Significance
Mammalian cells contain two RNA polymerase III isoforms that differ only in ubiquitous POLR3GL and developmentally regulated POLR3G subunits. Here, in contradiction to previous conclusions from POLR3G knockdown analyses, we show that POLR3G and POLR3GL are functionally redundant and, in the context of embryonic stem cell differentiation, can largely compensate for each other when expressed at appropriate levels. Moreover, whereas Polr3g knockout mice die at an early embryonic stage, Polr3gl knockout mice complete embryonic development but die at weaning with signs of both general growth defects and potential cerebellum-related neuronal defects. As interests grow in reported Pol III-related disorders, Polr3gl knockout mice provide a convenient model to study the physiological effect of variations in Pol III functions in more detail.
Keywords: RNA polymerase III, POLR3G, POLR3GL, ESC, development
Abstract
Mammalian cells contain two isoforms of RNA polymerase III (Pol III) that differ in only a single subunit, with POLR3G in one form (Pol IIIα) and the related POLR3GL in the other form (Pol IIIβ). Previous research indicates that POLR3G and POLR3GL are differentially expressed, with POLR3G expression being highly enriched in embryonic stem cells (ESCs) and tumor cells relative to the ubiquitously expressed POLR3GL. To date, the functional differences between these two subunits remain largely unexplored, especially in vivo. Here, we show that POLR3G and POLR3GL containing Pol III complexes bind the same target genes and assume the same functions both in vitro and in vivo and, to a significant degree, can compensate for each other in vivo. Notably, an observed defect in the differentiation ability of POLR3G knockout ESCs can be rescued by exogenous expression of POLR3GL. Moreover, whereas POLR3G knockout mice die at a very early embryonic stage, POLR3GL knockout mice complete embryonic development without noticeable defects but die at about 3 wk after birth with signs of both general growth defects and potential cerebellum-related neuronal defects. The different phenotypes of the knockout mice likely reflect differential expression levels of POLR3G and POLR3GL across developmental stages and between tissues and insufficient amounts of total Pol III in vivo.
Transcription of eukaryotic genomes is typically mediated by three essential RNA polymerases (Pols I, II, and III) and two plant-specific, specialized forms of Pol II (Pol IV and V). Among these, Pol III transcribes genes specifying short untranslated transcripts that include 1) cellular RNase P, RNase mitochondrial RNA processing (MRP), transfer RNAs (tRNAs), 5S RNA, 7SL RNA, 7SK RNA, U6 RNA, Vault RNAs, and Y RNAs and 2) viral VA-I, VA-II, and EBER RNAs (1). Pol III also transcribes short interspersed nuclear elements (SINES), including ALU genes that number over a million in humans (2). More recent studies have identified Pol III functions in the synthesis of other small noncoding regulatory RNAs and in the transcription of genomic loci that are positioned close to Pol II-transcribed loci and potentially involved in their regulation (3–9).
Pol III is the largest RNA polymerase with 17 subunits (1), 5 of which are specific to Pol III (10). Three of the five Pol III-specific subunits (POLR3C/RPC62, POLR3F/RPC39, and POLR3G/RPC32) form a stable subcomplex with a selective and critical function in transcription initiation (11). Mammalian cells actually contain two Pol III isoforms that differ in only a single subunit (12, 13), with POLR3G in one form (Pol IIIα) and the related POLR3GL in the other form (Pol IIIβ) (14). These paralogues present some differences in their interactions with POLR3C, suggesting possible differences in the functions of the two Pol III isoforms (15). Interestingly, POLR3G and POLR3GL are differentially expressed, with POLR3G expression being more highly expressed in undifferentiated embryonic stem cells (ESCs) and tumor cells relative to the broad expression of POLR3GL (14). However, POLR3G is also expressed in normal mouse liver and in untransformed human fibroblasts (16), suggesting that this subunit might not be strictly restricted to embryonic stem (ES) or cancer cells. A genome-wide analysis identified POLR3G as one of the most down-regulated genes during human ESC differentiation (17). In a related study, chromatin immunoprecipitation (ChIP) assays and small interfering RNA (siRNA)-mediated knockdown of NANOG or OCT4 in human ESCs indicated that POLR3G is a downstream target of these factors and is regulated by ERK1/2 signaling (18). Similarly, the oncoprotein c-MYC is found exclusively on the POLR3G promoter but not at the POLR3GL locus in human P493-6 Burkitt’s lymphoma cells (19). Altogether, these results indicate that POLR3G and POLR3GL expression levels are controlled by different mechanisms.
Despite the fact that both Pol III isoforms display different expression levels in cells and are subject to specific regulation, they have been reported to occupy largely the same loci in mouse liver and Hepa 1-6 cells (16). While these results suggested that POLR3G and POLR3GL are not involved in target gene selection, and that both Pol III isoforms transcribe the same genes, the lack of corresponding functional studies left open this question. Here, we show that both POLR3G and POLR3GL can be incorporated into Pol III complexes when overexpressed in mouse ESCs, with limited differences in the composition of the two types of Pol III complexes. Genome-wide ChIP-seq experiments further confirmed that both POLR3G- and POLR3GL-containing Pol III complexes bind the same targets in mouse embryonic fibroblasts (MEFs). We further generated individual Polr3g and Polr3gl knockout (KO) mice. As anticipated, Polr3g KO mice died early in development, between embryonic day (E3.5) and E6.5. Unexpectedly, Polr3gl KO mice were not only able to proceed through embryonic development but also survived up to weaning. More importantly, for each isoform the observed KO phenotype was accentuated by genetic reduction (in heterozygous mice) of the paralogous subunit, suggesting that these isoforms play redundant functions in vivo. Moreover, the differentiation defects of Polr3g knockout ESCs could be fully rescued by the exogenous expression of POLR3GL. Altogether, these experiments support a model in which POLR3G and POLR3GL indeed fulfill the same functions in ESCs and during mouse development. Hence, the phenotypic differences observed in mice lacking one or the other isoform likely result from the differential regulation of their expression during development and across tissues.
Results
Epitope-Tagged POLR3G and POLR3GL Are both Efficiently Incorporated into Pol III Complexes of Comparable Compositions in Mouse Embryonic Stem Cells.
The surprising enrichment and apparent specific functions fulfilled by POLR3G in ESCs (17, 18), combined with similar genomic occupancies of the two Pol III isoforms in liver cells (16), prompted us to evaluate whether POLR3G and its close paralogue POLR3GL exist in Pol III complexes with different compositions in ESCs. To this end, we established stable mouse E14TG2a cell lines expressing either FLAG-AviTag–tagged POLR3G or POLR3GL using a lentiviral-based delivery method. First, we found that neither POLR3G nor POLR3GL overexpression altered stem cell morphology or messenger RNA (mRNA) levels of the pluripotency marker Pou5f1 (Oct4) (SI Appendix, Fig. S1), which indicates that these cells are still in their pluripotent states. Furthermore, the two types of Pol III complexes, isolated by affinity purification from corresponding whole-cell extracts, displayed similar compositions as revealed by the band patterns obtained in Coomassie-stained gels (Fig. 1A). These results are in agreement with analyses of the natural Pol III isoforms purified from mouse plasmacytoma cells (13) and the POLR3G- and POLR3GL-tagged Pol III isoforms affinity-purified from HeLa cells (14). We further confirmed by mass spectrometry (MS) that both ESC preparations contain the same set of well-characterized Pol III subunits, with 15 of the 17 Pol III subunits detected in both complexes and the other 2, POLR2L (7.6 kDa) and POLR2K (7.0 kDa), missing due to their small sizes. Importantly, only a few additional proteins were identified in either preparation SI Appendix, Table S1) and most of these proteins, such as GAPDH and PPIA, are related to metabolic functions, expressed at very high levels and probable contaminants based on considerations of common contaminants identified in other mass spectrometric analyses (20). Other differences relate to proteins displaying low MS scores and coverage, suggesting that they are not stochiometric partners of the complexes. Therefore, and especially with respect to near-stoichiometric components, both ESC-derived Pol III isoforms most likely differ only with respect to the presence of POLR3G or POLR3GL. Furthermore, although we cannot formally exclude the possibility that some of the proteins differentially associated with the primary (17-subunit) POLR3G- and POLR3GL-containing Pol III complexes do not contribute to differential function, this is less likely based on results (below) indicating common target genes and functions.
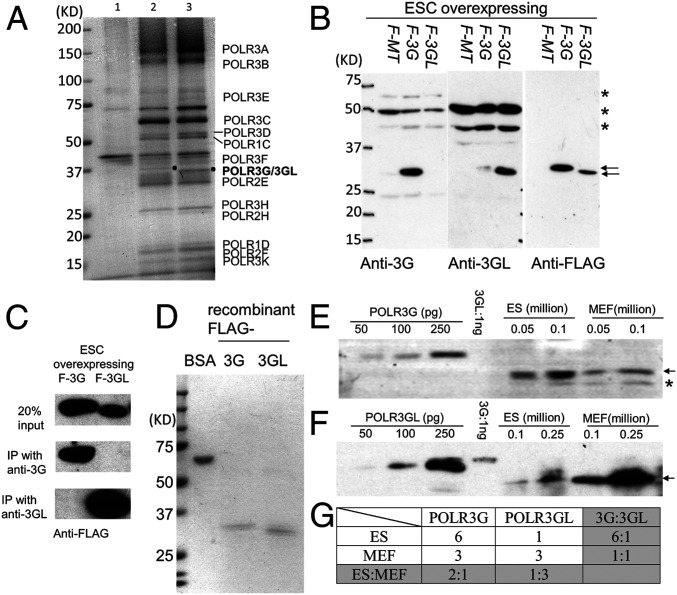
Fig. 1.
POLR3G and POLR3GL expression in mouse ESCs and MEFs. (A) Coomassie-stained gels showing minor differences in polypeptide composition of Pol III complexes immunoprecipitated (with anti-FLAG antibody and streptavidin beads) from the lysates of stable ESC lines harboring FLAG-AviTag-empty vector (lane 1), FLAG-AviTag-POLR3G (lane 2), or FLAG-AviTag-POLR3GL (lane 3). (B) Immunoblots of cell lysates from ESCs transduced with FLAG-AviTag-empty vector (F-MT), FLAG-AviTag-POLR3G (F-3G), or FLAG-AviTag-POLR3GL (F-3GL) showing the specificities of the raised antibodies. Immunoblots with anti-FLAG antibody revealed the relative amounts of overexpressed POLR3G and POLR3GL proteins in whole-cell lysates. Arrows denote the overexpressed proteins and asterisks indicate nonspecific bands. (C) Immunoprecipitation assay showing the specificities of anti-POLR3G and anti-POLR3GL antibodies for cognate Pol III subunits in cell lysates of ESCs. (D) Coomassie-stained gels showing the purities of recombinant FLAG-tagged POLR3G and POLR3GL proteins that were used as standards to determine the amounts of endogenous POLR3G and POLR3GL proteins in ESCs and MEFs. (E and F) Immunoblots showing the relative levels, to recombinant proteins, of POLR3G (E) or POLR3GL (F) in whole-cell lysates of ESCs and primary MEFs. The recombinant protein amounts (in pg) and the cell-number equivalents (1 million) of whole-cell lysate loaded in each lane are indicated. Based on the recombinant protein standards, the calculated values for total Pol III molecules per cell are ∼5 × 104 for ESCs and ∼4 × 104 for MEFs. Arrows indicate endogenous POLR3G and POLR3GL proteins, and the asterisk indicates a nonspecific band. (G) Calculated relative ratios of POLR3G and POLR3GL proteins in ESCs and primary MEFs.
Evaluation of POLR3G and POLR3GL Expression in ESCs and Primary MEFs and during Early Mouse Embryonic Development.
We next evaluated the precise levels of expression of POLR3G and POLR3GL in both ESCs (cultured E14TG2a line) and differentiated primary MEFs (isolated and cultured from mouse embryos at E12.5). For this purpose, we raised polyclonal antibodies against the first 146 residues of POLR3G and the first 151 residues of POLR3GL, respectively. Although these polyclonal antibodies cross-reacted with additional proteins in immunoblots of whole-cell extracts, they successfully distinguished the two proteins in ESC lines overexpressing the mouse POLR3G or POLR3GL proteins (Fig. 1B). Moreover, each antibody could specifically immunoprecipitate its cognate Pol III subunit (Fig. 1C).
To quantify the absolute protein expression levels in different cell lines, we compared the immunoblot signals obtained using whole-cell extracts prepared from the same number of E14TG2a ESCs and primary MEFs to the signals given by increasing amounts of highly purified FLAG-tagged POLR3G and POLR3GL (Fig. 1 D–F). Interestingly, we found that the POLR3G level was 6-fold higher than the POLR3GL level in E14TG2a cells. In contrast, POLR3G levels were significantly lower (∼2-fold) in MEFs relative to ESCs, whereas POLR3GL levels were ∼3-fold higher in MEFs relative to ESCs (Fig. 1G). Therefore, the ratio of POLR3G to POLR3GL is dramatically decreased from 6:1 in ESCs to 1:1 in MEFs, confirming previously reported results regarding the reduction of POLR3G-containing Pol III during differentiation.
We also evaluated whether POLR3G and POLR3GL had different expression patterns at the mRNA level during early mouse development. To this end, we measured mRNA levels in mouse embryos harvested from two-cell to blastula stages (SI Appendix, Fig. S2). The low expression levels (reflected by high cycle threshold [CT] numbers) of the two transcripts from the earliest stage reflect the presence of maternal transcripts in these embryos. These results indicate that, while POLR3G mRNA expression rapidly increases at the early eight-cell stage to reach its maximum at the blastula stage, the POLR3GL mRNA level remains unchanged throughout development to the blastocyst stage.
Genome-Wide POLR3C, POLR3G, and POLR3GL Occupancies in Mouse ESCs and MEFs.
Next, we identified target genes of POLR3G- and POLR3GL-containing Pol III complexes in mouse E14TG2a ESCs and MEFs using genome-wide ChIP-seq with antibodies raised against POLR3C, POLR3G, POLR3GL, and Pol II (RBP1) (21). We observed strong ChIP-seq signals of POLR3C, POLR3G, and POLR3GL binding at tRNA clusters, but no detectable levels of all Pol III subunits binding at most Pol II-transcribed genes, indicating the specificity of the Pol III antibodies for Pol III-transcribed genes (Fig. 2 A and B). Among these antibodies, the POLR3C antibody displayed the greatest specificity and sensitivity in ChIP-seq experiments. Therefore, we always observed more POLR3C-binding sites than POLR3G- or POLR3GL-binding sites (Fig. 2C and Dataset S1). Overall, except for the POLR3GL subunit that was not readily detected on ESC chromatin despite being expressed at a detectable level in ESCs, all Pol III subunits tested were found at the same genomic locations. The lack of POLR3GL ChIP-seq peaks in ESCs could reflect either a failure of POLR3GL-containing complexes to be used for Pol III gene transcription or, more likely, the lower level of POLR3GL expression relative to POLR3G expression (1:6 ratio in ESCs) and a consequent level of POLR3GL binding that falls below detection levels in ChIP-seq experiments.
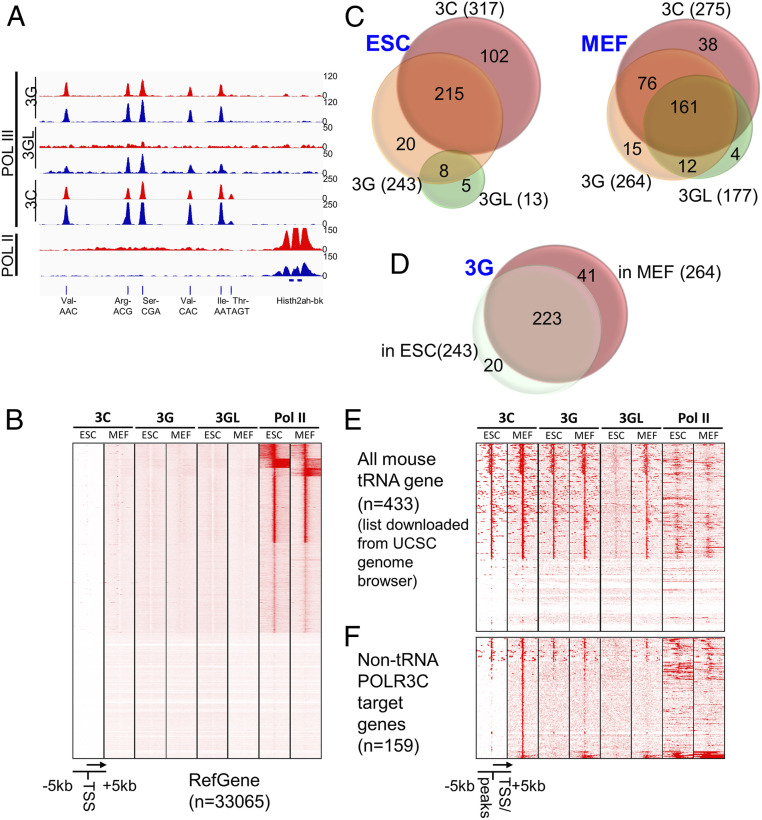
Fig. 2.
Genome-wide POLR3C, POLR3G, and POLR3GL occupancy in mouse ESCs and MEFs. (A) Representative Integrative Genomics Viewer view showing the ChIP-seq signals of POLR3C, POLR3G, POLR3GL, and Pol II at the tRNA cluster of Chr13. The binding profiles in ESCs are shown in red, while the profiles in MEFs are shown in blue. (B) Heatmap showing no detectable POLR3C, POLR3G, and POLR3GL occupancy at most Pol II-transcribed genes, thereby indicating the specificity of the antibodies for Pol III-transcribed genes. (C) Venn diagrams showing overlaps of POLR3C, POLR3G, and POLR3GL target sites in ESCs (Left) and MEFs (Right). (D) Venn diagram showing overlap of POLR3G target sites in ESCs and primary MEFs. (E and F) Heatmaps showing the ChIP-seq signals of POLR3C, POLR3G, POLR3GL, and Pol III at all murine tRNA loci (E) and non-tRNA targets of PLOR3C (F) in ESCs and MEFs.
POLR3C was detected at 317 loci in ESCs and 275 loci in MEFs, respectively (Fig. 2C). Most of these loci mapped to known Pol III target genes that include tRNA genes, 5S RNA genes, 7SK RNA genes, U6 small nuclear RNA genes, RNase MRP, Y RNA genes, vault RNA genes, and SINEs. We also observed a total of 243 and 264 loci that were occupied by POLR3G in ESCs and MEFs, respectively. Around 90% of these loci overlapped loci co-occupied by POLR3C (as expected for integral Pol III subunits) (Fig. 2C and Dataset S1), with the incomplete overlap likely reflecting the greater affinity of the POLR3C antibody (below). A total of 223 genes were commonly targeted by POLR3G in both ESCs and MEFs, while only 20 and 41 genes were specifically bound, respectively, in these cell types (Fig. 2D and Dataset S2) and could reflect cell-specific transcription of Pol III target genes regulated by other factors. In this regard, over half of the 61 outliers were tRNA genes that also were bound by POLR3C, consistent with previous reports of cell-type–specific expression of tRNA genes (22, 23). In contrast to POLR3G, POLR3GL was detected at only very few loci in ESCs (likely because of lower abundance and antibody affinity; but see also discussion of Fig. 2E data below) but showed a dramatically higher occupancy of 177 loci in MEFs (Fig. 2C). These POLR3GL loci presented a significant level of overlap with the POLR3G loci in MEFs (173 of 177 loci), indicating that both forms of polymerase were likely recruited to the same loci in these cells. The apparent differences may again reflect differences in antibody affinities (POLR3C > POLR3G > POLR3GL) and protein levels leading to differences in detection limits. As an example, when each of the apparent POLR3G-specific loci in MEFs was manually reviewed, many showed extremely low levels of POLR3GL binding that had been filtered out during the original analysis because the signals were closer to background.
The annotated mouse tRNA genes comprise 430 standard tRNA genes, 2 selenocysteine tRNA genes and 1 possible suppressor tRNA gene. More than half of these genes were occupied by POLR3C and POLR3G in both ESCs and MEFs (Fig. 2E), whereas POLR3GL exhibited strong binding to these tRNA loci only in MEFs. Although weak, the POLR3GL signals in ESCs nonetheless appear significant but, being closer to background, would have been filtered out in the analyses in Fig. 2C. Interestingly, these Pol III-dependent tRNA loci often displayed significant levels of Pol II binding, raising the possibility of active Pol II transcription units (Fig. 2E). Similar binding patterns were also observed in non-tRNA target genes of POLR3C (Fig. 2F).
Altogether, these results suggest that the two Pol III isoforms mediate expression of a very similar, if not the same, set of genes and that they have limited specificity, if any, other than that relating to levels of expression. These results are surprising in view of the earlier suggestion that the POLR3G-containing Pol III has a gene-specific role in the regulation of ESC differentiation (18) but are in line with results of genomic analyses in the mouse liver (16) and with current results of genetic analyses (below) that fail to show any differences in intrinsic functions.
Generation and Characterization of POLR3G and POLR3GL Conditional Knockout Mice.
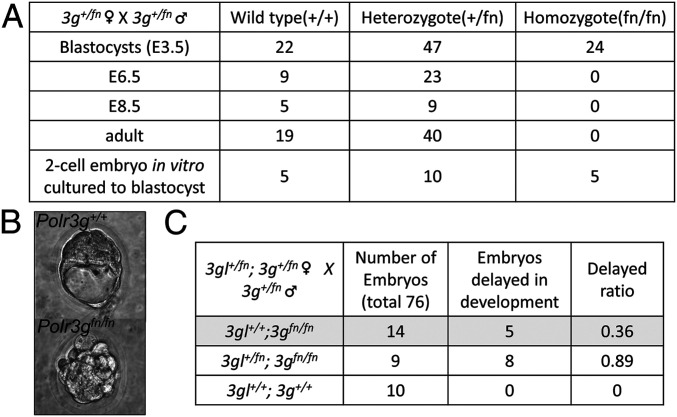
The apparent contradiction between the earlier report proposing different functions of the Pol III isoforms in regulating ESC differentiation and the current results indicating that both isoforms are found primarily on the same loci in MEFs prompted us to examine the functions of POLR3G and POLR3GL subunits in vivo. To understand the physiological function of POLR3G and POLR3GL during mouse development, we engineered, first, knockout mice harboring large insertions at Polr3g and Polr3gl regions. These insertions disrupt gene expression and generate functional null mutants (fn/fn). The insertions flanked by FRT/loxp sequences could later be removed to generate conditional (fl/fl) knockout mice (SI Appendix, Figs. S3 and S4). The initial crosses revealed that the complete loss of POLR3G is embryonic lethal. Therefore, we monitored early embryonic development more closely at specific time points (Fig. 3A). Indeed, we detected a Mendelian ratio of 0.26 (24 of 93) Polr3gfn/fn embryos at E3.5, although 8 embryos displayed delayed or arrested development as shown in Fig. 3B. In contrast, no Polr3gfn/fn embryos were observed at E6.5 from a total of 32 embryos, indicating that Polr3gfn/fn embryos die between E3.5 and E6.5. To confirm that Polr3gfn/fn embryos could survive to the blastocyst stage, we also collected 20 two-cell stage embryos from a Polr3g+/fn heterozygous cross and cultured them in vitro for 3 d. All embryos, including five Polr3gfn/fn homozygotes, were able to develop a blastocyst cavity, with one of five Polr3gfn/fn homozygotes arrested at the early blastocyst (Fig. 3A, last column).
Fig. 3.
Complete loss of POLR3G in mice is early embryonic lethal. (A) Summary of viability data indicating that Polr3gfn/fn embryos died between E3.5 and E6.5. (B) An example of a Polr3gfn/fn embryo, compared to a control embryo, arrested at the beginning of the blastula stage. (C) Summary of data indicating that a 50% reduction of POLR3GL in Polr3gfn/fn embryos results in an enhanced arrest (from 36 to 89%) of embryonic development at the early blastula stage.
The aforementioned results suggest either that POLR3G-containing Pol III complexes are not required until E3.5 or, more likely, that the levels of maternal Polr3g, maternal Polr3gl, and zygotic Polr3gl mRNAs were sufficient to maintain an appropriate level of Pol III up to E3.5. An alternative possibility is that POLR3GL is up-regulated (in embryos lacking POLR3G) to an extent sufficient to maintain viability up to E3.5, seemingly feasible in view of our demonstration (below) that cells can compensate, at least partially, for the loss of one subunit by overexpressing the other. Regarding the basis for the embryonic lethality post E3.5, we wondered if this arose because of a low level of POLR3GL-containing Pol III that was insufficient to compensate for the loss of POLR3G-containing Pol III or because POLR3G-containing Pol III was fulfilling a specific function at this developmental stage. To test these hypotheses, we conducted a genetic analysis to determine whether a 50% reduction in the POLR3GL level in Polr3gfn/fn embryos could aggravate the lethality phenotype. Indeed, our data show that the observed 36% fraction of delayed Polr3gfn/fn E3.5 embryos increased to 89% for Polr3gl+/fn; Polr3gfn/fn E3.5 embryos. Therefore, most Polr3gl+/fn; Polr3gfn/fn embryos were incapable of reaching the full blastocyst stage (Fig. 3C), whereas all wild-type embryos were able to develop normally to the full blastocyst stage. Notably, these results support the hypothesis that POLR3GL can compensate for the loss of POLR3G up to E3.5. Moreover, because POLR3GL is expressed at a low level during early embryonic development, it is likely that the much less abundant POLR3GL-containing Pol III complexes are not sufficient to fully compensate for the loss of POLR3G after the E3.5 blastula stage. The likelihood that the observed effects are due to differences in overall Pol III levels rather than isoform-selective functions is further supported by studies (below) showing functional equivalence of POLR3G and POLR3GL in driving ESC differentiation.
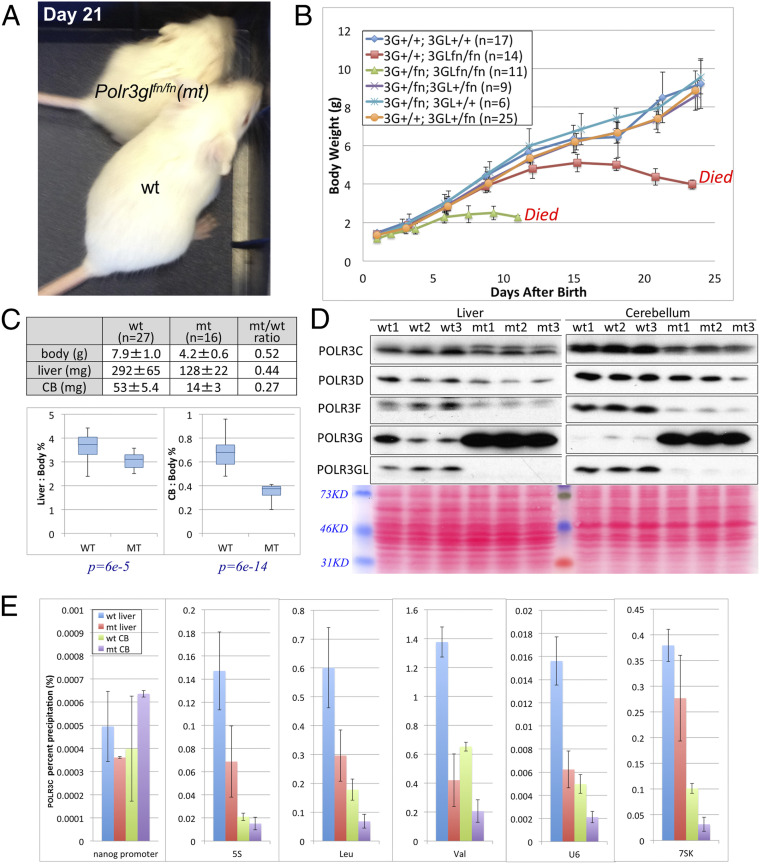
In stark contrast to Polr3gfn/fn mice, Polr3glfn/fn mice, to our surprise, were able to complete embryonic development up to birth without noticeable defects. However, postnatally, these mice grew more slowly than heterozygous or wild-type mice (Fig. 4A) and eventually died between postnatal day 19 (P19) to P25 with only half of the body weight of their wild-type siblings (Fig. 4B). Cell growth depends on protein production rates and, therefore, is tied to the synthesis of ribosomal RNAs and tRNAs by Pol I and Pol III (2). Hence, the observed phenotypes are consistent with a reduction of Pol III functions. Most importantly, however, the fact that Polr3glfn/fn mice could undergo development and even survive postnatally for a period of time indicates that POLR3G-containing Pol III can fulfill the same developmental functions as POLR3GL-containing Pol III. Similarly, we also performed a dosage effect experiment for POLR3G in Polr3glfn/fn mice. This analysis revealed that Polr3g+/fn; Polr3glfn/fn mice die much earlier than Polr3g+/+; Polr3glfn/fn mice, with an average life span of 10 d and more severe growth retardation phenotypes (the average weight at death was 2.3 ± 0.1 g; Fig. 4B, green line). These results confirmed that POLR3G can substitute partially for POLR3GL in Polr3glfn/fn mice.
Fig. 4.
Complete loss of POLR3GL in mice results in pup death at the weaning stage. (A) A Polr3glfn/fn mouse with about half the body size of its wild-type sibling at P21 after birth. (B) Growth curves of mouse pups with different dosages of Polr3g and Polr3gl genes from P1 to P25. Each mouse was weighed every 2 or 3 d in the morning. Genotypes and numbers of mice used for each genotype are described in the legend. Polr3glfn/fn mice are indicated by the red line and Polr3g+/fn;Polr3glfn/fn mice by the green line. (C) Graph indicating that Polr3glfn/fn mice show a statistically smaller cerebellum, relative to body size, than do control mice. (D) Immunoblots of liver and cerebellum protein extracts using POLR3C, POLR3D, POLR3F, POLR3G, and POLR3GL antibodies. Organs from three wild-type and three Polr3glfn/fn mice were used, and each lane was loaded with the same amount of protein extract as indicated by Ponceau S staining at the bottom. (E) POLR3C ChIP-qPCR assays were performed at several Pol III-targeted loci using liver and cerebellum tissues from wild-type and Polr3glfn/fn mice at P21.
In their final days (post P16), in addition to their smaller size, Polr3glfn/fn mice exhibited poor body balance and displayed locomotor ataxia indicative of putative cerebellar function defects. Indeed, whole-body phenotyping using eight Polr3glfn/fn mice and eight wild-type mice at P20 to P22 indicated that most organs in mutant mice were relatively normal morphologically and histologically, except for the cerebellum, which appeared statistically smaller than wild type when normalized to body weight (Fig. 4C). We next probed the levels of total Pol III in the cerebellum and, as a control, in liver in three wild-type and three Polr3gl knockout mice by immunoblot. In addition to confirming that these mice lacked POLR3GL expression in both tissues, these analyses revealed, interestingly, that POLR3G levels were significantly increased in all three mutant mice. This again suggests partial compensatory effects via the up-regulation of the other Pol III isoform in these tissues. However, the compensation was incomplete as revealed by the significantly decreased expression of POLR3C, POLR3D, and POLR3F subunits in both mutant organs (Fig. 4D). Next, we further validated these results by evaluating the recruitment of total Pol III to cognate target genes by performing ChIP-qPCR assays on pooled chromatin samples from multiple mice. These experiments revealed that the levels of total Pol III in the analyzed genes decreased by 50 to 75% in KO mice when compared to results obtained for wild-type mice (Fig. 4E).
Overexpressing Either POLR3G or POLR3GL Can Rescue the Differentiation Defect of POLR3G-Deficient ESCs.
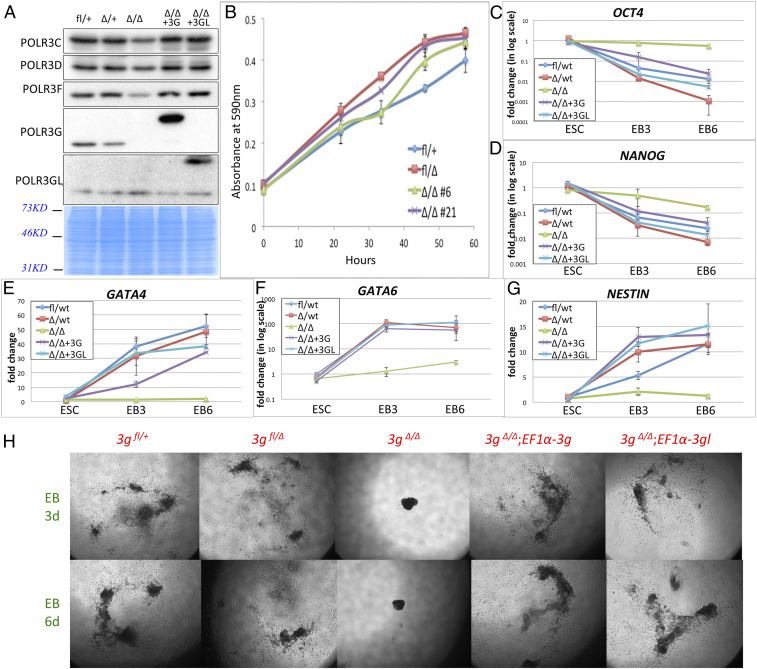
The dosage-effect experiments performed in either Polr3gfn/fn embryos or Polr3glfn/fn newborns suggested that POLR3G could partially substitute for POLR3GL and vice versa. To further test this hypothesis, we generated Polr3gfl/fl and Polr3gΔ/+ mice as described in SI Appendix, Figs. S3 and S4. The “Δ” in these Polr3g and Polr3gL mouse lines refers to the permanent deletion of the Polr3g or Polr3gl gene region by CRE recombinase, which eliminates all Polr3g or Polr3gl mRNA transcripts. We next established stable Polr3gfl/+ and Polr3gfl/Δ ESC lines (hereafter referred to, respectively, as 3gfl/+ and 3gfl/Δ) from E3.5 embryos by crossing Polr3gfl/fl male and Polr3gΔ/+ female mice. Two stable 3gΔ/Δ ESC lines (Δ/Δ #6 and #21) then were established by transient transfection of 3gfl/Δ ESCs with a plasmid expressing CRE recombinase. An immunoblot confirmed that neither of the 3gΔ/Δ ESC lines expressed any POLR3G at the protein level (Fig. 5A). To our surprise, both 3gΔ/Δ ESC lines displayed the same cell proliferation rates as Polr3g wild-type or heterozygous ESCs in vitro (Fig. 5B), suggesting that POLR3G is dispensable for ESC growth. Next, we stably expressed either POLR3G or POLR3GL under the control of an EF1a promoter in the 3gΔ/Δ ESC lines via lentivirus-based delivery (3gΔ/Δ; EF1a-3g and 3gΔ/Δ; EF1a-3gl ESC lines, respectively) to evaluate whether both Pol III isoforms could compensate for the loss of POLR3G in differentiation. Immunoblot analysis confirmed the high levels of exogenous POLR3G or POLR3GL overexpression in 3gΔ/Δ ESCs.
Fig. 5.
Overexpressed POLR3G or POLR3GL in Polr3gΔ/Δ ESCs equally rescue the phenotype of Polr3gΔ/Δ ESCs. (A) Immunoblots (Top five panels) of whole-cell extracts of ESCs with different genotypes (indicated at the top) with POLR3C, POLR3D, POLR3F, POLR3G, and POLR3GL antibodies (indicated at the left). Each lane was loaded with the same amount of protein extract as indicated by Coomassie blue staining at the bottom. (B) Cell proliferation assays of wild-type, Polr3gΔ/+, and Polr3gΔ/Δ ESCs. Two Polr3gΔ/Δ stable ESC lines were used in this assay as shown by the green and purple lines. (C–G) Differentiation assays performed with wild-type, Polr3gΔ/+, and Polr3gΔ/Δ ESC lines and with rescue lines Polr3gΔ/Δ;EF1a-3g and Polr3gΔ/Δ;EF1a-3gl. RT-PCR assays were performed on Oct4, Nanog, Gata4, Gata6, and Nestin genes using three time points (ESCs at the differentiation start point, 3 d after EB formation and 6 d after EB formation) individually. (H) Embryoid body differentiation analyses indicating that overexpression of either POLR3G or POLR3GL rescues the undifferentiated phenotype of Polr3gΔ/Δ ESCs.
To see whether the differentiation ability was affected in ESC lines lacking POLR3G and whether exogenous POLR3G or POLR3GL could rescue the defect in POLR3G-deficient (3gΔ/Δ) ESCs, we used the hanging drop method to form embryoid bodies (EBs) from 3gfl/+, 3gfl/Δ, 3gΔ/Δ, 3gΔ/Δ;EF1a-3g, and 3gΔ/Δ;EF1a-3gl ESC lines in vitro (24). After 3 and 6 d of differentiation, both wild-type (3gfl/+) and heterozygous (3gfl/Δ) ESCs showed full differentiation into embryoid bodies (Fig. 5H). In contrast, the POLR3G-deficient (3gΔ/Δ) ESCs could not achieve differentiation under these conditions. As expected, POLR3G fully rescued the differentiation defect of POLR3G-deficient (3gΔ/Δ) ESCs. Notably, however, POLR3GL overexpression could also achieve the same level of differentiation (Fig. 5H), confirming that both Pol III isoforms can support the same functions in cell-fate determination. We further confirmed the rescue efficiency by measuring the mRNA expression levels of several ESC pluripotency and differentiation marker genes that included Oct4, Nanog, Gata4, Gata6, and Nestin. The results indicate that both POLR3G and POLR3GL can fully rescue the differentiation defect of POLR3G-deficient (3gΔ/Δ) ESCs (Fig. 5 C–G). Altogether, our observations provide very strong evidence to support the conclusion that the total Pol III level, and not which Pol III isoforms are selectively expressed, is the main factor regulating the transition between ESC proliferation and differentiation.
Discussion
Redundant Functions and Differential Expression of the Two Pol III Isoforms.
Here, we have shown that the compositions of POLR3G- and POLR3GL-containing complexes in mouse ESCs are essentially the same, at least at the level of near stoichiometric components. In addition, both Pol III isoforms are found largely at the same loci in MEFs. These results suggest that, in these cell types and likely in many others (16), both types of complexes generally fulfill the same function. This hypothesis is further supported by genetic analyses indicating that the phenotype of either the Polr3g or the Polr3gl knockout mouse is further enhanced by loss of one allele of the other isoform. Most importantly, overexpression of either POLR3G or POLR3GL can rescue the differentiation defect of Polr3gΔ/Δ ESCs. This latter observation confirms that POLR3GL can functionally replace POLR3G in ESCs. Hence, the previous suggestions of a specific function of POLR3G in ESCs (14, 17) are more likely explained by the low expression of POLR3GL in these cells. Indeed, our data show that the ratio of POLR3G to POLR3GL in ESCs is about 6:1.
POLR3G and POLR3GL are highly similar in sequence, with 46% amino acid identities. Interestingly, invertebrates express only one isoform that is more closely related to POLR3GL (16). It therefore is possible that, during evolution, POLR3GL became limiting with respect to an increasing demand for Pol III during early embryogenesis, which could have favored gene duplication and fixation. The Polr3g gene is controlled by a promoter that can drive acute and massive expression of Polr3g mRNA during early development, whereas the Polr3gl gene is expressed more ubiquitously and more stably later in development. The difference in their regulation lies, at least in part, in the specific recruitment of MYC to the Polr3g promoter (16). The high level of MYC expression in ESCs probably accounts for the increased production of POLR3G in these rapidly dividing cells that present a high demand for Pol III activity. The latter hypothesis would also explain the increased levels observed in cancer cells that divide rapidly (14). The normal embryonic development of Polr3gl knockout mice indicates either that POLR3G is continuously expressed during embryogenesis or that a feedback mechanism monitoring Pol III activity can reactivate Polr3g alleles when needed. Very likely, POLR3G itself is not sufficient to meet the growth demands of mice after birth, thereby leading to the growth defect and early lethality phenotypes observed in Polr3gl KO mice.
Differential Phenotypes of Polr3g and Polr3gl Knockout Mice and Auto-Compensation of Two Forms In Vivo.
Polr3g knockout mice die between E3.5 and 6.5. To our surprise, however, it was possible to establish Polr3gΔ/Δ ESC lines in vitro via transient transfection of Polr3gfl/Δ cells with a CRE recombinase. Interestingly, Polr3gfn/fn ESC lines could also be established by crossing Polr3g+/fn mice and deriving ESCs from E3.5 embryos. In both cases, these Polr3g knockout cell lines were found to proliferate at rates similar to those of wild-type ESCs with no significant changes in the expression of pluripotency marker genes such as Nanog and Oct4. These results are contradictory to previous conclusions drawn from POLR3G knockdown analyses that suggested that this Pol III isoform was important for the proliferation and pluripotency maintenance of ESC cells (14, 18). However, the previous studies relied exclusively on either siRNA- or short hairpin RNA-mediated knockdowns, such that the differences compared to the current results with knockout cell lines could reflect either off-target effects of the interfering RNAs or an acute effect of temporarily reducing POLR3G. In our system, POLR3G was removed permanently and may have induced an increase in POLR3GL expression that partially compensated for the loss of POLR3G. Indeed, our immunoblot data showed that POLR3GL protein was increased about twofold in a Polr3gΔ/Δ ESC line (Fig. 5A). Although this amount appeared to be sufficient to maintain the proliferation of ESCs, it proved insufficient to match the higher Pol III demand during ESC differentiation, as evidenced by the inability of Polr3g knockout ESCs to differentiate into embryoid bodies. These results are also consistent with the early embryonic lethality observed in Polr3g knockout mice.
Unexpectedly, Polr3gl knockout mice displayed prenatal development without notable defects, which clearly demonstrates that POLR3G is not restricted to ESCs. However, Polr3gl knockout mice did present clear postnatal growth defects and eventually died at 3 wk after birth. Remarkably, compared to their wild-type siblings, these mice showed a massive (5- to 10-fold) induction of POLR3G expression at 3 wk of age. Nevertheless, this induction was insufficient to ensure long-term survival of Polr3gl knockout mice. The compensatory increase of one protein upon reduction or loss of function of an isoform is common in nature. For example, alpha 1-globin gene expression is increased in individuals heterozygous for the alpha-2 globin deletion (25). In mice heterozygous for a null mutation in Scn8a, Nav1.2 increases to keep total sodium channel expression approximately constant despite partial loss of Nav1.6 channels (26). The fact that the POLR3G and POLR3GL isoforms can compensate for one another strongly suggests that they have similar intrinsic functions in vivo.
Pol III/POLR3-Related Disorders.
Mutations in Pol III subunits POLR3A, POLR3B, POLR1C, or POLR3K in humans are causative for various pathologies of the central nervous system. Recently, because of their similarity and shared molecular function, these conditions have been designated POLR3-related leukodystrophies (27, 28). Among all of the neurologic impairments, cerebellar signs appear first and slowly worsen toward severe intention tremor, dysarthria, dysmetria, and gait ataxia (29). In addition to mutations in the previously mentioned Pol III subunits, mutations in CLP1 (a multifunctional kinase involved in tRNA splicing) and BRF1 (one of three subunits of the Pol III transcription initiation complex) also cause abnormalities in cerebellar development and neurodegeneration (30, 31). The phenotypes that we have observed in Polr3gl knockout mice are in line with the cerebellar abnormalities in humans carrying mutations in Pol III-related genes. Both immunoblot and ChIP analyses in Polr3gl knockout mice confirmed reductions both in the total amount of Pol III and in Pol III binding to target genes in vivo. There currently are no reported POLR3GL loss-of-function mutations in humans, which may be due to their early lethality. However, a very recent report described three individuals with biallelic Polr3gl variants in which exon 2 or exon 5 of Polr3gl was skipped (32). These three patients were characterized mainly by axial endosteal hyperostosis, oligodontia, short stature, and mild facial dysmorphisms. These phenotypes largely fit within the spectrum of Pol III-related disorders and are supported by our mouse model. It will be interesting to see if there are other Polr3gl mutations leading to reductions of POLR3GL, thereby giving similar phenotypes as other POLR3-related disorders. In this regard, Polr3gl mutations should be examined in early infant death cases and in patients with impaired cerebellar development. Our Polr3gl knockout mice could also constitute a convenient model to study these pathologies in more detail.
Materials and Methods
Generation of Polr3g and Polr3gl Conditional Knockout Mice.
The Polr3g targeting vector was obtained from EUCOMM (Polr3gtm1a [EUCOMM]Wtsi HTGR04004_Z_1_E09) (SI Appendix, Fig. S3). The Polr3gl-targeting Bac vector was generated by recombination (SI Appendix, Fig. S4). Both mouse lines were generated by the Rockefeller Gene Targeting Center in a C57BL/6 background. B6(Cg)-Tyrc-2J/J (#000058), Actin-Flp (#3005703), and EIIA-CRE (#003724) mice were obtained from The Jackson Laboratory. All mice were maintained according to institutional guidelines. Transgenic mice with single onsite recombination were confirmed by Southern blot analysis using 32P-labeled external probes and neoprobes. The “knockout-first” alleles (fn alleles) were later crossed to transgenic FLP mice to generate conditional knockout mice (fl allele). The fl allele mice were further crossed to transgenic EIIA-CRE mice to obtain heterozygous Δ alleles. All animal experiments were approved and performed in accordance with the Institutional Animal Care and Use Committee at The Rockefeller University.
Mouse Embryo Isolation, Culture, and Genotyping.
After mating, plugged females were euthanized at certain time points. Embryos were flushed out or isolated from oviducts or uteri as described in Manipulating the Mouse Embryo (33). Viagen DirectPCR solution (#102-T) was used to lyse embryos followed by regular PCR and agarose gel isolation. For embryos on and before 3.5 d, two-round nested PCR analyses were used to amplify the signal. A table listing all primers is provided in Dataset S3. To culture two-cell-stage embryos, 1.5-d embryos were flushed out using fresh EmbryoMax medium (Millipore #MR-121-D) and cultured in 20 μL EmbryoMax medium drops in a 37 °C, 5% CO2 incubator until they reached the blastocyst stage.
Cell Culture.
E14TG2a cells were cultured on 0.1% gelatinized tissue-culture plates in complete ESC growth medium (Gibco) supplemented with mouse leukemia inhibitory factor (LIF) (Millipore) and 15% fetal bovine serum (FBS). Primary MEFs were derived from E12.5 embryos, maintained in ESC growth medium supplemented with 10% FBS (without LIF), and passaged three to five times prior to immunoblot or ChIP-seq analyses. Mouse embryonic stem cell lines used for Fig. 5 were derived from E3.5 blastocyst embryos and cultured in Glasgow’s modified Eagle’s medium (Sigma) supplemented with 10% FBS, 0.1 mM 2-mercaptoethanol, 1 mM nonessential amino acids, 1 mM sodium pyruvate, 1% l-glutamine, 1,000 U/mL of mouse LIF, and 2 inhibitors (1 μM MEK inhibitor PD0325901 and 3 μM GSK3 inhibitor CHIR99021). Stable Polr3gΔ/Δ ESC lines were established by temporarily transfecting Polr3gfl/Δ ESCs with a plasmid expressing a CRE recombinase for 36 h (the Cre-CAG-Ires Puro plasmid was a kind gift from Hiromitsu Nakauchi, Institute of Medical Science, The University of Tokyo, Tokyo, Japan (34). After puromycin selection, single clones were picked and confirmed by PCR genotyping.
EB Formation and Spontaneous Differentiation.
EBs were formed using the hanging drop method (24, 35). EBs were later transferred to gelatin-coated plates to initiate spontaneous differentiation toward three germ lineages (endoderm, mesoderm, and ectoderm). The first day on which formed EBs were transferred to gelatin-coated plates was defined as EB 0 d. Medium was changed daily afterward. At EB-3 and EB-6 d, three EBs from each genotype were lysed and subjected to qRT-PCR assays.
Antibodies, Immunoblots, and Immunoprecipitation.
GST-tagged POLR3G (residues 1 to 146), GST-tagged POLR3GL (residues 1 to 151), and 6 His-tagged POLR3C (full length) were expressed in Escherichia coli and, after purification, used as antigens to generate polyclonal antibodies in rabbits (Covance). POLR3D (RPC53) and POLR3F (RPC39) antibodies were as described before (11). Corresponding sera were diluted 5,000-fold for immunoblot and 200-fold for immunoprecipitation (IP). Anti-FLAG M2 antibody and M2 beads were obtained from Sigma. For IP, samples were lysed, incubated with M2 beads, and washed in BC300+1% Triton X-100.
POLR3G and POLR3GL Pull-Down and Mass Spectrometric Assays.
E14TG2a cells were first transfected with a lentiviral vector expressing BirA-neomycin under the EF1α promoter pEF1a-BirA-IRES-neo. Neomycin-positive clones were selected and transfected with lentiviral vectors expressing FLAG-AviTag-POLR3G or -POLR3GL and puromycin-N-acetyltransferase under the EF1α promoter PL-SIN-EF1α-Fb-IRES-puro. Stable cell lines expressing exogenous POLR3G or POLR3GL were selected and confirmed by streptavidin-IP and FLAG immunoblot. For pull-down assays, whole-cell lysates were extracted with 0.42 M KCl + 0.2% Triton X-100 and washed four times with BC300 + 0.2% Triton X-100, followed by purification on M2-Agarose and streptavidin beads. Final samples were eluted with sodium dodecyl sulfate (SDS) buffer at room temperature and analyzed by SDS/polyacrylamide gel electrophoresis (PAGE). For MS, each gel lane was excised and cut into 16 pieces that were denatured and digested with trypsin at 37 °C overnight. Peptides were separated by liquid chromatography followed with tandem MS. All procedures were performed and analyzed by the Rockefeller University Mass Spectrometry facility.
Quantification of POLR3G and POLR3GL Levels in Cells.
DNA sequences encoding FLAG-tagged full-length POLR3G and POLR3GL proteins were cloned into the pFastBac expression vectors. Baculoviruses were generated and amplified in Sf9 cells and transduced to High Five cells for protein expression. POLR3G and POLR3GL proteins were purified on M2-Agarose beads and eluted in PBS+0.1% Triton X-100 buffer containing 150 μg/mL 3xFLAG peptide. Final concentrations of full-length POLR3G and POLR3GL were measured by SDS/PAGE with reference to bovine serum albumin standards (Sigma A4503) in the linear range. To prepare the whole-cell lysates, E14TG2a cells and primary MEFs were carefully counted and lysed directly in SDS loading buffer. Immunoblots were carried out with either anti-POLR3G or anti-POLR3GL. Signals were detected and analyzed with a LI-COR Odyssey imaging system. Protein amounts in cell lysates were determined by reference to purified recombinant POLR3G or POLR3GL proteins. The size differences between recombinant and endogenous proteins reflect the presence of the FLAG tag.
Cell Proliferation Assay.
A 96-well plate was precoated with 0.1% gelatin overnight. A total of 2,000 cells in 100 μL of ES medium were added to each well on the next day. A Promega CellTiter 96 AQueous cell proliferation assay kit was used to measure cell growth.
Quantitative RT-PCR.
Complementary DNA (cDNA) was synthesized using 1 μg of total RNA and the qScript cDNA synthesis kit (Quanta Biosciences #95047) as per manufacturer’s instructions. A table listing all primers is provided in Dataset S3. For qPCR, QuantiTect SYBR Green was used in triplicate for three biological repeats.
Single-Cell-to-CT.
Embryos at two-cell, four-cell, eight-cell, and blastula stages were lysed and subjected to single-cell-to-CT analyses using an Ambion single-cell-to-CT kit according to the prescribed protocol. The preamplification mix contained primer sets for Gapdh, Oct3/4, Nanog, Sox2, Klf4, Polr3g, Polr3gl, Rn5S, Rn7SK, and RnU6. Second-round qPCR assays were performed with Applied Biosystems Real-Time PCR Instruments 7100.
ChIP-Seq Assays.
For each ChIP assay, 30 to 50 million E14TG2a cells or primary MEFs were used. Cells were cross-linked in growth medium with 1% formaldehyde for 10 min at room temperature. Chromatin was prepared from isolated nuclei and sonicated to an average size of 200 to 700 bp in lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid [EDTA], 0.1% SDS, 0.1% Na-deoxycholate, and 1× complete Protease Inhibitor [Roche]). Soluble chromatin (∼200 μg DNA) was immunoprecipitated with 10 μg of rabbit polyclonal Pol II (N-20, Santa Cruz sr-899), POLR3G, POLR3GL, and POLR3C antibodies/100 μL Dynal protein G matrix at 4 °C for overnight. Bound materials were extensively washed, eluted, treated with RNase A and proteinase K, and reverse-cross-linked overnight at 65 °C. ChIP’ed DNA fragments were recovered using a PCR purification kit from Qiagen and submitted to the Epigenomic Core Facility at Weill Cornell Medical College for library construction (TruSeq preparation kit from Illumina) and for 50-bp single-read high-throughput sequencing in an Illumina HiSeq2000.
Analysis of ChIP-Seq Data.
ChIP-seq data were analyzed as previously described (36). Briefly, the 50-bp sequence tags generated for each ChIP-seq assay were aligned to mouse genome mm9 assembly by Bowtie pipeline 1.0.1 release using parameters -m 1, -n 2, and -best. The SAMtools was used to sort the aligned reads and remove PCR duplicates. Peak calling was performed using the MACS software (37) or HOMER package (38). The final numbers of peaks were manually determined by comparison to the input gene track in Integrative Genomics Viewer. Clustering heat maps of the ChIP-seq results were performed using the seqMINER program (39) with the transcription start site dataset of murine tRNA and RefGene from University of California at Santa Cruz genome table browser. The annotated mouse tRNA genes are previously described (40). Statistics of ChIP-seq experiments are in SI Appendix, Table S2.
ChIP.
Liver and cerebellum samples were collected from either wild-type or mutant mice and fixed in 1% formaldehyde for 5 min at room temperature. Fixed samples were lysed in 50 mM Hepes (pH 7.5), 140 mM NaCl, 1 mM EDTA, 10% glycerol, 05% Nonidet P-40, 0.25% Triton X-100 with proteinase inhibitors for 20 min at 4 °C. After centrifugation, pellets were washed two times with 10 mM Tris⋅HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA, and 0.5 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA). After washing, pellets were resuspended in 10 mM Tris⋅HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, and proteinase inhibitor and sonicated to fragments with an average size of 200 to 800 bp. For each IP, ∼30 μg of soluble chromatin was incubated overnight with 1.5 μL POLR3C antibody and 30 μL Dynal protein G beads. Dynal beads were then washed four times with 20 mM Tris⋅HCl (8.0), 1 mM EDTA, 0.1% SDS, 1% Triton-X100, and 500 mM NaCl. After cross-link reversal, ChIP’ed DNA fragments were recovered using a PCR purification kit from Qiagen. Final data were generated from three biological repeats.
Data Availability.
The ChIP-seq dataset has been deposited under accession no. GSE143969 in the National Center for Biotechnology Information Gene Expression Omnibus database on January 22, 2020 (21).
Supplementary Material
Acknowledgments
We thank Dr. Keiichi Ito for advice on ESC-related experiments, Dr. Martin Teichmann for discussions, Dr. Zhengxin Wang for POLR3C antibody preparation, Dr. Qingwen Yang (Rockefeller Gene Targeting Center) for designing targeting constructs and generating transgenic mice, and the Epigenomics Core Facility at Weill Cornell Medical College for the Next Generation Sequencing services. This work was supported by National Cancer Institute Grants CA129325 and CA202245 (to R.G.R.). A.G. was supported by a Swiss National Science Foundation Early Mobility Fellowship (P2GEP3_151952) and by a Human Frontier Science Program Long-Term Fellowship (LT001083/2014).
Footnotes
The authors declare no competing interest.
Data deposition: The ChIP-seq dataset has been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database on January 22, 2020 (accession no. GSE143969).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922821117/-/DCSupplemental.
References
- 1.Dieci G., Fiorino G., Castelnuovo M., Teichmann M., Pagano A., The expanding RNA polymerase III transcriptome. Trends Genet. 23, 614–622 (2007). [DOI] [PubMed] [Google Scholar]
- 2.White R. J., RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol. 6, 69–78 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Oler A. J.et al., Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat. Struct. Mol. Biol. 17, 620–628 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barski A.et al., Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat. Struct. Mol. Biol. 17, 629–634 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moqtaderi Z.et al., Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat. Struct. Mol. Biol. 17, 635–640 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canella D., Praz V., Reina J. H., Cousin P., Hernandez N., Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 20, 710–721 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrière L.et al., Genomic binding of Pol III transcription machinery and relationship with TFIIS transcription factor distribution in mouse embryonic stem cells. Nucleic Acids Res. 40, 270–283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alla R. K., Cairns B. R., RNA polymerase III transcriptomes in human embryonic stem cells and induced pluripotent stem cells, and relationships with pluripotency transcription factors. PLoS One 9, e85648 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeganeh M.et al.; CycliX consortium , Differential regulation of RNA polymerase III genes during liver regeneration. Nucleic Acids Res. 47, 1786–1796 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumay-Odelot H., Durrieu-Gaillard S., Da Silva D., Roeder R. G., Teichmann M., Cell growth- and differentiation-dependent regulation of RNA polymerase III transcription. Cell Cycle 9, 3687–3699 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z., Roeder R. G., Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 11, 1315–1326 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Sklar V. E., Schwartz L. B., Roeder R. G., Distinct molecular structures of nuclear class I, II, and III DNA-dependent RNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 72, 348–352 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sklar V. E. F., Roeder R. G., Purification and subunit structure of deoxyribonucleic acid-dependent ribonucleic acid polymerase III from the mouse plasmacytoma, MOPC 315. J. Biol. Chem. 251, 1064–1073 (1976). [PubMed] [Google Scholar]
- 14.Haurie V.et al., Two isoforms of human RNA polymerase III with specific functions in cell growth and transformation. Proc. Natl. Acad. Sci. U.S.A. 107, 4176–4181 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefèvre S.et al., Structure-function analysis of hRPC62 provides insights into RNA polymerase III transcription initiation. Nat. Struct. Mol. Biol. 18, 352–358 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Renaud M.et al., Gene duplication and neofunctionalization: POLR3G and POLR3GL. Genome Res. 24, 37–51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enver T.et al., Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum. Mol. Genet. 14, 3129–3140 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Wong R. C.-B.et al., A novel role for an RNA polymerase III subunit POLR3G in regulating pluripotency in human embryonic stem cells. Stem Cells 29, 1517–1527 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Lin C. Y.et al., Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151, 56–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellacheruvu D.et al., The CRAPome: A contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen W.-Y., Wang X., Roeder R. G., Genome-wide maps of RNA Pol III subunits POLR3C, POLR3G, and PLOR3GL in ES-E14TG2a and primary MEF cells. National Center for Biotechnology Information Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE143969. Deposited 22 January 2020.
- 22.Sagi D.et al., Tissue- and time-specific expression of otherwise identical tRNA genes. PLoS Genet. 12, e1006264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dittmar K. A., Goodenbour J. M., Pan T., Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2, e221 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X., Yang P., In vitro differentiation of mouse embryonic stem (mES) cells using the hanging drop method. J. Vis. Exp., 825 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liebhaber S. A., Cash F. E., Main D. M., Compensatory increase in alpha 1-globin gene expression in individuals heterozygous for the alpha-thalassemia-2 deletion. J. Clin. Invest. 76, 1057–1064 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vega A. V., Henry D. L., Matthews G., Reduced expression of Na(v)1.6 sodium channels and compensation by Na(v)1.2 channels in mice heterozygous for a null mutation in Scn8a. Neurosci. Lett. 442, 69–73 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Bernard G.et al., Mutations of POLR3A encoding a catalytic subunit of RNA polymerase Pol III cause a recessive hypomyelinating leukodystrophy. Am. J. Hum. Genet. 89, 415–423 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorboz I.et al., Mutation in POLR3K causes hypomyelinating leukodystrophy and abnormal ribosomal RNA regulation. Neurol. Genet. 4, e289 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghoumid J.et al., Cerebellar hypoplasia with endosteal sclerosis is a POLR3-related disorder. Eur. J. Hum. Genet. 25, 1011–1014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaffer A. E.et al., CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell 157, 651–663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borck G.et al., BRF1 mutations alter RNA polymerase III-dependent transcription and cause neurodevelopmental anomalies. Genome Res. 25, 155–166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terhal P. A.et al., Biallelic variants in POLR3GL cause endosteal hyperostosis and oligodontia. Eur. J. Hum. Genet. 28, 31–39 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Behringer R.et al., Manipulating the Mouse Embryo: A Laboratory Manual, (Cold Spring Harbor Laboratory Press, 4th Ed., 2014). [Google Scholar]
- 34.Kobayashi T., Kato-Itoh M., Nakauchi H., Targeted organ generation using Mixl1-inducible mouse pluripotent stem cells in blastocyst complementation. Stem Cells Dev. 24, 182–189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang H.et al., Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell 144, 513–525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimada M.et al., Gene-specific H1 eviction through a transcriptional activator→p300→NAP1→H1 pathway. Mol. Cell 74, 268–283.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y.et al., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinz S.et al., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye T.et al., seqMINER: An integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 39, e35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan P. P., Lowe T. M., GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, D184–D189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ChIP-seq dataset has been deposited under accession no. GSE143969 in the National Center for Biotechnology Information Gene Expression Omnibus database on January 22, 2020 (21).