Abstract
The Human Microbiome Initiative of NIH, begun in 2007, has opened the door to the power of the intestinal microbiome in health and disease. The 100 trillion gut microbes influence body function through three pathways: (1) via the neural route where 500 million neurons of the enteric nervous system (the body's second brain) connect to the brain and spinal cord, (2) via the immune route where the gut-immune capacity prevents infection and elicits immune response to vaccines, and (3) by the hormonal route wherein biologically active chemicals are released from enteroendocrine cells to control mood and body functions. Through research, the identification of diseases and disorders associated with abnormal microbiome (“dysbiosis”) has increased in number with potential for reversibility. Our team has developed an orally administered fecal microbiota transplantation product that is effective in reversing dysbiosis in recurrent Clostridioides difficile (C. difficile) and is being used to reverse abnormal microbiomes in chronic dysbiotic disorders.
INTRODUCTION
In 1901, Elie Metchnikoff remarked on the abundance of bacteria in the gastrointestinal tract, and he suggested they would be shown to have functional value in the future (1). The modern era of this science began in 2007 when the NIH-funded Human Microbiome Initiative (2) was begun and the European Metagenomics of the Human Intestinal Tract project was planned (3). The totality of microbiota in the human is referred to as the microbiome. Ninety-five percent of the body's microbiota reside in the gut, primarily in the colon where microbial cells outnumber the total cells in the human body (4). Using available sequencing methods, it is now possible to determine the presence of healthy or disarrayed microbiomes. Daily we are learning about new associations between the intestinal microbiome and human disease.
The gut has the largest surface area of any body tissue; it is estimated to be 344 ft2 (5). A majority of the microbial load in the intestine is in the free lumen of the colon. The body is protected by the physical barrier of the epithelial lining that is covered with a mucus coating that is densest in the distal colon where the greatest counts of microbes reside. Distanced by an inner layer of mucus, the mucosal-associated bacteria, residing in crypts and transverse folds of the proximal colon, are adjacent to and interact with the enteric nervous system (ENS) and lymphoid tissues of the gut.
In this review, we will examine what makes a healthy microbiome and how a pathologic microbiome contributes to the development of disease.
MATERIALS AND METHODS
The present review focuses on how the gut, the gut microbiome, and the brain communicate and how an abnormal reduction in intestinal microbiome diversity, or dysbiosis, contributes to disease. Finally, we describe a local program aimed at reversal of dysbiosis in disease and disorders of the intestinal microbiome by fecal microbiota -transplantation (FMT).
DEFINITIONS USED IN THIS REVIEW
Alpha-Diversity of Intestinal Microbiome
The α-diversity of species in the gut microbiome is measured by their number (richness) and relative proportions (evenness). In the measurements of α-diversity, the different organisms are counted as operational taxonomic units (OTU).
Beta-Diversity of Intestinal Microbiome
The proportion of specific taxa (phyla, family, species, or strains) in the gut microbiome simultaneously studied provides an analysis of β-diversity. Regarding genera (and family) in the intestinal microbiome of healthy persons, approximately 90% of the bacterial organisms are made up of anaerobic Gram-positive Firmicutes (Clostridiaceae) and anaerobic Gram-negative Bacteroidetes (Bacteroidaceae).
Healthy Microbiome
The composition of microbiome communities between people shows strikingly different proportions in taxa, which presents a challenge in defining a healthy microbiome. However, there are consistent features among healthy people that make the microbiome “healthy.” First, a rich and diverse microbiome can be observed when looking at the total number of bacterial species, measured as OTU (α-diversity). Second, β-diversity in a healthy microbiome contains less than 10% of the facultative anaerobe phylum, Proteobacteria (family, Enterobacteriaceae). Third, within the Firmicutes and Bacteroidetes, there is a high variation in species and stains among individuals.
Dysbiosis
Microbial imbalance or maladaptation of the intestinal microbiome is identified by reduced α-diversity or specific changes in taxa (β-diversity) and commonly an elevation (beyond 10%) in the proportion of the phylum Proteobacteria (Enterobacteriaceae) (6). Important anaerobic bacteria species associated with dysbiosis include variation in phyla (species) within the following: Firmicutes (Blautia spp., Ruminococcus spp., Coprococcus spp., Coprobacillus spp.); Bacteroidetes (Flavobacterium spp., Prevotella spp.); and Verrucomicrobia (Akkermansia spp.). Alteration in proportion of these species is frequently found in chronic disorders associated with dysbiosis.
IMPORTANCE OF DIET
The adage, “You are what you eat,” is true of the intestinal microbiome. Diet is the most important driver of the intestinal microbiome where human gut microbiota adjusts to food ingested. The most important constituent of a diet needed to obtain a healthy microbiome is fiber, which may be soluble or insoluble (Table 1). Soluble fiber is more beneficial than insoluble fiber to metabolism by microbiota and fermentation. Resistant starches (resistant to digestion in the upper gut), although insoluble, can be fermented by bacteria. Some foods (e.g., seeds and nuts) contain both soluble and insoluble fiber and -contribute significantly to the shaping of a healthy microbiome.
TABLE 1.
Foods That Shape a Healthy Microbiome
| Soluble Fiber1 | Insoluble Fiber2 |
|---|---|
| Soluble grains: oat bran (oatmeal), rye, barley | Whole grains: wheat and corn bran, brown rice |
| Fruits: figs, prunes, plums, -berries, apricots, apples, bananas, pears, guavas, avocados | Fruits: avocados, unripe bananas, kiwi, grape and tomato skins |
| Vegetables: beans (all types), peas, soybeans, broccoli, root vegetables (onions, leeks, carrots, lentils, sweet potatoes, yams, Jerusalem artichokes, turnips, garlic), Brussel sprouts | Vegetables: green beans, peas, potato skins, cauliflower, zucchini, cabbage, celery |
| Seeds: flax, Chia, sunflower, sesame | Seeds: flax, Chia, sunflower, sesame |
| Nuts: all nuts—almonds have highest fiber | Nuts: all nuts |
Soluble fiber (dissolves in aqueous colon and is readily fermented by indigenous microbiota producing organic acids that promote cardiovascular, metabolic health).
Insoluble fiber (provides bulk to stool and prevents diverticulosis; resistant starches (in bold above) can also be fermented which retains the value of soluble fiber).
FUNCTIONS OF A HEALTHY INTESTINAL MICROBIOME
The 100 trillion bacteria in the gut modulate gut-health structurally, biochemically, and immunologically. On a structural level, microbiota are essential for proper intestinal epithelial cell and tight junction formation and enteric nervous system development, based on data from numerous animal models (7,8). Enterocyte proliferation and wound healing are impaired in germ-free mice in the dextran sulfate sodium (DSS) colitis model (9). Decreased epithelial apoptosis and crypt cell proliferation in germ-free piglets can be corrected by orally delivered microbiota (7). In addition, the healthy human colon is lined by two layers of mucus. The outer layer, rich in oligosaccharides, serves as a nutrient source for commensal bacteria, whereas the densely packed inner layer lacks bacteria and shields the gut epithelium from bacteria (10,11). Commensal bacteria such as Lactobacillus stimulate mucin production, as do short-chain fatty acids (SCFAs) produced by anaerobic bacteria (12). Transfer of microbiota to germ-free mice, which lack an intact mucus layer, resulted in transfer of a healthy mucus phenotype (13), indicating the role of microbiota in generating the mucus barrier.
SCFAs, which include acetate, propionate, and butyrate, are produced upon fermentation of dietary fiber by anaerobic bacteria. In addition to stimulating mucin production, they serve as energy sources for colonic epithelial cells and accelerate tight junction formation. They are therefore thought to enhance gut barrier integrity (7). They also engage in anti-inflammatory activities, such as modulating neutrophil trafficking, inducing regulatory CD4 T cell (Treg) -generation, and decreasing pro-inflammatory cytokine production by monocytes and macrophages (14).
Gut bacteria convert primary bile acids, originating from the liver, into the secondary bile acids deoxycholic acid and lithocholic acid by 7 α-dehydroxylation. Bacteria from Lachnospiraceae and Ruminococcaceae families are key in this step (15). Bile acids can, in turn, shape the composition of the gut microbiota and damage bacterial cell membranes and DNA, which would kill the organism. Susceptibility to this condition varies across bacterial species. Secondary bile acids inhibit C. difficile germination and growth (16).
An intact microbiome also produces antimicrobial peptides. Bacillus thuringiensis produces thuricin CD, which is toxic to C. difficile (17). Numerous gut species produce nisin, which targets many Gram-positive bacteria, but may not be present in sufficient levels in vivo to have an effect. Lactobacillus reuteri produces reuterin, which can inhibit C. difficile growth (18).
The microbiome has pleiotropic effects on the mucosal and systemic immune system. A healthy microbiome suppresses IFNγ and TNF production by peripheral blood mononuclear cells while stimulating development of Treg and IL-10 production (19). Bacteroides spp. induces Tregs and stimulates IL-10 production (19,20). Clostridioides spp., especially clusters IV, XIVa, and XVIII, are also Treg inducers (21). Intestinal plasma cells secrete polyreactive IgA to coat and contain commensal microbiota (22), yet it is not known how this immune response affects the organisms gut physiology and function. Also unknown is whether epithelium-associated commensal bacteria such as Mucispirillum and segmented filamentous bacteria, in turn, stimulate IgA production and therefore modulate immune responses (23).
GUT-BRAIN AXIS
Central to health orchestrated by the intestinal microbiome is the functioning of the central nervous system. The gut and viscera communicate with the brain via three important general pathways that collectively constitute the gut-brain axis (Figure 1).
Fig. 1.
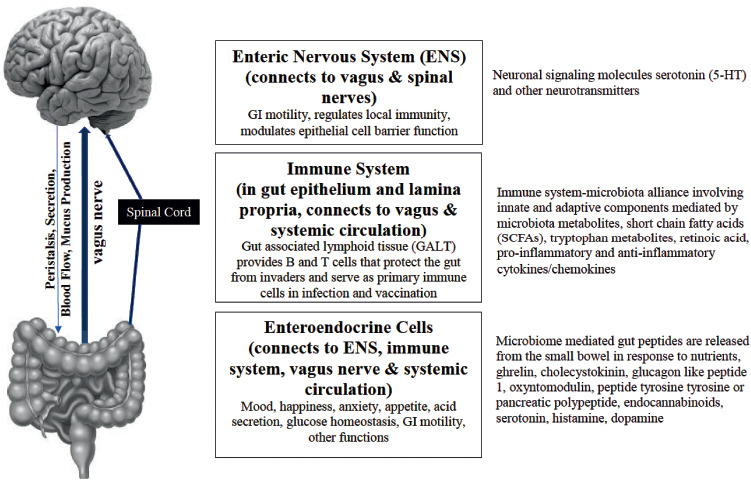
The Three Major Components of the Microbiota-Gut-Brain Axis.
Neural Pathway
In the first pathway, gut microbiota and the gastrointestinal tract communicate directly with the brain and spinal cord through -hardwired connections involving the approximately 500 million intrinsic afferent neurons in the enteric nervous system (ENS), often referred to as the body's second brain (24). Microbiota are required for this connection, and germ-free animals do not develop fully functioning brains (25). ENS activity depends upon microbiota-released neurotransmitters, including gamma-aminobutyric acid (GABA). The vagus nerve (cranial nerve X) is the parasympathetic channel through which information is transmitted from the gut to the brain, which then reciprocates via cognitive and emotional functions, and by modification of intestinal secretory systems, changes in peristalsis, and blood flow (26).
Immune Pathway
The second pathway whereby microbes affect body function including brain function is through effects on immune signals from gut--associated lymphoid tissue (GALT). These signals alter messages to the brain via sensory neurons of the ENS (27, 28). More than 70% of the body's immune cell population is contained in the GALT, which depends upon an intimate relationship with adjacent microbiota. GALT-microbiome activity defines innate immunity through toll-like receptors and NOD-like receptors (NLR) that recognize disruption in the microbiome and damaged tissues (29). GALT functions in harmony with a healthy microbiome mucosal-associated lymphoid tissue (MALT) to protect the body from potential intraluminal pathogens. In addition to secondary bile acids and SCFAs mentioned above, the microbiome influences the innate immune system through release of metabolites, folate, indole, trimethylamine-N-oxide, and neurotransmitters such as GABA and serotonin. Gram-negative bacterial lipopolysaccharides (LPS) from the microbiota trigger inflammation via the innate immune system (30).
The Endocrine Pathway
The third connection between the gut and brain is the endocrine system of the gut. While enteroendocrine cells (EECs) comprise less than 1% of gut epithelial cells, the large number of intestinal epithelial cells explains why the gut is the largest endocrine organ of the body (31). EECs detect signals from microbiota through toll-like receptors (TLR) which recognize LPS or SCFAs. EECs produce several hormones based on stimuli from intraluminal bacteria and fatty acid metabolites: cholecystokinin, peptide YY, glucagon-like peptide-1, ghrelin, oxyntomodulin, peptide tyrosine tyrosine, pancreatic polypeptide, and 5-hydroxytryptamine (5-HT). The gut hormones have access to the hypothalamus through the circulation and via the vagus nerve.
PATHOGENESIS OF MICROBIOME-MEDIATED DISEASES AND DISORDERS (TABLE 2)
TABLE 2.
Pathophysiology of Diseases and Disorders Associated with Dysbiosis
| Initiating Event | Factor Influencing | Disease, Disorder |
|---|---|---|
| GI dysmotility | Depending upon region of the gut affected by dysbiosis | Functional bowel disease with diarrhea or constipation, small bowel bacterial overgrowth, gastroparesis, pseudo-obstruction |
| Mucosal permeability | Increased permeability can lead to mucosal inflammation | Inflammatory bowel disease, irritable bowel syndrome |
| Translocation of bacterial products (e.g., LPS), release of peptides (amyloid β) and proteins (α-synuclein) can act as prions in the central nervous system | Metabolic alterations, obesity, neurodegeneration (Alzheimer's and Parkinson's disease and multiple sclerosis) | |
| Unhealthy diet | High-fat diet producing intestinal inflammation | Obesity, insulin-resistance, type-2 diabetes mellitus |
| Carcinogenic metabolites of food (fat contributing to formation of peroxides and sterol metabolites, salted fish contaminated during processing, red and processed meats, sugary beverages) | Colorectal cancer | |
| Dysbiosis can affect the GALT | Altered GALT responses | Allergic disorders, failure to react to exposure to infectious agents, vaccines, or immunotherapy |
| Enteroendocrine cells release of biologic chemicals and hormones | Serotonin and 5-HT signals to ENS | Sensory and motor activity of gut as part of -gut-brain axis |
| Release of hormones: histamine, somatostatin, motilin, cholecystokinin, neurotensin, vasoactive intestinal peptide, enteroglucagon, GLP-1, cholecystokinin, histamine | Effects on gut secretion and motility, regulation of glucose metabolism, activity on ENS as part of gut-brain axis, effects on GALT, some act as classical hormones affecting body organs | |
| Neurotransmitters: leptin, ghrelin, dopamine, GABA, serotonin, peptide YY, cholecystokinin | Alteration of mood, anxiety, depression, hunger, satiation | |
| Exposure to recurrent courses of antibiotics can lead to profound α- and β-diversity defects | Defenses of gut lumen are reduced allowing overgrowth of bacteria given a selective advantage for growth | Clostridioides difficile infection and colitis |
Abbreviations: LPS, lipopolysaccharide; GALT, gut-associated lymphoid tissue; 5-HT, 5-hydroxytryptophan; ENS, enteric nervous system; GLP-1, glucagon-like peptide 1; GABA, gamma-aminobutyric acid.
Abnormal distribution of microbiota in the gut alters intestinal function. Abnormal signals through the parasympathetic nervous system via the vagus nerve to the brain as seen in neurodegenerative diseases affect bowel motility and hormone secretion. Dysmotility depends upon the region of the gut affected by microbiota.
Alteration of the mucosal barrier and tight junctions of the gut lead to altered permeability of microbial products and metabolites of partially digested foods, as seen in inflammatory bowel disease, irritable bowel syndrome, celiac disease, and obesity with insulin resistance.
Gut permeability defects, abnormal immune responses, and migration of proteins and peptides from the gut through the vagus nerve or systemic circulation to the brain can cause neurodegeneration, which is discussed later.
A diet rich in animal fat and digestible carbohydrates (i.e., “Western diet”) leads to weight gain and metabolic changes associated with obesity and metabolic changes from released hormones and neurologic events through release of neurotransmitters that affect the central and peripheral nervous systems. Such a diet can also result in changes in appetite and satiation, acid secretion, glucose homeostasis, and level of insulin resistance, which is important in the metabolic syndrome of obesity and diabetes. Diet-influenced production of microbial metabolites and secondary bile acids can cause mucosal neoplastic changes in the pathogenesis of colorectal carcinoma.
Abnormal gut-associated lymphoid immune response associated with dysbiosis leads to allergic disorders, failure to develop protective immunity and immune response to vaccines, and lack of effectiveness of anticancer immunotherapy (32).
In antibiotic-induced dysbiosis, reduced gut microbiota diversity, lowered levels of organic acids normally produced by rich growth of anaerobic bacteria, and space made available through inhibition of indigenous microbiota allowing growth of foreign microbiota as seen in Clostridioides difficile infection (CDI).
More research is needed on gut viruses as mediators of a healthy microbiome. All bacteria are capable of being infected by bacteriophages, and diversity of the virome has been associated with microbiome recovery from dysbiosis (33,34) and may explain how FMT product filtered to remove bacteria retains its ability to restore the microbiome diversity in patients with recurrent Clostridioides difficile infection (CDI) (35).
DISEASES AND DISORDERS ASSOCIATED WITH DYSBIOSIS (FIGURE 2)
Fig. 2.
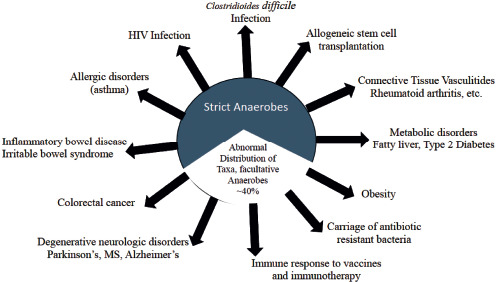
Conditions associated with Dysbiosis.
Clostridioides difficile Infection (CDI)
In CDI, antibiotic therapy typically alters the intestinal microbiome, reducing intestinal protection and allowing C. difficile spores to germinate and produce colonic mucosal inflammation. The subsequent antibiotics taken for CDI further impair gut microbiome defenses, and recurrent disease is common. In recurrent CDI, both α- and β-diversity of the microbiome are altered (36). Specifically, species of butyrate-producing Lachnospiraceae and Ruminococcaceae are depleted. High SCFA levels correlate with resistance to CDI and may even be bactericidal to C. difficile (37). Gut microbiota are also important for conversion of primary bile acids into secondary bile acids. Whereas primary bile acids stimulate germination of C. difficile spores, secondary bile acids block germination. Consistent with this finding, CDI is associated with high primary bile acid levels and low secondary bile acid levels (38).
Allogeneic Stem Cell Transplantation
In allogeneic stem cell transplantation, reduction in diversity of the intestinal microbiome contributes to morbidity and mortality, -graft-versus-host disease, and subsequent mucosal infection (39).
Connective Tissue Vasculitis Disorders
Dysbiosis involving the gut and mouth (dental and salivary) is -associated with the development of rheumatoid arthritis, representing targets of prevention or therapy (40). Preliminary data indicate that the microbiome may play a role in systemic lupus erythematosus, -systemic sclerosis, Sjögren's syndrome, and Behçet's disease (41).
Metabolic Disorders, Obesity and Nonalcoholic Fatty Liver Disease
The Western diet of excessive animal fats and digestible carbohydrates leads to obesity and dysbiosis. Patients with nonalcoholic fatty liver disease (NAFLD) have been shown to have alterations of their intestinal microbiome, including higher proportions of Proteobacteria, and some genera of Bacteroidetes (Prevotella spp.) and Firmicutes (Erysipelotrichia and Lactobacillus spp.) (42). Metabolic and microbiome changes can lead to secondary bile acid release, alteration of metabolites (SCFAs), and release of bacterial products (e.g., LPS), which can result in endothelial barrier permeability disruption, inflammation, small bowel bacterial overgrowth, and insulin resistance. These changes cause intrahepatic alteration of lipid metabolism, accumulation of fat, inflammation, and fibrosis (43-45). Both animal and human studies have shown that the intestinal microbiome is involved in the development of liver injury in steatosis. Germ-free mice fed high-fat diets are resistant to hepatic steatosis and dyslipidemia and have improved glucose tolerance and insulin sensitivity (46).
Carriage of Antimicrobial Resistance
In people who receive multiple courses of antibiotics, increased proportions of facultative anaerobes evolve in the gut microbiome (47). Improvement in microbiome diversity by fecal microbiota transplantation reduces antibiotic-resistance by narrowing the proportion of facultative anaerobes in the gut by replacing them with strict anaerobes (48).
Efficacy of Vaccine and Immunotherapeutic Agents
Vaccines that are effective in industrialized regions such as the United States, Canada, and Western Europe may not be helpful in developing countries where nutritional standards are lacking. This likely relates to the necessity of GALT in vaccine immune responses. Improvement of microbiome diversity may become an important objective in global vaccine programs (49).
Immunotherapy has revolutionized the treatment of many otherwise fatal cancers. Unleashing T-cell activity with these novel immune checkpoint inhibitor drugs results in high success rates in curing cancer. However, this success may depend upon the presence of an intact intestinal microbiome (50). One of the complications of immunotherapy for cancer patients is host-directed, T-cell-induced tissue responses and immune colitis. The microbiome seems to be important in this complication of immunotherapy, as shown in its reversal by FMT (51).
Neurodegenerative Disorders: Parkinson's Disease (PD), Multiple Sclerosis (MS), and Alzheimer's Disease (AD)
Gastrointestinal dysfunction, most commonly in the form of constipation, is observed in a majority of people destined for Parkinson's disease, and it tends to occur years to decades before their neurologic impairment (52). Neuroinflammation of the brain from pathologic immune system changes and increased mucosal permeability is postulated as an event that can complicate dysbiosis in PD (53,54). Bacterial metabolites such as SCFAs have been shown to affect microglia maturation (55).
The bidirectional gut-brain axis may be involved in the development of PD. Neurons of the ENS that contain aggregated and phosphorylated α-synuclein allow the spread of this protein from the gut to the central nervous system via the vagus nerve and olfactory tract. The protein is deposited in the substantia nigra as a prion-like molecule causing degeneration of dopamine neurons leading to motor deficits in PD (56-58). Indirect evidence that the gut and vagus nerve are involved in the development of PD was found in a Swedish study showing that truncal vagotomy five years or more before development of neurologic complaints reduced the risk of PD development (59). Abnormal microbiota seen in PD may further contribute to disease progression through enhanced metabolic degradation of anti-PD drugs, including levodopa (60).
Inflammation from dysbiosis in the intestine could contribute to altered blood brain barrier structures in PD (61). Abnormalities of intestinal microbiota with misfolding, aggregation, and deposition of proteins in the brain are observed in other forms of neurodegenerative disease (62), including Alzheimer's disease, where release of microbial β-amyloid can lead to plaque formation and neurofibrillary tangles in the brain (63). The microbiota in multiple sclerosis patients can lead to autoimmune reactivity (64), possibly through aberrant T--regulatory cell responses involving GALT (65). Abnormal gut microbiome composition in these neurodegenerative conditions is associated with increased permeability of the gut mucosa and immune activation that can lead to neuroinflammation, injury, and degeneration (66).
Colorectal Cancer
Diet and intestinal microbiome both appear to be important in the pathogenesis of colorectal cancer. Diet shapes the intestinal microbiome, and certain foods not only lead to a pathogenic microbiome but also encourage gut inflammation leading to cytotoxic changes; other foods are anti-tumorigenic (67). Patients with colorectal cancer characteristically show abnormal β-diversity of their intestinal microbiomes (68).
Inflammatory Bowel Disease (IBD)
Patients with Crohn's disease and ulcerative colitis show unique microbiomes with depletion of strict anaerobes compared to healthy people (69). Abundant species that generate metabolites important in disease pathogenesis reflect adaptation to oxidative stress in patients with IBD (70).
Irritable Bowel Syndrome (IBS)
IBS is a common chronic gastrointestinal disorder associated with visceral hypersensitivity, abdominal pain, and changes in bowel -pattern. The gut microbiome is abnormal with alteration in β-diversity compared with normal healthy persons (71). Microbiota alterations have been associated with changes in fecal levels of SCFAs compared with healthy controls (72).
Allergic Disorders and Asthma
The gut microbiome communicates with the respiratory microbiome, which affects the T helper 2 arm of the adaptive immune system in atopy and asthma (73), making the microbiome a target area for treatment in a variety of allergic disorders.
HIV/AIDS
The gut microbiota in people with HIV harbor unique microbiota that are associated with persistent inflammation and T-cell activation (74) Also, HIV therapy is associated with changes in the microbiome that influence response to treatment (75).
HOUSTON CENTER FOR MICROBIOME RESEARCH
The Kelsey Research Foundation, a 501(c)(3) Houston organization devoted to digestive disease research, and the University of Texas Health Science Center, Houston, have created the Houston Center for Microbiome Research. This collaborative research program also involves Baylor College of Medicine, MD Anderson Cancer Hospital, Memorial Hermann Hospital, Baylor St. Luke's Medical Center, and Houston Methodist Hospital. The Center is attempting to document diseases and disorders associated with dysbiosis and to look for ways to control or reverse disease through restoration of the microbiome. In the Center, a lyophilized fecal microbiota product has been derived from filtered fecal samples from well-screened healthy donors that is delivered orally in enteric-coated (acid-resistant) capsules. This product is as effective as delivering frozen FMT product by enema or colonoscopy (36,76) and is being used to treat multiple conditions under IND with the FDA, including CDI, Parkinson's disease, nonalcoholic fatty liver diseases, functional bowel disease, and HIV/AIDS. The Center is also exploring FMT augmentation of immunotherapy response in cancer and in treatment of immune colitis associated with anticancer immunotherapy (51). We have recently prepared a review of the current scientific literature exploring microbiome enhancement by FMT as a way to improve health in the broad range of conditions for which studies have been carried out (77).
CONCLUSIONS
The microbiota-gut-brain axis is a bidirectional communication system involving integrated neurological, immunological, and neuroendocrine pathways that are influenced by metabolites of microbiota and affect homeostasis and health. The vagus nerve, a component of the parasympathetic nervous system, senses metabolites of microbiota and relays information to the central nervous system.
The physiologic contributions of gut microbiome orchestrated by the 100 trillion bacteria residing in the gut either as mucosal-associated or free luminal bacteria relate to the large surface area of the gut, 100 times larger than the skin surface, with gut-associated lymphoid tissue containing 70% of the body's immune cells with plentiful enteroendocrine cells releasing large quantities of hormones that act locally and systemically.
Growing data support the idea that changes in the proportion of bacterial populations and overgrowth of some strains can cause and promote disease states rather than represent secondary events. Many groups, including our own, are exploring the value of reversal of dysbiosis as a way to improve health and treat or prevent diseases. The future will bring great advances in microbiome science, as researchers explore boundaries of the intestinal microbiome and the pharmaceutical industry plows hundreds of millions of dollars into new product development.
ACKNOWLEDGMENTS AND FINANCIAL SUPPORT
Funds for this review were provided by the Kelsey Research Foundation and the University of Texas Health Science Center, both in Houston, Texas. The review was supported by the Texas Medical Center, Digestive Disease Center (Public Health Service grant DK56338).
Footnotes
Potential Conflicts of Interest: Drs. DuPont and Jiang have a patent application for FMT product. Dr. DuPont has received a grant from Rebiotix.
DISCUSSION
Gravellese, Boston: Your work really looks at the microbiome as a bulk population of different strains of bacteria, and, in the literature, you see reports of one or another strain that's either protective or pathogenic. So, I wonder if you can walk us through how you think about this. Do you think they will prove to be specific strains that we should be particularly interested in, or do you think it's more about a balance in a bulk population?
DuPont, Houston: Great question. On the slide I showed you, I didn't emphasize this point. In looking at beta diversity, we find that certain organisms are indicated as protective, as you point out, and others are considered disease-associated. For Clostridium difficile infection, you look at the totality of microbes, the alpha diversity. In chronic disorders, it is beta diversity abnormalities that are important. It is this mix of organisms and abnormal, unhealthy bacteria and lack of healthy ones that causes concern. I'd be glad to send you the slide that lists the specific bacteria that we are interested in (health and disease), and you will see them when this paper is published in the Transactions.
Hook, Birmingham: Thanks for your talk. In dysbiosis and other sites, specifically the vagina, biofilms also play an important role. What about in the gut? Are biofilms active there, and are they part of this whole complex picture?
DuPont, Houston: I think biofilms are very much involved in the mucosal-associated microbiota that interact with the immune system and the enteric nervous system.
Hardin, New York: Great talk. I am interested in the oral microbiome and how it relates to diseases like rheumatoid arthritis. It might be a place that's even more easily manipulated than the gut. I wonder if you could update us on where we stand with the oral microbiome.
DuPont, Houston: Well, in studies on rheumatoid arthritis, the oral microbiome and the gut both show a dysfunctional dysbiosis. If you're a scatologist like I am, I focus on the stool rather than the organisms in the mouth. We are studying antibody-coated bacteria as a way to get to the gut-associated bacteria, and we find that the same IgA-coated bacteria in the gut can be found in the mouth in close proximity to immune cells. Gut microbiota contribute to the oral microbiota and vice versa. So it isn't a very far reach to see how the gut and the mouth relate. They're anatomically connected.
Tweardy, Houston: Excellent talk as always. You made an interesting comment early on in your talk that everybody's healthy microbiota are different, yet when you treat a patient you use presumably a single source of microbiota. Does one size fit all, or how does that difference in all of our healthy microbiota translate to a single donor actually being successful in remediating disease in the particular recipient?
DuPont, Houston: That's a wonderful question, David, and one size does not fit all. With Clostridium difficile infection recurrence, it is pure dysbiosis. There's abnormal alpha and beta. All donors work in FMT. When you deal with diseases where there's a primary disorder of beta diversity, all donors don't work. The best studies were with inflammatory bowel disease where one study found that a single donor successfully treated all the IBD patients but other donors didn't. With our studies of chronic disease like Parkinson's, we are combining stool product from three donors before putting product in capsules to maximize our possibility. So I would say all donors work for Clostridium difficile, but we don't know which donors work for chronic.
Tweardy, Houston: Thank you.
Rockey, Charleston: We'll welcome everybody to Charleston next year. I have two questions. The first one is similar to the previous one: How do you know which stool to give to which patient? It sounds like there's going to be some nuances to that, particularly in diseased states. The second question is Would you comment on the death associated with fecal transplantation and an FDA ruling or ban?
DuPont, Houston: We don't know which stool to use so we are combining fecal products for non-Clostridium difficile indications. In some sites where researchers are doing fecal microbiota transplantation (FMT), extended spectrum beta-lactamase-resistant E.coli were transplanted into the recipients with recurring Clostridium difficile and they have developed fatal antibiotic-resistant infections. The FDA sent a warning out, and they jumped on this like gangbusters. We have screened for antibiotic resistance since the moment we started our program, and we never had that problem. If you screen for antibiotic resistance, you should be able to prevent acquisition of antibiotic-resistant genes. We haven't had any trouble. The FDA did not stop FMT during that time. It just required everyone to have a protocol for preventing it by screening for antibiotic resistance.
Rockey, Charleston: What about my first question—do you think you're going to have to use different stool for different patients based on their disease?
DuPont, Houston: As I said, we're using stool products from three donors in each of our doses. We are isolating thousands of colonies of bacteria from our treated patients and looking for biologic activity. We hope to get rid of the F in FMT, and we're trying to look for microbes with biologic activity that can be given in pure form. In my opinion, stool is not destined for greatness in microbiome reversal. We need to develop purified non-fecal-derived products. Also, various diseases will require their own microbiome reversal formula.
REFERENCES
- 1.The Wilde Medal and Lecture of the Manchester Literary and Philosophical Society. Br Med J. 1901;1(2104):1027–8. [PMC free article] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32(8):834–41. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 4.Cani PD. Human gut microbiome: hopes, threats and promises. Gut. 2018;67(9):1716–25. doi: 10.1136/gutjnl-2018-316723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helander HF, Fandriks L. Surface area of the digestive tract—revisited. Scand J Gastroenterol. 2014;49(6):681–9. doi: 10.3109/00365521.2014.898326. [DOI] [PubMed] [Google Scholar]
- 6.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33(9):496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 7.Yu LC, Wang JT, Wei SC, Ni YH. Host-microbial interactions and regulation of intestinal epithelial barrier function: From physiology to pathology. World J Gastrointest Pathophysiol. 2012;3(1):27–43. doi: 10.4291/wjgp.v3.i1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ge X, Ding C, Zhao W, Xu L, Tian H, Gong J, et al. Antibiotics-induced depletion of mice microbiota induces changes in host serotonin biosynthesis and intestinal motility. J Transl Med. 2017;15(1):13. doi: 10.1186/s12967-016-1105-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci U S A. 2005;102(1):99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105(39):15064–9. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4659–65. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng S, Ma X, Geng S, Jiang X, Li Y, Hu L, et al. Fecal Microbiota Transplantation Beneficially Regulates Intestinal Mucosal Autophagy and Alleviates Gut Barrier Injury. mSystems. 2018;3(5) doi: 10.1128/mSystems.00137-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jakobsson HE, Rodriguez-Pineiro AM, Schutte A, Ermund A, Boysen P, Bemark M, et al. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 2015;16(2):164–77. doi: 10.15252/embr.201439263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Correa-Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MA. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunology. 2016;5(4):e73. doi: 10.1038/cti.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baktash A, Terveer EM, Zwittink RD, Hornung BVH, Corver J, Kuijper EJ, et al. Mechanistic Insights in the Success of Fecal Microbiota Transplants for the Treatment of Clostridium difficile Infections. Front Microbiol. 2018;9:1242. doi: 10.3389/fmicb.2018.01242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang JD, Myers CJ, Harris SC, Kakiyama G, Lee IK, Yun BS, et al. Bile Acid -7alpha-Dehydroxylating Gut Bacteria Secrete Antibiotics That Inhibit Clostridium difficile: Role of Secondary Bile Acids. Cell Chem Biol. 2019;26(1):27–34 e4. doi: 10.1016/j.chembiol.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rea MC, Sit CS, Clayton E, O'Connor PM, Whittal RM, Zheng J, et al. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc Natl Acad Sci U S A. 2010;107(20):9352–7. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spinler JK, Auchtung J, Brown A, Boonma P, Oezguen N, Ross CL, et al. Next-Generation Probiotics Targeting Clostridium difficile through Precursor-Directed Antimicrobial Biosynthesis. Infect Immun. 2017;85(10) doi: 10.1128/IAI.00303-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blander JM, Longman RS, Iliev ID, Sonnenberg GF, Artis D. Regulation of inflammation by microbiota interactions with the host. Nat Immunol. 2017;18(8):851–60. doi: 10.1038/ni.3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu H, Khosravi A, Kusumawardhani IP, Kwon AH, Vasconcelos AC, Cunha LD, et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352(6289):1116–20. doi: 10.1126/science.aad9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–6. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 22.Bunker JJ, Erickson SA, Flynn TM, Henry C, Koval JC, Meisel M, et al. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science. 2017;358(6361) doi: 10.1126/science.aan6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bunker JJ, Flynn TM, Koval JC, Shaw DG, Meisel M, McDonald BD, et al. Innate and Adaptive Humoral Responses Coat Distinct Commensal Bacteria with Immunoglobulin A. Immunity. 2015;43(3):541–53. doi: 10.1016/j.immuni.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forsythe P, Kunze W, Bienenstock J. Moody microbes or fecal phrenology: what do we know about the microbiota-gut-brain axis? BMC Med. 2016;14:58. doi: 10.1186/s12916-016-0604-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharon G, Sampson TR, Geschwind DH, Mazmanian SK. The Central Nervous System and the Gut Microbiome. Cell. 2016;167(4):915–32. doi: 10.1016/j.cell.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furness JB, Costa M. The adrenergic innervation of the gastrointestinal tract. Ergeb Physiol. 1974;69(0):2–51. [PubMed] [Google Scholar]
- 27.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8(6):411–20. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 28.Elenkov IJ, Wilder RL, Chrousos GP, Vizi ES. The sympathetic nerve—an integrative interface between two supersystems: the brain and the immune system. Pharmacol Rev. 2000;52(4):595–638. [PubMed] [Google Scholar]
- 29.O'Neill LA, Golenbock D, Bowie AG. The history of Toll-like receptors—redefining innate immunity. Nat Rev Immunol. 2013;13(6):453–60. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- 30.Hakansson A, Molin G. Gut microbiota and inflammation. Nutrients. 2011;3(6):637–82. doi: 10.3390/nu3060637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Latorre R, Sternini C, De Giorgio R, Greenwood-Van Meerveld B. Enteroendocrine cells: a review of their role in brain-gut communication. Neurogastroenterol Motil. 2016;28(5):620–30. doi: 10.1111/nmo.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lazar V, Ditu LM, Pircalabioru GG, Gheorghe I, Curutiu C, Holban AM, et al. Aspects of Gut Microbiota and Immune System Interactions in Infectious Diseases, Immunopathology, and Cancer. Front Immunol. 2018;9:1830. doi: 10.3389/fimmu.2018.01830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Draper LA, Ryan FJ, Smith MK, Jalanka J, Mattila E, Arkkila PA, et al. Long-term colonisation with donor bacteriophages following successful faecal microbial transplantation. Microbiome. 2018;6(1):220. doi: 10.1186/s40168-018-0598-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuo T, Wong SH, Lam K, Lui R, Cheung K, Tang W, et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut. 2018;67(4):634–43. doi: 10.1136/gutjnl-2017-313952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ott SJ, Waetzig GH, Rehman A, Moltzau-Anderson J, Bharti R, Grasis JA, et al. Efficacy of Sterile Fecal Filtrate Transfer for Treating Patients with Clostridium difficile Infection. Gastroenterology. 2017;152(4):799–811 e7. doi: 10.1053/j.gastro.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Jiang ZD, Jenq RR, Ajami NJ, Petrosino JF, Alexander AA, Ke S, et al. Safety and preliminary efficacy of orally administered lyophilized fecal microbiota product compared with frozen product given by enema for recurrent Clostridium difficile infection: A randomized clinical trial. PLoS One. 2018;13(11):e0205064. doi: 10.1371/journal.pone.0205064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rolfe RD. Role of volatile fatty acids in colonization resistance to Clostridium difficile. Infect Immun. 1984;45(1):185–91. doi: 10.1128/iai.45.1.185-191.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allegretti JR, Kearney S, Li N, Bogart E, Bullock K, Gerber GK, et al. Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Aliment Pharmacol Ther. 2016;43(11):1142–53. doi: 10.1111/apt.13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staffas A, Burgos da Silva M, van den Brink MR. The intestinal microbiota in allogeneic hematopoietic cell transplant and graft-versus-host disease. Blood. 2017;129(8):927–33. doi: 10.1182/blood-2016-09-691394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21(8):895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 41.Talotta R, Atzeni F, Ditto MC, Gerardi MC, Sarzi-Puttini P. The Microbiome in Connective Tissue Diseases and Vasculitides: An Updated Narrative Review. J Immunol Res. 2017;2017:6836498. doi: 10.1155/2017/6836498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wieland A, Frank DN, Harnke B, Bambha K. Systematic review: microbial dysbiosis and nonalcoholic fatty liver disease. Aliment Pharmacol Ther. 2015;42(9):1051–63. doi: 10.1111/apt.13376. [DOI] [PubMed] [Google Scholar]
- 43.Aragones G, Gonzalez-Garcia S, Aguilar C, Richart C, Auguet T. Gut Microbiota-Derived Mediators as Potential Markers in Nonalcoholic Fatty Liver Disease. Biomed Res Int. 2019;2019:8507583. doi: 10.1155/2019/8507583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bashiardes S, Shapiro H, Rozin S, Shibolet O, Elinav E. Nonalcoholic fatty liver and the gut microbiota. Mol Metab. 2016;5(9):782–94. doi: 10.1016/j.molmet.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Musso G, Cassader M, Paschetta E, Gambino R. Bioactive Lipid Species and Metabolic Pathways in Progression and Resolution of Nonalcoholic Steatohepatitis. Gastroenterology. 2018;155(2):282–302 e8. doi: 10.1053/j.gastro.2018.06.031. [DOI] [PubMed] [Google Scholar]
- 46.Rabot S, Membrez M, Bruneau A, Gerard P, Harach T, Moser M, et al. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 2010;24(12):4948–59. doi: 10.1096/fj.10-164921. [DOI] [PubMed] [Google Scholar]
- 47.Langdon A, Crook N, Dantas G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016;8(1):39. doi: 10.1186/s13073-016-0294-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Millan B, Park H, Hotte N, Mathieu O, Burguiere P, Tompkins TA, et al. Fecal Microbial Transplants Reduce Antibiotic-resistant Genes in Patients with Recurrent Clostridium difficile Infection. Clin Infect Dis. 2016;62(12):1479–86. doi: 10.1093/cid/ciw185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferreira RB, Antunes LC, Finlay BB. Should the human microbiome be considered when developing vaccines? PLoS Pathog. 2010;6(11):e1001190. doi: 10.1371/journal.ppat.1001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell. 2018;33(4):570–80. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nature Medicine. 2018 doi: 10.1038/s41591-018-0238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cersosimo MG, Raina GB, Pecci C, Pellene A, Calandra CR, Gutierrez C, et al. Gastrointestinal manifestations in Parkinson's disease: prevalence and occurrence before motor symptoms. J Neurol. 2013;260(5):1332–8. doi: 10.1007/s00415-012-6801-2. [DOI] [PubMed] [Google Scholar]
- 53.Keshavarzian A, Green SJ, Engen PA, Voigt RM, Naqib A, Forsyth CB, et al. Colonic bacterial composition in Parkinson's disease. Mov Disord. 2015;30(10):1351–60. doi: 10.1002/mds.26307. [DOI] [PubMed] [Google Scholar]
- 54.Scheperjans F, Aho V, Pereira PA, Koskinen K, Paulin L, Pekkonen E, et al. Gut microbiota are related to Parkinson's disease and clinical phenotype. Mov Disord. 2015;30(3):350–8. doi: 10.1002/mds.26069. [DOI] [PubMed] [Google Scholar]
- 55.Erny D, Hrabe de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci. 2015;18(7):965–77. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim S, Kwon SH, Kam TI, Panicker N, Karuppagounder SS, Lee S, et al. Transneuronal Propagation of Pathologic alpha-Synuclein from the Gut to the Brain Models Parkinson's Disease. Neuron. 2019 doi: 10.1016/j.neuron.2019.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pan-Montojo F, Schwarz M, Winkler C, Arnhold M, O'Sullivan GA, Pal A, et al. Environmental toxins trigger PD-like progression via increased alpha-synuclein release from enteric neurons in mice. Sci Rep. 2012;2:898. doi: 10.1038/srep00898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shannon KM, Keshavarzian A, Mutlu E, Dodiya HB, Daian D, Jaglin JA, et al. Alpha-synuclein in colonic submucosa in early untreated Parkinson's disease. Mov Disord. 2012;27(6):709–15. doi: 10.1002/mds.23838. [DOI] [PubMed] [Google Scholar]
- 59.Svensson E, Horvath-Puho E, Thomsen RW, Djurhuus JC, Pedersen L, Borghammer P, et al. Vagotomy and subsequent risk of Parkinson's disease. Ann Neurol. 2015;78(4):522–9. doi: 10.1002/ana.24448. [DOI] [PubMed] [Google Scholar]
- 60.Maini Rekdal V, Bess EN, Bisanz JE, Turnbaugh PJ, Balskus EP. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science. 2019;364(6445) doi: 10.1126/science.aau6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guan J, Pavlovic D, Dalkie N, Waldvogel HJ, O'Carroll SJ, Green CR, et al. Vascular degeneration in Parkinson's disease. Brain Pathol. 2013;23(2):154–64. doi: 10.1111/j.1750-3639.2012.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walker LC, Schelle J, Jucker M. The Prion-Like Properties of Amyloid-beta Assemblies: Implications for Alzheimer's Disease. Cold Spring Harb Perspect Med. 2016;6(7) doi: 10.1101/cshperspect.a024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Friedland RP, Chapman MR. The role of microbial amyloid in neurodegeneration. PLoS Pathog. 2017;13(12):e1006654. doi: 10.1371/journal.ppat.1006654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berer K, Gerdes LA, Cekanaviciute E, Jia X, Xiao L, Xia Z, et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci U S A. 2017;114(40):10719–24. doi: 10.1073/pnas.1711233114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kirby TO, Ochoa-Reparaz J. The Gut Microbiome in Multiple Sclerosis: A Potential Therapeutic Avenue. Med Sci (Basel) 2018;6(3) doi: 10.3390/medsci6030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kowalski K, Mulak A. Brain-Gut-Microbiota Axis in Alzheimer's Disease. J Neurogastroenterol Motil. 2019;25(1):48–60. doi: 10.5056/jnm18087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Almeida CV, de Camargo MR, Russo E, Amedei A. Role of diet and gut microbiota on colorectal cancer immunomodulation. World J Gastroenterol. 2019;25(2):151–62. doi: 10.3748/wjg.v25.i2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105(24):1907–11. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104(34):13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4(2):293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Enck P, Mazurak N. Dysbiosis in Functional Bowel Disorders. Ann Nutr Metab. 2018;72(4):296–306. doi: 10.1159/000488773. [DOI] [PubMed] [Google Scholar]
- 72.Sun Q, Jia Q, Song L, Duan L. Alterations in fecal short-chain fatty acids in patients with irritable bowel syndrome: A systematic review and meta-analysis. Medicine (Baltimore) 2019;98(7):e14513. doi: 10.1097/MD.0000000000014513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fujimura KE, Lynch SV. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe. 2015;17(5):592–602. doi: 10.1016/j.chom.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bandera A, De Benedetto I, Bozzi G, Gori A. Altered gut microbiome composition in HIV infection: causes, effects and potential intervention. Curr Opin HIV AIDS. 2018;13(1):73–80. doi: 10.1097/COH.0000000000000429. [DOI] [PubMed] [Google Scholar]
- 75.Ji Y, Zhang F, Zhang R, Shen Y, Liu L, Wang J, et al. Changes in intestinal microbiota in HIV-1-infected subjects following cART initiation: influence of CD4 + T cell count. Emerg Microbes Infect. 2018;7(1):113. doi: 10.1038/s41426-018-0117-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jiang ZD, Ajami NJ, Petrosino JF, Jun G, Hanis CL, Shah M, et al. Randomised clinical trial: faecal microbiota transplantation for recurrent Clostridum difficile infection—fresh, or frozen, or lyophilised microbiota from a small pool of healthy donors delivered by colonoscopy. Aliment Pharmacol Ther. 2017;45(7):899–908. doi: 10.1111/apt.13969. [DOI] [PubMed] [Google Scholar]
- 77.DuPont H, Jiang Z-D, DuPont A, Utay N. Abnormal intestinal microbiome in medical disorders and potential reversibility by fecal microbiota transplantation. Dig Dis Sci. 2020 doi: 10.1007/s10620-020-06102-y. doi.org/10.1007/s10620-020-06102-y. [DOI] [PubMed] [Google Scholar]


