Significance
Burkholderia pseudomallei is a bacterial pathogen that causes melioidosis, an infectious disease in the tropics with high morbidity and mortality. It has a unique property among bacteria: to fuse infected host cells. We found that our immune system detects bacterial- or chemical-induced host cell fusion as a danger signal. Abnormal cell fusion leads to genomic instability and formation of micronuclei. This triggers the host to activate a signaling pathway leading to a form of cell death known as autophagic death, which likely serves to limit abnormal cellular transformation.
Keywords: cell fusion, cGAS–STING, T6SS, autophagy, DNA damage
Abstract
Burkholderia pseudomallei is the causative agent of melioidosis, an infectious disease in the tropics and subtropics with high morbidity and mortality. The facultative intracellular bacterium induces host cell fusion through its type VI secretion system 5 (T6SS5) as an important part of its pathogenesis in mammalian hosts. This allows it to spread intercellularly without encountering extracellular host defenses. We report that bacterial T6SS5-dependent cell fusion triggers type I IFN gene expression in the host and leads to activation of the cGAMP synthase–stimulator of IFN genes (cGAS–STING) pathway, independent of bacterial ligands. Aberrant and abortive mitotic events result in the formation of micronuclei colocalizing with cGAS, which is activated by double-stranded DNA. Surprisingly, cGAS–STING activation leads to type I IFN transcription but not its production. Instead, the activation of cGAS and STING results in autophagic cell death. We also observed type I IFN gene expression, micronuclei formation, and death of chemically induced cell fusions. Therefore, we propose that the cGAS–STING pathway senses unnatural cell fusion through micronuclei formation as a danger signal, and consequently limits aberrant cell division and potential cellular transformation through autophagic death induction.
Melioidosis is an infectious disease endemic in Southeast Asia, Northern Australia, and increasingly reported in other tropical parts of the world (1, 2). The clinical spectrum of the disease can range from localized skin lesions to severe sepsis and death (1). The causative agent is Burkholderia pseudomallei, a gram-negative facultative intracellular bacterium capable of surviving within various eukaryotic host cells (3–5). One very unusual phenotypic signature of its intracellular lifecycle is the formation of multinucleated giant cells (MNGCs) in many different cell types, through the fusion of an infected cell with its neighboring cells (4, 6). Upon uptake by host cells, the bacterium escapes from endosomal vesicles by one of its type III secretion systems (T3SS3) into the cytosol (7). Through a two-component sensor-regulator system VirAG, it senses reduced glutathione in the cytosol to turn on the expression of T6SS5 (8). T6SS5 is involved in inducing cell–cell fusion leading to MNGC formation (9–12) and is a critical virulence factor for Burkholderia pathogenesis in mammalian hosts (9). Besides T6SS5, intracellular motility also contributes to MNGC formation (10). B. pseudomallei relies on the fla2 flagellar motility system and BimA protein for intracellular motility within the cytoplasm (13–15). BimA protein located on the tail-end of the bacterium allows B. pseudomallei to exploit host actin by polymerizing monomeric actin and propelling the bacterium through the cell (13–15). Induction of MNGCs is thought to facilitate localized dissemination of the bacterium and evasion from host defenses or antimicrobials (6). Cell fusion is potentially involved in the pathogenesis of melioidosis as MNGCs have been observed in the tissue samples of melioidosis patients and in a C57BL/6 chronic mouse model of B. pseudomallei infection (16, 17).
The T6SS is a specialized molecular machine in gram-negative bacteria for the export and delivery of effectors across bacterial membranes into the host. It is composed of 13 essential components that assembles to form a contractile bacteriophage tail-resembling structure anchored onto the bacterial envelope by a transmembrane complex (18–20). The T6SS tail is evolutionarily, structurally, and functionally related to the tail of contractile bacteriophages, which is composed of an inner tube enveloped by a contractile sheath (21–23). The T6SS inner tube is comprised of hexamers of Hcp stacked in a head-to-tail fashion with the VgrG trimer at its tip (21–23) and the contraction of the TssBC sheath propels the inner Hcp tube toward the target cell for delivery of the effectors (24–26). ClpV is a cytosolic AAA+ ATPase protein that functions to improve the efficiency of T6SS by recycling TssBC subunits after sheath contraction (27). T6SS effectors have diverse functions. In Burkholderia T6SS5, the VgrG5 effector has an evolved C-terminal extension domain that is responsible for cell fusion, although the exact mechanism of how it triggers host cell fusion is not known (11, 12).
Initially, we examined whether there were additional phenotypes conferred by T6SS5 on infected cells besides cell fusion. Using in vitro cell-infection models involving various T6SS5 and cell fusion-defective mutants, we found to our surprise that bacterial-induced cell fusion, not T6SS secreted effectors, triggers the innate immune response. Host pattern recognition receptors (PRRs) detect danger-associated molecular patterns that arise from the host and pathogen-associated molecular patterns present on microbes. cGAMP synthase (cGAS) is a PRR known to recognize cytosolic double-stranded DNA and catalyze the formation of 2′3′-cGAMP, a cyclic dinucleotide (CDN). CDNs associate with adaptor stimulator of IFN genes (STING) present in the endoplasmic reticulum, leading to type I IFN gene induction. cGAS is most often implicated in the recognition of self-DNA from tumor cells (28, 29) and DNA derived from pathogens, such as viruses (30–32) or cytosolic bacteria (33, 34). Intriguingly, we discovered that bacterial and chemical cell fusion triggers genomic instability that results in micronuclei formation and activation of cGAS and STING. This process leads to autophagic cell death instead of type I IFN production. Therefore, our study reveals a mechanism of how cell–cell fusion acts as a danger signal to the host to trigger the cGAS–STING pathway and cell death, potentially limiting cellular transformation.
Results
Burkholderia Defective in T6SS5 or Intracellular Motility Induce Significantly Lower Type I IFN Gene Expression.
To identify phenotypes dependent on T6SS5 during infection, we utilized various B. pseudomallei T6SS5 mutants, including ∆clpV5, ∆hcp5, and T6SS5 two-component sensor-regulator mutant ∆virAG (35) for infection in RAW264.7 macrophages. B. pseudomallei-infected cells had IFN-β (IFNB1) expression at levels ∼10-fold higher than T6SS5 mutant-infected cells at 16 h after infection (Fig. 1A). Burkholderia thailandensis—a closely related species with a homologous T6SS5 and similar intracellular life cycle—similarly led to 10-fold higher induction of IFNB1 gene expression as compared to T6SS5 mutants ∆clpV5- and ∆vgrG5-infected cells at both low and high multiplicities of infection (MOI) at 11 to 15 h but not at 4 to 8 h postinfection (Fig. 1B). Intracellular bacterial loads were comparable between cells infected with B. thailandensis and the T6SS5 mutants, indicating that the lower IFNB1 expression was not due to differential bacterial burden (SI Appendix, Fig. S1A). As B. pseudomallei is a risk group 3 agent requiring high containment, B. thailandensis was used in subsequent experiments for further mechanistic investigations.
Fig. 1.
Burkholderia defective in T6SS5 or intracellular motility induce significantly lower type I IFN gene expression. (A and B) Expression of IFN-β (IFNB1) in RAW264.7 macrophages infected with Burkholderia compared with various T6SS5 mutants. (A) RAW264.7 macrophages were infected with B. pseudomallei strains at an MOI of 1 for 16 h. An assay was performed in triplicates and the values are expressed as mean ± SD. (B) RAW264.7 macrophages were infected with B. thailandensis strains at an MOI of 10 for 8 and 15 h or MOI of 100 for 4 and 11 h. An assay was performed in duplicates and the values are expressed as mean ± SD. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; n.s., not significant. (C) Light micrographs of Giemsa-stained RAW264.7 macrophages infected with various B. thailandensis strains. Cells were infected at an MOI of 10 or 100 and were fixed for staining and visualization of MNGC formation under 10× objective magnification at 15 or 11 h postinfection respectively. (Scale bars, 100 µm.) Note: ∆vgrG5(a) is an insertional mutant while ∆vgrG5(b) is an in-frame deletion mutant.
The differential IFNB1 expression at late time points (11 to 15 h postinfection) when MNGC formation occurred (Fig. 1C) suggests that the induction of type I IFN gene expression could be dependent on the event of cell–cell fusion. Quantification of MNGCs is recorded in SI Appendix, Table S1. To explore this possibility, we utilized the ∆bimA∆motA2 double mutant, which exhibits the delayed cell fusion phenotype (10). BimA affects actin polymerization and MotA2 affects intracellular motility, both of which are not part of the T6SS5 and therefore should not have defective T6SS5 function. Infection by the ∆bimA∆motA2 mutant at an MOI of 10 for 12 h in RAW264.7 macrophages did not induce MNGC formation or increase IFNB1 gene expression, whereas the wild-type bacterial infection formed MNGCs and up-regulated IFNB1 (SI Appendix, Fig. S1 B and C). This is despite the two having comparable bacterial loads (SI Appendix, Fig. S1D). Under these conditions, ∆bimA∆motA2 expressed T6SS5 genes hcp5, vgrG5, and virA at levels higher than B. thailandensis (SI Appendix, Fig. S1E). Collectively, these present the possibility that cell fusion could be triggering type I IFN gene expression.
Bacterial-Driven Cell Fusion Induces Type I IFN Gene Expression Independent of Bacterial Ligands.
Some VgrGs that form the tip of the T6SS5 apparatus are considered “evolved” and have an extended carboxyl-terminal domain (CTD) that confers effector functions. The CTD of Burkholderia VgrG5 has been shown to possess fusogenic activity responsible for cell–cell fusion and MNGC formation and is separable from VgrG’s role in in T6SS secretion (12). As expected, in RAW264.7 cells infected with vgrG5-∆CTD, no MNGCs were formed (Fig. 2A and SI Appendix, Table S1). The fusion-defective vgrG5-∆CTD mutant also showed correspondingly reduced IFNB1 gene expression compared to B. thailandensis-infected cells (Fig. 2B), suggesting that cell–cell fusion induces up-regulation of type I IFN gene expression. However, it is unclear whether the vgrG5-∆CTD mutant exhibits defective binding of effectors at the VgrG tip as VgrG can act as specific adaptors for effector delivery (36, 37). Thus, when we infected sparsely seeded cells with wild-type bacteria so that they did not fuse (Fig. 2C), type I IFN gene expression was not induced even though T6SS5 was up-regulated (Fig. 2 D and E). This eliminates the role of any potential bacterial effectors bound to the CTD of VgrG5 in IFNB1 gene induction.
Fig. 2.
Bacterial-driven cell fusion induces type I IFN gene expression independent of bacterial ligands. (A) Light micrographs of Giemsa-stained RAW264.7 macrophages infected with B. thailandensis wild-type and cell fusion defective vgrG5-∆CTD. Cells infected with ∆clpV5 were included as a control. Cells were infected at an MOI of 10 and fixed for staining and visualization of MNGC formation under 10× objective magnification at 15 h postinfection. (Scale bars, 100 µm.) (B) Expression of IFNB1 in RAW264.7 macrophages infected with various B. thailandensis strains, including fusion dysfunctional vgrG5-∆CTD. RAW264.7 macrophages were infected at an MOI of 10 for 15 h. Assay was performed in triplicates and the values are expressed as mean ± SD. (C) Light micrographs of sparsely seeded RAW264.7 macrophages infected with B. thailandensis wild-type and cell fusion-defective vgrG5-∆CTD. Cells were infected at an MOI of 10 and were fixed for Giemsa staining and visualization of MNGC formation under 10× objective magnification at 15 h postinfection. (Scale bars, 100 µm.) (D) Expression of IFNB1 in sparsely seeded RAW264.7 macrophages infected with B. thailandensis and vgrG5-∆CTD. RAW264.7 macrophages were infected at an MOI of 10 for 15 h. Transcript levels of IFNB1 were measured by qRT-PCR and normalized to uninfected cells. GAPDH gene was used as a housekeeping gene control. An assay was performed in triplicates and the values are expressed as mean ± SD. (E) Expression of bacterial T6SS5 gene hcp5 in sparsely seeded RAW264.7 cells infected with B. thailandensis. Transcript levels of hcp5 was measured by qRT-PCR and normalized to that at 2 h postinfection. An assay was performed in triplicate and the values are expressed as mean ± SD. (F) Light micrographs of Giemsa-stained HepG2 cells treated with PEG-6000. HepG2 cells were treated with PEG-6000 and allowed a duration of 21 h for cell–cell fusion to occur. D-PBS–treated cells were included as a control. Cells were stained and visualized under 10× objective magnification. (Scale bar, 100 µm.) (G) Expression of IFNB1 in PEG-6000–treated cells. HepG2 cells were treated with PEG-6000 and allowed to fuse over 21 h. Transcripts were harvested and analyzed via qRT-PCR. Expression levels of genes were normalized to the control D-PBS–treated cells. An assay was performed in triplicate and the values are expressed as mean ± SD. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; n.s., not significant.
To further confirm that cell fusion drives type I IFN gene expression independent of bacterial ligands, we replaced bacterial infection with polyethylene glycol (PEG) to induce cell fusion so that there will be no interference from bacterial nucleic acids. We found that PEG-6000–induced cell fusion correlated with increased type I IFN gene expression in both HepG2 liver epithelial cells (Fig. 2 F and G and SI Appendix, Table S1) and primary human mesenchymal stem cells (hMSCs) (SI Appendix, Fig. S1 F and G and Table S1). These findings support the notion that cell fusion is the driver for type I IFN gene expression.
Activation of the cGAS–STING Pathway during Cell–Cell Fusion.
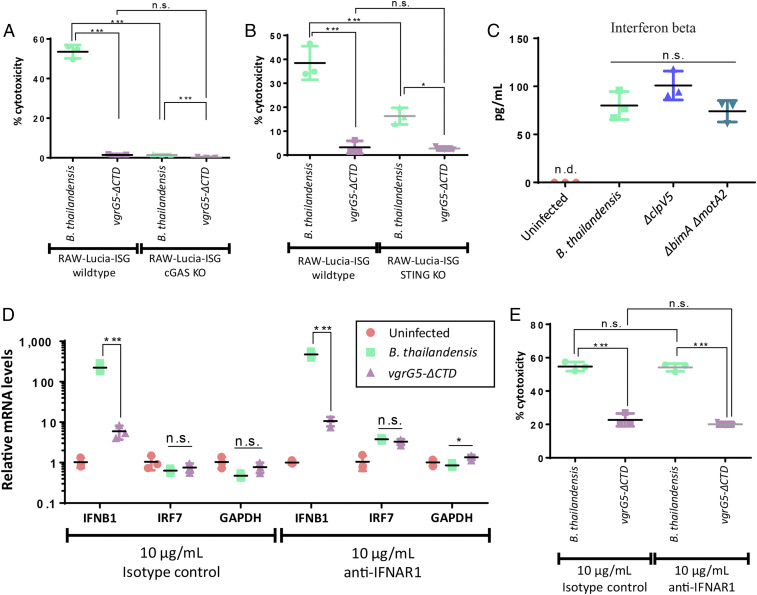
Induction of type I IFN gene expression can be driven by the activation of PRRs, including Toll-like receptors and other cytosolic nucleic acid-sensing receptors (38). Since T6SS5 expression and MNGC formation occurs after bacterial escape from the endosome into the cytosol (8), cytosolic PRRs may be involved in the up-regulation of type I IFN gene expression during cell–cell fusion. STING senses cyclic dinucleotides and has been reported to be involved in the induction of type I IFN through the sensing of membrane disturbance events, such as viral cell fusion (39, 40). STING knockout RAW macrophages infected with wild-type bacteria did not show up-regulation of IFNB1 gene expression compared to the infected wild-type macrophages although cell–cell fusion was extensive (Fig. 3 A and B). As STING also functions as an adaptor for cytosolic DNA sensors, we assessed the involvement of sequence-independent cytosolic DNA sensor cGAS, which functions upstream of STING. B. thailandensis infection in macrophages and their corresponding cGAS knockout cells led to extensive MNGC formation, whereas ∆clpV5 infection resulted in less MNGC formation and ∆vgrG5 was completely defective in cell–cell fusion (Fig. 3C). Significant up-regulation of IFNB1 gene expression was only observed in B. thailandensis infected wild-type cells but not B. thailandensis infected cGAS knockout cells (Fig. 3D).
Fig. 3.
cGAS and STING are required for cell fusion induction of type I IFN gene expression. (A) Light micrographs of Giemsa-stained RAW-Lucia-ISG wild-type and STING knockout cells infected with various B. thailandensis strains. Cells were infected at an MOI of 10 and were fixed for staining and visualization of MNGC formation under 10× objective magnification at 15 h postinfection. (Scale bar, 100 µm.) (B) Expression of IFNB1 in RAW-Lucia-ISG wild-type and STING knockout cells infected with B. thailandensis and various T6SS5 mutants. Cells were infected with B. thailandensis strains at an MOI of 10 for 15 h. An assay was performed in triplicate and the values are expressed as mean ± SD. (C) Light micrographs of Giemsa-stained RAW-Lucia-ISG wild-type and cGAS knockout cells infected with various B. thailandensis strains. Cells were infected at an MOI of 10 and were fixed for staining and visualization of MNGC formation under 10× objective magnification at 15 h postinfection. (Scale bar, 100 µm.) (D) Expression of IFNB1 in RAW-Lucia-ISG wild-type and cGAS knockout cells infected with B. thailandensis and various T6SS5 mutants. Cells were infected with B. thailandensis strains at an MOI of 10 for 15 h. An assay was performed in triplicate and the values are expressed as mean ± SD. Note: ∆vgrG5(a) is an insertional mutant while ∆vgrG5(b) is an in-frame deletion mutant. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; n.s., not significant.
We also utilized the TRAMP C2 prostate epithelial cell infection model with the corresponding STING knockout cells to determine the involvement of STING. B. thailandensis infection in wild-type and STING knockout cells resulted in the formation of MNGCs but not in T6SS5 ∆vgrG5-infected cells (SI Appendix, Fig. S2A and Table S1). The ∆clpV5 mutant-infected wild-type and STING knockout cells also formed MNGCs but to a lesser extent compared to those infected with B. thailandensis (SI Appendix, Fig. S2A and Table S1). This is expected, as ClpV is known to be nonessential for initial T6SS assembly and function but is required for disassembly and recycling of the T6SS sheath subunits after contraction, thereby showing an intermediate phenotype (24, 27, 41). The extent of cell–cell fusion corresponded to the up-regulation of IFNB1 gene expression in infected TRAMP C2 wild-type cells (SI Appendix, Fig. S2B). B. thailandensis-infected STING knockout cells had no significant up-regulation of IFNB1 gene expression despite forming MNGCs (SI Appendix, Fig. S2). Thus, cGAS and STING are stimulated downstream of MNGC formation and are required for cell–cell fusion-dependent IFNB1 gene expression.
Aberrant Cell Division and the DNA Damage Response after MNGC Formation.
To better understand factors that contribute to cGAS activation upon cell fusion, we employed time-lapse microscopy to image cell–cell fusion during B. thailandensis infection of HepG2 cells. We chose HepG2 cells as their larger size facilitated visualization. The extent of cell–cell fusion in infected HepG2 cells corresponded to the up-regulation of IFNB1 gene expression (SI Appendix, Fig. S3 A and B and Table S1). The bacteria had chromosomal insertion of the mApple gene encoding red fluorescence, whereas the cell periphery and nuclei were marked by CellMask and Hoechst 33342, respectively. At 14 h postinfection, we observed a huge MNGC containing highly condensed genetic material, some of which resembling mitotic chromosomes (Fig. 4A, orange asterisks, and Movie S1). Unlike regular mitosis, where mitotic chromosomes are drawn to the poles of the cell, the genetic content in the MNGC reshuffled without being pulled toward any direction. By 15 h 30 min postinfection, the genetic content of the MNGC became less condensed and three points of constriction at the membrane were observed (Fig. 4A, white arrowheads). The membrane constrictions, however, dissipated by 16 h 15 min and did not lead to the formation of daughter cells. These observations suggest that MNGCs were undergoing abortive cell division. Defective mitosis promotes DNA damage (42), which has been reported to lead to induction of type I IFN by the cGAS–STING pathway (43–46). Thus, we assessed indicators of the DNA damage response that are activated in response to DNA damage. Monoubiquitination of H2AX induced upon DNA double-strand breaks initiates DNA damage signaling through the recruitment of ataxia telangiectasia mutated (ATM). ATM in turn phosphorylates H2AX to form γ-H2AX, a biomarker for DNA damage (47). Higher levels of monoubiquitinated H2AX and γ-H2AX were detected in B. thailandensis-infected cells compared to cells infected with fusion defective vgrG5-∆CTD (Fig. 4B and SI Appendix, Fig. S3C). Monoubiquitinated γ-H2AX, a biomarker for double-stranded breaks, was more abundant in B. thailandensis-infected cells than vgrG5-∆CTD–infected cells (Fig. 4B and SI Appendix, Fig. S3C), suggesting that double-stranded breaks are the major form of DNA damage in the fused cells. We also treated cells with MitoQ, an inhibitor of mitochondrial stress, to determine whether mitochondria could be a source of DNA damage and leakage to trigger IFNB1 gene expression, but found no difference due to the treatment (SI Appendix, Fig. S3D). Together, these data demonstrate that cell–cell fusion triggers cGAS activation and subsequent type I IFN gene expression through DNA damage.
Fig. 4.
Burkholderia-induced MNGCs have increased genomic instability that can be sensed by cGAS. (A) Confocal micrographs of B. thailandensis tssA::mApple-infected HepG2 cells from 13 h 45 min to 17 h 30 min postinfection. Cell periphery and nuclei are labeled by CellMask and Hoechst, represented in magenta and blue respectively. Bacteria are represented in red. Orange asterisks indicate mitotic chromosomes, white arrows indicate points of constriction. Yellow arrows represent dissipation of membrane constriction. (Scale bar, 20 µm.) (B) Expression of DNA damage markers in fused cells. TRAMP C2 cells were infected with cell fusion proficient B. thailandensis and cell fusion-defective vgrG5-∆CTD at an MOI of 10. Total lysates were harvested at 18 h postinfection. Proteins were resolved on 4 to 15% SDS gel and analyzed via immunoblotting with antibodies against γ-H2AX and H2AX. Western blot against β-actin was included as a control. (C) Effect of cell–cell fusion induced by B. thailandensis infection on genomic instability. HepG2 cells were infected with B. thailandensis strains at an MOI of 50. Fused HepG2 cells infected with B. thailandensis and nonfused cells infected with vgrG5-∆CTD were examined for occurrence of micronuclei formation at 19 h postinfection. The occurrence of micronuclei was quantified as the number of micronuclei per nucleus examined and expressed as the mean from biological triplicates ± SD. Uninfected cells were included as a control. *P ≤ 0.05. (D–F) Confocal micrographs of B. thailandensis tssA::mApple infected HepG2 cells to visualize micronuclei. (D) Cells were infected with B. thailandensis tssA::mApple at an MOI of 50:1. Infected cells were fixed at 19 h postinfection for the labeling of host cell nuclei, nuclear lamin B1, and cell periphery. B. thailandensis express the fluorescent mApple protein. Cells were visualized under the confocal microscope at magnification of 63×. The nuclei are displayed in blue, nuclear lamin B1 in green, cell periphery in magenta, and bacteria in red. The overlay composite image presented was automatically generated by the ZEN software after image acquisition of a 5 × 5 tile scan. The zone of MNGC is highlighted by the white line. (Scale bar, 100 µm.) (E) Areas indicated by the yellow boxes in D were enlarged for the visualization of micronuclei. White arrows indicate ruptured micronuclei. Yellow asterisks indicate the absence of colocalization between DNA aggregates with nuclear lamin B1. (F) Areas indicated by the red box in the first panels of E were further enlarged for clearer visualization of micronuclei. White arrows indicate ruptured micronuclei. Absence of colocalization between DNA aggregates with nuclear lamin B1 are indicated with yellow asterisks. (G and H) Localization of cGAS in HepG2 cells infected with B. thailandensis and cell fusion-defective vgrG5-∆CTD. HepG2 cells were infected at an MOI of 50 and fixed at 19 h postinfection for immunostaining of cGAS, which are represented in green. Nuclei and cell periphery were labeled via Hoechst and CellMask, represented in blue and magenta, respectively. White arrows indicate colocalization between cGAS and micronuclei. Colocalization between DNA aggregates with cGAS are indicated with yellow asterisks. (G) Cells were visualized at a magnification of 63×. (Scale bar, 50 µm.) (H) Areas of colocalization of cGAS with micronuclei or DNA aggregates were enlarged from other fields of view of B. thailandensis infected HepG2 cells. (Scale bar, 20 µm.)
Cell Fusion Leads to Micronuclei Formation.
A hallmark of DNA damage and abortive cell division is genomic instability and formation of micronuclei (48–50). We examined the occurrence of micronuclei by staining B. thailandensis-infected HepG2 cells with nuclear DNA stain Hoechst 33342. Since cGAS has been reported to bind micronuclear DNA upon rupture and collapse of the nuclear envelope (28), we also labeled the infected cells with nuclear envelope marker, nuclear lamin B1, to assess nuclear envelope integrity. There was a significant increase in the number of micronuclei in B. thailandensis-infected MNGCs, compared to unfused vgrG5-∆CTD–infected or –uninfected cells (Fig. 4 C–F), indicating enhanced genomic instability in the fused cells likely due to mitotic errors. Immunofluorescence analysis with lamin B1 antibodies indicated that a subset of the micronuclei was ruptured, depicted by the lack of lamin B1 staining (Fig. 4 E and F). Interestingly, we also observed large DNA aggregates within the MNGCs that were not surrounded by a nuclear envelope (Fig. 4 E and F). Since cGAS can gain access to genetic material within micronuclei during nuclear membrane rupture, the lack of compartmentalisation of the large DNA aggregates in the cytosol may permit cGAS binding. Indeed, we observed that cGAS colocalized with a portion of the micronuclei as well as large DNA aggregates within B. thailandensis-infected cells that formed MNGCs (Fig. 4 G and H). We also observed cGAS colocalization with micronuclei and detected increased occurrence of micronuclei in PEG-6000–mediated fused cells compared to the unfused cells (SI Appendix, Fig. S3 E–G). Together, these data indicate that unnatural cell fusion leads to genomic instability and micronuclei formation.
cGAS–STING Activation Leads to Cell Death but Not Type I IFN Production.
cGAS–STING activation has been reported to lead to various modes of cell death (51–55). Hence, we examined the possibility of cell death in bacterial-induced fused cells. RAW cGAS and STING knockout cells infected with B. thailandensis had significantly less cell death as compared to B. thailandensis-infected wild-type cells (Fig. 5 A and B). This was also observed in TRAMP C2 cells, where B. thailandensis-infected STING knockout cells had lower cytotoxicity compared to B. thailandensis-infected wild-type cells (SI Appendix, Fig. S4A). Here, reinfection of the host cells was prevented by the addition of antibiotics in the media. Higher levels of cytotoxicity corresponded with lower intracellular bacterial loads, indicating a release of the bacteria into the media during cell death (SI Appendix, Fig. S4 B and C). To investigate whether the observed induction of cytotoxicity is dependent on type I IFN-mediated signaling, we examined the amount of IFN-β produced by the infected cells. Cells infected with B. thailandensis wild-type and cell fusion-defective strains ∆clpV5 and ∆bimA∆motA2 produced similar low concentrations of IFN-β (Fig. 5C) that did not correlate with the differential gene expression seen between wild-type and mutant infected cells. Levels of IFN regulatory factor 7 (IRF7) transcripts, which mediate type I IFN production and are up-regulated during type I IFN signaling, were also similar between B. thailandensis and vgrG5-∆CTD–infected cells despite the differential IFNB1 gene expression (Fig. 5D). These data show that cell fusion induces type I IFN gene expression but does not lead to downstream protein production or signaling. Blocking type I IFN receptor IFNAR1 during infection had no effect on cytotoxicity (Fig. 5E) and intracellular bacterial loads (SI Appendix, Fig. S4D). These data show that cell–cell fusion induces cell death via cGAS–STING signaling independent of type I IFN.
Fig. 5.
Cell fusion-triggered cGAS and STING activation leads to enhanced cytotoxicity independent of type I IFN signaling. (A and B) Effects of cell–cell fusion on cytotoxicity and its dependence on the cGAS–STING pathway. (A) RAW-Lucia-ISG wild-type and cGAS knockout macrophages were infected with B. thailandensis and cell fusion defective vgrG5-∆CTD at an MOI of 10. Cells were assessed for cytotoxicity via LDH assay at 18 h postinfection. (B) RAW-Lucia-ISG wild-type and STING knockout macrophages were infected with B. thailandensis and cell fusion defective vgrG5-∆CTD at an MOI of 10. Cells were assessed for cytotoxicity via LDH assay at 17 h postinfection. Assays were performed in triplicates and the values are expressed as mean ± SD. (C) IFN-β production in RAW264.7 macrophages infected with B. thailandensis and various T6SS5 mutants. Cells were infected at an MOI of 10 and assessed for IFN-β production via ELISA at 24 h postinfection. Assay was performed in triplicate and the values are expressed as mean ± SD. (D) Effects of IFNB1 gene induction on downstream IRF7 gene expression in RAW264.7 macrophages infected with B. thailandensis and cell fusion-defective vgrG5-∆CTD. Cells were infected at an MOI of 10:1 and subjected to type I IFN signaling blockade 8 h postinfection using anti-IFNAR1 receptor blocking antibody. Isotype-treated controls were included in the experiment. Transcripts were harvested 18 h postinfection. Transcript levels of IFNB1 and IRF7 were measured by qRT-PCR and normalized to uninfected cells under each treatment using the threshold cycle (2−ΔΔCt) method with β-actin as the reference gene. An assay was performed in triplicate and the values are expressed as mean ± SD. (E) Effects of type I IFN signaling on cytotoxicity in RAW264.7 macrophages infected with B. thailandensis and cell fusion-defective vgrG5-∆CTD. Cells were infected at an MOI of 10:1 and subjected to type I IFN signaling blockade 8 h postinfection using anti-IFNAR1 receptor blocking antibody. Isotype-treated controls were included in the experiment. Cells were assessed for cell death via LDH assay after 18 h of infection. Assay was performed in triplicate and the values are expressed as mean ± SD. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; n.s., not significant; n.d. not detected.
cGAS–STING-Dependent Autophagic Cell Death.
The cGAS–STING pathway has been shown to be essential for autophagic cell death during replicative crisis to limit genomic instability (51). The domain of STING responsible for TBK and IRF3 activation leading to type I IFN expression was also found to be decoupled from the region responsible for autophagy induction (55). Hence, we explored the possibility that cell–cell fusion triggers cytotoxicity through cGAS–STING-dependent autophagy. The conversion of LC3-I to LC3-II by phosphatidylethanolamine conjugation is accepted as a reliable measure for autophagosome formation, and therefore, indicative of autophagy activation (56). We found that B. thailandensis-infected cells displayed higher levels of LC3A-II/LC3A-I ratios than uninfected or vgrG5-∆CTD–infected cells (Fig. 6A). STING knockout cells infected with B. thailandensis did not exhibit higher LC3A-II/LC3AI ratios compared to wild-type cells (Fig. 6A), correlating with the much lower cytotoxicity in cGAS and STING knockout cells (Fig. 5 A and B and SI Appendix, Fig. S4A). In addition, PEG-induced cell fusion resulted in enhanced cytotoxicity, which corresponded with the increased ratio of lipidated LC3B-II to LC3B-I (SI Appendix, Fig. S4 E and F).
Fig. 6.
Cell–cell fusion leads to cGAS–STING-dependent autophagic cell death. (A) Induction of autophagy in TRAMP C2 cells infected with B. thailandensis and vgrG5-∆CTD. To investigate the induction of autophagy via STING pathway, TRAMP C2 and its corresponding STING KO cells were infected with cell fusion-proficient B. thailandensis and cell fusion-defective vgrG5-∆CTD at an MOI of 10. Total lysates were harvested at 22 h postinfection. Proteins were resolved on 4 to 15% SDS gel and analyzed via immunoblotting with antibodies against LC3A. Western blot against β-actin was included as a control. Intensity of the bands were assessed for biological triplicates via densitometric analysis and represented as mean LC3AII/LC3AI ratio or mean arbitrary units (AU) ± SD. (B) Effects of autophagy on cytotoxicity in HepG2 cells infected with B. thailandensis and cell fusion-defective vgrG5-∆CTD. HepG2 cells were transfected with NC-1 scrambled RNA or ATG-7 siRNA for knockdown of ATG-7. Transfected cells were infected at MOI of 50:1. Cells were assessed for cytotoxicity via LDH assay at 24 h postinfection. An assay was performed with biological triplicates and the values are expressed as mean ± SD. (C) Light micrographs of NC-1 scrambled RNA or ATG-7 siRNA-transfected HepG2 cells infected with B. thailandensis. Cells were infected at an MOI of 50 and were fixed for Giemsa staining and visualization of MNGC formation at 24 h postinfection. (Scale bars, 100 µm.) *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; n.s., not significant.
To examine if the induction of autophagy during cell fusion leads to cell death, we performed small-interfering RNA (siRNA) knockdown of an essential autophagy gene, ATG-7, and subjected the cells to B. thailandensis infection. Knockdown of ATG-7 resulted in a significant reduction in cytotoxicity during B. thailandensis infection as compared to controls transfected with scrambled RNA (Fig. 6B). Transfection with scrambled RNA or ATG-7 did not affect MNGC formation during B. thailandensis infection (Fig. 6C). Inhibition of autophagy with bafilomycin A1 in B. thailandensis-infected wild-type cells also resulted in reduced cytotoxicity as compared to negative-control DMSO-treated cells, indicating that autophagy contributes to fusion-dependent cell death (SI Appendix, Fig. S4G). These data confirm that autophagic cell death occurs during cell fusion.
cGAS KO Mice Showed Slightly Delayed Death from a Lower Dose Infection.
We assessed survival of cGAS knockout mice following acute and chronic infection. During acute infection, no differences in mortality were observed between wild-type and cGAS knockout mice (SI Appendix, Fig. S5). In contrast, cGAS knockout mice clearly survived longer during a more chronic infection, although statistical analyses via the Gehan–Breslow–Wilcoxon (P = 0.4471) test revealed the differences to be not significant. Our results suggest that the cGAS–STING pathway may paradoxically contribute to increased bacterial pathogenesis, perhaps by hastening cell death and bacterial spread through release of intracellular bacteria only in a low-dose infection, and even in this scenario, it is clearly still not the major pathway controlling disease.
Discussion
B. pseudomallei is a facultative intracellular pathogen responsible for causing melioidosis that is estimated to kill 89,000 people worldwide per year (1). After the bacteria have escaped from endosomal compartments to the cytosol, B. pseudomallei triggers infected cells to undergo fusion with its neighboring cells (10). The induction of cell–cell fusion by Burkholderia has been traditionally regarded as a Trojan horse-like strategy for the bacteria to hide from immune detection while allowing localized bacterial spread during chronic infection (6). Contrary to this well-accepted paradigm, we discovered that Burkholderia-induced MNGCs are not quiescent affairs that go undetected. They are in fact triggering the activation of the host DNA-sensing cGAS–STING pathway, leading to autophagic cell death. The activation of the cGAS–STING pathway by MNGCs occur through DNA damage and micronuclei formation. Our discovery of the molecular mechanisms pertaining to Burkholderia-induced cell fusion also extends to chemically induced cell fusion, as well as different cell types, including primary hMSCs, demonstrating that this is potentially a common mechanism triggered by substantial cell–cell fusion.
Prior to our work, membrane perturbation triggered by liposome and enveloped virus cell fusion have been suggested as a danger signal leading to STING activation (39, 40). However, the mechanism of STING activation by these modes of membrane disturbances is undefined. We found that fused cells have DNA damage and enhanced genomic instability that in recent years have been shown to be detected by cGAS–STING (28, 43, 48–51, 55, 57, 58). Our observations that MNGCs undergo abortive cell division (Fig. 4A and Movie S1) shows that DNA damage is introduced when cell–cell fusion occurs between nonsynchronized cells. As liposome-fusion and enveloped virus–cell fusion do not form heterokaryons, leading to deregulated cell cycle, it is improbable that these modes of membrane disturbances trigger STING via the same mechanism. While multinucleated cells within granulomas in Mycobacterium bovis Bacillus Calmette-Guerin (bacillus Calmette–Guérin) and Mycobacterium tuberculosis animal models have also been described to harbor double-stranded breaks, the MNGCs formed were due to failure of cytokinesis or endoreplication and not cell–cell fusion (59). Here, it is important to note the crucial difference that Mycobacteria have no inherent ability to fuse cells, whereas Burkholderia have. It remains to be investigated whether such polykaryons could trigger a similar cGAS–STING activation and their downstream responses.
There had been previous reports of chemically induced fused cells or virus-induced syncytia with genomic alterations including DNA breaks and chromosomal instability (60–62). MNGCs in human melioidosis patients were also observed to have abnormal genetic material including diffused chromatin and nuclei which appeared polylobated with jagged edges (16), similar to what we observed in our B. thailandensis in vitro HepG2 infection model. Literature in the 1960s and 1970s had reported premature chromosome condensation and pulverisation in asynchronously fused cells (63–65), events where DNA damage could possibly be introduced. Accumulation of DNA damage and genomic instability increases the chances of malignant transformation. In fact, cell fusion events have been proposed to facilitate the acquisition of malignant properties. This is supported by reports that the progeny of fused cells have enhanced metastatic capabilities and are more tumorigenic (66–71).
On the other hand, one could argue that cell fusions contribute to the natural defense against malignancies through cGAS–STNG activation. It is increasingly clear that cancer cells undergoing cell cycle arrest or replicative crisis harbor DNA damage and various chromosomal abnormalities and instability that could lead to cell death. The triggering of type I IFN by the cGAS–STING pathway has been reported to trigger tumor immunity (43, 57, 72–78). Transformed cells have also been recently described to undergo autophagic cell death via the cGAS–STING pathway through a mechanism distinct from type I IFN induction (55). Mirroring this finding, we demonstrated that cell–cell fusion also leads to activation of autophagy and corresponding cell death through the cGAS–STING pathway, independent of type I IFN. The extensive genomic instability likely shuts down protein synthesis as the cells undergo autophagic cell death. It is thus possible that Burkholderia-induced cell fusions increase the risk of malignant cellular transformation through our proposed mechanism and the cGAS–STING pathway functions to limit this process. Hence, defects in the cGAS–STING pathway could potentially predispose the host to cancer development in circumstances where unnatural cell–cell fusions occur, including virus-induced cell–cell fusions such as those mediated by HIV.
B. pseudomallei and B. thailandensis are environmental saprophytes and accidental human pathogens. What could be the evolutionary advantage of maintaining an energetically expensive secretion system that fuses mammalian cells? The bacterium’s natural habitat in the soil could expose it to unicellular organisms, like amoebas living in a colony, that would resemble a multicellular organism. Burkholderia’s T3SS3 and T6SS5 could have evolved with the adaptation to escape amoebic defenses. The ability to fuse a colony of unicellular amoebas to form a multinucleated giant amoeba would seem to be a clever solution to expand its infection radius and access the cytosolic compartments directly without multiple rounds of exit and entry through phagosomes. Although the cGAS–STING pathway is majorly involved in type I IFN responses in vertebrates, the C-terminal tail of STING critical for recruitment of type I IFN signaling components TBK1 and IRF3 is absent in invertebrates (79, 80). It is therefore postulated that type I IFN induction is a relatively recently emerged function of cGAS–STING signaling (79). The ancestral function of STING is believed to be the induction of autophagy as STING from the primitive invertebrate, sea anemone, has been shown to induce autophagic cell death but not type I IFN production (55). Multicellular mammalian hosts have evolved the cGAS–STING pathway to deal with cytosolic DNA incursions, whether from genomic instability or foreign DNA to induce type I IFN, but also preserved the initiation of autophagic cell death under catastrophic circumstances. Given that Burkholderia has not evolved and adapted to encounter with mammalian hosts, the subsequent cGAS–STING-triggered host cell death and outcome may not have adapted to either the bacterial or host’s advantage. This could also explain the relatively weak effect in pathogenesis in infected cGAS knockout mice compared to wild-type mice.
In summary, we have discovered an instance of a pathogen-triggering micronuclei formation through the induction of host cell fusion, thereby activating the cGAS–STING pathway and subsequent cell death (SI Appendix, Fig. S6). We propose that during infection or fusogenic chemical exposure, cell–cell fusion is a danger signal sensed by host innate immune sensors cGAS and STING. The body likely exhibits a graded response; smaller cytosolic incursions by pathogens result in an efficient type I IFN immune response and autophagy that could serve to remove cytosolic pathogens upon cGAS activation. However, extensive cell fusion is a catastrophic event resulting in aberrant and abortive mitosis, which leads to DNA damage and genomic instability. This in turn triggers autophagic cell death instead of a productive type I IFN response, with the end goal of limiting cellular transformation. It awaits to be seen whether chronic melioidosis patients show increased susceptibility to cancer and if so, whether these patients have mutations in the cGAS–STING or autophagy pathways that would attenuate the body’s natural defenses against unwelcomed cellular fusions.
Materials and Methods
Bacterial Strains and Construction of Mutants.
All bacterial strains used in this study are listed in SI Appendix, Table S2. Bacterial strains were routinely maintained on lysogeny broth (LB) agar. Mutants were derived from the E264 strain via the allelic exchange method. Plasmids and primers used for the generation of mutants are listed in SI Appendix, Tables S2 and S3, respectively. Details of mutant construction are in SI Appendix.
Cell Infection and Treatments.
See SI Appendix for details of cell culture and maintenance.
As T6SS5 expression levels are higher when the bacteria are grown in RPMI1640 (Gibco) as compared to DMEM, the media was changed to RPMI containing 10% FBS prior to the infection. Cells were infected with stationary overnight cultures of Burkholderia strains at the indicated MOIs and centrifuged at 250 × g for 5 min at room temperature. After 1 to 2 h of infection, kanamycin (Sigma) was added to kill off extracellular bacteria at final concentrations of 250 µg/mL and 300 µg/mL for B. pseudomallei and B. thailandensis, respectively. For infections involving HepG2 cells, the bacteria were opsonised with 50% human serum (Sigma) prior to infection for 1 h at 37 °C with shaking.
To assess the effect of late-phase type I IFN response, cells were treated with 10 μg/mL anti-IFNAR1 receptor-blocking antibody (eBioscience, 16−5945−85) or its corresponding isotype control (eBioscience, 16−4714−82) at 8 h postinfection.
For chemically induced cell–cell fusions, cells were incubated with 50% (wt/vol) PEG-6000 or D-PBS as a negative control for 2 min at 37 °C, 5% CO2. After the treatment, the cells were washed with growth media thrice to remove residual PEG-6000.
ATG-7 Knockdown in HepG2 Cells.
HepG2 cells were concurrently seeded and transfected with 30 nM ATG-7 siRNA (hs.Ri.ATG7.13.1, Integrated DNA Technologies) or scrambled RNA (NC-1 DsiRNA, Integrated DNA Technologies). After 3 d, transfected cells were trypsinized for reseeding and retransfection with 30 nM ATG-7 siRNA or NC-1 scrambled RNA. After 3 d, the cells were used for B. thailandensis infection at the indicated MOI.
Analysis of Gene Expression by Real-Time PCR.
Total RNA was isolated using PureZOL (Bio-Rad) and illustra RNAspin Mini Kit (GE Healthcare). cDNA was synthesized using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). Transcripts were quantified using iQ SYBR Green Supermix (Bio-Rad) in a CFX Connect real-time PCR detection system (Bio-Rad). PCR primers are listed in SI Appendix, Table S4. For host genes, relative RNA levels of a particular gene were normalized to D-PBS–treated or uninfected controls using the threshold cycle (2−ΔΔCt) method with β-actin as the reference gene. Housekeeping gene GADPH was used an internal control. For bacterial genes, relative RNA levels of the genes in mutant strains were normalized to the wild-type using the 2−ΔΔCt method with 16S as the reference gene. rpoB was used as an internal control.
Giemsa Staining for Viewing MNGCs.
Cells were fixed with 100% methanol (Fischer Scientific) for 2 min, washed once with distilled water, and stained with 20× diluted Giemsa stain (Sigma) for 15 min. After staining, cells were washed once with distilled water and air-dried. Stained cells were examined with light microscopes (Olympus Model CKX53 or IX51) and images taken using Lumenera InfinityX Microscope Camera and Infinity Capture software at an objective magnification of 10×.
Cytotoxicity Assay.
Cell cytotoxicity was measured by assaying for lactate dehydrogenase (LDH) release in the cell supernatants using an LDH assay kit (Clontech).
Live Confocal Imaging.
HepG2 cells were infected with mApple fluorescent B. thailandensis at an MOI of 50. Kanamycin was added to a final concentration of 300 µg/mL at 1 h postinfection. Prior to imaging, the media was replaced to RPMI containing 10% FBS, 10 µg/mL Hoechst 33342, and 0.1 µL/mL CellMask Deep Red Plasma membrane stain. Cells were visualized using LSM710 (Zeiss) at a magnification of 63× from 13 h 45 min to 17 h 30 min postinfection at intervals of 15 min.
Immunofluorescence.
Cells were fixed in 4% paraformaldehyde (Sigma) for 15 min at room temperature. Cells were then permeabilized in 0.25% (vol/vol) Triton-X for 10 min at room temperature and blocked in staining buffer, 5% FBS, 2% BSA (HyClone), and 0.1% (vol/vol) Triton-X (Merck) in 1× PBS for 30 min at room temperature. For visualization of nuclear envelope, cells were stained with rabbit anti-lamin B1 antibody (ab16048, Abcam) overnight at 4 °C and was subsequently stained with secondary antibody Alexa Fluor 488 anti-rabbit antibody (Invitrogen) at room temperature for 2 h. For cGAS localization to micronuclei, cells were stained with rabbit anti-cGAS antibody (D1D3G, Cell Signaling Technologies) overnight at 4 °C and followed by Alexa Fluor 555 anti-rabbit antibody (Invitrogen) at room temperature for 2 h. Cells were then stained with 10 μg/mL Hoechst 33342 (ThermoScientific) and 1:10,000 diluted CellMask Deep Red Plasma membrane stain (Invitrogen). In between each incubation step, the cells were washed with 0.1% Triton-X (Merck). The coverslips were mounted onto clean glass slides with ProLong Gold Antifade (Invitrogen) and imaged with Zeiss LSM710 Confocal microscope using a 63× objective lens.
Quantification of Micronuclei.
Confocal micrographs of fused cells were assessed for the occurrence of micronuclei. Only MNGCs comprising greater than five nuclei were examined. Uninfected or D-PBS–treated cells were included as a negative control. Micronuclei were counted manually according to these criteria: 1) micronuclei are less than one-third the size of nucleus but large enough to discern shape and color; 2) micronuclei are round in shape; 3) micronuclei are located within cell boundary as demarcated by cell mask stain; and 4) micronuclei boundary is distinguishable from nuclear boundary. Total number of nuclei assessed were enumerated for normalization across conditions. At least 150 nuclei were examined in each condition. Occurrence of micronuclei is represented as the number of micronuclei per nucleus examined. cGAS localization to micronuclei was determined visually by the complete overlay of cGAS signals with micronuclei.
ELISA.
Culture supernatants were obtained for determining IFN-β production by ELISA using the LEGEND MAX kit (Biolegend), according to manufacturer’s instructions in triplicates.
Immunoblotting.
Cells were lysed in 0.5% (vol/vol) Triton-X supplemented with 1× protease phosphatase inhibitor mixture (Thermo Scientific). Lysates were spun down at 6,500 × g for 10 min at 4 °C. Supernatant fractions were boiled in 1× SDS laemmli buffer (Bio-Rad). Histone acid extraction was conducted overnight at 4 °C using 0.2N HCl (Spectrum chemical) on the pellet, which contains intact nuclei. Nuclear debris were removed through centrifugation at 6,500 × g for 10 min at 4 °C. Histones were neutralized with 2M NaOH (Merck) before subjecting to boiling in 1× SDS laemmli buffer (Bio-Rad). Proteins were resolved on 4 to 15% SDS/PAGE gels (Bio-Rad) and transferred onto nitrocellulose membranes (Thermo Scientific). Membranes were blocked with 5% BSA at room temperature for 2 h and probed with the specific antibodies overnight at 4 °C. Anti-H2AX, anti–γ-H2AX, and anti–β-actin antibodies were obtained from Invitrogen, Biolegend, and Sigma, respectively. Secondary antibodies HRP-linked anti-mouse antibody and HRP-linked anti-rabbit antibody were obtained from Abcam and Cell Signaling Technology, respectively.
For LC3A detection in TRAMP C2-infected cells, cells were lysed and processed as described in SI Appendix.
All blots were developed using ECL plus reagent (Thermo Scientific) and bands were visualized using ChemiDoc MP Imaging System (Bio-Rad). Densitometric analysis was conducted using ImageJ.
Statistical Analysis.
Student’s two-tailed unpaired t test was conducted to determine statistical significance of 95% confidence between two samples compared. Statistical significance of 95% confidence between three or more groups were determined by ANOVA followed by Tukey’s test. All statistics were conducted using the GraphPad Prism software. Statistical significance is indicated as follows: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Ethical Statement.
Animal studies were ethically reviewed and approved by the National University of Singapore Institutional Animal Care and Use Committee, according to the National Advisory Committee for Laboratory Animal Research guidelines. The National University of Singapore is an American Association for Laboratory Animal Care International-accredited institution (Frederick, MD).
Data Availability.
All data are available in the main text and SI Appendix.
Supplementary Material
Acknowledgments
We thank Jeff Miller and Christopher Todd French (University of California, Los Angeles) for providing us with Burkholderia thailandensis strains; Lee Shu Ying (National University of Singapore) for providing support on the microscopy imaging; Mok Chee Keng (National University of Singapore BSL-3 Core Facility) for rendering assistance on the ABSL-3 experiments; Hayden Tan (National University of Singapore) for advising us on the siRNA knockdown procedure; and Heng-Phon Too (National University of Singapore) for the kind gift of human mesenchymal stem cells. This work was supported by the Singapore National Medical Research Council (NMRC/CBRG/0052/2013), Ministry of Education (MOE2018-T3-1-003) and Yong Loo Lin School of Medicine BSL-3 Core Facility and National Medical Research Council, Centre Grant MINE, Research Core no. 4 (NMRC/CG/013/2013). This research was also funded by the Singapore Ministry of Education under its Singapore Ministry of Education Academic Research Fund Tier 2 (MOE2018-T2-2-179) to K.C.C.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006908117/-/DCSupplemental.
Change History
June 9, 2021: The Acknowledgments have been updated.
References
- 1.Wiersinga W. J.et al., Melioidosis. Nat. Rev. Dis. Primers 4, 17107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Limmathurotsakul D.et al., Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat. Microbiol. 1, 15008 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Jones A. L., Beveridge T. J., Woods D. E., Intracellular survival of Burkholderia pseudomallei. Infect. Immun. 64, 782–790 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whiteley L.et al., Entry, intracellular survival, and multinucleated-giant-cell-forming activity of Burkholderia pseudomallei in human primary phagocytic and nonphagocytic cells. Infect. Immun. 85, e00468-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pruksachartvuthi S., Aswapokee N., Thankerngpol K., Survival of Pseudomonas pseudomallei in human phagocytes. J. Med. Microbiol. 31, 109–114 (1990). [DOI] [PubMed] [Google Scholar]
- 6.Kespichayawattana W., Rattanachetkul S., Wanun T., Utaisincharoen P., Sirisinha S., Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: A possible mechanism for cell-to-cell spreading. Infect. Immun. 68, 5377–5384 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevens M. P.et al., An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46, 649–659 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Wong J., Chen Y., Gan Y. H., Host cytosolic glutathione sensing by a membrane histidine kinase activates the type VI secretion system in an intracellular bacterium. Cell Host Microbe 18, 38–48 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Burtnick M. N.et al., The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 79, 1512–1525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.French C. T.et al., Dissection of the Burkholderia intracellular life cycle using a photothermal nanoblade. Proc. Natl. Acad. Sci. U.S.A. 108, 12095–12100 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwarz S.et al., VgrG-5 is a Burkholderia type VI secretion system-exported protein required for multinucleated giant cell formation and virulence. Infect. Immun. 82, 1445–1452 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toesca I. J., French C. T., Miller J. F., The type VI secretion system spike protein VgrG5 mediates membrane fusion during intercellular spread by pseudomallei group Burkholderia species. Infect. Immun. 82, 1436–1444 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevens M. P.et al., Identification of a bacterial factor required for actin-based motility of Burkholderia pseudomallei. Mol. Microbiol. 56, 40–53 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Breitbach K.et al., Actin-based motility of Burkholderia pseudomallei involves the Arp 2/3 complex, but not N-WASP and Ena/VASP proteins. Cell. Microbiol. 5, 385–393 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Benanti E. L., Nguyen C. M., Welch M. D., Virulent Burkholderia species mimic host actin polymerases to drive actin-based motility. Cell 161, 348–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong K. T., Puthucheary S. D., Vadivelu J., The histopathology of human melioidosis. Histopathology 26, 51–55 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Conejero L.et al., Low-dose exposure of C57BL/6 mice to burkholderia pseudomallei mimics chronic human melioidosis. Am. J. Pathol. 179, 270–280 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coulthurst S. J., The Type VI secretion system—A widespread and versatile cell targeting system. Res. Microbiol. 164, 640–654 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Cascales E., Cambillau C., Structural biology of type VI secretion systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1102–1111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zoued A.et al., Architecture and assembly of the type VI secretion system. Biochim. Biophys. Acta 1843, 1664–1673 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Brunet Y. R., Hénin J., Celia H., Cascales E., Type VI secretion and bacteriophage tail tubes share a common assembly pathway. EMBO Rep. 15, 315–321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leiman P. G.et al., Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 106, 4154–4159 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanamaru S., Structural similarity of tailed phages and pathogenic bacterial secretion systems. Proc. Natl. Acad. Sci. U.S.A. 106, 4067–4068 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basler M., Pilhofer M., Henderson G. P., Jensen G. J., Mekalanos J. J., Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunet Y. R., Espinosa L., Harchouni S., Mignot T., Cascales E., Imaging type VI secretion-mediated bacterial killing. Cell Rep. 3, 36–41 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Kostyuchenko V. A.et al., The tail structure of bacteriophage T4 and its mechanism of contraction. Nat. Struct. Mol. Biol. 12, 810–813 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Bönemann G., Pietrosiuk A., Diemand A., Zentgraf H., Mogk A., Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28, 315–325 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackenzie K. J.et al., cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo S. R.et al., STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 41, 830–842 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao D.et al., Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341, 903–906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lam E., Stein S., Falck-Pedersen E., Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade. J. Virol. 88, 974–981 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dansako H.et al., The cyclic GMP-AMP synthetase-STING signaling pathway is required for both the innate immune response against HBV and the suppression of HBV assembly. FEBS J. 283, 144–156 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Watson R. O.et al., The cytosolic sensor cGAS detects Mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host Microbe 17, 811–819 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen K.et al., Listeria monocytogenes induces IFNβ expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J. 33, 1654–1666 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y.et al., Regulation of type VI secretion system during Burkholderia pseudomallei infection. Infect. Immun. 79, 3064–3073 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pissaridou P.et al., The Pseudomonas aeruginosa T6SS-VgrG1b spike is topped by a PAAR protein eliciting DNA damage to bacterial competitors. Proc. Natl. Acad. Sci. U.S.A. 115, 12519–12524 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bondage D. D., Lin J. S., Ma L. S., Kuo C. H., Lai E. M., VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor-effector complex. Proc. Natl. Acad. Sci. U.S.A. 113, E3931–E3940 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu J., Chen Z. J., Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32, 461–488 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Holm C. K.et al., Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat. Immunol. 13, 737–743 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hare D. N.et al., Membrane perturbation-associated Ca2+ signaling and incoming genome sensing are required for the host response to low-level enveloped virus particle entry. J. Virol. 90, 3018–3027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng J., Ho B., Mekalanos J. J., Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One 6, e23876 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.S Pedersen R.et al., Profiling DNA damage response following mitotic perturbations. Nat. Commun. 7, 13887 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lam A. R.et al., RAE1 ligands for the NKG2D receptor are regulated by STING-dependent DNA sensor pathways in lymphoma. Cancer Res. 74, 2193–2203 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lan Y. Y., Londoño D., Bouley R., Rooney M. S., Hacohen N., Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Rep. 9, 180–192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahn J.et al., Inflammation-driven carcinogenesis is mediated through STING. Nat. Commun. 5, 5166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Härtlova A.et al., DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 42, 332–343 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Wu C. Y.et al., Critical role of monoubiquitination of histone H2AX protein in histone H2AX phosphorylation and DNA damage response. J. Biol. Chem. 286, 30806–30815 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harding S. M.et al., Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crasta K.et al., DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bakhoum S. F.et al., Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nassour J.et al., Autophagic cell death restricts chromosomal instability during replicative crisis. Nature 565, 659–663 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zierhut C.et al., The cytoplasmic DNA sensor cGAS promotes mitotic cell death. Cell 178, 302–315.e23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brault M., Olsen T. M., Martinez J., Stetson D. B., Oberst A., Intracellular nucleic acid sensing triggers necroptosis through synergistic type I IFN and TNF signaling. J. Immunol. 200, 2748–2756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaidt M. M.et al., The DNA inflammasome in human myeloid cells is initiated by a STING-cell death program upstream of NLRP3. Cell 171, 1110–1124.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gui X.et al., Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567, 262–266 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kabeya Y.et al., LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen Y. J.et al., Genome-derived cytosolic DNA mediates type I interferon-dependent rejection of B cell lymphoma cells. Cell Rep. 11, 460–473 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Le Bert N.et al., STING-dependent cytosolic DNA sensor pathways regulate NKG2D ligand expression. OncoImmunology 3, e29259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herrtwich L.et al., DNA damage signaling instructs polyploid macrophage fate in granulomas. Cell 167, 1264–1280.e18 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Sorimachi K., Naora H., Akimoto K., Niwa A., Naora H., Multinucleation and preservation of nucleolar integrity of macrophages. Cell Biol. Int. 22, 351–357 (1998). [DOI] [PubMed] [Google Scholar]
- 61.Duelli D. M.et al., A virus causes cancer by inducing massive chromosomal instability through cell fusion. Curr. Biol. 17, 431–437 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Nichols W. W., Levan A., Aula P., Norrby E., Chromosome damage associated with the measles virus in vitro. Hereditas 54, 101–118 (1965). [DOI] [PubMed] [Google Scholar]
- 63.Yanishevsky R. M., Prescott D. M., Late S phase cells (Chinese hamster ovary) induce early S phase DNA labeling patterns in G1 phase nuclei. Proc. Natl. Acad. Sci. U.S.A. 75, 3307–3311 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rao P. N., Johnson R. T., Mammalian cell fusion: Studies on the regulation of DNA synthesis and mitosis. Nature 225, 159–164 (1970). [DOI] [PubMed] [Google Scholar]
- 65.Takagi N., Aya T., Kato H., Sandberg A. A., Relation of virus-induce cell fusion and chromosome pulverization to mitotic events. J. Natl. Cancer Inst. 43, 335–347 (1969). [PubMed] [Google Scholar]
- 66.Goldenberg D. M.et al., Horizontal transmission and retention of malignancy, as well as functional human genes, after spontaneous fusion of human glioblastoma and hamster host cells in vivo. Int. J. Cancer 131, 49–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jacobsen B. M.et al., Spontaneous fusion with, and transformation of mouse stroma by, malignant human breast cancer epithelium. Cancer Res. 66, 8274–8279 (2006). [DOI] [PubMed] [Google Scholar]
- 68.Miller F. R., Mohamed A. N., McEachern D., Production of a more aggressive tumor cell variant by spontaneous fusion of two mouse tumor subpopulations. Cancer Res. 49, 4316–4321 (1989). [PubMed] [Google Scholar]
- 69.Lu X., Kang Y., Cell fusion as a hidden force in tumor progression. Cancer Res. 69, 8536–8539 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu X., Kang Y., Efficient acquisition of dual metastasis organotropism to bone and lung through stable spontaneous fusion between MDA-MB-231 variants. Proc. Natl. Acad. Sci. U.S.A. 106, 9385–9390 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou X.et al., Cell fusion connects oncogenesis with tumor evolution. Am. J. Pathol. 185, 2049–2060 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marcus A.et al., Tumor-derived cGAMP triggers a STING-mediated interferon response in non-tumor cells to activate the NK cell response. Immunity 49, 754–763.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deng L.et al., STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity 41, 843–852 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang H.et al., cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc. Natl. Acad. Sci. U.S.A. 114, 1637–1642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Demaria O.et al., STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc. Natl. Acad. Sci. U.S.A. 112, 15408–15413 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ohkuri T.et al., STING contributes to antiglioma immunity via triggering type I IFN signals in the tumor microenvironment. Cancer Immunol. Res. 2, 1199–1208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ho S. S.et al., The DNA structure-specific endonuclease MUS81 mediates DNA sensor STING-dependent host rejection of prostate cancer cells. Immunity 44, 1177–1189 (2016). [DOI] [PubMed] [Google Scholar]
- 78.Shen Y. J.et al., Genome-derived cytosolic DNA contributes to type I interferon expression and immunogenicity of B-cell lymphoma cells. Cytokine 76, 581–582 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Margolis S. R., Wilson S. C., Vance R. E., Evolutionary origins of cGAS-STING signaling. Trends Immunol. 38, 733–743 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Kranzusch P. J.et al., Ancient origin of cGAS-STING reveals mechanism of universal 2′,3′ cGAMP signaling. Mol. Cell 59, 891–903 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text and SI Appendix.