Abstract
Salmonella is an important genus of Gram-negative pathogens, treatment of which has become problematic due to increases in antimicrobial resistance. This is partly attributable to the overexpression of tripartite efflux pumps, particularly the constitutively expressed AcrAB-TolC. Despite its clinical importance, the structure of the Salmonella AcrB transporter remained unknown to-date, with much of our structural understanding coming from the Escherichia coli orthologue. Here, by taking advantage of the styrene maleic acid (SMA) technology to isolate membrane proteins with closely associated lipids, we report the very first experimental structure of Salmonella AcrB transporter. Furthermore, this novel structure provides additional insight into mechanisms of drug efflux as it bears the mutation (G288D), originating from a clinical isolate of Salmonella Typhimurium presenting an increased resistance to fluoroquinolones. Experimental data are complemented by state-of-the-art molecular dynamics (MD) simulations on both the wild type and G288D variant of Salmonella AcrB. Together, these reveal several important differences with respect to the E. coli protein, providing insights into the role of the G288D mutation in increasing drug efflux and extending our understanding of the mechanisms underlying antibiotic resistance.
Keywords: Salmonella, multidrug efflux pump, membrane proteins, multidrug resistance, AcrB, cryo-EM, molecular dynamics
1. Introduction
Bacterial multidrug resistance (MDR) is a growing global concern with many antibiotics now ineffective against major classes of pathogens. This is especially pertinent in Gram-negative organisms, which are intrinsically more resistant due to the presence of a double membrane. Salmonella is an important Gram-negative pathogen, the prevalence of which is increasing especially in nosocomial settings. While there are many adaptive routes for the development of MDR in Salmonella [1], the role of the multidrug efflux-pumps in the process has been identified as one of the key mechanisms for escaping the antibiotic and biocide selective pressures [2,3,4]. The action of the resistance-nodulation-division (RND) transporter AcrB provides the principal antimicrobial resistance efflux function in Salmonella enterica serovar Typhimurium (from here on, S. Typhimurium) [5]. In vivo AcrB forms a complex with the outer membrane factor (OMF) TolC and the periplasmic adaptor protein (PAP) AcrA, forming a functional tripartite complex with a 3:6:3 stoichiometry respectively, that spans both the inner and outer membrane [3,6,7,8,9]. Among the nine different tripartite efflux pumps involved in MDR in S. Typhimurium [5], seven form tripartite complexes with TolC [10], and AcrB is the primary transporter associated with MDR. Consistent with its major role, the loss of AcrB confers a loss of virulence in Salmonella [11] despite the considerable overlap of specificities and partial compensation from overexpression of homologous RND-based tripartite-pumps [12]. Furthermore, the presence of the AcrAB-TolC efflux pump is required for the development of ciprofloxacin resistance [13] and is essential for biofilm formation [14].
Most of our current knowledge of AcrB transporter function is based on the E. coli orthologue, which shares 94.7% sequence identity to the S. Typhimurium pump. AcrB is organised as a homotrimer [15], the functional rotation of which has been proposed as a structural mechanism of the efflux process with each protomer undergoing sequential conformational cycling of states known as access, binding and extrusion (A, B and E respectively) [16], or, alternatively, in parallel to the ATP-synthase cycle—loose, tight, open (L, T, O) [17]. Our understanding of this mechanism has been reinforced by both structural and mutagenesis approaches including conditional cross-linking [18,19,20], as well as by computer simulations [21,22]. The sequestering of substrates/drugs by the pump can occur through a number of different pathways—known as substrate channels, that allow for capture of the cargo from the periplasmic space (channel 2 and channel 3) or from the outer leaflet of the inner membrane (channel 1 and channel 4) [23,24,25]. In the L (A) protomer, the channels 1 and 2 converge on a “proximal” drug-binding pocket (PBP), also known as the “access” pocket [19,23,26], which is separated by a “switch-loop” from the deep- or distal-binding pocket (DBP). The latter becomes occupied in the T protomer. Channel 3, which originates from an interprotomer space known as the vestibule has been shown to bypass the PBP providing direct access to the DBP and a distinct selectivity [27]. Channel 4 was discovered very recently as the putative entry path for fusidic acid and other carboxylated antibiotics, and is directly connected to the DBP within the T protomer (from here on referred as DBPT) [25,28].
The electrochemical potential of the membrane is utilised for transport via a proton-relay located in the transmembrane (TM) portion of the protein, which induces a conformational change that is communicated to the drug binding pockets over > 50 Å via a piston-like action involving TM helix 2 [21,29]. Recent free energy calculations suggest that protonation of D408 in the TM domain of the drug-bound protomer drives the functional rotation [21], albeit all the residues forming the proton relay are essential.
Although the main mechanism of efflux-based resistance in Enterobacteriaceae is provided by overexpression of the efflux pumps via mutations of the regulatory regions of their operons or their respective repressors (e.g., in E. coli expression of the genes of the principal efflux pump acrAB/tolC is predominantly controlled by the interplay of the local and global transcription regulators AcrR and MarA, while in Salmonella, these genes are under the control of AcrR and RamA, respectively, for reference see [30,31,32]), a new mechanism of adaptive resistance has been reported, namely the selection of mutations altering the specificity of the RND-transporter itself [33]. The particular mutation, which arose during the antibiotic treatment of a patient with a complex Salmonella infection [34,35], has resulted in a substitution, G288D, within the Salmonella Typhimurium AcrB transporter (hereafter STmAcrBG288D). This mutation, is responsible for the occurrence of MDR, particularly to ciprofloxacin, increasing the MIC to the drug over 60 folds. Thus, unveiling the molecular structure of STmAcrB and the role of specific residues is critical to our understanding of the mechanism of antibiotic recognition and efflux that confers the clinically relevant MDR in Salmonella, yet to date there has not been an experimental structure of Salmonella AcrB available.
Here, we report the first structure of the Salmonella AcrB transporter carrying the clinically relevant G288D mutation, solved by cryogenic electron microscopy (cryo-EM). This provides an insight into the structural rearrangement AcrB and also the role of the G288D mutation that underlies its multidrug resistance. Molecular dynamics (MD) simulations were further employed to provide atomistic details on the alteration of protein functional dynamics due to the G288D substitution.
2. Materials and Methods
2.1. Cloning of AcrB G288D
DNA library from clinical isolate strain L18 [33], was used to amplify Salmonella Typhimurium AcrB G288D mutant (STmAcrBG288D) using the following primers:
Salm_acrB+: TAGCTTCATATGCCTAATTTCTTTATCGATCGCCCTATATTTGCGTGGGTG
Salm_acrB−: ATATTCCTCGAGGCGATGTTCTGTCGAATGACTATGCTCAATATCTTCGC
The PCR product was cloned into pET26b using the NdeI and XhoI restriction sites (underlined). BW25113 was used as the propagating strain for cloning. E. coli C43ΔacrB (DE3) cells were used for protein expression.
2.2. Protein Overexpression and Purification
The protein was expressed using auto-induction and membrane preparation, styrene maleic acid (SMA) solubilisation and protein purification were performed using the protocols previously reported [36,37]. Cells were disrupted with a cell disrupter (Constant Systems Ltd.) at 30 kpsi after resuspension in 20 mM Tris, 0.5mM EDTA pH 7.9 (4 °C) in a ratio of 4:1 buffer to pellet. The cell membranes were isolated by differential centrifugation as previously described [38]. AcrB was extracted in its native trimeric form (~340kDa) using 2.5% (w/v) SMA, incubated for 2 h at room temperature as previously described [36,39]. The insoluble material was removed by centrifugation (100,000× g, 1 h, 4 °C). The supernatant was mixed with 2 mL HisPur™ Cobalt Resin (Thermo scientific) and incubated in batch overnight at 4 °C with gentle agitation. Following packing, the resin was washed twice with wash buffer (50 mM Tris-HCl, 500 mM NaCl, 10% (w/v) glycerol). After elution (50 mM Tris-HCl, 500 mM NaCl, 10% (w/v) glycerol, 300 mM imidazole), the peak fractions were pooled out and dialysed against a buffer free of imidazole.
2.3. Negative Stain Electron Microscopy
The purified STmAcrBG288D sample was assessed for sample quality by negative stain microscopy and grids were prepared as previously described [39]. Briefly, carbon-coated grids were glow-discharged for 40 s, following which 3µl of the STmAcrBG288D (~20 µg/mL) was added to the grid and stained with 2% uranyl acetate. Grids were imaged on a Technai F20 microscope fitted with a field emission gun operating at 200kV with a nominal magnification of 50,000× on a CMOS detector.
2.4. Cryo-Electron Microscopy
For single particle cryo-EM analysis, gold quantifoil grids (2.1 mesh) were glow discharged (20 s) before applying 3µL of ~1 mg/mL STmAcrBG288D. Grids were prepared using a Vitrobot Mark IV and blotted with Ash-free Whatman filter paper (No. 50) for 6 s using a blot force of 6. Data were collected on a G2 Titan Krios fitted with a Gatan K2 direct electron detector operating in counting mode at the Astbury Biostructure facility, Leeds, UK. EPU was used to setup a 72-h data collection resulting in 3210 micrographs at 1.07 Å/pixel. Motion correction was carried out in MotionCor2 and CTF values determined using Gctf [40,41]. Initial two-dimensional (2D) references were generated in RELION from manually picked particles to generate reference free classes for autopicking the remaining data [42]. Autopicking resulted in a total of 965,863 particles which were subjected to 2D classification and after removal of those belonging to poorly resolved classes ~316,000 particles remained. After further rounds of 2D and three-dimensional (3D) classification the particle number was reduced to 105,901. The 3D refinement was conducted in RELION 2.1 using a previously published cryo-EM AcrB structure (Electron Microscopy Databank (EMD)-3887), filtered to 60 Å as a starting model. Following the 3D auto-refinement and particle polishing the resulting model had a global resolution of 4.6 Å, as determined by the 0.143 cut-off criteria. The final model had an automatically determined B-factor of –305 applied within RELION 2.1 [42]. To examine the effects of the G288D mutation on the oligomerization of STmAcrBG288D reconstructions were generated in both C1 and C3 symmetry with the C1 symmetry reconstruction reaching a similar resolution and showing the same overall architecture. For model fitting the E. coli AcrB crystal structure (Protein databank (PDB) ID: 4ZLJ) was put through “threading” in Phyre 2 [43] to produce a model of the STmAcrBWT structure. The G288D mutation was manually edited within Coot [44]. The high sequence similarity between E. coli AcrB and STmAcrBG288D resulted in a model of high confidence. The resulting STmAcrBG288D model was subsequently docked in Chimera followed by rigid body refinement using the MDFF program [45]. The model then underwent model refinement in Coot and real space refinement in PHENIX. Data collection and refinement statistics are given in Table S1. The STmAcrBG288D highest resolution C3 map and fitted structure have been deposited within the Electron Microscopy Database (EMDB) with accession number 4460 and 6Z12.
2.5. Methods for Structural Analysis
Bioinformatic analysis: Multiple sequence alignments were prepared using MAFFT as implemented in MAFFT v.7 server (https://mafft.cbrc.jp/) [46]. ESPript 3.0 was used for structural annotations of the alignments [47]. Model superposition and additional visualisation were performed in Coot [44] and PyMOL (PyMOL Molecular Graphics System, Version 1.71 Schrödinger, LLC).
2.6. Homology Modelling
Three homology models were built of STmAcrBWT and STmAcrBG288D using different X-ray crystal structures of wild type AcrB from E. coli (hereafter EcAcrBWT) (PDB IDs 4DX5, 4DX7 [19] and 2J8S [48]) as templates. The amino acid sequences of both EcAcrBWT and STmAcrBWT were obtained from the Uniprot database (Uniprot IDs: P31224 and Q8ZRA7, respectively). The absence of gaps was verified through a sequence alignment with ClustalOmega [49]. The homology models were generated using Modeller 9.3 [50], each having a MOLPDF score greater than 1.5× 105, and included the full range of residues (1–1033) in every template. The homology models of STmAcrBG288D were further energy-minimized into the experimental C1 cryo-EM map presented in this work with the program Flex-EM [51]. We performed structural optimization of the models for up to 40 iterations, and we ranked the final structures based on their cross-correlation function (hereafter CCF) (see Table S2).
2.7. Molecular Dynamics Simulations
The homology models of STmAcrBWT and STmAcrBG288D were used as starting structures to perform all-atom MD simulations. Following previous work [33,52], we simulated the truncated structure including only the porter domain and a few residues at the interface with the TM domain (namely, residue segments 32-335 and 564-870), imposing positional restraints on the Cα atoms of the residues found within 5 Å from the bottom of the structure (weight of the restraints: 1 kcal/mol). Those residues involved were in Subdomain PN1 (V32, A33, Q34, T37, I38, and A39), subdomain PN2 (A297, N298, A299, T330, P331, and F332), subdomain PC1 (L564, P565, D566, K632, D633, W634, P638, G639, E640, A670, I671, V672, T676, A677, and T678) and subdomain PC2 (P710, D711, L712, G838, E839, A840, Q865, E866, and R867). The selected portion of the protein was inserted in a truncated octahedron filled with 0.15 KCl aqueous solution, setting the minimum distance between the protein and the edge of the box to 16 Å. The topology and the initial coordinate files were created through the leap module of AMBER18 [53]. Protein and water were represented using the ff14SB force field [54] and the TIP3P model [55], respectively, while the parameters for the ions were retrieved from [56]. The system was enclosed in a truncated octahedron filled with 0.15 M KCl aqueous solution, and the minimum distance of the protein and the border of the box was set to 16 Å. The MD simulations of each system were done according to the following procedure. Firstly, we performed a multi-step structural relaxation combining steepest descent and conjugate gradient methods, using the pmemd module of AMBER18 [53], as described in previous publications [52,57,58,59]. The relaxation was followed by two MD simulations runs to heat the system from 0 to 310 K: i) from 0 to 100 K in 1 ns under constant-volume conditions and with harmonic restraints (k = 1 kcal·mol−1·Å−2) on the heavy atoms of both the protein and the lipids; ii) from 100 to 310 K in 5 ns under constant pressure (set to a value of 1 atm) and with restraints on the heavy atoms of the protein and on the z coordinates of the phosphorous atoms of the lipids to allow membrane rearrangement during heating. Next, we performed a series of 10 equilibration steps to equilibrate the box dimensions. Each step was of 100 ps in duration (total 1 ns) and was carried out under isotropic pressure scaling conditions through the Berendsen barostat. The Langevin thermostat was also used to maintain the temperature constant, with a collision frequency of 1 ps−1. Finally, for every system we performed three independent MD simulations, each with a production run of 150 ns in length. Time steps of 0.5 fs and 2 fs were used during the heating and equilibration stages, respectively. In the production run a time step of 4 fs was adopted under an isothermal-isobaric ensemble after hydrogen mass repartitioning [60]. Moreover, the lengths of all the R-H bonds were constrained with the SHAKE algorithm. Coordinates were saved every 100 ps. Long-range electrostatic forces were evaluated with the particle mesh Ewald (PME) algorithm, with a non-bonded cut-off of 9 Å.
2.8. Post-Processing of MD Trajectories
The MD trajectories of STmAcrBWT and STmAcrBG288D were firstly processed by performing a cluster analysis with the cpptraj module of AMBER18 [53]. For each trajectory, we considered only the last 140 ns of the production run, where the RMSD of the protein with respect to the first frame is fairly constant (Figure S1). Every trajectory was subjected to three clustering procedures, in each of which the distance-RMSD metric was applied to the DBP of a different monomer of AcrB, generating 100 clusters. In this way, we obtained 300 clusters per trajectory, divided in three equal subsets (1 subset per monomer). For each subset, the representative centroid structures of all clusters were used to perform several analyses aimed at assessing how the size and shape of the DBP are affected by the G288D mutation. To this end, we firstly estimated its volume of in the L, T, and O monomers of both STmAcrBWT and STmAcrBG288D.
The same analysis was then performed on 5 experimentally derived crystal structures of the EcAcrBWT, which were chosen as reference structures to identify variations between E. coli and STmAcrBWT. These structures have PDB IDs 4DX5, 4DX7 [19], 2J8S [48], 2I6W (the last being a symmetric LLL structure) [61] and 6BAJ (the structural model recently derived from cryo-EM data by Qiu et al.) [62]. The volume calculations were performed using the POVME 2.0 software [63], adopting a grid spacing of 0.5 Å. Additional analyses were conducted to better characterize of the impact of the G288D mutation. These included the calculation of the gyration radius of the DBP, the number of (pseudo)contacts between the PC1 and PC2 subdomains and the number of waters in the first and second solvation shell of residue 288. Such analyses were conducted on every protomer of AcrB. Calculations of the radius of gyration and of the number of (pseudo)contacts were carried out using in-house tcl scripts and performed on the cluster representatives of STmAcrBWT and STmAcrBG288D, as well as on the reference structures of the EcAcrBWT. The radius of gyration was computed for three different regions of the DBP: the whole DBP (S46, Q89, S128, E130, E134, F136, V139, Q176, L177, F178, S180, E273, N274, D276, Y327, L573, F610, V612, R620, F628), the hydrophobic trap (hereafter HP trap) (F136, V139, F178, Y327, L573, F610, V612, and F628) and the upper DBP (S46, Q89, S128, E130, Q176, L177, G179, S180, E273, N274, D276 and R620). As to the number of (pseudo)contacts, it was calculated by using a distance cut-off of 10 Å among the Cα carbons of selected regions of PC1 (segment 571-667) and PC2 (segments 679-721, 822-859). Regarding the first and second water shells of residue 288, these regions were defined using distance cut-offs of 3.4 Å and 5 Å, respectively. Calculations were performed on the last 140 ns of every MD trajectory of STmAcrBWT and STmAcrBG288D, using the cpptraj module of AMBER18 [53].
Moreover, we monitored the Loose/Tight/Open (LTO) asymmetry of STmAcrBG288D along the MD trajectories. To perform this analysis, we used as a reference the EcAcrBWT crystal structure with PDB ID 4DX7 [19], in which the protein is found in the LTO state. Thus, for each frame in the last 140ns of the MD production run, we calculated the RMSD of each conformer of the mutant with respect to every conformer of the E. coli reference structure. Only the Cα atoms were considered for this calculation.
3. Results
3.1. Oligomeric State and Overall Fold of S. Typhimurium AcrB
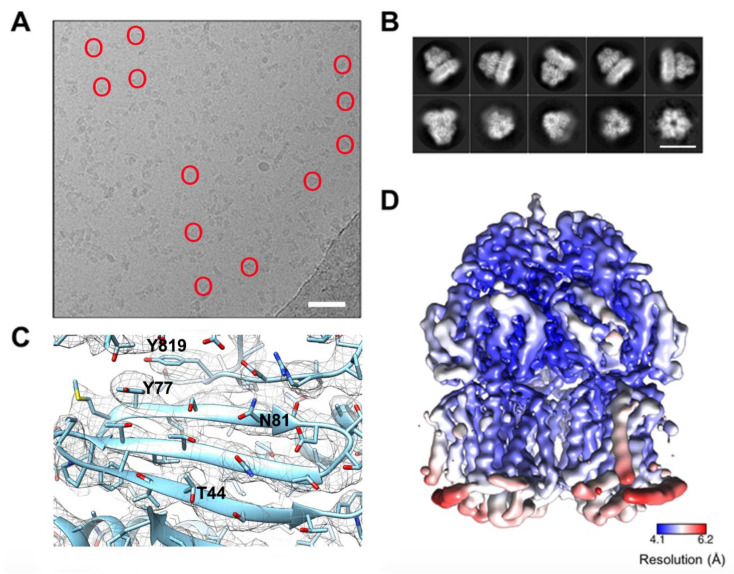
Here we report the first ever experimentally-derived structure of the AcrB from S. Typhimurium. STmAcrBG288D structure was determined by single particle cryo-EM to a global resolution of 4.6 Å. STmAcrBG288D was expressed in an E. coli strain deficient of the native AcrB transporter (C43ΔacrB(DE3)) to ensure only the Salmonella orthologue was expressed. For protein extraction a styrene maleic acid (SMA) co-polymer approach was used, which we have previously shown to be effective at isolating the E. coli AcrB in sufficient levels for both negative-stain and cryo-EM analysis [36,37,39]. Negative stain and the subsequent cryo-EM sample (Figure 1A) showed a mono-disperse sample with no significant aggregation or degradation.
Figure 1.
Single particle cryo-electron microscopy reconstruction of STmAcrB G288D processed in C3 space group. (A) Representative micrograph of STmAcrB G288D collected on a K2 camera, with a scale bar in white representing 50 nm. Some representative STmAcrB G288D particles are circled in red. (B) Two-dimensional (2D) classes of STmAcrB G288D, produced in RELION as seen from the side, high angle, and top views. The scale bar represents 15 nm. (C) Representative experimental electron map density processed in C3 space group (mesh), with the refined STmAcrB G288D model fitted (cyan). Reference residues labelled within the map include, T44, Y77, N81 and Y819. (D) The C3 derived map from STmAcrB G288D coloured by local resolution in Angstroms (Å) showing the core to be more highly resolved, while the transmembrane flanking regions display lower resolution.
In contrast to our previous studies of EcAcrB by single particle cryo-EM here we have collected significantly more data to improve the resulting resolution and quality of the map [36]. The micrographs showed a monodisperse sample with variation in the orientation within the ice (Figure 1A). The resultant classes revealed clear secondary structure detail for both side and top views (Figure 1B). As expected, based on the high sequence similarity, the overall architecture when processed in C3 symmetry is similar to that of E. coli AcrB and the functional biological unit of the protein is formed by three protomers displaying overall 3-fold symmetry [15] (Figure 1B). The transmembrane domain of each protomer is formed by 12 α-helices with a single α-helix flanking the base of the transmembrane domain. All the α-helices were well-defined in the cryo-EM map and could be placed with high confidence. Although additional density can be seen between the helices the resolution is not sufficient to assign any lipids or SMA-polymer with confidence. The periplasmic domain is well-resolved and shows features consistent with a local resolution of ~4.1 Å. Both α-helices and β-strands are well-resolved and bulky amino acid side-chains can be fitted within the map with confidence (Figure 1C). Analysis of the local resolution in C3 map shows the core to be more highly resolved with lower resolution features displayed on the exterior of the complex, especially within the transmembrane region (Figure 1D).
3.2. Comparison between Salmonella AcrB G288D Processed in the C3 Space Group and the WT E. coli Transporters
The overall structure of STmAcrB is closely related to that of the previously reported E. coli orthologue [15], which is consistent with the high level of sequence conservation between the two transporters. Sequence analysis reveals a 94.7% identity over the 1048 residues as calculated by the SIM Alignment tool (https://web.expasy.org/sim/) [64], (Figure S2).
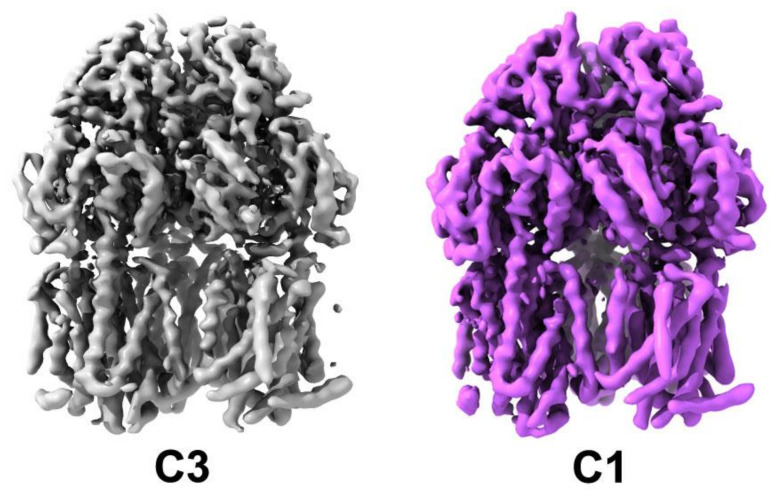
The position of the secondary structure elements, and correspondingly the (sub)domain organisation is preserved, as is to be expected given there are no significant additional residue inserts between the sequences (Figure 2; Figure S2). Consistently, the refined trimer preserves its central symmetry, but there are slight discrepancies between the individual protomers which amount to an RMSD of 0.35–0.38 Å over the C-alpha backbone. This small change results from each chain being fitted and refined within the map without the enforcement of threefold symmetry, reflective of the modest (~4.6 Å) resolution within the map. While symmetric structures in the R32 space group have been produced both in apo- (1IWG.pdb, [15]), and in a linezolid-bound form of EcAcrB (4K7Q.pdb, [65]), as mentioned in the Introduction, in its functional state AcrB presents a breakdown of the central-axis three-fold symmetry (with conformers referred to as either L, T, O (Loose, Tight, Open) [17]; or A, B, E (Access; Bound; or Extrusion) [16]), which is also observed in the associated X-ray structures. The related EcAcrB cryo-EM structure (6CSX.pdb, [62]) also displayed a clear breakdown in the 3-fold symmetry related to the presence of defined states within the complex. Therefore, we also refined STmAcrBG288D in the C1 space group corresponding to an asymmetric trimer resulting in a map which closely resembled that of the C3 but with small local changes, consistent with the change in state of each protomer (Figure 3).
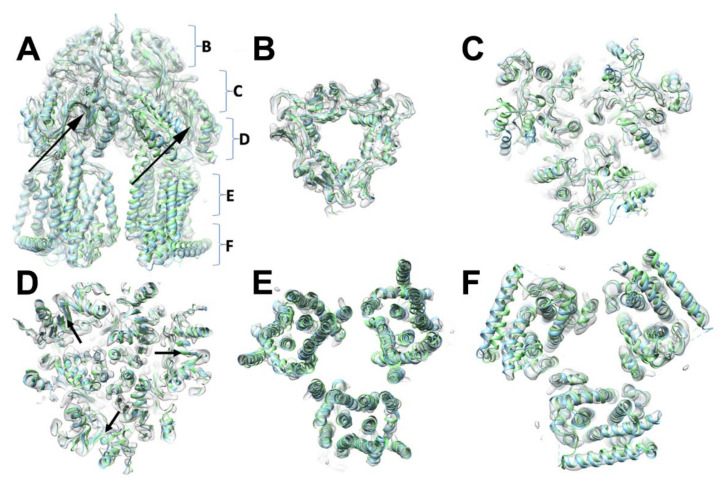
Figure 2.
Comparison of E. coli and S. Typhimurium AcrB three-dimensional (3D) structures. (A). A side-view of the trimer of the STmAcrB G288D refined model (cyan) superposed over the E. coli orthologue (EcAcrB) (light green) and fitted into the experimental electron density map (semi-transparent grey). (B–F) Horizontal slices through the STmAcrB C3 cryo-EM electron-density map from the top (B) to base (F). Both STmAcrB G288D and EcAcrB are fitted within the density. The relative positions of the slices are indicated in (A). The position of G288D mutation within the structure is indicated with black arrows in (A) and (D).
Figure 3.
Comparison of STmAcrB G288D cryo-EM maps refined in C3 and C1 with no difference in the overall architecture observed but slightly improved resolution in the C3 map.
To establish whether the functional rotation states are also preserved in the STmAcrBG288D we set out to analyse which of the reported EcAcrB protomer-states our structure resembled most closely. Pairwise superpositions using the high-resolution asymmetric structure 4DX7 [19], resulted in a RMSD over the STmAcrB backbone in the ranges of 1.69–1.75 to 1.54–1.64 and to 1.51–1.54 Å (against the L, T, and O protomers, respectively), while the superposition with the symmetric 4K7Q yielded a lower RMSD of 1.03–1.08 Å. Note that superposition of 4K7Q onto 4DX7 chains yielded RMSDs corresponding to 1.19, 1.26 and 1.40 Å (for the L, T, and O conformers, respectively). The above indicates that the while asymmetric, the conformation observed within our cryo-EM STmAcrB structure deviates only moderately from the symmetrised E. coli protomer state.
3.3. Implications for Drug Selectivity and Efflux Efficiency
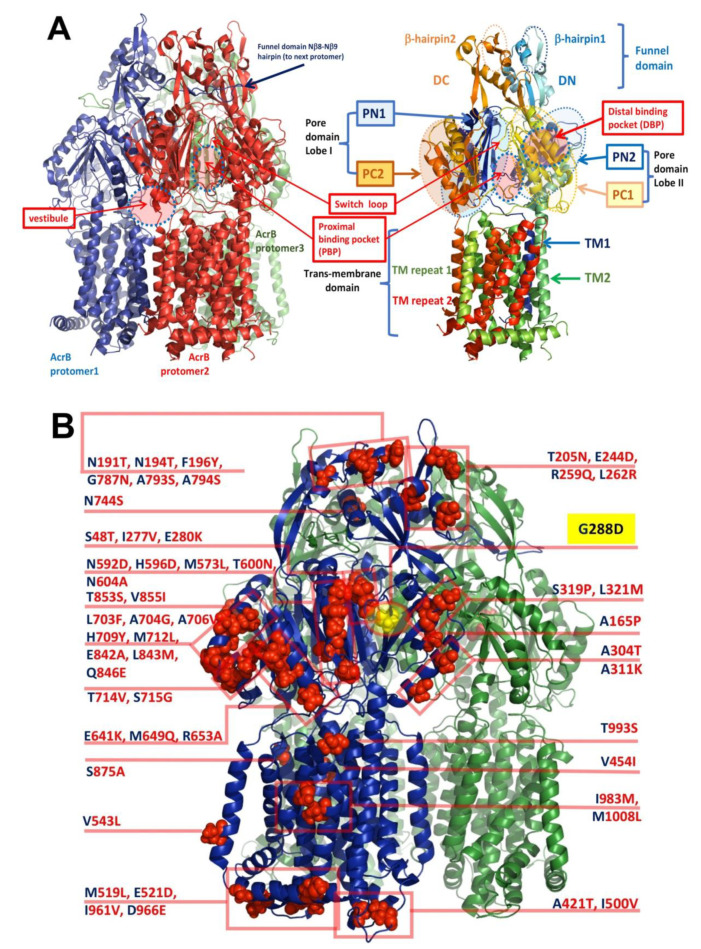
We next sought to understand how sequence differences between the E. coli and S. Typhimurium AcrB could translate into differential selectivity to antibiotics. Compared to the EcAcrB, the novel STmAcrB structure shows changes in the substrate pathway and the DBP, which is sandwiched between the PC1 and PN2 subdomains (Figure 4A) of the periplasmic “pore”- or porter-domain. The principal access for soluble drugs from the periplasmic space is via access-tunnel 2, which is located between the two principal lobes of the pore-domain, and is connected to the “access” or proximal binding pocket (PBP), which also receives the membrane-connected access-tunnel 1 [23]. As discussed in the Introduction, the DBP is separated from the PBP by the so-called “Glycine” or “switch”-loop, the flexibility of which affects the drug-passage through the latter [19,66]. As will be discussed in detail further below, the new STmAcrB structure refined in C1 space group, displays asymmetry of protomers, which allows them to be assigned to the specific conformational states (L, T, O conformers) described above, however the range of structural variation between them appears to be significantly lower than the corresponding conformers within EcAcrB; therefore, we analysed the sidechains based on the C3 map, which had a higher local resolution. Due to their high overall homology and lack of gaps in the alignment, for ease of presentation, below, when referring to the amino-acid differences between the E. coli and Salmonella AcrB, we refer to the divergent residues occupying the same position as “substitutions” (see Figure 4B).
Figure 4.
(A) General organisation of the STmAcrB G288D trimer and detailed sub-domain organisation of the single protomer. (B) Mapping of the sequence differences of STmAcrBG288D vs. EcAcrBWT shown on one of the three protomers as red spheres. The labels indicate single-letter residue code for the differing residues in E. coli AcrB (in blue), alongside with the equivalent positions and substituted side-chains in Salmonella AcrB (in red). The location of the mutation G288D is indicated with a yellow box.
Yet, despite the low overall percentage of sequence difference, the amino-acid differences cluster into several discrete groups which may have significant impact on the functional profile of the pump and its dynamics. These residue differences can be split into two broad classes based on their potential effects—one affecting the conformational dynamics of the protomer, while the second impacting the substrate recognition and processing (Figure 4B).
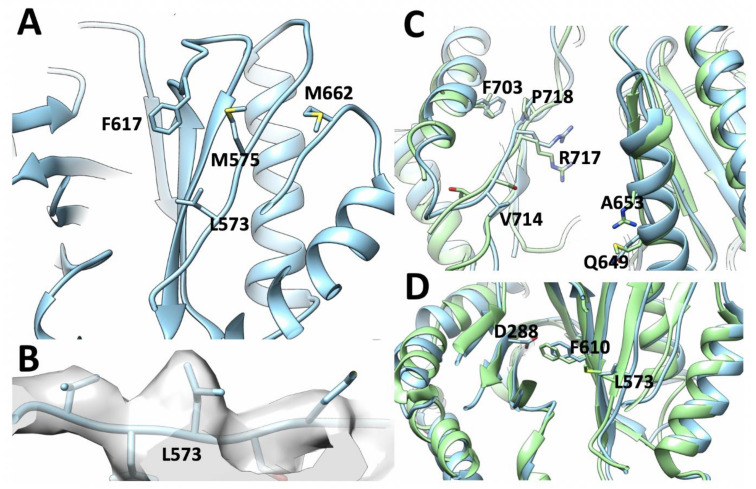
Of the latter, of particular interest is the M573L substitution in the primary substrate pathway. In EcAcrB M573 occupies a critical position between the proximal binding pocket and the switch loop, where it is part of a larger Met-cluster that includes M575 and M662; and, in conjunction with the F617 which belongs to the switch-loop, also provides a part of the hydrophobic barrier. The residue is well resolved in the electron-density map and the Leucine-substitution appears to cause local rearrangement of the hydrophobic residues in the so-called hydrophobic trap (HT) of the DBP, including a displacement of the ring of F617 (Figure 5A,B).
Figure 5.
Close look of the drug-binding pockets of STmAcrB G288D. (A) The PBP (access binding pocket) and the switch loop in STmAcrB G288D with those residues that make up the larger hydrophobic “Met-cluster” shown. (B) Representative map density for STmAcrB G288D showing the proximity of L573, which in EcAcrB has a bulkier methionine allowing unambiguous placement of this sidechain. (C) The outer rim of the cleft between the two lobes of the porter domain which provide entry to tunnel 2. (D) Comparison of the structural differences in the DBP in proximity of the G288D mutation. In all panels STmAcrB is shown in cyan and EcAcrB in green.
A double substitution T714V/S715G located on the outer left rim of the cleft between the two lobes of the porter domain, forming the entry of tunnel 2 leading to the proximal drug-binding pocket might plausibly impact the selectivity of some drugs. On the opposite rim of the tunnel 2 cleft there are two more neighbouring substitutions, namely M649Q plus R653A. While these latter substitutions are on the outer rim of the entrance leading to the PBP their nature suggests that they would result in a significant rearrangement of the electrostatic properties as well as water-coordination at the entrance of the pocket and thus may affect drug access (Figure 5C).
Furthermore, the double substitution L703F/A704G is able to provide a novel phenyl-ring stacking interaction with the P718, which could affect the local dynamics of the PBP entry, as these residues are located just above V714/G715 and immediately next to R717 which has been shown to participate in substrate coordination in the proximal pocket [19,23]. While the quality of the map at this region does not permit the unambiguous positioning of F703 its increased bulk must be accommodated within the structure (Figure 5C).
3.4. Structural Changes Attributable to G288D Mutation
An important aspect of our cryo-EM STmAcrB structure refined in a C3 is the presence of a G288D mutation which confers greater resistance to fluoroquinolones [33]. However, analysis of the novel cryo-EM structure reveals that the G288D mutation brings D288 in close contact with the well-resolved sidechain of F610. Unfortunately, since negatively charged side-chains are more susceptible to radiation damage their resulting density is often poor within the map [67]. Hence, while there is no-clear density for D288 side chain, there are also no preferred rotamers that could avoid this close contact, suggesting that D288 is either in a less-favoured conformation or maybe in multiple conformations between protomers, explaining the poor density for this region. It is interesting to note that the β-strand on which G288 is located does not show a significant shift to accommodate the extra bulk of aspartate sidechain in the mutated Salmonella structure compared to the EcAcrBWT DBP (Figure 5D). However, the residues around D288 show a notable shift, which may accommodate the increased bulk and the major change in the electrostatics of the pocket associated with the increased hydration detected in our previous work [33]. In particular, F610 is positioned further away from position 288 causing an increase in pocket size (Figure 5D). Through these changes the lower part of hydrophobic pocket is locally enlarged. This expansion occurs around Q176 and F178, which have both previously been shown to participate in the coordination of substrates within AcrB [19,26,33,68].
3.5. Mechanism of G288D Mutation Inferred from MD Simulations
Notably, the above observations were based on the static and medium-resolution cryo-EM structure; therefore, we sought to further validate and extend our predictions by performing MD simulations starting from several independent homology models. Many different all-atom structural models of STmAcrBG288D and STmAcrBWT were derived through homology modelling, and the models of the former were minimized against the C1 cryo-EM map presented in this work. Model accuracy was evaluated through the CCF (see Materials and Methods), which, for each model, slightly improved starting from every template (Table S2). Moreover, the optimization against the cryo-EM map did not cause significant structural changes from the initial models (last column in Table S2).
Starting from these structural models, we performed three independent all-atom MD simulations for each system in order to compare structural and dynamical features of the wild type and substituted STmAcrB. As described in Materials and Methods, we used a truncated model of STmAcrB (largely validated in previous studies; see [69] for a recent review) containing the periplasmic portion of the protein only, as it is primarily responsible for drug binding. In fact, despite this approximation the asymmetric LTO structure remained well-preserved throughout the MD simulations, consistent with the results described previously [33,52]. In particular, cross-RMSD calculations between each conformer of our models against each conformer of the reference structure (PDB ID 4DX7 [19], see Materials and Methods) revealed that the asymmetric LTO structure is similarly retained in STmAcrBG288D (Table 1, below).
Table 1.
Cross-RMSD of each chain of STmAcrBG288D, with respect to every conformer of the EcAcrB crystal structure 4DX7. For each model, the calculation was performed on the last 140 ns of the production run (see Materials and Methods); the reported values correspond to the average RMSD and its standard deviation in the Loose (L), Tight (T) and Open (O) conformer. Only the Cα atoms were considered for this calculation.
| Homology Model | Chain | RMSD (Reference Structure: EcAcrB 4DX7) | ||
|---|---|---|---|---|
| L-Conformer | T-Conformer | O-Conformer | ||
| 1 (template: 2J8S) | A | 2.1 (0.1) | 2.7 (0.1) | 3.2 (0.1) |
| B | 3.0 (0.1) | 2.3 (0.1) | 4.1 (0.1) | |
| C | 3.5 (0.1) | 3.9 (0.1) | 2.2 (0.1) | |
| 2 (template: 4DX5) | A | 2.4 (0.1) | 3.1 (0.1) | 3.0 (0.1) |
| B | 3.0 (0.1) | 2.2 (0.1) | 3.9 (0.1) | |
| C | 3.2 (0.1) | 3.9 (0.1) | 2.2 (0.1) | |
| 3 (template: 4DX7) | A | 2.1 (0.1) | 2.8 (0.1) | 3.4 (0.1) |
| B | 3.1 (0.1) | 2.3 (0.1) | 4.1 (0.1) | |
| C | 3.1 (0.1) | 3.5 (0.1) | 2.1 (0.1) | |
We first focused our analysis on the impact of the G288D substitution on the volume of the DPB (VDBP) across all AcrB conformers. The largest differences are seen in the volume of the distal binding pocket of monomer T (hereafter DBPT), which undergoes a significant expansion with respect to the WT Salmonella protein (Table 2). Note that while relative to the E. coli orthologue the VDBP of the STmAcrB WT is about 800 Å3 smaller, the effect of G288 mutant on the DBP results in an expansion of approximately 450 Å3 in the T-conformer bringing it closer to that of the E. coli orthologue.
Table 2.
Values of the volume of the deep- or distal-binding pocket (DBP) (standard deviations in parentheses) in each conformer of AcrB, measured on the E. coli AcrBWT reference structures and on the molecular dynamics (MD) trajectories of S. Typhimurium AcrBWT and AcrBG288D (see Materials and Methods).
| System | Volume of DBP [Å3] | ||
|---|---|---|---|
| L-Conformer | T-Conformer | O-Conformer | |
| EcAcrBWT1 | 763 (86) | 2315 (76) | 1094 (76) |
| STmAcrBWT2 | 957 (93) | 1534 (163) | 991 (96) |
| STmAcrBG288D2 | 807 (78) | 1979 (72) | 1151 (64) |
1 calculated on experimental reference structures; 2 calculated on representatives of each of the 100 clusters extracted from the MD trajectories.
The trend seen for VDBP is confirmed by the calculation of the gyration radius (Table 3), which increased significantly only for the DBPT, and in particular in the HP trap, for which the increment amounts to almost 1 Å with respect to EcAcrBWT (of less than 0.5 Å in the other monomers).
Table 3.
Radius of gyration of the DBP calculated for every AcrB conformer. The three regions of the DBP considered in this calculation are indicated as Whole (entire DBP), Upper (upper part of the binding site), and HP trap (hydrophobic trap) (see Materials and Methods for the definition of these regions).
| System | Radius of Gyration [Å] | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| L-Conformer | T-Conformer | O-Conformer | |||||||
| Whole | Upper | HP Trap | Whole | Upper | HP Trap | Whole | Upper | HP Trap | |
| EcAcrBWT1 | 10.2 (0.1) | 9.0 (0.1) | 6.4 (0.3) | 10.8 (0.1) | 9.4 (0.1) | 7.1 (0.3) | 10.6 (0.1) | 9.9 (0.1) | 6.2 (0.0) |
| STmAcrBWT2 | 10.5 (0.1) | 9.1 (0.1) | 6.3 (0.2) | 10.7 (0.2) | 8.7 (0.1) | 6.9 (0.2) | 10.9 (0.1) | 9.8 (0.1) | 6.3 (0.2) |
| STmAcrBG288D2 | 10.5 (0.1) | 9.2 (0.1) | 6.4 (0.1) | 11.2 (0.1) | 9.2 (0.2) | 8.0 (0.2) | 11.0 (0.1) | 9.9 (0.1) | 6.5 (0.2) |
1 calculated on experimental reference structures. 2 calculated on representatives of each of the 100 clusters extracted from the MD trajectories.
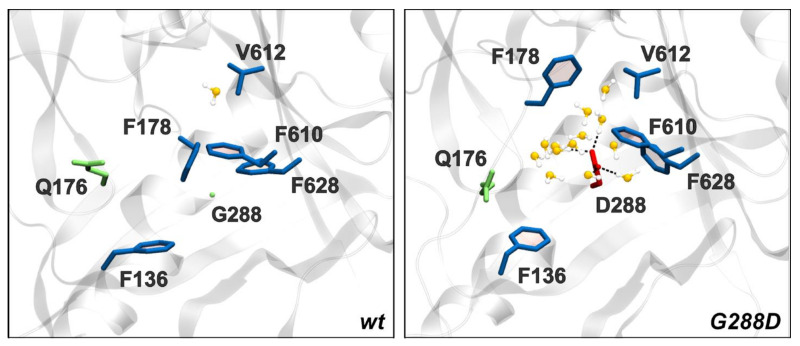
In addition, the analyses performed on multiple trajectories of both the WT and substituted STmAcrB transporter reveal that the number of waters in the 1st and 2nd solvation shells of residue 288 drastically increases upon mutation (Table 4 and Figure 6, below). The increased hydration of the DBPT “breaks” the hydrophobic character of the hydrophobic trap (HP trap), and is likely responsible for the increased volume and gyration radius of the DBPT and for the altered specificity of the transporter described previously [33]. Our findings are reasonable, as the mutation of a glycine into a charged and bulkier residue is expected to have the largest impact on the structure, dynamics, hydration of the surrounding (prevalently hydrophobic) region. Moreover, they agree with previously published results [33], although here we have extended the analyses to conformers other than T and we have increased confidence by using multiple and independent structural models of AcrB.
Table 4.
Number of waters in the first and second solvation shell around residue 288 (in DBP), in STmAcrBWT and STmAcrBG288D. The two solvation shells were defined by using a distance cut-off of 3.4 Å and 5.0 Å, respectively.
| System | AcrB Conformer | ||
|---|---|---|---|
| L-Conformer | T-Conformer | O-Conformer | |
| # 1st solv. shell waters | |||
| STmAcrBWT | - | 0.1 (0.3) | 0.0 (0.1) |
| STmAcrBG288D | - | 6.3 (0.6) | 3.1 (0.3) |
| # 2nd solv. shell waters | |||
| STmAcrBWT | 0.0 (0.1) | 0.5 (0.4) | 0.2 (0.2) |
| STmAcrBG288D | 0.2 (0.3) | 11.3 (1.2) | 5.7 (0.5) |
Figure 6.
Close-up of the residue 288 and surrounding residues of the HP trap in STmAcrBWT and STmAcrBG288D. Waters belonging to the first and second hydration shell of residue 288 (distance threshold: 5 Å, see Materials and Methods) are also shown, and hydrogen bonds involving residue 288 are represented as dashed lines. This image has been created using two representative frames of MD trajectories.
Along these lines, in addition to the impact of the mutation on DBPT, we also evaluated here the occurrence of structural changes in the other main binding pocket of AcrB, the PBP of the L conformer (hereafter PBPL). This site is found beneath the entrance cleft [24] and involves subdomains PC1 and PC2. The calculation of the volume of the PBP is less-straightforward due to its relatively open architecture and somewhat vague definition of boundaries. Therefore, to quantify the changes occurring in the region we evaluated the number of contacts between domains PC1 and PC2, which should reflect the opening/closing of the external cleft leading to the access pocket. Indeed, within representative E. coli structures, such as 4DX7, the number of average contacts increases from monomer L to T and to O. Rather surprisingly we found that these subdomains lose contact in the G288D STmAcrB variant compared to the WT protein (Table 5, below), and this especially affects the L-conformer. This results in L-conformer being more open towards the periplasm and thus providing easier entrance for effluxed compounds.
Table 5.
Number of contacts between the subdomains PC1 and PC2 in the three conformers of AcrB. Two residues have been considered in contact if the distance between their Cαs is below 10 Å (see Materials and Methods).
| System | Number of Contacts (PC1-PC2) | ||
|---|---|---|---|
| L-Conformer | T-Conformer | O-Conformer | |
| EcAcrBWT1 | 9 (5) | 10 (2) | 38 (2) |
| STmAcrBWT2 | 12 (4) | 6 (2) | 38 (7) |
| STmAcrBG288D2 | 1(1) | 7(2) | 31 (7) |
1 calculated on experimental reference structures. 2 calculated on representatives of each of the 100 clusters extracted from the MD trajectories.
4. Discussion
Despite its clinical importance, the structure of Salmonella AcrB has remained poorly understood until now. This could partially be due to the fact that while the E. coli AcrB readily crystallises and has been described as a persistent contaminant of both 2D and 3D crystallization screens even at picogram amounts [70,71], the Salmonella transporter proved rather challenging for crystallization via traditional approaches, which is unexpected considering the close homology between their sequences. This may partially be explained as EcAcrB crystallises in both the H32 or C121 space groups and in both these instances the regions which form the crystal contacts differ significantly between EcAcrB and STmAcrB.
To circumvent these issues, here we set-out to employ a cryo-electron microscopy approach to the solution of structure of STmAcrB G288D. Furthermore, to minimise structural artefacts we decided to pursue the determination of STmAcrB in a native-like membrane environment, by utilizing styrene maleic acid (SMA) copolymer [37] for membrane extraction and direct solubilisation of the protein from bacterial membranes. The SMA has many benefits over traditional detergents, the main one being that it allows for a one-step extraction from cell membrane allowing the retention of native structural lipids. We have shown previously the applicability of the SMA-approach, especially in relation to AcrB for both negative stain [39] and cryo-EM structure determination [36], the latter work reaching sub-9 Å resolution. Optimisation of sample preparation allowed us to push the resolution to near 4 Å (in the periplasmic porter domains) of STmAcrB, allowing confident placement of most side-chains. Our new STmAcrB structure adds to only a few available recent native-lipid membrane structures of RND-transporters including the Acinetobacter baumannii AdeB [72] and EcAcrB [62].
The transmembrane domain shows a high degree of similarity between the two models with no significant changes in helices position that could not be attributed to differences one would expect from a lower resolution model. The base of the transmembrane domain shows subtle movement of the helices at the base of the membrane (Figure 2E). This helix is clearly resolved within the EM map and can be placed with confidence although it is more poorly resolved than the core helices, consistent with some degree of mobility of this element, likely due to its position on the periphery of the complex.
On the DBP Pocket and the Effect of G288D
The resolution of the structures obtained by cryo-EM often does not allow to investigate subtle but possibly relevant conformational changes between the WT and G288D variant of STmAcrB, nor does it capture the dynamic properties of the respective molecules (and of their dynamic interaction with the solvent and the membrane) that ultimately translate into their functional characteristic.
These dynamic changes, which could be of particular interest at putative binding sites and entrance/exit gates for substrates and/or inhibitors, were thus investigated by means of full-atomic molecular dynamics simulations. The impact of the G288D substitution was expected to be most pronounced for the DBP, where it was predicted to localise. Indeed, several previous high-resolution structures of EcAcrB [19,48] demonstrate that G288 sits in the middle of a β-strand beneath F178, a residue which is critical for coordination of hydrophobic drugs in the DBP. Moreover, the side chains of neighbouring residues point away from the G288D substitution (as seen in Figure 5 and Figure 6), suggesting that the side chain of the substituted aspartate residue will protrude into the pocket cavity, thus having a major effect on the coordination of potential substrates [33].
The role of mutations within the drug-binding pockets of RND transporters have only recently been appreciated as a possible additional mechanism of drug resistance [33,73,74], although earlier reports exist [75]. The G288D substitution has been demonstrated to alter the antibiotic resistance profiles for clinical isolate of Salmonella, notably increasing resistance to fluoroquinolones. The new cryo-EM structure, alongside the computational data presented here sheds further light on the possible mechanism of action of G288D substitution. Building on the previous work on the G288D [33], here we expanded the scope and the statistical significance of the analysis and moreover, we describe the impact of the mutation on all protomers of the transporter by means of additional analyses not performed in previous publications. Interestingly, a significant compression of the DBPT was seen in STmAcrBWT as compared to the value calculated from the X-ray structures of EcAcrB orthologue in the MD simulations. The analysis of our MD simulations suggests that the DBP of STmAcrB is significantly less-voluminous than the E. coli orthologue, pointing to an increased steric hindrance within the pocket, which might translate into altered substrate-processing kinetics and specificities between the two transporters. However, it should be noted that such a compression was also seen in previous MD simulations of EcAcrB in the absence of any ligand within the pocket, pointing to the possibility of unresolved ligands within the DBPT of these experimental structures [58] although further analysis of this feature is beyond the scope the current study. Despite that, the MD analysis suggests that DBP of WT STmAcrB exhibits markedly different dynamic properties to either the STmAcrB D288G or EcAcrB, both of which show similar, larger volumes in the T-conformer (Table 2 and Table 3). This may translate into altered specificity and increased efflux of the substrates that bind in the DBP. Indeed, changes in the DBP characteristics have been reported to account for the discrepancies in the substrate specificity of other RND-transporters e.g., AcrB and AcrD; MexB and MexY [26,58,76]. Such effect may be further enhanced by the observed loss of contacts between PC1 and PC2 of the L-conformer (Table 5), resulting in a conformation of the PBP that is more-open to the periplasm and hence more accessible for potential substrates. Thus, the significant DBP and PBP rearrangements observed in the structure and in the MD simulations likely account for the discrepancies between the WT and G288D variants, and its increased pumping efficiency for ciprofloxacin. It is worth noting however, that concurrently to increased fluoroquinolone resistance the G288D causes an increased susceptibility to doxorubicin and minocycline [33].
While there are still open questions as to the role played by the G288 within the functional cycle of AcrB, and so far the G288D mutation is the only mutation affecting this codon resulting in multidrug resistance [33], it is notable that mutations at this position have been reported more than once, both from Salmonella and E. coli [33,68,77]. The study by Soparkar et al., [68] was searching for spontaneous gain-of-function revertants compensating for the critical substitution F610A [78] affecting the packing of the hydrophobic trap within the DBP, while the Schuster et al., [77] isolated a G288S mutation in response to exposure of cells to the efflux pump inhibitor (EPI) 1-(1-Naphthylmethyl)-piperazine (NMP), strongly suggesting the residue has an important structural role affecting the DBP. Our present data further reinforces the importance of G288, highlighting not only changes of the DBP volume and conformation, but also subtle yet tangible conformational rearrangements which radiate from the mutation point (Figure 6, Table 4) affecting the packing of subdomains PC1 and PC2 and by proxy the proximal binding pocket (PBP). As reported above, these might be communicated via different arrangements of the switch-loop [24] and of the loop connecting PC2 to the funnel domain, which move closer in STmAcrBG288D compared to STmAcrBWT. Thus, the G288D mutation appears to impact the plasticity of the drug-binding pocket and drug-transport pathway. These results are further corroborated by the recent analysis of STmAcrBG288D sensitivity to another EPI, namely phenylalanine-arginine-β-naphthylamide (PAβN) [79], which not only demonstrates that PAβN effectively inhibits both WT and G288D version of the Salmonella AcrB, but that that ciprofloxacin and PAβN can stably occupy the DBP at the same time. This implies that PAβN potentiates antibiotic activity by restraining drug-binding pocket dynamics, rather than preventing antibiotic binding. Hence, further analysis and docking utilising the new structure reported here would be invaluable to further elucidate the exact effects of the mutation on the efflux, and inform the design of new EPIs.
In addition to elucidating the direct effect of G288D mutation, the cryo-EM structure of STmAcrB also highlights several important differences in the residues lining the principal drug-entrance tunnel 2 access leading to the PBP affecting its electrostatics (Figure 4B), as well as identifies a possible role for the M573, a residue in the control of coordination of residues at the upper-side of the DBP. Taken together these modifications and rearrangements likely account for the subtle differences between the specificities and efflux properties of EcAcrB and STmAcrB.
5. Conclusions
AcrB is a key transporter contributing to multidrug resistance and recent studies have provided significant developments in our understanding of its function. Through the study of different species and sequence variants we can start to better understand how these mechanisms are conserved across the wider RND-transporter family.
Increases in MDR in Salmonella Typhimurium are transforming this microorganism into a growing health threat globally, which is progressively difficult to treat. Here we have used a single particle cryo-EM approach to solve the structure of the STmAcrB G288D mutant derived from a clinical isolate strain. Consistent with the high sequence similarity between the STmAcrB and its well-characterised E. coli orthologue, the structural similarity between the two proteins is high, without significant changes in overall architecture. However, there are subtle differences present within the structure that could account for the changes in substrate transport. Indeed, computational modelling followed by MD-simulations highlighted specific features of the two transporters at their respective putative substrate-interaction sites. We also further clarify the effect of G288D mutation on STmAcrB, and report that it results in a subtle expansion of the DBP pocket and may thus affect the dynamics of the pocket alongside the interprotomer-interactions within the Salmonella AcrB trimer. While the significance of the specific substitutions within the DBP of the STmAcrB might require further investigation, the availability of the experimental structure of this clinically important protein provides the platform to inform future structural analysis and structure-driven design of potential novel efflux-pump inhibitors.
Acknowledgments
The authors thank the Astbury Biostructure Laboratory for their assistance with EM data collection.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/2076-2607/8/6/943/s1, Figure S1. Protein RMSD calculated along the MD trajectories of STmAcrBG288D aligned to the STmAcrBWT; Figure S2. Multiple sequence alignment of STmAcrBG288D aligned to the STmAcrBWT and EcAcrBWT; Table S1. Data collection, processing and model fitting statistics.; Table S2 Values of the Cross-Correlation Function obtained through Flex-EM for the homology models of STmAcrBG288D, before and after the optimization inside the cryo-EM map.
Author Contributions
Conceptualisation and experimental design: S.P.M., V.L.G.P., and V.N.B.; Performed experiments and modelling: R.M.J., C.F., M.P., H.C., R.L.M., S.J.H., and A.V.V.; Analysis of data and model refinement: R.M.J., C.F., V.L.G.P., A.V.V., M.C.F., P.R., S.P.M., and V.N.B.; supervision V.L.G.P., A.V.V., P.R., S.P.M., and V.N.B.; Discussed the data and wrote the manuscript: R.M.J., C.F., A.V.V., S.P.M., and V.N.B. with contributions from all other authors; all authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a Biotechnology and Biological Sciences Research Council (BBSRC) (BB/N002776/1) and Wellcome Trust grant (108372/A/15/Z) to V.N.B. R.M.J., thanks the Wellcome Trust for PhD studentship support (109158/B/15/Z) and the Astbury Biostructure Laboratory is supported through the Wellcome Trust (108466/Z/15/Z). V.L.G.P., P.R., and A.V.V., thank the support of the National Institutes of Allergy and Infectious Diseases (project number AI136799). S.J.H was supported by a BBSRC White Rose Doctoral Training Fellowship (BB/M011151/1).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Curiao T., Marchi E., Grandgirard D., León-Sampedro R., Viti C., Leib S.L., Baquero F., Oggioni M.R., Martinez J.L., Coque T.M. Multiple adaptive routes of Salmonella enterica Typhimurium to biocide and antibiotic exposure. BMC Genomics. 2016;17:491. doi: 10.1186/s12864-016-2778-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Misra R., Bavro V.N. Assembly and transport mechanism of tripartite drug efflux systems. Biochim. Biophys. Acta. 2009;1794:817–825. doi: 10.1016/j.bbapap.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du D., Wang-Kan X., Neuberger A., van Veen H.W., Pos K.M., Piddock L.J.V., Luisi B.F. Multidrug efflux pumps: Structure, function and regulation. Nat. Rev. Microbiol. 2018;16:523–539. doi: 10.1038/s41579-018-0048-6. [DOI] [PubMed] [Google Scholar]
- 4.Kobylka J., Kuth M.S., Müller R.T., Geertsma E.R., Pos K.M. AcrB: A mean, keen, drug efflux machine. Ann. N. Y. Acad. Sci. 2019;1459:38–68. doi: 10.1111/nyas.14239. [DOI] [PubMed] [Google Scholar]
- 5.Nishino K., Latifi T., Groisman E.A. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2006;59:126–141. doi: 10.1111/j.1365-2958.2005.04940.x. [DOI] [PubMed] [Google Scholar]
- 6.Janganan T.K., Bavro V.N., Zhang L., Matak-Vinkovic D., Barrera N.P., Venien-Bryan C., Robinson C.V., Borges-Walmsley M.I., Walmsley A.R. Evidence for the assembly of a bacterial tripartite multidrug pump with a stoichiometry of 3:6:3. J. Biol. Chem. 2011;286:26900–26912. doi: 10.1074/jbc.M111.246595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Symmons M.F., Marshall R.L., Bavro V.N. Architecture and roles of periplasmic adaptor proteins in tripartite efflux assemblies. Front. Microbiol. 2015;6:513. doi: 10.3389/fmicb.2015.00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z., Fan G., Hryc C.F., Blaza J.N., Serysheva I.I., Schmid M.F., Chiu W., Luisi B.F., Du D. An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. Elife. 2017;6:e24905. doi: 10.7554/eLife.24905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNeil H.E., Alav I., Torres R.C., Rossiter A.E., Laycock E., Legood S., Kaur I., Davies M., Wand M., Webber M.A., et al. Identification of binding residues between periplasmic adapter protein (PAP) and RND efflux pumps explains PAP-pump promiscuity and roles in antimicrobial resistance. PLoS Pathog. 2019;15:e1008101. doi: 10.1371/journal.ppat.1008101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horiyama T., Yamaguchi A., Nishino K. TolC dependency of multidrug efflux systems in Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. 2010;65:1372–1376. doi: 10.1093/jac/dkq160. [DOI] [PubMed] [Google Scholar]
- 11.Wang-Kan X., Blair J.M.A., Chirullo B., Betts J., La Ragione R.M., Ivens A., Ricci V., Opperman T.J., Piddock L.J.V. Lack of AcrB Efflux Function Confers Loss of Virulence on Salmonella enterica Serovar Typhimurium. MBio. 2017;8:607–613. doi: 10.1128/mBio.00968-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blair J.M.A., Smith H.E., Ricci V., Lawler A.J., Thompson L.J., Piddock L.J. V Expression of homologous RND efflux pump genes is dependent upon AcrB expression: Implications for efflux and virulence inhibitor design. J. Antimicrob. Chemother. 2015;70:424–431. doi: 10.1093/jac/dku380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ricci V., Piddock L.J.V. Ciprofloxacin selects for multidrug resistance in Salmonella enterica serovar Typhimurium mediated by at least two different pathways. J. Antimicrob. Chemother. 2009;63:909–916. doi: 10.1093/jac/dkp054. [DOI] [PubMed] [Google Scholar]
- 14.Baugh S., Phillips C.R., Ekanayaka A.S., Piddock L.J.V., Webber M.A. Inhibition of multidrug efflux as a strategy to prevent biofilm formation. J. Antimicrob. Chemother. 2014;69:673–681. doi: 10.1093/jac/dkt420. [DOI] [PubMed] [Google Scholar]
- 15.Murakami S., Nakashima R., Yamashita E., Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 16.Murakami S., Nakashima R., Yamashita E., Matsumoto T., Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 17.Seeger M.A., Schiefner A., Eicher T., Verrey F., Diederichs K., Pos K.M. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 18.Seeger M.A., von Ballmoos C., Eicher T., Brandstätter L., Verrey F., Diederichs K., Pos K.M. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat. Struct. Mol. Biol. 2008;15:199–205. doi: 10.1038/nsmb.1379. [DOI] [PubMed] [Google Scholar]
- 19.Eicher T., Cha H., Seeger M.A., Brandstatter L., El-Delik J., Bohnert J.A., Kern W.V., Verrey F., Grutter M.G., Diederichs K., et al. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc. Natl. Acad. Sci. USA. 2012;109:5687–5692. doi: 10.1073/pnas.1114944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takatsuka Y., Nikaido H. Covalently linked trimer of the AcrB multidrug efflux pump provides support for the functional rotating mechanism. J. Bacteriol. 2009;191:1729–1737. doi: 10.1128/JB.01441-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsunaga Y., Yamane T., Terada T., Moritsugu K., Fujisaki H., Murakami S., Ikeguchi M., Kidera A. Energetics and conformational pathways of functional rotation in the multidrug transporter AcrB. Elife. 2018;7:e31715. doi: 10.7554/eLife.31715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vargiu A.V., Ramaswamy V.K., Malvacio I., Malloci G., Kleinekathöfer U., Ruggerone P. Water-mediated interactions enable smooth substrate transport in a bacterial efflux pump. Biochim. Biophys. Acta -Gen. Subj. 2018;1862:836–845. doi: 10.1016/j.bbagen.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 23.Nakashima R., Sakurai K., Yamasaki S., Nishino K., Yamaguchi A. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature. 2011;480:565–569. doi: 10.1038/nature10641. [DOI] [PubMed] [Google Scholar]
- 24.Husain F., Nikaido H. Substrate path in the AcrB multidrug efflux pump of Escherichia coli. Mol. Microbiol. 2010;78:320–330. doi: 10.1111/j.1365-2958.2010.07330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tam H.-K., Malviya V.N., Foong W.-E., Herrmann A., Malloci G., Ruggerone P., Vargiu A.V., Pos K.M. Binding and Transport of Carboxylated Drugs by the Multidrug Transporter AcrB. J. Mol. Biol. 2019;2019 432:861–877. doi: 10.1016/j.jmb.2019.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakashima R., Sakurai K., Yamasaki S., Hayashi K., Nagata C., Hoshino K., Onodera Y., Nishino K., Yamaguchi A. Structural basis for the inhibition of bacterial multidrug exporters. Nature. 2013;500:102–106. doi: 10.1038/nature12300. [DOI] [PubMed] [Google Scholar]
- 27.Zwama M., Yamasaki S., Nakashima R., Sakurai K., Nishino K., Yamaguchi A. Multiple entry pathways within the efflux transporter AcrB contribute to multidrug recognition. Nat. Commun. 2018;9:124. doi: 10.1038/s41467-017-02493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oswald C., Tam H.-K., Pos K.M. Transport of lipophilic carboxylates is mediated by transmembrane helix 2 in multidrug transporter AcrB. Nat. Commun. 2016;7:13819. doi: 10.1038/ncomms13819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eicher T., Seeger M.A., Anselmi C., Zhou W., Brandstätter L., Verrey F., Diederichs K., Faraldo-Gómez J.D., Pos K.M. Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB. Elife. 2014;3:e03145. doi: 10.7554/eLife.03145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atac N., Kurt-Azap O., Dolapci I., Yesilkaya A., Ergonul O., Gonen M., Can F. The Role of AcrAB-TolC Efflux Pumps on Quinolone Resistance of E. coli ST131. Curr. Microbiol. 2018;75:1661–1666. doi: 10.1007/s00284-018-1577-y. [DOI] [PubMed] [Google Scholar]
- 31.Sharma P., Haycocks J.R.J., Middlemiss A.D., Kettles R.A., Sellars L.E., Ricci V., Piddock L.J.V., Grainger D.C. The multiple antibiotic resistance operon of enteric bacteria controls DNA repair and outer membrane integrity. Nat. Commun. 2017;8:1444. doi: 10.1038/s41467-017-01405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weston N., Sharma P., Ricci V., Piddock L.J.V. Regulation of the AcrAB-TolC efflux pump in Enterobacteriaceae. Res. Microbiol. 2018;169:425–431. doi: 10.1016/j.resmic.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Blair J.M.A., Bavro V.N., Ricci V., Modi N., Cacciotto P., Kleinekathӧfer U., Ruggerone P., Vargiu A.V., Baylay A.J., Smith H.E., et al. AcrB drug-binding pocket substitution confers clinically relevant resistance and altered substrate specificity. Proc. Natl. Acad. Sci. USA. 2015;112:3511–3516. doi: 10.1073/pnas.1419939112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piddock L.J., Griggs D.J., Hall M.C., Jin Y.F. Ciprofloxacin resistance in clinical isolates of Salmonella typhimurium obtained from two patients. Antimicrob. Agents Chemother. 1993;37:662–666. doi: 10.1128/AAC.37.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piddock L.J., Whale K., Wise R. Quinolone resistance in salmonella: Clinical experience. Lancet (London, UK) 1990;335:1459. doi: 10.1016/0140-6736(90)91484-R. [DOI] [PubMed] [Google Scholar]
- 36.Parmar M., Rawson S., Scarff C.A., Goldman A., Dafforn T.R., Muench S.P., Postis V.L.G. Using a SMALP platform to determine a sub-nm single particle cryo-EM membrane protein structure. Biochim. Biophys. Acta. Biomembr. 2018;1860:378–383. doi: 10.1016/j.bbamem.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee S.C., Knowles T.J., Postis V.L.G., Jamshad M., Parslow R.A., Lin Y.-P., Goldman A., Sridhar P., Overduin M., Muench S.P., et al. A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc. 2016;11:1149–1162. doi: 10.1038/nprot.2016.070. [DOI] [PubMed] [Google Scholar]
- 38.Roach P.C.J., Postis V.L.G., Deacon S.E., Wright G.S.A., Ingram J.C., Xia X., McPherson M.J., Baldwin S.A. Large-scale preparation of bacterial cell membranes by tangential flow filtration. Mol. Membr. Biol. 2008;25:609–616. doi: 10.1080/09687680802530451. [DOI] [PubMed] [Google Scholar]
- 39.Postis V., Rawson S., Mitchell J.K., Lee S.C., Parslow R.A., Dafforn T.R., Baldwin S.A., Muench S.P. The use of SMALPs as a novel membrane protein scaffold for structure study by negative stain electron microscopy. Biochim. Biophys. Acta. 2015;1848:496–501. doi: 10.1016/j.bbamem.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng S.Q., Palovcak E., Armache J.-P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kimanius D., Forsberg B.O., Scheres S.H., Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife. 2016;5:18722. doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casanal A., Lohkamp B., Emsley P. Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data. Protein Sci. 2020;29:1069–1078. doi: 10.1002/pro.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 46.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sennhauser G., Amstutz P., Briand C., Storchenegger O., Grütter M.G. Drug Export Pathway of Multidrug Exporter AcrB Revealed by DARPin Inhibitors. PLoS Biol. 2006;5:e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webb B., Sali A. Protein Structure Modeling with MODELLER. Methods Mol. Biol. 2017;1654:39–54. doi: 10.1007/978-1-4939-7231-9_4. [DOI] [PubMed] [Google Scholar]
- 51.Topf M., Lasker K., Webb B., Wolfson H., Chiu W., Sali A. Protein structure fitting and refinement guided by cryo-EM density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vargiu A.V., Nikaido H. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 2012;109:20637–20642. doi: 10.1073/pnas.1218348109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Case D.A., Huang Y., Walker R.C., Lin C., Cheatham T.E., III, Mermelstein D.J., Simmerling C., Li P., Roitberg A., Onufriev A., et al. AMBER 2018. University of California; San Francisco, CA, USA: 2018. [Google Scholar]
- 54.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 56.Joung I.S., Cheatham T.E. Determination of Alkali and Halide Monovalent Ion Parameters for Use in Explicitly Solvated Biomolecular Simulations. J. Phys. Chem. B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vargiu A.V., Ruggerone P., Opperman T.J., Nguyen S.T., Nikaido H. Molecular Mechanism of MBX2319 Inhibition of Escherichia coli AcrB Multidrug Efflux Pump and Comparison with Other Inhibitors. Antimicrob. Agents Chemother. 2014;58:6224–6234. doi: 10.1128/AAC.03283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramaswamy V.K., Vargiu A.V., Malloci G., Dreier J., Ruggerone P. Molecular Rationale behind the Differential Substrate Specificity of Bacterial RND Multi-Drug Transporters. Sci. Rep. 2017;7:8075. doi: 10.1038/s41598-017-08747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sjuts H., Vargiu A.V., Kwasny S.M., Nguyen S.T., Kim H.-S., Ding X., Ornik A.R., Ruggerone P., Bowlin T.L., Nikaido H., et al. Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc. Natl. Acad. Sci. USA. 2016;113:3509–3514. doi: 10.1073/pnas.1602472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hopkins C.W., Le Grand S., Walker R.C., Roitberg A.E. Long-Time-Step Molecular Dynamics through Hydrogen Mass Repartitioning. J. Chem. Theory Comput. 2015;11:1864–1874. doi: 10.1021/ct5010406. [DOI] [PubMed] [Google Scholar]
- 61.Das D., Xu Q.S., Lee J.Y., Ankoudinova I., Huang C., Lou Y., DeGiovanni A., Kim R., Kim S.-H. Crystal structure of the multidrug efflux transporter AcrB at 3.1Å resolution reveals the N-terminal region with conserved amino acids. J. Struct. Biol. 2007;158:494–502. doi: 10.1016/j.jsb.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qiu W., Fu Z., Xu G.G., Grassucci R.A., Zhang Y., Frank J., Hendrickson W.A., Guo Y. Structure and activity of lipid bilayer within a membrane-protein transporter. Proc. Natl. Acad. Sci. USA. 2018;115:12985–12990. doi: 10.1073/pnas.1812526115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Durrant J.D., de Oliveira C.A.F., McCammon J.A. POVME: An algorithm for measuring binding-pocket volumes. J. Mol. Graph. Model. 2011;29:773–776. doi: 10.1016/j.jmgm.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang X., Miller W. A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 1991;12:337–357. doi: 10.1016/0196-8858(91)90017-D. [DOI] [Google Scholar]
- 65.Hung L.-W., Kim H.-B., Murakami S., Gupta G., Kim C.-Y., Terwilliger T.C. Crystal structure of AcrB complexed with linezolid at 3.5 Å resolution. J. Struct. Funct. Genomics. 2013;14:71–75. doi: 10.1007/s10969-013-9154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Müller R.T., Travers T., Cha H.-J., Phillips J.L., Gnanakaran S., Pos K.M. Switch Loop Flexibility Affects Substrate Transport of the AcrB Efflux Pump. J. Mol. Biol. 2017;429:3863–3874. doi: 10.1016/j.jmb.2017.09.018. [DOI] [PubMed] [Google Scholar]
- 67.Hryc C.F., Chen D.-H., Afonine P.V., Jakana J., Wang Z., Haase-Pettingell C., Jiang W., Adams P.D., King J.A., Schmid M.F., et al. Accurate model annotation of a near-atomic resolution cryo-EM map. Proc. Natl. Acad. Sci. USA. 2017;114:3103–3108. doi: 10.1073/pnas.1621152114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Soparkar K., Kinana A.D., Weeks J.W., Morrison K.D., Nikaido H., Misra R. Reversal of the Drug Binding Pocket Defects of the AcrB Multidrug Efflux Pump Protein of Escherichia coli. J. Bacteriol. 2015;197:3255–3264. doi: 10.1128/JB.00547-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vargiu A.V., Ramaswamy V.K., Malloci G., Malvacio I., Atzori A., Ruggerone P. Computer simulations of the activity of RND efflux pumps. Res. Microbiol. 2018;169:384–392. doi: 10.1016/j.resmic.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 70.Psakis G., Polaczek J., Essen L.-O. AcrB et al.: Obstinate contaminants in a picogram scale. One more bottleneck in the membrane protein structure pipeline. J. Struct. Biol. 2009;166:107–111. doi: 10.1016/j.jsb.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 71.Glover C.A.P., Postis V.L.G., Charalambous K., Tzokov S.B., Booth W.I., Deacon S.E., Wallace B.A., Baldwin S.A., Bullough P.A. AcrB contamination in 2-D crystallization of membrane proteins: Lessons from a sodium channel and a putative monovalent cation/proton antiporter. J. Struct. Biol. 2011;176:419–424. doi: 10.1016/j.jsb.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 72.Su C.-C., Morgan C.E., Kambakam S., Rajavel M., Scott H., Huang W., Emerson C.C., Taylor D.J., Stewart P.L., Bonomo R.A., et al. Cryo-Electron Microscopy Structure of an Acinetobacter baumannii Multidrug Efflux Pump. MBio. 2019;10:e01295-e19. doi: 10.1128/mBio.01295-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trampari E., Holden E.R., Wickham G.J., Ravi A., Prischi F., de Oliveira Martins L., Savva G.M., Bavro V.N., Webber M.A. Antibiotics select for novel pathways of resistance in biofilms. bioRxiv. 2019:605212. doi: 10.1101/605212. [DOI] [Google Scholar]
- 74.Yao H., Shen Z., Wang Y., Deng F., Liu D., Naren G., Dai L., Su C.-C., Wang B., Wang S., et al. Emergence of a Potent Multidrug Efflux Pump Variant That Enhances Campylobacter Resistance to Multiple Antibiotics. MBio. 2016;7:e01543-16. doi: 10.1128/mBio.01543-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bohnert J.A., Schuster S., Fähnrich E., Trittler R., Kern W. V Altered spectrum of multidrug resistance associated with a single point mutation in the Escherichia coli RND-type MDR efflux pump YhiV (MdtF) J. Antimicrob. Chemother. 2007;59:1216–1222. doi: 10.1093/jac/dkl426. [DOI] [PubMed] [Google Scholar]
- 76.Ramaswamy V.K., Vargiu A.V., Malloci G., Dreier J., Ruggerone P. Molecular Determinants of the Promiscuity of MexB and MexY Multidrug Transporters of Pseudomonas aeruginosa. Front. Microbiol. 2018;9:1144. doi: 10.3389/fmicb.2018.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schuster S., Kohler S., Buck A., Dambacher C., König A., Bohnert J.A., Kern W. V Random mutagenesis of the multidrug transporter AcrB from Escherichia coli for identification of putative target residues of efflux pump inhibitors. Antimicrob. Agents Chemother. 2014;58:6870–6878. doi: 10.1128/AAC.03775-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bohnert J.A., Schuster S., Seeger M.A., Fähnrich E., Pos K.M., Kern W. V Site-directed mutagenesis reveals putative substrate binding residues in the Escherichia coli RND efflux pump AcrB. J. Bacteriol. 2008;190:8225–8229. doi: 10.1128/JB.00912-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reading E., Ahdash Z., Fais C., Ricci V., Kan X.W., Grimsey E., Stone J., Malloci G., Lau A.M., Findlay H., et al. Perturbed structural dynamics underlie inhibition and altered specificity of the multidrug efflux pump AcrB. bioRxiv. 2020:063511. doi: 10.1101/2020.04.27.063511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.