Abstract
B cell activating factor (BAFF) is essential for B cells to develop and respond to Ags. Dysregulation of BAFF contributes to the development of some autoimmune diseases and malignancies. Little is known about when, where and how BAFF is produced in vivo and about which BAFF-producing cells contribute to B cell responses. To better understand BAFF functions, we created BAFF reporter mice (BAFF-RFP) and Baff floxed (Bafffl/fl) mice. Splenic and bone marrow (BM) neutrophils (Nphs) from BAFF-RFP mice expressed the highest constitutive levels of BAFF; other myeloid subsets including conventional dendritic cells (cDCs) and monocyte (MO) subsets expressed lower levels. Treatment of BAFF-RFP mice with Poly(I:C) increased BAFF expression in splenic Ly6Chi inflammatory MOs, CD11bhi activated NK subset, and in BM myeloid precursors. After infection with West Nile virus (WNV), BAFF increased in CD8− cDCs and Nphs, and BAFF+ CD11bhi NK cells expanded in draining lymph nodes. The cell- and tissue-specific increases in BAFF expression were dependent on type I IFN signaling. MAVS signaling also was required or contributed to BAFF expression in DC and MO subsets, respectively. Mice with deletion of Baff in either cDCs or Nphs had reduced Ab responses after NP-Ficoll immunization; thus, BAFF produced by both cDCs and Nphs contributes to T cell-independent Ab responses. Conversely, mice with a cDC Baff-deficiency had increased mortality after WNV infection and decreased WNV-specific IgG and neutralizing Ab responses. BAFF produced by Nphs and cDCs is regulated differently and has key roles in Ab responses and protective immunity.
Introduction
B-cell-activating factor (BAFF) (also known as BLyS or Tnfsf13b) is a member of the TNF family of ligands that binds to three different TNF receptor family members: BAFFR, TACI and BCMA (1–3). Baff−/− and Baffr−/− mice develop few or no mature B cells displaying a block in B cell development from the transitional 1 (T1) to T2 stage onward (4–6). BAFF signaling, like signaling through the BCR, functions as a key survival factor for maintaining mature B cell homeostasis (5, 6). BAFF is overexpressed in patients with autoimmune diseases such as Sjögren’s Syndrome, systemic lupus erythematosus (SLE), type I diabetes, B cell lymphoma and chronic lymphocytic leukemia (3, 7–9). In addition to BAFF’s contributing to the development of autoimmune diseases (10), we and others have shown that BAFF plays an important role in infectious diseases, including HIV and West Nile virus (WNV) (11–15). BAFF may also contribute to the pathology of HIV (16). Moreover, BAFF has been associated to inflammatory conditions, such as inflammatory bowel disease (17) and allergic airway inflammatory diseases (18).
Although BAFF dysregulation has been implicated in a number of human diseases, little is known about the regulation of BAFF expression and function in vivo. Most studies have focused on the effects of blocking BAFF receptors and the development of BAFF-blocking drugs (17, 19, 20). However, which BAFF-producing cells contribute to specific B cell responses and B cell-associated diseases is not entirely clear. Studies primarily done in vitro have established that BAFF can be expressed by different immune cell types including monocytes (MOs), macrophages (MΦs), dendritic cells (DCs), neutrophils (Nphs), and follicular dendritic cells (FDCs) as well as epithelial cells and stromal cells (1, 11, 21–23). In particular, BAFF produced by DCs, Nphs, epithelial cells, FDCs and stromal cells helps to regulate humoral immune responses (24). Several cytokines including IFN-γ, IL-10, G-CSF and GM-CSF upregulate BAFF expression in different cell types (1, 21, 25–28).
Type I IFN is a key regulator of BAFF. IFNα induces DCs to produce BAFF, which can play a role in class switch recombination (CSR) and drive B cells to become Ab producing cells (29–33). In particular, Type I IFN-dependent BAFF produced by DCs is likely to play a key role in T-independent type 2 (TI-2) Ab responses (29, 32–35). Both ssRNA and dsRNA viruses induce Type I IFN via RNA-sensing pathways, including TLR3, TLR7 and the RIG-I/MAVS pathway (36). Poly(I:C) promotes BAFF production in tonsillar mononuclear cells, which in turn triggers IgA CSR and aggravates IgA nephropathy (37). However which specific myeloid cell subset is responsible for this effect is not known. Infection with either ssRNA or dsRNA viruses also induces BAFF production in vitro in human salivary gland epithelial cells, DCs and MOs (11).
To define the BAFF-producing cells implicated in specific immune responses, we developed two novel transgenic (Tg) mouse lines, BAFF reporter (BAFF-RFP) mice and Baff floxed (Bafffl/fl) mice. Using BAFF-RFP reporter mice to monitor BAFF levels, we detected changes in BAFF expression in specific myeloid populations after in vivo activation of RNA-sensing pathways. Changes in BAFF production by MOs, cDCs and Nphs were mostly dependent on IFNAR but differentially affected by the absence of MAVS. Furthermore, we identified a new BAFF-expressing CD11bhi NK subset induced by either Poly(I:C) or WNV infection. Selective removal of Baff from either cDCs or Nphs revealed that BAFF produced from both DCs and Nphs is required for optimal TI-2 Ag-specific Ab responses. Mice lacking BAFF expression on cDCs were more susceptible to infection with WNV and had reduced WNV-specific IgG and neutralizing Abs as compared to controls. Thus, BAFF produced by cDCs is required to protect against a lethal viral infection. These findings underscore that BAFF produced by Nphs and cDCs is regulated differently and has distinct roles in Ab responses and protective immunity.
Materials and Methods
Mice
C57BL/6J, (B6) mice were purchased from Jackson Labs (Bar Harbor, ME). BAFF-RFP and Bafffl/fl mice (on the B6 background) were developed at the University of California at Davis Mouse Biology Program (UC Davis, CA) (see supplemental Fig. S1A). Mavs−/− (B6×129Sv/Ev) mice were kindly provided by Dr. S. Akira (Osaka University, Osaka, Japan) and backcrossed onto the B6 background (38). Ifnar−/− B6 mice were a kind gift from Dr. Michael Gale (University of Washington, Seattle, WA). Mrp8Cre and zDCCre B6 mice were purchased from Jackson Labs (Bar Harbor, ME). All mice were age- and sex-matched for experiments and used at 8–11 wks of age. Mice were housed in a specific pathogen free environment; all procedures were approved by the University of Washington Institutional Animal Care and Use Committee.
Generation of BAFF-RFP reporter and Bafffl/fl mice
A two-construct approach was used to generate BAFF reporter mice expressing a Tag-Red Fluorescence Protein-T (RFP) (39) under the Tnfsf13b/Baff promoter (BAFF-RFP) and Bafffl/fl mice to conditionally knockout the Tnfsf13b/Baff gene. First, the endogenous reporter BAFF-RFP mice were generated by replacing a Tnfsf13b allele with a targeting construct expressing IRES-RFP between exon 2 and exon 3 of Baff gene and containing loxP sites between exon 3 and exon 7. In the BAFF-RFP allele Baff is functionally knocked-out where the endogenous reporter expresses BAFF-IRES-RFP (BAFF-RFP), the RFP protein under the control of the BAFF promoter but with an internal ribosome entry site (IRES). In a second step, Bafffl/fl mice were generated by excising the RFP sequence and leaving wild type Baff gene with floxed exons 3–7 (for details about the construct and the generation of the transgenic mice see supplemental Fig. S1A, B). The Bafffl/fl mice express wild type Baff until Cre-mediated deletion of exons 3–7 render the Baff gene inactive. Bafffl/fl mice were crossed with Mrp8Cre mice (40, 41) and zDCCre mice (42, 43), to generate a conditional knockout (cKO) where Baff is selectively deleted in Nphs (Bafffl/fl Mrp8Cre) or cDCs (Bafffl/fl zDCCre).
For in vivo experiments we used heterozygous BAFF-IRES-RFP+/− (BAFF-RFP+/−) mice, which still have mature B cells and, therefore, are suitable for functional studies. Unless otherwise specified, “BAFF-RFP”, indicates heterozygous BAFF-RFP+/− mice. BAFF-RFP+/− mice were crossed with Ifnar−/− mice or Mavs−/− mice to generate BAFF-RFP Ifnar−/− mice and BAFF-RFP Mavs−/− mice, respectively.
Injection of TLR agonists and harvesting of PBMC, spleen and bone marrow (BM) cells
Mice were injected i.p. with 200μg/mouse Poly(I:C), 200μg/mouse R848 or 20nmol/mouse CpG; 6 h after injection of the TLR ligand, Brefeldin A (eBioscience, San Diego, CA, USA) was administered i.v. (44, 45). Brefeldin A was injected in vivo to optimize the retention of the BAFF-RFP signal inside the cells after TLR agonist administration; 24 h after TLR agonist injection, spleens and BMs were harvested for cell subsets analysis. Spleens from naïve or TLR-injected mice were harvested and dissociated into single cell suspensions as previously described with some modification (33). Briefly spleens were removed and dissociated by enzymatic digestion at 37 °C with liberase TL and Dnase I (Roche, Indianapolis, IN, USA), followed by mincing the tissue between the ends of two frosted microscope glass slides to obtain a single cell suspension. BM cells were isolated by cutting one end of the femur and flushing BM cells out of the bone by centrifugation. For PBMC harvesting, blood was collected by retroorbital eye bleeds using heparinized capillary tubes. After erythrocytes were lysed, PBMC, splenocytes and BM cell suspensions were processed for staining for flow cytometry. In initial experiments, BAFF-RFP mice were injected with Brefeldin A only for 18 h as controls, no significant differences in the BAFF-RFP signal/subset by flow cytometry, between naïve BAFF-RFP mice and Brefeldin A only injected BAFF-RFP mice were observed.
West Nile virus infections and inguinal LN (iLN) harvesting
The pathogenic lineage 1 WNV-TX infectious clone, derived from the Texas 2002-HC strain, was generously provided by Dr. Michael Gale (University of Washington, Seattle, WA) and prepared as described (13). For infections for iLN analysis, mice were inoculated under anesthesia with 1000 PFU WNV-TX subcutaneously into both footpads (f.p.) in a total volume of 40 μL; 20 μL (500 PFU) were injected in each f.p. (13). 24 h or 48 h after WNV infection iLNs were removed, minced into small fragments and digested for 40 min at 37 °C with liberase TL and Dnase I (Roche, Indianapolis, IN, USA), as described (46). Single cell suspensions were processed for staining for flow cytometry. For survival studies infected mice were inoculated under anesthesia with 100 PFU WNV-TX subcutaneously into one f.p. in a total volume of 20 μL. Mice were monitored at least once daily for survival. Serum was isolated from blood, collected via the retro-orbital route at d 4 and d 7, and stored at −80°C until use.
WNV RNA quantitation, WNV-specific Ab ELISA and FRNT50
Viral RNA was extracted from sera by using QiAMP viral RNA extraction kit (Qiagen, Valencia, CA, USA). Sequences for primers and TaqMan probes used, were as described (13). WNV envelope protein (WNVE)-specific IgG was quantitated by ELISA assay using a recombinant WNV-E protein (MyBioSource, San Diego, CA, USA) as described (13). Titers of WNV neutralizing Abs were quantitated by a focus forming reduction neutralization test (FRNT) using Vero cells as described (47). The FRNT50 titers were calculated as the lowest dilution of serum with less than 50% of the average number of spots in the virus only wells.
NP-Ficoll immunization and NP-Ab ELISA
Mice were administered i.p. with 20 ug/mouse of NP (4-Hydroxy-3-nitrophenylacetic) hapten conjugated to AminoEthylCarboxyMethyl-FICOLL (AECM-FICOLL), NP-Ficoll (Biosearch Technologies, Petaluma, CA, USA). At the indicated time points mice were bled and serum was isolated. Serum NP Ab titers were measured as described (33). Plates were developed with tetramethylbenzidine substrate and read at 450 nm absorbance. Values were compared with known dilutions of IgM or IgG to calculate Ab concentrations.
Flow cytometry
Splenocytes, PBMCs, BM cells and iLNs were processed into single cell suspensions as above and stained for flow cytometry analysis as described (13). Cells were incubated with an Aqua Live-Dead fixable viability dye (Molecular Probes, Life Technologies, Waltham, MA, USA) in the absence of FBS, to discriminate dead cells. Cells were then blocked using an anti-FC receptor Ab (anti-CD16/CD32) (2.4G2) (BioLegend, (San Diego, CA, USA) and stained for surface markers and then fixed in 1–2% paraformaldehyde. Cells were stained with mAbs conjugated to FITC, allophycocyanin, eFluor450, allophycocyanin-eFluor780, PerCPCy5.5, PE-Cy7, AlexaFluor647, BUV395, BV605, BV421, BV711, BV650 and BUV395. For analysis of splenic or BM cell subsets eleven- to twelve-colors flow cytometry was performed using combinations of mAbs against: CD19 (1D3), CD11b (M1/70) and CD11c (N418) from eBioscience (San Diego, CA, USA); B220 (RA3–6B2), CD93 (AA4.1) and Ly6C (AL-21) from BD Horizon/Biosciences (San Jose, CA, USA); CD19 (1D3), B220 (RA3–6B2), CD3 (17A2), NK1.1 (PK136), CD8α (53–6.7), Ly6G (1A8), SiglecH (440c), Ly49c (14B11), CD49b (DX5), NKp46/CD335 (29A1.4), CD127 (SB/199), CD21/35 (7E9), CD23 (B3B4) and CD24 (M1/69) from BioLegend (San Diego, CA, USA). The BAFF-RFP signal was detected in the PE channel. Cells were processed with an LSRII flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) using a FACSDiva software and data were analyzed using FlowJo (v.10, Tree Star). See supplemental Fig S2A and Fig. S2B for gating strategies for spleens (and iLNs) and BM, respectively. Splenic myeloid cell subsets were defined as described previously (13), as in supplemental Fig S2A. For BM we identified 5 populations of myeloid cells and precursors in the CD11b+B220−CD19−Ly6G−SSC− gate and defined them based on the literature (48–50) (See supplemental Fig. S2B and supplemental Table S1). A similar gating strategy as for spleen cells was used for PBMC. PBMC subsets were defined as follows: Nphs, CD11bhiLy6GhiLy6CintSSCint−NK1.1−; Ly6Chi MOs, CD11bhiLy6ChiCD11c−SSC−Ly6G−NK1.1−SiglecH−; DCs, CD11b+Ly6C−CD11c+SSC−Ly6G−NK1.1−SiglecH−; NK cells, NK1.1hiCD11bint; T cells, CD3+CD19−; B cells, CD3−CD19+.
Splenic B cells subsets were defined as described previously (13, 51). After gating out debris, doublets and dead cells, B cells subsets in the CD19+ B220+ gate were defined as follows: FO B cells, CD24midCD21/35midCD93−CD23−; Marginal zone (MZ) B cells, CD24hiCD21/35hiCD93−CD23−; MZ B cell precursors CD24hiCD21/35hiCD93loCD23+; T2 B cells, CD24hiCD21/35int/hiCD93+CD23+; T1 B cells CD24hiCD21/35loCD93+CD23−. For BM B cell precursors and long lived plasma cells (PCs) (52) we gated out NK cells, pDCs and CD11b+ cells. In the NK1.1−SiglecH−CD11b− gate we defined PreProB cells as B220−CD43+CD19−; ProB cells as B220loCD43+CD19+; PreB cells as B220+CD43−CD19+IgM−IgD−; newly formed B cells (NFB) as B220+CD43−CD19+IgM+IgD−; mature B cells (MatB) as B220+CD43−CD19+IgM+IgD+; PreBNFMatB (B220+CD43−). Splenic PCs and long lived PCs in the BM were defined as B220loCD138+. Splenic T cell subsets are defined as follows: CD8 T cells as CD3+ CD8+CD19−, CD4 T cells as CD3+ CD8−CD19−.
Sorting Strategy for BAFF-RFP mice and BAFF cKO mice verification and BAFF qPCR analysis
BM cells from WT and BAFF-RFP (+/−) mice were harvested as described above and cell subsets sorted with FACS Aria cell sorter (Becton Dickinson, Franklin Lakes, NJ, USA). Nphs were sorted from WT and BAFF-RFP mice as CD11b+Ly6GhiLy6CintSSCint−. B cells were sorted from WT mice as Ly6G−B220+ cells, and as Ly6G−B220+RFP+ B cells and Ly6G−B220+RFP− B cells from BAFF-RFP mice.
Splenocytes were harvested from Bafffl/fl, Bafffl/fl MRP8Cre or Bafffl/fl zDCCre mice as described above and enriched with CD11b+ CD11c+ magnetic bead positive selection (Milteny, CA, USA). Cell populations were sorted using a FACS Aria cell sorter (Becton Dickinson, Franklin Lakes, NJ, USA). For gating strategies see Supplemental Fig. S2A. Sorted cells were lysed and RNA extracted for qRT-PCR analysis as described (33). The primer sequences used were as follows: mBaff-F 5’-AGGCTGGAAGAAGGAGATGAG-3’ and mBaff-R 3’- CAGAGAAGACGAGGGAAGGG −5’. See Supplemental (Fig. S1C, S1D) for verification of BAFF Nph cKO (Bafffl/fl MRP8Cre) mice or BAFF cDCs cKO (Baff fl/fl zDCCre) mice. As additional controls, sorted cells from WT mice, MRP8Cre mice and zDCCre mice all showed similar levels of Baff mRNA as Bafffl/fl mice (not shown).
Statistical analyses
Survival data were analyzed by Mantel-Cox log-rank test. Timeline data were analyzed using a 2-way ANOVA with Tukey’s multiple comparison test. Analysis between more than two groups were performed using 1-way ANOVA with Holm-Sidak method for multiple comparisons. Analyses between two groups were performed using unpaired Student t test. GraphPad Prism 7 was used for statistical analyses. Differences of p<0.05 were considered significant.
Results
BAFF-RFP expression in BM and splenic cell populations
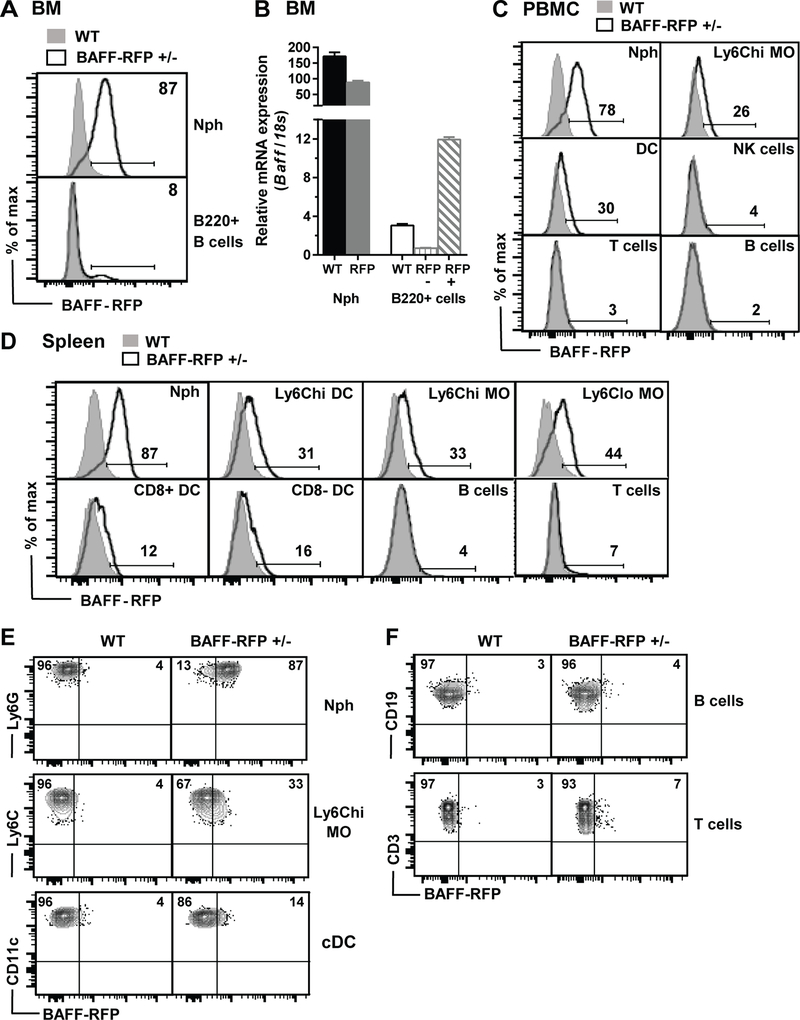
We developed mouse models to define how BAFF expression by cell subsets is regulated and what BAFF-producing cells are required for B cell responses. We used a two-construct approach to generate BAFF IRES-TagRFP-T (BAFF-RFP) reporter mice concurrently with Bafffl/fl mice (see supplemental Fig. S1A for details). In BM from naïve BAFF-RFP (+/−) mice most Nphs were BAFF-RFP+ and a small population of BAFF-RFP+ B220+ B cells was detectable (Fig. 1A). To determine whether RFP expression in BAFF-RFP mice correlated with Baff mRNA expression, we sorted BM Nphs, BAFF-RFP+ B220+ cells and BAFF-RFP− B220+ B cells and quantified Baff mRNA levels (Fig. 1B, for sorting strategy see Methods). As expected Baff mRNA levels in Nphs from heterozygous BAFF-RFP mice were about half the levels detected in WT mice. Baff mRNA expression was lower in B cells than in Nphs in both WT and BAFF-RFP+/− mice. Importantly, Baff mRNA levels were higher in BAFF-RFP+ B cells compared to BAFF-RFP− B cells or unsorted WT B cells (Fig. 1B). Therefore, BAFF-RFP detected by flow cytometry correlates with Baff mRNA expression.
Figure 1. BAFF-RFP detection in blood and splenic cell subsets.
A. BAFF-RFP by flow cytometry in BM Nph and a subset of B220+ B cells in BAFF-RFP+/− mice (See Methods for gating strategy). B. BAFF mRNA by qPCR in sorted Nphs, B220+ B cells from WT mice, B220+RFP+ B cells and B220+RFP− B cells from BAFF-RFP+/− mice. Baff mRNA expression in arbitrary units relative to 18S. A and B, data show one representative of two experiments. C and D, expression of BAFF-RFP in PBMC (C) and splenic (D, E and F) cell populations from naïve BAFF-RFP+/− mice. C and D, data show one representative of two (C) or more than five (D, E and F) experiments. In A, C and D, WT (C57BL/6): grey filled histograms; BAFF-RFP+/− mice: black empty histograms. E and F, dot plots of BAFF-RFP expression in myeloid (E), B cells and T cells (F). In A-E, Neutrophils, Nph; Ly6Chi monocytes, Ly6Chi MO; dendritic cells, DC, conventional DC, cDC. For gating strategy of cell populations in Spleen (D, E and F) see Fig. S2B; and for cell populations in BM (A) and PBMCs (C) see Methods PBMCs (C) see Methods.
Analysis of cell populations from spleens or PBMCs (see Methods and supplemental Fig. S2A for gating strategy) from naïve BAFF-RFP+/− mice revealed that BAFF is constitutively expressed at high levels in Nphs, expressed at lower levels in DCs and MOs and had very low expression or was undetectable in other leukocyte subsets (Fig. 1C and 1D, Fig. S3A and S3B). These data are consistent with previous reports (1, 11, 21). Although BAFF-RFP expression overall was very low in B cells, BAFF was detectable particularly in MZ B cells and MZ B cell precursors and T2 B cells and in small numbers of FO and T1 B cells (Fig. S3A), in agreement with Baff mRNA expression in B cells subsets (53).
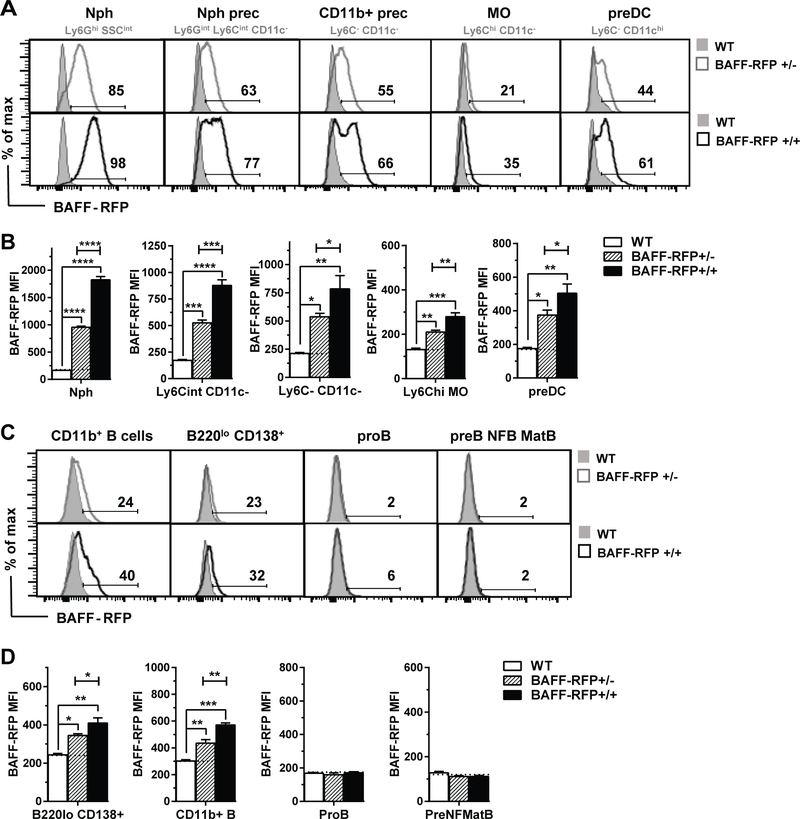
Interestingly, in the BM of naïve BAFF-RFP+/− mice, in addition to Nphs, other myeloid cell subsets and myeloid precursors constitutively expressed significant levels of BAFF-RFP (Fig. 2). Using mice with either one Rfp allele (RFP+/−) or two Rfp alleles (RFP+/+), we examined the CD11b+ BM population in detail and designated subsets based on their cell surface phenotypes (see Methods and supplemental Fig. S2B and Table S1). In particular, a high percentage of Nph precursors, CD11b+ myeloid precursors and preDCs expressed BAFF-RFP compared to MOs (Fig. 2A). BAFF-RFP expression levels were also greater in Nphs and other myeloid precursors compared to MOs, as shown by their higher levels of expression (Fig. 2B). The BAFF-RFP signals in both BAFF-RFP+/− and BAFF-RFP+/+ were significantly different from RFP background detection in WT mice (Fig. 2A–D). The higher levels of BAFF-RFP in BAFF-RFP+/+ mice vs BAFF-RFP+/− mice in all BM populations, confirmed that BAFF-RFP protein detection by flow cytometry correlates with the amount of Baff gene expression (Fig. 2A–D).
Figure 2. BAFF-RFP detection in BM cell populations.
A-D, BAFF-RFP detected by flow cytometry in BM myeloid cell populations (A-B) or B cell populations (C-D) from naïve WT mice, BAFF-RFP+/− mice and BAFF-RFP+/+ mice. For gating strategy of myeloid cell populations (A and B) in BM see Fig. S2B. For gating strategy of B cell populations (C and D) see Methods. Data shown are representative histograms (A and C) or mean ± SEM of RFP MFI (B and D) from one of two independent experiments performed with three mice per group each. B and D, dotted lines show RFP MFI background in WT control. Statistics were performed by one-way ANOVA with Holm-Sidak method for multiple comparisons; * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
We also analyzed BM B cell subsets and precursors. BAFF-RFP was expressed at low levels in B220lo CD138+ long-lived plasma cells (LLPCs) and in a subset of CD11b+ B220+ CD19+ B cells but not in ProB cells or more mature B cells precursors (Fig. 2C and 2D). As expected, BAFF-RFP+/− mice had fewer mature B cells precursors in the BM than WT mice, while BAFF-RFP+/+ mice had almost no mature B cells and normal numbers of earlier B cell precursors (Fig. S2C and S2D). Although the BAFF-RFP+/− mice had lower expression of BAFF, they were still capable of producing mature B cells and thus could be used to measure B cell functions and Ab responses in vivo. For subsequent studies we used BAFF-RFP+/− mice and refer to them simply as BAFF-RFP mice.
Cell and tissue specific upregulation of BAFF-RFP in myeloid subsets after in vivo administration of TLR agonists or WNV infection
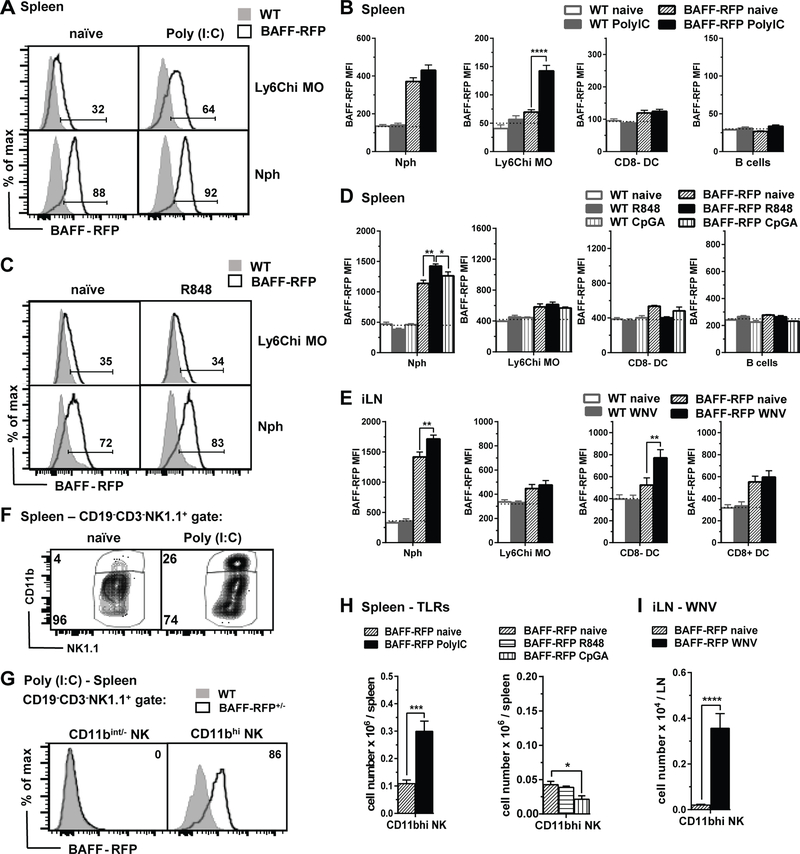
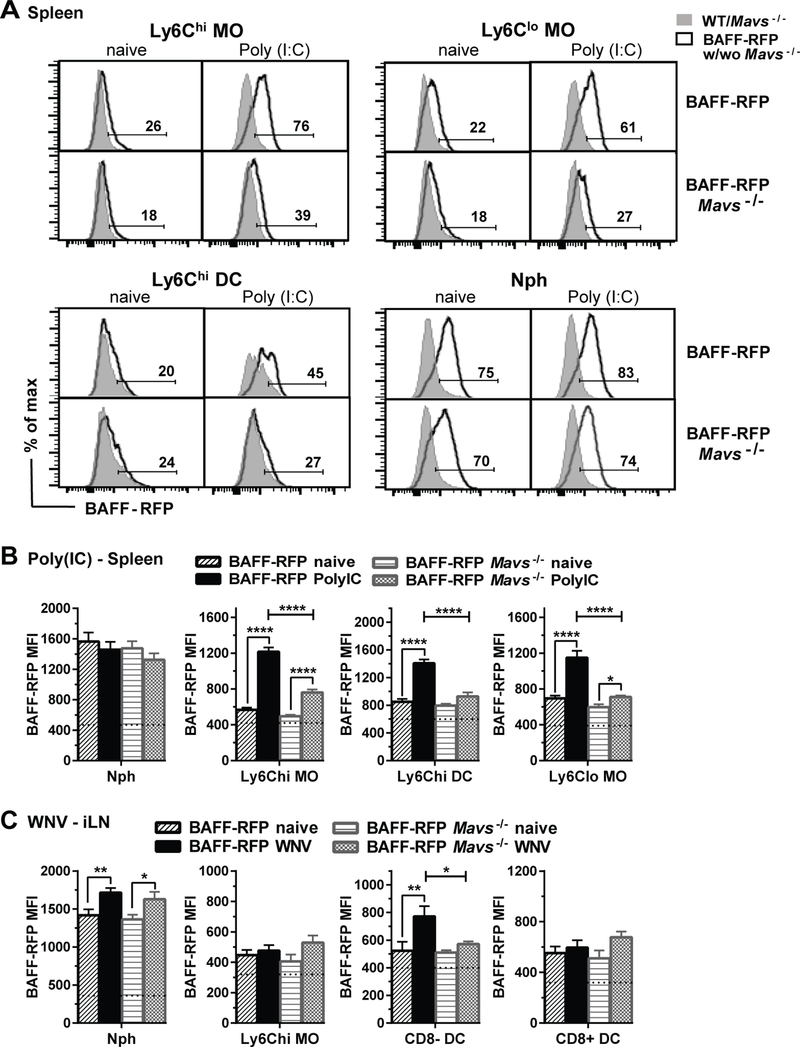
To address how the activation of RNA-sensing pathways regulate BAFF expression in specific cell subsets in vivo, we injected BAFF-RFP mice with TLR3 agonist Poly(I:C) or the TLR7/8 agonist R848 and analyzed BAFF-RFP expression in splenic cell populations 24 h later (Fig 3A–D). Poly(I:C) administration led to significant increases in BAFF-RFP MFI (Fig. 3A and 3B) and the percentage of BAFF-RFP+ cells (not shown) in inflammatory Ly6Chi MOs but not in cDCs, Nphs or B cells. In contrast, R848 administration had no effect on BAFF-RFP expression in splenic Ly6Chi MOs but slightly upregulated BAFF-RFP in Nphs (Fig. 3C and 3D). Furthermore, the TLR9 agonist, CpG did not affect BAFF-RFP expression in either Ly6Chi MOs or Nphs (Fig. 3D). Thus, changes in BAFF-RFP expression in myeloid subsets varied depending on the TLR-ligand used.
Figure 3. Cell and tissue specific up-regulation of BAFF-RFP in myeloid subsets after in vivo administration of TLR agonists or WNV infection.
A-D and F-G, WT and BAFF-RFP mice were injected i.p. or not for 24 h with 200μg/mouse Poly(I:C) (A,B,F and G), 200μg/mouse R848 (C,D and G), or 20nmol/mouse CpG (D and G). A-D and F-G, 6 h after i.p. injection of the TLR ligand, Brefeldin A was administered i.v. (see Methods). In E and H, WT and BAFF-RFP+/− mice were infected via f.p. with 1000pfu WNV. A-H, BAFF-RFP+ cell populations from spleens (A-D and F-G) or iLNs (E and H) were analyzed by flow cytometry and RFP MFI was compared to WT mice as controls. In F-H, CD11bhi NK cell subset upregulated in spleen after Poly(I:C) injection (F and G) or in iLN after WNV infection (H). A, F and C, data are shown as representative histograms from more than three independent experiments (A and F, Poly(I:C)), or two independent experiments (C, R848). B, D and E bar graphs show means ± SEM of RFP MFI. H and I, bar graphs show total cell numbers of BAFF-RFP+ CD11bhi NK cells in spleens from Poly(I:C) (H, left panel), R848 or CpG (H, right panel), or in iLN from WNV infected mice (I). Graphs summarize data from three independent experiments (B, E, H, left panel and I (N=9)); or are from one representative of two independent experiments (D and H right panel (N=3–4). In B, D and E, dotted lines show RFP MFI background in WT control mice. Statistics were performed by one-way ANOVA with Holm-Sidak method for multiple comparisons; except unpaired Student t test was used in G left panel and H; * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
Next we challenged BAFF-RFP mice with WNV-TX (WNV), a pathogenic ssRNA virus, and examined BAFF-RFP expression in myeloid cells within the draining iLNs 24 h and 48 h after infection. Similarly to the effect of the TLR3 agonist in the spleen, WNV infection slightly increased BAFF-RFP levels in Nphs from iLNs (Fig. 3E), and in addition, significantly up-regulated BAFF-RFP expression in CD8− cDCs 24 h post-infection (Fig. 3E). BAFF-RFP levels did not change in iLN CD8+ cDC, MO subsets, B cells or T cells after WNV infection (Fig. 3E and not shown).
Interestingly, after either Poly(I:C) administration or WNV infection, we detected increased numbers of an activated NK1.1 subset, which expressed high levels of both BAFF-RFP and CD11b (CD11bhi NK). In contrast, CD11bint− NK cells were BAFF-RFP− (Fig. 3F, 3G, 3H left panel and 3I). This BAFF-RFP+ CD11bhi NK subset was not upregulated after in vivo administration of R848 or CpG (Fig. 3H right panel). A phenotypic analysis revealed that the CD11bhi NK cells, like other NK cell subsets, express CD49b, NKp46 and Ly-49C, and in addition express Ly6G, a marker found on Nphs (Supplemental Fig. S3C). Furthermore, CD11bhi NK cells did not express CD127 suggesting they were not innate lymphoid cell 1 (ILC1) (54). In summary, by using BAFF-RFP reporter mice we detected cell- and tissue- specific up-regulation of BAFF after in vivo challenge with different RNA-sensing TLR agonists or WNV infection.
Requirement for Type I IFN and MAVS signaling pathways for BAFF-RFP up-regulation in myeloid subsets
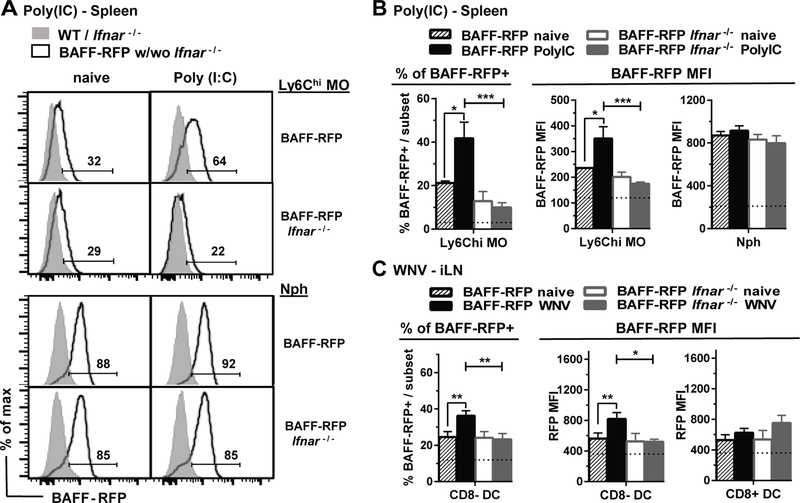
Both dsRNA and ssRNA viruses trigger three RNA sensing pathways: TLR3 (dsRNA), TLR7/TLR8 (ssRNA) and RIG-I/MDA5/MAVS (ssRNA, dsRNA), which induce type I IFN production in plasmacytoid DCs or other cells (36, 55). Type I IFN is a major inducer of BAFF. However, direct regulation of BAFF by TLR engagement also has been described (22). Thus, to test whether Type-1 IFN was required for BAFF upregulation, we crossed BAFF-RFP mice with Ifnar−/− mice and examined BAFF-RFP expression after either Poly(I:C) administration or WNV infection (Fig. 4). Unlike in BAFF-RFP WT mice, BAFF-RFP Ifnar−/− mice did not upregulate BAFF-RFP in Ly6Chi MOs and Ly6Chi DCs in response to Poly(I:C), while there was no change in the constitutive BAFF-RFP expression in Nphs (Fig. 4A and 4B, and not shown). Furthermore, BAFF-RFP Ifnar−/− mice didn’t upregulate BAFF in either iLN Nphs or CD8− cDC after infection with WNV (Fig. 4C and not shown). Thus, the increases in BAFF expression in splenic inflammatory monocyte subsets in response to Poly(I:C) immunization and in iLN Nphs and CD8− cDCs after WNV infection all require type I IFN signaling.
Figure 4. The type I IFN receptor (IFNAR) is required for BAFF up-regulation in MOs and cDCs induced by Poly(I:C) and WNV.
BAFF-RFP mice and BAFF-RFP Ifnar−/− mice were treated or not for 24 h with 200μg/mouse Poly(I:C) (A and B) by i.p. injection or f.p. infection with WNV (1000pfu) (C). Data show BAFF-RFP levels by flow cytometry in myeloid populations from spleens (A and B) and iLNs (C). A, data shown are representative histograms from two independent experiments, the numbers indicate % of BAFF-RFP+ cells. B and C bar graphs show means ± SEM of % of BAFF-RFP+ cells (left panels) or RFP MFI (right panels) and summarize data from two independent experiments (N=6–7). B and C, dotted lines show % of RFP+ cells or RFP MFI background in WT and Ifnar−/− mice. Statistics were determined with one-way ANOVA with Holm-Sidak method for multiple comparisons test; *** p<0.001, **** p<0.0001.
Since both dsRNA and ssRNA can induce type I IFN via the MAVS pathway, we next tested whether Poly(I:C)- or WNV-induced changes in BAFF-RFP expression required MAVS. Using BAFF-RFP Mavs−/− mice, we found that myeloid subsets differed in their requirement for MAVS to regulate BAFF expression. Poly(I:C)-induced increases in BAFF-RFP MFI in Ly6Chi DCs required MAVS, while the BAFF-RFP MFI increase in Ly6Chi MOs and Ly6Clo MOs was only partially inhibited in BAFF-RFP Mavs−/− mice (Fig. 5A and 5B). As expected, there was no change in the BAFF-RFP expression in Nphs. These data suggest that for MO subsets, type I IFN production that up-regulates BAFF, occurs through either the TLR3 pathway or the MAVS pathway. In contrast, the BAFF increase in inflammatory Ly6Chi DCs is dependent solely on type I IFN produced via the MAVS pathway.
Figure 5. Differential requirement of MAVS for BAFF changes induced by Poly(I:C) and WNV in different myeloid subsets.
A-C. BAFF-RFP mice and BAFF-RFP Mavs−/− mice were left untreated or treated i.p. with 200μg/mouse Poly(I:C) or infected with 1000pfu WNV via the f.p. for 24 h. Data show BAFF-RFP levels by flow cytometry in myeloid populations from spleens (A and B) and iLNs (C). In A, data shown are representative histograms from two independent experiments, the numbers indicate % of BAFF-RFP+ cells. In B and C bar graphs show means ± SEM of BAFF-RFP MFI and summarize data from two independent experiments (N=6–7). In B and C, dotted lines show RFP MFI background in WT and Mavs−/− mice that were included as controls in each experiment. Statistics were determined with one-way ANOVA with Holm-Sidak method for multiple comparisons test; ** p<0.01, *** p<0.001, **** p<0.0001.
We also analyzed BM cells from BAFF-RFP Ifnar−/− and BAFF-RFP Mavs−/− mice after Poly(I:C) immunization. Both the IFNAR and MAVS signaling pathways were required to induce increases in the percentage of inflammatory BAFF-RFP+ Ly6Chi MOs and BAFF-RFP+ Ly6Chi CD11c+ cells as well as in other myeloid precursors and BAFF+ CD11b+ B cells (Supplemental Fig. S3D and S3E). Thus, BAFF is differentially regulated in both a cell–specific manner and in a tissue-specific fashion. For example, BAFF regulation in Ly6Chi MOs is only partially dependent on MAVS in the spleen, but requires MAVS in the BM.
BAFF MFI was not significantly upregulated in CD8− cDCs in BAFF-RFP Mavs−/− mice after WNV infection; however, BAFF upregulation in Nphs after infection was independent of MAVS (Fig. 5C). Therefore, after WNV infection the BAFF increase in CD8− cDCs requires the activation of the MAVS pathway, while TLR7/8 pathway may be required for increasing BAFF expression in Nphs. In summary, the changes in BAFF expression by myeloid subsets induced after Poly(I:C) immunization or WNV infection require type I IFN signaling but only in some cases are dependent on MAVS.
The expansion of BAFF+ CD11bhi NK cells and BAFF+ Ly6Chi MOs and DCs is regulated by IFNAR and MAVS signaling
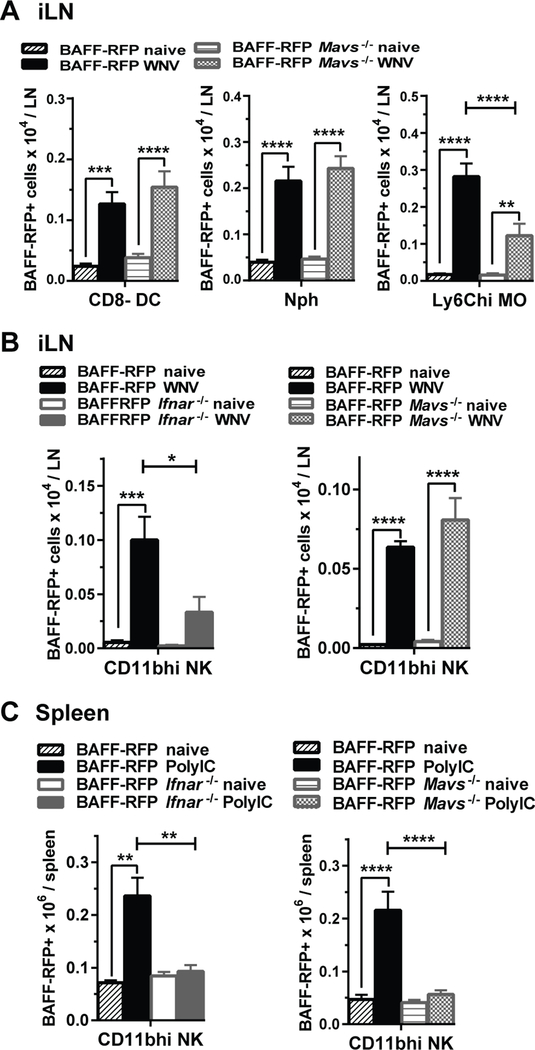
BAFF-RFP reporter mice allowed us not only to detect BAFF expression per cell but also to analyze changes in populations of BAFF-RFP producing cells. Several BAFF-RFP+ myeloid cell subsets including Nphs, inflammatory MOs and cDCs were expanded in the iLNs 24 h after WNV infection (Fig. 6A). A number of factors, including chemokines and chemokine receptors, could regulate migration and the expansion of myeloid cells upon infection. Therefore, we tested whether MAVS/IFNAR signaling pathways were required for the changes in cell numbers. Interestingly, MAVS signaling contributed to BAFF-RFP+ inflammatory Ly6Chi MO (Fig. 6A) and Ly6Chi DC (not shown) expansion but not to the increased numbers of BAFF+ CD8− cDCs and BAFF+ Nphs (Fig. 6A). Thus, even though BAFF-RFP expression per cell in CD8− cDC required MAVS signaling, the expansion of CD8− cDCs was MAVS independent. The requirements for IFNAR in the expansion of myeloid subsets were similar to the MAVS requirements (not shown). Taken together these results suggests that RIG-I/MAVS-Type I IFN signaling axis plays a role in the expansion of inflammatory Ly6Chi MOs and Ly6Chi DCs, while other factors are responsible for the increased numbers of Nphs and cDCs upon WNV infection.
Figure 6. Regulation by MAVS and IFNAR signaling of BAFF+ CD11bhi NK cells and BAFF+ Ly6Chi MO/DC expansion in response to WNV and Poly(I:C).
BAFF-RFP mice, BAFF-RFP Mavs−/− mice (A) BAFF-RFP Ifnar−/− mice (B and C) or WT mice were infected or not with 1000pfu WNV (f.p.) (A and B) or injected i.p with 200μg/mouse Poly(I:C) (C). Data show numbers of BAFF-RFP+ myeloid cell subsets from iLNs harvested 24 h post-infection (A and B), or from spleens 24 hrs post- Poly(I:C) immunization (C). Bar graphs show means ± SEM of BAFF-RFP+ cell numbers per iLN and summarize data in A and B right panel from three independent experiments (N=9); in B left panel from two independent experiments (N=6–9), in C left panel and C right panel from two independent experiments each (N=6–7). Statistics were performed using one-way ANOVA with Holm-Sidak method for multiple comparisons test; * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
The expansion of the BAFF-RFP+ NK CD11bhi cells in the iLN after WNV infection required the IFNAR signaling pathway, but was independent of MAVS signaling (Fig. 6B). In contrast, administration of Poly(I:C) to either BAFF-RFP Ifnar−/− or BAFF-RFP Mavs−/− mice failed to expand BAFF+ CD11bhi NK cells (Fig. 6C). Taken together these data suggest that type I IFN-dependent expansion of activated BAFF+ CD11bhi NK cells, upon WNV infection is mostly dependent on TLR7/8 signaling. In contrast, after dsRNA challenge, the type I IFN-dependent expansion of this BAFF+ NK subset, occurs via engagement of the MAVS pathway rather than TLR3 activation.
BAFF produced by DCs is required for protective immune responses to WNV infection
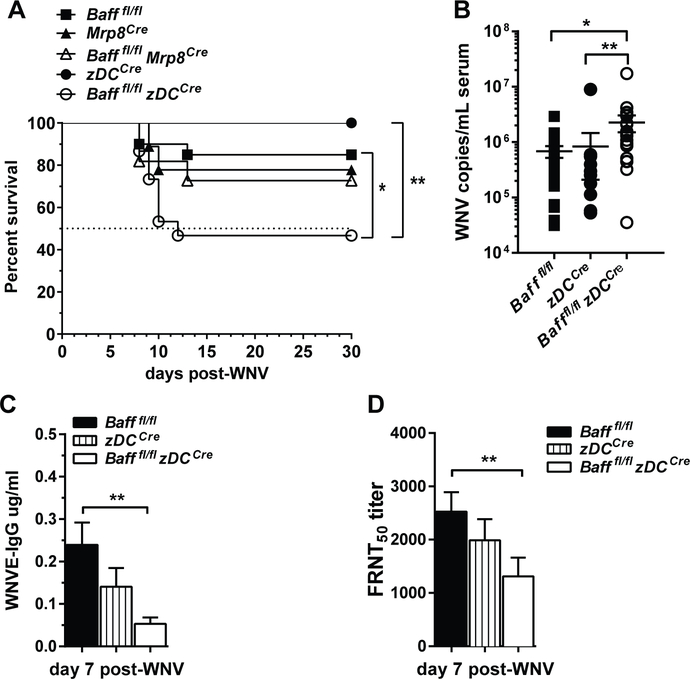
BAFF expression increases in cDCs and Nphs, and BAFF+ cDCs and BAFF+ Nphs expand after WNV infection (Fig. 3E, 6A). To examine the role of these sources of BAFF during infection, we generated Baff conditional knockout (BAFF cKO) mice, where we specifically deleted Baff in cDCs or Nphs, Bafffl/fl mice (See Supplemental Fig. S1A and Methods) were crossed with either zDCCre mice or Mrp8Cre mice to generate mice deficient in BAFF expression on cDCs (Bafffl/fl zDCCre), and mice deficient in BAFF expression on Nphs (Bafffl/fl Mrp8Cre). Analysis of the expression of Baff mRNA in purified cDCs or Nphs confirmed that BAFF expression was knocked out appropriately in the respective BAFF cKO mice (Fig. S1C, S1D). Baff mRNA reduction was restricted to cDCs in Bafffl/fl zDCCre mice. BAFF mRNA in Bafffl/fl Mrp8Cre mice was dramatically reduced in Nphs, but some reduction was evident in MOs. In agreement with previous findings suggesting that radiation-resistant but not hematopoietic cells were responsible for B cell homeostasis (56), we found that depleting BAFF from either Nphs or DCs did not alter the splenic B cell compartment in naïve mice (data not shown).
We infected Bafffl/fl zDCCre mice and Bafffl/fl Mrp8Cre mice and monitored their survival and Ab responses. Bafffl/fl zDCCre mice had significantly increased mortality compared to Bafffl/fl and zDCCre controls (Fig. 7A). In contrast, Bafffl/fl Mrp8Cre mice had similar survival rate as Bafffl/fl mice (Fig. 7A). Furthermore, Bafffl/fl zDCCre mice had increased virus titers (Fig. 7B) and decreased WNV-specific IgG and WNV neutralizing Abs (Fig. 7C and 7D) compared to controls. Thus, BAFF produced by DCs is required for optimal protective immune responses to WNV infection. Apparently, BAFF from cDCs helps to sustain or promote B cell humoral responses to WNV, since WNV-specific Ab responses are decreased in mice lacking BAFF expression on cDC.
Figure 7. BAFF produced by DCs is required for protective immune responses to WNV infection.
A-D, Bafffl/fl, Mrp8Cre, Bafffl/fl Mrp8Cre mice, zDCCre, Bafffl/fl zDCCre mice were infected via f.p. with WNV (100pfu). Mice were monitored daily for survival (A). Viral titers in sera were quantified using real time qPCR (B). WNVE-specific IgG were analyzed by ELISA (C). Neutralizing Abs were analyzed by FRNT50 in Vero cells (D). A-D, Data are combined from three independent experiments: Bafffl/fl, N=20; Mrp8Cre, N=9; Bafffl/fl Mrp8Cre, N=11; zDCCre, N=14; Bafffl/fl zDCCre, N=15. Statistics were performed using a log-rank test for significance (A), or one-way ANOVA corrected with Holm-Sidak method for multiple comparisons test (B-D); * p<0.05, ** p<0.01.
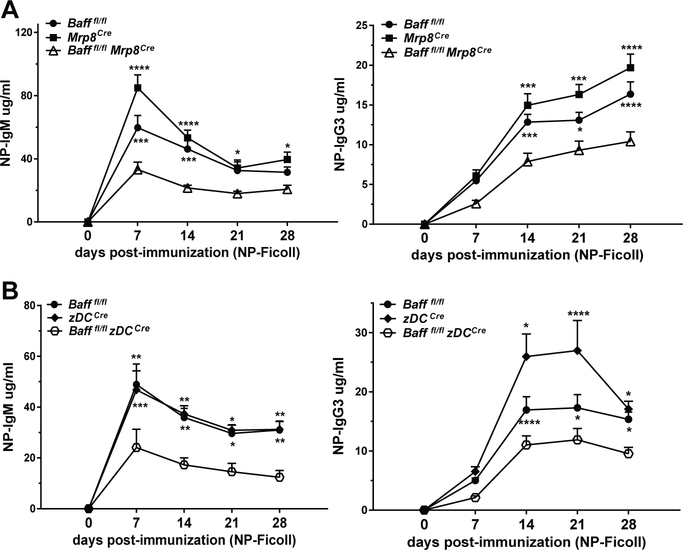
BAFF from Nphs and cDCs contributes to TI-2 IgG Ab responses
BAFF produced by DCs, Nphs or MOs has been reported to contribute to TI-2 Ab responses (25, 32, 33, 57). To examine further whether BAFF from Nphs or cDCs play a role in TI-2 Ab responses, we immunized Bafffl/fl Mrp8Cre mice and Bafffl/fl zDCCre mice with the TI-2 Ag, NP-Ficoll (Fig. 8). Both Bafffl/fl Mrp8Cre mice and Bafffl/fl zDCCre mice had reduced NP-specific IgM and IgG3 production (Fig. 8A, and 8B, respectively). Thus, both Nph and cDC BAFF sources are required for normal Ag-specific Ab responses to TI-2 Ags. Since BAFF depletion in Nph in Bafffl/fl Mrp8Cre mice was selective but not restricted to Nphs, we cannot formally exclude that BAFF from MOs may also contribute to the reduced Ab response to NP-Ficoll. However, Bafffl/fl Cx3cr1Cre mice, which have reduced BAFF expression in MOs, did not have reduced NP-specific Ab responses compared to control mice (Fig S1E).
Figure 8. BAFF from Nphs and cDCs contributes to TI-2 IgG Ab responses.
Bafffl/fl Mrp8Cre mice (A) or Bafffl/fl zDCCre mice (B) were immunized i.p. with 20 μg of NP-Ficoll, bled at the indicated time points, and NP-specific IgM and IgG3 were analyzed by ELISA. A and B, The data presented show means ± SEM of NP-IgM and NP-IgG3 and are combined from three independent experiments for each cKO genotype; A, Bafffl/fl mice, N=17; Mrp8Cre mice, N=7; Bafffl/fl Mrp8Cre mice, N=17. B, Bafffl/fl mice, N=17; zDCCre mice, N=8; Bafffl/fl zDCCre mice, N=15. Statistics were performed using 2-way ANOVA with Tukey’s multiple comparison test. Statistics shown are comparison between Bafffl/fl mice or Cre mice controls vs. BAFF cKO mice; * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
Discussion
BAFF is essential for the host response to pathogens and provides a critical link between innate and adaptive immune responses (1, 58). Although BAFF dysregulation has been implicated in several diseases, studies investigating BAFF producing cells have been limited either to in vitro experiments or used models where BAFF or its receptors were completely blocked or ablated (13, 29, 32, 33, 57, 59). Here we developed and used BAFF-RFP reporter mice and Bafffl/fl mice to better understand BAFF sources and their functions in B cell responses. We found that BAFF is expressed in several blood cell subsets as well as lymphoid tissues, including the spleen and LNs. The major cells expressing BAFF are Nphs, DCs and MO subsets. Furthermore, BAFF expression can undergo cell- and tissue-specific up-regulation upon in vivo activation of different RNA-sensing pathways. In addition, selective deletion of BAFF expression in cDCs or Nphs revealed that BAFF from both cDCs and Nphs contribute to TI-2 Ab responses, while cDC BAFF, unlike Nph BAFF, is required for optimal protective immunity against WNV.
Nphs express the highest BAFF levels in blood and in every tissue we tested, including the spleen, iLNs and BM. These data are consistent with previous reports describing Nphs as a major source of BAFF (21, 60, 61). BAFF production by Nphs has been reported in human autoimmune diseases and mouse models (62–64). However, BAFF-production by Nphs during viral infections has not been previously noted. BAFF expression by Nphs was slightly up-regulated after in vivo challenge with two ssRNA stimuli: R848 and WNV. WNV-induced BAFF increase in Nphs was IFNAR-dependent but MAVS-independent, suggesting that Type-1 IFN production that regulates BAFF in Nphs might be induced through the TLR7/8 pathway. Although, Nph BAFF production increased after viral infection, it is not required for protective immunity to WNV. In contrast, BAFF produced by Nphs is required for protective immunity against Salmonella typhimurium infection (Kuley et al. manuscript in preparation). Further investigation is required to elucidate the possible roles of BAFF produced by Nphs during viral and bacterial infections. Puga et al. (56) previously reported that BAFF produced by human Nphs may contribute to TI Ab responses (57). In support of this study, we demonstrated that selective depletion of BAFF from Nphs reduces Ag-specific Ab responses to a TI-2 Ag. Nphs strategically located in the peri-marginal zone of the spleen may help B cells to rapidly respond to blood-borne TI Ags (57, 65, 66).
Earlier studies also implicated DCs in TI Ab responses, and more recently, several cDC subsets were reported to be localized in the marginal zone (32, 67–69). In agreement with these findings, we found that BAFF from cDCs is required for optimal humoral responses to a TI-2 Ag. The reduction but not full ablation of Ab responses to NP-Ficoll in either BAFF Nph cKO or BAFF cDC cKO mice, suggests that both Nphs and cDCs contribute to TI-2 Ab responses. It is also possible that some BAFF-producing cells compensate for the absence of BAFF-production by another source. Further studies are needed to identify specific niches in lymphoid tissues where BAFF-producing cDCs and Nphs provide help to B cell responses (70, 71).
BAFF plays an important protective role during viral infections (13, 72). In particular, BAFF signaling is an important regulator of humoral responses and required for survival against lethal WNV infection (73). Here we found that cDC BAFF is essential for protection from WNV infection. In contrast to BAFF Nph cKO mice, BAFF cDC cKO mice after WNV infection, had increased mortality and increased virus titers. The reduced WNV-specific IgG and WNV neutralizing Abs in BAFF cDC cKO mice suggest that BAFF from cDCs is essential for the development of protective humoral responses to WNV. How BAFF-producing cells contribute to control WNV replication, whether it is through the regulation of B cells responses or other mechanism needs further investigation. Consistent with the protective role of cDC-derived BAFF, WNV infection up-regulated BAFF expression in CD8− cDCs. WNV induced up-regulation of BAFF in CD8− cDCs was dependent on MAVS, suggesting that this occurred through WNV activation of the RIG-I pathway (36, 55). Since both pDCs and cDCs express the RIG-I/MAVS pathway and can produce Type I IFN upon viral challenge (47, 55, 74), it remains to be determined if BAFF upregulation in cDCs after WNV infection is dependent on Type I IFN produced by cDCs or by pDCs.
The IFNAR-dependent and MAVS-dependent BAFF regulation in inflammatory MO subsets by Poly(I:C) most likely occurs via MDA5 triggered by long dsRNA (36, 55, 75). Our finding that Poly(I:C) increased BAFF levels in splenic inflammatory Ly6Chi MO and Ly6Chi DC subsets is consistent with previous reports. Poly(I:C) and virus-induced Type I IFN production by pDCs upregulates BAFF levels in MOs and myeloid DCs (12, 31, 76). Inflammatory MO/DCs also express TLR3 and produce IFNα in response to Poly(I:C) and dsRNA (74, 77–80). Furthermore, the MAVS signaling pathway has been implicated in type I IFN production by inflammatory MOs and MO-derived DCs (74, 79). Further studies are needed to establish whether the MAVS/IFNAR requirement for Poly(I:C)-induced BAFF up-regulation in inflammatory MO subsets occurs via type I production by pDCs, or in an autocrine fashion.
Using BAFF-RFP mice we identified a murine NK cell subset expressing BAFF induced by the activation of RNA-sensing pathways. The increased expression of CD11b, NKp46 and Ly49C in BAFF+ CD11bhi NK cells, compared to CD11bint/− NK cells, suggests that the BAFF+ CD11bhi NK cells are activated (54, 81). The fact that the expansion of BAFF+ CD11bhi NK cells is dependent on IFNAR suggests type I IFN plays a major role not only in the up-regulation of BAFF expression by NK cells but also in the induction of this activated NK subset. These findings are in agreement with previous reports implicating type I IFN in the activation of NK cells during viral infections (82–85). NK cells play an important role in antiviral immunity not only through the NK-mediated killing of virus infected cells but also by regulating adaptive immune responses (84, 86, 87). NK cells can enhance Ag presentation by B cells and can promote TI Ab responses against bacteria or in autoimmune settings (88–90). Also a human NK subset has been shown to produce BAFF (91). BAFF expression by human NK cells inhibits sensitivity to treatment of chronic lymphoid leukemia, and has been correlated to a SNP associated with susceptibility to autoimmune disease (92, 93). It would be interesting to investigate whether the activated BAFF expressing NK cells we have characterized play a role in regulating B cell responses during infections or during the development of autoimmune diseases.
Remarkably, in the BM not only Nphs, but also some CD11b+ myeloid precursors express high levels of BAFF. Interestingly, Poly(I:C)-dependent BAFF regulation in BM MO subsets was similar to their counterparts in the spleen, and required the MAVS/IFNAR pathway. These BAFF+ myeloid precursors are most likely late precursors since they express CD11b. Among the myeloid precursors expressing higher BAFF levels, the population we define as CD11b+ myeloid precursor lacking Ly6C may be an earlier progenitor of a precursor not committed to a specific granulocyte/monocyte lineage, as described by Yanez et al. (48, 50). Consistent with reports of BM-derived DCs being a source of BAFF (33), we found that BM preDCs expressed substantial BAFF levels. Despite the high BAFF expression in Nph and DC precursors in the BM and in spleens, these cell sources were not required to maintain the general mature B cell compartment. Our results are in agreement with earlier findings suggesting that BAFF produced by radiation-resistant cells, possibly stromal cells, are responsible for normal B cell homeostasis (56),
Interestingly, a subset of CD19+B220+CD11b+ B cells in the BM expressed BAFF; these cells were CD43+loCD5−CD11c−, and some were IgM+ and IgD+. Further analysis is required to define this subset better. They may belong to the B1 B cell lineage because since they express CD11b and BAFF (53, 94) or be a BM B cell subset providing BAFF to BM LLPCs as a survival factor (1, 5, 95). We also found that BAFF is expressed in some peripheral B cell subsets; further studies are required to define the functions of this source of BAFF.
Our new BAFF-RFP reporter and Bafffl/fl mouse lines may be useful for identifying BAFF sources and defining their in vivo functions. Localization of BAFF-RFP positive cells in the spleen and lymph nodes, would also be very helpful to identify specific niches of BAFF producing cells during immune responses. Given the major role of BAFF in the host response to pathogens (58) and in autoimmune diseases, more insights into the localization, timing and function of BAFF-producing cells regulating B cell responses may help in the development of more targeted vaccines and therapies.
Supplementary Material
Key points.
Cell- and tissue-specific type I IFN-dependent BAFF increases in myeloid subsets
Neutrophils and DCs BAFF is required for optimal T-independent 2 humoral responses
BAFF produced by DCs is required for protective immunity to West Nile virus
Acknowledgements
We thank Dr. Keith Elkon for helpful insights and discussion. We thank Michel Black and the Cell and Analysis Flow Cytometry and Imaging Core in the Department of Immunology at the University of Washington for their support. The authors declare they have no competing financial interests.
This work was supported by a grant from the National Institutes of Health (R01AI44257, EA Clark).
Abbreviations
- B6
C57BL/6
- BAFF
B cell activating factor
- BM
bone marrow
- cKO
conditional knockout
- cDC
conventional DC
- DC
dendritic cell
- FO
follicular
- f.p.
footpad
- IFNAR
Type I IFN receptor
- KO
knockout
- cKO
conditional knockout
- LLPC
long-lived plasma cell
- MAVS
mitochondrial activator of virus signaling
- MΦ
macrophage
- MO
monocyte
- MZ
marginal zone
- Nph
neutrophil
- NP
4-hyrody-3-nitrophenacetyl
- Poly(I:C)
polyinosinic:polycytidylic acid
- RIG-I
retinoic acid-inducible gene I
- Tg
transgenic
- WNV
West Nile Virus
- WNVE
West Nile Virus E protein
References
- 1.Mackay F, and Schneider P 2009. Cracking the BAFF code. Nat. Rev. Immunol. 9: 491–502. [DOI] [PubMed] [Google Scholar]
- 2.Rickert RC, Jellusova J, and Miletic AV 2011. Signaling by the tumor necrosis factor receptor superfamily in B-cell biology and disease. Immunol. Rev. 244: 115–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincent FB, Saulep-Easton D, Figgett WA, Fairfax KA, and Mackay F 2013. The BAFF/APRIL system: Emerging functions beyond B cell biology and autoimmunity. Cytokine Growth Factor Rev. 24: 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorelik L, Cutler AH, Thill G, Miklasz SD, Shea DE, Ambrose C, Bixler SA, Su L, Scott ML, and Kalled SL 2004. Cutting Edge: BAFF Regulates CD21/35 and CD23 Expression Independent of Its B Cell Survival Function. J. Immunol. 172: 762–766. [DOI] [PubMed] [Google Scholar]
- 5.Mackay F, Figgett WA, Saulep D, Lepage M, and Hibbs ML 2010. B-cell stage and context-dependent requirements for survival signals from BAFF and the B-cell receptor. Immunol. Rev. 237: 205–225. [DOI] [PubMed] [Google Scholar]
- 6.Smulski CR, and Eibel H 2018. BAFF and BAFF-Receptor in B Cell Selection and Survival. Front. Immunol. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang S, Li J-Y, and Xu W 2014. Role of BAFF/BAFF-R axis in B-cell non-Hodgkin lymphoma. Crit. Rev. Oncol. Hematol. 91: 113–122. [DOI] [PubMed] [Google Scholar]
- 8.Samy E, Wax S, Huard B, Hess H, and Schneider P 2017. Targeting BAFF and APRIL in systemic lupus erythematosus and other antibody-associated diseases. Int. Rev. Immunol. 36: 3–19. [DOI] [PubMed] [Google Scholar]
- 9.Shabgah AG, Shariati-Sarabi Z, Tavakkol-Afshari J, and Mohammadi M 2019. The role of BAFF and APRIL in rheumatoid arthritis. J. Cell. Physiol. 234: 17050–17063. [DOI] [PubMed] [Google Scholar]
- 10.Boneparth A, and Davidson A 2012. B-cell activating factor targeted therapy and lupus. Arthritis Res. Ther. 14: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ittah M, Miceli-Richard C, Lebon P, Pallier C, Lepajolec C, and Mariette X 2011. Induction of B Cell-Activating Factor by Viral Infection Is a General Phenomenon, but the Types of Viruses and Mechanisms Depend on Cell Type. J. Innate Immun. 3: 200–207. [DOI] [PubMed] [Google Scholar]
- 12.Gomez AM, Ouellet M, and Tremblay MJ 2015. HIV-1–Triggered Release of Type I IFN by Plasmacytoid Dendritic Cells Induces BAFF Production in Monocytes. J. Immunol. 194: 2300–2308. [DOI] [PubMed] [Google Scholar]
- 13.Giordano D, Draves KE, Young LB, Roe K, Bryan MA, Dresch C, Richner JM, Diamond MS, M. G. Jr, and Clark EA 2017. Protection of mice deficient in mature B cells from West Nile virus infection by passive and active immunization. PLOS Pathog. 13: e1006743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tarantino G, Marco VD, Petta S, Almasio PL, Barbaria F, Licata A, Bosco GL, Tripodo C, Stefano RD, and Craxì A 2009. Serum BLyS/BAFF predicts the outcome of acute hepatitis C virus infection. J. Viral Hepat. 16: 397–405. [DOI] [PubMed] [Google Scholar]
- 15.Alturaiki W, McFarlane AJ, Rose K, Corkhill R, McNamara PS, Schwarze J, and Flanagan BF 2018. Expression of the B cell differentiation factor BAFF and chemokine CXCL13 in a murine model of Respiratory Syncytial Virus infection. Cytokine 110: 267–271. [DOI] [PubMed] [Google Scholar]
- 16.Borhis G, Trovato M, Chaoul N, Ibrahim HM, and Richard Y 2017. B-Cell-Activating Factor and the B-Cell Compartment in HIV/SIV Infection. Front. Immunol. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uzzan M, Colombel J-F, Cerutti A, Treton X, and Mehandru S 2016. B Cell-Activating Factor (BAFF)-Targeted B Cell Therapies in Inflammatory Bowel Diseases. Dig. Dis. Sci. 61: 3407–3424. [DOI] [PubMed] [Google Scholar]
- 18.Kato A, and Schleimer RP 2007. Beyond inflammation: airway epithelial cells are at the interface of innate and adaptive immunity. Curr. Opin. Immunol. 19: 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferraccioli G, and Gremese E 2017. B cell activating factor (BAFF) and BAFF receptors: fakes and facts. Clin. Exp. Immunol. 190: 291–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou B, Zhang H, Su X, Luo Y, Li X, Yu C, Xie Q, Xia X, He G, and Yang L 2019. Therapeutic effects of a novel BAFF blocker on arthritis. Signal Transduct. Target. Ther. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scapini P, Bazzoni F, and Cassatella MA 2008. Regulation of B-cell-activating factor (BAFF)/B lymphocyte stimulator (BLyS) expression in human neutrophils. Immunol. Lett. 116: 1–6. [DOI] [PubMed] [Google Scholar]
- 22.Ittah M, Miceli-Richard C, Gottenberg J-E, Sellam J, Eid P, Lebon P, Pallier C, Lepajolec C, and Mariette X 2008. Viruses induce high expression of BAFF by salivary gland epithelial cells through TLR- and type-I IFN-dependent and -independent pathways. Eur. J. Immunol. 38: 1058–1064. [DOI] [PubMed] [Google Scholar]
- 23.Bombardieri M, Kam N-W, Brentano F, Choi K, Filer A, Kyburz D, McInnes IB, Gay S, Buckley C, and Pitzalis C 2011. A BAFF/APRIL-dependent TLR3-stimulated pathway enhances the capacity of rheumatoid synovial fibroblasts to induce AID expression and Ig class-switching in B cells. Ann. Rheum. Dis. 70: 1857–1865. [DOI] [PubMed] [Google Scholar]
- 24.Cerutti A, Puga I, and Cols M 2011. Innate control of B cell responses. Trends Immunol. 32: 202–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craxton A, Magaletti D, Ryan EJ, and Clark EA 2003. Macrophage- and dendritic cell—dependent regulation of human B-cell proliferation requires the TNF family ligand BAFF. Blood 101: 4464–4471. [DOI] [PubMed] [Google Scholar]
- 26.Kim H-A, Jeon S-H, Seo G-Y, Park J-B, and Kim P-H 2008. TGF-β1 and IFN-γ stimulate mouse macrophages to express BAFF via different signaling pathways. J. Leukoc. Biol. 83: 1431–1439. [DOI] [PubMed] [Google Scholar]
- 27.Woo S-J, Im J, Jeon JH, Kang S-S, Lee M-H, Yun C-H, Moon E-Y, Song MK, Kim H-H, and Han SH 2013. Induction of BAFF expression by IFN-γ via JAK/STAT signaling pathways in human intestinal epithelial cells. J. Leukoc. Biol. 93: 363–368. [DOI] [PubMed] [Google Scholar]
- 28.Ohata J, Zvaifler NJ, Nishio M, Boyle DL, Kalled SL, Carson DA, and Kipps TJ 2005. Fibroblast-Like Synoviocytes of Mesenchymal Origin Express Functional B Cell-Activating Factor of the TNF Family in Response to Proinflammatory Cytokines. J. Immunol. 174: 864–870. [DOI] [PubMed] [Google Scholar]
- 29.Litinskiy MB, Nardelli B, Hilbert DM, He B, Schaffer A, Casali P, and Cerutti A 2002. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat. Immunol. 3: 822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He B, Qiao X, and Cerutti A 2004. CpG DNA Induces IgG Class Switch DNA Recombination by Activating Human B Cells through an Innate Pathway That Requires TLR9 and Cooperates with IL-10. J. Immunol. 173: 4479–4491. [DOI] [PubMed] [Google Scholar]
- 31.Cerutti A, Qiao X, and He B 2005. Plasmacytoid dendritic cells and the regulation of immunoglobulin heavy chain class switching. Immunol. Cell Biol. 83: 554–562. [DOI] [PubMed] [Google Scholar]
- 32.Balázs M, Martin F, Zhou T, and Kearney JF 2002. Blood Dendritic Cells Interact with Splenic Marginal Zone B Cells to Initiate T-Independent Immune Responses. Immunity 17: 341–352. [DOI] [PubMed] [Google Scholar]
- 33.Giordano D, Draves KE, Li C, Hohl TM, and Clark EA 2014. Nitric Oxide Regulates BAFF Expression and T Cell–Independent Antibody Responses. J. Immunol. 193: 1110–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Bülow G-U, van Deursen JM, and Bram RJ 2001. Regulation of the T-Independent Humoral Response by TACI. Immunity 14: 573–582. [DOI] [PubMed] [Google Scholar]
- 35.Lindh E, Lind SM, Lindmark E, Hässler S, Perheentupa J, Peltonen L, Winqvist O, and Karlsson MCI 2008. AIRE regulates T-cell-independent B-cell responses through BAFF. Proc. Natl. Acad. Sci. 105: 18466–18471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chow KT, Gale M, and Loo Y-M 2018. RIG-I and Other RNA Sensors in Antiviral Immunity. Annu. Rev. Immunol. 36: 667–694. [DOI] [PubMed] [Google Scholar]
- 37.He L, Peng X, Wang J, Tang C, Zhou X, Liu H, Liu F, Sun L, and Peng Y 2015. Synthetic Double-Stranded RNA Poly(I:C) Aggravates IgA Nephropathy by Triggering IgA Class Switching Recombination through the TLR3-BAFF Axis. Am. J. Nephrol. 42: 185–197. [DOI] [PubMed] [Google Scholar]
- 38.Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, and Akira S 2006. Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med. 203: 1795–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, and Tsien RY 2008. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 5: 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Passegué E, Wagner EF, and Weissman IL 2004. JunB Deficiency Leads to a Myeloproliferative Disorder Arising from Hematopoietic Stem Cells. Cell 119: 431–443. [DOI] [PubMed] [Google Scholar]
- 41.Abram CL, Roberge GL, Hu Y, and Lowell CA 2014. Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. J. Immunol. Methods 408: 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loschko J, Schreiber HA, Rieke GJ, Esterházy D, Meredith MM, Pedicord VA, Yao K-H, Caballero S, Pamer EG, Mucida D, and Nussenzweig MC 2016. Absence of MHC class II on cDCs results in microbial-dependent intestinal inflammation. J. Exp. Med. 213: 517–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loschko J, Rieke GJ, Schreiber HA, Meredith MM, Yao K-H, Guermonprez P, and Nussenzweig MC 2016. Inducible targeting of cDCs and their subsets in vivo. J. Immunol. Methods 434: 32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu F, and Whitton JL 2005. Cutting Edge: Re-evaluating the In Vivo Cytokine Responses of CD8+ T Cells during Primary and Secondary Viral Infections. J. Immunol. 174: 5936–5940. [DOI] [PubMed] [Google Scholar]
- 45.Hosking MP, Flynn CT, and Whitton JL 2014. Antigen-Specific Naive CD8+ T Cells Produce a Single Pulse of IFN-γ In Vivo within Hours of Infection, but without Antiviral Effect. J. Immunol. 193: 1873–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giordano D, Li C, Suthar MS, Draves KE, Ma DY, Gale M, and Clark EA 2011. Nitric oxide controls an inflammatory-like Ly6ChiPDCA1+ DC subset that regulates Th1 immune responses. J. Leukoc. Biol. 89: 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roe K, Giordano D, Young LB, Draves KE, Holder U, Suthar MS, M. G. Jr, and Clark EA 2019. Dendritic cell-associated MAVS is required to control West Nile virus replication and ensuing humoral immune responses. PLOS ONE 14: e0218928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Auffray C, Sieweke MH, and Geissmann F 2009. Blood Monocytes: Development, Heterogeneity, and Relationship with Dendritic Cells. Annu. Rev. Immunol. 27: 669–692. [DOI] [PubMed] [Google Scholar]
- 49.Auffray C, Fogg DK, Narni-Mancinelli E, Senechal B, Trouillet C, Saederup N, Leemput J, Bigot K, Campisi L, Abitbol M, Molina T, Charo I, Hume DA, Cumano A, Lauvau G, and Geissmann F 2009. CX3CR1+ CD115+ CD135+ common macrophage/DC precursors and the role of CX3CR1 in their response to inflammation. J. Exp. Med. 206: 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yáñez A, Ng MY, Hassanzadeh-Kiabi N, and Goodridge HS 2015. IRF8 acts in lineage-committed rather than oligopotent progenitors to control neutrophil vs monocyte production. Blood 125: 1452–1459. [DOI] [PubMed] [Google Scholar]
- 51.Roe K, Shu GL, Draves KE, Giordano D, Pepper M, and Clark EA 2019. Targeting Antigens to CD180 but Not CD40 Programs Immature and Mature B Cell Subsets to Become Efficient APCs. J. Immunol. ji1900549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hardy RR, and Shinton SA 2004. Characterization of B Lymphopoiesis in Mouse Bone Marrow and Spleen In B Cell Protocols. Methods in Molecular Biology Gu H, and Rajewsky K, eds. Humana Press, Totowa, NJ: 1–24. [DOI] [PubMed] [Google Scholar]
- 53.Chu VT, Enghard P, Riemekasten G, and Berek C 2007. In Vitro and In Vivo Activation Induces BAFF and APRIL Expression in B Cells. J. Immunol. 179: 5947–5957. [DOI] [PubMed] [Google Scholar]
- 54.Annunziato F, Romagnani C, and Romagnani S 2015. The 3 major types of innate and adaptive cell-mediated effector immunity. J. Allergy Clin. Immunol. 135: 626–635. [DOI] [PubMed] [Google Scholar]
- 55.Szabo A, and Rajnavolgyi E 2013. Collaboration of Toll-like and RIG-I-like receptors in human dendritic cells: tRIGgering antiviral innate immune responses. Am. J. Clin. Exp. Immunol. 2: 195–207. [PMC free article] [PubMed] [Google Scholar]
- 56.Gorelik L, Gilbride K, Dobles M, Kalled SL, Zandman D, and Scott ML 2003. Normal B Cell Homeostasis Requires B Cell Activation Factor Production by Radiation-resistant Cells. J. Exp. Med. 198: 937–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puga I, Cols M, Barra CM, He B, Cassis L, Gentile M, Comerma L, Chorny A, Shan M, Xu W, Magri G, Knowles DM, Tam W, Chiu A, Bussel JB, Serrano S, Lorente JA, Bellosillo B, Lloreta J, Juanpere N, Alameda F, Baró T, de Heredia CD, Torán N, Català A, Torrebadell M, Fortuny C, Cusí V, Carreras C, Diaz GA, Blander JM, Farber C-M, Silvestri G, Cunningham-Rundles C, Calvillo M, Dufour C, Notarangelo LD, Lougaris V, Plebani A, Casanova J-L, Ganal SC, Diefenbach A, Aróstegui JI, Juan M, Yagüe J, Mahlaoui N, Donadieu J, Chen K, and Cerutti A 2012. B cell–helper neutrophils stimulate the diversification and production of immunoglobulin in the marginal zone of the spleen. Nat. Immunol. 13: 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakai J, and Akkoyunlu M 2017. The Role of BAFF System Molecules in Host Response to Pathogens. Clin. Microbiol. Rev. 30: 991–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nardelli B, Belvedere O, Roschke V, Moore PA, Olsen HS, Migone TS, Sosnovtseva S, Carrell JA, Feng P, Giri JG, and Hilbert DM 2001. Synthesis and release of B-lymphocyte stimulator from myeloid cells. Blood 97: 198–204. [DOI] [PubMed] [Google Scholar]
- 60.Scapini P, Nardelli B, Nadali G, Calzetti F, Pizzolo G, Montecucco C, and Cassatella MA 2003. G-CSF–stimulated Neutrophils Are a Prominent Source of Functional BLyS. J. Exp. Med. 197: 297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scapini P, Carletto A, Nardelli B, Calzetti F, Roschke V, Merigo F, Tamassia N, Pieropan S, Biasi D, Sbarbati A, Sozzani S, Bambara L, and Cassatella MA 2005. Proinflammatory mediators elicit secretion of the intracellular B-lymphocyte stimulator pool (BLyS) that is stored in activated neutrophils: implications for inflammatory diseases. Blood 105: 830–837. [DOI] [PubMed] [Google Scholar]
- 62.Assi LK, Wong SH, Ludwig A, Raza K, Gordon C, Salmon M, Lord JM, and Scheel-Toellner D 2007. Tumor necrosis factor α activates release of B lymphocyte stimulator by neutrophils infiltrating the rheumatoid joint. Arthritis Rheum. 56: 1776–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coquery CM, Wade NS, Loo WM, Kinchen JM, Cox KM, Jiang C, Tung KS, and Erickson LD 2014. Neutrophils Contribute to Excess Serum BAFF Levels and Promote CD4+ T Cell and B Cell Responses in Lupus-Prone Mice. PLOS ONE 9: e102284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Németh T, Mócsai A, and Lowell CA 2016. Neutrophils in animal models of autoimmune disease. Semin. Immunol. 28: 174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deniset JF, Surewaard BG, Lee W-Y, and Kubes P 2017. Splenic Ly6Ghigh mature and Ly6Gint immature neutrophils contribute to eradication of S. pneumoniae. J. Exp. Med. 214: 1333–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cerutti A, Cols M, and Puga I 2013. Marginal zone B cells: virtues of innate-like antibody-producing lymphocytes. Nat. Rev. Immunol. 13: 118–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caton ML, Smith-Raska MR, and Reizis B 2007. Notch–RBP-J signaling controls the homeostasis of CD8− dendritic cells in the spleen. J. Exp. Med. 204: 1653–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qiu C-H, Miyake Y, Kaise H, Kitamura H, Ohara O, and Tanaka M 2009. Novel Subset of CD8α+ Dendritic Cells Localized in the Marginal Zone Is Responsible for Tolerance to Cell-Associated Antigens. J. Immunol. 182: 4127–4136. [DOI] [PubMed] [Google Scholar]
- 69.Chappell CP, Draves KE, Giltiay NV, and Clark EA 2012. Extrafollicular B cell activation by marginal zone dendritic cells drives T cell–dependent antibody responses. J. Exp. Med. 209: 1825–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cerutti A, Puga I, and Cols M 2012. New helping friends for B cells. Eur. J. Immunol. 42: 1956–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vinuesa CG, and Chang P-P 2013. Innate B cell helpers reveal novel types of antibody responses. Nat. Immunol. 14: 119–126. [DOI] [PubMed] [Google Scholar]
- 72.Xu HC, Huang J, Khairnar V, Duhan V, Pandyra AA, Grusdat M, Shinde P, McIlwain DR, Maney SK, Gommerman J, Löhning M, Ohashi PS, Mak TW, Pieper K, Sic H, Speletas M, Eibel H, Ware CF, Tumanov AV, Kruglov AA, Nedospasov SA, Häussinger D, Recher M, Lang KS, and Lang PA 2015. Deficiency of the B Cell-Activating Factor Receptor Results in Limited CD169+ Macrophage Function during Viral Infection. J. Virol. 89: 4748–4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Samuel MA, and Diamond MS 2006. Pathogenesis of West Nile Virus Infection: a Balance between Virulence, Innate and Adaptive Immunity, and Viral Evasion. J. Virol. 80: 9349–9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Silva MC, Guerrero-Plata A, Gilfoy FD, Garofalo RP, and Mason PW 2007. Differential Activation of Human Monocyte-Derived and Plasmacytoid Dendritic Cells by West Nile Virus Generated in Different Host Cells. J. Virol. 81: 13640–13648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, and Akira S 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid–inducible gene-I and melanoma differentiation–associated gene 5. J. Exp. Med. 205: 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Asselin-Paturel C, and Trinchieri G 2005. Production of type I interferons: plasmacytoid dendritic cells and beyond. J. Exp. Med. 202: 461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsumoto M, Funami K, Tanabe M, Oshiumi H, Shingai M, Seto Y, Yamamoto A, and Seya T 2003. Subcellular Localization of Toll-Like Receptor 3 in Human Dendritic Cells. J. Immunol. 171: 3154–3162. [DOI] [PubMed] [Google Scholar]
- 78.Kadowaki N, Antonenko S, and Liu Y-J 2001. Distinct CpG DNA and Polyinosinic-Polycytidylic Acid Double-Stranded RNA, Respectively, Stimulate CD11c− Type 2 Dendritic Cell Precursors and CD11c+ Dendritic Cells to Produce Type I IFN. J. Immunol. 166: 2291–2295. [DOI] [PubMed] [Google Scholar]
- 79.Haist KC, Burrack KS, Davenport BJ, and Morrison TE 2017. Inflammatory monocytes mediate control of acute alphavirus infection in mice. PLOS Pathog. 13: e1006748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boyette LB, Macedo C, Hadi K, Elinoff BD, Walters JT, Ramaswami B, Chalasani G, Taboas JM, Lakkis FG, and Metes DM 2017. Phenotype, function, and differentiation potential of human monocyte subsets. PLOS ONE 12: e0176460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vivier E, Tomasello E, Baratin M, Walzer T, and Ugolini S 2008. Functions of natural killer cells. Nat. Immunol. 9: 503–510. [DOI] [PubMed] [Google Scholar]
- 82.Martinez J, Huang X, and Yang Y 2008. Direct Action of Type I IFN on NK Cells Is Required for Their Activation in Response to Vaccinia Viral Infection In Vivo. J. Immunol. 180: 1592–1597. [DOI] [PubMed] [Google Scholar]
- 83.Zhu J, Huang X, and Yang Y 2008. A Critical Role for Type I IFN–dependent NK Cell Activation in Innate Immune Elimination of Adenoviral Vectors In Vivo. Mol. Ther. 16: 1300–1307. [DOI] [PubMed] [Google Scholar]
- 84.Madera S, Rapp M, Firth MA, Beilke JN, Lanier LL, and Sun JC 2016. Type I IFN promotes NK cell expansion during viral infection by protecting NK cells against fratricide. J. Exp. Med. 213: 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Paolini R, Bernardini G, Molfetta R, and Santoni A 2015. NK cells and interferons. Cytokine Growth Factor Rev. 26: 113–120. [DOI] [PubMed] [Google Scholar]
- 86.Waggoner SN, Reighard SD, Gyurova IE, Cranert SA, Mahl SE, Karmele EP, McNally JP, Moran MT, Brooks TR, Yaqoob F, and Rydyznski CE 2016. Roles of natural killer cells in antiviral immunity. Curr. Opin. Virol. 16: 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ali A, Gyurova IE, and Waggoner SN 2019. Mutually assured destruction: the cold war between viruses and natural killer cells. Curr. Opin. Virol. 34: 130–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jennings P, and Yuan D 2009. NK Cell Enhancement of Antigen Presentation by B Lymphocytes. J. Immunol. 182: 2879–2887. [DOI] [PubMed] [Google Scholar]
- 89.Gao N, Jennings P, Guo Y, and Yuan D 2011. Regulatory role of natural killer (NK) cells on antibody responses to Brucella abortus. Innate Immun. 17: 152–163. [DOI] [PubMed] [Google Scholar]
- 90.Yuan D, Thet S, Zhou XJ, Wakeland EK, and Dang T 2011. The role of NK cells in the development of autoantibodies. Autoimmunity 44: 641–651. [DOI] [PubMed] [Google Scholar]
- 91.Cella M, Otero K, and Colonna M 2010. Expansion of human NK-22 cells with IL-7, IL-2, and IL-1β reveals intrinsic functional plasticity. Proc. Natl. Acad. Sci. 107: 10961–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Almeida E. R. A. de, and Petzl-Erler ML 2013. Expression of genes involved in susceptibility to multifactorial autoimmune diseases: estimating genotype effects. Int. J. Immunogenet. 40: 178–185. [DOI] [PubMed] [Google Scholar]
- 93.Wild J, Schmiedel BJ, Maurer A, Raab S, Prokop L, Stevanović S, Dörfel D, Schneider P, and Salih HR 2015. Neutralization of (NK-cell-derived) B-cell activating factor by Belimumab restores sensitivity of chronic lymphoid leukemia cells to direct and Rituximab-induced NK lysis. Leukemia 29: 1676–1683. [DOI] [PubMed] [Google Scholar]
- 94.Esplin BL, Welner RS, Zhang Q, Borghesi LA, and Kincade PW 2009. A differentiation pathway for B1 cells in adult bone marrow. Proc. Natl. Acad. Sci. 106: 5773–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thai L-H, Gallou SL, Robbins A, Crickx E, Fadeev T, Zhou Z, Cagnard N, Mégret J, Bole C, Weill J-C, Reynaud C-A, and Mahévas M 2018. BAFF and CD4+ T cells are major survival factors for long-lived splenic plasma cells in a B-cell–depletion context. Blood 131: 1545–1555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.