Abstract
This paper represents the first article in a series on Yunnanese microfungi. We herein provide insights into Magnolia species associated with microfungi. All presented data are reported from the Kunming Botanical Gardens. Final conclusions were derived from the morphological examination of specimens coupled with phylogenetic sequence data to better integrate taxa into appropriate taxonomic ranks and infer their relationships. Shearia formosa, the type species of Shearia, lacks type material, and its phylogenetic position accordingly remains unresolved. A fresh collection of Shearia formosa, obtained from Magnolia denudata and M. soulangeana in China, therefore, designated a neotype for stabilizing the application of the species and/or genus name. Phylogenetic analyses of a combined DNA data matrix containing SSU, LSU, RPB2 and TEF loci of representative Pleosporales revealed that the genera Crassiperidium, Longiostiolum and Shearia are a well-defined monophylum. It is recognized as the family Longiostiolaceae and strongly supported by Bayesian and Maximum Likelihood methods. Its members are characterized by immersed to semi-immersed, globose to subglobose ascomata with a central, periphysate ostiole, a peridium composed of rectangular to polygonal cells, cylindrical to clavate asci, broadly fusiform, hyaline to pale brown ascospores, a coelomycetous asexual morph with pycnidial conidiomata, enteroblastic, annellidic, ampulliform, doliiform or cylindrical conidiogenous cells and cylindrical to fusiform, transverse and sometimes laterally distoseptate conidia without a sheath or with a basal lateral sheath. Nigrograna magnoliae sp. nov. is introduced from Magnolia denudata with both asexual and sexual morphs. We observed the asexual morph of Brunneofusispora sinensis from the culture and therefore amended the generic and species descriptions of Brunneofusispora.
Introduction
Located in Southwestern China, Yunnan is renowned for harboring one of the botanically richest and most diverse terrestrial regions on Earth [1]. Yunnan represents >50% of China’s overall floristic diversity and accounts for 18,000 vascular plant species [2], with high levels of endemism [3]. A highly variable climate and flourishing vegetation facilitate rapid fungal growth, reproduction and speciation. Based on its high level of plant diversity, it is estimated that approximately 104,000 fungal species may exist in Yunnan [4].
Conversely, among these fungal encounters, ascomycetes are being neglected compared to the level of research conducted on basidiomycetes in Yunnan Province. Owing to their abundance in all ecosystems, ascomycetous taxa simply cannot be overlooked in any region. Most taxa of ascomycetes are plant-associated fungi that can be pathogens, endophytes, saprobes or epiphytes across a wide range of hosts in terrestrial as well as aquatic habitats. Microfungi contribute both positively and negatively to human and economic well-being. As pathogens, they pose a threat to agriculture [5], and the rapid identification of potentially problematic species and accurate prediction of their behavior will facilitate the adoption of proper mitigation and phytosanitary measures. Given their ubiquitous nature, additional taxonomic knowledge are prerequisites to understanding the biological and environmental significance of ascomycetes. Supporting this obligation, the CAS Key Laboratory for Plant Biodiversity and Biogeography of East Asia (KLPB) has begun to study plant-based ascomycetes in Yunnan Province. The current study represents the first in a series comprising a taxonomic circumscriptive project of microfungi in Yunnan Province. It accounts for a group of ascomycetes recovered from the twigs of Magnolia denudata and M. soulangeana in the East Garden of Kunming Botanical Garden (Kunming, Panlong District). This garden functions as an ex-situ conservation of plants from Southwest China, particularly focusing on the conservation of endangered, endemic and economically important plant species native to the Yunnan Plateau and the southern Hengduan Mountains [6]. Based on morphology and multi-gene phylogenetic evidences of the collected ascomycetes, we characterized a neotype, a new species and a new host record in the order Pleosporales.
Materials and methods
Isolates and specimens
Fresh fungal materials were collected from twigs of Magnolia denudata and M. soulangeana in the East Garden of Kunming Botanical Garden (Yunnan Province, China) during both dry (February) and wet (August) seasons. Kunming Botanical Garden is in the central part of Yunnan Province in the city of Kunming, located in a plateau basin. The wet and dry seasons are distinct in Kunming, and precipitation is concentrated from May to October, accounting for about 85% of the annual precipitation [7]. The dry seasons is from November to April, the rainfall of accounts for only about 15% of total annual rainfall [7]. The Garden is located at 25°07’004.9”–25°08’054.8”N, 102°44’015.2”–102°44’047.3”E at an elevation of 1914–1990 m above sea level, and has an annual average rainfall of 1006.5 mm, an annual average temperature of 14.7 °C and an annual average relative humidity of 73% [6]. The collected specimens were brought to the laboratory in Zip lock plastic bags. Samples were examined with an Olympus SZ61 Series microscope. Single spore isolation was carried out following the method described in [8]. Germinated spores were individually transferred to potato dextrose agar (PDA) plates and grown at 20 °C in the daylight. Isolates including accession numbers of gene sequences are listed in are listed in Suppl. material 1: Table 1. Isolates listed as MFLUCC and KUMCC are maintained in the Culture Collection of Mae Fah Luang University, Chiang Rai, Thailand and Kunming Institute of Botany Culture Collection, China, respectively. Specimens have been deposited in the Mae Fah Luang University (MFLU) Herbarium, Chiang Rai, Thailand. Faces of Fungi and Index Fungorum numbers are provided as outlined in [9] and [10].
Table 1. Genes/loci used in the study with PCR primers, references and protocols.
| Locus a | Primers | PCR: thermal cycles: b (Annealing temp. in bold) | References |
|---|---|---|---|
| ITS | ITS5 | (95 °C: 30 s, 55 °C:50 s, 72 °C: 90 s) × 35 cycles | [11] |
| ITS4 | |||
| LSU | LR0R | (95 °C: 30 s, 55 °C:50 s, 72 °C: 90 s) × 35 cycles | [12, 13] |
| LR5 | |||
| SSU | NS1 | (95 °C: 30 s, 55 °C:50 s, 72 °C: 90 s) × 35 cycles | [11] |
| NS4 | |||
| RPB2 | fRPB2-5f | (94 °C: 60 s, 58 °C: 60 s, 72 °C: 90 s) × 40 cycles | [14] |
| fRPB2-7cR | |||
| TEF | EF1-983F | (95 °C: 30 s, 55 °C:50 s, 72 °C: 90 s) × 35 cycles | [15, 16] |
| EF1-2218R |
a ITS: Part of rDNA 18S (3' end), the first internal transcribed spacer (ITS1), the 5.8S rRNA gene, the second ITS region (ITS2), and part of the 28S rRNA (5' end); LSU: Large subunit (28S); SSU: Small subunit rDNA (18S); RPB2: RNA polymerase II second largest subunit; TEF: translation elongation factor 1-alpha gene
b All the PCR thermal cycles include Initiation step of 95 °C: 5 min, and final elongation step of 72 °C: 10 min and final hold at 4 °C
Morphological observations
In hand sections of the ascomata/ conidiomata, which were mounted in distilled water, the following characteristics were evaluated: ascomata/ conidiomata diameter, height, colour and shape; width of peridium; height and diameter of ostioles. Length and width (at the widest point) of asci, ascospores, conidiophores and conidia were measured. Images were captured with a Canon EOS 600D digital camera fitted to a Nikon ECLIPSE Ni compound microscope. Measurements were made with the Tarosoft (R) Image Frame Work program and images used for figures processed with Adobe Photoshop CS5 Extended version 10.0 software (Adobe Systems, USA).
DNA extraction, PCR amplifications and sequencing
Mycelia for DNA extraction from each isolate were grown on PDA for 3–4 weeks at 20 °C and total genomic DNA was extracted from approximately 150 ± 50 mg axenic mycelium scraped from the edges of the growing culture. Mycelium was ground to a fine powder with liquid nitrogen and DNA extracted using the Biospin Fungus Genomic DNA Extraction Kit-BSC14S1 (BioFlux, P.R. China) following the instructions of the manufacturer. DNA to be used as template for PCR were stored at 4 °C for use in regular work and duplicated at -20 °C for long term storage.
DNA sequence data was obtained from the partial sequences of three ribosomal and two protein coding genes. The genes, primers, references and PCR protocols are summarized in Table 1. Polymerase chain reaction (PCR) was carried out in a volume of 25 μl which contained 12.5 μl of 2 × Power Taq PCR MasterMix (Bioteke Co., China), 1 μl of each primer (10 μM), 1 μl genomic DNA and 9.5 μl deionized water. The amplified PCR fragments were sent to a commercial sequencing provider (BGI, Ltd Shenzhen, P.R. China). The nucleotide sequence data acquired were deposited in GenBank.
Molecular phylogenetic analyses
Sequencing and sequence alignment
Sequences generated from different primers of the five genes were analysed with other sequences retrieved from GenBank (see S1 Table). Sequences with high similarity indices were determined from a BLAST search to find the closest matches with taxa in Pleosporales, and from recently published data [17, 18, 19]. The multiple alignments of all consensus sequences, as well as the reference sequences were automatically generated with MAFFT v. 7 (http://mafft.cbrc.jp/alignment/server/index.html; [20, 21]), and were improved manually when necessary using BioEdit v. 7.0.5.2 [22].
Phylogenetic analyses
The single-locus data sets were examined for topological incongruence among loci for members of the Pleosporales. The conflict-free alignments were concatenated into a multi-locus alignment that was subjected to maximum-likelihood (ML) and Bayesian (BI) phylogenetic analyses. The evolutionary models for Bayesian analysis and maximum-likelihood were selected independently for each locus using MrModeltest v. 2.3 [23] under the Akaike Information Criterion (AIC) implemented in both PAUP v. 4.0b10. GTR+I+G model is resulted in each locus for Bayesian analysis and maximum-likelihood by AIC in MrModeltest as the best-fit model.
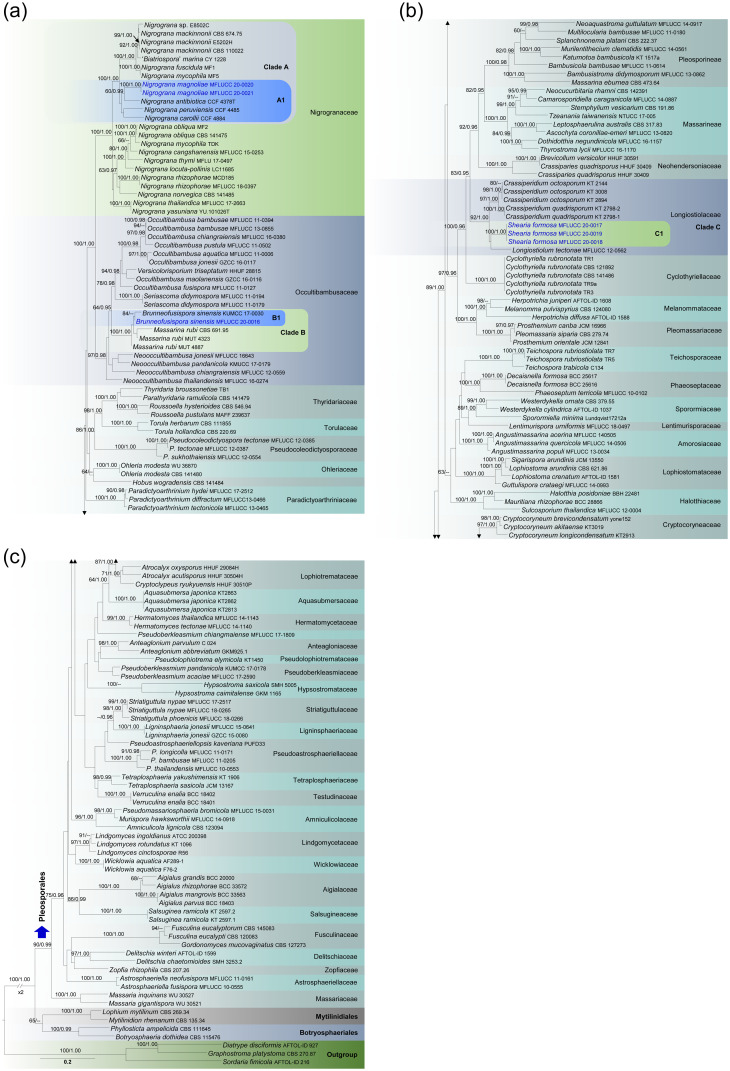
Bayesian analysis was conducted with MrBayes v. 3.1.2 [24] to evaluate Bayesian posterior probabilities (BYPP) [25, 26] by Markov Chain Monte Carlo sampling (BMCMC). GTR+I+G was used in the command. Six simultaneous Markov chains were run for 2,000,000 generations and trees were sampled every 200th generation. The distribution of log-likelihood scores was examined to determine stationary phase for each search and to decide if extra runs were required to achieve convergence, using the program Tracer 1.5 [27]. First 20% of generated trees were discarded and remaining 80% of trees were used to calculate posterior probabilities of the majority rule consensus tree. BYPP greater than 0.95 are given above each node (Fig 1).
Fig 1. RAxML tree based on a combined dataset of partial SSU, LSU, RPB2 and TEF sequence analyses.
Bootstrap support values for ML equal to or greater than 60% and Bayesian posterior probabilities (BYPP) equal to or greater than 0.95 are shown as ML/BI above the nodes. The new isolates are blue. The scale bar represents the expected number of nucleotide substitutions per site.
Maximum likelihood trees were generated using the RAxML-HPC2 on XSEDE (8.2.8) [28, 29] in the CIPRES Science Gateway platform [30] using GTR+I+G model of evolution. Maximum likelihood bootstrap values (ML) equal or greater than 70% are given above each node (Fig 1).
Phylograms were visualized with FigTree v1.4.0 program [31] and reorganized in Microsoft power point (2007) and Adobe Illustrator® CS5 (Version 15.0.0, Adobe®, San Jose, CA). The finalized alignment and tree were deposited in TreeBASE, submission ID: 25920 (http://purl.org/phylo/treebase/phylows/study/TB2:S25920).
Compliance with ethical standards
The authors declare that there is no conflict of interest (financial or non-financial) and agree to the submission of this paper. The authors also confirm that the fieldwork did not involve endangered or protected species and no specific permissions were required for these locations as the land belongs to the Kunming Institute of Botany, which is the first affiliated institution on this paper.
Nomenclature
The electronic version of this article in Portable Document Format (PDF) in a work with an ISSN or ISBN will represent a published work according to the International Code of Nomenclature for algae, fungi, and plants, and hence the new names contained in the electronic publication of a PLOS ONE article are effectively published under that Code from the electronic edition alone, so there is no longer any need to provide printed copies.
In addition, the new name contained in this work has been submitted to Index Fungorum from where they will be made available to the Global Names Index. The unique Index Fungorum number can be resolved and the associated information viewed through any standard web browser by appending the Index Fungorum number contained in this publication to the prefix www.indexfungorum.org/. The online version of this work is archived and available from the following digital repositories: PubMed Central, LOCKSS.
Results
Phylogenetic analysis
The concatenated dataset (SSU, LSU, RPB2 and TEF loci) consisted of 180 strains including the new taxa proposed in this study, with Diatrype disciformis (AFTOL-ID 927), Graphostroma platystoma (CBS 270.87) and Sordaria fimicola (AFTOL-ID 216) as the out-group taxa. Topologies of trees (ML and BI) derived from analyses of single gene dataset were compared and the overall tree topology was congruent to those obtained from the combined dataset.
The RAxML analysis of the combined dataset yielded a best scoring tree (Fig 1) with a final ML optimization likelihood value of -80641.514118. The matrix had 2357 distinct alignment patterns, with 24.7% of undetermined characters or gaps. Parameters for the GTR + I + G model of the combined LSU, SSU, RPB2 and TEF were as follows: Estimated base frequencies; A = 0.248843, C = 0.245092, G = 0.273138, T = 0.232926; substitution rates AC = 1.416663, AG = 4.473370, AT = 1.440148, CG = 1.224282, CT = 9.886557, GT = 1.000; proportion of invariable sites I = 0.46547; gamma distribution shape parameter α = 0.580812. Comparison of the alignment properties and nucleotide substitution models are provided in Table 2. The Bayesian analysis resulted in 10001 trees after 2000000 generations. The first 1000 trees, representing the burn-in phase of the analyses, were discarded, while the remaining 9001 trees were used for calculating posterior probabilities in the majority rule consensus tree.
Table 2. Comparison of alignment properties of genes and nucleotide substitution models used in Pleosporales phylogenetic analysis.
| SSU | LSU | RPB2 | TEF | |
|---|---|---|---|---|
| Alignment strategy (MAFFT v. 7) | G-INS-1 | G-INS-1 | G-INS-1 +manual | G-INS-1 +manual |
| Nucleotide substitution models for Bayesian analysis (determined by MrModeltest | GTR+I+G | GTR+I+G | GTR+I+G | GTR+I+G |
Forty-one families and two suborders (Massarineae, Pleosporineae) in Pleosporales are represented in the phylogenetic tree (Fig 1). The topologies resulting from the ML and BI analyses (Fig 1) are generally congruent with the results reported by [19] and other large-scale phylogenies of Dothideomycetes e.g. [32]. Multigenic (Fig 1) analyses agree to support significantly the monophyletic origin of every interleaved family and the two suborders in Pleosporales. Six newly isolated strains from this study (MFLUCC 20–0016, MFLUCC 20–0017, MFLUCC 20–0018, MFLUCC 20–0019, MFLUCC 20–0020 and MFLUCC 20–0021) constituted three lineages in Clade A, B and C (Fig 1). MFLUCC 20–0020 and MFLUCC 20–0021 grouped within Nigrogranaceae (Clade A, Fig 1) as a well-supported monophyletic clade with 100% ML 1.00 BYPP support (Fig 1). This is closely related to Nigrograna antibiotica (CCF 4378T), N. carollii (CCF 4884) and N. peruviensis (CCF 4485), but the corresponding affiliation is not statistically supported (Subclade A1, Fig 1). MFLUCC 20–0016 grouped in Occultibambusaceae (Clade B, Fig 1). MFLUCC 20–0016, Brunneofusispora sinensis (KUMCC 17 0030) and three strains labeled as ‘Massarina rubi’ (CBS 691.95 MUT 4323, MUT 4887) formed a strongly supported monophyletic association (Subclade B1, Fig 1) in Occultibambusaceae. Three of our newly isolated strains (MFLUCC 20–0017, MFLUCC 20–0018, MFLUCC 20–0019), Crassiperidium quadrisporum (KT 2798 1, KT 2798 2), C. octosporum (KT 3008, KT 2144, KT 2894) and Longiostiolum tectonae (MFLUCC 12 0562) formed an isolated, well-supported clade (92% ML and 1.00 PP, Clade C, Fig 1) within Pleosporales, which is regarded as Longiostiolaceae.
Taxonomy
Longiostiolaceae Phukhams., Doilom & K.D. Hyde, Fungal Diversity 101: 43 (2020) amended
[urn:lsid:indexfungorum.org:names: IF557086]
Facesoffungi number: FoF 07215.
Saprobic in terrestrial habitats. Sexual morph: Ascomata solitary to gregarious, immersed to semi-immersed, globose to subglobose, with a central ostiole. Ostiole long, circular, central, sometimes periphysate. Peridium composed of rectangular to polygonal, pale brown to dark brown cells. Hamathecium comprising numerous, cellular, septate pseudoparaphyses. Asci 4–8-spored, bitunicate, fissitunicate, cylindrical to clavate, pedicellate. Ascospores mostly overlapping biseriate to 3-seriate, broadly fusiform, thick-walled, 1–10-septate, hyaline to pale brown, smooth-walled. Asexual morph: Conidiomata pycnidial, sometimes pseudostromatic, solitary to gregarious, immersed, globose to conical, unilocular, dark brown, ostiolate. Ostiole papillate ostiole, central, circular. Conidiomata wall composed of angular, pale brown to brown cells. Conidiophores absent or reduced to conidiogenous cells. Conidiogenous cells enteroblastic, annellidic, ampulliform, doliiform or cylindrical. Conidia cylindrical to fusiform, base truncate, with several transverse and sometimes laterally distoseptate, continuous, without a sheath or with a basal lateral sheath.
Type genus: Longiostiolum Doilom, Ariyaw. & K.D. Hyde
Notes: Clade C (Fig 1) comprises Crassiperidium, Longiostiolum and Shearia, which are phylogenetically highly supported and belong to Longiostiolaceae in the Pleosporales (Fig 1). [33] recently introduced Longiostiolaceae to accommodate Crassiperidium and Longiostiolum. In this study, Shearia has also proven to be a genus in Longiostiolaceae (Clade C1, Fig 1), and we hereby amend the family description in order to accommodate its morphological characteristics.
Longiostiolaceae has a close phylogenetic affinity to Cyclothyriellaceae in large-scale multi-gene phylogenetic analyses of Pleosporales. But they are not grouped in a monophyletic clade, rather forming discrete clades adjacent to each other. The asexual characteristics of this new family distinguish it from Cyclothyriellaceae. Cyclothyriellaceae has 1-celled conidia, whereas Longiostiolaceae has multi-celled conidia. Morphologically, Crassiperidium is most similar to Pseudoasteromassaria [34] in its cylindrical, multi-septate, hyaline conidia. However, their ascospores are different, and phylogenetically, both of them are not closely associated. Shearia shares some morphological resemblances to camarosporium-like and stegonsporiopsis-like taxa by its pycnidial conidiomata, holoblastic annellidic conidiogenous cells and distoseptate, pale pigmented conidia [35]. However, neither camarosporium-like nor stegonsporiopsis-like taxa are phylogenetically closely related to Shearia.
Accepted genera in Longiostiolaceae
Crassiperidium M. Matsum. & Kaz. Tanaka, Mycosphere 9(6): 1259 (2018)
Type species: Crassiperidium octosporum M. Matsum. & Kaz. Tanaka (2018)
Notes: [18] established the genus Crassiperidium to accommodate two new species, C. octosporum and C. quadrisporum from Fagus crenata in Japan. Crassiperidium is characterized by ‘globose to depressed globose ascomata with a well-developed ascomatal wall at the sides, clavate asci, broadly fusiform, hyaline ascospores, pycnidial conidiomata, and cylindrical, multi-septate, hyaline conidia produced by annellidic conidiogenous cells’ [18]. Phylogenetic analyses of multi-genes show that Crassiperidium is closely affiliated with Cyclothyriellaceae (Pleosporales, Dothideomycetes), but the exact familial placement was uncertain. Morphologically, Crassiperidium is similar to Pseudoasteromassaria by its ascomatal, pycnidial and conidial characteristics [18, 34]. Also, both are recorded from the same host plant (Fagus crenata). However, phylogenetically, Pseudoasteromassaria belongs to Latoruaceae and is not closely related to Crassiperidium (Fig 1). In addition, their ascospores and conidiogenous cells differ [18, 36].
Longiostiolum Doilom, Ariyaw. & K.D. Hyde, Fungal Diversity 78: 55 (2016)
Type species: Longiostiolum tectonae Doilom, D.J. Bhat & K.D. Hyde, Fungal Diversity 78: 55 (2016)
Notes: Longiostiolum is introduced by [37] as a monotypic genus in the suborder Massarineae with L. tectonae as the type species. The genus is characterized by black, immersed to semi-immersed, uniloculate, globose to subglobose ascostromata with a long central ostiole and phragmosporous ascospores. Longiostiolum was an incertae sedis genus in Pleosporales and [38] listed this in Massarinaceae. In this study, Longiostiolum tectonae grouped sister to Shearia in Longiostiolaceae (Clade C, Fig 1).
Shearia Petr., Annales Mycologici 22 (1–2): 180 (1924)
Type species: Shearia formosa (Ellis & Everh.) Petr., Sydowia 15 (1–6): 216 (1962)
Notes: [39] introduced the genus Shearia with S. magnoliae (Shear) Petr. (basionym Camarosporium magnoliae Shear 1902) as the type species. However, [40] regarded that Stegonsporium formosa Ellis & Everh. (1883) is the oldest name for this taxon and thus introduced the new name, Shearia formosa (Ellis & Everh.) Petr. (1962). [41], however, erroneously listed Shearia magnoliae as the type species of Shearia, although [42] accepted Shearia formosa as the type species. [35] illustrated Shearia fusa (Berk. & M.A. Curtis) M.E. Barr based on HHUF 30474 (on twigs of Magnolia praecocissima var. borealis), Japan.
[42] mentioned that Pleomassaria magnoliae Shear (1902) is the sexual morph of Shearia formosa (as Camarosporium magnoliae). [43] reported Shearia fusa from putative culture of Pleomassaria maxima Ellis & Everh. (current name: Splanchnonema maximum (Ellis & Everh.) M.E. Barr fide [44]. However, the asexual morph of Pleomassaria maxima was reported as Shearia formosa [45]. There are no sequence data available for Pleomassaria maxima (= Splanchnonema maximum) in GenBank, thus it is not possible to check its phylogenetic relationship with other taxa in Pleosporales. With the availability of molecular data, we have included Pleomassaria siparia (CBS 279.74) and Splanchnonema platani (CBS 222.37) in our phylogenetic analyses to represent Pleomassaria and Splanchnonema. However, they are not closely related to each other (Fig 1), whereas CBS 279.74 grouped in Pleomassariaceae and CBS 222.37 grouped in Massarineae.
During this study of microfungi from Magnolia, we recollect Shearia fermosa from Kunming. Below we illustrate and re-describe our new collection as a neo-type. In multi-gene analyses (Fig 1), Shearia formosa (MFLUCC 20–0017, MFLUCC 20–0018, MFLUCC 20–0019) groups in Longiostiolaceae.
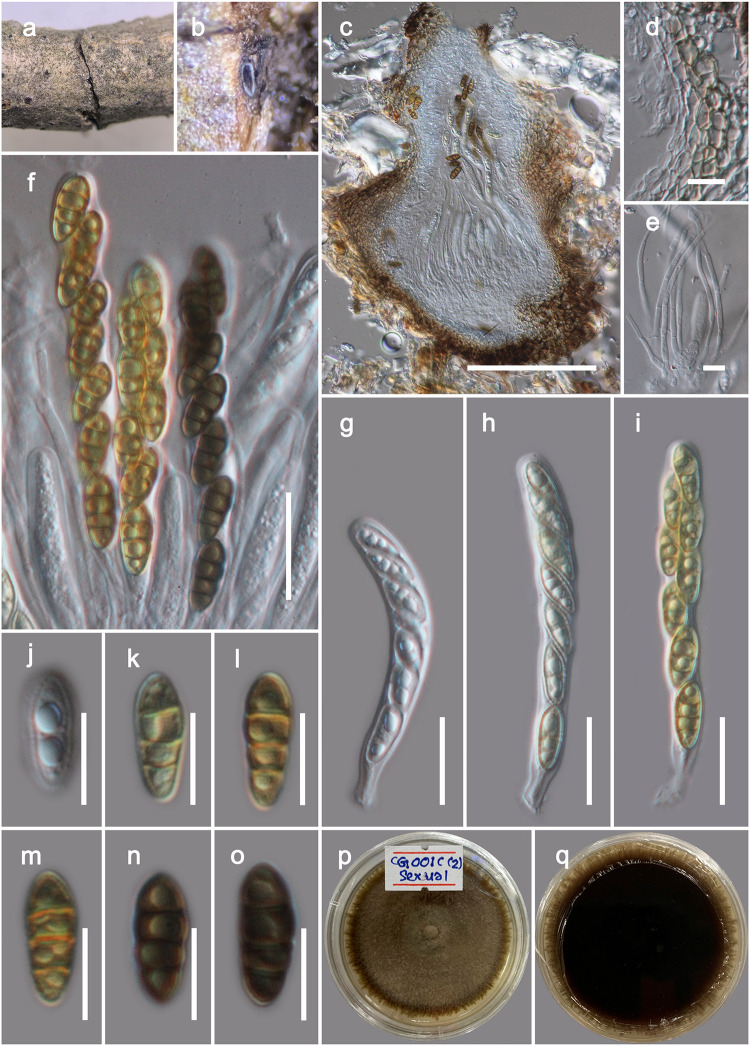
Shearia formosa (Ellis & Everh.) Petr., Sydowia 15 (1–6): 216 (1962) Fig 2.
Fig 2. Shearia formosa (Neotype, MFLU 19–2368).
a Habitat. b, c Conidiomata observed on host substrate. d, e Vertical section of conidiomata. f Cells of pycnidia wall. g–i Conidiogenous cells and developing conidia. j–q Conidia. r–v Colonies on PDA (t and v from the bottom). w, x Conidiomata on PDA. Scale bars: 200 μm (d, e), 20 μm (f, i, j–q), 10 μm (g, h).
≡ Stegonsporium formosa Ellis & Everh., Bull. Torrey bot. Club 10(7): 76 (1883)
= Shearia magnoliae (Shear) Petr., Annls mycol. 22(1/2): 180 (1924)
= Camarosporium magnoliae Shear, Bull. Torrey bot. Club 29: 455 (1902)
[urn:lsid:indexfungorum.org:names:339263]
Facesoffungi number: FoF 07747
Saprobic on dead and living twigs of Magnolia spp. Sexual morph: Undetermined. Asexual morph: Conidiomata 500–700 μm high, 600–900 μm diam. ( μm, n = 10), pseudostromatic, solitary to gregarious, immersed, peridermal to subperidermal, globose to conical, unilocular, dark brown, papillate ostiole, central, circular. Conidiomata wall 15–25 μm wide at the base, 40–70 μm wide at the sides outer layer composed of thick-walled, very dark brown occluded cells, lateral and basal walls composed of peridermal cells and thick-walled, brown cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 8–12 μm long, 5–8 μm wide ( μm, n = 20), holoblastic, annellidic, ampulliform, doliiform or cylindrical, discrete, indeterminate, hyaline, often thick and smooth-walled. Conidia 70–95 μm × 24–30 μm ( μm, n = 30), fusiform, base truncate, apex obtuse, with several transverse and laterally distoseptate, continuous, smooth and thick-walled; initially enveloped in a gelatinous sheath, depressed at the apex, at maturity remaining as a basal lateral sheath.
Culture characteristics: When cultured on PDA, colonies reached up to 40 mm diam after 10 d at 18 °C with smooth and undulate edge, whitish at the beginning, becoming pale brown in center, yellowish green in median area, creamy margin, slightly raised, circular appearance, with sparse to moderate aerial mycelium on the surface with black, scattered conidiomata. On reverse, dirty green or brownish orange, with distinct zonation.
Known Distribution: Magnolia sp.: China [46; this study], Magnolia kobus: Japan [43, 47], Magnolia grandiflora, Magnolia sp.: USA [42, 48]
Material examined: CHINA, Yunnan Province, Heilongtan, Kunming Institute of Botany, 25.137711° N, 102.745185° E, on living branches of Magnolia denudata (Magnoliaceae), 02 February, 2019, D.N. Wanasinghe, DWCG001 (neotype designated here, MFLU 19–2368), ex-neotype living cultures MFLUCC 20–0019, KUMCC 20–0181, ibid. DWCG001E (MFLU 19–2369), living cultures MFLUCC 20–0018, KUMCC 20–0180, ibid. 19 August 2019, on living branches of Magnolia soulangeana DWCG001B (MFLU 20–0091), living cultures MFLUCC 20–0017, KUMCC 20–0182.
Notes: We could not find any herbarium materials for Shearia formosa, Stegonsporium formosa or Camarosporium magnoliae. There are three herbarium specimens were located in the herbarium of Royal Botanic Gardens (Kew, HerbIMI) for Shearia viz. IMI 200150 (1947), IMI 200273 (1904) and IMI 365970 (1904) as S. magnoliae from Magnolia spp. However, all three are labelled as Shearia magnoliae and none of them is regarded as type. Since the holotype of Shearia formosa seems to be lost or not elected, a neotype specimen (MFLU 19–2368) and an ex-neotype culture (MFLUCC 20–0019) are designated here to fix the use of the name. The descriptions and illustrations in [42] for Shearia formosa (from IMI 200150) are similar to MFLU 19–2368 which we describe herein (Fig 2).
Nigrogranaceae Jaklitsch & Voglmayr, Hyde, Stud. Mycol. 85: 54 (2016)
Nigrograna Gruyter, Verkley & Crous, Stud. Mycol. 75: 31 (2012)
Type species: Nigrograna mackinnonii (Borelli) Gruyter, Verkley & Crous, Stud. Mycol. 75: 31 (2012)
Notes: The genus Nigrograna was introduced by [49] with Nigrograna mackinnonii (basionym: Pyrenochaeta mackinnonii Borelli) as the type species. Originally, Pyrenochaeta mackinnonii was reported from a mycetoma of a human. However, the later studies by [17, 50, 51, 52] supplemented more species associated with plants. [53] introduced Nigrograna antibiotica, N. carollii, N. peruviensis and N. yasuniana by synonymizing the endophytic Biatriospora species in [54]. There are 14 species listed in [10]. In this study, we introduce Nigrograna magnoliae as a new species which was collected from Magnolia denudata in Kunming, China.
Nigrograna magnoliae Wanas. sp. nov. Figs 3 and 4
Fig 3. Sexual morph of Nigrograna magnoliae (MFLU 20–0092, Holotype).
a, b Ascomata observed on host substrate. c Vertical sections through an ascoma. d Cells of peridium. e Pseudoparaphyses. f–i Asci. j–o Ascospores. p, q Colonies on PDA (q from the bottom). Scale bars: 100 μm (c), 20 μm (f–i), 10 μm (e, j–o).
Fig 4. Asexual morph of Nigrograna magnoliae (MFLU 20–0092, Holotype).
a, b Conidiomata observed on host substrate. c Vertical sections through a conidioma. d–f Conidiophores and phialides. g Conidia. h, i Colonies on PDA (i from the bottom). Scale bars: 100 μm (c), 10 μm (d–f), 5 μm (g).
[urn:lsid:indexfungorum.org:names: 557331]
Facesoffungi number: FoF 06278
Etymology: The name of the species refers to the host genus, Magnolia.
Saprobic on dead aerial twigs of Magnolia denudata. Sexual morph: Ascomata 200–300 μm high, 150–220 μm diam. ( μm, n = 10), perithecioid, solitary or gregarious, immersed to erumpent through host tissue, subglobose or obpyriform, brown to dark brown, with an ostiole. Ostiole mostly central, brittle. Peridium composed of angular cells, outer layer, dark brown, thick-walled cells, inner layer, hyaline with thin-walled cells. Hamathecium composed of numerous, 2–3 μm wide, filamentous, septate pseudoparaphyses. Asci 60–100 × 8–12 μm (x = 74.2 × 9.5 μm, n = 20), 8-spored, bitunicate, fissitunicate, clavate to cylindric-clavate, short pedicellate, apically rounded, with a minute ocular chamber. Ascospores 12–16 × 5–6.5 μm (x = 14.1 × 5.7 μm, n = 30), overlapping uni to bi-seriate, ellipsoid, yellowish-brown to brown, 3-septate, guttulate. Asexual morph (on the natural host): Coelomycetous. Conidiomata 300–500 μm high, 200–300 μm diam. ( μm, n = 10), pseudostromatic, solitary to gregarious, immersed, peridermal to subperidermal, globose to conical, unilocular, dark brown, papillate ostiole, central, circular. Conidiomata wall 15–30 μm wide, outer layer composed of thick-walled, very dark brown occluded cells, inner layers composed with hyaline, thin cells of textura angularis. Conidiophores, branched at the base, septate, hyaline, straight or sinuous to slightly curved, with pegs along one or two sides and solitary phialides terminally. Phialides 10–30 μm long, 2–4 μm wide ( μm, n = 30), variable in shape, ampulliform-lageniform-subcylindrical, hyaline, smooth-walled. Conidia 3.5–5 μm × 1.3–1.7 μm ( μm, n = 30), oblong to cylindrical or allantoid, subhyaline to hyaline, 1-celled, containing 2 guttules, smooth.
Culture characteristics: On PDA, colonies reached up to 40 mm diam after 12 d at 18 °C. Colony dense, circular, slightly raised, surface smooth, with serrate edge, floccose, greenish grey at the center and brown towards margin from the top and reverse dark brown.
Material examined: CHINA, Yunnan Province, Heilongtan, Kunming Institute of Botany, 25.137711° N, 102.745185° E, on living branches of Magnolia denudata (Magnoliaceae), 02 February, 2019, D.N. Wanasinghe, DWCG001C (MFLU 20–0092), ex-type living cultures MFLUCC 20–0020, KUMCC 20–0178 (asexual), MFLUCC 20–0021, KUMCC 20–0179 (sexual).
Notes: The new fungus was collected from Magnolia denudata in Kunming. It can be labeled a typical Nigrograna taxa based on its ascomata, asci and ascospore characteristics. Phylogenetically it has a close affinity to Nigrograna antibiotica, N. carollii and N. peruviensis, nonetheless this relationship is statistically not supported (Subclade A1, Fig 1). These three species are reported as endophytes from Peru in the phloem of living Ulmus laevis, on living sapwood of wild Hevea brasiliensis, and on living sapwood of wild Virola sp. respectively [54]. All of these species are known only from their culture characteristics, precluding comparing the morphology of either sexual or asexual morph with our new collection. We were able to isolate both ascospores and conidia from the fruiting structures on the natural host. Both single spore isolation of ascospore and conidia formed identical culture morphologies on PDA (Figs 3p, 3q, 4h and 4i) and the DNA based sequences derived from those cultures were also similar in comparisons. Therefore, we introduce Nigrograna magnoliae sp. nov. providing both asexual and sexual morphs.
Occultibambusaceae D.Q. Dai & K.D. Hyde, Fungal Diversity 82: 25 (2016)
Notes: [55] introduced family Occultibambusaceae and currently it comprises five genera viz. Brunneofusispora S.K. Huang & K.D. Hyde, Neooccultibambusa Doilom & K.D. Hyde, Occultibambusa D.Q. Dai & K.D. Hyde, Seriascoma Phook., D.Q. Dai & K.D. Hyde and Versicolorisporium Sat. Hatak., Kaz. Tanaka & Y. Harada [56]. Brunneofusispora was introduced by [57] to accommodate B. sinensis and it was known only from its sexual morph. We collected Brunneofusispora sinensis from Magnolia and in this study, we report the coelomycetous asexual morph from the culture. Hence, we amend Brunneofusispora in order to accommodate its asexual morph.
Brunneofusispora S.K. Huang & K.D. Hyde, Fungal Diversity 95: 36 (2019) amended
[urn:lsid:indexfungorum.org:names:555599]
Facesoffungi number: FoF04862
Saprobic on dead wood or twigs. Sexual morph. See [57]. Asexual morph. Coelomycetous. Conidiomata solitary, superficial, globose to sub-globose, unilocular, dark brown. Conidiomata wall composed of thick-walled, dark brown cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells enteroblastic, phialidic, ampulliform, doliiform or cylindrical, discrete, indeterminate, hyaline, smooth-walled. Conidia oblong to cylindrical, 1-celled, hyaline, smooth, guttulate.
Type species. Brunneofusispora sinensis S.K. Huang & K.D. Hyde
Notes: Based on the combined SSU, LSU, RPB2 and TEF sequence analyses of Pleosporales, one of our new strain (MFLUCC 20–0016) clusters with Brunneofusispora sinensis with strong bootstrap support (Subclade B1, Fig 1).
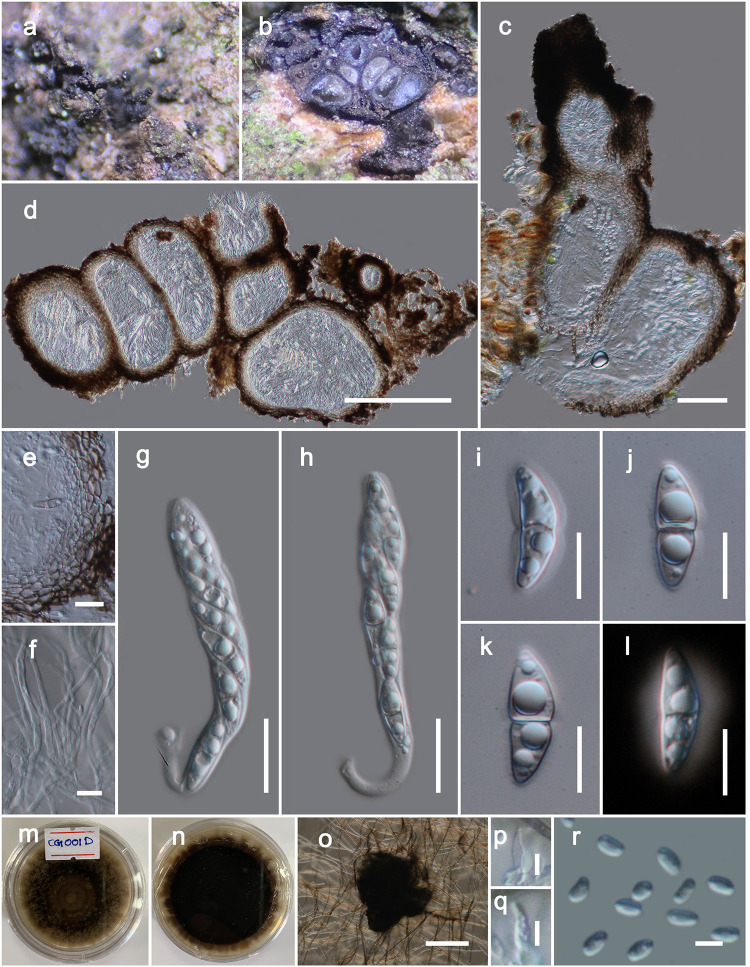
Brunneofusispora sinensis S.K. Huang & K.D. Hyde, Fungal Diversity 95: 38 (2019). Fig 5
Fig 5. Brunneofusispora sinensis (MFLU 20–0093).
a Ascomata observed on host substrate. b–d Vertical sections through ascomata. e Cells of peridium. f Pseudoparaphyses. g, h Asci. i–l Ascospores (Note the sheath stained with Indian Ink in l). m, n Colonies on PDA (n from the bottom). o Squashed conidiomata. p, q Conidiogenous cells. r Conidia. Scale bars: 50 μm (c), 200 μm (d), 20 μm (e, g, h), 10 μm (f, j–o), 100 μm (o), 5 μm (p–r).
[urn:lsid:indexfungorum.org:names:555600]
Facesoffungi number: FoF04863
Saprobic on dead wood or twigs. Sexual morph. Ascomata 200–300 μm high, 150–300 μm diam. ( μm, n = 10), perithecial, solitary to scattered, immersed, eventually erumpent, globose to subglobose, uni to multi-loculate, glabrous, dark brown to black, ostiolate. Ostioles 80–150 μm long, 40–70 μm diam. ( μm, n = 5), papillate, central. Peridium 20–30 μm wide, equally thick-walled, composed of blackened cells, arranged in a textura angularis. Hamathecium composed of numerous, 1.5–2.5 μm wide, filamentous, septate pseudoparaphyses. Asci 60–100 × 10–14 μm (x = 82.4 × 12.3 μm, n = 20), 8-spored, bitunicate, fissitunicate, cylindric-clavate to clavate, pedicellate, rounded at the apex, with an ocular chamber. Ascospores 16–20 × 6–8 μm (x = 18.5 × 6.4 μm, n = 30), overlapping biseriate, hyaline, broadly fusiform, 1-septate, deeply constricted at the septum, smooth-walled, with large guttules, surrounded by a thick mucilaginous sheath. Asexual morph. Coelomycetous. Conidiomata 120–160 μm high, 80–120 μm diam. ( μm, n = 5), solitary, superficial, globose to sub-globose, unilocular, dark brown. Conidiomata wall composed of thick-walled, very dark brown cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 6–7.5 μm long, 2.5–3 μm wide ( μm, n = 10), enteroblastic, phialidic, ampulliform, doliiform or cylindrical, discrete, indeterminate, hyaline, smooth-walled. Conidia 3–4 μm × 1.9–2.5 μm ( μm, n = 25), mostly cylindrical or ovoid, 1-celled, hyaline, smooth, guttulate.
Culture characteristics: On PDA, colonies reached up to 40 mm diam after 15 d at 18 °C. Colony dense, circular, slightly raised at the center and raised at the margin, surface smooth, with serrate edge, floccose to fluffy, greenish grey to brown from the top and reverse dark brown, sporulated after 15–18 weeks.
Material examined: CHINA, Yunnan Province, Heilongtan, Kunming Institute of Botany, 25.137711° N, 102.745185° E, on living branches of Magnolia denudata (Magnoliaceae), 02 February, 2019, Dhanushka N. Wanasinghe, DWCG001D (MFLU 20–0093), living cultures MFLUCC 20–0016, KUMCC 20–0183.
Notes: In this study we have acquired DNA from the mycelium of a sexual morph and in multi-gene phylogeny, our novel strain and the Brunneofusispora sinensis group in a monophyletic clade (subclade B1, Fig 1). Even though B1 is not strongly supported, there were only three bp differences (not including gaps) in the comparison of the 525 nucleotides across the ITS regions. Morphological characteristics i.e. asci and ascospores, are not significantly different to each other in shape or dimensions [57]. Therefore, we identify our collection as Brunneofusispora sinensis. We observed its asexual morph from the culture and therefore we amended the generic and species descriptions herein. Nevertheless, it is worthy to remark that we note the ascospores of our new isolate were remaining hyaline at maturity whereas brownish in the holotype of Brunneofusispora sinensis.
Discussion
The genus Shearia is one of the ‘most distinctive’ genera in coelomycetes [42]. Its morphology is highly conspicuous and easy to distinguish from other known dematiaceous coelomycetes [35, 42]. Several studies have discussed the morphology of this genus [35, 42]. [42, 43 and 45] reported the sexual morph of Shearia as Pleomassaria. However, Pleomassaria maxima, which was reported as the sexual morph of Shearia formosa [45] and Shearia fusa [43], has been transferred to Splanchnonema maximum (Ellis & Everh.) M.E. Barr (1993). [58] showed that Pleomassaria s. str. grouped in Prosthemium Kunze in their phylogenetic analyses. Based on this result, [59, 60] proposed to reduce Pleomassaria under Prosthemium. Furthermore, Splanchnonema s. str. has been accepted as a well-established genus in Pleomassariaceae [38, 56]. Hence, we assume that taxa linked to Shearia (Shear 1902) such as Camarosporium magnoliae [36, 42, 45], are not related to Prosthemium (= Pleomassaria) s. str. or Splanchnonema s. str., which are placed in Pleomassariaceae. This assumption has been confirmed by our phylogenetic analyses, placing Shearia s. str. groups in Longiostiolaceae, whereas Splanchnonema and Prosthemium strains are phylogenetically apart (Fig 1). Currently, Pleomassaria magnoliae (Shear 1902) and Splanchnonema maximum [36, 45] are lacking DNA sequencing to confirm their placements in Pleosporales. Consequently, doubts exist regarding the sexual morph of Shearia. Since both sexual morphs were collected from Magnola spp., we conclude that further collections from this host are necessary.
The distribution of Magnolia is variable but concentrated particularly around temperate and tropical South East and East Asia. They are treasured around the world as ornamental trees due to their attractive flowers and foliage and are used as timber and medicine by local and international communities. However, 48% of all Magnolia species are endangered and facing habitat loss [61]. The Kunming Botanical Garden functions as an ex-situ conservation for endangered, endemic and economically important plant species native to the Yunnan Plateau and the southern Hengduan Mountains [6]. Micro-fungi on Magnolia have been studied for decades around the world, and so far over 1000 fungal species have been reported on this host [62]. However, only 46 fungal species have been reported from China, across just 14 Magnolia species (M. alba, M. albosericea, M. candolii, M. coco, M. cylindrica, M. delavayi, M. denudata, M. grandiflora, M. liliiflora, M. officinalis, M. paenetalauma, M. soulangeana, M. stellata and M. zenii) [62]. This study highlights the Kunming Botanical Garden as an ideal locale to conduct diverse research on the micro-fungal occurrences on Magnolia as it is a repository of a great number of Magnolia species.
Supporting information
The newly generated sequences are indicated in bold.
(DOC)
Acknowledgments
Wanasinghe and Ratchadawan Cheewangkoon thank Chiang Mai University for partially supporting this research. Austin Smith is thanked for the English editing in the manuscript. Nimali I. de Silva and Danushka S. Tennakoon are thanked for the help with culture and herbarium deposits.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Peter E. Mortimer thanks the National Science Foundation of China and the Chinese Academy of Sciences for financial support under the following grant: 41761144055. Dhanushka Wanasinghe would like to thank CAS President’s International Fellowship Initiative (PIFI) for funding his postdoctoral research (number 2019PC0008) and the 64th batch of China Postdoctoral Science Foundation (grant no.: Y913083271). Nalin Wijayawardene thanks the National Natural Science Foundation of China (No. NSFC 31950410558).
References
- 1.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature 2000; 403:853–858. 10.1038/35002501 [DOI] [PubMed] [Google Scholar]
- 2.Yang Y, Tian K, Hao J, Pei S, Yang Y. Biodiversity and biodiversity conservation in Yunnan, China. Biodiver Conserv. 2004; 13:813–826. 10.1023/B:BIOC.0000011728.46362.3c [DOI] [Google Scholar]
- 3.Zhang LS, Fang XQ. Palaeogeography of China: the Formation of China's Natural Environment. Science Press, Beijing: 2012 [Google Scholar]
- 4.Feng B, Yang Z. Studies on diversity of higher fungi in Yunnan, southwestern China: A review Plant divers. 2018; 40:165–171. 10.1016/j.pld.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Q, Hou LW, Crous PW, Cai L. Didymellaceae revisited. Stud in Mycol. 2017; 87:105–159. 10.1016/J.SIMYCO.2017.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen G, Sun W. The role of botanical gardens in scientific research, conservation, and citizen science. Plant Divers. 2018; 40:181–188. 10.1016/j.pld.2018.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Shi Z, Ye L, Su B. Analysis on the Characteristics of Extreme Weather Events in Kunming City during Recent 20 Years. IOP Conf. Series: Environ Earth Sci. 2019; 252:042124. 10.1088/1755-1315/252/4/042124 [DOI]
- 8.Chomnunti P, Hongsanan S, Aguirre-Hudson B, Tian Q, Peršoh D, Dhami MK, et al. The sooty moulds. Fungal Divers. 2014; 66:1–36. 10.1007/s13225-014-0278-5 [DOI] [Google Scholar]
- 9.Jayasiri SC, Hyde KD, Ariyawansa HA, Bhat DJ, Buyck B, Cai L et al. The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015; 74:3–18. 10.1007/s13225-015-0351-8 [DOI] [Google Scholar]
- 10.Index Fungorum. 2020 http://www.indexfungorum.org/names/Names.asp (Accessed 1 February 2020).
- 11.White TJ, Bruns T, Lee SJWT, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR—Protocols and Applications. 1990; 18:315–322. 10.1016/b978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- 12.Rehner SA, Samuels GJ. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol Res. 1994; 98, 625–634. 10.1016/S0953-7562(09)80409-7 [DOI] [Google Scholar]
- 13.Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990; 172:4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sung G-H, Sung J-M, Hywel-Jones NL, Spatafora JW. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Mol Phylogenet Evol. 2007; 44, 1204–1223. 10.1016/j.ympev.2007.03.011 [DOI] [PubMed] [Google Scholar]
- 15.Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005; 97:84–98. 10.3852/mycologia.97.1.84 [DOI] [PubMed] [Google Scholar]
- 16.Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes evidence from an RNA polymerase II subunit. Mol Biol Evol. 1999; 16:1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- 17.Jaklitsch WM, Voglmayr H. Hidden diversity in Thyridaria and a new circumscription of the Thyridariaceae. Stud Mycol. 2016; 85:35–64. 10.1016/j.simyco.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumura M, Kato W, Hashimoto A, Takahashi YS, Shirouzu T, Tanaka K. Crassiperidium (Pleosporales, Dothideomycetes), a new ascomycetous genus parasitic on Fagus crenata in Japan. Mycosphere. 2018; 9:1256–1267. 10.5943/mycosphere/9/6/13 [DOI] [Google Scholar]
- 19.Zhang S-N, Hyde KD, Jones EBG, Jeewon R, Cheewangkoon R, Liu J-K. Striatiguttulaceae, a new pleosporalean family to accommodate Longicorpus and Striatiguttula gen. nov. from palms. MycoKeys. 2019; 49: 99–129. 10.3897/mycokeys.49.30886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuraku S, Zmasek CM, Nishimura O, Katoh K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013; 41:W22–W28. 10.1093/nar/gkt389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katoh k, Rozewicki J, Yamada KD.–MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization, Briefings in Bioinformatics, 2017; bbx108. Available from: 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acid Symp Ser. 1999; 41:95–98 [Google Scholar]
- 23.Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008, 24:581–583. 10.1093/bioinformatics/btm388 [DOI] [PubMed] [Google Scholar]
- 24.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001; 17:754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- 25.Rannala B, Yang Z. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J Mol Evol 1996; 43:304–311. 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- 26.Zhaxybayeva O, Gogarten JP. Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC genomics. 2002; 3:4 10.1186/1471-2164-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rambaut A, Drummond AJ. 2007 Tracer v1, 4. http://beast.bio.ed.ac.uk/Tracer. (Accessed: 15 February 2020)
- 28.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008; 57,758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 29.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014; 30:1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, 2010, pp. 1–8. 10.1109/GCE.2010.5676129 [DOI]
- 31.Rambaut A. 2012 FigTree version 1.4.0. http://tree.bio.ed.ac.uk/software/figtree/ (Accessed: 8 February 2020)
- 32.Liu JK, Hyde KD, Jeewon R, Phillips AJ, Maharachchikumbura SS, Ryberg M et al. Ranking higher taxa using divergence times: a case study in Dothideomycetes. Fungal Divers. 2017; 84:75–99. 10.1007/s13225-017-0385-1 [DOI] [Google Scholar]
- 33.Phukhamsakda C, McKenzie EHC, Phillips AJL, Jones EBG, Bhat DJ, Stadler M et al. Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 2020; 101:1–204. 10.1007/s13225-020-00448-4 [DOI] [Google Scholar]
- 34.Ariyawansa HA, Hyde KD, Jayasiri SC, Buyck B, Chethana KWT, Dai DQ, et al. Fungal diversity notes 111–252: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015; 75:27–274. 10.1007/s13225-015-0346-5 [DOI] [Google Scholar]
- 35.Wijayawardene NN, Hyde KD, Wanasinghe DN, Papizadeh M, Goonasekara ID, Camporesi E et al. Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Divers 2016; 77:1–316. 10.1007/s13225-016-0360-2 [DOI] [Google Scholar]
- 36.Tanaka K, Hirayama K, Yonezawa H, Sato G, Toriyabe A, Kudo H et al. Revision of the Massarineae (Pleosporales, Dothideomycetes). Stud Mycol. 2015; 82:75–136. 10.1016/j.simyco.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li GJ, Hyde KD, Zhao RN, Hongsanan S, Abdel-Aziz FA, Abdel-Wahab MA et al. Fungal Divers notes 253–366: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016; 78:1–237. 10.1007/s13225-016-0366-9 [DOI] [Google Scholar]
- 38.Wijayawardene NN, Hyde KD, Lumbsch HT, Liu JK, Maharachchikumbura SSN, Ekanayaka AH et al. Outline of Ascomycota: 2017. Fungal Divers. 2018; 88:167–263. 10.1007/s13225-018-0394-8 [DOI] [Google Scholar]
- 39.Petrak F. Mykologische Notizen. VII. Annales Mycologici. 1924; 22(1–2):1–182. [Google Scholar]
- 40.Petrak F. Ergebnisse einer Revision der Grundtypen verschiedener Gattungen der Askomyzeten und Fungi Imperfecti. Sydowia. 1962, 16(1–6):353–361 [Google Scholar]
- 41.Sutton BC. Coelomycetes VI. Nomenclature of generic names proposed for Coelomycetes Mycol Pap. 1977; 141:1–253. [Google Scholar]
- 42.Sutton BC. The Coelomycetes. fungi imperfecti with pycnidia, acervuli and stromata. Commonwealth Mycological Institute, Kew, Surrey, England. 1980.
- 43.Tanka K, Ooki Y, Hatakeyama S, Harada Y, Barr ME. Pleosporales in Japan (5): Pleomassaria, Asteromassaria, and Splanchnonema. Mycoscience. 2005; 46:248–260. 10.1007/S10267-005-0245-9 [DOI] [Google Scholar]
- 44.Species Fungorum. 2020 http://www.speciesfungorum.org/Names/Names.asp (Accessed 8 March 2020).
- 45.Tubaki K, Murata S, Nakagiri A. Materials for the fungus flora of Japan (33). Trans Mycol Soc Jpn. 1983; 24:121–125. 10.1007/BF02268632 [DOI] [Google Scholar]
- 46.Teng SC. Fungi of China. Mycotaxon, Ltd, Ithaca, NY, 1996. [Google Scholar]
- 47.Kobayashi T. Index of fungi inhabiting woody plants in Japan. Host, Distribution and Literature. Zenkoku-Noson-Kyoiku Kyokai Publishing Co., Ltd; 2007.
- 48.Miller JW. Bureau of Plant Pathology. Tri-ology Techn. Rep. Div. Pl. Indust., Florida. 1990; 29:6.
- 49.De Gruyter J, Woudenberg JHC, Aveskamp AA, Verkley GJM, Groenewald JZ, Crous PW. Redisposition of Phoma-like anamorphs in Pleosporales. Stud Mycol. 2013; 75:1–36. 10.3114/sim0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hyde KD, Norphanphoun C, Abreu VP, Bazzicalupo A, Chethana KWT, Clericuzio M et al. Fungal diversity notes 603–708: Taxonomic and phylogenetic notes on genera and species. Fungal Divers 2017; 87:1–235. 10.1007/s13225-017-0391-3 [DOI] [Google Scholar]
- 51.Tibpromma S, Hyde KD, Jeewon R, Maharachchikumbura SSN, Liu JK, Bhat DJ et al. Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2017; 83:1–261. 10.1007/s13225-017-0378-0 [DOI] [Google Scholar]
- 52.Zhao YZ, Zhang ZF, Cai L, Peng WJ, Liu F. Four new filamentous fungal species from newly-collected and hive-stored bee pollen. Mycosphere. 2018; 9:1089–1116. 10.5943/mycosphere/9/6/3 [DOI] [Google Scholar]
- 53.Kolařík M. New taxonomic combinations in endophytic representatives of the genus Nigrograna. Czech Mycol. 2018; 70:123–126. [Google Scholar]
- 54.Kolařík M, Spakowicz DJ, Gazis R, Shaw J, Kubátová A, Nováková A et al. Biatriospora (Ascomycota: Pleosporales) is an ecologically diverse genus including facultative marine fungi and endophytes with biotechnological potential. Plant Syst Evol. 2017; 303:35–50. 10.1007/s00606-016-1350-2 [DOI] [Google Scholar]
- 55.Dai DQ, Phookamsak R, Wijayawardene NN, Li WJ, Bhat DJ, Xu JC, et al. Bambusicolous fungi. Fungal Divers. 2017; 82:1–105. 10.1007/s13225-016-0367-8 [DOI] [Google Scholar]
- 56.Hongsanan S, Hyde KD, Phookamsak R, Wanasinghe DN, Mckenzie EHC, Sarma VV et al. Refined families of Dothideomycetes. Fungal Divers. 2020. Forthcoming [Google Scholar]
- 57.Phookamsak R, Hyde KD, Jeewon R, Bhat DJ, Gareth EB, Maharachchikumbura SSN et al. Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 2019; 95:1–273. 10.1007/s13225-019-00421-w [DOI] [Google Scholar]
- 58.Tanaka K, Mel’nik VA, Kamiyama M, Hirayama K, Shirouzu T. Molecular phylogeny of two coelomycetous fungal genera with stellate conidia, Prosthemium and Asterosporium, on Fagales trees. Botany. 2010; 88:1057–1071. 10.1139/B10-078 [DOI] [Google Scholar]
- 59.Wijayawardene NN, Crous PW, Kirk PM, Hawksworth DL, Boonmee S, Braun U et al. Naming and outline of Dothideomycetes–2014 including proposals for the protection or suppression of generic names. Fungal Divers. 2014; 69:1–55. 10.1007/s13225-014-0309-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rossman AY, Crous PW, Hyde KD, Hawksworth DL, Aptroot A, Bezerra JL et al. Recommended names for pleomorphic genera in Dothideomycetes. IMA Fungus 2015; 6:507–523. 10.5598/imafungus.2015.06.02.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rivers M, Beech E, Murphy L, Oldfield S. The Red List of Magnoliaceae—revised and extended. BGCI. Richmond, UK. 2016.
- 62.Farr DF, Rossman AY. Fungal Databases, U.S. National Fungus Collections, ARS, USDA. https://nt.ars-grin.gov/fungaldatabases/ (Accessed 9 March 2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The newly generated sequences are indicated in bold.
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.