StpA is normally considered a molecular backup of the nucleoid-structuring protein H-NS, which was reported as a transcriptional repressor of the type I-E CRISPR-Cas system in Escherichia coli. However, the role of StpA in regulating the type I-E CRISPR-Cas system remains elusive. Our previous work uncovered a new route for double-stranded DNA (dsDNA) entry during natural transformation of E. coli. In this study, we show that StpA plays a role opposite to that of its paralogue H-NS in regulating the type I-E CRISPR-Cas system against natural transformation of E. coli. Our work not only expands our knowledge on CRISPR-Cas-mediated adaptive immunity against extracellular nucleic acids but also sheds new light on understanding the complex regulation mechanism of the CRISPR-Cas system. Moreover, the finding that paralogues StpA and H-NS share a DNA binding site but play opposite roles in transcriptional regulation indicates that higher-order compaction of bacterial chromatin by histone-like proteins could switch prokaryotic transcriptional modes.
KEYWORDS: CRISPR-Cas system, Escherichia coli, histone-like proteins, natural transformation, transcriptional regulation
ABSTRACT
Working mechanisms of CRISPR-Cas systems have been intensively studied. However, far less is known about how they are regulated. The histone-like nucleoid-structuring protein H-NS binds the promoter of cas genes (Pcas) and suppresses the type I-E CRISPR-Cas system in Escherichia coli. Although the H-NS paralogue StpA also binds Pcas, its role in regulating the CRISPR-Cas system remains unidentified. Our previous work established that E. coli is able to take up double-stranded DNA during natural transformation. Here, we investigated the function of StpA in regulating the type I-E CRISPR-Cas system against natural transformation of E. coli. We first documented that although the activated type I-E CRISPR-Cas system, due to hns deletion, interfered with CRISPR-Cas-targeted plasmid transfer, stpA inactivation restored the level of natural transformation. Second, we showed that inactivating stpA reduced the transcriptional activity of Pcas. Third, by comparing transcriptional activities of the intact Pcas and the Pcas with a disrupted H-NS binding site in the hns and hns stpA null deletion mutants, we demonstrated that StpA activated transcription of cas genes by binding to the same site as H-NS in Pcas. Fourth, by expressing StpA with an arabinose-inducible promoter, we confirmed that StpA expressed at a low level stimulated the activity of Pcas. Finally, by quantifying the level of mature CRISPR RNA (crRNA), we demonstrated that StpA was able to promote the amount of crRNA. Taken together, our work establishes that StpA serves as a transcriptional activator in regulating the type I-E CRISPR-Cas system against natural transformation of E. coli.
IMPORTANCE StpA is normally considered a molecular backup of the nucleoid-structuring protein H-NS, which was reported as a transcriptional repressor of the type I-E CRISPR-Cas system in Escherichia coli. However, the role of StpA in regulating the type I-E CRISPR-Cas system remains elusive. Our previous work uncovered a new route for double-stranded DNA (dsDNA) entry during natural transformation of E. coli. In this study, we show that StpA plays a role opposite to that of its paralogue H-NS in regulating the type I-E CRISPR-Cas system against natural transformation of E. coli. Our work not only expands our knowledge on CRISPR-Cas-mediated adaptive immunity against extracellular nucleic acids but also sheds new light on understanding the complex regulation mechanism of the CRISPR-Cas system. Moreover, the finding that paralogues StpA and H-NS share a DNA binding site but play opposite roles in transcriptional regulation indicates that higher-order compaction of bacterial chromatin by histone-like proteins could switch prokaryotic transcriptional modes.
INTRODUCTION
To cope with environmental stresses, bacteria and archaea acquire exogenous genes from genomes and plasmids of other strains via horizontal gene transfer (HGT) (1–3). Simultaneously, prokaryotes are challenged by invasion of harmful genes. To reduce the risk of acquisition of exogenous DNA, prokaryotes have evolved both innate immunity that is mediated by restriction-modification systems (4) and adaptive immunity that is mediated by CRISPR-Cas systems (5–13). The CRISPR-Cas system, consisting of clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (cas) genes, is widespread in genomes of prokaryotes (5, 6, 14–17). According to the organization of effector modules, CRISPR-Cas systems are grouped into two classes, and each class is further divided into three types (5, 18). Class 1 CRISPR-Cas systems (including types I, III, and IV) are considered the ancestral system, whereas class II CRISPR-Cas systems (including types II, V, and VI) are most widely applied in genome editing of a wide range of organisms (6). In general, the CRISPR-Cas-mediated adaptive immunity involves three processes (adaptation, expression, and interference) (6). Two conserved proteins, Cas1 and Cas2, mediate the adaptation process during which new spacers are formed in the CRISPR region near the leader sequence (19–21). In the expression process, a group of Cas proteins are produced, accompanied by transcription of the CRISPR array into the precursor CRISPR RNA (pre-crRNA), which is subsequently processed into small mature crRNA by CasE or RNase III (10, 22). The interference process involves crRNA-directed cleavage of the targeted DNA by Cas nucleases within the ribonucleoprotein complex composed of Cas proteins and crRNA (12, 14, 23–27).
Escherichia coli K-12 is equipped with a type I-E CRISPR-Cas immune system that consists of eight cas genes (cas1, cas2, cas3, and casABCDE) and two CRISPR arrays (CRISPR I and CRISPR II) (10, 16, 28). Genes casABCDE encode Cas proteins that form the Cascade complex. The Cascade-Cas3 complex and the RecBCD nuclease generate single-stranded DNAs (ssDNAs) that are used for naive and primed adaptation (29–32). Protospacer-adjacent motif (PAM)-containing ssDNA strands are captured by Cas1 and Cas2, which then anneal complementary strands (20, 21, 33, 34). Prespacer precursors are trimmed for integration, with the mature PAM-derived end being integrated at the spacer side of the CRISPR, followed by duplicating repeats as a result of filling of gaps (35, 36). The CRISPR array is transcribed into pre-crRNA and further processed into mature crRNA by CasE (10). Together with crRNA, CasABCDE form a CRISPR ribonucleoprotein (crRNP) complex which can be directed to the targeted DNA region (10, 37, 38). The conformational changes in the Cascade complex lock the R loop, which is formed by hybridization of the crRNA and the target strand of the double-stranded DNA (dsDNA), leading to the nontarget strand being replaced (39–42). Then, the nuclease Cas3, which is stabilized by the heat shock protein 90 homologue HtpG (43), is recruited to the target strand to cleave DNA (39, 44).
The working mechanisms of CRISPR-Cas systems have been intensively studied (6–15, 19–27, 29–44) whereas less is known about the mechanisms regulating them (45). Normally, the type I-E CRISPR-Cas system of E. coli is suppressed (28, 46). It can be activated under certain physiological conditions (i.e., in medium supplemented with glucose) and confer immunity to bacteriophage P1 (47). Expression of the type I-E CRISPR-Cas system is regulated by multiple regulators in E. coli and Salmonella enterica (28, 46–48). H-NS, a histone-like nucleoid-structuring protein that inhibits gene transcription, represses expression of cas genes by binding to the upstream region of the promoter of the casABCDE operon (Pcas) (28). It also reduces the amount of crRNA through inhibiting transcription of the CRISPR array and limiting pre-crRNA processing due to insufficient CasE (28). H-NS also partially suppresses transcription of cas3 (49), which encodes the nuclease for DNA cleavage (50, 51). Temperature of incubation is important to maintain a sufficient amount of Cas3 for DNA interference. Incubating the hns mutant at 30°C improves the immunity conferred by the CRISPR-Cas system against λ phage infection (49). Overexpression of LeuO, the LysR-type regulator, can activate transcription of casABCDE through antagonizing H-NS-mediated transcriptional repression and increase the amount of crRNA (46). The carbon catabolism regulator cAMP-cAMP receptor protein (cAMP-CRP) complex negatively regulates type I-E CRISPR-Cas system by competing with LeuO for the binding site of Pcas (47) whereas CRP plays an opposite role in Pectobacterium atrosepticum (52). It activates the type I-F CRISPR-Cas system in that species.
The H-NS paralogue StpA, considered a backup for H-NS (53), can fill the role of H-NS in repressing transcription of genes when H-NS is absent (54, 55). At the transcriptional level, H-NS and StpA can suppress expression of each other (54). In some cases, StpA and H-NS can serve as transcriptional activators and stimulate transcription of the maltose regulon and crp via the stringent response regulator ppGpp (56, 57). Normally, H-NS and StpA bind to the same region of promoters, and StpA has a 4- to 6-fold-greater affinity for DNA than H-NS (58).
As a main driving force for evolution, natural transformation has been found widespread in prokaryotes (59). In general, during natural bacterial transformation, a single-stranded DNA (ssDNA) from extracellular dsDNA is pulled into the cytoplasm via a highly conserved DNA uptake machinery that is assembled in the cell membrane (59–61). CRISPR-Cas systems limit ssDNA uptake during natural transformation of both Streptococcus pneumoniae and Neisseria meningitidis (62, 63). Our previous work has shown that stationary-phase E. coli is able to develop natural competence and allows dsDNA to enter the cytoplasm (64–67). Natural transformation of E. coli is regulated by the transcriptional regulators RpoS and cAMP-CRP complex (66, 68). Although CRISPR-Cas-mediated immunity to bacteriophage infection has been well documented in E. coli (10, 47), the potential effect of the CRISPR-Cas system on limiting dsDNA uptake during natural transformation has not yet been investigated. It has been demonstrated that StpA is able to bind the promoter of the cas operon in E. coli (28). However, the role of StpA in regulating expression of the cas genes remains unknown. In this study, we explored the potential role of StpA in regulating the type I-E CRISPR-Cas system of E. coli and CRISPR-Cas-mediated immunity to dsDNA transfer during natural transformation. Our data showed that StpA played a role opposite to that of H-NS: it served as an activator in regulating the type I-E CRISPR-Cas system and enhanced immunity against DNA transfer during natural transformation of E. coli.
RESULTS
StpA is required for the immunity against plasmids that is conferred by the CRISPR-Cas system in an hns null mutant.
H-NS suppresses expression of the type I-E CRISPR-Cas system in E. coli (28). Although the H-NS paralogue StpA was shown to bind the promoter of the cas operon (28), its role in regulating the type I-E CRISPR-Cas system remains unexplored. We attempted to characterize the role of StpA in regulating the type I-E CRISPR-Cas system against natural transformation in E. coli. To this end, we constructed stpA null mutants in hns-deficient and hns+ E. coli strains. By using λ-Red-mediated recombineering, hns and stpA single- and double-deletion mutants were constructed (see Fig. S1 in the supplemental material). Construction and genotype examination of these mutants are shown in Fig. S1 and S2. A Western blot assay showed that StpA was indeed absent in the hns stpA null mutant (Fig. S1D). Morphologies of constructed strains were observed under a phase-contrast microscope. Similar to a previous observation (69), both hns and hns stpA null mutants were obviously longer than their wild-type (WT) parent and the stpA null mutant (Fig. S2). The hns stpA null mutant grew remarkably more slowly than the hns null mutant (Fig. S8A), in line with previous reports (54, 70).
To evaluate the DNA interference by the type I-E CRISPR-Cas system, we constructed the plasmid pCR1, which can be targeted by the CRISPR-Cas system of E. coli. To ensure the sensitivity of pCR1 to the CRISPR-Cas system, the DNA fragment (CR1) with four protospacer-adjacent motif (PAM)-containing DNA regions was chemically synthesized and cloned into pUC57 (Fig. S3). DNA interference by the type I-E CRISPR-Cas system was evaluated by levels of natural transformation (Fig. 1) and by the viability of transformants after transformation (Fig. S9). Natural transformation was performed by spreading the mixture of plasmid and the cell culture onto an LB plate containing 5% (wt/vol) agar (Fig. 1A). Levels of natural transformation were compared with those of the CRISPR-Cas-targeted plasmid (pCR1) and nontargeted plasmid (pDsRED) carrying a red fluorescence gene whose sequence cannot be recognized by the CRISPR-Cas system in E. coli. Both pCR1 (GenBank accession number MT437282 and Addgene plasmid repository catalog no. 154270 [https://www.addgene.org/154270/]) and pDsRED are derivatives of pUC19 (Table 1), and their sequences are available in the supplemental material. With pDsRED as the donor plasmid, transformation efficiencies of the stpA and hns null mutants and their WT parent were similar (∼100 CFU/μg) (Fig. 1B). In contrast, with pCR1 as the donor plasmid, although the transformation efficiency was not significantly reduced in the stpA null mutant with respect to that of the WT strain, it was reduced by more than 200-fold in the hns null mutant (Fig. 1B). The results demonstrate that activation of the type I-E CRISPR-Cas system in the hns null mutant strongly inhibits plasmid transfer during natural transformation. We observed that transformation of the hns stpA null mutant with pCR1 as the donor plasmid reduced the level of transformation by less than 3-fold (Fig. 1B). Although stpA inactivation restored transformation in the hns null mutant, the level of transformation in the hns stpA null mutant was still lower than that in the WT strain, indicating that StpA is partially responsible for activating CRISPR-Cas-mediated DNA interference in the hns mutant.
FIG 1.
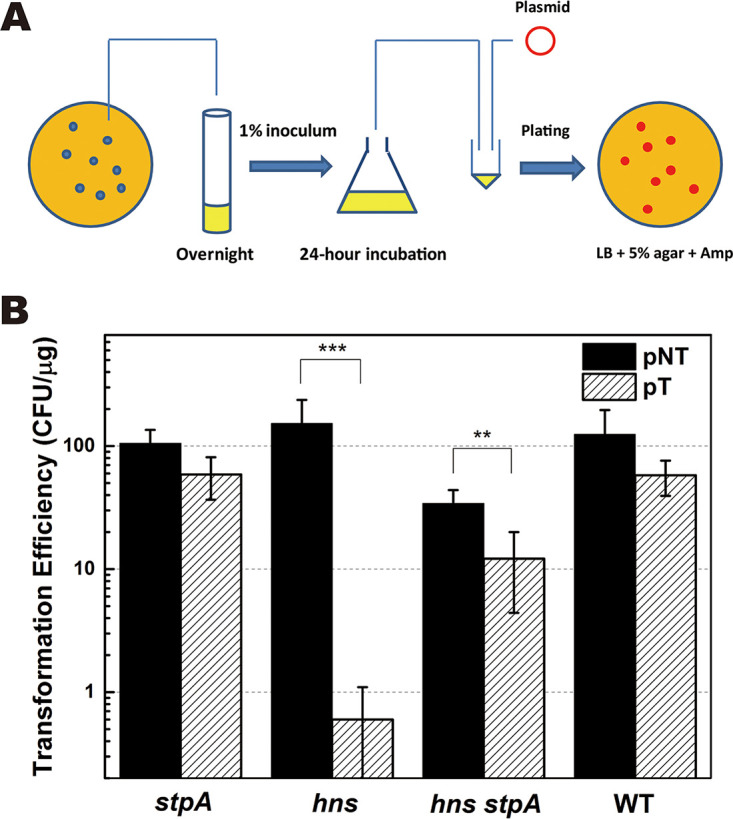
Effect of stpA inactivation on natural transformation with CRISPR-Cas-targeted plasmid. (A) Schematic representation of natural transformation of E. coli for evaluating the CRISPR-Cas-mediated immunity against extracellular DNA. (B) Natural transformation was performed by spreading the mixture of the 24-h-incubated E. coli culture and plasmid onto LB plates containing 5% (wt/vol) agar supplemented with ampicillin (100 μg ml−1). Levels of natural transformation with the CRISPR-Cas-targeted plasmid pCR1 (pT) or nontargeted plasmid pDsRED (pNT) were measured in Δhns, ΔstpA, Δhns ΔstpA, and WT strains. Cell viability of each transformant was measured (see Fig. S9 in the supplemental material). Data are shown as means ± standard deviations (n = 3). Statistical significance was determined using a two-tailed Student's t test (**, P ≤ 0.01; ***, P ≤ 0.005).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| MC4100 | F− λ− araD139 Δ(argF-lac)U169 rpsL150 relA deoC1 ptsF25 rbsR flbB5301 | 66 |
| ZJUTCBB0015 | MC4100 ΔstpA::cat, Cmr | This study |
| ZJUTCBB0016 | MC4100 Δhns::kan, Kanr | This study |
| ZJUTCBB0017 | MC4100 ΔstpA::cat Δhns::kan, Kanr Cmr | This study |
| ZJUTCBB0018 | MC4100 ΔstpA pGLO-Pcas-GFP | This study |
| ZJUTCBB0019 | MC4100 Δhns pGLO-Pcas-GFP | This study |
| ZJUTCBB0020 | MC4100 ΔstpA Δhns pGLO-Pcas-GFP | This study |
| ZJUTCBB0021 | MC4100 pGLO-Pcas-GFP | This study |
| ZJUTCBB0022 | MC4100 Δhns pSUStpA pGLO-Pcas-GFP | This study |
| ZJUTCBB0023 | MC4100 Δhns pSUHNS pGLO-Pcas-GFP | This study |
| ZJUTCBB0024 | MC4100 Δhns pSU19 pGLO-Pcas-GFP | This study |
| ZJUTCBB0025 | MC4100 ΔstpA Δhns pSUStpA pGLO-Pcas-GFP | This study |
| ZJUTCBB0026 | MC4100 ΔstpA Δhns pSUHNS pGLO-Pcas-GFP | This study |
| ZJUTCBB0027 | MC4100 ΔstpA Δhns pSU19 pGLO-Pcas-GFP | This study |
| ZJUTCBB0028 | MC4100 Δhns pSUStpA pGLO-Pcas*-GFP | This study |
| ZJUTCBB0029 | MC4100 Δhns pSUHNS pGLO-Pcas*-GFP | This study |
| ZJUTCBB0030 | MC4100 Δhns pSU19 pGLO-Pcas*-GFP | This study |
| ZJUTCBB0031 | MC4100 ΔstpA Δhns pSU-PBAD-stpA | This study |
| ZJUTCBB0032 | MC4100 ΔstpA Δhns pSU-PBAD-hns | This study |
| ZJUTCBB0033 | MC4100 ΔstpA pSU-PJ23119-GFP | This study |
| ZJUTCBB0034 | MC4100 Δhns pSU-PJ23119-GFP | This study |
| ZJUTCBB0035 | MC4100 ΔstpA Δhns pSU-PJ23119-GFP | This study |
| ZJUTCBB0036 | MC4100 pSU-PJ23119-GFP | This study |
| Plasmids | ||
| pKD46 | Expressing Red recombinase, repA101(Ts) oriR101, Ampr | 94 |
| pCP20 | Expressing FLP recombinase, repA101(Ts) pSC101 ori, Cmr, Ampr | 94 |
| pKD3 | R6K γ ori, Ts replicon, Cmr | 94 |
| pKD4 | R6K γ ori, Ts replicon, Kanr | 94 |
| pSU19 | p15A replicon, Cmr | Lab reserve |
| pSUStpA | pSU19 derivative, stpA expressed with the original promoter, Cmr | This study |
| pSUHNS | pSU19 derivative, hns expressed with the original promoter, Cmr | This study |
| pGLO-Pcas-gfp | pGLO derivative, gfp expressed with Pcas, Ampr | This study |
| pGLO-PrscA-gfp | pGLO derivative, gfp expressed with PrscA, Ampr | This study |
| pGLO-Pcas*-gfp | pGLO-Pcas-gfp with mutated Pcas | This study |
| pSU-PBAD-gfp | pSU19 derivative, gfp expressed with PBAD, Cmr | This study |
| pSU-PBAD-stpA | pSU19 derivative, stpA expressed with PBAD, Cmr | This study |
| pSU-PBAD-hns | pSU19 derivative, hns expressed with PBAD, Cmr | This study |
| pSU-PJ23119-gfp | pSU19 derivative, gfp expressed with PJ23119 which is a constitutive promoter, Cmr | Lab reserve |
| pUC57 | pUC19 derivative, Ampr | Lab reserve |
| pT/pCR1 | pUC19 derivative, carrying a DNA fragment targeted by the type I-E CRISPR-Cas system of E. coli, Ampr | This study |
| pNT/pDsRED | pUC19 derivative, expressing red fluorescence protein, Ampr | Lab reserve |
| pET28a-StpA-His | pET28a derivative, StpA expressed by T7/lac promoter, Kanr | This study |
| pET28a-HNS-His | pET28a derivative, HNS expressed by T7/lac promoter, Kanr | This study |
Ts, temperature sensitive.
We analyzed DNA interference in transformants of the hns and hns stpA null mutants after transformation. Growth rates of transformants of mutants and their wild-type parent were evaluated. In liquid LB broth, no significant difference in cell growth was observed in WT and the hns null mutant carrying either the CRISPR-Cas-targeted plasmid pCR1 or the nontargeted plasmid pDsRED (Fig. S9A) whereas a significant growth defect in LB broth was observed in the hns and hns stpA null mutants carrying pCR1 compared with those carrying pDsRED (Fig. S9A). A growth defect was also observed in the hns and hns stpA null mutants carrying pCR1 and grown on LB-agar plates. By using drop plating and plate streaking methods, we showed that both the hns and hns stpA null mutants carrying pDsRED grew well on plates, but the mutants carrying pCR1 grew poorly under the same conditions (Fig. S9B and C). To quantify DNA interference after natural transformation, plasmid loss was evaluated by counting the number of viable cells having (ampicillin positive [Amp+]) or losing (Amp−) the targeted plasmid. In both the hns and hns stpA null mutants carrying pCR1, the number of Amp+ viable counts in the overnight-grown culture was ∼1,000-fold smaller than that of Amp− viable counts (Fig. S9D), suggesting that the targeted plasmid could have suffered significant damage in these mutants. Together, these results clearly demonstrate that StpA is required for CRISPR-Cas-mediated DNA interference in plasmid transformation of E. coli.
To evaluate the potential effect of StpA on DNA interference during chemical transformation, we compared transformation efficiencies in the hns and the hns stpA null mutants. The CRISPR-Cas-targeted plasmid pCR1 transformed both the stpA null mutant and its wild-type parent with a high efficiency (1 × 105 to 2 × 105 CFU/μg). In contrast, it transformed the hns null mutant with an efficiency of ∼4 CFU/μg. Inactivating stpA in the hns null mutant increased transformation efficiency by more than 10-fold, reaching ∼50 CFU/μg. Nevertheless, the efficiency of transformation with pCR1 was more than 1,000-fold lower than that with the nontargeted plasmid pDsRED in the hns stpA mutant, reflecting that StpA weakly stimulated CRISPR-Cas-mediated DNA interference during chemical transformation of the hns mutant.
StpA is required for transcription of cas genes in an hns null mutant.
H-NS was reported to be able to both suppress and activate gene expression (55, 56, 71) while StpA serves as a backup for H-NS, fulfilling the role of H-NS when it is absent (54, 55). Since deleting stpA remarkably reduced the activity of the CRISPR-Cas system against natural transformation in the hns mutant (Fig. 1B), we suspected that H-NS and StpA played opposite roles in regulating the type I-E system in E. coli. To test this, we quantified transcription of the cas operon in E. coli strains with single or double deletions of hns and stpA. We observed that inactivating stpA in an hns null mutant further increased transcription of an H-NS-suppressed gene, bglG, by more than 5-fold (Fig. 2A), and further decreased transcription of an H-NS-activated gene, malE, by about 7-fold (Fig. 2B). These results are in good accordance with previous reports (55, 56, 71). Transcription of the cas operon was reported to be increased by inactivating hns (55, 56, 71). We also observed that transcription of a gene in the cas operon (casB) was increased by more than 6-fold in an hns null mutant with respect to the level in the wild type (Fig. 2C). However, transcription of casB was increased by only about 3-fold in the hns stpA null mutant, about 2-fold lower than that in the hns null mutant (Fig. 2C). The data clearly show that StpA is required for high-level transcription of the cas operon.
FIG 2.
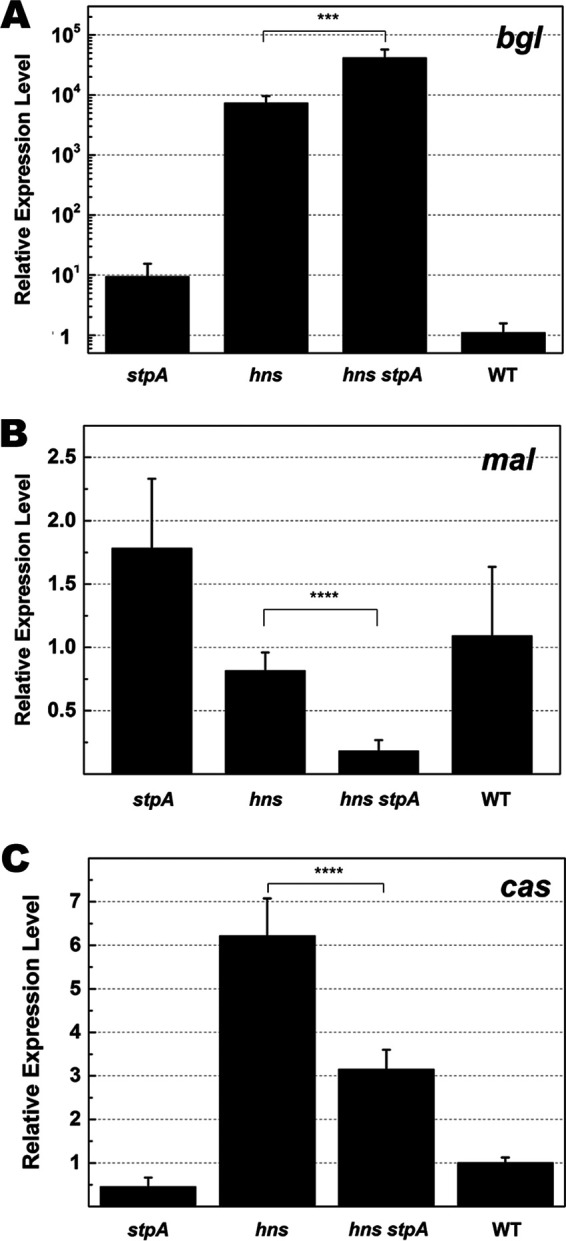
Quantification of transcriptional levels of the cas operon in the absence of H-NS and/or StpA. To quantify gene transcription, the cDNA was synthesized with RNA isolated from mutants (ΔstpA, Δhns, and Δhns ΔstpA strains) and their wild-type parent. Relative expression levels of bgl (A), mal (B), and cas (C) operons in these strains were measured with qPCR using 16S RNA as the reference. Primers used for qPCR are listed in Table 2. Each column represents results from an average of 4 samples. Error bars denote standard deviation. Statistical significance was determined using a two-tailed Student's t test (***, P ≤ 0.005; ****, P ≤ 0.001).
To further explore the role of StpA in regulating the type I-E CRISPR-Cas system, we monitored transcription of the cas operon during cell growth by fusing the promoter of the cas operon with the green fluorescent protein (GFP) gene (Pcas-gfp) (Fig. 3A and Fig. S4A). Inactivation of hns was reported to relieve transcriptional repression of cas genes (28). As expected, strong green fluorescence was observed in an hns null mutant whereas it was much weaker in the WT parent (Fig. 3B). The H-NS-suppressed promoter PrcsA was reported to be activated by mutating hns (72). We also observed that relative intensities of green fluorescence with the transcriptional fusion construct of PrcsA-gfp were remarkably higher in the hns and hns stpA null mutants than those in the stpA null mutant and the WT strain (Fig. S11). We noticed that transcription of the cas operon was low in the stpA null mutant (Fig. 3B), in line with the previous observation that inactivation of stpA did not significantly affect transcription of cas genes (28). After 12 h of incubation in LB medium, green fluorescence was ∼3-fold lower in the hns stpA null mutant than that in the hns null mutant (Fig. 3B), indicating that StpA played a positive role in regulating the cas operon. In contrast, expression levels of GFP with a constitutive promoter in the hns and hns stpA null mutant were not higher than those in the stpA null mutant and WT (Fig. S12). A stimulation effect of StpA on transcription of cas genes was more evident when cells were grown in M9 minimal medium. Throughout incubation, relative intensities of green fluorescence were ∼3-fold lower in the hns stpA null mutant than in the hns null mutant (Fig. 3C). Taken together, these results clearly show that StpA activates transcription of cas genes in the absence of hns.
FIG 3.
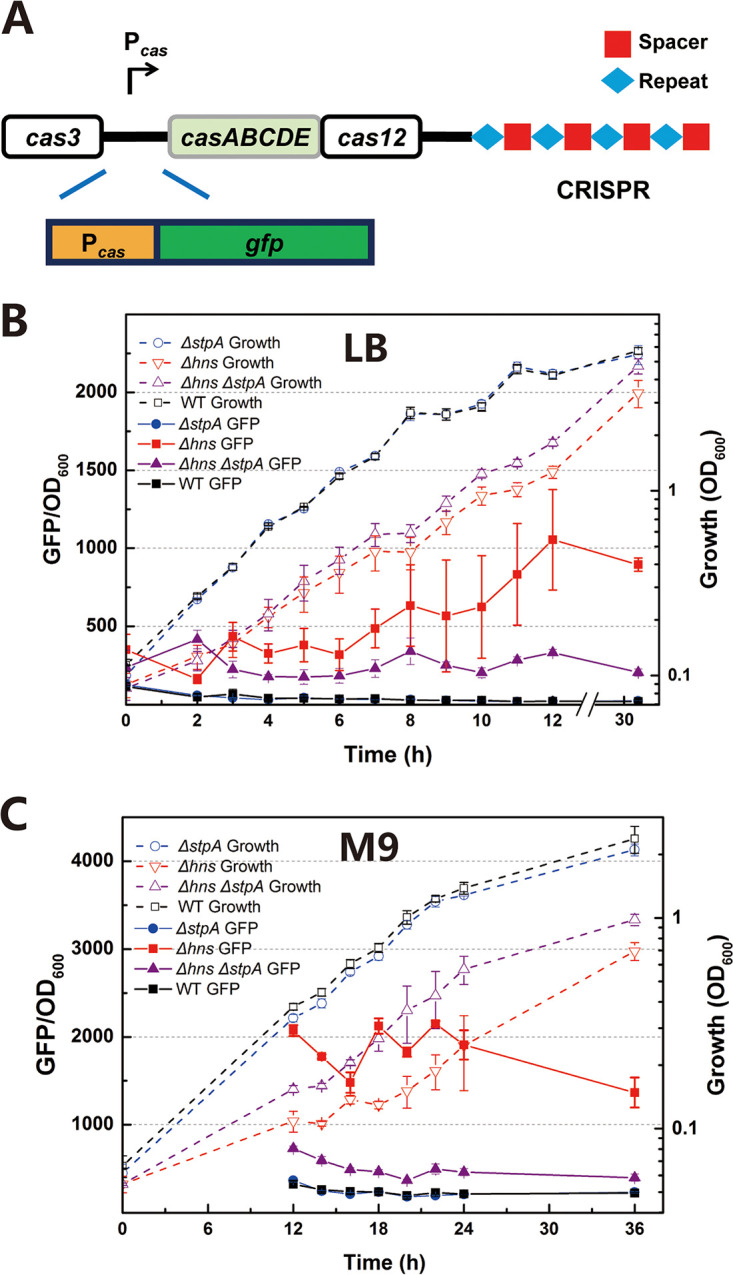
Effect of stpA inactivation on transcription of the cas operon. (A) The reporter plasmid pGLO-Pcas-gfp was constructed by fusing the promoter of the cas operon (Pcas) with gfp to quantify the transcriptional level of cas genes. (B) Expression levels of Pcas-gfp were measured in Δhns, ΔstpA, Δhns ΔstpA, and WT strains that were grown in LB broth. (C) Expression levels of Pcas-gfp were measured in Δhns, ΔstpA, Δhns ΔstpA, and WT strains that were grown in M9 minimal medium supplemented with 0.32% (wt/vol) fructose as the carbon source. Dashed lines represent optical growth, and solid lines represent activity of Pcas. All experiments were performed at 30°C with shaking at 180 rpm. Refer to supplemental material for details about construction of mutants (Δhns, ΔstpA, and Δhns ΔstpA strains) (see Fig. S1 and Table S1 in the supplemental material) and the plasmid pGLO-Pcas-gfp (Fig. S4A). Data are shown as means ± standard deviations (n = 3).
StpA activates transcription of cas genes by binding to the same site as H-NS in the promoter of the cas operon.
StpA often shares the same DNA binding site (DBS) as H-NS in the wild-type cell. But DNA binding profiles of StpA were different in wild-type and hns mutant cells (73). In vitro experiments showed that both H-NS and StpA bound Pcas, and the corresponding DBS of H-NS in Pcas was predicted (28). We asked whether H-NS and StpA bound to the same site in regulating the cas operon. To confirm that H-NS suppressed the cas operon by binding to the predicted DBS, we replaced the four conserved A/T residues with G/C in the putative DBS and constructed the promoter Pcas* (Fig. 4A and Fig. S5). Transcriptional activities of Pcas and Pcas* were compared when H-NS was expressed from a multicopy-number plasmid (Fig. S6). By using GFP as the reporter, we observed that activities of both Pcas and Pcas* were strong in an hns null mutant carrying the empty vector pSU19 (Fig. 4B). Although expressing H-NS on pSU19 reduced the activity of Pcas by 4.41-fold in the hns null mutant, it reduced the activity of Pcas* by only 1.84-fold (Fig. 4B). Moreover, the activity of Pcas* was higher than that of Pcas in both wild-type and stpA mutant cells (Fig. S15). These results demonstrated that mutations in Pcas* relieved transcriptional suppression by H-NS, indicating that H-NS indeed inhibited transcription of the cas operon by binding to the predicted DBS.
FIG 4.
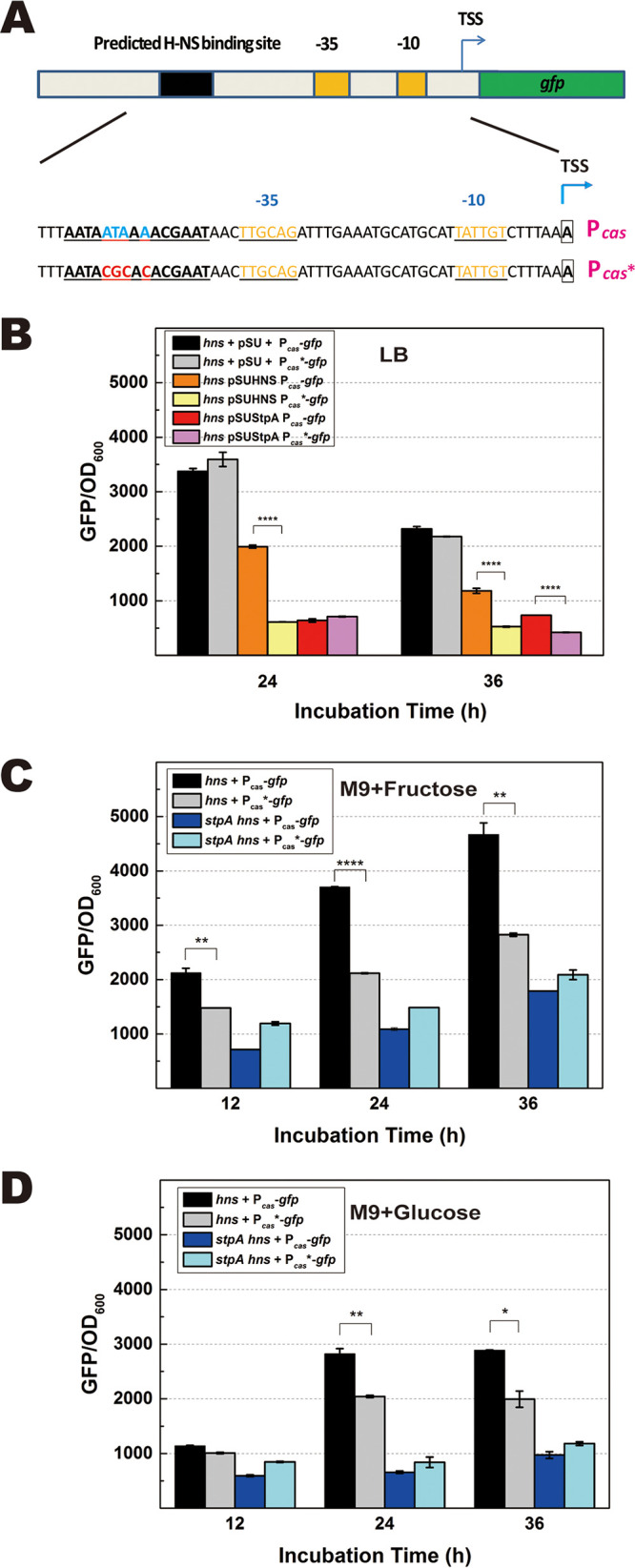
DNA binding site for transcriptional regulation by StpA and H-NS. Previous work has shown that H-NS and StpA can bind to Pcas, and the DNA binding site (DBS) for H-NS was predicted. (A) The putative DBS of H-NS/StpA was mutated by replacing the conserved sequence ATAAA to CGCAC. Pcas with a disrupted DBS was named Pcas*. Refer to Fig. S5 in the supplemental material for details about plasmid construction. (B) With the reporter GFP as an indicator, activities of Pcas and Pcas* were compared in the hns null mutant that expressed StpA or H-NS from the multicopy-number plasmid pSU19 in LB broth. Activities of Pcas-gfp and Pcas*-gfp were compared in hns and hns stpA null mutants that were grown in M9 minimal medium supplemented with 0.32% (wt/vol) fructose (C) or 1% (wt/vol) glucose (D) as the carbon source. TSS, transcriptional start site. Data are shown as means ± standard deviations (n = 4). Statistical significance was determined using a two-tailed Student's t test (*, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.001).
Although expressing H-NS on pSU19 increased the natural transformability of the hns stpA mutant to the level of the wild type, expressing StpA on pSU19 partially restored the natural transformability of the hns stpA mutant (Fig. S13). The results indicated that ectopic expression of H-NS fully, but overexpression of StpA slightly, suppressed the DNA interference activity of the CRISPR-Cas system. In accordance with the above observation, overexpression of StpA slightly suppressed the activity of Pcas after 36 h of incubation. This result contradicts our previous observation that StpA positively regulated the cas operon in the hns null mutant. A high concentration of StpA was reported to block the accessibility of DNA and interfere with gene transcription (74). We reasoned that the level of StpA expressed from both the genome and the multicopy-number plasmid pSUStpA in the hns null mutant should be very high and thus affect transcription of Pcas. To test whether StpA expressed at the physiological level promoted transcription of the cas operon by binding to the DBS of Pcas, we compared activities of Pcas and Pcas* in hns and hns stpA null mutants. In M9 medium supplemented with fructose, we observed that after 24 h of incubation the activity of Pcas was about 2-fold higher than that of Pcas* in the hns null mutant (Fig. 4C). Similar differential activities of Pcas and Pcas* in the hns null mutant were observed in M9 medium supplemented with glucose (Fig. 4D) whereas activities of both Pcas and Pcas* were similarly low in the hns stpA null mutant grown in M9 medium supplemented with either fructose or glucose (Fig. 4C and D). These results reflect that StpA indeed promotes the activity of Pcas by binding to the DBS of H-NS when it is expressed by a single chromosomal copy of the stpA in the hns null mutant.
Expression of StpA at a low level activates transcription of the cas operon.
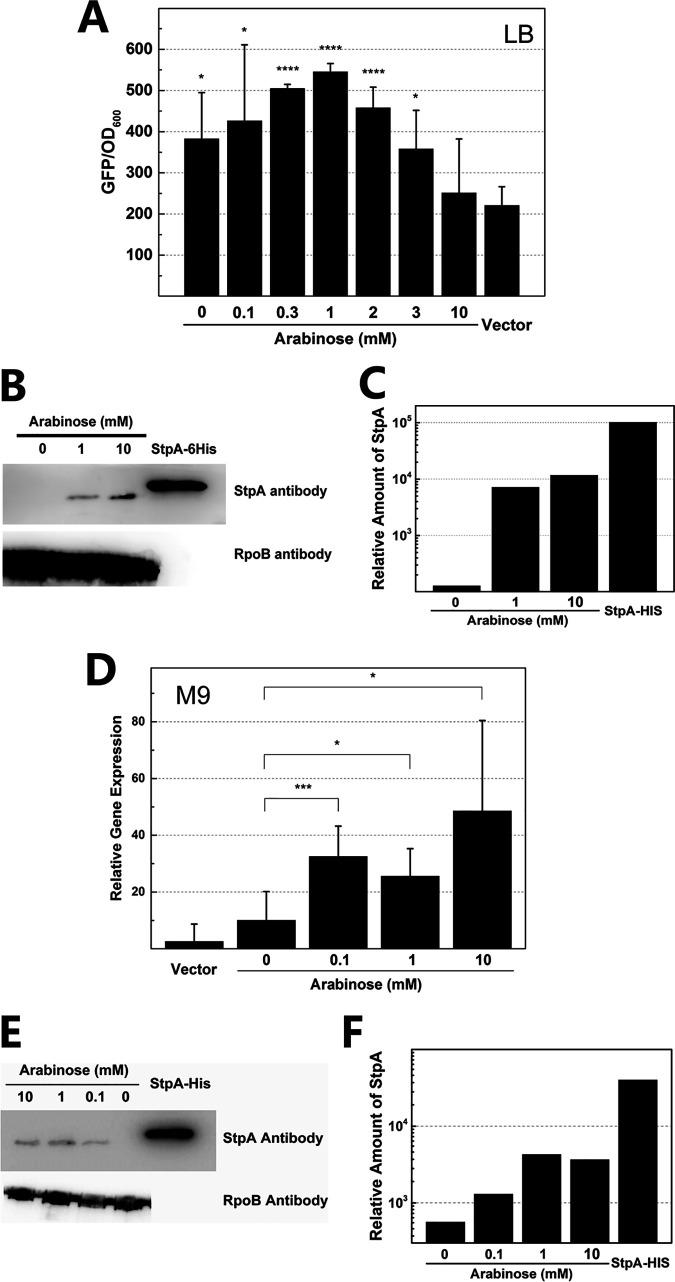
Low expression of StpA on the chromosome promoted the activity of Pcas (Fig. 2, 3, and 4C) whereas high expression of StpA on a multicopy-number plasmid failed to stimulate Pcas (Fig. 4B). This led us to consider whether expressing StpA at different levels yielded different effects on Pcas. To test this, we controlled expression of StpA by using an arabinose-inducible promoter (PBAD-stpA) on the plasmid (Fig. S7). The activity of Pcas was evaluated when StpA was expressed at different levels in the hns stpA null mutant grown in LB and M9 media (Fig. 5). We indeed observed that low concentrations of arabinose (0.1 mM to 3 mM) induced weak expression of StpA (Fig. 5B and C), which stimulated Pcas in LB medium (Fig. 5A). When 1 mM arabinose was added to the cell culture, the activity of Pcas in the hns stpA null mutant expressing StpA by PBAD reached the highest level, which was more than 2-fold higher than that in a strain carrying an empty vector (Fig. 5A). We also observed that even when no arabinose was added into the LB medium, the activity of Pcas in the strain containing PBAD-stpA on the vector was still obviously higher than that in the strain carrying the empty vector (Fig. 5A), indicating that leaky expression of StpA was sufficient for activating transcription of the cas operon in LB medium, whereas the activity of Pcas was gradually decreased by further increasing the concentration of arabinose (Fig. 5A). In contrast, a high concentration of arabinose (i.e., 10 mM) remarkably increased the amount of StpA expressed by PBAD (Fig. 5B and C) but reduced the activity of Pcas to a level similar to that with the empty vector (Fig. 5A). To test the effect of low expression of StpA on the CRISPR-Cas system-mediated DNA interference, levels of natural transformation with the CRISPR-Cas-targeted plasmid pCR1 and nontargeted plasmid pDsRED were evaluated in the hns stpA mutant containing PBAD-stpA. Considering that transformation occurred exclusively on agar plates (64, 65), expression of StpA was controlled by adding arabinose in both liquid form and on agar plates (see the supplemental material for details about the experimental procedure). When no arabinose was added, pCR1 transformed cells with an efficiency of ∼35 CFU/μg (Table S2) whereas no transformants were detected when 1 or 2 mM arabinose was added (Table S2). The result indicated that arabinose-induced expression of StpA activated CRISPR-Cas-mediated DNA interference during natural transformation of the E. coli hns stpA mutant.
FIG 5.
Complementation analysis of the effect of StpA on transcriptional regulation of Pcas. To modulate expression of StpA, the araBAD promoter (PBAD) was fused with stpA, and the construct was transformed into the hns stpA null mutant. The effect of StpA on transcription of the cas operon was monitored in cell cultures incubated in LB medium for 10 h (A) and in M9 medium for 24 h (D) supplemented with different concentrations of arabinose. A Western blot assay was performed to determine expression levels of StpA in cell cultures incubated in LB (B) and M9 (E) media. The StpA protein amounts from cell cultures grown in LB (C) and M9 (F) media were analyzed by using Quality One software. All experiments were performed in duplicate or triplicate. Statistical significance was determined using a two-tailed Student's t test (*, P ≤ 0.05; ***, P ≤ 0.005; ****, P ≤ 0.001).
The effect of StpA on the activity of Pcas was also evaluated in E. coli strains grown in M9 medium with glucose as the carbon source, which was reported to suppress the activity of PBAD (75, 76). Expressing StpA by PBAD in M9 medium supplemented with glucose indeed yielded a small amount of StpA (Fig. 5E and F). When no arabinose was added, the activity of Pcas in the hns stpA null mutant with PBAD-stpA on a vector was comparable to that of a strain carrying the empty vector (Fig. 5D), indicating remarkably reduced leaky expression of StpA in M9 medium. Amounts of StpA expressed by PBAD were equally low in cell cultures supplemented with 0.1 mM to 10 mM arabinose (Fig. 5E and F). Correspondingly, activities of Pcas in the above cultures were increased by 2- to 5-fold with respect to the level in the culture without arabinose (Fig. 5D). In contrast, expression of H-NS at either a high or a low level failed to stimulate transcription of the cas operon in the hns stpA null mutant (Fig. S14). Taken together, our results clearly demonstrate that expressing StpA at a low level stimulates transcription of the cas operon.
StpA is required for the formation of mature crRNA in an hns null mutant.
A functional CRISPR-Cas system consists of Cas proteins and mature CRISPR RNA (crRNA), which guides Cas proteins to the target DNA (5, 6, 14, 15). We have shown that StpA activated transcription of the cas genes in the hns null mutant. Finally, we examined the potential effect of StpA on the amount of mature crRNA. Mature crRNA is formed by processing pre-crRNA with CasE, encoded by a gene in the cas operon (10) (Fig. 6A). To remove long RNA (including pre-crRNA), we purified mature crRNA from total RNA by using a microRNA (miRNA) purification kit, in which the spin column contains resins that selectively bind small RNAs (sRNAs) (<200 nucleotides [nt]) with high affinity (Fig. 6B). An adaptor, which consisted of poly(A) and a primer for further quantitative PCR, was attached to the 3′ end of the sRNA fragment (Fig. 6B). Purified sRNA was analyzed by gel electrophoresis, which showed that sizes of most purified sRNAs were approximately 50 bp (Fig. S16A). With tRNA (encoded by glnU) as the internal control, quantitative PCR (qPCR) was performed to determine the amount of crRNA. Melt curve analysis showed single peaks for both crRNA and tRNA (Fig. S16B and C), indicating that the target cDNA was specifically amplified. PCR products of expected sizes were detected by gel electrophoresis (Fig. S16D). The qPCR data revealed that the relative amount of mature crRNA in the hns null mutant was more than 20-fold greater than that in the hns stpA null mutant (Fig. 6C), indicating that stpA was required for the formation of a large amount of cRNA in the hns mutant. Nevertheless, the amount of crRNA is still significantly higher than that in the wild type. Therefore, StpA is partially responsible for the increase in the amount of mature crRNA in the hns mutant whereas in the hns+ background, the relative amount of mature crRNA in the stpA mutant was ∼5-fold higher than that in its wild-type parent, showing that the effect of StpA on the amount of crRNA was influenced by H-NS.
FIG 6.
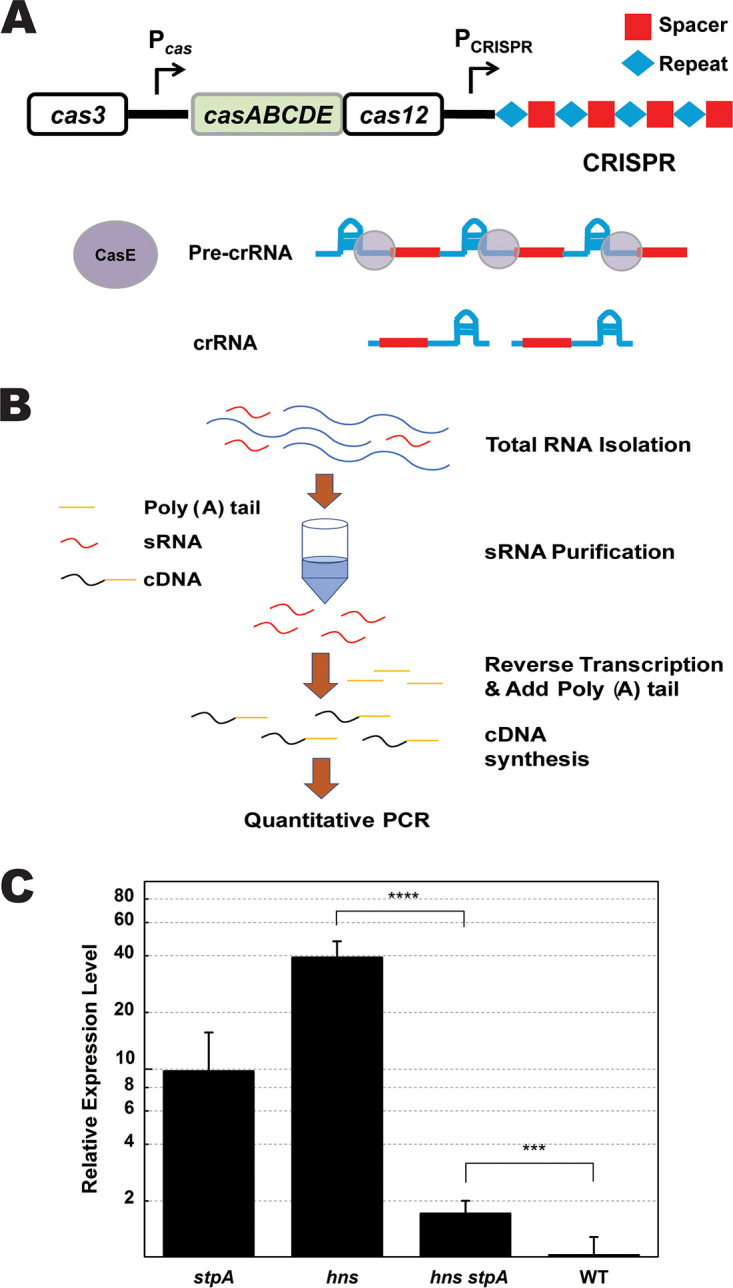
Effect of stpA inactivation on the amount of mature crRNA. (A) Schematic representation of the mechanism of crRNA formation. Followed by transcription of the pre-crRNA from CRISPR and expression of the cas operon, CasE cleaves the repeat sequence to form mature crRNA. (B) Schematic representation of the procedure of mature crRNA purification (refer to the supplemental material for details). (C) After 12 h of incubation in LB broth, total RNA was isolated, and a small RNA (containing crRNA) was purified for quantitative PCR assay. The relative amounts of mature crRNA from Δhns, ΔstpA, Δhns ΔstpA, and WT strains were quantified by real-time qPCR assay with tRNA as the reference. Data are shown as means ± standard deviations (n = 4). Statistical significance was determined using a two-tailed Student's t test (***, P ≤ 0.005; ****, P ≤ 0.001).
DISCUSSION
Previous work established that H-NS suppressed the type I-E CRISPR-Cas system in E. coli (28). In this study, we showed that the H-NS paralogue StpA played an opposite role in regulating the type I-E CRISPR-Cas system when H-NS was absent. It is able to promote both the transcriptional level of the cas operon and the amount of mature crRNA. We also document that the type I-E CRISPR-Cas system activated by StpA was able to defend against the transfer of DNA mediated by natural transformation in E. coli. CRISPR adaptation is mediated by Cas1 and Cas2, whose encoding genes are located in the cas operon of E. coli (16, 28). Considering that StpA activates transcription of the cas operon, we anticipate that it could also stimulate CRISPR adaptation during natural transformation of E. coli. The type I CRISR-Cas system is widely distributed in prokaryotes (5, 18). Particularly, homologues of H-NS and StpA have been found in both Salmonella and Klebsiella, which are also equipped with the type I-E CRISPR-Cas system (48, 77, 78). MvaT and MvaU play roles similar to those of H-NS and StpA in Pseudomonas (79), which also contains an active type I-E CRISPR-Cas system (80, 81). It would be interesting to investigate whether StpA homologues are involved in such regulatory roles in organisms other than E. coli.
CRISPR-Cas systems limit not only ssDNA uptake during natural transformation of S. pneumoniae and N. meningitidis (62, 63) but also dsDNA transferred via bacteriophage infection in E. coli (10, 47). Our previous work uncovered a new type of natural transformation, in which dsDNA was transferred across E. coli cell membranes (1). Here, we have shown that the type I-E CRISPR-Cas system in E. coli reduced levels of natural transformation with the CRISPR-Cas-targeted plasmid but did not affect transformation with the nontargeted plasmid (Fig. 1). We also observed that an StpA-activated CRISPR-Cas system reduced chemical transformation with the targeted plasmid (see Fig. S10 in the supplemental material). Therefore, transforming dsDNA is subject to CRISPR-Cas-mediated interference. It has been documented that inactivating cas3, which encodes the nuclease for DNA cleavage, reduced bacteriophage DNA interference by the type I CRISPR-Cas system in the hns mutant (46, 49). The StpA-activated type I-E CRISPR-Cas system could also provide immunity against “infecting” ssDNA (e.g., conjugative DNA and bacteriophage DNA) due to DNA cleavage on the target dsDNA formed by recombination or replication of ssDNA after its entry into the cell. With the hns stpA null mutant as the recipient, although we detected a number of transformants carrying the CRISPR-Cas-targeted plasmid, these transformants were unable to grow further (Fig. S9), suggesting that the type I-E CRISPR-Cas system in the hns stpA null mutant was unable to cleave all CRISPR-Cas-targeted DNA immediately after plasmid entry. Instead, dsDNA should be gradually degraded over a prolonged period of time in that strain (Fig. S9). Nevertheless, delayed DNA interference in the hns stpA null mutant but immediate degradation of the entering plasmid in the hns null mutant indicated that the efficiency of DNA interference was enhanced by StpA in the hns null mutant (Fig. S9). It is also noticeable that the level of transformation with the CRISPR-Cas-targeted plasmid was slightly lower than that with the nontargeted plasmid in the WT strain (Fig. 1B), indicating that the type I-E CRISPR-Cas system could be moderately activated during natural transformation of E. coli.
H-NS and StpA are xenogeneic silencing proteins for suppressing gene transcription from foreign-derived DNAs, which are widespread in the Gram-negative alpha-, beta-, and gammaproteobacteria as well as in the Gram-positive Actinobacteria (79). Although interaction between xenogeneic silencers and DNA have recently been intensively studied (74, 82–86), it remains unclear how the H-NS/StpA nucleoprotein structure observed in vitro affects gene transcription in vivo. It is not unprecedented to discover that StpA can act as a transcriptional activator, given that StpA normally functions as a backup for H-NS, which was shown to act as both activator and repressor, as revealed by transcriptome analysis (73). For example, both H-NS and StpA stimulate transcription of crp by changing the DNA topology to a form that favors the use of highly compacted DNA for transcription (57). We also observed that both H-NS and StpA positively regulated transcription of the mal operon (Fig. 2B). Nevertheless, data presented here strongly argue that H-NS and StpA play opposite roles in regulating the same promoter (i.e., the cas promoter): the former suppresses gene transcription but the latter stimulates gene transcription (Fig. 2 and 5). StpA and H-NS share DNA binding regions in both the gal and cas promoters (28, 54). In the promoter of the cas operon, a footprinting experiment showed that StpA and H-NS bound to the same region (28) in which the H-NS binding motif was predicted (28, 46, 47). In this study, we provided in vivo evidence supporting the finding that StpA and H-NS recognized the same DBS in Pcas but played different roles in regulating transcription of the cas operon (Fig. 4). Overexpressing H-NS strongly suppressed the activity of Pcas in the hns null mutant (Fig. 4B). In contrast, mutating the DBS derepressed transcription of the cas operon that was inhibited by H-NS (Fig. 4B), showing that H-NS suppresses the activity of Pcas by binding to the predicted DBS. With respect to the activity of Pcas containing an intact DBS, the activity of Pcas* containing a mutated DBS was remarkably reduced in the hns null mutant (Fig. 4C and D). Nevertheless, Pcas and Pcas* showed similar activity levels in the hns stpA null mutant (Fig. 4C and D). These facts clearly show that certain amounts of StpA promoted the activity of Pcas by acting on the DBS of H-NS. The reason for the different behaviors of H-NS and StpA on the activity of Pcas needs further exploration. Previous in vitro experiments documented that StpA and H-NS compact DNA in different ways (74). It is possible that differential compaction of DNA by H-NS and StpA may lead to different modes of transcriptional regulation in vivo.
StpA is more versatile than H-NS in transcriptional regulation. In the presence or absence of StpA, we observed that H-NS inhibited the activity of Pcas (Fig. 7). In contrast, a small amount of StpA induced the activity of Pcas (Fig. 5A and D and Fig. 7), but a large amount of StpA failed to stimulate Pcas (Fig. 5A and 7). Previous in vitro experiments revealed that linearized double-stranded DNA incubated with 1 StpA per 1 bp (1:1 StpA/DNA) showed rigid StpA-coated DNA hairpins that blocked DNA accessibility (74) whereas at ratios of both 1:10 and 1:100 StpA/DNA, StpA-induced DNA bridging was evident (74). We calculated the cellular concentration of StpA in cells grown in M9 medium supplemented with 1 mM arabinose and found that Pcas was activated when the ratio of StpA/DNA was ∼1:200 (see the results and discussion in the supplemental material and Table S3). Possibly, a low cellular concentration of StpA may induce DNA bridging, which could facilitate transcription initiation by altering the configuration of StpA/DNA. Nevertheless, the explanation for transcriptional activation by StpA awaits more direct evidence.
FIG 7.
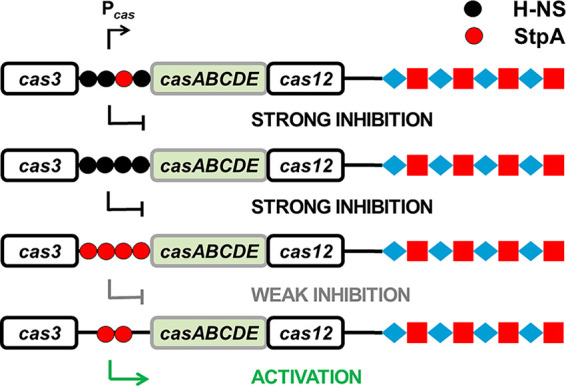
Schematic diagram of transcriptional regulation by StpA and H-NS. Transcription of the cas operon was suppressed by H-NS in the presence (first row) or absence of StpA (second row). Although StpA expressed at a high level weakly suppresses transcription of the cas operon (third row), a low concentration of StpA stimulated the activity of Pcas (fourth row).
We observed a transcriptional stimulation effect of StpA only in the hns null strain. In line with a previous study (28), inactivation of stpA did not affect transcription of cas genes in the hns+ strain (Fig. 2 and 3). Considering that transcription of H-NS can be upregulated by inactivating stpA (54), the effect of StpA on transcription of the type I-E CRISPR-Cas system could be masked by stronger repression due to the increased expression level of H-NS in the stpA null mutant. Similar to previous studies (54), we also observed that inactivating both hns and stpA caused a more serious growth defect than inactivating only hns (Fig. S8A). Interestingly, when a high-copy-number plasmid was introduced into E. coli strains, the hns null mutant grew remarkably more slowly than the hns stpA null mutant (Fig. S8B and C). Providing that H-NS alters plasmid and chromosomal DNA supercoiling (87), as well as chromosome partitioning and replication (69), it is possible that the aberrant topological structure of the plasmid could have an effect on cell growth in the hns null mutant.
Although StpA increased the amount of crRNA in the hns deletion mutant, it reduced the amount of crRNA in the hns+ strain (Fig. 6C). The result reflects that H-NS influenced the effect of StpA on the level of crRNA. Considering that StpA can function as both an RNA chaperon (88–90) and a transcriptional regulator of the gene encoding CasE for pre-crRNA processing (Fig. 6A), it could play pleiotropic roles in stabilizing/processing pre-crRNA and/or mature crRNA in the hns mutant. The complex interplay between these processes contributes to crRNA abundance in a nonlinear way (91). Alternatively, H-NS and StpA in the WT strain may form a heteromeric complex (92), which could have a stronger suppression effect on expression of crRNA than only H-NS in the stpA null mutant.
Of note, a recent report documented that ectopic expression of StpA on a multicopy-number plasmid suppressed transcription of casA in an hns cas1 double mutant of E. coli, resulting in a decrease of DNA interference during λ phage infection (93). The result is in accordance with our observation that ectopic expression of StpA at a high level suppressed transcription of cas genes in the hns null mutant (Fig. 4B) and moderately reduced DNA interference during natural transformation (Fig. S13). It is also documented that although inactivation of stpA further increased transcription of casA in the hns cas1 mutant, stpA seems to have no additional role to further improve immunity against bacteriophage infection in that strain (93). This finding is different from our observation in that stpA deletion in an hns null mutant reduced transcription of cas genes (Fig. 3) and decreased immunity against plasmid DNA in both natural and chemical transformations (Fig. 1 and Fig. S10).
In summary, we showed that expression of the type I-E CRISPR-Cas system is positively regulated by StpA which activates the promoter of the cas operon and augments the amount of crRNA in the hns null background. The type I-E CRISPR-Cas system activated by StpA increases DNA interference during natural transformation and thus improves the cellular immune response to horizontally transferred genetic elements. Moreover, we provided evidence showing that DNA-structuring protein paralogues StpA and H-NS bound on the same site in Pcas but played opposite roles in transcriptional regulation of the cas operon, implying that higher-order compaction of bacterial chromatin by histone-like proteins could switch prokaryotic transcriptional modes.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, growth conditions, and media.
Bacterial strains, plasmids, and primers used in this study are listed in Tables 1 and 2. Strains lacking hns and/or stpA were constructed with pKD46 (see Fig. S1 in the supplemental material), a temperature-sensitive plasmid carrying the λ-derived Red recombination system (94). Mutants were examined through PCR and Western blot assay (Fig. S1 and Table S1). Recombinant plasmids were constructed with a One-Step PCR Cloning kit (Fig. S3 to S7). Details of the construction of mutants and plasmids are described in supplemental material. Plasmids were isolated with a plasmid isolation kit according to the manufacturer’s instructions (Axygen Biotech Co., Ltd.). E. coli was grown at 30°C or 37°C as follows: in LB broth containing 1% tryptone (wt/vol), 0.5% (wt/vol) yeast extract, and 1% (wt/vol) NaCl; in 1.5× LB broth containing 1.5% tryptone (wt/vol), 0.75% (wt/vol) yeast extract, and 1.5% (wt/vol) NaCl; in M9 minimal medium containing 61 mM K2HPO4·3H2O, 38.21 mM KH2PO4, 4.15 mM MgSO4, 16.70 mM NH4Cl, 0.1% tryptone (wt/vol), and 0.32% (wt/vol) fructose or 0.8% (wt/vol) glucose as the carbon source, supplemented with an appropriate amount of arabinose for inducing transcription of genes when necessary or not supplemented; or on LB agar plates containing 1.5% (wt/vol) or 5% (wt/vol) agar. When necessary, the medium was supplemented with ampicillin (100 μg ml−1), kanamycin (50 μg ml−1), or chloramphenicol (25 μg ml−1), unless otherwise specified. Cell growth was measured in a Spectrumlab S23A spectrophotometer or in a Tecan Sunrise 96-well plate reader. All experiments were repeated independently at least three times.
TABLE 2.
Primers
| Primer no. | Name | Sequence (5′–3′)a |
|---|---|---|
| P01 | ΔstpA H1P1 | ATCGCTTACACTACGCGACGAAATACTTTTTTTGTTTTGGCGTTAAAAGGGTGTAGGCTGGAGCTGCTTC (−62, −12) |
| P02 | ΔstpA H2P2 | ATAAGATGCCGTGGAACCAACGAGCTTGAGAAGCGACGCCGGACGCGCCCCATATGAATATCCTCCTTAG (+25, +75) |
| P03 | Δhns H1P1 | TCTATTATTACCTCAACAAACCACCCCAATATAAGTTTGAGATTACTACAGTGTAGGCTGGAGCTGCTTC (−51, −1) |
| P04 | Δhns H2P2 | TTATTAAATTGTCTTAAACCGGACAATAAAAAATCCCGCCGCTGGCGGGCATATGAATATCCTCCTTAG (+443, +493) |
| P05 | ΔstpA-1500F | TATGTATTTACGACCAGAC |
| P06 | ΔstpA-999R | ATTTTAGCGGAGCCTGCC |
| P07 | Δhns-1500F | GCATTGCCCTTCTGGGGCCG |
| P08 | Δhns-999R | AGGAACCAGGATGTTGCCGG |
| P09 | gfp-F | AGGAGAACAATTTAAGAAGGAGATATACAT |
| P10 | gfp-R | AATAATGGTTTCTTAGACGTCAAGCTTGCATGCCTGCAGG |
| P11 | Pcas promoter-F | TCCCCGAAAAGTGCCACCTGCTTCGGGAATGATTGTTATC |
| P12 | Pcas promoter-R | CCTTCTTAAATTGTTCTCCTTCATATGCTC |
| P13 | gfp-rscA-F | CATTGAGTGAGGGTATGCTACTAGAAAGAGGAGAAATA |
| P14 | gfp-rscA-R | TTACGGAATTATATTAATGGGGTAACGAATCAGACAATTG |
| P15 | PrscA promoter-F | CAATTGTCTGATTCGTTACCGGGTCTGAATGCGACGTTAA |
| P16 | PrscA promoter-R | TATTTCTCCTCTTTCTAGTATTAAATTCTCCTGGACTG |
| P17 | pSU19-stpA-F | CGCTTCTCAAGCTCGTTGGGGTACCGAGCTCGAATTC |
| P18 | pSU19-stpA-R | GAAAACATCCATCACTGGTGCCTGCAGGCATGCAAGCTTG |
| P19 | stpA-pSU19-F | CAAGCTTGCATGCCTGCAGGCACCAGTGATGGATGTTTTC |
| P20 | stpA-pSU19-R | GAATTCGAGCTCGGTACCCCAACGAGCTTGAGAAGCG |
| P21 | hns-pSU19-F | CGTGGATAACACCGATACGGGGGTACCGAGCTCGAATTC |
| P22 | hns-pSU19-R | TACGAGAATTCCCTATCCTGCAGGCATGCAAGCTTG |
| P23 | pSU19-hns-F | CAAGCTTGCATGCCTGCAGGATAGGGAATTCTCGTA |
| P24 | pSU19-hns-R | GAATTCGAGCTCGGTACCCCCGTATCGGTGTTATCCACG |
| P25 | cas-gfp-ΔBS-F | CTTTAATAGCGAGACGAATAAC |
| P26 | cas-gfp-ΔBS-R | GTTATTCGTCTCGCTATTAAAG |
| P27 | tRNA-F | CGGTTTTTGATACCGGCAT |
| P28 | crRNA-F | CTCCCTGTCGGTTGTAATTG |
| P29 | sRNA-R | GATCGCCCTTCTACGTCGTAT |
| P30 | PBAD-gfp-pSU-F | TTATGACAACTTGACGGCTACA |
| P31 | PBAD-gfp-pSU-R | TTATTTGTAGAGCTCATCCATGC |
| P32 | pSU-PBAD-gfp-F | TGGATGAGCTCTACAAATAAGAATTCGAGCTCGGTACCCG |
| P33 | pSU-PBAD-gfp-R | TAGCCGTCAAGTTGTCATAAATTGCGTTGCGCTCACTGCC |
| P34 | pSU-PBAD-F | GAATTCGAGCTCGGTACCCG |
| P35 | pSU-PBAD-R | ATGTATATCTCCTTCTTAAAGTTAAACAAAATTATTTC |
| P36 | PBAD-stpA-pSU-F | TTTAAGAAGGAGATATACATATGTCCGTAATGTTACAAAG |
| P37 | PBAD-stpA-pSU-R | GGGTACCGAGCTCGAATTCTTAGATCAGGAAATCGTCGAG |
| P38 | PBAD-hns-pSU-F | TTTAAGAAGGAGATATACATATGAGCGAAGCACTTAAAATTCTG |
| P39 | PBAD-hns-pSU-R | GGGTACCGAGCTCGAATTCTTATTGCTTGATCAGGAAATCGTCG |
Boldface letters indicate nucleotide extensions, complementary to antibiotic resistance cassettes, that were introduced in the primers to amplify either the cat gene or the kan gene from plasmid pKD3 or pKD4, respectively. Following each primer sequence, the corresponding gene and the positions of the first and final nucleotides (in parentheses) are indicated (with respect to the ATG of the gene).
Quantification of transcription of cas genes with GFP.
To monitor transcription of cas genes by GFP, the promoter PBAD was replaced by the promoter of the cas operon (Pcas) in the plasmid pGLO-gfp (Fig. 3A and Fig. S4A). The recombinant plasmid was transformed into E. coli cells, and transformants were subsequently grown in LB or M9 medium supplemented with appropriate antibiotics. Cell growth was monitored spectrophotometrically at an optical density of 600 nm (OD600). Intensity of the culture fluorescence, as an indicator of transcription of Pcas, was measured by a SpectraMax Gemini EM microplate reader with excitation and emission wavelengths at 395 and 509 nm, respectively.
Quantification of gene transcripts and mature crRNA with quantitative PCR.
Total RNA was isolated from E. coli cells grown in LB broth at 30°C for 12 h. The cell density was adjusted to an OD600 of 1.0. To quantify transcription of chromosomal genes, the isolated RNA was reverse transcribed, and quantitative PCR (qPCR) was performed with a Bio-Rad iCycle iQ. Small RNA (sRNA) was purified from the isolated total RNA with an EasyPure miRNA kit (TransGen Biotech Co., Ltd.) which contains columns filled with resin with high affinity to sRNA (<200 bp) fragments (Fig. 2A). The 3′ terminally tagged cDNA was synthesized by reverse transcription (miRNA First-Strand cDNA Synthesis SuperMix; TransGen Biotech Co., Ltd.) and quantified by qPCR. Relative expression levels of genes and sRNAs were calculated according to the formula 2−(ΔCT target − ΔCT reference) (95), with 16S RNA and tRNA, respectively, as the internal references. Primers used for qPCR are listed in Table 2. Refer to supplemental material for detailed procedures.
Natural transformation of E. coli.
Natural transformation was performed according to a previously documented method (66, 67). A single E. coli colony grown on an LB-agar plate was inoculated into a glass tube containing 5 ml of LB broth and incubated with shaking at 30°C. An overnight-grown E. coli culture (14 to 18 h of incubation) was inoculated into a flask containing 1.5× LB broth at a ratio of 1:100 (vol/vol). The culture was incubated with shaking at a speed of 180 rpm at 30°C. After 24 h of incubation, the cell culture was precipitated by centrifugation at room temperature, and 90% (vol/vol) of the supernatant was discarded. The cell pellet was resuspended in the remaining 10% (vol/vol) of the supernatant and placed in a metal bath at 30°C. Plasmids pDsRED and pCR1 were added to the cell suspension solution to a final concentration of 40 μg ml−1. Fifty microliters of the mixture was spread on LB plates containing 5% (wt/vol) agar (BD Difco) and 200 μg ml−1 of ampicillin, which had been placed at room temperature (∼30°C) for 1 to 2 days before use. Numbers of transformants were counted on petri plates which were incubated at 30°C for 1 to 2 days. Transformation efficiency was calculated by dividing the number of transformants by the amount of DNA.
Protein purification and Western blot assay.
Protein with a His tag was purified from E. coli BL21 with a tagged-protein purification kit (soluble protein) (Beijing ComWin Biotech Co., Ltd.). The antibody against StpA, which can interact with both StpA and its paralogue H-NS (Fig. S1C), was used for evaluating the expression level of StpA. RpoB was set as the control, and the corresponding primary antibody was commercially obtained from BioLegend. Refer to the supplemental material for details about procedures for protein purification and Western blot assay.
Data availability.
The pCR1 sequence has been deposited in GenBank under accession number MT437282 and the Addgene plasmid repository under catalog no. 154270 (https://www.addgene.org/154270/).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Donald Morrison (University of Illinois at Chicago) for critical reading of the manuscript and anonymous reviewers for their efforts to improve the quality of the manuscript.
This work was supported by the National Natural Science Foundation of China (grants 31670084 and 31100071), the Key Research and Development Program of Zhejiang Province (grant 2020C02031), and the Zhejiang Provincial Natural Science Foundation of China (grants LY16C010003 and Y3110237).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Sun D. 2018. Pull in and push out: mechanisms of horizontal gene transfer in bacteria. Front Microbiol 9:2154. doi: 10.3389/fmicb.2018.02154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Claverys JP, Prudhomme M, Martin B. 2006. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol 60:451–475. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- 3.Dubnau D, Blokesch M. 2019. Mechanisms of DNA uptake by naturally competent bacteria. Annu Rev Genet 53:217–237. doi: 10.1146/annurev-genet-112618-043641. [DOI] [PubMed] [Google Scholar]
- 4.Wilson GG, Murray NE. 1991. Restriction and modification systems. Annu Rev Genet 25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 5.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, van der Oost J. 2016. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353:aad5147. doi: 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- 7.Xue C, Sashital DG. 6 February 2019, posting date. Mechanisms of type I-E and I-F CRISPR-Cas systems in Enterobacteriaceae. EcoSal Plus 2019. doi: 10.1128/ecosalplus.ESP-0008-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 9.Deveau H, Garneau JE, Moineau S. 2010. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol 64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 10.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 12.Marraffini LA, Sontheimer EJ. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karginov FV, Hannon GJ. 2010. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell 37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorek R, Kunin V, Hugenholtz P. 2008. CRISPR–a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol 6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 15.Sorek R, Lawrence CM, Wiedenheft B. 2013. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem 82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 16.Pougach K, Semenova E, Bogdanova E, Datsenko KA, Djordjevic M, Wanner BL, Severinov K. 2010. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol Microbiol 77:1367–1379. doi: 10.1111/j.1365-2958.2010.07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol 190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. 2015. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radovcic M, Killelea T, Savitskaya E, Wettstein L, Bolt EL, Ivancic-Bace I. 2018. CRISPR-Cas adaptation in Escherichia coli requires RecBCD helicase but not nuclease activity, is independent of homologous recombination, and is antagonized by 5’ ssDNA exonucleases. Nucleic Acids Res 46:10173–10183. doi: 10.1093/nar/gky799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amitai G, Sorek R. 2016. CRISPR-Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol 14:67–76. doi: 10.1038/nrmicro.2015.14. [DOI] [PubMed] [Google Scholar]
- 21.Sternberg SH, Richter H, Charpentier E, Qimron U. 2016. Adaptation in CRISPR-Cas systems. Mol Cell 61:797–808. doi: 10.1016/j.molcel.2016.01.030. [DOI] [PubMed] [Google Scholar]
- 22.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. 2011. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A 108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marraffini LA, Sontheimer EJ. 2010. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. 2010. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 28.Pul U, Wurm R, Arslan Z, Geissen R, Hofmann N, Wagner R. 2010. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol Microbiol 75:1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- 29.Musharova O, Klimuk E, Datsenko KA, Metlitskaya A, Logacheva M, Semenova E, Severinov K, Savitskaya E. 2017. Spacer-length DNA intermediates are associated with Cas1 in cells undergoing primed CRISPR adaptation. Nucleic Acids Res 45:3297–3307. doi: 10.1093/nar/gkx097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunne T, Kieper SN, Bannenberg JW, Vogel AI, Miellet WR, Klein M, Depken M, Suarez-Diez M, Brouns SJ. 2016. Cas3-derived target DNA degradation fragments fuel primed CRISPR adaptation. Mol Cell 63:852–864. doi: 10.1016/j.molcel.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 31.Levy A, Goren MG, Yosef I, Auster O, Manor M, Amitai G, Edgar R, Qimron U, Sorek R. 2015. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semenova E, Savitskaya E, Musharova O, Strotskaya A, Vorontsova D, Datsenko KA, Logacheva MD, Severinov K. 2016. Highly efficient primed spacer acquisition from targets destroyed by the Escherichia coli type I-E CRISPR-Cas interfering complex. Proc Natl Acad Sci U S A 113:7626–7631. doi: 10.1073/pnas.1602639113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arslan Z, Hermanns V, Wurm R, Wagner R, Pul U. 2014. Detection and characterization of spacer integration intermediates in type I-E CRISPR-Cas system. Nucleic Acids Res 42:7884–7893. doi: 10.1093/nar/gku510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yosef I, Goren MG, Qimron U. 2012. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res 40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S, Loeff L, Colombo S, Jergic S, Brouns SJJ, Joo C. 2020. Selective loading and processing of prespacers for precise CRISPR adaptation. Nature 579:141–145. doi: 10.1038/s41586-020-2018-1. [DOI] [PubMed] [Google Scholar]
- 36.Nunez JK, Lee AS, Engelman A, Doudna JA. 2015. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 519:193–198. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Erp PB, Jackson RN, Carter J, Golden SM, Bailey S, Wiedenheft B. 2015. Mechanism of CRISPR-RNA guided recognition of DNA targets in Escherichia coli. Nucleic Acids Res 43:8381–8391. doi: 10.1093/nar/gkv793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beloglazova N, Kuznedelov K, Flick R, Datsenko KA, Brown G, Popovic A, Lemak S, Semenova E, Severinov K, Yakunin AF. 2015. CRISPR RNA binding and DNA target recognition by purified Cascade complexes from Escherichia coli. Nucleic Acids Res 43:530–543. doi: 10.1093/nar/gku1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao Y, Luo M, Dolan AE, Liao M, Ke A. 2018. Structure basis for RNA-guided DNA degradation by cascade and Cas3. Science 361:eaat0839. doi: 10.1126/science.aat0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao YB, Luo M, Hayes RP, Kim J, Ng S, Ding F, Liao MF, Ke AL. 2017. Structure basis for directional R-loop formation and substrate handover mechanisms in type I CRISPR-Cas system. Cell 170:48–60. doi: 10.1016/j.cell.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao H, Sheng G, Wang J, Wang M, Bunkoczi G, Gong W, Wei Z, Wang Y. 2014. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature 515:147–150. doi: 10.1038/nature13733. [DOI] [PubMed] [Google Scholar]
- 42.Jackson RN, Golden SM, van Erp PBG, Carter J, Westra ER, Brouns SJJ, van der Oost J, Terwilliger TC, Read RJ, Wiedenheft B. 2014. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science 345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yosef I, Goren MG, Kiro R, Edgar R, Qimron U. 2011. High-temperature protein G is essential for activity of the Escherichia coli clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system. Proc Natl Acad Sci U S A 108:20136–20141. doi: 10.1073/pnas.1113519108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redding S, Sternberg SH, Marshall M, Gibb B, Bhat P, Guegler CK, Wiedenheft B, Doudna JA, Greene EC. 2015. Surveillance and processing of foreign DNA by the Escherichia coli CRISPR-Cas system. Cell 163:854–865. doi: 10.1016/j.cell.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patterson AG, Yevstigneyeva MS, Fineran PC. 2017. Regulation of CRISPR-Cas adaptive immune systems. Curr Opin Microbiol 37:1–7. doi: 10.1016/j.mib.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Westra ER, Pul U, Heidrich N, Jore MM, Lundgren M, Stratmann T, Wurm R, Raine A, Mescher M, Van Heereveld L, Mastop M, Wagner EG, Schnetz K, Van Der Oost J, Wagner R, Brouns SJ. 2010. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol Microbiol 77:1380–1393. doi: 10.1111/j.1365-2958.2010.07315.x. [DOI] [PubMed] [Google Scholar]
- 47.Yang CD, Chen YH, Huang HY, Huang HD, Tseng CP. 2014. CRP represses the CRISPR/Cas system in Escherichia coli: evidence that endogenous CRISPR spacers impede phage P1 replication. Mol Microbiol 92:1072–1091. doi: 10.1111/mmi.12614. [DOI] [PubMed] [Google Scholar]
- 48.Medina-Aparicio L, Rebollar-Flores JE, Gallego-Hernández AL, Vázquez A, Olvera L, Gutiérrez-Ríos RM, Calva E, Hernández-Lucas I. 2011. The CRISPR/Cas immune system is an operon regulated by LeuO, H-NS, and leucine-responsive regulatory protein in Salmonella enterica serovar Typhi. J Bacteriol 193:2396–2407. doi: 10.1128/JB.01480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Majsec K, Bolt EL, Ivančić-Baće I. 2016. Cas3 is a limiting factor for CRISPR-Cas immunity in Escherichia coli cells lacking H-NS. BMC Microbiol 16:28. doi: 10.1186/s12866-016-0643-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gong B, Shin M, Sun J, Jung CH, Bolt EL, van der Oost J, Kim JS. 2014. Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3. Proc Natl Acad Sci U S A 111:16359–16364. doi: 10.1073/pnas.1410806111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. 2011. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J 30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patterson AG, Chang JT, Taylor C, Fineran PC. 2015. Regulation of the type I-F CRISPR-Cas system by CRP-cAMP and GalM controls spacer acquisition and interference. Nucleic Acids Res 43:6038–6048. doi: 10.1093/nar/gkv517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang AX, Belfort M. 1992. Nucleotide-sequence of a newly-identified Escherichia coli gene, stpA, encoding an H-NS-like protein. Nucleic Acids Res 20:6735–6735. doi: 10.1093/nar/20.24.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonden B, Uhlin BE. 1996. Coordinated and differential expression of histone-like proteins in Escherichia coli: regulation and function of the H-NS analog StpA. EMBO J 15:4970–4980. doi: 10.1002/j.1460-2075.1996.tb00877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolf T, Janzen W, Blum C, Schnetz K. 2006. Differential dependence of StpA on H-NS in autoregulation of stpA and in regulation of bgl. J Bacteriol 188:6728–6738. doi: 10.1128/JB.00586-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johansson J, Dagberg B, Richet E, Uhlin BE. 1998. H-NS and StpA proteins stimulate expression of the maltose regulon in Escherichia coli. J Bacteriol 180:6117–6125. doi: 10.1128/.180.23.6117-6125.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johansson J, Balsalobre C, Wang SY, Urbonaviciene J, Jin DJ, Sonden B, Uhlin BE. 2000. Nucleoid proteins stimulate stringently controlled bacterial promoters: a link between the cAMP-CRP and the (p)ppGpp regulons in Escherichia coli. Cell 102:475–485. doi: 10.1016/S0092-8674(00)00052-0. [DOI] [PubMed] [Google Scholar]
- 58.Sonnenfield JM, Burns CM, Higgins CF, Hinton J. 2001. The nucleoid-associated protein StpA binds curved DNA, has a greater DNA-binding affinity than H-NS and is present in significant levels in hns mutants. Biochimie 83:243–249. doi: 10.1016/S0300-9084(01)01232-9. [DOI] [PubMed] [Google Scholar]
- 59.Johnston C, Martin B, Fichant G, Polard P, Claverys JP. 2014. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 60.Chen I, Dubnau D. 2004. DNA uptake during bacterial transformation. Nat Rev Microbiol 2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 61.Seitz P, Blokesch M. 2013. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol Rev 37:336–363. doi: 10.1111/j.1574-6976.2012.00353.x. [DOI] [PubMed] [Google Scholar]
- 62.Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. 2012. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe 12:177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Heidrich N, Ampattu BJ, Gunderson CW, Seifert HS, Schoen C, Vogel J, Sontheimer EJ. 2013. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell 50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun D, Zhang Y, Mei Y, Jiang H, Xie Z, Liu H, Chen X, Shen P. 2006. Escherichia coli is naturally transformable in a novel transformation system. FEMS Microbiol Lett 265:249–255. doi: 10.1111/j.1574-6968.2006.00503.x. [DOI] [PubMed] [Google Scholar]
- 65.Sun D, Zhang X, Wang L, Prudhomme M, Xie Z, Martin B, Claverys JP. 2009. Transforming DNA uptake gene orthologs do not mediate spontaneous plasmid transformation in Escherichia coli. J Bacteriol 191:713–719. doi: 10.1128/JB.01130-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, Shi C, Yu J, Ren J, Sun D. 2012. RpoS regulates a novel type of plasmid DNA transfer in Escherichia coli. PLoS One 7:e33514. doi: 10.1371/journal.pone.0033514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun D. 2016. Two different routes for double-stranded DNA transfer in natural and artificial transformation of Escherichia coli. Biochem Biophys Res Commun 471:213–218. doi: 10.1016/j.bbrc.2016.01.137. [DOI] [PubMed] [Google Scholar]
- 68.Guo M, Wang H, Xie N, Xie Z. 2015. Positive effect of carbon sources on natural transformation in Escherichia coli: role of low-level cyclic AMP (cAMP)-cAMP receptor protein in the derepression of rpoS. J Bacteriol 197:3317–3328. doi: 10.1128/JB.00291-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaidow A, Wachi M, Nakamura J, Magae J, Nagai K. 1995. Anucleate cell production by Escherichia coli delta hns mutant lacking a histone-like protein, H-NS. J Bacteriol 177:3589–3592. doi: 10.1128/jb.177.12.3589-3592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang A, Rimsky S, Reaban ME, Buc H, Belfort M. 1996. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J 15:1340–1349. doi: 10.1002/j.1460-2075.1996.tb00476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Free A, Williams RM, Dorman CJ. 1998. The StpA protein functions as a molecular adapter to mediate repression of the bgl operon by truncated H-NS in Escherichia coli. J Bacteriol 180:994–997. doi: 10.1128/JB.180.4.994-997.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sledjeski D, Gottesman S. 1995. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc Natl Acad Sci U S A 92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uyar E, Kurokawa K, Yoshimura M, Ishikawa S, Ogasawara N, Oshima T. 2009. Differential binding profiles of StpA in wild-type and hns mutant cells: a comparative analysis of cooperative partners by chromatin immunoprecipitation-microarray analysis. J Bacteriol 191:2388–2391. doi: 10.1128/JB.01594-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lim CJ, Whang YR, Kenney LJ, Yan J. 2012. Gene silencing H-NS paralogue StpA forms a rigid protein filament along DNA that blocks DNA accessibility. Nucleic Acids Res 40:3316–3328. doi: 10.1093/nar/gkr1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Katz L, Englesberg E. 1971. Hyperinducibility as a result of mutation in structural genes and self-catabolite repression in the ara operon. J Bacteriol 107:34–52. doi: 10.1128/JB.107.1.34-52.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gendron RP, Sheppard DE. 1974. Mutations in the l-arabinose operon of Escherichia coli B-r that result in hypersensitivity to catabolite repression. J Bacteriol 117:417–421. doi: 10.1128/JB.117.2.417-421.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin T, Pan Y, Hsieh P, Hsu C, Wu M, Wang J. 2016. Imipenem represses CRISPR-Cas interference of DNA acquisition through H-NS stimulation in Klebsiella pneumoniae. Sci Rep 6:31644. doi: 10.1038/srep31644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Medina-Aparicio L, Rebollar-Flores J, Beltrán-Luviano A, Vázquez A, Gutiérrez-Ríos R, Olvera L, Calva E, Hernández-Lucas I. 2017. CRISPR-Cas system presents multiple transcriptional units including antisense RNAs that are expressed in minimal medium and upregulated by pH in Salmonella enterica serovar Typhi. Microbiology 163:253–265. doi: 10.1099/mic.0.000414. [DOI] [PubMed] [Google Scholar]
- 79.Navarre WW, McClelland M, Libby SJ, Fang FC. 2007. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev 21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- 80.Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O'Toole GA. 2009. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol 191:210–219. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pawluk A, Bondy-Denomy J, Cheung V, Maxwell K, Davidson A. 2014. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. mBio 5:e00896-14. doi: 10.1128/mBio.00896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qin L, Bdira FB, Sterckx YGJ, Volkov AN, Vreede J, Giachin G, van Schaik P, Ubbink M, Dame RT. 2020. Structural basis for osmotic regulation of the DNA binding properties of H-NS proteins. Nucleic Acids Res 48:2156–2172. doi: 10.1093/nar/gkz1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shahul Hameed UF, Liao C, Radhakrishnan AK, Huser F, Aljedani SS, Zhao X, Momin AA, Melo FA, Guo X, Brooks C, Li Y, Cui X, Gao X, Ladbury JE, Jaremko L, Jaremko M, Li J, Arold ST. 2019. H-NS uses an autoinhibitory conformational switch for environment-controlled gene silencing. Nucleic Acids Res 47:2666–2680. doi: 10.1093/nar/gky1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van der Valk RA, Vreede J, Qin L, Moolenaar GF, Hofmann A, Goosen N, Dame RT. 2017. Mechanism of environmentally driven conformational changes that modulate H-NS DNA-bridging activity. Elife 6:e27369. doi: 10.7554/eLife.27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu Y, Chen H, Kenney LJ, Yan J. 2010. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev 24:339–344. doi: 10.1101/gad.1883510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Arold ST, Leonard PG, Parkinson GN, Ladbury JE. 2010. H-NS forms a superhelical protein scaffold for DNA condensation. Proc Natl Acad Sci U S A 107:15728–15732. doi: 10.1073/pnas.1006966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mojica FJ, Higgins CF. 1997. In vivo supercoiling of plasmid and chromosomal DNA in an Escherichia coli hns mutant. J Bacteriol 179:3528–3533. doi: 10.1128/jb.179.11.3528-3533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Doetsch M, Gstrein T, Schroeder R, Furtig B. 2010. Mechanisms of StpA-mediated RNA remodeling. RNA Biol 7:735–743. doi: 10.4161/rna.7.6.13882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grossberger R, Mayer O, Waldsich C, Semrad K, Urschitz S, Schroeder R. 2005. Influence of RNA structural stability on the RNA chaperone activity of the Escherichia coli protein StpA. Nucleic Acids Res 33:2280–2289. doi: 10.1093/nar/gki515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Waldsich C, Grossberger R, Schroeder R. 2002. RNA chaperone StpA loosens interactions of the tertiary structure in the td group I intron in vivo. Genes Dev 16:2300–2312. doi: 10.1101/gad.231302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Djordjevic M, Djordjevic M, Severinov K. 2012. CRISPR transcript processing: a mechanism for generating a large number of small interfering RNAs. Biol Direct 7:24. doi: 10.1186/1745-6150-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johansson J, Eriksson S, Sonden B, Wai SN, Uhlin BE. 2001. Heteromeric interactions among nucleoid-associated bacterial proteins: localization of StpA-stabilizing regions in H-NS of Escherichia coli. J Bacteriol 183:2343–2347. doi: 10.1128/JB.183.7.2343-2347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mitić D, Radovčić M, Markulin D, Ivančić-Baće I. 2020. StpA represses CRISPR-Cas immunity in H-NS deficient Escherichia coli. Biochimie 174:136–143. doi: 10.1016/j.biochi.2020.04.020. [DOI] [PubMed] [Google Scholar]
- 94.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The pCR1 sequence has been deposited in GenBank under accession number MT437282 and the Addgene plasmid repository under catalog no. 154270 (https://www.addgene.org/154270/).