Abstract
Study Objectives
Multisensor wearable consumer devices allowing the collection of multiple data sources, such as heart rate and motion, for the evaluation of sleep in the home environment, are increasingly ubiquitous. However, the validity of such devices for sleep assessment has not been directly compared to alternatives such as wrist actigraphy or polysomnography (PSG).
Methods
Eight participants each completed four nights in a sleep laboratory, equipped with PSG and several wearable devices. Registered polysomnographic technologist-scored PSG served as ground truth for sleep–wake state. Wearable devices providing sleep–wake classification data were compared to PSG at both an epoch-by-epoch and night level. Data from multisensor wearables (Apple Watch and Oura Ring) were compared to data available from electrocardiography and a triaxial wrist actigraph to evaluate the quality and utility of heart rate and motion data. Machine learning methods were used to train and test sleep–wake classifiers, using data from consumer wearables. The quality of classifications derived from devices was compared.
Results
For epoch-by-epoch sleep–wake performance, research devices ranged in d′ between 1.771 and 1.874, with sensitivity between 0.912 and 0.982, and specificity between 0.366 and 0.647. Data from multisensor wearables were strongly correlated at an epoch-by-epoch level with reference data sources. Classifiers developed from the multisensor wearable data ranged in d′ between 1.827 and 2.347, with sensitivity between 0.883 and 0.977, and specificity between 0.407 and 0.821.
Conclusions
Data from multisensor consumer wearables are strongly correlated with reference devices at the epoch level and can be used to develop epoch-by-epoch models of sleep–wake rivaling existing research devices.
Keywords: machine learning, big data, polysomnography, actigraphy, smartphone, wearable, artificial intelligence
Statement of Significance.
Inexpensive and accessible multisensor wearable devices that allow for the collection of data relevant to sleep assessment, such as triaxial accelerometer signals and heart rate, are increasingly common. Sleep–wake classifications from several wrist actigraph devices were compared against ground truth classifications derived from polysomnography. Machine learning was subsequently used to develop a novel classification algorithm of sleep–wake, using data from consumer multisensor wearables. The model that was developed using data from wearable devices generated more accurate epoch-by-epoch sleep–wake classifications than existing wrist actigraph devices, suggesting that such consumer devices may allow for the study of sleep among a more general population and in more ecologically valid scenarios than was previously possible.
Introduction
The use of wrist-worn actigraphy, or watch-like devices sensitive to motion, to distinguish sleep from wake has been explored for over 40 years. The performance of a device to distinguish sleep from wake can be considered at two levels of analysis: the epoch level, defined as the ability of a device to correctly classify each sleep epoch (typically 30 s) within the night, and the night level, i.e. the ability of the device to summarize the entire night of sleep.
Kupfer et al. [1] initially observed that at the night level, movement counts derived from a wrist-worn actigraphy device were highly correlated with movement counts derived from polysomnography (PSG). Wrist actigraphy has since emerged as a popular alternative to PSG because of the low obtrusiveness, more manageable cost, and adequate accuracy for monitoring sleep–wake states determined by PSG, the gold standard for monitoring sleep [2]. Despite actigraphy’s long history and excellent ecological monitoring utility, however, devices that focus on motion-sensing have common limitations in wake detection using current algorithms [3].
Human scorers of PSG or wrist actigraphy generate similar night-level summary statistics for sleep–wake, such as the total minutes of sleep within the night [4], and can discriminate sleep from wake based on actigraphy data for as narrow as 1-min sleep durations with high accuracy [5]. Automated staging algorithms using actigraphy data now classify sleep–wake, although with the continued need for a human scorer to define the “time in bed” period typically corroborated using sleep diaries [6–8].
Common algorithms using wrist actigraphy data appear biased toward the classification of sleep [3], with poorer accuracy for the detection of wakefulness. In terms of Signal Detection Theory treating sleep as the “target” to be detected within a sleep–wake classifier, actigraphy tends to be more accurate at correctly classifying periods of sleep (classifier sensitivity) than correctly classifying periods of wake (classifier specificity) [3]. In practice, this detection imbalance means that actigraphy-based sleep–wake classifiers misclassify a greater number of wake epochs as sleep than vice-versa, leading to an overclassification of sleep. As a result, overall classification performance will be poorer for nights or individuals with a greater amount of wakefulness. For example, among individuals with sleep disorders, the bias against detecting wake is especially pronounced, with wakefulness detection of only 35%–50% [3, 9–14]. As poor sleep impacts many aspects of daily life, including work productivity [15–20], the risk of accidents [21–25], chronic disease risk, and health care costs [26–30], improving the assessment of sleep–wake outside of a laboratory environment is critical.
Commonly used actigraphy-based sleep–wake algorithms include the “Cole–Kripke” [6] and “Sadeh” [31] approaches, which use statistics on the motion from both the epoch being classified and from epochs several minutes into the past and future. As wrist actigraphy is conventionally implemented on a device dedicated to monitoring motion, some of the limitations in detecting periods of wake may be attributable to the use of a single monitoring modality. Many consumer wearable devices such as “smartwatches” contain both triaxial accelerometers and other sensor modalities. With their increasing ubiquity, there is an opportunity to noninvasively assess the synergy of multi-modal or multi-device-informed algorithm performance in sleep–wake classification.
Although sleep stages are typically assessed via changes in the central nervous system (CNS), as measurable with electroencephalography (EEG), the activity of the autonomic nervous system (ANS) also varies by sleep stage and is measurable through changes in heart rate and heart rate variance over time. For example, de Zambotti et al. [32] describe in a recent review the links between CNS and ANS activity with respect to sleep. The newest generation of wearable devices confers monitoring capabilities that make an evaluation of ANS activity more accessible, primarily via the monitoring of cardiac activity via photoplethysmography (PPG) optical sensors. For the purpose of sleep–wake assessment, these PPG sensors provide a supplement to the accelerometers traditionally found in wrist actigraphs.
Some consumer wearables include the Oura Ring, Apple Watch, and Fitbit lines of devices. The Oura Ring, Apple Watch, and several models in the Fitbit line are capable of measuring both user motion and cardiac activity. In addition, some companies such as Oura and Fitbit have developed their own proprietary algorithms to generate measures of sleep quality from the raw data collected by their devices. Independent evaluations of these devices referenced against concurrently collected PSG have suggested accuracy for discriminating sleep–wake that is comparable to existing research-grade wrist actigraphy devices. For example, the Oura Ring has been reported to have a sensitivity to detect sleep of 96% and a specificity to detect wake of 48% [33], whereas the Fitbit Charge 2 has been reported to have a sensitivity to detect sleep of 96% and a specificity to detect wake of 61% [34]. We refer to these new types of devices, which contain both accelerometers suitable for motion actigraphy and technology to measure cardiac activity (such as a PPG sensor), as multisensor wearables.
Beyond the algorithms provided by device manufacturers, many wearable devices also allow users or software developers to access the data from some or all of the sensors included on the device, via (for example) an application user interface or software development kit. However, as noted by de Zambotti et al. [35], there is still room for improvement on the part of manufacturers. Specifically, device standardization, complete data access, and more algorithmic openness would benefit the research, clinical, and consumer communities. Allowing users and developers access to the raw sensor data permits the creation of novel algorithms and applications; for example, leveraging advances in machine learning to develop new classifiers beyond those provided by the manufacturer. In some cases, the sensor data from a device could be used by developers as it is collected to support “real-time” detection and innovative interventions designed to enhance sleep.
Combining actigraphy with cardiac data has the potential to improve consumer-friendly devices beyond those which utilize motion data alone. Existing techniques such as the Cole–Kripke sleep–wake algorithm use motion alone, necessitating the need for the development of new procedures to incorporate additional orthogonal data modalities. Supervised machine learning techniques are one method to flexibly incorporate new measurement modalities, such as the combination of motion and heart rate data, to distinguish sleep from the wake. In the present study, several analyses were conducted to develop and evaluate the performance of a machine learning model relative to other common devices.
Within analysis 1, we first examined the quality of sleep–wake classification from several wearable devices, including common research wrist actigraphy devices and the Oura Ring, versus registered polysomnographic technologist (RPSGT) scored PSG as ground truth. Each device was evaluated in terms of how well it corresponded to PSG scoring at both an epoch-by-epoch and night level. The Apple Watch was not included within this first analysis, as it does not natively output sleep–wake classifications. Within analysis 2, we evaluated the quality of data available from two multisensor wearable devices, the Oura Ring and Apple Watch, by comparing their data to those obtained from an electrocardiography (ECG) channel of a PSG montage and to triaxial motion data from a triaxial wrist actigraph, the ActiGraph Link. This analysis was performed to establish the suitability of these devices for the development of a sleep–wake classifier. Finally, within analysis 3, we used supervised machine learning techniques to develop and evaluate a series of sleep–wake classification algorithms informed by motion and cardiac data from the wearable devices. In order to evaluate how well similar models would compare using common laboratory data, models were also developed using data from the ECG channel of the PSG in conjunction with triaxial motion data from a common research-grade wrist actigraph, serving as a point of comparison.
Methods
Participants
Healthy adults between 35 and 50 years of age with normal color vision and normal hearing were eligible to enroll. Participants were excluded for chronic or acute medical conditions requiring the use of medication with a reasonable likelihood to interfere with sleep or circadian structure, smoking within the past year, nocturnal shift work within the last 6 months or travel through more than three time zones within the last 3 months, substance use (verified with 9-panel urine toxicology screening for amphetamines, cocaine, marijuana, opiates, phencyclidine, barbiturates, benzodiazepines, methadone, and propoxyphene at admission), and sleep disorders (screened during the first inpatient night). Participants were also asked to refrain from caffeine and alcohol for the 3 days immediately prior to inpatient admission. All participants provided written informed consent. All procedures were approved by the Institutional Review Board of the Pennsylvania State University and conducted in accordance with the Declaration of Helsinki.
Screening
Initial screening was conducted to evaluate typical participant sleep at home using a sleep diary and wrist actigraphy (Spectrum Plus; Philips Respironics, Murrysville, PA) worn for at least 4 days and nights. Average sleep duration during the at-home screening was determined according to a previously described algorithm using 30-s epochs of movement count [3]; consecutive low-movement epochs were used to approximate sleep onset and awakening and therefore sleep time each night. Each individual participant’s average sleep duration from actigraphy was used as an alternate criterion for their minimum average sleep opportunity (time in bed) before later inpatient admission: participants received instruction to be in bed for 8 h or their average, whichever was longer, for each of the three nights prior to lab admission.
Participants were confirmed to be free of sleep-related breathing disorders at home through pulse oximetry (Nonin Model 3150; Nonin Medical, Plymouth, MN); specifically, no nocturnal oxygenation fluctuations that resulted in substantial time spent below 88% peripheral oxygen saturation or five or more oxygen fluctuations per hour. Following sleep screening, participants underwent a physical examination and medical history by a physician or nurse practitioner.
Protocol
Participants in the study completed four consecutive days and nights in the laboratory. On the second and fourth night of participation, participants received auditory stimulation, designed to either enhance or disrupt sleep, referred to here as the Enhancing and Disruptive nights, respectively. The order of Enhancing and Disruptive stimulation was counterbalanced across participants. No auditory stimulation was presented on the first or third night, which is referred to as Habituation and Sham, respectively. The results of the auditory stimulation are outside the scope of this report, which is focused on the ability of various wearable devices to dissociate sleep from the wake, and have been discussed elsewhere [36]. None of the devices or classification algorithms in this report had knowledge of the experimental condition or the presence of auditory stimulation.
Participants were admitted to the Clinical Research Center at the Pennsylvania State University (University Park campus) and provided a biospecimen (urine) for toxicology screening. Participants were maintained in a light and sound-attenuated sleep laboratory suite and instrumented with PSG (TrackIt v2.8.0.8; Lifelines Ltd, Irvine, CA and Polysmith v10.0 build 7956; Nihon-Kohden, Tokyo), comprised of EEG, electrooculography, electromyography, and ECG, all sampled at 200 Hz. The EEG montage contained 11 channels: ground at location FPz, left mastoid (M1), right mastoid (M2), F3, Fz, F4, C3, Cz, C4, O1, and O2. Midline electrodes were referenced to the left mastoid, while lateral electrodes were referenced to the contralateral mastoid. ECG electrodes (two) were placed according to the American Academy of Sleep Medicine (AASM) standard [37].
In addition to PSG, participants were outfitted with several wearable devices, including an Apple Watch (wrist of nondominant hand; distal), Oura Ring (best fitting finger per participant), ActiGraph Link (wrist of dominant hand), and Philips Respironics Spectrum Plus (wrist of nondominant hand; proximal) (Table 1). Although the ActiGraph Link was placed on an alternate hand from the other two wrist devices, prior work has reported a high correspondence for wrist actigraphy concurrently collected from dominant and nondominant wrists [38].
Table 1.
Manufacturer, Device Name, and Data Details for Wearable Devices Used Within the Study
| Manufacturer | Device | Data | Reference |
|---|---|---|---|
| ActiGraph (Pensacola, FL) | Link | Wrist-worn device, triaxial accelerometer sampling at 80 Hz. | http://actigraphcorp.com/products/actigraph-link/ |
| Apple (Cupertino, CA) | Watch (Series 2) | Wrist-worn device, triaxial accelerometer sampling at 1.33 Hz, PPG sensor provides BPM estimates at approximately 0.2 Hz. | http://www.apple.com/watch/ |
| Oura (Oulu, Finland) | Ring (Version 1) | Finger worn device, accelerometer internally sampling at 50 Hz, providing motion counts at 30 s intervals, PPG sensor internally sampling at 250 Hz, providing R–R intervals. | http://ouraring.com/ |
| Philips Respironics (Murrysville, PA) | Spectrum Plus | Wrist-worn device, accelerometer internally sampling at 32 Hz, providing motion counts at 30-s intervals. | http://www.actigraphy.com/devices/actiwatch/actiwatch-pro.html |
Participants were scheduled for a 9-h time in bed sleep opportunity in darkness; due to technical and time constraints, actual sleep opportunity in the laboratory ranged from 8h 41m to 9h 13m (according to PSG, lights out to lights on).
Polysomnography processing and sleep staging
PSG data were staged in 30-s epochs according to the AASM standards by the RPSGT (author MMS). During staging, the RPSGT was blinded to the auxiliary data channel that included information regarding the timing presence of auditory stimulation. In some cases of discontinuous nocturnal recording due to unreadable EEG data (e.g. poor data quality or participant disconnected for restroom use), sections of data were considered “unscorable” and were excluded from epoch-level analyses.
Analysis 1: Evaluation of device classification
Sleep–wake classification from devices was compared to RPSGT-derived sleep staging to establish a point of reference for evaluating our machine learning model performance. Philips Actiware software v6.0.9 was used to export the Actiwatch Spectrum Plus sleep–wake output at 30-s intervals. When classifying each 30-s sleep epoch, the Actiware software computes a weighted sum of activity counts within the current epoch, the prior four epochs, and the following four epochs. If this sum exceeds a predefined threshold, the epoch is classified as a wake. Classifications were exported from the Actiware software using the default wake threshold of “Medium,” corresponding to an activity count sum threshold of 40. ActiGraph ActiLife software v6.11.8 was used to export the ActiGraph Link sleep–wake output at 60-s intervals. The ActiLife software allows the selection of one of two algorithms, which are based upon (but may not be implemented identically to) existing sleep–wake algorithms: “Cole–Kripke” [6] or “Sadeh” [31]. These two algorithm selections were separately output for comparison to our RPSGT staging. The Oura Ring (version 1) provides sleep staging with four categories (wake, “light” sleep, “deep” sleep, rapid eye movement) at 30-s intervals. To compare the Oura output to other sleep–wake output, the four-category staging was discretized into sleep and wake.
While the Actiwatch and ActiGraph devices score data continuously, the Oura Ring stores data when it determines the wearer is in bed. In some cases, the Oura Ring did not fully capture the period that the participant was in bed and thus did not begin to provide staging information by the time the experimental lights-out period began or stopped providing staging information before the experimental lights-out period ended. In these cases, those missing sleep epochs were filled to the bounds of the experimental in-bed period with the classification of “wake.” In addition to the device classifications, a naïve “model” was constructed that always predicts sleep (because the “time in bed” period is typically skewed toward sleep rather than wake), serving as a point of reference for the performance of a theoretical classifier that provides no information.
The intervals used by each device for classification are not necessarily aligned to the intervals which define a 30-s sleep epoch labeled by the RPSGT. For example, even if the device provides classification in 30-s time windows, the start of the 30-s period may not be aligned to the start of the epoch used by the RPSGT for staging. For the purposes of comparison, each RPSGT-staged epoch was compared to the epoch from each device with the closest timestamp. In the case of data from the ActiGraph Link, while the device epochs are at 60-s intervals, the nearest labeled epoch to each 30-s RPSGT-staged reference epoch was used. If a device lacked a staged epoch within 30 s of a given PSG epoch due to missing data, that epoch was not evaluated for that device. Any epochs that could not be staged by the RPSGT (labeled “unscorable”) were excluded from the comparison. In addition, in order to mitigate the influence of clock offsets between each device and the PSG, which can especially influence the epoch-by-epoch comparisons, the sleep–wake output from each device was shifted ±5 min relative to the PSG output, to identify the lag (if any) that optimized the sleep–wake correspondence between the device and PSG, for each night. This shifted version was subsequently used to compute both epoch-by-epoch and night-level statistics on data correspondence. A similar procedure (also with a ±5 min window) has been previously used to mitigate potential time offsets between actigraphy counts and sleep–wake staging [3].
Analysis 1A: Epoch-by-epoch correspondence between devices and PSG
Several metrics were computed using the Caret package for R [39] to evaluate the performance of the classifications from each device, treating the RPSGT-staged PSG as the correct or “ground truth” label for comparison, with sleep as the “positive” class to be detected. These comparisons were performed after restricting to the complete cases of epochs that were present for both a given device and for the PSG-derived staging; epochs that were missing for a given device or deemed unscorable by the RPSGT were not included in the comparison. The following metrics were evaluated:
Accuracy: the percentage of epochs correctly classified.
Balanced accuracy: the mean of wake epochs correctly classified and sleep epochs correctly classified.
Sensitivity: also known as recall, the percentage of sleep epochs correctly classified.
Specificity: the percentage of wake epochs correctly classified.
Precision: also known as a positive predictive value, the percentage of epochs classified as sleep that are correctly classified.
Cohen’s kappa (κ): classifier agreement with PSG, relative to chance [40]. Specifically, kappa is calculated via (po−pe)/ (1−pe), where po is the percentage of observed classifications with agreement, and is the percentage of classifications that would be expected by chance.
Signal detection theory d-prime (d′): the difference in standard deviation between theoretical signal and noise distributions [41]. Computed by first transforming the percentage of sleep epochs correctly classified as sleep (classifier sensitivity, or “hit rate”) and the percentage of wake epochs incorrectly classified as sleep (classifier “false alarm rate”) into z-scores via the inverse of the normal cumulative density function and then computing the difference.
Analysis 1B: Night-level correspondence between devices and PSG
For some applications, especially clinical, night-level summaries of sleep quality may be of greater interest than epoch-by-epoch sleep staging. For this reason, several metrics of sleep quality were also calculated, first using the ground truth values from the RPSGT-derived sleep staging and then using the classifications provided by each device. These metrics were computed after restricting to the complete cases of epochs that were present for both the device being evaluated and the PSG-derived sleep staging:
Sleep efficiency: the percentage of the in-bed period classified as sleep.
Sleep onset latency (SOL): the latency, in minutes, from Lights Out until the first epoch of sleep occurs.
Total sleep time (TST): the total number of minutes of staged sleep.
Wakefulness after sleep onset (WASO): the number of minutes spent awake following the first epoch staged sleep.
For each device and classifier, these metrics were computed for all epochs present in the dataset and compared against the PSG staging data with the same epochs present.
Analysis 2: Evaluation of motion and heart rate data obtained from wearables
PSG preprocessing
The ECG channel of the PSG was processed to serve as a point of comparison to the PPG data collected by the Apple Watch and Oura Ring, which each provide data that has already had some processing applied. The PSG data files were imported into MATLAB (Natick, MA), the ECG channel was selected, mean centered (removing any direct current offset), and bandpass filtered between 0.05 and 40 Hz, with a filter of order 20. The filtered time series was further decomposed using a symlet wavelet (sym4) to 5 levels, with the data at the fourth and fifth levels retained. In order to identify the R-peaks of the ECG QRS waveform, the square of the absolute value of the resulting wavelet-filtered time series was entered into a peak finding algorithm (the MATLAB function “findpeaks”) with a minimum peak distance of 100 samples (0.5 s) and a minimum peak height of 5 mV. Finally, the identified R-peaks were converted into R–R intervals by labeling each peak by the latency in seconds from the previous peak.
ActiGraph Link preprocessing
Triaxial accelerometer data from the ActiGraph Link, sampled at 80 Hz, were used as a point of comparison to actigraphy data derived from the Apple Watch and Oura Ring. In order to reduce the influence of gravity on accelerometer measurements, each dimension (x,y,z) in the accelerometer time series was high-pass filtered at 0.1 Hz with a third-order Butterworth filter. Following filtering, each three-element (x,y,z) sample was then converted to the magnitude of the three-dimensional acceleration vector ().
Apple Watch preprocessing
Although the Apple Watch contains a PPG sensor, access to the raw PPG signal was not available. Instead, an estimate of heart rate in beats per minute (BPM) is provided by the device approximately every 5 s (at a rate of 0.2 Hz). The degree of processing performed by the device to transform the PPG signal to heart rate, or the time window used to compute each heart rate sample, is not documented by Apple, though it may be presumed that some temporal filtering is performed. As the Apple Watch provides values corresponding to heart rate, and not interbeat interval (IBI), heart rate values were transformed to pseudo-IBI for consistency with the other devices, by dividing 60 s by the reported heart rate. Each Apple Watch sample was time-stamped as it was collected from the device for synchronization with other devices in the study.
Triaxial accelerometer data, corrected for the force of gravity, were additionally collected via the Apple Watch, sampled at 1.33 Hz (40 samples/30 s sleep epoch). Although the device can be configured to sample at a higher rate, a higher sampling rate also requires more power, reducing battery life. A lower rate was used to ensure that the battery in the device would last through each night of data collection. The Apple Watch contains a triaxial gyroscope, which allows the force of gravity to be separated from the raw accelerometer measurement, via knowledge of device orientation at each sample. The triaxial, gravity-corrected time series was converted into a time series of vector magnitude.
Oura Ring preprocessing
The Oura Ring contains a PPG sensor (raw sensor signal unavailable). This signal is processed on the device to extract the R-peaks of the QRS waveform. When an R-peak is identified, the device logs the interval from the previous peak (the R–R interval) along with a timestamp.
The Oura Ring additionally contains a triaxial accelerometer; however, the raw accelerometer data were not made available for the present study. Instead, a set of values summarizing the motion data were provided by the Oura device at 30-s intervals. Provided summary values included “motion seconds,” “motions low,” and “motions high.” Motion seconds is defined by Oura as the number of seconds in which an acceleration greater than 64 mg (where g is the acceleration due to gravity) occurs on any of the three accelerometer channels. Motions low and motions high are defined as counts, referencing the number of times the acceleration vector exceeds a predefined threshold within the epoch; however, the actual thresholds exceeded are not provided by Oura. For each staged epoch, the summary value “motion seconds” was used from the closest 30-s Oura provided epoch.
Temporal alignment of data
Data from each device were time-stamped during the time of recording. In order to reduce the possibility of poor temporal alignment between devices, an additional alignment step was performed to align the cardiac activity data between the ECG, Apple Watch, and Oura Ring. For each recording and data source, outlier samples were removed, defined as an IBI outside the range of 0.4286 and 2 s (corresponding to an instantaneous heart rate range of 30–140 BPM), or more than four SDs from the mean IBI for a given recording and data source.
Comparing device data using raw samples is problematic, as a single missed IBI could shift all subsequent samples in the time series. In order to directly compare the timing of the devices, the IBI values derived from each device were linearly interpolated to a common sampling interval of integer seconds within the recording. To reduce the influence of outliers on the timing evaluation, an 11-sample median filter was applied to each interpolated time series. Following the median filter, any chunk of data in which 10 or more contiguous seconds were missing (interpolated across) for either time series under comparison was removed from both time series. For the ECG and Apple Watch comparison, an average of 8.49 min of data was excluded per night, while for the ECG and Oura Ring comparison, an average of 18.93 min of data was excluded per night. Finally, the cross-correlation was performed between the ECG and device data at lags between 1000 and 1000 s to identify the timing offset that produced the highest correlation between the time series pair, separately for each recording.
A similar alignment process could not be performed for the actigraphy data, because unlike the ECG data, which is recorded as a channel within the PSG, no single source of actigraphy data could serve as ground truth with respect to PSG staging.
Data comparison
The data from the Apple Watch and Oura Ring were compared to the data derived from the ECG or ActiGraph Link in order to evaluate how accurately the devices could capture variations in heart rate or motion across the night. To determine the quality of heart rate measurement, the mean IBI, SD of IBI, and root mean square of successive differences of IBI within each 30-s sleep epoch were correlated within each night between the ECG and Apple Watch and separately between the ECG and Oura Ring. To determine the quality of motion actigraphy, the mean and SD of actigraphy vector magnitude within each 30-s sleep epoch were correlated within each night between the ActiGraph Link and the Apple Watch. The magnitude of the three-element vector was used to compare accelerometer data instead of the three-coordinate time series because the vector magnitude is invariant to coordinate rotation between the devices under comparison. As raw motion actigraphy was not available for the Oura Ring (version 1) used here, the “motion seconds” variable instead correlated with the mean and SD of vector magnitude as derived from the ActiGraph Link.
Analysis 3: Development and evaluation of machine learning approach
Feature extraction
Each 30-s sleep epoch was labeled with the RPSGT sleep–wake determination as well as a set of features for the supervised machine learning model. The term “features” refers to a set of computed values that are used as the input to the classifier. Features were extracted separately for each of three model variants constructed: (i) a model informed by PSG-derived ECG and actigraphy from the ActiGraph Link, (ii) a model informed by cardiac activity and actigraphy from the Apple Watch, and (iii) a model informed by cardiac activity and motion from the Oura Ring. In addition to the features derived from each device, a feature encoding the amount of time that has elapsed since the start of the “lights out” period was included, as the prior probability of a given epoch being sleep or wake was not assumed to be constant across the night.
Feature extraction began with the original data, not the interpolated data that was used to determine the cardiac temporal offsets. Temporal offsets for each recording (discussed earlier) were applied to both the Oura Ring and Apple Watch data. Outlier rejection was again performed according to the guidelines mentioned previously.
For the combination of PSG and Link, and for the Apple Watch, the features within each epoch include the number of seconds that have elapsed since the “lights out” period began, the mean IBI in the epoch, the SD of IBI in the epoch, and the mean of actigraphy vector magnitude in the epoch. For the data derived from the Oura Ring, the variable “motion seconds” as output by the device were used in place of the mean of actigraphy vector magnitude. In addition to data from the current epoch, the mean IBI, SD of IBI, and mean actigraphy vector magnitude from the prior eight epochs (4 min) were used as features within each current epoch, resulting in 27 cardiac or motion features and one time feature (the seconds elapsed since the in-bed period began) per epoch. The use of data from prior epochs when evaluating the current epoch was inspired in part by the Cole–Kripke actigraphy algorithm [6], which uses a weighted average of past and future epochs to determine the sleep–wake state of the current epoch. Here, only past and not future epochs are used in order to support the desired “real-time” prediction of sleep state.
Machine learning approach
Our machine learning approach used a gradient boosting classifier [42] as implemented within the Python package Scikit-learn [43]. Models were constructed and evaluated using nested leave-one-participant-out cross-validation. Within any machine learning paradigm, the data used to train the model should not be used to evaluate the model, motivating separate training and test partitions of the data [44]. In the case of grouped data, such as the present report where sleep epochs are grouped or clustered into nights and nights are grouped into participants, it is ideal if the test set contains more than a single or few participant instances, which may not be representative of the dataset as a whole. This leads to a leave-one-participant-out cross-validation structure, in which each of the n participants serves as the test set within one of n model runs, and each model is trained on the remaining n−1 participants. The mean and SD of performance across the n model runs are then collected, resulting in a value that is not biased toward any particular test participant.
However, many common machine learning methods contain parameters that are not adjusted as part of the model, which instead govern the structure of the model itself. For example, for gradient-boosted decision trees as used within the present report, such model structure parameters, or hyperparameters, include the number of decision trees within the ensemble, the number of observations within the final terminating nodes, and others. A particular set of hyperparameters may be optimal for a given dataset but are often unknown a priori. Iterating through model parameters to minimize the test set error generates a biased estimate of model performance, as test set performance is no longer outside of the training procedure [45, 46].
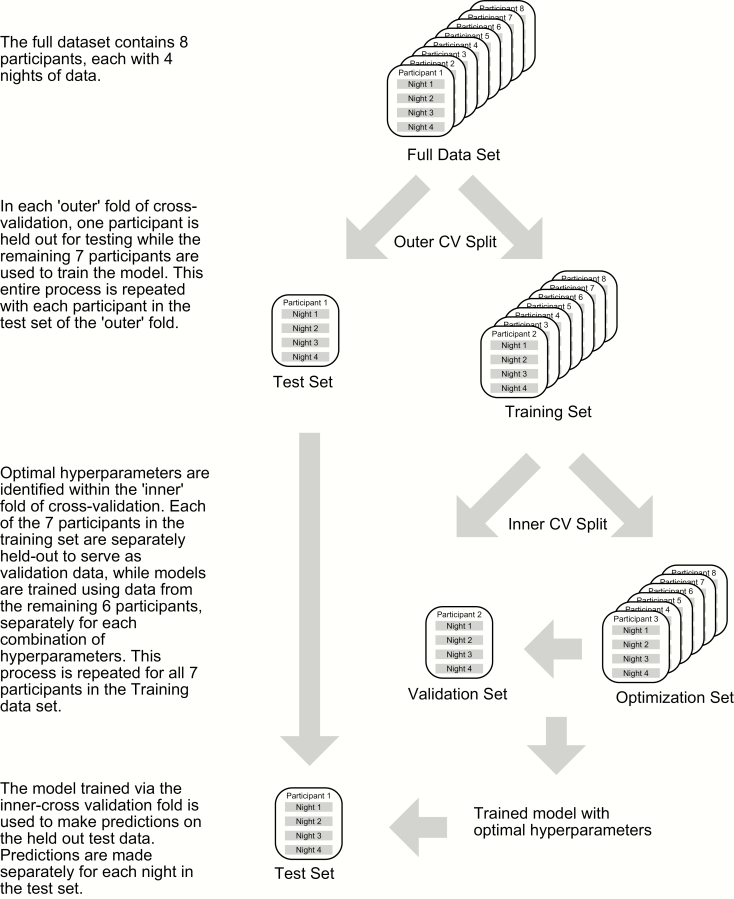
Within the nested leave-one-participant-out cross-validation procedure employed here (Figure 1), the selection of model parameters is implemented inside each outer cross-validation fold, via a separate series of (n−1) cross-validation folds per participant. Within each fold of the interior cross-validation, one of the (n−1) participants serves as a validation test for tuning the model hyperparameters. For each outer cross-validation fold, the best candidate model from the interior cross-validation is then evaluated on the held-out test data. The final results reported are the mean and SD of performance measures across the eight outer cross-validation folds. In the present report, hyperparameters and their options selected in the interior cross-validation include the learning rate (0.01, 0.1), the number of estimators (50, 100, 150), the maximum decision tree depth (1, 2, 3, 4), and the minimum number of samples per tree split (2, 10, 20, 30). Other model hyperparameters were fixed: loss, fixed at deviance, criterion, fixed at Friedman mean square error, and the minimum number of samples per leaf, fixed at 1. During training, models are optimized for the area under the curve (AUC).
Figure 1.
The nested cross-validation procedure used.
Models were separately trained and tested for each of the three datasets (PSG ECG channel in combination with ActiGraph Link accelerometer data, Oura Ring, and Apple Watch). For each dataset, gradient boosting classifier models were trained and tested with and without night-level normalization, and with and without class balance, totaling four variations per dataset. In all cases, the target of the classifier for each 30-s epoch was a binary classification of sleep or wake.
Normalizing data is a common preprocessing step within a machine learning paradigm, serving to place both features and datasets on a common scale. While scaling features is not required for a decision tree approach as described here, scaling between datasets can still provide a benefit when using physiological data such as heart rate, where dataset shift [47] may occur between training and test sets due to individual differences in mean heart rate. Here, night-level normalization refers to transforming each feature within each night by transforming to a z-score via subtracting the mean and dividing by the SD of that feature within the night. However, normalizing at the night level only allows classifications to be made once the full night is completed, as the distribution of values across the night is not known until the entire night of data is collected. Here, models were also run without normalization to simulate model performance when classifications are made in “real time,” epoch-by-epoch, prior to the completion of the night.
“Class balance” refers to the balancing of sleep and wake instances used for model training (but not evaluation). Models were developed either with or without balanced classes during training. Class balance was achieved via random oversampling over the minority class (Wake) during training, via the Python package imbalanced-learn [48].
Classifier evaluation
The output of each classifier was evaluated in the same manner as the device output previously described within sections 1A and, by restricting to complete cases between a given classifier and PSG-derived staging, at both the epoch-by-epoch and night level analyses 3A and 3B respectively. All of the epoch-by-epoch metrics previously described in the context of evaluating device output were calculated. In addition, classifier output probabilities were used to calculate the AUC of the receiver operating characteristic (ROC) curve. AUC could not be calculated for device output, such as from the Actiwatch Spectrum, etc. as these devices only provide discrete classifications, rather than class probabilities.
Results
Participants
Nine participants were enrolled in the protocol. One participant was excluded during the inpatient portion for medical attention not related to experimental interventions. The eight remaining participants completed the full, four-night protocol (five female, M = 40.75 years, SD = 4.84). Participants were adherent in maintaining their typical sleep schedule during the Pre-Inpatient period (mean TST = 514.2 min, SD = 59.8 min), relative to Screening (M = 496.7 min, SD = 44.4 min), according to actigraphy, t(7) = 0.95, p = .374, d = 0.33.
Missing data
Data from some of the 32 total nights of participation (eight participants with four nights each) were missing or corrupted for some datasets. Missing values were not imputed but were instead excluded from analyses. For data from the Apple Watch, three participants were each missing one night of data due to data corruption. In addition, the Apple Watch failed to accurately measure heart rate for two nights of a single participant, often oscillating between a value close to ECG-derived heart rate, and a value approximately twice the ECG-derived heart rate. These two nights were additionally removed from the analysis. For Oura Ring data, two participants were each missing one night due to data corruption. In addition, two nights were missing large amounts of data, possibly due to an issue with the device’s detection of the in-bed period, as described in the Methods section. For one night, the device did not begin collecting data until approximately 2.5 h following lights out. For a second night, the device was missing a contiguous chunk of approximately 2 h of data within the night. These two nights were excluded from the comparison of night-level metrics for the Oura Ring but were included in the epoch-by-epoch sleep–wake comparison, after excluding only the missing periods.
Epoch-by-epoch and night-level statistics were computed using the complete cases of epochs present for both a given device or classifier, and the PSG-derived sleep staging. For the Oura Ring staging output, no epochs were missing beyond the two datasets each missing 22%–27% of data as described above. As previously described, while the Oura Ring sometimes began staging late, or ended staging early relative to the in-bed period, these periods were considered to have an Oura Ring classification of wake and thus were not considered missing. No epochs were missing from the ActiGraph Link or Actiwatch Spectrum device output datasets. For the classification datasets, the Apple Watch datasets were missing a mean of 0.85% of epochs (SD = 2.15%), the ECG-Link datasets were missing a mean of 0.23% of epochs (SD = 0.34%), while the Oura Ring datasets were missing a mean of 4.01% of epochs (SD = 4.82%).
Data from one night of participation had a high percentage of epochs that were unscorable by the RPSGT (28%) due to a disconnected EEG electrode. This night was excluded from the comparison of night-level metrics. However, this night was included in epoch-by-epoch staging device comparisons and in our machine learning models, as these comparisons exclude any unscorable epochs from the analysis. For the remaining 31 nights, the mean percentage of epochs marked unscorable by the RPSGT was 0.41% (SD = 0.64%).
Analysis 1: Evaluation of device classification
Analysis 1A: Epoch-by-epoch correspondence between devices and PSG
The sleep–wake classifications provided by several devices were compared to the values scored by the RPSGT (Table 2). For healthy individuals without a sleep disorder, the in-bed period is dominated by periods of sleep. In this case, overall classification accuracy (% of epochs correctly classified) can be a misleading performance metric. As an illustration, Table 2 displays the epoch-by-epoch “performance” that can be obtained by simply classifying every epoch within the in-bed period as Sleep, the “naïve” model. The sleep–wake classification accuracy of this no-information model is near to each of the other devices used within the study. For metrics such as balanced accuracy or d-prime, the ActiGraph Link exported with the “Sadeh” algorithm produced the highest concordance with RPSGT-derived staging. All classifiers perform better on the detection of sleep (sensitivity) than the detection of wake (specificity), as has been previously observed [3]. This bias was most pronounced for classifications from the Actiwatch Spectrum Plus, which produced both the greatest sensitivity for sleep epochs and the poorest specificity for wake epochs. Despite this imbalance, as epochs within the in-bed period are predominantly sleep, the classifications from the Actiwatch Spectrum Plus produced the highest overall accuracy.
Table 2.
Comparison of Device Sleep–Wake Classification Relative to Ground Truth Sleep–Wake Classification Derived From PSG Staging
| Source | Accuracy | Balanced accuracy | d′ | Kappa | Precision | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|
| Naïve Model (result of always predicting Sleep, for reference) | 0.876 (0.064) | 0.500 (0.000) | 0.646 (0.204) | 0.000 (0.000) | 0.876 (0.064) | 1.000 (0.000) | 0.000 (0.000) |
| ActiGraph Link with “Cole–Kripke” algorithm | 0.891 (0.046) | 0.752 (0.070) | 1.807 (0.375) | 0.482 (0.120) | 0.940 (0.042) | 0.936 (0.050) | 0.568 (0.163) |
| ActiGraph Link with “Sadeh” algorithm | 0.880 (0.054) | 0.779 (0.071) | 1.874 (0.463) | 0.487 (0.146) | 0.949 (0.041) | 0.912 (0.064) | 0.647 (0.163) |
| Actiwatch Spectrum Plus algorithm | 0.904 (0.050) | 0.674 (0.065) | 1.831 (0.327) | 0.424 (0.121) | 0.914 (0.055) | 0.982 (0.012) | 0.366 (0.136) |
| Oura Ring | 0.899 (0.046) | 0.686 (0.075) | 1.771 (0.502) | 0.423 (0.150) | 0.923 (0.042) | 0.963 (0.039) | 0.410 (0.165) |
The “naïve” model is not a device but is instead the performance that is obtained by classifying every in-bed epoch as sleep, included here for reference. Values displayed within each cell are the mean and SD across nights.
Analysis 1B: Night-level correspondence between devices and PSG
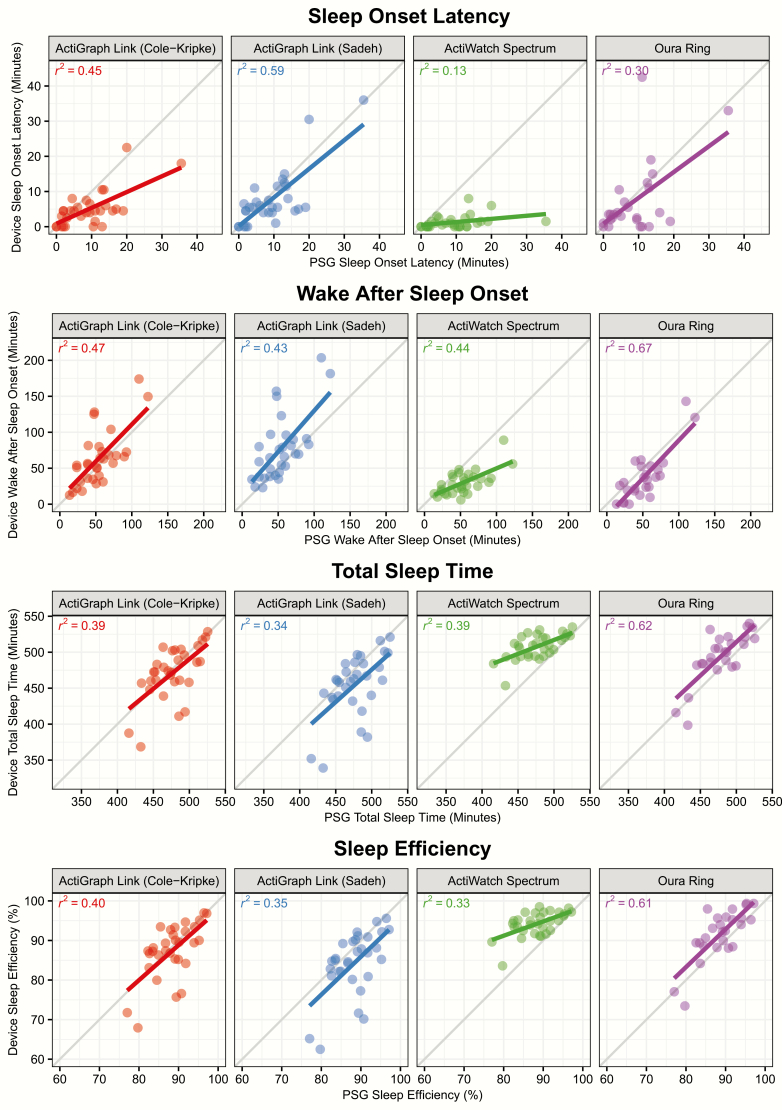
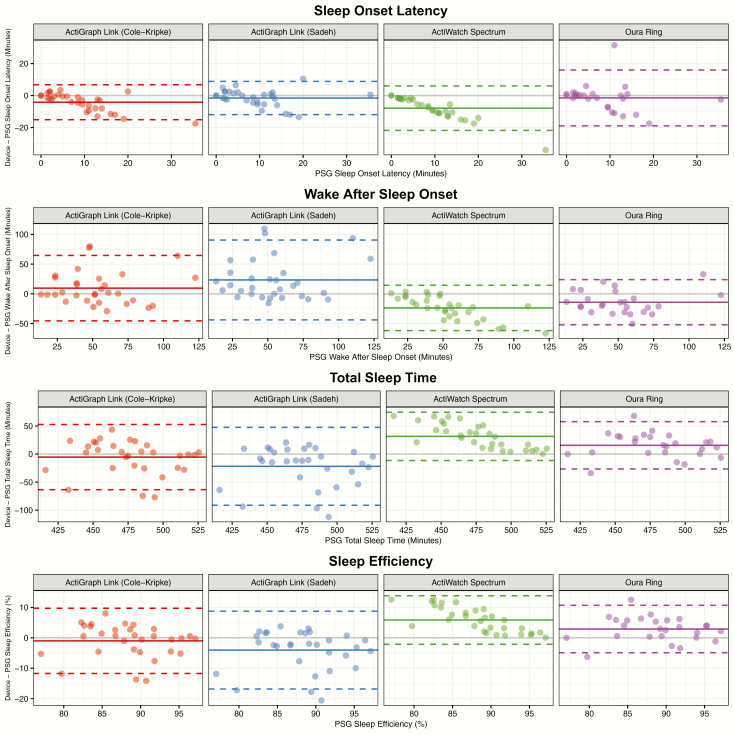
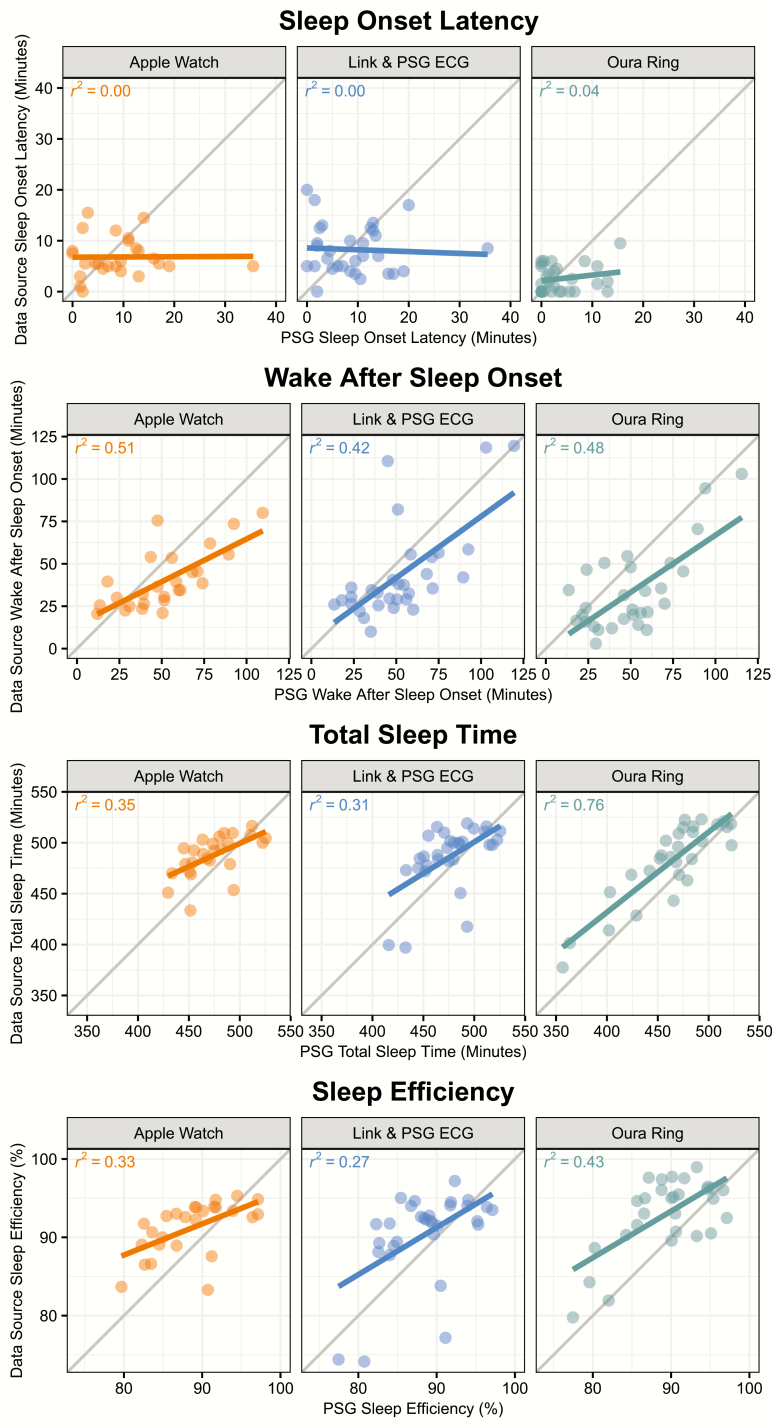
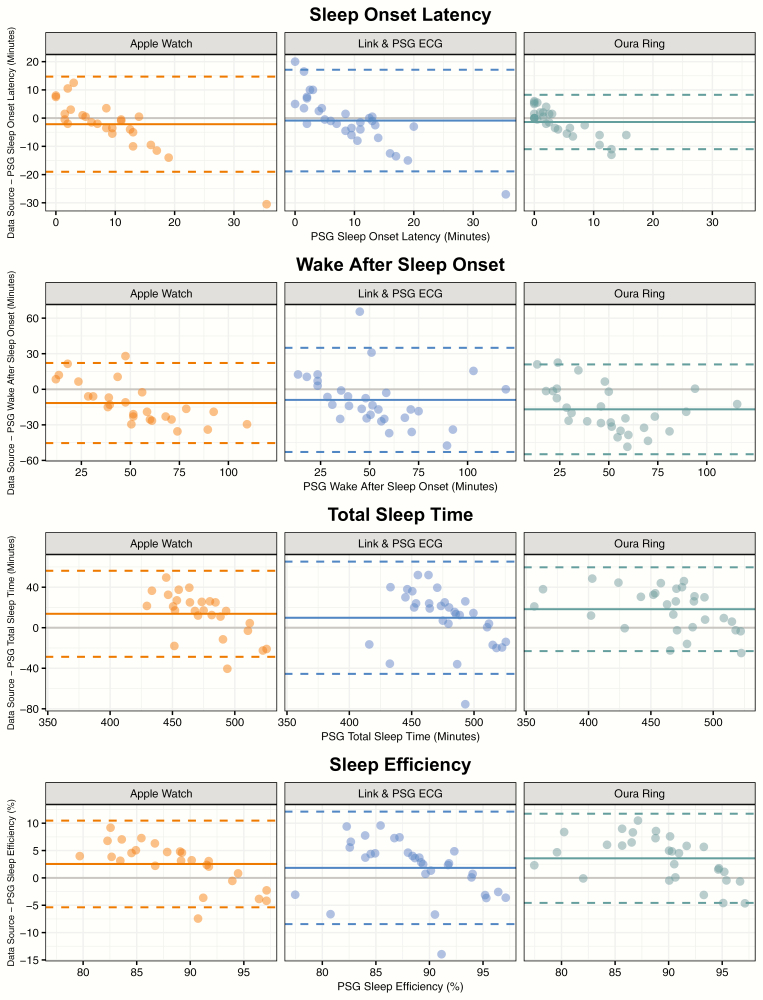
The sleep–wake classifications provided by each device were used to compute several night-level metrics, including sleep efficiency, SOL, WASO, and TST, which were compared to the same metrics computed from the PSG-derived staging. Measures of device error are displayed in Table 3. Figure 2 displays a scatterplot of the correspondence between the device and PSG-derived staging for each metric, while Figure 3 displays the bias of each device relative to PSG-derived staging for each metric. No device consistently outperforms the other across all metrics, and the best performing device in terms of root mean squared error for a given metric is not necessarily the same as the best performing device in terms of R2. The Actiwatch Spectrum Plus results demonstrate the device’s bias toward the classification of sleep. For example, Figure 2 illustrates that the Spectrum underpredicts SOL and WASO, while overpredicting TST and SE. The device biases within Figure 3 further demonstrate that this bias is not constant; for example, sleep efficiency is overpredicted to a greater extent for nights with lower sleep efficiency.
Table 3.
Comparison of Night-Level Metrics Derived From Device Data to Those Derived From RPSGT Staging
| Metric | Device | Mean error | MAE | RMSE | R 2 |
|---|---|---|---|---|---|
| Sleep efficiency (%) | ActiGraph Link (Cole–Kripke) | −0.99 | 4.04 | 5.47 | 0.40 |
| ActiGraph Link (Sadeh) | −4.03 | 5.25 | 7.58 | 0.35 | |
| Actiwatch Spectrum | 5.86 | 5.86 | 7.10 | 0.33 | |
| Oura Ring | 2.89 | 3.93 | 4.86 | 0.61 | |
| Sleep onset latency (min) | ActiGraph Link (Cole–Kripke) | −4.16 | 5.00 | 6.89 | 0.45 |
| ActiGraph Link (Sadeh) | −1.53 | 3.92 | 5.44 | 0.59 | |
| Actiwatch Spectrum | −7.87 | 7.87 | 10.52 | 0.13 | |
| Oura Ring | −1.46 | 5.31 | 8.86 | 0.30 | |
| WASO (min) | ActiGraph Link (Cole–Kripke) | 9.61 | 20.61 | 29.25 | 0.47 |
| ActiGraph Link (Sadeh) | 23.37 | 27.50 | 41.00 | 0.43 | |
| Actiwatch Spectrum | −23.65 | 24.13 | 30.42 | 0.44 | |
| Oura Ring | −14.10 | 20.75 | 23.67 | 0.67 | |
| Total sleep time (min) | ActiGraph Link (Cole–Kripke) | −5.45 | 21.84 | 29.68 | 0.39 |
| ActiGraph Link (Sadeh) | −21.84 | 28.39 | 41.10 | 0.34 | |
| Actiwatch Spectrum | 31.52 | 31.51 | 38.19 | 0.39 | |
| Oura Ring | 15.56 | 21.21 | 26.22 | 0.62 |
Displayed are the mean error, mean absolute error (MAE), root mean squared error (RMSE), and R2.
Figure 2.
Comparison of PSG-derived and device-derived night-level sleep metrics. Points depict the PSG and device-derived values for each night of data within the study (except nights previously described as excluded). Lines depict the linear fit between PSG value and device value for each device.
Figure 3.
Bland-Altman [49] plots comparing PSG-derived sleep metrics to the difference between each device-derived sleep metric and the PSG-derived sleep metric. Points depict difference values for each night of data within the study (except nights previously described as excluded).
Analysis 2: Evaluation of motion and heart rate data obtained from wearables
The Apple Watch failed to accurately measure the heart rate for two nights of a single participant. The Apple Watch data for those nights for that participant were excluded. Within these nights, the Apple Watch-derived heart rate fluctuated between a value that was either correct or twice the rate of the participant’s ECG-derived heart rate. For the remaining recordings, the heart rate data from each device was aligned based on the identified offsets from the ECG time series. For the Oura Ring, the offset relative to the ECG ranged between −2 and 4 s (mean = 1.53, SD = 1.69) where negative values represent the device time leading the ECG, and positive values represent the device time lagging the ECG. For the Apple Watch, the offset relative to the ECG ranged between 0 and 12 s (mean = 8.43, SD = 3.31). The greater timing offset on the part of the Apple Watch may result from temporal smoothing of PPG data performed on the device, though the details of any preprocessing performed are not provided by Apple.
IBIs (or pseudo-IBIs in the case of the Apple Watch) that were outliers or extreme values were removed prior to feature extraction. For the ECG datasets, an average of 0.06% of samples was rejected (SD = 0.19%). For the Oura Ring datasets, an average of 2.47% of samples was rejected (SD = 2.53%). For the Apple Watch datasets, an average of 0.01% of samples was rejected (SD = 0.03%).
The epoch-level feature values were compared between the ECG (for cardiac activity features) and ActiGraph Link (for motion feature) and the two wearable devices in order to determine the wearable device data quality. For the comparison of the Oura Ring with ActiGraph Link motion, the “motion seconds” provided by the Oura Ring within a given epoch was compared to the mean and SD of vector magnitude from the ActiGraph Link, as raw accelerometer data were not available from the Oura Ring. For both the Apple Watch and Oura Ring, both heart rate and motion data from the wearables were reasonably correlated with data from the reference devices at the epoch level (Table 4). For example, the correlation of IBI with the ECG channel of the PSG montage averaged 0.92 and 0.85 for the Apple Watch and Oura Ring, respectively.
Table 4.
Standardized Correlation Coefficients (r) for Correlation of PSG ECG or ActiGraph Link-Derived Features, With Features Derived From the Wearable Devices Under Study
| Device | Mean of vector magnitude (Apple Watch) or motion seconds (Oura Ring) | SD of vector magnitude (Apple Watch) or motion seconds (Oura Ring) | Mean of IBI | SD of IBI | RMSSD of IBI |
|---|---|---|---|---|---|
| Apple Watch | 0.83 (0.08) | 0.79 (0.08) | 0.92 (0.04) | 0.76 (0.09) | 0.58 (0.18) |
| Oura Ring | 0.53 (0.16) | 0.47 (0.13) | 0.85 (0.14) | 0.78 (0.15) | 0.62 (0.17) |
Values within each cell represent the mean and SD across nights. IBI, interbeat interval.
Analysis 3: Evaluation of machine learning approach
Analysis 3A: Epoch-by-epoch correspondence between classifier output and PSG
Epoch-by-epoch sleep–wake classifications collected from the outer loop of the nested cross-validation procedure were compared to PSG derived staging, for each classifier variation under evaluation. Classifiers varied in their dataset, whether data were normalized within a night, and whether class oversampling was performed, resulting in 12 separate models. Table 5 displays the epoch-by-epoch performance of all model variations on several performance metrics. Datasets using heart rate data from the ECG channel of the PSG in combination with actigraphy data from the ActiGraph Link generally outperform classifiers using data from either the Apple Watch or Oura Ring. Normalization of data within a night generally improves classifier performance. Oversampling of wake epochs during training reduces much of the bias toward the classification of sleep epochs, which improves specificity to detect wake epochs while decreasing sensitivity to detect sleep epochs.
Table 5.
Performance of Machine Learning Classifiers
| Dataset | Normalization | Oversampling | AUC | Accuracy | Balanced accuracy | d′ | Kappa | Precision | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|---|---|
| Apple Watch | FALSE | None | 0.918 (0.040) | 0.918 (0.040) | 0.753 (0.083) | 2.228 (0.319) | 0.544 (0.117) | 0.936 (0.050) | 0.974 (0.029) | 0.532 (0.187) |
| FALSE | Oversampling | 0.917 (0.037) | 0.872 (0.077) | 0.825 (0.060) | 2.154 (0.377) | 0.514 (0.145) | 0.962 (0.038) | 0.890 (0.095) | 0.760 (0.156) | |
| TRUE | None | 0.926 (0.040) | 0.928 (0.029) | 0.789 (0.063) | 2.347 (0.373) | 0.602 (0.102) | 0.943 (0.039) | 0.976 (0.022) | 0.602 (0.136) | |
| TRUE | Oversampling | 0.922 (0.041) | 0.882 (0.039) | 0.853 (0.048) | 2.237 (0.398) | 0.533 (0.154) | 0.967 (0.033) | 0.898 (0.049) | 0.807 (0.105) | |
| Oura Ring | FALSE | None | 0.897 (0.053) | 0.914 (0.045) | 0.692 (0.083) | 1.926 (0.428) | 0.441 (0.149) | 0.929 (0.050) | 0.977 (0.021) | 0.407 (0.176) |
| FALSE | Oversampling | 0.888 (0.063) | 0.839 (0.075) | 0.780 (0.079) | 1.855 (0.451) | 0.404 (0.135) | 0.960 (0.039) | 0.853 (0.096) | 0.707 (0.208) | |
| TRUE | None | 0.892 (0.063) | 0.906 (0.050) | 0.689 (0.082) | 1.827 (0.454) | 0.418 (0.135) | 0.925 (0.051) | 0.971 (0.031) | 0.407 (0.174) | |
| TRUE | Oversampling | 0.896 (0.062) | 0.844 (0.057) | 0.805 (0.067) | 1.876 (0.462) | 0.422 (0.153) | 0.964 (0.030) | 0.855 (0.071) | 0.755 (0.143) | |
| ECG + Link | FALSE | None | 0.924 (0.039) | 0.917 (0.049) | 0.755 (0.070) | 2.321 (0.330) | 0.560 (0.112) | 0.938 (0.047) | 0.970 (0.049) | 0.540 (0.170) |
| FALSE | Oversampling | 0.923 (0.037) | 0.870 (0.082) | 0.830 (0.055) | 2.191 (0.350) | 0.521 (0.140) | 0.966 (0.035) | 0.883 (0.105) | 0.776 (0.144) | |
| TRUE | None | 0.926 (0.041) | 0.922 (0.043) | 0.788 (0.065) | 2.383 (0.436) | 0.595 (0.113) | 0.943 (0.040) | 0.968 (0.045) | 0.607 (0.147) | |
| TRUE | Oversampling | 0.924 (0.042) | 0.878 (0.048) | 0.856 (0.043) | 2.253 (0.372) | 0.536 (0.156) | 0.970 (0.029) | 0.890 (0.059) | 0.821 (0.096) |
Values in each cell are the mean and SD across nights.
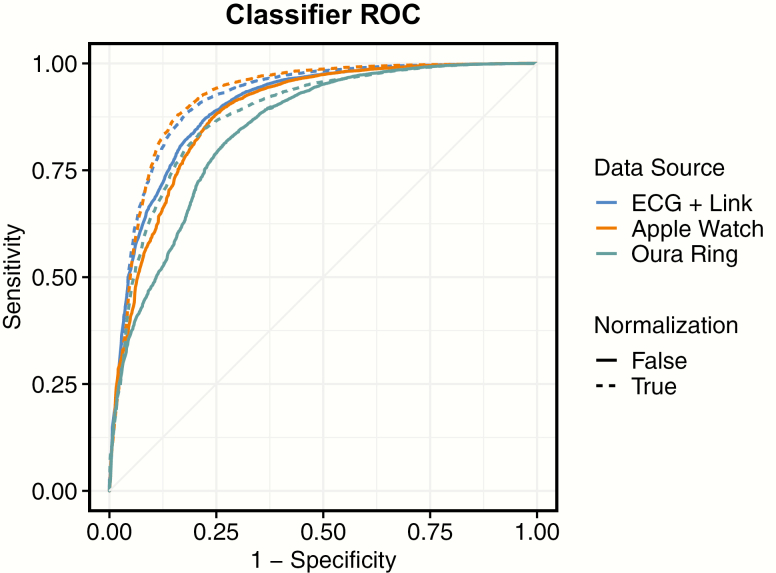
Figure 4 displays the mean ROC curve across nights, for the three datasets under study, both with and without night-level normalization. Model variations with class oversampling are not displayed, as AUC is largely unaffected by class oversampling.
Figure 4.
Mean receiver operating characteristic (ROC) curves, for each classifier, both with and without night-level normalization. Each line depicts the point-by-point average of ROC across nights classified. Classifiers depicted are without class oversampling.
Analysis 3B: Night-level metrics derived from classifier output
The sleep–wake output provided by each classifier was used to compute several night-level metrics, including sleep efficiency, SOL, WASO, and TST, which were compared to the same metrics computed from the PSG-derived staging. Measures of classifier error are displayed within Table 6. While the classifiers perform reasonably well at reproducing sleep efficiency, WASO, and TST, they perform poorly at reproducing SOL. Figure 5 displays a scatterplot of the correspondence between classifier and PSG-derived staging for each metric, most evident is the difficulty of capturing the variance of SOL across nights. Figure 6 displays the bias of each classifier relative to PSG-derived staging for each metric.
Table 6.
Estimation of Night-Level Metrics, Using Data From Classifiers Built on Various Data Sources
| Metric | Device | Mean error | MAE | RMSE | R 2 |
|---|---|---|---|---|---|
| Sleep efficiency (%) | Apple Watch | 2.55 | 4.24 | 4.72 | 0.33 |
| Oura Ring | 3.58 | 4.55 | 5.44 | 0.43 | |
| ECG + Link | 1.84 | 4.64 | 5.48 | 0.27 | |
| Sleep onset latency (min) | Apple Watch | −2.15 | 5.89 | 8.70 | 0.00 |
| Oura Ring | −1.38 | 3.79 | 5.02 | 0.04 | |
| ECG + Link | −0.87 | 6.52 | 9.08 | 0.00 | |
| WASO (min) | Apple Watch | −11.58 | 18.27 | 20.49 | 0.51 |
| Oura Ring | −16.90 | 21.51 | 25.42 | 0.48 | |
| ECG + Link | −8.95 | 19.11 | 23.79 | 0.42 | |
| Total sleep time (min) | Apple Watch | 13.73 | 22.69 | 25.29 | 0.35 |
| Oura Ring | 18.28 | 23.28 | 27.64 | 0.76 | |
| ECG + Link | 9.82 | 24.92 | 29.48 | 0.31 |
Here, the values reported are for the models computed with night-level normalization, but without class oversampling.
Figure 5.
Comparison of PSG- and classifier-derived night-level sleep metrics. Points depict the PSG- and classifier-derived values for each night of data within the study (except nights previously described as excluded). Lines depict the linear fit between PSG and classifier-derived values.
Figure 6.
Bland-Altman [49] plots comparing PSG-derived sleep metrics to the difference between each classifier-derived sleep metric and the PSG-derived sleep metric. Points depict difference values for each night of data within the study (except nights previously described as excluded). Solid lines depict the mean bias between PSG and device-derived values, while dashed lines depict the 95% confidence interval for the bias across nights.
Discussion
Both epoch-by-epoch and night-level measures of sleep–wake detection were evaluated for two wrist devices and one ring device, relative to “gold standard” RPSGT-scored polysomnography. Consistent with past work that evaluated actigraphy for sleep–wake detection [3], typical actigraphy and multisensor algorithms were of high sensitivity (% of sleep epochs correctly classified) but low specificity (% of sleep epochs correctly classified). Sleep, the more prevalent class within the in-bed period (approximately 88% of staged epochs in the present experiment), was overclassified, while wake was underclassified. This imbalance in performance is consistent with previous reports for the sleep–wake output of the Actiwatch Spectrum [3] and Oura Ring [33].
The night-level analysis, comparing how accurately data from each device can be used to derive measures of sleep efficiency, SOL, WASO, and TST, provides a complementary view of device performance. For example, although no single device was consistently optimal for all measures, the Oura Ring generated the highest R2 relative to PSG-derived staging for three of the four measures under study, despite not necessarily generating the lowest mean error, and having lower performance than the ActiGraph Link in terms of the epoch-by-epoch performance measures of balanced accuracy or d′. While poor time synchronization between a given device and PSG could contribute to a discrepancy between epoch-by-epoch and night-level performance, a posthoc synchronization process was performed within the present experiment to correct for any static time offsets between each device’s sleep–wake output and PSG. Specifically, the sleep–wake output from each device was shifted ±5 min relative to the PSG-derived staging, to identify the optimal correspondence.
Following analysis of the sleep–wake classifications output from the wrist actigraphy devices and Oura Ring, we evaluated the raw data quality from the Oura Ring and Apple Watch and developed our own novel machine learning models of sleep–wake. We also developed a classifier using data assumed to be of higher quality, using heart rate data derived from the ECG channel of the PSG in conjunction with triaxial actigraphy data from the ActiGraph Link, for comparison.
In terms of raw data quality, we observed moderate to strong correlations between the data from the wearables and data from clinical measurement tools at the level of the 30-s sleep epoch. For example, despite the Apple Watch providing heart rate estimates approximately every 5 s, the mean pseudo-IBI within a sleep epoch computed from the Apple Watch had an average correlation of 0.92 to the mean IBI derived from the ECG channel of the PSG. Similarly, the mean IBI from the Oura Ring has an average correlation of 0.85 to the mean IBI from the ECG channel of the PSG.
Actigraphy features were also reasonably correlated despite some differences in the underlying data collection parameters. For example, the mean vector magnitude within an epoch derived from the triaxial accelerometer collected from the Apple Watch at 1.33 Hz had an average correlation of 0.83 to the mean vector magnitude collected from the ActiGraph Link at 80 Hz. Similarly, the “motion seconds” value provided by the Oura Ring at 30-s intervals had an average correlation of 0.53 to the mean vector magnitude from the ActiGraph Link. The lower correspondence for the motion feature derived from the Oura Ring does not necessarily suggest that the Oura Ring is poorer at capturing motion than the Apple Watch, but likely reflects the lower granularity of the data that was available for export from the Oura Ring (“motion seconds” at 30-s intervals) during the time of the study. In general, the high correspondence of data from these multisensor devices relative to PSG or high-resolution triaxial actigraphy is promising with respect to using the devices when PSG is not available and is applicable to the goal of longitudinal, noninvasive measurement of the sleep.
We trained and tested variations of our classifier, using data from either the Apple Watch, Oura Ring, or for a point of comparison, data from the ECG channel of the PSG in combination with actigraphy data from the ActiGraph Link. In addition to the data features from each device, a temporal feature encoding the number of seconds that had elapsed into the in-bed period was included. Our temporal feature is linear, increasing in 30-s increments with each epoch. Other recent reports have also included temporal features in sleep staging models. For example, Fonseca et al. [50] used a linear time feature, while Walch et al. [51] used a mathematical model of the circadian clock specific to each participant. Each model was additionally trained with variations that incorporated class balancing via random oversampling of the minority class (wake), and/or normalization of datasets at the night level via transformation of each feature to a z-score within the night. Oversampling was intended to ameliorate the common behavior of sleep–wake classifiers in the performance imbalance between the sensitivity and specificity.
Oversampling to balance classes during training did improve specificity (20%–35%), but at a cost of reducing sensitivity (8%–12%). Whether sensitivity or specificity is more favored may depend on the application of the researcher. For example, because the oversampling procedure detects a greater percentage of wake epochs, it may be more appropriate to use in populations who have a sleep pathology. Practically, as oversampling did not affect AUC for the paradigm used here and our classifier natively outputs probabilities, a similar result could be achieved by shifting the probability threshold.
Night-level normalization resulted in a small improvement in model performance for all models with the exception of the model using Oura Ring data without oversampling. While normalizing data at the night level can only be performed once the night has ended and the distribution of values across the night is known, using the distribution of data for a given user from the prior session may allow for both data normalization and real-time classification.
The devices included in Analysis 1 vary along several dimensions, including the type of physiological data they use, their hardware implementation, and the algorithm internally used to generate sleep–wake classifications. However, despite these variations, they serve as a useful point of comparison for evaluating the performance of the alternative sleep–wake classifier we developed within Analysis 3.
The best performing research device for measuring epoch-by-epoch sleep–wake within the experiment as evaluated by d′ was the ActiGraph Link running the Sadeh algorithm (d′ = 1.874, sensitivity = 0.912, specificity = 0.647). For comparison, the classifier we developed, which was trained and tested using data from the Apple Watch with our normalization algorithm over the night achieved epoch-by-epoch sleep–wake classification performance (d′ = 2.347, sensitivity = 0.976, specificity = 0.602, AUC = 0.926). The same model with the addition of balancing classes prior to training results had a similar d′, but a shift in a bias away from sleep and toward wake (d′ = 2.237, sensitivity = 0.898, specificity = 0.807, AUC = 0.922).
Comparison of the performance of each device, and our models, on night-level summary measures such as sleep efficiency, SOL, WASO, and TST revealed that the best performing device in terms of epoch-by-epoch sleep–wake classification is not necessarily the best for each summary measure. This is especially the case when considering that these measures can be evaluated in terms of either absolute accuracy (mean error) or their ability to capture variance across nights (R2). For example, while the Oura Ring was not the most accurate device in terms of epoch-by-epoch accuracy among the research devices, it tended to capture the most variance relative to RPSGT-derived ground-truth metrics (R2), for all metrics except SOL. One caveat is that four nights were excluded from the Oura comparison; two nights were excluded due to corrupted recordings, and two were excluded due to a seeming failure of the device to detect the wearer was in bed and begin to provide classifications.
Similarly, while our models performed favorably in comparison to the research devices in terms of the epoch-by-epoch analysis, they were not more effective at deriving night-level summary values. As these summary values are not “known” by the classifier during training, the optimal epoch-by-epoch classifier may not generate the optimal night summary, though in theory a perfect epoch-by-epoch classifier would reproduce all summary values precisely. One illustration of this discrepancy is that the classifier we developed using data from the Oura Ring often captured more variance between nights in terms of R2 relative to the classifiers we developed using data from the Apple Watch or ECG-Link, despite having lower epoch-by-epoch performance. The data from our classifiers had particular trouble reproducing RPSGT staging-derived measures of SOL. As SOL according to AASM criteria is the time between “lights out” and the first epoch scored as sleep, misclassification of a single early epoch can have a large influence on the accuracy of the measure.
A recent report by Walch et al. [51] also used data collected from an Apple Watch to develop classifiers for sleep–wake and sleep stage. Additionally, as previously described, this recent report also used a feature that encoded information about time within the night. However, there are several differences between this prior report and the present result. While Walch et al. collected triaxial accelerometer data from the Apple Watch, accelerometer data were subsequently converted into movement counts using an algorithm developed to approximate Actiwatch movement counts from microelectromechanical systems accelerometers [52]. However, because the accelerometer contained within the Actiwatch is most sensitive to movements along a single axis (palmar–dorsal) [52], this approach utilizes only one axis of the original triaxial accelerometer data, in support of approximating Actiwatch style movement counts. In contrast, our approach uses the vector magnitude of the triaxial accelerometer data, the exception being the Oura Ring, for which we were only able to obtain count-level motion data from the device.
An additional distinction is that the approach described by Walch et al. [51] uses data from a 10-min window around each sleep epoch being scored, while our approach uses only historical data, namely, data from the 30-s epoch being scored in addition to data from the prior eight epochs (4 min). Using data from the near future is common for sleep–wake staging, for example, the Cole–Kripke [6] and Sadeh [31] algorithms utilize movement counts from both the recent past and near future when classifying a given epoch, and is a reasonable approach if sleep–wake state is going to be evaluated retrospectively, after the night has completed. However, our approach instead facilitates each 30-s epoch to be classified immediately, allowing for real-time applications.
Many multisensor wearable devices, such as the Apple Watch, allow data to be processed in real time, as opposed to conventional actigraphy devices, which are designed to log the data on the device, and make the raw data accessible only after the sleep session has occurred. This precludes utility to inform real-time interventions that are delivered during monitoring with PSG. For example, ECG data collected in PSG were combined with pulse oximetry to detect sleep stage and sleep apnea during sleep for the purpose of administering interventions to mitigate breathing obstruction [53–55]. The real-time availability of data in multisensor devices like the Apple Watch creates the possibility of delivering interventions during sleep for enhancing or manipulating sleep quality. Real-time sleep staging on accessible consumer wearables has several potential applications, including the treatment of sleep disorders [53–55], enhancement of sleep with deep sleep stimulation [56], and enhancement of memory through targeted memory reactivation [57–59].
There are also notable limitations to the use of consumer wearable devices, in their present form. The battery life on a device such as the Apple Watch, while sufficient to collect data throughout a night of sleep, is currently not capable of 24 h of continuous recording of high-resolution data without needing to be recharged. Full access to raw sensor data from consumer wearables is often not available but could further improve the accuracy and transparency of sleep detection models. For example, the Apple Watch is equipped with a PPG heart rate sensor, but Apple does not currently allow developers to access the PPG sensor data. Instead, the PPG data are processed by the device and an estimate of the instantaneous heart rate is provided at intervals of approximately 5 s. If the raw PPG sensor data were available from the Apple Watch, additional measures could be derived from the time series [60], such as arterial blood pressure [61], which could be used to improve the classification of sleep stage [62].
There are several limitations to the present study. It included a relatively small sample of eight healthy participants, all in early midlife, sleeping in a highly controlled laboratory environment. While it was important to include only healthy participants when investigating a small sample, additional work is required to identify how well sleep–wake models trained on healthy participants generalize to participants with sleep disorders and in outpatient settings. A potential solution is to add wearable devices to sleep studies already occurring, facilitating the relatively easy procurement of generalizing data to a broader cross-section of individuals.
Additionally, although several data sources were included in the present report to demonstrate the relative performance that can be obtained with different sources of data, models were trained and tested within a given data source. More effort on data standardization may be required, for example, to train a model on PSG data and implement it using a wearable like the Apple Watch. Similarly, some devices provided sleep–wake classifications at 60-s intervals rather than the 30-s intervals in which the RPSGT staged the data, requiring alignment. Another limitation of the present study was that models were developed over the course of the night, as opposed to over a 24-h period. A model developed on 24-h data which contains a greater number of periods of wakefulness may be useful to decrease the bias toward the classification of sleep that is common in sleep–wake classification. Furthermore, while the present report focuses on the classification of sleep–wake, a similar approach may be used to classify the sleep stage.
The results of the study highlight the potential for using multisensor wearables to measure sleep in not only research and clinical contexts, but also in ecologically valid in-home study settings. Relative to many research-grade and clinical-grade actigraphy devices, consumer devices like the Oura Ring and Apple Watch are more affordable and accessible. Such devices are capable of sleep monitoring in the outpatient environment, which may be more reimbursable by healthcare companies than the expensive inpatient setting. The longitudinal, repeated, and multisensor quality of these devices makes them ideal for evaluating insomnia or circadian rhythm disorders and can be further developed to provide users with tailored interventions that examine the cause of their sleep issues. Clinicians can then use these new tools to facilitate behavior change and administer more effective cognitive behavioral therapy for insomnia [63].
Funding
This work was supported by the National Science Foundation (NSF) under grant 1622766 awarded to DG (PI; CEO Mobile Sleep Technologies LLC/Proactive Life Inc. [DBA Sonic Sleep Coach]). Work was conducted at Pennsylvania State University (via subcontract) and further supported by the Pennsylvania State University Clinical and Translational Sciences Institute (funded by the National Center for Advancing Translational Sciences, National Institutes of Health, through grant UL1TR002014) and institutional funds from the College of Health and Human Development of the Pennsylvania State University to OMB. Collaboration also includes a separate project: National Institutes of Health/National Institute on Aging SBIR R43AG056250 to DG (PI; CEO Mobile Sleep Technologies LLC/Proactive Life Inc., DBA Sonic Sleep Coach) “Non-pharmacological improvement of sleep structure in midlife and older adults.” OMB also receives an honorarium from the National Sleep Foundation for his role as Editor in Chief (Designate) of Sleep Health (sleephealthjournal.org).
Acknowledgments
We would like to thank the participants for providing their data.
Financial Disclosures: DMR and DG are employed by Proactive Life, Inc. a for-profit company. Proactive Life has two patents granted or in application related to monitoring sleep: Sleep Stimulation and Monitoring (US patent 14302482A); Cyclical Behavior Modification (US Patent 8468115). OMB discloses that he received two subcontract grants to Penn State from Mobile Sleep Technologies (NSF/STTR 1622766, NIH/NIA SBIR R43AG056250), received honoraria/travel support for lectures from Boston University, Boston College, Tufts School of Dental Medicine, and Allstate, and receives an honorarium for his role as the Editor in Chief (designate) of Sleep Health (sleephealthjournal.org).
Nonfinancial Disclosures: None.
References
- 1. Kupfer DJ, et al.. The application of Delgado’s telemetric mobility recorder for human studies. Behav Biol. 1972;7(4):585–590. [DOI] [PubMed] [Google Scholar]
- 2. Ancoli-Israel S, et al.. The role of actigraphy in the study of sleep and circadian rhythms. Sleep. 2003;26(3):342–392. [DOI] [PubMed] [Google Scholar]
- 3. Marino M, et al.. Measuring sleep: accuracy, sensitivity, and specificity of wrist actigraphy compared to polysomnography. Sleep. 2013;36(11):1747–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kripke DF, et al.. Wrist actigraphic measures of sleep and rhythms. Electroencephalogr Clin Neurophysiol. 1978;44(5):674–676. [DOI] [PubMed] [Google Scholar]
- 5. Mullaney DJ, et al.. Wrist-actigraphic estimation of sleep time. Sleep. 1980;3(1):83–92. [DOI] [PubMed] [Google Scholar]
- 6. Cole RJ, et al.. Automatic sleep/wake identification from wrist activity. Sleep. 1992;15(5):461–469. [DOI] [PubMed] [Google Scholar]
- 7. Jean-Louis G, et al.. Sleep detection with an accelerometer actigraph: comparisons with polysomnography. Physiol Behav. 2001;72(1–2):21–28. [DOI] [PubMed] [Google Scholar]
- 8. Webster JB, et al.. An activity-based sleep monitor system for ambulatory use. Sleep. 1982;5(4):389–399. [DOI] [PubMed] [Google Scholar]
- 9. Blood ML, et al.. A comparison of sleep detection by wrist actigraphy, behavioral response, and polysomnography. Sleep. 1997;20(6):388–395. [PubMed] [Google Scholar]
- 10. de Souza L, et al.. Further validation of actigraphy for sleep studies. Sleep. 2003;26(1):81–85. [DOI] [PubMed] [Google Scholar]
- 11. Kushida CA, et al.. Comparison of actigraphic, polysomnographic, and subjective assessment of sleep parameters in sleep-disordered patients. Sleep Med. 2001;2(5):389–396. [DOI] [PubMed] [Google Scholar]
- 12. Paquet J, et al.. Wake detection capacity of actigraphy during sleep. Sleep. 2007;30(10):1362–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Signal TL, et al.. Sleep measurement in flight crew: comparing actigraphic and subjective estimates to polysomnography. Aviat Space Environ Med. 2005;76(11):1058–1063. [PubMed] [Google Scholar]
- 14. Sivertsen B, et al.. A comparison of actigraphy and polysomnography in older adults treated for chronic primary insomnia. Sleep. 2006;29(10):1353–1358. [DOI] [PubMed] [Google Scholar]
- 15. Chattu VK, et al.. Insufficient sleep syndrome: is it time to classify it as a major noncommunicable disease? Sleep Sci. 2018;11(2):56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hafner M, et al.. Why Sleep Matters — the Economic Costs of Insufficient Sleep: A Cross-Country Comparative Analysis. Santa Monica, CA: RAND Corporation; 2016: 101. [PMC free article] [PubMed] [Google Scholar]
- 17. Hillman D, et al.. The economic cost of inadequate sleep. Sleep. 2018;41(8). doi: 10.1093/sleep/zsy083 [DOI] [PubMed] [Google Scholar]
- 18. Rosekind MR, et al.. Insomnia risks and costs: health, safety, and quality of life. Am J Manag Care. 2010;16(8):617–626. [PubMed] [Google Scholar]
- 19. Rosekind MR, et al.. The cost of poor sleep: workplace productivity loss and associated costs. J Occup Environ Med. 2010;52(1):91–98. [DOI] [PubMed] [Google Scholar]
- 20. Sluiter JK. High-demand jobs: age-related diversity in work ability? Appl Ergon. 2006;37(4):429–440. [DOI] [PubMed] [Google Scholar]
- 21. Bhattacharyya N. Abnormal sleep duration is associated with a higher risk of accidental injury. Otolaryngol Head Neck Surg. 2015;153(6):962–965. [DOI] [PubMed] [Google Scholar]
- 22. de Mello MT, et al.. Sleep disorders as a cause of motor vehicle collisions. Int J Prev Med. 2013;4(3):246–257. [PMC free article] [PubMed] [Google Scholar]
- 23. Koppel S, et al.. Older drivers, crashes and injuries. Traffic Inj Prev. 2011;12(5):459–467. [DOI] [PubMed] [Google Scholar]
- 24. Pandi-Perumal SR, et al.. Sleep disorders, sleepiness and traffic safety: a public health menace. Braz J Med Biol Res. 2006;39(7):863–871. [DOI] [PubMed] [Google Scholar]
- 25. Tefft BC. Risks older drivers pose to themselves and to other road users. J Safety Res. 2008;39(6):577–582. [DOI] [PubMed] [Google Scholar]
- 26. Léger D, et al.. Societal costs of insomnia. Sleep Med Rev. 2010;14(6):379–389. [DOI] [PubMed] [Google Scholar]
- 27. Liu Y, et al.. Relationship between sleep duration and self-reported health-related quality of life among US adults with or without major chronic diseases, 2014. Sleep Health. 2018;4(3):265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stenholm S, et al.. Sleep duration and sleep disturbances as predictors of healthy and chronic disease–free life expectancy between ages 50 and 75: a pooled analysis of three cohorts. J Gerontol Ser A. February 2018. doi: 10.1093/gerona/gly016 [DOI] [PubMed] [Google Scholar]
- 29. Wang S, et al.. Relationship of sleep duration with sociodemographic characteristics, lifestyle, mental health, and chronic diseases in a large Chinese adult population. J Clin Sleep Med. 2017;13(03):377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Watson NF. Evolve sleep: optimized solutions to help your sleep medicine practice thrive. J Clin Sleep Med. 2015;11(8):841–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sadeh A, et al.. Activity-based sleep-wake identification: an empirical test of methodological issues. Sleep. 1994;17(3):201–207. [DOI] [PubMed] [Google Scholar]
- 32. de Zambotti M, et al.. Dynamic coupling between the central and autonomic nervous systems during sleep: a review. Neurosci Biobehav Rev. 2018;90:84–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Zambotti M, et al.. The sleep of the ring: comparison of the ŌURA sleep tracker against polysomnography. Behav Sleep Med. March 2017:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Zambotti M, et al.. A validation study of Fitbit Charge 2™ compared with polysomnography in adults. Chronobiol Int. 2018;35(4):465–476. [DOI] [PubMed] [Google Scholar]
- 35. de Zambotti M, et al.. The boom in wearable technology: cause for alarm or just what is needed to better understand sleep? Sleep. 2016;39(9):1761–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schade MM, et al.. Auditory stimulation during sleep transiently increases delta power and all-night proportion of NREM stage 3 sleep while preserving total sleep time and continuity. Presented at: Society for Neuroscience; November 4, 2018; San Diego, CA. https://www.abstractsonline.com/pp8/#!/4649/presentation/38903. Accessed August 14, 2019.
- 37. Berry RB, et al.. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications. Darien, IL: American Academy of Sleep Medicine; 2017. [Google Scholar]
- 38. Driller MW, et al.. What wrist should you wear your actigraphy device on? Analysis of dominant vs. non-dominant wrist actigraphy for measuring sleep in healthy adults. Sleep Sci. 2017;10(3):132–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wing MKC, et al.. Caret: Classification and Regression Training; 2019. https://CRAN.R-project.org/package=caret.
- 40. Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas. 1960;20(1):37–46. [Google Scholar]
- 41. Stanislaw H, et al.. Calculation of signal detection theory measures. Behav Res Methods Instrum Comput. 1999;31(1):137–149. [DOI] [PubMed] [Google Scholar]
- 42. Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Stat. 2001;29(5):1189–1232. [Google Scholar]
- 43. Pedregosa F, et al.. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825− 2830. [Google Scholar]
- 44. Stone M. Cross-validatory choice and assessment of statistical predictions. J R Stat Soc Ser B Methodol. 1974;36(2):111–147. [Google Scholar]
- 45. Cawley GC, et al.. On over-fitting in model selection and subsequent selection bias in performance evaluation. J Mach Learn Res. 2010;11:2079–2107. [Google Scholar]
- 46. Varma S, et al.. Bias in error estimation when using cross-validation for model selection. BMC Bioinformatics. 2006;7:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moreno-Torres JG, et al.. A unifying view on dataset shift in classification. Pattern Recognit. 2012;45(1):521–530. [Google Scholar]
- 48. Lemaître G, et al.. Imbalanced-learn: a python toolbox to tackle the curse of imbalanced datasets in machine learning. J Mach Learn Res. 2017;18(17):1–5. [Google Scholar]
- 49. Bland JM, et al.. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1(8476):307–310. [PubMed] [Google Scholar]
- 50. Fonseca P, et al.. A comparison of probabilistic classifiers for sleep stage classification. Physiol Meas. 2018;39(5):055001. [DOI] [PubMed] [Google Scholar]
- 51. Walch O, et al.. Sleep stage prediction with raw acceleration and photoplethysmography heart rate data derived from a consumer wearable device. Sleep. 2019;42(12). doi: 10.1093/sleep/zsz180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. te Lindert BHW, et al.. Sleep estimates using microelectromechanical systems (MEMS). Sleep. 2013;36(5):781–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Heneghan C, et al.. A portable automated assessment tool for sleep apnea using a combined Holter-oximeter. Sleep. 2008;31(10):1432–1439. [PMC free article] [PubMed] [Google Scholar]
- 54. Xie B, et al.. Real-time sleep apnea detection by classifier combination. IEEE Trans Inf Technol Biomed. 2012;16(3):469–477. [DOI] [PubMed] [Google Scholar]
- 55. Xie B, et al.. An improved approach for real-time detection of sleep apnea. In: proceedings of the International Conference on Bio-Inspired Systems and Signal Processing; Rome, Italy: SciTePress - Science and Technology Publications; 2011:169–175. doi: 10.5220/0003137101690175 [DOI] [Google Scholar]
- 56. Ngo HV, et al.. Auditory closed-loop stimulation of the sleep slow oscillation enhances memory. Neuron. 2013;78(3):545–553. [DOI] [PubMed] [Google Scholar]
- 57. Oudiette D, et al.. Upgrading the sleeping brain with targeted memory reactivation. Trends Cogn Sci. 2013;17(3):142–149. [DOI] [PubMed] [Google Scholar]
- 58. Portas CM, et al.. Auditory processing across the sleep-wake cycle: simultaneous EEG and fMRI monitoring in humans. Neuron. 2000;28(3):991–999. [DOI] [PubMed] [Google Scholar]
- 59. Rasch B, et al.. Odor cues during slow-wave sleep prompt declarative memory consolidation. Science. 2007;315(5817):1426–1429. [DOI] [PubMed] [Google Scholar]
- 60. Shelley KH. Photoplethysmography: beyond the calculation of arterial oxygen saturation and heart rate. Anesth Analg. 2007;105(6 Suppl):S31–S36, tables of contents. [DOI] [PubMed] [Google Scholar]
- 61. Teng XF, et al.. Continuous and noninvasive estimation of arterial blood pressure using a photoplethysmographic approach. In: proceedings of the 25th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (IEEE Cat. No.03CH37439); Cancun, Mexico: IEEE; 2003:3153–3156. doi: 10.1109/IEMBS.2003.1280811 [DOI] [Google Scholar]
- 62. Loredo JS, et al.. Sleep quality and blood pressure dipping in normal adults. Sleep. 2004;27(6):1097–1103. [DOI] [PubMed] [Google Scholar]
- 63. Avidan AY. Sleep and neurologic problems in the elderly. Sleep Med Clin. 2006;1(2):273–292. [Google Scholar]