Abstract
Background
Jails may facilitate spread of methicillin-resistant Staphylococcus aureus (MRSA) in urban areas. We examined MRSA colonization upon entrance to a large urban jail to determine if there are MRSA transmission networks preceding incarceration.
Methods
Males incarcerated in Cook County Jail (Chicago) were enrolled, with enrichment for people living with human immunodeficiency virus (PLHIV), within 72 hours of intake. Surveillance cultures assessed prevalence of MRSA colonization. Whole-genome sequencing (WGS) identified preincarceration transmission networks.
We examined methicillin-resistant Staphylococcus aureus (MRSA) isolates to determine if there are transmission networks that precede incarceration. A large proportion of individuals enter jail colonized with MRSA. Molecular epidemiology and colonization risk factors provide clues to community reservoirs for MRSA.
Results
There were 718 individuals (800 incarcerations) enrolled; 58% were PLHIV. The prevalence of MRSA colonization at intake was 19%. In multivariate analysis, methamphetamine use, unstable housing, current/recent skin infection, and recent injection drug use were predictors of MRSA. Among PLHIV, recent injection drug use, current skin infection, and HIV care at outpatient clinic A that emphasizes comprehensive care to the lesbian, gay, bisexual, transgender community were predictors of MRSA. Fourteen (45%) of 31 detainees with care at clinic A had colonization. WGS revealed that this prevalence was not due to clonal spread in clinic but rather to an intermingling of distinct community transmission networks. In contrast, genomic analysis supported spread of USA500 strains within a network. Members of this USA500 network were more likely to be PLHIV (P < .01), men who have sex with men (P < .001), and methamphetamine users (P < .001).
Conclusions
A large proportion of individuals enter jail colonized with MRSA. Molecular epidemiology and colonization risk factors provide clues to identify colonized detainees entering jail and potential community reservoirs of MRSA.
Keywords: MRSA, epidemiology, incarceration, whole genomic sequencing, community-associated MRSA
We examined methicillin-resistant Staphylococcus aureus (MRSA) isolates from detainees entering jail to determine if there are community transmission networks that precede incarceration. A large proportion of individuals enter jail colonized with MRSA. Molecular epidemiology and colonization risk factors provide clues to community reservoirs for MRSA.
Community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) has emerged as a significant pathogen in community [1, 2] and healthcare settings [3]. By pulsed-field gel electrophoresis, USA300 is the most common CA-MRSA strain in the United States [4], affecting several distinct populations, including illicit drug users [5], men who have sex with men (MSM) [6], military recruits [7], athletes [8], homeless individuals [9, 10], and those in correctional facilities [11, 12]. While USA300 remains the predominant CA-MRSA pathogen, USA500 MRSA has been reported as an important pathogen among people living with human immunodeficiency virus (PLHIV) [13].
Congregate settings, those characterized by close person-to-person contact, are hypothesized to facilitate spread of MRSA, for example, urban jails have had sustained outbreaks of CA-MRSA skin infections [14–16]. Jails are characterized by short lengths of stay (pending sentencing) and high detainee turnover and recidivism; it is unclear if these features impact downstream rates of MRSA in the community. In one study, it was observed that residence in a high detainee-release zip code in Chicago led to a nearly 2-fold increased risk of having CA-MRSA infection [17], suggesting there may be interplay between incarceration and particular geographic areas for MRSA risk.
In addition to geography, type of residence (eg, homeless, substance abuse center, public housing) has been associated with CA-MRSA [17, 18]. Illicit drug use has been a consistent risk factor for MRSA in inner-city populations [5, 19, 20] and also could lead to an increase in S. aureus infections given the current opioid epidemic. Uneven geographic distribution of MRSA in urban areas [6, 17] and association of specific community exposures with MRSA risk has led to speculation that community-based transmission networks for MRSA exist.
MRSA transmission dynamics before, during, and following incarceration at large inner-city jails, hypothesized to be a focal point of MRSA spread, are unclear. In addition, the extent and impact of community MRSA transmission networks on the importation of MRSA into jails is unknown. Our objectives in this study were to determine the prevalence of MRSA colonization at entrance to a large inner-city jail, identify epidemiologic predictors of MRSA colonization upon entrance to the jail, and use genomics to infer community networks of MRSA transmission that precede incarceration.
METHODS
Study Population and Patient Enrollment
The study setting was the Cook County Jail (Chicago, IL), one of the largest single-site US jails with roughly 250 incarcerations per day and an average daily census of 9000–10 000 detainees. From January 2016 through December 2017, incarcerated males were enrolled within 72 hours of jail intake. Given our prior work demonstrating the significant burden MRSA has had on PLHIV [17, 21] and that 1 in 7 (14%) PLHIV pass through correctional facilities annually [22], we enriched the study population with PLHIV by enrolling from the jail HIV clinic. Furthermore, enriching with PLHIV allowed us to better understand the social networks that could account for previously reported high rates of MRSA in this population [9, 21]. The estimated seroprevalence of PLHIV at the Cook County Jail is approximately 2%. For those not living with HIV, enrollment occurred in the early evening when newly arrested males were brought into the jail.
Individuals identified or known to be living with HIV upon entering the jail were typically seen in the jail HIV clinic within 24–48 hours from jail entrance. All PLHIV seen in the jail HIV clinic were eligible for enrollment. For those not living with HIV, enrollment occurred 1 to 2 evenings a week when newly arrested males were brought into the jail. To allow for enrollment throughout the year and given the large number of eligible individuals not living with HIV, we targeted enrollment at 10 males each week.
Swab Collection and Processing
Surveillance cultures from the anterior nares, throat, and bilateral inguinal area were collected at the initial enrollment visit to determine the prevalence of MRSA colonization. Specimens were obtained using the Copan ESwab for MRSA detection [23]. Nasal swabs were collected by swabbing both anterior nares; throat swabs were collected by swabbing the posterior pharynx; and inguinal swabs were collected from a 10-cm2 area of skin bilaterally. Sample sites were chosen to maximize identification of MRSA carriers [9, 24] and participant acceptability.
To increase culture sensitivity, swabs were inoculated into enrichment broth [25]. Aliquots of overnight broth cultures were inoculated on ChromID MRSA (bioMérieux, Durham, NC). Staphylococcus aureus was confirmed by standard biochemical tests; methicillin resistance was confirmed by cefoxitin disk. Confirmed MRSA isolates underwent DNA extraction.
Whole-genome Sequencing
Genomic DNA extracted from MRSA isolates was prepared for sequencing using a Nextera XT library XT library preparation kit (Illumina, San Diego, CA) according to the manufacturer’s instructions. Sequencing was performed on an Illumina NextSeq500 instrument using a high-output kit with paired-end 2 × 75 base reads. Library preparation and sequencing were performed at the University of Illinois Chicago Sequencing Core. Details on sequenced strains are available in Supplementary Table 1. Sequence data are available under Bioproject PRJNA530184.
Genomic Analysis
The quality of sequencing reads was assessed with FastQC [26]; Trimmomatic [27] was used for trimming adapter sequences and low-quality bases. Single-nucleotide variants (SNVs) were identified by mapping cleaned sequencing reads against a finished USA300 MRSA reference genome and applying a series of filters to retain high-quality variants (see Supplementary Methods). Phylogenetic analyses were performed using IQTREE v1.5.5 [28] (see Supplementary Methods). For individuals with multiple isolates sequenced (see Supplementary Figure 1), representative isolates were selected using a combination of phylogenetic clustering and an SNV distance threshold to include multiple isolates from an individual if they were deemed to represent independent acquisition events (see Supplementary Materials).
Risk Factors and Statistical Analyses
A survey to identify predictors of MRSA colonization was administered to study participants at the initial enrollment visit and included questions about demographics, drug use, sexual history, medical care, housing status, and incarceration history.
Prevalence of MRSA colonization at entrance to the jail was calculated using the total number of individuals sampled as the denominator. The prevalence of extranasal and exclusive extranasal colonization (colonization at a body site when the anterior nares were negative for MRSA) was also examined.
SAS software version 9.4 (SAS Institute, Cary, NC) was used for statistical analysis; χ2 analysis was used for categorical variables; and the Fisher exact test was used for low-frequency predictors. Statistically important (ie, P < .2) factors on univariate analysis were included in multivariate analysis, with backward elimination of covariates. Predictors with low frequencies (ie, n <5 cases) in any cell were excluded from multivariate analyses. Multivariate analyses were performed using a generalized estimating equation to account for multiple admissions for some individuals.
The study was approved by the Cook County Health Institutional Review Board, which oversees local approval for enrollment of Cook County Jail detainees, and by the Rush University Medical Center institutional review boards. Verbal consent was obtained. Approval from the Office for Human Research Protections was obtained in order to enroll current detainees.
RESULTS
Features of the Study Population
We enrolled 718 individuals (800 incarcerations). Recidivism was high in the population, with 91% of individuals having previously been in jail, 21% in the prior 6 months. Sixty-one percent of individuals had been in prison in the past, 7% in the prior 6 months. Of individuals, 653 (91%) had 1, 54 (7.5%) had 2, 8 (1.1%) had 3, and 3 (0.3%) had 4–6 incarcerations in which the individual was enrolled during the study period.
Due to our sampling strategy, 58% of the enrollment population was living with HIV; 82% of individuals were African-American and 9% were Hispanic. The mean age of enrolled individuals was 37.6 years (standard deviation, 12.3). Ninety-four percent of individuals reported ever using illicit drugs and 46% self-reported as homeless, residing in a shelter, living in a substance abuse center, or “couch surfing.” Among PLHIV, 69% were on antiretroviral therapy and 10% reported no care.
Prevalence of CA-MRSA Colonization
A total of 155 individuals (19.4%) were colonized with MRSA at any body site assessed at jail entrance; 137 (19.1%) at their first enrollment. Nares (11.1%), throat (11%), and inguinal (11.4%) colonization were all common. Exclusive extranasal colonization was present in 66 individuals (42.6% of colonized and 8.3% of all individuals). Colonization occurred as follows: 81 (52.3%) at 1 body site, 35 (22.5%) at 2 body sites, and 39 (25.2%) at all 3 body sites. MRSA colonization was not more common among individuals who were enrolled once (19%) vs more than once (21.1%). There was no significant difference in the prevalence of colonization at intake by HIV status (PLHIV, 20.69%; people not living with HIV, 17.56%; P = .27).
Predictors of MRSA Colonization at Admission to the Jail
On univariate analysis (Table 1), significant predictors of MRSA colonization at admission to the jail were methamphetamine use, injection drug use in the past year, homeless or unstable housing, current skin infection, and self-report of skin or MRSA infection in the past year. Among PLHIV (Table 1), recent incarceration, current skin infection, and receiving outpatient HIV care at clinic A, a clinic that emphasizes comprehensive care to the lesbian, gay, bisexual, transgender, and questioning (LGBTQ) community, were significant predictors of MRSA colonization. Reporting HIV care at clinic B was negatively associated with MRSA risk; other reported clinics for HIV care were not significant. Among those not living with HIV (Table 1), self-report of skin or MRSA infection in the past year was a significant predictor of MRSA.
Table 1.
Univariate Analysis of Predictors of Methicillin-Resistant Staphylococcus aureus Colonization at Admission to the Jail at the First Enrollment Visit in the Total Study Population and by Human Immunodeficiency Virus Status
| Total Study Population | Individuals Living With HIV | Individuals Not Living With HIV | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Epidemiologic Factor | MRSA (n = 137) | No MRSA (n = 581) | OR | 95% CI | P Value | MRSA (n = 80) | No MRSA (n = 306) | OR | 95% CI | P Value | MRSA (n = 57) | No MRSA (n = 275) | OR | 95% CI | P Value |
| Race/Ethnicity | |||||||||||||||
| African-American | 113 (82.5) | 475 (81.8) | 0.9 | .5, 1.7 | .70 | 63 (78.8) | 243 (79.4) | 1.0 | .5, 2.1 | .97 | 50 (87.7) | 232 (84.4) | 0.8 | .2, 2.4 | .63 |
| Hispanic | 10 (7.3) | 54 (9.3) | 0.7 | .3, 1.7 | .41 | 7 (8.8) | 25 (8.2) | 1.1 | .4, 3.2 | .91 | 3 (5.3) | 29 (10.5) | 0.4 | .1, 1.8 | .22 |
| White/Other | 14 (10.2) | 52 (9.0) | 10 (12.5) | 38 (12.4) | 4 (7.0) | 14 (5.1) | |||||||||
| Methamphetamine use | 28 (20.4) | 69 (11.9) | 1.9 | 1.2, 3.1 | <.01 | 22 (27.5) | 58 (19.0) | 1.6 | .9, 2.9 | .09 | 6 (10.5) | 11 (4.0) | 2.8 | 1.0, 8.0 | .09 |
| Ecstasy or psychedelic use | 55 (40.1) | 212 (36.5) | 1.2 | .8, 1.7 | .43 | 27 (33.8) | 94 (30.7) | 1.1 | .7, 1.9 | .60 | 28 (49.1) | 118 (42.9) | 1.3 | .7, 2.3 | .39 |
| Illicit inhalant use | 7 (5.1) | 41 (7.1) | 0.7 | .3, 1.6 | .41 | 5 (6.3) | 30 (9.8) | 0.6 | .2, 1.6 | .32 | 2 (3.5) | 11 (4.0) | 0.9 | .2, 4.0 | 1.00 |
| Marijuana use | 121 (88.3) | 517 (89.0) | 0.9 | .5, 1.7 | .82 | 68 (85.0) | 258 (84.3) | 1.1 | .5, 2.1 | .88 | 53 (93.0) | 259 (94.2) | 0.8 | .3, 2.5 | .76 |
| Cocaine use | 70 (51.1) | 275 (47.3) | 1.2 | .8, 1.7 | .43 | 51 (63.8) | 177 (57.8) | 1.3 | .8, 2.1 | .34 | 19 (33.3) | 98 (35.6) | 0.9 | .5, 1.7 | .74 |
| Heroin use | 37 (27.0) | 144 (24.8) | 1.1 | .7, 1.7 | .59 | 28 (35.0) | 92 (30.1) | 1.3 | .7, 2.1 | .40 | 9 (15.8) | 52 (18.9) | 0.8 | .4, 1.7 | .58 |
| Other narcotics (eg, codeine, oxycontin) | 27 (19.7) | 126 (21.7) | 0.9 | .6, 1.4 | .61 | 8 (10.0) | 55 (18.0) | 0.5 | .2, 1.1 | .09 | 19 (33.3) | 71 (25.8) | 1.4 | .8, 2.7 | .25 |
| Illicit benzodiazepine use | 33 (24.1) | 137 (23.6) | 1.0 | .7, 1.6 | .90 | 13 (16.3) | 59 (19.3) | 0.8 | .4, 1.6 | .54 | 20 (35.1) | 78 (28.4) | 1.4 | .7, 2.5 | .31 |
| Taking prescription drugs to get high | 14 (10.2) | 68 (11.7) | 0.9 | .5, 1.6 | .62 | 7 (8.8) | 39 (12.7) | 0.7 | .3, 1.5 | .33 | 7 (12.3) | 29 (10.5) | 1.2 | .5, 2.9 | .7 |
| Injection drug use in the past year | 16 (11.7) | 33 (5.7) | 2.2 | 1.2, 4.1 | .01 | 14 (17.5) | 31 (10.1) | 1.9 | .9, 3.7 | .07 | 2 (3.5) | 2 (0.7) | 5 | .7, 36.0 | .14 |
| Men who have sex with men | 41 (29.9) | 130 (22.4) | 1.5 | 1.0, 2.2 | .06 | 39 (48.8) | 126 (41.2) | 1.4 | .8, 2.2 | .22 | 2 (3.5) | 4 (1.5) | 2.5 | .4, 13.8 | .27 |
| Previous incarceration in jail | 129 (94.2) | 525 (90.4) | 1.7 | .8, 3.7 | .16 | 73 (91.3) | 273 (89.2) | 1.3 | .5, 3.0 | .60 | 56 (98.2) | 252 (91.6) | 5.1 | .7, 38.6 | .09 |
| Release from jail in the prior 3 months | 21 (15.3) | 66 (11.4) | 1.4 | .8, 2.4 | .20 | 13 (16.3) | 26 (8.5) | 2.1 | 1.0, 4.3 | .04 | 8 (14.0) | 40 (14.5) | 1 | .4, 2.2 | .92 |
| Release from jail in the prior 6 months | 32 (23.4) | 121 (20.8) | 1.2 | .7, 1.8 | .52 | 17 (21.3) | 48 (15.7) | 1.5 | .8, 2.7 | .24 | 15 (26.3) | 73 (26.5) | 1 | .5, 1.9 | .97 |
| Previous incarceration in prison | 84 (61.3) | 354 (60.9) | 1.0 | .7, 1.5 | .93 | 44 (55.0) | 183 (59.8) | 0.8 | .5, 1.3 | .44 | 40 (70.2) | 171 (62.2) | 1.4 | .8, 2.7 | .25 |
| Release from prison in the prior 3 months | 4 (2.9) | 7 (1.2) | 2.5 | .7, 8.5 | .24 | 3 (3.8) | 5 (1.6) | 2.3 | .5, 10.0 | .37 | 1 (1.8) | 2 (0.7) | 2.4 | .2, 27.3 | .43 |
| Release from prison in the prior 6 months | 8 (5.8) | 42 (7.2) | 0.8 | .4, 1.7 | .57 | 5 (6.3) | 24 (7.8) | 0.8 | .3, 2.1 | .63 | 3 (5.3) | 18 (6.5) | 0.8 | .2, 2.8 | 1.00 |
| Homeless or unstable housing in the past year | 74 (54.0) | 254 (43.7) | 1.5 | 1.0, 2.2 | .03 | 47 (58.8) | 158 (51.6) | 1.3 | .8, 2.2 | .26 | 27 (47.4) | 96 (34.9) | 1.7 | .9, 3.0 | .08 |
| Current skin infection | 11 (8.0) | 22 (3.8) | 2.2 | 1.0, 4.7 | .03 | 10 (12.5) | 16 (5.2) | 2.6 | 1.1, 6.0 | .02 | 1 (1.8) | 6 (2.2) | 0.8 | .1, 6.8 | 1.00 |
| Emergency room visit in the past year | 73 (53.3) | 284 (48.9) | 1.2 | .8, 1.7 | .35 | 46 (57.5) | 163 (53.3) | 1.2 | .7, 2.0 | .50 | 27 (47.4) | 121 (44.0) | 1.1 | .6, 2.0 | .64 |
| Skin or MRSA infection in the past year | 14 (10.2) | 16 (2.8) | 4.0 | 1.9, 8.5 | <.01 | 8 (10.0) | 14 (4.6) | 2.3 | .9, 5.7 | .10 | 6 (10.5) | 2 (0.7) | 16.1 | 3.2, 81.8 | <.01 |
| Hospitalized in the past year | 39 (28.5) | 165 (28.4) | 1.0 | .7, 1.5 | .99 | 25 (31.3) | 103 (33.7) | 0.9 | .5, 1.5 | .68 | 14 (24.6) | 62 (22.5) | 1.1 | .6, 2.2 | .74 |
| Living with HIV | 80 (58.4) | 306 (52.7) | 1.3 | .9, 1.8 | .23 | … | … | … | … | … | … | … | … | … | … |
| No HIV care in the past year | … | … | … | … | … | 9 (11.3) | 31 (10.1) | 1.1 | .5, 2.5 | .77 | … | … | … | … | … |
| Care at clinic A | … | … | … | … | … | 14 (17.5) | 17 (5.6) | 3.6 | 1.7, 7.7 | <.01 | … | … | … | … | … |
| Care at clinic B | … | … | … | … | … | 1 (1.3) | 27 (8.8) | 0.1 | .0, 1.0 | .02 | … | … | … | … | … |
| Care at clinic C | … | … | … | … | … | 30 (37.5) | 125 (40.8) | 0.9 | .5, 1.4 | .59 | … | … | … | … | … |
| Care at clinic D | … | … | … | … | … | 2 (2.5) | 9 (2.9) | 0.8 | .2, 4.0 | .1 | … | … | … | … | … |
| Taking antiretrovirals | … | … | … | … | … | 50 (62.5) | 215 (70.5) | 0.7 | .4, 1.2 | .17 | … | … | … | … | … |
| Taking trimethoprim- sulfamethoxazole | … | … | … | … | … | 9 (11.3) | 30 (9.8) | 1.2 | .5, 2.6 | .70 | … | … | … | … | … |
| Taking other antibiotics in the prior 2 weeks | … | … | … | … | … | 12 (15.0) | 41 (13.4) | 1.1 | 0.6, 2.3 | .71 | … | … | … | … | … |
Values represent no (%). As individuals could be enrolled more than once in the study, results in this table represent epidemiologic factors from an individual’s first enrollment.
Abbreviations: CI, confidence interval; HIV, human immunodeficiency virus; OR, odds ratio; MRSA, methicillin-resistant Staphylococcus aureus.
In multivariable analysis (Table 2), with adjustment for HIV status and race/ethnicity, methamphetamine use (odds ratio [OR], 2.0; P = .01), having a skin or MRSA infection in the past year (OR, 3.5; P < .01), and being homeless or having unstable housing (OR, 1.5; P = .03) were significant factors associated with MRSA colonization. Among PLHIV, after controlling for race/ethnicity, injection drug use in the past year (OR, 2.0; P = .04), having a current skin infection (OR, 2.7; P = .01), and receiving care at clinic A (OR, 3.4; P < .01) were significant predictors of MRSA (Table 2). Among individuals not living with HIV (Table 2), after controlling for race/ethnicity, methamphetamine use was a risk factor for MRSA (OR, 4.1; P = .03).
Table 2.
Multivariate Analysis of Predictors of Methicillin-Resistant Staphylococcus aureus Colonization at Admission to the Jail in the Total Study Population and Among Individuals Living With Human Immunodeficiency Virus
| Predictor | Odds Ratio | 95% Confidence Interval | P Value |
|---|---|---|---|
| Total population (n = 800) | |||
| Race/Ethnicity | |||
| African-American | 1.4 | .7, 2.9 | .32 |
| Hispanic | 1.0 | .4, 2.6 | .98 |
| White/Other | … | … | … |
| Living with HIV | 1.0 | .7, 1.5 | .97 |
| Methamphetamine use | 2.0 | 1.2, 3.4 | .01 |
| Homeless or unstable housing | 1.5 | 1.0, 2.2 | .03 |
| Previous incarceration in jail | 2.0 | 1.0, 4.3 | .06 |
| Skin or methicillin-resistant Staphylococcus aureus infection in past year | 3.5 | 1.6, 7.5 | <.01 |
| Individuals living with HIV (n = 464) | |||
| Race/Ethnicity | |||
| African-American | 1.6 | .7, 3.4 | .27 |
| Hispanic | 1.3 | .4, 4.0 | .65 |
| White/Other | … | … | … |
| Injection drug use in the past year | 2.0 | 1.0, 3.7 | .04 |
| Receives HIV care at clinic A | 3.4 | 1.6, 7.5 | <.01 |
| Current skin infection | 2.7 | 1.2, 6.1 | .01 |
| Individuals not living with HIV (n = 336) | |||
| Race/Ethnicity | |||
| African-American | 1.7 | .4, 7.2 | .47 |
| Hispanic | 0.8 | .1, 5.0 | .80 |
| White/Other | … | … | … |
| Methamphetamine use | 4.1 | 1.2, 14.3 | .03 |
The total study population was used in multivariate analysis with a generalized estimating equation used to account for multiple admissions for some individuals.
Abbreviation: HIV, human immunodeficiency virus.
Whole-genome Sequencing
There were 265 MRSA isolates from 145 individuals (Supplementary Figure 1). We used whole-genome phylogeny and an SNV cutoff to group each individual’s isolates into those plausibly linked back to a single acquisition event (see Supplementary Methods). Overall, 150 MRSA acquisition events were inferred for the 145 individuals, with the remaining intraindividual genetic variation hypothesized to reflect intrahost evolution (Supplementary Figures 2 and 3). Supporting our grouping of individual isolates, we observed a median core SNV difference of 3 and a mean of 5.4 (max = 38 SNVs), which is consistent with the magnitude of intrahost variation observed for MRSA in previous studies [29]. Among those individual isolates deemed to be the result of separate acquisition events (5 individuals), the median core SNV distance was 517 and the mean was 5676 (max = 13 610). Of the 150 independently acquired MRSA isolates, 101 were closely related to USA300 (median core SNV distance = 119), 13 to USA500 (median core SNV distance = 124.5), 25 were clustered with ST5 isolates (median core SNV distance = 420), and 10 other isolates were not clustered with common sequence types (Supplementary Figure 4). Among the 8 individuals who were enrolled in the study more than once and colonized with MRSA on multiple admissions, phylogenetic analysis demonstrated that 4 individuals had highly similar strains at each incarceration across body sites, while there were 4 instances where genomics demonstrated colonization with 2 distinct strains (mean time between enrollments, 154 days; Supplementary Figure 5).
Genomic Epidemiology of MRSA Isolates from Clinic A
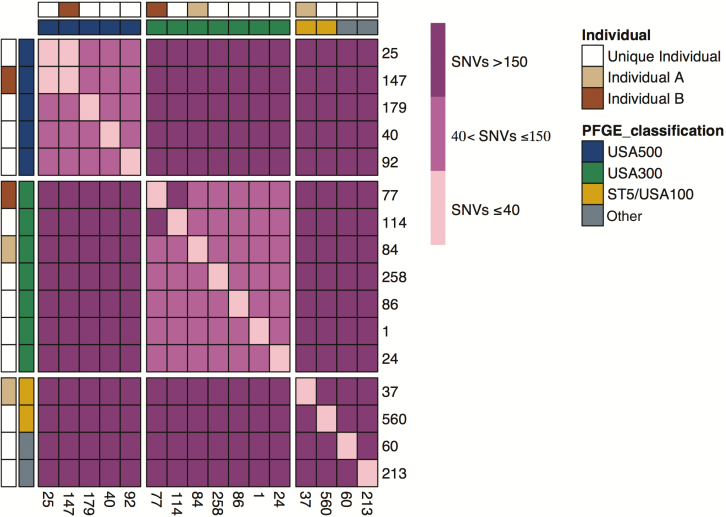
Among the 31 individuals who reported receiving HIV care at clinic A, 14 (45%) had MRSA colonization. We used whole-genome sequencing to determine if the high prevalence of MRSA at clinic A was due to dissemination of a single strain at that clinic or if individuals carried genetically unrelated strains. All major strain lineages were present among the 14 isolates (eg, USA300, USA500, USA100/ST5, and others). Moreover, even among strains of the same sequence type, there was extensive underlying variation with a median pairwise distance of 515 SNVs and only 1 pair of isolates within 40 SNVs of each other (Figure 1). These results demonstrate that the high prevalence of MRSA among individuals reporting receipt of care at clinic A was not due to clonal spread but instead due to an intermingling of distinct community MRSA transmission networks.
Figure 1.
Genomic epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) isolates from clinic A. The number of SNVs between all pairs of MRSA isolates from MRSA-positive individuals who received care at clinic A are displayed as a heat map. The rows and columns show the 16 MRSA isolates from the 14 individuals who received care at clinic A. Column and row annotations indicate the strain of the isolate (USA300, USA500, or ST5) and whether an isolate was from 1 of the 2 individuals colonized with multiple strains (individuals A and B). Abbreviations: PFGE, pulsed-field gel electrophoresis; SNV, single nucleotide variant.
Genomic Epidemiology of MRSA Strains Isolated at Entrance to the Jail
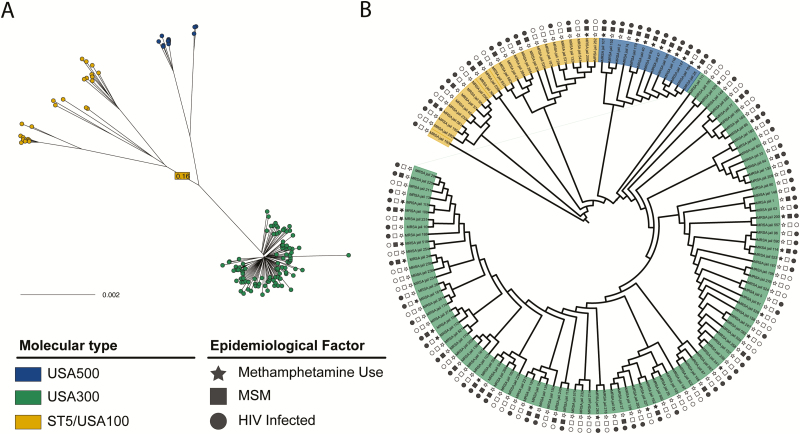
To determine if different strains of MRSA spread via distinct social or healthcare networks, we tested for an association between individual exposures and strain type (USA300, USA500, USA100/ST5; Figure 2). This analysis revealed a distinct epidemiologic profile for individuals harboring USA500 strains (Table 3). Relative to other intake-positive individuals, those colonized with USA500 were more likely to be living with HIV (P < .01), MSM (P < .001), and methamphetamine users (P < .001). There was no association of recent hospitalization with USA500 carriage. Five (38%) individuals colonized with USA500 reported receiving care at clinic A, suggesting that the social networks underlying spread of USA500 overlap with those seeking care at this clinic. Placing USA500 strains in this study in the context of previously sequenced USA500 isolates [30] (Supplementary Figure 6), we observed that our strains belonged to the C1 and C2 USA500 sublineages, with C1 being noted for its association with multidrug resistance [30].
Figure 2.
Genomic epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) isolates collected at intake to the jail. A, An unrooted maximum likelihood phylogenetic tree showing isolates from all study individuals colonized with MRSA on admission to Cook County Jail. Tips are colored according to inferred strain type (USA300, USA500, and ST5). The branch leading to ST5 is truncated for display purposes, with the length of the branch overlaid on the truncated branch. B, A rooted representation of the tree from panel A is shown, with branch lengths ignored for visualization purposes. Clades corresponding to the 3 strain types are indicated by color, and the shapes in the outer rings indicate the epidemiologic characteristics of MRSA-colonized individuals. Filled circles, squares, and stars indicate that the corresponding individual is living with HIV, reports being a MSM, or has a history of methamphetamine use, respectively. Abbreviations: HIV, human immunodeficiency virus; MSM, man who has sex with men.
Table 3.
Epidemiologic Factors Associated With Colonization With USA500 Methicillin-Resistant Staphylococcus aureus
| Epidemiologic Factora | Number (%) of Individuals Colonized With USA500 (n = 13)b | Number (%) of Individuals Colonized With USA300 or ST5 (n = 126) | Odds Ratio | 95% Confidence Interval | P Value |
|---|---|---|---|---|---|
| White (vs non-Hispanic white and black) | 4 (30.8%) | 10 (7.9%) | 5.2 | 1.3, 19.8 | .03 |
| Hispanic (vs non-Hispanic white and black) | 1 (7.7%) | 8 (6.3%) | 1.2 | .1, 10.7 | .6 |
| Black (vs white and non-Hispanic white) | 8 (61.5%) | 108 (85.7%) | 0.3 | .1, .9 | .04 |
| Methamphetamine use | 10 (76.9%) | 24 (19%) | 14.2 | 3.6, 55.5 | <.001 |
| Ecstasy or psychedelic use | 7 (53.8%) | 56 (44.4%) | 1.5 | .5, 4.6 | .57 |
| Illicit inhalant use | 2 (15.4%) | 9 (7.1%) | 2.4 | .5, 12.3 | .27 |
| Marijuana use | 12 (92.3%) | 111 (88.1%) | 1.6 | .2, 13.4 | 1 |
| Cocaine use | 7 (53.8%) | 70 (55.6%) | 1 | .3, 2.9 | 1 |
| Heroin use | 0 | 43 (34.1%) | … | … | .01 |
| Other narcotics (eg, codeine, oxycontin) | 2 (15.4%) | 28 (22.2%) | 0.6 | .1, 3 | .73 |
| Illicit benzodiazepine use | 3 (23.1%) | 32 (25.4%) | 0.9 | .2, 3.4 | 1 |
| Taking prescription drugs to get high | 1 (7.7%) | 16 (12.7%) | 0.6 | .1, 4.7 | 1 |
| Injection drug use in the past year | 5 (38.5%) | 17 (13.5%) | 4 | 1.2, 13.7 | .03 |
| Men who have sex with men | 12 (92.3%) | 33 (26.2%) | 33.8 | 4.2, 270 | <.001 |
| Living with human immunodeficiency virus | 13 (100%) | 77 (61.1%) | … | … | <.01 |
| Previous incarceration in jail | 11 (84.6%) | 120 (95.2%) | 0.3 | .05, 1.5 | .16 |
| Release from jail in the prior 3 months | 0 | 23 (18.3%) | … | … | .13 |
| Release from jail in the prior 6 months | 2 (15.4%) | 37 (29.4%) | 0.4 | .1, 2.1 | .35 |
| Previous incarceration in prison | 0 | 85 (67.5%) | … | … | <.001 |
| Homeless or unstable housing in the past year | 6 (46.2%) | 72 (57.1%) | 0.6 | .2, 2 | .56 |
| Current skin infection | 0 | 12 (9.5%) | … | … | .6 |
| Emergency room visit in the past year | 8 (61.5%) | 71 (56.3%) | 1.2 | .4, 4 | .78 |
| Skin or methicillin-resistant Staphylococcus aureus infection in the past year | 2 (15.4%) | 14 (11.1%) | 1.5 | .3, 7.2 | .65 |
| Hospitalized in the past year | 4 (30.8%) | 38 (30.2%) | 1 | .3, 3.5 | 1 |
Eleven individuals had methicillin-resistant Staphylococcus aureus strains that were not categorized as USA500, USA300, or ST5. Of the 126 non-USA500 isolates, 101 were USA300 and 25 were ST5. All 13 individuals colonized with USA500 strains were living with human immunodeficiency virus; therefore no odds ratio could be calculated. No individuals colonized with USA500 strains had the following epidemiologic factors: heroin use, release from jail in the prior 3 months, previous incarceration in prison, or current skin infection and therefore no odds ratio could be calculated.
aAll 13 individuals colonized with USA500 strains were living with human immunodeficiency virus; therefore no odds ratio could be calculated. No individuals colonized with USA500 strains had the following epidemiologic factors: heroin use, release from jail in the prior 3 months, previous incarceration in prison, or current skin infection.
bGiven our phylogenetic analysis, we wanted to further investigate predictors of USA500 and therefore conducted a 2 × 2 analysis of this category compared to the other 2 categories combined. Results of the 2 × 3 analyses are included in Supplementary Table 2.
DISCUSSION
A large proportion of males who entered jail were colonized with MRSA. Entrance risk factors for colonization suggest potential community reservoirs for MRSA. We observed a 45% colonization rate of MRSA among PLHIV who receive care at a clinic that emphasizes comprehensive services for the LGBTQ community, and 38% of patients colonized with USA500 received care at that clinic. Using phylogenetic analysis, we observed that USA500 MRSA strains appear to be associated with distinct social networks. Of note, individuals who harbor USA500 strains were more likely to be living with HIV, MSM, and methamphetamine users.
The intake prevalence of 19% observed in our study is dramatically higher than in the general population of nares MRSA carriage (1.5%) [31] and other nonincarcerated high-risk groups (6%–16%) [9, 21]. Prior estimates of colonization prevalence among detainees have been done in prisons [12, 32] and jails [33]. A study in Baltimore reported a 15.8% prevalence of nasal MRSA carriage at central booking among arrestees [34]. Our study extends this work by including extranasal sites and focusing on the prevalence of colonization upon entrance to a large inner-city jail. Establishing this intake prevalence is critical for understanding the high burden of MRSA that has been observed during incarceration. For example, Kajita et al [35] developed a mathematical model to estimate transmission rates and intervention effects during a large MRSA outbreak at the Los Angeles County Jail. Our calculated intake prevalence may be useful for updating this and similar models, especially to derive estimates that accurately reflect the contribution of intake MRSA colonization. In addition, a prior model predicted that increased contacts per day during incarceration are associated with increased release of individuals infected with MRSA into the community [36]. Updating that model with our estimated entrance colonization rate may help delineate the downstream effect of preincarceration colonization on risk for MRSA in the surrounding community.
Given our prior observations [9, 17, 21], we chose a priori to enrich our sample for PLHIV. We did not identify a difference in the prevalence for MRSA colonization at intake according to HIV status, suggesting that factors other than from HIV status may account for the elevated MRSA risk previously observed. In prior work by our group, we similarly observed that when one controlled for community exposures and incarceration, HIV status was no longer associated with MRSA colonization [21]. In addition, taking antiretrovirals or being on trimethoprim-sulfamethoxazole prophylaxis did not protect against MRSA colonization, suggesting that factors other than immune status [37] may be relevant. Our current study supports prior speculation that high-risk social networks outside of healthcare settings or correctional facilities may be important for MRSA risk and could be a focus of intervention.
We observed that methamphetamine use, unstable housing, and a history of skin or MRSA infection were risk factors for MRSA colonization upon entrance to the jail. Others have observed methamphetamine use to be a significant risk factor for MRSA infection, particularly among MSM living with HIV [38] and that methamphetamine use is associated with high-risk sexual behaviors [39], such as meeting sexual partners via the internet, bathhouses, or sex parties, all of which have also been associated with MRSA risk [38]. We speculate that the association of methamphetamine use with MRSA observed in our study may relate to high-risk sexual activity within distinct social networks. Among PLHIV, recent injection drug use was significantly associated with MRSA risk. The impact of the current opioid epidemic on rates of MRSA infection among at-risk patient populations warrants close monitoring and may need to become a basis for intervention. Finally, others have similarly noted that current skin infection or history of skin or MRSA infection in the prior year can be a significant risk factor for MRSA colonization [21, 40]. Such a finding could potentially be used in an intervention strategy, for example, to identify those at highest risk upon entrance to the jail.
We observed that receiving HIV care at clinic A was a significant predictor of MRSA colonization. By using phylogenetic analysis, we were able to extend traditional epidemiologic analysis to demonstrate that the high prevalence of MRSA among detainees with care at clinic A (45%) was not due to exposure to the clinic itself but more likely to high-risk individuals from different social networks seeking care at that clinic.
Most prior work for CA-MRSA has identified USA300 as the most common community strain [4]. While the majority of sequenced strains in this study were USA300, USA500 strains were isolated from 13 individuals who had distinct epidemiologic characteristics. All USA500 carriers were MSM living with HIV and 77% used methamphetamines, suggesting distinct social networks in the MSM community. Prior work in MSM has highlighted a high burden of MRSA infection [6]; our work extends this by utilizing genomics to highlight possible networks within the larger MSM community. As we examined only incarcerated MSM, further work is needed to determine the extent of MRSA carriage in the larger MSM community, particularly in populations where close contacts in tight social networks have previously been shown to facilitate spread of sexually transmitted diseases [41, 42].
Our study has limitations. We only examined a single large urban jail and did not factor in history of MRSA infections into our enrollment. However, we believe our results would be generalizable to other large urban jails in the United States and potentially other congregate settings. We focused our study on the prevalence and risk factors for MRSA; additional evaluation of methicillin-susceptible S. aureus would be of use. Recidivism was common in this study. Future work with mathematical modeling may tease out the impact of incarceration and recidivism on the downstream spread of MRSA in the community.
The prevalence of MRSA colonization entering a large inner-city jail is high. Whole-genome sequencing and phylogenic analysis extended traditional epidemiologic analysis and revealed possible community transmission networks for MRSA among males before incarceration. Genomics demonstrated that the high burden of MRSA among patients seeking care at clinic A is due to an intermixing of at-risk individuals in different MRSA transmission networks. Our study highlights preadmission colonization risk factors and community transmission networks that could be a potential focus for infection prevention interventions.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank Emma Richardson (Rush University Medical Center) and Jon Zelner (University of Michigan) for providing content expertise for the project and the Collaborative Research Unit at Cook County Health for their assistance with data collection.
Financial support. The project described was supported by the National Institute of Allergy and Infectious Diseases (grant R01AI114688; Principal Investigator: K. J. P.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Potential conflicts of interest. M. K. H. has served as a coinvestigator on projects for Sage, Molnlycke, OpGen, and Clorox. R. A. W. reports participation in clinical studies where participating hospitals or nursing homes received contributed product from Sage Products Inc, Molnlycke, or Medline. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Fridkin SK, Hageman JC, Morrison M, et al. ; Active Bacterial Core Surveillance Program of the Emerging Infections Program Network Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med 2005; 352:1436–44. [DOI] [PubMed] [Google Scholar]
- 2. Herold BC, Immergluck LC, Maranan MC, et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 1998; 279:593–8. [DOI] [PubMed] [Google Scholar]
- 3. Popovich KJ, Weinstein RA, Hota B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin Infect Dis 2008; 46:787–94. [DOI] [PubMed] [Google Scholar]
- 4. Tenover FC, McDougal LK, Goering RV, et al. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol 2006; 44:108–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jackson KA BM, Brooks JT, et al. Invasive methicillin-resistant Staphylococcus aureus infections among persons who inject drugs—six sites, 2005–2016. Morb Mortal Wkly Rep 2018; 67:625–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Diep BA, Chambers HF, Graber CJ, et al. Emergence of multidrug-resistant, community-associated, methicillin-resistant Staphylococcus aureus clone USA300 in men who have sex with men. Ann Intern Med 2008; 148:249–57. [DOI] [PubMed] [Google Scholar]
- 7. Ellis MW, Hospenthal DR, Dooley DP, Gray PJ, Murray CK. Natural history of community-acquired methicillin-resistant Staphylococcus aureus colonization and infection in soldiers. Clin Infect Dis 2004; 39:971–9. [DOI] [PubMed] [Google Scholar]
- 8. Kazakova SV, Hageman JC, Matava M, et al. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N Engl J Med 2005; 352:468–75. [DOI] [PubMed] [Google Scholar]
- 9. Popovich KJ, Hota B, Aroutcheva A, et al. Community-associated methicillin-resistant Staphylococcus aureus colonization burden in HIV-infected patients. Clin Infect Dis 2013; 56:1067–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Landers TF, Harris RE, Wittum TE, Stevenson KB. Colonization with Staphylococcus aureus and methicillin-resistant S. aureus among a sample of homeless individuals, Ohio. Infect Control Hosp Epidemiol 2009; 30:801–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Malcolm B. The rise of methicillin-resistant Staphylococcus aureus in U.S. correctional populations. J Correct Health Care 2011; 17:254–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lowy FD, Aiello AE, Bhat M, et al. Staphylococcus aureus colonization and infection in New York State prisons. J Infect Dis 2007; 196:911–8. [DOI] [PubMed] [Google Scholar]
- 13. Peters PJ, Brooks JT, McAllister SK, et al. Methicillin-resistant Staphylococcus aureus colonization of the groin and risk for clinical infection among HIV-infected adults. Emerg Infect Dis 2013; 19:623–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. CDC. Methicillin-resistant Staphylococcus aureus infections in correctional facilities—Georgia, California, and Texas, 2001–2003. MMWR Morb Mortal Wkly Rep 2003; 52:992–6. [PubMed] [Google Scholar]
- 15. Pan ES, Diep BA, Carleton HA, et al. Increasing prevalence of methicillin-resistant Staphylococcus aureus infection in California jails. Clin Infect Dis 2003; 37:1384–8. [DOI] [PubMed] [Google Scholar]
- 16. David MZ, Mennella C, Mansour M, Boyle-Vavra S, Daum RS. Predominance of methicillin-resistant Staphylococcus aureus among pathogens causing skin and soft tissue infections in a large urban jail: risk factors and recurrence rates. J Clin Microbiol 2008; 46:3222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Popovich KJ, Weinstein RA, Aroutcheva A, Rice T, Hota B. Community-associated methicillin-resistant Staphylococcus aureus and HIV: intersecting epidemics. Clin Infect Dis 2010; 50:979–87. [DOI] [PubMed] [Google Scholar]
- 18. Hota B, Ellenbogen C, Hayden MK, Aroutcheva A, Rice TW, Weinstein RA. Community-associated methicillin-resistant Staphylococcus aureus skin and soft tissue infections at a public hospital: do public housing and incarceration amplify transmission? Arch Intern Med 2007; 167:1026–33. [DOI] [PubMed] [Google Scholar]
- 19. Rhee Y, Aroutcheva A, Hota B, Weinstein RA, Popovich KJ. Evolving epidemiology of Staphylococcus aureus bacteremia. Infect Control Hosp Epidemiol 2015; 36:1417–22. [DOI] [PubMed] [Google Scholar]
- 20. Charlebois ED, Bangsberg DR, Moss NJ, et al. Population-based community prevalence of methicillin-resistant Staphylococcus aureus in the urban poor of San Francisco. Clin Infect Dis 2002; 34:425–33. [DOI] [PubMed] [Google Scholar]
- 21. Popovich KJ, Smith KY, Khawcharoenporn T, et al. Community-associated methicillin-resistant Staphylococcus aureus colonization in high-risk groups of HIV-infected patients. Clin Infect Dis 2012; 54:1296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spaulding AC, Seals RM, Page MJ, Brzozowski AK, Rhodes W, Hammett TM. HIV/AIDS among inmates of and releasees from US correctional facilities, 2006: declining share of epidemic but persistent public health opportunity. PLoS One 2009; 4:e7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saegeman V, Flamaing J, Muller J, Peetermans WE, Stuyck J, Verhaegen J. Clinical evaluation of the Copan ESwab for methicillin-resistant Staphylococcus aureus detection and culture of wounds. Eur J Clin Microbiol Infect Dis 2011; 30:943–9. [DOI] [PubMed] [Google Scholar]
- 24. Miller LG, Diep BA. Clinical practice: colonization, fomites, and virulence: rethinking the pathogenesis of community-associated methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis 2008; 46:752–60. [DOI] [PubMed] [Google Scholar]
- 25. Safdar N, Narans L, Gordon B, Maki DG. Comparison of culture screening methods for detection of nasal carriage of methicillin resistant Staphylococcus aureus: a prospective study comparing 32 methods. J Clin Microbiol. 2003; 41:3163–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andrews S. FastQC: a quality control tool for high throughput sequence data. Available at: www.bioinformatics.babraham.ac.uk/projects/fastqc. [Google Scholar]
- 27. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015; 32:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Golubchik T, Batty EM, Miller RR, et al. Within-host evolution of Staphylococcus aureus during asymptomatic carriage. PLoS One 2013; 8:e61319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frisch MB, Castillo-Ramirez S, Petit RA 3rd, et al. Invasive methicillin-resistant Staphylococcus aureus USA500 strains from the U.S. Emerging Infections Program constitute three geographically distinct lineages. 2018; 3: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gorwitz RJ, Kruszon-Moran D, McAllister SK, et al. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. J Infect Dis 2008; 197:1226–34. [DOI] [PubMed] [Google Scholar]
- 32. Mukherjee DV, Herzig CT, Jeon CY, et al. Prevalence and risk factors for Staphylococcus aureus colonization in individuals entering maximum-security prisons. Epidemiol Infect 2014; 142:484–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maree CL, Eells SJ, Tan J, et al. Risk factors for infection and colonization with community-associated methicillin-resistant Staphylococcus aureus in the Los Angeles County Jail: a case-control study. Clin Infect Dis 2010; 51:1248–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Farley JE, Ross T, Stamper P, Baucom S, Larson E, Carroll KC. Prevalence, risk factors, and molecular epidemiology of methicillin-resistant Staphylococcus aureus among newly arrested men in Baltimore, Maryland. Am J Infect Control 2008; 36:644–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kajita E, Okano JT, Bodine EN, Layne SP, Blower S. Modelling an outbreak of an emerging pathogen. Nat Rev Microbiol 2007; 5:700–9. [DOI] [PubMed] [Google Scholar]
- 36. Okano JT, Blower S. Are correctional facilities amplifying the epidemic of community-acquired methicillin-resistant Staphylococcus aureus? Nat Rev Microbiol 2010; 8:83. [DOI] [PubMed] [Google Scholar]
- 37. Shet A, Mathema B, Mediavilla JR, et al. Colonization and subsequent skin and soft tissue infection due to methicillin-resistant Staphylococcus aureus in a cohort of otherwise healthy adults infected with HIV type 1. J Infect Dis 2009; 200:88–93. [DOI] [PubMed] [Google Scholar]
- 38. Lee NE, Taylor MM, Bancroft E, et al. Risk factors for community-associated methicillin-resistant Staphylococcus aureus skin infections among HIV-positive men who have sex with men. Clin Infect Dis 2005; 40:1529–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Urbina A, Jones K. Crystal methamphetamine, its analogues, and HIV infection: medical and psychiatric aspects of a new epidemic. Clin Infect Dis 2004; 38:890–4. [DOI] [PubMed] [Google Scholar]
- 40. Hidron AI, Kourbatova EV, Halvosa JS, et al. Risk factors for colonization with methicillin-resistant Staphylococcus aureus (MRSA) in patients admitted to an urban hospital: emergence of community-associated MRSA nasal carriage. Clin Infect Dis 2005; 41:159–66. [DOI] [PubMed] [Google Scholar]
- 41. Morgan E, Skaathun B, Nikolopoulos GK, et al. A network intervention to locate newly HIV infected persons within MSM networks in Chicago. AIDS Behav 2019; 23:15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fujimoto K, Flash CA, Kuhns LM, Kim JY, Schneider JA. Social networks as drivers of syphilis and HIV infection among young men who have sex with men. Sex Transm Infect 2018; 94:365–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.