Abstract
Cetuximab, a monoclonal antibody targeting EGFR, is a standard of care for the treatment for locally advanced or metastatic head and neck squamous cell carcinoma (HNSCC). However, despite overexpression of EGFR in over 90% of HNSCC lesions, most HNSCC patients fail to respond to cetuximab treatment. In addition, there are no available biomarkers to predict sensitivity or resistance to cetuximab in the clinic. Here, we sought to advance precision medicine approaches for HNSCC by identifying PI3K-mTOR signaling-network-specific cetuximab resistance mechanisms. We first analyzed the frequency of genomic alterations in genes involved in the PI3K-mTOR signaling circuitry in the HNSCC TCGA dataset. Experimentally, we took advantage of CRISPR/Cas9 genome editing approaches to systematically explore the contribution of genomic alterations in each tumor suppressor gene (TSG) controlling the PI3K-mTOR pathway to cetuximab resistance in HNSCC cases that do not exhibit PIK3CA mutations. Remarkably, we found that many HNSCC cases exhibit pathway-specific gene copy number loss of multiple TSGs that normally restrain PI3K-mTOR signaling. Among them, we found that both engineered and endogenous PTEN gene deletions can mediate resistance to cetuximab. Our findings suggest that PTEN gene copy number loss, which is highly prevalent in HNSCC, may result in sustained PI3K/mTOR signaling independent of EGFR, thereby representing a promising mechanistic biomarker predictive of cetuximab resistance in this cancer type. Further prospective studies are needed to investigate the impact of PTEN loss on cetuximab efficacy in the clinic.
Keywords: PTEN, tumor suppressor gene, cetuximab, EGFR, mTOR, head and neck cancer, signal transduction
Introduction
Head and neck squamous cell carcinomas (HNSCC), which include cancers of the oral cavity, oropharynx, and larynx, are among the top 10 most common cancers worldwide, with over 65,000 estimated new cases per year in the United States alone, and accounting for about 15,000 estimated cancer deaths (1). Despite aggressive multimodality therapies and recent advances in treatment, the prognosis of patients with HNSCC is still poor, with 5-year survival estimates of approximately 65% (1), which are even lower if the cancers are detected at advanced stages. Thus, there is an urgent unmet need to develop new therapeutic options to treat patients with HNSCC. The recent deep sequencing of HNSCC has revealed that this malignancy harbors a remarkable multiplicity and diversity of genomic alterations, with particular emphasis on aberrant activation of the epidermal growth factor receptor (EGFR) and PI3K/Akt/mTOR signaling pathways (2–5).
Over 90% of HNSCC lesions overexpress EGFR, one of the upstream molecules of PI3K/mTOR signaling (6,7), and EGFR expression is associated with poorer survival of patients with HNSCC (8,9). Cetuximab, a chimeric IgG1 monoclonal antibody against the EGFR extracellular domain, was approved in 2006 by the US Food and Drug Administration (FDA) for the treatment of HNSCC patients based on the results of seminal clinical trials (10–12). However, the overall response rates of cetuximab as a single agent or the increased response rates observed when cetuximab is added to radiation or chemotherapy are less than 10 to 20% (10–12), much lower than initially expected considering the high level of EGFR expression in HNSCC. In addition, recent phase III clinical trials showed that cetuximab-based chemoradiation therapy (CRT) demonstrated inferior efficacy to cisplatin-based CRT in HPV-positive oropharyngeal cancer (13,14).
EGFR-targeted therapies have demonstrated improvement in clinical outcomes in several cancer types, including non-small cell lung cancer and colorectal cancer (15–18), where, unlike HNSCC, molecular biomarkers have been identified to determine which patients are most likely to benefit from these agents. In the case of HNSCC, there are no biomarkers available in the clinic to predict sensitivity or resistance to cetuximab despite the recent characterization of the molecular alterations of this malignancy. The upregulation and activation of multiple receptor tyrosine kinases (RTKs), including HER3, c-MET, and AXL have been reported to mediate intrinsic or acquired resistance to cetuximab in HNSCC (19–22). In addition, we have shown that mutation of the PIK3CA gene, the most commonly mutated oncogene in HNSCC (5), as well as mutant RAS gene can confer cetuximab resistance in HNSCC experimental models (23). However, ~75 % of the HNSCC lesions lack PIK3CA or RAS mutations. Additional biomarkers predictive of cetuximab resistance or sensitivity are warranted to further advance precision medicine in HNSCC.
In this study, we took advantage of the CRISPR/Cas9 genome editing approaches to systematically explore the contribution of genomic alteration in the PI3K/mTOR signaling network to cetuximab resistance in HNSCC cases that do not exhibit PIK3CA mutations. Remarkably, we found that many HNSCC cases exhibit pathway-specific gene copy number loss of multiple tumor suppressor genes (TSGs) involved in PI3K/mTOR signaling. Among them, we found that both engineered and endogenous PTEN loss can mediate resistance to cetuximab, due to sustained PI3K/mTOR signaling activity. Our findings suggest that PTEN gene copy number loss, which is highly prevalent in HNSCC, may represent a promising biomarker predictive of cetuximab resistance in this disease.
Materials and methods
Antibodies and reagents
Antibodies against pEGFRY1068 (#2236), pERK1/2T202/Y204 (#4370), ERK1/2 (#4696), pS6S235/S236 (#2211), S6 (#2217), α-tubulin (#3873), and GAPDH (#2118) were purchased from Cell Signaling Technology (Beverly, MA). Antibody against EGFR (#sc-03) was purchased from Santa Cruz Biotechnology (Dallas, TX). Erlotinib was purchased from Selleck Chemical (Houston, TX), and cetuximab was obtained from the pharmacy of UCSD Moores Cancer Center (La Jolla, CA).
Cell lines, culture condition, and transfection
The human HNSCC cell lines HN12, CAL27 and Detroit 562 were genetically characterized as part of NIH/NIDCR Oral and Pharyngeal Cancer Branch cell collection, and have been described previously (24,25). All cells were cultured in DMEM (D-6429, Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum (FBS) at 37°C in 5% CO2. All cell lines underwent DNA authentication by multiplex STR profiling (Genetica DNA Laboratories, Inc. Burlington, NC) prior to the described experiments to ensure consistency in cell identity. No presence of Mycoplasma was found according to Mycoplasma Detection Kit-QuickTest from Biomake. PTEN-, TSC2-, STK11-, and EIF4EBP1-knockout cells were engineered using the CRISPR/Cas9 system as described previously (26). LentiCRISPR v2 plasmid was purchased from Addgene (#52961). A single guide RNA (sgRNA) was designed according to Zhang Lab protocol (27). sgRNAs targeting sequences for PTEN, TSC2, STK11, and EIF4E-BP1 were 5’-ACCGCCAAATTTAATTGCAG-3’, 5’-GGTCGCGGATCTGTTGCAGC-3’, 5’-CAGGTGTCGTCCGCCGCGAA-3’, and 5’-GCACCACCCGGCGAGTGGCG-3’, respectively. These oligo were cloned into lentiCRISPR v2 plasmid, and packaged into lentivirus in HEK 293T cells. HN12 and CAL27 cells were infected with lentiviruses for 2 days. The infected cells were selected with puromycin (4 μg/ml) for 3 days. Single cell clones were obtained, and knockout of each gene was validated by western blot analysis. siRNA for EGFR was purchased from Sigma (MISSION siRNA human EGFR SIHK0659).
Western blot analysis and image quantifications
Cells and tissues were lysed on ice in RIPA buffer (0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 50 mM Tris/HCl, pH 7.5, 1.0% NP-40) containing Halt™ Protease and Phosphatase Inhibitor Cocktail (#78440, ThermoFisher Scientific). Protein concentrations were measured by Bio-Rad Protein Assay (Bio-Rad, Hercules, CA). Equal amounts of total proteins were subjected to SDS-polyacrylamide gel electrophoresis and transferred to PVDF membranes. Membranes were blocked with 3% BSA or 5% nonfat dry milk in TBS-T buffer (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.1% [v/v] Tween-20) for 1 h, and then incubated with primary antibodies in blocking buffer for 1 h at room temperature. Detection was conducted by incubating the membranes with horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit IgG secondary antibodies (Southern Biotech, Birmingham, AL) used at a dilution of 1:10,000 in 5% milk-TBS-T buffer for 1 h at room temperature, and visualized with Immobilon Western Chemiluminescent HRP substrate (Millipore, Billerica, MA). The image quantifications were performed with ImageJ.
Cellular proliferation and viability assay
Cells were cultured in 96-well plates and treated with drugs for 72 hours, then incubated with AlamarBlue (Invitrogen, Carlsbad, CA) for 2 h at 37°C. Absorbance was read at 570 nm, using 600 nm as a reference wavelength. Each experiment was repeated three times in triplicate.
Sphere formation assay
Cells were seeded in 96-well ultra-low attachment culture plates (Corning, Tweksbury, MA) at 100 cells per well. Medium consisted of serum free DMEM/F12 Glutamax supplement medium (#10565042, Thermo Fisher Scientific), basis fibroblast growth factor (bFGF: 20 ng/ml, #13256029, Thermo Fisher Scientific), epithelial growth factor (EGF: 20 ng/ml, #PHG0313, Thermo Fisher Scientific), B-27 (1:50 dilution, #17504044, Thermo Fisher Scientific), and N2 supplement (1:100 dilution, #17502–048, Thermo Fisher Scientific). Ten days after seeding, photographs were obtained, and the sizes and numbers of sphere colonies on each well were counted using a microscope. Each experiment was repeated three times in triplicate.
Animal work
All studies in mice were approved by the Institutional Animal Care and Use Committees (IACUC) of the University of California, San Diego (protocol #S15195) or the University of California, San Francisco (protocol # AN173372–02C). To establish tumor xenografts, 2.0 × 106 cells were transplanted into the both flanks of athymic nude mice (female, four to six weeks old) (Charles River Laboratories, Wilmington, MA), and when the tumor volume reached approximately 100 mm3, the mice were randomized into groups and treated by intraperitoneal injection (ip) with cetuximab (40 mg/kg, three times a week) or control diluent (10 tumors in 5 mice per each group). Tumor volume was calculated by using the formula length × width × width/2. The mice were euthanized at the indicated time points and tumors isolated for histologic and immunohistochemical evaluation.
Tissue analysis
All samples were fixed in zinc formalin (Z-Fix, Anatech) and embedded in paraffin; 5 μm sections were stained with Hematoxylin-Eosin for diagnostic purposes. For immunohistochemistry (IHC) studies, the sections were deparaffinized, and hydrated through graded ethanols. The slides were extensively washed with distilled water and antigen retrieval was performed by high temperature treatment with 10 mM citric acid in a microwave. After washing with water and PBS, the slides were successively incubated with the primary and secondary antibodies, and the ABC reagent (Vector Laboratories, Burlingame, CA). The reaction was developed with 3–3’-diamonobenzifdine under microscopic control.
Genomic data analysis
Gene mutation and copy number variation analyses where performed using publicly available data generated by The Cancer Genome Atlas consortium, accessed through cBio portal (www.cbioportal.org) (28,29).
Data and statistical analysis
Data were analyzed using GraphPad Prism version 8.02 for Windows (GraphPad Software, San Diego, CA). The overall survival and progression free survival time were assessed using the Kaplan-Meier method and compared using the log-rank test. Comparisons between experimental groups were made using unpaired t test. The correlation between PTEN mRNA and protein expression was evaluated using Pearson test. P < 0.05 was considered to be statistically significant.
Results
The landscape of genomic alterations in PI3K/mTOR pathway in HNSCC.
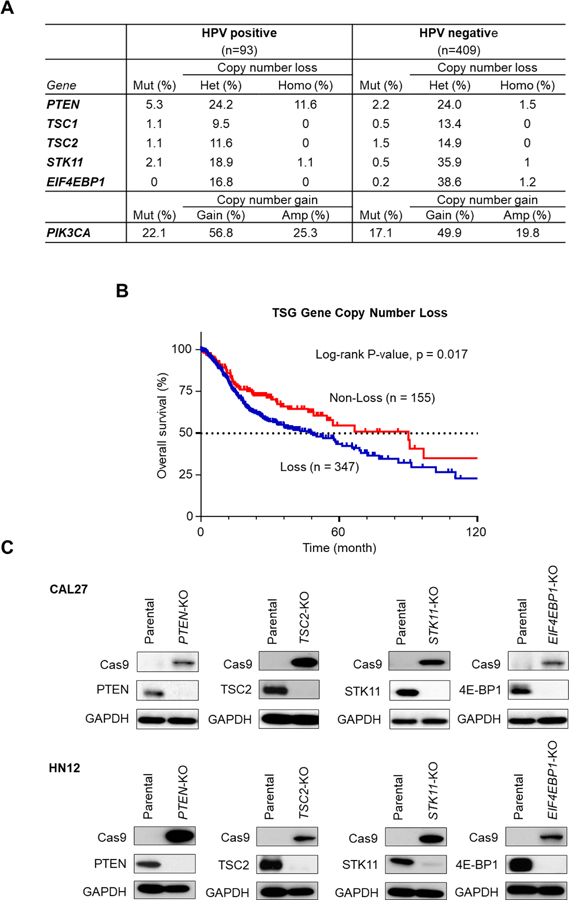
Pathway-specific analysis of the HNSCC oncogenome suggests that most genomic alterations are involved in aberrant mitogenic signaling, with particular emphasis on the PI3K/mTOR pathway (2). In this study, we focused on the TSGs in the PI3K/mTOR signaling circuitry, including PTEN, TSC1/2, STK11, and EIF-4EBP1, the loss of which are expected to result in persistent activation of mTOR signaling. We first analyzed the frequency of genomic alteration in the TCGA HNSCC dataset (n = 502). As shown in Figure 1A, frequent mutations of PTEN (5.4, 2.2%) and less frequent mutations in the TSC1 (1.1, 0.5%), TSC2 (1.1, 1.5%), STK11 (2.2, 0.5%), and EIF-4EBP1 (0, 0.2%) genes were observed in HPV-positive (n=93) and HPV-negative (n=409) HNSCC patients, respectively. On the other hand, the loss of copy number in PTEN (35.8, 25.5%), TSC1 (9.5, 13.4%), TSC2 (11.6, 14.9%), STK11 (18.9, 36.0%), and EIF-4EBP1 (16.8, 39.8%) were much more frequently observed in both HPV-positive (n=93) and HPV-negative HNSCC patients (n=409), respectively. In addition, the copy number loss of either PTEN, TSC1/2, STK11, or EIF-4EBP1 was associated with poorer survival of HNSCC patients in TCGA HNSCC cohort (log-rank test p=0.017) (Fig. 1B), although mutations of these genes individually was not significantly associated with overall and progression free survival (Supplementary Fig. 1). Thus, most of the alterations in TSGs in the PI3K/mTOR signaling pathway are not mutations, but losses of gene copy number, which are significantly associated with worse clinical outcome in HNSCC patients. To elucidate the role of loss of PI3K/mTOR TSGs in resistance to cetuximab, we sought to generate HNSCC isogenic cell line panels with PTEN, TSC2, STK11, or EIF4EBP1 gene knockouts using the CRISPR-Cas9 system. We employed CAL27 and HN12 cells, both of which are human HNSCC cell lines that harbor no genomic alterations in PI3K/mTOR and RAS pathway genes (25). Single guide RNAs (sgRNAs) targeting each gene were cloned into pLentiCRISPR-v2 vector, and packaged into lentiviruses using HEK 293T cells. The packaged lentiviruses were transduced into CAL27 and HN12 cells, and cells were selected with puromycin, (4 μg/ml) for 3 days. Single cell clones for each deleted gene were isolated and gene knockouts were validated by western blot of Cas9 expression and depletion of each protein expression (Fig. 1C).
Figure 1. Genomic alterations in genes involved in PI3K/Akt/mTOR signaling in HNSCC.
(A) Frequency of genomic alterations in genes involved in PI3K/Akt/mTOR signaling in the HNSCC TCGA dataset (n = 504), including HPV-positive (n = 95) and negative (n = 409) lesions. (B) Comparison of overall survival between patients with and without copy number loss of tumor suppressor genes (TSGs) in either PTEN, TSC1, TSC2, STK11, or EIF4E-BP1. (C) Isogenic cell panels of HNSCC cell lines, CAL27 and HN12, parental controls and with gene knockout in PTEN, TSC2, STK11, or EIF4E-BP1, as indicated.
Dependency of PI3K/mTOR signaling on EGFR activity in HNSCC cell lines: Knockout of PI3K/mTOR TSGs confers EGFR-independent signaling.
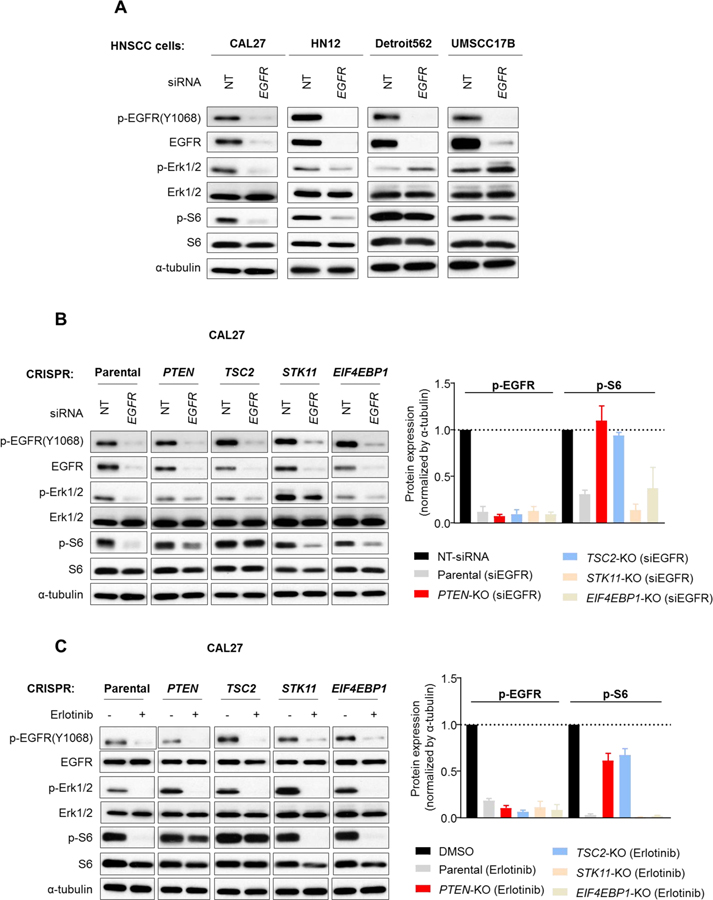
We next tested the impact of EGFR inhibition in a panel of different HNSCC cell lines, including cells that do not have obvious driver oncogene mutations (HN12, CAL27), using Detroit562 (PIK3CA mutant) and UM-SCC-17B (HRAS mutant) HNSCC cells (25), as controls. We performed knockdown of EGFR using siRNA, and analyzed Ser235/Ser236 phospho-S6 (p-S6S235/236), herein referred to as p-S6, and phospho-Erk1/2 (p-ErkT202/Y204), herein referred to as p-Erk1/2, as markers of PI3K/mTOR and RAS/RAF/MAPK signaling activity, respectively. We found that p-S6 and p-Erk1/2 were suppressed by EGFR knockdown in both of HN12 and CAL27 cells under serum free conditions. As expected, Detroit562 (PIK3CA mutant) and UM-SCC-17B (HRAS mutant) cells were resistant to p-S6 and p-Erk1/2 reduction by EGFR knockdown (Fig. 2A). This finding was validated by EGFR inhibition using a clinically relevant small molecule EGFR tyrosine kinase inhibitor, erlotinib (Supplementary Fig. 2A). These results suggest that PI3K/Akt/mTOR and RAS/RAF/MAPK signaling are mainly regulated under basal conditions by EGFR in both CAL27 and HN12, but not in Detroit562 and UM-SCC-17B cells.
Figure 2. Effects of knocking out PI3K/mTOR TSGs on the dependency of PI3K/mTOR signaling on EGFR activity in HNSCC cells.
(A) HNSCC cell lines were treated with control non-targeting (NT) or EGFR-siRNA under serum starvation. Cell lysates were analyzed for the indicated protein by western blotting. (B) Parental CAL27, and isogenic cells with the indicated gene knockouts were treated with control non-targeting (NT) or EGFR-siRNA under serum starvation. Cell lysates were analyzed for the indicated proteins by western blotting (Left panel). The band density was analyzed and normalized with control siRNA-treated samples. Data were from triplicate experiments (Right Panel). (C) Parental CAL27, and isogenic cells with indicated gene knockout were serum starved overnight, and then treated with 0.1% DMSO or erlotinib (3 μM) for 1h. Cell lysates were analyzed for the indicated proteins by western blot (left panel). The band density was analyzed and normalized with the vehicle control (0.1% DMSO)-treated samples. Data were from triplicate experiments (right Panel).
We next used these isogenic cell line panels to investigate which of the PI3K/mTOR pathway TSGs knockouts led to independence from EGFR activity with regard to downstream signaling. The isogenic CAL27 cells were treated with EGFR siRNA or erlotinib (3 μM), followed by immunoblotting for total and phosphorylated forms of Erk1/2 or S6. Remarkably, both PTEN- and TSC2-knockout CAL27 cells were resistant to PI3K/mTOR signaling inhibition as shown by failure to reduce p-S6 in both EGFR siRNA and erlotinib treated cells. By contrast, STK11- and EIF4EBP1-knockout CAL27 cells remained sensitive to p-S6 reduction following EGFR knockdown or erlotinib treatment (Figs. 2B and 2C). Similar experiments were also performed in isogenic HN12 cell panels, revealing that PTEN-knockout cells were resistant to EGFR knockdown as well as to erlotinib treatment, whereas partial p-S6 reduction was observed in TSC2-knockout HN12 cells under these EGFR inhibiting conditions (Supplementary Fig. 2B, C).
Effects of PTEN knockout on the response to cetuximab in cell proliferation and orosphere formation.
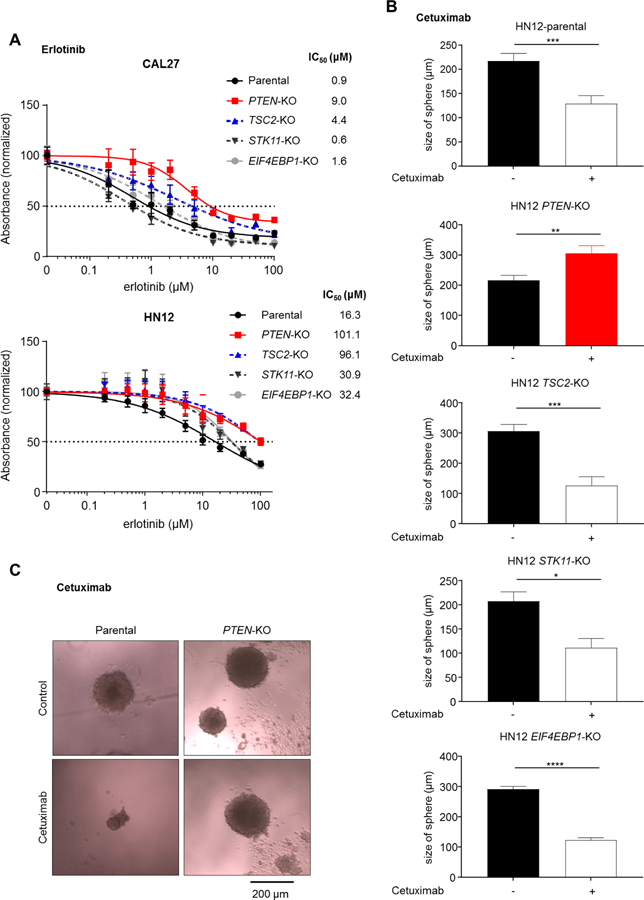
We next compared anti-proliferative efficacy of erlotinib between parental and PI3K/mTOR pathway TSG knockout cells. Cells were treated with serial concentrations of erlotinib for 72 h in 96-well plates. The IC50 values for erlotinib in parental, PTEN-, TSC2-, STK11-, and EIF4EBP1-knockout CAL27 cells were 0.9 μM, 9.0 μM, 4.4 μM, 0.6 μM, and 1.6 μM, respectively (Fig. 3A). The IC50 values in parental, PTEN-, TSC2-, STK11-, and EIF4EBP1-knockout HN12 cells were 16.3 μM, 101.1 μM, 96.1 μM, 30.9 μM, and 32.4 μM, respectively (Fig. 3A). Remarkably, among all of these changes the IC50 values for erlotinib in PTEN-KO cells were consistently 6 – 10 fold higher than that in parental cells.
Figure 3. Effects of PTEN knockout on the response to cetuximab in cell proliferation and orosphere formation assays.
(A) Parental and isogenic CAL27 and HN12 cells seeded in 96-well plates (2000 per well) were treated with the indicated concentrations of erlotinib for 72h. Cell viabilities were normalized with that of the corresponding vehicle control (0.1% DMSO)-treated cells. (B) Parental and isogenic HN12 cells were seeded in 96-well ultra-low attachment culture dishes at 100 cells per well (n = 10), and treated with vehicle control (0.9% NaCl) or cetuximab (10 μg/ml). 10 days after treatment, the size of spheres in each well were determined. (C) Representative spheres obtained from parental and PTEN knockout cells treated with vehicle control or cetuximab (10 μg/ml).
We next investigated whether the knockout of these PI3K/mTOR TSGs could interfere with the inhibition of the tumorigenic potential by cetuximab. For this, we compared the ability of sphere growth (orosphere) between parental and TSGs knockout cells after treatment with cetuximab at 10 μg/ml, which is equivalent to the serum trough concentration in patients treated with cetuximab (30). In parental HN12 cells, cetuximab significantly reduced the size of sphere formation (Fig. 3B). Notably, PTEN-knockout HN12 cells showed resistance to cetuximab-induced inhibition of sphere growth, while the other knockout cells were still partially or completely sensitive to cetuximab in terms of orosphere formation (Fig. 3B, C). Collectively, these results suggest that among all genes tested, knockout of PTEN showed the most robust phenotype, conferring independence from EGFR activity and resistance to EGFR inhibition in terms of downstream signaling, proliferation, and orosphere growth.
Effects of PTEN knockout on the response to cetuximab in HNSCC tumor xenografts.
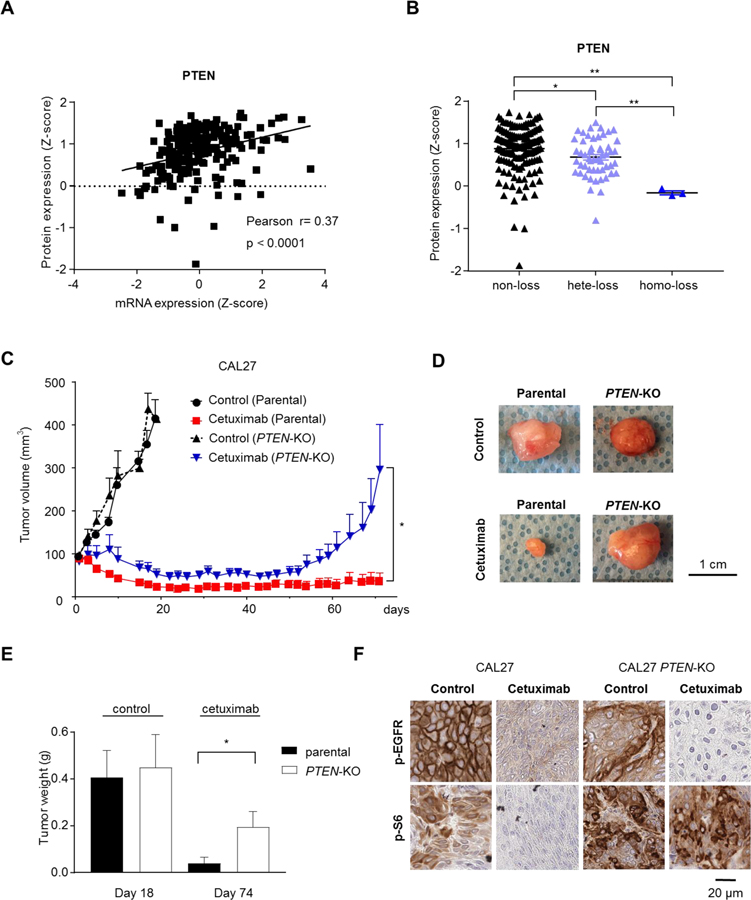
Given that PTEN knockout cells were the most robust in demonstrating resistance to EGFR inhibition, we first extended our genomics studies and analyzed the correlation between protein expression, mRNA expression, and PTEN copy number in the large TCGA HNSCC dataset. The protein expression of PTEN was significantly correlated with mRNA expression (Fig. 4A), and copy number (Fig. 4B), suggesting that PTEN gene copy loss results in reduced transcripts and PTEN protein levels. Experimentally, we compared the efficacy of cetuximab between parental and PTEN-knockout tumors in vivo to confirm the role of PTEN knockout in conferring resistance to cetuximab. We transplanted parental or PTEN-KO CAL27 cells into the flanks of nude mice, and then treated with either cetuximab (40 mg/kg, three times a week) or control diluent (10 tumors per each group). The growth of PTEN-KO tumors treated with control diluent was almost identical compared to that of control-treated parental tumors. The volume of PTEN-KO tumors decreased after the cetuximab treatment, while parental CAL27 tumors responded to greater extent. However, PTEN-KO tumors regrew rapidly, in contrast to wild-type CAL27 cells which exhibited prolonged tumor remission (Fig. 4C–E). These results suggest that cetuximab displayed greater efficacy in wild-type parental tumors compared with that achieved in PTEN-KO tumors. Immunohistochemistry analysis showed that cetuximab-treated PTEN-KO tumors exhibited sustained p-S6 staining (Fig. 4F), despite clear suppression of p-EGFR. In contrast, cetuximab treatment clearly suppressed p-S6 as well as p-EGFR in the wild-type parental tumors.
Figure 4. Effects of PTEN knockout on the response to cetuximab in HNSCC tumor xenografts.
Correlation of PTEN protein expression with PTEN mRNA (A) and PTEN gene copy number (B) in the TCGA dataset. (C) Parental and PTEN knockout CAL27 were transplanted into nude mice, and treated with vehicle control diluent or cetuximab (40 mg/kg), 3 times per week. Cetuximab treatment was continued until 6 weeks (D). Representative tumors treated with or without cetuximab are shown. (E) Tumor weight at the indicated day. Control diluent- and cetuximab-treated tumors were collected 18 days and 74 days after treatment, respectively. (F) Representative immunohistochemical analysis of pEGFR (Y1068) and pS6, as indicated.
Efficacy of cetuximab on naturally occurring PTEN-deficient HNSCCs
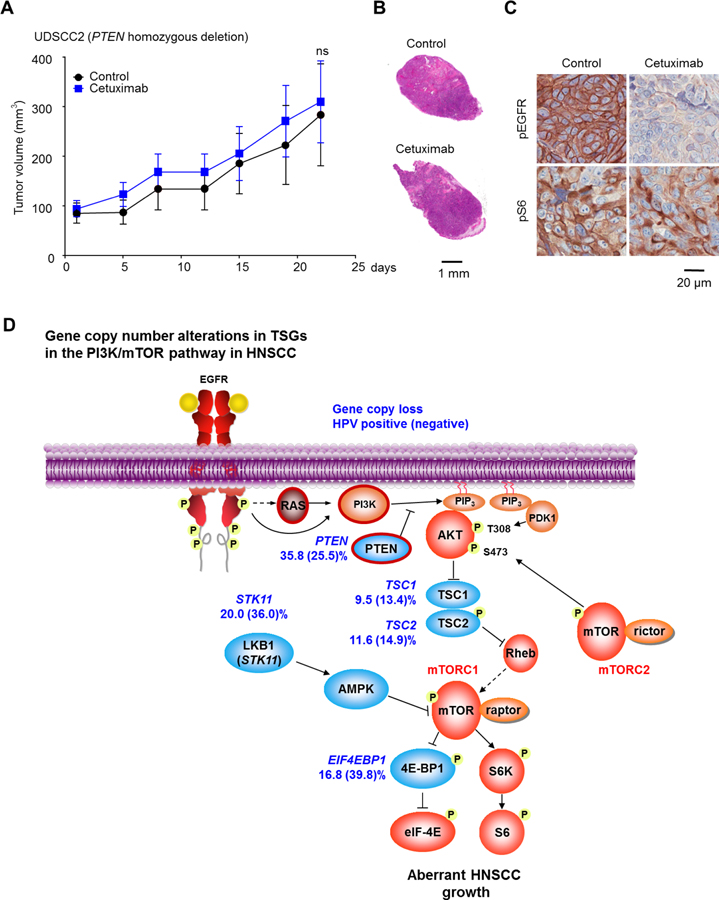
To further confirm our findings that PTEN loss promotes cetuximab resistance in HNSCC, we employed UDSCC2 cells, which harbors an endogenous homozygous PTEN-loss (25). We injected the UDSCC2 cells into the flank of nude mice, and treated UDSCC2-bearing mice with cetuximab (40 mg/kg, three times per week) or control solution (10 tumors per each treatment group) for 3 weeks. Consistent with our findings in isogenic genome-edited cells, UDSCC2 tumors were totally insensitive to cetuximab treatment (Fig. 5A, B). In the UDSCC2 tumors treated with cetuximab, persistent p-S6 staining was also observed, despite successful suppression of p-EGFR, strongly supporting the failure of cetuximab to perturb PI3K/mTOR signaling in the UDSCC2 xenograft model (Fig. 5C).
Figure 5. Resistance to cetuximab in HNSCC tumors with endogenous PTEN loss, and PI3K/mTOR network-based analysis of refractoriness to cetuximab.
(A) UDSCC2 cells were transplanted into nude mice, and treated with vehicle control diluent or cetuximab (40 mg/kg), 3 times per week. (B) Representative tumors treated with vehicle control or cetuximab (HE staining). (C) Representative immunohistochemical analysis of pEGFR (Y1068) and pS6, as indicated. (D), graphic depicting copy number variations in the PI3K/mTOR pathway in HNSCC. Resistance to cetuximab can be specifically conferred by PIK3CA and RAS mutations (from reference 23), as well as from frequent PTEN gene copy loss (red border).
Discussion
Although cetuximab displays antitumoral activity in HNSCC and is approved by the FDA for treatment of this disease, only a small minority of patients benefit clinically from cetuximab treatment. Thus, precise stratification of patients that are sensitive or resistant to cetuximab is needed to harness the full clinical potential of this therapeutic agent. Here, we show that the HNSCC cells that are originally sensitive to cetuximab become resistant when the PTEN gene is genetically disrupted. We also show that HNSCC cell lines that harbor endogenous PTEN loss are highly resistant to cetuximab treatment. These findings strongly indicate that PTEN loss, a frequent event in HNSCC, is sufficient to promote cetuximab resistance, and that this genomic alteration may be responsible for intrinsic cetuximab resistance.
Our analysis of TCGA data from 409 HPV-negative and 93 HPV-positive HNSCC samples revealed that a high percentage (69.3%) of HNSCC lesions exhibited the loss of at least one copy of candidate PI3K/mTOR pathway TSGs. Loss of these TSGs was observed to occur with much greater frequency than mutation of the genes. In addition, copy number loss of PI3K/mTOR TSGs seems to be clinically important, because they were found to be associated with poor survival in HNSCC patients. Among the TSGs tested, copy number loss in PTEN gene was comparably observed in both HPV-negative (24.2%) and HPV-positive (31.2%) HNSCC cases, which is consistent with the frequency of reduced PTEN protein expression in HNSCC (31,32).
Since EGFR is commonly overexpressed in HNSCC and HNSCC cells and tumors are often addicted to EGFR signaling for sustained survival and proliferation, EGFR targeting therapy including small molecule EGFR inhibitors (EGFRi), such as erlotinib, and targeting antibodies, e.g., cetuximab, would be expected to be effective in a broader subset of HNSCC patients. In fact, the HNSCC cell lines HN12 and CAL27, both of which do not have obvious driver oncogene mutations in PI3K/mTOR or RAS/RAF/MAPK signaling pathways, are dependent on EGFR signaling, and are highly sensitive to cetuximab treatment in vitro and in vivo. However, we have previously shown that genomic alterations in the PIK3CA or RAS genes that lead to aberrant signaling and EGFR-independent proliferation can confer cetuximab resistance in these HNSCC cell lines (23). Through the use of genome editing approaches we have now performed a systematic analysis of the contribution of PI3K/mTOR TSG copy loss to resistance to cetuximab. We found that HNSCC cells genetically engineered for PTEN loss or HNSCC cells with endogenous PTEN loss were resistant to cetuximab and erlotinib treatment, the latter as an example of EGFRi. Aligned with our observations, reduced expression, as judged by immune histochemistry (IHC) or mutation of PTEN, correlates with poor response to cetuximab treatment in retrospective studies in metastatic HNSCC and colorectal cancer, respectively (17,33). Collectively, these findings demonstrate that PTEN loss is sufficient to promote resistance to cetuximab and suggest that PTEN deficiency may play a role in the resistance to this agent in a large percentage of HNSCC cancer patients.
In addition to promoting resistance to EGFR targeting therapies, PTEN loss has previously been shown to confer resistance to clinically viable inhibitors of PI3K and CDK4/6 (34,35). Hence, restoration of cellular PTEN activity, particularly in cells retaining only one copy of the PTEN gene, could prove highly valuable for restoring sensitivity to cetuximab, as well as other molecular targeting anti-cancer agents. Recent studies have discovered that the PTEN protein is negatively regulated by the upstream E3 ubiquitin ligase WWP1, and genetic deletion of the Wwp1 gene led to upregulation of PTEN activity, and corresponding loss of PI3K/Akt signaling, in cells with only one copy of the PTEN gene (36). Moreover, treatment of cells characterized by heterozygous loss of PTEN with the WWP1 inhibitor indole-3-carbinol potently suppressed in vivo tumor growth and PI3K-mediated signaling. These findings suggest that targeted inhibition of WWP1 may be a promising strategy for reversing resistance to cetuximab, and other agents, resulting from genetic alteration of PTEN.
The use of EGFR targeting therapy for non-small cell lung cancer and metastatic colorectal cancer is restricted to selected patients based on molecular characteristics such as the presence of EGFR activating mutation and the absence of RAS/BRAF mutations, respectively. However, in HNSCC cetuximab is currently prescribed regardless of the presence or absence of these, or other, genetic alterations. Our findings suggest that the PTEN loss, which can result in activation of the PI3K/mTOR signaling pathway independently of EGFR, can be used as a mechanistic biomarker for the selection of patients that could be considered for exclusion from cetuximab treatment.
Our study includes several limitations. First, the impact of PTEN loss on cetuximab resistance is based on in vitro and experimental mouse models rather than based on clinical studies. Second, HNSCC cells with PTEN-KO respond less to cetuximab compared with parental HNSCC cells, however they still show partial responses followed by tumor relapse. Thus, cetuximab may inhibit yet to be identified growth promoting signals in addition to PI3K/mTOR in PTEN-KO tumors, until PTEN deficiency and aberrant PI3K/mTOR signaling may drive tumor regrowth and treatment failure. Therefore, further clinical studies are warranted to investigate the clinical utility of PTEN loss as a negative predictive biomarker of cetuximab sensitivity and for patient stratification.
Supplementary Material
Acknowledgments
J. Gutkind was supported by the National Institute of Dental and Craniofacial Research grants R01DE026870 and R01DE026644, and by the National Cancer Institute grant U54CA209891. J. Grandis was supported by NIH grants R01DE023685 and R35CA231998 and the American Cancer Society. D. Johnson was supported by NIH grants R01DE024728 and R01DE023685.
J.S. G. has received other commercial research support from Kura Oncology and Mavupharma, and is a consultant/advisory board member for Oncoceuitics Inc., Vividion Therapeutics, and Domain Therapeutics; D.E.J. and J.R.G are co-inventors of cyclic STAT3 decoy and have financial interests in STAT3 Therapeutics. STAT3 Therapeutics holds an interest in cyclic STAT3 decoy.
Abbreviations list
- CRT
Chemoradiation therapy
- EGFR
Epidermal growth factor receptor
- FDA
US Food and Drug Administration
- EGFRi
EGFR inhibitor
- HNSCC
Head and neck squamous cell carcinomas
- HPV
Human papillomavirus
- IHC
Histochemistry
- KO
Gene knock out
- NT
Non-targeting
- sgRNAs
Single guide RNAs
- TCGA
The Cancer Genome Atlas
- TSG
Tumor suppressor gene
Footnotes
Conflicts of interest: The remaining authors declare no conflicts.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin [Internet]. 2019;69:7–34. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30620402 [DOI] [PubMed] [Google Scholar]
- 2.Lui VWY, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, et al. Frequent Mutation of the PI3K Pathway in Head and Neck Cancer Defi nes Predictive Biomarkers. Cancer Discov. 2013;3:761–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pickering CR, Zhang J, Yoo SY, Bengtsson L, Moorthy S, Neskey DM, et al. Integrative Genomic Characterization of Oral Squamous Cell Carcinoma Identifi es Frequent Somatic Drivers. Cancer Discov. 2013;3:770–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iglesias-Bartolome R, Martin D, Silvio Gutkind J. Exploiting the head and neck cancer oncogenome: Widespread PI3K-mTOR pathway alterations and novel molecular targets. Cancer Discov. 2013;3:722–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grandis JR, Tweardy DJ. Elevated Levels of Transforming Growth Factor α and Epidermal Growth Factor Receptor Messenger RNA Are Early Markers of Carcinogenesis in Head and Neck Cancer. Cancer Res [Internet]. 1993;53:3579–84. Available from: http://cancerres.aacrjournals.org/content/53/15/3579.abstract [PubMed] [Google Scholar]
- 7.Kalyankrishna S, Grandis JR. Epidermal Growth Factor Receptor Biology in Head and Neck Cancer. J Clin Oncol. 2006;24:2666–72. [DOI] [PubMed] [Google Scholar]
- 8.Chung CH, Ely K, Mcgavran L, Varella-garcia M, Parker J, Parker N, et al. Increased Epidermal Growth Factor Receptor Gene Copy Number Is Associated With Poor Prognosis in Head and Neck Squamous Cell Carcinomas. J Clin Oncol. 2006;24:4170–6. [DOI] [PubMed] [Google Scholar]
- 9.Grandis JR, Melhem M, Gooding W. Levels of TGF-alfa and EGFR Protein in Head and Neck Squamous Cell Carcinoma and Patient Survival. J Natl Cancer Inst. 1998;90:824–32. [DOI] [PubMed] [Google Scholar]
- 10.Vermorken JB, Trigo J, Hitt R, Koralewski P, Diaz-Rubio E, Rolland F, et al. Open-label, uncontrolled, multicenter phase II study to evaluate the efficacy and toxicity of cetuximab as a single agent in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck who failed to respond to platinum-based the. J Clin Oncol. 2007;25:2171–7. [DOI] [PubMed] [Google Scholar]
- 11.Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, et al. Radiotherapy plus Cetuximab for Squamous- Cell Carcinoma of the Head and Neck. N Engl J Med. 2006;354:567–78. [DOI] [PubMed] [Google Scholar]
- 12.Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, et al. Platinum-Based Chemotherapy plus Cetuximab in Head and Neck Cancer. N Engl J Med. 2008;359:1116–27. [DOI] [PubMed] [Google Scholar]
- 13.Mehanna H, Robinson M, Hartley A, Kong A, Foran B, Fulton-Lieuw T, et al. Radiotherapy plus cisplatin or cetuximab in low-risk human papillomavirus-positive oropharyngeal cancer (De-ESCALaTE HPV): an open-label randomised controlled phase 3 trial. Lancet. 2019;393:51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillison ML, Trotti AM, Harris J, Eisbruch A, Harari PM, Adelstein DJ, et al. Radiotherapy plus cetuximab or cisplatin in human papillomavirus-positive oropharyngeal cancer (NRG Oncology RTOG 1016): a randomised, multicentre, non-inferiority trial. Lancet [Internet]. Elsevier Ltd; 2019;393:40–50. Available from: 10.1016/S0140-6736(18)32779-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol [Internet]. 2010;11:121–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20022809 [DOI] [PubMed] [Google Scholar]
- 16.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med [Internet]. 2010. [cited 2014 Aug 7];362:2380–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20573926 [DOI] [PubMed] [Google Scholar]
- 17.Cutsem EC, Koohne C-H, Hitre E, Zaluski J, Chien C-RC, Lim R, et al. Cetuximab and Chemotherapy as Initial Treatment for Metastatic Colorectal Cancer. N Engl J Med. 2009;360:1408–17. [DOI] [PubMed] [Google Scholar]
- 18.Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, de Braud F, et al. Fluorouracil , Leucovorin , and Oxaliplatin With and Without Cetuximab in the First-Line Treatment of Metastatic Colorectal Cancer. J Clin Oncol. 2009;27:663–71. [DOI] [PubMed] [Google Scholar]
- 19.Brand TM, Iida M, Stein AP, Corrigan KL, Braverman CM, Luthar N, et al. AXL mediates resistance to cetuximab therapy. Cancer Res. 2014;74:5152–64. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 20.Leonard B, Brand TM, O’Keefe RA, Lee ED, Zeng Y, Kemmer JD, et al. BET inhibition overcomes receptor tyrosine kinase-mediated cetuximab resistance in HNSCC. Cancer Res. 2018;78:4331–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang N, Wang D, Hu Z, Shin HJC, Qian G, Rahman MA, et al. Combination of Anti-HER3 Antibody MM-121/SAR256212 and Cetuximab Inhibits Tumor Growth in Preclinical Models of Head and Neck Squamous Cell Carcinoma. Mol Cancer Ther. 2014;13:1826–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madoz-Gúrpide J, Zazo S, Chamizo C, Casado V, Caramés C, Gavín E, et al. Activation of MET pathway predicts poor outcome to cetuximab in patients with recurrent or metastatic head and neck cancer. J Transl Med. BioMed Central; 2015;13:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Martin D, Molinolo AA, Patel V, Iglesias-Bartolome R, Degese MS, et al. mTOR co-targeting in cetuximab resistance in head and neck cancers harboring PIK3CA and RAS mutations. J Natl Cancer Inst. 2014;106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amornphimoltham P, Patel V, Sodhi A, Nikitakis NG, Sauk JJ, Sausville EA, et al. Mammalian Target of Rapamycin , a Molecular Target in Squamous Cell Carcinomas of the Head and Neck. Cancer Res. 2005;1:9953–62. [DOI] [PubMed] [Google Scholar]
- 25.Martin D, Abba MC, Molinolo AA, Vitale-Cross L, Wang Z, Zaida M, et al. The head and neck cancer cell oncogenome: A platform for the development of precision molecular therapies. Oncotarget. 2014;5:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science (80- ) [Internet]. 2014;343:84–7. Available from: https://www.bps.go.id/dynamictable/2018/05/18/1337/persentase-panjang-jalan-tol-yang-beroperasi-menurut-operatornya-2014.html [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov [Internet]. 2012;2:401–4. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22588877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal [Internet]. 2013;6:pl1 Available from: http://www.ncbi.nlm.nih.gov/pubmed/23550210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibata K, Naito T, Okamura J, Hosokawa S, Mineta H. Simple and rapid LC-MS / MS method for the absolute determination of cetuximab in human serum using an immobilized trypsin. J Pharm Biomed Anal [Internet]. 2017;146:266–72. Available from: 10.1016/j.jpba.2017.08.012 [DOI] [PubMed] [Google Scholar]
- 31.Squarize CH, Castilho RM, Abrahao AC, Molinolo A, Lingen MW, Gutkind JS. PTEN Deficiency Contributes to the Development and Progression of Head and Neck Cancer 1. Neoplasia. 2013;15:461–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bian Y, Hall B, Sun Z, Molinolo A, Chen W, Gutkind JS, et al. Loss of TGF- b signaling and PTEN promotes head and neck squamous cell carcinoma through cellular senescence evasion and cancer-related inflammation. Oncogene. 2011;31:3322–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.da Costa AABA, Costa FD, Araújo DV, Camandaroba MPG, de Jesus VHF, Oliveira A, et al. The roles of PTEN, cMET, and p16 in resistance to cetuximab in head and neck squamous cell carcinoma. Med Oncol [Internet]. Springer US; 2019;36:1–9. Available from: 10.1007/s12032-018-1234-0 [DOI] [PubMed] [Google Scholar]
- 34.Costa C, Wang Y, Ly A, Hosono Y, Ellen M, Walmsley CS, et al. PTEN loss mediates clinical cross-resistance to CDK4/6 and PI3Kα inhibitors in breast cancer. Cancer Discov. 2019;[Epub ahead of print]. [DOI] [PubMed]
- 35.Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kα inhibitor. Nature. 2015;518:240–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee YR, Chen M, Lee JD, Zhang J, Lin SY, Fu TM, et al. Reactivation of PTEN tumor suppressor for cancer treatment through inhibition of a MYC-WWP1 inhibitory pathway. Science (80- ). 2019;364:eaau0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.