Supplemental Figures S6 and S7).
Movie S2.
Example of image denoising of EB1 dynamics. Confocal image sequence of dynamic EB1 comets before (left) and after (right) image denoising. Images have been acquired at one frame every 0.65 s. Playback speed is 40 fps (∼ 20× real speed).
We next tested whether OF can pick up changes in the architecture of MT neworks in the oocyte and analyzed cells harboring a mutation in gurken (grk), which exhibit MT organization and cell polarity defects (Gonzalez-Reyes et al., 1995; Roth et al., 1995; Januschke et al., 2002). With our OF analysis we were able to detect an aberrant MT organization in grk oocytes, with EB1 comets showing a variable orientation from cell to cell (Supplemental Movie S3 and Supplemental Figure S6). Although, the number of tested cells is low, and thus the actual biological meaning of these findings needs further investigation, our results confirm that OF is able to detect aberrations in MT networks in complex systems like the oocyte.
Movie S3.
OF motion estimation in follicle cells. Confocal image sequence of dynamic EB1 comets in follicle cells and subsequent OF motion estimation. Images have been acquired at one frame every 0.65 s. Playback speed is 40 fps (∼ 20× real speed).
Aberrantly fast cytoplasmic flows change MT motion, bundling, and orientation
To investigate the relationship between cytoplasmic advection and the organization of the MT network, we monitored the bulk behavior of MTs in control cells and in oocytes with faster flows. Oocytes mutant for cappuccino (capu) or spire display faster flows at st9 (Supplemental Figure S9, A and B) and fail to localize posterior cargoes like the mRNA-binding protein Staufen (Figure 1, I, K, and M) (Theurkauf, 1994; Dahlgaard et al., 2007; Quinlan, 2013). Capu (a formin) and Spire are actin nucleators that are required for the formation of a cytoplasmic actin mesh that regulates the magnitude of flows by a yet unknown mechanism. It has been shown in fixed samples that the removal of the actin mesh seems to cause bundling of MTs in close proximity to the cell cortex. However, dynamic MT behavior in living mutant oocytes has not been addressed in detail.
To monitor the bulk movement of MTs, we imaged cells expressing the MT-associated protein Jupiter::GFP (Jup), which labels the entire MT filament. Compared with control cells, we observed that MTs in homozygous capuEY12344 or transheterozygous spire1/Df(2L)Exel6046 mutant oocytes appear as thick bundles, which dramatically bend and buckle (Figure 1, I–N’ and Supplemental Movie S5). Such higher “packing” of filaments does not necessarily indicate their cross-linking, but argues that the high hydrostatic pressure in fast-flowing oocytes is sufficient to bundle MT filaments. More importantly, and in contrast to previous descriptions, our data demonstrate that in capu and spire oocytes, MTs cluster even when they are not in close proximity to the cortex, and thus bundles can form even when the MTs are not pushed against the cortex. Furthermore, the observed MT bundles appear longer when compared with control cells, extending further into the posterior (red arrows in Figure 1, L and N). In summary, these results indicate that changing the regime of cytoplasmic advection impacts the bulk motion of MTs, as well as their cytoplasmic bundling state.
In capu and spire mutant oocytes, flows can reach speeds up to 200 nm/s, well within the regime of plus tip growth. In such mutant situation, the plus tip of single MTs might not be able to “outgrow” the effect of cytoplasmic advection, and thus flows might become a dominant contributor of MT orientation. To test whether faster flows alter the orientation of MTs, we quantified the directionality of EB1 signals in capu mutant cells with our OF approach and found substantial alterations (Figure 1, T–X). Compared with controls, capu mutants exhibit large amounts of comets in close proximity to each other, which move in the same direction. This suggets that capu mutant oocytes display arrays of parallel growing filaments (Figure 1T and Supplemental Movie S4). The motion of such EB1 “arrays” (which varies from cell to cell; Figure 1, T–X) is likely to correlate with the bundling of MT filaments described above and could be interpreted as a direct result of faster flows. Interestingly, the global posterior EB1 orientation bias was only mildly decreased in most capu mutant oocytes (Figure 1, V and W vs. Q and R, and Table 1). However, MTs are less focused toward the posterior tip in capu mutants, with only 25% of the posterior-directed EB1 signals growing with an angle between 30° and 330°. These findings are in good agreement with the determinant mislocalization defects in capu and spire mutant cells (Figure 1, X vs. K and M).
Movie S4.
Dynamic behaviour of EB1 comets in grk mutant oocytes. Denoised confocal image sequence of dynamic EB1 comets in a grk2B6/grk2E12 mutant oocyte. The nucleus (n) of the cell is detached and fails to migrate to the antero-lateral cortex. Images have been acquired at one frame every 0.65 s. Playback speed is 40 fps (∼ 20× real speed).
In summary, these findings together demonstrate that the lack of the actin mesh, and the resulting increase in flows speeds, impact the orientation of MTs and lead us to speculate that at midoogenesis, flows must be kept in a precise regime for molecular determinants to be localized correctly. However, how the actin mesh actually regulates the magnitude of flows in the oocyte is not known.
The cytoplasmic actin mesh counteracts the recruitment of Kinesin to MTs in vivo
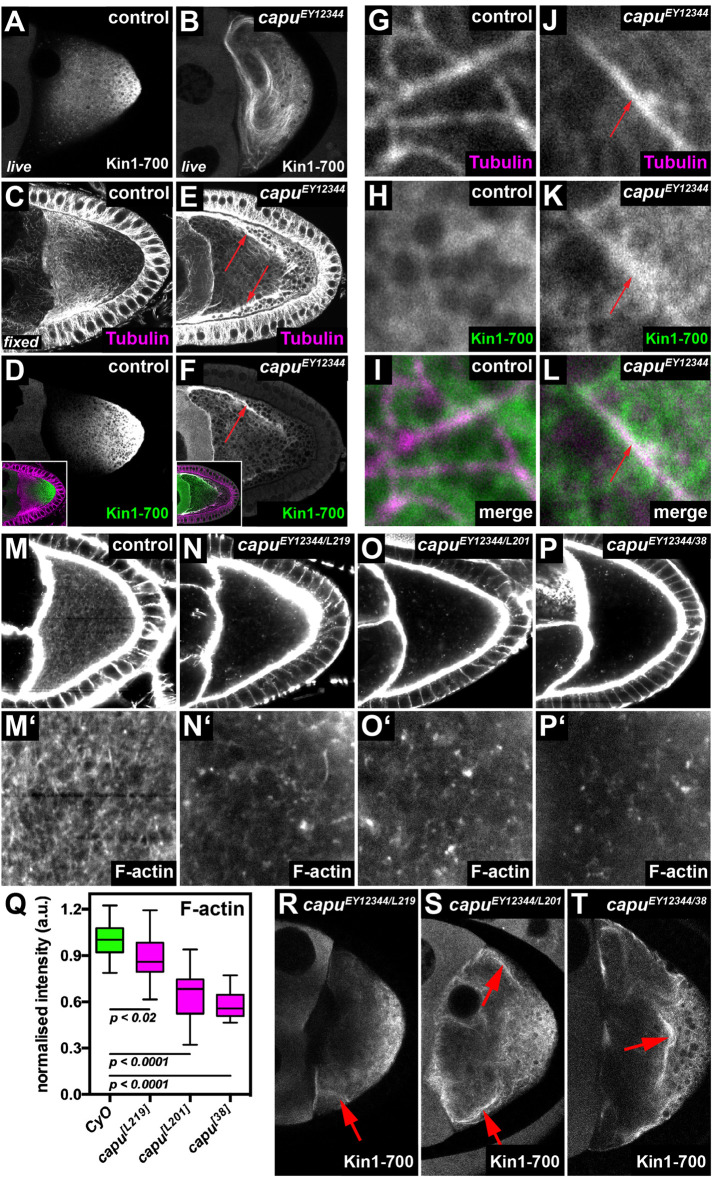
The MT plus tip-directed motor protein Kin constitutes the major driving force of cytoplasmic flows in the Drosophila oocyte (Palacios and St Johnston, 2002). To start investigating possible mechanisms by which the actin mesh regulates the magnitude of cytoplasmic flows, we monitored the distribution of Kin (Kin1-700-GFP) in control and homozygous capuEY12344 mutant oocytes. This truncated motor protein lacks all of its tail domains and is thus not autoinhibited and unable to transport cargo (Williams et al., 2014). In control oocytes, Kin1-700 predominantly localizes to the posterior of the cell, in a gradient of diminishing abundance from posterior to anterior (Figure 2, A and D; Williams et al., 2014). In stark contrast, Kin1-700 fails to accumulate at the posterior in capu mutant oocytes (Figure 2, B and F; Supplemental Movie S10) and is found instead on filamentous structures closely resembling MTs (compare Figure 2B to Figure 1L and Supplemental Movie S10 to Supplemental Movie S5). This result indicates that, in the absence of capu, Kin becomes strongly recruited to MTs. The same Kin behavior can be induced acutely by ex vivo treatment with F-actin destabilizing drugs like Cytochalasin D (unpublished data), indicating that the observed behavior of Kin is due to the lack of the actin mesh and not to potential developmental defects that might arise from the lack of capu during earlier stages of oogenesis. In fixed control cells, Kin1-700 accumulates to posterior regions of the cell (Figure 2D), where MTs are less abundant (Figure 2C), while the motor strongly colocalizes with MTs in fixed capu mutant oocytes (Figure 2, E, F, and J–L). This observation suggests that the actin mesh alters the ability of Kin to become recruited to MTs.
FIGURE 2:
The cytoplasmic actin mesh counteracts the recruitment of Kinesin to MTs. (A, B) Living control (capuEY12344/CyO in A) and capu mutant oocyte (capuEY12344/capuEY12344 in B) expressing Khc::GFP. While the fusion protein mainly localizes posteriorly in control cells (A), it strongly decorates MTs in capu mutant cells (B and Supplemental Movie S10). (C–F) Fixed control (capuEY12344/CyO) oocytes (C, D) and capu mutant cells (capuEY12344/capuEY12344) (E, F) expressing Khc::GFP. Cells were stained against αTubulin (magenta) and the Kin1-700::GFP fusion protein (green). In controls, Tubulin and Khc::GFP localize to opposed gradients and show little overlap (C, D). Conversely, in capu mutants, Khc::GFP strongly colocalizes with Tubulin (red arrows in E and F). Insets in D and F show merged channels. (G–L) Higher resolution images of fixed control (capuEY12344/CyO) (G–I) and capu mutant (J–L) cells stained against αTubulin (magenta) and GFP (green). While Khc::GFP localizes diffusely around MTs in control cells (G–I), it strongly colocalizes to MTs in capu mutants (red arrows in J–L). (M–P) The cytoplasmic actin mesh in control (M, capuEY12344/CyO) and transheterozygous capuEY12344/capuL219 (N), capuEY12344/capuL201(O), and capuEY12344/capu38 (P). (Q) Quantification of signal intesities of the actin mesh as shown in M–P. All cells harbor the strong hypomorphic allele capuEY12344 over CyO (n = 15), capuL219 (n = 21), capuL201 (n = 20), or capu38 (n = 21). (R–T) Khc::GFP expressed in living capuEY12344/capuL219 (R), capuEY12344/capuL201(S), and capuEY12344/capu38 (T) oocytes. Red arrows indicate Khc::GFP positive MT bundles.
Movie S5.
Dynamic behaviour of EB1 comets in control and capu mutant oocytes. Denoised confocal image sequence of dynamic EB1 comets in control (left) and capuEY12344 mutant oocytes (right). Images have been acquired at one frame every 0.65 s. Playback speed is 40 fps (∼ 20× real speed).
To further understand the relationship between the actin mesh and the interaction of Kin with MTs, we monitored the localization of the motor in different capu mutant alleles. For this, we utilized three mutant alleles that harbor distinct point mutations in the FH2 domain and that affect the in vitro actin assembly rate of Capu to different degress (capuL219, capuL201, and capu38 in increasing severity) (Yoo et al., 2015). To exclude effects from potential second site mutations, we generated trans-heterozygous animals between these alleles and the strong hypomorphic mutation capuEY12344 (Figure 2 and Supplemental Table S1). To estimate the amount of cytoplasmic F-actin, we measured relative signal intensities in phalloidin-stained control and mutant oocytes (Figure 2, M–Q). In agreement with the in vitro data (Yoo et al., 2015), capuL219 oocytes displayed a mild decrease in signal intensities, followed by capuL201 and capu38, resulting in the strongest reduction in fluorescence intensities (Figure 2Q). We next tested the localization of Kin1-700 in these three allelic combinations of capu and found that in capuL219 oocytes, Kin is found at the posterior region of the cell (Figure 2R), but also at small MT bundles (arrow in Figure 2R). This recruitment of Kin to MTs was further enhanced in capuL201 and capu38 to levels similar to what we observed in the homozygous capuEY12344 cells (Figure 2, S and T compared with B). Thus, the strength of Kin recruitment to MTs seems to follow the strength of the defects in the actin mesh. It is important to note that the speed of cytoplasmic flows is equally increased in capuL219, capuL201, and capu38 oocytes (Yoo et al., 2015 and unpublished data). Taken together, this indicates that the amount of Kin recruited to MTs mainly depends on the amount of cytoplasmic F-actin. Together with our previous finding that a higher number of active Kin results in faster flows (Ganguly et al., 2012), these findings point toward a model by which the actin mesh is regulating the magnitude of flows by limiting the efficient recruitment of Kin to MTs.
Cytoplasmic streaming is necessary for MTs to display a correct orientation
Our results so far show that Kin-dependent cytoplasmic flows need to be maintained at low speeds in st9 oocytes for MTs not to misorient. Consequently, we next asked whether cytoplasmic streaming might actually be necessary to sustain a proper organization of the MT network. In other words, is the motion direction of MT plus ends altered in st9 oocytes that lack Kin-dependent cytoplasmic streaming?
To address this question, we first analyzed MTs in oocytes lacking kinesin heavy chain (khc). The complete loss of the Kin motor unit (mutant allele khc27, from here on called khcnull) results in a fully penetrant absence of cytoplasmic flows (Supplemental Figure S8) (Palacios and St Johnston, 2002; Serbus et al., 2005). Compared with control cells, MTs in khcnull mutant oocytes display very little overall motion and appear rather “stiff” (Figure 3, A and A’ compared with Figure 1, J and J’ and Supplemental Movie S6). However, oocytes without Kin not only lack cytoplasmic streaming but also lack cargo transport toward the posterior tip and display an aberrant actin mesh (Figure 3D and Supplemental Figure S9) (Brendza et al., 2000; Drechsler et al., 2017). To test whether the altered bulk behavior of MTs is due to the lack of streaming or other aberrations in Kin mutant oocytes, we monitored MTs in st9 oocytes carrying distinct mutations in the Kin motor domain, resulting in a slower motor (two mutant alleles known as khc23 and khc17, hereafter summarized as khcslow) (Brendza et al., 1999). St9 khcslow oocytes display a normal cytoplasmic actin mesh (Supplemental Figure S9) and are able to transport a considerable amount of cargo toward the posterior (Figure 3, E and F) (Serbus et al., 2005; Loiseau et al., 2010). Importantly, and identical to khcnull cells, they lack any cytoplasmic advection at st9, allowing us to study the impact of flows on MT behavior in the presence of transport (Supplemental Figure S8) (Serbus et al., 2005). Similar to khcnull cells, MTs in khcslow oocytes appear stiff and no motion could be detected in kymographs (Figure 3. B–C’, and Supplemental Movie S6). These observations indicate that the altered motion of MTs in oocytes without Kin is indeed due to a lack of cytoplasmic streaming. Thus, Kin-dependent cytoplasmic advection is necessary for MTs to display a wild-type bulk motion.
Movie S6.
MT bulk motion in control and different Kin mutant oocytes. Dynamic bulk motion of Jup-labelled MTs in control (first from left), khc27 (second from left), khc23 (third from left), and khc17 (right) mutant oocyte. Images have been acquired at one frame every 10.4 s. Playback speed is 20 fps (∼ 200× real speed).
We next analyzed EB1 directionality in khcslow oocytes (Figure 3, G–K). As in control cells, we observed dynamic EB1 comets throughout the entire cytoplasm (Supplemental Movie S7). However, in stark contrast, the distribution of orientation angles displayed a more focused bias toward the posterior of the cell (Figure 3I vs . Figure 1Q). Consequently, khcslow mutant oocytes displayed an increased posterior plus tip bias, with 75% of EB1 signals directed toward posterior, compared with 66% in control cells (Figure 3J vs. Figure 1R and Table 1). Furthermore, 54% of these posterior-directed EB1 signals (the group consisting of 75% of all comets) displayed a posterior tip orientation, compared with 40% in controls (Figure 3K vs. Figure 1S and Table 1). These findings indicate that the orientation of the growing MT plus ends in st9 oocytes depends not only on nucleation or anchoring of minus ends but also on the presence of cytoplasmic flows. In other words, advection is necessary for MTs to display a correct orientation.
Movie S7.
Dynamic behaviour of EB1 comets in control and different Kin mutant oocytes. Denoised confocal time series of dynamic EB1 comets in control (left), khc23 (middle), and khc27 (right) mutant oocytes. Images have been acquired at one frame every 0.65 s. Playback speed is 40 fps (∼ 20× real speed).
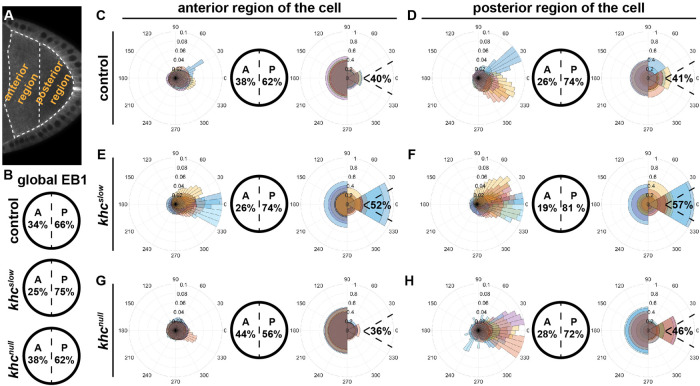
To further investigate the impact of cytoplasmic flows and Kin activity on MT orientation, we analyzed the regional organization of the MT network along the A-P axis in control, khcnull, and khcslow oocytes. We divided each oocyte into an anterior and a posterior region (Figure 4A) and analyzed EB1 directionality in each of these two regions (Table 1). As previously shown, the posterior EB1 bias increases along the A-P axis of st9 oocytes (Figure 4, B–D) (Parton et al., 2011). In the anterior region, we found 62% of signals directed toward the posterior (Figure 4C), while this bias was increased to 74% in the posterior region (Figure 4D). As already demonstrated for the global posterior EB1 bias (Figure 4B), khcslow oocytes also showed a dramatic change of MT orientation along the A-P axis, with a 74% posterior bias in the anterior region (Figure 4, E vs. C) and an even further increased 81% posterior bias in the posterior region (Figure 4, F vs. D). Additionally, the ratio of signals directed toward the posterior tip (Figure 1C) in both anterior and posterior regions of the khcslow cell was substantially increased compared with controls (Figure 4, E and F, and Table 1). This clearly demonstrates that oocytes with a slower Kin, and thus without cytoplasmic streaming, display a stronger polarization of the entire MT network toward the posterior pole, a key region in the establishment of the embryonic body axis. While the lack of Kin (khcnull) seemed to cause only minor defects in the global organization of the MT cytoskeleton (Figure 3, L–P, and Figure 4B), the regional analysis of EB1 directionality in khcnull oocytes revealed major differences compared with control cells. In the anterior region of khcnull cells, we detected an unexpected drop of the posterior bias (56% compared with 66% in controls), which indicates that the complete lack of Kin does indeed affect MT network organization (Figure 4, G vs. C). It needs to be mentioned here that oocytes lacking Kin also fail to localize the nucleus, which in turn is associated with MT minus ends (Williams et al., 2014). Therefore, this observed MT behavior in khcnull cells might primarily reflect the mislocalization of a certain subset of MTOCs in the cell. However, MTs in the posterior region of khcnull cells displayed a slightly more focused growth toward the key posterior tip (Figure 1C), similar to khcslow cells (Figure 4, H vs. D and F, right rosette panels, and Table 1).
FIGURE 4:
Anterior and posterior regional impact of Kin activity on MT orientation. (A) Single frame of an oocyte expressing EB1. Dashed lines represent the anterior and posterior regions of the oocyte used to extract orientation data. (B) Global EB1 signal directions within the entire oocyte (as shown in Figures 1 and 3). (C–H) In each case, from left to right: distribution of EB1 growth orientation visualized in a rose diagram, the anterior-posterior orientation, and the posterior tip orientation. (C, D) Data for control cells (n = 8). (E, F) Data for khcslow mutant cells (n = 10). (G, H) Data for khcnull mutant cells (n = 10).
Taken together, our data allow us to draw certain conclusions about the relationship of Kin activity, cytoplasmic streaming, and the organization of MTs. First, in st9 oocytes, cytoplasmic flows need to be in a defined regime to ensure proper MT orientation. Second, in the presence of Kin-mediated transport, but in the absence of cytoplasmic flows (khcslow condition), there are more plus ends growing toward the posterior, suggesting that cytoplasmic streaming is necessary for MTs to polarize in the precise pattern observed in control cells. And third, together with cytoplasmic flows, other Kin-mediated processes, such as cargo transport or nucleus anchoring, affect the organization of the MT network, supporting previously published findings (Zimyanin et al., 2008; Nieuwburg et al., 2017).
Reconstitution of cytoplasmic flows in st9 khc slow oocytes rescues MT orientation
Our data suggest that Kin-mediated cytoplasmic streaming is necessary for the MT network to completely adopt its wild-type organization in the st9 oocyte. To further investigate the connection between cytoplasmic advection and MT orientation, we analyzed EB1 directionality in capu,khcslow double mutant oocytes. With this experiment we aimed to test whether the re-establishment of cytoplasmic streaming in a mutant background is sufficient to rescue MT orientation.
It has been suspected that faster cytoplasmic flows in capu mutants can be slowed down again by introducing a khcslow mutation (Dahlgaard et al., 2007). Therefore, we generated a double mutant, harboring the alleles capuEY12344 and khc17. To verify the functionality of this double mutant, we first investigated posterior cargo localization in fixed cells. As expected, ∼85% of capu,khcslow/+ cells (which are essentially capu mutants; Supplemental Table S1) failed to correctly localize Staufen to the posterior pole of the cell (Figure 5A, n = 20). In comparison, ∼72% of capu,khcslow double mutant cells localized Staufen into a posterior crescent (n = 25). However, the majority of oocytes that displayed Staufen in a crescent also showed Staufen accumulation in posterior dots (n = 11/18). This constitutes a phenotype that is usually associated with the khcslow alleles (Figure 5B vs. Figure 3, E and F). Together, these data confirmed that our double mutant is comparable to the previously reported alleles (Dahlgaard et al., 2007), and that slow Kin is sufficient to rescue the major cargo localization defects seen in capu mutants. However, since cargo transport was not rescued to wild-type levels and was instead found to be similar to khcslow oocytes (Loiseau et al., 2010), it is obvious that the decreased cargo transport efficiency of slow Kin cannot be rescued by reintroducing cytoplasmic streaming by the lack of capu. It is furthermore unclear whether this is due to a reduced translocation speed of slow Kin or to a defect in cargo anchoring in capu mutant cells (Tanaka et al., 2011).
Next we tested whether premature fast cytoplasmic flows in capu mutants are indeed slowed down again in capu,khcslow oocytes. As expected, and similar to capu single mutants, capu,khcslow/+ oocytes displayed aberrant fast flows (Figure 5C; Supplemental Movie S8). In contrast, cytoplasmic flows in capu,khcslow double mutant oocytes were indeed slowed down again and appeared similar to those observed in control cells (Supplemental Figure S8). We then tested whether the rescue of cytoplasmic streaming in the double mutant also affected MT bulk movement. Compared with capu,khcslow/+ oocytes, which displayed fast flows and consequently MT bundling, the reduction of flow speeds in the double mutant caused a MT bulk movement similar to that observed in control cells (Figure 5, C–D’, and Supplemental Movie S8).
Movie S8.
MT bulk motion in capu,khcslow/+ and capu,khcslow double mutant oocytes. Dynamic bulk motion of Jup-labelled MTs in capuEY12344,khc17/capuRK,GFP (left, transheterozygous mutant for capuEY12344/capuRK and heterozygous for khc17) and capuEY12344,khc17/capuRT, khc17 (right, transheterozygous mutant for capuEY12344/capuRK and homozygous for khc17). Images have been acquired at one frame every 10.4 s. Playback speed is 20 fps (∼ 200× real speed).
To finally test whether the re-establishment of cytoplasmic streaming in a khcslow background (or the reduction of fast flows in capu background) rescues the MT orientation defects observed in either single mutant, we analyzed EB1 directionality in capu,khcslow/+ (Figure 5, E–I) and capu,khcslow double mutant oocytes (Figure 5, J–N, and Supplemental Movie S9). EB1 comets in capu,khcslow/+ oocytes exhibited values that are similar to what we observed in capu mutants (Figure 1, T–X), suggesting that the heterozygous presence of a khcslow mutant chromosome does not substantially affect the capu mutant phenotype (Supplemental Movie S9, 61% of EB1 signals pointing toward posterior, and 34% of this 61% are growing toward the posterior tip in capu,khcslow/+ cells; Figure 5, F–I, and Table 1). Conversely, EB1 comets in capu,khcslow double mutant cells displayed a posterior motion bias indistinguishable from that of control cells (Supplemental Movie S9, 66% posterior bias, Figure 5M vs. Figure 1R and Table 1). Finally, the fraction of all posterior comets pointing toward the posterior tip was also found to be similar in capu,khcslow double mutant cells (39%) and controls (40%) (Figure 5N vs. Figure 1S and Table 1), further strengthening the idea that cytoplasmic streaming constitutes a substantial contributor to a correct MT plus end focusing, and thus MT organization, in the oocyte.
Movie S9.
Dynamic behaviour of EB1 comets in capu,khcslow/+ and capu,khcslow double mutant oocytes. Denoised confocal time series of dynamic EB1 comets in capuEY12344,khc17/capuRK,GFP (left, transheterozygous mutant for capuEY12344/capuRK and heterozygous for khc17) and capuEY12344,khc17/capuRT, khc17 (right, transheterozygous mutant for capuEY12344/capuRK and homozygous for khc17). Images have been acquired at one frame every 0.65 s. Playback speed is 40 fps (∼ 20× real speed).
In summary, our results now clearly demonstrate that in the absence of capu and the actin mesh, a slower Kin motor protein is sufficient to restore the correct regime of cytoplasmic flows, resulting in the correct polarization of the MT cytoskeleton and an almost restored transport of cargo toward the posterior. Therefore, it seems that the actin mesh is essential in oocytes that exhibit normal Kin-mediated transport, likely by regulating the amount of Kin recruited to MTs in order to ensure the proper regulation of cytoplasmic streaming, which in turn is an important contributor to the observed MT orientation.
DISCUSSION
Due to its large size (80–100 µm along the A-P axis) and the fact that MT minus ends are nucleated and anchored along the antero-lateral cortices of the cell, the Drosophila st9 oocyte is arguably one of the most challenging in vivo systems in which to analyze the dynamic behavior of MTs. The use of confocal microscopy, together with the OF-based analysis tool described in this study, allowed us to quantify the motion direction of plus ends within a 2D focal plane along the entire length of this cell. Together with others, the present study supports the existence of a complex mechanical and/or biochemical relationship between motion of cytoplasmic components (whether by advection or by transport), cytoplasmic F-actin, and MTs in the Drosophila oocyte. This holds true for our model system, but is likely transferable to many other cell types. For example, experimental data and modeling of cytoplasmic motion in Characean algae (where cytoplasmic flows are acto-myosin dependent) suggest that parallel actin cables and ordered streaming patterns self-organize in an interdependent manner (Foissner and Wasteneys, 2000; Woodhouse and Goldstein, 2013).
Kin-mediated cargo transport through a highly viscous medium, such as the cytoplasm, inevitably induces bulk motion of such medium, which will cause lateral displacement forces on MT filaments and thus induce a feedback on their spatial orientation. As a consequence, cytoplasmic streaming needs to be kept in a defined speed regime and at a biased random pattern in order for the MT network to properly polarize. In the absence of the actin mesh (as in capu or spire mutants), there is an increased persistence and speed of flows, resulting in parallel alignment and bundling of MTs, as well as strong defects on MT orientation. The actin mesh—which itself requires Kin activity for its proper organization (Drechsler et al., 2017)—is required for the maintenance of this correct regime of advection.
With our work, we show that our image analysis approach—based on a rigorous image analysis framework—is able to infer and quantify directionality of EB1 motion from confocal image sequences. The use of an efficient iterative optimization algorithm enabled us to analyze entire image sequences at once, as compared with a frame-by-frame analysis. In comparison to more commonly used techniques, such as particle tracking or PIV, OF allows us to perform motion estimation on pixel level in challenging circumstances, such as low signal-to-noise ratios and small particle size. Most importantly, this image analysis approach can handle standard confocal microscopy data and the method does not require demanding imaging techniques or costly computing hardware. Moreover, the image analysis depends only on a few parameters that can be easily adjusted. However, two limitations of the developed methodology need to be pointed out. First, due to the use of a variational framework for the image analysis, both the denoised sequences and the estimated velocities exhibit a loss of contrast, leading to underestimated speeds of EB1 comets. This is particularly due to the temporal regularization required in both steps to overcome the above-mentioned challenges. Second, our statistical analyses of directions of EB1 comets are based on velocities computed for all pixels within each segmented oocyte. We are aware that not every image pixel portrays exactly one EB1 comet. Furthermore, EB1 comets in the oocyte are subjected to different forces. In summary, however, our results demonstrate that our approach is able to monitor general plus end direction in most, if not all, cell types and thus constitutes an efficient and reliable analytical framework for MT polarity studies. Therefore, OF is absolutely suitable to efficiently describe the orientation of MTs in crowded intracellular environments.
It is unknown how the actin mesh slows down Kin-dependent cytoplasmic flows or how it might affect MT organization. It has been proposed that the presence of a viscoelastic actin network can increase the effective viscosity of the cytoplasm and counteract the viscous drag of cargo transport by Kin (Quinlan, 2016). Furthermore, actin-MT cross-linking proteins have been demonstrated to allow a potent cross-talk between both filament species (Preciado Lopez et al., 2014), presumably coordinating cytoskeletal organization in vivo. However, our data from capu,khcslow double mutant oocytes, which do not form an actin mesh, suggest that the mesh itself is not essential to allow a correct MT orientation when Kin is slower. In this “artificial” mutant situation, the correct regime of flows seems sufficient to allow proper MT network organization. It is thus possible that the actin mesh regulates the activity of Kin more directly, for example by tethering the motor to its filaments (directly or indirectly through cargo). Such a model is indeed supported by our finding that Kin becomes efficiently recruited to MTs when the actin mesh is absent (Supplemental Movie S10 and Figure 2). This observation could explain how the mesh regulates not only flows but also the higher degree of MT bundling, as a larger number of Kin molecules recruited to MTs is likely to result in faster flows (Ganguly et al., 2012). Furthermore, this could also result in a higher degree of effective cross-linking of adjacent filaments (Lu et al., 2016). However, the recruitment of Kin to MTs depends on various regulatory mechanisms, including interactions with cargo and MT-associated proteins like Ensconsin (Metivier et al., 2019). For this reason we used the truncated form Khc1-700 in our analyses. Whether and how the actin mesh influences the interaction of full-length Kin with MTs—in the presence of other MT-binding regulatory mechanisms—need to be further addressed in the future. Nevertheless, our findings indicate a novel potential mechanism of how the actin and MT cytoskeletons can regulate each other.
Movie S10.
Recruitment of Kin towards MTs in the absence of the actin mesh. A truncated KHC fusion protein (aa 1-700, C-terminal GFP fusion), localised in a posterior cloud in control oocytes (left), and was detected to weakly associate with filamentous MTs in the anterior region of the oocyte. Loss of capu caused a re-localisation of the protein, which is could now be found to heavily decorate MTs in the entire oocyte (right). Images have been acquired at one frame every 10.4 s. Playback speed is 20 fps (∼ 200× real speed).
Our data establish cytoplasmic streaming in st9 oocytes as a contributing factor for the correct organization of the MT cytoskeleton. If and how a stronger posterior polarization of the MT network may affect cargo delivery to the posterior are unknown. Tracking oskar mRNA particles in khcslow oocytes did not reveal a stronger orientation bias of cargo movement (Zimyanin et al., 2008). However, it is unclear whether trajectories of oskar transport always reflect the organization of the underlying MT network and whether all MTs would be equally used by slow Kin motor/cargo complexes.
Previous studies suggested that the pattern of nucleation and minus end anchoring along the cortex are sufficient to organize the MT network and to allow correct cargo transport to define the A-P axis of the animal. Consequently, mutant oocytes harboring defects in nucleation and/or anchoring of minus ends also display polarity defects (Doerflinger et al., 2006; Nashchekin et al., 2016). Furthermore, mathematical modeling suggested that cytoplasmic streaming at midoogenesis is negligible to explain the correct localization of posterior cargoes like oskar mRNA (Khuc Trong et al., 2015). However, our study demonstrates that cytoplasmic streaming is involved in the orientation of MT plus ends, and we conclude that the localization of the minus ends alone is not sufficient to define the precise overall organization of the network. This was most obvious in khcslow oocytes, which in our hands lack cytoplasmic streaming entirely and displayed an increased posterior orientation MT bias (Figure 3). The advantage of analyzing slow Kin mutants is that other Kin-dependent processes, like cargo transport and formation of the actin mesh, do take place. Despite showing only mild defects in the distribution of developmental determinants, khcslow mutant oocytes fail to give rise to a healthy offspring, suggesting that oocyte polarity is affected (Moua et al., 2011). Consistently, we found an increased posterior bias of EB1 signals in khcslow mutant oocytes, strongly suggesting that the lack of cytoplasmic streaming was causative for this observation.
In summary, the combination of various forces produced within living cells demands a complex set of biochemical and biomechanical regulatory mechanisms for cytoskeletal networks to organize correctly. Our results show that, in Drosophila st9 oocytes, advection by cytoplasmic streaming contributes to the polarization of MTs by affecting the directional motion of MT plus ends. All these observations further stress the need for a combination of different experimental approaches in order to fully understand the dynamic organization of cytoskeletons, from simplified in vitro systems to complex in vivo situations.
MATERIALS AND METHODS
Fly stocks and genetics
Flies were kept at standard corn meal agar and raised at room temperature (21°C). Detailed genotypes of all fly stocks can be found in Supplemental Table S1. Germline clones for the analysis of khc mutant alleles have been induced by the FLP/FRT ovoD system (Chou and Perrimon, 1996). Germline clones in Figure 5 were identified by the absence of nuclear GFP in germline cells.
Live imaging
Female flies of the desired genotypes were collected and fattened on dry yeast for 12–16 h prior to imaging. Ovaries were dissected in a small drop of halocarbon oil (Voltalef S10, VWR) on a glass coverslip and single egg chambers were separated using fine tungsten needles. Images were acquired on a Leica SP5 inverted confocal microscope, using a 40×/1.3 Oil DIC Plan-Neofluar (Jup) or a 100×/1.4 Oil DIC objective (EB1). Signals were detected using a Leica HyD Hybrid Detector. For MT bulk movement, a single plane from the middle of the oocyte was imaged at a scan speed of 100 Hz and at an image resolution of 1024 × 1024 pixels (corresponding to one image every 10.4 s). For EB1 imaging, the oocyte was fitted and oriented within a 512 × 256 pixel frame and a single plane image was taken every 0.65 s. Image sequences of at least 100 frames (65 s) were taken, inspected visually, and bleach corrected using Fiji (Schindelin et al., 2012). In all experiments, the middle of the oocyte was defined as the focal plane in which the oocyte had its largest area.
Immunostainings
Egg chambers were dissected in phosphate-buffered saline (PBS) + 0.1% Tween 20 and fixed in 10% formaldehyde in PBS + 0.1% Tween 20 for 10 min. Fixed ovaries were incubated with anti-αTubulin (MAB1864, Sigma-Aldrich, cloneYL1/2, 1:100) or anti-Staufen (a gift from. D. St Johnston, 1:3,000) in PBS + 2% Tween 20 overnight at 4°C. After four consecutive washes Alexa-coupled secondary antibodies (1:100) were incubated for 2 h at room temperature. Native fluorescence of GFP was imaged without amplification. Images were acquired on a Leica SP5 inverted confocal microscope, using a 40×/1.3 Oil DIC Plan-Neofluar objective. For phalloidin staining, egg chambers were dissected and fixed in 10% methanol free formaldehyde in PBS containing 0.1% Tween 20 (PBT0.1) for a maximum of 10 min. Fixed cells were washed 4× in PBT0.1 and incubated with 1 µM TRITC-coupled phalloidin (in PBT0.1, Sigma-Aldrich) overnight at 4 °C. The stained samples were washed 4× in PBT0.1, mounted, and imaged imediately under identical conditions using a Zeiss LSM800 equipped with a 40×/1.3 Oil DIC Plan-Neofluar objective. Normalized fluorescence intensities were acquired using Fiji.
Image denoising and optical flow-based motion estimation
Motion analysis of the recorded 2D image sequences was performed using a two-step procedure. The first step aimed to remove noise contamination from the unprocessed sequences, while the goal of the second step was to estimate motion in terms of displacement vector fields from the so-improved sequences. As noise may significantly disturb the motion estimation (Ruhnau et al., 2005; Burger et al., 2018), this two-step procedure is crucial for obtaining robust estimation results. In the first step, we recovered from each recorded image sequence uδ an improved version u by solving a variational image denoising problem with spatio-temporal regularization. It reads
While the first two terms on the right-hand side resemble standard image denoising with total variation regularization (Rudin et al., 1992) in space, the third term connects subsequent frames by penalizing temporal changes within the recovered solution. As EB1 comets typically appear in several subsequent frames at similar positions, it allows us to effectively remove randomly distributed noise from a sequence (see Figure 1, D and E, Supplemental Movie S2, and Supplemental Information). Here, α,β > 0 are regularization parameters that balance the three terms and need to be chosen appropriately.
The result u served as input to the motion estimation step, in which we estimated a displacement vector field 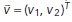 by solving
by solving
Here, 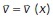 is defined for every pixel x of the image u and is an estimate of the motion that u experiences at x from one image frame to the subsequent one in time. That is, the vector
is defined for every pixel x of the image u and is an estimate of the motion that u experiences at x from one image frame to the subsequent one in time. That is, the vector 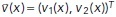 points in the direction of motion at x, and
points in the direction of motion at x, and 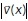 is an estimate of the speed of the motion at x. The first term on the right-hand side in the above equation aims to approximately solve the OF equation (Horn and Schunck, 1981), while the second and third terms incorporate spatio-temporal regularization of the vector-valued unknown (for further details, see Supplemental Information). The use of the vector-valued total variation allows for spatial discontinuities in the displacement vector field. We found that in both steps the temporal regularization was key and the analysis of individual frames did not yield satisfactory results. Both minimization problems were approximately solved using the primal-dual hybrid gradient method (Chambolle, 2011) and graphics-processing unit (GPU) acceleration. An in-depth description of both models, their numerical solution, and parameter choices as well as implementation details can be found in the Supplemental Information.
is an estimate of the speed of the motion at x. The first term on the right-hand side in the above equation aims to approximately solve the OF equation (Horn and Schunck, 1981), while the second and third terms incorporate spatio-temporal regularization of the vector-valued unknown (for further details, see Supplemental Information). The use of the vector-valued total variation allows for spatial discontinuities in the displacement vector field. We found that in both steps the temporal regularization was key and the analysis of individual frames did not yield satisfactory results. Both minimization problems were approximately solved using the primal-dual hybrid gradient method (Chambolle, 2011) and graphics-processing unit (GPU) acceleration. An in-depth description of both models, their numerical solution, and parameter choices as well as implementation details can be found in the Supplemental Information.
Supplementary Material
Acknowledgments
We are greatful to Fabio Giavazzi (Milan) for help with the PIV analysis in some of our experiments. We thank D. St Johnston and I. Davis for reagents and M. Wayland for assistance with imaging. We also thank Lena Frerking and Sujoy Ganguly for fruitful discussions during the initial phase of the project and MD Martin Bermudo and D. St Johnston for comments on the manuscript and discussions.
M.D. and I.M.P. were supported by the BBSRC, the University of Cambridge, and Queen Mary University of London. M.D. was also supported by an Isaac Newton Trust fellowship and acknowledges funding by the DFG/SFB944 (Physiology and Dynamics of Cellular Microcompartments), the University of Osanbrück, and the State of Lower Saxony. L.F.L. and C.B.S. acknowledge support from the Leverhulme Trust (Breaking the non-convexity barrier), EPSRC (grant EP/M00483X/1), the EPSRC Centre (EP/N014588/1), the RISE projects CHiPS and NoMADS, the Cantab Capital Institute for the Mathematics of Information, and the Alan Turing Institute. L.A.K. was supported by Queen Mary University of London. M.B. and H.D. acknowledge support by the European Research Council (EU FP7-ERC Consolidator Grant 615216 LifeInverse). We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Quadro P6000 GPU used for this research.
Abbreviations used:
- capu
cappuccino
- EB1
EB1::GFP
- khc
kinesin heavy chain
- GPU
graphics-processing unit
- Kin
Kinesin-1
- grk
gurken
- MT
microtubule
- Jup
Jupiter::GFP
- MTOC
MT organizing center
- OF
optical flow
- PBS
phosphate-buffered saline
- PIV
particle image velocimetry
- st9
stage 9.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-08-0440) on April 8, 2020.
REFERENCES
- Akhmanova A, Steinmetz MO. (2015). Control of microtubule organization and dynamics: two ends in the limelight. Nat Rev Mol Cell Biol , 711–726. [DOI] [PubMed] [Google Scholar]
- Amat F, Myers EW, Keller PJ. (2013). Fast and robust optical flow for time-lapse microscopy using super-voxels. Bioinformatics , 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S, Scharstein D, Lewis JP, Roth S, Black MJ, Szeliski R. (2011). A database and evaluation methodology for optical flow. Int J Comput Vision , 1–31. [Google Scholar]
- Boquet-Pujadas A, Lecomte T, Manich M, Thibeaux R, Labruyere E, Guillen N, Olivo-Marin JC, Dufour AC. (2017). BioFlow: a non-invasive, image-based method to measure speed, pressure and forces inside living cells. Sci Rep , 9178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boric K, Orio P, Vieville T, Whitlock K. (2013). Quantitative analysis of cell migration using optical flow. PLoS One , e69574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza KM, Rose DJ, Gilbert SP, Saxton WM. (1999). Lethal kinesin mutations reveal amino acids important for ATPase activation and structural coupling. J Biol Chem , 31506–31514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza RP, Serbus LR, Duffy JB, Saxton WM. (2000). A function for Kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science , 2120–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger M, Dirks H, Schönlieb C-B. (2018). A Variational Model for Joint Motion Estimation and Image Reconstruction. SIAM J Imaging Sci , 94–128. [Google Scholar]
- Chambolle APT. (2011). A first-order primal-dual algorithm for convex problems with applications to imaging. J Math Imaging Vis , 120–145. [Google Scholar]
- Chou TB, Perrimon N. (1996). The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster . Genetics , 1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgaard K, Raposo AA, Niccoli T, St Johnston D. (2007). Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila Oocyte. Dev Cell , 539–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpiano J, Jara J, Scheer J, Ramírez OA, Ruiz-del-Solar J, Steffen H. (2012). Performance of optical flow techniques for motion analysis of fluorescent point signals in confocal microscopy. Mach Vis Appl , 675–689. [Google Scholar]
- Doerflinger H, Benton R, Torres IL, Zwart MF, St Johnston D. (2006). Drosophila anterior-posterior polarity requires actin-dependent PAR-1 recruitment to the oocyte posterior. Curr Biol , 1090–1095. [DOI] [PubMed] [Google Scholar]
- Drechsler M, Giavazzi F, Cerbino R, Palacios IM. (2017). Active diffusion and advection in Drosophila oocytes result from the interplay of actin and microtubules. Nat Commun , 1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassier C, Freal A, Gasmi L, Delphin C, Ten Martin D, De Gois S, Tambalo M, Bosc C, Mailly P, Revenu C, Peris L, et al. (2018). Motor axon navigation relies on Fidgetin-like 1-driven microtubule plus end dynamics. J Cell Biol , 1719–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissner I, Wasteneys GO. (2000). Microtubule disassembly enhances reversible cytochalasin-dependent disruption of actin bundles in characean internodes. Protoplasm , 33–44. [Google Scholar]
- Forrest KM, Gavis ER. (2003). Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila . Curr Biol , 1159–1168. [DOI] [PubMed] [Google Scholar]
- Frerking L, Burger M, Vestweber DCB. (2014). TGV-based flow estimation for 4D leukocyte transmigration. In: Proceedings of the Inverse Problems from Theory to Applications Conference, ed. Louis AK, Arridge S, Rundell B, Bristol, UK: IOP Publishing, 79–83. [Google Scholar]
- Ganguly S, Williams LS, Palacios IM, Goldstein RE. (2012). Cytoplasmic streaming in Drosophila oocytes varies with kinesin activity and correlates with the microtubule cytoskeleton architecture. Proc Natl Acad Sci USA , 15109–15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Reyes A, Elliott H, St Johnston D. (1995). Polarization of both major body axes in Drosophila by gurken-torpedo signalling. Nature , 654–658. [DOI] [PubMed] [Google Scholar]
- Guo VDV, Zhou X. (2014). Red blood cell tracking using optical flow methods. IEEE J Biomed Health Inform , 991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn BKP, Schunck BG. (1981). Determining optical flow. Artif Intell , 185–203 [Google Scholar]
- Huang Y, Hao L, Li H, Liu Z, Wang P. (2017). Quantitative analysis of intracellular motility based on optical flow model. J Healthc Eng . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd TR, Herrmann B, Sauerwald J, Sanny J, Grosch M, Lehmann R. (2016). Long oskar controls mitochondrial inheritance in Drosophila melanogaster . Dev Cell , 560–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J, Gervais L, Dass S, Kaltschmidt JA, Lopez-Schier H, Johnston DS, Brand AH, Roth S, Guichet A. (2002). Polar transport in the Drosophila oocyte requires Dynein and Kinesin I cooperation. Curr Biol , 1971–1981. [DOI] [PubMed] [Google Scholar]
- Jiang K, Toedt G, Montenegro Gouveia S, Davey NE, Hua S, van der Vaart B, Grigoriev I, Larsen J, Pedersen LB, Bezstarosti K, et al. (2012). A Proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr Biol , 1800–1807. [DOI] [PubMed] [Google Scholar]
- Kent IA, Rane PS, Dickinson RB, Ladd AJ, Lele TP. (2016). Transient pinning and pulling: a mechanism for bending microtubules. PLoS One , e0151322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuc Trong P, Doerflinger H, Dunkel J, St Johnston D, Goldstein RE. (2015). Cortical microtubule nucleation can organise the cytoskeleton of Drosophila oocytes to define the anteroposterior axis. Elife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama A, Karakesisoglou I, Wong E, Vaezi A, Fuchs E. (2003). ACF7. An essential integrator of microtubule dynamics. Cell , 343–354. [DOI] [PubMed] [Google Scholar]
- Loiseau P, Davies T, Williams LS, Mishima M, Palacios IM. (2010). Drosophila PAT1 is required for Kinesin-1 to transport cargo and to maximize its motility. Development , 2763–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Lakonishok M, Serpinskaya AS, Kirchenbuechler D, Ling SC, Gelfand VI. (2018). Ooplasmic flow cooperates with transport and anchorage in Drosophila oocyte posterior determination. J Cell Biol , 3497–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Winding M, Lakonishok M, Wildonger J, Gelfand VI. (2016). Microtubule-microtubule sliding by kinesin-1 is essential for normal cytoplasmic streaming in Drosophila oocytes. Proc Natl Acad Sci USA , E4995–E5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier M, Monroy BY, Gallaud E, Caous R, Pascal A, Richard-Parpaillon L, Guichet A, Ori-McKenney KM, Giet R. (2019). Dual control of Kinesin-1 recruitment to microtubules by Ensconsin in Drosophila neuroblasts and oocytes. Development . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. (1984). Dynamic instability of microtubule growth. Nature , 237–242. [DOI] [PubMed] [Google Scholar]
- Monteith CE, Brunner ME, Djagaeva I, Bielecki AM, Deutsch JM, Saxton WM. (2016). A mechanism for cytoplasmic streaming: kinesin-driven alignment of microtubules and fast fluid flows. Biophys J , 2053–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moua P, Fullerton D, Serbus LR, Warrior R, Saxton WM. (2011). Kinesin-1 tail autoregulation and microtubule-binding regions function in saltatory transport but not ooplasmic streaming. Development , 1087–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashchekin D, Fernandes AR, St Johnston D. (2016). Patronin/Shot cortical foci assemble the noncentrosomal microtubule array that specifies the Drosophila anterior-posterior axis. Dev Cell , 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwburg R, Nashchekin D, Jakobs M, Carter AP, Khuc Trong P, Goldstein RE, St Johnston D. (2017). Localised dynactin protects growing microtubules to deliver oskar mRNA to the posterior cortex of the Drosophila oocyte. Elife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios IM, St Johnston D. (2002). Kinesin light chain-independent function of the Kinesin heavy chain in cytoplasmic streaming and posterior localisation in the Drosophila oocyte. Development , 5473–5485. [DOI] [PubMed] [Google Scholar]
- Parton RM, Hamilton RS, Ball G, Yang L, Cullen CF, Lu W, Ohkura H, Davis I. (2011). A PAR-1-dependent orientation gradient of dynamic microtubules directs posterior cargo transport in the Drosophila oocyte. J Cell Biol , 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preciado Lopez M, Huber F, Grigoriev I, Steinmetz MO, Akhmanova A, Koenderink GH, Dogterom M. (2014). Actin-microtubule coordination at growing microtubule ends. Nat Commun , 4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purro SA, Ciani L, Hoyos-Flight M, Stamatakou E, Siomou E, Salinas PC. (2008). Wnt regulates axon behavior through changes in microtubule growth directionality: a new role for adenomatous polyposis coli. J Neurosci , 8644–8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan ME. (2013). Direct interaction between two actin nucleators is required in Drosophila oogenesis. Development , 4417–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan ME. (2016). Cytoplasmic streaming in the drosophila oocyte. Annu Rev Cell Dev Biol , 173–195. [DOI] [PubMed] [Google Scholar]
- Roth S, Neuman-Silberberg FS, Barcelo G, Schupbach T. (1995). cornichon and the EGF receptor signaling process are necessary for both anterior-posterior and dorsal-ventral pattern formation in Drosophila . Cell , 967–978. [DOI] [PubMed] [Google Scholar]
- Rudin LI, Osher S, Fatemi E. (1992). Nonlinear total variation based noise removal algorithms. Physica D: Nonlinear Phenomena , 259–268. [Google Scholar]
- Ruhnau P, et al. (2005). Variational optical flow estimation for particle image velocimetry. Expe Fluids , 21–32. [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbus LR, Cha BJ, Theurkauf WE, Saxton WM. (2005). Dynein and the actin cytoskeleton control kinesin-driven cytoplasmic streaming in Drosophila oocytes. Development , 3743–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Kato Y, Matsuda K, Hanyu-Nakamura K, Nakamura A. (2011). Drosophila Mon2 couples Oskar-induced endocytosis with actin remodeling for cortical anchorage of the germ plasm. Development , 2523–2532. [DOI] [PubMed] [Google Scholar]
- Theurkauf WE. (1994). Premature microtubule-dependent cytoplasmic streaming in cappuccino and spire mutant oocytes. Science , 2093–2096. [DOI] [PubMed] [Google Scholar]
- Vig DK, Hamby AE, Wolgemuth CW. (2016). On the quantification of cellular velocity fields. Biophys J , 1469–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LS, Ganguly S, Loiseau P, Ng BF, Palacios IM. (2014). The auto-inhibitory domain and ATP-independent microtubule-binding region of Kinesin heavy chain are major functional domains for transport in the Drosophila germline. Development , 176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse FG, Goldstein RE. (2013). Cytoplasmic streaming in plant cells emerges naturally by microfilament self-organization. Proc Natl Acad Sci USA , 14132–14137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H, Roth-Johnson EA, Bor B, Quinlan ME. (2015). Drosophila Cappuccino alleles provide insight into formin mechanism and role in oogenesis. Mol Biol Cell , 1875–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D. (2008). In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell , 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.